Abstract
Objective
The effectiveness of bariatric surgery in restoring β-cell function has been described in type-2 diabetes (T2D) patients and animal models for years, whereas the mechanistic underpinnings are largely unknown. The possibility of vertical sleeve gastrectomy (VSG) to rescue far-progressed, clinically-relevant T2D and to promote β-cell recovery has not been investigated on a single-cell level. Nevertheless, characterization of the heterogeneity and functional states of β-cells after VSG is a fundamental step to understand mechanisms of glycaemic recovery and to ultimately develop alternative, less-invasive therapies.
Methods
We performed VSG in late-stage diabetic db/db mice and analyzed the islet transcriptome using single-cell RNA sequencing (scRNA-seq). Immunohistochemical analyses and quantification of β-cell area and proliferation complement our findings from scRNA-seq.
Results
We report that VSG was superior to calorie restriction in late-stage T2D and rapidly restored normoglycaemia in morbidly obese and overt diabetic db/db mice. Single-cell profiling of islets of Langerhans showed that VSG induced distinct, intrinsic changes in the β-cell transcriptome, but not in that of α-, δ-, and PP-cells. VSG triggered fast β-cell redifferentiation and functional improvement within only two weeks of intervention, which is not seen upon calorie restriction. Furthermore, VSG expanded β-cell area by means of redifferentiation and by creating a proliferation competent β-cell state.
Conclusion
Collectively, our study reveals the superiority of VSG in the remission of far-progressed T2D and presents paths of β-cell regeneration and molecular pathways underlying the glycaemic benefits of VSG.
Keywords: Vertical sleeve gastrectomy, Type-2 diabetes, Beta-cell dedifferentiation, Beta-cell redifferentiation, Beta-cell function, scRNA-sequencing
Highlights
-
•
Vertical sleeve gastrectomy (VSG) rapidly improves late-stage diabetes in db/db mice.
-
•
VSG recovers β-cell function and induces intrinsic changes in the β-cell transcriptome.
-
•
VSG improves markers of β-cell dedifferentiation.
-
•
VSG regenerates functional β-cell mass by re-differentiation and proliferation.
1. Introduction
Pancreatic β-cells secrete insulin with precision to regulate glucose uptake by peripheral tissues and maintain normoglycaemia. Obesity-linked T2D is characterized by a condition of insulin resistance and progressive β-cell failure. The latter results from an overall decline of functional β-cell mass while compensating for metabolic demand and insulin resistance [1,2]. Reportedly, functional loss of β-cell mass seems to be a consequence of glucolipotoxicity and oxidative, inflammatory and ER stress [3]. Among different mechanisms proposed for β-cell failure, β-cell dedifferentiation has been identified as a pathogenic mechanism in rodent and human forms of T2D. In this process, β-cells lose their identity and acquire a characteristic signature distinct from functional β-cells that includes the induction of Aldehyde-dehydrogenase 1 A3 (Aldh1a3) [[4], [5], [6], [7], [8]]. The exact causes of the loss of β-cell identity are not fully understood and remain an active and promising area of investigation, especially with respect to exploring therapeutic interventions aimed at inducing β-cell redifferentiation to a functional state. In fact, there are reports showing that the dedifferentiated state of β-cells can be partly reversed by strict dietary intervention [9,10] and combined insulin and glucagon-like peptide-1 (GLP-1)/estrogen conjugate-mediated targeted delivery to β-cells [11]. Redifferentiation, however, cannot be accomplished by common glucose-lowering agents [10]. Classic insulin substitutive therapies may involve adverse complications, including life-threatening hypoglycaemia, and are often insufficient for controlling glycaemia in advanced stages of T2D, whereas alternative glucose-regulating pharmacotherapies are currently limited to a few anti-diabetic pharmaceuticals of varying effect [[12], [13], [14]]. Bariatric surgery has risen in reputation for its lasting weight-lowering effects in severely obese patients as well as achieving high T2D remission rates [[15], [16], [17]]. Though it was initially attributed to solely calorie-restrictive mechanisms, it has become clear during the last decade that the profound benefits of bariatric surgery can most likely be explained by the combined actions of a multitude of physiological adaptations [18,19]. Strikingly, studies of human and animal models show that glycaemic control is rapidly improved after bariatric surgery and prior to its primary effect on weight loss [[20], [21], [22], [23], [24]], suggesting that factors independent of decreased obesity cause changes in glucose regulation and diabetes remission. VSG is the most commonly performed bariatric surgical procedure [25] and exerts remarkable metabolic effects, including islet intrinsic effects and improved peripheral insulin sensitivity [21]. VSG has recently been shown to enhance β-cell function independent of body weight, paralleled by marked intrinsic adaptations in the islet transcriptome in pre-diabetic high fat diet (HFD)-fed mice [20], and improve β-cell diabetes markers in early diabetic (8- to 10-week-old and 4-week-old) db/db mice [21,26]. The exact molecular complement of T2D after VSG is not yet fully resolved. Current literature has mainly focused on models resembling early insulin-compensatory stages of T2D, such as diet-induced obesity (DIO) models and young db/db mice. However, studies investigating the glucoregulatory mechanisms of VSG in models mirroring clinical features of extreme obesity and severe T2D are still missing. Db/db mice have an inbuilt adaptive flexibility that enables them to adjust insulin production rates and β-cell mass relative to the metabolic demand in early stages [27,28] while showing progressive β-cell failure and dedifferentiation [10,29] and a decline in proliferative capacity and β-cell mass after 12 weeks of age [28]. Here, we employed 16- to 18-week-old db/db mice that mirror clinical features of obesity-linked T2D [30] to investigate whether VSG is able to reverse far-progressed T2D. Unlike in previous studies, we took advantage of single-cell transcriptomics to investigate 1) how VSG induces β-cell–intrinsic glucoregulatory mechanisms for diabetes remission, 2) whether VSG affects heterogeneous β-cell states and 3) whether VSG supports β-cell redifferentiation to a functional glucose-sensing state.
2. Results
2.1. VSG restores glycaemia and improves β-cell function in overtly diabetic db/db mice
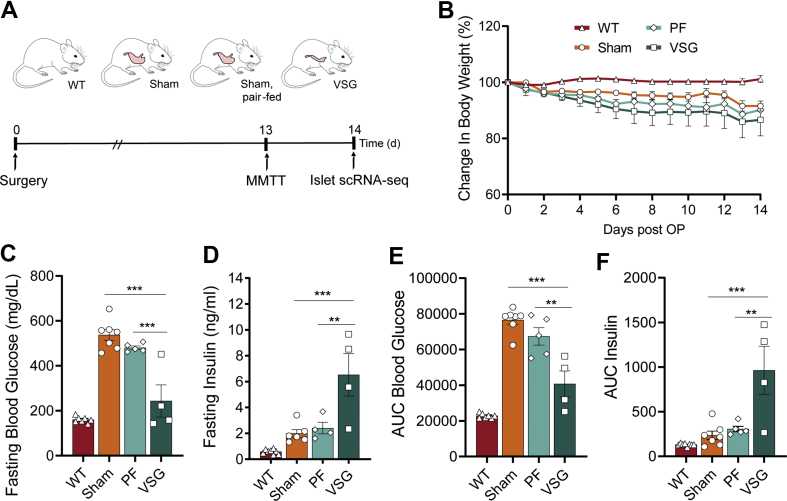
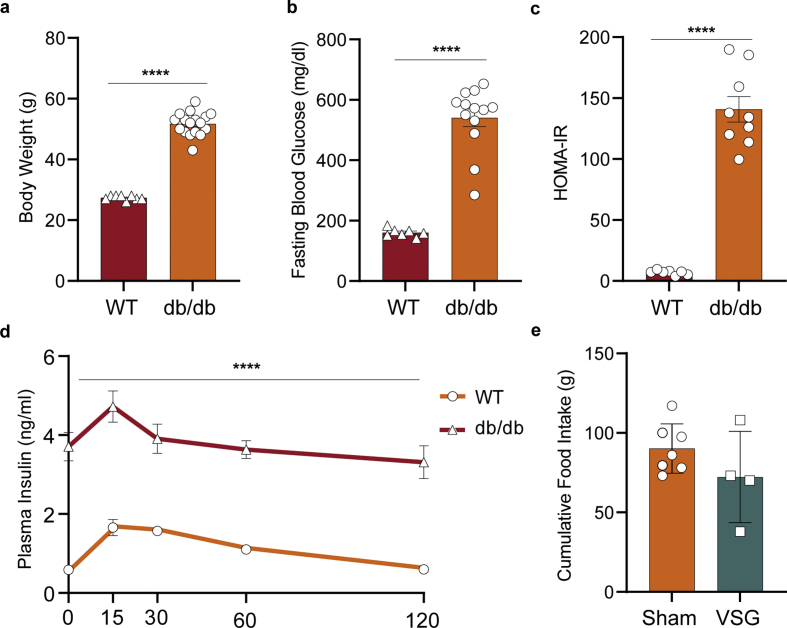
Preoperatively, db/db mice presented a strong obese-diabetic phenotype with insulin resistance. Mice achieved an average body weight (BW) of 51.7 g and fasting glycaemia of 540 mg/dL while lacking a regulated insulin response during a mixed-meal tolerance test (MMTT, Figure S1). Mice of this cohort were homogeneously allocated to VSG and sham surgery groups. An additional sham-operated, pair-fed (PF) group was calorically restricted to match the postoperative food intake of the VSG group (Figure 1A). Previous studies showed that VSG both improved glucoregulation in pre-diabetes HFD models [20,23] and reversed glycaemia in early-onset T2-diabetic db/db mice [21], while PF accomplished the former but only partly succeeded in the latter. However, the effectiveness of VSG in overt T2D mice, in which regenerative capacity declines, remains to be addressed. To test the effects of VSG in 18- to 20-week-old db/db mice on glucose regulation dependently of gastrointestinal functions, VSG and control mice underwent an MMTT. Despite similar weight profiles and food intake in all groups, arising from genetically-driven hyperphagia in db/db mice, glycaemic control markedly improved from VSG within just two weeks of intervention (Figure 1B, Figure S1). These findings indicate that VSG is capable of rapidly reversing long-term T2D independently of body weight in aged db/db mice, similar to reports in early T2-diabetic (8- to 10-week-old) db/db mice [21]. We show that VSG rescued fasting glycaemia almost to the levels of WT mice, increased fasting insulin levels, and improved glycaemia during an MMTT in comparison to Sham and PF (Figure 1C–E). VSG mice secreted 3-fold more insulin upon oral application of a liquid diet than Sham and PF (Figure 1F). This suggests that VSG is most likely able to restore glycaemic control by increasing basal and postprandial insulin secretion. Interestingly, unlike in pre-diabetes models, PF glucose and insulin levels did not differ from Sham, suggesting that a solely calorie restriction–based intervention is no longer sufficient to restore glycaemic control after prolonged and severe T2D.
Figure 1.
VSG restores glycaemia and improves β-cell function in overtly diabetic db/db mice. A, Schematic of single-cell experiment. 16- to 18-week-old db/db mice were subjected to VSG, sham surgery (Sham) and sham surgery with subsequent pair-feeding. Animals were challenged with a mixed-meal tolerance test (MMTT) two weeks prior to surgery and one day before the end of experiment at day 14, followed by islet scRNA-seq (age 18–20 weeks). B, Postsurgical changes in BW (WT, n = 8; Sham, n = 6, PF, n = 6; VSG, n = 4; two-way ANOVA with Bonferroni's post hoc test; differences in VSG versus Sham and VSG versus PF were not statistically significant; VSG versus WT (d4, p = 0.099; d5, p = 0.0045; d6-d14, p < 0.0001)). C, Fasting blood glucose (WT, n = 7; Sham, n = 7, PF, n = 5; VSG, n = 4; one-way ANOVA with Bonferroni's post hoc test; WT versus Sham (p < 0.0001), WT versus PF (p < 0.0001), Sham versus VSG (p < 0.0001), PF versus VSG (p = 0.0003)) and D, fasting insulin levels (WT, n = 8; Sham, n = 6; PF, n = 4; VSG, n = 4; one-way ANOVA with Bonferroni's post hoc test; WT versus VSG (p < 0.0001), Sham versus VSG (p = 0.0008), PF versus VSG (p = 0.0047)). E, Area under the curve (AUC) of blood glucose levels during an MMTT (WT, n = 7; Sham, n = 7, PF, n = 5; VSG, n = 4; one-way ANOVA with Bonferroni's post hoc test; Sham versus VSG (p < 0.0001); PF versus VSG (p = 0.0011) F, AUC of insulin levels during an MMTT (WT, n = 8; Sham, n = 7, PF, n = 5; VSG, n = 4; one-way ANOVA with Bonferroni's post hoc test; Sham versus VSG (p = 0.0002); PF versus VSG (p = 0.0016). All data are presented as mean ± s.e.m.
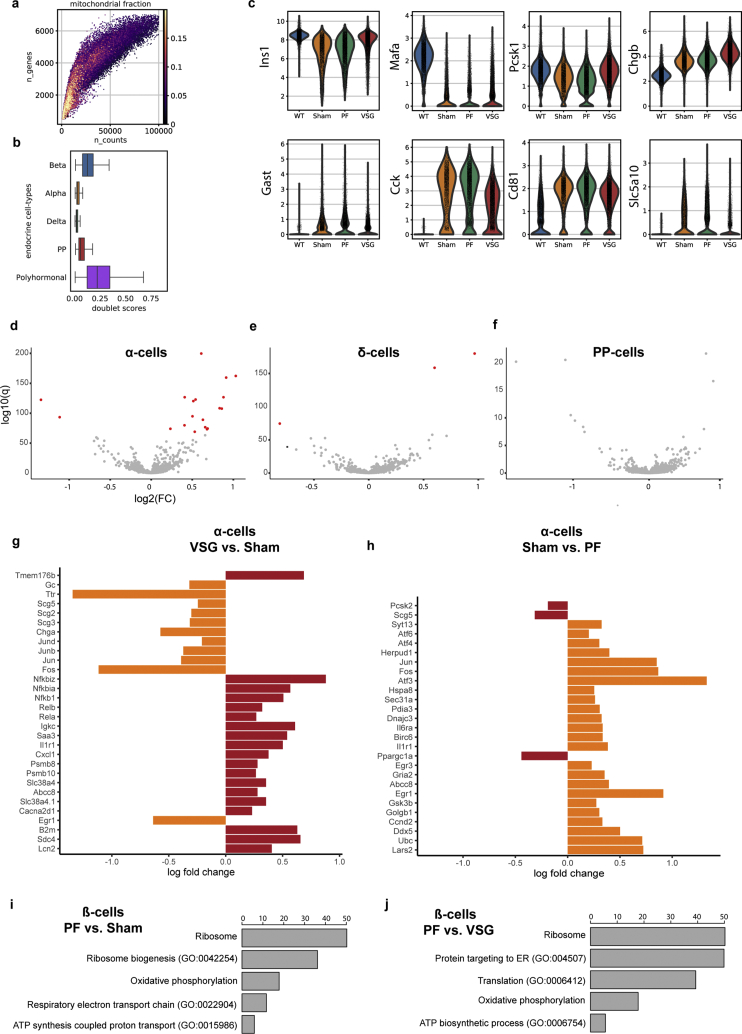
2.2. Islets of extreme obese-diabetic mice show massive β-cell dedifferentiation
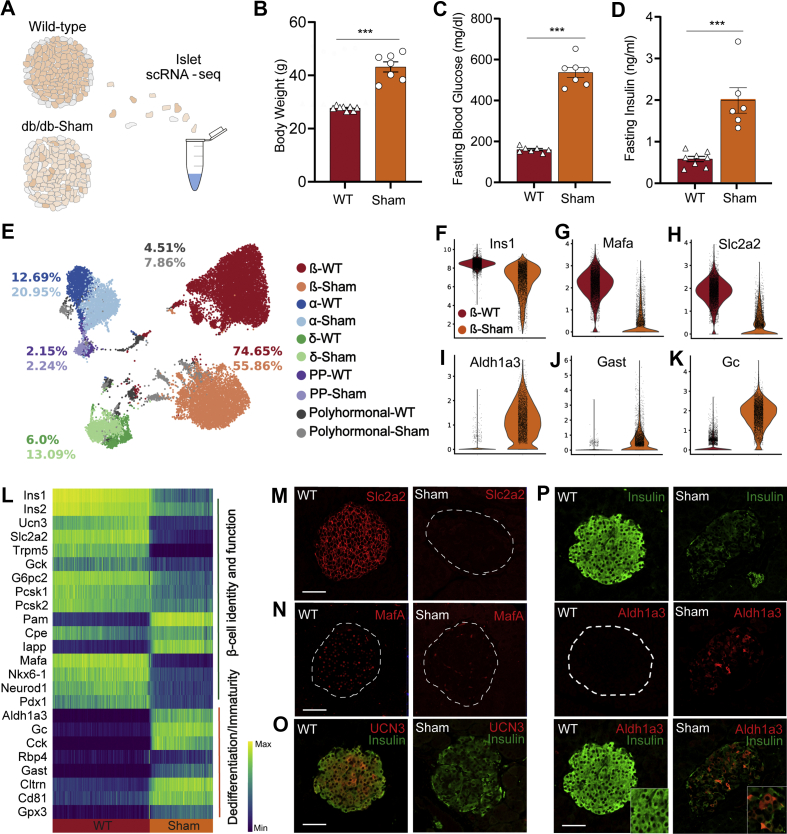
While the metabolic phenotype of the db/db mouse is well described [28], there are no reports on how islets and β-cells respond to sustained T2D. To this end, we used single-cell RNA sequencing (scRNA-seq) to analyze the islet transcriptomes of 18- to 20-week-old wild-type (WT) (n = 2) and sham-operated db/db mice (n = 2) (Figure 2A). Sham mice had developed a severe state of obesity (Figure 2B) and showed markedly enhanced levels of fasting blood glucose (Figure 2C) and fasting insulin (Figure 2D), permitting the investigation of the underlying molecular mechanisms of islet and β-cell dysfunction in an overt T2D model.
Figure 2.
Islets of extreme obese-diabetic mice reveal massive β-cell dedifferentiation. A, Experimental design of islet scRNA-seq of wildtype and 18- to 20-week-old Sham db/db mice. B, Sham mice showing markedly enhanced body weight (WT, n = 8; Sham, n = 7; p < 0.0001) C, fasting insulin (WT, n = 8; Sham, n = 6; p = 0.0002) and D, fasting blood glucose levels (WT, n = 7; Sham, n = 7; p < 0.0001). Data was analyzed by unpaired student's t-tests. E, UMAP plot of 19,878 endocrine cells from WT and Sham mice. Colours indicate the five main endocrine cell types, and colour tone differentiates between WT (darker tones) and Sham (lighter tones). Numbers highlight the proportion of each cell cluster in WT or Sham, respectively. F–K, Violin plots indicating gene expression of selected β-cell maturity (Ins1, Mafa, Slc2a2) and dedifferentiation (Aldh1a3, Gast, Gc) markers, highlighting key differences between WT and Sham β-cells. Violin plots show the distribution of expression as a kernel density estimate (kde) fit. Points within the violin indicate individual cell points. L, Changes in gene expression of major markers of β-cell identity, function, dedifferentiation and immaturity along a diffusion trajectory path from WT to Sham. Cells are ordered using a random-walk–based distance metric computed using diffusion pseudotime (DPT). A starting point or root cell is randomly selected within the WT β-cells and cell transition from the origin is studied thereafter. Expression shown is the running average along the inferred trajectory, scaled to the maximum observed level per gene. Smaller clusters were removed to ensure smooth transition. M–P, Immunohistochemical analysis of Slc2a2, MafA, UCN3, Insulin and Aldh1a3 in β-cells of WT and Sham mice at study end. Scale is 50 μm. All data are presented as mean ± s.e.m.
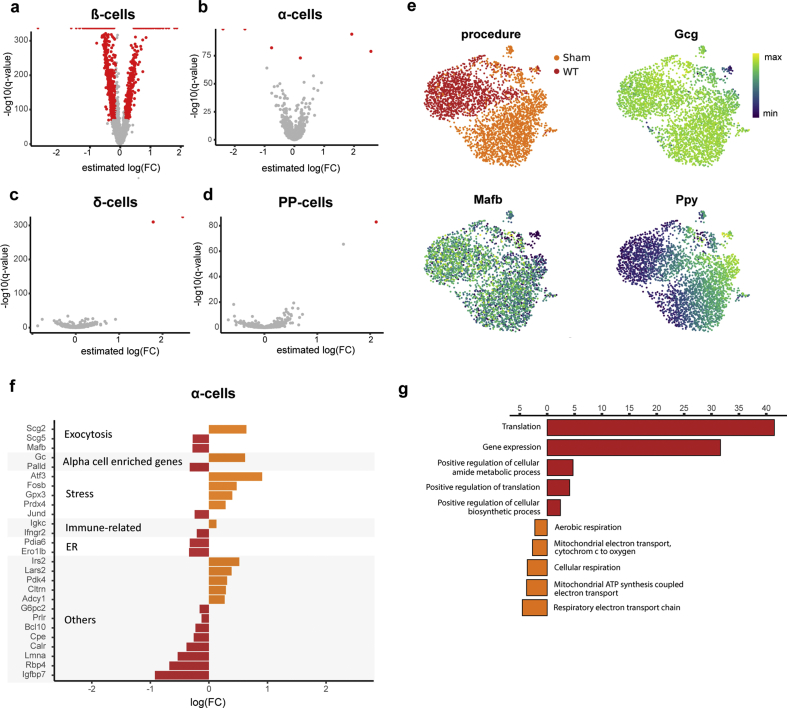
In order to generate a highly resolved profile of pancreatic islets upon overt diabetes, we analyzed 19,878 endocrine cells (expressing Chromogranin A, Chga) using scRNA-seq (WT: n = 2, 10,725 cells; db/db: n = 2, 9153 cells). Single cells were clustered and visualized based on their transcriptomic similarity using an unbiased graph-based approach. Specifically, we used Uniform Manifold Approximation and Projection [31] (UMAP) and Louvain clustering [32] to assign cells into five main endocrine populations: α-cells, β-cells, δ-cells, pancreatic polypeptide (PP) cells and polyhormonal cells. Clusters were manually annotated based on the predominant expression of specific marker genes, namely, glucagon (Gcg), insulin (Ins1), somatostatin (Sst) and pancreatic polypeptide (Ppy) or a combination of hormones that defined polyhormonal cells. Our scRNA-seq data revealed an altered cell composition in diabetic islets as well as intrinsic changes in the gene expression profiles of endocrine clusters (specifically β-cells), indicated by a shift in cell cluster location in the UMAP space. Indeed, the Sham mice showed a proportional decline in the percentage of β-cells, from 74.65% in WT to 55.86%, while α-, δ-, PP- and polyhormonal cells increased compared to WT (Figure 2E). Differential gene expression analysis in β-, α-, δ-, and PP-cells revealed that Sham-derived β-cells were transcriptionally more strongly modulated than α-, δ-, and PP-cells in the db/db model (Figure S2, Supplementary Table 5). This suggests that β-cells are more sensitive to the hostile metabolic environment than other islet cell types. Of note, Sham-derived α-cells exclusively co-expressed Ppy but showed fewer differentially-expressed genes, including adaptive response genes [[33], [34], [35], [36]] (Figure S2). Gene Ontology (GO) analysis revealed an upregulation of the translation machinery and downregulation of cellular respiration in Sham-derived diabetic α-cells (Figure S2). Because of these findings, we next sought to transcriptionally characterize the identity and functionality of β-cells. Interestingly, Sham-derived β-cells showed reduced expression levels of β-cell identity (Insulin-1, Ins1; transcription factor Mafa [37], Mafa; Pancreatic and duodenal homeobox 1, Pdx1; Urocortin 3 [38], Ucn3; Neuronal differentiation 1 [39], Neurod1) and β-cell functional markers (Solute carrier family 2, Slc2a2, also known as Glut2; Transient receptor potential cation channel subfamily M member 5, Trpm5; Proprotein convertase 1, Pcsk1; Figure 2F–H and L). β-cell failure in T2D is characterized by gradual loss of identity and function, which involves β-cell dedifferentiation in both rodents [4,40] and humans [7]. Aldh1a3 is the most prominent and established marker of β-cell dedifferentiation during β-cell failure [40], found to be robustly increased in the majority of db/db Sham-derived β-cells. Other markers of β-cell dedifferentiation, including Gastrin [41,42] (Gast), Vitamin D binding protein [43] (Gc) and Cluster of differentiation 8144 (Cd81), Collectrin [11] (Cltrn) and Glutathione peroxidase 3 [11] (Gpx3) were similarly increased in db/db β-cells (Figure 2I-L). The changes observed in our scRNA-seq data were further corroborated by immunohistochemistry (Figure 2M–P). Consequently, our data imply a transcriptional reprogramming of β-cells during disease progression in db/db mice involving the loss of identity and key functional markers and a progressive increase in characteristic β-cell dedifferentiation markers. Collectively, we underscore the prominence of the db/db mouse to better understand the process of β-cell failure and thereby provide an opportunity to examine β-cell protective and regenerative mechanisms through therapeutic interventions.
2.3. VSG distinctly improves β-cell identity and function
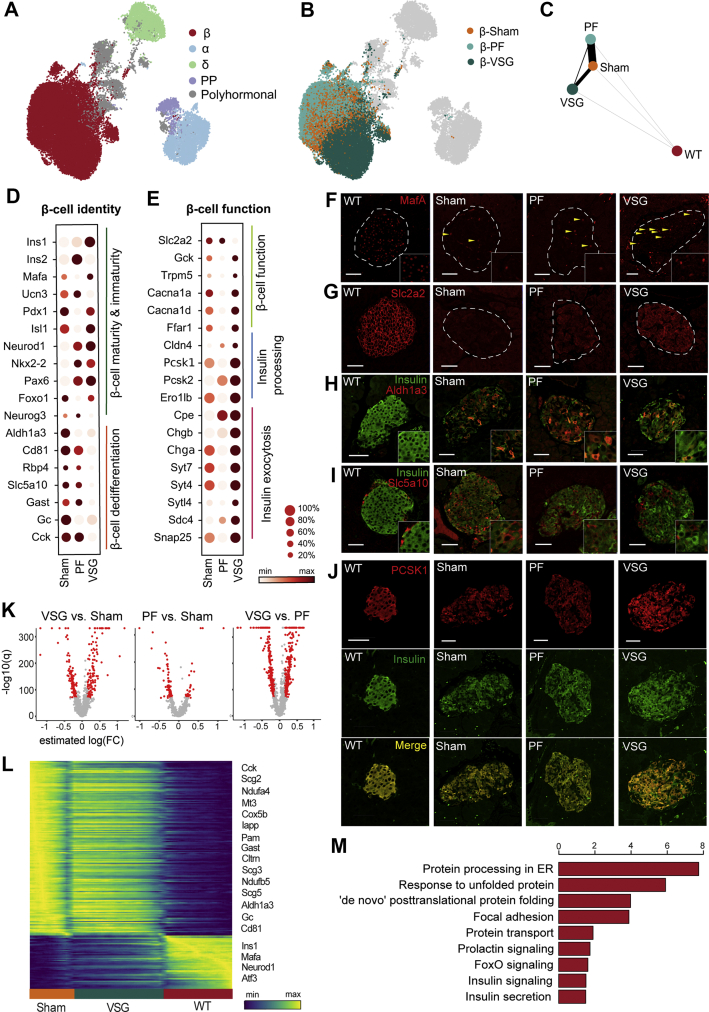
In order to gain insights into the β-cell mechanisms governing the fast recovery of glycaemia by VSG, we analyzed the single-cell transcriptomes of Sham, PF controls and VSG islets. Prior studies have either focused on pre-diabetes models and used bulk islet transcriptomics [20] or early stage T2D models [21,26] addressing selected β-cell markers. However, the impact of VSG on islets has not been dealt with at a single-cell resolution to address cell type and cell state–specific gene expression profiles after VSG. To fill this knowledge gap, we profiled 35,941 single pancreatic endocrine cells from Sham, PF and VSG groups (Sham: n = 2, 9153 cells; PF: n = 1, 13,184 cells; VSG: n = 2, 13,604 cells) 14 days post-surgery, which formed five major clusters corresponding to distinct cell-types (α-cells, β-cells, δ-cells, PP-cells and polyhormonal cells) based on canonical marker expression (Figure 3A). β-cells revealed a large number of differentially regulated genes, while α-, δ- and PP-cells showed fewer transcriptional differences after VSG when compared to Sham, with only a small portion of genes showing differential expression (Figure S3, Supplementary Table 5). However, it should be noted here that scRNA-seq is less sensitive in capturing differences in small cell clusters, such as α-, δ- or PP-cells. Collectively, our data not only captures the fact that β-cells are mostly affected in T2D (Figure 2), but also that β-cells are modulated by VSG intervention and may likely play a crucial role in the islet intrinsic glucoregulatory mechanisms of VSG. In order to unravel specific mRNA changes underlying VSG-mediated glycaemic recovery, we then compared the β-cell transcriptome of VSG with that of diabetic Sham and PF controls. β-cells from the VSG group clustered separately from those of Sham and PF groups in the UMAP space, emphasizing that VSG invokes distinct transcriptional changes that may underlie the recovery of β-cells. The PF β-cells clustered more closely with Sham β-cells, suggesting shared transcriptional programs and divergent effects of PF and VSG on the β-cell transcriptome (Figure 3B). These observations were further supported by Partition-based graph abstraction (PAGA) analysis, which is based on a statistical framework that measures the connectivity and relatedness of single-cell clusters [45]. The nodes of the abstracted graph represent β-cell clusters from the four distinct groups, with the edges implicating the expected trajectory. PAGA analysis highlighted a strong connectivity between Sham- and PF-derived β-cells and a weaker connectivity between VSG and Sham- or PF-derived β-cells (Figure 3C). These findings suggest that VSG induces a distinct β-cell signature compared to Sham and PF controls, while PF intervention does not induce great alterations in the β-cell transcriptome. Consequently, we charted expression profiles of key β-cell identity and function markers and found that VSG rapidly improved the expression of β-cell transcription factors and maturation markers (Ins1, Mafa, Pdx1, Isl1, Nkx2-2, Pax6, Foxo1 and Neurod1) within only two weeks of intervention. Ucn3 (maturation marker) expression was unaffected by VSG, suggesting that long-term intervention may be required. Concomitantly, the VSG-derived β-cells had a significantly decreased expression of dedifferentiation markers Aldh1a3, Cd81, Retinol binding protein 4 [46] (Rbp4), Sodium/glucose cotransporter 5 [11] (Slc5a10), Gast, Gc and Cholecystokinin [11] (Cck, Figure 3D). Immunohistochemical stainings corroborated that the transcriptional effects observed after VSG majorly translated to protein levels (Figure 3F, H and I). Genes identified as playing a crucial role in glucose sensing, such as Glucokinase (Gck), Trpm5, Calcium voltage gated channel subunit alpha 1 A (Cacna1a) and Free fatty acid receptor 1 (Ffar1), were robustly increased in VSG (Figure 3E). The major glucose sensor in β-cells, Slc2a2, was found to be partly restored by VSG (shown by immunofluorescence) but was surprisingly not mirrored at the mRNA-level (Figure 3G), potentially owing to dietary regulation of Slc2a2 protein levels [47,48]. The critical gene for proinsulin processing Pcsk1 was upregulated at both the mRNA and protein expression levels (Figure 3E and J). Additional insulin processing genes, such as Proprotein convertase 2 (Pcsk2) and Carboxypeptidase E (Cpe), were also expressed at higher levels in VSG. Genes involved in insulin secretion, such as Chromogranin B [49] (Chgb), Synaptotagmin 7 [50] (Syt7) and Synaptosomal-associated protein 25 kDa [51] (Snap25), were significantly upregulated in VSG (Figure 3E). To assess gene expression changes with regard to WT, a selection of genes from all groups is shown as violin plots (Figure S3). In line with the lack of metabolic effects of PF, the β-cell signature was only mildly altered. PF did not greatly impact β-cell identity, dedifferentiation and functionality markers (Figure 3D and E) and enrichment of upregulated genes in PF compared to Sham, and VSG solely revealed terms related to ribosomal and mitochondrial functions (Figure S3).
Figure 3.
VSG distinctly improves the identity and function of β-cells. A,B, UMAP plots of 35,941 endocrine cells from Sham, VSG and PF. Colours in (a) indicate the five main endocrine cell clusters and in (b) distinguish β-cells specifically according to the three groups. C, Abstracted graph of cluster connectivity assessing transcriptomic similarity between VSG-, PF-, Sham- and WT-derived β-cells, inferred using PAGA. PAGA is based on a statistical method that measures relatedness between single-cell clusters, with edge weights indicating link significance. D,E, Gene expression for key genes involved in β-cell identity and function changed upon VSG. (d) Dot plot showing that VSG-derived β-cells upregulated essential maturity markers and downregulated dedifferentiation markers. (e) Dot plot showing genes critical for β-cell function and metabolism, insulin processing and exocytosis. Color intensity shows normalized mean expression in a group; dot size indicates the percentage of cells that express the corresponding gene. Expression is scaled per gene. F–J, Immunohistochemical analysis of MafA, Slc2a2, Insulin, Aldh1a3, Slc5a10 and Pcsk1 in β-cells of WT, Sham, PF and VSG mice at study end. Scale is 50 μm K, Volcano plots indicating differences in the number of genes that were differentially expressed when comparing β-cells from Sham and VSG versus β-cells from Sham and PF, obtained from differential analysis testing using limma-trend. Red dots denote significantly regulated genes thresholded on significance levels (absolute estimated log (fold change (FC))>0.15 and B > 150). p-values were corrected for multiple testing using BH. L, Heatmap showing expression changes in genes that were differentially regulated across Sham, VSG and WT β-cells. As a first step, a trajectory of β-cell dedifferentiation upon treatment was estimated using diffusion pseudotime (DPT) and randomly assigning a root cell within the dedifferentiated cells. β-cells were further grouped according to Sham, VSG and WT and gene expression trends were visualized along this one-dimensional axis. Y-axis shows normalised gene expression as the running average along the inferred trajectory, scaled to the maximum observed level per gene. M, Enrichment analysis of GO terms and KEGG pathways of upregulated genes at estimated log (FC) > 0.15 and B > 150 in β-cells between VSG and PF. Method limma-trend was used to identify differentially expressed genes and gene-set enrichment was done with EnrichR webtool using Fisher's exact test to find enriched terms and pathways. X-axis indicates –log10 (adjusted p-value).
To further examine the extent to which VSG was able to improve β-cell identity and function, β-cells were first ordered along a trajectory using diffusion pseudotime [52] (DPT), followed by grouping according to Sham, VSG and WT and reconstructing expression changes along PAGA paths for a given set of genes (Supplementary Table 1 and Methods). VSG ameliorated the expression of several prominent identity and functional markers and simultaneously decreased the expression of genes involved in dedifferentiation. A multitude of markers, including Ins1, Mafa, Neurod1, Cck, Gast, Cltrn, Aldh1a3, Gc and Cd81, displayed a gene expression pattern trending towards WT levels (Figure 3L, Supplementary Table 1). This observation is in line with the previous PAGA analysis and shows that VSG, but not PF, inched towards a WT β-cell–like identity. To understand the unique signaling and functional pathways altered by VSG, we analyzed the VSG β-cell transcriptome through GO and the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses of significantly upregulated genes. Interestingly, differential gene expression analysis revealed a larger set of genes being significantly regulated in the VSG group compared to Sham and PF. Furthermore, a high transcriptional similarity was again observed between PF- and Sham-derived β-cells, supporting a discrete β-cell phenotype in the VSG group (Figure 3K). We identified pathways associated with the unfolded protein response (UPR), endoplasmic reticulum protein processing and insulin secretion to be upregulated in VSG versus PF, suggesting that VSG enhances insulin synthesis and pathways that ensure proper folding of insulin (Figure 3M). In summary, our findings reveal that VSG, but not PF, induces significant changes in the β-cell transcriptome shortly after surgery that likely mirror the glycaemic correction and recovery of β-cell function after VSG.
2.4. VSG augments UPR and enhances ERAD to counteract ER stress
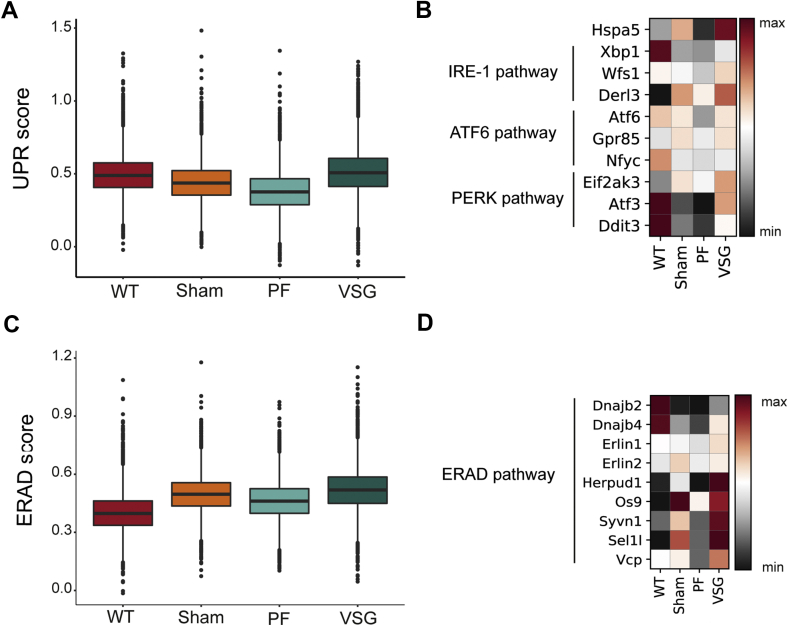
The UPR is a critical adaptive response to endoplasmic reticulum stress (ER stress), restoring cell homeostasis and, when exceeding manageable levels, contributing to loss of β-cell mass in T1D and T2D [53,54]. We scored canonical UPR genes (Supplementary Table 2 and Methods) in β-cells and observed that VSG significantly increased the overall expression score compared to WT, Sham and PF (Figure 4A). Heat shock protein family A member 5 (Hspa5, also known as 78-kDa glucose-regulated protein, GRP78), a major ER chaperone and upstream regulator of the UPR with anti-apoptotic properties [55], was found upregulated on the mRNA level upon VSG. Additionally, VSG increased the expression of numerous other UPR genes, such as X-box binding protein 1 (Xbp1), Wolfram syndrome 1 (Wfs1), Activating transcription factor 6 (Atf6) and Eukaryotic translation initiation factor 2 alpha kinase 3 (Eif2ak3, also known as Perk, Figure 4B), which play a role in β-cell homeostasis [56,57], adaptive β-cell proliferation [58] and insulin processing [59]. ER-associated protein degradation (ERAD) serves to mitigate ER stress and has recently also been attributed an indispensable role in the maintenance of β-cell identity and function [60,61]. Our data reveal that VSG specifically increased the score of characteristic ERAD markers in β-cells (Supplementary Table 2 and Methods) (Figure 4C). In particular, the expression of the major components of the ERAD complex, SEl1L adaptor subunit of ERAD E3 ubiquitin ligase (Sel1l) and Synoviolin 1 [62] (Syvn1, also known as Hrd1), were increased upon VSG (Figure 4D). Additionally, we found the expression of other ERAD-related genes, such as DnaJ Heat Shock Protein Family (Hsp40) Member B2 (Dnajb2), Dnajb4, ER lipid raft associated 1 (Erlin1), Homocysteine-responsive endoplasmic reticulum–resident ubiquitin-like domain member 1 protein (Herpud1) and Osteosarcoma amplified 9 (Os9) increased in the β-cells of VSG-treated mice (Figure 4D). Collectively, our results reveal that VSG enhances adaptive-response mechanisms to ER stress in β-cells, which likely parallels the extreme insulin secretion upon VSG and indicates a VSG-specific protective role of ERAD during ER stress in β-cells.
Figure 4.
VSG augments UPR and enhances ERAD to counteract ER stress. A, Box plots showing the distribution of UPR scores per group, calculated using characteristic UPR markers. Box plots show median (WT = 0.489, Sham = 0.437, PF = 0.376, VSG = 0.508), quartile and whisker values. Points within the box plot indicate individual data points or cells in that corresponding group. VSG showed an increased UPR score compared to WT (p < 0.0001), Sham (p < 0.0001) and PF (p < 0.0001) using Welch's Test (one way) with Bonferroni's post-hoc test. B, Matrix plot showing expression of selected genes involved in the three main signalling branches of the UPR. Color gradient indicates mean z-score scaled gene expression. C, Box plots showing the distribution of ERAD scores per group, calculated using canonical ERAD genes. Box plots show median (WT = 0.398, Sham = 0.497, PF = 0.461, VSG = 0.519), quartile and whisker values. Points within the box plot indicate individual data points or cells in that corresponding group. VSG showed a higher ERAD score compared to WT (p < 0.0001), Sham (p < 0.0001) and PF (p < 0.0001) using Welch's Test (one way) with Bonferroni's post-hoc test. D, Matrix plot showing expression of selected genes involved in ERAD. Color gradient indicates mean z-score scaled gene expression.
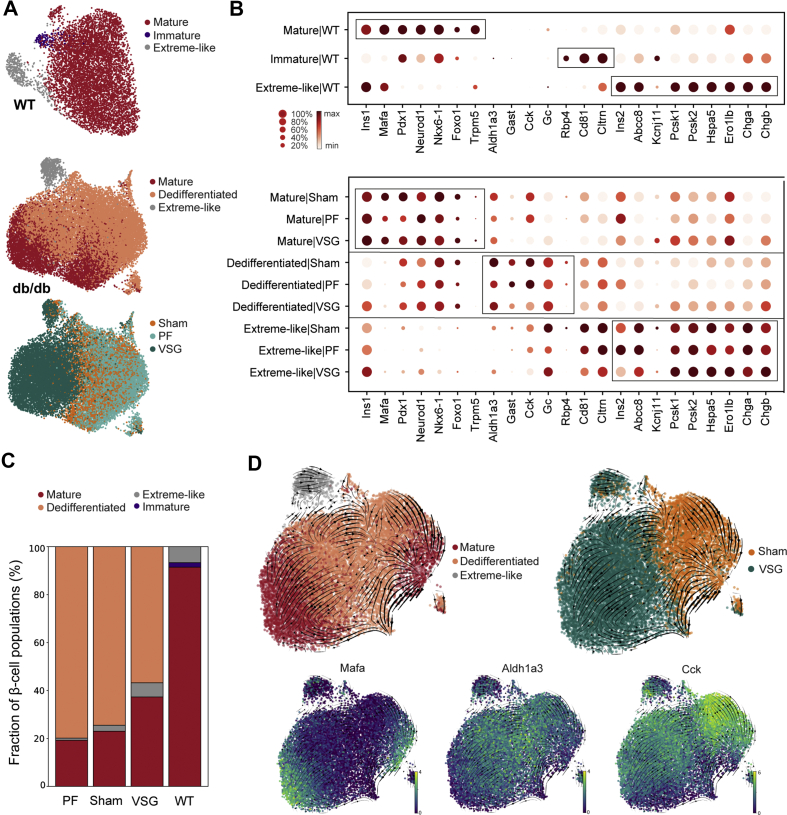
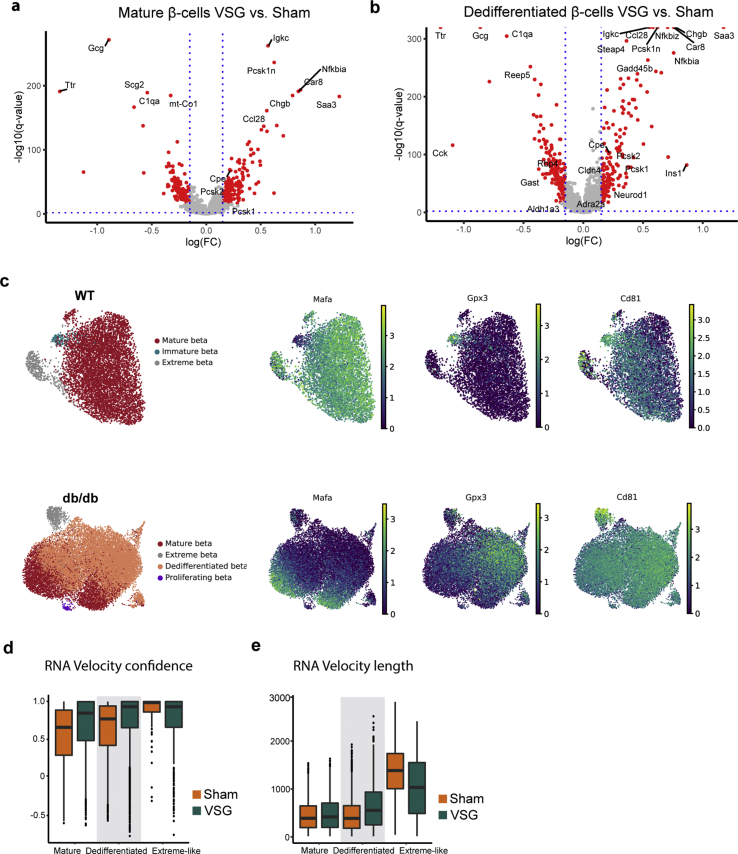
2.5. VSG shifts β-cells towards mature and extreme-like states
β-cell heterogeneity has now been well described in healthy and diabetic conditions as well as during the process of de- and redifferentiation [11,[63], [64], [65], [66], [67], [68], [69], [70], [71]]. However, it has not yet been established how VSG affects β-cell subpopulations in terms of cell composition and cell-specific marker expression. In order to catalogue heterogeneity of β-cell states after VSG, we performed refined clustering on 28,230 β-cells from WT (n = 2), Sham (n = 2), PF (n = 1) and VSG (n = 2). Markers were shown separately for WT- and db/db-derived β-cells (Sham, PF, VSG), as their expression profiles varied significantly. We identified four major β-cell subpopulations and classified them as mature, immature, dedifferentiated and extreme-like (Figure 5A). The mature β-cell subpopulation exhibited the highest expression of β-cell maturation markers and transcription factors (Mafa, Pdx1, Neurod1 and Nkx6-1) as well as functional markers (Ins1, Forkhead box protein O1 (Foxo1) and Trpm5) in all groups (Figure 5B). With the purpose of delineating group-specific proportional shifts in the subpopulations, we estimated the relative fractional change in cell numbers. As anticipated, Sham and PF mice depicted a reduced fraction of mature β-cells (23% and 19%, respectively) compared to WT (91%), while VSG increased the fraction of mature β-cells (37%, Figure 5C). We next performed differential expression testing within mature and dedifferentiated β-cells in order to investigate the cluster-specific transcriptional changes arising upon VSG intervention. Interestingly, we found that the VSG-derived mature β-cells had significantly higher levels of β-cell functionality genes (Pcsk1, Pcsk2, Cpe and Chgb) compared to mature β-cells of Sham, implying that VSG is able to boost the function of mature β-cells (Figure S4, Supplementary Table 5). The molecular identity of immature β-cells is increasingly being investigated. Recently, a scRNA-seq study characterized an immature β-cell subset based on low expression levels of Mafa and high expression levels of Cltrn and Gpx3 [11]. Another study by our laboratory has recently identified CD81 as a marker for immature and dedifferentiated β-cells [44]. In our scRNA-seq dataset, only the WT β-cells presented a clearly distinguishable immature subset (2%) with high expression levels of Cltrn, Gpx3 and Cd81 and low expression of Mafa (Figure 5A and B, Figure S4). In db/db β-cells, immature β-cells could not be distinguished from dedifferentiated β-cells (Figure S4), which highly expressed Aldh1a3 and Cd81 and comprised the majority of the β-cells (Figure 5A–C). The highest proportions of dedifferentiated β-cells were present in Sham and PF (74% and 80% respectively), while levels were decreased in VSG (57%, Figure 5C). Dedifferentiated β-cells were not present in WT. Yet displaying a dedifferentiated signature, VSG improved the transcriptome of dedifferentiated β-cells. Markers of β-cell dedifferentiation (Aldh1a3, Gast, Cck and Rbp4) were reduced while β-cell identity and functionality markers (Ins1, Neurod1, Cpe, Pcsk1, Pcsk2, Adrenoreceptor alpha A [72] (Adra2a), Claudin 4 [73] (Cldn4) and Chgb) were increased in VSG compared to Sham (Figure S4, Supplementary Table 5). In order to investigate the dynamics of mature and dedifferentiated β-cells, we performed an RNA velocity analysis that included VSG and Sham groups [74]. We observed clear transitions from mature β-cells towards a dedifferentiated state in Sham, whereas VSG-derived dedifferentiated β-cells were distinctively directed towards maturity (Figure 5D). These observations of cell movements are indicative of dedifferentiation processes occurring in Sham and promotion of redifferentiation processes by VSG. Furthermore, we computed velocity length and confidence in order to gain insights about the rate of differentiation and coherence of the velocity field, respectively, for each individual cell. Dedifferentiated β-cells from VSG depicted a substantially faster pace of transition as well as higher measure of confidence in RNA velocity compared to Sham, suggesting more active cell profile changes after VSG (Figure S4; Supplementary Table 3). Taken together, our findings 1) indicate that VSG improves function in both mature and dedifferentiated β-cells and 2) reveal a role of VSG in promoting β-cell redifferentiation, implied by a proportional and transcriptional shift from dedifferentiated to more mature β-cells.
Figure 5.
VSG shifts β-cells towards mature and extreme-like states. A, UMAP plot showing refined clustering of 7183 β-cells from WT and 21,047 β-cells from db/db (VSG + PF + Sham) into four main β-cell subpopulations. Colours indicate the subpopulations identified: mature, immature, dedifferentiated and extreme-like β-cells, as well as control and intervention groups. B, Dot plot showing expression changes of key genes involved in β-cell maturity, immaturity, dedifferentiation and extreme β-cell identity in WT β-cell subclusters (top panel) and db/db (VSG + PF + Sham) β-cell subclusters (bottom panel). Color intensity shows normalized mean expression in a cell cluster; dot size indicates the percentage of cells in a cluster that express the corresponding gene. Expression is scaled per gene. C, Proportion of cells per β-cell subpopulation in WT, Sham, PF and VSG. D, RNA velocity estimates of cell transitions applied to Sham and VSG β-cells and direction of cell movement indicated as streamlines projected onto UMAP embedding (top panel). UMAP plots showing expression of key genes (Mafa, Aldh1a3, Cck) to delineate dedifferentiation from mature cells.
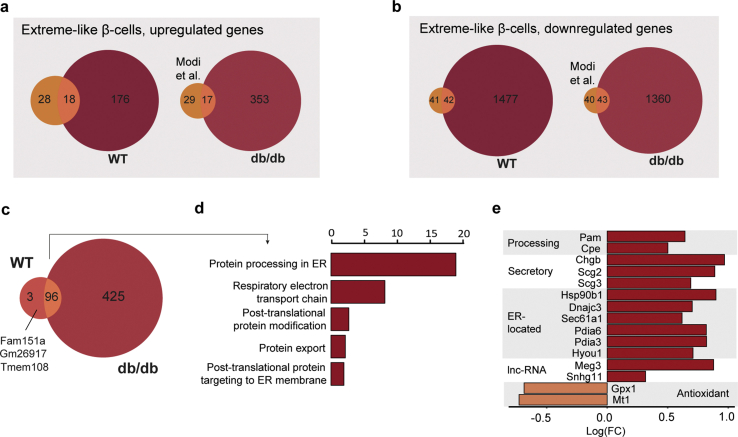
Using single-molecule fluorescence in situ hybridization (smFISH), Farack et al. recently reported a unique β-cell subpopulation termed “extreme” β-cells based on their explicitly high expression of Ins2 and key β-cell function genes with normal to low levels of mature insulin [75]. Thus, extreme β-cells were attributed a specialized role in constant basal insulin secretion, which is necessary throughout the day. Due to the fact that the proportions of extreme β-cells rise in early T2-diabetic db/db mice, extreme β-cells were speculated to counteract constant hyperglycaemia present in T2D [75]. Whether extreme β-cells also play a role in long-term T2D and whether this subpopulation can be targeted by therapeutic interventions remain open questions. In our scRNA-seq dataset, we identified an extreme-like β-cell population in WT and db/db mice based on the smFISH markers described by Farack and colleagues (Ins2, Ins1, Pcsk1, Pcsk2, Chga, Chgb, Abcc8 and Kcnj11) (Figure 5B). Although all markers described by Farack et al. were upregulated in our extreme-like β-cells, not all markers were as pronounced as reported (e.g., Ins1), which is possibly inherent to the different techniques being used (scRNA-seq versus smFISH). Using single-cell transcriptomics, a recent study by the Johnson laboratory revealed a β-cell state exhibiting explicitly high Ins2 that likely reflects the extreme β-cell state described by Farack and colleagues [68]. Corroborating this, we found a great overlap of genes enriched in our extreme-like β-cell population with those reported by the Johnson laboratory (Figure S5, Supplementary Table 4 and Methods). Surprisingly, we also observed that the proportion of extreme-like β-cells was lowered in Sham (3%) and PF (1%) compared to WT (7%) and was restored to WT-level in VSG (6%, Figure 5C). These findings suggest that, unlike in early-diabetic compensatory conditions, extreme-like β-cells are lost in late-stage T2D; however, they can be recovered by VSG intervention. To better understand the functional relevance of extreme-like β-cells, we analyzed overexpressed genes in extreme-like β-cells of WT and db/db mice (Supplementary Table 5). Most genes were common in extreme-like β-cells of WT and db/db mice, indicating that they share functional features (Figure S5). GO and KEGG enrichment analyses revealed terms related to cell respiration, protein processing, ER stress and protein export ()Figure S5. Extreme-like β-cells of db/db mice overexpressed insulin processing and secretory genes as well as genes linked to ER stress and protein synthesis (Figure 5B; Figure S5), supporting the role of extreme-like β-cells in increasing insulin secretion. Collectively, our data suggest that the proportion of extreme-like β-cells changes in number in late-stage diabetes, eventually contributing to T2D progression. Nevertheless, we show that the extreme-like state of β-cells can rapidly be regained within two weeks of VSG intervention and thus might play a role in glycaemic normalization post-VSG.
2.6. VSG rapidly expands β-cell mass
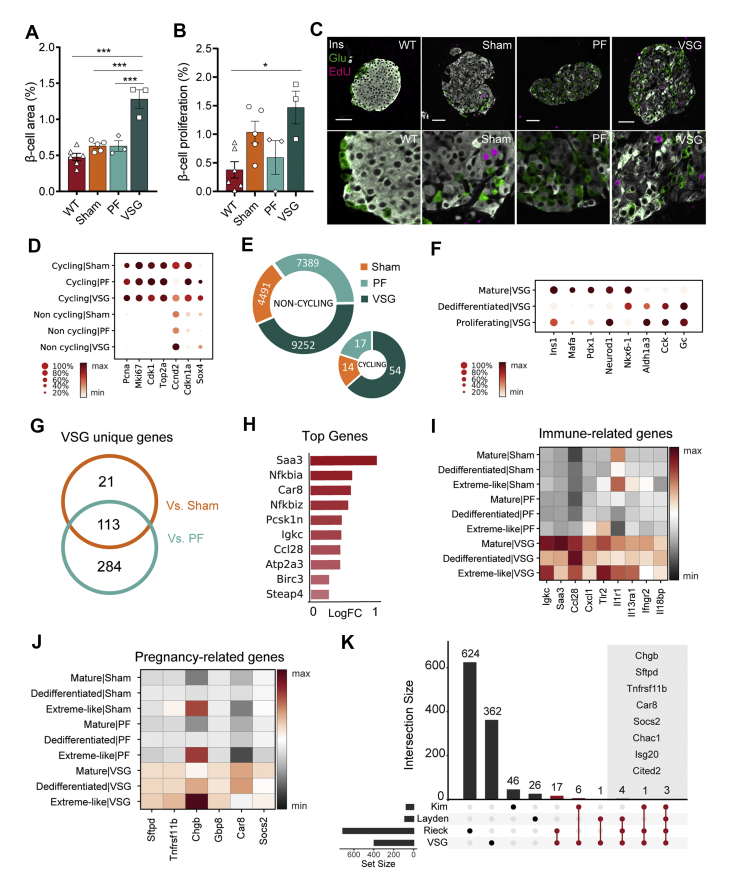
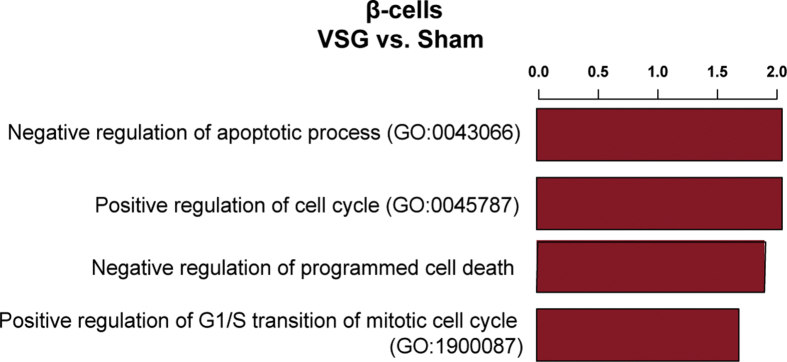
Loss and dysfunction of β-cells are the primary determinants of diabetes pathophysiology. Based on the findings from the MMTT, VSG intervention greatly restores glycaemic control. To understand the source of increased insulin secretion, we assessed islet mass and morphology quantitatively. VSG rapidly triggered a pronounced expansion of β-cell area within two weeks of intervention, compared to all other groups (Figure 6A). We reasoned this change to not only correlate with β-cell redifferentiation, but also expansion of dedifferentiated β-cells. A previous report shows that db/db mice increase β-cell proliferation in the early and intermediate stages of the disease to compensate for the rising metabolic demand, whereas the proliferation rate declines in late-stage diabetic db/db mice instead [28]. Here, we detected a modest, albeit statistically non-significant, increase in β-cell proliferation in VSG compared to Sham and PF (Figure 6B and C). Notwithstanding, we cannot rule out the possibility that the major β-cell proliferative activity occurred during the first week of VSG, since we labelled the proliferating cells in mice 72 h before sacrifice. Supporting the proliferative β-cell phenotype, we identified a cycling β-cell population in our scRNA-seq dataset, marked by high expression of cell cycle genes (Mki67, Top2a and Cdk1), which originated from VSG (54/9252, 0.6%) and to a lesser extent from Sham (14/4491, 0.31%) and PF controls (17/7389, 0.23%, Figure 6D and E). Pathway analysis of VSG-versus PF-derived β-cells revealed an enrichment of cell cycle–related terms, such as positive regulation of cell cycle and negative regulation of apoptosis (Figure S6). Interestingly, the non-cycling β-cell populations of VSG showed a higher expression of Cyclin D2 (Ccnd2), the essential cell-cycle progression gene in adult β-cells [76], compared to controls. Additionally, we observed an upregulation of SRY-box transcription factor 4 (Sox4) and concomitant downregulation of Cyclin-dependent kinase inhibitor 1 A (Cdkn1a) in non-cycling β-cells of the VSG group, which has been shown to allow adult β-cell replication [77] (Figure 6D). These findings suggest that VSG could trigger a proliferation-competent β-cell state. We next addressed whether the VSG-derived proliferating β-cell cluster presents a mature or dedifferentiated identity and found that the proliferating β-cells from VSG exhibited dedifferentiation features by expressing the highest levels of Aldh1a3, Cck and Gc. Concomitantly, there was a lower expression of maturity markers (Ins1, Mafa and Pdx1, Figure 6F), suggesting that dedifferentiated β-cells are more likely to enter the cell cycle.
Figure 6.
VSG rapidly expands β-cell mass. A, Pancreas morphometry analysis reveals that VSG doubled the relative β-cell area (WT, n = 6; Sham, n = 5, PF, n = 3; VSG, n = 3; WT versus VSG (p < 0.0001); Sham versus VSG (p < 0.0001), PF vs VSG (p = 0.0003)). B, Quantification of β-cell proliferation at study end (WT, n = 6; Sham, n = 5, PF, n = 3; VSG, n = 3; WT versus VSG (p = 0.019), VSG versus Sham (p > 0.999), VSG versus PF (p = 0.156)). All data are presented as mean ± s.e.m and were analyzed by one-way ANOVA with Bonferroni's post-hoc test. C, Representative images of immuno-stainings against insulin, glucagon and EdU. Scale is 50 μm D, Dot plot showing expression of cell cycle–related genes in cycling and non-cycling β-cells per group. Color intensity shows normalized mean expression in a cell cluster. Expression is scaled per gene. E, Doughnut plots indicating fraction of proliferating β-cells per group (VSG-54/9252, PF-17/7389, Sham-14/4491). F, Dot plot showing expression of β-cell identity genes in mature, dedifferentiated and proliferating β-cells of VSG, implying the dedifferentiated signature of proliferating β-cells. Expression is scaled per gene. G, Venn diagram showing uniquely upregulated β-cell genes in VSG versus control groups. H, Top 10 unique VSG genes (estimated log (FC) > 0.25, B > 150). Log (FC) plotted for VSG versus Sham. I–J, Matrix plots showing expression of (i) immune-related and (j) pregnancy-related genes in β-cells per group resolved per subcluster. Color gradient indicates mean z-score scaled gene expression. K, Upset plot showing the overlap between VSG-specific genes derived from our study and literature curated genes that are differentially expressed in islets during pregnancy (Layden et al. 2010, Rieck et al. 2009, Kim et al. 2010). Upset plot is used to visualize intersection of sets. The set size bar plot on the left indicates the total number of genes populating each of our genesets. The intersection size bar plot shows the total number of genes that overlap between two or more sets. The bars highlighted in red refer to the genes that are uniquely shared among VSG and the participating sets. Limma-trend method was used to find differentially expressed genes for VSG versus Sham and PF at absolute estimated log (FC) > 0.15 and B > 150. Venn diagrams aided in identifying the VSG-regulated genes.
Underscoring the impact of VSG on β-cell mass, we next examined the uniquely regulated genes that could elucidate the potential proliferative phenotype in the β-cell population of VSG mice. We found 418 genes to be upregulated in VSG mice versus control groups (Figure 6G and H), some of which were related to immune function (Figure 6I). Surprisingly, a number of key genes, remarkably upregulated in VSG, are documented to be highly expressed during pregnancy, a condition in which profound adaptive functional and proliferative mechanisms occur in β-cells. These genes included Sftpd, coding for Surfactant-associated protein D (SP-D); Carbonic anhydrase 8 (Car8); Tnfrsf11b, coding for Osteoprotegerin (OPG) and Chgb (Figure 6J). OPG (Tnfrsf11 b) is a soluble decoy receptor of the TNFR superfamily, best known to be induced by lactogens and act as a β-cell mitogen by preventing the interaction of Receptor activator of NF-kB Ligand (RANKL) and RANK (Figure 6J). OPG robustly activates β-cell proliferation under various conditions, such as pregnancy and young, aged and diabetic STZ-treated mice [78,79]. Here, we present OPG for the first time as a possible mediator of β-cell replication in the context of VSG. In order to further outline the pregnancy-associated genes induced by VSG, we compared our β-cell dataset with published islet transcriptome datasets of pregnant mice [[80], [81], [82]] (Figure 6K). We found 32 common genes induced by both VSG and pregnancy. Strikingly, these included most key pregnancy genes (Sftpd, Tnfrsf11b, Car8 and Socs2), immune-modulatory genes (Il13ra1 and Ifngr2) and β-cell identity and functionality genes (Mafa, Pdx1, Ins1 and Chgb).
Taken together, we show that VSG increased the β-cell area within only two weeks and suggest that this increase is attributed to two additive mechanisms, β-cell redifferentiation and proliferation. More precisely, our findings indicate that VSG triggers a proliferation-competent β-cell state characterized by up- and downregulated expression levels of the cell cycle regulator gene Ccnd2 and the cell cycle inhibitor gene Cdkn1a, respectively, and induces adaptive pregnancy-associated factors.
3. Discussion
Bariatric surgeries such as VSG improve glucose control and lead to high rates of T2D remission in patients [16,83]. Despite the high interest in a global understanding of bariatric surgical mechanisms, with the overarching objective of finding novel, less-invasive therapeutic strategies, the exact complement of glucoregulatory mechanisms after VSG remains elusive.
This study investigates the mechanistic role of VSG in restoring glycaemic control after overt T2D, with special regard to β-cell redifferentiation and novel VSG-driving factors in β-cells using single-cell profiling. In contrast to previous studies, we chose a clinically more-relevant model of overt late-stage diabetic 16- to 18-week-old db/db mice. Although the db/db model does not fully recapitulate human obesity-linked diabetes (amongst other reasons, because it relies on a rare Lepr mutation and starts very early in life, before β-cell maturation is fully completed), the db/db model seems to be closer to human β-cell pathophysiology than DIO models [84]. It mirrors features of morbid obesity-linked diabetes, representing a common indication for bariatric surgical interventions, and resembles human T2D β-cell pathophysiology, including β-cell dedifferentiation. VSG outcomes were studied 14 days post-surgery to give sufficient time for recovery but also capture the earliest dynamics where critical metabolic events would take effect. Clinically, preoperative markers that can predict T2D remission upon surgery are constantly subject to investigations [[85], [86], [87], [88]] and raise concerns of the effectiveness of bariatric surgery performed on patients with highly progressed T2D. Strikingly, we demonstrate that VSG is indeed capable of reversing severe T2D within only two weeks of intervention, while a mild calorie-restrictive intervention is insufficient to improve T2D in this model. This was reflected not only on the physiological level, but also the single-cell transcriptome level, as we did not observe relevant differences between the PF and Sham β-cell transcriptomes. This finding expands our knowledge of the superiority of VSG over calorie restriction. In contrast to previous reports showing that calorie restriction is effective in prediabetes models [20,89], our data indicates that PF becomes inefficacious in highly progressed T2D.
The only study that addresses the effects of VSG on islet transcriptome demonstrates a unique regulation by VSG, but could not decipher cell-type specific contributions [20]. To advance our mechanistic understanding of the glycaemic rescue induced by VSG in specific islet cell types, we conducted a comprehensive single-cell transcriptome analysis of the islets of Langerhans. Our data revealed that obesity-driven T2D primarily affected the transcriptome of β-cells and not that of α-, δ- or PP-cells. In particular, Sham-derived β-cells revealed features of extreme dedifferentiation and compromised β-cell identity and function. Likewise, we demonstrate that β-cells majorly benefitted while α-, δ- and PP-cells only benefitted to a low extent from surgical intervention, indicating a superior role of β-cells in the glycaemic rescue by VSG in our model. Furthermore, we suggest that the intrinsic molecular changes in β-cells, to an extent, account for the marked glucoregulatory improvements after VSG intervention. However, we cannot rule out the possibilities that the transcriptional changes reflect secondary outcomes of preceding peripheral metabolic improvements driven by VSG, or that the limited number of VSG-treated mice used for metabolic assessment and scRNA-seq falsely captured physiological outliers. Additionally, our conclusions on β-cell function were made based on transcriptomic data and MMTTs assessing the systemic regulation of glycaemia, including gastro-intestinal functions, instead of pure β-cell function.
Given the remarkable increase in insulin secretion after VSG, possibly driven by extrinsic signals such as GLP-1 and adaptations inherent to β-cells, we reasoned that there must be intrinsic compensatory mechanisms in VSG-derived β-cells that ensure proper insulin synthesis, processing and secretion. Indeed, our data revealed uniquely upregulated levels of insulin secretion and processing machinery as well as UPR, which likely mirrors the parallelly increased ER stress and insulin secretion levels present in VSG. Furthermore, our findings put forward ERAD to mitigate ER stress and maintain β-cell homeostasis in VSG-treated mice.
Talchai et al. [4] demonstrated that the loss of functional β-cells in T2D mainly emerges due to β-cell dedifferentiation and correlates with the increased expression of Aldh1a3 and decreased levels of mature β-cell markers. In fact, a few studies address β-cell dedifferentiation or maturation markers after bariatric surgery. It was shown that two forms of bariatric surgery, Roux-en-Y gastric bypass (RYGB) and VSG, reduce β-cell dedifferentiation in diabetic Goto-Kakizaki (GK) rats [90] or improve β-cell identity markers in young (4 weeks and 8–10 weeks old) diabetic db/db mice [21,26]. However, a complete single-cell mRNA profile of the changes occurring on the β-cell level to dissect the cellular and molecular changes occurring during surgical intervention and the resulting physiological phenotyping remained elusive. VSG triggered fast β-cell recovery to a more functional state, characterized by improved expression profiles of β-cell identity and functionality markers as well as a remarkable decline in dedifferentiation markers. Furthermore, we identified a route of redifferentiation, evidenced by unique cell transitions from the dedifferentiated to a more mature β-cell state using RNA velocity as well as by a higher proportion of mature and lower number of dedifferentiated β-cells. Of note, the β-cell maturity marker Ucn3 did not recover after VSG in our late-stage T2D db/db mice (16–18 weeks). Differences in UCN3 levels likely depend on the stage of T2D as shown in a study by the Ben-Zvi laboratory [21], in which VSG progressively improved UCN3 levels between one week and one month after VSG in early diabetic (8–10 weeks) db/db mice. Nevertheless, our data shows that even in the absence of Ucn3, β-cells recover within two weeks of VSG intervention, which is in line with a study revealing that UCN3 is functionally not required in β-cells [91]. Furthermore, based on the findings by the Ben-Zvi laboratory [21], we speculate that VSG would likely further enhance β-cell redifferentiation on a long–term scale. It has been reported that Gastrin expression is turned on in a subset of β-cells after severe hyperglycaemia [42]. Interestingly, lowering of glycaemia post-VSG decreased the expression of Gast and Cck, indicating that hyperglycaemia drastically compromises the β-cell identity.
Reports on β-cell mass after bariatric surgery are highly inconsistent. Two studies show an increase in β-cell mass upon VSG in GK rats [92] and upon RYGB in pigs [93], whereas others report no change in β-cell mass after RYGB in GK rats [94] or lowered β-cell proliferation in db/db mice [21] and decreased β-cell area in DIO mice [95] upon VSG. In our model of late-stage T2D, we show that VSG increased β-cell area and that this expansion is most likely driven by additive mechanisms of β-cell redifferentiation and proliferation. We found that VSG induces a proliferation-competent β-cell state by unique changes in cell-cycle regulator genes and, surprisingly, an induction of pregnancy-associated factors. Maternal β-cells compensate for physiological insulin resistance by proliferating and expanding during pregnancy to maintain maternal euglycaemia [96]. These adaptations have been reported to be mainly driven by increased levels of prolactin and placental lactogens [[97], [98], [99]] and proven to be distinct from those activated by nutritional and injury stress [80,100]. A study reported that male islets can also acquire a pregnancy-like signature through exposure to prolactin [101]. Indeed, a possible link was provided by a recent study showing that VSG increased blood prolactin levels in male patients [102]. Future research will need to validate this observation and investigate the mechanistic link between VSG and prolactin-levels as well as the exact downstream consequences on β-cells in a setting of VSG. The role of the commonly upregulated genes in β-cells during pregnancy and upon VSG, including Sftpd, Car8 and Tnfrsf11b (OPG), is largely unknown but may be causally linked to compensatory β-cell function and proliferation, both of which are well-known phenomena during pregnancy [80,82,103]. Sftpd has been shown to be overexpressed in newly formed β-cells during the perinatal period and during pregnancy, and has been suggested to confer anti-inflammatory properties to β-cells [81,104]. Car8 expression in adult β-cells was found to be weak, although it is upregulated in mice during pregnancy [82,101]. In contrast, OPG is well known for its mitogenic effects in β-cells under various settings, including during pregnancy and when pharmaceutically administered [78,[105], [106], [107]]. In this study, we present OPG as a potential new β-cell mitogen induced by VSG that could contribute to β-cell expansion and thus to the restoration of glycaemia after VSG. Cytokines present after VSG or increased prolactin levels might be upstream drivers, inducing OPG expression upon VSG [78,106,107]; however, this needs to be conclusively proven in future experiments. Taken together, we provide evidence for the superiority of VSG in a clinically-relevant, morbidly obese–T2D mouse model and give insights into the diverse β-cell mechanisms that underlie VSG-mediated T2D remission. Additionally, this understanding serves as a basis for the potential discovery of novel therapeutic targets, including pregnancy pathways, that might be able to mimic bariatric surgery for T2D therapy.
4. Methods
4.1. Animals
Male Lepr-deficient mice, homozygous for Lepr-db (db/db mice), and corresponding control mice, WT for Lepr (both stock #000642, BKSCg-Dock7m+/+Leprdb/J), were purchased from The Jackson Laboratory (Maine, US) and maintained under standard controlled conditions. Db/db mice were single-housed one week prior to surgery. WT mice were housed in groups of 2–4 per cage. Before surgery, mice were matched for body weight and metabolic parameters and assigned to VSG, Sham and pair-fed (PF) groups for surgery at the age of 16–18 weeks. All animal experiments were carried out in compliance with the German Animal Protection Act and with the approved guidelines of the Society of Laboratory Animals (GV-SOLAS) and the Federation of Laboratory Animal Science Associations (FELASA). This study was approved by the Animal Use and Care Committee of Bavaria, Germany.
4.2. Surgical procedures
VSG and sham surgeries were conducted as described previously [108] (PMID: 23554454). Briefly, VSG surgery included the lateral excision of 70% of the stomach, leaving a tubular gastric remnant in continuity with the esophagus superiorly and the pyloric sphincter and duodenum inferiorly. For VSG sham surgery (Sham), the stomach was isolated, followed by manual application of pressure with blunt forceps at a vertical line, starting at the esophagus superiorly and ending at the duodenum inferiorly. Surgeries were performed on 12-h fasted mice after administration of Buprenorphin (0.1 mg/kg body weight) and isoflurane anaesthesia. WT mice were considered a healthy control group and did not undergo surgery.
4.3. Postoperative care and dietary regimen
Postoperatively, all surgical groups received subcutaneous injections of Meloxicam (1 mg/kg body weight daily for 2–3 days) and warm saline (1 mL daily for 2–3 days). Mice had access to ad libitum Osmolite HiCal liquid diet (Abbott) for 3 days after surgery before being returned to a solid diet on day 4. Henceforth, VSG and Sham mice received ad libitum chow. A subgroup of Sham mice was pair-fed to match the food intake of VSG mice. To this end, PF mice received pre-weighed amounts of chow once daily, calculated based on the mean food consumption by the VSG group during the previous 24 h.
4.4. Mixed meal tolerance test
Mixed-meal tolerance tests (MMTTs) were performed 14 days prior to and 13 days post-surgery. Mice were fasted for 6 h and tail vein blood was collected for basal blood glucose and insulin measurements. Subsequently, mice were gavaged with 200 μL of Osmolite HiCal liquid diet enriched with 20% dextrose. Tail vein blood was collected after 15, 30, 60, and 120 min to assess glucose and circulating insulin levels during the MMTT. Blood glucose was quantified in plasma using mouse glucose assay kit (CrystalChem, #81692). Levels of circulating insulin were assayed in plasma using mouse-specific ELISA kits (CrystalChem, #90080).
4.5. HOMA insulin resistance (HOMA-IR) index
The HOMA-IR index was calculated by multiplying fasting blood glucose (mg/dL) with fasting plasma insulin (μIU/mL) and dividing by 405.
4.6. Proliferative cell labeling in mice
To label proliferative cells, mice were injected with 10 mg/g body weight 5-ethynyl-2′-deoxyuridine (EdU, Thermo, #E−10187) intraperitoneally once a day for 3 consecutive days before the end of experiment.
4.7. Islet isolation and single cell preparation
Pancreata were excised and digested using collagenase P (Roche, #11213857001) followed by centrifugation using OptiPrep™ density gradient medium (Sigma, #D1556). Isolated islets were handpicked under a stereomicroscope and cultured in RPMI 1640 medium (Invitrogen, #21875091) supplemented with 10% FCS (GIBCO) and 1% penicillin-streptomycin (Life Technologies, #15140122) for 1 h at 37 °C. For single cell suspension, islets of different sizes were dissociated by incubation with TrypLE (Life technologies, #12605) for 12–15 min at 37 °C and gentle repeated pipetting. Cells were washed twice with FACS buffer (2% FCS in PBS) before continuing with library generation for single-cell RNA sequencing.
4.8. Single-cell RNA sequencing: RNA preparation, library generation and sequencing
Single-cell samples from islets derived from WT (n = 2), Sham (n = 2), PF (n = 1) and VSG mice (n = 2) were prepared as described above. Number of dead cells was estimated by trypan blue staining. Single-cell libraries were generated using the Chromium™ Single Cell 3′ GEM, Library & Gel Bead Kit v3 (10x Genomics, #1000075) according to the manufacturer's instructions. Briefly, to obtain a target cell number of 10,000 cells per sample, approximately 16,000 cells per sample were loaded onto a single channel of the 10x chip to generate Gel Bead-in-Emulsions (GEMs). These underwent reverse transcription to barcode RNA before cleanup and cDNA amplification. Afterwards, enzymatic fragmentation and attachment of the 5′ adaptor and sample index was performed. Libraries were sequenced on a NovaSeq6000 (Illumina) with 150 bp paired-end sequencing.
4.9. Immunohistochemistry and EdU assay
Pancreata were fixed in 4% (w/v) neutrally buffered formalin and entirely cross-sectioned into 3–5 parallel, equidistant slices per case. Maintaining their orientation, the tissue slices were vertically embedded in paraffin to ensure complete coverage of the entire pancreas, including head, neck, body and tail. Sections of 3-μm thickness were cut. The following primary antibodies were used: Monoclonal rabbit anti-insulin, #3014, Cell Signaling, 1:400; Polyclonal guinea pig anti-insulin, PA1-26938, Thermo Scientific, 1:200, Polyclonal guinea pig anti-glucagon, M182, Takara, 1:2500; Monoclonal mouse anti-GRP78, BD Bioscience, 610,979, 1:200; Polyclonal goat anti-Sel1L, Novus Biologicals, NB100-93463, 1:200; Polyclonal rabbit anti-MafA N-terminal region, ARP47760_P050, Aviva systems biology, 1:200; polyclonal rabbit anti-UCN3, H-019-29, Phoenix Pharmaceuticals, 1:500; Polyclonal goat anti-Slc2a2, ab111117, abcam, 1:300; Polyclonal rabbit anti-Aldh1a3, LS-C498339-100, LSBio, 1:300; Polyclonal rabbit anti-Slc5a10, ab167156, abcam, 1:300 and Monoclonal mouse anti-Pcsk1, sc-100578, Santa Cruz, 1:200. The following secondary antibodies were used: AlexaFluor750-conjugated goat anti-rabbit, A21039, Invitrogen, 1:2500; Donkey anti-rabbit IgG 488, Invitrogen, A21206, 1:800; Donkey anti-rabbit IgG 555, Invitrogen, A31572, 1:800; Biotinylated goat anti-guinea pig, BA7000, Vector, 1:100; Cy3-conjugated streptavidin, SA1010, Invitrogen 1:100; Donkey anti-guinea pig Alexa 488, 706-545-148, Dianova, 1:800; Donkey anti-mouse IgG 555, Invitrogen, A31570, 1:800 and Donkey anti-goat IgG 555, Invitrogen, A21432, 1:800. EdU staining was performed as per manufacturer's protocol (Click-IT EdU Alexa Fluor 647 Imaging Kit, Thermo Fisher Scientific). Nuclei were labelled with Hoechst33342 (H1399, Thermo Fischer) or 4′,6-diamidino-2-phenylindole (DAPI; Life Technologies, 1:500). Slides were mounted with VectaShield and imaged using confocal microscopy (Zeiss LSM Airy Scan).
4.10. Pancreas morphometry and quantification
For pancreas quantification, 6 tissue sections per animal were analyzed. The stained tissue sections were scanned with an AxioScan.Z1 digital slide scanner (Zeiss, Jena, Germany) equipped with a 40× magnification objective. Subsequently, islets were automatically detected (100 islets on average per animal) and insulin-, glucagon- and EdU-positive cells were quantified using the image analysis software Definiens Developer XD2 (Definiens AG, Germany). To evaluate α- and β-cell replication, the respective ratio of glucagon/EdU/Hoechst or insulin/EdU/Hoechst co-positive cells to the total glucagon- or insulin-positive cells were quantified.
4.11. Statistical analysis of metabolic data
The results are expressed as mean ± s.e.m. (Standard Error Mean). For statistical comparison of longitudinal data, two-way ANOVA corrected by Bonferroni's multiple comparison test was used. Any other data were compared by one-way ANOVA corrected by Bonferroni's multiple comparison test or student's t-test. A value of p<0.05% was considered statistically significant. Analyses were performed using GraphPad Prism 9 Software (GraphPad Software, USA).
4.12. Preprocessing of 10x scRNA-seq data
The CellRanger analysis pipeline (v3.1.0) provided by 10x Genomics was used for demultiplexing the binary base call (BCL) files, aligning reads to the mm10 genome build and reading filtering, barcode and unique molecular identifier (UMI) counting to generate a digital gene expression matrix (DGE). The standard cell detection algorithm of CellRanger was used to select high quality barcodes based on the overall distribution of total UMI counts per cell. All downstream analyses, unless stated otherwise, were performed on Python 3.7.7 using the Scanpy package [109] (v1.4.6). Next, the cell-by-gene count matrices for each sample were iteratively concatenated into one merged dataset. Standard preprocessing and quality control (QC) measures were adopted to filter noisy and unreliable cells and genes from the raw data obtained after sequencing. These included filtering cells based on number of detected genes, count depth and fraction of counts originating from mitochondrial genes, which is indicative of potential cellular stress. This ensured that downstream analysis results were obtained only with viable cells and not technical artifacts, apoptotic cells, doublets, etc. On the basis of the joint distribution of these QC metrics, we investigated cells for anomalous behavior, followed by filtering using thresholding decisions. Briefly, genes expressed in fewer than 10 cells were excluded. Additionally, low-quality cells were removed if they 1) showed a high fraction (>20%) of counts due to mitochondrial transcripts, 2) expressed more than 7,000 genes, 3) had more than 100,000 UMI counts and 4) expressed fewer than 500 genes (the argument being that below this threshold, potentially dying cells or empty droplets contaminated by ambient RNA expression would be picked up for analysis). Five non-endocrine clusters, identified using bona fide markers, including ductal cells (expressing Krt19), acinar cells (expressing Prss2), endothelial cells (expressing Plvap), stellate cells (expressing Col1a2) and immune cells (expressing Cd74), were removed from further analysis. Furthermore, we applied Scrublet pipeline [110] (v0.2.1, with manually adjusted parameters, see Code) to mark potential technical artifacts of cell doublets that can arise due to arbitrary co-encapsulation of two cells with differing expressions. Predicted doublet-like cells that co-localized in distinct clusters and co-expressed endocrine and non-endocrine markers were not considered for analysis. Examining the distribution of cell counts and genes and thresholding on outlier peaks, we measured 19,878 endocrine cells from WT and Sham with a median number of transcripts per cell of approximately 3962 and a mean of 33,206 counts per cell. Additionally, we detected 35,941 endocrine cells from Sham, PF and VSG collectively and detected a median number of transcripts per cell of approximately 3621 and a mean of 30,251 counts per cell. Cell filtering was followed by normalization of the read counts using the scaling normalization function computeSumFactors (with min.mean = 1) from scran available through R. This was done to account for cell-specific biases due to differences in sequencing depth and capture efficiency between cells and the values that were subsequently log-transformed. Additionally, highly variable genes (HVGs) to be used as input for dimensionality reduction were identified using function pp.highly_variable_genes from Scanpy with default parameters. 4000 top HVGs were retained for use in principal component analysis (PCA), followed by clustering and visualization via UMAP.
4.13. Dimensionality reduction, clustering and cell-type annotation
The single-cell RNA-seq data was analyzed for transcriptomic similarities between cells in two-dimensional embedding by computing a k-nearest-neighborhood (KNN) graph using the function pp.neighbors from Scanpy. Prior to this, 50 principal components were computed with principal component analysis (PCA) on the list of 4000 highly variable genes using the function pp.pca. The size of the local neighborhood for the KNN graph was set to 30 and the top 30 principal components were used. This was followed by computation of a Uniform Manifold Approximation and Projection (UMAP) embedding (v0.4.0) and Louvain-based clustering (louvain: v0.6.1, igraph: v0.8.0) using Scanpy functions tl.umap and tl.louvain, respectively. Important to note here is that no fixed resolution parameter was used; instead, Louvain clustering was repeated at different resolutions within subclusters to account for specific differences in the data manifold. Additionally, each UMAP visualization in the study was plotted after re-computing the KNN graph and UMAP on the cell subset under study using the first 30 principal components and the highly variable genes as initially determined. Significant endocrine clusters were identified on the basis of mRNA expression of well-known canonical markers, mainly, Ins1, Gcg, Sst and Ppy. We also identified cells expressing a combination of hormones that could be indicative of polyhormonal cells or potential doublets. As studying cells expressing multiple hormones was beyond the scope of the current study, we did not further resolve the identification of polyhormonal singlets from doublet-like islet clusters. For the identification of β-cell subpopulations, we computed a new KNN graph followed by Louvain clustering at varied resolutions using a combination of literature-based gene sets, as well as enriched gene ontologies and pathways using the web interface EnrichR [111]. The gene expression profile of each cluster was compared to the profiles of all other cells using a variance-overestimated t-test, implemented through Scanpy using tl.rank_genes_groups, in order to identify key marker genes for each cluster that additionally assisted in annotating the more unconventional and transient β-cell–states.
4.14. Differential expression analyses between treatment interventions and subpopulations
In order to explore the divergent transcriptional programs in β-cells accompanying VSG, differentially expressed genes were determined in our dataset by the comparison of β-cells between treatment groups and control viz. WT, Sham, PF and VSG. Moreover, β-cell subclusters were also described in more detail by exploring VSG-specific effects. For this purpose, differential analysis tests were performed on the log-normalized counts (output from scran method, which accounts for library size differences) using limma-trend from R package limma [112] (v3.42.2) and edgeR [113] (v3.28.1) through an rpy2 (v3.2.7) interface. Additionally, to exclude spurious results, genes expressed in fewer than 1% cells in either of the two groups tested were also removed. Genes with a false discovery rate (FDR) < 0.01 were used for subsequent analysis unless stated otherwise. Briefly, a design matrix was constructed with cell type and batch information. Using the lmFit function, linear models were fit to the expression of each gene. The genes were subsequently ranked with enhanced statistical power using an empirical Bayes method, eBayes, to detect significant changes in expression. Finally, the top-ranked genes were corrected for multiple testing and FDR using the adjustment method (Benjamini-Hochberg or BH) and extracted from the linear model fit. Gene set enrichment analysis was subsequently performed using the web-based tool EnrichR [111] (https://maayanlab.cloud/Enrichr/), which uses a standard hypergeometric test or Fisher's exact test to report enrichment results, and significantly regulated KEGG pathways and GO terms/ontologies were reported. The results were visualized using custom plotting functions.
4.15. Inferring pseudotime, cluster connectivity and cell transitions
In order to assess transcriptomic similarity of β-cells between treatment groups, we applied Partition-based graph abstraction (PAGA) to the KNN-graph using the Scanpy function tl.paga, including low-connectivity edges. PAGA is a measure of cluster connectivity wherein edge weights indicate significance of a connection.
To infer pseudotime of β-cell dedifferentiation upon treatment, we ordered cells along a trajectory using a random-walk based distance metric, employing diffusion pseudotime (DPT) computed with tl.dpt in Scanpy. For this purpose, we randomly assigned a root cell within the dedifferentiated β-cells and followed the direction cells transitioned from after this starting point. Prior to this step, all β-cells were batch-corrected using BBKNN [114] (v1.4.1) from the Scanpy external package (using external.pp.bbknn with default settings) in order to augment pseudotime inference by minimizing the variation added by the different sample batches. This resulted in a modified neighborhood graph that could be directly used to infer the maturation trajectory using DPT. β-cells were grouped according to Sham, VSG and WT and visualized by reconstruction of expression changes in genes that were robustly and gradually regulated along this one-dimensional axis. Extreme-like β-cells were excluded from this analysis, as they were not part of the linear trajectory and formed a separate cluster by expressing few very specific genes.
In order to identify direction of potential cell movements upon VSG treatment, we applied RNA velocity to our single-cell data using the default stochastic model as implemented in the scVelo python package (v0.2.2). Model fitting and subsequent velocity estimation was performed in VSG-treated and Sham control β-cells. The data was prepared following standard protocol and using default parameters. Briefly, 2000 top genes were retained after filtering genes that did not pass a minimum threshold of 20 counts simultaneously for spliced and unspliced mRNA. This was followed by computing the first and second order moments for each cell using 30 principal components with the 30 nearest neighbors. Two additional metrics, namely velocity confidence and velocity length, were also estimated, as our key focus was to affirm the extent to which VSG improved re-differentiation (Supplementary Table 4). The length was indicative of the pace at which differentiation occurs; thereby, higher values suggested faster transition. Confidence, on the other hand, was a measure of correlation within the velocity vector fields, with higher values indicating higher coherence.
4.16. Scoring single-cell identity with ERAD and UPR markers
In order to assign to each cell ERAD and UPR scores, we extracted from literature several genes that have been implicated in ERAD and UPR and serve as markers for the two critical quality-control machineries within a cell (Supplementary Table 2). Scores were then computed using the tl.score_genes function in Scanpy (with default settings) for each of the groups (WT, Sham, PF and VSG) using curated ERAD and UPR gene signatures. Statistical differences of the cell scores among the four groups were further assessed using Welch's Heteroscedastic F Test, which does not assume variance homogeneity like ANOVA, and using Bonferroni's correction to adjust the significance level for pairwise comparisons (p value < 0.01, welch.test from R package onewaytests (v2.4)).
4.17. Comparison of β-cell extreme markers to those published by Modi et al., 2019 and Farack et al., 2019
To compare our extreme-like β-cells to the extreme β-cells previously described in literature, we referred to a publicly available single-cell RNA sequencing study by Modi et al. [68], who characterized Ins2(GFP)high β-cells as synonymous to the extreme β-cells reported by Farack et al. [75]. Briefly, genes that were differentially expressed along the GFP gradient were indicated as specific markers distinguishing extreme β-cells from the rest and were subsequently used as a signature for our study (Supplementary Table 3). Differentially expressed genes between extreme-like vs non–extreme-like β-cells in WT and, additionally, across all db/db cells collectively (Sham, PF and VSG-derived β-cells) were used for the comparison. Differential expression analysis was performed using limma-trend from R package limma (v 3.42.2) and edgeR (v3.28.1) through an rpy2 (v3.2.7) interface. Genes that were expressed in fewer than 1% cells in either of the two groups tested were removed, and genes with a false discovery rate (FDR) < 0.01, B > 50 and an absolute estimated log(fold change) > 0.2 were used as input. The B scores represent the log-odds that a gene is significantly differentially expressed. Following this, Venn diagrams were plotted to reveal the intersection between extreme-like β-cells identified in this study with those from Modi et al.
4.18. Comparison between VSG-specific β-cell markers and literature curated differentially expressed genes in pancreatic islets during pregnancy
To assess similarities between VSG- and pregnancy-specific β-cell adaptations, we compared published islet transcriptome datasets from pregnant versus non-pregnant mice [[80], [81], [82]] with our VSG dataset. To obtain genes unique to VSG, we compared genes from all groups versus control groups (Sham and PF, respectively) using limma-trend as mentioned above, thresholded with an absolute estimated log(fold change) > 0.15 and B > 150. We used Venn diagrams to identify the VSG-unique intersection, including the overlapping VSG/WT intersection (representing genes with expression levels restored to WT) for up- and downregulated genes separately. The list of genes obtained from this analysis was subsequently compared to differentially expressed genes in islets during pregnancy, derived from Rieck et al., Layden et al. and Kim et al. The intersection was then visualized using an upset plot. The set size bar plot indicates the total number of genes in each set, while the intersection size bar plot shows the total number of genes that overlap between two or more sets.
4.19. Data and code availability
The single-cell RNA sequencing dataset generated in this study has been deposited at the National Center for Biotechnology Information Gene Expression Omnibus (GEO) under accession number GSE174194, along with the code and custom scripts for analyzing scRNA-seq data generated in this study which is available at GitHub (https://github.com/theislab/2021_Oppenlaender_Palit_Pancreas_VSG).
Author contributions
L.O., A., K.S. and H.L. conceived and designed the study; L.O., K.S., A., M.S., A.F., T.G., C.S., A.B.P. and A.B. conducted the experiments; S.P. designed and performed all bioinformatics analysis; L.O., S.P. and A. interpreted the scRNA-seq data; L.O., S.P., A. and H.L. co-wrote the manuscript; F.J.T., H.L., A. supervised the study and H.L. acquired funding.
Acknowledgements
The authors wish to thank Ines Kunze, Jessica Jaki, Jacqueline Haag, Julia Beckenbauer, Kaiyung Yang and Silvia Schirge for their excellent technical assistance, as well as Stephan Sachs for discussion. The study was supported by the Helmholtz Foundation and German Center for Diabetes Research (DZD e.V.).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.molmet.2021.101330.
Contributor Information
Ansarullah, Email: ansar.aman@jax.org.
Fabian J. Theis, Email: fabian.theis@helmoltz-muenchen.de.
Heiko Lickert, Email: heiko.lickert@helmholtz-muenchen.de.
Conflict of interest
F.J.T. reports consulting fees from ImmunAI and ownership interest in Dermagnostix.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
FigS1.
Preoperative physiological parameters of db/db mice. a–d, Preoperative values of (a) BW (WT, n = 8; db/db, n = 17; unpaired student's t-test, p < 0.0001), (b) fasting blood glucose (WT, n = 7; db/db, n = 13; unpaired student's t-test; p < 0.0001), (c) HOMA-IR (WT, n = 7; db/db, n = 9; unpaired student's t-test; p < 0.0001) and (d) plasma insulin during a mixed-meal tolerance test (WT, n = 8; db/db, n = 17; two-way ANOVA; p(group) < 0.0001, p(time) = 0.0007). e, Postsurgical cumulative food intake (Sham, n = 7; VSG, n = 4; unpaired student's t-test; p = 0.2).
FigS2.
Diabetic stress mainly affects the β-cells transcriptome and, to a low extent, that of α-, δ- and PP-cells. a–d, Volcano plots showing numbers of significantly differentially expressed genes, comparing Sham versus WT in (a) β-cells, (b) α-cells, (c) δ-cells and (d) PP-cells with a threshold at an absolute estimated logFC > 0.15, B > 150, adjusted p < 0.01. e, UMAP plot showing WT- and Sham-derived α-cells (3279 cells in total) based on the expression of hormone glucagon (Gcg) and depicting co-expression of Mafb and Ppy. f, Bar plot showing the log(FC) differences between WT- and Sham-derived α-cells for a selection of genes. g, Selected Gene Ontology (GO) terms from enrichment analysis of significantly up- and downregulated genes in α-cells of Sham compared to WT at absolute estimated log(FC) > 0.15 and B > 20.
FigS3.
VSG distinctly modulates the β-cells transcriptome and, to a lesser extent, that of α-, δ- and PP-cells. a, Scatter plot showing the distribution of total counts (x-axis), number of genes expressed (y-axis) and fraction of counts in mitochondrial genes (color scale) in all endocrine cells. b, Boxplot showing the distribution of doublet scores in endocrine cell types. All groups display low median scores. Boxplot shows the extreme values and the interquartile range (IQR). Whiskers indicate 1.5 IQR of the lower and upper quartile. Outliers not shown. c, Violin plots of selected β-cell maturity, function and dedifferentiation in the β-cell cluster per group. d–f, Volcano plots showing very few significantly differentially expressed genes in (d) α-cells, (e) δ-cells and (f) PP-cells of VSG-treated mice compared to Sham (absolute estimated log(FC) > 0.15, B > 150 and FDR < 0.01). g–h, Bar plot depicting a selection of genes and their corresponding log(FC) differences between (g) VSG- and Sham- and (h) PF- and Sham-derived α-cells. i–j, GO terms and KEGG pathways derived from genes specifically upregulated in PF β-cells compared to (i) Sham and (j) VSG. Threshold at absolute estimated logFC > 0.15, B > 20, adjusted p < 0.01. X-axis indicates –log10(adjusted p-value).
FigS4.
Refined clustering shows distinct β-cell states. a–b, Differentially expressed genes in (a) mature β-cells and (b) dedifferentiated β-cells in VSG versus Sham at absolute estimated logFC > 0.15, B > 30, p(adj) < 0.01. c, Expression of selected markers defining immature β-cells (absence of Mafa, expression of Gpx3, Cd81). d–e, Velocity confidence and length of single-cell RNA velocity in VSG- and Sham-derived β-cells per subcluster. Velocity confidence indicates coherence within the neighboring velocity vector fields and length is a measure of speed of differentiation. (d) Box plots showing median (Sham(dediff)=0.768, VSG(dediff)=0.931), quartile and whisker values of velocity confidence. VSG showed a significantly higher velocity confidence compared to Sham (p < 0.0001, Welch-Test (One-Way) with Bonferroni's post-hoc test). (e) Box plots showing median (Sham(dediff)=385.41, VSG(dediff)=552.51), quartile and whisker values of velocity length. VSG showed a significantly higher velocity length compared to Sham (p < 0.0001, Welch-Test (One-Way) with Bonferroni's post-hoc test).
FigS5.
Transcriptional characterization of extreme-like β-cells in T2D. a–b, Comparison of (a) upregulated and (b) downregulated genes from the Ins2-high β-cell population described by Modi et al. (2019) with db/db extreme-like β-cells and WT extreme-like β-cells from our dataset, thresholded at estimated logFC > 0.2 or logFC < –0.2, B > 50, p(adj) < 0.01. c, Venn diagram showing the overlap of enriched genes of WT- and db/db-derived (Sham, PF, VSG) extreme-like β-cells. Limma-trend method was used to find differentially expressed genes at absolute log(fold change) > 0.15 and B > 100. d, GO and KEGG pathway enrichment analysis using EnrichR webtool with Fisher's exact test revealed significantly upregulated ontologies for genes commonly overexpressed in both WT and db/db extreme-like β-cells. Venn diagrams aided in identifying the overlapping genes that were group invariant. X-axis indicates –log10(adjusted p-value). e, Bar plot showing selected regulated genes in db/db-derived (Sham, PF, VSG) extreme-like β-cells.
FigS6.
Proliferative phenotype in VSG-derived β-cells. GO and KEGG analysis of significantly upregulated genes in VSG compared to PF revealed cell cycle–related terms. Upregulated genes were obtained from limma-trend test comparing VSG- and Sham-derived β-cells at logFC > 0.15, B > 150, adj(p) < 0.01. X-axis indicates –log10(adjusted p-value).
References
- 1.Rhodes C.J. Type 2 diabetes-a matter of ß-cell life and death? Science. 2005;307(5708):380–384. doi: 10.1126/science.1104345. [DOI] [PubMed] [Google Scholar]
- 2.Nolan C.J., Prentki M. Insulin resistance and insulin hypersecretion in the metabolic syndrome and type 2 diabetes: time for a conceptual framework shift. Diabetes and Vascular Disease Research. 2019;16(2):118–127. doi: 10.1177/1479164119827611. [DOI] [PubMed] [Google Scholar]
- 3.Halban P., Polonsky K., Bowden D.W., Hawkins M.A., Ling C., Mather K.J. Beta-cell failure in type 2 diabetes: postulated mechanisms and prospects for prevention and treatment. Diabetes Care. 2014;37:1751–1758. doi: 10.2337/dc14-0396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Talchai C., Xuan S., Lin H.V., Sussel L., Accili D. Pancreatic β cell dedifferentiation as a mechanism of diabetic β cell failure. Cell. 2012;150(6):1223–1234. doi: 10.1016/j.cell.2012.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weir G.C., Aguayo-Mazzucato C., Bonner-Weir S. β-cell dedifferentiation in diabetes is important, but what is it? Islets. 2014 doi: 10.4161/isl.27494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang Z., York N.W., Nichols C.G., Remedi M.S. Pancreatic β-cell dedifferentiation in diabetes and redifferentiation following insulin therapy. Cell Metabolism. 2015;19(5):872–882. doi: 10.1016/j.cmet.2014.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cinti F., Bouchi R., Kim-Muller J.Y., Ohmura Y., Sandoval P.R., Masini M. Evidence of β-cell dedifferentiation in human type 2 diabetes. The Journal of Cinical Endocrinology and Metabolism. 2016;101(3):1044–1054. doi: 10.1210/jc.2015-2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Butler A.E., Dhawan S., Hoang J., Cory M., Zeng K., Fritsch H. β-Cell deficit in obese type 2 diabetes, a minor role of β-cell dedifferentiation and degranulation. The Journal of Cinical Endocrinology and Metabolism. 2016;101(2):523–532. doi: 10.1210/jc.2015-3566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sheng C., Li F., Lin Z., Zhang M., Yang P., Bu L. Reversibility of β -Cell-Specific transcript factors expression by long-term caloric restriction in db/db mouse. Journal of Diabetes Research. 2016 doi: 10.1155/2016/6035046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ishida E., Kim-Muller J.Y., Accili D. Pair feeding, but not insulin, phloridzin, or rosiglitazone treatment, curtails markers of β-cell dedifferentiation in db/db mice. Diabetes. 2017;66(8):2092–2101. doi: 10.2337/db16-1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sachs S., Bastidas-Ponce A., Tritschler S., Bakhti M., Böttcher A., Sánchez-Garrido M.A. Targeted pharmacological therapy restores β-cell function for diabetes remission. Nature Metabolism. 2020;2(2):192–209. doi: 10.1038/s42255-020-0171-3. [DOI] [PubMed] [Google Scholar]
- 12.Capehorn M., Ghani Y., Hindsberger C., Johansen P., Jódar E. Once-weekly semaglutide reduces HbA1c and body weight in patients with type 2 diabetes regardless of background common OAD: a subgroup Analysis from SUSTAIN 2–4 and 10. Diabetes Therapy. 2020;11(5):1061–1075. doi: 10.1007/s13300-020-00796-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wadden T., Bailey T., Billings L., Davies M., Frias J., Koroleva A. Effect of subcutaneous semaglutide vs placebo as an adjunct to intensive behavioral therapy on body weight in adults with overweight or obesity: the STEP 3 randomized clinical trial. Journal of the American Medical Association. 2021;325(14):1403–1413. doi: 10.1001/jama.2021.1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tahrani A.A., Barnett A.H., Bailey C.J. Pharmacology and therapeutic implications of current drugs for type 2 diabetes mellitus. Nature Reviews Endocrinology. 2016;12(10):566–592. doi: 10.1038/nrendo.2016.86. [DOI] [PubMed] [Google Scholar]
- 15.Rubino F., Nathan D.M., Eckel R.H., Schauer P.R., Alberti K.G.M.M., Zimmet P.Z. Metabolic surgery in the treatment algorithm for type 2 diabetes: a joint statement by international diabetes organizations. Diabetes Care. 2016;39(6):861–877. doi: 10.2337/dc16-0236. [DOI] [PubMed] [Google Scholar]
- 16.Schauer P.R., Bhatt D.L., Kirwan J.P., Wolski K., Aminian A., Brethauer S.A. Bariatric surgery versus intensive medical therapy for diabetes — 5-year outcomes. New England Journal of Medicine. 2017;376(7):641–651. doi: 10.1056/NEJMoa1600869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Buchwald H., Estok R., Fahrbach K., Banel D., Jensen M.D., Pories W.J. Weight and type 2 diabetes after bariatric surgery: systematic review and meta-analysis. Americas Journal of Medicine. 2009;122(3) doi: 10.1016/j.amjmed.2008.09.041. [DOI] [PubMed] [Google Scholar]
- 18.Evers S.S., Sandoval D.A., Seeley R.J. The physiology and molecular underpinnings of the effects of bariatric surgery on obesity and diabetes. Annual Review of Physiology. 2017;79 doi: 10.1146/annurev-physiol-022516-034423. [DOI] [PubMed] [Google Scholar]
- 19.Seeley R.J., Chambers A.P., Sandoval1 D.A. The role of gut adaptation in the potent effects of multiple bariatric surgeries on obesity and diabetes. Cell Metabolism. 2015;21(3):369–378. doi: 10.1016/j.cmet.2015.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Douros J.D., Niu J., Sdao S., Gregg T., Fisher-Wellman K., Bharadwaj M. Sleeve gastrectomy rapidly enhances islet function independently of body weight. JCI Insight. 2019;4(6) doi: 10.1172/jci.insight.126688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Abu-Gazala S., Horwitz E., Schyr R.B.H., Bardugo A., Israeli H., Hija A. Sleeve gastrectomy improves glycemia independent of weight loss by restoring hepatic insulin sensitivity. Diabetes. 2018;67(6):1079–1085. doi: 10.2337/db17-1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jørgensen N.B., Jacobsen S.H., Dirksen C., Bojsen-Møller K.N., Naver L., Hvolris L. Acute and long-term effects of Roux-en-Y gastric bypass on glucose metabolism in subjects with Type 2 diabetes and normal glucose tolerance. American Journal of Physiology. Endocrinology and Metabolism. 2012;303(1):122–131. doi: 10.1152/ajpendo.00073.2012. [DOI] [PubMed] [Google Scholar]
- 23.Chambers A.P., Jessen L., Ryan K.K., Sisley S., Wilson-Pérez H.E., Stefater M.A. Weight-Independent changes in blood glucose homeostasis after gastric bypass or vertical sleeve gastrectomy in rats. Gastroenterology. 2011;141(3):950–958. doi: 10.1053/j.gastro.2011.05.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Laferrère B., Teixeira J., McGinty J., Tran H., Egger J.R., Colarusso A. Effect of weight loss by gastric bypass surgery versus hypocaloric diet on glucose and incretin levels in patients with type 2 diabetes. The Journal of Cinical Endocrinology and Metabolism. 2008;93(7):2479–2485. doi: 10.1210/jc.2007-2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Welbourn R., Hollyman M., Kinsman R., Dixon J., Liem R., Ottosson J. Bariatric surgery worldwide: baseline demographic description and one-year outcomes from the fourth IFSO global registry report 2018. Obesity Surgery. 2019;29:782–795. doi: 10.1007/s11695-018-3593-1. [DOI] [PubMed] [Google Scholar]
- 26.Li F., Sheng C., Song K., Zhang M., Bu L., Yang P. Preventative sleeve gastrectomy contributes to maintaining β cell function in db/db diabetic mouse. Obesity Surgery. 2016;26(10):2402–2410. doi: 10.1007/s11695-016-2112-5. [DOI] [PubMed] [Google Scholar]
- 27.Kang T., Boland B.B., Jensen P., Alarcon C., Nawrocki A., Grimsby J.S. Characterization of signaling pathways associated with pancreatic β-cell adaptive flexibility in compensation of obesity-linked diabetes in db-db mice. Molecular & Cellular Proteomics. 2020;19(6):971–993. doi: 10.1074/mcp.RA119.001882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dalbøge L.S., Almholt D.L.C., Neerup T.S.R., Vassiliadis E., Vrang N., Pedersen L. Characterisation of age-dependent beta cell dynamics in the male db/db mice. PloS One. 2013;8(12):1–10. doi: 10.1371/journal.pone.0082813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Neelankal John A., Ram R., Jiang F.X. RNA-seq analysis of islets to characterise the dedifferentiation in type 2 diabetes model mice db/db. Endocrine Pathology. 2018;29(3):207–221. doi: 10.1007/s12022-018-9523-x. [DOI] [PubMed] [Google Scholar]
- 30.Hummel K.P., Dickie M.M., Coleman D.L. Diabetes, a new mutation in the mouse. Science. 1966;153(3740):1127–1128. doi: 10.1126/science.153.3740.1127. [DOI] [PubMed] [Google Scholar]
- 31.Becht E., McInnes L., Healy J., Dutertre C.A., Kwok I.W.H., Ng L.G. Dimensionality reduction for visualizing single-cell data using UMAP. Nature Biotechnology. 2019;37(1):38–47. doi: 10.1038/nbt.4314. [DOI] [PubMed] [Google Scholar]
- 32.Blondel V.D., Guillaume J.L., Lambiotte R., Lefebvre E. Fast unfolding of communities in large networks. Journal of Statistical Mechanics: Theory and Experiment. 2008;2008(10):1–12. [Google Scholar]
- 33.Zmuda E.J., Qi L., Zhu M.X., Mirmira G.R., Montminy M.R., Hai T. The roles of ATF3, an adaptive-response gene, in high-fat-diet-induced diabetes and pancreatic β-cell dysfunction. Molecular Endocrinology. 2019;24:1423–1433. doi: 10.1210/me.2009-0463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li D., Yin X., Zmuda E.J., Wolford C.C., Dong X., White M.F. The repression of IRS2 gene by ATF3, a stress-inducible gene, contributes to pancreatic β-Cell apoptosis. Diabetes. 2008;57(3):635–644. doi: 10.2337/db07-0717. [DOI] [PubMed] [Google Scholar]
- 35.Cantley J., Choudhury A.I., Asare-Anane H., Selman C., Lingard S., Heffron H. Pancreatic deletion of insulin receptor substrate 2 reduces beta and alpha cell mass and impairs glucose homeostasis in mice. Diabetologia. 2007;50(6):1248–1256. doi: 10.1007/s00125-007-0637-9. [DOI] [PubMed] [Google Scholar]
- 36.Hennige A.M., Burks D.J., Ozcan U., Kulkarni R.N., Ye J., Park S. Upregulation of insulin receptor substrate-2 in pancreatic β cells prevents diabetes. Journal of Clinical Investigation. 2003;112:1521–1532. doi: 10.1172/JCI18581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Blum B., Hrvatin S., Schuetz C., Bonal C., Rezania A., Melton D.A. Functional beta-cell maturation is marked by an increased glucose threshold and by expression of urocortin 3. Nature Biotechnology. 2012;30(3):261–264. doi: 10.1038/nbt.2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.van der Meulen T., Xie R., Kelly O.G., Vale W.W., Sander M., Huising M.O. Urocortin 3 marks mature human primary and embryonic stem cell-derived pancreatic alpha and beta cells. PloS One. 2012;7(12):1–12. doi: 10.1371/journal.pone.0052181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oliver-Krasinski J.M., Stoffers D.A. On the origin of the beta cell. Genes & Development. 2008;22:1998–2021. doi: 10.1101/gad.1670808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim-Muller J.Y., Fan J., Kim Y.J.R., Lee S.A., Ishida E., Blaner W.S. Aldehyde dehydrogenase 1a3 defines a subset of failing pancreatic β cells in diabetic mice. Nature Communications. 2016;7:2016. doi: 10.1038/ncomms12631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gittes G.K., Rutter W.J., Debas H.T. Initiation of gastrin expression during the development of the mouse pancreas. Americas Journal of Surgery. 1993;165(1):23–26. doi: 10.1016/s0002-9610(05)80399-x. [DOI] [PubMed] [Google Scholar]
- 42.Dahan T., Ziv O., Horwitz E., Zemmour H., Lavi J., Swisa A. Pancreatic β-cells express the fetal islet hormone gastrin in rodent and human diabetes. Diabetes. 2017;66(2):426–436. doi: 10.2337/db16-0641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kuo T., Lazar M.A., Accili D., Kuo T., Damle M., González B.J. vol. 4. 2019. (Induction of α cell – restricted Gc in dedifferentiating β cells contributes to stress-induced β cell dysfunction). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Salinno C., Büttner M., Cota P., Tritschler S., Tarquis-Medina M., Bastidas-Ponce A. CD81 marks immature and dedifferentiated pancreatic β-cells. Molecular Metabolism. 2021;49 doi: 10.1016/j.molmet.2021.101188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wolf F.A., Hamey F.K., Plass M., Solana J., Dahlin J.S., Göttgens B. PAGA: graph abstraction reconciles clustering with trajectory inference through a topology preserving map of single cells. Genome Biology. 2019;20(1):1–9. doi: 10.1186/s13059-019-1663-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Camunas-Soler J., Dai X.-Q., Hang Y., Bautista A., Lyon J., Suzuki K. Patch-seq links single-cell transcriptomes to human islet dysfunction in diabetes. Cell Metabolism. 2020;31(5):1017–1031. doi: 10.1016/j.cmet.2020.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gremlich S., Roduit R., Thorens B. Dexamethasone induces posttranslational degradation of GLUT2 and inhibition of insulin secretion in isolated pancreatic β cells. Comparison with the effects of fatty acids. Journal of Biological Chemistry. 1997;272(6):3216–3222. doi: 10.1074/jbc.272.6.3216. [DOI] [PubMed] [Google Scholar]
- 48.Ohtsubo K., Takamatsu S., Minowa M.T., Yoshida A., Takeuchi M., Marth J.D. Dietary and genetic control of glucose transporter 2 glycosylation promotes insulin secretion in suppressing diabetes. Cell. 2005;123(7):1307–1321. doi: 10.1016/j.cell.2005.09.041. [DOI] [PubMed] [Google Scholar]
- 49.Bearrows S.C., Bauchle C.J., Becker M., Haldeman J.M., Swaminathan S., Stephens S.B. Chromogranin B regulates early-stage insulin granule trafficking from the Golgi in pancreatic islet β-cells. Journal of Cell Science. 2019;132(13) doi: 10.1242/jcs.231373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dolai S., Xie L., Zhu D., Liang T., Qin T., Xie H. Synaptotagmin-7 functions to replenish insulin granules for exocytosis in human islet β-cells. Diabetes. 2016;65:1962–1976. doi: 10.2337/db15-1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sadoul K., Lang J., Montecucco C., Weller U., Regazzi R., Catsicas S. SNAP-25 is expressed in islets of Langerhans and is involved in insulin release. The Journal of Cell Biology. 1995;128(6):1019–1028. doi: 10.1083/jcb.128.6.1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Haghverdi L., Büttner M., Wolf F.A., Buettner F., Theis F.J. Diffusion pseudotime robustly reconstructs lineage branching. Nature Methods. 2016;13(10):845–848. doi: 10.1038/nmeth.3971. [DOI] [PubMed] [Google Scholar]
- 53.Fonseca S.G., Gromada J., Urano F. Endoplasmic reticulum stress and pancreatic β-cell death. Trends in Endocrinology and Metabolism. 2011;22(7):266–274. doi: 10.1016/j.tem.2011.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bilekova S., Sachs S., Lickert H. Pharmacological targeting of endoplasmic reticulum stress in pancreatic beta cells. Trends in Pharmacological Sciences. 2021;42(2):85–95. doi: 10.1016/j.tips.2020.11.011. [DOI] [PubMed] [Google Scholar]
- 55.Lee A.S. The ER chaperone and signaling regulator GRP78/BiP as a monitor of endoplasmic reticulum stress. Methods. 2005;35(4):373–381. doi: 10.1016/j.ymeth.2004.10.010. [DOI] [PubMed] [Google Scholar]
- 56.Hassler J.R., Scheuner D.L., Wang S., Han J., Kodali V.K., Li P. The IRE1α/XBP1s pathway is essential for the glucose response and protection of β cells. PLoS Biology. 2015;13(10) doi: 10.1371/journal.pbio.1002277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Abreu D., Asada R., Revilla J.M.P., Lavagnino Z., Kries K., Piston D.W. Wolfram syndrome 1 gene regulates pathways maintaining beta-cell health and survival. Laboratory Investigation. 2020;100(6):849–862. doi: 10.1038/s41374-020-0408-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sharma R.B., O’Donnell A.C., Stamateris R.E., Ha B., McCloskey K.M., Reynolds P.R. Insulin demand regulates β cell number via the unfolded protein response. Journal of Clinical Investigation. 2015;125(10):3831–3846. doi: 10.1172/JCI79264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sowers C.R., Wang R., Bourne R.A., McGrath B.C., Hu J., Bevilacqua S.C. The protein kinase PERK/EIF2AK3 regulates proinsulin processing not via protein synthesis but by controlling endoplasmic reticulum chaperones. Journal of Biological Chemistry. 2018;293(14):5134–5149. doi: 10.1074/jbc.M117.813790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shrestha N., Liu T., Ji Y., Reinert R.B., Torres M., Li X. Sel1L-Hrd1 ER-associated degradation maintains β cell identity via TGF-β signaling. Journal of Clinical Investigation. 2020;130(7):3499–3510. doi: 10.1172/JCI134874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hu Y., Gao Y., Zhang M., Deng K.Y., Singh R., Tian Q. Endoplasmic reticulum–associated degradation (ERAD) has a critical role in supporting glucose-stimulated insulin secretion in pancreatic B-cells. Diabetes. 2019;68(4):733–746. doi: 10.2337/db18-0624. [DOI] [PubMed] [Google Scholar]
- 62.Christianson J.C., Olzmann J.A., Shaler T.A., Sowa M.E., Bennett J., Richter C.M. Defining human ERAD networks through an integrative mapping strategy. Nature Cell Biology. 2012;14(1):93–105. doi: 10.1038/ncb2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bader E., Migliorini A., Gegg M., Moruzzi N., Gerdes J., Roscioni S.S. Identification of proliferative and mature β-cells in the islets of Langerhans. Nature. 2016;535:430–434. doi: 10.1038/nature18624. [DOI] [PubMed] [Google Scholar]
- 64.Roscioni S.S., Migliorini A., Gegg M., Lickert H. Impact of islet architecture on β-cell heterogeneity, plasticity and function. Nature Reviews Endocrinology [Internet] 2016;12:695–709. doi: 10.1038/nrendo.2016.147. https://www-nature-com.are.uab.cat/articles/nrendo.2016.147 Available from: [DOI] [PubMed] [Google Scholar]
- 65.Baron M., Veres A., Wolock S.L., Faust A.L., Gaujoux R., Vetere A. A single-cell transcriptomic map of the human and mouse pancreas reveals inter- and intra-cell population structure. Cell Systems. 2016;3(4):346–360. doi: 10.1016/j.cels.2016.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Segerstolpe Å, Athanasia Palasantza P.E., Andréasson E.-M.A.A.-C., Sun X., Picelli S., Sabirsh A. Single-cell transcriptome profiling of human pancreatic islets in health and type 2 diabetes. Cell Metabolism. 2016;24(4):593–607. doi: 10.1016/j.cmet.2016.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Xin Y., Gutierrez G.D., Okamoto H., Kim J., Lee A.H., Adler C. Pseudotime ordering of single human B-cells reveals states of insulin production and unfolded protein response. Diabetes. 2018;67:1783–1794. doi: 10.2337/db18-0365. [DOI] [PubMed] [Google Scholar]
- 68.Modi H., Skovsø S., Ellis C., Krentz N.A.J., Zhao Y.B., Cen H. Ins2 gene bursting activity defines a mature β-cell state. bioRxiv. 2019 [Google Scholar]
- 69.Fang Z., Weng C., Li H., Tao R., Mai W., Liu X. Single-cell heterogeneity analysis and CRISPR screen identify key β-cell-specific disease genes. Cell Reports [Internet] 2019;26(11):3132–3144.e7. doi: 10.1016/j.celrep.2019.02.043. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Feng Y., Qiu W.L., Yu X.X., Zhang Y., He M.Y., Li L.C. Characterizing pancreatic β-cell heterogeneity in the streptozotocin model by single-cell transcriptomic analysis. Molecular Metabolism. 2020;37 doi: 10.1016/j.molmet.2020.100982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yu X.X., Xu C.R. Understanding generation and regeneration of pancreatic β cells from a single-cell perspective. Development (Cambridge) 2020;147 doi: 10.1242/dev.179051. [DOI] [PubMed] [Google Scholar]
- 72.Ganic E., Singh T., Luan C., Fadista J., Johansson J.K., Cyphert H.A. MafA-controlled nicotinic receptor expression is essential for insulin secretion and is impaired in patients with type 2 diabetes. Cell Reports. 2016;14(8):1991–2002. doi: 10.1016/j.celrep.2016.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Li H., Neelankal John A., Nagatake T., Hamazaki Y., Jiang F.X. Claudin 4 in pancreatic β cells is involved in regulating the functional state of adult islets. FEBS Open Bio. 2020;10(1):28–40. doi: 10.1002/2211-5463.12735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bergen V., Lange M., Peidli S., Wolf F.A., Theis F.J. Generalizing RNA velocity to transient cell states through dynamical modeling. Nature Biotechnology [Internet] 2020;38(12):1408–1414. doi: 10.1038/s41587-020-0591-3. Available from: [DOI] [PubMed] [Google Scholar]
- 75.Farack L., Golan M., Egozi A., Dezorella N., Bahar Halpern K., Ben-Moshe S. Transcriptional heterogeneity of beta cells in the intact pancreas. Developmental Cell. 2019;48(1):115–125. doi: 10.1016/j.devcel.2018.11.001. [DOI] [PubMed] [Google Scholar]
- 76.Kushner J.A., Ciemerych M.A., Sicinska E., Wartschow L.M., Teta M., Long S.Y. Cyclins D2 and D1 are essential for postnatal pancreatic β-cell growth. Molecular and Cellular Biology. 2005;25(9):3752–3762. doi: 10.1128/MCB.25.9.3752-3762.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Xu E.E., Sasaki S., Speckmann T., Nian C., Lynn F.C. SOX4 allows facultativeβ-cellproliferation through repression of cdkn1a. Diabetes. 2017;66(8):2213–2219. doi: 10.2337/db16-1074. [DOI] [PubMed] [Google Scholar]
- 78.Kondegowda N.G., Fenutria R., Pollack I.R., Orthofer M., Garcia-Ocaña A., Penninger J.M. Osteoprotegerin and denosumab stimulate human beta cell proliferation through inhibition of the receptor activator of NF-κB ligand pathway. Cell Metabolism. 2015;22(1):77–85. doi: 10.1016/j.cmet.2015.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Schmitz F., Roscioni S., Lickert H. Repurposing an osteoporosis drug for β cell regeneration in diabetic patients. Cell Metabolism [Internet] 2015;22(1):58–59. doi: 10.1016/j.cmet.2015.05.024. Available from: [DOI] [PubMed] [Google Scholar]
- 80.Rieck S., White P., Schug J., Fox A.J., Smirnova O., Gao N. The transcriptional response of the islet to pregnancy in mice. Molecular Endocrinology. 2009;23(10):1702–1712. doi: 10.1210/me.2009-0144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Layden B.T., Durai V., Newman M.V., Marinelarena A.M., Ahn C.W., Feng G. Regulation of pancreatic islet gene expression in mouse islets by pregnancy. Journal of Endocrinology. 2010;207(3):265–279. doi: 10.1677/JOE-10-0298. [DOI] [PubMed] [Google Scholar]
- 82.Kim H., Toyofuku Y., Lynn F.C., Chak E., Uchida T., Mizukami H i. Serotonin regulates pancreatic beta cell mass during pregnancy. Nature Medicine. 2010;16(7) doi: 10.1038/nm.2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Schauer P.R., Bhatt D.L., Kirwan J.P., Wolski K., Brethauer S.A., Navaneethan S.D. Bariatric surgery versus intensive medical therapy for diabetes — 3-year outcomes. New England Journal of Medicine. 2014;370(21):2002–2013. doi: 10.1056/NEJMoa1401329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Burke S.J., Batdorf H.M., Burk D.H., Noland R.C., Eder A.E., Boulos M.S. Db/db mice exhibit features of human type 2 diabetes that are not present in weight-matched C57BL/6J mice fed a western diet. Journal of Diabetes Research. 2017;2017 doi: 10.1155/2017/8503754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lee W.J., Chong K., Ser K.H., Chen J.C., Lee Y.C., Chen S.C. C-peptide predicts the remission of type 2 diabetes after bariatric surgery. Obesity Surgery. 2012;22(2):293–298. doi: 10.1007/s11695-011-0565-0. [DOI] [PubMed] [Google Scholar]
- 86.Souteiro P., Belo S., Neves J.S., Magalhães D., Silva R.B., Oliveira S.C. Preoperative beta cell function is predictive of diabetes remission after bariatric surgery. Obesity Surgery [Internet] 2017;27(2):288–294. doi: 10.1007/s11695-016-2300-3. Available from: [DOI] [PubMed] [Google Scholar]
- 87.Luo Y., Guo Z., He H., Yang Y., Zhao S., Mo Z. Predictive model of type 2 diabetes remission after metabolic surgery in Chinese patients. The Internet Journal of Endocrinology. 2020 doi: 10.1155/2020/2965175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Elgenaied I., El Ansari W., Elsherif M.A., Abdulrazzaq S., Qabbani A.S., Elhag W. Factors associated with complete and partial remission, improvement, or unchanged diabetes status of obese adults 1 year after sleeve gastrectomy. Surgery for Obesity and Related Diseases [Internet. 2020;16(10):1521–1530. doi: 10.1016/j.soard.2020.05.013. Available from: [DOI] [PubMed] [Google Scholar]
- 89.Corezola do Amaral M.E., Kravets V., Dwulet J.M., Farnsworth N.L., Piscopio R., Schleicher W.E. Caloric restriction recovers impaired β-cell-β-cell gap junction coupling, calcium oscillation coordination, and insulin secretion in prediabetic mice. American Journal of Physiology. Endocrinology and Metabolism. 2020;319(4):E709–E720. doi: 10.1152/ajpendo.00132.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Qian B., Zhou X., Li B., Li B., Liu Z., Wu J. Reduction of pancreatic b -cell dedifferentiation after gastric bypass surgery in diabetic rats. Journal of Molecular Cell Biology. 2014;6:531–534. doi: 10.1093/jmcb/mju042. [DOI] [PubMed] [Google Scholar]
- 91.Huang J.L., Lee S., Hoek P., van der Meulen T., Van R., Huising M.O. Genetic deletion of Urocortin 3 does not prevent functional maturation of beta cells. Journal of Endocrinology. 2020;246(1):69–78. doi: 10.1530/JOE-19-0535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Grong E., Kulseng B., Arbo I.B., Nord C., Eriksson M., Ahlgren U. Sleeve gastrectomy, but not duodenojejunostomy, preserves total beta-cell mass in Goto-Kakizaki rats evaluated by three-dimensional optical projection tomography. Surgical Endoscopy. 2016;30(2):532–542. doi: 10.1007/s00464-015-4236-4. [DOI] [PubMed] [Google Scholar]
- 93.Lindqvist A., Spégel P., Ekelund M., Garcia Vaz E., Pierzynowski S., Gomez M.F. Gastric bypass improves β-cell function and increases β-cell mass in a porcine model. Diabetes. 2014;63(5):1665–1671. doi: 10.2337/db13-0969. [DOI] [PubMed] [Google Scholar]
- 94.Miskelly M., Shcherbina L., Fischer A.-H., Abels M., Lindqvist A., Wierup N. GK-rats respond to gastric bypass surgery with improved glycemia despite unaffected insulin secretion and beta cell mass. Peptides. 2021:136. doi: 10.1016/j.peptides.2020.170445. [DOI] [PubMed] [Google Scholar]
- 95.McGavigan A.K., Garibay D., Henseler Z.M., Chen J., Bettaieb A., Haj F.G. TGR5 contributes to glucoregulatory improvements after vertical sleeve gastrectomy in mice. Gut. 2017;66(2):226–234. doi: 10.1136/gutjnl-2015-309871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ernst S., Demirci C., Valle S., Velazquez-Garcia S., Ocaña A.G.- Mechanisms in the adaptation of maternal β-cells during pregnancy. Diabetes Management. 2011;1(2):239–248. doi: 10.2217/dmt.10.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Brelje T.C., Scharp D.W., Lacy P.E., Ogren L., Talamantes F., Robertson M. Effect of homologous placental lactogens, prolactins, and growth hormones on islet B-cell division and insulin secretion in rat, mouse, and human islets: implication for placental lactogen regulation of islet function during pregnancy. Endocrinology. 1993;132(2):879–887. doi: 10.1210/endo.132.2.8425500. [DOI] [PubMed] [Google Scholar]
- 98.Vasavada R.C., Gonzalez-Pertusa J.A., Fujinaka Y., Fiaschi-Taesch N., Cozar-Castellano I., Garcia-Ocaña A. Growth factors and beta cell replication. The International Journal of Biochemistry & Cell Biology. 2005;38(5):931–950. doi: 10.1016/j.biocel.2005.08.003. [DOI] [PubMed] [Google Scholar]
- 99.Kondegowda N.G., Zhang X., Williams K., Mozar A., Vasavada R.C. Growth factor mediated regulation of beta cell survival. The Open Endocrinology Journal. 2011;4(1):78–93. [Google Scholar]
- 100.Pepin M.E., Bickerton H.H., Bethea M., Hunter C.S., Wende A.R., Banerjee R.R. Prolactin receptor signaling regulates a pregnancy-specific transcriptional program in mouse islets. Endocrinology. 2019;160(5):1150–1163. doi: 10.1210/en.2018-00991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Goyvaerts L., Lemaire K., Arijs I., Auffret J., Granvik M., Van Lommel L. Prolactin receptors and placental lactogen drive male mouse pancreatic islets to pregnancy-related mRNA changes. PloS One. 2015;10(3) doi: 10.1371/journal.pone.0121868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Wang X., Ma B., Li G., Sheng C., Yang P., Gao J. Glucose-lipid metabolism in obesity with elevated prolactin levels and alteration of prolactin levels after laparoscopic sleeve gastrectomy. Obesity Surgery. 2020;30(10):4004–4013. doi: 10.1007/s11695-020-04771-2. [DOI] [PubMed] [Google Scholar]
- 103.Schraenen A., Lemaire K., De Faudeur G., Hendrickx N., Granvik M., Van Lommel L. Placental lactogens induce serotonin biosynthesis in a subset of mouse beta cells during pregnancy. Diabetologia. 2010;53(12):2589–2599. doi: 10.1007/s00125-010-1913-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Aye T., Toschi E., Sharma A., Sgroi D., Bonner-Weir S. Identification of markers for newly formed β-cells in the perinatal period: a time of recognized β-cell immaturity. Journal of Histochemistry and Cytochemistry. 2010;58(4):369–376. doi: 10.1369/jhc.2009.954909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Tang S., Xin Y., Yang M., Zhang D., Xu C. Osteoprotegerin promotes islet β cell proliferation in intrauterine growth retardation rats through the PI3K/AKT/FoxO1 pathway. International Journal of Clinical and Experimental Pathology. 2019;12(6):2324–2338. [PMC free article] [PubMed] [Google Scholar]
- 106.Kuroda Y., Maruyama K., Fujii H., Sugawara I., Ko S.B.H., Yasuda H. Osteoprotegerin regulates pancreatic β-cell homeostasis upon microbial invasion. PloS One. 2016;11(1) doi: 10.1371/journal.pone.0146544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Schrader J., Rennekamp W., Niebergall U., Schoppet M., Jahr H., Brendel M.D. Cytokine-induced osteoprotegerin expression protects pancreatic beta cells through p38 mitogen-activated protein kinase signalling against cell death. Diabetologia. 2007;50(6):1243–1247. doi: 10.1007/s00125-007-0672-6. [DOI] [PubMed] [Google Scholar]
- 108.Stefater M.A., Pérez-tilve D., Chambers A.P., Wilson-Pérez H.E., Sandoval D.A., Berger J. Sleeve gastrectomy induces loss of weight and fat mass in obese rats, but does not affect leptin sensitivity. Gastroenterology. 2010;138(7):2426–2436. doi: 10.1053/j.gastro.2010.02.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Wolf F.A., Angerer P., Theis F.J. SCANPY: large-scale single-cell gene expression data analysis. Genome Biology. 2012;19(15) doi: 10.1186/s13059-017-1382-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Wolock S.L., Lopez R., Klein A.M. Scrublet: computational identification of cell doublets in single-cell transcriptomic data. Cell Systems [Internet] 2019;8(4):281–291. doi: 10.1016/j.cels.2018.11.005. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Kuleshov M.V., Jones M.R., Rouillard A.D., Fernandez N.F., Duan Q., Wang Z. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Research. 2016;44(W1):90–W97. doi: 10.1093/nar/gkw377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Research. 2015;43(7):e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Robinson M.D., McCarthy D.J., Smyth G.K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2009;26(1):139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Polański K., Young M.D., Miao Z., Meyer K.B., Teichmann S.A., Park J.E. BBKNN: fast batch alignment of single cell transcriptomes. Bioinformatics. 2020;36(3):964–965. doi: 10.1093/bioinformatics/btz625. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The single-cell RNA sequencing dataset generated in this study has been deposited at the National Center for Biotechnology Information Gene Expression Omnibus (GEO) under accession number GSE174194, along with the code and custom scripts for analyzing scRNA-seq data generated in this study which is available at GitHub (https://github.com/theislab/2021_Oppenlaender_Palit_Pancreas_VSG).