Significance
Necroptosis has been shown to be involved in mediating pathology of major human inflammatory and degenerative diseases. Necroptosis is mediated by the activation of death receptors in response to the stimulation of cognate ligands such as the activation of TNFR1 by TNF. Here we provide unexpected insight that necroptosis promotes the activation of ER-localized transmembrane unfolded protein response (UPR) signal sensors, including protein kinases IRE1α and PERK in an unusual manner without disrupting their binding with GRP78. Thus, our results demonstrate that necroptosis promotes an unconventional ER stress response by directly disrupting the ER membrane without inducing the accumulation of misfolded proteins and the role of RNA as the content of extracellular vesicles released by necroptotic cells.
Keywords: necroptosis, ER stress, UPR sensors, PERK, IRE1α
Abstract
Necroptosis is a form of regulated necrosis mediated by the formation of the necrosome, composed of the RIPK1/RIPK3/MLKL complex. Here, we developed a proximity ligation assay (PLA) that allows in situ visualization of necrosomes in necroptotic cells and in vivo. Using PLA assay, we show that necrosomes can be found in close proximity to the endoplasmic reticulum (ER). Furthermore, we show that necroptosis activates ER stress sensors, PERK, IRE1α, and ATF6 in a RIPK1-RIPK3-MLKL axis–dependent manner. Activated MLKL can be translocated to the ER membrane to directly initiate the activation of ER stress signaling. The activation of IRE1α in necroptosis promotes the splicing of XBP1, and the subsequent incorporation of spliced XBP1 messenger RNA (mRNA) into extracellular vesicles (EVs). Finally, we show that unlike that of a conventional ER stress response, necroptosis promotes the activation of unfolded protein response (UPR) sensors without affecting their binding of GRP78. Our study reveals a signaling pathway that links MLKL activation in necroptosis to an unconventional ER stress response.
Necroptosis is a regulated necrotic cell death mechanism that can be activated upon stimulation of death receptors, such as TNFR1, by their cognate ligands (1–3). Activation of TNFR1 promotes the formation of an intracellular complex, known as complex I, which controls the activation of receptor-interacting protein kinase 1 (RIPK1). Activated RIPK1 interacts with RIPK3 to promote the activation of RIPK3, which in turn recruits and phosphorylates mixed lineage kinase-like protein (MLKL) to form complex IIb (the necrosome) (4–8). Phosphorylation of MLKL by RIPK3 unleashes its N-terminal four-helix bundle (4HB) domain to promote its oligomerization into oligomers, which can permeabilize the cell membrane to promote cell lysis (9, 10). The phosphorylated MLKL localizes not only to the plasma membrane (PM) but also to internal membranes (11). However, while the mechanism that promotes the loss of cytoplasmic membrane integrity during necrosis has been extensively characterized, we know very little about the mechanism and consequence that mediates the damage to intracellular organelles during necroptosis.
The endoplasmic reticulum (ER) is an intracellular organelle where all proteins in transit through the secretory pathway are folded, modified, and assembled into multisubunit complexes. The state of protein folding in the ER is monitored by three ER-localized transmembrane unfolded protein response (UPR) signal sensors, including two protein kinases, IRE1 (inositol requiring kinase 1) (12, 13) and PERK (double-stranded RNA-activated protein kinase-like ER kinase) (14), and the transcription factor ATF6 (activating transcription factor 6) (15). Under normal homeostatic conditions, UPR sensors, including IRE1, PERK, and ATF6, are known to be kept inactive by the interaction of their luminal domains with protein chaperone GRP78 (BiP). In response to ER stress, accumulated unfolded proteins sequester GRP78 from the UPR sensors, which promotes the activation of IRE1, PERK, and ATF6 and the induction of GRP78. Activation of IRE1α, an evolutionarily conserved ER sensor, promotes its endoribonuclease activity, which leads to the splicing of a 26-nucleotide intron from XBP1 messenger RNA (mRNA). Thus, disrupting the binding of GRP78 with IRE1α and PERK and transcriptional induction of GRP78 has been recognized as the hallmarks of the ER stress response (16).
Here we investigated the dynamics of necrosomes during necroptosis using proximity ligation assay (PLA). We found an enriched association of necrosomes with ER, which led us to study the involvement of the ER stress response during necroptosis. We found a robust activation of UPR sensors, including the phosphorylation of IRE1α, PERK, and the cleavage of ATF6 during necroptosis. Activation of IRE1α by necroptosis promoted Xbp-1 splicing and loading of sXbp-1 in extracellular vesicles (EVs). We investigated the mechanism by which necroptosis promotes the activation of UPR sensors. We found that oligomerization of MLKL could promote the disruption of the ER membrane and activation of UPR sensors without disrupting their binding with GRP78. Our study suggests that necroptosis promotes an unconventional ER stress response by directly disrupting the ER membrane without inducing the accumulation of misfolded proteins.
Results
Detection of the RIPK1-RIPK3 Necrosome In Situ by PLA.
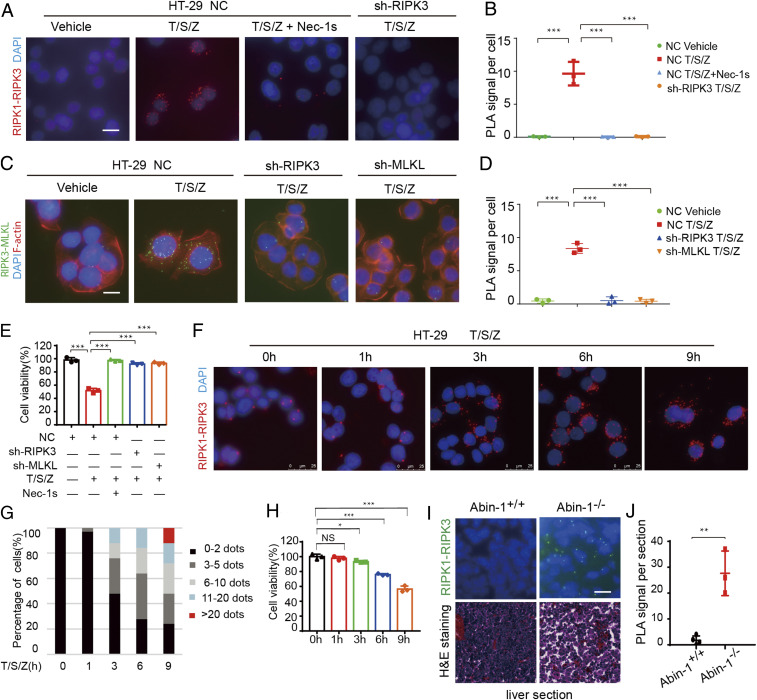
We developed a PLA for detecting the interaction of RIPK1-RIPK3 during necroptosis in situ. Necroptosis of HT-29 cells was induced by treatment with TNF (T), SM-164 (S), and pan-caspase inhibitor z-VAD.fmk (Z) (T/S/Z) (17). Interaction of RIPK1 and RIPK3 was detected specifically in T/S/Z treated HT-29 cells (Fig. 1 A and B). The PLA-RIPK1/RIPK3 interaction signal was inhibited upon the treatment with RIPK1 kinase inhibitor Nec-1s or knockdown of RIPK3 (Fig. 1 A and B). Furthermore, the PLA-RIPK3/MLKL interaction signal was inhibited upon knockdown of RIPK3 or MLKL (Fig. 1 C and D). The levels of PLA-RIPK1/RIPK3 and RIPK3/MLKL signals per cell correlated with viability (Fig. 1 B, D, and E), suggesting the specificity of the assay in determining the interaction of RIPK1 and RIPK3 and that of RIPK3 and MLKL in necroptotic cells. A PLA assay was also developed for detecting in situ RIPK1-RIPK3 interacting signal in necroptosis of mouse cells, and the specificity of signal was demonstrated by its absence in RIPK1 or RIPK3 knockout cells (SI Appendix, Fig. S1 A and B). Thus, this PLA assay can be used to detect in situ specific interaction of RIPK1-RIPK3 and RIPK3-MLKL, which are the key components of the necrosome during necroptosis of human and mouse cells.
Fig. 1.
Detection of the RIPK1-RIPK3 necrosome in situ by PLA. (A and B) HT-29 cells were treated with human TNF (50 ng/mL), SM-164 (100 nM), z-VAD.fmk (25 μM), and Nec-1s (10 μM) as indicated. The RIPK1-RIPK3 complex was detected by in situ PLA. (Scale bar, 10 μm.) RIPK1-RIPK3 in situ PLA signals per cell were quantified (>100 cells per group from three independent experiments were assessed) (B). (C and D) HT-29 cells were treated with human TNF (50 ng/mL), SM-164 (100 nM), z-VAD.fmk (25 μM), and Nec-1s (10 μM) as indicated. The RIPK3-MLKL complex was detected by in situ PLA. (Scale bar, 10 μm.) RIPK3-MLKL in situ PLA signals per cell were quantified (>100 cells per group from three independent experiments were assessed) (D). (E) The viability of the HT-29 cells used in A–D was determined by Cell-Titer-Glo assay. (F) HT-29 cells were treated with human TNF (50 ng/mL), SM-164 (100 nM), and z-VAD.fmk (25 μM) for the indicated periods of time. The RIPK1-RIPK3 complex was detected by in situ PLA assay. (Scale bar, 25 μm.) (G) RIPK1-RIPK3 PLA signals per cell were quantified. Percentages of cells with different numbers of RIPK-RIPK3 PLA signals in the population were calculated at each time point (n = 2 independent experiments; >100 cells per group were assessed). (H) The viability of the HT-29 cells (G) was determined by Cell-Titer-Glo assay. (I) H&E staining and RIPK1-RIPK3 in situ PLA assays were conducted on liver sections of Abin-1+/+ and Abin-1−/− E16.5 embryos. (Scale bar, 10 μm.) (J) RIPK1-RIPK3 PLA signals per section were quantified (n = 3 independent experiments). All data are shown as mean ± SEM; *P < 0.05; **P < 0.01; ***P < 0.001; NS, not significant; based on two-tailed Student’s t test (J) and one-way ANOVA followed by Tukey’s test (B, D, E, and H).
Next, we evaluated the dynamics of PLA-necrosome signals during necroptosis progression. We treated HT-29 cells with T/S/Z and performed the PLA-RIPK1-RIPK3 assay at different time points. The numbers of necrosomes per cell and the numbers of cells with more necrosomes started to increase from 3 h after the addition of T/S/Z and increased over time until reaching high levels at 9 h (Fig. 1 F and G). The dynamic increases in necrosomes per cell correlated with the loss of cell viability (Fig. 1H), suggesting that the in situ PLA assay of RIPK1-RIPK3 represents a measurement for the progression of necroptosis. These results suggest that different cells in the populations die with different kinetics. While the cell viability assay determines the average viability of a cell population, the number of necrosomes detected by in situ PLA in each cell profiles necroptosis progression at a single-cell level.
We next used the PLA assay to characterize the necrosome in vivo. Knockout of ABIN-1, an important ubiquitin binding protein that controls the activation of RIPK1, leads to embryonic lethality (E18.5) due to necroptosis of the liver (18). The PLA-RIPK1-RIPK3 assay detected the presence of necrosomes in the livers of E16.5 Abin-1−/− mice, but not that of wild-type (WT) mice (Fig. 1 I and J and SI Appendix, Fig. S1C). Thus, our PLA assay made it possible to observe endogenous necrosomes in vivo.
Association of the Necrosome with the ER during Necroptosis.
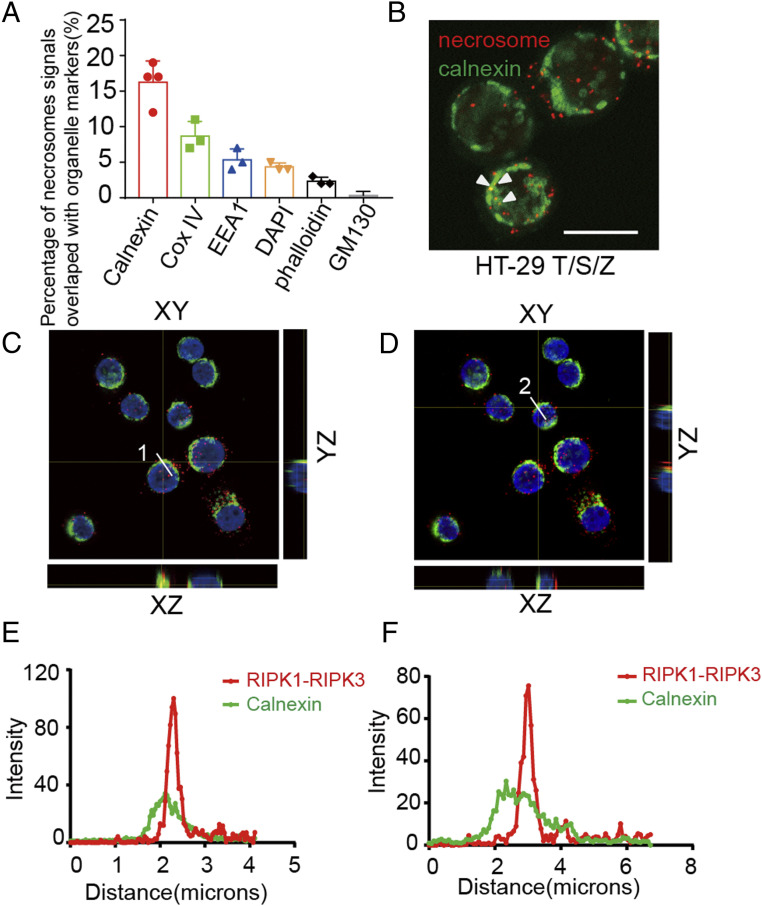
We next used the PLA assay to characterize the subcellular localization of the necrosome. Necrosomes in necroptotic cells were colabeled with PLA-RIPK1/RIPK3 and different organelle markers, including ER membrane protein calnexin, mitochondrial inner membrane protein COX4, early endosome marker EEA1, nuclear dye DAPI, phalloidin for F-actin, and Golgi protein GM130. We determined the percentages of PLA signals that overlapped with different organelle markers, which revealed the association of necrosomes with different subcellular compartments during necroptosis. We found that PLA signals of necrosomes were partially overlapped with markers of ER, mitochondria, early endosome, nucleus, and cytoplasm, but rarely with Golgi (Fig. 2A). Interestingly, we noted that ∼17% of necrosomes were found to be associated with ER (Fig. 2 A–F).
Fig. 2.
Association of the necrosome with ER during necroptosis. (A) HT-29 cells were treated with human TNF (50 ng/mL), SM-164 (100 nM), and z-VAD.fmk (25 μM), and RIPK1-RIPK3 complex was detected by in situ PLA assay. The subcellular markers were detected by immunofluorescence or specific fluorescent dye staining. Quantification of the percentages of RIPK1-RIPK3 in situ PLA signals overlapped with subcellular organelle marker, calnexin (ER), COX IV (mitochondria), EEA1 (early endosome), DAPI (nucleus), phalloidin (F-actin), and GM-130 (Golgi). At least 100 cells per group were calculated. (B) A representative image of RIPK1-RIPK3 PLA signals overlapped with calnexin immunofluorescence signals. (Scale bar, 10 μm.) (C and D) Orthogonal view of RIPK1-RIPK3 PLA signals overlapped with calnexin immunofluorescence signals. (E and F) Quantification of the intensity along the line shown in C and D. (Scale bars, 10 μm.)
We next examined whether the morphology of the ER was changed during necroptosis. We used GRP78, an ER-localized chaperone protein, as a biomarker of ER integrity. While GRP78 is normally excluded from the nucleus, we detected nuclear GRP78 in rounded necroptotic cells, suggesting that the ER is damaged during necroptosis, which leads to the leakage of GRP78 into the nucleus (SI Appendix, Fig. S2 A and B). The nuclear GRP78 could be inhibited in cells treated with Nec-1s and GSK′872 (SI Appendix, Fig. S2 C and D). We also used ER-Tracker Red dye, a cell-permeable live cell stain that is highly selective for the ER to confirm the observation. The ER-Tracker staining displayed a condensed ER pattern in rounded necroptotic cells. Thus, we conclude that the morphology of the ER was dramatically changed during necroptosis.
Necroptosis Activates a Noncanonical ER Stress Response.
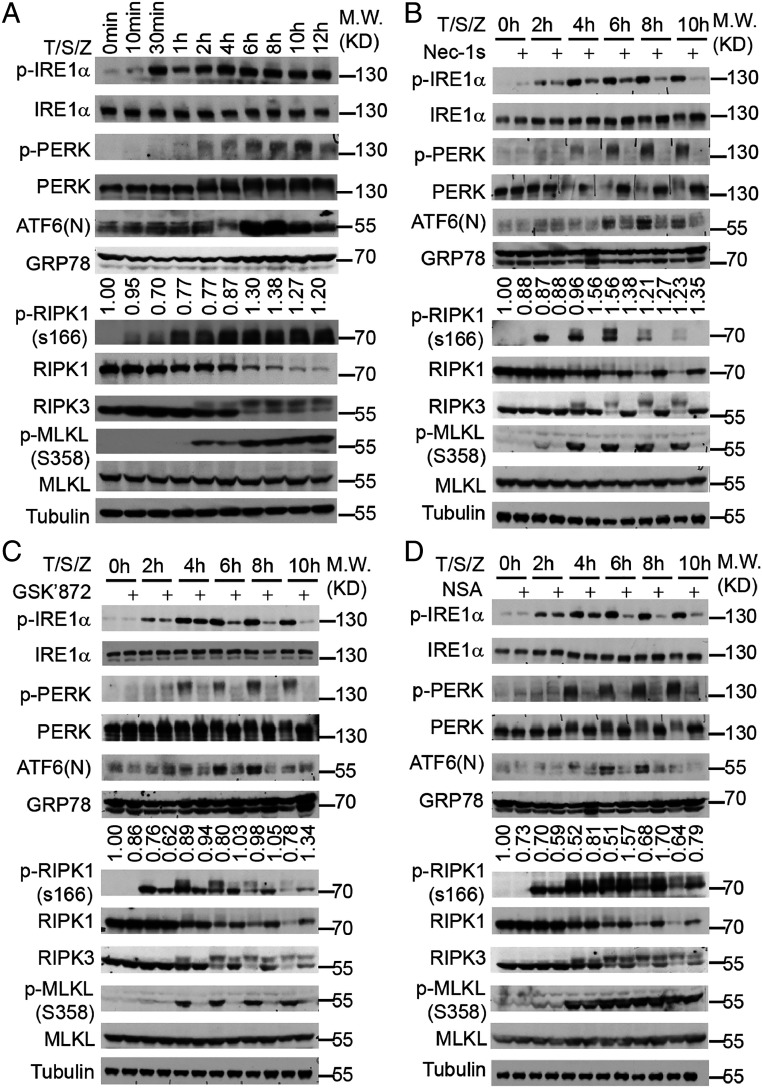
Disturbance of ER homeostasis is known to trigger an ER stress response (19–21). To determine whether necroptosis may activate an ER stress response, we examined the phosphorylation of protein kinase RNA-like endoplasmic reticulum kinase (PERK), inositol-requiring enzyme 1α (IRE1α), the cleavage of activating transcription factor 6 (ATF6), which are the hallmarks of ER stress signaling activation, during necroptosis progression. In HT-29 cells treated with T/S/Z, p-S166 RIPK1 was detected first at 1 h, which was followed by the appearance of p-RIPK3 and p-MLKL starting after 2 h. We detected phosphorylation of IRE1α as early as 30 min (Fig. 3A). Both upshifted and phosphorylated PERK could be detected after 2 h (Fig. 3A). The cleavage of ATF6 was significantly increased at 6 h, 8 h, and 10 h (Fig. 3A). As expected, the phosphorylation of IRE1α and PERK, and the cleavage of ATF6, were also detected in HT-29 cells treated with the classical ER stress–inducing models, thapsigargin (Tg) and tunicamycin (Tm) (SI Appendix, Fig. S3 A and B). In contrast, the phosphorylation of IRE1α was also detected but weaker and more transient in TNF(T) and TNF/SM-164(T/S) model than that in the necroptosis model (SI Appendix, Fig. S3 C and D). Neither PERK phosphorylation nor XBP1 splicing was detectable in the T and T/S models (SI Appendix, Fig. S3 E and F). ATF6 cleavage was detected in T/S, but not in the T model (SI Appendix, Fig. S3 C and D). Thus, the upstream sensors of ER stress signaling were most robustly activated in the necroptosis model.
Fig. 3.
Necroptosis activates a noncanonical ER stress response. (A–D) HT-29 cells were treated with human TNF (50 ng/mL), SM-164 (100 nM), z-VAD.fmk (25 μM), Nec-1s (10 μM), GSK′872 (10 μM), and NSA (2.5 μM) as indicated. The cell lysate was collected and analyzed by immunoblotting using indicated antibodies. The values under the blots indicate quantification of relative density of GRP78 bands.
Up-regulation of GRP78 is a robust downstream readout in a canonical unfolded protein response (21). Under normal homeostatic conditions, GRP78 is in complex with IRE1α, PERK, and ATF6 to inactivate these UPR transmembrane stress sensors. Accumulation of misfolded proteins releases GRP78 from these UPR sensors, which in turn promote the activation of the ER stress response. Treatment with canonical ER stress inducers, including SERCA inhibitor thapsigargin and the N-linked glycosylation inhibitor tunicamycin, induced GRP78 as expected (SI Appendix, Fig. S3 A and B). In contrast, we found only very weak induction of GRP78 during necroptosis (Fig. 3A). Thus, the activation of UPR during necroptosis deviates from what occurs under canonical ER stress conditions.
Next, we tested whether the RIPK1-RIPK3-MLKL axis was required for the activation of ER stress signaling during necroptosis. We found that treatment with RIPK1 inhibitor Nec-1s, RIPK3 inhibitor GSK′872, and MLKL membrane translocation inhibitor NSA (4) effectively inhibited the activation of PERK and ATF6 at all time points and phosphorylation of IRE1α from the 2-h time point (Fig. 3 B–D). Thus, the kinase activities of RIPK1 and RIPK3, as well as MLKL translocation to the membrane, are essential for the activation of ER stress signaling activation during necroptosis. We also detected the phosphorylation of IRE1α, upshift of PERK in mouse embryonic fibroblasts (MEFs) treated with T/S/Z (SI Appendix, Fig. S3G). In contrast, Nec-1s, GSK′872, and NSA cannot inhibit the phosphorylation of PERK and IRE1α, or the cleavage of ATF6 in the conventional ER stress response induced by thapsigargin (SI Appendix, Fig. S3 H–J). Thus, the ER stress response can be activated by necroptotic signaling but may be mediated by a mechanism distinct from that conventional ER stress response.
Incorporation of Spliced XBP1 mRNA into Extracellular Vesicles during Necroptosis.
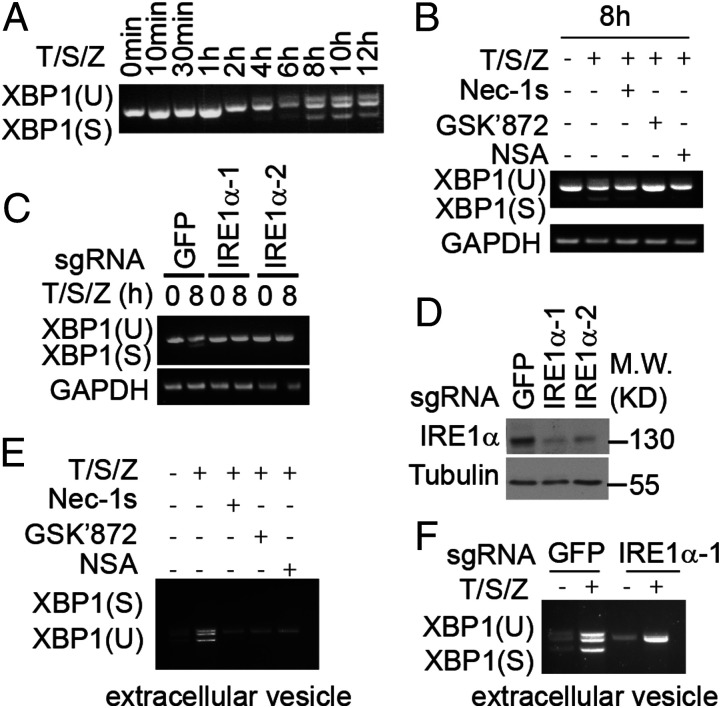
Activation of IRE1α during ER stress promotes its endoribonuclease activity, which in turn mediates unconventional splicing of XBP1 mRNA (13). The cleavage of XBP1 was detected 4 h after activation of necroptosis by T/S/Z (Fig. 4A). Furthermore, the splicing of XBP1 activated in necroptotic cells was blocked by Nec-1s, GSK′872, and NSA, and by knockout of IRE1α (Fig. 4 B–D). Emerging evidence suggests that the spliced form of XBP1 mRNA can be incorporated into extracellular vesicles known as the exosomes during ER stress (22). EVs, with the capacity to transfer proteins, lipids, and nucleic acids, have been increasingly recognized to play an important role in intercellular communication (23). MLKL could regulate extracellular vesicle generation during necroptosis to promote cytokine release (24, 25). But the potential presence of RNA cargo in the extracellular vesicles generated by necroptosis has not been investigated. Thus, we next examined whether the activation of IRE1α could promote the loading of spliced sXBP1 in the extracellular vesicles during necroptosis. We detected the spliced form of XBP1 mRNA in the extracellular vesicles isolated from T/S/Z-treated HT-29 cells, which was inhibited by treatment with Nec-1s, GSK′872, NSA, and silencing of IRE1α (Fig. 4 C–E). IRE1α knockout did not affect cell viability, the phosphorylation of RIPK1, RIPK3, and MLKL in HT-29 cells treated with T/S/Z (SI Appendix, Fig. S4 A and B), or the particle number, size, and protein marker level of extracellular vesicles during necroptosis (SI Appendix, Fig. S4 C and D). Thus, we conclude that activation of IRE1α not only regulates XBP1 splicing in the necroptotic cells but also promotes the secretion of spliced XBP1 mRNA by extracellular vesicles.
Fig. 4.
Incorporation of spliced XBP1 mRNA into extracellular vesicles during necroptosis. (A and B) HT-29 cells were treated with human TNF (50 ng/mL), SM-164 (100 nM), z-VAD.fmk (25 μM), Nec-1s (10 μM), GSK′872 (10 μM), and NSA (2.5 μM) as indicated. The cell lysate was collected and analyzed by RT-PCR. (C) Sg-GFP, sg-IRE1α-1, and sg-IRE1α-2 HT-29 cells were treated with human TNF (50 ng/mL), SM-164 (100 nM), and z-VAD.fmk (25 μM). The cell lysate was collected and analyzed by RT-PCR. (D) The knockout efficiency of sg-IRE1α-1, sg-IRE1α-2 HT-29 cells. (E) HT-29 cells were treated with human TNF (50 ng/mL), SM-164 (100 nM), z-VAD.fmk (25 μM), Nec-1s (10 μM), GSK′872 (10 μM), and NSA (2.5 μM) for 24 h. The exosome was isolated from culture medium and the RNA was analyzed by RT-PCR. (F) Sg-GFP, sg-IRE1α-1, and sg-IRE1α-2 HT-29 cells were treated with human TNF (50 ng/mL), SM-164 (100 nM), and z-VAD.fmk (25 μM) as indicated for 24 h. The exosome was isolated from culture medium and the RNA was analyzed by RT-PCR.
Oligomerization of MLKL Triggers ER Stress Signaling during Necroptosis.
Since ER stress signaling during necroptosis could be blocked by treatment with NSA, which inhibits the membrane translocation of oligomerized MLKL (4), a downstream event in necroptosis, we wondered whether MLKL oligomerization in necroptosis could directly trigger ER stress signaling. We used the expression construct of MLKL fused with the FV domain, which was induced to undergo oligomerization by the addition of AP20187 (Fig. 5A). MLKL oligomerization induced by AP20187 triggered necroptosis, which was inhibited by NSA but not by Nec-1s and GSK′872 (26) (SI Appendix, Fig. S5A), suggesting that this system was able to bypass the upstream regulation of RIPK1 and RIPK3 to activate MLKL directly. Induction of MLKL oligomerization rapidly induced ER stress signaling as demonstrated by the phosphorylation of both PERK and IRE1α as early as 0.5 h after the addition of AP20187 (Fig. 5B). XBP1 splicing also occurred at 2 h and 3 h after the induction of MLKL dimerization (Fig. 5C). The membrane translocation of oligomerized MLKL was important, as the addition of NSA effectively inhibited the phosphorylation of PERK and IRE1α, as well as the splicing of XBP1 (Fig. 5 B and C). While thapsigargin induced up-regulation of eIF2α phosphorylation and ATF4, T/S/Z and AP20187-mediated dimerization of MLKL did not significantly up-regulate GRP78, eIF2α phosphorylation, or ATF4 (SI Appendix, Fig. S5C).
Fig. 5.
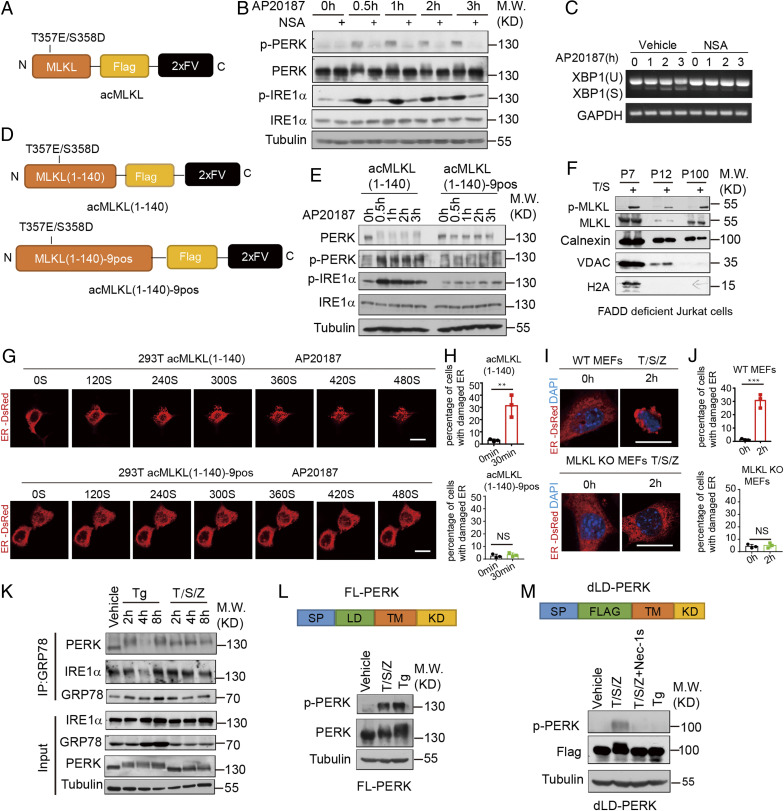
Oligomerized MLKL can activate UPR sensors without disrupting their binding with GRP78. (A) A schematic diagram of the oligomerizable MLKL (acMLKL). T357E/S358D MLKL mutations mimic the phosphorylation by RIPK3. (B and C) acMLKL HT-29 cells were treated with AP20187 (100 nM) and NSA (2.5 μM) as indicated. The cell lysate was collected and analyzed by immunoblotting (B). The cell lysate was collected and analyzed by RT-PCR (C). (D) Schematic representation of the oligomerizable N-terminal part of MLKL (acMLKL-140) and 9 positively charged amino acid MLKL mutants (acMLKL-140–9pos). (E) acMLKL-140 HT-29 cells and acMLKL-140–9pos HT-29 cells were treated with AP20187 (100 nM). The cell lysate was collected and analyzed by immunoblotting. (F) FADD-deficient Jurkat cells were treated with human TNF (50 ng/mL) and SM-164 (100 nM), and the sequential fractions were analyzed by immunoblotting. (G) 293T cells were transfected with the expression constructs of acMLKL (1 to 140), acMLKL (1 to 140)-9pos and ER-DsRed. The cells were treated with AP20187 (100 nM) and ER morphology was detected by time-lapsed imaging. (Scale bar, 10 μm.) (H) Quantification of the percentage of cells with ER puncta in G (>100 cells per group from three independent experiments were assessed). (I) WT MEFs and MLKL KO cells stably expressed with ER-DsRed were treated with mouse TNF (10 ng/mL), SM-164 (100 nM), and z-VAD.fmk (25 μM). The ER morphology was detected by confocal microscopy. (Scale bar, 10 μm.) (J) Quantification of the percentage of cells with ER puncta in I (>100 cells per group from three independent experiments were assessed). (K) HT-29 cells were treated with thapsigargin (500 nM) and human TNF (50 ng/mL), SM-164 (100 nM), and z-VAD.fmk (25 μM) as indicated. The level of IRE1α and PERK binding with GRP78 was detected by coimmunoprecipitation. (L) A schematic diagram of the full-length PERK (FL-PERK) construct. PERK KO HT-29 cells reconstituted with FL-PERK were treated with thapsigargin (500 nM) and human TNF (50 ng/mL), SM-164 (100 nM), z-VAD.fmk (25 μM), and the cell lysate was collected and analyzed by immunoblotting. (M) A schematic diagram of the luminal domain defect PERK (dLD-PERK) construct. PERK KO HT-29 cells reconstituted with dLD-PERK were treated with thapsigargin (500 nM) and human TNF (50 ng/mL), SM-164 (100 nM), z-VAD.fmk (25 μM), and Nec-1s (10 μM). The cell lysate was collected and analyzed by immunoblotting. All data are shown as mean ± SEM; **P < 0.01; ***P < 0.001; NS, not significant; based on two-tailed unpaired Student’s t test (H and J).
Homooligomerization of the N-terminal part of MLKL (acMLKL 1 to 140) was found to be necessary and sufficient for mediating the plasma membrane translocation of MLKL to mediate necrosis (27). To explore whether the oligomerization of MLKL 1 to 140 aa might be sufficient to activate PERK and IRE1α, we constructed an expression plasmid for oligomerizable WT acMLKL and mutant acMLKL-9PosA that were fused with the 2xFV domain for inducing oligomerization by binding with AP20187 (28). As reported, oligomerization of WT acMLKL could induce necroptosis while acMLKL-9PosA could not (SI Appendix, Fig. S5B). Induction of oligomerization of acMLKL by the addition of AP20187 rapidly induced phosphorylation of PERK and IREα, while oligomerization of the mutant acMLKL-9PosA, which mutated the 9 crucial amino acids for binding to the membrane, could not activate PERK and IREα (Fig. 5 D and E). These data suggest that oligomerization of the MLKL N terminus and the binding to the membrane are important for activation of ER stress signaling.
Oligomerized MLKL Can Activate UPR Sensors without Disrupting Their Binding with GRP78.
Translocation of trimerized MLKL to the cytoplasmic membrane is known to be a critical execution event in necroptosis (27, 29). Thus, we hypothesized that MLKL might translocate to the ER membrane to trigger the activation of UPR sensors. We performed an ER fraction assay by ultracentrifugation. We found that phosphorylated MLKL could be detected in the ER membrane fraction during necroptosis (Fig. 5F and SI Appendix, Fig. S5D), suggesting that MLKL may translocate to the ER membrane during necroptosis.
We next used time-lapsed microscopy to visualize the dynamic change of the ER upon inducing MLKL oligomerization. Interestingly, we found that the network of the ER was rapidly broken into puncta-like structures upon induction of acMLKL(1 to 140) oligomerization (Fig. 5 G and H). Similar ER-puncta generation was observed in necroptosis of WT MEFs induced by treatment with T/S/Z (Fig. 5 I and J). Taken together, our results suggest that oligomerized MLKL is translocated to the ER membrane to promote ER membrane damage during necroptosis.
Since the induction of protein chaperone GRP78 expression is a hallmark of a conventional ER stress response, while we observed the activation of IRE1α, PERK, and ATF6 with the minimum induction of GRP78 during necroptosis, we next investigated whether the binding of GRP78 with IRE1α might be affected by necroptosis. Interestingly, while the binding of GRP78 and IRE1α was reduced in cells treated with thapsigargin to induce a conventional ER stress, the binding of GRP78 and IRE1α was minimally affected during necroptosis (Fig. 5K), which is consistent with a minimum effect of necroptosis on the expression of GRP78 (Fig. 3A and SI Appendix, Fig. S3A). Thus, our results suggest that necroptosis is able to promote the activation of IRE1α without significantly affecting its binding with GRP78. We constructed the expression plasmid for full-length PERK (FL-PERK) and luminal domain deleted PERK (dLD-PERK). Our results showed that while both FL-PERK and dLD-PERK could be activated by T/S/Z and inhibited by Nec-1s, thapsigargin could only activate FL-PERK and not dLD-PERK (Fig. 5 L and M and SI Appendix, Fig. S5E). Thus, we conclude that necroptosis can activate an unconventional ER stress response without disrupting the binding of GRP78 with the luminal domains of UPR sensors.
Discussion
IRE1α, PERK, and ATF6 in complex with GRP78 are UPR sensors in the ER lumen that monitor for the levels of misfolded proteins (16, 21). In a conventional ER stress response, the abnormal accumulation of improperly folded proteins within the ER lumen competes for binding with GRP78 that normally binds with the luminal domains of IRE1α, PERK, and ATF6, which in turn promotes the activation of these UPR sensors (16). The induction of GRP78 is a hallmark of conventional ER stress responses. Interestingly, we found that the activation of ER sensors, including IRE1α, PERK, and ATF6 during necroptosis, was neither associated with a dramatic induction of GRP78 as that in thapsigargin-treated cells nor was the interaction of IRE1α and PERK with GRP78 affected by necroptosis. Thus, necroptosis can activate an unconventional ER stress response.
MLKL can facilitate endosomal function and generation of EVs, independently of RIPK1 or RIPK3, and also the phosphorylation of MLKL by RIPK3 can further enhance EV generation (24). Our results demonstrate secretion of spliced XBP1 mRNA by extracellular vesicles generated by necroptosis. The role of RNA in mediating extracellular signaling has been increasingly recognized (30). Our results suggest that the RNA content of extracellular vesicles from necroptotic cells may transmit signals to the nearby cellular environment about the impending danger.
We show that oligomerization of MLKL is sufficient to promote the activation of UPR sensors in necroptosis. While the induction of GRP78 is an established hallmark of the ER stress response (31), we show that oligomerization of MLKL can also disrupt the ER membrane to promote the activation of UPR sensors independent of their luminal domains and with minimum effect on the levels of GRP78. MLKL is an indispensable mediator of necroptosis (32). The oligomerization of MLKL induced by T/S/Z or artificial dimerization promotes its association with lipid rafts in the plasma membrane (27). MLKL has been shown to promote Ca2+ influx by generating plasma membrane “bubbles” in a process involving the ESCRT-III machinery during necroptosis, which precedes the loss of PM integrity. Necroptosis strongly induces the transcription of proinflammatory cytokines and chemokines (26). Thus, the lack of GRP78 induction is unlikely due to the inability of necroptotic cells to launch transcription. IRE1α and PERK can be activated by perturbation of cellular lipid composition independently of their luminal domains, which has led to the suggestion that the UPR sensors can also monitor the lipid composition of the ER membrane (33). Our results suggest that the UPR sensors can sense the integrity of the ER membrane, and furthermore, necroptosis provides a unique ER stress paradigm that can activate ER sensors, including IRE1α and PERK with minimum effect on their bindings with GRP78. Thus, necroptosis activates ER stress sensors, including IRE1α and PERK, in an unconventional manner.
Materials and Methods
Reagents.
The reagents used in this study included: Recombinant human TNFα (Cat. No. C008) from Novoprotein; z-VAD.fmk (Cat. No. S7023) from Selleck; GSK′872 (Cat. No. 530389) and necrosulfonamide (NSA) (Cat. No. 480073) from Calbiochem; thapsigargin (Cat. No. ab120286) and tunicamycin (Cat. No. ab120296) from Abcam; AP20187 (Cat. No. HY-13992) from MedChemExpress; R-7-Cl-O-Nec-1 (Nec-1s) and SM-164 were custom synthesized; DAPI nucleic acid stain (Cat. No. D1306) and ER-Tracker Red (Cat. No. E34250) from Invitrogen; Actin-stain 555 fluorescent phallodin (Cat. No. PHDH1) from Cytoskeleton; puromycin (Cat. No. S7417) from Selleck; Endoplasmic Reticulum Isolation Kit (Cat. No. ER0100); Duolink In Situ PLA Probe Anti-Rabbit MINUS (Cat. No. DUO92005); Duolink In Situ PLA Probe Anti-Goat PLUS (Cat. No. DUO92003); Duolink In Situ Detection Reagents Green (Cat. No. DUO92014); Duolink In Situ Detection Reagents Red (Cat. No. DUO92008); Duolink In Situ Mounting Medium with DAPI (Cat. No. DUO82040) from Sigma-Aldrich; and CellTiter-Glo Luminescent Cell Viability Assay (Cat. No. G7572) from Promega.
PLA Assay.
PLA Duolink kit instructions were followed with minor modifications. Briefly, cells were plated on chamber slides and treated as indicated. Cells were fixed in 4% paraformaldehyde and permeabilized with 0.1% Triton X-100. The cells were then blocked with Duolink block buffer and incubated with the primary antibody overnight at 4 °C. Cells were washed with washing buffer A three times, each for 5 min. Then the cells were incubated with the Duolink PLUS and MINUS secondary antibodies for 1 h at room temperature. The slides were then washed with washing buffer A three times for 5 min each. The Duolink ligation-ligase solution was added to each sample, incubated at 37 °C for 45 min, and then washed with washing buffer A two times for 2 min each. The Duolink amplification-polymerase solution was added to each sample and incubated at 37 °C for 100 min. The slides were washed with washing buffer B two times for 10 min each. The slide was mounted with a coverslip using a minimal volume of Duolink In Situ Mounting Medium with DAPI. Washing buffer A contained 0.01 M Tris, 0.15 M NaCl and 0.05% Tween 20, pH 7.4; and washing buffer B contained 0.2 M Tris and 0.1 M NaCl, pH 7.4.
The PLA assay protocol on mouse embryo sections was the same as on culture cells except for fixation steps. E16.5 mouse embryos were dissected from pregnant mice and directly embedded in OCT (optimal cutting temperature compound) at −40 °C. Sagittal sections of the whole embryo at the thickness of 10 μm were prepared and stored at −80 °C before use. The slices were fixed by precold methanol for 10 min and the methanol was washed out by PBS three times. Antibodies used in proximity ligation assay include: Anti-RIPK1 (Cat. No. 3493, 1:2,000) from CST; anti-MLKL(human) (Cat. No. ab183770, 1:800) anti-RIPK3(mouse)(C-16) (sc-47364, 1:400); anti-RIPK3(human)(N-14) (sc-47368, 1:400) from Santa Cruz. The signals of PLA dots were quantified by ImageJ software (analyzing particles).
Live Cell Imaging.
Cells were plated at 24-well glass bottom plates (Cellvis, P24-1.5H-N) and transfected as described. Cells were maintained in complete media at 37 °C and 5% CO2. As indicated, 100 nM AP20187 was added into medium to stimulate the cells before the image was captured (Leica Lightsheet Confocal System). Images were processed using ImageJ.
Quantification and Statistical Analysis.
All statistical analyses were performed using GraphPad Prism 7.00. All data were shown as mean ± SEM. All experiments were repeated two to three times.
Supplementary Material
Acknowledgments
This work was supported in part by the China National Natural Science Foundation (Grant 21837004, Grant 91849204, and Grant 92049303), the Science and Technology Commission of Shanghai Municipality (Grant 18JC1420500), the National Key R&D Program of China (Grant 2016YFA0501900), the Shanghai Municipal Science and Technology Major Project (Grant 2019SHZDZX02), and the Chinese Academy of Sciences (Grant XDB39030200).
Footnotes
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2110476118/-/DCSupplemental.
Data Availability
All study data are included in the article and/or SI Appendix.
References
- 1.Shan B., Pan H., Najafov A., Yuan J., Necroptosis in development and diseases. Genes Dev. 32, 327–340 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wallach D., Kang T. B., Dillon C. P., Green D. R., Programmed necrosis in inflammation: Toward identification of the effector molecules. Science 352, aaf2154 (2016). [DOI] [PubMed] [Google Scholar]
- 3.Fulda S., The mechanism of necroptosis in normal and cancer cells. Cancer Biol. Ther. 14, 999–1004 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sun L., et al., Mixed lineage kinase domain-like protein mediates necrosis signaling downstream of RIP3 kinase. Cell 148, 213–227 (2012). [DOI] [PubMed] [Google Scholar]
- 5.Degterev A., et al., Identification of RIP1 kinase as a specific cellular target of necrostatins. Nat. Chem. Biol. 4, 313–321 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cho Y. S., et al., Phosphorylation-driven assembly of the RIP1-RIP3 complex regulates programmed necrosis and virus-induced inflammation. Cell 137, 1112–1123 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.He S., et al., Receptor interacting protein kinase-3 determines cellular necrotic response to TNF-alpha. Cell 137, 1100–1111 (2009). [DOI] [PubMed] [Google Scholar]
- 8.Zhang D. W., et al., RIP3, an energy metabolism regulator that switches TNF-induced cell death from apoptosis to necrosis. Science 325, 332–336 (2009). [DOI] [PubMed] [Google Scholar]
- 9.Hildebrand J. M., et al., Activation of the pseudokinase MLKL unleashes the four-helix bundle domain to induce membrane localization and necroptotic cell death. Proc. Natl. Acad. Sci. U.S.A. 111, 15072–15077 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang D., et al., The MLKL channel in necroptosis is an octamer formed by tetramers in a dyadic process. Mol. Cell. Biol. 37, e00497-16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang H., et al., Mixed lineage kinase domain-like protein MLKL causes necrotic membrane disruption upon phosphorylation by RIP3. Mol. Cell 54, 133–146 (2014). [DOI] [PubMed] [Google Scholar]
- 12.Sidrauski C., Walter P., The transmembrane kinase Ire1p is a site-specific endonuclease that initiates mRNA splicing in the unfolded protein response. Cell 90, 1031–1039 (1997). [DOI] [PubMed] [Google Scholar]
- 13.Yoshida H., Matsui T., Yamamoto A., Okada T., Mori K., XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell 107, 881–891 (2001). [DOI] [PubMed] [Google Scholar]
- 14.Harding H. P., Zhang Y., Ron D., Protein translation and folding are coupled by an endoplasmic-reticulum-resident kinase. Nature 397, 271–274 (1999). [DOI] [PubMed] [Google Scholar]
- 15.Haze K., Yoshida H., Yanagi H., Yura T., Mori K., Mammalian transcription factor ATF6 is synthesized as a transmembrane protein and activated by proteolysis in response to endoplasmic reticulum stress. Mol. Biol. Cell 10, 3787–3799 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hetz C., Zhang K., Kaufman R. J., Mechanisms, regulation and functions of the unfolded protein response. Nat. Rev. Mol. Cell Biol. 21, 421–438 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Degterev A., et al., Chemical inhibitor of nonapoptotic cell death with therapeutic potential for ischemic brain injury. Nat. Chem. Biol. 1, 112–119 (2005). [DOI] [PubMed] [Google Scholar]
- 18.Dziedzic S. A., et al., ABIN-1 regulates RIPK1 activation by linking Met1 ubiquitylation with Lys63 deubiquitylation in TNF-RSC. Nat. Cell Biol. 20, 58–68 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Boyce M., Yuan J., Cellular response to endoplasmic reticulum stress: A matter of life or death. Cell Death Differ. 13, 363–373 (2006). [DOI] [PubMed] [Google Scholar]
- 20.Schröder M., Kaufman R. J., The mammalian unfolded protein response. Annu. Rev. Biochem. 74, 739–789 (2005). [DOI] [PubMed] [Google Scholar]
- 21.Ron D., Walter P., Signal integration in the endoplasmic reticulum unfolded protein response. Nat. Rev. Mol. Cell Biol. 8, 519–529 (2007). [DOI] [PubMed] [Google Scholar]
- 22.Hosoi T., Nakashima M., Ozawa K., Incorporation of the endoplasmic reticulum stress-induced spliced form of XBP1 mRNA in the exosomes. Front. Physiol. 9, 1357 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yáñez-Mó M., et al., Biological properties of extracellular vesicles and their physiological functions. J. Extracell. Vesicles 4, 27066 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yoon S., Kovalenko A., Bogdanov K., Wallach D., MLKL, the protein that mediates necroptosis, also regulates endosomal trafficking and extracellular vesicle generation. Immunity 47, 51–65.e7 (2017). [DOI] [PubMed] [Google Scholar]
- 25.Gong Y. N., et al., ESCRT-III acts downstream of MLKL to regulate necroptotic cell death and its consequences. Cell 169, 286–300.e16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhu K., et al., Necroptosis promotes cell-autonomous activation of proinflammatory cytokine gene expression. Cell Death Dis. 9, 500 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen X., et al., Translocation of mixed lineage kinase domain-like protein to plasma membrane leads to necrotic cell death. Cell Res. 24, 105–121 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dondelinger Y., et al., MLKL compromises plasma membrane integrity by binding to phosphatidylinositol phosphates. Cell Rep. 7, 971–981 (2014). [DOI] [PubMed] [Google Scholar]
- 29.Cai Z., et al., Plasma membrane translocation of trimerized MLKL protein is required for TNF-induced necroptosis. Nat. Cell Biol. 16, 55–65 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dinger M. E., Mercer T. R., Mattick J. S., RNAs as extracellular signaling molecules. J. Mol. Endocrinol. 40, 151–159 (2008). [DOI] [PubMed] [Google Scholar]
- 31.Lee A. S., The ER chaperone and signaling regulator GRP78/BiP as a monitor of endoplasmic reticulum stress. Methods 35, 373–381 (2005). [DOI] [PubMed] [Google Scholar]
- 32.Wu J., et al., Mlkl knockout mice demonstrate the indispensable role of Mlkl in necroptosis. Cell Res. 23, 994–1006 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Volmer R., van der Ploeg K., Ron D., Membrane lipid saturation activates endoplasmic reticulum unfolded protein response transducers through their transmembrane domains. Proc. Natl. Acad. Sci. U.S.A. 110, 4628–4633 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or SI Appendix.