Abstract
We previously identified a novel TATA-binding protein (TBP)-interacting protein (TIP120) from the rat liver. Here, in an RNA polymerase II (RNAP II)-reconstituted transcription system, we demonstrate that recombinant TIP120 activates the basal level of transcription from various kinds of promoters regardless of the template DNA topology and the presence of TFIIE/TFIIH and TBP-associated factors. Deletion analysis demonstrated that a 412-residue N-terminal domain, which includes an acidic region and the TBP-binding domain, is required for TIP120 function. Kinetic studies suggest that TIP120 functions during preinitiation complex (PIC) formation at the step of RNAP II/TFIIF recruitment to the promoter but not after the completion of PIC formation. Electrophoretic mobility shift assays showed that TIP120 enhanced PIC formation, and TIP120 also stimulated the nonspecific transcription and DNA-binding activity of RNAP II. These lines of evidence suggest that TIP120 is able to activate basal transcription by overcoming a kinetic impediment to RNAP II/TFIIF integration into the TBP (TFIID)-TFIIB-DNA-complex. Interestingly, TIP120 also stimulates RNAP I- and III-driven transcription and binds to RPB5, one of the common subunits of the eukaryotic RNA polymerases, in vitro. Furthermore, in mouse cells, ectopically expressed TIP120 enhances transcription from all three classes (I, II, and III) of promoters. We propose that TIP120 globally regulates transcription through interaction with basal transcription mechanisms common to all three transcription systems.
The efficiency of transcription is regulated mainly by two steps, initiation and elongation (42–44). In the case of RNA polymerase II (RNAP II)-dependent genes, in vitro studies revealed that the initiation of transcription from TATA-containing promoters requires multiple general transcription factors (GTFs) in addition to RNAP II, and early studies with isolated factors reveal an assembly of these components into a functional preinitiation complex (PIC) (8, 44, 65). The initial step involves TFIID binding to the TATA box, which may be stimulated by TFIIA. TFIIB, TFIIF, and RNAP II then bind to the TFIID-TATA box complex to form a minimal PIC that, in some case, is active on supercoiled templates. Further activation and complete PIC assembly involve recruitment of TFIIE and TFIIH. More recent studies have described the isolation of RNAP II complexes (RNAP II holoenzyme) containing associated GTFs and mediators, suggesting a mechanism that can bypass several steps in PIC assembly (26, 29, 63).
Although the assembly of RNAP II and GTFs on the promoter is indispensable for transcription, regulation of the assembly through modulation in the levels of these factors is not apparent. Instead, gene-specific DNA-binding regulatory factors have been considered to play a predominant role in this regulation. These factors may interact directly or indirectly with RNAP II and GTFs to regulate either the assembly of the PIC or its subsequent function. Potential mechanisms include increasing the local concentration of GTFs and RNAP II on a promoter, activating these factors via stoichiometric interactions (44), enzymatic modification (52), and modifying chromatin and its associated proteins by the activities of enzymes such as histone acetyltransferases and deacetylases (51). In some cases, transcriptional mediators, which do not bind DNA directly, may serve as bridging factors between gene-specific regulators and GTFs and RNAP II (22, 44). Apart from functions at the level of initiation, DNA-binding activators and mediators may also stimulate transcriptional elongation. RNAP II elongation factors include SII, SIII, ELL, p-TEFb, DSIF, and TFIIF are thought to act on most class II genes through binding to RNAP II (42, 56).
The entire gene expression program in a cell is thought to be systematically and generally controlled in a manner dependent on various cell activities. For example, if a resting cell enters the proliferation mode, a number of genes must be simultaneously activated. Such concerted gene regulation could involve, in part, a sequence-independent transcriptional regulator that acts on wide spectrum of regulated genes transcribed by RNAP I, II, and III. In such a case, the regulatory protein would be expected to interact with a component common to the RNAP I, II, and III general transcriptional mechanisms. Such components include common RNA polymerase subunits (47) and the TATA-binding protein (TBP) (11, 16, 21).
TBP was initially identified as a component of TFIID (TBP plus TBP-associated factors [TAFs]) in the RNAP II system (9, 14). TBP can bind to the TATA box to nucleate PIC assembly either in isolated form or in the context of TFIID (43). TBP was also shown to be an essential component of the RNAP I accessory factor SL1/TIF and the RNAP III accessory factor TFIIIB and thus is regarded as a universal transcription factor (7, 53). TBP binds to numerous proteins that include other GTFs, viral transactivators, tumorigenesis-related factors, and other transcriptional activators and mediators (4, 18, 44). These interactions are presumed to effect transcriptional regulation through modulations of the efficiency of PIC formation or function.
TBP-interacting protein 120 (TIP120) was originally identified as one of several rat liver proteins that bind to a histidine-tagged TBP (64). Although TIP120 has none of the motifs (leucine zipper, zinc finger, etc.) usually found in transcription factors, its N-terminal region has a sequence that is related to a region of Drosophila TAF80 (10, 28). Indeed, TIP120 interacts directly with TBP in vitro and is associated with TBP in nuclear extracts (64), thus suggesting a role in transcriptional regulation. In this study, we investigated the effect of TIP120 on basal transcription both in vitro and in vivo, and we found that TIP120 stimulates the basal level of transcription via enhancement of RNAP II-containing PIC formation. It is likely that TIP120 plays an important role in PIC formation at the entry of RNAP II into a template DNA.
MATERIALS AND METHODS
Expression, purification, and antibody for recombinant TIP120.
The entire open reading frame of TIP120 was linked to an N-terminal polyhistidine tag (20), subcloned into a baculovirus vector (His-pBlueBacIII) (Invitrogen), and expressed in Spodoptera frugiperda Sf9 cells. Sf9 cells expressing TIP120 were harvested, resuspended in a lysis buffer containing 20 mM Tris-HCl (pH 7.9), 100 mM KCl, 0.1% NP-40, 1 mM 2-mercaptoethanol, 1 mM phenylmethylsulfonyl fluoride (PMSF), and 10% glycerol, and disrupted by sonication. After centrifugation, the supernatant was applied onto a Ni-agarose column (Qiagen) equilibrated with lysis buffer, and the column was washed with BC100 buffer (20 mM Tris-HCl [pH 7.9], 100 mM KCl, 1 mM 2-mercaptoethanol, 1 mM PMSF, 10% glycerol) supplemented with 20 mM imidazole-HCl (pH 7.9). The bound proteins were eluted with BC100 buffer containing 300 mM imidazole-HCl (pH 7.9), and the peak fractions were dialyzed against BC50 buffer (BC100 with 50 rather than 100 mM KCl). The Ni column eluate was loaded onto a MonoQ (Pharmacia) column, and the column was eluted with a linear gradient from 50 to 500 mM KCl in BC buffer. The 0.45 M KCl fraction was collected and dialyzed against BC100 buffer.
For antibody production, TIP120 was further purified through a preparative sodium dodecyl sulfate (SDS)-polyacrylamide gel, and the gel was stained with Coomassie brilliant blue. The band corresponding to TIP120 was excised, and the protein was electroeluted from the gel to immunize rabbits. The serum was affinity purified through TIP120-immobilized HiTrap N-hydroxysuccinimide-activated beads (Pharmacia). The bound antibody was eluted from the column with 0.2 M glycine-HCl (pH 2.5) and neutralized with a 1/10 volume of 1 M Tris-HCl (pH 8.0).
Reconstituted in vitro transcription. (i) For RNAP II promoters.
Polyhistidine-tagged mouse TBP (24), human TFIIB, and human TFIIE (15, 41) were expressed in Escherichia coli and purified as described previously. The α and β-γ subunits of the human TFIIA coexpressed in E. coli were as reported by Ma et al. (33). Human TFIIF was purified from Sf9 cells coinfected with recombinant baculoviruses encoding the RAP74 and RAP30 subunits as described elsewhere (1). TFIIH was prepared from HeLa cells as previously described (34). For purification of human TFIID, HeLa cell nuclear extracts were fractionated through phosphocellulose (Whatman) and MonoQ (Pharmacia) columns as previously described (54). RNAP II was purified from calf thymus as described by Hodo and Blatti (19).
Reconstituted in vitro transcription reactions were performed as previously described with the promoters of the adenovirus major late (AdML) and adenovirus E4 (E4) genes followed by a G-free cassette (40, 46). Twenty microliters of reaction mixture containing 10 mM HEPES-KOH (pH 7.6), 25 mM KCl, 6 mM MgCl2, 3% glycerol, 45 ng of TBP (or 5 μl of TFIID fraction plus 0.1 μg of TFIIA), 50 ng of TFIIB, 120 ng of TFIIF, 0.2 μg of calf thymus RNAP II, and 400 ng of supercoiled DNA template (AdML and E4 promoters) was preincubated for 20 min on ice, and the reaction was initiated by the addition of ribonucleoside triphosphates (NTPs) to final concentrations of 620 μM ATP, 620 μM UTP, 25 μM CTP, and 5 μCi of [α-32P]CTP. The standard runoff transcription was carried out with TFIIA, TFIIB, TBP, TFIIE, TFIIF, TFIIH, RNAP II (34), and AdML and rabbit β-globin (61) promoters as described. Transcription was performed for 45 min at 30°C. Transcripts were resolved on a 5% sequencing gel. When necessary, transcripts were quantified with a BAS 1500 RI-image analyzer (Fuji Film).
(ii) For RNAP III promoters.
TFIIIB, TIIIC, and RNAP III were immunopurified from the nuclear extracts made from FLAG-tagged cell lines (6, 58–60). The partially purified TFD were prepared as described previously (59). The adenovirus VA1 gene from −96 to +212 (AdVA1) and human methionine tRNA gene from −131 to +148 (htRNA[Met]) were used to yield runoff products. Reconstituted in vitro runoff transcription was performed for 60 min at 30°C in a reaction cocktail similar to that previously described (58, 59).
(iii) For the mouse rRNA promoter.
The mouse ribosomal RNA gene (mrDNA) promoter sequence from −330 to +291 in mrDNA (37) linearized to yield a 320-base transcript was used for the runoff transcription assay. Reconstituted in vitro transcription was performed as previously reported (23), using fraction A′ (RNAP I stimulatory factor) (63a), C (RNAP I), and D (SL1 and UBF) prepared from FM3A mouse ascites cells. The reaction mixture was incubated at 30°C for 60 min in the presence of 100 μg of α-amanitin per ml (23).
EMSA.
Electrophoretic mobility shift assay (EMSA) was performed with an AdML TATA box probe from −45 to +20 as previously described (3, 32). The binding mixture (10 μl) consisting of 50 mM KCl, 12.5 mM HEPES-KOH (pH 7.6), 6.3 mM MgCl2, 0.05 mM EDTA, 0.05% NP-40, 0.5 mM dithiothreitol, 5% glycerol, 80 of ng poly(dG-dC), 100 ng of bovine serum albumin (BSA), 1 ng of end-labeled probe, TBP (10 ng), TFIIB (10 ng), TFIIF (50 ng), purified RNAP II (200 ng), and TIP120 (200 ng) was incubated for 30 min at room temperature. The reaction products were analyzed by polyacrylamide gel electrophoresis (PAGE) through a 4% polyacrylamide gel containing 5% glycerol and running buffer consisting of 25 mM Tris-base (pH 8.3), 190 mM glycine, and 5% glycerol.
Nonspecific transcription of RNAP II.
Nonspecific RNA synthesis was carried out by the method of Sekimizu et al. (48). The reaction mixture (50 μl) contained 50 mM Tris-HCl (pH 7.9), 0.02 mM EDTA, 100 mM (NH4)2SO4, 3 mM MnSO4, 2 mM MgCl2, 5 mM 2-mercaptoethanol, 1 mM PMSF, 0.5 mM GTP, 0.5 mM UTP, 0.5 mM ATP, 0.1 mM [α-32P]CTP (0.5 μCi), 10% glycerol, 20 μg of activated calf thymus DNA, and 0.2 pmol of purified calf thymus RNAP II. When necessary, 2 to 16 pmol of TIP120 or 16 pmol of GTFs was added to the reaction. Where indicated, α-amanitin (1 μg/ml) was included. The mixture was incubated at 30°C for 10 min, and a 15-μl aliquot was spotted on Whatman DE52 paper. The paper was rinsed five times with 5% Na2HPO4 solution, and radioactivities on the paper were measured by a liquid scintillation counter.
In vitro binding for TIP120 and RNA polymerase subunits.
TIP120 carrying FLAG tag at its N terminus was purified from Sf9 cells by passage through M2-agarose and MonoQ columns as described above. cDNAs of RPB5, RPB6, RPB8, and RPB10α, common subunits for RNA polymerases, were cloned by PCR-mediated techniques. The histidine-tagged RNA polymerase subunits were expressed in E. coli and purified by using Ni-agarose as described above. Interaction with TIP120 and each RPB protein was analyzed by affinity chromatography and pull-down experiments. TIP120 affinity columns (50 μl) were prepared by immobilizing FLAG-TIP120 fusion protein (5 μg) on M2-agarose beads as described above. The columns were equilibrated with BC100 buffer. Control column contained no TIP120 protein. Purified RPB proteins (0.5 μg) were loaded onto the columns. After extensive washing of the columns with BC100 buffer, bound proteins were eluted with FLAG peptide, analyzed by SDS-PAGE (15% gel), and detected by silver staining.
Cells and transient luciferase assay.
P19 mouse embryonal carcinoma cells and HEp-2 cells were cultured in alpha minimal essential medium and Dulbecco’s modified Eagle’s medium (Gibco), respectively, supplemented with 10% fetal bovine serum. For retinoic acid treatment, all-trans retinoic acid (Sigma) was added into the medium at a final concentration of 0.5 mM. For the transient luciferase assay, P19 cells were transfected by the use of Lipofectamine Plus (Gibco) with each luciferase reporter plasmid (150 ng) together with various amounts of TIP120-expressing effector plasmid (pRcCMV-HA120). After incubation for 5 h, the cells were transferred to new dishes at a dilution of 1:2 and further cultured for 40 h. The cells were harvested, and cell extracts (1 ml) were prepared. The luciferase activity in 20 ml of reaction cocktail was measured with a luciferase assay system (Promega) and TD20/20 luminometer (Terner Designs). Obtained values were normalized by protein concentration.
For construction of the effector plasmid, the entire TIP120 cDNA coding sequence having a hemagglutinin tag at the N terminus was inserted downstream from the cytomegalovirus (CMV) enhancer-promoter regulatory unit of pRc/CMV DNA (Invitrogen). Reporter plasmids were constructed from pGV-B vector DNA (Toyo Ink Co., Ltd.) by inserting a promoter sequence into the multicloning site of the vector. Promoter sequences included in each plasmid were as follows: pGV-rDNA, positions −330 to +291 of the mrDNA promoter; pGV-ML33, positions −33 to +33 of the AdML promoter; and pGV-ML677, positions −677 to +33 of the AdML promoter.
Nucleotide sequence accession number.
Amino acid and nucleotide sequences of rat TIP120 appears in the GenBank, EMBL, and DDBJ databases with accession no. D87671.
RESULTS
Activation of basal transcription of RNAP II genes by TIP120.
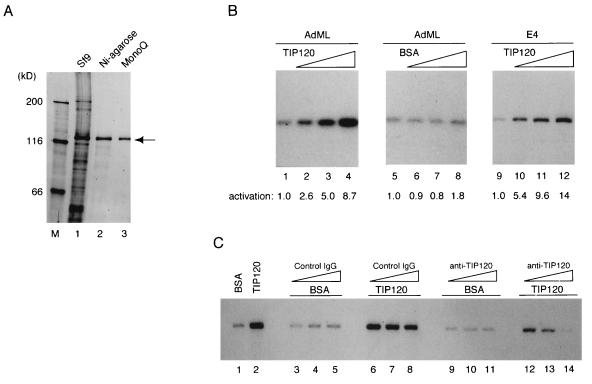
To perform biochemical studies on TIP120, we expressed polyhistidine-tagged TIP120 in Sf9 cells and purified the recombinant protein to near homogeneity by Ni-agarose and MonoQ chromatography (Fig. 1A). To test the effect of TIP120 on transcriptional regulation, we established a minimal system containing TBP, TFIIB, TFIIF, RNAP II, and supercoiled DNA templates with AdML and E4 promoters followed by a G-free cassette. The use of supercoiled templates abrogated the need for TFIIE and TFIIH (39, 40, 55). Both AdML and E4 promoters were transcribed accurately under our conditions (Fig. 1B, lanes 1, 5, and 9), and the addition of recombinant TIP120 to the reaction resulted in activation of transcription from both promoters in a dose-dependent manner (Fig. 1B, lanes 2 to 4 and 10 to 12). A twofold molar excess of TIP120 per TBP molecule increased the level of transcription from AdML and E4 promoters 8.7- and 14-fold, respectively (Fig. 1B; compare lanes 1 and 4 and lanes 9 and 12), whereas the addition of equivalent amounts of BSA had little effect (lanes 6 to 8). To further establish that TIP120 itself causes transcriptional activation, we added affinity purified anti-TIP120 antibody to the reaction. As shown in Fig. 1C, anti-TIP120 antibody repressed TIP120-stimulated transcription (lanes 12 to 14) but not basal transcription (lanes 9 to 11), whereas an equivalent amount of control immunoglobulin G had little effect on basal or TIP120-activated transcription (lanes 3 to 5 and 6 to 8, respectively). These results further indicate that TIP120 itself was responsible during the enhanced transcription.
FIG. 1.
Stimulation of basal transcription in vitro by TIP120. (A) Expression and purification of recombinant TIP120. Histidine-tagged TIP120 expressed in Sf9 cells (lane 1) was purified by Ni-agarose (lane 2) and then by MonoQ (lane 3). The proteins were resolved by SDS-PAGE (7.5% gel) and stained with silver. The arrow indicates the position of the recombinant TIP120. (B) Effect of TIP120 on in vitro reconstituted transcription. Transcription reactions were performed as described in Materials and Methods, using supercoiled AdML (lanes 1 to 8) and E4 (lanes 9 to 12) templates. Reaction mixtures contained 100 ng (lanes 2 and 10), 200 ng (lanes 3 and 11), or 400 ng (lanes 4 and 12) of the recombinant TIP120. Equivalent amounts of BSA were added to the reactions (lanes 5 to 8). TIP120-mediated stimulation is presented as fold activation relative to controls (lanes 1, 5, and 9) at the bottom. (C) Anti-TIP120 antibody suppressed the TIP120-mediated transcriptional stimulation. In vitro transcription was performed with the AdML promoter as for panel B. Each reaction contained 200 ng of TIP120 or BSA as indicated. Control immunoglobulin G (IgG) (lanes 3 to 8) and affinity-purified anti-TIP120 antibody (lanes 9 to 14) were added to the reaction mixture.
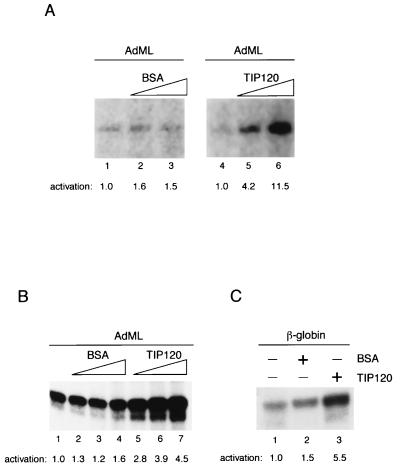
Since TFIID, rather than the derived TBP, is thought to function in RNAP II transcription in vivo, we reconstituted a transcription system in which TBP was replaced by purified TFIID and TFIIA. In this case, TIP120 again stimulated (up to 11.5-fold) transcription from the AdML promoter in a dose-dependent manner (Fig. 2A). We also investigated the effects of TIP120 on linearized templates in a complete transcription system that contained TFIIA, TFIIE, and TFIIH in addition to the minimal components (Fig. 2B and C). As observed in the minimal system with supercoiled templates (Fig. 1B), transcription from a linear AdML template in the complete system was enhanced (up to 4.5-fold) by TIP120 in a dose-dependent manner (Fig. 2B). Further studies indicated that TIP120 also significantly stimulated transcription from β-globin promoter (Fig. 2C). Some promoters (e.g., the conalbumin promoter) were stimulated only weakly (data not shown), implying that TIP120 may have a promoter preference. Thus, the combined results from Fig. 1 and 2 indicate that TIP120 can stimulate transcription from a variety of different promoters, independently of TAFs, TFIIE, and TFIIH or DNA topology.
FIG. 2.
TIP120 stimulates the basal transcription in other in vitro transcription systems. (A) TIP120-stimulated transcription in the presence of TFIID. The supercoiled AdML G-free template was transcribed in a transcription system with the minimal set of GTFs including TFIID and TFIIA instead of TBP. Reaction mixtures contained 200 and 400 ng of BSA (lanes 2 and 3) and TIP120 (lanes 5 and 6). TIP120-mediated stimulation is presented as fold activation relative to controls (lanes 1 and 4) at the bottom. (B) TIP120 enhances transcription from linearized AdML promoter in the complete transcription system with TFIIB, TBP, TFIIE, TFIIF, TFIIH, and RNAP II. Reaction mixtures contained 100, 200, and 400 ng of the recombinant TIP120 (lanes 5 to 7, respectively). Equivalent amounts of BSA were added to the reactions (lanes 2 to 4). TIP120-mediated stimulation is presented as fold activation relative to the control (lane 1) at the bottom. (C) TIP120 enhances transcription from linearized β-globin promoter in the complete transcription system. The β-globin promoter was transcribed in the complete transcription system as described above. Reaction mixtures in lanes 2 and 3 contained 200 ng of BSA and TIP120, respectively. TIP120-mediated stimulation is presented as fold activation relative to the control (lane 1) at the bottom.
Domain for transcriptional activation and TBP binding.
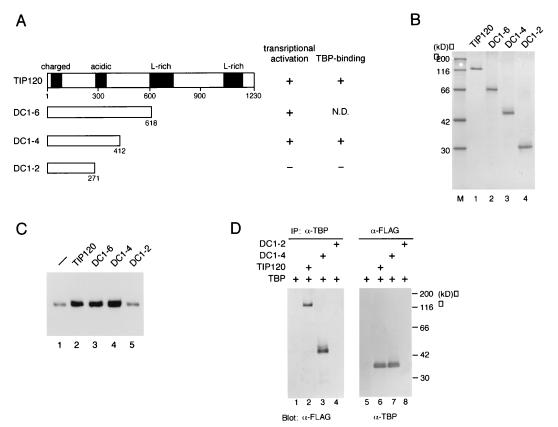
To determine the domain of the TIP120 molecule responsible for transcriptional activation, we constructed a series of C-terminal deletion mutants as depicted in Fig. 3A. The full-length TIP120 and the deletion mutants (Fig. 3A) carrying both FLAG and histidine tags at their N termini were expressed in Sf9 cells and purified by Ni-agarose, MonoQ, and M2-agarose chromatography to near homogeneity (Fig. 3B). These highly purified proteins were analyzed for the ability to activate basal transcription in the reconstituted system. Mutants DC1-6 (amino acids [aa] 1 to 618) and DC1-4 (aa 1 to 412), as well as full-length TIP120 (Fig. 3C, lanes 2 to 4), enhanced transcription from AdML promoter, whereas mutant DC1-2 (aa 1 to 271) failed to do so (lane 5). These results suggest that the N-terminal one-third (upstream from aa 412) that includes charged and acidic regions is sufficient for transcription activation.
FIG. 3.
The N-terminal one-third of TIP120 is sufficient for transcriptional stimulation and TBP binding. (A) Structures of deletion mutants. Several characteristic sequences such as charged, acidic, and leucine-rich regions are indicated. These four proteins contain both FLAG and histidine tags at their N termini. Numbers indicate amino acid positions of TIP120 from the N terminus. Results for transcriptional stimulation and TBP binding for each construct are summarized. N.D., not determined. (B) The proteins depicted in panel A were expressed in Sf9 cells and purified. One hundred nanograms of the purified proteins was analyzed by SDS-PAGE and Coomassie brilliant blue staining. (C) Effects of deletion mutants on basal transcription. Transcription reactions were performed as described in the legend to Fig. 1B, using supercoiled AdML template. Equimolar amounts (2.4 pmol) of full-length TIP120 (lane 2) and the C-terminal deletion mutants (lanes 3 to 5) were added to the reaction. Lane 1, without TIP120 protein. (D) Mapping of the TBP-binding region of TIP120. Equimolar amounts (7.5 pmol) of TBP and the deletion mutants were incubated as indicated. The mixtures were immunoprecipitated (IP) with anti-TBP antibody (lanes 1 to 4) or anti-FLAG antibody (lanes 5 to 8). The precipitated proteins were resolved by SDS-PAGE (10% gel) and analyzed by Western blotting with anti-FLAG antibody (lanes 1 to 4) or anti-TBP antibody (lanes 5 to 8).
TIP120 interacts directly with TBP in solution (64) but is not stably incorporated into a TBP-DNA complex (data not shown). We next examined the ability of truncation versions to interact with TBP. Equimolar amounts of full-length TIP120 and truncated mutants DC1-2 and DC1-4 were incubated with TBP and immunoprecipitated with anti-TBP antibody or anti-FLAG antibody. Anti-TBP antibody efficiently coimmunoprecipitated full-length TIP120 and DC1-4 (Fig. 3D, lanes 2 and 3) but not DC1-2 (lane 4). Equivalent results were obtained in the case of coimmunoprecipitation with anti-FLAG antibody (Fig. 3D, lanes 6 to 8). We also observed that anti-TBP antibody and anti-FLAG antibody were not able to immunoprecipitate DC1-4 (data not shown) and TBP (Fig. 3D, lane 5), respectively. These results (summarized in Fig. 3A) clearly demonstrate the correlation between the abilities of TBP binding and transcriptional stimulation. This correlation supports the idea that TBP binding is one of the essential requirements for TIP120-mediated transcriptional stimulation.
TIP120 contributes to PIC formation.
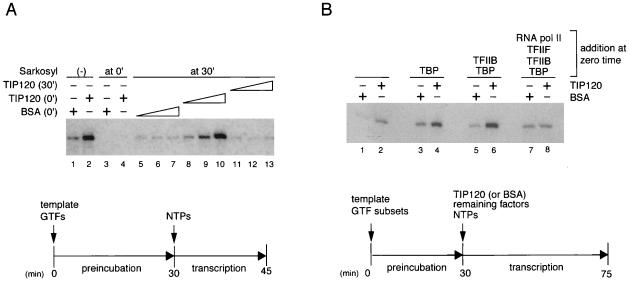
The primary event of transcription entails the assembly of GTFs and RNAP II on a promoter to form the PIC (17). To investigate whether the TIP120-mediated stimulation occurs prior to or subsequent to PIC formation, we used a previously described preincubation-Sarkosyl addition protocol (17). In this assay, transcription factors and DNA are first incubated in the absence of NTPs to allow PIC formation, and transcription is thus allowed to proceed in the presence of NTPs and a Sarkosyl concentration that inhibits the formation but not function of the PIC. When 0.01% Sarkosyl was present during PIC formation, both basal and TIP120-stimulated transcription were inhibited (Fig. 4A; compare lanes 3 and 4 to lanes 1 and 2). As expected, Sarkosyl did not abolish basal transcription when added after a time (30 min) sufficient for PIC formation (Fig. 4A, lanes 5 to 7). TIP120 had no effect when added with Sarkosyl at 30 min, consistent either with TIP120 function during PIC formation or with a general inhibitory effect of Sarkosyl on TIP120 function (Fig. 4A, lanes 11 to 13). However, TIP120 did enhance transcription when present during PIC assembly (0-min addition) and prior to Sarkosyl addition (Fig. 4A, lanes 8 to 10). Since the level of TIP120-enhanced transcription was comparable to that observed in standard assay, these results suggest that TIP120 functions mainly during PIC assembly and not at subsequent initiation or elongation steps.
FIG. 4.
TIP120 functions during PIC formation. (A) Effects of Sarkosyl on TIP120-mediated transcriptional stimulation. Transcription reactions were performed with the AdML promoter. Template DNA, GTFs, and RNAP II were mixed at zero time (0′; the time at which preincubation at 30°C started). After preincubation for 30 min (30′), transcription was initiated by the addition of nucleotide substrates. TIP120 was added to the reaction at zero time (lanes 2, 4, and 8 to 10) or after 30 min (lanes 11 to 13). Sarkosyl (0.01%) was also added at zero time (lanes 3 and 4) or at 30 min (lanes 5 to 13). The reaction mixtures contained 100 ng (lanes 8 and 11), 200 ng (lanes 9 and 12), or 400 ng (lanes 2, 10, and 13) of TIP120. Lanes 1 and 2 did not contain Sarkosyl. Equivalent amounts of BSA were added at zero time (lanes 1, 3, and 5 to 7). (B) Completion of PIC including the three GTFs and RNAP II impairs the TIP120 effect. The AdML DNA was preincubated with different subsets of GTFs for 30 min as indicated, and then transcription was initiated by addition of nucleotide substrates and remaining factors. Recombinant TIP120 (400 ng) or BSA was added as indicated.
To study the mechanism of TIP120-mediated activation in more detail, we preincubated the DNA template with various combinations of GTFs and RNAP II before addition of TIP120, as shown in Fig. 4B. The absence of any GTFs during the preincubation resulted in a weak basal activity (Fig. 4B, lane 1), whereas addition of TBP alone or with other factors during the preincubation period considerably increased basal transcription (lanes 3, 5, and 7). This observation is consistent with previous reports that TBP binding to the TATA box is a rate-limiting step for transcription initiation (8, 66). Addition of TIP120 after template preincubation with no GTFs (Fig. 4B, lane 2), with TBP (lane 4), or with TBP plus TFIIB (lane 6) significantly enhanced the basal level of transcription. In striking contrast, TIP120 had no effect on the basal level of transcription when added after the template had been preincubated with all of the components (TBP, TFIIB, TFIIF, and RNAP II) necessary for formation of a minimal stable PIC (Fig. 4B, lane 8 versus lane 7). Inactivation of GTFs and/or RNAP II during the preincubation time was negligible (Fig. 4B, lanes 3, 5, and 7). These results, together with those of Fig. 4A, show that TIP120 functions during PIC formation at a step that precedes or is coincident with the entry of RNAP II/TFIIF.
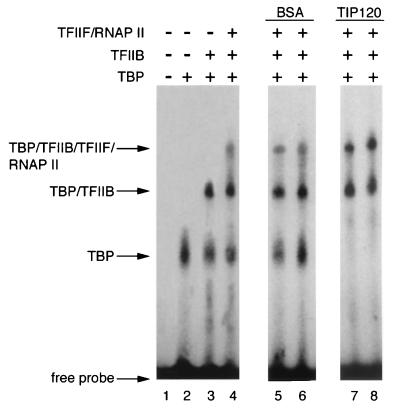
Stimulation of PIC formation by TIP120.
Next, we examined the effect of TIP120 on complex formation with the AdML TATA box by EMSA. TIP120 alone showed no stable binding to promoter DNA in EMSA (data not shown). This finding suggests that TIP120 is not a conventional DNA-binding protein, which is consistent with the homology search data (64). An EMSA with TBP, TFIIB, TFIIF, and RNAP II resulted in three distinct bands (Fig. 5, lanes 2 to 4) which were not observed in the absence of the GTFs and RNAP II (Fig. 5, lane 1). The formation of those complexes was dependent on TBP (Fig. 5, lane 2), TFIIB (lane 3), and TFIIF-RNAP II (lane 4), as previously reported (3, 32). In addition, the binding of RNAP II to the TBP-TFIIB complex was dependent on TFIIF (data not shown). The addition of TIP120 significantly reduced the amount of TBP-DNA complex, whereas the amount of the completed PIC (TBP-TFIIB-TFIIF-RNAP II) was significantly increased (Fig. 5; compare lanes 5 and 6 to 7 and 8; the experiment was done in duplicate). The completed complex found in Fig. 5 (lanes 7 and 8) was not supershifted by TIP120 and did not contain immunologically detectable TIP120 (data not shown). Thus, TIP120 evidently is not integrated into the TBP-DNA-complex even though it can bind to TBP in solution. These observations suggest that TIP120 acts to increase the amount of the RNAP II-containing transcription-competent complex and are consistent with the order of addition experiments of Fig. 4B. However, since it is unlikely that TIP120 is stably incorporated into the PIC, we anticipate that TIP120 transiently interacts with the components of the PIC.
FIG. 5.
TIP120 facilitates the formation of RNAP II-containing PIC. DNA-protein complexes were formed with various GTFs and RNAP II and analyzed by EMSA as described in Materials and Methods. A 5′-end-labeled DNA fragment containing the AdML TATA box was used in the DNA binding assay. Lane 1, no protein. Various combinations of GTFs and RNAP II, as indicated, were added to the DNA binding assay (lanes 2 to 8). PIC formation was performed in the absence (lanes 5 and 6) or presence (lanes 7 and 8) of TIP120. An equivalent amount of BSA was used (lanes 5 and 6). The reactions were done in duplicate. Positions of the specific complexes are indicated.
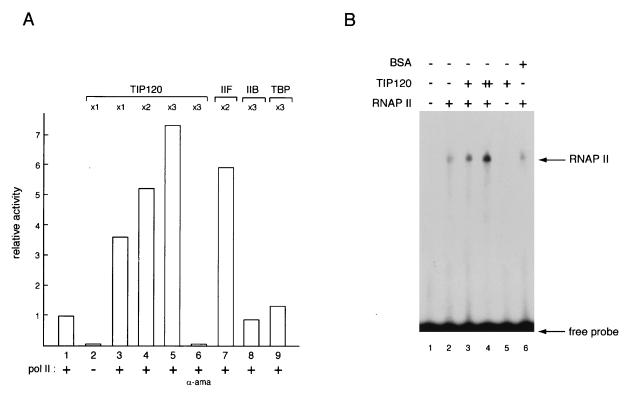
TIP120 stimulates nonspecific transcription by RNAP II.
The results of Fig. 4 and 5 imply that TIP120 can functionally interact with RNAP II as well as TBP. To investigate the effect of TIP120 on RNAP II itself, we assayed TIP120 function in a nonspecific RNAP II transcription reaction. In experiments shown in Fig. 6A, RNAP II was incubated with template DNA in the presence or absence of TIP120, and then transcription was initiated by adding nucleotides. TIP120 dramatically (>7-fold) increased RNAP II activity in a dose-dependent manner (Fig. 6A, lanes 3 to 5). Neither TBP nor TFIIB affected the RNAP II activity (Fig. 6A, lanes 8 and 9), whereas TFIIF, which is known to promote the elongation rate as well as the initiation frequency of RNAP II (2, 12), facilitated RNAP II activity as expected (lane 7). The TIP120 preparation had no RNAP II activity, and TIP120-stimulated transcription was sensitive to α-amanitin (Fig. 6A, lanes 2 and 6). These findings clearly demonstrated that TIP120 can stimulate RNAP II activity. We also found that TIP120 was able to increase the activity of RNAP II of HeLa and mouse cells prepared by different purification protocols (data not shown). These findings emphasized our supposition that TIP120 targets RNAP II for transcriptional activation as well as TBP.
FIG. 6.
Effect of TIP120 on RNAP II activity. (A) Stimulation of enzyme activity by TIP120. Each RNAP II reaction contains 4 pmol (×1), 8 pmol (×2), or 12 pmol (×3) of TIP120. Lanes 7 to 9 contain 8 pmol (×2) of TFIIF and 16 pmol (×3) of TFIIB and TBP, respectively. Lane 6 contains α-amanitin. The reactions were done in duplicate. (B) TIP120 stimulates the DNA binding of RNAP II. DNA-protein complexes were formed with the DNA fragment used in the previous experiment (Fig. 5) and RNAP II and analyzed by EMSA in the absence of a carrier DNA. Lane 1, no protein. Various combinations of RNAP II (200 ng), TIP120, and BSA, as indicated, were added to the DNA binding assay. Reaction mixtures contained 100 ng (lanes 3 and 5) and 200 ng (lane 4) of the recombinant TIP120. Lane 6 contained 100 ng of BSA. Positions of the specific complexes are indicated.
The above observations suggest the existence of a mechanism by which TIP120 stimulates RNAP II recruitment to a template DNA or the RNAP II elongation rate. To examine these possibilities, we tested the effect of TIP120 on DNA-binding activity of RNAP II. It is known that RNAP II is able to bind nonspecifically to a DNA fragment in vitro (25, 27). The purified RNAP II was incubated with the radiolabeled oligonucleotide used in the previous experiment (Fig. 5) in the absence of a carrier DNA. As expected, an EMSA with only RNAP II resulted in one distinct band (Fig. 6B, lane 2) which was not detected in the absence of RNAP II (lane 1). The addition of TIP120 significantly increased the amount of RNAP II-DNA complex in a dose-dependent manner (Fig. 6B, lanes 3 and 4), whereas TIP120 itself did not form a clear retarded band (lane 5), and an equivalent amount of BSA had little effect on the formation of the RNAP II-DNA complex (lane 6). Moreover, we found that TIP120 had little effect on the elongation rate of RNAP II (data not shown), which agrees with the results of the Sarkosyl and kinetic experiments (Fig. 4). These results clearly demonstrate that TIP120 facilitates the binding of RNAP II to DNA. Thus, it is likely that the stimulation of RNAP II activity by TIP120 is a consequence of the ability of TIP120 to enhance DNA-binding activity of RNAP II.
Effects of TIP120 on other classes of promoters.
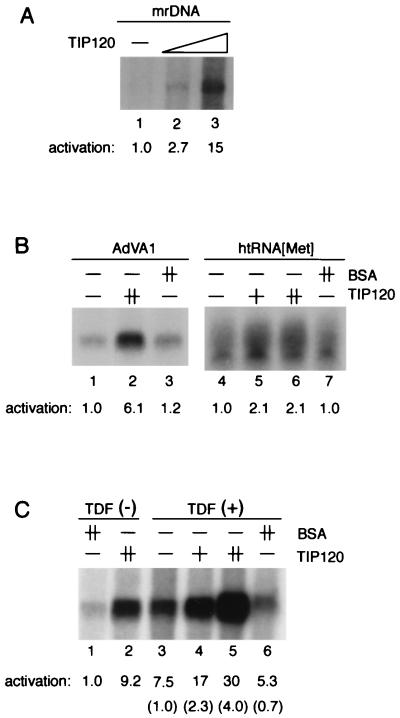
To link the above observations to our assumption that a target(s) of TIP120 is some general component or common reaction for transcriptional regulation, we next investigated the effect of TIP120 on two other transcription systems, i.e., those utilizing RNAP I and RNAP III. First, mrDNA was examined in a reconstituted transcription system composed of mouse factors (Materials and Methods), since in vitro transcription of RNAP I exhibits a species specificity (14, 45). As shown in Fig. 7A, mrDNA was weakly but significantly transcribed in this assay system (lane 1), and TIP120 stimulated the specific transcription (lanes 2 and 3). The activation index obtained with 500 ng of TIP120 was 15 (compare lanes 1 and 3), nearly the same as that obtained in the RNAP II system (Fig. 1B).
FIG. 7.
TIP120 stimulates RNAP I- and III-directed in vitro transcription. (A) TIP120 enhances transcription from mrDNA. mrDNA was transcribed in the presence of mouse factors as described in Materials and Methods). Reaction mixtures contained 250 and 500 ng of the recombinant TIP120 (lanes 2 and 3, respectively). TIP120-mediated stimulation is presented as fold activation relative to the control below the panel. (B) Effect of TIP120 on transcription from class III promoters. The AdVA1 and htRNA[Met] promoters were transcribed. Reaction mixtures contained 200 ng (lane 5) or 800 ng (lanes 2 and 6) of TIP120. Lanes 3 and 7 contained 800 ng of BSA. TIP120-mediated stimulation is presented as fold activation relative to the controls (lanes 1 and 4) at the bottom. (C) TIP120-mediated transcriptional stimulation of the AdVA1 promoter is independent of TDF. The AdVA1 promoter was transcribed as shown in panel B. Transcription reactions were performed in the absence (lanes 1 to 4) or presence (lanes 3 to 6) of TDF. Reactions contained 200 ng (lane 4) or 800 ng (lanes 2 and 5) of TIP120. Lanes 1 and 6 contained 800 ng of BSA. TIP120-mediated stimulation is presented as fold activation relative to the control (lane 1) at the bottom. Parentheses indicate fold activation relative to lane 3.
We further examined TIP120 effects on two different RNAP III genes, AdVA1 and htRNA[Met], in a system reconstituted with human TFIIIB, TFIIIC, and RNAP III (57, 59). Transcription from the AdVA1 promoter was stimulated by TIP120, with a stimulation index of 6.1 (Fig. 7B; compare lanes 1 and 2). htRNA[Met] transcription was also weakly stimulated by TIP120 (Fig. 7B, lane 6). As TDF (translation-dependent factor) has been demonstrated to enhance RNAP III-driven in vitro transcription (59), the effect of TIP120 was also examined in the presence of added TDF. As seen in Fig. 7C, TIP120 stimulated AdVA1 transcription 9.2-fold (compare lanes 1 and 2) and 4.0-fold (compare lanes 3 and 5) in the absence and presence of TDF, respectively. Thus, TIP120-mediated stimulation is TDF independent. Significantly, the combined stimulation index for AdVA1 gene transcription by both TDF and TIP120 was exceptionally high, 30-fold (Fig. 7C; compare lanes 1 and 5).
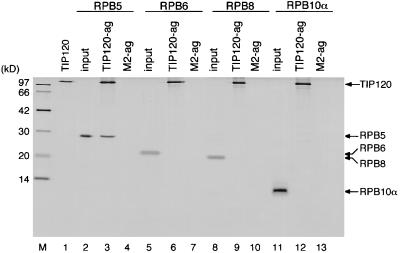
To elucidate a mechanism of TIP120-mediated stimulation of three classes of RNA polymerases, we investigated whether TIP120 interacts with subunits common to all three RNA polymerases. RPB5, RPB6, RPB8, RPB10α, and RPB10β are common subunits of three RNA polymerases, whose structures and functions are highly conserved in the eukaryotes (49). Affinity columns were prepared by immobilizing the FLAG-TIP120 fusion proteins on M2-agarose beads. Equivalent amounts of human RPB5, RPB6, RPB8, and RPB10α were loaded onto the columns, and the adsorbed proteins were detected. As shown in Fig. 8, the TIP120 column significantly retained RPB5 (lane 3) but not RPB6, RPB8, and RPB10α (lanes 6, 9, and 12). No binding of those four subunits to M2-agarose was observed (Fig. 8, lanes 4, 7, 10, and 13), suggesting that binding between TIP120 and RPB5 is specific. These results imply that at least RPB5 is a possible target for TIP120 in three RNA polymerases.
FIG. 8.
TIP120 binds to RPB5 in vitro. FLAG-TIP120 fusion proteins were immobilized on M2-agarose beads (TIP120-ag). As a control, M2-agarose beads (M2-ag) were used. Recombinant RPB5 (lanes 3 and 4), RPB6 (lanes 6 and 7), RPB8 (lanes 9 and 10), or RPB10α (lanes 12 and 13) was loaded onto the columns. Proteins retained the columns were analyzed as described in Materials and Methods. Lanes 2, 5, 8, and 11 show 20% of each input subunit. Lane 1, recombinant FLAG-tagged TIP120. M, molecular weight markers. Note that RPB6 and RPB8 were scarcely stained with silver (lanes 5 and 8) even though equivalent amounts of subunits were loaded.
TIP120 stimulates transcription from three different classes of promoters in P19 cells.
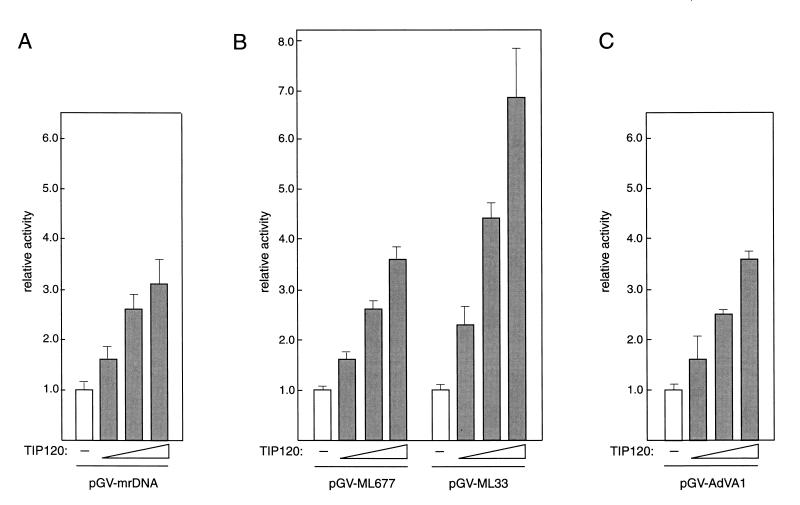
The above studies have shown that TIP120 stimulates in vitro transcription from three classes of promoters. To further assess the physiological relevance of these observations, we studied the contribution of TIP120 to transcriptional regulation in vivo. P19 mouse cells (35) were used for transient transfection assays in which various promoter-carrying luciferase reporter plasmids were introduced into the cells together with the TIP120 expression plasmid. P19 cells constitutively expressed a low level of TIP120 protein (see below). As observed in Fig. 9, increased amounts of effector plasmids enhanced the relative luciferase activity from all of the promoters used. When 540 ng of effector DNA was used, stimulation indices in transcription efficiency of pGV-mrDNA for RNAP I, pGV-ML677 (positions −677 to +33 of the AdML promoter), and pGV-ML33 (positions −33 to +33 of the AdML promoter) for RNAP II and of pGV-AdVA1 for RNAP III were 3.1, 3.6, 6.8, and 3.6, respectively. The failure of the control (empty) effector plasmid to exhibit any stimulation indicates that ectopically expressed TIP120 is responsible for the observed stimulation, whereas the failure to see high levels of stimulation (as observed in vitro) may reflect contributions of endogenous TIP120. Interestingly, TIP120 exhibited a higher stimulatory effect on the enhancerless AdML core promoter (pGV-ML33) than on the enhancer-containing promoter (Fig. 9B). These results are consistent with the view stated above that TIP120 acts on the basal transcription apparatus. Taken together, these data indicate that TIP120 enhances transcription from all three classes of promoters in vivo.
FIG. 9.
TIP120 stimulates transcription of three classes of promoters in vivo. Promoter fragments from mrDNA, AdML (−677 to +33), AdML (−33 to +33), and AdVA1 were inserted into the promoterless luciferase vector to construct pGV-mrDNA (A), pGV-ML677 (B), pGV-ML33 (B), and pGV-AdVA1 (C), respectively. Each reporter plasmid (150 ng) was used to transfect to P19 cells in the presence of the CMV promoter-driven TIP120-expressing effector plasmid (180, 360, and 540 ng). Enzyme activities are normalized with protein concentration. The data are presented as activity relative to that without effector plasmid. The height of each column represents the mean of three independent transfections performed in duplicate. Standard deviations are indicated by vertical lines.
DISCUSSION
TIP120 commonly stimulates transcription in vitro.
TIP120 stimulated transcription from multiple RNAP II-driven genes in the reconstituted cell-free systems. Because the recombinant TIP120 was highly pure (Fig. 1A) and a parallel preparation from mock-infected Sf9 cells did not exhibit an activating property (data not shown), this stimulatory effect must be due to TIP120 itself and not to contaminating proteins. In support of this conclusion, an antibody directed against recombinant TIP120 specifically suppressed the TIP120-mediated activated transcription (Fig. 1C).
In vitro transcription might be elevated in a nonspecific manner by proteins that are not true transcription factors but somehow stabilize bona fide transcription factors or suppress inhibitory agents. However, the following observations indicate that the effect of TIP120 is specific. First, TIP120 shows some promoter specificity, in that chicken conalbumin (data not shown) and htRNA[Met] (Fig. 7B) promoters, in marked contrast to others in the cognate class, showed low stimulation by TIP120. Second, TIP120 function was apparent only when TIP120 was added at a particular time during PIC formation, being essentially ineffective when added after PIC formation (Fig. 4). Third, high TIP120-dependent stimulation levels were seen when TIP120 was included at nearly equimolar amounts with GTFs, implying a specific interaction between TIP120 and a component(s) thereof. Finally, ectopic TIP120 resulted in enhanced transcription from all three classes of promoters in P19 cells (Fig. 9). In these cases, TIP120-mediated stimulation was much more pronounced for the AdML core promoter (6.8-fold) than for the enhancer-carrying counterpart (3.6-fold) (Fig. 9B), in good agreement with the observed effect of TIP120 on AdML core promoter function in vitro (2.6- to 8.7-fold [Fig. 1B]).
Our analysis showed that a stimulatory effect of TIP120 on transcription by RNAP II was independent of DNA topology, the type of class II promoter, or the specific GTF requirement (Fig. 2). In the latter case, the stimulatory effect was independent of TFIIE, TFIIH, TFIIA, and TAFs. Moreover, since the degrees of stimulation were similar under various RNAP II reaction conditions, TIP120 appears to act on a common process in RNAP II-mediated transcription. Importantly, TIP120 also stimulated transcription by RNAP I (Fig. 7A) and by RNAP III, most notably on the AdVA1 gene in the latter case (Fig. 7B). In the case of the RNAP III system, the effect of TIP120 was demonstrated to be independent of the stimulatory factor TDF. These observations indicate that TIP120 generally stimulates all three transcription systems in vitro. To data, no other nonessential transcription factors have been clearly demonstrated to activate transcription by all three RNA polymerases at the basal level. Thus, TIP120 appear to be unique among known transcriptional regulators, and the present study may provide new insights into mechanisms of gene regulation. The situation for TIP120 may be analogous to that of PC4 and topoisomerase I (Topo I), earlier-described RNAP II coactivators (13, 30, 31, 36, 50) that more recently were found in a TFIIIC-containing complex and to function in RNAP III transcription (13, 36, 60). It was further argued that PC4 and Topo I might function in transcription by all three RNA polymerases (60). In contrast to TIP120, PC4 and Topo I affected only the activated or multiple round transcription, not basal or single-round transcription.
Mechanism of TIP120-mediated stimulation.
Since TIP120 could stimulate transcription by RNAP II only when it was added at a specific stage in PIC assembly (i.e., before TFIIF-RNAP II is recruited into the TBP-TFIIB-DNA complex) (Fig. 4), TIP120 may act at a specific time or affect a particular process in PIC assembly. EMSA clearly indicated that TIP120 shifted the balance between TBP-DNA complexes (decreased) and TBP-TFIIB-TFIIF-RNAP II-DNA complexes (increased) (Fig. 5). Hence, we suggest that TIP120 facilitates PIC assembly and that it must act at a specific step prior to completion of PIC formation. Consistent with this idea, TIP120 had little effect on the elongation rate of RNAP II (data not shown).
What is the target of TIP120, and how does TIP120 facilitate PIC formation? Since the stimulatory effect of TIP120 was seen in all RNA polymerase transcription systems, the logical hypothetical target would be a general process or/and a molecule common to all three reaction systems. Based on this aspect, TBP is a more likely target molecule. First, TBP is included in SL1, TFIID, and TFIIIB, which are involved in core promoter recognition events on genes transcribed by RNAP I, II, and III, respectively (7, 9, 53). Second, TIP120 can specifically bind to TBP in solution (64), and deletion analysis demonstrated a correlation between the ability of TIP120 to bind TBP and TIP120-mediated stimulation (Fig. 3). NC2 is a good contrast to TIP120, because NC2 also binds to TBP but represses the basal transcription by RNAP II and III (62) and perhaps RNAP I (56a). However, even though TIP120 can bind to TBP in solution, no evidence was obtained for the stable binding of TIP120 to TBP-DNA or to the completed PIC. This suggests an unstable or transient association of TIP120 with DNA-bound TBP and higher-order complexes during PIC assembly.
Additionally, since several polypeptides (RPB5, RPB6, RPB8, and RPB10α) are shared by all three classes of RNA polymerases (47, 49), we expected that such a common RNA polymerase subunit(s) would be another target for TIP120. In this study, we presented evidence for specific interaction of TIP120 with RPB5 (Fig. 8). RPB5 was found to interact with human hepatitis B virus X protein to modulate the function of RNA polymerase (5). Recent work also demonstrated RPB5 plays a role in activated transcription (38). The protein-protein interaction between TIP120 and RPB5 can explain the results in Fig. 6. We also found that TIP120 could stimulate enzymatic activity of RNAP I and III (our unpublished observations). TIP120 may modify RNAP II function by inducing a conformational change in RNAP II through interaction with RPB5 subunit. Eventually, such a mechanism is likely to stimulate the entry of RNAP II to PIC. Alternatively, TIP120 could also bind to DNA and function as a mediator between RNAP II and DNA.
We propose a model in which TIP120 transiently interacts with both TBP (or TBP-TFIIB-DNA) and RNAP II to facilitate specific integration of RNAP II into the PIC. TIP120 may dissociate from the PIC soon after RNA polymerases are incorporated at the specific region. Hence, TIP120 might exhibit some chaperone-like activity toward the RNA polymerases. Consequently, TIP120 can be categorized as a novel type of transcriptional modulator that communicates with RNA polymerase and general transcription factor TBP.
Relevance of TIP120 to in vivo gene regulation.
Since the transcriptional stimulation function of TIP120 was initially found in the in vitro reconstituted transcription systems, the question arises as to whether TIP120 works in gene regulation in the cell. That TIP120 plays a role in vivo is suggested by the following findings. First, TIP120 was effective in an RNAP II reaction system containing the more physiological TATA-binding factor TFIID rather than the derived TBP (Fig. 2A). Second, TIP120 copurified with TFIIIC (data not shown), which as described above also contains the general coactivators PC4 and Topo I (60). Third, TIP120 can directly bind to TBP (64) and a particular subunit of RNA polymerases (Fig. 8). Both TBP and TIP120 were found in the same nuclear protein complex (64). Fourth, the TIP120-mediated transcriptional stimulation by all three RNA polymerases was demonstrated in cotransfection experiments (Fig. 9).
When cells are exposed to various inducible agents that elicit proliferation and differentiation, a number of genes must be regulated simultaneously at the transcription step. Moreover, growth stimuli are thought to influence transcriptional regulation for multiple sets of genes that include not only class II genes but also class I and III genes. Neither conventional gene-specific regulators nor GTFs can account for the occurrence of such total gene regulation in a cell. We thus hypothesize the existence of a global activator which allows amplification of the expression of multiple genes, and TIP120 appears to serve as such a regulator. If this is the case, analysis of TIP120 will provide new insight into global gene expression.
ACKNOWLEDGMENTS
Y.M. and S.Y. contributed equally to this study.
We thank R. C. Conaway, D. Reinberg, and H. Handa for their generous gifts of cDNA clones of GTFs. We also thank H. Handa and T. Fukasawa for helpful discussions. This work was supported in part by grants from the program Grants-in-Aid for Scientific Research on Priority Areas of The Japanese Ministry of Education, Science, Sports, and Culture.
REFERENCES
- 1.Aso T, Conaway J W, Conaway R C. Role of core promoter structure in assembly of the RNA polymerase II preinitiation complex. A common pathway for formation of preinitiation intermediates at many TATA and TATA-less promoters. J Biol Chem. 1994;269:26575–26583. [PubMed] [Google Scholar]
- 2.Bengal E, Flores O, Krauskopf A, Reinberg D, Aloni Y. Role of the mammalian transcription factors IIF, IIS, and IIX during elongation by RNA polymerase II. Mol Cell Biol. 1991;11:1195–1206. doi: 10.1128/mcb.11.3.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buratowski S, Hahn S, Guarente L, Sharp P A. Five intermediate complexes in transcription initiation by RNA polymerase II. Cell. 1989;56:549–561. doi: 10.1016/0092-8674(89)90578-3. [DOI] [PubMed] [Google Scholar]
- 4.Burley S K, Roeder R G. Biochemistry and structural biology of transcription factor IID (TFIID) Annu Rev Biochem. 1996;65:769–799. doi: 10.1146/annurev.bi.65.070196.004005. [DOI] [PubMed] [Google Scholar]
- 5.Cheong J, Yi M, Lin Y, Murakami S. Human RPB5, a subunit shared by eukaryotic nuclear RNA polymerases, binds human hepatitis B virus X protein and may play a role in X transactivation. EMBO J. 1995;14:143–150. doi: 10.1002/j.1460-2075.1995.tb06984.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chiang C M, Ge H, Wang Z, Hoffmann A, Roeder R G. Unique TATA-binding protein-containing complexes and cofactors involved in transcription by RNA polymerase II and III. EMBO J. 1993;12:2749–2762. doi: 10.1002/j.1460-2075.1993.tb05936.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Comai L, Zomerdijk J C, Beckmann H, Zhou S, Admon A, Tjian R. Reconstitution of transcription factor SL1: exclusive binding of TBP by SL1 or TFIID subunits. Science. 1995;266:1966–1972. doi: 10.1126/science.7801123. [DOI] [PubMed] [Google Scholar]
- 8.Conaway R C, Conaway J W. General initiation factors for RNA polymerase II. Annu Rev Biochem. 1993;62:161–190. doi: 10.1146/annurev.bi.62.070193.001113. [DOI] [PubMed] [Google Scholar]
- 9.Dynlacht B D, Hoey T, Tjian R. Isolation of coactivators associated with the TATA-binding protein that mediate transcriptional activation. Cell. 1991;55:563–576. doi: 10.1016/0092-8674(81)90019-2. [DOI] [PubMed] [Google Scholar]
- 10.Dynlacht B D, Weinzierl R O J, Admon A, Tjian R. The dTAFII80 subunit of Drosophila TFIID contains β-transducin repeats. Nature. 1993;363:176–179. doi: 10.1038/363176a0. [DOI] [PubMed] [Google Scholar]
- 11.Eisenmann D M, Dollard C, Winston F. SPT15, the gene encoding the yeast TATA binding factor TFIID, is required for normal transcription initiation in vivo. Cell. 1989;58:1183–1191. doi: 10.1016/0092-8674(89)90516-3. [DOI] [PubMed] [Google Scholar]
- 12.Flores O, Maldonado E, Reinberg D. Factors involved in specific transcription by mammalian RNA polymerase II. Factors IIE and IIF independently interact with RNA polymerase II. J Biol Chem. 1989;264:8913–8921. [PubMed] [Google Scholar]
- 13.Ge H, Roeder R G. Purification, cloning, and characterization of a human coactivator, PC4, that mediates transcriptional activation of class II genes. Cell. 1994;78:513–523. doi: 10.1016/0092-8674(94)90428-6. [DOI] [PubMed] [Google Scholar]
- 14.Grummt I, Roth E, Paule M R. Ribosomal RNA transcription in vitro is species specific. Nature. 1982;296:173–174. doi: 10.1038/296173a0. [DOI] [PubMed] [Google Scholar]
- 15.Ha I, Lane W, Reinberg D. Cloning a human gene encoding the general transcription initiation factor TFIIB. Nature. 1991;352:689–695. doi: 10.1038/352689a0. [DOI] [PubMed] [Google Scholar]
- 16.Hahn S, Buratowski S, Sharp P A, Guarente L. Isolation of the gene encoding the yeast TATA binding protein TFIID: a gene identical to the SPT15 suppressor of Ty element insertions. Cell. 1989;58:1173–1181. doi: 10.1016/0092-8674(89)90515-1. [DOI] [PubMed] [Google Scholar]
- 17.Hawley D, Roeder R G. Functional steps in transcription initiation and reinitiation from the major late promoter in a HeLa nuclear extract. J Biol Chem. 1987;262:3452–3461. [PubMed] [Google Scholar]
- 18.Hernandez N. TBP, a universal eukaryotic transcription factor? Genes Dev. 1993;7:1291–1308. doi: 10.1101/gad.7.7b.1291. [DOI] [PubMed] [Google Scholar]
- 19.Hodo H G, Blatti S P. Purification using polyethylenimine precipitation and low molecular weight subunit analyses of calf thymus and wheat germ DNA-dependent RNA polymerase II. Biochemistry. 1977;16:2334–2343. doi: 10.1021/bi00630a005. [DOI] [PubMed] [Google Scholar]
- 20.Hoffmann A, Roeder R G. Purification of his-tagged proteins in non-denaturing conditions suggests a convenient method for protein interaction studies. Nucleic Acids Res. 1991;19:6337–6338. doi: 10.1093/nar/19.22.6337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Horikoshi M, Wang C K, Fujii H, Cromlish J A, Weil P A, Roeder R G. Cloning and structure of a yeast gene encoding a general transcription initiation factor TFIID that binds to the TATA box. Nature. 1989;341:299–303. doi: 10.1038/341299a0. [DOI] [PubMed] [Google Scholar]
- 22.Kaiser K, Meisterernst M. The human general co-factor. Trends Biochem Sci. 1996;21:342–345. [PubMed] [Google Scholar]
- 23.Kato H, Nagamine M, Kominami R, Muramatsu M. Formation of the transcription initiation complex on mammalian rDNA. Mol Cell Biol. 1986;6:3418–3427. doi: 10.1128/mcb.6.10.3418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kato K, Makino Y, Kishimoto T, Yamauchi J, Kato S, Muramatsu M, Tamura T. Multimerization of the mouse TATA binding protein (TBP) driven by its C-terminal conserved domain. Nucleic Acids Res. 1994;22:1179–1185. doi: 10.1093/nar/22.7.1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Killeen M T, Greenblatt J F. The general transcription factor RAP30 binds to RNA polymerase II and prevents it from binding nonspecifically to DNA. Mol Cell Biol. 1992;12:30–37. doi: 10.1128/mcb.12.1.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim Y-J, Bjorklund S, Li Y, Sayre M H, Kornberg R D. A multiprotein mediator of transcriptional activation and its interaction with the C-terminal repeat domain of RNA polymerase II. Cell. 1994;77:599–608. doi: 10.1016/0092-8674(94)90221-6. [DOI] [PubMed] [Google Scholar]
- 27.Kimura M, Ishiguro A, Ishihama A. RNA polymerase II subunits 2, 3, and 11 form a core subassembly with DNA binding activity. J Biol Chem. 1997;272:25851–25855. doi: 10.1074/jbc.272.41.25851. [DOI] [PubMed] [Google Scholar]
- 28.Kokubo T, Gong D-W, Yamashita S, Takada R, Roeder R G, Horikoshi M, Nakatani Y. Molecular cloning, expression, and characterization of the Drosophila 85-kilodalton TFIID subunit. Mol Cell Biol. 1993;13:7859–7863. doi: 10.1128/mcb.13.12.7859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Koleske A, Young R A. An RNA polymerase II holoenzyme responsive to activators. Nature. 1994;368:466–469. doi: 10.1038/368466a0. [DOI] [PubMed] [Google Scholar]
- 30.Kretzschmar M, Kaiser K, Lottspeich F, Meisterems M. A novel mediator of class II gene transcription with homology to viral immediate early transcriptional regulators. Cell. 1994;78:525–534. doi: 10.1016/0092-8674(94)90429-4. [DOI] [PubMed] [Google Scholar]
- 31.Kretzschmar M, Meisteremst M, Roeder R G. Identification of human DNA topoisomerase I as a cofactor for activator-dependent transcription by RNA polymerase II. Proc Natl Acad Sci USA. 1994;90:11508–11512. doi: 10.1073/pnas.90.24.11508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lu H, Zawel L, Fisher L, Egly J-M, Reinberg D. Human general transcription factor IIH phosphorylates the C-terminal domain of RNA polymerase II. Nature. 1992;358:641–645. doi: 10.1038/358641a0. [DOI] [PubMed] [Google Scholar]
- 33.Ma D, Watanabe H, Mermelstein F, Admon A, Oguri K, Sun X, Wada T, Imai T, Shiroya T, Reinberg D, Handa H. Isolation of a cDNA encoding the largest subunit of TFIIA reveals functions important for activated transcription. Genes Dev. 1993;7:2246–2257. doi: 10.1101/gad.7.11.2246. [DOI] [PubMed] [Google Scholar]
- 34.Marinoni J-C, Roy R, Vermeulen W, Miniou P, Lutz Y, Weeda W, Seroz T, Gomez D M, Hoeijmakers J H J, Egly J-M. Cloning and characterization of p52, the fifth subunit of the core of the transcription/DNA repair factor TFIIH. EMBO J. 1997;16:1093–1102. doi: 10.1093/emboj/16.5.1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McBurney M, Rogers B J. Isolation of male embryonal carcinoma cells and their chromosome replication patterns. Dev Biol. 1982;89:503–508. doi: 10.1016/0012-1606(82)90338-4. [DOI] [PubMed] [Google Scholar]
- 36.Merino A, Madden K R, Lane W S, Champoux J J, Reinberg D. DNA topoisomerase I is involved in both repression and activation of transcription. Nature. 1993;365:227–232. doi: 10.1038/365227a0. [DOI] [PubMed] [Google Scholar]
- 37.Mishima Y, Yamamoto O, Kominami R, Muramatsu M. In vitro transcription of a mouse ribosomal RNA gene. Nucleic Acids Res. 1981;9:6773–6785. doi: 10.1093/nar/9.24.6773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miyao T, Woychik N A. RNA polymerase RPB5 plays a role in transcriptional activation. Proc Natl Acad Sci USA. 1998;95:15281–15286. doi: 10.1073/pnas.95.26.15281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Parvin J D, Sharp P A. DNA topology and a minimal set of basal factors for transcription by RNA polymerase II. Cell. 1993;73:533–540. doi: 10.1016/0092-8674(93)90140-l. [DOI] [PubMed] [Google Scholar]
- 40.Parvin J D, Shykind B M, Meyers R E, Kim J, Sharp P. Multiple sets of basal factors initiate transcription by RNA polymerase II. J Biol Chem. 1994;269:18414–18421. [PubMed] [Google Scholar]
- 41.Peterson M G, Inostroza J, Maxon M E, Flores O, Admon A, Reinberg D, Tjian R. Structure and function of the recombinant subunits of human TFIIE. Nature. 1991;354:369–373. doi: 10.1038/354369a0. [DOI] [PubMed] [Google Scholar]
- 42.Reines D, Conaway J W, Conaway R C. The RNA polymerase II general elongation factors. Trends Biochem Sci. 1996;21:351–355. [PMC free article] [PubMed] [Google Scholar]
- 43.Roeder R G. The complexities of eukaryotic transcription initiation: regulation of preinitiation complex assembly. Trends Biochem Sci. 1991;16:402–408. doi: 10.1016/0968-0004(91)90164-q. [DOI] [PubMed] [Google Scholar]
- 44.Roeder R G. The role of general initiation factors in transcription by RNA polymerase II. Trends Biochem Sci. 1996;21:327–335. [PubMed] [Google Scholar]
- 45.Safrany G, Tanaka N, Kishimoto T, Ishikawa Y, Kato H, Kominami R, Muramatsu M. Structural determinant of the species-specific transcription of the mouse rRNA gene promoter. Mol Cell Biol. 1989;9:349–353. doi: 10.1128/mcb.9.1.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sawadogo M, Roeder R G. Factors involved in specific transcription by human RNA polymerase II: analysis by a rapid and quantitative in vitro assay. Proc Natl Acad Sci USA. 1985;82:4394–4398. doi: 10.1073/pnas.82.13.4394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sawadogo M, Sentenac A. RNA polymerase B (II) and general transcription factors. Annu Rev Biochem. 1990;59:711–754. doi: 10.1146/annurev.bi.59.070190.003431. [DOI] [PubMed] [Google Scholar]
- 48.Sekimizu K, Kobayashi N, Mizuno D, Natori S. Purification of a factor from Ehrlich ascites tumor cells specifically stimulating RNA polymerase II. Biochemistry. 1976;15:5064–5070. doi: 10.1021/bi00668a018. [DOI] [PubMed] [Google Scholar]
- 49.Shpakovski G V, Acker J, Wintzerith M, Lacroix J-F, Thuriaux P, Vigneron M. Four subunits that shared by the three classes of RNA polymerase are functional interchangeable between Homo sapiens and Saccharomyces cerevisiae. Mol Cell Biol. 1995;15:4702–4710. doi: 10.1128/mcb.15.9.4702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shykind B M, Kim J, Steward L, Champoux J J, Sharp P A. Topoisomerase I enhances TFIID-TFIIA complex assembly during activation of transcription. Genes Dev. 1997;11:397–407. doi: 10.1101/gad.11.3.397. [DOI] [PubMed] [Google Scholar]
- 51.Struhl K. Histone acetylation and transcriptional regulatory mechanisms. Genes Dev. 1998;12:599–606. doi: 10.1101/gad.12.5.599. [DOI] [PubMed] [Google Scholar]
- 52.Svejstrup J Q, Vichi P, Egly J-M. The multiple roles of transcription/repair factor TFIIH. Trends Biochem Sci. 1996;21:346–350. [PubMed] [Google Scholar]
- 53.Taggart A K, Fisher T S, Pugh B F. The TATA-binding protein and associated factors are components of pol III transcription factor TFIIIB. Cell. 1992;71:1015–1028. doi: 10.1016/0092-8674(92)90396-t. [DOI] [PubMed] [Google Scholar]
- 54.Tamura T, Sumita K, Hirose S, Mikoshiba K. Core promoter of the mouse myelin basic protein gene governs brain-specific transcription in vitro. EMBO J. 1990;9:3101–3108. doi: 10.1002/j.1460-2075.1990.tb07507.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tyree C M, George C P, Lira-DeVito L M, Wampler S L, Dahmus M E, Zawel L, Kadonaga J. Identification of a minimal set of proteins that is sufficient for accurate initiation of transcription by RNA polymerase II. Genes Dev. 1993;7:1254–1265. doi: 10.1101/gad.7.7a.1254. [DOI] [PubMed] [Google Scholar]
- 56.Wada T, Takagi T, Yamaguchi Y, Ferdous A, Imai T, Hirose S, Sugimoto S, Yano K, Hartzog G A, Winston F, Buratowski S, Handa H. DSIF, a novel transcription elongation factor that regulates RNA polymerase II processivity, is composed of human Spt4 and Spt5 homologs. Genes Dev. 1998;12:343–356. doi: 10.1101/gad.12.3.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56a.Wang, Z. Unpublished observation.
- 57.Wang Z, Roeder R G. Structure and function of a human transcription factor TFIIIB subunit that is evolutionarily conserved and contains both TFIIB- and high-mobility-group protein 2-related domains. Proc Natl Acad Sci USA. 1995;92:7026–7030. doi: 10.1073/pnas.92.15.7026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang Z, Roeder R G. TFIIIC acts through a downstream region to stabilize TFIIIC2 binding to RNA polymerase III promoters. Mol Cell Biol. 1996;16:6841–6850. doi: 10.1128/mcb.16.12.6841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang Z, Luo T, Roeder R G. Identification of an autonomously initiating RNA polymerase III holoenzyme containing a novel factor that is selectively inactiated during protein synthesis inhibition. Genes Dev. 1997;11:2371–2382. doi: 10.1101/gad.11.18.2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang Z, Roeder R G. DNA topoisomerase I and PC4 can interact with human TFIIIC to promote both accurate termination and transcription reinitiation by RNA polymerase III. Mol Cell. 1998;1:749–757. doi: 10.1016/s1097-2765(00)80074-x. [DOI] [PubMed] [Google Scholar]
- 61.Wasylyk B, Wasylyk C, Augereau P, Chambon P. The SV40 72 bp repeat preferentially potentiates transcription starting from proximal natural or substitute promoter elements. Cell. 1983;32:503–514. doi: 10.1016/0092-8674(83)90470-1. [DOI] [PubMed] [Google Scholar]
- 62.White R J, Khoo B C, Inostroza J A, Reinberg D, Jackson S P. Differential regulation of RNA polymerases I, II, and III by the TBP-binding repressor Dr1. Science. 1994;265:448–450. doi: 10.1126/science.7939686. [DOI] [PubMed] [Google Scholar]
- 63.Wilson C J, Chao D M, Imbalzano A N, Schnitzler G R, Kingston R E, Young R A. RNA polymerase II holoenzyme contains SWI/SNF regulators involved in chromatin remodeling. Cell. 1996;84:235–244. doi: 10.1016/s0092-8674(00)80978-2. [DOI] [PubMed] [Google Scholar]
- 63a.Yamamoto, K. Unpublished results.
- 64.Yogosawa S, Makino Y, Yoshida T, Kishimoto T, Muramatsu M, Tamura T. Molecular cloning of a novel 120-kDa TBP-interacting protein. Biochem Biophys Res Commun. 1996;229:612–617. doi: 10.1006/bbrc.1996.1852. [DOI] [PubMed] [Google Scholar]
- 65.Zawel L, Reinberg D. Advances in RNA polymerase II transcription. Curr Opin Cell Biol. 1992;4:488–495. doi: 10.1016/0955-0674(92)90016-6. [DOI] [PubMed] [Google Scholar]
- 66.Zawel L, Reinberg D. Initiation of transcription by RNA polymerase II: a multi-step process. Prog Nucleic Acid Res Mol Biol. 1993;44:67–108. doi: 10.1016/s0079-6603(08)60217-2. [DOI] [PubMed] [Google Scholar]