Abstract
Purpose:
CACNA1C encodes the alpha-1-subunit of a voltage-dependent L-type calcium channel expressed in human heart and brain. Heterozygous variants in CACNA1C have previously been reported in association with Timothy syndrome and Long QT syndrome. Several case reports have suggested that CACNA1C variation may also be associated with a primarily neurological phenotype.
Methods:
We describe 25 individuals from 22 families with heterozygous variants in CACNA1C, who present with predominantly neurological manifestations.
Results:
Fourteen individuals have de novo, non-truncating variants and present variably with developmental delays, intellectual disability, autism, hypotonia, ataxia, and epilepsy. Functional studies of a subgroup of missense variants via patch-clamp experiments demonstrated differential effects on channel function in vitro, including loss-of-function (p.Leu1408Val), neutral effect (p.Leu614Arg), and gain-of-function (p.Leu657Phe, p.Leu614Pro). The remaining 11 individuals from eight families have truncating variants in CACNA1C. The majority of these individuals have expressive language deficits, and half have autism.
Conclusion:
We expand the phenotype associated with CACNA1C variants to include neurodevelopmental abnormalities and epilepsy, in the absence of classic features of Timothy syndrome or Long QT syndrome.
Introduction:
CACNA1C (MIM:114205) encodes the alpha-1 subunit of a voltage-dependent L-type calcium channel that is broadly expressed and best studied in human heart, brain, smooth muscle, and endocrine tissue1. Pathogenic variants in CACNA1C are associated with various cardiac rhythm disorders. Specific heterozygous gain-of-function variants are associated with Timothy syndrome (MIM:601005), a multisystem disorder characterized by long QT interval, structural cardiac defects, dysmorphic facial features, syndactyly, intellectual disability, autism, hypotonia, and, less commonly, epilepsy1. Heterozygous gain-of-function variants have also been reported in association with non-syndromic Long QT syndrome2. Loss-of-function variants in CACNA1C were previously implicated as a cause of Brugada syndrome (MIM:611875), but this association has not been firmly established3.
In addition to the role of CACNA1C in cardiac rhythm disorders, this gene has also been implicated in diverse neurological phenotypes. Polymorphisms in CACNA1C are associated with increased risk of schizophrenia, major depressive disorder, bipolar disorder, and autism4. One individual presenting with neonatal epileptic encephalopathy has been previously reported with a de novo, rare missense variant in CACNA1C; functional studies were not performed5. Individuals with de novo variants in CACNA1C have been detected in large-scale sequencing studies for neurodevelopmental disorders, including two individuals with missense variants and an individual with a truncating variant6.7. In addition, two individuals with intragenic deletions in CACNA1C in association with neurodevelopmental and behavioral abnormalities have been previously reported8. There is also a report of a de novo, intragenic duplication within CACNA1C in a child with learning disabilities9. There have also been numerous reports of individuals with neurodevelopmental phenotypes associated with deletions at 12p, both interstitial and terminal, disrupting CACNA1C 10. This has led to speculation that haploinsufficiency of this gene may largely account for the neurodevelopmental phenotype of 12p13.33 interstitial deletion syndrome8. Finally, one large Chinese kindred was reported with a heterozygous intronic variant in CACNA1C segregating with an adult-onset autosomal dominant cerebellar ataxia phenotype11.
Here we describe 25 new individuals from 22 families with novel variants in the CACNA1C gene presenting with a predominantly neurological phenotype, variably including developmental delay, intellectual disability, autism, hypotonia, epilepsy, and ataxia, in the absence of significant cardiac manifestations or additional features of Timothy syndrome. Broad phenotypic differences were evident between those with non-truncating and truncating variants. We functionally characterize a subgroup of these variants with patch clamping.
Patients and Methods:
Ascertainment of cases
Cases were ascertained from GeneMatcher through the Matchmaker Exchange Network between July 2018 and March 202012,13. In addition, 2 cases were ascertained through the ClinVar database. CACNA1C variants in index cases were detected on exome sequencing or intellectual disability gene panel. Additional genetic findings for individual patients are summarized in the supplement. Signed consent was obtained from parents/legal guardians for publication of photographs, in accordance with the institutional review boards/ethics committees at each institution.
Selection of variants for functional studies:
Whole-cell patch clamp analysis to assess functionality of the Cav1.2 calcium channels was conducted for a small number of missense variants: (p.Leu614Pro), (p.Leu614Arg), (p.Leu657Phe) and (p.Leu1408Val). These variants were specifically selected because they are located near channel regions where there appeared to be a clustering of variants in the cohort, and are likely of functional importance. For our selection of variants to functionally characterize, we also considered intolerance to variation of the residue, and whether variants at homologous residues in related channels have been reported as pathogenic.
Whole-cell patch clamp electrophysiology of p.Leu614Arg variant
Human CACNA1C cDNA (Accession no. NM_000719) was mutagenized to generate p.Leu614ARg using site-directed mutagenesis (Agilent), following the manufacturer’s instructions using the following primers: 5’ CAAGATCATGTCCCCAAGAGGCATCTCCGTGCTCAG 3’ (forward) and 5’ CTGAGCACGGAGATGCCTCTTGGGGACATGATCTTG 3’ (reverse). The underlined and bolded nucleotides indicate the variant. Wild type or p.L614R CACNA1C (3 μg) along with CACNB2 (1.5 μg) and CACNA2D (1.5 μg) and EGFP (0.3 μg) were transfected into HEK293 cells using Lipofectamine 2000 (ThermoFisher Scientific, MA). The HEK293 cells were grown in Dulbecco’s modified Eagle’s culture medium with 10% fetal bovine serum in 60-mm culture dishes in a 37°C incubator with 5% CO2. Whole-cell voltage-gated calcium currents were measured as previously described14.
Whole-cell patch clamp electrophysiology and Western blot of p.Leu614Pro, p.Leu657Phe and p.Leu1408Val variants
Molecular Biology and Mutagenesis:
The mouse Cav1.2 (Cacna1c; clone #26572), the rat Cavβ3 (Cacnb3; clone#26574) and the rat Cavα2δ1 (Cacna2d1; clone #26575), as previously described15, were obtained from Addgene. The p.Leu614Pro, p.Leu657Phe, and p.Leu1408Val variants of Cav1.2 were generated by site-directed mutagenesis.
Cell Culture:
HEK293 cells were used for patch clamp and Western blot experiments. HEK cells were cultured and maintained using standard cell culture approaches as previously described16. Transfection was performed using jetPRIME (Polypus) according to the manufacturer’s instructions. Calcium currents were recorded 72 hours following transfection. Western blots were performed 72 hours after transfection.
Whole-cell patch clamp electrophysiology:
Patch clamp experiments were performed as previously described16. Solutions used for Whole cell patch clamp recordings were: Bath solution (in mM): 135 choline-Cl, 1 MgCl2, 2 CaCl2, and 10 HEPES, adjusted to pH 7.4 with CsOH; Pipette solution: 135 CsCl, 4 MgATP, 1 EGTA, 1 EDTA, and 10 HEPES, adjusted to pH 7.4 with CsOH. Experiments were conducted at room temperature (22 ± 1°C). Currents were adjusted with P/4 leak subtraction during data acquisition.
Western blot:
Cell lysates were collected in NP-40 lysis buffer (1% NP-40, 150 mM NaCl, 50 mM Tris-HCl) supplemented with 1% protease inhibitor, 72 hours after transfection. Western blots were performed as previously described16. Cav1.2 was detected using a mouse monoclonal Cav1.2 antibody (1:1000 dilution, clone #L57/46; NeuroMab) and HRP-conjugated goat anti-mouse antibody (1:20,000 dilution, SH023; Applied Biological Materials).
Results:
Demographics, molecular data, and clinical characteristics for our cohort are summarized in tables 1 and 2 (More detailed clinical information for each individual, including variants of uncertain significance identified in additional genes, is provided in the Supplementary Data). Thirteen individuals are male, and 12 are female. Age at most recent follow-up ranges from 10 months to 40 years (median age 6.8 years).
Table 1.
Clinical features of individuals with nontruncating CACNA1C variants.
| Individual | P1 | P2 | P3 | P4 | P5 | P6 | P7 | P8 | P9 | P10 | P11 | P12 | P13 | P14 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Variants | p.Phe166Leu | p.Lys177Arg | p.Arg324Trp | p.Val403Met | p.Leu601Arg | p.Met611Thr | p.Leu614Arg | p.Leu614Pro | p.Leu657Phe | p.Ile743del | p.Val1187Ala | p.Leu1408Val | p.Val1411Leu | p.Val1411Leu |
| c.496T>C | c.530A>G | c.970C>T | c.1207G>A | c.1802T>G | c.1832T>C | c.1841 T>G | c.1841T>C | c.1969C>T | c.2227_2229delATC | c.3560T>C | c.4222C>G | c.4231G>C | c.4231G>T | |
| Inheritance | de novo | de novo | de novo | de novo | de novo | de novo | de novo | de novo | de novo | de novo | de novo | de novo | de novo | de novo |
| Age at most recent exam | 23 years | 7.6 years | 9 years | 2.3 years | 4 years | 4 years | 2.5 years | 24 years | 15 years | 3.5 years | 1.5 years | 6.8 years | 7.8 years | 10 months |
| Sex | Female | Female | Female | Female | Female | Male | Male | Male | Female | Male | Male | Female | Female | Male |
| Development | ||||||||||||||
| Motor delay | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| Ambulatory (age started walking) | + (16 months) | + (15 months) | + (21 months) | − | + (28 months) | + (17 months) | + (3 years) | − | + (2 years) | − | − | − | − | − |
| Abnormal Muscle tone | Hypotonia | − | Hypotonia | Hypotonia | Hypotonia | Hypotonia | Hypotonia | Hypotonia | − | Hypotonia, Leg spasticity | Hypotonia | Hypotonia | Hypotonia | Hypotonia |
| Abnormal balance/coordination | − | + | − | − | + | + | + | N/A | − | N/A | N/A | N/A | N/A | N/A |
| Tremor | − | − | − | Action tremor in arms in first 2 years, subsequently resolved | Intention tremor in arms, head tremor | − | − | − | − | − | − | − | − | − |
| Language delay | + | − | − | + | + | + | + | + | + | + | + | + | + | + |
| Verbal Ability | − | + | + | − | − | + | − | − | + | − | − | − | − | N/A |
| Clinical Diagnosis of Autism | − | − | − | − | − | − | − | + | − | − | − | − | − | − |
| ADHD | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| Cognition | Severe intellectual disability | Learning disability | Learning disability, dyslexia | Mild-moderate global developmental delay | Mild intellectual disability | Mild global developmental delay | Severe developmental delay | Intellectual disability, severity unspecified | moderate-severe intellectual disability | Severe global developmental delay | Unknown | Severe intellectual disability | Severe intellectual disability | Severe global developmental delay |
| Epilepsy | ||||||||||||||
| Seizures | + | + | + | − | − | + | + | + | + | − | + | + | + | + |
| Age of onset | 6 years | 11 months | 8 years | N/A | N/A | 7 months | 10 months | 1 year | 6 years | N/A | 1 year | 6 months | 11 months | 2 months |
| Seizure types/Epilepsy syndrome | Absence, generalized tonic-clonic | Focal motor, absence. Seizures fever sensitive. | Focal seizures | N/A | N/A | West syndrome/ Infantile spasms | Myoclonic seizures; generalized tonic-clonic | Generalized epilepsy. Seizures are fever sensitive. | Behavioral arrest spells +/- head or limb jerking | N/A | Seizures provoked by fever, diagnosed as febrile seizures | Myoclonic seizures àmultiple generalized seizure types/ generalized epilepsy syndrome. | West syndrome/Infantile spasms à Lennox-Gastaut syndrome | Generalized epilepsy |
| Cardiac abnormalities | ||||||||||||||
| Abnormal ECG/Holter | N/A | − | − | − | N/A | − | − | − | − | − | N/A | Possible ventricular conduction delay | Prolonged QT | − (borderline QTc 450msec) |
| Abnormal ECHO | N/A | N/A | − | Small interatrial communication in the upper part of the atrial septum with left-right shunt | N/A | − | N/A | − | N/A | − | − | Trace AI, trivial TR, trace PR | − | − (previously known PDA/PFO have since closed) |
| Orthopedic abnormalities | Pes plagovalgus | Congenital hip dysplasia | Hypermobility | Left hip dislocation and right hip subluxation, joint hypermobility | Pes planovalgus, joint hypermobility, toe-walking | − | Bilateral clubfoot, and adducted thumbs | Kyphosis | Small appearing feet, tapered fingers | Bilateral clubfoot, prominent heels in small appearing feet, adducted thumb, and camptodactyly of fingers, tapering fingers | Cutaneous 2–3 toe syndactyly and mild 3–4 finger syndactyly, congenital torticollis | Kyphoscoliosis, Coxa valga | − | Bilateral congenital hip dysplasia, toe 2–3 syndactyly bilaterally, bilateral adducted thumbs |
| Ophthalmological abnormalities | Unknown | − | Strabismus, hyperopia | Strabismus (non paralytic) | Myopia | − | Strabismus | Hyperopia, astigmatism | − | Severe hyperopia | Hyperopia, astigmatism, esotropia | Strabismus | Cortical visual impairment | − |
| Dysmorphic features | + | − | − | − | + | − | − | + | + | + | − | − | − | + |
Table 2.
Clinical features of individuals with truncating CACNA1C variants.
| Individual | P15 | P16a | P16b | P17 | P18 | P19 | P20 | P21 | P22a | P22b | P22c |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Variants | p.Gln80Leufs*49 | p.Arg161* | p.Arg161* | p.Trp528* | splice | p.Val996Trpfs*19 | p.Arg1377Profs*61 | p.Leu1518Profs*3 | p.Arg1989* | p.Arg189* | p.Arg1989* |
| c.239_244delAGCGGAinsTTGCAGCTCC | c.481C>T | c.481C>T | c.1584G>A | c.2663+1G>C | c.2926delG | c.4129dup | c.4553del | c.5860C>T | c.5860C>T | c.5860C>T | |
| Inheritance | de novo | Paternal | de novo | Paternal | not paternal, maternal sample unavailable for testing | de novo | de novo | de novo | Paternal | Paternal | Unknown |
| Age at most recent exam | 15 years | 6 years | 33 years | 14.5 years | 16 years | 6 years | 4 years | 40 years | 2 years | 3 years | 32 years |
| Sex | Female | Male | Male | Female | Male | Male | Female | Female | Male | Male | Male |
| Development | |||||||||||
| Motor delay | − | + | − | − | − | + | − | + | − | − | Unknown |
| Ambulatory (age started walking) | + (15 months) | + (14 months) | + | + (12 months) | + (12 months) | + (18 months) | + (12 months) | + (26 months) | + (10 months) | + | + |
| Abnormal Muscle tone | − | Hypotonia | − | − | − | Unknown | − | Unknown | Hypotonia | Hypotonia | Unknown |
| Abnormal balance/coordination | − | + | − | − | + | + | − | − | − | + | − |
| Tremor | − | High frequency tremor | − | − | − | − | − | − | − | Intention tremor | − |
| Language delay | + | + | + | + | + | + | + | + | + | + | Unknown |
| Verbal Ability | + | − (2 words only) | + | + | + | + | + | + | − (Couple words only) | − (Several words only) | + |
| Clinical Diagnosis of Autism | + | + | − | − | − | − | + | − | + | + | − |
| ADHD | + | - | − | - | + | + | _ | _ | − | − | − |
| Cognition | IQ in low-normal range | Mild -moderate intellectual disability | Learning disability with normal IQ. | Borderline intellectual disability | Moderate intellectual disability | Normal | Moderate global developmental delay | Moderate intellectual disability | Unknown | Unknown | Learning disability |
| Epilepsy | |||||||||||
| Seizures | − | − | − | − | − | − | − | − | − | − | + |
| Abnormal Brain MRI | Chiari 1 malformation | N/A | N/A | − | N/A | − | N/A | N/A | N/A | N/A | N/A |
| Cardiac abnormalities | |||||||||||
| Abnormal ECG/Holter | − | − | − | N/A | N/A | − | N/A | N/A | N/A | − | N/A |
| Abnormal ECHO | N/A | N/A | N/A | N/A | N/A | − | N/A | N/A | N/A | N/A | N/A |
| Orthopedic abnormalities | Scoliosis | − | Unknown | − | Pes planus, genu valgum | − | − | − | Metatarsus adductus | − | Unknown |
| Ophthalmological abnormalities | − | − | − | Unknown | − | Unknown | − | Strabismus | − | − | Unknown |
| Dysmorphic features | − | + | − | + | + | + | + | − | + | + | − |
Thirteen unrelated individuals have missense variants in CACNA1C, one individual has an in-frame deletion, and 11 individuals from 8 families have predicted truncating variants, including one canonical splice variant. All the non-truncating variants and all but three of the truncating variants occurred de novo. The three truncating variants that were inherited segregated with neurodevelopmental abnormalities in the families. None of the variants in our cohort are found in public databases, including the Broad Genome Aggregation Database (gnomAD) (https://www.biorxiv.org/content/10.1101/531210v4), 1000 genomes17, or the Exome Variant Server (Exome Variant Server, NHLBI GO Exome Sequencing Project (ESP), Seattle, WA (URL: http://evs.gs.washington.edu/EVS/). All missense variants occur at highly conserved positions and are predicted by in silico models to be damaging (Mutation Taster, Poly-Phen2, SIFT). CADD scores are listed in table 1. One truncating variant we describe occurs in exon 45/45, potentially escaping nonsense-mediated decay. All other reported truncating variants are predicted to be subject to nonsense-mediated decay.
Non-truncating variants:
For individuals with missense variants and the in-frame deletion (i.e. non-truncating variants), prenatal history was remarkable for reduced fetal movement (n=3/14), bilateral clubfoot detected on prenatal ultrasound (n=2/14), and oligohydramnios (n=2/14). All growth parameters at birth were normal except one individual with IUGR. Two individuals had neonatal feeding difficulties.
All 14 individuals with non-truncating variants have a history of developmental delays or intellectual disability. Two individuals, after early delays, currently have a learning disability. Nine individuals are non-verbal at most recent follow-up, and six are non-ambulatory (age at last evaluation ranging from 2 years to 24 years). This does not include one individual that is still an infant (10 months at last evaluation), who is manifesting severe global developmental delays and is unable to babble, roll, or sit. One individual has been formally diagnosed with autism, and six additional individuals have autistic features, including stereotypies and abnormal socialization. The overall developmental trajectory in our cohort is consistent with a static neurodevelopmental disorder. Regression was reported in one individual within the context of developing infantile spasms.
Twelve of 14 individuals in this group are hypotonic, and four are described as having abnormal balance and coordination (ataxia). One individual was diagnosed with bilateral vestibular areflexia.
Ten of 14 individuals in our cohort with non-truncating variants have epilepsy, with onset ranging from 2 months to 8 years. Clinical details are summarized in table 1. Both generalized and focal epilepsy disorders were reported. Fever sensitivity was reported in 3 individuals. Epilepsy was medically refractory in 4 individuals. On EEG, both generalized and focal epileptiform abnormalities were seen across the cohort, without consistent patterns. One individual was reported to have excessive diffuse fast (beta) activity. MRI brain was normal in all individuals that underwent imaging, with the exception of an incidentally detected arachnoid cyst in one individual and prominent cerebellar folia in another individual.
Eleven of the individuals with non-truncating variants had at least one ECG. One individual had mildly prolonged QTc on routine ECG at 7 years of age. Holter monitoring in this individual found significant QT prolongation, especially at higher heart rates (decreased repolarization reserve), and evidence of repolarization instability with macroscopic T wave alternans. Another individual had a possible ventricular conduction delay on routine ECG. Holter monitoring in this individual demonstrated only premature ventricular contractions. The other 9 individuals had normal ECGs. Nine individuals had echocardiograms; seven were entirely normal; one individual had trace aortic insufficiency, trivial tricuspid regurgitation, and trace pulmonary regurgitation; and one individual had a small interatrial communication in the upper part of the atrial septum with a left-right shunt.
Orthopedic abnormalities were common in individuals with non-truncating variants (n = 12/14). Two of the individuals had bilateral clubfoot and congenital finger contractures. Two additional individuals had persistently adducted thumbs. Additional orthopedic abnormalities include kyphosis or scoliosis (n=2/14); congenital hip dysplasia (n=3/14); syndactyly (n=2/14); and joint hypermobility (n=3/14).
Additional variable features in the individuals with non-truncating variants include leg spasticity (n=1/14), tremor (n=2/14), hyperopia (n=4/14), strabismus (n=4/14).
For the most part, growth parameters are normal in individuals with non-truncating variants. Two individuals are microcephalic, and the remainder are normocephalic. Three individuals have short stature (<3rd %ile for height), including the two with history of clubfoot. Minor facial dysmorphisms are seen in a number of individuals (see figure 1), but there are no consistent features and only two of the individuals have the syndactyly characteristic of Timothy syndrome.
Figure 1.
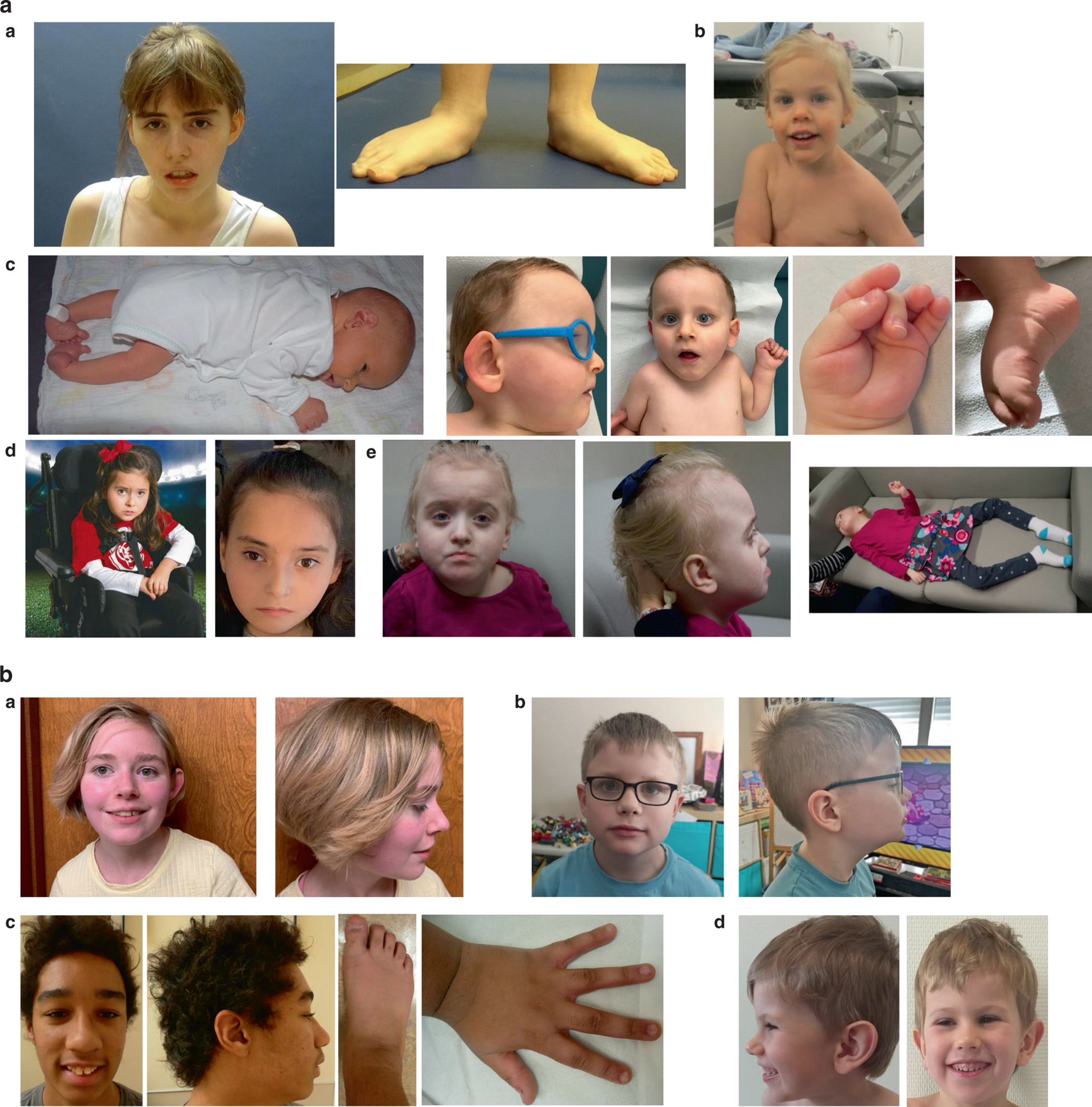
a: Spectrum of dysmorphology seen in individuals with CACNA1C non-truncating variants. A) Individual P1 at age 23 years. Note pes planovalgus B) Individual P5 at age 4 years C) Individual P10. Note clubfoot in infancy, and small appearing feet with prominent heels, adducted thumb, and camptodactyly later in childhood. D) Individual P12 at age 6 years of age. Note strabismus and kyphoscoliosis. E) Individual 13 at age 6 years old.
b: Spectrum of dysmorphology seen in individuals with CACNA1C truncating variants: A) Individual 15 at 15 years old. B) Individual 16a at 6 years old. Note large ears. C) Individual P18. Note mildly broad thumbs, hypoplastic 5th fingernails, broad halluces, and hypoplastic fifth toes. D) Individual P19 at age.
Whole-cell patch clamp analysis was conducted to assess functionality of the Cav1.2 calcium channels in individuals with missense variants (p.Leu614Pro), (p.Leu614Arg), (p.Leu657Phe) and (p.Leu1408Val). These variants were initially selected because they are located in or near regions where there appeared to be a clustering of variants in our cohort and also channel regions with likely functional importance. L614 is a recurrently affected residue, and located in close physical proximity to other variants (L601R and M611T) in the apex of the Domain II voltage sensor. L657F (Domain II, S5) is located in close proximity to V401M (Domain I, S6) in a neighboring domain, near the cytoplasmic gate. Lastly L1408V is nearly adjacent to V1411 (Domain IV, S5), also near the cytoplasmic gate. Review of the variants using MetaDome also indicates that the L657 and L1408 residues are located in regions highly intolerant to variation, and variants at homologous residues in related channels have been reported as pathogenic18. Missing from the functional characterization thus far are clusters of variants in S1 and outer pore of Domain I. Based on our initial characterization of 4 missense variants, as described below, we recognized that there were disparate effects and a more comprehensive study would be required as data from more patients and variants is collected. Cav1.2 (p.Leu1408Val) displays an overall reduction in current density (Figure 2) with unaltered activation kinetics compared WT Cav1.2. Missense variants (p.Leu614Pro and p.Leu657Phe) appear to transmit gain-of-function properties to the channel. Modest current increases were observed with the p.Leu614Pro variant, but were not statistically significant (Figure 2). A more pronounced, statistically significant enhancement of calcium current was observed with the p.Leu657Phe variant (Figure 2). The larger currents were also accompanied by shifts in the voltage-dependence of activation, in a hyperpolarizing direction for these variants. Western blot analysis was employed to identify whether the gain- or loss- of -function effects were a result of altered protein expression (Figure 2). Compared to WT Cav1.2, the p.Leu657Phe variant was associated with elevated proteins levels (Figure 2), providing explanation for the drastic increase in calcium current with this variant. Protein levels were similar to WT with the p.Leu614Pro and p.Leo1408Val variants. The p.Leu614Arg variant did not appear to confer any statistically significant change in current or properties of voltage-dependent gating (see Figure 2).
Figure 2: Characterization of Cav1.2 missense variants (L614P, L657F, L1408V, and L614R):
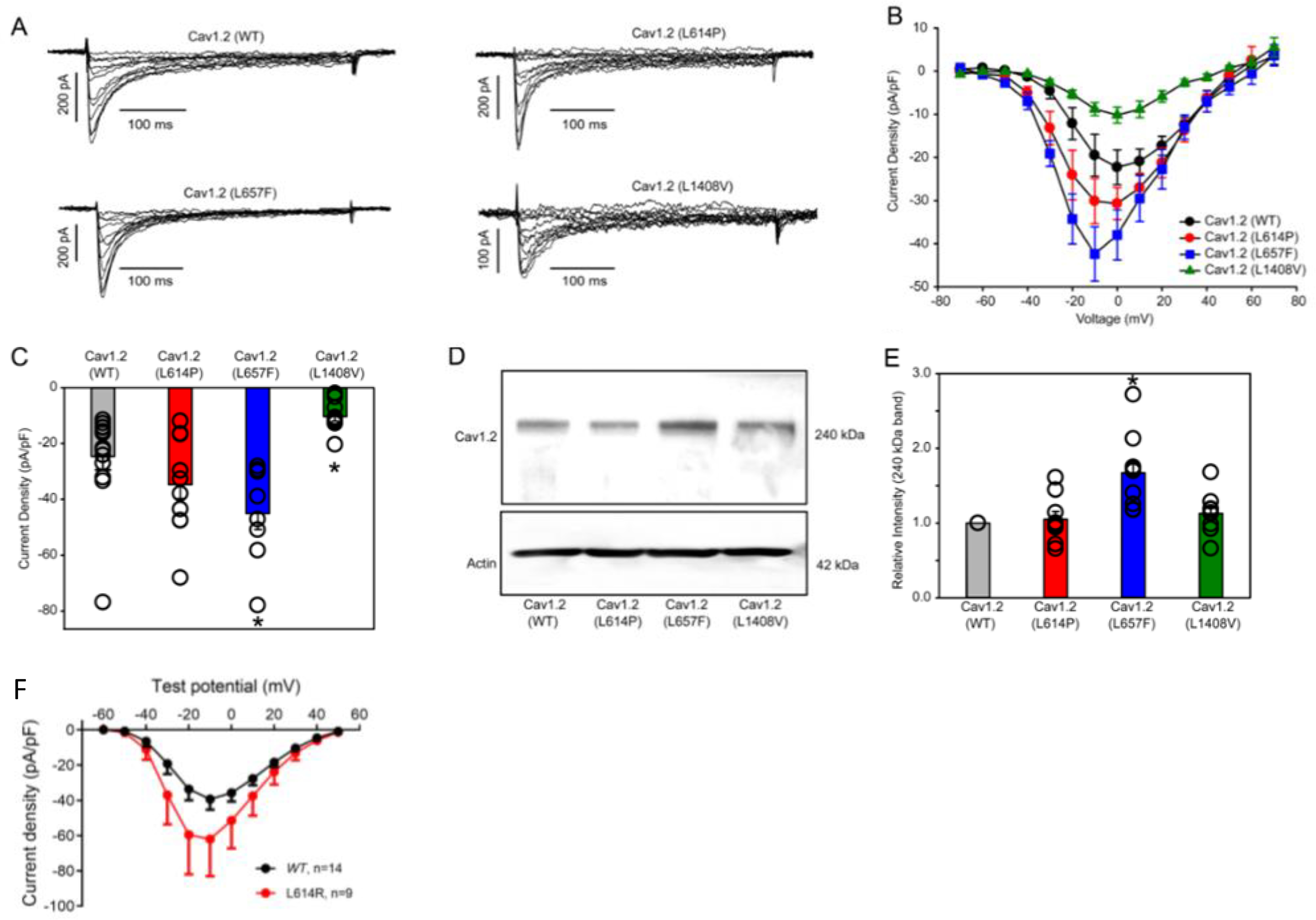
A), Representative whole-cell Ca2+ current traces after heterologous expression of Cavβ3, Cavα2δ1 and either Cav1.2 WT or Cav1.2 variants L614P, L657F, and L1408V in HEK293 cells. Solutions and voltage protocols used to elicit Ca2+ currents are described in the methods section. B), Averaged current-voltage (I-V) curves from whole-cell patch clamp measurements of HEK293 cells expressing Cavβ3, Cavα2δ1 and either Cav1.2 WT (n=14) or Cav1.2 variants L614P (n=11), L657F (n=8), and L1408V (n=10). C), Current density (pA/pF) analysis of the peak current of Cav1.2 WT (n=14) or Cav1.2 variants L614P (n=11), L657F (n=8), and L1408V (n=10) from cell recordings in A and B. Individual cells (open circles) are overlapped with the average current density (pA/pF), depicted as bars in the graph. Statistical analyses were carried out with a one-way ANOVA test. * p < 0.05 vs. Cav1.2 WT. D), Representative Western blot of whole-cell lysate from HEK293 cells heterologously expressed with Cavβ3, Cavα2δ1 and either Cav1.2 WT or Cav1.2 variants L614P, L657F, and L1408V. Actin was used as loading control. Antibodies used for immunoblotting of proteins are described in the methods section. E), Densitometry analysis of Cav1.2 WT (n=9) or Cav1.2 variants (L614P (n=9), L657F (n=9), and L1408V (n=9)) protein band intensities relative to the respective actin band intensities. Individual cells (open circles) are overlapped with the average relative Cav1.2 band intensities (240 kDa), depicted as bars in the graph. Statistical analyses were carried out with a one-way ANOVA test. * p < 0.05 vs. Cav1.2 WT. F) Averaged current-voltage (I-V) curves from whole-cell patch clamp measurements of HEK293 cells expressing CACNB2, CACNA2D, EGFP and either Cav1.2 WT (n=14) or Cav1.2 variant L614R (n=9). Differences in current density do not meet statistical significance.
To account for the possibility that a variant may create a cryptic splice site, the Transcript-inferred Pathogenicity (TraP) score was reviewed for all missense variants (Supplementary Data). The TraP score utilizes an algorithm that can detect creations and disruptions of cryptic splice sites by a variant19. The TraP scores reported for each missense variant in the cohort fell between 0–0.359, suggesting that none of the variants would be strongly predicted to affect splicing.
Truncating variants:
For individuals with truncating variants, prenatal history was unremarkable, other than one individual with maternal history of gestational diabetes and one individual with maternal history of cholestasis of pregnancy. All growth parameters at birth were normal. The neonatal period was uncomplicated in most individuals, with the exception of perinatal respiratory difficulties in one individual and feeding difficulties in another individual.
In contrast to the individuals with non-truncating variants, most of the individuals with truncating variants in our cohort had essentially normal early motor development and achieved independent ambulation. Five of 11 individuals with truncating variants have been diagnosed with autism, and 3 of these individuals are considered non-verbal. An additional individual was previously diagnosed with pervasive developmental disorder not otherwise specified. Only 1/11 of these individuals has a history of seizures, and this individual is currently seizure-free without treatment. Only three of the individuals in this group have hypotonia (tone is reported as unknown for 3/11). Four individuals are described as having poor balance or coordination, and two individuals have a tremor. One individual with a de novo truncating variant is described as having paroxysmal dyskinesia, and another with a paternally inherited truncating variant had a paroxysmal movement disorder in infancy that subsequently resolved.
Only 5/11 of individuals with truncating variants had an ECG, which was normal in all cases. One individual has undergone an echocardiogram that was normal. Seven of the individuals are reported to have minor dysmorphic features. Recurrent infections, including otitis media, were reported in 4/11 individuals; one of these individuals was diagnosed with hypogammaglobulinemia. One individual has a history of cryptorchidism.
Discussion:
We ascertained 25 individuals from 22 families with heterozygous variants in the CACNA1C gene presenting with a neurological phenotype variably associated with developmental delay, intellectual disability, autism, motor abnormalities, and epilepsy. While our sample size is limited, there were apparent phenotypic differences between individuals with non-truncating (missense variants, in-frame deletion) and truncating variants in our cohort.
Non-truncating variants in our cohort all occurred de novo. As a group, the individuals with non-truncating variants had more severe global developmental delay/intellectual disability and motor impairment compared to those with truncating variants. About half were non-verbal and non-ambulatory at most recent follow-up. The majority had hypotonia. One of the individuals in this group was formally diagnosed with autism, although six additional had autistic behaviors.
Epilepsy was much more common in the group with non-truncating variants, present in more than two-thirds of individuals. A spectrum of epilepsy syndromes and phenotypes were found, ranging from medically refractory infantile epileptic encephalopathy to later onset focal or generalized seizures with good response to anticonvulsants. Seizures were fever-sensitive in a number of individuals. There were no consistent EEG findings across the cohort. One individual was found to have excess fast activities similar to the previously reported infant with CACNA1C-related epileptic encephalopathy, but this is generally viewed as a non-specific finding and may be a medication effect5.
Two of the individuals in our cohort with non-truncating variants have a history of bilateral clubfoot and congenital finger contractures. Bilateral clubfoot and camptodactyly was also reported in the previously reported individual with neonatal epileptic encephalopathy associated with a de novo missense variant in CACNA1C5. This may represent a less common manifestation of the CACNA1C-related neurological disorder.
In contrast, three of the truncating variants in our cohort were familial (paternally inherited). These individuals as a group appeared to have more selective developmental abnormalities involving expressive language; 10/11 individuals in this group had delayed expressive language, whereas early motor development was essentially normal in all but one patient. Autism was more common in this group, present in 5/11 individuals. Finally, there appeared to be variable penetrance and expressivity of features amongst different family members with truncating variants.
Only one of the individuals in our cohort (P13; p.Val1411Leu) is known to have cardiac disease (prolonged QT), and none has physical features of Timothy syndrome, highlighting the novel, CNS-predominant clinical presentation of the reported individuals.
The CACNA1C channel subunit is broadly expressed in brain, heart, smooth muscle, endocrine tissue, and connective tissue, where it regulates cellular excitability, calcium signaling, and has downstream effects on gene expression20. In the central nervous system, CACNA1C shows the highest expression in the cerebral cortex, hippocampi, thalami, cerebellum, and suprachiasmatic nucleus5. It has also been implicated in the development of the serotonergic system21. These neuroanatomic substrates can account for the reported neurological phenotypes in our cohort.
The CACNA1C channel consists of 4 homologous domains (DI-DIV), each comprised of 6 transmembrane segments, cytoplasmic segments, and transmembrane linker segments1,22. Variants reported in association with Timothy syndrome cluster in the C-terminal end of the S6 transmembrane segment of domain I or in the DIII-DIV loop, and cause a gain-of-function either by decreasing voltage-dependent inactivation or increasing “window” current, respectively1,22. Similarly, non-syndromic Long QT syndrome has been associated with gain-of-function variants in the DI-DII loop and elsewhere23. The reported protein alterations in our cohort are spread throughout the protein, although there is a clustering of missense variants in one of the linker segments of DII (exon 13) and DIV (exon 35), suggesting the potential significance of these regions of the protein in the function of CACNA1C in the central nervous system (Figure 3).
Figure 3: Location of CACNA1C variants in 25 individuals.
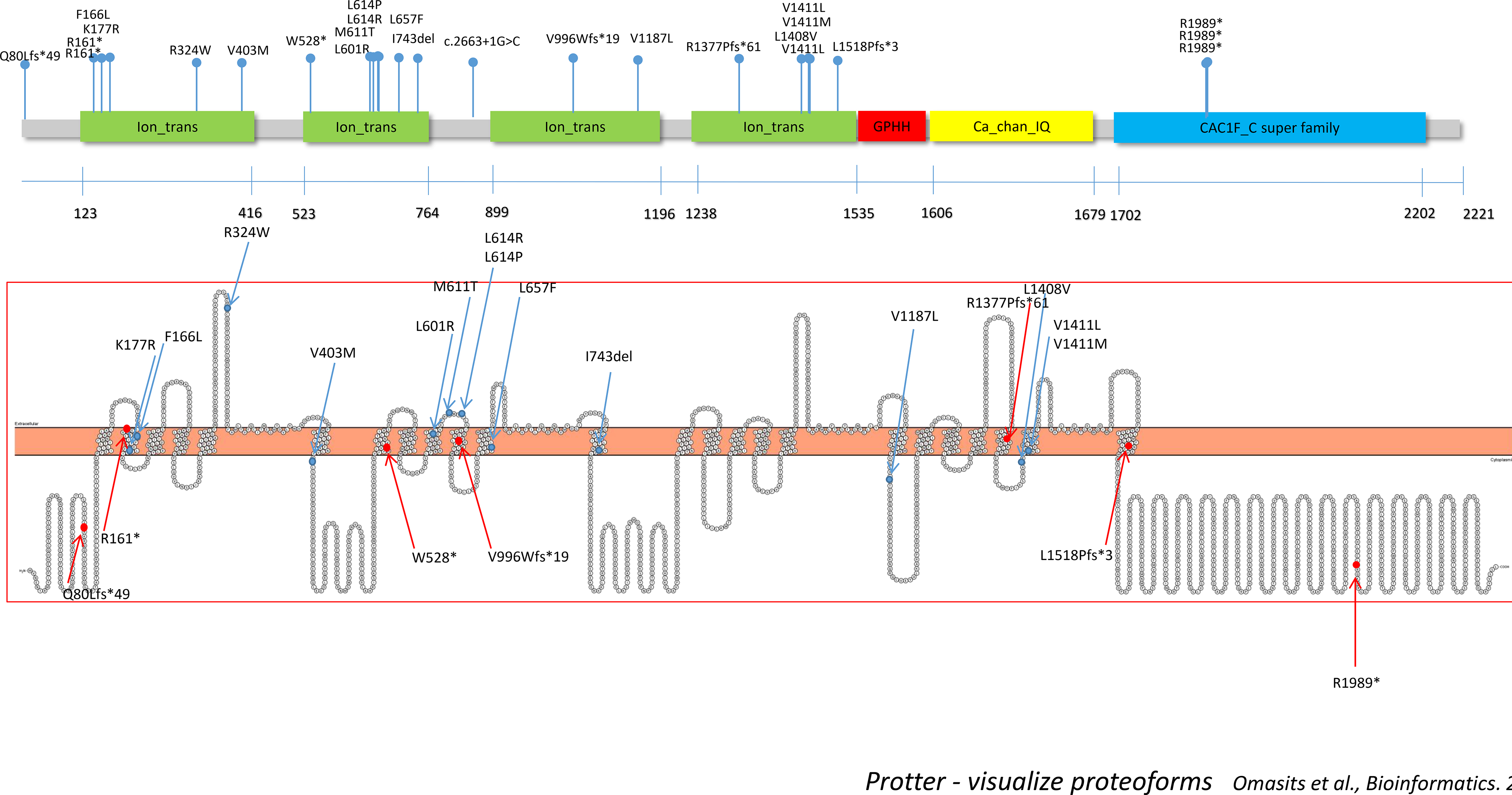
Blue dots indicate missense variants or in-frame deletion, and red dots indicate truncating variants. Protein alterations are spread throughout the protein, although there is a clustering of missense variants in one of the linker segments of domain II (exon 13) and domain IV (exon 35), suggesting the potential significance of these regions of the protein in the function of CACNA1C in the central nervous system. Figure was created using open-source tool, Protter30.
CACNA1C functions as a component of a larger complex with accessory subunits, including auxiliary β and α2δ subunits20. There are tissue-specific differences in these accessory subunits between brain and heart that allow for modifications in channel function. In addition, CACNA1C undergoes extensive alternative splicing, with some transcripts undergoing additional RNA editing24,25. It has recently been suggested that there are 241 unique CACNA1C transcripts in the human brain, and that these may encode channels with altered function and different kinetics24. Further, different brain regions may themselves exhibit different CACNA1C splicing profiles24. These complexities in the interaction of CACNA1C with different accessory subunits, and plethora of CACNA1C isoforms in the central nervous system make interpretation of the effects of rare variants in this gene challenging.
To provide further insight into the molecular mechanism of disease in our cohort, we performed patch clamping on a subgroup of variants in our cohort. These studies demonstrated diverse effects on channel current associated with specific variants, including loss-of-function (p.Leu1408Val), neutral effect (p.Leu614Pro, p.Leu614Arg), and gain-of-function alterations (p.Leu657Phe). It is important to note that these functional studies were conducted only in the canonical transcript, and so it is possible that employing alternate transcripts could yield different results. Also, the patch clamp experiments of the p.Leu1408Val, p.Leu657Phe, and p.Leu614Pro variants yielding abnormal results were conducted using the CACNB3 accessory subunit, whereas the experiments for the p.Leu614Arg variant yielding normal results utilized CACNB2. The reason for this discrepancy is that the work evolved in multiple centers, as contributing clinical groups were simultaneously identifying CACNA1C variants and characterizing their effects with tools available to them. The landscape of potential combinations of beta subunits (each with multiple splice variants) and CACNA1C variants is broad, and it is uncertain yet which conditions are best to reveal relevant functional disruptions. The CACNB3 accessory subunit has a relatively higher expression in brain versus heart compared to CACNB2. The specific accessory subunits used for the patch clamp experiments could potentially affect the results and may also have implications regarding why the particular variants we report have predominant CNS manifestations in the relative absence of cardiac findings.
Overall, the missense variants tested thus far did not cause pronounced changes in the voltage-dependence or kinetics of gating. Variants differed in terms of overall current density and protein expression. The gain-of-function effect of the Leu657Phe variant is most likely explained by an increase in protein expression, as both the current density and protein level increased by approximately 2-fold. The Leu1408Val variant appears to have a clear effect on overall channel activity, as current density associated with this variant was significantly reduced to approximately 30% of control, but the protein expression was unchanged.
The finding that both CACNA1C gain of function and loss of function can result in a neurological phenotype is not unexpected, and there is a precedent for other CNS-expressed calcium channels to be dosage sensitive (eg. CACNA1A)26. Timothy syndrome is caused by CACNA1C gain of function, and frequently has a neurological phenotype, including intellectual disability, autism, hypotonia, and rarely epilepsy. Conversely, truncation/deletion of CACNA1C has previously been reported in association with developmental delay/intellectual disability, and it has been suggested that CACNA1C is haploinsufficient10. This is further supported by constraint analysis on the gnomAD dataset, demonstrating an observed/expected ratio of for predicted loss-of-function variants of 0.05 and probability of loss-of-function intolerance (pLI) score of 1.027. It is still not clear how loss-of-function variants in CACNA1C result in an epilepsy syndrome, since reduced channel activity in excitatory pyramidal neurons would theoretically be protective against seizures. It is possible that the CACNA1C channel is also expressed in GABAergic inhibitory interneurons, analogous to the mechanism seen with loss-of-function SCN1A pathogenic variants that result in Dravet syndrome28.
In individuals with missense variants in our cohort associated with a loss-of-function on patch clamp studies, it remains unknown whether the basis is haploinsufficiency or a dominant negative effect, potentially by encoding non-functioning channels that compete with wild-type for accessory subunits, or preventing the correct folding of wild type protein. The more severe neurodevelopmental and epilepsy phenotype in patients with missense, predicted loss-of-function variants compared to those with whole gene/exonic deletions would better support a dominant-negative effect.
In terms of the variant with an apparent neutral effect on patch clamp experiments, there are a number of possible explanations. As previously stated, it is possible that employing alternate transcripts or accessory subunits for patch clamp studies for these variants could have yielded different results. Splice-variant dependent gating changes have previously been demonstrated in another calcium channel subunit gene, CACNA1A29. Another possibility is differential effects of a variant in vivo due to protein-protein interactions. Finally, one could hypothesize that specific CACNA1C transcripts serve non-canonical roles in the central nervous system that were not captured on our patch clamp studies. Further exploration of these variants in more complex systems such as iPSC-derived neurons, and a more comprehensive study of additional variants, may reveal trends in underlying functional outcomes.
Our study was underpowered to correlate phenotypic differences in patients with missense variants between those with predicted gain or loss-of-function on patch clamp studies. Further, not all missense variants were studied with patch clamp experiments.
In conclusion, we delineate the phenotypic spectrum of a CACNA1C-related neurological disorder variably associated with developmental delay, intellectual disability, autism, epilepsy, hypotonia, and ataxia. Significant cardiac abnormalities were absent in all but one of the individuals in our cohort at most recent follow-up. Although our sample size is limited, there appear to be phenotypic differences between individuals with truncating and non-truncating variants. While it is tempting to assume that these truncating variants result in a loss-of-function through haploinsufficiency, given the complexity of alternate splicing of this gene, this requires further validation. Further efforts to determine the precise molecular mechanism(s) for the neurological-predominant symptoms in some individuals with variants in the CACNA1C gene will provide additional insight into pathophysiology and potentially inform more targeted treatment.
Supplementary Material
Acknowledgements:
Research reported in this manuscript was supported by the NIH Common Fund, through the Office of Strategic Coordination/Office of the NIH Director under Award Number(s) [U01HG007709 (Baylor College of Medicine) and U01HG007672 (Duke University to Dr. Vandana Shashi)]. Additional funding for this project was under Award Number 1RO1HD090132-01A1 (Cornell University to Dr. Geoffrey Pitt). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Additional funding was provided by a Canadian Institutes of Health Research Grant (MOP-97988 to Harley T. Kurata), and a Rowland and Muriel Haryett Fellowship (University of Alberta, to Shawn M. Lamothe) and a fellowship from the Ministry of Education and Research of the Community of Madrid to Marta Pacio Míguez (B2017/BMD-3721), and microgrant from the Rare Disease Foundation (P.Y. Billie Au and Harley T. Kurata).
Sequencing and analysis was supported by the National Human Genome Research Institute grants UM1 HG008900 and R01 HG009141. K.A.D., B.C., and E.O. were supported the National Institute of Mental Health U01 MH119689.
Footnotes
Ethics Declaration:
Signed consent was obtained from parents/legal guardians for publication of photographs, in accordance with the institutional review boards/ethics committees at each individual institution. Clinical (de-identified and identifiable) and genetic data from all cases was analyzed with IRB approval under the UDN Study protocol (15-HG-0130) approved by the NIH IRB.
Data Availability:
The data for P12 (VCV000190670.1) and P13 (VCV00052282.1) have been submitted to ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/) in accordance with the requirements of the Undiagnosed Diseases Network. The remaining de-identified data and variant information are available individually upon request.
References:
- 1.Splawski I, Timothy KW, Sharpe LM, Decher N, Kumar P, Bloise R, Napolitano C, Schwartz PJ, Joseph RM, Condouris K, et al. Ca(V)1.2 calcium channel dysfunction causes a multisystem disorder including arrhythmia and autism. Cell, 119(1) (2004), pp. 19–31. [DOI] [PubMed] [Google Scholar]
- 2.Zhang Q, Chen J, Qin Y, Wang J, Zhou L. Mutations in voltage-gated L-type calcium channel: implications in cardiac arrhythmia. Channels (Austin), 12(1) (2018), pp. 201–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hosseini SM, Kim R, Udupa S, Costain G, Jobling R, Liston E, Jamal SM, Szybowska M, Morel CF, Bowdin S, et al. Reappraisal of Reported Genes for Sudden Arrhythmic Death: Evidence-Based Evaluation of Gene Validity for Brugada Syndrome. Circulation, 138(12) (2018), pp. 1195–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Green EK, Grozeva D, Jones I, Jones L, Kirov G, Caesar S et al. The bipolar disorder risk allele at CACNA1C also confers risk of recurrent major depression and of schizophrenia. Mol Psychiatry, 15 (2009), pp. 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bozarth X, Dines JN, Cong Q, Mirzaa GM, Foss K, Merritt JL II, Thies J, Mefford HC, Novotny E. Expanding clinical phenotype in CACNA1C related disorders: From neonatal onset severe epileptic encephalopathy to late-onset epilepsy. American Journal of Medical Genetics, 176(12)(2018), pp. 2733–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Iossifov I, O’Roak BJ, Sanders SJ, Romemus M, Krumm N, Levy D, Stressman HA, Witherspoon KT, Vives L, Patterson KE, et al. The contribution of de novo coding mutations to autism spectrum disorder. Nature, 515(7526) (2014), pp. 216–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deciphering Developmental Disorders Study. Prevalence and architecture of de novo mututations in developmental disorders. Nature, 542(7642) (2017), pp. 433–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mio C, Passon N, Baldan F, Bregant E, Monaco E, Mancini L, Demori E, Damante G. CACNA1C haploinsufficiency accounts for the common features of interstitial 12p13.33 deletion carriers. Eur J. Med Genet, 63(4)(2020),pp. 103843. [DOI] [PubMed] [Google Scholar]
- 9.Roberts JL, Hovanes K, Dasouki M, Manzardo AM, Butler MG. Chromosomal microarray analysis of consecutive individuals with autism spectrum disorders or learning disability presenting for genetic services. Gene, 535(1)(2014), pp. 70–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Quintela I, Eirís J, Gómez-Lado C, Pérez-Gay L, Dacruz D, Cruz R, Castro-Gago M, Míguez L, Carracedo Á, Barros F. Copy number variation analysis of patients with intellectual disability from North-West Spain. Gene, 626 (2017), pp. 189–199. [DOI] [PubMed] [Google Scholar]
- 11.Chen J, Sun Y, Liu X, Li J. Identification of a novel mutation in the CACNA1C gene in a Chinese family with autosomal dominant cerebellar ataxia. BMC Neurol, 19(1)(2019), pp. 157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sobreira N, Schiettecatte F, Valle D, Hamosh A. GeneMatcher: A Matching Tool for Connecting Investigators with an Interest in the Same Gene. Hum Mutat. 36(10)(2015), pp. 928–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Philippakis AA, Azzariti DR, Beltran S, Brookes AJ, Brownstein CA, Brudno M, Brunner HG, Buske OJ, Carey K, Doll C, et al. The Matchmaker Exchange: A Platform for Rare Disease Gene Discovery. Hum Mutat. 36(10)(2015), pp.915–921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hennessey JA, Boczek NJ, Jiang Y, Miller JD, Patrick W, Pfeiffer R, Sutphin BS, Tester DJ, Barajas-Martinez H, Ackerman MJ, et al. Voltage-dependent inactivation, increased CaV1.2 channel window current, and arrhythmogenesis. PLos One. 9(9)(2014), pp. e106982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lin Y, McDonough S, Lipscombe D. Alternative splicing in the voltage-sensing region of N-Type CaV2.2 channels modulates channel kinetics. J Neurophysiol. 92(5)(2004):2820–30. [DOI] [PubMed] [Google Scholar]
- 16.Wang AW, Yau MC, Wang CK, Sharmin N, Yang RY, Pless SA, Kurata HT. Four drug-sensitive subunits are required for maximal effect of a voltage sensor-targeted KCNQ opener. J Gen Physiol. 150(10)(2018),pp.1432–1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Auton A, Abecasis G, Altshuler D, et al. A global reference for human genetic variation. Nature. 526 (2015), pp.68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wiel L, Baakman C, Gilissen D, Veltman JA, Vriend G, Gilissen C. MetaDome: Pathogenicity analysis of genetic variants through aggregation of homologous human protein domains. Hum Mutat. 40(8)(2019), pp.1030–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gelfman S, Wang Q, McSweeney KM, Ren Z, La Carpia F, Halvorsen M, Schoch K, Ratzon F, Heinzen EL, Boland MJ, Petrovski S, Goldstein DB. Annotating pathogenic non-coding variants in genic regions. Nat Commun. 8(1)(2017), pp. 236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Striessnig J, Pinggera A, Kaur G, Bock G, Tuluc P. L-type Ca2+ channels in heart and brain. Wiley Interdisciplinary Reviews: Membrane Transport and Signaling, 3(2)(2014), pp. 15–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ehlinger DG, Commons KG. Cav1.2 L-type calcium channels regulate stress coping behavior via serotonin neurons. Neuropharmacology, 144 (2019), pp.282–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Betzenhauser MJ, Pitt GS, Antzelevitch C. Calcium Channel Mutations in Cardiac Arrhythmia Syndromes. Curr Mol Pharmacol, 8(2)(2015), pp.133–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wmhoner K, Friedrich C, Stallmeyer B, Coffey AJ, Grace A, Zumhagen S, Seebohm G, Ortiz-Bonnin B, Rinne S, Sachse FB, et al. Gain-of-function mutations in the calcium channel CACNA1C (Cav1.2) cause non-syndromic long-QT but not Timothy syndrome. J. Mol. Cell Cardio, 80(2015), pp.186–195. [DOI] [PubMed] [Google Scholar]
- 24.Clark MB, Wrzesinski T, Garcia AB, Hall NAL, Kleinman JE, Hyde T, Weinberger DR, Harrison PJ, Haerty W, Tunbridge EM. Long-read sequencing reveals the complex splicing profile of the psychiatric risk gene CACNA1C in human brain. Molecular Psychiatry, 25(2020), pp.37–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lipscombe D, Andrade A. Calcium Channel Cava1 Splice Isoforms – Tissue Specificty and Drug Action. Curr Mol Pharmacol, 8(1)(2015), pp. 22–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sintas C, Carreno O, Fernandez-Castillo N, Corominas R, Vila-Pueyo M, Toma C, Cuenca-Leon E, Barroeta I, Roig C, Volpini V, et al. Mutation Spectrum in the CACNA1A Gene in 49 Patients with intolEpisodic Ataxia. Sci Rep. 31(7)(1)(2017), pp.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Karczewski KJ, Francioli LC, Tiao G, Cummings BB, Alfoldi J, Wang Q, Collins RL, Larichia KM, Ganna A, Birnbaum DP, et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 581 (2020), pp. 434–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yu FH, Mantegazza M, Westenbroek RE, Robbins CA, Kalume F, Burton KA, Spain WJ, McKnight GS. Scheuer T, Catterall WA. Reduced sodium current in GABAergic interneurons in a mouse model of severe myoclonic epilepsy in infancy. Nature Neuroscience. 9(9)(2006),pp.1142–1149 [DOI] [PubMed] [Google Scholar]
- 29.Pinggeraa A, Negroa G, Tuluc a P, Brownb MJ, Lieb a A, Striessni J. Gating defects of disease-causing de novo mutations in Cav1.3 Ca2+ channels. Channels. 12(1)(2018), pp.388–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Omasits U, Ahrens CH, Müller S, Wollscheid B. Protter: interactive protein feature visualization and integration with experimental proteomic data. Bioinformatics. 15;30(6)(2014):884–886. doi: 10.1093/bioinformatics/btt607. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data for P12 (VCV000190670.1) and P13 (VCV00052282.1) have been submitted to ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/) in accordance with the requirements of the Undiagnosed Diseases Network. The remaining de-identified data and variant information are available individually upon request.


