ABSTRACT
The collective polarization of cellular structures and behaviors across a tissue plane is a near universal feature of epithelia known as planar cell polarity (PCP). This property is controlled by the core PCP pathway, which consists of highly conserved membrane-associated protein complexes that localize asymmetrically at cell junctions. Here, we introduce three new mouse models for investigating the localization and dynamics of transmembrane PCP proteins: Celsr1, Fz6 and Vangl2. Using the skin epidermis as a model, we characterize and verify the expression, localization and function of endogenously tagged Celsr1-3xGFP, Fz6-3xGFP and tdTomato-Vangl2 fusion proteins. Live imaging of Fz6-3xGFP in basal epidermal progenitors reveals that the polarity of the tissue is not fixed through time. Rather, asymmetry dynamically shifts during cell rearrangements and divisions, while global, average polarity of the tissue is preserved. We show using super-resolution STED imaging that Fz6-3xGFP and tdTomato-Vangl2 can be resolved, enabling us to observe their complex localization along junctions. We further explore PCP fusion protein localization in the trachea and neural tube, and discover new patterns of PCP expression and localization throughout the mouse embryo.
KEY WORDS: Celsr1, Frizzled 6, Vangl2, Epidermis, Planar cell polarity, Super resolution
Summary: This paper presents three endogenously-tagged mouse models for investigating the localization and dynamics of transmembrane planar cell polarity proteins Celsr1, Frizzled6 and Vangl2.
INTRODUCTION
Planar cell polarity (PCP) describes the coordinated alignment of cell polarity across a tissue plane, and is exemplified by polarized protrusions that emanate from epithelial surfaces such as bristles, hairs, scales and motile cilia (Apodaca, 2018; Devenport, 2016; Goodrich and Strutt, 2011; Iwasaki et al., 2018; Schweisguth, 2015; Wallingford and Mitchell, 2011). The PCP pathway, a highly conserved set of membrane-associated proteins, orients these diverse structures and directs a range of polarized behaviors including cell rearrangements and oriented cell divisions (Butler and Wallingford, 2017; Carroll and Yu, 2012; Segalen and Bellaïche, 2009; Wallingford, 2012). Landmark studies in Drosophila identified the ‘core’ PCP components through mutations that cause cuticular bristles and wing hairs to form disordered patterns (Adler, 2012; Carvajal-Gonzalez and Mlodzik, 2014; Goodrich and Strutt, 2011; Maung and Jenny, 2011). The core PCP components were later shown to have multiple orthologs in vertebrates and new, vertebrate-specific PCP functions including convergent extension, ciliogenesis and neural tube closure (Adler and Wallingford, 2017; Butler and Wallingford, 2017; Jussila and Ciruna, 2017; Nikolopoulou et al., 2017; Sutherland et al., 2020; Wang et al., 2019). Importantly, human mutations in PCP genes are associated with neural tube defects, ciliopathies and congenital heart defects, linking the PCP pathway to structural birth defects in humans (Cai and Shi, 2014; De Marco et al., 2014; Wang et al., 2019). Given the expansion of PCP functions in vertebrates and the implications of PCP in human disease, it is important to define the features that are both conserved and unique to vertebrate PCP. A necessary step toward this goal is the development of tools to investigate the cell biology of core PCP components in vertebrate model systems.
The core PCP pathway consists of a set of transmembrane and cytoplasmic proteins that asymmetrically localize to cell junctions. On one side of the cell, transmembrane protein Frizzled (Fz) colocalizes with the cytoplasmic protein Dishevelled (Dvl), while Vangl [Van Gogh (Vang) in Drosophila] and its cytoplasmic partner prickle (Pk), localize to the opposite side. A third transmembrane protein, Celsr [Flamingo (also known as Stan) in Drosophila], localizes to both sides of the cell, forms homophilic adhesions and helps to bridge opposing complexes of Fz-Dvl and Vangl-Pk across cell interfaces (Fig. 1A) (Butler and Wallingford, 2017; Devenport, 2014; Harrison et al., 2020; Strutt and Strutt, 2009; Yang and Mlodzik, 2015). Whether all vertebrate tissues exhibit these asymmetries is still unclear, in part due to limitations of the tools used for visualizing PCP distributions (Jones and Chen, 2007; Wang and Nathans, 2007). Antibodies for immunofluorescence vary in quality, and overexpression of PCP components often perturbs their localization and function. Further, PCP proteins must be expressed clonally or mosaically to determine whether their distribution is unipolar because conventional light microscopy cannot resolve the two sides of a junction (Olofsson and Axelrod, 2014). Recent advances in CRISPR-Cas9 technology, however, provide an efficient means to introduce fluorescent proteins into endogenous loci, helping to overcome the issues arising from transgenic overexpression (Gu et al., 2018). Moreover, super-resolution microscopy can resolve asymmetries across junctional interfaces without the need for clones or mosaics (Ezan and Montcouquiol, 2013; Stahley et al., 2021; Wegel et al., 2016). These technological advances provide an opportunity to clarify ambiguities and conflicting reports regarding PCP protein localization in the vertebrate literature.
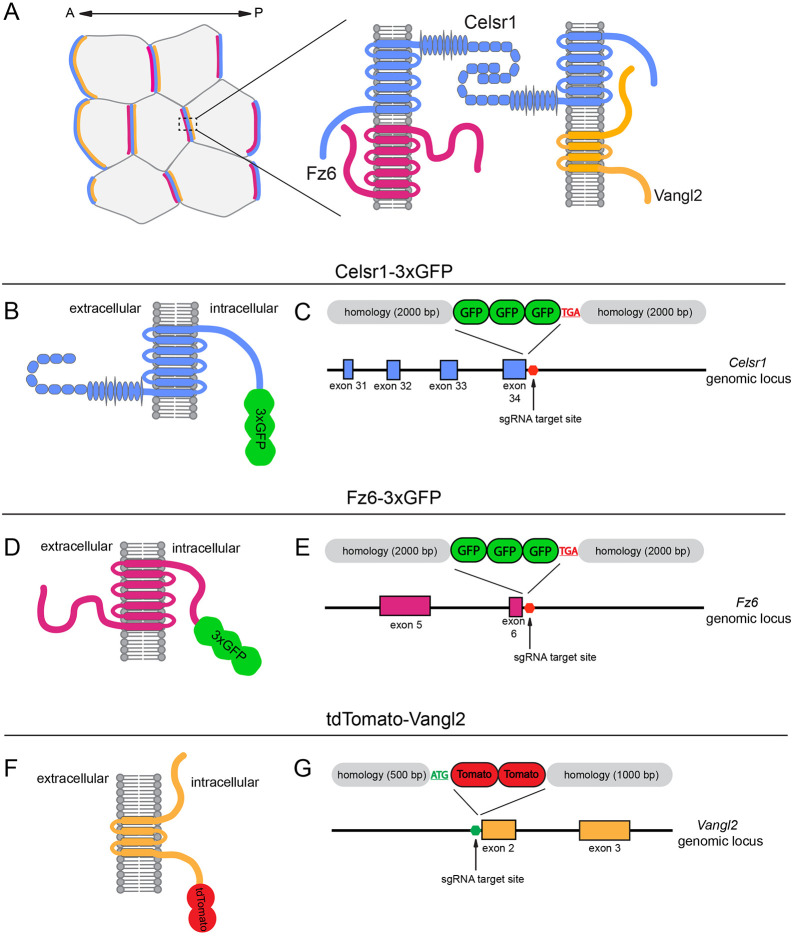
Fig. 1.
Strategy for endogenously tagging Celsr1, Fz6 and Vangl2. (A) Schematic of the proposed localization of the core transmembrane PCP proteins at the cellular and junctional level. (B,D,F) Diagram of Celsr1-3xGFP (B), Fz6-3xGFP (D) and tdTomato-Vangl2 (F) fusion proteins illustrating domain structure and the position of insertion. (C,E) Targeting arm design for the C-terminal Celsr1-3xGFP (C) and Fz6-3xGFP (E) insertion. The Celsr1 and Fz6 stop codons targeted by the sgRNA and the C-terminal exons of the Celsr1 (blue) and Fz6 (magenta) genomic region are shown. (G) Targeting arm design for N-terminal fusion of tdTomato-Vangl2. The Vangl2 start codon targeted by the sgRNA and the N-terminal exons (orange) of the Vangl2 genomic region are shown.
During polarization, PCP proteins redistribute from initially uniform distributions (Devenport, 2014). Live imaging studies in Drosophila were the first to monitor PCP protein dynamics over the course of polarization. These studies revealed directed transport of Fz and Dvl-containing membrane vesicles toward the proximal side of wing epithelial cells (Matis et al., 2014; Shimada et al., 2006). Using fluorescence recovery after photobleaching (FRAP) in the Drosophila wing, PCP proteins were shown to assemble into highly stable punctate assemblies at polarized junctions, whereas non-punctate regions displayed higher mobility and turnover (Strutt et al., 2016, 2011). Differential stability of PCP proteins along the axis of asymmetry has also been observed in Xenopus epidermis (Butler and Wallingford, 2015; Chien et al., 2015) and neural plate (Butler and Wallingford, 2018), as well as in the mouse oviduct (Shi et al., 2016), suggesting that the regulation of clustering and aggregation of PCP complexes is an important, conserved mechanism in establishing PCP. Studies of PCP protein dynamics in mouse are much more limited, in part because of the difficulties associated with live imaging embryonic development in utero and because of a lack of mouse models suitable for visualizing PCP proteins in living tissue.
Following polarization, asymmetrically localized PCP proteins act on downstream, cytoskeletal factors to polarize epithelial protrusions such as wing hairs, bristles or cilia (Devenport, 2016). In vertebrates, PCP asymmetry also induces large-scale convergent extension movements through cell intercalation driven by polarized cell crawling and junctional remodeling (Huebner and Wallingford, 2018; Shindo et al., 2018; Sutherland et al., 2020; Tada and Heisenberg, 2012). In the Xenopus neuroectoderm, for example, enrichment of PCP proteins at cell junctions correlates with actomyosin recruitment and junction shrinkage to drive convergent extension (Butler and Wallingford, 2018). The impact of cell rearrangements, where junctions must dissolve and reform, on PCP asymmetry is unknown. Is asymmetry continually refined and reestablished during cell rearrangements? Furthermore, how is the long-range coordinated asymmetry of PCP proteins preserved in highly proliferative tissues as cells round up, divide and establish new junctions with their neighbors?
Here, we use 2C-HR-CRISPR (Gu et al., 2018) to introduce fluorescent tags in the endogenous loci of the three core transmembrane PCP components, Celsr1, Fz6 and Vangl2. We show that Celsr1-3xGFP, Fz6-3xGFP and tdTomato-Vangl2 strains are viable and display bright PCP protein labeling from their endogenous loci. We use live imaging of the mouse epidermis to show how, despite extensive cell divisions and cell rearrangements throughout the tissue, the average planar polarity of epidermal progenitor cells is maintained through continuous, local repolarization events. We further show, using super-resolution imaging, how these tools allow us to resolve the asymmetric partitioning of Fz6-3xGFP and tdTomato-Vangl2 to opposite sides of cell junctions without the use of chimeras or mosaic expression. Finally, we discover new patterns of PCP gene expression and protein localization throughout the mouse embryo.
RESULTS
Generating endogenously tagged Celsr1-3xGFP, Fz6-3xGFP and tdTomato-Vangl2 mouse lines
To generate mouse strains that express fluorescently-tagged PCP proteins from their endogenous loci, we used 2C-HR-CRISPR to insert 3xGFP at the intracellular C-terminus of Celsr1 and Fz6 (Fig. 1B,D), and tdTomato at the N-terminus of Vangl2 (Fig. 1F) (Gu et al., 2020b). All three are multipass transmembrane proteins but, unlike Celsr1 and Fz6, Vangl2 is a four-pass transmembrane protein with cytosolic domains at both N- and C-termini. The N-terminus of Vangl2 was chosen for tagging to avoid interfering with a highly conserved C-terminal PDZ-binding motif (Bailly et al., 2018; Kibar et al., 2001b; Murdoch et al., 2001; Park and Moon, 2002; Torban et al., 2004; Katoh and Katoh, 2005). We inserted 3xGFP in frame and upstream of Celsr1 and Fz6 stop codons (Fig. 1C,E), and tdTomato in frame and downstream of the Vangl2 start codon (Fig. 1G). Founder mice were screened for GFP or tdTomato insertion, outcrossed and sequenced to ensure correct and mutation-free targeting of the fluorescent tag. Celsr1-3xGFP and Fz6-3xGFP knock-in lines were homozygous viable and fertile, and did not display any known PCP phenotypes. By contrast, although heterozygous tdTomato-Vangl2 mice were viable and fertile, homozygous animals at postnatal stages displayed curly tails, hermaphroditism and head shaking behaviors. At embryonic stages, tdTomato-Vangl2 homozygotes presented variably and incompletely penetrant neural tube closure defects including curly tails, spina bifida, anencephaly or, most severely, craniorachischisis. These phenotypes were less severe than those caused by loss-of-function mutations in the Vangl2 gene (Kibar et al., 2001a,b; Murdoch et al., 2001; Torban et al., 2008), but nevertheless they indicate that fusion of tdTomato to the N-terminal cytoplasmic domain does impair some Vangl2 functions.
Celsr1-3xGFP, Fz6-3xGFP and tdTomato-Vangl2 show proper localization and function in the embryonic epidermis
To verify that Celsr1-3xGFP, Fz6-3xGFP, and tdTomato-Vangl2 fusion proteins were functional, we characterized their expression, localization and function in the skin epidermis where the PCP pathway directs the orientation of hairs across the skin surface. PCP proteins are expressed in basal cells of the interfollicular epidermis (IFE), the progenitors that give rise to hair follicles and the outer stratified skin layers (Chang et al., 2016; Devenport and Fuchs, 2008). As early as embryonic day (E) 11.5, PCP proteins become asymmetrically localized and, over the course of development, asymmetry increases in magnitude and alignment, peaking at around E15.5 (Aw et al., 2016; Devenport and Fuchs, 2008). The polarized distribution of PCP proteins specifies the direction of hair follicles, which bud from polarized basal cells and point in an anterior orientation (Fig. 2). Hair follicle polarity is lost or randomized in PCP mutants, and PCP proteins become uniformly distributed within cells (Cetera et al., 2017; Chang et al., 2016; Devenport and Fuchs, 2008; Guo et al., 2004; Ravni et al., 2009; Wang et al., 2006b, 2010). If Celsr1-3xGFP, Fz6-3xGFP and tdTomato-Vangl2 fusion proteins impair PCP function, we expect to observe reduced asymmetric localization and hair follicle misalignment.
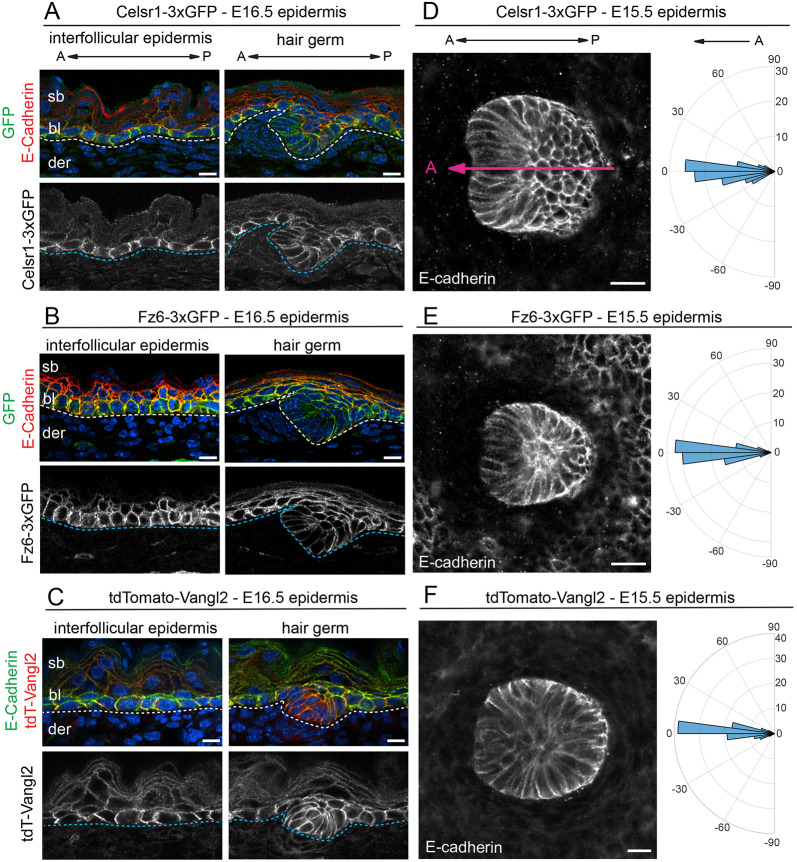
Fig. 2.
Expression and function of Celsr1-3xGFP, Fz6-3xGFP and tdTomato-Vangl2 in the embryonic skin. (A-C) Expression and localization of PCP fusion proteins in the interfollicular epidermis and developing hair follicles (HF) in E16.5 homozygous embryos. Sagittal cryosections were labeled with E-cadherin and Hoechst to mark nuclei (blue). (A) Celsr1-3xGFP (green), E-cadherin (red). (B) Fz6-3xGFP (green), E-cadherin (red). (C) tdTomato-Vangl2 (red), E-cadherin (green). Dotted lines indicate position of the epidermal-dermal boundary. Note that Celsr1-3xGFP, Fz6-3xGFP and tdTomato-Vangl2 are expressed primarily in the basal layer. (D-F) HF orientations at E15.5. Planar views of single HFs of flat-mount epidermis with E-cadherin are shown. Quantification of HF alignment is shown in circular histograms. Anterior is to the left (at 0°). Vectors point toward the anterior, growing end of HFs. (D) Celsr1-3xGFP, n=184, three embryos. Magenta line overlaid on the HF indicates orientation, where the anterior is marked by low E-cadherin expression. (E) Fz6-3xGFP homozygous, n=361, three embryos. (F) tdTomato-Vangl2 homozygotes with a curly tail and closed neural tube phenotype (CNT), n=434, three embryos. Note that in all three homozygous lines, HFs point anteriorly and align along the A-P axis, similar to their orientations in wild-type epidermis (Fig. S1). bl, basal layer; der, dermis; sb, suprabasal layer. Scale bars: 10 µm (A-C); 20 µm (D-F).
In sagittal views of E16.5 epidermis, Celsr1-3xGFP Fz6-3xGFP, and tdTomato-Vangl2 fusion proteins were expressed in the basal layer of the IFE and developing hair follicles (Fig. 2A-C), similar to their previously reported expression in wild-type epidermis using immunofluorescence (Devenport and Fuchs, 2008). Nascent hair follicles emerged from the epithelium with an anterior-directed tilt, and displayed normal growth and morphology (Fig. 2A-C). To quantify hair follicle polarization, flat-mounted E15.5 backskins were imaged and tiled to capture hundreds of hair follicles across the skin surface and the angle of hair follicle growth was measured relative to the anterior-posterior (A-P) axis. In all three lines, developing hair follicles pointed anteriorly and collectively aligned along the A-P axis, and their distributions were indistinguishable from wild-type epidermis (Fig. 2D-F; Fig. S1). Notably, in homozygous tdTomato-Vangl2 embryos, hair follicles were properly aligned even in embryos with neural tube closure defects, indicating that although the fusion protein is partially impaired in the neural tube, it is largely functional in the skin (Fig. S1D,E).
To characterize PCP protein localization and asymmetry in the epidermis, we compared the distributions of Celsr1-3xGFP, Fz6-3xGFP and tdTomato-Vangl2 at E15.5 to the distribution of their corresponding untagged proteins in control embryos. We used automated segmentation (see Materials and Methods) to detect basal cell edges using E-cadherin, and calculated the nematic order of Celsr1-3xGFP, Fz6-3xGFP and tdTomato-Vangl2 distributions. Compared with E-cadherin, which was localized uniformly at basal cell edges, endogenously-tagged PCP proteins were enriched along A-P junctions (Fig. 3A-E). Importantly, their polarized distributions were similar in magnitude and collective alignment to their untagged counterparts in control embryos (Fig. 3F-H). Moreover, the distribution of other PCP components was unaffected in each of the endogenously-tagged mouse lines (Fig. S2A-D). For example, Vangl2, the asymmetric localization of which relies on Celsr1 (Devenport and Fuchs, 2008; Stahley et al., 2021), was correctly polarized in Celsr1-3xGFP epidermis (Fig. S2A) and was comparable with Vangl2 localization in wild-type controls (Fig. 3H). Again, despite the variable neural tube defects in tdTomato-Vangl2 homozygous embryos, we did not observe a difference in tdTomato-Vangl2 distribution in the epidermis of embryos with open or closed neural tubes (Fig. 3C,D) and the distribution in both was comparable with the untagged protein in control embryos (Fig. 3H).
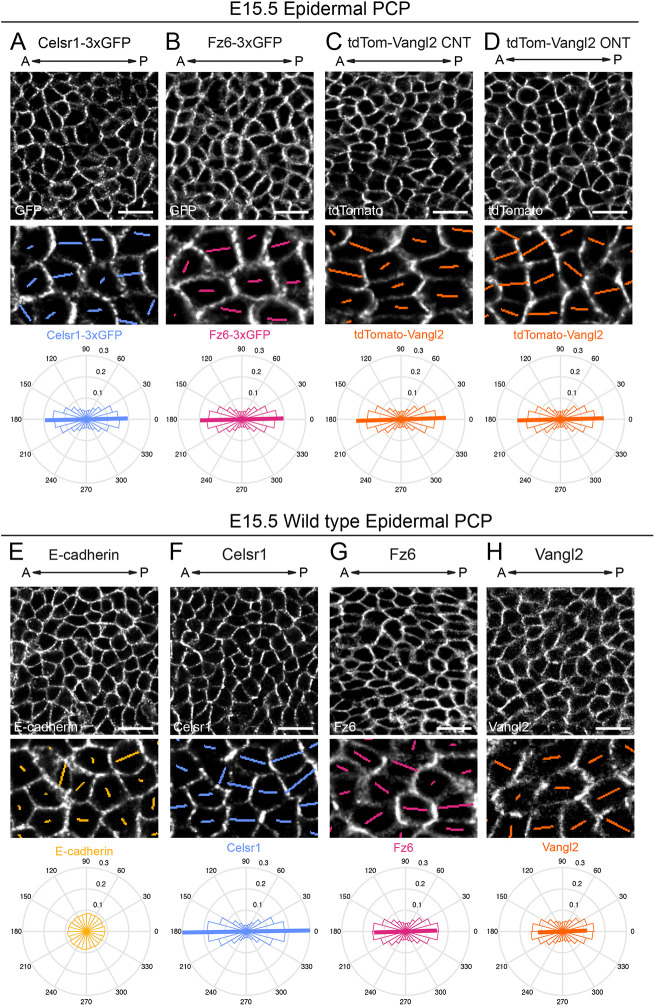
Fig. 3.
Planar polarized distribution of Celsr1-3xGFP, Fz6-3xGFP and tdTomato-Vangl2 in the epidermis. (A-H) Representative planar views of the basal layer of the interfollicular epidermis in E15.5 epidermis showing fluorescently-tagged PCP protein distributions in homozygous embryos (A-D) and Celsr1, Fz6, Vangl2 and E-cadherin localization in wild-type epidermis as detected by immunofluorescence against the endogenous untagged proteins (E-H). Anterior is to the left. Magnified areas below are overlaid with colored lines representing the axis (line angle) and magnitude (line length) of polarity. Quantification of polarity distributions are displayed below on circular histograms. (A) Celsr1-3xGFP, n=13,605 basal cells, three embryos. (B) Fz6-3xGFP, n=18,665 basal cells, three embryos. (C) tdTomato-Vangl2 in embryos displaying a CNT phenotype (CNT), n=10,775 basal cells, three embryos. (D) tdTomato-Vangl2 in embryos displaying an open neural tube (ONT) phenotype, n=9884 basal cells, three embryos. (E) E-cadherin, n=12,669 basal cells, three embryos. Note that compared with endogenous and tagged PCP proteins, E-cadherin is uniformly distributed at cell junctions. (F) Celsr1 distribution, n=12,669 basal cells, three embryos. (G) Fz6 distribution, n=8162 basal cells, three embryos. (H) Vangl2 distribution, n=9300 basal cells, three embryos. Scale bars: 20 µm.
Overall, these data indicate that fusion of 3xGFP and tdTomato onto the transmembrane PCP components does not impair their expression, localization or function in the epidermis. Moreover, proper expression and localization of tdTomato-Vangl2 in the basal layer, along with the lack of hair follicle phenotypes, indicates that although the tdTomato insertion interferes with Vangl2 function in the neural tube, it does not appear to affect its function in the epidermis, even when homozygous.
Resolving Fz6-3xGFP and tdT-Vangl2 asymmetry across cell junctions using super-resolution imaging
We next sought to determine whether our endogenously tagged PCP lines could be used to improve resolution of Fz6 and Vangl2 asymmetry across junctions with super-resolution microscopy. We have recently shown that endogenous Fz6 and Vangl2 labeled with commercially available antibodies could be resolved using structured illumination microscopy (SIM) (Stahley et al., 2021). However, the 100 nm maximum lateral resolution of SIM was insufficient to consistently resolve the two proteins at all junctions, especially in thicker skin samples (later than E14.5) where background and out-of-plane fluorescence interfered. Therefore, we turned to stimulated emission depletion (STED) super-resolution microscopy, which can achieve less than 50 nm resolution (Wegel et al., 2016). In addition, as the Fz6 antibody epitope lies in its extracellular domain, we reasoned that fluorescent tags inserted at the cytoplasmic termini of Fz6 and Vangl2 would increase the distance between fluorescent signals and improve their resolution.
Focusing on A-P junctions that are enriched for PCP proteins (Fig. 4A), we compared standard confocal and STED microscopy on dual-labeled Fz6-3xGFP; tdTomato-Vangl2 epidermis. Even with conventional confocal imaging, some separation between the GFP and td-Tomato signal was detectable, and this was significantly improved with STED (Fig. 4B; Fig. S3A). Using STED, we observed Fz6-3xGFP and tdTomato-Vangl2 organized into punctate assemblies along A-P junctions (Fig. 4B,C; Fig. S3A,B), consistent with their organization in Drosophila and with our previous observations using SIM (Cho et al., 2015; Stahley et al., 2021; Strutt et al., 2016, 2011). We generated surface plots of Fz6-3xGFP and tdTomato-Vangl2 intensity along a junction and identified puncta as peaks of fluorescence intensity (Fig. 4D; Fig. S3C; Movie 1, peaks labeled with black and gray arrowheads). Fz6-3xGFP and tdTomato-Vangl2 puncta were often paired along A-P junctions and, depending on the position of Fz6-3xGFP (F) and tdTomato-Vangl2 (V) relative to the A-P axis, we categorized puncta pairs as having an F-V or V-F orientation. We found that the mean fraction of paired puncta in the F-V orientation was higher (69.4%) than puncta in the V-F orientation (30.6%) (Fig. 4E). To test whether the F-V directional bias of puncta pairs was specific to A-P oriented junctions, we performed similar analysis on horizontally aligned mediolateral (M-L) junctions, where PCP proteins accumulate at lower levels (see Fig. 3B-D; Aw et al., 2016). Fz6-3xGFP and tdTomato-Vangl2 were more diffusely localized along M-L junctions, but punctate assemblies could still be observed (Fig. 4F-H). Puncta pairs were identified (Fig. 4H; Movie 2) and categorized as having an F-V or V-F orientation relative to the M-L axis. However, we found no bias in puncta orientation relative to the M-L axis (Fig. 4I). Together, these results demonstrate that super-resolution microscopy coupled with endogenously-tagged PCP protein reporters can resolve Fz6 and Vangl2 unipolar asymmetric localizations. Moreover, these results show that although PCP protein localization is often depicted as smooth and unipolar along a junction, their submicron organization is more complex.
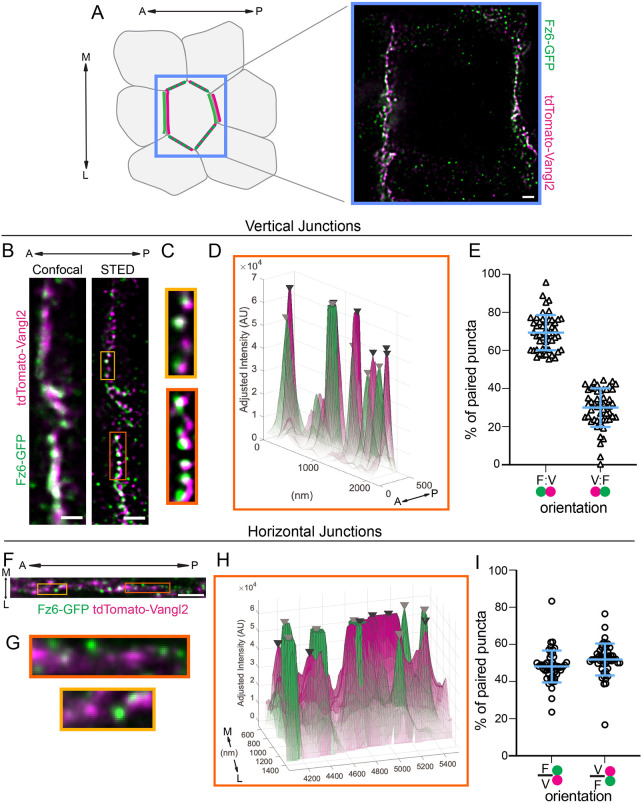
Fig. 4.
Super-resolution (STED) imaging of Fz6-3xGFP and tdTomato-Vangl2 resolves their asymmetric localization. (A-I) Whole-mount E15.5 Fz6-3xGFP/Fz6-3xGFP; tdTomato-Vangl2/+ skins labeled for GFP and tdTomato and imaged using stimulated emission depletion microscopy (STED). Anterior is oriented to the left. (A) Schematic of PCP localization in basal epidermal cells (left). Anterior-posterior (A-P; vertical) and mediolateral (M-L; horizontal) junctions of a single cell are highlighted. STED image of a full cell with Fz6-3xGFP (green) and tdTomato-Vangl2 (magenta) (right). Note that horizontal junctions appear less bright and punctate than vertical junctions. (B-I) STED imaging and analysis of vertical (B-E) and horizontal (F-I) junctions. (B,F) Representative planar view of Fz6-3xGFP (green) and tdTomato-Vangl2 (magenta) localization at a vertical junction imaged using standard confocal imaging (left) and STED (right) (B) and at a horizontal junction (F). (C,G) Magnified regions of the junction corresponding to the boxed areas in B and F. (D,H) Corresponding surface plots of GFP and tdTomato fluorescence intensity along the junction of the magnified region outlined in orange in B and F. Black and gray arrowheads mark tdTomato-Vangl2 and Fz6-3xGFP peaks, respectively. Opacity of the plot corresponds to pixel intensity, with low opacity indicating low pixel intensity and high opacity indicating high pixel intensity. Note in horizontal junctions (H) that the non-peak signal is more diffuse with higher opacity compared with vertical junctions (D). (E) Quantification of the orientation of puncta pairs in vertical junctions. The percent of puncta pairs along a junction in the F-V orientation (where Fz6-3xGFP localizes to the anterior of each puncta pair) versus the V-F orientation, n=50 junctions, three embryos. (I) Quantification of the orientation of puncta pairs in horizontal junctions. The percent of puncta pairs along a junction in the F-V orientation (where Fz6-3xGFP localizes to the medial side of the pair) versus V-F orientation, n=51 junctions, three embryos. Blue bar overlay indicates the mean±s.d. across all junctions. Scale bars: 1 µm.
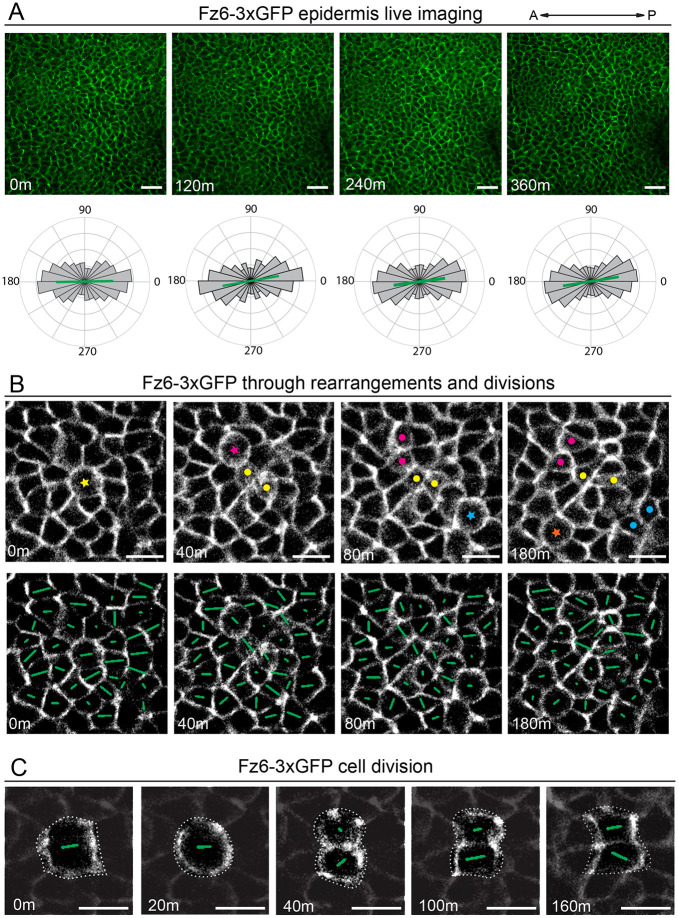
Live imaging in the epidermis reveals PCP asymmetry is maintained through extensive repolarization events
Basal progenitor cells of the epidermis rapidly proliferate to expand the progenitor pool and to produce the skin suprabasal stratified layers (Hardman et al., 1998; Muroyama and Lechler, 2012). Despite the cell shape changes in mitosis and the rearrangements accompanying daughter-cell cleavage, the asymmetry and collective alignment of PCP proteins is preserved across the tissue (Devenport et al., 2011; Shrestha et al., 2015). Using the fusion PCP reporters generated here, we were able to monitor the dynamics of PCP asymmetry through epithelial divisions and rearrangements.
To capture the dynamics of epidermal PCP, live skin explants from E15.5 Fz6-3xGFP or tdTomato-Vangl2 embryos were cultured at an air-liquid interface and z-stacks were captured every 20 min for ∼6 h (Fig. 5; Fig. S4; Movies 3 and 4) (Cetera et al., 2018). Both reporters displayed bright junctional labeling and minimal photobleaching. Focusing our analysis on Fz6-3xGFP, we quantified asymmetry over the entire field of view at 0, 2, 4 and 6 h time points. Over this time period, the average orientation and magnitude of polarity across the field of view was unchanged (Fig. 5A). However, when smaller, local regions of polarity were monitored over shorter time intervals, it became clear that, although the global polarity of the tissue is preserved through time, the polarity of individual cells and connected groups of cells is highly dynamic (Fig. 5B; Fig. S5; Movie 5). Fluctuations in junction length and formation of multicellular vertices were associated with shifts in the direction and/or the magnitude of polarity (Fig. S5A). Cell divisions displayed the largest changes in polarity, which occurred with high frequency. Fig. 5B shows a time course of Fz6-3xGFP localization in a group of about 50 cells during which four cell divisions occur. Each cell division is associated with local shifts in Fz6-3xGFP polarity within the dividing cell, its daughters and in connected neighbors. Consistent with our previous observations in fixed tissue, the magnitude of polarity was diminished in mitosis and cytokinesis (Devenport et al., 2011; Heck and Devenport, 2017). Following abscission, Fz6-3xGFP asymmetry in daughter cells steadily increased (Fig. 5B,C; Fig. S5B,C). We monitored Fz6-GFP distribution across individual mitoses and found that, although the angle of polarity was sometimes perfectly preserved from mother to daughter cells, other divisions resulted in significant deviation from the original polarity axis (Fig. 5C; Fig. S5B,C; Movie 6). Over time, as the polarity of some cells rotated away from the A-P axis, the polarity of other cells became better aligned with it, thereby preserving global Fz6-GFP polarity and alignment across the tissue.
Fig. 5.
Fz6-3xGFP localization is highly dynamic during cell rearrangements and divisions yet the global axis of asymmetry is maintained. (A) Still frames from live imaging of Fz6-3xGFP homozygous E15.5 skin explants at 0, 2, 4 and 6 h time points. Circular histograms below show quantification of Fz6-3xGFP polarity across the entire field of view. Note the orientation and magnitude of the polarity is maintained over the time course. (B) Still frames of Fz6-3xGFP live imaging during cell divisions and rearrangements. Top panels: a colored star indicates cells in mitosis and a pair of dots after cytokinesis. Bottom panels: same still frames as above with polarity nematics overlaid. Note that Fz6-3xGFP polarity within individual cells is not fixed through time, rather polarity changes in magnitude and orientation, especially with division. (C) Representative example of Fz6-3xGFP during cell division. Polarity nematics are overlaid on the dividing cell (dashed outline), and neighboring cells are darkened to emphasize Fz6-3xGFP in the dividing cell. Note, in this example daughter cells share the same axis of asymmetry as the mother cells before mitosis. Scale bars: 40 µm (A); 20 µm (B,C).
Characterization of Celsr1-3xGFP, Fz6-3xGFP and tdTomato-Vangl2 in a multiciliated tissue: the adult mouse trachea
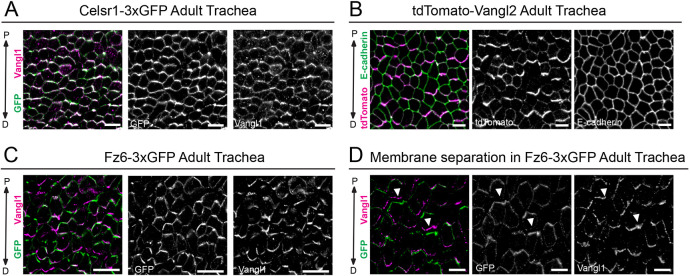
One of the best characterized functions for PCP in vertebrate systems is to direct the polarized beating of cilia in multiciliated cells (MCCs) (Meunier and Azimzadeh, 2016; Ohata and Alvarez, 2016; Wallingford and Mitchell, 2011). In mouse, MCCs produce directional fluid flow in the upper airways, the oviduct and brain ventricles. PCP is essential for the rotational alignment of cilia basal bodies within individual MCCs, as well as for the coordinated alignment of MCC polarity across the tissue (Boutin et al., 2014; Guirao et al., 2010; Mitchell et al., 2009; Shi et al., 2014; Usami et al., 2021; Vladar et al., 2012, 2015, 2016). In PCP mutants, loss of directed ciliary beating results in fluid buildup in the brain (hydrocephalus), impaired airway clearance and infertility (Ohata et al., 2014; Shi et al., 2014; Tissir et al., 2010; Vladar et al., 2012). The polarized localization of core PCP components has been well documented in the mouse trachea and in primary mouse trachea epithelial cells in culture (MTECs) (Vladar et al., 2012, 2015, 2016). These studies showed asymmetric localization of Vangl1 and Fz6 along the proximal-distal (P-D) axis. They further inferred from the position of Pk2 that Vangl2 is positioned on the distal side, whereas Fz6 localizes to the proximal side (Vladar et al., 2012). Celsr1 and Vangl2 localization have not been reported in the trachea in vivo, but their localization in MTECs is consistent with core PCP protein distributions described in the Drosophila wing: Celsr1 shows bipolar asymmetry along the P-D axis, whereas Vangl2 is unipolar and on the same side of the cell as Pk2 (Vladar et al., 2012).
Confirming previous findings, we found that endogenously tagged Fz6-3xGFP was strongly polarized along the P-D axis in the adult trachea (Fig. 6C). In addition, Celsr1-3xGFP showed collective alignment at P-D cell edges where it overlapped with polarized Vangl1 (Fig. 6A; Fig. S6A,B). The localization of Celsr1-3xGFP and Fz6-3xGFP was similar to their endogenous counterparts in wild-type adult trachea (Fig. S6C,D). tdTomato-Vangl2 was also planar polarized and aligned with the P-D axis (Fig. 6B), but we were unable to label Vangl2 in wild-type trachea to compare endogenous protein localization. E-cadherin by comparison was uniformly distributed around cell edges (Fig. 6B; Fig. S6E). Thus, similar to the polarized distribution previously reported for Fz6 and Vangl1, these data show that Celsr1 and Vangl2 are polarized in airway epithelial cells in vivo.
Fig. 6.
Polarized localization of Celsr1-3xGFP, and mutually exclusive localization of tdTomato-Vangl2 and Fz6-3xGFP to opposite cell edges in the adult trachea. (A-C) Representative planar view of flat-mounted trachea. (A) Homozygous Celsr1-3xGFP adult labeled with antibodies against GFP (green) and Vangl1 (magenta). Note that Celsr1-3xGFP and Vangl1 are asymmetrically localized at proximal-distal (P-D) junctions. Proximal is oriented up. (B) Heterozygous tdTomato-Vangl2 adult labeled with tdTomato (green) and E-cadherin (magenta). tdTomato-Vangl2 is asymmetrically localized along the P-D axis, whereas E-cadherin is uniform around cell edges. (C) Fz6-3xGFP homozygous adult labeled with GFP (green) and Vangl1 (magenta). Fz6-3xGFP and Vangl1 are polarized along the P-D axis. (D) Representative image showing mutually exclusive localization of Fz6-3xGFP (green) and Vangl1 (magenta) to opposing sides of P-D junctions in cells where membranes have separated due to methanol fixation (arrowheads indicate areas showing opposing localization across cell junctions). Scale bars: 10 µm (A,C,D); 5 µm (B).
Fortuitously, in some trachea samples, our methanol fixation protocol caused the membranes of neighboring cells to separate, allowing us to clearly distinguish the proximal and distal sides of cell junctions that are normally too close together to be resolved with conventional microscopy (Fig. 6D). In trachea from Fz6-3xGFP homozygotes co-labeled with Vangl1, the two proteins showed mutually exclusive localization to opposite sides of cell junctions. Whereas Fz6-3xGFP accumulated at the proximal edges, Vangl1 localized to the distal sides. The partitioning of Fz6-3xGFP of Vangl1 was such that the two proteins displayed almost no overlap in areas where membranes had clearly separated (Fig. 6D). These findings not only confirm previous accounts of Vangl1 and Fz6 asymmetry in the trachea, they provide conclusive evidence for the unipolar localizations of Vangl1 and Fz6 to distal and proximal cell edges, respectively.
Characterization of Celsr1-3xGFP, Fz6-3xGFP and tdTomato-Vangl2 in the early embryo: neural tube and somites
Across vertebrate species, defects in the PCP pathway severely impair neural tube closure (NTC), the process by which the neural plate bends, folds and seals into a tube that gives rise to the brain and spinal cord. In mouse, PCP mutants fail to initiate NTC resulting in craniorachischisis, the most severe form of neural tube defect (NTD) (Curtin et al., 2003; Greene et al., 1998; Hamblet et al., 2002; Kibar et al., 2001b; Murdoch et al., 2001; Wang et al., 2006a,c). Human NTD patients have been shown to carry mutations in PCP genes, including in Vangl2, Celsr1 and Fz6, closely linking the PCP pathway with human NTDs (Allache et al., 2012; De Marco et al., 2012; Juriloff and Harris, 2012; Kibar et al., 2011; Lei et al., 2010; Merello et al., 2015; Qiao et al., 2016; Wang et al., 2019). Despite the clear genetic link between PCP and NTC in vertebrates, the subcellular localization of PCP components during neurulation is only partially understood. In Xenopus, Vangl2 and Prickle2 strongly enrich on the anterior side of neuroepithelial cell junctions, where their localization correlates with junction shrinkage during convergent extension movements (Butler and Wallingford, 2018). Celsr1 localizes asymmetrically to A-P cell borders in the neural-plate of chick embryos (Nishimura et al., 2012). In mouse, Vangl2 is broadly expressed in the neuroepithelium throughout neurulation, but whether it is localized asymmetrically is unclear (Galea et al., 2018; Kibar et al., 2001b; Torban et al., 2008).
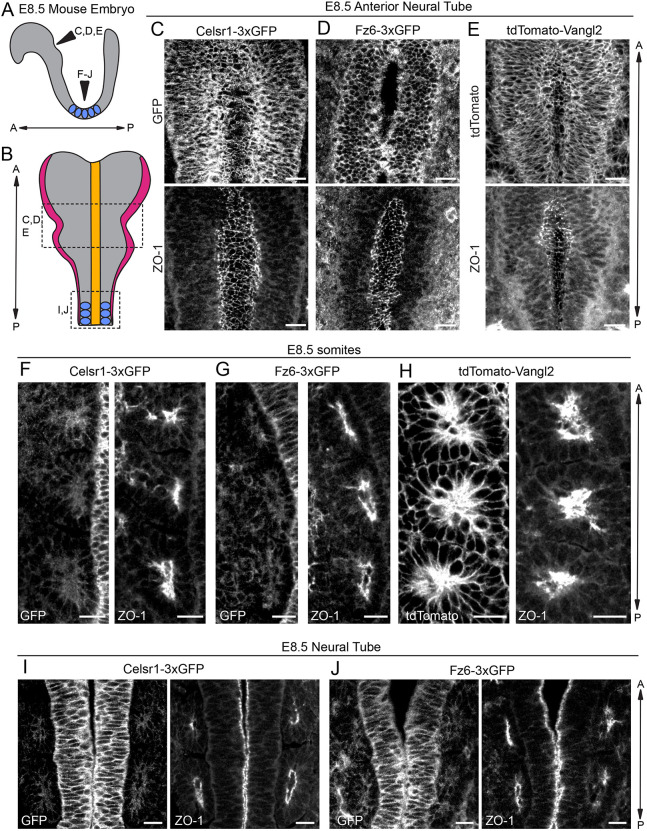
To address these questions, we examined Celsr1-3xGFP, Fz6-3xGFP and tdTomato-Vangl2 localization in E8.5 embryos at different A-P positions of the neural tube which close at different rates (Fig. 7A,B). In wild-type embryos stained for the endogenous PCP proteins, similar expression patterns to the tagged proteins were observed (Fig. S7). All three fusion proteins were expressed throughout the neural tube, and were broadly localized along the lateral edges of the pseudostratified neuroepithelial cells. Notably, Celsr1-3xGFP, Fz6-3xGFP and tdTomato-Vangl2 were not enriched at the apical surface, which was marked by co-labeling with the apical, tight junction marker ZO-1 (Tjp1). Rather, all three proteins showed reduced levels at positions of high ZO-1 accumulation (Fig. 7C-E). Focusing on the ventral surface of still open regions of the neural tube to capture PCP protein distribution along the epithelial plane, we were unable to detect clear asymmetric localization of any of the PCP fusion proteins. Based on the extensive cell rearrangements and divisions that drive NTC, we suspect PCP asymmetries are highly dynamic and transient and may only be detectable by live imaging. Alternatively, mosaic expression of the fusion proteins may be required to reveal asymmetries of PCP proteins during neural tube development. This could be achieved by generating embryo chimeras between endogenously-tagged PCP reporter and wild-type embryos to produce mosaic expression of the tagged PCP proteins.
Fig. 7.
Celsr1-3xGFP, Fz6-3xGFP and tdTomato-Vangl2 in the early embryo: neural tube and somites. (A) Schematic of lateral view of E8.5 embryo showing positions of neural tube imaged in C-J and somites (blue). (B) Schematic of dorsal view of E8.5 embryo showing neural folds and somites. Adapted from Brooks et al. (2020). (C-E) Representative images of endogenously-tagged PCP protein localization in the rostral neural tube of E8.5 homozygous embryos labeled with ZO-1 to mark the apical positions of neural epithelial cells. Maximum intensity projections of 5-8 µm are shown. (C) Celsr1-3xGFP (top), ZO-1 (bottom). (D) Fz6-3xGFP (top), ZO-1 (bottom). Note that a single plane was chosen for the Fz6-3xGFP channel to more clearly display its localization. (E) tdTomato-Vangl2 (top), ZO-1 (bottom). (F-H) Representative images showing endogenously-tagged PCP protein localization in the somites of E8.5 homozygous embryos labeled with ZO-1. Three somites from one side of the midline are shown. (F) Celsr1-3xGFP (left) and ZO-1 (right). (G) Fz6-3xGFP (left) and ZO-1 (right). (H) tdTomato-Vangl2 (left) and ZO-1 (right). Note the strong expression of tdTomato-Vangl2 in somites compared with Celsr1-3xGFP and Fz6-3xGFP. (I,J) Representative images of Celsr1-3xGFP and Fz6-3xGFP localization at a more caudal position of the neural tube that has already closed at E8.5. (I) Celsr1-3xGFP (left) and ZO-1 (right). (J) Fz6-3xGFP (left) and ZO-1 (right). Scale bars: 20 µm (C-E); 10 µm (F-J).
In imaging E8.5 embryos, we also noted that tdTomato-Vangl2 was highly expressed in the somites. Similar to its distribution in neural epithelial cells, tdTomato-Vangl2 was localized more broadly around the cell surface than ZO-1 (Fig. 7F-H). By contrast, Celsr1-3xGFP and Fz6-3xGFP were expressed at low levels in somites, especially compared with their levels in the neural tube midline (Fig. 7I-J). We saw similar expression patterns of endogenous proteins in the midline and somites of wild-type embryos, including more widespread expression of Vangl2 (Fig. S7). Thus, the neural tube defects observed in the tdTomato-Vangl2 line do not appear to be due to aberrant expression or localization of the fusion protein (Fig. S7F-H). Another marked difference between the three PCP proteins was their relative expression levels in the non-neural surface ectoderm. Fz6-3xGFP levels were notably elevated in the surface ectoderm compared with other tissues at E8.5 and E9 (Fig. S7I-J). This observation is consistent with the predominant phenotype in Fz6 mutants, in which PCP defects are mainly observed in the skin epidermis. Overall, these data highlight overlapping and distinct expression patterns of the three transmembrane core PCP proteins in the early embryo, prompting us to explore their localization patterns at later stages of organogenesis.
Whole-embryo imaging of Celsr1-3xGFP, Fz6-3xGFP and tdTomato-Vangl2: new patterns of expression and localization
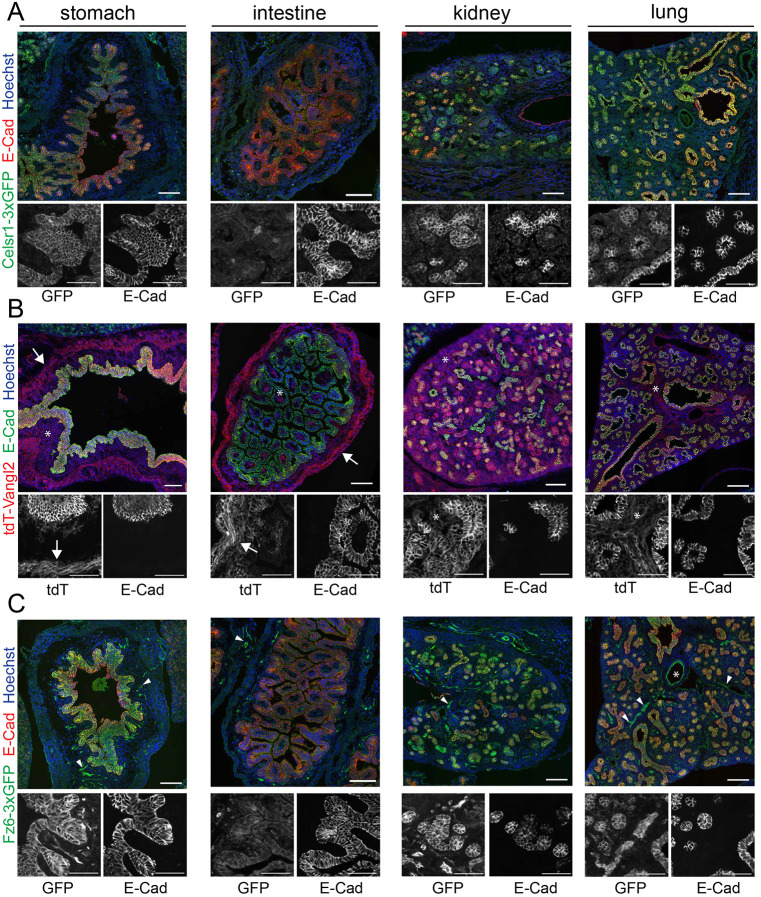
The PCP pathway functions broadly in mammalian development in a wide range of tissues, organs and cell types (Butler and Wallingford, 2017; Wang and Nathans, 2007; Yang and Mlodzik, 2015). The insertion of fluorescent reporters into endogenous PCP loci allows investigation of the localization and dynamics of Celsr1, Fz6 and Vangl2 in any tissue in which they are endogenously expressed. To explore and compare expression patterns, we surveyed Celsr1-3xGFP, Fz6-3xGFP and tdTomato-Vangl2 localization across a range of organs in sagittal sections of E16.5 embryos (Fig. 8). Overall, Celsr1-3xGFP, Fz6-3xGFP and tdTomato-Vangl2 were expressed in most epithelial tissues where they often, but not always, overlapped with E-cadherin localization. For example, PCP fusions were co-expressed and largely colocalized with E-cadherin in the lung, stomach and kidney epithelia (Fig. 8A-C). By contrast, in the intestinal epithelium, PCP protein expression was more restricted. Vangl2 and Fz6 were found at the base of intestinal villi but were only weakly expressed in or absent from the rest of the epithelium. Celsr1 was detectable only in a small number of cells (of unknown identity) scattered across the intestine (Fig. 8A-C). In certain regions of the kidney tubules, the PCP fusion proteins and E-cadherin were differentially expressed. In the liver, Fz6 was abundant and diffusely localized whereas Celsr1 and Vangl2 were only weakly expressed (Fig. 8A-C).
Fig. 8.
Celsr1-3xGFP, Fz6-3xGFP and tdTomato-Vangl2 expression and localization across embryonic organs. (A-C) Sagittal sections from Celsr1-3xGFP, tdTomato-Vangl2 and Fz6-3xGFP embryos at E16.5 labeled with E-cadherin to mark epithelia and Hoechst (blue) to mark nuclei. Composite images of E16.5 sagittal sections showing Celsr1-3xGFP (A, green), tdTomato-Vangl2 (B, red) and Fz6-3xGFP (C, green) expression in the stomach, intestine, kidney and lung. Below each merged image are zoomed-in regions of individual channels in grayscale. In B, arrows indicate expression in smooth muscle, asterisks denote expression in stromal cells. In C, arrowheads point to expression in vasculature, asterisk marks the lumen of a blood vessel in the lung. Scale bars: 100 µm (merged images); 50 µm (grayscale images).
We also observed expression patterns unique to each transmembrane PCP protein. Celsr1 appeared to be more restricted to epithelial cell types than either Fz6 or Vangl2. By contrast, Fz6 was highly expressed in the vascular endothelium. Throughout the embryo bright Fz6-3xGFP labeling was associated with the vasculature in every organ we surveyed, including in the brain and heart (arrowheads in Fig. 8C; Fig. S8). Perhaps most surprising was the widespread expression of Vangl2, especially in non-epithelial cell types (Fig. 8B). tdTomato-Vangl2 was abundant in the heart, smooth muscle surrounding the digestive tract (arrows in Fig. 8B; Fig. S8), the brain, and in stromal cells surrounding epithelial tubes like the kidney and intestine (asterisks in Fig. 8B; Fig. S8). This broad expression pattern of Vangl2 may explain why loss-of-function mutations in Vangl2 give rise to some of the strongest embryonic phenotypes of all PCP mutants. In summary, these results demonstrate the broad utility of our endogenously-tagged, fluorescent PCP reporters to investigate PCP protein localization and function across the mouse embryo. They also suggest possible functions for PCP in the vasculature, smooth muscle and stroma of numerous organs.
DISCUSSION
The PCP pathway regulates many facets of mammalian development, but the tools to investigate the cell biology of its core components have relied on transgenic overexpression and immunolabeling, with variable success. The three new mouse models described here, which express fluorescent Celsr1, Fz6 and Vangl2 fusion proteins from their endogenous loci, significantly improve upon existing tools for PCP protein localization and overcome many of the shortcomings associated with overexpression and immunofluorescence. We demonstrate that Celsr1-3xGFP, Fz6-3xGFP and tdTomato-Vangl2 mice are viable and show proper function and localization in the embryonic epidermis. These models enable direct visualization of PCP protein dynamics during development, and the Fz6-3xGFP and tdTomato-Vangl2 lines can be combined with super-resolution imaging to observe localization at cell junctions. Overall, these new tools will allow for visualization of Celsr1, Fz6 and Vangl2 localization and dynamics in virtually any mouse tissue or organ where they are normally expressed.
Using endogenously tagged Fz6-3xGFP, we were able to resolve some discrepancies in the literature regarding its localization. Fz6 was initially reported to localize asymmetrically to A-P junctions in the embryonic epidermis (Devenport and Fuchs, 2008), but this was brought into question in experiments in which Fz6 was overexpressed or deleted mosaically (Dong et al., 2018). Under these conditions, Dong et al. observed uniform Fz6 distribution. However, non-endogenous levels of PCP proteins have a well-documented impact on PCP signaling and mosaic overexpression or depletion of Fz can disrupt neighboring wild-type cell polarity non-autonomously (Goodrich and Strutt, 2011). Our data clearly demonstrate that endogenous Fz6-3xGFP is asymmetrically localized in epidermal cells along the A-P axis. More significantly, we show in both the epidermis and the trachea that Fz6 localization is unipolar. Using STED in the epidermis, we observed complex punctate organization of Fz6, where Fz6 was predominantly enriched on the anterior side of individual puncta pairs where it opposed Vangl2. In addition, using methanol fixation to cause membrane separation in the trachea, we observed mutually exclusive, unipolar localization of Fz6 and Vangl1 to opposing sides of junctions. Thus, our data resolves any ambiguity in the asymmetric and opposing localization of Fz6 and Vangl1/2 in two different epithelial mammalian tissues.
Performing live imaging over 6 h of epidermal development, we demonstrated that endogenously-tagged PCP reporters can be used to visualize PCP protein dynamics in living tissue and provided new insights into how polarity is maintained in the highly proliferative epidermis. Movies Movies 3,5,6 revealed that the local polarity of Fz6-3xGFP is constantly in flux, shifting in both orientation and magnitude as cells divide and rearrange. And yet, the average, global polarity of the tissue is preserved. We have previously shown that PCP is preserved during mitosis by removal of Celsr1, with associated Fz6 and Vangl2, from the cell surface via bulk internalization into membrane vesicles (Devenport et al., 2011; Shrestha et al., 2015). The proteins are held in endosomes until cytokinesis when their membrane localization and asymmetry are restored (Heck and Devenport, 2017). Our data capture the temporary loss and restoration of Fz6-3xGFP asymmetry through mitosis and cytokinesis, which had previously only been observed at fixed time points. We found that the polarity of daughter cells does not always perfectly align with that of the mother cell before division, suggesting that interactions with neighboring cells may be more important in directing polarity restoration after division, as opposed to the dividing cell carrying an intrinsic memory of polarity. Although the conditions required to perform long-term live imaging of the epidermis were insufficient to track endosome movements over the course of mitosis (explants are grown at an air-liquid interface, and images are captured using air objectives at 10-20 min intervals), we believe high speed imaging of the tagged lines with immersion objectives will be possible for shorter time periods and enable high resolution imaging of PCP trafficking during mitosis.
A considerable drawback of our mouse models is the tissue-specific phenotype caused by fusing tdTomato to Vangl2. When homozygous, tdTomato-Vangl2 embryos display curly tails and neural tube closure (NTC) defects with incomplete penetrance. Yet we show that tdTomato-Vangl2 is properly localized and functional in the epidermis. One possible explanation is that the tdTomato tag interferes with the rate of Vangl2 maturation, transport or turnover, which might be crucial in highly dynamic morphogenetic processes like NTC. Alternatively, the N-terminal tag may interfere with a neural tube-specific Vangl2 protein interaction. Our favored hypothesis, however, stems from the recent discovery of an alternative N-terminally extended form of Vangl2 which was reported during the preparation of this article. Vangl2-Long is generated through an alternative translational start site and appears to be conserved in all vertebrates (Walton et al., 2020 preprint). Selective depletion of Vangl2-Long in Xenopus embryos resulted in severe NTC defects, potentially explaining the phenotypes we observe in our tdTomato-Vangl2 line. The N-terminal tdTomato tag is predicted to selectively block production of Vangl2-Long, which would result in loss-of-function phenotypes specifically where the Vangl2-Long isoform is expressed. Future experiments to determine where Vangl2-Long is expressed in mouse embryos will be important for determining the utility of our tdTomato-Vangl2 line. We can say with confidence that tdTomato-Vangl2 functions properly in the skin epidermis, and based on the normal morphology of other tissues that abundantly express Vangl2 it is likely that the tdTomato fusion does not interfere with Vangl2 functions in several other tissues.
Finally, using fluorescent PCP reporters to explore Celsr1, Fz6 and Vangl2 expression and localization in several embryonic tissues and organs, we identified both overlapping and unique expression patterns for each of the three transmembrane proteins, raising new questions about PCP function in mammals. For example, Fz6, but not Celsr1 or Vangl2, is highly expressed in the vasculature, suggesting it either works with other Celsr and Vang homologs in endothelial cells, or that it may have functions that are independent of PCP. Vangl2 expression is surprisingly widespread in non-epithelial tissues including smooth muscle, cardiac muscle and stromal fibroblasts. Significantly, Celsr1 and Fz6 are not expressed in these cell types, suggesting that Vangl2 could be functioning independently of the core PCP pathway. There is some evidence that Vangl2 functions separately from the other PCP proteins in neurons (Dos-Santos Carvalho et al., 2020). Given that much of what we know about PCP function comes from epithelia, it will be important to decipher to what extent the principles of epithelial PCP, such as asymmetric localization and intercellular interactions at cell junctions, apply to non-epithelial tissues. Overall, these expression patterns indicate there are many unexplored functions for core PCP components in mammals. Our fluorescent PCP reporters should serve as broadly useful tools for investigating PCP localization and dynamics in a wide range of mouse tissues and organs.
MATERIALS AND METHODS
Generation of Celsr1-3xGFP, Fz6-3xGFP and tdTomato-Vangl2 targeting constructs
To generate the Celsr1-3xGFP and Fz6-3xGFP repair constructs, a 4 kb area surrounding the Celsr1 or Fz6 stop codons (2 kb upstream and 2 kb downstream of the stop codons) was amplified from genomic DNA prepared from keratinocytes derived from CD1 mice. This sequence was inserted into the pAAV backbone to serve as homology arms (Addgene plasmid #131777, gift from the Toettcher lab, Princeton University, NJ, USA) (Wilson et al., 2017) via InFusion cloning (Takara). 3xGFP was amplified (from plasmid fat2-3xGFP floxDsRed, gift from Sally Horne-Badovinac, University of Chicago, IL, USA) (Barlan et al., 2017) and inserted before the stop codon in the homology arm constructs of Celsr1 and Fz6 through InFusion cloning.
To generate the tdTomato-Vangl2 repair constructs, homology arms of 500 bp upstream and 1 kb downstream surrounding the Vangl2 start codon were amplified from genomic DNA from keratinocytes derived from CD1 mice. The homology arms were inserted into the pUC backbone (gift from Toettcher lab) (Ravindran et al., 2020) via InFusion cloning (Takara). tdTomato was amplified (Addgene #26771, gift from Bradley Joyce, Princeton University, NJ, USA) and inserted immediately following the start codon in the homology arm constructs of Vangl2 through InFusion cloning.
sgRNA target sequences were designed around the stop codon for C-terminal tagging of Celsr1 and Fz6 and the start codon for N-terminal tagging of Vangl2. Design of sgRNA and evaluation of off-target sites was carried out using the optimized CRISPR design tool (crispr.mit.edu). Two sgRNAs lacking predicted off-target sites in coding regions were selected for each line. If the sgRNA did not overlap with the insertion site, silent point mutations were introduced in the repair constructs. Synthetic gRNAs were either purchased from Synthego or produced in house as previously described (Gu et al., 2018). For sequences see Table S1.
Generation of mouse lines and breeding
Mice were housed in an AAALAC-accredited facility following the Guide for the Care and Use of Laboratory Animals. Animal maintenance and husbandry followed the laboratory Animal Welfare Act. Princeton University's Institutional Animal Care and Use Committee (IACUC) approved all animal procedures.
The Celsr-3xGFP, Fz6-3xGFP and tdTomato-Vangl2 repair constructs (30 ng/µl), Cas9 mRNA (100 ng/µl) and sgRNA (50 ng/µl) were prepared and injected into two-cell stage CD1 mouse embryos by E.P. as previously described (Gu et al., 2020a,b). Positive founders were determined by two sets of PCRs (described below). Founder mice were outcrossed to C57Bl/6J. N1 mice generated from these crosses were genotyped by PCR and the genomic regions spanning the homology arms were cloned into TOPO pCR4 vector (Thermo Fisher Scientific) and sequenced to ensure correct targeting and lack of mutations. N1 Celsr1-3xGFP and Fz6-3xGFP mice were outcrossed to C57Bl/6J multiple times (up to five times) and N2/N3 mice were intercrossed to generate homozygotes. tdTomato-Vangl2 mice were maintained in CD1 and C57BL/6J mixed backgrounds. Although heterozygous tdTomato-Vangl2 mice were viable and fertile, phenotypes were observed with variable penetrance when backcrossed into different backgrounds. In BL6, heterozygous mice displayed curly tails, whereas in the CD1 background, heterozygous mice displayed curly tails and hermaphroditism. Homozygous animals at postnatal stages were viable and fertile, but displayed curly tails, hermaphroditism and head shaking behaviors.
Genotyping PCRs were designed to discriminate between wild-type, heterozygous, and homozygous mice for all lines. To genotype for the 3xGFP insertion, a PCR using the forward primer upstream of the 3xGFP insert and a reverse primer within the 3xGFP were used to amplify a region only in mice with the 3xGFP insertion. Similarly, a forward primer in the homology arm upstream of the insertion and a reverse primer in the tdTomato, amplified a fragment in homozygous and heterozygous knock-in animals. For all three lines, a PCR using a primer upstream of the insert and downstream of the insert, amplified a large fragment in knock-in animals and a small wild-type fragment when the insert was absent. Heterozygous animals have both large and small fragments. For primer sequences see Table S2.
Immunofluorescence and image acquisition
For studying the embryonic epidermis, E15.5 embryos were dissected in PBS and fixed in 4% paraformaldehyde (PFA) for 1 h at room temperature. Backskins dissected from fixed embryos were blocked overnight at 4°C in 2% normal goat serum, 2% normal donkey serum (or 4% normal donkey serum and no goat serum when staining for Fz6), 1% bovine serum albumin (BSA) and 1% fish gelatin in PBT2 (PBS with 0.2% Triton X-100). Samples were incubated in primary antibody in PBT2 overnight at 4°C, including antibodies against GFP and tdTomato, as PFA fixation weakens endogenous signals. Following incubation with primary antibody, samples were washed three times in PBT2 for 30 min at room temperature and then incubated with secondary antibodies and Hoechst (Invitrogen, H1399, 1:1000) for a minimum of 2 h at room temperature or overnight at 4°C. Samples were washed in PBT2 and PBS at room temperature and mounted in Prolong Gold.
The following primary antibodies were used: guinea pig anti-Celsr1 (Devenport and Fuchs, 2008; 1:1000), rat anti-E-cadherin (1:1000, DECMA-1, Thermo Fisher Scientific, MA1-25160), rabbit anti-E-cadherin (1:250, Cell Signaling Technology, 3195), rat anti-Vangl2 (1:100, Millipore, MABN750), goat anti-Fz6 (1:400, R&D Biosystems, AF1526), chicken anti-GFP (1:2000, Abcam, ab6556) and rabbit anti-RFP (1:200, Rockland, 600-4010379). Alexa Fluor-488, -555 and -647 secondary antibodies were used at 1:2000 (Invitrogen or Jackson ImmunoResearch). For full details on antibodies and reagents see Table S3.
Images were acquired on either an inverted Nikon A1 or Nikon A1R-Si confocal microscope controlled by NIS Elements software. Plan Apo 40/1.3 NA oil immersion (Nikon) objectives were used. NIS elements software, ImageJ and Photoshop were used for image processing.
Live imaging and movie processing
Live imaging of E15.5 epidermal explants was performed as previously described (Cetera et al., 2018). Briefly, E15.5 dorsal flank skin explants were dissected in PBS and mounted dermal side down onto a 1% agarose gel with F-media containing 10% fetal bovine serum. A 35-mm air permeable Lummox membrane dish (Sarstedt) was placed on the epidermal side of the skin explants. To image, explants were cultured in a humidified imaging chamber with 5% CO2. Images were acquired using a Nikon TiE Spinning Disc with Plan Apo 20/0.75 NA air objective. Z-stacks with 2 µm step sizes were acquired at 20 min intervals for 7-8 h. Cells close to the edge of the explants were not imaged to avoid differences in planar cell polarity that could occur as a result of a wound healing response. ImageJ was used for movie processing. Drift that occurred during time-lapse imaging was corrected using the ImageJ plugin, MultiStackReg.
Image segmentation and polarity analysis
Image segmentation for the whole-mount embryonic epidermis of epithelial cell edges was performed using an implementation of U-Net (Ronneberger et al., 2015), a convolutional neural network (CNN) architecture that has been shown to be very effective for image segmentation problems. For training the CNN we used six images of whole-mount embryonic epidermis generated as described above. We separated the channels into 8-bit images of E-cadherin+Vangl2, E-cadherin, Vangl2 and GFP to train the CNN on 24 total images. Post-training, one cell segmentation mask was generated using the Ecadherin marker and hand corrected using Tissue Analyzer (Aigouy et al., 2016). The masks produced from the CNN were post-processed in a MATLAB-based GUI application to produce binary masks. The post-processing included user-controlled thresholding for generating a binary mask, trimming of incomplete edges and removing small objects left over from thresholding. The same mask was applied as the output label for all other channels in the image.
Segmentation of live imaged Fz6-3xGFP embryonic epidermis was carried out using Tissue Analyzer. Segmentation of tracheal cells was carried out using Tissue Analyzer and E-cadherin to mark cell edges (Aigouy et al., 2010).
Polarity was calculated using Packing Analyzer V2 software as previously described (Aigouy et al., 2010). The software calculates the axis and magnitude (nematic order) of PCP protein distribution along cell edges, which are defined by binary masks generated by automated segmentation as described above. Data were plotted in circular histograms using MATLAB. The average polarity magnitude, as defined in Aigouy et al. (2016), was plotted on the circular histograms to indicate both angle and strength of polarity. Before polarity analysis, images were rotated up to 15° to align them with the A-P axis.
STED imaging and image analysis
E15.5 backskins were dissected, fixed and stained as described above. Primary antibodies used were chicken anti-GFP (1:2000, Abcam, ab6556) and rabbit anti-RFP (1:200, Rockland, 600-4010379). Samples were incubated in secondary antibodies in PBT2 overnight at 4°C. Secondary antibodies used were goat anti-chicken STAR ORANGE (1:100, Abberior: STORANGE-1005) and goat anti-rabbit STAR RED (1:100, Abberior: STRED-1002). Skins were mounted in Prolong Gold antifade reagent (Molecular Probes).
Images were acquired using a Nikon A1R-HD25 confocal microscope coupled to an inverted Ti-2 stand with a STEDYCON stimulated emission depletion (STED) module. STED images were acquired with a Plan Apo 100×1.49 NA oil-immersion objective (Nikon). Z-stacks were acquired with 0.25 µm steps. NIS elements software, NIS-Elements AR analysis, ImageJ and Adobe Photoshop were used for image deconvolution and processing.
The x-y resolution for the STARRED and STARORANGE antibodies was acquired by imaging single molecules of the secondary antibodies smeared on a slide and mounted in Prolong Gold, similar to our epidermis samples. We determined the point spread function of the signal from single molecules of antibody. We found a mean full width at half maximum (FWHM) across 12 molecules of 48.7 nm in the x-axis and a mean of 51.1 nm in the y-axis for the STARRED antibody. The STARORANGE antibody showed a reduction in resolution, with a mean of 57.8 nm in the x-axis and 59.1 nm in the y-axis across 17 molecules. Based on these values, our following analyses assume an x-y resolution of 65 nm.
Binary masks were generated in Fiji in order to identify the junctions for analysis and exclude any obvious cytoplasmic signal. These masks, along with images of full junctions, were acquired and imported into MATLAB 2021a for analysis, with masks being applied to full junction images by image multiplication. To identify the center of puncta, the imregionalmax function was used to identify local maxima in the image. Local maxima which were below three standard deviations of the mean intensity of the image were filtered out. Peaks within 3.25 pixels (resolution divided by pixel size) of another peak of the same channel were combined by averaging their x- and y-coordinates and intensity values. Puncta of Fz6-3xGFP and tdTomato-Vangl2 peaks were then paired, with a cut off of 220 nm, to account for peaks with no obvious corresponding peak of the other channel. A given peak can be paired with any number of peaks from the other channel; however, pairs are unique. All MATLAB scripts are available at https://github.com/PrincetonUniversity/DevenportLab-2DSegmentation.
Orientation of pairs relative to the junction orientation were then calculated. This orientation was defined by either the relative x-coordinate for vertical junctions or y-coordinate for horizontal junctions. For vertical junctions, if the x-coordinate of the Fz6-3xGFP peak was less than or equal to that of the tdTomato-Vangl2, it was defined as being in the F-V orientation. If greater than, then the pair was defined as being in the V-F orientation. For horizontal junctions, if the y-coordinate of the Fz6-3xGFP peak was greater than or equal to that of the tdTomato-Vangl2, it was defined as being in the V-F orientation. If less than, then the pair was defined as being in the F-V orientation.
To further visualize this STED microscopy data, for both stained channels, image intensity values were stretched to saturate the bottom and top 1% of pixel values using the imadjust function in MATLAB, to increase contrast. These adjusted image data were then used to generate a surface plot for each image using the surf function in MATLAB. The previously calculated peaks were then plotted on the surface plots using the scatter3 function in MATLAB. To aid in visualization, each point on the surface plot was assigned a transparency based on its corresponding pixel intensity: low values had high transparency and high values had low transparency. Rotating videos of these plots were generated using the CaptureFigVid package (Jennings, 2021).
Trachea dissection and immunostaining
Tracheas were dissected from adult mice and fixed in 100% methanol at −20°C for 10 min or in 4% PFA for 1 h at 4°C. Tracheas were then washed in PBS, blocked overnight in 4% normal donkey serum, 1% BSA and 1% fish gelatin in PBT2. Tracheas were incubated with primary antibody in blocking buffer overnight at 4°C, washed with PBT2 at room temperature, incubated with secondary antibody overnight, and then washed in PBT2 followed by PBS and mounted in Prolong Gold. The following primary antibodies were used: guinea pig anti-Celsr1 (Devenport and Fuchs, 2008; 1:1000), rabbit anti-E-cadherin (1:250, Cell Signaling Technology, 3195), goat anti-Fz6 (1:400, R&D Biosystems, AF1526), rabbit anti-Vangl1 (1:500, Sigma-Aldrich, HPA025235-10), chicken anti-GFP (1:2000, Abcam, ab6556) and rabbit anti-RFP (1:200, Rockland, 600-4010379). Alexa Fluor-488, -555 and -647 secondary antibodies were used at 1:2000 (Invitrogen or JacksonImmunoResearch). For full details of antibodies and reagents see Table S3. GFP and tdTomato were stained against as trachea fixation in methanol and PFA weakens the endogenous signal of both GFP and tdTomato.
Images were acquired on either an inverted Nikon A1 or Nikon A1R-Si confocal microscope controlled by NIS Elements software. Plan Apo 40× 1.3 NA oil immersion (Nikon) and Plan Apo 60×1.40 NA oil immersion (Nikon) objectives were used. NIS elements software, ImageJ and Photoshop were used for image processing.
Neural tube E8.5 immunostaining
E8.5 embryos were dissected in M2 media as previously described (Xiao et al., 2018). Briefly, the pregnant female was sacrificed, E8.5 embryos were removed from the abdomen, and uterine muscles were removed to free the decidua. A longitudinal incision through the decidua was made to separate the embryo from the future placenta. A portion of the yolk sac was removed for genotyping. E8.5 embryos were fixed in 4% paraformaldehyde for 45 min at room temperature and blocked overnight at 4°C in 2% normal goat serum, 2% normal donkey serum (or 4% normal donkey serum and no goat serum when staining for Fz6), 1% BSA and 1% fish gelatin in PBT2. Samples were incubated in primary antibody in PBT2 overnight at 4°C. Following incubation with primary antibody, samples were washed in PBT2 at room temperature and then incubated with secondary antibodies and Hoechst (Invitrogen, H1399, 1:1000) overnight at 4°C. Samples were washed in PBT2 and PBS at room temperature and mounted in Prolong Gold.
The following primary antibodies were used: chicken anti-GFP (1:2000, Abcam, ab6556), rabbit anti-RFP (1:200, Rockland, 600-4010379) and rat anti-ZO-1 (1:100, Developmental Studies Hybridoma Bank, R26.4C). The following secondary antobodies were used: Alexa Fluor-488, -555 and -647 at 1:2000 (Invitrogen or Jackson ImmunoResearch).
Images were acquired on an inverted Nikon A1 confocal microscope controlled by NIS Elements software. Plan Apo 40×1.3 NA oil immersion (Nikon) objectives were used. Images were acquired using 1 µm z-steps. NIS Elements software, ImageJ and Photoshop were used for image processing.
E16.5 embedding, sectioning, and staining
E16.5 embryos were dissected in PBS by removing the tail for genotyping and making 3-5 mm incisions on the ventral side of the embryo to expose internal organs. Embryos were fixed in 4% PFA for 1.5 h at 4°C, washed three times in PBS for at least 15 min at 4°C, and then washed additionally in PBS overnight at 4°C. Embryos were infused with 20% sucrose in PBS for 1 h at 4°C, in 30% sucrose in PBS for 1 h at 4°C, in 1:1 30% sucrose:OCT for 1 h at 4°C, and lastly in 1:1 30% sucrose:OCT overnight at 4°C. Embryos were embedded in OCT on dry ice and stored at −80°C.
Frozen OCT-embedded embryos were cryosectioned on a Leica CM3050 cryostat. Full embryo sagittal sections were cut at 10 µm thickness, collected on Superfrost Plus-coated slides and allowed to dry for at least 30 min before being stored at −20°C.
For immunostaining, sections were rehydrated for 10 min in PBS at room temperature in a humidified chamber on a flat surface, permeabilized in PBT (PBS with 0.1% Triton X-100) for 10 min at room temperature, and blocked in 2% normal goat serum, 2% normal donkey serum and 2% BSA in PBT for 1 h at room temperature. Slides were incubated with primary antibody in block for 1 h at room temperature: rat anti-E-cadherin (1:2000, DECMA-1, Thermo Fisher Scientific, MA1-25160), chicken anti-GFP (1:2000, Abcam, ab6556) and rabbit ant-RFP (1:500, Rockland, 600-4010379). Slides were washed three times for 10 min with PBT and incubated with secondary antibodies in PBT for 1 h at room temperature. Alexa Fluor-488, -555 and -647 secondary antibodies were used at 1:2000 (Invitrogen). Slides were washed in PBT with Hoechst (Invitrogen, H1399, 1:2000) for 10 min, followed by two 10 min washes in PBT. A final wash in PBS for 10 min was carried out before mounting in glycerol-based mounting media (90% glycerol in PBS) with anti-fade reagent and sealed with nail polish. Slides were stored at −20°C.
Images were acquired on either an inverted Nikon A1R-Si or a Nikon A1R-HD25 confocal microscope controlled by NIS Elements software. Plan Apo 60×1.40 NA oil immersion (Nikon) objectives were used. When acquiring large tissue areas, 5×5 or larger x-y regions were collected using resonance scanning with 16× averaging. Images were denoised and stitched together with 15% overlap using NIS elements software. ImageJ and Photoshop were used for further image processing.
Supplementary Material
Acknowledgements
The authors thank all those who provided resources, technical support and valuable discussions that contributed to this work. We thank all Devenport lab members for their feedback and advice, especially Liliya Leybova for her help with dissections and experimental set up. We particularly acknowledge technical support from the Model Production Core staff led by M. Gertsenstein at the Centre for Phenogenomics, Toronto. We thank Pavithran T. Ravindran for his help in construct design and cloning assistance. We thank Gary Laevsky in the Princeton University Confocal Imaging Facility, a Nikon Center of Excellence, for imaging support, expertise and assistance.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: L.P.B., D.D.; Methodology: L.P.B., S.V.P., M.C., E.P., D.D.; Software: R.S., A.B.; Validation: L.P.B., M.H.-O.; Formal analysis: L.P.B., R.S., A.G.; Investigation: L.P.B., M.H.-O., S.V.P., M.C., K.A.L., E.P., D.D.; Resources: A.B.; Writing - original draft: L.P.B., D.D.; Writing - review & editing: L.P.B., M.H.-O., R.S., D.D., S.V.P., K.A.L.; Visualization: L.P.B., S.V.P., R.S., D.D.; Supervision: D.D.; Funding acquisition: L.P.B., D.D.
Funding
This work was supported by the National Institute of Arthritis and Musculoskeletal and Skin Diseases under grant numbers R01 AR066070, R01 AR068320 and F31 AR077407 and the National Institute of General Medical Sciences of the National Institutes of Health under grant number T32GM007388. Deposited in PMC for release after 12 months.
Data availability
All MATLAB scripts are available at https://github.com/PrincetonUniversity/DevenportLab-2DSegmentation
Peer review history
The peer review history is available online at https://journals.biologists.com/dev/article-lookup/doi/10.1242/dev.199695
References
- Adler, P. N. (2012). The frizzled/stan pathway and planar cell polarity in the Drosophila wing. Curr. Top. Dev. Biol. 101, 1-31. 10.1016/B978-0-12-394592-1.00001-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adler, P. N. and Wallingford, J. B. (2017). From planar cell polarity to ciliogenesis and back: the curious tale of the PPE and CPLANE proteins. Trends Cell Biol. 27, 379-390. 10.1016/j.tcb.2016.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aigouy, B., Farhadifar, R., Staple, D. B., Sagner, A., Röper, J.-C., Jülicher, F. and Eaton, S. (2010). Cell flow reorients the axis of planar polarity in the wing epithelium of Drosophila. Cell 142, 773-786. 10.1016/j.cell.2010.07.042 [DOI] [PubMed] [Google Scholar]
- Aigouy, B., Umetsu, D. and Eaton, S. (2016). Segmentation and quantitative analysis of epithelial tissues. Methods Mol. Biol. 1478, 227-239. 10.1007/978-1-4939-6371-3_13 [DOI] [PubMed] [Google Scholar]
- Allache, R., De Marco, P., Merello, E., Capra, V. and Kibar, Z. (2012). Role of the planar cell polarity gene CELSR1 in neural tube defects and caudal agenesis. Birth Defects Res. A Clin. Mol. Teratol. 94, 176-181. 10.1002/bdra.23002 [DOI] [PubMed] [Google Scholar]
- Apodaca, G. (2018). Role of polarity proteins in the generation and organization of apical surface protrusions. Cold Spring Harb. Perspect. Biol. 10, a027813. 10.1101/cshperspect.a027813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aw, W. Y., Heck, B. W., Joyce, B. and Devenport, D. (2016). Transient tissue-scale deformation coordinates alignment of planar cell polarity junctions in the mammalian skin. Curr. Biol. 26, 2090-2100. 10.1016/j.cub.2016.06.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailly, E., Walton, A. and Borg, J. (2018). The planar cell polarity Vangl2 protein: from genetics to cellular and molecular functions. Semin. Cell Dev. Biol. 81, 62-70. 10.1016/j.semcdb.2017.10.030 [DOI] [PubMed] [Google Scholar]
- Barlan, K., Cetera, M. and Horne-Badovinac, S. (2017). Fat2 and Lar define a basally localized planar signaling system controlling collective cell migration. Dev. Cell 40, 467-477.e5. 10.1016/j.devcel.2017.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutin, C., Labedan, P., Dimidschstein, J., Richard, F., Cremer, H., André, P., Yang, Y., Montcouquiol, M., Goffinet, A. M. and Tissir, F. (2014). A dual role for planar cell polarity genes in ciliated cells. Proc. Natl. Acad. Sci. USA 111, E3129-E3138. 10.1073/pnas.1404988111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks, E. R., Islam, M. T., Anderson, K. V. and Zallen, J. A. (2020). Sonic hedgehog signaling directs patterned cell remodeling during cranial neural tube closure. eLife 9, e60234. 10.7554/eLife.60234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler, M. T. and Wallingford, J. B. (2015). Control of vertebrate core planar cell polarity protein localization and dynamics by Prickle 2. Development 142, 3429-3439. 10.1242/dev.121384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler, M. T. and Wallingford, J. B. (2017). Planar cell polarity in development and disease. Nat. Rev. Mol. Cell Biol. 18, 375-388. 10.1038/nrm.2017.11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler, M. T. and Wallingford, J. B. (2018). Spatial and temporal analysis of PCP protein dynamics during neural tube closure. eLife 7, e36456. 10.7554/eLife.36456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai, C. and Shi, S. (2014). Genetic evidence in planar cell polarity signaling pathway in human neural tube defects. Front. Med. 8, 68-78. 10.1007/s11684-014-0308-4 [DOI] [PubMed] [Google Scholar]
- Carroll, T. J. and Yu, J. (2012). The kidney and planar cell polarity. Curr. Top. Dev. Biol. 101, 185-212. 10.1016/B978-0-12-394592-1.00011-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvajal-Gonzalez, J. M. and Mlodzik, M. (2014). Mechanisms of planar cell polarity establishment in Drosophila. F1000prime Rep. 6, 98. 10.12703/P6-98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cetera, M., Leybova, L., Woo, F. W., Deans, M. and Devenport, D. (2017). Planar cell polarity-dependent and independent functions in the emergence of tissue-scale hair follicle patterns. Dev. Biol. 428, 188-203. 10.1016/j.ydbio.2017.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cetera, M., Leybova, L., Joyce, B. and Devenport, D. (2018). Counter-rotational cell flows drive morphological and cell fate asymmetries in mammalian hair follicles. Nat. Cell Biol. 20, 541-552. 10.1038/s41556-018-0082-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, H., Smallwood, P. M., Williams, J. and Nathans, J. (2016). The spatio-temporal domains of Frizzled6 action in planar polarity control of hair follicle orientation. Dev. Biol. 409, 181-193. 10.1016/j.ydbio.2015.10.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien, Y.-H., Keller, R., Kintner, C. and Shook, D. R. (2015). Mechanical strain determines the axis of planar polarity in ciliated epithelia. Curr. Biol. 25, 2774-2784. 10.1016/j.cub.2015.09.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho, B., Pierre-Louis, G., Sagner, A., Eaton, S. and Axelrod, J. D. (2015). Clustering and negative feedback by endocytosis in planar cell polarity signaling is modulated by ubiquitinylation of prickle. PLoS Genet. 11, e1005259. 10.1371/journal.pgen.1005259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtin, J. A., Quint, E., Tsipouri, V., Arkell, R. M., Cattanach, B., Copp, A. J., Henderson, D. J., Spurr, N., Stanier, P., Fisher, E. M.et al. (2003). Mutation of Celsr1 disrupts planar polarity of inner ear hair cells and causes severe neural tube defects in the mouse. Curr. Biol. 13, 1129-1133. 10.1016/S0960-9822(03)00374-9 [DOI] [PubMed] [Google Scholar]
- De Marco, P., Merello, E., Rossi, A., Piatelli, G., Cama, A., Kibar, Z. and Capra, V. (2012). FZD6 is a novel gene for human neural tube defects. Hum. Mutat. 33, 384-390. 10.1002/humu.21643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Marco, P., Merello, E., Piatelli, G., Cama, A., Kibar, Z. and Capra, V. (2014). Planar cell polarity gene mutations contribute to the etiology of human neural tube defects in our population. Birth Defects Res. A Clin. Mol. Teratol. 100, 633-641. 10.1002/bdra.23255 [DOI] [PubMed] [Google Scholar]
- Devenport, D. (2014). The cell biology of planar cell polarity. J. Cell Biol. 207, 171-179. 10.1083/jcb.201408039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devenport, D. (2016). Tissue morphodynamics: translating planar polarity cues into polarized cell behaviors. Semin. Cell Dev. Biol. 55, 99-110. 10.1016/j.semcdb.2016.03.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devenport, D. and Fuchs, E. (2008). Planar polarization in embryonic epidermis orchestrates global asymmetric morphogenesis of hair follicles. Nat. Cell Biol. 10, 1257-1268. 10.1038/ncb1784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devenport, D., Oristian, D., Heller, E. and Fuchs, E. (2011). Mitotic internalization of planar cell polarity proteins preserves tissue polarity. Nat. Cell Biol. 13, 893-902. 10.1038/ncb2284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong, B., Vold, S., Olvera-Jaramillo, C. and Hao, C. (2018). Functional redundancy of frizzled 3 and frizzled 6 in planar cell polarity control of mouse hair follicles. Development 145, dev168468. 10.1242/dev.168468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dos-Santos Carvalho, S., Moreau, M. M., Hien, Y. E., Garcia, M., Aubailly, N., Henderson, D. J., Studer, V., Sans, N., Thoumine, O. and Montcouquiol, M. (2020). Vangl2 acts at the interface between actin and N-cadherin to modulate mammalian neuronal outgrowth. eLife 9, e51822. 10.7554/eLife.51822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezan, J. and Montcouquiol, M. (2013). Revisiting planar cell polarity in the inner ear. Semin. Cell Dev. Biol. 24, 499-506. 10.1016/j.semcdb.2013.03.012 [DOI] [PubMed] [Google Scholar]
- Galea, G. L., Nychyk, O., Mole, M. A., Moulding, D., Savery, D., Nikolopoulou, E., Henderson, D. J., Greene, N. D. E. and Copp, A. J. (2018). Vangl2 disruption alters the biomechanics of late spinal neurulation leading to spina bifida in mouse embryos. Dis. Models Mech. 11, dmm032219. 10.1242/dmm.032219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodrich, L. V. and Strutt, D. (2011). Principles of planar polarity in animal development. Development 138, 1877-1892. 10.1242/dev.054080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene, N. D. E., Gerrelli, D., Van Straaten, H. W. M. and Copp, A. J. (1998). Abnormalities of floor plate, notochord and somite differentiation in the loop-tail (Lp) mouse: a model of severe neural tube defects. Mech. Dev. 73, 59-72. 10.1016/S0925-4773(98)00029-X [DOI] [PubMed] [Google Scholar]
- Gu, B., Posfai, E. and Rossant, J. (2018). Efficient generation of targeted large insertions by microinjection into two-cell-stage mouse embryos. Nat. Biotechnol. 36, 632-637. 10.1038/nbt.4166 [DOI] [PubMed] [Google Scholar]
- Gu, B., Gertsenstein, M. and Posfai, E. (2020a). Generation of large fragment knock-in mouse models by microinjecting into 2-cell stage embryos. Methods Mol. Biol. 2066, 89-100. 10.1007/978-1-4939-9837-1_7 [DOI] [PubMed] [Google Scholar]
- Gu, B., Posfai, E., Gertsenstein, M. and Rossant, J. (2020b). Efficient generation of large-fragment knock-in mouse models using 2-Cell (2C)-Homologous Recombination (HR)-CRISPR. Curr. Protoc. Mouse Biol. 10, e67. 10.1002/cpmo.67 [DOI] [PubMed] [Google Scholar]
- Guirao, B., Meunier, A., Mortaud, S., Aguilar, A., Corsi, J.-M., Strehl, L., Hirota, Y., Desoeuvre, A., Boutin, C., Han, Y. G.et al. (2010). Coupling between hydrodynamic forces and planar cell polarity orients mammalian motile cilia. Nat. Cell Biol. 12, 341-350. 10.1038/ncb2040 [DOI] [PubMed] [Google Scholar]
- Guo, N., Hawkins, C. and Nathans, J. (2004). Frizzled6 controls hair patterning in mice. Proc. Natl. Acad. Sci. 101, 9277-9281. 10.1073/pnas.0402802101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadley Wickham, D. N. and Thomas Lin, P. (2016). ggplot2: Elegant Graphics for Data Analysis, 1st edn. New York, USA: Springer International Publishing. [Google Scholar]
- Hamblet, N. S., Lijam, N., Ruiz-Lozano, P., Wang, J., Yang, Y., Luo, Z., Mei, L., Chien, K. R., Sussman, D. J. and Wynshaw-Boris, A. (2002). Dishevelled 2 is essential for cardiac outflow tract development, somite segmentation and neural tube closure. Development 129, 5827-5838. 10.1242/dev.00164 [DOI] [PubMed] [Google Scholar]
- Hardman, M. J., Sisi, P., Banbury, D. N. and Byrne, C. (1998). Patterned acquisition of skin barrier function during development. Development 125, 1541-1552. 10.1242/dev.125.8.1541 [DOI] [PubMed] [Google Scholar]
- Harrison, C., Shao, H., Strutt, H. and Strutt, D. (2020). Molecular mechanisms mediating asymmetric subcellular localisation of the core planar polarity pathway proteins. Biochem. Soc. Trans. 48, 1297-1308. 10.1042/BST20190404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heck, B. W. and Devenport, D. (2017). Trans-endocytosis of planar cell polarity complexes during cell division. Curr. Biol. 27, 3725-3733.e4. 10.1016/j.cub.2017.10.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huebner, R. J. and Wallingford, J. B. (2018). Coming to consensus: a unifying model emerges for convergent extension. Dev. Cell 46, 389-396. 10.1016/j.devcel.2018.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki, M., Kuroda, J., Kawakami, K. and Wada, H. (2018). Epidermal regulation of bone morphogenesis through the development and regeneration of osteoblasts in the zebrafish scale. Dev. Biol. 437, 105-119. 10.1016/j.ydbio.2018.03.005 [DOI] [PubMed] [Google Scholar]
- Jennings, A. (2021). Create Video of Rotating 3D Plot. MATLAB Central File Exchange. https://www.mathworks.com/matlabcentral/fileexchange/41093-create-video-of-rotating-3d-plot
- Jones, C. and Chen, P. (2007). Planar cell polarity signaling in vertebrates. BioEssays 29, 120-132. 10.1002/bies.20526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juriloff, D. M. and Harris, M. J. (2012). A consideration of the evidence that genetic defects in planar cell polarity contribute to the etiology of human neural tube defects. Birth Defects Res. A Clin. Mol. Teratol. 94, 824-840. 10.1002/bdra.23079 [DOI] [PubMed] [Google Scholar]
- Jussila, M. and Ciruna, B. (2017). Zebrafish models of non-canonical Wnt/planar cell polarity signalling: fishing for valuable insight into vertebrate polarized cell behavior. Wiley Interdiscip. Rev. Dev. Biol. 6, e267. 10.1002/wdev.267 [DOI] [PubMed] [Google Scholar]
- Katoh, Y. and Katoh, M. (2005). Comparative genomics on Vangl1 and Vangl2 genes. Int. J. Oncol. 26, 1435-1440. 10.3892/ijo.26.5.1435 [DOI] [PubMed] [Google Scholar]
- Kibar, Z., Underhill, D. A., Canonne-Hergaux, F., Gauthier, S., Justice, M. J. and Gros, P. (2001a). Identification of a new chemically induced allele (Lpm1Jus) at the loop-tail locus: morphology, histology, and genetic mapping. Genomics 72, 331-337. 10.1006/geno.2000.6493 [DOI] [PubMed] [Google Scholar]
- Kibar, Z., Vogan, K. J., Groulx, N., Justice, M. J., Underhill, D. A. and Gros, P. (2001b). Ltap, a mammalian homolog of Drosophila Strabismus/Van Gogh, is altered in the mouse neural tube mutant Loop-tail. Nat. Genet. 28, 251-255. 10.1038/90081 [DOI] [PubMed] [Google Scholar]
- Kibar, Z., Salem, S., Bosoi, C. M., Pauwels, E., De Marco, P., Merello, E., Bassuk, A. G., Capra, V. and Gros, P. (2011). Contribution of VANGL2 mutations to isolated neural tube defects. Clin. Genet. 80, 76-82. 10.1111/j.1399-0004.2010.01515.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei, Y.-P., Zhang, T., Li, H., Wu, B.-L., Jin, L. and Wang, H.-Y. (2010). VANGL2 mutations in human cranial neural-tube defects. N Engl. J. Med. 362, 2232-2235. 10.1056/NEJMc0910820 [DOI] [PubMed] [Google Scholar]
- Matis, M., Russler-Germain, D. A., Hu, Q., Tomlin, C. J. and Axelrod, J. D. (2014). Microtubules provide directional information for core PCP function. eLife 3, e02893. 10.7554/eLife.02893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maung, S. M. T. W. and Jenny, A. (2011). Planar cell polarity in Drosophila. Organogenesis 7, 165-179. 10.4161/org.7.3.18143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merello, E., Mascelli, S., Raso, A., Piatelli, G., Consales, A., Cama, A., Kibar, Z., Capra, V. and De Marco, P. (2015). Expanding the mutational spectrum associated to neural tube defects: literature revision and description of novel VANGL1 mutations. Birth Defects Res. A Clin. Mol. Teratol. 103, 51-61. 10.1002/bdra.23305 [DOI] [PubMed] [Google Scholar]
- Meunier, A. and Azimzadeh, J. (2016). Multiciliated cells in animals. Cold Spring Harb. Perspect. Biol. 8, a028233. 10.1101/cshperspect.a028233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell, B., Stubbs, J. L., Huisman, F., Taborek, P., Yu, C. and Kintner, C. (2009). The PCP pathway instructs the planar orientation of ciliated cells in the Xenopus larval skin. Curr. Biol. 19, 924-929. 10.1016/j.cub.2009.04.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murdoch, J. N., Doudney, K., Paternotte, C., Copp, A. J. and Stanier, P. (2001). Severe neural tube defects in the loop-tail mouse result from mutation of Lpp1, a novel gene involved in floor plate specification. Hum. Mol. Genet. 10, 2593-2601. 10.1093/hmg/10.22.2593 [DOI] [PubMed] [Google Scholar]
- Muroyama, A. and Lechler, T. (2012). Polarity and stratification of the epidermis. Semin. Cell Dev. Biol. 23, 890-896. 10.1016/j.semcdb.2012.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolopoulou, E., Galea, G. L., Rolo, A., Greene, N. D. and Copp, A. J. (2017). Neural tube closure: cellular, molecular and biomechanical mechanisms. Development 144, 552-566. 10.1242/dev.145904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura, T., Honda, H. and Takeichi, M. (2012). Planar cell polarity links axes of spatial dynamics in neural-tube closure. Cell 149, 1084-1097. 10.1016/j.cell.2012.04.021 [DOI] [PubMed] [Google Scholar]
- Ohata, S. and Alvarez-Buylla, A. (2016). Planar organization of multiciliated ependymal (E1) cells in the brain ventricular epithelium. Trends Neurosci. 39, 543-551. 10.1016/j.tins.2016.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohata, S., Nakatani, J., Herranz-Pérez, V., Cheng, J., Belinson, H., Inubushi, T., Snider, W. D., García-Verdugo, J. M., Wynshaw-Boris, A. and Alvarez-Buylla, A. (2014). Loss of Dishevelleds disrupts planar polarity in ependymal motile cilia and results in hydrocephalus. Neuron 83, 558-571. 10.1016/j.neuron.2014.06.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olofsson, J. and Axelrod, J. D. (2014). Methods for studying planar cell polarity. Methods 68, 97-104. 10.1016/j.ymeth.2014.03.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, M. and Moon, R. T. (2002). The planar cell-polarity gene stbm regulates cell behaviour and cell fate in vertebrate embryos. Nat. Cell Biol. 4, 20-25. 10.1038/ncb716 [DOI] [PubMed] [Google Scholar]
- Qiao, X., Liu, Y., Li, P., Chen, Z., Li, H., Yang, X., Finnell, R. H., Yang, Z., Zhang, T., Qiao, B.et al. (2016). Genetic analysis of rare coding mutations of CELSR1-3 in congenital heart and neural tube defects in Chinese people. Clin. Sci. 130, 2329-2340. 10.1042/CS20160686 [DOI] [PubMed] [Google Scholar]
- Ravindran, P. T., Wilson, M. Z., Jena, S. G. and Toettcher, J. E. (2020). Engineering combinatorial and dynamic decoders using synthetic immediate-early genes. Commun. Biol. 3, 436. 10.1038/s42003-020-01171-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravni, A., Qu, Y., Goffinet, A. M. and Tissir, F. (2009). Planar cell polarity cadherin Celsr1 regulates skin hair patterning in the mouse. J. Invest. Dermatol. 129, 2507-2509. 10.1038/jid.2009.84 [DOI] [PubMed] [Google Scholar]
- Ronneberger, O., Fischer, P. and Brox, T. (2015). U-Net: convolutional networks for biomedical image segmentation springerlink. In Medical Image Computing and Computer-Assisted Intervention – MICCAI 2015, pp. 234-241. Cham: Springer. [Google Scholar]
- Schweisguth, F. (2015). Asymmetric cell division in the Drosophila bristle lineage: from the polarization of sensory organ precursor cells to Notch-mediated binary fate decision. Wiley Interdiscip. Rev. Dev. Biol. 4, 299-309. 10.1002/wdev.175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segalen, M. and Bellaïche, Y. (2009). Cell division orientation and planar cell polarity pathways. Semin. Cell Dev. Biol. 20, 972-977. 10.1016/j.semcdb.2009.03.018 [DOI] [PubMed] [Google Scholar]
- Shi, D., Komatsu, K., Hirao, M., Toyooka, Y., Koyama, H., Tissir, F., Goffinet, A. M., Uemura, T. and Fujimori, T. (2014). Celsr1 is required for the generation of polarity at multiple levels of the mouse oviduct. Development 141, 4558-4568. 10.1242/dev.115659 [DOI] [PubMed] [Google Scholar]
- Shi, D., Usami, F., Komatsu, K., Oka, S., Abe, T., Uemura, T. and Fujimori, T. (2016). Dynamics of planar cell polarity protein Vangl2 in the mouse oviduct epithelium. Mech. Dev. 141, 78-89. 10.1016/j.mod.2016.05.002 [DOI] [PubMed] [Google Scholar]
- Shimada, Y., Yonemura, S., Ohkura, H., Strutt, D. and Uemura, T. (2006). Polarized transport of Frizzled along the planar microtubule arrays in Drosophila wing epithelium. Dev. Cell 10, 209-222. 10.1016/j.devcel.2005.11.016 [DOI] [PubMed] [Google Scholar]
- Shindo, A., Audrey, A., Takagishi, M., Takahashi, M., Wallingford, J. B. and Kinoshita, M. (2018). Septin-dependent remodeling of cortical microtubule drives cell reshaping during epithelial wound healing. J. Cell Sci. 131, jcs212647. 10.1242/jcs.212647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrestha, R., Little, K. A., Tamayo, J. V., Li, W., Perlman, D. H. and Devenport, D. (2015). Mitotic control of planar cell polarity by polo-like Kinase 1. Dev. Cell 33, 522-534. 10.1016/j.devcel.2015.03.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahley, S. N., Basta, L. P., Sharan, R. and Devenport, D. (2021). Celsr1 adhesive interactions mediate the asymmetric organization of planar polarity complexes. elife 10, e62097. 10.7554/eLife.62097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strutt, H. and Strutt, D. (2009). Asymmetric localisation of planar polarity proteins: mechanisms and consequences. Semin. Cell Dev. Biol. 20, 957-963. 10.1016/j.semcdb.2009.03.006 [DOI] [PubMed] [Google Scholar]
- Strutt, H., Warrington, S. J. and Strutt, D. (2011). Dynamics of core planar polarity protein turnover and stable assembly into discrete membrane subdomains. Dev. Cell 20, 511-525. 10.1016/j.devcel.2011.03.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strutt, H., Gamage, J. and Strutt, D. (2016). Robust asymmetric localization of planar polarity proteins is associated with organization into signalosome-like domains of variable stoichiometry. Cell Rep. 17, 2660-2671. 10.1016/j.celrep.2016.11.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutherland, A., Keller, R. and Lesko, A. (2020). Convergent extension in mammalian morphogenesis. Semin. Cell Dev. Biol. 100, 199-211. 10.1016/j.semcdb.2019.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tada, M. and Heisenberg, C.-P. (2012). Convergent extension: using collective cell migration and cell intercalation to shape embryos. Development 139, 3897-3904. 10.1242/dev.073007 [DOI] [PubMed] [Google Scholar]
- Tissir, F., Qu, Y., Montcouquiol, M., Zhou, L., Komatsu, K., Shi, D., Fujimori, T., Labeau, J., Tyteca, D., Courtoy, P.et al. (2010). Lack of cadherins Celsr2 and Celsr3 impairs ependymal ciliogenesis, leading to fatal hydrocephalus. Nat. Neurosci. 13, 700-707. 10.1038/nn.2555 [DOI] [PubMed] [Google Scholar]
- Torban, E., Wang, H.-J., Groulx, N. and Gros, P. (2004). Independent mutations in mouse Vangl2 that cause neural tube defects in looptail mice impair interaction with members of the Dishevelled family. J. Biol. Chem. 279, 52703-52713. 10.1074/jbc.M408675200 [DOI] [PubMed] [Google Scholar]
- Torban, E., Patenaude, A.-M., Leclerc, S., Rakowiecki, S., Gauthier, S., Andelfinger, G., Epstein, D. J. and Gros, P. (2008). Genetic interaction between members of the Vangl family causes neural tube defects in mice. Proc. Natl. Acad. Sci. USA 105, 3449-3454. 10.1073/pnas.0712126105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trichas, G., Begbie, J. and Srinivas, S. (2008). Use of the viral 2A peptide for bicistronic expression in transgenic mice. BMC Biol. 6, 40. 10.1186/1741-7007-6-40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usami, F. M., Arata, M., Shi, D., Oka, S., Higuchi, Y., Tissir, F., Takeichi, M. and Fujimori, T. (2021). Intercellular and intracellular cilia orientation is coordinated by CELSR1 and CAMSAP3 in oviduct multi-ciliated cells. J. Cell Sci. 134, jcs257006. 10.1242/jcs.257006 [DOI] [PubMed] [Google Scholar]
- Vladar, E. K., Bayly, R. D., Sangoram, A. M., Scott, M. P. and Axelrod, J. D. (2012). Microtubules enable the planar cell polarity of airway cilia. Curr. Biol. 22, 2203-2212. 10.1016/j.cub.2012.09.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vladar, E. K., Lee, Y. L., Stearns, T. and Axelrod, J. D. (2015). Observing planar cell polarity in multiciliated mouse airway epithelial cells. Methods Cell Biol. 127, 37-54. 10.1016/bs.mcb.2015.01.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vladar, E. K., Nayak, J. V., Milla, C. E. and Axelrod, J. D. (2016). Airway epithelial homeostasis and planar cell polarity signaling depend on multiciliated cell differentiation. JCI Insight 1, e88027. 10.1172/jci.insight.88027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallingford, J. B. (2012). Planar cell polarity and the developmental control of cell behavior in vertebrate embryos. Annu. Rev. Cell Dev. Biol. 28, 627-653. 10.1146/annurev-cellbio-092910-154208 [DOI] [PubMed] [Google Scholar]
- Wallingford, J. B. and Mitchell, B. (2011). Strange as it may seem: the many links between Wnt signaling, planar cell polarity, and cilia. Genes Dev. 25, 201-213. 10.1101/gad.2008011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walton, A., Revinski, D., Sergé, A., Audebert, S., Camoin, L., Puvirajesinghe, T. M., Isnardon, D., Marchetto, S., Kodjabachian, L., Bailly, E.et al. (2020). A novel Golgi-associated Vangl2 translational variant required for PCP regulation in vertebrates. bioRxiv 10.1101/2020.10.20.347385 [DOI] [Google Scholar]
- Wang, Y. and Nathans, J. (2007). Tissue/planar cell polarity in vertebrates: new insights and new questions. Development 134, 647-658. 10.1242/dev.02772 [DOI] [PubMed] [Google Scholar]
- Wang, J., Hamblet, N. S., Mark, S., Dickinson, M. E., Brinkman, B. C., Segil, N., Fraser, S. E., Chen, P., Wallingford, J. B. and Wynshaw-Boris, A. (2006a). Dishevelled genes mediate a conserved mammalian PCP pathway to regulate convergent extension during neurulation. Development 133, 1767-1778. 10.1242/dev.02347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y., Badea, T. and Nathans, J. (2006b). Order from disorder: Self-organization in mammalian hair patterning. Proc. Natl. Acad. Sci. USA 103, 19800-19805. 10.1073/pnas.0609712104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y., Guo, N. and Nathans, J. (2006c). The role of Frizzled3 and Frizzled6 in neural tube closure and in the planar polarity of inner-ear sensory hair cells. J. Neurosci. 26, 2147-2156. 10.1523/JNEUROSCI.4698-05.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y., Chang, H. and Nathans, J. (2010). When whorls collide: the development of hair patterns in frizzled 6 mutant mice. Development 137, 4091-4099. 10.1242/dev.057455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, M., de Marco, P., Capra, V. and Kibar, Z. (2019). Update on the role of the non-canonical Wnt/planar cell polarity pathway in neural tube defects. Cells 8, 1198. 10.3390/cells8101198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegel, E., Göhler, A., Lagerholm, B. C., Wainman, A., Uphoff, S., Kaufmann, R. and Dobbie, I. M. (2016). Imaging cellular structures in super-resolution with SIM, STED and localisation microscopy: a practical comparison. Sci. Rep. 6, 27290. 10.1038/srep27290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson, M. Z., Ravindran, P. T., Lim, W. A. and Toettcher, J. E. (2017). Tracing information flow from Erk to target gene induction reveals mechanisms of dynamic and combinatorial control. Mol. Cell 67, 757-769.e5. 10.1016/j.molcel.2017.07.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao, C., Nitsche, F. and Bazzi, H. (2018). Visualizing the node and notochordal plate in gastrulating mouse embryos using scanning electron microscopy and whole mount immunofluorescence. J. Vis. Exp. 6, e58321. 10.3791/58321 [DOI] [PubMed] [Google Scholar]
- Yang, Y. and Mlodzik, M. (2015). Wnt-Frizzled/planar cell polarity signaling: cellular orientation by facing the wind (Wnt). Annu. Rev. Cell Dev. Biol. 31, 623-646. 10.1146/annurev-cellbio-100814-125315 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.