Significance
Superantigens are proteins that induce a vast activation of the immune system and are typically found in pathogenic microbes. Recently, proteins with superantigen-like properties were discovered in a commensal bacterium inhabiting the human gut, but little is known about the impact of this protein on host/commensal interaction. Here, we characterize the superantigen-like protein immunoglobulin-binding protein (Ibp) expressed by Ruminococcus gnavus, which has broad specificity to human and mouse immunoglobulins. We present the crystal structure of the Ibp/Fab complex and show that one of Ibp’s domains alone is sufficient to activate B cells. This work provides an understanding of superantigen-like molecules potentially playing a role in the maintenance of host/microbiome homeostasis.
Keywords: superantigen, antibody, immunoglobulin-binding protein, Ruminococcus gnavus, commensal microbe
Abstract
Microorganisms have coevolved diverse mechanisms to impair host defenses. A major one, superantigens, can result in devastating effects on the immune system. While all known superantigens induce vast immune cell proliferation and come from opportunistic pathogens, recently, proteins with similar broad specificity to antibody variable (V) domain families were identified in a commensal microbiota. These proteins, identified in the human commensal Ruminococcus gnavus, are called immunoglobulin-binding protein (Ibp) A and B and have been shown to activate B cells in vitro expressing either human VH3 or murine VH5/6/7. Here, we provide molecular and functional studies revealing the basis of this Ibp/immunoglobulin (Ig) interaction. The crystal structure and biochemical assays of a truncated IbpA construct in complex with mouse VH5 antigen-binding fragment (Fab) shows a binding of Ig heavy chain framework residues to the Ibp Domain D and the C-terminal heavy chain binding domain (HCBD). We used targeted mutagenesis of contact residues and affinity measurements and performed studies of the Fab-IbpA complex to determine the stoichiometry between Ibp and VH domains, suggesting Ibp may serve to cluster full-length IgA antibodies in vivo. Furthermore, in vitro stimulation experiments indicate that binding of the Ibp HCBD alone is sufficient to activate responsive murine B cell receptors. The presence of these proteins in a commensal microbe suggest that binding a broad repertoire of immunoglobulins, particularly in the gut/microbiome environment, may provide an important function in the maintenance of host/microbiome homeostasis contrasting with the pathogenic role of structurally homologous superantigens expressed by pathogens.
The relationship between host and microbiota can be viewed as a spectrum between commensalism and pathogenicity (1, 2). Commensal microbes coevolved many strategies to manipulate and protect the host immune system from pathogens (3). Whereas most microbial antigens stimulate only a small fraction of lymphocytes, certain pathogenic organisms evolved molecules to hijack the host system by harnessing and redirecting the T or B cell stimulatory potential of hosts against themselves. These broadly reactive proteins are called superantigens (SAgs); until recently, this term was reserved for mostly pathogenic Staphylococci or Streptococci species and some viruses (4). For T cell SAgs, their predominant mechanism of activation is through crosslinked binding to a germline region of a variable beta chain of the T cell receptors and outside of the peptide-binding groove of a class II major histocompatibility complex (5, 6). Similarly, B cell SAgs, that is, Staphylococcal protein A (SpA) and Streptoccoccal protein G, bind outside of the complementarity-determining region (CDR) of the variable heavy (VH) chain of an antigen-binding fragment (Fab) as well as antibody-crystallizable fragment (Fc) (7, 8). Additional examples of superantigenic-like broadly reactive interactions have been uncovered that expand on the classical definition of conventional superantigens, e.g., protein Fv (a human liver sialoprotein), protein L (a coat protein of Peptostreptococcus magnus), and protein M (a human mycoplasma protein) (9–11). More recently, in our studies of host-microbiota interactions, we identified proteins exhibiting superantigen-like binding characteristics, called immunoglobulin-binding proteins (Ibp) A and B (12).
Previously, we observed that in mice colonized with human microbiota, a large portion of the microbiota are coated with mouse microbiota-reactive monoclonal antibodies derived from small intestinal IgA plasma cells (12). This study also revealed that a subset of the commensal Lachnospiracae family can also bind to antibodies expressing particular immunoglobulin variable gene regions. Specifically, we focused on the Ruminococcus gnavus species, which is a Gram-positive anaerobe that inhabits the majority of humans yet represents a small fraction of the gut microbiota (13, 14). The species is well adapted to the gut by using a unique sialic acid metabolism pathway, which provides a competitive strategy to colonize a niche in the gut mucosal layer (15, 16). Despite being a small percentage of the healthy gut microbiota, R. gnavus is disproportionately represented in inflammatory conditions and diseases such as inflammatory bowel disease (IBD), lupus nephritis, or atopy (17–21). Notably, R. gnavus represents a genetically heterogeneous species that can be grouped in distinct functionally relevant clusters, and IBD flare-ups were associated with transient expansions of a genetically and functionally related R. gnavus clade (18, 22, 23). However, only limited molecular associations have been described, and it is currently unknown how, or if at all, R. gnavus is contributing to either disease or dysbiosis or if it is just expanding due to a competitive advantage during inflammation. There are several potential reasons for this disproportionate correlation with inflammatory diseases, including factors such as a higher capacity in utilization of mucin glycans, expression of unique proinflammatory polysaccharides by R. gnavus, and expression of IbpA and IbpB (12, 24, 25), the subjects of this study. Considering the need for understanding these host/microbe interactions, our discovery of these new protein effectors, IbpA and IbpB, led us to investigate the molecular and structural features of these proteins.
IbpA and IbpB have 83% amino acid sequence similarity and are located directly adjacent to one another in the R. gnavus genome, suggesting they arose via gene duplication. Even though the consequences of Ibp expression for commensal and host are poorly understood, these proteins include several features reminiscent of other bacterial immunoglobulin-binding proteins, including common B cell SAgs such as SpA or Streptoccoccal protein G (26). For example, IbpA/B contain four 67 amino acid long repeat domains that are reminiscent of the repeat domains in SpA and protein G that were shown to mediate binding to Fc or Fab fragments of immunoglobulins. In humans, SpA binds to VH3 Igs, inducing a vast supraclonal expansion of VH3 plasmablasts. In addition, in VH7 (previously T15) transgenic mice, high-dose injections of SpA induce VH7+-targeted B cells depletion by an activation-induced cell-death (AICD) pathway (27–30). A survey of Ibp binding to a large panel of monoclonal mouse and human antibodies revealed a broad Ibp-reactive repertoire with a bias toward murine VH5/6/7 and human VH3 families, which in humans is the largest subset of total B cells and suggests similar recognition as SpA (12). Interestingly, both IbpA and IbpB were also found to potently stimulate human and mouse B cells in vitro, presumably by B cell receptor (BCR) crosslinking through the series of repeat domains. Finally, an N-terminal signal peptide and C-terminal SPKTG sortase motif indicate cell wall anchoring, reminiscent of SpA tethering on the surface of virulent strains of Staphylococcus aureus (12, 31). Despite functional and structural similarity of Ibp to B cell SAgs, the expression of Ibp in human commensal bacteria in healthy humans suggest they may be involved in host/microbe homeostasis instead of pathogenicity; therefore, we propose the use of the term superantigen-like to describe these novel proteins.
In this work, we provide detailed molecular and functional information about R. gnavus Ibp, defining the in vitro properties of full-length Ibp and single Ibp domains. We have solved the crystal structure of a truncated IbpA construct in complex with two mouse VH5 Fab fragments showing association mediated mostly through Ig framework residues with Domain D and the C-terminal heavy chain binding domain (HCBD). We show Ibp can bind to a broad repertoire of human and mouse B cells that exceed the repertoire of antibodies bound to SpA. We utilized individual IbpA contact domains to study their ability to activate B cells in vitro and could observe B cell responses with even single IbpA domains. Overall, our results give new insights of how R. gnavus interacts with the host and utilizes its surface proteins to attract a wide range of antibodies.
Results
IbpA and IbpB Bind to a Broad Repertoire of Human and Mouse B Cells and Initiate BCR Signaling.
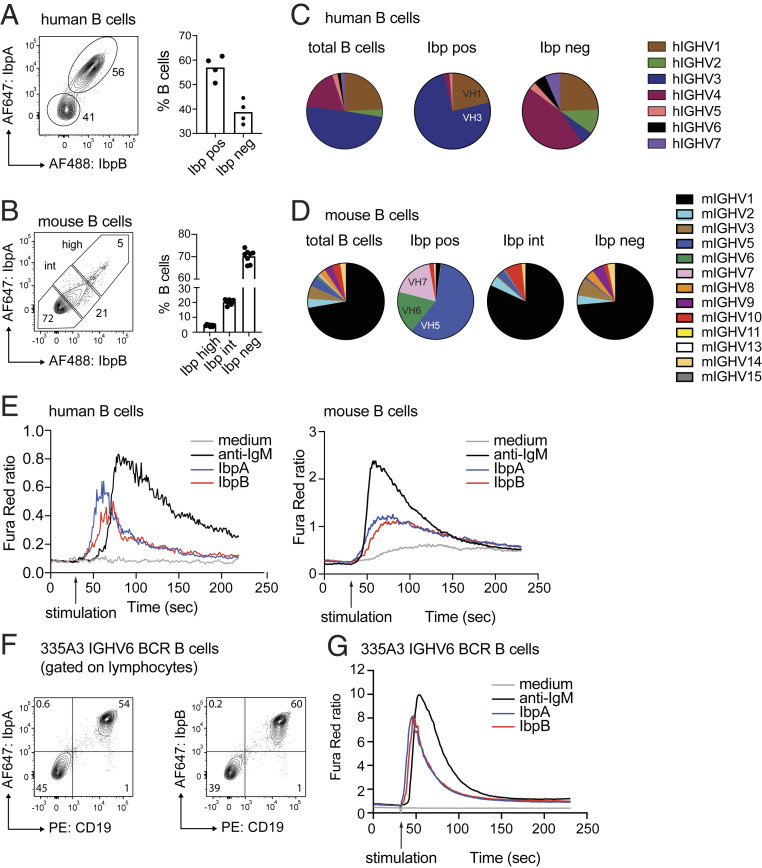
We previously tested an archive of mouse and human-derived monoclonal antibodies for Ibp reactivity and observed preferential binding of mouse VH5/6/7 and human VH3 variable regions. However, we also detected a considerable fraction of antibodies expressing other VH families that were bound by Ibp (12). To test if IbpA and IbpB could also bind to membrane-associated immunoglobulins and to more precisely define the repertoire of Ibp-reactive mouse and human B cells, we labeled full-length IbpA and IbpB with fluorophores and used them as staining reagents for flow cytometry. Fifty percent to 60% of human peripheral blood B cells were costained with IbpA and IbpB (Fig. 1A), whereas the frequency of brightly labeled splenic mouse B cells was lower (Ibphigh, 5%) (Fig. 1B). No discernable difference was observed between IbpA and IbpB staining, and incubation with either IbpA or IbpB or both prevented IbpA/IbpB double staining, suggesting binding to a shared repertoire and epitopes (SI Appendix, Fig. S1A). In addition, we detected that a dose-dependent population of mouse B cells with an intermediate binding pattern could be detected (Ibpint, Fig. 1B and SI Appendix, Fig. S1B). We next sorted Ibp-bound, Ibp-unbound, and total human and mouse B cell populations and performed repertoire sequencing. The majority of Ibp-positive human B cells (75%) expressed VH3 gene families, whereas 25% showed expression of VH1 genes (Fig. 1C). Ibphigh murine B cells exclusively expressed VH5/6/7 genes (Fig. 1D). The repertoire of Ibpint B cells was more heterogenous but mostly consisted of VH1 families. There was a partial overlap between the heavy chain repertoire of Ibp negative (Ibpneg) and Ibpint B cells, but some families, such as mouse VH3, VH8, and VH9, were only found within the Ibpneg fraction (Fig. 1D). A restricted light-chain repertoire was not detectable in either dataset, suggesting Ibp binding to antibodies is solely mediated through the heavy chain variable regions (SI Appendix, Fig. S1 C and D). These data confirm our previous findings and indicate that IbpA and IbpB both bind a shared and broad repertoire of human and mouse antibodies (12).
Fig. 1.
IbpA and IbpB activate a defined repertoire of human and mouse BCRs. (A) Flow cytometry plots show human CD19pos B cells stained with fluorescently labeled IbpA and IbpB (Left) and bar graph quantification collected from three individuals (Right). (B) Analogous flow cytometry plots for mouse CD19pos B cells stained with fluorescently labeled IbpA or IbpB (Left) and bar graph quantification (Right). (C) Immunoglobulin heavy chain repertoire of sorted human B cells. Each symbol represents the repertoire of the corresponding sorted B cell population. Total B cells indicate repertoire of all CD19pos B cells regardless of Ibp binding. (D) Analogous immunoglobulin heavy chain repertoire of sorted mouse B cells and mouse B cells. (E) Ca2+ flux analysis of human peripheral blood CD19pos B cells (Left) and mouse CD19pos splenocytes (Right) after in vitro stimulation with IbpA, IbpB, or anti-IgM. (F) Representative contour plots of transgenic B cells expressing a VH6 BCR show bright staining of CD19 B cells with IbpA (Left) or IbpB (Right). (G) Ca2+ flux analysis mouse CD19pos splenocytes expressing a transgenic VH6 BCR after in vitro stimulation with IbpA, IbpB, or anti-IgM.
We then sought to determine whether recombinant IbpA and IbpB also initiate BCR signaling. We therefore stimulated B cells in vitro and examined intracellular calcium flux by flow cytometry. A strong calcium flux signal could be detected in bulk human B cells (Fig. 1E, Left), whereas the signal of bulk mouse B cells was lower, consistent with Ibp’s engagement of a more restricted segment of the mouse Ig repertoire (Fig. 1E, Right). However, in transgenic B cells expressing an IGHV6 BCR, both IbpA and IbpB induced calcium flux considerably faster and stronger than in wild-type B cells (Fig. 1 F and G). Thus, recombinant Ibp binds to a broad repertoire of human and mouse BCRs and, in a soluble form, can activate BCR signaling in vitro.
A Truncated IbpA Construct Is Functional and Shows Higher Affinity to Antibodies than Wild-Type IbpA.
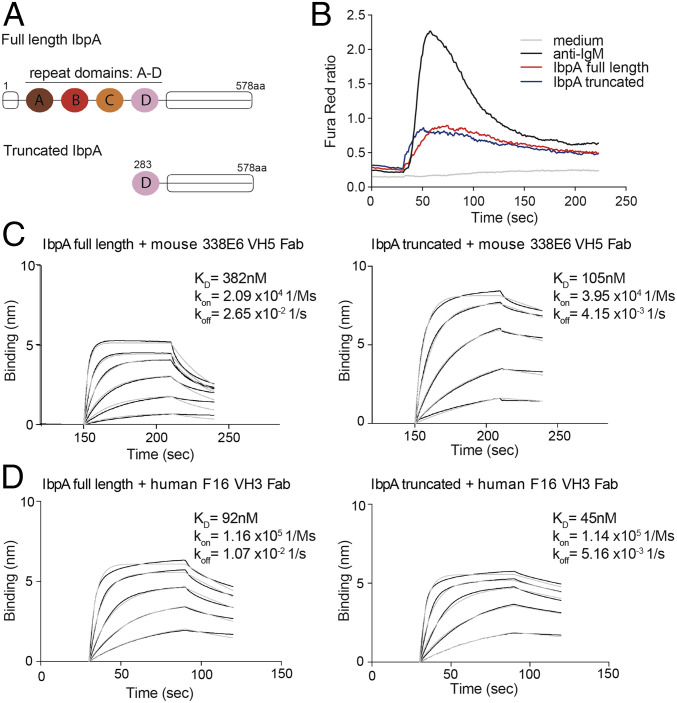
To analyze the molecular details of how Ibp binds to antibodies and maintains its B cell repertoire specificity, we pursued structural studies of full-length Ibp in complex with a mouse VH5 Fab. Structural trials with full-length IbpA or IbpB in complex with Fab either by X-ray crystallography or by negative stain electron microscopy were unsuccessful. We therefore generated an optimized IbpA (truncated IbpA) construct that bears only one of the four 67 amino acid repeat domains, which in complex with Fab will reduce binding heterogeneity. This construct contains the fourth repeat (Domain D) and extends to the C terminus of the full-length IbpA (Fig. 2A and SI Appendix, Fig. S2A). The size-exclusion chromatography profile of the truncated IbpA in complex with Fab shows a peak that shifts to longer retention times compared to the full-length IbpA and Fab complex, consistent with smaller hydrodynamic radius and reduced molecular mass (SI Appendix, Fig. S2 B and C). We also tested the ability of truncated IbpA to bind and activate mouse BCRs in vitro and found that the truncated Ibp activated as well as the full length (Fig. 2B). Interestingly, direct affinity measurement by biolayer interferometry (BLI) showed three times higher affinity (lower dissociation constant, KD) and slower dissociation rate (koff) for the truncated IbpA construct compared to the full length when tested for binding with mouse VH5 338E6 Fab (Fig. 2C). We also observed similar trends in KD and koff for a human VH3 IgG F16 clone (Fig. 2D). Altogether, these data demonstrate that truncated IbpA is as effective as full-length IbpA in activating mouse splenic B cells; it additionally binds to specific mouse and human Fabs with higher affinity and a slower dissociation rate than full-length IbpA, suggesting that the full-length IbpA might adopt a conformation that modifies its interaction with the Fab portion of antibodies.
Fig. 2.
Truncated IbpA maintains binding to Fab fragment with higher affinity. (A) Cartoon representation shows domains of full-length IbpA protein (Top) in comparison with truncated IbpA construct (Bottom) which was used for crystallization and in vitro B cell activation assays. (B) Ca2+ flux analysis comparing full-length and truncated IbpA after in vitro stimulation of mouse splenic B cells. (C) Binding studies using BLI comparing affinity of full-length (Left) versus truncated IbpA (Right) to mouse 338E6 VH5 Fab. (D) Binding studies using BLI comparing affinity of full-length (Left) versus truncated IbpA (Right) to human F16 VH3 Fab.
A Truncated IbpA Structure Reveals Two Superantigen-Like Fold Domains that Engage a Fab: The Domain D and the C-Terminal HCBD.
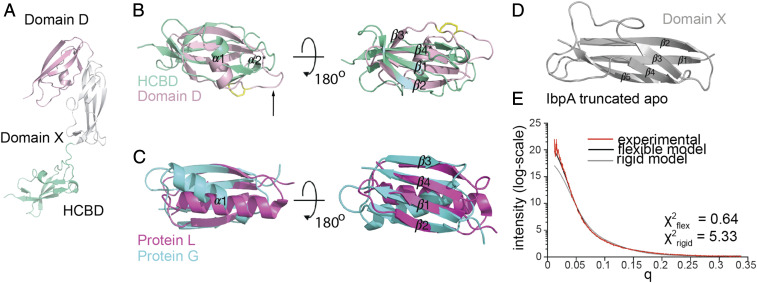
In contrast to full-length IbpA, the truncated Ibp construct proved successful in our crystallization attempts in complex with an engineered mouse VH5 Fab (clone 338E6), where the variable domains derive from a mouse-naïve splenic B2 Fab, and the constant CL and CH1 domains are from a mutagenized human IgG1 scaffold. We solved the resulting structure and refined to a 2.1A resolution with Rwork/Rfree 0.2297/0.2658 (SI Appendix, Table S1). The truncated IbpA structure reveals three equal size domains: Domain D (light pink), middle Domain X (gray), and a C-terminal domain, from now on referred to as HCBD (teal) (Fig. 3A). Domain D comprises one of the four repeat domains (A, B, C, and D) present in the full-length structure of Ibp, which are highly similar to one another.
Fig. 3.
Crystal structure of truncated IbpA reveals two superantigen-like domains and one adhesion-like novel domain. (A) Truncated IbpA structure from complex crystal structure showing three equal-size domains: repeat Domain D (light pink) with disulfide bridge (yellow) and black arrow pointing at the unstructured loop, middle Domain X (gray), and HCBD (green-teal). (B) Close-up analysis of superimposed structures of Domain D (light pink) and HCBD (green-teal). (C) Close-up analysis of superimposed structure of related superantigen fold of protein L (magenta) and protein G (cyan). (D) Structure of novel adhesion-like domain of middle Domain X (gray). (E) Experimental fit of SAXS data of truncated IbpA apo analyzed with rigid model by FoXS and flexible multistate modeling with MultiFoXS servers and plotted by I(q).
Strikingly, despite having no sequence homology and low DALI server z score = 2.4, both Domain D and HCBD adopt a fold reminiscent of that of protein G and L (Fig. 3 B and C and SI Appendix, Fig. S3A). Domain D has three instead of four complete β strands (β1, β2, and β4), with a truncated β3* likely due to a disulfide bond between Cys317 and Cys338, bringing the two loops closer to each other and away from the β4 strand (Fig. 3A, yellow). An equivalent of Cys317 forming this S-S bond is present in all repeat domains of full-length IbpB and only in the last repeat of four in exchange of Y317 in the full-length IbpA. The alpha helix between β2 and β3 is shorter and turned ∼40° to 60° compared to protein G and L, respectively, and accommodates a long loop not present in protein G/L (Fig. 3A, black arrow and SI Appendix, Fig. S3B). HCBD also resembles a superantigen-like fold despite being located outside of the repeat region of IbpA, whereas no additional antibody binding sites were reported for other known superantigens (SI Appendix, Fig. S3C). While composed of two beta (and shorter β4*) strands, HCBD additionally has a metal binding site mediated by three Asp in a DGDGDG motif, the role of which is unclear (SI Appendix, Fig. S3D). The middle Domain X fold (Fig. 3D) distantly resembles other adhesion-type proteins, which link domains with unstructured regions (mucus-binding proteins, PDB ID: 3LYY and 4MT5) according to the DALI server (32), suggesting a lack of structural coordination between Domain D and HCBD.
To evaluate potential flexibility between the three domains of the truncated IbpA, we used small-angle X-ray scattering (SAXS) analysis to investigate the flexibility of truncated IbpA when not bound to Fab. The initial fit of the experimental data with a rigid body IbpA apo model from our crystal structure did not yield a good score using the FoXS server (χ2 = 5.33), suggesting that the solution structure may adopt additional conformations to that of our crystal structure (Fig. 3E) (33). Using MultiFoXS server, which allows for conformational sampling and multistate modeling, we stipulated flexibility in the two linker regions connecting the three domains. Using this analysis, we determined six distinct topologies from that of our crystal structure with an average χ2 = 0.64 (Fig. 3E and SI Appendix, Fig. S3E) (33). Taken together, our structure of truncated IbpA reveals three new domains, two of which (Domain D and HCBD) adopt a fold similar to protein G and L, despite no sequence conservation. The middle domain serves as a linker between these two independent superantigen-like domains, allowing truncated IbpA to sample multiple conformations.
Truncated IbpA in Complex with Mouse VH5 Fab Shows Contacts between Two Distinct IbpA Domains and Both Chains of the Fab.
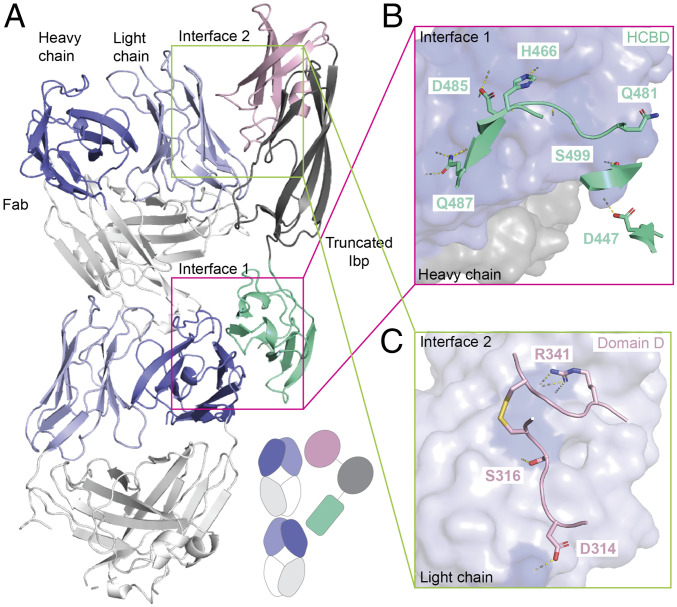
The asymmetric unit of our crystal contains two Fabs and two IbpAs, where each IbpA establishes contacts with the heavy chain of one Fab and the light chain of the other (Fig. 4A), producing a 2:2 stoichiometric complex (SI Appendix, Fig. S4 A and B). The primary Fab/IbpA interface is between the heavy chain and the HCBD (Fig. 4B) and is composed of 44 contacts between the HCBD and the D, C’’, and E β-strands of the VH Fab with 15 H-bonds between 7 residues of the HCBD and 12 residues of the Fab (SI Appendix, Table S2). This interface is discussed further in the following section. A secondary interface was also noted between the variable region of the light chain of the Fab and Domain D of IbpA (Fig. 4C and SI Appendix, Table S2). In contrast to protein L/G and HCBD, which maintain interactions with the Fab through formation of strong backbone-mediated beta-zippers with their β2 or β3 strands, the Domain D contact site in this interface maps to two unstructured loops between β2 and β4 (SI Appendix, Fig. S4C). The middle Domain X, between Domain D and the HCBD, only maintains three van der Waals contacts with the Fab (SI Appendix, Table S2). Taken together, both interfaces dock on the side of the variable domain of the Fab, outside of the conventional CDR binding region, reminiscent of the binding of known B cell superantigens, that is, protein A and protein L (SI Appendix, Fig. S4D). Our complex structure revealed that the VH domain of the Fab is contacted exclusively through the HCBD, while a smaller contact exists between the light-chain variable domain (VL) and Domain D of IbpA. These interfaces are investigated in more detail as follows.
Fig. 4.
Complex crystal structure of functional, truncated IbpA shows unconventional binding to mouse VH5 Fab clone 338E6 involving heavy and light chains. (A) Crystal structure showing two distinct interfaces between truncated IbpA and heavy and light chains of two mouse VH5 Fab fragments. Simplified cartoon present in lower right corner. (B) Close-up view of interface 1 between heavy chain (blue, surface representation) and participating residues of the HCBD (green-teal, cartoon representation); BSA ∼670 Å. (C) Close-up view of interface 2 between light chain (light blue, surface representation) and participating residues of Domain D (pink, cartoon representation); BSA ∼380 Å.
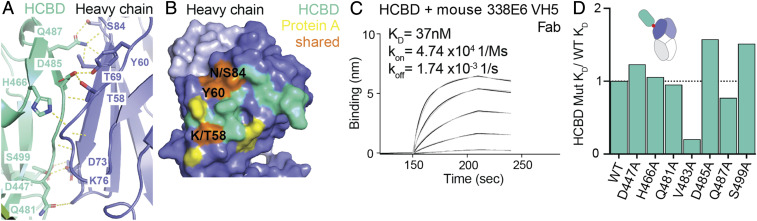
The C-Terminal HCBD of IbpA Confers VH Specificity.
The primary interface between IbpA and Fab revealed in the crystal structure involves interactions between the C-terminal HCBD and the VH domain of the Fab. This result is consistent with our B cell repertoire sequencing studies, where IbpA binding was heavily biased toward Fabs containing particular VH family members. This interface includes one salt bridge held by K76 and seven backbone H-bonds involving G55 in the CDR2 and R67, I70, R72, A75, and E82 in the framework region 3 (FR3) (Fig. 5A and SI Appendix, Table S2). Notably, the contact area of this primary interface is ∼633 Å2 and not only spans FR3, as typically seen for protein A superantigen, but also involves the CDR2 region of the Fab (SI Appendix, Fig. S5A). Despite SpA and HCBD sharing a similar VH germline bias, only three contact residues are commonly recognized on Ig (K/T58, Y60 and N/S84), suggesting a differential molecular recognition of the VH (Fig. 5B). To understand how this Ig binding interface is recognized by HCBD, we have mutated individual contact residues on HCBD to alanine and tested for binding by BLI. No individual alanine mutation on HCBD site was able to eliminate binding, and only a few had a slight effect. Interestingly, the V483A mutation shows a ∼5-fold higher affinity than wild-type HCBD. (Fig. 5 C and D and SI Appendix, Fig. S5B). These observations are likely attributable to the 37 nM affinity between HCBD and Fab, their slow dissociation, and their vast contact interface.
Fig. 5.
IbpA HCBD interacts with framework region of mouse VH5 heavy chain through extensive contact sites. (A) Close-up view of interface 1 between HCBD and mouse VH5. (B) Binding footprint of HCBD (green-teal), protein A (yellow), and three shared sites between the two (orange), mapped on the surface representation of heavy (dark blue) and light chains (light blue) of the Fab. (C) BLI binding plot between immobilized wild-type HCBD and analyzed mouse VH5 Fab versus time. (D) Alanine mutational analysis of measured KD of HCBD contact residues using bar graph, normalized to HCBD WT.
To investigate how this interaction translates across the repertoire of VH family members, we selected representative mouse and human Fabs from different VH families and tested for binding with HCBD. Consistent with our repertoire sequencing studies, binding to only mouse VH5 but not VH3 or VH14 and only human VH1 and VH3 but not VH4 germline specificities could be observed (SI Appendix, Fig. S5 C and D). Intriguingly, there were no discernable differences in key contact residues in amino acid sequences from other VH families (SI Appendix, Fig. S5A), therefore we investigated the possibility that these VH families might have intrinsic domain structural differences (i.e., Cα backbone differences) that might explain the biases in binding. We therefore compared the overall backbone structures of available atomic models of these Fab families. Although no major structural differences are seen between binders (slate) and nonbinders (black) Ig (SI Appendix, Fig. S5E), we hypothesize IbpA germline bias could be attributed to interface complementarity in this high-affinity interaction as many interactions are mediated by backbone atoms (slate circles). Taken together, we show that the HCBD is able to mediate strong Fab interactions with a clear heavy chain preference, although we cannot identify precisely how this specificity is conferred.
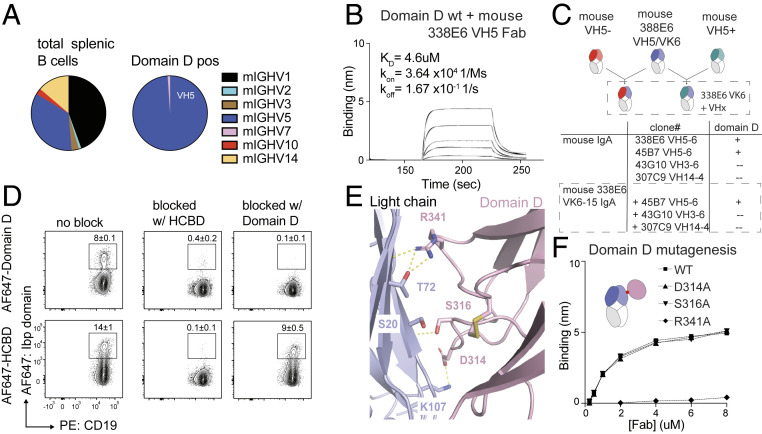
IbpA Domain D Contacts the Light Chain of the Fab in the Complex Structure.
All previously described B cell superantigens mediate their binding to antibodies through 50 to 70 amino acid long domains that are repeated in their structures, which for protein L was shown to confer an enhanced avidity toward their targets (34). To explore the role of Domain D in Fab binding, which has three other highly homologous repeats in the full-length IbpA (Domains A, B, and C), we further examined its interface with the Fab in our crystal structure. While our repertoire data with full-length IbpA suggest the bias in binding is mediated solely through the VH contacts, we wanted to establish whether this interface was real or a product of a crystal-packing artifact.
First, to determine any repertoire preference for Domain D, we sorted mouse B cells stained with fluorescently labeled Domain D and again observed a restricted binding to VH5 and VH7 gene families (Fig. 6A and SI Appendix, Fig. S6A) with no light-chain bias, again reiterating the original heavy chain bias of the full-length and truncated Ibp. Notably, our method for repertoire analysis used in this experiment had limitations with regard to detection of the whole spectrum of variable gene regions compared to next-generation sequencing (NGS) shown in Fig. 1D; however, a clear bias toward the heavy chain VH5/7 families was nevertheless observed. We have also validated Domain D VH specificity to selected mouse and human Fab clones by direct binding measurement by BLI, and, similarly to HCBD, only mouse VH5 and human VH1 and VH3 binding was observed (Fig. 6B and SI Appendix, Fig. S6 B and C).
Fig. 6.
Characterization of IbpA repeat Domain D contacts with Fab. (A) Heavy chain repertoire of sorted mouse CD19pos B cells binding to single repeat Domain D. (B) BLI binding plot between immobilized wild-type Domain D and analyzed mouse VH5 Fab versus time. (C) Experimental scheme of mouse Fabs with constrained light chain and swapped heavy chains (Top), table reflecting measure binding between wild-type Domain D and swapped Fabs; “+” = 10−6, “−” = no binding (Bottom). (D) Representative contour plots show wild-type mouse B cells stained with HCBD or domain with and without preincubation with unlabeled HCBD (Middle column) or Domain D (Right column). (E) Close-up view of crystal contacts between IbpA Domain D (light pink) and VL of a Fab (light blue). (F) Comparison plot of alanine substitutions in Domain D, directly involved residues in binding to Fab fragment versus Fab concentration determined by BLI.
Finally, to rule out the molecular functionality of this Domain D/light-chain interface, we generated hybrid Fabs that carried the Vk6-15 light chain from Fab 338E6 used in the complex crystal structure (where this interface was observed) and swapped in heavy chains from other VH families. Consistent with our repertoire and binding studies, Domain D bound only to Fabs containing the VH5 domain (Fig. 6C and SI Appendix, Fig. S6D). This observation was also confirmed with another repeat Ibp domain, Domain A, which also binds specifically to 338E6 VH5 Fab and not the hybrid Fab (SI Appendix, Fig. S6E). These data in conjunction with the lack of k and l light-chain bias in IbpA binding (SI Appendix, Fig. S1 C and D) strongly suggest that the interface between Domain D and the light chain in our structure is a crystal-packing artifact.
Next, to investigate whether Domain D competes with the HCBD for binding to VH, we tested if staining of polyclonal mouse B cells with labeled Domain D or HCBD could be blocked by preincubation with either purified Domain D or HCBD. If Domain D indeed binds light chains, we would expect a signal even after blocking with heavy chain–specific HCBD. However, Domain D staining could be blocked by both HCBD and Domain D, indicating binding to a shared heavy chain–specific binding site (Fig. 6D). HCBD staining, on the other hand, was merely reduced after preincubation with Domain D, consistent with the lower affinity (KD) and faster dissociation (koff) of Domain D than HCBD to Fab (Fig. 6D). In addition, we saw that Domain D exclusively binds to IbpA-positive but not IbpA-negative human B cells (SI Appendix, Fig. S6F). Finally, we mutated three Domain D residues apparent in the crystal-packing interface in the crystal structure: D314, S316, and R341 to A, and observed that both D314 and S316 are presumptive to bind to the heavy chain; however, the R341A mutant abrogated that binding (Fig. 6 E and F and SI Appendix, Fig. S6G). Altogether, these data suggest that Domain D binds the same heavy chain repertoire as HCBD but with lower affinity. Exact identification of the residues involved in that binding would require a dedicated study of the heavy chain–Domain D complex, but the observation that binding is abrogated with a R341A mutated construct suggests that at least R341 is involved.
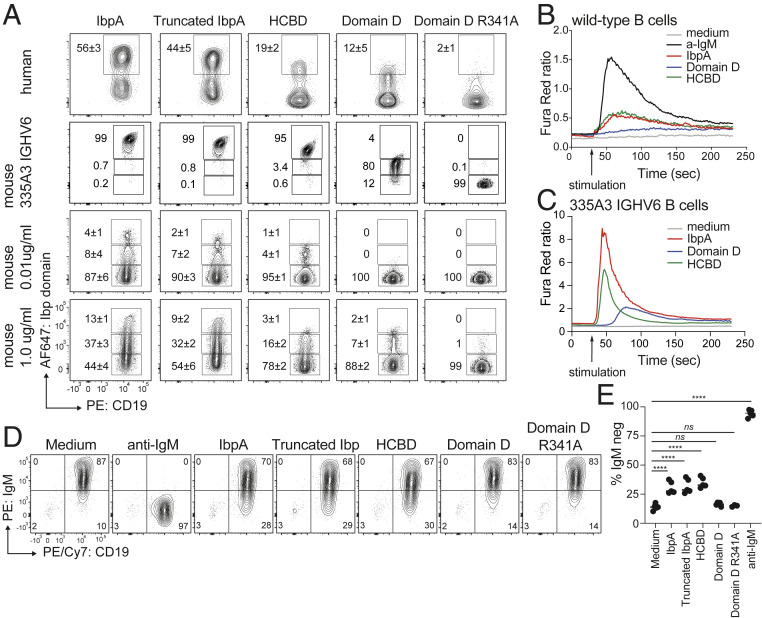
Stimulation with Individual IbpA Domains Is Sufficient to Induce BCR Signaling.
Next, we sought to compare the frequency of human and mouse B cells binding full-length and truncated IbpA versus single IbpA domains. The frequency of wild-type mouse and human B cells labeled with truncated IbpA was only slightly diminished compared to wild-type IbpA (Fig. 7A). However, even at higher concentrations, Domain D stained only 5% to 10% of splenic B cells. Furthermore, staining was almost completely abrogated with the R341A mutated construct (Fig. 7A). The same was true when a homogenous transgenic mouse VH6-expressing B cell population was examined. B cells could be brightly stained with full-length and truncated IbpA, but the signal decreased from HCBD to Domain D and was completely gone in samples stained with Domain D R341 (Fig. 7A). Furthermore, HCBD not only showed binding to a larger fraction of mouse B cells compared to Domain D but also induced calcium flux in wild-type and transgenic B cells (Fig. 7 B and C), whereas Domain D only induced a detectable calcium flux response when VH6 B cells were analyzed. The signal, however, was slower and diminished compared to HCBD (Fig. 7C). This could be the result of a longer dwell time of HCBD on the BCR, as suggested by the higher affinity and slower dissociation in BLI studies (Fig. 5C). In addition, surface IgM down-regulation occurred to a similar degree on wild-type B cells stimulated with full-length IbpA, truncated IbpA, and HCBD but not with single Domain D (Fig. 7 D and E).
Fig. 7.
Functional characterization of single IbpA domains. (A) Contour plots show frequency of human and mouse B cells binding to fluorescently labeled full-length, truncated, and single IbpA domains. (B and C) Ca2+ flux analysis of wild-type (B) or VH6 transgenic (C) mouse B cell after in vitro stimulation with full-length IbpA versus single IbpA domains. (D) Analysis of BCR down-regulation of MACS-purified mouse B cells after 2 h in vitro stimulation with single IbpA domains compared with full-length and truncated IbpA. (E) Quantification of B cell down-regulation with tested IbpA constructs.
Role of Domain D and HCBD in IbpA Interaction with Immunoglobulins.
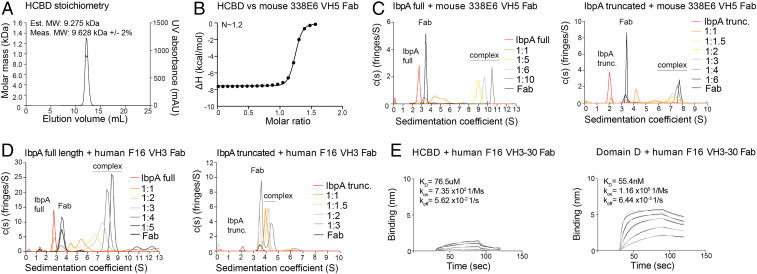
The convention for most receptor signaling models involves crosslinking of receptors through engagement of either a multivalent or multimerized ligand. Here, we have shown that a single domain of IbpA, the HCBD, can activate B cells through their BCR. To explore whether the HCBD itself can exist as a multimer, we investigated the multimeric states of this domain as well as for truncated and full-length IbpA. Using multi-angle light scattering (MALS), we show that the HCBD exists exclusively in a monomeric state, even at high concentrations, at least 1,000-fold above that used in the activation experiments (Fig. 8A). To explore the stoichiometry of HCBD binding to Fab, we employed isothermal titration calorimetry (ITC), which directly measures the thermodynamics of binding but can also estimate the stoichiometry of binding. Our results approximate a 1:1 binding stoichiometry (n = 1.2, Fig. 8B) between the HCBD and 338E6 VH5 Fab from our structure. This shows how soluble HCBD is monomeric yet is sufficient to induce signaling in B cells.
Fig. 8.
Role of multiple binding sites in full-length and truncated IbpA. (A) Stoichiometry of individual HCBD by MALS. (B) Stoichiometry of HCBD and Fab complex by ITC. (C) Stoichiometry of full-length (Left) or truncated (Right) IbpA and mouse 338E6 VH5 Fab complex by AUC. (D) Stoichiometry of full-length (Left) or truncated (Right) IbpA and human F16 VH3 Fab complex by AUC. (E) BLI binding plot between immobilized wild-type HCBD (Left) or Domain D (Right) and analyzed human VH3-30 Fab clone F16 versus time.
To further explore the stoichiometry of Fab binding to the full-length IbpA, we performed a series of sedimentation velocity analytical ultracentrifugation (AUC) experiments at increasing molar ratios between full-length IbpA and 338E6 VH5 Fab (Fig. 8C, Left). Each protein, when characterized alone, provided accurate molecular weight (MW) estimates for monomers and reasonable shape information based on SAXS and crystallography data (SI Appendix, Fig. S7C). Our titration experiments reveal the formation of large oligomers, with essentially a single species (9 S peak) present when full-length IbpA was mixed with a 5-fold molar excess of the Fab. The MW estimate of the 9 S complex is 343 kDa, similar to the sequence-based MW of 314 kDa for a 1:5 IbpA:Fab complex (SI Appendix, Fig. S7C). Both a 6-fold and 10-fold excess resulted in excess monomeric Fab at 3.5 S (Fig. 8C, Left). To better determine the stoichiometry of the IbpA:Fab complex at 10.5 S, we collected the data for the full-length IbpA:Fab (1:6 molar ratio) at 301 nm. At this wavelength, we only see an absorbance signal from the Fab rather than from both IbpA and Fab. This allows us to attribute all of the absorbance signal in the resulting c(s) distribution to Fab (SI Appendix, Fig. S7A). Therefore, the integration under the c(s) distribution can define a ratio for the absorbance per micromolar of Fab based on the intended loading concentration. We observed ∼4.7 molar equivalents of Fab in the complex peak, with ∼0.83 molar equivalent of the Fab sedimenting as a monomer, supporting the conclusion that the primary complex is a 1:5 IbpA:Fab complex (SI Appendix, Fig. S7A, Right). A 1:5 complex is consistent with Fab binding to four available repeat domains and the HCBD binding site.
By contrast, the truncated IbpA preincubated with 3-fold molar excess of the Fab yields essentially a single species at ∼7.2 S, with an estimated MW of 190 kDa, similar to the sequence-based MW of 182.3 kDa for a 1:3 complex (Fig. 8C, Right and SI Appendix, Fig. S7C). Using a similar approach as with full-length IbpA, when the 1:4 loading ratio is examined, we see ∼0.83 and ∼3.2 molar equivalent of monomeric Fab and Fab in the complex, respectively (SI Appendix, Fig. S7B). Thus, truncated IbpA may potentially have an additional Fab binding site not apparent in the full-length IbpA or form a mixture of different complex configurations. We have additionally tested the stoichiometry of full-length and truncated IbpA with human F16 VH3 Fab. Conversely to our results with mouse Fab, we observed 1:4 and 1:1 stoichiometries of F16 Fab to full-length and truncated IbpA, respectively (Fig. 8D). This result supports our BLI binding studies, where we observed Domain D and not HCBD binding to F16 Fab (SI Appendix, Figs. S5D and S6C). Taken together, we hypothesize that both Domain D and HCBD are complementary in binding to mouse and human Fabs and, given the potential flexibility of truncated IbpA, the observed higher stoichiometry for mouse Fab could be indicative of crosslinked binding rather than formation of a homogenous complex.
Due to the confounding results generated by the AUC measurements, we sought to investigate, at low-resolution, the solution structure of the apo full-length IbpA by SAXS. Similar to earlier studies of truncated IbpA, we used MultiFoXS, stipulating flexibility in the six linker regions between all domains (SI Appendix, Fig. S7D, average χ2 = 0.49). Although some flexibility was present in the more external domains (the unknown N-term domain in black and HCBD in green-teal) of the full-length IbpA, SAXS analysis of the entire IbpA supported mostly extended conformations (f/f0 ∼1.7) with a more arched architecture, suggesting a cooperative binding to Fabs between the linked domains (SI Appendix, Fig. S7E). Only one of the predicted full-length IbpA conformations was able to accommodate all five Fab molecules concurrently, forming a tightly coordinated complex. Taken together, both truncated and full-length IbpA structures show some level of flexibility, but the truncated construct has more structural freedom while binding to Fabs.
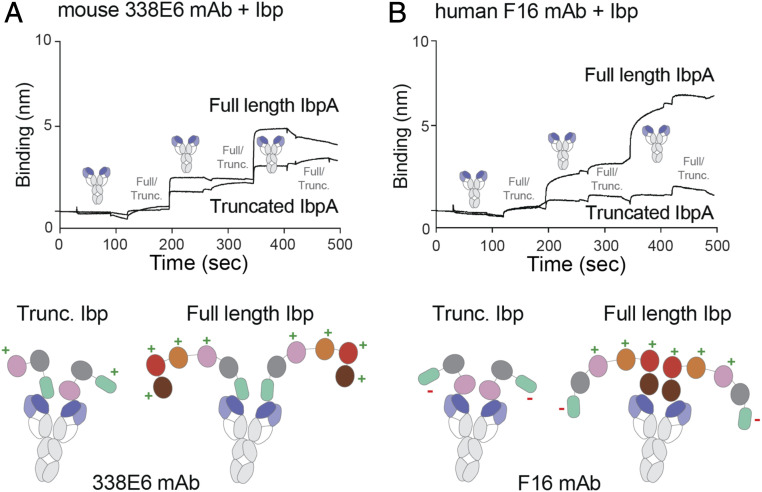
We sought to investigate whether differences in affinities between Domain D and HCBD affect Ibp binding to a full-length antibody (mAb). We tested for oligomerization between Ibp variants and two studied mAb, mouse 338E6 and human F16, by BLI. We immobilized mAb on a protein A tip and followed it by alternating analytes, Ibp (either truncated or full length) and mAb, for three rounds. Our previous BLI and AUC binding studies with 338E6 Fab showed that HCBD dominates the binding over Domain D, but still some low affinity participation from Domain D is present. We have therefore observed a formation of a continuous oligomer between truncated IbpA and 338E6 mAb, but only initial oligomerization events followed by saturation for the full-length IbpA (Fig. 9A). This suggests that truncated IbpA allows for binding more mAbs, whereas full-length IbpA adopts conformations that limit its oligomerization. In contrast to 338E6, Domain D is a dominant binding site for F16 Fab without any participation from HCBD according to our BLI and AUC data. Therefore, no oligomerization was observed for truncated IbpA and F16 mAb, consistent with a singular binding site coming from Domain D. Additionally, F16 rather than 338E6 mAb seems to form larger oligomers with full-length IbpA (Fig. 9B). Our structural modeling simplified with cartoons in Fig. 9 shows our proposed models of full-length antibodies binding to IbpA variants and forming varying degrees of oligomerization that is dependent on binding strengths of Domain D (and other repeats) and HCBD.
Fig. 9.
Proposed model of binding of truncated and full-length Ibp to two discrete clones. (A) Multimerization experiment by BLI of mouse 338E6 full-length antibody binding to either truncated or full-length Ibp (Top); proposed cartoon model of 338E5 mAb shows high levels of binding to both Ibp variants (Bottom), “+” indicates available binding site, and “−” indicates no binding site. (B) Multimerization experiment by BLI of human F16 full-length antibody binding to either truncated or full-length Ibp (Top); proposed cartoon model of F16 mAb shows high levels of binding only to full-length but not truncated Ibp (Bottom), “+” indicates available binding site, and “−” indicates no binding site.
Discussion
In this work, we molecularly characterize a novel Ibp expressed by a commensal R. gnavus which activates mouse and human B cells in vitro. We provide detailed repertoire analysis of sorted IbpA-bound mouse and human B cells confirming high affinities to mouse VH5/6/7 and human VH1/3 families and do not detect any associated light-chain bias. However, we also detected a range of mouse BCRs, mostly expressing VH1 families, that bound Ibps with lower affinities.
To uncover the molecular basis for this binding, we determined the structure of IbpA bound to Fab, revealing that truncated IbpA primarily engages mouse VH5 Fab with two separate domains. First, a C-terminal domain called HCBD is able to strongly bind to the VH region. Second, Domain D binds weakly to a site on the VH domain that likely overlaps with the binding site of the HCBD. Even though the crystal structure suggested an interface between Domain D and the Fab light chain, further investigations confirmed a heavy chain–restricted binding pattern, leading us to conclude that the interface between Domain D and the light chain in the crystal structure is a crystal-packing artifact. In concert with the number of contacts at each interface, Fab binding was abrogated by mutating a single Domain D residue R314A, whereas binding of HCBD was only moderately affected by targeted mutagenesis of individual HCBD residues, suggesting a distribution of binding energy across the interface and no “hot-spot” residues. The exact molecular basis for VH germline recognition could not be determined in this study, as no clear sequence or structural differences between different VH families were apparent. Additionally, as shown for 338E6 and F16 clones, Domain D and HCBD may exhibit different affinities for various mouse and human clones, which makes sequence and structure-based analysis of Ibp germline bias complicated. These domains may therefore demonstrate either redundant or nonredundant binding depending on the precise variable region structure of the targeted antibody. Interestingly, our findings for the mouse system emphasize that the HCBD, with its stronger affinity and slower dissociation rate, outcompetes Domain D binding and elicits vast mouse BCR activation in vitro without self-multimerization. This remarkable monovalent ligation of BCR without crosslinking has been also observed for a certain HEL-specific BCR (35); however, we cannot entirely exclude other Ibp-binding partners on the surface.
Having identified two binding sites on a single truncated IbpA molecule in our structure, we examined its binding capabilities by BLI and AUC and compared it with wild-type IbpA. We observed that truncated IbpA has higher affinity and, based on AUC, shows stoichiometry of three mouse Fab molecules per truncated IbpA. This is more than its number of identified binding sites; one explanation for this greater valency for truncated IbpA and not the full-length IbpA would be the possibility of a third binding site in truncated IbpA that is somehow sequestered in the full-length IbpA. Indeed, the full-length IbpA is more rigid in structure, as determined by our SAXS data, whereas the conformational flexibility of the truncated IbpA is much greater.
The same experiments performed using human Fab gave 1:1 stoichiometry with truncated IbpA and 1:4 for the full length, consistent with the engagement of repeat domains and not HCBD. Whether there is a possible coordination between all repeat domains that constrains movements of full-length IbpA remains to be investigated. Our oligomerization experiments with mAbs suggest varying degrees of daisy-chaining that are tuned by the binding strength of Domain D (and other repeats) and HCBD in Ibp. These subtle but interesting observations could potentially enable R. gnavus to affect the magnitude of its IgA coating and in turn its balance with other commensal bacteria. Further studies should assess metagenomic heterogeneity of R. gnavus Ibp genes, especially for mutations within repeat and HCBD domains and their functional consequences (36). However, whether intentional IgA coating through Ibp is consequently also beneficial for R. gnavus still needs to be determined. Furthermore, we hypothesize IbpA/B to be expressed on the cell surface via a sortase-mediated mechanism, but the presence of secreted forms of IbpA/B cannot be excluded (12). Soluble IbpA could possibly reach and stimulate immune cells typically excluded from mucosal surfaces in healthy individuals.
The in vivo functional significance of having a different number of binding sites for human Igs is currently unclear. Whether truncated IbpA has a hidden site for mouse 338E6 Fab or higher propensity to crosslink that could also apply to human Igs should be further investigated. Having multiple weaker binding domains could be potentially beneficial for R. gnavus. Unlike T cell SAgs, BCR-targeting SAgs have to additionally accommodate high levels of circulating immunoglobulins. Paradoxically, SpA, with its ability to also bind the Fc-regions of soluble IgG, has been attributed a critical role for efficient BCR crosslinking and induction of AICD (37). Despite the single Ibp HCBD being able to stimulate B cells in vitro, our data suggest that having multiple available binding sites enhances avidity and B cell activation, as shown by our binding and cellular studies. Therefore, in a physiological environment where IgA dominates as dimers, IbpA/B having the potential to bind up to five antibodies is consistent with high IgA coating of R. gnavus in vivo (12).
Besides IbpA being expressed by a commensal microbiota and utilizing similar mechanism to common B cell SAgs, it is also unique in its ability to bind to a VH with two distinct domains. Despite Domain D and HCBD having a superantigenic-like fold, they have little sequence similarity or superantigenic potential when compared with either protein G or L. This study opens the need to search for other commensals expressing similar Ibps as a novel mechanism of host-microbiota communication.
Materials and Methods
Crystal Structure Determination and Analysis.
Crystals of the truncated IbpA-mouse VH5 Fab complex were grown at room temperature by hanging drop vapor diffusion. Equal molar volumes of a protein solution containing 15 mg/mL complex was mixed with a reservoir solution containing 0.2 M calcium chloride and 18% PEG 4000. Crystals were cryoprotected in 0.2 M calcium chloride, 18% PEG 3350, and 20% ethylene glycol and flash frozen in liquid nitrogen. One dataset was collected from a single region of a single crystal at 100 K at advanced photon source (APS) beamline 24ID-E (λ = 0.9791 Å) on an Eiger detector. The data were processed, scaled, and merged using iMosfilm with diffraction limits of 2.1 Å (38). Data collection and processing statistics are listed in SI Appendix, Table 1.
VH5 Fab, with its CDR and V-C hinges omitted, was used as the search model using PHASER as molecular replacement tool (39). Two VH5 Fabs packed against each other were determined per asymmetric unit. Since no search model was known for IbpA, the remaining electron density was fitted with secondary structures using Buccaneer, an auto-build package in CCP4 i2 (40). Cycles of manual model building and refinement (torsional noncrystallographic symmetry restraints applied throughout) with Coot and PHENIX allowed us to connect helices and strands and trace the backbone of truncated IbpA (41, 42). Electron density for the most C-terminal ∼40 amino acids of IbpA is not visible, which suggest disordered structure, which was also confirmed by structure prediction with the Phyre server (43). Refinement and validation statistics are listed in SI Appendix, Table 1. Structure figures were generated with PyMOL (https://pymol.org/2/).
Sizing Analysis.
For stoichiometry analysis, 1 mg of HCBD was injected onto a preconditioned size-exclusion chromatography column (Superdex 200 10/300 GL, GE Healthcare) with Hepes buffer saline (10 mM Hepes pH 7.2, 150 mM NaCl) at a flow rate of 0.5 mL/min. The effluent of the SEC column flowed through an inline ultraviolet/visible detector (Waters Corporation), the MALS/dynamic light scattering (DLS) detector, and an Optilab UT-rEX dRI detector (Wyatt Technology Corporation) at room temperature. Data collection and analysis was performed with ASTRA software, version 7.1.1 (Wyatt Technology Corporation). ASTRA software utilizes a patented method for the interdetector delay and band broadening correction (44). First-order fit Zimm formalism was used for all molar mass calculation.
Binding Studies.
All the binding studies were carried out using BLI (either ForteBio or Octet K2 from Pall Life Sciences) at room temperature (22 °C). Concentrations of different IbpA constructs were individually immobilized on a Ni-NTA sensor to reach ∼2 to 3 RU for binding with mouse Fab clones at different concentrations and responses (in nanometer) were recorded. Hepes buffer saline (10 mM Hepes pH 7.2, 150 mM NaCl) was used in all the measurements, and sensor tips were recharged with 350 mM imidazole after each round. For binding studies with human Fab clones, Ibp constructs were nonspecifically labeled with NHS-SS-Biotin at 1:1 ratio. Excess of biotin was removed by size-exclusion chromatography, and the level of biotinylation was assessed by sodium dodecyl sulfate–polyacrylamide gel electrophoresis shift assay with Trap protein. Biotinylated Ibp constructs were immobilized with Streptavidin sensor tips and recharged with 0.1 M glycine pH 2.5. Traces were reference subtracted, and kinetic parameters (KD, kon, koff) were calculated by fitting the data with ForteBio built-in analysis software.
Mice.
C57BL/6J mice were used for in vitro stimulation experiments, labeling studies, and sequencing analysis. IgH335A3/wt Igk335A3/wt VH6 BCR transgenic mice were generated as part of a separate study. Mice were crossed on Rag1−/− background (JAX) before analysis to avoid BCR expression from wild-type alleles.
Repertoire Analysis of Sorted Ibp-Binding Mouse and Human B Cells.
Peripheral blood mononuclear cells were obtained from healthy human donors; all subjects provided written informed consent for participation. This study was approved by the University of Chicago Institutional Review Board (IRB) under IRB #12623B. Cells were stained with labeled full-length IbpA and IbpB, and indicated B cell populations were sorted using an AriaIIIu cell sorter (BD). Postsort analysis showed purity of ≥95%. Cells were washed and RNA purified using RNeasy mini kits (Qiagen) following the manufacturer’s recommendations. Samples were sent to iRepertoire, Inc., for sequencing heavy and light-chain repertoires by NGS. Alternatively, repertoire analysis in Fig. 6A was performed by sorting Domain D labeled and total splenic CD19 B cells. After RNA purification (RNeasy mini kit, Qiagen), complementary DNA (cDNA) was synthesized using the SuperScript IV First Strand cDNA Synthesis kit (Thermo Fisher). Heavy chain variable regions were amplified using the forward primer MsVHE (GGGAATTCGAGGTGCAGCTGCAGGAGTCTGG) and CM reverse primer (AGGGGGCTCTCGCAGGAGACGAGG). After agarose gel electrophoresis, PCR products were excised and purified using the QIAquick Gel Extraction Kit (Qiagen). PCR fragments were cloned into a TOPO vector using the TOPO TA Cloning Kit for Sequencing, with One Shot TOP10 Chemically Competent E. coli (Thermo Fisher). After transformation, bacteria were plated on Luria–Bertani agar containing carbenicillin, 192 individual colonies were picked and miniprepped, and inserts were Sanger sequenced using M13 forward primers. Sequences were mapped to mouse-variable region genes using IMGT (www.imgt.org).
Ibp HCBD and Repeat Domain Blocking Experiments.
To block mouse B cell staining with labeled repeat domain, cells were incubated with high-dose (100 μg/mL) unlabeled HCBD for 20 min at 4 to 8 °C and washed and stained with labeled repeat domain at the indicated concentrations for 20 min at 4 to 8 °C. Next, cells were stained with fluorophore-labeled mAbs before acquisition on an LSR II cytometer (BD).
Mouse B Cell In Vitro Stimulation.
MACS-purified untouched B cells were counted using a Neubauer counting chamber, and cells in Roswell Park Memorial Institute (RPMI) medium supplemented with 10% fetal calf serum (Gibco) were transferred to a 96-well U-bottom plate (150,000 per well). For analysis of BCR, down-regulation cells were stimulated for 2 h at 37 °C with indicated Ibp constructs at 10 μg/mL or F(ab’)2 -goat anti-mouse (10 μg/mL). Before stimulation with Ibp constructs, possible endotoxin contamination was minimized using High Capacity Endotoxin Removal Spin Columns (Thermo Fisher).
ITC.
HCBD and VH5 Fab pure protein samples in matching buffers and protein concentrations measured by BCA were analyzed for binding stoichiometry using Malvern MicroCal iTC200. The 200 μL of 30 μM HCBD was injected into the measured cell kept at 30 °C, and then 40 μL of 1 mM of VH5 Fab was titrated into the temperature-regulated cell in one preinjection of 0.4 μL and 20 subsequent injections of 2 μL. Data were analyzed via the MicroCal ITC Origin Analysis software to extract the binding stoichiometry.
AUC.
Experiments were performed using a Beckman Coulter XL-I analytical ultracentrifuge with an An-60 Ti rotor. Data were collected using both interference and absorbance optics (wavelength was varied depending on sample). Experiments were run at a speed of 36,000 rpm for 16 to 20 h or until sedimentation became insignificant. Proteins were purified by size-exclusion chromatography into 10 mM Hepes pH 7.2 and 150 mM NaCl and diluted to desired concentrations in the same buffer prior to loading the samples. Volumes of 450 μL were loaded into two-sector epon-charcoal 1.2-cm centerpieces with sapphire windows. Samples were equilibrated at 20 °C for at least 1 h before beginning experiments. Data were analyzed using the continuous c(s) distribution model in SEDFIT version 16.1 (45). Partial specific volumes, buffer density, and buffer viscosity were estimated using SEDNTERP (46) (20130813 beta release).
The S20,w value is the sedimentation coefficient of a single species corrected for protein partial specific volume, buffer density, viscosity, and temperature. The S20,w value, frictional ratio (f/f0), and experimental MW (MWexp, kDa) were reported by SEDFIT’s c(s) distribution analysis. The sequence-based MW (MWseq) was calculated using ProtParam.
SAXS Analysis.
SAXS data were collected at the SIBYLS 12.3.1 beamline at the Advanced Light Source using High Throughput Mail-In program. Scattering measurements were performed on 20-μL samples at concentrations ranging between 0.1 and 2 mg/mL of truncated or full-length IbpA. Collected datasets were processed using ATSAS and Scatter software by trimming low/high scattering angles and adjusting Rg and dmax based on Guinier peak analysis and real space plots. Multistate modeling was done in MultiFoXS by indicating flexible regions in the models, analyzing scattering profiles, and fitting them to the experimental data (33, 47). Full-length IbpA models were generated in Coot by using a combination of duplicating repeat domains and Phyre server predictions (43). We defined the linker residues in truncated IbpA models (79-83, 157-163) and full-length IbpA models (85-89, 151-155, 218-222, 285-289, 352-356, 430-436) as flexible. The server sampled over 10,000 conformations, calculated their SAXS profiles, and enumerated and scored multistate models. The quality of fit between models and experimental SAXS data was assessed by the χ2 value <1 based on I(q) plots. Frictional ratio (f/f0) of models were calculated using HullRad (48).
Supplementary Material
Acknowledgments
This work is based upon research conducted at the Northeastern Collaborative Access Team (NE-CAT) beamlines, which are funded by the National Institute of General Medical Sciences from NIH grant (P30 GM124165). The Eiger 16M detector on the 24-ID-E beamline is funded by a NIH-ORIP HEI grant (S10OD021527). This research used resources of the Advanced Photon Source, a US Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract No. DE-AC02-06CH11357. We thank the staff of the APS at NE-CAT (24ID-E) for their use and assistance with X-ray beamlines and for help and advice during data collection. We thank Dr. Steven Erickson for providing 335A3 VH6 BCR transgenic mice. We thank Dr. Ahmed Rohaim and Dr. Sobhan Roy for help and advice on solving the crystal structure, Dr. Kristof Nolan for help and advice on solving the crystal structure and processing SAXS data, and Dr. Peter Chung for help and advice with ITC. We thank the staff of SIBYLS, which is supported by the award DOE-BER IDAT DE-AC02-05CH11231 and NIGMS ALS-ENABLE grants (P30 GM124169 and S10OD018483), where SAXS data were collected and preprocessed. We thank the University of Chicago Flow Cytometry Core for assistance with cell sorting and the University of Chicago DNA Sequencing core for assistance with plasmid minipreps and sequencing. This work was supported by NIH grants R01A115471 (to E.J.A); U01AI125250, R01AI144094, and R01AI038339 (to A.B.); R01GM094363 (to A.B.H.); and T32GM007281 and F30AI124476 (to J.J.B.) as well as a fellowship by the German Research Foundation (to C.D.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2023898118/-/DCSupplemental.
Data Availability
The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.wwpdb.org (PDB ID code 7KPJ).
References
- 1.Round J. L., Palm N. W., Causal effects of the microbiota on immune-mediated diseases. Sci. Immunol. 3, eaao1603 (2018). [DOI] [PubMed] [Google Scholar]
- 2.Hooper L. V., Gordon J. I., Commensal host-bacterial relationships in the gut. Science 292, 1115–1118 (2001). [DOI] [PubMed] [Google Scholar]
- 3.Geva-Zatorsky N., et al., Mining the human gut microbiota for immunomodulatory organisms. Cell 168, 928–943.e11 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Proft T., Fraser J. D., Bacterial superantigens. Clin. Exp. Immunol. 133, 299–306 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Choi Y. W., et al., Residues of the variable region of the T-cell-receptor β-chain that interact with S. aureus toxin superantigens. Nature 346, 471–473 (1990). [DOI] [PubMed] [Google Scholar]
- 6.Dellabona P., et al., Superantigens interact with MHC class II molecules outside of the antigen groove. Cell 62, 1115–1121 (1990). [DOI] [PubMed] [Google Scholar]
- 7.Graille M., et al., Crystal structure of a Staphylococcus aureus protein A domain complexed with the Fab fragment of a human IgM antibody: Structural basis for recognition of B-cell receptors and superantigen activity. Proc. Natl. Acad. Sci. U.S.A. 97, 5399–5404 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Derrick J. P., Wigley D. B., Crystal structure of a streptococcal protein G domain bound to an Fab fragment. Nature 359, 752–754 (1992). [DOI] [PubMed] [Google Scholar]
- 9.Patella V., Bouvet J. P., Marone G., Protein Fv produced during vital hepatitis is a novel activator of human basophils and mast cells. J. Immunol. 151, 5685–5698 (1993). [PubMed] [Google Scholar]
- 10.Grover R. K., et al., A unique human mycoplasma protein that generically blocks antigen-antibody union. Science 343, 656–661 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Graille M., et al., Complex between Peptostreptococcus magnus protein L and a human antibody reveals structural convergence in the interaction modes of Fab binding proteins. Structure 9, 679–687 (2001). [DOI] [PubMed] [Google Scholar]
- 12.Bunker J. J., et al., B cell superantigens in the human intestinal microbiota. Sci. Transl. Med. 11, eaau9356 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Qin J., et al., A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464, 59–65 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Arumugam M., et al., Enterotypes of the human gut microbiome. Nature 473, 174–180 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bell A., et al., Elucidation of a sialic acid metabolism pathway in mucus-foraging Ruminococcus gnavus unravels mechanisms of bacterial adaptation to the gut. Nat. Microbiol. 4, 2393–2404 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tailford L. E., et al., Discovery of intramolecular trans-sialidases in human gut microbiota suggests novel mechanisms of mucosal adaptation. Nat. Commun. 6, 7624 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lloyd-Price J., et al., Multi-omics of the gut microbial ecosystem in inflammatory bowel diseases. Nature 569, 655–662 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hall A. B., et al., A novel Ruminococcus gnavus clade enriched in inflammatory bowel disease patients. Genome Med. 9, 103 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Azzouz D., et al., Lupus nephritis is linked to disease-activity associated expansions and immunity to a gut commensal. Ann. Rheum. Dis. 78, 947–956 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chua H.-H., et al., Intestinal dysbiosis featuring abundance of Ruminococcus gnavus associates with allergic diseases in infants. Gastroenterology 154, 154–167 (2018). [DOI] [PubMed] [Google Scholar]
- 21.Zheng H., et al., Altered gut microbiota composition associated with eczema in infants. PLoS One 11, e0166026 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Arevalo P., VanInsberghe D., Elsherbini J., Gore J., Polz M. F., A reverse ecology approach based on a biological definition of microbial populations. Cell 178, 820–834.e14 (2019). [DOI] [PubMed] [Google Scholar]
- 23.Sorbara M. T., et al., Functional and genomic variation between human-derived isolates of Lachnospiraceae reveals inter- and intra-species diversity. Cell Host Microbe 28, 134–146.e4 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Crost E. H., et al., Utilisation of mucin glycans by the human gut symbiont Ruminococcus gnavus is strain-dependent. PLoS One 8, e76341 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Henke M. T., et al., Ruminococcus gnavus, a member of the human gut microbiome associated with Crohn’s disease, produces an inflammatory polysaccharide. Proc. Natl. Acad. Sci. U.S.A. 116, 12672–12677 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Silverman G. J., Goodyear C. S., Confounding B-cell defences: Lessons from a staphylococcal superantigen. Nat. Rev. Immunol. 6, 465–475 (2006). [DOI] [PubMed] [Google Scholar]
- 27.Goodyear C. S., Silverman G. J., Death by a B cell superantigen: In vivo VH-targeted apoptotic supraclonal B cell deletion by a Staphylococcal toxin. J. Exp. Med. 197, 1125–1139 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Silverman G. J., et al., A B cell superantigen-induced persistent “Hole” in the B-1 repertoire. J. Exp. Med. 192, 87–98 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pauli N. T., et al., Staphylococcus aureus infection induces protein A-mediated immune evasion in humans. J. Exp. Med. 211, 2331–2339 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim H. K., Falugi F., Missiakas D. M., Schneewind O., Peptidoglycan-linked protein A promotes T cell-dependent antibody expansion during Staphylococcus aureus infection. Proc. Natl. Acad. Sci. U.S.A. 113, 5718–5723 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schneewind O., Missiakas D. M., Protein secretion and surface display in Gram-positive bacteria. Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci. 367, 1123–39 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Holm L., Park J., DaliLite workbench for protein structure comparison. Bioinformatics 16, 566–567 (2000). [DOI] [PubMed] [Google Scholar]
- 33.Schneidman-Duhovny D., Hammel M., Tainer J. A., Sali A., FoXS, FoXSDock and MultiFoXS: Single-state and multi-state structural modeling of proteins and their complexes based on SAXS profiles. Nucleic Acids Res. 44 (W1), W424-9 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kastern W., Sjöbring U., Björck L., Structure of peptostreptococcal protein L and identification of a repeated immunoglobulin light chain-binding domain. J. Biol. Chem. 267, 12820–12825 (1992). [PubMed] [Google Scholar]
- 35.Kim Y.-M., et al., Monovalent ligation of the B cell receptor induces receptor activation but fails to promote antigen presentation. Proc. Natl. Acad. Sci. U.S.A. 103, 3327–3332 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Palm N. W., et al., Immunoglobulin A coating identifies colitogenic bacteria in inflammatory bowel disease. Cell 158, 1000–1010 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ulloa-Morales A. J., Goodyear C. S., Silverman G. J., Essential domain-dependent roles within soluble IgG for in vivo superantigen properties of staphylococcal protein A: Resolving the B-cell superantigen paradox. Front. Immunol. 9, 2011 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Battye T. G., Kontogiannis L., Johnson O., Powell H. R., Leslie A. G., iMOSFLM: A new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallogr. D Biol. Crystallogr. 67, 271–281 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McCoy A. J., et al., Phaser crystallographic software. J. Appl. Cryst. 40, 658–674 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cowtan K., The Buccaneer software for automated model building. 1. Tracing protein chains. Acta Crystallogr. D Biol. Crystallogr. 62, 1002–1011 (2006). [DOI] [PubMed] [Google Scholar]
- 41.Emsley P., Lohkamp B., Scott W. G., Cowtan K., Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Adams P. D., et al., PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kelley L. A., Mezulis S., Yates C. M., Wass M. N., Sternberg M. J., The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 10, 845–858 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wyatt P. J., Submicrometer particle sizing by multiangle light scattering following fractionation. J. Colloid Interface Sci. 197, 9–20 (1998). [DOI] [PubMed] [Google Scholar]
- 45.Schuck P., Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and Lamm equation modeling. Biophys. J. 78, 1606–1619 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ribaric S., Peterec D., Sketelj J., Computer aided data acquisition and analysis of acetlycholinesterase velocity sedimentation profiles. Comput. Methods Programs Biomed. 49, 149–156 (1996). [DOI] [PubMed] [Google Scholar]
- 47.Dyer K. N., et al., High-throughput SAXS for the characterization of biomolecules in solution: A practical approach. Methods Mol. Biol. 1091, 245–258 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fleming P. J., Fleming K. G., HullRad: Fast calculations of folded and disordered protein and nucleic acid hydrodynamic properties. Biophys. J. 114, 856–869 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.wwpdb.org (PDB ID code 7KPJ).