Abstract
Mutations in the isocitrate dehydrogenase 1 (IDH1) and IDH2 genes are frequently observed in a wide variety of hematologic malignancies, including myeloid and T-cell leukemias. In this study, we generated Idh2R140Q transgenic mice to examine the role of the Idh2R140Q mutation in leukemia. No leukemia developed in Idh2R140Q transgenic mice, suggesting a need for additional genetic events for leukemia development. Since myeloid cells from NUP98-HOXD13 fusion (NHD13) transgenic mice frequently acquire somatic Idh mutations when they transform to AML, we generated Idh2R140Q/NHD13 double transgenic mice. Idh2R140Q/NHD13 transgenic mice developed an immature T cell leukemia with an immunophenotype similar to double-negative 1 (DN1) or DN2 thymocytes. Idh2R140Q/NHD13 leukemic cells were enriched for an early thymic precursor transcriptional signature, and the gene expression profile for Idh2R140Q/NHD13 DN1/DN2 T-ALL closely matched that of human early/immature T cell precursor (EITP) ALL. Moreover, recurrent mutations found in EITP ALL patients, including KRAS, PTPN11, JAK3, SH2B3, and EZH2 were also found in Idh2R140Q/NHD13 DN1/DN2 T-ALL. In vitro treatment of Idh2R140Q/NHD13 thymocytes with enasidenib, a selective inhibitor of mutant IDH2, led to a marked decrease in leukemic cell proliferation. These findings demonstrate that Idh2R140Q/NHD13 mice can serve as a useful in vivo model for the study of EITP ALL development and therapy.
Introduction
IDH genes code for isocitrate dehydrogenase enzymes. These homo-dimeric enzymes are part of the tricarboxylic acid cycle and play an important role in cellular metabolism. For both isocitrate dehydrogenase 1(IDH1) (located in cytoplasm and peroxisomes) or its isoform isocitrate dehydrogenase 2 (IDH2) (located in mitochondria), the primary function of IDH enzymes is to convert isocitrate to alpha ketoglutarate in a nicotinamide adenine dinucleotide (NAD) phosphate dependent manner(1).
Mutations in IDH genes are associated with numerous forms of human cancer. IDH mutations have been reported in human gliomas, chondrosarcoma(2), cholangiocarcioma(3) and hematopoietic malignancies, including angioimmunoblastic T cell lymphoma(4), AML and immature T-lineage leukemias(5–8). IDH1 and IDH2 have been shown to be frequently mutated in acute myeloid leukemia (up to 33% reported in one study), with IDH2 mutations reported in 19% of AML cases(6). The majority of IDH1/2 mutations occur at arginine residue p.R132 for IDH1 and p.R140 and p.R172 for IDH2 in AML. The IDH2 p.R140 mutation(9, 10) has regularly been reported to be the most frequent IDH mutation among IDH mutations identified in AML(6, 11, 12).
As a consequence of the p.R140 point mutation, IDH2 acquires neomorphic enzymatic activity resulting in NADPH dependent reduction of alpha-ketoglutarate to 2-hydroxyglutarate (2-HG). This new functional activity is a common feature for all of the common IDH1 and IDH2 mutations, and leads to accumulation of 2-HG in patients with IDH2 p.R140 mutation at levels 10– 100 fold higher compared to wild type IDH AML samples(10). 2-HG is a competitive inhibitor of alpha-ketoglutarate dependent dioxygenases including histone demethylases and TET family methylcytosine dioxygenase enzymes, leading to aberrant histone and DNA methylation, resulting in epigenetic modification which is suggested to contribute to tumorigenesis(13). To investigate the role of IDH2 mutation in leukemia transformation we generated mice that expressed an Idh2R140Q mutant in the hematopoietic compartment of mice.
Materials and Methods
Transgenic mice
The NHD13 transgenic mouse was previously described(14) and has been maintained in NIH intramural animal facilities. To generate the Vav- Idh2R140Q transgenic mouse, a murine Idh2 cDNA (Cat# MMM1013–9201034) was purchased from Thermo Fisher Scientific, and site-directed mutagenesis (QuikChange, Agilent Technologies, part # 200519) was used to generate a p.R140Q mutation. A full length Idh2 cDNA was PCR amplified with primers containing a SfiI restriction site at the 5’ end and a NotI site at the 3’ end. This PCR fragment was cloned into pGEM-T Easy (Promega, Cat# A1360) and sequenced to verify that no sequence error had been generated by PCR. This plasmid was digested with SfiI and NotI and a 1677bp cDNA fragment was cloned into the HS 21/45-vav vector which contained 5’ and 3’ Vav regulatory elements(15).
The Vav-Idh2R140Q construct was linearized and separated from the plasmid backbone and microinjected into zygotes from FVB/N mice; mice that had incorporated the transgene were identified by Southern blot, and offspring from two founders (L3 and K6) were followed. To generate Idh2R140Q /NHD13 double transgenic mice, Idh2R140Q mice (L3 line) were crossed with NHD13 mice. Genotyping of tail tissue genomic DNA was performed by PCR using primers designated in supplementary Table S1.
Leukemia evaluation
Mice were monitored for signs of illness such as tachypnea, weight loss, lethargy, and kyphosis. In addition, complete blood counts (CBC) were obtained at 2–4month intervals beginning at age 6 months. Moribund mice were euthanized and evaluated for leukemia using established guidelines(16). These evaluations included routine clinical studies, such as a CBC, peripheral blood smear, bone marrow cytospin, and necropsy. Single cell suspensions of BM, spleen, thymus, and enlarged lymph nodes were analyzed by multicolor flow cytometry (Supplemental Table S2)and formalin fixed hematopoietic (BM, spleen, thymus) and non-hematopoietic (liver, kidney, lung) organs were analyzed by immunohistochemistry. In addition, selected samples were assessed by whole exome sequencing, RNA-seq, or oligonucleotide-based gene expression arrays.
For the Idh2/NHD13 transgenic survival study all male and female mice were followed from birth to 22 months of age by animal caretakers. The animal caretakers were blinded as to experimental group. The study was powered to be able to detect a 20% increase in frequency of leukemia.
Flow cytometry, histology and immunohistochemistry
Flow cytometry analysis was performed as described previously(17), using a FACScan (BD Pharmingen) instrument. The antibodies used to characterize leukemic cells are indicated in supplementary Table S2. 4′,6-diamidino-2-phenylindole (DAPI) (BD Pharmingen, Cat#564907) or propidium iodide (PI) (Invitrogen, Cat# P3566) were used for viability staining.
For sorting DN cells; DN thymocytes were enriched by staining with CD4 (L3T4)(Cat# 130–117-043) and CD8a (Ly-2)(Cat#130–117-044) Microbeads (Miltenyi biotec), and depletion of the stained cells using MACS LD column (Cat# 130–042-901, Miltenyi biotec), according to the manufacturer’s recommended protocol. The depleted cells were then stained with mouse anti CD4, CD8, CD25, CD44 specific antibodies and sorted with an Aria FACS Aria II, FACS instrument.
Tissues were fixed in 10% Formalin, paraffin-embedded, and sections stained with hematoxylin and eosin, CD3 (MCA1477, AbD Serotec, Raleigh, NC, USA), B220 (553086, BD Pharmingen), or Myeloperoxidase (1:1000, DAKO-A0398). Tissue histology was analyzed as described previously(18).
Tcrb VDJ and DJ gene rearrangements assay
Tcrb VDJ and DJ gene rearrangements assay was performed as described previously(18, 19) using leukemic and control RNA and genomic DNA respectively.
Transplantation and engraftment of leukemic cells
Eight- to 10-week-old C57BL/6 recipient mice that express the CD45.1 allotype of CD45 (purchased from Charles River) were lethally or sub-lethally irradiated (900cGy or 600cGy respectively) and transplanted through tail vein injections with donor leukemia cells harvested from spleen. The lethal radiation recipients were also transplanted with a life-sparing dose of 2 × 105 WT BMNC as described previously(20). Peripheral blood of transplanted mice was examined for donor leukemic cell engraftment at 2, 4, 6 and 10 weeks for primary leukemia transplantation and 2 and 3 weeks for secondary transplantation as described previously(18).
OP9-DL1 co-culture and AG-221 treatment
Pooled DN1/DN2 thymocytes were purified from whole thymus as described above from Idh2R140Q/NHD13, NHD13, Idh2R140Q and wild type mice for OP9-DL1 coculture. CD4 and CD8 double negative (DN) thymocytes (1X10E5) were plated onto an OP9-DL1 monolayer and cultured in a MEM media with 20% fetal bovine serum (FBS), 5 ng/mL Flt3L and 1 ng/mL interleukin (IL) 7 (PeproTech, Rockynd Hill, NJ, USA). Media was changed every 2 days and passaged every 4 days as previously described(21, 22). Three independent replicates done for this study.
For experiments with AG-221 (Selleck, USA), an immortalized Idh2R140Q/NHD13 cell line was developed by continuous growth on OP9-DL1 with Flt3/IL7 supplement as described above. AG-221 was dissolved in DMSO at a stock concentration of 10mM, divided into aliquots, stored at −20 degree Celsius, and used at a final concentration of 5 or 10uM.
Whole exome sequencing
Whole exome sequencing was performed on Idh2R140Q/NHD13 DN1/DN2 T-ALL and NHD13 non-DN1/DN2 T-ALL (CD4/CD8 single or double positive) mice samples as described previously(23). Briefly, libraries were prepared by tagmentation using the Nextera DNA Library Kit (Illumina, Inc.) according to the manufacturer’s instructions with minor modifications. Exome capture was carried out using the SureSelect XT Mouse All Exon, 49.6Mb Kit (Agilent Technologies, Inc.) and libraries were quantified by qPCR (KAPA Biosystems) and sequenced on a HiSeq 2500 System (Illumina, Inc.). Data processing and variant calling procedure followed the Best Practices Workflow for Somatic short variant discovery recommended by the Broad Institute. The sequences were aligned to mouse genome build mm10. Somatic variant calling was performed by comparison of a tumor sample to WT samples using the MuTect2(24) somatic variant caller developed the Broad Institute. Single nucleotide variants (SNV) were first filtered with the filtering criteria recommended by the GATK (https://software.broadinstitute.org/gatk/documentation/article.php?id=3225) and then filtered with the following additional filters: (1)Minimum fraction of altered reads in a tumor is 0.3; (2) Minimum number of altered reads in a tumor is 2; (3) Minimum log fisher is 2; (4) Impact effect is ‘High’ or ‘Moderate’ for Tier 1 indels and missense mutation; (5) Exclude SNPs reported in dbSNP build 137 or previously identified as germline variants in the NIH C57Bl6 colony.
For validation of mutations we performed PCR using primers specific for amplification of exonic regions with mutations (identified by WES). PCR product was then sequenced by conventional PCR-based Sanger sequencing and the sequencing reads were analyzed for mutations using Sequencher software.
Gene expression array analysis
Total RNA was isolated from leukemic Idh2R140Q/NHD13 spleen, leukemic SCL/LMO1 thymi and WT thymi using TRIzol reagent (Cat# 15596026, Invitrogen) following the manufacturer’s recommended protocol. Genomic DNA was removed from Total RNA using Qiagen DNase kit (Cat No./ID: 79254). 1ug of total RNA was reverse transcribed, labelled, and hybridized to GeneChip array (GeneChip Mouse Genome 430 2.0) for gene expression array experiment performed by the NCI Gene Expression Core.
Gene set enrichment analysis
Gene set enrichment analysis (GSEA) was performed using GSEA software (v 4.0.3). The 100 most highly upregulated and down regulated genes identified from fold-change of the differential gene expression analysis study for mice Idh2R140Q/NHD13 vs WT samples were used for enrichment analysis with publicly available data GSE24142(25), comparing adult ETP(DN1) with adult DN3 cells.
For GSEA enrichment analysis comparing mice leukemia with human leukemia, we used the 150 probesets with the greatest fold-change from patients with ETP ALL compared to non-ETP T-ALL(26) and generated gene signatures of upregulated and downregulated genes. This gene signature was then compared to gene expression arrays from Idh2R140Q/NHD13 DN1/DN2 T-ALL or SCL/LMO1 non-DN1/DN2 T-ALL using GSEA. Similar analysis was performed using an independent set of early immature, ETP-like T-ALL vs mature T-ALL(8).
2-hydroxyglutarate (2-HG) detection
30 ± 0.05 mg Tissue was placed in 0.600 ml acetonitrile:water (70:30 v/v) and 20 μl of DL-Norleucine (DLN-IS,10μM) added, followed by homogenization using a Precellys homogenizer (Bertin Instruments, Montigny-le-Bretonneux, France), utilizing 1.0 mm zirconia/silica beads for 30 sec at 6500 rpm. The samples were centrifuged at 20,000g for 15 min at 4˚C and 500 μl of supernatant was taken and dried in a SpeedVac concentrator at room temperature. The dried residue was derivatized by adding 60 μl BSTFA and sonicated for 30 min at room temperature. The samples were diluted with 60 μl acetonitrile, briefly vortexed for 10 s and 1.00 μl was injected into the GCMS using an autosampler.
Calibration standards solutions of 2-HG (5.0 mM) were prepared in acetonitrile. Further working solutions were prepared using intermediate solutions of 500.0 μM and 50.0 μM in acetonitrile:water (50:50 v/v). Calibration curve standards were made at 0.5, 5.0, 10, 40, 80 and 100 μM. while the quality control samples were prepared at four levels, that is, 85 μM (HQC, high quality control), 30 μM (MQC, middle quality control), 15.0 μM (LQC, low quality control), 0.5 μM (LLOQ QC, lower limit of quantitation quality.
2-HG chromatography carried out on a HP-5MS capillary column (30 m × 0.250 mm, 0.25 μm; Agilent Technologies, Foster City, CA) with 17.0 min run time. Analyte was separated at 10.4 min retention time, m/z 247.1 (qualifier ions m/z 203.1, 349.2) TMS derivatives on SIM mode of 40–500 m/z. Analyses were performed with an Agilent 6890N gas chromatograph coupled to an Agilent 5973 mass-selective detector (MSD) with following chromatographic conditions: Initial temperature 100˚C for 2 min, increasing to 200˚C at 10˚C/min for 17 min. The front inlet temperature was 230˚C operating with a split less mode. MSD ion source and interface temperature was 230˚C. The MSD operated in EI mode at 70 eV. Carrier gas was He (1.0 ml/min). GCMS data were acquired and processed using Agilent MassHunter WorkStation software.
Reduced Representation Bisulfite Sequencing (RRBS)
Three WT thymus, three non-DN1/DN2 NHD13 T-ALL, and eight DNA/DN2 NHD13/Idh2R140Q samples were analyzed by RRBS, as previously described(27). Briefly Illumina TruSeq libraries were generated from 500 ng of genomic DNA(28) and 36 bp single-end sequenced on an Illumina HiSeq 2000. Reads were mapped (mm9) with BSMAP 2.74. Quantification and differential analyses were performed with methylKit 0.9.2.(29). Analyses focused on 500,789 CpG sites interrogated 3 or more times in all 14 experiments. Methylation data were mapped both to 5’ regulatory regions of genes of interest as described in the text as well as hematopoietic enhancer regions(30). Differential methylation between groups was calculated by logistic regression and p-values were adjusted to q-values using the SLIM method. Differentially methylated regions were selected based on q-value and percent methylation difference cutoffs of 0.05 and 25%, respectively. Gene-enhancer assignments were predicted with the GREAT 2.0 Web server. Functional enrichment analyses of hyper- and hypo-methylated enhancer associated genes was performed with DAVID.
Statistics
Log-rank (Mantel-Cox) test was used to determine statistical significance in survival studies. Mann Whitney test was used for calculating p value for difference in 2-HG concentration between WT thymocytes and Idh2R140Q thymocytes. Student t test was used to determine statistical significance for the WBC, hemoglobin and platelet counts from CBC analysis between the four mouse genotypes and for 2-HG concentration between WT splenocytes and Idh2R140Q/NHD13 splenocytes. A Fisher exact test was used for the Notch1 mutation comparison. The GraphPad Prism statistical package (v 8.4.3) was used.
Study approval
All animal studies were approved by the National Cancer Institute (NCI) Intramural Animal Care and Use Committee, and all experiments were performed in accordance with the relevant guidelines and regulations.
Results
Generation of Idh2R140Q transgenic mice
We used Vav1 regulatory elements(15) to express an Idh2R140Q mutant cDNA (Supplemental Figure S1A) in all hematopoietic tissues (Supplemental Figure S1B). We followed the progeny of two founder lines (L3 and K6) and observed Mendelian ratios of inheritance (Supplemental Figure S1D). 2-HG levels were significantly higher in Idh2R140Q thymocytes compared to wildtype thymocytes, indicating that the Idh2R140Q allele was functional and produced the 2-HG oncometabolite (Supplemental Figure S1E). However, the overall survival of Idh2R140Q mice was not significantly different than that of wildtype mice (n=33 WT and 40 Idh2R140Q mice; p=0.201) (Supplemental Figure S1F). These findings are in agreement with other studies which showed that expression of mutant IDH1/2 was not sufficient for development of leukemia in mice(31, 32).
Idh2R140Q/NHD13 mice die of an immature T cell leukemia
We recently identified recurrent, acquired Idh1 p.R132H mutations in NUP98-HOXD13-driven AML(33), suggesting a possible collaboration of these two aberrations during leukemic transformation. To address this hypothesis, we crossed Idh2R140Q mice with NUP98-HOXD13 (NHD13) mice and monitored the progeny for signs of disease. As shown in Figure 1A, all Idh2R140Q/NHD13 mice died by the age of one year (median survival of 315 days), exhibiting a significantly shorter overall survival compared with all other genotypes.
Figure 1. Idh2R140Q/NHD13 mice develop a highly penetrant, lethal DN1/DN2 T-ALL.
A, survival of Idh2R140Q/NHD13(n=31), Idh2R140Q(n=30), NHD13(n=34) and WT(n=20) mice. Log-rank (Mantel-Cox) p value <0.0001 for Idh2R140Q/NHD13 vs WT, <0.0001 for Idh2R140Q/NHD13 vs Idh2R140Q, and <0.05 for Idh2R140Q/NHD13 vs NHD13. B, leukocytosis and blast morphology in peripheral blood from Idh2R140Q/NHD13 leukemia C, H&E and CD3 staining from Idh2R140Q/NHD13 leukemic mouse. D, flow cytometry of leukemic spleen stained for CD44, CD25, CD90 and cKit to identify DN1 or DN2 cells.
Complete blood counts (CBC) revealed anemia, thrombocytopenia and leukocytosis in Idh2R140Q/NHD13 mice, consistent with development of acute leukemia (Supplemental Figure S2A; Supplemental Table S3). NHD13 mice showed anemia, thrombocytopenia, and variable leukopenia or leukocytosis, consistent with prior studies of NHD13 mice(14, 34).
Leukemic Idh2R140Q/NHD13 mice consistently exhibited splenomegaly (Supplemental Figure S2B). As expected, the level of 2-HG in leukemic Idh2R140Q/NHD13 spleens was significantly higher than in wild type splenocytes (Supplemental Figure S2C). Peripheral blood smears from Idh2R140Q/NHD13 leukemic mice showed leukocytosis and cells with blast morphology (Figure 1B) and tissue histology showed infiltration of leukemic blasts into the bone marrow and perivascular regions of the liver, consistent with disseminated leukemia (Figure 1C). We focused our detailed studies on the initial ten leukemic Idh2R140Q/NHD13 mice, and characterized the leukemias with a panel of flow cytometry antibodies (CD3, CD4, CD8, B220, CD19, Mac1, Gr1), immunohistochemistry (IHC; CD3), and Tcrb gene rearrangements to classify the developmental stage of the Idh2R140Q/NHD13 leukemia.
All ten leukemias were negative or dim for B220 and CD19, ruling out the possibility that these were B-cell leukemia. Two of the ten Idh2R140Q/NHD13 leukemias (numbers 570 and 590) were positive for Mac1, demonstrating that these mice developed AML, consistent with the AML previously identified in NHD13+ mice(14, 34) (Table 1). The remaining eight samples were CD4- CD8- (Double Negative or DN) by flow cytometry (Table 1), ruling out a mature T-cell leukemia. All eight of these were positive for CD3 by IHC (Figure 1C and Table 1), suggesting that these Idh2R140Q/NHD13 mice might have developed a leukemia of immature precursor T-cell origin(26, 35–39). Surprisingly, six of eight Idh2R140Q/NHD13 leukemias that stained positive for CD3 by IHC were negative for CD3 by flow cytometry.
Table.1.
Summary of the immunophenotype of Idh2R140Q/NHD13 mice
| Mouse ID | IHC | Flow surface markers | Flow DN1/DN2 markers | TCRB V-DJ rearrangements | TCRB D-J rearrangements | Diagnosis | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CD4 | CD8 | CD3 | CD19 | B220 | Mac1 | cKit | CD25 | CD44 | CD90 | |||||
| 524 | CD3+ | - | - | - | - | - | - | low | + | + | + | - | D1J1S3 | DN2 ALL |
| 525 | CD3+ | - | - | - | - | - | - | + | - | + | - | - | D1J1S3, D1J1S6, D2J2S4 | DN1 ALL |
| 526 | CD3+ | - | - | - | - | - | - | low | + | + | + | - | D1J1S1 | DN2 ALL |
| 531 | CD3+ | - | - | - | - | - | - | low | - | + | - | - | GL | DN1 ALL |
| 546 | CD3+ | - | - | + | NT | - | - | low | low | + | low | - | D1J1S3 | DN1/DN2 ALL |
| 550 | CD3+ | - | - | + | - | - | - | - | - | + | + | - | GL | DN1 ALL |
| 555 | CD3+ | - | - | - | - | - | - | low | - | + | - | - | D1J1S2, D1J1S5 | DN1 ALL |
| 559 | CD3+ | - | - | - | - | - | - | + | - | + | + | - | D1J1S2 | DN1 ALL |
| 570 | NT | - | - | - | - | low | + | - | - | + | - | - | GL | Immature AML |
| 590 | NT | - | - | - | - | - | + | - | - | + | - | - | GL | AML |
| 662 | NT | - | - | - | - | - | + | low | low | + | + | NT | Thymus-D1J1S3, D1J1S6, D2J2S4, Spleen- D2J2S4 | DN1/DN2 lymphoma |
| 664 | NT | - | - | - | - | - | + | low | low | low | + | - | Thymus- D1J1S1, D1J1S3, Spleen- D1J1S1 | DN2 lymphoma |
| 668 | NT | - | - | - | - | - | - | low | + | low | + | NT | Thymus-D1J1S4 (major clone), D1J1S3, D1J1S6, D2J2S2 (minor clone) | DN2 lymphoma |
NT: not tested
The discordant IHC and flow cytometry results for CD3 were initially puzzling, however, since cytoplasmic CD3 expression precedes surface CD3 expression during normal thymocyte development(40–42), IHC, which uses formalin fixed tissue, will detect cytoplasmic proteins, whereas flow cytometry will only detect surface proteins (unless the cells have been permeabilized prior to staining). This finding was also consistent with the possibility that these leukemias, which expressed cytoplasmic CD3 but lacked cell surface CD3 (as well as CD4 and CD8), were leukemias of immature, DN thymocyte precursors. We therefore evaluated these leukemias using additional immunophenotypic and genetic markers of thymocyte differentiation.
Supplementary Figure S3 depicts the pattern of cell surface marker staining and T cell receptor beta rearrangement during normal T cell development. As shown in Table 1 and Figure 1D, Idh2R140Q/NHD13 leukemias exhibited an immunophenotype consistent with either DN1 thymocytes (Lin-CD44+CD25-CD90-Kit+) (mouse #525, 531), DN2 thymocytes, (Lin-CD44+CD25+CD90+Kit-) (#524), or an immunophenotype intermediate between DN1 and DN2 (#559, 526).
We used an established assay for clonal Tcrb VDJ rearrangement based on amplification of Tcrb mRNA using a degenerate V primer and a C primer which anneals to both Tcrb constant regions (CB1 and CB2)(17, 19, 43). Consistent with a DN1 or DN2 immunophenotype, the Idh2R140Q/NHD13 leukemias did not have evidence for clonal Tcrb VDJ rearrangement (Table 1, Supplemental Figure S4A). However, D-J rearrangement precedes V-DJ rearrangement during thymocyte development (Fig S3), and the Tcrb VDJ rearrangement assay will not detect Tcrb DJ rearrangements. Therefore, we developed an assay for clonal DJ rearrangements as outlined in Fig S4B(18, 19). The Idh2R140Q/NHD13 leukemias either showed clonal Tcrb DJ rearrangements (#524, 525, 526, 546, 555, 559) or were germline for Tcrb (Table 1, Supplemental Figure S4C). These results further support the assertion that the Idh2R140Q/NHD13 leukemias originate from an immature thymocyte precursor.
Idh2R140Q/NHD13 leukemia is transplantable.
In addition to clonality, metastatic spread, and block to differentiation, transplantation to wild-type organisms is another characteristic of malignant disease. We injected 1 × 10E05 cells from a primary Idh2R140Q/NHD13 leukemia (M#555) into non-irradiated, sublethal irradiated (600 cGy), or lethally irradiated (900 cGy) congenic recipients; the lethally irradiated mice received a radiation sparing dose of 2 × 10E05 WT bone marrow cells. The leukemic donors expressed the CD45.2 allele of CD45, whereas recipients and WT bone marrow expressed CD45.1. Survival of primary and secondary transplant recipients is shown in Figure 2A-B, and CBC at or near the time of death are shown in supplementary Table S4. In the first experiment, all mice transplanted with primary leukemia cells (from mouse # 555; immunophenotype CD4-, CD8-, sCD3-, CD19-, B220-, Mac1-, Gr1-, CD44+, CD25-, Kitdim, CD90-) without irradiation survived, three of four transplanted with sublethal irradiation (600 cGy) survived, and one of three transplanted with myeloablative, lethal irradiation (900 cGy) survived. CBC and engraftment data indicated that these mice died of acute leukemia. All secondary transplant recipients, both sublethal and lethal radiation recipients, died of leukemia (Figure 2B), suggesting that a more aggressive Idh2R140Q/NHD13 leukemic subclone had been selected in vivo.
Figure 2. Idh2R140Q/NHD13 leukemia is transplantable.
A,survival of lethal (900rads, n=3), sub-lethal (600rads, n=4) and non-irradiated (n=5) recipients transplanted with primary Idh2R140Q/NHD13 leukemic splenocytes. p values by Mantel-Cox test. B, survival of lethal (n=10), sub-lethal (n=5), non-irradiated (n=8) and WT BM control (n=3) recipients transplanted with secondary Idh2R140Q/NHD13 leukemic splenocytes. p values by Mantel-Cox test. C & D, flow cytometry analysis showing engraftment of Idh2R140Q/NHD13 donor cells in primary and secondary recipients. E, histology showing infiltration of CD3 and myeloperoxidase positive cells in spleen and perivascular regions of the liver. F, Tcrb DJ rearrangement assay of donor and recipient mice.
Flow cytometry of the primary transplant recipients demonstrated that BM and spleen were almost completely replaced with leukemic donor cells (Figure 2C). Additional immunophenotyping (Figure 2C) indicated that the recipient leukemic cells were CD4-CD8-CD44+CD25-B220-Kitdim. Of note, a majority of the leukemic cells in the BM had acquired Mac1 or Gr1 expression; the degree of expression seemed to be dependent on the tissue, suggesting a potential effect of the microenvironment on expression of these antigens.
Characterization of secondary transplant recipients demonstrated a majority of the leukemic cells in the BM had acquired Mac1 or Gr1 (Figure 2D). Immunohistochemistry demonstrated that the leukemic cells infiltrated parenchymal organs, such as the liver, and stained for both CD3 and MPO (Figure 2E). Figure 2F shows that the primary and secondary leukemic recipients had clonal Tcrb gene rearrangements that were identical to the parental cells, demonstrating that despite the immunophenotypic differentiation, the leukemic clone in the transplant recipients was clearly derived from the primary leukemia.
Idh2R140Q/NHD13 leukemia originates from an early T cell precursor in the thymus
The hypothesis that the Idh2R140Q/NHD13 leukemia originated from an immature T precursor was further supported by three additional Idh2R140Q/NHD13 mice (#662, 664, 668) that were moribund and euthanized at a relatively young age (4, 7.5 and 8.5 months). These mice had normal CBCs and no evidence of splenomegaly (Supplemental Table S3). However, flow cytometry revealed a profound block in T cell maturation in these mice as shown by an expansion of DN thymocytes (Table 1, Supplemental Figure S5). In addition, PCR analysis revealed oligoclonal thymic Tcrb DJ rearrangements (Table 1), indicating an expansion of immature T cell precursors. Moreover, in two samples (662 and 664), one of the Tcrb DJ rearrangements that was detected in the thymus was also found in the spleen (Table 1). Taken together, these observations suggest that the DN1/DN2 T cell leukemia originates in the thymus, and subsequently metastasizes and invades other tissues including BM and spleen.
At six weeks of age, the Idh2R140Q/NHD13 thymus was populated primarily by DP cells, similar to wild type mice (Supplemental Figure S6). A subtle expansion of DN cells was noted (4.65% in Idh2R140Q/NHD13 vs 2.5% in WT), with a decrease in DN3 cells, suggesting a relative block at the DN2-DN3 transition. There was no evidence of a clonal expansion, as the Idh2R140Q/NHD13 thymocytes showed predominantly polyclonal Tcrb DJ and VDJ rearrangements at this time point. However, at 12 weeks of age, a marked expansion of DN cells was detected (33.4%), accompanied by a clonal expansion of DN2 cells (Supplemental Figure S6). Similar results were obtained with a 15-week-old Idh2R140Q/NHD13 mouse (Supplemental Figure S6). These results support the hypothesis that the clonal Idh2R140Q/NHD13 leukemia originates from a DN thymocyte.
Both Idh2R140Q and NHD13 are required for severe impairment of thymocyte differentiation in vitro
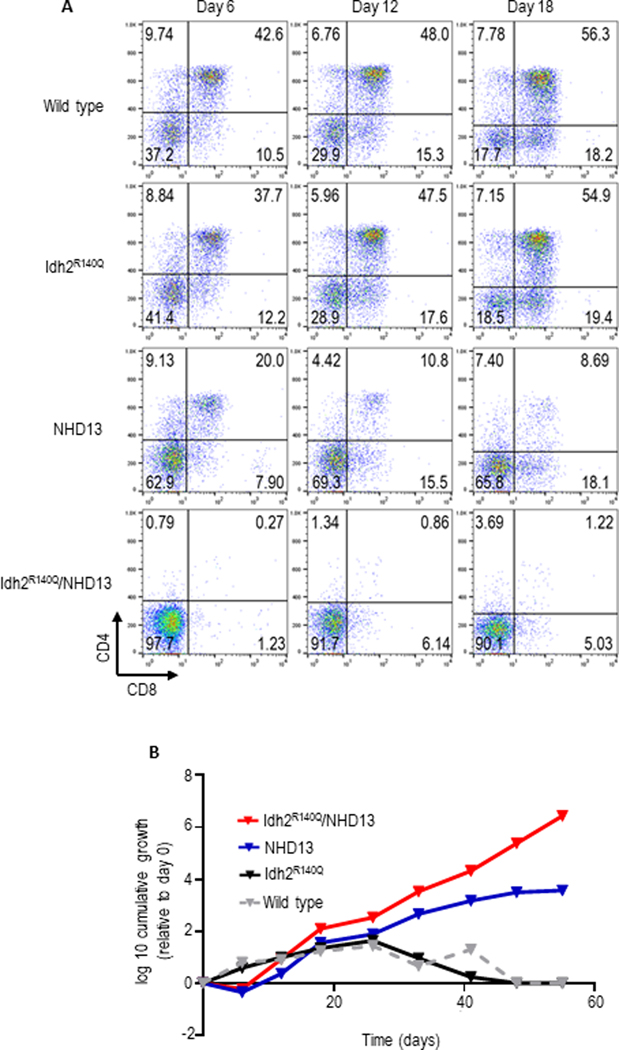
To better understand the role of each mutation in the generation of DN1/DN2 leukemia, we examined in vitro thymocyte differentiation of pooled DN1 and DN2 thymocytes (CD4-CD8-CD44+CD25+/−) purified from 5 week old mice of the four possible genotypes using a standard in vitro assay for thymocyte differentiation(44). After six days in culture, approximately 40% of WT DN1–2 thymocytes had matured to DP (CD4+CD8+) cells and 20% had progressed to the single positive stage (primarily CD8+, Figure 3A). By day 18, only ~18% of the WT cells remained DN (Figure 3A). Similar results were obtained with Idh2R140Q DN1–2 thymocytes, indicating that Idh2R140Q alone does not disrupt T cell differentiation. A partial block in differentiation was detected in NHD13 DN1–2 culture at day 18 (Figure 3A). However, Idh2R140Q/NHD13 culture exhibited an almost complete block in T cell differentiation; at day 6, 98% of the culture remained DN and at day 18, 90% remained DN (Figure 3A).
Figure 3. In vitro co-culture of Idh2R140Q/NHD13 thymocytes on OP9-DL1 stromal cells reveals block in DN to DP differentiation.
A,DN1/2 thymocytes (CD4-CD8-CD44+CD25+/−) from 5-week-old mice were cocultured with OP9-DL1 stromal cells. Suspension cells were obtained from the coculture at indicated time points and assessed for CD4 and CD8 expression. B, growth curve of suspension cells; Idh2R140Q/NHD13 cells display continued exponential growth.
At day 18, the DN population in WT, Idh2R140Q and NHD13 cultures was composed primarily of DN3 cells while most of the Idh2R140Q/NHD13 cells were DN2, again consistent with a partial block at the DN2-DN3 transition (Supplemental Figure S7). In addition, whereas the WT and Idh2R140Q cultures stopped proliferating after 5 weeks in culture, and the NHD13 proliferated for ~ 6 weeks before becoming quiescent, the Idh2R140Q/NHD13 culture remained highly proliferative for at least eight weeks (Figure 3B). These results demonstrate that although an NHD13 transgene alone leads to impaired thymocyte differentiation, the combination of NHD13 and Idh2R140Q leads to a more severe block at the DN2-DN3 transition, accompanied by immortalization.
Idh2R140Q/NHD13 DN1/DN2 T-ALL acquire additional mutations similar to those associated with human early/immature thymocyte precursor acute lymphoblastic leukemia (EITP ALL)
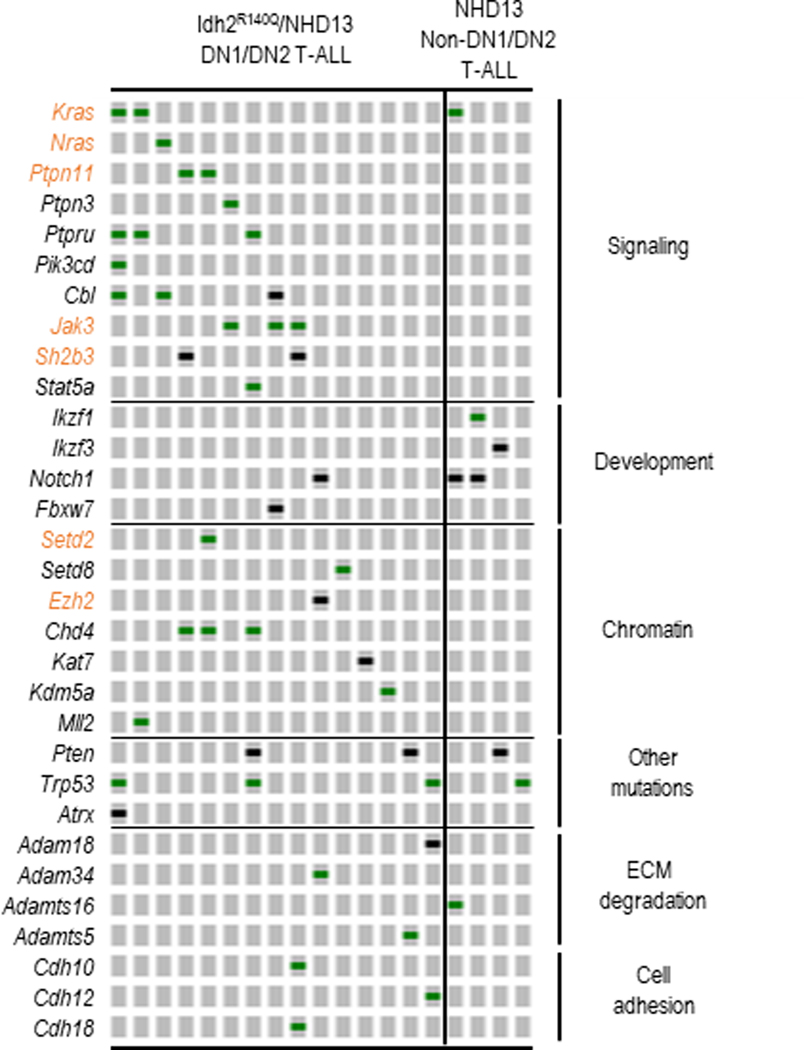
Because of the variable time for leukemia development in Idh2R140Q/NHD13 mice (Figure 1A), we suspected that these two mutations were not sufficient to induce DN1/DN2 T-ALL, but that additional mutations in cancer driver genes would be required. Human leukemias of immature T cell precursor origin with similar immunophenotypes have been variably designated as early immature or early T cell precursor ALL(8, 26); we have elected to group these together as “early immature thymocyte precursor ALL (EITP)” as previously suggested(45). Recent studies on the genomic landscape of human T-ALL demonstrated that acquired mutations in EITP ALL were distinct from non-EITP ALL and included frequent mutations in cytokine receptor (FLT3, JAK3, IL7R), Ras signaling (NRAS, KRAS, SH2B3, PTPN11), and chromatin modifier (EZH2, SETD2) genes(8, 46–48). Therefore, we used whole exome sequencing (WES) to identify acquired mutations in 15 samples of Idh2R140Q/NHD13 DN1/DN2 T-ALL and four samples of non-DN1/DN2 T-ALL (DP or SP) T- ALL that arose in NHD13 only mice (Supplemental Table S5 and S6). A subset of these mutations is validated by Sanger sequencing in supplementary Figure S8. Remarkably, recurrent mutations that occurred predominantly in EITP ALL patients were also found primarily in Idh2R140Q/NHD13 DN1/DN2 T-ALL compared to NHD13 non-DN1/DN2 T-ALL. These include mutations in Kras, Nras, Ptpn11, Jak3, Sh2b3, Setd2 and Ezh2 (Figure 4, highlighted in orange). Both samples with Sh2b3 mutations (Figure 4) had biallelic mutations, suggesting that Sh2b3 functioned as a tumor suppressor gene. In total, all Idh2R140Q/NHD13 DN1/DN2 T-ALL samples had at least one acquired mutation of a gene known to be involved in malignant transformation, consistent with the hypothesis that Idh2R140Q and NHD13 alleles were not sufficient to produce leukemia but required additional mutations for leukemic transformation. Finally, only one out of 15 Idh2R140Q/NHD13 DN1/DN2 T-ALL had acquired a mutation in Notch1 compared with two out of four NHD13 T-ALLs (Figure 4, Fisher exact test p=0.0970). Of note, mutations in NOTCH1 are frequent in patients with non-EITP ALL but less common in EITP ALL patients(8, 46), highlighting the similarity between murine Idh2R140Q/NHD13 DN1/DN2 T-ALL and human EITP ALL.
Figure 4. Recurrent mutations in Idh2R140Q/NHD13 DN1/DN2 T-ALL are similar to those in human EITP ALL.
15 Idh2R140Q/NHD13 DN1/DN2 T-ALL and 4 NHD13 non-ETP T-ALL (CD4/CD8 single or double positive) samples were assessed by WES. Recurrent mutations occurring predominantly in ETP ALL patients are highlighted in orange. Green bars represent missense mutations and black bars represent truncation mutations.
The expression profile of Idh2R140Q/NHD13 DN1/DN2 T-ALL is consistent with an arrest in T cell development at the DN2-DN3 transition.
We used gene expression arrays to compare the global gene expression profiles of Idh2R140Q/NHD13 DN1/DN2 T-ALL (N=9) to non-DN1/DN2 cortical T-ALL (CD4+CD8+) (N=6) from SCL-LMO1 mice(49) and wild-type thymus (N=3). Unsupervised hierarchical clustering of expression data showed separation into the three anticipated sample groups (Figure 5A). Gene set enrichment analysis (GSEA) using the top 100 genes upregulated in Idh2R140Q/NHD13 DN1/DN2 T-ALL compared to published array data (GSE24142)(25) showed a enrichment in genes that were upregulated in ETP (DN1) versus DN3 thymocytes (Figure 5B). The converse was also true; genes downregulated in Idh2R140Q/NHD13 DN1/DN2 T-ALL were downregulated in ETP (DN1) compared to DN3 thymocytes (Figure 5C). Similar to the in vitro co-culture and in vivo thymocyte development findings, these observations were consistent with a block in differentiation at the DN2 to DN3 transition. Comparison of genes that were differentially expressed in Idh2R140Q/NHD13 DN1/DN2 T-ALL compared to cortical T-ALL yielded very similar results (Supplemental Figure S9) with the Idh2R140Q/NHD13 DN1/DN2 T-ALL enriched in DN1 (ETP) signature compared to DN3 signature genes.
Figure 5. Idh2R140Q/NHD13 DN1/DN2 T-ALL gene expression is similar to murine ETP (DN1) thymocytes.
Global gene expression from 9 Idh2R140Q/NHD13 DN1/DN2 T-ALL, 6 SCL/LMO1 non-DN1/DN2 T-ALL, and 3 WT thymi was assessed by oligonucleotide microarrays. A, dendrogram of unsupervised hierarchical clustering using centered correlation and average linkage. B & C, the top 100 upregulated and down regulated genes between the Idh2R140Q/NHD13 DN1/DN2 T-ALL and WT thymus were compared to mouse ETP (DN1) and DN3 thymocytes (GSE24142)(25).
The transcriptional signature of human EITP ALL is similar to that of murine Idh2R140Q/NHD13 DN1/DN2 T-ALL.
The initial characterization of human EITP leukemia identified a set of genes that discriminated between human EITP and non-EITP ALL(26). We assessed expression of this 32 gene-set in the murine Idh2R140Q/NHD13 DN1/DN2 T-ALL and SCL/LMO1 cortical (non-DN1/DN2) T-ALL; again the data separated into DN1/DN2 T-ALL and cortical (non-DN1/DN2) T-ALL group (Figure 6A). Genes found to be upregulated in the Idh2R140Q/NHD13 DN1/DN2 T-ALL are enriched in hematopoietic stem and progenitor cells (such as Nfe2, Tal1, Kit and Gata2) and committed myeloid progenitors (Spi1) whereas downregulated genes include those known to increase as thymocytes differentiate (Lat, Cd3e/d, Lck, Zap70, Tcf7, Gata3 and Notch1/3).
Figure 6. The gene expression signature of murine Idh2R140Q/NHD13 DN1/DN2 T-ALL is similar to human EITP ALL.
A, expression of 32 gene set defined by Coustan-Smith et al. in murine Idh2R140Q/NHD13 DN1/DN2 T-ALL and murine non-DN1/DN2 T-ALL. B, 100 most differentially expressed probe sets from patients with EITP ALL (n=9) compared to non-EITP T-ALL (26) were used to generate gene signatures of upregulated and downregulated genes. This gene signature was then compared to gene expression arrays from Idh2R140Q/NHD13 DN1/DN2 T-ALL (n=9) or SCL/LMO1 non-DN1/DN2 T-ALL (n=6) using Gene Set Enrichment Analysis (GSEA). Upper panel, up regulated gene comparison; lower panel, down-regulated gene comparison. C, an analogous comparison was performed using a gene signature derived from an independent set of EITP ALL vs mature T-ALL(8).
GSEA analysis using the 100 most differentially expressed probesets in EITP ALL patients vs non-EITP T-ALL patients(26) demonstrated that the upregulated genes in EITP ALL patients are highly enriched in the mouse DN1/DN2 T-ALL and a strong negative enrichment was detected using the down regulated genes (Figure 6B). A similar pattern of enrichment was obtained using an independent data set (GSE33469; Figure 6C). These results demonstrate that a common “early immature T cell precursor” transcriptional program is activated in both human EITP ALL and murine DN1/DN2 T-ALL.
Differential methylation of Idh2R140Q/NHD13 DN1/DN2 T-ALL.
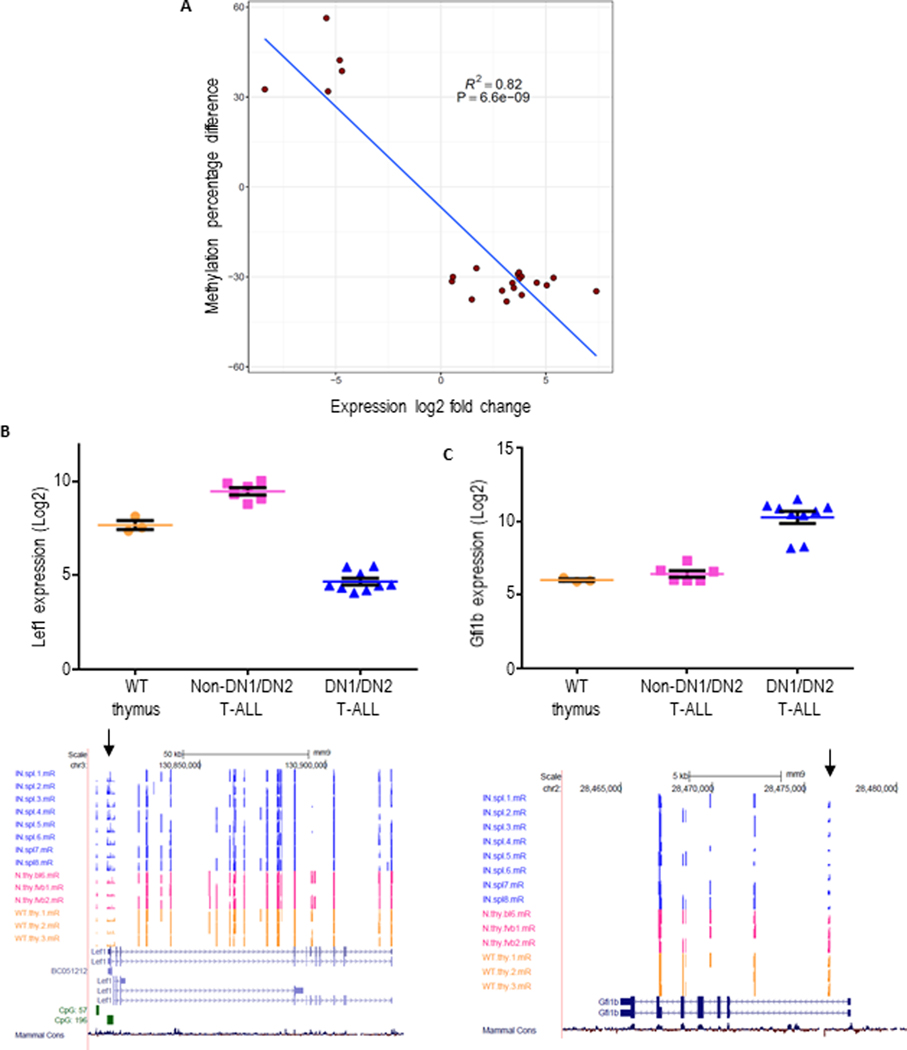
Since the production of 2-HG by Idh2 p.R140 mutations leads to aberrant cytosine methylation, we used Reduced Representation Bisulfite Sequencing (RRBS) to identify regions of the genome that were differentially methylated between Idh2R140Q/NHD13 DN1/DN2 T-ALL, NHD13 CD4+/CD8+ (DP) T-ALL, and WT thymus. Supplemental Figure S10A-B shows the distribution of methylation patterns observed among 5106 previously reported hematopoietic enhancer enhancer regions(30). The Idh2R140Q/NHD13 DN1/DN2 T-ALL possessed an enhancer methylation profile distinct from that of NHD13 T-ALL and WT thymus. Compared to NHD13 T-ALL, we observed hypermethylation in Idh2R140Q/NHD13 DN1/DN2 T-ALL of 130 enhancers and hypomethylation of 522 enhancers. Differential methylation of enhancers specifying T cell development and maturation was not stochastic. Hypermethylation of active or poised CD4 or CD8 lineage enhancers was significantly enriched compared to enhancers not involved in these lineages (OR = 5.64 P = 7.23e-06, two-tailed Fisher’s Exact Test). In an analysis of all potentially functional hematopoietic enhancers, hypermethylation was enriched for genes involved in embryonic development and protein tyrosine kinase signaling whereas enhancer hypomethylation was associated with genes involved in transcriptional regulation as well as cell fate commitment (Supplemental Table S7). We next generated a set of genes which were important for early thymocyte differentiation by combining 37 genes that were differentially expressed in both Idh2R140Q/NHD13 DN1/DN2 T-ALL and ETP (DN1) thymocytes based on the GSEA analysis in Figure 5 with 36 genes reported from a literature review(39, 50) (Supplemental Table S8). Of these genes, 22 were at least 25% differentially methylated between Idh2R140Q/NHD13 DN1/DN2 T-ALL and NHD13 DP T-ALL. Figure 7A shows a strong correlation (R2=0.82 P = 6.6 × 10−9, Pearson Correlation Test) between enhanced expression and decreased methylation in this gene set. Figure 7B-C shows examples of differential methylation and expression involving genes well known to be involved in early thymocytes differentiation such as Lef1 and Gfi1b.
Figure 7. Inverse correlation between methylation and expression.
A, mRNA expression of genes important for early thymic development which showed differential methylation of at least 25% at 5’ regulatory regions in NHD13/Idh2R140Q DN1/DN2 T- ALL compared to NHD13 non-DN1/DN2 T-ALL was determined by expression array as in Figure 6. Methylation percentage difference is plotted on the y axis and log2 fold change expression ratio is plotted on the x axis. B, upper panel, Lef1 expression in WT thymus, NHD13/Idh2R140Q DN1/DN2 T-ALL and non-DN1/DN2 T-ALL. Lower panel, individual cytosine methylation tracks for NHD13/Idh2R140Q DN1/DN2 T-ALL (blue), NHD13 non-DN1/DN2 T-ALL (pink) and WT thymus (yellow) samples. CpG island are indicated in green. Arrow shows hypermethylated region in NHD13/Idh2R140Q EITP ALL samples. C, upper panel, Gfi1b expression in WT thymus, NHD13/Idh2R140Q DN1/DN2 T-ALL and non-DN1/DN2 T-ALL. Lower panel, individual cytosine methylation tracks for NHD13/Idh2R140Q DN1/DN2 T- ALL (blue), NHD13 non-DN1/DN2 T-ALL (pink) and WT thymus (yellow) samples. Arrow shows hypomethylated region in NHD13/Idh2R140Q DN1/DN2 T-ALL samples.
In-vitro treatment of Idh2R140Q/NHD13 DN cells with a selective IDH2 inhibitor led to marked decrease in cell proliferation
Enasidenib (AG-221) is a selective IDH2 inhibitor used as a targeted therapeutic for relapsed or refractory (R/R) AML patients with IDH2 mutations(51–53). To examine the effect of AG-221 on Idh2R140Q/NHD13 DN cells in vitro, we established immortalized Idh2R140Q/NHD13 DN cells using the OP9-DL1 co-culture system (Supplemental Figure S11A). These immortalized cells were then treated with AG-221, leading to a dose-dependent decrease in cell proliferation (Supplemental Figure S11B). Examination of clonality for the in vitro immortalized Idh2R140Q/NHD13 cells demonstrated emergence of a clonal population of Idh2R140Q/NHD13 cells with time (Supplemental Figure S11C). This in vitro data suggests that targeting an IDH2 mutation may be an effective treatment of EITP leukemia as well as AML.
Discussion
In this report we examined the role of Idh2 p.R140Q mutation in leukemia development using a genetically engineered mouse model that expressed an Idh2 p.R140Q mutation in hematopoietic tissues. Idh2R140Q transgenic mice produced the 2-HG oncometabolite but did not result in a leukemic phenotype. Lack of leukemic transformation due to mutant Idh2 expression in murine models has been documented by others(31, 54) highlighting the need for additional genetic events for leukemia development in Idh2 mouse models. Idh2R140Q/NHD13 double transgenic mice, generated by crossing NHD13 transgenic mice with Idh2R140Q transgenic mice developed an immature T cell leukemia with median survival age of 10 months. The malignant immature T cells were most commonly at a stage of thymocyte differentiation similar to DN1 or DN2 thymocytes. These results stand in contrast to those obtained with NHD13 only transgenic mice, which most commonly develop AML, precursor thymocyte lymphoblastic leukemia/lymphoma (pre-T LBL), or, less commonly, B-cell precursor ALL(14, 55)
Using GSEA, we demonstrated that the expression profile of gene sets expressed in DN1 thymocytes was enriched in the Idh2R140Q/NHD13 leukemia samples, whereas gene sets enriched in the more differentiated DN3 thymocytes were downregulated in the Idh2R140Q/NHD13 leukemia samples. The presence of clonal Tcrb DJ and absence of clonal VDJ rearrangements in young (12–15 week) Idh2R140Q/NHD13 mice further reinforces the notion that the Idh2R140Q/NHD13 leukemias originate from a DN1/DN2 thymocyte. Finally, the transcriptional signature of human EITP T-ALL is consistent with a thymocyte differentiation arrest, and is distinct from the transcriptional signature identified in non-EITP T-ALL(45).
WES of Idh2R140Q/NHD13 leukemia samples identified additional acquired mutations associated with leukemic transformation. Mutations involving signaling pathways, such as NRAS, KRAS, PTPN11, SH2B3, and PTEN are prevalent in EITP T-ALL(46) and were also recurrently mutated in Idh2R140Q/NHD13 samples.
Both NUP98 fusions and IDH mutations are recurrent events in EITP ALL. One of the initial studies of EITP ALL, identified IDH1/2 mutations in 14% of adult EITP ALL patients(8), and a more recent survey of 1085 T-ALL patients identified IDH1 or IDH2 mutations in 47 patients (4%)(56). The IDH1/2 mutations were associated with an EITP immunophenotype, and acquired mutations such as RAS, ETV6, or DNMT3A that have also been associated with an EITP ALL. In this study, the most common IDH mutation was an IDH2 p.R140Q mutation, which conferred a poor prognosis, leading the authors to speculate that a subset of EITP patients may be candidates for therapy with a mutant IDH2 inhibitor.
A comprehensive study of pediatric T-ALL mutations identified NUP98 translocations in 16% of EITP-ALL patients(47). In addition, NUP98 fusion genes, including the NHD13 fusion, commonly activate HOXA/B genes (34), which are associated with stem cell self-renewal signatures, and are downstream targets of a wide spectrum of mutations associated with leukemia, including MLL fusions, CALM-AF10 fusion, monosomy 7, and NPM1 truncation mutations(57). In the clinic, overexpression of HOXA cluster genes was strongly associated with both poor overall survival as well as an EITP ALL immunophenotype when compared to non-EITP T-ALL patients(58). These observations highlight the fact that NUP98 and IDH mutations are relevant for human EITP ALL and suggest that deregulation of Hoxa/b cluster genes mediated by a NUP98 fusion gene may be an important step during leukemic transformation of Idh2R140Q/NHD13 mice.
Previous reports of EITP ALL models based either on Zeb2 gain of function or a combination of Cdkn2a inactivation and constitutive Il7r activation (Arf −/−/Il7r) displayed an immunophenotype similar to that observed in the Idh2R140Q/NHD13 DN1/DN2 T-ALL(59, 60). This immunophenotype was characterized by surface markers typical of DN1/2 thymocytes (cKit and CD44), and the absence of mature thymocyte markers (CD4 and CD8). Some mice reported in the Zeb2 model developed AML, similar to findings with Idh2R140Q/NHD13 mice, again suggesting that pre-malignant DN1/DN2 thymocytes may have the potential for myeloid lineage transformation. Furthermore, an additional study showed that Lmo2 overexpression could inhibit normal murine thymocyte differentiation at the DN2 stage (60, 61); consistent with those findings, Lmo2 mRNA was overexpressed in Idh2R140Q/NHD13 DN1/DN2 T-ALL. These observations, together with the finding that Idh2R140Q/NHD13 leukemias frequently stained positive for myeloperoxidase, highlight a differentiation block at the DN2-DN3 transition and an accumulation of DN1/DN2 thymocytes that possess both myeloid and T-lymphocyte developmental potential (62).
In summary we have shown that Idh2R140Q/NHD13 mice develop a highly penetrant DN1/DN2 T-ALL leukemia. The immunophenotype, gene expression profile, and pattern of spontaneous acquired mutations seen in Idh2R140Q/NHD13 DN1/DN2 T-ALL closely resemble those seen in human EITP T-ALL. Finally, effective treatment of immortalized Idh2R140Q/NHD13 DN thymocytes with AG-221 suggests that this model can be used to study EITP biology and identify therapies for EITP leukemia patients.
Supplementary Material
Statement of Significance.
T-cell leukemia induced in Idh2R140Q/NUP98-HOXD13 mice is immunophenotypically, transcriptionally and genetically similar to human EITP ALL, providing a model for studying disease development and treatment.
Acknowledgements
The authors thank current and former members of the Aplan lab, Avinash Bhandoola, Terry Fry, and Warren Pear for insightful discussions. We thank the NCI Sequencing Minicore for Sanger sequencing, the NCI Transgenic Core for generation of transgenic mice, the NCI Genomics Core for Next Generation Sequencing, the NCI Flow cytometry core for cell sorting, the NCI Pathology/Histotechnology Lab (PHL) for immunohistochemistry, Shelley Hoover and Mark Simpson of the NCI Molecular Pathology Unit for assistance with slide imaging, and Maria Jorge for excellent animal husbandry. This work was supported by the Intramural Research Program of the National Cancer Institute, National Institutes of Health (grant numbers ZIA SC 010378 and BC 010983).
Footnotes
Disclosure of Conflicts of Interest:
Peter D. Aplan receives royalties from the NIH Technology Transfer Office for the invention of NUP98-HOXD13 mice.
References
- 1.Medeiros BC, Fathi AT, DiNardo CD, Pollyea DA, Chan SM, Swords R. Isocitrate dehydrogenase mutations in myeloid malignancies. Leukemia. February2017;31(2):272–281. doi: 10.1038/leu.2016.275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amary MF, Bacsi K, Maggiani F, et al. IDH1 and IDH2 mutations are frequent events in central chondrosarcoma and central and periosteal chondromas but not in other mesenchymal tumours. J Pathol. July2011;224(3):334–43. doi: 10.1002/path.2913 [DOI] [PubMed] [Google Scholar]
- 3.Jusakul A, Cutcutache I, Yong CH, et al. Whole-Genome and Epigenomic Landscapes of Etiologically Distinct Subtypes of Cholangiocarcinoma. Cancer Discov. October2017;7(10):1116–1135. doi: 10.1158/2159-8290.Cd-17-0368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cairns RA, Iqbal J, Lemonnier F, et al. IDH2 mutations are frequent in angioimmunoblastic T-cell lymphoma. Blood. February232012;119(8):1901–3. doi: 10.1182/blood-2011-11-391748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yan H, Parsons DW, Jin G, et al. IDH1 and IDH2 mutations in gliomas. N Engl J Med. February192009;360(8):765–73. doi: 10.1056/NEJMoa0808710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marcucci G, Maharry K, Wu YZ, et al. IDH1 and IDH2 gene mutations identify novel molecular subsets within de novo cytogenetically normal acute myeloid leukemia: a Cancer and Leukemia Group B study. J Clin Oncol. May102010;28(14):2348–55. doi: 10.1200/jco.2009.27.3730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mardis ER, Ding L, Dooling DJ, et al. Recurring mutations found by sequencing an acute myeloid leukemia genome. N Engl J Med. September102009;361(11):1058–66. doi: 10.1056/NEJMoa0903840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Van Vlierberghe P, Ambesi-Impiombato A, Perez-Garcia A, et al. ETV6 mutations in early immature human T cell leukemias. J Exp Med. December192011;208(13):2571–9. doi: 10.1084/jem.20112239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Green A, Beer P. Somatic mutations of IDH1 and IDH2 in the leukemic transformation of myeloproliferative neoplasms. N Engl J Med. January282010;362(4):369–70. doi: 10.1056/NEJMc0910063 [DOI] [PubMed] [Google Scholar]
- 10.Ward PS, Patel J, Wise DR, et al. The common feature of leukemia-associated IDH1 and IDH2 mutations is a neomorphic enzyme activity converting alpha-ketoglutarate to 2-hydroxyglutarate. Cancer Cell. March162010;17(3):225–34. doi: 10.1016/j.ccr.2010.01.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Green CL, Evans CM, Zhao L, et al. The prognostic significance of IDH2 mutations in AML depends on the location of the mutation. Blood. July142011;118(2):409–12. doi: 10.1182/blood-2010-12-322479 [DOI] [PubMed] [Google Scholar]
- 12.Ok CY, Loghavi S, Sui D, et al. Persistent IDH1/2 mutations in remission can predict relapse in patients with acute myeloid leukemia. Haematologica. February2019;104(2):305–311. doi: 10.3324/haematol.2018.191148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu W, Yang H, Liu Y, et al. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of α-ketoglutarate-dependent dioxygenases. Cancer Cell. January182011;19(1):17–30. doi: 10.1016/j.ccr.2010.12.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin YW, Slape C, Zhang Z, Aplan PD. NUP98-HOXD13 transgenic mice develop a highly penetrant, severe myelodysplastic syndrome that progresses to acute leukemia. Blood. July12005;106(1):287–95. doi: 10.1182/blood-2004-12-4794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ogilvy S, Metcalf D, Gibson L, Bath ML, Harris AW, Adams JM. Promoter elements of vav drive transgene expression in vivo throughout the hematopoietic compartment. Blood. September151999;94(6):1855–63. [PubMed] [Google Scholar]
- 16.Morse HC 3rd, Anver MR, Fredrickson TN, et al. Bethesda proposals for classification of lymphoid neoplasms in mice. Blood. July12002;100(1):246–58. doi: 10.1182/blood.v100.1.246 [DOI] [PubMed] [Google Scholar]
- 17.Beachy SH, Onozawa M, Silverman D, Chung YJ, Rivera MM, Aplan PD. Isolated Hoxa9 overexpression predisposes to the development of lymphoid but not myeloid leukemia. Experimental hematology. 2013;41(6):518–529.e5. doi: 10.1016/j.exphem.2013.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kundu S, Park ES, Chung YJ, et al. Thymic precursor cells generate acute myeloid leukemia in NUP98-PHF23/NUP98-HOXD13 double transgenic mice. Sci Rep. November202019;9(1):17213. doi: 10.1038/s41598-019-53610-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Choi CW, Chung YJ, Slape C, Aplan PD. A NUP98-HOXD13 fusion gene impairs differentiation of B and T lymphocytes and leads to expansion of thymocytes with partial TCRB gene rearrangement. J Immunol. November152009;183(10):6227–35. doi: 10.4049/jimmunol.0901121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chung YJ, Choi CW, Slape C, Fry T, Aplan PD. Transplantation of a myelodysplastic syndrome by a long-term repopulating hematopoietic cell. Proc Natl Acad Sci U S A. September162008;105(37):14088–93. doi: 10.1073/pnas.0804507105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Beachy SH, Onozawa M, Silverman D, Chung YJ, Rivera MM, Aplan PD. Isolated Hoxa9 overexpression predisposes to the development of lymphoid but not myeloid leukemia. Exp Hematol. June2013;41(6):518–529 e5. doi: 10.1016/j.exphem.2013.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Holmes R, Zuniga-Pflucker JC. The OP9-DL1 system: generation of T-lymphocytes from embryonic or hematopoietic stem cells in vitro. Cold Spring Harb Protoc. February2009;2009(2):pdb.prot5156. doi: 10.1101/pdb.prot5156 [DOI] [PubMed] [Google Scholar]
- 23.Yin M, Baslan T, Walker RL, et al. A unique mutator phenotype reveals complementary oncogenic lesions leading to acute leukemia. JCI Insight. December52019;4(23)doi: 10.1172/jci.insight.131434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cibulskis K, Lawrence MS, Carter SL, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol. March2013;31(3):213–9. doi: 10.1038/nbt.2514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Belyaev NN, Biro J, Athanasakis D, Fernandez-Reyes D, Potocnik AJ. Global transcriptional analysis of primitive thymocytes reveals accelerated dynamics of T cell specification in fetal stages. Immunogenetics. August2012;64(8):591–604. doi: 10.1007/s00251-012-0620-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Coustan-Smith E, Mullighan CG, Onciu M, et al. Early T-cell precursor leukaemia: a subtype of very high-risk acute lymphoblastic leukaemia. Lancet Oncol. February2009;10(2):147–56. doi: 10.1016/s1470-2045(08)70314-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mayle A, Yang L, Rodriguez B, et al. Dnmt3a loss predisposes murine hematopoietic stem cells to malignant transformation. Blood. January222015;125(4):629–38. doi: 10.1182/blood-2014-08-594648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Boyle P, Clement K, Gu H, et al. Gel-free multiplexed reduced representation bisulfite sequencing for large-scale DNA methylation profiling. Genome Biol. October32012;13(10):R92. doi: 10.1186/gb-2012-13-10-r92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Akalin A, Kormaksson M, Li S, et al. methylKit: a comprehensive R package for the analysis of genome-wide DNA methylation profiles. Genome Biol. October32012;13(10):R87. doi: 10.1186/gb-2012-13-10-r87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lara-Astiaso D, Weiner A, Lorenzo-Vivas E, et al. Immunogenetics. Chromatin state dynamics during blood formation. Science. August222014;345(6199):943–9. doi: 10.1126/science.1256271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sasaki M, Knobbe CB, Munger JC, et al. IDH1(R132H) mutation increases murine haematopoietic progenitors and alters epigenetics. Nature. August302012;488(7413):656–9. doi: 10.1038/nature11323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ogawara Y, Katsumoto T, Aikawa Y, et al. IDH2 and NPM1 Mutations Cooperate to Activate Hoxa9/Meis1 and Hypoxia Pathways in Acute Myeloid Leukemia. Cancer Res. May152015;75(10):2005–16. doi: 10.1158/0008-5472.Can-14-2200 [DOI] [PubMed] [Google Scholar]
- 33.Goldberg L, Gough SM, Lee F, et al. Somatic mutations in murine models of leukemia and lymphoma: Disease specificity and clinical relevance. Genes Chromosomes Cancer. June2017;56(6):472–483. doi: 10.1002/gcc.22451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gough SM, Slape CI, Aplan PD. NUP98 gene fusions and hematopoietic malignancies: common themes and new biologic insights. Blood. December82011;118(24):6247–57. doi: 10.1182/blood-2011-07-328880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bell JJ, Bhandoola A. The earliest thymic progenitors for T cells possess myeloid lineage potential. Nature. April102008;452(7188):764–7. doi: 10.1038/nature06840 [DOI] [PubMed] [Google Scholar]
- 36.Campana D, Behm FG. Immunophenotyping of leukemia. J Immunol Methods. September212000;243(1–2):59–75. doi: 10.1016/s0022-1759(00)00228-3 [DOI] [PubMed] [Google Scholar]
- 37.Shortman K, Wu L. Early T lymphocyte progenitors. Annu Rev Immunol. 1996;14:29–47. doi: 10.1146/annurev.immunol.14.1.29 [DOI] [PubMed] [Google Scholar]
- 38.Yui MA, Feng N, Rothenberg EV. Fine-scale staging of T cell lineage commitment in adult mouse thymus. J Immunol. July12010;185(1):284–93. doi: 10.4049/jimmunol.1000679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yui MA, Rothenberg EV. Developmental gene networks: a triathlon on the course to T cell identity. Nat Rev Immunol. August2014;14(8):529–45. doi: 10.1038/nri3702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Levelt CN, Carsetti R, Eichmann K. Regulation of thymocyte development through CD3. II. Expression of T cell receptor beta CD3 epsilon and maturation to the CD4+8+ stage are highly correlated in individual thymocytes. The Journal of experimental medicine. 1993;178(6):1867–1875. doi: 10.1084/jem.178.6.1867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chetty R, Gatter K. CD3: structure, function, and role of immunostaining in clinical practice. J Pathol. August1994;173(4):303–7. doi: 10.1002/path.1711730404 [DOI] [PubMed] [Google Scholar]
- 42.Staal FJ, Weerkamp F, Langerak AW, Hendriks RW, Clevers HC. Transcriptional control of t lymphocyte differentiation. Stem Cells. 2001;19(3):165–79. doi: 10.1634/stemcells.19-3-165 [DOI] [PubMed] [Google Scholar]
- 43.Danska JS, Pflumio F, Williams CJ, Huner O, Dick JE, Guidos CJ. Rescue of T cell-specific V(D)J recombination in SCID mice by DNA-damaging agents. Science. October211994;266(5184):450–5. doi: 10.1126/science.7524150 [DOI] [PubMed] [Google Scholar]
- 44.Schmitt TM, Zúñiga-Pflücker JC. Induction of T cell development from hematopoietic progenitor cells by delta-like-1 in vitro. Immunity. December2002;17(6):749–56. doi: 10.1016/s1074-7613(02)00474-0 [DOI] [PubMed] [Google Scholar]
- 45.Haydu JE, Ferrando AA. Early T-cell precursor acute lymphoblastic leukaemia. Curr Opin Hematol. July2013;20(4):369–73. doi: 10.1097/MOH.0b013e3283623c61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang J, Ding L, Holmfeldt L, et al. The genetic basis of early T-cell precursor acute lymphoblastic leukaemia. Nature. January112012;481(7380):157–63. doi: 10.1038/nature10725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu Y, Easton J, Shao Y, et al. The genomic landscape of pediatric and young adult T-lineage acute lymphoblastic leukemia. Nat Genet. August2017;49(8):1211–1218. doi: 10.1038/ng.3909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Neumann M, Heesch S, Schlee C, et al. Whole-exome sequencing in adult ETP-ALL reveals a high rate of DNMT3A mutations. Blood. June62013;121(23):4749–52. doi: 10.1182/blood-2012-11-465138 [DOI] [PubMed] [Google Scholar]
- 49.Chervinsky DS, Zhao XF, Lam DH, Ellsworth M, Gross KW, Aplan PD. Disordered T-cell development and T-cell malignancies in SCL LMO1 double-transgenic mice: parallels with E2A-deficient mice. Mol Cell Biol. July1999;19(7):5025–35. doi: 10.1128/mcb.19.7.5025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rothenberg EV, Moore JE, Yui MA. Launching the T-cell-lineage developmental programme. Nat Rev Immunol. January2008;8(1):9–21. doi: 10.1038/nri2232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kim ES. Enasidenib: First Global Approval. Drugs. October2017;77(15):1705–1711. doi: 10.1007/s40265-017-0813-2 [DOI] [PubMed] [Google Scholar]
- 52.Stein EM, DiNardo CD, Pollyea DA, et al. Enasidenib in mutant IDH2 relapsed or refractory acute myeloid leukemia. Blood. August102017;130(6):722–731. doi: 10.1182/blood-2017-04-779405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yen K, Travins J, Wang F, et al. AG-221, a First-in-Class Therapy Targeting Acute Myeloid Leukemia Harboring Oncogenic IDH2 Mutations. Cancer Discov. May2017;7(5):478–493. doi: 10.1158/2159-8290.Cd-16-1034 [DOI] [PubMed] [Google Scholar]
- 54.Kats LM, Reschke M, Taulli R, et al. Proto-oncogenic role of mutant IDH2 in leukemia initiation and maintenance. Cell Stem Cell. March62014;14(3):329–41. doi: 10.1016/j.stem.2013.12.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Slape C, Lin YW, Hartung H, Zhang Z, Wolff L, Aplan PD. NUP98-HOX translocations lead to myelodysplastic syndrome in mice and men. J Natl Cancer Inst Monogr. 2008;(39):64–8. doi: 10.1093/jncimonographs/lgn014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Simonin M, Schmidt A, Bontoux C, et al. Oncogenetic landscape and clinical impact of IDH1 and IDH2 mutations in T-ALL. J Hematol Oncol. May32021;14(1):74. doi: 10.1186/s13045-021-01068-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Collins CT, Hess JL. Deregulation of the HOXA9/MEIS1 axis in acute leukemia. Curr Opin Hematol. July2016;23(4):354–61. doi: 10.1097/moh.0000000000000245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bond J, Marchand T, Touzart A, et al. An early thymic precursor phenotype predicts outcome exclusively in HOXA-overexpressing adult T-cell acute lymphoblastic leukemia: a Group for Research in Adult Acute Lymphoblastic Leukemia study. Haematologica. June2016;101(6):732–40. doi: 10.3324/haematol.2015.141218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Goossens S, Radaelli E, Blanchet O, et al. ZEB2 drives immature T-cell lymphoblastic leukaemia development via enhanced tumour-initiating potential and IL-7 receptor signalling. Nature Communications. 2015/01/July 2015;6(1):5794. doi: 10.1038/ncomms6794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Treanor LM, Zhou S, Janke L, et al. Interleukin-7 receptor mutants initiate early T cell precursor leukemia in murine thymocyte progenitors with multipotent potential. J Exp Med. April72014;211(4):701–13. doi: 10.1084/jem.20122727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.McCormack MP, Shields BJ, Jackson JT, et al. Requirement for Lyl1 in a model of Lmo2-driven early T-cell precursor ALL. Blood. September192013;122(12):2093–103. doi: 10.1182/blood-2012-09-458570 [DOI] [PubMed] [Google Scholar]
- 62.Bhandoola A, Sambandam A. From stem cell to T cell: one route or many? Nat Rev Immunol. February2006;6(2):117–26. doi: 10.1038/nri1778 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.