Significance
The ability of immune cells to distinguish self tissue from nonself pathogens is a key characteristic of immunity, allowing responses to be targeted against invading pathogens while protecting against self-directed immune damage. The recognition of nonself by innate immune cells has been extensively characterized, but the mechanisms that allow for self recognition and self-tolerance remain largely unexplored. Here, we uncover a self-tolerance system in Drosophila that relies on the N-glycosylation of extracellular matrix proteins: immune activity is restrained by recognition of a self signal and proceeds when encountering self tissues missing the self signal. This allows the host to recognize and protect self tissues, destroy aberrant tissue, and, perhaps, respond to pathogens that evade nonself recognition systems.
Keywords: innate immunity, self-recognition, self-tolerance, autoimmunity, protein N-glycosylation
Abstract
In order to respond to infection, hosts must distinguish pathogens from their own tissues. This allows for the precise targeting of immune responses against pathogens and also ensures self-tolerance, the ability of the host to protect self tissues from immune damage. One way to maintain self-tolerance is to evolve a self signal and suppress any immune response directed at tissues that carry this signal. Here, we characterize the Drosophila tuSz1 mutant strain, which mounts an aberrant immune response against its own fat body. We demonstrate that this autoimmunity is the result of two mutations: 1) a mutation in the GCS1 gene that disrupts N-glycosylation of extracellular matrix proteins covering the fat body, and 2) a mutation in the Drosophila Janus Kinase ortholog that causes precocious activation of hemocytes. Our data indicate that N-glycans attached to extracellular matrix proteins serve as a self signal and that activated hemocytes attack tissues lacking this signal. The simplicity of this invertebrate self-recognition system and the ubiquity of its constituent parts suggests it may have functional homologs across animals.
Immune systems have evolved to protect organisms from infection by a range of pathogens (1). One of the defining characteristics of host immunity is the ability to discriminate healthy self tissue from nonself pathogens. There are at least two mechanisms by which a host can distinguish its own tissue from that of a pathogen (2, 3): In nonself recognition, the host deploys receptors that recognize pathogen-specific molecules; when these receptors bind their ligands, the pathogen is identified and host immune mechanisms are triggered. In self recognition, the host deploys receptors that recognize host-specific molecules; when these receptors bind the corresponding self ligand, a self tissue is identified and host immune mechanisms are repressed, a process known as self-tolerance. Self-recognition is important in protecting self tissue during an immune response and for disposing of diseased or damaged self tissue.
Blood cells, the unique mobile immune cells of animals, often act as sentinels of infection and control whether immune responses are mounted against different tissues (2, 4). Our understanding of immune cell function is dominated by examples of nonself recognition (5–7). For example, invertebrate blood cells and most cells from the vertebrate myeloid lineage (i.e., cells responsible for innate immunity) identify nonself pathogens using pattern recognition receptors (PRRs), which recognize conserved pathogen-associated molecular patterns (PAMPs) such as viral double-stranded RNAs (dsRNAs), bacterial lipopolysaccharides and peptidoglycans, and fungal β-glucans (8–12). However, these cells also contribute to self-recognition (3), allowing hosts to recognize self tissues and suppress autoimmunity. Although much of the research uncovering mechanisms of recognition and self-tolerance has focused on adaptive immune mechanisms (5, 7), a growing body of work has demonstrated the importance of innate immune mechanisms in the suppression of autoimmunity (13–18).
Successful innate-mediated self-tolerance requires that nonimmune tissues produce and display specific self-associated molecular patterns (SAMPs; ref. 19) and that blood cells recognize SAMPs in these tissues and suppress proimmune signaling. A loss of self-tolerance occurs when innate immune cells can no longer recognize SAMPs, when target tissues are unable to display SAMPs, or when the recognition of self is insufficient to restrain immunity. The self-tolerance mechanism can also act as an initiator of immunity itself if failure to recognize a self signal on the pathogen surface triggers an immune response, in what is known as “missing-self recognition” (20, 21). It has been proposed that glycans (complex sugar groups) are likely candidates for SAMP signals given that they dominate cell surfaces and extracellular matrices and can be very diverse (19), although the general mechanisms underlying innate immune-mediated self-tolerance and missing-self recognition remain largely unexplored.
The fruit fly Drosophila melanogaster has proven an excellent model for the study of conserved innate immune mechanisms (22–27). Flies are infected by a wide range of pathogens and, being invertebrates, lack a classical adaptive immune response, allowing for the study of innate immune mechanisms in isolation. In Drosophila, circulating hemocytes (blood cells) known as plasmatocytes actively surveil the hemocoel (body cavity) for damaged tissues and invading pathogens (4), showing they are capable of self/nonself discrimination. These macrophage-like immune cells are responsible for phagocytosis of micropathogens (e.g., bacteria) and encapsulation of macroparasites, during which thousands of blood cells attack large foreign objects (e.g., parasitoid wasp eggs). Drosophila phagocytic responses are genetically conserved with those of vertebrates (28, 29), and the insect encapsulation process is functionally related to the production of granulomas in vertebrates, during which natural killer (NK) cells and other blood cells attack, and occasionally melanize, large foreign tissues (30–34). Thus, Drosophila might also serve as a model for immune self-tolerance if potential self-recognition systems (including SAMP production or SAMP recognition) could be targeted and studied.
Here, we focus on Drosophila mutants that mount misdirected immune responses against their own tissues. Surprisingly, numerous classical Drosophila mutant strains known as melanotic tumor mutants, in which blood cells attack and encapsulate aberrant self tissues due to a deficiency in self-tolerance, were never fully genetically characterized (35–40). Of those still available in public stock collections (SI Appendix, Table S1), we decided to characterize the D. melanogaster tumor(1)Suzuki (tuSz) melanotic tumor strain, which was isolated in a large-scale temperature-sensitive mutagenesis screen (41, 42). This mutant displays a temperature-sensitive self-encapsulation phenotype directed at its own posterior fat body tissue (37). We demonstrate that the tuSz1 mutant phenotype is the result of two distinct mutations: 1) a loss of function mutation in the GCS1 gene, which disrupts the protein N-glycosylation pathway in the posterior fat body, and 2) a gain-of-function mutation in the hopscotch gene (hop), the Drosophila Janus Kinase (JAK) ortholog, which causes precocious activation of immune cells. Our results demonstrate that N-glycosylated extracellular matrix (ECM) proteins serve as SAMPs and that activated innate immune cells attack tissues that lack these SAMPs.
Results
The tuSz1 Phenotype Results from a Self-Directed Immune Response.
In accordance with previous work (37), we find that tuSz1 mutants display a temperature-sensitive phenotype characterized by the melanization of posterior fat body tissue. This phenotype first appears during late larval development (third instar), and the melanized tissue persists throughout the life of the fly (Fig. 1 A–C). This is morphologically reminiscent of the melanotic encapsulation of a parasitoid wasp egg (Fig. 1D). Drosophila larvae and pupae are regularly infected by parasitoid wasps in nature (43–45), and their blood cells attack the wasp eggs (46, 47). In this encapsulation response, plasmatocytes become activated and adhere to the surface of the parasitoid egg (48, 49). Immune stimulation also triggers the production of specialized flattened immune cells known as lamellocytes (50, 51), which adhere to the plasmatocyte cell layer to form a multicellular, multilayered capsule around the parasitoid egg. The capsule is consolidated and melanized, and free radicals are released into the capsule, leading to parasitoid death (49, 52, 53). The morphological similarity between melanization of self fat body tissue and wasp eggs suggested that the tuSz1 mutant phenotype may represent a self-directed immune response.
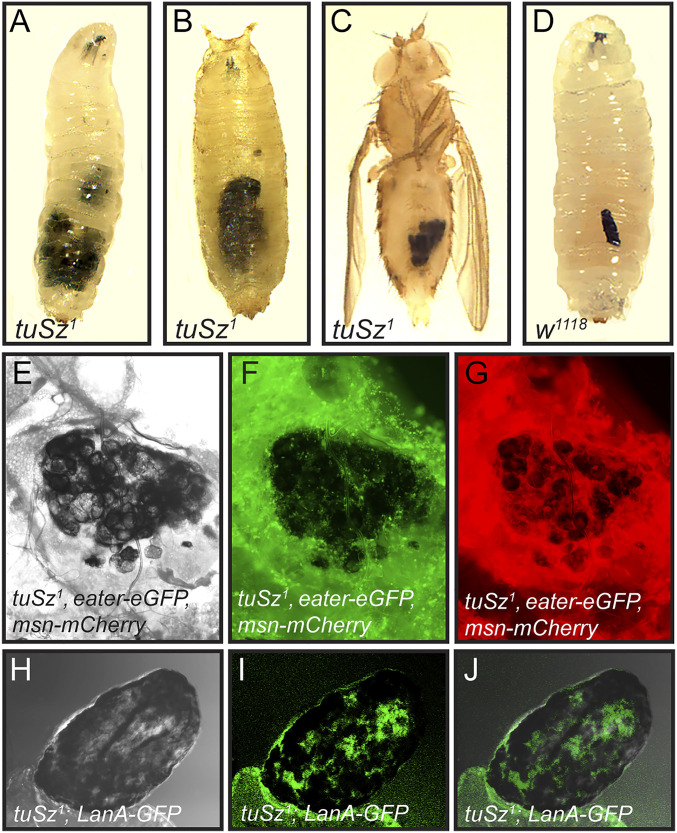
Fig. 1.
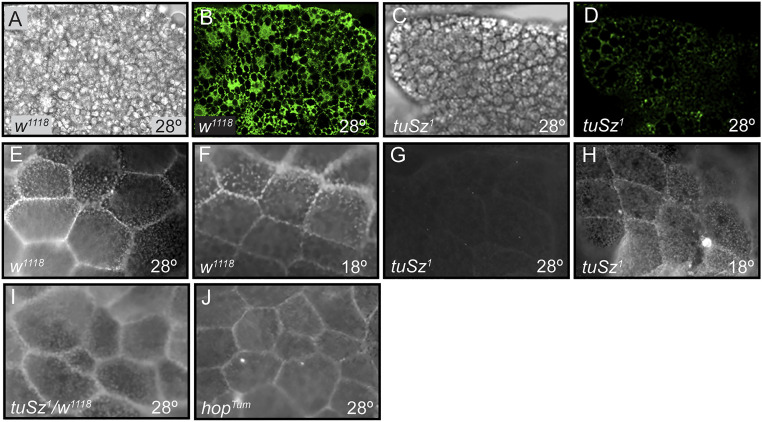
The tuSz self-encapsulation phenotype. (A–C) In tuSz1 mutants at 28 °C, posterior fat body tissues are melanized during the third-instar larval stage (A), and the melanization persists through the pupal (B) and adult stages (C). The self-encapsulation phenotype is morphologically similar to the encapsulation of a Leptopilina clavipes (parasitoid wasp) egg by a w1118 control larva (D). Brightfield (E), eGFP (F), and mCherry (G) images of fat bodies dissected from tuSz1, eater-eGFP, msn-mCherry larvae demonstrate that plasmatocytes (marked by eGFP expression) and lamellocytes (marked by mCherry expression) interact with fat body tissue that is undergoing melanization. Brightfield (H), GFP (I), and merged (J) images of fat bodies dissected from tuSz1; LanA-GFP larvae show that self-encapsulated fat body tissue nevertheless maintains intact ECM (marked by GFP expression).
To test whether the tuSz1 melanization phenotype results from a melanotic encapsulation of self tissue, we used the eater-eGFP and msn-mCherry reporter lines to mark plasmatocytes and lamellocytes, respectively, in control and tuSz1 mutant backgrounds. We find that both plasmatocytes (Fig. 1F) and lamellocytes (Fig. 1G) adhere to, and form a capsule around, fat body tissue that is undergoing melanization in tuSz1 mutant larvae, mirroring their role in the encapsulation of parasitoid wasp eggs (52, 53). By contrast, there are minimal interactions between plasmatocytes and fat body tissue in the control background (SI Appendix, Fig. S1), and lamellocytes are rarely observed in naïve wild-type larvae (54). This suggests that the tuSz1 phenotype arises from a loss of self-tolerance leading to the autoimmune targeting of self tissue by the encapsulation response.
A similar self-encapsulation response in Drosophila was previously linked to the loss of ECM (55), leading to the hypothesis that self-tolerance is mediated by a SAMP associated with the ECM. To test the presence of the ECM in tuSz1 mutants, we expressed GFP fused to either Laminin A (LanA) or Viking (Vkg) in the tuSz1 background. LanA and Vkg are Drosophila orthologs of subunits of the major structural ECM proteins laminin and collagen, respectively (56), so imaging GFP fusions with either protein allows us to assay the state of the ECM in tuSz1 mutants. In the mutants, the posterior fat body, including sites of self-encapsulation, is covered by laminin (Fig. 1 H–J) and collagen (SI Appendix, Fig. S2), demonstrating that the ECM is intact in the mutant background. This is also observed in the melanotic tumor mutant tu(2)bw (57), suggesting that, while necessary, ECM integrity may not be sufficient for self-tolerance. We therefore hypothesize that the self-tolerance signal may be a specific posttranslational modification of one or more ECM proteins and that this modification is disrupted in tuSz1 mutants.
To understand the mechanism mediating self-tolerance in Drosophila, we began by further characterizing the tuSz1 mutant phenotype. The tuSz1 mutation was previously mapped to the X chromosome (37, 58), so we first tested whether the phenotype was influenced by the sex of the fly. We find no difference in the penetrance of the self-encapsulation (melanized posterior fat body) phenotype between hemizygous male and homozygous female flies (SI Appendix, Table S2), so we pooled the sexes for further analyses. As previously described (37), we find that the tuSz1 phenotype is temperature sensitive, with homozygous mutant flies displaying a high penetrance of self-encapsulations at the restrictive temperature of 28 °C but not at the permissive temperature of 18 °C (Fig. 2A and SI Appendix, Table S2). We additionally find that heterozygous female flies do not display the self-encapsulation phenotype at 28 °C (Fig. 2A and SI Appendix, Table S2), demonstrating that this phenotype is recessive.
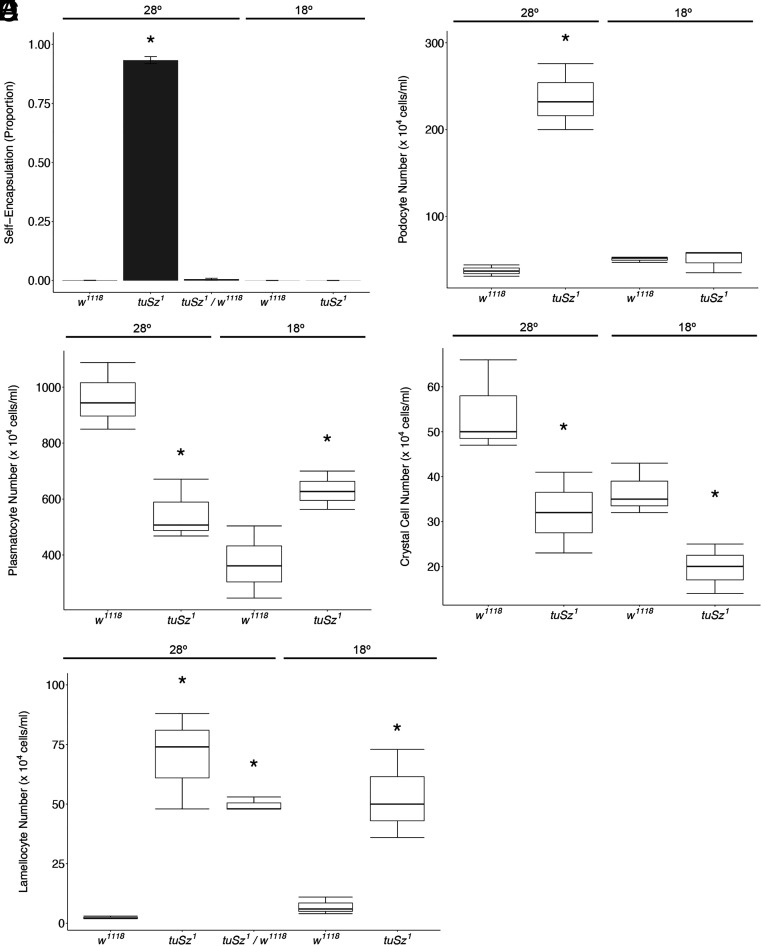
Fig. 2.
Phenotypic characterization of tuSz flies. (A) Penetrance of the self-encapsulation phenotype of w1118 (control), tuSz1, and tuSz1/w1118 heterozygous flies raised at the indicated temperature. *P < 0.05 compared to w1118 at 28 °C. (B–E) Plasmatocyte number (B), lamellocyte number (C), podocyte number (D), and crystal cell number (E) of the indicated genotypes at the indicated temperatures. *P < 0.05 compared to w1118 at each temperature. P values for A and C were determined by Dunnett’s test. P values for B, D, and E were determined by Welch’s two-sample t test.
The Hemocyte Population Is Altered in tuSz1 Mutants.
This self-encapsulation phenotype is morphologically similar to the Drosophila encapsulation response against wasp eggs. The encapsulation response is characterized by an increase in hemocyte numbers and the differentiation of lamellocytes (46, 59–61), so we tested whether the self-encapsulating tuSz1 mutants showed similar alterations in the numbers or ratios of different hemocyte populations. We counted the circulating hemocyte subtypes in tuSz1 and control w1118 (both strains derived from the wild-type Oregon R strain) larvae at 18 °C and 28 °C. We find that plasmatocyte counts are elevated in tuSz1 mutant larvae raised at 18 °C relative to w1118 controls raised at 18 °C (Fig. 2B), suggesting that tuSz1 mutants have a significantly increased plasmatocyte population. However, circulating plasmatocyte counts are significantly decreased in tuSz1 mutants relative to w1118 controls at 28 °C (Fig. 2B). This difference is likely due to the fact that, in tuSz1 mutants at 28 °C, the immune cells are actively participating in the self-encapsulation response (Fig. 1F) and are sequestered out of circulation as they adhere to the developing capsule.
Lamellocytes are Drosophila immune cells that are usually only produced during an encapsulation response, and their numbers are significantly increased in tuSz1 mutant larvae at both temperatures (Fig. 2C), in agreement with previous findings (37). Notably, these data functionally separate the production of lamellocytes (occurring at both 18 °C and 28 °C) from the self-encapsulation response (28 °C only) and suggest that immune cell alterations and self-encapsulation penetrance are genetically separable aspects of the tuSz1 phenotype. To further test the genetic basis of the immune cell phenotype, we assayed lamellocyte number in heterozygous (tuSz1/w1118) flies at 28 °C and found that heterozygous flies have significantly more lamellocytes than control flies (Fig. 2C), demonstrating that this phenotype is dominant. Lamellocytes can be produced directly from hemocyte precursors in the lymph gland (the Drosophila hematopoietic organ) or via the transdifferentiation of circulating or sessile plasmatocyte populations (50, 59, 60). The production of lamellocytes within the lymph gland leads to the dispersal of the lymph gland lobes (50, 61), and we find that lymph glands are dispersed in tuSz1 mutant larvae at both temperatures (SI Appendix, Fig. S3). Additionally, podocytes, also known as prelamellocytes, are plasmatocyte-like cells with fine cytoplasmic filaments extending outward from the cell body and are hypothesized to represent an intermediate step when circulating plasmatocytes transdifferentiate into lamellocytes during a macroparasite immune challenge (54, 59, 62, 63). There is a significant increase in the production of podocytes in tuSz1 mutants, but only at 28 °C (Fig. 2D), perhaps contributing to the production of additional lamellocytes in the restrictive condition. These findings suggest that lamellocytes are produced via both routes in tuSz1 mutants.
Finally, crystal cells are a Drosophila immune cell type involved in the melanization of wounds, and their numbers are significantly decreased in tuSz1 mutant larvae at both temperatures (Fig. 2E). The decrease in crystal cells in the absence of self-encapsulation at 18 °C suggests that this decrease is not due to the participation of crystal cells in melanotic encapsulation. We speculate that the loss of crystal cells may result from changes in signal transduction during the specification of the shared hemocyte precursor cells in the lymph gland, away from crystal cell production and to lamellocyte production, a process known to be regulated by the Notch and JAK-STAT signal transduction pathways (64–67).
The tuSz1 Phenotype Is Caused by Two Distinct Mutations.
Overall, these phenotypic characterization data lead to the hypothesis that the tuSz1 mutant phenotype is dependent on two distinct mutations: One is a nonconditional, dominant mutation that results in an alteration in the population of circulating hemocytes, including the production of lamellocytes, which are a hallmark of the Drosophila encapsulation response. The other is a temperature-sensitive, recessive mutation that results in an alteration of the posterior fat body tissue, making it a target of this activated immune cell response. This second mutation is likely to affect a gene required for production of a Drosophila SAMP that serves as a self-tolerance signal.
The tuSz1 mutation was isolated in a temperature-sensitive mutagenesis screen (41) and mapped to the X chromosome (42). Subsequent mapping efforts using meiotic recombination mapping and complementation testing with large genomic deletions mapped the tuSz1 mutation to a 572-kb region of the X chromosome containing 80 predicted genes, but despite strong efforts, the genetic basis of the phenotype was not identified (37, 58). Since that time, the Drosophila genome has been sequenced and annotated, the protein products of many genes in the mapped region have been characterized, and mutant strains have been developed for many of these genes. Furthermore, numerous molecularly defined chromosomal aberration collections have become available in Drosophila (68–72), which enables fine scale mapping of many classic mutations that had not been previously localized to the genome (73). We took a candidate gene approach as well as a mapping approach that makes use of complementation testing using defined genomic deletions and duplications. Along with targeted genomic DNA sequencing, we were able to more precisely map and identify the mutations leading to the loss of self-tolerance in tuSz1 mutants.
The Dominant Mutation in tuSz1 Is a Gain-of-Function Mutation in hop.
The tuSz1 mutant locus includes hop, the Drosophila homolog of JAK, a central member of the JAK-STAT pathway. Gain-of-function mutations in hop have been demonstrated to increase hemocyte numbers and cause differentiation to the lamellocyte type (74, 75). Additionally, JAK-STAT pathway activity is linked to the melanotic encapsulation immune response (76, 77). We therefore hypothesized that a mutation in hop may be the dominant mutation causing constitutive hemocyte production as part of the self-encapsulation phenotype in tuSz1 mutant flies.
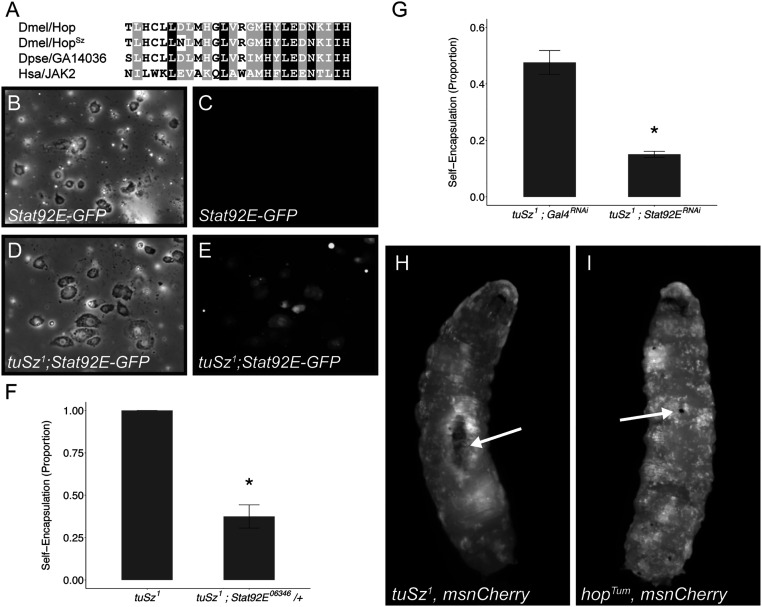
Sequencing the hop gene from the tuSz1 strain reveals that the hopSz allele contains a missense mutation in the JH2 pseudokinase domain, converting the aspartic acid at position 682 to an asparagine (D682N; Fig. 3A). The JH2 domain is conserved across species and is required for the autoinhibition of JAK when the pathway is inactive (78). Mutations in the JH2 domain lead to gain-of-function JAK activity and are linked to human diseases including multiple forms of leukemia and other cancers (79–82). We predict that the D682N missense mutation may disrupt JH2 domain function and result in ectopic pathway signaling, as this residue only shows conservative substitutions between species, with glutamic acid found at the corresponding position in the human JAK2 protein (Fig. 3A). To test this prediction, we assayed JAK-STAT pathway activity using the Stat92E-GFP reporter (83). This reporter expresses GFP under the control of the Stat92E transcription factor, and GFP fluorescence intensity gives a relative readout of pathway activity. The reporter is activated in hemocytes from tuSz1; Stat92E-GFP mutants but not control Stat92E-GFP larvae (Fig. 3 B–E), suggesting that the hopSz allele encodes a gain-of-function Hop protein. To test the hypothesis that the hopSz allele is dominant, we imaged the Stat92E-GFP reporter in heterozygous tuSz1/+; Stat92E-GFP larvae. The reporter is activated in this heterozygous background (SI Appendix, Fig. S4), supporting the dominant nature of the hopSz allele.
Fig. 3.
The role of JAK-STAT signaling in tuSz1 mutants. (A) An amino acid sequence alignment within the JH2 domain of Hop and the Hop orthologs GA14036 from Drosophila pseudoobscura and JAK2 from humans shows the missense D. melanogaster HopSz allele. (B–E) Brightfield (B) and GFP (C) images of immune cells from Stat92E-GFP larvae raised at 28 °C show no JAK-STAT pathway activity. Brightfield (D) and GFP (E) images of immune cells from tuSz1; Stat92E-GFP larvae raised at 28 °C show JAK-STAT pathway activity. (F) Penetrance of the self-encapsulation phenotype in tuSz1 and tuSz1; Stat92E06346/+ flies raised at 28 °C. *P < 0.05 compared to tuSz1. (G) Penetrance of the self-encapsulation phenotype in tuSz1; msn-GAL4; UAS-GAL4RNAi and tuSz1; msn-GAL4; UAS-Stat92ERNAi flies raised at 28 °C. *P < 0.05 compared to tuSz1; msn-GAL4; UAS-GAL4RNAi. P values were determined by generalized linear models. (H and I) Lamellocytes (marked by mCherry expression) aggregate around large self-encapsulations in tuSz1, msn-mCherry larvae (arrow in H) but are dispersed throughout hopTum, msn-mCherry larvae (I). Tissue self-encapsulations are not seen in hopTum, msn-mCherry larvae, but small melanized nodules are common (arrow in I).
To test whether this ectopic activation of the JAK-STAT pathway is an important contributor to the tuSz1 phenotype, we genetically decreased the dosage of Stat92E in tuSz1 mutants and assayed the self-encapsulation phenotype. Loss of one copy of Stat92E in tuSz1 mutant larvae is sufficient to suppress the mutant phenotype in tuSz1; Stat92E06346/+ larvae (Fig. 3F and SI Appendix, Table S2). To validate this finding and test the tissue specificity of the role of Stat92E in the tuSz1 phenotype, we crossed UAS-Stat92ERNAi with tuSz1; msn-GAL4 flies to knock down Stat92E specifically in lamellocytes in the tuSz1 mutant background. Once again, we observe a significant decrease in penetrance of the self-encapsulation phenotype relative to the UAS-GAL4RNAi control cross (Fig. 3G and SI Appendix, Table S2), suggesting that ectopic JAK-STAT activation in immune cells plays a role in the production of the self-encapsulation phenotype.
Importantly, gain-of-function JAK-STAT signaling alone is insufficient for the self-encapsulation phenotype. The encapsulation of self tissue is not observed in larvae carrying the widely studied hopTum gain-of-function mutation (Fig. 3 H and I), even though the hopTum and hopSz mutations both lead to the production of excess plasmatocytes and lamellocytes (Fig. 2, refs. 74 and 75). Whereas in tuSz1 mutants the lamellocytes are mainly observed on the surface of posterior fat body tissue (arrow in Fig. 3H), the excess lamellocytes in hopTum are dispersed in circulation, occasionally forming small melanized nodules by adhering to each other (arrow in Fig. 3I). This suggests that the hopSz allele may serve as a potentiating factor for the loss of self-tolerance, but there is likely a second mutation responsible for targeting of hopSz lamellocytes to the posterior fat body.
A Conditional, Recessive Loss-of-Function Allele of GCS1 Disrupts Protein N-glycosylation in tuSz1 Mutants.
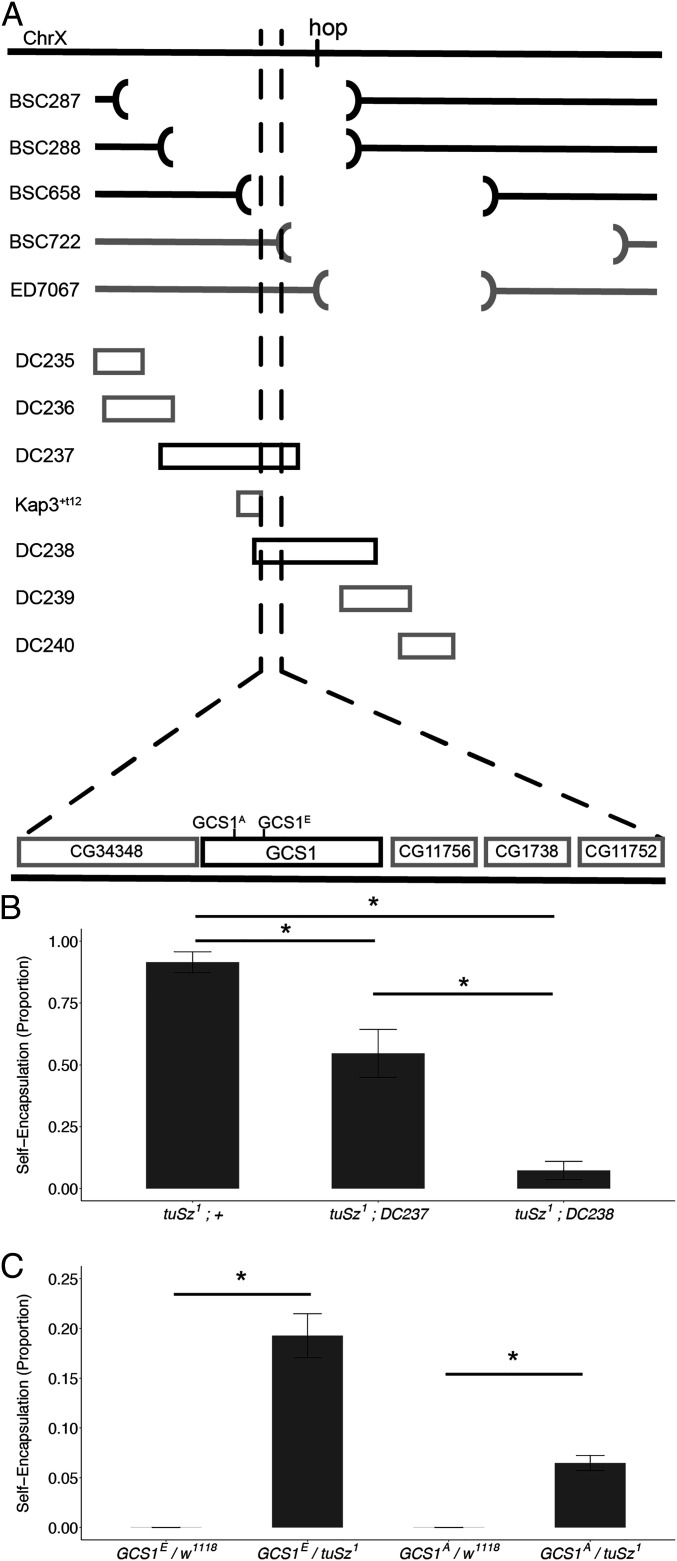
To uncover the genetic basis of the loss of self-tolerance phenotype observed in tuSz1 mutants, we used molecularly defined chromosomal aberrations to map the conditional recessive mutation within the previously defined X chromosome region. Five X chromosome deletion strains that, in combination, uncover the entirety of the previously mapped 572 kb tuSz1 locus were tested for their ability to complement the tuSz1 self-encapsulation phenotype (Fig. 4A). Three of these deletions fail to complement tuSz1, while two deletions do complement tuSz1, with the overlap defining a 24.4-kb chromosomal region responsible for the self-encapsulation phenotype (Fig. 4A). To further narrow down the location of the self-tolerance locus, we tested whether defined X chromosome genomic duplications (with small regions of the X chromosome translocated to the third chromosome) (71) within the mapped region could rescue the self-encapsulation phenotype in homozygous mutant tuSz1 flies. Of the seven X chromosome duplication strains tested, two rescue the self-encapsulation phenotype, while five fail to rescue, and together, these narrowed the self-tolerance locus to an even smaller 10.2-kb region containing five candidate genes (Fig. 4A).
Fig. 4.
Mapping the hypothesized tuSz1 self-tolerance mutation. (A) Schematic demonstrating the mapping of the tuSz1 recessive mutation on the X chromosome. Lines are listed by strain name. The hop gene locus is indicated on the chromosome. (Top) Interrupted segments illustrate the location of genome deletions in the indicated Drosophila strains. Black lines indicate deletions that fail to complement the tuSz1 self-encapsulation phenotype. Gray lines indicate complementing deletions. (Middle) Blocks represent duplicated regions of the X chromosome. Black blocks indicate duplications that rescue the tuSz1 self-encapsulation phenotype, and gray blocks indicate failure to rescue. (Bottom) The deletion and duplication lines indicate a small locus for the tuSz1 SAMP mutation. Candidate genes are shown. (B) Penetrance of the self-encapsulation phenotype of tuSz1 flies in combination with the indicated duplication raised at 28 °C. *P < 0.05 for the indicated comparisons. (C) Penetrance of the self-encapsulation phenotype of tuSz1 heterozygous flies in combination with w1118 controls or the indicated GCS1 allele raised at 28 °C. *P < 0.05 for the indicated comparisons. P values for B and C were determined by Dunnett’s test.
Note that the hop gene is located within 15 kb of the tuSz1 locus and that in our mapping experiments, we observed that the DC237 duplication, which includes the newly defined tuSz locus but excludes hop, was only able to partially rescue the self-encapsulation phenotype. However, the DC238 duplication, which includes both the tuSz and hop loci, rescued the phenotype nearly to control levels (Fig. 4B). This provided additional experimental evidence that hopSz, along with one other distinct mutation, are both required for the full tuSz1 self-encapsulation phenotype.
We find that two EMS-generated recessive lethal mutations in the GCS1 gene (84), one of the five genes found within the newly defined tuSz1 locus, fail to complement the tuSz1 self-encapsulation phenotype (Fig. 4C). Sequencing of the GCS1 locus from tuSz1, including 2 kb up- and downstream (Dataset S1), reveals 30 sequence variants compared to the D. melanogaster reference genome (SI Appendix, Table S3). Of these sequence changes, 27 are observed in the Drosophila Genetics Reference Panel (85), suggesting they may reflect standing variation in natural Drosophila populations. To further assess the potential functional significance of these sequence variants, we sequenced the GCS1 locus from the w1118 control strain (Dataset S1), which complements the tuSz1 self-encapsulation phenotype (Fig. 2A). Except for one mutation that is unique to tuSz1, the tuSz1 and w1118 GCS1 sequences are identical, suggesting that this lone mutation may be the causative mutation in tuSz1.
The identified mutation in tuSz1 (referred to as GCS1Sz) is a C-to-T transition 35 bp upstream of the transcription start site of GCS1. This sequence change may alter transcription factor binding sites identified in high-throughput transcription factor mapping projects (86–88) (Dataset S2) and falls within an identified transcription factor hotspot (89), suggesting that gene expression may be affected in tuSz1 mutants. To test this, we stained fat body tissues from early third-instar control w1118 and tuSz1 mutant larvae raised at 28 °C with an α-GCS1 antibody. We find α-GCS1 strongly stains control posterior fat body tissues (Fig. 5 A and B); however, α-GCS1 staining is largely undetectable in posterior fat body tissue from early third-instar tuSz1 mutants (Fig. 5 C and D). Importantly, α-GCS1 staining is maintained in anterior fat body tissue in tuSz1 mutants at 28 °C (SI Appendix, Fig. S5 A and B), mirroring the restriction of the self-encapsulation phenotype to posterior fat body tissue. We hypothesize that the GCS1Sz mutation alters an upstream enhancer that regulates GCS1 expression in the posterior fat body and that the mutation results in the temperature-sensitive disruption of gene expression. Interestingly, while plasmatocytes are responsible for coating other fly larval tissues with ECM, the fat body secretes its own ECM (90–92), which is consistent with the idea that the GCS1 locus might contain a fat-body–specific enhancer that is mutated in GCS1Sz.
Fig. 5.
Fat body GCS1 expression and protein N-glycosylation. (A–D) Posterior fat body tissue dissected from early third-instar w1118 (A and B) and tuSz1 (C and D) larvae raised at 28 °C prior to tissue melanization. (A and C) Brightfield images of dissected fat body tissue demonstrating the absence of tissue melanization. (B and D) Corresponding fluorescent images of α-GCS1 antibody staining. (E–J) Magnified posterior fat body tissue dissected from early third-instar larvae and stained by FITC-WGA to assay protein N-glycosylation. w1118 fat body from larvae raised at 28 °C (E) or 18 °C (F) is positive for FITC-WGA staining. tuSz1 fat body tissue is negative for FITC-WGA at 28 °C (G) but positive at 18 °C (H). tuSz1/w1118 heterozygous (I) and hopTum (J) larvae raised at 28 °C are positive for FITC-WGA staining, indicating that hop mutations do not affect fat body ECM protein N-glycosylation.
GCS1 encodes a mannosyl-oligosaccharide glucosidase, an enzyme required in the protein N-glycosylation pathway (93, 94). To test whether decreased GCS1 expression has a functional outcome in tuSz1 mutants, we incubated fat body tissues dissected from early third-instar w1118 and tuSz1 larvae with fluorescein isothiocyanate (FITC)-conjugated wheat germ agglutinin (WGA), a lectin that specifically recognizes N-glycosylated proteins (95). The magnified surfaces of fat body tissue from w1118 control larvae grown at both 18 °C and 28 °C show WGA binding displaying the characteristic honeycomb pattern of neighboring fat body cells (Fig. 5 E and F), indicating that fat body ECM proteins are N-glycosylated. tuSz1 mutants display a temperature-dependent loss of this WGA binding, with binding lost from posterior (but not anterior) fat body tissue in larvae grown at 28 °C (Fig. 5 G and H and SI Appendix, Fig. S5C). The loss of α-GCS1 staining and WGA binding are observed in tuSz1 fat body tissue from younger fly larvae prior to the onset of melanization. This suggests that loss of ECM protein N-glycosylation renders tissues susceptible to self-encapsulation, not that loss of staining is a consequence of tissue melanization. Furthermore, the loss of WGA reactivity is distinct from the effects of hop gain-of-function mutations: WGA staining is observed in heterozygous tuSz1/w1118 larvae (Fig. 5I) and hopTum mutant larvae (Fig. 5J), both of which have activated immune cells with no self-encapsulation. These findings are consistent with the maintenance of self-tolerance in these genotypes (Figs. 2A and 3I) and suggest that N-glycosylated ECM proteins may be recognized as SAMPs to promote self-tolerance.
ECM Protein N-glycosylation Is a Self-Tolerance Mark in Drosophila.
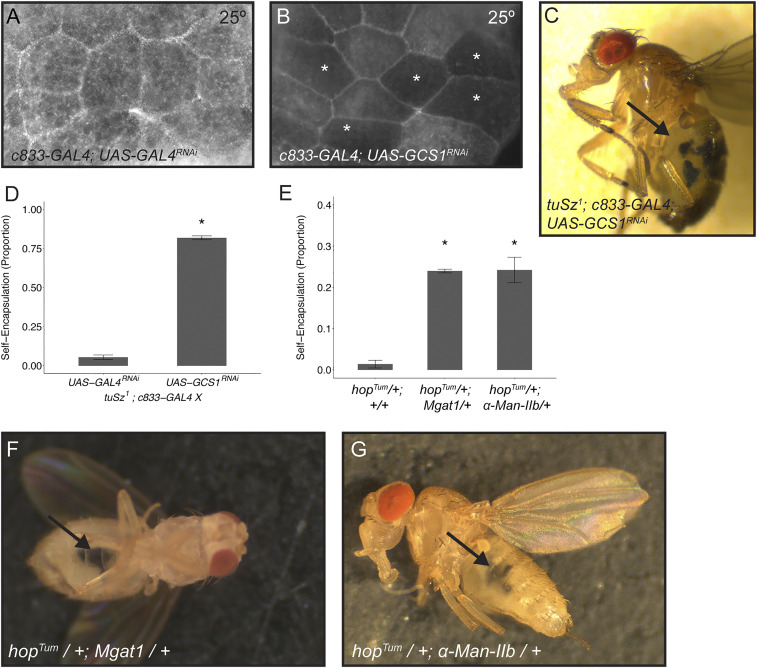
To test the hypothesis that ECM protein N-glycosylation plays a role in self-tolerance in Drosophila, we disrupted protein N-glycosylation in gain-of-function hop backgrounds and assayed for self-encapsulation. First, we used a UAS- GCS1RNAi construct in the tuSz1 background to knock down GCS1 levels specifically in fat body tissue by crossing to the c833-GAL4 driver. As previously discussed, the tuSz1 self-encapsulation phenotype is temperature sensitive and only observed at the restrictive temperature of 28 °C (Fig. 2A), whereas the JAK-STAT gain-of-function phenotype was seen at all temperatures. Taking into account the temperature-dependent nature of the GAL4/UAS expression system (96), in which knockdown is more efficient at higher temperatures, we raised larvae at 25 °C, at which we predict that tuSz1 larvae should have activated JAK-STAT signaling but no self-encapsulation response. We find that knockdown of GCS1 in c833-GAL4; UAS-GCS1RNAi larvae at 25 °C results in decreased fat body WGA binding compared to the c833-GAL4; UAS-GAL4RNAi knockdown control (Fig. 6 A and B), although to a lesser degree than tuSz1 mutants at 28 °C (compare Figs. 5E and 6B). We do not, however, observe the self-encapsulation phenotype when GCS1 is knocked down in fat body tissue in this otherwise wild-type background (n >100), which is consistent with our data suggesting that the hopSz gain-of-function allele is required for the self-encapsulation phenotype. We also do not observe the self-encapsulation phenotype in tuSz1; c833-GAL4; UAS-GAL4RNAi control knockdown flies raised at 25 °C, confirming that 25 °C is a permissive temperature for the temperature-sensitive self-encapsulation phenotype. However, we do see a strong self-encapsulation phenotype in tuSz1; c833-GAL4; UAS-GCS1RNAi flies raised at 25 °C (Fig. 6 C and D), providing further support for our hypothesis that GCS1 is required for self-tolerance. Given that c833-GAL4 is a pan–fat body driver, we expected and observed self-encapsulation of both anterior and posterior fat body tissues in this genotype.
Fig. 6.
Loss of GCS1 leads to loss of self-tolerance. (A and B) Posterior fat body tissue dissected from larvae raised at 25 °C and stained with FITC-WGA to assay protein N-glycosylation. Control c833-GAL4; UAS-GAL4RNAi tissue is positive for FITC-WGA staining (A). c833-GAL4; UAS-GCS1RNAi shows a mosaic loss of FITC-WGA staining, where * marks fat body cells with decreased staining (B). (C) tuSz1; c833-GAL4; UAS-GCS1RNAi flies raised at 25 °C show melanized self-encapsulations (indicated by the arrow). (D) Penetrance of the self-encapsulation phenotype in control tuSz1; c833-GAL4; UAS-GAL4RNAi and tuSz1; c833-GAL4; UAS-GCS1RNAi flies raised at 25 °C. *P < 0.05 compared to tuSz1; c833-GAL4; UAS-GAL4RNAi. (E) Penetrance of the self-encapsulation phenotype in hopTum/+, hopTum/+; Mgat1KG02444/+, and hopTum/+; α-Man-IIbMI09613/+ flies raised at 28 °C. *P < 0.05 compared to hopTum/+. (F) hopTum/+; Mgat1KG02444/+ flies raised at 28 °C show melanized self-encapsulations (indicated by the arrow). (G) hopTum/+; α-Man-IIbMI09613/+ flies raised at 28 °C show melanized self-encapsulations (indicated by the arrow). P values (D and E) were determined by generalized linear models.
Second, we tested whether enzymes in the protein N-glycosylation pathway that function downstream of GCS1 also play a role in the maintenance of self-tolerance. Specifically, we assayed if mutations in either of the N-glycan–processing enzymes Mgat1 or α-Man-IIb are sufficient to cause a loss of self-tolerance in a second hop gain-of-function mutant background, hopTum. hopTum is a temperature-sensitive mutant, and at 28 °C, the mutants display an activated immune response (75, 97). However, unlike tuSz1, this immune activation does not lead to the encapsulation of self tissue (Fig. 3I), suggesting that self-tolerance is maintained. As expected, we find that adult hopTum/+ flies raised at 28 °C display the characteristic melanotic nodule phenotype (44.6 ± 4.5% of flies have melanized nodules) but very rarely encapsulate self tissues (Fig. 6E). However, the removal of a single copy of either Mgat1 (Fig. 6 E and F) or α-Man-IIb (Fig. 6 E and G) in the hopTum/+ background in flies raised at 28 °C leads to a significant increase in the proportion of adults with the self-encapsulation phenotype. These findings suggest that the disruption of the N-glycosylation pathway leads to a loss of self-tolerance, supporting our hypothesis that ECM protein N-glycosylation serves as a self-tolerance signal. Interestingly, though the hopTum and Mgat1/α-Man-IIb mutant combinations might have led to melanotic encapsulation of other host tissues, the self-encapsulations we observed in these genotypes are predominantly localized to fat body tissue, which might be more sensitive to N-glycosylation processing enzyme dosage than other tissues given that it secretes its own ECM (90–92).
Discussion
A Two-Step Model for Maintaining Self-Tolerance in Drosophila.
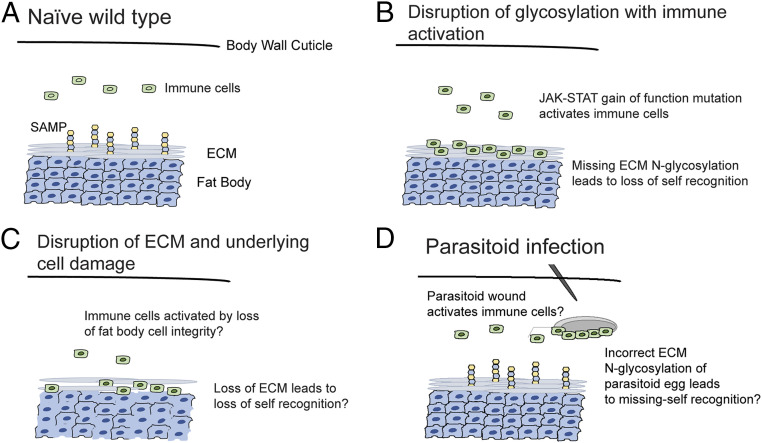
In this work, we have investigated the Drosophila tuSz1 mutant strain. tuSz1 is a temperature-sensitive mutant, and at the restrictive temperature, posterior fat body tissue is melanotically encapsulated by hemocytes in a reaction similar to the antiparasitoid immune response. We found that the tuSz1 phenotype is caused by two tightly linked mutations: a nonconditional, dominant gain-of-function mutation in hop that leads to ectopic immune activation and a temperature-sensitive, recessive mutation in GCS1 that leads to loss of protein N-glycosylation of the ECM overlaying the posterior fat body. These data lead us to propose a two-step model in which immune activation and the loss of SAMP presentation/recognition are both necessary for the breakdown of self-tolerance (Fig. 7). In a naïve wild-type larva, neither condition is met and self-tolerance is maintained (Fig. 7A). In the case of the tuSz1 mutant, the posterior fat body lacks appropriate ECM protein N-glycosylation and is targeted by constitutively activated hemocytes for encapsulation (Fig. 7B). This two-step model is also reflected in the hopTum mutant background, in which the simultaneous disruption of N-glycosylation in this immune-activated background results in tissue self-encapsulation similar to the tuSz1 mutant (Fig. 6 E–G).
Fig. 7.
Models describing the necessity for two independent signals in fly encapsulation responses. (A) Homeostasis is maintained in naïve wild-type larvae. (B) In tuSz1 mutant larvae, immune cells are inappropriately activated by JAK-STAT pathway activation due to the hopSz gain-of-function mutation. The loss of protein N-glycosylation in posterior fat body tissue due to the GCS1Sz mutation leads to loss of self-tolerance and tissue encapsulation. (C) In the model of self-tolerance described by Kim and Choe (55), the coupled phenotypes of loss of cell integrity and loss of ECM integrity are sufficient to disrupt self-tolerance. (D) Immune cells are activated following parasitoid wasp infection, presumably due to the wound-mediated activation of JAK-STAT signaling. SAMP-presenting host tissues are protected from encapsulation, and wasp eggs may be targeted for encapsulation because they are missing the ECM N-glycosylation SAMP.
Interestingly, previous work (55) also documented the necessity of at least two signals for self-encapsulation in Drosophila. In that case, both the loss of the ECM (with its glycosylated proteins) and the disrupted positional integrity of the underlying fat body cells (potentially mimicking a wound) were required for immune cells to become activated and encapsulate the self tissue (Fig. 7C). A similar loss of ECM and underlying cell integrity was also found in the classical melanotic tumor mutant tu(2)W (98, 99). This model, in which at least two factors are required for self-encapsulation, may explain why the several classically described self-encapsulation mutants (SI Appendix, Table S1), unlike virtually all other types of visible Drosophila mutants, were never successfully mapped (38).
The disruption of either factor in the two-step model in isolation is not sufficient to cause self-encapsulation. This can be seen in parasitoid wasp infected larvae; the wounding associated with parasitoid infection leads to immune activation (59, 60), but in the absence of SAMP disruption, the fly is able to specifically encapsulate the parasitoid egg while protecting against self-encapsulation (Figs. 1D and 7D). Conversely, while internal tissue damage in a naïve larva does attract hemocyte interactions, in the absence of an immune stimulus, this does not lead to self-encapsulation, but rather the hemocytes attempt to repair the damaged tissue (100). That blood cells err on the side of fixing disrupted self tissue rather than treating it as pathogenic and encapsulating it unless another stress signal is also present suggests that flies may have evolved a multi-input system to safeguard against spurious encapsulation (100, 101).
A Conserved Mechanism of Innate Self-Tolerance?
D. melanogaster immune responses have proven to be an excellent model for understanding the mechanisms underlying conserved innate immune responses, including those of humans (23, 102). Our findings on Drosophila self-tolerance may also be relevant to human innate self-tolerance. Indeed, data from a range of studies are consistent with the idea that protein glycosylation is a mediator of vertebrate immune responses, and cell-surface glycans have been proposed as candidate SAMPs for the innate immune response to distinguish healthy self tissues from aberrant or foreign tissues even if the mechanisms are not entirely understood (3, 19, 103–105). Protein-linked sugar groups should presumably fit this role well, as they can take on diverse combinations of sugar residues and branching patterns (19, 106).
Protein N-glycosylation is a complex multistep process (106, 107) that begins with the addition of a presynthesized glycosyl precursor to the protein at an asparagine residue. This nascent glycan is then trimmed back to a core glycan structure, a process that is initiated by the activity of GCS1 (108). The core glycan is then elaborated with the addition of multiple carbohydrate groups to give rise to a variety of final structures, with hybrid and complex type N-glycans among the most prevalent (109–111). Glycan elaboration begins with the activity of Mgat1, which leads to the production of hybrid type N-glycans (112). These hybrid N-glycans can be further processed by the α-mannosidase-I and -II family of enzymes to produce paucimannose N-glycans, which can serve as complex N-glycan precursors and are further elaborated by downstream enzymes to give rise to the final complex N-glycan structure (113). Our data suggest that disruption of any of the genes encoding key N-glycan–processing enzymes will be associated with the loss of self-tolerance in Drosophila. Similarly, the loss of the α-mannosidase-II (αM-II) gene in mice is linked with the development of an autoimmune phenotype that is likened to systemic lupus erythematosus (114–116). Like the tuSz1 mutant, the αM-II mouse phenotype arises due to alterations in protein N-glycosylation in nonimmune tissues and is mediated by innate immune cells (115). Altered patterns of protein N-glycosylation are also observed in additional mouse models of autoimmune disease (117, 118) and have been linked to autoimmune disease in human patients (119–123).
The ECM is a conserved structure made up of numerous proteins, many of which are N-glycosylated, including laminin and collagen (124–127). A role for the ECM in mediating self-tolerance has been previously proposed: The encapsulation of self tissues in D. melanogaster is also observed following RNA interference (RNAi) knockdown of genes encoding the ECM proteins laminin and collagen, supporting the idea that SAMPs reside in the ECM (55). The role of the ECM in self-tolerance is further emphasized by tissue transplantation studies in Drosophila. Drosophila larvae are largely tolerant of conspecific tissue transplants, but this tolerance is abolished when tissues are first treated with collagenase to disrupt the ECM, leading to the specific encapsulation of treated tissues (128). Reactivity to ECM proteins is also associated with various forms of human autoimmune disease (129–131). Based on these data, we propose that the N-glycosylation of ECM proteins may serve as a conserved self-tolerance signal for innate immune mechanisms and that loss of ECM protein N-glycosylation may lead to loss of self-tolerance and, consequently, autoimmune disease in a diverse range of species.
Glycosylation and Missing-Self Recognition.
An alternative means by which hosts can recognize pathogens is missing-self recognition. Instead of tracking pathogen diversity with numerous recognition receptors, as in nonself recognition, missing-self recognition does not rely on tracking pathogens at all, but only on specifically recognizing self and attacking tissues that lack the self signal. Further, unlike germ line–encoded forms of nonself recognition, missing-self recognition systems allow host species to respond to novel pathogen types that they have never encountered in their evolutionary history. Protein glycosylation plays an important role in the handful of identified missing-self recognition systems of vertebrates (3, 19, 132–135). The most well-known case of missing-self recognition involves the interaction between vertebrate NK cells and host MHC class I (MHCI) proteins (136). All vertebrate cells produce MHCI to display any possible antigens present in their cytoplasm to T cells, but intracellular pathogens often suppress host cell MHCI expression to prevent their molecules from being displayed (137). NK cells are lymphoid-type cells that induce cytolysis in infected host cells (138). In an uninfected state, recognition of properly glycosylated MHCI inhibits NK cell cytolysis of host cells, but in an infected state in which host cells are missing the MHCI self signal, the NK cell inhibitory receptors fail to recognize “self,” and the infected host cells are lysed, an effective means of killing intracellular pathogens that have suppressed host MHC signaling (21, 132, 139, 140).
As of yet, there are no examples of missing-self immune recognition systems in invertebrates (141, 142), and it has been hypothesized that invertebrate immune systems rely largely on PRRs for nonself recognition of pathogens (143). Still, invertebrates do mount immune responses against a variety of inanimate objects like oil droplets, sterile nylon, and charged chromatography beads as well as tissue transplants from other insect species (128, 144–147). All of these foreign bodies presumably lack distinct PAMPs, suggesting that invertebrates have some sort of missing-self recognition system. Additionally, while multiple antimicrobial PRRs have been identified in the Drosophila genome (11), PRRs targeting macroparasites like parasitoid wasps have not yet been discovered (52). Our model of self-recognition suggests that following parasitoid infection, activated immune cells assess all exposed tissue surfaces for the self-tolerance glycan signal and that the absence of this Drosophila SAMP on parasitoid wasp eggs might be the cue that targets them for melanotic encapsulation (Fig. 7D).
Materials and Methods
Insect Strains.
D. melanogaster used in this study were maintained on standard Drosophila medium on a 12-h light–dark cycle. The D. melanogaster strains and sequence analysis are described in more detail in SI Appendix, Supplementary Materials and Methods.
Imaging.
To determine GCS1 expression in fat body tissue, fat bodies were dissected from early third-instar larvae in phosphate-buffered saline (PBS), fixed in 4% paraformaldehyde for 10 min, and permeabilized in PBS + 0.1% Triton-X100 (PBSTx). Tissues were then blocked in 5% normal goat serum in PBSTx and stained with the α-GCS1/MOGS antibody (Sigma-Aldrich, Prestige Antibodies HPA011969) diluted to 1:100 in blocking solution.
To assay tissue protein N-glycosylation, we dissected fat body tissues from early third-instar larvae in PBS and stained them with 100 mg/mL FITC-conjugated wheat germ agglutinin (FITC-WGA; Vector Laboratories, FL-1021) for 3 min at room temperature (53). Stained tissues were washed three times with Drosophila Ringer’s solution prior to imaging.
To assay hemocyte localization, whole mount larvae carrying the fluorescent hemocyte marker strain msn-mCherry (lamellocytes) were imaged in PBS. To assay hemocyte–fat body interactions, fat body tissue from larvae carrying the eater-eGFP (plasmatocytes) and msn-mCherry markers were dissected and imaged under paraffin oil.
Hemocyte Counts.
To determine the numbers of different hemocyte subtypes, five late third-instar larvae were rinsed in Drosophila Ringer’s solution and bled into 20 μL PBS + 0.01% phenylthiourea. Hemocytes were transferred to a disposable hemocytometer (Incyto C-Chip DHC-N01) and allowed to adhere for 30 min. Hemocytes from each sample were counted from sixteen 0.25 × 0.25 × 0.1 mm squares. Experiments were performed in triplicate, as described in ref. 53. The different hemocyte types were identified by their stereotypical morphologies (148).
Statistical Analysis.
To analyze phenotypic penetrance data, we used generalized linear models with quasibinomial errors for parametric data, and ANOVA of aligned rank transformed data for comparisons involving genotypes with a penetrance proportion of 0 or 1. For experiments with multiple comparisons, Dunnett’s test was used for pairwise comparisons to the w1118 control genotype.
Welch’s two-sample t tests were used to compare immune cell count data between two genotypes. For multiple comparisons, immune cell count data were analyzed by ANOVA followed by Dunnett’s test for pairwise comparisons. All statistics were done in the R statistical computing environment (149) using the multcomp (150) and ARTool (151) packages. Graphs were produced using the ggplot2 package (152).
Supplementary Material
Acknowledgments
We thank two anonymous reviewers, as well as members of the N.T.M. and T.A.S. laboratories, for detailed and constructive feedback that greatly improved this manuscript. Research reported in this publication was supported by NIH Awards R35GM133760 to N.T.M. and R01AI081879 to T.A.S. Stocks obtained from the Bloomington Drosophila Stock Center (NIH P40OD018537) were used in this study. The Illinois State University Confocal Microscopy Facility was funded by NSF Grant DBI-1828136.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2017460118/-/DCSupplemental.
Data Availability
All study data are included in the article and/or supporting information.
References
- 1.Danilova N., The evolution of immune mechanisms. J. Exp. Zoolog. B Mol. Dev. Evol. 306, 496–520 (2006). [DOI] [PubMed] [Google Scholar]
- 2.Janeway C. A. Jr, Medzhitov R., Innate immune recognition. Annu. Rev. Immunol. 20, 197–216 (2002). [DOI] [PubMed] [Google Scholar]
- 3.Medzhitov R., Janeway C. A. Jr, Decoding the patterns of self and nonself by the innate immune system. Science 296, 298–300 (2002). [DOI] [PubMed] [Google Scholar]
- 4.Babcock D. T., et al., Circulating blood cells function as a surveillance system for damaged tissue in Drosophila larvae. Proc. Natl. Acad. Sci. U.S.A. 105, 10017–10022 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cooper M. D., Alder M. N., The evolution of adaptive immune systems. Cell 124, 815–822 (2006). [DOI] [PubMed] [Google Scholar]
- 6.Kumar H., Kawai T., Akira S., Pathogen recognition in the innate immune response. Biochem. J. 420, 1–16 (2009). [DOI] [PubMed] [Google Scholar]
- 7.Romagnani S., Immunological tolerance and autoimmunity. Intern. Emerg. Med. 1, 187–196 (2006). [DOI] [PubMed] [Google Scholar]
- 8.Berke I. C., Li Y., Modis Y., Structural basis of innate immune recognition of viral RNA. Cell. Microbiol. 15, 386–394 (2013). [DOI] [PubMed] [Google Scholar]
- 9.Brown G. D., Gordon S., Immune recognition of fungal beta-glucans. Cell. Microbiol. 7, 471–479 (2005). [DOI] [PubMed] [Google Scholar]
- 10.Kang D., Liu G., Lundström A., Gelius E., Steiner H., A peptidoglycan recognition protein in innate immunity conserved from insects to humans. Proc. Natl. Acad. Sci. U.S.A. 95, 10078–10082 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lu Y., et al., Pattern recognition receptors in Drosophila immune responses. Dev. Comp. Immunol. 102, 103468 (2020). [DOI] [PubMed] [Google Scholar]
- 12.Park B. S., Lee J.-O., Recognition of lipopolysaccharide pattern by TLR4 complexes. Exp. Mol. Med. 45, e66 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Toubi E., Vadasz Z., Innate immune-responses and their role in driving autoimmunity. Autoimmun. Rev. 18, 306–311 (2019). [DOI] [PubMed] [Google Scholar]
- 14.Waldner H., The role of innate immune responses in autoimmune disease development. Autoimmun. Rev. 8, 400–404 (2009). [DOI] [PubMed] [Google Scholar]
- 15.Bentham J., et al., Genetic association analyses implicate aberrant regulation of innate and adaptive immunity genes in the pathogenesis of systemic lupus erythematosus. Nat. Genet. 47, 1457–1464 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cavalcante P., et al., Etiology of myasthenia gravis: Innate immunity signature in pathological thymus. Autoimmun. Rev. 12, 863–874 (2013). [DOI] [PubMed] [Google Scholar]
- 17.Kramer J. M., Early events in Sjögren’s Syndrome pathogenesis: The importance of innate immunity in disease initiation. Cytokine 67, 92–101 (2014). [DOI] [PubMed] [Google Scholar]
- 18.O’Reilly S., Innate immunity in systemic sclerosis pathogenesis. Clin. Sci. (Lond.) 126, 329–337 (2014). [DOI] [PubMed] [Google Scholar]
- 19.Varki A., Since there are PAMPs and DAMPs, there must be SAMPs? Glycan “self-associated molecular patterns” dampen innate immunity, but pathogens can mimic them. Glycobiology 21, 1121–1124 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yokoyama W. M., The search for the missing ‘missing-self’ receptor on natural killer cells. Scand. J. Immunol. 55, 233–237 (2002). [DOI] [PubMed] [Google Scholar]
- 21.Kärre K., Natural killer cell recognition of missing self. Nat. Immunol. 9, 477–480 (2008). [DOI] [PubMed] [Google Scholar]
- 22.Brennan C. A., Anderson K. V., Drosophila: The genetics of innate immune recognition and response. Annu. Rev. Immunol. 22, 457–483 (2004). [DOI] [PubMed] [Google Scholar]
- 23.Hoffmann J. A., Kafatos F. C., Janeway C. A., Ezekowitz R. A., Phylogenetic perspectives in innate immunity. Science 284, 1313–1318 (1999). [DOI] [PubMed] [Google Scholar]
- 24.Lemaitre B., Hoffmann J., The host defense of Drosophila melanogaster. Annu. Rev. Immunol. 25, 697–743 (2007). [DOI] [PubMed] [Google Scholar]
- 25.Parsons B., Foley E., Cellular immune defenses of Drosophila melanogaster. Dev. Comp. Immunol. 58, 95–101 (2016). [DOI] [PubMed] [Google Scholar]
- 26.Williams M. J., Drosophila hemopoiesis and cellular immunity. J. Immunol. 178, 4711–4716 (2007). [DOI] [PubMed] [Google Scholar]
- 27.Howell L., et al., A directed miniscreen for genes involved in the Drosophila anti-parasitoid immune response. Immunogenetics 64, 155–161 (2012). [DOI] [PubMed] [Google Scholar]
- 28.Rämet M., Manfruelli P., Pearson A., Mathey-Prevot B., Ezekowitz R. A. B., Functional genomic analysis of phagocytosis and identification of a Drosophila receptor for E. coli. Nature 416, 644–648 (2002). [DOI] [PubMed] [Google Scholar]
- 29.Stroschein-Stevenson S. L., Foley E., O’Farrell P. H., Johnson A. D., Identification of Drosophila gene products required for phagocytosis of Candida albicans. PLoS Biol. 4, e4 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Anthony R. M., Rutitzky L. I., Urban J. F. Jr, Stadecker M. J., Gause W. C., Protective immune mechanisms in helminth infection. Nat. Rev. Immunol. 7, 975–987 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Koppang E. O., Haugarvoll E., Hordvik I., Aune L., Poppe T. T., Vaccine-associated granulomatous inflammation and melanin accumulation in Atlantic salmon, Salmo salar L., white muscle. J. Fish Dis. 28, 13–22 (2005). [DOI] [PubMed] [Google Scholar]
- 32.Mukhopadhyay S., et al., Causes of pulmonary granulomas: A retrospective study of 500 cases from seven countries. J. Clin. Pathol. 65, 51–57 (2012). [DOI] [PubMed] [Google Scholar]
- 33.Richards D. T., Hoole D., Lewis J. W., Ewens E., Arme C., Adherence of carp leucocytes to adults and cercariae of the blood fluke Sanguinicola inermis. J. Helminthol. 70, 63–67 (1996). [DOI] [PubMed] [Google Scholar]
- 34.Swartz J. M., et al., Plasminogen activator inhibitor-2 (PAI-2) in eosinophilic leukocytes. J. Leukoc. Biol. 76, 812–819 (2004). [DOI] [PubMed] [Google Scholar]
- 35.Rizki R. M., Rizki T. M., Cell interactions in the differentiation of a melanotic tumor in Drosophila. Differentiation 12, 167–178 (1979). [DOI] [PubMed] [Google Scholar]
- 36.Watson K. L., Johnson T. K., Denell R. E., Lethal(1) aberrant immune response mutations leading to melanotic tumor formation in Drosophila melanogaster. Dev. Genet. 12, 173–187 (1991). [DOI] [PubMed] [Google Scholar]
- 37.Rizki T. M., Rizki R. M., Developmental analysis of a temperature-sensitive melanotic tumor mutant inDrosophila melanogaster. Wilhelm Roux Arch. Dev. Biol. 189, 197–206 (1980). [DOI] [PubMed] [Google Scholar]
- 38.Sparrow J. C., “Melanotic ‘tumours’” in The Genetics and Biology of Drosophila, Ashburner M., Wright T., Eds. (Academic Press, London, 1978), pp. 277–313. [Google Scholar]
- 39.Burnet B., Sang J. H., Physiological genetics of melanotic tumors in Drosophila melanogaster. II. the genetic basis of response to tumorigenic treatments in the tu K and tu bw; st su-tu strains. Genetics 49, 223–235 (1964). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Minakhina S., Steward R., Melanotic mutants in Drosophila: Pathways and phenotypes. Genetics 174, 253–263 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Suzuki D. T., Temperature-sensitive mutations in Drosophila melanogaster. Science 170, 695–706 (1970). [DOI] [PubMed] [Google Scholar]
- 42.Suzuki D. T., et al., Temperature-sensitive mutations in Drosophila melanogaster. I. Relative frequencies among gamma-ray and chemically induced sex-linked recessive lethals and semilethals. Proc. Natl. Acad. Sci. U.S.A. 57, 907–912 (1967). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Driessen G., Alphen J. J. M. V., Hemerik L., Drosophila species, breeding in the stinkhorn (Phallus impudicus pers.) and their larval parasitoids. Neth. J. Zool. 40, 409–427 (1989). [Google Scholar]
- 44.Fleury F., et al., Ecological and genetic interactions in Drosophila-parasitoids communities: A case study with D. melanogaster, D. simulans and their common Leptopilina parasitoids in south-eastern France. Genetica 120, 181–194 (2004). [DOI] [PubMed] [Google Scholar]
- 45.Janssen A., Driessen G., De Haan M., Roodbol N., The impact of parasitoids on natural populations of temperate woodland Drosophila. Neth. J. Zool. 38, 61–73 (1987). [Google Scholar]
- 46.Honti V., Csordás G., Kurucz É., Márkus R., Andó I., The cell-mediated immunity of Drosophila melanogaster: Hemocyte lineages, immune compartments, microanatomy and regulation. Dev. Comp. Immunol. 42, 47–56 (2014). [DOI] [PubMed] [Google Scholar]
- 47.Vlisidou I., Wood W., Drosophila blood cells and their role in immune responses. FEBS J. 282, 1368–1382 (2015). [DOI] [PubMed] [Google Scholar]
- 48.Mortimer N. T., et al., Parasitoid wasp venom SERCA regulates Drosophila calcium levels and inhibits cellular immunity. Proc. Natl. Acad. Sci. U.S.A. 110, 9427–9432 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Russo J., Dupas S., Frey F., Carton Y., Brehelin M., Insect immunity: Early events in the encapsulation process of parasitoid (Leptopilina boulardi) eggs in resistant and susceptible strains of Drosophila. Parasitology 112, 135–142 (1996). [DOI] [PubMed] [Google Scholar]
- 50.Lanot R., Zachary D., Holder F., Meister M., Postembryonic hematopoiesis in Drosophila. Dev. Biol. 230, 243–257 (2001). [DOI] [PubMed] [Google Scholar]
- 51.Rizki T. M., Rizki R. M., Lamellocyte differentiation in Drosophila larvae parasitized by Leptopilina. Dev. Comp. Immunol. 16, 103–110 (1992). [DOI] [PubMed] [Google Scholar]
- 52.Carton Y., Poirié M., Nappi A. J., Insect immune resistance to parasitoids. Insect Sci. 15, 67–87 (2008). [Google Scholar]
- 53.Mortimer N. T., Kacsoh B. Z., Keebaugh E. S., Schlenke T. A., Mgat1-dependent N-glycosylation of membrane components primes Drosophila melanogaster blood cells for the cellular encapsulation response. PLoS Pathog. 8, e1002819 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rizki M. T. M., Alterations in the haemocyte population of Drosophila melanogaster. J. Morphol. 100, 437–458 (1957). [Google Scholar]
- 55.Kim M. J., Choe K.-M., Basement membrane and cell integrity of self-tissues in maintaining Drosophila immunological tolerance. PLoS Genet. 10, e1004683 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Isabella A. J., Horne-Badovinac S., “Building from the ground up: Basement membranes in Drosophila development” in Basement Membranes, Miner J. H., Ed., Current Topics in Membranes (Academic Press, 2015), pp. 305–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nappi A. J., Hemocyte reactions and early cellular changes during melanotic tumor formation in Drosophila melanogaster. J. Invertebr. Pathol. 43, 395–406 (1984). [Google Scholar]
- 58.Zhimulev I., et al., Fine cytogenetical analysis of the band 10A1-2 and the adjoining regions in the Drosophila melanogaster X-chromosome. V. Genetic characteristics of the loci in the 9E-10B region. Biol. Zent. Bl. 106, 699–720 (1987). [Google Scholar]
- 59.Anderl I., et al., Transdifferentiation and proliferation in two distinct hemocyte lineages in Drosophila melanogaster larvae after wasp infection. PLoS Pathog. 12, e1005746 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Márkus R., et al., Sessile hemocytes as a hematopoietic compartment in Drosophila melanogaster. Proc. Natl. Acad. Sci. U.S.A. 106, 4805–4809 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sorrentino R. P., Carton Y., Govind S., Cellular immune response to parasite infection in the Drosophila lymph gland is developmentally regulated. Dev. Biol. 243, 65–80 (2002). [DOI] [PubMed] [Google Scholar]
- 62.Shrestha R., Gateff E., Ultrastructure and cytochemistry of the cell-types in the tumorous hematopoietic organs and the hemolymph of the mutant Lethal (1) Malignant Blood Neoplasm (l(1)mbn) of Drosophila melanogaster. Dev. Growth Differ. 24, 83–98 (1982). [DOI] [PubMed] [Google Scholar]
- 63.Rizki T. M., Rizki R. M., “The cellular defense system of Drosophila melanogaster” in Insect Ultrastructure, King R. C., Akai H., Eds. (Springer US, 1984), vol. 2, pp. 579–604. [Google Scholar]
- 64.Duvic B., Hoffmann J. A., Meister M., Royet J., Notch signaling controls lineage specification during Drosophila larval hematopoiesis. Curr. Biol. 12, 1923–1927 (2002). [DOI] [PubMed] [Google Scholar]
- 65.Krzemien J., Oyallon J., Crozatier M., Vincent A., Hematopoietic progenitors and hemocyte lineages in the Drosophila lymph gland. Dev. Biol. 346, 310–319 (2010). [DOI] [PubMed] [Google Scholar]
- 66.Mukherjee T., Kim W. S., Mandal L., Banerjee U., Interaction between Notch and Hif-α in development and survival of Drosophila blood cells. Science 332, 1210–1213 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Small C., et al., An unexpected link between notch signaling and ROS in restricting the differentiation of hematopoietic progenitors in Drosophila. Genetics 197, 471–483 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cook R. K., et al., The generation of chromosomal deletions to provide extensive coverage and subdivision of the Drosophila melanogaster genome. Genome Biol. 13, R21 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Roote J., Russell S., Toward a complete Drosophila deficiency kit. Genome Biol. 13, 149 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cook R. K., et al., A new resource for characterizing X-linked genes in Drosophila melanogaster: Systematic coverage and subdivision of the X chromosome with nested, Y-linked duplications. Genetics 186, 1095–1109 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Venken K. J. T., et al., A molecularly defined duplication set for the X chromosome of Drosophila melanogaster. Genetics 186, 1111–1125 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ryder E., et al., The DrosDel deletion collection: A Drosophila genomewide chromosomal deficiency resource. Genetics 177, 615–629 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Spana E. P., et al., speck, first identified in Drosophila melanogaster in 1910, is encoded by the Arylalkalamine N-Acetyltransferase (AANAT1) gene. G3 (Bethesda) 10, 3387–3398 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Harrison D. A., Binari R., Nahreini T. S., Gilman M., Perrimon N., Activation of a Drosophila Janus kinase (JAK) causes hematopoietic neoplasia and developmental defects. EMBO J. 14, 2857–2865 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Luo H., Hanratty W. P., Dearolf C. R., An amino acid substitution in the Drosophila hopTum-l Jak kinase causes leukemia-like hematopoietic defects. EMBO J. 14, 1412–1420 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sorrentino R. P., Melk J. P., Govind S., Genetic analysis of contributions of dorsal group and JAK-Stat92E pathway genes to larval hemocyte concentration and the egg encapsulation response in Drosophila. Genetics 166, 1343–1356 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yang H., Kronhamn J., Ekström J.-O., Korkut G. G., Hultmark D., JAK/STAT signaling in Drosophila muscles controls the cellular immune response against parasitoid infection. EMBO Rep. 16, 1664–1672 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Saharinen P., Vihinen M., Silvennoinen O., Autoinhibition of Jak2 tyrosine kinase is dependent on specific regions in its pseudokinase domain. Mol. Biol. Cell 14, 1448–1459 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hammarén H. M., Virtanen A. T., Raivola J., Silvennoinen O., The regulation of JAKs in cytokine signaling and its breakdown in disease. Cytokine 118, 48–63 (2019). [DOI] [PubMed] [Google Scholar]
- 80.Levine R. L., et al., The JAK2V617F activating mutation occurs in chronic myelomonocytic leukemia and acute myeloid leukemia, but not in acute lymphoblastic leukemia or chronic lymphocytic leukemia. Blood 106, 3377–3379 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lupardus P. J., et al., Structure of the pseudokinase-kinase domains from protein kinase TYK2 reveals a mechanism for Janus kinase (JAK) autoinhibition. Proc. Natl. Acad. Sci. U.S.A. 111, 8025–8030 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Springuel L., Renauld J.-C., Knoops L., JAK kinase targeting in hematologic malignancies: A sinuous pathway from identification of genetic alterations towards clinical indications. Haematologica 100, 1240–1253 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bach E. A., et al., GFP reporters detect the activation of the Drosophila JAK/STAT pathway in vivo. Gene Expr. Patterns 7, 323–331 (2007). [DOI] [PubMed] [Google Scholar]
- 84.Yamamoto S., et al., A drosophila genetic resource of mutants to study mechanisms underlying human genetic diseases. Cell 159, 200–214 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Mackay T. F. C., et al., The Drosophila melanogaster genetic reference panel. Nature 482, 173–178 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Junion G., et al., A transcription factor collective defines cardiac cell fate and reflects lineage history. Cell 148, 473–486 (2012). [DOI] [PubMed] [Google Scholar]
- 87.MacArthur S., et al., Developmental roles of 21 Drosophila transcription factors are determined by quantitative differences in binding to an overlapping set of thousands of genomic regions. Genome Biol. 10, R80 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Nègre N., et al., A cis-regulatory map of the Drosophila genome. Nature 471, 527–531 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.modENCODE Consortium et al., Identification of functional elements and regulatory circuits by Drosophila modENCODE. Science 330, 1787–1797 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Banerjee U., Girard J. R., Goins L. M., Spratford C. M., Drosophila as a genetic model for hematopoiesis. Genetics 211, 367–417 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Fessler J. H., Fessler L. I., Drosophila extracellular matrix. Annu. Rev. Cell Biol. 5, 309–339 (1989). [DOI] [PubMed] [Google Scholar]
- 92.Matsubayashi Y., et al., A moving source of matrix components is essential for de novo basement membrane formation. Curr. Biol. 27, 3526–3534.e4 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Boisson M., et al., Arabidopsis glucosidase I mutants reveal a critical role of N-glycan trimming in seed development. EMBO J. 20, 1010–1019 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Katoh T., et al., Deficiency of α-glucosidase I alters glycoprotein glycosylation and lifespan in Caenorhabditis elegans. Glycobiology 23, 1142–1151 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Burger M. M., Goldberg A. R., Identification of a tumor-specific determinant on neoplastic cell surfaces. Proc. Natl. Acad. Sci. U.S.A. 57, 359–366 (1967). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Duffy J. B., GAL4 system in Drosophila: A fly geneticist’s Swiss army knife. Genesis 34, 1–15 (2002). [DOI] [PubMed] [Google Scholar]
- 97.Luo H., et al., Mutation in the Jak kinase JH2 domain hyperactivates Drosophila and mammalian Jak-Stat pathways. Mol. Cell. Biol. 17, 1562–1571 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Rizki R. M., Rizki T. M., Basement membrane abnormalities in melanotic tumor formation of Drosophila. Experientia 30, 543–546 (1974). [DOI] [PubMed] [Google Scholar]
- 99.Rizki T. M., Rizki R. M., Topology of the caudal fat body of the tumor-w mutant of Drosophila melanogaster. J. Invertebr. Pathol. 24, 37–40 (1974). [DOI] [PubMed] [Google Scholar]
- 100.Pastor-Pareja J. C., Wu M., Xu T., An innate immune response of blood cells to tumors and tissue damage in Drosophila. Dis. Model. Mech. 1, 144–154 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Tsai C.-R., Wang Y., Galko M. J., Crawling wounded: Molecular genetic insights into wound healing from Drosophila larvae. Int. J. Dev. Biol. 62, 479–489 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Medzhitov R., Toll-like receptors and innate immunity. Nat. Rev. Immunol. 1, 135–145 (2001). [DOI] [PubMed] [Google Scholar]
- 103.Maverakis E., et al., Glycans in the immune system and the altered glycan theory of autoimmunity: A critical review. J. Autoimmun. 57, 1–13 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Paulson J. C., Macauley M. S., Kawasaki N., Siglecs as sensors of self in innate and adaptive immune responses. Ann. N. Y. Acad. Sci. 1253, 37–48 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Mahla R. S., Reddy M. C., Prasad D. V. R., Kumar H., Sweeten PAMPs: Role of sugar complexed PAMPs in innate immunity and vaccine biology. Front. Immunol. 4, 248 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Schwarz F., Aebi M., Mechanisms and principles of N-linked protein glycosylation. Curr. Opin. Struct. Biol. 21, 576–582 (2011). [DOI] [PubMed] [Google Scholar]
- 107.Gagneux P., Aebi M., Varki A., “Evolution of glycan diversity” in Essentials of Glycobiology, Varki A., Ed. et al. (Cold Spring Harbor Laboratory Press, 3rd Ed, 2017), pp. 253–264. [Google Scholar]
- 108.Helenius A., Aebi M., Roles of N-linked glycans in the endoplasmic reticulum. Annu. Rev. Biochem. 73, 1019–1049 (2004). [DOI] [PubMed] [Google Scholar]
- 109.Moremen K. W., Tiemeyer M., Nairn A. V., Vertebrate protein glycosylation: Diversity, synthesis and function. Nat. Rev. Mol. Cell Biol. 13, 448–462 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Vandenborre G., et al., Diversity in protein glycosylation among insect species. PLoS One 6, e16682 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Walski T., et al., Protein N-glycosylation and N-glycan trimming are required for postembryonic development of the pest beetle Tribolium castaneum. Sci. Rep. 6, 35151 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Schachter H., Narasimhan S., Gleeson P., Vella G., Control of branching during the biosynthesis of asparagine-linked oligosaccharides. Can. J. Biochem. Cell Biol. 61, 1049–1066 (1983). [DOI] [PubMed] [Google Scholar]
- 113.Moremen K. W., Trimble R. B., Herscovics A., Glycosidases of the asparagine-linked oligosaccharide processing pathway. Glycobiology 4, 113–125 (1994). [DOI] [PubMed] [Google Scholar]
- 114.Chui D., et al., Genetic remodeling of protein glycosylation in vivo induces autoimmune disease. Proc. Natl. Acad. Sci. U.S.A. 98, 1142–1147 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Green R. S., et al., Mammalian N-glycan branching protects against innate immune self-recognition and inflammation in autoimmune disease pathogenesis. Immunity 27, 308–320 (2007). [DOI] [PubMed] [Google Scholar]
- 116.Van Dyken S. J., Locksley R. M., Autoimmunity: Altered self-N-glycans trigger innate-mediated autoimmunity. Immunol. Cell Biol. 85, 572–574 (2007). [DOI] [PubMed] [Google Scholar]
- 117.Hashii N., et al., Alteration of N-glycosylation in the kidney in a mouse model of systemic lupus erythematosus: Relative quantification of N-glycans using an isotope-tagging method. Immunology 126, 336–345 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Alavi A., Axford J. S., Sweet and sour: The impact of sugars on disease. Rheumatology (Oxford) 47, 760–770 (2008). [DOI] [PubMed] [Google Scholar]
- 119.Grigorian A., et al., Pathogenesis of multiple sclerosis via environmental and genetic dysregulation of N-glycosylation. Semin. Immunopathol. 34, 415–424 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Ząbczyńska M., et al., Altered N-glycan profile of IgG-depleted serum proteins in Hashimoto’s thyroiditis. Biochim. Biophys. Acta, Gen. Subj. 1864, 129464 (2020). [DOI] [PubMed] [Google Scholar]
- 121.Labasque M., et al., Specific contactin N-glycans are implicated in neurofascin binding and autoimmune targeting in peripheral neuropathies. J. Biol. Chem. 289, 7907–7918 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Reily C., Stewart T. J., Renfrow M. B., Novak J., Glycosylation in health and disease. Nat. Rev. Nephrol. 15, 346–366 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Cvetko A., et al., Glycosylation alterations in multiple sclerosis show increased proinflammatory potential. Biomedicines 8, 410 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Jürgensen H. J., et al., A novel functional role of collagen glycosylation: Interaction with the endocytic collagen receptor uparap/ENDO180. J. Biol. Chem. 286, 32736–32748 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Morita A., Sugimoto E., Kitagawa Y., Post-translational assembly and glycosylation of laminin subunits in parietal endoderm-like F9 cells. Biochem. J. 229, 259–264 (1985). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Sasaki T., Fässler R., Hohenester E., Laminin: The crux of basement membrane assembly. J. Cell Biol. 164, 959–963 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Vandenborre G., et al., Glycosylation signatures in Drosophila: Fishing with lectins. J. Proteome Res. 9, 3235–3242 (2010). [DOI] [PubMed] [Google Scholar]
- 128.Rizki R. M., Rizki T. M., Hemocyte responses to implanted tissues in Drosophila melanogaster larvae. Wilhelm Roux Arch. Dev. Biol. 189, 207–213 (1980). [DOI] [PubMed] [Google Scholar]
- 129.Medina C. O., Nagy N., Bollyky P. L., Extracellular matrix and the maintenance and loss of peripheral immune tolerance in autoimmune insulitis. Curr. Opin. Immunol. 55, 22–30 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Nagy N., et al., Hyaluronan in immune dysregulation and autoimmune diseases. Matrix Biol. 78–79, 292–313 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Bei R., et al., Long-lasting tissue inflammatory processes trigger autoimmune responses to extracellular matrix molecules. Int. Rev. Immunol. 27, 137–175 (2008). [DOI] [PubMed] [Google Scholar]
- 132.Parham P., NK cell receptors: Of missing sugar and missing self. Curr. Biol. 10, R195–R197 (2000). [DOI] [PubMed] [Google Scholar]
- 133.Lian R. H., Freeman J. D., Mager D. L., Takei F., Role of conserved glycosylation site unique to murine class I MHC in recognition by Ly-49 NK cell receptor. J. Immunol. 161, 2301–2306 (1998). [PubMed] [Google Scholar]
- 134.Crocker P. R., Paulson J. C., Varki A., Siglecs and their roles in the immune system. Nat. Rev. Immunol. 7, 255–266 (2007). [DOI] [PubMed] [Google Scholar]
- 135.Blaum B. S., et al., Structural basis for sialic acid-mediated self-recognition by complement factor H. Nat. Chem. Biol. 11, 77–82 (2015). [DOI] [PubMed] [Google Scholar]
- 136.Kärre K., NK cells, MHC class I molecules and the missing self. Scand. J. Immunol. 55, 221–228 (2002). [DOI] [PubMed] [Google Scholar]
- 137.Lodoen M. B., Lanier L. L., Viral modulation of NK cell immunity. Nat. Rev. Microbiol. 3, 59–69 (2005). [DOI] [PubMed] [Google Scholar]
- 138.Trinchieri G., Biology of natural killer cells. Adv. Immunol. 47, 187–376 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Ljunggren H. G., Kärre K., In search of the ‘missing self’: MHC molecules and NK cell recognition. Immunol. Today 11, 237–244 (1990). [DOI] [PubMed] [Google Scholar]
- 140.Yokoyama W. M., Kim S., Licensing of natural killer cells by self-major histocompatibility complex class I. Immunol. Rev. 214, 143–154 (2006). [DOI] [PubMed] [Google Scholar]
- 141.Mushegian A., Medzhitov R., Evolutionary perspective on innate immune recognition. J. Cell Biol. 155, 705–710 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Williams M. J., Regulation of antibacterial and antifungal innate immunity in fruitflies and humans. Adv. Immunol. 79, 225–259 (2001). [DOI] [PubMed] [Google Scholar]
- 143.Royet J., Infectious non-self recognition in invertebrates: Lessons from Drosophila and other insect models. Mol. Immunol. 41, 1063–1075 (2004). [DOI] [PubMed] [Google Scholar]
- 144.Eslin P., Doury G., The fly Drosophila subobscura: A natural case of innate immunity deficiency. Dev. Comp. Immunol. 30, 977–983 (2006). [DOI] [PubMed] [Google Scholar]
- 145.Lavine M. D., Strand M. R., Surface characteristics of foreign targets that elicit an encapsulation response by the moth Pseudoplusia includens. J. Insect Physiol. 47, 965–974 (2001). [DOI] [PubMed] [Google Scholar]
- 146.Sadd B. M., Siva-Jothy M. T., Self-harm caused by an insect’s innate immunity. Proc. Biol. Sci. 273, 2571–2574 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Cooper E. L., Transplantation immunity in annelids. I. Rejection of xenografts exchanged between Lumbricus terrestris and Eisenia foetida. Transplantation 6, 322–327 (1968). [DOI] [PubMed] [Google Scholar]
- 148.Rizki T. M., Rizki R. M., Properties of the larval hemocytes of Drosophila melanogaster. Experientia 36, 1223–1226 (1980). [Google Scholar]
- 149.R Core Team , R: A Language and Environment for Statistical Computing (R Core Team, 2021). [Google Scholar]
- 150.Hothorn T., Bretz F., Westfall P., Simultaneous inference in general parametric models. Biom. J. 50, 346–363 (2008). [DOI] [PubMed] [Google Scholar]
- 151.Wobbrock J. O., Findlater L., Gergle D., Higgins J. J., The aligned rank transform for nonparametric factorial analyses using only anova procedures in Proceedings of the SIGCHI Conference on Human Factors in Computing Systems. (Association for Computing Machinery, 2011), pp. 143–146. [Google Scholar]
- 152.Wickham H., ggplot2: Elegant Graphics for Data Analysis (Springer-Verlag, 2009). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or supporting information.