Abstract
Ewing sarcoma (EwS) is characterized by pathognomonic translocations, most frequently fusing EWSR1 with FLI1. An estimated 30% of EwS tumors also display genetic alterations in STAG2, TP53, or CDKN2A (SPC). Numerous attempts to develop relevant EwS models from primary human cells have been unsuccessful in faithfully recapitulating the phenotypic, transcriptomic and epigenetic features of EwS. In this study, by engineering the t(11;22)(q24;q12) translocation together with a combination of SPC mutations, we generated a wide collection of immortalized cells (EWIma cells) tolerating EWSR1-FLI1 expression from primary mesenchymal stem cells (MSC) derived from an EwS patient. Within this model, SPC alterations strongly favored EwS oncogenicity. Xenograft experiments with independent EWIma cells induced tumors and metastases in mice, which displayed bona fide features of EwS. EWIma cells presented balanced but also more complex translocation profiles mimicking chromoplexy, which is frequently observed in EwS and other cancers. Collectively, these results demonstrate that bone marrow-derived MSCs are a source of origin for EwS and also provide original experimental models to investigate Ewing sarcomagenesis.
SIGNIFICANCE
These findings demonstrate that Ewing sarcoma can originate from human bone marrow-derived mesenchymal stem cells and that recurrent mutations support EWSR1-FLI1 translocation-mediated transformation.
INTRODUCTION
Ewing sarcoma (EwS) is the second most frequent bone or soft-tissue cancer of children, adolescents, and young adults. It is characterized by a chromosomal translocation between EWSR1 and members of the ETS (E26 transforming-specific) family of transcription factors, most frequently with FLI1 (t(11;22)(q24;q12)) (1). EWSR1-FLI1 exerts a strong oncogenic role but also cytotoxic effects when expressed in various primary cells (2,3). Only few additional recurrent genetic alterations are observed in EwS, primarily including inactivating mutations of STAG2 (~15–20%) and TP53 (~5–10%), as well as CDKN2A deletions (~9–22%) (4-6). Co-occurrence of STAG2 and TP53 mutations was reported to be associated with poor outcome (6). However, the pro-oncogenic role of these additional mutations in EwS origin and progression remains elusive.
Despite numerous efforts to generate murine EwS models, none of them faithfully recapitulated phenotypic, transcriptomic and epigenetic features of EwS (7-11). This may be partially explained by the poor conservation of cis-regulatory enhancers containing GGAA-microsatellites (mSats) that are uniquely bound by EWSR1-FLI1, and that appear critical for Ewing sarcomagenesis (12). However, several factors such as the exact nature of the cell(s) of origin, the timing and the (co)-occurrence of oncogenic events involved in Ewing sarcomagenesis are still poorly characterized (12). Although EwS histogenesis has been a long-lasting debate, experimental evidence has converged on either a neural crest origin, as neural crest-derived cells appeared to be permissive to EWSR1-FLI1 expression (13), or a mesenchymal origin, as for instance EWSR1-FLI1 inhibition in EwS cells induced features of mesenchymal stem cells (MSCs) (14). However, none of the attempts succeeded to model bona fide EwS tumors in vivo from any types of primary human cells.
To better mimic the pathophysiological context of EwS, we and others engineered the EWSR1-FLI1 translocation using genome editing technologies in human stem cells (15,16). These approaches lead to formation of the specific t(11;22)(q24;q12) translocation, starting from two double-strand breaks (17), one in EWSR1 and the other in FLI1. In contrast to models with ectopic EWSR1-FLI1 expression, the precise generation of the translocation at the endogenous loci enables faithful and ‘natural’ oncogene regulation, and reproduces heterozygosity at the EWSR1 locus resulting from the translocation of one allele. However, the specific isolation of immortalized/transformed EWSR1-FLI1 translocated clones remained unsuccessful (15,16). To overcome this issue, we reasoned that the genetic background of the starting cells could be of strong relevance. Indeed, the incidence of EwS is much higher in Europeans or European-Americans than in Africans or Afro-Americans (18-20). Genome-wide association studies (GWAS) suggest that a specific genetic germline background may be more permissive to EWSR1-ETS translocation and may favor EWSR1-ETS activity (21), as reported for the EGR2 susceptibility locus (22). In addition, we reasoned that additional recurrent mutations identified in this cancer may also contribute to Ewing sarcomagenesis. Here, we describe a model generated from primary human MSCs of a European EwS patient by introducing the t(11;22)(q24;q12) translocation and additional alterations in STAG2, TP53 and CDKN2A. Strikingly, this model displays molecular and phenotype features of EwS tumor, including expression of EwS-associated markers (including membranous CD99), and efficiently generated EwS tumors and metastases in immunodeficient mice. The bona fide genetically engineered EwS model generated in this study provides novel insights in Ewing sarcomagenesis and highlights the role of additional somatic mutations in this transformation.
MATERIALS AND METHODS
Primary cell and cell line culture
MSCPat cells (MSC7-BJ), human primary BMSCs, derived from bone marrow aspirates previously described in (23) and hMPCs described in (24) were cultured in αMEM supplemented with 10% MSC-FBS (12662029; Life Technologies), Glutamine 10mM (Life Technologies) and 2 ng/mL Recombinant Human FGF (233-FB-025; R&D Systems). For these primary cells, written informed consent was obtained according to the Declaration of Helsinki and studies were approved by the ethics committees of the contributing institutions. A673 were cultured in DMEM medium supplemented with 10% FBS (Life Technologies) and cultured at 37 °C in a humidified atmosphere with 5% CO2 and 20% O2. MSCPat and hMPCs were cultured in hypoxic-like conditions (3% O2). Culture cells were tested monthly for mycoplasma contamination (with the VenorGeM qEP kit (11-9250, Minerva Biolabs), and if positive, cells were treated with the mycoplasma treatment kit (Myco-1&2 set A8360.0010, VWR). MSCPat were culture at low passages (up to 10). EWIma clones were kept in culture from passage 1 and up to 100 days in culture. Cells used in the study are fully described in Supplementary Materials.
CRISPR/Cas9 transfections
Cells were transfected by 4D Nucleofector Amaxa technology (Lonza) using the cell line nucleofector (solution P1, FF-104) with 1 μg of plasmid pCAS9-GFP (44719; Addgene) and 1 μg of each plasmid (MLM3636, 43860; Addgene) encoding for the different gRNAs (2 μg for gRNAEWS). For Cas9/gRNA RNP complexes, MSCPat and hMPCs were transfected directly with the different combination of gRNAs and Cas9-GFP protein (ratio 2:1). gRNA sequences are listed in the Supplementary material.
PCR-based translocation detection and frequency
For detection of translocations from bulk cells, DNA (E.Z.N.A. Tissue DNA Kit, Omega Bio-Tek, GA, USA) was amplified by PCR or Nested PCR, 6 to 8 days post-transfection. Serial dilutions of DNA enable the assessment of translocation frequency as previously described in (25). Primer sequences are reported in Supplementary Material.
Soft-agar colony formation assays
A first agar layer was placed in 10 cm plates at 0.8% (w/v) of low melting temperature agarose (50101; Life) in αMEM-10%FBS. Once solidified, a second layer of 0.48% agar containing 4 x 104 cells was added. The plates were maintained at 4 °C for 5 min and 10mL of fresh culture medium was subsequently deposited as a top layer. The plates were incubated in hypoxia conditions and colonies were isolated after 3 to 4 weeks and further analyzed. Counting of colonies was performed using ImageJ/Fiji software and T-test analysis was applied.
Cell proliferation and siRNA assay
hMPCs were seeded in 6-well plates (40.000 or 60.000 cells/well) and maintained in hypoxic like or standard conditions for proliferation assay. Cell growth was monitored and analyzed by the IncuCyte Live Cell Analysis system (IncuCyte S3, Essen Bioscience) every 24 hours for 4 to 10 days. For knock-down experiments, 40nM of siRNA against EWSR1-FLI1 (5’-GGC AGC AGA ACC CUU CUU A-3’) (Eurofins) (or control siRNA (D-001810-01-50; Dharmacon) was transfected using Lipofectamine RNAiMAX Transfection Reagent (ThermoFisher Scientific) following manufacturer’s instructions. Cell growth was monitored and analyzed by the IncuCyte system every 4 hours during 3 days. All experiments were independently performed in triplicate, and T-test analysis was applied. SiRNA sequences are reported in Supplementary Material.
Fluorescence In Situ Hybridization (FISH), multicolor FISH analysis and conventional cytogenetic analysis
Fresh cells with few passages were harvested after 1-6 hours with 10μl/mL of KaryoMAX colcemid (Gibco) treatment, resuspended in 0.075M KCl at 37 °C for 30 minutes and fixed in methanol/acetic acid (3:1). Cells were dropped onto glass slides and dried. FISH was performed on the metaphases using EWSR1 and FLI1 probes (LPS007, Cytocell) to detect the t(11;22) chromosomal translocation. Cell images were captured with the Zeiss Spinning Disk Confocal microscopy 63x. Alternatively, for multicolor FISH imaging metaphase spreads were stained with 24XCyte, Multicolor Painting mFISH Probe Kit (MetaSystems), which was prepared following supplier’s instructions. Metaphases were imaged using a ZEISS AxioImager.Z2 microscope and the Metafer automated capture system (MetaSystems). Karyotyping was performed using Isis software (MetaSystems). For conventional karyotypes, metaphase spreads, R-banded chromosomes were analyzed by standard procedures.
Flow cytometry
Immunostaining of cells for CD99 marker was performed by incubating cells with FITC Mouse Anti-Human CD99 (BD Pharmingen, 555688) or FITC Mouse IgG2a, κ Isotype Ctrl Antibody (Biolegend, 400208) for 30 minutes at 4 °C prior to flow cytometry analysis based on SS-A/FS-H gating on alive cells (BD FACS Aria II- BD bioscience, and FlowJo software). Cells labelled with unspecific FITC Mouse IgG2a were used as negative cells for CD99 expression.
Western blotting
Whole-cell extracts were prepared with protein lysis buffer (50mM Tris-HCl at pH 7.4, 1% Triton X-100, 0.1% SDS, 150mM NaCl, 1mM EDTA, and 1mM DTT), with addition of cocktail protease inhibitor tablets (Complete, Roche). Membranes were stained with FLI1 (ab133485; Abcam) (used to detect the EWSR1-FLI1 translocation), STAG2 (sc81852; Santa Cruz), p53 (sc126; Santa Cruz) or p16 (554079; BD Pharmingen) antibodies. ACTIN (sc1616; Santa Cruz) and VINCULIN (sc73614; Santa Cruz) antibodies were used as loading controls. Membranes were visualized with Odyssey CLx Imaging System (LI-COR Biosciences).
qRT-PCR
RNA was extracted with the RNeasy Plus Mini Kit (Qiagen) and reverse-transcribed using the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems). qRT-PCRs were performed using PowerSYBR green Mastermix (Applied Biosystems). Oligonucleotides were purchased from MWG Eurofins Genomics (Oligonucleotides and Primers, see Supplementary Materials). Reactions were run on CFX384 Touch Real-Time PCR instrument (Bio-Rad) and analyzed using the CFX Manager Software (Bio-Rad). Relative expression level was assessed with the ddCT method using RPLP0 as a housekeeping gene. Primer sequences are reported in Supplementary Material.
Telomerase Repeat Amplification Protocol (TRAP)
Fresh cells were resuspended in 100μL of CHAPS Lysis Buffer and TRAP assay was performed following the manufacturer’s instructions of TRAPeze Telomerase Detection Kit (S7700, Millipore). PCR products were run on a TBE/acrylamide:bisacrylamide (19:1) gel, stained with SYBR Gold Nucleic Acid Gel Stain (Invitrogen) and visualized with a FLA-3000 Phosphorimager (Fujifilm).
Telomere Restriction Fragment (TRF)
DNA was isolated and digested overnight with HinfI and RsaI enzymes. DNA samples were run in a 0.7% agarose gel overnight and transferred to a membrane. The [γ32P]ATP-labeled telomere probe (CCCTAAA)4 was subsequently hybridized by using the Easy Hyb reagent (Roche). Membranes were exposed to Phosphorimager screens and screens were scanned with a FLA-3000 Phosphorimager (Fujifilm).
SNP array
Infinium Core-24 Chip (Illumina Inc. San Diego, USA) containing more than 300.000 SNPs that were hybridized with genomic DNA.
RNA-seq and bioinformatics analysis
Sequencing was carried out using 2x100 cycles (paired-end reads 100 nucleotides) for all samples on Illumina HiSeq2500 or NovaSeq6000 instruments. Reads were aligned with STAR 2.5.3 (Supplementary Reference 47) to the human genome (GRCh37/hg19 version). We used the count matrix generated by STAR with the human gene annotation v19 of GENCODE as reference. DESEQ2 (Supplementary Reference 48) was used to normalize data and performed differential analysis with the Wald test. The p-value was adjusted using the Benjamini-Hochberg method. We considered a gene expressed if the normalized expression was higher than 10. STAR-Fusion v1.4.0 (Supplementary Reference 49) was applied to predict fusion transcripts. No statistical methods were used to predetermine sample size. RNAseq experiments were performed in duplicates or triplicates.
ChIP-Seq
Chromatin Immunoprecipitation (ChIP) experiments were performed following manufacturer instructions using iDeal ChIP-seq kit for transcription factors and for histones (Diagenode) with respectively rabbit polyclonal anti-FLI1 antibody (ab15289, Abcam), rabbit polyclonal anti-H3K4me3 (C15410003, Diagenode) and rabbit polyclonal anti-H3K27ac (ab4729, Abcam). For ChIP sequencing, libraries were generated using TruSeq ChIP library preparation kit (Illumina) and sequenced on Illumina HiSeq 2500 (single end, 100 bp). Reads were aligned to human reference genome (GRCh37/hg19) with bowtie2 2.2.9 (Supplementary Renference 50). Peaks were called with MACS2 2.1.1 (Supplementary Reference 51) with the option narrow for FLI1 ChIP-seq and broad for H3K27ac histone mark. ChIP-seq data were normalized according to their respective input DNA sample. The ChIP-seq signal tracks were generated by macs2 with bdgcmp option (and m FE to compute fold enrichment between the ChIP and the control). Then, we run bedGraphToBigWig to convert the file to a binary format (BigWig). To define enhancers and super-enhancers, we used the ROSE tool (Supplementary Reference 52) with the parameter –t 2500 in order to exclude H3K27ac peaks which overlap the theoretical TSS (<2.5kb) regions. Here defined enhancers were then stitched and therefore some enhancers or super-enhancers can contain active promoters. We annotate them by associating the closest expressed genes. Control-FREEC (29) on input DNA was used to determine Copy Number Variants in EWIma1 (MSCPat was used as reference).
Primer sequences are reported in Supplementary Material.
Mice
Animal care and use for this study were performed in accordance with the recommendations of the European Community (2010/63/UE) for the care and use of laboratory animals. Experimental procedures were specifically approved by the ethics committee of the Institut Curie CEEA-IC #118 (Authorization APAFIS#11206-2017090816044613-v2 given by National Authority) in compliance with the international guidelines. The tumorigenic and metastatic potential of hMPC clones (4-7 mice) or EWIma1, 5 and 7 (2-4 mice per clone) cells was investigated by injecting 1 million cells in an orthotopic intra-osseous model as described previously (Supplementary Reference 53). Seven-week-old NSG (NOD.Cg-Prkdcscid IL2rgtm1Wjl/SzJ) female mice were purchased from Charles Rivers (France). Mice were anesthetized by inhalation of a combination isoflurane/air (1.5%, 1 L/min) and followed up for tumor growth. The tumor volume was calculated by using the formula L × (l2)/2, where L and l represent respectively the longest and the smallest perpendicular diameter. Tumor samples were fixed for 24 hours in AFA solution and processed for paraffin embedding and sectioning.
Immunohistochemistry
Xenograft sections (4μm) were cut and stained with hematoxylin and eosin. Immunohistochemistry was performed using the following antibodies and dilutions: FLI1 1:50 (Abcam, ab15289), STAG2 1:25 (Santa Cruz, sc81852), Ki67 1:500 (Abcam, Ab15580), cleaved CASP3 1:250 (Cell Signaling, #9661) and CD99 1:1 ready-to-use (Agilent, IS057).
Data availability
ChIP-seq and RNA-seq data have been deposited in NCBI’s Gene Expression Omnibus under accession number: GSE150783.
Raw data are available at Mendeley. Reserved DOI: doi:10.17632/fx29by5k43.1 https://data.mendeley.com/datasets/fx29by5k43/draft?a=63bd5c8f-f13b-4100-b9bf-988ddb0c9131
RESULTS
Generation of stable cell lines expressing EWSR1-FLI1 (EWIma cells) starting from wild type MSCs of an EwS patient
Aiming at recapitulating the t(11;22;)(q24;q12) in situ and knowing that the genetic background is a potential factor of incidence in Ewing sarcoma (18), we derived primary normal MSCs of a patient (MSCPat) (23) who was affected by an EwS of the ulna (Supplementary Fig. 1A to C). The EwS tumor from this patient exhibited four large deletions (chr3q; chr9p comprising CDKN2A; chr16q; and chr17p comprising TP53), one gain (chr1q), and an isodisomy (chr5p) at time of diagnosis (Supplementary Fig. 1A and 2A). This tumor also exhibited altered expression of the 3’ end of STAG2 (4.90 ratio of average coverage of [Exons 26–30]/[Exons 1–25], compared to 1.62 for MSCPat cells suggesting the existence of 3’ truncated transcripts) (Supplementary Fig. 2A), however, without detectable genetic alteration in the coding region of this gene. MSCPat were derived from a bone marrow aspirate at the time of diagnosis and exhibit no copy number alterations (Supplementary Fig. 1B). Genomic analyses of these cells confirmed the absence of mutations or alterations in oncogene or tumor suppressor genes. As comparison, we also used mesenchymal stem/precursor cells (hMPCs) derived from human embryonic stem cells which are proficient for multi-lineage differentiation (fat, cartilage, bone, and skeletal muscle)(24). We noticed a growth advantage of hypoxia on cell morphology and proliferation of hMPCs (Supplementary Fig. 2B and C). This is consistent with the observation that the center of solid tumors and the niche of MSCs are mostly hypoxic environments and that hypoxia enhances growth of MSCs (also reported in 25). Based on these results, we cultured MSCPat and hMPC cells in hypoxic conditions (3% O2) throughout this study and used the CRISPR-Cas9 technology to engineer the t(11;22)(q24;q12) translocation (26)(Fig. 1A). Upon transfection of Cas9, gRNAEWSR1 and gRNAFLI1 (EF) coding plasmids in MSCPat, EWSR1-FLI1 translocation positive cells were readily detected at 12 and up to 61 day post-transfection (Supplementary Fig. 2D), although at a lower frequency for the latter time point. However, we could not recover any viable clones in these conditions.
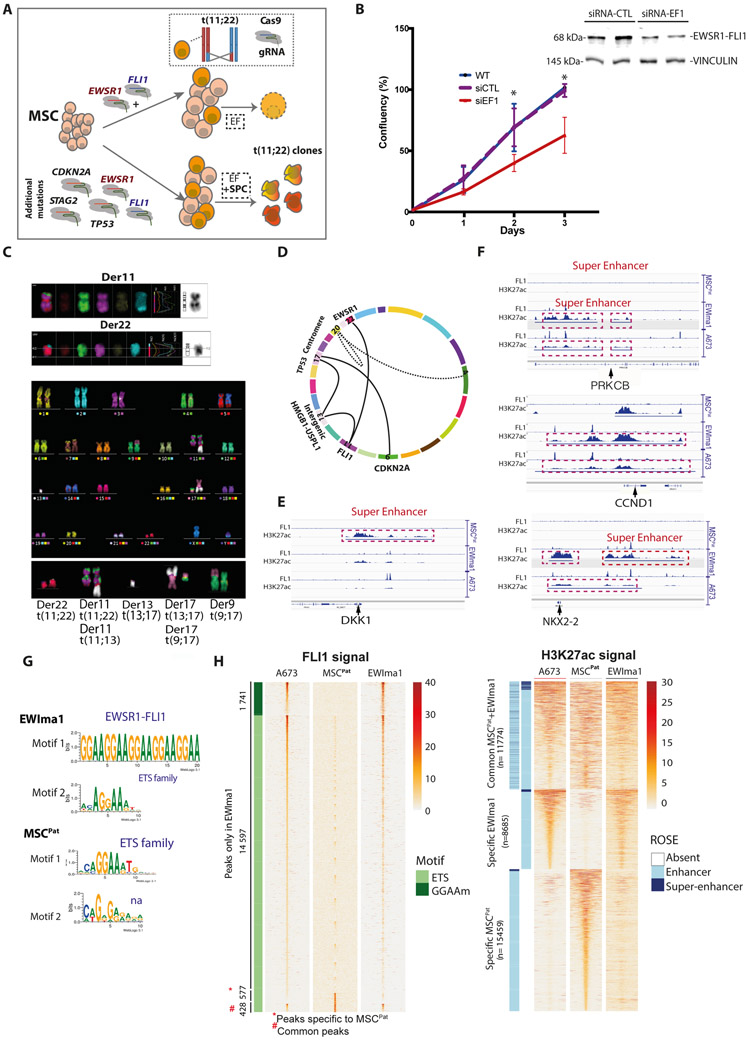
Figure 1. Generation of EWIma1 cells derived from MSCPat cells, recapitulating molecular and epigenetic features of EwS.
A- CRISPR/Cas9 based strategy to obtain EWSR1-FLI1 translocated clones from MSCs with or without inducing STAG2, TP53 and CDKN2A additional mutations. EF: gRNAEWS and gRNAFLI1. EF+SPC: gRNAEWS, gRNAFLI1, gRNATP53, gRNACDKN2A and gRNASTAG2.
B- Cumulative cell counts over time in EWIma1 cells transfected with siRNA targeting EWSR1-FLI1 (si-EF1) compared to wild type (WT) and siRNA control (si-CTL). Top, western blot against FLI1, shown at 4 days post transfection. Results represent the mean ± SD from three independent experiments. * p<0.05.
C- Representative image of spectral karyotype (SKY multi-colored fluorescent FISH analysis) obtained from EWIma1 cells with a chromoplectic like pattern (reciprocal translocations t(11;22)(q24;q12), t(13;17), t(9;17)) and derivative chromosome 11 (der11) of t(11;13)). N= 22 metaphases. See also Supplementary Table 1.
D- Schematic circos plot of the main rearrangements seen in EWIma1 cells (dot lines: rearrangements not found in all cells).
E- Integrative Genomics Viewer representation for FLI1 and H3K27ac ChIP-seq profiles at DKK1 locus showing the disappearance of super-enhancer in EWIma1, and A673 cells compared to MSCPat. Super-enhancers are framed in red.
F- Integrative Genomics Viewer representation for FLI1 and H3K27ac ChIP-seq profiles at PRKCB, CCND1 and NKX2-2 loci showing the appearance of super-enhancers in EWIma1, and A673 cells compared to MSCPat. Super-enhancers are framed in red.
G- Top two motifs predicted by ChIPMunk corresponding to known motifs in Jaspar database identified in EWIma1 and MSCPat FLI1 ChIP-seq data.
H- Left: Heatmap representation of FLI1 ChIP-seq peaks ranked by intensity at GGAA microsatellite (GGAAm) or ETS sites in EWIma1 only or MSCPat common sites. Right: Heatmap representation of H3K27ac ChIP-seq peaks sorted by ROSE SE rank in EWIma1 and MSCPat specific and common sites. A673 data are shown as positive control. Read density is displayed within a 5kb (H3K27ac or FLI1) window around peak center and color scale intensities are shown in normalized coverage (scale is shown on the right of each panel).
We hypothesized that additional somatic mutations found in EwS may facilitate the transformation potential of EWSR1-FLI1 fusion protein. We focused on the three most recurrently mutated genes identified in EwS: STAG2 (S), TP53 (P) and CDKN2A (C), knowing that their expression is also altered in the tumor of origin of this EwS patient (Supplementary Fig. 2A). Using CRISPR-Cas9 and gRNA plasmid transfection, we simultaneously induced the translocation with EF gRNAs with SPC gRNAs in MSCPat cells (Fig. 1A), and could recover numerous clones with SPC mutations. One clone (hereafter termed EWIma1), among hundred isolated clones, displayed morphologic changes with rounder cells, indicative of a gradual acquisition of the classical small-round-cell morphology of EwS cells (2-3 weeks after its isolation, Supplementary Fig. 2E). Three additional independent experiments using the same approach allowed recovering two additional EWIma clones (termed EWIma1* and EWIma1#). SNP arrays of EWIma1 cells did not show any of the copy number changes found in the patient EwS cells, excluding a hypothetical initial contamination of the MSCPat with patient tumor cells (Supplementary Fig. 1A and 1C). In contrast to the original MSCPat, but similarly to the prototypic A673 EwS cell line, EWIma1 cells stably expressed EWSR1-FLI1 oncoprotein (Supplementary Fig. 2F). As expected, these cells did not express STAG2 nor p16 and expressed a truncated form of p53 (Supplementary Fig. 2G). Inhibition of EWSR1-FLI1 by RNA interference led to a significant decrease of their proliferation (Fig. 1B) and reverted EWIma1 cells to a more mesenchymal spread-like morphology, as previously described in EwS cell lines (14)(Supplementary Fig. 2H and 2I).
Karyotypes and PCR analysis of EWIma1 cells revealed three additional translocations involving chromosomes 9, 11, 13, and 17 (Fig. 1C and 1D, Supplementary Fig. 2J, 2K and 2L and Supplementary Table 1). Most of the breakpoint junctions corresponded to the CRISPR/Cas9 targeted loci or to a predicted off-target site of gRNACDKN2A located on chr13 (in the promoter region of USPL1) (Supplementary Fig. 2K). Only t(9;17) breakpoints, which involve the centromere of chr17, could not be amplified by PCR but probably implicates the CDKN2A on chr9 as only one allele of CDKN2A can be amplified. An additional duplication of chr20q, potentially involved in a derivative chromosome 20, was detected in 73% (16/22) of the metaphases (Supplementary Fig. 2L and Supplementary Table 1). Similarly to EWIma1, we identified translocations involving chromosomes 9, 11 and 13, 17 in EWIma1* and EWIma1# (Supplementary Fig. 2J). Strikingly, these particular chromosomal rearrangements are reminiscent of concomitant intricate genetic events previously described as chromoplexy, which is characterized by a sudden burst of complex, loop-like rearrangements (27), found in more than 35% of EwS tumors (28).
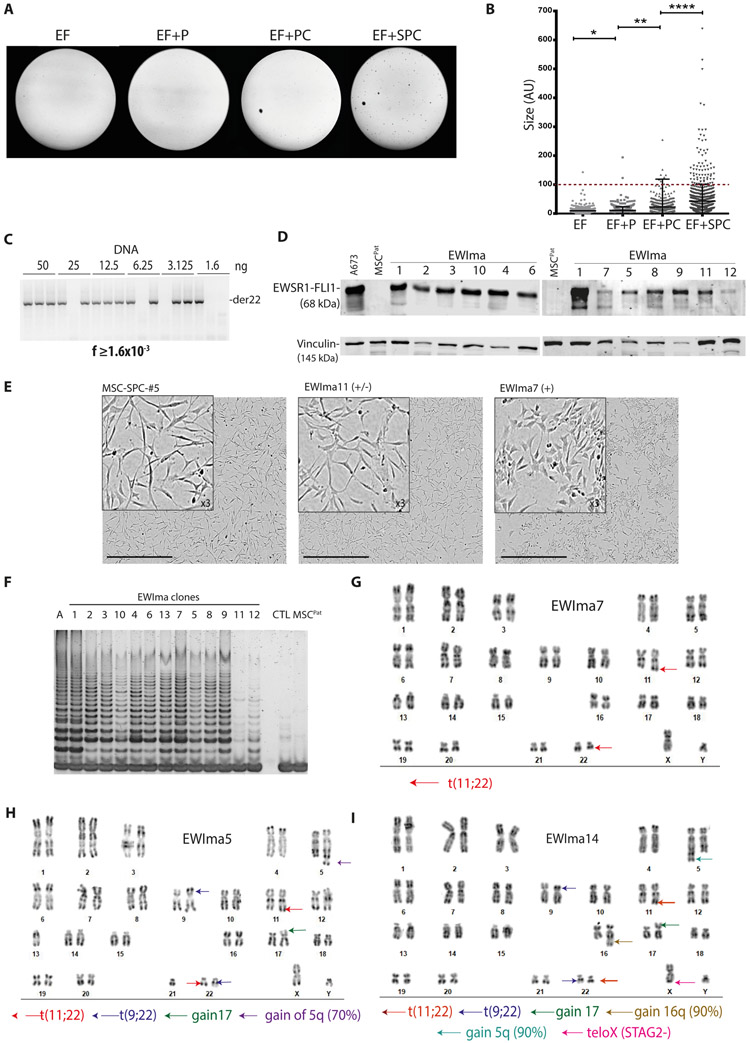
Figure 2. Molecular characterization of a collection of EWIma cells derived from MSCPat cells.
A- Representative images of agar colony formation assays using combinations of gRNAEWS (E), gRNAFLI1 (F), gRNATP53 (P), gRNACDKN2A (C), and gRNASTAG2 (S).
B- Colony size quantification mean +/− SD, p value (* p= 0.0186; ** p=0.0029; ****p<0.0001); n>600 colonies per condition.
C- Nested PCR to detect the translocated chromosome derivative 22 (der22) on serial dilutions from a DNA pool of MSCPat cells transfected with EF+SPC gRNAs (from 50 to 1.6 ng in triplicates). Translocation frequency (f) is calculated as described in (26) using the assumption that a human diploid cell contains ~6 pg of DNA.
D- Western blot against FLI1 in a panel of EWIma clones compared to positive (A673) and negative (MSCPat) controls. Vinculin is used as loading control.
E- Representative images of cellular morphology for the negative EWSR1-FLI1 translocated MSC-SPC-#5 clone (SPC mutated and with typical MSC morphology), for a positive EWSR1-FLI1 translocated EWIma11 (intermediate EwS morphology) and EWIma7 (classical EwS morphology) clones. Scale bar: 800 μm.
F- Telomerase Repeat Amplification Protocol (TRAP) assay showing telomerase activity in the different EWIma clones compared to A673 Ewing cells (A), MSC-SPC-#5 (CTL) and MSCPat cells.
G- Karyotype analysis of EWIma7.
H- Karyotype analysis of EWIma5.
I- Karyotype analysis of EWIma14.
To further characterize the EW1ma1 model and to assess whether it faithfully recapitulates EwS properties, we performed ChIP-seq experiments against FLI1 and H3K27ac in MSCPat and EWIma1 cells. Chromatin patterns for these marks at known EWSR1-FLI1 targets genes in EWIma1 cells strongly resembled those of established A673 EwS cells and noticeably differed from the one of MSCPat (Fig. 1E and 1F).
Importantly, the canonical EWSR1-FLI1 GGAA-mSat and ETS binding motifs (29) were identified as the first and second motifs among all known transcription factor motifs in EWIma1 FLI1 ChIP-seq data (Fig. 1G). Conversely, ETS site was the most prominent identified motif in ChIP-seq peaks in MSCPat which express FLI1 (Fig. 1G, Supplementary Fig. 3A). In EWIma1 cells, 16,338 specific EWSR1-FLI1 peaks were identified (1,741 at GGAA-mSats and 14,597 at ETS binding sites). These specific EWSR1-FLI1 peaks were highly similar to those identified in the A673 cells (Fig. 1H). Strikingly, peaks at GGAA-mSat regions were completely absent in MSCPat. Using H3K27ac ChIP-seq, we further identified 8,685 regions that were specific for EWIma1 compared to MSCPat (Fig. 1H). Again, these regions were highly conserved in A673. Furthermore, we performed ROSE analysis in EWIma1 and MSCPat cells and identified super-enhancers (SEs) associated to known EwS-specific genes such as BCL11B, CCND1, GLG1, NKX2-2, and SOX6 (30-32) as top hits in EWIma1 cells (Supplementary Fig. 3B).
De novo Ewing sarcomagenesis models display heterogeneous morphologies and immortalization patterns, simple and chromoplectic-like phenotypes and are favored by additional mutations.
Aiming at obtaining a broad collection of EWIma models by increasing the translocation frequency, we transfected MSCPat with the ribonucleic protein (RNP)/Cas9 complexes (replacing the above plasmid based approach) (33). Very few small colonies grew in agar after transfection with gRNAEWSR1 and gRNAFLI1 (EF) or together with gRNATP53 (EF+P). In contrast, additional combinations with gRNACDKN2A (EF+PC) and even more strikingly with gRNASTAG2 (EF+SPC) significantly increased the size of these soft agar grown colonies (Fig. 2A and 2B). Using EF+SPC conditions, we reached a translocation frequency of 1.6x10−3 (Fig. 2C), representing a ~30-fold improvement as compared to plasmid based approach (Supplementary Fig. 2D). Remarkably, EF+SPC combination allowed for the identification of 116 PCR positive clones for EWSR1-FLI1 out of a total of 274 isolated clones from these agar plates (Supplementary Fig. 3C). We further kept in culture 30 clones (Supplementary Table 2). Of these, we first randomly selected 13 clones (EWIma2 to EWIma14) for further molecular and cellular characterization, and confirmed expression of EWSR1-FLI1 fusion protein (Fig. 2D) and mutation in SPC genes for all of them (Table 1). Using flow cytometry, we also confirmed higher CD99 expression levels in EWIma clones as compared to MSCPat mutated in SPC (Supplementary Fig. 3D). Most EWIma clones displayed classical EwS cell morphology (Fig. 2E, Supplementary Fig. 3E). Using TRAP assay, most EWIma clones showed strong telomerase activity and grew past 100 days, in agreement with a full immortalized phenotype (Fig. 2F). Interestingly, we observed a more mesenchymal intermediate morphology in some clones (EWIma 11, 12, but also for two additional clones named EWIma 30 and 31) (Fig. 2E, Supplementary Fig. 3E). These intermediate clones appeared to express lower levels of EWSR1-FLI1 transcript (Supplementary Table 2) and protein (Fig. 2D), as compared to EWIma models displaying a clear EwS morphology, and also exhibited a lower telomerase activity (Fig. 2F). Karyotype analysis of immortalized EWIma models revealed that 7 out of 13 analyzed clones (EWIma 2 to 4 and 6 to 8, 12) displayed a rather simple and stable karyotype with t(11;22)(q24;q12) (Fig. 2G and Supplementary Fig. 4A). A few additional somatic alterations could be also detected by simple karyotype, including a loss of 16q (EWIma2), which is also recurrently identified in EwS tumors (Supplementary Fig. 4A). EWIma1, 1*, 1#, 5, 11 and 14 displayed chromoplectic-like translocation patterns (Fig. 1C, 1D, 2H, 2I, Supplementary Fig. 4A). This result is strikingly representative of the recently reported 30-40% incidence of chromoplexy in EwS tumors (28). Whereas the chromoplectic-like pattern of experimental independent EWIma1, EWIma1* and EWIma1# clones is almost identical, (Supplementary Fig. 2J), differences in the translocation patterns were observed in the other complex models. For instance, EWIma5 and EWIma14 showed a chromoplectic-like karyotype with t(11;22) but also the additional translocation t(9;22) (between CDKN2A and EWSR1), addition of chr17p (probably from the TP53 gene DNA break as only one allele can be detected) and loss of STAG2 in all metaphases (Fig. 2H and 2I, Supplementary Fig. 4A and 4B). Altogether, immortalized EWIma1 cells display stable EWSR1-FLI1 expression and faithfully recapitulate EwS cells features.
Table 1: Representative panel of genetic and morphologic features of EWIma model.
Sequences of STAG2, TP53 and CDKN2A mutations and cellular morphology (M) (+ classical EwS morphology, +/− intermediate EwS morphology or - MSC morphology) are indicated. Genomic sequences of der11 and der22 at breakpoints are indicated. MSC-SPC-#5 clone does not contain the EWSR1-FLI1 translocation and is used as negative control. EWINGness Score* for each EWIma model is displayed in the last column (see also Supplementary Table 2). “No seq” means that no sequence was PCR amplified for these clones probably due to large DNA deletions or presence of translocation implicated the gene.
| Name | STAG2 | TP53 | CDKN2A | M | Der11 (chr11,chr22) |
Der22 (chr22, chr11) |
Score * |
|---|---|---|---|---|---|---|---|
| EWIma2 | c.661_662insT | c.831delG | c.512_513insG/ c.512_514insTT | + | TCCAGCTA-CTTCACAC | TTTCCTAT-TAAACATCT | 70,0 |
| EWIma3 | c.661_662insT | no seq | c.512_513insG/ c.512_514insTT | + | TCCAGCTA-CTTCACAC | TTTCCTAT-TAAACATCT | 66,6 |
| EWIma13 | c.661_662insT | c.831delG | c.512_513insG/ c.512_514insTT | + | TCCAGCTA-CTTCACAC | TTTCCTAT-TAAACATCT | 66,5 |
| EWIma10 | c.661_662insT | c.831delG / c.817_838del | c.513delT/ c.512_513del | + | Del204 - Del24 | Del70 - Del15 | 66,2 |
| EWIma14 | c.661_662insT | c.811_830del | c.512_513insT | + | TCCAGCTA-CTTCACAC | TTTCCTAT-TAAACATCT | 65,4 |
| EWIma4 | c.661_662insT | c.831delG | c.512_513insG | + | TCCAGCTA-CTTCACAC | TTTCCTAT-TAAACATCT | 65,4 |
| EWIma6 | c.661_662insT | c.831delG/ c.832_833insCC | c.512delG/ c.512_513insT | + | TCCAGCTA-CTTCACAC | TTTCCTAT-TAAACATCT | 64,5 |
| EWIma7 | c.661_662insT | c.831_832insG/ c.831_837del | c.512_513insT | + | TCCAGCTA-CTTCACAC | TTTCCTAT-TAAACATCT | 61,4 |
| EWIma9 | c.661_662insT | c.831delG | c.512_513insT/ c.511_512del | + | TCCAGCTA-CTTCACAC | TTTCCTAT-TAAACATCT | 60,8 |
| EWIma5 | no seq | c.811_830del | c.512_513insT | + | TCCAGCTA-CTTCACAC | TTTCCTAT-TAAACATCT | 60,4 |
| EWIma8 | c.661_662insT | c.831_832insG/ c.831_837del | c.512_513insT | + | TCCAGCTA-CTTCACAC | TTTCCTAT-TAAACATCT | 60,1 |
| EWIma1 | c.661_662insT | c.831delG | c.511_519del | + | TCCAGCTA-CTTCACAC | TTTCCTAT-TAAACATCT | 58,3 |
| EWIma11 | c.661_662insT | c.832_833insC/ c.831delG + c.834C>T | c.512_515del / c.512_513insT | +/− | TCCAGCTA-CTTCACAC | TTTCCTAT-TAAACATCT | 40,9 |
| EWIma12 | c.661_662insT | c.832_833insC/ c.831delG + c.834C>T | c.512_513insG | +/− | TCCAGCTA-CTTCACAC | TTTCCTAT-TAAACATCT | 33,5 |
| MSC-SPC#5 | c.660_662del | c.831_836del/ c.815_836del | c.509_516del / c.512_513insT | - | - | - | 0,0 |
Engineering of t(11;22)(q24;q12) positive cells from multipotent mesenchymal precursor cells
Since MSCPat cells are extremely limited resources to generate these EwS models, we attempted to reproduce these results using published multipotent mesenchymal precursor cells (hMPCs)(24). An initial translocation frequency above 10−3 independently of the presence or absence of SPC gRNA could be achieved in these cells (Supplementary Fig. 5A). As reported for MSCPat, transfection of the unique pair of gRNAEWS and gRNAFLI1 in hMPCs resulted in a progressive loss of the t(11;22)(q24;q12) after two weeks of culture (Supplementary Fig. 5B). However, addition of SPC gRNAs increased the proliferation rate of bulk EF gRNAs transfected hMPCs cells (Supplementary Fig. 5C) and led to longer detection (up to 27 days) of EF fusion transcript (Supplementary Fig. 5B). Numerous clones grown in soft agar could be isolated from gRNA EF+SPC hMPC transfected cells and 0.9% contained the EwS translocation (3/336 clones). A similar frequency (0.7%, 3/408 isolated clones) of EWSR1-FLI1 positive clones was obtained if only additional gRNAs targeting TP53 and CDKN2A (but not STAG2) were used. Small colonies were obtained in soft agar when gRNAEWS and gRNAFLI1 were transfected alone, but they could ultimately not be recovered after isolation.
We further analyzed EWSR1-FLI1 translocated clones, with and without STAG2 mutations (Supplementary Fig. 5D to 5K). As in EwS cells, we could detect the EWSR1-FLI1 fusion transcript in all clones, with STAG2 WT clone 1 exhibiting a very low level of transcript (Supplementary Fig. 5F and 5G). Expression of EWSR1-FLI1 was detected in 3 clones for which we collected sufficient protein extract, confirming the low level of fusion protein in STAG2 WT clone 1 (Supplementary Fig. 5H). TP53 was mutated in these clones which expressed a truncated p53 protein (Supplementary Fig. 5E and 5I). CDKN2A mutations were also present in all four clones (Supplementary Fig. 5E). While no p16 expression was detected for the STAG2 knockout (KO) clones, the STAG2 WT clone 1 showed p16 expression (related to the induced mutation that leads to a late stop codon) (Supplementary Fig. 5E and 5I). All clones expressed the EwS-specific cell surface marker CD99 (Supplementary Fig. 5J). However, and in contrast to EWIma clones obtained from MSCPat, none of these EWSR1-FLI1 translocated clones was fully immortalized in vitro and they all stopped growing after 2–3 months in culture (after agar selection). During that time, we observed a progressive telomere shortening associated with a weak telomerase activity, factors known to be deleterious for long-term culture (Supplementary Fig. 5K).
Transcriptomic analysis of t(11;22)(q24;q12) engineered mesenchymal precursor models revealed a palette of EWSR1-FLI1 activation signature in these cells
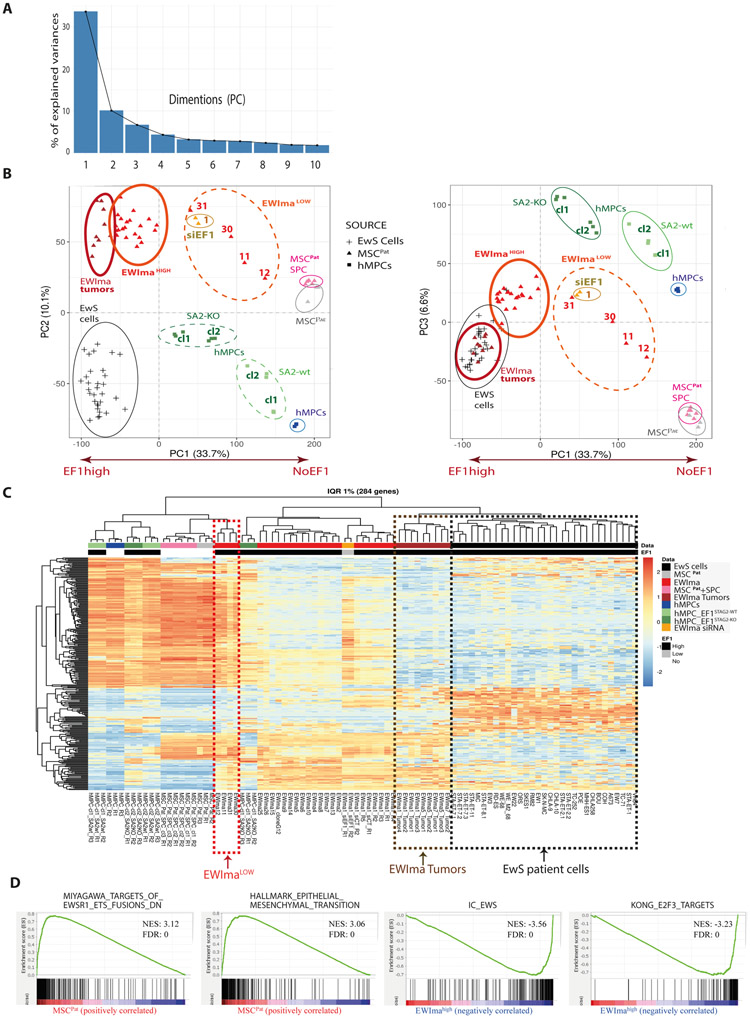
To further characterize the different models generated so far, we performed transcriptomic analyses in hMPCs, MSCPat cells, and the derived models containing either the EWSR1-FLI1 translocation and/or SPC mutations. These results were compared to transcriptomic profiles of 30 established EwS cell lines. Principal component analysis (PCA) revealed a striking weight of the first component (PC1) as compared to the other ones (Fig. 3A). PC1 was clearly associated with EWSR1-FLI1 expression (Fig. 3B). Importantly, most EWIma1 cells clustered close to a collection of EwS cells but far apart from their parental MSCPat. Conversely, MSCPat with or without SPC mutations co-localized on PC1 axis and were moderately segregated by PC2 and PC3. Short-term silencing of EWSR1-FLI1 in EWIma1 cells reverted their PC1 component (Fig. 2B, Supplementary Fig. 2H, 2I and Supplementary Fig. 6A). Interestingly, EWIma 11, 12, 30 and 31 models (termed hereafter EWImalow models), which displayed a more mesenchymal intermediate morphology (Fig. 2E and Supplementary Fig. 3E) and a moderate telomerase activity (Fig. 2F) were clearly segregated from the other EWIma models (EWImahigh, defined hereafter as all EWIma models except EWImalow) on the PC1 axis (Fig. 3B). Similarly, using an unsupervised hierarchical clustering analysis (HCA), all EWImahigh in vitro clones emerged distinctly of the original MSCPat, hMPCs and EWImalow, from a slightly distant branch compared to EwS patient cells (Fig. 3C). In contrast, engineered clones obtained from hMPCs cells clustered in between EwS cell lines and their parental cells (Fig. 3C). Quite remarkably, EWSR1-FLI1 expression appeared as the key factor driving the segregation of mesenchymal cells from EwS populations in both PCA and HCA. MSCPat carrying SPC alterations or hMPC-derived models displaying lower EWSR1-FLI1 levels (in particular clones 1 and 2 from hMPCs WT STAG2) (Fig. 3B and 3C), clustered close to WT MSCPat or hMPCs (Fig. 3C). To measure PC1 activity and evaluate the transition state of a broader collection of EWIma models without having to perform RNA-seq for each clone, we wondered if a small panel of known EWSR1-FLI1 transcriptional activated (EGR2, NKX2-2, PRKCB) (21,26,27) and repressed (TNC, DKK1, IGFBP3) target genes could be used as a surrogate marker using an RT-QPCR approach. For this, we defined an “EWINGness” score as the sum of log2FC (EGR2 + NKX2-2 + PRKCB) - (TNC + DKK1 + IGFBP3). Quite remarkably, a strong correlation (R2=0.93) between PC1 and EWINGness scores was observed in MSCPat and EWIma models (Supplementary Fig. 6B). A similar observation was made when all data were considered (R2=0.85) and not surprisingly, this correlation was poor among EwS cell lines (R2=0.08) (Supplementary Fig. 6B). Using this approach, we were able to evaluate the “EWINGness" of 30 EWIma clones (Supplementary Table 2) emphasizing a broad palette of EWSR1-FLI1 activation signature in these models (Supplementary Fig. 6C). Indeed, clones that were negative for the EWSR1-FLI1 translocation but mutated for SPC typically displayed a EWINGness score below 25. Independently of an analysis on their morphological aspect, EWImalow model had an intermediate EWINGness score which was comparable to EWIma1 cells silenced for EWSR1-FLI1. Similarly, hMPC-derived models, which were not fully transformed, clearly scored in the intermediate 25-50 window. All other EWIma models presenting an EWSR1-FLI1 translocation had a score > 50, which was also observed in a panel of 22 EwS cell lines (Supplementary Fig. 6C). We further explored the transcriptomic signature between MSCPat and EWIma models using gene-set enrichment analysis (GSEA). Heatmap of the top50 features for each phenotype highlighted again the intermediate signature of EWImalow models (Supplementary Fig. 6D). These last were removed from further GSEA to identify gene set signatures correlated with MSCPat or EWImahigh models. Quite remarkably, published EWSR1-FLI1 activation signatures ranked among the top50 EWIma correlated signatures (among 18580 investigated signatures) (Supplementary Table 3). Similarly, four E2F family member signatures were also identified in this top50 (Fig. 3D, Supplementary Table 3). Among those and of particular interest, E2F3 was previously shown to co-localize with EWSR1-FLI1 and to participate in the deregulation of cell cycle control of EwS (34). Conversely, signatures associated with a mesenchymal state were strongly enriched in MSCPat GSEA analysis (Fig. 3D, Supplementary Table 3).
Figure 3. EWSR1-FLI1 transcriptional signature is predominant in the de novo models.
A- Histogram for weight percentage of top 10 dimensions of principal component analysis (PCA).
B- PCA representation for parental and hMPC or MSCPat derived models (including in vitro, in vivo and EWSR1-FLI1 silenced EWIma1 models) compared to a collection of EwS cell lines. Left: PC1 vs PC2 and right: PC1 vs PC3.
C- Unsupervised Hierarchical clustering and heatmap based on top 1% inter-quartile range gene expression values. EWImalow models (EWIma12, 11, 31 and 30) are framed in red. EWIma orthotopic tumors (EWIma1, 5 and 7) are framed in brown. EwS patient cell lines are framed in black. Rn: biological replicates numbering.
D- GSEA enrichment plots from top signatures upon MSCPat versus EWIma models comparison. Top MSCPat correlated signatures included commonly down-regulated genes in mesenchymal progenitors upon EWSR1-FLI1 and EWSR1-ERG expression (Myagawa targets of EWSR1-ETS fusions DN) and the hallmark epithelial to mesenchymal transition gene sets. Top EWIma correlated signatures included the cell cycle independent EWSR1-FLI1 activation signature (IC_EWS from (23)) and up-regulated genes in embryonic fibroblasts upon serum stimulation and E2F3 knockdown (KONG_E2F3_TARGETS). In this analysis, EWImalow models were excluded.
EWIma cells display tumorigenic and metastatic properties in mice
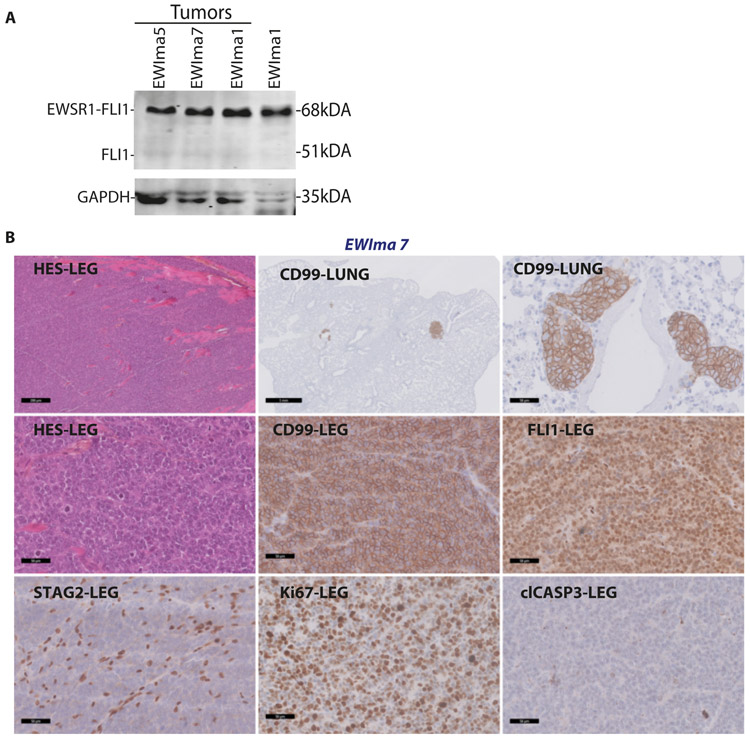
Ultimately, to evaluate the ability of the above described models to give rise to tumors in vivo, we performed orthotopic intra-femoral xenograft experiments. When injecting four hMPC derived models (STAG2 KO clone 1, n=4; STAG2 KO clone 2, n=4; STAG2 WT clone1 hMPC, n=4; STAG2 WT clone2 1, n=3), no tumors were detected in any of these mice after 5.5 months. Remarkably, when injecting two chromoplectic-like EWIma models (EWIma1, n=4 and EWIma5, n=2) and one simple rearranged model (EWIma7, n=4), all mice developed tumors that reached ethical endpoint criteria within 44-60 days (EWIma1), 84-90 days (EWIma7) and 90 days (EWIma5) post injection. These tumors were positive for t(11;22;)(q24;q12) and expressed EWSR1-FLI1 (Fig. 4A). Necropsy revealed distant metastases to the lungs and liver with all 3 models. Histological analysis evidenced a typical EwS small-round-cell morphology at primary and metastatic sites (Fig. 4B, Supplementary Fig. 7A and 7B). Immunohistochemistry (IHC) experiments confirmed a strong and homogeneous CD99 membrane staining, a nuclear FLI1 pattern and absence of nuclear STAG2 expression (Fig. 4B, Supplementary Fig. 7A and 7B). These tumors were highly proliferative and displayed a non-apoptotic pattern as revealed respectively by Ki67 and cleaved caspase 3 staining (Fig. 4B, Supplementary Fig. 7A and 7B). Finally, we profiled these EWIma tumors using RNA-seq. Notably, using unsupervised hierarchical clustering analysis (HCA), transformed EWIma1, 5 and 7 tumors clustered together with EwS cells, whereas their in vitro respective models emerged from a more distant branch (Fig. 3C). Similarly, these tumors were clearly more left shifted on PC1 axis as compared to their respective in vitro counterparts (Fig. 3B). All together, these EWIma models likely represent a novel and large panel of de novo generated EwS cellular models with immortalized and transforming properties.
Figure 4. In vivo tumors obtained from engineered EWIma7 cells.
A- Western blot against FLI1 in EWIma 5, 7 and 1 tumor cells and parental EWIma1 cells.
B- Histology of EWIma7 tumors (representative images). Top panel row: left, H&E staining in tumors at primary orthotopic implantation site (scale bar 200 μm) and anti-CD99 IHC staining at lung metastatic sites (scale bar, middle panel: 1 mm, right panel: 50 μm). Middle row left panel: magnification of H&E in the primary orthotopic tumor displaying a classical small round tumor cell morphology (scale bar: 50 μm). Additional IHC stainings against CD99, FLI1, STAG2, Ki67 and cleaved CASP3 (clCASP3) in primary orthotopic tumors are shown in middle and bottom panel rows (scale bar: 50 μm).
DISCUSSION
In the present work, we successfully and efficiently generated EWSR1-FLI1 transformed cells starting from “normal” non-cancerous MSCs of a EwS patient. These models (EWIma) faithfully recapitulated bona fide EwS characteristics, including cell morphology, transcriptomic, epigenetic, metastatic and plasticity aspects that have been previously reported in EwS cells lines and tumors (12). In addition, we defined here a EWINGness score as a simple surrogate marker to evaluate transformation potential of mesenchymal stem cells towards Ewing sarcoma. Our results further support that these MSCPat are permissive to EWSR1-FLI1 expression under its EWSR1 endogenous promoter and ultimately leads to their transformation in vivo. This demonstrates that Ewing sarcoma can originate from human bone marrow-derived MSC as previously anticipated but never demonstrated so far (8, 9, 11, 14).Whereas this cell is the only permissive one remains to be formally elucidated. For instance, repeating our experimental approach in neural-crest-derived or other stem/progenitor populations would be complementary to define if various cells of origin in EwS exist (13, 35, 36). The EwS tumor generated in this study combines endogenous EWSR1-FLI1 translocation together with most recurrent mutations found in EwS. How STAG2, TP53, and CDKN2A alterations specifically contribute to transformation in our model remains to be clarified in future studies. However, even if the SPC mutations appear to confer a growth advantage to mesenchymal stem cells (Supplementary Fig. 5C), they do not appear to confer a “primed” Ewing transcriptomic signature (MSCPat vs MSCPat_SPC), which is clearly mediated by the EWSR1-FLI1 transcription factor in our MSCPat or hMPC derived models (Fig. 3B and 3C). STAG2 and TP53 mutations can co-occur in EwS at diagnosis and appear to define an aggressive subtype (6). We also recently demonstrated that STAG2 loss-of-function (LOF) mutations reduced the cis-mediated activity of EWSR1-FLI1 (37). In that respect, we can speculate that STAG2 LOF in EWIma models may attenuate the known EWSR1-FLI1 toxicity and therefore favors the emergence of these clones. We also showed that STAG2 LOF increased migratory properties of EwS cells, including in EWIma1 cells (37), which was also previously reported at the clinical level to be associated with metastasis (5) and poor outcome (4). In possible support of this notion, orthotopically engrafted EWIma1, 5 and 7 cells also grew at distant sites such as in the lungs and liver. In addition, the original EwS tumor cells of this patient displayed two chromosomal deletions containing CDKN2A and TP53 at the time of the diagnosis, and gene expression data showed a transcript alteration of STAG2 (Supplementary Fig. 2A). These findings raise the possibility that the simultaneous alteration of p16, p53 and STAG2 expression had a direct “boosting” effect on Ewing sarcomagenesis in this particular patient. Whereas individual or combined SPC mutations in this particular patient tumor or more generally in EwS are concomitant to the translocation in EwS tumors or appear as secondary events remains to be elucidated, and both scenarios may occur. Since SPC mutations are absent from many EwS tumors at the time of diagnosis, it is likely that other combinations of more private mutations together with the pathognomic translocation may also allow to successfully transform MSC into faithful EwS models. Indeed, on average, ten coding variants per tumor were detected in EwS tumors at the time of diagnosis (6) and 120 unique genes were involved in chromoplectic breakpoints in Ewing sarcoma (28). Finally, the time scale and the in vitro aspects of our approach may also explain why we only transformed few MSCPat with the EF+SPC cocktail as several years and/or microenvironmental factors may be necessary to fully transform a cell of origin. For instance, using clock-like mutation signatures in primary and relapse EwS tumors, it was estimated that the cell that would give rise to the relapse existed years before diagnosis (28). The over-proportional weight of the first PCA component and the GSEA signatures (Fig. 3A and 3D) highlighted that the EWSR1-FLI1 transcriptional signature is the predominant feature in our model. Notably, the EWSR1-FLI1 binding pattern at both GGAA-mSats and canonical ETS-like binding sites in EWIma1 were strikingly overlapping with that of the established A673 EwS cell line but highly divergent from the FLI1 binding pattern observed in MSCPat (Fig. 1G and 1H). Acquisition of well-known SEs, reminiscent of a specific EwS identity, clearly demonstrates that EWIma1 cells also display bona fide (neo)-enhancer properties for EWSR1-FLI1 (Fig. 1H). Hierarchical clustering showed that EWIma tumors cluster within a large panel of EwS cell lines. Yet, PC2 which mostly discriminated hMPC and MSCPat derived models, also slightly segregated EWIma1 from EwS cell lines (Fig. 3B). Interestingly, all EWIma tumors display additional ‘EWINGness’ (similar PC1 values to EwS cell lines) as compared to their respective EWIma in vitro models (Fig. 3B and 3C). Exogenous signaling from the microenvironment may account for this difference but remains to be determined.
In this study, we suggested the existence of a ‘permissive milieu’ that could alleviate the potential toxicity of EWSR1-FLI1 expression while favoring its appropriate regulation. We recently demonstrated using GWAS that at least 6 loci were significantly associated with Ewing sarcoma (21). Future experiments, using MSCs with different genotypes at susceptibility loci will enable to more precisely decipher the key genetic elements that are required for EWSR1-FLI1-induced transformation. Besides, recent single cell RNA-seq study of EwS patient-derived xenograft (PDX) tumors highlighted a window in which EwS cells can proliferate (23). Low levels of EWSR1-FLI1 were associated with mesenchymal and apoptotic phenotypes (14, 38), while EwS cells displaying very high EWSR1-FLI1 activity led to absence of proliferation and HIF1α pathway activation (23). In that respect, the collection of EWSR1-FLI1 positive clones generated in this study display a broad and heterogeneous range of EWINGness scores, possibly recapitulating various levels of EWSR1-FLI1 transcriptional activity. However, although all EWIma models generated from MSCPat carried EWSR1-FLI1 translocation and SPC mutations, not all displayed fully immortalized patterns indicating that additional factors (e.g. stemness, cell cycle, oxphos status…) may contribute to the transformation of the EwS cell of origin. Additional investigation with these valuable models will be necessary to answer these key questions.
Here, we also attempted but did not succeed to engineer a transformed EwS model with hMPC cells. Whereas hMPC cells display a bona fide multipotent differentiation potential (24), we anticipated that MSC derived from healthy adolescent bone marrow (match of MSCPat conditions) would have been a better control. However, we could not collect such controls due to the very limited occurrence of such pediatric samples.
Besides modelling Ewing sarcomagenesis, we genetically engineered cells with different karyotypes, including some that are reminiscent of chromoplexy (EWIma 1, 5 and 14), which has been described in ~35% of EwS tumors. Chromoplexy comprises multiple chromosomal translocations that reshuffles chromosomes in a new and scrambled configuration instead of creating simple reciprocal translocations. Recently, chromoplexy has been described in 17.8% of 2,648 whole-cancer genomes from 38 tumor types (39,40) and plausibly as the source of their oncogenic transformation. However, if the exact mechanism of chromoplexy remains to be fully elucidated, modelling this event, as made possible with our gRNA cocktail approach, is of major interest for cancer research. Indeed, in our EWIma models, most chromoplectic-like rearrangements were proved to be initiated at gRNA target sites (and off-target sites for gRNACDKN2A), indicating that they originated from a single burst in MSCPat at the time of EWSR1-FLI1 translocation formation as suggested by genomic data on EwS tumors (28). In addition, genomic regions implicated in chromoplexy are often found in early replicating regions, rich in expressed genes (27, 28, 41). Remarkably, all loci implicated in chromoplectic-like events in EWIma1 cells (including the intergenic off-target site of gRNACDKN2A) were located within early replicating domains of the human MSC genome (Supplementary Fig. 7C)(42). In that respect, our approach may represent an attractive model to investigate how ‘normal’ cells adapt to such a catastrophic burst of rearrangements. In addition to chromoplexy, chromosomal alterations that have been observed in EwS are also present in some of our EWIma models. For instance, a duplication of chr20q is observed in EWIma1. In EwS, trisomy or focal amplifications of chr20 have been described in up to 15% of these tumors (43, 44). Similarly, deletion of chr16q observed in EwS tumors (43, 44) is particularly obvious in EWIma2 cells (Supplementary Fig. 4A).
Recent studies allowed to reconstruct clonal and temporal evolution of tumors using mutational signatures, multiple spatio-temporal tumor sampling and/or single cell sequencing approaches. These top-down approaches allow to speculate about the timing of genetic lesions in the cell of origin without, however, achieving this original stage (40, 45). Our bottom-up strategy is very complementary to these approaches that were also used in EwS to speculate about the timing of the translocation and of the additional alterations. In addition, since EwS genetic susceptibility loci have been identified (21), it would be interesting to expand this collection when starting from additional untransformed cell of origin collections (possibly derived from EwS and non-EwS patients) which may ultimately allow to determine how eQTL related genes affect Ewing sarcomagenesis. Combining top-down and bottom-up strategies may ultimately allow to answer these complex questions, especially in sarcoma where many new entities are presumed to be driven by candidate gene fusion oncogenes (46).
In conclusion, this work demonstrates that EwS can originate from bone marrow derived mesenchymal stem cells. It further provides evidence of the necessity to reach a minimal level of EWSR1-FLI1-mediated transcriptional activity, within a defined genomic susceptibility context, to achieve full immortalization and transformation of this cell of origin. All together, we successfully bypassed here the challenge of modeling EwS ab initio. Our model mimics a rather aggressive form of EwS with SPC mutations displaying single balanced EWSR1-FL1 translocation but also chromoplectic-like events and transforming properties in mice. Our successful approach to generate bona fide EwS cells opens broad avenues to gain important insights into Ewing sarcomagenesis but also into mechanisms related to chromoplexy. More generally, this transposable approach shall allow to investigate sarcomagenesis in the highly heterogeneous family of sarcoma tumors.
Supplementary Material
Acknowledgements
We thank the iPS (Nathalie Lefort), Imagine cytometry (Olivier Pellé), and microscopy (Meriem Garfa-Traore) platforms for help with SNP karyotyping, cell analysis and image analysis, Lina El Kassar (CECS/I-STEM), Dr. Chloé Lescale and Dr. Ludovic Deriano (Institut Pasteur) for help with the multicolor FISH analysis and Dr. Marion Piganeau for technical support and helpful discussions. We thank Rosalie Borry and each member of the ‘Genome Dynamics in the Immune System’ lab for scientific discussions and technical support. We thank E. Lapouble, G. Pierron, C. Thirant, R. Leclerc, V. Raynal, S. Baulande, P. Legoix, C. Kamoun, M-M. Aynaud and E. Barillot and all members of the Genetics and Biology of Pediatric Cancers laboratory for helpful discussions and/or technical or bioinformatics assistance. We thank H. Kovar for providing Ewing sarcoma cell lines and COG Childhood Cancer Repository for the CHLA cell line.
Funding:
This work was supported by the Institut National du Cancer (PLBIO16-291), by La Ligue Nationale Contre le Cancer (P.R/E.B. and O.D/D.S teams: Équipes Labellisées), the Fondation ARC (L.B.); the AIDA association (M.H.), by grants from the Institut Curie; the INSERM; the Canceropôle Ile-de-France; the projet de Recherche ‘Enfants, Adolescents et Cancer’; the Agence Nationale de la Recherche (ANR-10-EQPX-03, Institut Curie Génomique d’Excellence (ICGex) and the société française de lutte contre les cancers de l’enfant et de l’adolescent. This project also received support from European funding: ERA-NET TRANSCAN JTC 2014 (TRAN201501238) and TRANSCAN JTC 2017 (TRANS201801292). D.S. is supported by the Institut Curie–SIRIC (Site de Recherche Intégrée en Cancérologie) program. M.J is supported by NIH (NCI) R35 CA253174. T.G.P.G. is supported by grants from the Gert and Susanna Mayer foundation, the Barbara and Wilfried Mohr foundation, and the SMARCB1 association. We are indebted to the following associations for providing essential support: L’Etoile de Martin, Aida, la Course de l'Espoir, M la vie avec Lisa, ADAM Couleur Jade, Dans les pas du Géant, Courir pour Mathieu, Marabout de Ficelle, Olivier Chape, Les Bagouzamanon, Enfants et Santé, and les Amis de Claire.
Footnotes
The authors declare no potential conflicts of interest.
References
- 1.Delattre O, Zucman J, Plougastel B, Desmaze C, Melot T, Peter M, et al. Gene fusion with an ETS DNA-binding domain caused by chromosome translocation in human tumours. Nature 1992;359:162–5 [DOI] [PubMed] [Google Scholar]
- 2.Lessnick SL, Dacwag CS, Golub TR. The Ewing's sarcoma oncoprotein EWS/FLI induces a p53-dependent growth arrest in primary human fibroblasts. Cancer Cell 2002;1:393–401 [DOI] [PubMed] [Google Scholar]
- 3.Sohn EJ, Li H, Reidy K, Beers LF, Christensen BL, Lee SB. EWS/FLI1 oncogene activates caspase 3 transcription and triggers apoptosis in vivo. Cancer Res 2010;70:1154–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brohl AS, Solomon DA, Chang W, Wang J, Song Y, Sindiri S, et al. The genomic landscape of the Ewing Sarcoma family of tumors reveals recurrent STAG2 mutation. PLoS Genet 2014;10:e1004475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Crompton BD, Stewart C, Taylor-Weiner A, Alexe G, Kurek KC, Calicchio ML, et al. The genomic landscape of pediatric Ewing sarcoma. Cancer Discov 2014;4:1326–41 [DOI] [PubMed] [Google Scholar]
- 6.Tirode F, Surdez D, Ma X, Parker M, Le Deley MC, Bahrami A, et al. Genomic landscape of Ewing sarcoma defines an aggressive subtype with co-association of STAG2 and TP53 mutations. Cancer Discov 2014;4:1342–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Minas TZ, Surdez D, Javaheri T, Tanaka M, Howarth M, Kang HJ, et al. Combined experience of six independent laboratories attempting to create an Ewing sarcoma mouse model. Oncotarget 2017;8:34141–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Riggi N, Cironi L, Provero P, Suva ML, Kaloulis K, Garcia-Echeverria C, et al. Development of Ewing's sarcoma from primary bone marrow-derived mesenchymal progenitor cells. Cancer Res 2005;65:11459–68 [DOI] [PubMed] [Google Scholar]
- 9.Castillero-Trejo Y, Eliazer S, Xiang L, Richardson JA, Ilaria RL Jr., Expression of the EWS/FLI-1 oncogene in murine primary bone-derived cells Results in EWS/FLI-1-dependent, ewing sarcoma-like tumors. Cancer Res 2005;65:8698–705 [DOI] [PubMed] [Google Scholar]
- 10.El Beaino M, Liu J, Wasylishen AR, Pourebrahim R, Migut A, Bessellieu BJ, et al. Loss of Stag2 cooperates with EWS-FLI1 to transform murine Mesenchymal stem cells. BMC Cancer 2020;20:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Riggi N, Suva ML, De Vito C, Provero P, Stehle JC, Baumer K, et al. EWS-FLI-1 modulates miRNA145 and SOX2 expression to initiate mesenchymal stem cell reprogramming toward Ewing sarcoma cancer stem cells. Genes Dev 2010;24:916–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grünewald TGP, Cidre-Aranaz F, Surdez D, Tomazou EM, de Alava E, Kovar H, et al. Ewing sarcoma. Nat Rev Dis Primers 2018;4:5. [DOI] [PubMed] [Google Scholar]
- 13.von Levetzow C, Jiang X, Gwye Y, von Levetzow G, Hung L, Cooper A, et al. Modeling initiation of Ewing sarcoma in human neural crest cells. PLoS One 2011;6:e19305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tirode F, Laud-Duval K, Prieur A, Delorme B, Charbord P, Delattre O. Mesenchymal stem cell features of Ewing tumors. Cancer Cell 2007;11:421–9 [DOI] [PubMed] [Google Scholar]
- 15.Piganeau M, Ghezraoui H, De Cian A, Guittat L, Tomishima M, Perrouault L, et al. Cancer translocations in human cells induced by zinc finger and TALE nucleases. Genome research 2013;23:1182–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Torres-Ruiz R, Martinez-Lage M, Martin MC, Garcia A, Bueno C, Castano J, et al. Efficient Recreation of t(11;22) EWSR1-FLI1(+) in Human Stem Cells Using CRISPR/Cas9. Stem Cell Reports 2017;8:1408–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brunet E, Simsek D, Tomishima M, DeKelver R, Choi VM, Gregory P, et al. Chromosomal translocations induced at specified loci in human stem cells. Proc Natl Acad Sci U S A 2009;106:10620–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fraumeni JF Jr., Glass AG. Rarity of Ewing's sarcoma among U.S. Negro children. Lancet 1970;1:366–7 [DOI] [PubMed] [Google Scholar]
- 19.Jawad MU, Cheung MC, Min ES, Schneiderbauer MM, Koniaris LG, Scully SP. Ewing sarcoma demonstrates racial disparities in incidence-related and sex-related differences in outcome: an analysis of 1631 cases from the SEER database, 1973-2005. Cancer 2009;115:3526–36 [DOI] [PubMed] [Google Scholar]
- 20.Randall RL, Lessnick SL, Jones KB, Gouw LG, Cummings JE, Cannon-Albright L, et al. Is There a Predisposition Gene for Ewing's Sarcoma? J Oncol 2010;2010:397632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Machiela MJ, Grünewald TGP, Surdez D, Reynaud S, Mirabeau O, Karlins E, et al. Genome-wide association study identifies multiple new loci associated with Ewing sarcoma susceptibility. Nat Commun 2018;9:3184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grünewald TG, Bernard V, Gilardi-Hebenstreit P, Raynal V, Surdez D, Aynaud MM, et al. Chimeric EWSR1-FLI1 regulates the Ewing sarcoma susceptibility gene EGR2 via a GGAA microsatellite. Nat Genet 2015;47:1073–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aynaud MM, Mirabeau O, Gruel N, Grossetete S, Boeva V, Durand S, et al. Transcriptional Programs Define Intratumoral Heterogeneity of Ewing Sarcoma at Single-Cell Resolution. Cell Rep 2020;30:1767–79.e6 [DOI] [PubMed] [Google Scholar]
- 24.Barberi T, Willis LM, Socci ND, Studer L. Derivation of multipotent mesenchymal precursors from human embryonic stem cells. PLoS Med 2005;2:e161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fehrer C, Brunauer R, Laschober G, Unterluggauer H, Reitinger S, Kloss F, et al. Reduced oxygen tension attenuates differentiation capacity of human mesenchymal stem cells and prolongs their lifespan. Aging Cell 2007;6:745–57 [DOI] [PubMed] [Google Scholar]
- 26.Renouf B, Piganeau M, Ghezraoui H, Jasin M, Brunet E. Creating Cancer Translocations in Human Cells Using Cas9 DSBs and nCas9 Paired Nicks. Methods in enzymology 2014;546:251–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Baca SC, Prandi D, Lawrence MS, Mosquera JM, Romanel A, Drier Y, et al. Punctuated evolution of prostate cancer genomes. Cell 2013;153:666–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Anderson ND, de Borja R, Young MD, Fuligni F, Rosic A, Roberts ND, et al. Rearrangement bursts generate canonical gene fusions in bone and soft tissue tumors. Science 2018;361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Boeva V, Surdez D, Guillon N, Tirode F, Fejes AP, Delattre O, et al. De novo motif identification improves the accuracy of predicting transcription factor binding sites in ChIP-Seq data analysis. Nucleic Acids Res 2010;38:e126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kennedy AL, Vallurupalli M, Chen L, Crompton B, Cowley G, Vazquez F, et al. Functional, chemical genomic, and super-enhancer screening identify sensitivity to cyclin D1/CDK4 pathway inhibition in Ewing sarcoma. Oncotarget 2015;6:30178–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Baldauf MC, Orth MF, Dallmayer M, Marchetto A, Gerke JS, Rubio RA, et al. Robust diagnosis of Ewing sarcoma by immunohistochemical detection of super-enhancer-driven EWSR1-ETS targets. Oncotarget 2018;9:1587–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marchetto A, Ohmura S, Orth MF, Knott MML, Colombo MV, Arrigoni C, et al. Oncogenic hijacking of a developmental transcription factor evokes vulnerability toward oxidative stress in Ewing sarcoma. Nat Commun 2020;11:2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hendel A, Bak RO, Clark JT, Kennedy AB, Ryan DE, Roy S, et al. Chemically modified guide RNAs enhance CRISPR-Cas genome editing in human primary cells. Nature biotechnology 2015;33:985–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bilke S, Schwentner R, Yang F, Kauer M, Jug G, Walker RL, et al. Oncogenic ETS fusions deregulate E2F3 target genes in Ewing sarcoma and prostate cancer. Genome research 2013;23:1797–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Staege MS, Hutter C, Neumann I, Foja S, Hattenhorst UE, Hansen G, et al. DNA microarrays reveal relationship of Ewing family tumors to both endothelial and fetal neural crest-derived cells and define novel targets. Cancer Res 2004;64:8213–21 [DOI] [PubMed] [Google Scholar]
- 36.Tanaka M, Yamazaki Y, Kanno Y, Igarashi K, Aisaki K, Kanno J, et al. Ewing’s sarcoma precursors are highly enriched in embryonic osteochondrogenic progenitors. The Journal of clinical investigation 2014;124:3061–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Surdez D, Zaidi S, Grossetête S, Laud-Duval K, Ferre AS, Mous L, et al. STAG2 mutations alter CTCF-anchored loop extrusion, reduce cis-regulatory interactions and EWSR1-FLI1 activity in Ewing sarcoma. Cancer Cell 2021 [DOI] [PubMed] [Google Scholar]
- 38.Stoll G, Surdez D, Tirode F, Laud K, Barillot E, Zinovyev A, et al. Systems biology of Ewing sarcoma: a network model of EWS-FLI1 effect on proliferation and apoptosis. Nucleic Acids Res 2013;41:8853–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Campbell PJ, Getz G, Korbel JO. Pan-cancer analysis of whole genomes. Nature 2020;578:82–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gerstung M, Jolly C, Leshchiner I, Dentro SC, Gonzalez S, Rosebrock D, et al. The evolutionary history of 2,658 cancers. Nature 2020;578:122–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee JJ, Park S, Park H, Kim S, Lee J, Lee J, et al. Tracing Oncogene Rearrangements in the Mutational History of Lung Adenocarcinoma. Cell 2019;177:1842–57.e21 [DOI] [PubMed] [Google Scholar]
- 42.Rivera-Mulia JC, Buckley Q, Sasaki T, Zimmerman J, Didier RA, Nazor K, et al. Dynamic changes in replication timing and gene expression during lineage specification of human pluripotent stem cells. Genome research 2015;25:1091–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jahromi MS, Jones KB, Schiffman JD. Copy Number Alterations and Methylation in Ewing's Sarcoma. Sarcoma 2011;2011:362173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ozaki T, Paulussen M, Poremba C, Brinkschmidt C, Rerin J, Ahrens S, et al. Genetic imbalances revealed by comparative genomic hybridization in Ewing tumors. Genes Chromosomes Cancer 2001;32:164–71 [DOI] [PubMed] [Google Scholar]
- 45.Alexandrov LB, Jones PH, Wedge DC, Sale JE, Campbell PJ, Nik-Zainal S, et al. Clock-like mutational processes in human somatic cells. Nat Genet 2015;47:1402–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Watson S, Perrin V, Guillemot D, Reynaud S, Coindre JM, Karanian M, et al. Transcriptomic definition of molecular subgroups of small round cell sarcomas. The Journal of pathology 2018;245:29–40 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
ChIP-seq and RNA-seq data have been deposited in NCBI’s Gene Expression Omnibus under accession number: GSE150783.
Raw data are available at Mendeley. Reserved DOI: doi:10.17632/fx29by5k43.1 https://data.mendeley.com/datasets/fx29by5k43/draft?a=63bd5c8f-f13b-4100-b9bf-988ddb0c9131