ABSTRACT
Flagellar assembly depends on intraflagellar transport (IFT), a bidirectional motility of protein carriers, the IFT trains. The trains are periodic assemblies of IFT-A and IFT-B subcomplexes and the motors kinesin-2 and IFT dynein. At the tip, anterograde trains are remodeled for retrograde IFT, a process that in Chlamydomonas involves kinesin-2 release and train fragmentation. However, the degree of train disassembly at the tip remains unknown. Here, we performed two-color imaging of fluorescent protein-tagged IFT components, which indicates that IFT-A and IFT-B proteins from a given anterograde train usually return in the same set of retrograde trains. Similarly, concurrent turnaround was typical for IFT-B proteins and the IFT dynein subunit D1bLIC–GFP but severance was observed as well. Our data support a simple model of IFT turnaround, in which IFT-A, IFT-B and IFT dynein typically remain associated at the tip and segments of the anterograde trains convert directly into retrograde trains. Continuous association of IFT-A, IFT-B and IFT dynein during tip remodeling could balance protein entry and exit, preventing the build-up of IFT material in flagella.
KEY WORDS: Cilia, Dynein1b, Kinesin-2, BBSome, Flagella
Summary:In vivo imaging in Chlamydomonas supports a simple model of intraflagellar transport (IFT) turnaround at the tip, in which IFT-A, -B, and IFT dynein remain associated and anterograde trains convert directly into retrograde trains.
INTRODUCTION
Intraflagellar transport (IFT) is a motor-based protein shuttle dedicated to the assembly and maintenance of cilia and eukaryotic flagella (Rosenbaum and Witman, 2002). IFT trains are composed of 22 IFT particle proteins organized into IFT-A, -B1 and -B2 subcomplexes and the two IFT motors, the heterotrimeric kinesin-2 and IFT dynein (Cole et al., 1998; Kozminski et al., 1995; Piperno and Mead, 1997; Taschner and Lorentzen, 2016; Taschner et al., 2016; Walther et al., 1994). The trains assemble near the basal bodies, enter the flagellum and move anterogradely to the flagellar tip in a kinesin-2-dependent manner. The retrograde motor IFT dynein is a cargo on the anterograde trains and transported in an autoinhibited configuration with its microtubule-binding domain pointing away from the axonemal tracks, ensuring highly processive transport (Dentler, 2005; Jordan et al., 2018; Kozminski et al., 1993; Pazour et al., 1999; Porter et al., 1999; Toropova et al., 2017). In Chlamydomonas, most kinesin-2 is released from IFT near the tip, probably due to its phosphorylation by a Ca2+-dependent kinase, and diffuses back to the cell body (Chien et al., 2017; Engel et al., 2009; Liang et al., 2014). Anterograde and retrograde trains, while distinct in size, ultrastructure and velocity, are widely assumed to consist of the same proteins but for the absence of kinesin-2 in the latter (Jordan et al., 2018; Stepanek and Pigino, 2016). In Chlamydomonas, the number of retrograde trains exceeds that of anterograde trains, suggesting that trains disintegrate at the tip into smaller retrograde trains (Dentler, 2005; Reck et al., 2016). Indeed, in vivo imaging of fluorescent protein-tagged IFT-B proteins in T. brucei has shown that an anterograde train splits into three retrograde trains (Buisson et al., 2012). In Chlamydomonas, trains dwell for ∼2 s after arriving at the tip before splitting into an average of 2.4 fragments (Chien et al., 2017; Qin et al., 2007; Wren et al., 2013). Concomitantly, many IFT cargoes, including most axonemal proteins such as tubulin, are unloaded (Lechtreck, 2015). Then, IFT material from a given anterograde train mixes with complexes derived from other trains to form retrograde trains (Chien et al., 2017). However, the processes leading to the release of kinesin-2 and the cargoes, the activation of IFT dynein and the formation of retrograde trains at the flagellar tip are incompletely understood.
It is widely thought that anterograde trains dissolve into individual IFT-A, IFT-B, kinesin-2 and IFT dynein complexes at the flagellar tip (Chien et al., 2017; Cole and Snell, 2009; Dawson and House, 2010; Ishikawa and Marshall, 2011; Liang et al., 2014; Morga and Bastin, 2013; Nakayama and Katoh, 2018; Pedersen et al., 2006; Soares et al., 2019; Taschner and Lorentzen, 2016). Then, retrograde trains reassemble and IFT dynein becomes active (Chien et al., 2017). Single protein tracking in C. elegans cilia showed distinct dwell times for the different IFT subcomplexes, suggesting that IFT-A and -B proteins separate at the tip (Mijalkovic et al., 2018). Recent ultrastructural analysis revealed that anterograde IFT trains in Chlamydomonas are not built from IFT particles encompassing equimolar amounts of the IFT-A, -B and -motor subcomplexes but instead are organized into IFT-A, -B and IFT dynein layers, each with its distinct periodicity (Jordan et al., 2018). Thus, a disassembly of anterograde trains into individual IFT particles encompassing one each of IFT-A, -B and IFT dynein seems unlikely and other models of IFT reorganization at the flagellar tip need to be considered. For example, anterograde IFT trains could split more or less perpendicular through the layers into a few, somewhat irregular ‘carts’ consisting of multiple IFT-A, IFT-B, and probably IFT dynein complexes, which then return together to the flagellar base. Also, the trains could separate into oligomers of each IFT-A and IFT-B with or without associated dynein motors, which then recombine into a retrograde train. Finally, trains could disassemble into mostly individual IFT subcomplexes or even IFT proteins, which then reassemble with similar kinetics into retrograde trains.
To analyze the degree of IFT train disassembly at the tip, we used FRAP assays of strains expressing fluorescent protein-tagged IFT particle and motor subunits. All IFT proteins studied, including those of the IFT-A subcomplex, dwelled at the tip for ∼2 s, and proteins derived from one anterograde train distributed into approximately three retrograde trains. IFT-A and IFT-B subcomplexes derived from a given anterograde train returned in the same set of retrograde trains. Similarly, IFT dynein and IFT-B from a given anterograde train mostly returned in the same set of retrograde trains but recombination with material from other trains occurred as well. Our data indicate that IFT-A, -B, and IFT dynein typically remain associated with each other during tip turnaround. We propose a model in which segments of the anterograde trains convert directly into retrograde trains without major disassembly.
RESULTS
IFT-A and IFT-B proteins dwell at the flagellar tip
To analyze how anterograde IFT trains reorganize for retrograde travel at the flagellar tip, we employed Chlamydomonas strains expressing fluorescent protein (FP)-tagged versions of the IFT-A proteins IFT43 and IFT140, the IFT-B1 proteins IFT46, IFT81 and IFT74, the IFT-B2 protein IFT54, and the retrograde IFT motor subunit D1bLIC (Table S1). Expression of these proteins restored IFT and flagellar assembly in the corresponding mutants (see Fig. S5E and Materials and Methods). For our analysis, we focused on cells with high-frequency IFT and little stationary IFT.
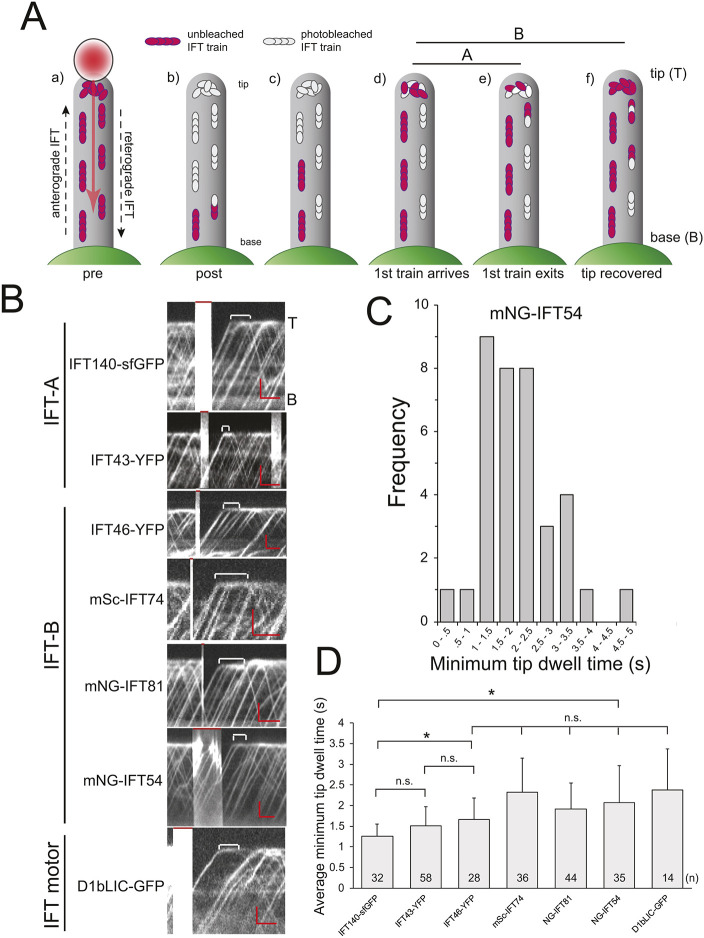
In Chlamydomonas, FP-tagged versions of IFT-B proteins IFT20 and IFT27 and the motor subunit D1bLIC pause for ∼2 s at the tip between their arrival by anterograde IFT and the onset of retrograde IFT (Chien et al., 2017; Qin et al., 2007; Reck et al., 2016; Wren et al., 2013). To analyze whether tip dwelling is a common feature of IFT proteins including those of the IFT-A subcomplex, we bleached FP-tagged IFT proteins in the tip region of flagella with a focused laser beam and analyzed the time elapsed between the arrival of the first unbleached train and the departure of the first unbleached retrograde train; we refer to this approach as tip-FRAP (Fig. 1A; Movie 1). For all IFT proteins analyzed, we observed a pause between their arrival at the tip and their departure (Fig. 1B). Pauses of less than 1 s or more than 4 s were rarely observed (Fig. 1C; Fig. S1A). For the different IFT protein analyzed, the average length of the pause varied from ∼1.3 s for IFT140–sfGFP to ∼2.3 s for mSc–IFT74 and D1bLIC–GFP (Fig. 1D). Although it provides a measure for the time needed to assemble retrograde trains, the tip-FRAP approach does not allow us to determine with certainty whether proteins in the first unbleached retrograde train are indeed derived from the first post-bleach anterograde train or from subsequent trains or a mixture of both. To determine the time an IFT protein dwells at the tip more directly, we expressed 2×Dendra–IFT54 in the ift54-2 background (Fig. S1B). A small portion of the inactive green-emitting Dendra was autoactivated into the red-emitting form. The behavior of these IFT54–DendraRED speckles deviated considerably from that of the bulk flow of IFT54–DendraGREEN supporting the notion that we imaged small clusters of IFT54–DendraRED or individual proteins (Fig. S1Ba–c, Movie 2). The average dwell time of the IFT54–DendraRED speckles was 2.2 s (s.d. 1.2, n=32, Fig. S1C). This value is similar to that observed for IFT54–mNG using tip-FRAP (Fig. 1C,D) and those previously reported for the IFT-B protein IFT27 based on the analysis of individual IFT trains (Chien et al., 2017). To summarize, IFT proteins including those of the IFT-A subcomplex pause at the tip and require ∼1.5 s or longer to transition from anterograde into retrograde trains.
Fig. 1.
IFT-A, IFT-B and IFT dynein proteins dwell for ∼2 s at the flagellar tip. (A) Schematic of the tip-FRAP approach. (a) The bleaching laser was moved from the flagellar tip toward the base, photobleaching all trains in the distal region of the flagellum. Alternatively, we used a stationary larger-diameter beam and flash-bleached the flagellar tip. (b,c) Fluorescent anterograde trains re-enter the distal flagellum. (d) The first unbleached anterograde train arrives at the tip. (e) The first retrograde train containing unbleached IFT material exits the tip. (f) The signal of the IFT pool at the tip is fully recovered. Line A, pause between arrival of the first unbleached anterograde train and departure of the first unbleached retrograde train. Line B, time between the arrival of the first unbleached anterograde train and the recovery of the tip signal. (B) Kymograms from tip-FRAP photobleaching experiments using tagged IFT strains. The time between the arrival of the first post-bleach anterograde train and the departure of the first unbleached retrograde train is marked by brackets; red bars at the top indicate the bleaching steps. Images are representative of three or more experiments for each strain. T, tip; B, base. Scale bars: 2 µm (vertical) and 2 s (horizontal). (C) Distribution of dwell times at the tip for mNG–IFT54 (n=37 particles from three experiments). (D) Mean±s.d. dwell time of the various tagged IFT proteins at the tip. *P<0.001; n.s., not significant (paired two-tailed t-test).
During the pause, the IFT54–DendraRED speckles remained mostly stationary but particles undergoing a slow diffusion were observed as well (Fig. S1Bd–i). Thus, we wondered whether IFT proteins are confined to the tip region during turnaround. First, we analyzed the behavior of BBS4, a subunit of the BBSome (Nachury et al., 2007). This octameric complex cycles via anterograde and retrograde IFT through flagella but also detaches from the IFT machinery along the length of the flagella and near the tip (Lechtreck et al., 2009; Liew et al., 2014). In Chlamydomonas, BBS proteins are substoichiometric to IFT proteins and FP-tagged versions of BBS4 are typically expressed at even lower levels (Lechtreck et al., 2009). Here, we used a mNG-tagged BBS4 expressed at low levels, so that only one or two tagged BBSomes are present on a subset of IFT trains allowing us to study the behavior of single BBSomes at the tip (Fig. S1D). BBS4–mNG dwelled for an average of 2.65 s (s.d. 2 s, n=31) and, in comparison to the IFT proteins, its behavior was more variable, ranging from remaining almost stationary at the tip to escaping from the tip region by diffusion, often for later pick-up by retrograde trains (Fig. S1D,E). Similarly, KAP–GFP (a subunit of kinesin-2) and IFT cargoes diffuse into the flagellar shaft after being released from IFT at the tip (Chien et al., 2017; Wren et al., 2013). In contrast, tagged IFT particle and dynein subunits largely remained sequestered within the tip region during turnaround and exit from this region by diffusion was uncommon.
The velocity of train conversion at the tip is temperature dependent
The variability in the length of the pause recorded for the different FP-tagged IFT proteins could indicate distinct behaviors at the tip. Alternatively, it could reflect differences between the various rescue strains and/or differences in the experimental conditions. Indeed, when cells were imaged at 16, 21 and 27°C, we observed that both velocity and dwell time at the tip of the FP-tagged proteins were temperature dependent (Fig. S2A–D). Generally, a temperature increase of 10°C increased anterograde velocity by a factor of ∼1.6 whereas the velocity of retrograde IFT and the pace of retrograde train departure from the tip increased by up to 2.7-fold. Thus, Chlamydomonas IFT shows a pronounced temperature sensitivity, similar to previous observations on IFT velocity and frequency in Trypanosoma brucei (Buisson et al., 2012). These observations were expected since the temperature dependency of motor protein activity and cellular transports is well documented (Yadav and Kunwar, 2021). In double-mutant double-rescue strains expressing combinations of two IFT proteins, the variation of the dwell times reported above for the single-tagged strains were not observed (see below). In conclusion, the dwell time variations observed in the single strains likely result from temperature fluctuations during imaging rather than presenting characteristic features of the IFT proteins analyzed.
Anterograde IFT trains typically fragment into several retrograde trains
Previous studies revealed that IFT27 and D1bLIC derived from a single anterograde train return to the flagellar base in several retrograde trains indicating that anterograde trains fragment at the tip (Chien et al., 2017). IFT27–GFP was expressed in wild-type Chlamydomonas cells also expressing endogenous IFT27. Furthermore, it has been proposed that IFT27 dissociates from the IFT-B complex at the tip of mammalian primary cilia (Liew et al., 2014). Thus, it is unclear whether the behavior of IFT27–GFP is representative of that of the other IFT proteins (Chien et al., 2017).
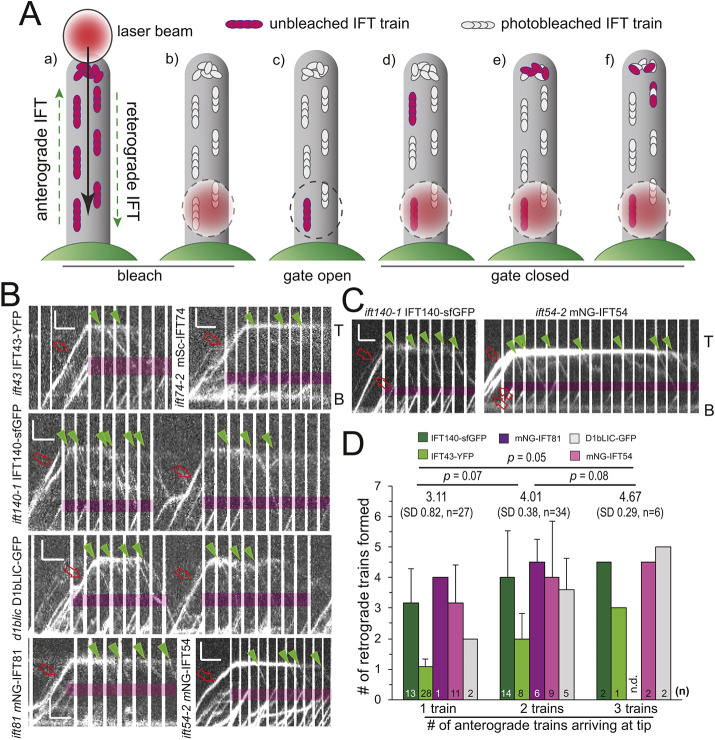
To answer this question, we used the photogate approach to analyze the behavior of individual fluorescent IFT trains at the tip (Chien et al., 2017). In brief, a focused laser beam was used to bleach the flagellum from the tip down (Fig. 2A). Once near the flagellar base, the bleaching laser is transiently blocked to allow one or a few unbleached IFT trains to enter the flagellum and continue their journey toward the tip. Then, the photogate is closed again, that is, the beam blinks on and off with a ratio, diameter and strength ensuring that subsequent IFT trains passing through the beam are bleached but allowing for the recording of the total internal reflection fluorescence (TIRF) signals while the laser is off. Upon reaching the tip, anterograde trains typically split into several retrograde trains for all IFT proteins studied (Fig. 2B; Movie 3). However, trains returning without fragmentation (Fig. S3A), trains splitting along the length of the flagellum (Fig. S3B) and/or trains returning prior to reaching the tip (Fig. S3C) were observed as well, testifying to the flexibility of IFT (Chien et al., 2017; Dentler, 2005; Reck et al., 2016). A single fluorescent anterograde train resulted in an average of 3.2 retrograde trains for IFT140–sfGFP (s.d. 1.1, n=11) and mNG–IFT54 (s.d. 1.2, n=10), 4 retrograde trains for mNG–IFT81 (n=1), 2 for D1bLIC–GFP (n=4) and 1.1 for IFT43–YFP (n=28) (Fig. 2B,D). The low number of retrograde trains observed for IFT43–YFP could result in part from the lower photostability of YFP, causing a loss of the signal at the tip. In addition, IFT43 has been reported to be a substoichiometric component of the IFT-A complex (Mukhopadhyay et al., 2010).
Fig. 2.
Fragmentation of IFT trains at the tip involves all IFT subcomplexes. (A) Schematic of the photogate technique (after Chien et al., 2017). (a) Tagged IFT proteins in the flagellum are photobleached by moving a focused laser beam from the tip and toward the base. (b) Once near the proximal end, the laser blinked, typically with a 0.9 s off and 0.1 s on pattern. (c) The photogate is opened by blocking the laser beam to allow one or a few unbleached trains to enter the distal flagellum. (d) The photogate is closed again and subsequent trains are bleached. (e) The unbleached train reaches the tip. (f) The first unbleached retrograde train departs from the tip. (B) Kymograms of photobleaching experiments for the strains indicated. Red open arrows, anterograde trains; green arrowheads, retrograde trains. T, tip; B, base. Scale bars: 2 µm (vertical) and 2 s (horizontal). The magenta boxes indicate the approximate position of the gating laser. (C) Kymograms showing two IFT140–sfGFP (left) and or three mNG–IFT54 (right) anterograde trains entering the bleached tip region and fragmenting into retrograde trains. Scale bars: 2 µm (vertical) and 2 s (horizontal). (D) Mean±s.d. (s.d. shown where n>2) number of retrograde trains derived from 1, 2 or 3 anterograde trains in photogate experiments. The number of anterograde trains analyzed for each IFT protein is indicated (n). The average number of retrograde trains formed was 3.11, 4.01, and 4.67 retrograde trains for one, two or three arriving anterograde trains, respectively. For this analysis the outlying data for IFT43–YFP were not considered. P indicates the result of a paired two-tailed t-test. n.d., no data collected.
In experiments where two or three fluorescent trains passed the gate into the cilium, the number of departing retrograde trains increased to an average of ∼4.0 and ∼4.7, respectively; the outlying data for IFT43–YFP were not considered for this analysis (Fig. 2C,D). The sub-proportional increase in the number of departing trains compared to that of arriving anterograde trains could indicate that several anterograde train fragments combine into a single retrograde train (Chien et al., 2017). While such fusion events have not yet been directly observed, they are supported by the widely observed ratio of ∼1.3 between anterograde and retrograde trains (Chien et al., 2017; Dentler, 2005; Reck et al., 2016). However, it should be noted that the photogate approach likely introduces a bias by analyzing mostly larger and brighter trains, which could lead to an overestimation of the average number of fragments formed per anterograde train.
We noticed that the return of retrograde trains originating from large anterograde trains as assessed by signal strength and the number of resulting retrograde trains, was spread over a longer time compared to retrograde trains derived from apparently smaller anterograde trains (Fig. 2B,C; Fig. S3D). For mNG–IFT54, for example, we observed the formation of up to eight retrograde trains from one anterograde train (Movie 3). Thus, the size of the incoming anterograde trains, which ranges from ∼100 nm to ∼700 nm (Jordan et al., 2018), appears to impact the kinetics of tip turnaround.
The size of the IFT protein pool at the tip is equivalent to approximately four anterograde trains
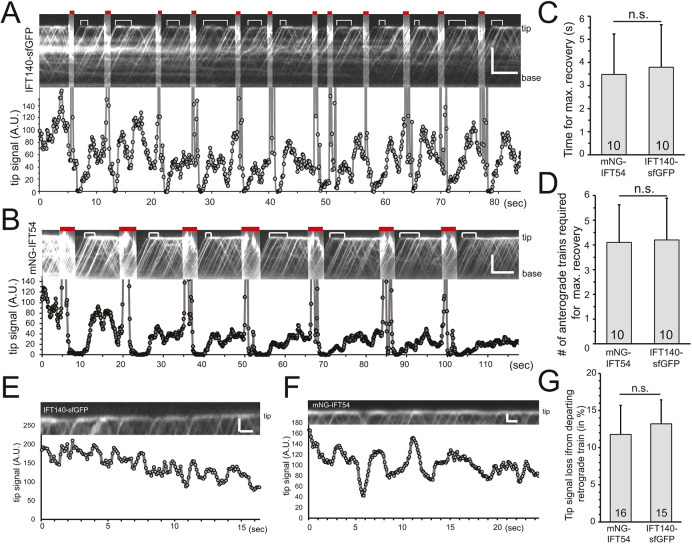
IFT proteins dwell for ∼1.6 s or longer at the flagellar tip and, during this time, additional anterograde trains arrive, indicating the presence of an IFT pool at the tip. To determine the size of this pool, we used the tip-FRAP assay and analyzed the time and the number of anterograde trains required to restore the fluorescent signal at the tip (Fig. 3A,B). For this analysis, we focused on the IFT-A protein IFT140–sfGFP and IFT-B protein mNG–IFT54. Often, the signal at the tip reached or approached pre-bleach intensity indicating a near complete exchange of the bleached IFT proteins with unbleached ones derived from the arriving anterograde trains (Fig. 3A). However, in other experiments, the recovery after the first bleaching step remained incomplete in comparison to the pre-bleach signal, indicating the presence of a stationary or slow-exchanging pool of IFT protein at the tip (Fig. 3B). Regardless of whether the recovery after the initial bleaching step was partial or near complete, a similar extent of signal recovery was observed after each of several subsequent bleaching steps, but for a slow progressive loss of signal caused by the prolonged observation (Fig. 3A,B). This suggests the presence of an active IFT pool and, occasionally, a slowly exchanging or standing IFT pool at the flagellar tip.
Fig. 3.
The size of the IFT protein pool at the tip is equivalent to approximately four anterograde trains. (A,B) Kymograms of IFT140–sfGFP (A, top panel) and mNG–IFT54 (B, top panel) and the corresponding quantification of fluorescence intensities (A.U., arbitrary units) at the tip (bottom panels). Red bars, photobleaching steps; white brackets, time required for the signal to plateau. Scale bars: 5 µm (vertical) and 5 s (horizontal). (C,D) Time in seconds (C) and the number of anterograde trains (D) required for the recovery of the IFT140–sfGFP or mNG–IFT54 tip signals. Results are mean±s.d. (n=10). (E,F) Kymograms of sfGFP–IFT140 (E) and mNG–IFT54 (F) and the corresponding quantification of the intensity decline at the tip induced by the departure of retrograde trains. Scale bars: 1 µm (vertical) and 1 s (horizontal). (G) Mean±s.d. loss of tip signal resulting from the departure of a retrograde train. The number of events analyzed is indicated. n.s., not significant (paired two-tailed t-test).
After the bleaching step, the signal at the tip recovered in 3.8±1.8 s and 3.5±1.8 s for IFT-A and IFT-B, respectively (n=10, mean±s.d.; Fig. 3C). During this time an average of 4.2±1.7 and 4.1±1.5 unbleached trains arrived for IFT140–sfGFP and mSc–IFT54, respectively (n=10; Fig. 3D). After a brief pause, retrograde trains continuously left the pool causing a transient decrease of the signal at the tip by an average of ∼12% with departing large trains causing a decay of up to 35% (Fig. 3E–G). Based on these data, we estimate that the IFT pool at the tip is equivalent to approximately three to four anterograde trains.
Evidence for continued association of IFT-A and IFT-B during train remodeling
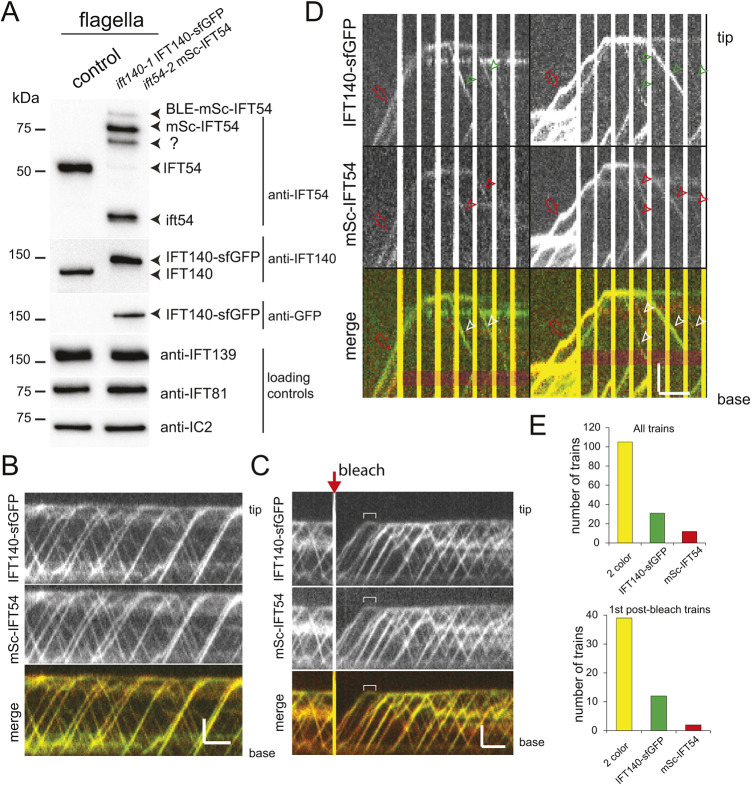
At the flagellar tip, IFT trains fragment but the level of fragmentation remains unclear. To test whether IFT-A and -B proteins separate or remain associated during train remodeling, we generated a double-tagged ift140-1 IFT140–sfGFP ift54-2 mSc–IFT54 strain (Fig. 4A). Both fluorescent proteins were present in all anterograde and retrograde trains identified and the ratio between the two proteins in trains was similar; for example, bright IFT140–sfGFP trains were also bright for mSc–IFT54 (Fig. 4B). The strength of the fluorescent pool at the tip for both proteins fluctuated similarly as trains arrived and departed resulting in a Pearson's correlation coefficient of 0.834 (s.d. 0.08, n=5; Fig. S4A–C). During tip-FRAP, the tip signals of mSc–IFT54 and IFT140–sfGFP recovered concurrently, and proteins arriving by anterograde IFT dwelled for the same time (∼2.5 s; Fig. 4C; Fig. S4D–F, Movie 4). The first and ensuing post-bleach retrograde trains generally displayed the signals of both proteins (Fig. 4C). Of 121 such post-bleach retrograde trains, 93% showed both sfGFP and mSc signals and 6% displayed only a sfGFP signal; the latter is probably due to the lower photostability of mSc.
Fig. 4.
IFT140–sfGFP and mSc–IFT54 from one anterograde train return in the same set of retrograde trains. (A) Western blot of flagella isolated from the control (g1) and the ift140-1 IFT140–sfGFP ift54-2 mSc–IFT54 strains probed with the antibodies as indicated. Anti-IFT54 stained endogenous IFT54 in wild type; in the experimental strain, this antibody stained mSc–IFT54, the unprocessed BLE-mSc–IFT54, a likely mSc–IFT54 degradation product (?), which could have formed during boiling of the samples (Gross et al., 2000), and IFT54, the non-functional truncated product of the ift54-2 mutant. Blot shown is representative of two biological replicates. (B) Kymograms showing IFT in an unbleached flagellum of the ift140-1 IFT140–sfGFP ift54-2 mSc–IFT54 strain. The two individual kymograms of sfGFP and mSc and the merged kymogram are shown. Scale bars: 2 µm (vertical) and 2 s (horizontal). (C) Kymogram from a tip-FRAP experiment using the ift140-1 IFT140–sfGFP ift54-2 mSc–IFT54 strain. The brackets indicate minimum tip dwell time. Scale bars: 2 µm (vertical) and 2 s (horizontal). (D) Kymograms from two photogate experiments showing the presence of both IFT140-sfGFP and mSc-IFT54 in the retrograde trains. Red open arrows, anterograde trains; arrowheads, retrograde trains. Scale bars: 2 µm (vertical) and 2 s (horizontal). (E) Distribution of the presence of only IFT14–sfGFP, only mSc-IFT54 or both unbleached proteins in all retrograde trains (top) and in only first departing trains (bottom) formed in photogate experiments. Because mSc bleached faster than sfGFP, we only considered those retrograde trains for this analysis that were formed while a signal of both proteins was visible at the tip. Kymograms shown are representative of three or more experiments, and results in E are for n=148 and 54 trains in the top and bottom graphs, respectively.
To analyze remodeling of individual trains, we used a two-color photogate approach (Fig. 4D; Movie 5). In 54 photogate experiments, we observed the formation of 148 retrograde trains of which 108 trains (71%) contained both proteins, 31 trains (21%) showed only an sfGFP signal and 12 trains (8%) were only visible in the mSc channel (Fig. 4E). Similarly, 74% of the first post-bleach retrograde trains contained both proteins, whereas 23% and 4% only showed an sfGFP or mSc signal (n=54, Fig. 4E). The photogate approach uses a strong 488-nm gating beam in addition to the light used for two-color TIRF imaging. Occasionally, some of the mSc signal survived in anterograde trains hit by the gating laser. This, together with the progressive loss of emitting fluorophores, especially of mSc, could explain the infrequent occurrence of retrograde trains showing only an IFT140–sfGFP or mSc–IFT54 signal. However, it is also possible that mSc–IFT54 and IFT140–sfGFP derived from the same anterograde train occasionally separate and end up in different retrograde trains.
We also generated a strain expressing the IFT-B1 protein mNG–IFT81 and the IFT-B2 protein mSc–IFT54 in the corresponding double mutant (Fig. S5). Typically, we observed a near perfect match of mNG–IFT81 and mSc–IFT54 presence in the trains during standard TIRF and in tip-FRAP; in the latter, the signal of the less photostable mSc was often diminished (Fig. S5B,C). This suggests similar kinetics and probably persistent association of these two IFT-B proteins during train remodeling at the tip. To summarize, an IFT-B1 and -B2 protein (i.e. mNG–IFT81 and mSc–IFT54) and an IFT-A and an IFT-B2 protein (i.e. IFT140–sfGFP and mSc–IFT54) from given anterograde trains typically returned to the cell body in the same set of retrograde trains and displayed matching intensity fluctuations at the tip. A simple explanation is that IFT-A and IFT-B proteins remain associated during train remodeling, suggesting that trains fragment into ‘carts’ composed of several IFT-A and -B complexes, which then commence retrograde movement.
Expression of mSc–IFT54 reduces IFT dynein entry into cilia
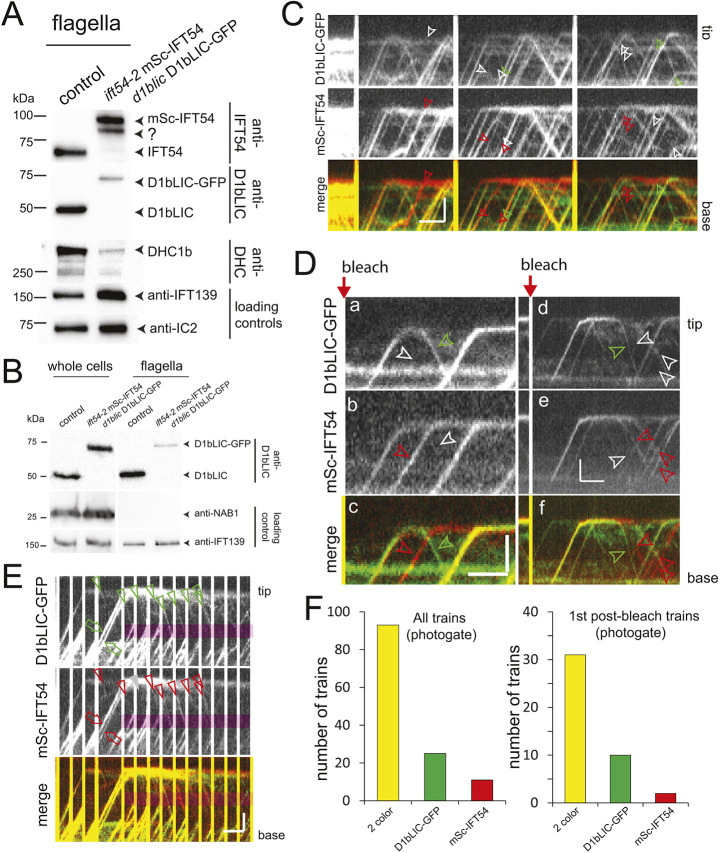
IFT dynein is a cargo during anterograde IFT before it is activated during train remodeling at the tip (Jordan et al., 2018). To explore the question of whether IFT dynein is released from the IFT particle backbone at the tip, we generated a d1blic D1bLIC–GFP ift54-2 mSc–IFT54 strain (Fig. 5A). Western blot analyses showed reduced levels of D1bLIC–GFP as well as of the untagged IFT dynein heavy chain DHC1b (also known as DHC2 in mammals) in flagella isolated from the double-mutant double-rescue strain (Fig. 5A,B). Despite the reduction of IFT dynein, flagellar length and the velocity and frequency of anterograde and retrograde IFT were essentially normal in the d1blic D1bLIC–GFP ift54-2 mSc–IFT54 strain, indicating a robustness of the IFT system despite reduced IFT dynein levels in the flagella (Fig. S5E and data not shown). In whole-cell samples of the d1blic D1bLIC–GFP ift54-2 mSc–IFT54 strain, the levels of D1bLIC and D1bLIC–GFP were similar to those in controls (Fig. 5B). Previous analysis of the d1bLIC D1bLIC–GFP single rescue strain revealed near wild-type levels of the tagged protein in both the cell bodies and flagella (Reck et al., 2016). Thus, FP-tagging of IFT54 interferes with the flagellar levels of IFT dynein, in particular when combined with GFP-tagged D1bLIC.
Fig. 5.
Concomitant and separated return of D1bLIC–GFP and mSc–IFT54 during IFT U-turns. (A) Western blot of flagella isolated from control (g1) and the d1blic D1bLIC–GFP ift54-2 mSc–IFT54 strain. Note reduced levels of D1bLIC–GFP and DHC1b in the double mutant double rescue strain. ?, putative degradation product of mSc-IFT54. (B) Western blot comparing whole-cell and flagella samples of control and d1blic D1bLIC-GFP ift54-2 mSc-IFT54. Antibodies to IFT139 and the cell body protein nucleic acid binding protein 1 (NAB1) were used as loading controls. Blots shown are representative of two biological replicates. (C,D) Kymograms of tip-FRAP experiments using the d1blic D1bLIC-GFP ift54-2 mSc-IFT54 strain. Red and green arrowheads, train trajectories composed mostly of mSc–IFT54 and D1bLIC–GFP, respectively; white arrowheads, position of such trains in the corresponding kymograms lacking clear trajectories. In Dd–f, the return of D1bLIC–GFP-positive trains (green arrowheads) precedes that of the first mSc–IFT54 trains. Scale bars: 2 µm (vertical) and 2 s (horizontal). (E) Kymograms from a photogate experiment show similar fragmentation patterns for D1bLIC–GFP and mSc–IFT54 derived from the two gated trains. Red arrows, anterograde trains; arrowheads, retrograde trains; gray arrowhead in mSc-IFT54 panel, train lacking a D1bLIC-GFP signal. Scale bars: 2 µm (vertical) and 2 s (horizontal). (F) Left-hand panel, distribution of the presence of only D1bLIC-GFP, only mSc-IFT54 or both unbleached proteins in the retrograde trains formed in photogate experiments. Right-hand panel, presence of the unbleached proteins in the first departing trains in photogate experiments. Kymograms shown are representative of nine sets of experiments, and results in F are for n=129 and 43 trains in the left- and right-hand graphs, respectively.
In agreement with the reduction of IFT dynein in flagella revealed by western blotting, we observed anterograde trains with reduced levels of fluorescent D1bLIC–GFP or lacking the signal entirely (Fig. 5C,D). In detail, of 393 anterograde trains analyzed from tip-FRAP experiments, 31 (∼8%) lacked an D1bLIC–GFP signal and ten showed a strongly reduced signal. We also observed seven trains (<2%) lacking an mSc–IFT54 signal and six trains (<2%) with a reduced mSc signal; however, these trains typically appeared after prolonged illumination, suggesting that the mSc–IFT54 signal could have been lost by photobleaching. The data suggest that co-expression of D1bLIC–GFP and mSc–IFT54 interferes with the binding of IFT dynein to anterograde trains and its import into flagella.
In tip-FRAP experiments, we occasionally observed a clear separation of the GFP and mSc signals in the post-bleach retrograde trains (Fig. 5D). Of 183 retrograde trains analyzed, 12 (∼7%) displayed only an mSc–IFT54 signal suggesting that mSc–IFT54 associates with bleached D1bLIC–GFP. Furthermore, nine (∼5%) of the retrograde trains only showed an D1bLIC–GFP signal. Such D1bLIC–GFP-only trains mostly departed the tip briefly after a bleaching step (Fig. 5D). Thus, during the assembly of retrograde trains, D1bLIC–GFP appears to occasionally associate with bleached mSc–IFT54 derived from earlier anterograde trains. However, in 43 two-color photogate experiments with one or a few unbleached anterograde trains arriving at the tip, the majority of the retrograde trains possessed both fluorescent IFT proteins (Fig. 5E,F). Both signals were detected in 31 of the 43 of the first-departing trains and in 93 of all 129 retrograde trains analyzed (Fig. 5F). As described above, differences in the photostability of the two employed fluorescent proteins could explain the occurrence of some single-color retrograde trains. Alternatively, IFT dynein could occasionally return together with IFT-B complexes derived from earlier trains. To summarize, the data suggest that D1bLIC–GFP and mSc–IFT54 from a given anterograde train typically end up in the same set of retrograde IFT trains, similar to our observations on IFT140–sfGFP and mSc–IFT54. However, an occasional separation of D1bLIC–GFP and mSc–IFT54 during the formation of retrograde trains was also apparent, suggesting some flexibility during train remodeling. Indeed, IFT dyneins apparently unconnected to IFT trains have been observed near the tip by cryo-ET (Jordan et al., 2018).
Expression of FP-tagged IFT74 destabilizes IFT dynein on anterograde trains
Recently, the coiled-coil domain of IFT54 was identified as a putative binding side for IFT dynein during anterograde traffic (Zhu et al., 2021). To test whether the expression of mSc–IFT54, an IFT-B2 protein, uniquely interferes with IFT dynein import into cilia, we generated a strain co-expressing D1bLIC–GFP and the IFT-B1 protein IFT74 fused at its N-terminus with mSc in the d1blic ift74-2 double mutant. Similar to our observations in the mSc–IFT54 D1bLIC–GFP strain, the level of D1bLIC–GFP was strongly reduced in flagella isolated from the d1blic D1bLIC–GFP ift74-2 mSc–IFT74 strain (Fig. 6A). We noticed that some cells of this strain had shorter than normal flagella and accumulated IFT material at the tip; for TIRF analysis, we focused on cells with near normal IFT and flagellar length. Anterograde trains as visualized by mSc–IFT74 occasionally lacked an D1bLIC–GFP signal (Fig. 6B,C). Such mSc–IFT74 trains often entered the flagella in conjunction with D1bLIC–GFP and then lost the protein while moving to the flagellar tip, indicating that mSc–IFT74 destabilizes IFT dynein binding to anterograde trains (Fig. 6B; Fig. S6Ba–c, Movie 6). In detail, 15% of all anterograde trains analyzed (n=472) lacked, showed a reduced presence or lost D1bLIC–GFP whereas only ∼4% of the trains lacked or showed a reduced presence of mSc–IFT74 (Fig. 6C). D1bLIC–GFP dissociating from the mSc–IFT74 trains diffused in the flagella as discrete particles (Fig. S6B). Occasionally, such fragments joined passing retrograde trains, similar to what was seen in single-molecule observations on XBX-1::EGFP, the D1bLIC homologue in C. elegans (Fig. S6Bd–f; Mijalkovic et al., 2017). In tip-FRAP experiments, a dissociation of the two proteins was observed more frequently in cells with low frequency IFT and sluggish conversion into retrograde trains, suggesting that separation of the two could be linked to low-fidelity IFT (Fig. 6D versus E; Fig. S6A,B). In cells with high frequency IFT, however, the two proteins mostly remained associated (Fig. 6D); 80% of the first retrograde trains in tip-FRAP experiments encompassed both fluorescent proteins and ∼18% showed only the more stable D1bLIC–GFP signal (Fig. 6F). In conclusion, D1bLIC–GFP and mSc–IFT74 from a given anterograde train typically return on the same set of retrograde trains but numerous exceptions were observed. A separation of the two markers was probably promoted by the reduced interaction between IFT-B and IFT dynein in the d1blic D1bLIC–GFP ift74-2 mSc–IFT74 strain.
Fig. 6.
Co-expression of mSc–IFT74 and D1bLIC–GFP weakens IFT-B and IFT dynein binding. (A) Western blot analysis comparing control and the ift74-2 mSc–IFT74 d1blic D1bLIC–GFP flagella. Anti-IFT74 stained IFT74 in control and mSc–IFT74, uncleaved BLE-mSc–IFT74 and a putative degradation product (?) in the tagged strain. Blot shown is representative of two biological replicates. (B) Kymogram of IFT in the d1bLIC D1bLIC–GFP ift74-2 mSc–IFT74 strain. Open arrowheads and white circles mark events of D1bLIC–GFP dissociation from mSc–IFT74 trains. The filled red and green arrowheads indicate trains composed mostly of mSc–IFT74 or D1bLIC–GFP, respectively. Scale bars: 2 µm (vertical) and 2 s (horizontal). (C) Distribution of anterograde IFT trains carrying D1bLIC–GFP and mSc–IFT74, only mSc–IFT74 or only D1bLIC–GFP. The data were collected from the control and experimental flagella from tip-FRAP experiments. (D,E) Kymograms from tip-FRAP experiments. White brackets indicate the time between the unbleached first anterograde and first retrograde train. Solid green arrowheads, D1bLIC–GFP train lacking a clear mSc–IFT74 signal (white arrowheads). Green and red open arrowheads, first postbleach train with GFP and mSc signals. Scale bars: 2 μm (vertical) and 2 s (horizontal). See Fig. S6 for additional examples of tip-FRAP experiments. (F) Analysis of the presence of D1bLIC–GFP, mSc–IFT54 or both unbleached proteins in the first postbleach retrograde trains during tip-FRAP. Kymograms shown are representative of four experiments and results in C,F are for n=472 and 35 trains, respectively.
FP-tagged IFT54 alters anterograde train structure causing reduced binding of IFT dynein
As the precise position of the individual IFT proteins in the trains remains unknown, we attempted to localize the FP tags in anterograde trains of an ift140-1 IFT140–mC (mCherry) ift54-2 mNG–IFT54 strain using cryo-electron microscopy. While this endeavor was not successful, we noticed an increased number of anterograde trains lacking IFT dynein (Fig. 7; Fig. S7). In detail, 28% (n=98) of anterograde trains of ift140-1 IFT140–mC ift54-2 mNG–IFT54 did not carry any dynein, in contrast to the 6% (n=245) of such trains observed in wild-type cilia (Jordan et al., 2018). Interestingly, the dynein coverage of the dynein-carrying trains of the double-mutant double-rescue strain showed an IFT-B:IFT dynein ratio of 1:0.20 (n=70), which is close to the ratio of 1:0.25 (n=58) observed in wild type. This agrees with the live imaging data showing both IFT dynein-carrying and IFT dynein-devoid anterograde trains in the d1blic D1bLIC–GFP ift54-2 mSc–IFT54 flagella. Western blot analysis of a similar ift140-1 IFT140–GFP ift54-2 mSc–IFT54 strain showed moderately reduced amounts of D1bLIC and DHC1b, supporting the conclusion that the presence of these tags interferes with dynein loading onto anterograde trains (Fig. S5D). Analysis of the contact sites between IFT-B and dynein in subtomogram averages of ift140-1 IFT140-mC ift54-2 mNG-IFT54 trains showed a reduced connection between neighboring IFT-B particles compared to that seen in the wild-type structure (Fig. 7A,B). The missing connecting density between IFT-B particles of the ift140-1 IFT140–mC ift54-2 mNG–IFT54 strain also corresponds to one of the contact points between IFT-B and IFT dynein in wild-type trains (Fig. 7B). Generally, the IFT-B average obtained from the tagged strain was not as well resolved as in wild type, indicating distortions introduced by the FP tags. Thus, the tags alter the structure of the IFT-B polymer in the ift140-1 IFT140–mC ift54-2 mNG–IFT54 strain affecting the binding of IFT dynein to anterograde trains and its import into flagella.
Fig. 7.
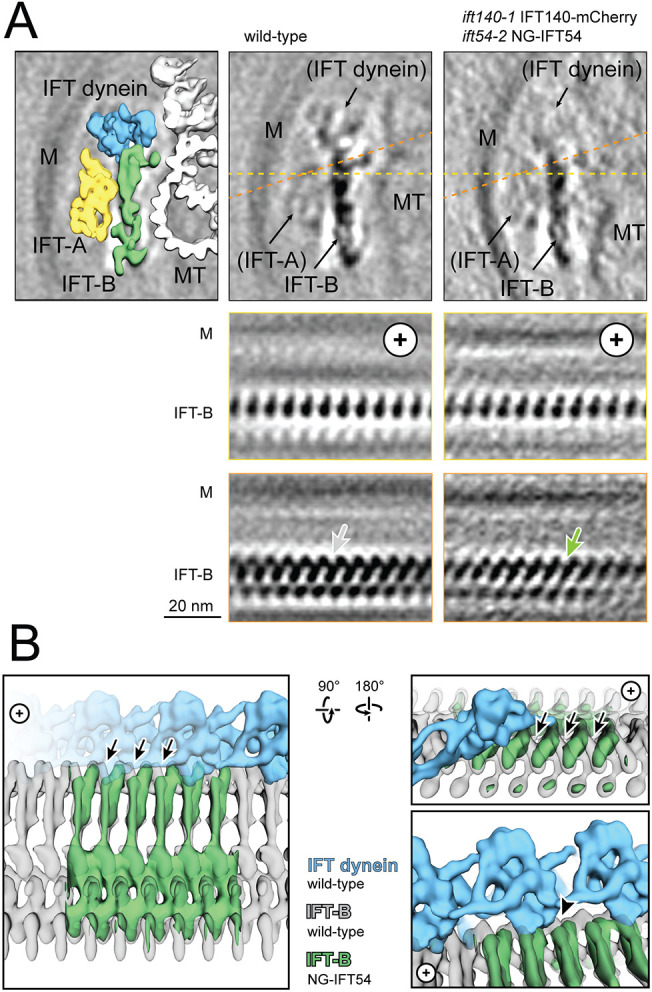
Reduction of IFT dynein in strains expressing tagged IFT particle proteins. (A) Comparison of IFT-B particles in control and tagged strains by cryo-EM. The top panels show cross sections and the orange and yellow lines indicate the position for sections shown in the lower panels. In the yellow sections, the IFT-B particles share overall similarity. Close to the IFT dynein binding side, the connection between neighboring particles is weakened in the mutant (green arrow). M, membrane; MT, microtubule; +, direction of the tip. Images are based on one experiment, analyzing 4508 mutant particles (from 98 IFT trains from 33 tomograms) and 2172 wild-type particles (from 57 IFT trains from 16 tomograms). (B) 3D Illustration of the missing connection (arrows) between IFT-B particles in the mutant compared to wild type (green, IFT-B from the mutant; gray, IFT-B from wild-type; blue, IFT dynein from wild-type). The density that connects neighboring IFT-B particles is also a contact side to IFT dynein (arrowhead), which could explain the reduced binding of IFT dynein. Left panel, view from the membrane towards the axoneme; upper right panel, longitudinal view as in A (orange); lower right panel, close up view of IFT dynein and IFT-B interaction, view as on the left; IFT-A is not shown.
DISCUSSION
FP-tagging of IFT proteins often causes subtle changes in IFT
To study the conversion of IFT trains at the flagellar tip, we employed live imaging of FP-tagged IFT proteins in Chlamydomonas. Expression of these tagged proteins restored IFT and flagellar assembly in corresponding loss-of-function mutants, indicating the principal functionality of the fusion proteins (Brown et al., 2015; Picariello et al., 2019; Reck et al., 2016; Wingfield et al., 2017; Zhu et al., 2017). However, expression of the N-terminally FP-tagged IFT54 and IFT74 resulted in the loss of IFT dynein from a subset of anterograde trains, reducing the amount of IFT dynein in flagella. In anterograde trains, IFT dynein is tightly attached as a cargo to the layer of IFT-B particles with the large IFT dynein complexes stretching over seven to eight IFT-B complexes forming numerous heterogenous IFT-B–IFT dynein contacts (Jordan et al., 2018; Toropova et al., 2019). The FP tags could sterically interfere with IFT dynein binding to IFT-B, reducing its presence on the trains. Indeed, IFT dynein import into flagella was particularly affected when FP tags on IFT54 or IFT74 were combined with a tag on D1bLIC, perhaps indicating that these proteins neighbor each other. Notably, our data are in line with a recent study showing that IFT54 directly interacts with IFT dynein and that small deletions in the coiled-coil domain of IFT54 reduce IFT dynein entry into flagella (Zhu et al., 2021). Furthermore, our cryo-EM analysis indicates that the FP tags cause a more general distortion of the IFT-B layer, probably weakening IFT-B–IFT dynein interaction. Reck et al. previously noted a reduced frequency of retrograde IFT as visualized by DIC imaging in the d1blic D1bLIC–GFP rescue strain compared to wild-type, also suggesting that FP tagging can cause subtle changes in IFT (Reck et al., 2016). Considering the dense construction of the IFT trains (Jordan et al., 2018), the positions that allow for the addition of large tags without interfering with train architecture could be limited, putting a cautionary tale on the use of multiple such tags to study the interactions between IFT proteins and complexes (Katoh et al., 2015). Placement of fluorescent protein tags informed by protein and train structure, longer linkers between the fluorescent tag and the target protein, and smaller tags such as the cysteine-tag in combination with a fluorescent payload or the split GFP technique are alternatives approaches, which could reduce tag-induced IFT anomalies (Backer et al., 2007; Kamiyama et al., 2016).
Anterograde trains lacking IFT dynein have also been observed in wild-type cells (Jordan et al., 2018) and, in C. elegans, IFT dynein frequently dissociates from moving anterograde trains (Mijalkovic et al., 2017). Thus, the expression of FP-tagged IFT-B proteins might merely reinforce a phenomenon already occurring in wild-type cells. Interestingly, FP tagging of IFT54 increased the share of trains lacking IFT dynein entirely whereas the other trains retained a near normal complement of motors. This suggests both interference of FP–IFT54 with the initiation of the IFT dynein layer during train assembly and cooperativity of IFT dynein binding to the IFT-B backbone (Toropova et al., 2019).
Persisting association of IFT-A, -B and IFT dynein during tip turnaround could balance IFT
IFT dynein needs to be activated at the flagellar tip while the large, highly structured anterograde trains morph into the smaller, less conspicuous retrograde trains (Stepanek and Pigino, 2016). Previous studies have established that kinesin-2, or at least its KAP subunit, is released from IFT at the tip, a process possibly involving phosphorylation of the KIF3B (also known as FLA8) subunit (Engel et al., 2009; Liang et al., 2014). It has also been shown that IFT-B1 and IFT dynein subunits dwell for several seconds at the tip, before distributing onto several retrograde trains, which consecutively return to the cell body (Chien et al., 2017; Engel et al., 2009; Qin et al., 2007; Wren et al., 2013). Here, similar dwell periods and fragmentation patterns were observed for IFT-A and IFT-B2 proteins revealing that in Chlamydomonas, the IFT-B, -A and IFT dynein complexes behave alike during tip turnaround. In C. elegans, IFT dynein, the anterograde motor OSM-3 and IFT-A U-turn almost instantaneously after arriving at the tip whereas the IFT-B protein OSM-6 pauses considerably longer, tentatively indicating that IFT-B dissociates from the other IFT complexes and mixes with material derived from later arriving trains during the formation of retrograde trains (Mijalkovic et al., 2017). This observation adds to the already known differences in IFT between C. elegans and Chlamydomonas; in the former, anterograde IFT uses two distinct anterograde motors, the BBSome participates in IFT train assembly, stabilization and tip turnaround, and the kinesin-2 motors move by retrograde IFT. Retrograde IFT of kinesin-2 and, in Bbs4-/- mutants, asynchronous movements of IFT-A and IFT-B have also been observed in mammalian cilia (Ou et al., 2005; Uytingco et al., 2019; Wei et al., 2012; Williams et al., 2014).
In Chlamydomonas, pairwise two-color imaging of subunits of the IFT-B1 and -B2 complex, the IFT-A and -B complex and IFT dynein and IFT-B1 or -B2 proteins revealed that tagged proteins from a given anterograde train typically return on the same set of retrograde trains. The most parsimonious explanation for this observation is that these IFT subcomplexes remain associated during tip turnaround. A similar pattern, however, could also result from the different complexes separating and mixing with the pool after arrival at the tip followed by reassociation with similar kinetics during the formation of retrograde trains (Chien et al., 2017). Based on the following observations, we favor a model of persistent association between multiple IFT complexes during remodeling at the tip. First, in photogate experiments, retrograde trains with stronger and weaker signals appear to emerge in apparently random order and, in two-color experiments, the strength of the IFT-A and IFT-B protein signals in the emerging retrograde trains often correlated (e.g. Fig. 4D). In photogate experiments, unbleached proteins entering the pool of bleached IFT material at the tip will be gradually diluted by the arrival of subsequent bleached trains as well as departing trains. If bleached and unbleached IFT complexes freely mingle in the pool, the signal strength of the emerging retrograde trains should progressively decline, which was not observed. Second, the mobility of the tagged IFT particle proteins and D1bLIC during tip remodeling was limited. This sets them apart from the kinesin-2 motor, which quickly diffuses into the flagellar shaft after its release from IFT (Chien et al., 2017; Engel et al., 2009). Also, IFT proteins dwelling at the tip were less mobile than the BBSome, a large protein complex, which is known to dissociate from and re-associate to IFT (Lechtreck et al., 2009; Liew et al., 2014). Finally, IFT dynein released from trains along the length of flagella quickly spreads by diffusion (Fig. S6), but remains near stationary during the tip U-turn. The diffusional escape of kinesin-2, the BBSome and IFT cargoes from the tip reveals that the tip is not an enclosed compartment in which the IFT proteins are restrained. Rather, the largely immobilized behavior of the IFT proteins at the tip suggests that the proteins remain associated in large fragments, likely encompassing several of each IFT-A, -B, and perhaps IFT dynein complexes and probably that such fragments remain attached to the axoneme or the flagellar membrane. Third, a subset of trains turned around at the tip without fragmentation, indicating that anterograde trains can morph directly into retrograde trains and that fragmentation is not an absolute prerequisite for conversion and turnaround. To summarize, our observations support a model in which anterograde trains convert directly into retrograde trains without major disassembly but that there is fragmentation into large train fragments encompassing all subcomplexes but kinesin-2 (Fig. 8). However, our observation that D1bLIC–GFP and IFT dynein occasionally separates from the IFT-A and -B backbone, especially in the presence of GFP-tagged IFT-B proteins, indicates some flexibility of the conversion process. A persistent association of the three IFT complexes would provide a simple mechanism of how cells maintain a balance of IFT material in the flagellum.
Fig. 8.
A model for IFT turnaround at the flagellar tip. This simple model suggests that (a) the compact configuration of the anterograde trains is mediated by the IFT-B backbone and requires kinesin-2 to be active and engaged with axonemal microtubules, (b) kinesin-2 is inactivated as the trains stall and/or run of the microtubular tracks. This allows the IFT-B backbone to assume a more expanded configuration. (c) The expanded configuration of the IFT-B backbone reduces inhibitory contacts with IFT dynein and/or rotates IFT dynein initiating its activation. Inactive kinesin-2 is released from the trains. (d,e) Additional portions of the IFT train expand and activated dynein engages with the A-tubule tracks. The expanded configuration could facilitate fragmentation of IFT trains. (f) Retrograde IFT commences.
A stop-and-turn model for IFT train conversion
Our data support a simple model in which IFT-A, IFT-B and, typically, IFT dynein remain mostly associated during the turnaround of IFT trains. How then is the conversion of anterograde into retrograde trains triggered and regulated? In an elegant recent study, Nievergelt and co-workers used a micro-mechanical device to compress flagella at some point along their length interrupting IFT (Nievergelt et al., 2021 preprint). Remarkably, anterograde trains reaching the site of compression paused briefly and ectopically converted into retrograde trains. This suggests that the specific structural and biochemical environment of the flagellar tip region is not essential for train conversion (Chaya and Furukawa, 2021; Louka et al., 2018). A common feature of the flagellar tip and a compression site along the flagellar shaft is that IFT trains will stall. Stalled IFT trains, often derived from anterograde trains, are commonly present in standard Chlamydomonas flagella (Stepanek and Pigino, 2016). Interestingly, such pausing trains assume an extended zigzag configuration resembling the structure of retrograde trains (Jordan et al., 2018; Stepanek and Pigino, 2016). Thus, stalling of anterograde trains at the tip or a compression site could initiate conversion into retrograde trains. Furthermore, conversion could be triggered by the trains running partially or completely off their microtubular tracks (Nievergelt et al., 2021 preprint). Such trains have been described by standard transmission electron microscopy (TEM) (Dentler, 2005; Pedersen et al., 2006) and, in tomographic analyses, such trains appear to lose the highly defined structure of anterograde trains in their distal regions, which have lost contact to the underlying doublet microtubules (Jordan et al., 2018; Nievergelt et al., 2021 preprint). Both stalling and losing contact with the track will prevent kinesin-2 from stepping and, in a hypothetical scenario, kinesin-2 activity could ensure a compressed configuration of the anterograde trains. Without kinesin-2 activity, trains will relax and assume the extended configuration of retrograde trains (Fig. 8). Perhaps, the more expanded zigzag configuration reduces the number of inhibitory IFT-B–IFT dynein contacts present in anterograde trains contributing to IFT dynein activation (Toropova et al., 2019). As the leading part of the trains will ‘derail’ first, it might convert earlier than the rest of the train, and then break off potentially explaining the observed staggered return of train fragments and the drawn-out return of fragments derived from large anterograde trains.
MATERIALS AND METHODS
Strains and culture conditions
The Chlamydomonas reinhardtii Dangeard rescue strains expressing IFT140–superfolderGFP (sfGFP), IFT43–YFP, IFT46–YFP, mNeonGreen (mNG)–IFT54, BBS4–GFP and D1bLIC–GFP (CC-4488) were previously described; the corresponding strain numbers of the Chlamydomonas Resource Center (http://www.chlamycollection.org/; RRID:SCR_014960) are added in parenthesis, when available (Lechtreck et al., 2009; Lv et al., 2017; Picariello et al., 2019; Reck et al., 2016; Wingfield et al., 2017; Zhu et al., 2017). The bald (i.e. lacking flagella) ift140-1 (CC-5490) and ift46-1 (CC-4375), have been previously described and were rescued by expressing IFT140–sfGFP or IFT46–YFP, respectively (Hou et al., 2007; Picariello et al., 2019; Zhu et al., 2017). The short flagella phenotype of ift43 and d1blic (CC-4053) mutants have also been described previously and were rescued by expressing IFT43–YFP or D1bLIC–GFP, respectively (Reck et al., 2016; Zhu et al., 2017). The bbs4-1 non-phototactic mutant was previously described and was rescued with BBS4–GFP or BBS4–mNG (Lechtreck et al., 2009; Liu and Lechtreck, 2018). The bald ift81-1 and ift74-2 mutants have been previously described (Brown et al., 2015; Kubo et al., 2016). ift81-1 was rescued as assessed by the restoration of flagellar assembly and IFT using a bicistronic construct consisting of the aphVIII selectable marker gene, conferring resistance to zeocin, and the genomic region of IFT81 fused at its N-terminus to an mNG that was codon-adapted for Chlamydomonas (Rasala et al., 2013). ift74-2 was rescued using a similar construct consisting of the aphVIII selectable marker gene and the genomic region of IFT74 fused at its N-terminus to an mScarlet-I (mSc) that was codon-adapted for mammals. As previously reported, the ift54-2 mutant lacked flagella and was obtained by insertional mutagenesis using the aph7″ cassette; it was rescued using a bicistronic construct consisting of the aphVIII selectable marker gene and the genomic region of IFT54 fused at its N-terminus to either mNG (Chlamydomonas codon usage), mSc or 2×Dendra with mammalian codon usage (Wingfield et al., 2017). For expression of FP-tagged IFT54, IFT71 and IFT81, we used a previously described expression vector in which the aphVIII selectable marker gene is separated from the transgenes by a cleavable 2A sequence (Rasala et al., 2013). As previously described, cleavage of the 2A sequence is not always complete, resulting in two or more bands of the tagged protein in western blots (Lechtreck et al., 2018). Furthermore, boiling of red fluorescence protein SDS-samples can lead to fragmentation and formation of additional bands (Gross et al., 2000). The ift140-1 IFT140–sfGFP ift54-2 mSc–IFT54 strain was obtained by mating the ift140-1 IFT140–sfGFP and the ift54-2 mSc–IFT54 strains. Motile progeny was analyzed by TIRF microscopy and the strains expressing IFT140–sfGFP and mSc–IFT54 were analyzed by western blotting using antibodies to C. reinhardtii IFT140 (Picariello et al., 2019) and IFT54 (Wingfield et al., 2017). The ift74-2 mSc–IFT74 ift81-1 mNG–IFT81 strain was obtained by mating the ift74-2 mSc-IFT74 and the ift81-1 mNG–IFT81 strain. Motile progeny was analyzed by TIRF microscopy and the strains expressing mSc–IFT74 and mNG–IFT81 were analyzed by western blotting using antibodies to C. reinhardtii IFT74 (Qin et al., 2004) and IFT81 (Cole et al., 1998). The ift54-2 mSc–IFT54 ift81-1 mNG–IFT81 strain was generated by mating the ift54-2 mSc-IFT54 and the ift81-1 mNG–IFT81 strain. Motile progeny was analyzed by TIRF microscopy and the strains expressing mSc–IFT54 and mNG–IFT81 were analyzed by western blotting using antibodies to C. reinhardtii IFT54 and IFT81. The d1blic D1bLIC–GFP ift54-2 mSc-IFT54 strain was obtained by mating the d1blic D1bLIC–GFP and the ift54-2 mSc–IFT54 strains. Motile progeny were analyzed by TIRF microscopy and strains expressing D1bLIC–GFP and mSc-IFT54 were analyzed by western blotting using antibodies to C. reinhardtii D1bLIC (Perrone et al., 2003) and IFT54. For antibody dilutions, see section on western blotting.
TIRF microscopy
For TIRF imaging, we used an Eclipse Ti-U microscope (Nikon) equipped with 60× NA1.49 TIRF objective and through-the-objective TIRF illumination provided by 40-mW 488-nm and 75-mW 561-nm diode lasers (Spectraphysics) as previously described (Lechtreck, 2013, 2016). The excitation lasers were cleaned up with a Nikon GFP/mCherry TIRF filter cube, and the emission was separated using an Image Splitting Device (Photometrics DualView2 with filter cube 11-EM). Imaging was mostly undertaken at 10 frames per second (fps) using an iXON3 (Andor) and the NIS-Elements Advanced Research software (Nikon).
To obtain a focused laser beam, the 488-nm laser beam was split using a 488 nm zero-order half-wave plate and a broad band polarized beam splitter; one of the beams was used for TIRF illumination. The other beam was expanded using a 3× beam expander, focused using 200 mm plano-convex lens and a 35 mm plano-convex lens and recombined with the TIRF laser beam using polarized beam splitter (all parts from Thorlabs Inc.). A motorized mirror connected to a joystick (Newfocus) was used to move the bleaching laser and the size of the laser spot was altered manually by moving the 35-mm lens. For Photogate experiments, the shutter and shutter driver (Uniblitz) for the bleaching laser were controlled via an Arduino Uno device and a custom-written macro for µ-Manager (https://micro-manager.org/) as described by Wingfield et al. (2017).
Observation chambers for C. reinhardtii were constructed by applying a ring of vacuum grease or petroleum jelly to a 24×60 mm no. 1.5 coverslip; 10 μl of cell suspension were applied and allowed to settle for ∼1 min. Then, the chamber was closed by inverting a 22×22 mm no. 1.5 cover glass with ∼5–10 μl of 5 mM HEPES, pH 7.3 supplemented with 3–5 mM EGTA onto the larger coverslip. Cells were imaged through the large cover glass at room temperature. FIJI (National Institutes of Health) software was used to generate kymograms using either the built-in Multi Kymogram tool or the KymoResliceWide plugin (https://imagej.net/KymoResliceWide). The Plot Profile tool was used to analyze signal intensity and Microsoft Excel was used for statistical analysis. For FRAP analysis of the IFT pool at the tip, kymograms were generated from videos showing the cells before, during and after the application of the laser pulse or bleaching phase. Grayscale profiles were plotted along a line covering the tip signal, the data were converted to Excel, and the fluorescence strength prior to the photobleaching step was set to 100%. Adobe Photoshop was used to adjust image contrast and brightness, and figures were prepared in Adobe Illustrator.
Flagellar isolation and western blotting
For western blot analyses, cells in 10 mM HEPES pH 7.4, 5 mM MgSO4 and 4% sucrose (w/v) were deflagellated by the addition of dibucaine. After removing the cell bodies by two differential centrifugations (1150 g for 3 min and 1600 g for 10 min), flagella were sedimented at 40,000 g, 20 min, 4°C as previously described (Witman, 1986). Flagella were dissolved by boiling in Laemmli SDS sample buffer, separated on Mini-Protean TGX gradient gels (BioRad), and transferred electrophoretically to PVDF membrane. After blocking, the membranes were incubated overnight in the primary antibodies; secondary antibodies were applied for 90-120 min at room temperature. After addition of the substrate (Femtoglow by Michigan Diagnostics or ECL Prime Western Blotting Detection Reagent by GE Healthcare), chemiluminescent signals were documented using a BioRad Chemi Doc imaging system. The following primary antibodies were used in this study. Polyclonals were rabbit anti-GFP (1:1000, A-11122; Thermo Fisher Scientific), anti-IFT54 (1:500; Wingfield et al., 2017), anti-IFT140 (1:1000; Picariello et al., 2019), anti-D1bLIC (1:2000; Hou et al., 2004), anti-IFT74 (1:2000; Qin et al., 2004), anti-DHC1b (1:2000; Pazour et al., 1999) and anti-NAB1 (1:5000; cat no. AS08 333, Agrisera), and monoclonals were anti-IC2 (1:1000; King and Witman, 1990), anti-IFT139 (1:200; Cole et al., 1998) and anti-IFT172 (1:100; Cole et al., 1998).
Cryo-electron tomography
Cells were applied on glow-discharged TEM grids with holey carbon support film (Quantifoil Micro Tools GmbH, R 3.5/1). 10-nm colloidal gold particles were added to the sample as fiducial markers during tomogram reconstruction. The samples were blotted from the back with Whatman filter paper, rapidly frozen in liquid ethane at −182°C using a Leica EM Grid Plunger and stored in liquid nitrogen until image acquisition.
Data were acquired with a FEI Titan Halo transmission electron microscope operated at 300 kV equipped with a FEG, a Gatan energy filter using a slit width of 20 eV, and a Gatan K2 direct detector. SerialEM software was used for the automated tomographic tilt series acquisition (Mastronarde, 2005). Full grid montages at low magnification were acquired to find suitable cilia, connected to the cell body. The nominal image magnification was 30,000×, resulting in a calibrated pixel size of 2.36 Å in super-resolution mode of the camera.
Tilt series were recorded with 2° increments with a bidirectional tilt scheme from −20° to 64° and −64° (when possible). The defocus range was −4.5 to −6 µm, and the cumulative dose was 130 to 150 e/Å2 per tomogram. Images were acquired in dose fractionation mode with frame times between 0.10 and 0.25 s.
Image processing and subtomogram averaging
The frames were aligned using K2Align software, which is based on the MotionCorr algorithm (Li et al., 2013). Tomogram reconstruction was performed with Etomo from IMOD version 4.9.3 (Kremer et al., 1996) using weighted back projection. CTF curves were estimated with CTFPLOTTER and corrected by phase-flipping with the software CTFPHASEFLIP, both implemented in IMOD (Xiong et al., 2009). The tomograms were binned six times, resulting in a pixel size of 1.41 nm. For particle picking and visualization of tomograms, a non-linear anisotropic diffusion filter by IMOD (Kremer et al., 1996) was applied to enhance the contrast of macromolecular structures.
Subtomogram averaging was performed on the unfiltered tomograms with PEET version 1.11.0 from the IMOD package (Heumann et al., 2011). IFT-B particles were picked with 6 nm spacing. The IFT-B complex and inhibited IFT dynein structure EMD-4303 (Jordan et al., 2018) was used as an initial reference for particle alignment. The references for the analysis of wild-type and ift140-1 IFT140-mC ift54-2 mNG-IFT54 structures were refined in subsequent iterations by adding more particles. To reduce the influence of the microtubule doublet, the membrane or the other repeating parts of the train structure on the alignment, loose binary masks that contained six IFT-B repeats were applied to the reference. The final averages of the wild-type structure contained 2172 particles for IFT-B. The structure of IFT-B of the ift140-1 IFT140–mC ift54-2 mNG–IFT54 strain was calculated from 4508 particles.
Visualization of tomograms and average densities was done in 3dmod from IMOD and rendering of isosurfaces was performed with UCSF Chimera (Pettersen et al., 2004).
Supplementary Material
Acknowledgements
We thank George B. Witman (University of Massachusetts Medical School, Worcester, MA, USA) for the kind gift of antibodies to IFT proteins and the Electron Microscopy facility of the MPI-CBG for expert technical support.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: K.L., G.P.; Formal analysis: J.L.W., M.J., G.P., K.L.; Investigation: J.L.W., B.M., I.M., P.L., M.J., D.D., K.L.; Data curation: J.L.W., M.J., K.L.; Writing - original draft: J.L.W., M.J., G.P., K.L.; Writing - review & editing: G.P., K.L.; Supervision: K.L.; Project administration: K.L.; Funding acquisition: G.P., K.L.
Funding
This study was supported by a grant by the National Institutes of Health (R01GM110413 to K.L.). M.J., D.D., and G.P. are supported by the Max Planck Society, the Human Technopole, and the European Research Council (ERC) under the European Union's Horizon 2020 Framework Programme research and innovation program (grant agreement No. 819826 to G.P.). B.M. was supported by an University of Georgia (UGA) CURO Research Assistantship for undergraduate researchers. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. Deposited in PMC for release after 12 months.
Peer review history
The peer review history is available online at https://journals.biologists.com/jcs/article-lookup/doi/10.1242/jcs.259010.
References
- Backer, M. V., Levashova, Z., Patel, V., Jehning, B. T., Claffey, K., Blankenberg, F. G. and Backer, J. M. (2007). Molecular imaging of VEGF receptors in angiogenic vasculature with single-chain VEGF-based probes. Nat. Med. 13, 504-509. 10.1038/nm1522 [DOI] [PubMed] [Google Scholar]
- Brown, J. M., Cochran, D. A., Craige, B., Kubo, T. and Witman, G. B. (2015). Assembly of IFT trains at the ciliary base depends on IFT74. Curr. Biol. 25, 1583-1593. 10.1016/j.cub.2015.04.060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buisson, J., Chenouard, N., Lagache, T., Blisnick, T., Olivo-Marin, J. C. and Bastin, P. (2012). Intraflagellar transport proteins cycle between the flagellum and its base. J. Cell Sci. 126, 327-338. 10.1242/jcs.117069 [DOI] [PubMed] [Google Scholar]
- Chaya, T. and Furukawa, T. (2021). Post-translational modification enzymes as key regulators of ciliary protein trafficking. J. Biochem., [Epub] mvab024. 10.1093/jb/mvab024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien, A., Shih, S. M., Bower, R., Tritschler, D., Porter, M. E. and Yildiz, A. (2017). Dynamics of the IFT machinery at the ciliary tip. eLife 6, e28606. 10.7554/eLife.28606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole, D. G. and Snell, W. J. (2009). SnapShot: intraflagellar transport. Cell 137, 784-784.e1. 10.1016/j.cell.2009.04.053 [DOI] [PubMed] [Google Scholar]
- Cole, D. G., Diener, D. R., Himelblau, A. L., Beech, P. L., Fuster, J. C. and Rosenbaum, J. L. (1998). Chlamydomonas kinesin-II-dependent intraflagellar transport (IFT): IFT particles contain proteins required for ciliary assembly in Caenorhabditis elegans sensory neurons. J. Cell Biol. 141, 993-1008. 10.1083/jcb.141.4.993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson, S. C. and House, S. A. (2010). Life with eight flagella: flagellar assembly and division in Giardia. Curr. Opin. Microbiol. 13, 480-490. 10.1016/j.mib.2010.05.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dentler, W. (2005). Intraflagellar transport (IFT) during assembly and disassembly of Chlamydomonas flagella. J. Cell Biol. 170, 649-659. 10.1083/jcb.200412021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel, B. D., Lechtreck, K.-F., Sakai, T., Ikebe, M., Witman, G. B. and Marshall, W. F. (2009). Total internal reflection fluorescence (TIRF) microscopy of Chlamydomonas flagella. Methods Cell Biol. 93, 157-177. 10.1016/S0091-679X(08)93009-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross, L. A., Baird, G. S., Hoffman, R. C., Baldridge, K. K. and Tsien, R. Y. (2000). The structure of the chromophore within DsRed, a red fluorescent protein from coral. Proc. Natl. Acad. Sci. USA 97, 11990-11995. 10.1073/pnas.97.22.11990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heumann, J. M., Hoenger, A. and Mastronarde, D. N. (2011). Clustering and variance maps for cryo-electron tomography using wedge-masked differences. J. Struct. Biol. 175, 288-299. 10.1016/j.jsb.2011.05.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou, Y., Pazour, G. J. and Witman, G. B. (2004). A dynein light intermediate chain, D1bLIC, is required for retrograde intraflagellar transport. Mol. Biol. Cell 15, 4382-4394. 10.1091/mbc.e04-05-0377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou, Y., Qin, H., Follit, J. A., Pazour, G. J., Rosenbaum, J. L. and Witman, G. B. (2007). Functional analysis of an individual IFT protein: IFT46 is required for transport of outer dynein arms into flagella. J. Cell Biol. 176, 653-665. 10.1083/jcb.200608041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa, H. and Marshall, W. F. (2011). Ciliogenesis: building the cell's antenna. Nat. Rev. Mol. Cell Biol. 12, 222-234. 10.1038/nrm3085 [DOI] [PubMed] [Google Scholar]
- Jordan, M. A., Diener, D. R., Stepanek, L. and Pigino, G. (2018). The cryo-EM structure of intraflagellar transport trains reveals how dynein is inactivated to ensure unidirectional anterograde movement in cilia. Nat. Cell Biol. 20, 1250-1255. 10.1038/s41556-018-0213-1 [DOI] [PubMed] [Google Scholar]
- Kamiyama, D., Sekine, S., Barsi-Rhyne, B., Hu, J., Chen, B., Gilbert, L. A., Ishikawa, H., Leonetti, M. D., Marshall, W. F., Weissman, J. S.et al. (2016). Versatile protein tagging in cells with split fluorescent protein. Nat. Commun. 7, 11046. 10.1038/ncomms11046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh, Y., Nozaki, S., Hartanto, D., Miyano, R. and Nakayama, K. (2015). Architectures of multisubunit complexes revealed by a visible immunoprecipitation assay using fluorescent fusion proteins. J. Cell Sci. 128, 2351-2362. 10.1242/jcs.168740 [DOI] [PubMed] [Google Scholar]
- King, S. M. and Witman, G. B. (1990). Localization of an intermediate chain of outer arm dynein by immunoelectron microscopy. J. Biol. Chem. 265, 19807-19811. 10.1016/S0021-9258(17)45444-5 [DOI] [PubMed] [Google Scholar]
- Kozminski, K. G., Johnson, K. A., Forscher, P. and Rosenbaum, J. L. (1993). A motility in the eukaryotic flagellum unrelated to flagellar beating. Proc. Natl. Acad. Sci. USA 90, 5519-5523. 10.1073/pnas.90.12.5519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozminski, K. G., Beech, P. L. and Rosenbaum, J. L. (1995). The Chlamydomonas kinesin-like protein FLA10 is involved in motility associated with the flagellar membrane. J. Cell Biol. 131, 1517-1527. 10.1083/jcb.131.6.1517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremer, J. R., Mastronarde, D. N. and McIntosh, J. R. (1996). Computer visualization of three-dimensional image data using IMOD. J. Struct. Biol. 116, 71-76. 10.1006/jsbi.1996.0013 [DOI] [PubMed] [Google Scholar]
- Kubo, T., Brown, J. M., Bellve, K., Craige, B., Craft, J. M., Fogarty, K., Lechtreck, K. F. and Witman, G. B. (2016). Together, the IFT81 and IFT74 N-termini form the main module for intraflagellar transport of tubulin. J. Cell Sci. 129, 2106-2119. 10.1242/jcs.187120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechtreck, K. F. (2013). In vivo imaging of IFT in Chlamydomonas Flagella. Methods Enzymol. 524, 265-284. 10.1016/B978-0-12-397945-2.00015-9 [DOI] [PubMed] [Google Scholar]
- Lechtreck, K. F. (2015). IFT-cargo interactions and protein transport in cilia. Trends Biochem. Sci. 40, 765-778. 10.1016/j.tibs.2015.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechtreck, K. F. (2016). Methods for Studying Movement of Molecules Within Cilia. Methods Mol. Biol. 1454, 83-96. 10.1007/978-1-4939-3789-9_6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechtreck, K.-F., Johnson, E. C., Sakai, T., Cochran, D., Ballif, B. A., Rush, J., Pazour, G. J., Ikebe, M. and Witman, G. B. (2009). The Chlamydomonas reinhardtii BBSome is an IFT cargo required for export of specific signaling proteins from flagella. J. Cell Biol. 187, 1117-1132. 10.1083/jcb.200909183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechtreck, K. F., Mengoni, I., Okivie, B. and Hilderhoff, K. B. (2018). In vivo analyses of radial spoke transport, assembly, repair and maintenance. Cytoskeleton (Hoboken) 75, 352-362. 10.1002/cm.21457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, X., Mooney, P., Zheng, S., Booth, C. R., Braunfeld, M. B., Gubbens, S., Agard, D. A. and Cheng, Y. (2013). Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. Nat. Methods 10, 584-590. 10.1038/nmeth.2472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang, Y., Pang, Y., Wu, Q., Hu, Z., Han, X., Xu, Y., Deng, H. and Pan, J. (2014). FLA8/KIF3B phosphorylation regulates kinesin-II interaction with IFT-B to control IFT entry and turnaround. Dev. Cell 30, 585-597. 10.1016/j.devcel.2014.07.019 [DOI] [PubMed] [Google Scholar]
- Liew, G. M., Ye, F., Nager, A. R., Murphy, J. P., Lee, J. S., Aguiar, M., Breslow, D. K., Gygi, S. P. and Nachury, M. V. (2014). The intraflagellar transport protein IFT27 promotes BBSome exit from cilia through the GTPase ARL6/BBS3. Dev. Cell 31, 265-278. 10.1016/j.devcel.2014.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, P. and Lechtreck, K. F. (2018). The Bardet-Biedl syndrome protein complex is an adapter expanding the cargo range of intraflagellar transport trains for ciliary export. Proc. Natl. Acad. Sci. USA 115, E934-E943. 10.1073/pnas.1713226115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louka, P., Vasudevan, K. K., Guha, M., Joachimiak, E., Wloga, D., Tomasi, R. F.-X., Baroud, C. N., Dupuis-Williams, P., Galati, D. F., Pearson, C. G.et al. (2018). Proteins that control the geometry of microtubules at the ends of cilia. J. Cell Biol. 217, 4298-4313. 10.1083/jcb.201804141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv, B., Wan, L., Taschner, M., Cheng, X., Lorentzen, E. and Huang, K. (2017). Intraflagellar transport protein IFT52 recruits IFT46 to the basal body and flagella. J. Cell Sci. 130, 1662-1674. 10.1242/jcs.200758 [DOI] [PubMed] [Google Scholar]
- Mastronarde, D. N. (2005). Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 152, 36-51. 10.1016/j.jsb.2005.07.007 [DOI] [PubMed] [Google Scholar]
- Mijalkovic, J., Prevo, B., Oswald, F., Mangeol, P. and Peterman, E. J. G. (2017). Ensemble and single-molecule dynamics of IFT dynein in Caenorhabditis elegans cilia. Nat. Commun. 8, 14591. 10.1038/ncomms14591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mijalkovic, J., van Krugten, J., Oswald, F., Acar, S. and Peterman, E. J. G. (2018). Single-molecule turnarounds of intraflagellar transport at the C. elegans Ciliary Tip. Cell Rep. 25, 1701-1707.e2. 10.1016/j.celrep.2018.10.050 [DOI] [PubMed] [Google Scholar]
- Morga, B. and Bastin, P. (2013). Getting to the heart of intraflagellar transport using Trypanosoma and Chlamydomonas models: the strength is in their differences. Cilia 2, 16. 10.1186/2046-2530-2-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay, S., Wen, X., Chih, B., Nelson, C. D., Lane, W. S., Scales, S. J. and Jackson, P. K. (2010). TULP3 bridges the IFT-A complex and membrane phosphoinositides to promote trafficking of G protein-coupled receptors into primary cilia. Genes Dev. 24, 2180-2193. 10.1101/gad.1966210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nachury, M. V., Loktev, A. V., Zhang, Q., Westlake, C. J., Peränen, J., Merdes, A., Slusarski, D. C., Scheller, R. H., Bazan, J. F., Sheffield, V. C.et al. (2007). A core complex of BBS proteins cooperates with the GTPase Rab8 to promote ciliary membrane biogenesis. Cell 129, 1201-1213. 10.1016/j.cell.2007.03.053 [DOI] [PubMed] [Google Scholar]
- Nakayama, K. and Katoh, Y. (2018). Ciliary protein trafficking mediated by IFT and BBSome complexes with the aid of kinesin-2 and dynein-2 motors. J. Biochem. 163, 155-164. 10.1093/jb/mvx087 [DOI] [PubMed] [Google Scholar]
- Nievergelt, A. P., Zykov, I., Diener, D., Buchholz, T.-O., Delling, M., Diez, S., Jug, F., S̆tĕpánek, L. and Pigino, G. (2021). Intraflagellar transport trains can turn around without the ciliary tip complex. bioRxiv, 2021.03.19.436138. 10.1101/2021.03.19.436138 [DOI] [Google Scholar]
- Ou, G., Blacque, O. E., Snow, J. J., Leroux, M. R. and Scholey, J. M. (2005). Functional coordination of intraflagellar transport motors. Nature 436, 583-587. 10.1038/nature03818 [DOI] [PubMed] [Google Scholar]
- Pazour, G. J., Dickert, B. L. and Witman, G. B. (1999). The DHC1b (DHC2) isoform of cytoplasmic dynein is required for flagellar assembly. J. Cell Biol. 144, 473-481. 10.1083/jcb.144.3.473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen, L. B., Geimer, S. and Rosenbaum, J. L. (2006). Dissecting the molecular mechanisms of intraflagellar transport in chlamydomonas. Curr. Biol. 16, 450-459. 10.1016/j.cub.2006.02.020 [DOI] [PubMed] [Google Scholar]
- Perrone, C. A., Tritschler, D., Taulman, P., Bower, R., Yoder, B. K. and Porter, M. E. (2003). A novel dynein light intermediate chain colocalizes with the retrograde motor for intraflagellar transport at sites of axoneme assembly in chlamydomonas and mammalian cells. Mol. Biol. Cell 14, 2041-2056. 10.1091/mbc.e02-10-0682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen, E. F., Goddard, T. D., Huang, C. C., Couch, G. S., Greenblatt, D. M., Meng, E. C. and Ferrin, T. E. (2004). UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605-1612. 10.1002/jcc.20084 [DOI] [PubMed] [Google Scholar]
- Picariello, T., Brown, J. M., Hou, Y., Swank, G., Cochran, D. A., King, O. D., Lechtreck, K., Pazour, G. J. and Witman, G. B. (2019). A global analysis of IFT-A function reveals specialization for transport of membrane-associated proteins into cilia. J. Cell Sci. 132, jcs220749. 10.1242/jcs.220749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piperno, G. and Mead, K. (1997). Transport of a novel complex in the cytoplasmic matrix of Chlamydomonas flagella. Proc. Natl. Acad. Sci. USA 94, 4457-4462. 10.1073/pnas.94.9.4457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter, M. E., Bower, R., Knott, J. A., Byrd, P. and Dentler, W. (1999). Cytoplasmic dynein heavy chain 1b is required for flagellar assembly in Chlamydomonas. Mol. Biol. Cell 10, 693-712. 10.1091/mbc.10.3.693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin, H., Diener, D. R., Geimer, S., Cole, D. G. and Rosenbaum, J. L. (2004). Intraflagellar transport (IFT) cargo: IFT transports flagellar precursors to the tip and turnover products to the cell body. J. Cell Biol. 164, 255-266. 10.1083/jcb.200308132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin, H., Wang, Z., Diener, D. and Rosenbaum, J. (2007). Intraflagellar transport protein 27 is a small G protein involved in cell-cycle control. Curr. Biol. 17, 193-202. 10.1016/j.cub.2006.12.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasala, B. A., Barrera, D. J., Ng, J., Plucinak, T. M., Rosenberg, J. N., Weeks, D. P., Oyler, G. A., Peterson, T. C., Haerizadeh, F. and Mayfield, S. P. (2013). Expanding the spectral palette of fluorescent proteins for the green microalga Chlamydomonas reinhardtii. Plant J. 74, 545-556. 10.1111/tpj.12165 [DOI] [PubMed] [Google Scholar]
- Reck, J., Schauer, A. M., VanderWaal Mills, K., Bower, R., Tritschler, D., Perrone, C. A. and Porter, M. E. (2016). The role of the dynein light intermediate chain in retrograde IFT and flagellar function in Chlamydomonas. Mol. Biol. Cell 27, 2404-2422. 10.1091/mbc.e16-03-0191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbaum, J. L. and Witman, G. B. (2002). Intraflagellar transport. Nat. Rev. Mol. Cell Biol. 3, 813-825. 10.1038/nrm952 [DOI] [PubMed] [Google Scholar]
- Soares, H., Carmona, B., Nolasco, S., Viseu Melo, L. and Goncalves, J. (2019). Cilia distal domain: diversity in evolutionarily conserved structures. Cells 8, 160. 10.3390/cells8020160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepanek, L. and Pigino, G. (2016). Microtubule doublets are double-track railways for intraflagellar transport trains. Science 352, 721-724. 10.1126/science.aaf4594 [DOI] [PubMed] [Google Scholar]
- Taschner, M. and Lorentzen, E. (2016). The intraflagellar transport machinery. Cold Spring Harb. Perspect. Biol. 8, a028092. 10.1101/cshperspect.a028092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taschner, M., Weber, K., Mourão, A., Vetter, M., Awasthi, M., Stiegler, M., Bhogaraju, S. and Lorentzen, E. (2016). Intraflagellar transport proteins 172, 80, 57, 54, 38, and 20 form a stable tubulin-binding IFT-B2 complex. EMBO J. 35, 773-790. 10.15252/embj.201593164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toropova, K., Mladenov, M. and Roberts, A. J. (2017). Intraflagellar transport dynein is autoinhibited by trapping of its mechanical and track-binding elements. Nat. Struct. Mol. Biol. 24, 461-468. 10.1038/nsmb.3391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toropova, K., Zalyte, R., Mukhopadhyay, A. G., Mladenov, M., Carter, A. P. and Roberts, A. J. (2019). Structure of the dynein-2 complex and its assembly with intraflagellar transport trains. Nat. Struct. Mol. Biol. 26, 823-829. 10.1038/s41594-019-0286-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uytingco, C. R., Williams, C. L., Xie, C., Shively, D. T., Green, W. W., Ukhanov, K., Zhang, L., Nishimura, D. Y., Sheffield, V. C. and Martens, J. R. (2019). BBS4 is required for intraflagellar transport coordination and basal body number in mammalian olfactory cilia. J. Cell. Sci. 132, jcs222331. 10.1242/jcs.222331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther, Z., Vashishtha, M. and Hall, J. L. (1994). The Chlamydomonas FLA10 gene encodes a novel kinesin-homologous protein. J. Cell Biol. 126, 175-188. 10.1083/jcb.126.1.175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, Q., Zhang, Y., Li, Y., Zhang, Q., Ling, K. and Hu, J. (2012). The BBSome controls IFT assembly and turnaround in cilia. Nat. Cell Biol. 14, 950-957. 10.1038/ncb2560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams, C. L., McIntyre, J. C., Norris, S. R., Jenkins, P. M., Zhang, L., Pei, Q., Verhey, K. and Martens, J. R. (2014). Direct evidence for BBSome-associated intraflagellar transport reveals distinct properties of native mammalian cilia. Nat. Commun. 5, 5813. 10.1038/ncomms6813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wingfield, J. L., Mengoni, I., Bomberger, H., Jiang, Y.-Y., Walsh, J. D., Brown, J. M., Picariello, T., Cochran, D. A., Zhu, B., Pan, J.et al. (2017). IFT trains in different stages of assembly queue at the ciliary base for consecutive release into the cilium. eLife 6, e26609. 10.7554/eLife.26609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witman, G. B. (1986). Isolation of Chlamydomonas flagella and flagellar axonemes. Methods Enzymol. 134, 280-290. 10.1016/0076-6879(86)34096-5 [DOI] [PubMed] [Google Scholar]
- Wren, K. N., Craft, J. M., Tritschler, D., Schauer, A., Patel, D. K., Smith, E. F., Porter, M. E., Kner, P. and Lechtreck, K. F. (2013). A differential cargo-loading model of ciliary length regulation by IFT. Curr. Biol. 23, 2463-2471. 10.1016/j.cub.2013.10.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong, Q., Morphew, M. K., Schwartz, C. L., Hoenger, A. H. and Mastronarde, D. N. (2009). CTF determination and correction for low dose tomographic tilt series. J. Struct. Biol. 168, 378-387. 10.1016/j.jsb.2009.08.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav, S. and Kunwar, A. (2021). Temperature-dependent activity of motor proteins: energetics and their implications for collective behavior. Front. Cell Dev. Biol. 9, 610899. 10.3389/fcell.2021.610899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, B., Zhu, X., Wang, L., Liang, Y., Feng, Q. and Pan, J. (2017). Functional exploration of the IFT-A complex in intraflagellar transport and ciliogenesis. PLoS Genet. 13, e1006627. 10.1371/journal.pgen.1006627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, X., Wang, J., Li, S., Lechtreck, K. and Pan, J. (2021). IFT54 directly interacts with kinesin-II and IFT dynein to regulate anterograde intraflagellar transport. EMBO J. 40, e105781. 10.15252/embj.2020105781 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.