Summary:
Many TRP channels respond to diverse stimuli and conditionally conduct small and large cations. Such functional plasticity is presumably enabled by a uniquely dynamic ion selectivity filter that is regulated by physiological agents. What’s currently missing is a ‘photo series’ of intermediate structural states that directly address this hypothesis and reveal specific mechanisms behind such dynamic channel regulation. Here, we exploit cryo-EM to visualize conformational transitions of the capsaicin receptor, TRPV1, as a model to understand how dynamic transitions of the selectivity filter in response to algogenic agents, including protons, vanilloid agonists, and peptide toxins, permit permeation by small and large organic cations. These structures also reveal mechanisms governing ligand binding substates, as well as allosteric coupling between key sites that are proximal to the selectivity filter and cytoplasmic gate. These insights suggest a general framework for understanding how TRP channels function as polymodal signal integrators.
Graphical Abstract
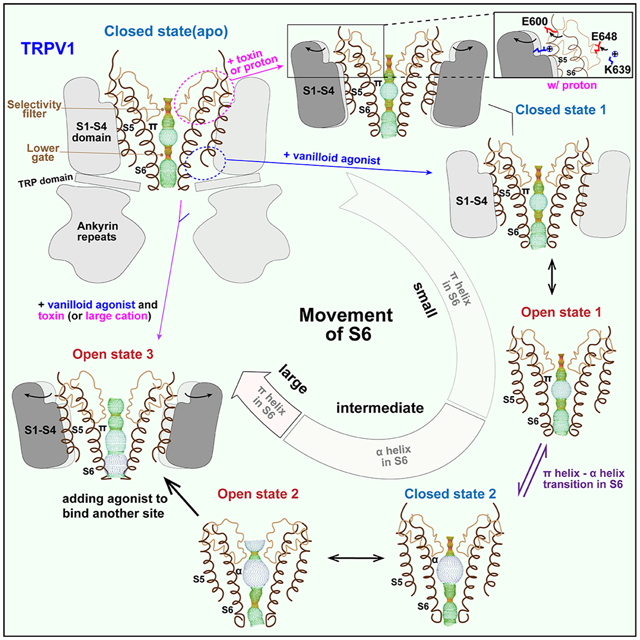
In Brief:
Visualizing substates of the TRPV1 channel reveals mechanisms underlying selectivity filter plasticity, ligand binding, and allosteric coupling.
INTRODUCTION
TRP channels are polymodal signal integrators that detect and respond to distinct physiologic stimuli. This is perhaps best exemplified by TRPV1 (Caterina et al., 1997), a heat-activated channel whose sensitivity to inflammatory agents, including extracellular protons (Jordt et al., 2000; Tominaga et al., 1998) and bioactive lipids (Rohacs, 2015; Starowicz et al., 2007) underlies its critical role in acute and persistent pain (Julius, 2013). Functional and structural studies have identified sites on the channel that confer sensitivity to these and other modulatory agents, and that lie in close proximity to restriction points along the ion permeation pathway - one in the ‘upper’ extracellular half of pathway near the selectivity filter, and another within the ‘lower’ half of the pathway, near the cytoplasmic gate (Cao et al., 2013).
Some TRP subtypes, such as TRPV5 and V6, are highly selective for divalent cations and have rigid selectivity filters resembling those of voltage-gated potassium channels (Dang et al., 2019; Hughes et al., 2019; McGoldrick et al., 2018). Others, such as TRPV1, V2, V3, and TRPA1, which accommodate monovalent, divalent, and large organic cations (Binshtok et al., 2007; Munns et al., 2015; Puopolo et al., 2013), possess outer pore regions that are plastic and undergo substantial widening during channel activation (Cao et al., 2013; Deng et al., 2020; Huynh et al., 2016; Paulsen et al., 2015; Pumroy et al., 2019; Zhao et al., 2020; Zubcevic et al., 2018a; Zubcevic et al., 2018b), presumably allowing for dynamic ion selectivity (Chung et al., 2008; Jara-Oseguera et al., 2019). While the physiological consequences of such structural plasticity are not fully understood, for TRPV1 or TRPA1 this may facilitate calcium-dependent release of peptide transmitters from sensory nerve endings to promote neurogenic inflammation and pain hypersensitivity (Lin King et al., 2019). Plasticity of the upper restriction may also allow permeation by cationic lidocaine derivatives, which can be exploited to deliver local anesthetics to TRPV1-expressing nociceptors (Binshtok et al., 2007). But whether and how the upper restriction accommodates both small and large cations remain unclear.
Aside from effects on ion permeation, conformational changes in the outer pore region influence TRPV1 gating. Indeed, algogenic gating modifiers such as spider toxins and extracellular protons interact with residues in the outer pore region to facilitate opening of the cytoplasmic gate. However, while pharmacological and biophysical studies strongly suggest that the upper and lower restrictions are allosterically coupled (Bohlen et al., 2010), structural mechanisms underlying such communication remain unclear. Also at issue is the relative contribution of evolutionarily conserved structural elements, such as S1–S4 voltage sensor-like domain (VSLD) or S4–S5 linker region, to gating of TRPs when compared with their topologically similar voltage-gated ion channel cousins. What role, if any, do such regions play in the allosteric coupling between upper and lower restrictions, and how do physiological modulators, such as inflammatory agents, natural products, or drugs exploit these interactions to regulate channel activity? Furthermore, many TRP channels are homo-tetrameric, raising questions about the functional equivalency of ligand binding sites and whether drugs or physiological modulators interact in a cooperative, sequential, or random manner to trigger gating.
Among TRP channels, TRPV1 enjoys the richest pharmacology, which we leverage here to capture snapshots of nanodisc-reconstituted channels using single particle cryo-electron microscopy (cryo-EM). These substates represent key structural intermediates in which the status of small and large permeating cations, stepwise engagement of ligands with TRPV1, and stoichiometry of ligand binding can be directly visualized at key regulatory points. Furthermore, these substates highlight transitions between channel conformations, revealing a two-step mechanism whereby extracellular protons, a key algogenic component of the inflammatory milieu (Bevan and Geppetti, 1994), alter the structure of the upper restriction to facilitate opening of the lower gate. These findings provide a framework for understanding the uniquely dynamic behavior of this extended family of excitatory ion channels.
RESULTS
A dynamic upper restriction regulates ion permeability
Previously, we stabilized the open state of TRPV1 with a combination of agents, including a peptide spider toxin (double-knot toxin, DkTx) and a vanilloid agonist (resiniferatoxin, RTX), that target upper and lower restrictions, respectively (Cao et al., 2013; Gao et al., 2016). Here, we sought to trap channel states using DkTx alone to focus in on conformational transitions that primarily target the outer pore and upper restriction. DkTx triggers transient single channel events preceding conversion to a stable open state (Bohlen et al., 2010), suggesting that the channel samples intermediate states before it is fully opened by the toxin. We reconstituted a minimal functional TRPV1 channel (Liao et al., 2013) into lipid nanodiscs and applied DkTx prior to freezing cryo-EM grids. Remarkably, we identified a range of substates within the same cryo-EM dataset that are characterized by distinct toxin interactions, pore diameters, coordination of ions by G643 backbone carbonyls (which form the upper restriction / selectivity filter), and accumulation of waters above and below this point (Figures 1, S1 to S4, and Table S1). As detailed below, these states reveal a series of presumptive sequential events that highlight the dynamic nature of this site, as well as stepwise engagement between DkTx and the channel (Movie S1).
Figure 1. Conformational transitions in the selectivity filter.
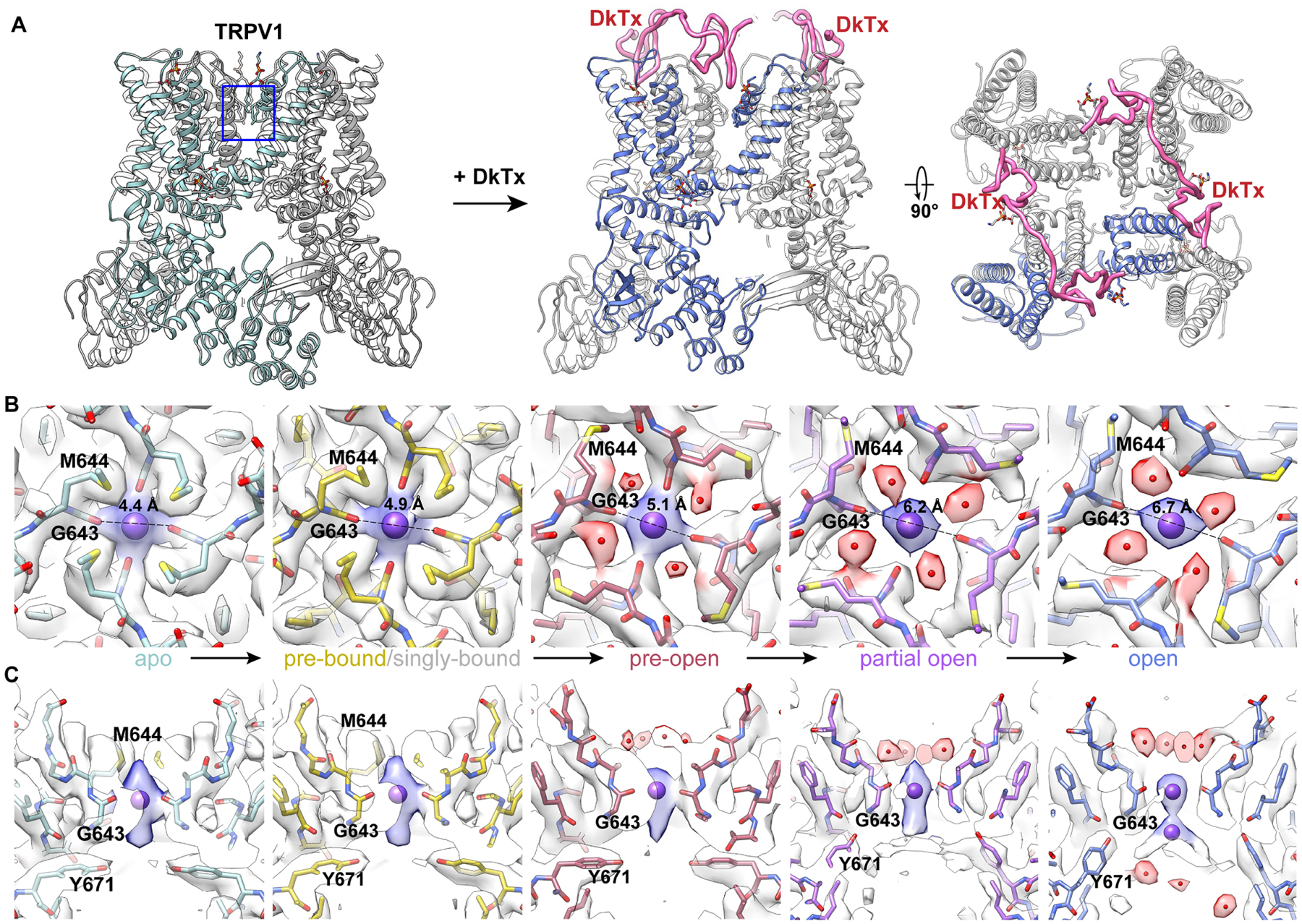
(A) Ribbon diagram of minimal TRPV1, illustrating location of the selectivity filter and binding of DkTx.
(B) and (C) Top and side views of the selectivity filter formed by G643 carbonyls with each panel representing a specific substate, as labeled. The distance between opposing carbonyls is marked in (B). From left to right, panels are arranged in a presumptive order of conformational transitions from apo (left) to open (right) via intermediate substates induced by DkTx. The density in between carbonyls is interpreted as a Na+, which elongates as coordination loosens and finally splits in two in the open pore when the selectivity filter diameter is the largest.
First, we observed a ‘pre-bound’ state in which there is evidence of DkTx interaction before a clear toxin density is seen, including movement of elements constituting the known toxin footprint, such as the loop between the pore helix and S5 and the position of specific lipids within the outer membrane leaflet. In the next presumptive state we observe engagement of a single DkTx molecule to one of two symmetric binding sites on the homo-tetrameric channel, albeit with relatively weak density, suggesting transient attachment to a channel otherwise resembling the pre-bound state (Figure S3A, S3B). Next, we see a ‘pre-open’ conformation containing two well-resolved, fully bound DkTx molecules in which the main change involves rotation of M644 away from the central axis (Figure 1B), as previously described (Cao et al., 2013). Finally, with two fully bound DkTx molecules, we observe two states (‘partial open’ and ‘open’) in which the distance between opposing G643 carbonyls increases from 4.4Å in the apo state to 6.2 and 6.7Å, respectively. This is accompanied by a range of poses of the resident regulatory phosphatidylinositol (PI) lipid within the vanilloid binding pocket, where the head group and one aliphatic chain of the lipid becomes increasingly mobile as the lower gate diameter increases (Figure S3C), and an annular lipid inserts its tail into a hydrophobic cleft between S5 and S6 helices of neighboring subunits (Figure S3D).
A particularly interesting feature of these transitional substates is the identification of densities corresponding to presumptive ions and waters within the pore (Figure 1). In apo and pre-bound structures, we see a well-resolved density in the middle of the upper restriction, which we interpret as a fully or partially dehydrated Na+ coordinated by four backbone carbonyls of G643 (Figures 1B). Water begins to appear in the pre-open state, where it is coordinated by backbone carbonyls of G645, coinciding with rotation of M644 side chains away from the central axis (Figure 1B). Transitioning to partial open and open states, coordination of the ion by G643 becomes weaker as the density progressively elongates along the vertical axis to eventually split into two discrete entities (Figure 1C), likely reflecting changes in probability distributions of cations within the filter at different conformations. When correlated with opening of the lower gate, the observed increase in water accessible volume in the cavity below (Figure S4A) should facilitate rehydration of cations emerging from the selectivity filter (Figure 1C, S4A and S4B).
Allosteric opening of the lower gate
DkTx engages the outer pore region of TRPV1, raising the question of how it allosterically opens the lower gate to elicit membrane currents. In examining the transition from the pre-bound to open, the VSLD is seen to tilt backward progressively (Figure 2A), consequently moving S5 (which connects the VSLD with the lower gate) and S6 away from the pore in partial open and open states (Figure 2B). By comparing the relative positions of two points (V469 at the top of VSLD and I679 at the lower gate) in apo and substate structures identified above, we see a sequential movement beginning with backward tilting of VSLD at pre-bound and pre-open states that does not move I679. Further sideward tilting or twisting of VSLD at partial open and open states is then accompanied by backward movement of S6 and a π- to α-helix transition that introduces a kink at the bottom of S6 and rotates the lower half of S6 by one residue, replacing I679 with L678 (Figure 2C, S4C). Furthermore, M682 is rotated from a hydrophobic nest, where its side chain density is fully resolved (Figure S4D), to a position facing the pore, where it now forms the narrowest point in the lower gate (Figure S4A, S4B). The widest distance between sulfur atoms of two opposing M682 in the open conformation is 6.5Å, producing a pore radius of 1.4Å, which is sufficient to permeate water and partially dehydrated cations (Figure S4J). When facing the pore, M682 looses its hydrophobic interaction with neighboring residues and becomes flexible (as evidenced from the less well-resolved side chain densities in these two substates; Figure S4E–I), likely making the pore radius larger than that measured from the built model. We thus designate these two substates as ‘partial open’ and ‘open.’ Additionally, S6 shifts continually backward (Figure S4C), consistent with progressive gating transitions.
Figure 2. Allosteric opening of the lower gate.
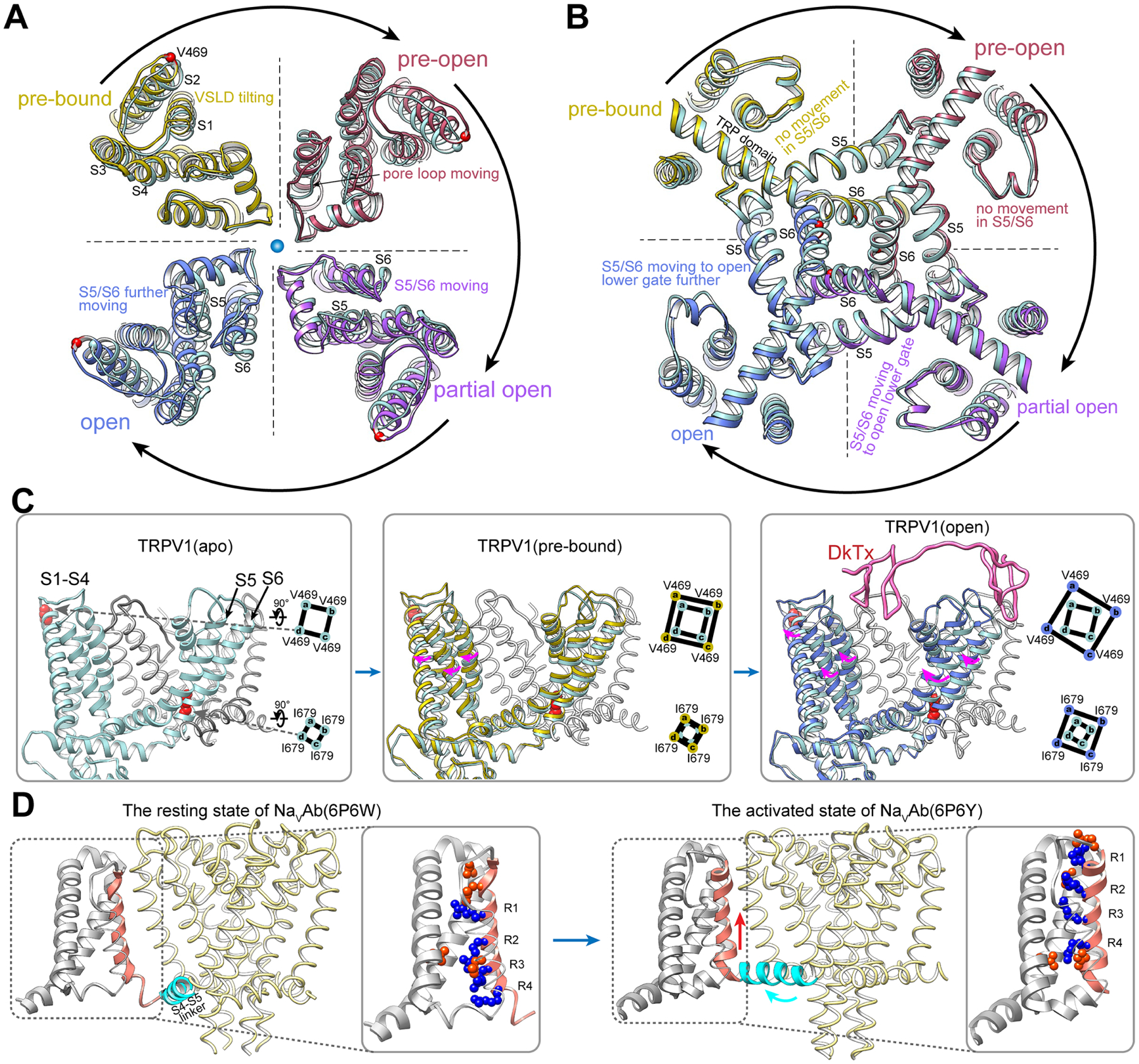
(A) and (B) Slab views of the selectivity filter (A, viewed from extracellular side) and lower gate (B, viewed from cytoplasmic side) of the channel. In clockwise order, each quarter shows overlay of the apo structure (light blue) with an intermediate observed upon DkTx binding.
(C) V469 and I679 are taken as reference points (left panel) to reveal the relationship between VSLD tilting and lower gate opening.
(D) In voltage-gated sodium channels (NavAb, PDB code 6P6W), the VSD gates the channel in response to membrane voltage via up and down movements of the arginine-containing S4 helix that move the S4–S5 linker.
Only when DkTx is fully engaged with the channel (in the open state), is the tilt and twist of VSLD sufficient to allosterically open the lower gate. Thus, we propose that the VSLD is a main module that allosterically couples changes in the outer pore to the lower gate, and does so in a ‘tunable’ manner, as further illustrated below in the case of proton modulation. Our observation of VSLD movement relative to the pore domain is reminiscent of previously described rotation of VSLD relative to the pore region in TRPV3 upon activation by 2-APB (Singh et al., 2018). In contrast this region moves differently in voltage-gated channels, where sliding of the S4 helix in response to changes in membrane voltage promotes backward movement of S5 to open the gate (Figure 2D) (Wisedchaisri et al., 2019), highlighting a mechanistically distinct manner in which this architecturally conserved domain regulates channel gating.
An adaptive filter accounts for TRPV1 non-selectivity
When activated by vanilloid agonists, TRPV1 is permeable to mono- and divalent cations, as well as large organic cations (Binshtok et al., 2007; Munns et al., 2015; Puopolo et al., 2013). However, our previous studies did not reveal changes in selectivity filter conformation unless DkTx was present to stabilize the outer pore region (Cao et al., 2013). To ask whether this same dynamic change in pore radius accounts for vanilloid-evoked permeation by large cations, we determined structures of the RTX-activated channel in the presence of both N-methyl-d-glucamine (NMDG) and the lidocaine derivative QX-314 (Figure S1G – S1I), each of which are reported to act as permeating organic cations (Jara-Oseguera et al., 2019; Puopolo et al., 2013). The first snapshot captures the channel in a conformation resembling that of the open state seen with DkTx, and in which only a Na+ ion is loosely coordinated within the filter (Figure 3A). In the next snapshot, we see a density that can be modeled as NMDG just as it engages with the upper restriction, displacing the Na+ ion further down the pore. In the last snapshot, NMDG has entered completely into the restriction site. In each case, the overall conformation of the outer pore resembles that stabilized by DkTx in which M644 is rotated away from the pore, consistent with prior mutagenesis studies implicating the outer pore domain as a key determinant of dynamic ion selectivity (Munns et al., 2015). Furthermore, the pore loop is shifted, the VSLD is tilted backward (Figure 3B), and the channel is fully open (Figure 3C and 3F). The surprising similarity between the RTX/NMDG and RTX/DkTx structures (Figure 3D and E) suggests that dynamic behavior of the selectivity filter is enhanced by the vanilloid agonist and stabilized by DkTx or a large cation (Figure 3G, Movie S2).
Figure 3. Permeation by NMDG illustrates selectivity filter plasticity.
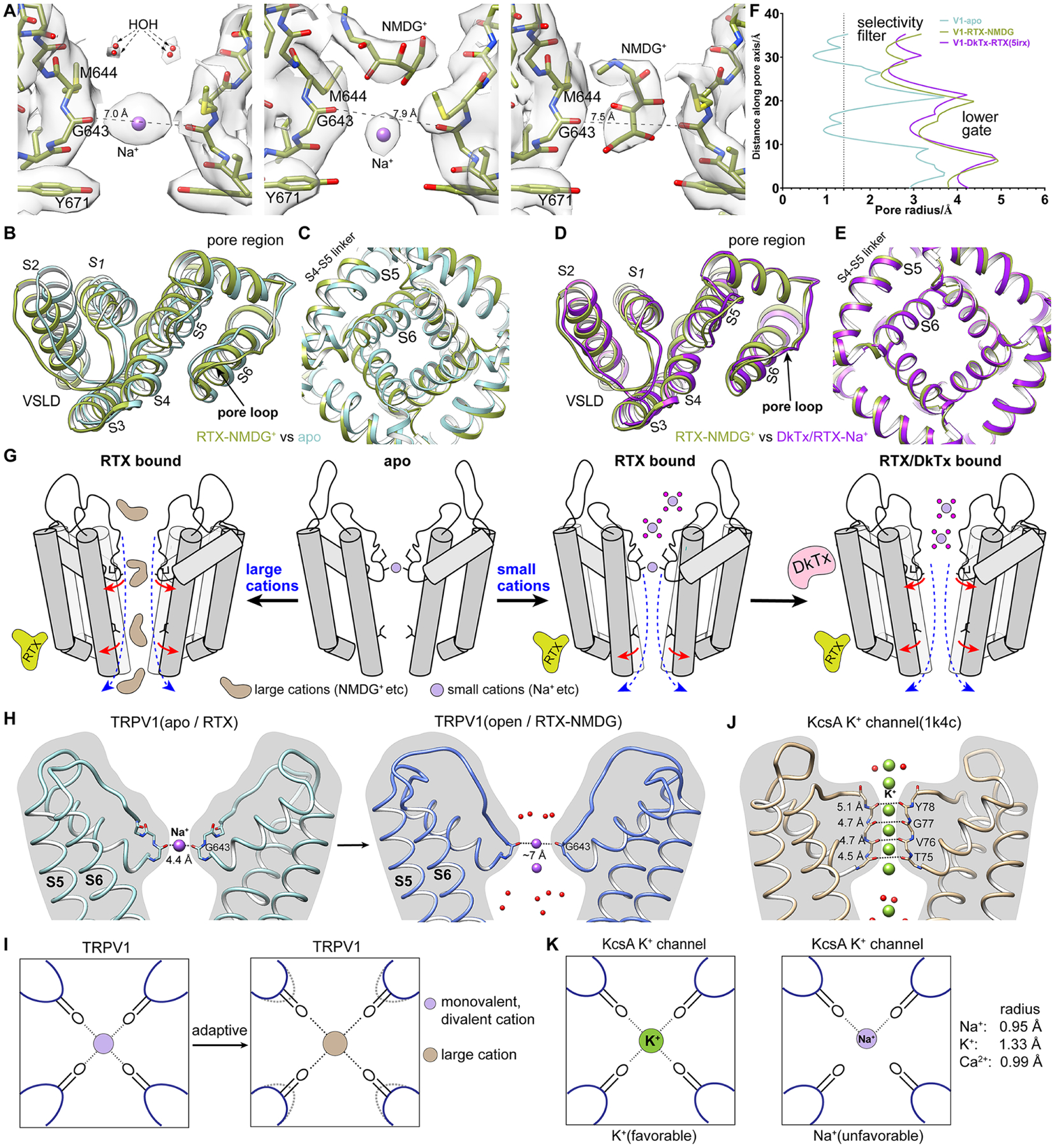
(A) In the presence of NMDG and QX-314, the selectivity filter of RTX-activated TRPV1 is fully open with M644 rotated away from the pore. Left: Na+ is loosely coordinated by G643 carbonyl with water molecules coordinated above. Middle: NMDG molecule is engaged above the selectivity filter. Right: NMDG is captured within the selectivity filter.
(B) Overlay with the apo structure reveals that, when permeating a large organic cation, the pore loop is shifted and VSLD is tilted backward.
(C) Bottom slab view shows that the lower gate is open in the presence of both RTX and NMDG.
(D) and (E) Same views as in (B) and (C) showing that the conformation of RTX-activated TRPV1 permeating NMDG resembles the fully open channel activated by both RTX and DkTx.
(F) Comparison of pore radii shows that TRPV1 exhibits a fully open ion permeation pathway when both RTX and NMDG are present.
(G) Schematic model showing how the channel transitions from apo to fully open by RTX and DkTx (right) or by RTX alone and large organic cation (left), illustrating adaptivity of the selectivity filter.
(H) The selectivity filter of TRPV1 is shallow and can be dilated in response to DkTx binding or permeation of NMDG.
(I) Positions of opposing carbonyls in the TRPV1 selectivity filter are adaptive in response to permeating cations of different sizes.
(J) The selectivity filter of KcsA is long and stable, providing tight coordination of K+.
(K) The stable configuration of the selectivity filter in KcsA provides tight coordination for K+ but is unfavorable to other cations.
The selectivity filter in TRPV1, defined by backbone carbonyls of G643 alone, is shallow and dynamic (Figure 3H). Adaptation of the pore diameter, captured above in the presence of DkTx or NMDG, allows for coordination of cations of different sizes within the filter (Figure 3I), even asymmetrically. Likely, the presence of different extracellular cations biases the selectivity filter to adopt conformations suitable for their permeation. This is in sharp contrast with the rigid selectivity filter of highly selective cation channels, such as voltage gated K+ channels (Zhou et al., 2001), in which backbone carbonyls of 4~5 pore-lining residues coordinate 3~4 fully dehydrated K+ ions (Figure 3J), a stable configuration that disfavors larger or smaller cations (Figure 3K). Our results are also consistent with biophysical studies (Jara-Oseguera et al., 2019) suggesting that the upper restriction of TRPV1 is permeable to Na+ in the apo state and does not function as a classic ‘open-and-closed’ gate.
Stepwise competition between regulatory lipids and vanilloid agonists
To favor permeation by large organic cations, we purposefully lowered sodium concentration. While this did not facilitate visualization of QX-314 or YO-PRO-1 in the upper restriction site, it fortuitously allowed us to capture distinct substates of RTX binding, providing an opportunity to resolve questions pertaining to stoichiometry and cooperativity of vanilloid binding and the competitive relationship with regulatory PI lipids. Thus, in an RTX-containing sample with 9 mM Na+ with QX-314, we observed several configurations of the resident PI lipid ranging from fully intact densities to pockets with weakened aliphatic lipid tails or with the tail but not inositol head group displaced by RTX. In a sample containing 60 mM Na+ with YO-PRO-1, intact RTX densities are seen in all pockets with no trace of the resident PI lipid (Figure 4A). Together, these intermediates suggest a two-step process by which vanilloid agonists displace the regulatory lipid, first competing off the aliphatic tail followed by displacement of the headgroup.
Figure 4. Sub-stoichiometric binding of RTX.
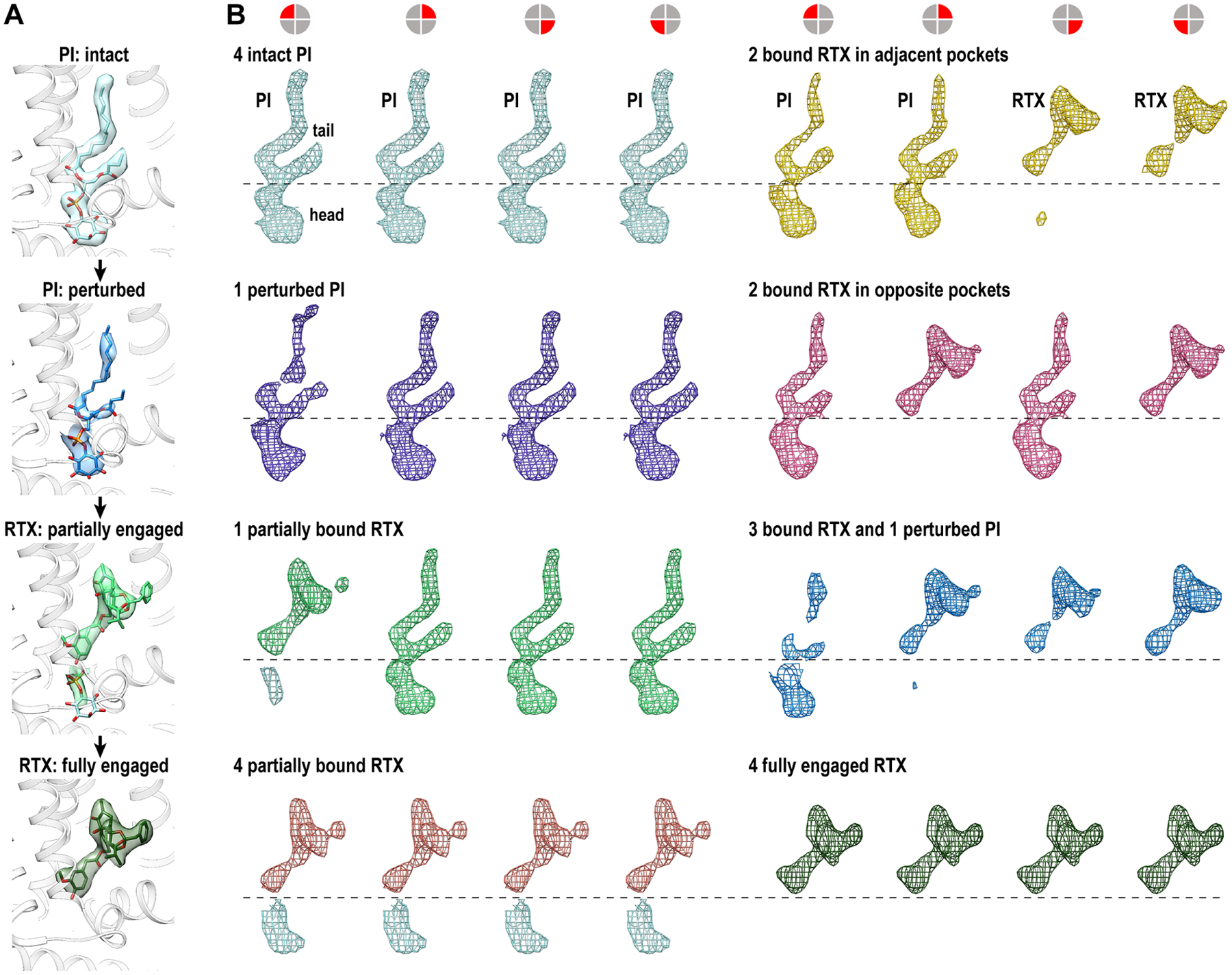
(A) Transitions from an intact PI lipid to weakened aliphatic tail, densities of an inositol headgroup and a less well-defined RTX, to the well-defined RTX density indicates, indicate a stepwise engagement of RTX binding to the vanilloid pocket.
(B) Engagement of RTX molecules with the four equivalent vanilloid pockets of TRPV1 is random without apparent order or coordination. Each panel shows densities within each of the four pockets in the same sequential order (as indicated by symbols atop the panel). Except for the first group (top left, 4 intact PI) and the last group (bottom right, 4 fully engaged RTX, found in 60 mM Na+ sample), the panels representing other substates are arranged without a specific order. RTX binding status in all panels is labeled.
See also Figure S5.
In the low sodium (9 mM) sample we observed all possible tetrameric intermediates with no apparent preference for relative binding position (Figure 4B). For example, in substates containing two RTX molecules, binding is observed in para and ortho positions with almost equal prevalence. Only once the PI lipid is completely displaced from a given subunit do we see obvious movement of S5, which is restricted to that subunit and insufficient to induce movement of the cognate S6 helix (Figure S5). Together with the higher sodium sample (60 mM), we see that all four endogenous PI lipids must be fully displaced by RTX to induce sufficient S5 movement in all subunits. In this case, the lower gate assumes two different “closed” conformations (C1 and C2); in C1, the S5 movement is still incremental and S6 does not move, while in C2, S5 moves sufficiently to enable S6 movement (Figure S5). Overall, the level of movement of S5 correlates closely with the level of RTX engagement in the vanilloid pocket: only fully engaged RTX causes sufficient S5 movement to allow S6 movement, a necessity to open the lower gate.
Channel gating by RTX
The two substates captured above have different closed conformations: C1 has a π-helix in S6 and the pore is closed by I679 (5.7Å), while C2 has an α-helix in S6 with the pore closed by M682 (5.0Å). To confirm that both are relevant to channel gating, we further determined structures of full-length TRPV1 with RTX bound under normal buffer conditions. As endpoint references, we also determined structures of full-length TRPV1 at neutral pH in apo and RTX/DkTx bound state. These are largely indistinguishable from their minimal channel counterparts except for an unstructured density above the outer pore contributed by the turret (Figures S6A–E).
We captured three different substates of the full-length channel bound to RTX (labeled as C1, O1 and C2) (Figure S6F–S6N). In each case, the selectivity filter remains unchanged from the apo state, RTX is fully engaged in all four vanilloid pockets, and S5 moves backwards to a similar position, with the main difference seen in S6, which moves progressively backwards from substates C1 to C2 (Figure 5A, 5B, Movie S3). C1 resembles the RTX C1 of the minimal channel with a closed gate (Figure S6F, S6H–S6J). While the distance of opposite gating residue I679 is increased from 5.1Å in apo state to 5.8Å and pore radius from 0.8Å to 1.1Å, it is insufficient to permeate water or partially dehydrated cations (Figure 5C, S6I–S6K). O1 has an open gate, in which the distance between opposite I679 is increased to 6.8Å and pore radius to 1.7Å, sufficient to permeate partially or fully dehydrated cation (Figure 5C, S6I, S6J and S6L). C2 resembles RTX C2 of the minimal channel (Figure S6G–S6J), in which the π-helix in S6 transitions to α-helix. Although the S6 has moved further backwards, M682 becomes the new gating residue. The pore profile calculated from the model shows a closed gate (Figure 5C). However, we observe a clear density that appears to be a cation coordinated by four sulfur atoms of M682 (Figure S6M, S6N). Although very rare, sulfur from methionine has been found to coordinate Na+ (Guskov et al., 2016), with an atom-to-atom distance of ~2.7Å (Zhou et al., 2021). If this density corresponds to a coordinated Na+, then the C2 conformation is permeable to partially dehydrated cations. Capturing three distinct conformations from the same dataset suggest that the channel can switch between these substates upon activation by a vanilloid agonist, consistent with the characteristic “flickery” behavior observed in single channel recording of RTX or capsaicin activated TRPV1 (Bohlen et al., 2010). Furthermore, these structures reveal multiple substates upon agonist binding while validating the minimal or full-length channel as valid models to dissect structural elements of channel gating.
Figure 5. RTX-mediated channel gating.
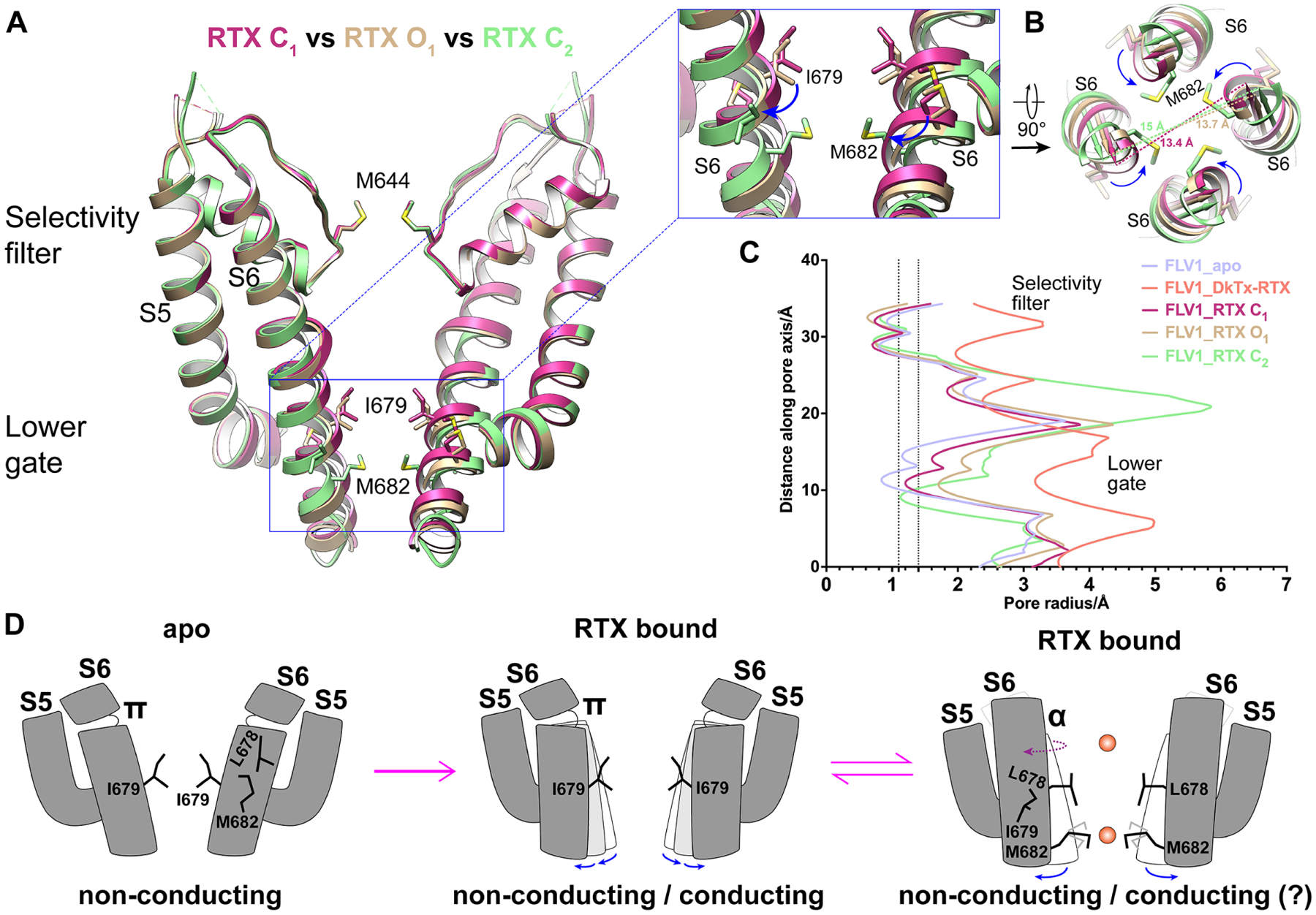
(A) Overlay of three RTX bound full-length structures reveals progressive backward movement in the lower part of S6 from C1 to C2 while the selectivity filter remains static. Enlarged view shows that transition in S6 from π- to α-helix in C2 rotates M682 into the pore.
(B) Progressive backward movement of S6 is viewed along symmetry axis.
(C) Pore radii of three RTX bound structures, apo (closed) and fully open with DkTx/RTX bound. Vertical dashed lines marks 1.1Å (radius of dehydrated Na+ and Ca2+) and 1.4Å (radius of water).
(D) Schematic model illustrates RTX-evoked channel gating. Channel is closed in apo (left). After RTX binding, both closed and conducting substates are seen with S6 π-helix configuration (middle). After π- to α-helix transition, cation permeation is regulated by M682 (right).
See also Figure S6.
Structural mechanisms of proton action
Local tissue acidosis is a driver of pain hypersensitivity, which is mediated, in part, through TRPV1 (Caterina et al., 2000; Davis et al., 2000). Structure-function studies have identified two negatively charged residues (E600 and E648) as key sites underlying sensitivity of TRPV1 to extracellular protons (Ahern et al., 2005; Jordt et al., 2000). Using the ‘minimal’ TRPV1 construct, we have previously shown that E600 resides in an extracellular loop that undergoes substantial movement upon channel opening, suggesting that protonation of E600 disrupts hydrogen bonding interactions that stabilize the apo configuration (Cao et al., 2013). In light of evidence that the turret region (which is missing from the minimal channel) influences proton-mediated potentiation (Liao et al., 2013), we used the full-length TRPV1 to further probe structural mechanisms underlying proton modulation. We first examined the channel at pH 6, which represents a threshold proton concentration for TRPV1 activation at room temperature (Hazan et al., 2015; Jordt et al., 2000; Tominaga et al., 1998). From this one dataset, 3D classification revealed three distinct conformations, which we refer to as pH6a, pH6b and pH6c (Figures 6A, 6B, S7C, S7D). In the apo structure at neutral pH, E600 engages in a salt bridge interaction with R455 in the VSLD of the neighboring subunit (Figures 6A, left panel, and 6D left panel), restricting motion around this locale. In all three pH6 structures, the E600 side chain rotates away from R455 (Figures 6A, second to fourth panels), consistent with protonation of E600 and a marked increase in calculated pKa from 5.2 to 8.4 (Figure 6C). Rotation also weakens hydrogen bonding between the backbone nitrogen of E600 and backbone carbonyl of N652, as surmised from an increased distance (2.8 to 4.4Å) between these points in apo versus pH6a structures, which is greater in pH6b and pH6c conformers (6.4 and 8.6Å, respectively). Interestingly, Y653, which exhibits distinct orientations in open and closed conformations, is also differentially oriented in pH6a and b versus pH6c configurations (Figure 6A), as expected for transitional substates. Furthermore, disruption of the E600-R455 interaction is accompanied by backward tilting of the VSLD and progressive movement of the pore loop towards the open channel conformation (Figure 6D), producing an outer pore cavity with a slightly more negatively charged surface (Figure S7C).
Figure 6. Proton-induced conformational changes.
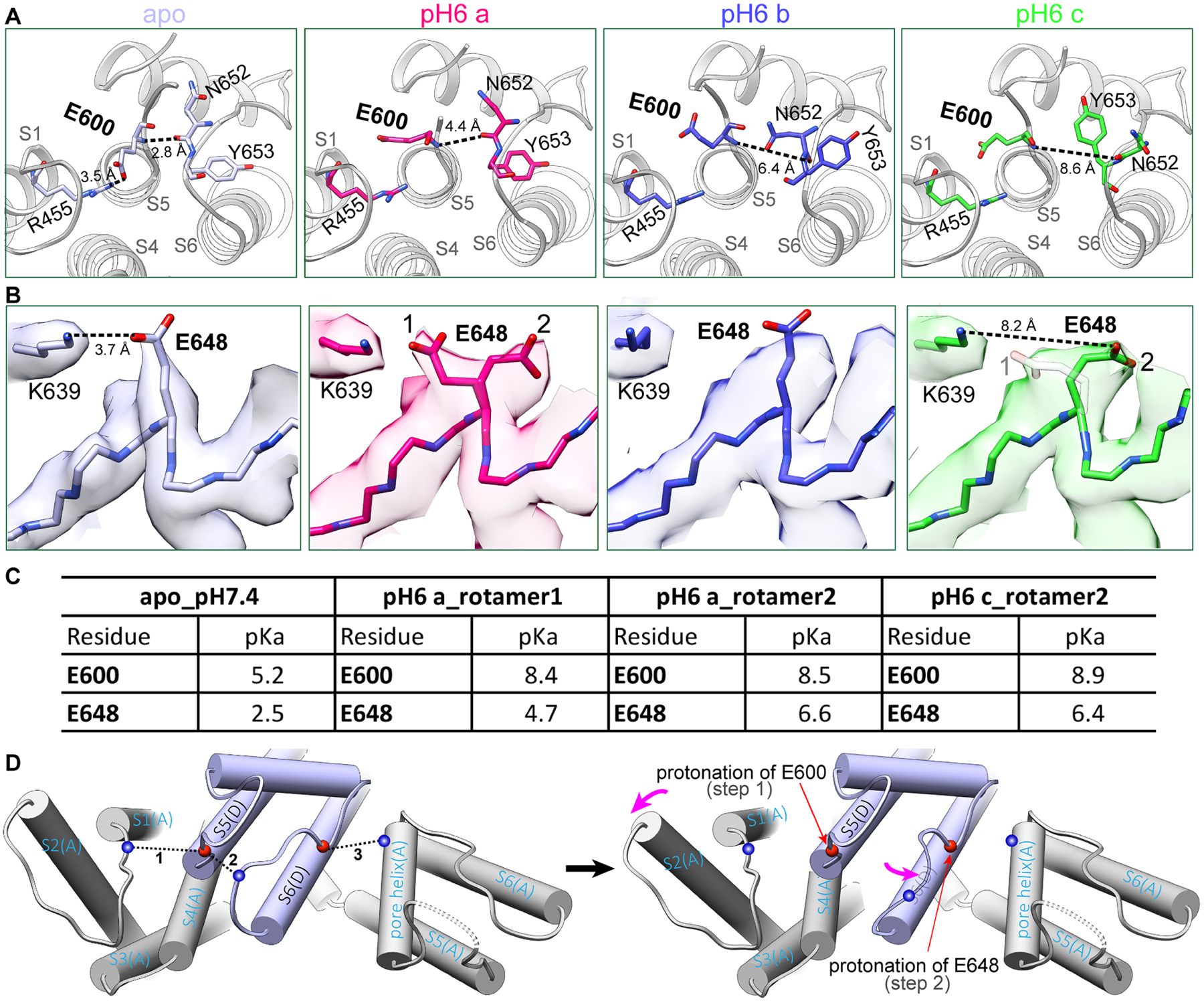
(A) At neutral pH apo state, E600 forms a salt bridge with R455 at VSLD of the neighboring subunit. At pH 6, protonation of E600 releases this salt bridge, breaks hydrogen bonds between the pore loop and S5 helix, moves upper pore loop progressively from pH6a to pH6c in the direction toward the fully open conformation induced by DkTx and RTX.
(B) Similarly, E648 density is well defined, forming a salt bridge with K639 in the pore helix of the neighboring subunit at neutral pH. At pH 6, side chain density of E648 is split, adopting two alternative rotamer conformations, pointing towards and away from K639, respectively. From pH6a to pH6c, the density of rotamer 1 becomes progressively weaker as 2 becomes stronger.
(C) Calculated pKa values for E600 and E648 in apo, pH6a and pH6c states. For E648, rotamer 2 is the protonated conformation.
(D) Schematic representation of a two-step model of extracellular proton-induced conformational changes: Left: E600 at the top of S5 helix and E648 in the middle of the pore loop stabilize the pore configuration at neutral pH. Right: E600 protonation followed by E648 protonation allows VSLD to tilt backwards and the pore loop to move.
See also Figure S7.
These changes alter the local environment and suggest that protonation of E600 proceeds, and possibly drives subsequent protonation at E648 (Figure 6B). Indeed, density maps at pH 6 show two possible rotameric side chain positions for E648, which are close to or pointing away from K639 (separated by ~3.7 and 8.2Å, respectively) (Figure 6B). Calculated pKa values for these two rotamers are also markedly different (4.7 versus 6.6, respectively) (Figure 6C), suggesting that the one pointing away from K639 predominates as the protonated species. Indeed, the density distribution between these rotameric positions shifts towards the species with a higher pKa value (i.e. further from K639) in the pH6c conformation compared to pH6a. Interestingly, the side chain density is less well-resolved for the pH6b species, suggesting that it is intermediate between pH6a and pH6c conformers (Figure 6B).
While the pH6c conformation shows hallmarks of the open state, the outer restriction / selectivity filter is not as fully dilated as DkTx/RTX or NMDG stabilized structures, the S5 movement is visible but insufficient to move S6 (Figure S7D), and the pore profile is not as wide as that seen in the fully open state (Figure S7D). To ask whether further acidification can promote a complete transition, we lowered the pH even further to 5.5, which was the practical limit for maintaining protein stability. However, this structure showed no further conformational changes beyond that seen in the pH6c structure, even when the lower gate was stabilized by RTX (Figure S7). Consistent with this, only one conformation similar to pH6c was observed at pH 5.5, suggesting that we reached the limit of proton-evoked transitions. While mutation of residues other than E600 and E648 have been shown to affect pH sensitivity, our results support the conclusion that they are not directly involved in proton detection (Aneiros et al., 2011; Ryu et al., 2007).
Two-way allosteric coupling between upper and lower restrictions
Protons and DkTx target the outer pore region but also alter status of the lower gate (Figure 7A). Our structures suggest that protonation of E600 and E648 affects discrete regions that can be roughly divided into the outer VSLD and central pore module, respectively. The former induces movement of the S4–S5 linker and generates space to allow for movement of S6, poising the lower gate for opening, whereas the latter destabilizes the upper restriction to facilitate transition to a fully protonated conformation, as noted above. DkTx binding also alters the ionic interaction between E600 and R455 and induces VSLD to undergo a similar outward tilt concomitant with outward movement of S5, thereby facilitating opening of the lower gate. Interestingly, the lower gate is more dilated in the presence of DkTx than at pH 6. The distortion of the bilayer by DkTx leads to insertion of a lipid tail into a hydrophobic cleft between S5 and S6 helices of neighboring subunits, thereby serving as a wedge to stabilize the lower gate. In the case of RTX, we see neither backward tilting of the VSLD nor appreciable widening of the upper restriction, indicating that the primary action of vanilloid agonists is to open the lower gate, which is sufficient to elicit ion flux through the channel. At the same time, our results with NMDG suggest that allosteric coupling works in both directions since binding of RTX favors the most open conformation of the upper restriction (as revealed under these stabilizing conditions), consistent with more rapid action of DkTx in the presence of vanilloid agonists (Bohlen et al., 2010).
Figure 7. Allosteric coupling and agonists induced channel activation.
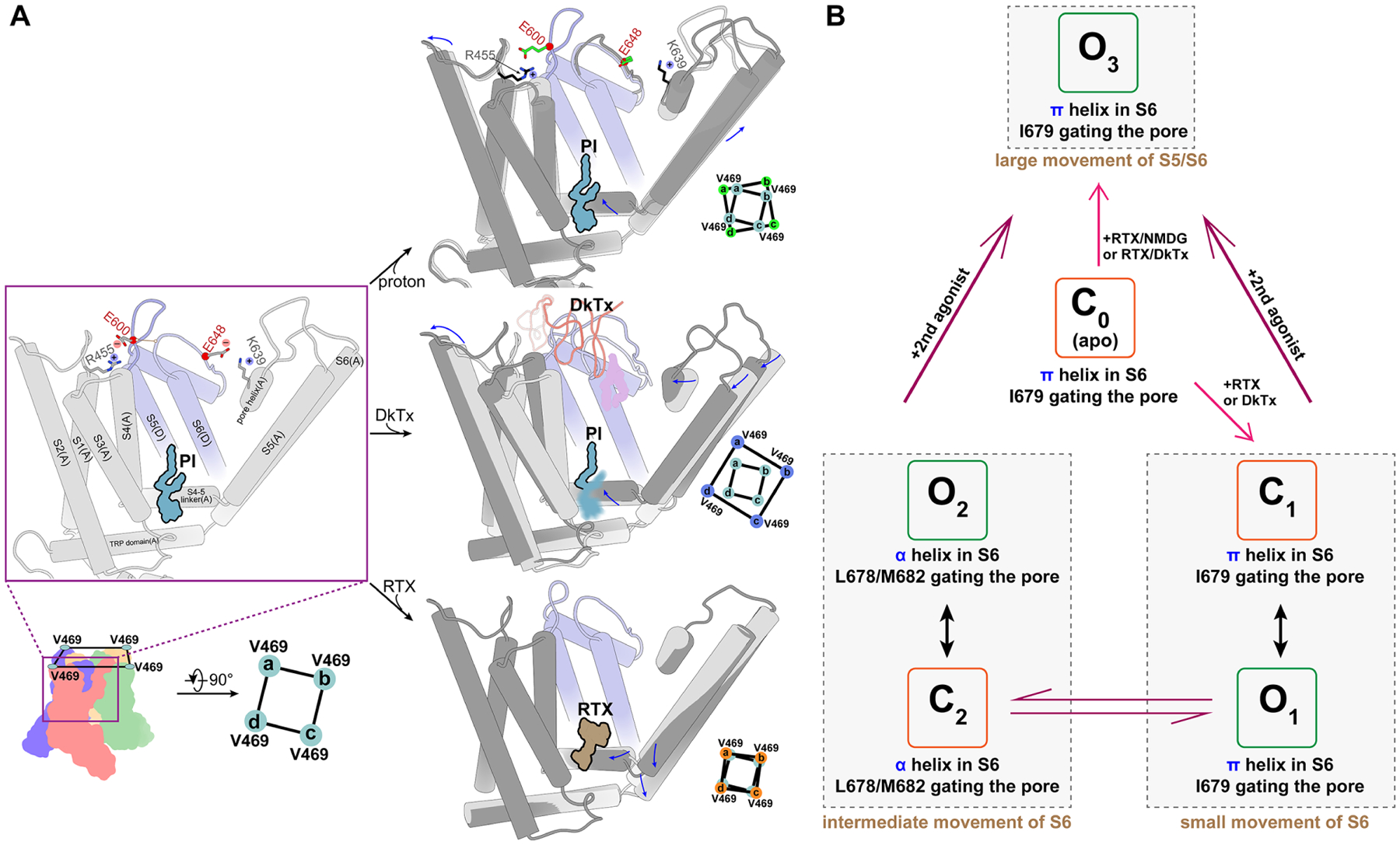
(A) Allosteric coupling elicited by different ligands illustrates the polymodal nature of TRPV1 gating. Key structural features and residues are shown in the left panel. Upper: Protonation of E600 allows backward tilting of VSLD, which allosterically moves the S4–S5 linker and S5 to prepare the lower gate for opening. E648 protonation loosens constraints on the upper restriction site to potentiate the channel. Middle: DkTx binding induces a larger backward tilting of VSLD, sufficient to allosterically move S5/S6 (via S4–S5 linker) and open the lower gate. Insertion of a lipid tail into the central cavity likely stabilizes coupling between the upper restriction site and lower gate in the open conformation. S4–S5 linker movement destabilizes the resident PI lipid in the vanilloid binding pocket. Bottom: RTX binding acts directly on S4–S5 linker to move S5 and S6, allosterically enhancing dynamic behavior of the upper restriction site. Permeating large organic cations bias the selectivity filter towards a wide-open conformation.
(B) Actions of different agonists transition TRPV1 into different close (C) and open (O) substates. C0 represents the closed channel in the apo state. C1 and O1 represent closed and open substates with a π-helix in S6. C2 and O2 represent closed and open substates with an α-helix in S6; in both cases, a single agonist binds to the channel, acting either on the lower gate or the outer pore. O3 represents the widest open channel, with two ligands acting on both lower gate and upper restriction sites simultaneously. The degree of S5/S6 movement increases from C0 to O3.
We notice that when the channel is activated by a single agonist, either DkTx or RTX alone, movement of S6 is coupled with a π-helix to α-helix transition (Movie S1 and S3) resembling that seen in the full-length squirrel TRPV1 channel (Nadezhdin et al., 2021). Interestingly, this transition is not seen when an agonist is bound in the vanilloid pocket and the upper restriction is stabilized by either DkTx (Cao et al., 2013; Gao et al., 2016) or NMDG (Movie S2), in which case S5 and S6 movements are the largest among all structures described in this study. Interestingly, movements of S6 can be roughly characterized as small, intermediate, or large (Figure 7B). When the movement is small, such as in going from the apo to O1 of RTX bound full-length structure, the π-helix is retained and the channel gate is formed by I679. In the intermediate category, exemplified by DkTx partial open and open (O2), C2 of minimal or full-length channel, the π-helix transitions to an α-helix and the gate is formed by M682. When the S5/S6 movement is large, as seen in RTX/DkTx or RTX/NMDG structures (O3), a π-helix configuration is observed and the open gate (9.8Å) is formed by I679. Transitions between π and α have also been seen in TRPV2 and V3 structure (Singh et al., 2018; Zubcevic et al., 2016; Zubcevic et al., 2018a), but the functional consequences of such transitions remain to be fully appreciated. Our more extensive observations with TRPV1 suggest that they may account for multiple closed and open substates and/or rapid transitions between substates typically observed following activation by capsaicin (Hui et al., 2003).
DISCUSSION
Visualizing plasticity of the ion selectivity filter
Permeability of P2X or TRP channels to large organic cations has been proposed to occur through a controversial process known as ‘pore dilation’ whereby repetitive or prolonged channel activation leads to dramatic changes in size selectivity (Bean, 2015; Chung et al., 2008; Khakh et al., 1999). Irrespective of whether or to what extent this involves a use- and time-dependent mechanism, high resolution TRP channel structures described here and elsewhere now show that dynamic changes in the outer pore and selectivity filter can accommodate small and large cations, upholding at least one intriguing aspect of the pore dilation hypothesis – namely, that pore size need not be limited to a fixed open state, but can be adaptive.
QX-314 and NMDG behave as permeant blockers that impede the flow of smaller Na+ ions, likely reflecting their relatively tight, but transient binding within the pore, perhaps explaining why we see NMDG poised within the outer pore region, but not deeper in the pathway. Future studies using various organic cations and channel activation parameters may reveal a more complete series of intermediates that detail movement along the length of the pore. In any event, elucidating dynamic transitions associated with selectivity filter flexibility is pertinent to understanding structural mechanisms that may be exploited to develop modality-specific analgesics or target drug delivery to nociceptive neurons in the pain pathway (Binshtok et al., 2007; Puopolo et al., 2013).
Exploiting cryo-EM to reveal ligand binding substates
Improvements in cryo-EM data collection and analysis have transformed our ability to identify structural subclasses of protein complexes that represent functional substates. These advances promise to revolutionize molecular pharmacology by providing direct visual insight into key parameters of ligand-receptor interaction, such as binding site competition, stoichiometry, cooperativity and so forth. This may be especially advantageous for receptors containing multiple ligand binding subunits, or where specific binding of unlabeled, low affinity or hydrophobic ligands is difficult to measure biochemically. By segmenting substates of TRPV1-ligand interactions, we can now visualize and define precise steps through which endogenous modulators or natural products interact with key regulatory sites to facilitate channel gating.
For example, functional studies implicated E600 and E648 as key sites mediating acid-evoked channel potentiation and activation, respectively. However, a structural understanding of the distinct contribution of these residues to acid sensitivity was lacking. The ‘snap shots’ shown here now resolve this question by revealing a progressive, stepwise process in which protonation of E600 initiates structural rearrangements within the outer pore region that prepare E648 for subsequent protonation. The E600-mediated transition initiates pre-gating movements in the VSLD and pore loop, providing a cogent explanation for how moderate acidosis potentiates sensitivity to other stimuli (heat or vanilloids), and why TRPV1 E648 mutants are sensitized, but not activated by protons (Jordt et al., 2000).
The other important interaction pertains to the interplay between vanilloids and resident PI lipids. Our structures reveal a dynamic competition in which vanilloids first displace the aliphatic PI lipid tail followed by the inositol headgroup. This may be consequential for understanding how different endogenous TRPV1 ligands, such as distinct PI lipid species, endocannabinoids or endovanilloids, bind to and modulate the channel, which is likely relevant to other TRPV subtypes that harbor lipids in the same location near the lower gate (van Goor et al., 2020).
Our structures also suggest that the four vanilloid sites are equivalent, with each contributing incrementally to gating transitions. This is in general agreement with functional studies of the cold-activated TRPM8 channel, where menthol binds to four independent and energetically equivalent sites, eliciting stepwise stabilization of the open state (Janssens and Voets, 2011). A conceptually similar analysis using concatenated TRPV1 channels suggested that occupancy of only one vanilloid site is sufficient to activate the channel, but this analysis may be complicated by the fact that a mutation (Y511A) introduced to render subunits capsaicin-insensitive may also affect PI lipid binding at the vanilloid site (Hazan et al., 2015). In any case, our findings and those of (Janssens and Voets, 2011) suggest that TRP channels differ from classic models exemplified by hetero-pentameric nicotinic acetylcholine receptors in which two non-equivalent agonist sites are occupied sequentially and with positive cooperativity to elicit gating (Sine and Claudio, 1991).
Communication between dynamic constrictions
The existence of two restrictions along the TRPV1 ion permeation pathway reflects distinct points of action for water soluble versus lipophilic agents that modulate upper and lower regions, respectively. Despite their distinct locations, these control points must communicate with one another to ultimately affect ion flux. A comparison of proton- and DkTx-mediated effects reveals a common framework for how modulation of the upper restriction is structurally coupled to the cytoplasmic gate through subtle, but consequential rigid body-like tilting of the VSLD to provoke movement of the S4–S5 linker and S5. While the specific structural movements are quite different, this is broadly reminiscent of the connection between VSD movements and gating in voltage-gated potassium channels, perhaps in keeping with their common evolutionary roots. Furthermore, our structures are consistent with recent biophysical studies suggesting that the upper restriction is not a true gate with clearly defined open and closed states, but rather a highly dynamic filter that enables permeation by small and large cations while also contributing to modulation of the lower gate (Jara-Oseguera et al., 2019). Interestingly, neither protons nor DkTx enhance capsaicin-evoked unitary currents or permeability to divalent cations (Bohlen et al., 2010; Geron et al., 2018; Liao et al., 2013), suggesting that their main physiologic effect is to facilitate opening of the lower gate. Conversely, vanilloids enhance adaptivity of the upper restriction, demonstrating the duality of allosteric communication between these control points.
Role of methionine in the pore
In TRPV1 and other TRPV subtypes, side chains of M644 in the upper restriction and M682 in lower gate can switch between two different positions, facing into or away from the pore. When the selectivity filter is not dilated by DkTx or NMDG, M644 faces the pore forming a restriction above backbone carbonyls of G643. In some structures, there is a cation-like density coordinated by the four sulfur atoms of M644. Moreover, mutation of M644 influences ion permeability ratios (Munns et al., 2015), but the exact role of M644 remains to be fully explored. In the lower gate, M682 forms a new restriction when the S6 π-helix transitions to an α-helix. Again, mutation of M682 (Susankova et al., 2007) has not provided clear insight into the role of this residue in channel gating. Our structures now suggest that, when facing the pore without a hydrophobic interaction network, methionine side chains are more flexible than those of other residues that form the selectivity filter (G643) or lower gate (I679). This is more pronounced as the distance between opposing M682 residues increases. We therefore propose that neither M644 nor M682 forms a rigid barrier for cation permeation and that a combination of changes in S6 position and π- to α-helix transitions generates multiple open and closed substates in response to different agonists (Figure 7B), as observed in single channel recordings (Hui et al., 2003).
Limitations of the study
To study structural mechanisms of proton action, both the intra and extracellular side of the channel were exposed to the same low pH environment. To evaluate the potential impact, we calculated pKa values of all potentially protonatable residues facing the cytosol. None showed a significant change of calculated pKa value at low pH conformations. While we therefore think that the low pH structures are not directly impacted by non-physiologically low intracellular pH, it remains a possibility. Another potential limitation is use of the minimal functional TRPV1 channel for structural studies. As demonstrated here and previously by us and others, we believe that the minimal channel is a valid model system for elucidating gating mechanism. Nonetheless, we do recognize that the electrophysiological behavior of the minimal and full-length channel is not identical.
STAR★METHODS
LEAD CONTACT AND MATERIALS AVAILABILITY
Lead contact
Further information and requests for reagents may be addressed to the Lead Contact, Yifan Cheng (Yifan.Cheng@ucsf.edu).
Materials availability
Cell lines and plasmids used in this manuscript will be available from the Lead Contact upon execution of a materials transfer agreement.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Cell lines
Sf9 cells were cultured in ESF 921 (Expression Systems) at 27 °C. These cells were used for Baculovirus generation. HEK293 GnTI− cells were cultured in FreeStyle 293 Expression Medium + 2% (v/v) fetal bovine serum at 37 °C with 8 % CO2. These cells are female in origin.
METHOD DETAILS
Protein purification and nanodisc reconstitution
Recombinant minimal functional (amino acids 110 to 603 and 627 to 764; miniV1) and full-length (amino acids 2 to 838; FLV1) rat TRPV1 protein were expressed and purified as previously described (Liao et al., 2013). For nanodisc reconstitution, membrane scaffold protein MSP2N2 was expressed and purified from Escherichia coli. Lipids were prepared as previously described (Gao et al., 2016). Purified TRPV1 protein in detergent (DDM) was mixed with MSP2N2 and soybean polar lipid extract (Avanti) using the ratio TRPV1 monomer:MSP2N2:soybean lipid = 1:1.8:80 and incubated on ice for 10–30 min. 2–3 batches of Bio-beads SM2 (Bio-Rad) were added to remove detergents from the mixture with constant rotation for overnight. After adding TEV protease and incubation for 4 hours to cleave the fused MBP tag, the reconstituted channels were cleared by moderate centrifugation via spin filter microtubes (Merck Millipore Ltd.) followed by size exclusion chromatography using a Superdex 200 Increase 10/300 GL column (GE) equilibrated with buffer containing 20 mM HEPES-NaOH, pH 7.4, 150 mM NaCl and 100 μM TCEP for the samples miniV1-apo at pH 7.4, miniV1-DkTx at pH 7.4, FLV1-apo at pH7.4, pH 6, FLV1-DkTx/RTX, FLV1-RTX at pH 7.4, pH 5.5. The peak fractions corresponding to tetrameric TRPV1 in lipid nanodisc were pooled, concentrated to around 3.2 mg ml−1 using Amicon Ultra filter device (50-kDa MWCO, Millipore) and analyzed through SDS-PAGE and negative-stain EM followed by immediate cryo-EM grid preparation. Nanodisc reconstituted TRPV1 were used throughout this study. The pH adjustments (to pH 6 or 5.5) or ligands addition (RTX or DkTx) were performed 30 min before cryo-sample preparation.
To prepare sample of channel with RTX, NMDG and QX-314, the reconstituted channels were subjected to size exclusion chromatography equilibrated with buffer containing 20 mM HEPES, pH 7.1, 6.3 mM NMDG, 100 μM TCEP and 375 μM NaOH. QX-314 and RTX were added into the sample 30 min before cryo-sample preparation. The final buffer contains 12.5 mM HEPES, pH 6.8, 28 mM NMDG, 23 mM QX-314, 64 μM TCEP, 240 μM NaOH and 40 μM RTX. To prepare sample of channel with RTX and QX-314 under 9 mM Na+ concentration, the reconstituted channels were subjected to size exclusion chromatography equilibrated with buffer containing 20 mM HEPES, pH 7.3, 5 mM NaCl, 100 μM TCEP and 6 mM NaOH. QX-314 and RTX were added into the sample 20 min before cryo-sample preparation. The final buffer contains 13.3 mM HEPES, pH 6.85, 25 mM QX-314, 66.7 μM TCEP, 5.6 mM NaOH, 3.3 mM NaCl and 41.7 μM RTX. To prepare sample of channel with RTX and YO-PRO-1 under 60 mM Na+ concentration, the reconstituted channels were subjected to size exclusion chromatography equilibrated with buffer containing 20 mM HEPES, pH 7.3, 5 mM NaCl, 100 μM TCEP and 6 mM NaOH. YO-PRO-1 in DMSO and RTX were added into the sample 20 min before cryo-sample preparation. The final buffer contains 17.6 mM HEPES, pH 7.3, 10.3 μM YO-PRO-1, 1 % DMSO, 87.9 μM TCEP, 5.3 mM NaOH, 54.5 mM NaCl and 51.7 μM RTX.
EM sample preparation and data acquisition
For negative-stain EM, 2.5 μl of TRPV1 samples at ~40 μg ml−1 were applied to a glow discharged Cu grid covered by continuous carbon film, and then stained with 0.75% (w/v) uranyl formate (Ohi et al., 2004). A Tecnai T12 microscope (ThermoFisher FEI Company) operated at 120 kV was employed to check these negatively stained grids. Micrographs were recorded at a nominal magnification of 52,000× using an UltraScan 4000 camera (Gatan), corresponding to a pixel size of 2.21 Å on the specimen.
To prepare cryo-EM grids of nanodisc reconstituted TRPV1 in neutral pH, channel was mixed with RTX at final concentration of 40–55 μM in a molar ratio of 1:6.9–9.5, or with DkTx at final concentration of 20 μM in a molar ratio of 1:3.5 for 20–30 min before vitrification. To prepare cryo-EM grids at low pH, nanodisc reconstituted full-length apo channel or preformed complex with RTX at pH 7.4 buffer (20 mM HEPES-NaOH, 150 mM NaCl and 100 μM TCEP) was titrated by 1 M Na acetate buffer (pH 4.6) to adjust the final pH to 6 for apo samples or to pH 5.5 for TRPV1-RTX samples. 3 μl samples at 1.5–2.5 mg/ml were applied onto a glow-discharged gold grids covered with holey carbon film (Quantifoil, 300 mesh 1.2/1.3) and blotted using a Vitrobot Mark IV (FEI Company) with 4.5-s blotting time and 100% humidity at 4 °C for FLV1-apo at neutral pH, pH 6 and FLV1-RTX at pH 5.5, or at 20 °C for miniV1-apo at pH 7.4, miniV1-DkTx at pH 7.4, miniV1-RTX/NMDG at pH 6.8, miniV1-RTX/QX-314/9mM Na+ at pH 6.85 and miniV1-RTX/ YO-PRO-1/60mM Na+ at pH 7.3, FLV1-DkTx/RTX at pH 7.4, FLV1-RTX at pH 7.4 and plunge frozen in liquid ethane cooled by liquid nitrogen.
Full-length apo channel at pH 6, in complex with RTX at pH 5.5 and in complex with DkTx/RTX at pH 7.4 were imaged with Talos Arctica transmission electron microscope (ThermoFisher FEI) operated at 200 keV. Full-length apo channel at neutral pH, in complex with RTX at pH 7.4, minimal apo channel at pH 7.4, in complex with DkTx, in complex with RTX/NMDG, in complex with RTX when QX-314 added under 9 mM Na+ concentration and in complex with RTX when YO-PRO-1 added under 60 mM Na+ concentration were imaged with a Titan Krios microscope (ThermoFisher FEI) operated at 300keV, equipped with a Bio Quantum post-column energy filter with zero-loss energy selection slit set to 20 eV. Both microscopes are equipped with a K2 or K3 camera (Gatan Inc), operating in super-resolution counting mode. Data collections were carried out by using SerialEM (Mastronarde, 2005). The data acquisition parameters, including dose rate, total dose and total frames per movie stack, etc. are summarized in the Table S1. For minimal channel with DkTx, two separate datasets were collected using the same microscope under the same condition. They were processed separately following the same protocol and combined together in the final stage of image processing.
Imaging processing
Motion correction of movie stacks were processed on-the-fly by using MotionCor2 (Zheng et al., 2017), which also bins motion corrected sum 2 × 2 by Fourier cropping. Sums without dose-weighting were used for contrast transfer function (CTF) determination using Gctf (Zhang, 2016) or Patch-Based CTF Estimation in cryoSPARC (Punjani et al., 2017). Dose-weighted sums were used for all other image processing. Particles were picked using a Gaussian template in cisTEM (Grant et al., 2018) or template selected from previous 2D averages in cryoSPARC followed by reference-free 2D classification (particles binned 4 × 4 during extraction). Particles within junk classes were removed entirely. An ab initio 3D reference model was generated based on the remaining particles by using cryoSPARC, which is then used for 3D classification in RELION-3. Particles within bad classes were further removed and remaining particles were subjected to further analysis. Obvious conformational heterogeneities were seen in the resulting 3D classes of minimal channel in complex with DkTx, with RTX / NMDG, in complex with RTX under 9 mM Na+ concentration and in complex with RTX under 60 mM Na+ concentration or full-length apo channel at pH 6 and in complex with RTX at pH 7.4. These datasets were subjected to further classification, as described below. No obvious conformational heterogeneity was observed in minimal apo channel, full-length apo channel, full-length channel in complex with RTX at pH 5.5 and in complex with DkTx/RTX at pH 7.4.
For datasets of minimal channel with DkTx, RTX/NMDG or RTX under 9 mM Na+ concentration, 3D classification of whole complex with global masks separated all particles into different classes with different conformations. To further improve the resolution and quality of these maps, particles within each conformational class was further C1 symmetry expanded using the program relion_particle_symmetry_expand implemented in RELION-3. The quadrupled particle stack was first subjected to the 3D classification without imposing symmetry using a mask of a single subunit. The bad particles (corresponding to bad subunit) were removed to obtain a cleaner particle stack. The remaining expanded particles were subjected to another round of 3D classification (Relion options: --tau2_fudge 20 –skip align) without local mask for minimal channel with DkTx, which separated symmetry expanded particles into two classes with the orientation of DkTx perpendicular to each other, with outer pore focused mask for minimal channel with RTX/NMDG and with vanilloid pocket focused mask for minimal channel with RTX under 9 mM Na+ concentration. For each original particle in minimal channel with DkTx, the orientational parameter of only one expansion belonging to the same DkTx bound class is kept. For each particle in minimal channel with RTX under 9 mM Na+ concentration, the presence of endogenous phosphatidylinositol lipid or RTX, their relative orientations and number of occurrences in four subunits are analyzed for distinguishing between different states. The quality of final density maps was further improved with local refinement without symmetry in RELION-3. As a validation control, we purposely calculated maps that contain different percentages of particles from apo states and fully bound RTX state, simulating densities in the vanilloid binding pocket contributed by misclassified particles. As shown in Figure S5B, these simulated densities are very different from what presented in Figure 4, supporting that it is largely free of particle misclassification.
For all other datasets, including full-length channel in complex DkTx/RTX at pH 7.4, C4 symmetry was applied in the 3D classification and refinement procedures. The symmetry expansion approach was also used to improve the density quality of Ankyrin repeat domain of apo channel at neutral pH. Directional Fourier Shell Correlation (dFSC) curves are calculated as described (Dang et al., 2017), and the nominal resolution is estimated from the averaged FSC using FSC=0.143 criterion (Rosenthal and Henderson, 2003). Local resolution maps were calculated in RELION-3. Conversion of star files from cryoSPARC to RELION-3 was performed using UCSF pyem (https://zenodo.org/record/3576630#.XuczyFVKjIU).
Model building and refinement
Atomic model (PDB accession 3J5P) were initially docked into the Relion-sharpened density maps with UCSF Chimera (Pettersen et al., 2004) and manually adjusted in Coot (Emsley et al., 2010). The models for ligands and associated lipids and their geometric constraints were generated using elBOW module in PHENIX (Adams et al., 2010) or JLigand module (Lebedev et al., 2012) in CCP4 package. All models were refined over multiple rounds using the module ‘phenix.real_space_refine’ (Afonine et al., 2018) in PHENIX and ISOLDE (Croll, 2018) implemented in ChimeraX (Goddard et al., 2018). The quality of all refined models was assessed using the ‘comprehensive model validation’ function in PHENIX and wwPDB validation server (Berman et al., 2003).
Pore radii, pKa and electrostatics calculation
pKa values were calculated in H++ (Anandakrishnan et al., 2012) as pK_(1/2), which in most cases is equivalent to pKa. For all pKa calculation, pH values are set to the same as in the experiments. HOLE program (Smart et al., 1996) was used to calculate all pore radii. Surface charges of the outer pore region were calculated using the latest version of the program (APBS and pdb2pqr) at pH 7, 6 and 5.5 respectively (Baker et al., 2001). UCSF Chimera (Goddard et al., 2018) were used to make figures.
QUANTIFICATION AND STATISTICAL ANALYSIS
The relative values of temperature factor were obtained using the same reference as indicated in figure legend and the generation of dot plots were performed using the program Graphpad Prism.
DATA AND CODE AVAILABILITY
Data Atomic coordinates and cryo-EM maps are deposited in EMDB and PDB as follows: Minimal TRPV1 in apo: EMD-23136, PDB-7L2P; Minimal TRPV1 with DkTx bound in pre-bound: EMD-23139, PDB-7L2S; with single DkTx bound: EMD-23161; pre-open: EMD-23138, PDB-7L2R; partial open: EMD-23140, PDB-7L2T; open: EMD-23141, PDB-7L2U; Minimal TRPV1 with RTX and NMDG, in state a: EMD-23143, PDB-7L2W; in state b: EMD-23142, PDB-7L2V; and in state c: EMD-23144, PDB-7L2X; minimal TRPV1 with RTX and QX-315 in buffer of 9 mM NaCl: with 1 perturbed PI: EMD-24084, PDB-7MZ6; with 1 partial bound RTX: EMD-24086, PDB-7MZ9; with 4 partial bound RTX: EMD-24085, PDB-7MZ7; with 2 bound RTX in adjacent subunits: EMD-24087, PDB-7MZA; with 2 bound RTX in opposite subunit: EMD-24091, PDB-7MZE; with 3 bound RTX and 1 perturbed PI: EMD-24088, PDB-7MZB; minimal TRPV1 with fully engaged RTX in buffer of 60mM NaCl and YO-PRO-1, in state C1: EMD-24089, PDB-7MZC; in state C2: EMD-24090, PDB-7MZD; Full-length TRPV1 in apo and neutral pH: EMD-23128, PDB-7L2H; Full-length TRPV1 with DkTx and RTX bound: EMD-23133, PDB-7L2M; Full-length TRPV1 with RTX bound, in state C1: EMD-23134, PDB-7L2N, in state O1: EMD-23132, PDB-7L2L; in state C2: EMD-24083, PDB-7MZ5; Full-length TRPV1 in pH 6, state a: EMD-23129, PDB-7L2I; in state b: EMD-23131, PDB-7L2K; in state c: EMD-23130, PDB-7L2J; Full-length TRPV1 in pH 5.5 and with RTX bound: EMD-23135, PDB-7L2O.
Code UCSF pyem is available at https://zenodo.org/record/3576630#.XuczyFVKjIU
Additional information None
Supplementary Material
Figure S1. Single particle cryo-EM structure determination, related to Figure 1, 3, 4, 5 and 6.
For each structure presented in this study, panels are arranged from left to right in the same order: directional Fourier Shell Correlation (dFSC) plots and averaged FSC (green curve) with resolution estimated on FSC=0.143 marked, side and top views of the density map color code with local resolution. (A) minimal TRPV1 apo; (B) with DkTx pre-bound; (C) single DkTx bound; (D) – (F) with double DkTx bound in pre-open (D), partial open (E) and open (F); (G) – (I) three conformations with RTX and NMDG bound; (J) – (O) six conformations of TRPV1 with RTX, QX-314 and 9 mM Na+, showing various sub-stoichiometric binding states of RTX; (P) and (Q) two conformations of TRPV1 with RTX, YO-PRO-1 and 60 mM Na+ with fully and stoichiometrically bound RTX; (R) Full length apo TRPV1 at neutral pH; (S) - (U) three conformations of full-length TRPV1 at pH6, pH6a, pH6b and pH6c; (V) full-length TRPV1 at pH5.5 with RTX bound. (W) – (Y) three conformations of full-length TRPV1 at neutral pH with RTX bound. (Z) Full-length TRPV1 with DkTx and RTX bound.
Figure S2. Classification scheme of TRPV1 with DkTx bound, related to Figure 1 and 2.
The scheme illustrates the classification processes that identified five different conformations in the dataset. Symmetry expansion applied to each conformation further improves the conformational homogeneity within each conformational class. For data processing efficiency, the entire dataset was separated into two groups that each went through the same classification scheme. Once a clean class was obtained after symmetry expansion, particles of the same conformation were recombined to a final reconstruction. The particle numbers of each final reconstruction are indicated.
Figure S3. Binding of DkTx to TRPV1, related to Figure 1 and 2.
(A) and (B) Side and top view of DkTx densities bound to the channel, shown as ribbon diagram, in four DkTx bound substates. From left to right, panels are single bound, pre-open, partial open and open.
(C) Segmented density of the phosphatidylinositol lipid residing in the vanilloid binding pocket. Densities of the lipid head group and one acyl chain become progressively weaker as the lower gate shifts towards opening.
(D) Segmented density of an annular lipid. In the open conformation, one of the lipid tails is seen inserting into a hydrophobic cleft between S5 and S6 of neighboring subunits.
Figure S4. Conformational dynamics upon DkTx binding, related to Figure 1 and 2.
(A) Solvent accessible pathway along the ion permeation pore of minimal TRPV1 in apo, pre-bound, pre-open, partial open and open substates. Residues that form the upper restriction and lower gate are labeled. Note that the water accessible volume of the central cavity is enlarged in partial open and open.
(B) Side views of ion permeation pathway with the distance between critical residues (M644, G643, Y671, I679 and M682) of two opposing subunits marked.
(C) Overlay of the lower part of S6 in apo, partial open and open states. Transition of S6 from π-helix in apo to α-helix in partial open and open substates rotates I679 away and M682 into the pore.
(D) – (H) M682 is well resolved in pre-open, but less well-resolved partial open and open substates.
(I) Temperature factor of M682 in partial open is higher than in other substates. Temperature factor of neighboring residue A680 of the same structure is used as a reference point.
(J) Pore radii of partial open and open are compared with that of apo and fully open (5IRX) states. Dashed line in the profile of partial open substate indicating the uncertainty in the calculation caused by less well-resolved M682 in one subunit. Two vertical dashed line marks the radii of dehydrated Ca2+ (1.1Å) and water (1.4Å).
Figure S5. Conformational changes at the lower gate induced by sub-stoichiometric binding of RTX, related to Figure 4.
(A) A bottom slab view of the lower gate in which the apo structure is overlaid with RTX binding substates (number and state of RTX binding are labeled as in Figure 5B). Perturbation of endogenous PI lipid does not induce noticeable changes in the lower gate. Partial binding of RTX induces movement of S5 within the same subunit. With RTX fully engaged in all binding sites (seen only in 60 mM Na+ sample), the lower gate assumes two different conformations: in one, the S5 movement is small and S6 does not move, while in the other, both S5 and S6 move and the lower gate is open. Overall, the level of movement of S5 correlates closely with the level of RTX engagement in the vanilloid pocket such that only fully engaged RTX causes sufficient S5 movement to move S6 and to open the lower gate.
(B) Ligand density within the vanilloid binding pocket calculated from combining different ratio of PI-containing and RTX-containing particles. These simulations show how the ligand density would appear if the PI containing and RTX containing vanilloid pockets were not cleanly separated by classification before calculating 3D reconstructions.
Figure S6. Binding of RTX to full-length TRPV1, related to Figure 5.
(A) Solvent accessible pathway along the ion permeation pore of minimal and full-length channels in apo and DkTx and RTX bound states.
(B) Overlay of full and minimal TRPV1 structures determined at neutral pH shows that the two structures are largely indistinguishable, except for the turret region in the enlarged view. The windowed regions are enlarged in the adjacent panel. The location of E600 is marked.
(C) The turret density appears as an unstructured density atop the pore.
(D) Pore radii of minimal and full-length channels in apo and DkTx and RTX bound states.
(E) Overlay of outer pore (left) and lower gate (right) for minimal and full-length DkTx/RTX bound channels.
(F) and (G) With RTX bound, overlay of substates C1 of minimal with C1 of full-length channel (F), substate C2 of minimal with C2 of full-length channel (G).
(H) Solvent accessible ion permeation pathway along the ion permeation pore of minimal TRPV1 with fully engaged RTX in substates C1 and C2.
(I) Solvent accessible ion permeation pathway along the ion permeation pore of full-length TRPV1 with RTX in substates C1, O1 and C2.
(J) Overlay of full-length TRPV1 structures with RTX in substates C1, O1 or C2 with apo structure.
(K) and (L) Bottom view of lower gate in substates C1 or O1.
(M) and (N) side and bottom view of substate C2 show a cation density in the vicinity of M682.
Figure S7. Proton-evoked conformational changes, related to Figure 6.
(A) and (B) Comparison between the full-length structures captured at low and neutral pH. Each panel shows the overlay of a low pH with neutral pH structure (as labeled), in either top view (A) of VSLD and pore domain, or bottom view of the lower gate (B). From the left to right, panels are arranged such that pH6a (red), pH6b (blue), pH6c (green) and pH5.5/RTX (fuchsia pink) are compared with the structure of neutral pH (light blue). The VSLD is tilted backwards in pH6a. Upper and lower parts of the pore loop are moved in pH6b, pH6c and pH5.5/RTX.
(C) Surface charges of the outer pore region calculated at neutral pH (left), pH6c (middle), and pH5.5 with RTX (right).
(D) Comparison of pore radii of the channel at pH6 and pH5.5 with that at neutral pH and fully open states.
Supplementary Movie 1, related to Figure 2
A morphing movie in ribbon diagram shows the conformational transition of TRPV1 upon DkTx binding, following the presumptive sequence of events as labeled. Key structural elements, such as π- to α-helix transition, are marked.
Supplementary Movie 2, related to Figure 3
A morphing movie in ribbon diagram shows the conformational transition of TRPV1 from apo to RTX-bound with NMDG in the pore. Note that there is no π- to α-helix transition.
Supplementary Movie 3, related to Figure 5
A morphing movie in ribbon diagram shows the conformational transition of full-length TRPV1 from apo to RTX bound C1, O1 to C2 conformations. Note that π- to α-helix transition only happen when S6 moves to open the lower gate.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and virus strains | ||
| BL21 Competent Cells | NEB | Cat#C2527H |
| DH10Bac Competent Cells | invitrogen | Cat#10361012 |
| Experimental Models: Cell Lines | ||
| Human: HEK293 GnTI− cells | ATCC | CRL-3022 |
| Insect: Sf9 cells | Expression Systems | Cat#94-001S |
| Chemicals, peptides, and recombinant proteins | ||
| Double-knot toxin (DkTx) | This paper | N/A |
| Resiniferatoxin (RTX) | Sigma | Cat#R8756 |
| N-Methyl-D-glucamine (NMDG) | Sigma | Cat#M2004 |
| QX 314 chloride | Tocris | Cat#2313 |
| YO-PRO-1 iodide | invitrogen | Cat#Y3603 |
| Freestyle 293 Expression Medium | gibco | Cat#12338-018 |
| Insect cell culture media | Expression Systems | Cat#96-001-01 |
| FETAL BOVINE SERUM | PEAK | Cat#PS-FB1 |
| BOVINDE CALF SERUM | HyClone | Cat#SH30072.03 |
| Deposited data | ||
| minimal TRPV1, apo | This paper | EMD-23136; PDB-7L2P |
| minimal TRPV1, DkTx pre-bound | This paper | EMD-23139; PDB-7L2S |
| minimal TRPV1, DkTx singly-bound | This paper | EMD-23161 |
| minimal TRPV1, DkTx, pre-open | This paper | EMD-23138; PDB-7L2R |
| minimal TRPV1, DkTx, partial open | This paper | EMD-23140; PDB-7L2T |
| minimal TRPV1, DkTx, open | This paper | EMD-23141; PDB-7L2U |
| minimal TRPV1, RTX/NMDG a | This paper | EMD-23143; PDB-7L2W |
| minimal TRPV1, RTX/NMDG b | This paper | EMD-23142; PDB-7L2V |
| minimal TRPV1, RTX/NMDG c | This paper | EMD-23144; PDB-7L2X |
| minimal TRPV1, 1 perturbed PI | This paper | EMD-24084; PDB-7MZ6 |
| minimal TRPV1, 1 partially bound RTX | This paper | EMD-24086; PDB-7MZ9 |
| minimal TRPV1, 4 partially bound RTX | This paper | EMD-24085; PDB-7MZ7 |
| minimal TRPV1, 2 bound RTX in adjacent pockets | This paper | EMD-24087; PDB-7MZA |
| minimal TRPV1, 2 bound RTX in opposite pockets | This paper | EMD-24091; PDB-7MZE |
| minimal TRPV1, 3 bound RTX and 1 perturbed PI | This paper | EMD-24088; PDB-7MZB |
| minimal TRPV1, 4 fully engaged RTX C1 | This paper | EMD-24089; PDB-7MZC |
| minimal TRPV1, 4 fully engaged RTX C2 | This paper | EMD-24090; PDB-7MZD |
| full-length TRPV1, apo, neutral pH | This paper | EMD-23128; PDB-7L2H |
| full-length TRPV1, DkTx/RTX | This paper | EMD-23133; PDB-7L2M |
| full-length TRPV1, RTX C1 | This paper | EMD-23134; PDB-7L2N |
| full-length TRPV1, RTX O1 | This paper | EMD-23132; PDB-7L2L |
| full-length TRPV1, RTX C2 | This paper | EMD-24083; PDB-7MZ5) |
| full-length TRPV1, pH6a | This paper | EMD-23129; PDB-7L2I |
| full-length TRPV1, pH6b | This paper | EMD-23131; PDB-7L2K |
| full-length TRPV1, pH6c | This paper | EMD-23130; PDB-7L2J |
| full-length TRPV1, pH5.5-RTX | This paper | EMD-23135; PDB-7L2O |
| TRPV1 in complex with DkTx and RTX | Gao et al., 2016 | PDB: 5irx |
| NavAb N49K/L109A/M116V/G94C/Q150C disulfide crosslinked mutant | Wisedchaisri et al., 2019 | PDB: 6P6W |
| NavAb V100C/Q150C disulfide crosslinked mutant | Wisedchaisri et al., 2019 | PDB: 6P6Y |
| Potassium Channel KcsA-Fab complex | Zhou et al., 2001 | PDB: 1k4c |
| Recombinant DNA | ||
| Plasmid: pET28a_MSP2N2 | (Grinkova et al., 2010) |
http://n2t.net/addgene:29520; RRID:Addgene_29520 |
| Plasmid: pBH_His_DkTx | Bohlen et al., 2010 | N/A |
| Plasmid: pFastBac1-CMV_His-MBP_mini-TRPV1 | This paper | N/A |
| Plasmid: pFastBac1-CMV_His-MBP_FL-TRPV1 | This paper | N/A |
| Software and algorithms | ||
| SerialEM | Mastronarde, 2005 |
http://bio3d.colorado.edu/SerialEM/; RRID:SCR_017293 |
| MotionCor2 | Zheng et al., 2017 |
https://msg.ucsf.edu/software; RRID:SCR_016499 |
| GCTF | Zhang, 2016 |
https://cam.ac.uk/kzha; RRID:SCR_016500 |
| Relion 3.0 | (Zivanov et al., 2018) |
https://cam.ac.uk/relion; RRID:SCR_016274 |
| Relion 3.1 | (Scheres, 2020) |
https://cam.ac.uk/relion; RRID:SCR_016274 |
| cryoSPARC | (Punjani et al., 2017) |
https://cryosparc.com; RRID:SCR_016501 |
| cisTEM | Grant et al., 2018 |
https://cistem.org/; RRID:SCR_016502 |
| PyEM | Daniel Asarnow, Yifan Cheng Lab |
https://github.com/asarnow/pyem; https://doi.org/10.5281/zenodo.3576630 |
| UCSF Chimera | Pettersen et al., 2004 |
https://www.cgl.ucsf.edu/chimera; RRID:SCR_004097 |
| COOT | Emsley et al., 2010 |
https://www2.mrclmb.cam.ac.uk/personal/pemsley/coot/; RRID:SCR_014222 |
| PHENIX | Adams et al., 2010 |
http://www.phenixonline.org; RRID:SCR_014224 |
| ISOLDE | Croll, 2018 | https://isolde.cimr.cam.ac.uk/ |
| Prism 7 | (GraphPad Software, San Diego, CA). |
https://www.graphpad.com/scientificsoftware/prism/; RRID:SCR_002798 |
| Other | ||
| Amylose Resin | NEB | Cat#E8021S |
| QUANTIFOIL R1.2/1.3 300 mesh Au holey carbon grids | quantifoil | https://www.quantifoil.com/ |
Highlight.
Transitional TRPV1 substates reveal two-step mechanism of proton modulation
Substates reveal PI lipid and vanilloid agonist binding stoichiometry and competition
Adaptive selectivity filter accommodates permeation by small and large organic cations
Conformational snapshots reveal allosteric mechanisms connecting key regulatory sites
Acknowledgments:
We thank Dr. Alexander Myasnikov for help in optimizing cryo-EM data collection during the initial stage of this project, Dr. Junrui Li for computational support, Dr. Kenton Swartz and members of the Cheng and Julius laboratories for helpful discussions. The UCSF cryo-EM facility is managed by Dr. David Bulkley and Mr. Glenn Gilbert. Cryo-EM equipment at UCSF is partially supported by NIH grants S10OD020054, S10OD021741. This work was supported by a Human Frontier Science Program Postdoctoral Fellowship (K.Z), and NIH grants (R35NS105038 to D.J. and 1R35GM140847 to Y.C). Y.C. is an Investigator of Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of interests: The authors declare no competing interests.
REFERENCES:
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. (2010). PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66, 213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afonine PV, Poon BK, Read RJ, Sobolev OV, Terwilliger TC, Urzhumtsev A, and Adams PD (2018). Real-space refinement in PHENIX for cryo-EM and crystallography. Acta Crystallogr D Struct Biol 74, 531–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahern GP, Brooks IM, Miyares RL, and Wang XB (2005). Extracellular cations sensitize and gate capsaicin receptor TRPV1 modulating pain signaling. J Neurosci 25, 5109–5116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anandakrishnan R, Aguilar B, and Onufriev AV (2012). H++ 3.0: automating pK prediction and the preparation of biomolecular structures for atomistic molecular modeling and simulations. Nucleic Acids Res 40, W537–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aneiros E, Cao L, Papakosta M, Stevens EB, Phillips S, and Grimm C (2011). The biophysical and molecular basis of TRPV1 proton gating. EMBO J 30, 994–1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker NA, Sept D, Joseph S, Holst MJ, and McCammon JA (2001). Electrostatics of nanosystems: application to microtubules and the ribosome. Proc Natl Acad Sci U S A 98, 10037–10041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bean BP (2015). Pore dilation reconsidered. Nat Neurosci 18, 1534–1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman H, Henrick K, and Nakamura H (2003). Announcing the worldwide Protein Data Bank. Nat Struct Biol 10, 980. [DOI] [PubMed] [Google Scholar]
- Bevan S, and Geppetti P (1994). Protons: small stimulants of capsaicin-sensitive sensory nerves. Trends Neurosci 17, 509–512. [DOI] [PubMed] [Google Scholar]
- Binshtok AM, Bean BP, and Woolf CJ (2007). Inhibition of nociceptors by TRPV1-mediated entry of impermeant sodium channel blockers. Nature 449, 607–610. [DOI] [PubMed] [Google Scholar]
- Bohlen CJ, Priel A, Zhou S, King D, Siemens J, and Julius D (2010). A bivalent tarantula toxin activates the capsaicin receptor, TRPV1, by targeting the outer pore domain. Cell 141, 834–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao E, Liao M, Cheng Y, and Julius D (2013). TRPV1 structures in distinct conformations reveal activation mechanisms. Nature 504, 113–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caterina MJ, Leffler A, Malmberg AB, Martin WJ, Trafton J, Petersen-Zeitz KR, Koltzenburg M, Basbaum AI, and Julius D (2000). Impaired nociception and pain sensation in mice lacking the capsaicin receptor. Science 288, 306–313. [DOI] [PubMed] [Google Scholar]
- Caterina MJ, Schumacher MA, Tominaga M, Rosen TA, Levine JD, and Julius D (1997). The capsaicin receptor: a heat-activated ion channel in the pain pathway. Nature 389, 816–824. [DOI] [PubMed] [Google Scholar]
- Chung MK, Guler AD, and Caterina MJ (2008). TRPV1 shows dynamic ionic selectivity during agonist stimulation. Nat Neurosci 11, 555–564. [DOI] [PubMed] [Google Scholar]
- Croll TI (2018). ISOLDE: a physically realistic environment for model building into low-resolution electron-density maps. Acta Crystallogr D Struct Biol 74, 519–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang S, Feng S, Tien J, Peters CJ, Bulkley D, Lolicato M, Zhao J, Zuberbuhler K, Ye W, Qi L, et al. (2017). Cryo-EM structures of the TMEM16A calcium-activated chloride channel. Nature 552, 426–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang S, van Goor MK, Asarnow D, Wang Y, Julius D, Cheng Y, and van der Wijst J (2019). Structural insight into TRPV5 channel function and modulation. Proc Natl Acad Sci U S A 116, 8869–8878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis JB, Gray J, Gunthorpe MJ, Hatcher JP, Davey PT, Overend P, Harries MH, Latcham J, Clapham C, Atkinson K, et al. (2000). Vanilloid receptor-1 is essential for inflammatory thermal hyperalgesia. Nature 405, 183–187. [DOI] [PubMed] [Google Scholar]
- Deng Z, Maksaev G, Rau M, Xie Z, Hu H, Fitzpatrick JAJ, and Yuan P (2020). Gating of human TRPV3 in a lipid bilayer. Nat Struct Mol Biol 27, 635–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, and Cowtan K (2010). Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66, 486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y, Cao E, Julius D, and Cheng Y (2016). TRPV1 structures in nanodiscs reveal mechanisms of ligand and lipid action. Nature 534, 347–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geron M, Kumar R, Zhou W, Faraldo-Gomez JD, Vasquez V, and Priel A (2018). TRPV1 pore turret dictates distinct DkTx and capsaicin gating. Proc Natl Acad Sci U S A 115, E11837–E11846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goddard TD, Huang CC, Meng EC, Pettersen EF, Couch GS, Morris JH, and Ferrin TE (2018). UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Protein Sci 27, 14–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant T, Rohou A, and Grigorieff N (2018). cisTEM, user-friendly software for single-particle image processing. Elife 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grinkova YV, Denisov IG, and Sligar SG (2010). Engineering extended membrane scaffold proteins for self-assembly of soluble nanoscale lipid bilayers. Protein Eng Des Sel 23, 843–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guskov A, Jensen S, Faustino I, Marrink SJ, and Slotboom DJ (2016). Coupled binding mechanism of three sodium ions and aspartate in the glutamate transporter homologue GltTk. Nat Commun 7, 13420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazan A, Kumar R, Matzner H, and Priel A (2015). The pain receptor TRPV1 displays agonist-dependent activation stoichiometry. Sci Rep 5, 12278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes TE, Del Rosario JS, Kapoor A, Yazici AT, Yudin Y, Fluck EC 3rd, Filizola M, Rohacs T, and Moiseenkova-Bell VY (2019). Structure-based characterization of novel TRPV5 inhibitors. Elife 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui K, Liu B, and Qin F (2003). Capsaicin activation of the pain receptor, VR1: multiple open states from both partial and full binding. Biophys J 84, 2957–2968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynh KW, Cohen MR, Jiang J, Samanta A, Lodowski DT, Zhou ZH, and Moiseenkova-Bell VY (2016). Structure of the full-length TRPV2 channel by cryo-EM. Nat Commun 7, 11130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssens A, and Voets T (2011). Ligand stoichiometry of the cold- and menthol-activated channel TRPM8. J Physiol 589, 4827–4835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jara-Oseguera A, Huffer KE, and Swartz KJ (2019). The ion selectivity filter is not an activation gate in TRPV1–3 channels. Elife 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordt SE, Tominaga M, and Julius D (2000). Acid potentiation of the capsaicin receptor determined by a key extracellular site. Proc Natl Acad Sci U S A 97, 8134–8139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julius D (2013). TRP channels and pain. Annu Rev Cell Dev Biol 29, 355–384. [DOI] [PubMed] [Google Scholar]
- Khakh BS, Bao XR, Labarca C, and Lester HA (1999). Neuronal P2X transmitter-gated cation channels change their ion selectivity in seconds. Nat Neurosci 2, 322–330. [DOI] [PubMed] [Google Scholar]
- Lebedev AA, Young P, Isupov MN, Moroz OV, Vagin AA, and Murshudov GN (2012). JLigand: a graphical tool for the CCP4 template-restraint library. Acta Crystallogr D Biol Crystallogr 68, 431–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao M, Cao E, Julius D, and Cheng Y (2013). Structure of the TRPV1 ion channel determined by electron cryo-microscopy. Nature 504, 107–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin King JV, Emrick JJ, Kelly MJS, Herzig V, King GF, Medzihradszky KF, and Julius D (2019). A Cell-Penetrating Scorpion Toxin Enables Mode-Specific Modulation of TRPA1 and Pain. Cell 178, 1362–1374 e1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastronarde DN (2005). Automated electron microscope tomography using robust prediction of specimen movements. J Struct Biol 152, 36–51. [DOI] [PubMed] [Google Scholar]
- McGoldrick LL, Singh AK, Saotome K, Yelshanskaya MV, Twomey EC, Grassucci RA, and Sobolevsky AI (2018). Opening of the human epithelial calcium channel TRPV6. Nature 553, 233–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munns CH, Chung MK, Sanchez YE, Amzel LM, and Caterina MJ (2015). Role of the outer pore domain in transient receptor potential vanilloid 1 dynamic permeability to large cations. J Biol Chem 290, 5707–5724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadezhdin KD, Neuberger A, Nikolaev YA, Murphy LA, Gracheva EO, Bagriantsev SN, and Sobolevsky AI (2021). Extracellular cap domain is an essential component of the TRPV1 gating mechanism. Nat Commun 12, 2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohi M, Li Y, Cheng Y, and Walz T (2004). Negative Staining and Image Classification - Powerful Tools in Modern Electron Microscopy. Biol Proced Online 6, 23–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulsen CE, Armache JP, Gao Y, Cheng Y, and Julius D (2015). Structure of the TRPA1 ion channel suggests regulatory mechanisms. Nature 520, 511–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, and Ferrin TE (2004). UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem 25, 1605–1612. [DOI] [PubMed] [Google Scholar]
- Pumroy RA, Samanta A, Liu Y, Hughes TE, Zhao S, Yudin Y, Rohacs T, Han S, and Moiseenkova-Bell VY (2019). Molecular mechanism of TRPV2 channel modulation by cannabidiol. Elife 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punjani A, Rubinstein JL, Fleet DJ, and Brubaker MA (2017). cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat Methods 14, 290–296. [DOI] [PubMed] [Google Scholar]
- Puopolo M, Binshtok AM, Yao GL, Oh SB, Woolf CJ, and Bean BP (2013). Permeation and block of TRPV1 channels by the cationic lidocaine derivative QX-314. J Neurophysiol 109, 1704–1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohacs T (2015). Phosphoinositide regulation of TRPV1 revisited. Pflugers Arch 467, 1851–1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenthal PB, and Henderson R (2003). Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J Mol Biol 333, 721–745. [DOI] [PubMed] [Google Scholar]
- Ryu S, Liu B, Yao J, Fu Q, and Qin F (2007). Uncoupling proton activation of vanilloid receptor TRPV1. J Neurosci 27, 12797–12807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres SHW (2020). Amyloid structure determination in RELION-3.1. Acta Crystallogr D Struct Biol 76, 94–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sine SM, and Claudio T (1991). Gamma- and delta-subunits regulate the affinity and the cooperativity of ligand binding to the acetylcholine receptor. J Biol Chem 266, 19369–19377. [PubMed] [Google Scholar]
- Singh AK, McGoldrick LL, and Sobolevsky AI (2018). Structure and gating mechanism of the transient receptor potential channel TRPV3. Nat Struct Mol Biol 25, 805–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smart OS, Neduvelil JG, Wang X, Wallace BA, and Sansom MS (1996). HOLE: a program for the analysis of the pore dimensions of ion channel structural models. J Mol Graph 14, 354–360, 376. [DOI] [PubMed] [Google Scholar]
- Starowicz K, Nigam S, and Di Marzo V (2007). Biochemistry and pharmacology of endovanilloids. Pharmacol Ther 114, 13–33. [DOI] [PubMed] [Google Scholar]
- Susankova K, Ettrich R, Vyklicky L, Teisinger J, and Vlachova V (2007). Contribution of the putative inner-pore region to the gating of the transient receptor potential vanilloid subtype 1 channel (TRPV1). J Neurosci 27, 7578–7585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tominaga M, Caterina MJ, Malmberg AB, Rosen TA, Gilbert H, Skinner K, Raumann BE, Basbaum AI, and Julius D (1998). The cloned capsaicin receptor integrates multiple pain-producing stimuli. Neuron 21, 531–543. [DOI] [PubMed] [Google Scholar]
- van Goor MK, de Jager L, Cheng Y, and van der Wijst J (2020). High-resolution structures of transient receptor potential vanilloid channels: Unveiling a functionally diverse group of ion channels. Protein Sci 29, 1569–1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisedchaisri G, Tonggu L, McCord E, Gamal El-Din TM, Wang L, Zheng N, and Catterall WA (2019). Resting-State Structure and Gating Mechanism of a Voltage-Gated Sodium Channel. Cell 178, 993–1003 e1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K (2016). Gctf: Real-time CTF determination and correction. J Struct Biol 193, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Lin King JV, Paulsen CE, Cheng Y, and Julius D (2020). Irritant-evoked activation and calcium modulation of the TRPA1 receptor. Nature 585, 141–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng SQ, Palovcak E, Armache JP, Verba KA, Cheng Y, and Agard DA (2017). MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat Methods 14, 331–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W, Trinco G, Slotboom DJ, Forrest LR, and Faraldo-Gomez JD (2021). On the Role of a Conserved Methionine in the Na(+)-Coupling Mechanism of a Neurotransmitter Transporter Homolog. Neurochem Res. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Morais-Cabral JH, Kaufman A, and MacKinnon R (2001). Chemistry of ion coordination and hydration revealed by a K+ channel-Fab complex at 2.0 A resolution. Nature 414, 43–48. [DOI] [PubMed] [Google Scholar]
- Zivanov J, Nakane T, Forsberg BO, Kimanius D, Hagen WJ, Lindahl E, and Scheres SH (2018). New tools for automated high-resolution cryo-EM structure determination in RELION-3. Elife 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zubcevic L, Herzik MA Jr., Chung BC, Liu Z, Lander GC, and Lee SY (2016). Cryo-electron microscopy structure of the TRPV2 ion channel. Nat Struct Mol Biol 23, 180–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zubcevic L, Herzik MA Jr., Wu M, Borschel WF, Hirschi M, Song AS, Lander GC, and Lee SY (2018a). Conformational ensemble of the human TRPV3 ion channel. Nat Commun 9, 4773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zubcevic L, Le S, Yang H, and Lee SY (2018b). Conformational plasticity in the selectivity filter of the TRPV2 ion channel. Nat Struct Mol Biol 25, 405–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Single particle cryo-EM structure determination, related to Figure 1, 3, 4, 5 and 6.
For each structure presented in this study, panels are arranged from left to right in the same order: directional Fourier Shell Correlation (dFSC) plots and averaged FSC (green curve) with resolution estimated on FSC=0.143 marked, side and top views of the density map color code with local resolution. (A) minimal TRPV1 apo; (B) with DkTx pre-bound; (C) single DkTx bound; (D) – (F) with double DkTx bound in pre-open (D), partial open (E) and open (F); (G) – (I) three conformations with RTX and NMDG bound; (J) – (O) six conformations of TRPV1 with RTX, QX-314 and 9 mM Na+, showing various sub-stoichiometric binding states of RTX; (P) and (Q) two conformations of TRPV1 with RTX, YO-PRO-1 and 60 mM Na+ with fully and stoichiometrically bound RTX; (R) Full length apo TRPV1 at neutral pH; (S) - (U) three conformations of full-length TRPV1 at pH6, pH6a, pH6b and pH6c; (V) full-length TRPV1 at pH5.5 with RTX bound. (W) – (Y) three conformations of full-length TRPV1 at neutral pH with RTX bound. (Z) Full-length TRPV1 with DkTx and RTX bound.
Figure S2. Classification scheme of TRPV1 with DkTx bound, related to Figure 1 and 2.
The scheme illustrates the classification processes that identified five different conformations in the dataset. Symmetry expansion applied to each conformation further improves the conformational homogeneity within each conformational class. For data processing efficiency, the entire dataset was separated into two groups that each went through the same classification scheme. Once a clean class was obtained after symmetry expansion, particles of the same conformation were recombined to a final reconstruction. The particle numbers of each final reconstruction are indicated.
Figure S3. Binding of DkTx to TRPV1, related to Figure 1 and 2.
(A) and (B) Side and top view of DkTx densities bound to the channel, shown as ribbon diagram, in four DkTx bound substates. From left to right, panels are single bound, pre-open, partial open and open.
(C) Segmented density of the phosphatidylinositol lipid residing in the vanilloid binding pocket. Densities of the lipid head group and one acyl chain become progressively weaker as the lower gate shifts towards opening.
(D) Segmented density of an annular lipid. In the open conformation, one of the lipid tails is seen inserting into a hydrophobic cleft between S5 and S6 of neighboring subunits.
Figure S4. Conformational dynamics upon DkTx binding, related to Figure 1 and 2.
(A) Solvent accessible pathway along the ion permeation pore of minimal TRPV1 in apo, pre-bound, pre-open, partial open and open substates. Residues that form the upper restriction and lower gate are labeled. Note that the water accessible volume of the central cavity is enlarged in partial open and open.
(B) Side views of ion permeation pathway with the distance between critical residues (M644, G643, Y671, I679 and M682) of two opposing subunits marked.
(C) Overlay of the lower part of S6 in apo, partial open and open states. Transition of S6 from π-helix in apo to α-helix in partial open and open substates rotates I679 away and M682 into the pore.
(D) – (H) M682 is well resolved in pre-open, but less well-resolved partial open and open substates.
(I) Temperature factor of M682 in partial open is higher than in other substates. Temperature factor of neighboring residue A680 of the same structure is used as a reference point.
(J) Pore radii of partial open and open are compared with that of apo and fully open (5IRX) states. Dashed line in the profile of partial open substate indicating the uncertainty in the calculation caused by less well-resolved M682 in one subunit. Two vertical dashed line marks the radii of dehydrated Ca2+ (1.1Å) and water (1.4Å).
Figure S5. Conformational changes at the lower gate induced by sub-stoichiometric binding of RTX, related to Figure 4.
(A) A bottom slab view of the lower gate in which the apo structure is overlaid with RTX binding substates (number and state of RTX binding are labeled as in Figure 5B). Perturbation of endogenous PI lipid does not induce noticeable changes in the lower gate. Partial binding of RTX induces movement of S5 within the same subunit. With RTX fully engaged in all binding sites (seen only in 60 mM Na+ sample), the lower gate assumes two different conformations: in one, the S5 movement is small and S6 does not move, while in the other, both S5 and S6 move and the lower gate is open. Overall, the level of movement of S5 correlates closely with the level of RTX engagement in the vanilloid pocket such that only fully engaged RTX causes sufficient S5 movement to move S6 and to open the lower gate.
(B) Ligand density within the vanilloid binding pocket calculated from combining different ratio of PI-containing and RTX-containing particles. These simulations show how the ligand density would appear if the PI containing and RTX containing vanilloid pockets were not cleanly separated by classification before calculating 3D reconstructions.
Figure S6. Binding of RTX to full-length TRPV1, related to Figure 5.
(A) Solvent accessible pathway along the ion permeation pore of minimal and full-length channels in apo and DkTx and RTX bound states.
(B) Overlay of full and minimal TRPV1 structures determined at neutral pH shows that the two structures are largely indistinguishable, except for the turret region in the enlarged view. The windowed regions are enlarged in the adjacent panel. The location of E600 is marked.
(C) The turret density appears as an unstructured density atop the pore.
(D) Pore radii of minimal and full-length channels in apo and DkTx and RTX bound states.
(E) Overlay of outer pore (left) and lower gate (right) for minimal and full-length DkTx/RTX bound channels.
(F) and (G) With RTX bound, overlay of substates C1 of minimal with C1 of full-length channel (F), substate C2 of minimal with C2 of full-length channel (G).
(H) Solvent accessible ion permeation pathway along the ion permeation pore of minimal TRPV1 with fully engaged RTX in substates C1 and C2.
(I) Solvent accessible ion permeation pathway along the ion permeation pore of full-length TRPV1 with RTX in substates C1, O1 and C2.
(J) Overlay of full-length TRPV1 structures with RTX in substates C1, O1 or C2 with apo structure.
(K) and (L) Bottom view of lower gate in substates C1 or O1.
(M) and (N) side and bottom view of substate C2 show a cation density in the vicinity of M682.
Figure S7. Proton-evoked conformational changes, related to Figure 6.
(A) and (B) Comparison between the full-length structures captured at low and neutral pH. Each panel shows the overlay of a low pH with neutral pH structure (as labeled), in either top view (A) of VSLD and pore domain, or bottom view of the lower gate (B). From the left to right, panels are arranged such that pH6a (red), pH6b (blue), pH6c (green) and pH5.5/RTX (fuchsia pink) are compared with the structure of neutral pH (light blue). The VSLD is tilted backwards in pH6a. Upper and lower parts of the pore loop are moved in pH6b, pH6c and pH5.5/RTX.
(C) Surface charges of the outer pore region calculated at neutral pH (left), pH6c (middle), and pH5.5 with RTX (right).
(D) Comparison of pore radii of the channel at pH6 and pH5.5 with that at neutral pH and fully open states.
Supplementary Movie 1, related to Figure 2
A morphing movie in ribbon diagram shows the conformational transition of TRPV1 upon DkTx binding, following the presumptive sequence of events as labeled. Key structural elements, such as π- to α-helix transition, are marked.
Supplementary Movie 2, related to Figure 3
A morphing movie in ribbon diagram shows the conformational transition of TRPV1 from apo to RTX-bound with NMDG in the pore. Note that there is no π- to α-helix transition.
Supplementary Movie 3, related to Figure 5
A morphing movie in ribbon diagram shows the conformational transition of full-length TRPV1 from apo to RTX bound C1, O1 to C2 conformations. Note that π- to α-helix transition only happen when S6 moves to open the lower gate.
Data Availability Statement
Data Atomic coordinates and cryo-EM maps are deposited in EMDB and PDB as follows: Minimal TRPV1 in apo: EMD-23136, PDB-7L2P; Minimal TRPV1 with DkTx bound in pre-bound: EMD-23139, PDB-7L2S; with single DkTx bound: EMD-23161; pre-open: EMD-23138, PDB-7L2R; partial open: EMD-23140, PDB-7L2T; open: EMD-23141, PDB-7L2U; Minimal TRPV1 with RTX and NMDG, in state a: EMD-23143, PDB-7L2W; in state b: EMD-23142, PDB-7L2V; and in state c: EMD-23144, PDB-7L2X; minimal TRPV1 with RTX and QX-315 in buffer of 9 mM NaCl: with 1 perturbed PI: EMD-24084, PDB-7MZ6; with 1 partial bound RTX: EMD-24086, PDB-7MZ9; with 4 partial bound RTX: EMD-24085, PDB-7MZ7; with 2 bound RTX in adjacent subunits: EMD-24087, PDB-7MZA; with 2 bound RTX in opposite subunit: EMD-24091, PDB-7MZE; with 3 bound RTX and 1 perturbed PI: EMD-24088, PDB-7MZB; minimal TRPV1 with fully engaged RTX in buffer of 60mM NaCl and YO-PRO-1, in state C1: EMD-24089, PDB-7MZC; in state C2: EMD-24090, PDB-7MZD; Full-length TRPV1 in apo and neutral pH: EMD-23128, PDB-7L2H; Full-length TRPV1 with DkTx and RTX bound: EMD-23133, PDB-7L2M; Full-length TRPV1 with RTX bound, in state C1: EMD-23134, PDB-7L2N, in state O1: EMD-23132, PDB-7L2L; in state C2: EMD-24083, PDB-7MZ5; Full-length TRPV1 in pH 6, state a: EMD-23129, PDB-7L2I; in state b: EMD-23131, PDB-7L2K; in state c: EMD-23130, PDB-7L2J; Full-length TRPV1 in pH 5.5 and with RTX bound: EMD-23135, PDB-7L2O.
Code UCSF pyem is available at https://zenodo.org/record/3576630#.XuczyFVKjIU
Additional information None


