Abstract
Early embryonic development in Xenopus laevis is characterized by transcriptional repression which is relieved at the midblastula stage (MBT). Here we show that the relative abundance of TATA-binding protein (TBP) increases robustly at the MBT and that the mechanism underlying this increase is translation of maternally stored TBP RNA. We show that TBP is rate-limiting in egg extract under conditions that titrate nucleosome assembly. Precocious translation of TBP mRNA in Xenopus embryos facilitates transcription before the MBT, without requiring TBP to be prebound to the promoter before injection. This effect is transient in the absence of chromatin titration and is sustained when chromatin is titrated. These data show that translational regulation of TBP RNA contributes to limitations on the transcriptional capacity before the MBT. Second, we examined the ability of trans-acting factors to contribute to promoter activity before the MBT. Deletion of cis-acting elements does not affect histone H2B transcription in egg extract, a finding indicative of limited trans-activation. Moreover, in the context of the intact promoter, neither the transcriptional activator Oct-1, nor TBP, nor TFIID enable transcriptional activation in vitro. HeLa cell extract, however, reconstitutes activated transcription in mixed extracts. These data suggest a deficiency in egg extract cofactors required for activated transcription. We show that the capacity for activated H2B transcription is gradually acquired at the early gastrula transition. This transition occurs well after the blastula stage when the basal transcription machinery can first be complemented with TBP.
The early embryonic development of vertebrate and many invertebrate species is characterized by a period during which the embryonic genome is transcriptionally silent. The developmental stage at which the major transcriptional activity starts ranges from the 2-cell stage in the mouse to the cycle 9 to 14 blastoderm stage in Drosophila melanogaster. In nematodes and amphibians the major activation of the embryonic genome starts after 7 cell cycles (in C. elegans, i.e., 90 to 125 cells) and 12 cell cycles (in Xenopus laevis, i.e., ca. 4,000 cells), respectively (for reviews, see references 4, 36, and 62). In Xenopus, the gradual activation of the embryonic genome coincides with the acquisition of cell motility and the loss of cell cycle synchrony. These three coinciding transitions are collectively referred to as the midblastula transition (MBT) (33, 34).
Newport and Kirschner (33, 34) suggested that both cell cycle lengthening and the relief of transcriptional repression at the MBT are regulated by stoichiometric titration of an inhibitor by the exponentially increasing amount of DNA in the embryo. In partial constriction experiments, only half of the egg inherits a nucleus. Only the nucleated side divides until a nucleus incidentally moves to the uncleaved half of the embryo. This results in twin embryos, of which the retarded-half embryo has a delayed MBT. This timing suggests that the nucleus/cytoplasm ratio regulates the MBT. Experiments with polyspermic eggs, containing an increased number of nuclei, were in line with this hypothesis (33). Similarly, transcriptional repression of a class III gene was found to be mediated by a low DNA/cytoplasm ratio, while microinjection of nonspecific DNA activated the promoter precociously (34). Cell cycle lengthening is not causal for transcription activation, since cell cycle arrest by aphidicolin, by cycloheximide, or by a proteolysis-resistant mutant of cyclin B does not relieve transcriptional repression (38).
Replication-coupled chromatin assembly correlates tightly with and is causal for repression of transcription in oocytes (2), suggesting that the stoichiometrically titrated inhibitor proposed by Newport and Kirschner (33, 34) involves one or more components of chromatin. In Drosophila, however, it has been shown that titration of a specific transcriptional repressor contributes to activation of the embryonic genome (39, 61). In addition, enhancers are inactive prior to zygotic gene activation in the mouse, which, besides other mechanisms involving chromatin remodeling, is attributed to a coactivator not being available until the activation of the genome (reviewed in references 26 and 36). This prompts the question whether repression of the embryonic genome in Xenopus is regulated at different levels, perhaps involving chromatin-mediated repression, deficiencies in the transcription machinery, and the presence of specific repressors.
TATA-binding protein (TBP) has been identified as a molecule that is able to facilitate transcription before the MBT if two conditions are met (3, 37, 38): (i) recombinant TBP protein is preincubated with the promoter template prior to injection into embryos, and (ii) a state of incomplete chromatin assembly is present. Such an immature chromatin structure is observed at early time points after injection of a promoter template and after coinjection of a large amount of nonspecific DNA that titrates chromatin assembly. If recombinant TBP protein is injected into the embryo separately from the promoter template, no transcription is observed, suggesting that a dynamic competition between chromatin and TBP accounts for the regulation of transcription before and after the MBT. In this view, chromatin prevents the access of transcription factors, such as TBP, to promoter DNA (38).
In addition, transcriptional activators seem to be impaired in their function. The artificial activator GAL4-VP16 was found to depress chromatin and activate transcription under conditions when endogenous activators could not (3), whereas in another study the same activator could bind its cis-acting element but failed to activate transcription (37), thus behaving more like endogenous activators under these circumstances.
In this report we explore the mechanisms underlying global repression of transcription before the MBT and its relief thereafter. We report that TBP protein is strongly upregulated at the MBT and that this is mediated by translation of previously masked maternal TBP RNA. Employing egg extracts to assay the pre-MBT basal transcription machinery under conditions that titrate nucleosome assembly effectively, we found that TBP stimulates transcription robustly, providing evidence that TBP is rate limiting before the MBT. Moreover, we found that if TBP RNA is translated precociously in Xenopus embryos, preincubation of TBP protein with the promoter template is not necessary for stimulation of transcription before the MBT. In addition, we explore the mechanistic basis of the absence of activated transcription before the MBT. Using the egg extract in vitro transcription system, we found that the inactivity of trans-acting transcription factors is due to a deficiency in the transcription machinery. We show here that this deficiency exists in vivo during a window of time from the onset of basal transcription at the MBT to the subsequent start of gastrulation a few hours later.
MATERIALS AND METHODS
Constructs.
The pH2B-Luc plasmid was described by Schilthuis (42) and contains the histone H2B.1 promoter of the Xenopus histone gene cluster Xl-hi-118 (12), fused to the luciferase reporter gene. pH2B-TATA-Luc was obtained by digestion of pH2B-Luc with SphI and religation of the plasmid. For microinjection experiments, capped xTBP (16) RNA was synthesized from linearized pSP64A-xTBP by using an in vitro RNA synthesis kit (Ambion).
DNA topology assays.
In vitro transcription reaction samples (see below), and embryo or oocyte homogenates were deproteinized for 2 h at 55°C with proteinase K (100 μg/ml, final concentration). The DNA was phenol extracted, ethanol precipitated, and resuspended in 10 μl of TE containing 100 μg of RNase A per ml. After incubation at 37°C for 1 h, 2 μl of 80% glycerol was added, and the DNA was loaded on a 1% agarose gel containing 1× TPE (40 mM Tris base, 30 mM NaH2PO4, 1 mM EDTA) and chloroquine. One-dimensional chloroquine gels were run for 18 h at 3 V/cm in 90 μg of chloroquine per ml. Two-dimensional chloroquine gels were run in 4 and 30 μg of chloroquine per ml for the first and second dimensions, respectively. After capillary transfer of the DNA to a Hybond N-Plus membrane (Amersham), the Southern blot was probed with radiolabeled (Rediprime; Amersham) plasmid-specific probes according to standard procedures.
Density gradient centrifugation and Northern blotting.
Extract from 200 oocytes or embryos was fractionated on a Nycodenz (Nycomed, Oslo, Norway) density gradient, as described previously (31, 50, 52). Fractions were deproteinized by phenol-chloroform extraction. The RNA was recovered by ethanol precipitation and analyzed by Northern blotting by using Hybond N-Plus (Amersham) membranes and Hybrisol I (Oncor) for blocking and hybridization solution. RNA isolated from a single fraction was loaded on the gel for the Nycodenz gradient blots, whereas 30 μg of total RNA per sample was loaded for the developmental Northern blot.
Extracts for Western blotting and luciferase assays.
Embryos were homogenized in 4 volumes of low-salt whole-cell extract buffer (25 mM Tris-HCl, pH 7.5; 70 mM KCl; 1 mM EDTA; 20% glycerol; 5 mM dithiothreitol (DTT); 1 μg of leupeptin, pepstatin A, and aprotinin per ml). Homogenates were centrifuged at 15,000 × g for 5 to 10 min at 4°C. Subsequently, the supernatant was frozen on dry ice and stored at −80°C for later use.
Western blot analysis and antibodies.
Two embryo equivalents of extract were loaded per lane. For early embryonic extracts this results in even amounts of total protein loaded per lane. Western blot analysis was performed by using Hybond-ECL (Amersham) membranes and an enhanced chemiluminescence (ECL) detection kit (Pierce). The following antibodies were used: anti-TBP (58C9 [Santa Cruz Biotechnologies] at 1:4,000), anti-RNAPolII (8WG16 [BabCO] at 1:2,000), anti-TFIIB (C18 [Santa Cruz Biotechnologies] at 1:500), and anti-TFIIF RAP74 (C18 at 1:500).
Luciferase assays.
Luciferase assays were performed by using a luciferase detection kit (Pharmingen) and a Monolight 2010 luminometer (Analytical Luminescence Laboratory). In all luciferase assays, one embryo equivalent of extract was assayed, representing the average activity of 20 to 30 embryos in the homogenate. Experiments were performed in triplicate with different batches of embryos derived from different frogs. For assaying the fold activation, the activity of the full-length histone H2B promoter (H2B) was normalized against that of the H2B core promoter (“TATA”). DNA loading per embryo was checked by Southern blot analysis.
In vitro transcription extracts.
Egg extracts (low-speed supernatants) were prepared as previously described by Blow (7). In vitro transcription experiments were performed by using either activated or unactivated low-speed egg extracts; similar results were obtained, irrespective of the type of egg extract used. HeLa cell extracts were prepared as described previously (30).
In vitro transcription analysis and primer extension.
Primer extension of chloramphenicol acetyltransferase (CAT) RNA and histone H4 RNA isolated from embryos or oocytes was described previously (27). Conditions for in vitro transcription analysis were essentially as described by Toyoda and Wolffe (55). For standard 25-μl reactions, 600 ng (160 fmol) of pH2B-Luc or pH2B-TATA-Luc and 1 μl of egg extract (60 μg of protein) or 1 to 5 μl of HeLa cell extract (30 to 150 μg of protein) was used. Reactions contained 0.1 to 1 ng (0.2 to 2 fmol) of synthetic LucΔ RNA, which served as internal standard for primer extension. Transcription reactions were incubated at 23°C for 60 min. The purification of HeLa TFIID (D-TFIID) and the expression, purification, and activity of human recombinant TBP, TFIIB, TFIIE, and TFIIF were as described before (53, 54). To verify the RNA polymerase II (Pol II) dependence of histone H2B transcription in egg extract, 0.25 μl of α-amanitin (1 mg/ml in dimethyl sulfoxide [DMSO]) was included in the 25-μl transcription reactions (final concentration, 10 μg/ml). Control reactions in these cases contained 0.25 μl of DMSO. The primer used for primer extension of luciferase RNA was a 32P-end-labeled oligonucleotide complementary to Luc RNA (Luc2, 5′-ATG TTC ACC TCG ATA TGT GCA TCT GTA AA-3′). This primer was annealed to the RNA for 45 min at 55°C in 12 μl of annealing buffer (5 mM Tris-HCl, pH 8.0; 0.2 mM EDTA; 160 mM KCl). The tubes were chilled on ice for 10 min. Subsequently, 28 μl of ice-cold RT-Mix (containing 10 μl of 4× SRT buffer [80 mM Tris-HCl, pH 8.4; 10 mM MgCl2; 400 μg of bovine serum albumin per ml; 40 mM DTT], 1.65 μl of actinomycin D [1 mg/ml], 6 μl of deoxynucleoside triphosphate mix [2 mM concentrations of dATP, dCTP, dGTP, and dTTP], 0.25 μl of RNase inhibitor [Boehringer Mannheim; 40 U/ml], 0.5 μl of Superscript reverse transcriptase [Gibco BRL; 200 U/ml]) was added, and the tubes were incubated for 60 min at 42°C. The single-stranded DNA was phenol extracted, precipitated with ethanol, dissolved in 5 μl of formamide loading buffer, and loaded onto a 5% acrylamide (19:1) sequencing gel containing 8 M urea. Gels were run for 75 min at 70 W, fixed (10% methanol, 8% acetic acid), and dried under vacuum at 80°C. Quantitation of primer extension products was performed with a PhosphorImager (Molecular Dynamics). Normalization of cytomegalovirus (CMV) promoter activity with H4 signals was performed by calculating the average H4 signal for each developmental stage. The relative normalization factor for each lane is the ratio of H4 signal to the stage-specific H4 average. This corrects for the developmental regulation of histone H4 mRNA.
RESULTS
TBP abundance increases at the MBT.
To examine the possibility that developmental regulation of components of the basal transcription machinery contributes to activation of the embryonic genome, extracts from staged embryos were prepared and subjected to Western blot analysis (Fig. 1A). TBP is relatively abundant during gastrulation and neurulation (stages 10 to 17), whereas the protein is barely detectable during early cleavage (stage 2) or in oocytes. TBP levels increase between cleavage and early blastula stages (stage 6½) and increase further at the midblastula stage (between stage 8 and 9), peaking at stage 9 at the onset of zygotic transcription, well before gastrulation commences.
FIG. 1.
Regulation of TBP during embryonic development. (A) Western blot analysis of TBP, the large subunit of RNA Pol II, the RAP74 subunit of TFIIF, and TFIIB. Developmental stages are described by Nieuwkoop and Faber (35). The MBT occurs between stages 8 and 9. (B) Northern blot analysis of TBP RNA. The position of TBP RNA is indicated with an arrowhead; the position of the 18S RNA is indicated with an asterisk. The lower panel shows 28S RNA as detected with methylene blue staining of the Northern blot. (C) Northern blot analysis of TBP RNA in fractions of oocyte (top panel) and embryonic (middle panel) extracts subjected to density gradient centrifugation. The fractions containing rRNA (fractions 8 to 12) were identified by methylene blue staining of the blot (bottom panel). Fraction 7 does not contain functional ribosomes, as indicated by the absence of 28S RNA (compare references 31, 50, and 52). (D) Quantitation of TBP protein and TBP RNA abundance during early embryogenesis (derived from panels A and B). TBP protein abundance was quantitated by using multiple ECL exposures of the Western blot to ensure quantitation in the linear range of film. Due to the limited linearity of the ECL, the magnitude of regulation of TBP abundance may be larger than what is shown here. Quantitation of Northern blot signals was performed by using a PhosphorImager.
We compared the expression profile exhibited by TBP with that of other components of the basal transcription machinery. The large subunit of RNA Pol II (Pol IIa) is hardly detectable in oocytes; however, protein levels are markedly higher in embryos, presumably reflecting translation of maternally stored RNA upon maturation of the oocyte. During embryogenesis RNA Pol II levels show only a moderate increase at the MBT. Coinciding with the MBT, however, is the appearance of the hyperphosphorylated form of Pol II (IIo), a finding consistent with the unphosphorylated and phosphorylated forms of RNA Pol II being the enzymes involved in preinitiation and transcript elongation respectively (reviewed by Dahmus [11] and Shilatifard [43]).
We asked the question whether the temporal regulation of TBP (and to a lesser extent Pol II) is a general phenomenon for basal transcription factors during embryogenesis. This is not the case. Two other basal transcription factors, TFIIB and TFIIF RAP74, appear to be expressed constitutively during embryogenesis (Fig. 1A). These results suggest that the ratio of TBP and, to a lesser degree Pol II, to other basal transcription factors undergoes major transitions during early embryonic development.
Translational regulation of maternal TBP RNA.
TBP protein starts accumulating at stage 6½, i.e., before the onset of embryonic transcription (Fig. 1A). We therefore hypothesized that this protein is translated from maternal TBP mRNA. To examine this possibility, RNA was isolated from staged embryos and subjected to Northern blot analysis. Using a Xenopus TBP probe, a single TBP mRNA of approximately 2,000 nucleotides was detected (Fig. 1B). This mRNA is relatively abundant in oocytes and declines in abundance during early embryogenesis, thereby exhibiting a temporal profile complementary to that of the protein it encodes (Fig. 1D), which suggests a shorter half-life for the TBP message when TBP protein is accumulating.
To examine the possibility that TBP is translated from maternal stores of RNA, ribonucleoproteins from oocytes and staged embryos were fractionated by density gradient centrifugation. Fractions were collected from these gradients, and the RNA isolated from these fractions was subjected to Northern blot analysis. A significant fraction of the TBP RNA from stage 9 embryos is associated with ribosomes (Fig. 1C), a finding indicative of TBP RNA being translated. In contrast, TBP RNA from oocytes is predominantly found in the nonribosomal messenger RNP (mRNP) fraction, indicative of translational masking (Fig. 1C [52]). The total signal corresponding to TBP RNA is less in stage 9 embryos compared to oocytes, which is consistent with the relative RNA levels observed in oocytes and embryos (Fig. 1B). Fractionation of RNPs from cleavage-stage embryos (stage 4) shows an intermediate level of association with mRNPs and ribosomes (data not shown). This is consistent with the gradual increase in TBP protein levels observed during embryogenesis (Fig. 1A; see also reference 6). We therefore conclude that TBP is translated from maternal stores of RNA. This is a process that starts before the MBT and that results in maximal TBP levels at the onset of transcription at the MBT.
TBP is rate limiting for basal transcription in egg extract.
TBP stimulates RNA Pol II-dependent transcription in pre-MBT embryos (3, 38). However, this stimulation was found to be dependent on two experimental conditions: (i) preincubation of recombinant TBP protein with the promoter template prior to injection into embryos and (ii) a state of incomplete chromatin assembly. Such an immature chromatin structure is observed at early time points after injection of the promoter template and after coinjection of a large amount of nonspecific DNA that titrates chromatin assembly.
The requirement to have TBP protein prebound to the promoter before injection into embryos suggested that a potential mechanism for this phenomenon involves a dynamic competition between TBP and histones. Our finding that TBP is regulated at the level of its translation from maternally stored RNA, such that TBP protein levels are very low before the MBT (Fig. 1), raised the possibility that TBP may be rate limiting for the basal transcription machinery. In the latter model, the requirement for interfering with pre-MBT chromatin assembly reflects a separate repression mechanism that is superimposed on top of the TBP regulation.
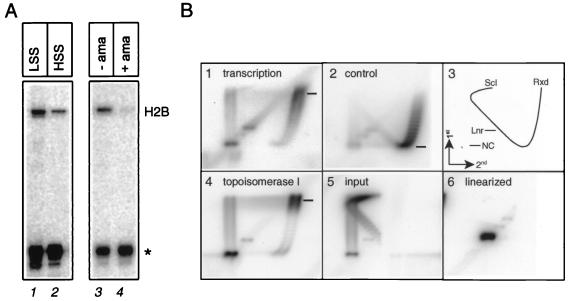
To examine this possibility, we employed a developmentally relevant in vitro transcription extract. Extracts from Xenopus eggs appear to support low levels of transcription that is RNA Pol II dependent (Fig. 2A). This reveals the presence of a complete and active basal transcription machinery, a result consistent with published data (38). The difference with regard to transcription between the egg extract and the egg from which the extract is made is explained by the titration of chromatin assembly by excess DNA template in the in vitro transcription assay. Consistent with this, two-dimensional chloroquine gels, which allow the resolution of DNA topoisomers according to the extent to which they are supercoiled, reveal that the plasmid containing the promoter template is in a relaxed state comparable to plasmid relaxed in vitro by recombinant topoisomerase I. This confirms the absence of detectable chromatin assembly under these reaction conditions (Fig. 2B).
FIG. 2.
RNA Pol II-dependent transcription in egg extract. (A) Both low-speed supernatant (LSS) and high-speed supernatant (HSS) egg extract contain a functional transcription machinery (lanes 1 and 2) that is dependent on RNA Pol II as assessed by sensitivity to low levels (10 μg/ml) of α-amanitin (lanes 3 and 4). Except for lane 2, all in vitro transcription analysis in this study was performed with LSS extract. “H2B” indicates the position primer extension products corresponding to transcripts specifically initiated from a Xenopus histone H2B promoter, and the asterisk indicates the position of the primer extension product of synthetic LucΔ RNA, which was used as internal standard. Incubation of 0.6 μg of pH2B-Luc (160 fmol of DNA) with 1 μl of LSS egg extract under the conditions described in Materials and Methods results in the synthesis of ca. 0.2 to 0.5 fmol of correctly initiated transcript. (B) Two-dimensional gel electrophoresis in the presence of chloroquine. Panel B1 shows the DNA topology under standard egg extract in vitro transcription conditions, panel B2 shows a control sample containing nucleosomes, panel B3 explains the hybridization signals (“Rxd,” “Scl,” “Lnr,” and “NC” represent, respectively, relaxed, supercoiled, linearized, and nicked pH2B-Luc plasmid), panel B4 shows the positions of plasmid DNA treated with recombinant topoisomerase I (resulting in nicked and relaxed DNA), panel B5 shows the topology of plasmid as isolated from Escherichia coli, and panel B6 shows the position of linearized pH2B-Luc plasmid. The estimated average number of supercoils of the DNA in the in vitro transcription reaction, based on quantitated PhosphorImager area profiles of resolved DNA isomers, is 3.9. The uncertainty in the number of supercoils as a result of a dynamic equilibrium between supercoiling (writhe) and winding (twist) is estimated to be ±2 (see panel B4).
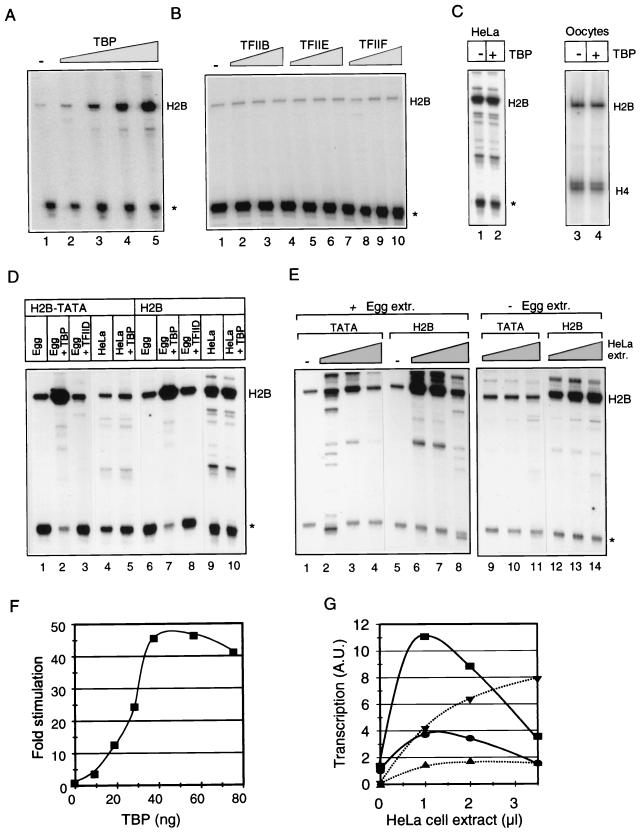
We investigated whether TBP or other basal transcription factors can stimulate transcription in this system. Addition of purified TBP to egg extract appears to stimulate transcription from a Xenopus histone H2B promoter 40- to 50-fold (Fig. 3A and F). We examined whether only TBP could stimulate H2B transcription in egg extract by the addition of several other human basal transcription factors (i.e., TFIIB, TFIIE, and TFIIF) in amounts up to twice the amount which saturates basal transcription in a reconstituted system (23). TFIIB, TFIIE, and TFIIF, however, do not affect the levels of transcript synthesized by egg extract (Fig. 3B). This stimulation of transcription by TBP is not an artifact of in vitro transcription, since transcription driven by HeLa cell extract is not affected by exogenous TBP (Fig. 3C). Likewise, no stimulation of transcription from a Xenopus histone H2B promoter is observed in oocytes upon TBP overexpression (Fig. 3C), providing additional evidence that TBP rate limitation is not a general phenomenon. To rule out the possibility that TBP was competing with an undetectable number of nucleosomes, 3 μg of nonspecific DNA was added to each reaction containing two egg equivalents of egg extract, and the effect of TBP was tested on transcription. Nonspecific DNA does not affect transcription in the presence or absence of TBP (data not shown).
FIG. 3.
TBP is rate limiting for basal transcription, but exogenous TBP is not sufficient to support activated transcription in egg extract. (A) Transcription from the histone H2B promoter in egg extract in the absence of exogenous TBP (lane 1) and in the presence of 9.5, 19, 28.5, or 38 ng of TBP (lanes 2 to 5). (B) The basal factors TFIIB, TFIIE, and TFIIF do not stimulate egg extract transcription. Transcription in the absence of exogenous basal factors (lane 1), in the presence of 12.5, 25, or 50 ng of TFIIB (lanes 2 to 4), in the presence of 22.5, 45, or 90 ng of TFIIE (lanes 5 to 7), and in the presence of 50, 100, or 200 ng of TFIIF (lanes 8 to 10). A total of 25 ng of TFIIB, 45 ng of TFIIE, and 100 ng of TFIIF is sufficient to saturate basal transcription in a reconstituted system (23). (C) TBP does neither stimulate nor inhibit transcription in HeLa cell extract or Xenopus oocytes. Lane 1, histone H2B transcription in the absence of TBP; lane 2, histone H2B transcription in the presence of 38 ng of TBP (larger amounts of TBP also do not stimulate [data not shown]); lanes 3 and 4, transcription from the H2B promoter in oocytes with or without 0.5 ng of TBP RNA injected into the cytoplasm 3 h before injection of the DNA (compare with Fig. 4). Primer extension of endogenous histone H4 was used as a control for RNA loading in this experiment. (D) trans-Activation was assayed under various circumstances by comparing the activities of the core histone H2B promoter (TATA) and the full-length histone H2B promoter. Lanes 1 to 5, transcription from basal H2B promoter; lanes 6 to 10, transcription from full-length H2B promoter. (E) Functional deficiency in egg extract affecting activated transcription. trans-Activation was assayed under various circumstances by comparing the activities of the core histone H2B promoter (H2B-TATA, lanes 1 to 4 and lanes 9 to 11) and the full-length histone H2B promoter (H2B, lanes 5 to 8 and lanes 12 to 14). Transcription driven by 1 μl of egg extract mixed with 0 μl (lanes 1 and 5), 1 μl (lanes 2 and 6), 2 μl (lanes 3 and 7), and 3.5 μl (lanes 4 and 8) of HeLa cell extract. Lanes 9 to 14 show the extent to which transcription is supported by the respective amounts of HeLa cell extract alone. Mixed extract and HeLa cell extract, but not egg extract alone, support activated transcription. (F) Quantitation of three TBP titration experiments, including the experiment shown in panel A. (G) Graphic representation of the experiment show in panel E. The levels of transcription obtained were related to the level obtained with egg extract, which was arbitrarily set at 1. Addition of 0.5 μl of HeLa cell extract to 1 μl of egg extract resulted in levels of transcription that were approximately twofold lower than those observed when 1 μl of HeLa extract was added to 1 μl of egg extract; activation was less than threefold when 0.5 μl of HeLa cell extract was used. Optimal, activated transcription is achieved when 1 μl of HeLa extract is added to 1 μl of egg extract, as shown in the graph. Symbols: ●, basal transcription driven by egg extract mixed with increasing amounts of HeLa extract; ■, transcriptional activity of the full-length promoter in the presence of egg extract mixed with increasing amounts of HeLa extract; ▴, basal transcription driven by increasing amounts of HeLa extract; ▾, transcriptional activity of the full-length promoter in the presence of increasing amounts of HeLa extract.
In conclusion, addition of TBP stimulates a rate-limiting step for transcription in egg extract in the absence of detectable chromatin assembly and independent of the titration of potentially undetectable low amounts of nucleosome assembly. This indicates that titration of chromatin assembly and the complementation of the basal transcription machinery with exogenous TBP represent two separate steps in relieving pre-MBT constraints on basal transcription.
TBP does not relieve a deficiency for activated transcription in vitro.
As complementation of the transcription machinery of the egg with TBP protein boosts the transcriptional capacity, we wondered whether this experimental situation would allow cross-talk between transcriptional activators and the basal transcription machinery in the egg extract in vitro transcription system.
Histone H2B promoters contain an Oct-1 binding site (14, 28, 46, 48, 59) directly upstream of the TATA box that contributes to the regulation of this promoter in vivo (14, 17, 22, 28). Oct-1 is one of the founding members of the POU domain family of transcriptional regulators (20, 41, 58), and is maternally derived in Xenopus (21, 22). To determine whether or not Oct-1 contributes to histone H2B promoter activity in egg extracts, Oct-1 was depleted from egg extracts by using a monoclonal antibody (56), and the effect of Oct-1 depletion on H2B promoter activity was tested in the absence or presence of exogenous TBP. Depletion of more than 90% of Oct-1, however, does not affect histone H2B promoter activity in egg extracts (data not shown). Therefore, the contribution of upstream promoter sequences was determined by using a core promoter and a full-length histone H2B promoter construct (designated, respectively, TATA and H2B). The TATA construct contains an initiator element and a TATA box but lacks the Oct-1 and CCAAT motifs present in the full-length histone H2B promoter. Since the TATA construct lacks any cis-acting elements except those recognized by the basal transcription machinery, we operationally define the activity of the TATA construct as basal transcription. The degree to which transcriptional activators contribute to overall promoter activity is given by the ratio of H2B to TATA promoter activity (fold activation).
The TATA and H2B constructs appear to be equally active in egg extract (Fig. 3D, lanes 1 and 6), indicating that activated transcription is not supported by egg extract whereas it is supported by HeLa cell extract (lanes 4 and 9). TBP stimulates only basal transcription in egg extract and does not influence basal and activated transcription in HeLa cell extract (lanes 5 and 10). Activated transcription is known to involve TBP-associated factors (reviewed by Burley and Roeder [15] and Goodrich et al. [10]), and since HeLa cell extract supports activated transcription, we addressed the question whether HeLa TFIID (54), a functional complex containing TBP and TBP-associated factors, would confer activated transcription to egg extract. HeLa TFIID, in contrast to TBP, moderately stimulates basal egg extract transcription, about two-fold (Fig. 3D, lane 3). In part this is explained by the relatively low concentration of TBP in the HeLa TFIID fraction (Western blot analysis [not shown]). However, the addition of still more TFIID did not result in a further increase of transcription. The stimulation of egg extract transcription by TFIID by using a full-length promoter was slightly stronger than the stimulation of basal transcription but never exceeded 1.5-fold activation, whereas HeLa cell extract, containing similar levels of TBP as assessed by Western blot analysis, supports 3- to 5-fold activation (Fig. 3D, compare lanes 3 to 5 and lanes 8 to 10).
The transcription machinery of the egg, as recruited by the histone H2B promoter, appears to be restricted in its ability to support activated transcription. Since the inability to support activated H2B transcription is not relieved by either TBP or TFIID, it represents a second constraint on the transcription machinery, one distinct from the constraint on basal transcription by limiting amounts of TBP protein.
We wanted to test whether egg extract is permissive to transcriptional activation in the presence of all the factors required for full activation or whether the lack of activation is a dominant property of egg extract, one mediated by a repressor of activated transcription. For instance, NAT (negative regulator of activated transcription), a complex containing the human Srb10 and Med6 proteins, represses activated transcription, while basal transcription is facilitated (51). However, egg extract does not dominantly repress activation; HeLa cell extract confers the ability to activate transcription to egg extract when these extracts are mixed (Fig. 3E and G). Transcript synthesis is most efficient in a 1:1 mixed extract, which supports levels of transcription that are substantially higher than that of either extract alone. The addition of more HeLa extract results in loss of synergy and in repression of mainly basal transcription. When increasing amounts of egg extract are added to a fixed amount of HeLa extract, it appears that egg extract inhibits basal HeLa transcription without affecting the level of activation (not shown). Therefore, the lack of transcription activation in egg extract is not dominant over the activation which is supported by HeLa cell extract but rather is caused by a functional deficiency in egg extract (Fig. 3E and G). This functional deficiency may involve either the physical absence or a lack of activity of a transcription cofactor(s). In addition, the observed capacity of the human HeLa cell extract to stimulate transcription in the frog egg extract indicates that this putative human cofactor also functions with the basal transcription factors from X. laevis. We conclude that transcription activation of the histone H2B promoter is impaired in vitro, independent of the levels of TBP that affect basal transcription.
Precocious translation of TBP mRNA stimulates transcription before the MBT in vivo.
If exogenous TBP stimulates transcription before the MBT by relieving a rate-limiting step, as suggested by the chromatin-independent stimulation by TBP in vitro, one might expect that precocious translation of otherwise masked TBP mRNA in the embryo will actually stimulate transcription in the embryo. This would not only provide evidence for the functional relevance of the translational regulation of TBP but would also show that TBP does not necessarily have to be prebound to the promoter for transcriptional stimulation in this system.
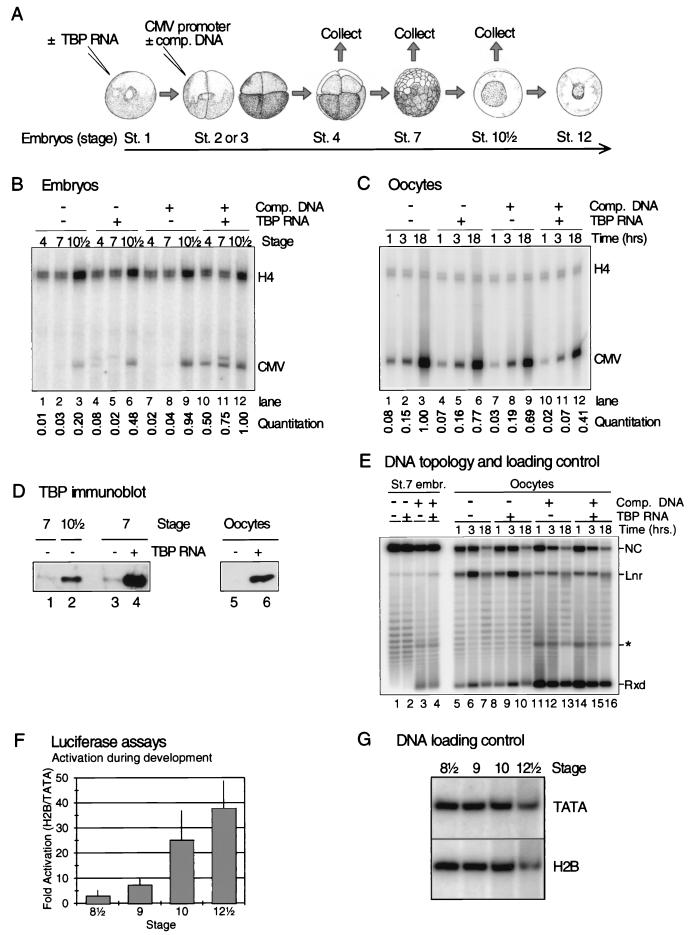
Our initial attempts to inject a CMV promoter template, with or without translationally competent TBP RNA and with or without nonspecific DNA, failed to reveal any stimulation of transcription by TBP before the MBT. However, it appeared that nonspecific DNA not only titrates chromatin assembly but also nonspecifically inhibits the translation of otherwise translationally competent TBP mRNA. Therefore, we redesigned the experiment (Fig. 4A) in such a way that TBP mRNA is injected into embryos at the one-cell stage, whereas 1 h later (at the two- to four-cell stages) the promoter template is injected into one blastomere with or without nonspecific DNA. This allows for a 1-h time window in which the TBP RNA is translated into protein. Subsequently, total RNA was isolated from cleavage stage embryos (stage 4, ca. 1 h after injection of DNA), early blastula embryos (stage 7, ca. 3 h after injection of DNA), and early gastrula embryos (stage 10½, post-MBT). CMV promoter activity was assayed by using primer extension, whereas primer extension of maternally loaded histone H4 mRNA served as a control.
FIG. 4.
Precocious translation of TBP RNA in embryos facilitates transcription before the MBT, but embryos do not fully activate an H2B promoter until well after TBP complementation of the basal transcription machinery. (A) Synthetic TBP RNA (0.5 ng) was injected into stage 1 embryos. Unlike endogenous TBP RNA, synthetic RNA is actively translated before the MBT. After 1 h 0.5 ng of double-stranded pCMV-CAT was injected into one blastomere of two-cell-stage embryos, with or without 20 ng of nonspecific competitor DNA (plasmid without eukaryotic promoter). Embryos were collected for RNA isolation after 1-h, 3-h (25°C), or overnight (14°C) incubation. A similar experiment was performed with oocytes. (B) Primer extension of RNA isolated from embryos subjected to the experiment described in panel A. “CMV” indicates the position of the primer extension product of RNA initiated at the injected CMV promoter; “H4” indicates the position of the primer extension product of endogenous histone H4 RNA used as a primer extension control. A quantitation of the primer extension products from the CMV promoter is shown. The CMV promoter activity was normalized by using the relative level of histone H4 RNA (see Materials and Methods). The results are reproducible between experiments. Two bands are visible for CMV transcription in the lanes containing TBP (lanes 4, 5, and 11); only the lower band represents accurately initiated transcription. The origin of the upper band is unknown. (C) Primer extension of RNA isolated from oocytes subjected to an experiment comparable to the one shown in panel B. At 3 h after injection of TBP RNA into the cytoplasm, 0.5 ng of double-stranded pCMV-CAT was injected into the nucleus, with or without 20 ng of nonspecific competitor DNA (plasmid without eukaryotic promoter). Oocytes were collected for RNA isolation after 1, 3, or 18 h. A quantitation is shown as in panel B. (D) Western blot analysis of TBP protein levels after overexpression in embryos (lanes 3 and 4) or oocytes (lanes 5 and 6). Endogenous TBP levels are shown in lanes 1, 2, and 5 (compare with Fig. 1). (E) Southern blot analysis of a chloroquine gel loaded with DNA isolated from embryos and oocytes subjected to the experiment described in panel A (compare panels B and C). This serves as a loading control (comparable amounts of DNA in all experimental samples) and reveals the DNA topology. The blot was labeled with a CMV-CAT-specific probe. The asterisk shows the position of a cross-hybridizing band corresponding to competitor DNA with a relaxed topology. (F) Activation of the Xenopus H2B promoter as assayed with luciferase assays by using the core promoter (TATA) and full-length promoter (H2B) constructs (0.2 ng of DNA per embryo). Errors ± the standard error of the mean are indicated. The difference between stage 9 and stage 12½ is statistically significant (P < 0.025). (G) DNA loading control of the experiments shown in panel F. This shows that no significant differences exist in extrachromosomal replication of the constructs during embryonic development.
The CMV promoter exhibits the normal developmental regulation observed for class II genes when injected without TBP RNA or nonspecific DNA (Fig. 4B). Correctly initiated transcripts derived from this promoter first appear after the MBT (lane 3), whereas these transcripts are not detectable at stage 4 and 7 (lanes 1 and 2). If TBP mRNA is injected prior to the DNA injection, a weak and temporary signal corresponding to correctly initiated transcripts is detected at stage 4 (lane 4). This signal, however is lost at stage 7, before the normal post-MBT transcriptional signal is detected at stage 10½ (lane 6). The weak signal observed at stage 4 apparently combines elevated TBP levels (Fig. 4D) with the relatively accessible promoter structure observed at early time points after injection of naked double-stranded DNA. However, a much more robust transcription before the MBT is observed when both TBP RNA was injected prior to DNA injection and nonspecific DNA was coinjected with the promoter template (Fig. 4B, lanes 10 to 12). Under these circumstances it is not necessary for the CMV promoter to have TBP bound to it before injection into embryos in order to be transcribed. Without TBP RNA injected, nonspecific DNA is not sufficient to allow transcription from the CMV promoter before the MBT (Fig. 4B, lanes 7 and 8).
Because the levels of endogenous TBP protein are not only low in early embryos but also in oocytes (Fig. 1A), we wondered whether the transcriptionally active oocyte would behave in the same way as the embryo does with respect to stimulation of transcription by TBP and nonspecific DNA. We therefore performed a similar experiment with oocytes. TBP RNA was injected into stage VI oocytes, and the CMV promoter template was injected with or without nonspecific DNA 3 h after RNA injection. RNA was isolated from oocytes after 1 or 3 h or after overnight incubation of the injected oocytes. Elevation of TBP levels does not affect transcription in oocytes, whereas nonspecific DNA causes a mildly inhibitory—rather than stimulatory—effect on transcription (Fig. 4C). This result shows that the strong correlation that exists in early embryos between transcriptional activity, TBP rate limitation, and TBP protein abundance comes into existence after the transition from oocyte to egg. If the stage VI oocyte is matured with progesterone into an egg, then repression of transcription is established (29). Another observation is that transcripts accumulate linearly with time in the oocyte, whereas in the embryo (in the presence of TBP and competitor DNA) transcript levels reach a steady state, suggesting a greater role for RNA turnover in the embryo compared to the oocyte.
As a control the DNA topology of the promoter template after injection was examined by using chloroquine gel analysis (Fig. 4E). Nonspecific DNA does titrate chromatin assembly as expected, which manifests itself as an increase in the relaxed fraction of DNA when competitor DNA was coinjected. TBP protein does not affect the topology of the promoter. A significant observation is that the topology of the promoter template in embryos in the presence of nonspecific DNA (Fig. 4E, lanes 3 to 4) is similar to that of promoter template in oocytes without nonspecific DNA (Fig. 4E, lanes 5 and 8), whereas the promoter template in embryos without nonspecific DNA shows a relatively strong supercoiling without a significant fraction of the DNA being in the relaxed state. This reinforces the fact that chromatin assembly in the embryo is very efficient and more repressive than that observed in oocytes (29). In addition, the differences in transcriptional activity cannot be attributed to differences in the amount of DNA injected in embryos or oocytes.
In conclusion, precocious translation of TBP mRNA in embryos but not in oocytes greatly enhances transcription in the presence of nonspecific DNA, and this stimulation of transcription does not require TBP to be prebound to the promoter prior to injection.
Acquisition of transcription activation in pregastrulation embryos.
Our experiments with the egg in vitro transcription system suggest that activated transcription is regulated in a fashion that is mechanistically independent of translational unmasking of TBP mRNA. It is known that some genes are transcribed ubiquitously at low levels right after the MBT, before developmental stage and cell type-specific expression is observed at the time gastrulation starts (8, 40, 60). We therefore asked whether TBP complementation of basal transcription and competence for activated transcription are regulated independently in vivo between the onset of zygotic transcription at the MBT and the onset of gastrulation. To address this question, the TATA and H2B promoter constructs were injected into fertilized eggs, and the promoter activity, as represented by the luciferase activity, was measured at different time points after the MBT (from stage 8½ onwards). We used luciferase assays because these are more sensitive than primer extension mediated detection of promoter activity and because we wanted to examine transcription activation very early after the MBT, when overall transcription levels are very low.
Between stage 8½ and stage 12½, luciferase expression driven by the H2B promoter increases by about 5 orders of magnitude. Luciferase activity increases from an average of about 500 light units per embryo at stage 8½ (integrated over the first 10 s) to 4 × 106 light units per embryo at stage 12½. Between stages 8½ and 9 the increase in activity of the TATA and H2B promoters is about the same at each stage, which means that the ability of the embryonic transcription machinery to activate transcription is limited (Fig. 4F), as it is in egg extract (Fig. 3D). Between stages 9 and 10, however, the activity of the full-length H2B promoter increases dramatically (ca. 170-fold), whereas the basal promoter construct increases significantly less (ca. 50-fold), representing a marked increase in transcriptional activation just before gastrulation commences.
As a control, the injected DNA was isolated from the embryos at different stages of embryonic development to ensure that the different activities observed were not due to differential replication of the constructs. Identical amounts of DNA were recovered from the injected embryos at each stage (Fig. 4G). Therefore, we conclude that a significant increase in transcriptional activation occurs between the MBT and the onset of gastrulation (Fig. 4F), the developmental timing of which is different from that of TBP accumulation between early and late blastula stages (compare Fig. 1A and 4F).
DISCUSSION
We have studied the molecular mechanisms that are underlying transcriptional repression during early embryogenesis and relief of that repression at the MBT. Our main conclusions are as follows. (i) TBP is translated from maternally stored RNA just before the MBT. The ratio of TBP to other basal transcription factors undergoes a major transition during early embryonic development as a result of this regulation. (ii) TBP is rate limiting for transcription before the MBT both in vitro and in vivo, and precocious translation of synthetic TBP RNA in the embryo facilitates transcription before the MBT in the presence of nonspecific DNA. (iii) The transcription machinery of the egg does not support activation of a H2B promoter above basal levels of transcription, even though transcriptional activators are present in the egg and are able to bind DNA. This lack of activated transcription is also observed in the embryo and is only alleviated between the MBT and gastrulation.
Developmental regulation of the embryonic transcription machinery.
The data presented here shed new light on the earlier work by Prioleau et al. (37, 38) and Almouzni and Wolffe (3). From the present study it seems quite clear that major deficiencies exist with regard to the early embryonic transcription machinery, even independent of chromatin-mediated repression mechanisms. Nevertheless, these chromatin-mediated repression mechanisms are quite important for repression of the genome before the MBT. Even in this study, we have not been able to see transcription before the MBT without interfering with chromatin assembly in some way. In our in vitro experiments this was obtained by high template concentrations, such that nucleosome assembly by egg extract was virtually completely abolished. In the embryo this was, as in previous studies, achieved by coinjection of competitor DNA and by assaying at early time points after injection when chromatin assembly is far from complete.
An interesting question is how the repressive effects of chromatin and the deficiencies in TBP and coactivators work together to achieve full repression before the MBT. One possible unifying model that historically has been considered entails a dynamic competition between histones and transcription factors for binding to DNA. The observations that seemed to suggest that this is true were as follows. Preincubation of the promoter template with recombinant TBP protein results in a transient basal transcription if the promoter subsequently is injected into the embryo (37, 38). This transient transcription by TBP alone is sustained if nonspecific competitor DNA (ca. 25 ng) is coinjected. If large amounts of competitor DNA are injected (ca. 80 ng per embryo) transcription is observed without any exogenous TBP (38). In these studies, TBP and the promoter template needed to be preincubated in order to stimulate transcription. If recombinant TBP protein and promoter template are injected separately, TBP fails to facilitate transcription, suggesting that it needs to be prebound in order to compete with histones for binding DNA. However, the current study suggests a different interpretation of these data. If the exclusion of nucleosomes by TBP was the driving force of the TBP-mediated stimulation before the MBT, two predictions could be made. The first is that, in the absence of nucleosomes, additional TBP would have no role in stimulating transcription because the endogenous TBP available in the embryo would be able to sustain high levels of basal transcription. The second prediction is that TBP translated from injected RNA would fail to stimulate transcription in the embryo because in this experimental regime TBP is not prebound to DNA before injection into the embryo, which in the experiments with recombinant protein was found to be required for transcription. It appears that either prediction fails. If nucleosome assembly is titrated by high amounts of DNA in egg extract, such that transcription is observed without addition of exogenous TBP (Fig. 2), TBP still potently stimulates basal transcription (Fig. 3) in a fashion that is independent of additional competitor DNA. The other prediction fails as well. As shown in Fig. 4, TBP translated in the embryo can stimulate basal transcription efficiently in vivo, without being bound to the template prior to injection. So the requirement to have TBP protein prebound to the template, which provided an argument in favor of the dynamic competition model, seems to be a peculiarity of the use of recombinant TBP rather than TBP RNA to achieve overexpression. This may be explained by differential mosaicisms of injected RNA and protein, respectively. In comparison with RNA, protein tends to be more localized at the site of injection (9, 56a), such that if the promoter template is injected separately the injected protein might not be close to the DNA. Taking these data together, repression by chromatin and constraints imposed by TBP represent distinct and independent levels of regulation. Although the effects of these different levels of regulation are superimposed on each other, the mechanisms themselves are not interdependent and can be uncoupled both in vitro and in vivo (Fig. 3 and 4).
In addition to the insights in the nature of stimulation of transcription by TBP provided by these functional studies, we have characterized the regulation of TBP mRNA, TBP protein, and TBP translation during early embryonic development. Western blot analysis establishes a clear underrepresentation of TBP within the early embryonic transcription machinery (Fig. 1), a machinery which is functional in the absence of chromatin-mediated repression (Fig. 2) and yet severely constrained, as revealed by the addition of exogenous TBP (Fig. 3 and 4). Therefore, TBP is rate limiting before the MBT, a condition alleviated by translation of stored maternal TBP RNA just preceding the MBT (Fig. 1). Currently, we have no information regarding the sequence of the Xenopus TBP mRNA outside the coding region (16). A specific dodecauridine element in the 3′ untranslated regions (UTRs) of Cl2 and activin receptor mRNA has been implicated in the embryonic polyadenylation and translation of these RNAs (44, 45). The accumulation of TBP from masked maternal mRNA occurs well before that of components of chromatin such as histone H1 and Polycomb accumulate (8, 13, 50). Comparison of regulatory elements in the 3′ UTRs of these masked mRNAs will be informative. The large subunit of Pol II is also translationally regulated (Fig. 1A). We have no information, however, on the expression of other subunits of RNA Pol II. In addition it is unclear whether limitation of components of the basal transcription machinery other than TBP also contributes to transcriptional constraint.
Not only is basal transcription regulated during early embryonic development. We demonstrate that the ability to activate a H2B promoter is constrained in the embryo at the MBT, after which this constraint is substantially alleviated by gastrulation (Fig. 4). Our in vitro characterization of the pre-MBT transcription machinery suggests that a functional deficiency of cofactor(s) is responsible for the absence of activation before the MBT (Fig. 3). This deficiency is likely to affect multiple trans-acting factors. It has been shown that Xenopus oocytes and embryos contain multiple maternally derived transcription factors capable of binding a histone H2B promoter (21, 22). Furthermore, in at least one study, Gal4-VP16 has been found to bind promoter DNA while failing to activate transcription in early embryos (37). We have been able to restore the ability to activate a histone H2B promoter at the MBT by injecting RNA encoding a human cofactor (58a). Although we have no data on the expression of the endogenous coactivator in Xenopus embryos and although this particular cofactor may not restore activation to all promoters under these conditions, the observation that expression of a single coactivator causes a robust and reproducible stimulation of activated but not of basal transcription in stage 9 embryos is in concordance with the in vitro data shown in Fig. 3. Evidently, the embryonic transcription machinery is regulated, and basal and activated transcription are independently constrained and potentiated, by TBP and (potentially multiple) coactivators.
Transcriptional regulation accompanying the early gastrula transition.
Our studies indicate that transcriptional activation of the histone H2B promoter initially is absent after the MBT; however, activation progressively increases through gastrulation. A number of genes are known to be transiently transcribed at the MBT, irrespective of their normal spatial or temporal regulation (8, 40, 60). The mechanism underlying this developmentally regulated phenomenon most likely is the appearance of the counteracting effects of chromatin-mediated repression of basal transcription and selective activation overruling such chromatin-mediated repression. For example, the histone deacetylase inhibitor trichostatin A fails to stimulate transcription before gastrulation, while it potently derepresses transcription after gastrulation (1, 50). In the case of the 5S rRNA genes, the counteracting effects of repression and activation are known to be mediated by histone H1 and the TFIIIA transcription factor, respectively (5, 8). Another example is the Xenopus MyoD gene, which is ubiquitously expressed after the MBT before a positive autoregulatory loop is established in myogenic cells at the onset of gastrulation (between stages 10 and 10¼), whereas in nonmyogenic cells MyoD is repressed by that time (40). The accumulation of histone H1 contributes to that repression, since ablation of H1 accumulation induces ectopic MyoD expression (49).
Interestingly, the developmental stage at which robust activation is observed correlates with early gastrula transition (24), between stages 10 and 10½, which represents a switch of maternal to zygotic control of cell cycle regulation (24, 25, 32), the switch of maternal to zygotic control of programmed cell death (18, 47), along with the earliest detectable endogenous apoptosis (19, 57), and the onset of gastrulation itself. It is conceivable that it is not just coincidence that acquisition by the embryo of full regulatory control of a variety of developmental processes occurs at a developmental stage at which the embryo for the first time is able to regulate gene expression from complete silencing to full activation. Our studies suggest that the translation of maternal TBP mRNA contributes to the basal levels of transcription observed for the first time at the MBT, while full control of gene expression may require the embryo to complement its transcription machinery with coactivators.
ACKNOWLEDGMENTS
We thank H. T. Marc Timmers, Peter C. van der Vliet, and J. Julian Blow for invaluable advice, suggestions, and reagents during early stages of this work.
This work was supported by a grant from the Dutch Organization for Scientific Research (NWO).
REFERENCES
- 1.Almouzni G, Khochbin S, Dimitrov S, Wolffe A P. Histone acetylation influences both gene expression and development of Xenopus laevis. Dev Biol. 1994;165:654–669. doi: 10.1006/dbio.1994.1283. [DOI] [PubMed] [Google Scholar]
- 2.Almouzni G, Wolffe A P. Replication-coupled chromatin assembly is required for the repression of basal transcription in vivo. Genes Dev. 1993;7:2033–2047. doi: 10.1101/gad.7.10.2033. [DOI] [PubMed] [Google Scholar]
- 3.Almouzni G, Wolffe A P. Constraints on transcriptional activator function contribute to transcriptional quiescence during early Xenopus embryogenesis. EMBO J. 1995;14:1752–1765. doi: 10.1002/j.1460-2075.1995.tb07164.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Andéol Y. Early transcription in different animal species: implication for transition from maternal to zygotic control in development. Roux’s Arch Dev Biol. 1994;204:3–10. doi: 10.1007/BF00189062. [DOI] [PubMed] [Google Scholar]
- 5.Andrews M T, Brown D D. Transient activation of oocyte 5S RNA genes in Xenopus embryos by raising the level of the trans-acting factor TFIIIA. Cell. 1987;51:445–453. doi: 10.1016/0092-8674(87)90640-4. [DOI] [PubMed] [Google Scholar]
- 6.Bell P, Scheer U. Developmental changes in RNA polymerase I and TATA box-binding protein during early Xenopus embryogenesis. Exp Cell Res. 1999;248:122–135. doi: 10.1006/excr.1999.4411. [DOI] [PubMed] [Google Scholar]
- 7.Blow J J. Preventing re-replication of DNA in a single cell cycle: evidence for a replication licensing factor. J Cell Biol. 1993;122:993–1002. doi: 10.1083/jcb.122.5.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bouvet P, Dimitrov S, Wolffe A P. Specific regulation of Xenopus chromosomal 5S rRNA gene transcription in vivo by histone H1. Genes Dev. 1994;8:1147–1159. doi: 10.1101/gad.8.10.1147. [DOI] [PubMed] [Google Scholar]
- 9.Brown D D, Schlissel M S. A positive transcription factor controls the differential expression of two 5S RNA genes. Cell. 1985;42:759–767. doi: 10.1016/0092-8674(85)90272-7. [DOI] [PubMed] [Google Scholar]
- 10.Burley S K, Roeder R G. Biochemistry and structural biology of transcription factor IID (TFIID) Annu Rev Biochem. 1996;65:769–799. doi: 10.1146/annurev.bi.65.070196.004005. [DOI] [PubMed] [Google Scholar]
- 11.Dahmus M E. Phosphorylation of the C-terminal domain of RNA polymerase II. Biochim Biophys Acta. 1995;1261:171–182. doi: 10.1016/0167-4781(94)00233-s. [DOI] [PubMed] [Google Scholar]
- 12.Destrée O H J, Bendig M M, De Laaf R T M, Koster J G. Organisation of Xenopus histone gene variants within clusters and their transcriptional expression. Biochim Biophys Acta. 1984;782:132–141. doi: 10.1016/0167-4781(84)90016-2. [DOI] [PubMed] [Google Scholar]
- 13.Dimitrov S, Almouzni G, Dasso M, Wolffe A P. Chromatin transitions during early Xenopus embryogenesis: changes in histone H4 acetylation and in linker histone type. Dev Biol. 1993;160:214–227. doi: 10.1006/dbio.1993.1299. [DOI] [PubMed] [Google Scholar]
- 14.Fletcher C, Heintz N, Roeder R G. Purification and characterization of OTF-1, a transcription factor regulating cell cycle expression of a human histone H2b gene. Cell. 1987;51:773–781. doi: 10.1016/0092-8674(87)90100-0. [DOI] [PubMed] [Google Scholar]
- 15.Goodrich J A, Cutler G, Tjian R. Contacts in context: promoter specificity and macromolecular interactions in transcription. Cell. 1996;84:825–830. doi: 10.1016/s0092-8674(00)81061-2. [DOI] [PubMed] [Google Scholar]
- 16.Hashimoto S, Fujita H, Hasegawa S, Roeder R G, Horikoshi M. Conserved structural motifs within the N-terminal domain of TFIID tau from Xenopus, mouse and human. Nucleic Acids Res. 1992;20:3788. doi: 10.1093/nar/20.14.3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heintz N. The regulation of histone gene expression during the cell cycle. Biochim Biophys Acta. 1991;1088:327–339. doi: 10.1016/0167-4781(91)90122-3. [DOI] [PubMed] [Google Scholar]
- 18.Hensey C, Gautier J. A developmental timer that regulates apoptosis at the onset of gastrulation. Mech Dev. 1997;69:183–195. doi: 10.1016/s0925-4773(97)00191-3. [DOI] [PubMed] [Google Scholar]
- 19.Hensey C, Gautier J. Programmed cell death during Xenopus development: a spatio-temporal analysis. Dev Biol. 1998;203:36–48. doi: 10.1006/dbio.1998.9028. [DOI] [PubMed] [Google Scholar]
- 20.Herr W, Sturm R A, Clerc R G, Corcoran L M, Baltimore D, Sharp P A, Ingraham H A, Rosenfeld M G, Finney M, Ruvkun G, Horvitz H R. The POU domain: a large conserved region in the mammalian pit-1, oct-1, oct-2, and Caenorhabditis elegans unc-86 gene products. Genes Dev. 1988;2:1513–1516. doi: 10.1101/gad.2.12a.1513. [DOI] [PubMed] [Google Scholar]
- 21.Hinkley C, Perry M. A variant octamer motif in a Xenopus H2b histone gene promoter is not required for transcription in frog oocytes. Mol Cell Biol. 1991;11:641–654. doi: 10.1128/mcb.11.2.641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hinkley C, Perry M. Histone H2b gene transcription during Xenopus early development requires functional cooperation between proteins bound to the CCAAT and octamer motifs. Mol Cell Biol. 1992;12:4400–4411. doi: 10.1128/mcb.12.10.4400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Holstege F C P, Tantin D, Carey M, Van der Vliet P C, Timmers H T M. The requirement for the basal transcription factor IIE is determined by the helical stability of promoter DNA. EMBO J. 1995;14:810–819. doi: 10.1002/j.1460-2075.1995.tb07059.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Howe J A, Howell M, Hunt T, Newport J W. Identification of a developmental timer regulating the stability of embryonic cyclin A and a new somatic A-type cyclin at gastrulation. Genes Dev. 1995;9:1164–1176. doi: 10.1101/gad.9.10.1164. [DOI] [PubMed] [Google Scholar]
- 25.Howe J A, Newport J W. A developmental timer regulates degradation of cyclin E1 at the midblastula transition during Xenopus embryogenesis. Proc Natl Acad Sci USA. 1996;93:2060–2064. doi: 10.1073/pnas.93.5.2060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jacobson M D, Weil M, Raff M C. Programmed cell death in animal development. Cell. 1997;88:347–354. doi: 10.1016/s0092-8674(00)81873-5. [DOI] [PubMed] [Google Scholar]
- 27.Jones P L, Veenstra G J C, Wade P A, Vermaak D, Kass S U, Landsberger N, Strouboulis J, Wolffe A P. Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nat Genet. 1998;19:187–191. doi: 10.1038/561. [DOI] [PubMed] [Google Scholar]
- 28.LaBella F, Sive H L, Roeder R G, Heintz N. Cell-cycle regulation of a human histone H2b gene is mediated by the H2b subtype-specific consensus element. Genes Dev. 1988;2:32–39. doi: 10.1101/gad.2.1.32. [DOI] [PubMed] [Google Scholar]
- 29.Landsberger N, Wolffe A P. Remodeling of regulatory nucleoprotein complexes on the Xenopus hsp70 promoter during meiotic maturation of the Xenopus oocyte. EMBO J. 1997;16:4361–4373. doi: 10.1093/emboj/16.14.4361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Manley J L, Fire A, Samuels M, Sharp P A. In vitro transcription: whole cell extract. Methods Enzymol. 1983;101:568–582. doi: 10.1016/0076-6879(83)01038-1. [DOI] [PubMed] [Google Scholar]
- 31.Meric F, Matsumoto K, Wolffe A P. Regulated unmasking of in vivo synthesized maternal mRNA at oocyte maturation. A role for the chaperone nucleoplasmin. J Biol Chem. 1997;272:12840–12846. doi: 10.1074/jbc.272.19.12840. [DOI] [PubMed] [Google Scholar]
- 32.Newport J, Dasso M. On the coupling between DNA replication and mitosis. J Cell Sci Suppl. 1989;12:149–160. doi: 10.1242/jcs.1989.supplement_12.13. [DOI] [PubMed] [Google Scholar]
- 33.Newport J, Kirschner M. A major developmental transition in early Xenopus embryos. I. Characterization and timing of cellular changes at the midblastula stage. Cell. 1982;30:675–686. doi: 10.1016/0092-8674(82)90272-0. [DOI] [PubMed] [Google Scholar]
- 34.Newport J, Kirschner M. A major developmental transition in early Xenopus embryos. II. Control of the onset of transcription. Cell. 1982;30:687–696. doi: 10.1016/0092-8674(82)90273-2. [DOI] [PubMed] [Google Scholar]
- 35.Nieuwkoop P D, Faber J. Normal table of Xenopus laevis (Daudin): a systematical and chronological survey of the development from the fertilized egg till the end of metamorphosis. 2nd ed. Amsterdam, The Netherlands: North Holland Publishing Company; 1967. [Google Scholar]
- 36.Nothias J-Y, Majumder S, Kaneko K J, DePamphilis M. Regulation of gene expression at the beginning of mammalian development. J Biol Chem. 1995;270:22077–22080. doi: 10.1074/jbc.270.38.22077. [DOI] [PubMed] [Google Scholar]
- 37.Prioleau M-N, Buckle R S, Méchali M. Programming of a repressed but committed chromatin structure during early development. EMBO J. 1995;14:5073–5084. doi: 10.1002/j.1460-2075.1995.tb00189.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Prioleau M-N, Huet J, Sentenac A, Méchali M. Competition between chromatin and transcription complex assembly regulates gene expression during early development. Cell. 1994;77:439–449. doi: 10.1016/0092-8674(94)90158-9. [DOI] [PubMed] [Google Scholar]
- 39.Pritchard D K, Schubiger G. Activation of transcription in Drosophila embryos is a gradual process mediated by the nucleocytoplasmic ratio. Genes Dev. 1996;10:1131–1142. doi: 10.1101/gad.10.9.1131. [DOI] [PubMed] [Google Scholar]
- 40.Rupp R A W, Weintraub H. Ubiquitous MyoD transcription at the midblastula transition precedes induction-dependent MyoD expression in presumptive mesoderm of X. laevis. Cell. 1991;65:927–937. doi: 10.1016/0092-8674(91)90545-a. [DOI] [PubMed] [Google Scholar]
- 41.Ryan A K, Rosenfeld M G. POU domain family values: flexibility, partnerships, and developmental codes. Genes Dev. 1997;11:1207–1225. doi: 10.1101/gad.11.10.1207. [DOI] [PubMed] [Google Scholar]
- 42.Schilthuis J G. Regulation of expression of histone genes during early development of Xenopus laevis. Ph.D. thesis. Utrecht, The Netherlands: Utrecht University; 1990. [Google Scholar]
- 43.Shilatifard A. The RNA polymerase II general elongation complex. Biol Chem. 1998;379:27–31. doi: 10.1515/bchm.1998.379.1.27. [DOI] [PubMed] [Google Scholar]
- 44.Simon R, Tassan J P, Richter J D. Translational control by poly(A) elongation during Xenopus development: differential repression and enhancement by a novel cytoplasmic polyadenylation element. Genes Dev. 1992;6:2580–2591. doi: 10.1101/gad.6.12b.2580. [DOI] [PubMed] [Google Scholar]
- 45.Simon R, Wu L, Richter J D. Cytoplasmic polyadenylation of activin receptor mRNA and the control of pattern formation in Xenopus development. Dev Biol. 1996;179:239–250. doi: 10.1006/dbio.1996.0254. [DOI] [PubMed] [Google Scholar]
- 46.Singh H, Sen R, Baltimore D, Sharp P A. A nuclear factor that binds to a conserved sequence motif in transcriptional control elements of immunoglobulin genes. Nature. 1986;319:154–158. doi: 10.1038/319154a0. [DOI] [PubMed] [Google Scholar]
- 47.Stack J H, Newport J W. Developmentally regulated activation of apoptosis early in Xenopus gastrulation results in cyclin A degradation during interphase of the cell cycle. Development. 1997;124:3182–3195. doi: 10.1242/dev.124.16.3185. [DOI] [PubMed] [Google Scholar]
- 48.Staudt L M, Singh H, Sen R, Wirth T, Sharp P A, Baltimore D. A lymphoid-specific protein binding to the octamer motif of immunoglobulin genes. Nature. 1986;323:640–643. doi: 10.1038/323640a0. [DOI] [PubMed] [Google Scholar]
- 49.Steinbach O C, Wolffe A P, Rupp R A W. Somatic linker histones cause loss of mesodermal competence in Xenopus. Nature. 1997;389:395–399. doi: 10.1038/38755. [DOI] [PubMed] [Google Scholar]
- 50.Strouboulis J, Damjanovski S, Vermaak D, Meric F, Wolffe A P. Transcriptional repression by XPc1, a new polycomb homolog in Xenopus laevis embryos, is independent of histone deacetylase. Mol Cell Biol. 1999;19:3958–3968. doi: 10.1128/mcb.19.6.3958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sun X, Zhang Y, Cho H, Rickert P, Lees E, Lane W, Reinberg D. NAT, a human complex containing Srb polypeptides that functions as a negative regulator of activated transcription. Mol Cell. 1998;2:213–222. doi: 10.1016/s1097-2765(00)80131-8. [DOI] [PubMed] [Google Scholar]
- 52.Tafuri S R, Wolffe A P. Selective recruitment of masked maternal mRNA from messenger ribonucleoprotein particles containing FRGY2 (mRNP4) J Biol Chem. 1993;268:24255–24261. [PubMed] [Google Scholar]
- 53.Timmers H T M. Transcription initiation by RNA polymerase II does not require hydrolysis of the β-Γ phosphoanhydride bond of ATP. EMBO J. 1994;13:391–399. doi: 10.1002/j.1460-2075.1994.tb06273.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Timmers H T M, Sharp P A. The mammalian TFIID protein is present in two functionally distinct complexes. Genes Dev. 1991;5:1946–1956. doi: 10.1101/gad.5.11.1946. [DOI] [PubMed] [Google Scholar]
- 55.Toyoda T, Wolffe A P. Characterization of RNA polymerase II-dependent transcription in Xenopus extracts. Dev Biol. 1992;153:150–157. doi: 10.1016/0012-1606(92)90099-3. [DOI] [PubMed] [Google Scholar]
- 56.Veenstra G J C, Beumer T L, Peterson-Maduro J, Stegeman B I, Karg H A, Van der Vliet P C, Destrée O H J. Dynamic and differential Oct-1 expression during early Xenopus embryogenesis: persistence of Oct-1 protein following down-regulation of the RNA. Mech Dev. 1995;50:103–117. doi: 10.1016/0925-4773(94)00328-k. [DOI] [PubMed] [Google Scholar]
- 56a.Veenstra, G. J. C., and O. H. J. Destree. Unpublished data.
- 57.Veenstra G J C, Peterson-Maduro J, Mathu M T, Van der Vliet P C, Destrée O H J. Non-cell autonomous induction of apoptosis and loss of posterior structures by activation domain-specific interactions of Oct-1 in the Xenopus embryo. Cell Death Differ. 1998;5:774–784. doi: 10.1038/sj.cdd.4400416. [DOI] [PubMed] [Google Scholar]
- 58.Veenstra G J C, Van der Vliet P C, Destrée O H J. POU domain transcription factors in embryonic development. Mol Biol Rep. 1997;24:139–155. doi: 10.1023/a:1006855632268. [DOI] [PubMed] [Google Scholar]
- 58a.Veenstra, G. J. C., and A. P. Wolffe. Unpublished data.
- 59.Verrijzer C P, Alkema M J, Van Weperen W W, Van Leeuwen H C, Strating M J J, Van der Vliet P C. The DNA binding specificity of the bipartite POU domain and its subdomains. EMBO J. 1992;11:4993–5003. doi: 10.1002/j.1460-2075.1992.tb05606.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wormington W M, Brown D D. Onset of 5 S RNA gene regulation during Xenopus embryogenesis. Dev Biol. 1983;99:248–257. doi: 10.1016/0012-1606(83)90273-7. [DOI] [PubMed] [Google Scholar]
- 61.Yasuda G K, Baker J, Schubiger G. Temporal regulation of gene expression in the blastoderm Drosophila embryo. Genes Dev. 1991;5:1800–1812. doi: 10.1101/gad.5.10.1800. [DOI] [PubMed] [Google Scholar]
- 62.Yasuda G K, Schubiger G. Temporal regulation in the early embryo: is MBT too good to be true? Trends Genet. 1992;8:124–127. doi: 10.1016/0168-9525(92)90369-F. [DOI] [PubMed] [Google Scholar]