Abstract
Background:
Although the gastrointestinal stromal tumor (GIST) genotype is not currently included in risk-stratification systems, a growing body of evidence shows that the pathogenic variant (PV) type and codon location hold a strong prognostic influence on recurrence-free survival (RFS). This information has particular relevance in the adjuvant setting, where an accurate prognostication could help to better identify high-risk tumors and guide clinical decision-making.
Materials and Methods:
Between January 2005 and December 2020, 96 patients with completely resected GISTs harboring a KIT proto-oncogene receptor tyrosine kinase (KIT) exon 11 PV were included in the study. We analyzed the type and codon location of the PV according to clinicopathological characteristics and clinical outcome; the metastatic sites in relapsed patients were also investigated.
Results:
Tumors harboring a KIT exon 11 deletion or deletion/insertion involving the 557 and/or 558 codons, showed a more aggressive clinical behavior compared with tumors carrying deletion/deletion/insertion in other codons, or tumors with duplication/insertion/single-nucleotide variant (SNV) (7-year RFS: 50% versus 73.1% versus 88.2%, respectively; p < 0.001). Notably, among 18 relapsed patients with 557 and/or 558 deletion or deletion/insertion, 14 patients (77.8%) harbored deletions simultaneously involving 557 and 558 codons, while only 4 patients (22.2%) harbored deletions involving only 1 of the 557/558 codons. Thus, when 557 or 558 deletions occurred separately, the tumor showed a prognostic behavior similar to the GIST carrying deletions outside the 557/558 position. Remarkably, patients with GISTs stratified as intermediate risk, but carrying the 557/558 deletion, showed a similar outcome to the high-risk patients with tumors harboring deletions in codons other than 557/558, or duplication/insertion/SNV.
Conclusion:
Our data support the inclusion of the PV type and codon location in routine risk prediction models, and suggest that intermediate-risk patients whose GISTs harbor 557/558 deletions may also need to be treated with adjuvant imatinib like the high-risk patients.
Keywords: 557/558 deletion, gastrointestinal stromal tumor, KIT proto-oncogene receptor tyrosine kinase, platelet-derived growth factor receptor alpha, prognostic biomarker
Introduction
Gastrointestinal stromal tumors (GISTs) are a subgroup of mesenchymal tumors originating from the interstitial cells of Cajal, which can arise from any part of the gastrointestinal tract, most frequently from the stomach and small intestine, characterized by the expression of the cell-surface transmembrane receptor KIT with tyrosine kinase activity in approximately 95% of tumors.1,2
Tumor mutational status is biologically and clinically important in GISTs and make this tumor a paradigmatic model of oncogene addiction. Constitutively activating mutations in the gene coding for KIT proto-oncogene receptor tyrosine kinase (KIT) or in platelet-derived growth factor receptor alpha (PDGFRA) oncogene are alternative and mutually exclusive, highlighting their important role in the pathogenesis of GISTs.3,4 KIT mutations can be identified in 70–80% of GISTs: they are found most commonly (70% of cases) in exon 11, encoding the intracellular juxtamembrane domain of the receptor, followed in the frequency of incidence by exon 9 (12–15%), encoding the extracellular ligand-binding domain. With decreasing frequency, a subset of KIT gene mutations occurs in exons 13 or 17. 1 Around 5–10% of GISTs present molecular alterations in PDGRFA, often in exon 18 (80%) and more rarely in exon 12 (10–15%) or 14 (1–5%). 3 Finally, 10–15% of GISTs in the adult population and 85% of pediatric GISTs are ‘PDGRFA/KIT wild type (WT)’, tumors that lack detectable PDGRA and KIT mutations but often harbor genomic or epigenetic aberrations in subunits of the succinate dehydrogenase (SDH) complex. 4 SDH is an enzyme complex characterized by four subunits (SDH A,B,C,D), involved in the Krebs cycle and electron transport of oxidative phosphorylation. The SDH-deficient GISTs, characterized by the loss of SDHB protein, are mostly associated with genetic syndromes such as the Carney triad syndrome and Carney–Stratakis syndrome: 5 the SDH-competent subgroup, instead, could be associated with alterations in neurofibromatosis type 1 (NF1) gene, BRAF, or RAS, referred to as RAS-pathway mutant GISTs. 6 The remaining cases, usually referred to as KIT/PDGFRA/SDH/RAS-P WT or quadruple WT GISTs, can show rare mutational events in Fibroblast Growth Factor Receptor (FGFR) or Neurotrophic Tyrosine Receptor Kinase (NRTK).4,7 –9
KIT and PDGFRA mutations represent known prognostic and predictive biomarkers for GISTs, and are useful in driving the choice of therapy in the adjuvant and metastatic setting.10,11 Patients with GISTs with a tumor harboring the KIT exon 11 mutation make up the most frequent subgroup and, in the metastatic setting, are usually highly sensitive to the standard dose of imatinib; 12 GISTs with KIT exon 9 mutation are, instead, more sensitive to the increased dose of imatinib 800 mg/day.13,14 Regarding PDGFRA, despite most of the mutations being imatinib-sensitive (D846Y, N848K, Y849K, and HDSN845-848P), the D842V mutation in exon 18 is known to be responsible for primary resistance to the targeted agents currently available, and clinical trials with novel tyrosine kinase inhibitors (TKIs) are ongoing to find therapeutic agents able to overcome this resistance. 15 Finally, in WT GISTs, the absence of any molecular targetable alteration makes these tumors less responsive to standard TKIs. 5
In recent years, there has been a special interest in exploring the prognostic influence of types and position of KIT exon 11 mutations on recurrence-free survival (RFS). The commonly used risk-stratification model did not take into account the genotyping; nevertheless, evidence to support the prognostic role of mutational status is growing. In patients with localized GISTs completely resected in a previous study, predominantly retrospective, showed that KIT exon 11 deletions, and deletions affecting KIT exon 11 codons 557 and/or 558, were associated with poor outcome.15 –17 In the metastatic setting, clinical studies reported that GISTs with mutations in KIT exon 11 exhibited a high predisposition to liver metastasis18,19 and previous research showed that 557/558 deletions facilitated the preferential spread to the liver, compared with other KIT exon 11 pathogenic variants (PVs), explaining the consequent more aggressive clinical behavior. 20
This information has particular relevance in the adjuvant setting, where an efficient therapy is available and an accurate prognostication could help clinicians to better identify high-risk tumors and to guide clinical decision-making.
Based on this background, we conducted a population-based, single-institution study. Our research aimed to investigate the effect of the exact type and codon location of KIT exon 11 critical mutations on clinical outcome, and the potential association between PV and metastatic sites in relapsed patients.
Materials and methods
Study population
Clinicopathological variables and mutational status data were analyzed from a GIST system database prospectively collected in an Italian referral center for the diagnosis and treatment of soft tissue sarcoma and GIST: the ‘Sicilian Regional Center for the Prevention, Diagnosis and Treatment of Rare and Heredo-Familial Tumors’ of the Section of Medical Oncology at the University Hospital Policlinico Paolo Giaccone of Palermo. In this retrospective study all consecutive patients with localized GISTs completely resected, referred to the center between January 2005 and December 2020, were included.
Patients with metastatic or inoperable primary GISTs, or patients treated with prior neoadjuvant imatinib or adjuvant imatinib therapy, were excluded. To study the clinicopathological characteristics of patients with GISTs according to mutational status, information was collected on the gender, age, site of origin of primary tumors, primary tumor diameter and mitosis, Miettinen–Lasota risk categories classification, 21 and KIT or PDGFRA PVs. The effect of KIT exon 11 mutations type on RFS was evaluated.
Staging was performed after surgery with contrast-enhanced computed tomography of the chest, abdomen, and pelvis. Blood biochemical analyses, blood cell counts, and abdominal computed tomography were assessed periodically. In KIT exon 11-mutated patients with relapsed tumors after curative surgery, tumor metastatic sites (i.e. liver, peritoneum, or either liver and peritoneum) were described.
All the patients provided written informed consent for the collection of the clinical, pathological, and genetic information required by the study. All data were anonymously recorded. All clinical data were available during follow-up. This study (G-Land 2017, approval number 01032017) was conducted in accordance with the Declaration of Helsinki and good clinical practice guidelines, and was approved by the ethics committee (Comitato Etico Palermo 1) of the university-affiliated hospital University Hospital Policlinico Paolo Giaccone
Mutation analysis
The GIST diagnosis was made based on histopathologic assessment and immunohistochemical staining for CD117 antigen expression by local pathologists with special expertise in GIST and soft tissue sarcoma. The pathologists also reported tumor mitoses from 50 high-power fields (HPFs) and diameter lesions. All GISTs were centrally examined for somatic mutations in KIT (exons 9, 11, 13, and 17) and PDGFRA (exons 12, 14, and 18) by polymerase chain reaction amplification, and direct Sanger sequencing. Samples scoring negative were further profiled by using a targeted next-generation sequencing panel for the presence of hot spot mutations in H/K/N RAS, BRAF, NTRK; according to the aim of the study, only the KIT exon 11 PVs were included in the outcome analyses.
Statistical analysis
Clinicopathological variables and PV type and codon location were evaluated for KIT exon 11 genetic subgroups of localized patients, according to critical mutations. The comparison between subgroups was made with Pearson’s chi-square test and ANOVA test.
RFS was measured between the date of surgery and the date of first documentation of GIST recurrence or death, censoring patients who were alive without recurrence on the date of the last follow-up. RFS between groups was compared using the Kaplan–Meier method and log-rank test. To identify independent prognostic factors for RFS, univariate and multivariate Cox proportional hazard regression models were built.
All tests were performed with a significance level of p = 0.05. Statistical analyses were conducted using IBM SPSS Statistics for Windows Version 27.0 (IBM Corporation, Armonk, NY, USA).
Results
Clinicopathological characteristics of KIT exon 11-mutated patients according to critical mutations
We chose to investigate KIT exon 11-mutated patients because they represent the largest molecular subgroup of GISTs, and are characterized by wide variability in PV types and clinical behavior.
Between January 2005 and December 2020, 96 consecutive patients with GISTs with localized disease, harboring a KIT exon 11 PV, were included in the study.
The patients were classified based on the KIT exon 11 PV as follows: (a) KIT exon 11 deletion or deletion/insertion involving 557 and/or 558 codons (named ‘Del-557/558’); (b) deletion or deletion/insertion in codons other than 557 and/or 558 (named ‘Del-No-557/558’); (c) duplication, insertion, or SNV (named ‘No-Del’).
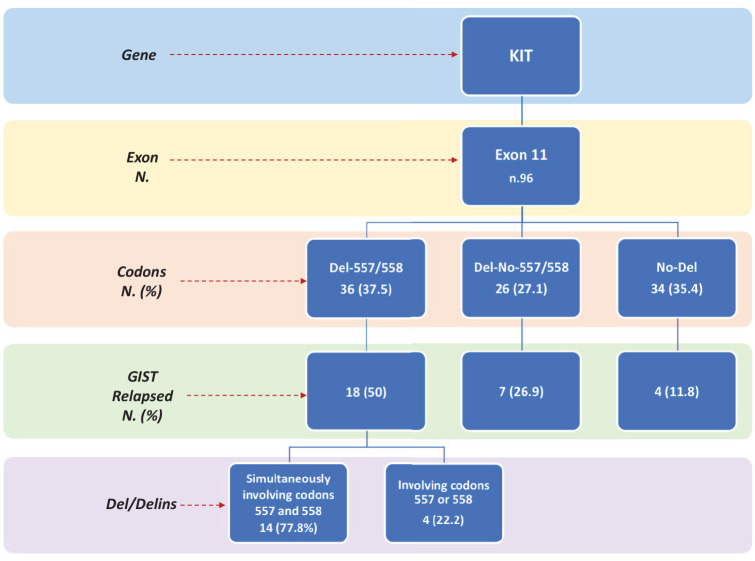
A total of 36 patients were in the Del-557/558 group (37.5%), 26 patients in the Del-No-557/558 group (27.1%), and 34 patients in the No-Del group (35.4%) (Figure 1).
Figure 1.
Diagram of mutational status and outcomes of the patients with KIT exon 11 GISTs included in the study.
Del, deletions; Delins, deletion/insertions; GIST, gastrointestinal stromal tumor; KIT, KIT proto-oncogene receptor tyrosine kinase.
To highlight the prognostic impact of mutational status on the natural history of GISTs, we analyzed the clinicopathological characteristics of patients with GISTs according to KIT exon 11 critical mutations (Table 1).
Table 1.
Clinical and pathological characteristics of patients with localized GISTs according to the type and location of KIT exon 11 critical mutations.
| Total KIT exon 11 patients | KIT exon 11 subgroups | p value * | |||
|---|---|---|---|---|---|
| Del-557/558 n (%) | Del-No-557/558 n (%) | No-Del n (%) | – | ||
| N.patients | 96 | 36 (37.5) | 26 (27.1) | 34 (35.4) | – |
| Gender | |||||
| Male | 56 (58.3) | 24 (60.6) | 13 (50) | 19 (55.9) | NS |
| Female | 40 (41.7) | 12 (31.4) | 13 (50) | 15 (44.1) | |
| Age at diagnosis (years) | |||||
| Median | 60 | 59 | 56.5 | 63.5 | NS |
| Mean | 59 | 58 | 53.9 | 61.1 | |
| Range | 33–82 | 33–76 | 33–67 | 39–82 | |
| Site of origin | |||||
| Gastric | 58 (60.4) | 27 (75) | 12 (46.1) | 19 (55.9) | 0.03 |
| Small bowel/other | 38 (39.6) | 9 (25) | 14 (53.9) | 15 (44.1) | |
| Baseline diameter | |||||
| ⩽5 cm | 41 (42.7) | 10 (27.8) | 16 (61.5) | 15 (44.1) | 0.02 |
| >5 cm | 55 (57.3) | 26 (72.2) | 10 (38.5) | 19 (55.9) | |
| Baseline mitosis | |||||
| ⩽5/50 HPF | 51 (53.1) | 13 (36.1) | 15 (57.7) | 23 (67.6) | 0.02 |
| >5/50 HPF | 45 (46.9) | 23 (63.9) | 11 (42.3) | 11 (32.4) | |
| M–L risk categories | |||||
| Very low/low | 22 (22.9) | 3 (8.3) | 10 (38.5) | 9 (26.5) | 0.008 |
| Intermediate | 35 (36.5) | 13 (36.2) | 12 (46.1) | 10 (29.4) | |
| High | 39 (40.6) | 20 (55.5) | 4 (15.4) | 15 (44.1) | |
Comparison Del-557/558 versus Del-No-557/558 versus No-Del.
Del, deletion; Del-557/558, KIT exon 11 deletion or deletion/insertions involving 557 and/or 558 codons; Del-No-557/558, KIT exon 11 deletion or deletion/insertions in codons other than 557 and/or 558; Delin, deletion/insertion; GIST, gastrointestinal stromal tumor; HPF, high-power field; KIT, KIT proto-oncogene receptor tyrosine kinase; M–L, Miettinen–Lasota; No-Del, KIT exon 11 duplication, insertion, or single nucleotide variant; NS, not significant.
The patients with 557 and/or 558 codon deletion or deletion/insertion, were mainly men (60.6%), more frequently with gastric GISTs (75%), primitive tumor diameter >5 cm (72.2%), and mitotic index >5/50 HPF (63.9%). The patients with other KIT exon 11 PVs, compared with the Del-557/558 group, less frequently showed GISTs of gastric origin (Del-No-557/558: 46.1%; No-Del: 55.9%; p = 0.03), but also a lower number of large primitive tumors with baseline diameter >5 cm (Del-No-557/558: 38.5%; No-Del: 55.9%; p = 0.02), and high median mitotic rate (Del-No-557/558: 42.3%; No-Del: 32.4%; p = 0.02). Therefore, patients with tumors carrying 557 and/or 558 codon deletion or deletion/insertion were stratified as high-risk GISTs in 55.5% of cases, while only 15.4% of patients with deletions in other KIT exon 11 codons, and 44.1% of patients with duplication, insertion, or SNV, were included in the high-risk category (p = 0.008) (Table 1).
Outcome analysis
The outcome investigated was RFS. The median follow-up time of the 96 KIT exon 11-mutated patients was 92 months (range, 10–190 months). RFS rate at 7 years was 69.8% [median 134.7 months; Confidence Interval (CI), 118.6–150.8]. During follow up, a total of 29 RFS events (recurrence or death) were observed (30.2%): 18 events in 36 Del-557/558 patients (50%); 7 events in 26 Del-No-557/558 (26.9%), and 4 events in 34 No-Del patients (11.8%).
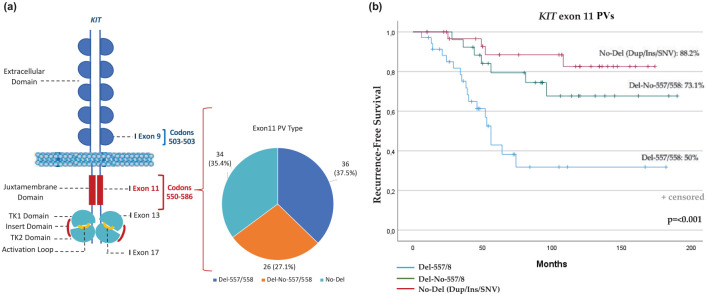
When RFS between the three groups was compared, patients with GISTs with deletion or deletion/insertion of codons 557 and/or 558 showed a worse outcome than any other KIT exon 11-mutated patients. At the same time, however, patients whose GISTs harbored a KIT exon 11 deletion or deletion/insertion in codons other than 557 and/or 558, had less favorable outcomes than patients with duplication, insertion, or SNV [Del-557/558 versus Del-No-557/558 versus No-Del: 7-year RFS, 50% versus 73.1% versus 88.2%; median RFS (mRFS), 86.9 months (95% CI, 60.1–13.7) versus 148.02 months (95% CI, 121.8–74.2) versus 155 months (95% CI, 137.7–172.2), respectively; p < 0.001] (Figure 2).
Figure 2.
(a) Schematic KIT receptor structure and locations of activating mutational hotspots. The KIT functional domains include five immunoglobulin-like domains (extracellular domain), juxtamembrane domain, TK1 domain, insert domain, TK2 domain, and activation loop. Number of patients in the three groups of KIT exon 11 PVs: (i) deletion or deletion/insertion in codons 557 and/or 558 (‘Del-557/558’); (ii) deletion or deletion/insertion in codons other than 557 and/or 558 (‘Del-No-557/558’); (iii) duplication, insertion, or SNV (‘No-Del’). (b) RFS in KIT exon 11 patients according to PV type and location.
KIT, KIT proto-oncogene receptor tyrosine kinase; PV, pathogenic variant; RFS, recurrence-free survival; SNV, single nucleotide variant; TKI, tyrosine kinase 1; TK2, tyrosine kinase 2.
Univariate and multivariate Cox proportional hazard regression models were built to identify the independent prognostic factors for RFS.
The following factors were found to be statistically significantly associated with RFS in univariable analyses: diameter of primary tumor >5 cm [Hazard Ratio (HR): 4.72; 95% CI, 1.91–11.66; p < 0.001]; mitosis >5/50 HPF (HR: 3.34; 95% CI, 1.57–7.10; p = 0.002); no gastric site of origin (HR: 2.13; 95% CI, 1.03–4.43; p = 0.03); risk categories (HR: 4.67; 95% CI, 1.89–11.54; p = 0.001), and KIT exon 11 deletions or deletion/insertion involving codons 557 and/or 558 (HR: 0.19; 95% CI, 0.08–0.42; p < 0.001).
In the final multivariable Cox regression model, tumor diameter (HR: 2.76; 95% CI, 1.01–7.57; p = 0.04), site of origin (HR: 2.49; 95% CI, 1.13–5.48; p = 0.02), and KIT exon 11 PV type (HR: 0.23; 95% CI, 0.1–0.53; p = 0.001) remained statistically significant. Table 2 summarizes the results of the univariable and multivariable prognostic factor analysis for RFS.
Table 2.
Univariate and multivariate analysis of prognostic factors for RFS in KIT exon 11-mutated patients.
| RFS | Univariate Cox regression | Multivariable Cox regression | ||
|---|---|---|---|---|
| HR (95% CI) | p value | HR (95% CI) | p value | |
| Gender (female versus male) | 0.65 (0.31–1.39) | NS | ||
| Age (>50 years versus ⩽50 years) | 0.75 (0.33–1.69) | NS | ||
| Primitive tumor diameter (⩽5 cm versus >5 cm) | 4.72 (1.91–11.66) | < 0.001 | 2.76 (1.01–7.57) | 0.04 |
| Mitosis (⩽5/50 HPF versus >5/50 HPF) | 3.34 (1.57–7.10) | 0.001 | 1.50 (0.60–3.73) | NS |
| Gastric site of origin (yes versus no) | 2.13 (1.03–4.43) | 0.03 | 2.49 (1.13–5.48) | 0.02 |
| M–L risk categories (very low/low versus intermediate/high) | 4.67 (1.89–11.54) | 0.001 | 2.34 (0.77–7.18) | NS |
| Exon 11 Del or Delins 557 and/or 558 no versus yes) | 0.19 (0.08–0.42) | < 0.001 | 0.23 (0.1–0.53) | 0.001 |
CI, confidence interval; Del, deletion; Delin, deletion/insertion; HPF, high-power field; HR, hazard ratio; KIT, KIT proto-oncogene receptor tyrosine kinase; M–L, Miettinen–Lasota; NS, not significant; RFS, recurrence-free survival.
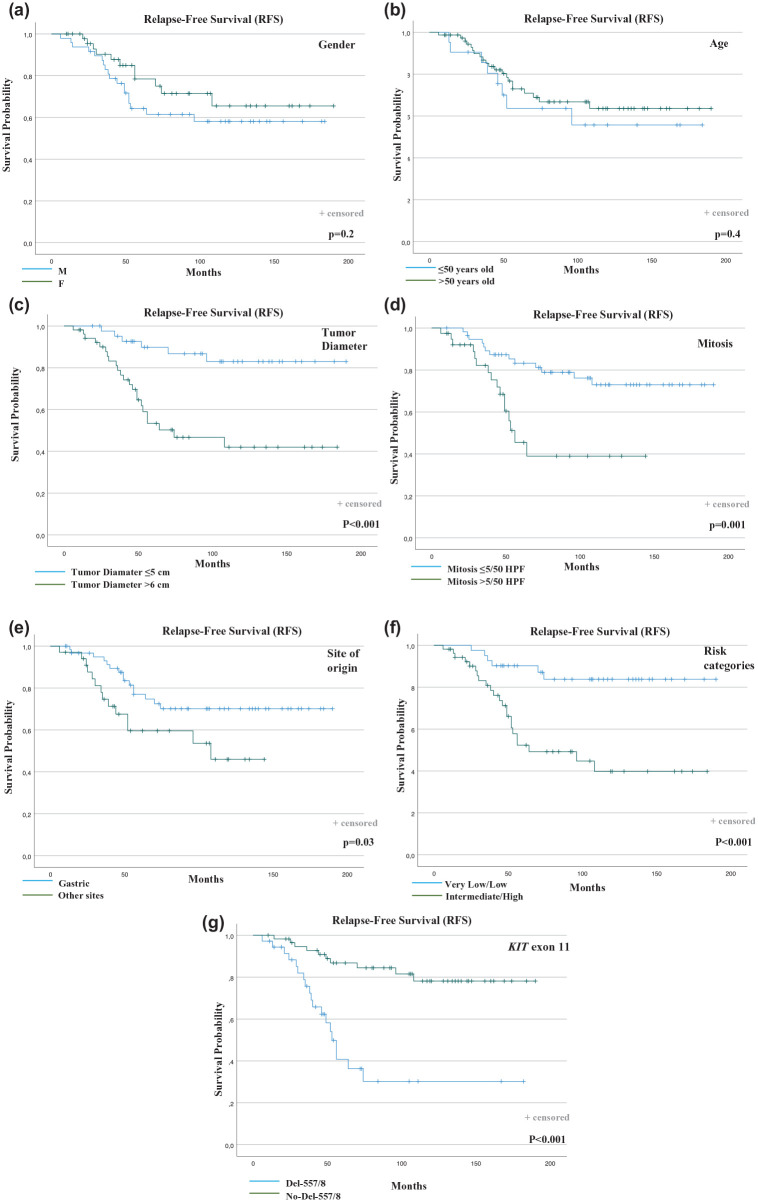
RFS curves were plotted according to each independent prognostic factor (Figure 3).
Figure 3.
Outcome analysis in KIT exon 11-mutated patients according to clinical and biological factors. (a) RFS according to gender. (b) RFS according to age. (c) RFS according to tumor diameter. (d) RFS according to mitosis. (e) RFS according to the site of origin. (f) RFS according to risk categories. (g) RFS according to KIT exon 11 PV type.
Del-557/558, KIT exon 11 deletion or deletion/insertion involving 557 and/or 558 codons; Del-No-557/558, KIT exon 11 deletion or deletion/insertion in codons other than 557 and/or 558; HPF, high-power field; KIT, KIT proto-oncogene receptor tyrosine kinase; PV, pathogenic variant; RFS, recurrence-free survival.
Metastatic sites and PV classification in KIT exon 11-mutated patients with relapsed tumors after curative surgery
At the median follow-up time of 92 months, a total of 29 out 96 KIT exon 11-mutated patients (30.2%) showed a tumor recurrence. In this population, tumor metastatic sites (i.e. liver, peritoneum, or either liver and peritoneum) were described. The patients with metastatic sites were classified on the basis of the presence/absence of deletion or deletion/insertion in codons 557 and/or 558.
In the group of relapsed patients with GISTs harboring 557 and/or 558 deletion or deletion/insertion, 72.2% of metastatic spread involved the peritoneum (13/18 patients), 16.7% the liver (3/18 patients), and 2 patients showed peritoneal and liver metastasis at tumor recurrence (11.1%). In the relapsed patients harboring tumor exon 11 PV not involving 557/558 deletions, 54.5% had peritoneal metastasis (6/11 patients), 27.3% peritoneal and liver metastasis (3/11 patients), and 18.2% only hepatic spread (2/11 patients) (p = 0.5) (Table 3).
Table 3.
Metastatic sites in KIT exon 11-mutated patients with relapsed tumors after curative surgery. The patients were classified as having GISTs harboring deletion or deletion/insertion involving codons 557/558 or other KIT exon 11 PVs.
| Metastatic sites | Total patients n (%) | Deletion or deletion/insertion codons 557/558 * n. (%) | Other exon 11 PVs * n (%) |
|---|---|---|---|
| Liver metastasis | 5 (17.2) | 3 (16.7) | 2 (18.2) |
| Peritoneal metastasis | 19 (65.6) | 13 (72.2) | 6 (54.5) |
| Liver and peritoneal metastasis | 5 (17.2) | 2 (11.1) | 3 (27.3) |
Pearson’s chi-square test: comparison deletion or deletion/insertion codons 557/558 versus other exon 11 PVs; p = 0.5.
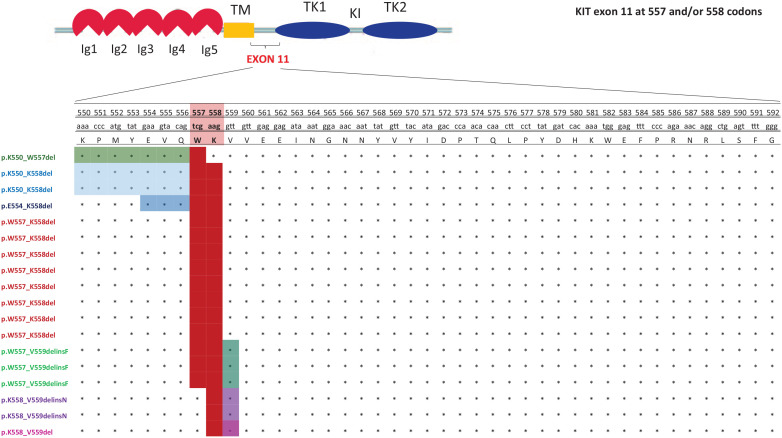
In the population of patients with tumors carrying 557 and/or 558 deletion or deletion/insertion, an exact PV classification was reported to investigate the rate of metastatic recurrence in patients with a deletion involving only one or both 557 and 558 codons. Among 18 relapsed patients, 14 (77.8%) showed GISTs harboring deletions simultaneously involving codons 557 and 558. The PVs were mainly KIT p.W557_K558del (n.8), deletion/insertions (n.3, p.W557_V559delinsF), or intron 10/exon 11 junction deletions resulting in p.K550_K558 deletion (n.2), plus n.1 patient with p.E554_K558del. When 557 and 558 deletions were analyzed separately, only 4 out of 18 relapsed patients (22.2%) showed a tumor relapse (n.2, p.K558_V559delinsN; n.1 p.K550_W557del; n.1 p.K558_V559del) (Figure 4).
Figure 4.
The exact KIT exon 11 PV classification of relapsed patients with GISTs with a deletion involving only one or both 557 and 558 codons (in red).
GIST, gastrointestinal stromal tumor; Ig, immunoglobulin; KIT, KIT proto-oncogene receptor tyrosine kinase; PV, pathogenic variant; TKI, tyrosine kinase 1; TK2, tyrosine kinase 2.
The impact of PV in the intermediate-risk versus high-risk KIT exon 11-mutated patients
We investigated the prognostic impact of the PV on the outcome of the high-risk versus intermediate-risk patients with GISTs. A total of 74 patients were included in the analysis: 35 patients in the intermediate-risk group and 39 patients in the high-risk group. During follow-up, a total of 6 RFS events (recurrence or death) were observed in the intermediate group (25%), and a total of 16 events were observed in the high-risk group (75%). RFS rate at 7 years was 82.8% for the intermediate-risk group (mRFS 149 months; 95% CI, 79.3–192.9), and 59% for the high-risk group (mRFS 98.5 months; 95% CI, 36.1–184.3).
The intermediate-risk and high-risk patients were grouped on the basis of the presence/absence of deletion or deletion/insertion, of the codons 557/558. When RFS between the four groups was compared, patients with GISTs with intermediate-risk and Del-557/558 showed a similar worst outcome compared with high-risk patients with tumors carrying no-Del-557/558 PV (duplication, insertion, or SNV). Indeed, the RFS between the two groups was not statistically significant, showing a lower RFS in the intermediate-risk Del-557/558 (7-year RFS, 69.2.2%; mRFS 117.9 months; 95% CI, 79.3–156.6) than in the high-risk No-Del-557/558 group (7-year RFS, 84.2%; mRFS 153.6; 95% CI, 122.8–184.3; p = 0.4) (Figure 5).
Figure 5.
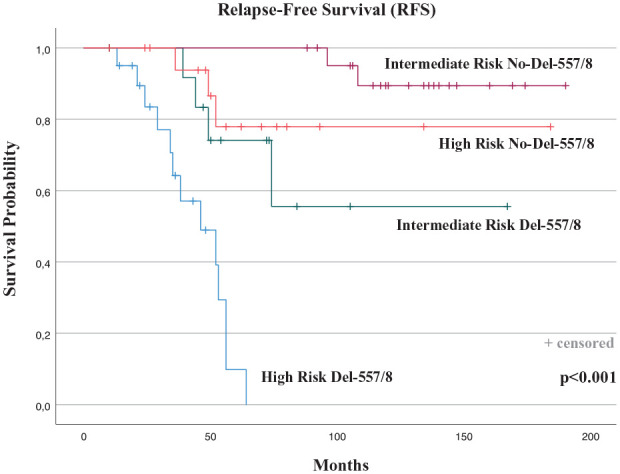
Outcome analysis in the intermediate-risk versus high-risk KIT exon 11-mutated patients according to the presence/absence of deletion or deletion/insertion of the codons 557/558.
KIT, KIT proto-oncogene receptor tyrosine kinase.
Discussion
The ability to identify high-risk tumors is a key element for the management of patients with GISTs and adjuvant treatment selection. Several studies have investigated the contribution of prognostic factors in predicting disease recurrence to better drive clinical decision-making in GISTs after curative surgery.22,23 Despite tumor size, mitotic index and anatomical tumor origin are the clinicopathological features currently included in risk-stratification systems. There is a consistent body of evidence that the mutational profiles of KIT or PDGFRA genes hold strong clinical prognostic value in patients with completely resected GISTs. 24 It is known that GISTs are composed of many different genetic subtypes. 25 Patients with GISTs with a tumor harboring KIT exon 11 PV are the most frequent and represent a heterogeneous genetic and clinical subgroup showing variable clinical outcomes. 13 Further subclassification of exon 11 tumors, based on exact mutation type and codon location, has highlighted, in previous studies, the clinical and prognostic impact of PV on RFS, in addition to well-established prognostic factors of the validated Miettienen–Lasota prediction model.15,18,26 –28
Regarding the clinical and pathological characteristics of patients according to specific tumor genotype, in the present study, KIT exon 11 deletions or deletion/insertion that involve codons 557 and/or 558, compared with other exon 11 PVs, were more frequent in gastric GISTs and characterized by larger primitive tumors and higher mitotic index, and thus 55.5% of localized GISTs with 557/558 deletions are stratified as high-risk tumors. Conversely, the KIT exon 11 SNVs (such as the p.W557R) and duplications (such as the p.D572_D579dup), classically associated with favorable outcomes, 15 were mostly related, in our cohort, with lower mitotic count and low/intermediate risk (68.4%) than KIT 557/558 deletions or deletion/insertion, indicating a more indolent clinical behavior. These data are consistent with the results of previous studies and confirm that KIT exon 11 557/558 deletions are associated with malignant tumor behavior.15,18,26 –28
Our data regarding the impact of PV type and location on RFS showed the worst outcome for patients with GISTs harboring deletion or deletion/insertion of the codons 557 and/or 558 compared with patients with any other KIT exon 11 PV (7-year RFS of 50% versus 73.1%); KIT exon 11 SNVs and duplications showed a better 7-year RFS rate (88.2%). Multivariate analysis confirmed the independent prognostic value of this finding. Remarkably, 14 out of 18 relapsed patients in the group of Del-557/558 harbored deletions simultaneously involving codons 557 and 558. The PVs were mainly KIT p.W557_K558del, deletion/insertions (p.W557_V559delinsF), or intron 10/exon 11 junction deletions resulting in p.K550_K558 deletion. These data underline a significantly higher risk of recurrence in patients with deletions involving both codons.15 –17 Interestingly, when 557 and 558 codon deletions were analyzed separately, only 22.2% of patients (4/18 patients) showed a tumor relapse, miming the prognostic behavior of tumor-carrying exon 11 deletion outside W557_K558. This result partially differs from previous evidence, which described similar deleterious RFS between patients with deletions affecting only codons 557 or 558 and patients harboring tumor PVs of either codon. 28 The reasons for the lower aggressive biology of deletions not simultaneously involving codons 557 and 558 can be explained by the critical autoinhibitory role on the process of tyrosine kinase activation exerted by 557 and 558 codon regions that, when both delete, results in a considerably increased spontaneous receptor phosphorylation and activation of the downstream pathway.29,30
Notably, when we examined the prognostic significance of the PV in the high-risk versus intermediate-risk patients with GISTs, the outcome of intermediate-risk patients with Del-557/558 and the high-risk patients without Del-557/558 (deletion or deletion/insertion in codons other than 557/558, or duplication, insertion, or SNV) was similar in terms of RFS. The two groups of patients showed the same worse prognosis. These current findings suggest that the patients with GISTs with intermediate-risk features and KIT exon 11 deletion or deletion/insertion in the 557/558 codons show a significant risk of relapse and may need to be treated with the adjuvant imatinib.
According to the aggressiveness and metastatic potential of 557/558 tumor deletions, 31 an organ-specific metastatic involvement of the liver has been proposed in these patients. In a previous study, patients with GISTs with KIT exon 11 deletions involving codons 557/558 reported a significant association with hepatic metastasis, suggesting an involvement of these mutations in the development of an organ-selective pattern of tumor metastasis in GISTs. 20 The study proposed that KIT exon 11 codons 557/558 deletion promote liver metastasis through CXC chemokine ligand (CXCL) 12/CXC chemokine receptor (CXCR) 4 axis, chemotactic factors implicated in cancer progression. According to this data, PVs of 557/558 critical codons increased CXCR4 expression by the upregulation of the transcription factor ETS Variant Transcription Factor 1 (ETV1) and the enhanced CXCL12-mediated GIST cell migration and invasion, ultimately promoting liver colonization. 20 In contrast to these findings, our clinical results showed that in GISTs with 557/558 tumor deletions, the peritoneum, not the liver, is the most frequent metastatic site in relapsed patients (72.2%). The reasons for these results remain speculative and represent the rationale to better investigate if other factors of clinical or biological interest32 –39 could have a further impact on tumor recurrence in patients with homogeneous intrinsic molecular features.
The strength of the current study is the comprehensive clinicopathological and molecular data from a population-based analysis with a long follow-up. Despite the relatively small cohort of relapsed patients for prognostic evaluation of PV on metastatic sites, these preliminary reports could encourage future studies in a larger patient population.
Conclusion
In patients with localized GISTs completely resected we showed that KIT exon 11 deletions or deletion/insertion involving both codons 557 and 558 are genotypes indicative of more aggressive tumor behavior and higher risk of recurrence: the KIT exon 11 deletion or deletion/insertion, when involving only one of 557 or 558 codons, resembles the more indolent prognostic behavior of tumor-carrying deletion outside 557 and 558 codons, supporting the importance of considering the PV type and codon location in routine risk stratification. Differing from other series, in relapsed patients with 557/558 deletions or deletion/insertion, the peritoneum and not the liver is the most frequent metastatic site. Finally, we found that intermediate-risk patients whose GISTs harbor KIT exon 11 deletion or deletion/insertion in the 557/558 codons showed a similar outcome to high-risk patients without Del-557/558 (GIST carrying deletion or deletion/insertion in codons other than 557/558, or duplication/insertion/SNV), suggesting that they may need to be treated with the adjuvant imatinib as with high-risk patients.
Footnotes
Author contributions: Conceptualization LI, GB, DF, AR, and VB; molecular analysis NB, CB, MB, MC, DC, AF, EP and AP; clinical data GB, LA, AB, LRC, GG, GP, and DC; data curation and analysis LI, BV, CB, IDL, AG and MC; writing LI, GB and DF; supervision AR, GB, and VB. All authors have read and agreed to the published version of the manuscript.
The authors declare that there is no conflict of interest.
Funding: The authors received no financial support for the research, authorship, and/or publication of this article.
ORCID iDs: Lorena Incorvaia  https://orcid.org/0000-0002-1199-7286
https://orcid.org/0000-0002-1199-7286
Marco Bono  https://orcid.org/0000-0001-7169-3463
https://orcid.org/0000-0001-7169-3463
Antonio Russo  https://orcid.org/0000-0002-4370-2008
https://orcid.org/0000-0002-4370-2008
Contributor Information
Lorena Incorvaia, Department of Biomedicine, Neuroscience and Advanced Diagnostics (Bi.N.D.), Section of Medical Oncology, University of Palermo, Via del Vespro 129, Palermo, 90127, Italy.
Giuseppe Badalamenti, Department of Surgical, Oncological and Oral Sciences, Section of Medical Oncology, University of Palermo, Palermo, Italy.
Daniele Fanale, Department of Surgical, Oncological and Oral Sciences, Section of Medical Oncology, University of Palermo, Palermo, Italy.
Bruno Vincenzi, Department of Medical Oncology, Biomedical Campus, University of Rome, Rome, Italy.
Ida De Luca, Department of Surgical, Oncological and Oral Sciences, Section of Medical Oncology, University of Palermo, Palermo, Italy.
Laura Algeri, Department of Surgical, Oncological and Oral Sciences, Section of Medical Oncology, University of Palermo, Palermo, Italy.
Nadia Barraco, Department of Surgical, Oncological and Oral Sciences, Section of Medical Oncology, University of Palermo, Palermo, Italy.
Chiara Brando, Department of Surgical, Oncological and Oral Sciences, Section of Medical Oncology, University of Palermo, Palermo, Italy.
Annalisa Bonasera, Department of Surgical, Oncological and Oral Sciences, Section of Medical Oncology, University of Palermo, Palermo, Italy.
Marco Bono, Department of Surgical, Oncological and Oral Sciences, Section of Medical Oncology, University of Palermo, Palermo, Italy.
Marta Castiglia, Department of Surgical, Oncological and Oral Sciences, Section of Medical Oncology, University of Palermo, Palermo, Italy.
Daniela Cancelliere, Department of Surgical, Oncological and Oral Sciences, Section of Medical Oncology, University of Palermo, Palermo, Italy.
Massimiliano Cani, Department of Surgical, Oncological and Oral Sciences, Section of Medical Oncology, University of Palermo, Palermo, Italy.
Lidia Rita Corsini, Department of Surgical, Oncological and Oral Sciences, Section of Medical Oncology, University of Palermo, Palermo, Italy.
Alessia Fiorino, Department of Surgical, Oncological and Oral Sciences, Section of Medical Oncology, University of Palermo, Palermo, Italy.
Antonio Galvano, Department of Surgical, Oncological and Oral Sciences, Section of Medical Oncology, University of Palermo, Palermo, Italy.
Erika Pedone, Department of Surgical, Oncological and Oral Sciences, Section of Medical Oncology, University of Palermo, Palermo, Italy.
Alessandro Perez, Department of Surgical, Oncological and Oral Sciences, Section of Medical Oncology, University of Palermo, Palermo, Italy.
Alessia Pivetti, Department of Surgical, Oncological and Oral Sciences, Section of Medical Oncology, University of Palermo, Palermo, Italy.
Giuseppa Graceffa, Department of Surgical, Oncological and Oral Sciences, Section of General and Oncological Surgery, University of Palermo, Palermo, Italy.
Gianni Pantuso, Department of Surgical, Oncological and Oral Sciences, Section of General and Oncological Surgery, University of Palermo, Palermo, Italy.
Daniela Cabibi, Department of Health Promotion, Mother and Child Care, University of Palermo, Palermo, Italy.
Antonio Russo, Department of Surgical, Oncological and Oral Sciences, Section of Medical Oncology, University of Palermo, Palermo, Italy.
Viviana Bazan, Department of Biomedicine, Neuroscience and Advanced Diagnostics (Bi.N.D.), Section of Medical Oncology, University of Palermo, Palermo, Italy.
References
- 1. Corless CL, Barnett CM, Heinrich MC. Gastrointestinal stromal tumours: origin and molecular oncology. Nat Rev Cancer 2011; 11: 865–878. [DOI] [PubMed] [Google Scholar]
- 2. Pantuso G, Macaione I, Taverna A, et al. Surgical treatment of primary gastrointestinal stromal tumors (GISTs): management and prognostic role of R1 resections. Am J Surg 2020; 220: 359–364. [DOI] [PubMed] [Google Scholar]
- 3. Corless CL, Schroeder A, Griffith D, et al. PDGFRA mutations in gastrointestinal stromal tumors: frequency, spectrum and in vitro sensitivity to imatinib. J Clin Oncol 2005; 23: 5357–5364. [DOI] [PubMed] [Google Scholar]
- 4. Wu CE, Tzen CY, Wang SY, et al. Clinical diagnosis of Gastrointestinal Stromal Tumor (GIST): from the molecular genetic point of view. Cancers (Basel) 2019; 11: 679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Boikos SA, Pappo AS, Killian JK, et al. Molecular subtypes of KIT/PDGFRA wild-type gastrointestinal stromal tumors: a report from the National Institutes of Health Gastrointestinal Stromal Tumor Clinic. JAMA Oncol 2016; 2: 922–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Vanden Bempt I, Vander Borght S, Sciot R, et al. Comprehensive targeted next-generation sequencing approach in the molecular diagnosis of gastrointestinal stromal tumor. Genes Chromosomes Cancer 2020; 60: 239–249. [DOI] [PubMed] [Google Scholar]
- 7. Miettinen M, Killian JK, Wang ZF, et al. Immunohistochemical loss of succinate dehydrogenase subunit A (SDHA) in gastrointestinal stromal tumors (GISTs) signals SDHA germline mutation. Am J Surg Pathol 2013; 37: 234–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cocco E, Scaltriti M, Drilon A. NTRK fusion-positive cancers and TRK inhibitor therapy. Nat Rev Clin Oncol 2018; 15: 731–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Li F, Huynh H, Li X, et al. FGFR-mediated reactivation of MAPK signaling attenuates antitumor effects of imatinib in gastrointestinal stromal tumors. Cancer Discov 2015; 5: 438–451. [DOI] [PubMed] [Google Scholar]
- 10. Hirota S, Isozaki K, Moriyama Y, et al. Gain-of-function mutations of c-kit in human gastrointestinal stromal tumors. Science 1998; 279: 577–580. [DOI] [PubMed] [Google Scholar]
- 11. Nannini M, Nigro MC, Bruno V, et al. Personalization of regorafenib treatment in metastatic gastrointestinal stromal tumours in real-life clinical practice. Ther Adv Med Oncol 2017; 9: 731–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Vincenzi B, Nannini M, Badalamenti G, et al. Imatinib rechallenge in patients with advanced gastrointestinal stromal tumors following progression with imatinib, sunitinib and regorafenib. Ther Adv Med Oncol 2018; 10: 1758835918794623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Debiec-Rychter M, Sciot R, Le Cesne A, et al. KIT mutations and dose selection for imatinib in patients with advanced gastrointestinal stromal tumours. Eur J Cancer 2006; 42: 1093–1103. [DOI] [PubMed] [Google Scholar]
- 14. Serrano C, George S. Gastrointestinal stromal tumor: challenges and opportunities for a new decade. Clin Cancer Res 2020; 26: 5078–5085. [DOI] [PubMed] [Google Scholar]
- 15. Wozniak A, Rutkowski P, Schöffski P, et al. Tumor genotype is an independent prognostic factor in primary gastrointestinal stromal tumors of gastric origin: a European multicenter analysis based on ConticaGIST. Clin Cancer Res 2014; 20: 6105–6116. [DOI] [PubMed] [Google Scholar]
- 16. Joensuu H, Wardelmann E, Sihto H, et al. Effect of KIT and PDGFRA mutations on survival in patients with gastrointestinal stromal tumors treated with adjuvant imatinib: an exploratory analysis of a randomized clinical trial. JAMA Oncol 2017; 3: 602–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Martin-Broto J, Gutierrez A, Garcia-Del-Muro X, et al. Prognostic time dependence of deletions affecting codons 557 and/or 558 of KIT gene for relapse-free survival (RFS) in localized GIST: a Spanish Group for Sarcoma Research (GEIS) Study. Ann Oncol 2010; 21: 1552–1557. [DOI] [PubMed] [Google Scholar]
- 18. Andersson J, Bümming P, Meis-Kindblom JM, et al. Gastrointestinal stromal tumors with KIT exon 11 deletions are associated with poor prognosis. Gastroenterology 2006; 130: 1573–1581. [DOI] [PubMed] [Google Scholar]
- 19. Cho S, Kitadai Y, Yoshida S, et al. Deletion of the KIT gene is associated with liver metastasis and poor prognosis in patients with gastrointestinal stromal tumor in the stomach. Int J Oncol 2006; 28: 1361–1367. [PubMed] [Google Scholar]
- 20. Wang HC, Li TY, Chao YJ, et al. KIT Exon 11 Codons 557-558 deletion mutation promotes liver metastasis through the CXCL12/CXCR4 axis in gastrointestinal stromal tumors. Clin Cancer Res 2016; 22: 3477–3487. [DOI] [PubMed] [Google Scholar]
- 21. Miettinen M, Lasota J. Gastrointestinal stromal tumors: pathology and prognosis at different sites. Semin Diagn Pathol 2006; 23: 70–83. [DOI] [PubMed] [Google Scholar]
- 22. Joensuu H. Predicting recurrence-free survival after surgery for GIST. Lancet Oncol 2009; 10: 1025. [DOI] [PubMed] [Google Scholar]
- 23. Rutkowski P, Nowecki ZI, Michej W, et al. Risk criteria and prognostic factors for predicting recurrences after resection of primary gastrointestinal stromal tumor. Ann Surg Oncol 2007; 14: 2018–2027. [DOI] [PubMed] [Google Scholar]
- 24. Bachet JB, Hostein I, Le Cesne A, et al. Prognosis and predictive value of KIT exon 11 deletion in GISTs. Br J Cancer 2009; 101: 7–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Badalamenti G, Rodolico V, Fulfaro F, et al. Gastrointestinal stromal tumors (GISTs): focus on histopathological diagnosis and biomolecular features. Ann Oncol 2007; 18(Suppl. 6): vi136–vi140. [DOI] [PubMed] [Google Scholar]
- 26. Singer S, Rubin BP, Lux ML, et al. Prognostic value of KIT mutation type, mitotic activity, and histologic subtype in gastrointestinal stromal tumors. J Clin Oncol 2002; 20: 3898–3905. [DOI] [PubMed] [Google Scholar]
- 27. Wardelmann E, Losen I, Hans V, et al. Deletion of Trp-557 and Lys-558 in the juxtamembrane domain of the c-kit protooncogene is associated with metastatic behavior of gastrointestinal stromal tumors. Int J Cancer 2003; 106: 887–895. [DOI] [PubMed] [Google Scholar]
- 28. Martín J, Poveda A, Llombart-Bosch A, et al. Deletions affecting codons 557-558 of the c-KIT gene indicate a poor prognosis in patients with completely resected gastrointestinal stromal tumors: a study by the Spanish Group for Sarcoma Research (GEIS). J Clin Oncol 2005; 23: 6190–6198. [DOI] [PubMed] [Google Scholar]
- 29. Bauer S, Duensing A, Demetri GD, et al. KIT oncogenic signaling mechanisms in imatinib-resistant gastrointestinal stromal tumor: PI3-kinase/AKT is a crucial survival pathway. Oncogene 2007; 26: 7560–7568. [DOI] [PubMed] [Google Scholar]
- 30. Ma Y, Cunningham ME, Wang X, et al. Inhibition of spontaneous receptor phosphorylation by residues in a putative alpha-helix in the KIT intracellular juxtamembrane region. J Biol Chem 1999; 274: 13399–13402. [DOI] [PubMed] [Google Scholar]
- 31. Incorvaia L, Fanale D, Vincenzi B, et al. Type and gene location of KIT mutations predict progression-free survival to first-line imatinib in gastrointestinal stromal tumors: a look into the exon. Cancers (Basel) 2021; 13: 993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. McDonald B, Kubes P. Cellular and molecular choreography of neutrophil recruitment to sites of sterile inflammation. J Mol Med (Berl) 2011; 89: 1079–1088. [DOI] [PubMed] [Google Scholar]
- 33. Lazennec G, Richmond A. Chemokines and chemokine receptors: new insights into cancer-related inflammation. Trends Mol Med 2010; 16: 133–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Incorvaia L, Badalamenti G, Rini G, et al. MMP-2, MMP-9 and activin A blood levels in patients with breast cancer or prostate cancer metastatic to the bone. Anticancer Res 2007; 27: 1519–1525. [PubMed] [Google Scholar]
- 35. Dumitru CA, Lang S, Brandau S. Modulation of neutrophil granulocytes in the tumor microenvironment: mechanisms and consequences for tumor progression. Semin Cancer Biol 2013; 23: 141–148. [DOI] [PubMed] [Google Scholar]
- 36. Psaila B, Lyden D. The metastatic niche: adapting the foreign soil. Nat Rev Cancer 2009; 9: 285–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Rutkowski P, Teterycz P, Klimczak A, et al. Blood neutrophil-to-lymphocyte ratio is associated with prognosis in advanced gastrointestinal stromal tumors treated with imatinib. Tumori 2018; 104: 415–422. [DOI] [PubMed] [Google Scholar]
- 38. Badalamenti G, Fanale D, Incorvaia L, et al. Role of tumor-infiltrating lymphocytes in patients with solid tumors: can a drop dig a stone? Cell Immunol 2019; 343: 103753. [DOI] [PubMed] [Google Scholar]
- 39. Fanale D, Incorvaia L, Badalamenti G, et al. Prognostic role of plasma PD-1, PD-L1, pan-BTN3As and BTN3A1 in patients affected by metastatic gastrointestinal stromal tumors: can immune checkpoints act as a sentinel for short-term survival? Cancers (Basel) 2021; 13: 2118. [DOI] [PMC free article] [PubMed] [Google Scholar]