Abstract
5-Deoxy(iso)flavonoids are structural representatives of phenylpropanoid-derived compounds and play critical roles in plant ecophysiology. Recently, 5-deoxy(iso)flavonoids gained significant interest due to their potential applications as pharmaceuticals, nutraceuticals, and food additives. Given the difficulties in their isolation from native plant sources, engineered biosynthesis of 5-deoxy(iso)flavonoids in a microbial host is a highly promising alternative approach. However, the production of 5-deoxy(iso)flavonoids is hindered by metabolic flux imbalances that result in a product profile predominated by non-reduced analogues. In this study, GmCHS7 (chalcone synthase from Glycine max) and GuCHR (chalcone reductase from Glycyrrhizza uralensis) were preliminarily utilized to improve the CHR ratio (CHR product to total CHS product). The use of this enzyme combination improved the final CHR ratio from 39.7% to 50.3%. For further optimization, a protein-protein interaction strategy was employed, basing on the spatial adhesion of GmCHS7:PDZ and GuCHR:PDZlig. This strategy further increased the ratio towards the CHR-derived product (54.7%), suggesting partial success of redirecting metabolic flux towards the reduced branch. To further increase the total carbon metabolic flux, 15 protein scaffolds were programmed with stoichiometric arrangement of the three sequential catalysts GmCHS7, GuCHR and MsCHI (chalcone isomerase from Medicago sativa), resulting in a 1.4-fold increase in total flavanone production, from 69.4 mg/L to 97.0 mg/L in shake flasks. The protein self-assembly strategy also improved the production and direction of the lineage-specific compounds 7,4′-dihydroxyflavone and daidzein in Escherichia coli. This study presents a significant advancement of 5-deoxy(iso)flavonoid production and provides the foundation for production of value-added 5-deoxy(iso)flavonoids in microbial hosts.
Keywords: Self-assembled enzymes, Protein-protein interaction, Enzyme scaffolds, 5-Deoxyflavonoid, Chalcone reductase
Highlights
-
•
5-Deoxyflavonoids synthetic pathways were constructed and engineered in E. coli.
-
•
CHR ratio was optimized by orthologous enzyme selection, and CHS-CHR interaction.
-
•
Artificial scaffold P4S2G1 was programmed to improve the total metabolic flux.
-
•
Daidzein and genistein were synthesized from L-tyr in E. coli for the first time.
1. Introduction
Flavonoids represent a class of polyphenolic plant secondary metabolites. Compounds belonging to this class are composed of a C6-C3-C6 skeleton and harbour hydroxy groups at characteristic positions of the three ring system (Dooner et al., 1991; Koes et al., 1994). To date, more than ten flavonoid subclasses have been identified in plants including chalcones, dihydrochalcones, aurones, flavanones, flavones, isoflavones, dihydroflavonols, flavonols, coumestans, petrocarpans, anthocyanidins, flavans, etc. (Ververidis et al., 2007; Winkel-Shirley, 2001). Subsequent enzyme-catalyzed modification reactions, such as, hydroxylation, methylation, acylation, glycosylation, and prenylation, decorate the flavan-skeleton, giving rise to more than 9000 known flavonoids (Arita and Suwa, 2008; Williams and Grayer 2004). Despite this tremendous number of flavonoids, compounds belonging to this class can be simply grouped into two groups, hydroxy- and 5-deoxyflavonoids, depending on the presence/absence of the C5-stable hydroxy group. Interestingly, the former group members are widespread in the plant kingdom, ranging from mosses to angiosperms, whereas the members of the latter group are restricted, predominantly to legumes (Koes et al., 1994; Naoumkina et al., 2010).
5-Deoxy(iso)flavonoids (with lead structures liquiritigenin, daidzein and formonoetin) are characteristic metabolites of Leguminosae. Such compounds are typically phytoalexins, naturally acting as an early metabolic defense strategy against pathogen attacks. Members of this class are also function as symbiotic signals toward Rhizobium bacteria involved in nitrogen fixation (Welle et al., 1991; Graham, 1991). In recent years, 5-deoxyflavonoids have been extensively investigated due to their potential applications as pharmaceuticals, nutraceuticals, and food additives for humans (Chouhan et al., 2017; Zhao et al., 2018). For example, liquiritigenin, a 5-deoxyflavanone mainly isolated from the roots of Glycyrrhiza sp., exhibits antitumor activity and a cytoprotective effect against oxidative stress (Xie et al., 2012; Kim et al., 2004). The well-known 5-deoxyisoflavones daidzein and its glycoconjungates, mainly found in soybeans, play an important role in the prevention of breast cancer (Choi and Kim, 2008), osteoporosis (Strong et al., 2014), and postmenopausal syndromes (Chen et al., 2019).
Traditionally, the major source of 5-deoxy(iso)flavonoids is a direct extraction from plant materials. However, this approach suffers from several drawbacks, such as the long maturity periods, unreliability due to the unexcepted seasonal changes, and a huge variation in the flavonoid content from different plant varieties (Tsai et al., 2008; Zhu et al., 2017). Some value-added flavonoids can also be obtained through chemical synthesis, which often requires the use of hazardous chemical catalysts, or extreme reaction conditions (Jha et al., 1981). Besides, there are also critical challenges in the accurate substitute position and chiral synthesis (Lim et al., 2001; Mavel et al., 2006). Engineered microbial cells represent an attractive alternative production platform for flavonoids in terms of the low energy requirements, low waste emissions, environmentally friendly feedstocks, and the potential for large-scale production (Chemler et al., 2006; Katsuyama et al., 2008).
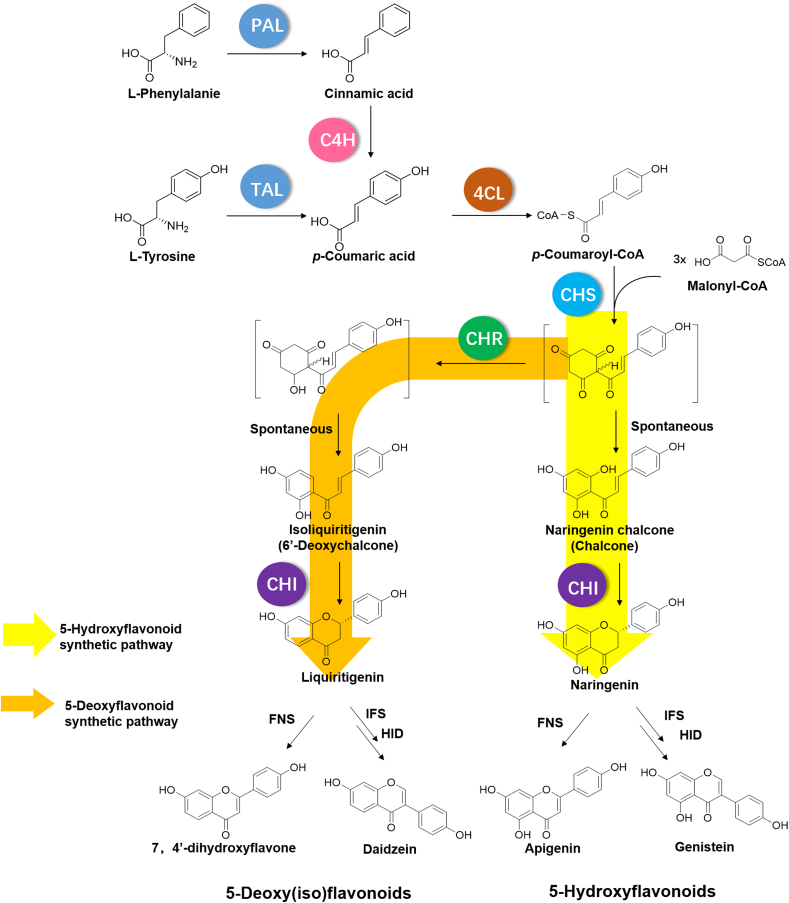
In plants, flavonoids are synthesized via parallel phenylpropanoid routes, starting from either phenylalanine or tyrosine (Weisshaar and Jenkins, 1998; Dixon and Steele, 1999) (Fig. 1). The initial steps are catalyzed by phenylalanine ammonia lyase (PAL)/tyrosine ammonia lyase (TAL) to form the phenylpropanoids cinnamic acid or p-coumaric acid, respectively. Cinnamate 4-hydroxylase (C4H) is required for the hydroxylation of cinnamic acid to yield p-coumaric acid, which is subsequently converted to its CoA thioester by the activity of 4-coumaryl:CoA ligase (4CL). Subsequently, chalcone synthase (CHS, a polyketide synthase), catalyzes the condensation of p-coumaryl-CoA as the starter unit and three molecules of the extension unit to form 4,2′,4′,6′-tetrahydroxychalcone (naringenin chalcone), which is then converted to naringenin in the presence of chalcone isomerase (CHI). Naringenin serves as the common precursor to generate either 5-hydroxyflavone by the activity of flavone synthase (FNS), or to form 5-hydroxyisoflavone by the catalytic activity of isoflavone synthase (IFS) and 2-hydroxyisoflavanone dehydratase (HID). Interestingly, in legumes, an additional polyketide reductase (chalcone reductase, CHR) is involved in the condensation step that leads to formation of 4,2′,4′-trihydroxychalcone (isoliquiritigenin), the common precursor of 5-deoxy(iso)flavonoids (Fig. 1).
Fig. 1.
Overview of the flavonoid biosynthesis pathway in plants. PAL: phenylalanine ammonia lyase; TAL: tyrosine ammonia lyase; C4H: cinnamate 4-hydroxylase; 4CL: 4-coumarate:CoA ligase; CHS: chalcone synthase; CHR: chalcone reductase; CHI: chalcone isomerase; FNS: flavone synthase; IFS: 2-hydroxyisoflavone synthase; HID: 2-hydroxyisoflavanone dehydratase. The yellow arrow represents the predominant 5-hydroxyflavonoid pathway, and the orange arrow represents the 5-deoxy(iso)flavonoid synthetic pathway. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Thanks to the full elucidation of the flavonoid biosynthetic pathway and the rapid development in the fields of genetic and metabolic engineering, significant progress has been made in the heterologous synthesis of flavonoids in engineered hosts (Fowler and Koffas, 2009; Leonard et al., 2007, 2008; Lyu et al., 2017; Wu et al., 2014a, 2014b). Complete flavonoid biosynthesis pathways, e.g. for the production of naringenin, pinocembrin, apigenin, quercetin, kaempferol, scutellarin, and baicalein have been reconstructed in microbial host systems in last decades (Cao et al., 2016; Jones et al., 2015, 2017; Li et al., 2019; Liu et al., 2018, 2020; Miyahisa et al., 2006). Various strategies have also been successfully employed to improve the engineered production of flavonoids, and several flavonoids have reached industrially relevant titers (Leonard et al., 2008; Leonard and Koffas, 2007; Wu et al., 2014b). In constrast to these successes, to date, the heterologous synthesis of 5-deoxy(iso)flavonoids in microbial hosts has been only sporadically investigated (Yan et al., 2007; Rodriguez et al., 2017; Stahlhut et al., 2015). An engineered E. coli strain expressing genes coding for Pc4CL2 (4CL-2 from Petroselinum crispum), MsCHR (CHR from M. sativa), PhCHS (CHS from P. crispum), and MsCHI (CHI from M. sativa) led to the production of liquiritigenin starting from p-coumaric acid (Yan et al., 2007). However, the titer of liquiritigenin was only one quarter of that of naringenin, indicating an imbalanced metabolic flux ratio of favoring production of the 5-hydroxyflavonoid over the 5-deoxyflavonoid. Currently, this challenge of ratios of competing products makes available engineering approaches unsuitable for the large-scale production of 5-deoxyflavonoids.
In the present work, we set efforts to redirect flavanoid metabolic flux into the reduced branch for the production of 5-deoxyflavonoids. A pair of preferred enzymes GmCHS7/GuCHR were selected from five plant species, and subsequently used for the reconstitution of liquiritigenin synthetic pathway in E. coli. By introducing a protein-peptide interaction system PDZ-PDZlig to gather GmCHS7 and GuCHR, the ratio of liquiritigenin in final product was promoted. Stoichiometric arrangement of GmCHS7, GuCHR, and MsCHI was designed by protein assembly scaffolds, but failed to further improve the CHR ratio. What's more, the protein self-assembly strategy was also used in de novo synthesis of linear 5-deoxyflavone and 5-deoxyisoflavone metabolites 7,4′-dihydroxyflavone, daidzein in E. coli.
2. Materials and methods
2.1. Strains, plasmids and chemicals
Escherichia coli DH10B was used for plasmid propagation and all plasmid constructions. E. coli BL21(DE3) was selectively used for the expression of heterologous genes and production of flavonoids. The compatible vectors pCDFDuet-1, and pET28a (Novagen, Germany) were used for the expression of multiple genes. The vector pET21c (Novagen, Germany) was used for protein scaffold assembly. All restriction enzymes and T4 ligases were purchased from NEB. All constructs were either amplified with Primerstar Max DNA Polymerase (Takara Bio Inc., Beijing) or the ClonExpress II One Step Cloning Kit (Vazyme Biotech Co., Ltd, Nanjing). Authentic chemical standards of naringenin chalcone, naringenin, 7,4′-dihydroflavone and apigenin were purchased from Yuanye Bio-Technology Co., Ltd (Shanghai). Isoliquiritigenin, liquiritigenin, daidzein, and genistein were purchased from Meilun Biotechnology Co., Ltd (Dalian).
2.2. cDNA isolation and gene cloning
RNAs were isolated from leaves of Glycine max, roots of Glycyrrhiza uralensis, and Arabidopsis thaliana. Full length cDNAs were obtained from total RNA by reverse transcription and PCR using PrimerStar Max mix (Takara). Primers were designed based on the sequences with Genebank accession numbers. GuCHS (ARH02397), and GuCHR (D86559) were amplified from G. uralensis, GmCHS7 (NP_001340309), GmCHR5 (LC309095), GmIFS (AAF45142), and GmHID (AB154415) were isolated from G. max, and AtCHS (NP_196897) was obtained from A. thaliana. CrCPR(X69791, cytochrome P450 reductase from Catharanthus roseus) was synthesized by GenScript (Nanjing) with codon optimization for E. coli. GuCHS and GuCHR were cloned into the pET21c vector between the NdeI/BamHI sites. GmCHS7, GmCHR5, and AtCHS were cloned into pET21c between the NdeI/XhoI sites. GmIFS, CcCPR and GmHID were cloned into the pRSFDuet-1 vector between the NcoI/XhoI sites. All primers are listed in Table S1.
2.3. Genetic reconstruction of the 5-deoxyflavonoid pathway
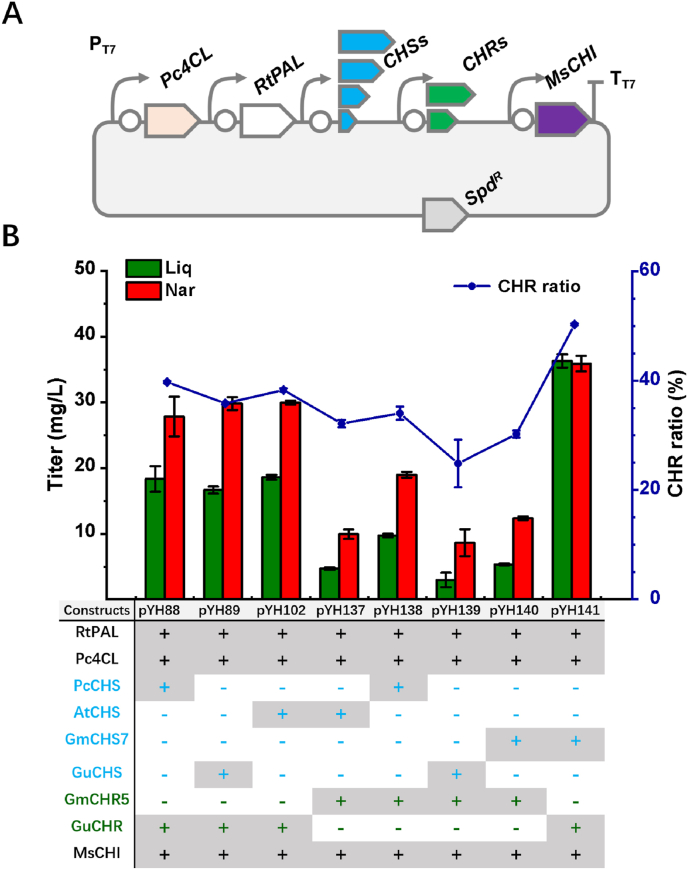
The plasmids pYH51 and pYH55 were used as the original plasmids for assembly of the 5-deoxyflavonoid pathway(Li et al., 2019). The plasmid pYH51 was first linearized with restriction endonucleases BamHI and EcoRI. The DNA fragment BamHI-T7-RBS-GuCHR-EcoRI, containing the GuCHR coding sequence, T7 promoter region and RBS (ribosome binding site), was amplified with primers BamHI-T7GuCHR-F and EcoRI-GuCHR-R using pET21c-GuCHR as a template. The obtained fragment BamHI-T7-RBS-GuCHR-EcoRI was then inserted into the linearized pYH51 to generate pYH70. Meanwhile, the DNA fragment HindIII-T7-RBS-GuCHR-NotI was amplified with primers HindIII-GuCHR-F and NotI-GuCHR-R, and inserted into the HindIII/NotI digested pYH55 to result in pYH88. In a similar way, the T7-RBS-GmCHR5 sequence was inserted into the linearized pYH55 to form pYH137. The DNA fragment T7-RBS-PcCHS of pYH88 was replaced by T7-RBS-GuCHS, T7-RBS-AtCHS, and T7-RBS-GmCHS7 to give pYH89, pYH102, and pYH141, respectively. The DNA fragment T7-RBS-PcCHS of pYH137 was replaced by the aforementioned sequences to yield pYH138, pYH139, and pYH140.
For protein engineering of GmCHS7 and GuCHR, the fusion protein GmCHS7:GuCHR was achieved by overlap extension PCR (Hilgarth and Lanigan, 2020). The fragment coding for the fusion protein GmCHS7-(GGGGS)2-GuCHR was obtained by two rounds of PCR. The first round was performed by using NotI-GmCHS-F/GmCHS7-GuCHR-R and GmCHS7-GuCHR-F/XhoI-GuCHR-R as primers, and the second round by using NotI-GmCHS-F/XhoI-GuCHR-R. The final fragment was inserted into the NotI/XhoI digested pYH55 to generate pYH152.
For construction of the two-protein complex of GmCHS7 and GuCHR, genes encoding for PDZ and PDZlig were cloned in a way that the proteins are fused to the C terminal of GuCHR and GmCHS7, respectively, via (GGGGS)2 as a (Table S2, S3). The obtained PCR products GmCHS7:PDZ and GuCHR:PDZlig were successively assembled into the pCDF-Duet-1 harboring Pc4CL (Petroselium crispum 4CL, KF765780), RtPAL (Rhodotorula toruloides, AAA33883), and MsCHI (Medicago sativa, KF765782) between HindIII/NotI/XhoI. The plasmid pYH153 was finally obtained. Another plasmid pYH149, which was contransformed with synthetic scaffolds to recuit GmCHS7, GuCHR and MsCHI, was also generated in the same way. Three separated protein ligands, PDZlig, SH3lig, and GBDlig were infused at the C terminus of the aforementioned individual pathway enzymes, respectively. All constructed plasmids were further verified by colony PCR and Sanger sequencing (Sangon Biotech).
2.4. Construction of synthetic scaffolds
Scaffolds were built by tethering multiple protein-protein interaction domains, including PDZ (α-synthrophin domain PSD95/DlgA/Zo-1 from Mus musculus), SH3 Crk domain from M. musculus), GBD (the GTPase binding domain from N-WASP from Rattus rattus) (Table S2), via fifteen-amino acid (GGGGS)3 linkers using the BioBrick strategy (Shetty et al., 2008) (Table S2, S3). The PDZ fragment was initially introduced into pET21c within the NdeI and SpeI/XhoI restriction enzyme sites. The fragment consisting of a DNA sequence encoding the N-terminal linker (GGGGS)3, and the interaction domain, was cloned into pEASY leaving it with 5′ flanking XbaI and 3′ SpeI/XhoI restriction enzyme sites. These basic parts were combined in desired arrangements by digesting the ultimate upstream part with SpeI and XhoI and the downstream part with XbaI and XhoI, followed by ligation. The cohesive ends created by SpeI and XbaI are compatible for ligation and destroy both of these recognition sites after ligation. Thus, the resulting composite part maintains the original unique restriction enzyme pattern for further addition of other parts. In this manner, synthetic scaffolds were constructed by iterative addition of parts.
2.5. Bacterial media and cultivation conditions
Lysogeny broth (LB) (10 g/L tryptone, 5 g/L yeast extract, and 10 g/L NaCl), Terrific broth (TB)(12 g/L tryptone, 24 g/L yeast extract, 5 g/L glycerol, 17 mmol KH2PO4, 72 mmol K2HPO4), M9 minimal medium (11.3 g/L M9 salts, 5 g/L D-glucose, 2 μM MgSO4, 0.1 μM CaCl2) supplemented with 1000 × trace metal mix (0.03 g/L H3BO3, 1 g/L Thiamine, 0.94 g/L ZnCl2, 0.5 g/L CoCl2, 0.38 g/L CuCl2, 1.6 g/L MnCl2, 3.6 g/L FeCl2), and MOPS minimal medium (Neidhardt et al., 1974) supplemented with 5 g/L glucose and an additional 4 g/L NH4Cl were used for bacterial fermentation. E. coli cells used for gene cloning, plasmid propagation, and inoculation preparations were cultured at 37 °C in LB medium supplemented with appropriate antibiotics. The working concentrations of antibiotics were kanamycin (50 μg/mL), chloramphenicol (34 μg/mL), ampicillin (50 μg/mL), and streptomycin (40 μg/mL). Various combinations of antibiotics were added into cultures of E. coli strains harbouring different plasmids for production of individual flavonoids.
For the production of flavonoids in recombinant E. coli strains, 100 μL of an overnight seed culture was inoculated into 10 mL of LB, TB, M9, and MOPS. After the OD600 reached 0.4–0.5, 0.3 mM isopropyl-β-D-thiogalactoside (IPTG), and 0.5 g/L of L-tyrosine (final concentration) were added to the cultures. 0.3 mM p-coumaric acid was added when producer did not express PAL. All fermentation was performed at 22 °C for 48 h. At the end of fermentation, 1 mL culture aliquot was disrupted by sonication and extracted twice with an equal volume of ethyl acetate. After centrifugation, the supernatant was evaporated to dryness and dissolved in 200 μL methanol for HPLC analysis.
2.6. Analysis of flavonoids
20 μL of each sample was analyzed by high-performance liquid chromatography (HPLC) on an Ultimate 3000 HPLC system (ThermoFisher Scientific) equipped with a photodiode-array (PDA) detector. Naringenin and liquiritigenin were measured at 280 nm using a Thermo syncronis C18 column (250 mm × 4.6 mm; particle size, 5 μm) maintained at 30 °C. Naringenin chalcone, isoliquiritigenin, 7,4′-dihydroflavone, and apigenin were measured at 340 nm, daidzein and genistein were measured at 260 nm. Obtained products were separated by acetonitrile (solvent A) and water (containing 0.1% formic acid, solvent B) at a flow rate of 1.0 mL/min under the following conditions: 20–55% solvent A for 20 min, 55–100% A for 10 min, 100% A for 5 min, 100-20% A for 2 min and 20% A for 4 min. For the analysis of daidzein and genistein, the conditions were slightly modified: 10–30% solvent A for 20 min, 35–50% A for 10 min, 50–100% A for 2 min, 100% A for 5 min, 100-10% A for 2 min and 10% A for 4 min. Identification of the target compounds was achieved via matching their retention time and their UV spectrum with authentic standards. The compounds were quantified based on calibration curves of various concentrations of standards using peak area. The data shown in this study were generated from three independent experiments.
The obtained products were also analyzed by Ultra performance liquid chromatography (UPLC)-electrospray ionization (ESI)-high-resolution (HR) mass spectrometry (MS). Data were acquired using a Q Exactive hybrid quadrupole Orbitrap mass spectrometer (Thermo Scientific) equipped with an Acquity UPLC BEH C18 column (2.1 × 50 mm, 1.7 μm). The mobile phase consisted of methanol (0.1% formic acid, solvent A) and H2O (solvent B). A linear gradient was set as follows: 0.0–10.0 min, 5%–100% A; 10.0–12.5 min, 100% A; 12.5–15.0, re-equilibrate to the initial condition. The flow was 0.25 mL/min. The mass acquisition was performed in negative and positive ionization mode over a range of 100–1000 m/z.
3. Results
3.1. Reconstruction of the liquiritigenin biosynthesis pathway in E. coli
Yan et al.,(2007) reported that the titer of liquiritigenin was only 1/4 of that of naringenin when gene coding for Pc4CL, MsCHR isolated from Medicago sativa, PhCHS and MsCHI were expressed in an engineered E. coli strain. To construct a more efficient pathway for liquiritigenin production, we first focused on searching for a preferred CHR enzyme with high substrate affinity or high turnover. Liquiritigenin represents a characteristic 5-deoxyflavanone and substantially accumulates in Glycyrrhiza sp. We speculated that a substrate-specific CHR with high activity, and involved in the synthesis of liquiritigenin, exists in this species. Thus, we used MsCHR for G. uralensis whole-genome searching (https://www.ncbi.nlm.nih.gov/bioproject/350202). We obtained only one CHR sequence hit (named GuCHR), which showed high amino acid similarity (99% identity) with a reported polyketide reductase (GGPKR2, BAA13114) from G. glabra (Hayashi et al., 1996).
Subsequently, the gene encoding GuCHR was amplified from G. uralensis cDNA and ligated into the plasmid pYH51 (Li et al., 2019), already containing Pc4CL, PhCHS, and MsCHI, to generate pYH70. E. coli BL21(DE3) harboring pYH70 was used for liquiritigenin production. HPLC analysis showed that two new products were observed in all culture media (LB, TB, MOPS, M9) with the addition of 0.3 mM p-coumaric acid (Fig. 2A and C). A product was eluted at 11.78 min, which had a retention time identical to that of authentic liquiritigenin (Fig. 2C). The identity of the product was further confirmed by a high resolution mass spectrum with a m/z value of 257.0800 [M+H]+ (Fig. 2D, S1C), consistent with the authentic liquiritigenin standard (Fig. S1A). An accompanying product, eluting at 13.75 min (Fig. 2C), also accumulated alongside liquiritigenin and was identified as the unreduced product narigenin by HPLC-MS analysis (Fig. S1B, S1D). The identity of the products were also confirmed by comparing their UV absorption spectra, and mass spectrum with the authentic standard (Fig. S2).
Fig. 2.
Reconstruction of the liquiritigenin pathway in E. coli. (A) The engineered strain E. coli BL21(DE3) carrying pYH70 (pCDF-Pc4CL-PcCHS-GuCHR-MsCHI) was cultivated with 0.3 mM p-coumaric acid after 48 h fermentation in LB, TB, MOPS, and M9 medium. The green and red bars indicate the yield of liquiritigenin (Liq) and naringenin (Nar), respectively. The blue lines indicate the CHR ratio (the yield of CHR product to total CHS product) in the final cultures. Error bars indicate the standard deviations of three replicates. (B) The engineered strain E. coli BL21(DE3) carrying pYH88 (pCDF-Pc4CL-RtPAL-PcCHS-GuCHR-MsCHI), was cultivated with 0.5 g/L L-tyrosine (final concentration), after 48 h fermentation in LB, TB, MOPS and M9 medium. (C) HPLC analysis of the fermentation product with E. coli BL21 (DE3) harbouring pYH70 in different media. The samples were detected at 280 nm. (D) The MS spectrum of synthetic liquiritigenin with E. coli BL21 (DE3) harbouring pYH70 in M9 medium. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
To reduce the cost of the fermentation, we further explored L-tyrosine as the precursor for liquiritigenin production. To achieve this goal, RtPAL was introduced into the aforementioned vector pYH70 to generate a final construct pYH88. The strain E. coli BL21(DE3) harboring pYH88 produced a maximum level of 21.2 mg/L liquiritigenin and 23.5 mg/L naringenin in M9 medium after incubation for 2 days with supplementation of 0.5 g/L tyrosine (Fig. 2B, S3). The final product gave the highest CHR ratio of 47.4% (Fig. 2B). Based on these preliminary results, M9 medium was employed for further fermentation processes.
3.2. Optimize synthetic pathway via orthologous CHSs and CHRs selection
Although the introduction of GuCHR successfully led to the production of liquiritigenin, the competing pathway leading to naringenin was still predominant in the recombinant host. However, compared to previous work (Yan et al., 2007), our utilization of GuCHR herein instead of MsCHR highly improved the CHR ratio. This result indicated that targeting the branch point of reduced and non-reduced direction through bioprospecting of CHR from various plant sources is an important strategy for fine-tuning 5-deoxyflavonoid biosynthesis. CHS is another rate-limiting enzyme that mediates the condensation of phenylpropanoic acid-CoA and malonyl-CoA to form the respective chalcone, hence, the adaptability of CHS and CHR would affect reduced and unreduced flavonoid distribution.
Based on this speculation, our next metabolic engineering strategy focused on the selection of CHS orthologous enzymes in addition to CHRs from individual plant species. Thus, except for PcCHS and GuCHR, three additional CHS genes, AtCHS from A. thaliana, GmCHS7 from G. max, GuCHS from G. uralensis, alongside GmCHR5 were carefully chosen and orthogonally used for the reduced branch pathway optimization. Constructs pYH89, pYH102 and pYH141 were assembled with GuCHR, alternatively combining it with the aforementioned CHSs, and RtPAL, Pc4CL, MsCHI to yield different combinations (Fig. 3). The results showed that the constructs pYH88, pYH89 and pYH102 exhibited nearly equal ability to synthesize liquiritigenin and naringenin (Fig. 3). However, pYH141 led to a significant increase in the titers of both liquiritigenin (36.3 mg/L) and naringenin (35.9 mg/L), and in addition, exhibited an improved CHR ratio of 50.3%. The other combinations including the replacement of GuCHR with GmCHR5 (resulting in the four constructs pYH137-pYH140), gave extremely low titers of liquiritigenin (range from 3.0 to 9.8 mg/L), indicating the weak catalytic activity of GmCHR5 (Fig. 3).
Fig. 3.
Orthologous screening of CHSs and CHRs. (A) Schematic diagrams of constructions with four CHSs and two CHRs from four plant species; (B) The engineering 5-deoxyflavanone producers, carrying orthologous CHSs and CHRs, were cultivated with 0.5 g/L L-tyrosine (final concentration). The samples were detected at 280 nm. Error bars indicate the standard deviations of three replicates.
3.3. Effects of CHS and CHR protein complexes
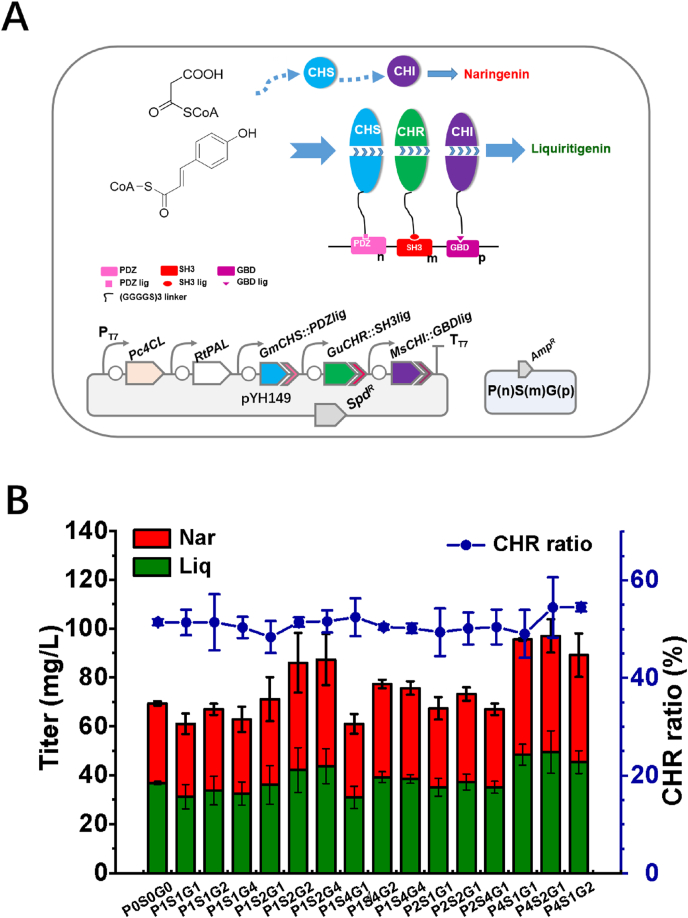
It is well known that the shorter the spatial distance between CHS and CHR, the shorter the transient time and distance of the unstable intermediate, and the higher the synthetic efficiency. To achieve this goal, two strategies were adopted in this study. Firstly, translational fusion of multiple enzymes in successive steps is an efficient method for improving enzyme performance (Yan et al., 2008). Hence, a fusion protein GmCHS7:GuCHR was constructed with a flexible linker (GGGGS)2 between the two proteins that is encoded by the final plasmid pYH152 (Fig. 4A). The second strategy involved the construction of protein complexes using engineered interactions between well-characterized interaction domains and their specific ligands. This strategy has been used to improve the catalytic efficiency of enzymes in vivo/in vitro (Dueber et al., 2009; Kang et al., 2019; Yang et al., 2017). A pair of protein-peptide interaction domains (PDZ, PSD95/Dlg1/zo-1) and its ligand (PDZ ligand) from metazoan cells (Dueber et al., 2009) was selected for the self-assembly of GuCHR and GmCHS7. A short peptide (GGGGS)2 was used to link target GuCHR to the PDZ domain yielding GuCHR:PDZ and the same linker (GGGGS)2 was used to connect GmCHS7 and the PDZ ligand yielding GmCHS7:PDZlig. Through the interaction of the protein-peptide (PDZ-PDZlig), GuCHR:PDZ and GmCHS7:PDZlig were designed to self-assemble and form a double enzyme complex that enables the substrate to be directly transferred from GmCHS7 to GuCHR. The GuCHR:PDZ and GmCHS7:PDZlig, together with Pc4CL, RtPAL, and MsCHI were assembled into pCDFDuet-1 yielding the plasmid pYH153 (Fig. 4A).
Fig. 4.
Enzymatic engineering of CHS and CHR by protein self-assembly to improve liquiritigenin production. (A) Schematic diagrams of plasmids pYH141, pYH152, and pYH153; (B) Protein-protein interaction between GmCHS and GuCHR, assayed by BiFC in N. benthamiana by co-expression of translational fusions with N-terminal (NeGFP) or C-terminal (CeGFP) fragments of eGFP. (a, b) GuCHR:PDZ interacts weakly with GmCHS7:PDZlig; (c,d) There is no significant interaction between GuCHR and GmCHS7. (C) The engineered 5-deoxyflavanone producer cultivated with 0.5 g/L L-tyrosine (final concentration) (D) HPLC chromatogram of the obtained products. The samples were detected at 280 nm.
In advance, to assess whether GuCHR:PDZ and GmCHS7:PDZlig were successfully assembled, interaction between these two proteins was assayed by bimolecular fluorescence complementation (BiFC). GuCHR:PDZ and GmCHS7:PDZlig were fused with the N-terminus (1–155) and C-terminus (156–273) of an eGFP (enhanced green fluorescent protein) fragment, yielding GuCHR:PDZ-NeGFP and GmCHS7:PDZlig-CeGFP, respectively. As shown in Fig.4B and Fig. S4, the co-expression of GuCHR:PDZ-NeGFP and GmCHS7:PDZlig-CeGFP in N. benthamiana led to a visible green fluorescence, suggesting the direct interaction between GuCHR:PDZ and GmCHS7:PDZlig. In contrast, no interaction was observed between GmCHS7 and GuCHR without PDZ-PDZlig. The aforementioned results indicated the successful association of GuCHR and GmCHS7 in the presence of PDZ and its ligand.
Compared with the production in E. coli BL21(DE3)-pYH141, the yield of liquiritigenin and naringenin were reduced to 21.6 mg/L and 20.4 mg/L, respectively, in E. coli BL21(DE3)-pYH152, in which GmCHS7 and GuCHR were attached via a fusion protein. On the contrary, their yields were dramatically boosted to 44.5 mg/L, and 31.8 mg/L, respectively, in E. coli BL21(DE3)-pYH153, in which GmCHS7 and GuCHR were connected via protein-peptide interaction. The CHR ratio was improved to be 54.7%, suggesting a successful re-direction of the carbon metabolic flux to the 5-deoxyflavanone branch to a certain extent.
3.4. Enzyme stoichiometric arrangement by protein assembly scaffolds
The designed double protein complex of GmCHS7 and GuCHR not only improved the metabolic flux, but also diverted the flavonoid pathway towards the reduced branch. However, the GmCHS7:PDZ and GuCHR:PDZlig only provided a rigid 1:1 ratio of individual enzymes. In order to explore the effect of modulating enzyme stoichiometry in the synthetic complex of more enzymes, tethered scaffolds, in various arrangements, were constructed to optimize the pathway. Three pairs of protein-protein interaction domains PDZ, SH3, GBD (Table S2) and their ligands (PDZ ligand, SH3 ligand, and GBD ligand) were applied to design a regulation machinery (Table S3). The separated PDZ ligand, SH3 ligand, and GBD ligand were correspondingly fused to the C-termini of GmCHS7, GuCHR, and MsCHI via a flexible linker (GGGGS)2, respectively (Fig. 5A). Finally, the plasmid pYH149 (pCDF-Pc4CL-RtPAL-GmCHS7:PDZlig-GuCHR:SH3lig-MsCHI:GBDlig) was constructed.
Fig. 5.
Engineered 5-deoxyflavonoid metabolon protein scaffolds to boost the yield of Liquiritigenin/Naringenin. (A) Schematic diagram showing the architectured protein scaffolds for Liq and Nar production in E. coli. (B) The engineering 5-deoxyflavanone producers harboring pYH149 and individual protein scaffolds. Production titers obtained with supplement of 0.5 g/L L-tyrosine.
A matrix of scaffolds was test with the architecture (N-terminus to C-terminus) of PDZ(n)SH3(m)GBD(p). The binding domains PDZ, SH3, and GBD were linked together by a fifteen amino acid peptide (GGGGS)3 and were used to target GmCHS7, GuCHR, and MsCHI, respectively. Fifteen designed scaffolds were generated and E. coli BL21(DE3) was individually co-transformed with pYH149 (Table S4) into BL21(DE3). Strain BL21(DE3) harboring pYH149 and pET21c (P0S0G0) was set as a control. Compared to the control, the scaffold P1S1G1 slightly reduced the titer of total flavanones, suggesting a possible metabolic burden as a consequence of scaffold proteins overexpression under a strong promoter. Production improvements depended on the number of single domain repeats in the scaffolds (Fig. 5B). For example, the scaffolds P1S1G1 and P1S2G1 that differ by one SH3 domain resulted in production of different flavonoid titers of 60.0 mg/L and 71.1 mg/L, respectively. However, the addition of two repeated SH3 domains in P1S4G1 reduced the flavanone production to 61.0 mg/L. In the same way, scaffolds P1S1G2 led to a higher titer than P1S1G1, indicating the positive effect of the GBD domain, whereas, the P1S1G4 slightly reduced the final titer. Details of the mechanisms through which scaffolding increases metabolic efficiency are still not known. Curiously, the PDZ domain played the most important role in flavonoid production in view of 60.0 mg/L from P1S1G1, 67.4 mg/L from P2S1G1, and 95.6 mg/L from P4S1G1. As expected, scaffold P4S2G1, with a 4:2:1 ratio of PDZ domain: SH3 domain: GBD domain, improved the flavanone titer 1.4-fold compared to the last strain E. coli BL21(DE3) harbouring pYH149, showing a maximum production level of 97.0 mg/L (49.5 mg/L of liquiritigenin and 47.5 mg/L of naringenin). However, none of the scaffolds significantly increased the CHR ratio.
3.5. Production of 7,4′-dihydroxyflavone and apigenin
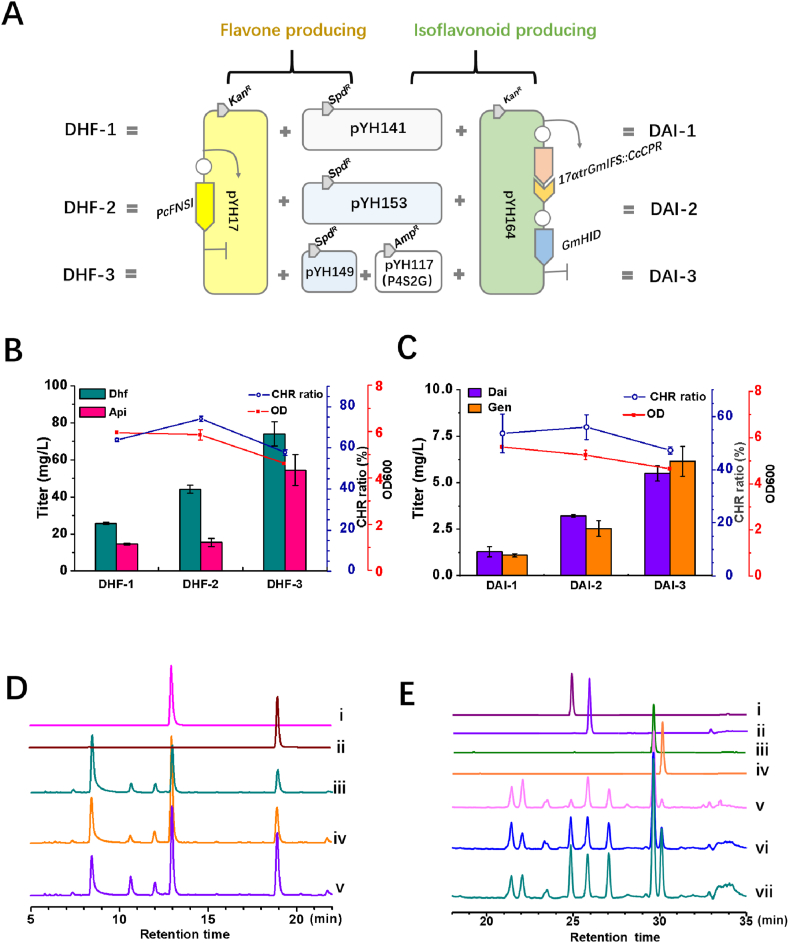
To further determine whether the designed protein scaffolds can improve 5-deoxyflavone production, FNSI, catalyzing the enzymatic step from flavanone to flavone, was introduced into the liquiritigenin/narigenin producer. The liquiritigenin producing strains were further engineered for the production of 7,4′-dihydroxyflavone by introducing pYH17, harboring a PcFNSI gene. The E. coli BL21(DE3) harboring pYH141 and pYH17 was named DHF-1, E. coli BL21(DE3) harboring pYH153 and pYH17 was named DHF-2, and a strain carrying pYH149, pYH117 and pYH17 was named DHF-3 (all strains are listed in Table 1). After 2 days of incubation, the cultures were analyzed by HPLC-UV and HPLC-ESI-MS. The product eluting at 12.98 min was identified to be 7,4′-dihydroxyflavone. The retention time and UV spectra were identical to those of authentic standard (Fig. 6A, Fig. S6). The identity of the product was further confirmed by high resolution mass spectrometry with an m/z value of 253.0511 [M-H]-, which was consistent with the authentic standard (Fig. S5A, S5C). Another product, eluting at 19.04 min, was identified to be apigenin by LC-MS analysis, UV absorption spectra and high-resolution MS compared with authentic apigenin (Fig. S5B, S5D, S6B, S6D). The titer of 7,4′-dihydroxyflavone and apigenin was boosted 1.7-fold, and 1.1-fold by direct interaction of GmCHS7 and GuCHR in DHF-2, compared to the strain DHF-1. The scaffold engineering of GmCHS7, GuCHR and MsCHI in DHF-3 dramatically increased the production of 7,4′-dihydroxyflavone to 74.3 mg/L and that of apigenin to 52.1 mg/L. However, the R ratio was not obviously improved by assembly of GmCHS7, GuCHR, and MsCHI through scaffold protein engineering.
Table 1.
Strains and plasmids used in the present work.
| Plasmids | Description | Source or reference | |
|---|---|---|---|
| pET21c | T7 promoter, pBR322 ori, AmpR | Novagen | |
| pRSFDuet-1 | Double T7 promoters, pBR322 ori, KanR | Novagen | |
| pCDFDuet-1 | Double T7 promoters, CloDF13 ori, SmR | Novagen | |
| pYH51 | pCDF-4CL-PcCHS-CHI | pCDF-Duet-1 carrying 4CL, PcCHS, and CHI | Li et al. (2019) |
| pYH55 | pCDF-4CL-PAL-PcCHS-CHI | pCDF-Duet-1 carrying PAL, 4CL, PcCHS, and CHI | Li et al. (2019) |
| pYH70 | pCDF-4CL-PcCHS-GuCHR-CHI | pCDF-Duet-1 carrying 4CL, GuCHR, PcCHS, and CHI | This study |
| pYH88 | pCDF-4CL-PAL-PcCHS-GuCHR-CHI | pCDF-Duet-1 carrying 4CL, PAL, PcCHS, GuCHR, and CHI | This study |
| pYH89 | pCDF-4CL-PAL-GuCHS-GuCHR-CHI | pCDF-Duet-1carrying 4CL, PAL, GuCHS, GuCHR, and CHI | This study |
| pYH102 | pCDF-4CL-PAL-AtCHS-GuCHR-CHI | pCDF-Duet-1 carrying 4CL, PAL, AtCHS, GuCHR, and CHI | This study |
| pYH137 | pCDF-4CL-PAL-PcCHS-GmCHR5-CHI | pCDF-Duet-1 carrying 4CL, PAL, AtCHS, GmCHR5, and CHI | This study |
| pYH138 | pCDF-4CL-PAL-GuCHS-GmCHR5-CHI | pCDF-Duet-1 carrying 4CL, PAL, GuCHS, GmCHR5, and CHI | This study |
| pYH139 | pCDF-4CL-PAL-AtCHS-GmCHR5-CHI | pCDF-Duet-1 carrying 4CL, PAL, AtCHS, GmCHR, and CHI | This study |
| pYH140 | pCDF-4CL-PAL-GmCHS7-GmCHR5-CHI | pCDF-Duet-1 carrying 4CL, PAL, GmCHS7, GmCHR5, and CHI | This study |
| pYH141 | pCDF-4CL-PAL-GmCHS7-GuCHR-CHI | pCDF-Duet-1 carrying 4CL, PAL, GmCHS, GuCHR, and CHI | This study |
| pYH149 | pCDF-4CL-PAL-GmCHS7::PDZlig-GuCHR::SH3lig-CHI::GBDlig | pCDF-Duet-1 carrying 4CL, PAL, GmCHS7::PDZlig, GuCHR::SH3lig, and CHI::GBDlig | This study |
| pYH152 | pCDF-4CL-PAL-GmCHS7::GuCHR-CHI | pCDF-Duet-1 carrying 4CL, PAL, GmCHS7::GuCHR, and CHI | This study |
| pYH153 | pCDF-4CL-PAL-GmCHS7::PDZlig-GuCHR::PDZ-CHI | pCDF-Duet-1 carrying 4CL, PAL, GmCHS7::PDZlig, GuCHR::PDZ, and CHI | This study |
| pYH17 | pET28a-FNS I | pET28a carrying FNS I | Li et al. (2019) |
| pYH164 | pRSF-17αtrGmIFS::CrCPR-GmHID | pRSFDuet-1 carrying 17αtrGmIFS::CrCPR, and GmHID | This study |
| Strains | |||
| E. coli BL21(DE3) | |||
| DHF-1 | E. coli BL21(DE3) harboring pYH141 and pYH17 | This study | |
| DHF-2 | E. coli BL21(DE3) harboring pYH153 and pYH17 | This study | |
| DHF-3 | E. coli BL21(DE3) harboring pYH149, pYH117, and pYH17 | This study | |
| DAI-1 | E. coli BL21(DE3) harboring pYH141, and pYH164 | This study | |
| DAI-2 | E. coli BL21(DE3) harboring pYH153, and pYH164 | This study | |
| DAI-3 | E. coli BL21(DE3) harboring pYH149, pYH117 and pYH164 | This study | |
Fig. 6.
Production of a flavone and an isoflavone in E. coli via protein self-assembly engineering. (A) Overviews of diagrammatic sketch of engineered stains for flavone and isoflavonoid production. (B) The engineered 5-deoxyflavone producer cultivated with 0.5 g/L L-tyrosine (final concentration). The cyan and pink bars indicate the yield of Dhf and Api, respectively. The blue line indicates the CHR ratio (the yield of 7,4′-dihydroxyflavone to total flavone) in the final cultures; the red line indicates the biomass at 600 nm. (C) The engineered 5-deoxyisoflavonoid producer cultivated with 0.5 g/L L-tyrosine (final concentration). The blue line indicates the CHR ratio (the yield of daidzein to total isoflavone) in the final cultures; the red line indicates the biomass at 600 nm. (D) HPLC analysis of the fermentation products with the flavone producer. i, Dhf (7,4′-dihydroxyflavone); ii, Api (apigenin); iii, product obtained with DHF-1; iv product obtained with DHF-2; v, product obtained with DHF-3. The samples were detected at 340 nm; (E) HPLC analysis of the fermentation products with the isoflavonoid producer. i, Dai (daidzein); ii, Liq; iii, Nar; iv, Gen (genistein); v, product obtained with DAI-1; vi, product obtained with DAI-2; vii, product obtained with DAI-3. The samples were detected at 260 nm. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
3.6. Production of daidzein and genistein
To validate our strategy for the improvement of 5-deoxyisoflavone production, an isoflavonoid synthase (IFS), a cytochrome P450 reductase, together with a 2-hydroxyisoflavanone dehydratase (HID) were introduced into the liquiritigenin producer. To achieve this, we chose GmIFS, GmHID from G. max, and CrCPR from Catharanthus roseus based on previous works (Akashi et al., 2005; Kim et al., 2009; Leonard and Koffas, 2007). The infused chimera 17αGmIFS:CcCPR, constructed by mutation of 17αGmIFS, a gene encoding for an N-terminal peptide substituted with 17α, and a truncated CcCPR was cloned into pRSFDuet-1 between the NcoI and EcoRI sites. Next, GmHID was also ligated into the vector between the NdeI and XhoI to create pYH164. The final plasmid pYH164 was then introduced into the liquiritigenin/naringenin producer for isoflavanone production. The E. coli BL21(DE3) harboring pYH141 and pYH164 was named DAI-1, E. coli BL21(DE3) harboring pYH153 and pYH164 was named DAI-2, and the strain carrying pYH149, pYH117 and pYH164 was named DHF-3. After 2 days of incubation, the cultures were analyzed by HPLC-UV and HPLC-ESI-MS. The product eluting at 24.88 min was identified to be daidzein, and the product eluting at 30.13 min was identified to be genistein, by comparing the retention time, UV spectrum and high-resolution mass spectrum with the authentic standards (Fig. S7 and S8). The intermediates liquiritigenin and narigenin were still be detected owing to the insufficient biotransformation of flavanones by the catalytic activity of GmIFS and GmHID. However, the production of daidzein and genistein were still observed to be improved in DAI-2 and DAI-3. The titer of daidzein and genistein were 3.2 mg/L and 2.5 mg/L in DAI-2, which is almost 2.5-fold and 2.3-fold higher than that in DAI-1 (1.3 mg/L of daidzein and 1.1 mg/L of genistein). Their titers were further improved to be 5.5 mg/L and 6.2 mg/L in DAI-3. The results indicated that the double or multiple protein complex remarkably increased the metabolic flux for the total 5-deoxyisoflavones production. The daidzein to genistein ratio was measured to be 1.2, 1.3 and 0.9 in DAI-1, DAI-2, and DAI-3, respectively. The physical proximity of GmCHS7 and GuCHR can slightly divert the flux towards the reduced branch, however, the triple protein complex engineering had a negative effect on flux branch flux control.
4. Discussion
5-Deoxy(iso)flavonoids are a distinct subclass of flavonoids that are predominantly distributed in Leguminosae (Aoki et al., 2000). Biosynthesis of these valuable natural products in a microbial host is an attractive alternative to chemical synthesis and extraction from plant tissues. However, regulation of carbon flux through the phenylpropanoid pathway is complicated and biased towards the non-reduced branch, rendering production of 5-deoxy(iso)flavonoid extremely challenging.
CHR sits on the branch point of the flavonoid pathway and plays a vital role in controlling the ratio of CHR product to CHS product. However, the branch point is natively biased towards non-reduced flavonoids while only a small fraction of flux is directed towards reduced flavonoids. An engineered E. coli expressing Medicago sativa MsCHR could produce 7.0 mg/L of liquiritigenin, that was only one quarter of naringenin (Yan et al., 2008). Whereas, in a more recent study, an A. mongholicus CHR was selected for the production of six flavonoids in yeast, resulting in 3.4-fold higher titer of liquiritigenin compared to that of naringenin (1.55 mg/L) (Rodriguez et al., 2017). Inspired by these previous results, we adopted a genomic search analysis targeting species that are known to accumulate reduced flavonoids, resulting in a positive hit, GuCHR from G. uralensis. An initial cloning of this gene into construct pYH88 led to the production of 16.7 mg/L liquiritigenin, which was much higher than in the previous reports (Fig. 2B). A comparison of the products of this GuCHR to that of GmCHR5 from G. max that achieved a lower liquirtigenin titer (9.8 mg/L in pYH138) (Fig. 3) highlights the critical role that enzymes from different species play in synthetic biology endeavors. Although most CHRs have been isolated from Leguminosae, some CHR-like enzymes have been identified in some non-leguminous plant species (Ballance and Dixon, 1995; Sallaud et al., 1995; Goormachtig et al., 1999; Manning 1998), suggesting extensive CHR mining from diverse plant species followed by activity screening as promising strategies in improving the toolbox for 5-deoxyflavonoid production.
For tailoring of enzymes to improve the titer of desired products, directed evolution based on the enzyme structure is usually the method of choice (Xiong et al., 2017). The generation of mutants with specific substrate affinity or high turnover rate has been widely applied for an improved production of valuable metabolites. Rational design and site-directed mutagenesis of CHSs was undertaken to improve their activity, which led to positive effects on pinocembrin production (Cao et al., 2016). Although the three-dimensional structure of chalcone reductases have been well resolved, it is still difficult to identify whether the linear polyketide-CoA intermediates or p-coumaryl-trione is the real substrate of CHR (Bomati et al., 2005). The proposed intermediate, with passive diffusion or spontaneous aromatization, greatly influences the final product distribution, including the ratio of deoxychalcone to chalcone. Due to these unresolved mechanistic issues, rational protein engineering on CHRs to fine-tune substrate specificity or catalytic efficiency remains inaccessible.
Improving the concentration of CHR would be an alternative choice for diversion of flux toward the reduced products. The expression of an extra copy of the chalcone synthase and chalcone reductase genes not only improved the titer of fisetin, but also slightly improved the ratio of fisetin to quercetin in an engineered yeast (Rodriguez et al., 2017). However, the upper limit of deoxychalcone to chalcone, which are the common precursors of 5-deoxyflavonoid and 5-hydroxyflavonoid compounds, respectively, have been experimentally observed to be ∼1:1 even in the presence of a great excess of CHR (Oguro et al., 2004). The reason was predominantly attributed to either reloading of polyketide-CoA intermediates onto the CHS catalytic center or the spontaneous aromatization of the coumaryl-trione in the surrounding aqueous solution that may compete with CHR binding and reduction and thus limit the yield of deoxychalcone (Bomati et al., 2005).
Based on the characteristic 5-deoxyflavonoids founded in legumes, it is highly speculative that a CHS-CHR enzymatic complex exists, which makes the reduced pathway efficient. However, neither GmCHS7-GmCHR5 nor GuCHS-GuCHR exhibited preferable ratio of CHR product or promotional effect on carbon fluxes. Thus, we carried out orthogonal screening of CHS and CHR from five different plant species, including M. sativa (legume), G. uralensis (legume), G. max (legume), A. thaliana (Brassicaceae), P. hybrida (Solanaceae) to optimize the liquiritigenin production. The best performance enzymes GmCHS7-GuCHR not only improved the CHR ratio, but also the final flavonoid titer. Although it is not clear how the selected enzymes benefited the production of liquiritigenin, this result highlights the importances of bioprospecting of genetic elements from various plant sources as an important strategy for developing optimal biocatalysts. Oftentimes there are several putative orthologous encoding enzymes with uncharacterized biochemical properties. In some cases, there are even several homologues in a plant species. For example, there are at least nine paralogs encoding CHS (Schmutz et al., 2010; Shimomura et al., 2015), 11 encoding CHR (Mameda et al., 2018), and 12 encoding CHI (Ralston et al., 2005) in the soybean genome. Under these circumstances, a library of constructs with different gene combinations can be achieved and screened for the most efficient pathway.
It is generally accepted that a short transient time or delivery distance can reduce the diffusion of unstable intermediates, thus conferring catalytic advantages. These conditions can be achieved through two approaches: protein fusion engineering or protein-protein interaction assembly. Fusions maximize the local concentrations of substrates for individual enzymes, allowing the reaction to proceed efficiently while minimizing degradation of unstable intermediates or reducing the negative effects of toxic intermediates. For example, FPPS (farnesyl diphosphate synthase) and PTS (patchoulol synthase) were fused via a short three amino acid linker, which resulted in a 2-fold increased production of patchoulol in E. coli (Albersen, et al., 2011). However, this strategy is not always useful, e.g. in case that the fused protein adversely affects the expression levels or disrupts the enzyme structure or prevent natural multimerization. For example, the fusion of CHS and CHR clearly did not provide any additional advantages when compared to free-floating enzymes in the production of fisetin (Rodriguez et al., 2017), and neither did we realize any significant gains in liquiritigenin production from a GmCHS7: GuCHR chimera in this work.
Another efficient tool to facilitate artificial multi-protein complexes is via protein-protein interaction (Liu et al., 2013). The sequential pathway enzymes are colocalized into synthetic complexes using engineered interactions between well-characterized protein-protein interaction domains and their specific ligands. These specifically organized complexes can shorten the transient time and space between enzymes and their substrates can prevent loss of intermediates through diffusion or to competing pathways. The net result is improved catalytic efficiency or tailored direction of the pathway towards the desired product. Protein-protein interaction domains have been shown to improve production of itaconic acid in E. coli and to enhance the biosynthesis of resveratrol in yeast (Wang and Yu, 2012; Yang et al., 2017). We adopted a similar strategy and engineered interaction of CHS and CHR through GmCHS7:PDZ and GuCHR:PDZlig in the strain BL21(DE3)-pYH153, which improved the production of liquiritigenin to 44.6 mg/L. Compared to non-colocalized producers, this strategy improved total titers to 76.4 mg/L and CHR ratio to 54.7%.
Recently, the sequential actions of a number of flavonoid enzymes that are recruited onto the cytoplasmic side of ER to form metabolons have been investigated (Stafford, 1974; Waki et al., 2016; Saito et al., 2013; Watkinson et al., 2018). The amount of 5-deoxy(iso)flavonoids found in soybean is mainly attributed to the isoflavonoid metabolons (Nakayama et al., 2019). The formation of metabolons is regarded crucial in flavonoid structural diversity and flavonoid synthetic efficiency. To date, specific flavonoid metabolons have been studied in multiple plant species. When using a heterologous platform, specific metabolons can also be constructed by protein self-assembly to mimic the flavonoid metabolons. Our programmed protein scaffolds are designed to construct a deoxyflavonoid metabolon CHS-CHR-CHI with stoichiometric control over enzymes to regulate pathway direction regulation. However, all 15 scaffolds failed to show an achieve significant positive effect on the CHR ratio of the final product. This could be caused by several factors such as dynamics of the association/dissociation events of the equilibrium state (a net effect of the systems’ conditions such as pH and ionic strength), that had a bearing on protein-protein affinity and interactions. Another possible explanation could be that the protein scaffolds only increased the concentration of the co-localized enzymes, but not the NADPH pool, a required cofactor for CHR. All in all, one of our artificial deoxyflavonoid metabolon, P4S2G1 significantly improved metabolic flux to yield a high titer of flavonoids.
5. Conclusion
This work presents a proof-of-concept on redirecting biosynthetic flux towards 5-deoxy(iso)flavonoids via orthologous gene screening and a protein self-assembly strategy. The engineered GmCHS7 and GuCHR complexes improved CHR ratio to 54.7%. In addition to a substantial overall proportion of 5-deoxyflavanone, our best strain carrying a self-assembly enzyme scaffold, P4S2G1, also enhanced the production of flavanones by 1.4-fold (97.0 mg/L). These strategies were validated by the production of lineage-specific flavones and isoflavones. The final titers of flavones and isoflavones were enhanced by 3.2-fold (126.4 mg/L), and 4.9-fold (11.7 mg/L), respectively. This work also achieved de novo biosynthesis of 7,4′-dihydroxyflavone, daidzein and genistein from L-tyrosine in an E. coli host for the first time. Our study presents a promising strategy for valuable molecule production in a heterologous platform, and highlights the utility of enzyme self-assembly for pathway efficiency improvement.
Author statement
J.L. and Y.W. designed the experiments. J.L., F.X., D.J., and C.T. conducted all experiments. J.L., S.Y., and I.M analyzed the data. J.L., I. M., and Y.W. wrote the manuscript, and all authors read and approved the final manuscript.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was funded by the National Key R&D Program of China (2018YFA0900600), the National Natural Science Foundation of China (Grant nos. 22077129, 32070328, 41876084) and Research Program of State Key Laboratory of Bioreactor Engineering, the Tianjin Synthetic Biotechnology Innovation Capacity Improvement Project (TSBICIP-KJGG-002-15), the Program of Shanghai Academic Research Leader (20XD1404400), the Strategic Priority Research Program ‘Molecular mechanism of Plant Growth and Development’ of CAS (XDB27020202), the Construction of the Registry and Database of Bioparts for Synthetic Biology of the Chinese Academy of Science (No. ZSYS-016), the International Partnership Program of Chinese Academy of Science (No. 153D31KYSB20170121) and the National Key Laboratory of Plant Molecular Genetics, SIPPE, CAS. Y. S. is supported by the Fundation of Youth Innovation Promotion Association of the Chinese Academy of Sciences.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.mec.2021.e00185.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- Akashi T., Aoki T., Ayabe S. Molecular and biochemical characterization of 2-hydroxyisoflavanone dehydratase. Involvement of carboxylesterase-like proteins in leguminous isoflavone biosynthesis. Plant Physiol. 2005;137:882–891. doi: 10.1104/pp.104.056747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki T., Akashi T., Ayabe S. Flavonoids of leguminous plants: structure, biological activity, and biosynthesis. J. Plant Res. 2000;113:475–488. doi: 10.1007/PL00013958. [DOI] [Google Scholar]
- Arita T., Suwa K. Search extension transforms wiki into a relational system: a case for flavonoid metabolite database. BioData Min. 2008;1:7. doi: 10.1186/1756-0381-1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballance G.M., Dixon R.A. Medicago sativa cDNAs encoding chalcone synthesis. Plant Physiol. 1995;107:1027–1028. doi: 10.1104/pp.107.3.1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bomati E.K., Austin M.B., Bowman M.E., Dixon R.A., Noel J.P. Structural elucidation of chalcone reductase and implications for deoxychalcone biosynthesis. J. Biol. Chem. 2005;280:30496–30503. doi: 10.1074/jbc.M502239200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao W., Ma W., Wang X., Zhang B., Cao X., Chen K., Li Y., Ouyang P. Enhanced pinocembrin production in Escherichia coli by regulating cinnamic acid metabolism. Sci. Rep. 2016;6:1–9. doi: 10.1038/srep32640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chemler J.A., Yan Y., Koffas M.A.G. Biosynthesis of isoprenoids, polyunsaturated fatty acids and flavonoids in Saccharomyces cerevisiae. Microb. Cell Factories. 2006;5:20. doi: 10.1186/1475-2859-5-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Ko N., Chen K. Isoflavone supplements for menopausal women: a systematic review. Nutrients. 2019;11:2649. doi: 10.3390/nu11112649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi E.J., Kim G.H. Daidzein causes cell cycle arrest at the G1 and G2/M phases in human breast cancer MCF-7 and MDA-MB-453 cells. Phytomedicine. 2008;15:683–690. doi: 10.1016/j.phymed.2008.04.006. [DOI] [PubMed] [Google Scholar]
- Chouhan S., Sharma K., Zha J., Guleria S., Kottas M.A.G. Recent advances in the recombinant biosynthesis of polyphenols. From. Microbiol. 2017;8:2259. doi: 10.3389/fmicb.2017.02259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon R.A., Steele C.L. Flavonoids and isoflavonoids – a goldmine for metabolic engineering. Trends Plant Sci. 1999;4:394–400. doi: 10.1016/s1360-1385(99)01471-5. [DOI] [PubMed] [Google Scholar]
- Dooner H.K., Robbins T.P., Jorgensen R.A. Genetic and developmental control of anthocyanin biosynthesis. Annu. Rev. Genet. 1991;25:173–199. doi: 10.1146/annurev.ge.25.120191.001133. [DOI] [PubMed] [Google Scholar]
- Dueber J.E., Wu G.C., Malmirchegini G.R., Moon T.S., Petzold C.J., Ullal A.V., Prather K.L.J., Keasling J.D. Synthetic protein scaffolds provide modular control over metabolic flux. Nat. Biotechnol. 2009;27:753–759. doi: 10.1038/nbt.1557. [DOI] [PubMed] [Google Scholar]
- Fowler Z.L., Koffas M.A.G. Biosynthesis and biotechnological production of flavanones: current state and perspectives. Appl. Microbiol. Biotechnol. 2009;83 doi: 10.1007/s00253-009-2039-z. [DOI] [PubMed] [Google Scholar]
- Goormachtig S., Lievens S., Herman S., Van Montagu M., Holsters M. Chalcone reductase-homologous transcripts accumulate during development of stem-borne nodules on the tropical legume Sesbania rostrata. Planta. 1999;209:45–52. doi: 10.1007/s004250050605. [DOI] [PubMed] [Google Scholar]
- Graham T.L. Flavonoid and isoflavonoid distribution in developing soybean seedling tissues and in seed and root exudates. Plant Physiol. 1991;95:594–603. doi: 10.1104/pp.95.2.594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi H., Murayama K., Hiraoka N., Ikeshiro Y. Plant Gene Register PGR 96-095: nucleotide sequences of cDNAs (accession nos. D86558 and D86559) for Glycyrrhiza glabra polyketide reductase. Plant Physiol. 1996;112:861–864. doi: 10.1104/pp.112.2.861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilgarth R.S., Lanigan T.S. Optimization of overlap extension PCR for efficient transgene construction. MethodX. 2020;7 doi: 10.1016/j.mex.2019.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jha H.G., Ziliken F., Breitmaier E. Isoflavone synthesis with 1,3,5-triazine. Angew. Chem. 1981;93:129–131. doi: 10.1002/anie.198101021. [DOI] [Google Scholar]
- Jones J.A., Collins S.M., Vernacchio V.R., Lachance D.M., Koffas M.A.G. Optimization of naringenin and p-coumaric acid hydroxylation using the native E. coli hydroxylase complex. HpaBC. Biotechnol. Prog. 2015;32:21–25. doi: 10.1002/btpr.2185. [DOI] [PubMed] [Google Scholar]
- Jones J.A., Vernacchio V.R., Collins S.M., Shirke A.N., Xiu Y., Englaender J.A., Cress B.F., McCutcheon C.C., Linhardt R.J., Gross R.A., Koffas M.A.G. Complete biosynthesis of anthocyanins using E.coli polycultures. mBio. 2017;8 doi: 10.1128/mBio.00621-17. e00621-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang W., Ma T., Liu M., Qu J., Liu Z., Zhang H., Shi B., Fu S., Ma J., Lai L.T.F., He S., Qu J., Au S.W.-N., Kang B.H., Lau W.C.Y., Deng Z., Xia J., Liu T. Modular enzyme assembly for enhanced cascade biocatalysis and metabolic flux. Nat. Commun. 2019;10:4248. doi: 10.1038/s41467-019-12247-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsuyama Y., Matsuzawa M., Funa N., Horinouchi S. Production of curcuminoids by Escherichia coli carrying an artifical biosynthesis pathway. Microbiology. 2008;74:641–645. doi: 10.1099/mic.0.2008/018721-0. [DOI] [PubMed] [Google Scholar]
- Kim D.H., Kim B.-G., Jung N.R., Ahn J.-H. Production of genistein from naringenin using Escherichia coli containing isoflavone synthase-cytochrome P450 reductase fusion protein. J. Microbiol. Biotechnol. 2009;19:1612–1616. doi: 10.4014/jmb.0905.05043. [DOI] [PubMed] [Google Scholar]
- Kim S.C., Byun S.H., Yang C.H., Kim C.Y., Kim J.W., Kim S.G. Cytoprotective effects of Glycyrrhiza radix extract and its active component liquiritigenin against cadmium-induced toxicity (effects on bad translocation and cytochrome c-mediated PARP cleavage) Toxicology. 2004;197:239–251. doi: 10.1016/j.tox.2004.01.010. [DOI] [PubMed] [Google Scholar]
- Koes R.E., Quattrocchio F., Mol J.N.M. The flavonoid biosynthetic pathway in plant: function and evolution. Bioessays. 1994;16:123–132. doi: 10.1002/bies.950160209. [DOI] [Google Scholar]
- Leonard E., Koffas M.A.G. Engineering of artificial plant cytochrome P450 enzymes for synthesis of isoflavones by Escherichia coli. Appl. Environ. Microbiol. 2007;73:7246–7251. doi: 10.1128/AEM.01411-07. http://doi:10.1128/AEM.01411-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard E., Lim K.H., Saw P.N., Koffas M.A.G. Engineering central metabolic pathways for high-level flavonoid production in Escherichia coli. Appl. Environ. Microbiol. 2007;73:3877–3886. doi: 10.1128/AEM.00200-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard E., Yan Y., Fowler Z.L., Li Z., Lim C.-G., Lim K.-H., Koffas M.A.G. Strain improvement of recombinant Escherichia coli for efficient production of plant flavonoids. Mol. Pharm. 2008;5:257–265. doi: 10.1021/mp7001472. [DOI] [PubMed] [Google Scholar]
- Li J., Tian C., Xia Y., Mutanda I., Wang K., Wang Y. Production of plant-specific flavones baicalein and scutellarin in an engineered E. coli from available phenylalanine and tyrosine. Metab. Eng. 2019;52:124–133. doi: 10.1016/j.ymben.2018.11.008. [DOI] [PubMed] [Google Scholar]
- Lim S.S., Jung S.H., Ji J., Shin K.H., Keum S.R. Synthesis of flavonoids and their effects on aldose reductase and sorbitol accumulation in streptozotocin-induced diabetic rat tissues. J. Pharm. Pharmacol. 2001;53:653–668. doi: 10.1211/0022357011775983. [DOI] [PubMed] [Google Scholar]
- Liu F., Banta S., Chen W. Functional assembly of a multi-enzyme methanol oxidation cascade on a surface-displayed trifunctional scaffold for enhanced NADH production. Chem. Commun. 2013;49:3766–3768. doi: 10.1039/c3cc40454d. [DOI] [PubMed] [Google Scholar]
- Liu X., Cheng J., Zhang G., Ding W., Duan L., Yang J., Kui L., Cheng X., Ruan J., Fan W., Chen J., Long G., Zhan Y., Cai J., Wang W., Ma Y., Dong Y., Yang S., Jiang H. Engineering yeast for the berviscapine by genomic analysis and synthetic biology approaches. Nat. Commun. 2018;9:448. doi: 10.1038/s41467-018-02883-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Cheng J., Zhu X., Zhang G., Yang S., Guo X., Jiang H., Ma Y. De Novo biosynthesis of multiple pinocembrin derivatives in Saccharomyces cerevisiae. ACS Synth. Biol. 2020;9:3042–3051. doi: 10.1021/acssynbio.0c00289. [DOI] [PubMed] [Google Scholar]
- Lyu X., Ng K.R., Lee J.L., Mark R., Chen W.N. Enhancement of naringenin biosynthesis from tyrosine by metabolic engineering of Saccharomyces cerevisiae. J. Agric. Food Chem. 2017;65:6638–6646. doi: 10.1021/acs.jafc.7b02507. [DOI] [PubMed] [Google Scholar]
- Mameda R., Waki T., Kawai Y., Takahashi S., Nakayama T. Involvement of chalcone reductase in the soybean isoflavone metabolon: identification of GmCHR5, which interacts with 2-hydroxyisoflavanone synthase. Plant J. 2018;96:56–74. doi: 10.1111/tpj.14014. [DOI] [PubMed] [Google Scholar]
- Manning K. Isolation of a set of ripening-related genes from strawberry: their identification and possible relationship to fruit quality traits. Planta. 1998;205:622–631. doi: 10.1007/s004250050365. [DOI] [PubMed] [Google Scholar]
- Mavel S., Dikic B., Palakas S., Emond P., Greguric I., de Gracia A.G., Mattner F., Garrigos M., Guilloteau D., Katsifis A. Synthesis and biological evaluation of a series of flavone derivatives as potential radioligands for imaging the multidrug resistance-associated protein 1(ABCC1/MRP1) Bioorg. Med. Chem. 2006;14:1599–1607. doi: 10.1016/j.bmc.2005.10.009. [DOI] [PubMed] [Google Scholar]
- Miyahisa I., Funa N., Ohnishi Y., Martens S., Moriguchi T., Horinouchi S. Combinatorial biosynthesis of flavones and flavonols in Escherichia coli. Appl. Microbiol. Biotechnol. 2006;71:53–58. doi: 10.1007/s00253-005-0116-5. [DOI] [PubMed] [Google Scholar]
- Nakayama T., Takahashi S., Waki T. Formation of flavonoid metabolons: functional significance of protein-protein interactions and impact on flavonoid chemodiversity. Front. Plant Sci. 2019 doi: 10.3389/fpls.2019.00821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naoumkina M.A., Zhao Q., Gallego-Giraldo L., Dai X., Zhao P.X., Dixon R.A. Genome-wide analysis of phenylpropanoid defence pathways. Mol. Plant Pathol. 2010;11:829–846. doi: 10.1111/j.1364-3703.2010.00648.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neidhardt F.C., Bloch P.L., Smith D.F. Culture medium for enterobacteria. J. Bacteriol. 1974 doi: 10.1128/jb.119.3.736-747.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oguro S., Akashi T., Ayabe S., Noguchi H., Abe I. Probing biosynthesis of plant polytides with synthetic N-acetylcysteamine thioesters. Biochem. Biophys. Res. Commun. 2004;325:561–567. doi: 10.1016/j.bbrc.2004.10.057. [DOI] [PubMed] [Google Scholar]
- Rodriguez A., Strucko T., Stahlhut S.G., Kristensen M., Svenssen D.K., Forster J., Nielsen J., Borodina I. Metabolic engineering of yeast for fermentative production of flavonoids. Bioresour. Technol. 2017;245:1645–1654. doi: 10.1016/j.biortech.2017.06.043. [DOI] [PubMed] [Google Scholar]
- Saito K., Yonekura-Sakakibara K., Nakabayashi R., Higashi Y., Yamazaki M., Tohge T., Fernie A. The flavonoid biosynthetic pathway in Arabidopsis: structural and genetic diversity. Plant Physiol. Biochem. 2013;72:21–34. doi: 10.1016/j.plaphy.2013.02.001. [DOI] [PubMed] [Google Scholar]
- Sallaud C., El-Turk J., Bigarré L., Sevin H., Welle R., Esnault R. Nucleotide sequences of three chalcone reductase genes from Alfalfa. Plant Physiol. 1995;108:869–870. doi: 10.1104/pp.108.2.869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmutz J., Cannon S.B., Schlueter J., Ma J., Mitros T., Nelson W. Genome sequence of the palaeopolyploid soybean. Nature. 2010;463:178–183. doi: 10.1038/nature08670. [DOI] [PubMed] [Google Scholar]
- Shetty R.P., Endy D., Knight T.F., JR. Engineering biobrick vectors from biobrick parts. J. Biol. Eng. 2008;2:5. doi: 10.1186/1754-1611-2-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimomura M., Kanamori H., Komatsu S., Namiki N., Mukai Y., Kurita K., Kamatsuki K., Ikawa H., Yano R., Ishimoto M., Kaga A., Katayose Y. The Glycine max cv. Enrei genome for improvement of Japanese soybean cultivars. Int. J. Genom. 2015;215 doi: 10.1155/2015/358127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stafford H.A. The metabolism of aromatic compounds. Annu. Rev. Plant Physiol. 1974;25:459–486. doi: 10.1146/annurev.pp.25.060174.002331. [DOI] [Google Scholar]
- Stahlhut S.G., Siedler S., Malla S., Harrison S.J., Maury J., Neves A.R., Forster J. Assembly of a novel biosynthetic pathway for production of the plant flavonoid fisetin in Escherichia coli. Metab. Eng. 2015;31:84–93. doi: 10.1016/j.ymben.2015.07.002. [DOI] [PubMed] [Google Scholar]
- Strong A.L., Ohlstein J.F., Jiang Q., Zhang Q., Zheng S., Boun S.M., Elliott S., Gimble J.M., Burow M.E., Wang G., Bunnell B.A. Novel daidzein analogs enhance osteogenic activity of bone marrow-derived mesenchymal stem cells and adipose-derived stromal/stem cells through estrogen receptor dependent and independent mechanisms. Stem Cell Res. Ther. 2014;5:105. doi: 10.1186/scrt493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai P.J., Wu S.C., Cheng Y.K. Role of polyphenyols in antioxidant capacity of napiergrass from different grow seasons. Food Chem. 2008;106:27–32. doi: 10.1016/j.foodchem.2007.05.037. [DOI] [Google Scholar]
- Ververidis F., Trantas E., Douglas C., Vollmer G., Kretzschmar G., Panopoulos N. Biotechnology of flavonoids and other phenylpropanoid-derived natural products. Part I: chemical diversity, impacts on plant biology and human health. Biotechnol. J. 2007;2:1214–1234. doi: 10.1002/biot.200700084. [DOI] [PubMed] [Google Scholar]
- Waki T., Yoo D., Fujino N., Mameda R., Denessiouk K., Yamashita S., Motohashi R., Akahi T., Aoki T., Ayabe S., Takahashi S., Nakayama T. Identification of protein-protein interactions of isoflavonoid biosynthetic enzymes with 2-hydroxyisoflavanone synthase in soybean (Glycine max(L.) Merr.) Biochem. Biophys. Res. Commun. 2016;469:546–551. doi: 10.1016/j.bbrc.2015.12.038. [DOI] [PubMed] [Google Scholar]
- Wang Y., Yu O. Synthetic scaffolds increased resveratrol biosynthesis in engineered yeast cells. J. Biotechnol. 2012;157:258–260. doi: 10.1016/j.jbiotec.2011.11.003. [DOI] [PubMed] [Google Scholar]
- Weisshaar B., Jenkins G.I. Phenylpropanoid biosynthesis and its regulation. Curr. Opin. Plant Biol. 1998;1:251–257. doi: 10.1016/S1369-5266(98)80113-1. [DOI] [PubMed] [Google Scholar]
- Welle R., Schröder G., Schiltz E., Grisebach H., Schröder J. Induced responds to pathogen attack, analysis and heterologous expression of the key enzyme in the biosynthesis of phytoalexins in soybean (Glycine max L. Merr. cv. Harosoy 63) Eur. J. Biochem. 1991;196:423–430. doi: 10.1111/j.1432-1033.1991.tb15833.x. [DOI] [PubMed] [Google Scholar]
- Winkel-Shirley B. Flavonoid Biosynthesis. A colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiol. 2001;126:485–493. doi: 10.1104/pp.126.2.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams C.A., Grayer R.J. Anthocyanins and other flavonoids. Nat. Prod. Rep. 2004;21:539–573. doi: 10.1039/b311404j. [DOI] [PubMed] [Google Scholar]
- Wu J., Du G., Zhou J., Chen J. Systems metabolic engineering of microorganisms to achieve large-scale production of flavonoid scaffolds. J. Biotechnol. 2014;188:72–80. doi: 10.1016/j.jbiotec.2014.08.016. [DOI] [PubMed] [Google Scholar]
- Wu J., Zhou T., Du G., Zhou J., Chen J. Modular optimization of heterologous pathways for de novo synthesis of (2S)-naringenin in Escherichia coli. PloS One. 2014;9 doi: 10.1371/journal.pone.0101492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie S., Wang Y., Liu C., Luo K., Cai Y. Liquiritigenin inhibits serum-induced HIF-1α and VEGF expression via the AKT/mTOR-p70S6K signalling pathway in Hela cells. Phytother Res. 2012;26:1133–1141. doi: 10.1002/ptr.3696. [DOI] [PubMed] [Google Scholar]
- Xiong D., Lu S., Wu J., Liang C., Wang W., Wang W., Jin J., Tang S. Improving key enzyme activity in phenylpropanoid pathway with a designed biosensor. Metab. Eng. 2017;40:115–123. doi: 10.1016/j.ymben.2017.01.006. [DOI] [PubMed] [Google Scholar]
- Yan Y., Huang L., Koffas M.A.G. Biosynthesis of 5-deoxyflavanones in microorganisms. Biotechnol. J. 2007;2:1250–1262. doi: 10.1002/biot.200700119. [DOI] [PubMed] [Google Scholar]
- Yan Y., Li Zhen, Koffas M.A.G. High-yield anthocyanin biosynthesis in engineered Escherichia coli. Biotechnol. Bioeng. 2008;100:126–140. doi: 10.1002/bit.21721. [DOI] [PubMed] [Google Scholar]
- Yang Z., Gao X., Xie H., Wang F., Ren Y., Wei D. Enhanced itaconic acid production by self-assembly of two biosynthetic enzymes in Escherichia coli. Biotechnol. Bioeng. 2017;114:457–462. doi: 10.1002/bit.26081. [DOI] [PubMed] [Google Scholar]
- Zhao C., Wang F., Lian Y., Xiao H., Zheng J. Biosynthesis of citrus flavonoids and their health effects. Crit. Rev. Food Sci. Nutr. 2018;60:566–583. doi: 10.1080/10408398.2018.1544885. [DOI] [PubMed] [Google Scholar]
- Zhu L., Huang Y., Zhang Y., Xu C., Lu J., Wang Y. The growing season impacts the accumulation and composition of flavonoids in grape skins in two-crop-a-year viticulture. J. Food Sci. Technol. MYS. 2017;54:2861–2870. doi: 10.1007/s13197-017-2724-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.