Abstract
Background
Allergies are on the rise globally, with an enormous impact on affected individuals’ quality of life as well as health care resources. They cause a wide range of symptoms, from slightly inconvenient to potentially fatal immune reactions. While allergies have been described and classified phenomenologically, there is an unmet need for easily accessible biomarkers to stratify the severity of clinical symptoms. Furthermore, biomarkers marking the success of specific immunotherapy are urgently needed.
Objectives
Plasma extracellular vesicles (pEV) play a role in coordinating the immune response and may be useful future biomarkers. A pilot study on differences in pEV content was carried out between patients with type I allergy, suffering from rhinoconjunctivitis with or without asthma, and voluntary non-allergic donors.
Methods
We examined pEV from 38 individuals (22 patients with allergies and 16 controls) for 38 chemokines, cytokines, and soluble factors using high-throughput data mining approaches.
Results
Patients with allergies had a distinct biomarker pattern, with 7 upregulated (TNF-alpha, IL-4, IL-5, IL-6, IL-17F, CCL2, and CCL17) and 3 downregulated immune mediators (IL-11, IL-27, and CCL20) in pEV compared to controls. This reduced set of 10 factors was able to discriminate controls and allergic patients better than the total array.
Conclusions
The content of pEV showed potential as a target for biomarker research in allergies. Plasma EV, which are readily measurable via blood test, may come to play an important role in allergy diagnosis. In this proof-of-principle study, it could be shown that pEV's discriminate patients with allergies from controls. Further studies investigating whether the content of pEVs may predict the severity of allergic symptoms or even the induction of tolerance to allergens are needed.
Keywords: Immune mediators, Allergy, Biomarkers, Extracellular vesicles-exosomes
Introduction
For decades, scientific efforts have been made to distinguish patients with allergies from non-allergic individuals and to detect routinely applicable biomarkers identifying patients successfully treated with allergen immunotherapy (AIT).1,2 There is still a need for specific quantifiable factors discriminating sensitized individuals with no clinically relevant allergic symptoms from patients with allergy and predicting the severity of allergic symptoms. Hence, additional parameters are necessary. Furthermore, single biomarkers may not be sufficient for reliable clinical predictions. Instead, patterns of biomarkers may have a higher diagnostic and predictive value.
Recent findings have pointed towards extracellular vesicles (EV) as players in the induction and suppression of allergic inflammation.3 EV are secreted by virtually all cells and circulate in the bloodstream in high concentrations. They, therefore, carry the potential to act as mediators between distant tissues and organs. Notably, they carry numerous immunological effectors, including chemokines, cytokines, other soluble ligands, microRNAs, and proteases, all of which may influence inflammation.4, 5, 6 As was recently shown by Sullivan and colleagues, EV and their cargo can exert regulatory functions through a combination of mechanisms, including cell surface receptor engagement, post-uptake secondary autocrine effects, and even long-term propagation of the stimulus to passers-by cells.7 In the specific context of that study, these interlocking mechanisms directly modulate a very restricted T cell subset, yet they appear to have the property to induce tolerance in allograft models. Thus, even small-scale EV-mediated effects may produce large-scale alterations.
Intrigued by the abundance of factors found in EV and their role in the immune system, we wanted to determine whether EV could serve as a diagnostic tool in distinguishing patients with allergies. As their extraction is minimally invasive, biomarker patterns in plasma EV (pEV) may serve as an ideal monitoring device to assess and supervise the course of allergies. This study tested the hypothesis that patients with a type I allergy may present a different biomarker pattern in pEV compared to participants with no allergies.
Materials and methods
Blood sample collection and processing
Six healthy individuals and 22 patients with allergies were recruited from the pool of participants of an ongoing study following approval by the responsible ethics committee. Patients with allergies were identified by a positive history of symptomatic allergy and an enzyme allergosorbent test (EAST) class >3 (Allergozyme® specific IgE test, Omega Diagnostics, Reinbek, Germany); a negative history of allergy was used to select non-allergic participants as controls. Blood samples of 10 additional healthy controls were included following the second ethics approval. Here, specific IgE testing was performed with ImmunoCAP™ System (Thermo Fisher Scientific Inc, USA). Pregnant candidates and those with severe or infectious diseases were excluded based on the eligibility criteria of the German Red Cross for blood donors. Participants were 18–67 years old at the time of blood collection, received no corticosteroids in the 4 weeks before the blood donation, and were not on antihistamines.
Fifty milliliters of blood were drawn into EDTA-containing syringes, of which 1 ml was used to screen for infection with HIV-1/2, HBV, and HCV. Within 2 h after blood collection, plasma was obtained by centrifugation and stored at −80 °C. pEV were purified from 5 ml plasma by standard differential centrifugation as described previously.6 pEV preparation was resuspended in 130 μl phosphate-buffered saline (Sigma®, lot RNBC0787) and stored in eight aliquots of 15 μl at −80 °C.
Particle tracking, protein concentration determination, and Western blot
pEV particle count per ml was determined by dynamic light scattering with a ZetaView® particle tracker according to the manufacturer's instructions.
Protein concentration was determined with a bicinchoninic acid (BCA) assay.
Western blots were performed with the primary antibodies anti-human CD9 (BD Pharmingen, Becton Dickinson, Heidelberg, Germany, #555370), anti-human CD63 (BD Pharmingen, Becton Dickinson, Heidelberg, Germany, #556019), and the secondary antibody anti-mouse horseradish peroxidase-linked antibody (Cell Signaling Technology, #7076) according to an established protocol. Western blots were developed using ECL detection reagent (Thermo Scientific, #34094) and chemiluminescence detected with Amersham Imager 600 (GE Healthcare Europa GmbH) according to an established protocol.
Quantification of cytokines
Cytokines were quantified according to the manufacturer's instructions using the following BioLegend LEGENDplex™ kits: Human pro-inflammatory Chemokine panel (Cat No. 740003, lot B 254807), Human Cytokine Panel 2 (Cat. No 74010 2, Lot B274719), and Human Th Cytokine Panel (Cat. No 740722, Lot B260858). The absolute concentration of each factor was determined using the LEGENDplex™ data analysis software based on a standard curve recorded for each factor and run.
The LEGENDplex™ assay captures molecules through beads coated with specific antibodies. Specific biotinylated antibodies then bind to the captured molecules and are recognized by dye-coupled streptavidin. The dye signal intensities are quantified in a flow cytometer.
Statistical analyses
Statistical evaluations and plotting were performed in R.8
A robust statistical analysis requires the replacement of missing and censored values. We decided to randomly impute values that failed to clear the limit of quantification (LOQ) as determined from the standard curves of the LEGENDplex™ measurements. Shapiro-Wilk tests of normality9 were conducted on the log-transformed data to assess whether a log-normal distribution could be assumed for each of the 39 measured factors. After p-value adjustment according to the method proposed by Benjamini and Hochberg,10 only one of the factors (MIP-3a/CCL20) was found to deviate significantly from the expected shape of a normal distribution. For the sake of simplicity and consistency, we decided to assume a log-normal distribution for all measured factors in the imputation procedure. For imputation, the parameters of each factor's assumed log-normal distribution were estimated from the empirical values in the corresponding measurement batch.11 Measurements below the LOQ were then replaced by sampling random values from the parametrized distribution and selecting the necessary number of those smaller than the defined LOQ.
Differential abundance analysis was performed with limma,12 which allowed simultaneous analysis, significance tests, and p-value correction for all included factors. The limma pipeline was employed on log-transformed and imputed data without any filtering. Sample batches were treated as blocking variables, including correction for consensus correlation.
For visualization, the R packages ggplot2, ggfortify, and pheatmap were used,13, 14, 15 and batch effects in the data were removed beforehand with limma's utility function removeBatchEffects. Hierarchical clustering in heat maps estimated sample (dis)similarity based on Euclidean distance. Principal component analysis was performed with function prcomp after log and z-score transformation.
Results
The study population (Table 1) included 22 Caucasian patients (8 female and 14 male, median age 49 years), and 16 non-allergic Caucasian donors served as controls (9 female and 7 male, median age 35 years). No notable immunologically relevant comorbidities were reported at the time of sampling in either group of the study population, except for atopic disease, ie, allergic asthma and atopic eczema in the group of participants with allergies.
Table 1.
Characterization of study participants. AIT – allergen immunotherapy; Het. FVLM – heterozygous Factor V-Leiden Mutation; discr. vWS – discrete von Willebrand Syndrome.
| Characteristics | Individuals w/o allergies N = 16 | Individuals with allergies N = 22 |
|---|---|---|
| Gender N (%) | ||
| M | 7 (44) | 14 (64) |
| F | 9 (56) | 8 (36) |
| Age (years) | ||
| Median (IQR) | 35 (26–49) | 49 (38–54) |
| Mean ± SD | 38 ± 12 | 46 ± 12 |
| Range | 22–57 | 21–67 |
| Total Serum IgE (kU/l) | ||
| N, Median (IQR) | 16, 32.7 (11.7–58.7) | 16, 179 (126–359) |
| Range | 4.56–199 | 22–5000 |
| Specific IgEa (kU/l) | ||
| N, Median (IQR) | ||
| Phleum | 16, 0.035 (.01 - .495) | 22, 12.5 (5.5–21.2) |
| Birch | 16, 0.02 (.0175 - .745) | 14, 19.2 (9.0–23.9) |
| Bet v1 | – | 10, 13.6 (6.4–21.6) |
| Alder | – | 2, 5.4 (2.8–7.9) |
| Hazel | – | 6, 6.6 (5.9–10.4) |
| House Dust Mites | 16, 0.11 (.06 - .14) | 6, 27.1 (17.5–82.8) |
| Mugwort | – | 5, 0.47 (0.45–1.02) |
| English Plantain | – | 2, 0.94 (0.91–0.97) |
| Cat | – | 3, 3.6 (1.8–24.4) |
| Rye | – | 3, 3.7 (2.5–6.6) |
| Apple | – | 1, 11.86 |
| Dog | – | 1, 0.87 |
| Short Ragweed | – | 1, 1.38 |
| Yellow Jacket | – | 1, 0.26 |
| Comorbiditiesb N (%) | ||
| Diabetes Mellitus | 1 (6.3) | |
| Hypertension | 1 (6.3) | 2 (9.1) |
| Het. FVLM, discr. vWS | 1 (6.3) | |
| Hyperthyroidism | 1 (6.3) | |
| Asthma | 1 (6.3) | 2 (9.1) |
| Old pulmonary embolism | 1 (4.5) | |
| Prostate Ca in remission | 1 (4.5) | |
| Atrial fibrillation | 1 (4.5) | |
| Atopic eczema | 1 (4.5) | |
| No comorbidity | 11 (69) | 14 (64) |
| Medicationb N (%) | ||
| AIT (completed in the past) | 0 (0) | 5 (23) |
| Metformin | 1 (6.3) | |
| Ramipril | 1 (6.3) | |
| L-Thyroxine | 1 (6.3) | |
| Rivaroxaban | 1 (4.5) | |
| Simvastatin, Flecainide | 1 (4.5) | |
| Metamizole | 1 (4.5) | |
| Not on chronic medication | 12 (75) | 16 (73) |
| no data | 1 (6.3) | 3 (14) |
| Blood collection (by quarter of year) | 0–0 – 15–1 | 6–12 – 1–2 |
One case of fish allergy was reported but not followed up with specific IgE measurement.
Some patients had more than one comorbidity and/or were on more than one medication
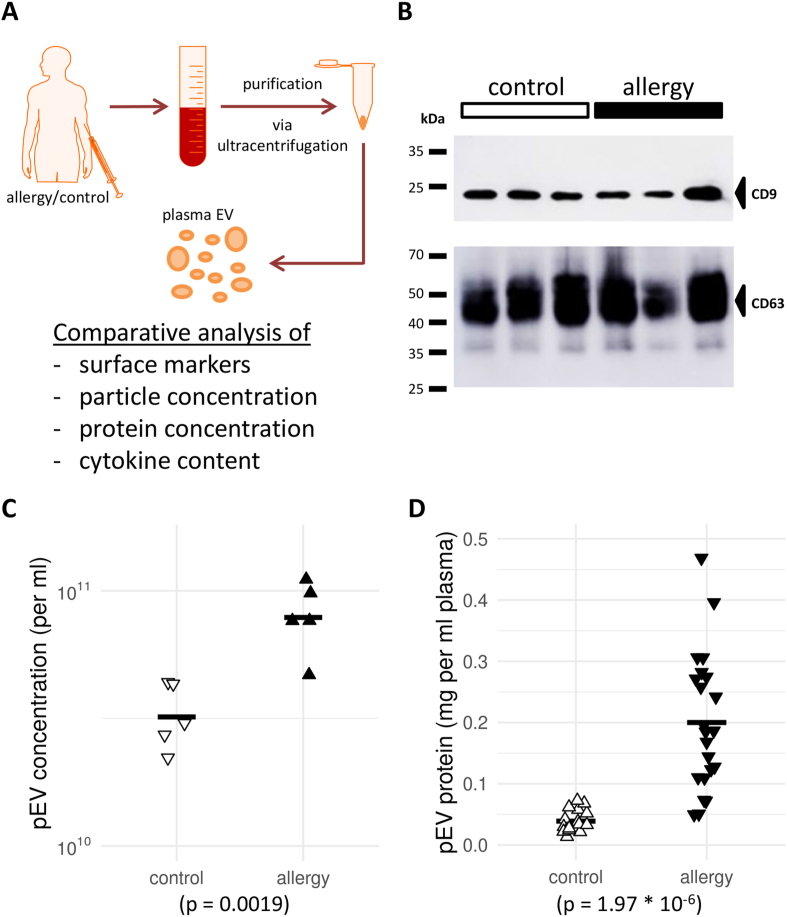
After purification and processing of the plasma samples (Fig. 1A) using differential ultracentrifugation, a Western blot for CD9 and CD63 (Fig. 1B) confirmed the presence of canonical pEV markers in the isolated fractions. Furthermore, a single-particle tracking analysis detected EV-sized particles as well as significant differences in their concentration between patients with allergies and controls (Fig. 1C). Supporting this finding, the average amount of protein obtained after pEV extraction was significantly higher in patients with allergies (Fig. 1D). This observation was robust even when accounting for assumed effects of gender and age in the model (data not shown). In summary, patients with allergies showed a 2- to 3-fold increase in pEV numbers and an up to 6-fold increase in pEV protein content compared to controls.
Fig. 1.
Characterization of plasma extracellular vesicles (pEV) from patients with allergies and controls reveals differences in total protein abundance. (A) Workflow chart for the study. Donor blood was obtained, and plasma-derived extracellular vesicles (pEV) were extracted by ultracentrifugation. The isolated pEV characteristics were then determined in multiple assays to compare patients with allergies and controls. (B) Western blot of the pEV markers CD9 and CD63 in the pEV fraction isolated by gradient ultracentrifugation. Three representative samples from either cohort are shown. (C) + (D) Scatter plots of (C) EV concentration in plasma or (D) total protein concentration in pEV from controls and patients with allergies. In (C), a sample of five individuals from each cohort is shown. Horizontal bars indicate sample or cohort averages, respectively. The observed differences were significant for both characteristics (see p-value below panels)
Subsequently, the presence of 39 selected cytokines was analyzed in a batched manner (list of factors in Suppl. Table S1). Random imputation from modeled empirical distributions was performed to replace data points outside the range of the limits of quantification. (see Suppl. Fig. S1 for an example of the largest measured batch and Methods in detail). Due to unrecoverably missing values, one factor (CCL4) was removed at this point.
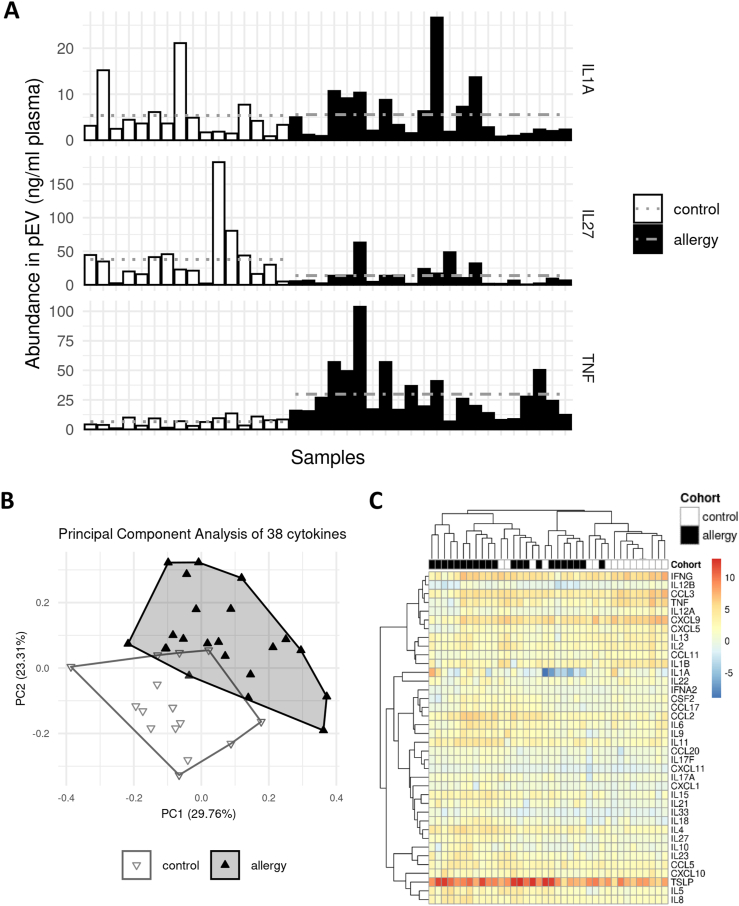
Pre-analysis inspection of the cytokine abundancies revealed a mixed pattern of increases and decreases. We found that some factors like IL-1A were unchanged while others were up to 5-fold more (TNF) or less (IL-27) abundant in the allergy cohort (Fig. 2A). To assess the data set's ability to discriminate between patients with allergies and controls, we performed a principal component analysis (PCA) using all cytokines after removing batch effects (Fig. 2B). We found a mix of separation and overlap in the result, with three controls and one allergy sample, respectively, crossing into the opposite cluster's territory. A heat map and hierarchical clustering of the full data set (Fig. 2C) partially replicated the findings from the PCA plot, with an obvious cluster structure; however, poor high-level separation of clusters. This observed distribution suggested a limited ability of the full set of measured cytokines to separate the two cohorts.
Fig. 2.
The cytokine profile of pEV is associated with allergies. (A) Illustration of the observed per-sample abundances of 3 specific measured cytokines: IL-1A (top, the least different among the cohorts), IL-27 (middle, highest gross decrease in patients with allergies), and TNF (bottom, highest gross increase in patients with allergies). Horizontal grey lines indicate cohort averages. (B) Dimensional reduction of the full data set via principal component analysis (PCA). Approximately half of the observed variance can be explained by the first 2 principal components. The control and the allergy cohorts occupy distinct but partially overlapping regions in the plot, arguing for an allergy-specific pattern in the measured cytokine profiles. (C) Heat map and hierarchical clustering of the full data set. While some subgroups within both cohorts cluster together, there is no high-level separation between allergy and control samples
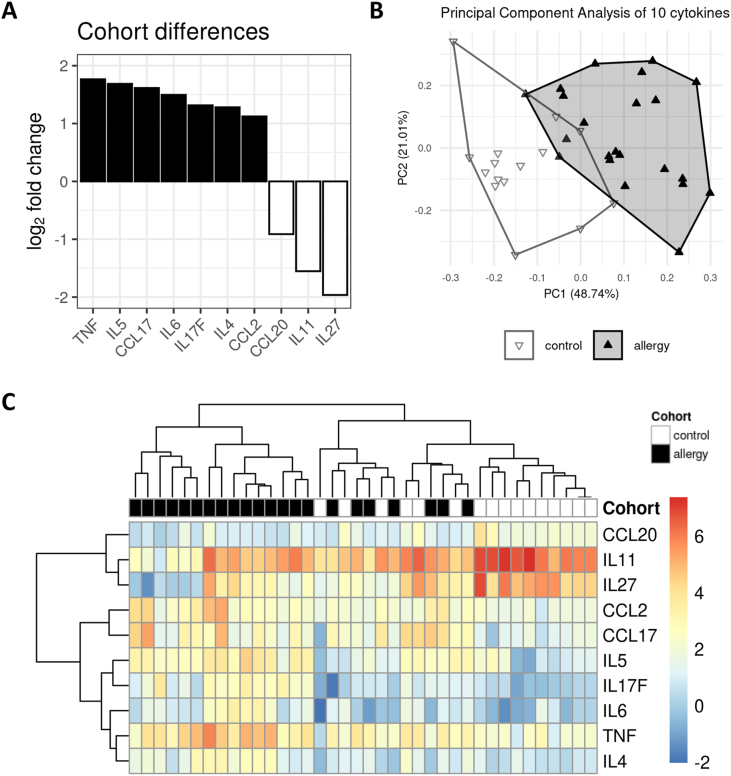
In the next step, we asked whether a subset of the measured cytokines would allow proper discrimination between patients with allergies and those with no allergies. We applied a method from high-throughput analysis workflows to our data set and identified ten cytokines that were significantly different between the two cohorts at an alpha level of .05 (Suppl. Table S1). The 10 cytokines and their calculated log2 fold changes are shown in Fig. 3A. While TNF-α, IL-4, IL-5, IL-6, IL-17F, CCL2, and CCL17 were increased in patients with allergies, CCL20, IL-11, and IL-27 were decreased.
Fig. 3.
Bioinformatics analysis reveals a subset of ten cytokines whose discriminatory power surpasses that of the full data set, yielding a pEV allergy signature. (A) Waterfall plot illustrating the observed abundance differences (shown as log2 fold change) of the nine cytokines selected as significantly different between the cohorts by a high-throughput analysis method that corrects for multiple testing. Upregulation in the allergy cohort (seven cytokines) is indicated by solid black, while downregulation (three cytokines) is shown as a black outline with white fill. (B) PCA with the allergy signature cytokines. The explained variance of the first two principal components is around 17% points higher than in the PCA on the full data set (Fig. 2B). Although there is no obvious improvement in separation of the control and allergy clusters, apart from less overlap along the first principal component. (C) Heat map and hierarchical clustering of the allergy signature cytokines. Compared to Fig. 2C, the reduced cytokine set improves the separation between allergy and control samples even though it fails to disentangle two mixed clusters with 13 out of 38 samples
We then performed a PCA on this reduced data set with only 10 cytokines (Fig. 3B). While the relative cohort positions are nearly identical to Fig. 2B, there is an increase in explained variance in the first 2 dimensions from 53% to 70%, suggesting that the reduced set yielded a cleaner internal structure than the full data set.
In the corresponding heat map (Fig. 3C), we found an improvement in the high-level structure, with 1 pure allergy cluster on the left, 1 pure control cluster on the right, and 2 mixed clusters in between.
In summary, we found changes in gross protein concentration and cytokine content in pEV that could help to discriminate patients with allergies from non-allergic controls and may serve as biomarkers to select and monitor therapeutic interventions.
Discussion
In this proof-of-principle study with a limited number of samples and factors, we demonstrated that biomarker patterns in pEV might be able to discriminate allergic from non-allergic individuals. EVs are involved in the upregulation and downregulation of numerous inflammatory and defense mechanisms. They comprise various kinds of bioactive molecules, such as cytokines, RNAs, and others. Numerous sources of their release, including immunonologically active cells, epithelial cells, vascular endothelium, and secretory organs (ie, liver), must be considered during interpretation of pEV pattern in special conditions of the immune response.16
Consistent with the knowledge of measurements of interleukins in sera of individuals with allergies, the abundances of pro-allergic cytokines IL-4 and IL-5 in pEV of patients with allergies were higher than in healthy controls combined with the pro-inflammatory cytokines IL-6 and TNFalpha. Furthermore, the concentration of MCP-1 (monocyte chemoattractant protein 1, CCL2) was higher in pEV of patients with allergies than in healthy controls. The chemokine CCL2 and its main chemokine receptor CCR2 have been implicated in the pathogenesis of several different disease processes, including vascular permeability and attraction of immune cells during metastasis, a number of different neurological disorders, autoimmune diseases, obesity, and atherosclerosis.17 Interestingly, a recent study on the definition of biomarkers predicting symptoms in patients with or without allergy upon seasonal pollen exposure found that nasal MCP-1 was higher in patients with seasonal allergic rhinitis than in individuals without symptoms.18
Moreover, the pattern of elevated factors in patients with allergies involves CCL17. CCL17 is present on dermal blood vessels in a pro-inflammatory microenvironment and interacts with Th2-activated helper cells, promoting their migration into the skin.19,20 Additionally, IL-17 F was increased in patients with allergies in our study, which aligned with its known involvement in allergic asthma and its abundance in upper airway nasal epithelia.21,22
Three factors were reduced in pEV of patients with allergies compared to healthy controls: CCL20, IL-27, and IL-11. The effect of IL-27 in airway allergies depends on the balance of its signaling pathways on different target cells of innate immunity and adaptive immunity and the stage of allergic response.23 Many inconsistencies exist when evaluating its role in allergic airway disease, as IL-27 may have pro-inflammatory and attenuating effects. Nevertheless, it is known that tolerance induction during AIT is supported by dendritic cells influencing naïve T-cells with a cocktail of IL-10, IL-12, and IL-27 to either provoke Th1 deviation or induction of regulatory T-cells (Tregs)- both promoting a Th1 dominant milieu.24
The association of lower levels of IL-11 in pEV of patients with allergies compared to controls is interesting. IL-11 is involved in inflammation, osteogenesis, hematopoiesis, fertility, and cancer.25,26 It is almost absent from the body fluids of healthy individuals, and it is not clear which cells produce IL-11 in vivo.27 Additionally, its effect has been described as pro-and anti-inflammatory.28 IL-11 receptors are present on numerous cells, including T cells, osteoclasts, megakaryocytes, macrophages, endothelial cells, and epithelial cells. However, the precise role of IL-11 in allergic contexts remains to be elucidated.
Further studies will be needed to address the reproducibility of the signature before it can be evaluated and its application as a predictor in clinical context can be warranted. In particular, larger cohorts in conjunction with intelligent data mining approaches may increase the diagnostic power of the signature. Biomarker data sets, rather than individual factors, may constitute a next-generation test system that could recognize and monitor abnormal immune conditions like allergies.
Aside from their diagnostic value, the question arises whether such findings match and extend the knowledge on the pathogenesis of allergies. The ten-cytokine signature that we found brings together factors with an established mechanistic role in allergy and factors whose impact is not yet clear. The nature of these factors suggests a vesicle secretion by cells of the immune system or a secretory organ like the liver.29 The inflammatory content of pEV may increase the reactivity of innate immune cells, hence, lowering the threshold for an allergic reaction. In line with this assumption, it has been observed that inflammatory pEV contributes to a pro-allergic immune response.30, 31, 32, 33 While pro-allergic signaling cascades are thought to occur mainly via soluble factors engaging membrane receptors,33,34 ligand-receptor interactions or cellular signaling may also occur following EV internalization on the level of endosomal compartments.35 Recent research demonstrated that the opposite is also possible, and pEV may coat target immune cells to induce immune tolerance.7
Plasma EV may also differentiate cells, such as monocytes, towards inflammatory dendritic cells, which may lead to secondary secretion events. Together this could lead to a pro-allergic microenvironment, e.g., characterized by a Th2 predominance.33,34 The increased abundance of TNF, IL-4, IL-5, IL-6, IL-17 F, CCL-2, and CCL-17 in pEV of our allergy cohort would support such a view.
From cytokine studies, we know, immune regulatory mechanisms change very fast in AIT. Therefore, in a future project, a defined group of patients with clinical risk markers and early changes in pEV could help identify patterns of markers that could be monitored during AIT. Such markers could allow identifying individuals who benefited from an AIT or who would profit from this therapeutic option.
Conclusion
In summary, this is a proof-of-principle study raising hope that by measuring additional factors in pEV and with the use of more sophisticated data mining algorithms, a better and more comprehensive diagnosis of allergic reactions might be possible. This may even help to predict the severity of clinical symptoms or discriminate responders and non-responders before or after allergen immunotherapy.
Abbreviations
AIT: allergen immunotherapy; IL: interleukin, LOQ: limit of quantification; pEV: plasma extracellular vesicles; PBS: phosphate-buffered saline; PCA: principal component analysis; TNF: tumor necrosis factor; Th1: type 1 helper T-cell; Th2: type 2 helper T-cell; T regs: regulatory T cells.
Funding
JV acknowledges support from the German Ministry of Education and Research (BMBF-Bundesministerium für Bildung und Forschung) through grants MelEVIR (031L0073A), MelAutim (01ZX1905A) KI-VesD (161L0244A), and from the Bavarian Ministry of Economic Affairs, Regional Development and Energy through Gaminfection-UKER (07 03/686 68/201/19/24/20/25/21). AB acknowledges support from the BMBF through grants MelEVIR (031L0073A) and the IZKF (Interdisziplinäres Zentrum für Klinische Forschung), Erlangen. Sample collection and annotation was sponsored by Allergopharma GmbH & Co. KG.
Ethics approval
Following approval of a study according to article 15 Berufsordnung (professional code for physicians) by the ethics committee of Schleswig-Holstein, Germany (reference no.064/14(II), 21.11.2016), candidates for this study were recruited from the pool of participants in an ongoing long-term trial by Allergopharma which included blood sampling between 11-2016 and 08–2018. Blood samples from further healthy donors were recruited by the Department of Dermatology in collaboration with the Department of Transfusion Medicine and Hemostaseology, Universitätsklinikum Erlangen, Friedrich-Alexander University Erlangen-Nürnberg (FAU), Erlangen, Germany upon approval by the FAU's ethics committee (reference no. 242_18B).
Author contributions
The study was initiated by AB, BL, and GS and conceptually designed by AB, BL and NW; sample collection and annotation: HK, BL and SA; sample processing: KB, FC, SG; data processing and analysis: ME; data interpretation: ME, NW, AB, JV, CB, GS; manuscript preparation: NW, ME, AB, CB, GS; funding acquisition: GS, CB, AB, JV. All authors have read and approved the final manuscript and consent for publication.
Declaration of competing interest
BL and HK are employees of Allergopharma GmbH & Co. KG.
Acknowledgements
We acknowledge the measurements of specific IgE by Sabine Schüpferling, Department of Dermatology, Universitätsklinikum Erlangen, Germany.
Footnotes
Full list of author information is available at the end of the article https://doi/org/10.1016/j.waojou.2021.100583
Supplementary data to this article can be found online at https://doi.org/10.1016/j.waojou.2021.100583.
Contributor Information
Nicola Wagner, Email: Nicola.Wagner@uk-erlangen.de.
Martin Eberhardt, Email: Martin.Eberhardt@uk-erlangen.de.
Julio Vera, Email: Julio.Vera@uk-erlangen.de.
Federica Cuomo, Email: Federica.Cuomo@icloud.com.
Katja Blume, Email: Katja.Blume@uk-erlangen.de.
Silvia Galster, Email: Silva.Galster@uk-erlangen.de.
Susanne Achenbach, Email: Susanne.Achenbach@uk-erlangen.de.
Bernd Laffert, Email: Bernd.Laffert@allergopharma.com.
Helga Kahlert, Email: Helga.Kahlert@web.de.
Gerold Schuler, Email: Gerold.Schuler@uk-erlangen.de.
Carola Berking, Email: Carola.Berking@uk-erlangen.de.
Andreas Baur, Email: Andreas.Baur@uk-erlangen.de.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Shamji M.H., Kappen J.H., Akdis M. Biomarkers for monitoring clinical efficacy of allergen immunotherapy for allergic rhinoconjunctivitis and allergic asthma: an EAACI Position Paper. Allergy. 2017;72:1156–1173. doi: 10.1111/all.13138. [DOI] [PubMed] [Google Scholar]
- 2.Pfaar O., Agache I., de Blay F. Perspectives in allergen immunotherapy: 2019 and beyond. Allergy. 2019;74(Suppl 108):3–25. doi: 10.1111/all.14077. [DOI] [PubMed] [Google Scholar]
- 3.Nazimek K., Bryniarski K., Askenase P.W. Functions of exosomes and microbial extracellular vesicles in allergy and contact and delayed-type hypersensitivity. Int Arch Allergy Immunol. 2016;171(1):1–26. doi: 10.1159/000449249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Konoshenko M.Y., Lekchnov E.A., Vlassov A.V., Laktionov P.P. Isolation of extracellular vesicles: general methodologies and latest trends. BioMed Res Int. 2018;30:8545347. doi: 10.1155/2018/8545347. eCollection 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Abels E.R., Brakefiled X.O. Introduction to extracellular vesicles: biogenesis, RNA cargo selection, content, release, and uptake in cellular and. Mol Neurobiol. 2016;36:301–312. doi: 10.1007/s10571-016-0366-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee J.H., Dindorf J., Eberhardt M. Innate extracellular vesicles from melanoma patients suppress beta-catenin in tumor cells by miRNA-34a. Life Sci Alliance. 2019;2 doi: 10.26508/lsa.201800205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sullivan J.A., Tomita Y., Jankowska-Gan E. Treg-cell-derived IL-35-coated extracellular vesicles promote infectious tolerance. Cell Re. 2020;30:1039–1051. doi: 10.1016/j.celrep.2019.12.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.R Core Team. R . R Foundation for Statistical Computing; Vienna, Austria: 2019. A Language and Environment for Statistical Computing.https://www.R-project.org/ URL. [Google Scholar]
- 9.Shapiro S.S., Wilk M.B. An analysis of variance test for normality (for complete samples) Biometrika. 1965;52(3/4):591–611. doi: 10.1093/biomet/52.3-4.591. [DOI] [Google Scholar]
- 10.Benjamini Y., Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Roy Stat Soc B. 1995;57:289–300. MR 1325392. [Google Scholar]
- 11.Millard S.P. Springer; New York: 2013. EnvStats: An R Package for Environmental Statistics. ISBN 978-1-4614-8455-4. [Google Scholar]
- 12.Ritchie M.E., Phipson B., Wu D. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015 Apr 20;43(7):e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wickham H. Springer; New York: 2016. ggplot2: Elegant Graphics for Data Analysis. [Google Scholar]
- 14.Tang Yuan, Horikoshi Masaaki, Li Wenxuan. Ggfortify: unified interface to visualize statistical result of popular R packages. The R Journal. 2016;8(2):478–489. [Google Scholar]
- 15.Kolde B Raivo. 2019. Pheatmap: Pretty Heatmaps. R Package.https://CRAN.R-project.org/package=pheatmap version 1.0.12. [Google Scholar]
- 16.Kalluri R., LeBleu V.S. The biology, function, and biomedical applications of exosomes. Science. 2020;367(6478) doi: 10.1126/science.aau6977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.O'Connor T., Borsig L., Heikenwalder M. CCL2-CCR2 signaling in disease pathogenesis. Endocr Metab Immune Disord - Drug Targets. 2015;15(2):105–118. doi: 10.2174/1871530315666150316120920. [DOI] [PubMed] [Google Scholar]
- 18.Gökkaya M., Damialis A., Nussbaumer T. Defining biomarkers to predict symptoms in subjects with and without allergy under natural pollen exposure. J Allergy Clin Immunol. 2020;146(3):583–594. doi: 10.1016/j.jaci.2020.02.037. e6. [DOI] [PubMed] [Google Scholar]
- 19.Rapp M., Wintergerst M.W.M., Kunz W.G. CCL22 controls immunity by promoting regulatory T cell communication with dendritic cells in lymph nodes. J Exp Med. 2019;216(5):1170–1181. doi: 10.1084/jem.20170277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Campbell J.J., O'Connell D.J., Wurbel M.A. Cutting Edge: chemokine receptor CCR4 is necessary for antigen-driven cutaneous accumulation of CD4 T cells under physiological conditions. J Immunol. 2007;178(6):3358–3362. doi: 10.4049/jimmunol.178.6.3358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sorbello V., Ciprandi G., Di Stefano A. Nasal IL-17F is related to bronchial IL-17F/neutrophilia and exacerbations in stable atopic severe asthma. Allergy. 2015;70(2):236–240. doi: 10.1111/all.12547. [DOI] [PubMed] [Google Scholar]
- 22.Chang S.H., Dong C. IL-17F: regulation, signaling and function in inflammation. Cytokine. 2009;46(1):7–11. doi: 10.1016/j.cyto.2008.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jafarzadeh A., Nemati M., Jafarzadeh S., Chauhan P., Saha B. The immunomodulatory potentials of interleukin-27 in airway allergies. Scand J Immunol. 2021;93(2) doi: 10.1111/sji.12959. [DOI] [PubMed] [Google Scholar]
- 24.Kirtland M.E., Tsitoura D.C., Durham S.R., Shamji M.H. Toll-like receptor agonists as adjuvants for allergen immunotherapy. Front Immunol. 2020;12:11. doi: 10.3389/fimmu.2020.599083. 599083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nishina T., Komazawa-Sakon S., Yanaka S. Interleukin-11 links oxidative stress and compensatory proliferation. Sci Signal. 2012;5(207) doi: 10.1126/scisignal.2002056. ra5. [DOI] [PubMed] [Google Scholar]
- 26.Nguyen P.M., Abdirahman S.M., Putoczki T.L. Emerging roles for Interleukin-11 in disease. Growth Factors. 2019;37(1-2):1–11. doi: 10.1080/08977194.2019.1620227. [DOI] [PubMed] [Google Scholar]
- 27.Lokau J., Agathe M., Flynn C.M., Garbers C. Proteolytic control of interleukin-11 and interleukin-6 biology. Biochim Biophys Acta Mol Cell Res. 2017;1864(11 Pt B):2105–2117. doi: 10.1016/j.bbamcr.2017.06.008. [DOI] [PubMed] [Google Scholar]
- 28.Cook S.A., Schafer S. Hiding in plain sight: interleukin-11 emerges as a master regulator of fibrosis, tissue integrity, and stromal inflammation. Annu Rev Med. 2020 27;71:263–276. doi: 10.1146/annurev-med-041818-011649. [DOI] [PubMed] [Google Scholar]
- 29.Schierer S., Ostalecki C., Zinser E. Extracellular vesicles from mature dendritic cells (DC) differentiate monocytes into immature DC. Life Sci Alliance. 2018;1(6) doi: 10.26508/lsa.201800093. eCollection 2018 Dec. PMID: 30519676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sastre B., Canas J.A., Rodrigo-Munoz J.M., Del Pozo V.7. Novel modulators of asthma and allergy: exosomes and MicroRNAs. Front Immunol. 2017;8:826. doi: 10.3389/fimmu.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Matthieu M., Martin-Jaular L., Lavieu G., Thery C. Specificities of secretion and uptake of exosomes and other extracellular vesicles for cell-to-cell communication. Nat Cell Biol. 2019;21:9–17. doi: 10.1038/s41556-018-0250-9. [DOI] [PubMed] [Google Scholar]
- 32.Wahlund C.J.E., Gucluler G., Hiltbrunner S., Veerman R.E., Naslund T.I., Gabrielsson S. Exosomes from antigen-pulsed dendritic cells induce stronger antigen-specific immune responses than microvesicles in vivo. Sci Rep. 2017;7(1):17095. doi: 10.1038/s41598-017-16609-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hough K.P., Deshane J.S. Exosomes in allergic airway diseases. Curr Allergy Asthma Rep. 2019;22:26. doi: 10.1007/s11882-019-0857-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Breiteneder H., Diamant Z., Eiwegger T. Future research trends in understanding the mechanisms underlying allergic diseases for improved patient care. Allergy. 2019;74(12):2293–2311. doi: 10.1111/all.13851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Admyre C., Bohle B., Johansson S.M. B cell-derived exosomes can present allergen peptides and activate allergen-specific T cells to proliferate and produce TH2-like cytokines. J Allergy Clin Immunol. 2007;120(6):1418–1424. doi: 10.1016/j.jaci.2007.06.040. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.