Abstract
B.1.351 is the SARS-CoV-2 variant most resistant to antibody neutralization. We demonstrate how the dose and number of immunizations influence protection. Nonhuman primates (NHP) received two doses of 30 or 100 μg of Moderna’s mRNA-1273 vaccine, a single immunization of 30 μg, or no vaccine. Two doses of 100 μg of mRNA-1273 induced reciprocal ID50 mean neutralizing antibody titers against live SARS-CoV-2 D614G and B.1.351 of 3,300 and 240, respectively. Higher neutralizing responses against B.1.617.2 were also observed after two doses compared to a single dose. Following challenge with B.1.351, there was ~4–5−log10 reduction of viral subgenomic RNA (sgRNA) and low to undetectable replication in bronchoalveolar lavages in the two-dose vaccine groups, with a 1−log10 reduction in nasal swabs (NS) in the 100 μg dose group. These data establish that a two-dose regimen of mRNA-1273 will be critical for providing upper and lower airway protection against major variants of concern.
INTRODUCTION
The emergence of SARS-CoV-2 variants of concern (VOC) that show reduced neutralization by sera from Wu-1 strain convalescent subjects or vaccinees1, 2, 3 has created uncertainty about the efficacy of current SARS-CoV-2 vaccines against VOC infection. To date, the most concerning variants contain combinations of mutations and deletions in the S receptor-binding domain (RBD) and N-terminal domain (NTD), respectively. Acquisition of amino acid substitutions in the S RBD- namely K417N, E484K, and N501Y—and in the NTD, such as L18F, D80A, D215G, and Δ242–244, is associated with increased transmissibility and reduction in neutralization sensitivity4, 5, 6, 7, 8, 9, 10, 11, 12. Variants containing these substitutions originally isolated in the United Kingdom (UK) (B.1.1.7, Alpha), Republic of South Africa (B.1.351, Beta), Brazil (P.1 lineage, Gamma), New York (B.1.526), and California (B.1.427/B.1.429), have shown varying reduction in neutralization by convalescent and vaccine serum, and are resistant to some monoclonal antibodies11, 13, 14, 15, 16, 17, 18, 19. Moreover, the B.1.617.2 (Delta) variant is now the most prevalent variant circulating globally and shows some resistance to neutralization by sera from vaccinated subjects20, 21.
Among these variants, B.1.351 contains the most mutations in the RBD and NTD subdomains22 and has been shown to have the largest fold-reduction in neutralizability by potent RBD-specific monoclonal antibodies, including LY-CoV55523, and convalescent serum from individuals infected with ancestral SARS-CoV-2 strains24, 25, 26. Additionally, it was reported that sera from mRNA-1273-immunized human and nonhuman primates (NHP) showed the greatest reduction of neutralization against B.1.351 compared to B.1.1.7, P.1, B.1.427/B.1.429, and B.1.1.7+E484K variants4, 5, 6, 7, 8, 9, 10, 11, 12, 27, 28, 29. In UK- or US-based clinical studies, NVX-CoV2373 (Novavax), AZD1222 (University of Oxford/AstraZeneca), and Ad26.COV2.S (Janssen/Johnson & Johnson) vaccines show between ~70 and 90% protection against the circulating D614G or B.1.1.7 variants8, 30, 31, 32, and vaccine efficacy against mild symptomatic COVID-19 caused by B.1.351 was up to 60% for Ad26.CoV232 and NVX-CoV237333 and ~10% for AZD1228, 30, 31, 32. A recent report showed BNT162b2, Pfizer’s mRNA vaccine, conferred ~75% protection against confirmed B.1.351 infection in Qatar34. While immunological assessments for all vaccine trials are underway and correlates of protection are not yet determined, these data highlight the potential impact that reduced neutralization capacity to B.1.351 may have on protection against mild symptomatic COVID-19 across various platforms. Though comparable to BNT162b2 in other settings, human efficacy trials with mRNA-1273 have not been conducted in regions where B.1.351 circulates as a dominant variant. 35.
Vaccine development for COVID-19 has benefitted from clinically translatable data from the NHP model36, 37, 38, 39, 40, 41, 42. As there have been no published studies on vaccine protection in NHP challenged with the B.1.351 variant, we evaluated the impact of the dose and number of immunizations with mRNA-1273 on immunogenicity and protection against B.1.351 challenge in NHP. Here, we show a correlation between high mRNA-1273 induced antibody responses and decreased viral replication following B.1.351 challenge to establish that a two-dose regimen of mRNA-1273 will be critical for providing upper and lower airway protection against major variants of concern.
RESULTS
Antibody responses following mRNA-1273 vaccination
In prior studies, vaccination of NHP with 10–100 μg of mRNA-1273 at weeks 0 and 4 conferred rapid and complete control of detectable viral replication in both the upper and lower airways following SARS-CoV-2 USA/Washington-1 (WA-1) challenge36, 41. In the current study, to assess the influence of dose and number of immunizations on immunogenicity and protection against B.1.351, NHP were immunized with 30 or 100 μg in the standard 0- and 4-week vaccine regimen (x2) or a single dose (x1) of 30 μg (Extended Data Fig. 1).
We first performed a temporal analysis of serum neutralizing antibody responses after single immunization or prime and boost with mRNA-1273. Consistent with a prior study41, there was a ~100-fold increase in D614G-specific lentiviral pseudovirus neutralizing antibodies following a boost with 30 or 100 μg of mRNA-1273 (Extended Data Fig. 2A). All 8 of the 100 μg x2 immunized NHP and 7 of 8 receiving 30 μg x2 had >103 reciprocal ID50 titers 7–8 weeks after boost, and 13/16 animals from both 2-dose regimens had detectable neutralizing activity against B.1.351 (Extended Data Figs. 2B). By contrast, only 6/8 NHP that received a single dose of 30 μg had detectable neutralizing responses against D614G at that same timepoint, and none (0/8) had detectable neutralizing antibodies against B.1.351 (Extended Data Figs. 2A,B).
Focusing immune analysis at the time of SARS-CoV2 B.1.351 challenge, ~8 weeks post-boost or 12 weeks after the single immunization, S-specific binding and neutralizing antibody responses were assessed. Using a 10-plex MULTI-ARRAY ELISA, we assessed WA-1 (Figs. 1A,E) and B.1.351 (Figs. 1D,H) S- and RBD- specific antibody binding responses, which represent the vaccine and challenge strains, respectively. B.1.351 S-specific (Figs. 1A,D) and RBD-specific (Figs. 1E,H) antibody responses were reduced less than 2–3 fold for each vaccine group as compared to WA-1 S. There was a vaccine dose-dependent increase in S- and RBD-specific antibody responses against WA-1 (Figs. 1A,E) and B.1.351 (Figs. 1D,H), B.1.1.7 (Figs. 1B,F) and P.1 (Figs. 1C,G).
Fig. 1. Serum antibody responses following mRNA-1273 immunization.
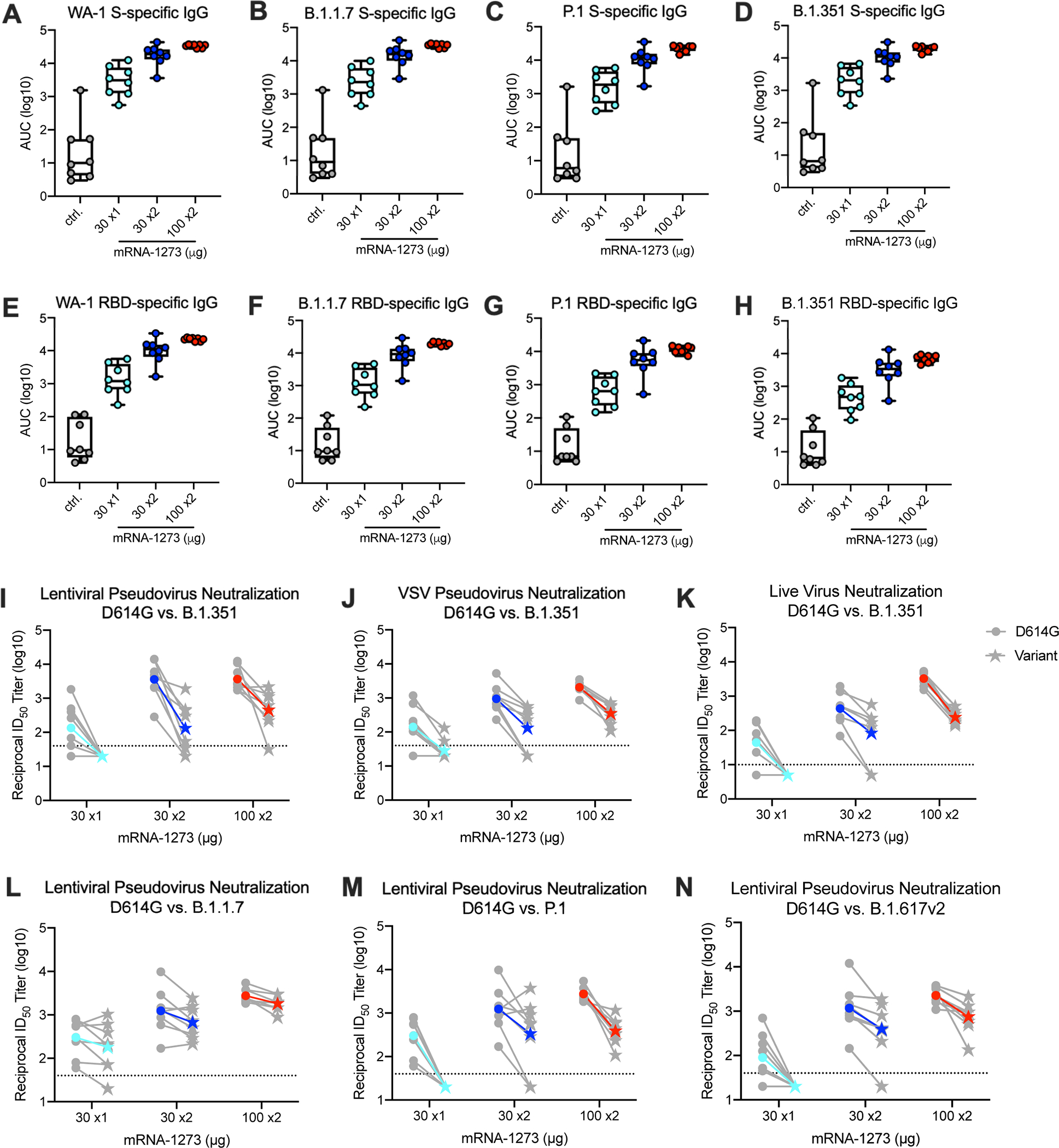
Twenty-four rhesus macaques were immunized with mRNA-1273 (30 μg, one dose – light blue; 30 μg, two doses – dark blue; or 100 μg, two doses – red), according to Extended Data Fig. 1. Eight aged-matched naïve NHP (gray) were used as controls. Sera collected at week 12, immediately before challenge, were assessed for SARS-CoV-2 USA/Washington1 (WA-1) (A, E), B.1.1.7 (B, F), P.1 (C, G), and B.1.351 (D, H) S- (A-D) and RBD-specific (E-D) IgG by MULTI-ARRAY ELISA. Lentiviral-based pseudovirus neutralization (I, L-N) was conducted on SARS-CoV-2 D614G and B.1.351 (I), B.1.1.7 (L), P.1 (M), and B.1.617.2 (N). D614G and B.1.351 VSV-based pseudovirus neutralization (J) and focus reduction live-virus neutralization (K) were also assessed. Data represents one independent experiment. (A-H) Circles represent individual NHP. Boxes and horizontal bars denote the IQR and medians, respectively; whisker end points are equal to the maximum and minimum values. (I-N) Gray lines represent individual NHP, and colored lines represent geometric mean titers (GMT). Dotted lines indicate neutralization assay limits of detection. Symbols represent individual NHP and may overlap for equal values. Data represents one independent experiment.
In vitro neutralizing activity was next determined using two pseudovirus neutralization assays and a live virus assay. Neutralizing titers against B.1.351 were compared to D614G, the benchmark strain. Using a D614G lentiviral-based pseudovirus neutralization assay, the reciprocal ID50 geometric mean titer (GMT) was ~3,600 following two doses of 100 μg (Fig. 1 I and Extended Data Fig. 2A). Consistent with the 8-fold reduction reported by us and others using human vaccine or convalescent serum3, 21, 29, 43, the reciprocal ID50 GMT against B.1.351 was ~450 (Fig. 1I and Extended Data Fig. 2B). Notably, in NHP that received a single 30 μg dose of mRNA, the reciprocal ID50 GMT against D614G was ~130, but there were no detectable neutralizing antibodies against B.1.351 (Fig. 1I). We observed similar outcomes comparing D614G to B.1.351 using VSV-based pseudovirus (Fig. 1J) and live virus (Fig. 1K) neutralization assays. To extend the analysis, there was little change in neutralization in any vaccine group comparing D614G to B.1.1.7 (Fig. 1L) but the reduction in neutralization against the P.1 variant (Fig.1M) was similar to that observed with B.1.351 (Fig. 1I). Last, there was an average 3-fold reduction in GMT (EC-50) in serum neutralizing activity against B.1.617.2 compared to D614G in animals that received two doses of 30 or 100 μg of mRNA-1273 and undetectable responses in the 30 μg single dose group. Nevertheless, reciprocal ID50 neutralizing responses were ~400 and ~750 in the 30 and 100 μg two dose groups respectively. Antibody binding and neutralization responses were highly correlated with one another (Extended Data Fig. 3).
To extend the antibody analyses to the mucosal sites of infection, S- and RBD-specific IgG and IgA in BAL and nasal wash samples were assessed at ~3 weeks post-boost or 7 weeks after the single 30 μg immunization. Consistent with systemic humoral responses, there was a dose-dependent increase in BAL and nasal wash WA-1 or B.1.351 S-specific IgG and IgA (Figs. 2A–H). BAL WA-1 (Fig. 2A) or B.1.351 (Fig. 2B). S-specific IgA responses in nasal wash trended higher only in the two-dose vaccine groups compared to control NHP (Figs. 2G,H). RBD-specific antibody responses followed similar trends as S-specific responses (Figs. 2I–P). Overall, mRNA-1273 vaccination elicits WA-1 and B.1.351 S-specific IgG and IgA antibodies in serum and lower and upper airways as previously shown41.
Fig. 2. Mucosal antibody responses following mRNA-1273 immunization.
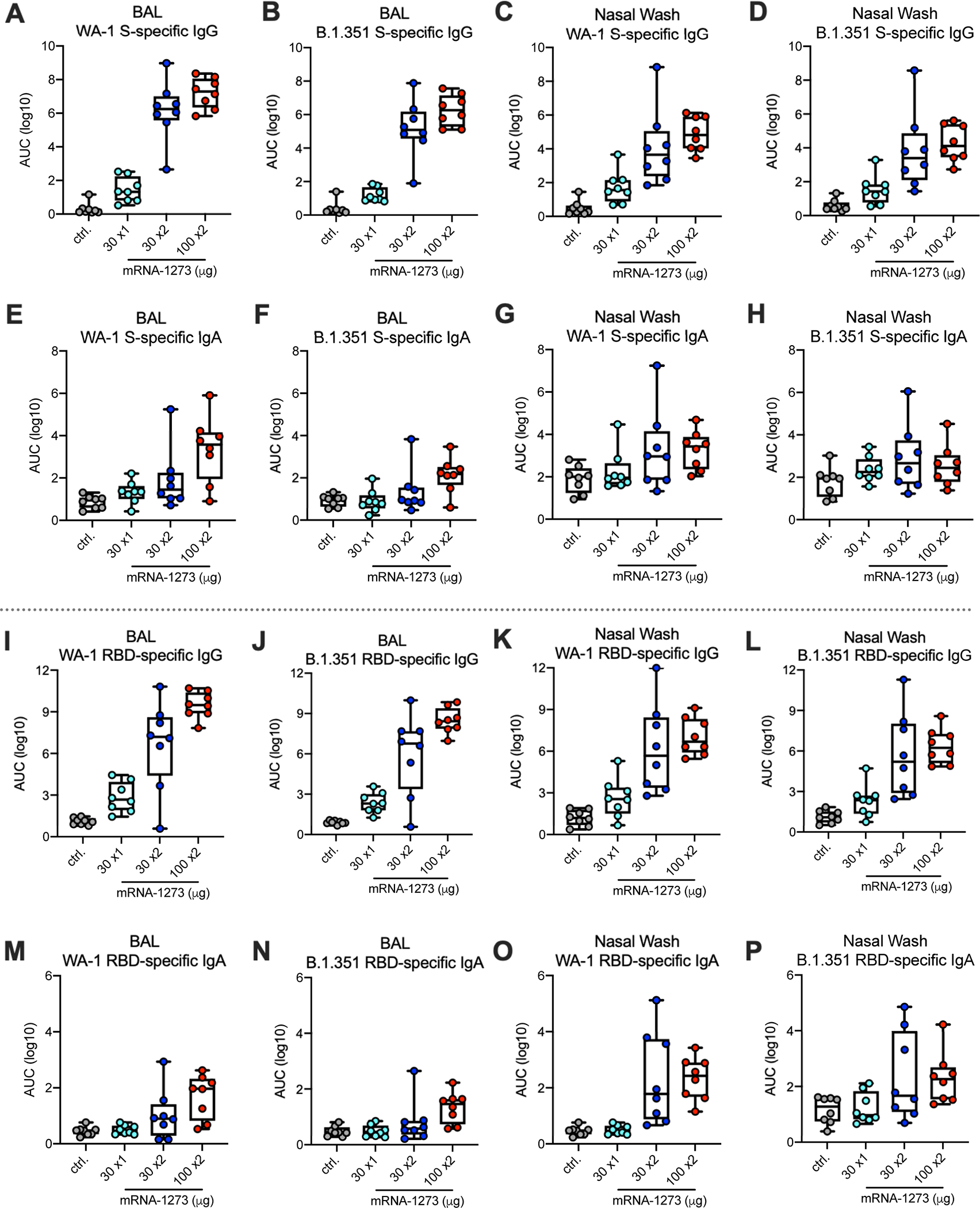
Twenty-four rhesus macaques were immunized according to Extended Data Fig. 1, and compared to eight age-matched controls. BAL (A-B, E-F, I-J, M-N) and nasal washes (C-D, G-H, K-L, O-P) collected at week 7 were assessed for SARS-CoV-2 WA-1 (A, C, E, G, I, K, M, O) and B.1.351 (B, D, F, H, J, L, N, P) S- (A-H) and RBD-specific (I-P) IgG (A-D, I-L) and IgA (E-H, M-P) by MULTI-ARRAY ELISA. Data represents one independent experiment. Circles represent individual NHP. Boxes and horizontal bars denote the IQR and medians, respectively; whisker end points are equal to the maximum and minimum values.
T cell responses following mRNA-1273 vaccination
mRNA-1273 induces Th1, CD4 T follicular helper (Tfh) responses and CD8 T cells in NHP and humans36, 44, 45. Consistent with these data, S-specific Th1 responses were induced in a dose-dependent manner with higher responses in the 100 μg dose group (Fig. 3A). There were low to undetectable Th2 responses in all vaccine groups (Fig. 3B). There was also dose-dependence in the frequency of S-specific Tfh responses expressing the surface marker CD40L (Fig. 3C) or the canonical cytokine IL-21 (Fig. 3D), which are critical for improving antibody responses. S-specific CD8 T cell responses were observed in 5/8 NHP that received two doses of 100 μg mRNA-1273 (Fig. 3E). These data show that mRNA-1273 induces Th1- and Tfh-skewed CD4 responses and CD8 T cells at the highest dose.
Fig. 3. T cell responses following mRNA-1273 immunization.
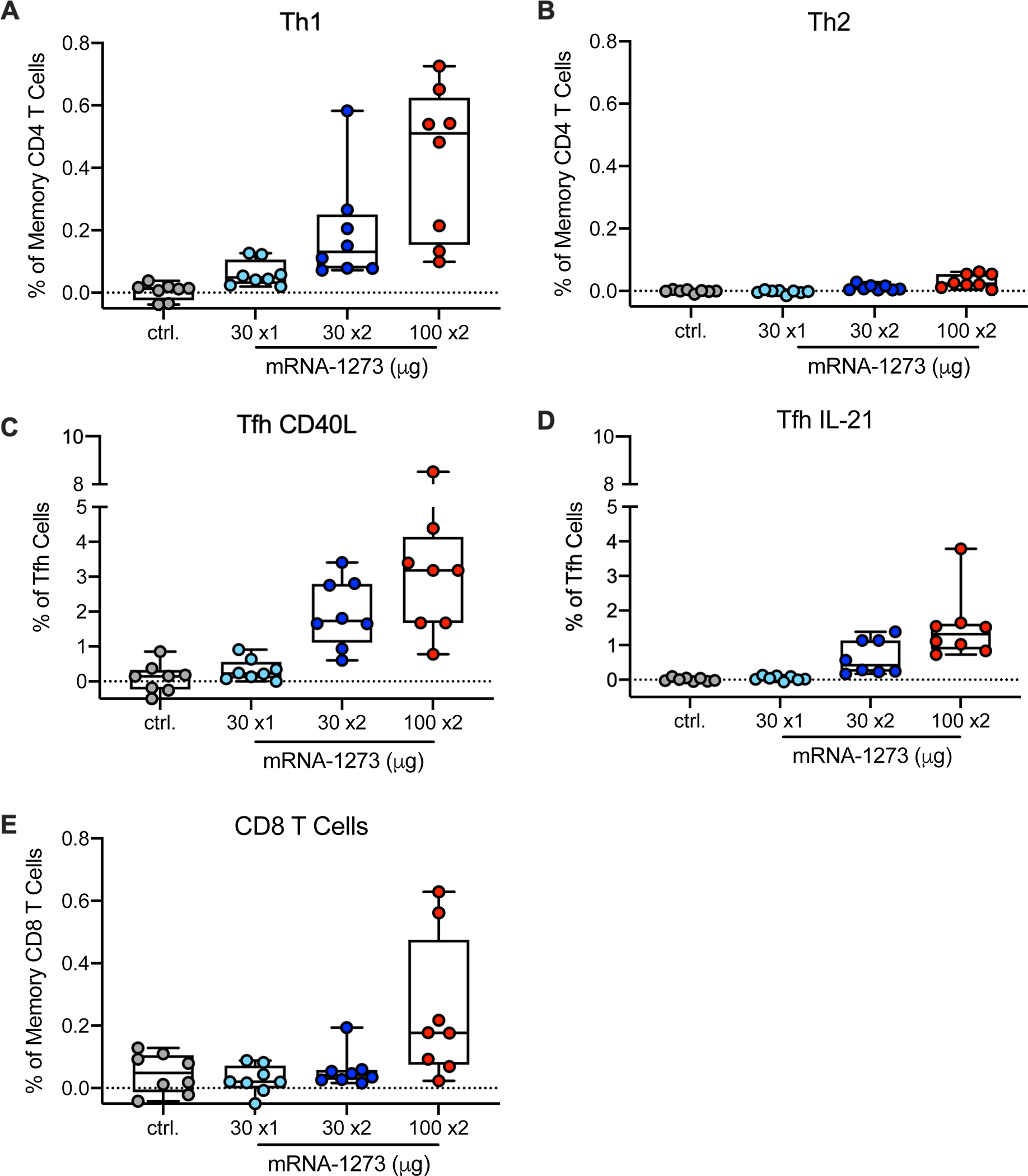
Twenty-four rhesus macaques were immunized according to Extended Data Fig. 1, and compared to eight age-matched controls. Intracellular staining was performed on PBMCs at week 7 to assess T cell responses to SARS-CoV-2 S protein peptide pools, S1 and S2. Responses to S1 and S2 individual peptide pools were summed. (A) Th1 responses (IFNg, IL-2, or TNF), (B) Th2 responses (IL-4 or IL-13), (C) Tfh CD40L upregulation (peripheral follicular helper T cells (Tfh) were gated on central memory CXCR5+PD-1+ICOS+ CD4 T cells), (D) Tfh IL-21, (E) CD8 T cells. Data represents one independent experiment. Boxes and horizontal bars denote IQR and medians, respectively; whisker end points are equal to the maximum and minimum values. Circles represent individual NHP. Dotted lines are set to 0%.
Protection against SARS-CoV-2 replication in the airways
An extensive analysis was performed to characterize the nucleotide sequence and in vivo pathogenicity. Deep sequencing was performed after each passage (P) of the B.1.351 strain and the challenge stock retained all mutations in the RBD and NTD sites on the spike protein (Extended Data Fig. 4A). The JHU B.1.351 P2 stock was then administered to Golden Syrian hamsters, a highly pathogenic SARS-CoV-2 animal model, at three different concentrations to characterize weight loss (Extended Data Fig. 4B) and in NHP to measure upper and lower airway viral replication by qRT-PCR for sgRNA (Extended Data Figs. 4C,D). Based on these data, a B.1.351 challenge dose of 5×105 PFU was selected for the vaccine study; this dose would induce sgRNA levels similar to the higher values obtained from nasal secretions of humans following SARS-CoV-2 infection46, 47.
To assess the protective efficacy of mRNA-1273 against the B.1.351 SARS-CoV-2 variant, the NHP were challenged with a total dose of 5×105 PFU of B.1.351 by intratracheal (IT) and intranasal (IN) routes 7–8 weeks post-boost for the two-dose regimens and 12 weeks after the single dose regimen. Two days post-challenge, only 2 of 8 NHP that received 100 μg x2 of mRNA-1273 had detectable SARS-CoV-2 envelope (E) sgRNA (sgRNA _E) in BAL compared to 8 of 8 in the control group (Fig. 4A). By qRT-PCR of sgRNA_E in BAL (Fig. 4A) or nucleocapsid (N) sgRNA (sgRNA _N) in BAL (Fig. 4C), the 100 μg x2 group had a significant decrease in viral load compared to NHP that received a single dose of 30 μg (p = 0.0054) or to the control NHP (p = 0.0009). NHP that received 30 μg x2 also had a significant decrease in viral load compared to control NHP (p = 0.0054) and showed a trend toward reduced viral replication compared to a single immunization with 30 μg (Figs. 4A,C). NHP that received a single vaccination with 30 μg showed a trend toward reduced viral replication compared to control NHP (Figs. 4A,C). On day 4 post-challenge, the pattern was the same, with a significant reduction in viral replication in BAL for all vaccine groups compared to control NHP, and for the 100 μg group compared to 30 μg x1 (Figs. 4A,C). By day 7, while 7 of 8 control NHP still had ~4 logs of sgRNA_E, there was no detectable sgRNA_E in 6 of 8 NHP in all vaccine groups (Fig. 4A), consistent with control of viral replication in the lower airway. To extend the analysis, we assessed viral titers in BAL and NS following challenge (Figs. 4E,H). There was low to undetectable virus cultured from BAL in the majority of NHP immunized with 30 and 100 μg x2 two days post-challenge (Fig. 4E). Moreover, on day 2 post-challenge, BAL viral titers and sgRNA were highly correlated (Figs. 4F,G), where no virus was culturable from BAL of all NHP with BAL sgRNA_N <1.2×104 RNA copies/mL (Fig. 4G).
Fig. 4. Efficacy of mRNA-1273 against upper and lower respiratory B.1.351 viral replication.
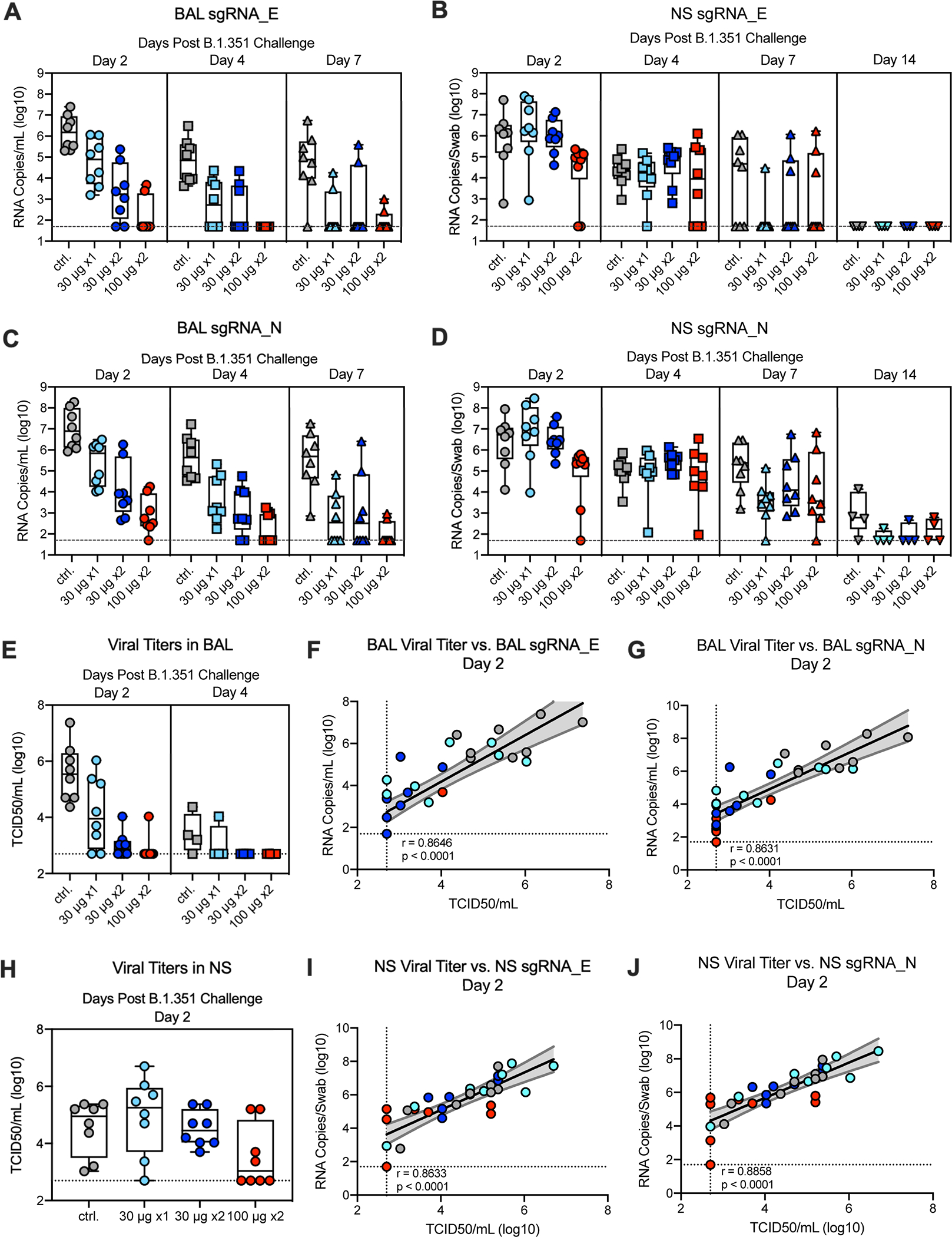
Twenty-four rhesus macaques were immunized and challenged as described in Extended Data Fig. 1, and compared to eight age-matched controls. BAL (A, C) and nasal swabs (NS) (B, D) were collected on days 2 (circles), 4 (squares), and 7 (triangles), and 14 (inverted triangles) post-challenge, where applicable, and viral replication was assessed by detection of SARS-CoV-2 E- (A-B) and N-specific (C-D) sgRNA. Viral titers were assessed by TCID50 assay for BAL collected on days 2 and 4 post-challenge (E) and for NS on day 2 (H). Boxes and horizontal bars denote the IQR and medians, respectively; whisker end points are equal to the maximum and minimum values. (F-G, I-J) Plots show correlations between viral titers and sgRNA_E (F, I) and sgRNA_N (G, J) in BAL (F-G) and NS (I-J) 2 days post-challenge. Data represents one independent experiment. Black and gray lines indicate linear regression and 95% confidence interval, respectively. ‘r’ and ‘p’ represent Spearman’s correlation coefficients and corresponding two-sided p-values, respectively; all p-values were <0.0001. Symbols represent individual NHP and may overlap for equal values.
In contrast to the significant reduction of viral replication in the lower airway, at day 2 post-challenge, the only significant reduction in viral replication for sgRNA_E (Fig. 4B) or sgRNA _N (Fig. 4D) in NS was in the 100 μg dose group. The differences in NHP that received 100 μg x2 were marginally lower compared to the groups that received 30 μg x2 or 30 μg x1 (p = 0.0273 and 0.0350, respectively) with no other significant pairwise differences (Figs. 4B,D). The groups were not significantly different at the later time points. A notable finding was that 5 of 8 control NHP still had ~4 log10 of sgRNA_E and all 8 NHP had sgRNA_N at day 7 in the NS (Figs. 4B, D), highlighting persistence of sgRNA following B.1.351 through day 7 post-challenge. With that, there was ~4 log10 of sgRNA_E at day 7 in the NS of 3 of 8 NHP in the 100 μg x2 group (Fig. 4B). Of note, 6 of 8 animals had low to undectable detection of virus in NS on day 2 post-challenge in the 100 μg x 2 group (Fig. 4H). NS viral titers and sgRNA were highly correlated (Figs. 4I,J). Overall, these data show significant reduction and rapid control of B.1.351 viral replication in the lower airways following vaccination with mRNA-1273 with more limited control in the upper airway and only in the 100 μg x 2 dose group.
Inflammation and viral load in lung tissue post-challenge
To provide a further assessment of protection following vaccination, NHP in each of the dose groups were assessed for virus-related pathology and the detection of viral antigen (VAg) in the lung 8 days post B.1.351 challenge. The severity of inflammation, which ranged from minimal to moderate, was similar across lung samples from NHP that received the vaccine in doses of 100 μg x2, 30 μg x2, or 30 μg x1 (Fig. 5). The inflammatory lesions in the lung were characterized by a mixture of lymphocytes, histiocytes, and fewer polymorphonuclear cells associated with variably expanded alveolar capillaries, occasional areas of perivascular inflammation, and Type II pneumocyte hyperplasia. Two out of four NHP that received 30 μg x1 of mRNA-1273 vaccine had trace amounts of virus detected in the lung. There was no detection of VAg in any lung sample from NHP that received two doses of 30 or 100 μg. All 4 NHP in the control group had variable amounts of VAg detected in the lung (Fig. 5 and Supplementary Table 1). These data corroborate viral replication data suggesting two doses of mRNA-1273 elicits substantial protection against lower airway disease following B.1.351 challenge.
Fig. 5. Post-challenge lung histopathological analysis and viral detection.
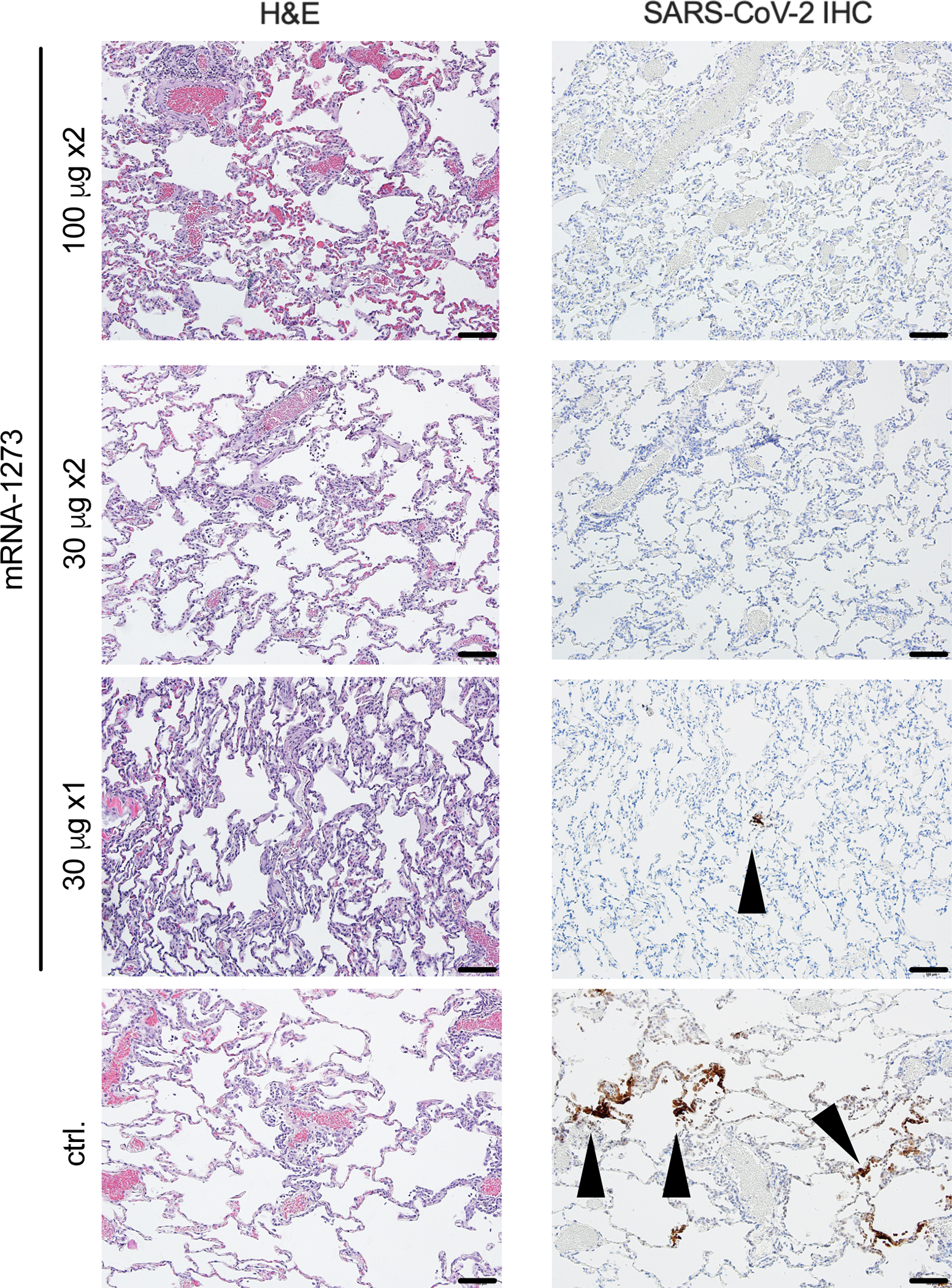
Rhesus macaques were immunized and challenged as described in Figure S1. Eight days post-challenge, lung samples (n = 4/group) were evaluated for the presence of inflammation by hematoxylin and eosin (H & E) staining (left) and evidence of virus infection by immunohistochemistry (IHC) for SARS-CoV-2 viral antigen (right). Representative images show the location and distribution of SARS-CoV-2 viral antigen in serial lung tissue sections. Arrows indicate areas positive for viral antigen. Each image is taken at 10x magnification; scale bars represent 100 microns.
Post-challenge antibody and T cell responses in the airways
The assessment of antibody responses post-challenge has been useful for determining whether viral replication in the BAL or NS is sufficient to boost vaccine-induced anamnestic S-specific antibody responses in these mucosal tissues41, 48. In BAL, WA-1 and B.1.351 S-specific IgG (Extended Data Figs. 5A,B) or IgA (Extended Data Figs. 5C,D) responses did not increase post-challenge in NHP that received two immunizations of 30 or 100 μg of mRNA-1273. However, by 14 days post-challenge, there was an increase in WA-1 and B.1.351 S-specific IgG responses in NHP that received 30 μg x1 of mRNA-1273, to titers that were similar to the NHP that received 30 or 100 μg x2 and higher than the unvaccinated controls (Extended Data Figs. 5A,B). In NS, there was an increase in WA-1 and B.1.351 S-specific IgG (Extended Data Figs. 5E,F) or IgA (Extended Data Figs. 5G,H) responses in unvaccinated NHP and those immunized with 30 μg of mRNA-1273 once or twice; however, there were no anamnestic S-specific antibody responses in 100 μg dose group. Overall, these data show that the increase in S-specific anamnestic antibody responses in both the BAL and NS are associated with viral replication in these mucosal sites and may explain the relatively rapid clearance of virus from NS in the 30 μg x1 dose group seen from day 4 to 7 (Figs. 4B,D). We also assessed anamnestic S-specific T cell responses in BAL following challenge. CD8 T cell responses, and not CD4 T cells, were increased post-challenge in BAL in 30 μg x1 mRNA-1273 immunized NHP and control NHP (Extended Data Fig. 6). These data are consistent with their increased viral load in BAL compared to the two-dose vaccine groups.
Antibody correlates of protection
Assessing immune correlates of protection following vaccination is a critical aspect of vaccine development. It was reported that mRNA-1273 induced antibody responses are a mechanistic correlate for reducing viral replication against WA-1 challenge in NHP41. Here, B.1.351 S-specific IgG antibody titers at week 12, the time of challenge, also correlated strongly with reduction of sgRNA in both BAL (Fig. 6A) and NS (Fig. 6D) at day 2 post-challenge. In addition, both pseudovirus and live viral neutralization correlate significantly with reduction of sgRNA in both BAL (Figs. 6B,C) and NS (Figs. 6E,F).
Fig. 6. Antibody correlates of protection.
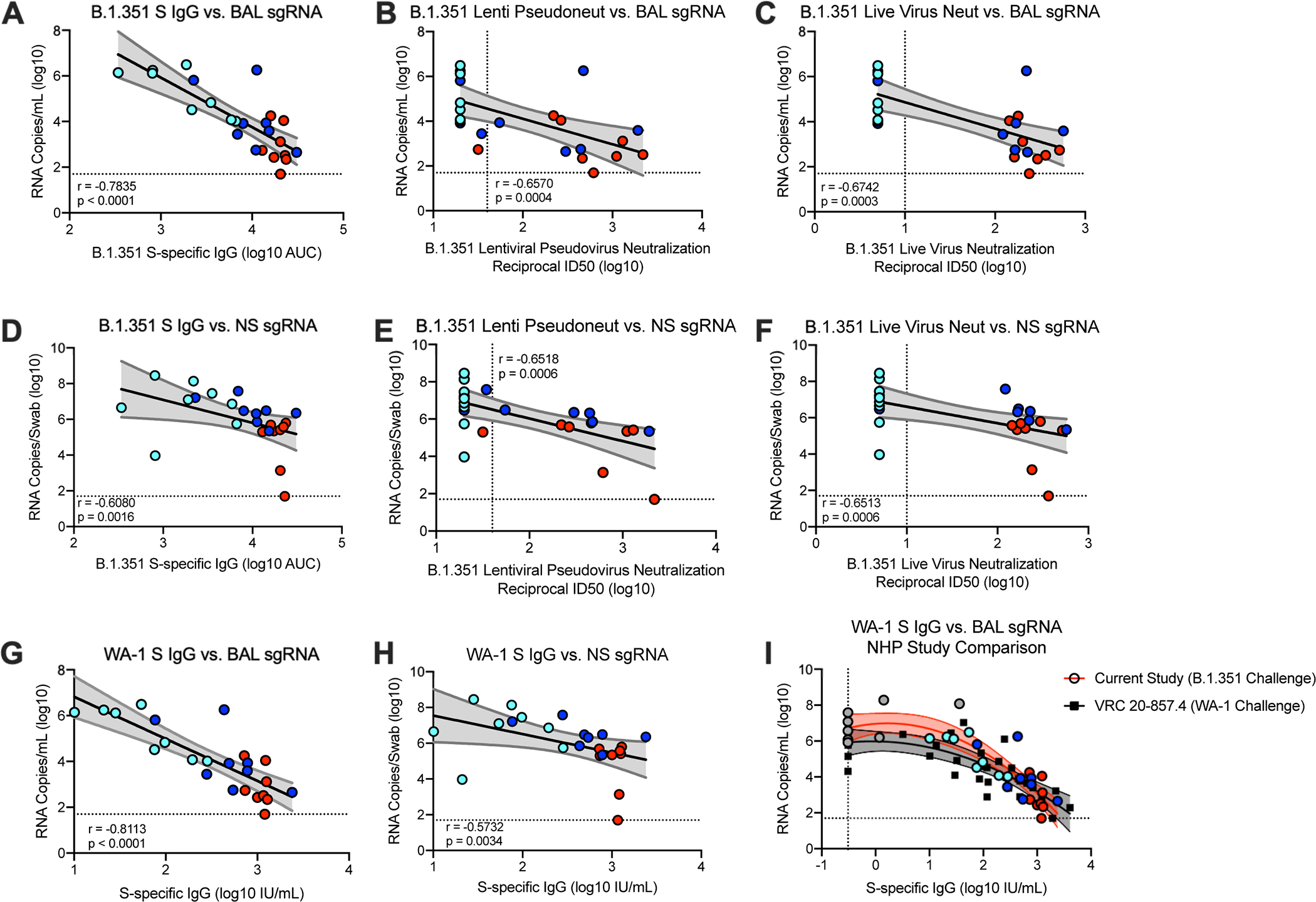
Twenty-four rhesus macaques were immunized and challenged as described in Extended Data Fig. 1, and compared to eight age-matched controls. (A-F) Plots show correlations and the corresponding two-sided p-values between week 12 SARS-CoV-2 B.1.351 S-specific IgG (A, D), lentiviral-based pseudovirus neutralization (B, E), and focus reduction live-virus neutralization (C, F) with N-specific sgRNA in BAL (A-C) and NS (D-F) at day 2 post-challenge. (G-H) Plots show correlations and the corresponding two-sided p-values between week 12 SARS-CoV-2 WA-1 S-specific IgG, converted to IU/mL, with N-specific sgRNA in BAL (G) and NS (H) at day 2 post-challenge. Circles represent individual NHP, where colors indicate mRNA-1273 dose. Dotted lines indicate assay limits of detection. (I) The relationship between pre-challenge WA-1 S-specific IgG and day 2 BAL sgRNA_N, with data from the current study using a B.1.351 challenge (filled circles, red curve fit) superimposed on data from our previous study with a WA-1 challenge (black squares, black curve fit); lines indicate quadratic curve fit and 95% confidence intervals. Data represents one independent experiment.
Symbols represent individual NHP and may overlap for equal values.
In a recent report on correlates of protection following mRNA-1273 immunization in NHP, we established a linear relationship between WA-1 S-specific antibody titers as defined by international units (IU) and subsequent sgRNA 2 days after SARS-CoV-2 WA-1 challenge. Here, we similarly converted pre-challenge WA-1 S-specific antibody titers to IU (Supplementary Table 2). Consistent with our prior NHP study, pre-challenge WA-1 S-specific IgG titers and sgRNA in BAL (Fig. 6G) and NS (Fig. 6H) at day 2 post-challenge were negatively correlated. Based on a linear model, vaccinated animals that had S-specific IgG of ~100 and 120 IU/mL were predicted to have sgRNA_N and sgRNA_E, respectively, in BAL of approximately 2 log10 lower than the average for PBS controls. Serum neutralizing activity was associated with control of viral replication; 13/14 of the NHP with detectable pseudovirus neutralizing activity against B.1.351 had BAL sgRNA_N <105 (Fig. 6B). A similar linear relationship between S-specific antibody titers and viral replication in nasal swabs was apparent (Fig. 6E), although few of the NHP had NS sgRNA_N below 105.
As the antibody titers of S-specific IgG are very similar to those we had previously reported using NHP challenged with the SARS-CoV-2 WA-141, we conducted an additional exploratory analysis of the data from both studies to investigate the consistency of the relationship between S-specific antibodies and BAL sgRNA, across the two different viruses. For low S-specific IgG levels, sgRNA was higher for animals challenged with B.1.351, but for animals with titers of S-specific IgG greater than approximately 100 IU/mL, the estimated regression curve slopes are similar as are the levels that correspond to 2–4 log10 reductions in sgRNA in BAL compared to controls (Fig. 6I). These data suggest a similar relationship between S-specific IgG titers and lower airway protection against WA-1, which contains S protein that is homologous to the mRNA-1273 insert, and the B.1.351 variant SARS-CoV-2 strain in NHP.
DISCUSSION
mRNA-1273 and BNT162b2 vaccines had ~95% efficacy in clinical trials performed in the US, when WA-1 and D614G variants circulated most widely49, 50. A critical issue is whether these and other vaccines will mediate protection against rapidly emerging variants. The B.1.351 variant is one of greatest concern compared to WA-1, D614G, or B.1.1.7 based on the higher reduction in neutralization using vaccine sera4, 5, 6, 7, 8, 9, 10, 11, 12, 27, 28 and clinical trials showing lower efficacy against symptomatic infection32, 33, 35. However, recent data show the B.1.617.2 variant is now the predominant circulating strain worldwide, although is less resistant to neutralization than B.1.351. Here, we present evidence that vaccine dose and number of immunizations had a significant effect on protection against B.1.351 challenge. These data are consistent with a recent report showing vaccine effectiveness against PCR-confirmed infection with the B.1.351 variant is 16% after 1 dose of BNT162b2 and 75% after two doses34.
Antibodies play a critical role in mediating vaccine-elicited protection against SARS-CoV-2 in NHP models36, 40, 42, 48, 51. Here, we show there was a dose-dependent increase in WA-1 and B.1.351 S- and RBD-specific antibody titers and D614G and B.1.351 neutralization titers. Neutralizing activity against B.1.351 and B.1.617.2 were both significantly increased following the second immunization. These data highlight the importance of a two-dose regimen for optimizing neutralization antibody responses, particularly against B.1.351 and likely for other variants of concern with increased neutralization resistance. The frequency of S-specific Th1 and Tfh cell responses were also dose-dependent, and CD8 T cell responses detected only in NHP receiving the 100 μg dose. These data are consistent with prior studies by us and others showing mRNA is a vaccine platform that induces Tfh cells and robust binding and neutralizing antibodies 36, 41, 44, 52.
As this was one of the first studies to use the B.1.351 variant for challenge in NHP, extensive sequence analyses were performed to propagate a challenge stock with an S sequence matched to the reference isolate. Naïve NHP infected with the B.1.351 stock notably had peak sgRNA levels of ~107 copies/mL in BAL, which is higher than reported by us and others for challenge studies using the WA-1 strain36, 40, 42, 48, 51. At 7 days post-challenge, most of the control NHP still had ~105 RNA copies/mL and copies/swab present in BAL and NS, respectively. This contrasts with the more rapid and complete reduction of viral replication following challenge with the WA-1 strain36, 40, 42, 48, 51 and is higher than viral loads typically seen in human infections. Whether the amount and persistence of B.1.351 virus in vivo in this study relates to challenge dose or suggests that this variant has inherent properties that make it more difficult to control and clear compared to the WA-1 strain53, 54 is a focus of ongoing analyses. 36, 40, 42, 48, 51 and is higher than viral loads typically seen in human infections. Whether the amount and persistence of the B.1.351 virus in vivo in this study relates to the challenge dose or suggests that this variant has inherent properties that make it more difficult to control and clear compared to the WA-1 strain53, 54 is a focus of ongoing analyses.
Based on the high levels of sgRNA in BAL and NS, the dose of B.1.351 used here provided a stringent challenge for vaccine-elicited protection. We observed significant reduction in viral replication in BAL at day 2 post-challenge in groups that received 30 and 100 μg twice; 6 of 8 NHP that received 100 μg vaccine regimen had no detectable sgRNA_E in BAL. Of note, while there was no detectable serum neutralizing activity against B.1.351 virus with a single immunization of 30 μg, there was still significant reduction in viral replication in BAL by day 4 post-challenge compared to controls with limited inflammation or viral antigen in lungs at day 8. These data are consistent with our recent study in which NHP immunized with only 1 or 3 μg of mRNA-1273 twice demonstrated reduction in viral replication in BAL and limited lung pathology following WA-1 challenge despite absence of detectable neutralizing activity41. Potential mechanisms for lower airway protection in the absence of detectable serum neutralizing antibodies include Fc effector functions and anamnestic responses post challenge. It is also possible that T cells contribute to control of viral replication in lower airways; however, there is no current evidence for viral control by T cells following vaccination or primary infection55 in the SARS-CoV-2 NHP model.
In contrast to high-level or complete protection observed in lower airways against B.1.351 challenge for all doses, control of viral replication in upper airway was only observed in animals receiving 2 doses of 100 μg. These data are consistent with prior studies in NHP showing higher amounts of antibody are required to reduce viral replication in upper than lower airways following mRNA-1273 vaccination36, 41. The implications of these findings on transmission of B.1.351 in vaccinated people are unknown. Of note, recent results from human vaccine trials show that there is greater protection against severe disease than mild disease against the B.1.351 variant following immunization with Ad26.CoV232 or BNT162b234. Serum antibody titers were strong predictors of reduced viral load in BAL and NS. These data are consistent with studies by us and others showing antibodies induced by vaccination41 or infection55 of NHP protect against reinfection36, 41. The implications of these findings on the transmission of B.1.351 in vaccinated people are unknown. Of note, recent results from human vaccine trials show that there is greater protection against severe disease than mild disease against the B.1.351 variant following immunization with Ad26.CoV232 or BNT162b234. Serum antibody titers were strong predictors of reduced reduction of viral load in BAL and NS. These data are consistent with studies by us and others showing that antibodies induced by can be a mechanistic correlate of protection in NHP following vaccination41 or infection55 of NHP protect against reinfection. Our modeling suggests animals with S-specific IgG titers of approximately 100 IU/mL reduce sgRNA levels in BAL by approximately 2 log10, with further decrease of approximately 2 log10 sgRNA for every 1 log10 increase in IgG IU/mL. We also noted that 13/14 animals with any detectable neutralizing antibodies against B.1.351 had BAL sgRNA_N <105 suggesting that vaccines eliciting low to undetectable B.1.351-specific neutralizing antibodies may still control viral replication in lower airways.
The NHP model has been critical for guiding vaccine development against COVID-19 in humans. This report provides evidence that a two-dose regimen with mRNA-1273 effectively protects against B.1.351. Importantly, these data show a two-dose regimen induces high neutralizing activity against B.1.617.2 variant-of-concern and ongoing studies will determine if it is sufficient to protect NHPs. A final issue is durability of immunity and protection following vaccination. Ongoing studies assess how additional boosting with WA-1 or variant strain spike antigens influence protection against B.1.351 and other variants in NHPs and humans. These data in conjunction with real-world clinical data inform public health decisions on future vaccine boost requirements.
METHODS
Propagation and Characterization of Viral Stocks
VeroE6 cells were obtained from ATCC (clone E6, ATCC, #CRL-1586). SARS-CoV-2/human/USA/GA-EHC-083E/2020) (referred to as the D614G variant) was derived from a residual nasopharyngeal swab collected from an Emory Healthcare patient in March 2020. The isolation and sequencing was previously described3. The B.1.351 variant isolate, kindly provided by A. Pekosz [John Hopkins University (JHU), Baltimore, MD], was propagated once in Vero-TMPRSS2 cells (generated by VRC/NIH) to generate P2 viral stocks. Viral titers were determined by plaque assay on Vero-TMPRSS2 cells. Viral stocks were sequenced as described below and stored at −80C until use.
Deep Sequencing of Virus Isolate
Illumina-ready libraries were generated using NEBNext Ultra II RNA Prep reagents (New England BioLabs) as previously described48. Briefly, we fragmented RNA, followed by double-stranded cDNA synthesis, end repair, and adapter ligation. The ligated DNA was then barcoded and amplified by a limited cycle PCR and the barcoded Illumina libraries were sequenced by using paired-end 150-base protocol on a NextSeq 2000 (Illumina). Demultiplexed sequence reads were analyzed in the CLC Genomics Workbench v.21.0.3 by (i) trimming for quality, length, and adaptor sequence, (ii) mapping to the Wuhan-Hu-1 SARS-CoV-2 reference (GenBank accession number: NC_045512), (iii) improving the mapping by local realignment in areas containing insertions and deletions (indels), and (iv) generating both a sample consensus sequence and a list of variants. Default settings were used for all tools.
Titration of B.1.351 in Golden Syrian Hamsters
Hamster experiments were carried out in compliance with US National Institutes of Health regulations and approval from the Animal Care and Use Committee of Bioqual, Inc. (Rockville, MD). Challenge studies were conducted at Bioqual, Inc. Golden Syrian hamsters (Envigo, 089), aged 8–9 weeks old, were randomized into groups of 10 based on weight, with each group containing a 1:1 male:female ratio. Hamsters were inoculated IN with total doses of 1×103 – 1×105 PFU of SARS-CoV-2 JHU B.1.351 P2 in a final volume of 100 μL split between each nostril. Body weight observations were made daily post-infection.
Titration of B.1.351 in Rhesus Macaques
Three year-old male and female Indian-origin rhesus macaques were sorted by age and weight and then stratified into groups. NHP were challenged with total doses of 2.4×105 and 2.4×106 PFU of JHU SARS-CoV-2 B.1.351 P2; the viral inoculum was administered in 3 mL IT and 1 mL IN (0.5 mL into each nostril). On days 2, 4, and 6 post-challenge, BAL and nasal swabs were collected and assessed for sgRNA as detailed in the below methods.
Pre-clinical mRNA-1273 mRNA and Lipid Nanoparticle Production Process
A sequence-optimized mRNA encoding prefusion-stabilized SARS-CoV-2 S-2P56, 57 protein was synthesized in vitro. The mRNA was purified by oligo-dT affinity purification and encapsulated in a lipid nanoparticle through a modified ethanol-drop nanoprecipitation process described previously58.
Rhesus Macaque Model
NHP experiments were carried out in compliance with US National Institutes of Health regulations and approval from the Animal Care and Use Committee of the Vaccine Research Center and Bioqual, Inc. (Rockville, MD). Challenge studies were conducted at Bioqual, Inc. Male and female, 3–12 year-old, Indian-origin rhesus macaques were sorted by sex, age and weight and then stratified into groups of 8. NHP were immunized intramuscularly (IM) at week 0 and week 4–5 with either 30 or 100 μg mRNA-1273 in 1 mL of 1X PBS into the right hind leg or 30 μg at week 0. Naïve aged-matched NHP were included as controls. At week 12 (7–8 weeks post-boost or 12 weeks after the single vaccination), all NHP were challenged with a total dose of 5×105 PFU of SARS-CoV-2 B.1.351 strain. The viral inoculum was administered as 3.75×105 PFU in 3 mL intratracheally (IT) and 1.25×105 PFU in 1 mL intranasally (IN) in a volume of 0.5 mL into each nostril. Pre- and post-challenge sample collection is detailed in Extended Data Fig. 1.
Quantification of SARS-CoV-2 RNA and sgRNA
At the time of collection, NS were frozen in 1 mL of 1X PBS containing 1 μL of SUPERase-In RNase Inhibitor (Invitrogen) and BAL was mixed with 1 mL of RNAzol BD containing 10 μL acetic acid and both were frozen at −80°C until extraction. Nasal specimens were thawed at 55°C, and the swab removed. The remaining PBS was mixed with 2 mL of RNAzol BD (Molecular Research Center) and 20 μL acetic acid. At the time of collection, 1 mL of BAL fluid was mixed with 1 mL of RNAzol BD containing 10 μL acetic acid and frozen at −80°C until extraction. BAL specimens were thawed at 20–22°C and mixed with an additional 1 mL of RNAzol BD containing 10 μL acetic acid. Total RNA was extracted from nasal specimens and BAL fluid using RNAzol BD Column Kits and eluted in 65 μL water. Subgenomic SARS-CoV-2 E and N mRNA was quantified via reverse transcription-polymerase chain reaction (PCR) as previously described41. Reactions were conducted with 5 μL RNA and TaqMan Fast Virus 1-Step Master Mix (Applied Biosystems) with 500 nM primers and 200 nM probes. Primers and probes were as follows: sgLeadSARSCoV2_F: 5’-CGATCTCTTGTAGATCTGTTCTC-3’, E subgenomic mRNA - E_Sarbeco_P: 5’-FAM-ACACTAGCCATCCTTACTGCGCTTCG-BHQ1–3’ and E_Sarbeco_R: 5’-ATATTGCAGCAGTACGCACACA-3’, N subgenomic mRNA - wtN_P: 5’-FAM-TAACCAGAATGGAGAACGCAGTGGG-BHQ1–3’and wtN_R: 5’-GGTGAACCAAGACGCAGTAT-3’. Reactions were run on a QuantStudio 6 Pro Real-Time PCR System (Applied Biosystems) at the following conditions: 50°C for 5 min, 95°C for 20 sec, and 40 cycles of 95°C for 15 sec and 60°C for 1 min. Absolute quantification was performed in comparison to a standard curve. For the standard curve, the E or N subgenomic mRNA sequence was inserted into a pcDNA3.1 vector (Genscript) and transcribed using MEGAscript T7 Transcription Kit (Invitrogen) followed by MEGAclear Transcription Clean-Up Kit (Invitrogen). The lower limit of quantification was 50 copies.
TCID50 Quantification of SARS-CoV-2 from BAL
Vero-TMPRSS2 cells (VRC/NIH) were plated at 25,000 cells/well in Dulbecco’s Modified Eagle Medium (DMEM) + 10% FBS + Gentamicin and the cultures were incubated at 37°C, 5.0% CO2. Cells should be 80–100% confluent the following day. Medium was aspirated and replaced with 180 μL of DMEM + 2% FBS + gentamicin. Twenty (20) μL of BAL sample was added to top row in quadruplicate and mixed using a P200 pipettor 5 times. Using the pipettor, 20 μL was transferred to the next row, and repeated down the plate (columns A-H) representing 10-fold dilutions. The tips were disposed for each row and repeated until the last row. Positive (virus stock of known infectious titer in the assay) and negative (medium only) control wells were included in each assay set-up. The plates were incubated at 37°C, 5.0% CO2 for 4 days. The cell monolayers are visually inspected for cytopathic effect (CPE). TCID50 value wass calculated using the Read-Muench formula. For optimal assay performance, the TCID50 value of the positive control should test within 2-fold of the expected value.
Histopathology and Immunohistochemistry (IHC)
As previously described 36, NHP lung tissue sections were stained with hematoxylin and eosin (H&E) for routine histopathology and a rabbit polyclonal SARS-CoV-2 (GeneTex, GTX135357) for detection of SARS-CoV-2 virus antigen. All samples were evaluated by a boarded-certified veterinary pathologist.
10-plex Meso Scale ELISA
Multiplexed plates (96-well) precoated with SARS-CoV-2 S-2P57 and RBD proteins from the following strains: WA-1, B.1.351, B.1.1.7, and P.1., SARS-CoV-2 N protein, and Bovine Serum Albumin (BSA) are supplied by the manufacturer [Meso Scale Display (MSD)]. See Supplementary Table 3 for reagent details. Determination of antibody binding was performed as previously described21. Briefly, on the day of the assay, the plate is blocked for 60 minutes with MSD Blocker A (5% BSA). The blocking solution is washed off and test samples are applied to the wells at 4 dilutions (1:100, 1:500, 1:2500 and 1:10,000) unless otherwise specified and allowed to incubate with shaking for two h. Plates are washed and SULFO-TAG™ labeled anti-IgG antibody is applied to the wells and allowed to associate with complexed coated antigen – sample antibody within the assay wells. Plates are washed to remove unbound detection antibody. A read solution containing ECL substrate is applied to the wells, and the plate is entered into the MSD Sector instrument. A current is applied to the plate and areas of well surface where sample antibody has complexed with coated antigen and labeled reporter will emit light in the presence of the ECL substrate. The MSD Sector instrument quantitates the amount of light emitted and reports this ECL unit response as a result for each sample and standard of the plate. Magnitude of ECL response is directly proportional to the extent of binding antibody in the test article. All calculations are performed within Excel and the GraphPad Prism software, v7.0. Readouts are provided as Area Under Curve (AUC).
4-plex Meso Scale ELISA
MSD SECTOR® plates are precoated by the manufacturer with SARS-CoV-2 proteins (S-2P57, RBD, and N) and a BSA control in each well in a specific spot-designation for each antigen. See Supplementary Table 3 for reagent details. Determination of antibody binding was performed as previously described41. In detail, serum samples will be heat-inactivated for 30 min at 56°C prior to assay. Plates are blocked for 60 min at RT with MSD blocker A solution without shaking. Plates are washed and MSD reference standard (calibrator), QC test sample (pool of COVID-19 convalescent sera) and human serum test samples are added to the precoated wells in duplicates in an 8-point dilution series. Reference standard is added in triplicates. MSD control sera (low, medium and high) are added undiluted in triplicates as per validated assay format. Additional assay controls might be added in triplicates. Samples are incubated at RT for 4 h with shaking on a Titramax Plate shaker (Heidolph) at 1500 rpm. SARS-CoV-2 specific antibodies present in the sera or controls bind to the coated antigens. Plates are washed to remove unbound antibodies. Antibodies bound to the SARS-CoV-2 viral proteins are detected using an MSD SULFO-TAG™ anti-human IgG detection antibody incubated for 60 min at RT and with shaking. Plates are washed and a read solution (MSD GOLD™ read buffer) containing electrochemiluminescence (ECL) substrate is applied to the wells, and the plate is entered into the MSD MESO Sector S 600 detection system. An electric current is applied to the plates and areas of well surface which form antigen-anti human IgG antibody SULFO-TAG™ complex will emit light in the presence of the ECL substrate.
The MSD MESO Sector S 600 detection system quantitates the amount of light emitted and reports the ECL unit response as a result for each test sample, control sample and reference standard of each plate. Analysis is performed with the MSD Discovery Workbench software, Version 4.0. Calculated ECLIA parameters to measure binding antibody activities will include interpolated concentrations or assigned arbitrary units (AU/mL) read from the standard curve.
Conversion to 4-plex Readout to International Units
Recently the arbitrary units were bridged to the WHO International Standard [provided in international units/mL (IU/mL)], and a conversion factor was calculated and confirmed. Parallelism was established for all three antigens (SARS-CoV S-2P, RBD, and N) between the MSD provided reference standard and the WHO provided international standard. Concentration assignments were performed and then confirmed both at MSD and as part of a multi-site confirmation study.
Meso Scale ELISA for Mucosal Antibody Responses
Using previously described methods48, total S-specific IgG and IgA were determined by MULTI-ARRAY ELISA using Meso Scale technology (Meso Scale Discovery, MSD).
Lentiviral Pseudovirus Neutralization Assay
As previously described59, pseudotyped lentiviral reporter viruses were produced by the co-transfection of plasmids encoding S proteins from Wuhan-1 strain (Genbank #: MN908947.3) with a D614G mutation, a luciferase reporter, lentivirus backbone, and human transmembrane protease serine 2 (TMPRSS2) genes into HEK293T/17 cells (ATCC CRL-11268). Similarly, pseudoviruses containing S from B.1.351, P.1, and B.1.1.7 were produced. See Supplementary Table 3 for reagent details. Sera, in duplicate, were tested for neutralizing activity against the pseudoviruses by quantification of luciferase activity [in relative light units (RLU)]. Percent neutralization was normalized considering uninfected cells as 100% neutralization and cells infected with pseudovirus alone as 0% neutralization. IC50 titers were determined using a log(agonist) vs. normalized-response (variable slope) nonlinear regression model in Prism v9.0.2 (GraphPad). For samples that do not neutralize at the limit of detection at 50%, a value of 20 was plotted and used for geometric mean calculations.
VSV Pseudovirus Neutralization Assay
To make SARS-CoV-2 pseudotyped recombinant VSV-ΔG-firefly luciferase virus, BHK21/WI-2 cells (Kerafast, EH1011) were transfected with the Wuhan-1 strain (Genbank #: MN908947.3) S plasmid expressing full-length S with D614G mutation or S of B.1.351. See Supplementary Table 3 for reagent details. After S plasmid transfection, cells were subsequently infected with VSV∆G-firefly-luciferase as previously described60. Neutralization assays were completed on A549-ACE2-TMPRSS2 cells, generated using lentivirus encoding hACE2-P2A-TMPRSS2, with serially diluted serum samples as previously described29. For samples that do not neutralize at the limit of detection at 50%, a value of 20 was plotted and used for geometric mean calculations.
Focus Reduction Neutralization Test (FRNT)
Viruses were propagated in Vero-TMPRSS2 cells (VRC/NIH) (to generate viral stocks. See Supplementary Table 3 for reagent details. Viral titers were determined by focus-forming assay on VeroE6 cells (ATCC). Viral stocks were stored at −80°C until use. FRNT assays were performed as previously described 61. In detail, samples were diluted at 3-fold in 8 serial dilutions using DMEM (VWR, #45000–304) in duplicates with an initial dilution of 1:10 in a total volume of 60 ml. Serially diluted samples were incubated with an equal volume of SARS-CoV-2 (100–200 foci per well) at 37° C for 1 h in a round-bottomed 96-well culture plate. The antibody-virus mixture was then added to Vero cells and incubated at 37° C for 1 h. Post-incubation, the antibody-virus mixture was removed and 100 μL of prewarmed 0.85% methylcellulose (Sigma-Aldrich, #M0512–250G) overlay was added to each well. Plates were incubated at 37° C for 24 h. After 24 h, methylcellulose overlay was removed, and cells were washed three times with PBS. Cells were then fixed with 2% paraformaldehyde in PBS (Electron Microscopy Sciences) for 30 min. Following fixation, plates were washed twice with PBS and 100 μl of permeabilization buffer (0.1% BSA [VWR, #0332], Saponin [Sigma, 47036–250G-F] in PBS), was added to the fixed Vero cells for 20 min. Cells were incubated with an anti-SARS-CoV S primary antibody directly conjugated to biotin (CR3022-biotin) for 1 h at RT. Next, the cells were washed 3x in PBS and avidin-HRP was added for 1 h at RT followed by three washes in PBS. Foci were visualized using TrueBlue HRP substrate (KPL, # 5510–0050) and imaged on an ELISPOT reader (CTL). Antibody neutralization was quantified by counting the number of foci for each sample using the Viridot program62. The neutralization titers were calculated as follows: 1 - (ratio of the mean number of foci in the presence of sera and foci at the highest dilution of respective sera sample). Each specimen was tested in duplicate. The FRNT-50 titers were interpolated using a 4-parameter nonlinear regression in GraphPad Prism v9.0.2.4.3. For samples that do not neutralize at the limit of detection at 50%, a value of 5 was plotted and used for geometric mean calculations.
Intracellular Cytokine Staining (ICS)
Cryopreserved PBMC were thawed and rested overnight in a 37C°/5% CO2 incubator. The next morning, cells were stimulated with SARS-CoV-2 Spike protein peptide pools (S1 and S2) that were matched to the vaccine insert (comprised of 158 and 157 individual peptides, respectively, as 15mers overlapping by 11 aa in 100% DMSO, JPT Peptides) at a final concentration of 2 μg/ml in the presence of 3 μM monensin for 6 hours. Negative controls received an equal concentration of DMSO instead of peptides (final concentration of 0.5%). Intracellular cytokine staining was performed as described63. The following monoclonal antibodies were used: CD3 APC-Cy7 (clone SP34.2, BD Biosciences), CD4 PE-Cy5.5 (clone S3.5, Invitrogen), CD8 BV570 (clone RPA-T8, Biolegend), CD45RA PE-Cy5 (clone 5H9, BD Biosciences), CCR7 BV650 (clone G043H7, Biolegend), CXCR5 PE (clone MU5UBEE, Thermo Fisher), CXCR3 BV711 (clone 1C6/CXCR3, BD Biosciences), PD-1 BUV737 (clone EH12.1, BD Biosciences), ICOS Pe-Cy7 (clone C398.4A, Biolegend), CD69 ECD (cloneTP1.55.3, Beckman Coulter), IFN-g Ax700 (clone B27, Biolegend), IL-2 BV750 (clone MQ1–17H12, BD Biosciences), IL-4 BB700 (clone MP4–25D2, BD Biosciences), TNF-FITC (clone Mab11, BD Biosciences), IL-13 BV421 (clone JES10–5A2, BD Biosciences), IL-17 BV605 (clone BL168, Biolegend), IL-21 Ax647 (clone 3A3-N2.1, BD Biosciences), and CD154 BV785 (clone 24–31, BioLegend). Aqua live/dead fixable dead cell stain kit (Thermo Fisher Scientific) was used to exclude dead cells. All antibodies were previously titrated to determine the optimal concentration. Samples were acquired on an BD FACSymphony flow cytometer and analyzed using FlowJo version 9.9.6 (Treestar, Inc., Ashland, OR) following the gating scheme in Extended Data Fig. 7.
Statistical Analysis
Graphs show data from individual NHP with dotted lines indicating assay limits of detection. Groups were compared by Kruskal-Wallis test, followed by pairwise two-sided Wilcoxon Rank-sum tests with Holm’s adjustment on the set of pairwise tests if the Kruskal-Wallis was significant, for the primary analysis of viral load at day 2 in the BAL and NS, as well as other comparisons between dose groups. Correlations were estimated and tested using Spearman’s nonparametric method. Linear regression was used to explore the relationship between antibody levels and sgRNA, including quadratic terms for comparing the newly generated data and that previously published 63, with likelihood ratio tests to compare models and assess interaction effects.
Extended Data
Extended Data Fig. 1. Study design: Ability of mRNA-1273 to protect NHP against B.1.351 challenge.
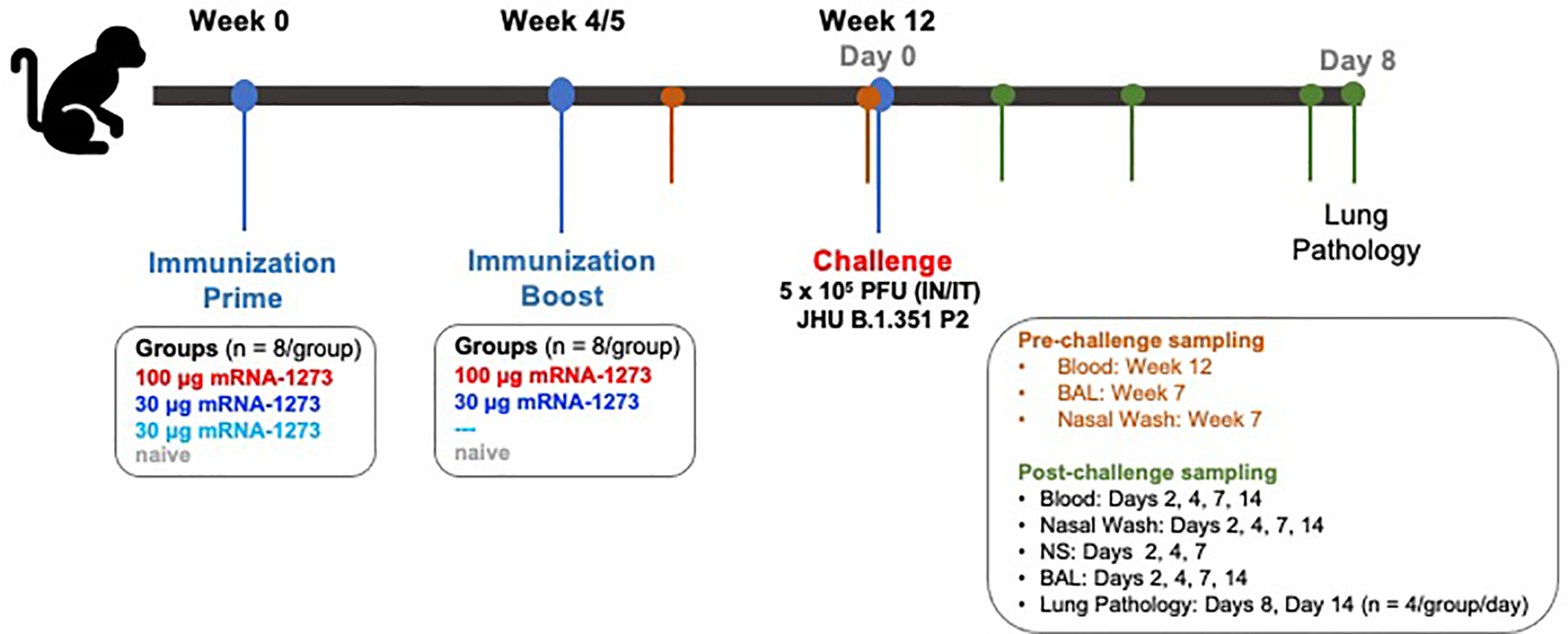
Nonhuman primates (NHP), Rhesus macaques (n = 8/group), were immunized with mRNA-1273 on the following schedule: Group 1: 0 and 4 weeks, 100 μg; Group 2: 0 and 5 weeks, 30 μg; Group 3: week 0, 30 μg. Naïve aged-matched NHP were included as controls. At week 12, animals were challenged with a total of 5×105 PFU of SARS-CoV-2 B.1.351. The viral inoculum was administered as 3.75×105 PFU in 3 mL intratracheally (IT) and 1.25×105 PFU in 1 mL intranasally (IN) in a volume of 0.5 mL into each nostril. Sera were collected at weeks 7 and week 12. Bronchoalveolar lavages (BAL) and nasal washes were also collected at week 7. Sera, BAL, and nasal washes were collected post-challenge on days 2, 4, 7, and 14, as indicated. Lung pathology was assessed on day 8 post-challenge in a subset of animals (n = 4/group)
Extended Data Fig. 2. Temporal neutralizing antibody responses following mRNA-1273 immunization.
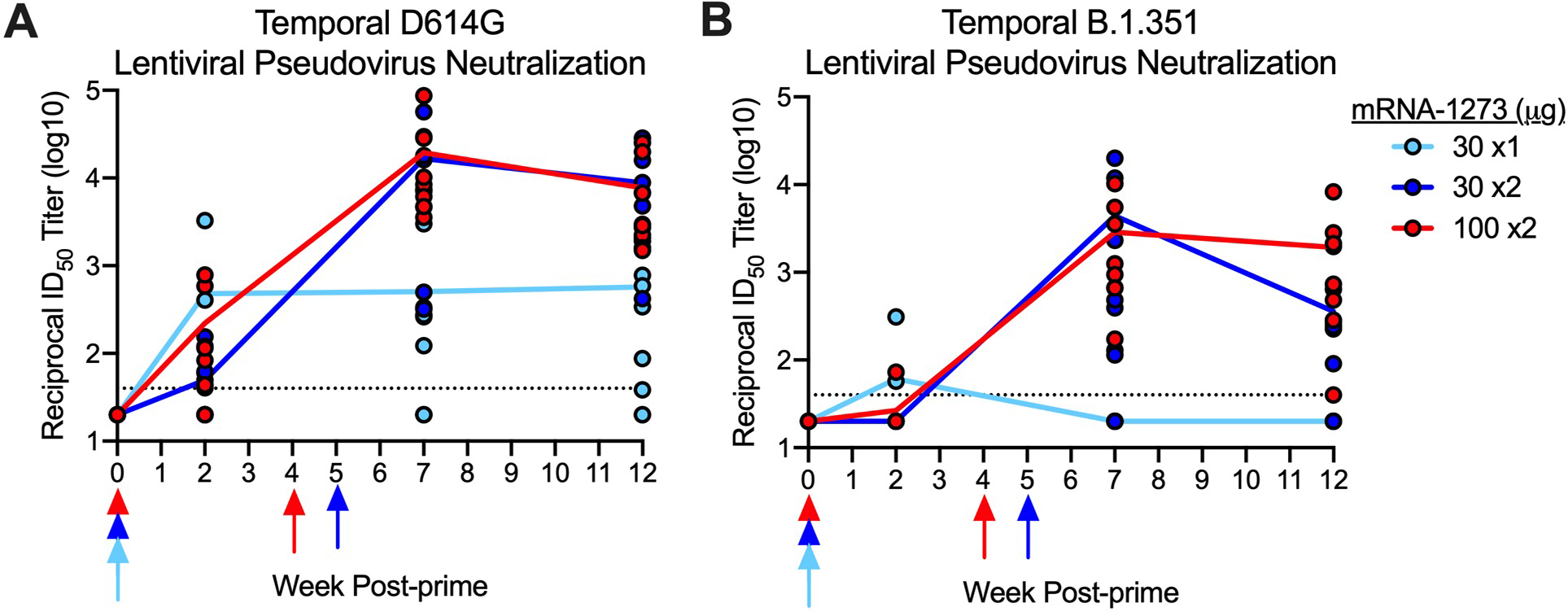
Rhesus macaques were immunized according to Figure S1. Sera collected at weeks 0, 2, 7, and 12 were assessed for SARS-CoV-2 D614G (A), and B.1.351 (B) lentiviral-based pseudovirus neutralization. Data represents one independent experiment. Circles represent individual NHP and may overlap where values are equal; lines represent geometric mean titers (GMT). Dotted lines indicate neutralization assay limits of detection. Arrows point to immunization weeks.
Extended Data Fig. 3. Correlations of humoral antibody analyses.
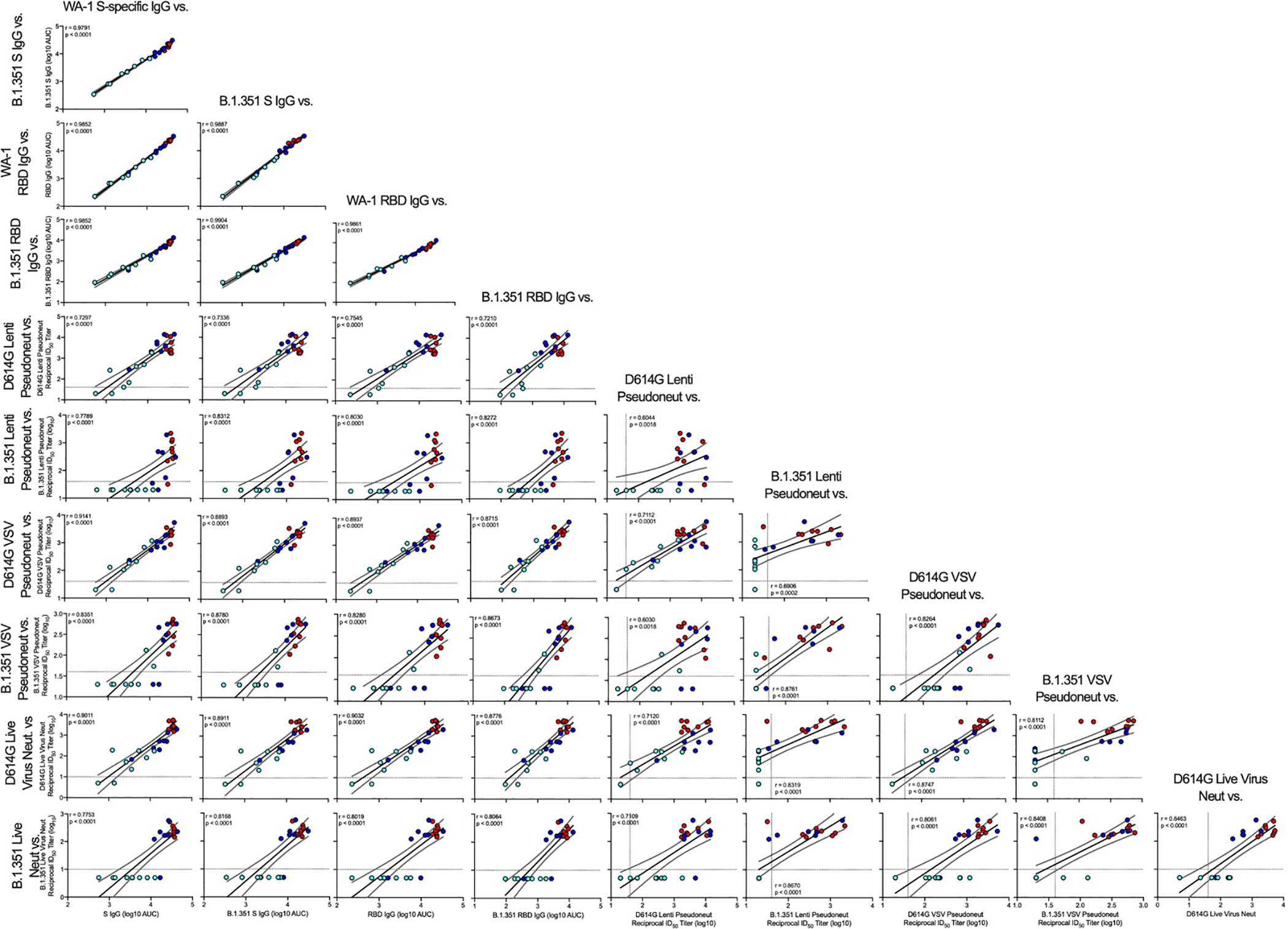
Rhesus macaques were immunized according to Figure S1C. Plots show correlations between SARS-CoV-2 WA-1 S-specific IgG, B.1.351 S-specific IgG, WA-1 RBD-specific IgG, B.1.351 RBD-specific IgG, D614G lentiviral-based pseudovirus neutralization, B.1.351 lentiviral-based pseudovirus neutralization, D614G VSV-based pseudovirus neutralization, B.1.351 VSV-based pseudovirus neutralization, D614G focus reduction neutralization, and B.1.351 focus reduction neutralization at week 12. Data represents one independent experiment. Circles represent individual NHP, where colors indicate mRNA-1273 dose as defined in Figure S1. Dotted lines indicate assay limits of detection. Black and gray lines indicate linear regression and 95% confidence interval, respectively. ‘r’ and ‘p’ represent Spearman’s correlation coefficients and corresponding two-sided p-values, respectively.
Extended Data Fig. 4. Characterization of B.1.351 viral isolate.
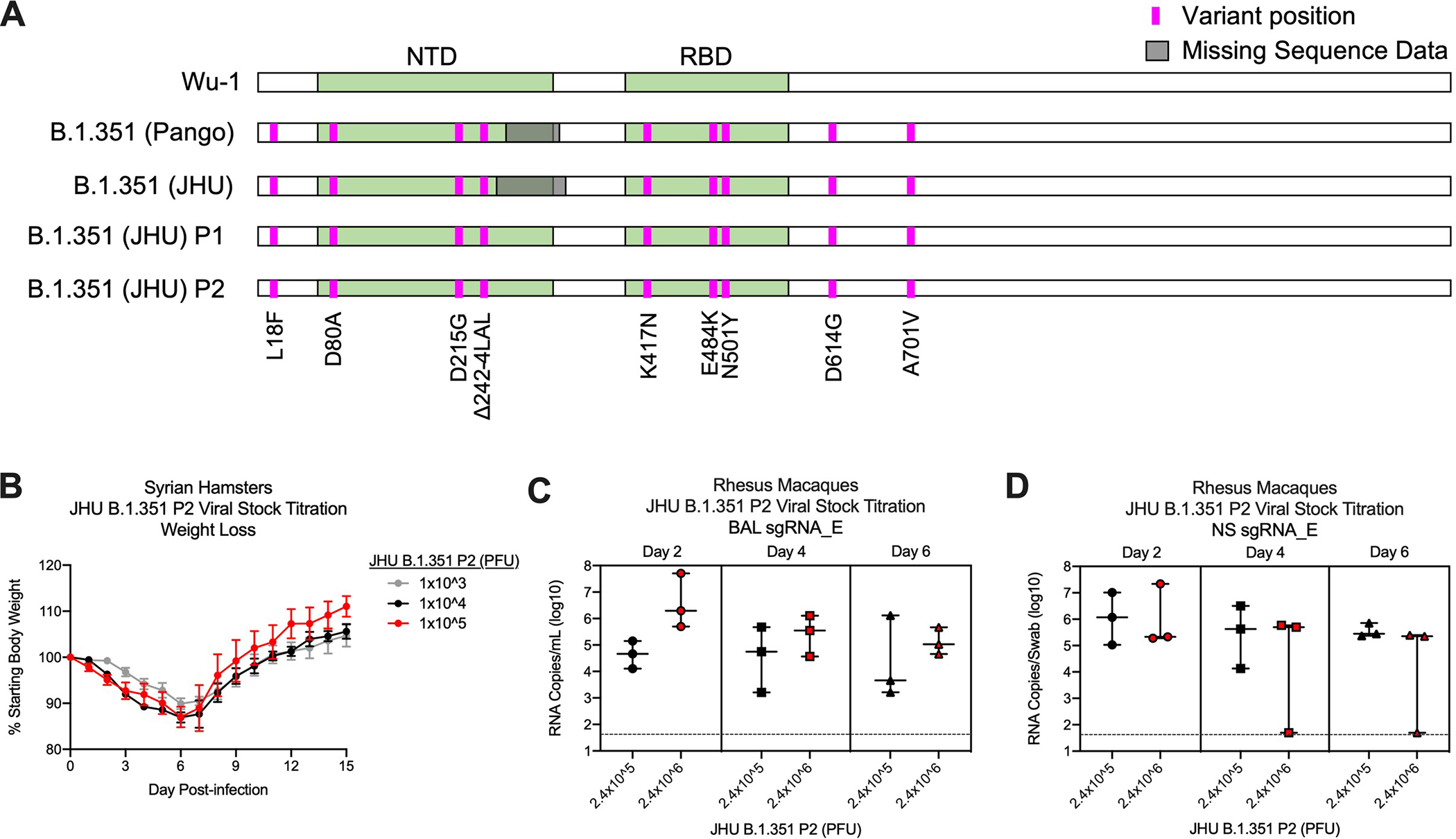
A SARS-CoV-2 B.1.351 clinical isolate was first passaged (P1) at Johns Hopkins University (JHU) on Vero cells then passaged again (P2) on Vero/TMPRSS2 cells. P1 and P2 underwent shotgun deep sequencing. (A) Alignment of S protein consensus sequence, where pink and gray indicate variant amino acid position and missing sequence data, respectively. (B) Syrian hamsters (n = 10/group) were infected with 1×103 (gray), 1×104 (black), or 1×105 (red) dilution of JHU B.1.351 P2 and monitored for weight loss for 15 days post-infection. Circles and error bars represent means and SEM, respectively. (C-D) Rhesus macaques (n = 3/group) were infected with 2.4×105 (black) or 2.4×106 (red) PFU of JHU B.1.351 P2 and viral replication was assessed by detection of SARS-CoV-2 E-specific sgRNA in BAL (C) and NS (D) on days 2, 4, and 6 post-infection. Data represents one independent experiment. Boxes and horizontal bars denote the interquartile ranges (IQR) and medians, respectively; whisker end points are equal to the maximum and minimum values. Dotted lines indicate assay limits of detection.
Extended Data Fig. 5. Mucosal antibody responses following SARS-CoV-2 challenge in mRNA-1273-immunized NHP.
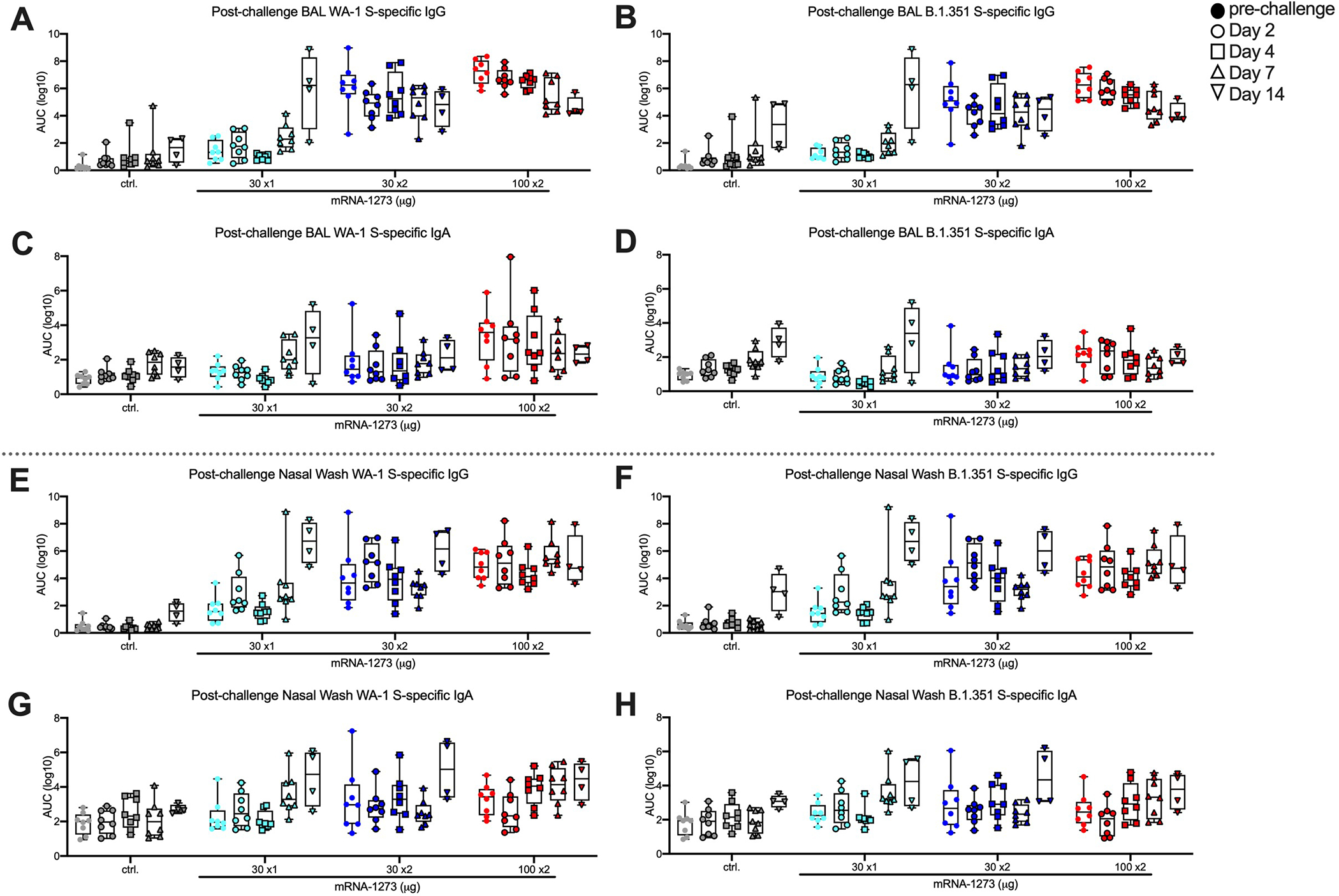
Rhesus macaques were immunized and challenged according to Figure S1. BAL (A-D) and nasal washes (E-H) collected at week 7 (filled circles) and days 2 (circles), 4 (squares), 7 (triangles), and 14 (inverted triangles) post-challenge were assessed for SARS-CoV-2 WA-1 (A, C, E, G) and B.1.351 (B, D, F, H) S-specific IgG (A-B, E-F) and IgA (C-D, G-H) by MULTI-ARRAY ELISA. Data represents one independent experiment. Boxes and horizontal bars denote the IQR and medians, respectively; whisker end points are equal to the maximum and minimum values. Symbols represent individual NHP.
Extended Data Fig. 6. T cell responses following SARS-CoV-2 challenge in mRNA-1273-immunized.
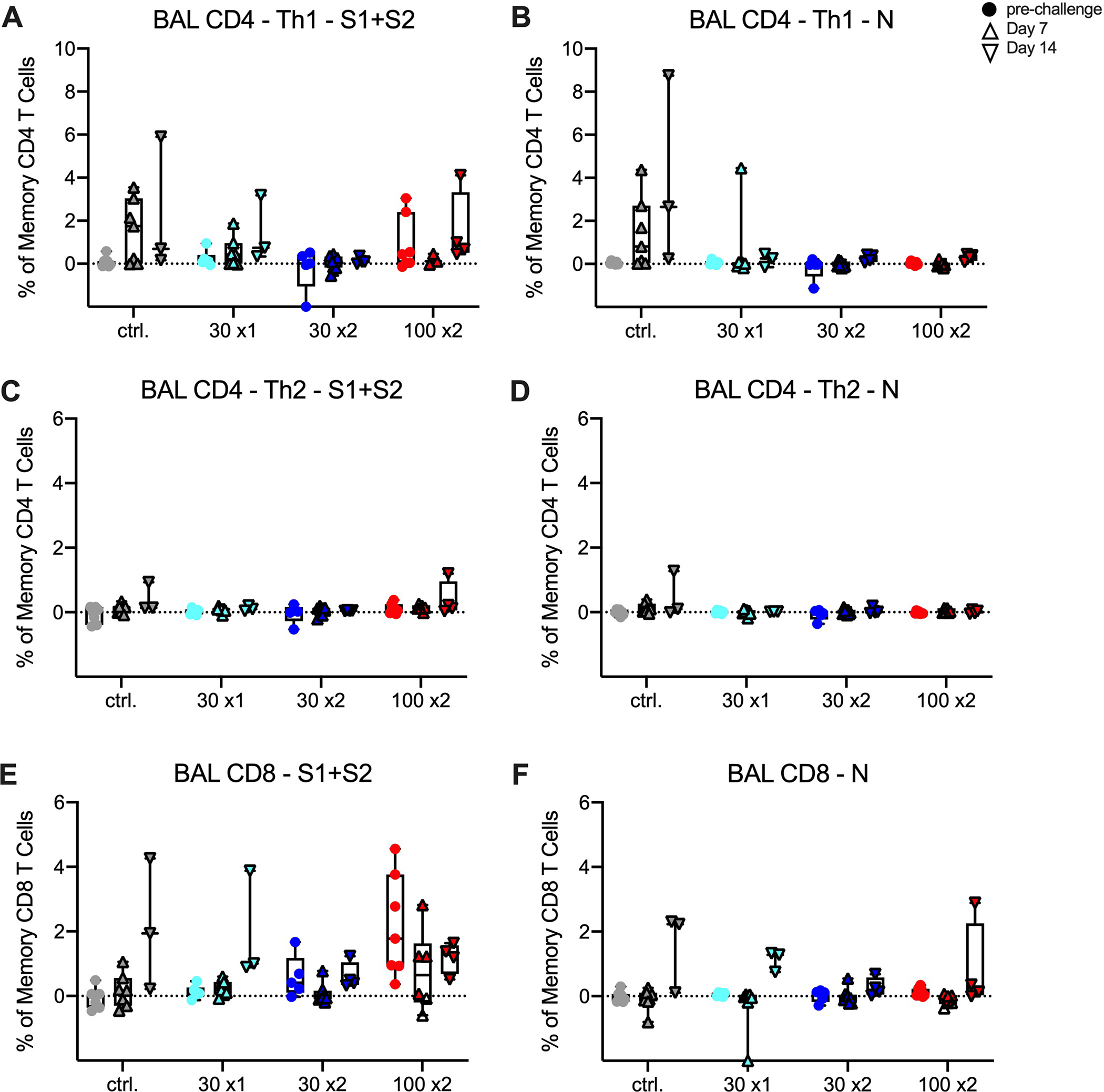
Rhesus macaques were immunized according to Extended Data Fig. 1. Intracellular cytokine staining was performed on BAL cells at week 27 (filled circles) and days 7 (triangles) and 14 (inverted triangles) post-challenge to assess T cell responses to SARS-CoV-2 S protein peptide pools, S1 and S2 (A, C, E) and N (B, D, F). Responses to S1 and S2 individual peptide pools were summed. Cytokine frequencies were measured from memory T cells as defined by CD45RA and CCR7. (A-B) Th1 responses (IFNg, IL-2, or TNF), (C-D) Th2 responses (IL-4 or IL-13), (E-F) CD8 T cell responses (IFNg, IL-2, or TNF). Data represents one independent experiment. Boxes and horizontal bars denote IQR and medians, respectively; whisker end points are equal to the maximum and minimum values. Circles represent individual NHP. Dotted lines are set to 0%.
Extended Data Fig. 7. Flow cytometry gating tree.
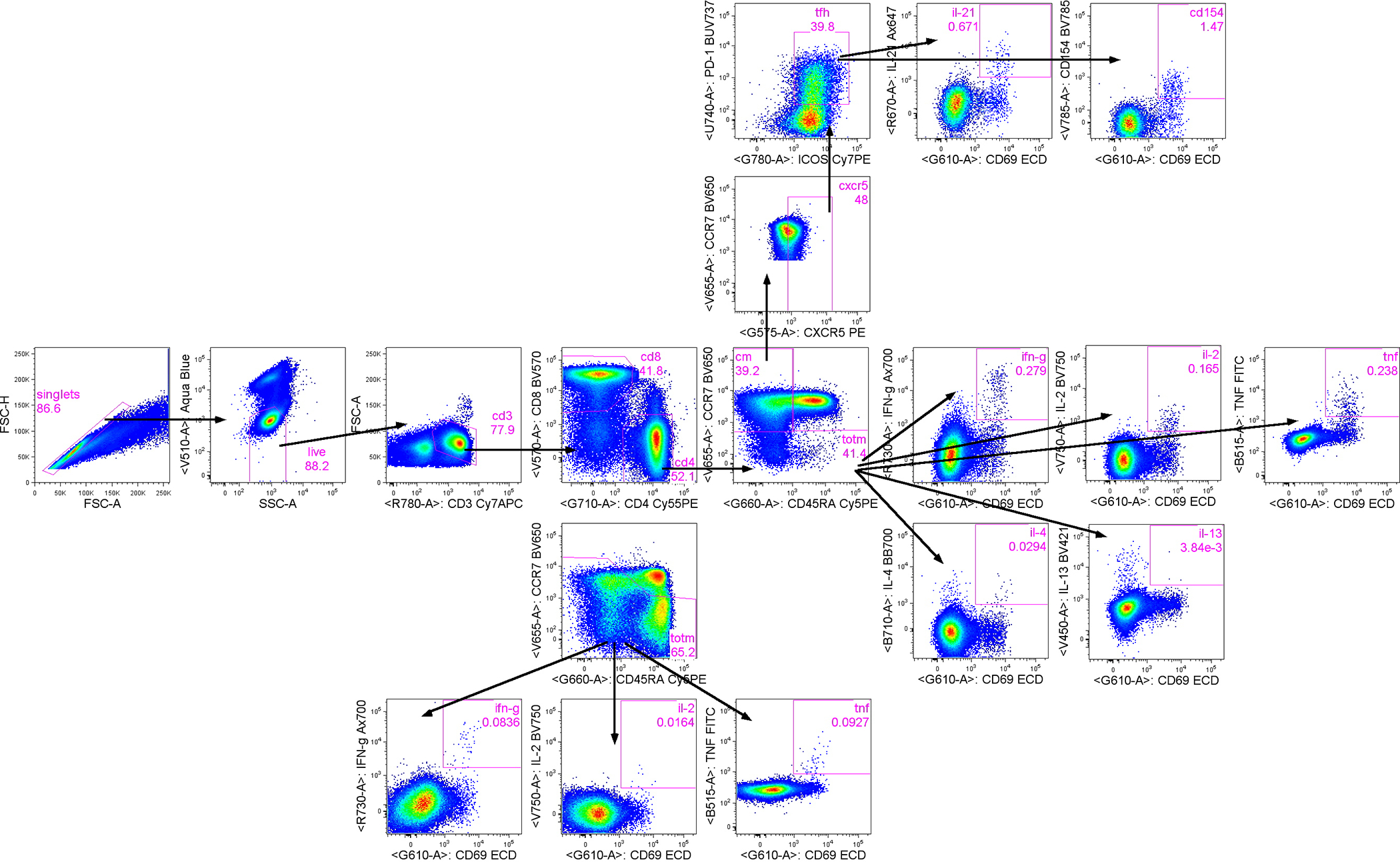
T cell populations were selected by subsequent gating: first single cells, followed by live/SSC low cells, then CD3+/FSC low cells, and finally CD4+ and CD8+ T cells. To measure IL-21 production and CD40L expression from Tfh, CD4 T cells were gated first on central memory (CM) T cells (CCR7+CD45RA−) then CXCR5+ CM cells followed by PD-1+ICOS+ cells to identify Tfh. Subsequently, from Tfh, CD69+IL-21+ and CD69+CD154+ cells were gated. Cytokine production from total memory (TotM) CD4 and CD8 T cells was measured by first gating on total memory cells (central memory CCR7+CD45RA− plus effector memory CCR7−CD45RA− plus terminal effector memory CCR7-CD45RA+ T cells) while excluding naïve CCR7+CD45RA+ T cells, then CD69+cytokine+ gates were drawn on total memory T cells.
Supplementary Material
Acknowledgements
We thank any additional members of all included laboratories for critical discussions and advice pertaining to experiments included in the manuscript. We thank J. Stein and M. Young for technology transfer and administrative support, respectively. We thank members of the NIH NIAID VRC Translational Research Program, including C. Case, H. Bao, E. McCarthy, J. Noor, A. Taylor, and R. Woodward, for technical and administrative assistance with animal experiments. We thank H. Mu and M. Farzan, from The Scripps Research Institue, for the ACE2-overexpressing 293 cells. We thank A. Creanga and M. Kanekiyo for the Vero-TMPRSS2 cells. We thank the laboratory of P. Kwong for providing protein for use in ELISA assays for detection of mucosal antibodies. We thank M. Brunner and M. Whitt, from the University of Tennessee Health Science Center College of Medicine, for kind support on recombinant VSV-based SARS-CoV-2 pseudovirus production. This work was supported by the Intramural Research Program of the VRC, NIAID, NIH. mRNA-1273 has been funded in part with Federal funds from the Department of Health and Human Services, Office of the Assistant Secretary for Preparedness and Response, Biomedical Advanced Research and Development Authority, under Contract 75A50120C00034. K.S.C.’s research fellowship was partially funded by the Undergraduate Scholarship Program, Office of Intramural Training and Education, Office of the Director, NIH. Virus propagation and live virus neutralization assays were funded by Emory Executive Vice President for Health Affairs Synergy Fund Award, Pediatric Research Alliance Center for Childhood Infections and Vaccines and Children’s Healthcare of Atlanta, Woodruff Health Sciences Center 2020 COVID-19 CURE Award, and NIAID Centers of Excellence for Influenza Research and Surveillance (CEIRS) contract HHSN272201400004C (M.S.S). M.S.S. is on the Advisory Board of Moderna.
Footnotes
Competing Interest Declaration
K.S.C. and B.S.G. are inventors on U.S. Patent No. 10,960,070 B2 and International Patent Application No. WO/2018/081318 entitled “Prefusion Coronavirus Spike Proteins and Their Use.” K.S.C. and B.S.G. are inventors on US Patent Application No. 62/972,886 entitled “2019-nCoV Vaccine”. A.C., M.K., S.E.3, K.W., D.K.E. and A.C. are employees of Moderna. A.V.R., Z.F., B.C., M.P., J.W., B.B., A.C., A.D., L.P., K.S., H.A., S.K., and M.G.L. are employees of Bioqual. No other authors declare any competing interests.
Data Availability Statement
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
References
- 1.Supasa P et al. Reduced neutralization of SARS-CoV-2 B.1.1.7 variant by convalescent and vaccine sera. Cell 184, 2201–2211.e2207 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen RE et al. Resistance of SARS-CoV-2 variants to neutralization by monoclonal and serum-derived polyclonal antibodies. Nat Med 27, 717–726 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Edara VV et al. Infection- and vaccine-induced antibody binding and neutralization of the B.1.351 SARS-CoV-2 variant. Cell Host & Microbe 29, 516–521.e513 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li Q et al. The Impact of Mutations in SARS-CoV-2 Spike on Viral Infectivity and Antigenicity. Cell 182, 1284–1294 e1289 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tegally H et al. Detection of a SARS-CoV-2 variant of concern in South Africa. Nature, doi: 10.1038/s41586-021-03402-9 (2021). [DOI] [PubMed] [Google Scholar]
- 6.Shen X et al. SARS-CoV-2 variant B.1.1.7 is susceptible to neutralizing antibodies elicited by ancestral spike vaccines. Cell Host Microbe, doi: 10.1016/j.chom.2021.03.002 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wibmer CK et al. SARS-CoV-2 501Y.V2 escapes neutralization by South African COVID-19 donor plasma. Nat Med, doi: 10.1038/s41591-021-01285-x (2021). [DOI] [PubMed] [Google Scholar]
- 8.Voysey M et al. Safety and efficacy of the ChAdOx1 nCoV-19 vaccine (AZD1222) against SARS-CoV-2: an interim analysis of four randomised controlled trials in Brazil, South Africa, and the UK. Lancet 397, 99–111 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Voysey M et al. Single-dose administration and the influence of the timing of the booster dose on immunogenicity and efficacy of ChAdOx1 nCoV-19 (AZD1222) vaccine: a pooled analysis of four randomised trials. Lancet 397, 881–891 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Andreano E et al. SARS-CoV-2 escape in vitro from a highly neutralizing COVID-19 convalescent plasma. bioRxiv, doi: 10.1101/2020.12.28.424451 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hoffmann M et al. SARS-CoV-2 variants B.1.351 and P.1 escape from neutralizing antibodies. Cell (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garcia-Beltran WF et al. Multiple SARS-CoV-2 variants escape neutralization by vaccine-induced humoral immunity. Cell, doi: 10.1016/j.cell.2021.03.013 (2021).. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tada T et al. Decreased neutralization of SARS-CoV-2 global variants by therapeutic anti-spike protein monoclonal antibodies. bioRxiv, doi: 10.1101/2021.02.18.431897 (2021). [DOI] [Google Scholar]
- 14.Tada T et al. Neutralization of viruses with European, South African, and United States SARS-CoV-2 variant spike proteins by convalescent sera and BNT162b2 mRNA vaccine-elicited antibodies. bioRxiv, doi: 10.1101/2021.02.05.430003 (2021). [DOI] [Google Scholar]
- 15.Deng X et al. Transmission, infectivity, and antibody neutralization of an emerging SARS-CoV-2 variant in California carrying a L452R spike protein mutation. medRxiv, doi: 10.1101/2021.03.07.21252647 (2021). [DOI] [Google Scholar]
- 16.Greaney AJ et al. Comprehensive mapping of mutations in the SARS-CoV-2 receptor-binding domain that affect recognition by polyclonal human plasma antibodies. Cell Host Microbe 29, 463–476 e466 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McCallum M et al. N-terminal domain antigenic mapping reveals a site of vulnerability for SARS-CoV-2. Cell, doi: 10.1016/j.cell.2021.03.028 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang P et al. Antibody resistance of SARS-CoV-2 variants B.1.351 and B.1.1.7. Nature, doi: 10.1038/s41586-021-03398-2 (2021). [DOI] [PubMed] [Google Scholar]
- 19.Zhou H et al. B.1.526 SARS-CoV-2 variants identified in New York City are neutralized by vaccine-elicited and therapeutic monoclonal antibodies. bioRxiv, doi: 10.1101/2021.03.24.436620 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu C et al. Reduced neutralization of SARS-CoV-2 B.1.617 by vaccine and convalescent serum. Cell, doi: 10.1016/j.cell.2021.06.020 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pegu A et al. Durability of mRNA-1273-induced antibodies against SARS-CoV-2 variants. bioRxiv, 2021.2005.2013.444010 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tegally H et al. Emergence and rapid spread of a new severe acute respiratory syndrome-related coronavirus 2 (SARS-CoV-2) lineage with multiple spike mutations in South Africa. medRxiv, 2020.2012.2021.20248640 (2020). [Google Scholar]
- 23.Hoffmann M et al. SARS-CoV-2 variants B.1.351 and P.1 escape from neutralizing antibodies. Cell 184, 2384–2393.e2312 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li Q et al. SARS-CoV-2 501Y.V2 variants lack higher infectivity but do have immune escape. Cell 184, 2362–2371.e2369 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wibmer CK et al. SARS-CoV-2 501Y.V2 escapes neutralization by South African COVID-19 donor plasma. Nat Med 27, 622–625 (2021). [DOI] [PubMed] [Google Scholar]
- 26.Zhou D et al. Evidence of escape of SARS-CoV-2 variant B.1.351 from natural and vaccine-induced sera. Cell 184, 2348–2361.e2346 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Starr TN et al. Deep Mutational Scanning of SARS-CoV-2 Receptor Binding Domain Reveals Constraints on Folding and ACE2 Binding. Cell 182, 1295–1310 e1220 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Starr TN et al. Deep mutational scanning of SARS-CoV-2 receptor binding domain reveals constraints on folding and ACE2 binding. bioRxiv, doi: 10.1101/2020.06.17.157982 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu K et al. Serum Neutralizing Activity Elicited by mRNA-1273 Vaccine. New England Journal of Medicine 384, 1468–1470 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Novavax (2021). Novavax COVID-19 Vaccine Demonstrates 89.3% Efficacy in UK Phase 3 Trial. Press Release January 28, 2021: https://ir.novavax.com/node/15506/pdf [Google Scholar]
- 31.K E et al. Efficacy of ChAdOx1 nCoV-19 (AZD1222) vaccineagainst SARS-CoV-2 variant of concern 202012/01(B.1.1.7): an exploratory analysis of a randomisedcontrolled trial. Lancet, doi: 10.1016/S0140-6736(21)00628-0 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sadoff J et al. Safety and Efficacy of Single-Dose Ad26.COV2.S Vaccine against Covid-19. New England Journal of Medicine, doi: 10.1056/NEJMoa2101544 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shinde V et al. Efficacy of NVX-CoV2373 Covid-19 Vaccine against the B.1.351 Variant. New England Journal of Medicine 384, 1899–1909 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Abu-Raddad LJ, Chemaitelly H & Butt AA Effectiveness of the BNT162b2 Covid-19 Vaccine against the B.1.1.7 and B.1.351 Variants. New England Journal of Medicine (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Madhi SA et al. Efficacy of the ChAdOx1 nCoV-19 Covid-19 Vaccine against the B.1.351 Variant. New England Journal of Medicine 384, 1885–1898, doi: 10.1056/NEJMoa2102214 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Corbett KS et al. Evaluation of the mRNA-1273 Vaccine against SARS-CoV-2 in Nonhuman Primates. New England Journal of Medicine 383, 1544–1555 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guebre-Xabier M et al. NVX-CoV2373 vaccine protects cynomolgus macaque upper and lower airways against SARS-CoV-2 challenge. Vaccine 38, 7892–7896 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mercado NB et al. Single-shot Ad26 vaccine protects against SARS-CoV-2 in rhesus macaques. Nature 586, 583–588 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van Doremalen N et al. ChAdOx1 nCoV-19 vaccination prevents SARS-CoV-2 pneumonia in rhesus macaques. bioRxiv : the preprint server for biology, 2020.2005.2013.093195 (2020). [Google Scholar]
- 40.Yu J et al. DNA vaccine protection against SARS-CoV-2 in rhesus macaques. Science 369, 806 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Corbett KS et al. Immune Correlates of Protection by mRNA-1273 Immunization against SARS-CoV-2 Infection in Nonhuman Primates. bioRxiv, 2021.2004.2020.440647 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Klasse PJ, Nixon DF & Moore JP Immunogenicity of clinically relevant SARS-CoV-2 vaccines in nonhuman primates and humans. Science Advances 7, eabe8065 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shen X et al. Neutralization of SARS-CoV-2 Variants B.1.429 and B.1.351. New England Journal of Medicine, doi: 10.1056/NEJMc2103740 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pardi N et al. Nucleoside-modified mRNA vaccines induce potent T follicular helper and germinal center B cell responses. The Journal of experimental medicine 215, 1571–1588 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Corbett KS et al. SARS-CoV-2 mRNA Vaccine Development Enabled by Prototype Pathogen Preparedness. Nature 586(7830), 567–571 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zou L et al. SARS-CoV-2 Viral Load in Upper Respiratory Specimens of Infected Patients. New England Journal of Medicine 382, 1177–1179 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wölfel R et al. Virological assessment of hospitalized patients with COVID-2019. Nature 581, 465–469 (2020). [DOI] [PubMed] [Google Scholar]
- 48.Francica JR et al. Vaccination with SARS-CoV-2 Spike Protein and AS03 Adjuvant Induces Rapid Anamnestic Antibodies in the Lung and Protects Against Virus Challenge in Nonhuman Primates. bioRxiv, 2021.2003.2002.433390 (2021). [Google Scholar]
- 49.Baden LR et al. Efficacy and Safety of the mRNA-1273 SARS-CoV-2 Vaccine. New England Journal of Medicine 384, 403–416 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Polack FP et al. Safety and Efficacy of the BNT162b2 mRNA Covid-19 Vaccine. New England Journal of Medicine 383, 2603–2615 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gao Q et al. Development of an inactivated vaccine candidate for SARS-CoV-2. Science 369, 77–81 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pardi N, Hogan MJ, Porter FW & Weissman D mRNA vaccines - a new era in vaccinology. Nat Rev Drug Discov 17, 261–279 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Munster VJ Subtle differences in the pathogenicity of SARS-CoV-2 variants of concern B.1.1.7 and B.1.351 in rhesus macaques. bioRxiv, doi: 10.1101/2021.05.07.443115 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Radvak PB 1.1.7 and B.1.351 variants are highly virulent in K18-ACE2 transgenic mice and show different pathogenic patterns from early SARS-CoV-2 strains. bioRxiv, doi: 10.1101/2021.06.05.447221 (2021). [DOI] [Google Scholar]
- 55.McMahan K et al. Correlates of protection against SARS-CoV-2 in rhesus macaques. Nature 590, 630–634 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
References (Methods)
- 56.Pallesen J et al. Immunogenicity and structures of a rationally designed prefusion MERS-CoV spike antigen. Proceedings of the National Academy of Sciences 114, E7348–E7357 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wrapp D et al. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 367, 1260–1263 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hassett KJ et al. Optimization of Lipid Nanoparticles for Intramuscular Administration of mRNA Vaccines. Molecular Therapy - Nucleic Acids 15, 1–11 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jackson LA et al. An mRNA Vaccine against SARS-CoV-2 - Preliminary Report. N Engl J Med 383, 1920–1931 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Whitt MA Generation of VSV pseudotypes using recombinant ΔG-VSV for studies on virus entry, identification of entry inhibitors, and immune responses to vaccines. J Virol Methods 169, 365–374 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vanderheiden A et al. Development of a Rapid Focus Reduction Neutralization Test Assay for Measuring SARS-CoV-2 Neutralizing Antibodies. Curr Protoc Immunol 131, e116 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Katzelnick LC et al. Viridot: An automated virus plaque (immunofocus) counter for the measurement of serological neutralizing responses with application to dengue virus. PLoS Negl Trop Dis 12, e0006862 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Donaldson MM, Kao SF & Foulds KE OMIP-052: An 18-Color Panel for Measuring Th1, Th2, Th17, and Tfh Responses in Rhesus Macaques. Cytometry A 95, 261–263 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Finak G et al. Mixture models for single-cell assays with applications to vaccine studies. Biostatistics 15, 87–101 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.


