Abstract
The WNT signaling pathway is a critical regulator of development and adult tissue homeostasis and becomes dysregulated in many cancer types. While hyperactivation of WNT signaling is common, the type and frequency of genetic WNT pathway alterations can vary dramatically between different cancers, highlighting possible cancer-specific mechanisms for WNT-driven disease. In this review, we discuss how WNT pathway disruption contributes to tumorigenesis in different organs and how WNT impacts the tumor cell and immune microenvironment. Finally, we describe recent and ongoing efforts to target oncogenic WNT signaling as a therapeutic strategy.
Introduction
WNT signaling is a critical molecular rheostat that guides a range of physiological processes including embryonic development, lineage commitment, adult stem cell homeostasis, and tissue regeneration. The first member of the WNT family was identified more than 30 years ago as the Int-1 proto-oncogene in a mouse mammary tumor virus model (1). Int-1 was later identified as the homolog of the Drosophila melanogaster segment polarity gene wingless, and thus, the term ‘WNT’ was born from the fusion of both gene names (2). While first described in a cancer setting, much of our fundamental understanding of WNT biology has come from studying development in model organisms such as Drosophila, Xenopus laevis, and the mouse. Indeed, these systems continue to be a critical resource in efforts to define all WNT signaling components, their functions, and how they serve to control normal WNT signaling. In this review we will focus on abnormal or dysregulated WNT signaling, how it drives cancer, its role in stemness and immune evasion, and the progress and challenges of targeting the WNT pathway as a therapeutic strategy.
β-catenin-dependent WNT signaling
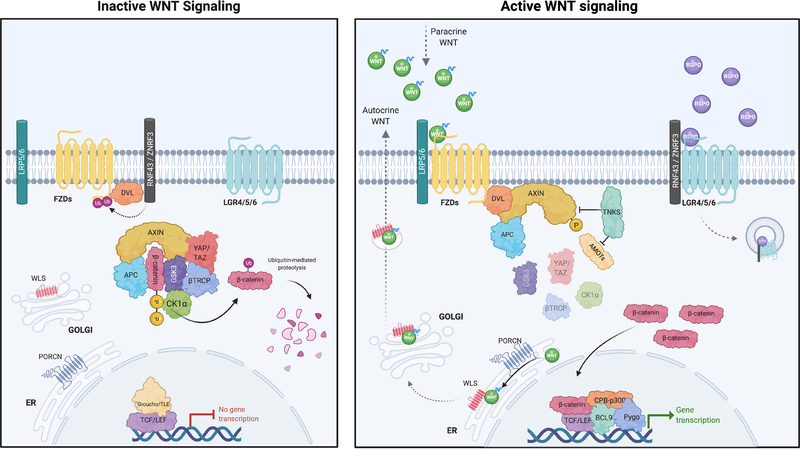
The WNT family consists of 19 secreted glycoproteins, which orchestrate cell fate specification, cell proliferation, cell migration, dorsal axis formation, asymmetric cell division, and many more functions, depending on cell and tissue context (3,4). The downstream effects of WNTs have traditionally been separated into two, sometimes overlapping categories: Canonical (β-catenin dependent) and non-canonical (β-catenin-independent). The β-catenin dependent pathway is induced by WNT ligands binding to Frizzled (FZD) and LRP5/6 co-receptor complexes, which initiate intracellular signaling and membrane recruitment of scaffold proteins (AXIN1/2 and DVL). This induces disruption of the core destruction complex [(AXIN, APC, casein kinase 1α (CK1α), and glycogen synthase kinase-3 (GSK3)], resulting in stabilization of β-catenin and its subsequent nuclear localization (3) (Figure 1). Numerous other proteins can interface with and modulate the core WNT pathway, including the Tankyrase enzymes (TNKS and TNKS2) that elevate WNT signaling by targeting AXIN1/2 for degradation (5). In the nucleus, β-catenin binds to members of the T-cell factor/lymphoid enhancer factor (TCF/LEF) family, recruit transcriptional co-activators including p300 and or CREB binding proteins (CBP) to drive a WNT transcriptional program. In the absence of WNT ligands, β-catenin is tagged for degradation via sequential phosphorylation by CK1α and GSK3 on serine and threonine residues (S45/T41/S37/S33) at the N-terminus. Phosphorylated β-catenin is recognized by a ubiquitin ligase complex which includes Beta-Transducin Repeat Containing E3 Ubiquitin Protein Ligase (β-TrCP), leading to poly-ubiquitination and proteasomal degradation. While not usually considered core members of the complex, Hippo pathway regulators YAP and TAZ (WWTR1) have been shown to play an important role in β-TrCP recruitment and β-catenin inactivation (6). Interestingly, YAP/TAZ downstream activity is also modulated by the TNKS enzymes through regulation of Angiomotin proteins (7).
Figure 1. Overview of the WNT signaling pathway.
In the absence of WNT ligand (inactive WNT signaling), accumulated β-catenin bound in the destruction complex by AXIN and APC is phosphorylated by CK1α, and GSK3 leading to its ubiquitination and proteasomal degradation by β-TrCP/YAP/TAZ. In the presence of WNT ligands (active WNT signaling), LRP5/6 and FZD co-receptors associate leading to activation and recruitment of AXIN1/2 and DVL to the membrane, disrupting the destruction complex. This results in stabilization and nuclear localization of β-catenin. In the nucleus, β-catenin binds to the TCF/LEF, recruiting co-activators p300 and CBP to induce WNT target gene target transcription. Created with BioRender.com
Before any intracellular pathway activation occurs, WNT ligands must be secreted from a WNT-producing cell to activate signaling in a WNT-responsive cell. The production of WNT ligands is tightly controlled and requires both post-translational modification by a serine O-palmitoleoyltransferase (PORCN) and association with Wntless (WLS or GPR177) in the endoplasmic reticulum (ER) to be secreted into the extracellular space (8). The enzymatic processing of WNTs by PORCN is an Achilles’ heel of the pathway that has been exploited to target WNT signaling pharmacologically, as discussed later.
In addition to the family of WNT ligands, there are a range of other extracellular modulators of WNT signaling. R-spondins1–4 (RSPO1–4) are small, secreted proteins that bind to leucine-rich repeat-containing G-protein-coupled-receptors (LGR4–6) to enhance WNT ligand-driven activation (9,10). Despite their name, LGR4–6 do not actually function as G-protein-coupled receptors. Rather, RSPO-bound LGR receptors bind to and sequester the transmembrane E3 ligases RNF43 and ZNRF3, preventing them from marking WNT-FZD receptors for lysosomal degradation (11,12). This results in accumulation of FZD receptors on the cell surface and amplification of the response to WNT ligand stimulation. The importance of the RSPO/LGR5/RNF43 module in controlling WNT pathway activation is evident across several cancer types with chromosome rearrangements driving overexpression of RSPO2 and RSPO3 in colorectal cancer (CRC), while inactivating mutations in RNF43 are observed in CRC, pancreatic ductal adenocarcinoma (PDAC), and endometrial cancers (13–15). Canonical WNT signaling is antagonized by several secreted proteins, including Dickkopf (DKK1) and members of the secreted frizzled-related proteins (SFRPs) that bind and sequester WNT ligands (16,17).
The general paradigm of WNT/β-catenin signaling has been recognized for more than a decade and is strongly supported by experimental evidence; yet the picture is far from complete. The order and kinetics of phosphorylation and protein-protein interactions at the cell membrane ‘signalosome’ remain an area of active investigation (18–20), as do the precise interactions that govern destruction complex binding, protein degradation, and β-catenin nuclear translocation (21–24). The dynamics of protein complex interactions are beginning to be revealed through the in vitro reconstruction of the destruction complex, while newer tools such as real-time pathway reporters provide insight into the kinetics of response, particularly in complex in vivo tissues (25,26). Further elucidation of how WNT signaling is tuned at each step of the process will provide an important ‘ground truth’ for deciphering and interpreting WNT dysregulation in cancer.
β-catenin-independent WNT signaling
The β-catenin-independent pathway is comparatively more diverse and less characterized than that of the canonical WNT pathway. By definition, the β-catenin-independent pathway operates without a β-catenin-mediated transcriptional response and regulates different signaling outputs, including cell polarity and migration (27,28). Like WNT/β-catenin signaling, the β-catenin-independent pathway is initiated by WNT ligands (e.g. WNT11 and WNT5A) binding to a panel of receptors including FZD, ROR2, ROR1 or RYK, resulting in activation of downstream effectors (29). WNT ligands are often grouped into canonical and non-canonical classes, but recent evidence suggests that WNT ligands once considered ‘canonical’ WNTs, like WNT3A, can activate β-catenin independent signaling (30). Given this crosstalk, it is difficult to disentangle how the β-catenin-independent pathway individually contributes to cancer phenotypes, though there is evidence for both pro and anti-tumorigenic roles (31). For instance, the Wnt5a-Ror2 axis is a prominent ligand receptor pair in the β-catenin-independent pathway that regulates planar cell polarity and tissue patterning. This signal exerts a tumor suppressive role in CRC while conferring invasiveness in other cancer cell types (32). Through the regulation of migration and cell polarity β-catenin-independent signaling likely influences tumorigenic behavior, though it is not clear that this arm of the pathway acts as a primary disease driver.
WNT signaling and tissue homeostasis
WNT is not only critical for the development of many organ systems, but it also plays a fundamental role in the maintenance of actively self-renewing tissues and in regeneration post-injury. Sustained WNT pathway activity is essential for homeostasis of the intestine, hair follicles, and hematopoietic system (3,33–35), while the induction of high WNT signaling is important for wound repair in a wide range of tissues, including skin, lung, pancreas, and liver (36–39).
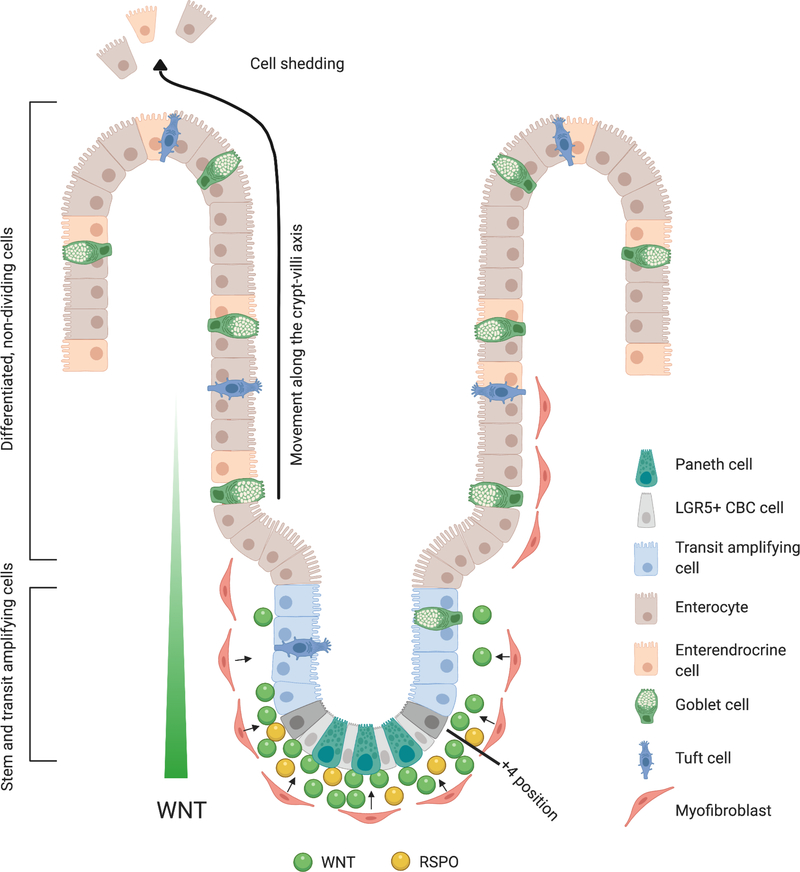
The intestinal mucosa is an archetypal example of WNT dependence in both normal tissue function and wound repair, and due to its unique cellular arrangement, provides a clear picture of how spatio-temporal WNT activity controls organ function. The small and large intestine are the most rapidly renewed tissues in adult mammals with the average life cycle of an individual epithelial cell less than a week. In mice, up to 200 new cells are generated per intestinal crypt each day and the entire epithelium is turned over within 3–5 days (40). The engine that drives this incredible flux is the LGR5-positive crypt-base columnar (CBC) stem cell that resides at the base of the crypt (Figure 2). Self-renewal of CBC cells is maintained by high levels of WNT ligand, produced and secreted by underlying stromal cells and interdigitated ‘niche cells’, such as the Paneth cell in the small intestine (41–43) (Figure 2). The differential regulation of β-catenin by transcriptional co-factors preserves the narrow window of WNT activity to govern the fate of intestinal stem cells (44). Recently, Borrelli et al revealed that C-terminal co-activators of β-catenin act as a binary on/off switch for β-catenin transcription of WNT target genes while N-terminal co-activators fine tune β-catenin transcriptional output to the exact level required for proliferation and self-renewal of intestinal stem cells (44). Rapid proliferation in the crypt results in a continuous movement of cells in an upward motion pushing cells out of the crypt where they begin to differentiate into all intestinal epithelial lineages and finally end their life cycle by undergoing apical extrusion or shedding at the tip of the villus (45). This cellular ‘conveyor belt’ leading to physical separation of cells from the WNT-high crypt niche is a key factor driving intestinal differentiation. The various processes that control lineage specification during exit from the crypt are complex and have been well-covered elsewhere (46).
Figure 2– WNT signaling in the Small intestine.
WNT ligand produced and secreted by predominantly by underlying stromal cells drives continuous proliferation in the intestinal crypt via LGR5+ crypt-base columnar (CBC) cells, interdigitated with Paneth cells. Proliferation drives the upwards motion of cells, through the transit amplifying (TA) zone where they continue to rapidly divide, and ultimately into the villus region where committed cells differentiate into all intestinal epithelial lineages. Cells are shed from the monolayer at the tip of the villus. Created with BioRender.com
The curious case of WNT pathway mutations in cancer
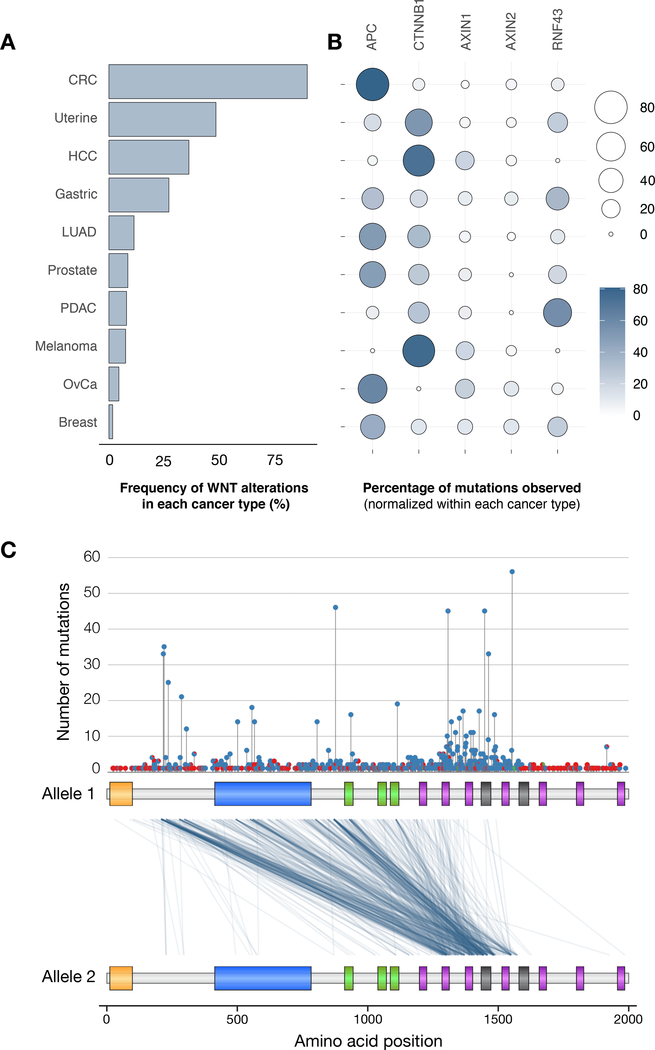
Oncogenic activation of the WNT pathway is observed to varying degrees across a range of cancers. Cancer-associated WNT hyperactivation can be WNT ligand-dependent or downstream of the ligand-receptor interface. Most mutations in RNF43 or RSPO result in ligand dependent WNT signaling and are sensitive to drugs that block WNT production (see below). Alterations, particularly truncating mutations in APC and AXIN1 disrupt the negative regulation of β-catenin by the destruction complex, whereas direct missense mutations or small in-frame deletions in β-catenin (CTNNB1) promote signaling by rendering the protein insensitive to proteosomal degradation (47). Though the outcome of each type of WNT pathway alteration is increased downstream transcriptional response, the level of pathway induction and, intriguingly, the pattern of genetic alterations that drive WNT activation in each cancer type is different (Figure 3A–B).
Figure 3. WNT pathway mutation distribution across cancer types.
A. Bar graph representing the frequency of alterations in core WNT regulators (APC, CTNNB1, AXIN1, AXIN2, RNF43) across different tumor types shown as the percentage of all tumors analyzed. B. Bubble plot showing the frequency of mutations in each WNT regulator, within the WNT-mutant subset of each cancer type. C. Lollipop plot showing number and position of truncating (blue) and missense (red) mutations in APC in CRCs. Plot below shows the position of mutations on each of two APC alleles in a given tumor, relative to the N-terminal portion of the APC protein (total length is 2843 amino acids). Many CRCs show early APC truncations, but most cancers contain at least one allele truncated within the Mutation Cluster Region (MCR) between 1200–1600 amino acids. All data derived from publicly available TCGA PanCan database (cbioportal.org).
In CRC, WNT hyperactivation is almost exclusively driven by truncating mutations in APC, whereas alterations in CTNNB1, AXIN1/2, RNF43, and RSPO2/3 (not shown) combined account for less than 15% of all WNT pathway changes. In other tumor types such as gastric, lung, prostate, ovarian and breast cancer, APC mutations are also common, but account for only half of all WNT pathway disruptions. In contrast, hepatocellular carcinoma (HCC), melanoma, uterine, and pancreatic ductal adenocarcinoma (PDAC) rarely present with APC alterations, but instead frequently harbor β-catenin (HCC, melanoma and uterine) and RNF43 (PDAC) mutations. Each of these mutations, and in particular, APC and β-catenin mutations, are potent drivers of WNT signaling; so why then, is there such a strong bias in mutation pattern between different cancer types?
The first and most obvious possibility is that environmental carcinogens or cell-intrinsic mutational signatures unique to each tissue type led to cancer-specific genetic changes. Indeed, there is some evidence for this in HCC. Through analysis of whole genome mutational patterns, Letouze et al reported that a liver cancer-specific mutational processes account for the majority CTNNB1 hotspot mutations in this disease (48). Moreover, despite their different patterns in human cancers, in animal models, engineered mutations in APC and β-catenin drive very similar disease progression in the liver, lung and colon (49–55), suggesting that functional differences may not be the dominant factor in defining cancer-selective WNT pathway alterations.
Although mutational processes are clearly important, they are likely not the whole story. By comparing observed and expected frequencies of mutations in different cancer types (which harbor distinct underlying mutational processes), Temko et al argue that biological selection can be dominant to mutational processes within a given cancer type (56). In particular, they compare APC and CTNNB1 mutations in liver, uterine and colorectal cancers, and note that selection for APC mutations is observed only in CRC. Consistent with the notion that APC mutations are favored in CRC, germline mutations in APC strongly predispose patients to the development of benign and malignant tumors in the colon (57), while other organs are less dramatically affected.
Exactly what might drive selection for APC disruption (over β-catenin mutation) is not clear, though it is worth noting that APC is a large multi-domain, multi-functional protein whose truncation or loss may cause pleiotropic effects in different cell types. APC has WNT/β-catenin-independent roles in DNA repair, apoptosis, spindle assembly, chromosome segregation, and cytoskeletal regulation through interaction with microtubules (58–60). In fact, through detailed analysis of intestinal crypts, Näthke and colleagues propose that APC truncation disrupts the orientation and asymmetry of cell division and may contribute to early tumor development by delaying cell transit from the crypt base (61). Like APC, β-catenin is also a multi-functional protein that, in addition to acting as a WNT transcription factor, interacts with E-cadherin at the epithelial adherens junction, which is essential for cell-cell contacts and tissue remodeling (62). However, unlike APC, in most cancers CTNNB1 mutations are present on only a single allele, with rare cases of loss of heterozygosity. Thus, it is likely the remaining wildtype β-catenin protein can support any lacking normal functions of mutant β-catenin at the membrane. To our knowledge the impact of loss of heterozygosity has not been investigated in CTNNB1 mutant tumors, so this point remains speculation.
The ‘Just Right’ or ‘Goldilocks’ hypothesis
In addition to mutation signatures and non-WNT related functions of key proteins, the relative level of WNT/β-catenin activation may have a strong impact on the selection of specific of WNT alterations in cancer. The ‘Goldilocks’ theory posits that WNT hyperactivation at an intermediate level (not too cold, and not too hot) is ideal for cell transformation (63). The precise ‘ideal’ level has been difficult to define experimentally, however, perhaps reflecting a varying set point across different tissues or cell types (64,65). It is noteworthy that distinct APC mutations can produce different levels of canonical WNT pathway activation for tumorigenesis in CRC and have been associated with tumor type (microsatellite instable vs. stable), location and response to targeted therapies (64,66,67). Further, while many types of truncating APC mutations are observed in colorectal (and other) cancers, most tumor cells carry at least one allele truncated within the Mutation Cluster Region (MCR) located between amino acids 1200–1600 (Figure 3C). Experimental evidence suggests that such mutations provide a hyperactivated, but not maximal, WNT response (67,68).
Thus, while there is clear bias in the types and frequency of different WNT pathway alterations in distinct cancer types, the underlying determinants of this are unclear, and so a key facet of our understanding of WNT as a cancer driver remains elusive.
WNT across different cancer types
Colorectal cancer
The WNT signaling pathway is the most dominant regulator of stem cell maintenance and proliferation in the gastrointestinal tract, driving complete renewal of the intestinal epithelium every 3–5 days (69,70). Given this, it is not surprising that alterations in the WNT signaling pathway are a near-universal feature of CRC, with more than 90% of CRCs harboring mutations in APC, CTNNB1, RNF43, AXIN1, or RSPO genes (Figure 3) (71). APC mutations were recognized as a likely initiating event in CRC around 30 years ago following the parallel identification of APC as the gene mutated in human familial adenomatous polyposis (FAP) (72) and in the intestinal cancer prone ApcMin (Multiple Intestinal Neoplasias) mouse (73). Development of engineered animal models that enabled timed disruption of Apc or activation of Ctnnb1 directly showed that WNT activation is sufficient to trigger hyperproliferation and block differentiation in the intestinal crypt (53–55,74). In particular, oncogenic WNT activation within Lgr5-positive stem cells drives rapid hyperplastic growth (74,75). Similarly, WNT induction in Bmi1+, Lrig1+, and Dclk1+ stem cell compartments can also initiate tumor growth, although the kinetics of adenoma development vary, and in the case of Dclk1, requires tissue injury and/or inflammation to instigate tumor growth (76–78). These data support a “bottom up” model whereby WNT mutations in normal crypt base stem cells drive tumor growth. In contrast, Schwitalla et al demonstrated that coincident activation of KRAS and NFkB can induce dedifferentiation and transformation of enterocytes in a “top down” model of tumor development (79). Whatever the path to tumor development, in mice and in humans, CRCs show high WNT-associated stem-cell like signatures (80). In fact, lineage tracing experiments in vivo in mice show that Lgr5-positive cells represent about 5–10% of cells in adenomas and give rise to all cancer cell lineages as well as to additional Lgr5-positive cells (81). Consistent with a key role for stem cell-like tumor propagation, ablation of cells expressing stem markers such as Bmi1 (82), or direct elimination of Lgr5-expressing cells (83,84) can reduce intestinal tumor burden.
As demonstrated in the case of KRAS and NFkB, contribution of ‘non-WNT’ factors can play a major role in determining the outcome of oncogenic WNT mutations in individual cells. For example, the homeobox transcription factor HOXA5, an important repressor of intestinal stem cell fate is suppressed by WNT but can attenuate WNT-driven cancer phenotypes if overexpressed or induced by retinoids (85). Alternatively, some proteins play a more indirect role in WNT regulation, such as the histone H3K36 methyltransferase SETD2, which restricts the WNT-dependent expansion of the stem cell compartment by influencing splicing of the WNT regulator DVL (86). WNT effects can also be bolstered by synergy with other oncogenic pathways. In particular, mutations in WNT and RAS/MAPK pathway genes are frequently observed in the same tumors (71), and in animal models show clear cooperativity in driving the early stages of tumorigenesis (87–89). While phenotypically, WNT and KRAS are cooperative drivers, the signaling mechanics between them is complicated. APC and KRAS cooperate to drive activation of cancer stem cells and tumor growth in vivo (90), while inhibition of MEK can show potent activation of the WNT pathway with increased stem cell plasticity (91). Similarly, Kabiri et al propose that WNT and RAS/MAPK pathways are mutually repressive in order to maintain the pool of intestinal stem cells at the crypt base (92). Fully elucidating the molecular details of how these two key pathways intersect and in what context may help determine future treatment strategies.
However, WNT is activated or augmented in CRC, it is usually a disease driver. Inhibiting WNT and/or β-catenin directly essentially eliminates Lgr5-positive cells, suppresses proliferation, and drives cell differentiation (55,67,93,94). In human CRC cells, overexpression of wildtype APC is sufficient to downregulate WNT signaling, induce expression of differentiation markers, and reduce tumor growth (95). Similarly, restoration of endogenous Apc expression in an in vivo APC-silencing CRC model, is sufficient to cause rapid disease regression, even in the presence of oncogenic Kras and p53 mutations (55). In this murine example, lineage tracing revealed that tumor cells could reintegrate within the normal epithelial monolayer and produce functional differentiated epithelial cells, highlighting the key role of WNT in controlling the switch between normal and transformed behavior in CRC.
Like APC and CTNNB1, RSPO fusions also act as tumor initiators and cancers drivers. Recurrent chromosome rearrangements creating EIF3E-RSPO2 and PTPRK-RSPO3 fusions are mutually exclusive with other WNT alterations in CRC, supporting a redundant role in oncogenic WNT activation (96). In clinical samples, RSPO fusions appear to be enriched in traditional serrated adenomas (TSAs) (97), though mouse models have not supported a direct link between RSPO fusions and this distinct adenoma subtype (98,99). This may reflect a difference in the cell of origin, mutational processes, and/or other cooperating genetic events absent in the ‘RSPO only’ animal models. Creation of RSPO fusions in the murine intestine using inducible in vivo CRISPR-Cas9 (99) or via cDNA expression (98) provided the first evidence that these events are tumor initiating, while multiple studies have shown that blocking WNT secretion via PORCN inhibitors, or directly inhibiting RSPO itself can block tumor growth (93,99,100). Similar to RSPO fusions, inactivating mutations in RNF43 are largely exclusive to APC mutations, but unlike other WNT alterations, are enriched in the microsatellite instable (MSI-H) tumors (13). Cancer-associated changes are predominantly nonsense and frameshift truncating mutations, spread throughout the coding sequence. Loss of the RNF43 locus or early truncations disrupt negative regulation of WNT receptor complexes and drive ligand-dependent activation of the pathway. As predicted, murine or patient-derived organoids and PDX tumors with these lesions are sensitive to PORCN inhibitors (101–103) (104) (103,105). Similar sensitivity is also seen in other tumor types, including pancreatic PDX models (106).
Two other types of RNF43 mutations reveal the complexity of how this protein controls WNT output and highlight the importance of understanding if and how specific mutations promote cancer growth. The most frequently observed RNF43 mutation is a frameshift over a short poly-guanine tract at G658-G659 (G659Vfs*41). Despite its recurrence in CRCs, this alteration has only minor effects on RNF43 activity and does not confer WNT hyperactivation. The lack of functional impact in CRC and gastric cancers suggests that G659Vfs*41 is likely a passenger mutation associated with microsatellite instability (101,107,108). Recently, Spit et al described a third category of RNF43 truncations within a 50 amino acid region (K514-Q563) in the C-terminal half of the protein (103). These truncations act to sequester or ‘trap’ CK1 at the plasma membrane, depleting it from the destruction complex and promoting β-catenin accumulation. Consequently, cells with RNF43 trapping mutations show WNT ligand independent activation of the pathway and are insensitive to PORCN inhibition.
The dominant role of WNT signaling in CRC makes it a favorable target of therapeutic interventions. As discussed further below, a variety of approaches and drugs have been developed to target hyperactive WNT and are in early phase clinical trials. We do not yet know the outcome of many of these studies, but it is evident from pre-clinical model systems that a deep understanding of recurrent WNT pathway mutations and the dependencies they create will be important for the development and deployment of effective treatments.
Liver cancer
Similar to CRC, the WNT signaling pathway is hyperactivated in a high proportion of hepatocellular carcinomas (HCCs), with frequent hotspot mutations in CTNNB1 (~30%) and inactivating alterations in AXIN1 (15%) or less commonly, APC (1.6%) (109). The frequency of CTNNB1 mutations differs based on etiology. Hepatitis C Virus (HCV)-associated tumors have a significantly higher frequency of CTNNB1 mutations (28%) compared to Hepatitis B virus (HBV)-associated disease (11%) (110). However, unlike CRC and gastric cancer, the role of WNT signaling as a driver or cooperating event in HCC pathogenesis is unclear. Work by Nejak-Bowen et al. supports WNT as a cooperative event, showing that overexpression of degradation-resistant β-catenin alone is not enough to drive HCC initiation in vivo (111). Further, transcriptomics suggests activation of the WNT/β-catenin pathway is restricted to mid-late stages of hepatocarcinogenesis (112). The role of β-catenin as a functional promoter of tumor progression is further supported by the observation that nuclear β-catenin is associated with late-stage HCC, even in the absence of oncogenic mutation (113). In addition to β-catenin mutation, WNT pathway activation is also associated upregulation of WNT ligands (WNT-1/3/5a/10b) or co-receptors (FZD3/6/7, LRP6) and downregulation of antagonist of the Wnt pathway (sFRP1/4/5 and DKK3/4) (114). The WNT-TGFβ class of HCC is linked with a more aggressive phenotype, whereas those containing CTNNB1 mutations tend to be less aggressive, more differentiated tumors (115).
While activating WNT mutations may be a mid-to-late-stage event in HCC, the story may be different in pediatric liver cancer. Hepatoblastoma (HB) is the most common pediatric liver cancer, most often diagnosed in young, pre-school age children (<3yo). Given the early onset, HBs carry relatively few genomic alterations, however CTNNB1 mutations have been observed in up to 75% of cases (116,117). Similarly, HBs have been associated with APC-mutation-linked FAP, suggesting WNT has a major driving role in this disease (118). What underlies the difference in WNT mutation prevalence and susceptibility to WNT stimuli in these distinct but related cancers is unclear, but it seems reasonable to assume that cell or origin / cell identity play a key role in WNT-driven tumorigenesis in the liver.
Lung cancer
Lung adenocarcinoma (LUAD) is the most common lung cancer subtype and the leading cause of cancer-related death, globally. In normal lung, WNT signaling is critical for both lung development and regeneration (119–121). Likewise, LUAD is frequently associated with increased expression of WNT pathway-activating genes such as WNT ligands and FZD receptors, as well as down-regulation of negative regulators of the pathway like APC, AXIN1, and DKKs (122). Depending on the assay used to quantify, it is estimated that 35–70% of LUADs have active WNT signaling (123,124), though unlike colorectal and liver cancers, only ~10% of tumors carry canonical oncogenic mutations in APC, CTNNB1 or RNF43.
These genomic data imply that downstream activation of WNT is important in LUAD, but that it can be mutationally acquired or ligand dependent. Consistent with the observation of APC and CTNNB1 mutations in some human LUAD, forced activation of the WNT pathway using engineered genetic alleles promotes progression of Kras or Braf mutant lung tumors (50,125,126). Moreover, recent work suggests that, like human LUAD, murine models carrying oncogenic Kras and p53 mutations, but without genetic WNT alterations, also rely on WNT ligands for tumor progression (127). Interestingly, in this setting, WNT-high Lgr5+ cancer cells reside in close proximity to Porcn+ cancer cells capable of secreting WNT ligand, thus forming a cancer stem cell niche, similar to those observed in normal tissues, but derived entirely of cancer cells. Indeed, inactivation of PORCN in LUAD cells (but not in the stroma) blocks tumor progression (127). Collectively, these data support the notion that cancer-cell intrinsic WNT signaling is an important component of lung cancer initiation and progression, but that WNT activation may be ligand dependent or independent. Additionally, the work in murine models implies that the creation of a WNT-dependent cancer cell derived niche is a critical step in driving ligand-dependent tumor growth.
Wnt signaling in metastasis
Metastasis is a hallmark of late stage cancer which presents as a therapeutic challenge responsible for more than 90% of cancer related mortality (128). Direct genetic evidence linking WNT signaling with metastatic progression in human cancers is scarce, though there are abundant examples in preclinical models that WNT signaling promotes stemness, proliferation, and cell motility (115,129–139), which may contribute to metastatic phenotypes.
In cases such as colorectal and gastric cancer, WNT activating mutations are early events and so common that they are rarely associated only with metastatic behavior. Indeed, in animal models that mimic tumor progression through adenoma-adenocarcinoma-metastasis, elevated WNT signaling is required for tumor cell survival at all sites (140). This is in agreement with the notion that metastasis is driven by plasticity and/or non-genetic alterations triggered by environmental cues (141). De Sousa et al first described a critical role for Lgr5+ CSC in the establishment of CRC-derived liver metastasis (83), and recently, Fumagalli et al built on this model, revealing that it is highly plastic Lgr5-negative stem cells drive dissemination prior to consolidation and growth through an emergent Lgr5+ stem cell population in the metastatic site (142). Though not directly tested, these data propose a model in which WNT does not drive the metastatic process per se but is required in both the primary tumor and metastatic site to establish and maintain tumorigenesis. Similar to CRC, once breast cancer cells form a metastatic niche, WNT ligands are recruited to reestablish signaling and maintain tumor growth (143,144). Yet, timed suppression of WNT may also be important for tumor survival. Mallardi and colleagues showed that transient suppression of WNT signaling via DKK1 induces a slow cycling state, driving immune evasion and long-term survival of latent, metastatic-initiating cells (145). As discussed below, WNT may also contribute to metastatic spread indirectly, by promoting an immunosuppressive microenvironment, allowing the establishment and growth of distant lesions.
One setting in which WNT seems to be preferentially involved in driving metastatic progression is prostate cancer. A recent comparison of localized and metastatic prostate cancers revealed that APC mutations are enriched specifically in metastatic tumors (146). This association was independent of androgen sensitivity, suggesting it is more strongly linked to metastatic progression. Using organoids and in situ genetically engineered mouse models, Leibold et al confirmed that activation of WNT by disruption of APC is sufficient to promote metastatic spread, and that WNT suppression may be an effective strategy to target disseminated disease (146).
WNT signaling and anti-tumor immunity
Since the introduction and success of immune checkpoint inhibitors and immune cell-based therapies for cancer treatment, understanding how developing cancers adapt to evade immune detection has become the most immediate goal in cancer research. In the effort to identify strategies to further improve immunotherapy outcomes and achieve long-term cancer remission, the WNT signaling pathway has emerged as a possible key to modulating immune cell function in cancers.
WNT signaling is a known regulator of immune cell function, notably suppressing the maturation and differentiation of T cells and dendritic cells (34). Active WNT signaling can promote increased survival of regulatory T cells, alter differentiation of CD4+ cells to adopt a pro tumorigenic Th17 subtype, impairs differentiation of CD8+ effector T cells, and drive dendritic cells into a more tolerogenic regulatory state (147–149). For instance, in melanoma, WNT signaling supports an immunosuppressive microenvironment by enhancing production of IL-10 and IL-12, which results in impaired dendritic cell and effector T cell function (150). Moreover, using an immune competent murine melanoma model Spranger et al highlighted a direct role for WNT activation in driving T cell exclusion and resistance to immune checkpoint inhibitors (151). They showed that WNT suppressed the recruitment of Batf3+ dendritic cells, resulting in a failure to prime CD8+ T cells in the tumor-draining lymph node (151). Near identical results were seen in HCC, where induction of mutant β-catenin in in situ derived MYC/p53−/− tumors resulted in immune evasion and resistance to αPD-1 treatment (152). Given the frequency of CTNNB1 mutations in HCC, these data highlight the potential therapeutic benefit of targeting WNT in combination with approved immunotherapies.
Beyond these two clear functionally validated examples, correlative analyses across 31 cancer types, shows that active WNT signaling is associated with non-T-cell inflamed tumors (153). Further, in CRC, where WNT is a clear driver, Grasso et al identified a correlation between high WNT transcriptional signatures and low T-cell infiltration, irrespective of CRC subtype and mutational load (154). Whether WNT signaling explains the current poor clinical response of CRCs to immunotherapies remains to be tested, but recent pre-clinical work with syngeneic CRC models suggests that WNT targeted agents may enhance anti-tumor immunity (155,156).
It is important to note that WNT may not be universally immunosuppressive. As mentioned, suppression of WNT via DKK1 induces latency associated with downregulation of cell surface immune sensors. This allows latent metastatic cells to persist long term by evading immune surveillance (157). Together, these data highlight key roles for WNT signaling in mediating anti-tumor immunity, and while most evidence suggest that WNT suppression would be an attractive therapeutic strategy further understanding the impact of WNT regulation on anti-tumor immune function will be critical to implement such approaches safely.
Therapeutic targeting of the WNT signaling pathway: inhibitors, clinical trials and resistance
Targeting the Wnt pathway
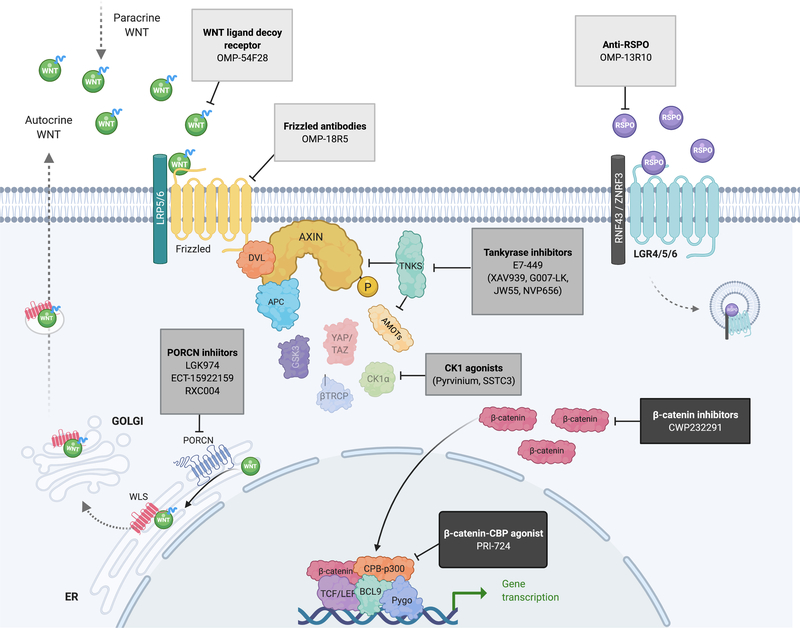
Given the high frequency of WNT pathway activation in cancer, and clear role in driving tumor progression and immunosuppression, there is immense interest in targeting the WNT pathway for cancer therapy. Current strategies to target WNT signaling can be grouped into three categories: Ligand or receptor-targeting agents, agents that promote degradation of β-catenin, and antagonists of β-catenin mediated transcription. An overview of inhibitors in completed or ongoing clinical trials are presented in Table 1 and Figure 4.
Table 1:
Wnt inhibitors used in the clinic
| Compound | Cancer type | Trial identifier |
|---|---|---|
| WNT ligand or receptor targeting agents | ||
| OMP-54F28 (Ipafricept) | Ovarian Pancreatic HCC |
NCT01608867
NCT02092363 NCT02050178 NCT02069145 |
| OMP-18R5/ Vanticumab | NSCLC Pancreatic Metastatic HER2-negative breast cancer |
NCT01345201
NCT01957007 NCT02005315 NCT01973309 |
| WNT974 (LGK974) | Pancreatic BRAF mutant and metastatic CRC Melanoma TNBC Head and Neck squamous cell cancer Cervical squamous cell cancer Esophageal squamous cell cancer Lung squamous cell cancer |
NCT01351103
NCT02278133 |
| ETC-1922159 | Advanced sold tumors | NCT02521844 |
| RXC004 | Solid tumors | NCT03447470 |
| Compounds that promote β-catenin degradation | ||
| CWP232291 | AML CML Myelodysplastic syndrome Myelofibrosis Multiple Myeloma |
NCT03055286
NCT01398462 NCT02426723 |
| E7–447 (2X-121) | Ovarian Breast Solid tumors TNBC Melanoma |
NCT03878849
NCT03562832 NCT01618136 |
| Antagonist of β-catenin mediated expression | ||
| PRI-724 | Pancreatic AML CML CRC |
NCT01764477
NCT01606579 NCT02413853 |
| E7386 | Solid neoplasms CRC neoplasms |
NCT03833700
NCT03264664 |
| SM08502 | Solid tumors | NCT0335066 |
Figure 4: Targeting the WNT pathway- inhibitors in clinical trial.
Schematic representation of the canonical WNT signaling pathway with inhibitors at various points along the pathway which are currently in clinical trials. Other validated inhibitors not yet in clinical trials are noted in parentheses. See also Table 1. Created with BioRender.com
Wnt ligand and receptor targeting agents
Cancers driven by RSPO fusions, RNF43 mutations, and autocrine/paracrine WNT activation rely on the engagement of the WNT ligand with cell surface receptors (Figure 1). Multiple pharmacologic strategies have been developed to intercept this upstream WNT trigger, including the direct inhibition of WNT ligand secretion, WNT-FZD antagonists, or RSPO3 neutralizing antibodies.
WNT-FZD binding can be blocked through direct inhibition of the receptor interface, or by sequestration of the WNT ligand. OMP-54F28 is a fusion protein comprising the WNT-binding domain of Fzd8 fused to a human IgG1 Fc domain and following evidence of efficacy in pre-clinical models is now in Phase I clinical trials for advanced HCC, ovarian, and pancreatic cancers (158). Vantictumab (OMP18R5) is a humanized monoclonal antibody that recognizes the extracellular domain of multiple FZD proteins (FZD1, 2, 5, 7, 8) and has shown anti-tumor efficacy in solid tumors (159) and trialed in combination with cytotoxic chemotherapies in lung, breast and pancreatic cancers, although dose escalation studies revealed significant bone-related toxicities (see below).
Following the identification of RSPO2 and RSPO3 fusion CRCs, neutralizing RPSO antibodies were developed to directly target these oncogenic drivers. Anti-RSPO3 antibody treatment has shown promise in RSPO fusion CRC PDX models (100) and an independently developed RSPO3 antibody (OMP-131R10) completed Phase I testing in 2018 (NCT02482441). Despite the clear rationale for this approach and evidence that RSPO3 is a disease driver (100,102), to date RSPO-focused treatments have not progressed further clinically.
LGK974 (now known as WNT974) (160), ETC-1922159 and RXC004 are PORCN inhibitors that suppress WNT secretion by preventing O-linked palmitoleoylation of WNT ligands. These inhibitors have been validated thoroughly in pre-clinical rodent models and have shown activity in RSPO fusion (93,102,161), RNF43 mutant (93,104) and WNT-ligand driven cancers (160). ETC-1922159 is also reported to synergize with PI3K/mTOR inhibitors in preclinical RNF43-mutant pancreatic cancer models in vivo (162). While preliminary results suggest WNT974 has a “manageable toxicity” profile (163), both WNT974, ETC-1922159, and vantictumab (anti-FZD) have shown dose limiting toxicities related to loss of bone mass (e.g. fractures). This is mostly likely due to the essential role of WNT signaling in osteoblast differentiation and regulation of bone homeostasis (3,164–166). Recently, Madan et al reported that blocking the resorption of bone during PORCN inhibitor treatment can ameliorate the negative consequences of WNT blockade in the bone while maintaining on target WNT inhibition (167); ETC-1922159 is currently in phase 1B testing in combination with the RANKL inhibitor denosumab to prevent bone loss (NCT02521844).
Compounds that promote β-catenin degradation
For cancers that activate WNT downstream of the receptor, signaling must be blocked at or below the regulation of β-catenin. For example, CWP232291 is a peptidomimetic drug that drives degradation of β-catenin via activation of caspases and is currently in phase I clinical trials as a single agent in acute myeloid leukemia (AML) and in chronic myeloid leukemia (CML), or in combination with lenalidomide and dexamethasone in multiple myeloma (MM).
Other strategies aim to re-engage endogenous tumor suppression by hijacking known regulators of β-catenin turnover. One such approach is via the inhibition of TNKS enzymes (TNKS/TNKS2) which control the abundance of AXIN1 protein (Figure 1) and can suppress WNT signaling, even in cells carrying truncated alleles of APC (see: “Resistance to WNT inhibition”). In preclinical studies, TNKS inhibitors demonstrate robust inhibition of WNT signaling, proliferation and synergistic effects with other targeted agents including CDK4/6, EGFR, MEK inhibitors or anti-PD-L1 (168–172). While PARP/TNKS inhibitors effectively suppress WNT signaling, there is considerable toxicity due to the dependence of normal epithelial and hematopoietic stem cells on WNT(173,174). Such on-target toxicities remain an issue in targeting the WNT signaling pathway. One dual-PARP/TNKS inhibitor that has not displayed overt intestinal toxicity in pre-clinical studies, E7449 (2X-121) (175), is currently in phase 2 clinical trials for advanced ovarian cancer and metastatic breast cancer. While E7449 shows TNKS inhibitor activity in cells, it is 50–100 times more potent against PARP1/PARP2 (IC50 1nM, and 1.2nM respectively) than TNKS/TNKS2 (50–100nM) (175), and early clinical data shows better responses in patients with BRCA mutant cancers (176), a known predictive biomarker of PARP inhibitor response. Thus, it remains unclear what role TNKS inhibition plays in the biological activity of this particular drug.
An alternate approach to re-engage DC-mediated β-catenin degradation is via direct activation of the kinases controlling β-catenin stability. Pyrvinium, an approved treatment for helminth (pinworm) infection, enhances CK1α activity and reduces WNT signaling, even in the context of stabilizing β-catenin mutations (177,178). Pyrvinium is poorly bioavailable, though newer derivatives exhibit efficacy against several WNT-driven cancer cell lines and organoids and may hold more therapeutic promise (179).
Antagonist of β-catenin mediated transcription
If β-catenin abundance cannot be controlled, its function as an oncogenic transcription factor could be targeted by interfering with the engagement of essential transcriptional co-factors. PRI-724 is a β-catenin-CBP antagonist that specifically inhibits the β-catenin-CBP interaction while promoting the formation of β-catenin and p300 interactions, thus inhibiting the self-renewal capacity of stem cells in vitro (180). E7386 is a selective inhibitor which also targets the β-catenin-CBP interaction, but was developed with improved microsomal stability, membrane permeability and solubility of C-82 which is an active form of PRI-724 (181). Similarly, antagonists of β-catenin responsive transcription (iCRT) specifically target the activity of β-catenin at the TCF transcriptional complex to inhibit WNT signaling (182). While not directly targeting β-catenin, a small molecule inhibitor of CDC-like kinase (CLK) was recently shown to inhibit a range of WNT pathway genes, including downstream transcriptional targets (e.g. AXIN2) and direct regulators of the pathway (e.g. LRP5, CTNNB1) at least in part by modifying mRNA splicing (183). This drug is currently in phase I trials for patients with advanced solid tumors.
Exploiting WNT activation to treat WNT-driven cancers
Direct and potent inhibition of WNT signaling carries significant deleterious consequences for normal tissues, but there may be opportunities to exploit WNT dependence without blocking pathway activity. Using a genome-wide CRISPR-based screen, Hinze et al recently showed that activation of the WNT pathway sensitized leukemias to treatment with asparaginase by preventing GSK3a-mediated proteosome degradation and catabolic production of asparagine (184). The same approach provided dramatic survival benefit in mice transplanted with RSPO3 fusion intestinal tumor cells, and in APC mutant cancers when co-treated with selective GSK3a inhibitors (185).
Resistance to WNT inhibition
For most molecularly targeted therapies, the emergence of drug resistance is a major hurdle to long-term clinical efficacy. To date, the minimal clinical use of WNT targeting drugs has limited the analysis of human tumor response and resistance. However, the use of cell lines and pre-clinical murine models has provided some insight into potential mechanisms of tumor escape.
We and others recently showed that sensitivity to TNKS inhibition can be dictated by type of truncating APC mutations present in a tumor or cell line (67,186). Surprisingly, the types of APC mutations that predict sensitivity or resistance to TNKS inhibition in these two studies was not the same, suggesting that there might be other factors that influence this response. Consistent with this idea, Menon et al, suggest that activation of KRAS may be associated to TNKS inhibitor resistance (169). This has not been formally demonstrated, but it is worth noting that treatment of cancer cell lines and xenografts with MEK or EGFR inhibitors can re-sensitize cells to treatment with TNKS inhibitors (170,172,187,188).
Using two human PTPRK-RSPO3 fusion cell lines, Picco et al showed that while RSPO fusion CRCs are sensitive to PORCN inhibitors, downstream activation of WNT signaling via disruptive mutations in AXIN1 can lead to the emergence of drug resistant cells (189). Such mutation-driven pathway reactivation is reminiscent of drug resistance in other settings such as treatment of EGFR mutant lung cancers with EGFR inhibitors. In the RSPO3 example, the VACO6 cells that developed AXIN1 mutations are microsatellite unstable (MSI-high) and thus have a hypermutation phenotype. Most RSPO fusion human tumors are microsatellite stable, so it is unclear whether this would be the dominant outcome in a clinical setting (96,190). Nevertheless, it provides a clear example of the need to accurately profile tumor mutations and ensure there are no downstream WNT alterations present when initiating treatment with receptor or ligand based WNT inhibitors.
Even in the case of effective tumor-intrinsic suppression of WNT production, it is possible that niche-supporting cells in the microenvironment may supplement WNT production. While it has not yet been demonstrated in tumors, Virshup and colleagues showed that normal intestinal stem cells can bypass the toxic effect of PORCN inhibitors due to juxtaposed WNT-producing stroma that avoids PORCN inhibitions via efficient drug efflux (191). Similarly, Seino et al identified a subtype of pancreatic cancer organoids that depend on WNT production from cancer-associated stroma as well as tumors cells that produce their own WNT ligands (106). In both of these contexts, tumor cells remain sensitive to PORCN inhibitors, but there was a third subtype that was at least partially resistant to PORCN inhibitor treatment, though the mechanism for this was not clear.
We recently described a genotype-dependent, but non-genetic mechanism of WNT inhibitor resistance (102), whereby induction of a YAP/TAZ-driven transcriptional program downstream of TGFβ signaling induced lineage reversion to a WNT-independent embryonic state (192). A similar phenomenon has been reported during tissue regeneration in the intestine (193,194). Interestingly, in AXIN1-mutant HCC, transformation can occur in the absence of WNT hyperactivation, associated with oncogenic signatures of Notch and YAP/TAZ (195). Recently, Kawasaki et al described a second form of lineage plasticity whereby transition to a neuroendocrine-like state, termed gastroenteropancreatic neuroendocrine neoplasms (GEP-NEN), can bypass the dependence on WNT pathway activity (196). Reminiscent of neuroendocrine transitions observed in prostate and lung cancers (197–199), the emergence of GEP-NENs is strongly correlated with disruption of both TP53 and RB1. As clinical trials of WNT inhibitors expand, it will be important to monitor lineage transitions as a possible driver of therapy failure. A mechanistic understanding of how to prevent or reverse such effects could be critical for achieving the greatest impact of WNT-targeted treatments.
Concluding remarks
Since its discovery more than 30 years ago, investigation of WNT pathway signaling in normal development and cancer has revealed an array of fascinating biological mechanisms. Our ever-expanding understanding of how WNT is activated and promotes cancer progression has highlighted opportunities to tackle the deadliest malignancies. We must now exploit what we have learned and explore new approaches to WNT-focused therapy, including broadening our gaze beyond the tumor cell. Defining the way in which WNT activation interacts with and is modulated by the tumor microenvironment (TME) and the immune system is a major challenge for the next decade and may usher a new wave of efforts to drive WNT targeted treatments into the clinic.
Statement of significance.
WNT signaling is a fundamental regulator of tissue homeostasis and oncogenic driver in many cancer types. In this review, we highlight recent advances in our understanding of WNT signaling in cancer, in particular the complexities of WNT activation in distinct cancer types, its role in immune evasion, and the challenge of targeting the WNT pathway as a therapeutic strategy.
Acknowledgements
We thank Alyna Katti for discussion and critical reading of the manuscript. L.E.D. received support from the American Cancer Society under a Research Scholar Grant (131461-RSG-17-202-01-TBG) and the National Cancer Institute (1R01CA222517-01A1). L.E.D. and T.T. received support from the Starr Cancer Consortium (SCC I14-0023).
Footnotes
Conflict of Interest Statement
L.E.D. holds equity in and is a scientific advisor for Mirimus Inc. (unrelated to this work)
References
- 1.Nusse R, Varmus HE. Many tumors induced by the mouse mammary tumor virus contain a provirus integrated in the same region of the host genome. Cell 1982;31(1):99–109 doi 10.1016/0092-8674(82)90409-3. [DOI] [PubMed] [Google Scholar]
- 2.Baker NE. Molecular cloning of sequences from wingless, a segment polarity gene in Drosophila: the spatial distribution of a transcript in embryos. Embo j 1987;6(6):1765–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clevers H, Nusse R. Wnt/β-catenin signaling and disease. Cell 2012;149(6):1192–205 doi 10.1016/j.cell.2012.05.012. [DOI] [PubMed] [Google Scholar]
- 4.McCartney BM, Näthke IS. Cell regulation by the Apc protein Apc as master regulator of epithelia. Curr Opin Cell Biol 2008;20(2):186–93 doi 10.1016/j.ceb.2008.02.001. [DOI] [PubMed] [Google Scholar]
- 5.Yang E, Tacchelly-Benites O, Wang Z, Randall MP, Tian A, Benchabane H, et al. Wnt pathway activation by ADP-ribosylation. Nat Commun 2016;7:11430 doi 10.1038/ncomms11430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Azzolin L, Panciera T, Soligo S, Enzo E, Bicciato S, Dupont S, et al. YAP/TAZ incorporation in the β-catenin destruction complex orchestrates the Wnt response. Cell 2014;158(1):157–70 doi 10.1016/j.cell.2014.06.013. [DOI] [PubMed] [Google Scholar]
- 7.Wang W, Li N, Li X, Tran MK, Han X, Chen J. Tankyrase Inhibitors Target YAP by Stabilizing Angiomotin Family Proteins. Cell Rep 2015;13(3):524–32 doi 10.1016/j.celrep.2015.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Takada R, Satomi Y, Kurata T, Ueno N, Norioka S, Kondoh H, et al. Monounsaturated fatty acid modification of Wnt protein: its role in Wnt secretion. Dev Cell 2006;11(6):791–801 doi 10.1016/j.devcel.2006.10.003. [DOI] [PubMed] [Google Scholar]
- 9.de Lau W, Barker N, Low TY, Koo BK, Li VS, Teunissen H, et al. Lgr5 homologues associate with Wnt receptors and mediate R-spondin signalling. Nature 2011;476(7360):293–7 doi 10.1038/nature10337. [DOI] [PubMed] [Google Scholar]
- 10.Carmon KS, Gong X, Lin Q, Thomas A, Liu Q. R-spondins function as ligands of the orphan receptors LGR4 and LGR5 to regulate Wnt/beta-catenin signaling. Proc Natl Acad Sci U S A 2011;108(28):11452–7 doi 10.1073/pnas.1106083108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Koo BK, Spit M, Jordens I, Low TY, Stange DE, van de Wetering M, et al. Tumour suppressor RNF43 is a stem-cell E3 ligase that induces endocytosis of Wnt receptors. Nature 2012;488(7413):665–9 doi 10.1038/nature11308. [DOI] [PubMed] [Google Scholar]
- 12.Hao HX, Xie Y, Zhang Y, Charlat O, Oster E, Avello M, et al. ZNRF3 promotes Wnt receptor turnover in an R-spondin-sensitive manner. Nature 2012;485(7397):195–200 doi 10.1038/nature11019. [DOI] [PubMed] [Google Scholar]
- 13.Giannakis M, Hodis E, Jasmine Mu X, Yamauchi M, Rosenbluh J, Cibulskis K, et al. RNF43 is frequently mutated in colorectal and endometrial cancers. Nat Genet 2014;46(12):1264–6 doi 10.1038/ng.3127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jiang X, Hao HX, Growney JD, Woolfenden S, Bottiglio C, Ng N, et al. Inactivating mutations of RNF43 confer Wnt dependency in pancreatic ductal adenocarcinoma. Proc Natl Acad Sci U S A 2013;110(31):12649–54 doi 10.1073/pnas.1307218110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bond CE, McKeone DM, Kalimutho M, Bettington ML, Pearson SA, Dumenil TD, et al. RNF43 and ZNRF3 are commonly altered in serrated pathway colorectal tumorigenesis. Oncotarget 2016;7(43):70589–600 doi 10.18632/oncotarget.12130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mao B, Wu W, Davidson G, Marhold J, Li M, Mechler BM, et al. Kremen proteins are Dickkopf receptors that regulate Wnt/beta-catenin signalling. Nature 2002;417(6889):664–7 doi 10.1038/nature756. [DOI] [PubMed] [Google Scholar]
- 17.Cruciat CM, Niehrs C. Secreted and transmembrane wnt inhibitors and activators. Cold Spring Harb Perspect Biol 2013;5(3):a015081 doi 10.1101/cshperspect.a015081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schaefer KN, Peifer M. Wnt/Beta-Catenin Signaling Regulation and a Role for Biomolecular Condensates. Dev Cell 2019;48(4):429–44 doi 10.1016/j.devcel.2019.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gerlach JP, Jordens I, Tauriello DVF, van ‘t Land-Kuper I, Bugter JM, Noordstra I, et al. TMEM59 potentiates Wnt signaling by promoting signalosome formation. Proc Natl Acad Sci U S A 2018;115(17):E3996–e4005 doi 10.1073/pnas.1721321115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee H, Evans T. TMEM88 Inhibits Wnt Signaling by Promoting Wnt Signalosome Localization to Multivesicular Bodies. iScience 2019;19:267–80 doi 10.1016/j.isci.2019.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van Kappel EC, Maurice MM. Molecular regulation and pharmacological targeting of the β-catenin destruction complex. Br J Pharmacol 2017;174(24):4575–88 doi 10.1111/bph.13922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schaefer KN, Pronobis MI, Williams CE, Zhang S, Bauer L, Goldfarb D, et al. Wnt regulation: exploring Axin-Disheveled interactions and defining mechanisms by which the SCF E3 ubiquitin ligase is recruited to the destruction complex. Mol Biol Cell 2020;31(10):992–1014 doi 10.1091/mbc.E19-11-0647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Griffin JN, Del Viso F, Duncan AR, Robson A, Hwang W, Kulkarni S, et al. RAPGEF5 Regulates Nuclear Translocation of β-Catenin. Dev Cell 2018;44(2):248–60.e4 doi 10.1016/j.devcel.2017.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lybrand DB, Naiman M, Laumann JM, Boardman M, Petshow S, Hansen K, et al. Destruction complex dynamics: Wnt/β-catenin signaling alters Axin-GSK3β interactions in vivo. Development 2019;146(13) doi 10.1242/dev.164145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Naik S, Piwnica-Worms D. Real-time imaging of beta-catenin dynamics in cells and living mice. Proc Natl Acad Sci U S A 2007;104(44):17465–70 doi 10.1073/pnas.0704465104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van de Moosdijk AAA, van de Grift YBC, de Man SMA, Zeeman AL, van Amerongen R. A novel Axin2 knock-in mouse model for visualization and lineage tracing of WNT/CTNNB1 responsive cells. Genesis 2020;58(9):e23387 doi 10.1002/dvg.23387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Anastas JN, Moon RT. WNT signalling pathways as therapeutic targets in cancer. Nat Rev Cancer 2013;13(1):11–26 doi 10.1038/nrc3419. [DOI] [PubMed] [Google Scholar]
- 28.Zhan T, Rindtorff N, Boutros M. Wnt signaling in cancer. Oncogene 2017;36(11):1461–73 doi 10.1038/onc.2016.304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang Y. Wnt/Planar cell polarity signaling: a new paradigm for cancer therapy. Mol Cancer Ther 2009;8(8):2103–9 doi 10.1158/1535-7163.Mct-09-0282. [DOI] [PubMed] [Google Scholar]
- 30.Flores-Hernández E, Velázquez DM, Castañeda-Patlán MC, Fuentes-García G, Fonseca-Camarillo G, Yamamoto-Furusho JK, et al. Canonical and non-canonical Wnt signaling are simultaneously activated by Wnts in colon cancer cells. Cell Signal 2020;72:109636 doi 10.1016/j.cellsig.2020.109636. [DOI] [PubMed] [Google Scholar]
- 31.Scheel C, Eaton EN, Li SH, Chaffer CL, Reinhardt F, Kah KJ, et al. Paracrine and autocrine signals induce and maintain mesenchymal and stem cell states in the breast. Cell 2011;145(6):926–40 doi 10.1016/j.cell.2011.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Endo M, Nishita M, Fujii M, Minami Y. Insight into the role of Wnt5a-induced signaling in normal and cancer cells. Int Rev Cell Mol Biol 2015;314:117–48 doi 10.1016/bs.ircmb.2014.10.003. [DOI] [PubMed] [Google Scholar]
- 33.Heath JP. Epithelial cell migration in the intestine. Cell Biol Int 1996;20(2):139–46 doi 10.1006/cbir.1996.0018. [DOI] [PubMed] [Google Scholar]
- 34.Staal FJ, Luis TC, Tiemessen MM. WNT signalling in the immune system: WNT is spreading its wings. Nat Rev Immunol 2008;8(8):581–93 doi 10.1038/nri2360. [DOI] [PubMed] [Google Scholar]
- 35.Lowry WE, Blanpain C, Nowak JA, Guasch G, Lewis L, Fuchs E. Defining the impact of beta-catenin/Tcf transactivation on epithelial stem cells. Genes Dev 2005;19(13):1596–611 doi 10.1101/gad.1324905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ito M, Yang Z, Andl T, Cui C, Kim N, Millar SE, et al. Wnt-dependent de novo hair follicle regeneration in adult mouse skin after wounding. Nature 2007;447(7142):316–20 doi 10.1038/nature05766. [DOI] [PubMed] [Google Scholar]
- 37.Boulter L, Govaere O, Bird TG, Radulescu S, Ramachandran P, Pellicoro A, et al. Macrophage-derived Wnt opposes Notch signaling to specify hepatic progenitor cell fate in chronic liver disease. Nat Med 2012;18(4):572–9 doi 10.1038/nm.2667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang Y, Goss AM, Cohen ED, Kadzik R, Lepore JJ, Muthukumaraswamy K, et al. A Gata6-Wnt pathway required for epithelial stem cell development and airway regeneration. Nat Genet 2008;40(7):862–70 doi 10.1038/ng.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huch M, Bonfanti P, Boj SF, Sato T, Loomans CJ, van de Wetering M, et al. Unlimited in vitro expansion of adult bi-potent pancreas progenitors through the Lgr5/R-spondin axis. Embo j 2013;32(20):2708–21 doi 10.1038/emboj.2013.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reya T, Clevers H. Wnt signalling in stem cells and cancer. Nature 2005;434(7035):843–50 doi 10.1038/nature03319. [DOI] [PubMed] [Google Scholar]
- 41.Sato T, van Es JH, Snippert HJ, Stange DE, Vries RG, van den Born M, et al. Paneth cells constitute the niche for Lgr5 stem cells in intestinal crypts. Nature 2011;469(7330):415–8 doi 10.1038/nature09637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shoshkes-Carmel M, Wang YJ, Wangensteen KJ, Tóth B, Kondo A, Massasa EE, et al. Subepithelial telocytes are an important source of Wnts that supports intestinal crypts. Nature 2018;557(7704):242–6 doi 10.1038/s41586-018-0084-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Greicius G, Kabiri Z, Sigmundsson K, Liang C, Bunte R, Singh MK, et al. PDGFRα(+) pericryptal stromal cells are the critical source of Wnts and RSPO3 for murine intestinal stem cells in vivo. Proc Natl Acad Sci U S A 2018;115(14):E3173–e81 doi 10.1073/pnas.1713510115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Borrelli C, Valenta T, Handler K, Vélez K, Gurtner A, Moro G, et al. Differential regulation of β-catenin-mediated transcription via N- and C-terminal co-factors governs identity of murine intestinal epithelial stem cells. Nat Commun 2021;12(1):1368 doi 10.1038/s41467-021-21591-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Blander JM. Death in the intestinal epithelium-basic biology and implications for inflammatory bowel disease. Febs j 2016;283(14):2720–30 doi 10.1111/febs.13771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gehart H, Clevers H. Tales from the crypt: new insights into intestinal stem cells. Nat Rev Gastroenterol Hepatol 2019;16(1):19–34 doi 10.1038/s41575-018-0081-y. [DOI] [PubMed] [Google Scholar]
- 47.Yaeger R, Chatila WK, Lipsyc MD, Hechtman JF, Cercek A, Sanchez-Vega F, et al. Clinical Sequencing Defines the Genomic Landscape of Metastatic Colorectal Cancer. Cancer Cell 2018;33(1):125–36.e3 doi 10.1016/j.ccell.2017.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Letouzé E, Shinde J, Renault V, Couchy G, Blanc JF, Tubacher E, et al. Mutational signatures reveal the dynamic interplay of risk factors and cellular processes during liver tumorigenesis. Nat Commun 2017;8(1):1315 doi 10.1038/s41467-017-01358-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Juan J, Muraguchi T, Iezza G, Sears RC, McMahon M. Diminished WNT -> β-catenin -> c-MYC signaling is a barrier for malignant progression of BRAFV600E-induced lung tumors. Genes Dev 2014;28(6):561–75 doi 10.1101/gad.233627.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rogers ZN, McFarland CD, Winters IP, Seoane JA, Brady JJ, Yoon S, et al. Mapping the in vivo fitness landscape of lung adenocarcinoma tumor suppression in mice. Nat Genet 2018;50(4):483–6 doi 10.1038/s41588-018-0083-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Molina-Sánchez P, Ruiz de Galarreta M, Yao MA, Lindblad KE, Bresnahan E, Bitterman E, et al. Cooperation Between Distinct Cancer Driver Genes Underlies Intertumor Heterogeneity in Hepatocellular Carcinoma. Gastroenterology 2020;159(6):2203–20.e14 doi 10.1053/j.gastro.2020.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Colnot S, Decaens T, Niwa-Kawakita M, Godard C, Hamard G, Kahn A, et al. Liver-targeted disruption of Apc in mice activates beta-catenin signaling and leads to hepatocellular carcinomas. Proc Natl Acad Sci U S A 2004;101(49):17216–21 doi 10.1073/pnas.0404761101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sansom OJ, Reed KR, Hayes AJ, Ireland H, Brinkmann H, Newton IP, et al. Loss of Apc in vivo immediately perturbs Wnt signaling, differentiation, and migration. Genes Dev 2004;18(12):1385–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Harada N, Tamai Y, Ishikawa T, Sauer B, Takaku K, Oshima M, et al. Intestinal polyposis in mice with a dominant stable mutation of the beta-catenin gene. EMBO J 1999;18(21):5931–42 doi 10.1093/emboj/18.21.5931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dow LE, O’Rourke KP, Simon J, Tschaharganeh DF, van Es JH, Clevers H, et al. Apc Restoration Promotes Cellular Differentiation and Reestablishes Crypt Homeostasis in Colorectal Cancer. Cell 2015;161(7):1539–52 doi 10.1016/j.cell.2015.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Temko D, Tomlinson IPM, Severini S, Schuster-Böckler B, Graham TA. The effects of mutational processes and selection on driver mutations across cancer types. Nat Commun 2018;9(1):1857 doi 10.1038/s41467-018-04208-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Groden J, Thliveris A, Samowitz W, Carlson M, Gelbert L, Albertsen H, et al. Identification and characterization of the familial adenomatous polyposis coli gene. Cell 1991;66(3):589–600 doi 10.1016/0092-8674(81)90021-0. [DOI] [PubMed] [Google Scholar]
- 58.Näthke IS, Adams CL, Polakis P, Sellin JH, Nelson WJ. The adenomatous polyposis coli tumor suppressor protein localizes to plasma membrane sites involved in active cell migration. J Cell Biol 1996;134(1):165–79 doi 10.1083/jcb.134.1.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hanson CA, Miller JR. Non-traditional roles for the Adenomatous Polyposis Coli (APC) tumor suppressor protein. Gene 2005;361:1–12 doi 10.1016/j.gene.2005.07.024. [DOI] [PubMed] [Google Scholar]
- 60.Nelson S, Näthke IS. Interactions and functions of the adenomatous polyposis coli (APC) protein at a glance. J Cell Sci 2013;126(Pt 4):873–7 doi 10.1242/jcs.100479. [DOI] [PubMed] [Google Scholar]
- 61.Quyn AJ, Appleton PL, Carey FA, Steele RJ, Barker N, Clevers H, et al. Spindle orientation bias in gut epithelial stem cell compartments is lost in precancerous tissue. Cell Stem Cell 2010;6(2):175–81 doi 10.1016/j.stem.2009.12.007. [DOI] [PubMed] [Google Scholar]
- 62.Valenta T, Hausmann G, Basler K. The many faces and functions of β-catenin. Embo j 2012;31(12):2714–36 doi 10.1038/emboj.2012.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Albuquerque C, Breukel C, van der Luijt R, Fidalgo P, Lage P, Slors FJ, et al. The ‘just-right’ signaling model: APC somatic mutations are selected based on a specific level of activation of the beta-catenin signaling cascade. Hum Mol Genet 2002;11(13):1549–60. [DOI] [PubMed] [Google Scholar]
- 64.Buchert M, Athineos D, Abud HE, Burke ZD, Faux MC, Samuel MS, et al. Genetic dissection of differential signaling threshold requirements for the Wnt/beta-catenin pathway in vivo. PLoS Genet 2010;6(1):e1000816 doi 10.1371/journal.pgen.1000816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Leedham SJ, Rodenas-Cuadrado P, Howarth K, Lewis A, Mallappa S, Segditsas S, et al. A basal gradient of Wnt and stem-cell number influences regional tumour distribution in human and mouse intestinal tracts. Gut 2013;62(1):83–93 doi 10.1136/gutjnl-2011-301601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Christie M, Jorissen RN, Mouradov D, Sakthianandeswaren A, Li S, Day F, et al. Different APC genotypes in proximal and distal sporadic colorectal cancers suggest distinct WNT/β-catenin signalling thresholds for tumourigenesis. Oncogene 2013;32(39):4675–82 doi 10.1038/onc.2012.486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Schatoff EM, Goswami S, Zafra MP, Foronda M, Shusterman M, Leach BI, et al. Distinct Colorectal Cancer-Associated APC Mutations Dictate Response to Tankyrase Inhibition. Cancer Discov 2019;9(10):1358–71 doi 10.1158/2159-8290.Cd-19-0289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gaspar C, Franken P, Molenaar L, Breukel C, van der Valk M, Smits R, et al. A targeted constitutive mutation in the APC tumor suppressor gene underlies mammary but not intestinal tumorigenesis. PLoS Genet 2009;5(7):e1000547 doi 10.1371/journal.pgen.1000547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Schepers A, Clevers H. Wnt signaling, stem cells, and cancer of the gastrointestinal tract. Cold Spring Harb Perspect Biol 2012;4(4):a007989 doi 10.1101/cshperspect.a007989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Krausova M, Korinek V. Wnt signaling in adult intestinal stem cells and cancer. Cell Signal 2014;26(3):570–9 doi 10.1016/j.cellsig.2013.11.032. [DOI] [PubMed] [Google Scholar]
- 71.Comprehensive molecular characterization of human colon and rectal cancer. Nature 2012;487(7407):330–7 doi 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Galiatsatos P, Foulkes WD. Familial adenomatous polyposis. Am J Gastroenterol 2006;101(2):385–98 doi 10.1111/j.1572-0241.2006.00375.x. [DOI] [PubMed] [Google Scholar]
- 73.Su LK, Kinzler KW, Vogelstein B, Preisinger AC, Moser AR, Luongo C, et al. Multiple intestinal neoplasia caused by a mutation in the murine homolog of the APC gene. Science 1992;256(5057):668–70 doi 10.1126/science.1350108. [DOI] [PubMed] [Google Scholar]
- 74.Barker N, Ridgway RA, van Es JH, van de Wetering M, Begthel H, van den Born M, et al. Crypt stem cells as the cells-of-origin of intestinal cancer. Nature 2009;457(7229):608–11 doi 10.1038/nature07602. [DOI] [PubMed] [Google Scholar]
- 75.Barker N, van Es JH, Kuipers J, Kujala P, van den Born M, Cozijnsen M, et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature 2007;449(7165):1003–7 doi 10.1038/nature06196. [DOI] [PubMed] [Google Scholar]
- 76.Yanai H, Atsumi N, Tanaka T, Nakamura N, Komai Y, Omachi T, et al. Intestinal cancer stem cells marked by Bmi1 or Lgr5 expression contribute to tumor propagation via clonal expansion. Sci Rep 2017;7:41838 doi 10.1038/srep41838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Powell AE, Vlacich G, Zhao ZY, McKinley ET, Washington MK, Manning HC, et al. Inducible loss of one Apc allele in Lrig1-expressing progenitor cells results in multiple distal colonic tumors with features of familial adenomatous polyposis. Am J Physiol Gastrointest Liver Physiol 2014;307(1):G16–23 doi 10.1152/ajpgi.00358.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Westphalen CB, Asfaha S, Hayakawa Y, Takemoto Y, Lukin DJ, Nuber AH, et al. Long-lived intestinal tuft cells serve as colon cancer-initiating cells. J Clin Invest 2014;124(3):1283–95 doi 10.1172/jci73434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Schwitalla S, Fingerle AA, Cammareri P, Nebelsiek T, Göktuna SI, Ziegler PK, et al. Intestinal tumorigenesis initiated by dedifferentiation and acquisition of stem-cell-like properties. Cell 2013;152(1–2):25–38 doi 10.1016/j.cell.2012.12.012. [DOI] [PubMed] [Google Scholar]
- 80.Merlos-Suárez A, Barriga FM, Jung P, Iglesias M, Céspedes MV, Rossell D, et al. The intestinal stem cell signature identifies colorectal cancer stem cells and predicts disease relapse. Cell Stem Cell 2011;8(5):511–24 doi 10.1016/j.stem.2011.02.020. [DOI] [PubMed] [Google Scholar]
- 81.Schepers AG, Snippert HJ, Stange DE, van den Born M, van Es JH, van de Wetering M, et al. Lineage tracing reveals Lgr5+ stem cell activity in mouse intestinal adenomas. Science 2012;337(6095):730–5 doi 10.1126/science.1224676. [DOI] [PubMed] [Google Scholar]
- 82.Maynard MA, Ferretti R, Hilgendorf KI, Perret C, Whyte P, Lees JA. Bmi1 is required for tumorigenesis in a mouse model of intestinal cancer. Oncogene 2014;33(28):3742–7 doi 10.1038/onc.2013.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.de Sousa e Melo F, Kurtova AV, Harnoss JM, Kljavin N, Hoeck JD, Hung J, et al. A distinct role for Lgr5(+) stem cells in primary and metastatic colon cancer. Nature 2017;543(7647):676–80 doi 10.1038/nature21713. [DOI] [PubMed] [Google Scholar]
- 84.Shimokawa M, Ohta Y, Nishikori S, Matano M, Takano A, Fujii M, et al. Visualization and targeting of LGR5(+) human colon cancer stem cells. Nature 2017;545(7653):187–92 doi 10.1038/nature22081. [DOI] [PubMed] [Google Scholar]
- 85.Ordóñez-Morán P, Dafflon C, Imajo M, Nishida E, Huelsken J. HOXA5 Counteracts Stem Cell Traits by Inhibiting Wnt Signaling in Colorectal Cancer. Cancer Cell 2015;28(6):815–29 doi 10.1016/j.ccell.2015.11.001. [DOI] [PubMed] [Google Scholar]
- 86.Yuan H, Li N, Fu D, Ren J, Hui J, Peng J, et al. Histone methyltransferase SETD2 modulates alternative splicing to inhibit intestinal tumorigenesis. J Clin Invest 2017;127(9):3375–91 doi 10.1172/jci94292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Haigis KM, Kendall KR, Wang Y, Cheung A, Haigis MC, Glickman JN, et al. Differential effects of oncogenic K-Ras and N-Ras on proliferation, differentiation and tumor progression in the colon. Nat Genet 2008;40(5):600–8 doi 10.1038/ng.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Boutin AT, Liao WT, Wang M, Hwang SS, Karpinets TV, Cheung H, et al. Oncogenic Kras drives invasion and maintains metastases in colorectal cancer. Genes Dev 2017;31(4):370–82 doi 10.1101/gad.293449.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.D’Abaco GM, Whitehead RH, Burgess AW. Synergy between Apc min and an activated ras mutation is sufficient to induce colon carcinomas. Mol Cell Biol 1996;16(3):884–91 doi 10.1128/mcb.16.3.884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Moon BS, Jeong WJ, Park J, Kim TI, Min do S, Choi KY. Role of oncogenic K-Ras in cancer stem cell activation by aberrant Wnt/β-catenin signaling. J Natl Cancer Inst 2014;106(2):djt373 doi 10.1093/jnci/djt373. [DOI] [PubMed] [Google Scholar]
- 91.Zhan T, Ambrosi G, Wandmacher AM, Rauscher B, Betge J, Rindtorff N, et al. MEK inhibitors activate Wnt signalling and induce stem cell plasticity in colorectal cancer. Nat Commun 2019;10(1):2197 doi 10.1038/s41467-019-09898-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kabiri Z, Greicius G, Zaribafzadeh H, Hemmerich A, Counter CM, Virshup DM. Wnt signaling suppresses MAPK-driven proliferation of intestinal stem cells. J Clin Invest 2018;128(9):3806–12 doi 10.1172/jci99325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Madan B, Ke Z, Harmston N, Ho SY, Frois AO, Alam J, et al. Wnt addiction of genetically defined cancers reversed by PORCN inhibition. Oncogene 2016;35(17):2197–207 doi 10.1038/onc.2015.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Huang WS, Wang JP, Wang T, Fang JY, Lan P, Ma JP. ShRNA-mediated gene silencing of beta-catenin inhibits growth of human colon cancer cells. World J Gastroenterol 2007;13(48):6581–7 doi 10.3748/wjg.v13.i48.6581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Faux MC, Ross JL, Meeker C, Johns T, Ji H, Simpson RJ, et al. Restoration of full-length adenomatous polyposis coli (APC) protein in a colon cancer cell line enhances cell adhesion. J Cell Sci 2004;117(Pt 3):427–39 doi 10.1242/jcs.00862. [DOI] [PubMed] [Google Scholar]
- 96.Seshagiri S, Stawiski EW, Durinck S, Modrusan Z, Storm EE, Conboy CB, et al. Recurrent R-spondin fusions in colon cancer. Nature 2012;488(7413):660–4 doi 10.1038/nature11282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Sekine S, Yamashita S, Tanabe T, Hashimoto T, Yoshida H, Taniguchi H, et al. Frequent PTPRK-RSPO3 fusions and RNF43 mutations in colorectal traditional serrated adenoma. J Pathol 2016;239(2):133–8 doi 10.1002/path.4709. [DOI] [PubMed] [Google Scholar]
- 98.Hilkens J, Timmer NC, Boer M, Ikink GJ, Schewe M, Sacchetti A, et al. RSPO3 expands intestinal stem cell and niche compartments and drives tumorigenesis. Gut 2017;66(6):1095–105 doi 10.1136/gutjnl-2016-311606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Han T, Schatoff EM, Murphy C, Zafra MP, Wilkinson JE, Elemento O, et al. R-Spondin chromosome rearrangements drive Wnt-dependent tumour initiation and maintenance in the intestine. Nat Commun 2017;8:15945 doi 10.1038/ncomms15945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Storm EE, Durinck S, de Sousa e Melo F, Tremayne J, Kljavin N, Tan C, et al. Targeting PTPRK-RSPO3 colon tumours promotes differentiation and loss of stem-cell function. Nature 2016;529(7584):97–100 doi 10.1038/nature16466. [DOI] [PubMed] [Google Scholar]
- 101.van de Wetering M, Francies HE, Francis JM, Bounova G, Iorio F, Pronk A, et al. Prospective derivation of a living organoid biobank of colorectal cancer patients. Cell 2015;161(4):933–45 doi 10.1016/j.cell.2015.03.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Han T, Goswami S, Hu Y, Tang F, Zafra MP, Murphy C, et al. Lineage Reversion Drives WNT Independence in Intestinal Cancer. Cancer Discov 2020;10(10):1590–609 doi 10.1158/2159-8290.Cd-19-1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Spit M, Fenderico N, Jordens I, Radaszkiewicz T, Lindeboom RG, Bugter JM, et al. RNF43 truncations trap CK1 to drive niche-independent self-renewal in cancer. Embo j 2020;39(18):e103932 doi 10.15252/embj.2019103932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Koo BK, van Es JH, van den Born M, Clevers H. Porcupine inhibitor suppresses paracrine Wnt-driven growth of Rnf43;Znrf3-mutant neoplasia. Proc Natl Acad Sci U S A 2015;112(24):7548–50 doi 10.1073/pnas.1508113112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Yu J, Yusoff PAM, Woutersen DTJ, Goh P, Harmston N, Smits R, et al. The Functional Landscape of Patient-Derived RNF43 Mutations Predicts Sensitivity to Wnt Inhibition. Cancer Res 2020;80(24):5619–32 doi 10.1158/0008-5472.Can-20-0957. [DOI] [PubMed] [Google Scholar]
- 106.Seino T, Kawasaki S, Shimokawa M, Tamagawa H, Toshimitsu K, Fujii M, et al. Human Pancreatic Tumor Organoids Reveal Loss of Stem Cell Niche Factor Dependence during Disease Progression. Cell Stem Cell 2018;22(3):454–67 e6 doi 10.1016/j.stem.2017.12.009. [DOI] [PubMed] [Google Scholar]
- 107.Li S, Lavrijsen M, Bakker A, Magierowski M, Magierowska K, Liu P, et al. Commonly observed RNF43 mutations retain functionality in attenuating Wnt/β-catenin signaling and unlikely confer Wnt-dependency onto colorectal cancers. Oncogene 2020;39(17):3458–72 doi 10.1038/s41388-020-1232-5. [DOI] [PubMed] [Google Scholar]
- 108.Tu J, Park S, Yu W, Zhang S, Wu L, Carmon K, et al. The most common RNF43 mutant G659Vfs*41 is fully functional in inhibiting Wnt signaling and unlikely to play a role in tumorigenesis. Sci Rep 2019;9(1):18557 doi 10.1038/s41598-019-54931-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Guichard C, Amaddeo G, Imbeaud S, Ladeiro Y, Pelletier L, Maad IB, et al. Integrated analysis of somatic mutations and focal copy-number changes identifies key genes and pathways in hepatocellular carcinoma. Nat Genet 2012;44(6):694–8 doi 10.1038/ng.2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ding SL, Yang ZW, Wang J, Zhang XL, Chen XM, Lu FM. Integrative analysis of aberrant Wnt signaling in hepatitis B virus-related hepatocellular carcinoma. World J Gastroenterol 2015;21(20):6317–28 doi 10.3748/wjg.v21.i20.6317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Nejak-Bowen KN, Thompson MD, Singh S, Bowen WC Jr., Dar MJ, Khillan J, et al. Accelerated liver regeneration and hepatocarcinogenesis in mice overexpressing serine-45 mutant beta-catenin. Hepatology 2010;51(5):1603–13 doi 10.1002/hep.23538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Marquardt JU, Seo D, Andersen JB, Gillen MC, Kim MS, Conner EA, et al. Sequential transcriptome analysis of human liver cancer indicates late stage acquisition of malignant traits. J Hepatol 2014;60(2):346–53 doi 10.1016/j.jhep.2013.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Kim E, Lisby A, Ma C, Lo N, Ehmer U, Hayer KE, et al. Promotion of growth factor signaling as a critical function of β-catenin during HCC progression. Nat Commun 2019;10(1):1909 doi 10.1038/s41467-019-09780-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Bengochea A, de Souza MM, Lefrançois L, Le Roux E, Galy O, Chemin I, et al. Common dysregulation of Wnt/Frizzled receptor elements in human hepatocellular carcinoma. Br J Cancer 2008;99(1):143–50 doi 10.1038/sj.bjc.6604422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Lachenmayer A, Alsinet C, Savic R, Cabellos L, Toffanin S, Hoshida Y, et al. Wnt-pathway activation in two molecular classes of hepatocellular carcinoma and experimental modulation by sorafenib. Clin Cancer Res 2012;18(18):4997–5007 doi 10.1158/1078-0432.Ccr-11-2322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Koch A, Denkhaus D, Albrecht S, Leuschner I, von Schweinitz D, Pietsch T. Childhood hepatoblastomas frequently carry a mutated degradation targeting box of the beta-catenin gene. Cancer Res 1999;59(2):269–73. [PubMed] [Google Scholar]
- 117.Eichenmüller M, Trippel F, Kreuder M, Beck A, Schwarzmayr T, Häberle B, et al. The genomic landscape of hepatoblastoma and their progenies with HCC-like features. J Hepatol 2014;61(6):1312–20 doi 10.1016/j.jhep.2014.08.009. [DOI] [PubMed] [Google Scholar]
- 118.Giardiello FM, Petersen GM, Brensinger JD, Luce MC, Cayouette MC, Bacon J, et al. Hepatoblastoma and APC gene mutation in familial adenomatous polyposis. Gut 1996;39(6):867–9 doi 10.1136/gut.39.6.867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Clevers H, Loh KM, Nusse R. Stem cell signaling. An integral program for tissue renewal and regeneration: Wnt signaling and stem cell control. Science 2014;346(6205):1248012 doi 10.1126/science.1248012. [DOI] [PubMed] [Google Scholar]
- 120.Hogan BL, Barkauskas CE, Chapman HA, Epstein JA, Jain R, Hsia CC, et al. Repair and regeneration of the respiratory system: complexity, plasticity, and mechanisms of lung stem cell function. Cell Stem Cell 2014;15(2):123–38 doi 10.1016/j.stem.2014.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Herriges M, Morrisey EE. Lung development: orchestrating the generation and regeneration of a complex organ. Development 2014;141(3):502–13 doi 10.1242/dev.098186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Stewart DJ. Wnt signaling pathway in non-small cell lung cancer. Journal of the National Cancer Institute 2014;106(1):djt356 doi 10.1093/jnci/djt356. [DOI] [PubMed] [Google Scholar]
- 123.Licchesi JD, Westra WH, Hooker CM, Machida EO, Baylin SB, Herman JG. Epigenetic alteration of Wnt pathway antagonists in progressive glandular neoplasia of the lung. Carcinogenesis 2008;29(5):895–904 doi 10.1093/carcin/bgn017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Xu X, Sun PL, Li JZ, Jheon S, Lee CT, Chung JH. Aberrant Wnt1/β-catenin expression is an independent poor prognostic marker of non-small cell lung cancer after surgery. J Thorac Oncol 2011;6(4):716–24 doi 10.1097/JTO.0b013e31820c5189. [DOI] [PubMed] [Google Scholar]
- 125.Juan J, Muraguchi T, Iezza G, Sears RC, McMahon M. Diminished WNT -> beta-catenin -> c-MYC signaling is a barrier for malignant progression of BRAFV600E-induced lung tumors. Genes Dev 2014;28(6):561–75 doi 10.1101/gad.233627.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Pacheco-Pinedo EC, Durham AC, Stewart KM, Goss AM, Lu MM, Demayo FJ, et al. Wnt/beta-catenin signaling accelerates mouse lung tumorigenesis by imposing an embryonic distal progenitor phenotype on lung epithelium. The Journal of clinical investigation 2011;121(5):1935–45 doi 10.1172/JCI44871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Tammela T, Sanchez-Rivera FJ, Cetinbas NM, Wu K, Joshi NS, Helenius K, et al. A Wnt-producing niche drives proliferative potential and progression in lung adenocarcinoma. Nature 2017;545(7654):355–9 doi 10.1038/nature22334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Gupta GP, Massagué J. Cancer metastasis: building a framework. Cell 2006;127(4):679–95 doi 10.1016/j.cell.2006.11.001. [DOI] [PubMed] [Google Scholar]
- 129.Sundqvist A, Morikawa M, Ren J, Vasilaki E, Kawasaki N, Kobayashi M, et al. JUNB governs a feed-forward network of TGFβ signaling that aggravates breast cancer invasion. Nucleic Acids Res 2018;46(3):1180–95 doi 10.1093/nar/gkx1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Talbot LJ, Bhattacharya SD, Kuo PC. Epithelial-mesenchymal transition, the tumor microenvironment, and metastatic behavior of epithelial malignancies. Int J Biochem Mol Biol 2012;3(2):117–36. [PMC free article] [PubMed] [Google Scholar]
- 131.Zhang H, Xue Y. Wnt pathway is involved in advanced gastric carcinoma. Hepatogastroenterology 2008;55(84):1126–30. [PubMed] [Google Scholar]
- 132.Katoh M. Frequent up-regulation of WNT2 in primary gastric cancer and colorectal cancer. Int J Oncol 2001;19(5):1003–7 doi 10.3892/ijo.19.5.1003. [DOI] [PubMed] [Google Scholar]
- 133.Kurayoshi M, Oue N, Yamamoto H, Kishida M, Inoue A, Asahara T, et al. Expression of Wnt5a is correlated with aggressiveness of gastric cancer by stimulating cell migration and invasion. Cancer Res 2006;66(21):10439–48 doi 10.1158/0008-5472.Can-06-2359. [DOI] [PubMed] [Google Scholar]
- 134.Bo H, Gao L, Chen Y, Zhang J, Zhu M. Upregulation of the expression of Wnt5a promotes the proliferation of pancreatic cancer cells in vitro and in a nude mouse model. Mol Med Rep 2016;13(2):1163–71 doi 10.3892/mmr.2015.4642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Wu DJ, Jiang YS, He RZ, Tao LY, Yang MW, Fu XL, et al. High expression of WNT7A predicts poor prognosis and promote tumor metastasis in pancreatic ductal adenocarcinoma. Sci Rep 2018;8(1):15792 doi 10.1038/s41598-018-34094-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Pai P, Rachagani S, Lakshmanan I, Macha MA, Sheinin Y, Smith LM, et al. The canonical Wnt pathway regulates the metastasis-promoting mucin MUC4 in pancreatic ductal adenocarcinoma. Mol Oncol 2016;10(2):224–39 doi 10.1016/j.molonc.2015.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Peng YY, He YH, Chen C, Xu T, Li L, Ni MM, et al. NLRC5 regulates cell proliferation, migration and invasion in hepatocellular carcinoma by targeting the Wnt/β-catenin signaling pathway. Cancer Lett 2016;376(1):10–21 doi 10.1016/j.canlet.2016.03.006. [DOI] [PubMed] [Google Scholar]
- 138.Cai J, Xiong Q, Jiang X, Zhou S, Liu T. RNF6 facilitates metastasis and radioresistance in hepatocellular carcinoma through ubiquitination of FoxA1. Exp Cell Res 2019;374(1):152–61 doi 10.1016/j.yexcr.2018.11.019. [DOI] [PubMed] [Google Scholar]
- 139.Wang X, Wang X, Liu Y, Dong Y, Wang Y, Kassab MA, et al. LGR5 regulates gastric adenocarcinoma cell proliferation and invasion via activating Wnt signaling pathway. Oncogenesis 2018;7(8):57 doi 10.1038/s41389-018-0071-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.O’Rourke KP, Loizou E, Livshits G, Schatoff EM, Baslan T, Manchado E, et al. Transplantation of engineered organoids enables rapid generation of metastatic mouse models of colorectal cancer. Nat Biotechnol 2017;35(6):577–82 doi 10.1038/nbt.3837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Brabletz T. To differentiate or not--routes towards metastasis. Nat Rev Cancer 2012;12(6):425–36 doi 10.1038/nrc3265. [DOI] [PubMed] [Google Scholar]
- 142.Fumagalli A, Oost KC, Kester L, Morgner J, Bornes L, Bruens L, et al. Plasticity of Lgr5-Negative Cancer Cells Drives Metastasis in Colorectal Cancer. Cell Stem Cell 2020;26(4):569–78.e7 doi 10.1016/j.stem.2020.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Kim SJ, Garcia-Recio S, Creighton CJ, Perou CM, Rosen JM. Alterations in Wnt- and/or STAT3 signaling pathways and the immune microenvironment during metastatic progression. Oncogene 2019;38(31):5942–58 doi 10.1038/s41388-019-0852-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Eyre R, Alférez DG, Santiago-Gómez A, Spence K, McConnell JC, Hart C, et al. Microenvironmental IL1β promotes breast cancer metastatic colonisation in the bone via activation of Wnt signalling. Nat Commun 2019;10(1):5016 doi 10.1038/s41467-019-12807-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Malladi S, Macalinao DG, Jin X, He L, Basnet H, Zou Y, et al. Metastatic Latency and Immune Evasion through Autocrine Inhibition of WNT. Cell 2016;165(1):45–60 doi 10.1016/j.cell.2016.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Leibold J, Ruscetti M, Cao Z, Ho YJ, Baslan T, Zou M, et al. Somatic tissue engineering in mouse models reveals an actionable role for WNT pathway alterations in prostate cancer metastasis. Cancer Discov 2020. doi 10.1158/2159-8290.Cd-19-1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Hong Y, Manoharan I, Suryawanshi A, Majumdar T, Angus-Hill ML, Koni PA, et al. β-catenin promotes regulatory T-cell responses in tumors by inducing vitamin A metabolism in dendritic cells. Cancer Res 2015;75(4):656–65 doi 10.1158/0008-5472.Can-14-2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Fu C, Liang X, Cui W, Ober-Blöbaum JL, Vazzana J, Shrikant PA, et al. β-Catenin in dendritic cells exerts opposite functions in cross-priming and maintenance of CD8+ T cells through regulation of IL-10. Proc Natl Acad Sci U S A 2015;112(9):2823–8 doi 10.1073/pnas.1414167112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Ding Y, Shen S, Lino AC, Curotto de Lafaille MA, Lafaille JJ. Beta-catenin stabilization extends regulatory T cell survival and induces anergy in nonregulatory T cells. Nat Med 2008;14(2):162–9 doi 10.1038/nm1707. [DOI] [PubMed] [Google Scholar]
- 150.Yaguchi T, Goto Y, Kido K, Mochimaru H, Sakurai T, Tsukamoto N, et al. Immune suppression and resistance mediated by constitutive activation of Wnt/β-catenin signaling in human melanoma cells. J Immunol 2012;189(5):2110–7 doi 10.4049/jimmunol.1102282. [DOI] [PubMed] [Google Scholar]
- 151.Spranger S, Bao R, Gajewski TF. Melanoma-intrinsic β-catenin signalling prevents anti-tumour immunity. Nature 2015;523(7559):231–5 doi 10.1038/nature14404. [DOI] [PubMed] [Google Scholar]
- 152.Ruiz de Galarreta M, Bresnahan E, Molina-Sánchez P, Lindblad KE, Maier B, Sia D, et al. β-Catenin Activation Promotes Immune Escape and Resistance to Anti-PD-1 Therapy in Hepatocellular Carcinoma. Cancer Discov 2019;9(8):1124–41 doi 10.1158/2159-8290.Cd-19-0074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Luke JJ, Bao R, Sweis RF, Spranger S, Gajewski TF. WNT/β-catenin Pathway Activation Correlates with Immune Exclusion across Human Cancers. Clin Cancer Res 2019;25(10):3074–83 doi 10.1158/1078-0432.Ccr-18-1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Grasso CS, Giannakis M, Wells DK, Hamada T, Mu XJ, Quist M, et al. Genetic Mechanisms of Immune Evasion in Colorectal Cancer. Cancer Discov 2018;8(6):730–49 doi 10.1158/2159-8290.Cd-17-1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Waaler J, Leenders RGG, Sowa ST, Alam Brinch S, Lycke M, Nieczypor P, et al. Preclinical Lead Optimization of a 1,2,4-Triazole Based Tankyrase Inhibitor. Journal of Medicinal Chemistry 2020;63(13):6834–46 doi 10.1021/acs.jmedchem.0c00208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Wang C, Yan J, Yin P, Gui L, Ji L, Ma B, et al. β-Catenin inhibition shapes tumor immunity and synergizes with immunotherapy in colorectal cancer. Oncoimmunology 2020;9(1):1809947 doi 10.1080/2162402x.2020.1809947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Malladi S, Macalinao DG, Jin X, He L, Basnet H, Zou Y, et al. Metastatic Latency and Immune Evasion through Autocrine Inhibition of WNT. Cell 2016;165(1):45–60 doi 10.1016/j.cell.2016.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Pete Yeung LB, Cancilla Belinda, Dee-Hoskins Cristina, Evans James W., Fischer Marcus M., Yen Wan-Ching, Gurney Austin, Lewicki John, Hoey Timothy and Kapoun Ann M.. Abstract 1907: Wnt pathway antagonist OMP-54F28 (FZD8-Fc) inhibits tumor growth and reduces tumor-initiating cell frequency in patient-derived hepatocellular carcinoma and ovarian cancer xenograft models. 2014. DOI: 10.1158/1538-7445.AM2014-1907. [DOI] [Google Scholar]
- 159.Gurney A, Axelrod F, Bond CJ, Cain J, Chartier C, Donigan L, et al. Wnt pathway inhibition via the targeting of Frizzled receptors results in decreased growth and tumorigenicity of human tumors. Proc Natl Acad Sci U S A 2012;109(29):11717–22 doi 10.1073/pnas.1120068109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Liu J, Pan S, Hsieh MH, Ng N, Sun F, Wang T, et al. Targeting Wnt-driven cancer through the inhibition of Porcupine by LGK974. Proc Natl Acad Sci U S A 2013;110(50):20224–9 doi 10.1073/pnas.1314239110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Woodcock S, Bhamra I, Jones C, Cook AE, Eagle C, Phillips C. Abstract 3874: Efficacy of the Wnt/Beta-Catenin pathway inhibitor RXC004 in genetically-defined models of cancer. Cancer Research 2019;79(13 Supplement):3874- doi 10.1158/1538-7445.Am2019-3874. [DOI] [Google Scholar]
- 162.Zhong Z, Sepramaniam S, Chew XH, Wood K, Lee MA, Madan B, et al. PORCN inhibition synergizes with PI3K/mTOR inhibition in Wnt-addicted cancers. Oncogene 2019;38(40):6662–77 doi 10.1038/s41388-019-0908-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Janku F, Connolly R, LoRusso P, de Jonge M, Vaishampayan U, Rodon J, et al. Abstract C45: Phase I study of WNT974, a first-in-class Porcupine inhibitor, in advanced solid tumors. Molecular Cancer Therapeutics 2015;14(12 Supplement 2):C45–C doi 10.1158/1535-7163.Targ-15-c45. [DOI] [Google Scholar]
- 164.Glass DA 2nd, Bialek P, Ahn JD, Starbuck M, Patel MS, Clevers H, et al. Canonical Wnt signaling in differentiated osteoblasts controls osteoclast differentiation. Dev Cell 2005;8(5):751–64 doi 10.1016/j.devcel.2005.02.017. [DOI] [PubMed] [Google Scholar]
- 165.Houschyar KS, Tapking C, Borrelli MR, Popp D, Duscher D, Maan ZN, et al. Wnt Pathway in Bone Repair and Regeneration - What Do We Know So Far. Front Cell Dev Biol 2018;6:170 doi 10.3389/fcell.2018.00170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Kato M, Patel MS, Levasseur R, Lobov I, Chang BH, Glass DA 2nd, et al. Cbfa1-independent decrease in osteoblast proliferation, osteopenia, and persistent embryonic eye vascularization in mice deficient in Lrp5, a Wnt coreceptor. J Cell Biol 2002;157(2):303–14 doi 10.1083/jcb.200201089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Madan B, McDonald MJ, Foxa GE, Diegel CR, Williams BO, Virshup DM. Bone loss from Wnt inhibition mitigated by concurrent alendronate therapy. Bone Res 2018;6:17 doi 10.1038/s41413-018-0017-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Waaler J, Leenders RGG, Sowa ST, Alam Brinch S, Lycke M, Nieczypor P, et al. Preclinical Lead Optimization of a 1,2,4-Triazole Based Tankyrase Inhibitor. J Med Chem 2020. doi 10.1021/acs.jmedchem.0c00208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Menon M, Elliott R, Bowers L, Balan N, Rafiq R, Costa-Cabral S, et al. A novel tankyrase inhibitor, MSC2504877, enhances the effects of clinical CDK4/6 inhibitors. Sci Rep 2019;9(1):201 doi 10.1038/s41598-018-36447-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Schoumacher M, Hurov KE, Lehar J, Yan-Neale Y, Mishina Y, Sonkin D, et al. Inhibiting Tankyrases sensitizes KRAS-mutant cancer cells to MEK inhibitors via FGFR2 feedback signaling. Cancer Res 2014;74(12):3294–305 doi 10.1158/0008-5472.CAN-14-0138-T. [DOI] [PubMed] [Google Scholar]
- 171.Mizutani A, Yashiroda Y, Muramatsu Y, Yoshida H, Chikada T, Tsumura T, et al. RK-287107, a potent and specific tankyrase inhibitor, blocks colorectal cancer cell growth in a preclinical model. Cancer Sci 2018;109(12):4003–14 doi 10.1111/cas.13805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Foronda M, Tarumoto Y, Schatoff EM, Leach BI, Diaz BJ, Zimmerman J, et al. Tankyrase inhibition sensitizes cells to CDK4 blockade. PLoS One 2019;14(12):e0226645 doi 10.1371/journal.pone.0226645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Zhong Y, Katavolos P, Nguyen T, Lau T, Boggs J, Sambrone A, et al. Tankyrase Inhibition Causes Reversible Intestinal Toxicity in Mice with a Therapeutic Index < 1. Toxicol Pathol 2016;44(2):267–78 doi 10.1177/0192623315621192. [DOI] [PubMed] [Google Scholar]
- 174.Lau T, Chan E, Callow M, Waaler J, Boggs J, Blake RA, et al. A novel tankyrase small-molecule inhibitor suppresses APC mutation-driven colorectal tumor growth. Cancer Res 2013;73(10):3132–44 doi 10.1158/0008-5472.Can-12-4562. [DOI] [PubMed] [Google Scholar]
- 175.McGonigle S, Chen Z, Wu J, Chang P, Kolber-Simonds D, Ackermann K, et al. E7449: A dual inhibitor of PARP1/2 and tankyrase1/2 inhibits growth of DNA repair deficient tumors and antagonizes Wnt signaling. Oncotarget 2015;6(38):41307–23 doi 10.18632/oncotarget.5846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Plummer R, Dua D, Cresti N, Drew Y, Stephens P, Foegh M, et al. First-in-human study of the PARP/tankyrase inhibitor E7449 in patients with advanced solid tumours and evaluation of a novel drug-response predictor. Br J Cancer 2020;123(4):525–33 doi 10.1038/s41416-020-0916-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Thorne CA, Hanson AJ, Schneider J, Tahinci E, Orton D, Cselenyi CS, et al. Small-molecule inhibition of Wnt signaling through activation of casein kinase 1α. Nat Chem Biol 2010;6(11):829–36 doi 10.1038/nchembio.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Cui L, Zhao J, Liu J. Pyrvinium Sensitizes Clear Cell Renal Cell Carcinoma Response to Chemotherapy Via Casein Kinase 1α-Dependent Inhibition of Wnt/β-Catenin. Am J Med Sci 2018;355(3):274–80 doi 10.1016/j.amjms.2017.11.017. [DOI] [PubMed] [Google Scholar]
- 179.Li B, Orton D, Neitzel LR, Astudillo L, Shen C, Long J, et al. Differential abundance of CK1α provides selectivity for pharmacological CK1α activators to target WNT-dependent tumors. Sci Signal 2017;10(485) doi 10.1126/scisignal.aak9916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Gang EJ, Hsieh YT, Pham J, Zhao Y, Nguyen C, Huantes S, et al. Small-molecule inhibition of CBP/catenin interactions eliminates drug-resistant clones in acute lymphoblastic leukemia. Oncogene 2014;33(17):2169–78 doi 10.1038/onc.2013.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Yamada K, Hori Y, Inoue S, Yamamoto Y, Iso K, Kamiyama H, et al. E7386, a selective inhibitor of the interaction between β-catenin and CBP, exerts antitumor activity in tumor models with activated canonical Wnt signaling. Cancer Res 2021. doi 10.1158/0008-5472.Can-20-0782. [DOI] [PubMed] [Google Scholar]
- 182.Gonsalves FC, Klein K, Carson BB, Katz S, Ekas LA, Evans S, et al. An RNAi-based chemical genetic screen identifies three small-molecule inhibitors of the Wnt/wingless signaling pathway. Proc Natl Acad Sci U S A 2011;108(15):5954–63 doi 10.1073/pnas.1017496108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Tam BY, Chiu K, Chung H, Bossard C, Nguyen JD, Creger E, et al. The CLK inhibitor SM08502 induces anti-tumor activity and reduces Wnt pathway gene expression in gastrointestinal cancer models. Cancer Lett 2020;473:186–97 doi 10.1016/j.canlet.2019.09.009. [DOI] [PubMed] [Google Scholar]
- 184.Hinze L, Pfirrmann M, Karim S, Degar J, McGuckin C, Vinjamur D, et al. Synthetic Lethality of Wnt Pathway Activation and Asparaginase in Drug-Resistant Acute Leukemias. Cancer Cell 2019;35(4):664–76.e7 doi 10.1016/j.ccell.2019.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Hinze L, Labrosse R, Degar J, Han T, Schatoff EM, Schreek S, et al. Exploiting the Therapeutic Interaction of WNT Pathway Activation and Asparaginase for Colorectal Cancer Therapy. Cancer Discov 2020. doi 10.1158/2159-8290.Cd-19-1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Tanaka N, Mashima T, Mizutani A, Sato A, Aoyama A, Gong B, et al. APC Mutations as a Potential Biomarker for Sensitivity to Tankyrase Inhibitors in Colorectal Cancer. Mol Cancer Ther 2017;16(4):752–62 doi 10.1158/1535-7163.Mct-16-0578. [DOI] [PubMed] [Google Scholar]
- 187.Solberg NT, Waaler J, Lund K, Mygland L, Olsen PA, Krauss S. TANKYRASE Inhibition Enhances the Antiproliferative Effect of PI3K and EGFR Inhibition, Mutually Affecting β-CATENIN and AKT Signaling in Colorectal Cancer. Mol Cancer Res 2018;16(3):543–53 doi 10.1158/1541-7786.Mcr-17-0362. [DOI] [PubMed] [Google Scholar]
- 188.Moon JH, Hong SW, Kim JE, Shin JS, Kim JS, Jung SA, et al. Targeting β-catenin overcomes MEK inhibition resistance in colon cancer with KRAS and PIK3CA mutations. Br J Cancer 2019;120(9):941–51 doi 10.1038/s41416-019-0434-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Picco G, Petti C, Centonze A, Torchiaro E, Crisafulli G, Novara L, et al. Loss of AXIN1 drives acquired resistance to WNT pathway blockade in colorectal cancer cells carrying RSPO3 fusions. EMBO Mol Med 2017;9(3):293–303 doi 10.15252/emmm.201606773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Seeber A, Kocher F, Xiu J, Spizzo G, Puccini A, Swensen J, et al. Molecular landscape of colorectal cancers harboring R-spondin fusions. Journal of Clinical Oncology 2019;37(15_suppl):3588- doi 10.1200/JCO.2019.37.15_suppl.3588. [DOI] [Google Scholar]
- 191.Kabiri Z, Greicius G, Madan B, Biechele S, Zhong Z, Zaribafzadeh H, et al. Stroma provides an intestinal stem cell niche in the absence of epithelial Wnts. Development 2014;141(11):2206–15 doi 10.1242/dev.104976. [DOI] [PubMed] [Google Scholar]
- 192.Mustata RC, Vasile G, Fernandez-Vallone V, Strollo S, Lefort A, Libert F, et al. Identification of Lgr5-independent spheroid-generating progenitors of the mouse fetal intestinal epithelium. Cell Rep 2013;5(2):421–32 doi 10.1016/j.celrep.2013.09.005. [DOI] [PubMed] [Google Scholar]
- 193.Yui S, Azzolin L, Maimets M, Pedersen MT, Fordham RP, Hansen SL, et al. YAP/TAZ-Dependent Reprogramming of Colonic Epithelium Links ECM Remodeling to Tissue Regeneration. Cell Stem Cell 2018;22(1):35–49 e7 doi 10.1016/j.stem.2017.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Ayyaz A, Kumar S, Sangiorgi B, Ghoshal B, Gosio J, Ouladan S, et al. Single-cell transcriptomes of the regenerating intestine reveal a revival stem cell. Nature 2019;569(7754):121–5 doi 10.1038/s41586-019-1154-y. [DOI] [PubMed] [Google Scholar]
- 195.Abitbol S, Dahmani R, Coulouarn C, Ragazzon B, Mlecnik B, Senni N, et al. AXIN deficiency in human and mouse hepatocytes induces hepatocellular carcinoma in the absence of β-catenin activation. J Hepatol 2018;68(6):1203–13 doi 10.1016/j.jhep.2017.12.018. [DOI] [PubMed] [Google Scholar]
- 196.Kawasaki K, Toshimitsu K, Matano M, Fujita M, Fujii M, Togasaki K, et al. An Organoid Biobank of Neuroendocrine Neoplasms Enables Genotype-Phenotype Mapping. Cell 2020;183(5):1420–35.e21 doi 10.1016/j.cell.2020.10.023. [DOI] [PubMed] [Google Scholar]
- 197.Mu P, Zhang Z, Benelli M, Karthaus WR, Hoover E, Chen CC, et al. SOX2 promotes lineage plasticity and antiandrogen resistance in TP53- and RB1-deficient prostate cancer. Science 2017;355(6320):84–8 doi 10.1126/science.aah4307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Ku SY, Rosario S, Wang Y, Mu P, Seshadri M, Goodrich ZW, et al. Rb1 and Trp53 cooperate to suppress prostate cancer lineage plasticity, metastasis, and antiandrogen resistance. Science 2017;355(6320):78–83 doi 10.1126/science.aah4199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Niederst MJ, Sequist LV, Poirier JT, Mermel CH, Lockerman EL, Garcia AR, et al. RB loss in resistant EGFR mutant lung adenocarcinomas that transform to small-cell lung cancer. Nat Commun 2015;6:6377 doi 10.1038/ncomms7377. [DOI] [PMC free article] [PubMed] [Google Scholar]