Abstract
The osteogenic effect of a composite electrospun core-shell nanofiber membrane encapsulated with Emdogain® (EMD) was evaluated. The membrane was developed through coaxial electrospinning using polycaprolactone as the shell and polyethylene glycol as the core. The effects of the membrane on the osteogenic differentiation of periodontal ligament stem cells (PDLSCs) were examined using Alizarin Red S staining and qRT-PCR. Characterization of the nanofiber membrane demonstrated core-shell morphology with a mean diameter of ~1µm. Examination of the release of fluorescein isothiocyanate-conjugated bovine serum albumin (FITC-BSA) from core-shell nanofibers over a 22-day period showed improved release profile of encapsulated proteins as compared to solid nanofibers. When cultured on EMD-containing core-shell nanofibers, PDLSCs showed significantly improved osteogenic differentiation with increased Alizarin Red S staining and enhanced osteogenic gene expression, namely OCN, RUNX2, ALP, and OPN. Core-shell nanofiber membranes may improve outcomes in periodontal regenerative therapy through simultaneous mechanical barrier and controlled drug delivery function.
Keywords: Guided tissue regeneration, Bone regeneration, Enamel matrix proteins, Periodontal regeneration, Nanofibers
1. INTRODUCTION
Periodontal diseases encompass those pathologic processes affecting the periodontium, including gingivitis and periodontitis. The public health concern is staggering, with the most recent NHANES data reporting that nearly half (46%) of American adults over the age of 30 have some form of periodontitis.1 This is an infectious disease caused by a primarily Gram-negative bacterial biofilm, which in turn results in an inflammatory response mediated by cytokines and molecules such as IL-1β, IL-6, PGE2, TNF-α, RANKL, and MMPs.2 Clinically, this manifests as progressive attachment and bone loss, associated with the presence of bacterial plaque and calculus.3
The goals of periodontal therapy are to reduce and/or eliminate inflammation caused by bacterial plaque, correct defects caused by periodontal disease, and to regenerate periodontal tissues. After periodontal therapy, such as scaling and root planing or osseous surgery, healing occurs via repair, typically in the form of a long junctional epithelium or connective tissue adhesion/attachment.4,5 In contrast, healing by regeneration results in complete restoration of the lost periodontium.6 Effective regeneration requires a proper combination of signaling molecules, progenitor/stem cells, and/or extracellular matrix mimicking scaffolds.7
Signaling molecules include proteins that stimulate the differentiation of stem cells, such as those found in the periodontal ligament (PDL). One such group of proteins are the enamel matrix proteins, which include amelogenin (90%), ameloblastin, enamelin, tuft protein, proteases, and albumin.8 Enamel matrix proteins are commercially available as Emdogain® (Straumann, Basel, Switzerland), which contains enamel matrix derivative (EMD) from porcine tooth buds in a propylene glycol alginate carrier. Its mechanism of action is a form of biological mimicry to stimulate periodontal regeneration, since enamel matrix proteins play a pivotal role in the events of root development that lead to cementogenesis and subsequent bone formation.8 Other beneficial effects of EMD include anti-inflammatory effects, with decreased production of IL-1β and IL-8, diminished osteoclastic activity by interfering with the OPG/RANKL balance, improved angiogenesis, and anti-bacterial activity.8–11 Histologic evidence has shown that treatment with EMD can result in periodontal regeneration, but not in 100% of cases.12 Nevertheless, several clinical studies using EMD have shown significant results in probing depth reduction, clinical attachment level gain, and bone fill.13–15
Guided tissue regeneration aims to restore the lost bone and periodontium in areas of attachment loss by excluding epithelium and gingival connective tissue from the root surface, allowing cells from the periodontal ligament (PDL) and bone to repopulate the space adjacent to the root. This is achieved by using a barrier membrane over the bony defect, such as non-resorbable polytetrafluoroethylene (PTFE) or resorbable synthetic and collagen-based membranes. Limitations for currently available membranes, however, do exist. Non-resorbable membranes provide mechanical strength but require an additional surgery for removal. Resorbable collagen-based membranes are biocompatible but have insufficient mechanical properties and unpredictable degradation profiles.7 Moreover, these membranes do not offer drug delivery function or antibacterial properties, and complications such as microbial colonization and post-operative infection may occur upon premature membrane exposure.7,16
Electrospinning has emerged as one of the simplest, most cost-effective methods to produce fibrous membranes that closely mimic the extracellular matrix of native tissue.17–19 Endowed with topographical and biological signals, electrospun nanofiber membranes have potential use in guided tissue regeneration to provide an optimal microenvironment for cell differentiation and proliferation.20,21 Electrospinning uses electrostatic forces to draw a charged polymer solution into a mesh of fibers in nanometer dimensions. The conventional type of electrospinning utilizes a single polymer, while newer methods can create fibers with multiple components.21 One such technique is coaxial electrospinning, which produces “core-shell” fibers made of an inner “core” containing a drug, growth factor, and/or protein, and an outer “shell” polymer which provides mechanical properties.22,23 These core-shell nanofibers allow for direct incorporation of biologically active molecules into the core component, enabling controlled delivery of a desired drug or protein.22,23
The goal of our current study is to develop a multifunctional core-shell electrospun nanofiber membrane capable of delivering encapsulated EMD to periodontal ligament stem cells (PDLSCs). We hypothesize that this core-shell nanofiber membrane could be used for periodontal regeneration to provide a mechanical barrier for epithelial downgrowth, and furthermore act as a delivery vehicle for biologically active molecules (such as EMD) to induce stem cell differentiation and bone formation in periodontal defects.
2. MATERIALS AND METHODS
2.1. Materials.
Polycaprolactone (PCL) (80 kDa MW), fluorescein isothiocyanate-conjugated bovine serum albumin (FITC-BSA), polyethylene glycol (PEG) (35 kDa MW), ascorbic acid (AA), β-glycerophosphate (BGP), and Alizarin Red S were purchased from Sigma Aldrich (St. Louis, MO, USA). Solvent 1,1,1,3,3,3-hexafluoro-2-isopropanol (HFIP) was purchased from Oakwood Chemical (Estill, SC, USA). Emdogain® (enamel matrix derivative, EMD) (30 mg/mL) was obtained from Straumann (Basel, Switzerland).
2.2. Coaxial Electrospinning.
The electrospinning is based on previously published methodology from our group.23 The shell fluid consisted of PCL (12% w/v) dissolved in HFIP. The core fluid was prepared with 1 mg of FITC-BSA and 100 mg of PEG (10% w/v) dissolved in 1 mL of deionized (DI) water. Core-shell nanofibers were electrospun using a coaxial needle and a 10-kV voltage applied between the needle and a grounded copper collecting sheet, separated by 11 cm. Electrospinning was performed at room temperature with a relative humidity of 40% using shell/core flow rates of 2.0/0.2, 2.0/0.4, 4.0/0.2, and 4.0/0.4 mL/h. For EMD-releasing nanofibers, the core fluid was prepared by dissolving 0.3 mL of EMD and 50 mg of PEG in 1 mL of DI water, and a shell/core flow rate of 4.0/0.4 mL/h was used.
2.3. Characterization of Core-Shell Nanofibers.
Core-shell nanofibers were imaged via Scanning Electron Microscopy (SEM), Transmission Electron Microscopy (TEM) and epifluorescent microscopy. For cross-sectional imaging, the membranes were freeze-fractured in liquid nitrogen and oriented to have broken ends perpendicular to the SEM sample stub.
2.4. Release from Core-Shell Nanofibers.
Solid nanofibers were fabricated via conventional electrospinning using a solution of PCL and FITC-BSA. Negative controls included solid PCL nanofibers and PCL/PEG core-shell nanofibers. The nanofibers were placed into wells of a 24-well plate, to which PBS (1 mL) was added and then incubated at 4°C. At designated time intervals, PBS was extracted and replenished. The PBS extracts were placed in a 48-well plate and analyzed via a microplate reader (Synergy Mx, Biotek, Winooski, VT, USA) along with blank and standard solutions of FITC-BSA, ranging from 0–25,000 ng/mL. Fluorescence intensity was quantified according to the fluorescence emission maximum (528nm for FITC-BSA). Using the resulting linear calibration curve, FITC-BSA concentrations were calculated at each time interval. At the end of the release period, the nanofibers were dissolved in a 1:1 HFIP:DI-water solution, and fluorescence intensity was calculated. Accumulation of release was determined and divided against the sum of the total amount of FITC-BSA in the nanofibers (i.e. the sum of FITC-BSA released and remaining FITC-BSA after dissolving the nanofibers).
2.5. PDLSC Isolation and Culture.
Collection of teeth for use in this study was approved by the Research Subjects Review Board of the University of Rochester (Rochester, NY, USA). Extracted teeth with healthy periodontal tissues were placed in Dulbecco’s Modified Eagle’s Medium (DMEM) containing 15% fetal bovine serum (FBS) and 1% penicillin/streptomycin (P/S). The PDL tissues were removed and incubated in cell medium in a humidified atmosphere of 5% CO2 at 37°C. The cells were allowed to migrate from the PDL tissues for 1 week, then filtered using a 70 µm cell strainer (BD Falcon, USA) and seeded onto a new culture dish with cell media change every 2 days. The cells were expanded to passage 5.
2.6. PDLSC Characterization.
PDLSCs from passage 3 were delivered to the Pryhuber Lab (University of Rochester, Rochester, NY, USA) for antibody staining and flow cytometric analysis. The following antibodies and fluorochromes were used and purchased from BD Biosciences (San Jose, CA, USA) unless otherwise specified: CD105(BV 605), CD146(BV 711), CD44(BV 786), CD34(BV 510, BioLegend, San Diego, CA, USA), CD29(AF 700, BioLegend, San Diego, CA, USA), CD73(PE-CF594), CD45(PE-Cy5), CD90(Pe-Cy7), Ki67(FITC), and 7 AAD (Live/Dead, Invitrogen). CD31 (PE), CD45 (V450), CD14 (BV 510) and CD19 (BV510) were used as exclusion markers. The cells were first washed in Dulbecco’s PBS (DPBS) and stained with 7 AAD for 10 minutes at room temperature. The cells were then washed with DPBS with 2% BSA and blocked with 4% normal mouse serum for 10 minutes at 4°C. Next, the cells were stained for 90 minutes at 4°C in a cocktail containing anti-CD105, CD31, CD146, CD44, CD34, CD29, CD73, CD45, CD14, CD19 and CD90. The stained cells were resuspended and passed through a 100 μm filter and analyzed on an LSR II flow cytometer (BD Biosciences, San Jose, CA, USA), along with compensation beads (Bangs Laboratories, Fishers, IN, USA). Gating was applied to isolate single cells and to eliminate dead cells. The cells were further gated to eliminate CD19, CD14, CD31 or CD45 positive cells. Once these gates were applied, cells were examined for the stem cell markers of interest, namely CD105, CD90, CD146, and CD44.
2.7. PDLSC Culture with EMD.
12-well plates were coated overnight with 0.05 mL of EMD per well. Human PDLSCs from passage 3 were seeded at a density of 1.5 × 106 cells/well and incubated in a humidified atmosphere of 5% CO2 at 37°C with cell media change every 2–3 days. Appropriate test and control groups were used, including cells seeded with or without EMD and/or osteogenic enhancers. Osteogenic differentiation medium consisting of 15% FBS, 1% P/S, 50 µg/mL AA, and 10 mM BGP in DMEM was added at 5 days after initial culture.
2.8. Evaluation of PDLSC Differentiation in the Presence of EMD.
Alizarin Red S staining was done 21 days after addition of osteogenic enhancers by dissolving 2 g Alizarin Red S in 100 mL DI-water and adjusted to pH 4.1–4.3 using 19% hydrochloric acid. The cells were rinsed with PBS, fixed with 10% formalin (1 mL), and rinsed with DI-water (1 mL). The Alizarin Red S solution (1 mL) was added to each well to cover the cellular monolayer and incubated at room temperature for 30 minutes.
2.9. Cell Seeding onto Core-Shell Nanofibers.
The nanofiber membranes were sterilized with 75% ethanol and UV irradiation. The membranes were then rinsed in sterile PBS three times, soaked in medium, and incubated in a humidified atmosphere of 5% CO2 at 37°C overnight prior to cell seeding. Human PDLSCs from passage 5 were seeded onto the nanofiber membranes at a density of 1.2 × 105 cells/well. The well plates were then incubated in a humidified atmosphere of 5% CO2 at 37°C. Cell media change was done every 2–3 days. Osteogenic differentiation medium in DMEM was added at 5 days after initial culture. In the experimental group, the PCL/PEG-EMD nanofiber membranes were placed on the well plate onto which the PDLSCs were seeded. PCL/PEG nanofiber membranes were used as controls.
2.10. Evaluation of PDLSC Differentiation on Core-Shell Nanofibers.
At day 19 after initial cell culture in osteogenic medium, Alizarin Red S staining and osteogenic gene expression via total RNA isolation and qRT-PCR analyses were determined. The primer sequences for osteogenic marker genes osteocalcin (OCN), runt related transcription factor 2 (RUNX2), alkaline phosphatase (ALP), and osteopontin (OPN) are listed in Table 1. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) and beta actin (H-ACTIN) were used as controls. Three samples were collected from each group, which consisted of PDLSCs cultured on the control or test membranes in the presence of osteogenic enhancers. Total RNA was isolated with the RNeasy plus mini-kit (Qiagen) according to the manufacturer’s recommendations. RNA purity and quantity were determined with NanoDrop1000 (ThermoFisher, Waltham, MA, USA) and quality was assessed using Bioanalyzer 2100 (Agilent Technologies, Santa Clara, CA, USA). RNA samples were reverse transcribed into cDNA using the High Capacity Reverse Transcription kit (ThermoFisher, Waltham, MA, USA) following manufacturer’s recommendations. cDNA from three samples of each group were pooled and used for qRT-PCR analyses. qRT-PCR was performed in triplicate for each sample with QuantStudio 12K FLEX (ThermoFisher, Waltham, MA, USA) with PowerSYBR Green Master Mix reagent (ThermoFisher, Waltham, MA, USA). Comparative ΔΔCt was performed to determine relative mRNA expression, where relative abundance of each gene was first internally normalized to the mean expression of GAPDH and H-ACTIN.
Table 1.
Primer sequences for qRT-PCR analyses of gene expression in PDLSCs cultured on electrospun core-shell nanofibers.
| Gene | Full Name | Primer | Sequence |
|---|---|---|---|
| GAPDH | Glyceraldehyde phosphate dehydrogenase | Forward | 5'-TGCCACCCAGAAGACCGT-3′ |
| Reverse | 5'-GGATGACCTTGCCCACAGC-3′ | ||
| H-ACTIN | Beta actin | Forward | 5'-AGAGATGGCCACGGCTGCTT-3′ |
| Reverse | 5'-ATTTGCGGTGGACGATGGAG-3′ | ||
| OCN | Osteocalcin | Forward | 5′-CCCAGGCGCTACCTGTATCAA-3′ |
| Reverse | 5′-GGTCAGCCAACTCGTCACAGTC-3′ | ||
| RUNX2 | Runt-related transcription factor 2 | Forward | 5'-CACTGGCGCTGCAACAAGA-3′ |
| Reverse | 5'-CATTCCGGAGCTCAGCAGAATAA-3′ | ||
| ALP | Alkaline phosphatase | Forward | 5'-CCTGGACCTCGTTGACACCT-3′ |
| Reverse | 5'-GTCCCCTGGCTCGAAGAGA-3′ | ||
| OPN | Osteopontin | Forward | 5'-CGAAGTAAGTCCAACGAAAG-3′ |
| Reverse | 5'-GGTGATGTCCTCGTCTGTA-3′ |
2.11. Statistical Analyses.
All measurements are expressed as mean ± standard deviation. Statistical analyses for the fiber diameter and release data were conducted using a one-way and two-way Analysis of Variance (ANOVA), respectively, with Bonferroni post-tests to compare groups. Statistical analyses for the qPCR data were conducted using a two-way sample t-test. A p value <0.05 was considered statistically significant.
3. RESULTS
To obtain core-shell nanofiber membranes, PEG and PCL solutions were used to produce electrospun nanofibers with a core-shell structure. Based on prior published results, 10% PEG (w/v, core) and 12% PCL (w/v, shell) were optimal concentrations for generating core-shell nanofibers with desired uniformity and reproducibility (Figure 1).23 The effects of the flow rates on fiber diameter have also been previously published by our group.23 Analyses using SEM showed bead-free fibers with a hollow structure upon freeze-fracturing (Figure 1C and D). TEM further demonstrated the core-shell structure in about 60% of the spun nanofibers (Figure 1E). When encapsulated with FITC-BSA, green fluorescence could be observed in the inner core of the nanofibers (Figure 1F), demonstrating the successful encapsulation of the protein within the nanofibers. While the 4.0/0.2 mL/h shell/core flow rate combination produced fibers that were slightly larger than those produced by the other three flow rates, the diameters of all core-shell fibers were similar, ranging from 0.5~1.5 µm (Figure 2).
Figure 1.
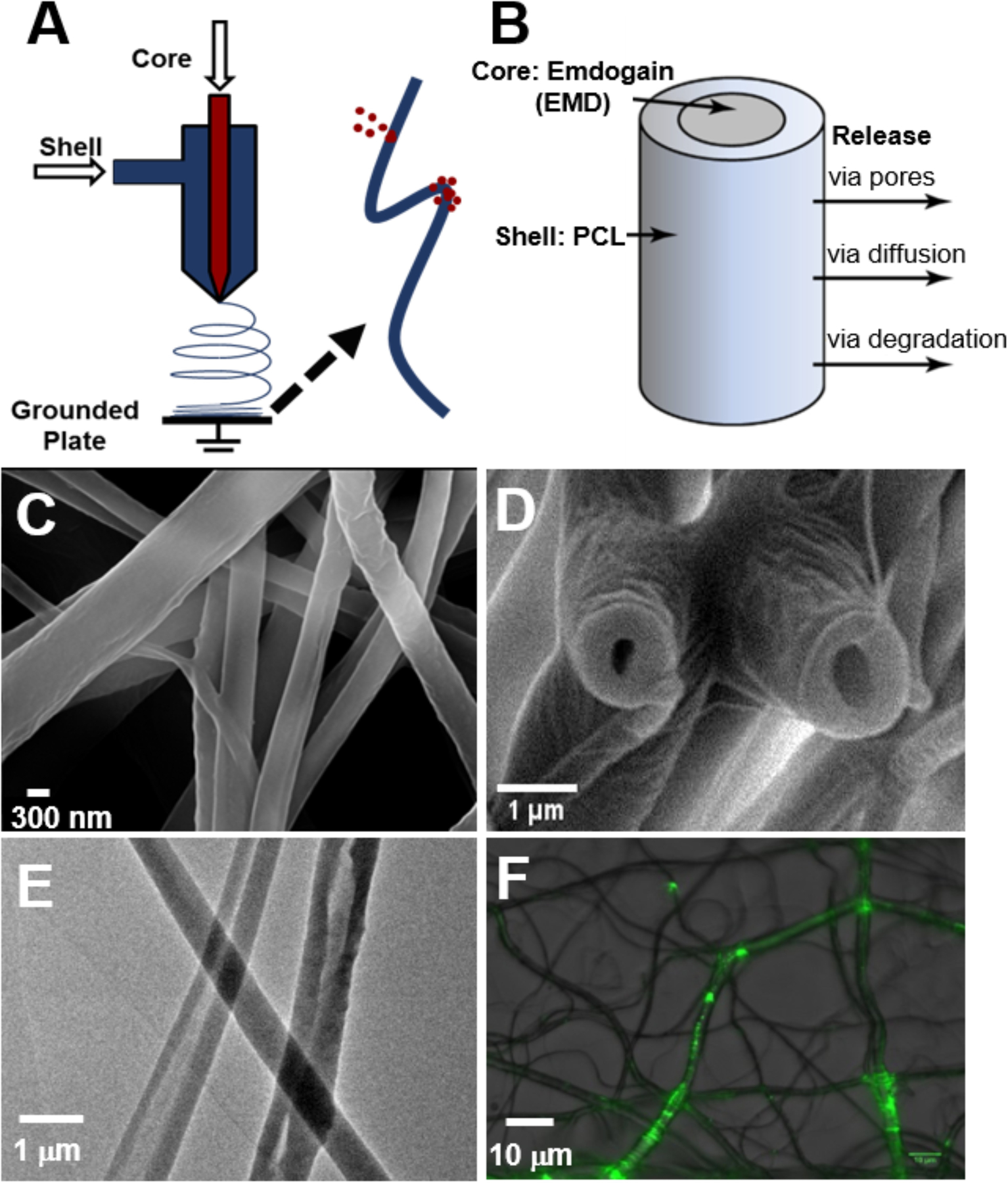
(A) Schematic illustration of coaxial electrospinning. (B) Schematic representation of a core-shell nanofiber. (C) SEM images of core-shell nanofibers spun at 2.0/0.4 mL/h (shell/core flow rate). (D) SEM cross-section image of core-shell nanofibers upon freeze-fracturing. (E) TEM image shows hollow structure and core-shell composition of nanofibers. (F) Epifluorescent images show encapsulated FITC-BSA in nanofibers.
Figure 2.
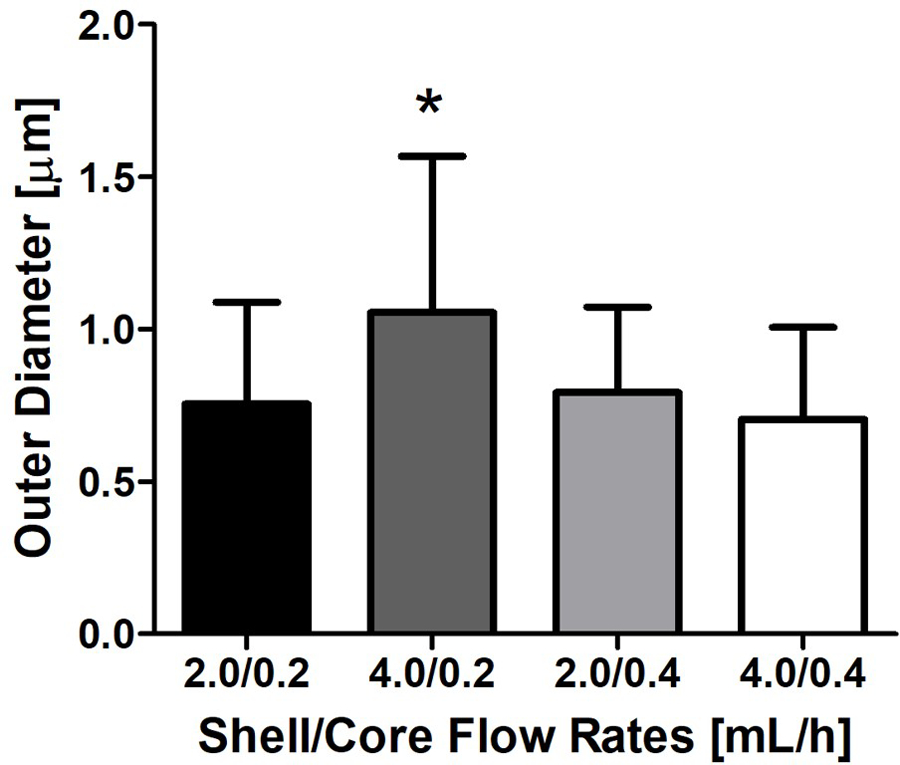
Outer diameter of core-shell nanofibers at varying flow rates. * indicates p<0.05.
To determine the payload and the kinetics of the macromolecular release from the core-shell nanofibers, FITC-BSA was encapsulated in nanofibers during coaxial electrospinning. FITC-BSA encapsulation in core-shell fibers has been previously performed by several groups in order to establish release rates and mechanisms.24–27 FITC-BSA consists of FITC covalently conjugated to BSA protein, which allows for detection and subsequent quantification of the release of BSA from the core-shell nanofibers. The release curves for FITC-BSA from the core-shell nanofibers were found to be significantly different from those of the solid counterpart (Figure 3A, p<0.05). The solid nanofibers showed a burst release of FITC-BSA within the first 12 hours, with 90% of the payload released by 22 days. By contrast, all four groups of core-shell nanofibers showed a delayed release at early time points and a sustained release (i.e., a non-zero slope) over weeks, such that only about 40–60% of the payload was released after 22 days. When comparing the loading capacity for nanofibers corresponding to the four flow rates indicated above, the group of 2.0/0.4 mL/h shell/core flow rates showed a higher level of FITC-BSA encapsulation than all the other groups (Figure 3B, p<0.05). The negative control based on solid PCL nanofibers and negative control based on PCL/PEG core-shell nanofibers both showed no detectable fluorescence.
Figure 3.
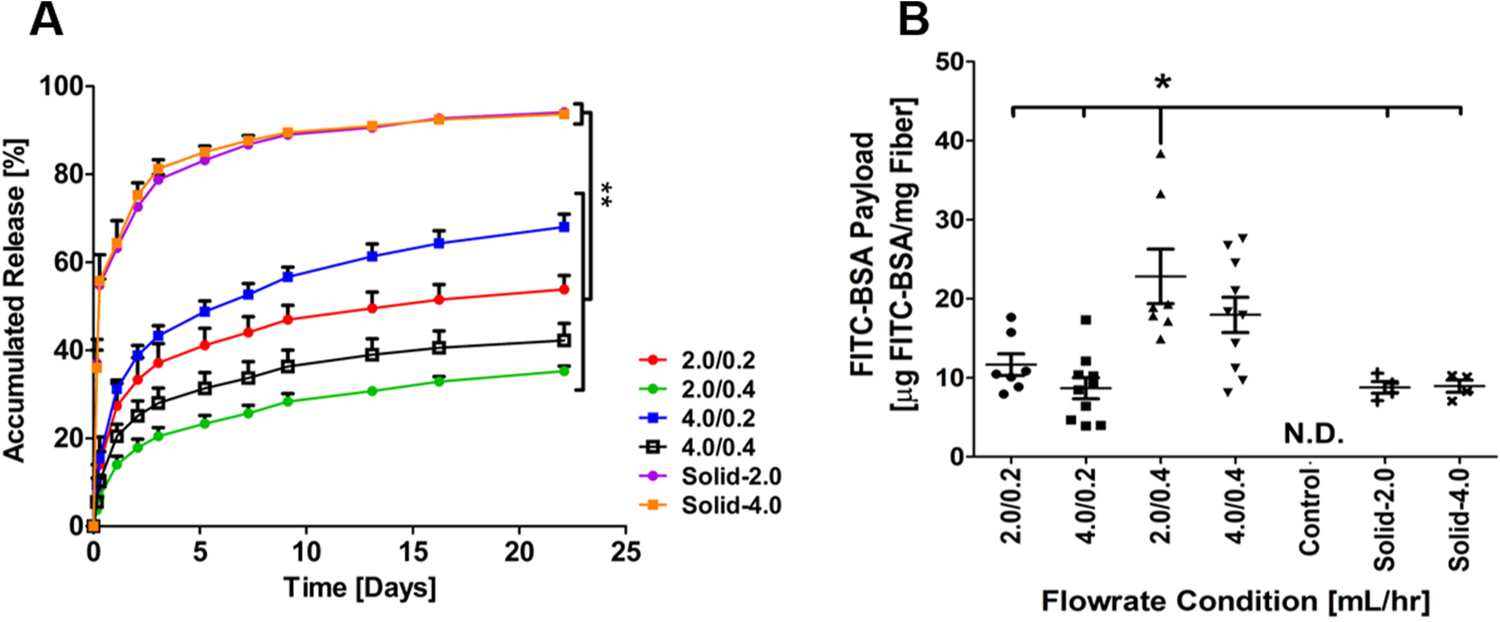
Payload and release of FITC-BSA from core-shell nanofibers. (A) The accumulated release from both core-shell and solid nanofibers spun with FITC-BSA over 22 days at 37°C in PBS (n=3 for solid groups and n=4 for core-shell groups). (B) Conventional and coaxial electrospinning nanofiber payload (N.D. = Not Detectable). * indicates p<0.05.
To further characterize the release kinetics of FITC-BSA from core-shell nanofibers, the Ritger-Peppas model was used to analyze the release mechanisms from various fibers.27–29 The FITC-BSA in core-shell nanofibers produced by the four different flow rate combinations showed significantly slower release during the first few hours followed by a more sustained phase of release that lasted for more than 20 days. When applying the Peppas equation of fit, core-shell nanofiber membrane release exponents (n) from all four groups were close to 0.45 (p>0.05; R2>0.9), indicating Fickian diffusion via cylinders.30 In contrast, the release exponents were much smaller than 0.45(~0.25) from FITC-BSA-encapsulating solid nanofibers, indicating non-Fickian or quasi-Fickian diffusion (Table 2).
Table 2.
FITC-BSA release kinetic analyses.
| Sample | Flow Rate | Peppas Model | ||
|---|---|---|---|---|
| KP | n | R2 | ||
| FITC-BSA | Solid-2.0 | 0.290 ± 0.053 | 0.251 ± 0.052 | 0.92 |
| Solid-4.0 | 0.278 ± 0.108 | 0.271 ± 0.086 | 0.88 | |
| 2.0/0.2 | 0.037 ± 0.013 | 0.440 ± 0.036 | 0.93 | |
| 4.0/0.2 | 0.065 ± 0.011 | 0.424 ± 0.036 | 0.94 | |
| 2.0/0.4 | 0.027 ± 0.004 | 0.438 ± 0.022 | 0.96 | |
| 4.0/0.4 | 0.038 ± 0.005 | 0.407 ± 0.023 | 0.95 | |
The EMD-containing nanofiber membrane was subsequently made with a goal to deliver adequate EMD initially with sustained long-term release to maximize its efficacy. Accordingly, a shell/core flow rate of 4.0/0.4 mL/h was chosen based off the release profile of FITC-BSA, where it demonstrated greater initial and overall release of FITC-BSA as compared to the 2.0/0.4 mL/h shell/core flow rate, while also providing greater encapsulation as compared to the remaining groups.
The EMD-containing nanofiber membranes were used to culture PDLSCs isolated from extracted human teeth. Flow cytometry analysis was used to establish the purity and identity of the primary cells isolated from extracted human teeth using a panel of known stem cell markers.31,32 As shown (Figure 4), isolated and culture expanded cells were mostly negative for hematopoietic and endothelial cell markers CD45 and CD31 (99.8%), respectively. The cells were also negative for CD14 and CD19, demonstrating lack of monocyte and/or B cell contamination. Among CD31−CD45− cells, 98.6% of cells were CD105+CD90+, and 95.1% of were CD146+CD44+, suggesting that greater than 95% of the culture-expanded PDL cells expressed markers of human mesenchymal stem cells.31,32
Figure 4.
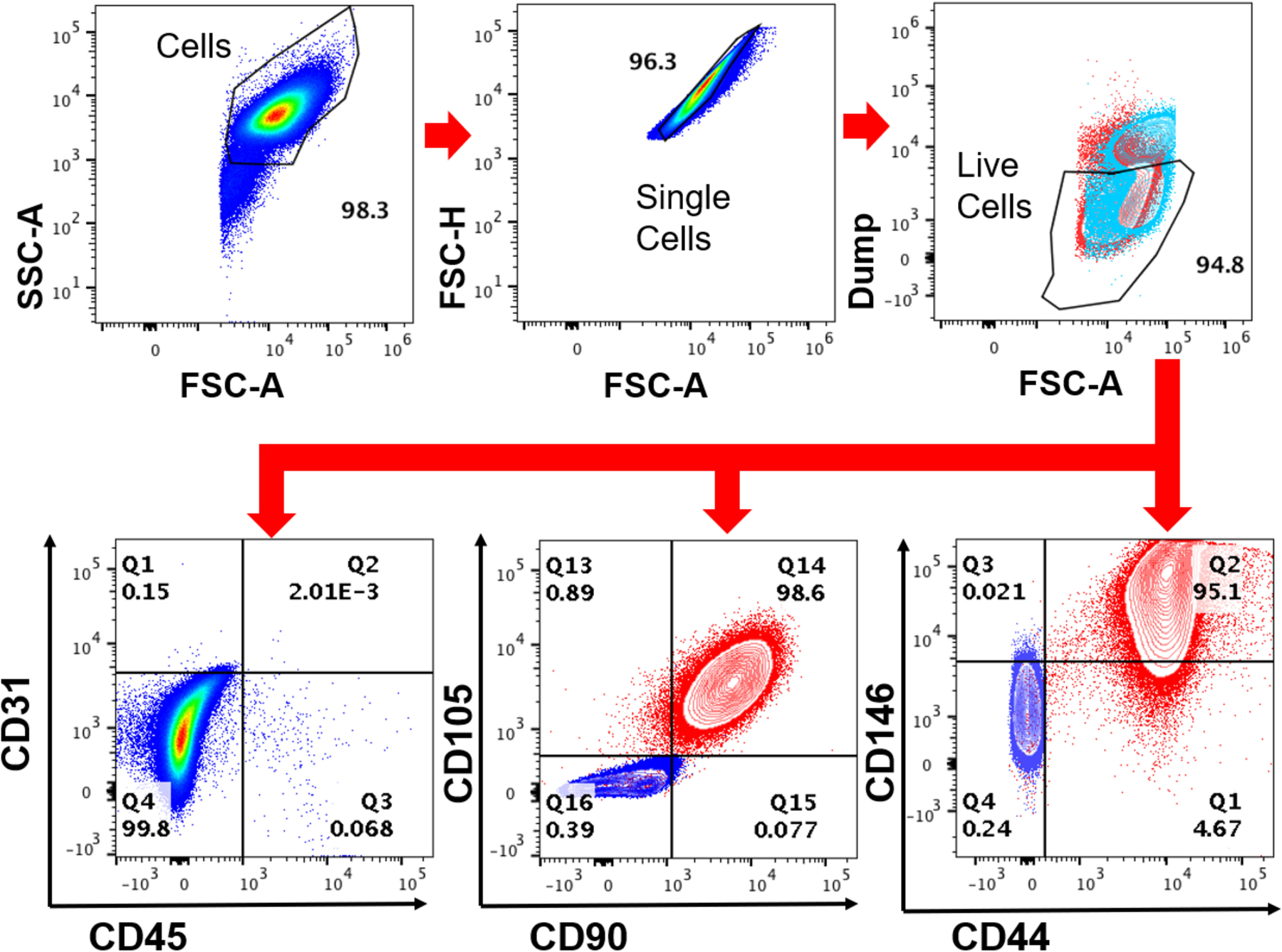
Phenotyping PDLSCs by flow cytometry analysis. PDLSCs were isolated and expanded as described. Flow cytometry analysis showed that the PDLSCs were predominantly negative for hematopoietic and endothelial markers CD45 and CD31 but strongly positive for mesenchymal markers CD105, CD90, CD146, and CD44.
Alizarin Red S staining was used to determine the effects of EMD-releasing nanofibers on PDLSC differentiation. EMD had a slight positive effect on PDLSCs in culture with normal medium in the absence of osteogenic enhancers (Figure 5A and C). Addition of EMD in osteogenic medium strongly induced osteogenic differentiation and mineralization of the PDLSCs (Figure 5B and D). On EMD-containing nanofibers, Alizarin Red S staining showed greater mineralization as compared to controls in both media, with and without osteogenic enhancers (Figure 5E–H). Further quantitative qRT-PCR results showed a significant difference (p<0.05) in gene expression of osteogenic markers OCN, RUNX2, ALP, and OPN (Figure 6). Upregulation of all osteogenic marker genes of interest were noted in the PDLSCs seeded onto EMD-containing membranes as compared to the control group.
Figure 5.
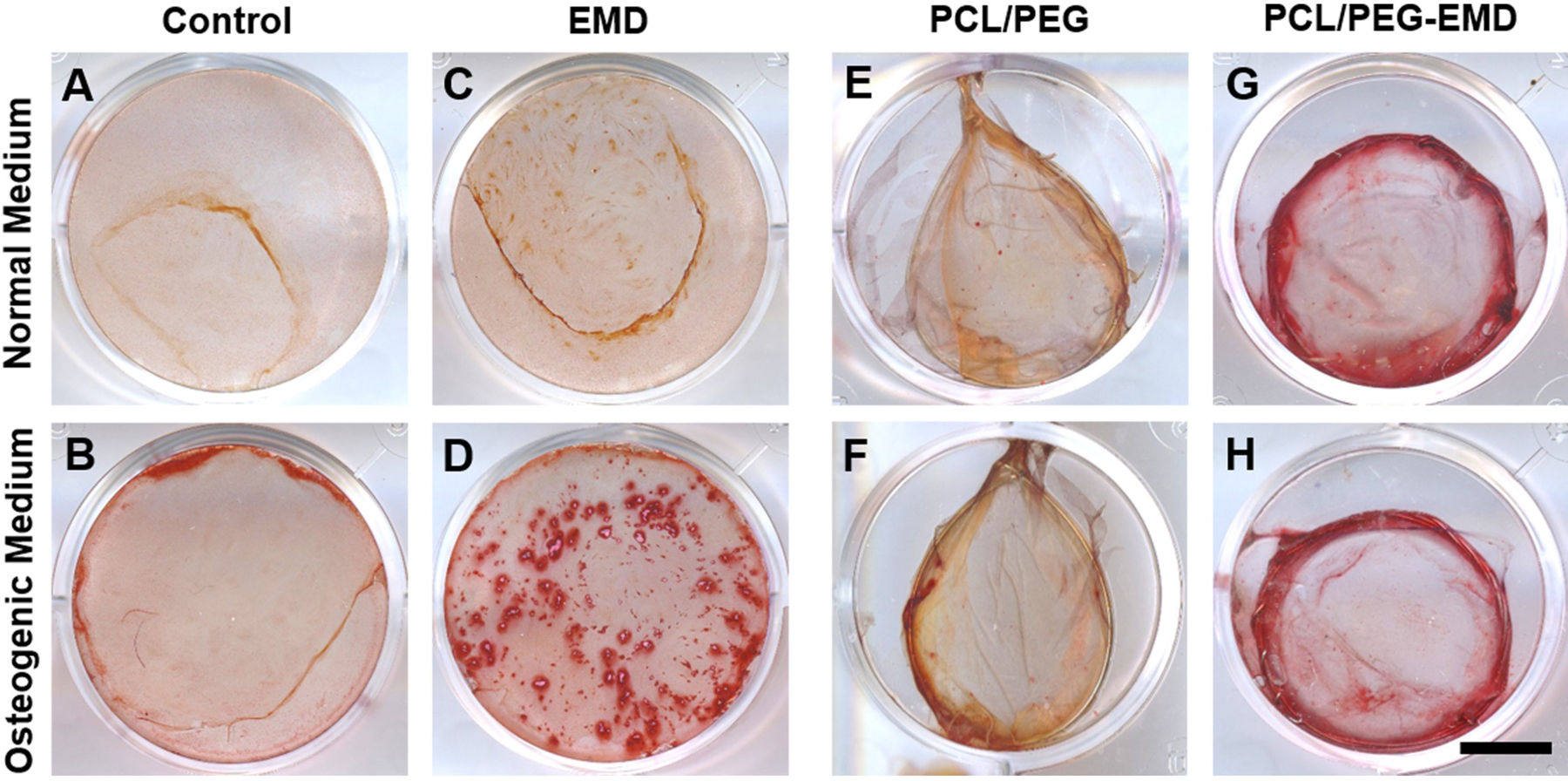
Alizarin Red S staining in PDLSC culture on a plastic surface (A–D) and directly on nanofibers (E–H). EMD was coated on a plastic surface (C, D) or encapsulated into nanofibers (G, H). PDLSC mineralization is shown in cultures with normal medium (A, C, E, G) or with osteogenic differentiation medium (B, D, F, H). Scale bar = 10 mm.
Figure 6.
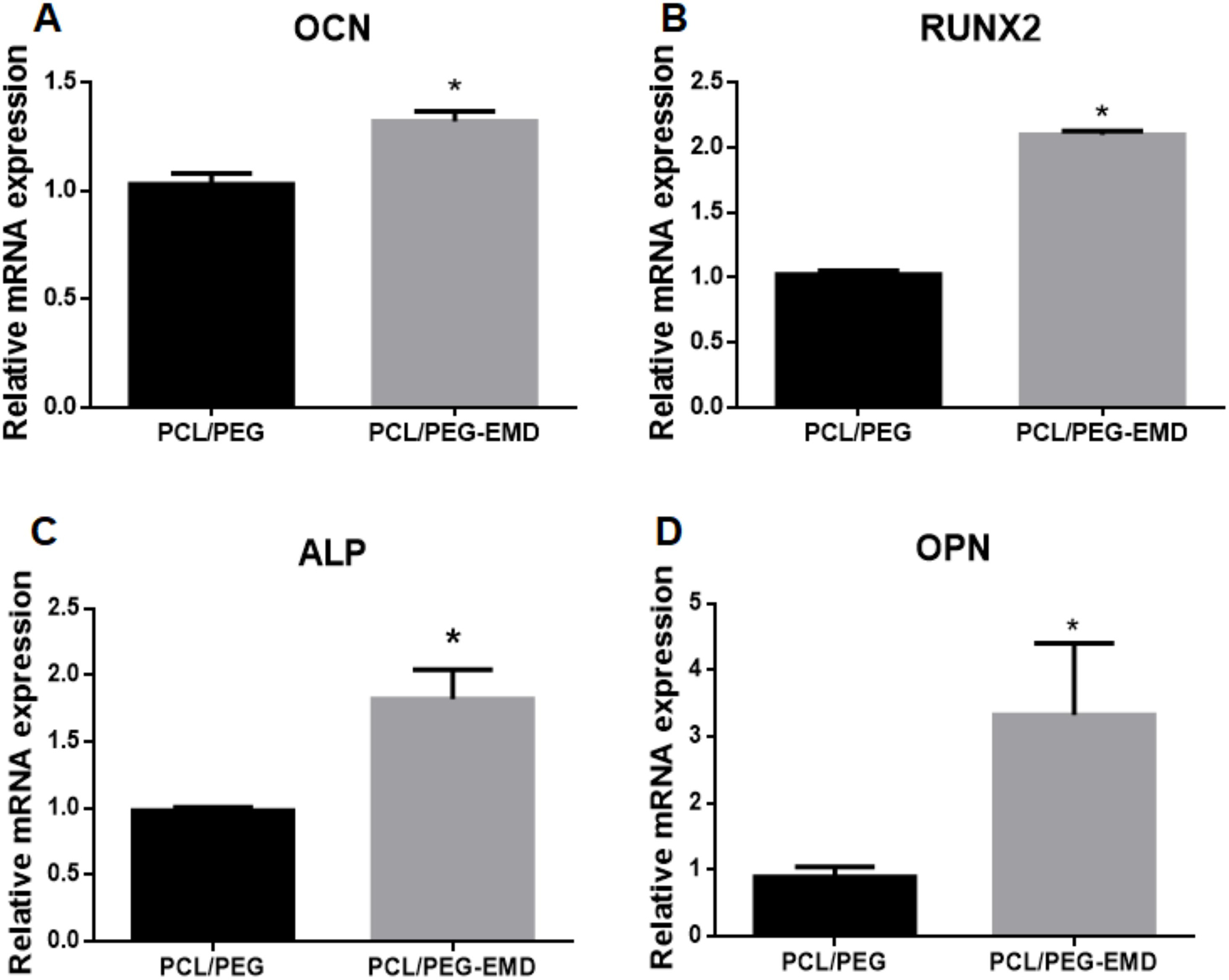
qRT-PCR analyses of gene expression in PDLSCs cultured on the control (PCL/PEG) and EMD-containing (PCL/PEG-EMD) nanofibers. Relative mRNA expression of osteogenic markers OCN, RUNX2, ALP, and OPN are shown. * indicates p<0.05.
4. DISCUSSION
With a goal to create a multifunctional barrier membrane capable of delivery of therapeutic agents for regeneration of periodontal tissue and bone, a core-shell nanofiber membrane was created via coaxial electrospinning. Compared with conventional electrospun fibers, the core-shell nanofiber membrane demonstrated sustained release of the core component over 22 days. Experiments utilizing PDLSCs isolated from extracted human teeth further showed that core-shell nanofiber membranes supported PDLSC culture and differentiation. The encapsulation of EMD in nanofibers promoted osteogenic differentiation of PDLSCs as evidenced by enhanced mineralization and increased expression of osteogenic marker genes.
Previous studies have shown that a variety of therapeutic agents including proteins can be encapsulated and released from PCL.25,33,34 In most cases, the release of these agents exhibits two phases: a relatively rapid release followed by a slow release that occurs over a period of days or months. Since PCL is relatively hydrophobic and does not swell significantly in aqueous solutions, leaching of molecules from PCL must occur by slow diffusion through the polymer during the initial release stage. The slower long-term release phase has been attributed to material degradation. Since PCL does not significantly degrade within days, FITC-BSA release from a PCL carrier must be dominated by diffusion. Indeed, it has been shown that PCL, although hydrophobic by itself, is porous and permits water diffusion into the matrix which could subsequently affect the release and diffusion of hydrophilic molecules.35 By modeling the release kinetics using the Ritger-Peppas model, our study demonstrated that the release of hydrophilic FITC-BSA from core-shell nanofibers is significantly different from that of solid fibers. FITC-BSA from core-shell nanofibers was diffused more slowly via a mechanism of Fickian diffusion, whereas FITC-BSA in solid nanofibers was released rapidly through mixed mechanisms that may involve irregular protein transport.36
Electrospun nanofiber membranes have many potential applications due to their ability to mimic the extracellular matrix, provide mechanical strength, and simultaneously deliver drugs or therapeutic agents.17–21 Specifically, core-shell nanofibers allow for controlled release of the inner “core” component to the cellular environment.22,23 Previous studies have demonstrated the advantages of using electrospun nanofibers for periodontal tissue regeneration.7,37–41 In a study using solid PCL nanofibers containing metronidazole, the authors observed sustained release over 19 days with low burst release.38 In another study that incorporated tetracycline into core–shell nanofibers, there was sustained release for 75 days with only 19% of burst release within the first two hours.39 Bone formation was observed in another study that cultured human bone marrow stromal cells on core-shell nanofibers containing recombinant human bone morphogenetic protein 2 (rhBMP-2) and a PCL shell.40 Using an in vivo rat model, it was demonstrated that an electrospun membrane composed of poly(D,L-lactic acid) and poly(D,L-lactic-co-glycolic acid) could prevent cellular infiltration up to 13 weeks.41 The available research suggests that electrospun nanofibers can provide prolonged drug release, biocompatibility, antibacterial effects, and mechanical properties.7,20–23,37–41
While multiple clinical studies have demonstrated beneficial effects of EMD in the healing of periodontal defects, the concept of delivering encapsulated EMD via core-shell nanofiber membranes to human PDLSCs is novel. EMD-containing nanofibers provide a biomimetic barrier membrane that promotes PDLSC recruitment and differentiation. Our study is the first to incorporate EMD in biomimetic core-shell nanofibers and demonstrate a positive effect on human PDLSCs. While EMD has similar regenerative effects as barrier membranes in clinical studies, significant benefits to support the adjunctive use of EMD in addition to barrier membranes has yet to be clearly shown.42–44 This may be in part due to its gelatinous nature: EMD alone does not have space maintenance properties and is best suited for isolated, well-contained intraosseous defects.45,46 In this study, we also attempted to coat electrospun nanofiber membranes with EMD; however the EMD coating did not adhere in the presence of cell medium. This is akin to the challenges of intraoral EMD application in the presence of saliva and/or blood, especially in wide defects. By enclosing EMD within core-shell nanofibers, we created a barrier membrane with desired rigidity and controlled, sustained drug release.
The rationale for utilizing EMD in this study stems from its well-documented, significant biologic effects.8–11,47,48 This includes enhanced cell attachment, proliferation, and growth of PDL fibroblasts, as well as accelerated wound fill rates through a chemotactic and proliferative effect on endothelial cells.8,9,47 EMD also upregulates genes for growth factors such as BMP-2, BMP-7, PDGF, TGF-ß, and VEGF, while modulating the OPG-RANKL axis to indirectly favor bone formation.10,17,48
Limitations of this present study include its short duration and small dosage of EMD within a membrane. Using a rat model, it has been found that EMD applied directly to a periodontal defect is detectable up to 4 weeks.49 Thus, it is anticipated that EMD may persist for a longer period when encapsulated and released via core-shell nanofibers. The results of this 19-day study show that EMD can be released slowly from nanofibers. Subsequently, its peak effects may occur at an even later timepoint than observed here. To increase the loading dose of EMD, layering of multiple membranes could be utilized in future in vivo models. In this study, we also found that relative gene expression of RUNX2 and OPN was greater than that of OCN, suggesting that the PDLSCs were in an early stage of osteogenic differentiation since these genes are upstream of OCN.50 Longer follow-up is needed to determine the long-term effects of using such membranes for bone regeneration. Further studies using other cell types such as osteoblasts or pulpal cells may also be of interest.
A novel idea for future research is the development of a multilayered, multifunctional, and spatially designed bioactive core-shell nanofiber membrane. The membrane can be functionally oriented with multiple layers containing different drugs or proteins. For instance, on the PDL-facing side of an osseous defect, the layer could consist of encapsulated EMD nanofibers. An inner layer may contain other encapsulated growth factors such as rhBMP-2. On the tissue facing side, a layer containing encapsulated antibiotics to prevent infection may be appropriate. By tailoring the function of the membrane to its location in the defect, one could achieve more directed and predictable outcomes.
5. CONCLUSIONS
We have developed a method to create a core-shell nanofiber membrane containing encapsulated EMD. These nanofibers promoted the upregulation of osteogenic gene expression in PDLSCs in vitro. Further studies exploring other processing parameters or core components, as well as in vivo models are warranted. Coaxial electrospinning may have potential use in periodontal regeneration by creating core-shell nanofiber membranes that can simultaneously act as mechanical barriers and deliver bioactive molecules, such as EMD, to induce stem cell differentiation and reconstitute a lost periodontium.
ACKNOWLEDGEMENTS:
The authors would like to acknowledge Drs. David Fraser and Gloria Pryhuber (University of Rochester, Rochester, NY, USA) for their assistance with the experiments in this manuscript. The University of Rochester Genomics Research Center (Rochester, NY, USA) performed the PCR experiments for this work. This study is supported by grants from the National Institute of Health RO1AR067859, RO1DE019902, R21DE026256, RO1DE029790, R21AR076056, and P30AR069655 awarded to XZ, R01 diversity supplement awarded to KS, and U01-HL122700 awarded to GP. Further support was granted from the Eastman Institute for Oral Health Basil G. Bibby Fellowship awarded to LWL. The authors declare no conflicts of interest.
REFERENCES
- 1.Eke PI, Dye BA, Wei L, Slade GD, Thornton-Evans GO, Borgnakke WS, et al. Update on prevalence of periodontitis in adults in the United States: NHANES 2009 to 2012. J Periodontol. 2015;86(5):611–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hajishengallis G Immunomicrobial pathogenesis of periodontitis: keystones, pathobionts, and host response. Trends Immunol. 2014;35(1):3–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Page RC, Schroeder HE. Pathogenesis of inflammatory periodontal disease. A summary of current work. Lab Invest. 1976;34(3):235–249. [PubMed] [Google Scholar]
- 4.Caton JG, Zander HA. The attachment between tooth and gingival tissues after periodic root planing and soft tissue curettage. J Periodontol. 1979;50(9):462–466. [DOI] [PubMed] [Google Scholar]
- 5.Caton J, Nyman S, Zander H. Histometric evaluation of periodontal surgery. II. Connective tissue attachment levels after four regenerative procedures. J Clin Periodontol. 1980;7(3):224–231. [DOI] [PubMed] [Google Scholar]
- 6.Gottlow J, Nyman S, Lindhe J, Karring T, Wennström J. New attachment formation in the human periodontium by guided tissue regeneration. Case reports. J Clin Periodontol. 1986;13(6):604–616. [DOI] [PubMed] [Google Scholar]
- 7.Bottino MC, Thomas V, Schmidt G, Vohra YK, Chu TM, Kowolik MJ, et al. Recent advances in the development of GTR/GBR membranes for periodontal regeneration--a materials perspective. Dent Mater. 2012;28(7):703–721. [DOI] [PubMed] [Google Scholar]
- 8.Miron RJ, Sculean A, Cochran DL, Froum S, Zucchelli G, Nemcovsky C, et al. Twenty years of enamel matrix derivative: the past, the present and the future. J Clin Periodontol. 2016;43(8):668–783. [DOI] [PubMed] [Google Scholar]
- 9.Gestrelius S, Andersson C, Lidström D, Hammarström L, Somerman M. In vitro studies on periodontal ligament cells and enamel matrix derivative. J Clin Periodontol. 1997;24(9 Pt 2):685–792. [DOI] [PubMed] [Google Scholar]
- 10.Rodrigues TL, Marchesan JT, Coletta RD, Novaes AB Jr, Grisi MF, Souza SL, et al. Effects of enamel matrix derivative and transforming growth factor-beta1 on human periodontal ligament fibroblasts. J Clin Periodontol. 2007;34(6):514–522. [DOI] [PubMed] [Google Scholar]
- 11.Galli C, Macaluso GM, Guizzardi S, Vescovini R, Passeri M, Passeri G. Osteoprotegerin and receptor activator of nuclear factor-kappa B ligand modulation by enamel matrix derivative in human alveolar osteoblasts. J Periodontol. 2006;77(7):1223–1228. [DOI] [PubMed] [Google Scholar]
- 12.Yukna R, Mellonig J.T. Histologic evaluation of periodontal healing in humans following regenerative therapy with enamel matrix derivative. A 10-case series. J Periodontol. 2000;71(5):752–759. [DOI] [PubMed] [Google Scholar]
- 13.Trombelli L, Farina R. Clinical outcomes with bioactive agents alone or in combination with grafting or guided tissue regeneration. J Clin Periodontol. 2008;35(Suppl 8):117–135. [DOI] [PubMed] [Google Scholar]
- 14.Froum SJ, Weinberg MA, Rosenberg E, Tarnow D. A comparative study utilizing open flap debridement with and without enamel matrix derivative in the treatment of periodontal intrabony defects: a 12-month re-entry study. J Clin Periodontol. 2001;72(1):25–34. [DOI] [PubMed] [Google Scholar]
- 15.Tonetti MS, Lang NP, Cortellini P, Suvan JE, Adriaens P, Dubravec D, et al. Enamel matrix proteins in the regenerative therapy of deep intrabony defects. A multicentre randomized controlled clinical trial. J Clin Periodontol. 2002;29(4): 317–325. [DOI] [PubMed] [Google Scholar]
- 16.Chen YT, Wang HL, Lopatin DE, O’Neal R, MacNeil RL. Bacterial adherence to guided tissue regeneration barrier membranes exposed to the oral environment. J Periodontol. 1997;68(2):172–179. [DOI] [PubMed] [Google Scholar]
- 17.Liu W, Thomopoulos S, Xia Y. Electrospun nanofibers for regenerative medicine. Adv Healthc Mater. 2012;1(1):10–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gao X, Song J, Zhang Y, Xu X, Xu X, Zhang S, Ji P, et al. Bioinspired design of polycaprolactone composite nanofibers as artificial bone extracellular matrix for bone regeneration application. ACS Appl Mater Interfaces. 2016;8(41):27594–27610. [DOI] [PubMed] [Google Scholar]
- 19.Dahlin RL, Kasper FK, Mikos AG. Polymeric nanofibers in tissue engineering. Tissue Eng Part B Rev 2011;17(5):349–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang T, Zhai Y, Nuzzo M, Yang X, Yang Y, Zhang X. Layer-by-layer nanofiber-enabled engineering of biomimetic periosteum for bone repair and reconstruction. Biomaterials. 2018;182:279–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sill TJ, von Recum HA. Electrospinning: applications in drug delivery and tissue engineering. Biomaterials. 2008;29(13):1989–2006. [DOI] [PubMed] [Google Scholar]
- 22.Sultanova Z, Kaleli G, Kabay G, Mutlu M. Controlled release of a hydrophilic drug from coaxially electrospun polycaprolactone nanofibers. Int J Pharm. 2016;505:133–8. [DOI] [PubMed] [Google Scholar]
- 23.Schilling K, El Khatib M, Plunkett S, Xue J, Xia Y, Vinogradov SA, et al. Electrospun fiber mesh for high-resolution measurements of oxygen tension in cranial bone defect repair. ACS Appl Mater Interfaces. 2019;11(37):33548–33558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang YZ, Wang X, Feng Y, Li J, Lim CT, Ramakrishna S. Coaxial electrospinning of (fluorescein isothiocyanate-conjugated bovine serum albumin)-encapsulated poly(epsilon-caprolactone) nanofibers for sustained release. Biomacromolecules. 2006April;7(4):1049–57. [DOI] [PubMed] [Google Scholar]
- 25.Li X, Su Y, Liu S, Tan L, Mo X, Ramakrishna S. Encapsulation of proteins in poly(L-lactide-co-caprolactone) fibers by emulsion electrospinning. Colloids Surf B Biointerfaces. 2010February1;75(2):418–24. [DOI] [PubMed] [Google Scholar]
- 26.Baek J, Lee E, Lotz MK, D’Lima DD. Bioactive proteins delivery through core-shell nanofibers for meniscal tissue regeneration. Nanomedicine. 2020January;23:102090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ji W, Yang F, van den Beucken JJJP, Bian Z, Fan M, Chen Z, et al. Fibrous scaffolds loaded with protein prepared by blend or coaxial electrospinning. Acta Biomaterialia. 2010;6:4199–207. [DOI] [PubMed] [Google Scholar]
- 28.Peppas NA. 1. Commentary on an exponential model for the analysis of drug delivery: Original research article: a simple equation for description of solute release: I II. Fickian and non-Fickian release from non-swellable devices in the form of slabs, spheres, cylinders or discs, 1987. J Control Release. 2014September28;190:31–2. [PubMed] [Google Scholar]
- 29.Maleki H, Gharehaghaji AA, Toliyat T, Dijkstra PJ. Drug release behavior of electrospun twisted yarns as implantable medical devices. Biofabrication. 2016September16;8(3):035019. [DOI] [PubMed] [Google Scholar]
- 30.Peppas NA. 1. Commentary on an exponential model for the analysis of drug delivery: Original research article: a simple equation for description of solute release: I II. Fickian and non-Fickian release from non-swellable devices in the form of slabs, spheres, cylinders or discs, 1987. J Control Release. 2014September28;190:31–2. [PubMed] [Google Scholar]
- 31.Unpublished observations. (Manuscript in preparation, Misra R.)
- 32.Nagatomo K, Komaki M, Sekiya I, Sakaguchi Y, Noguchi K, Oda S, et al. Stem cell properties of human periodontal ligament cells. J Periodont Res. 2006;41(4): 303–310. [DOI] [PubMed] [Google Scholar]
- 33.Hu J, Tian L, Prabhakaran MP, Ding X, Ramakrishna S. Fabrication of nerve growth factor encapsulated aligned poly(ε-caprolactone) nanofibers and their assessment as a potential neural tissue engineering scaffold. Polymers (Basel). 2016February19;8(2):54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chaparro FJ, Presley KF, Coutinho da Silva MA, Lannutti JJ. Sintered electrospun polycaprolactone for controlled model drug delivery. Mater Sci Eng C Mater Biol Appl. 2019June;99:112–120. [DOI] [PubMed] [Google Scholar]
- 35.Rosenberg R, Devenney W, Siegel S, Dan N. Anomalous release of hydrophilic drugs from poly(epsilon-caprolactone) matrices. Mol Pharm. 2007Nov-Dec;4(6):943–8. [DOI] [PubMed] [Google Scholar]
- 36.Huang X, Brazel CS. On the importance and mechanisms of burst release in matrix-controlled drug delivery systems. J Control Release. 2001June15;73(2–3):121–36. [DOI] [PubMed] [Google Scholar]
- 37.Zafar M, Najeeb S, Khurshid Z, Vazirzadeh M, Zohaib S, Najeeb B, et al. Potential of electrospun nanofibers for biomedical and dental applications. Materials. 2016;9(2):73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zamani M, Morshed M, Varshosaz J, Jannesari M. Controlled release of metronidazole benzoate from poly epsilon-caprolactone electrospun nanofibers for periodontal diseases. Eur J Pharm Biopharm. 2010;75(2):179–185. [DOI] [PubMed] [Google Scholar]
- 39.Ranjbar-Mohammadi M, Zamani M, Prabhakaran MP, Bahrami SH, Ramakrishna S. Electrospinning of PLGA/gum tragacanth nanofibers containing tetracycline hydrochloride for periodontal regeneration. Mater Sci Eng C Mater Biol Appl. 2016;58:521–31. [DOI] [PubMed] [Google Scholar]
- 40.Zhu H, Yu D, Zhou Y, Wang C, Gao M, Jiang H, et al. Biological activity of a nanofibrous barrier membrane containing bone morphogenetic protein formed by core-shell electrospinning as a sustained delivery vehicle. J Biomed Mater Res B Appl Biomater. 2013;101(4):541–52. [DOI] [PubMed] [Google Scholar]
- 41.Zhang E, Zhu C, Yang J, Sun H, Zhang X, Li S, et al. Electrospun PDLLA/PLGA composite membranes for potential application in guided tissue regeneration. Mater Sci Eng C Mater Biol Appl. 2016;58:278–85. [DOI] [PubMed] [Google Scholar]
- 42.Pontoriero R, Wennström J, Lindhe J. The use of barrier membranes and enamel matrix proteins in the treatment of angular bone effects. A prospective controlled clinical study. J Clin Periodontol. 1999;26(12):833–840. [DOI] [PubMed] [Google Scholar]
- 43.Sanz M, Tonetti MS, Zabalegui I, Sicilia A, Blanco J, Rebelo H, et al. Treatment of intrabony defects with enamel matrix proteins or barrier membranes: results from a multicenter practice-based clinical trial. J Periodontol. 2004;75(5):726–33. [DOI] [PubMed] [Google Scholar]
- 44.Sculean A, Kiss A, Miliauskaite A, Schwarz F, Arweiler NB, Hannig M. Ten-year results following treatment of intra-bony defects with enamel matrix proteins and guided tissue regeneration. J Clin Periodontol. 2008;35(9):817–24. [DOI] [PubMed] [Google Scholar]
- 45.Cortellini P, Tonetti MS. Clinical performance of a regenerative strategy for intrabony defects: scientific evidence and clinical experience. J Periodontol. 2005;76(3):341–50. [DOI] [PubMed] [Google Scholar]
- 46.Froum S, Lemler J, Horowitz R, Davidson B. The use of enamel matrix derivative in the treatment of periodontal osseous defects: a clinical decision tree based on biologic principles of regeneration. Int J Periodontics Restorative Dent. 2001;21(5):437–49. [PubMed] [Google Scholar]
- 47.Rincon JC, Xiao Y, Young WG, Bartold PM. Enhanced proliferation, attachment and osteopontin expression by porcine periodontal cells exposed to Emdogain. Arch Oral Biol. 2005;50(12):1047–54. [DOI] [PubMed] [Google Scholar]
- 48.Parkar MH, Tonetti M. Gene expression profiles of periodontal ligament cells treated with enamel matrix proteins in vitro: analysis using cDNA arrays. J Periodontol. 2004;75(11):1539–46. [DOI] [PubMed] [Google Scholar]
- 49.Hamamoto Y, Kawasaki N, Jarnbring F, Hammarström L. Effects and distribution of the enamel matrix derivative Emdogain in the periodontal tissues of rat molars transplanted to the abdominal wall. Dent Traumatol. 2002;18(1):12–23. [DOI] [PubMed] [Google Scholar]
- 50.Miron RJ, Zhang YF. Osteoinduction: a review of old concepts with new standards. J Dent Res. 2012;91(8):736–44. [DOI] [PubMed] [Google Scholar]


