Abstract
The RHO1 gene encodes a yeast homolog of the mammalian RhoA protein. Rho1p is localized to the growth sites and is required for bud formation. We have recently shown that Bni1p is one of the potential downstream target molecules of Rho1p. The BNI1 gene is implicated in cytokinesis and the establishment of cell polarity in Saccharomyces cerevisiae but is not essential for cell viability. In this study, we screened for mutations that were synthetically lethal in combination with a bni1 mutation and isolated two genes. They were the previously identified PAC1 and NIP100 genes, both of which are implicated in nuclear migration in S. cerevisiae. Pac1p is a homolog of human LIS1, which is required for brain development, whereas Nip100p is a homolog of rat p150Glued, a component of the dynein-activated dynactin complex. Disruption of BNI1 in either the pac1 or nip100 mutant resulted in an enhanced defect in nuclear migration, leading to the formation of binucleate mother cells. The arp1 bni1 mutant showed a synthetic lethal phenotype while the cin8 bni1 mutant did not, suggesting that Bni1p functions in a kinesin pathway but not in the dynein pathway. Cells of the pac1 bni1 and nip100 bni1 mutants exhibited a random distribution of cortical actin patches. Cells of the pac1 act1-4 mutant showed temperature-sensitive growth and a nuclear migration defect. These results indicate that Bni1p regulates microtubule-dependent nuclear migration through the actin cytoskeleton. Bni1p lacking the Rho-binding region did not suppress the pac1 bni1 growth defect, suggesting a requirement for the Rho1p-Bni1p interaction in microtubule function.
The Rho family (Rho) belongs to the small G-protein superfamily and regulates various cell functions, such as cell adhesion, cell motility, and cytokinesis, through the reorganization of the actin cytoskeleton (18, 57). Many potential downstream target molecules of Rho have been identified (19), but it has not yet been thoroughly clarified how Rho regulates the reorganization of the actin cytoskeleton through these target molecules.
The actin cytoskeleton plays a pivotal role in the budding processes of the yeast Saccharomyces cerevisiae (4). This yeast has several Rho family members, including Rho1p, Rho2p, Rho3p, Rho4p, and Cdc42p, which are involved in the budding processes (4, 58). We have isolated Bni1p as a potential downstream target molecule of Rho1p that regulates the reorganization of the actin cytoskeleton (29). Bni1p has subsequently been shown to be a potential downstream target molecule of Cdc42p, Rho3p, and Rho4p (11, 23). We have also found that Bnr1p is a Bni1p-related protein that is a potential downstream target molecule of Rho4p (24). Bni1p and Bnr1p are members of the FH protein family, which is defined by the presence of “formin homology” domains, the proline-rich FH1 domain and the FH2 domain. The FH proteins play important roles in actin cytoskeleton-dependent processes, including cytokinesis and the establishment of cell polarity (12, 62). Bni1p and Bnr1p, at their FH1 domains, bind to profilin, an actin monomer-binding protein that is implicated in actin polymerization (11, 24). Bni1p and Bnr1p also interact with Bud6p (Aip3p), an actin-binding protein (1, 11, 28). Moreover, Bni1p interacts with elongation factor 1α (EF1α), which binds to and bundles actin filaments (60). Bni1p is known to localize at sites of bud growth, where the actin cytoskeleton is actively reorganized (25). We have found that Spa2p is a Bni1p-binding protein and that this interaction is required for the localization of Bni1p to the bud tip (13).
The cytoplasmic microtubule system is believed to functionally and physically interact with the actin system, although the molecular mechanisms of this interaction remain to be clarified (2, 9, 27, 34, 43). Genetic and cell biological studies with S. cerevisiae tub2 mutants have shown that cytoplasmic microtubules are not required for budding but are required for the migration of a daughter nucleus into the bud (22, 56). A minus-end-directed, microtubule-based motor protein, dynein, is involved in this nuclear migration process (50, 68). Cell biological studies with act1 mutants have revealed that the actin system is involved in the control of spindle position and nuclear migration (7, 43), but it remains to be clarified how the actin system interacts with the cytoplasmic microtubule system.
In this study, we have shown that Bni1p regulates nuclear migration. A bni1 mutation shows a synthetic lethal interaction with mutations in PAC1 or NIP100 (14, 26). PAC1 encodes a protein similar to a human lissencephaly gene product, LIS1 (45), and to an Aspergillus nidulans gene product, NUDF, which is implicated in dynein function (65, 66). NIP100 encodes a protein similar to the largest polypeptide component of the dynein-activating dynactin complex, p150Glued of rat and Neurospora crassa (46, 59, 64). Both pac1 bni1 and nip100 bni1 mutant cells show a binucleate phenotype, probably due to the misorientation of cytoplasmic microtubules. Our results suggest that Bni1p is a cortical marker that guides cytoplasmic microtubules to the bud tip.
MATERIALS AND METHODS
Strains, media, and yeast transformations.
The yeast strains used in this study are listed in Table 1. Yeast strains were grown on the rich media YPDAU and YPGalAU. YPDAU contained 2% Bacto Peptone (Difco Laboratories, Detroit, Mich.), 1% Bacto Yeast Extract (Difco), 0.04% adenine sulfate, 0.02% uracil, and 2% glucose. YPGalAU was the same except that 3% galactose plus 0.2% sucrose replaced the glucose. A medium that contained 2% Bacto Peptone, 1% Bacto Yeast Extract, 0.02% uracil, and 4% glucose (YPDDU) was used to isolate bsl mutants. Yeast transformations were performed by the lithium acetate method (16). Transformants were selected on SD medium, which contained 2% glucose and 0.7% yeast nitrogen base without amino acids. SG medium contained 3% galactose, 0.2% sucrose, and 0.7% yeast nitrogen base without amino acids. SD or SG medium was supplemented with amino acids or bases when required. SDAAU or SGalAAU contained 0.5% Casamino Acids, 0.025% adenine sulfate, and 0.025% uracil, in addition to SD or SG medium. Standard yeast genetic manipulations were performed as described previously (52). Where indicated, benomyl (Wako, Osaka, Japan), dissolved at 10 mg/ml in dimethyl sulfoxide, was added to YPDAU to a final concentration of 10, 20, or 30 μg/ml. Escherichia coli DH5α was used for construction and propagation of plasmids.
TABLE 1.
Yeast strains used in this studya
| Strain | Genotype | Reference | |
|---|---|---|---|
| OHNY1 | MATa ura3 leu2 trp1 his3 ade2 | 41 | |
| OHNY2 | MATα ura3 leu2 trp1 his3 ade2 | 41 | |
| OHNY3 | MATa/MATα ura3/ura3 leu2/leu2 trp1/trp1 his3/his3 ade2/ade2 | 42 | |
| KY4 | MATa/MATα ura3/ura3 leu2/leu2 trp1/trp1 his3/his3 ade2/ade2 bni1::HIS3/bni1::HIS3 | 29 | |
| STFY1 | MATα ura3 leu2 trp1 his3 ade2 ade3 bni1::HIS3 | ||
| BTY3 | MATa ura3 leu2 trp1 his3 ade2 bni1::HIS3 | 13 | |
| HIY2 | MATα ura3 leu2 trp1 his3 ade2 bnr1::TRP1 | 24 | |
| HIY11 | MATa ura3 leu2 trp1 his3 ade2 bni1::HIS3 bnr1::TRP1 | 24 | |
| PFIY1 | MATα ura3 leu2 trp1 his3 ade2 pac1::URA3 | ||
| PFIY11 | MATα ura3 leu2 trp1 his3 ade2 nip100::URA3 | ||
| AFIY1 | MATα ura3 leu2 trp1 his3 ade2 arp1::URA3 | ||
| CFIY1 | MATα ura3 leu2 trp1 his3 ade2 cin8::URA3 | ||
| PFIY5 | MATα ura3 leu2 trp1 his3 ade2 pac1::URA3 bni1::HIS3 [pRS314-GAL1-myc-BNI1] | ||
| PFIY6 | MATα ura3 leu2 trp1 his3 ade2 pac1::URA3 bni1::HIS3 [pRS315-GAL1-HA-BNI1] | ||
| PFIY15 | MATa ura3 leu2 trp1 his3 ade2 nip100::URA3 bni1::HIS3 [pRS314-GAL1-myc-BNI1] | ||
| AMFY1 | MATa ura3 leu2 trp1 his3 ade2 act1-4 | ||
| APFY1 | MATa ura3 leu2 trp1 his3 ade2 pac1::URA3 act1-4 |
Strains used in this study are isogenic.
Molecular biological techniques.
Standard molecular biological techniques were used for construction of plasmids, DNA sequencing, and PCR (48). Plasmids used in this study are listed in Table 2. DNA sequences were determined with an ALFexpress DNA sequencer (Amersham Pharmacia Biotech, Inc., Little Chalfont, United Kingdom), and PCRs were performed with the GeneAmp PCR System 2400 (Perkin-Elmer, Norwalk, Conn.).
TABLE 2.
Plasmids used in this study
| Plasmid | Characteristicsa | Reference |
|---|---|---|
| YEp351 | LEU2 2μm | 20 |
| YEp351-BNI1 | BNI1 LEU2 2μm; made by inserting the 6.5-kb BamHI-SmaI fragment from pBS-BNI1 into the corresponding sites of YEp351 | 29 |
| YEp351-BNI1-ADE3 | BNI1 ADE3 LEU2 2μm; made by inserting the 5.4-kb SmaI-SalI (filled-in) ADE3 fragment (a gift from S. Nomoto) into the SmaI site of YEp351-BNI1 | |
| pRS315 | LEU2 CEN6 | 54 |
| pRS316 | URA3 CEN6 | 54 |
| YCp50-PAC1 | PAC1 URA3 CEN6; isolated from a genomic library in YCp50 as a clone containing a 9.6-kb genomic DNA fragment of PAC1 | 41 |
| YCp50-NIP100 | NIP100 URA3 CEN6; isolated from a genomic library in YCp50 as a clone containing a 14-kb genomic DNA fragment of NIP100 | 41 |
| pRS316-BNI1 | BNI1 URA3 CEN6; made by inserting the 6.5-kb BamHI-SmaI fragment from pBS-BNI1 into the corresponding sites of pRS316 | 29 |
| pRS316-PAC1 | PAC1 URA3 CEN6; made by inserting the 3.1-kb BamHI-EcoRV fragment from YCp50-PAC1 into the BamHI-SmaI sites of pRS316 | |
| pRS316-NIP100 | NIP100 URA3 CEN6; made by inserting the 4.2-kb EcoRI-KpnI fragment from YCp50-NIP100 into the corresponding sites of pRS316 | |
| pRS314-GAL1-myc | TRP1 CEN6 | 13 |
| pRS314-GAL1-myc-BNI1 | TRP1 CEN6 GAL1-myc-BNI1 | 13 |
| pRS315-GAL1 | LEU2 CEN6; made by inserting the 0.74-kb ScaI-HindIII fragment, containing the GAL1 promoter, a multicloning site, and the TDH1 terminator, into the SpeI (filled-in)-HindIII sites of pRS315 | 13 |
| pRS315-GAL1-HA | LEU2 CEN6; made by inserting the synthetic oligonucleotide encoding two copies of the HA epitope, YPYDVPDYA, which is derived from the influenza virus hemagglutinin (HA) protein, into the EcoRI-KpnI sites in the multicloning site of pRS315-GAL1 | |
| pRS315-GAL1-HA-BNI1 | LEU2 CEN6 GAL1-HA-BNI1; made by inserting the 5.9-kb BamHI-SmaI fragment from pRS314-GAL1-myc-BNI1 containing the BNI1 open reading frame into the corresponding sites of the multicloning site of pRS315-GAL1-HA, to place BNI1 downstream of the two HA epitopes | 29 |
| pRS314-PBNI1-myc | TRP1 CEN6; made by inserting the 0.62-kb PCR fragment upstream of the BNI1 open reading frame into the EcoRI-SacI sites of pRS314-GAL1-myc to replace the GAL1 promoter with the BNI1 promoter; the 0.62-kb BNI1 promoter region was amplified by PCR with upstream primer 1 (5′-GGCCGAGCTCAGTTGGTATGGATAGAGCCAGAATGTAAAACAAGGTGGCA) and downstream primer 2 (5′-GGCCGAATTCTTCCTTTCCTTCTCTTCTTTTCTTGCTTCTCG) | |
| pRS314-PBNI1-myc-BNI1 | TRP1 CEN6 PBNI1-myc-BNI1; made by inserting the 5.9-kb BamHI-SmaI fragment from pRS314-GAL1-myc-BNI1 containing the BNI1 open reading frame into the corresponding sites of the multicloning site of pRS314-PBNI1-myc, to place BNI1 downstream of the three myc epitopes | 13 |
| pRS314-PBNI1-myc-BNI1 (490–1954) | TRP1 CEN6 PBNI1-myc-BNI1 (490–1954); made by inserting the 4.4-kb BamHI-SmaI fragment from pRS314-GAL1-myc-BNI1 (490–1954) into the corresponding sites of the multicloning site of pRS314-PBNI1-myc, to place BNI1 (490–1954) downstream of the three myc epitopes | 13 |
| pRS314-PBNI1-myc-BNI1′-1 | TRP1 CEN6 PBNI1-myc-BNI1 (Δ826–987); made by inserting the 2.5-kb BamHI site-bounded PCR fragment encoding amino acids 1 to 825 of Bni1p and the 2.9-kb BamHI-SmaI site-bounded PCR fragment encoding amino acids 988 to 1954 of Bni1p into the BamHI-SmaI sites of pRS314-PBNI1-myc, to place BNI1 (Δ826–987) downstream of the three myc epitopes; the 2.5-kb DNA fragment was amplified by PCR with upstream primer 3 (5′-TATAGGATCCAGTTGGTATGGATAGAGCCAGAATGTAAAACAAGGTGGCA) and downstream primer 4 (5′-GGCCGGATCCCGAGTTCATACCTCTGGTGCCTTC); the 2.9-kb DNA fragment was amplified by PCR with upstream primer 5 (5′-GGCCGGATCCGACGAGATTATGGACAGTCCAAAG) and downstream primer 6 (5′-CTCTCCCGGGCTAGTGCTTGTTTGGATGTTTGTTTTGGTATTACTGTTGT) | |
| pRS314-PBNI1-myc-BNI1′-2 | TRP1 CEN6 PBNI1-myc-BNI1 (Δ1239–1328); made by inserting the 3.7-kb BamHI site-bounded PCR fragment encoding amino acids 1 to 1238 of Bni1p and the 1.9-kb BamHI-SmaI site-bounded PCR fragment encoding amino acids 1329 to 1954 of Bni1p into the BamHI-SmaI sites of pRS314-PBNI1-myc, to place BNI1 (Δ1239–1328) downstream of the three myc epitopes; the 3.7-kb DNA fragment was amplified by PCR with upstream primer 3 and downstream primer 7 (5′-GGCCGGATCCCTGTGAGGAGAGTACAGATGATTG); the 1.9-kb DNA fragment was amplified by PCR with upstream primer 8 (5′-GGCCGGATCCGCATCGCAAATCAAATCAGCTGTAAC) and the downstream primer 6 | |
| pRS314-PBNI1-myc-BNI1′-3 | TRP1 CEN6 PBNI1-myc-BNI1 (1–1750); made by inserting the 5.3-kb BamHI-SacII (filled-in) fragment from pRS314-GAL1-myc-BNI1 containing the BNI1 open reading frame into the BamHI-SmaI sites of the multicloning site of pRS314-PBNI1-myc, to place BNI1 (1–1750) downstream of the three myc epitopes | 13 |
| pBS-PAC1 | PAC1; made by inserting the 3.1-kb BamHI-EcoRV fragment from YCp50-PAC1 into the corresponding sites of pBluescript KS(+) | |
| pUC19-NIP100 | NIP100; made by inserting the 4.2-kb EcoRI-KpnI fragment from pRS316-NIP100 into the corresponding sites of pUC19 | |
| pBS-pac1::URA3 | A derivative of pBS-PAC1; made by replacing the 0.85-kb SphI-HpaI internal fragment of PAC1 corresponding to amino acids 38 to 321 with the 1.2-kb URA3 fragment | |
| pUC19-nip100::URA3 | A derivative of pUC19-NIP100; made by replacing the 4.4-kb EcoRV-SacI internal fragment of NIP100 corresponding to amino acids 67 to 731 with the 1.2-kb URA3 fragment | |
| pUC19-ARP1 | ARP1; made by inserting the 2.2-kb BamHI-SalI site-bounded coding region plus the fragment including the ARP1 0.5-kb upstream and 0.5-kb downstream noncoding regions into the corresponding sites of pUC19; the 2.2-kb DNA fragment was amplified by PCR with upstream primer 9 (5′-GGCCGGATCCCATAATACAGTATTTATTGATCAACG) and downstream primer 10 (5′-GGCCGTCGACTCTGAAAGGTATGTCTTCCTATTAGAC) | |
| pUC19-arp1::URA3 | A derivative of pUC19-ARP1; made by replacing the 0.97-kb ClaI-StyI internal fragment of ARP1 corresponding to amino acids 1 to 306 with the 1.2-kb URA3 fragment | |
| pUC19-CIN8 | CIN8; made by inserting the 4.1-kb BamHI-SalI site-bounded coding region plus the fragment including the CIN8 0.5-kb upstream and 0.5-kb downstream noncoding regions into the corresponding sites of pUC19; the 4.1-kb DNA fragment was amplified by PCR with upstream primer 11 (5′-GGCCGGATCCATCTGGTTTCAAGCAAGAATTGAAC) and downstream primer 12 (5′-GGCCGTCGACAAGACCATCAAAGGTTTCATTTG) | |
| pUC19-cin8::URA3 | A derivative of pUC19-CIN8; made by replacing the 2.57-kb MunI internal fragment of CIN8 corresponding to amino acids 158 to 1016 with the 1.2-kb URA3 fragment |
Underlined sequences are portions of BNI1, ARP1, or CIN8.
Screening for bsl mutants.
To obtain mutations (bsl) that cause synthetic lethality with the bni1 mutation, strain STFY1 containing plasmid YEp351-BNI1-ADE3 was plated on YPDDU and subsequently treated with UV for 30 s. Colonies that showed a red nonsectoring phenotype were isolated and subsequently transformed with plasmid pRS316-BNI1 to test whether the isolated clones showed a white sectoring phenotype. Clones that showed a sectoring phenotype were retained as bsl mutants.
Tetrad analysis of bsl mutants.
To determine whether the bsl mutants contained single mutations, the bsl bni1/YEp351-BNI1-ADE3 haploid cells were crossed to the parental bni1 cells (BTY3) and diploid clones that had lost YEp351-BNI1-ADE3 were selected. Sporulation and tetrad analyses were done by standard methods (51), and at least 10 tetrads were analyzed for each cross. The bsl mutants that showed 2:2 segregation in terms of the growth phenotypes were characterized further.
Cloning of PAC1 and NIP100.
Each bsl bni1/YEp351-BNI1-ADE3 mutant was transformed with a yeast genomic library made in centromeric vector YCp50 (41) and screened for white colonies on SD plates lacking uracil but containing 0.00125% adenine sulfate. Cells of single white colonies were replica-plated onto SD plates lacking leucine. Library plasmids were recovered through E. coli transformation from clones that did not grow on these plates. The recovered plasmids were transformed again into the bsl mutant to identify clones that reproducibly conferred sectoring activity. The recovered plasmids were sequenced with primer 5′-GCTACTTGGAGCCACTATCGAC and primer 5′-AGGCGCCAGCAACCGCACCTGT, and the partial nucleotide sequences of the cloned genomic DNAs were determined. The plasmids were mapped by restriction enzyme digestion, and several different and overlapping regions of the genomic fragment were subcloned into pRS316 (54) and tested for colony-sectoring activity. PAC1 and NIP100 were isolated from the bsl1 and bsl2 mutants, respectively, and these genes were characterized further in this study. To confirm whether the bsl1 and bsl2 mutations were located in PAC1 and NIP100, respectively, the 0.85-kb SphI-HpaI fragment in the PAC1 open reading frame and the 2.0-kb EcoRV-SacI fragment in the NIP100 open reading frame were deleted and the resulting genomic fragments were tested for colony-sectoring activity. Neither genomic fragment showed any sectoring activity (data not shown). Moreover, the pac1 and nip100 disruption mutants were crossed with the bsl1 bni1 and bsl2 bni1 mutants, respectively, and tetrad analysis indicated that the bsl1 and bsl2 mutations occurred in PAC1 and NIP100, respectively (data not shown).
Disruption of PAC1, NIP100, ARP1, and CIN8.
To construct pac1 and nip100 disruption mutants, plasmids pBS-pac1::URA3 and pUC19-nip100::URA3 were cut with PvuII or with EcoRI and SalI, respectively, and the digested DNA was introduced into strain OHNY3. To construct the arp1 and cin8 disruption mutants, plasmids pUC19-arp1::URA3 and pUC19-cin8::URA3 were cut with PvuII and the digested DNA was introduced into strain OHNY2. Genomic DNA was isolated from each transformant, and the proper disruption of each gene was verified by PCR (data not shown). These pac1, nip100, arp1, and cin8 mutant strains were used for further genetic studies.
Cytological techniques.
Actin and DNA were stained with rhodamine-phalloidin (Molecular Probes, Eugene, Oreg.) and 4′,6-diamidino-2-phenylindole dihydrochloride (DAPI) (Sigma Chemical Co., St. Louis, Mo.), respectively, as described previously (67). Indirect immunofluorescence of microtubules was performed as described previously (67). Microtubules were stained with a rat anti-α-tubulin YOL1/34 monoclonal antibody (Harlan Sera-lab, Loughborough, England). Stained cells were observed with an Axiophoto microscope (Carl Zeiss, Oberkochen, Germany) and photographed with a peltier cooling 3CCD color camera (C5810-01; Hamamatsu Photonics KK., Hamamatsu, Japan).
RESULTS
Deficiency in nuclear migration in the bni1 mutant.
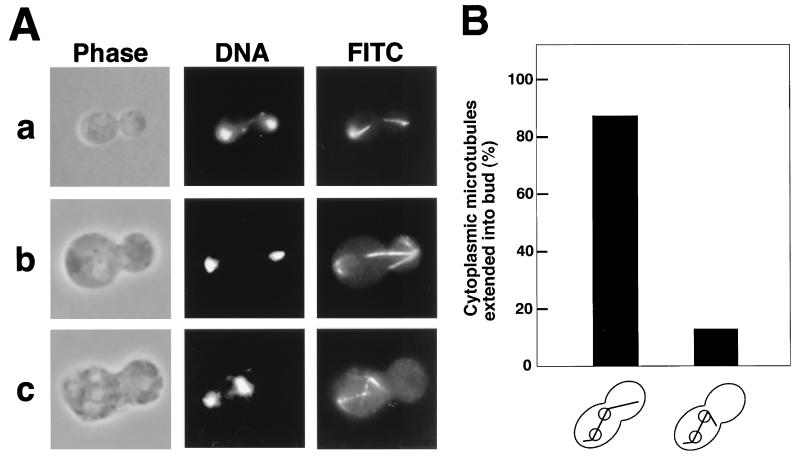
Disruption of BNI1 does not produce a strongly deleterious effect on cell growth, although the bni1 mutant grows slowly at higher temperatures (29). A diploid strain homozygous for the bni1 mutation is partially deficient in cytokinesis due to a wide bud neck and shows a random budding pattern (29, 69). Recently, evidence has emerged suggesting that there is a functional linkage between the actin cytoskeleton and cytoplasmic microtubules (33, 43). To investigate whether the bni1 mutation has any effect on the microtubule system, cells of the diploid bni1 mutant were grown in YPDAU at 30°C for 12 h. Because the diploid bni1 mutant shows a cytokinesis phenotype, demonstrated by, for example, cells with many unseparated large buds (29), we focused on cells with one large bud and a wide bud neck. We found that 22% showed this phenotype, and of the 503 cells counted that had this phenotype, 72% showed normal nuclear migration and microtubule orientation as in wild-type cells (Fig. 1A, panels a and b). In contrast, 28% of the cells showed a nuclear migration defect (panel c). In 141 cells with a nuclear migration defect, the frequency of the entry of cytoplasmic microtubules into the bud was measured (Fig. 1B). Of these cells, 87% showed cytoplasmic microtubules extending into the bud. These results suggest that Bni1p is involved in the regulation of microtubule-regulated nuclear migration but not in the entry of cytoplasmic microtubules into the bud.
FIG. 1.
Morphological phenotype of the bni1 mutant. Cells of diploid strains OHNY3 (wild type) and KY4 (bni1) were incubated at 30°C in YPDAU for 12 h, fixed, and doubly stained with DAPI and an anti-α-tubulin monoclonal antibody for DNA and cytoplasmic microtubules (FITC), respectively. All fields were photographed at the same magnification. (A) Nuclear migration defect in the bni1 mutant. Normal nuclear migration is seen in a wild-type cell (a) and a bni1 mutant cell (b). Nuclear migration defect is seen in a bni1 mutant cell (c). (B) Quantitation of cytoplasmic microtubules extended into the bud in cells of the bni1 mutant with nuclear migration defects.
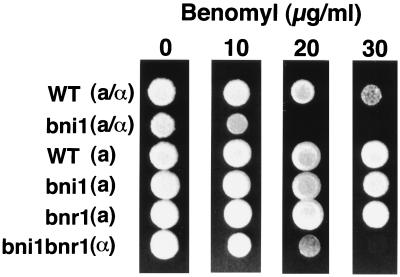
To investigate further whether microtubule function is compromised in cells of the diploid bni1 mutant, the sensitivity of the mutant to the microtubule-destabilizing drug benomyl was tested. Haploid bni1 and bnr1 mutants showed benomyl sensitivities similar to that of the wild type (Fig. 2). The haploid bni1 bnr1 mutant showed a slightly increased sensitivity to benomyl. Moreover, the diploid bni1 mutant showed more severe sensitivity to benomyl than did either the wild type or the haploid bni1 bnr1 mutant. These results suggest that BNI1 function is involved in the stability of microtubules, at least in diploids.
FIG. 2.
Increased benomyl sensitivity of the diploid bni1 mutant. Approximately 105 cells from diploid strains OHNY3 (wild type [WT]) and KY4 (bni1) and haploid strains OHNY1 (WT), BTY3 (bni1), HIY2 (bnr1), and HIY11 (bni1 bnr1) were plated in 3-μl spots on YPDAU containing 0, 10, 20, and 30 μg of benomyl per ml. The cells were incubated at 30°C for 3 days.
Identification of pac1 and nip100 as mutations that are synthetically lethal with a bni1 mutation.
To identify genes that interact with bni1, we screened for mutations that were lethal in combination with a bni1 mutation. A synthetic lethality screen by the color-sectoring assay (30) was set up with the haploid bni1 strain STFY1. A total of 14,000 colonies from UV-mutagenized cells (about 13% viability) were screened for the nonsectoring red phenotype at 24°C. Three nonsectoring bsl (bni1 synthetic lethal) mutants were recovered and crossed to a bni1 mutant, BTY3, to determine if they contained single mutations. All three mutants contained nonlinked single mutations (data not shown). To clone the corresponding genes, a genomic library was screened for plasmids that rescued the synthetic lethality. BSL1 and BSL2 were cloned and proved to be PAC1 and NIP100, respectively (Fig. 3). (see Materials and Methods).
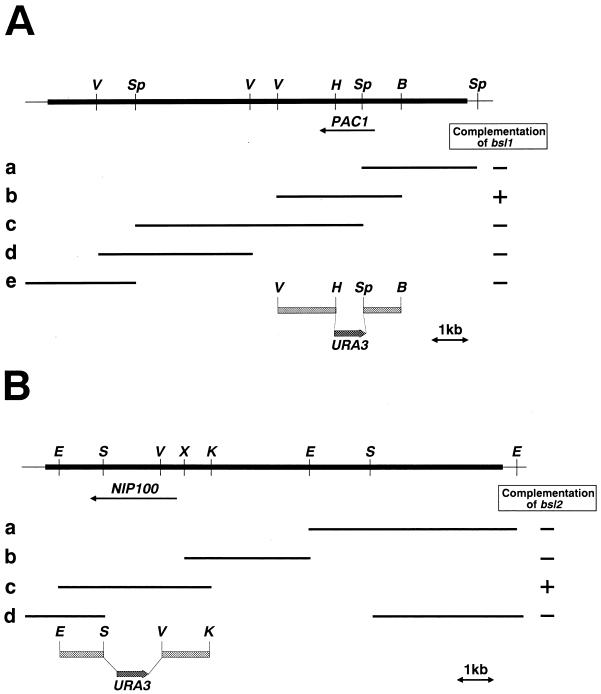
FIG. 3.
Restriction enzyme map and mapping of PAC1 and NIP100. (A) The bsl1 mutation lies in PAC1. The thick line represents the 9.6-kb original genomic DNA fragment, whereas the thin lines represent portions of the vector YCp50. B, BamHI; Sp, SphI; V, EcoRV; H, HpaI. The HpaI site is not unique in the insert DNA. Various fragments of the insert DNA were subcloned into YCp50 or pRS316 and tested for their ability to suppress the synthetic lethality. The complementing DNA was present in fragment b. (B) The bsl2 mutation lies in NIP100. The thick line represents the 14-kb original genomic DNA fragment, whereas the thin lines represent portions of the vector YCp50. E, EcoRI; S, SacI; V, EcoRV; X, XbaI; K, KpnI. The EcoRV, XbaI, and KpnI sites are not unique in the insert DNA. Various fragments of the insert DNA were subcloned into YCp50 or pRS316 and tested for their ability to suppress the synthetic lethality. The complementing fragment was present in fragment c. The constructs used to disrupt PAC1 and NIP100 are shown below the original genomic fragment in panels A and B, respectively.
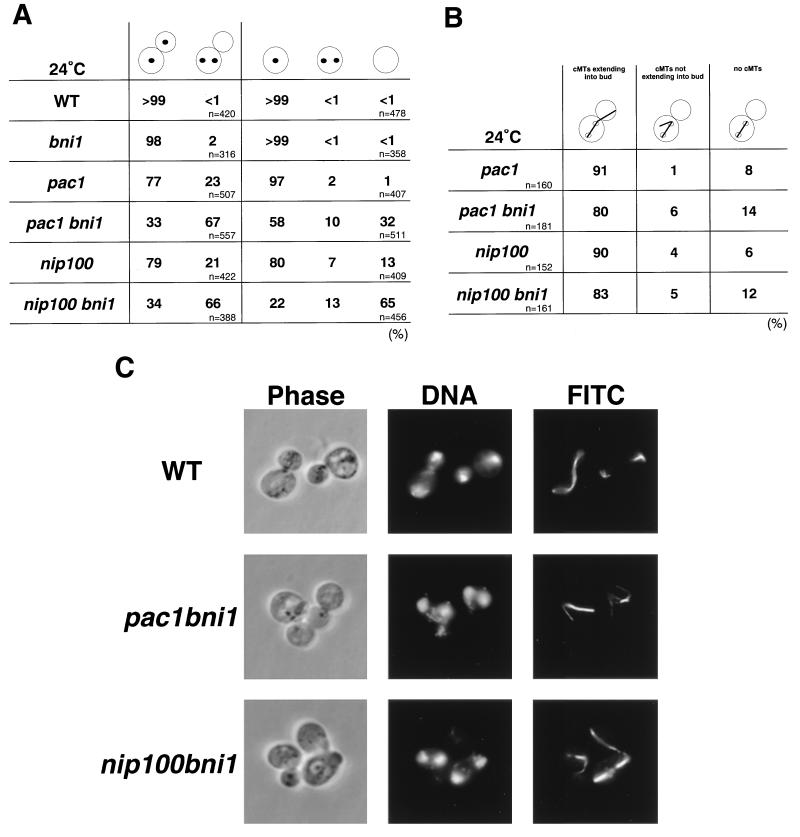
Disruption of either PAC1 or NIP100 causes a defect in nuclear migration that becomes particularly evident at lower temperatures (14, 26). This phenotype is similar to that of the dynein heavy-chain mutant dyn1 (10, 32). The pac1 bni1 and nip100 bni1 mutants were synthetically lethal (Table 3), and the phenotypes of both mutants were analyzed cytologically by using double-mutant strains that contained a plasmid with a GAL1 promoter-regulated BNI1 gene. Cells of the bni1, pac1, pac1 bni1, nip100, and nip100 bni1 mutants were grown in YPGalAU, transferred to YPDAU, and incubated for 13 h. Cells of the pac1 and nip100 mutants showed a binucleate phenotype in 23 and 21%, respectively, of the large-budded cells, whereas cells of the bni1 mutant showed this phenotype in only 2% of cases (Fig. 4A). Cells of both double mutants showed an increased proportion of binucleate cells (Fig. 4A and C). Cells of the pac1 bni1 and nip100 bni1 mutants showed a binucleate phenotype in 67 and 66%, respectively, of the large-budded cells. In the double mutants, cells with a cytokinesis defect, containing one large bud and a wide bud neck, were observed in less than 1% of the large-budded cells, indicating that the lethal phenotype was not due to an enhancement of the bni1 cytokinesis defect (data not shown). In binucleate cells of the pac1 bni1 and nip100 bni1 mutants, 80 and 83%, respectively, exhibited cytoplasmic microtubules that extended into the bud (Fig. 4B). These results indicate that BNI1 is involved in the regulation of microtubule function required for proper nuclear migration and that the PAC1 and NIP100 functions are related to that of BNI1.
TABLE 3.
Viability of mutants carrying mutations in combination with bni1 mutationa
| Genotypeb | No. of tetrads analyzed | No. of predicted mutants | % of lethal mutants | Viability of double mutant |
|---|---|---|---|---|
| pac1 bni1 | 19 | 23 | 100 | Synthetic lethal |
| nip100 bni1 | 20 | 26 | 100 | Synthetic lethal |
| arp1 bni1 | 29 | 22 | 100 | Synthetic lethal |
| cin8 bni1 | 25 | 26 | 19 | Viable |
PFIY1 (pac1::URA3), PFIY11 (nip100::URA3), AFIY1 (arp1::URA3), and CFIY1 (cin8::URA3) were crossed to BTY3 (bni1::HIS3), and the resulting diploids were subjected to tetrad analysis.
Genotypes of the inviable segregants were inferred from those of the viable segregants.
FIG. 4.
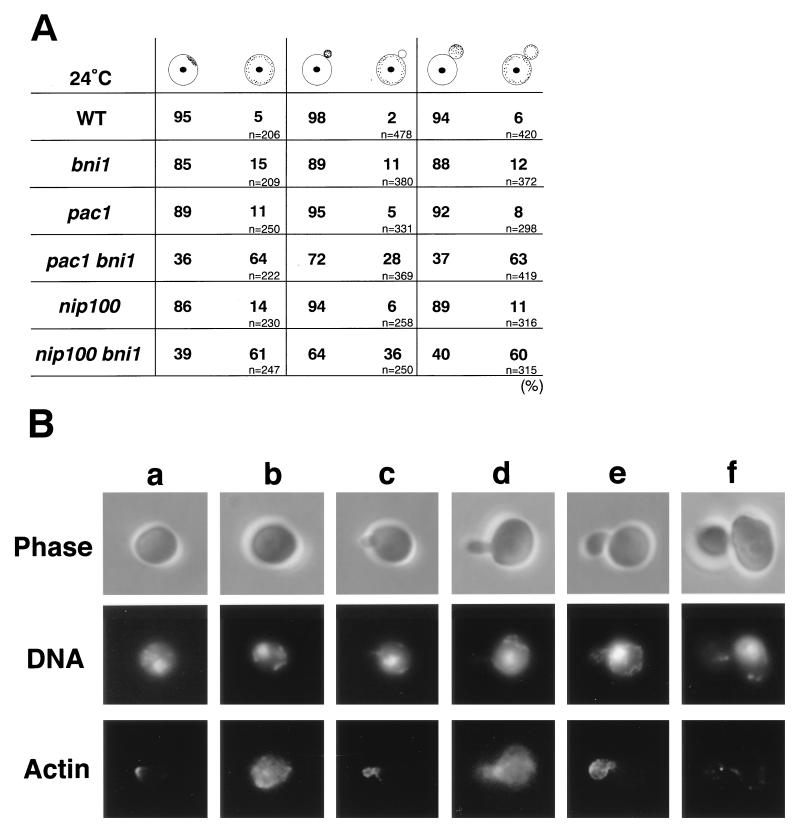
Morphological phenotypes of the pac1 bni1 and nip100 bni1 mutants. Cells of haploid strains OHNY1 (wild type [WT]), BTY3 (bni1), PFIY1 (pac1), PFIY5 (pac1 bni1), PFIY11 (nip100), and PFIY15 (nip100 bni1) were incubated at 24°C in YPDAU for 13 h, fixed, and doubly stained with DAPI and an anti-α-tubulin monoclonal antibody for DNA and cytoplasmic microtubules (FITC), respectively. (A) Quantitation of the nuclear migration defect. The percentage of cells with nuclear migration defects was determined in large-budded and unbudded cells. (B) Cytoplasmic microtubule orientation in cells with two nuclei within the mother cell of the pac1 bni1 and nip100 bni1 mutants. (C) Nuclear migration defect in large-budded cells of the pac1 bni1 and nip100 bni1 mutants. All fields were photographed at the same magnification.
In unbudded cells of the double mutants, the proportions of anucleate cells increased >30- and ∼5-fold, respectively, compared to the pac1 and nip100 single mutants (Fig. 4A). These results suggest that the binucleate cells in the pac1 bni1 and nip100 bni1 mutants go through cell division to produce anucleate daughter cells.
We investigated whether the genetic interactions between BNI1 and PAC1 or NIP100 are specific to the dynein system. Arp1p appears to be one of the components of the dynactin complex that operates in nuclear migration in cooperation with the dynein heavy chain (36, 40). CIN8 is a kinesin-related gene required for the separation of the spindle pole bodies (21, 47). The bni1 mutant BTY3 was crossed to the arp1 and cin8 mutants AFIY1 and CFIY1, respectively. Tetrad analysis indicated that the bni1 mutant is synthetically lethal with the arp1 mutant but not with the cin8 mutant (Table 3). We conclude that the synthetic lethal interaction between bni1 and pac1 or nip100 is specific to the dynein system.
Abnormal distribution of cortical actin patches in the pac1 bni1 and nip100 bni1 mutants.
We have shown previously that the diploid bni1 mutant has abnormal morphology and distribution of cortical actin patches (29). We examined the distribution of cortical actin patches in the pac1 bni1 and nip100 bni1 cells. Cells of the bni1, pac1, pac1 bni1, nip100, and nip100 bni1 mutants were grown in YPGalAU and shifted to YPDAU for 15 h. Of the cells that contained a single nucleus within the mother cell, only 5 to 11% of the pac1 and 6 to 14% of the nip100 single-mutant cells showed a nonpolarized localization of cortical actin patches in unbudded, tiny-budded, and small-budded cells, as also observed in the wild type and the bni1 single mutant (Fig. 5). In contrast, 64 and 63% of the pac1 bni1 and 61 and 60% of the nip100 bni1 double-mutant cells showed a nonpolarized distribution of cortical actin patches in unbudded and small-budded cells, respectively. We found that 28 and 36% of the tiny-budded cells in the pac1 bni1 and nip100 bni1 mutants, respectively, showed an abnormal actin distribution. We also observed the distribution of cortical actin patches in both double-mutant cells after 8.5 h of Bni1p depletion. More than 53% of unbudded and small-budded cells and more than 31% of tiny-budded cells showed a nonpolarized distribution of cortical actin patches (data not shown). These results suggest that the distribution of cortical actin patches is somewhat compromised before the nuclear migration defect becomes apparent in both the pac1 bni1 and nip100 bni1 mutants.
FIG. 5.
Abnormalities in the distribution of cortical actin patches in cells of the pac1 bni1 and nip100 bni1 mutants with one nucleus within the mother cell. Cells of haploid strains OHNY1 (wild type [WT]), BTY3 (bni1), PFIY1 (pac1), PFIY5 (pac1 bni1), PFIY11 (nip100), and PFIY15 (nip100 bni1) were incubated at 24°C in YPDAU for 15 h, fixed, and doubly stained with DAPI and rhodamine-phalloidin for DNA and actin, respectively. (A) Quantitation of defects in the distribution of cortical actin patches in unbudded, tiny-budded, and small-budded cells. (B) Distributions of actin patches in unbudded (a and b), tiny-budded (c and d), and small-budded (e and f) cells of the pac1 and pac1 bni1 mutants. a, c, e, the pac1 mutant; b, d, f, the pac1 bni1 mutant. All fields were photographed at the same magnification.
Involvement of actin function in PAC1-regulated nuclear migration.
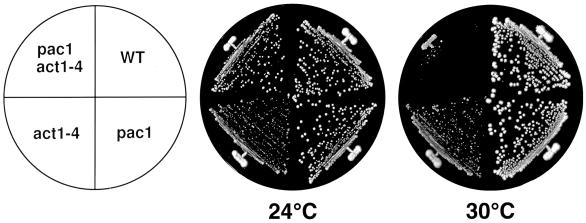
Previous studies have shown that disruption of actin function results in improper spindle orientation and nuclear migration defects (7, 43). However, the molecular mechanism of the linkage between nuclear migration and actin function has not yet been clarified. The synergistic effects of bni1 and pac1 and of bni1 and nip100 mutations on the polarized actin cytoskeleton suggested that Pac1p and Nip100p might be involved in this linkage. To investigate this possibility further, we used the act1-4 mutation, which has been shown previously to affect microtubule function (43). The act1-4 mutant AMFY1 was crossed with the pac1 mutant PFIY1 to construct the pac1 act1-4 mutant APFY1. Cells of this strain showed no detectable growth defect at 24°C but had a severe growth defect when shifted to 30°C (Fig. 6). The nip100 act1-4 mutant also had a severe growth defect when shifted to 30°C (data not shown). These results suggest that the actin function genetically interacts with the PAC1 and NIP100 functions.
FIG. 6.
The temperature-sensitive growth phenotype of the pac1 act1-4 mutant. Cells of haploid strains OHNY1 (wild type [WT]), PFIY1 (pac1), AMFY1 (act1-4), and APFY1 (pac1 act1-4) were streaked onto YPDAU plates, which were subsequently incubated at 24 or 30°C for 4 days.
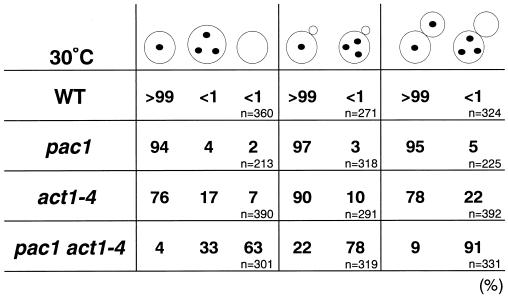
The phenotypes of the pac1 act1-4 mutant cells were observed after growth in YPDAU at 30°C for 13 h. In large-budded cells, 22 and 5% of the act1-4 and pac1 single mutants, respectively, contained three or more nuclei within the mother cell, and this phenotype was enhanced in the pac1 act1-4 double mutant to 91% of the large-budded cells (Fig. 7), a phenotype much more severe than that observed in the pac1 bni1 and nip100 bni1 mutants (Fig. 4A and C). Similarly, in tiny-budded cells, the proportion of cells with three nuclei within the mother cell increased to 78% in the double mutant compared to 10 and 3% in the act1-4 and pac1 single mutants, respectively. In unbudded cells, an increased proportion of anucleate cells was observed, consistent with the results for the pac1 bni1 and nip100 bni1 mutants. Enlarged multinucleate cells, a phenotype observed in the act1-4 mutant, also were twice as common in the pac1 act1-4 double mutant as in the act1-4 mutant. These results indicate that loss of actin function in the pac1 mutant causes a defect in nuclear migration that is similar to, but more severe than, that observed in the pac1 bni1 and nip100 bni1 mutants.
FIG. 7.
Morphological phenotypes of the pac1 act1-4 mutant. Cells of haploid strains OHNY1 (wild type [WT]), PFIY1 (pac1), AMFY1 (act1-4), and APFY1 (pac1 act1-4) were incubated at 30°C in YPDAU for 13 h, fixed, and stained with DAPI for DNA. The percentages of cells with nuclear migration defects were determined in unbudded, tiny-budded, and large-budded cells.
Determination of the region in Bni1p required for suppression of the pac1 bni1 lethality.
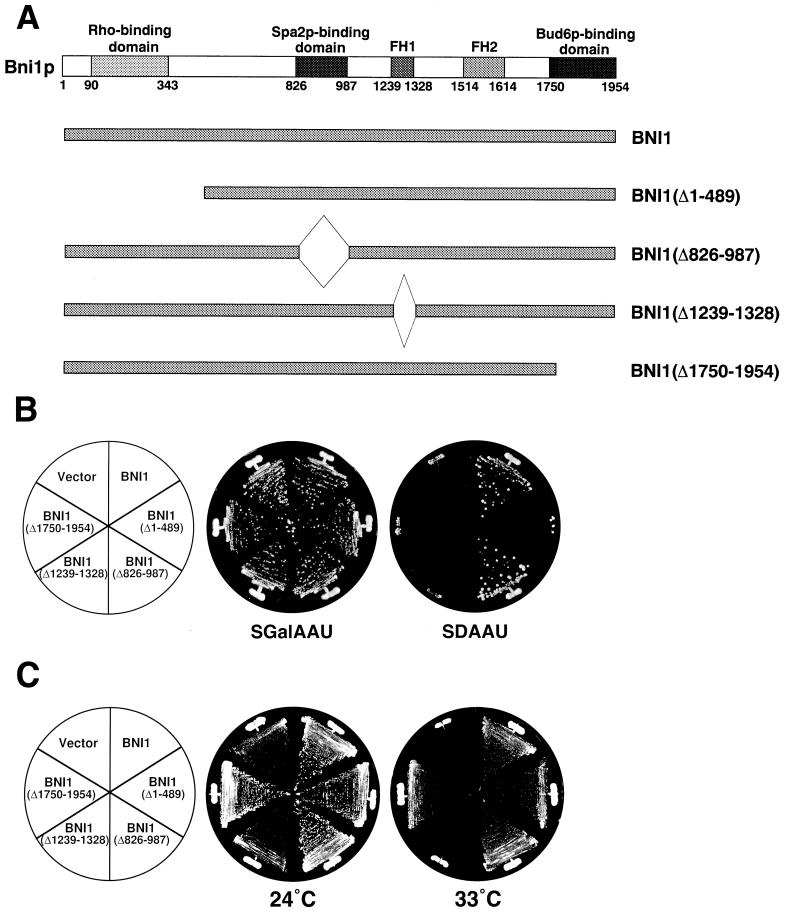
We constructed several deletion mutants of myc-tagged Bni1p to identify the Bni1p-binding protein required for the microtubule-related Bni1p function. Bni1p mutants lacking the Rho- (11, 23), Spa2p- (13), profilin- (24), or Bud6p (11)-binding region were constructed to be expressed under the control of the BNI1 promoter (Fig. 8A). These mutant proteins did not interact with the corresponding partner proteins as judged by the two-hybrid method (data not shown). The expression of these mutant proteins was confirmed by Western blot analysis with an anti-myc monoclonal antibody (data not shown). The lethality of the pac1 bni1 mutation was suppressed in the Bni1p mutant lacking the Spa2p-binding region but not in those lacking the Rho-, profilin-, or Bud6p-binding region (Fig. 8B). On the other hand, we also tested whether these mutations could suppress the temperature-sensitive growth defect of the bni1 bnr1 mutant which shows deficiency in the actin cytoskeleton (24). The Bni1p mutant lacking the Rho-, Spa2p-, or Bud6p-binding region showed suppression of the growth defect at 33°C, but the mutant lacking the profilin-binding region did not (Fig. 8C). We furthermore investigated genetically the possible requirement for the Bni1p-Bud6p and Bni1p-Spa2p interactions in nuclear migration. Neither the bud6 nor the spa2 mutation was synthetically lethal with the pac1 mutation (data not shown). These results suggest that the interactions of Bni1p with the Rho family members and profilin may be required for the microtubule-related functions of Bni1p.
FIG. 8.
Suppression of the synthetic lethality of the pac1 bni1 double mutation by BNI1 (Δ826–987) but not by BNI1 (Δ1–489), BNI1 (Δ1239–1328), or BNI1 (Δ1750–1954). (A) Structures of the deletion mutants of Bni1p. (B) Cells of strain PFIY6 (pac1 bni1) transformed with pRS314-PBNI1-myc-BNI1, pRS314-PBNI1-myc-BNI1 (490–1954), pRS314-PBNI1-myc-BNI1′-1, pRS314-PBNI1-myc-BNI1′-2, pRS314-PBNI1-myc-BNI1′-3, or the pRS314-PBNI1-myc vector were streaked onto SGalAAU and SDAAU plates and incubated at 30°C for 4 days. (C) Cells of strain HIY11 (bni1 bnr1) transformed with plasmids described in panel B were streaked onto SDAAU plates and incubated at 24 or 33°C for 3 days.
DISCUSSION
In this study we have provided evidence that a diploid bni1 mutant shows a nuclear migration defect. Because nuclear migration is known to be regulated by cytoplasmic microtubules (22, 56), our results thus suggest that Bni1p, known to be involved in cytokinesis and the establishment of cell polarity (69), also serves as a regulator of microtubule function. In Drosophila, the genes which encode proteins related to BNI1, cappuccino and diaphanous, are implicated in the regulation of microtubules (8, 15), suggesting a conserved role for proteins containing the FH1 and FH2 domains in microtubule function.
To clarify the molecular mechanism of how Bni1p regulates the microtubule system, we screened for mutations that resulted in lethality in combination with a bni1 mutation. This identified two genes, PAC1 and NIP100, which are involved in dynein function (10, 14, 26, 32). Dynein plays an important role in nuclear migration, probably by pulling the spindle pole body into the bud via cytoplasmic microtubules (68). Our results indicate that the inviability of the pac1 bni1 and nip100 bni1 double mutants is mainly due to the reduced function of cytoplasmic microtubules. This phenotype is consistent with the results observed with the diploid bni1 mutant.
Cytoplasmic microtubules in yeast are elongated from the spindle pole body and are oriented toward the site of bud growth (50, 55). It has recently been reported that the kar9 mutation is synthetically lethal with the dyn1 mutation (37). In the kar9 dyn1 mutant, cytoplasmic microtubules are misoriented, resulting in the binucleate phenotype. Kar9p is a novel protein implicated in the regulation of microtubule orientation and is localized at the bud tip, as is Bni1p. Kar9p may serve as a cortical marker to which plus ends of cytoplasmic microtubules are oriented. The present results, together with the fact that Bni1p is localized to the bud tip (13, 25), strongly suggest that Bni1p is another marker for cytoplasmic microtubules at the bud tip. The functional and physical interactions between Kar9p and Bni1p in regulating microtubule orientation to the bud tip should be investigated in future studies.
The molecular mechanism of spindle positioning and its functional relationship with cytoplasmic microtubules are not thoroughly understood (3, 6, 49, 50, 68). Recently, two kinesin-related proteins, Kip2p and Kip3p, were shown to be involved in nuclear migration (39). The kip3 mutation shows a synthetic lethal interaction with the dyn1 mutation, suggesting that Kip3p and dynein function in a redundant pathway. On the other hand, Kip2p has been suggested to function partially in the dynein pathway (39). It is important to clarify whether Bni1p functionally interacts with Kip3p and regulates spindle positioning. During the preparation of this paper, Miller et al. (38) showed that the cortical localization of Kar9p is strongly dependent on the actin cytoskeleton and on Bni1p and Lee et al. (31) showed that a bni1 mutation results in abnormal spindle positioning and suggested that Bni1p functions in the kinesin pathway. These findings are consistent with each other and with our present results. Thus, it can be concluded from these three different series of experiments that Bni1p, Kar9p, and Kip3p function in the same pathway and genetically interact with the dynein function.
Bni1p is a downstream target molecule of the Rho family members and regulates the reorganization of the actin cytoskeleton (11, 29). Therefore, it is possible that Bni1p regulates cytoplasmic microtubules through the actin cytoskeleton. Bni1p interacts with at least three actin-binding proteins, profilin, Bud6p, and EF1α (11, 24, 60). Genetic and functional analyses of Bni1p suggest that its interaction with profilin is important for the regulation of microtubule function. The findings that binucleate large-budded cells mostly showed depolarized cortical actin patches and that the act1-4 mutation strengthened the pac1 and nip100 phenotypes are also consistent with the idea that Bni1p regulates cytoplasmic microtubules through the actin cytoskeleton. In contrast, the Bni1p-Bud6p interaction does not seem to be important for the regulation of microtubule function. This result suggests that there exists a Bni1p-binding protein other than Bud6p that is required for the regulation of cytoplasmic microtubules. Bni1p may interact with an unknown protein involved in the regulation of cytoplasmic microtubules at the Bud6p-bindindg region. A rat dynactin component, p150Glued, binds both microtubules and the actin-related protein centractin (64). This result suggests that Nip100p might directly interact with some kind of actin-related protein which is under the regulation of Bni1p and that this might lead to the functional relationship between cytoplasmic microtubules and the actin cytoskeleton. NUDF, an Aspergillus nidulans homolog of Pac1p, has been suggested to affect nuclear migration by acting on the dynein motor system (65). From this result, Pac1p may function upstream of the dynein-dynactin system and functionally interact with the Bni1p-regulated actin cytoskeleton. Recently, a yeast homolog of Coronin, Crn1p, has been isolated by microtubule affinity chromatography (17). Crn1p has actin filament-bundling and microtubule-binding activities. Crn1p, or a protein with similar properties, may functionally interact with Pac1p, Nip100p, and/or Bni1p and link cytoplasmic microtubules with the actin cytoskeleton. EF1α possesses a microtubule-severing activity (53). The Bni1p-EF1α interaction also may be involved in the regulation of cytoplasmic microtubules.
Analysis of the functional domains of Bni1p furthermore suggests that the Rho1p-Bni1p interaction is involved in microtubule function. It has previously been shown that Rom2p, a GDP/GTP exchange factor specific for Rho1p and Rho2p (35, 42), and Bem2p, a GTPase-activating protein specific for Rho1p (44), are involved in the regulation of the microtubule system (35, 61). Therefore, the Rho1p-Bni1p interaction may be involved in microtubule function. We examined whether the pac1 and nip100 mutations enhance the phenotypes of the rho1-104, rho1 (F44Y), and rho1 (E45I) mutations (41, 67). However, no enhanced phenotypes were observed (data not shown). Other Rho family members which interact with Bni1p, such as Cdc42p, Rho3p, or Rho4p (11, 23), may play a role in the Bni1p-regulated microtubule function.
In mammalian cells, mDia has been isolated as a mammalian counterpart of Bni1p and/or Bnr1p and has been shown to be a downstream target molecule of RhoA (63), a mammalian homolog of Rho1p (67). It has been shown that the activation of Rho causes stabilization of microtubules in starved cells (5) and that microtubules are targeted and stabilized at focal adhesion sites where reorganization of the actin cytoskeleton is spatially regulated (27). By analogy to our present studies, mDia may also be involved in the regulation of the microtubule system in mammalian cells.
ACKNOWLEDGMENTS
We thank Y. Ohya for the yeast strains DBY2326 and DJTDZ16A.
This investigation was supported by grants-in-aid for Scientific Research and for Cancer Research from the Ministry of Education, Science, Sports, and Culture, Japan (1998), and by grants from the Human Frontier Science Program (1998).
REFERENCES
- 1.Amberg D C, Zahner J E, Mulholland J W, Pringle J R, Bostein D. Aip3/Bud6, a yeast actin-interacting protein that is involved in morphogenesis and the selection of bipolar budding sites. Mol Biol Cell. 1997;8:729–753. doi: 10.1091/mbc.8.4.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bershadsky A, Chausovsky A, Becker E, Lyubimova A, Geiger B. Involvement of microtubules in the control of adhesion-dependent signal transduction. Curr Biol. 1996;6:1279–1289. doi: 10.1016/s0960-9822(02)70714-8. [DOI] [PubMed] [Google Scholar]
- 3.Carminati J L, Stearns T. Microtubules orient the mitotic spindle in yeast through dynein-dependent interactions with the cell cortex. J Cell Biol. 1997;138:629–641. doi: 10.1083/jcb.138.3.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cid V J, Durán A, del Ray F, Snyder M P, Nombela C, Sánchez M. Molecular basis of cell integrity and morphogenesis in Saccharomyces cerevisiae. Microbiol Rev. 1995;59:345–386. doi: 10.1128/mr.59.3.345-386.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cook T A, Nagasaki T, Gundersen G G. Rho guanosine triphosphatase mediates the selective stabilization of microtubules induced by lysophosphatidic acid. J Cell Biol. 1998;141:175–185. doi: 10.1083/jcb.141.1.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.DeZwaan T M, Ellingson E, Pellman D, Roof D M. Kinesin-related KIP3 of Saccharomyces cerevisiae is required for a distinct step in nuclear migration. J Cell Biol. 1997;138:1023–1040. doi: 10.1083/jcb.138.5.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Drubin D G, Jones H D, Wertman K F. Actin structure and function: roles in mitochondrial organization and morphogenesis in budding yeast and identification of the phalloidin-binding site. Mol Biol Cell. 1993;4:1277–1294. doi: 10.1091/mbc.4.12.1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Emmons S, Phan H, Calley J, Chen W, James B, Manseau L. cappucino, a Drosophila maternal effect gene required for polarity of the egg and embryo, is related to the vertebrate limb deformity locus. Genes Dev. 1995;9:2482–2494. doi: 10.1101/gad.9.20.2482. [DOI] [PubMed] [Google Scholar]
- 9.Enomoto T. Microtubule disruption induces the formation of actin stress fibers and focal adhesions in cultured cells: possible involvement of the rho signal cascade. Cell Struct Funct. 1996;21:317–326. doi: 10.1247/csf.21.317. [DOI] [PubMed] [Google Scholar]
- 10.Eshel D, Urrestarazu L A, Vissers S, Jauniaux J C, van Vliet-Reedijk J C, Planta R J, Gibbons I R. Cytoplasmic dynein is required for normal nuclear segregation in yeast. Proc Natl Acad Sci USA. 1993;90:11172–11176. doi: 10.1073/pnas.90.23.11172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Evangelista M, Blundell K, Longtine M S, Chow C J, Adames N, Pringle J R, Peter M, Boone C. Bni1p, a yeast formin linking Cdc42p and the actin cytoskeleton during polarized morphogenesis. Science. 1997;276:118–122. doi: 10.1126/science.276.5309.118. [DOI] [PubMed] [Google Scholar]
- 12.Frazier J A, Field C M. Actin cytoskeleton: are FH proteins local organizers? Curr Biol. 1997;7:R414–R417. doi: 10.1016/s0960-9822(06)00205-3. [DOI] [PubMed] [Google Scholar]
- 13.Fujiwara T, Tanaka K, Mino A, Kikyo M, Takahashi K, Shimizu K, Takai Y. Rho1p-Bni1p-Spa2p interactions: implication in localization of Bni1p at the bud site and regulation of the actin cytoskeleton in Saccharomyces cerevisiae. Mol Biol Cell. 1998;9:1221–1233. doi: 10.1091/mbc.9.5.1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Geiser J R, Schott E J, Kingsbury T J, Cole N B, Totis L J, Bhattacharyya G, He L, Hoyt M A. Saccharomyces cerevisiae genes required in the absence of the CIN8 encoded spindle motor act in functionally diverse mitotic pathways. Mol Biol Cell. 1997;8:1035–1050. doi: 10.1091/mbc.8.6.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Giansanti M G, Bonaccorsi S, Williams B, Williams E L, Santolamazza C, Goldberg M L, Gatti M. Cooperative interactions between the central spindle and the contractile ring during Drosophila cytokinesis. Genes Dev. 1998;12:396–410. doi: 10.1101/gad.12.3.396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gietz D, Jean A S, Woods R A, Schiestl R H. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 1992;20:1425. doi: 10.1093/nar/20.6.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goode B L, Wong J J, Butty A C, Peter M, McCormack A L, Yates J R, Drubin D G, Barnes G. Coronin promotes the rapid assembly and cross-linking of actin filaments and may link the actin and microtubule cytoskeletons in yeast. J Cell Biol. 1999;144:83–98. doi: 10.1083/jcb.144.1.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hall A. Small GTP-binding proteins and the regulation of the actin cytoskeleton. Annu Rev Cell Biol. 1994;10:31–54. doi: 10.1146/annurev.cb.10.110194.000335. [DOI] [PubMed] [Google Scholar]
- 19.Hall A. Rho GTPases and the actin cytoskeleton. Science. 1998;279:509–514. doi: 10.1126/science.279.5350.509. [DOI] [PubMed] [Google Scholar]
- 20.Hill J E, Myers A M, Koerner T J, Tzagoloff A. Yeast/E. coli shuttle vectors with multiple unique restriction sites. Yeast. 1986;2:163–167. doi: 10.1002/yea.320020304. [DOI] [PubMed] [Google Scholar]
- 21.Hoyt M A, He L, Loo K K, Saunders W S. Two Saccharomyces cerevisiae kinesin-related gene products required for mitotic spindle assembly. J Cell Biol. 1992;118:109–120. doi: 10.1083/jcb.118.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huffaker T C, Thomas J H, Botstein D. Diverse effects of β-tubulin mutations in microtubule formation and function. J Cell Biol. 1988;106:1997–2010. doi: 10.1083/jcb.106.6.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Imamura, H., and Y. Takai. Unpublished data.
- 24.Imamura H, Tanaka K, Hihara T, Umikawa M, Kamei T, Takahashi K, Sasaki T, Takai Y. Bni1p and Bnr1p: downstream targets of the Rho family small G-proteins which interact with profilin and regulate actin cytoskeleton in Saccharomyces cerevisiae. EMBO J. 1997;16:2745–2755. doi: 10.1093/emboj/16.10.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jansen R-P, Dowzer C, Michaelis C, Galova M, Nasmyth K. Mother cell-specific HO expression in budding yeast depends on the unconventional myosin Myo4p and other cytoplasmic proteins. Cell. 1996;84:687–697. doi: 10.1016/s0092-8674(00)81047-8. [DOI] [PubMed] [Google Scholar]
- 26.Kahana J A, Schlenstedt G, Evanchuk D M, Geiser J R, Hoyt M A, Silver P A. The yeast dynactin complex is involved in partitioning the mitotic spindle between mother and daughter cells during anaphase B. Mol Biol Cell. 1998;9:1741–1756. doi: 10.1091/mbc.9.7.1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kaverina I, Rottner K, Small J V. Targeting, capture, and stabilization of microtubules at early focal adhesions. J Cell Biol. 1998;142:181–190. doi: 10.1083/jcb.142.1.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kikyo, M., K. Tanaka, T. Kamei, K. Ozaki, T. Fujiwara, E. Inoue, Y. Takita, Y. Ohya, and Y. Takai. An FH domain-containing Bnr1p is a multifunctional protein interacting with a variety of cytoskeletal proteins in Saccharomyces cerevisiae. Oncogene, in press. [DOI] [PubMed]
- 29.Kohno H, Tanaka K, Mino A, Umikawa M, Imamura H, Fujiwara T, Fujita Y, Hotta K, Qadota H, Watanabe T, Ohya Y, Takai Y. Bni1p implicated in cytoskeletal control is a putative target of Rho1p small GTP-binding protein in Saccharomyces cerevisiae. EMBO J. 1996;15:6060–6068. [PMC free article] [PubMed] [Google Scholar]
- 30.Koshland D, Kent J C, Hartwell L H. Genetic analysis of the mitotic transmission of minichromosomes. Cell. 1985;40:393–403. doi: 10.1016/0092-8674(85)90153-9. [DOI] [PubMed] [Google Scholar]
- 31.Lee L, Saskia K K, Evangelista M, Boone C, Pellman D. Control of mitotic spindle position by the Saccharomyces cerevisiae formin Bni1p. J Cell Biol. 1999;144:947–961. doi: 10.1083/jcb.144.5.947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li Y Y, Yeh E, Hays T, Bloom K. Disruption of mitotic spindle orientation in a yeast dynein mutant. Proc Natl Acad Sci USA. 1993;90:10096–10100. doi: 10.1073/pnas.90.21.10096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lillie S H, Brown S S. Suppression of a myosin defect by a kinesin-related gene. Nature. 1992;356:358–361. doi: 10.1038/356358a0. [DOI] [PubMed] [Google Scholar]
- 34.Lloyd C W, Smith C G, Woods A, Rees D A. Mechanism of cellular adhesion. II. The interplay between adhesion, the cytoskeleton and morphology in substrate-attached cells. Exp Cell Res. 1977;110:427–437. doi: 10.1016/0014-4827(77)90309-3. [DOI] [PubMed] [Google Scholar]
- 35.Manning B D, Padmanabha R, Snyder M. The Rho-GEF Rom2p localizes to sites of polarized cell growth and participates in cytoskeletal functions in Saccharomyces cerevisiae. Mol Biol Cell. 1997;8:1829–1844. doi: 10.1091/mbc.8.10.1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McMillan J N, Tatchell K. The JNM1 gene in the yeast Saccharomyces cerevisiae is required for nuclear migration and spindle orientation during the mitotic cell cycle. J Cell Biol. 1994;125:143–158. doi: 10.1083/jcb.125.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Miller R K, Rose M D. Kar9p is a novel cortical protein required for cytoplasmic microtubule orientation in yeast. J Cell Biol. 1998;140:377–390. doi: 10.1083/jcb.140.2.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miller R K, Matheos D, Rose M D. The cortical localization of the microtubule orientation protein, Kar9p, is dependent upon actin and proteins required for polarization. J Cell Biol. 1999;144:963–975. doi: 10.1083/jcb.144.5.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miller R K, Heller K K, Frisen L, Wallack D L, Loayza D, Gammie A E, Rose M D. The kinesin-related proteins, Kip2p and Kip3p, function differently in nuclear migration in yeast. Mol Biol Cell. 1998;9:2051–2068. doi: 10.1091/mbc.9.8.2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Muhua L, Karpova T S, Cooper J A. A yeast actin-related protein homologous to that in vertebrate dynactin complex is important for spindle orientation and nuclear migration. Cell. 1994;78:669–679. doi: 10.1016/0092-8674(94)90531-2. [DOI] [PubMed] [Google Scholar]
- 41.Nonaka H, Tanaka K, Hirano H, Fujiwara T, Kohno H, Umikawa M, Mino A, Takai Y. A downstream target of RHO1 small GTP-binding protein is PKC1, a homolog of protein kinase C, which leads to activation of the MAP kinase cascade in Saccharomyces cerevisiae. EMBO J. 1995;14:5931–5938. doi: 10.1002/j.1460-2075.1995.tb00281.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ozaki K, Tanaka K, Imamura H, Hihara T, Kameyama T, Nonaka H, Hirano H, Matsurra Y, Takai Y. Rom1p and Rom2p are GDP/GTP exchange proteins (GEPs) for the Rho1p small GTP binding protein in Saccharomyces cerevisiae. EMBO J. 1996;15:2196–2207. [PMC free article] [PubMed] [Google Scholar]
- 43.Palmer R E, Sullivan D S, Huffaker T, Koshland D. Role of astral microtubules and actin in spindle orientation and migration in the budding yeast, Saccharomyces cerevisiae. J Cell Biol. 1992;119:583–593. doi: 10.1083/jcb.119.3.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Peterson J, Zheng Y, Bender L, Myers A, Cerione R, Bender A. Interactions between the bud emergence proteins Bem1p and Bem2p and Rho-type GTPases in yeast. J Cell Biol. 1994;127:1395–1406. doi: 10.1083/jcb.127.5.1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Reiner O, Carrozzo R, Shen Y, Wehnert M, Faustinella F, Dobyns W B, Caskey C T, Ledbetter D H. Isolation of a Miller-Dieker lissencephaly gene containing G protein β-subunit-like repeats. Nature. 1993;364:717–721. doi: 10.1038/364717a0. [DOI] [PubMed] [Google Scholar]
- 46.Riehemann K, Sorg C. Sequence homologies between four cytoskeleton-associated proteins. Trends Biochem Sci. 1993;18:82–83. doi: 10.1016/0968-0004(93)90159-k. [DOI] [PubMed] [Google Scholar]
- 47.Roof D M, Meluh P B, Rose M D. Kinesin-related proteins required for assembly of the mitotic spindle. J Cell Biol. 1992;118:95–108. doi: 10.1083/jcb.118.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 49.Shaw S L, Maddox P, Skibbens R V, Yeh E, Salmon E D, Bloom K. Nuclear and spindle dynamics in budding yeast. Mol Biol Cell. 1998;9:1627–1631. doi: 10.1091/mbc.9.7.1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shaw S L, Yeh E, Maddox P, Salmon E D, Bloom K. Astral microtubule dynamics in yeast: a microtubule-based searching mechanism for spindle orientation and nuclear migration into the bud. J Cell Biol. 1997;139:985–994. doi: 10.1083/jcb.139.4.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sherman F. Getting started with yeast. Methods Enzymol. 1991;194:3–21. doi: 10.1016/0076-6879(91)94004-v. [DOI] [PubMed] [Google Scholar]
- 52.Sherman F, Fink G R, Hicks J B. Methods in yeast genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1986. [Google Scholar]
- 53.Shiina N, Gotoh Y, Kubomura N, Iwamatsu A, Nishida E. Microtubule severing by elongation factor 1α. Science. 1994;266:282–285. doi: 10.1126/science.7939665. [DOI] [PubMed] [Google Scholar]
- 54.Sikorski R S, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Snyder M, Gehrung S, Page B D. Studies concerning the temporal and genetic control of cell polarity in Saccharomyces cerevisiae. J Cell Biol. 1991;114:515–532. doi: 10.1083/jcb.114.3.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sullivan D S, Huffaker T C. Astral microtubules are not required for anaphase B in Saccharomyces cerevisiae. J Cell Biol. 1992;119:379–388. doi: 10.1083/jcb.119.2.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Takai Y, Sasaki T, Tanaka K, Nakanishi H. Rho as a regulator of the cytoskeleton. Trends Biochem Sci. 1995;20:227–231. doi: 10.1016/s0968-0004(00)89022-2. [DOI] [PubMed] [Google Scholar]
- 58.Tanaka K, Takai Y. Control of reorganization of the actin cytoskeleton by Rho family small GTP-binding proteins in yeast. Curr Opin Cell Biol. 1998;10:112–116. doi: 10.1016/s0955-0674(98)80093-8. [DOI] [PubMed] [Google Scholar]
- 59.Tinsley J H, Minke P F, Bruno K S, Plamann M. P150Glued, the largest subunit of the dynactin complex, is nonessential in Neurospora but required for nuclear distribution. Mol Biol Cell. 1996;7:731–742. doi: 10.1091/mbc.7.5.731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Umikawa M, Tanaka K, Kamei T, Shimizu K, Imamura H, Sasaki T, Takai Y. Interaction of Rho1p target Bni1p with F-actin-binding elongation factor 1α: implication in Rho1p-regulated reorganization of the actin cytoskeleton in Saccharomyces cerevisiae. Oncogene. 1998;16:2011–2016. doi: 10.1038/sj.onc.1201724. [DOI] [PubMed] [Google Scholar]
- 61.Wang T, Bretscher A. The rho-GAP encoded by BEM2 regulates cytoskeletal structure in budding yeast. Mol Biol Cell. 1995;6:1011–1024. doi: 10.1091/mbc.6.8.1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wasserman S. FH proteins as cytoskeletal organizers. Trends Cell Biol. 1998;8:111–115. doi: 10.1016/s0962-8924(97)01217-8. [DOI] [PubMed] [Google Scholar]
- 63.Watanabe N, Madaule P, Reid T, Ishizaki T, Watanabe G, Kakizuka A, Saito Y, Nakao K, Jockusch B M, Narumia S. p140mDia, a mammalian homolog of Drosophila diaphanous, is a target protein for Rho small GTPase and is a ligand for profilin. EMBO J. 1997;16:3044–3056. doi: 10.1093/emboj/16.11.3044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Waterman-Storer C M, Karki S, Holzbaur E L. The p150Glued component of the dynactin complex binds to both microtubules and the actin-related protein centractin (Arp1) Proc Natl Acad Sci USA. 1995;92:1634–1638. doi: 10.1073/pnas.92.5.1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Willins D A, Liu B, Xiang X, Morris N R. Mutations in the heavy chain of cytoplasmic dynein suppress the nudF nuclear migration mutation in Aspergillus nidulans. Mol Gen Genet. 1997;255:194–200. doi: 10.1007/s004380050489. [DOI] [PubMed] [Google Scholar]
- 66.Xiang X, Osmani A H, Osmani S A, Xin M, Morris N R. NudF, a nuclear migration gene in Aspergillus nidulans, is similar to the human LIS-1 gene required for neuronal migration. Mol Biol Cell. 1995;6:297–310. doi: 10.1091/mbc.6.3.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yamochi W, Tanaka K, Nonaka H, Maeda A, Musha T, Takai Y. Growth site localization of Rho1 small GTP-binding protein and its involvement in bud formation in Saccharomyces cerevisiae. J Cell Biol. 1994;125:1077–1093. doi: 10.1083/jcb.125.5.1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yeh E, Skibbens R V, Cheng J W, Salmon E D, Bloom K. Spindle dynamics and cell cycle regulation of dynein in the budding yeast, Saccharomyces cerevisiae. J Cell Biol. 1995;130:687–700. doi: 10.1083/jcb.130.3.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zahner J E, Harkins H A, Pringle J R. Genetic analysis of the bipolar pattern of bud sites selection in the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:1857–1870. doi: 10.1128/mcb.16.4.1857. [DOI] [PMC free article] [PubMed] [Google Scholar]