Abstract
CDK4/6 inhibitors are approved to treat breast cancer and are in trials for other malignancies. We examined CDK4/6 inhibition in mouse and human CD8 T cells during early stages of activation. Mice receiving tumor-specific CD8 T cells treated with CDK4/6 inhibitors displayed increased T cell persistence and immunologic memory. CDK4/6 inhibition upregulated Mxd4, a negative regulator of Myc, in both mouse and human CD8 T cells. Silencing of Mxd4 or Myc in mouse CD8 T cells demonstrated the importance of this axis for memory formation. We used single cell transcriptional profiling and TCR clonotype tracking to evaluate recently activated human CD8 T cells in breast cancer patients before and during treatment with either palbociclib or abemaciclib. CDK4/6 inhibitor therapy in humans increases the frequency of CD8 memory precursors and downregulates their expression of MYC target genes, suggesting that CDK4/6 inhibitors in cancer patients may augment long-term protective immunity.
Keywords: memory CD8 T cells, CDK4/6, cell cycle, palbociclib, abemaciclib, ribociclib, Myc
Introduction
Cancer immunotherapy has made remarkable progress, with its signature achievement being extension of overall survival and long-term durable remissions in a minority of patients. So-called “tail of the curve” 5-year survivors are touted as the most significant benefit of immunotherapeutics and rely on memory CD8 T cells to provide long-term immune surveillance (1). Yet, the question of how memory CD8 T cells form in cancer patients is not well understood. Effector CD8 T cells, including those re-invigorated by checkpoint blockade, can lyse tumor cells and produce cytokines like IFNγ to control tumor growth; however, these are not the same cells that confer long-term memory (2). Anti-CTLA4 may influence both effector and memory responses, although the mechanism by which memory cells form in the context of checkpoint blockade is unclear (3, 4). No therapies are specifically designed to improve memory formation in cancer patients.
CD8 T cell fate decisions occur soon after naive T cells first encounter peptide-MHC on activated dendritic cells (5). The majority of CD8 T cells become effector cells, although a few become memory cells. Memory T cell subtypes include tissue-resident memory cells that seed the tissues and become sentinel cells for subsequent infections, as well as central and peripheral memory cells that patrol blood, lymph, and secondary lymphoid organs. In total, memory cells represent 2–5% of the T cell response given their lower rates of proliferation compared to effector cells(6). Factors associated with augmented memory include lower affinity, lower abundance, or temporally interrupted contacts between the T cell receptor (TCR) and peptide-MHC complexes (7, 8). Memory cells undergo metabolic changes distinct from short-lived effector cells, relying on oxidative phosphorylation rather than glycolysis (9, 10). Inhibition of mTOR during immunization can skew a larger fraction of cells toward a memory phenotype (11). These metabolic changes are associated with altered mitochondrial morphology and signaling downstream of IL-15 (12).
Single cell analysis of activated murine T cells reveals a bifurcation of transcriptional phenotypes in the first cell division, with approximately half of the cells expressing transcripts consistent with effector cells and half expressing transcripts consistent with naïve/memory cells (13, 14). These differences are diminished in later cell divisions due to the highly proliferative nature of effector cells and their precursors, which rapidly dilute out the memory cell precursors resulting in a ratio of 1:20 to 1:50 memory to effector cells at the peak of an immune response. The hypothesis that memory cell fate decisions are made prior to the first cell division is supported by observations that the first cell division is asymmetric (14–17). Both MYC and mTOR segregate unevenly into daughter cells, with the cell receiving more copies of MYC and mTOR becoming an effector cell, whereas the daughter cell receiving less of these protein is fated to become a memory cell (15, 17).
Cellular proliferation is a hallmark of T cell activation, with rare antigen-stimulated clones dividing exponentially. The rate of cell division is heterogenous, and CD4 T cells arrested in G1 phase have been shown to produce higher levels of effector cytokines than more rapidly dividing cells (8). Central memory precursors divide at slower rates in vivo than their effector cell counterparts (18) although this slower division speed is not apparent in the initial cell divisions (19). Genetic deletion of the CDK regulator p27kip1 results in augmented effector and memory T cells responses, but these results may not extrapolate to acute drug-mediated interference with cell cycle progression (20). We hypothesized that pharmacologic inhibition of the cell cycle might affect T cell fate decisions, in particular that slowly dividing cells, or cells arrested in the G1 phase of the cell cycle, might be skewed toward a long-lived memory phenotype.
CDK4/6 inhibitors are approved for treatment of metastatic hormone-receptor positive breast cancer and are in clinical trials for other tumor types, either as single agents or in combination therapies. Although these drugs were developed based on their ability to cause cytostatic G1 arrest in tumor cells, they also have profound effects on anti-tumor immunity (21–24); however, the mechanisms proposed are divergent, resulting in an incomplete understanding of how best to apply CDK4/6 inhibitors as immunotherapy agents. Combination of CDK4/6 inhibitors with PD-1 blockade (with and without endocrine therapy) unfortunately led to high rates of hepatitis and several events of fatal pneumonitis in clinical trials for both early and late stage hormone-receptor positive breast cancer (25). These trials have largely been stopped for toxicity, and in light of these devastating results, the clinical path forward for use of CDK4/6 inhibitors in combination with immunotherapy is not clear.
To determine the precise effect of CDK4/6 inhibition on tumor-specific CD8 T cells, we examined the influence of these drugs on CD8 T cells during their initial activation and examined how CDK4/6 inhibitor exposed cells expanded and controlled tumor growth in both the primary and rechallenge setting. We demonstrate that CDK4/6 inhibition skews CD8 T cells toward a memory cell fate in both mice and humans; however, this occurs by regulation of the Myc/Mxd4 axis and is unrelated to the rate of cell division. We propose that a short course of CDK4/6 inhibition could be used to establish a tumor-specific memory CD8 T cell pool, thereby allowing for subsequent administration of checkpoint blockade.
Results
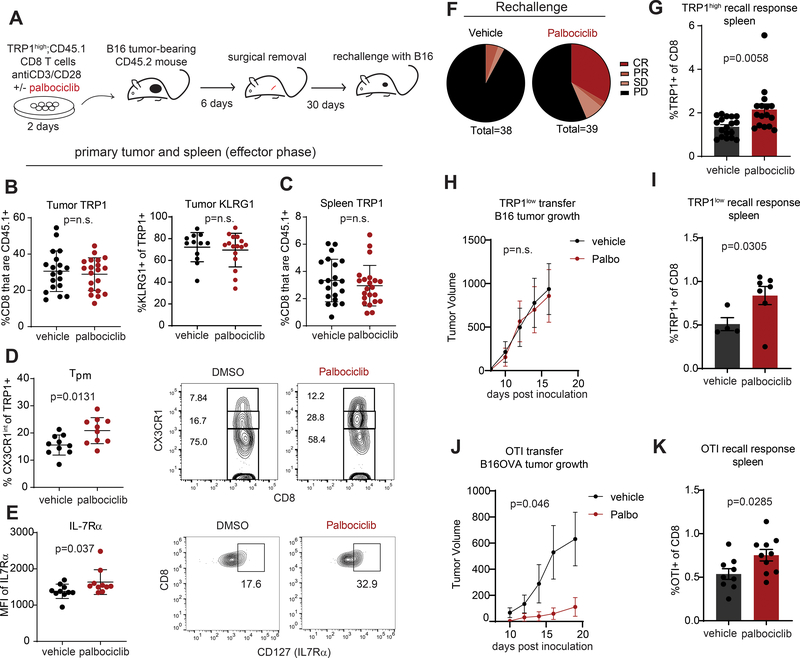
To track the fate of tumor-specific CD8 T cells in vivo, we used TRP1 transnuclear mice cloned from the nuclei of CD8 T cells specific for the melanoma antigen tyrosinase related protein 1 (TRP1). Both TRP1high and TRP1low effector CD8 T cells can delay growth of B16F10 melanoma in vivo to similar extents, despite a 100-fold difference in affinity of the TCRs for native TRP1 peptide presented on MHC class I (26, 27). We isolated TRP1high CD8 T cells and activated them ex vivo in the presence of the CDK4/6 inhibitor palbociclib (500nM) or vehicle. After 48 hours, CD8 T cells were adoptively transferred into host mice bearing palpable B16F10 tumors and congenically mismatched at the CD45 locus to facilitate identification of transferred cells by flow cytometry. Six days later, tumors were surgically excised, and mice were rested for an additional 30 days to allow for contraction of effector CD8 T cells, leaving a pool of memory TRP1high cells. To test the quality of the memory response induced, mice were challenged again with twice the original dose of B16F10 cells on the opposite flank (Figure 1A). At the time of surgery, TRP1 frequencies in both the tumor and spleen were similar between vehicle and palbociclib groups (Figure 1B–C). The transferred TRP1 cells efficiently became effector cells as evidenced by similarly high rates of KLRG1 expression in the tumor infiltrates (Figure 1B). During the peak of the primary response, more palbociclib-exposed CD8 T cells became memory precursor cells, as defined by intermediate expression of CX3CR1 (Figure 1D) and expression of IL7Rα (Figure 1E) (28). Upon rechallenge with B16 tumors in the memory phase, mice receiving palbociclib-treated CD8 T cells fared better than their vehicle counterparts with a higher proportion of animals fully rejecting their tumors (Figure 1F). Consistent with an improved memory response, mice in the palbociclib group had higher frequencies of TRP1high CD8 T cells in their spleens after rechallenge (Figure 1G). To determine whether the effects of palbociclib were consistent across a range of TCR clonotypes, we performed the same experimental protocol shown in Figure 1A using either low affinity CD8 T cells stimulated with TRP1 peptide (TRP1low, Figure 1H–I) or OT-I CD8 T cells stimulated with anti-CD3/CD28 beads (Figure 1J–K). Regardless of the TCR clonotype examined, previous exposure to palbociclib augmented the frequency of T cells observed in the memory phase. However, the degree of tumor control greatly differed among the different clonotypes, with OT-I T cells slightly outperforming TRP1high cells, while TRP1low cells had no ability to control tumor growth. Thus, palbociclib augments immunologic memory to a similar extent regardless of the TCR expressed; tumor control at rechallenge is determined by the affinity of the TCR.
Figure 1: Treatment of tumor-specific CD8 T cells ex vivo with CDK4/6i induces long-term protective memory in mice.
A) Diagram of experimental protocol. CD8 T cells were isolated by magnetic beads from pooled spleen and LNs of TRP1high CD45.1+ mice and activated with anti-CD3/28 beads in the presence of vehicle (DMSO) or 500nM palbociclib for 48 hours. Cells were washed and transferred into CD45.2+ mice bearing 100mm3 B16 tumors. Tumors were surgically removed after 6 days, and the mice recovered for 30 days prior to inoculation with 500,000 B16 cells subcutaneously on the opposite flank. B) Surgically excised tumors were digested and analyzed by flow cytometry for the frequency of CD45.1+ TRP1 cells and their expression of KLRG1. C) Spleens were analyzed by flow cytometry from mice 6 days after transfer of activated TRP1high cells. D) Surgically excised tumors from A were digested and analyzed by flow cytometry. Peripheral memory (Tpm) cells were defined as CD45.1+ CX3CR1intermediate as shown in the representative flow plots. Data pooled from 2 independent experiments. E) IL-7Rα expression on tumor-infiltrating TRP1high cells at the time of surgery. Representative flow plots are shown. Data pooled from 2 independent experiments. F) Pie charts show the response to B16 rechallenge pooled from 5 independent experiments by defining tumor response on the day that the first tumor reached 1000mm3 (day 12–16 post inoculation). complete response (CR, <10 mm3); partial response (PR, <200 mm3); stable disease (SD, <800 mm3); and progressive disease (<800 mm3). G) Frequency of TRP1high CD45.1+ cells as a fraction of total CD8 T cells in spleens of mice 10 days after tumor rechallenge. Data pooled from 3 independent experiments. H) TRP1low CD45.2 CD8 T cells were activated in vitro for 48 hours with peptide-pulsed antigen presenting cells in the presence of palbociclib or vehicle and transferred into CD45.1+ mice bearing B16 tumors according to the scheme in panel A. Upon tumor rechallenge, tumor growth was similar between vehicle and palbociclib groups. N=10 per group. I) Frequency of TRP1low CD45.2+ cells as a fraction of total CD8 T cells in spleens of mice 17 days after tumor rechallenge. J) OT-I CD8 T cells were activated in vitro for 48 hours with anti-CD3/CD28 beads in the presence of palbociclib or vehicle and transferred into C57BL/6 mice bearing B16OVA tumors according to the scheme in panel A. Tumor growth during the rechallenge phase is shown. N=10 per group. K) Frequency of SIINFEKL tetramer+ CD8 T cells as a fraction of total CD8 T cells in spleens of mice 19 days after tumor rechallenge. Error bars are SEM throughout.
To determine whether augmentation of immunologic memory could be observed in mice treated systemically with palbociclib, we adoptively transferred OT-I T cells into naïve mice and vaccinated with SIINFEKL peptide conjugated to an alpaca antibody fragment targeting MHC class II that we have previously shown is an effective vaccine platform (Supplemental Figure 1A) (29). Oral dosing of palbociclib during the 3 days post-vaccination increased the expression of IL-7Rα on antigen-specific CD8 T cells detected in the spleen at day 7, indicating increased formation of memory precursors (Supplemental Figure 1B–D). To determine whether systemic palbociclib increased memory formation of antigen-specific T cells in the context of a tumor-bearing mouse, we adoptively transferred naïve TRP1high CD8 T cells into congenically mismatched B16-tumor bearing hosts. Mice were treated with vehicle or palbociclib for 6 days prior to surgical removal of the tumors and establishment of immunologic memory (Supplemental Figure 1E). Upon rechallenge with tumor 30 days later, mice that had previously been treated with palbociclib showed slightly better tumor control and increased TRP1high cells in the spleen, consistent with augmented immunologic memory (Supplemental Figure 1F–G).
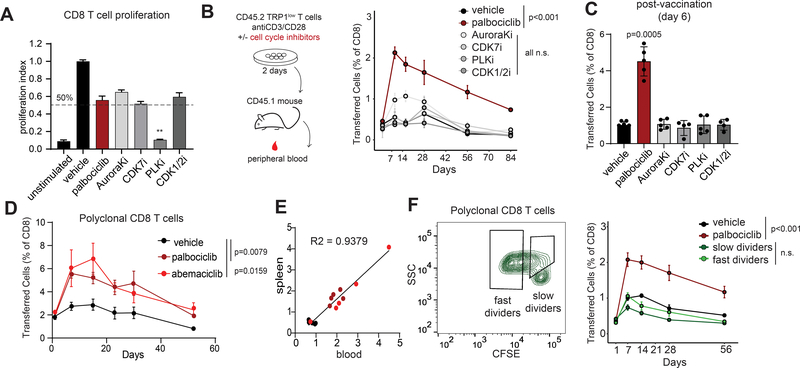
Activated T cells expand and contract over the course of an immune response. To track how CDK4/6 inhibition affects the kinetics of T cell responses, CD8 T cells were activated with anti-CD3/CD28 in vitro for 48 hours in the presence of palbociclib or other cell cycle inhibitors (Figure 2A). Each compound, with the exception of Polo-like kinase inhibitor (PLKi), was titrated and used at a final concentration that inhibited CD8 T cell proliferation by approximately 50% as measured by dilution of the dye CFSE 4 days after stimulation (Figure 2A). PLK inhibition was highly effective at blocking cell division, although CD8 T cells were able to start dividing upon removal of the drug and transfer into mice. CD8 T cells were washed, transferred into congenically-mismatched host mice, and their frequencies in peripheral blood were monitored over time. CD8 T cells activated with anti-CD3/CD28 in the presence of palbociclib expanded to a higher degree and contracted to a higher baseline level than CD8 T cell primed in the presence of vehicle or inhibitors of any of the other cell cycle components tested (CDK7, Aurora kinase, PLK, CDK1/2) (Figure 2B and Supplemental Figure 2A). After 84 days, mice were vaccinated with irradiated B16-GVAX mixed with TRP1 peptide. This vaccine elicited a more robust post-vaccination response in the palbociclib group, consistent with a higher frequency of memory cells (Figure 2C). Increased persistence and recall responses were also observed when CD8 T cells were activated in the presence of two other CDK4/6 inhibitors abemaciclib or ribociclib (Figure 2D). This occurred whether T cells were activated with anti-CD3/CD28 or with peptide-pulsed antigen presenting cells and occurred across multiple TCR clonotypes including polyclonal CD8 T cells (Figure 2D and Supplemental Figure 2A–B). Transferred T cell frequencies were tracked longitudinally in blood and were found to correlate well with frequencies in the spleen (Figure 2E). CDK4 inhibition has been shown to lead to increased surface PD-L1 expression (21), a finding we confirmed on CD8 T cells activated in the presence of CDK4/6i (Supplemental Figure 2C). This increased surface PD-L1 did not affect the ability of CDK4/6i to enhance memory formation as increased persistence of transferred CD8 T cell was observed in the presence of PD-1 blocking antibodies (Supplemental Figure 2D). To determine whether the augmented memory response was a general property of slowly cycling cells, we took advantage of the natural heterogeneity in T cell activation to sort cells that were initially either slow or fast dividers. Polyclonal CD8 T cells were labeled with the dye CFSE and activated in vitro for 48 hours in the absence of drug treatment. We then sorted cells that had divided only once during this period (“slow dividers”) versus cells that had divided three or more times (“fast dividers”) and transferred these into congenically mismatched host mice. Unfractionated cells activated in the presence of palbociclib or vehicle were included as positive and negative controls. Although palbociclib-treated CD8 T cells showed increased expansion and persistence, no differences were observed between slow and fast cyclers (Figure 2F). These findings demonstrate that slow-dividing cells are not intrinsically more capable of memory formation. Given that augmented memory formation was observed with three different CDK4/6 inhibitors but not with multiple other means of slowing the cell cycle, we concluded that the augmentation of memory formation was not solely dependent on cell cycle arrest. Although we did not observe augmented memory with the several other cell cycle inhibitors tested, CDK4/6i may be one of several routes to achieving augmented memory formation.
Figure 2: CDK4/6 inhibitors induce long term persistence of CD8 T cells independent of cell cycle.
A) Naïve CD8 T cells were labeled with CFSE and activated with anti-CD3/CD28 beads in the presence of 500nM palbociclib, 1μM AuroraKi, 100nM CDK7i, 150nM PLKi, 4.5 μM CDK1/2i, or vehicle. Proliferation indexes were calculated based on number of cell divisions determined by flow cytometry after 4 days. B) Diagram of experimental protocol. TRP1low CD45.2+ CD8 T cells were activated in the presence of the indicated cell cycle inhibitors at the concentrations used in A and transferred into CD45.1 recipients. Frequency of transferred cells in peripheral blood was measured over time. Representative of 3 independent experiments. C) At 85 days post transfer, the mice in B were immunized with irradiated B16-GVAX admixed with TRP1 peptide. Frequency of TRP1low cells was analyzed in the spleen 6 days later. Representative of 3 independent experiments. D) Polyclonal CD45.2+ CD8 T cells were activated in the presence of the indicated CDK4/6 inhibitors and transferred into CD45.1 recipients. Frequency of transferred cells in peripheral blood was measured over time. Representative of 3 independent experiments. E) After 56 days, mice from D were analyzed for frequencies of transferred cells in spleen and in blood. Plot shows correlation between the two compartments. Individual mice are represented by colored dots representing the treatment groups. F) Polyclonal CD45.2+ CD8 T cells were stained with CFSE and activated in vitro with anti-CD3/28 beads. Fast or slow cycling cells were sorted by FACS using the indicated gates, transferred into CD45.1+ recipients and monitored over time in peripheral blood. Error bars are SEM throughout.
Several mechanisms of action linking CDK4/6 inhibition to anti-tumor immunity have been proposed including increased phosphorylation of NFAT4 leading to increased IL-2 production from human CD4 T cells (24). NFAT is a family of transcription factors that in response to increased intracellular calcium become phosphorylated, dimerize, and rapidly translocate to the nucleus. NFAT activity is then regulated by its rate of nuclear export, which can occur over minutes to hours (30). We evaluated NFAT1, the primary isoform in mouse T cells (31), in CD8 T cells with and without palbociclib and found no differences in the rate of nuclear export, indicating that at least in mice, NFAT phosphorylation does not account for the effects of CDK4/6 inhibition on T cell fate (Supplemental Figure 3A–B). CD4 and to a lesser extent CD8 T cells have been reported to increase production of other cytokines such as IFNγ upon activation and exposure to CDK4/6 inhibitors (24). To examine whether effector cells generated in the presence of palbociclib displayed long-term potentiation of effector cell functions in the context of restimulation through the TCR, we activated TRP1high CD8 T cells in the presence of palbociclib and then rested them for 6 days to generate effector cells. Upon restimulation of these CD8 T cell effectors, we found that the presence of palbociclib during activation had no discernable effect on IFNγ production or the ability to kill melanoma tumor cells (Supplemental Figure 4A–E), which is consistent with our findings of equivalent effector cell frequencies in tumor infiltrates from primary tumors of mice receiving adoptively transferred TRP1high cells (Figure 1B). We therefore conclude that CDK4/6 inhibition augments memory responses in CD8 T cells while minimally affecting effector capacity upon re-engagement with antigen.
CD8 T cell fate decisions occur early during activation, usually prior to the first cell division (13, 26). Surface markers such as IL7Rα are shared between memory precursors and naïve cells. To distinguish between true memory precursors and cells that failed to become activated, we labeled CD8 T cells with CFSE and activated them in the presence of palbociclib or the CDK4/6 degrader molecule BSJ-02–162 (Supplemental Figure 5A–E) (32). Both palbociclib and BSJ-02–162 induced long-term persistence of CD8 T cell in vivo (Supplemental Figure 5B–C). Both compounds also delayed cell division rates in vitro (Supplemental Figure 5D); however, gating on CFSE-diluted cell as an indicator of activation revealed increased expression of IL7Rα and Bcl-2 on non-naïve cells after 48 hours (Supplemental Figure 5E). These results indicate that both enzymatic inhibition and targeted degradation of CDK4/6 result in similar skewing of activated CD8 T cells toward a memory precursor fate.
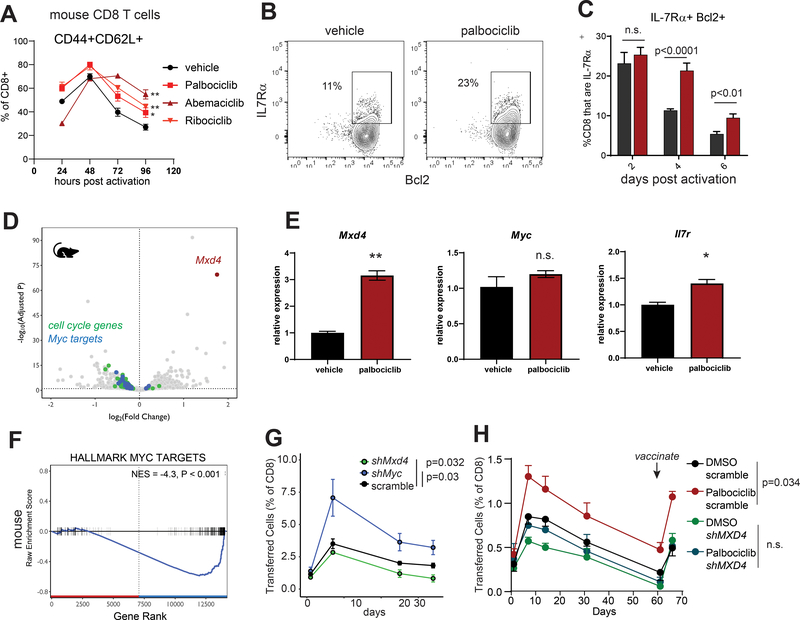
Murine CD8 T cells activated in the presence of three different CDK4/6 inhibitors showed persistent expression of CD62L (Figure 3A and Supplemental Figure 5F–G) as well as IL7Rα and Bcl-2 (Figure 3B–C), consistent with a memory precursor state. Having established that 48 hours of CDK4/6 inhibition during CD8 T cell activation was sufficient to confer long-term differences in immunologic memory, we proceeded to characterize the cellular changes in the first 48 hours correlating with altered cell fates. Transcriptional analysis of bulk RNA from mouse CD8 T cells activated in the presence of vehicle or palbociclib revealed Mxd4 as a top differentially expressed gene (Figure 3D–E). We examined Mxd4 expression by qPCR at 48 hours post-activation in the presence of our panel of cell cycle inhibitors. Mxd4 was significantly higher in the palbociclib condition than in any of the other groups, suggesting that the increased Mxd4 is specific to CDK4/6 inhibition and not replicated across the other cell cycle inhibitors tested (Supplemental Figure 6A). Mxd4 belongs to a family of transcription factors that associate with Max, the obligate heterodimeric partner of Myc (33). Binding of Mxd4 to Max sequesters Max away from Myc and represses the transcription of Myc target genes (33). We observed a distinct downregulation of Myc target genes in CD8 T cells activated in the presence of CDK4/6 inhibitors despite Myc transcript levels remaining unchanged (Figure 3E–F). To determine whether this Mxd4/Myc axis regulates memory formation in CD8 T cells, we transduced anti-CD3/CD28 activated TRP1high CD8 T cells with shRNAs targeting Mxd4, Myc, or scrambled control (Supplemental Figure 6B–C). Mxd4 silenced CD8 T cells showed increased transcription of several Myc target genes, as expected given the known role of Mxd4 as a negative regulator of Myc activity (Supplemental Figure 6D). We performed transcriptional profiling on control versus Mxd4-silenced CD8 T cells and examined differentially expressed genes at 72 hours post-activation with anti-CD3/CD28 beads. In this context, control cells with normal levels of Mxd4 had increased expression of genes associated with memory cells (Cd28, Il7r) and with cell cycle regulation (Ckdn1a, Ccng1) while Mxd4-silenced cells had increased expression of genes associated with effector cells (Ccl5, Eomes, Klf2, Gzma, Ifng) (Supplemental Figure 6E). Upon transfer into host mice, Myc-silenced cells expanded to greater frequencies and persisted at higher levels than the scrambled shRNA control cells (Figure 3G). By contrast, Mxd4-silenced cells failed to persist, and 2 out of 5 mice had undetectable TRP1high cells at the final time point. To determine whether Mxd4 is required for the palbociclib-induced memory formation, control or Mxd4-silenced CD8 T cells were activated with anti-CD3/CD28 in the presence of vehicle or palbociclib and transferred into congenically mismatched host mice. Whereas control CD8 T cells exposed to palbociclib showed greater expansion, persistence and recall response to vaccination, Mxd4-silenced cells showed no increase in persistence when exposed to palbociclib (Figure 3H). These results indicate that silencing Myc confers increased memory potential in CD8 T cells, and Mxd4-mediated suppression of Myc is responsible for the augmentation of memory potential in CD8 T cells treated with CDK4/6 inhibitors.
Figure 3: CDK4/6 inhibition acts through the Mxd4/Myc transcription factor axis to inducea memory precursor phenotype mouse CD8 T cells.
A) CD8 T cells were isolated by magnetic beads from pooled spleen and LNs of a C57BL/6 mouse and activated in vitro with anti-CD3/28 beads in the presence of the indicated compounds. Cells were analyzed by flow cytometry for expression of CD44 and CD62L at the indicated times. Representative of 3 independent experiments. B) CD8 T cells were isolated by negative selection on magnetic beads from pooled spleen and lymph nodes of a C57BL/6 mouse. Cells were labeled with CFSE and activated with anti-CD3/CD28 beads in the presence of the indicated compounds. 72 hours later cells were analyzed by flow cytometry. C) Quantification of Bcl-2 and IL7Rα by flow cytometry after gating on cells experiencing at least one cell division as shown in B. D) Murine TRP1high CD8 T cells were activated for 48 hours with anti-CD3/28 beads in the presence of vehicle or 500nM palbociclib. Differentially expressed transcripts were identified by bulk RNAseq. Hallmark cell cycle genes are shown in green; hallmark Myc targets are shown in blue. E) Quantitative PCR validation of the indicated transcript levels in mouse CD8 T cells at 48 hours post activation. Representative of 2 independent experiments. *p<0.05, **p<0.01. Error bars are SEM. F) Gene-set enrichment analysis of MYC target genes in mouse palbociclib treated CD8 T cells. G) TRP1high CD45.1+ CD8 T cells were transduced with retroviruses encoding RFP and scrambled, Mxd4 or Myc shRNAs. Cells were transferred into CD45.2+ recipient mice, and their frequency in peripheral blood was monitored over time. Representative of 2 independent experiments. Area under the curve values were calculated for individual mice and p values determined versus the scrambled control group using a Mann-Whitney test. H) Similar to G, TRP1 CD8 T cells were silenced and activated in the presence of vehicle or palbociclib. After 60 days, mice were vaccinated with TRP1 peptide mixed with irradiated B16-GVAX.
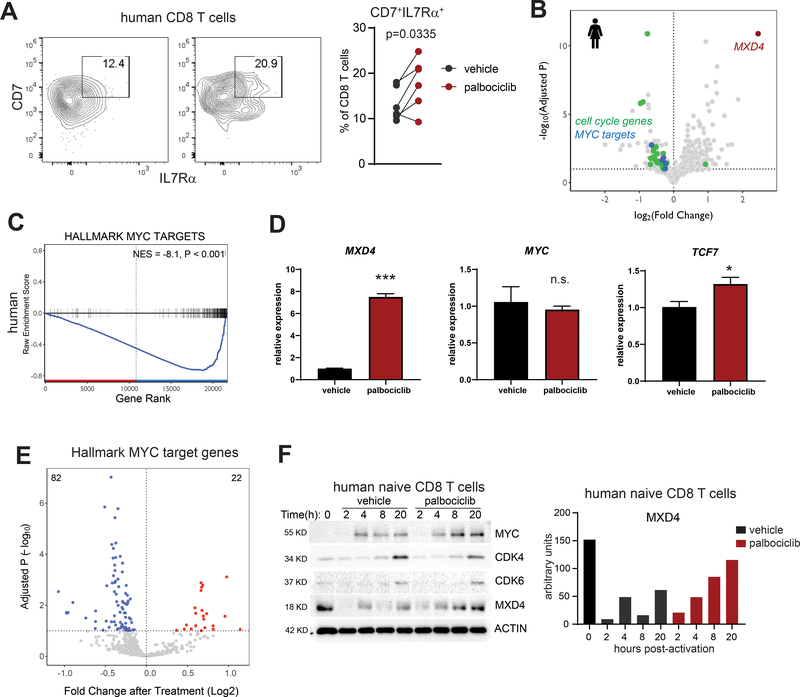
To determine whether similar effects occurred in humans, we isolated CD8 T cells from healthy donor peripheral blood and activated them with anti-CD3/28 beads in the presence of vehicle or palbociclib. CDK4/6 inhibitor treatment increased the frequency of IL7Rα+CD7+ memory precursors in five of six individual donors (Figure 4A). Upon transcriptional profiling of CD8 T cells activated in the presence of palbociclib, MXD4 was the top upregulated gene (Figure 4B). As we had observed in mouse cells, palbociclib exposure resulted in decreased transcription of MYC target genes despite no change in MYC transcript levels (Figure 4C–D). CDK4/6 inhibition imprinted an epigenetic signature consistent with decreased MYC activity. ATACseq analysis revealed that accessibility throughout MYC target gene loci was decreased with CDK4/6 inhibition, consistent with decreased target gene transcription (Figure 4E). MXD4 and MYC protein levels are dynamically regulated following T cell activation: MYC increases post-activation, while MXD4 decreases immediately post-activation and increases slowly thereafter. Palbociclib treatment results in retention of MXD4 protein even at early time points (Figure 4F).
Figure 4: CDK4/6 inhibition and MXD4-Myc transcription factors regulate memory cell fate decisions in human CD8 T cells.
A) CD8 T cells were isolated by magnetic beads from peripheral blood of healthy donors and activated in vitro with anti-CD3/28 beads in the presence of vehicle or palbociclib. Cells were analyzed by flow cytometry for expression of IL7Rα and CD7 at 48 hours. Matched pairs are from six individual donors. Data pooled from 2 independent experiments. B) Human CD8 T cells were isolated by magnetic beads from healthy blood donors and activated in vitro with anti-CD3/28 beads in the presence of vehicle, abemaciclib or palbociclib. RNAseq was performed on 48-hour samples. Differential expression analysis was performed between vehicle treated and combined CDK4/6i treated cells. C) Gene-set enrichment analysis of MYC target genes in human CDK4/6i treated CD8 T cells. D) Quantitative PCR validation of the indicated transcript levels in human CD8 T cells at 48 hours post activation. Representative of 2 independent experiments. *p<0.05, ***p<0.001. Error bars are SEM. E) CD8 T cells were isolated by negative selection from the blood of healthy donors and treated for 48 hours with palbociclib, abemaciclib, or vehicle. Areas of open, transcriptionally active chromatin were identified and grouped into consensus peaks. Peaks corresponding to open chromatin were observed in 184 genes (92%) from the hallmark MYC target gene set. The change in accessibility at each peak after treatment with palbociclib is shown as a volcano plot. P values were FDR-controlled. F) Human naïve CD8 T cells were isolated by magnetic beads and activated with anti-CD3/CD28 beads for the indicated times. Protein lysates were prepared from 3 healthy donors, pooled, and analyzed by immunoblot for the indicated proteins. MXD4 protein levels were quantified. Representative of 2 independent experiments.
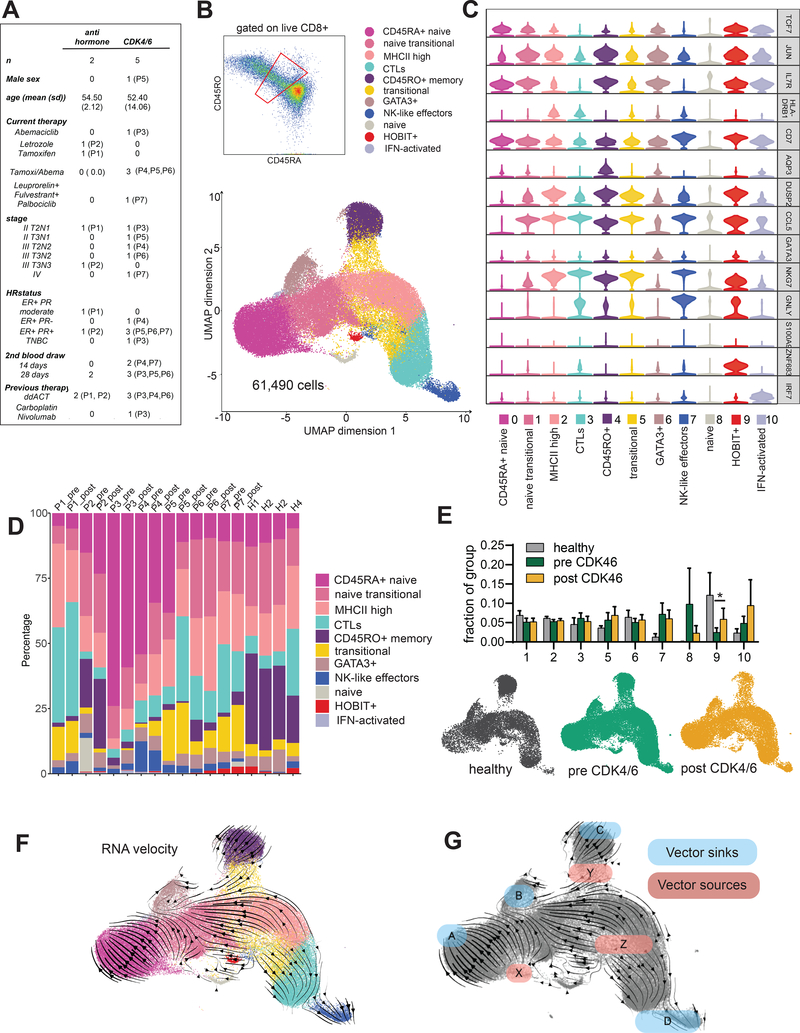
CDK4/6 inhibitors are used to treat metastatic hormone receptor positive breast cancer and are in trials in the adjuvant setting (34). To determine whether newly activated CD8 T cells in humans treated with CDK4/6 inhibitors are skewed toward a memory fate, we obtained peripheral blood mononuclear cells from 7 breast cancer patients (4 patients on CDK4/6 inhibitor with endocrine therapy, one on CDK4/6 inhibitor monotherapy, and 2 patients on endocrine therapy alone) and 4 healthy donor controls (Table S1). Five of the 7 cancer patients were receiving therapy in the adjuvant setting for breast cancer and had no evidence of disease during this study. Two patients (one in the CDK4/6 inhibitor group and one in the endocrine therapy group) were receiving therapy for metastatic breast cancer. For cancer patients, we collected blood prior to the initiation of CDK4/6 inhibitor treatment, and again at 2 or 4 weeks on treatment (Figure 5A). To identify recently activated cells, we sorted CD8+ cells that were CD45RO/RA intermediate. We used a generous sorting gate such that for each patient we would sequence some true naïve (CD45RA+) and memory (CD45RO+) cells, thus ensuring 1) that we obtained naïve and memory cell signatures for comparison to recently activated cells and 2) that we captured all transitional populations between these two poles (Figure 5B). We performed droplet-based single cell transcriptional profiling on more than 61,000 cells from 18 different samples and grouped these into 11 clusters (Figure 5B). Clusters were in smooth contiguity with each other when visualized by UMAP, a technique geared toward authentic representation of the global topology of complex data, in line with our goal of comprehensively capturing the full spectrum of T cell transitional transcriptional states. IL7R and TCF7 identified naïve and memory cells, while CCL5 reliably identified all antigen-experienced cells (Figure 5C and Table S2). Cytolytic effector cells expressed key defining genes GNLY, PRF1, GZMB. The highest levels of cytotoxic effector transcripts were found in cluster 7 and co-expressed with NK cell ligands suggesting an NK-like highly cytolytic population. Naïve cells (cluster 0) and memory cells (cluster 4) were clearly identifiable in each sample, indicating that we had captured cells from the highly abundant CD45RA and RO single positive cell clusters at the edge of our sorting gate (Figure 5D). Since we had intentionally captured only a small fraction of the total naïve and memory cells, clusters 0 and 4 were not counted as part of the frequency distribution. The remaining cells showed a similar distribution pattern among healthy and cancer patients, and between pre- and post- CDK4/6 inhibitor treatment, indicating that CD8 T cell activation occurs normally in patients with breast cancer and is not obstructed by treatment with CDK4/6 inhibitors (Figure 5E).
Figure 5: Memory cell precursors can be found by single cell transcriptional analysis of recently activated CD8 T cells from human peripheral blood.
A) Clinical characteristics of the patient cohort. B) Peripheral blood mononuclear cells were enriched for CD8 T cells using magnetic beads and stained with antibodies to CD8, CD45RO, and CD45RA. Cells were gated on live CD8+ prior to sorting the population gated in red. Single cell transcriptional profiling was performed using the 10X Genomics platform. 61,889 cells were analyzed from a combined 4 healthy donors and paired pre- and on-treatment samples from the 7 breast cancer patients shown in A. Cells were projected onto 2 dimensions by UMAP and grouped into 11 clusters. C) Violin plots of select cluster defining genes. D) Cluster representation per sample. E) Fractional representation of healthy, pre CDK4/6 and post CDK4/6 cells represented in each cluster. CD45RA+ naïve cells (cluster 0) and CD45RO+ memory cells (cluster 4) were excluded from the analysis. Error bars are SEM. Significance was evaluated using paired analysis of pre-CDK4/6 and post-CDK4/6 values for each patient; only cluster 9 was significant, *p=0.0403. UMAP projections of cells from healthy, pre and post CDK4/6 samples are shown. P1 and P2 post samples are included in the pre-CDK4/6 plot. F) RNA velocity analysis. G) Diagram of sinks and sources based on RNA velocity.
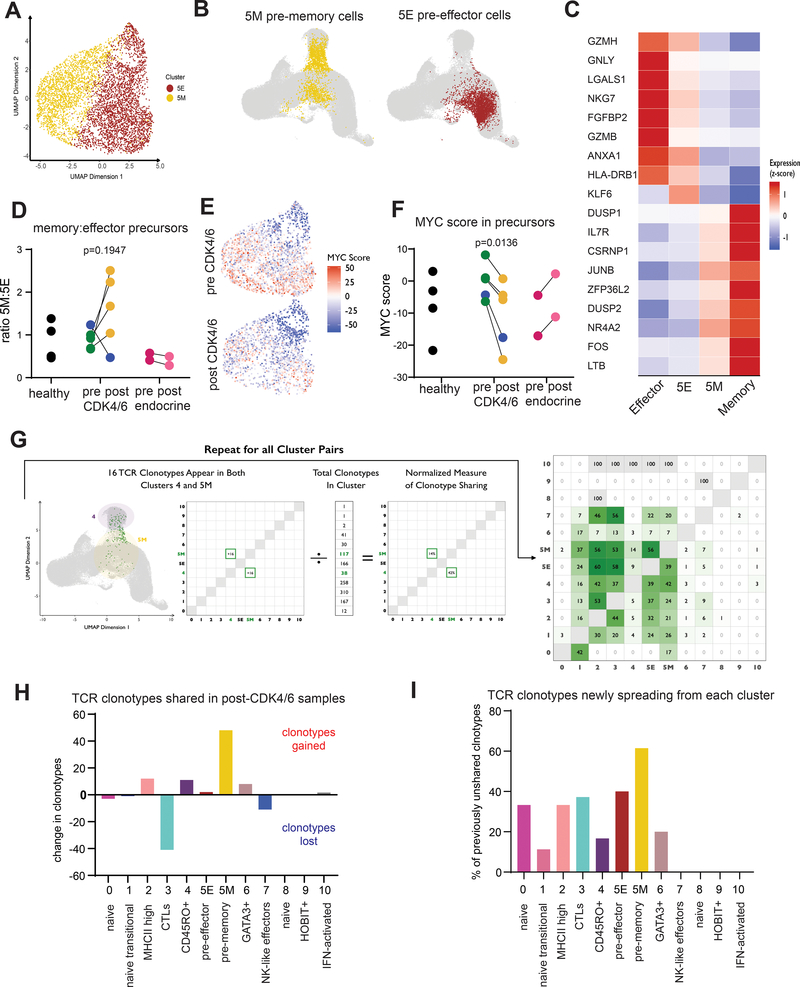
We explored whether we could identify central memory precursors in our scRNAseq dataset of recently activated CD8 T cells. Although we could assign identities to most of the clusters in our dataset, cluster 5 proved difficult since it expressed few globally distinct markers and appeared scattered in two places on the UMAP plot. We performed RNA velocity analysis, which infers the near-future states of cells from their unspliced mRNA (35). By this approach, two of the three velocity vector sources were in cluster 5 emanating toward sinks in the effector, memory and GATA3+ populations, clearly indicating that cells in cluster 5 are engaged in fate decision making (Figure 5F–G). We performed a subclustering analysis and found that cluster 5 separated into two clusters with distinct differentially expressed genes that paralleled effector and memory cells (Figure 6A). Remapping of cluster 5 onto the original UMAP also showed subcluster 5M near the memory cell cluster and subcluster 5E near the effector cell cluster, consistent with these being pre-memory and pre-effector cell populations, respectively (Figure 6B). The top differentially expressed transcripts between 5M and 5E correlated with higher levels of those same transcripts in more terminally differentiated memory and effector cell populations (Figure 6C). The ratio of cells in 5M to 5E reflects the proportion of newly activated CD8 T cells becoming pre-memory versus pre-effector cells. In healthy donors and in patients prior to CDK4/6 inhibitor treatment, this ratio is approximately 1. However, in 4 out of 5 post-CDK4/6 treatment samples, the ratio increased approximately 2-fold, indicating skewing toward memory precursors (Figure 6D). Of note, in the single patient whose 5M:5E ratio did not increase, the top expanded clonotype matched an Influenza-A specific TCR Vα sequence from the TCR3d database (36), suggesting that this patient may have been in the contraction phase of an acute viral infection at the start of CDK4/6 inhibitor treatment. Given our previous in vitro data connecting suppression of the MYC pathway to CD8 T cell memory augmentation, we also calculated an aggregated MYC target gene expression score for the transitional cells in cluster 5 pre- and on-treatment with CDK4/6 inhibitors. We found a marked decrease in MYC scores post-treatment, suggesting that CDK4/6 inhibitors indeed suppress the MYC axis in recently activated transitional CD8 T cells in vivo (Figure 6E–F).
Figure 6: TCR clonotype tracing reveals a transitional population of pre-effector and pre-memory cells in peripheral blood whose fractional composition and MYC signature are affected by CDK4/6i treatment.
A) Cluster 5 (transitional cells) was further divided into 2 sub-clusters. B) UMAP projections of sub-clusters 5M and 5E. C) Top differentially expressed genes among cluster 5 sub-clusters are displayed as a heatmap. Expression scores for those same genes are shown for memory cells (Cluster 4) and cytolytic effector cells (Cluster 3). D) Ratio of cells in sub-clusters 5M:5E analyzed in pairwise fashion. Blue dots indicate P5 (dominant clonotype has influenza-specific TCR). E) MYC scores for individual cells in Cluster 5. F) MYC hallmark gene signature was defined across the entire dataset and analyzed in pairwise comparison between pre and on CDK4/6 inhibitor treatment and between pre- and on-treatment samples from control patients receiving endocrine therapy. G) TCR clonotypes that were shared across clusters in pre-CDK4/6 samples are plotted in the matrix shown in green as percent of total clonotypes in that cluster. H) For each cluster, the total number of shared clonotypes gained was subtracted from the total number of shared clonotypes lost to quantify the total flux of TCR clonotypes occurring for each cluster. I) For each cluster, the number of unshared clonotypes in the pre-treatment samples was identified. Of these, clonotypes that became shared in the on-treatment samples are plotted as a percentage of total previously unshared for each cluster.
TCRs from single cell sequencing data can be used as molecular barcodes to track cell fates over time (37). We obtained paired αβ TCR sequences and identified clonally expanded T cells as cells whose TCR was found more than once in our dataset and was not one of the canonical MAIT TCRs (Supplemental Figure 7A–C). We then asked which TCR clonotypes were shared across multiple cell clusters, indicating movement of cells between multiple populations. The clusters with the greatest fraction of shared clonotypes were 5M and 5E, consistent with these being transitional populations (Figure 6G). We next compared the flux of shared clonotypes through each cluster in the pre- versus post-CDK4/6 samples. Cluster 5M showed a net gain of more than 40 clonotypes, far more than any other cluster (Figure 6H). Cluster 5M also showed the highest percentage of previously unshared clonotypes that then become shared in the post-treatment samples (Figure 6I). Our analyses suggest that CDK4/6 inhibition leads to an increase in both the number and diversity of peripheral CD8 T cells entering the memory precursor pool in breast cancer patients. We therefore conclude that breast cancer patients newly started on CDK4/6 inhibitor therapy skew CD8 T cells toward a memory cell fate.
Discussion
Here we show that inhibition of CDK4/6 in CD8 T cells at the time of T cell activation has little effect on cytolytic function or cytokine production, but skews the CD8 T cells toward a memory cell fate. Mice administered tumor-specific CD8 T cells activated in the presence of CDK4/6 inhibitors were protected from subsequent tumor challenge. Memory CD8 T cells tracked longitudinally for up to three months were consistently elevated if the T cells had been activated in the presence of a CDK4/6 inhibitor, but this effect was not seen with other means of inhibiting or slowing the cell cycle. We traced the mechanism to increased expression of Mxd4 in both mouse and human CD8 T cells and a resultant downregulation in Myc activity.
Myc is dynamically regulated during T cell development both at the RNA and post-transcriptional level. Myc downregulation during the double-positive stage of thymic development is associated with upregulation of Mxd family members and marks the end of the post TCRβ selection proliferative burst (38). In mature T cells, Myc is upregulated upon stimulation through the TCR and controls the temporal window of proliferation, with loss of Myc protein immediately preceding transition to quiescence in activated T cells regardless of how many cell divisions had occurred (39). Myc is tightly linked to cellular proliferation in many cell types, underscoring its importance as an oncogene in multiple cancer types including T and B cell lymphomas. Myc has also been linked to immunologic memory, with asymmetrically dividing cells generating high Myc pre-effector cells and low Myc central memory precursor cells (15).
Mxd4 is part of a family of Max-binding transcription factors that include the transcriptional activator Myc and transcriptional repressors Mxd1–4 and Mnt (33). Mxd4 protein levels increase upon T cell activation and are enhanced by costimulation through the TNFR family member OX40 (40). OX40 engagement prevents cells from Myc-induced apoptosis, and confers long-lived persistence in vitro and in vivo, consistent with our observations that increased Mxd4 and resultant down regulation of Myc target genes lead to increased memory CD8 T cells in vivo. We observed dynamic regulation of Mxd4 transcripts in both mouse and human CD8 T cells, suggesting that although Mxd4 protein stability is regulated by serine phosphorylation (40), Mxd4 is unlikely to be a direct phosphorylation target of CDK4 or CDK6. CDK4 phosphorylates and inactivates the transcription factor FoxO1, which has been linked to memory formation through its regulation of Tcf7 and linked to negative regulation of MYC (41, 42). The promoter region of Mxd4 contains forkhead transcription factor binding motifs, and thus FoxO1 may be a direct transcriptional regulator of Mxd4, thereby providing feedback inhibition of MYC target genes, a key part of promoting the memory lineage. Alternatively, Lelliot et al found differential phosphorylation of the transcription factor host cell factor 1 (Hcf1) in mouse and human cells treated with palbociclib suggesting that Hcf1 might be phosphorylated by CDK4/6. Hcf1 forms the repressive half of a heterodimeric complex with Miz-1 that binds to the Mxd4 promoter, and loss of Hcf1 from this complex enables transcriptional activation of Mxd4 (43, 44). We hypothesize that CDK4/6 inhibition and prolongation of the G1-S transition results in increased Mxd4 transcription and increased competition with Myc for Max binding.
The T cell receptor is a unique molecular barcode that can be used to track clonally expanded cells and movement of those clones over time (37). Our analysis of paired pre- and on-treatment samples allowed a dynamic analysis of the flux of clonotypes over time and identified memory precursor cells as showing the highest degree of sharing with other cell clusters, indicating the transitional nature of this population. Both the flux of new clonotypes into, and newly originating from, the memory precursor population increases dramatically with CDK4/6 inhibitor treatment. These findings were corroborated by RNA velocity analysis, an orthogonal approach that relies on unspliced mRNA rather than on TCR sequences. Importantly, we also observed an overall decrease in MYC target gene expression in transitional CD8 T cells from patients receiving CDK4/6 inhibitor therapy, suggesting that the MXD4/MYC axis may regulate this fate decision in vivo, as well as in vitro. Thus, in human breast cancer patients, we found that treatment with either palbociclib or abemaciclib skews newly activated CD8 T cells toward a memory precursor fate.
Our single cell dataset comprises greater than 60,000 recently activated CD8 T cells from both healthy donors and cancer patients. We provide a comprehensive landscape of the transitional states that CD8 T cells undergo post-activation, as well as bona fide naïve and memory CD8 T cells from the same set of patients. These analyses revealed several populations of interest in addition to the central memory precursors that we have highlighted herein. Recently activated effector cell populations included cytolytic effector cells (cluster 3) as well as a highly cytolytic population reminiscent of NK cells (cluster 7). Unexpectedly, we also found that GATA3 expressing CD8 T cells in a relatively quiescent state (cluster 6) represented a terminal sink. The functional importance of these cells is still unknown. Finally, cluster 9 included cells with high levels of the transcription factor HOBIT (ZNF683) and other markers of tissue-resident memory cells (Trms). Trms are typically found in tissues and are the predominant CD8 T cell population in breast cancer correlating with good prognosis (45). Trms are not typically found in peripheral circulation, although they have been reported in HIV+ patient blood and lymph nodes (46). The fraction of HOBIT+ cells in our dataset is very low, although intriguingly the frequency of these rare cells increases with CDK4/6 inhibitor therapy and may be worth further investigation.
Abemaciclib, palbociclib, and ribociclib all increase overall survival in breast cancer patients, with the longest reported data extending to 4 years (47–52). Of note, in the analysis of ribociclib, overall survival curves did not separate until 24 months, a pattern more consistent with an immunotherapy than a cytotoxic agent (47). We conjecture that the increase in overall survival may reflect a dynamic equilibrium with adaptive immunity, suggesting that the greatest benefit to CDK4/6 inhibitors may occur early in treatment. Our findings suggest that CDK4/6 inhibitors may have broad utility outside of breast cancer, and importantly, that these drugs would have the highest utility in the neoadjuvant setting when CD8 T cell activation in response to tumor antigens may occur. Early treatment with CDK4/6 inhibitors to establish a memory CD8 T cell pool followed by checkpoint blockade would allow for non-concurrent administration of these agents, thereby likely avoiding the synergistic toxicity observed when CDK4/6 inhibitors are given simultaneously with PD-1 blockade (25). Neoadjuvant CDK4/6 inhibition followed by checkpoint blockade is currently being explored (NCT04075604), potentially paving the way for treatment of a range of resectable cancers.
Materials and Methods
Animal care
Animals were housed at the Dana-Farber Cancer Institute and were maintained according to protocols approved by the DFCI Institutional Animal Care and Use Committee (IACUC) (#14–019 and #14–037). TRP1high and TRP1low transnuclear mouse lines were generated by us as previously reported and maintained in house (27). Both lines are now available through Jackson labs (IMSR Cat# JAX:30957, RRID:IMSR_JAX:30957 and IMSR Cat# JAX:30958, RRID:IMSR_JAX:30958). TRP1high mice were also crossed to CD45.1+ mice from Jackson labs (B6.SJL-Ptprca Pepcb/BoyJ, IMSR Cat# JAX:002014, RRID:IMSR_JAX:002014). C57BL/6-Tg(TcraTcrb)1100Mjb/J (OT-I) (IMSR Cat# JAX:003831, RRID:IMSR_JAX:003831) and C57BL/6 (IMSR Cat# JAX:000664, RRID:IMSR_JAX:000664) mice were purchased from Jackson Labs. Female mice aged 6–8 weeks and 18–25g were used throughout for both T cell donors and recipients to avoid immunogenicity of Y-chromosome encoded genes. In vitro experiments were performed on T cells isolated from pooled male and female mice.
Patient samples
For breast cancer patients, we obtained written informed consent from the patients, and the studies were conducted in accordance with the Declaration of Helsinki and the Belmont Report. Patient peripheral blood samples were obtained via Dana-Farber Cancer Institute Institutional Review Board protocols 18–258, 13–364, and/or 17–024. Anonymous healthy donor leukopacks were obtained from the Kraft Family Blood Donor Center at Dana-Farber Cancer Institute and Brigham and Women’s Hospital, protocol T0363. Breast cancer patients included both female and male patients. Age range 33–67. Inclusion criteria were diagnosis of breast cancer and availability of a pre-treatment blood sample. No patients were excluded. No patients were removed from the analysis. Healthy donors also included both males and females.
Cell lines
B16-F10 cells were purchased from the American Type Culture Collection (ATCC Cat# CRL-6475, RRID:CVCL_0159). B16-GVAX and B16-OVA cells (RRID:CVCL_WM78) were a gift from Glenn Dranoff. Cells were cultured in RPMI 1640 medium (Gibco) supplemented with 10% heat-inactivated FBS (Omega Scientific catalogue # FB-11), 2 mM L-glutamine (Gibco), penicillin G sodium (100 U/mL, Gibco), streptomycin sulfate (100 μg/mL, Gibco), 1 mM sodium pyruvate (Gibco), 0.1 mM nonessential amino acids (Gibco), and 0.1 mM β-mercaptoethanol (Sigma). Cells were passaged 2–6 times prior to use and were used for experiments at 80–90% confluency. Mycoplasma testing was performed by Charles River Laboratories every 6 months and was negative for the entire course of this study. No further authentication was performed. HEK 293T cells (ATCC Cat# CRL-3216, RRID:CVCL_0063) were purchased from ATCC and cultured in DMEM medium (Gibco) supplemented with 10% heat inactivated FBS, 2mM L-glutamine and 100 U/ml penicillin G.
Chemicals
All cell cycle inhibitors were titrated on CFSE labeled CD8 T cells to identify a dose that inhibited proliferation by approximately 50%. Mouse and human CD8 T cells showed similar responses to each drug; therefore, the same concentrations were used for both species. Palbociclib (PD 0332991, Sigma product #PZ0383) was used at 500nM. Abemaciclib (LY2835219, obtained from Eli Lilly Pharmaceuticals) was used at 100 nM. Ribociclib (LEE011, Selleckchem product #S7440) was used at 200 nM. CDK1/2i (RO-3306, Selleckchem product #S7747) was used at 4.5 μM. Aurora kinase inhibitor (MLN-8054, Selleckchem product #S1100) was used at 1 μM. Polo-like kinase inhibitor (GSK-461364, Selleckchem product #S2193) was used at 0.15 μM. CDK7i (YKL-5124) was developed by Nathanael Gray’s lab as reported (53) and used at 100 nM. CDK4/6 degrader (BJS 2–162) was developed by Nathanael Gray’s lab as reported (32) and used at 2 μM.
In vivo experiments
In vivo experiments were performed as described (54). Briefly, 3×105 B16-F10 cells (RRID:CVCL_0159) were inoculated by subcutaneous injection into the flank in 100μL of phosphate buffered saline (PBS). Tumors were allowed to grow for 6–7 days and then mice were randomized to different adoptive cell therapy treatment groups. Tumors were allowed to grow for an additional 6 days. Tumors were then surgically excised, minced with scissors, and incubated in RPMI containing digestion enzymes (Miltenyi tumor dissociation kit, catalogue # 130–096-730) at 37°C for 30 minutes. Tumors were filtered through a 40-micron cell strainer, washed with PBS, and centrifuged at 300g for 5 min. The resulting cell pellet containing tumor debris and infiltrating immune cells was resuspended in FACS buffer (PBS with 2% fetal calf serum) and stained with a master mix of antibodies described below. Spleens were crushed through a 40-micron cell strainer and erythrocytes removed with ACK lysis buffer (150mM NH4Cl, 10mM KHCO3, 0.1mM Na2EDTA). In some cases, mice were reinoculated after 30 days with B16-F10 (RRID:CVCL_0159) or B16-OVA (RRID:CVCL_WM78) cells (3×105) and tumor size was measured over time. Measurements were not blinded.
Flow cytometry
Cells from spleen, total tumor draining lymph nodes, and approximately 50mg tumor were incubated with extracellular staining mix including 2% fetal calf serum (FCS) for 30 minutes at 4°C, washed once in PBS, and either resuspended in 2% FCS and 1% EDTA in PBS for extracellular analysis only or were fixed, permeabilized, and stained with intracellular antibodies against specific cytokines (intracellular cytokine buffer kit from Biolegend catalogue # 421002). Analysis was performed on a BD Fortessa flow cytometer. Analysis was performed on CD45+ cells after gating out doublets (via SSC-A SSC-H gating) and dead cells (Zombie NIR Fixable Viability Kit, Biolegend catalogue # 423105). Flow cytometry antibodies were purchased from Biolegend: CD8 (clone 53–6.7 BioLegend Cat# 100728, RRID:AB_493426); CD25 (clone 3C7 BioLegend Cat# 101910, RRID:AB_2280288); CD44 (clone IM7 BioLegend Cat# 103028, RRID:AB_830785); CD45 (clone 30-F11 BioLegend Cat# 103147, RRID:AB_2564383); CD45.1 (clone A20 BioLegend Cat# 110706, RRID:AB_313495); granzyme B (clone QA16A02 BioLegend Cat# 372214, RRID:AB_2728381); IFNγ (clone XMG1.2 BioLegend Cat# 505829, RRID:AB_10897937); CD45.2 (clone Ly5–2 BioLegend Cat# 109806, RRID:AB_313443); CX3CR1 (clone SA011F11 BioLegend Cat# 149006, RRID:AB_2564315); human IL7Rα (clone A019D5 BioLegend Cat# 351332, RRID:AB_2562304); mouse IL7Rα (clone A7R34 BioLegend Cat# 135033, RRID:AB_2564576); human CD7 (clone 4H9 BioLegend Cat# 395604, RRID:AB_2820049); CD45RO (clone UCHL1 BioLegend Cat# 304206, RRID:AB_314422); CD45RA (clone HI100 BioLegend Cat# 304148, RRID:AB_2564157). Analysis was performed using Flow Jo software (RRID:SCR_008520).
Cell culturing, proliferation assay, and cytokine analysis
Primary cells were cultured in RPMI 1640 medium (Gibco) supplemented with 10% heat-inactivated FBS (Omega Scientific catalogue # FB-11), 2 mM L-glutamine (Gibco), penicillin G sodium (100 U/mL, Gibco), streptomycin sulfate (100 μg/mL, Gibco), 1 mM sodium pyruvate (Gibco), 0.1 mM nonessential amino acids (Gibco), and 0.1 mM β-mercaptoethanol (Sigma), further referred to as RPMI complete. CD8+ T cells were isolated from spleen and lymph nodes of TRP1high and TRP1low mice using EasySep™ Mouse CD8+ T Cell Isolation Kit (negative selection kit, StemCell catalogue # 19853) T cells were cultured in RPMI complete containing human IL-2 (100 U/mL Peprotech catalogue # 200–02-250UG; day 0) in the presence of CD3/CD28 beads (Dynabeads Mouse T Activator, Gibco catalogue # 11456). After coculture and treatment for 2 days, T cells were either adoptively transferred via tail vein injection, stained and analyzed by flow cytometry or RNA was isolated for downstream analyses (qPCR and RNAseq). For assessment of proliferation, cells were stained with Celltrace CFSE (Invitrogen catalogue # C34554) according to the manufacturer’s instructions. Proliferation index was calculated by dividing the mitotic events by the progenitors after 48h. CD8 effector T cells and cytotoxicity assays with cocultured B16-F10 cells were performed as previously reported (54).
Adoptive Transfer
CD8+ T cells were isolated from TRP1high CD45.1+ / TRP1low CD45.2+ or OTI CD45.2+ mice and treated with different cell cycle inhibitors in the presence of IL2 and CD3/CD28 beads as already described. After 48 hours, 2×106 CD45.1+CD8+ T cells were transferred by intravenous injection (150μL volume, diluted in sterile PBS) into sex matched CD45.1+ or CD45.2+ recipient mice respectively. For the longitudinal monitoring of persistence after transfer, mice were bled in the indicated intervals and flow cytometry was performed after red blood cell lysis and staining as described above. For the tumor rechallenge experiments, adoptive transfer was performed into B16 tumor-bearing mice at either 2 days or 5 days post tumor-inoculation as indicated in the figure legend. For some experiments, tumor size was monitored daily by precision calipers. For other experiments, tumor size was not measured and mice were euthanized 6 days post adoptive transfer, and spleens, tumor draining lymph nodes, and tumors were dissociated and analyzed by flow cytometry as described above. Transferred T cells were identified as CD45+CD8+CD45.1+ cells or CD45+CD8+CD45.2+ cells respectively.
Quantitative PCR
RNA from CDK4/6 inhibitor treated T cells was isolated as described above. RNA was transcribed to cDNA with the iScriptTM Reverse Transcription Supermix (BioRad catalogue # 10020178). Quantitative PCR was performed using the Sybr Green method (SsoAdvanced™ Universal SYBR® Green Supermix, BioRad cataloque # 1725271). All experiments were performed on a BioRad CF96 Cycler. Actin was used as the housekeeping gene. Results are presented as 2^-Delta Delta Ct values.
Mammalian vectors and RNA interference
Templates for shRNA sequences for targets Myc and MXD4 were designed on the Broad Institute Genetic Perturbation Platform (https://portals.broadinstitute.org/gpp/public/) and cloned into a retroviral vector expressing RFP (pMSCVURP-U6-(xx)-UbiC-RFP-2A-Puro, RRID:Addgene_28289) under control of the U6 promoter. Successful cloning was confirmed by Sanger sequencing. Retrovirus was produced in HEK 293T cells (RRID:CVCL_0063) in DMEM media. Cells were transfected with cloned vector and packaging vector (pCl Eco, RRID:Addgene_12371) at equal concentrations using the TransIT293™ (Mirus, cataloque # MIR2704) protocol according to manufactureŕs recommendations. Virus was harvested after 48h, filtered through a .45um filter and concentrated in a PEG8000 based buffer. After resuspension in DMEM, concentrated virus was flash frozen in liquid nitrogen and stored at −80°C or used directly for transduction.
Transduction of murine T cells
CD8 T cells were isolated and activated as described above. After 24h of activation, cells were transferred to 1.5 ml microcentrifuge tubes and resuspended in RPMI with 10% FCS containing 12μg/ml DEAE (1*10^6 cells/ml) and 100 μl of concentrated retrovirus was added. Spin transduction was performed at 6000g at 25°C for 90 minutes. Afterwards, supernatant was removed and cells were plated for another 48h with IL-2 and anti-CD3/CD28 beads. RFP positivity was assessed via flow cytometry on a BD Fortessa flow cytometer after 48hr and subsequently after adoptive transfer into recipient mice (see above) at the indicated timepoints.
Immunoblot
Protein was isolated using lab-made RIPA buffer. Protein content was then quantified using the Micro BCA™ Protein Assay Kit (Thermo Fisher catalogue #23235). Equal amounts of protein were then resolved on a 10% SDS PAGE after 5 minutes at 95°C in Laemmli buffer. After transfer the PVDF membrane was blocked in 5% BSA for 1 hour at room temperature, then exposed to primary antibody in 5% BSA over night at 4°C. Membranes were imaged after exposure to HRP-conjugated secondary antibody and enhanced chemiluminescence substrate (Perkin Elmer catalogue # NEL104001EA).
NFAT Signaling Index
Murine CD8 T cells were isolated as described above and activated with 50 ng/mL PMA and 1 μg/mL ionomycin in the presence of vehicle or 0.5 μM palbociclib. After 5 minutes of incubation at 37°C, an aliquot was deposited on a coverslip coated with poly-L-lysine as Time 0. After a further 5 minutes of incubation, cyclosporin A was added to the remaining cells to a final concentration of 1 μM. After 15, 35, and 55 minutes, an aliquot of cells were plated on the poly-L-lysine-coated coverslips. Slides were incubated for 5 minutes in a 37°C water bath then fixed with 4% formaldehyde for 15 minutes, at which point they were covered with PBS. Free aldehydes were quenched with 50 mM ammonium chloride in PBS before the slides were blocked and stained for NFAT1 (clone D43B1, secondary Anti-rabbit IgG AF633) and DAPI for immunofluorescence. Images were analyzed using Fiji (RRID:SCR_002285) to calculate the NFAT Signal Index as described previously (30).
Single-Cell RNA Sequencing
Peripheral blood monocytic cells (PBMC) were isolated from blood from healthy human donors (Kraft Family Blood Donor Center at Dana-Farber Cancer Institute and Brigham and Women’s Hospital, protocol #T0363) and matched samples from breast cancer patients before and on CDK4/6-inhibitor treatment or endocrine therapy using Ficoll™ gradient centrifugation. PBMCs were stained for CD8, CD45RA and CD45RO. The CD45RO+ CD45RA+ CD8+ population was sorted on a BD FACS Aria™ III after gating out doublets and dead cells as described above. A library was constructed from each sample using the Chromium Single Cell 5’ Kit (10x Genomics PN-1000006). In addition to the standard single-cell library, a VDJ-enriched library was created for each sample using a specialized Chromium kit (PN-1000005) for TCR sequencing. Libraries were sequenced on an Illumina HiSeq system generating paired-end 150bp reads. The 10x CellRanger pipeline (v3.0.2) was used to align reads to the GRCh38 reference genome and generate a single-cell feature count matrix for each library using default parameters. The count matrices were imported for downstream analysis into R using the “Seurat” package (v3.1.4). Genes expressed in fewer than 3 cells were discarded from further analysis. Barcodes were classified as cells if they satisfied the following criteria: reads detected in greater than 100 distinct genes, percentage of mitochondrial reads less than 2 MADs from the median, and total reads within 2 SDs of the mean. To focus our analysis on T cells, any cell with reads in the genes IGHM, IGHD, or CSF3R was removed from analysis. Data from each sample were log-normalized and combined into one batch-corrected expression matrix by Canonical Correlation Analysis (CCA). Counts were then scaled and subject to dimensionality reduction using Principal Component Analysis (PCA). Uniform Manifold Approximation and Projection (UMAP) embedding was generated from the top 14 dimensions of the PCA. Clusters were identified first by constructing a Shared Nearest Neighbor (SNN) graph based on each cell’s 20-nearest neighbors and then applying modularity refinement with the Louvain algorithm. Markers for each cluster were identified by comparing expression using the Wilcoxon rank sum test. For each cell, a “Myc Score” was calculated as follows: the established gene set “HALLMARK_MYC_TARGETS_V1” was obtained from the Molecular Signatures Database, its 200 members were narrowed down to the 193 genes that were measured by in our sequencing experiment, then for each gene the cell received an expression z-score based on the global mean and variance of that gene’s expression, and finally the single-gene z-scores were summed to yield the final score for that cell; cluster-level scores were computed as means of their constituent single-cell scores.
Fractional representation
To make the number of cells from different patients in each cluster comparable, we normalized the number of cells for each patient to the median of all patients’ cells in that cluster. Then the percentages of cells from different patients were calculated in each cluster and the two-sided Wilcoxon test was applied to obtain p values between pre-CDK4/6 and post-CDK4/6 groups and between healthy and post-CDK4/6 groups. No significant differences were observed across these comparisons.
RNA Velocity
Alignment products (BAM files) obtained from the single-cell RNA libraries described above were analyzed by the Python (RRID:SCR_001658) tool “velocyto” (v0.17.17) to generate spliced and unspliced expression matrices. These data were then processed using the package “scVelo” (v0.1.25): genes with less than 30 counts in both spliced and unspliced reads were excluded, the top 2,000 most variable genes were selected, filtered counts were log-normalized, per-cell velocities were estimated, and finally averaged vector fields were visualized as a stream plot overlaid on the UMAP generated as described above.
T-Cell Clonotype Analysis
T-Cell Receptor sequencing was performed per the 10x Genomics protocol described above. Cells expressing the V gene TRAV1–2 and any of the three J genes TRAJ33, TRAJ12, or TRAJ20 were labeled as Mucosal-Associated Invariant T (MAIT) cells and excluded from further analysis. Of the set of all unique CDR3 sequences (i.e. clonotypes), we considered only those that appeared in more than one cell. We then focused our analysis on clonotypes that appeared in both pre-treatment and on-treatment samples. To generate the clonotype sharing grid, we first counted the number of unique clonotypes that appeared in common between all possible cluster pairs for each CDK4/6 inhibitor treated patient, then summed the five resulting matrices together. Finally, each row was normalized by converting each of its values to a percentage of the total number of clonotypes in the cluster represented by that row.
Mouse Bulk RNA Sequencing
One million TRP1high or TRP1low CD8+ T cells were treated with CDK4/6 inhibitors or vehicle in the presence of CD3/CD28 beads and IL-2. Total RNA was prepared after 48h of treatment (Qiagen RNeasy Plus Mini Kit catalogue #74134). Library preparation using the KAPA mRNA HyperPrep Kit and sequencing on an Illumina NextSeq 500 was performed by the Dana-Farber Molecular Biology Core Facility. The resulting 75bp reads were trimmed for adapter sequences using the tool “cutadapt” (v2.9) then aligned to the GRCm38 genome with the corresponding ENSEMBL 97 annotation using “STAR” (v2.7.0f) (RRID:SCR_004463). Feature counting was performed using the R package “Rsubread” (v1.32.4), allowing for multi-mapping and multi-overlapping reads. Differential expression analysis was done using the package “DESeq2” (v1.22.2) (RRID:SCR_000154) with effective size moderation using “apeglm” (v1.8.0) (55). Gene sets for Gene Set Enrichment Analysis (GSEA) were obtained from the Molecular Signatures Database (MSigDB), loaded into R using the package “GSEABase” (v1.48.0), and analyzed using the package “fgsea” (v1.12.0).
Human Bulk RNA Sequencing
CD8 T-cells were isolated by negative selection from the blood of healthy donors and treated for 48 hours with palbociclib, abemaciclib, or vehicle. Total RNA was prepared after 48h of treatment (Qiagen RNeasy Plus Mini Kit catalogue #74134). Library preparation using the KAPA mRNA HyperPrep Kit and sequencing on an Illumina NextSeq 500 was performed by the Dana-Farber Molecular Biology Core Facility. The resulting 75bp reads were trimmed for adapter sequences using the tool “cutadapt” (v2.9) then aligned to the GRCh38 genome with the corresponding ENSEMBL 97 annotation using “STAR” (v2.7.0f) (RRID:SCR_004463). The remainder of the analysis proceeded identically to the mouse bulk RNA experiment described above.
ATAC Sequencing
CD8 T cells were isolated by negative selection from the blood of healthy donors and treated for 48 hours with palbociclib, abemaciclib, or vehicle. Library preparation and paired-end sequencing on an Illumina NextSeq 500 was performed by the Dana-Farber Center for Functional Cancer Epigenetics. The resulting 35bp reads were aligned to the GRCh38 genome using “Bowtie2” (v2.3.4.3) configured with the “very-sensitive” flag. Peaks were called for each sample using “Genrich” (v0.6), a custom peak-caller developed by the Harvard FAS Informatics group, with flags to discount mitochondrial reads and reads arising from PCR duplication. Consensus peaks were computed using the R package “GenomicRanges” (v1.38.0), and peaks that occurred in only 1 sample were discarded. Reads were assigned to each peak region using the package “Rsubread” (v1.32.4). Differential expression using “DESeq2” (v1.22.2) (RRID:SCR_000154) was performed on the resulting count matrix to identify differentially accessible regions (DARs) of chromatin between treatment conditions. Analysis for transcription factor motifs that are over- and underrepresented in DARs was done with HOMER (v4.10.3) (RRID:SCR_010881).
Statistics
Unpaired student’s t test with Welch’s correction was used for experiments with 2 groups, one-way ANOVA with Tukey’s post-hoc was used for experiments with 3 or more groups. Error bars are SEM, p values <0.05 were considered significant. Data were analyzed using Graphpad Prism software (RRID:SCR_002798) or R version 3.3.0. Experiments were replicated 3 times unless otherwise indicated in the figure legends.
Power calculation
From previous mouse experiments, the lowest difference between control and CDK4/6i treated groups AUC values had means of 35.1 vs. 50.5 with a SD of 8.65. Using a probability (power) of 0.95, and a 0.05 probability we will reject the null hypothesis, we estimated a sample size of 8 mice per group. In cases where mice underwent surgical removal of tumors, 10 mice per group were included to account for any losses related to surgery.
Data and code availability Bulk and single cell RNAseq data sets along with code used to analyze data are available here: doi:10.5281/zenodo.4781889
Supplementary Material
Statement of Significance.
CDK4/6 inhibition skews newly activated CD8 T cells toward a memory phenotype in mice and humans with breast cancer. CDK4/6 inhibitors may have broad utility outside breast cancer, particularly in the neoadjuvant setting to augment CD8 T cell priming to tumor antigens prior to dosing with checkpoint blockade.
Acknowledgements:
SKD and NSG were funded by the Hale Center for Pancreatic Cancer Research. SKD was funded by Eli Lilly, the Ludwig Center at Harvard, NIH U01 CA224146-01, NIH R01 AI158488-01, and is a Pew-Stewart Scholar in Cancer Research. MD was funded by a Mentored Clinical Scientist Development Award 1K08DK114563 – 01. KWW, GCY, MD, and SKD were funded by the Melanoma Research Alliance and American Cancer Society. SG was supported by the NHMRC of Australia (Investigator Grant GNT 1177357), Susan G. Komen (Career Catalyst Award CCR18547966), and the NIH SPORE in Breast cancer to the Dana-Farber/Harvard Cancer Centre (P50 CA168504). MH was funded by the German Research Foundation (DFG; project number: 398222819). LQ was funded by a SITC-Bristol Myers Squibb Postdoctoral Cancer Immunotherapy Translational Fellowship. E.C-T. and PL were funded by NIH T32CA207021. PL was funded by the Medical Scientist Training Program at Harvard. We thank Michael Manos and the DFCI Center for Immune-Oncology for sample processing. We thank the DFCI Center for Functional Cancer Epigenetics for ATACseq, and the DFCI Center for Cancer Immunology Research for the 10x Genomics pipeline.
Conflict of interest: SKD received research funding from Eli Lilly and Company for this project. SKD received unrelated research funding from Novartis Pharmaceuticals and Bristol-Myers Squibb and is a founder, science advisory board member (SAB) and equity holder in Kojin. MD is a science advisory board member for Neoleukin. NSG is a founder, science advisory board member (SAB) and equity holder in Gatekeeper, Syros, Petra, C4, B2S, Aduro and Soltego (board member). The Gray lab receives or has received research funding from Novartis, Takeda, Astellas, Taiho, Janssen, Kinogen, Voronoi, Her2llc, Deerfield and Sanofi. SG received research funding from Eli Lilly and G1 Therapeutics and has served on advisory boards for Eli Lilly, G1 Therapeutics, Pfizer, and Novartis. SMT reported receiving institutional research support from Merck, Bristol-Myers Squibb, Exelixis, Eli Lilly, Pfizer, Novartis, AstraZeneca, Eisai, Nektar, Odonate, Sanofi, and Genentech and has served on advisory boards for Genentech, Eli Lilly, Novartis, Pfizer, Nektar, Immunomedics, Nanostring, Daiichi-Sankyo, Bristol-Meyers Squibb, Sanofi, Athenex, AstraZeneca, Gilead, Eisai, Puma, and Merck.
References
- 1.Dougan MD,G; Dougan SK Cancer Immunotherapy: Beyond Checkpoint Blockade. Annual Review of Cancer Biology. 2019;3(1):55–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huang AC, Postow MA, Orlowski RJ, Mick R, Bengsch B, Manne S, et al. T-cell invigoration to tumour burden ratio associated with anti-PD-1 response. Nature. 2017;545(7652):60–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pedicord VA, Montalvo W, Leiner IM, and Allison JP. Single dose of anti-CTLA-4 enhances CD8+ T-cell memory formation, function, and maintenance. Proc Natl Acad Sci U S A. 2011;108(1):266–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hodi FS, O’Day SJ, McDermott DF, Weber RW, Sosman JA, Haanen JB, et al. Improved survival with ipilimumab in patients with metastatic melanoma. N Engl J Med. 2010;363(8):711–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kaech SM, and Ahmed R. Memory CD8+ T cell differentiation: initial antigen encounter triggers a developmental program in naive cells. Nat Immunol. 2001;2(5):415–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Backer RA, Hombrink P, Helbig C, and Amsen D. The Fate Choice Between Effector and Memory T Cell Lineages: Asymmetry, Signal Integration, and Feedback to Create Bistability. Adv Immunol. 2018;137:43–82. [DOI] [PubMed] [Google Scholar]
- 7.Henrickson SE, Perro M, Loughhead SM, Senman B, Stutte S, Quigley M, et al. Antigen availability determines CD8(+) T cell-dendritic cell interaction kinetics and memory fate decisions. Immunity. 2013;39(3):496–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Munitic I, Ryan PE, and Ashwell JD. T cells in G1 provide a memory-like response to secondary stimulation. J Immunol. 2005;174(7):4010–8. [DOI] [PubMed] [Google Scholar]
- 9.Sukumar M, Liu J, Ji Y, Subramanian M, Crompton JG, Yu Z, et al. Inhibiting glycolytic metabolism enhances CD8+ T cell memory and antitumor function. J Clin Invest. 2013;123(10):4479–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van der Windt GJ, O’Sullivan D, Everts B, Huang SC, Buck MD, Curtis JD, et al. CD8 memory T cells have a bioenergetic advantage that underlies their rapid recall ability. Proc Natl Acad Sci U S A. 2013;110(35):14336–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Araki K, Turner AP, Shaffer VO, Gangappa S, Keller SA, Bachmann MF, et al. mTOR regulates memory CD8 T-cell differentiation. Nature. 2009;460(7251):108–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van der Windt GJ, Everts B, Chang CH, Curtis JD, Freitas TC, Amiel E, et al. Mitochondrial respiratory capacity is a critical regulator of CD8+ T cell memory development. Immunity. 2012;36(1):68–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kakaradov B, Arsenio J, Widjaja CE, He Z, Aigner S, Metz PJ, et al. Early transcriptional and epigenetic regulation of CD8(+) T cell differentiation revealed by single-cell RNA sequencing. Nat Immunol. 2017;18(4):422–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Arsenio J, Kakaradov B, Metz PJ, Kim SH, Yeo GW, and Chang JT. Early specification of CD8+ T lymphocyte fates during adaptive immunity revealed by single-cell gene-expression analyses. Nat Immunol. 2014;15(4):365–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Verbist KC, Guy CS, Milasta S, Liedmann S, Kaminski MM, Wang R, et al. Metabolic maintenance of cell asymmetry following division in activated T lymphocytes. Nature. 2016;532(7599):389–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chang JT, Palanivel VR, Kinjyo I, Schambach F, Intlekofer AM, Banerjee A, et al. Asymmetric T lymphocyte division in the initiation of adaptive immune responses. Science. 2007;315(5819):1687–91. [DOI] [PubMed] [Google Scholar]
- 17.Pollizzi KN, Sun IH, Patel CH, Lo YC, Oh MH, Waickman AT, et al. Asymmetric inheritance of mTORC1 kinase activity during division dictates CD8(+) T cell differentiation. Nat Immunol. 2016;17(6):704–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kretschmer L, Flossdorf M, Mir J, Cho YL, Plambeck M, Treise I, et al. Differential expansion of T central memory precursor and effector subsets is regulated by division speed. Nat Commun. 2020;11(1):113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kinjyo I, Qin J, Tan SY, Wellard CJ, Mrass P, Ritchie W, et al. Real-time tracking of cell cycle progression during CD8+ effector and memory T-cell differentiation. Nat Commun. 2015;6:6301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Singh A, Jatzek A, Plisch EH, Srinivasan R, Svaren J, and Suresh M. Regulation of memory CD8 T-cell differentiation by cyclin-dependent kinase inhibitor p27Kip1. Mol Cell Biol. 2010;30(21):5145–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang J, Bu X, Wang H, Zhu Y, Geng Y, Nihira NT, et al. Cyclin D-CDK4 kinase destabilizes PD-L1 via cullin 3-SPOP to control cancer immune surveillance. Nature. 2018;553(7686):91–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schaer DA, Beckmann RP, Dempsey JA, Huber L, Forest A, Amaladas N, et al. The CDK4/6 Inhibitor Abemaciclib Induces a T Cell Inflamed Tumor Microenvironment and Enhances the Efficacy of PD-L1 Checkpoint Blockade. Cell Rep. 2018;22(11):2978–94. [DOI] [PubMed] [Google Scholar]
- 23.Goel S, DeCristo MJ, Watt AC, BrinJones H, Sceneay J, Li BB, et al. CDK4/6 inhibition triggers anti-tumour immunity. Nature. 2017;548(7668):471–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Deng J, Wang ES, Jenkins RW, Li S, Dries R, Yates K, et al. CDK4/6 Inhibition Augments Antitumor Immunity by Enhancing T-cell Activation. Cancer Discov. 2018;8(2):216–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rugo HSKP, Beck JT, Chisamore MJ, Hossain A, Chen Y, Tolaney SM. A phase Ib study of abemaciclib in combination with pembrolizumab for patients with hormone receptor positive (HR+), human epidermal growth factor receptor 2 negative (HER2-) locally advanced or metastatic breast cancer (MBC) (NCT02779751): Interim results. Journal of Clinical Oncology. 2020;38(15):1.31682550 [Google Scholar]
- 26.Clancy-Thompson E, Devlin CA, Tyler PM, Servos MM, Ali LR, Ventre KS, et al. Altered Binding of Tumor Antigenic Peptides to MHC Class I Affects CD8(+) T Cell-Effector Responses. Cancer Immunol Res. 2018;6(12):1524–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dougan SK, Dougan M, Kim J, Turner JA, Ogata S, Cho HI, et al. Transnuclear TRP1-specific CD8 T cells with high or low affinity TCRs show equivalent antitumor activity. Cancer Immunol Res. 2013;1(2):99–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gerlach C, Moseman EA, Loughhead SM, Alvarez D, Zwijnenburg AJ, Waanders L, et al. The Chemokine Receptor CX3CR1 Defines Three Antigen-Experienced CD8 T Cell Subsets with Distinct Roles in Immune Surveillance and Homeostasis. Immunity. 2016;45(6):1270–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Crowley SJ, Bruck PT, Bhuiyan MA, Mitchell-Gears A, Walsh MJ, Zhangxu K, et al. Neoleukin-2 enhances anti-tumour immunity downstream of peptide vaccination targeted by an anti-MHC class II VHH. Open Biol. 2020;10(2):190235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marangoni F, Murooka TT, Manzo T, Kim EY, Carrizosa E, Elpek NM, et al. The transcription factor NFAT exhibits signal memory during serial T cell interactions with antigen-presenting cells. Immunity. 2013;38(2):237–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martinez GJ, Pereira RM, Aijo T, Kim EY, Marangoni F, Pipkin ME, et al. The transcription factor NFAT promotes exhaustion of activated CD8(+) T cells. Immunity. 2015;42(2):265–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jiang B, Wang ES, Donovan KA, Liang Y, Fischer ES, Zhang T, et al. Development of Dual and Selective Degraders of Cyclin-Dependent Kinases 4 and 6. Angew Chem Int Ed Engl. 2019;58(19):6321–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Conacci-Sorrell M, McFerrin L, and Eisenman RN. An overview of MYC and its interactome. Cold Spring Harb Perspect Med. 2014;4(1):a014357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pernas S, Tolaney SM, Winer EP, and Goel S. CDK4/6 inhibition in breast cancer: current practice and future directions. Ther Adv Med Oncol. 2018;10:1758835918786451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.La Manno G, Soldatov R, Zeisel A, Braun E, Hochgerner H, Petukhov V, et al. RNA velocity of single cells. Nature. 2018;560(7719):494–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gowthaman R, and Pierce BG. TCR3d: The T cell receptor structural repertoire database. Bioinformatics. 2019;35(24):5323–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Luoma AM, Suo S, Williams HL, Sharova T, Sullivan K, Manos M, et al. Molecular Pathways of Colon Inflammation Induced by Cancer Immunotherapy. Cell. 2020;182(3):655–71 e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mingueneau M, Kreslavsky T, Gray D, Heng T, Cruse R, Ericson J, et al. The transcriptional landscape of alphabeta T cell differentiation. Nat Immunol. 2013;14(6):619–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Heinzel S, Binh Giang T, Kan A, Marchingo JM, Lye BK, Corcoran LM, et al. A Myc-dependent division timer complements a cell-death timer to regulate T cell and B cell responses. Nat Immunol. 2017;18(1):96–103. [DOI] [PubMed] [Google Scholar]
- 40.Vasilevsky NA, Ruby CE, Hurlin PJ, and Weinberg AD. OX40 engagement stabilizes Mxd4 and Mnt protein levels in antigen-stimulated T cells leading to an increase in cell survival. Eur J Immunol. 2011;41(4):1024–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Riddell M, Nakayama A, Hikita T, Mirzapourshafiyi F, Kawamura T, Pasha A, et al. aPKC controls endothelial growth by modulating c-Myc via FoxO1 DNA-binding ability. Nat Commun. 2018;9(1):5357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lu Y, Wu Y, Feng X, Shen R, Wang JH, Fallahi M, et al. CDK4 deficiency promotes genomic instability and enhances Myc-driven lymphomagenesis. J Clin Invest. 2014;124(4):1672–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kime L, and Wright SC. Mad4 is regulated by a transcriptional repressor complex that contains Miz-1 and c-Myc. Biochem J. 2003;370(Pt 1):291–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Piluso D, Bilan P, and Capone JP. Host cell factor-1 interacts with and antagonizes transactivation by the cell cycle regulatory factor Miz-1. J Biol Chem. 2002;277(48):46799–808. [DOI] [PubMed] [Google Scholar]
- 45.Savas P, Virassamy B, Ye C, Salim A, Mintoff CP, Caramia F, et al. Single-cell profiling of breast cancer T cells reveals a tissue-resident memory subset associated with improved prognosis. Nat Med. 2018;24(7):986–93. [DOI] [PubMed] [Google Scholar]
- 46.Buggert M, Nguyen S, Salgado-Montes de Oca G, Bengsch B, Darko S, Ransier A, et al. Identification and characterization of HIV-specific resident memory CD8(+) T cells in human lymphoid tissue. Sci Immunol. 2018;3(24). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Slamon DJ, Neven P, Chia S, Fasching PA, De Laurentiis M, Im SA, et al. Overall Survival with Ribociclib plus Fulvestrant in Advanced Breast Cancer. N Engl J Med. 2020;382(6):514–24. [DOI] [PubMed] [Google Scholar]
- 48.Sledge GW Jr., Toi M, Neven P, Sohn J, Inoue K, Pivot X, et al. MONARCH 2: Abemaciclib in Combination With Fulvestrant in Women With HR+/HER2- Advanced Breast Cancer Who Had Progressed While Receiving Endocrine Therapy. J Clin Oncol. 2017;35(25):2875–84. [DOI] [PubMed] [Google Scholar]
- 49.Goetz MP, Toi M, Campone M, Sohn J, Paluch-Shimon S, Huober J, et al. MONARCH 3: Abemaciclib As Initial Therapy for Advanced Breast Cancer. J Clin Oncol. 2017;35(32):3638–46. [DOI] [PubMed] [Google Scholar]
- 50.Sledge GW Jr., Toi M, Neven P, Sohn J, Inoue K, Pivot X, et al. The Effect of Abemaciclib Plus Fulvestrant on Overall Survival in Hormone Receptor-Positive, ERBB2-Negative Breast Cancer That Progressed on Endocrine Therapy-MONARCH 2: A Randomized Clinical Trial. JAMA Oncol. 2019; September29;6(1):116–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Johnston S, Martin M, Di Leo A, Im SA, Awada A, Forrester T, et al. MONARCH 3 final PFS: a randomized study of abemaciclib as initial therapy for advanced breast cancer. NPJ Breast Cancer. 2019;5:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Turner NC, Slamon DJ, Ro J, Bondarenko I, Im SA, Masuda N, et al. Overall Survival with Palbociclib and Fulvestrant in Advanced Breast Cancer. N Engl J Med. 2018;379(20):1926–36. [DOI] [PubMed] [Google Scholar]
- 53.Zhang H, Christensen CL, Dries R, Oser MG, Deng J, Diskin B, et al. CDK7 Inhibition Potentiates Genome Instability Triggering Anti-tumor Immunity in Small Cell Lung Cancer. Cancer Cell. 2020;37(1):37–54 e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tyler PM, Servos MM, de Vries RC, Klebanov B, Kashyap T, Sacham S, et al. Clinical Dosing Regimen of Selinexor Maintains Normal Immune Homeostasis and T-cell Effector Function in Mice: Implications for Combination with Immunotherapy. Molecular cancer therapeutics. 2017;16(3):428–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhu A, Ibrahim JG, and Love MI. Heavy-tailed prior distributions for sequence count data: removing the noise and preserving large differences. Bioinformatics. 2019;35(12):2084–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.