Abstract
Determining the processes that drive the evolution of pathogen host range can inform our understanding of disease dynamics and the potential for host-shifts. In natural populations, patterns of host range could be driven by genetically based differences in pathogen infectivity or ecological differences in host availability. In northwestern Italy, four reproductively isolated lineages of the fungal plant-pathogen Microbotryum have been shown to co-occur on several species in the genus Dianthus. We carried out cross-inoculation experiments to determine whether patterns of realized host range in these four lineages were driven by differences in infectivity and to test whether there was evidence of a trade-off between host range and within-host reproduction. We found strong concordance between field patterns of host range and pathogen infectivity on different Dianthus species using experimental inoculation, indicating that infection ability is a major driving force of host range. However, we found no evidence of a trade-off between the ability to infect a wider range of host species and spore production on a shared host.
INTRODUCTION
The host range of a pathogen, defined broadly here by the number of host species that a given pathogen species infects, is a critically important pathogen trait that can impact epidemiology (Cleaveland et al. 2001; Poulin and Keeney 2008), host community composition (Power and Mitchell 2004; Maloney et al. 2005; Alexander 2010; Mordecai 2011) and the risk of pathogen spillover into crop, livestock and human populations (Cleaveland et al. 2001; Woolhouse et al. 2005; Gilbert and Webb 2007). A distinction can be made between the ‘realized host range’ defined as the range of host species the pathogen is observed to infect in nature, and the ‘potential host range’ defined as the range of host species that a given pathogen can infect under experimental conditions. The latter is particularly critical for assessing risks of host shifts and novel disease emergence (Woolhouse et al. 2005; Gilbert and Webb 2007, Antonovics et al. 2013). Determining the processes that drive the evolution of pathogen host range in natural populations has the potential to inform predictions of disease dynamics and the development of control measures.
Modern molecular phylogenetics has reshaped our understanding of the realized host range of many pathogens. Many pathogens with a similar morphology and which were once thought to be generalists on a wide range of host species, have been shown to be species complexes, composed of multiple cryptic species each specialized to a small number of hosts (Szabo 2006; Le Gac et al. 2007; Malenke et al. 2009; Rouxel et al. 2013; Vialle et al. 2013). However, phylogenetic methods alone may be insufficient for determining a pathogen’s potential host range if sampling of host diversity is limited. For example, a pathogen may appear to be specialized if only one host species is well sampled. Experimental cross-inoculation studies can help bridge the gap between pattern and process by teasing apart infection ability from host availability (Poulin and Keeney 2008). Indeed, in most cases, inoculation studies have found that the potential host range of pathogens is greater than that observed in nature (Ebert 1998; King and Cable 2007; de Vienne et al. 2009; Parker et al. 2015). However, realized host ranges can be constrained if contact between host species is limited at fine spatial scales (Pedersen and Davies 2009, Stephens et al. 2019).
A key issue in the evolution of host range has been determining the factors that favor the evolution of specialists vs. generalists. One widely hypothesized mechanism is that a trade-off occurs in which pathogen traits that increase transmission within one species come at cost of reduced transmission on the other species (Fry 1990; Jaenike 1990; Kawecki 1994; Joshi and Thompson 1995; Ebert 1998). Evidence of such costs are exemplified by experimental evolution studies, that show a reduction in parasite fitness following several generations of selection on a novel host (Ebert et al. 1998, Bono et al. 2016). In natural populations, support for the trade-off hypothesis comes from observations of higher abundance of specialists compared to generalists on their shared hosts (Straub et al. 2011; Mederios et al. 2014). However, trade-offs are not always evident (Hellgren et al. 2009, Bedhomme et al. 2012). Moreover, the magnitude of trade-offs required to facilitate the evolution of host specialization will also depend on host community composition (Poisot et al. 2011; Asplen et al. 2012) and environmental heterogeneity (Bono et al. 2016). Thus, the trade-off approach to host breadth is best utilized when measures of trade-offs can be combined with knowledge of relative host abundance.
Anther-smut (caused by Basidiomycete fungi in the genus Microbotryum) is a pollinator-transmitted sterilizing disease that has become a model system for disease ecology (Bernasconi et al. 2009) and pathogen speciation (Giraud et al. 2008, 2010; Refrégier et al. 2008; Devier et al. 2010; Büker et al. 2013). Hosts infected with anther-smut produce fungal spores in place of pollen, and the ovary is also sterilized. The infection is systemic and can only be sustained on perennial hosts (Thrall et al. 1993; Hood et al. 2010). The disease is found on a wide range of plant species, most commonly in the Caryophyllaceae (Hood et al. 2010; Kido and Hood 2019). While the disease was initially described as being caused by a single widespread species, Microbotryum violaceum (=Ustilago violacea), this is now known to be a species complex with multiple host-specific pathogen lineages (Lutz et al. 2005; Le Gac et al. 2007; Kemler et al. 2013). Microbotryum species infecting hosts in the genus Silene are highly specialized; each pathogen species is generally found on just a single host species (Le Gac et al. 2007). Moreover, this specificity is generally maintained even in sympatry with other closely related host species (Hood et al. 2019; Tang et al. 2019), although occasional host shifts have been identified (Antonovics et al. 2002; Refregier et al. 2008; Tyson et al. 2018; Hood et al. 2019).
Here, we measured the infectivity and disease-related traits associated with pathogen fecundity (i.e., number of diseased flowers and spore production) for anther-smut pathogens on Dianthus species that vary in their realized host range in nature. The anther-smuts found on Dianthus are an exception to the general rule of strict specificity of Microbotryum; we studied the infection potential of four distinct putative species (hereafter referred to as ‘lineages’) of the anther-smut pathogen associated with Dianthus host species in the Mediterranean Alps (Petit et al. 2017). These four lineages have highly overlapping geographic and realized host ranges but are maintained through substantial post-zygotic isolation mechanisms in the form of post-meiotic F1 sterility (Petit et al. 2017). Three of the four lineages were found on two or more host species, indicative of some generalism, while one lineage occurred only on a single host species, D. pavonius, indicative of specialization. However, such patterns of host range could also be affected by host geography and variation in ecological opportunity for cross-species transmission, as there is large variation in the degree of overlap of host species (Bruns et al. 2018).
In this study, we investigate three research questions about the realized and potential host range of Microbotryum lineages on Dianthus:
Is there concordance between the experimental infectivity of each pathogen lineage and its observed distribution on Dianthus host species in nature?
Do generalist pathogen lineages have lower infectivity and spore production than specialist lineages on their shared host species?
Does the infection ability of generalist and specialist pathogen lineages vary across host populations within a host species?
To answer these questions, we carried out two greenhouse cross-inoculation experiments. The first experiment tested the infection ability and sporulation capacity of the four fungal lineages described by Petit et al. (2017) on three different Dianthus host species, to determine the potential host range and fitness of each pathogen (Questions 1 and 2). The second experiment tested the infection ability of the most specialized and most generalized lineage on multiple different populations of a single host species, Dianthus seguieri, to determine the ability of generalists and specialist to infect variable populations within a single host species (Question 3).
METHODS
Study system
The Western Mediterranean Alps are a hot spot of diversity for Dianthus (Valente et al. 2010). We recently surveyed host and pathogen abundance across 59 elevational transects in this region of the Alps (Bruns et al. 2018) and found anther-smut disease on seven perennial species: D. pavonius, D. furcatus, D. seguieri, D. sylvestris, D. superbus, D. carthusianorum, D. deltoides. Here we focus on the three most abundant species, D. pavonius, D. furcatus, and D. seguieri.
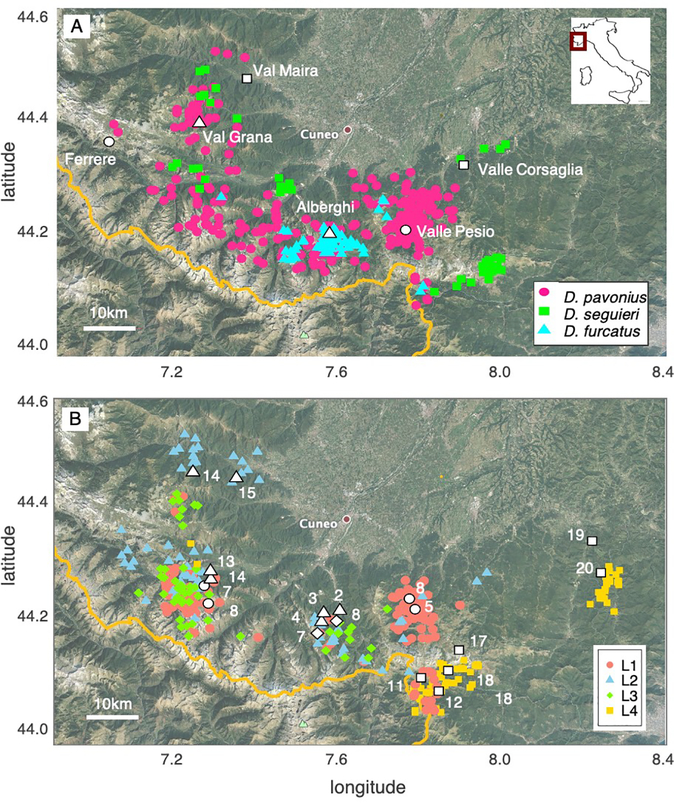
Dianthus pavonius is common in alpine meadow habitats from 1500m-2300m. In this region, it is the most abundant of the three species: it was present in 80% of transects surveyed in the large-scale study of the distribution of hosts and anther-smut disease (Fig. 1A; Bruns et al. 2018). Dianthus furcatus, has an elevational range (1400m-2100m) that overlaps with D. pavonius, and these species are frequently found in sympatry, in some cases, only a few meters apart (e.g., see the Alberghi population in Fig. 1A). However, D. furcatus has a more limited range than D. pavonius and was present in just 20% of surveyed transects (Bruns et al. 2018). Dianthus furcatus is largely absent from the easternmost populations including the Valle Pesio population.
Fig. 1.
Distribution and sampling locations for pathogen and host accessions used in Experiment 1. A) Distribution of three Dianthus host species (data from Bruns et al. 2018). Colors and shapes indicate host species identity. Points were jittered randomly to minimize overlap. The six locations where seeds were collected are shown in the brighter outlined shapes and labeled by locality. The yellow line is the border with France (where we do not have distribution data). The inset shows the general location in Italy. The closest city, Cuneo, is marked for reference. Inset: the general location in Italy. B) Distribution of the four Microbotryum lineages (L1-L4) in the Mediterranean Alps (data from Petit et al. 2017). The location of the 19 Microbotryum isolates tested in Experiment 1 are shown in white (see Table 1).
The third host species, D. seguieri is found at lower elevations between 500–1200m. We have only encountered close sympatry between D. seguieri and D. pavonius once, but they are frequently found in the same valley systems, separated by ca. 3–5km. We have never found sympatry between D. seguieri and D. furcatus (closest known locations 5km apart).
Petit et al. (2017) identified four unique genetic lineages of Microbotryum infecting these focal Dianthus species in this same region, based on 11 microsatellite markers and sequence data from 2 nuclear genes. One of these (‘Lineage 1’) was only ever found on D. pavonius plants in the field and thus appeared to be highly specialized. The other pathogen lineages showed a wider host range: Lineage 2 was found on all three Dianthus species, Lineage 3 was found on D. pavonius and D. furcatus, and Lineage 4 was found on D. pavonius and D. seguieri. Petit et al. (2017) also found that the distributions of the lineages varied in space: with Lineage 2 was more common in the northwest, while Lineage 4 was only found in the southeast (Fig. 1B).
Overview of experiments:
We carried out two cross-inoculation experiments. In Experiment 1 (Table 1A), we tested the infection ability and sporulation capacity of the four different pathogen lineages (with 2–7 pathogen isolates per lineage) against three different Dianthus host species (with 2 populations per host species). Due to a low sample size in the host D. seguieri, this experiment included a second round of inoculations on this host in a follow up inoculation trial (Table S1), using a subset of isolates from each of the four lineages, to test repeatability (We did not have enough D. furcatus seeds for a repeat inoculation). In Experiment 2 (Table 1B) we aimed to determine whether the specificity of the tested pathogen lineages was consistent across host populations within a species. We inoculated an additional eight populations of a single host species, D. seguieri, with a single isolate from pathogen Lineage 1 and a single isolate from Lineage 2.
Table 1.
Design and sample size (number of plants that survived and flowered) of the two cross-inoculation experiments. A) Experiment 1. B) Experiment 2, two pathogen lineages inoculated onto eight different populations of D. seguieri.
| A) Experiment 1 | ||||||||
|
| ||||||||
| Host population locations | Number of surviving plants that flowered | |||||||
|
| ||||||||
| Line age 1 | Lineage 2 | Line age 3 | Lineage 4 | |||||
|
Host
species |
Host pop | Lat. |
Lon
g. |
Num. Families sampled | (6,7,8)1 | (2,3,4,13,14,15,16) | (9,10) | (11,12,17,18,19,20) |
|
| ||||||||
| Dianthus pavonius | Valle | 44.18 | 7.68 | 20 | 30 | 81 | 17 | 53 |
| Pesio | 986 | 991 | ||||||
| Ferrere | 44.36 | 6.92 | 8 | 15 | 34 | 8 | 32 | |
| 022 | 607 | |||||||
|
| ||||||||
| Dianthus seguieri | Val | 44.48 | 7.28 | 5 | ||||
| Maira | 215 | 111 | 6 | 27 | 3 | 27 | ||
| Valle | 44.31 | 7.83 | 3 | |||||
| Corsaglia | 532 | 958 | 2 | 24 | 7 | 7 | ||
|
| ||||||||
| Dianthus furcatus | Alberghi | 44.18 | 7.49 | 4 | 3 | 5 | 2 | 8 |
| 235 | 404 | |||||||
| Valle | 44.39 | 7.15 | 6 | 3 | 4 | 4 | 8 | |
| Grana | 608 | 869 | ||||||
|
| ||||||||
| B) Experiment 2 | ||||||||
|
| ||||||||
| Dianthus seguieri host population locations | Number of surviving plants that flowered | |||||||
|
| ||||||||
| Population number | Location | Latitude | Longitude | Lineag e 12 | Lineag e 2 | |||
|
| ||||||||
| 24 | Val Maira | 44.4447 | 7.1784 | 33 | 12 | |||
| 32 | Valle Grana | 44.3638 | 7.2077 | 40 | 19 | |||
| 42 | Val Maira | 44.4121 | 7.2633 | 63 | 66 | |||
| 30 | Valle Gesso | 44.2689 | 7.4075 | 80 | 79 | |||
| 6 | San Bernardo di Mendatica | 44.1128 | 7.9199 | 39 | 16 | |||
| 7 | San Bernardo di Mendatica | 44.1078 | 7.923 | 27 | 16 | |||
| 36 | Valle Corsaglia | 44.3397 | 7.9434 | 60 | 70 | |||
| 38 | Valle Corsaglia | 44.3395 | 7.9434 | 65 | 48 | |||
The identity of replicate Microbotryum isolates within each lineage are listed in parenthesis. Information on these isolates is found in Table 2
Only a single isolate of each Lineage 1 and Lineage 2 were used in Experiment 2 and these differed from the ones used in Experiment 1 above. See text for details.
Experiment 1: Host species by pathogen lineage
Pathogen isolates
It would have been ideal to obtain all 12 combinations of host-of-origin and pathogen lineage, but a number of these were unavailable due to field specificities; for example, Lineage 1 was only found on D. pavonius (Petit et al. 2017). We eventually selected 19 pathogen isolates (Table 2; Fig. 1B) collected in July 2011 from the three focal host species in the Mediterranean Alps (10 from D. pavonius, 1 from D. furcatus, and 8 from D. seguieri). Microsatellite cluster analysis assigned these isolates to four distinct fungal lineages (Petit at al. 2017), all except two of these isolates (12 and 15) had <80% cluster assignment (Table 2). We therefore ran analyses with and without the inclusion of these two isolates (Supplemental Material S2). In total, there were 7 combinations of host-of-origin by fungal lineage out of a possible 12.
Table 2.
Lineage identity, host of origin, and collection location for 19 Microbotryum isolates used in the cross-inoculation studies on three host species: Dianthus pavonius, D. seguieri, and D. furcatus.
| Path. Lineage | Isolate1 | Lineage assignment score2 | Host of origin | Location name | Host collected from same location | Lat | Lon |
|---|---|---|---|---|---|---|---|
| Lineage 1 | 5 | 0.982 | D. pavonius | Valle Pesio | Yes | 44.2085 | 7.7182 |
| 6* | 0.979 | D. pavonius | Valle Pesio | Yes | 44.2288 | 7.7035 | |
| 7* | 0.824 | D. pavonius | Rio Freddo | No | 44.2196 | 7.1855 | |
| 8 | 0.979 | D. pavonius | Rio Freddo | No | 44.2538 | 7.1751 | |
| Lineage 2 | 2* | 0.976 | D. furcatus | Alberghi | Yes | 44.1745 | 7.4955 |
| 3* | 0.96 | D. pavonius | Alberghi | Yes | 44.1705 | 7.4957 | |
| 4 | 0.917 | D. pavonius | Alberghi | Yes | 44.1705 | 7.4957 | |
| 13* | 0.975 | D. seguieri | Val Maira | Yes | 44.4822 | 7.2811 | |
| 14 | 0.983 | D. seguieri | Val Maira | Yes | 44.5087 | 7.1289 | |
| 15* | 0.747* | D. seguieri | Rio Freddo | No | 44.2960 | 7.1571 | |
| 16 | 0.965 | D. seguieri | Rio Freddo | No | 44.2960 | 7.1592 | |
| Lineage 3 | 9* | 0.885 | D. pavonius | Alberghi | Yes | 44.1677 | 7.4953 |
| 10 | 0.9479 | D. pavonius | Alberghi | Yes | 44.1677 | 7.4953 | |
| Lineage 4 | 11* | 0.872 | D. pavonius | San Bernardo di Mendatica | No | 44.0511 | 7.7472 |
| 12 | 0.789* | D. pavonius | San Bernardo di Mendatica | No | 44.0566 | 7.7360 | |
| 17* | 0.98 | D. seguieri | San Bernardo di Mendatica | No | 44.0936 | 7.8278 | |
| 18 | 0.976 | D. seguieri | San Bernardo di Mendatica | No | 44.0953 | 7.8035 | |
| 19 | 0.975 | D. seguieri | Lago di Osiglia | No | 44.3106 | 8.2013 | |
| 20* | 0.988 | D. seguieri | Lago di Osiglia | No | 44.3106 | 8.2013 |
Asterisks indicate isolates that were also used in Experiment 2
Probability of belonging to the assigned lineage in Structure analysis (Petit et al. 2107).
Asterisks indicate isolates with scores <80%, that may be hybrids.
Host species
Seeds were collected in late August 2011, from six different geographic locations including two locations (“populations”) per host species (Table 2; Fig. 1A). Where possible, we tried to collect seed from the same populations where spores were collected earlier in the season, however, this was not always possible since some populations had already dispersed their seeds. Within each population seeds were collected from 4 to 20 plants and mixed together to form a bulk population. The number of available seeds varied considerably, due to a combination of host density, temporal variation in seed set, and seed predation.
To ensure even germination, seeds were started in 150mm culture dishes filled with 0.75% Agar media, amended with a 10% Murishage and Skoog plant nutrient medium. Prior to starting, seeds were surface sterilized for two 3-minute washes in 20% commercial bleach, 10% ethanol solution and rinsed 5 times in sterile distilled water. Each seed was then nicked with a sterile razor blade to break the seed coat. Plates were incubated in a growth chamber with 12 h days at 20°C day and 18°C night temperatures.
Inoculations
To standardize inoculation, teliospores from each pathogen isolate were placed on potato dextrose agar media and germination rate was scored after incubation at room temperature for 24 hours. We used these germination rates to make spore suspensions that contained the equivalent of 500 viable spores/uL for each of the 19 isolates. Solutions were made with sterile water and a drop of surfactant (Triton X-100) to break up spore clusters. Seedlings were inoculated 7 days after planting by placing a 2uL drop of spore suspension solution onto the apical meristem of the seedling. Only seedlings that had fully expanded cotyledons were inoculated. In total, we inoculated 140 D. furcatus, 431 D. pavonius, and 164 D. seguieri seedlings. Several seedlings germinated but were not ready for the inoculation and were left as uninoculated controls. (See Supplemental Material).
Following inoculations, seedlings were incubated in the growth chamber at 20°C for an additional 7 days. Each seedling was then transferred to a 4 × 13.5cm ‘conetainer’ (Stuewe and Sons Inc, Tangent, Oregon) filled with PromixBX, and moved to a greenhouse with 12 hours light/dark cycle, 20° days and 18° nights. Treatments were completely randomized. Plants were transferred outside in November for vernalization, and were moved back inside the greenhouse in April, prior to flowering.
Data collection
We recorded the date of first flowering, disease status, and number of inflorescences for each plant. To quantify spore production, we collected a single bud from each diseased plant. Each bud was dried and weighed and then cut open and put into vials with 1mL of 70% ethanol. Vials were vortexed and spores counted with an automatic cell counter (Cellometer Mini, Nexcelom Bioscience, Lawrence, Massachusetts). Inoculated plants that did not flower the first year were maintained and treated similarly for a second winter exposure and scored for disease the following spring.
Repeat inoculation trial with D. seguieri
Final samples sizes for the D. seguieri and D. furcatus host treatments were much smaller than anticipated due to lower starting seed numbers, reduced survival, and low flowering rates. We did not have additional seeds of D. furcatus to run a repeat, but were able to carry out a partial replicate of Experiment 1 on D. seguieri using a subset of 10 of the tested pathogen isolates (Table 2, asterisk, Table S1). We inoculated each isolate onto 10-day old D. seguieri plants using greenhouse-reared seed from the two original D. seguieri populations. We used the same inoculation method detailed above, and recorded infection rate at flowering.
Experiment 2: Host population by pathogen lineage
To determine whether the pathogen lineage specificity measured in Experiment 1 was consistent across a broader range of host populations we collected seed from eight additional D. seguieri populations from across the study region in 2013 (Table 1B, Fig. S1) and inoculated these with a single isolate of Lineage 1 originating from D. pavonius (44.2529, 7.1151) and a single isolate of Lineage 2 originating from D. seguieri (44.2960, 7.1572). Both isolates were collected from the Rio Freddo valley. We focused on D. seguieri in this second experiment because this host species showed strong lineage specific susceptibility in Experiment 1; in particular, Lineage 1 was unable to infect D. seguieri, whereas Lineage 2 infected at a high rate.
Data analysis
A total of 57% of the inoculated plants in Experiment 1 flowered within 2 years and were scored for infection (N=420; Table 1). Isolate 5 did not produce a single infection and was removed from all subsequent analyses.
We used generalized linear mixed models, with a binomial link function, to determine the effect of host species and pathogen lineage on infection rate (‘glmer’ function in ‘lme4’ package, R 3.6.1). Host species and pathogen lineage were fixed effects in the model, with pathogen isolate nested as a random effect within lineage. Since several combinations of host population by pathogen isolate had fewer than 10 flowering individuals (Table 1A), we pooled host populations from the same species. We confirmed that host population did not have a significant effect within each lineage-by-host species combination using Chi-squared and Fisher exact tests (Table S3). Fisher tests also confirmed that replicate isolates from the same lineage and host-of-origin did not differ significantly in infectivity (Table S4).
To examine the effect of pathogen host-of-origin on infectivity, we ran binomial glm models with host species and host-of-origin within the two lineages (Lineage 2 and 4) that had replicate isolates collected from at least two different hosts.
To test the hypothesis that specialist and generalist pathogens differ in fitness-related traits on their shared host, we used hierarchical random effects models to determine the effect of pathogen lineage on flowering date (phenology), inflorescence number, and spore production on the most commonly shared host. We used the ‘lme’ function in R 3.6.1 to estimate the variance components of pathogen lineage and pathogen isolate (nested in lineage) and used likelihood ratio tests to determine the effect of pathogen lineage. We carried out a similar analysis to determine the effect of host-of-origin. Pathogen fitness-related traits were analyzed using only the set of plants that flowered in the first year. We also used linear regression, weighted by sample size, to examine the relationship between infection ability of individual Microbotryum isolates on different host species and spore production.
A total of 650 D. seguieri plants flowered and were scored in the repeat inoculation trial (Table S1). To test for repeatability between the two replicate runs on D. seguieri we used a glm model with experimental replicate, pathogen lineage and the interaction of experiment and lineage as fixed effects. We used a quasibinomial error structure, with an estimated dispersion parameter because there was evidence of significant overdispersion in a strict binomial model. For this analysis we only included the subset of Experiment 1 isolates that were tested in both inoculation trials (Table 2, asterisks).
For Experiment 2 we used a glm model with binomial error structure to examine the main effects of pathogen lineage and host population. To determine whether there was an interaction effect between pathogen lineage and population: we multiplied the infection rate of each population when inoculated with Lineage 2 by the overall average reduction in infection by Lineage 1, to generate the expected infection rate of each population by Lineage 1. We multiplied the expected infection rate by the observed sample size for each population and used a Chi-squared contingency test to evaluate whether the expected number of infected Lineage 1 plants deviated from the observed number. We did not do the test in other direction (e.g., predicting Lineage 2 infections from Lineage 1 infection rates) because several populations inoculated with Lineage 1 had zero infections.
RESULTS
Experiment 1.
A total of 370 inoculated plants (out of 735 transplanted individuals) flowered in the first year of the experiment. An additional 50 inoculated plants flowered and were scored in the two subsequent years. Inoculation had a significant, positive effect on flowering rate (See Supplemental Results, S5).
Infectivity:
Host specificity of pathogen lineages:
Infection rate was significantly affected by pathogen lineage (X2=9.801, df=3, p=0.0203) and host species (X2=169.48, df=2, p<0.0001). There was also a significant pathogen lineage by host interaction effect (X2=40.8, df=6, p<0.000l). Lineage 1 showed the highest host specificity and was only able to infect D. pavonius (Fig. 2A). Lineage 4 was able to infect both D. pavonius and D. seguieri, but the latter to a lesser degree. Lineages 2 and 3 caused disease on all three host species. Exclusion of the two isolates with lineage assignment scores below 80% (isolates 12 and 15) had little effect on the results (Table S2).
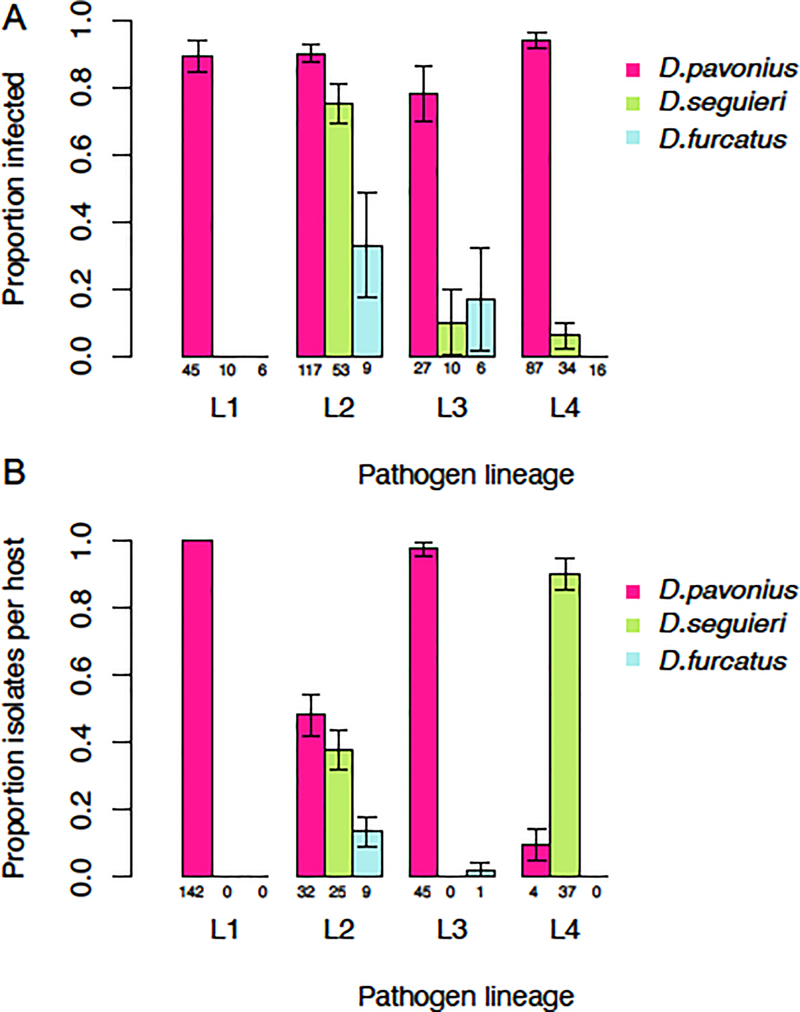
Fig. 2.
Comparison of the A) potential and B) realized host range of four Microbotryum lineages (L1-L4). A) Infection results from Experiment 1. Bars show the proportion of each host species infected by pathogen lineages L1-L4. Colors indicate host species: Dianthus pavonius (pink), D. seguieri (green), or D. furcatus (blue). Numbers indicate the total number that flowered and were scored. B) Field collections of the distribution of Microbotryum lineages on Dianthus species are shown for reference (data redrawn from Petit et al. 2017). Bars show the proportion of isolates from each lineage that were found on each host. Numbers of isolates in each category are indicated at the bottom. Error bars are 1 SEM.
Dianthus pavonius was the most susceptible of the three host species (Fig. 2A, pink bars) and infection rates did not differ significantly among the four lineages on this host species (X2=5.547, df = 3, p-value = 0.135). Dianthus seguieri and D. furcatus were infected at a lower rate, and only then by a subset of lineages (Fig. 2A, green and blue bars). The three pathogen lineages that were able to infect D. seguieri differed significantly in their infectivity on that host (X2= 56.415, df = 3, p<0.0001). Lineage 2 had the highest infection rate on D. seguieri (75%), while the other three lineages infected at much lower rates (0, 6, and 13%). In D. furcatus, only Lineage 2 and 3 were able to cause disease and there was no significant difference in infectivity between these two lineages (X2= 0.014205, df = 1, p-value = 0.9051).
Overall, there was a strong concordance of the experimental infection rates (Fig. 2A) with the realized host range of these four lineages in the field (Fig. 2B). Lineage 1, which was only found on D. pavonius in the field (Fig. 2B), was only able to infect this host species in Experiment 1 (Fig. 2A). In addition, Lineage 2, which was found on all three host species in the field, was able to infect all three species in Experiment 1.
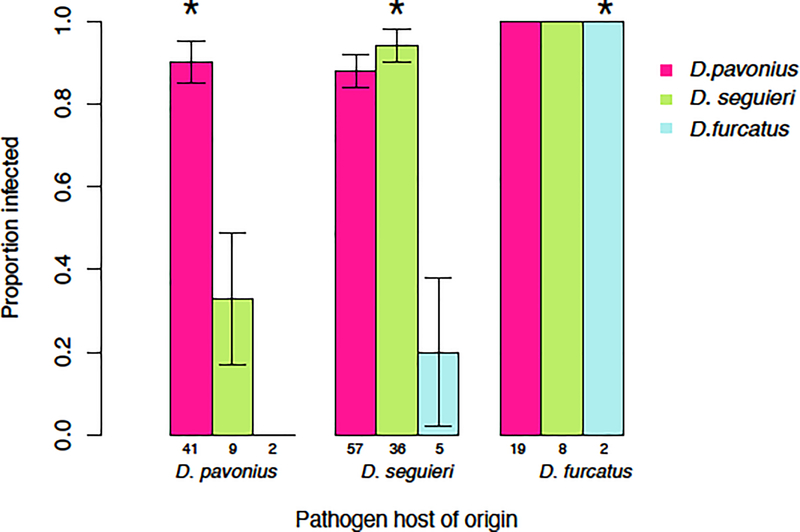
Host-of-origin of the pathogen:
Within the most generalist Lineage 2, where isolates were available from each of the three host species, the host-of-origin had a significant main effect on infection rate (X2=8.37, df=2, p=0.0152). There was also a significant host-of-origin by host species interaction effect (X2= 18.41, df=4, p=.0010 ). Pathogen isolates performed the best, or equal to the best on their host-of-origin (Fig. 3). The case was most extreme for Lineage 2 isolates originating from D. pavonius; these infected D. pavonius at a high rate but were unable to infect D. furcatus and had low infectivity on D. seguieri (Fig. 3). Lineage 2 isolates originating from D. seguieri had the highest infectivity on D. seguieri and D. pavonius, but low infectivity on D. furcatus. The single Lineage 2 isolate collected from D. furcatus had high infectivity on D. furcatus and D. pavonius, but lower infectivity on D. seguieri (Fig. 3).
Fig. 3.
Infectivity of Lineage 2 isolates collected from three different hosts-of-origin in Experiment 1. Asterisks indicate combinations where the pathogen was tested on its own host-of-origin. Numbers indicate the total number of inoculated plants that flowered and were scored for disease. Error bars 1 SEM.
Isolates from Lineage 4 were collected from two different host species in the field (D. pavonius, and D. seguieri; Fig. 1A). However, within this lineage there was no significant effect of host-of-origin (X2=0.002, df=1, p=0.964). Isolates of Lineage 4 collected from D. pavonius and D. seguieri had equally high infection on D. pavonius (93% and 80%, respectively), and equally low infection on D. seguieri (4% and 5%, respectively).
Effects of host population and isolate locality:
We did not detect significant population differences among host populations within any of the three tested species in Experiment 1 (Table S3). Nor did we find evidence of variation in infectivity among pathogen isolates from the same lineage and host-of origin (Table S4).
Repeatability of D. seguieri results.
When we compared the infection rate of the same subset of isolates on D. seguieri in both experiments, we found a significant main effect of pathogen lineage (χ2=68.2, df=3, p=0.034), and differences between experiments that approached significance (χ2=23.7, df=1, p=0.061); the infection rate was lower in this repeat experiment (12%, N=432) than in Experiment 1 (40%, N= 67). However, there was no significant interaction between pathogen lineage and experiment (χ2 =11.78, df=3, p<0.684) and the rank order of infectivity among the four pathogen lineages was consistent across the experiments: pathogen Lineages 2 and 3 were better at infecting D. seguieri than Lineages 1 and 4 (Fig. S2A). However, within the replicate experiment, there was a significant interaction between pathogen lineage and host population (X2=8.71, df=3, p=0.0334; Fig. S2B), which was not detected in the original experiment.
Floral and spore traits
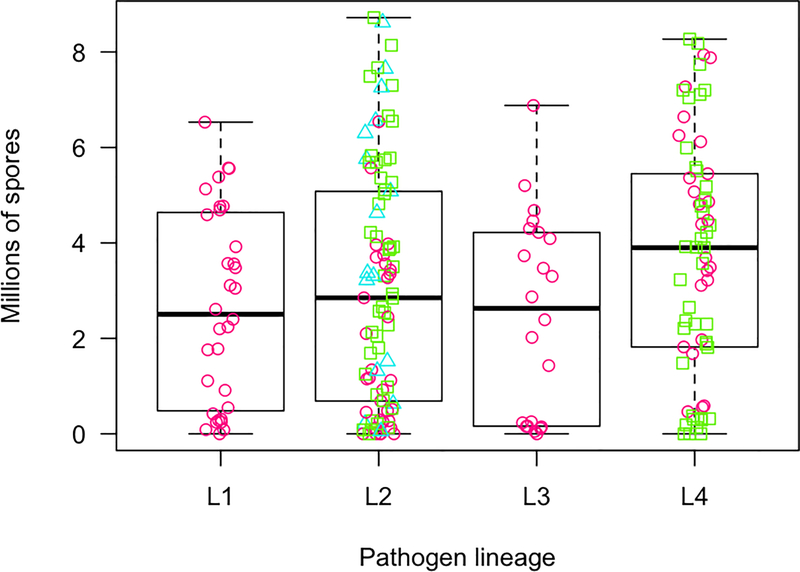
We measured spore production, flowering time and inflorescence number on 212 D. pavonius plants that flowered diseased in the first year. Among these diseased plants, pathogen isolate explained a significant amount of the total variation in flowering date (5.7%), inflorescence number (15.8%), and spore production (11%). However, there was no significant effect of pathogen lineage, or host-of-origin on any of these traits (Table 3). Lineage 1, which is specialized to D. pavonius, did not have higher spore production on this host species than the other pathogen lineages (Fig. 4).
Table 3.
Proportion of the variation in flowering and spore traits on Dianthus pavonius explained by either pathogen lineage or host of origin, accounting for the variation among isolates. Likelihood ratio tests and p-values show the overall significance of including lineage or host origin in the model.
| Among lineage or origin | Among isolate | LR.test | p | |
|---|---|---|---|---|
|
| ||||
| Days to first flower | ||||
| Pathogen lineage | <0.01% | 14.60% | 1.8E-07 | 0.9997 |
| Host of origin | 5% | 10.60% | 1.9E+00 | 0.1630 |
| Log inflorescence number | ||||
| Pathogen lineage | <0.01% | 5.70% | 1.3E-07 | 0.9997 |
| Host of origin | 0.2% | 5.54% | 1.3E-07 | 0.9997 |
| Total spores | ||||
| Pathogen lineage | <0.01% | 11.30% | 6.9E-07 | 0.9993 |
| Host of origin | 2.8% | 9.40% | 0.4654 | 0.4951 |
Fig. 4.
Spore production of four pathogen lineages on Dianthus pavonius of infected plants in Experiment 1 that flowered in the first year. Colors and shapes indicate host-of-origin: Pink circles-Dianthus pavonius, Green squares- D. seguieri, Blue triangles-D. furcatus. Neither lineage nor host-of-origin was a significant predictor of spore production (Table 3).
We did not find any evidence of negative correlation between infection ability of individual Microbotryum isolates and spore production on D. pavonius (Fig. S3). In weighted linear models, neither infection rate on D. pavonius (t=0.363, p=0.722), nor infection rate on other Dianthus hosts pavonius (t=1.590, p= 0.133) was a significant predictor of spore production on D. pavonius. However, we urge caution interpreting this result, as the sample size for estimating infection rates of individual isolates was low.
Experiment 2: Variation among Dianthus seguieri populations
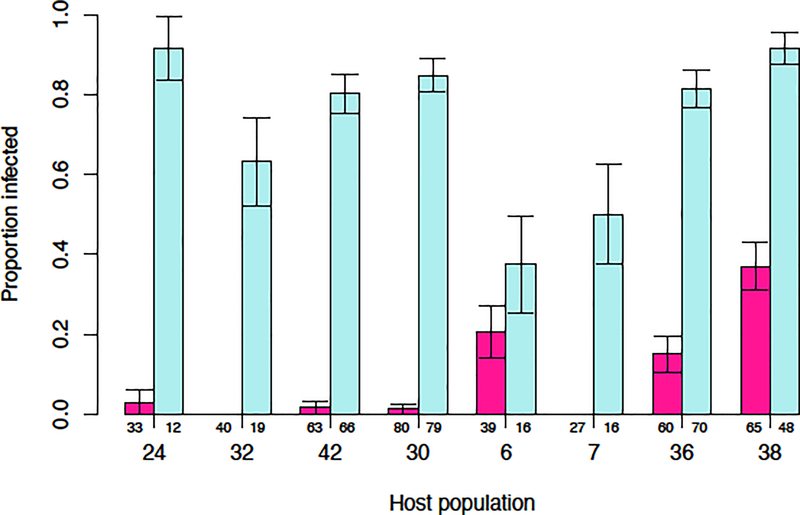
In the assessment of lineage specificity on multiple D. seguieri populations (Experiment 2), a total of 733 plants flowered. There was a significant effect of pathogen lineage on infection rate (X2=389.26, df=1, p<0.0001). The Lineage 2 isolate (from D. seguieri) had a much higher overall infection rate (72.5%) on D. seguieri than the Lineage 1 isolate (from D. pavonius) (9.8%), similar to the results obtained in Experiment 1. There was also significant variation in infection rate among host populations by both the Lineage 2 isolate (X2=35.49, df=7, p<0.0001) and the Lineage 1 isolate (X2=74.2, df=7, p<0.0001; Fig. 5). There was also a significant interaction effect between pathogen lineage and host population; observed infection rates of Lineage 1 across populations differed from expected values based on Lineage 2 values (X2=70.79, df=7, p<0.0001). Only the three D. seguieri populations, all from the eastern region (Populations 6, 36, 38) showed substantial susceptibility to the pathogen Lineage 1 (Fig. 5).
Fig. 5.
Infectivity of a single isolate of Lineage 1 (pink) and a single isolate of Lineage 2, (blue) on eight different D. seguieri populations in Experiment 2. Large numbers indicate the population ID number (Table 1B), small numbers indicate the sample size of each treatment. Error bars are 1 SEM.
DISCUSSION
Variation in Pathogen Host range
We found that the infectivity of the four Microbotryum lineages on Dianthus was highly concordant with observed patterns of their distributions among natural populations of the host species. Lineage 1, which has only ever been found on D. pavonius in nature (Petit et al. 2017), does indeed appear to be a true specialist to this host. Replicate isolates from this lineage were able to infect D. pavonius at high rate (89%) but had very low infection rates (0–5%) on the other Dianthus species. In contrast, Lineage 2 which Petit et al. (2017) found on all three Dianthus species in the field, was also able to infect all three host species at high levels in the lab, indicating that it is indeed a true generalist. Lineage 3 was also able infect all three Dianthus species, although it had only previously been reported on D. pavonius and D. seguieri. Lineage 4 was found on D. pavonius and D. seguieri in the field, and was able to infect only these two species, but not D. furcatus, in the lab. Moreover, the lineage-specific infectivity that we found here was generally robust to variation among pathogen isolates within lineages, and among host populations within a species. These results therefore show that the wide host range of these Microbotryum lineages on Dianthus are not simply the result of occasional host-shifts or spillover events, and that these lineages are likely maintained as viable populations on multiple Dianthus host species.
These results with anther-smut fungi of Dianthus differ from patterns of host specificity of Microbotryum on host species in the genus Silene where phylogeographic studies have shown that most species of Microbotryum are specialized to a single host species (Lutz et al. 2005; Le Gac et al. 2007). While host shifts can and do occur between closely related species (Antonovics et al. 2002; Refrégier et al. 2008; Tyson et al. 2018), there has been little evidence to suggest a true generalist species in Silene that is actively maintained on two or more hosts. Indeed, patterns of host-specificity among Silene-infecting Microbotryum are present even in communities with sympatric host species (Van Putten et al. 2005; Gladieux et al. 2010; Hood et al. 2019) and overlapping floral phenologies (Tang et al. 2019). One possible explanation for the contrast in host breadth between Microbotryum on Silene and those pathogen lineages on Dianthus is that the Dianthus species studied here were part of very recent radiation that occurred during the early Pleistocene (Valente et al. 2010). Consequently, Dianthus species are less differentiated than older Silene species (a phenomenon noted by Darwin 1869, p. 311). More generally, phylogenetic distance is known to be an important predictor of host-pathogen specificity (Gilbert and Webb 2007; Refrégier et al. 2008; de Vienne et al. 2009; Streicker et al. 2010; Antonovics et al. 2013; Parker et al. 2015).
The discovery that there are Microbotryum lineages that are actively maintained on multiple host species of Dianthus raises new questions about the ecology of this group of anther-smut pathogens. The dynamics of multi-host pathogens can fundamentally differ from those of single-host pathogens (Woolhouse et al. 2001; Dobson 2004). For example, transmission from highly susceptible host species can maintain the disease on less susceptible species (Power and Mitchell 2004). Generalist pathogens can also affect host community structure through apparent competition (Tompkins et al. 2003; Mordecai 2011) and are more likely to drive hosts to extinction than specialist pathogens (Woolhouse et al. 2001; Dobson 2004). However, the extent to which the disease dynamics of these multi-host lineages of Microbotryum may differ from the dynamics of specialist lineages is currently unknown. The only population where we have studied the transmission dynamics in depth contains a single host (D. pavonius) and a single pathogen lineage, Lineage 1 (Bruns et al. 2017). However, Dianthus species do occur in sympatry in other regions of the broader Western Alps (Bruns et al. 2018) and harbor multiple pathogen lineages (Petit et al. 2017), indicating the potential for multi-host interactions in nature.
Trade-offs
We expected to find evidence of a cost to generalism in the form of lower infectivity or spore production of the generalist lineages relative to the specialists on their shared host species, consistent with results from several other host-pathogen systems (Straub et al. 2011; Medeiros et al. 2014). However, we found no evidence of such trade-offs: Lineage 1, which was restricted to D. pavonius was not significantly better than any of the generalist lineages at infecting D. pavonius, nor did it have higher spore production following infection. This result was consistent across the two tested populations of D. pavonius, as well as replicate isolates of Microbotryum within each lineage. In contrast to the present study, Microbotryum species from Silene have been shown to have lower fitness on closely-related novel hosts than on their host-of-origin, producing fewer spores (Biere and Honders 1996) and abnormal flowers (Antonovics et al. 2002), and resulting in lower levels of sterility (Sloan et al. 2008).
The lack of strong trade-offs in the Microbotryum lineages on Dianthus may be one reason why generalist lineages are maintained in Dianthus-associated but not in Silene-associated Microbotryum lineages. Without trade-offs, generalist pathogens should have higher fitness than specialist through access to a broader range of transmission opportunities (Jaenike 1990; Joshi and Thompson 1995; Fry 1990). Why then is the specialist Lineage 1 so abundant on D. pavonius? One potential explanation is geographic variation in host species distributions. Petit et al. (2017) showed that the highest incidence of the D. pavonius specialist Lineage 1 was found within the eastern region where D. pavonius is largely allopatric from D. furcatus (See Fig. 1A), even though they grow sympatrically in other parts (Fig. 1B). Dianthus seguieri is generally not found in direct sympatry with D. pavonius, as it grows at lower elevations (Bruns et al. 2018). The specialist lineage could easily be maintained in these allopatric populations of D. pavonius. However, a lack of opportunity for cross-species transmission is unlikely to be the only explanation since Lineage 1 is also found throughout other parts of the hosts’ range, including sympatrically with the other four lineages (Petit et al. 2017).
Another possibility is that there are trade-offs between host breadth and other pathogen traits we did not measure in this experiment, such as competitive ability or infection ability at later host life-stages. Previous work has shown that pathogen isolates vary in their ability to colonize, and drive out other Microbotryum isolates within a host (Hood 2003). If the specialist lineage is a superior competitor on D. pavonius, this could help explain its high abundance. It is also possible that costs of generalism are only evident when hosts are older. For example, in birds, Medeiros et al. (2014) showed that specialist blood parasites were more common on the adults of shared host species than generalist parasites, but did not differ in relative abundance on juveniles. In this study, we only measured infection ability at the seedling stage, but in D. pavonius resistance to anther-smut increases substantially with age (Bruns et al. 2017; Bruns 2019). It could be that the specialist lineage has an infection advantage over the generalist on older D. pavonius plants.
Variation in susceptibility among host species
The three host species differed substantially in their overall susceptibility. Dianthus pavonius was a universally susceptible host to all the pathogen lineages and was infected at much higher rates than the other two species. This finding is consistent with observations from nature where we have found all four Microbotryum lineages on D. pavonius (Petit et al. 2017). Moreover, the high relative susceptibly of D. pavonius was consistent across two tested host populations, despite significant differences in disease prevalence observed in the field (the Valle Pesio population had a prevalence of 41%, while the Ferrere population was completely healthy). The potential epidemiological consequences of this variation in susceptibility among Dianthus species are unclear. In nature, all three host species have similar average disease incidence (59%, 55%, and 44%) and prevalence (17%, 20%, and 18% for D. furcatus, D. pavonius, and D. seguieri, respectively; Bruns et al. 2018).
There was substantial asymmetry in the species-specific susceptibility and infectivity of host and pathogen species. The pathogen with the most constrained host range (Lineage 1) was only found on the most susceptible host species (D. pavonius). A similar pattern was found by de Vienne et al. (2009) in a cross-species inoculation experiment with six Microbotryum species associated with: Silene latifolia, S. dioica, S. vulgaris, S. paradoxa, Saponaria officinalis, and Dianthus carthusianorum. They showed that host species varied in their susceptibility, and that the pathogens originating from more susceptible hosts had more restricted hosts ranges. For example, D. carthusianorum was susceptible to all six pathogen species, but the pathogen originating from D. carthusianorum was only able to infect D. carthusianorum. A similar pattern has been found in animals; Vázquez et al. (2005) showed that specialist parasites of fish and small mammals tended to infect hosts species that harbored the highest parasite richness. De Vienne et al. (2009) hypothesize that this type of asymmetric pattern could be caused by differences in strength of coevolution among different host-pathogen pairs. Strong coevolutionary interactions within one pair should lead to the evolution of greater host defenses and pathogen counter-defenses. Pathogens from these coevolutionary ‘hot-spot’ hosts may be better equipped to infect other, less well defended host species. In contrast, pathogens co-evolving with highly susceptible hosts, such as D. pavonius, may not evolve as many counter-offenses, making host shifts less likely.
The high abundance and universal susceptibility of D. pavonius may make it an open target for host shifts and lead to subsequent specialization. Indeed, our data shows that within the generalist Lineage 2, isolates from D. pavonius had lower infectivity on the other two host species compared with pathogens from the same lineage that were collected from either D. furcatus or D. seguieri. The phylogeny of the four Microbotryum lineages studied here has not been fully resolved, so we cannot infer the evolutionary history of the lineages. Gene trees from the two nuclear loci indicate that Lineage 3 is monophyletic and basal, but fail to fully resolve Lineages 1, 2, and 4 (Petit et al. 2017). Further genomic work resolving these evolutionary relationships could help determine whether the specialist Lineage 1 is indeed recently derived from generalists, or vice versa.
Variation in infectivity among D. seguieri populations
The host-species specificity of Microbotryum lineages varied among populations within a host species, in that the eight D. seguieri populations differed in their infection rate by two pathogen lineages. As expected, Lineage 2 (the generalist pathogen) was consistently better than Lineage 1 (the D. pavonius specialist) at infecting D. seguieri populations, but there were some interesting deviations. Lineage 2 had lower infectivity on host populations 6 and 7 from the San Bernardo di Mendatica locality in the southeast, and population 6 was also more susceptible to Lineage 1 (Fig. 4). Interestingly, Petit et al. (2017) found that Lineage 2 is rare in this region and Lineages 1 and 4 account for most of the Microbotryum samples from this locality. One possibility is that higher levels of host resistance in the southern D. seguieri populations has prevented the spread of Lineage 2 into this locality. Another possibility is that Lineage 2 has recently arrived in the south and has not yet had time to locally adapt. In either case, further information on the phylogeographic history of these Microbotryum lineages is needed.
CONCLUSIONS
The results of this study illustrate the complexity of host-parasite interactions during their “early” evolution, when host and pathogen species are young. Most macro-evolutionary models of host-parasite divergence assume specialization is an inherent trait of the pathogen (Brooks and McLennan 1991, de Vienne et al. 2007), and a shift onto a new host is followed by pathogen diversification and speciation. Indeed, this type of host-shift driven diversification has been clearly demonstrated for Microbotryum on Silene as evidenced by discordant phylogenies (Refrégier et al. 2008). However, in the Dianthus-Microbotryum system, the concordance between the experimental infectivity of each pathogen lineage and its observed distribution on several Dianthus host species in nature, has shown that Microbotryum lineages on Dianthus vary in host range from specialists on a single species to generalists capable of infecting several species. Specialist pathogen lineages can often co-occur with generalists, and do not necessarily have higher infectivity or spore production on their specialized host species. The infection ability of generalist and specialist pathogen lineages even varies across host populations within a host species.
This variation in host range is likely maintained by a combination of low costs of generalism, and heterogeneity in the degree of sympatry between different host species in different regions, which may allow the persistence of both specialist and generalist lineages. A further issue we have not addressed is that both the host species and the pathogen lineages are capable of hybridizing, with the latter appearing to be common (Petit et al. 2017). Therefore, an important future goal will be to integrate these newly described host range patterns into a broader understanding of speciation processes in host-pathogen systems.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Tim Park, who helped with the planting and inoculation, and Wendy Cranage, at the University of Virginia greenhouse who cared for the plants. We also thank Prima Vithoontien, Maggie Berrigan, and Megan Wu who assisted with spore counts and the follow up D. seguieri experiment. We also thank the editors and two anonymous reviewers who helped improve the final manuscript. We gratefully acknowledge funding support from the NSF (DEB-1936334 to EB), and the NIH ( R01GM122061 to JA and R01GM140457 to MH).
Footnotes
CONFLICT OF INTEREST
The authors have no conflicts of interest to report
Publisher's Disclaimer: This article has been accepted for publication and undergone full peer review but has not been through the copyediting, typesetting, pagination and proofreading process, which may lead to differences between this version and the Version of Record.
DATA ACCESSIBILITY
All the data described in the manuscript has been archived in the Dryad repository: https://doi.org/10.5061/dryad.q2bvq83jj
REFERENCES
- Alexander HM 2010. Disease in natural plant populations, communities, and ecosystems: insights into ecological and evolutionary processes. Plant Disease 94:492–503. [DOI] [PubMed] [Google Scholar]
- Antonovics J, Hood M, and Partain J. 2002. The ecology and genetics of a host Shift: Microbotryum as a model system. The American Naturalist 160: S40–S53. [DOI] [PubMed] [Google Scholar]
- Antonovics J, Boots M, Ebert D, Koskella B, Poss M, Sadd BM 2013. The Origin of Specificity by Means of Natural Selection: Evolved and Nonhost Resistance in Host–Pathogen Interactions. Evolution 67: 1–9 [DOI] [PubMed] [Google Scholar]
- Asplen MK, Bruns E, David AS, Denison RF, Epstein B, Kaiser MC, Kaser JM, Lacroix C, Mohl EK, Quiram G, Prescott K, Stanton-Geddes J, Vincent JB, Wragg PD, and May G. 2012. Do trade-offs have explanatory power for the evolution of organismal interactions? Evolution 66:1297–1307. [DOI] [PubMed] [Google Scholar]
- Bedhomme S, Lafforgue G, and Elena SF. 2012. Multihost Experimental Evolution of a Plant RNA virus reveals local adaptation and host-specific mutations. Mol Biol Evol 29:1481–1492. [DOI] [PubMed] [Google Scholar]
- Bernasconi G, Antonovics J, Biere A, Charlesworth D, Delph LF, Filatov D, Giraud T, Hood ME, Marais GAB, McCauley D, Pannell JR, Shykoff JA, Vyskot B, Wolfe LM, and Widmer A. 2009. Silene as a model system in ecology and evolution. Heredity 103:5–14. [DOI] [PubMed] [Google Scholar]
- Biere A, and Honders S. 1996. Host adaptation in the anther smut fungus Ustilago violacea (Microbotryum violaceum): infection success, spore production and alteration of floral traits on two host species and their f1-hybrid. Oecologia 107:307–320. [DOI] [PubMed] [Google Scholar]
- Bruns E (2019) Effects of host lifespan on the evolution of age-specific resistance: a case study of anther-smut disease on wild carnations. Chapter for ‘Wildlife Disease Ecology: Linking Theory to data and application’ Eds. Wilson K, Fenton A, and Tompkins D Cambridge University Press, Cambridge. [Google Scholar]
- Bruns EL, Antonovics J, Carasso V, and Hood M. 2017. Transmission and temporal dynamics of anther-smut disease ( Microbotryum) on alpine carnation (Dianthus pavonius ). Journal of Ecology 105:1413–1424. [Google Scholar]
- Bruns EL, Antonovics J, and Hood M. 2018. Is there a disease-free halo at species range limits? The co-distribution of anther-smut disease and its host species. Journal of Ecology 107:1–11. [Google Scholar]
- Büker B, Petit E, Begerow D, and Hood ME. 2013. Experimental hybridization and backcrossing reveal forces of reproductive isolation in Microbotryum. BMC Evolutionary Biology 13:224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleaveland S, Laurenson MK, and Taylor LH. 2001. Diseases of humans and their domestic mammals: pathogen characteristics, host range and the risk of emergence. Philosophical Transactions of the Royal Society of London. Series B: Biological Sciences 356:991–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darwin CR 1869. On the origin of species by means of natural selection, or the preservation of favoured races in the struggle for life. London: John Murray. 5th edition. [PMC free article] [PubMed] [Google Scholar]
- de Vienne DM, Hood ME, and Giraud T. 2009. Phylogenetic determinants of potential host shifts in fungal pathogens. Journal of Evolutionary Biology 22:2532–2541. [DOI] [PubMed] [Google Scholar]
- Devier B, Aguileta G, Hood ME, and Giraud T. 2010. Using phylogenies of pheromone receptor genes in the Microbotryum violaceum species complex to investigate possible speciation by hybridization. Mycologia 102:689–696. [DOI] [PubMed] [Google Scholar]
- Dobson A 2004. Population dynamics of pathogens with multiple host species. The American Naturalist 164:S64–S78. [DOI] [PubMed] [Google Scholar]
- Ebert D 1998. Experimental Evolution of Parasites. Science 282: 1432–1436. [DOI] [PubMed] [Google Scholar]
- Fellous S, Angot G, Orsucci M, Migeon A, Auger P, Olivieri I, and Navajas M. 2014. Combining experimental evolution and field population assays to study the evolution of host range breadth. Journal of Evolutionary Biology 27:911–919. [DOI] [PubMed] [Google Scholar]
- Fry JD 1990. Trade-Offs in fitness on different hosts: evidence from a selection experiment with a phytophagous mite. The American Naturalist 136:569–580. [Google Scholar]
- Gilbert GS, and Webb CO. 2007. Phylogenetic signal in plant pathogen–host range. PNAS 104:4979–4983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraud T, Gladieux P, and Gavrilets S. 2010. Linking the emergence of fungal plant diseases with ecological speciation. Trends in Ecology & Evolution 25:387–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraud T, Refrégier G, Le Gac M, de Vienne DM, and Hood ME. 2008. Speciation in fungi. Fungal Genetics and Biology 45:791–802. [DOI] [PubMed] [Google Scholar]
- Gladieux P, Vercken E, Fontaine MC, Hood ME, Jonot O, Couloux A and Giraud T, 2010. Maintenance of fungal pathogen species that are specialized to different hosts: allopatric divergence and introgression through secondary contact. Molecular Biology and Evolution, 28:459–471. [DOI] [PubMed] [Google Scholar]
- Hellgren O, Pérez-Tris J, and Bensch S. 2009. A jack-of-all-trades and still a master of some: prevalence and host range in avian malaria and related blood parasites. Ecology 90:2840–2849. [DOI] [PubMed] [Google Scholar]
- Hood ME 2003. Dynamics of multiple infection and within-host competition by the anther-smut pathogen. The American Naturalist 162:122–133. [DOI] [PubMed] [Google Scholar]
- Hood ME, Antonovics J, Wolf M, Stern ZL, Giraud T, and Abbate JL. 2019. Sympatry and interference of divergent Microbotryum pathogen species. Ecology and Evolution 9:5457–5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hood ME, Mena-Alí JI, Gibson AK, Oxelman B, Giraud T, Yockteng R, Arroyo MTK, Conti F, Pedersen AB, Gladieux P, and Antonovics J. 2010. Distribution of the anther-smut pathogen Microbotryum on species of the Caryophyllaceae. New Phytologist 187:217–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaenike J 1990. Host specialization in phytophagous insects. Annual Review of Ecology and Systematics 21:243–273. [Google Scholar]
- Joshi A, and Thompson JN. 1995. Trade-offs and the evolution of host specialization. Evol Ecol 9:82–92. [Google Scholar]
- Kawecki TJ 1994. Accumulation of deleterious mutations and the evolutionary cost of being a generalist. The American Naturalist 144:833–838. [Google Scholar]
- Kemler M, Martín MP, Telleria MT, Schäfer AM, Yurkov A, and Begerow D. 2013. Contrasting phylogenetic patterns of anther smuts (Pucciniomycotina: Microbotryum) reflect phylogenetic patterns of their caryophyllaceous hosts. Organisms Diversity & Evolution 13:111–126. [Google Scholar]
- Kido A and Hood ME 2020. Mining new sources of natural history observations for disease interactions. American Journal of Botany. 107: 3–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King TA, and Cable J. 2007. Experimental infections of the monogenean Gyrodactylus turnbulli indicate that it is not a strict specialist. International Journal for Parasitology 37:663–672. [DOI] [PubMed] [Google Scholar]
- Le Gac M, Hood ME, Fournier E, and Giraud T. 2007. Phylogenetic evidence of host-specific cryptic species in the anther smut fungus. Evolution 61:15–26. [DOI] [PubMed] [Google Scholar]
- Lutz M, Göker M, Piatek M, Kemler M, Begerow D, and Oberwinkler F. 2005. Anther smuts of Caryophyllaceae: Molecular characters indicate host-dependent species delimitation. Mycol Progress 4:225–238. [Google Scholar]
- Malenke JR, Johnson KP, and Clayton DH. 2009. Host specialization differentiates cryptic species of feather-feeding lice. Evolution 63:1427–1438. [DOI] [PubMed] [Google Scholar]
- Maloney PE, Lynch SC, Kane SF, Jensen CE, and Rizzo DM. 2005. Establishment of an emerging generalist pathogen in redwood forest communities. Journal of Ecology 93:899–905. [Google Scholar]
- Medeiros MCI, Ellis VA, and Ricklefs RE. 2014. Specialized avian Haemosporida trade reduced host breadth for increased prevalence. Journal of Evolutionary Biology 27:2520–2528. [DOI] [PubMed] [Google Scholar]
- Mordecai E 2011. Pathogen impacts on plant communities: unifying theory, concepts, and empirical work. Ecological Monographs 81:429–441. [Google Scholar]
- Parker IM, Saunders M, Bontrager M, Weitz AP, Hendricks R, Magarey R, Suiter K, and Gilbert GS. 2015. Phylogenetic structure and host abundance drive disease pressure in communities. Nature 520:542–544. [DOI] [PubMed] [Google Scholar]
- Pedersen AB, and Davies TJ. 2009. Cross-species pathogen transmission and disease emergence in primates. EcoHealth 6:496–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petit E, Silver C, Cornille A, Gladieux P, Rosenthal L, Bruns E, Yee S, Antonovics J, Giraud T, and Hood ME. 2017. Co-occurrence and hybridization of anther-smut pathogens specialized on Dianthus hosts. Molecular Ecology 26:1877–1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poisot T, Bever JD, Nemri A, Thrall PH, and Hochberg ME. 2011. A conceptual framework for the evolution of ecological specialisation. Ecology Letters 14:841–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulin R, and Keeney DB. 2008. Host specificity under molecular and experimental scrutiny. Trends in Parasitology 24:24–28. [DOI] [PubMed] [Google Scholar]
- Power AG, and Mitchell CE. 2004. Pathogen spillover in disease epidemics. The American Naturalist 164:S79–S89. [DOI] [PubMed] [Google Scholar]
- Refrégier G, Le Gac M, Jabbour F, Widmer A, Shykoff JA, Yockteng R, Hood ME, and Giraud T. 2008. Cophylogeny of the anther smut fungi and their Caryophyllaceous hosts: Prevalence of host shifts and importance of delimiting parasite species for inferring cospeciation. BMC Evolutionary Biology 8:100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouxel M, Mestre P, Comont G, Lehman BL, Schilder A, and Delmotte F. 2013. Phylogenetic and experimental evidence for host-specialized cryptic species in a biotrophic oomycete. New Phytologist 197:251–263. [DOI] [PubMed] [Google Scholar]
- Sloan DB, Giraud T, and Hood ME. 2008. Maximized virulence in a sterilizing pathogen: the anther-smut fungus and its co-evolved hosts. Journal of Evolutionary Biology 21:1544–1554. [DOI] [PubMed] [Google Scholar]
- Stephens PR, Altizer S, Ezenwa VO, Gittleman JL, Moan E, Han B, Huang S, and Pappalardo P. 2019. Parasite sharing in wild ungulates and their predators: Effects of phylogeny, range overlap, and trophic links. Journal of Animal Ecology 88:1017–1028. [DOI] [PubMed] [Google Scholar]
- Straub CS, Ives AR, and Gratton C. 2011. Evidence for a trade-off between host-range breadth and host-use efficiency in aphid parasitoids. The American Naturalist 177:389–395. [DOI] [PubMed] [Google Scholar]
- Streicker DG, Turmelle AS, Vonhof MJ, Kuzmin IV, McCracken GF, and Rupprecht CE. 2010. Host phylogeny constrains cross-species emergence and establishment of rabies virus in bats. Science 329:676–679. [DOI] [PubMed] [Google Scholar]
- Szabo LJ 2006. Deciphering species complexes: Puccinia andropogonis and Puccinia coronata, examples of differing modes of speciation. Mycoscience 47:130–136. [Google Scholar]
- Tang H, Hood ME, Ren Z-X, Li H-D, Zhao Y-H, Wolfe LM, Li D-Z, and Wang H. 2019. Specificity and seasonal prevalence of anther smut disease Microbotryum on sympatric Himalayan Silene species. Journal of Evolutionary Biology 32: 451–462. [DOI] [PubMed] [Google Scholar]
- Thrall PH, Biere A, and Antonovics J. 1993. plant life-history and disease susceptibility--The occurrence of Ustilago violacea on different species within the Caryophyllaceae. The Journal of Ecology 81:489. [Google Scholar]
- Tompkins DM, White AR, and Boots M. 2003. Ecological replacement of native red squirrels by invasive greys driven by disease. Ecology Letters 6:189–196. [Google Scholar]
- Tyson DA, Antonovics J and Bruns EL 2018. Anther smut disease caused by Microbotryum on berry campion Silene baccifera : endemic pathogen or host shift?. Plant Pathology 67: 1850–1856. [Google Scholar]
- Van Putten WF, Biere A and Van Damme JMM, 2005. Host-related genetic differentiation in the anther smut fungus Microbotryum violaceum in sympatric, parapatric and allopatric populations of two host species Silene latifolia and S. dioica. Journal of Evolutionary Biology, 18:203–212. [DOI] [PubMed] [Google Scholar]
- Valente LM, Savolainen V, and Vargas P. 2010. Unparalleled rates of species diversification in Europe. Proceedings of the Royal Society B: Biological Sciences 277:1489–1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vázquez DP, Poulin R, Krasnov BR, and Shenbrot GI. 2005. Species abundance and the distribution of specialization in host–parasite interaction networks. Journal of Animal Ecology 74:946–955. [Google Scholar]
- Vialle A, Feau N, Frey P, Bernier L, and Hamelin RC. 2013. Phylogenetic species recognition reveals host-specific lineages among poplar rust fungi. Molecular Phylogenetics and Evolution 66:628–644. [DOI] [PubMed] [Google Scholar]
- Woolhouse MEJ, Haydon DT, and Antia R. 2005. Emerging pathogens: the epidemiology and evolution of species jumps. Trends in Ecology & Evolution 20:238–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolhouse MEJ, Taylor LH, and Haydon DT. 2001. Population biology of multihost pathogens. Science 292:1109–1112. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the data described in the manuscript has been archived in the Dryad repository: https://doi.org/10.5061/dryad.q2bvq83jj