Abstract
Background and Purpose
The alarming increase in the prevalence of CTX-M-15 extended-spectrum β-lactamase (ESBL) producing E. coli has been significantly linked to the clonal expansion of emerging sequence type (ST131). This study aimed to screen for the O16/O25-ST131 clones among different phylogenetic types of E. coli strains isolated from urinary and diarrhoeal samples.
Methods
A total of 205 E. coli strains isolated from patients with UTI and acute diarrhoea were investigated by phenotypic and genotypic methods for ESBL identification. Molecular methods were used for identification of O25/O16-ST131 clone and phylogenetic typing of E. coli isolates.
Results
O25-ST131 clone was detected in 89/105 (84.8%) and 47/100 (47%) of urinary and intestinal E. coli isolates, respectively, with a significant difference (P-value<0.001). There was a significant high rate of occurrence of ESBLs, MDR, and antibiotic resistance to most antibiotic classes among O25-ST131 than non-O25-ST131 isolates. CTX-M-15 gene was detected in 64/71 (90%) of ESBLs producing intestinal isolates and 54/79 (68.4%) of urinary ESBLs producing isolates. The O25-ST131 clone was reported among all phylogenetic groups. The O16-ST131 clone serotype was not detected in the study isolates.
Conclusion
High prevalence of the O25-ST131 clone was reported among extraintestinal and intestinal E. coli isolates. First detection of the O25-ST131 clone among phylogenetic groups other than group B2 draws attention of the ability of this clone to transfer among commensal groups. An increasing in the prevalence of CTX-M-15 among E. coli strains especially of intestinal origin is alarming as the intestine is the main reservoir for ExPEC strains causing UTI.
Keywords: E. coli, O25-ST131 clone, ESBLs, UTI, diarrhoea
Introduction
Escherichia coli (E. coli) is one of the common causes of bacterial infections in both digestive and urinary tracts.1 E. coli is a member of the gut flora. Different species of bacteria interact in the gut and can gain virulence and antibiotic resistance genes developing pathogenic E. coli strains.2 Pathogenic E. coli strains cause either diarrhoea by intestinal pathogenic E. coli (IPEC) or extra-intestinal infections by extra-intestinal pathogenic E. coli (ExPEC). Urinary tract infection (UTI) is the commonest extra-intestinal infection as uropathogenic E. coli (UPEC) was found to be associated with more than 80% of UTI.3,4
The emerging E. coli ST131 was identified in 2008 as a multidrug resistant clone producing ESBLs. CTX-M-15 is the most dominant ESBL reported among E. coli ST131 strains.5 The worldwide alarming increase in CTX-M-15 ESBL producing Enterobacteriaceae, especially E. coli strains, represents a major challenge to public health.6 This increased isolation of CTX-M-15 producing E. coli strains was linked to the rapid clonal expansion of ST131 during the last 20 years.2 The success of dissemination of E. coli ST131 clone is explained by carrying ESBL resistance genes and different virulence factors7 and also by its predominance in the human gut.1 E. coli O25-ST131 is the predominant clone serotype, while O16-ST131 was reported in a very small percentage in some studies.2 Misuse of antibiotics in Middle East countries due to lack of policies controlling for sale of antibiotics plays an important role in the emergence of resistant bacterial strains. The relationships between use of third generation cephalosporins and increasing ESBL producing strains have been reported in several studies.8
E. coli is classified phylogenetically into four main group; A, B1, D, and B2.9 Most previous studies restrict the E. coli ST131 clone to phylogenetic group B2 but recently some studies reported ST131 isolates in other phylogenetic groups which may be an index of risk.10,11 Increasing rates of intestinal colonization with CTX-M-15 ESBL producing E. coli ST131 is particularly worrisome as the intestine is the main reservoir for ExPEC strains causing UTI and it was found that most UTIs were caused by E. coli in feces of the patients.12 The information about the E. coli ST131 clone in Egypt is very sparse. Therefore, the current study aimed to identify the prevalence of O16/O25-ST131 clones among different phylogenetic types of E. coli strains isolated from UTIs and diarrhoeal samples. The study also aimed to identify the association between this clone and antibiotic resistance patterns, ESBL production, and the presence of the CTX-M-15 gene.
Patients and Methods
Study Design
This study was conducted in the period from July 2020 to February 2021 in the department of Microbiology and Immunology, Faculty of medicine, Minia University, Egypt. A total of 205 E. coli clinical isolates (104 from urine of UTI patients and 100 from stool of patients with acute diarrhoea) were included in this study. All study participants were adults attending Outpatient clinics of Minia University Hospitals. UTI was diagnosed by dysuria, frequency, urgency with pyuria, and significant bacteriuria E. coli >105 CFU/ mL. Acute diarrhoea was diagnosed clinically by passage of loose or watery stools more than 3-times daily with any of the symptoms vomiting, abdominal cramps, or dysentery. Patients with any history of antibiotic use or hospitalization 2 weeks before specimen collection or any history of chronic disease were excluded. The study was approved by the Medical Ethics of Minia University hospital and written informed consent was taken from each participant for the use of the samples. This study was performed in agreement with the guidelines of the Declaration of Helsinki.
Bacterial Isolation and Identification
Both urine and stool samples were collected under aseptic precautions in sterile containers and transported within 2 hours to the microbiology laboratory for immediate examination. Urine samples with pus >5 cells/HPF were cultured on chromogenic media (CHROMagarTM Orientation, Paris, France). Stool samples were inoculated into enrichment broth and then sub-cultured on MacConkey and Eosin Methylene Blue (EMB) agar (Oxoid, UK). Isolated colonies were then identified biochemically by indole, methyl red, citrate, Voges-Proskauer, urease, and sugar fermentation tests. Strains identified and confirmed as E. coli were grown in Trypticase soy broth (Oxoid, UK), mixed with sterilized glycerol 20% and stored at −20°C for further testing.
Antimicrobial Susceptibility Testing
All isolated E. coli strains were subjected to antimicrobial susceptibility testing using Kirby Bauer disc diffusion method and identified according to Clinical and Laboratory Standards Institute (CLSI).13 The following antibiotic discs were used; amoxicillin/clavulanic acid (AMC, 30 μg), ceftriaxone (CRO, 30 μg), ceftazidime (CAZ, 30 μg), imipenem (IPM, 10 μg), amikacin (AMK, 30 μg), sulphamethoxazole/trimethoprim (SXT, 25 μg), and Ciprofloxacin (CIP, 5 μg) (Thermo Scientific™ Oxoid, UK). Resistance to three or more different antimicrobial groups is identified as multiple drug resistance (MDR).14 ESBL-production is suggested in isolates giving inhibition zone size ≤22 mm with ceftazidime and ≤25 mm with ceftriaxone. Double disk synergy test (DDST) was used to confirm ESBL producers.15 E. coli isolate from a previous study was used as quality control in antimicrobial susceptibility testing.16
DNA Extraction
The DNA of 205 E. coli isolates was extracted using Gene JET genomic DNA purification kit (Thermo Scientific, USA) according to manufacturer’s manual procedures.
Phylogenetic Analysis
E. coli isolates were classified into four phylogenetic groups using the multiplex PCR procedure described by Doumith and his colleagues for gadA, chuA, yjaA genes, and the TSPE4.C2 DNA fragment.16 The glutamate decarboxylase-alpha (gadA) gene of E. coli was used as an internal control. It was performed with the following optimized cycling conditions: initial denaturation at 94°C for 4 minutes; then 30 cycles (94°C for 30 seconds, annealing at 65°C for 30 seconds, and extension at 72°C for 30 seconds) and a final extension step of 5 minutes at 72°C. On the basis of the results of multiplex PCR, detection of chuA– yjaA – TSPE4.C2–, chuA– yjaA – TSPE4.C2+, chuA+ yjaA – TSPE4.C2+, and chuA+ yjaA+ TSPE4.C2+ were belonging to groups A, B1, D, and B2, respectively, with gadA detected in all isolates as internal control.17 E. coli isolate from a previous study, belonging to group B2 with four amplified bands, was used as a positive control in each PCR run.16
Detection of blaCTX-M-15 Gene and ST131 Clones
Screening of E. coli isolates from urine and stool specimens were first done for CTX-M-15. The thermal cycling conditions were: initial denaturation at 95°C for 10 minutes, then 35 cycles (95°C for 1 minute, annealing at 55°C for 45 seconds, extension at 72°C for 1 minute) and final extension at 72°C for 10 minute.18 Secondly, isolates were screened for detection of E. coli ST131 clones using primers in Table 1 to amplify pabB and trpA genes for O25-ST131 and O16-ST131 clades, respectively. The following amplification conditions: Initial denaturation for 4 minutes at 94°C then 30 cycles of denaturation for 5 seconds at 94°C, annealing for 20 seconds at 63°C and extension for 30 seconds at 72°C) and lastly final extension at 72°C for 5 minutes.19,20 The PCR products were identified by 1.5% Agarose gel electrophoresis (Biometra Goettingen, Germany). Positive O25 and O16-ST131 E. coli isolates, from previous studies done in the same laboratory, were used as positive control in PCR reactions for the ST131 clone.21 All primers sequences used in this study and amplification band sizes are described in Table 1.
Table 1.
PCR Primers Used in the Study
| Primer | Primers (5_ to 3_) | Amplicon Size (bp) | Annealing Temperature (°C) | Reference |
|---|---|---|---|---|
| gadA | F-GATGAAATGGCGTTGGCGCAAG R-GGCGGAAGTCCCAGACGATATCC |
373 | 65 | [17] |
| chuA | F- ATGATCATCGCGGCGTGCTG R-AAACGCGCTCGCGCCTAAT |
281 | ||
| yjaA | F-TGTTCGCGATCTTGAAAGCAAACGT R-ACCTGTGACAAACCGCCCTCA |
216 | ||
| TSPE4.C2 | F-GCGGGTGAGACAGAAACGCG R- TTGTCGTGAGTTGCGAACCCG |
152 | ||
| CTX-M-15 | F-CACACGTGGAATTTAGGGACT R-GCCGTCTAAGGCCATAAACA |
996 | 55 | [18] |
| pabBST131-O25b | F- TCCAGCAGGTGCTGGATCGT R- GCGAAATTTTTCGCCGTACTGT |
347 | 63 | [19] |
| trpAST131-O16 | F- AAAACCGCGCCGCGTTACCT R- CCAGAAATCGCGCCCGCATT |
145 | 63 | [20] |
Statistical Analysis
Demographic, clinical, and laboratory data analyses were carried out using SPSS program for windows version 20.0 (IBM, USA). P≤0.05 were considered statistically significant.
Results
Demographic Characteristics of the Study Subjects
A total of 205 E. coli strains were isolated from patients with UTI and acute diarrhoea. Out of 105 urinary isolates, 71 (67.6%) were from female patients and 34 (32.4%) were from male patients. The mean age of all UTI patients was 36.9±11.8. The mean age of female patients was 33.8±11.00, but the mean age of males was 43.2±10.9. Clinically, 89/105 (84.7%) of UTI patients complained from dysuria and increased frequency and only 41/105 (39%) suffered from fever.
One hundred E. coli strains were isolated from the stool of patients with acute diarrhoea. The age range of patients was from 20–63 years old (mean=43.2±12). Out of 100 patients with diarrhoea, 53 were males and 47 were females. Clinically, all cases passed more than three loose stools daily. Blood and mucus were detected in 16/100 and 69/100 of samples, respectively.
Antimicrobial Susceptibility Testing
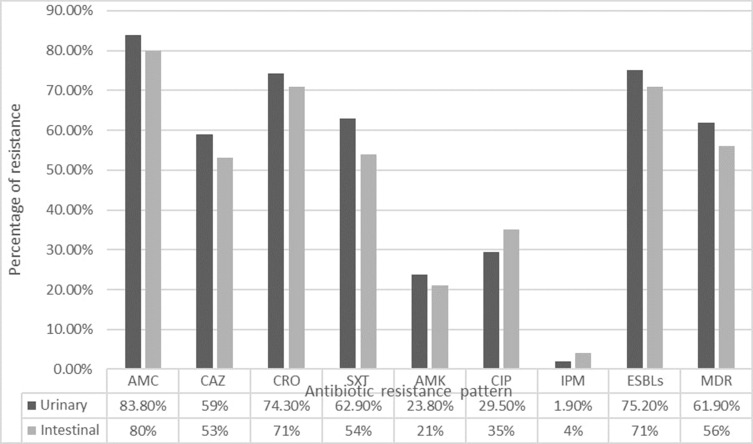
All E. coli isolates were tested for their resistance to seven antibiotics belonging to different classes (Figure 1). High resistance rates were detected against AMC, CRO, SXT, and CAZ in both urinary and intestinal E. coli isolates and the rates were higher in urinary isolates with no significant difference. Moderate resistance rates against CIP and AMK were detected in both groups. Imipenem showed very low resistance (1.9 and 4% in urinary and intestinal isolates, respectively).
Figure 1.
Antibiotic resistance, ESBLs, and MDR distributions among urinary and intestinal isolates.
Abbreviations: AMC, Amoxicillin-clavulanate; CAZ, Ceftazidime; CRO, Ceftriaxone; SXT, sulfamethoxazole-Trimethoprim; AMK, Amikacin; IPM, Imipenem; CIP, Ciprofloxacin.
ESBLs production was identified phenotypically using DDST in 75.2% of urinary isolates and 71% of intestinal isolates. MDR was found in 65/105 (61.9%) and 56/100 (56%) of urinary and intestinal E. coli isolates, respectively (Table 2).
Table 2.
Differentiation between E coli Isolates from Urinary and Intestinal Origin
| No. (%) of Strains | P-value | |||
|---|---|---|---|---|
| Urinary Isolates (n=105) | Intestinal Isolates (n=100) | |||
| Phylogenetic group | B2 | 32 (30.5%) | 5 (5%) | <0.001* |
| B1 | 30 (28.6%) | 39 (39%) | 0.114 | |
| A | 26 (24.8%) | 44 (44%) | 0.004* | |
| D | 17 (16.2%) | 12 (12%) | 0.398 | |
| O25-ST131 (pabB positive) | 89 (84.8%) | 47 (47%) | <0.001* | |
| MDR | 65 (61.9%) | 56 (56%) | 0.39 | |
| ESBL positive (synergy test) | 79 (75.2%) | 71 (71%) | 0.494 | |
| CTX-M-15 positive | 57 (54.3%) | 66 (66%) | 0.087 | |
| O16-ST131 (trpA) | 0 | 0 | – | |
Note: *P-value is significant.
Abbreviation: n, number.
Phylogenetic Grouping
The phylogenetic group distribution among the urinary isolates showed that 30.5%, 28.6%, 24.8, and 16.2% of isolates belonged to group B2, B1, A, and D, respectively. The B2 group was the predominant among urinary isolates. However, the phylogenetic group distribution among IPEC isolates was different, with 44, 39, 12, and 5% belonging to groups A, B1, D, and B2, respectively. There was a predominance of phylogenetic groups A and B2 among intestinal isolates. There was a significant difference between the urinary and intestinal isolates for phylogenetic groupings B2 & A (Table 2).
Detection of O25-ST131, O16-ST131 Clones
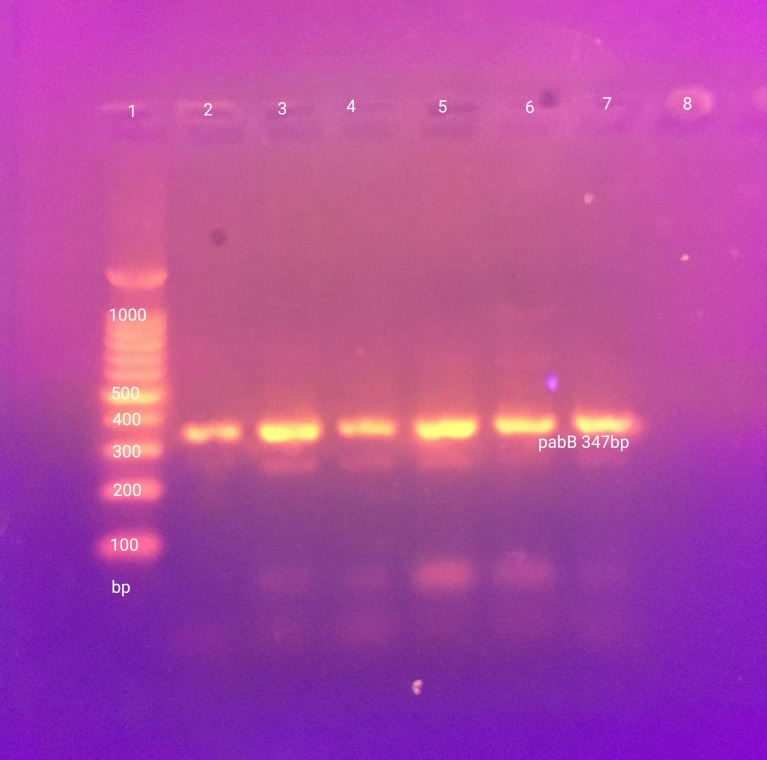
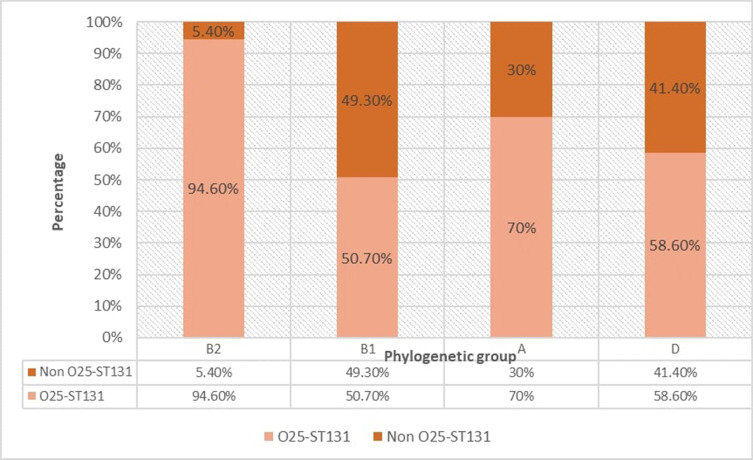
The pabB gene for the O25-ST131 clone (Figure 2) was detected in 89/105 (84.8%) and 47/100 (47%) of urinary and intestinal E. coli isolates. There was a significantly higher rate of this clone among urinary than intestinal isolates (Table 2). The distribution of phylogenetic groups among all O25-ST131 and non O25-ST131 isolates was as shown in Figure 3 and its distribution among urinary and intestinal isolates is shown in Table 3. There was a significant difference between O25-ST131 and non-O25-ST131 isolates in the percentage of ESBLs, MDR, and antibiotic resistance to most antibiotic classes (Table 3). No O16-ST131 clone was detected as the trpA gene was negative in all studied isolates.
Figure 2.
Screening of pabB gene for O25-ST131 in E. coli isolates. Lane 1, 100 bp molecular marker (iNtRON Biotechnology, South Korea); Lane 2–6 show (pabB gene) 347 bp; Lane 7 (positive control); Lane 8 (negative control).
Figure 3.
Phylogenetic distribution of O25-ST 131 and non O25-ST131 E. coli isolates.
Table 3.
Differentiation between O25-ST131 and Non-O25-ST131 E. coli Isolates
| n= (Urinary+Intestinal) | O25-ST131 | Non O25-ST131 | P-value | |||
|---|---|---|---|---|---|---|
| Urinary Isolates | Intestinal Isolates | Urinary Isolates | Intestinal Isolates | |||
| Phylogenetic group | B2, n=(32+5) | 30/32 (93.8%) | 5/5 (100%) | 2/32 (6.2%) | 0/5 0 | 1.000 |
| B1, n=(30+39) | 23/30 (76.7%) | 12/39 (30.8%) | 7/30 (23.3%) | 27/39 (69.2%) | <0.001* | |
| A, n=(26+44) | 24/26 (92.3%) | 25/44 (56.8%) | 2/26 (7.7%) | 19/44 (43.2%) | 0.002* | |
| D, n=(17+12) | 12/17 (70.6%) | 5/12 (41.7%) | 5/17 (29.4%) | 7/12 (58.3%) | 0.148 | |
| MDR n= (65+56) | 58 (89.2%) | 42 (75%) | 7 (10.8%) | 14 (25%) | 0.039* | |
| ESBLs positive (synergy test) n=(79+71) | 66 (83.5%) | 41 (57.7%) | 13 (16.5%) | 30 (42.3%) | <0.001* | |
| CTX-M-15, n=(57+66) | 45 (78.9%) | 35 (53%) | 12 (21.1%) | 31 (47%) | 0.004* | |
| Antibiotic resistance | AMC (88+80) | 74 (84.1%) | 45 (56.2%) | 14 (15.9%) | 35 (43.8%) | <0.001* |
| CAZ (62+53) | 53 (85.5%) | 38 (71.7%) | 9 (14.5%) | 15 (28.3%) | 0.07 | |
| CRO (78+71) | 66 (84.6%) | 42 (59.2%) | 12 (15.4%) | 29 (40.8%) | 0.001* | |
| STX (66+54) | 59 (89.4%) | 41 (75.9%) | 7 (10.6%) | 13 (24.1%) | 0.049* | |
| AMK (25+21) | 24 (96%) | 14 (66.7%) | 14 (4%) | 7 (33.3%) | 0.016* | |
| CIP (31+35) | 25 (80.6%) | 29 (82.9%) | 6 (19.4%) | 6 (17.1%) | 0.816 | |
| IPM (2+4) | 2 (100%) | 4 (100%) | 0 | 0 | ||
Note: *P-value is significant.
Abbreviations: MDR, Multi-Drug Resistant; ESBLs, Extended Spectrum B-Lactamases; AMC, Amoxicillin-clavulanate; CAZ, Ceftazidime; CRO, Ceftriaxone; SXT, sulfamethoxazole -Trimethoprim; AMK, Amikacin; IPM, Imipenem; CIP, Ciprofloxacin; n, number.
Detection of blaCTX-M-15 Gene
All E. coli isolates were examined for blaCTX-M-15 gene and it was positive in 66 and 54.3% of intestinal and urinary isolates, respectively, with a higher rate among intestinal over urinary isolates that was not significant (Table 2). CTX-M-15 was positive in 64/71 (90%) of ESBLs producing intestinal isolates. However, 54/79 (68.4%) of urinary ESBLs producing isolates were positive for CTX-M-15 and this difference was significant (Table 4).
Table 4.
Distribution of Phenotypic ESBLs Producing E coli Isolates According to O25-ST131, CTX-M-15, Phylogenetic Group, and Antibiotypes
| E coli Pattern (EC) | Source | No of Isolates | O25-ST131 | Non O25-ST131 | CTX-M-15 +ve | CTX-M-15 -ve | Phylogenetic Group | Antibiotype | |||
|---|---|---|---|---|---|---|---|---|---|---|---|
| A | B1 | D | B2 | ||||||||
| EC1 | Urinary | 21 | 15 | 6 | 12 | 9 | 2 | 9 | 4 | 6 | AMC, CAZ, CRO, SXT, CIP |
| Intestinal | 17 | 15 | 2 | 15 | 2 | 8 | 7 | 1 | 1 | ||
| EC2 | Urinary | 18 | 18 | 0 | 13 | 5 | 3 | 4 | 4 | 7 | AMC, CAZ, CRO, SXT, AMK |
| Intestinal | 5 | 3 | 2 | 5 | 0 | 4 | 1 | 0 | 0 | ||
| EC3 | Urinary | 7 | 3 | 4 | 7 | 0 | 0 | 1 | 4 | 2 | AMC, CRO |
| Intestinal | 12 | 0 | 12 | 12 | 0 | 3 | 9 | 0 | 0 | ||
| EC4 | Urinary | 9 | 9 | 0 | 6 | 3 | 2 | 4 | 0 | 3 | AMC, CAZ, CRO, SXT |
| Intestinal | 11 | 7 | 4 | 11 | 0 | 8 | 1 | 2 | 0 | ||
| EC5 | Urinary | 9 | 7 | 2 | 8 | 1 | 4 | 0 | 0 | 5 | AMC, CAZ, CRO |
| Intestinal | 5 | 1 | 4 | 4 | 1 | 4 | 1 | 0 | 0 | ||
| EC6 | Urinary | 4 | 4 | 0 | 2 | 2 | 1 | 1 | 1 | 1 | AMC, CRO, SXT |
| Intestinal | 4 | 3 | 1 | 4 | 0 | 4 | 0 | 0 | 0 | ||
| EC7 | Urinary | 3 | 3 | 0 | 2 | 1 | 2 | 1 | 0 | 0 | AMC, CRO, CIP |
| Intestinal | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ||
| EC8 | Urinary | 3 | 3 | 0 | 1 | 2 | 2 | 0 | 0 | 1 | AMC, CRO, SXT, AMK |
| Intestinal | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 0 | 0 | ||
| EC9 | Urinary | 1 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | AMC, CAZ, AMK |
| Intestinal | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ||
| EC10 | Urinary | 1 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | AMC, CAZ, CRO, CIP |
| Intestinal | 2 | 1 | 1 | 2 | 0 | 0 | 1 | 1 | 0 | ||
| EC11 | Urinary | 1 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | AMC, CAZ, SXT, AMK |
| Intestinal | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ||
| EC12 | Urinary | 1 | 1 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | AMC, CAZ, CRO, SXT, AMK, IPM |
| Intestinal | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ||
| EC13 | Urinary | 1 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | AMC, CAZ, CRO, SXT, CIP, IPM |
| Intestinal | 3 | 3 | 0 | 3 | 0 | 0 | 0 | 0 | 3 | ||
| EC14 | Urinary | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | AMC, CAZ. CRO, SXT, AMK, CIP |
| Intestinal | 7 | 6 | 1 | 5 | 2 | 0 | 4 | 2 | 1 | ||
| EC15 | Urinary | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | AMC, CAZ, CRO, AMK, CIP |
| Intestinal | 2 | 1 | 1 | 0 | 2 | 0 | 1 | 1 | 0 | ||
| EC16 | Urinary | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | AMC, CAZ, CRO, AMK, CIP |
| Intestinal | 1 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | 0 | ||
| EC17 | Urinary | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | AMC, CAZ, CRO, SXT, AMK, CIP, IPM |
| Intestinal | 1 | 1 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | ||
| Total (percent to ESBLs) | Urinary | 79 | 66 (83.5%) | 13 (16.5%) | 54 (68.4%) | 25 (31.1%) | 16 (20.3%) | 20 (25.3%) | 14 (17.7%) | 29 (36.7%) | |
| Intestinal | 71 | 41 (57.7%) | 30 (43.3%) | 64 (90.1%) | 7 (9.9%) | 32 (45.1%) | 26 (36.6%) | 8 (11.3%) | 5(7%) | ||
| P-value | <0.001* | 0.001* | <0.001* | ||||||||
Note: *P-value is significant.
Abbreviations: AMC, Amoxicillin-clavulanate; CAZ, Ceftazidime; CRO, Ceftriaxone; SXT, sulfamethoxazole-Trimethoprim; AMK, Amikacin; IPM, Imipenem; CIP, Ciprofloxacin.
Determination of Different E. coli Patterns among ESBLs Producing Strains According to Source, Antibiotype, Phylogenetic Group, CTX-M-15, and O25-ST131 Clone
All isolated ESBLs producing E. coli strains were classified according to source, antibiotype pattern, phylogenetic group, CTX-M-15, and O25-ST131 clone detection into 17 different patterns. Nine patterns “EC 1, 2, 3, 4, 5, 6, 8, 10, & 13” were found in both urinary and intestinal studied groups. Four patterns “EC 7, 9, 11, & 12” were only found among intestinal isolates only and four different patterns “EC 14, 15, 16, & 17” were found among urinary isolates only. EC1 is the most prevalent pattern in both studied groups and its antibiotype is MDR to AMC, CAZ, CRO, SXT, and CIP (Table 4).
Discussion
There is a dramatic increase in the prevalence of ESBL producing E. coli, where CTX-M is the commonest detected type.22 This increased prevalence was significantly linked to the clonal expansion of E. coli ST131.23 Most previous studies reported this emerging clone in extraintestinal infections, especially UTI.2 However, intestinal colonization by the E. coli ST131 clone has been reported by recent studies.1 Data about the E. coli ST131 clone in Egypt is still scarce. To the best of our knowledge, this study is the first to investigate the prevalence of E. coli ST131 clone in both extra-intestinal (urinary) and intestinal isolates in Upper Egypt. The study demonstrated that the prevalence of the O25-ST131 clone was very high in urinary isolates (83.4%) and the prevalence among intestinal isolates was less (47%), with a significant difference (P-value<0.001). Our results were similar to a study from India that detected the O25-ST131 clone in 87% of extra-intestinal clinical E. coli isolates.11 However, our results were high compared to other studies from different countries in the Middle East region and across the world: Iraq (15%), Iran (24.7%), Turkey (22%), Pakistan (46%), China (12.5%), and Mexico (11%).8,10,24–27 This low prevalence of the ST131 clone in most of these studies can be explained by using different molecular techniques, but the main reason of this variation is that most of these studies investigated only phylogenetic group B2 so the percentage of the clone was dependent on the distribution of phylogenetic group B2. Lately, different studies investigated the ST131 clone among the different phylogenetic groups.10,11
No isolates of the O16-ST131 clone were detected among the studied isolates. Our finding agrees with the worldwide prevalence of O16-ST131 serotype, where it was reported in a very small percentage in some countries (1% in Australia, 4.3% in Spain, 8% in France).2
Regarding the phylogenetic distribution of the studied isolates, the current study showed that the phylogenetic group B2 was more predominant among the urinary isolates (30.5%) than intestinal isolates (5%). However, most of the studied intestinal isolates were belonging to phylogenetic groups A and B1 (45%, 39%). These findings were in agreement with many previous reports, considered groups B2 and D as the commonest in UPEC isolates, while groups A and B1 are commensal phylogenetic groups.28,29 For the prevalence of O25-ST131 clone among different phylogenetic groups, we found a high prevalence of this clone among group B2 (94.6%) that agrees with several previous studies.11,30,31 The current study is the first study in Egypt to report the ST131 clone among phylogenetic groups other than group B2. There was a significant difference of O25-ST131 clone prevalence in different phylogenetic groups among urinary and intestinal isolates. Most of the positive O25-ST131 clone isolates in phylogenetic groups A, B1, and D were from urinary origin and most intestinal isolates were non-O25-ST131. However, all intestinal isolates of thephylogenetic group B2 carry this clone gene. The first study of this clone in Egypt was by Fam and his colleagues in 2011, who found the E. coli ST131 clone was representing 75% of phylogenetic group B2 E. coli clinical isolates and this group was representing 26% of all isolates.30 We found a high prevalence of ST131 clones among group B2 (94.6%). This indicated that there is an increasing rate of prevalence of ST131 clones in the last decade and it is in agreement with results of studies from Saudi Arabia that reported increasing the ST131 clone prevalence from 17.3 to 61.7% from UPEC isolates in the last few years.4,32 The detection of the ST131 clone in different groups other than B2 was reported also in other studies in India and Mexico.10,11 These findings draw attention about the ability of this clone to transfer among commensal groups and so the increased risk of this clone dissemination.
ESBL producing E. coli strains were detected among clinical and commensal isolates worldwide.33 Globally, CTX-M-15 is now the most prevalent ESBL type in both community and hospital setting.23 In Egypt, CTX-M-15 is the most predominant ESBL producing type among uropathogenic and diarrheagenic E. coli strains.22,30,34 In this study, the prevalence of ESBL production among both urinary and intestinal isolates were 75.2% and 71%, respectively. These results are higher than reports of previous studies in Egypt, with ESBL prevalence range from 52–69.6%.16,22,34,35 In the same context as our results, Valverdi et al36 documented that acute diarrhoea is a frequent reason for intestinal colonization by ESBL producers that reached to 55.5% in acute diarrhoea. This can be explained by the change in the gut flora that occurs in acute diarrhoea, causing predominance of pathogenic bacterial strains.
Our results demonstrated that 90% of ESBL producing isolates from patients with acute diarrhoea carried the blaCTX-M-15 gene. However, only 68.4% of ESBL producing urinary isolates carried this gene with a significant difference between the two studied groups. These results showed that there is an increasing in the prevalence of CTX-M-15 among E. coli, especially of intestinal origin, compared to previous results in Egypt, where the CTX-M-15 gene was detected in only 37.5% of ESBL producing diarrhoeagenic E. coli.16 This was worrisome as the intestine is the main reservoir for ExPEC strains causing UTI. Also, the blaCTX-M-15 gene was detected in 48.8% of ESBL producing Enterobacteriaceae including E. coli from patients with UTI in Egypt.34 A study in Riyadh in Saudi Arabia detected CTX-M-15 in 96.7% of ESBL producing E. coli from clinical isolates with the majority of samples from urine.37 This was higher than our results among urinary isolates, which indicates that some of the phenotypically ESBL producing urinary isolates carry other ESBLs gene types.
About 83.5% of ESBL producing urinary isolates identified as O25-ST131 clone, while only 57.7% of intestinal isolates were O25-ST131 positive with a significant difference between the studied groups. This is in agreement with several previous studies that reported a significant association of ESBL production with this clone.4,27,38 A study done in 2015 on the intestinal colonization by ESBL producing enterobacteria in Egyptian patients with liver disease found that all E. coli isolates of the phylogenetic group B2 were belonging to the ST131 clone and were O25-ST131 positive.31 This is similar to our results that all intestinal isolates of group B2 were ESBL producing and positive O25-ST131. However, another study in Egypt in 2017 detected the ST131 clone in 92.3% of ESBL producing E. coli from clinical isolates and 43.8% of them were 025b positive serotype.39
The association between emerging E. coli ST131 clones with CTX-M-15 production was reported in strains isolated from the stool and urine of patients from different countries.40 In this study, 45/57 (78.9%) of urinary and 35/66 (53%) of intestinal CTX-M-15 positive isolates were identified as O25-ST131 clone. Similar results were reported in a study in Bangladesh, where 71% of positive blaCTX-M-15 strains isolated from UTI patients were belonging to the O25-ST131 clone.41 Although the frequency of O25-ST131 was lower in intestinal than urinary study isolates, it was high compared to a previous Jordanian study investigated the intestinal colonization by CTX-M-15 producing the E. coli ST131 clone in infants.42
Regarding antibiotic resistant patterns of the study isolates, most ESBL producing isolates were found to be MDR. Classifying E. coli isolates into different patterns shows that the EC1 pattern (resistant to AMC, CAZ, CRO, SXT, and CIP) was the commonest pattern in both urinary and intestinal isolates. The association of ESBL production with quinolones and Trimethoprim/Sulfamethoxazole resistance can be explained by dissemination of multidrug resistant ST131 clones and by horizontal gene transfer by large plasmids carrying resistance genes to these antibiotic classes.43 More than 80% of isolates resistant to ciprofloxacin were belonging to the ST131 clone and this is similar to other studies that associate between fluoroquinolone resistance and ST131 clone in Saudi Arabia,4 Pakistan,26 and Taiwan.38
The important predisposing factor of this high prevalence of ESBL producing isolates in our country is misuse of antibiotics as there are no policies controlling purchasing these drugs and high self-medication without prescription, especially to third generation cephalosporins, and this led to selective pressure for resistant strains and dissemination of ESBLs producing bacteria.
A limitation of this study was choosing only patients with acute diarrhoea for studying intestinal colonization of the E. coli ST131 clone and ESBL production and not including a healthy group. But it was preferred to compare extraintestinal pathogenic strains of urinary origin with intestinal potentially pathogenic strains in patients with acute diarrhoea.
In conclusion, we detected high prevalence of the O25-ST131 clone among E. coli isolates in Egypt. It was higher in isolates from patients with UTI than those with acute diarrhoea. We provided the first detection of ST131 clone in different phylogenetic groups other than B2 in Egypt. An alarming increase in prevalence of CTX-M-15 ESBL producing E. coli in intestinal and urinary isolates was found. O25-ST131 E. coli clone isolates showed high antibiotic resistance rates, ESBL, and MDR compared to non-ST131 isolates. No isolates of the O16-ST131 clone were detected in both studied groups. We recommend the need to optimize the antibiotic use by establishing antimicrobial stewardship programs and taking these results of studies as guidance in empirical antibiotic selection in treatment. Also, more additional studies are needed to evaluate this emerging clone and risk factors in our country.
Acknowledgments
We thank the health care workers in Minia university hospitals for their cooperation.
Funding Statement
There is no funding to report.
Abbreviations
CFU, colony forming unit; DDST, double-disc synergy test; ESBL, extended-spectrum beta-lactamase; ExPEC, extra-intestinal pathogenic E. coli; HPF, high power field; IPEC, intestinal pathogenic E. coli; MDR, multiple drug resistance; ST131, Escherichia coli sequence type 131; UPEC, uropathogenic E. coli; UTI, urinary tract infection.
Data Sharing Statement
All data generated or analysed during this study are included in this article.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Morales Barroso I, Lopez-Cerero L, Navarro MD, Gutierrez-Gutierrez B, Pascual A, Rodriguez-Bano J. Intestinal colonization due to Escherichia coli ST131: risk factors and prevalence. Antimicrob Resist Infect Control. 2018;7(1):135. doi: 10.1186/s13756-018-0427-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nicolas-Chanoine MH, Bertrand X, Madec JY. Escherichia coli ST131, an intriguing clonal group. Clin Microbiol Rev. 2014;27(3):543–574. doi: 10.1128/CMR.00125-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ronald A. The etiology of urinary tract infection: traditional and emerging pathogens. Am J Med. 2002;113(Suppl 1A):14S–19S. doi: 10.1016/s0002-9343(02)01055-0 [DOI] [PubMed] [Google Scholar]
- 4.Alqasim A, Abu Jaffal A, Alyousef AA. Prevalence and molecular characteristics of sequence type 131 clone among clinical uropathogenic Escherichia coli isolates in Riyadh, Saudi Arabia. Saudi J Biol Sci. 2020;27(1):296–302. doi: 10.1016/j.sjbs.2019.09.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nicolas-Chanoine MH, Blanco J, Leflon-Guibout V, et al. Intercontinental emergence of Escherichia coli clone O25: H4-ST131 producing CTX-M-15. J Antimicrob Chemother. 2008;61(2):273–281. doi: 10.1093/jac/dkm464 [DOI] [PubMed] [Google Scholar]
- 6.Brolund A. Overview of ESBL-producing Enterobacteriaceae from a Nordic perspective. Infect Ecol Epidemiol. 2014;4. doi: 10.3402/iee.v4.24555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Can F, Azap OK, Seref C, Ispir P, Arslan H, Ergonul O. Emerging Escherichia coli O25b/ST131 clone predicts treatment failure in urinary tract infections. Clin Infect Dis. 2015;60(4):523–527. doi: 10.1093/cid/ciu864 [DOI] [PubMed] [Google Scholar]
- 8.Al-Guranie D, Al-Mayahie S. Prevalence of E. coli ST131 among Uropathogenic E. coli isolates from Iraqi patients in Wasit Province, Iraq. Int J Microbiol. 2020;Article ID 8840561:1–9. doi: 10.1155/2020/8840561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clermont O, Bonacorsi S, Bingen E. Rapid and simple determination of the Escherichia coli phylogenetic group. Appl Environ Microbiol. 2000;66(10):4555–4558. doi: 10.1128/aem.66.10.4555-4558.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Belmont-Monroy L, Ribas-Aparicio RM, Navarro-Ocaña A, et al. Characterization of Escherichia coli causing community acquired urinary tract infections in Mexico City. Diagn Microbiol Infect Dis. 2017;87(2):193–195. doi: 10.1016/j.diagmicrobio.2016.11.006 [DOI] [PubMed] [Google Scholar]
- 11.Abdullah AH, Lakshmidevi N. Prevalence of Escherichia coli sequence type 131 (ST131) among extra-intestinal clinical isolates in different phylogenetic groups. Health Sci. 2016;5:90–94. [Google Scholar]
- 12.Nielsen KL, Dynesen P, Larsen P, Frimodt-Møller N. Faecal Escherichia coli from patients with E. coli urinary tract infection and healthy controls who have never had a urinary tract infection. J Med Microbiol. 2014;63(4):582–589. doi: 10.1099/jmm.0.068783-0 [DOI] [PubMed] [Google Scholar]
- 13.CLSI. Performance Standards for Antimicrobial Disk Susceptibility Tests. M012 -A12. Wayne: Clinical and Laboratory Standards Institute; 2015. [Google Scholar]
- 14.Magiorakos AP, Srinivasan A, Carey RB, et al. Multidrug-resistant, extensively drug-resistant and pan drug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. 2012;18(3):268. doi: 10.1111/j.1469-0691.2011.03570.x [DOI] [PubMed] [Google Scholar]
- 15.Jacoby GA, Medeiros AA. More extended-spectrum beta-lactamases. Antimicrob Agents Chemother. 1991;35(9):1697–1704. doi: 10.1128/aac.35.9.1697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Khairy RMM, Fathy ZA, Mahrous DM, Mohamed ES, Abdelrahim SS. Prevalence, phylogeny, and antimicrobial resistance of Escherichia coli pathotypes isolated from children less than 5 years old with community acquired- diarrhea in Upper Egypt. BMC Infect Dis. 2020;20(1):908. doi: 10.1186/s12879-020-05664-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Doumith M, Day MJ, Hope R, Wain J, Woodford N. Improved multiplex PCR strategy for rapid assignment of the four major Escherichia coli phylogenetic groups. J Clin Microbiol. 2012;50(9):3108–3110. doi: 10.1128/JCM.01468-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Paterson DL, Hujer KM, Hujer AM, et al. Extended-spectrum beta-lactamases in Klebsiella pneumoniae bloodstream isolates from seven countries: dominance and widespread prevalence of SHV- and CTX-M-type beta-lactamases. Antimicrob Agents Chemother. 2003;47(11):3554–3560. doi: 10.1128/aac.47.11.3554-3560.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Clermont O, Dhanji H, Upton M, et al. Rapid detection of the O25b-ST131 clone of Escherichia coli encompassing the CTX-M-15-producing strains. J Antimicrob Chemother. 2009;64(2):274–277. doi: 10.1093/jac/dkp194 [DOI] [PubMed] [Google Scholar]
- 20.Johnson JR, Clermont O, Johnston B, et al. Rapid and specific detection, molecular epidemiology, and experimental virule`nce of the O16 subgroup within Escherichia coli sequence Type 131. J Clin Microbiol. 2014;52:1358–1365. doi: 10.1128/JCM.03502-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hefzy EM, Hassuna NA. Fluoroquinolone-resistant sequence Type 131 Subgroups O25b and O16 among extraintestinal Escherichia coli isolates from community-acquired urinary tract infections. Microb Drug Resist. 2017;23(2):224–229. doi: 10.1089/mdr.2016.0040 [DOI] [PubMed] [Google Scholar]
- 22.Hassuna NA, Khairalla AS, Farahat EM, Hammad AM, Abdel-Fattah M. Molecular characterization of Extended-spectrum β lactamase- producing E. coli recovered from community-acquired urinary tract infections in Upper Egypt. Sci Rep. 2020;10(1):2772. doi: 10.1038/s41598-020-59772-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Muller A, Gbaguidi-Haore H, Cholley P, Hocquet D, Sauget M, Bertrand X. Hospital-diagnosed infections with Escherichia coli clonal group ST131 are mostly acquired in the community. Sci Rep. 2021;11(1):5702. doi: 10.1038/s41598-021-85116-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Namaei MH, Yousefi M, Ziaee M, et al. First report of prevalence of CTX-M-15-producing Escherichia coli O25b/ST131 from Iran. Microb Drug Resist. 2017;23(7):879–884. doi: 10.1089/mdr.2016.0272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Demirci M, Ö Ü, Istanbullu Tosun A. Detection of O25b-ST131 clone, CTX-M-1 and CTX-M-15 genes via real-time PCR in Escherichia coli strains in patients with UTIs obtained from a university hospital in Istanbul. J Infect Public Health. 2019;12(5):640–644. doi: 10.1016/j.jiph.2019.02.017 [DOI] [PubMed] [Google Scholar]
- 26.Ali I, Rafaque Z, Ahmed I, et al. Phylogeny, sequence-typing and virulence profile of uropathogenic Escherichia coli (UPEC) strains from Pakistan. BMC Infect Dis. 2019;19:620. doi: 10.1186/s12879-019-4258-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhong YM, Liu WE, Meng Q, Li Y. Escherichia coli O25b-ST131 and O16-ST131 causing urinary tract infection in women in Changsha, China: molecular epidemiology and clinical characteristics. Infect Drug Resist. 2019;12:2693–2702. doi: 10.2147/IDR.S212658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sabaté M, Moreno E, Pérez T, Andreu A, Prats G. Pathogenicity Island markers in commensal and uropathogenic Escherichia coli isolates. Clin Microbiol Infect. 2006;12(9):880–886. doi: 10.1111/j.1469-0691.2006.01461.x [DOI] [PubMed] [Google Scholar]
- 29.Lee JH, Subhadra B, Son YJ, et al. Phylogenetic group distributions, virulence factors and antimicrobial resistance properties of uropathogenic Escherichia coli strains isolated from patients with urinary tract infections in South Korea. Lett Appl Microbiol. 2016;62(1):84–90. doi: 10.1111/lam.12517 [DOI] [PubMed] [Google Scholar]
- 30.Fam N, Leflon-Guibout V, Fouad S, et al. CTX-M-15-producing Escherichia coli clinical isolates in Cairo (Egypt), Including Isolates of clonal complex ST10 and clones ST131, ST73, and ST405 in both community and hospital settings. Microb Drug Resist. 2011;17(1):67–73. doi: 10.1089/mdr.2010.0063 [DOI] [PubMed] [Google Scholar]
- 31.Fam NS, Defasque S, Bert F, et al. Faecal carriage of extended-spectrum β-lactamase (ESBL)-producing enterobacteria in liver disease patients from two hospitals in Egypt and France: a comparative epidemiological study. Epidemiol Infect. 2015;143(6):1247–1255. doi: 10.1017/S0950268814001812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Alghoribi MF, Gibreel TM, Farnham G, Al Johani SM, Balkhy HH, Upton M. Antibiotic-resistant ST38, ST131 and ST405 strains are the leading uropathogenic Escherichia coli clones in Riyadh, Saudi Arabia. J Antimicrob Chemother. 2015;70(10):2757–2762. doi: 10.1093/jac/dkv188 [DOI] [PubMed] [Google Scholar]
- 33.Nairoukh YR, Mahafzah AM, Irshaid A, Shehabi AA. Molecular characterization of multidrug resistant uropathogenic E. Coli isolates from Jordanian patients. Open Microbiol J. 2018;12:1–7. doi: 10.2174/1874285801812010001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mohamed ES, Khairy RMM, Abdelrahim SS. Prevalence and molecular characteristics of ESBL and AmpC β -lactamase producing Enterobacteriaceae strains isolated from UTIs in Egypt. Antimicrob Resist Infect Control. 2020;9:198. doi: 10.1186/s13756-020-00856-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Abdel-Moaty MM, Mohamed WS, Abdel-All SM, El-Hendawy HH. Prevalence and molecular epidemiology of extended spectrum β-lactamase producing Escherichia coli from hospital and community settings in Egypt. J App Pharm Sci. 2016;6(01):042–047. doi: 10.7324/JAPS.2016.600107 [DOI] [Google Scholar]
- 36.Valverde A, Turrientes MC, Norman F, et al. CTX-M-15-non-ST131 Escherichia coli isolates are mainly responsible of faecal carriage with ESBL-producing Enterobacteriaceae in travellers, immigrants and those visiting friends and relatives. Clin Microbiol Infect. 2015;21(3):252.e1–252.e2524. doi: 10.1016/j.cmi.2014.09.021 [DOI] [PubMed] [Google Scholar]
- 37.Al-Agamy MH, Shibl AM, Hafez MM, Al-Ahdal MN, Memish ZA, Khubnani H. Molecular characteristics of extended-spectrum β-lactamase-producing Escherichia coli in Riyadh: emergence of CTX-M-15-producing E. coli ST131. Ann Clin Microbiol Antimicrob. 2014;13:4. doi: 10.1186/1476-0711-13-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang IF, Lee WY, Wang JL, et al. Fecal carriage of multidrug-resistant Escherichia coli by community children in southern Taiwan. BMC Gastroenterol. 2018;18(1):86. doi: 10.1186/s12876-018-0807-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.El-Badawy MF, Tawakol WM, Maghrabi IA, Mansy MS, Shohayeb MM, Ashour MS. Iodometric and molecular detection of ESBL production among clinical isolates of E. coli Fingerprinted by ERIC-PCR: the first egyptian report declares the emergence of E. coli O25b-ST131clone Harboring blaGES. Microb Drug Resist. 2017;23(6):703–717. doi: 10.1089/mdr.2016.0181 [DOI] [PubMed] [Google Scholar]
- 40.Dandachi I, Chaddad A, Hanna J, Matta J, Daoud Z. Understanding the epidemiology of multi-drug resistant gram-negative Bacilli in the middle east using a one health approach. Front Microbiol. 2019;10:1941. doi: 10.3389/fmicb.2019.01941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Begum N, Shamsuzzaman SM. Emergence of CTX-M-15 producing E. coli O25b-ST131 clone in a tertiary care hospital of Bangladesh. Malays J Pathol. 2016;38(3):241–249. [PubMed] [Google Scholar]
- 42.Badran EF, Din RAQ, Shehabi AA. Low intestinal colonization of Escherichia coli clone ST131 producing CTX-M-15 in Jordanian infants. J Med Microbiol. 2016;65(2):137–141. doi: 10.1099/jmm.0.000210 [DOI] [PubMed] [Google Scholar]
- 43.Bennett PM. Plasmid encoded antibiotic resistance: acquisition and transfer of antibiotic resistance genes in bacteria. Br J Pharmacol. 2008;153(1):S347–S357. doi: 10.1038/sj.bjp.0707607 [DOI] [PMC free article] [PubMed] [Google Scholar]