Abstract
Objectives
As early prediction of severe illness and death for patients with coronavirus disease 2019 (COVID-19) is important, we aim to explore the clinical value of laboratory indicators in evaluating the progression and prognosis of patients with COVID-19.
Design
Retrospective cohort study.
Setting
Hospital-based study in China.
Participants
Adult patients with COVID-19 from December 15, 2019 to March 15, 2020.
End point
Disease severity and mortality.
Methods
Clinical data of 638 patients with COVID-19 were collected and compared between severe and non-severe groups. The predictive ability of laboratory indicators in disease progression and prognosis of COVID-19 was analysed using the receiver operating characteristic curve. The survival differences of COVID-19 patients with different levels of laboratory indicators were analysed utilising Kaplan-Meier analysis.
Results
29.8% (190/638) of patients with COVID-19 progressed to severe. Compared with patients with no adverse events, C reactive protein (CRP), neutrophil-to-lymphocyte ratio (NLR) and D-dimer were significantly higher in severe patients with adverse events, such as acute myocardial injury, respiratory failure, acute kidney injury, mechanical ventilation, intensive care unit admission, multiple organ dysfunction syndromes and death (all p<0.05). The multivariate logistic analysis suggested that CRP, NLR and D-dimer were independent risk factors for the disease progression of COVID-19 (all p<0.05). The model combining all of them owned the highest area under the receiver operating characteristic curve (AUC) predicting disease progression and death of COVID-19, with AUC of 0.894 (95% CI 0.857 to 0.931) and 0.918 (95% CI 0.873 to 0.962), respectively. Survival analysis suggested that the patients with a high level of CRP, NLR or D-dimer performed shorter overall survival time (all p<0.05).
Conclusions
The combination of CRP, NLR and D-dimer could be an effective predictor for the aggravation and death in patients with COVID-19. The abnormal expression of these indicators might suggest a strong inflammatory response and multiple adverse events in patients with severe COVID-19.
Keywords: COVID-19, infection control, virology
Strengths and limitations of this study.
We used laboratory indicators to predict both disease severity and mortality in patients with COVID-19.
We combined inflammatory and coagulation indicators that were the important mechanisms for predicting the prognosis of patients with COVID-19.
Our study detailed the incidence of adverse events in patients with severe COVID-19 and recorded the levels of C reactive protein (CRP), neutrophil-to-lymphocyte ratio (NLR) and D-dimer in different adverse events.
Our study provided a simple way to determine the poor prognosis of patients with COVID-19, which was conducive to the allocation of medical resources.
Our study lacked dynamic monitoring of CRP, NLR and D-dimer.
Introduction
A pneumonia of unknown cause was first reported in December 2019 and rapidly spread throughout the world.1 It was found that the pathogen was a beta coronavirus, similar to SARS-CoV and Middle East respiratory syndrome coronavirus.2 The International Committee on Taxonomy of Viruses named the virus SARS-CoV-2 and the WHO named the disease COVID-19. SARS-CoV-2 could enter human cells by binding to ACE2 receptors in various organs of the human body, which was one of the important pathogeneses of COVID-19 disease.3 As of 22 August 2020, at least 22 812 529 people have been infected, and 795 133 people have died worldwide. Patients with mild and moderate COVID-19 accompany with a good prognosis, while severe patients may suffer from acute respiratory distress syndrome, multiple organ dysfunction syndromes (MODS) and acute kidney injury (AKI) or even death.4 Therefore, early identification of severe patients and active intervention are particularly important.
Several studies have shown that laboratory indicators can effectively predict disease progression and poor prognosis in patients with COVID-19. A study has reported that C reactive protein (CRP), a sensitive biomarker for inflammation, infection and tissue injury, is an independent risk factor for the progression of COVID-19.5 Neutrophil-to-lymphocyte ratio (NLR) has been reported to have a high accuracy in predicting severe COVID-19.6 7 The inflammatory response can cause a significant increase in D-dimer, and studies have reported that the D-dimer level can predict disease severity and death in patients with COVID-19.8–10 However, the current models for predicting disease progression and prognosis of COVID-19 are not ideal. Therefore, in this study, we explored the relationship between relevant laboratory indicators and disease progression and adverse events, aiming to provide a theoretical basis for early identification and intervention for patients with severe COVID-19.
Methods
Study design and participants
All 638 confirmed COVID-19 patients were enrolled according to the New Coronavirus Pneumonia Prevention and Control Guideline (seventh edition).11 All of them were admitted to Wuhan No.7 Hospital, which was temporarily taken over by Zhongnan Hospital of Wuhan University, China from 15 December 2019 to 15 March 2020. We followed up the clinical outcomes such as severity and mortality until 30 March 2020. This study was conducted following the Declaration of Helsinki. This study was approved by the Ethics Committee of Zhongnan Hospital of Wuhan University, China (grant numbers 2020082K).
Data collection
The electronic medical records data of patients with COVID-19 were carefully reviewed by three independent researchers, including demographics (gender and age), underlying comorbidities (hypertension, cardiovascular disease, chronic respiratory disease and chronic liver disease), clinical symptoms (fever, cough, dyspnoea, chest distress, fatigue, myalgia, stuffy nose, pharyngalgia, dizziness or headache, diarrhoea, nausea or vomiting and anorexia), radiological images (unilateral pneumonia and bilateral pneumonia), treatment measures (antiviral treatment, antibiotics, corticosteroids, intravenous immunoglobin and traditional Chinese medicine), complications (mechanical ventilation, intensive care unit (ICU) admission, MODS, acute myocardial injury, AKI and respiratory failure) and time of hospitalisation. Routine laboratory results included oxygenation index (OI), arterial oxygen partial pressure (PaO2), CRP, serum amyloid A (SAA), erythrocyte sedimentation rate (ESR), NLR, platelet-to-lymphocyte ratio (PLR), white cell count, lymphocyte count, monocyte cell count, neutrophil count, haemoglobin, platelet count, red cell distribution width, alanine aminotransferase (ALT), aspartate aminotransferase (AST), total bilirubin, albumin, cardiac troponin I, creatine kinase MB isoenzyme (CK-MB), glomerular filtration rate (GFR), serum creatinine, blood urea nitrogen, D-dimer, fibrinogen, activated partial thromboplastin time, prothrombin time (PT), thrombin time and systemic immune-inflammation index (SII), which was defined as platelet count*neutrophil count/lymphocyte count ratio.
Definition
Patients with severe COVID-19 were diagnosed if one of the following criteria was met: (1) respiratory rate ≥30/min; (2) oxygen saturation ≤93% at rest; (3) OI ≤300 mm Hg; (4) >50% lesion progression on pulmonary imaging within 24~48 hours; (5) respiratory failure requiring mechanical ventilation; (6) shock; (7) multiple organ failure requiring intensive care unit admission.11 Acute myocardial injury was defined as a hypersensitive troponin level above the reference upper limit of the 99th percentile, regardless of new abnormalities in electrocardiograms and echocardiography.12 AKI was defined as followed: (1) increased serum creatinine ≥0.3 mg/dL (≥26.5 umol/L) within 48 hours; (2) serum creatinine ≥1.5 times baseline within the first 7 days; and (3) urine volume ≤0.5 mL/kg/hour for 6 hours.13 Respiratory failure was defined as PaO2 below 60 mm Hg.14 MODS was defined if simultaneous or sequential dysfunction was observed in two or more organs or systems in the course of an acute disease so that the internal environment could not be maintained steady.15
Patient and public involvement
No identifying or personal information of patients was included in our study, and all information was anonymously analysed and processed, so clients and the public were not directly involved in the study, and all patients and the public were not involved in the design, or conduct, or reporting, or dissemination plans of this research.
Statistical analysis
SPSS V.25.0 was used for statistical analysis. Continuous variables were judged by the normality test. Normal data were analysed by t-test of two independent samples and described as mean±SD (X±S). Non-normal data were analysed by the Mann-Whitney test and described by the median and quartile range (IQR). Categorical variables, described by frequency and percentage (%), were statistically analysed using the χ2 test or the Fisher’s exact test. Univariate logistic regression was adopted to analyse risk factors of severe COVID-19, and the important factors whose p value were lower than 0.05 in univariate logistic regression and might influence the degree of COVID-19 disease were taken into multivariate logistic regression. ORs and 95% CIs were also calculated. We drew the receiver operating characteristic curve (ROC) and calculated area under the receiver operating characteristic curve (AUC) to evaluate the predictive efficiency of laboratory indicators. Kaplan-Meier curve was used to compare the survival of COVID-19 patients with different levels of laboratory indicators. Bilateral tests were used for all tests. P<0.05 was considered statistically significant.
Results
Demographic and clinical characteristics
A total of 638 confirmed COVID-19 patients were included in this study, including 190 severe patients (29.8%) and 448 non-severe patients (70.2%). The median age was 59 (47-69) years, and 301 (47.2%) patients were male. The median time of hospitalisation (IQR) was 15 (9–24) days. More than half of the patients accompanied with underlying comorbidities, with hypertension (207, 32.4%) and cardiovascular disease (70, 11.0%) being regarded as the most common comorbidities. In comparison with the non-severe group, the aggravated patients were older (66 vs 56, p<0.001), had more male patients (60.5% vs 41.5%, p<0.001) and higher ratios of comorbidity than non-severe cases, which included hypertension (43.7% vs 27.7%, p<0.001), cardiovascular disease (15.8% vs 8.9%, p=0.011) and chronic respiratory disease (12.1% vs 6.0%, p=0.009). Besides, fever (453, 71.0%) and cough (388, 60.8%) were the most common symptoms, followed with anorexia (237, 37.1%), fatigue (230, 36.1%), dyspnoea (208, 32.6%) and chest distress (167, 26.2%), and the severe group was more prone to have fever (83.7% vs 65.6%, p<0.001), dyspnoea (46.3% vs 26.8%, p<0.001), fatigue (45.3% vs 32.1%, p=0.002) and anorexia (46.8% vs 33.0%, p=0.001) than the non-severe group. According to chest computed tomography, most patients with COVID-19 manifested bilateral pneumonia (481, 75.4%), while a small number of them presented unilateral pneumonia (71, 11.1%), and the proportion of patients with bilateral pneumonia (86.3% vs 70.8%, p<0.001) was higher in patients with aggravated COVID-19 (table 1).
Table 1.
Baseline characterisitcs of the patients with COVID-19
| Variables | Total (n=638) | Severe (n=190) | Non-severe (n=448) | P value |
| Age (years), median (IQR) | 59 (47–69) | 66 (56–73) | 56 (45–66) | <0.001 |
| ≥65, no. (%) | 227 (35.6) | 105 (55.3) | 122 (27.2) | <0.001 |
| <65, no. (%) | 411 (64.4) | 85 (44.7) | 326 (72.8) | |
| Gender | ||||
| Male, no. (%) | 301 (47.2) | 115 (60.5) | 186 (41.5) | <0.001 |
| Female, no. (%) | 337 (52.8) | 75 (39.5) | 262 (58.5) | |
| Time of hospitalisation (days), median (IQR) | 15 (9–24) | 19 (11–27) | 14 (9–22) | 0.002 |
| Comorbidity | ||||
| Hypertension, no. (%) | 207 (32.4) | 83 (43.7) | 124 (27.7) | <0.001 |
| Cardiovascular disease, no. (%) | 70 (11.0) | 30 (15.8) | 40 (8.9) | 0.011 |
| Chronic respiratory disease, no. (%) | 50 (7.8) | 23 (12.1) | 27 (6.0) | 0.009 |
| Chronic liver disease, no. (%) | 37 (5.8) | 13 (6.8) | 24 (5.4) | 0.463 |
| Clinical symptoms | ||||
| Fever, no. (%) | 453 (71.0) | 159 (83.7) | 294 (65.6) | <0.001 |
| Cough, no. (%) | 388 (60.8) | 126 (66.3) | 262 (58.5) | 0.064 |
| Dyspnoea, no. (%) | 208 (32.6) | 88 (46.3) | 120 (26.8) | <0.001 |
| Chest distress, no. (%) | 167 (26.2) | 52 (27.4) | 115 (25.7) | 0.655 |
| Fatigue, no. (%) | 230 (36.1) | 86 (45.3) | 144 (32.1) | 0.002 |
| Myalgia, no. (%) | 60 (9.4) | 17 (8.9) | 43 (9.6) | 0.797 |
| Stuffy nose, no. (%) | 9 (1.4) | 0 (0.0) | 9 (2.0) | 0.109 |
| Pharyngalgia, no. (%) | 68 (10.7) | 14 (7.4) | 54 (12.1) | 0.079 |
| Dizziness or headache, no. (%) | 49 (7.7) | 12 (6.3) | 37 (8.3) | 0.399 |
| Diarrhoea, no. (%) | 78 (12.2) | 29 (15.3) | 49 (10.9) | 0.127 |
| Nausea or vomiting, no. (%) | 62 (9.7) | 18 (9.5) | 44 (9.8) | 0.892 |
| Anorexia, no. (%) | 237 (37.1) | 89 (46.8) | 148 (33.0) | 0.001 |
| Treatment | ||||
| Antiviral treatment, no. (%) | 466 (73.0) | 151 (79.5) | 315 (70.3) | 0.017 |
| Antibiotics, no. (%) | 452 (70.8) | 172 (90.5) | 280 (62.5) | <0.001 |
| Corticosteroids, no. (%) | 197 (30.9) | 107 (56.3) | 90 (20.1) | <0.001 |
| Intravenous immunoglobin, no. (%) | 120 (18.8) | 66 (34.7) | 54 (12.1) | <0.001 |
| Traditional Chinese medicine, no. (%) | 419 (65.7) | 102 (53.7) | 317 (70.8) | <0.001 |
| Imaging features | ||||
| Unilateral pneumonia, no. (%) | 71 (11.1) | 10 (5.3) | 61 (13.6) | 0.002 |
| Bilateral pneumonia, no. (%) | 481 (75.4) | 164 (86.3) | 317 (70.8) | <0.001 |
P values indicate differences between patients with severe and non-severe COVID-19. P<0.05 was considered statistically significant. Continuous variables were described as median with IQR and analysed by Mann-Whitney test. Categorical variables were described as percentages and analysed by the χ2 test or Fisher’s exact test.
Laboratory findings
On admission, many patients with COVID-19 tended to have lymphopenia, decreased levels of OI and albumin, elevated levels of CRP, SAA, ESR and D-dimer, and abnormal levels of NLR, PLR and SII. Compared with patients with non-severe COVID-19, severe patients had higher levels of CRP (p<0.001), SAA (p<0.001), ESR (p<0.001), NLR (p<0.001), PLR (p<0.001), SII (p<0.001), D-dimer (p<0.001), white cell count (p<0.001), neutrophil count (p<0.001), ALT (p<0.001), AST (p<0.001), total bilirubin (p<0.001), cardiac troponin I (p<0.001), CK-MB (p<0.001), serum creatinine (p=0.027), fibrinogen (p<0.001) and PT (p<0.001) and lower levels of OI, PaO2 (p<0.001), albumin (p<0.001), lymphocyte count (p<0.001), platelet count (p=0.004) and GFR (p=0.014). However, although the differences of some laboratory indicators between the two groups were statistically significant, the changes were all within the normal range, so the value of these indicators was limited, for example, white cell count, neutrophil count, ALT, AST, total bilirubin, cardiac troponin I, CK-MB, serum creatinine, fibrinogen, platelet count and GFR (table 2).
Table 2.
Laboratory findings of the patients with COVID-19 on admission
| Variables | Normal range | Total (n=638) | Severe (n=190) | Non-severe (n=448) | P value |
| Oxygenation index (mm Hg), median (IQR) | 400–500 | 376.2 (219.5–485.7) | 209.10 (128.1–281.0) | 449.6 (390.5–558.3) | <0.001 |
| Arterial oxygen partial pressure (mm Hg), median (IQR) | 70–107 | 87(64–120) | 62(49–79) | 107(88–139) | <0.001 |
| Creactive protein (mg/L), median (IQR) | 0–3 | 18.1 (2.4–58.4) | 71.9 (32.9–131.5) | 6.4 (1.2–27.9) | <0.001 |
| Serum amyloid A (mg/L), median (IQR) | 0–10 | 60.92 (11.82–168.82) | 92.94 (58.23–168.82) | 33.12 (7.68–168.60) | <0.001 |
| Erythrocyte sedimentation rate (mm/hour) | 0–15 | 22.96±16.50 | 33.40±14.33 | 19.14±15.56 | <0.001 |
| NLR, median (IQR) | NA | 3.0 (1.9–6.4) | 9.0 (4.0–15.0) | 2.4 (1.6–3.9) | <0.001 |
| PLR, median (IQR) | NA | 184.5 (128.4–280.2) | 262.6 (160.2–375.9) | 165.8 (119.4–233.4) | <0.001 |
| SII, median (IQR) | NA | 583.6 (348.9–1265.2) | 1433.0(654.0–2657.7) | 446.1 (304.6–782.6) | <0.001 |
| White cell count (×10∧9/L), median (IQR) | 3.5–9.5 | 5.17 (3.94–7.17) | 7.26 (4.85–9.98) | 4.74 (3.82–6.14) | <0.001 |
| Lymphocyte count (×10∧9/L), median (IQR) | 1.1–3.2 | 1.02 (0.65–1.50) | 0.66 (0.48–1.01) | 1.22 (0.81–1.64) | <0.001 |
| Monocyte cell count (×10∧9/L) | 0.1–0.6 | 0.42±0.32 | 0.45±0.52 | 0.40±0.15 | 0.191 |
| Neutrophil count (×10∧9/L), median (IQR) | 1.8–6.3 | 3.43 (2.40–5.22) | 5.90 (3.62–8.24) | 3.00 (2.22–3.91) | <0.001 |
| Haemoglobin (g/L) | 130–175 | 125.40±17.36 | 126.72±18.06 | 124.79±17.03 | 0.212 |
| Platelet count (×109/L), median (IQR) | 125–350 | 193(149–249) | 181(134–234) | 197(158–254) | 0.004 |
| Red cell distribution width (fl) | 39.0–51.5 | 38.92±3.30 | 39.15±3.08 | 38.81±3.39 | 0.247 |
| Alanine aminotransferase (IU/L), median (IQR) | 9–50 | 21(14–35) | 28(18–47) | 19(13–30) | <0.001 |
| Aspartate aminotransferase (IU/L), median (IQR) | 15–40 | 26(18–39) | 39(26–55) | 23(17–31) | <0.001 |
| Total bilirubin (μmol/L), median (IQR) | 2–23 | 8.0 (5.7–11.2) | 9.6 (6.4–13.1) | 7.5 (5.5–10.3) | <0.001 |
| Albumin (g/L) | 40–55 | 37.18±6.17 | 33.54±5.03 | 38.85±5.92 | <0.001 |
| Cardiac troponin I (ng/mL), median (IQR) | 0–0.014 | 0.008 (0.006–0.014) | 0.013 (0.008–0.024) | 0.007 (0.005–0.011) | <0.001 |
| Creatine kinase MB (ng/mL), median (IQR) | 0–6.22 | 1.21 (0.75–2.20) | 1.80 (0.92–3.80) | 1.05 (0.68–1.66) | <0.001 |
| Glomerular filtration rate (mL/min) | 66–143 | 115.96±36.28 | 109.93±42.37 | 118.72±32.79 | 0.014 |
| Serum creatinine (μmol/L) | 57–111 | 74.27±88.06 | 90.48±140.08 | 66.82±46.31 | 0.027 |
| Blood urea nitrogen (mmol/L), median (IQR) | 3.1–8.0 | 4.39 (3.44–5.79) | 5.40 (3.57–7.80) | 4.23 (3.43–5.37) | 0.139 |
| D-dimer (μg/mL), median (IQR) | 0–0.243 | 0.212 (0.105–0.617) | 0.755 (0.274–5.999) | 0.164 (0.081–0.303) | <0.001 |
| Fibrinogen (g/L), median (IQR) | 2–5 | 3.49 (2.64–4.49) | 4.23 (3.23–4.96) | 3.19 (2.39–4.11) | <0.001 |
| Acivated partial thromboplastin time (s) | 24.6–35.4 | 33.76±20.07 | 33.74±23.98 | 33.76±17.62 | 0.989 |
| Prothrombin time (s), median (IQR) | 9.3–12.9 | 12.3 (11.6–13.4) | 13.3 (12.2–14.2) | 12.1 (11.3–12.9) | <0.001 |
| Thrombin time (s) | 11.6–16.8 | 15.25±6.30 | 15.59±10.45 | 15.07±1.43 | 0.390 |
P values indicate differences between patients with severe and non-severe COVID-19. P<0.05 was considered statistically significant. Continuous normal variables were described as mean with SD and anaysed by t test. Continuous variables that were not normal were described as median with IQR and analysed by Mann-Whitney test.
NLR, neutrophil-to-lymphocyte ratio; PLR, platelet-to-lymphocyte ratio; SII, systematic immune-inflammatory index.
Adverse events in severe patients
Among the patients with severe COVID-19, 170 had respiratory failure (89.5%), 98 presented MODS (51.6%), 77 observed acute myocardial injury (44.8%), 70 needed mechanical ventilation (36.8%), 61 died (32.1%), 46 were admitted to ICU (24.2%) and 43 emerged AKI (23.5%). Compared with patients with no adverse events, CRP, NLR and D-dimer were significantly higher in patients with adverse events such as acute myocardial injury, respiratory failure, AKI, mechanical ventilation, ICU admission, MODS and death (all p<0.05) (table 3).
Table 3.
The adverse events and laboratory findings in patients with severe COVID-19
| Adverse event | Counts | CRP | NLR | D-dimer | ||||||
| Median (IQR, mg/L) |
U value | P value | Median (IQR) |
U value | P value | Median (IQR, μg/mL) |
U value | P value | ||
| Clinical outcomes | 4.026 | <0.05 | NLR | 4.097 | <0.05 | D-dimer | 4.279 | <0.05 | ||
| Death | 61 (32.1%) | 97.7 (63.6–179.1) | 13.5 (7.3–20.8) | 7.979 (0.495–25.253) | ||||||
| Alive | 129 (67.9%) | 61.4 (26.7–101.5) | 6.7 (3.7–12.4) | 0.604 (0.169–2.877) | ||||||
| Mechanical ventilation | 4.302 | <0.05 | 3.897 | <0.05 | 4.904 | <0.05 | ||||
| Yes | 70 (36.8%) | 92.2 (64.4–168.2) | 12.9 (7.0–19.8) | 5.580 (0.512–22.018) | ||||||
| No | 120 (63.2%) | 59.9 (22.7–99.8) | 6.6 (3.6–11.5) | 0.436 (0.151–2.618) | ||||||
| Intensive care unit admission | 2.955 | <0.05 | 2.925 | <0.05 | 2.688 | <0.05 | ||||
| Yes | 46 (24.2%) | 97.7 (64.4–168.2) | 14.8 (6.2–21.8) | 2.357 (0.498–15.210) | ||||||
| No | 144 (75.8%) | 62.9 (30.2–121.9) | 7.3 (4.0–13.0) | 0.613 (0.210–4.354) | ||||||
| Multiple organ dysfunction syndrome | 4.563 | <0.05 | 4.774 | <0.05 | 5.160 | <0.05 | ||||
| Yes | 98 (51.6%) | 85.5 (59.9–146) | 12.3 (6.2–18.5) | 2.493 (0.495–13.105) | ||||||
| No | 92 (48.4%) | 53.3 (16.9–92.5) | 5.6 (2.9–11.0) | 0.347 (0.144–1.027) | ||||||
| Acute myocardial injury | 3.621 | <0.05 | 3.497 | <0.05 | 4.946 | <0.05 | ||||
| Yes | 77 (44.8%) | 92.2 (59.5–145.8) | 11.7 (5.9–18.6) | 4.596 (0.615–14.730) | ||||||
| No | 95 (55.2%) | 60.8 (22.7–99.8) | 6.7 (3.1–11.9) | 0.392 (0.150–1.611) | ||||||
| Acute kidney injury | 2.892 | <0.05 | 2.506 | <0.05 | 2.709 | <0.05 | ||||
| Yes | 43 (23.5%) | 92.2 (58.4–195.5) | 13.1 (5.8–23.4) | 4.651 (0.495–15.858) | ||||||
| No | 140 (76.5%) | 64.7 (29.7–108.8) | 7.6 (3.8–13.4) | 0.613 (0.232–3.476) | ||||||
| Respiratory failure | 3.052 | <0.05 | 2.272 | <0.05 | 4.640 | <0.05 | ||||
| Yes | 170 (89.5%) | 76.0 (40.8–135.2) | 9.7 (4.4–15.4) | 4.651 (0.495–19.155) | ||||||
| No | 20 (10.5%) | 17.1 (6.7–68.5) | 4.9 (3.0–8.9) | 0.509 (0.151–2.461) | ||||||
Continuous variables were described as median with IQR and analysed by Mann-Whitney test. P values indicate differences of laboratory indicators in severe COVID-19 patients with and without adverse events. P<0.05 was considered statistically significant.
CRP, C reactive protein; NLR, neutrophil-to-lymphocyte ratio.
The risk factors for disease progression
The univariate logistic regression analysis indicated that the progression of COVID-19 was associated with age (OR=3.301; 95% CI 2.318 to 4.701; p<0.001), gender (OR=2.160; 95% CI 1.528 to 3.054; p<0.001), hypertension (OR=2.085; 95% CI 1.419 to 3.062; p<0.001), cardiovascular disease (OR=2.027; 95% CI 1.423 to 2.887; p<0.001), chronic respiratory disease (OR=1.648; 95% CI 1.036 to 2.621; p=0.035), OI (OR=0.985; 95% CI 0.983 to 0.988; p<0.001), PaO2 (OR=0.958; 95% CI 0.949 to 0.967; p<0.001), CRP (OR=1.031; 95% CI 1.025 to 1.0381; p=0.001), ESR (OR=1.057; 95% CI 1.035 to 1.080; p<0.001), NLR (OR=1.282; 95% CI 1.219 to 1.348; p<0.001), PLR (OR=1.005; 95% CI 1.004 to 1.006; p<0.001), SII (OR=1.001; 95% CI 1.001 to 1.001; p<0.001), albumin (OR=0.841; 95% CI 0.809 to 0.874; p<0.001), lymphocyte count (OR=0.922; 95% CI 0.765 to 0.972; p=0.043), D-dimer (OR=1.757; 95% CI 1.403 to 2.201; p<0.001), and PT (OR=1.138; 95% CI 1.023 to 1.265; p=0.017). The multivariate logistic regression analysis indicated that after the adjustment of potential factors including age, gender, hypertension, cardiovascular disease and chronic respiratory disease and so on, the results of multivariate logistic regression analysis showed that CRP (OR=1.015, 95% CI 1.001 to 1.029, p=0.034), NLR (OR=1.105, 95% CI 1.003 to 1.219, p=0.044) and D-dimer (OR=1.286, 95% CI 1.022 to 1.618, p=0.032) were independent risk factors and PaO2 (OR=0.958, 95% CI 0.943 to 0.974, p<0.001) was an independent protective factor for the progression of COVID-19 (table 4)
Table 4.
Logistic analysis of factors related to the progression of patients with COVID-19
| Variables | Univariate | Multivariate | ||
| OR (95% CI) | P value | OR (95% CI) | P value | |
| Age (>65 years) | 3.301 (2.318 to 4.701) | <0.001 | ||
| Gender (male) | 2.160 (1.528 to 3.054) | <0.001 | ||
| Hypertension | 2.085 (1.419 to 3.062) | <0.001 | ||
| Cardiovascular disease | 2.027 (1.423 to 2.887) | <0.001 | ||
| Chronic respiratory disease | 1.648 (1.036 to 2.621) | 0.035 | ||
| Oxygenation index | 0.985 (0.983 to 0.988) | <0.001 | ||
| Arterial oxygen partial pressure | 0.958 (0.949 to 0.967) | <0.001 | 0.958 (0.943 to 0.974) | <0.001 |
| C reactive protein | 1.031 (1.025 to 1.038) | 0.001 | 1.015 (1.001 to 1.029) | 0.034 |
| Serum amyloid A | 1.002 (1.000 to 1.005) | 0.108 | ||
| Erythrocyte sedimentation rate | 1.057 (1.035 to 1.080) | <0.001 | ||
| NLR | 1.282 (1.219 to 1.348) | <0.001 | 1.105 (1.003 to 1.219) | 0.044 |
| PLR | 1.005 (1.004 to 1.006) | <0.001 | ||
| SII | 1.001 (1.001 to 1.001) | <0.001 | ||
| Albumin | 0.841 (0.809 to 0.874) | <0.001 | ||
| Lymphocytes count | 0.922 (0.765 to 0.972) | 0.043 | ||
| D-dimer | 1.757 (1.403 to 2.201) | <0.001 | 1.286 (1.022 to 1.618) | 0.032 |
| Prothrombin time | 1.138 (1.023 to 1.265) | 0.017 | ||
Univariate and multivariate analyses were carried out using a logistic regression model. P<0.05 was considered statistically significant.
NLR, neutrophil-to-lymphocyte ratio; PLR, platelet-to-lymphocyte ratio; SII, systematic immune-inflammatory index.
Predictive performance of laboratory indicators
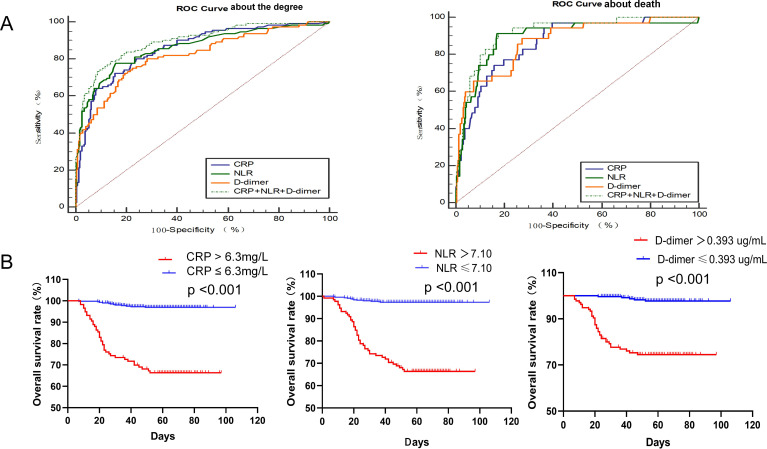
To predict the disease progression of patients with COVID-19, the ROC indicated that CRP, NLR and D-dimer had better accuracy than other laboratory indicators. The AUC of CRP was 0.856 (95% CI 0.813 to 0.898, p<0.001), and 40.5 mg/L was the optimal threshold value with 72.3% as sensitivity and 84.6% as specificity. The AUC of NLR was 0.861 (95% CI 0.816 to 0.906, p<0.001), and the optimal threshold value was 4.65 with 77.7% as sensitivity and 84.2% as specificity. The AUC of D-dimer was 0.819 (95% CI 0.769 to 0.869, p<0.001), and 0.335 µg/mL was taken as the optimal threshold value with 72.3% as sensitivity and 80.4% as specificity. The model that was combined with CRP, NLR and D-dimer exhibited a maximum AUC of 0.894 (95% CI 0.857 to 0.931, p<0.001), and the diagnostic sensitivity was 78.6% and the specificity was 86.3% (table 5 and figure 1).
Table 5.
Diagnostic performance of laboratory indicators in patients with COVID-19
| Model | AUC (95% CI) | SE | P value | Cut-off | Sensitivity | Specificity | Youden index |
| Severe patients | |||||||
| C reactive protein | 0.856 (0.813 to 0.898) | 0.022 | <0.001 | 40.5 | 0.723 | 0.846 | 0.569 |
| Neutrophil count | 0.787 (0.744 to 0.831) | 0.022 | <0.001 | 4.24 | 0.687 | 0.810 | 0.497 |
| Serum amyloid A | 0.658 (0.582 to 0.733) | 0.038 | <0.001 | 34.96 | 1.000 | 0.413 | 0.413 |
| Erythrocyte sedimentation rate | 0.756 (0.683 to 0.829) | 0.037 | <0.001 | 23.84 | 0.782 | 0.687 | 0.469 |
| NLR | 0.861 (0.816 to 0.906) | 0.023 | <0.001 | 4.65 | 0.777 | 0.842 | 0.619 |
| PLR | 0.694 (0.645 to 0.743) | 0.025 | <0.001 | 239.65 | 0.545 | 0.684 | 0.229 |
| SII | 0.789 (0.747 to 0.832) | 0.022 | <0.001 | 969.35 | 0.650 | 0.828 | 0.478 |
| White cell count | 0.721 (0.673 to 0.769) | 0.024 | <0.001 | 6.79 | 0.555 | 0.827 | 0.382 |
| Prothrombin time | 0.730 (0.682 to 0.778) | 0.025 | <0.001 | 13.3 | 0.503 | 0.841 | 0.344 |
| Acivated partial thromboplastin time | 0.425 (0.367 to 0.482) | 0.029 | 0.007 | 40.15 | 0.067 | 0.983 | 0.050 |
| Thrombin time | 0.464 (0.409 to 0.519) | 0.028 | 0.203 | 16.95 | 0.105 | 0.940 | 0.045 |
| Fibrinogen | 0.687 (0.636 to 0.738) | 0.026 | <0.001 | 3.77 | 0.646 | 0.679 | 0.325 |
| D-dimer | 0.819 (0.769 to 0.869) | 0.025 | <0.001 | 0.335 | 0.723 | 0.804 | 0.527 |
| CPR+NLR+D-dimer | 0.894 (0.857 to 0.931) | 0.019 | <0.001 | 0.26 | 0.786 | 0.863 | 0.649 |
| Death of patients | |||||||
| C reactive protein | 0.865 (0.806 to 0.923) | 0.030 | <0.001 | 63.3 | 0.743 | 0.842 | 0.585 |
| Neutrophil count | 0.782 (0.710 to 0.853) | 0.036 | <0.001 | 4.53 | 0.768 | 0.731 | 0.499 |
| Serum amyloid A | 0.606 (0.505 to 0.708) | 0.052 | 0.237 | 50.47 | 0.857 | 0.583 | 0.440 |
| Erythrocyte sedimentation rate | 0.738 (0.619 to 0.858) | 0.061 | 0.002 | 33.50 | 0.688 | 0.741 | 0.429 |
| NLR | 0.894 (0.832 to 0.956) | 0.032 | <0.001 | 7.10 | 0.914 | 0.830 | 0.744 |
| PLR | 0.618 (0.538 to 0.699) | 0.041 | 0.004 | 244.15 | 0.567 | 0.786 | 0.353 |
| SII | 0.752 (0.679 to 0.824) | 0.037 | <0.001 | 1171.40 | 0.691 | 0.770 | 0.461 |
| White cell count | 0.723 (0.643 to 0.803) | 0.041 | <0.001 | 7.46 | 0.571 | 0.808 | 0.379 |
| Prothrombin time | 0.728 (0.652 to 0.804) | 0.039 | <0.001 | 13.3 | 0.615 | 0.761 | 0.376 |
| Acivated partial thromboplastin time | 0.360 (0.275 to 0.445) | 0.043 | <0.001 | 42.70 | 0.020 | 0.988 | 0.008 |
| Thrombin time | 0.554 (0.465 to 0.644) | 0.046 | <0.001 | 17.05 | 0.240 | 0.952 | 0.192 |
| Fibrinogen | 0.545 (0.461 to 0.629) | 0.043 | 0.295 | 3.32 | 0.667 | 0.451 | 0.118 |
| D-dimer | 0.872 (0.810 to 0.934) | 0.032 | <0.001 | 0.393 | 0.886 | 0.726 | 0.612 |
| CPR+NLR+D-dimer | 0.918 (0.873 to 0.962) | 0.023 | <0.001 | 0.07 | 0.914 | 0.823 | 0.737 |
Receiver operating characteristic curve and AUC were carried out to evaluate the value of laboratory indicators predicting severe disease and death of COVID-19. P<0.05 was considered statistically significant.
AUC, area under the receiver operating characteristic curve; NLR, neutrophil-to-lymphocyte ratio; PLR, platelet to lymphocyte ratio; SII, systemic immune-inflammation index.
Figure 1.
The relationship and predictive effect of CRP, NLR and D-dimer on severity and death for patients with COVID-19. (A) Receiver operating characteristics curve of CRP, NLR and D-dimer for the diagnosis of severity and death of patients with COVID-19. (B) The time-dependent risk of COVID-19 patients with lower or higher levels of CRP, NLR, D-dimer using Kaplan-Meier method. CRP, C reactive protein; NLR, neutrophil-to-lymphocyte ratio; ROC, receiver operating characteristics.
To predict death of patients with COVID-19, the ROC indicated that CRP, NLR and D-dimer also possessed better accuracy than other laboratory indicators. The AUC of CRP was 0.865 (95% CI 0.806 to 0.923, p<0.001), and 63.3 mg/L was the optimal threshold value with 74.3% as sensitivity and 84.2% as specificity. The AUC of NLR was 0.894 (95% CI 0.832 to 0.956, p<0.001), and the optimal threshold value was 7.10 with 91.4% as sensitivity and 83.0% as specificity. The AUC of D-dimer was 0.872 (95% CI 0.810 to 0.934, p<0.001), and 0.393 ug/mL was taken as the optimal threshold value with 88.6% as sensitivity and 72.6% as specificity. The model that was combined with CRP, NLR and D-dimer reflected the largest AUC of 0.918 (95% CI 0.873 to 0.962, p<0.001), and the diagnostic sensitivity and specificity were 91.4% and 82.3%, respectively. These results indicated that CRP, NLR and D-dimer were of high diagnostic value in predicting disease severity and death in patients with COVID-19, especially when all of them were combined (table 5 and figure 1).
Prognosis analysis
The Kaplan-Meier curve and log-rank test demonstrated that the survival rate of COVID-19 patients with high CRP levels (66.7%) was significantly lower than that of COVID-19 patients with low CRP levels (97.1%), and the difference was statistically significant (p<0.001). The survival rate of COVID-19 patients with a high NLR level (66.7%) was significantly lower than that of COVID-19 patients with a low NLR level (97.3%), and the difference was statistically significant (p<0.001). The survival rate of COVID-19 patients with high D-dimer levels (74.8%) was significantly lower than that of COVID-19 patients with low D-dimer level (98.1%), and the difference was statistically significant (p<0.001). These results indicated that the patients with high levels of CRP, NLR and D-dimer had a worse prognosis (figure 1).
Discussion
SARS-CoV-2 is one of the three coronaviruses known to cause serious diseases among humans. It emerged in the pandemic of COVID-19, which has been prevalent in China, the USA, Japan, South Korea and other places worldwide, seriously threatening the health of humans.16–18 When the spike protein of SARS-CoV-2 is tightly bound to ACE2 of human, the protein of TMPRSS2 increases the activity of the spike protein to promote the virus to enter into the tissues and organs where ACE2 is highly expressed, such as the alveolar, heart, intestinal tract and blood vessel and so on, and then different symptoms may occur.19 20 Most patients are mild performance; however, in some patients, severe pulmonary oedema, coagulation dysfunction and MODS may occur within a short period, eventually leading to death. Early identification of severe patients can effectively allocate medical resources and provide adequate interventions to improve the survival chances of patients. Laboratory indicators can effectively anticipate disease progression and adverse events in patients with COVID-19.
In our study, we compared the patients with and without severe COVID-19, and the results indicated that more elderly patients with comorbidity were found in severe cases. A research of 1000 cases in the USA showed that patients who required ICU monitoring and treatment were older and predominantly male, and usually had baseline comorbidities, including hypertension, cardiovascular disease and so on, which was consistent with our results.21 Besides, most studies found that male patients with COVID-19 were more likely to experience severe symptoms and death than female patients, but this phenomenon was rarely discussed. Takahashi et al22 indicated that male patients had more poor T cell activation than female patients during SARS-CoV-2 infection. Sama et al23 reported that plasma ACE2 concentration in male patients was significantly higher than that in females, thus increasing the COVID-19 infection probability in male patients. Acharya et al24 attributed it to the absence of the X chromosome, high levels of testosterone that inhibit antibody production and terrible habits in men such as increased smoking and alcohol consumption and low levels of handwashing. Therefore, all of these results indicated that elderly men with underlying diseases were more likely to develop severe COVID-19, and their condition is more serious and needs more attention and treatment.
Excessive inflammation and imbalanced immune status are associated with COVID-19 disease progression and poor prognosis.25 Under normal physiological conditions, inflammatory and anti-inflammatory responses are in a balanced state to protect the body against pathogen invasion and maintain normal physiological functions. When the inflammatory response is too strong and beyond our bodies’ capacity, it will cause extensive damage to cells and tissue. CRP is an acute reactive protein that increases rapidly in conditions such as inflammation and infection. In particular, the increase in CRP is more obvious when the body has a bacterial infection, while the CRP generally does not increase or slightly increases when the body has a viral infection. However, Xu et al26 analysed 187 cases of patients with COVID-19 in Wuhan, reporting that CRP in severe patients was about 10 times higher than that in non-severe patients. Yarza et al27 reported that COVID-19 patients with respiratory failure presented higher levels of CRP than patients without respiratory failure. Our study also showed that the CRP level of the severe COVID-19 group was significantly higher than that of the non-severe COVID-19 group. The reasons for the significant increase of CRP in patients with severe COVID-19 may be as follows: first, patients with severe COVID-19 have higher levels of inflammatory factors such as interleukin-6 (IL-6), which can lead to the synthesis of CRP by hepatocytes. Also, severe patients are often accompanied by tissue and cell damage, which also increases CRP levels. Finally, severe patients often have bacterial infections and ultimately increase the level of CRP. Neutrophils are the most abundant part of white blood cells. In general, when the body is infected, it can rapidly increase and move to the infected site, killing pathogens by releasing reactive oxygen species and using neutrophil extracellular traps, which is a spider web structure outside the neutrophil with a variety of enzymes attached to it.28 However, Xie et al29 showed that when compared with patients with non-severe COVID-19, neutrophils in patients with severe COVID-19 were slightly elevated at 4.96×10∧9/L on admission but still within the normal range. Our research also found that neutrophils in patients with severe COVID-19 were not significantly increased. The reasons may be as follows: first, the virus can inhibit the haematopoietic function of bone marrow through direct or indirect action. Second, when neutrophils kill the virus, they will cause damage, destruction and excessive consumption of neutrophils. Thus, neutrophils were not significantly elevated in patients with severe COVID-19. Lymphocytes are important cells that kill SARS-CoV-2, and they significantly reduce in patients with severe COVID-19. When the inflammatory response is too strong, neutrophils can damage our tissues and inhibit the function of lymphocytes and induce T cell apoptosis. The number of blood lymphocytes can continue to decline when SARS-CoV-2 invades the alveoli because this process can cause tissue damage and chemokines release that induces lymphocytes to transfer from blood to lung tissue. Thus, the significant decrease of lymphocytes in the blood may be related to the apoptosis and redistribution of lymphocytes. NLR is a systemic inflammatory indicator derived from the blood count, reflecting the balance between the number of neutrophils and lymphocytes. The inflammatory response caused by SARS-CoV-2 can promote an increase in neutrophils and a decrease in lymphocytes, but sometimes their changes are not significant. However, NLR could significantly increase in COVID-19 disease by calculating the ratio of neutrophils to lymphocytes. NLR, which is similar to the albumin–globulin ratio, is an indirect laboratory indicator of inflammatory. Many studies have shown that NLR is an excellent indicator of inflammation in judging the disease progression and poor prognosis of COVID-19, and it can better reflect the inflammation situation in patients with COVID-19. Yang et al6 showed that NLR had a high accuracy in predicting patients with severe COVID-19, and a high level of NLR indicated severe disease and poor prognosis. Qin et al30 reported that the level of NLR in patients with severe COVID-19 was significantly higher than that in non-severe patients, which was consistent with our study. Besides, inflammatory indicators such as CRP, SAA, ESR, PLR and SII were significantly higher and lymphocyte levels were lower in patients with severe COVID-19 in our study. The above-mentioned studies indicated that patients with severe COVID-19 have a more dramatic inflammatory response in vivo, which aggravate the damage of cell and tissue to exacerbate the disease. D-dimer is derived from the cross-linked fibrin clot dissolved by plasmin. The inflammatory response can significantly increase the level of D-dimer. Excessive systemic inflammation can destroy the microvascular system, thus activating the coagulation system abnormally, which is manifested as systemic microvasculitis and microthrombus formation.25 31 Wu et al4 reported that the increased D-dimer was a predictor of severe disease and death in patients with COVID-19. Tang et al32 analysed 183 patients with COVID-19 and found that D-dimer was increased in patients died by COVID-19. Chen et al33 found that patients with severe COVID-19 had a higher level of D-dimer than non-severe patients. Our study found a significant increase of D-dimer in patients with severe COVID-19, which was consistent with the previous conclusions, suggesting that increasing D-dimer played an important predictive role in disease progression and death in patients with COVID-19.
To further clarify the factors leading to the progression of severe COVID-19, the multivariate logistic analysis was conducted. The results showed that CRP, NLR and D-dimer were still independent risk factors after adjusting for age, gender and underlying diseases and so on. The ROC showed that CRP, NLR and D-dimer were highly accurate in predicting severe COVID-19, and the AUC was the highest when the three were combined, with excellent sensitivity and specificity. This study shows that the above-mentioned laboratory indicators could effectively help clinicians to judge the severity of patients with COVID-19 quickly and then take corresponding measures. Besides, age is one of the important factors affecting the progression of patients with COVID-19, the univariate logistic regression analysis was conducted and showed that age over 65 years was a risk factor for progression to severe COVID-19 (OR=3.301, p<0.001), suggesting that older age is associated with severity of COVID-19 disease. However, the results of the multivariate analysis did not show age as an independent risk factor for the progression of COVID-19. This could be the result of the influence of multiple confounding factors and the limited sample size in our study. In terms of adverse events, patients with severe COVID-19 had a high incidence of mortality, mechanical ventilation, ICU admission, MODS, acute myocardial injury, respiratory failure and AKI. Compared with patients with no adverse events, CRP, NLR and D-dimer were significantly higher in patients with adverse events. These results suggested that there were more organ failures in patients with severe COVID-19, which might be associated with an inflammatory response. The ROC showed that CRP, NLR and D-dimer were highly accurate in predicting the death of COVID-19, and the AUC was the highest when the three were combined. Kaplan-Meier analysis showed that the survival time and survival rate of COVID-19 patients with a high level of CRP, NLR or D-dimer were significantly reduced. These results indicated that high levels of CRP, NLR and D-dimer could effectively predict the poor prognosis of patients with COVID-19; hence, we need to pay more attention to COVID-19 patients with abnormally high levels of these indicators. And active control of inflammatory storms might be effective in reducing the incidence and mortality of severe patients.
There were still some limitations. First, this study was a single-centre retrospective cohort study, and more multicentre prospective studies were needed to verify our conclusions. Second, this study lacked dynamic monitoring of CRP, NLR and D-dimer. In addition, inflammatory markers such as IL-1, IL-6, tumor necrosis factor alpha (TNF-α) and interferon gamma (IFN-γ) were not included in this study. Finally, the majority of patients in our study were ordinary, severe or critically ill patients but less mild patients, so the selection bias might still exist.
Conclusion
In summary, the combination of CRP, NLR, and D-dimer could be an effective predictor for the aggravation and death in patients with COVID-19. The abnormal expression of these indicators might suggest a strong inflammatory response and multiple adverse events in patients with severe COVID-19. Therefore, inflammatory responses in patients with severe COVID-19 required special attention and active treatment.
Supplementary Material
Acknowledgments
The authors would like to thank the patients from the Wuhan No.7 Hospital who participated in the study.
Footnotes
QW, JC and JS contributed equally.
Contributors: QW, JC and JS conceived the study and designed the trial. JL obtained institutional review board approval and financial support to carry out this research. QW, JC and JS collected the data, interpreted results, drafted, revised and wrote the final manuscript. Please note cofirst authorship of QW, JC and JS given the equal contributions of these authors. JW and Y-qY designed and performed statistical analysis. YW and W-bL collected data, contributed to writing the manuscript and helped with revising the manuscript. H-PZ and J-pW contributed to interpreting results, writing and revising the manuscript. X-yW and Z-aL contributed to designing the trial, obtained funding for the project, interpreting results, writing and revising the manuscript.
Funding: This work was supported by the National Natural Science Foundation of China (grant numbers 81800494), the National Major Scientific and Technological Special Project for Significant New Drugs Development (grant numbers 2020Z×09201007) and Hubei central government guides local science and technology development special project (grant numbers 2020ZYYD012).
Competing interests: None declared.
Patient and public involvement: Patients and/or the public were not involved in the design, or conduct, or reporting, or dissemination plans of this research.
Provenance and peer review: Not commissioned; externally peer reviewed.
Data availability statement
Data are available from the corresponding author on reasonable request.
Ethics statements
Patient consent for publication
Not applicable.
Ethics approval
This study was approved by the Ethics Committee of Zhongnan Hospital of Wuhan University, China (grant numbers 2020082K).
References
- 1.Li Q, Guan X, Wu P, et al. Early transmission dynamics in Wuhan, China, of novel coronavirus-infected pneumonia. N Engl J Med 2020;382:1199–207. 10.1056/NEJMoa2001316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cui J, Li F, Shi ZL. Origin and evolution of pathogenic coronaviruses. Nat Rev Microbiol 2019;17:181–92. 10.1038/s41579-018-0118-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lu R, Zhao X, Li J, et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet 2020;395:565–74. 10.1016/S0140-6736(20)30251-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu C, Chen X, Cai Y, et al. Risk factors associated with acute respiratory distress syndrome and death in patients with coronavirus disease 2019 pneumonia in Wuhan, China. JAMA Intern Med 2020;180:934–43. 10.1001/jamainternmed.2020.0994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang G, Wu C, Zhang Q, et al. C-Reactive protein level may predict the risk of COVID-19 aggravation. Open Forum Infect Dis 2020;7:ofaa153. 10.1093/ofid/ofaa153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang AP, Liu JP, Tao WQ, et al. The diagnostic and predictive role of NLR, d-NLR and PLR in COVID-19 patients. Int Immunopharmacol 2020;84:106504. 10.1016/j.intimp.2020.106504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Paliogiannis P, Zinellu A, Scano V, et al. Laboratory test alterations in patients with COVID-19 and non COVID-19 interstitial pneumonia: a preliminary report. J Infect Dev Ctries 2020;14:685–90. 10.3855/jidc.12879 [DOI] [PubMed] [Google Scholar]
- 8.Zhou F, Yu T, Du R, et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet 2020;395:1054–62. 10.1016/S0140-6736(20)30566-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Léonard-Lorant I, Delabranche X, Séverac F, et al. Acute pulmonary embolism in patients with COVID-19 at CT angiography and relationship to D-dimer levels. Radiology 2020;296:E189–91. 10.1148/radiol.2020201561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Paliogiannis P, Mangoni AA, Dettori P, et al. D-Dimer concentrations and COVID-19 severity: a systematic review and meta-analysis. Front Public Health 2020;8:432. 10.3389/fpubh.2020.00432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.National Health Commission of the People’s Republic of China . The new coronavirus pneumonia prevention and control guideline (7th edition). Available: http://www.nhc.gov.cn/yzygj/s7653p/202003/46c9294a7dfe4cef80dc7f5912eb1989.shtml
- 12.Shi S, Qin M, Shen B, et al. Association of cardiac injury with mortality in hospitalized patients with COVID-19 in Wuhan, China. JAMA Cardiol 2020;5:802–10. 10.1001/jamacardio.2020.0950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Khwaja A. KDIGO clinical practice guidelines for acute kidney injury. Nephron Clin Pract 2012;120:c179–84. 10.1159/000339789 [DOI] [PubMed] [Google Scholar]
- 14.Chiumello D, Chevallard G, Gregoretti C. Non-Invasive ventilation in postoperative patients: a systematic review. Intensive Care Med 2011;37:918–29. 10.1007/s00134-011-2210-8 [DOI] [PubMed] [Google Scholar]
- 15.Murray MJ, Coursin DB. Multiple organ dysfunction syndrome. Yale J Biol Med 1993;66:501–10. [PMC free article] [PubMed] [Google Scholar]
- 16.Holshue ML, DeBolt C, Lindquist S, et al. First case of 2019 novel coronavirus in the United States. N Engl J Med 2020;382:929–36. 10.1056/NEJMoa2001191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nishiura H, Kobayashi T, Yang Y, et al. The rate of Underascertainment of novel coronavirus (2019-nCoV) infection: estimation using Japanese passengers data on evacuation flights. J Clin Med 2020;9:419. 10.3390/jcm9020419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lin X, Gong Z, Xiao Z, et al. Novel coronavirus pneumonia outbreak in 2019: computed tomographic findings in two cases. Korean J Radiol 2020;21:365–8. 10.3348/kjr.2020.0078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hoffmann M, Kleine-Weber H, Schroeder S, et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 2020;181:271–80. 10.1016/j.cell.2020.02.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dai Y-J, Hu F, Li H, et al. A profiling analysis on the receptor ACE2 expression reveals the potential risk of different type of cancers vulnerable to SARS-CoV-2 infection. Ann Transl Med 2020;8:481. 10.21037/atm.2020.03.61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Argenziano MG, Bruce SL, Slater CL, et al. Characterization and clinical course of 1000 patients with coronavirus disease 2019 in New York: retrospective case series. BMJ 2020;369:m1996. 10.1136/bmj.m1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Takahashi T, Ellingson MK, Wong P, et al. Sex differences in immune responses that underlie COVID-19 disease outcomes. Nature 2020;588:315–20. 10.1038/s41586-020-2700-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sama IE, Ravera A, Santema BT, et al. Circulating plasma concentrations of angiotensin-converting enzyme 2 in men and women with heart failure and effects of renin-angiotensin-aldosterone inhibitors. Eur Heart J 2020;41:1810–7. 10.1093/eurheartj/ehaa373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Acharya Y, Pant S, Gyanwali P, et al. Gender disaggregation in COVID-19 and increased male susceptibility. J Nepal Health Res Counc 2020;18:345–50. 10.33314/jnhrc.v18i3.3108 [DOI] [PubMed] [Google Scholar]
- 25.Xu Z, Shi L, Wang Y, et al. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir Med 2020;8:420–2. 10.1016/S2213-2600(20)30076-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu B, Fan CY, Wang AL, et al. Suppressed T cell-mediated immunity in patients with COVID-19: a clinical retrospective study in Wuhan, China. J Infect 2020;81:e51–60. 10.1016/j.jinf.2020.04.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yarza R, Bover M, Paredes D, et al. SARS-CoV-2 infection in cancer patients undergoing active treatment: analysis of clinical features and predictive factors for severe respiratory failure and death. Eur J Cancer 2020;135:242–50. 10.1016/j.ejca.2020.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu J, Dong Z. Neutrophil extracellular traps in ischemic AKI: new way to kill. Kidney Int 2018;93:303–5. 10.1016/j.kint.2017.09.031 [DOI] [PubMed] [Google Scholar]
- 29.Xie L, Wu Q, Lin Q, et al. Dysfunction of adaptive immunity is related to severity of COVID-19: a retrospective study. Ther Adv Respir Dis 2020;14:1753466620942129. 10.1177/1753466620942129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Qin C, Zhou L, Hu Z, et al. Dysregulation of immune response in patients with coronavirus 2019 (COVID-19) in Wuhan, China. Clin Infect Dis 2020;71:762–8. 10.1093/cid/ciaa248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Levi M, van der Poll T. Coagulation and sepsis. Thromb Res 2017;149:38–44. 10.1016/j.thromres.2016.11.007 [DOI] [PubMed] [Google Scholar]
- 32.Tang N, Li D, Wang X, et al. Abnormal coagulation parameters are associated with poor prognosis in patients with novel coronavirus pneumonia. J Thromb Haemost 2020;18:844–7. 10.1111/jth.14768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen G, Wu D, Guo W, et al. Clinical and immunological features of severe and moderate coronavirus disease 2019. J Clin Invest 2020;130:2620–9. 10.1172/JCI137244 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data are available from the corresponding author on reasonable request.