Abstract
There is momentum towards implementing patient-derived xenograft models (PDX) in cancer research to reflect the histopathology, tumor behavior, and metastatic properties observed in the original tumor. To study PDX cells preclinically, we used both bioluminescence imaging (BLI) to evaluate cell viability and magnetic particle imaging (MPI), an emerging imaging technology to allow for detection and quantification of iron nanoparticles. The goal of this study was to develop the first successful iron labeling method of breast cancer cells derived from patient brain metsastases and validate this method with imaging during tumor development. The overall workflow of this labeling method is as follows: adherent and non-adherent luciferase expressing human breast cancer PDX cells (F2-7) are dissociated and concurrently labeled after incubation with micron-sized iron oxide particles (MPIO; 25 μg Fe/ml), with labeling validated by cellular imaging with MPI and BLI. In this study, NOD/SCID/ILIIrg-/- (n = 5) mice Received injections of 1 × 106 iron-labeled F2-7 cells into the fourth mammary fat pad (MFP). BLI was performed longitudinally to day 49 and MPI was performed up to day 28. In vivo BLI revealed that signal increased over time with tumor development. MPI revealed decreasing signal in the tumors over time. Here, we demonstrate the first application of MPI to monitor the growth of a PDX MFP tumor and the first successful labeling of PDX cells with iron oxide particles. Imaging of PDX cells provides a powerful system to better develop personalized therapies targeting breast cancer brain metastasis.
Keywords: bioluminescence imaging, breast cancer, cellular imaging, magnetic particle imaging, patient-derived xenografts
INTRODUCTION
Breast cancer is one of the most common cancers seen in women, currently affecting 1 in 8 women in North America [1]. Mortality associated with this disease is caused most frequently by metastasis, which is the spread of cancer from the primary tumor to other distant locations in the body. In breast cancer, these locations often include the brain, bone, lungs, and lymph nodes [2]. For brain metastases in particular, prognosis is poor, with mean 1-year survival rates of only 20% [3], and 2-year survival rates of < 2% [4]. Additionally, the incidence of brain metastasis (BM) is increasing, as neuroimaging techniques improve and treatments that allow for longer patient survival permits more time for cells to metastasize to the central nervous system [5]. Insight into the mechanisms and pathophysiology of breast cancer brain metastasis have long relied on the use of immortalized cell lines that have been injected intracardially into the left ventricle of the mouse heart in order to selectively grow tumors within the brain. While these cell lines have been well characterized, they do not represent the tumor heterogeneity, metastatic behaviours, and histopathology seen clinically, and are unsuitable for evaluating therapies due to their fast progression in vivo [6].
Patient-derived xenografts, or PDXs, have begun to supplant traditional cell lines due to their retention of clinical biomarkers and heterogeneity from the original tumor [7]. In recent years, PDXs have been developed to grow in the mammary fat pad of NOD/SCID/ILIIrg-/- (NSG) mice, with a long-term objective to advance personalized medicine. This strategy has shown exciting progress for the development of novel PDXs from brain metastases in breast cancer patients. In 2017, Contreras-Zárate et al. developed BM-PDXs to study the biology of brain metastasis and to serve as tools for testing novel therapeutic approaches [8]. These novel BM-PDXs retained intratumoral heterogeneity and metastatic potential, providing a clinically relevant model to study mechanisms of brain metastatic colonization and slower progression to allow for therapeutic testing. Currently, most PDX models are typically studied using methods such as histology, immunohistochemistry, and fluorescent microscopy, limiting the ability to study these models before an experimental endpoint has been reached. Tools that enable the longitudinal study of the fate of BM-PDXs would provide valuable information to characterize to the mechanisms and metastatic patterns in vivo.
Cellular imaging and cell tracking can be used to study cancer cell populations and metastatic processes in vivo. Bioluminescence imaging (BLI) with the reporter firefly luciferase (Fluc) has been widely utilized for tracking preclinical models of cancer. BLI is a valuable imaging modality as it allows for the longitudinal study of cell viability. Fluc BLI requires adenosine triphosphate (ATP) as a co-factor, and thus, the signal measured with BLI is directly proportional to the number of viable cells in a region of interest [9]. This modality is exceptionally useful when measuring treatment and therapeutic response, as cell viability may change but tumor volumes can remain unaltered.
Our group has previously shown that BLI can be complemented with iron-based cellular MRI technology to provide longitudinal measures of cancer cell viability in preclinical models [10]. Iron-based cellular MRI requires cells to be loaded with superparamagnetic iron oxide nanoparticles (SPIONs) and has shown to provide single cell sensitivity [11]. However, a limitation of this modality is that SPIONs create regions signal loss where iron-labeled cells are present in images, and so, determining the quantitation of signal loss is challenging and it is not possible to determine cell number [12]. Magnetic particle imaging (MPI) is an emerging imaging modality that directly detects SPIONs [13], resulting in a positive signal that appears as a “hot spot” in images. The signal strength detected is linearly proportional to the number of SPIONs, allowing for quantitation [14]. Presently, MPI has been used as a cell tracking modality for immortalized cancer cell lines [15], stem cells [16-18], pancreatic islets [19], T-cells [20] and macrophages [21,22]. However, no studies exist studying the growth of patient-derived xenografts labeled efficiently with an SPION in vivo.
Efficient iron labeling of a patient-derived xenograft cell line presents a challenge due to the mixed and heterogeneous population of cells. In this work, we report the first iron-labeling method for a patient-derived xenograft cell line and validate its utility for cell tracking with MPI and BLI.
MATERIALS AND METHODS
F2-7 cell line origin and cell culture
A firefly luciferase and enhanced green fluorescent protein (eGFP) expressing patient-derived xenograft cell line (F2-7) was obtained from the Cittely Lab (UC Denver), and was originally developed from a triple-negative brain metastatic patient-derived xenograft [8]. F2-7 cells were maintained in T75cm2 flasks coated with collagen-I (1 mg/ml) for 2 h to encourage attachment. 12 ml of Dulbecco’s modified Eagle’s medium (DMEM)-F12 supplemented with 10% of fetal bovine serum (FBS), 1 μg/ml hydrocortisone, 100 ng/ml of cholera toxin, and 1 nM of insulin was added to flasks after collagen coating. Cells grew under 37°C and 5% CO2. Since cells grow as both non-adherent mammospheres and adherent single cells, both populations require maintenance with each passage.
MPIO labeling procedure
Day 1
DMEM-F12, PBS, and trypsin were heated at 37°C for 30 min in a water bath. Conditioned media and non-adherent cells were collected into 15 ml conical tubes from confluent flasks of F2-7 cells. Conical tubes were centrifuged with a ThermoFisher Cytospin 4 for 5 min at 900 rpm, and the supernatant was removed and stored for future use as conditioned media. 1 ml of 0.25% Trypsin-EDTA was added to the cell pellet within the conical tube and was placed in the water bath for 2 min to trypsinize cells. In parallel, 1 ml of 0.25% Trypsin-EDTA was added to the flask with remaining adherent cells and incubated for approximately 5 min. Once adherent cells and mammospheres were dissociated, cell populations were mixed and resuspended in 5 ml of fresh media. After, the mixed population of cells were centrifuged again at 900 rpm for 5 min. The supernatant with trypsin was removed, and the cell pellet was resuspended in 2–5 ml. Cells were assessed for viability with Trypan blue exclusion assay and counted to achieve the correct volume of media to plate 2 × 106 cells. Cells were plated in uncoated T75 cm2 flasks to discourage attachment with 50% fresh media and 50% conditioned media. To iron label these cells, 3 methods of iron labeling were performed to determine the most efficient labeling technique. In the first trial, cells were supplemented with 25 μg Fe/ml of micron-sized iron oxide particles (MPIO) (0.9 μm diameter, ~63% magnetite, labeled with Flash Red; Bangs Laboratory, Fishers, IN, USA), and swirled in the flask to mix. The second trial used the 50 ug Fe/ml of the same MPIO, and the third used 25 ug Fe/ml with a magnetic plate under the flask to allow for magnetofection [23]. Following this, all flasks were incubated for 24 h.
Day 2
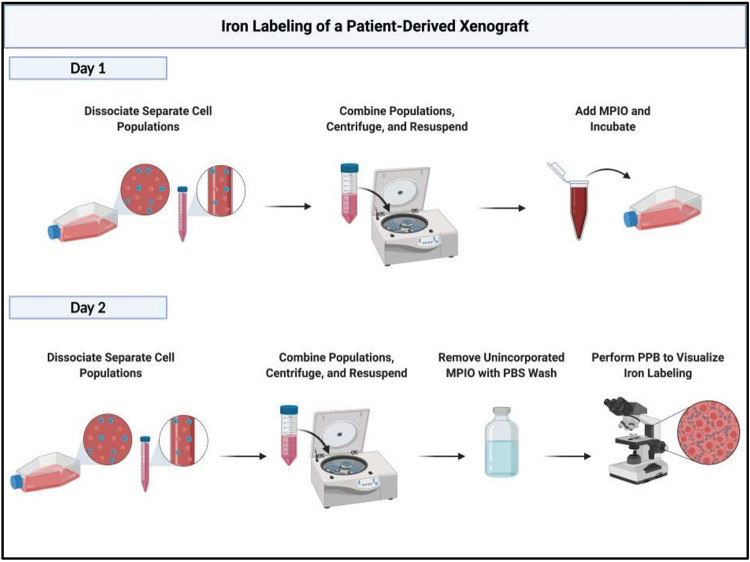
After 24 h, cells from the flasks were harvested and dissociated in accordance to the protocol in Day 1. After, cells were harvested and washed with 10 ml PBS in the flask to remove unincorporated MPIO. Cells were centrifuged at 900 rpm for 5 min. This process was repeated thrice to thoroughly remove unincorporated MPIO prior to cell injections. Cell viability was assessed and calculated using the Trypan blue exclusion assay. A simplified visual of this protocol is shown in Figure 1.
Figure 1.
A simplified illustration of the workflow to efficiently label patient-derived xenograft cells with iron-oxide particles. Created with BioRender.com.
Assessing iron labeling
To visualize MPIO labeling, labeled cells were affixed to a glass slide with a ThermoFisher Cytospin 4 cytocentrifuge and fixed with a Methanol/Acetic acid solution. Slides were then stained with a Perl’s Prussian Blue (PPB) solution and counterstained with Nuclear Fast Red. Slides were dehydrated with increasing concentrations of ethanol, cleared with xylene, and coverslipped with a xylene-based mounting medium. These PPB-stained slides were examined to assess the localization of MPIO within the cell to determine the labeling efficiency using a Zeiss AXIO Imager A1 Microscope (Zeiss Canada, Toronto, ON, Canada). Iron oxide particles appear dark blue, and cells appear light pink in color.
Animal model and workflow
All animals were cared for in accordance with the standards of the Canadian Council on Animal Care, under an approved protocol of the University of Western Ontario’s Council on Animal Care and housed in a pathogen-free barrier facility. Female NSG mice (n = 5) (NOD.Cg-PrkdcscidIl2rgtm1Wjl/SzJ, 6–8 weeks, Humanized Mouse and Xenotransplantation Facility, Robarts Research Institute, University of Western Ontario, London, ON) were first anesthetized with isoflurane (2% in 100% oxygen). NSG mice were then injected into the fourth mammary fat pad (MFP) with a suspension of 1 × 106 MPIO-labeled F2-7luc/eGFP+ cells in 50 μl of sterile saline and 50 ul of Matrigel. Mice were imaged with BLI (n = 2) out to day 49 and MPI (n = 3) out to day 28. At endpoint, mice were sacrificed by isoflurane overdose. Tumors were excised and placed in paraformaldehyde. Ex vivo MFP tumor volumes were estimated using the following formula = 0.52 (width)2 (length) to approximate the volume of an ellipsoid (mm3). Fixed MFP tumors were then processed, paraffin embedded and then cut into 5 μm sections with 5 sections placed on each slide. Immunohistochemistry was performed on contiguous slides for epidermal growth factor receptor (EGFR) and cytokeratin 5 (CK5) and the subsequent slide was stained with hematoxylin and eosin (H&E),
In vitro BLI procedure
To evaluate the relationship between cell number and BLI signal, cells were seeded in 24-well plates in 0.5 ml of growth medium at concentrations of 2 × 105, 6 × 105, 1 × 106 cells per well. Cells were allowed to adhere for 24 h and then 5 μl of D-luciferin (30 mg/ml) was added to the cell media 2 min prior to imaging using a hybrid optical/X-ray scanner (IVIS Lumina XRMS In Vivo Imaging System, PerkinElmer). Region-of-interest (ROI) analysis was performed for each well using Living Image Software (IVIS Imaging Systems, PerkinElmer) and data is expressed as the average radiance (photons/sec/cm2/steradian).
In vivo BLI procedure
BLI was performed on NSG mice (n = 2) on days 4, 28, 35, 42, and 49 using a hybrid optical/X-ray scanner (IVIS Lumina XRMS In Vivo Imaging System, PerkinElmer). Mice were anesthetized with isoflurane (2% in 100% oxygen) using a nose cone attached to an activated carbon charcoal filter. Mice Received a 100 ul intraperitoneal injection of D-luciferin (30 mg/ml), and luminescent images were captured for approximately 35 min to obtain the peak signal at each imaging session. Region-of-interest (ROI) analysis was performed for each mouse using Living Image Software (IVIS Imaging Systems, PerkinElmer) and data is expressed as the average radiance (photons/sec/cm2/steradian).
MPI acquisition
Full body MPI images of mammary fat pad tumor bearing NSG mice (n = 3) were acquired at days 0 (30 min post-injection), 7, 14, and 28. Images were collected on a Momentum™ scanner (Magnetic Insight Inc., Alameda, CA, USA) using the 3D high sensitivity isotropic (multichannel) scan mode. In this mode, images were acquired using a 5.7 T/m gradient, 21 projections and a FOV (field of view) of 12 × 6 × 6 cm, for a total scan time ~18 mins per mouse. Mice were anesthetized with 2% isoflurane in 100% oxygen for the entirety of the scan. 3D high sensitivity isotropic images of ex vivo tumors (n = 2) removed on Day 40 were acquired using the same parameters and a FOV of 4 × 6 × 6 cm, for a total scan time of ~12 min.
MPI calibration and signal quantification
To generate a calibration curve for converting MPI signal to iron content, samples were made with 2 μl aliquots of MPIO and 2 μl PBS and were imaged using the 3D high sensitivity isotropic parameters. The FOV was 12 × 6 × 6 cm. The following samples were scanned separately: 0.175 μg, 0.35 μg, 0.70 μg, 1.40 μg, 2.80 μg, 5.6 μg. Images were analyzed using the open-source Horos imaging software, version 3.3.5 (Annapolis, MD USA). To quantify the MPI signal in each image set, signal intensities were set to full dynamic range in order to represent the full range of signal in each specific ROI, such as the calibration samples and mammary fat pad tumors. Areas of interest were then segmented manually, and 3D volumes were reconstructed and calculated using the Horos volume algorithm. The total MPI signal was calculated using the equation mean signal × volume (mm3).
Hematoxylin and eosin staining
Sections were washed briefly in distilled water, stained in Harris hematoxylin solution for 5 min and differentiated in 1% acid alcohol for 30 s. After washing in 0.2% ammonia for 5 min, sections were counterstained in eosin-phloxine solution for 30 s; dehydrated through 95% alcohol, 2 changes of absolute alcohol, and 5 min each. Sections were then dehydrated and cleared through 95% ethyl alcohol, absolute alcohol and xylene, and mounted with resinous medium.
Immunohistochemistry
EGFR and cytokeratin 5 immunostaining were performed using a rabbit anti-EGFR antibody (1:200 dilution; Cat. # ab32077, Abcam) and a rabbit anti-cytokeratin 5 antibody (1:200 dilution; Cat. # ab64081, Abcam). All sections were rinsed in PBS, incubated for 10 min in PBS containing 3% hydrogen peroxide, and immersed for 30 min in blocking reagent (Dako Protein Block, S0909, Dako). Sections were then incubated with primary antibody at 4°C for 24 h followed by biotinylated horse anti-rabbit IgG secondary antibody (1:300 dilution; BA1100, Vector Laboratories) for 30 min at room temperature and rinsed in PBS for 3 changes for 5 min each. Tissue sections were processed using the (ABC) solution for 30 min (Vector Laboratories, Burlington, ON), and visualized with 3,3’-diaminobenzidine (0.5 mg/ml; Sigma-Aldrich Canada, Oakville, ON). Negative control staining (without addition of primary antibody) was performed on adjacent sections. Counterstaining of EGFR and cytokeratin 5 stained sections was performed using hematoxylin. Slides were then dehydrated through 70–100% alcohol, cleared with xylene, and cover slipped for microscopic examination.
Statistical analysis
Statistical analysis was performed using GraphPad Prism 8 Software (GraphPad, San Diego, CA, SA). Pearson’s rank correlation was used to determine the relationship between total MPI signal and iron content. In vivo data was expressed as mean ± SD and analyzed with a one-way ANOVA. Differences were considered statistically significant at P < 0.05.
RESULTS
Cell labeling
For each trial of labeling, F2-7 cells had varying labeling efficiencies demonstrated by a PPB stain shown in Figure 2, with cancer cells appearing pink, and intracellular iron in blue. For trial 1 with 25 μg Fe/ml of MPIO, F2-7 cells were efficiently labeled with labeling efficiency of 81.80 ± 10.14% (Fig. 1A) and viability of over 90%. This labeling efficiency was deemed successful and the 25 ug Fe/ml of MPIO labeling procedure was used for the remainder of the study. The second trial, using the 50 ug Fe/ml of the same MPIO, resulted in a labeling efficiency of 27.51 ± 1.19% (Fig. 1B). Finally, the third trial which used 25 ug Fe/ml with a magnetic plate resulted in a labeling efficiency of 5.55 ± 1.65% (Fig. 1C).
Figure 2.
Perl’s Prussian Blue (PPB) stains of F2-7 cells labeled with MPIO.A. PPB stain showing F2-7 cells labeled with 25 μg Fe/ml of MPIO, with labeling efficiency of 81.80 ± 10.14%. B. PPB stain showing F2-7 cells labeled with 50 μg Fe/ml of MPIO, with a labeling efficiency of 27.51 ± 1.19%. C. PPB stain showing F2-7 cells labeled with 25 ug Fe/ml and a magnetic plate, resulting in a labeling efficiency of 5.5 5 ± 1.65%.
Bioluminescence imaging
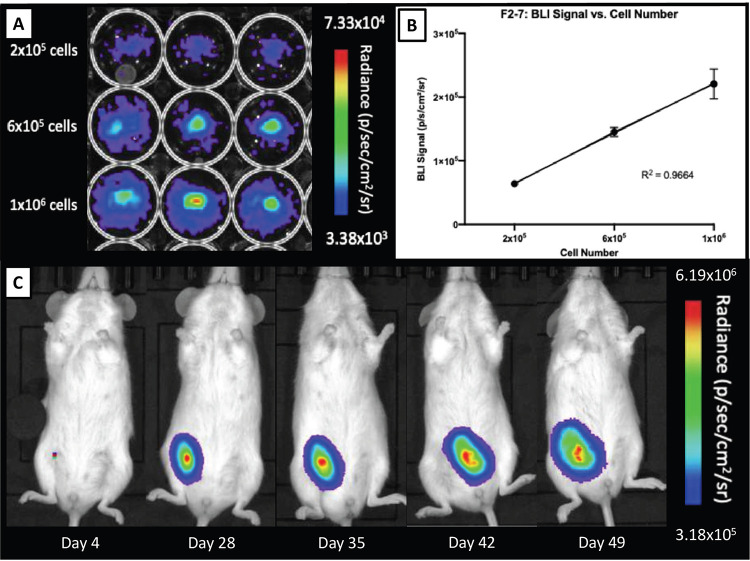
F2-7/eGFP-luc cells were seeded at concentrations of 2 × 105, 6 × 105, 1 × 106 cells per well and in vitro BLI was performed (Fig. 3A). A significant positive correlation was found between the number of F2-7/eGFP-luc cells and BLI signal (R2 = 0.9664). Specifically, as seeded cell number increased, BLI signal also increased (Fig. 3B). In Figure 3C, the BLI signal from a representative tumor is shown to increase over time with tumor development. No BLI signal was detected in any other region of the mouse body.
Figure 3.
In vitro and in vivo bioluminescent imaging of F2-7luc/eGFP+ cells. A. F2-7luc/eGFP+ cells seeded at various concentrations. B. A strong linear correlation is seen between cell number and BLI signal; R2 = 0.9664. C. BLI signal from a representative tumor is shown to increase over time with tumor development from day 4 to day 49.
Magnetic particle imaging
A calibration line was generated to determine iron content for a given MPI signal based on the 3D, high sensitivity, isotropic parameters used to image MPIO. An example of the images of samples measured to generate calibration curves are shown in Figure 4A for MPIO and the calibration line generated from this data is shown in 4B. Based on this data, we determined here was a strong linear relationship between iron content and MPI signal (arbitrary units, A.U.) for MPIO (R2 = 0.9836, P < 0.001). The equation of the line was: MPI Signal = 69.559 (Iron Content) for MPIO. Using this relationship, iron content could be determined for a given MPI signal.
Figure 4.
MPI calibration line generation and signal quantification. A. Images of MPIO samples measured to generate calibration curves. B. A strong linear relationship is seen between iron content and MPI signal. The asterisks in the equation represents a multiplication sign. R2 = 0.9836.
Figure 5A shows representative MPI of a tumor bearing mouse on day 0, 7, 14, and 28 post-injection of 1 × 106 MPIO-labeled F2-7 cells. MPI signal is clearly visualized in the lower, right MFP tumor, with signal decreasing over time (box). The mean iron content in the MFP tumors decreased significantly between day 0 (M = 4.06 ± 2.09 ug) and day 28 (M = 0.41 ± 0.25 ug) (P = 0.0095) (Fig. 5B). Note that some MPI signal can also be detected elsewhere in the body. Additionally, ex vivo MPI was performed on tumors removed on day 40 (Fig. 5C). A representative ex vivo tumor is shown in Figure 5D. The tumors measured 152.88 mm3 and 178.36 mm3, and the iron content was 0.05 ug and 0.75 ug, respectively.
Figure 5.
In vivo and ex vivo MPI of mammary fat pad tumors. Representative MPI of a tumor bearing mouse on day 0, 7, 14, and 28 post-injection of 1x106 MPIO-labeled F2-7 cells, with images window-leveled to the tumor (orange box). B. Quantification of mean iron content over time in MFP tumors decreasing significantly between day 0 (M = 4.06 ± 2.09 ug) and day 28 (M = 0.41 ± 0.25 ug) (P = 0.0095). The double asterisks represents statistical significance, indicating (P = 0.0095). C. Representative MPI of an ex vivo tumor removed on day 40. D. A representative ex vivo tumor of F2-7 cells.
Histology and immunohistochemistry
Tumors that formed from both the MPIO labeled and the unlabeled F2-7 cells retained expression of both EGFR and CK5, H&E stained sections confirmed the presence of tumor growth in the MFP (Fig. 6).
Figure 6.
Expression of cytokeratin 5 (CK5) and estrogen growth factor receptor (EGFR) in MPIO-labeled and unlabeled F2-7 MFP tumors and corresponding hematoxylin and eosin (H&E) staining.
DISCUSSION
The increasing incidence of breast cancer brain metastases and its poor prognosis has highlighted the critical need for clinically relevant models to develop new therapeutic strategies and to understand the mechanisms underlying its progression. PDX models have been used increasingly to understand the role of tumor heterogeneity in the development of novel drugs and metastatic progression, and while other groups have made exciting progress in this area, most methods employed have been performed ex vivo, through histology, or with immunohistochemistry. Alone, these do not allow for the longitudinal study of cancer progression in vivo, and therefore, tools such as cellular imaging would be extremely valuable in further investigation and characterization of these models. Additionally, only a handful of studies exist that validate the engraftment and growth patterns of PDX models with imaging. At this time, only four studies have used MRI to image PDX models of breast cancer brain metastasis. In 2016, Ni et al. monitored therapeutic response to combination therapies by both MRI and BLI of an orthoptic PDX brain metastatic model [24]. In 2017, two novel brain metastatic PDX cell lines, including F2-7, as well as BM-E22-1, were imaged to determine successful engraftment and detect metastases within the mouse brain [8]. Sharma et al. also confirmed the tumor establishment of two PDX cell lines (PDX2147 and PDX1435) with MRI after 30 d [25]. In 2020, Oshi et al. developed new implantation techniques for higher engraftment and growth rates of a PDX model and monitored tumor development with MRI [26]. Including the mentioned study by Ni et al., only three studies have used BLI to monitor cell viability in PDX models of breast cancer brain metastasis. Turner et al. used BLI to monitor the response of two basal-like, triple-negative PDX cell lines (WHIM2 and WHIM30) to treatment [27]. Liu et al. described establishment of orthotopic mouse models of BCBM-PDXs and monitored their engraftment with BLI [28]. To address this, we developed a novel method to label luciferase expressing PDX cells with iron-oxide particles to allow for the in vivo, longitudinal cell tracking with two imaging modalities for the first time.
Our group has previously demonstrated that BLI is a valuable tool for cell tracking preclinical models of breast cancer brain metastasis, as is it is able to provide longitudinal measures of tumor growth and indicates cell viability [10]. MPI is an emerging, highly sensitive imaging modality that can be used for cell tracking and offers the benefits of directly detecting iron-oxide nanoparticles to allow for the quantification of iron in a region of interest. These modalities are complementary, as BLI allows for real-time monitoring of viable cells longitudinally. However, experiences tissue attenuation of emitted light. MPI addresses this by providing a quantitative method to directly measure iron content within cells longitudinally but does not provide measures of viability or proliferation. Additionally, while MPI has been used to detect cell lines of breast cancer [15,29], to date, no studies exist demonstrating the labeling of PDX breast cancer cells with iron-oxide nanoparticles to allow for MPI cell tracking.
In this work, we visualized the iron labeling of the luciferase expressing F2-7-eGFP with PPB stains and demonstrated that 25 μg Fe/ml was the most successful labeling trial. The addition of the magnetic plate for labeling was likely ineffective due to the fact that the majority of these cells are in suspension during labeling, which differs from most studies which have labeled adherent cells. Previous work by our group has shown that this concentration of MPIO effectively labels immortalized cancer cells for the purposes of cell tracking [30]. However, the methods of labeling described in this work varies from the traditional labeling protocol of adherent cells due to the differences in homogeneity and complex cell culture required to grow PDX cells. Consequently, a limitation of this method is that labeling efficiency may vary due to the mixed cell populations within established PDX cell lines. Future work will investigate labeling trials with different iron particles and whether this changes the visualization of these cells in vivo with MPI or MRI.
In this model, F2-7 mammary fat pad tumors were monitored out to day 42 with BLI. BLI signal increased over time, demonstrating that the PDX cells had successfully engrafted and were proliferating over time. In contrast, the MPI signal from the MFP decreased over time during the 28-day period. This may be related to the clearance of iron released from dead labeled cancer cells. We observed MPI signal elsewhere in the body, which may be signal from the gut and attributed to iron in the mouse feed, or signal from the liver, which may result from clearance of iron from the tumor. Wang et al. have shown that when iron-labeled islets were transplanted under the kidney capsule MPI signal is detected in the kidney on day 1 post-transplant but on day 14 MPI signal also appears in the liver, where the released iron particles accumulate [19]. These observations require more study. Future work will investigate this signal located in elsewhere in the body through ex vivo imaging and iron quantification with MPI.
Our group and others have used either BLI or MPI to detect iron-labeled cancer cells in vivo. However, only one study has used both modalities [29], although they performed imaging at a single timepoint in a single mouse. We have previously shown that labeling cancer cells with iron-oxide nanoparticles does not significantly affect cell viability, proliferation, apoptosis, or metastatic efficiency, demonstrating that this labeling agent is an effective technique to track cells in vivo [11]. With the increased use of PDX models as a platform to study cancer metastasis and develop novel drugs and therapeutics, this method provides a reliable and efficient method to determine the fate of these cells in vivo and their response to therapeutic treatments.
Acknowledgments
The authors would like to thank the Canadian Institutes of Health Research and the Breast Cancer Society of Canada for their funding.
References
- 1.DeSantis C, Ma J, Bryan L, Jemal A. Breast cancer statistics, 2013. CA Cancer J Clin. 2014Jan-Feb;64(1):52–62. https://doi.org/10.3322/caac.21203 10.3322/caac.21203PMID: [DOI] [PubMed] [Google Scholar]
- 2.Kennecke H, Yerushalmi R, Woods R, Cheang MC, Voduc D, Speers CH, et al. Metastatic behavior of breast cancer subtypes. J Clin Oncol. 2010July;28(20):3271–7. https://doi.org/10.1200/JCO.2009.25.9820 10.1200/JCO.2009.25.9820PMID: [DOI] [PubMed] [Google Scholar]
- 3.Shaffrey ME, Mut M, Asher AL, Burri SH, Chahlavi A, Chang SM, et al. Brain metastases. Curr Probl Surg. 2004August;41(8):665–741. https://doi.org/10.1067/j.cpsurg.2004.06.001 10.1067/j.cpsurg.2004.06.001PMID: [DOI] [PubMed] [Google Scholar]
- 4.Engel J, Eckel R, Aydemir U, Aydemir S, Kerr J, Schlesinger-Raab A, et al. Determinants and prognoses of locoregional and distant progression in breast cancer. Int J Radiat Oncol Biol Phys. 2003April;55(5):1186–95. https://doi.org/10.1016/S0360-3016(02)04476-0 10.1016/S0360-3016(02)04476-0PMID: [DOI] [PubMed] [Google Scholar]
- 5.Barnholtz-Sloan JS, Sloan AE, Davis FG, Vigneau FD, Lai P, Sawaya RE. Incidence proportions of brain metastases in patients diagnosed (1973 to 2001) in the Metropolitan Detroit Cancer Surveillance System. J Clin Oncol. 2004July;22(14):2865–72. https://doi.org/10.1200/JCO.2004.12.149 10.1200/JCO.2004.12.149PMID: [DOI] [PubMed] [Google Scholar]
- 6.Hidalgo M, Amant F, Biankin AV, Budinská E, Byrne AT, Caldas C, et al. Patient-derived xenograft models: an emerging platform for translational cancer research. Cancer Discov. 2014September;4(9):998–1013. https://doi.org/10.1158/2159-8290.CD-14-0001 10.1158/2159-8290.CD-14-0001PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dobrolecki LE, Airhart SD, Alferez DG, Aparicio S, Behbod F, Bentires-Alj M, et al. Patient-derived xenograft (PDX) models in basic and translational breast cancer research. Cancer Metastasis Rev. 2016December;35(4):547–73. https://doi.org/10.1007/s10555-016-9653-x 10.1007/s10555-016-9653-xPMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Contreras-Zárate MJ, Ormond DR, Gillen AE, Hanna C, Day NL, Serkova NJ, et al. Development of novel patient-derived xenografts from breast cancer brain metastases. Front Oncol. 2017November;7:252. https://doi.org/10.3389/fonc.2017.00252 10.3389/fonc.2017.00252PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Prescher JA, Contag CH. Guided by the light: visualizing biomolecular processes in living animals with bioluminescence. Curr Opin Chem Biol. 2010February;14(1):80–9. https://doi.org/10.1016/j.cbpa.2009.11.001 10.1016/j.cbpa.2009.11.001PMID: [DOI] [PubMed] [Google Scholar]
- 10.Parkins KM, Hamilton AM, Makela AV, Chen Y, Foster PJ, Ronald JA. A multimodality imaging model to track viable breast cancer cells from single arrest to metastasis in the mouse brain. Sci Rep. 2016October;6(1):35889. https://doi.org/10.1038/srep35889 10.1038/srep35889PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heyn C, Ronald JA, Ramadan SS, Snir JA, Barry AM, MacKenzie LT, et al. In vivo MRI of cancer cell fate at the single-cell level in a mouse model of breast cancer metastasis to the brain. Magn Reson Med. 2006November;56(5):1001–10. https://doi.org/10.1002/mrm.21029 10.1002/mrm.21029PMID: [DOI] [PubMed] [Google Scholar]
- 12.Makela AV, Murrell DH, Parkins KM, Kara J, Gaudet JM, Foster PJ. Cellular imaging with MRI. Top Magn Reson Imaging. 2016October;25(5):177–86. https://doi.org/10.1097/RMR.0000000000000101 10.1097/RMR.0000000000000101PMID: [DOI] [PubMed] [Google Scholar]
- 13.Bulte JW. Superparamagnetic iron oxides as MPI tracers: A primer and review of early applications. Adv Drug Deliv Rev. 2019January;138(138):293–301. https://doi.org/10.1016/j.addr.2018.12.007 10.1016/j.addr.2018.12.007PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu LC, Zhang Y, Steinberg G, Qu H, Huang S, Cheng M, et al. A Review of Magnetic Particle Imaging and Perspectives on Neuroimaging. AJNR Am J Neuroradiol. 2019February;40(2):206–12. https://doi.org/10.3174/ajnr.A5896 10.3174/ajnr.A5896PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Parkins KM, Melo KP, Chen Y, Ronald JA, Foster PJ. Visualizing tumour self-homing with magnetic particle imaging. Nanoscale. 2021March;13(12):6016–23. https://doi.org/10.1039/D0NR07983A 10.1039/D0NR07983APMID: [DOI] [PubMed] [Google Scholar]
- 16.Nejadnik H, Pandit P, Lenkov O, Lahiji AP, Yerneni K, Daldrup-Link HE. Ferumoxytol Can Be Used for Quantitative Magnetic Particle Imaging of Transplanted Stem Cells. Mol Imaging Biol. 2019June;21(3):465–72. https://doi.org/10.1007/s11307-018-1276-x 10.1007/s11307-018-1276-xPMID:30194566 [DOI] [Google Scholar]
- 17.Zheng B, von See MP, Yu E, Gunel B, Lu K, Vazin T, et al. Quantitative Magnetic Particle Imaging Monitors the Transplantation, Biodistribution, and Clearance of Stem Cells In Vivo. Theranostics. 2016January;6(3):291–301. https://doi.org/10.7150/thno.13728 10.7150/thno.13728PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bulte JW, Walczak P, Janowski M, Krishnan KM, Arami H, Halkola A, et al. Quantitative “Hot Spot” Imaging of Transplanted Stem Cells using Superparamagnetic Tracers and Magnetic Particle Imaging (MPI). Tomography. 2015December;1(2):91–7. https://doi.org/10.18383/j.tom.2015.00172 10.18383/j.tom.2015.00172PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang P, Goodwill PW, Pandit P, Gaudet J, Ross A, Wang J, et al. Magnetic particle imaging of islet transplantation in the liver and under the kidney capsule in mouse models. Quant Imaging Med Surg. 2018March;8(2):114–22. https://doi.org/10.21037/qims.2018.02.06 10.21037/qims.2018.02.06PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rivera-Rodriguez A, Hoang-Minh LB, Chiu-Lam A, Sarna N, Marrero-Morales L, Mitchell DA, et al. Tracking adoptive T cell immunotherapy using magnetic particle imaging. Nanotheranostics. 2021April;5(4):431–44. https://doi.org/10.7150/ntno.55165 10.7150/ntno.55165PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Makela AV, Gaudet JM, Schott MA, Sehl OC, Contag CH, Foster PJ. Magnetic Particle Imaging of Macrophages Associated with Cancer: Filling the Voids Left by Iron-Based Magnetic Resonance Imaging. Mol Imaging Biol. 2020August;22(4):958–68. https://doi.org/10.1007/s11307-020-01473-0 10.1007/s11307-020-01473-0PMID: [DOI] [PubMed] [Google Scholar]
- 22.Gaudet J, Mansfield J, Goodwill P. Imaging Cancer Immunology: Tracking Immune Cells in vivo with Magnetic Particle Imaging. J Immunol. 2019May1;202(1 Supplement):130.7-130.7. [Google Scholar]
- 23.Arbab AS, Yocum GT, Kalish H, Jordan EK, Anderson SA, Khakoo AY, et al. Efficient magnetic cell labeling with protamine sulfate complexed to ferumoxides for cellular MRI. Blood. 2004August;104(4):1217–23. https://doi.org/10.1182/blood-2004-02-0655 10.1182/blood-2004-02-0655PMID: [DOI] [PubMed] [Google Scholar]
- 24.Ni J, Ramkissoon SH, Xie S, Goel S, Stover DG, Guo H, et al. Combination inhibition of PI3K and mTORC1 yields durable remissions in mice bearing orthotopic patient-derived xenografts of HER2-positive breast cancer brain metastases. Nat Med. 2016July;22(7):723–6. https://doi.org/10.1038/nm.4120 10.1038/nm.4120PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sharma S, Wu SY, Jimenez H, Xing F, Zhu D, Liu Y, et al. Ca2+ and CACNA1H mediate targeted suppression of breast cancer brain metastasis by AM RF EMF. EBioMedicine. 2019June;44:194–208. https://doi.org/10.1016/j.ebiom.2019.05.038 10.1016/j.ebiom.2019.05.038PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oshi M, Okano M, Maiti A, Rashid OM, Saito K, Kono K, et al. Novel Breast Cancer Brain Metastasis Patient-Derived Orthotopic Xenograft Model for Preclinical Studies. Cancers (Basel). 2020February;12(2):444. https://doi.org/10.3390/cancers12020444 10.3390/cancers12020444PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Turner TH, Alzubi MA, Sohal SS, Olex AL, Dozmorov MG, Harrell JC. Characterizing the efficacy of cancer therapeutics in patient-derived xenograft models of metastatic breast cancer. Breast Cancer Res Treat. 2018July;170(2):221–34. https://doi.org/10.1007/s10549-018-4748-4 10.1007/s10549-018-4748-4PMID: [DOI] [PubMed] [Google Scholar]
- 28.Liu Z, Wang Y, Kabraji S, Xie S, Pan P, Liu Z, et al. Improving orthotopic mouse models of patient-derived breast cancer brain metastases by a modified intracarotid injection method. Sci Rep. 2019January;9(1):622. https://doi.org/10.1038/s41598-018-36874-3 10.1038/s41598-018-36874-3PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yu EY, Bishop M, Zheng B, Ferguson RM, Khandhar AP, Kemp SJ, et al. Magnetic Particle Imaging: A Novel in Vivo Imaging Platform for Cancer Detection. Nano Lett. 2017March;17(3):1648–54. https://doi.org/10.1021/acs.nanolett.6b04865 10.1021/acs.nanolett.6b04865PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Knier NN, Hamilton AM, Foster PJ. Comparing the fate of brain metastatic breast cancer cells in different immune compromised mice with cellular magnetic resonance imaging. Clin Exp Metastasis. 2020August;37(4):465–75. https://doi.org/10.1007/s10585-020-10044-0 10.1007/s10585-020-10044-0PMID: [DOI] [PubMed] [Google Scholar]