A new model of human neuroblastoma provides ways to track tumor formation and expansion in living animals, allowing identification of CD4+ T-cell and macrophage functions required for oncogenesis.
Abstract
Immune cells regulate tumor growth by mirroring their function as tissue repair organizers in normal tissues. To understand the different facets of immune–tumor collaboration through genetics, spatial transcriptomics, and immunologic manipulation with noninvasive, longitudinal imaging, we generated a penetrant double oncogene–driven autochthonous model of neuroblastoma. Spatial transcriptomic analysis showed that CD4+ and myeloid populations colocalized within the tumor parenchyma, while CD8+ T cells and B cells were peripherally dispersed. Depletion of CD4+ T cells or CCR2+ macrophages, but not B cells, CD8+ T cells, or natural killer (NK) cells, prevented tumor formation. Tumor CD4+ T cells displayed unconventional phenotypes and were clonotypically diverse and antigen independent. Within the myeloid fraction, tumor growth required myeloid cells expressing arginase-1. Overall, these results demonstrate how arginine-metabolizing myeloid cells conspire with pathogenic CD4+ T cells to create permissive conditions for tumor formation, suggesting that these protumorigenic pathways could be disabled by targeting myeloid arginine metabolism.
Significance:
A new model of human neuroblastoma provides ways to track tumor formation and expansion in living animals, allowing identification of CD4+ T-cell and macrophage functions required for oncogenesis.
Graphical Abstract

Introduction
Despite the burgeoning interest in tumor immunology and inflammation, and its importance to understanding and treating human cancer, many conceptual and technical limitations are restraining the basic understanding of the immune–tumor interplay and the practical outcomes of new knowledge about cancer. These restrictions encompass widespread use of orthotopic transplantation of tumor cell lines of dubious relevance to human cancer, transplantation to anatomic sites of limited relevance to the natural origins of a given cancer, inability to track and predict immune cell infiltration patterns, and interlaboratory variance in models. A limitation of mouse cancer models is that they do not accurately reflect the time frame of human malignancies, or the accumulation of mutations. Neuroblastoma is an ideal “laboratory” to explore the immune–tumor interface. Neuroblastoma arises in the neuroectoderm and mainly forms from the adrenal medulla. The driver oncogenes for neuroblastoma are well understood (MYCN, LIN28, ALK) and form the basis of neuroblastoma mouse models, which reflect the rate of incidence, location and pathology of human disease (1,2,3). The predictable location and incidence of neuroblastoma allows focus across a window of tumor growth, as opposed to conventional cell line or xenograft systems where a bolus of cells is introduced into an animal.
Immune cells, and especially myeloid cells such as different types of macrophages, dendritic cells, and myeloid-derived suppressor cells (MDSC), invade tumors from the earliest stages of tumor growth. Large numbers of macrophages and other myeloid cell types, as well as regulatory T cells (Treg) within tumors, are correlated with poor outcomes in most childhood and adult cancers, spurring a multitude of clinical approaches to target these pathways (4, 5). Macrophage infiltration of tumors is inseparable from the process of “cancer inflammation”, a “hallmark” of cancer. More specifically, cancer is a type of nonresolving inflammation (6, 7). Other examples of nonresolving inflammation include leprosy, latent tuberculosis, asbestosis, the foreign body reaction to implanted medical devices, and chronic diseases such as Crohn's Disease and lupus. While the “rules” governing immune cell behavior in resolving inflammation in different organs are rapidly emerging, nonresolving inflammation is more complex because the resolving phase of inflammation is absent or aberrant. That the tumor microenvironment is immune “suppressive” is an accepted facet of most cancers because: (i) since tumors are “self” the numbers of antigen-specific lymphocytes are low, and (ii) overcoming tolerance to self-antigens is a key strategy for clinical development of agents to provoke an antitumor response. In this regard, attempts to bypass self-tolerance are key goals of cancer therapy.
Poorly vascularized and hypoxic inflammatory microenvironments are deficient in key nutrients necessary to support expansion of effector immune functions. Examples of these phenomena include infections associated with hypoxic granulomas such as tuberculosis and schistosomiasis (8, 9). In these diseases, arginine consumption by myeloid cells is required to suppress tissue destructive T-cell proliferation as part of the tissue repair and resolution process (8, 10). Within the immune system and especially the myeloid compartment, regulated enzymes metabolize amino acids (11), including Arginase-1 (Arg1), a cytoplasmic hydrolase that metabolizes arginine to ornithine, and Arg2, a closely related arginase to Arg1 that performs the same biochemical reaction, but is restricted to mitochondria. Both enzymes have been reported in multiple tumor types (12, 13) and are under complex regulatory control by cytokines and microbial products. The notion that T-cell responses can be suppressed by local myeloid cells expressing amino acid–consuming enzymes is hypothesized to be a component of the regulatory network of “infectious tolerance”, where different immune cells propagate to suppress neighboring cells (14, 15).
In cancer, a disease generally associated with aberrant vascularization and hypoxia, both resident myeloid cells and the tumor cells themselves can consume amino acids (16). Indeed, in pancreatic cancer models, arginine and tryptophan are specifically depleted from the tumor microenvironment relative to all other amino acids (17). Tumors may have therefore harnessed a natural immunoregulatory program as a mechanism to aid in immune evasion. Arg1 is closely associated with cancer immunosuppression and is a recent clinical target (18,19,20,21). The overall goal of targeting myeloid amino acid–consuming enzymes is to enhance local antitumor T-cell responses, or to overcome a “negative” signal that blocks antitumor T-cell reactivity. So far, genetic tests of the role of myeloid amino acid–metabolizing enzymes in autochthonous tumors have been limited to a single study where the loss of IDO1 on a KRAS-driven lung cancer model had limited effect in prolonging survival (22). We hypothesized that amino acid–dependent pathways are corrupted in the tumor microenvironment, and this phenomenon is linked to the inability of the host to control tumor proliferation and to changes in T-cell phenotypic states. To test this, we constructed a genetic system to measure the contribution of a key cancer-associated myeloid amino acid–metabolizing enzyme Arg1 to cancer development and progression.
Materials and Methods
Mice
C57BL/6, Ccr2−/−, Stat6−/−, Rag1−/−, Argflox, Tie2-Cre and OT-II mice were purchased from The Jackson Laboratory. Generation of Alk-mutant strains is described in Supplementary Materials and Methods. All mice used in this study were bred and housed within a single cubicle and maintained in a 12-hour day–night cycle with constant temperature and humidity. The SJCRH Institutional Animal Care and Use Committee approved all studies performed.
Murine neuroblastoma study criteria and study design
Inclusion criteria were presence of the TH-MYCN allele, and heterozygosity for one Alk-mutant allele and an age range from 1 to 3 weeks postweaning. Criteria for analysis were the presence or absence of an ultrasound-detectable tumor at any point within a 7-week period, where mice were imaged once per week. End point criteria are listed in Supplementary Materials and Methods. All raw data are available in Supplementary Tables S1 and S2.
Immune cell isolation
Tumors were excised, mechanically dissociated and transferred to basal medium containing 0.1% Collagenase Type IV and 150 mg/mL DNase I, incubated for 30-minute shaking at 37°C, then passed through a 70-μm nylon mesh. Cells were centrifuged for 5 minutes at 400 × g, resuspended in PBS, and overlaid on a 35%/60% Percoll gradient. Gradients were centrifuged at 2,000 × g for 20 minutes at 4°C without brake. Cells were collected from the 35%/60% interface and washed with PBS, prior to further analysis.
Flow cytometry
Tumor-derived cells were stained for cell surface markers and transcription factors as described in Supplementary Materials and Methods. Data were acquired with a BD LSRFortessa X-20 and Cytek Aurora spectral flow cytometer, and analyzed in FlowJo v10 (Treestar).
Human IHC
Patient sample analysis was approved by the Institutional Review Board at SJCRH and informed written consent was obtained, where applicable, for all human sample analysis. IHC studies were performed on archival formalin-fixed paraffin-embedded tissue blocks from 5 primary human neuroblastoma (3 with MYCN amplification; 2 without MYCN amplification). A 4-μm section from a representative block of each tumor was subjected to IHC. Antibodies are listed in Supplementary Materials and Methods.
In vivo depletions
In vivo depletions of CD4+, CD8+, B-cell, and natual killer (NK)–cell populations are described in Supplementary Materials and Methods. Successful depletion was confirmed by flow cytometric analysis.
TCR transgenic CD4 transfer
OT-II spleens and lymph nodes were manually dissociated through a 70-μm nylon mesh, centrifuged for 5 minutes at 400 × g, and subjected to red cell lysis. Cells were then surface stained and cell sorted. AlkF1178L; TH-MYCN were intravenously administered with 5 × 106 CD4+ Vβ5.1, 5.2+ 1 week prior to study enrollment. After detection by ultrasound, tumors were processed as detailed above for downstream processing.
In vitro amino acid starvation assays
Amino acid starvation experiments were performed as described previously (23). 2 ng/mL recombinant murine TGFβ (eBioscience) was added to cultures where indicated.
Bulk transcriptome analysis
Human primary neuroblastoma microarray data from Gene Expression Omnibus (GEO) sources (GSE12460, GSE13136) were RMA normalized, summarized and batch corrected in Partek Genomics Suite 6.6. These data were merged to RMA summarized public human Ewing sarcoma, retinoblastoma, and rhabdomysarcoma samples (E-TABM-1202 ArrayExpress, GSE29683, GSE37372). The detailed bioinformation analyses are described in Supplementary Materials and Methods. The data were then matched by gene symbols to deduplicated mouse neuroblastoma model data (GSE98763, GSE27516). The data for 70 selected genes were then z transformed, hierarchically clustered, and a heatmap produced using Partek Genomics Suite 6.6. For murine gene expression profiles, 100 ng of total RNA was processed using the Affymetrix Whole Transcript (WT) Plus assay according to the manufacturer's protocol. The resulting cDNA was labeled, fragmented hybridized to the Mouse Gene 2.0 assay at 60 rpm for 16 hours at 45°C. The arrays were then washed and stained using the Affymetrix Model 450 Fluidics Station and scanned with the Affymetrix Model 3000 7G scanner according to the manufacturer's protocols. Detailed bioinformation analyses are described in Supplementary Materials and Methods. Expression measures were background-corrected, normalized, and summarized from scanned intensity files using the oligo R package. Probes were annotated using the annotate package with the mogene20sttranscriptcluster database, and predicted genes, pseudogenes, and genes on the Y chromosome were excluded from downstream analyses. Differential expression across treatments was assessed using limma, with a false discovery rate (FDR) of 0.05 as the cutoff for statistical significance. Heatmaps were generated using the NMF package. Microarray files have been deposited in the GEO (accession GSE126024).
Single-cell gene expression
Tumor-resident CD45+ cells were distinguished from circulating CD45+ cells as described in Supplementary Materials and Methods. Viable CD45− and tumor-resident CD45+ populations were sorted, centrifuged at 400 × g for 5 minutes and resuspended in 0.04% BSA in PBS. CD45− and CD45+ populations were combined and approximately 20,000 cells were loaded on the Chromium controller (10X Genomics) with the single-cell 3′ kit (V2) to capture approximately 10,000 cells. The remaining steps were performed according to the manufacturer's protocol. Sequencing was performed on the Illumina Novaseq to generate 500M clusters per library. Detailed data analysis methods are outlined in Supplementary Materials and Methods. Transcriptional clusters were annotated manually by considering expression of various cell type–specific genes (Supplementary Fig. 1C; ref. 24), as well as with SingleR (25).
Spatial transcriptomics
Two tumors between 100 and 200 mm3 were harvested from distinct mice and were processed as described in Supplementary Materials and Methods. Cryosectioned tumors were used in the Visium Spatial Transcriptomics kit (10X Genomics) and libraries were sequenced on the Illumina NovaSeq platform at 28 × 120 bp. The detailed bioinformatic analyses are described in Supplementary Materials and Methods.
TCR repertoire analyses
CD4+ CD25+ cells were sorted into 384-well plates, where a single cell is in an individual well, with index sorting enabled. Two columns of each 384-well plate were left unsorted to serve as a negative control. Reverse transcription was completed using SuperScript VILO (Invitrogen) based on manufacturer's protocol. TCR αβ cDNA were amplified and sequencing with previously described methods (26). TCR sequencing output was analyzed via TCRdist (27).
Statistical analysis
All mice included into the study were analyzed sequentially. At the cessation of the study, all primary data was independently checked against the original database by two people and entered into GraphPad Prism v7 for survival curve comparison. The log-rank test was used as the primary statistic to estimate significance or nonsignificance. Exact P values for curve comparisons are reported in the figures. Two-tailed t tests or Kruskal–Wallis tests corrected for multiple comparisons were used as appropriate. Additional survival analyses were conducted using Cox proportional hazard (CoxPH) regression models in the R survival package after verification of model assumptions and visualized using survminer. The R ggplot2 package was used for the customized forest plot after conducting pairwise comparisons; FDR was used to adjust for multiple comparisons within each study group, with error bars representing the SE of the coefficients as determined by the respective CoxPH model.
Data availability
Single-cell gene expression (scGEX) and spatial transcriptomics data are accessioned in the Short Read Archive in association with BioProject PRJNA662418. Microarray files have been deposited in the GEO (accession GSE126024).
Results
Development of a penetrant neuroblastoma model
Neuroblastoma is the most prevalent solid tumor of children and originates from the sympathetic nervous system that gives rise to the catecholamine-producing adrenal medulla (28). Consequently, neuroblastoma predominantly arise in the kidney-aorta region. MYCN amplification is the most common genetic abnormality in pediatric neuroblastoma (28). Tyrosine hydroxylase (TH, the rate-limiting enzyme for catecholamine generation) promoter-driven MYCN transgenic mice are an established model of murine neuroblastoma that parallel aspects of the childhood disease (1, 2, 29) with tumors exhibiting predictable kinetics, quantifiable by noninvasive means. However, in our vivarium, tumor formation in TH-MYCN mice occurred in less than 10% of screened animals. To improve our understanding of neuroblastoma biology and intratumoral immunology, we created a highly penetrant autochthonous neuroblastoma model that builds upon the previous genetic approach. To accelerate tumor formation in the low penetrance TH-MYCN background, we introduced point mutations in Alk, a tyrosine kinase oncogene, which, after MYCN, is the most commonly associated gene with human neuroblastoma. Two gain-of-function mutations were created: AlkF1178L (equivalent to human ALKF1174L, which is the predominant ALK mutation in human neuroblastoma; ref. 29), and AlkY1282S (equivalent to human ALKY1278S, also found in human neuroblastoma; Fig. 1A; Supplementary Fig. S1A and S1B; refs. 30, 31). The F1178 L or Y1282S mutations were crossed into the TH-MYCN background in heterozygosity. We longitudinally tracked neuroblastoma formation and growth by ultrasound imaging (Supplementary Table S1) within a design appropriate to end point comparison of individual animals across time (Fig. 1B). Mice with animal welfare issues unrelated to a tumor were excluded (Supplementary Table S2). Mice with AlkF1178L on a TH-MYCN background developed neuroblastoma with an incidence of approximately 50% within the study window, and approximately 40% of mice achieved a study end point within the study period. The AlkY1282S mutation on the TH-MYCN background produced approximately half the tumor incidence and survival rate of the AlkF1174L mutation (Fig. 1C). Consistent with other studies (29, 32, 33), we found that the transcriptomes of the tumor samples were concordant with human neuroblastoma and distinct from other childhood solid tumors (Fig. 1D; Supplementary Tables S3 and S4). We observed significant expression of the current drug targets GD2 (Fig. 1E) and B7-H3 (CD276; Fig. 1F), and low amounts of PD-L1 and PD-L2 (Fig. 1G), replicating expression patterns seen in human neuroblastoma (34,35,36) and demonstrating the utility of this animal model as a preclinical tool. For all remaining longitudinal studies, we used the high penetrance AlkF1178L; TH-MYCN mice to investigate how manipulating amino acid metabolic pathways contributes to tumor growth. Unlike the widely used xenograft transplantation models, our autochthonous, genetics-based model system allows for precise identification of the conditions in which malignancy is established and permits detailed analysis of the earliest stages of the antitumor immune response.
Figure 1.
Point mutations in the endogenous Alk locus and neuroblastoma formation. A, Diagram of the two ALK mutations introduced into the endogenous Alk locus. B, Experimental design of the longitudinal study for animals reported herein. C, Incidence and survival curves comparing tumor formation of AlkF1178L; TH-MYCN or AlkY1282S; TH-MYCN mice. Representative image of an AlkF1178L; TH-MYCN mouse with a large abdominal neuroblastoma. D, Clustered heat map of “signature” gene expression comparing murine neuroblastoma (NB) models and other human solid tumors. E, GD2 expression in CD45− tumor cells. Purple and green histograms show GD2 expression from two individual tumors compared with isotype control (gray histogram). F, CD276 (B7-H3) expression in tumor cells. Purple and green histograms show CD276 expression from CD45− cells from two individual tumors compared with isotype control (gray histogram). G, Frequency of PD-L1– and PD-L2–expressing cells in CD45+ or CD45− intratumoral populations. In C, data were analyzed by the log-rank test. Criteria for scoring tumor incidence and inclusion and exclusion criteria are described in Materials and Methods. In D, the complete transcriptional signature is available in Supplementary Table S3 and available in full using accession numbers GSE12460, GSE13136, GSE16237, GSE37372, GSE29683, E-TABM-1202, GSE27516, GSE98763, and GSE27516. Red font, canonical neuroblastoma-associated mRNAs.
Single-cell sequencing identified transcripts encoding amino acid–metabolizing enzymes
We next sought to characterize the immune infiltrate of our neuroblastoma tumor model using single-cell transcriptomics. We collected scGEX data on our AlkF1178L; TH-MYCN tumors by isolating tumor cells and parenchymal immune cells, which were identified by injecting an anti-CD45 antibody intravenously prior to euthanizing the animals. After tumor processing, we stained with a CD45 antibody conjugated with a different fluorophore, allowing us to segregate vascular and parenchymal immune cells. Parenchymal immune cells and tumor were mixed at equal ratios for scGEX profiling (Fig. 2A; Supplementary Fig. S1C).
Figure 2.
Single-cell gene expression and spatial transcriptomic analysis of the neuroblastoma landscape characterize tumor-infiltrating immune populations and tumor cell subsets. A, t-SNE dimensionality reduction of mouse AlkF1178L; TH-MYCN CD45− and CD45+ tumor cells based on scGEX data, with cell clusters identified based on marker gene expression (Supplementary Fig. S1C). DC, dendritic cell; Tfh, T follicuar helper. B, Feature plot of tumor cells indicated by module scoring of Ncam1, Th, and Mycn expression. C, Box plots representing the proportion of cell types among the tumor-infiltrating CD45+ cells using the clusters identified in the scGEX analysis. Colors indicate replicate tumor identity within and across plots. D, Feature plots showing expression of gene markers of specific myeloid populations found in the tumor sample. E, Spatial transcriptomics feature plots from a representative section (A1), where expression of individual genes is overlaid on tumor section. Capture area points are enlarged for ease of visualization and are transparent for values below 0.1 on the accompanying color scale. Tumor cluster signatures represent classification scores derived from integration of scGEX data with spatial transcriptomics data. F, Violin plots of genes enriched in tumor cluster 3 compared with tumor cluster 7, including neural-like genes and cancer-associated genes. G–I, Spatial transcriptomics feature plots from a representative neuroblastoma section (A1) for B cells, CD8+ T cells, CD4+ T cells, and macrophages. Cd8a and Cd4 values are from section-specific log normalization. J, Box plots showing scGEX signature classification scores from spatial transcriptomics section A1 for capture spots with a macrophage classification score of 0 (no macs) versus spots with a positive macrophage classification score (with macs). Statistical comparisons were made with Wilcoxon rank sum tests.
Tumor cells were identified as those expressing known neuroblastoma gene hallmarks including those encoding Mycn, Th, and Ncam1, (Fig. 2B; Supplementary Fig. S1D) with no expression of Ptprc (the gene encoding the CD45 antigen, a marker of hematopoietic origin). This signature identified three tumor clusters that comprised 16.9% of the total cells captured. Unsurprisingly, the main gene expression pattern enriched in these clusters compared to nontumor immune cells included those encoding products known to be associated with tumor biology, including Tuba1a (37), Stmn2 (38), Uchl1 (39), and Tubb3 (Supplementary Fig. S1D; ref. 40). Known neuronal genes were upregulated in the tumor clusters, including Pirt, Tmeff2, and Dst, which have been previously identified as upregulated genes in neuroblastoma tumors (Supplementary Fig. S1D; ref. 41). Overall, the transcriptional profiling of tumor cells substantiated the complex nature of neuroblastoma, with the observation of neuronal signatures and various tumor biology signatures including growth, adhesion, proliferation, invasion, and metastasis.
Among the nontumor immune cells, B cells dominated the cellular enumeration (32%), followed by CD4+ T (26%) cells, and CD8+ T (14.5%) cells (Fig. 2C; Supplementary Fig. S1E). Flow cytometric analysis broadly confirmed these findings (Supplementary Fig. S1F). Within the innate compartment, discrete populations of macrophages and monocytes encompassing various states of activation and development were present, with smaller populations of neutrophils, granulocytes, NK cells, and innate-like lymphocytes also observed. Among the myeloid populations, we distinguished macrophages, inflammatory monocytes, neutrophils, and dendritic cells/immature monocytes based on gene expression (Fig. 2D). The largest component of this compartment was macrophages, which made up over 13% of the total immune cell population, with only a few percent of cells in the other compartments.
While scGEX analyses are useful for resolving the cellular heterogeneity of the tumor, the spatial organization of these cells can significantly impact their role within the tumor microenvironment. To explore this, we performed spatial transcriptomic analysis on sections from multiple tumors. We leveraged the scGEX data by integrating the single-cell and spatial molecular signals and identifying overlaps in expression patterns. Unsurprisingly, the presence of the scGEX tumor signature was nearly ubiquitous throughout the spatial transcriptomics data, though notably absent in large areas of presumptive vasculature. The distinct transcriptional clusters identified within this broader tumor signature appeared to correspond with spatial features within the tumor environment, where the most prevalent tumor scGEX cluster (cluster 3, Supplementary Fig. S1C) was most enriched at the tumor core, whereas the second most prevalent tumor cluster (cluster 7) was more spatially dispersed (Fig. 2E), indicating distinct tumor microenvironments that correspond with density and immune infiltration. Further analysis to compare tumor clusters 3 and 7 demonstrated that cluster 3 was more enriched in neural-like genes (Fig. 2F). Although both tumor clusters were enriched with many cancer-associated genes, cluster 3 in general expressed numerous genes known to be involved in multiple facets of tumor biology at higher levels than cluster 7 (Fig. 2F; Supplementary Fig. S1D).
We next examined the distribution of the innate and adaptive immune cell subsets in the tumor. Although B cells constituted a large proportion of the lymphocytes identified in the scGEX analyses, their corresponding expression signature was notably absent from the inner areas of the tumor and instead were enriched around the tumor periphery (Fig. 2G). Similarly, CD8+ T cells were not evenly distributed within the tumor, instead mostly accruing in peripheral areas (Fig. 2H). In contrast, both CD4+ T cells and macrophages were widely distributed throughout the tumor (Fig. 2I). Notably, CD4+ T-cell classification scores were significantly higher among areas with a positive macrophage classification score (Fig. 2J), indicating colocalization between these two cell subsets.
CD4+ T cells are pathogenic in neuroblastoma
Given the significant presence of adaptive immune cells in the tumor parenchyma, we next wanted to determine their contribution to tumor restraint or growth. To address the role of both B cells and T cells, we first crossed AlkF1178L; TH-MYCN mice with Rag1−/− animals to generate a strain lacking B and T cells from birth. Strikingly, we have never observed a single tumor in this line, indicating that that an intact adaptive immune compartment is crucial for neuroblastoma formation in this model (Fig. 3A). We therefore undertook experiments to identify the effectors of this protumorigenic response. The role of B lymphocytes in the antitumor immune response to solid tumors remains poorly understood, with both anti- and protumor roles reported (42,43,44), and no studies to date have systematically investigated the role of B cells in neuroblastoma. To determine the overall contribution of B cells to tumorigenesis in our model, we treated AlkF1178L; TH-MYCN mice with dual anti-CD19 and anti-B220–depleting antibodies. We observed no significant differences in tumor incidence or survival rates (Fig. 3B), suggesting that B lymphocytes were not required for tumor formation.
Figure 3.
CD4+, but not CD8+ or B cells, are pathogenic in AlkF1178L; TH-MYCN neuroblastoma. A, Incidence and survival curves comparing tumor formation between AlkF1178L; Rag1−/−; TH-MYCN mice and AlkF1178L; TH-MYCN mice. B, Incidence and survival curves of AlkF1178L; TH-MYCN mice treated with dual anti-CD19 and anti-B220 antibodies or control. C, Incidence and survival curves in AlkF1178L; TH-MYCN mice treated with anti-CD8 depleting antibody or control. D, Flow cytometric analysis of MHC class I expression on intratumoral CD45+ and CD45− populations. E, Incidence and survival curves in AlkF1178L; TH-MYCN mice treated with anti-CD4 depleting antibody or control. F, Successful depletion of CD4+ T cells in the tumor (first panel). Presence of Foxp3+ CD8+ cells in AlkF1178L; TH-MYCN mice treated with anti-CD4 depleting antibody. Data are representative of n = 2 mice. G, Dot plot of genes enriched across the CD4+ T-cell clusters in these tumor samples with genes binned in functional groups “Conventional CD4 genes,” “Cancer-related,” “Unconventional lineage markers,” and “Residency genes.” Color corresponds to expression of each gene relative to the average among the four focal populations, and the size of the dot represents the proportion of cells from the cluster expressing each gene. H, Donut plot of the frequency (freq) of Foxp3+ cells within each cluster. Clusters with 0% cells expressing Foxp3 are not shown. I, Flow cytometric analysis of tumor-resident CD4+ cells. CD4+ CD62L− cells express higher amounts of the “unconventional” markers Foxp3, Helios, PD1, and ICOS than CD4+ CD62L+ cells. J, t-SNE of only CD4+ T cells subsetted from the full dataset, with subclusters identified on the basis of marker gene expression. Dotted line indicates CD4 cluster 7. K, CD4 t-SNE from J, with colors indicating the originating tumor. Dotted line indicates CD4 cluster 7. L, Dot plot of “Unconventional lineage markers.” In A–C and E, data were analyzed by the log-rank test.
We next measured the effect of depleting CD8+ T cells, though in our spatial transcriptomic data these cells did not appear to be well infiltrated in the tumor. CD8+ T cells were dispensable for tumor formation, with depleted animals demonstrating similar rates of tumor development as control animals (Fig. 3C). CD8+ T cells exert their antitumor function in part by recognizing tumor-associated antigens on MHC class I. scGEX and spatial transcriptomic data indicated that the tumor cells failed to express significant amounts of β2-microglobulin (Supplementary Fig. S2A). Similarly, human pediatric neuroblastoma expresses extremely low amounts of β2-microglobulin, especially when compared with other tumor types (Supplementary Fig. S2B). Staining for MHC class I expression, we found that the tumor cells were largely class I negative, confirming our transcriptional data (Fig. 3D) and suggesting that neuroblastoma downregulate these recognition molecules to blunt an effective tumor-specific cytotoxic CD8+ T-cell response. Although MHC class I–independent tumor control can occur through NK-cell–mediated cytotoxic activity, the depletion of NK cells also had no effect on tumor incidence or survival (Supplementary Fig. S2C).
As both B cells and CD8+ T cells were dispensable for tumor formation, we turned to CD4+ T cells as the likely candidate for the established protumorigenic effect of adaptive immunity. We depleted total CD4+ cells via monoclonal antibody treatment in AlkF1178L; TH-MYCN mice starting 1 week prior to study enrollment. CD4+ depletion, which depletes both conventional and regulatory CD4+ populations, prevented tumor formation (Fig. 3E). We noted a small number of CD4+-depleted animals still developed tumors. In two of these mice, we analyzed their tumor microenvironment to confirm successful CD4+ depletion and observe the phenotype of any residual CD4+ T cells that might account for the tumor formation. While no CD4+ cells were observed in the blood or spleen, an exceptionally small number were observed within the tumor and appeared to be conventional CD4+ T cells, with no expression of Foxp3, which would be indicative of a regulatory phenotype (Fig. 3F). Surprisingly, a population of Foxp3+ CD8+ cells was enriched in these tumors. These populations were not observed in nondepleted mice (Supplementary Fig. S3), suggesting that this CD8+ regulatory phenotype emerges as a compensatory mechanism in the absence of CD4+ T cells, and that, at minimum, T cells with the capacity for regulatory activity are required for tumor formation.
The spatial transcriptomic data showing infiltration of CD4+ T cells along with these depletion data establish a critical role for CD4+ T cells in promoting tumor growth. This prompted us to define the features of these cells within the tumor. In our scGEX analysis, three clusters of CD4+ T cells were identified. Analysis of distinguishing genes allowed us to characterize them as: “conventional”, expressing standard markers of naïve and memory T cells and tissue residency (including Il7r, Ccr7, Ifngr1, Vps37b); “cancer-related,” expressing genes known to be enriched in cancers, including Fxyd5, Hspa1a, Tcf7, and Igfp4; and “unconventional”, expressing an array of genes representative of unconventional T-cell populations (including innate-like T cells and Tregs), such as Icos, Ikzf2, Ctla4, Pcdc1, and Tnfrsf4 (Fig. 3G). Importantly, the “unconventional” cluster was the only cluster with appreciable Foxp3 expression (Fig. 3H). scGEX analysis revealed that Sell expression was critical in distinguishing “unconventional” CD4 cells from “cancer-related” and “conventional” CD4 cells, where “unconventional” cells did not express Sell (Fig. 3G). Using CD62 L expression (encoded by Sell), we assessed the expression of “unconventional” markers on tumor-resident CD4+ cells by flow cytometry. Confirming our scGEX data, CD62L− cells expressed higher amounts of Helios, Foxp3, ICOS, and PD1 compared with CD62L+ cells (Fig. 3I).
Our scGEX analysis was based on a pooled dataset from multiple tumors. Given the potential for intertumoral heterogeneity, we analyzed the scGEX clustering of the CD4+ T cells only, identifying nine subclusters by k-means clustering (Fig. 3J). Using this finer discrimination, we observed one cluster common to all tumors (Fig. 3K), which displayed an “unconventional” gene expression pattern (Fig. 3L). Given the absolute requirement of CD4+ cells for tumor formation, these data suggest that CD4+ cells with a distinct “unconventional” phenotype may be responsible for this tumor-promoting effect.
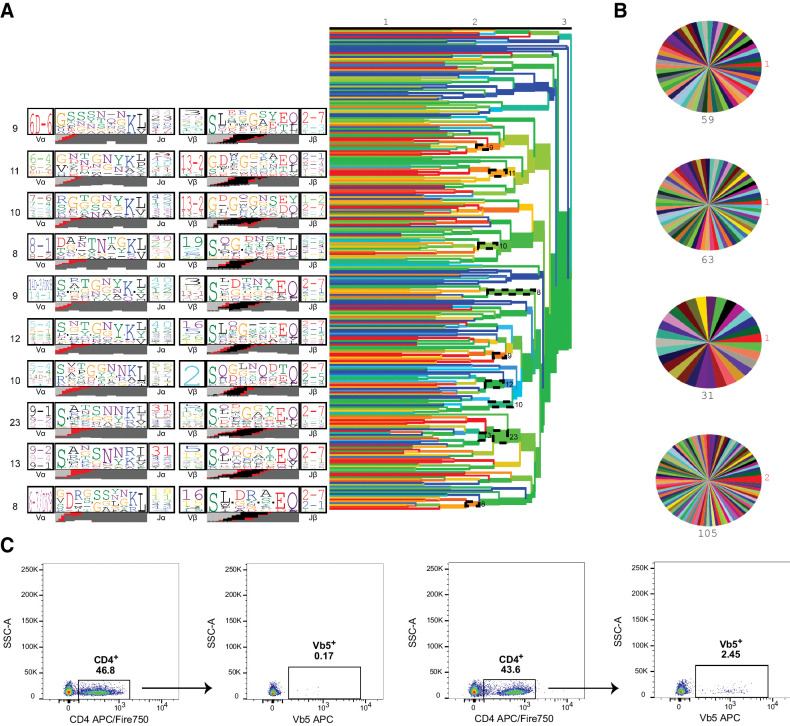
CD4+ T cells, like all T cells, are defined by the presence of a T-cell receptor conferring specificity to an epitope (usually peptide in complex with MHC class II). However, T cells can also perform more “innate” like functions, analogous to TCR-deficient innate lymphocytes, in a TCR-independent manner. Along with nuclear expression of the transcription factor Foxp3, CD4+ T cells with a regulatory phenotype highly express IL2rα, or CD25, on their cell surface (45). Because T cells with regulatory potential appear to be essential to tumorigenesis, we examined whether the CD4+ CD25+ T cells in our mouse neuroblastoma shared an epitope specificity. We performed single-cell TCR sequencing on parenchymal CD4+ CD25+ T cells isolated from multiple animals. We and others (27) have previously shown that T cells that share specificity for an epitope target usually carry TCRs with convergent motifs across individuals. Applying the TCRdist algorithm to detect the presence of such motifs, we found that there were no consistent motifs either within or between tumors that would indicate clonal expansion or shared specificity (Fig. 4A and B).
Figure 4.
Tumor-resident CD4+ CD25+ cells are not antigen specific. A, CD4+ CD25+ cells were isolated from AlkF1178L; TH-MYCN mice for TCR repertoire analysis. TCRdist trees (right) with dashed ellipses indicating groups of similar TCRs, or neighbors, as a cluster. Representative TCR logos (left) depict most highly used V and J genes, CDR3 amino acid sequences, and predicted VDJ rearrangement of each cluster (n = 4 mice, 262 TCR clones). B, Clone pie charts for four independent mice. Each wedge represents a unique TCR clone. The size of top clone is in red, and total number of sequences is in black. C, CD4 cells migrate to the tumor in an antigen-independent manner. AlkF1178L; TH-MYCN mice were intravenously injected with 5 × 106 CD4+ cells from OT-II mice prior to tumor formation. Tumors from mice receiving OT-II donor cells show increased numbers of Vβ5+ CD4 cells (right) compared with control mice (left).
To further confirm that the tumor infiltrating CD4+ T cells were acting in an antigen-independent manner, we transferred ovalbumin-specific OT-II CD4+ T cells to mice 1 week prior to study enrollment and the onset of detectable tumors. We then analyzed whether these cells would be recruited into the tumor parenchyma, despite the animals lacking expression of cognate antigen. Indeed, we observed an increased frequency of intratumoral CD4+ cells expressing Vβ5 TCR from mice transferred with OT-II CD4+ T cells compared with wild-type mice (Fig. 4C), suggesting that the phenotype of tumor-resident CD4+ cells can be obtained by a mechanism that does not require a specific antigen.
Bone marrow–derived CCR2+ myeloid cells drive neuroblastoma
Our spatial transcriptomic data indicated that in CD4 T cells, macrophage and monocyte clusters were the only immune cell types infiltrating throughout the tumor. We considered the cooccurrence of the distinct immune populations across all spatial transcriptomics sections. On the basis of probabilistic classification scoring of 6,729 spatial transcriptomics gene expression spots, we found a clear enrichment in the cooccurrence of moderate CD4+ T-cell and macrophage classification scores that was not present for other cell subsets (Supplementary Fig. S4A). In solid tumors, most macrophages originate from monocytes, whose egress from the bone marrow is largely controlled by the chemokine CCL2 and its receptor CCR2 (46). We found AlkF1178L; TH-MYCN; Ccr2−/− mice had >80% depletion of circulating monocytes compared with cognate controls (Supplementary Fig. S4B), and these mice had a trend to reduced tumor incidence and significantly increased survival compared with AlkF1178L; TH-MYCN mice (Fig. 5A). In the tumors arising in AlkF1178L; TH-MYCN; Ccr2−/− mice, we observed a relative depletion of monocytes and macrophages compared with controls, as expected (Supplementary Fig. S4C and S4D). Consistent with the “leaky” phenotype of Ccr2−/− mice, it is possible some residual monocytes can seed tumors and potentially interact with embryonic macrophages present in the adrenal gland, but nevertheless, these data demonstrate that bone marrow–derived CCR2-dependent monocytes and macrophages contribute to neuroblastoma development and growth.
Figure 5.
Myeloid Arg1 is pathogenic in neuroblastoma. A, Incidence and survival curves comparing tumor formation of AlkF1178L; TH-MYCN; Ccr2−/− mice. B, Feature plots showing cell-specific expression of Arg1. C, Dot plot of enzymes and known markers of monocyte, macrophage, and granulocyte populations. Color corresponds to expression of each gene relative to the average among the four focal populations, and the size of the dot represents the proportion of cells from the cluster expressing each gene. D, CD45− tumor cells express negligible Arg1 as detected by flow cytometry. E, Arg1 expression is predominantly expressed in CD45+ cells that also express CD68 and CCR2. Data are representative of three independent tumors. F, Heat map and volcano plot of gene expression in amino acid–starved CD4+ T cells. Data are from three biological replicates. Complete microarray data available in GEO (accession number GSE126024). G, Amino acid–starved CD4+ T cells reversibly increase Foxp3 expression. T cells were stimulated in the presence of APC in complete RPMI containing 100% (1200 μmol/L) or 1% (12 μmol/L) l-arginine with or without TGFβ and Foxp3 expression measured after 72 hours. Data are representative of n = 5 biologically independent experiments. H, Amino acid–starved CD4+ cells have increased expression of the “unconventional” markers Foxp3, Helios, and ICOS (but not PD1) compared with nonstarved CD4+ cells. I, Incidence and survival curves comparing tumor formation between AlkF1178L; TH-MYCN; Stat6−/−mice and AlkF1178L; TH-MYCN mice. J, Incidence and survival curves comparing tumor formation of AlkF1178L; TH-MYCN mice lacking Arg1 in all hematopoietic cells. K, Pairwise HRs as determined by CoxPH regression modeling. Reported P values are adjusted for multiple comparisons. F, female; M, male; WT, wild-type. Error bars represent the SE of the coefficients. In A, I, and J, data were analyzed by the log-rank test.
Myeloid Arg1 is a tumor driver
CCR2-dependent monocytes and macrophages that reside within tumor microenvironments have diverse functions that provide growth and survival advantages to the tumor cells, and importantly, suppress antitumor immune responses. A major means by which this occurs is via the production of the amino acid–metabolizing enzyme Arg1. Arg1 is expressed in a variety of tumor environments (19,20,21), both by resident myeloid cells and the tumor cells themselves (47,48,49). Arg1 depletes l-arginine from the local environment and hydrolyzes it to ornithine and urea. T cells are exquisitely sensitive to the deprivation of L-arginine, resulting in proliferative arrest (50) and, in the case of CD4+ cells, the promotion of Th2 and Treg phenotypes (51). Our scGEX data revealed that in our model of neuroblastoma, Arg1 expression was restricted to monocytic cells, with minimal amounts detected within the tumor cells (Fig. 5B and C). This finding was confirmed through flow cytometric analysis, where Arg1 expression was predominantly observed in CD68+ CCR2+ immune cells and not CD45− tumor cells (Fig 5D and E; Supplementary Fig. 5A). While CD68 is widely considered a pan-macrophage marker, nonmyeloid cells such as tumor cells have been reported to also express CD68 (52). While a small proportion of CD45− cells expressed CD68, they expressed negligible Arg1 (Supplementary Fig. 5B). Similarly, IHC analysis of human neuroblastoma revealed Arg1 expression only in interspersed myeloid-like cells that overlapped partly with CD68 and CD163 staining (Supplementary Fig. 5C). Our data thus far demonstrated spatial relationships between resident CCR2-dependent myeloid cells and non–antigen-specific CD4+ cells, and that both populations were required for tumor formation. Considering the conserved “unconventional” phenotype in intratumoral CD4+ cells, characterized by the expression of a suite of genes associated with suppressive activity, and prior links between amino acid metabolism and Treg induction, we sought to investigate the role of Arg1 within our model.
To determine the effects of arginine deprivation on CD4+ populations, we performed transcriptome profiling of CD4+ cells stimulated in arginine-depleted media. Compared to cells in nutritionally complete conditions, arginine-starved CD4+ cells exhibited increased expression of a suite of mRNAs encoding proteins associated with regulatory activity, including PD-L1, CD30L (Tnfsf8), RANKL (Tnfsf11), TROY (Tnfrsf19), and IL2, in addition to Foxp3 (Fig. 5F). Cytometric analysis revealed that arginine restriction alone was sufficient to promote the expression of Foxp3 in CD4+ cells, and this effect was potentiated with the addition of TGFβ (Fig. 5G). Foxp3 expression was further enhanced when cells remained in arginine-depleted media, and was reversed upon replenishment of arginine and subsequent entry into the cell cycle, consistent with a model where nutrient deprivation provokes plastic Foxp3 expression. Increased expression of other “unconventional” markers including Helios and ICOS were observed in arginine starved CD4+ cells compared with controls (Fig. 5H). Interestingly, PD1 expression was not increased in arginine-starved CD4+ cells, which may reflect the influence of multiple signaling pathways on surface PD1 in vivo.
Within the tumor microenvironment, Arg1 expression can be linked to multiple functional groups of myeloid cells. Tissue-infiltrating monocytes can differentiate into macrophages with distinct effector phenotype, including broadly defined “M1” and “M2”-like cells. Most tumor-associated monocytes and macrophages (TAM) are considered to be M2-like, which are key components of tissue repair and wound healing, and actively promote tumor growth through multiple mechanisms (4, 53). The canonical pathway for M2 macrophage polarization employs STAT6 downstream of the IL4 and IL13 receptors, and in coordination with C/EBPβ, PPARγ, KLF4 and other transcription factors, induces the expression of M2-related genes, including Arg1 (54), which potently suppress the antitumor immune response. To test whether the monocytic contribution to tumor formation depended on these M2 pathways, we crossed our AlkF1178L; TH-MYCN mice to mice lacking Stat6. We found no evidence that IL4/IL13 M2 macrophages were drivers of disease, as tumor formation in AlkF1178L; TH-MYCN; Stat6−/− were similar to their cognate controls (Fig. 5I). Thus, while monocyte-differentiated macrophages contributed to neuroblastoma formation, their effects were not mediated through STAT6-dependent, M2-dependent pathways. These data are consistent with previous studies demonstrating that Arg1 can be induced in monocytes and macrophages in response to hypoxia and the high lactate conditions of the tumor microenvironment (55,56,57).
MDSCs are a functional heterogeneous subset of myeloid cells that develop in response to cancer and other inflammatory conditions, and act as potent suppressors of T-cell activity. MDSC are defined by a number of immunoregulatory molecules, including Arg1 (58). However, our single-cell gene expression data did not detect significant numbers of Arg1-expressing cells that coexpressed mRNAs encoding S100A9 and IL4Rα, key markers of MDSC (Supplementary Fig. S5D; ref. 59). Flow cytometric analysis confirmed our transcriptomic data, demonstrating no IL4Rα-expressing monocytic populations (Supplementary Fig. S5E). In the absence of these critical markers required for MDSC identification, these data suggested the tumorigenic effects of myeloid cells are due to TAM rather than MDSC.
Although Arg1 is consistently linked with malignancy, it is important to note that there have been no loss-of-function studies to test the role of Arg1 in a genetic model of cancer. To evaluate the requirement of Arg1 to the development of neuroblastoma, we used conditional deletion of Arg1 in AlkF1178L; TH-MYCN mice. In this genetic strategy, the Arg1flox allele was crossed to Tek-Cre mice (Tg(Tek-cre)1Ywa, referred to as Tie2-Cre hereafter) to generate a complete knockout in all hematopoietic cells and some endothelial cells. Our scGEX data and other cancer-related studies using implantable models demonstrate Arg1 is primarily expressed in myeloid cells (21, 60). As other known cells, such as ILC2 and endothelia, which express Arg1, were not detected by scGEX, nor by flow cytometric methods (Supplementary Fig. S5F), we are confident that this genetic approach targets myeloid Arg1, as we have demonstrated previously (61). The use of Tie2-Cre is also necessary to achieve complete deletion of the conditional Arg1 locus, as the commonly used LysM-Cre (Lyz2-Cre) strain provides incomplete deletion (61). Using this approach, we found that loss of myeloid Arg1 was protective against tumor formation (Fig. 5J). Pairwise comparisons demonstrate that Arg1 and CCR2 deficiency in AlkF1178L; TH-MYCN mice provide protection against tumor formation compared with wild-type mice, and while bone marrow–derived macrophages are required for tumorigenesis, conventional STAT6-mediated M2-polarized macrophages were dispensable (Fig. 5K). Taken together, our findings indicated that a regulatory loop of Arg1-positive myeloid cells and non–antigen-specific unconventional CD4+ promoted a highly tumorigenic environment in this model of neuroblastoma.
Discussion
Our results indicate that a major protumor effect of myeloid cells is through pathways that deplete amino acids. We rescued the lethal effects of AlkF1178L; TH-MYCN neuroblastoma by depleting macrophages, CD4+ T cells, or myeloid Arg1; any one of these manipulations was sufficient to block tumor formation. By contrast, neither the STAT6-regulated pathways involved in M2 macrophage polarization, a pathway linked to tumor formation, nor CD8+ T cells were required in this model. We demonstrate the colocalization of CD4+ T cells and myeloid cells within the tumor microenvironment, suggesting functional relationships between these cell types. These findings were made possible by a highly penetrant model of neuroblastoma, which combines tyrosine hydroxylase–driven MYCN amplification with a known activating mutation in the proto-oncogene Alk, resulting in tumors that replicate the gene expression profiles and immune infiltration patterns of human pediatric neuroblastoma. Given the stark downregulation of MHC class I and β2-microglobulin expression on both human and murine tumor cells, we anticipated NK cells would accelerate tumor formation; they are not required during these early stages of tumor formation and growth.
The adrenal medulla is populated with sparse numbers of macrophages derived from the fetal liver, but it is possible that some or all macrophages in the adrenal gland at the time of tumor formation may be bone marrow derived. Without a clear genetic approach to eliminate embryonic-derived adrenal macrophages, we cannot determine their exact role in tumor formation. Through the use of Ccr2−/− mice, which have substantial (but not complete) depletion of bone marrow–derived macrophage and monocyte populations, we determined that Ccr2−/−; AlkF1178L; TH-MYCN mice had a trend to lower tumor incidence and significantly longer survival rates. Further work involving lineage-tracing would be necessary to determine the exact contribution of adrenal embryonic and bone marrow–derived macrophages to tumor formation. Nevertheless, we have demonstrated the importance of bone marrow macrophages to neuroblastoma development and disease progression.
An important finding of the depletion studies is that CD4+ T cells, collectively, are pathogenic in neuroblastoma, while B cells, CD8+ T cells, and NK cells are neutral for tumor formation and growth. This is supported by the finding that Rag1−/−; AlkF1178L; TH-MYCN mice, which lack an adaptive immune system, did not develop tumors. We speculate that an unconventional population of CD4+ cells, potentially including Tregs, is likely the pathogenic component of the CD4+ compartment. This requirement cannot be tested directly in our tumor model because depletion of Treg cells either by antibodies or genetic ablation would result in the development of autoimmunopathology during the study window. Further studies are required to elucidate the exact mechanisms by which the deprivation of arginine triggers the development of these non–antigen-specific unconventional cells, and precisely how these cells facilitate the tumorigenic process. However, from infection models, arginine depletion by myeloid cells has a critical role in suppressing excessive inflammation, and is linked to the development of a tolerogenic state (7, 10, 11, 14). In our neuroblastoma model, the interplay between myeloid amino acid depletion, CD4+ T cells and the tumor cells is probably highly dynamic, emphasizing the need for temporal quantification live animal imaging methods to measure specific metabolites, along with the tumor growth and immune infiltration.
The gene expression profile of these unconventional CD4+ cells shared features with resident CD4+ populations, some of which are only recently described in antigen-specific models. A curious feature of our data was that there was no evidence of any antigen-dependent enrichment, potentially indicating a non-TCR–based mechanism for T-cell activation. The transcriptional state of the cells clearly indicated a nonnaïve population, but the lack of any clonal expansion or TCR clustering argues against antigen-dependent activation and expansion. While this is speculative, the transcriptional state is consistent with other tissue repair–associated lymphocyte populations such as innate lymphoid cells (ILC) and γδ T cells, both of which can be activated by direct cytokine signaling (62, 63). Additional studies are required to identify the signals that activate CD4 cells within the tumor environment, and what function(s) they perform in the tumorigenic process, be it direct immune cell suppression, tissue repair, or a combination of both.
The conclusion that at least two immune populations play a critical role for tumor formation in MYCN-amplified neuroblastoma is somewhat surprising given the characterization of these tumors as largely immunologic quiescent (64). It is important to emphasize that immune populations make up a very small fraction of the overall tumor cellularity of our neuroblastoma tumors. Our characterization of the tumor populations was performed on material that was enriched for immune cells, where RNA was obtained from CD45− and parenchymal (nonvascularly accessible) CD45+ populations and mixed at equal ratios to provide sufficient depth of coverage within our analyses. Still, our data clearly demonstrate these cells have an immunoregulatory potency belied by their numbers.
Our results suggest that inhibitors of Arg1 and other immune modulatory interventions may have significant value in cancer therapy. Immunotherapy thus far has been focused on promoting antitumor effects of NK or antigen-specific T cells. The paradigm suggested here is that disruption of the protumorigenic, tissue-forming effects of immune cells, including T cells and macrophages, may be therapeutically beneficial in neuroblastoma, and perhaps in other low mutation burden developmental tumors.
Authors' Disclosures
P.G. Thomas reports grants from NIH-National Institute of Allergy and Infectious Diseases (NIAID), NIH-NCI, and Key For a Cure Foundation, and grants and other support from ALSAC during the conduct of the study; personal fees and nonfinancial support from 10X Genomics and Illumina, and personal fees from Immunoscape and PACT Pharma outside the submitted work; and a patent for WO US US20190040381A1 pending, a patent for WO WO2021003114A2 pending, a patent for WO WO2020257575A1 pending, and a patent for WO US US20170304293A1 issued. P.J. Murray reports other support from Max Planck Gesellschaft, American Lebanese Syrian Associated Charities, and grants from Key For A Cure Foundation, NIH, and Deutsche Forschungsgemeinschaft during the conduct of the study, and is on the scientific advisory boards for Palleon Pharma and ImCheck Therapeutics. No activities for these advisory boards are related to the manuscript. P.J. Murray also has a research contract with Boehringer Ingelheim (Biberach an der Ries, Germany) concerning inflammation research, which is unrelated to the manuscript. No disclosures were reported by the other authors.
Supplementary Material
Acknowledgments
The authors thank the staff of the St. Jude Center for In Vivo Imaging and Therapeutics for their dedication and expertise; the St. Jude Transgenic Core Unit for zygote and ES cell injections; Stefan Schattgen, Jessica Haverkamp, Dorian Obino, Crystal Neely, and Lidija Barbaric for assistance; and the St. Jude Department of Immunology Flow Cytometry facility for cell sorting. This work was supported by the NIH grant CA189990, the Max Planck Gesellschaft, DFG Grants FOR 2599 and TRR 127, the Key For A Cure Foundation, the American Lebanese Syrian Associated Charities, and NIH Cancer Center P30 CA21765.
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked advertisement in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
Note: Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
#E.K. Allen and J.C. Crawford contributed equally to this article.
Cancer Res 2021;81:5047–59
Authors' Contributions
L.-A. Van de Velde: Resources, data curation, formal analysis, validation, investigation, visualization, methodology, writing–original draft, project administration, writing–review and editing. E.K. Allen: Resources, data curation, software, formal analysis, investigation, visualization, writing–original draft, writing–review and editing. J.C. Crawford: Resources, data curation, software, formal analysis, investigation, visualization, writing–review and editing. T.L. Wilson: Formal analysis, investigation, writing–review and editing. C.S. Guy: Resources, investigation. M. Russier: Investigation. L. Zeitler: Investigation. A. Bahrami: Resources. D. Finkelstein: Formal analysis. S. Pelletier: Methodology. S. Schultz-Cherry: Resources. P.G. Thomas: Conceptualization, resources, supervision, funding acquisition, methodology, writing–original draft, project administration, writing–review and editing, co-corresponding author. P.J. Murray: Conceptualization, resources, formal analysis, supervision, funding acquisition, validation, investigation, visualization, methodology, writing–original draft, project administration, writing–review and editing.
References
- 1. Weiss WA. Targeted expression of MYCN causes neuroblastoma in transgenic mice. EMBO J 1997;16:2985–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Molenaar JJ, Domingo-Fernández R, Ebus ME, Lindner S, Koster J, Drabek K, et al. LIN28B induces neuroblastoma and enhances MYCN levels via let-7 suppression. Nat Genet 2012;44:1199–206. [DOI] [PubMed] [Google Scholar]
- 3. Cazes A, Lopez-Delisle L, Tsarovina K, Pierre-Eugène C, De Preter K, Peuchmaur M, et al. Activated Alk triggers prolonged neurogenesis and Ret upregulation providing a therapeutic target in ALK-mutated neuroblastoma. Oncotarget 2014;5:2688–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Biswas SK, Mantovani A. Macrophage plasticity and interaction with lymphocyte subsets: cancer as a paradigm. Nat Immunol 2010;11:889–96. [DOI] [PubMed] [Google Scholar]
- 5. Qian BZ, Pollard JW. Macrophage diversity enhances tumor progression and metastasis. Cell 2010;141:39–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Murray PJ. Nonresolving macrophage-mediated inflammation in malignancy. FEBS J 2018;285:641–53. [DOI] [PubMed] [Google Scholar]
- 7. Nathan C, Ding A. Nonresolving inflammation. Cell 2010;140:871–82. [DOI] [PubMed] [Google Scholar]
- 8. Duque-Correa MA, Kuhl AA, Rodriguez PC, Zedler U, Schommer-Leitner S, Rao M, et al. Macrophage arginase-1 controls bacterial growth and pathology in hypoxic tuberculosis granulomas. Proc Natl Acad Sci U S A 2014;111:e4024–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Araújo AP, Frezza TF, Allegretti SM, Hypoxia GS. Hypoxia-inducible factor-1α and vascular endothelial growth factor in a murine model of Schistosoma mansoni infection. Exp Mol Path 2010;89:327–33. [DOI] [PubMed] [Google Scholar]
- 10. Pesce JT, Ramalingam TR, Mentink-Kane MM, Wilson MS, El Kasmi KC, Smith AM, et al. Arginase-1-expressing macrophages suppress Th2 cytokine-driven inflammation and fibrosis. PLoS Pathog 2009;5:e1000371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Murray PJ. Amino acid auxotrophy as a system of immunological control nodes. Nat Immunol 2016;17:132–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Singla S, Bhojnagarwala PS, O'Brien S, Moon EK, Garfall AL, Rao AS, et al. Origin and role of a subset of tumor-associated neutrophils with antigen-presenting cell features in early-stage human lung cancer. Cancer Cell 2016;30:120–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Giatromanolaki A, Harris AL, Koukourakis MI. The prognostic and therapeutic implications of distinct patterns of argininosuccinate synthase 1 (ASS1) and arginase-2 (ARG2) expression by cancer cells and tumor stroma in non-small-cell lung cancer. Cancer Metab 2021;9:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cobbold SP, Adams E, Farquhar CA, Nolan KF, Howie D, Lui KO, et al. Infectious tolerance via the consumption of essential amino acids and mTOR signaling. Proc Natl Acad Sci U S A 2009;106:12055–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Andersson J, Tran DQ, Pesu M, Davidson TS, Ramsey H, O'Shea JJ, et al. CD4+ FoxP3+ regulatory T cells confer infectious tolerance in a TGF-β–dependent manner. J Exp Med 2008;205:1975–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gajewski TF, Schreiber H, Fu YX. Innate and adaptive immune cells in the tumor microenvironment. Nat Immunol 2013;14:1014–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sullivan MR, Danai LV, Lewis CA, Chan SH, Gui DY, Kunchok T, et al. Quantification of microenvironmental metabolites in murine cancers reveals determinants of tumor nutrient availability. eLife 2019;8:e44235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Steggerda SM, Bennett MK, Chen J, Emberley E, Huang T, Janes JR, et al. Inhibition of arginase by CB-1158 blocks myeloid cell-mediated immune suppression in the tumor microenvironment. J Immunother Cancer 2017;5:101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Katzenelenbogen Y, Sheban F, Yalin A, Yofe I, Svetlichnyy D, Jaitin DA, et al. Coupled scRNA-Seq and intracellular protein activity reveal an immunosuppressive role of TREM2 in cancer. Cell 2020;182:872–85. [DOI] [PubMed] [Google Scholar]
- 20. Molgora M, Esaulova E, Vermi W, Hou J, Chen Y, Luo J, et al. TREM2 modulation remodels the tumor myeloid landscape enhancing anti-PD-1 immunotherapy. Cell 2020;182:886–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Arlauckas SP, Garren SB, Garris CS, Kohler RH, Oh J, Pittet MJ, et al. Arg1 expression defines immunosuppressive subsets of tumor-associated macrophages. Theranostics 2018;8:5842–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Smith C, Chang MY, Parker KH, Beury DW, DuHadaway JB, Flick HE, et al. IDO Is a nodal pathogenic driver of lung cancer and metastasis development. Cancer Dis 2012;2:722–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Van de Velde LA, Murray PJ. Proliferating helper T cells require rictor/mTORC2 complex to integrate signals from limiting environmental amino acids. J Biol Chem 2016;291:25815–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Franzén O, Gan LM, Björkegren JLM. PanglaoDB: a web server for exploration of mouse and human single-cell RNA sequencing data. Database (Oxford). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Aran D, Looney AP, Liu L, Wu E, Fong V, Hsu A, et al. Reference-based analysis of lung single-cell sequencing reveals a transitional profibrotic macrophage. Nature Immunol 2019;20:163–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Dash P, Wang GC, Thomas PG. Single-cell analysis of T-cell receptor αβ repertoire. Methods Mol Biol 2015;1343:181–97. [DOI] [PubMed] [Google Scholar]
- 27. Dash P, Fiore-Gartland AJ, Hertz T, Wang GC, Sharma S, Souquette A, et al. Quantifiable predictive features define epitope-specific T cell receptor repertoires. Nature 2017;547:89–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Matthay KK, Maris JM, Schleiermacher G, Nakagawara A, Mackall CL, Diller L, et al. Neuroblastoma. Nature Rev Dis Primers 2016;2:16078. [DOI] [PubMed] [Google Scholar]
- 29. Berry T, Luther W, Bhatnagar N, Jamin Y, Poon E, Sanda T, et al. The ALKF1174L mutation potentiates the oncogenic activity of MYCN in neuroblastoma. Cancer Cell 2012;22:117–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Janoueix-Lerosey I, Lequin D, Brugières L, Ribeiro A, de Pontual L, Combaret V, et al. Somatic and germline activating mutations of the ALK kinase receptor in neuroblastoma. Nature 2008;455:967–70. [DOI] [PubMed] [Google Scholar]
- 31. Pugh TJ, Morozova O, Attiyeh EF, Asgharzadeh S, Wei JS, Auclair D, et al. The genetic landscape of high-risk neuroblastoma. Nat Genet 2013;45:279–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Heukamp LC, Thor T, Schramm A, Preter KD, Kumps C, Wilde BD, et al. Targeted expression of mutated ALK induces neuroblastoma in transgenic mice. Science Transl Med 2012;4:141ra91. [DOI] [PubMed] [Google Scholar]
- 33. Durbin AD, Zimmerman MW, Dharia NV, Abraham BJ, Iniguez AB, Weichert-Leahey N, et al. Selective gene dependencies in MYCN-amplified neuroblastoma include the core transcriptional regulatory circuitry. Nat Genet 2018;50:1240–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Castriconi R, Dondero A, Augugliaro R, Cantoni C, Carnemolla B, Sementa AR, et al. Identification of 4Ig-B7-H3 as a neuroblastoma-associated molecule that exerts a protective role from an NK cell-mediated lysis. Proc Natl Acad Sci USA 2004;101:12640–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Majzner RG, Simon JS, Grosso JF, Martinez D, Pawel BR, Santi M, et al. Assessment of programmed death-ligand 1 expression and tumor-associated immune cells in pediatric cancer tissues. Cancer 2017;123:3807–15. [DOI] [PubMed] [Google Scholar]
- 36. Schulz G, Cheresh DA, Varki NM, Yu A, Staffileno LK, Reisfeld RA. Detection of ganglioside GD2 in tumor tissues and sera of neuroblastoma patients. Cancer Res 1984;44:5914–20. [PubMed] [Google Scholar]
- 37. Wang D, Jiao Z, Ji Y, Zhang S. Elevated TUBA1A might indicate the clinical outcomes of patients with gastric cancer, being associated with the infiltration of macrophages in the tumor immune microenvironment. J. Gastrointestin Liver Dis 2020;29:509–22. [DOI] [PubMed] [Google Scholar]
- 38. Byrne FL, Yang L, Phillips PA, Hansford LM, Fletcher JI, Ormandy CJ, et al. RNAi-mediated stathmin suppression reduces lung metastasis in an orthotopic neuroblastoma mouse model. Oncogene 2014;33:882–90. [DOI] [PubMed] [Google Scholar]
- 39. Kwan SY, Au-Yeung CL, Yeung TL, Rynne-Vidal A, Wong KK, Risinger JI, et al. Ubiquitin carboxyl-terminal hydrolase L1 (UCHL1) promotes uterine serous cancer cell proliferation and cell cycle progression. Cancers 2020;12:118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Person F, Wilczak W, Hube-Magg C, Burdelski C, Möller-Koop C, Simon R, et al. Prevalence of βIII-tubulin (TUBB3) expression in human normal tissues and cancers. Tumour Biol 2017;39:1010428317712166. [DOI] [PubMed] [Google Scholar]
- 41. Brady SW, Liu Y, Ma X, Gout AM, Hagiwara K, Zhou X, et al. Pan-neuroblastoma analysis reveals age- and signature-associated driver alterations. Nature Comm 2020;11:5183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Helmink BA, Reddy SM, Gao J, Zhang S, Basar R, Thakur R, et al. B cells and tertiary lymphoid structures promote immunotherapy response. Nature 2020;577:549–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Petitprez F, de Reyniès A, Keung EZ, Chen TWW, Sun CM, Calderaro J, et al. B cells are associated with survival and immunotherapy response in sarcoma. Nature 2020;577:556–60. [DOI] [PubMed] [Google Scholar]
- 44. Sharonov GV, Serebrovskaya EO, Yuzhakova DV, Britanova OV, Chudakov DM. B cells, plasma cells and antibody repertoires in the tumour microenvironment. Nat Rev Immunol 2020;20:294–307. [DOI] [PubMed] [Google Scholar]
- 45. Sakaguchi S, Sakaguchi N, Shimizu J, Yamazaki S, Sakihama T, Itoh M, et al. Immunologic tolerance maintained by CD25+ CD4+ regulatory T cells: their common role in controlling autoimmunity, tumor immunity, and transplantation tolerance. Immunol Rev 2001;182:18–32. [DOI] [PubMed] [Google Scholar]
- 46. Qian BZ, Li J, Zhang H, Kitamura T, Zhang J, Campion LR, et al. CCL2 recruits inflammatory monocytes to facilitate breast-tumour metastasis. Nature 2011;475:222–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Singh R, Pervin S, Karimi A, Cederbaum S, Chaudhuri G. Arginase activity in human breast cancer cell lines: N(omega)-hydroxy-L-arginine selectively inhibits cell proliferation and induces apoptosis in MDA-MB-468 cells. Cancer Res 2000;60:3305–12. [PubMed] [Google Scholar]
- 48. Krzystek-Korpacka M, Szczęśniak-Sięga B, Szczuka I, Fortuna P, Zawadzki M, Kubiak A, et al. L-arginine/nitric oxide pathway is altered in colorectal cancer and can be modulated by novel derivatives from oxicam class of non-steroidal anti-inflammatory drugs. Cancers 2020;12:2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Jang TJ, Kim SA, Kim MK. Increased number of arginase 1-positive cells in the stroma of carcinomas compared to precursor lesions and nonneoplastic tissues. Pathol Res Pract 2018;214:1179–84. [DOI] [PubMed] [Google Scholar]
- 50. Van de Velde LA, Subramanian C, Smith AM, Barron L, Qualls JE, Neale G, et al. T cells encountering myeloid cells programmed for amino acid-dependent immunosuppression use Rictor/mTORC2 protein for proliferative checkpoint decisions. J Biol Chem 2017;292:15–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Castellano F, Molinier-Frenkel V. Control of T-cell activation and signaling by amino-acid catabolizing enzymes. Front Cell Dev Biol 2020;8:613416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Gottfried E, Kunz-Schughart LA, Weber A, Rehli M, Peuker A, Müller A, et al. Expression of CD68 in non-myeloid cell types. Scand J Immunol 2008;67:453–63. [DOI] [PubMed] [Google Scholar]
- 53. Eming SA, Wynn TA, Martin P. Inflammation and metabolism in tissue repair and regeneration. Science 2017;356:1026–30. [DOI] [PubMed] [Google Scholar]
- 54. Murray PJ. Macrophage polarization. Annu Rev Physiol 2017;79:541–66. [DOI] [PubMed] [Google Scholar]
- 55. Carmona-Fontaine C, Deforet M, Akkari L, Thompson CB, Joyce JA, Xavier JB. Metabolic origins of spatial organization in the tumor microenvironment. Proc Natl Acad Sci USA 2017;114:2934–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Colegio OR, Chu N-Q, Szabo AL, Chu T, Rhebergen AM, Jairam V, et al. Functional polarization of tumour-associated macrophages by tumour-derived lactic acid. Nature 2014;513:559–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Zhang D, Tang Z, Huang H, Zhou G, Cui C, Weng Y, et al. Metabolic regulation of gene expression by histone lactylation. Nature 2019;574:575–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Gabrilovich DI. Myeloid-derived suppressor cells. Cancer Immunol Res 2017;5:3–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Bronte V, Brandau S, Chen SH, Colombo MP, Frey AB, Greten TF, et al. Recommendations for myeloid-derived suppressor cell nomenclature and characterization standards. Nature Comm 2016;7:12150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Kratochvill F, Neale G, Haverkamp JM, Van de Velde LA, Smith AM, Kawauchi D, et al. TNF counterbalances the emergence of M2 tumor macrophages. Cell Reports 2015;12:1902–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. El Kasmi KC, Qualls JE, Pesce JT, Smith AM, Thompson RW, Henao-Tamayo M, et al. Toll-like receptor–induced arginase 1 in macrophages thwarts effective immunity against intracellular pathogens. Nat Immunol 2008;9:1399–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. McGinty JW, von Moltke J. A three course menu for ILC and bystander T cell activation. Curr Op Immunol 2020;62:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Shiromizu CM, Jancic CC. γδ T lymphocytes: an effector cell in autoimmunity and infection. Front Immunol 2018;9:2389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Vareki SM. High and low mutational burden tumors versus immunologically hot and cold tumors and response to immune checkpoint inhibitors. J Immunother Cancer 2018;6:157. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Single-cell gene expression (scGEX) and spatial transcriptomics data are accessioned in the Short Read Archive in association with BioProject PRJNA662418. Microarray files have been deposited in the GEO (accession GSE126024).