Abstract
Zinc Finger Protein 217 (ZNF217), a transcription factor and oncogene product, has been found to dysregulate Bone Morphogenetic Protein (BMP) signaling and induce invasion in breast tumors. In this study, the effect of BMP-2 or an active BMP-2 peptide, AISMLYLDEN, on the expression of ZNF217, BMP4 and CDK-inhibitor p21 gene, CDKN1A, was investigated in MCF-7 breast cancer cells. In parallel, the entire protein (BMP-2) as well as the aforementioned peptide were investigated in hDPSCs during osteogenic differentiation. The treatment of MCF-7 cancer cells with different concentrations of peptide AISMLYLDEN showed that the addition of 22.6 ng/ml was more effective in comparison to the other used concentrations. In particular, 48 h after treatment, CDKN1A and BMP4 mRNA levels were substantially increased in contrast to ZNF217 mRNA levels which were decreased. These results are strongly supported by BrdU assay that clearly indicated inhibition of cancer cell proliferation. Taken together, these results open ways for a concurrent use, at appropriate concentrations, of the peptide AISMLYLDEN during conventional therapeutic treatment in breast tumors with a metastatic tendency to the bones. Regarding the effect of the entire protein as well as its peptide on hDPSCs differentiation into osteocytes, the mRNA levels of osteocalcin, an osteogenic marker, showed that the peptide enhanced osteogenesis at a higher degree in comparison to the entire BMP-2 without however altering ZNF217, CDKN1A and BMP4 expression levels, which remained as expected of non-cancer cells.
Keywords: Bone Morphogenetic Protein-2, Breast cancer, Osteogenic differentiation, CDK-inhibitor p21, Zinc Finger Protein 217
Highlights
-
•
BMP-2 and BMP-2 peptide reduce proliferation of MCF-7 cells.
-
•
BMP-2 and peptide increase CDKN1A and BMP4 mRNA levels in MCF-7 cells.
-
•
BMP-2 peptide, at specific concentration, reduces ZNF217 expression in MCF-7 cells.
-
•
ZNF217, BMP4 and CDKN1A expressions remain at levels expected of non-cancer cells in peptide-treated osteocytes.
1. Introduction
Bone Morphogenetic Proteins (BMPs) are growth factors that belong to the TGF-β superfamily. Initially, they were found to induce the formation of bone and cartilage in animal models (Wozney et al., 1988; Urist, 1965; Reddi, 1997). However, in recent years, it has been discovered that they regulate growth, differentiation and apoptosis of mesenchymal, epithelial and neuronal cells as well as monocytes (Hogan, 1996). In addition, it has been found that they are involved in the morphogenesis of several tissue types and organs (Hogan, 1996; Kawabata et al., 1998). BMPs bind to surface receptors of target cells, BMPRs, and activate multiple signaling pathways, mainly the pathway of SMAD-1, -5 and -8 transcription factors (Bragdon et al., 2011).
BMP-2 can trigger the differentiation of human mesenchymal stem cells (hMSCs) into osteoblasts (Beederman et al., 2013). In addition, it has been found to be involved in the proliferation and apoptosis of tumor cells (Ghosh-Choudhury et al., 2000; Arnold et al., 1999; Wang et al., 2012; Wang et al., 2011). Through its interaction with receptors BMPRIa/BMPRII, BMP-2 can trigger the SMAD pathway, which is the main pathway that leads to pre-osteoblast and osteoblast differentiation of hMCSs, as well as SMAD-independent pathways that involve MAP kinases (Bragdon et al., 2011; Nohe et al., 2002). In the SMAD-dependent pathway, when BMP-2 forms a complex with its receptors, the constitutively active tyrosine kinase BMPRII phosphorylates and activates BMPRIa. The activated receptor BMPRIa phosphorylates SMAD-1, -5 or -8, which then forms a complex with co-SMAD, SMAD-4 (Attisano and Wrana, 2000; Massagué and Chen, 2000). The complex is transported to the nucleus, where it activates the expression of several transcription factors that promote osteogenesis (Bragdon et al., 2011; Sieber et al., 2009). Subsequently, genes necessary for osteogenesis, such as ALPL (which encodes the tissue non-specific alkaline phosphatase isoenzyme), are expressed and, a few weeks later, the mineralization of the extracellular matrix commences. BMP-2, also, activates MAPK pathways, which lead to the expression of ALPL, SPP1 (osteopontin) and COL1A1/2 (type I collagen) (Sieber et al., 2009; Katagiri et al., 2002; Ryoo et al., 2006).
ZNF217 is a transcription factor that belongs to the C2H2-type zinc-finger protein family. The ZNF217 gene has been recognized as a proto-oncogene with important role in the occurrence and progression of breast, colorectal, stomach, cervical, prostate and lung cancer (Krig et al., 2007; Collins et al., 2001). It is already known that ZNF217 is a component of a complex that contains CtBP1, which is a co-suppressor for many transcriptional suppressors in histone modifying complexes (Cowger et al., 2007; Shi et al., 2003). Although ZNF217 was initially recognized as a transcriptional suppressor, recent studies also indicate its role in transcriptional activation. It has been found that in mouse embryonic stem cells, ZFP217, the murine homolog of ZNF217, binds to promoters and enhancers of pluripotency genes, such as NANOG and SOX2, and activates their expression, thus maintaining the cells in their un-differentiated state (Lee et al., 2016).
In recent studies, it was demonstrated that ZNF217 triggers BMP signaling leading to metastasis of breast cancer cells to bone (Bellanger et al., 2017). Increased ZNF217 expression was observed in breast tumors that metastasized to the bones, in comparison to non-metastatic tumors and tumors that metastasized to other tissues. Ιn a breast cancer cell line, MDA-MB-231, high levels of ZNF217 mRNA were also found to be correlated with the deregulation of several genes that are involved in osteogenesis. Among them, BMP-2 and antagonists of the BMP pathway were downregulated while BMP-4 and BMPRIa, BMPRIb and BMPRII were upregulated (Bellanger et al., 2017). These observations exhibit a possible positive feedback loop between ZNF217 and BMP-4 expression, and a negative feedback loop between ZNF217 and BMP-2 expression. BMP-4 is overexpressed in a variety of human tumors and cancer cells lines. Interestingly, the addition of BMP-4 in breast cancer cell lines or the overexpression of its gene has been shown to inhibit their proliferation, but increase their invasive and metastatic potential (Kallioniemi, 2012). Treatment of MCF-7 breast cancer cells with BMP-2 has also been found to cause cell cycle arrest in G1 phase by increasing the production the cell cycle inhibitor p21 (Ghosh-Choudhury et al., 2000). Also, ZNF217 has been found to form a complex with the ubiquitin-ligase that regulates the stability of p53, Mouse Double Minute homolog-2 (MDM2), and, as a result, the transcription of the CDKN1A gene. Possible ZNF217 binding sites have also been found in silico in the promoter of CDKN1A (Mantsou et al., 2016).
The aim of this study was to investigate the effect of active dimeric BMP-2 or a short active decapeptide, derived from its carboxyterminal domain, on the proliferation of MCF-7 breast cancer cells and on the expression of ZNF217 in human dental pulp stem cells (hDPSCs) during induced osteogenesis. The BMP-2-derived peptide AISMLYLDEN has been previously found to induce differentiation of hMSCs more effectively than the active dimeric BMP-2 at concentration 50 ng/ml (Karoulias et al., 2021). It is interesting that, although the correlation between components of the BMP pathway and ZNF217 or CDKN1A has been studied in cancer cells (Ghosh-Choudhury et al., 2000; Bellanger et al., 2017; Chapellier et al., 2015a; Davis et al., 2008), there have not been studies in healthy cells so far. Investigating the relationship between BMP-2 and genes ZNF217, CDKN1A and BMP4 in mesenchymal stem cells will help to understand how these genes are affected during osteogenesis, with and without the addition of BMP-2 or its peptide.
2. Materials and methods
2.1. Materials
The following reagents were used in cell culture, in cell differentiation and in the staining assays: Dulbecco's Modified Eagle's Medium (Thermo Fisher Scientific, USA), StemPro Osteogenesis Kit (Thermo Fisher Scientific, USA), Fetal Bovine Serum (Thermo Fisher Scientific, USA), Penicillin/streptomycin solution (Thermo Fisher Scientific, USA), Phosphate-Buffered Saline (Thermo Fisher Scientific, USA), Trypsin/EDTA solution (BIOSERA, France), Recombinant active human Bone Morphogenetic Protein-2, BMP-2 (CUSABIO, USA) and BMP-2 peptide, AISMLYLDEN (Biomatik, Canada). Additionally, the following chemicals reagents were used in these assays: BCIP (C8H6BrClNOP4), NBT (C40H30N10O·2Cl6), Alizarin Red S (C14H8O4), Formaldehyde (CH2O), Tris (C4H11NO3), Hydrogen chloride (HCl), Sodium chloride (NaCl), Disodium hydrogen phosphate (Na2HPO4) and Sodium dihydrogen phosphate dihydrate (NaH2PO4·2H2O). The cell proliferation assay was conducted with a BrdU cell proliferation kit (MilliporeSigma, USA).
The following reagents were used in gene expression analysis: NucleoSpin RNA Kit (MACHEREY-NAGEL, Germany), M-MuLV Reverse Transcriptase (MINOTECH Biotechnology, Greece), oligo(dT)20 (Jena Bioscience, Germany), dNTP mix (MINOTECH biotechnology, Greece), Dithiothreitol (DTT) solution (MINOTECH biotechnology, Greece) and KAPA SYBR FAST qPCR Kit Master Mix (2×) ABI Prism (MilliporeSigma, Germany).
Disposable supplies were purchased from Thermo Fisher Scientific (USA), KISKER BIOTECH (Germany) and SARSTEDT (Germany).
2.2. Culture and differentiation of hDPSCs
Human Dental Pulp Stem Cells (hDPSCs) are a valuable source of multipotent stem cells and studies highlight their capacity to in vitro differentiate into active osteoblasts (Mortada and Mortada, 2018). For the purposes of this study, human hDPSCs were kindly provided by Assistant Professor A. Bakopoulou from the School of Dentistry, Aristotle University of Thessaloniki. The cells had been established from third molars of young healthy donors, aged 18–24, with the enzymatic dissociation method described in Bakopoulou et al. (2015). The samples had been collected in accordance to all the relevant guidelines and regulations and had been approved by the Institutional Review Board of the Aristotle University of Thessaloniki (Nr. 66/18-06-2018). All the donors signed an informed consent form.
The cells were cultured in Dulbecco's Modified Eagle's Medium (DMEM) supplemented with 10% v/v Fetal Bovine Serum (FBS), 100 U/ml penicillin and 100 μg/ml streptomycin, at 37 °C and 5% CO2. When the cells reached around 80% confluency, they were detached from the culture dish with 0.05% w/v Trypsin/EDTA solution, counted and seeded in 6-well and 12-well plates at a density of 20,000 cells per well. The cells were incubated overnight until they formed monolayers, then two groups of cells were treated with either 50 ng/ml active dimeric human BMP-2 or 50 ng/ml BMP2 peptide in the culture medium (Day 0). Three days later, DMEM was substituted by StemPro Osteogenesis Complete Medium in all groups except for a negative control group which was cultured in DMEM. The medium was refreshed every three days. RNA was extracted on the 7th, the 14th and the 21st day after treatment.
2.3. Culture and treatment of MCF-7 with ΒΜP-2 and BMP-2 peptide
MCF-7 cells were cultured in Dulbecco's Modified Eagle's Medium (DMEM) supplemented with 10% v/v Fetal Bovine Serum (FBS), 100 U/ml penicillin and 100 μg/ml streptomycin, at 37 °C and 5% CO2 until they reached around 70–80% confluency. Then they were seeded in 6-well plates at a density of 105 cells per well and were incubated overnight to form monolayers. Once attached, the cells were treated with 100 ng/ml active dimeric BMP-2 or different concentrations of BMP-2 peptide (45.2 ng/ml, 22.6 ng/ml and 4.52 ng/ml). RNA was extracted 6 h, 12 h, 24 h and 48 h after treatment. For each time point, there was also a negative control group, which was not treated with BMP-2 or BMP-2 peptide (untreated).
2.4. Total RNA extraction
Total RNA was extracted from hDPSCs, osteoblasts and MCF-7 using the NucleoSpin RNA kit, according to instructions. Briefly, the cells were washed with 1× PBS and incubated with 350 μl of lysis buffer containing 1% v/v β-mercaptoethanol for 4–5 min at room temperature. Then, they were scraped from the bottom, collected into 1.5 ml tubes and vortexed vigorously. To reduce viscosity, the lysed cells were transferred into appropriate filters and centrifuged at 13,000 rpm for 1 min. The lysates were mixed with cold 70% v/v ethanol at 1:1 volume ratio and, then, the RNA was bound to NucleoSpin RNA silica-membrane columns by centrifugation at 13,000 rpm for 30 s. Τhe silica membrane was desalted by addition of appropriate buffer and centrifugation at 13,000 rpm for 30 s to allow subsequent DNA digestion by incubation with rDNase solution at room temperature for 15 min. After the digestion of DNA, the membrane-bound RNA was washed twice with buffer containing 80% v/v ethanol and dried by centrifugation at 13,000 rpm for 2 min. RNA was eluted in 60 μl RNase-free H2O, by centrifugation at 13,000 rpm.
The integrity of the purified RNA was checked by electrophoresis in 2% w/v agarose gel and staining with 1× Gel Red Nucleic Acid Gel Stain (Biotium, USA). The bands corresponding to the most abundant RNA molecules, 18S (approximately 2 kb) and 28S rRNA (approximately 5 kb), were intact, indicating that the total RNA had not been hydrolysed during or after the isolation process (Suppl. Fig. 1) (Scientific and Methods to check RNA integrity, n.d.). The concentration of RNA in each sample was determined by measuring the optical density at 260 nm.
Fig. 1.
Evaluation of proliferation of MCF-7 breast cancer cells 24 h and 48 h after treatment with BMP-2 (100 ng/ml) or with BMP-2 peptide (45.2 ng/ml or 22.6 ng/ml) as assessed by the BrdU assay. The optical density (OD) was measured against blank (cell-free and BrdU-free wells) at 450 nm with a reference filter at 690 nm.
2.5. Detection of alkaline phosphatase activity (Alkaline Phosphatase Assay)
The Alkaline Phosphatase Assay (ALP assay) was performed on the 14th day of osteogenesis, on hDPSCs that had been seeded on 12-well plates at a density of 20,000 cells/well and treated with BMP-2 or BMP-2 peptide. The medium was removed and the cell monolayers were washed with 1× PBS and then fixed in 10% formalin solution (10% v/v formaldehyde, 0.4% w/v NaH2PO4.H2O, 0.65% w/v Na2HPO4) at room temperature for a maximum of 60 s (to avoid irreversible inactivation of alkaline phosphatase). Immediately afterwards, formalin was removed and the cells were washed twice with TWEEN 20-PBS solution (1× PBS, 0.05% v/v TWEEN-20). TWEEN 20-PBS was removed and the monolayers were incubated in 1× Alkaline Phosphatase Solution (100 mM Tris-HCl pH 9.5, 100 mM NaCl and 5 mM MgCl2) that contained 0.45% v/v of each of the substrates NBT and BCIP, at 37 °C until areas with blue NBT precipitate started to appear on the monolayers. The enzymatic activity was terminated by washing the cells with TWEEN 20-PBS solution. Finally, TWEEN 20-PBS was removed and 1× PBS was added in all the wells. The cells were observed with a Nikon ECLIPSE TS-100 inverted optical microscope (objective lenses: 10× and 40×, ocular lens: 10×) and photographs of the stained monolayers were taken at 100× and 400× magnification using a Nikon COOLPIX P5100 camera.
2.6. Staining of extracellular calcium deposits (Alizarin Red S assay)
The Alizarin Red staining assay was performed on the 21st day of differentiation, on hDPSCs that had been seeded on 12-well plates at a density of 20,000 cells/well and treated with BMP-2 or BMP-2 peptide. The medium was removed and the cells were washed with 1× PBS and then fixed in 10% formalin solution (10% v/v formaldehyde, 0.4% w/v NaH2PO4.H2O, 0.65% w/v Na2HPO4) at room temperature for 30 min. Afterwards, formalin was removed, the monolayers were washed twice with sterile distilled H2O and incubated in 2% w/v Alizarin Red S solution at room temperature for 25–45 min. Then, the cells were washed multiple times with sterile distilled water to remove excess dye. The cells were observed with a Nikon ECLIPSE TS-100 inverted optical microscope (objective lenses: 10× and 40×, ocular lens: 10×) and photographs of the stained monolayers were taken at 100× and 400× magnification using a Nikon COOLPIX P5100 camera.
2.7. cDNA synthesis
First-strand cDNA was synthesized from each total RNA sample with M-MuLV Reverse Transcriptase (MINOTECH biotechnology, Greece). Each reaction was set up with 0.5 ng/ml RNA, 5 μΜ Oligo(dT)20 (Jena Bioscience, Germany) and 500 μΜ of each dNTP (MINOTECH biotechnology, Greece). The mixture was heated to 65 °C for 5 min to denature secondary structures of the RNA and the Oligo(dT)20 and, immediately, cooled on ice for 1 min. 200 U of Reverse transcriptase (RT), 1× RT Buffer and 5 μΜ DTT (MINOTECH biotechnology, Greece) were added and the mixture was incubated at 25 °C for 5 min for efficient annealing of Oligo(dT)20 on the RNA templates. First-strand cDNA synthesis was performed at 37 °C for 1 h, then the enzyme was inactivated at 70 °C for 15 min.
2.8. Real time qPCR
Relative quantification of gene expression against reference gene RPLP0 was performed on a StepOne Real time PCR System (Thermo Fisher Scientific, USA), using KAPA SYBR FAST qPCR Kit Master Mix (2Χ) ABI PRISM. RPLP0 has been found to be an optimal housekeeping gene for studying the differentiation of hMSCs into osteoblasts and one of the genes with the most stable expression in breast tumors, therefore it was selected as the reference gene for all the experiments (Ragni et al., 2013; Wang et al., 2015). Primers for ZNF217, BMP4, CDKN1A, BGLAP and RPLP0 were purchased from Eurofins Genomics (Germany) and Thermo Fisher Scientific (USA). Each 10 μl-reaction was prepared with cDNA (1 μl), gene-specific forward and reverse primers (1 pmol of each) and 1× Master Mix. The annealing temperature that was used for all the primer pairs was 60 °C. The qPCR program that was selected for all the reactions consisted of the following stages: an initial denaturation stage at 95 °C for 20 s, followed by 40 cycles of amplification (denaturation at 95 °C for 3 s, annealing and extension at 60 °C for 20 s). The primer sequences are listed in Table 1.
Table 1.
Sequences of the primers designed for qPCR.
| Primer | Sequence (5′ to 3′) |
|---|---|
| ZNF217 forward | CAGCGAGGTCGATTCTCCAA |
| ZNF217 reverse | GGCCTTTTTCCTTCTAACGTCG |
| CDKN1A forward | GCAGACCAGCATGACAGATTTC |
| CDKN1A reverse | ATGTAGAGCGGGCCTTTGAG |
| BMP4 forward | GGAGGAGGAGGAAGAGCAGA |
| BMP4 reverse | CCAGATGTTCTTCGTGGTGGA |
| BGLAP forward | CCTCACACTCCTCGCCCTAT |
| BGLAP reverse | TGCTTGGACACAAAGGCTGC |
| RPLP0 forward | AATGTGGGCTCCAAGCAGAT |
| RPLP0 reverse | TGAGGCAGCAGTTTCTCCAG |
2.9. Cell proliferation assay
The proliferation rate of MCF-7 cells was investigated by the BrdU (5-bromo-2-deoxyuridine) assay (MilliporeSigma, USA). MCF-7 cells were seeded in 96-well plates at a density of 10,000 cells/well and were allowed to attach overnight at 37 °C and 5% CO2. Then the culture medium was renewed (DMEM supplemented with 10% FBS, 100 U/ml penicillin and 100 μg/ml streptomycin) and cells were treated with 100 ng/ml BMP-2, 45.2 ng/ml BMP-2 peptide or 22.6 ng/ml BMP-2 peptide, for 24 h and 48 h. After these time points, 1× BrdU labeling solution was added to each well, the cells were incubated for 4 h at 37 °C and 5% CO2 and then the incorporated BrdU was detected according to user protocol. Briefly, the plate contents were removed completely, and the cells were fixed in Fixative/Denaturing solution for 30 min at room temperature. Then, the solution was removed and the cells were incubated with 1× anti-BrdU antibody solution for 1 h at room temperature. The wells were washed thoroughly with solution containing PBS and surfactant and blotted on paper gently, before addition of 1× anti-mouse IgG-HRP conjugate and incubation for 30 min at room temperature. The wells were washed again and blotted on paper to remove secondary antibody and incubated in substrate solution (tetra-methylbenzidine) in the dark at room temperature for 7–9 min. Afterwards, the enzymatic reaction was terminated with 2.5 N sulfuric acid (Stop solution) and the absorbance was measured in a spectrophotometric plate reader at a wavelength of 450 nm and a reference filter of 690 nm. Cell-free and BrdU-free wells served as internal controls for this assay. The resulting OD values of those wells were used as blank (negative control) and background control (positive control), respectively. The experiment was conducted in 6-plicates.
2.10. Data analysis
The real-time PCR reactions were performed in 3-plicates and the Ct values were analyzed on Microsoft Excel 365. Relative quantification of the expression of the genes ΖΝF217, BMP4, CDKN1A and BGLAP was performed with the 2−ΔΔCt method (Livak method) (Livak and Schmittgen, 2001). The mean Ct values of these genes were normalized against the mean Ct value of the housekeeping gene RPLP0 in each sample and specific samples in each experiment were used as calibrators. These samples were the 7-day untreated (negative control) hDPSCs in the experiment that involved the osteogenic differentiation of HDPSCs and the 6-hour untreated MCF-7 cells in the experiment in which the effect of BMP-2 and the BMP-2 peptide was studied on the expression of ΖΝF217, BMP4 and CDKN1A in MCF-7 cells. The bar charts that depict the fold changes in gene expression and the mean absorbance values in the BrdU cell proliferation assay were constructed on GraphPad Prism 8.
3. Results and discussion
3.1. Effect of BMP-2 and the BMP-2 peptide on the proliferation of MCF-7 breast cancer cells and on the expression of ZNF217, BMP4 and CDKN1A
To assess the effect of BMP-2 and the peptide AISMLYLDEN on the proliferation of MCF-7 cells, a Bromodeoxyuridine (BrdU) labeling assay was performed. BrdU, a thymidine analog, is incorporated into newly synthesized DNA strands of proliferating cells and can be determined immunohistochemically. The colored product of a reaction catalyzed by horseradish peroxidase which is conjugated to the secondary antibody, can be determined by measuring the absorbance at 450 nm. The absorbance is proportional to the amount of incorporated BrdU, therefore to the number of actively proliferating cells (Millipore Sigma, BrdU cell proliferation assay user protocol, n.d.). The assay was performed 24 h and 48 h after the addition of BMP-2 (100 ng/ml) or the BMP-2 peptide (45.2 ng/ml or 22.6 ng/ml) in MCF-7 cultures. At 24 h, no apparent effect on cell proliferation was observed in BMP-2-treated MCF-7, but 27–28% reduction in the number of actively proliferating cells was observed in the peptide treated groups, compared to untreated MCF-7 (Fig. 1). At 48 h, 30% reduction of proliferation was detected in BMP-2-treated cells, 23% reduction in cells treated with 45.2 ng/ml of BMP-2 peptide and 42% reduction in cells treated with 22.6 ng/ml of BMP-2 peptide, compared to untreated MCF-7 (Fig. 1). Therefore, both BMP-2 and the peptide showed significant efficiency in inhibiting the proliferation of MCF-7 cells when added exogenously. 48 h after it had been added in the culture medium, BMP-2 had strong effect on cell proliferation The peptide had more immediate effect than BMP-2 and its effect on the cells increased overtime, from 24 h to 48 h, when it was added at the concentration of 22.6 ng/ml.
The mRNA levels of ZNF217, BMP4 and CDKN1A were monitored by real-time qPCR 6, 12, 24 and 48 h after treatment of MCF-7 cells with 100 ng/ml BMP-2 or different concentrations of BMP-2 peptide (45.2 ng/ml, 22.6 ng/ml and 4.52 ng/ml). The genes' expression was studied at multiple time points within a 48-hour span according to previous studies (Ghosh-Choudhury et al., 2000; Wang et al., 2012; Wang et al., 2011; Chen et al., 2012; Clement et al., 2000) and the concentration of 100 ng/ml BMP-2 was selected according to studies on the effect of BMP-2 on gene expression in MCF-7 cells (Ghosh-Choudhury et al., 2000; Steinert et al., 2008). The effect of the BMP-2 peptide on MCF-7 was studied at a concentration of 4.52 ng/ml which was equimolar to BMP-2 (3.87 nM), for comparison, and at concentrations 5 times (22.6 ng/ml) and 10 times higher (45.2 ng/ml), to observe if there are concentration-dependent effects on the genes' expression. The mRNA levels of ZNF217 in untreated MCF-7 at 6 h of culture were set as the “reference levels” for ZNF217. Similarly, the mRNA levels of BMP4 and CDKN1A in untreated MCF-7 at 6 h of culture were set as the reference levels for each of these genes, respectively. In conjunction with the findings of the BrdU assay, the gene expression results are analyzed and discussed at 24 h and 48 h, and at the most effective peptide concentrations, (45.2 ng/ml and 22.6 ng/ml).
In untreated MCF-7 cells, ZNF217 mRNA levels were 1.5–2-fold higher than reference levels at 24 h and 48 h (Fig. 2a). In BMP-2-treated cells, ZNF217 expression was 5-fold higher than the reference level at 24 h, but equal to the untreated group at 48 h. MCF-7 treated with 45.2 ng/ml BMP-2 peptide had low ZNF217 mRNA levels at 24 h (equal to reference levels) but increased levels at 48 h. On the other hand, cells treated with 22.6 ng/ml BMP-2 peptide had decreased ZNF217 expression at both time points (Fig. 2a). Therefore, at a molar concentration ten times that of BMP-2, the peptide had an upregulatory effect on ZNF217 expression at 48 h. However, an intermediate concentration of 22.6 ng/ml BMP-2 peptide (5 times the molar concentration of BMP-2) was associated with ZNF217 expression lower than (24 h) or almost equal to (48 h) the untreated group.
Fig. 2.
Fold change of ZNF217 (a), BMP-4 (b) and CDKN1A (c) mRNA levels at 6 h, 12 h, 24 h and 48 h after treatment of MCF-7 cultures with 100 ng/ml BMP-2 or different concentrations (45.2 ng/ml, 22.6 ng/ml and 4.52 ng/ml) BMP-2 peptide. The mRNA levels of untreated MCF-7 at 6 h were determined as the reference levels in all of the cases, a, b and c. The concentration of 4.52 ng/ml BMP-2 peptide is equimolar to 100 ng/ml BMP-2. “Un”: Undetected.
BMP-4 has been associated with decreased cell proliferation and increased migration in vitro. Treatment with recombinant BMP-4 decreased cell count in 16 breast cancer cell lines, including MCF-7, after 2 days, by inducing cell cycle arrest in G1 (Ketolainen et al., 2010). Also, MCF-7 and MDA-MB-231 transfected with BMP-4-expressing adenoviral vector showed decreased cell population after 2 to 5 days and increased migration after 24 to 36 h, as determined by MTT assay and wound healing and transwell assays, respectively (Guo et al., 2012). These cell lines had high expression of the genes that encode Matrix Metalloproteinase-1 (MMP1) and C-X-C chemokine receptor type 4 (CXCR4). Ιn this study, untreated MCF-7, BMP4 mRNA levels decreased 3-fold at 24 h compared to the 6-h reference level and, then, increased 2-fold the reference levels at 48 h (Fig. 2b). In BMP-2-treated cells, BMP4 mRNA was undetected at 24 h and increased at 48 h reaching equal levels to the untreated group. MCF-7 that were treated with 45.2 ng/ml or 22.6 ng/ml BMP-2 peptide had higher BMP-4 mRNA levels at 24 h and 48 h than untreated and BMP-2-treated cells.
BMP-2 has been associated with cell cycle arrest and inhibition of invasion of breast cancer cells (Davis et al., 2008; Chapellier et al., 2015b). MTT and immunoblotting assays have demonstrated that BMP-2 inhibits estradiol-induced proliferation of MCF-7 cells, by inducing the production of p21, which leads to inactivation of CDK-2 and hypophosphorylation of Rb (Ghosh-Choudhury et al., 2000). p21 is a cell-cycle inhibitor belonging to the Cip/Kip family of Cyclin-dependent kinase inhibitors. It is the product of the CDKN1A gene and it is known to bind to and inhibit cyclin- CDK2 complexes during G1 and S phases, causing cell cycle arrest in G1, in case of DNA damage and other types of cellular stress. However, it has also been shown that p21 increases normally in response to mitogenic signaling, through the ERK pathway, to form complexes with D-CDK-4/6 and activate them. In response to cellular stress, CDKN1A expression is induced in a p53-dependent manner and high levels of the protein accumulate, thus inhibiting CDK2 complexes and preventing the progression of S-phase (DNA replication) and Mitosis (Barnum and O'Connell, 2014; Karimian et al., 2016; Fischer et al., 2016). In this study, in untreated MCF-7, CDKN1A mRNA levels dropped 334-fold at 24 h and returned to reference levels at 48 h (Fig. 2c). However, treatment with 100 ng/ml BMP-2 or the higher concentrations of BMP-2 peptide lessened this downregulatory phenomenon. BMP-2-treated MCF-7 showed only 36-fold reduction in CDKN1A expression at 24 h, MCF-7 treated with 45.2 ng/ml BMP-2 peptide showed 42-fold and, those treated with 22.6 ng/ml BMP-2 peptide, 54-fold reduction in CDKN1A mRNA levels at 24 h. Additionally, the MCF-7 cells that had been treated with 22.6 ng/ml BMP-2 peptide had a 3-fold increase in CDKN1A expression at 48 h, compared to reference levels, which was not observed in the aforementioned groups (Fig. 2c). Therefore, treatment of MCF-7 with 100 ng/ml BMP-2 was associated with higher CDKN1A mRNA levels at 24 h after treatment, but no significant change was observed before or after this time point, compared to untreated cells. Treatment with the BMP-2 peptide, at a molar concentration 5 times that of BMP-2 (22.6 ng/ml), was associated with higher CDKN1A mRNA levels at 24 h but also with 3-fold increase at 48 h, compared to the untreated group. In most of the MCF-7 groups, including the untreated, it was observed that ZNF217 mRNA levels peaked 12 h before CDKN1A mRNA levels dropped (Fig. 2a and c), indicating the existence of a possible negative correlation between the two genes, regardless of the presence or absence of the BMP-2 peptide in their medium.
3.2. Effect of the BMP-2 peptide on osteogenic differentiation of hDPSCs and on the expression of ZNF217, BMP4 and CDKN1A
In parallel, the effect of BMP-2 and the peptide AISMLYLDEN on osteogenic differentiation of hDPSCs and on the expression of genes ZNF217, BMP4 and CDKN1A was investigated. Four groups were tested: a) a negative control group, consisting of untreated hDPSCs cultured in DMEM, b) a positive control group, consisting of hDPSCs that were induced to differentiate with osteogenesis medium, c) hDPSCs that were induced to differentiate with osteogenesis medium and were also treated with 50 ng/ml active dimeric human BMP-2, and d) hDPSCs that were induced to differentiate with osteogenesis medium and were also treated with 50 ng/ml BMP-2 peptide.
During the formation of their extracellular matrix (approximately 7th to 14th day of osteogenesis), proliferating osteoblasts exhibit high expression of the enzyme Alkaline Phosphatase (ALP). ALP activity can be detected by chromogenic reaction (NBT-BCIP staining) (Kunimatsu et al., 2018; Kim et al., 2019; Hoemann et al., 2009; Yuan et al., 2019; Wang et al., 2018). On the 14th day, higher alkaline phosphatase activity was observed in hDPSCs that had been treated with BMP-2 or BMP-2 peptide, compared to hDPSCs that differentiated only in the presence of osteogenesis medium, as indicated by the NBT-formazan deposits (Fig. 3a–d). Purple staining was observed in the negative control hDPSCs, which probably indicates that proliferating hMSCs produced oxidative compounds that reduced NBT (Chen et al., 2008; Atashi et al., 2015). The morphology of the positive control and the treated groups became polygonal, while negative control cells maintained the spindle-shaped, fibroblast-like morphology that is typical of hDPSCs (Fig. 3) (Suchanek et al., 2009; Awais et al., 2020). At the final stage of osteogenic differentiation, osteoblasts form hydroxyapatite deposits in the extracellular matrix (mineralization). On the 21st day of osteogenesis, bone ECM mineralization was verified by staining the extracellular hydroxyapatite deposits with Alizarin Red S, a dye that binds to calcium cations (Kunimatsu et al., 2018; Kim et al., 2019; Hoemann et al., 2009; Yuan et al., 2019; Wang et al., 2018). The Alizarin Red Assay showed that both the BMP-2-treated and BMP-2 peptide-treated cells had higher concentration of calcium deposits compared to positive control cells. The highest concentration was observed in cells treated with BMP-2 peptide (Fig. 3e–h). Additionally, the mRNA levels of the osteocalcin gene, BGLAP, were monitored by qPCR (Fig. 4). The production of osteocalcin mRNA was high in all the groups that had been induced to differentiate. Treatment with 50 ng/ml BMP-2 increased the expression of osteocalcin mRNA by 50-fold on Day 21 and treatment with 50 ng/ml BMP-2 peptide increased the expression by almost 500-fold, compared to reference levels (Fig. 4). According to the results of these assays, osteogenic differentiation was enhanced on the 21st day.
Fig. 3.
Staining assays for the verification of osteogenesis. (a–d): Alkaline Phosphatase Assay performed on the 14th day of osteogenesis, for the verification of successful differentiation of hDPSCs into osteoblasts. a) Untreated hDPSCs in DMEM (negative control), b) hDPSCs in osteogenesis medium (positive control), c) BMP-2-treated hDPSCs in osteogenesis medium, d) BMP-2 peptide-treated hDPSCs in osteogenesis medium. Left: 100× magnification, right: 400× magnification. (e–h): Alizarin Red Assay performed on the 21st day of osteogenesis, for the verification of successful mineralization of the extracellular matrix. e) Untreated hDPSCs in DMEM (negative control), f) hDPSCs in osteogenesis medium (positive control), g) BMP-2-treated hDPSCs in osteogenesis medium, h) BMP-2 peptide-treated hDPSCs in osteogenesis medium. Left: 100× magnification, right: 400× magnification. Dashed-line boxes indicate polygonal cells in the differentiated groups.
Fig. 4.
Fold change (FC) of BGLAP mRNA levels of osteoblasts compared to Day 7 Untreated hDPSCs. Untreated: hDPSCs cultured in DMEM, without factors that promote differentiation, Osteo: hDPSCs treated with osteogenesis medium, Osteo + BMP-2: hDPSCs treated with osteogenesis medium and 50 ng/ml BMP-2, Osteo + peptide: hDPSCs treated with osteogenesis medium and 50 ng/ml BMP-2 peptide.
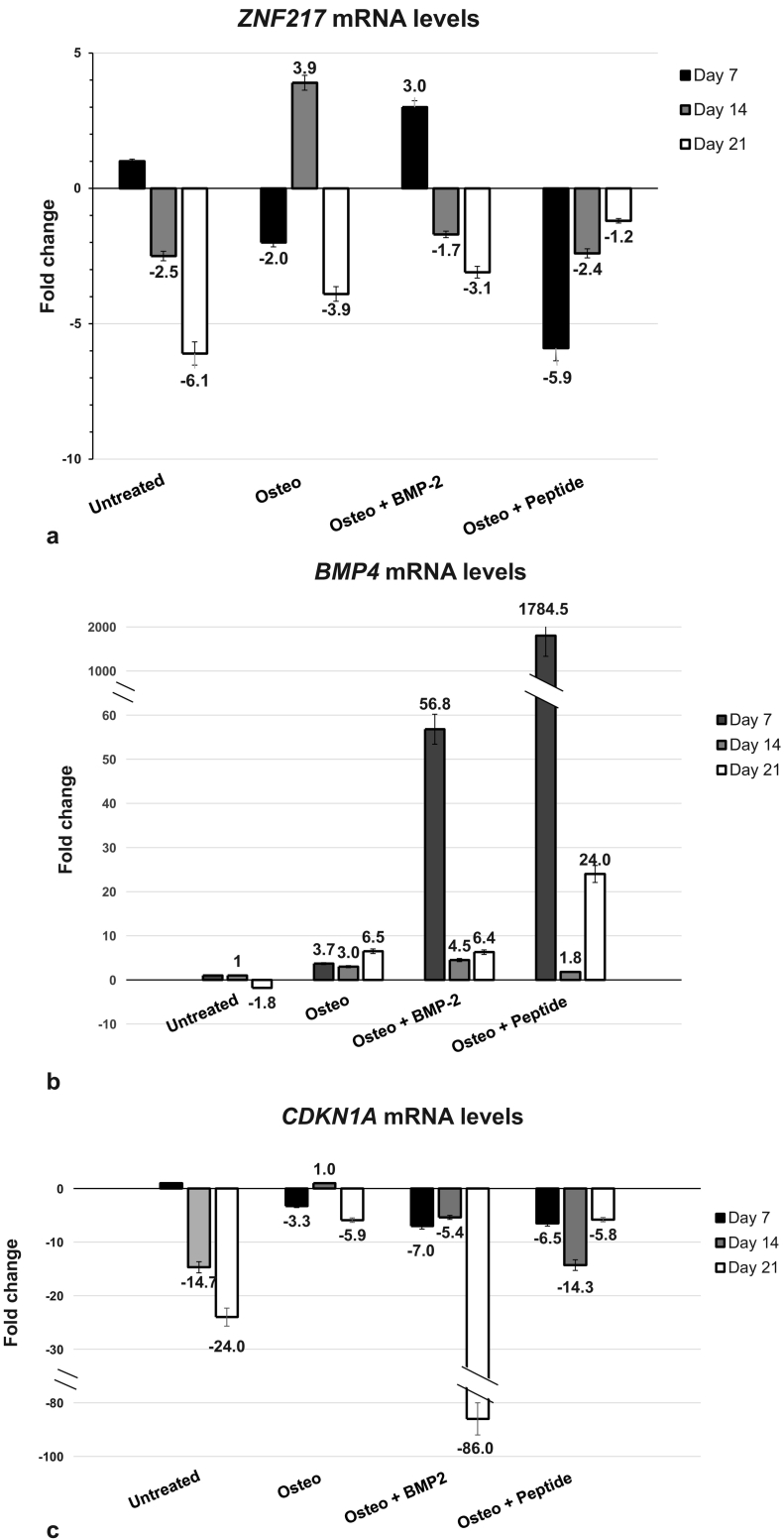
The mRNA levels of ZNF217, BMP4 and CDKN1A (p21) were quantified by real-time qPCR. On the 21st day of osteogenic differentiation, in the positive control group, ZNF217 mRNA levels decreased 4-fold below reference level (Fig. 5a). hDPSCs treated with 50 ng/ml recombinant human BMP-2 or BMP-2 peptide also had low ZNF217 levels on the 21st day.
Fig. 5.
Fold change of ZNF217 (a), BMP-4 (b) and CDKN1A (c) mRNA levels in hDPSCs and osteoblasts at 7, 14 and 21 days after induction of osteogenesis. Untreated: hDPSCs cultured in DMEM, without factors that promote differentiation, Osteo: hDPSCs treated with osteogenesis medium, Osteo + BMP-2: hDPSCs treated with osteogenesis medium and 50 ng/ml BMP-2, Osteo + peptide: hDPSCs treated with osteogenesis medium and 50 ng/ml BMP-2 peptide. The mRNA levels of untreated hDPSCs at 7 days were determined as the reference levels in all the cases, a, b and c.
BMP-4 is a major regulator of osteoblastic differentiation and is known to act through the same receptors as BMP-2 (Miyazono et al., 2010; Yang et al., 2014; Kang et al., 2004; Bandyopadhyay et al., 2006). Induction of differentiation with osteogenesis medium was accompanied by 6.5-fold increase in BMP4 mRNA levels on the 21st day (Fig. 5b). On the same day, BMP-2-treated osteocytes had equal BMP4 mRNA levels to the positive control and the peptide-treated osteocytes had about 3.5 times higher levels. Evidently, the short peptide, AISMLYLDEN, is a potent promoter of osteogenesis, increasing the production of another bone morphogenetic factor, BMP-4, early in the differentiation process and keeping it at relatively high levels even after the mineralization process has begun.
In the positive control group, CDKN1A mRNA levels decreased 6-fold below the reference level on the 21st day (Fig. 5c). In BMP-2-treated and in peptide-treated cells, reduction in CDKN1A mRNA was observed throughout the osteogenic differentiation process. On the 21st day, in peptide-treated osteocytes, CDKN1A mRNA levels were equal to the positive control's levels.
4. Conclusions
Altogether, these results show that BMP-2 and the BMP-2 peptide, AISMLYLDEN, can reduce the proliferation of MCF-7 cells. The peptide at 22.6 ng/ml, which was the most effective concentration at reducing cell proliferation, was associated with increase in CDKN1A and BMP4 and decrease in ZNF217 mRNA levels. In particular, the upregulation of genes CDKN1A and BMP4, respectively, as described in detail within the text above is of great importance due to their implication in cell cycle arrest. In addition, ZNF217 mRNA levels that are associated with promotion of cancer cell proliferation are downregulated. Taken together, the assessments regarding the regulation of the above described factors clearly indicate the desired influence of peptide AISMLYLDEN on their cell proliferation implication. Thus, these results open a “possible road” for concurrent use of the peptide AISMLYLDEN, at appropriate concentrations, as a novel treatment during conventional therapy, in patients with breast tumors with a metastatic tendency to the bones.
In addition, the treatment of hDPSCs with 50 ng/ml BMP-2 peptide promoted the osteogenic differentiation of hDPSCs effectively and did not alter significantly the expression of genes ZNF217, CDKN1A and BMP4, as observed on the 21st day of differentiation, when both the differentiation and the bone extracellular matrix mineralization processes had begun.
5. Future work
In the future, it would be interesting to extend the study to other breast cancer cell lines and primary breast cancer cells with metastatic tendency to the bones. An important step towards the clarification of how BMP-2 and the peptide affect cell proliferation and the genes in the present study is the investigation of the induced signaling pathways (ERK/p38- or SMAD-1/5/8-pathways) in each case, in MCF-7 and other breast cancer cell lines.
The following is the supplementary data related to this article.
Agarose gel electrophoresis of total RNA from (a) hDPSCs during osteogenic differentiation and (b) MCF-7 cells. a) Lanes 1, 5, 9: untreated hDPSCs in DMEM at 7, 14 and 21 days. Lanes 2, 6, 10: hDPSCs in osteogenesis medium at 7, 14 and 21 days. Lanes 3, 9, 11: hDPSCs in osteogenesis medium and treated with 50 ng/ml BMP-2. Lanes 4, 8, 12: hDPSCs in osteogenesis medium and treated with 50 ng/ml BMP-2 peptide. b): Lanes 1, 6, 11, 16: untreated MCF-7 at 6 h, 12 h, 24 h and 48 h. Lanes 2, 7, 12, 17: MCF-7 treated with 100 ng/ml BMP-2 at 6 h, 12 h, 24 h and 48 h. Lanes 3, 8, 13, 18: MCF-7 treated with 45.2 ng/ml BMP-2 peptide at 6 h, 12 h, 24 h and 48 h. Lanes 4, 9, 14, 19: MCF-7 treated with 22.6 ng/ml BMP-2 peptide at 6 h, 12 h, 24 h and 48 h. Lanes 5, 10, 15, 20: MCF-7 treated with 4.52 ng/ml BMP-2 peptide at 6 h, 12 h, 24 h and 48 h. The bands corresponding to the 28S rRNA and 18 S rRNA are visible.
CRediT authorship contribution statement
Aglaia Mantsou: Conceptualization, Methodology, Investigation, Formal analysis, Writing – original draft, Writing – review & editing, Visualization, Funding acquisition. Maria Pitou: Writing – review & editing. Eleni Papachristou: Investigation, Validation. Rigini M. Papi: Resources, Writing – review & editing, Funding acquisition. Paraskevas Lamprou: Validation, Supervision, Funding acquisition. Theodora Choli-Papadopoulou: Conceptualization, Methodology, Validation, Resources, Writing – original draft, Writing – review & editing, Supervision, Project administration, Funding acquisition.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was supported by the European Regional Development Fund of the European Union and Greek national funds through the Operational Program Competitiveness, Entrepreneurship and Innovation, under the call RESEARCH – CREATE – INNOVATE (Project code: T1EDK-04567) and by the Hellenic Foundation for Research and Innovation (Fellowship number: 371). We would like to thank Assistant Professor A. Bakopoulou of the Laboratory of Fixed Prosthesis and Implant Prosthodontics of the School of Dentistry, Aristotle University of Thessaloniki, for providing the hDPSCs. Also, we thank Dr. S. Poulios for his assistance in the qPCR reactions which were performed at the Department of Genetics, Development and Molecular Biology of the School of Biology, Aristotle University of Thessaloniki.
References
- Arnold S.F., Tims E., Mcgrath B.E. Identification of bone morphogenetic proteins and their receptors in human breast cancer cell lines: importance of BMP-2. Cytokine. 1999;11:1031–1037. doi: 10.1006/cyto.1999.0508. [DOI] [PubMed] [Google Scholar]
- Atashi F., Modarressi A., Pepper M.S. The role of reactive oxygen species in mesenchymal stem cell adipogenic and osteogenic differentiation: a review. Stem Cells Dev. 2015;24:1150–1163. doi: 10.1089/scd.2014.0484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attisano L., Wrana J.L. Smads as transcriptional co-modulators. Curr. Opin. Cell Biol. 2000;12:235–243. doi: 10.1016/S0955-0674(99)00081-2. [DOI] [PubMed] [Google Scholar]
- Awais S., Balouch S.S., Riaz N., Choudhery M.S. Human dental pulp stem cells exhibit osteogenic differentiation potential. Open Life Sci. 2020;15:229–236. doi: 10.1515/biol-2020-0023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakopoulou A., Leyhausen G., Volk J., Papachristou E., Koidis P., Geurtsen W. Wnt/ß-catenin signaling regulates dental pulp stem cells’ responses to pulp injury by resinous monomers. Dent. Mater. 2015;31:542–555. doi: 10.1016/j.dental.2015.02.004. [DOI] [PubMed] [Google Scholar]
- Bandyopadhyay A., Tsuji K., Cox K., Harfe B.D., Rosen V., Tabin C.J. Genetic analysis of the roles of BMP2, BMP4, and BMP7 in limb patterning and skeletogenesis. PLoS Genet. 2006;2:2116–2130. doi: 10.1371/journal.pgen.0020216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnum K.J., O’Connell M.J. Cell cycle regulation by checkpoints. Methods Mol. Biol. 2014;1170:29–40. doi: 10.1007/978-1-4939-0888-2_2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beederman M., Lamplot J.D., Nan G., Wang J., Liu X., Yin L., Li R., Shui W., Zhang H., Kim S.H., Zhang W., Zhang J., Kong Y., Denduluri S., Rogers M.R., Pratt A., Haydon R.C., Luu H.H., Angeles J., Shi L.L., He T.-C. BMP signaling in mesenchymal stem cell differentiation and bone formation. J. Biomed. Sci. Eng. 2013;6:32–52. doi: 10.4236/jbise.2013.68a1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellanger A., Donini C.F., Vendrell J.A., Lavaud J., Machuca-Gayet I., Ruel M., Vollaire J., Grisard E., Gyorffy B., Bièche I., Peyruchaud O., Coll J.L., Treilleux I., Maguer-Satta V., Josserand V., Cohen P.A. The critical role of the ZNF217 oncogene in promoting breast cancer metastasis to the bone. J. Pathol. 2017;242:73–89. doi: 10.1002/path.4882. [DOI] [PubMed] [Google Scholar]
- Bragdon B., Moseychuk O., Saldanha S., King D., Julian J., Nohe A. Bone morphogenetic proteins: a critical review. Cell. Signal. 2011;23:609–620. doi: 10.1016/j.cellsig.2010.10.003. [DOI] [PubMed] [Google Scholar]
- Chapellier M., Bachelard-Cascales E., Schmidt X., Clément F., Treilleux I., Delay E., Jammot A., Ménétrier-Caux C., Pochon G., Besançon R., Voeltzel T., Caron De Fromentel C., Caux C., Blay J.Y., Iggo R., Maguer-Satta V. Disequilibrium of BMP2 levels in the breast stem cell niche launches epithelial transformation by overamplifying BMPR1B cell response. Stem Cell Rep. 2015;4:239–254. doi: 10.1016/j.stemcr.2014.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapellier M., Bachelard-Cascales E., Schmidt X., Clément F., Treilleux I., Delay E., Jammot A., Ménétrier-Caux C., Pochon G., Besançon R., Voeltzel T., Caron De Fromentel C., Caux C., Blay J.Y., Iggo R., Maguer-Satta V. Disequilibrium of BMP2 levels in the breast stem cell niche launches epithelial transformation by overamplifying BMPR1B cell response. Stem Cell Rep. 2015;4:239–254. doi: 10.1016/j.stemcr.2014.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C.-T., Shih Y.-R.V., Kuo T.K., Lee O.K., Wei Y.-H. Coordinated changes of mitochondrial biogenesis and antioxidant enzymes during osteogenic differentiation of human mesenchymal stem cells. Stem Cells. 2008;26:960–968. doi: 10.1634/stemcells.2007. [DOI] [PubMed] [Google Scholar]
- Chen A., Wang D., Liu X., He S., Yu Z., Wang J. Inhibitory effect of BMP-2 on the proliferation of breast cancer cells. Mol. Med. Rep. 2012;6:615–620. doi: 10.3892/mmr.2012.962. [DOI] [PubMed] [Google Scholar]
- Clement J.H., Marr N., Meissner A., Schwalbe M., Sebald W., Kliche K.-O., Hoè ken K., Woè S. Bone morphogenetic protein 2 (BMP-2) induces sequential changes of Id gene expression in the breast cancer cell line MCF-7. J. Cancer Res. Clin. Oncol. 2000;126:271–279. doi: 10.1007/s004320050342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins C., Volik S., Kowbel D., Ginzinger D., Ylstra B., Cloutier T., Hawkins T., Predki P., Martin C., Wernick M., Kuo W.-L., Alberts A., Gray J.W. Comprehensive genome sequence analysis of a breast cancer amplicon. Genome Res. 2001;11:1034–1042. doi: 10.1101/gr.174301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowger J.J.M., Zhao Q., Isovic M., Torchia J. Biochemical characterization of the zinc-finger protein 217 transcriptional repressor complex: identification of a ZNF217 consensus recognition sequence. Oncogene. 2007;26:3378–3386. doi: 10.1038/sj.onc.1210126. [DOI] [PubMed] [Google Scholar]
- Davis S.R., Watkins G., Douglas-Jones A., Mansel R., Jiang W.G. Bone morphogenetic proteins 1 to 7 in human breast cancer, expression pattern and clinical/prognostic relevance. J. Exp. Ther. Oncol. 2008;7:327–338. [PubMed] [Google Scholar]
- Fischer M., Quaas M., Steiner L., Engeland K. The p53–p21-DREAM-CDE/CHR pathway regulates G2/M cell cycle genes. Nucleic Acids Res. 2016;44:164–174. doi: 10.1093/nar/gkv927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh-Choudhury N., Ghosh-Choudhury G., Celeste A., Ghosh P.M., Moyer M., Abboud S.L., Kreisberg J. Bone morphogenetic protein-2 induces cyclin kinase inhibitor p21 and hypophosphorylation of retinoblastoma protein in estradiol-treated MCF-7 human breast cancer cells. Biochim. Biophys. Acta. 2000;1497:186–196. doi: 10.1016/s0167-4889(00)00060-4. [DOI] [PubMed] [Google Scholar]
- Guo D., Huang J., Gong J. Bone morphogenetic protein 4 (BMP4) is required for migration and invasion of breast cancer. Mol. Cell. Biochem. 2012;363:179–190. doi: 10.1007/s11010-011-1170-1. [DOI] [PubMed] [Google Scholar]
- Hoemann C.D., El-Gabalawy H., McKee M.D. In vitro osteogenesis assays: influence of the primary cell source on alkaline phosphatase activity and mineralization. Pathol. Biol. 2009;57:318–323. doi: 10.1016/j.patbio.2008.06.004. [DOI] [PubMed] [Google Scholar]
- Hogan B.L. Bone morphogenetic proteins: multifunctional regulators of vertebrate development. Genes Dev. 1996;10:1580–1594. doi: 10.1101/gad.10.13.1580. [DOI] [PubMed] [Google Scholar]
- Kallioniemi A. Bone morphogenetic protein 4-a fascinating regulator of cancer cell behavior. Cancer Genet. 2012;205:267–277. doi: 10.1016/j.cancergen.2012.05.009. [DOI] [PubMed] [Google Scholar]
- Kang Q., Sun M.H., Cheng H., Peng Y., Montag A.G., Deyrup A.T., Jiang W., Luu H.H., Luo J., Szatkowski J.P., Vanichakarn P., Park J.Y., Li Y., Haydon R.C., He T.C. Characterization of the distinct orthotopic bone-forming activity of 14 BMPs using recombinant adenovirus-mediated gene delivery. Gene Ther. 2004;11:1312–1320. doi: 10.1038/sj.gt.3302298. [DOI] [PubMed] [Google Scholar]
- Karimian A., Ahmadi Y., Yousefi B. Multiple functions of p21 in cell cycle, apoptosis and transcriptional regulation after DNA damage. DNA Repair (Amst) 2016;42:63–71. doi: 10.1016/j.dnarep.2016.04.008. [DOI] [PubMed] [Google Scholar]
- Karoulias S.-Z., Pitou M., Papi R., Lamprou P., Choli-Papadopoulou T. Specific amino acids from the broad C-terminal region of BMP-2 are crucial for osteogenesis. Bone Rep. 2021;14 doi: 10.1016/j.bonr.2021.101092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katagiri T., Imada M., Yanai T., Suda T., Takahashi N., Kamijo R. Identification of a BMP-responsive element in Id1, the gene for inhibition of myogenesis. Genes Cells. 2002;7:949–960. doi: 10.1046/j.1365-2443.2002.00573.x. [DOI] [PubMed] [Google Scholar]
- Kawabata M., Imamura T., Miyazono K. Signal transduction by bone morphogenetic proteins. Cytokine Growth Factor Rev. 1998;9:49–61. doi: 10.1016/S1359-6101(97)00036-1. [DOI] [PubMed] [Google Scholar]
- Ketolainen J.M., Alarmo E.L., Tuominen V.J., Kallioniemi A. Parallel inhibition of cell growth and induction of cell migration and invasion in breast cancer cells by bone morphogenetic protein 4. Breast Cancer Res. Treat. 2010;124:377–386. doi: 10.1007/s10549-010-0808-0. [DOI] [PubMed] [Google Scholar]
- Kim Y.H., Jang W.G., Oh S.H., Kim J.W., Lee M.N., Song J.H., Yang J.W., Zang Y., Koh J.T. Fenofibrate induces PPARa and BMP2 expression to stimulate osteoblast differentiation. Biochem. Biophys. Res. Commun. 2019;520:459–465. doi: 10.1016/j.bbrc.2019.10.048. [DOI] [PubMed] [Google Scholar]
- Krig S.R., Jin V.X., Bieda M.C., O’Geen H., Yaswen P., Green R., Farnham P.J. Identification of genes directly regulated by the oncogene ZNF217 using chromatin immunoprecipitation (ChIP)-chip assays. J. Biol. Chem. 2007;282:9703–9712. doi: 10.1074/jbc.M611752200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunimatsu R., Nakajima K., Awada T., Tsuka Y., Abe T., Ando K., Hiraki T., Kimura A., Tanimoto K. Comparative characterization of stem cells from human exfoliated deciduous teeth, dental pulp, and bone marrow–derived mesenchymal stem cells. Biochem. Biophys. Res. Commun. 2018;501:193–198. doi: 10.1016/j.bbrc.2018.04.213. [DOI] [PubMed] [Google Scholar]
- Lee D.-F., Walsh M.J., Aguiló F. ZNF217/ZFP217 meets chromatin and RNA. Trends Biochem. Sci. 2016;41:986–988. doi: 10.1016/j.tibs.2016.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2-??CT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Mantsou A., Koutsogiannouli E., Haitoglou C., Papavassiliou A.G., Papanikolaou N.A. Regulation of expression of the p21CIP1 gene by the transcription factor ZNF217 and MDM2. Biochem. Cell Biol. 2016;94:560–568. doi: 10.1139/bcb-2016-0026. [DOI] [PubMed] [Google Scholar]
- Massagué J., Chen Y.-G. Controlling TGF-signaling. Genes Dev. 2000;14:627–644. doi: 10.1101/gad.14.6.627. [DOI] [PubMed] [Google Scholar]
- Millipore Sigma, BrdU cell proliferation assay user protocol, (n.d.). https://www.merckmillipore.com/INTL/en/product/BrdU-Cell-Proliferation-Assay,EMD_BIO-QIA58?ReferrerURL=https%3A%2F%2Fwww.google.com%2F&bd=1#anchor_USP (accessed August 20, 2021).
- Miyazono K., Kamiya Y., Morikawa M. Bone morphogenetic protein receptors and signal transduction. J. Biochem. 2010;147:35–51. doi: 10.1093/jb/mvp148. [DOI] [PubMed] [Google Scholar]
- Mortada I., Mortada R. Dental pulp stem cells and osteogenesis: an update. Cytotechnology. 2018;70:1479–1486. doi: 10.1007/s10616-018-0225-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nohe A., Hassel S., Ehrlich M., Neubauer F., Sebald W., Henis Y.I., Knaus P. The mode of bone morphogenetic protein (BMP) receptor oligomerization determines different BMP-2 signaling pathways. J. Biol. Chem. 2002;277:5330–5338. doi: 10.1074/jbc.M102750200. [DOI] [PubMed] [Google Scholar]
- Ragni E., Viganò M., Rebulla P., Giordano R., Lazzari L. What is beyond a qRT-PCR study on mesenchymal stem cell differentiation properties: how to choose the most reliable housekeeping genes. J. Cell. Mol. Med. 2013;17:168–180. doi: 10.1111/j.1582-4934.2012.01660.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddi A.H. Bone morphogenetic proteins: an unconventional approach to isolation of first mammalian morphogens. Cytokine Growth Factor Rev. 1997;8:249–376. doi: 10.1016/S1359-6101(96)00049-4. [DOI] [PubMed] [Google Scholar]
- Ryoo H.M., Lee M.H., Kim Y.J. Critical molecular switches involved in BMP-2-induced osteogenic differentiation of mesenchymal cells. Gene. 2006;366:51–57. doi: 10.1016/j.gene.2005.10.011. [DOI] [PubMed] [Google Scholar]
- ThermoFisher Scientific, Methods to check RNA integrity, (n.d.). https://www.thermofisher.com/gr/en/home/references/ambion-tech-support/rna-isolation/tech-notes/is-your-rna-intact.html (accessed August 20, 2021).
- Shi Y., Sawada J.-I., Sui G., Bachir Affar E., Whetstine J.R., Lan F., Ogawa H., Po M., Luke S., Nakatani Y., Shi Y. Coordinated histone modifications mediated by a CtBP co-repressor complex. Nature. 2003;422:735–738. doi: 10.1038/nature01550. [DOI] [PubMed] [Google Scholar]
- Sieber C., Kopf J., Hiepen C., Knaus P. Recent advances in BMP receptor signaling. Cytokine Growth Factor Rev. 2009;20:343–355. doi: 10.1016/j.cytogfr.2009.10.007. [DOI] [PubMed] [Google Scholar]
- Steinert S., Kroll T.C., Taubert I., Pusch L., Hortschansky P., Höffken K., Wölfl S., Clement J.H. Differential expression of cancer-related genes by single and permanent exposure to bone morphogenetic protein 2. J. Cancer Res. Clin. Oncol. 2008;134:1237–1245. doi: 10.1007/s00432-008-0396-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suchanek J., Soukup T., Visek B., Ivancakova R., Kucerova L., Mokry J. Dentall pulp stem cells and their characterization. Biomed. Pap. 2009;153:31–36. doi: 10.5507/bp.2009.005. [DOI] [PubMed] [Google Scholar]
- Urist M.R. Bone: formation by autoinduction. Science (80-.) 1965;150 doi: 10.1126/science.150.3698.893. [DOI] [PubMed] [Google Scholar]
- Wang L., Park P., Zhang H., La Marca F., Claeson A., Valdivia J., Lin C.Y. BMP-2 inhibits the tumorigenicity of cancer stem cells in human osteosarcoma OS99-1 cell line. Cancer Biol. Ther. 2011;11:457–463. doi: 10.4161/cbt.11.5.14372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Park P., Zhang H., La Marca F., Claeson A., Than K., Rahman S., Lin C.Y. BMP-2 inhibits tumor growth of human renal cell carcinoma and induces bone formation. Int. J. Cancer. 2012;131:1941–1950. doi: 10.1002/ijc.27444. [DOI] [PubMed] [Google Scholar]
- Wang H., Yang B., Geng T., Li B., Dai P., Chen C. Tissue-specific selection of optimal reference genes for expression analysis of anti-cancer drug-related genes in tumor samples using quantitative real-time RT-PCR. Exp. Mol. Pathol. 2015;98:375–381. doi: 10.1016/j.yexmp.2014.10.014. [DOI] [PubMed] [Google Scholar]
- Wang X., Zhang Y., Choukroun J., Ghanaati S., Miron R.J. Effects of an injectable platelet-rich fibrin on osteoblast behavior and bone tissue formation in comparison to platelet-rich plasma. Platelets. 2018;29:48–55. doi: 10.1080/09537104.2017.1293807. [DOI] [PubMed] [Google Scholar]
- Wozney J., Rosen V., Celeste A., Mitsock L., Whitters M., Kriz R., Hewick R., Wang E. Novel regulators of bone formation: molecular clones and activities. Science (80-.) 1988;242:1528–1534. doi: 10.1126/science.3201241. [DOI] [PubMed] [Google Scholar]
- Yang J., Shi P., Tu M., Wang Y., Liu M., Fan F., Du M. Bone morphogenetic proteins: relationship between molecular structure and their osteogenic activity. Food Sci. Hum. Wellness. 2014;3:127–135. doi: 10.1016/j.fshw.2014.12.002. [DOI] [Google Scholar]
- Yuan Y., Duan R., Wu B., Huang W., Zhang X., Qu M., Liu T., Yu X. Gene expression profiles and bioinformatics analysis of insulin-like growth factor-1 promotion of osteogenic differentiation. Mol. Genet. Genomic Med. 2019;7 doi: 10.1002/mgg3.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Agarose gel electrophoresis of total RNA from (a) hDPSCs during osteogenic differentiation and (b) MCF-7 cells. a) Lanes 1, 5, 9: untreated hDPSCs in DMEM at 7, 14 and 21 days. Lanes 2, 6, 10: hDPSCs in osteogenesis medium at 7, 14 and 21 days. Lanes 3, 9, 11: hDPSCs in osteogenesis medium and treated with 50 ng/ml BMP-2. Lanes 4, 8, 12: hDPSCs in osteogenesis medium and treated with 50 ng/ml BMP-2 peptide. b): Lanes 1, 6, 11, 16: untreated MCF-7 at 6 h, 12 h, 24 h and 48 h. Lanes 2, 7, 12, 17: MCF-7 treated with 100 ng/ml BMP-2 at 6 h, 12 h, 24 h and 48 h. Lanes 3, 8, 13, 18: MCF-7 treated with 45.2 ng/ml BMP-2 peptide at 6 h, 12 h, 24 h and 48 h. Lanes 4, 9, 14, 19: MCF-7 treated with 22.6 ng/ml BMP-2 peptide at 6 h, 12 h, 24 h and 48 h. Lanes 5, 10, 15, 20: MCF-7 treated with 4.52 ng/ml BMP-2 peptide at 6 h, 12 h, 24 h and 48 h. The bands corresponding to the 28S rRNA and 18 S rRNA are visible.