Abstract
The Saccharomyces cerevisiae HOP2 gene is required to prevent formation of synaptonemal complex between nonhomologous chromosomes during meiosis. The HOP2 gene is expressed specifically in meiotic cells, with the transcript reaching maximum abundance early in meiotic prophase. The HOP2 coding region is interrupted by an intron located near the 5′ end of the gene. This intron contains a nonconsensus 5′ splice site (GUUAAGU) that differs from the consensus 5′ splice signal (GUAPyGU) by the insertion of a nucleotide and by a single nucleotide substitution. Bases flanking the HOP2 5′ splice site have the potential to pair with sequences in U1 small nuclear RNA, and mutations disrupting this pairing reduce splicing efficiency. HOP2 pre-mRNA is spliced efficiently in the absence of the Mer1 and Nam8 proteins, which are required for splicing the transcripts of two other meiosis-specific genes.
RNA splicing is required to generate an mRNA containing a functional open reading frame (ORF) from a pre-mRNA containing one or more introns. Splicing is a two-step reaction (54). During the first step, the 5′ end of the intron is cleaved and joined to a specific nucleotide within the intron, resulting in formation of a lariat. In the second reaction, the two exons are ligated together and the intron is released.
RNA splicing is catalyzed by a protein-RNA complex called the spliceosome, which assembles in a highly ordered manner. Many of the steps in spliceosome assembly involve base pairing between conserved pre-mRNA sequences and the small nuclear RNA (snRNA) components of the spliceosome (46, 54). In Saccharomyces cerevisiae, most introns contain three highly conserved splicing signals (53): the 5′ splice site, GUAPyGU; the 3′ splice site, PyAG; and the UACUAAC sequence that lies upstream of the 3′ splice site and functions as the branch point for lariat formation. Previous studies have demonstrated that the 5′ splice site pairs sequentially with sequences in U1 and U6 snRNAs. Exon sequences immediately adjacent to the 5′ and 3′ splice sites pair with U5 snRNA, while the branch point sequence pairs with U2 snRNA. Proper pairing between splicing signal sequences and snRNAs is important for the efficiency and accuracy of splicing (54).
In some cases, splicing serves to regulate gene expression (28, 52). Regulated splicing often requires specialized factors, in addition to components of the general splicing machinery (28, 59). The MER2 gene provides one of few examples of regulated splicing in budding yeast. The MER2 gene has a nonconsensus 5′ splice site (16), and splicing of MER2 pre-mRNA requires two proteins, Mer1 and Nam8 (also known as Mre2), that are not required for general splicing (11, 16, 32, 38). MER2 pre-mRNA is present in both mitotic and meiotic cells (16), but the transcript is spliced efficiently only in meiotic cells because the Mer1 protein is produced only during meiosis (14). Compared to the consensus 5′ splice site in yeast, the MER2 5′ splice site has one less potential base pair with U1 snRNA (16). If pairing between the MER2 5′ splice site and U1 snRNA is increased by mutating nucleotides in either the MER2 intron or U1 snRNA, the requirement for Mer1 is alleviated (34). These observations suggest that the function of Mer1 is to promote or stabilize the interaction between the MER2 5′ splice site and U1 snRNA. Like MER2, the MER3 gene contains an intron with a nonconsensus 5′ splice site and requires the Mer1 and Nam8 proteins for splicing. However, no obvious regulatory role can be ascribed to MER3 splicing because the MER3 gene is transcribed only in meiotic cells (33).
Meiosis is a special form of cell division that produces haploid gametes from diploid parental cells. During meiotic prophase, homologous chromosomes pair with each other, undergo genetic recombination and engage in synaptonemal complex formation (44). These interactions between homologous chromosomes are essential for proper chromosome segregation at the first meiotic division. In S. cerevisiae, the meiosis-specific HOP2 gene plays an important role in promoting interhomolog interactions (26). In the hop2 mutant, chromosomes engage in nearly wild-type amounts of synaptonemal complex formation, but most synapsis involves nonhomologous chromosomes. In addition, the hop2 mutant sustains approximately the wild-type number of meiotic double-strand breaks (the initiators of meiotic recombination), but these breaks remain unrepaired. hop2 cells arrest at pachytene due to a checkpoint triggered by the failure to complete recombination and/or by the aberration in synaptonemal complex assembly.
In this study, we have identified the HOP2 ORF and an intron in the HOP2 coding region. This intron contains an unusual 5′ splice site that differs from the consensus sequence by the insertion of an extra nucleotide and by a single nucleotide substitution. Nonetheless, splicing of HOP2 transcripts does not require the Nam8 protein, nor does it require Mer1 or any other meiosis-specific factors. There is potential for extended base pairing between U1 snRNA and sequences flanking the HOP2 5′ splice site; these flanking sequences are important for efficient splicing of HOP2 pre-mRNA.
MATERIALS AND METHODS
Strains and genetic procedures.
Yeast strain genotypes are listed in Table 1. Yeast manipulations were performed and media were prepared as specified by Sherman et al. (49). Substitutive and integrative transformations (45) were carried out by the lithium acetate procedure (23). In diploid strains carrying the HOP2-H2, hop2-M, hop2-H3, hop2-M3, and hop2-M4 mutations, both copies of the chromosomal HOP2 gene were replaced with the indicated hop2 mutant allele by the two-step transplacement method (45). The nam8::TRP1 disruption was generated by the method of Baudin et al. (3). Oligonucleotides with homology to the NAM8 gene were used to amplify TRP1 from pR314 (50); the resulting PCR product, containing a TRP1 gene flanked by 50 bp of sequences upstream and downstream of the NAM8 coding region, was then transformed into yeast.
TABLE 1.
Yeast strains used
| Strain | Genotypea |
|---|---|
| BR2171-7B | MATa leu2ura3-1HO trp1-1arg4-8 thr1-4ura3-1ade2-1 |
| MATα leu2 ura3-1 HO trp1-1 arg4-8 thr1-4 ura3-1 ade2-1 | |
| YAB36 | MATa leu2-27 his4-280ura3-1trp1-1 spo13::ura3-1 arg4-9 thr1-4cyh10 LYS2ade2-1 |
| MATα leu2-3,112 his4-260 ura3-1 trp1-289 SPO13 arg4-8 thr1-1 CHY10 lys2-1 ade2-1 | |
| YAB27 | YAB36 but homozygous hop2::URA3 |
| YAB97 | YAB36 but homozygous HOP2-H2 |
| YAB98 | YAB36 but homozygous hop2-M |
| YAB99 | YAB36 but homozygous hop2-M3 |
| YAB100 | YAB36 but homozygous hop2-M4 |
| YAB103 | YAB36 but homozygous hop2-H3 |
| YAB274 | YAB36 but hop2-H2::lacZ |
| HOP2 | |
| YAB275 | YAB36 but hop2::lacZ |
| HOP2 | |
| YAB265 | YAB27 + pL44 (HOP2) |
| YAB266 | YAB27 + pL45 (HOP2-H2) |
| YAB71 | YAB36 but homozygous mer1::ADE2 |
| YAB276 | YAB71 but hop2-H2::lacZ |
| HOP2 | |
| YAB277 | YAB71 but hop2::lacZ |
| HOP2 | |
| YAB68 | YAB36 but homozygous hop2::URA3 mer1::ADE2 |
| YAB262 | YAB68 + pL44 (HOP2) |
| YAB263 | YAB68 + pL45 (HOP2-H2) |
| YAB230 | YAB36 but homozygous nam8::TRP1 |
| YAB278 | YAB230 but hop2-H2::lacZ |
| HOP2 | |
| YAB279 | YAB230 but hop2::lacZ |
| HOP2 | |
| YAB231 | YAB36 but homozygous hop2::URA3 nam8::TRP1 |
| YAB267 | YAB231 + pL44 (HOP2) |
| YAB268 | YAB231 + pL45 (HOP2-H2) |
| YAB270 | YAB36 + pL52 (ADH1::hop2::lacZ) |
| YAB269 | YAB36 + pL53 (ADH1::hop2-H2::lacZ) |
| YAB280 | YAB27 + pL62 (hop2-M2) |
| YAB281 | YAB27 + pL66 (hop2-M5) |
| YAB248 | YAB27 + pL81 (hop2-5P) |
| YAB247 | YAB27 + pL82 (hop2-3P) |
| YAB249 | YAB27 + pL84 (hop2-35P) |
| S168 | MATα ura3 lys2 ho::LYS2 arg4-bgl |
| NKY291 | MATa leu2::his4G ura3 lys2 ho::LYS2 |
| AS4 | MATα trp1 arg4 tyr7 ade6 ura3 MAL2 |
| Y260 | MATa his4-580 leu2-3,112 ura3-52 pep4-3 |
| BR1373-6D | MATa leu2-27 his4-280 ura3-1 spo13::ura3-1 arg4-8 thr1-1 trp1-1 ade2-2 cyh 10 |
The relevant HOP2 alleles present on replicating plasmids are indicated in parentheses following the plasmid designations.
In sporulation assays, diploid cells were grown to saturation in YPAD, or synthetic medium lacking leucine or uracil (to select for plasmids), and then diluted two- to fourfold into YPAD and grown for 12 h. Cells were then washed, diluted fivefold into 2% potassium acetate, and incubated at 30°C with vigorous shaking.
Escherichia coli XL1-Blue (Stratagene) was used for plasmid constructions. Bacterial strains used for transposon mutagenesis have been described by Hoekstra et al. (22). pL14 (25) was subjected to transposon mutagenesis in E. coli, using derivatives of the Tn3 transposon containing the bacterial lacZ coding region and the yeast LEU2 or URA3 gene (22). The transposon insertions were mapped by sequence analysis and introduced into a homothallic strain (BR2171-7B [42]) after digestion with NotI. The resulting transformants (heterozygous for the transposon) were sporulated, and tetrads were dissected; cells from Leu+ or Ura+ (depending on the transposon) diploid spore colonies were then tested for the ability to sporulate.
Plasmids.
Plasmids were constructed by standard methods (47). Gene disruptions were introduced into yeast by using pME39 for mer1::ADE2 (16) and pL21 for hop2::URA3 (25).
Plasmids carrying mutant HOP2 genes were constructed as follows. To generate pL68, the 2.2-kb SphI-EcoRI fragment containing the wild-type HOP2 gene from pL14 (25) was subcloned into pUN50 (12); the 2.2-kb HOP2-containing HindIII fragment from the pUN50 derivative was then subcloned into the integrating vector, pRS306 (50). The hop2-M, hop2-H3, hop2-M3, and hop2-M4 mutants were engineered by recombinational PCR (21). The PCR fragments were digested with NheI and BglII and substituted for the wild-type NheI-BglII fragment in pL68 to make the integration plasmids pL69 (hop2-M), pL73 (hop2-H3), pL70 (hop2-M3), and pL71 (hop2-M4). The HOP2-H2 integration plasmid, pL72, was made by replacing the wild-type NheI-BglII fragment of pL68 with the cDNA fragment obtained by reverse transcription-PCR (RT-PCR). All integration plasmids described above were cut with NheI before transformation into yeast. pL36 was derived from pL13 (containing CEN4, ARS1, LEU2, and the wild-type HOP2 gene) (25) by deleting the 0.5-kb SpeI-HindIII fragment of HOP2-adjacent DNA and the EcoRI-SalI segment of the polylinker. The hop2-M2, hop2-M5, hop2-3P, hop2-5P, and hop2-35P mutations were engineered by recombinational PCR. The PCR fragments were digested with BsiWI and BamHI and substituted for the wild-type BsiWI-BamHI fragment in pL36 to make pL62 (hop2-M2), pL66 (hop2-M5), pL82 (hop2-3P), pL81 (hop2-5P), and pL84 (hop2-35P).
Plasmids used to generate probes for RNase protection assays were constructed as follows. To create pL59 (which was used to generate wild-type HOP2 probe 1 [Fig. 1]), a 0.7-kb PCR product was amplified by using primers P10 (5′-GAACTGACCAAGCTTTATTTGAAAGATATG-3′, corresponding to bases −27 to +3) and P5 (5′-GGTCTCGAAAAAGCTTCAAAATACATACCA-3′, corresponding to bases +717 to +688). (Nucleotides are numbered as in Fig. 2C, where +1 indicates the first nucleotide in the HOP2 ORF.) The resulting PCR product was digested with HindIII and HincII and inserted between the HindIII and HincII sites of pBS M13+ (Stratagene). To generate HOP2 probe 2 for wild-type and mutant genes, 134-bp PCR products were amplified from pL68, pL73, pL70, pL82, pL81, and pL84 by using primers P20 (5′-GCTCTATTTGAATTCTATGGCACC-3′, corresponding to bases −16 to +9) and P11 (5′-AATACCCGGGGTACAATTACTAGTAATGGC-3′, corresponding to bases +118 to +89). The PCR fragments were then digested with EcoRI and SpeI and inserted between the EcoRI and XbaI sites of pBS M13+ to generate pL107 (HOP2), pL108 (hop2-H3), pL109 (hop2-M3), pL110 (hop2-3P), pL111 (hop2-5P), and pL118 (hop2-35P).
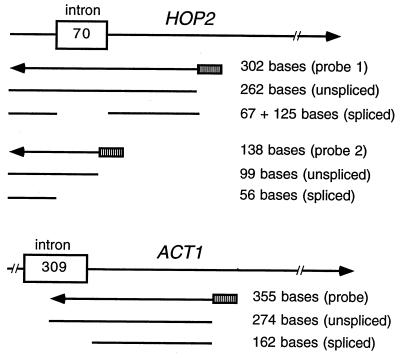
FIG. 1.
HOP2 and ACT1 probes used for RNase protection experiments. The HOP2 and ACT1 primary transcripts are represented diagrammatically; indicated below each diagram are the probes used for RNase protection and the protection products expected from spliced and unspliced RNA. The hatched boxes indicate the promoter and polylinker regions of the vector.
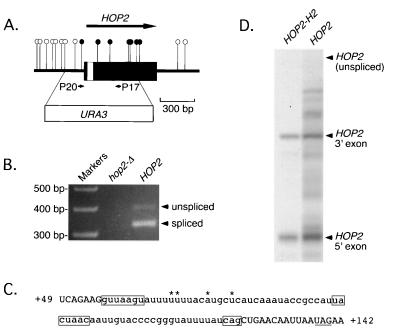
FIG. 2.
Identification and sequence of the HOP2 intron. (A) Diagram of the HOP2 gene, with exon sequences indicated in black and the intron indicated in white. The arrow depicts the direction and extent of the HOP2 coding region. Closed circles represent transposon insertions that disrupt HOP2 function, while open circles represent those do not disrupt gene function. P20 and P17 are the primers that were used for RT-PCR. Indicated below the HOP2 diagram are the endpoints of the hop2::URA3 deletion/disruption mutation used for panel B. (B) Products of RT-PCR derived from wild-type (YAB36) and hop2::URA3 (YAB27) cells. Molecular weight markers are indicated on the left. (C) RNA sequence of the HOP2 intron (lowercase) and flanking regions (uppercase). Numbers refer to the RNA sequence, with +1 indicating the first base of the HOP2 initiation codon. The boxed regions represent the splicing signal sequences. An alternative 3′ splice site that may be used in the hop2-M mutant is underlined. The nucleotides that vary among yeast strains are indicated by asterisks. (D) RNase protection of RNA from strains carrying the wild-type HOP2 gene (YAB36) or the HOP2-H2 cDNA allele (YAB97). RNA isolated from diploid cells harvested after 14 h in sporulation medium was subjected to RNase protection using the antisense HOP2 probe 1 (Fig. 1). Unspliced HOP2 RNA is detectable only after prolonged exposure.
Plasmids containing HOP2::lacZ fusion genes were engineered as follows. pL105 and pL106 were constructed by subcloning the 3.2-kb BamHI-BglII fragment containing lacZ from RP370 (60) into the BglII sites of pL72 and pL68. pL105 and pL106 were cut with NheI and integrated into the yeast genome to generate hop2-H2::lacZ and hop2::lacZ strains, respectively, for β-galactosidase assays.
Plasmids in which HOP2::lacZ genes are fused to the ADH1 promoter were constructed as follows. First, a HindIII site was introduced just before the HOP2 ORF by recombinational PCR. The PCR fragment was then digested with BamHI and NheI and substituted for the wild-type BamHI-XbaI fragment in pL36 to generate pL44. pL45 was constructed in a similar manner except that the intronless HOP2 cDNA was used as the template for amplification by PCR. The 1.5-kb BamHI-HindIII fragment containing the ADH1 promoter from pAAH5 (2) was inserted between the HindIII site and the BamHI site upstream of HOP2 in pL44 and pL45 to generate pL50 and pL51, respectively. The 2.1- and 2.0-kb BamHI fragments from pL50 and pL51 were then cloned into RP370, which contains lacZ, URA3, and the 2μm circle origin of DNA replication (60), to generate pL52 (hop2::lacZ) and pL53 (hop2-H2::lacZ), respectively.
To clone the HOP2 gene from different strains, total yeast genomic DNA was prepared from BR1373-6D (41), AS4 (10), S168 (20), NKY291 (1), and Y260 (obtained from Michael Snyder), and the HOP2 ORF was then amplified by PCR using primers P10 and P5. The products of PCR were purified and subcloned into pBS M13+ prior to sequencing.
RNase protection analysis and PCR amplification of HOP2 cDNA.
RNA isolation was carried out as described by Engebrecht et al. (16). RNase protection assays were carried out by using an RPAII kit from Ambion, Inc. (Austin, Tex.) as described by Engebrecht et al. (16). The antisense ACT1 probe was generated by cutting pL85 (26) with SspI and transcribing with T7 RNA polymerase (Fig. 1). The antisense HOP2 probe 1 (Fig. 1) was synthesized by in vitro transcription (24) of pL59 linearized with HindIII, using T7 RNA polymerase. The antisense HOP2 probe 2 (Fig. 1) for wild-type and mutant HOP2 genes was generated by cutting pL107, pL108, pL109, pL110, pL111, and pL112 with EcoRI and transcribing with T3 RNA polymerase. The levels of spliced and unspliced RNAs were quantitated using Multi-Analyst software (Bio-Rad) to scan autoradiograms and analyze the resulting data.
PCR analysis was performed from RNA templates with a GeneAmp Thermostable rTth reverse transcriptase RNA PCR kit (Perkin-Elmer Cetus) as instructed by the supplier. The upstream primer was P20, and the downstream primer was P17 (5′-CATTTCTCAGTTGCAATACAG-3′, corresponding to bases +383 to +363).
β-Galactosidase assays.
β-Galactosidase assays were performed as described by Chua and Roeder (6). β-Galactosidase activity units are defined as nanomoles of o-nitrophenyl-β-d-galactopyranoside cleaved per minute per milligram of protein.
RESULTS
The HOP2 intron contains a unique 5′ splice site.
The wild-type HOP2 gene was cloned from a yeast genomic library based on complementation of the hop2-1 sporulation defect (25). Complementing activity was localized to a region of 0.8 kbp by subcloning followed by transposon mutagenesis (Fig. 2A). Sequence analysis revealed that the 0.8-kb fragment does not contain an obvious ORF but does include a long coding region that lacks an in-frame initiation codon. These observations suggested that the HOP2 transcript might be spliced. A potential branch point sequence (UACUAAC) is found near the 5′ end of the 0.8-kbp fragment, and several consensus 3′ splice sites are located downstream of the UACUAAC box. However, no consensus 5′ splice site sequence is located upstream of the UACUAAC box.
To determine whether the HOP2 transcript is spliced and, if it is, to define the ends of the intron, HOP2 RNA from meiotic cells was reverse transcribed and the resulting cDNA was then amplified by PCR. Two PCR products that differ in size by less than 100 bp were amplified from total RNA of wild-type cells but not from cells carrying a HOP2 deletion mutation (Fig. 2B). Cloning and sequencing of the smaller RT-PCR product revealed that this fragment lacks 70 bp present in the genomic sequence. The UACUAAC box is located within the putative intron (nucleotides [nt] 98 to 105 [Fig. 2C]), and the 3′ end of the intron is defined by a consensus 3′ splice site (nt 123 to 135 [Fig. 2C]). However, the sequence present at the 5′ end of the intron (GUUAAGU, nt 56 to 62 [Fig. 2C]) differs from the consensus 5′ splice site (GUAPyGU) by the insertion of a U residue adjacent to the first U in the consensus to create a UU dinucleotide. In addition, the pyrimidine residue present at the fourth position of the consensus sequence is a purine (at the fifth position) in the HOP2 5′ splice site.
Diploids homozygous for the intronless version of HOP2 recovered by RT-PCR sporulate efficiently and make viable spores (Fig. 3C), indicating that this version of the gene encodes a functional Hop2 protein. To determine whether the HOP2 mRNA amplified by RT-PCR is the major product of HOP2 splicing, total RNA isolated from meiotic cells was analyzed by RNase protection assays using a probe spanning the intron (Fig. 1). Diploids in which both copies of the wild-type HOP2 gene have been replaced by the intronless HOP2 gene (HOP2-H2) recovered by RT-PCR generate two RNase protection products (Fig. 2D). One fragment is the size expected for sequences upstream of the putative intron, while the other corresponds to sequences downstream of the intron. The two predominant RNase protection products derived from wild type are the same size as those derived from the HOP2-H2 gene (Fig. 2D), indicating that the spliced product recovered by RT-PCR is indeed the predominant spliced mRNA generated from the wild-type gene. In addition to the two major products, we detected a number of other fragments that may represent minor splicing products or RNAs that have been degraded. Little or no unspliced HOP2 pre-mRNA was detected in wild-type meiotic cells.
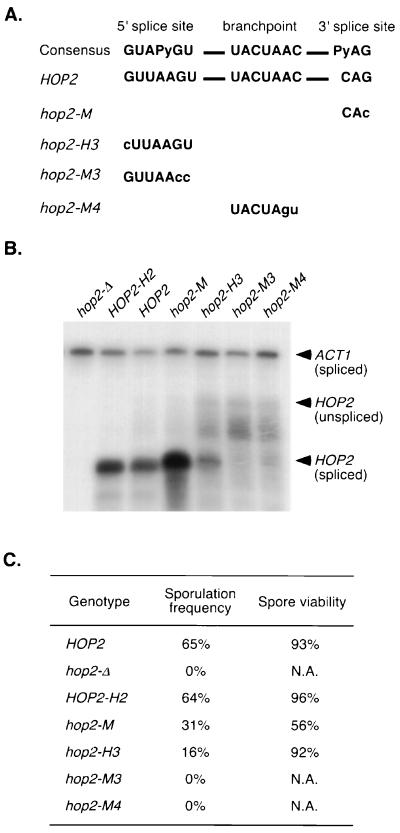
FIG. 3.
Mutational analysis of HOP2 splicing signal sequences. (A) Sequences of HOP2 mutations in the branch point sequence, 5′ splice site, and 3′ splice site. The nucleotides altered by mutation are presented in lowercase. (B) RNase protection of RNA from the wild type and hop2 mutants. RNAs isolated from diploid cells harvested after 14 h in sporulation medium were assayed by RNase protection using the antisense HOP2 probe 2 (Fig. 1). RNAs derived from strains YAB27 (hop2::URA3), YAB36 (HOP2), YAB97 (HOP2-H2), YAB98 (hop2-M), and YAB100 (hop2-M4) were hybridized with probe derived from pL107. RNAs from strains YAB103 (hop2-H3) and YAB99 (hop2-M3) were hybridized with probes derived from pL108 and pL109, respectively. All HOP2 probes are similar except that 1 or 2 nt have been changed, such that each probe is complementary to the corresponding mutant transcript. Transcription of the ACT1 gene is constitutive; therefore, ACT1 mRNA serves as a loading control. Use of the alternative 3′ splice site postulated for the hop2-M mutant (see Results) would not change the size of the protection product obtained with HOP2 probe 2 (Fig. 1), because this probe does not extend to the 3′ end of the intron. (C) Sporulation efficiency and spore viability of the wild type and hop2 mutants. Spore viability was determined from dissected tetrads; at least 200 spores were scored for each strain.
The sequence of the spliced HOP2 transcript contains an ORF that is 203 codons in length, with 55 nt of coding sequence located upstream of the intron and 554 nt of coding sequence downstream (25). To confirm that the HOP2 ORF has been correctly identified, sequences near the putative ATG initiation codon were mutated and examined for their effects on HOP2 gene function by monitoring sporulation efficiencies. Insertion of a nucleotide immediately 5′ of the initiation codon will not change the ORF and therefore should not affect gene function. In contrast, insertion of a single nucleotide immediately 3′ of the initiation codon will create a frameshift mutation and thus should result in a failure to make functional protein. In the hop2-M5 mutant, a T residue was inserted before the predicted initiation codon; this mutant sporulates as efficiently as the wild type (65% sporulation in both mutant [YAB281] and wild type [YAB267]) and displays the wild-type level of spore viability type (92% in the mutant and 93% in the wild type). In the hop2-M2 mutant strain YAB280, a G residue was inserted after the initiation codon; this mutant fails to sporulate (0% spore formation). These data indicate that translation of the Hop2 protein does indeed initiate at the predicted ATG codon.
Mutational analysis of HOP2 splice site signals.
To confirm that splicing is required for HOP2 gene expression and to identify the signal sequences required for splicing, a series of mutations were constructed by site-directed mutagenesis and then examined for their effects on splicing and sporulation. In the hop2-M4 mutant, the UACUAAC box is changed to UACUAGU (Fig. 3A), which should prevent the first step in splicing (54). hop2-M4 cells make very little spliced HOP2 mRNA; some unspliced transcript is detected, as well as a number of other RNAs that may be degradation products (Fig. 3B). Diploids homozygous for the hop2-M4 mutation fail to sporulate (Fig. 3C), as expected if proper splicing is required for HOP2 gene expression.
In the hop2-M3 mutant, the HOP2 5′ splice site GUUAAGU is changed to GUUAACC (Fig. 3A). The hop2-M3 mutant does not sporulate and makes little or no spliced RNA of the appropriate size (Fig. 3B and C), suggesting that the 5′ splice site identified by RT-PCR is the only 5′ splice site used to make functional HOP2 mRNA. Alternative (potential) 5′ splice sites are not utilized and/or lead to the production of mRNAs that cannot be translated to generate functional protein. (For example, the potential 5′ splice site [GUAUUU] at positions 61 to 66 [Fig. 2C] could not be used to make protein since there is a stop codon [UAA, nt 56 to 58 [Fig. 2C] in frame with the HOP2 coding region immediately upstream of this potential splice site.) In the hop2-H3 mutant, the first nucleotide of the 5′ splice site is changed from a G to a C. This mutation reduces both the efficiency of splicing and the level of sporulation, but not as dramatically as the hop2-M3 mutation (Fig. 3B and C). Thus, the presence of a G residue at the first position in the HOP2 5′ splice site appears to be important for splicing, but it is not absolutely essential.
In the hop2-M mutant, the HOP2 3′ splice site (bases 123 to 125 [Fig. 2C]) is changed from CAG to CAC, but the resulting HOP2 pre-mRNA appears nevertheless to be spliced efficiently (Fig. 3B). Given the unusual HOP2 5′ splice site and known interactions between the 5′ and 3′ splice site, it is possible that the mutant 3′ splice site can still be utilized. Alternatively, it is possible that splicing of hop2-M mutant pre-mRNA depends on a consensus 3′ splice site (UAG, corresponding to bases 138 to 140 in Fig. 2C) located 15 bp downstream of the 3′ splice site identified by RT-PCR. Use of the normally cryptic 3′ splice site would result in an mRNA that is 15 bases smaller than its wild-type counterpart and a protein that differs from the wild-type Hop2 protein by the deletion of five amino acids. In the hop2-M mutant, both sporulation efficiency and spore viability are decreased about twofold. If the mutant 3′ splice site (CAC) is used, then this meiotic defect suggests a reduction in efficiency of the second step in splicing (the RNase protection assay in Fig. 3B monitors only the first step in splicing). If the cryptic 3′ splice site (UAG) is used, then the meiotic phenotype suggests that the five amino acids deleted from the mutant protein influence the function or stability of the Hop2 protein.
Sequences flanking the HOP2 5′ splice site are important for splicing.
Alignment of the HOP2 5′ splice site with sequences in U1 snRNA indicates that the potential for base pairing between U1 snRNA and HOP2 pre-mRNA extends outside the 5′ splice site, to include 3 nt upstream of the 5′ splice site and 2 nt downstream of the splice site (Fig. 4A). In yeast introns, sequences flanking the 5′ splice site per se are not very conserved (27), suggesting that pairing between the 6-nt 5′ splice site and the complementary sequence in U1 snRNA is normally sufficient to stabilize the interaction between these RNA molecules. However, since pairing between U1 snRNA and the 7-nt HOP2 5′ splice site is predicted to be less stable due to the extra nucleotide, pairing between snRNA and nucleotides flanking the 5′ splice site proper may be important for a stable interaction. To investigate this possibility, mutants that reduce or eliminate the potential for pairing of U1 snRNA with HOP2 nucleotides outside the 5′ splice site were constructed and analyzed.
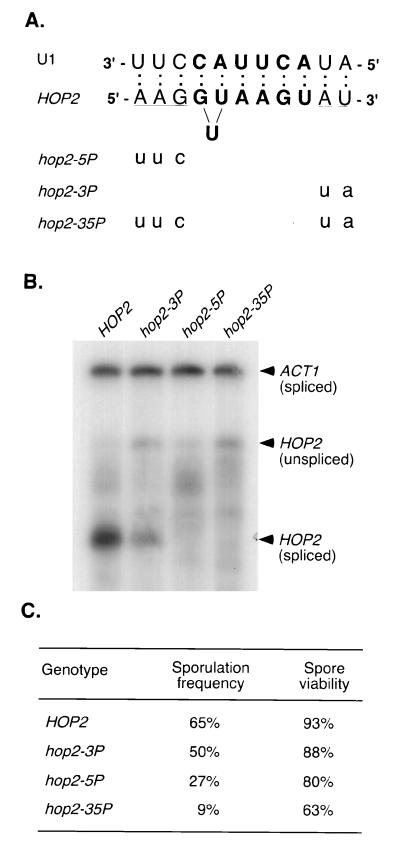
FIG. 4.
Mutational analysis of sequences flanking the HOP2 5′ splice site. (A) Mutations in sequences flanking the 5′ splice site. The sequences flanking the wild-type 5′ splice site (indicated in bold) are underlined, and potential base pairing between U1 snRNA and HOP2 RNA is indicated by colons. The exact nature of pairing between the HOP2 5′ splice site the U1 snRNA is uncertain, since the nucleotide that fails to pair could be either one of the two adjacent U residues in the splice site. Nucleotides altered by mutation are presented in lowercase. (B) RNase protection assays of RNA from the wild type and hop2 mutants. RNA isolated from diploid cells after 14 h in sporulation medium was assayed by RNase protection using antisense HOP2 probe 2 and the ACT1 probe (Fig. 1). RNAs from strains YAB265 (HOP2), YAB247 (hop2-3P), YAB248 (hop2-5P), and YAB249 (hop2-35P) were hybridized with HOP2 probes derived from pL107, pL110, pL111, and pL112, respectively. All HOP2 probes are similar except that 2 to 5 nt have been changed to match the mutant transcripts. (C) Sporulation efficiency and spore viability of the wild type (YAB265) and hop2 mutants (YAB247, YAB248, and YAB249). Spore viability was determined from dissected tetrads; at least 200 spores were scored for each strain.
In the hop2-3P mutant, the potential for base pairing between U1 snRNA and sequences downstream of the HOP2 5′ splice site was destroyed by changing the eighth and ninth nucleotides (bases 63 and 64) of the intron from AU to UA (Fig. 4A). The efficiency of splicing of the hop2-3P transcript is reduced to 35% of the wild-type level (Fig. 4B), and the sporulation frequency of hop2-3P cells is also slightly decreased (Fig. 4C). In the hop2-5P mutant, the last 3 nt of the upstream exon (bases 53 to 55) were changed to eliminate the potential for pairing between U1 snRNA and sequences upstream of the 5′ splice site (Fig. 4A). Splicing of the hop2-5P transcript is reduced severely (Fig. 4B). Although 27% of hop2-5P cells eventually form asci (Fig. 4C), sporulation is delayed by about 24 h and many asci contain fewer than four spores. (This defect in sporulation may be due in part to the two amino acids substitutions effected by the hop2-5P mutation, E18V and A19P.) In the hop2-35P mutant, sequences both upstream and downstream of the 5′ splice site were mutated to eliminate the potential for pairing with U1 snRNA (Fig. 4A). In this mutant, sporulation efficiency and spore viability are even lower than those of the hop2-3P and hop2-5P mutants (Fig. 4B and C). Furthermore, no spliced product is detected (Fig. 5B), indicating that hop2-3P and hop2-5P have additive or synergistic effects on splicing. These results indicate that nucleotides flanking the HOP2 5′ splice site are important for splicing, presumably because they pair with nucleotides in U1 snRNA.
FIG. 5.
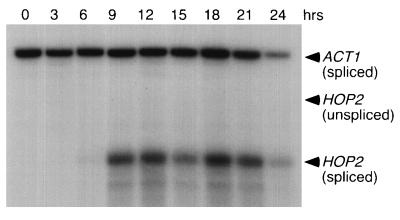
Transcription of the HOP2 gene is induced during meiotic prophase. RNA prepared from wild-type cells (YAB36) at various times after transfer to sporulation medium was subjected to RNase protection using HOP2 probe 2 and the ACT1 probe (Fig. 1). Unspliced HOP2 RNA is detectable only after prolonged exposure.
Transcription of the HOP2 gene is induced in meiotic prophase.
Previous studies have shown that the abundance of the Hop2 protein increases during meiosis (25). In principle, the elevated level of Hop2 protein might be achieved through either transcriptional or posttranscriptional regulation. Since RNA splicing is required for HOP2 gene expression, we considered the possibility that the HOP2 gene is transcribed in both vegetative and meiotic cells, but the transcript is spliced efficiently only in meiotic cells (as found for MER2).
To examine the regulation of HOP2 gene expression, RNase protection assays were used to analyze total RNA isolated from wild-type cells at various times after the introduction into sporulation medium (Fig. 5). The HOP2 transcript was first detected after 6 h in sporulation medium; a maximum level of transcript was reached by 9 h and then maintained for several hours thereafter. Unspliced HOP2 RNA was barely detectable in meiotic cells, indicating that the HOP2 transcript is efficiently spliced. Neither spliced nor unspliced HOP2 RNA was detected in vegetative cells, suggesting that the HOP2 gene is transcribed only during meiosis. Consistent with this result, a sequence (TCGGCGGCTA) that matches the consensus URS1 sequence (YCGGCGGCTA) is located at bases −130 to −121 in the HOP2 upstream region. The URS1 sequence is responsible for both repressing transcription in vegetative cells and inducing transcription in meiotic cells (5).
In the strain used for this analysis, most cells reach the pachytene stage of meiotic prophase (when chromosomes are fully engaged in synaptonemal complex formation) after 15 to 16 h in sporulation medium. Thus, the timing of HOP2 mRNA accumulation is consistent with the requirement for Hop2 specifically for meiotic interhomolog interactions and with the observed localization of the Hop2 protein to chromosomes prior to and during the pachytene stage (25).
HOP2 splicing does not require Nam8 or meiosis-specific factors.
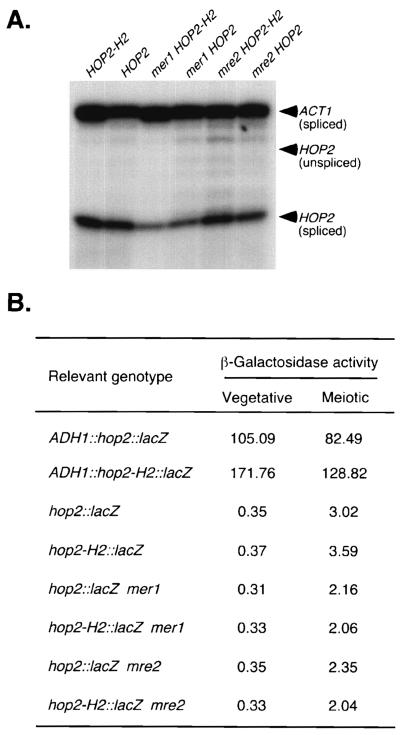
The meiosis-specific Mer1 protein is required for efficient splicing of the MER2 and MER3 transcripts, both of which have nonconsensus 5′ splice sites (16, 33). To determine whether Mer1 is required for splicing of HOP2 pre-mRNA, RNase protection assays were used to analyze total RNA from meiotic cells of mer1 mutant strains carrying either the wild-type HOP2 gene or the intronless HOP2-H2 gene. Similar levels of spliced products were observed in both strains (Fig. 6A), indicating that the Mer1 protein is not essential for splicing of HOP2 transcripts. To assay the levels of Hop2 protein produced by the wild-type and intronless HOP2 genes in the absence of Mer1, a lacZ coding domain was fused in frame to sequences in the second exon and β-galactosidase activity was measured in strains carrying these constructs. mer1 mutants carrying the hop2::lacZ or the hop2-H2::lacZ translational fusion gene exhibit similar levels of β-galactosidase activity (Fig. 6B), indicating that the HOP2 gene is expressed normally in the absence of the Mer1 protein.
FIG. 6.
Analysis of HOP2 splicing in the wild type and mer1 and nam8 mutants. (A) RNase protection of RNA from wild-type, mer1, or nam8 cells carrying either the wild-type or intronless version of the HOP2 gene. RNA isolated from cells after 13 h in sporulation medium was assayed by RNase protection using the antisense HOP2 probe 2 and the ACT1 probe (Fig. 1). Unspliced HOP2 RNA is detectable only after prolonged exposure. The level of HOP2 mRNA is lower in mer1 cells than wild-type cells because mer1 cells sporulate poorly (15). Strains analyzed are (from left to right) YAB266, YAB265, YAB263, YAB262, YAB268, and YAB267. (B) β-Galactosidase assays. Production of the Hop2–β-galactosidase fusion protein was monitored in strains carrying either the hop2::lacZ or the intronless hop2-H2::lacZ gene. Strains analyzed were (from top to bottom) YAB270, YAB269, YAB275, YAB274, YAB277, YAB276, YAB279, and YAB278. All fusion genes are controlled by the original HOP2 promoter except in strains YAB270 (ADH1::hop2::lacZ) and YAB269 (ADH1::hop2-H2::lacZ), in which HOP2 is fused to the constitutive ADH1 promoter. β-Galactosidase assays were performed on vegetative cells (0 h of sporulation) and meiotic cells (15 h of sporulation). Values given are the averages of three independent cultures for each strain.
To determine whether splicing of the HOP2 transcript requires any other meiosis-specific factor(s), the hop2::lacZ and hop2-H2::lacZ genes were fused to the ADH1 promoter such that the fusion genes are expressed in both vegetative and meiotic cells (2). The wild-type ADH1::hop2::lacZ and intronless ADH1::hop2-H2::lacZ fusion genes were introduced into wild-type cells, and β-galactosidase activity was measured both prior to the introduction into sporulation medium and after 15 h of sporulation. In both vegetative and meiotic cells, β-galactosidase activity was somewhat higher in the strain carrying the intronless ADH1::hop2-H2::lacZ fusion gene than in the strain carrying the intron-containing fusion gene (Fig. 6B), raising the possibility that the HOP2 transcript is not correctly spliced with 100% efficiency when the gene is overexpressed. However, the levels of β-galactosidase activity in meiotic cells relative to vegetative cells are similar for the two constructs (ADH1::hop2::lacZ and ADH1::hop2-H2::lacZ), indicating that none of the factors required for HOP2 splicing is specific to meiosis.
The Nam8 protein also is required for splicing of the MER2 and MER3 transcripts; however, unlike Mer1, the Nam8 protein is present in both vegetative and meiotic cells (11, 32, 33). To determine whether Nam8 is required for HOP2 splicing, RNase protection and β-galactosidase assays were performed in the nam8 mutant. The hop2::lacZ and hop2-H2::lacZ constructs produce equivalent levels of β-galactosidase activity both in the wild type and in the nam8 mutant, indicating that the Nam8 protein is not necessary for splicing of HOP2 pre-mRNA (Fig. 6).
Sequences in the HOP2 intron are less conserved than exon sequences.
The HOP2 gene was cloned from five different yeast strains, and the sequences of the cloned genes were compared to that of the original hop2-complementing clone (25). The sequences derived from BR1373-6D (41) and AS4 (10) are identical to that of the original clone. In the Y260 strain (obtained from Michael Snyder), the nucleotide at position 73 (in the intron) is changed from an A to a G. In the SK1 strain, NKY291 (1), the intron nucleotides at positions 67 and 68 are changed from TT to CC, and the nucleotide at position 221 in the 3′ exon is changed from an A to a T. The exon mutation changes a threonine residue to a similar amino acid (serine) and thus might not affect the function of the Hop2 protein. Another SK1-related strain, S168 (20), carries the same three mutations as NKY291, but the nucleotide at position 77 in the intron is also changed from a T to a C. The HOP2 sequence present in the Saccharomyces genome database (46a) differs from that of the original hop2-complementing clone (25) by the deletion of an A residue at position +634. The sequences downstream of the site of the mutation encode 31 amino acids in the case of the database sequence and 16 (different) amino acids in the case of the published sequence (25).
Overall, this comparative analysis revealed variation at a total of 6 nucleotide positions, including 4 (out of 70) in the intron (Fig. 2C) and 2 (out of 609) in exon sequences. These results suggest, as expected, that there is greater selective pressure for maintenance of HOP2 exon sequences than there is for intron sequences.
DISCUSSION
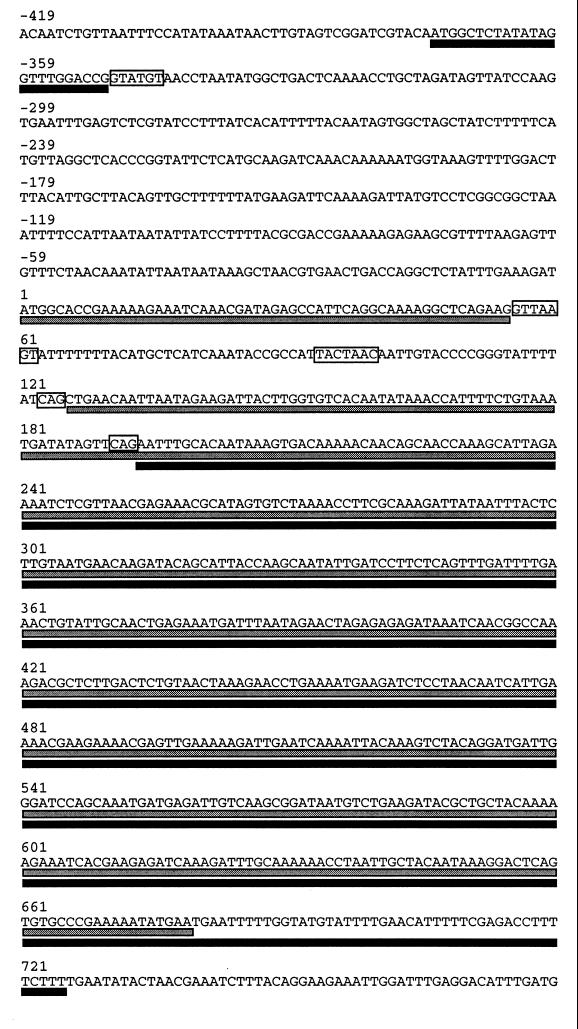
In the Saccharomyces genome database (46a) there is a hypothetical ORF called YGL033w (57) that overlaps extensively with the HOP2 ORF (Fig. 7). Expression of YGL033w requires the removal of an intron that contains a consensus 5′ splice (Fig. 7). However, several observations indicate that YGL033w, as annotated, is not the HOP2 gene. First, the 5′ exon of YGL033w is positioned at bases −374 to −351 relative to the HOP2 start codon, but the results of our transposon mutagenesis demonstrate that insertions in the putative exon or a few hundred base pairs downstream do not disrupt hop2-complementing activity. Second, the results of site-specific mutagenesis of splice site signals indicate that the HOP2-H2 mRNA is the only spliced product that encodes a fully functional Hop2 protein. Third, a cDNA corresponding to YGL033w was not detected by RT-PCR using a pair of primers that ought to have amplified both spliced and unspliced RNA corresponding to this ORF. This pair of primers did generate small amounts of a product in which the 5′ exon of YGL033w is fused to the 3′ exon of HOP2-H2 (data not shown). However, because the coding regions in the two exons are not fused in frame, this apparent cDNA has the capacity to encode only a very short peptide. Furthermore, when this intronless cDNA is introduced back into yeast (with appropriate 5′ and 3′ flanking sequences restored), it does not complement the hop2 defect. Finally, although a conserved 5′ splice site is used to generate the hypothetical spliced product encoding YGL033w, splicing depends on a 3′ splice site (bases 191 to 193) that is far from the branch point sequence. The distance between the branch point and the 3′ splice site is 90 nt, compared to 20 to 50 nt in most yeast introns. In addition, the 3′ splice site predicted to result in expression of YGL033w would need to compete with three other 3′ splice signals located further upstream. Based on these observations, it seems unlikely that the ORF designated as YGL033w is expressed in vivo.
FIG. 7.
Comparison of the HOP2 and YGL033w ORFs. Protein-coding sequences in the HOP2 gene are indicated by the gray bars directly below the nucleotide sequence. Coding sequences for YGL033w are indicated by the black bars. The splice site signals for both HOP2 and YGL033w are boxed; the two introns utilize the same branch point sequence but different 5′ and 3′ splice sites. The sequence present in the Saccharomyces genome database differs from that of the hop2-complementing clone (presented in the figure) by the deletion of an A residue from the stretch of A residues present at positions 629 to 634.
The HOP2 gene utilizes a unique 5′ splice site (GUUAAGU). In fact, it is the only known 5′ splice site in yeast in which the region of base pairing with U1 snRNA is interrupted by an extra nucleotide. Two factors may compensate for this interruption in pairing. First, the HOP2 5′ splice site, unlike the consensus, contains an A residue at the nucleotide corresponding to position 4 in the consensus; this A residue can pair with the appropriately positioned U residue in U1 snRNA. In contrast, the nucleotide present at the fourth position in the consensus 5′ splice site (U or C) cannot pair with the U residue in U1 snRNA. In addition, there are 3 nt immediately upstream of the HOP2 5′ splice site and 2 nt downstream of the splice site that are capable of base pairing with U1 snRNA. Mutational analysis demonstrates that these flanking sequences are indeed important for efficient splicing of HOP2 pre-mRNA. In the case of the hop-3P mutation, the reduction in splicing could result from reduced base pairing with U1 and/or U6 snRNA. The wild-type HOP2 intron has the potential to form 5 bp with U6 snRNA; 5′-GUAUU-3′ (positions 61 to 65 [Fig. 2C]) can pair with 3′-CAUAA-5′ in U6. Two of these base pairs are destroyed by the hop3-3P mutation. Similarly, the hop2-5P mutation may affect splicing due to its effect on pairing with U1 and/or U5 snRNAs (27, 37). In the wild-type HOP2 gene, nucleotides at the second and third positions upstream of the intron (nt 53 and 54 [Fig. 2C]) have the ability to base pair with nucleotides in U5; both of these base pairs are destroyed by the hop2-5P mutation.
Recently, Staley and Guthrie (55) have reported that increasing base pairing between U1 snRNA and the ACT1 5′ splice site (from 6 bp interrupted by one mismatch to 10 or 12 continuous bp) decreases splicing efficiency. A number of observations indicate that this effect is due to hyperstabilization of the interaction between U1 snRNA and the 5′ splice site. Thus, it is perhaps surprising that the HOP2 transcript is spliced with nearly 100% efficiency, despite an extensive interaction between U1 and the 5′ splice site (11 bp interrupted by one unpaired base). However, the interaction of U1 with the HOP2 5′ splice site is not predicted to be as strong as its interactions with the extended ACT1 5′ splice sites. The free energy of the U1 interaction with the HOP2 5′ splice site is −9.8 kcal/mol, compared to −12.3 kcal/mol for the 10-bp ACT1 site and −13.8 kcal/mol for the 12-bp ACT1 site (48). Also, it is important to note that the extended ACT1 5′ splice sites have significant effects only at low temperature. For example, the 10-bp ACT1 5′ splice site reduced splicing efficiency 16-fold when cells were grown at 16°C but only 3-fold when cells were grown at 30°C (the temperature used to study HOP2). Also, Staley and Guthrie (55) showed that the deficiency in splicing conferred by hyperstabilization of the interaction between U1 and the 5′ splice site reflects a competition between U1 and U6 snRNAs for base pairing with the 5′ splice site. The defect in splicing conferred by hyperstabilization of the interaction between U1 and the 5′ splice site can be suppressed by increasing the number of base pairs between U6 and the 5′ splice site. In fact, as noted above, sequences in or near the HOP2 5′ splice site have the potential to form 5 bp with U6 snRNA, compared to 3 bp between U6 and the consensus 5′ splice site. Thus, hyperstabilization of the interaction between U1 and the 5′ splice site might be compensated for by an increase in the strength of the interaction between the splice site and U6 snRNA.
The meiosis-specific MER2 and MER3 genes also contain nonconsensus 5′ splice sites. The MER2 5′ splice site (GUUCGU) differs from the consensus sequence by a single base substitution (A to U at the third position), while the MER3 5′ splice site (GUA_GU) is missing the fourth nucleotide of the consensus sequence (16, 33). The MER2 intron is also unusual in terms of its position within the gene; the intron starts at position +319 of the coding region, whereas the vast majority of yeast introns are located within a few codons of the ATG start codon (46). In addition, the MER2 and MER3 introns are unusual with respect to the distance between the branch point sequence and the 3′ splice site. This distance is 20 to 50 nt in most yeast introns (46), but it is 78 nt in the case of MER3 and only 10 nt in the case of MER2 (16, 33).
In contrast to the situation in the HOP2 gene, the presumed defect in the interaction between U1 snRNA and the MER2 and MER3 5′ splice sites is not compensated for by pairing in regions flanking the 5′ splice site. In both cases, the potential for pairing outside the splice site is limited to a single residue; an A residue located immediately downstream of the splice site in both introns could pair with the opposing U residue in U1 snRNA. Instead, in the case of MER2 and MER3, the defect in splicing is compensated for by the Mer1 and Nam8 proteins, which apparently promote or stabilize the interaction between U1 snRNA and the impaired 5′ splice site signals. The molecular basis for this enhancement of pairing is poorly understood. The Mer1 protein contains the K-protein-homologous motif characteristic of some RNA-binding proteins (51), and this protein has been demonstrated to bind specifically to sequences in and near the MER2 intron (35). The Nam8 protein contains three copies of an RNA-binding domain and is a component of the U1 snRNP (19). Both in vivo and in vitro, the Nam8 protein is required for efficient 5′ splice site recognition only when this process is impaired due to a nonconsensus 5′ splice site or the absence of a methylated cap at the 5′ end of the transcript (40).
Despite the fact that the HOP2 transcript, like the MER2 and MER3 transcripts, contains a nonconsensus 5′ splice site, the Mer1 and Nam8 proteins are not necessary for HOP2 gene expression. Our data also indicate that no meiosis-specific factors are required for HOP2 splicing. However, the possibility cannot be excluded that splicing of HOP2 transcripts depends on specialized splicing factors (present in both vegetative and meiotic cells) that have not yet been identified. When the HOP2 gene is fused to the ADH1 promoter and therefore overexpressed, the intronless hop2-H2::lacZ fusion gene generates a higher level of β-galactosidase activity than the wild-type HOP2 gene fused to lacZ. It is possible that the lower level of activity produced by the wild-type gene is due to the limited abundance or activity of a specialized splicing factor. Another possibility is that splicing of HOP2 transcripts is negatively regulated by the Hop2 protein, leading to inhibition of splicing in the presence of excess Hop2. Such a regulatory mechanism has been demonstrated to operate in the yeast RPL32 gene, which also contains a nonconsensus 5′ splice site (7, 13).
Unexpectedly, we found that mutation of the first nucleotide in the HOP2 5′ splice site from a G to a C reduces splicing, but it does not completely prevent splicing. This result is surprising since the same mutation completely abolishes splicing in the ACT1 gene (18, 58). Analysis of this mutation and others at the first position in the intron has led to the conclusion that the 5′ splice site sequence performs two distinct and important functions (18, 36, 58). First, it is involved in defining the 5′ splice site. Second, if the first step in splicing is executed and a lariat is formed, then the presence of a G at the branch junction is necessary for the next step in splicing (i.e., cutting at the 3′ splice site and exon ligation). Perhaps the less stringent requirement for a G at the first position in the HOP2 intron (compared to other introns) reflects the operation of a specialized splicing factor in the case of HOP2. It is also possible that a C at the first position in the intron is tolerated in the HOP2 gene (though not in other genes) because of the extended base pairing between the 5′ splice site and U1 snRNA.
Only 3.5% of all genes in the S. cerevisiae genome contain introns (2a, 43, 46, 53). Many of these introns are in genes encoding ribosomal proteins; in this class, about 60% of all genes have introns. Exclusive of the genes encoding ribosomal proteins; only 2.5% of genes contain introns (2a, 43, 46). It is therefore surprising that introns are found in a large fraction of genes expressed specifically in meiotic cells, especially in the early class of meiotic genes preferentially expressed during meiotic prophase (31). Approximately 21 early genes have been identified and examined for expression; of these, six have introns. This frequency (29%) is almost 10 times higher than the frequency of intron-containing genes in the genome as a whole. The meiosis-specific early genes that contain introns include HOP2, MER2, and MER3 (discussed above), as well as the REC114 (29), ME14 (30), and DMC1 (4) genes. The MEI4 and DMC1 genes are similar to the bulk of intron-containing genes with respect to intron position, intron size, and consensus splice signals. However, the intron in REC114 is unusual in two respects (29): (i) it is located a very long distance (1,242 nt) from the start of the coding region, and (ii) the 3′ splice site (AAG) does not conform to the consensus. Splicing of the REC114 transcript does not require the Mer1 protein, which is not surprising if one assumes (as the data suggest) that Mer1 is involved specifically in promoting the utilization of nonconsensus 5′ splice sites.
The reason why so many of the early meiotic genes contain introns remains a mystery. It has been suggested that these introns play an essential role in the proper regulation of meiotic gene expression (43). However, in the five genes for which an intronless version of the gene was constructed and introduced into yeast, the gene lacking the intron was found to complement the corresponding null mutant (16, 25, 29, 30, 33). Thus, the introns in meiotic genes do not appear to have essential functions. Nevertheless, we cannot exclude the possibility that regulation of splicing of the transcripts of the early meiotic genes is required under conditions that induce meiosis and sporulation in nature, even though it is not important under laboratory conditions. A second possible explanation for the high frequency of introns in meiotic genes is based on the assumption that there is selection for a small and compact genome (and therefore for intron loss) in a rapidly dividing organism such as yeast. In this case, meiotic genes would be exposed to less selection pressure than genes expressed in vegetative cells since many cycles of vegetative growth intervene between meioses in nature.
Fink (17) has suggested that introns have been lost from the yeast genome due to reverse transcription of mRNAs followed by homologous recombination between the resulting intronless cDNAs (17). This model also explains why most introns are located near the 5′ ends of genes. Removal of an intron by homologous recombination requires that crossing over take place on both sides of the intron; thus, introns near the ends of pre-mRNAs would be less likely to be removed. Also, introns near the 5′ ends of genes would be less likely to be reverse transcribed, since reverse transcription starts at the 3′ end of the transcript but does not always proceed all the way to the 5′ end. Experimental evidence demonstrates that yeast introns can be removed by reverse transcription followed by homologous recombination. Artificially engineered intron-containing genes undergo intron loss in vivo, and this loss depends on an active reverse transcriptase encoded by the transposable element Ty (8, 9). Thus, as suggested previously (39), the high frequency of meiotic genes containing introns might be attributable to the low level of reverse transcriptase present in meiotic cells. Transcription of Ty elements is repressed about 20-fold in MATa/MATα cells grown in a nonfermentable carbon source (one of the conditions required to induce meiosis), and so genes expressed only under these conditions would be exposed to much lower concentrations of reverse transcriptase (56).
ACKNOWLEDGMENTS
We are grateful to Manuel Ares, Jr., Sean Burgess, Janet Novak, Beth Rockmill, and Pedro San-Segundo for helpful comments on the manuscript. The Howard Hughes Biopolymer/Keck Foundation Biotechnology Resource Laboratory at Yale University provided oligonucleotides and performed DNA sequence analysis.
This work was supported by National Institutes of Health grant GM28904 to G.S.R. and by the Howard Hughes Medical Institute.
REFERENCES
- 1.Alani E, Cao L, Kleckner N. A method for gene disruption that allows repeated use of URA3 selection in the construction of multiply disrupted yeast strains. Genetics. 1987;116:541–545. doi: 10.1534/genetics.112.541.test. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ammerer G. Expression of genes in yeast using the ADC1 promoter. Methods Enzymol. 1983;101:192–201. doi: 10.1016/0076-6879(83)01014-9. [DOI] [PubMed] [Google Scholar]
- 2a.Ares Lab Intron Site. 13 August 1999, revision date. [Online.] http://www.cse.ucsc.edu/research/compbio/yeast_introns.html. [13 August 1999, last date accessed.]
- 3.Baudin A, Ozier-Kalogeropoulos O, Denouel A, Lacroute F, Cullin C. A simple and efficient method for direct gene deletion in Saccharomyces cerevisiae. Nucleic Acids Res. 1993;21:3329–3330. doi: 10.1093/nar/21.14.3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bishop D K, Park D, Xu L, Kleckner N. DMC1: a meiosis-specific yeast homolog of E. coli recA required for recombination, synaptonemal complex formation, and cell cycle progression. Cell. 1992;69:439–456. doi: 10.1016/0092-8674(92)90446-j. [DOI] [PubMed] [Google Scholar]
- 5.Buckingham L E, Wang H-T, Elder R T, McCarroll R M, Slater M R, Esposito R E. Nucleotide sequence and promoter analysis of SPO13, a meiosis-specific gene of Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1990;87:9406–9410. doi: 10.1073/pnas.87.23.9406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chua P R, Roeder G S. Zip2, a meiosis-specific protein required for the initiation of chromosome synapsis. Cell. 1998;93:349–359. doi: 10.1016/s0092-8674(00)81164-2. [DOI] [PubMed] [Google Scholar]
- 7.Dabeva M D, Post-Beittenmiller M A, Warner J R. Autogenous regulation of splicing of the transcript of a yeast ribosomal protein gene. Proc Natl Acad Sci USA. 1986;83:5854–5857. doi: 10.1073/pnas.83.16.5854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Derr L K, Strathern J N. A role for reverse transcripts in gene conversion. Nature. 1993;361:170–173. doi: 10.1038/361170a0. [DOI] [PubMed] [Google Scholar]
- 9.Derr L K, Strathern J N, Garfinkel D J. RNA-mediated recombination in S. cerevisiae. Cell. 1991;67:355–364. doi: 10.1016/0092-8674(91)90187-4. [DOI] [PubMed] [Google Scholar]
- 10.Detloff P, White M A, Petes T D. Analysis of a gene conversion gradient at the HIS4 locus in Saccharomyces cerevisiae. Genetics. 1992;132:113–123. doi: 10.1093/genetics/132.1.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ekwall K, Kermorgant M, Dujardin G, Groudinsky O, Slominski P P. The NAM8 gene in Saccharomyces cerevisiae encodes a protein with putative RNA binding motifs and acts as a suppressor of mitochrondrial splicing deficiencies when overexpressed. Mol Gen Genet. 1992;233:136–144. doi: 10.1007/BF00587571. [DOI] [PubMed] [Google Scholar]
- 12.Elledge S J, Davis R W. A family of versatile centromeric vectors designed for use in the sectoring-shuffle mutagenesis assay in Saccharomyces cerevisiae. Gene. 1988;70:303–312. doi: 10.1016/0378-1119(88)90202-8. [DOI] [PubMed] [Google Scholar]
- 13.Eng F G, Warner J R. Structural basis of splicing of a yeast messenger RNA. Cell. 1991;65:797–804. doi: 10.1016/0092-8674(91)90387-e. [DOI] [PubMed] [Google Scholar]
- 14.Engebrecht J, Roeder G S. MER1, a yeast gene required for chromosome pairing and genetic recombination, is induced in meiosis. Mol Cell Biol. 1990;10:2379–2389. doi: 10.1128/mcb.10.5.2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Engebrecht J, Roeder G S. Yeast mer1 mutants display reduced levels of meiotic recombination. Genetics. 1989;121:237–247. doi: 10.1093/genetics/121.2.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Engebrecht J, Voelkel-Meiman K, Roeder G S. Meiosis-specific RNA splicing in yeast. Cell. 1991;66:1257–1268. doi: 10.1016/0092-8674(91)90047-3. [DOI] [PubMed] [Google Scholar]
- 17.Fink G R. Pseudogenes in yeast? Cell. 1987;49:5–6. doi: 10.1016/0092-8674(87)90746-x. [DOI] [PubMed] [Google Scholar]
- 18.Fouser L A, Friesen J D. Mutations in a yeast intron demonstrate the importance of specific conserved nucleotides for the two stages of nuclear mRNA splicing. Cell. 1986;45:81–93. doi: 10.1016/0092-8674(86)90540-4. [DOI] [PubMed] [Google Scholar]
- 19.Gottschalk A, Tang J, Puig O, Salgado J, Neubauer G, Colot H V, Mann M, Seraphin B, Rosbash M, Luhrmann R, Fabrizio P. A comprehensive biochemical and genetic analysis of the yeast U1 snRNP reveals five novel proteins. RNA. 1998;4:374–393. [PMC free article] [PubMed] [Google Scholar]
- 20.Goyon C, Lichten M. Timing of molecular events in meiosis in Saccharomyces cerevisiae: stable heteroduplex DNA is formed late in meiotic prophase. Mol Cell Biol. 1993;13:373–382. doi: 10.1128/mcb.13.1.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Higuchi R. Recombination PCR. In: Innis M A, Gelfand D H, Sninsky J J, White T J, editors. PCR protocols. New York, N.Y: Academic Press, Inc.; 1990. pp. 177–183. [Google Scholar]
- 22.Hoekstra M F, Burbee D, Singer J, Mull E, Chiao E, Heffron F. A Tn3 derivative that can be used to make short in-frame insertions within genes. Proc Natl Acad Sci USA. 1991;88:5457–5461. doi: 10.1073/pnas.88.12.5457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ito H, Fukada Y, Murata K, Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Krieg P A, Melton D A. In vitro RNA synthesis with SP6 RNA polymerase. Methods Enzymol. 1987;155:397–415. doi: 10.1016/0076-6879(87)55027-3. [DOI] [PubMed] [Google Scholar]
- 25.Leu J-Y, Chua P R, Roeder G S. The meiosis-specific Hop2 protein of S. cerevisiae ensures synapsis between homologous chromosomes. Cell. 1998;94:375–386. doi: 10.1016/s0092-8674(00)81480-4. [DOI] [PubMed] [Google Scholar]
- 26.Leu, J.-Y., and G. S. Roeder. The pachytene checkpoint in S. cerevisiae depends on Swe1-mediated phosphorylation of the cyclin-dependent kinase Cdc28. Mol. Cell, in press. [DOI] [PubMed]
- 27.Long M, de Souza S J, Gilbert W. The yeast splice site revisited: new exon consensus from genomic analysis. Cell. 1997;91:739–740. doi: 10.1016/s0092-8674(00)80462-6. [DOI] [PubMed] [Google Scholar]
- 28.Lopez A J. Alternative splicing of pre-mRNA: developmental consequences and mechanisms of regulation. Annu Rev Genet. 1998;32:279–305. doi: 10.1146/annurev.genet.32.1.279. [DOI] [PubMed] [Google Scholar]
- 29.Malone R E, Pittman D L, Nau J J. Examination of the intron in the meiosis-specific recombination gene REC114 in Saccharomyces. Mol Gen Genet. 1997;225:410–419. doi: 10.1007/s004380050513. [DOI] [PubMed] [Google Scholar]
- 30.Menees T M, Ross-Macdonald P B, Roeder G S. MEI4, a meiosis-specific yeast gene required for chromosome synapsis. Mol Cell Biol. 1992;12:1340–1351. doi: 10.1128/mcb.12.3.1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mitchell A P. Control of meiotic gene expression in Saccharomyces cerevisiae. Microbiol Rev. 1994;58:56–70. doi: 10.1128/mr.58.1.56-70.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nakagawa T, Ogawa H. Involvement of the MRE2 gene of yeast in formation of meiosis-specific double-strand breaks and crossover recombination through RNA splicing. Genes Cells. 1997;2:65–79. doi: 10.1046/j.1365-2443.1997.d01-283.x. [DOI] [PubMed] [Google Scholar]
- 33.Nakagawa, T., and H. Ogawa.MER3, a novel helicase-like gene, regulates meiotic crossover recombination in meiosis. EMBO J., in press. [DOI] [PMC free article] [PubMed]
- 34.Nandabalan K, Price L, Roeder G S. Mutations in U1 snRNA bypass the requirement for a cell type-specific RNA splicing factor. Cell. 1993;73:407–415. doi: 10.1016/0092-8674(93)90239-m. [DOI] [PubMed] [Google Scholar]
- 35.Nandabalan K, Roeder G S. Binding of a cell type-specific RNA splicing factor to its target regulatory sequence. Mol Cell Biol. 1995;15:1953–1960. doi: 10.1128/mcb.15.4.1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Newman A J, Lin R-J, Cheng S-C, Abelson J. Molecular consequences of specific intron mutations on yeast mRNA splicing in vivo and in vitro. Cell. 1985;42:335–344. doi: 10.1016/s0092-8674(85)80129-x. [DOI] [PubMed] [Google Scholar]
- 37.Newman A J, Norman C. U5 snRNA interacts with the exon sequences at 5′ and 3′ splice sites. Cell. 1992;68:743–754. doi: 10.1016/0092-8674(92)90149-7. [DOI] [PubMed] [Google Scholar]
- 38.Ogawa H, Johzuka K, Nakagawa T, Leem S-H, Hagihara A H. Functions of the yeast meiotic recombination genes, MRE11 and MRE2. Adv Biophys. 1995;31:67–76. doi: 10.1016/0065-227x(95)99383-z. [DOI] [PubMed] [Google Scholar]
- 39.Pittman D, Lu W, Malone R E. Genetic and molecular analysis of REC114, an early meiotic recombination gene in yeast. Curr Genet. 1993;23:295–304. doi: 10.1007/BF00310890. [DOI] [PubMed] [Google Scholar]
- 40.Puig O, Gottschalk A, Fabrizio P, Seraphin B. Interaction of the U1 snRNP with nonconserved intronic sequences affects 5′ splice site selection. Genes Dev. 1999;13:569–580. doi: 10.1101/gad.13.5.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rockmill B, Roeder G S. Meiosis in asynaptic yeast. Genetics. 1990;126:563–574. doi: 10.1093/genetics/126.3.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rockmill B, Roeder G S. A meiosis-specific protein kinase homolog required for chromosome synapsis and recombination. Genes Dev. 1991;5:2392–2404. doi: 10.1101/gad.5.12b.2392. [DOI] [PubMed] [Google Scholar]
- 43.Rodriguez-Medina J, Rymond B C. Prevalence and distribution of introns in non-ribosomal protein genes of yeast. Mol Gen Genet. 1994;243:532–539. doi: 10.1007/BF00284201. [DOI] [PubMed] [Google Scholar]
- 44.Roeder G S. Meiotic chromosomes: it takes two to tango. Genes Dev. 1997;11:2600–2621. doi: 10.1101/gad.11.20.2600. [DOI] [PubMed] [Google Scholar]
- 45.Rothstein R. Targeting, disruption, replacement and allele rescue: integrative DNA transformation in yeast. Methods Enzymol. 1991;194:281–301. doi: 10.1016/0076-6879(91)94022-5. [DOI] [PubMed] [Google Scholar]
- 46.Rymond R C, Rosbash M. Yeast pre-mRNA splicing. In: Jones E W, Pringle J R, Broach J R, editors. The molecular biology of the yeast Saccharomyces cerevisiae: gene expression. II. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1992. pp. 143–192. [Google Scholar]
- 46a.Saccharomyces Genome Database. 25 August 1999, revision date. [Online.] http://genome-www.stanford.edu/Saccharomyces/. [25 August 1999, last date accessed.]
- 47.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 48.Serra M J, Turner D H. Predicting thermodynamic properties of RNA. Methods Enzymol. 1995;259:242–261. doi: 10.1016/0076-6879(95)59047-1. [DOI] [PubMed] [Google Scholar]
- 49.Sherman F, Fink G R, Hicks J B. Methods in yeast genetics: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1986. [Google Scholar]
- 50.Sikorski R, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Siomi H, Matunis M J, Michael W M, Dreyfuss G. The pre-mRNA binding K protein contains a novel evolutionarily conserved motif. Nucleic Acids Res. 1993;21:1193–1198. doi: 10.1093/nar/21.5.1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Smith C W J, Patton J G, Nadal-Ginard B. Alternative splicing in the control of gene expression. Annu Rev Genet. 1989;23:527–577. doi: 10.1146/annurev.ge.23.120189.002523. [DOI] [PubMed] [Google Scholar]
- 53.Spingola M, Grate L, Haussler D, Ares M., Jr Genome-wide bioinformatic and molecular analysis of introns in Saccharomyces cerevisiae. RNA. 1999;5:221–234. doi: 10.1017/s1355838299981682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Staley J P, Guthrie C. Mechanical devices of the spliceosome: motors, clocks, springs, and things. Cell. 1998;92:315–326. doi: 10.1016/s0092-8674(00)80925-3. [DOI] [PubMed] [Google Scholar]
- 55.Staley J P, Guthrie C. An RNA switch at the 5′ splice site requires ATP and the DEAD box protein Prp28p. Mol Cell. 1999;3:55–64. doi: 10.1016/s1097-2765(00)80174-4. [DOI] [PubMed] [Google Scholar]
- 56.Taguchi A K, Ciriacy M, Young E T. Carbon source dependence of transposable element associated gene activation in Saccharomyces cerevisiae. Mol Cell Biol. 1984;4:61–68. doi: 10.1128/mcb.4.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tettelin H, Carbone M L A, Albermann K, Albers M, Arroyo J, Backes U, Barreiros T, Bertani I, Bjourson A J, Bruckner M, Bruschi C V, Carignani G, Castagnoli L, Cerdan E, Clemente M L, Coblenz A, Coglievina M, Coissac E, Defoor E, Bino S D, Delius H, Delneri D, de Wergifosse P, Dujon B, Kleine K. The nucleotide sequence of Saccharomyces cerevisiae chromosome VII. Nature. 1997;387(6632 Suppl.):81–84. [PubMed] [Google Scholar]
- 58.Vijayraghavan U, Parker R, Tamm J, Ilmura Y, Rossi J, Abelson J, Guthrie C. Mutations in conserved intron sequences affect multiple steps in the yeast splicing pathway, particularly assembly of the spliceosome. EMBO J. 1986;5:1683–1695. doi: 10.1002/j.1460-2075.1986.tb04412.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang J, Manley J L. Regulation of pre-mRNA splicing in metazoa. Curr Opin Genet Dev. 1997;7:205–211. doi: 10.1016/s0959-437x(97)80130-x. [DOI] [PubMed] [Google Scholar]
- 60.Yocum R R, Hanley S, R. W Jr, Ptashne M. Use of lacZ fusions to delimit regulatory elements of the inducible divergent GAL1-GAL10 promoter in Saccharomyces cerevisiae. Mol Cell Biol. 1984;4:1985–1998. doi: 10.1128/mcb.4.10.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]