Abstract
The loss of FMR1 expression due to trinucleotide repeat expansion leads to fragile X syndrome, a cause of mental retardation. The encoded protein, FMRP, is a member of a gene family that also contains the fragile X-related proteins, FXR1P and FXR2P. FMRP has been shown to be a nucleocytoplasmic shuttling protein that selectively binds a subset of mRNAs, forms messenger ribonucleoprotein (mRNP) complexes, and associates with translating ribosomes. Here we describe a cell culture system from which we can isolate epitope-tagged FMRP along with mRNA, including its own message, and at least six other proteins. We identify two of these proteins as FXR1P and FXR2P by using specific antisera and identify a third protein as nucleolin by using mass spectrometry. The presence of nucleolin is confirmed by both reactivity with a specific antiserum as well as reverse coimmunoprecipitation where antinucleolin antiserum immunoprecipitates endogenous FMRP from both cultured cells and mouse brain. The identification of nucleolin, a known component of other mRNPs, adds a new dimension to the analysis of FMRP function, and the approach described should also allow the identification of the remaining unknown proteins of this FMRP-associated mRNP as well as the other bound mRNAs.
Fragile X syndrome is a common form of inherited mental retardation. It is caused by a loss of expression of the FMR1 gene, most often due to an expansion of a CGG repeat in the first exon (reviewed in references 2 and 41). Although this region is untranslated, repeat expansion leads to abnormal methylation and chromatin deacetylation, which results in transcriptional silencing of FMR1 (9, 18, 28, 30, 39). The FMR1 gene encodes an approximately 78-kDa protein, FMRP, although multiple isoforms exist due to alternate splicing (1). FMRP contains two hnRNP K-homologous (KH) domains and an RGG box, motifs thought to mediate interactions with mRNA (13). Indeed, FMRP has been shown to bind its own mRNA, homopolymer RNA in vitro, and a subset of brain mRNAs (3, 7, 35). In addition, FMRP is associated with ribosomes in an RNA-dependent manner (12, 40). When lysates were treated with EDTA to dissociate the ribosomal subunits, FMRP was released as a large (greater than 669-kDa) messenger ribonucleoprotein (mRNP) particle containing both poly(A)+ mRNA and protein (12, 14).
Such mRNP complexes are thought to be formed in the cytoplasm after the hnRNP proteins, which associate with the mRNA in transit from the nucleus to the cytoplasm, are released and exchanged for cytoplasmic proteins (11). Some cytoplasmic RNA binding proteins, however, are identical to those found in the nucleus (17). Thus, some proteins seem to remain associated with mRNAs regardless of where the complex is located in the cell. FMRP contains both a functional nuclear localization signal (NLS) and a nuclear export signal (12, 38), and although it is primarily cytoplasmic at steady state, about 5% of the cellular FMRP is nuclear (15). FMRP is therefore believed to shuttle between the nucleus and cytoplasm, compartmentalizing to the cytoplasm through ribosome association. Since FMRP is found in both the nucleus and cytoplasm, it is not clear where FMRP becomes a part of the mRNP particle.
The proteins that makeup the FMRP-containing mRNP are largely unknown. However, certain candidate proteins exist, such as the autosomal homologs of FMRP, namely, the fragile X-related proteins encoded by the FXR1 and FXR2 genes, FXR1P and FXR2P, respectively. Both proteins are similar to FMRP in overall structure, each having two KH domains and conservation of the NLS and nuclear export signal found in FMRP (36, 37, 46). FXR1P and FXR2P have also been shown to bind RNA and associate with ribosomes (34). FXR2 was first identified in a yeast two-hybrid screen using FMRP as the bait. FXR2P was then shown to associate with FMRP in vivo in HeLa cells (46). FXR1 was identified by screening a Xenopus laevis cDNA library with the FMR1 cDNA (36). FXR1P has since been shown to interact with FMRP in the yeast two-hybrid system (46). Moreover, glutathione S-transferase fusion proteins of FXR1P FXR2P and FMR1P have been shown to each associate with one another in vitro as well as form homodimers. Based on these data, FXR1P and FXR2P may well be components of the FMRP mRNP, although this has not yet been established.
Besides the possibility of FXR1P and FXR2P constituting the FMRP mRNP, it is likely, given a mass in excess of 669 kDa (14), that additional proteins are involved. However, progress on answering this question has been hampered by the lack of suitable immunoprecipitating antibodies against FMRP. We show below that an epitope-tagged FMRP can be immunoprecipitated from stably transfected mouse L-M(TK−) cells and that the immunoprecipitation contains at least six other proteins and RNA. We identify two of these proteins as the FXR1P and FXR2P by using specific antibodies and identify a third as nucleolin by using matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-MS) as well as a specific antiserum. Finally, we provide the first in vivo evidence that FMRP-mRNP associates with the FMR1 mRNA.
MATERIALS AND METHODS
Cell lines, DNA constructs, and transfection studies.
The murine cell line L-M(TK−) was obtained from the American Type Culture Collection (Rockville, Md.) and was grown at 37° in 8% CO2 in Dulbecco’s minimal essential medium containing 10% fetal calf serum supplemented with 10 mM HEPES and 100 U of penicillin-streptomycin per ml (complete medium). All media and supplements were purchased from GIBCO-BRL unless otherwise noted. We transfected the amino-terminal, Flag epitope-tagged FMR1 cDNA (7), which contains a truncated 3′ untranslated region (UTR) with only the first 153 nucleotides of the 2,130 nucleotides of the 3′ UTR. This construct was subcloned into the BamHI site of the mammalian expression vector, RSV.5(gpt), kindly provided by Eric Long, National Institutes of Health (25). Either the Flag-FMR1 construct or the RSV.5(gpt) vector alone was introduced into L-M(TK−) cells by the calcium phosphate precipitation method as described elsewhere (32). Transfected cells were selected for guanine phosphoribosyltransferase (GPT) expression in mycophenolic acid (6 μg/ml) and xanthine (252 μg/ml) (Sigma, St. Louis, Mo.). After 10 to 14 days, the 100-mm-diameter tissue culture dishes were trypsinized and the drug-resistant cells were replated at limiting dilution to obtain independent clones. Each clone was tested for Flag-FMRP expression by Western blotting as described below.
Metabolic labeling.
The day before the labeling, 5 × 105 to 10 × 105 vector-only or Flag-FMRP-expressing cells from a clonal culture were plated in complete medium in 60-mm-diameter tissue culture dishes. The following day, the cells were labeled in leucine-free Dulbecco’s minimal essential medium (Cellgro) supplemented with 5% dialyzed fetal calf serum to which [3H]leucine (250 μCi/ml; Amersham Pharmacia Biotech) was added. After 16 h, the dishes were washed twice in ice-cold phosphate-buffered saline (PBS) and lysed in lysis buffer (1 ml per dish; 50 mM Tris-HCl, 150 mM NaCl, 30 mM EDTA, 0.5% Triton X-100 [pH 7.6]) with protease inhibitor tablets (Boehringer Mannheim) added as instructed. All subsequent manipulations were carried out at 4°C or on ice. After 20 to 30 min, the plates were scraped into an Eppendorf tube, which was then spun at 20,000 × g for 5 min to remove the nuclei. The lysates were sequentially precleared for 1 h with protein G-agarose (100 μl per sample; Boehringer Mannheim) and then for 1 to 2 h with M1-coupled matrix (100 μl per sample; Sigma). M1 is an anti-Flag antibody that does not recognize the epitope-tagged construct that we use. The lysate was then immunoprecipitated for 2 to 3 h with 50 μl of the anti-Flag M2 matrix per 1 ml of sample. The matrix was washed five times with 1 to 1.5 ml of lysis buffer over a 1-h period. To elute the Flag-FMRP complexes, the matrix was resuspended in 90 μl of lysis buffer to which 10 μl of Flag peptide (5 mg/ml; Sigma) was added. The mixture was rotated for 1.5 h and then spun twice at 20,000 × g for 5 min to completely remove the matrix. The supernatants were removed, and 30-μl aliquots were resolved on a sodium dodecyl sulfate (SDS)–5 to 20% polyacrylamide gradient gel at 65 V overnight. The following day, the gel was fixed in 10% acetic acid–30% methanol for 20 to 30 min, soaked in 15 to 20 volumes of water, and then treated with Fluoro-Hance (Research Products International Corp.) for 30 min before drying at 75° C for 2 h. The dried gel was subjected to autoradiography at −90°C.
Protein purification.
To obtain enough protein for the identification of peptide masses by MALDI-MS, we adapted the transfected L-M(TK−) cells expressing either the vector-only or Flag-FMRP to spinner flasks, where they were grown nonadherently. One-liter volumes of cells were harvested weekly when they reached cell densities between 5 × 105 to 10 × 105 cells/ ml. The cells were spun down, washed twice in PBS, and resuspended to less than 108 cells/ml in ice-cold lysis buffer as described above. The nuclei were removed by spinning for 15 min at 2,200 × g, and the lysates were frozen at −90°C until 6 × 109 to 8 × 109 cell equivalents were obtained. The combined lysates were thawed and precleared for 2.5 h with 5-ml packed volume of the M1 matrix (Sigma) and then immunoprecipitated with 1-ml packed volume of the anti-Flag M2 matrix for 3 to 5 h. The matrix was then washed in 50 ml of lysis buffer followed by three washes over a 1-h period with 10 to 50 ml of lysis buffer. Bound Flag-FMRP-containing complexes were eluted for 1.5 h in 0.5 ml of lysis buffer and 0.5 ml of Flag peptide (5 mg/ml) synthesized by the Emory University Microchemical Facility. The peptide elution was combined with a 2-ml wash of the matrix, and the total volume was precipitated with 10% trichloroacetic acid. The protein pellet was washed once with 0.5% trichloroacetic acid and then twice with acetone, resuspended in SDS sample buffer, and resolved on an SDS–7.5% polyacrylamide minigel (7.7% of the sample [5 of 65 μl] was set aside for later Western analysis). After staining with Coomassie brilliant blue, the band at 100 kDa was cut out and sent to the Howard Hughes Medical Institute (HHMI) Biopolymer Facility at Yale University, where in-gel tryptic digestions were carried out and the peptides were purified by microbore high-pressure liquid chromatography. MALDI-MS was used to determine the molecular mass/charge ratios of the peptides. The primary program used for searching a database of predicted masses is ProFound, which relies on the OWL database. PeptideSearch was also performed because this algorithm uses the EMBL nonredundant database.
Antibodies, immunoprecipitation, and Western blotting analysis.
Cells were lysed in the lysis buffer described above at the cell numbers given in the figure legends. The nuclei were removed by spinning at 20,800 × g for 5 min. Lysates were immunoprecipitated either with the anti-Flag M2 matrix as described above or with the antinucleolin antiserum generously provided by Renato Aguilera (University of California, Los Angeles) as follows. For each immunoprecipitation, 1 to 3 μl of antiserum was prebound for >1 h to 60 μl of a 50% solution of protein A-Sepharose (Amersham Pharmacia Biotech) in 1 ml of PBS at 4°C. The cytoplasmic lysates were then added to the washed, antinucleolin-bound beads and immunoprecipitated for at least 2 h. The immunoprecipitates were then washed two to three times and boiled in SDS sample buffer for electrophoresis and Western analysis. The antinucleolin antibody was also used for Western blot staining and visualized with an anti-rabbit horseradish peroxidase (HRP) conjugate (Amersham Pharmacia Biotech) as described. The anti-FXR2P antibody (A42) and the anti-hnRNP A1 antibody (4B10) were provided by Gideon Dreyfuss (HHMI, University of Pennsylvania). The anti-FXR1P antiserum was provided by Andre Hoogeveen (Erasmus University, Rotterdam, The Netherlands). Anti-Flag antibody M2 was purchased from Sigma, and anti-FMRP antibody 1FM.1AC.484A.1 was obtained from Jean-Louis Mandel (Institute of Genetics, Illkirch, France). The Western blots probed with murine antibodies were visualized with anti-mouse HRP conjugates obtained from either Amersham Pharmacia Biotech or Kirkegaard & Perry Laboratories.
Isolation and labeling of mRNA.
Approximately 1.7 × 109 mouse L-M cells expressing Flag-FMRP or vector alone were harvested from nonadherent cultures growing at 106 cells/ml. Cells were washed three times in 50 ml of PBS and lysed gently on ice for 45 min at 7.7 × 107 cells/ml in lysis buffer (described above) with rRNAsin (100 U/ml; Promega) and 2× protease inhibitors (Complete tabs; Boehringer Mannheim). The nuclei were pelleted at 3,300 × g for 15 min at 4°C. The cytoplasmic supernatant was precleared for 3 h with 1 ml of the anti-Flag M2 matrix that was preblocked with 1 mg of its ligand, Flag peptide, by cross-linking in 20 μM dimethyl pimelimidate-2 HCl (DMP; Pierce). After preclearing, the cross-linked M2-Flag preclearing matrix was pelleted at 1,000 × g, and the supernatant was precleared a second time with 1 ml of the anti-Flag M1 matrix for 45 min. The M1 matrix was pelleted, and the final precleared lysate supernatant was immunoprecipitated with 470 μl of fresh, anti-Flag M2 matrix for 2.5 h, rotating at 4°C. The immunoprecipitated material was washed twice with 10 ml of lysis buffer for 15 min at 4°C. The third wash contained 50 U of RNase-free DNase (Promega) and 200 U rRNAsin, and the matrix was allowed to settle by gravity for 2 h. The fourth wash contained 200 U rRNAsin, and the matrix was pelleted by gravity overnight. The next day, protein-RNA complexes were eluted with 200 μl lysis buffer–200 μl of Flag peptide (5-mg/ml stock) for 45 min, and the matrix was washed with 500 μl of lysis buffer for 45 min. The elution and elution-wash were pooled, and 1/10 of the eluted material was saved for protein analysis. The remaining 9/10 of eluted material was treated with 100 μg of proteinase K (RNase free; Sigma) and 200 U rRNAsin at 37°C for 15 min. After phenol-chloroform extraction, the RNA was isolated by ethanol precipitaton. One-twentieth of the RNA yield was used in a first-strand cDNA synthesis reaction using 30 μCi of [α-32P]dATP (Amersham), 50 U of Moloney murine leukemia virus reverse transcriptase (Clontech), and 50 pmol of oligo(dT)18 primer. The reaction mixtures were heat inactivated at 99°C and run out on a 1% SeaKem GTG ethidium-agarose gel. The gel was dried, and the molecular weight standards were visualized by UV light photography. The dried gel was exposed to film (Biomax MS; Kodak) for 1 h at −70°C.
RNA analysis by reverse transcription-PCR.
One-tenth of the mRNA purified from the immunoprecipitated protein complexes was reverse transcribed by using an anchored oligo(dT) primer, (T)16VN (Gibco BRL), at 70°C for 2 min, 37°C for 3 min, 25°C for 1 min, 42°C for 20 min, 48°C for 10 min, 99°C for 5 min, and 4°C for 5 min. One-fourth of the reaction product was used to amplify the mouse FMR1 gene with primers E9f (AAAGCTAGAAGCTTTCTCG) and El1r (CCCTTGAATTATTGGAAGG), using an RNA PCR kit (Perkin-Elmer). Thermocycling was carried out at 95°C for 1 min followed by 35 cycles of 95°C for 30 s, 52°C for 45 s, and 72°C for 45 s. Forty percent of the yield was analyzed via agarose gel electrophoresis and visualized with ethidium bromide.
Mouse brain preparations.
Two wild-type littermates and two FMR1 knockout mice (10) were asphyxiated with CO2, and their brains were harvested into 2 ml of lysis buffer. The brains were disrupted by 10 strokes with a Dounce homogenizer. The lysates were then spun at 90,000 × g at 4°C in an ultracentrifuge for 0.5 h. The supernatant was removed and precleared with 360 μl of protein A-Sepharose (Amersham Pharmacia Biotech) and immunoprecipitated with the antinucleolin antibody as described above.
RNase treatment.
Immunoprecipitations of L-M(TK−) cells transfected with either the vector only or Flag-FMRP were carried out essentially as described above. Approximately 8 × 107 cells were lysed, enucleated, and then immunoprecipitated with 50 μl of the anti-Flag M2 matrix overnight at 4°C. The following day, the immunoprecipitates were washed twice in 1 ml of lysis buffer at 4°C. The third wash was for 15 min at 37°C, rotating in either lysis buffer alone with 2× protease inhibitors (Boehringer Mannheim) (mock treatment) or lysis buffer containing RNase T1 (50 U/ml; Sigma) and RNase A (120 μg/ml; Sigma) as described elsewhere (14). The immunoprecipitates were washed again at 4°C, pelleted and boiled in sample buffer, and resolved on a 7.5% gel, which was blotted to nitrocellulose and probed with the antinucleolin or anti-hnRNP A1 antibody.
RESULTS
Flag-FMRP can be stably expressed in murine L-M(TK−) cells.
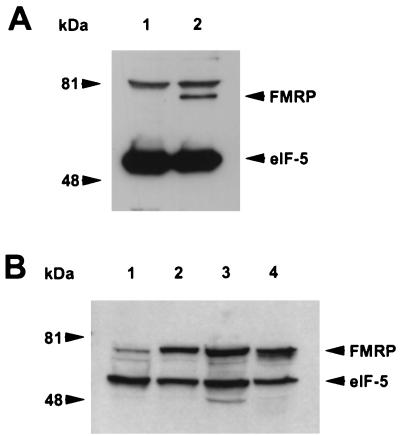
To identify proteins that interact with FMRP, we developed a cellular system expressing N-terminal Flag-tagged FMR1 cDNA. Such an epitope-tagged strategy was required since the available antibodies against FMRP immunoprecipitate poorly, and due to the highly conserved nature of FMRP (31), development of additional antibodies has been problematic. We attempted to express this transgene in a number of cultured cell lines, including murine fibroblasts [L-M(TK−)], mouse embryonic carcinoma cells (P19), human B-lymphoblasts (J1), and African green monkey kidney cells (COS). Although we were able to isolate numerous drug-resistant colonies of P19, J1, and COS cells, none expressed the Flag-FMRP by Western analysis using an anti-Flag antibody. In contrast, approximately 40% of the GPT-positive murine L-M(TK−) clones expressed Flag-FMRP. One of these clones was used in all of the subsequent studies, although similar results were obtained with other independently derived, Flag-FMRP-expressing clones (data not shown). As a control, a clone derived from transfection of the empty expression vector into L-M(TK−) was used. As shown in Fig. 1A, Western analysis using an anti-Flag antibody reveals a protein of the expected molecular weight in the lane containing transfectant lysate that is absent in the control clone transfected with just the parental plasmid (RSV.5). Probing with an antibody against eukaryotic initiation factor 5 (eIF-5) showed an expected 49-kDa protein, indicating equal loading between the lanes. An 81-kDa band was observed in both lysates due to reactivity of an unknown protein with the secondary antibody.
FIG. 1.
Transfected murine L-M(TK−) cells express epitope-tagged FMRP at levels comparable to that observed in a transformed B-cell line. (A) Approximately 2 × 105 cell equivalents from L-M(TK−) cells expressing either vector only (lane 1) or Flag-FMRP were loaded (lane 2). Positions of molecular weight markers are shown on the left; positions of Flag-FMRP and eIF-5 are indicated by arrowheads on the right. The cytoplasmic proteins were resolved on a 7.5% gel, transferred to nitrocellulose, and probed simultaneously with anti-Flag monoclonal antibody M2 and with a monoclonal antibody that recognizes eIF-5 to show that equal amounts of cytoplasmic lysate were loaded. The upper band (>81 kDa) present in both lanes is detected by this particular goat anti-mouse HRP-conjugated antibody alone (data not shown). The secondary antibodies used for panel A are different from those used for panel B. (B) Approximately 5 × 105 cell equivalents of cytoplasmic lysates from untransfected L-M(TK−) cells (lane 1), two independently derived clones expressing Flag-tagged FMRP (lanes 2 and 3), and an Epstein-Barr virus-transformed B-cell line (J1; lane 4) were loaded per lane of a 12% gel. After transfer, the blot was probed simultaneously with anti-FMRP monoclonal antibody 1FM.1AC.484A.1 and eIF-5 to show equal loading.
Interestingly, none of the Flag-FMRP-expressing clones appeared to express more FMRP than the endogenous levels of FMRP observed in J1 (Fig. 1B) or P19 and COS (data not shown) cells. In addition, untransfected L-M(TK−) cells appeared to have the lowest level of endogenous FMRP expression among all cell types examined (Fig. 1B and data not shown). Thus, it is possible that the inability to express the transfected FMR1 in any of the other cell types is due to toxic overexpression of FMRP, whereas L-M(TK−) cells tolerate the transgene since their endogenous levels are already low. In any event, these data show the construction of a mammalian cell system expressing epitope-tagged FMRP that would now be amenable to immunoprecipitation.
Coimmunoprecipitation of FMRP, FXR1P, FXR2P, and mRNA.
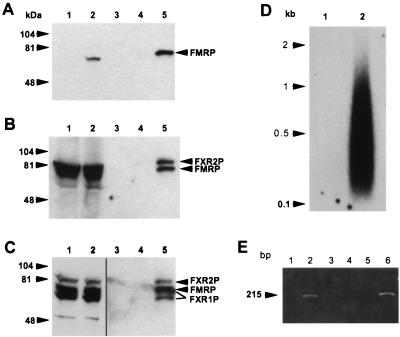
To determine whether the Flag-FMRP was able to form complexes in L-M(TK−) cells with proteins known to interact with FMRP, we immunoprecipitated with anti-Flag antibody M2 coupled to matrix and did a series of sequential probings with different antibodies. As shown in Fig. 2A, Western analysis using an anti-FMRP monoclonal antibody shows the overexpressed FMRP in the transfected cell lysate. Longer exposure of this blot revealed the endogenous FMRP in the L-M(TK−) cells (data not shown), similar to results in Fig. 1B. Immunoprecipitation using the M2 anti-Flag matrix followed by Flag peptide elution showed FMRP being captured in the transfected cell lysate of 107 cells, while no signal was observed in the peptide eluate of the M2 matrix alone or from the immunoprecipitation of vector-alone control cells. Examination of the matrix flowthrough of the transfected cell lysate showed that nearly all of the Flag-FMRP was captured (data not shown). Thus, under relatively nondenaturing conditions (150 mM salt and 0.5% detergent), FMRP can be effectively immunoprecipitated.
FIG. 2.
FXR1P and FXR2P assemble with Flag-FMRP in transfected L-M(TK−) cells to form an mRNP particle that binds mRNA. (A) Cytoplasmic lysates from approximately 5 × 105 L-M(TK−) cells expressing vector alone and expressing Flag-FMRP were loaded into lanes 1 and 2, respectively; lanes 3 to 5 contain Flag peptide elutions from the anti-Flag antibody M2 alone (lane 3) or from immunoprecipitations of 107 cells expressing the vector -only (lane 4) or Flag-FMRP (lane 5). The immunoprecipitated FMRP in lane 5 appears to run slower than the FMRP detected in the cytoplasmic lysates, probably because there is much less protein in the lanes containing the peptide elutions than in the lanes containing cytoplasmic lysates. The gel was blotted and sequentially probed with a monoclonal antibody recognizing FMRP (A), then with both anti-FMRP and anti-FXR2P antibodies (B), and finally with anti-FMRP, anti-FXR2P and anti-FXR1P antibodies (C). Lanes 1 and 2 in panel C are shown as separate because they are a lighter exposure of the same blot. Positions of the molecular weight standards are shown on the left, and positions of the proteins are shown on the right. The long and short isoforms of FXR1P are indicated by lines. (D) mRNA was purified from L-M(TK−) cells expressing either the vector only (lane 1) or Flag-FMRP (lane 2) as described in Materials and Methods. The mRNA was recovered, and the polyadenylated species were labeled by priming with oligo(dT) and synthesizing first-strand cDNA with reverse transcriptase. (E) MRNA obtained from immunoprecipitations of L-M(TK−) cells expressing either Flag-FMRP (lanes 1 and 2) or vector alone (lanes 3 and 4) or from mouse brain (lanes 5 and 6) was reverse transcribed with an oligo(dT) primer in either the presence (lanes 2, 4, and 6) or the absence (lanes 1, 3, and 5) of reverse transcriptase. A fraction of each reaction mixture was then added to a PCR mixture with mouse FMR1 primers. The PCR products were resolved on an agarose gel and stained with ethidium bromide.
Since FXR2P has previously been shown to associate with FMRP in vivo (46), we reprobed the same blot with anti-FXR2P to determine if FXR2P coimmunoprecipitated with FMRP. As shown in Fig. 2B, FXR2P was present at high levels in lysates of the L-M(TK−) cells. There appears to be more FXR2P than FMRP, which likely reflects the lower levels of FMRP than of FXR2P in these cells, as well as different affinities of the relevant antibodies to these proteins. In addition, FXR2P coimmunoprecipitates with FMRP, thus confirming the in vivo association of FMRP with FXR2P described before (46). This result establishes that our coimmunoprecipitation system is capable of isolating FMRP-associated proteins. Next we examined, again by reprobing the same blot, if FXR1P associates with FMRP in vivo. While FXR1P has been shown to interact with FMRP in vitro, there are no published data indicating that such interaction occurs in vivo in mammalian cells. As shown in Fig. 2C, FXR1P is detected in the immunoprecipitate. These data indicate that cellular FXR1P and FXR2P are associated with FMRP in vivo.
Because the lysis buffer used above also contained 30 mM EDTA to disrupt the ribosomes, it is likely that the immunoprecipitate consists of the large mRNP particle containing FMRP that we have previously observed (14). Accordingly, we tested whether mRNA was also present in the anti-Flag immunoprecipitate by performing first-strand cDNA synthesis on the immunoprecipitated material with reverse transcriptase and oligo(dT) priming. As shown in Fig. 2D, mRNAs are immunoprecipitated from the Flag-FMRP-expressing cells, while very little, if any, is eluted from the anti-Flag immunoprecipitation of the empty vector-expressing cells. Thus, immunoprecipitation of Flag-FMRP under these conditions is able to bring down mRNA as well as the FXR-encoded proteins, which may, therefore, represent components of an FMRP-associated mRNP.
Previously, we showed that purified FMRP bound its own mRNA in vitro (7). To determine whether the FMR1 mRNA was found in our immunoprecipitated complexes, we performed PCR using primers for the FMR1 cDNA. We show in Fig. 2D that the FMR1 mRNA was present in the immunoprecipitations from Flag-FMRP-expressing L-M(TK−) cells but not from those expressing the vector only. This is the first evidence that the FMRP-containing mRNP associates in vivo with the FMR1 mRNA.
Novel proteins assemble with FMRP mRNP.
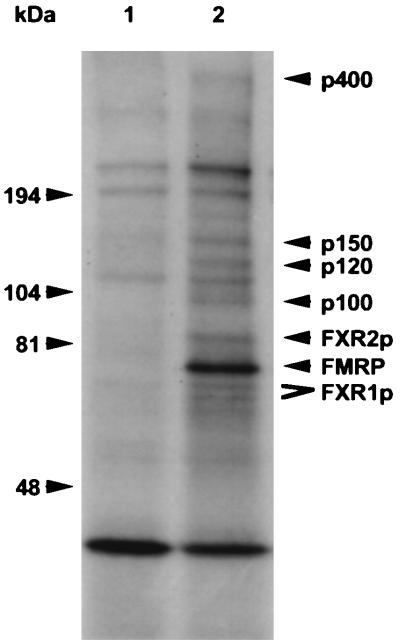
To identify additional proteins in the mRNP complex containing FMRP, transfected cells were cultured in the presence of [3H]leucine to metabolically label the cellular proteins. Following lysis and mRNP particle capture, as described above, the protein components were resolved by gel electrophoresis and visualized by autoradiography. As shown in Fig. 3, at least six proteins are observed in the immunoprecipitation of the Flag-FMRP-transfected cell lysate that are not present in the immunoprecipitate of cells transfected with the vector alone (which shows at least four proteins eluted from the M2 antibody). FMRP appears as the most intense band and was verified by Western analysis (data not shown). FMRP also appears more abundant in the eluate than the other proteins, most of which appear in approximately equivalent levels. Since the M2 matrix specifically captures Flag-FMRP, it is possible that this is due to the extensive washing of the bound complex, which may deplete associated proteins. Alternatively, FMRP could be differentially labeled or a significant portion of the Flag-FMRP may not be associated with the mRNP, although sucrose gradient fractionation has shown that the majority of FMRP is found to migrate with a faster mobility than free protein, suggesting that FMRP exists largely as a complex (data not shown). Therefore, these data indicate that at least seven proteins may compose the mRNP complex that coimmunoprecipitates with FMRP. Since one of the proteins is FMRP and two were identified as FXR1P and FXR2P, four unknown proteins, designated p100, p120, p150, and p400, remain to be further characterized.
FIG. 3.
Novel proteins in addition to FXR1P and FXR2P assemble with Flag-FMRP in L-M(TK−) cells. L-M(TK−) cells transfected with either the eukaryotic expression vector alone (lane 1) or with Flag-FMRP (lane 2) were labeled overnight with [3H]leucine and then immunoprecipitated with matrix coupled to anti-Flag antibody M2. After extensive washing, the Flag matrix was eluted with Flag peptide and the proteins were resolved on a 5 to 20% gradient gel. Migration of molecular weight markers is indicated on the left. Known proteins are indicated on the right, and the new proteins are indicated by their molecular sizes. Both the long and short isoforms of FXR1P are indicated, and FMRP is highlighted.
Large-scale purification and subsequent identification of p100.
To identify the unknown proteins of the mRNP containing FMRP, large-scale cultures of both L-M(TK−) cells expressing the vector alone or L-M(TK−) cells expressing Flag-FMRP were established. We adapted these normally adherent cells to spinner flasks to ease large-scale culturing. Based on pilot experiments, we determined that lysate from at least 5 × 109 L-M(TK−) cells would need to be immunoprecipitated to obtain enough protein for MALDI-MS identification.
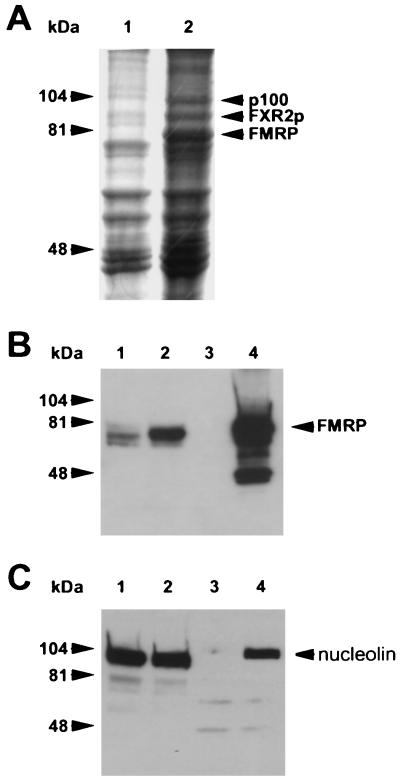
A Coomassie brilliant blue stain of Flag peptide elutions from a large scale purification is shown in Fig. 4A. The position of FMRP, the most abundant protein observed, was confirmed by Western analysis shown in Fig. 4B. FXR2P was identified by its position relative to FMRP and was confirmed by both Western analysis and MALDI-MS identification (data not shown). Of the four unknown proteins, p100 was chosen for characterization because it was the most clearly resolved on a 7.5% minigel. The 100-kDa band indicated in Fig. 4A was excised from the gel and sent to the HHMI Biopolymer Facility at Yale University, where it was subjected to a tryptic digest, of which 5% was analyzed by MALDI-MS. The peptide masses obtained from p100 matched 27% of the predicted peptide masses of mouse nucleolin by using the ProFound search program. With a different program, PeptideSearch, the peptide masses obtained from p100 also matched mouse nucleolin. An additional ProFound search was performed on the p100-derived masses after deletion of those masses which matched mouse nucleolin, and no additional proteins were identified in the sample.
FIG. 4.
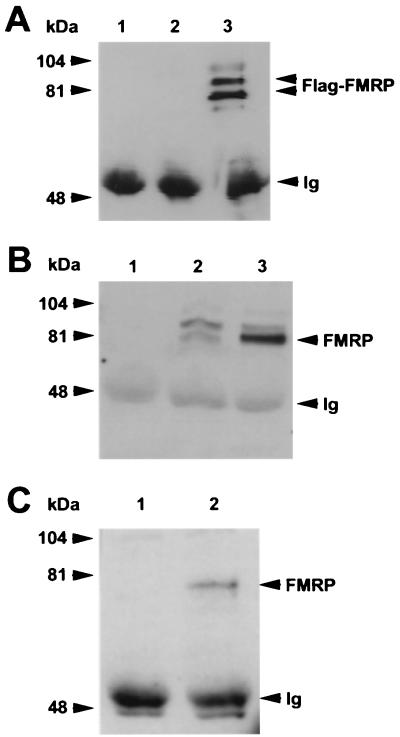
Nucleolin coimmunoprecipitates with Flag-FMRP. (A) Scanned image of the Coomassie brilliant blue-stained gel from which p100 was harvested. Lanes 1 and 2 are the Flag peptide elutions from the large-scale purifications of L-M(TK−) cells expressing vector alone and Flag-FMRP, respectively. The proteins were resolved on 7.5% minigels. The position of FMRP was determined by Western blotting of a gel run in parallel (B). The position of FXR2 was determined by both Western blotting and mass analysis (data not shown). The p100 band was cut out and analyzed as described in the text. (B) FMRP Western analysis of the large-scale purification. Lanes 1 and 2 contain a fraction of the pooled lysates from L-M(TK−) cells expressing the vector and Flag-FMRP before immunoprecipitation, respectively; lanes 3 and 4 show 7.5% of the peptide elutions from each of the large-scale purifications. (C) A rabbit antiserum derived against murine nucleolin was used to reprobe the Western blot shown in panel B. Positions of the molecular weight markers are shown on the left; the position of nucleolin is shown on the right.
To confirm the MALDI-MS identification of p100 as nucleolin, we obtained an antiserum (from Renato Aguilera) that recognizes murine nucleolin (27). We reprobed the Western blot shown in Fig. 4B with the antinucleolin antibody and showed that although nucleolin is abundant in L-M(TK−) cells, it is significantly immunoprecipitated only from the Flag-FMRP-expressing cells compared to the vector-only cells (Fig. 4C). Thus, nucleolin coimmunoprecipitates with Flag-FMRP, suggesting that p100 is indeed nucleolin.
FMRP coimmunoprecipitates with nucleolin.
To further establish and confirm that nucleolin coimmunoprecipitates with FMRP, the reverse immunoprecipitation was carried out with antinucleolin as the precipitating antibody. As shown in Fig. 5A, the antinucleolin antibody coimmunoprecipitates the epitope-tagged FMRP from the FMR1-transfected cells when analyzed with the anti-Flag antibody. We do not know why two prominent, anti-Flag-reactive proteins are immunoprecipitated with the antinucleolin antibody. Since they are not present in the immunoprecipitation of vector-only-expressing cells, they may represent posttranslational modifications of FMRP. In general, FMRP does not nonspecifically associate with irrelevant antibody because we can immunoprecipitate unrelated proteins with rabbit antiserum from L-M(TK−) cells and not see FMRP association (data not shown). Therefore, we believe that the interaction between FMRP and nucleolin is specific. To determine whether the endogenous FMRP in L-M(TK−) cells associates with nucleolin, we immunoprecipitated both the vector-only and the Flag-FMRP-expressing cells with an antinucleolin antibody and then probed with an anti-FMRP antibody. FMRP is observed in immunoprecipitations from both the vector-only and the Flag-FMRP-expressing cells, and as expected, there is much more FMRP brought down in the transgene-expressing cells (Fig. 5B). Thus, nucleolin is associated with the complex containing endogenous FMRP in mouse L-M(TK−) cells. This observation indicates that the association of nucleolin with the FMRP complex is not an artifact of the transfection model system since nucleolin associates with both endogenous and epitope-tagged FMRP.
FIG. 5.
FMRP coimmunoprecipitates with nucleolin. (A) Lane 1 contains the antinucleolin antibody alone; lanes 2 and 3 show antinucleolin immunoprecipitation of cytoplasmic lysates from 107 L-M(TK−) cells expressing the vector alone and from 107 L-M(TK−) cells expressing Flag-FMRP, respectively. The proteins were resolved on a 7.5% gel, blotted to nitrocellulose, and probed with anti-Flag antibody M2. Positions of the molecular weight markers are shown on the left, and positions of FMRP and the heavy chain of the antinucleolin antibody (immunoglobulin [Ig]) are shown on the right. (B) An experiment similar to that shown in panel A except that transferred proteins were probed with the anti-FMRP antibody. Lanes 1 to 3 are as described above: the antinucleolin antibody alone, an immunoprecipitation of vector-only-containing L-M(TK−) cells, and an immunoprecipitation of Flag-FMRP-expressing L-M(TK−) cells with the antinucleolin antibody. In lane 2, the endogenous murine FMRP is immunoprecipitated with nucleolin in addition to the Flag-tagged FMRP observed in lane 3. Positions of the heavy chain of the antinucleolin antibody, which reacts with the second-step goat anti-mouse HRP conjugate, and Flag-FMRP are indicated on the right. (C) Total brain homogenates were prepared from either FMR1 knockout mice (lane 1) or their wild-type, FMRP-positive littermates (lane 2). The cytoplasmic lysates were immunoprecipitated with the antinucleolin antibody, washed extensively, and boiled. The samples were resolved on a 7.5% gel, transferred to nitrocellulose, and then probed with a monoclonal antibody that recognizes FMRP. Positions of FMRP and the heavy chain of the antinucleolin antibody are shown on the right.
The association of nucleolin with the FMRP complex was demonstrated above by using murine fibroblast L-M(TK−) cells. While there are subtle connective tissue abnormalities in patients with fragile X syndrome (41), the major phenotypic consequence of the absence of FMRP is neuronal. Accordingly, we next prepared homogenates from the brains of male FMR1 knockout mice (10) and from their normal male littermates. With the antinucleolin antibody as the precipitating antibody and anti-FMRP for Western analysis, FMRP coimmunoprecipitates, with nucleolin from the normal mouse brain (Fig. 5C). That this band is truly FMRP is indicated by its absence from the antinucleolin immunoprecipitation of knockout brains. Hence, the association of nucleolin with FMRP appears to occur in the brain as well.
RNase treatment does not disrupt association of nucleolin with FMRP.
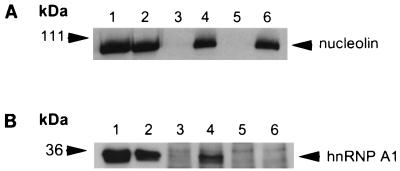
Because both nucleolin and FMRP contain RNA binding domains, it is possible that the association between nucleolin and FMRP occurs through independent binding of a common RNA molecule. To test this hypothesis, we treated anti-Flag immunoprecipitations of both the vector-only-expressing cells and the Flag-FMRP-expressing cells with RNases under conditions previously shown to disrupt the FMRP-associated mRNP particle (14). Figure 6 shows that the same amount of nucleolin is found associated with FMRP, regardless of treatment with RNases. In addition, a longer treatment with 10 times the amount of RNase for a longer period of time yielded the same result (data not shown). As a control for RNase digestion, we used an antibody directed against hnRNP A1 protein. hnRNP A1 protein binds poly(A)+ mRNA and, like FMRP, contains RNA binding domains and shuttles between the nucleus and cytoplasm (reviewed in reference 11). As shown in Fig. 6B, hnRNP A1 protein was found in the immunoprecipitated FMRP complex. However, unlike nucleolin, this association was lost following RNase treatment. Thus, it is likely that the association of hnRNP A1 protein with the complex containing FMRP is mediated by independent and separate interactions with the same RNA molecules. These data, therefore, serve as a control verifying the activity of the RNase treatment. Thus, we conclude that the association of nucleolin with the FMRP complex involves a close association, not separated by exposed RNA, and may well include protein-protein interaction. Since the FXR proteins also are not lost from the complex by RNase treatment (data not shown), it remains to be determined which protein nucleolin may directly interact with.
FIG. 6.
Treatment with RNase does not affect the association of nucleolin with FMRP. (A) Lanes 1 and 2 contain cytoplasmic lysates from 2.5 × 105 L-M(TK−) cells expressing the vector alone and from L-M(TK−) cells expressing Flag-FMRP, respectively; lanes 3 and 4 contain mock-treated, anti-Flag antibody immunoprecipitations from L-M(TK−) cells expressing the vector alone and from L-M(TK−) cells expressing Flag-FMRP, respectively; lanes 5 and 6 contain RNase-treated anti-Flag antibody immunoprecipitations from L-M(TK−) cells expressing the vector alone and from L-M(TK−) cells expressing Flag-FMRP, respectively. The proteins were resolved on a 7.5% gel, blotted to nitrocellulose, and probed with an antinucleolin antibody. Positions of molecular weight markers are shown on the left, and the position of nucleolin is shown on the right. (B) An experiment similar to that shown in panel A except that the transferred proteins were probed with an antibody to hnRNP A1. Lanes 1 and 2 are as described above except that lysates from 5 × 104 cells were loaded. Lanes 3 and 4 contain mock-treated anti-Flag antibody immunoprecipitations from L-M(TK−) cells expressing the vector alone and from L-M(TK−) cells expressing Flag-FMRP; lanes 5 and 6 contain RNase-treated anti-Flag antibody immunoprecipitations from L-M(TK−) cells expressing the vector alone and from L-M(TK−) cells expressing Flag-FMRP. The proteins were resolved on a 10% gel, blotted to nitrocellulose, and probed with an antibody that recognizes hnRNP A1. Positions of molecular weight markers are shown on the left, and the position of hnRNP A1 is shown on the right.
DISCUSSION
We have developed a cell transfection system to characterize the proteins and nucleic acids that associate with FMRP by expressing Flag-tagged FMRP in cultured murine fibroblasts from which we could perform efficient immunoprecipitations. The epitope-tagged FMRP in mouse L-M(TK−) cells was able to assemble with at least six other proteins and mRNAs, one of which is the FMR1 mRNA. Although we do not know for certain whether all of these molecules assemble into one large mRNP particle, as opposed to several smaller complexes, our previous results showing that FMRP was found as a >669-kDa particle indicate that this is a likely explanation (14). Using specific antisera, we confirmed that FXR2P is a part of this complex and provided in vivo evidence that FXR1P is also a component of the complex. Metabolic labeling revealed four other, unidentified proteins in the complex, and poly(T)-primed, first-strand cDNA synthesis demonstrated a mixture of mRNAs associated with the immunoprecipitate. Thus, this system establishes a method to identify both the protein and mRNA components of the FMRP complex. Accordingly, we identified by mass peptide analysis and confirmed by coimmunoprecipitation that one of the unknown proteins of the FMRP complex, designated p100, is nucleolin. Finally, we show that this association is resistant to treatment with RNases.
Nucleolin was first identified as spot C23 in preparations of nucleolar proteins resolved by two-dimensional gel electrophoresis (29). In addition to being very abundant in the nucleolus, comprising 5 to 10% of the total nucleolar protein, nucleolin was also detected in the cytoplasm (5, 8), where its function has only recently been studied (44, 45). Like FMRP and the FXR proteins, nucleolin contains an NLS, which enables it to shuttle between the nucleolus and cytoplasm (4, 16, 26, 33). Nucleolin has four RNA binding domains in the central portion of the molecule (8) and a carboxy-terminal RGG domain that binds RNA, as well as some proteins (6, 16, 20). Although the central RNA binding domains differ from the KH domains of the FMRP family of proteins, the RGG domains are shared among all four proteins. The identification of nucleolin and the FXR proteins as components of the FMRP complex indicates that the mRNAs of the complex could be directly interacting with any of the protein components. Hence, subsequent identification and functional studies of the mRNAs of the FMRP complex need to be evaluated as complex-associated mRNAs rather than as mRNAs associating with a single purified protein. Similarly, it remains to be established which of the protein components of the complex are in direct association.
The identification of nucleolin as a component of the FMRP complex provides some insight into possible FMRP function. Although nucleolin is normally localized to the nucleolus, a significant pool of it is cytoplasmic (5, 8, 45). Conversely, FMRP, while largely cytoplasmic, has been found in the nucleolus (42). Thus, only a fraction of the cellular nucleolin may be associated with FMRP. Recent studies that have shown that nucleolin is a part of other mRNP particles. For example, Zaidi and Malter identified nucleolin as one of the proteins bound to the 3′ UTR of amyloid precursor protein mRNA (45).
Nucleolin was also identified as one of the components of a 320-kDa mRNP particle isolated from X. laevis oocytes. Preincubation of this complex with two different mRNAs resulted in their translational suppression in a wheat germ extract as well as in a rabbit reticulolysate (44). Incubation of a smaller mRNP particle, without nucleolin and seven other polypeptides, did not suppress translation, suggesting that at least one of these components is important for mediating translational inhibition. It has been speculated that FMRP can play a role as a masking protein (13), preventing the translation of associated mRNAs until a specific signal is received. Indeed, recent studies have shown that FMRP can suppress translation of bound messages in an in vitro translation assay (23).
One of the reasons to identify the components of the FMRP complex is that mutations in the corresponding genes could lead to neuropsychiatric disease. While mental retardation has not yet been associated with mutations in FXR1 or FXR2 (24), it is unlikely that a similar search has been conducted for the nucleolin locus since this is the first correlation, albeit indirect, with such a disorder. No human mutations in nucleolin have been reported, nor have nucleolin knockout mice been created. However, the loss of nucleolin may not be lethal, based on studies of the yeast homolog Nsr1, which shares many structural and functional similarities with mammalian nucleolin (21, 43). When this gene is disrupted, the yeast survive but with a severe growth defect (19, 21, 22). Thus, null mutations of nucleolin in humans may present with a much more severe phenotype than fragile X syndrome but could include mental retardation as part of the phenotype.
In conclusion, we demonstrate a system by which the apparent mRNP particle associated with epitope-tagged FMRP in murine fibroblasts was isolated. From such isolations, several proteins can be found to copurify with FMRP as well as a complex mixture of mRNAs, including the FMR1 mRNA. Thus, this system can be used for future identification of those mRNAs as well as associated proteins. For the latter use, we have identified three of the associated proteins. Two of the proteins, FXR1P and FXR2P, were previously shown or suspected to be part of the FMRP mRNP. This confirms the interaction of the FMRP family proteins and validates the system as a method to isolate the mRNP containing FMRP. Finally, we identify p100, one of the four remaining unknown associated proteins, as nucleolin. This is confirmed by reverse coimmunoprecipitation using antinucleolin antibody and validated by demonstrating that endogenous FMRP can coimmunoprecipitate with nucleolin both in mouse L-M(TK−) cells and in mouse brain lysate. The identification of nucleolin, a known component of other mRNPs, adds a new dimension to the analysis of FMRP function and fragile X syndrome, as will the ongoing studies aimed at identifying the remaining associated proteins and mRNAs.
ACKNOWLEDGMENTS
We thank Dave Pallas, Keith Wilkinson, and members of the Warren lab for assistance and thoughtful comments and Cathy Alden for editorial assistance. We also thank Renato Aguilera, Andre Hoogeveen, Gideon Dreyfuss, and Jean-Louis Mandel for generously providing antibodies, as well as Ben Oostra and Patrick Willems for providing FMR knockout mice.
This project was supported in part by grants R37HD20521 and PO1HD35576. S.C. is an associate and S.T.W. is an investigator with the Howard Hughes Medical Institute.
REFERENCES
- 1.Ashley C T, Sutcliffe J S, Kunst C B, Leiner H A, Eichler E E, Nelson D L, Warren S T. Human and murine FMR-1: alternative splicing and translational initiation downstream of the CGG repeat. Nat Genet. 1993;4:244–251. doi: 10.1038/ng0793-244. [DOI] [PubMed] [Google Scholar]
- 2.Ashley C T, Warren S T. Trinucleotide repeat expansion and human disease. Annu Rev Genet. 1995;29:703–728. doi: 10.1146/annurev.ge.29.120195.003415. [DOI] [PubMed] [Google Scholar]
- 3.Ashley C T, Wilkinson K D, Reines D, Warren S T. FMR1 protein: conserved RNP family domains and selective RNA binding. Science. 1993;262:563–566. doi: 10.1126/science.7692601. [DOI] [PubMed] [Google Scholar]
- 4.Borer R A, Lehner C F, Eppenberger H M, Nigg E A. Major nucleolar proteins shuttle between nucleus and cytoplasm. Cell. 1989;56:379–390. doi: 10.1016/0092-8674(89)90241-9. [DOI] [PubMed] [Google Scholar]
- 5.Bouche G, Amalric F, Caizergues-Ferrer M, Zalta J P. Effects of heat shock on gene expression and subcellular protein distribution in Chinese hamster ovary cells. Nucleic Acids Res. 1979;7:1739–1747. doi: 10.1093/nar/7.7.1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bouvet P, Diaz J J, Kindbeiter K, Madjar J J, Amalric F. Nucleolin interacts with several ribosomal proteins through its RGG domain. J Biol Chem. 1998;273:19025–19029. doi: 10.1074/jbc.273.30.19025. [DOI] [PubMed] [Google Scholar]
- 7.Brown V, Small K, Lakkis L, Feng Y, Gunter C, Wilkinson K D, Warren S T. Purified recombinant Fmrp exhibits selective RNA binding as an intrinsic property of the fragile X mental retardation protein. J Biol Chem. 1998;273:15521–15527. doi: 10.1074/jbc.273.25.15521. [DOI] [PubMed] [Google Scholar]
- 8.Bugler B, Caizergues-Ferrer M, Bouche G, Bourbon H, Amalric F. Detection and localization of a class of proteins immunologically related to a 100-kDa nucleolar protein. Eur J Biochem. 1982;128:475–480. doi: 10.1111/j.1432-1033.1982.tb06989.x. [DOI] [PubMed] [Google Scholar]
- 9.Coffee B, Zhang F, Warren S T, Reines D. Acetylated histones are associated with FMR1 in normal but not fragile X-syndrome cells. Nat Genet. 1999;22:98–101. doi: 10.1038/8807. [DOI] [PubMed] [Google Scholar]
- 10.Consortium D-B F X. FMR1 knockout mice: a model to study fragile X mental retardation. Cell. 1994;78:23–33. [PubMed] [Google Scholar]
- 11.Dreyfuss G, Matunis M J, Pinol-Roma S, Burd C G. hnRNP proteins and the biogenesis of mRNA. Annu Rev Biochem. 1993;62:289–321. doi: 10.1146/annurev.bi.62.070193.001445. [DOI] [PubMed] [Google Scholar]
- 12.Eberhart D E, Malter H E, Feng Y, Warren S T. The fragile X mental retardation protein is a ribonucleoprotein containing both nuclear localization and nuclear export signals. Hum Mol Genet. 1996;5:1083–1091. doi: 10.1093/hmg/5.8.1083. [DOI] [PubMed] [Google Scholar]
- 13.Eberhart D E, Warren S T. The molecular basis of fragile X syndrome. Cold Spring Harbor Symp Quant Biol. 1996;61:679–687. [PubMed] [Google Scholar]
- 14.Feng Y, Absher D, Eberhart D E, Brown V, Malter H, Warren S T. FMRP associates with polyribosomes as an mRNP, and the I304N mutation of severe fragile X syndrome abolishes this association. Mol Cell. 1997;1:109–118. doi: 10.1016/s1097-2765(00)80012-x. [DOI] [PubMed] [Google Scholar]
- 15.Feng Y, Gutekunst C-A, Eberhart D E, Yi H, Warren S T, Hersch S M. Fragile X mental retardation protein: nucleocytoplasmic shuttling and association with somatodendritic ribosomes. J Neurosci. 1997;17:1539–1547. doi: 10.1523/JNEUROSCI.17-05-01539.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ghisolfi L, Kharrat A, Joseph G, Amalric F, Erard M. Concerted activities of the RNA recognition and the glycine-rich C-terminal domains of nucleolin are required for efficient complex formation with pre-ribosomal RNA. Eur J Biochem. 1992;209:541–548. doi: 10.1111/j.1432-1033.1992.tb17318.x. [DOI] [PubMed] [Google Scholar]
- 17.Hamilton B J, Nagy E, Malters J S, Arrick B A, Rigby W F C. Association of heterogeneous nuclear ribonucleoprotein A1 and C proteins with reiterated AUUUA sequences. J Biol Chem. 1993;268:8881–8887. [PubMed] [Google Scholar]
- 18.Hornstra I K, Nelson D L, Warren S T, Yang T P. High resolution methylation analysis of the FMR1 gene trinucleotide repeat region in fragile X syndrome. Hum Mol Genet. 1993;2:1659–1665. doi: 10.1093/hmg/2.10.1659. [DOI] [PubMed] [Google Scholar]
- 19.Kondo K, Kowalski L R, Inouye M. Cold shock induction of yeast NSR1 protein and its role in pre-rRNA processing. J Biol Chem. 1992;267:16259–16265. [PubMed] [Google Scholar]
- 20.Lapeyre B, Amalric F, Ghaffari S H, Rao S V, Dumbar T S, Olson M O. Protein and cDNA sequence of a glycine-rich, dimethylarginine-containing region located near the carboxyl-terminal end of nucleolin (C23 and 100 kDa) J Biol Chem. 1986;261:9167–9173. [PubMed] [Google Scholar]
- 21.Lee W C, Xue Z X, Melese T. The NSR1 gene encodes a protein that specifically binds nuclear localization sequences and has two RNA recognition motifs. J Cell Biol. 1991;112:1–12. doi: 10.1083/jcb.113.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee W C, Zabetakis D, Melese T. NSR1 is required for pre-rRNA processing and for the proper maintenance of steady-state levels of ribosomal subunits. Mol Cell Biol. 1992;12:3865–3871. doi: 10.1128/mcb.12.9.3865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li, Z., K. Wilkinson, S. T. Warren, and Y. Feng. Selective translation inhibition in vitro by the fragile X mental retardation protein. Submitted for publication.
- 24.Limprasert P, Zhong N, Dobkin C, Brown W T. Polymorphism of FXR1 showing lack of association with autism. Am J Med Genet. 1997;74:453–454. [PubMed] [Google Scholar]
- 25.Long E O, Rosen-Bronson S, Karp D R, Malnati M, Sekaly R P, Jaraquemada D. Efficient cDNA expression vectors for stable and transient expression of HLA-DR in transfected fibroblast and lymphoid cells. Hum Immunol. 1991;31:229–235. doi: 10.1016/0198-8859(91)90092-n. [DOI] [PubMed] [Google Scholar]
- 26.Martin M, Garcia-Fernandez L F, Diaz de la Espina S M, Noaillac-Depeyre J, Gas N, Javier Medina F. Identification and localization of a nucleolin homologue in onion nucleoli. Exp Cell Res. 1992;199:74–84. doi: 10.1016/0014-4827(92)90463-i. [DOI] [PubMed] [Google Scholar]
- 27.Miranda G A, Chokler I, Aguilera R J. The murine nucleolin protein is an inducible DNA and ATP binding protein which is readily detected in nuclear extracts of lipopolysaccharide-treated splenocytes. Exp Cell Res. 1995;217:294–308. doi: 10.1006/excr.1995.1090. [DOI] [PubMed] [Google Scholar]
- 28.Oberle I, Heilig R, Moisan J P, Kloepfer C. Fragile-X mental retardation syndrome with two flanking polymorphic DNA markers. Proc Natl Acad Sci USA. 1986;83:1016–1020. doi: 10.1073/pnas.83.4.1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Orrick L R, Olson M O, Busch H. Comparison of nucleolar proteins of normal liver and Novikoff hepatoma ascites cells by two-dimensional polyacrylamide gel electrophoresis. Proc Natl Acad Sci USA. 1973;70:1316–1320. doi: 10.1073/pnas.70.5.1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pieretti M, Zhang F, Fu Y H, Warren S T, Oostra B A, Caskey C T, Nelson D L. Absence of expression of the FMR-1 gene in fragile X syndrome. Cell. 1991;66:817–822. doi: 10.1016/0092-8674(91)90125-i. [DOI] [PubMed] [Google Scholar]
- 31.Price D K, Zhang F, Ashley C T, Warren S T. The chicken FMR1 gene is highly conserved with a CCT 5′ untranslated repeat and encodes an RNA-binding protein. Genomics. 1996;31:3–12. doi: 10.1006/geno.1996.0002. [DOI] [PubMed] [Google Scholar]
- 32.Sant A J, Braunstein N S, Germain R N. Predominant role of amino-terminal sequences in dictating efficiency of class II major histocompatibility complex alpha beta dimer expression. Proc Natl Acad Sci USA. 1987;84:8065–8069. doi: 10.1073/pnas.84.22.8065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schmidt-Zachmann M S, Nigg E A. Protein localization to the nucleolus: a search for targeting domains in nucleolin. J Cell Sci. 1993;105:799–806. doi: 10.1242/jcs.105.3.799. [DOI] [PubMed] [Google Scholar]
- 34.Siomi H, Choi M, Siomi M C, Nussbaum R L, Dreyfuss G. Essential role for KH domains in RNA binding: impaired RNA binding by a mutation in the KH domain of FMR1 that causes fragile X syndrome. Cell. 1994;77:33–39. doi: 10.1016/0092-8674(94)90232-1. [DOI] [PubMed] [Google Scholar]
- 35.Siomi H, Siomi M C, Nussbaum R L, Dreyfuss G. The protein product of the fragile X gene, FMR1, has characteristics of an RNA binding protein. Cell. 1993;74:291–298. doi: 10.1016/0092-8674(93)90420-u. [DOI] [PubMed] [Google Scholar]
- 36.Siomi M, Siomi H, Sauer W H, Srinivasan S, Nussbaum R L, Dreyfuss G. FXR1, an autosomal homolog of the fragile X mental retardation gene. EMBO J. 1995;14:2401–2408. doi: 10.1002/j.1460-2075.1995.tb07237.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Siomi M C, Zhang Y, Siomi H, Dreyfuss G. Specific sequences in the fragile X syndrome protein FMR1 and the FXR proteins mediate their binding to 60S ribosomal subunits and the interactions among them. Mol Cell Biol. 1996;16:3825–3832. doi: 10.1128/mcb.16.7.3825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sittler A, Devys D, Weber C, Mandel J-L. Alternative splicing of exon 14 determines nuclear or cytoplasmic localisation of fmr1 protein isoforms. Hum Mol Genet. 1996;5:95–102. doi: 10.1093/hmg/5.1.95. [DOI] [PubMed] [Google Scholar]
- 39.Sutcliffe J S, Nelson D L, Zhang F, Pieretti M, Caskey C T, Saxe D, Warren S T. DNA methylation represses FMR-1 transcription in fragile X syndrome. Hum Mol Genet. 1992;1:397–400. doi: 10.1093/hmg/1.6.397. [DOI] [PubMed] [Google Scholar]
- 40.Tamanini F, Meijer N, Verheij C, Willems P J, Galjaard H, Oostra B A, Hoogeveen A T. FMRP is associated to the ribosomes via RNA. Hum Mol Genet. 1996;5:809–813. doi: 10.1093/hmg/5.6.809. [DOI] [PubMed] [Google Scholar]
- 41.Warren, S. T., and S. L. Sherman. The fragile X syndrome. In C. R. Scriver, A. L. Beaudet, W. S. Sly, and D. Valle (ed.), The metabolic and molecular basis of inherited disease, 8th ed., in press. McGraw-Hill Book Co., New York, N.Y.
- 42.Willemsen R, Bontekoe C F T, Galjaard H, Hoogeveen A, Oostra B. Association of FMRP with ribosomal precursor particles in the nucleolus. Biochem Biophys Res Commun. 1996;225:27–33. doi: 10.1006/bbrc.1996.1126. [DOI] [PubMed] [Google Scholar]
- 43.Xue Z, Shan X, Lapeyre B, Melese T. The amino terminus of mammalian nucleolin specifically recognizes SV40 T-antigen type nuclear localization sequences. Eur J Cell Biol. 1993;62:13–21. [PubMed] [Google Scholar]
- 44.Yurkova M S, Murray M T. A translation regulatory particle containing the Xenopus oocyte Y box protein mRNP3+4. J Biol Chem. 1997;272:10870–10876. doi: 10.1074/jbc.272.16.10870. [DOI] [PubMed] [Google Scholar]
- 45.Zaidi S H, Malter J S. Nucleolin and heterogeneous nuclear ribonucleoprotein C proteins specifically interact with the 3′-untranslated region of amyloid protein precursor mRNA. J Biol Chem. 1995;270:17292–17298. doi: 10.1074/jbc.270.29.17292. [DOI] [PubMed] [Google Scholar]
- 46.Zhang Y, O’Connor J P, Siomi M C, Srinivasan S, Dutra A, Nussbaum R L, Dreyfuss G. The fragile X mental retardation syndrome protein interacts with novel homologs FXR1 and FXR2. EMBO J. 1995;14:5358–5366. doi: 10.1002/j.1460-2075.1995.tb00220.x. [DOI] [PMC free article] [PubMed] [Google Scholar]