Abstract
Chromosomal translocations generate oncogenic fusion proteins in approximately one-third of sarcomas, but how these proteins promote tumorigenesis is not well understood. Interestingly, some translocation-driven cancers exhibit dramatic clinical responses to therapy, such as radiotherapy, though the precise mechanism has not been elucidated. Here we reveal a molecular mechanism by which the fusion oncoprotein FUS-CHOP promotes tumor maintenance that also explains the remarkable sensitivity of myxoid liposarcomas to radiation therapy. FUS-CHOP interacted with chromatin remodeling complexes to regulate sarcoma cell proliferation. One of these chromatin remodelers, SNF2H, co-localized with FUS-CHOP genome-wide at active enhancers. Following ionizing radiation, DNA damage response kinases phosphorylated the prion-like domain of FUS-CHOP to impede these protein-protein interactions, which are required for transformation. Therefore, the DNA damage response after irradiation disrupted oncogenic targeting of chromatin remodelers required for FUS-CHOP-driven sarcomagenesis. This mechanism of disruption links phosphorylation of the prion-like domain of an oncogenic fusion protein to DNA damage after ionizing radiation and reveals that a dependence on oncogenic chromatin remodeling underlies sensitivity to radiation therapy in myxoid liposarcoma.
Significance
Prion-like domains, which are frequently translocated in cancers as oncogenic fusion proteins that drive global epigenetic changes, confer sensitivity to radiation via disruption of oncogenic interactions.
Keywords: Radiation, Cancer, chromatin, sarcoma, translocation, phase separation
Introduction
Translocation-driven sarcomas, such as Ewing sarcoma, comprise around one third of soft tissue sarcomas (1). In approximately half of these sarcomas, the N-terminal domain of the chimeric oncoprotein is derived from one of the FET proteins (FUS, EWSR1, TAF15). For example, in Ewing sarcoma, the EWSR1 N-terminus prion-like domain (PrLD) is fused to the carboxyl terminal domain of an ETS family transcription factor, typically FLI1, to generate EWS-FLI1. EWS-FLI1 acts as a neomorphic transcription factor by targeting GGAA-containing microsatellite repeats and altering chromatin accessibility mediated through interactions with the BAF (BRG1/BRM-associated factors) complex (2-4). Importantly, the interactions between EWS-FLI1 and the BAF complex are mediated through the PrLD of EWSR1 (2).
In a subset of Ewing sarcomas, FUS (fused in sarcoma) constitutes the N-terminal domain of the fusion protein (5). FUS is also the N-terminus partner of other fusion proteins that drive sarcomagenesis. FUS-CHOP (also known as FUS-DDIT3) results from a t(12;16)(q13;p11) translocation that drives 95% of myxoid liposarcomas (MLPS) (6-9). Exome analysis of MLPS has revealed few other recurrent mutations in coding sequences (10). FUS-CHOP is specific to MLPS and has not been detected in other liposarcoma subtypes or other cancers (11). These data suggest that the FUS-CHOP translocation is the predominant driver in MLPS tumorigenesis, though its precise mechanism is unknown. Over a century ago, James Ewing recognized that, like Ewing sarcoma, these sarcomas with a myxoid background were exquisitely sensitive to radiation therapy (12), but the mechanism underlying this radiosensitivity remains unknown. Indeed, MLPS is distinguished by its remarkable clinical response to radiation therapy compared to most other sarcoma subtypes (13-15). We hypothesized that the PrLD of FUS-CHOP interacts with chromatin remodeling complexes and identified new interactions with imitation switch (ISWI) chromatin remodeling complexes. Furthermore, we hypothesized that disruption of these interactions contributes to the radiosensitivity of MLPS and determined that phosphorylation of the PrLD mediates these interactions, but impedes transformation by FUS-CHOP.
Materials and Methods
Radiation treatments
Cells were cultured at least 24 hours prior to irradiation experiments. The X-RAD 160 (Precision X-Ray) cell irradiation system was used at 160 kVp x 18 mA energy. An F1 filter (2 mm aluminum) was used for beam conditioning. Sample distance was set to 40 cm. For a dose of 10 Gy, calculations based on annual dosimetry determined a treatment duration of 230 seconds.
Immunoblotting and antibodies
Samples were lysed in radioimmunoprecipitation assay buffer (RIPA) buffer (50 mM Tris-HCl, pH 8.0 with 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulfate (SDS)) for 30 min on ice (Sigma-Aldrich, R0278), sonicated briefly, then centrifuged at 10,000x g for 20 min at 4°C. Protein concentration was determined for the lysate supernatant by bicinchoninic acid assay (BCA) assay (Pierce, 23225). Samples was boiled in 4X Laemmli sample buffer (Bio-Rad, 1610747) at 95°C for 5 min, then cooled to room temperature before loading in a 4-20% Tris-Bis polyacrylamide gel. Samples were electrophoresed at 200 V for 30 min before transfer to nitrocellulose. Membranes were blocked in 5% non-fat dry milk or 5% bovine serum albumin (BSA) in Tris-buffered saline (TBS, Corning, 46-012-CM). Next, membranes were incubated overnight at 4°C with primary antibodies diluted in TBS-T (0.1% Tween-20) with 3% BSA: CHOP 1:1,000 (Cell Signaling Technology, 2895S); CHD4 1:1000 (D8B12, Cell Signaling Technology); BRG1 1:1000 (D1Q7F, Cell Signaling Technology); SNF2H 1:5000 (ABE1026, EMD Millipore); BAF170 1:1000 (D8O9V, Cell Signaling Technology); ACF1 1:1000 (ab187670, abcam); HDAC1 1:1000 (10E2, Cell Signaling Technology); TBP 1:1000 (D5C9H, Cell Signaling Technology). Membranes were washed three times in TBS-T for 5 min before secondary antibody incubation with goat anti-rabbit IRDye800 (Li-Cor Biosciences, P/N 925- 32211) and goat anti-mouse IRDye680 (Li-Cor Biosciences, P/N 925-68070) both at 1:10,000 dilutions in TBS-T for 1 h at room temperature. The membranes were washed three times in TBS-T for 5 min and imaged using an Odyssey CLx (Li-Cor Biosciences). Image analysis for normalization and quantification were performed using Image Studio (Version 5.2, Li-Cor Biosciences, P/N 9140-500).
Co-immunoprecipitation (Co-IP)
The co-IP protocol was adapted from Boulay et al. (2). Briefly, media was aspirated and washed twice with cold PBS. On ice, 1 mL of cytoplasmic lysis buffer (Buffer A: 10 mM Tris-HCl pH 8.0, 10 mM KCl, 1.5 mM MgCl2, 0.15% NP-40, 1 mM PMSF, 1 mM DTT, 1 tablet PhosSTOP and 1 tablet cOmplete protease inhibitor per 10 mL buffer) was added per 10 cm dish and scraped into a tube. Cells were incubated on ice for 5 minutes in Buffer A with periodic pipetting to mix and shear cells. Nuclei were pelleted at 1200 rpm for 5 minutes, and supernatant was aspirated carefully. The nuclei were washed with Buffer A without NP-40 and pelleted again via centrifugation at 1000 x g for 5 minutes. The supernatant was aspirated and nuclei were resuspended in nuclear lysis buffer (Buffer B: 50 mM Tris-HCl pH 8.0, 150 mM NaCl, 5 mM EDTA, 0.5% NP-40, 10% glycerol, 1 mM PMSF, 1 tablet PhosSTOP and 1 tablet cOmplete protease inhibitor per 10 mL buffer), approximately 5-6 volumes for adequate lysis. Gentle sonication with a QSONICA 800 R instrument at 20 Amp, 1 sec on/1 sec off, for 60 seconds was used to help lyse nuclei. After incubation for 30 minutes, nuclear lysate was centrifuged for 15 minutes at 14000 rpm at 4°C. The nuclear fraction was collected and quantified, then the final concentration was adjusted to 1 mg/mL with Buffer B. Each IP fraction was incubated with 3 ug of antibody (IgG rabbit, Cell Signaling Technology; CHD4 (D8B12), Cell Signaling Technology; BRG1 (D1Q7F), Cell Signaling Technology; SNF2H (ABE1026), EMD Millipore) or (1:50 dilution CHOP (L63F7), Cell Signaling Technology) overnight at 4°C on a tube rotator. Dynabeads were resuspended in the vial by vortexing >30 seconds and enough beads for 20 uL of beads for each IP reaction were transferred to a new tube. The tube was placed on a magnet to separate the beads from the solution, and the beads were washed once with Buffer B. The IP samples were then incubated with 20 uL of protein G Dynabeads for 2 hours at 4°C on the tube rotator. Samples were then washed 5 times at 4°C with 500 uL of Buffer B with gentle pipetting or inversion on a magnetic rack. After the final wash, the beads were resuspended in 30 uL 2x gel loading buffer with DTT, boiled, and processed using a standard western blotting protocol.
Urea denaturation assay
Urea denaturation was performed exactly as the co-IP protocol above with one exception. Before incubation with the primary antibody overnight, lysate was incubated at room temperature for 15 minutes with various concentration of urea (0M, 0.5M, 1M, 2M).
RNase/DNase assay
RNase/DNase assays were performed exactly as the co-IP protocol above with two changes. Before incubation with the primary antibody overnight, lysate was incubated at 37°C for 15 minutes with RNase A in Buffer B at 100 ug/mL then incubated with IP antibody overnight at 4°C for the RNase assay. Sample lysate was incubated at 37°C for 15 minutes with 100 units/mL DNase in Buffer B then incubated with IP antibody overnight at 4°C.
Proximity ligation assay (PLA)
The Duolink PLA red mouse/rabbit kit was used to perform proximity ligation assay per manufacturer’s protocols. Briefly, cells were plated in 8-well chamber slides overnight. The next day, media was aspirated, cells were fixed with 4% formaldehyde with methanol permeabilization. Cells were then blocked with Duolink blocking solution and incubated with primary antibody overnight. Next, Duolink PLA probes were incubated with the samples followed by ligation and amplification. Finally, the slides were mounted with a coverslip using Duolink in situ mounting medium with DAPI, and the slides were either imaged 15 minutes later or kept at −20°C until imaging using a Leica SP5 Inverted Confocal Microscope (DMI6000CS) with the 405 nm and 647 nm lasers for excitation.
Chromatin Immunoprecipitation Sequencing (ChIP-seq)
Chromatin was prepared using a SimpleChIP plus Sonication Chromatin IP Kit from Cell Signaling Technology according to manufacturer’s instructions following optimization of crosslinking, sonication and antibody conditions for each cell line. Briefly, one chromatin preparation for each cell line was prepared from approximately 20 million cells. Cells were crosslinked in 1% formaldehyde (methanol-free) for 9 minutes at room temperature. Glycine neutralization followed for 5 minutes at room temperature followed by 2x wash with cold PBS. Next, cells were scraped into cold 1x PBS with protease inhibitor, then pelleted at 1000 x g for 5 minutes at 4C. PBS was removed and cells were resuspended in 1X cell lysis buffer with protease inhibitor for 10 minutes prior to nuclei preparation. Nuclei were pelleted at 5000 x g for 5 minutes at 4C and the pellet was resuspended in 1X cell lysis buffer with protease inhibitor for 5 minutes before pelleting at 5000 x g for 5 minutes at 4C. Pelleted nuclei were then resuspended in cold nuclear lysis buffer with protease inhibitor and incubated on ice for 10 minutes. 1 mL of the nuclear lysate was transferred to appropriate tubes for sonication. Chromatin was sonicated using a Qsonica Q700 sonicator with Cup Horn and chiller (70 Amp, 15 sec on, 45 sec off) for the following durations per cell line: SW872, 15 minutes; DL221, 12 minutes; MLS402, 20 minutes. Following sonication, lysates were clarified by centrifugation at 21000 x g for 10 minutes at 4C and supernatant was transferred to a new tube. 50 uL of each lysate was taken for analysis of fragmentation and concentration determination. 10 ug ChIP DNA was used per IP sample. Samples were incubated with primary antibody overnight at 4C on a tube rotator. The next day, samples were incubated with protein G magnetic beads for 2 hours at 4C. Beads were isolated and washed 3 times in low salt wash buffer and 1 time in high salt wash buffer. Finally, chromatin was eluted from magnetic beads, protein was reverse crosslinked, and DNA was purified and pooled for library prep and sequencing.
CUT&RUN
CUT&RUN was performed as published and as on protocols.io (16,17). Briefly, cells were bound to conacavalin beads, permeabilized with digitonin and incubated with primary antibody overnight. Protein A/G-MNase was used to digest chromatin for 30 minutes and soluble DNA was purified via phenol-chloroform extraction. DNA libraries were prepped and barcoded using NEBNext adapters and barcodes before pooling and sequencing at Genewiz.
RNA-seq
Cells were washed with PBS twice and lysed with TRIzol reagent. RNA was purified following manufacturer’s instructions using a DirectZol RNA miniprep kit (Zymo Research). RNA samples were spiked with ERCC for normalization and sent to Genewiz for sequencing.
Protein production and purification
Target sequences for MBP-FUS-CHOP WT, MBP-FUS-CHOP 12A, and MBP-FUS-CHOP 12E cDNA sequences were synthesized by Genscript and subcloned into an E. coli expression vector pET28a-MBP that contains 6xHis-MBPtag. A TEV sequence was inserted between the MBP tag and FUS-CHOP cDNAs for MBP tag release via cleavage by TEV protease. BL21 star (DE3) E. coli cells were transformed with the recombinant plasmids and a single colony was inoculated into Terrific Broth (TB) medium containing the appropriate antibiotic. A 2 L culture was incubated at 37°C and when the OD600 reached about 1.2, protein expression was induced with IPTG at 15C for 16 hours. Cells were harvested by centrifugation. Following centrifugation cell pellets were resuspended with lysis buffer followed by sonication. The supernatant was kept for further purification. The target proteins were purified via one-step purification on a Ni column followed by sterilization through a 0.22 μm filter. No visible precipitation was observed after purification and two cycles of freeze-thaw testing was performed to ensure no visible precipitation after freezing. pH 7.4 PBS with 10% glycerol was used as the final vehicle for the proteins. Proteins were analyzed via SDS-PAGE and western blot using anti-His and anti-MBP antibodies that detected a strong band approximately 110 kDa in size. Protein concentration was determined by Bradford assay using a BSA standard.
Dynamic light scattering (DLS)
Aliquots of purified MBP-FUS-CHOP WT, MBP-FUS-CHOP 12A, and MBP-FUS-CHOP 12E were thawed on ice and diluted to 0.34 mg/mL. 176 μL of protein was aliquoted into two separate 0.5 mL low-bind Eppendorf tubes per protein. 20 μL of TEV protease buffer was added to all tubes. Either 4 μL of TEV protease (New England Biolabs) or 4 μL of PBS was added to each protein sample. Samples were incubated at room temperature overnight and transferred into a black wall, clear bottom 96-well plate. The plate was analyzed using a Wyatt DynaPro Plate Reader at 25°C.
Soft agar transformation assays
1.8% Bactoagar was made with diH2O and autoclaved. 2x and 1x RPMI were prepared using RPMI powder (ThermoFisher Scientific, 12100046) fetal bovine serum (ThermoFisher Scientific, 16000044), and antibiotic-antimycotic (ThermoFisher Scientific, 10091148). 0.6% agar was made by diluting 1.8% agar with DMEM and kept in a 42°C water bath. 3 mL of 0.6% agar was poured per well of 6-well plates and allowed to solidify in a hood for 10 minutes before transferring to an incubator. Cells were trypsinized and counted using Trypan Blue solution (ThermoFisher Scientific, 15250061). 500 μL of cells and 1 mL of 0.6% agar were mixed and gently pipetted onto the bottom agar in each well to create the 0.4% agar top layer. 15000 cells were plated per well in duplicate for each cell line. After plating, plates were placed in an incubator and allowed to grow for 2-3 weeks. Media were supplemented each week by adding 250 μL of RPMI per well to prevent drying. Plates were stained with 0.05% crystal violet in 10% ethanol for 1 hour, then destained with distilled water rinses. Colonies were imaged using a Leica inverted light microscope, and colonies were counted using ImageJ.
Clonogenic assays
Cells were plated at various densities in triplicate in a 6-well plate. Cells were allowed to attach overnight before irradiation at 2 Gy, 4 Gy, and 8 Gy using parameters described under ‘Radiation treatments’ in these methods. After formation and visualization of colonies, cells were fixed with 70% ethanol for 10 minutes, stained with Coomassie Brilliant Blue (Bio-Rad), rinsed with deionized water, and dried overnight. Surviving fractions were calculated relative to unirradiated controls. Colonies were imaged and counted using ImageJ.
Study material and analysis details are available in the Supplemental Materials.
Study Approval and Data Availability.
All data generated or analyzed in this study are included in this published article and its supplementary materials. All sequencing data are deposited at http://bit.ly/fuschopGEO and may be accessed using the token = ovyxuiyebjepvaz
Results
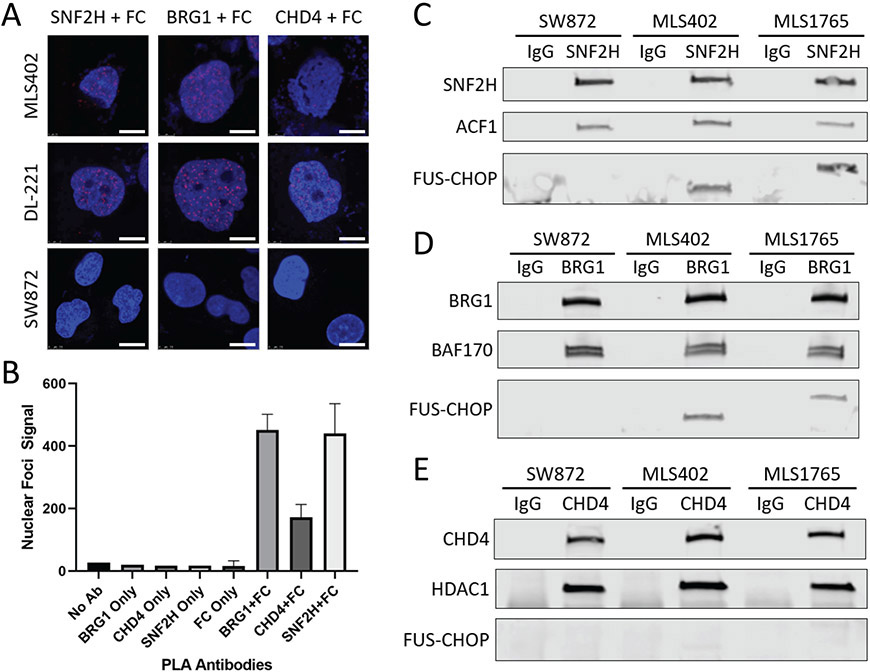
To identify chromatin remodeling complexes that interact with FUS-CHOP, we performed proximity ligation assays (PLA) for three of the major chromatin remodeler complex families: BAF, ISWI, and nucleosome remodeling deacetylase (NuRD). We found that all three chromatin remodeler families were in close proximity to FUS-CHOP in the nuclei of human MLPS cells (MLS402(6), DL221(18)), in contrast to fusion-negative liposarcoma cells (SW872) (Fig. 1). SNF2H and BRG1 PLA foci had relatively stronger signal compared to CHD4 PLA foci (Fig. 1, A and B). To confirm these interactions we performed co-IPs in the three human sarcoma cell lines (Fig. 1C). MLS402 expresses the 7-2 translocation FUS-CHOP variant (approx. 75 kDa); MLS1765 expresses the 13-2 translocation FUS-CHOP variant (approx. 100 kDa); and SW872 (negative control) (19). We detected an interaction between FUS-CHOP and BRG1 as previously reported (3). We also discovered a new interaction between FUS-CHOP and SNF2H of the ISWI complex (Fig. 1, C and D). CHD4 did not interact with FUS-CHOP in either fusion-expressing cell line (Fig. 1E). The controls ACF1, BAF170, and HDAC1 are known interactors with the chromatin remodelers ISWI, BAF, and NuRD, respectively (Fig. 1).
Fig. 1. FUS-CHOP interacts with multiple chromatin remodeling complexes.
(A) Confocal images of fusion-positive (DL-221, MLS402) and fusion-negative (SW872) nuclei stained with DAPI (blue). Red foci mark proteins that are in close proximity to FUS-CHOP (FC). SNF2H, BRG1, and CHD4 are ATPase subunits of their respective chromatin remodeling complexes. Scale bars = 10 μm. (B) Quantification of red foci in DL-221 nuclei with either single or paired PLA antibodies. (C) SNF2H co-IP for FUS-CHOP and the ACF1 subunit of ISWI complexes. (D) BRG1 co-IP for FUS-CHOP and the BAF170 subunit of the BAF complex. (E) CHD4 co-IP for FUS-CHOP and the HDAC1 subunit of the NuRD complex. Experiments were performed in duplicate.
To further validate these interactions in a different organism, we performed co-IPs in mouse cell lines and mouse tumor cell lines with and without FUS-CHOP (fig. S1, A-C) (20,21). The FUS-CHOP-expressing tumor cell lines used for these experiments were derived from the conditional mouse model of FUS-CHOP-driven sarcoma that we generated using Cre and CRISPR/Cas9 technology (21). FUS-CHOP co-immunoprecipitated with Snf2h and Brg1 in these mouse cell lines (fig. S1, A-C). As observed in the human tumor cells, FUS-CHOP was not detected in the Chd4 co-IP. Therefore, the mouse co-IP validated the results of the human co-IP in support of this interaction in multiple different cell lines and across species.
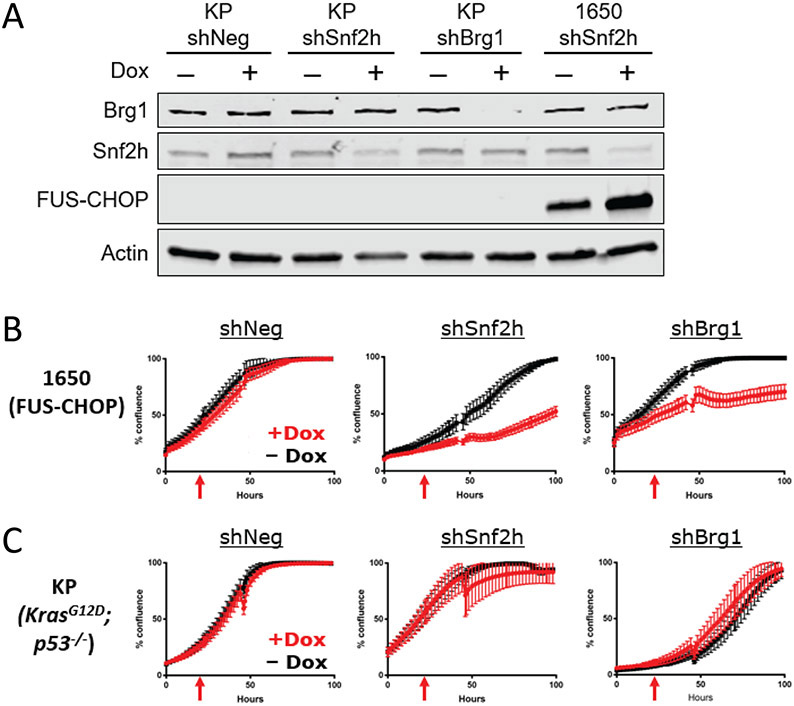
To determine if SNF2H and BRG1 impact the proliferation of FUS-CHOP-expressing cells, we stably knocked down Snf2h and Brg1 in KP (KrasG12D; p53−/−) (20) and 1650 (FUS-CHOP positive) (21) mouse sarcoma cells (Fig. 2A). Silencing Snf2h or Brg1 decreased proliferation of 1650 FUS-CHOP-driven mouse sarcoma cells (Fig. 2, B and C). However, proliferation was unaffected in KP murine sarcoma cell lines. These data indicate that Snf2h and Brg1 are selectively important for the proliferation of FUS-CHOP-driven murine sarcomas.
Fig. 2. Snf2h and Brg1 are required for proliferation of FUS-CHOP-driven tumor cells.
(A) Knockdown of Snf2h and Brg1 protein expression in 1650 (fusion-positive) and KP (fusion-negative) murine sarcoma cell lines. (B) Effect of Snf2h and Brg1 knockdown on proliferation of FUS-CHOP-driven murine sarcoma cell lines and (C) KP (fusion-negative) murine sarcoma cell lines. Red arrows denote addition of doxycycline (Dox) 24 hours after plating to induce shRNAs. At least two technical replicates were performed for knockdown experiments.
We then explored the stability of the interaction between SNF2H and FUS-CHOP and its dependence on nucleic acids (fig. S1, D-G). Interactions between SNF2H and FUS-CHOP were less stable than those between ISWI complex members SNF2H and ACF1 (fig. S1, D and E). The decreased stability of FUS-CHOP with the ISWI complex suggests a transient interaction, similar to EWS-FLI1 interaction with the BAF complex (2). Although FUS-CHOP interacts with SNF2H independently of RNA or DNA, the interaction seems to be enhanced in the absence of RNA suggesting that RNA may modulate this interaction (fig. S1, F and G)(22).
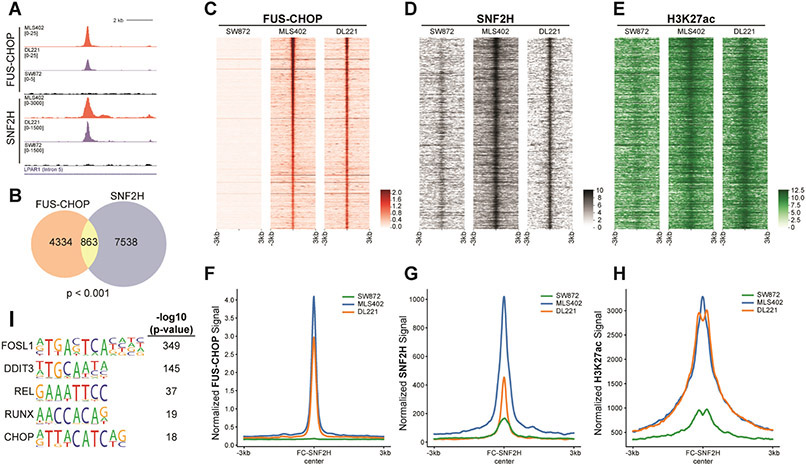
Because of the importance of the relationship between FUS-CHOP and SNF2H, we investigated the mechanism by which FUS-CHOP and SNF2H contribute to sarcomagenesis in MLPS. We first explored the genomic targeting of FUS-CHOP. To map FUS-CHOP binding sites genome-wide, we performed ChIP-seq in human MLPS cells as well as the FUS-CHOP-negative human liposarcoma cell line as a control (Fig. 3, fig S2). Distinct FUS-CHOP binding sites were detected in fusion-positive, but not fusion-negative chromatin (Fig. 3A, fig. S2A). 14,146 and 11,395 FUS-CHOP binding sites were identified in the MLS402 and DL221 cell lines, respectively (fig. S2, B and C). To increase the stringency of the analysis, we focused on those shared 5399 FUS-CHOP-bound sites detected in both MLS402 and DL221 cells. The FUS-CHOP binding sites were primarily located in intronic and distal intergenic regions with 5,197 FUS-CHOP binding sites at enhancers (fig. S2D). Since EWS-FLI1 is known to target GGAA repeats, we searched for repetitive sequence motifs, but no such repeat sequences were identified through our analysis. To determine whether SNF2H colocalized with FUS-CHOP on chromatin, we performed SNF2H CUT&RUN (Fig. 3, A and C, fig. S2, F-H). We identified 8,344 sites of SNF2H binding that were shared by the FUS-CHOP positive MLPS cell lines (but not in the fusion-negative cells) (Fig. 3B, fig. S2, G and H). Of these sites, we identified about 10% that were also bound by FUS-CHOP (Fig. 3B). H3K27 acetylation at the sites targeted by FUS-CHOP and SNF2H in MLS402 and DL221 cell lines indicates that they represent active enhancer elements (Fig. 3C-E). Interestingly, modest H3K27ac in the FUS-CHOP-negative SW872 cells suggests that other transcription factors may mediate acetylation at some of these sites. FUS-CHOP bound at these enhancers correlates with the abundance of SNF2H and H3K27ac and supports a model of recruitment of SNF2H to these enhancers defined by FUS-CHOP (Fig. 3F-H). DNA motif analysis of shared FUS-CHOP and SNF2H binding sites revealed significant enrichment of FOSL1, REL, and RUNX motifs, relative to genomic background (Fig. 3I). Interestingly, despite detection of the DDIT3 (CHOP) motif, it was less enriched than FOSL1. SNF2H/FUS-CHOP shared sites demonstrate similarly enriched DNA-binding motifs as those bound by FUS-CHOP (Fig. 3I, fig. S2E). Interestingly, the DDIT3 (or CHOP) motif was a prominent shared motif, but not a prominent motif in FUS-CHOP binding alone suggesting that some less prevalent motifs (such as DDIT3) are enriched at shared FUS-CHOP and SNF2H binding sites (fig. S2E). Wild type CHOP is implicated in the unfolded protein response (UPR), and under stress conditions, CHOP and ATF4 bind to very similar sequences (23). However, we observed a different set of DNA-binding motifs in our analysis. Differences in DNA binding between FUS-CHOP and wild type CHOP may explain how the fusion protein is oncogenic while the wild type CHOP is pro-apoptotic and initiates ER stress-induced cell death (24). Yet, the binding of FUS-CHOP to “classical” DDIT3 sequence motifs also supports a model where FUS-CHOP displaces or prevents wild type CHOP from binding its transcriptional targets important for growth arrest or apoptosis. Further experiments are required to elucidate if this “dominant negative” effect occurs either at the DNA/chromatin level or through protein-protein interactions, though evidence in the literature suggests the former mechanism is more likely (7,24,25).
Fig. 3. FUS-CHOP colocalizes with SNF2H at active enhancers.
(A) Representative tracks of a FUS-CHOP SNF2H shared binding site. (B) Venn diagram showing the overlap of FUS-CHOP ROE at enhancers and SNF2H ROE (Permutation test, p < 0.001). (C-E) Heatmaps of FUS-CHOP, SNF2H, and H3K27ac signal densities in human fusion-positive (DL221, MLS402) and fusion-negative (SW872) liposarcoma cell lines. 6-kb windows in each panel are centered on FUS-CHOP SNF2H shared binding sites (n = 863), ranked by significance of overlapping peak calls. (F-H) Average line plots of FUS-CHOP, SNF2H, and H3K27ac signal at ROE bound by both FUS-CHOP and SNF2H. SNF2H signal is increased at these enhancers in the presence of FUS-CHOP. H3K27ac signal is also increased at these enhancers in the presence of FUS-CHOP and SNF2H. The x-axis represents a 6-kb window centered on FUS-CHOP SNF2H binding sites. (I) Enriched motifs found at overlapping FUS-CHOP and SNF2H regions of enrichment. Two biological replicates were used for MLPS cell lines in all experiments.
To determine how the shared occupancy of SNF2H and FUS-CHOP impacted gene expression, we performed RNA-seq on human MLPS and liposarcoma cell lines and associated the transcriptomic differences between these cell lines with regions of shared occupancy (fig. S3). Our analysis revealed that regions of shared FUS-CHOP and SNF2H enrichment were associated with 711 genes, of which 338 genes were upregulated and 83 genes were downregulated in the DL221 cell line relative to SW872 (fig. S3A). These regions were associated with 212 upregulated and 144 downregulated genes in the MLS402 MLPS cell line compared to SW872 (fig. S3B). There was a strong correlation between the magnitude of both gene activation and repression in DL221 and MLS402 cell lines at these genes (fig. S3, C and D). These data show that the enhancers occupied by FUS-CHOP and SNF2H upregulate a common set of genes. Activated genes are enriched for those involved in cell proliferation, suggesting a direct role for that FUS-CHOP function in sarcomagenesis (fig. S3, E and F ) (26,27). Analyzing publicly available RNA-seq data for adipocyte progenitor cells and myxoid liposarcoma samples, we found that BRG1 and CHD4 expression was in the upper quartile of all expressed genes in both sets. While SNF2H expression levels were also in the upper quartile of expressed genes in adipocyte progenitor cells, expression levels in the MLS tumors were near the median abundance (fig. S3G). These data suggest that transcripts of SNF2H, BRG1, and CHD4 are present in MLS tumors at average to above average abundance. However, our model neither proposes nor is dependent on changes in SNF2H expression.
The FUS-CHOP PrLD is required for transformation of NIH-3T3 cells suggesting that the PrLD may mediate transactivation in the fusion protein. Substitution of the PrLD by other transactivation domains does not strongly transform cells (25). In contrast, substitution of FUS with EWS in the fusion oncoprotein results in similar transformation (25) and the EWS-CHOP fusion is also associated with MLPS in patients (28,29). These results indicate that FUS and EWS confer specific activities necessary for transformation. To investigate a potential link between the role of FUS PrLD as a transactivator and the exquisite clinical response of MLPS to radiotherapy, we hypothesized that the interaction of FUS-CHOP with binding partners, such as SNF2H, might be regulated through DNA damage-induced phosphorylation of the FUS-CHOP PrLD, which could interfere with chromatin remodeler interactions and contribute to the sensitivity of MLPS to ionizing radiation.
Immunoblotting with antibodies that recognize phospho-SQ/TQ (pSQ/TQ) sites, we explored the phosphorylation state of FUS-CHOP immunoprecipitated from cells treated with ATM and DNA-PK inhibitors. We found that FUS-CHOP was phosphorylated after irradiation by DNA-PK and ATM (fig. S4A). pSQ/TQ phosphorylation levels following irradiation in the presence of the DNA-PK inhibitor NU-7026 were similar to unirradiated cells (fig. S4B). Using a human FUS phospho-Ser42 specific antibody, we observed that sites in the PrLD were phosphorylated in FUS-CHOP driven mouse sarcoma cells (fig. S4C). These data show that, like wild type FUS, FUS-CHOP is phosphorylated by DNA-PK and ATM after irradiation (30).
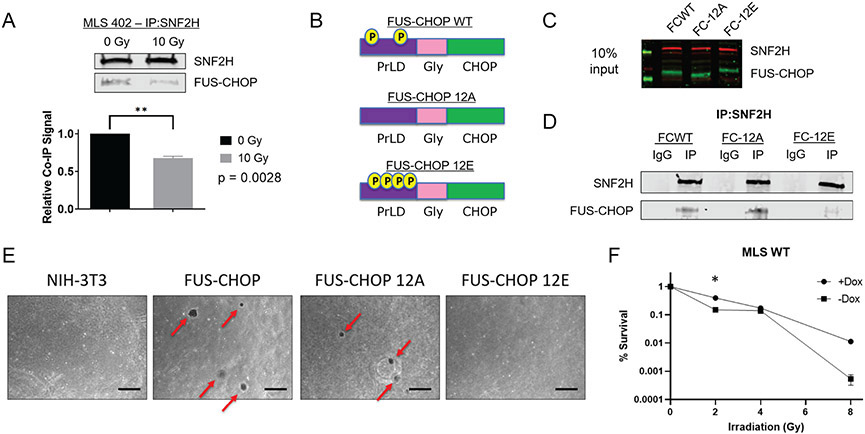
Next, we investigated if radiation affects the interaction of FUS-CHOP with SNF2H. 45 minutes after 10 Gy X-ray irradiation, we observed a significant decrease in the FUS-CHOP interaction with SNF2H (Fig. 4A). To directly test if the decreased interaction of FUS-CHOP with SNF2H was due to phosphorylation of the PrLD, we generated wild type (FCWT), phospho-dead (FC-12A) or phospho-mimic (FC-12E) FUS-CHOP mutants at the twelve SQ/TQ sites in the FUS-CHOP PrLD. These FUS-CHOP variants were stably expressed in NIH-3T3 cells (Fig. 4, B and C) (31). In contrast to the wild type FUS-CHOP and the phospho-dead 12A mutant, SNF2H interaction with the phospho-mimic 12E mutant was diminished (Fig. 4D). We then tested the transformation activity of these mutants by soft agar colony formation. Wild type and the 12A mutant, but not the 12E mutant, transformed NIH-3T3 cells (Fig. 4E). These data suggest that phosphorylation of the FUS-CHOP PrLD disrupts functions of the fusion protein necessary for transformation and proliferation, including interactions with SNF2H and potentially other nuclear proteins.
Fig. 4. Ionizing radiation decreases protein-protein interactions between SNF2H and FUS-CHOP by DNA damage-induced phosphorylation of the PrLD.
(A) Co-IP of SNF2H and FUS-CHOP 45 minutes after either sham or 10 Gy X-ray irradiation. ** p = 0.0028, unpaired t-test. (B) Schematic of FUS-CHOP wild type, phospho-dead (12A), and phospho-mimic (12E) cDNA constructs. (C) NIH-3T3 cells stably expressing FUS-CHOP and its phosphomutant forms. (D) Co-IP of SNF2H in NIH-3T3 cell lines stably expressing FUS-CHOP (FCWT), FUS-CHOP 12A (FC-12A), and FUS-CHOP 12E (FC-12E). (E) Soft agar colony formation assay to evaluate cellular transformation using NIH-3T3 cells that stably express either wild type FUS-CHOP or phosphomutant forms of FUS-CHOP. Representative images from two technical replicates are shown. Scale bars = 500 μm. (F) Clonogenic assay of human MLS 402 cells with and without dox-inducible shRNA targeting endogenous FUS-CHOP at 2, 4, and 8 Gy X-ray irradiation. *, p < 0.05
A recently appreciated property of proteins containing PrLDs, including the FET proteins, is the ability to form phase-separated compartments, or biomolecular condensates. Indeed, it is likely that FUS-CHOP activates enhancers and gene targets by forming nuclear condensates that contain chromatin remodelers and transcriptional machinery (32,33). We demonstrated that specific protein-protein interactions between the PrLD of FUS-CHOP and SNF2H can be disrupted by using a phospho-mimic mutant of FUS-CHOP (Fig. 4D). This decreased interaction between FUS-CHOP and SNF2H following irradiation may result from increased solubility of the phosphorylated form of FUS-CHOP mediated by DNA damage. Using purified proteins, we observed that the FUS-CHOP 12E phospho-mimetic mutant remains soluble after cleavage of an MBP solubility tag (fig. S4D). In contrast, wild type and the 12A mutant proteins aggregate into micron-scale particles.
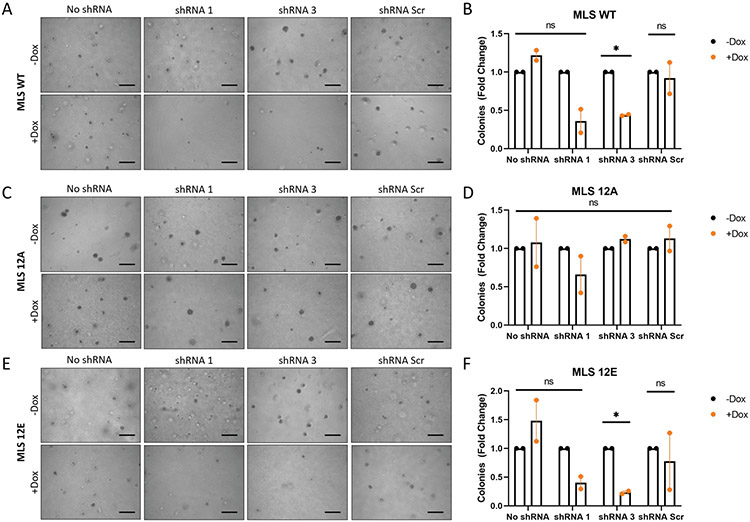
To further explore the role of FUS-CHOP in MLPS response to radiation, we performed clonogenic assays using MLS WT cells containing shRNA 1 (sh1), which showed the best knockdown based on Western blot (fig. S5). Interestingly, we observed MLS WT cells survive better with endogenous FUS-CHOP knockdown at 2 Gy, but there was no statistically significant difference at 4 Gy or 8 Gy (Fig. 4F). Therefore, to directly and more rigorously test the hypothesis that phosphorylation of the FUS-CHOP PrLD acts as a switch for tumorigenesis, we performed a rescue experiment using the human MLPS cell line, MLS 402. First, we generated MLS 402 cell lines that were either wild type (MLS WT), or expressed the 12A (MLS 12A) or 12E (MLS 12E) mutant. Next, we generated stable MLS WT, MLS 12A, and MLS 12E cell lines containing dox-inducible shRNAs targeting the 3’ UTR of FUS-CHOP (fig. S5). This strategy allowed us to specifically knockdown endogenous FUS-CHOP, but not the exogenous 12A or 12E mutants. Using these cell lines, we performed an in vitro soft agar assay to determine the effect phosphorylation of the FUS-CHOP PrLD had on cellular transformation as measured by colony formation (fig. 5). With knockdown of FUS-CHOP in MLS WT cells, fewer colonies formed (fig. 5A-B). Similarly, with knockdown of FUS-CHOP in MLS 12E cells, colony formation was diminished (fig. 5E-F). However, we observed that the 12A mutant was able to rescue cellular transformation as measured by increased colony formation (fig. 5C-D). These data indicate that the 12A FUS-CHOP mutant can rescue transformation in human cells, while the phospho-mimic FUS-CHOP mutant (12E) is not able to rescue transformation. Importantly, these data suggest that the 12E mutant disrupts interactions between FUS-CHOP and other proteins or nucleic acids required for transformation, but the 12A mutant retains these interactions required for transformation. Taken together, these data support the notion that the phosphorylation status of the FUS-CHOP PrLD can act as a switch for interactions between FUS-CHOP and proteins or nucleic acids required for transformation.
Fig. 5. Phosphorylation of the FUS-CHOP PrLD abrogates cellular transformation.
Soft agar assay for colony formation of FUS-CHOP cell line MLS WT (A-B), MLS 12A (C-D), and MLS 12E (E-F) with and without dox-inducible shRNA knockdown of endogenous FUS-CHOP. Representative images from two replicates are shown. Scale bars = 500 μm. *, p < 0.05; ns, not significant.
Discussion
Our work demonstrated that retargeting chromatin remodeling complexes by FET fusion proteins may be a more broadly applicable mechanism beyond EWS-FLI1 in Ewing sarcomagenesis (2). Importantly, we showed that in addition to the BAF complex, FET proteins can interact with other chromatin remodeling complexes such as the ISWI complex, which contains the SNF2H ATPase subunit (Fig. 1, fig. S1). We also showed that the SNF2H and BRG1 ATPase subunits were important for proliferation of FUS-CHOP-driven sarcomas (Fig. 2). We hypothesized that proliferation of FUS-CHOP-driven sarcomas may be dependent on these chromatin remodelers because recruitment of chromatin remodeling complexes to DNA regulatory elements by FUS-CHOP was important for sarcomagenesis. To test this model of oncogenesis, we first mapped FUS-CHOP and SNF2H binding sites genome-wide (Fig. 3, fig S2). These two proteins were found to co-localize at active enhancers in human MLPS cell lines that upregulated a common set of genes implicated in cell proliferation, migration, apoptosis, and several other processes (Fig. S3). This general mechanism of FET fusion-mediated chromatin remodeling may be a model for sarcomagenesis of an entire class of translocation-positive sarcomas that contain FET protein translocation partners. Additionally, to address lessons from modeling sarcomas in other organisms, this mechanism may explain why fusion-positive sarcomas are difficult to model in mice where the specific DNA-binding motifs and enhancers required for sarcomagenesis in humans differ from those required for these processes in mice (21,34).
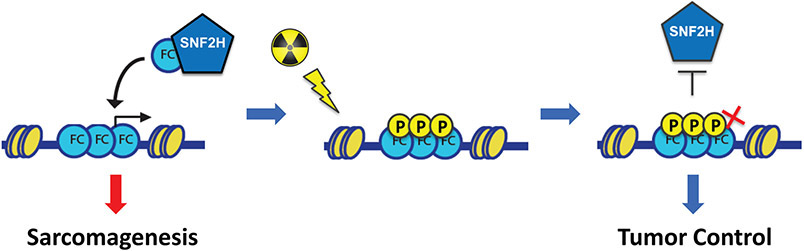
An intriguing and emerging property of proteins containing PrLDs, such as FET proteins, is the ability to form phase-separated biomolecular condensates (22,35-37). As such, FET proteins can phase separate under physiological conditions, and perhaps most interestingly, these intrinsically disordered regions (IDRs) can be post-translationally modified to promote or prevent phase separation and other intermolecular interactions. Given that the IDR of FUS is translocated in FUS-CHOP, we began to view “retargeting of chromatin remodelers” through the lens of biomolecular condensates and phase separation. Indeed, FUS-CHOP may be forming intranuclear condensates that contain chromatin remodelers and transcriptional machinery to activate enhancers and gene targets (32). Although our work is limited to the level of chromatin and does not directly explore formation of biomolecular condensates, we demonstrate specific protein-protein interactions between the PrLD of FUS-CHOP and SNF2H can be disrupted by using a phospho-mimic mutant of FUS-CHOP (Fig. 4). Post-translational modifications of FET PrLDs can change the charge of these proteins and either promote or prevent phase separation. In the case of FUS, phosphorylation of the domain at twelve phospho-SQ/TQ sites in the PrLD has been shown to increase the solubility of FUS and to disrupt phase separation (31,38). Taken together, our results suggest a model linking oncogenic activity and radiosensitivity in MLPS supporting a critical role for the PrLD of FUS-CHOP to mediate oncogenic retargeting of SNF2H (Fig. 6). Phosphorylation of the PrLD by activated DNA damage response kinases alters the solubility of FUS-CHOP, which destabilizes FUS-CHOP/SNF2H interactions necessary for MLPS maintenance. An important point to consider is the role of the FUS-CHOP PrLD in sarcoma maintenance in addition to promoting tumor cell proliferation. We discovered that a phospho-dead FUS-CHOP PrLD could rescue transformation in human MLS cells, but that the phospho-mimic PrLD mutant could not (Fig. 5). Considering these results with the data that radiation decreases interactions between FUS-CHOP and SNF2H (Fig. 4), and the recent demonstration that FUS-CHOP undergoes phase separation (32), we propose that FUS-CHOP not only drives sarcomagenesis in MLPS, but also mediates the radiosensitivity observed in the clinic by radiation-induced disruption of the phase separation properties of FUS-CHOP that diminishes protein-protein interactions required for tumor maintenance. Small molecules that promote phosphorylation of the PrLD in FET proteins like FUS that disrupt interactions with chromatin remodeling complexes may be effective treatments for FET fusion positive sarcomas.
Fig. 6. Model of FUS-CHOP mediated oncogenic activity and radiosensitivity in MLPS.
FUS-CHOP and SNF2H interact to target enhancers and activate genes leading to sarcomagenesis. Irradiation induces phosphorylation of the FUS-CHOP PrLD, which interferes with SNF2H interactions and MLPS sarcomagenesis leading to tumor control.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| CHOP/DDIT3 | Cell Signaling Technology | #2895 |
| CHD4 | Cell Signaling Technology | #11912 |
| BRG1 | Cell Signaling Technology | #49360 |
| SNF2H | EMD Millipore | ABE1026 |
| BAF170 | Cell Signaling Technology | #12760 |
| ACF1 | Abcam | ab187670 |
| HDAC1 | Cell Signaling Technology | #5356 |
| TBP | Cell Signaling Technology | #44059 |
| H3K27me3 | Cell Signaling Technology | #9733 |
| IgG - rabbit | Cell Signaling Technology | #2729 |
| H3K27ac | Sigma | MABE647 |
| H3K27ac | Active Motif | 39133 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| MBP-FUS-CHOP WT | This paper | |
| MBP-FUS-CHOP 12A | This paper | |
| MBP-FUS-CHOP 12E | This paper | |
| Critical Commercial Assays | ||
| Duolink Proximity Ligation Assay | Sigma-Aldrich | DUO92101-1KT |
| Deposited Data | ||
| ChIP-seq and RNA-seq raw data | NCBI GEO | GSE145678 |
| Experimental Models: Cell Lines | ||
| NIH-3T3 | ATCC | CRL-1658 |
| SW-872 | ATCC | HTB-92 |
| MLS 402 | Gift from Pierre Aman | |
| MLS 1765 | Gift from Pierre Aman | |
| DL-221 | Gift from Keila Torres and Alexander Lazar | |
| Recombinant DNA | ||
| N174(Puro)-FUS-CHOP WT | this paper | |
| N174(Puro)-FUS-CHOP 12A | this paper | |
| N174(Puro)-FUS-CHOP 12E | this paper | |
| N174-MCS (Puro) backbone | Addgene | Addgene plasmid # 81068 ; RRID:Addgene_81068 |
| Tet-pLKO-Puro | Addgene | Addgene plasmid # 21915 ; RRID:Addgene_21915 |
Acknowledgements
We thank Pierre Åman (University of Gothenberg) for cell lines MLS402 and MLS1765, and thank Keila Torres and Alexander Lazar (MD Anderson) for cell line DL221. We thank Steven Henikoff, Derek Janssens, and Nan Liu for their advice, shared reagents, and protocols for CUT&RUN. We also thank Andrea Ventura (Memorial Sloan Kettering) for helpful suggestions and discussion.
Financial Support:
This work was supported by the National Cancer Institute of the US, NIH (awards R35 CA197616 to DGK, F30 CA206424 to MC) and the T32 GM007171 MSTP training grant (Duke University).
Footnotes
Conflict of interest disclosure statement: The authors declare no competing interests for this manuscript. DGK is on the scientific advisory board and owns stock in Lumicell, which is commercializing intraoperative imaging technology. DGK is a co-founder of XRAD Therapeutics, which is developing radiosensitizers and has received research support from XRAD, Merck, Bristol Myers Squibb, and Eli Lilly.
References
- 1.Mertens F, Antonescu CR, Mitelman F. Gene fusions in soft tissue tumors: Recurrent and overlapping pathogenetic themes. Genes Chromosomes Cancer. 2016;55:291–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boulay G, Sandoval GJ, Riggi N, Iyer S, Buisson R, Naigles B, et al. Cancer-Specific Retargeting of BAF Complexes by a Prion-like Domain. Cell. 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lindén M, Thomsen C, Grundevik P, Jonasson E, Andersson D, Runnberg R, et al. FET family fusion oncoproteins target the SWI/SNF chromatin remodeling complex. EMBO Rep. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Patel M, Simon JM, Iglesia MD, Wu SB, McFadden AW, Lieb JD, et al. Tumor-specific retargeting of an oncogenic transcription factor chimera results in dysregulation of chromatin and transcription. Genome Res. 2012;22:259–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shing DC, McMullan DJ, Roberts P, Smith K, Chin S-F, Nicholson J, et al. FUS/ERG Gene Fusions in Ewing’s Tumors. Cancer Res. American Association for Cancer Research; 2003;63:4568–76. [PubMed] [Google Scholar]
- 6.Aman P, Ron D, Mandahl N, Fioretos T, Heim S, Arheden K, et al. Rearrangement of the transcription factor gene CHOP in myxoid liposarcomas with t(12;16)(q13;p11). Genes Chromosomes Cancer. 1992;5:278–85. [DOI] [PubMed] [Google Scholar]
- 7.Crozat A, Aman P, Mandahl N, Ron D. Fusion of CHOP to a novel RNA-binding protein in human myxoid liposarcoma. Nature. 1993;363:640–4. [DOI] [PubMed] [Google Scholar]
- 8.Panagopoulos I, Mandahl N, Ron D, Höglund M, Nilbert M, Mertens F, et al. Characterization of the CHOP breakpoints and fusion transcripts in myxoid liposarcomas with the 12;16 translocation. Cancer Res. 1994;54:6500–3. [PubMed] [Google Scholar]
- 9.Antonescu CR, Tschernyavsky SJ, Decuseara R, Leung DH, Woodruff JM, Brennan MF, et al. Prognostic impact of P53 status, TLS-CHOP fusion transcript structure, and histological grade in myxoid liposarcoma: a molecular and clinicopathologic study of 82 cases. Clin Cancer Res. 2001;7:3977–87. [PubMed] [Google Scholar]
- 10.Joseph CG, Hwang H, Jiao Y, Wood LD, Kinde I, Wu J, et al. Exomic analysis of myxoid liposarcomas, synovial sarcomas, and osteosarcomas. Genes Chromosomes Cancer. 2014;53:15–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wylie J. Pathology and Genetics of Tumours of Soft Tissue and Bone. Published 2002, 1st edition, ISBN 92 832 24132. Surg Oncol. 2004;13:43. [Google Scholar]
- 12.Ewing J. FASCIAL SARCOMA AND INTERMUSCULAR MYXOLIPOSARCOMA. Arch Surg. American Medical Association; 1935;31:507–20. [Google Scholar]
- 13.Chung PWM, Deheshi BM, Ferguson PC, Wunder JS, Griffin AM, Catton CN, et al. Radiosensitivity translates into excellent local control in extremity myxoid liposarcoma: a comparison with other soft tissue sarcomas. Cancer. 2009;115:3254–61. [DOI] [PubMed] [Google Scholar]
- 14.Guadagnolo BA, Zagars GK, Ballo MT, Patel SR, Lewis VO, Benjamin RS, et al. Excellent local control rates and distinctive patterns of failure in myxoid liposarcoma treated with conservation surgery and radiotherapy. Int J Radiat Oncol Biol Phys. 2008;70:760–5. [DOI] [PubMed] [Google Scholar]
- 15.Pitson G, Robinson P, Wilke D, Kandel RA, White L, Griffin AM, et al. Radiation response: an additional unique signature of myxoid liposarcoma. Int J Radiat Oncol Biol Phys. Elsevier; 2004;60:522–6. [DOI] [PubMed] [Google Scholar]
- 16.Skene PJ, Henikoff JG, Henikoff S. Targeted in situ genome-wide profiling with high efficiency for low cell numbers. Nat Protoc. 2018;13:1006–19. [DOI] [PubMed] [Google Scholar]
- 17.Meers MP, Bryson TD, Henikoff JG, Henikoff S. Improved CUT&RUN chromatin profiling tools. Elife. 2019;8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de Graaff MA, Yu JSE, Beird HC, Ingram DR, Nguyen T, Juehui Liu J, et al. Establishment and characterization of a new human myxoid liposarcoma cell line (DL-221) with the FUS-DDIT3 translocation. Lab Invest. 2016;96:885–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stratford EW, Castro R, Daffinrud J, Skårn M, Lauvrak S, Munthe E, et al. Characterization of liposarcoma cell lines for preclinical and biological studies. Sarcoma. 2012;2012:148614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kirsch DG, Dinulescu DM, Miller JB, Grimm J, Santiago PM, Young NP, et al. A spatially and temporally restricted mouse model of soft tissue sarcoma. Nat Med. 2007;13:992–7. [DOI] [PubMed] [Google Scholar]
- 21.Chen M, Xu ES, Leisenring NH, Cardona DM, Luo L, Ma Y, et al. The Fusion Oncogene FUS-CHOP Drives Sarcomagenesis of High-Grade Spindle Cell Sarcomas in Mice. Sarcoma. Hindawi; 2019;2019:1340261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maharana S, Wang J, Papadopoulos DK, Richter D, Pozniakovsky A, Poser I, et al. RNA buffers the phase separation behavior of prion-like RNA binding proteins. Science. American Association for the Advancement of Science; 2018;360:eaar7366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Han J, Back SH, Hur J, Lin Y-H, Gildersleeve R, Shan J, et al. ER-stress-induced transcriptional regulation increases protein synthesis leading to cell death. Nat Cell Biol. 2013;15:481–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barone MV, Crozat A, Tabaee A, Philipson L, Ron D. CHOP (GADD153) and its oncogenic variant, TLS-CHOP, have opposing effects on the induction of G1/S arrest. Genes Dev. 1994;8:453–64. [DOI] [PubMed] [Google Scholar]
- 25.Zinszner H, Albalat R, Ron D. A novel effector domain from the RNA-binding protein TLS or EWS is required for oncogenic transformation by CHOP. Genes Dev. 1994;8:2513–26. [DOI] [PubMed] [Google Scholar]
- 26.Chen EY, Tan CM, Kou Y, Duan Q, Wang Z, Meirelles GV, et al. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics. 2013;14:128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kuleshov MV, Jones MR, Rouillard AD, Fernandez NF, Duan Q, Wang Z, et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016;44:W90–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Panagopoulos I, Höglund M, Mertens F, Mandahl N, Mitelman F, Aman P. Fusion of the EWS and CHOP genes in myxoid liposarcoma. Oncogene. 1996;12:489–94. [PubMed] [Google Scholar]
- 29.Dal Cin P, Sciot R, Panagopoulos I, Aman P, Samson I, Mandahl N, et al. Additional evidence of a variant translocation t(12;22) with EWS/CHOP fusion in myxoid liposarcoma: clinicopathological features. J Pathol. 1997;182:437–41. [DOI] [PubMed] [Google Scholar]
- 30.Gardiner M, Toth R, Vandermoere F, Morrice NA, Rouse J. Identification and characterization of FUS/TLS as a new target of ATM. Biochem J. 2008;415:297–307. [DOI] [PubMed] [Google Scholar]
- 31.Monahan Z, Ryan VH, Janke AM, Burke KA, Rhoads SN, Zerze GH, et al. Phosphorylation of the FUS low-complexity domain disrupts phase separation, aggregation, and toxicity. EMBO J. 2017;36:2951–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Owen I, Yee D, Wyne H, Perdikari TM, Johnson V, Smyth J, et al. The oncogenic transcription factor FUS-CHOP can undergo nuclear liquid-liquid phase separation. bioRxiv. 2021. [cited 2021 May 10]. page 2021.02.24.432743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Boija A, Klein IA, Sabari BR, Dall’Agnese A, Coffey EL, Zamudio AV, et al. Transcription Factors Activate Genes through the Phase-Separation Capacity of Their Activation Domains. Cell. 2018;175:1842–55.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Minas TZ, Surdez D, Javaheri T, Tanaka M, Howarth M, Kang H-J, et al. Combined experience of six independent laboratories attempting to create an Ewing sarcoma mouse model. Oncotarget. 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Banani SF, Lee HO, Hyman AA, Rosen MK. Biomolecular condensates: organizers of cellular biochemistry. Nat Rev Mol Cell Biol. 2017;18:285–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ryan JJ, Sprunger ML, Holthaus K, Shorter J, Jackrel ME. Engineered protein disaggregases mitigate toxicity of aberrant prion-like fusion proteins underlying sarcoma. J Biol Chem. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shin Y, Berry J, Pannucci N, Haataja MP, Toettcher JE, Brangwynne CP. Spatiotemporal Control of Intracellular Phase Transitions Using Light-Activated optoDroplets. Cell. Elsevier; 2017. page 159–71.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rhoads SN, Monahan ZT, Yee DS, Shewmaker FP. The Role of Post-Translational Modifications on Prion-Like Aggregation and Liquid-Phase Separation of FUS. Int J Mol Sci. 2018;19. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.