Abstract
Sentrin/small ubiquitin-like modifier (SUMO)-specific protease 2 (SENP2)-deficient mice develop spontaneous seizures in early life because of a marked reduction in M currents, which regulate neuronal membrane excitability. We have previously shown that hyper-SUMOylation of the Kv7.2 and Kv7.3 channels is critically involved in the regulation of the M currents conducted by these potassium voltage-gated channels. Here, we show that hyper-SUMOylation of the Kv7.2 and Kv7.3 proteins reduced binding to the lipid secondary messenger PIP2. CaM1 has been shown to be tethered to the Kv7 subunits via hydrophobic motifs in its C termini and implicated in the channel assembly. Mutation of the SUMOylation sites on Kv7.2 and Kv7.3 specifically resulted in decreased binding to CaM1 and enhanced CaM1-mediated assembly of Kv7.2 and Kv7.3, whereas hyper-SUMOylation of Kv7.2 and Kv7.3 inhibited channel assembly. SENP2-deficient mice exhibited increased acetylcholine levels in the brain and the heart tissue because of increases in the vagal tone induced by recurrent seizures. The SENP2-deficient mice develop seizures followed by a period of sinus pauses or atrioventricular conduction blocks. Chronic administration of the parasympathetic blocker atropine or unilateral vagotomy significantly prolonged the life of the SENP2-deficient mice. Furthermore, we showed that retigabine, an M-current opener, reduced the transcription of SUMO-activating enzyme SAE1 and inhibited SUMOylation of the Kv7.2 and Kv7.3 channels, and also prolonged the life of SENP2-deficient mice. Taken together, the previously demonstrated roles of PIP2, CaM1, and retigabine on the regulation of Kv7.2 and Kv7.3 channel function can be explained by their roles in regulating SUMOylation of this critical potassium channel.
Keywords: SENP2, Kv7, PIP2, CaM1, retigabine
Abbreviations: AV, atrioventricular; CaM, calmodulin; co-IP, coimmunoprecipitation; PA, phosphatidic acid; PE, phosphatidylethanolamine; PIP2, phosphatidylinositol 4,5-bisphosphate; PS, phosphatidylserine; SENP2, sentrin/SUMO-specific protease 2; SUDEP, sudden unexplained death in epilepsy; SUMO, small ubiquitin-like modifier
Potassium channels located in the heart and brain were the first to be linked to sudden unexplained death in epilepsy (SUDEP) (1, 2), the most common cause of epilepsy-related death (3). SUDEP occurs in many patients at a relatively young age and accounts for a disproportionate number of lives lost (4). Isolating the underlying genes and mechanisms for this potentially preventable form of epilepsy mortality is essential to detect and treat those at high risk of SUDEP (5). Our previous studies showed that hyper-SUMOylation of the Kv7 potassium channel diminishes the M current and causes seizures and sudden death (6); however, deciphering the molecular mechanism has remained elusive.
The Kv7.2 and Kv7.3 subunits assemble to form a heterotetrameric channel that mediates the M current, and the C terminus is critical for the assembly, gating, and trafficking of Kv7.2 and Kv7.3 (7, 8). Calmodulin (CaM) is critical modulator that regulates the activation of Kv7 channels (9). It was reported that mutations in the CaM-binding domain of Kv7.2 interfered with phosphatidylinositol 4,5-bisphosphate (PIP2) activation of the channel (10), indicating that CaM might be competitive with PIP2. Mutations in the KCNQ2 and KCNQ3 genes account for epileptic encephalopathies and benign familial neonatal seizures. Some of these mutations have been shown to affect the binding of CaM to specific C-terminal motifs of KCNQ2 and KCNQ3 subunits, including the A and B helices (11). CaM has been shown to be tethered to the Kv7 subunits via hydrophobic motifs in their C termini and has been implicated in the channel assembly. The binding of CaM to Kv7.3 subunits is required for their heteromeric association with Kv7.2 (12). It was reported that CaM binding is crucial for the assembly and function of potassium channel subunits, which mediate one of the main components of the M current (13). Both CaM and PIP2 regulate Kv7 channel opening and activation (14, 15); however, whether SUMOylation is involved in the regulation process remains unclear.
Retigabine is a potent and selective Kv7 channel modulator that is used in the clinic to treat epilepsy (16). Retigabine was shown to induce the M current and hyperpolarize CHO cells expressing Kv7.2 and Kv7.3 channels. Retigabine activates the Kv7.2 and Kv7.3 channels via an increase in the maximal opening probability of the channels and a hyperpolarization shift of the voltage dependence of channel activation (17). We have recently shown that retigabine restored the M current, inhibited spontaneous firing, and prevented seizures and atrioventricular (AV) block in sentrin/small ubiquitin-like modifier (SUMO)-specific protease 2 (SENP2)-deficient mice (6); however, whether the treatment of epilepsy with retigabine has any relationship with Kv7 SUMOylation needs to be determined.
SUMO is a ubiquitin-like protein that can be conjugated to lysine residues of target proteins by utilizing SUMO-specific E1, E2, and E3 enzymes for SUMO conjugation (SUMOylation) and a family of SENPs (SUMO/sentrin-specific proteases) for SUMO removal (deSUMOylation) (18, 19). SUMOylation and deSUMOylation are versatile regulatory systems and have been implicated in various biological and pathological processes, including cancer, heart disease, and neurodegenerative disorders (20, 21, 22). There are six SENPs with different substrate specificities (19), among which SENP2 is involved in the binding of the polycomb complex to H3K27me3, and SENP2 KO mouse embryos exhibit cardiac development defects and do not survive to birth (23). SENP2 is also involved in regulating myostatin expression and myogenesis (24).
We have previously shown that mice deficient in SENP2 develop spontaneous seizures and sudden death and that hyper-SUMOylation of Kv7.2 and Kv7.3 could directly contribute to the development of seizures in SENP2-deficient mice (6, 25, 26). However, it is still unknown how SUMOylation modulates Kv7.2 and Kv7.3 activity and whether CaM and PIP2 are involved in the SUMOylation process. In this report, we show that mutation of Kv7.2 and Kv7.3 SUMOylation sites decreases the binding of Kv7.2 and Kv7.3 with CaM1, which affects the assembly of Kv7.2 and Kv7.3. The SENP2-deficient mice died between 6 and 8 weeks after birth, whereas the chronic administration of retigabine prolonged their life by 1 month and significantly promoted the deSUMOylation of Kv7.2 and Kv7.3 via decreased SAE1 levels. Taken together, our findings identified a critical physiological role of SENP2 in the regulation of Kv7.2 and Kv7.3 function.
Results
SENP2 mediates deSUMOylation of Kv7.2 and Kv7.3
Our previous study showed that SENP2 deficiency leads to hyper-SUMOylation of Kv7.2 and Kv7.3 in hippocampal neurons (6). To further define the role of SENP2 in the regulation of Kv7.2 and Kv7.3 SUMOylation, we determined whether SENP2 interacted with Kv7.2 and Kv7.3 in the HEK293T cell overexpression system, and the coimmunoprecipitation (co-IP) assay showed that SENP2 strongly interacted with both Kv7.2 and Kv7.3 (Fig. 1A). To explore the potential role of SENP2 in the deSUMOylation of Kv7.2 and Kv7.3, Kv7.2 or Kv7.3 plasmids were transfected with WT or catalytic mutant SENP2 plasmid into HEK293T cells. The results showed that Kv7.2 and Kv7.3 were modified by SUMO2, and WT SENP2 deconjugated their SUMOylation, whereas the SENP2 catalytic mutant did not affect their SUMOylation (Fig. 1B). Thus, SENP2 interacts with Kv7.2 and Kv7.3 and deconjugates their SUMO modifications.
Figure 1.
SENP2 specifically deconjugates the SUMOylation of Kv7.2 and Kv7.3.A, SENP2 interacts with Kv7.2 and Kv7.3. Indicated plasmids were transfected into HEK293T cells, and the immunoprecipitates (IP) with Flag from cell lysates were detected by immunoblotting (IB) with the anti-GFP antibody. The whole-cell lysates (WCLs) were detected by IB with anti-Flag or anti-GFP antibodies (∗ nonspecific band). B, SENP2 deconjugates SUMO2 from Kv7.2 and Kv7.3. Indicated plasmids were transfected into HEK293T cells, and the IP with Flag from cell lysates were detected by IB with the anti-HA antibody. The WCLs were detected by IB with anti-HA, anti-Flag, or anti-GFP antibodies (bottom). The results of quantitative analysis of Western blot are shown in the upper panel (n = 3 repeats/group, one-way ANOVA, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001). C, the predicted SUMOylation sites of Kv7.2 and Kv7.3 by online software (www.abgent.com/sumoplot). D, K634 is the major SUMO site of Kv7.2. Indicated plasmids were transfected into HEK293T cells, and the IP with Flag or HA from cell lysates were detected by IB with anti-HA or anti-Flag antibodies (bottom). The results of quantitative analysis of Western blot are shown in the upper panel (n = 3 repeats/group, one-way ANOVA, ∗∗p < 0.01, ∗∗∗p < 0.001). E, K624 is the major SUMO site of Kv7.3. Indicated plasmids were transfected into HEK293T cells, and the IP with Flag or HA from cell lysates was detected by IB with anti-HA or anti-Flag antibodies (bottom). The results of quantitative analysis of Western blot are shown in the upper panel (n = 3 repeats/group, one-way ANOVA, ∗∗p < 0.01). F, the predicted SUMO site K634 of Kv7.2 and K624 of Kv7.3 were conserved in various species. IP, immunoprecipitates; SENP2, sentrin/SUMO-specific protease 2; SUMO, small ubiquitin-like modifier.
As predicted by SUMOplot (www.abcepta.com.cn/sumoplot), K21 and K645 are highly scored SUMOylation sites of Kv7.2, while K298 and K625 are highly scored SUMOylation sites of Kv7.3 (Fig. 1C). We generated plasmids bearing WT or predicted SUMOylation site mutants of Kv7.2 or Kv7.3 and transfected them into HEK293T cells. Co-IP analysis showed that K634 and K624 were the major SUMOylation sites of Kv7.2 and Kv7.3, respectively (Fig. 1, D and E), and these two sites were both conserved in various species (Fig. 1F). Taken together, these results indicate that both SUMO2 and SENP2 dynamically regulate the balance of Kv7.2 and Kv7.3 SUMOylation.
Mutation of the SUMOylation site of Kv7.2 and Kv7.3 increased M currents
The subcellular localization and supramembrane assembly of Kv7.2 and Kv7.3 channels are essential for the regulation of channel function (27). To determine whether SUMOylation has any effect on the subcellular localization of Kv7.2 and Kv7.3, we transfected plasmids bearing WT or SUMO mutant Kv7.2 or Kv7.3 into HEK293T cells. IF staining showed that both WT and SUMO mutant Kv7.2 and Kv7.3 were localized in the cell membrane (Fig. 2A). We further performed the surface biotinylation assay to confirm the role of SUMOylation in subcellular localization of Kv7.2 and Kv7.3, and the results showed that both WT and SUMO mutant Kv7.2 and Kv7.3 had similar surface expression levels under normal or hyper-SUMOylation conditions (Fig. 2B). SUMOylation had no significant effect on the subcellular localization of Kv7.2 or Kv7.3. These results indicated that SUMOylation has no significant effect on the subcellular localization of Kv7.2 and Kv7.3.
Figure 2.
SUMOylation site mutant Kv7.2 and Kv7.3 enhanced M currents.A, SUMOylation site mutation has no significant effect on the subcellular localization of Kv7.2 or Kv7.3. Indicated plasmids were transfected into HEK293T cells and harvested for immunofluorescent staining with anti-Kv7.2 or anti-Kv7.3 antibodies (green). DAPI (blue) was used to show nuclei. The scale bar is 10 μm. B, SUMOylation has no significant effect on the subcellular localization of Kv7.2 and Kv7.3. Indicated plasmids were transfected into HEK293T cells, and harvested for surface biotinylation assay, and the surface Kv7.2 and Kv7.3 was detected by IB with indicated antibodies (bottom). The results of quantitative analysis of Western blot are shown in the upper panel (n = 3 repeats/group, one-way ANOVA, p > 0.05). C, stimulation program of electrophysiological recordings. Cell membrane voltage is clamped at −80 mV with 10 mV increment from −80 mV to +30 mV, and the duration is 200 ms to induce M currents. D and E, representative traces and time course of M currents in HEK293T cells transfected with WT (D) and mutant (E) Kv7.2 and Kv7.3. The holding potential is −80 mV, followed by a series of depolarization steps from −80 mV to +30 mV in 10 mV increment, followed by a 50 ms hyperpolarization step to −120 mV to record the tail current. F, representative traces of WT or mutant Kv7.2 and Kv7.3 channels in HEK293T cells. The peak current and the current amplitude of mutant Kv7.2 and Kv7.3 were significantly increased when compared with WT Kv7.2 and Kv7.3. G, the comparison diagram of I-V curve of WT or mutant Kv7.2 and Kv7.3 channels in HEK293T cells. When the activation voltage is about −80 mV, the average total potassium current of mutant Kv7.2 and Kv7.3 was significantly increased when compared with WT Kv7.2 and Kv7.3 (n = 5 cells/group, one-way ANOVA, ∗∗p < 0.01, ∗∗∗∗p < 0.0001). H, the comparison diagram of G-V curve of WT or mutant Kv7.2 and Kv7.3 channels in HEK293T cells. When the activation voltage is about −80 mV, the average total potassium current of mutant Kv7.2 and Kv7.3 was significantly increased when compared with WT Kv7.2 and Kv7.3 (n = 5 cells/group, one-way ANOVA, ∗p < 0.05, ∗∗∗∗p < 0.0001). IB, immunoblotting; SUMO, small ubiquitin-like modifier.
Our previous study demonstrated that hyper-SUMOylation of Kv7 potassium channels diminished M currents and resulted in seizures and sudden death (6). To explore the roles of SUMOylation in Kv7.2 and Kv7.3 channel activity in vitro, we examined the M currents with patch clamp assay in HEK293T cells. WT or SUMO site mutant Kv7.2 and Kv7.3 plasmids were cotransfected into HEK293T cells, and the M currents were recorded with standard deactivation voltage protocol (Fig. 2, C–E). The results showed that the traces, peak current, and conductance of M currents in mutant Kv7.2 and Kv7.3 were significantly enhanced when compared with WT channels (Fig. 2, F–H). However, there was no significant difference in the tail currents between two groups, which might be because the mutant Kv7.2 and Kv7.3 had no functional effect on the closure of potassium channels, and the different activation constants could also lead to similar changes in the tail currents (28). In addition, the negative shift of the tail traces may be related to the changes of voltage-gated function in mutant Kv7.2 and Kv7.3. The negative shift of I-V and G-V curves showed that the potassium channel of the mutant Kv7.2 and Kv7.3 was more easily activated and the cell membrane was more easily repolarized. Taken together, these results indicated that SUMOylation of Kv7.2/7.3 negatively regulated M currents without effect on subcellular localization.
SUMOylation of Kv7.2 and Kv7.3 decreases their interaction with PIP2
The C termini of Kv7.2 and Kv7.3 play an important role in mediating their interaction with PIP2 and thus modulate the pore opening and activation of Kv7 (29, 30). We found that the major SUMOylation sites of Kv7.2 and Kv7.3 were localized in the C-terminal domain, and we hypothesized that SUMOylation might affect the binding of the C termini of Kv7.2 and Kv7.3 with PIP2. The protein–lipid overlay assay enables the identification of the lipid ligands that interacts and binds with lipid, providing qualitative information on the relative affinity between lipid and other protein ligands. We generated prokaryotic expression vectors of the C terminus of WT or SUMO2-fusion Kv7.2 (pMAL-Kv7.2-C-SUMO2, 313–844) and Kv7.3 (pMAL-Kv7.3-C-SUMO2, 358–872, Fig. 3A) and expressed and purified the proteins from Escherichia coli (Fig. 3B) for the protein–lipid overlay assay with PIP strips including sphingosine 1-phosphate, PI(3,4)P2, PI(3,5)P2, PI(4,5)P2, PI(3,4,5)P3, phosphatidic acid (PA), phosphatidylserine (PS), lysophosphatidic acid, lysophosphatidylcholine, PI, PI(3)P, PI(4)P, PI(5)P, phosphatidylethanolamine (PE), and phosphatidylcholine. The protein–lipid overlay assay showed that the SUMO2-fusion of the Kv7.2 C terminus decreased its binding with PI(3,4)P2, PI(3,5)P2, PI(4,5)P2 PI(3,4,5)P3, PI(3)P, PI(4)P, and PI(5)P but had no significant effect on its binding with PA, PS, and PE (Fig. 3, C and D). On the other hand, the SUMO2-fusion of the Kv7.3 C terminus decreased its binding with PI(3,4)P2, PI(3,5)P2, PI(4,5)P2, PI(3,4,5)P3, PA, PS, PI(3)P, PI(4)P, and PI(5)P but had no significant effect on its binding with PE (Fig. 3, C and D). Taken together, these results showed that SUMOylation decreased the binding of Kv7.2 and Kv7.3 with PIP2, indicating that SUMOylation of Kv7.2 and Kv7.3 diminished its binding with PIP2 and blocked pore opening, ultimately affecting the function of Kv7.2 and Kv7.3 channels.
Figure 3.
SUMOylation of Kv7.2 affects its binding with PIP2.A, schematic diagram of the prokaryotic expression plasmid for recombinant protein expression. B, the purification of C-terminal of WT or SUMO2-fusion MBP-Kv7.2 or MBP-Kv7.3 protein. SDS-PAGE showing purification of MBP-Kv7.2-C, MBP-Kv7.2-C-SUMO2, MBP-Kv7.3w, and MBP-Kv7.3-C-SUMO2 (D) fusion protein. C, protein–lipid overlay assay with WT or SUMO mutant Kv7.2 or Kv7.3 C-terminal MBP proteins. PIP strips bound with WT or mutant Kv7.2 or Kv7.3 MBP fusion proteins. D, quantitative analysis of WT or SUMO2-fusion Kv7.2 or Kv7.3 C-terminal MBP proteins bound PIP strips (n = 3 repeats/group, one-way ANOVA, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001). E, eluted protein; I, induced cells; LPA, lysophosphatidic acid; LPC, lysophosphatidylcholine; PA, phosphatic acid; PC, phosphatidylcholine; PE, phosphatidylethanolamine; PIP2, phosphatidylinositol 4,5-bisphosphate; PS, phosphatidylserine; S1P, sphingosine-1-phosphate; SUMO, small ubiquitin-like modifier; U, uninduced cells.
SUMOylation site mutation of Kv7.2 and Kv7.3 decreases their binding with CaM1
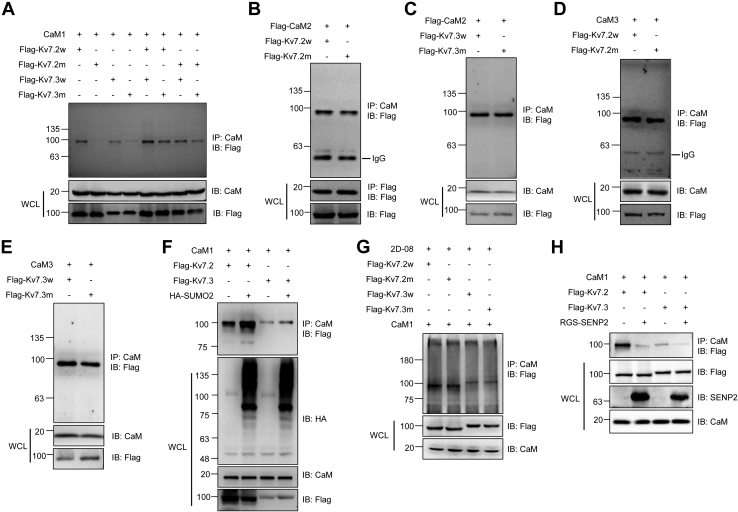
CaM is another critical modulator that binds the C-terminal motifs of KCNQ subunits and regulates the activation of Kv7 channels (11). Kv7.2 and Kv7.3 are SUMO targets, and we hypothesized that SUMOylation might affect their binding with CaM. WT and SUMO mutant Kv7.2 or Kv7.3 were transfected with CaM1 into HEK293T cells, and the co-IP assay showed that both Kv7.2 and Kv7.3 interacted with CaM1, whereas the mutant Kv7.2 and Kv7.3 showed dramatically decreased binding with CaM1 (Fig. 4A). Because there are three CaM proteins in mammalian cells, we further detected the binding of mutant Kv7.2 and Kv7.3 with CaM2 and CaM3. We found that the SUMOylation site mutations had no significant effect on the binding of Kv7.2 or Kv7.3 with CaM2 or CaM3 (Fig. 4, B–E). These results indicated that SUMOylation of Kv7.2 and Kv7.3 specifically regulates their binding with CaM1.
Figure 4.
SUMOylation of Kv7.2 and Kv7.3 affects their interaction with CaM1.A, Kv7.2 and Kv7.3 SUMO mutation diminished their interaction with CaM1. Indicated plasmids were transfected into HEK293T cells, and the IP with CaM1 from cell lysates were detected by IB with anti-Flag antibody. The WCLs were detected by IB with anti-CaM1 or anti-Flag antibodies. B and C, Kv7.2 (B) and Kv7.3 (C) SUMO mutation had no significant effect on their interaction with CaM2. Indicated plasmids were transfected into HEK293T cells, and the IP with CaM or Flag from cell lysates were detected by IB with anti-Flag antibody. The WCLs were detected by IB with anti-CaM or anti-Flag antibodies. D and E, Kv7.2 (D) and Kv7.3 (E) SUMO mutation had no significant effect on their interaction with CaM3. Indicated plasmids were transfected into HEK293T cells, and the IP with CaM from cell lysates were detected by IB with anti-Flag antibody. The WCLs were detected by IB with anti-CaM or anti-Flag antibodies. F, Kv7.2 and Kv7.3 SUMO mutation decreased the interaction between Kv7.2/Kv7.3 and CaM1. Indicated plasmids were transfected into HEK293T cells, and the IP with CaM from cell lysates were detected by IB with the anti-Flag antibody. The WCLs were detected by IB with anti-HA, anti-CaM, or anti-Flag antibodies. G, SUMO inhibition weakened the interaction of Kv7.2 and Kv7.3 with CaM1. Indicated plasmids were transfected into HEK293T cells, followed by 2D-08 treatment, and the IP with CaM from cell lysates were detected by IB with the anti-Flag antibody. The WCLs were detected by IB with anti-CaM or anti-Flag antibodies. H, SENP2 decreased the interaction of Kv7.2 and Kv7.3 with CaM1. Indicated plasmids were transfected into HEK293T cells, and the IP with CaM from cell lysates were detected by IB with the anti-Flag antibody. The WCLs were detected by IB with anti-Flag, anti-SENP2, or anti-CaM antibodies. CaM, calmodulin; IB, immunoblotting; IP, immunoprecipitates; SENP2, sentrin/SUMO-specific protease 2; SUMO, small ubiquitin-like modifier; WCLs, whole-cell lysates.
To verify the role of SUMOylation of Kv7.2 and Kv7.3 in their binding with CaM1, these plasmids were transfected with SUMO2, and the co-IP results showed that SUMO2 significantly enhanced the binding of both Kv7.2 and Kv7.3 with CaM1 (Fig. 4F). To further investigate the role of SUMOylation in the binding of Kv7.2 and Kv7.3 with CaM1, we detected whether loss of SUMOylation of Kv7.2 and Kv7.3 affects the interaction between Kv7.2 or Kv7.3 and CaM1. The SUMOylation inhibitor 2D-08 was used to block the SUMOylation of Kv7.2 and Kv7.3, and the results showed that there was no significant difference in the interaction of CaM1 with the WT or SUMO mutant Kv7.2 or Kv7.3 after SUMO inhibition (Fig. 4G). Moreover, we then detected whether SENP2 has any effect on the interaction of Kv7.2 and Kv7.3 with CaM1. The co-IP results showed that SENP2 significantly decreased the interaction of Kv7.2 and Kv7.3 with CaM1 (Fig. 4H). Taken together, these results showed that SUMOylation of Kv7.2 and Kv7.3 significantly enhanced the interaction of Kv7.2 and Kv7.3 with CaM1.
SUMOylation decreases the assembly of Kv7.2 and Kv7.3
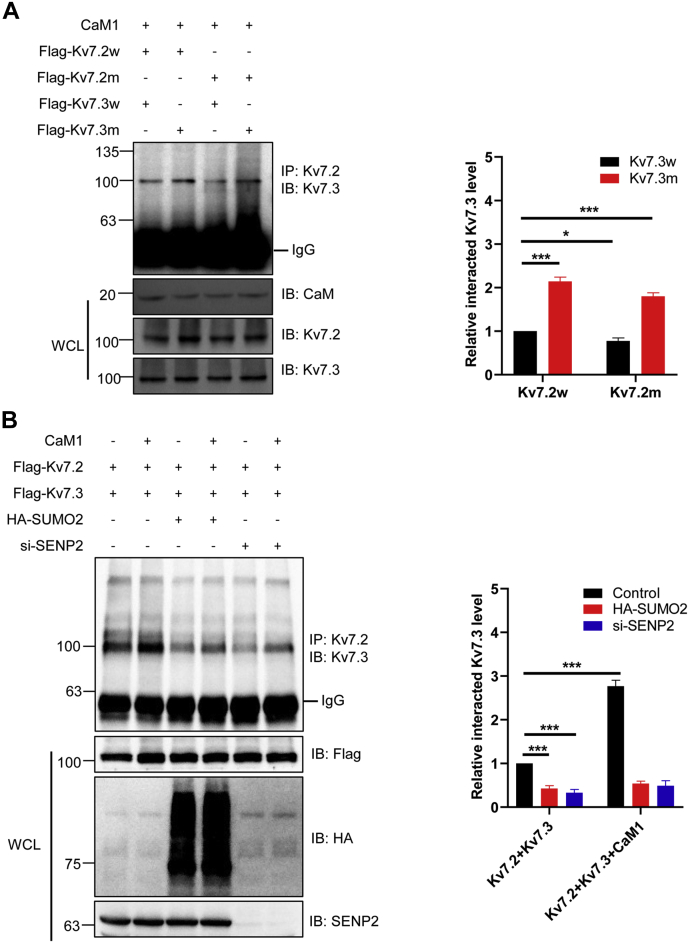
It was reported that the assembly of Kv7.2 and Kv7.3 is crucial for the function of potassium channel subunits, and we next detected whether SUMOylation has any effect on the assembly of Kv7.2 and Kv7.3. Indicated plasmids were transfected into HEK293T cells, and the co-IP results showed that in the presence of CaM1, mutant Kv7.2 diminished the assembly of Kv7.2 and Kv7.3, whereas mutant Kv7.3 significantly enhanced the assembly of Kv7.2 and Kv7.3, ultimately both mutant Kv7.2 and mutant Kv7.3 promoted the assembly of Kv7.2 and Kv7.3 (Fig. 5A). Moreover, we then detected the assembly of Kv7.2 and Kv7.3 under hyper-SUMOylation conditions induced by SUMO2 overexpression or SENP2 knockdown. Plasmids were transfected into HEK293T cells, and the co-IP results showed that SUMOylation of Kv7.2 and Kv7.3 induced by SUMO2 overexpression or SENP2 knockdown dramatically decreased Kv7 channel assembly (Fig. 5B). As expected, CaM1 promoted channel assembly in normal condition, whereas the effect of CaM1 was abolished by SUMOylation of Kv7.2 and Kv7.3 in hyper-SUMOylation status (Fig. 5B). Taken together, these results showed that SUMOylation plays a critical role in the interaction of Kv7.2 and Kv7.3 with CaM1 and the assembly of Kv7.2 and Kv7.3.
Figure 5.
SUMOylation of Kv7.2 and Kv7.3 interrupted their assembly.A, SUMO mutation of Kv7.2 or Kv7.3 enhanced the assembly of Kv7.2 and Kv7.3 at the presence of CaM1. Indicated plasmids were transfected into HEK293T cells, and the IP with Kv7.2 from cell lysates were detected by IB with anti-Kv7.3 antibody. The WCLs were detected by IB with anti-CaM, anti-Kv7.2, or anti-Kv7.3 antibodies (left). The results of quantitative analysis of Western blot are shown in the right panel (n = 3 repeats/group, two-way ANOVA, ∗p < 0.05, ∗∗∗p < 0.001). B, SUMOylation of Kv7.2 and Kv7.3 diminished the assembly of Kv7.2 and Kv7.3 at the presence of CaM1. Indicated plasmids were transfected into HEK293T cells, and the IP with Kv7.2 from cell lysates were detected by IB with the anti-Kv7.3 antibody (left). The results of quantitative analysis of Western blot are shown in the right panel (n = 3 repeats/group, two-way ANOVA, ∗∗∗p < 0.001). CaM, calmodulin; IB, immunoblotting; IP, immunoprecipitates; SUMO, small ubiquitin-like modifier; WCLs, whole-cell lysates.
Parasympathetic nervous system mediates SENP2 deficiency–induced sudden death
Our previous study showed that chronic injection of atropine decreased the parasympathetic tone and therefore prevented sinus pauses and AV blocks after seizures in SENP2-deficient mice (6). To further identify the effect of atropine in SENP2-deficient mice, we chronically injected SENP2-deficient mice with atropine daily until sudden death occurred. The results showed that atropine prolonged the mouse life span from less than 60 days to more than 70 days; thus, pretreatment with atropine prolonged the life span of SENP2-deficient mice by more than 2 weeks (Fig. 6A). Because atropine selectively blocks the parasympathetic nervous system, we wondered whether the secretion of neurotransmitters was affected in SENP2-deficient mice. The ELISA results indicated that the level of the parasympathetic neurotransmitter acetylcholine was significantly increased in the brain and heart tissue of SENP2-deficient mice (Fig. 6, B and C). Epinephrine and norepinephrine, neurotransmitters of the sympathetic nervous system, were not significantly different among SENP2-deficient, heterozygous, and control mice (Fig. 6, D and E). To further verify the role of the parasympathetic nervous system in SENP2 deficiency–induced sudden death, we performed unilateral vagotomy to rescue the mice from sudden death. The results showed that vagotomy significantly prolonged the life span of SENP2-deficient mice (Fig. 6F). These results indicated that the parasympathetic nervous system has an important role during SENP2 deficiency-induced sudden death.
Figure 6.
Vagus nerve–mediated SENP2 deficiency induced sudden death.A, chronic treatment of atropine prolonged the life span of SENP2-deficient mice. SENP-deficient and WT mice were chronically injected with atropine, and the life span was analyzed (n = 20 mice/group, Log-rank and Mantel–Cox test, p < 0.0001). The red dotted line represents the survival index when the survival rate is 50%. B and C, the acetylcholine level was increased in the brain (B) and heart (C) tissue of SENP2-deficient mice. The brain and heart tissues were isolated from SENP2-deficient, SENP2 heterozygous, and WT mice, and the acetylcholine level was measured by the ELISA kit (n = 5 mice/group, one-way ANOVA, ∗p < 0.05). D and E, the epinephrine (D) and norepinephrine (E) levels have no significant difference between SENP2-deficient and WT mice. The urine was collected from SENP2-deficient, SENP2 heterozygous, and WT mice, and the epinephrine and norepinephrine levels were measured by the ELISA kit (n = 5 mice/group, one-way ANOVA, p > 0.05). F, vagotomy prolonged the life span of SENP2-deficient mice. Vagotomy surgery was performed in SENP2-deficient and WT mice, and the life span was analyzed (n = 20 mice/group, Log-rank and Mantel–Cox test, p < 0.0001). The red dotted line represents the survival index when the survival rate is 50%. SENP2, sentrin/SUMO-specific protease 2.
Retigabine-regulated de-SUMOylation of Kv7 contributes to Kv7 channel opening
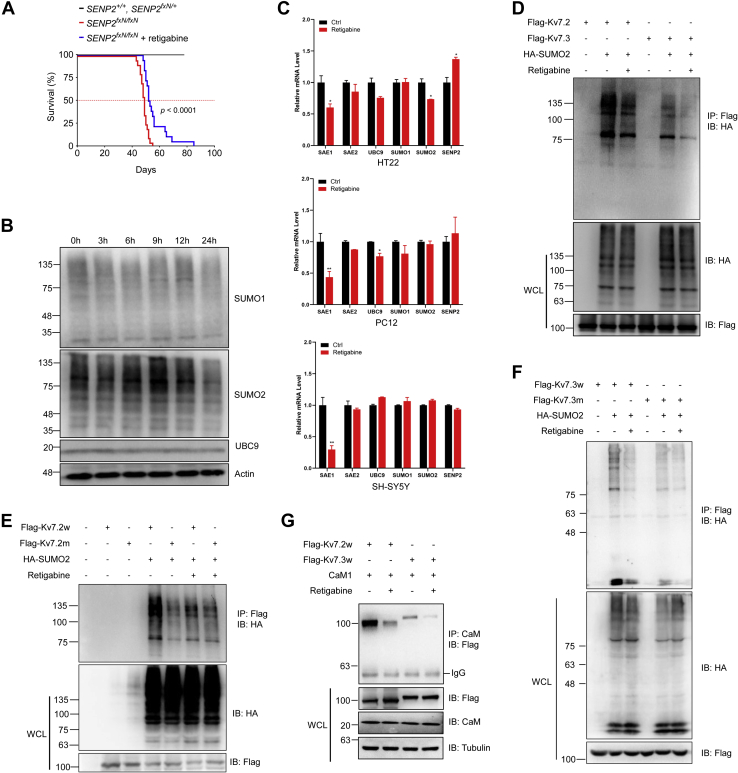
We previously showed that seizure-induced AV block could be prevented by atropine and that both seizures and AV blocks could be prevented by retigabine (6). However, it is not known whether sudden death is preventable by pharmacologic intervention with retigabine. We applied chronic administration of retigabine to determine whether sudden death can be prevented. Retigabine (10 mg/kg) was administrated until the last day of life, and the results showed that retigabine prolonged mouse life span from less than 60 days to almost 80 days; thus, retigabine prolonged the life span by more than 3 weeks (Fig. 7A).
Figure 7.
Retigabine decreased SUMOylation of Kv7.3 to rescue sudden death in SENP2-deficient mice.A, retigabine prolonged the life span of SENP2-deficient mice. SENP2-deficient, SENP2 heterozygous, and WT mice were chronically injected with retigabine, and the life span was analyzed (n = 20 mice/group, Log-rank and Mantel-Cox test, p < 0.0001). The red dotted line represents the survival index when the survival rate is 50%. B, retigabine decreased the SUMOylation level of SUMO2 but not SUMO1. COS7 cells were treated with retigabine for indicated times and harvested for IB with anti-SUMO1, anti-SUMO2, anti-UBC9, and anti-actin antibodies. C, the expression level of SAE1 transcripts was significantly decreased after retigabine treatment in SHSY5Y, PC12, and HT22 cells. The cells were treated with retigabine, and the transcript level of SUMO-related genes was measured by real-time PCR, normalized to control (n = 3 repeats/group, one-way ANOVA, ∗p < 0.05, ∗∗p < 0.01). D, retigabine diminished the SUMOylation of Kv7.2 and Kv7.3. Indicated plasmids were transfected into HEK293T cells, followed by retigabine treatment, and the IP with anti-Flag from cell lysates were detected by IB with the anti-HA antibody. The WCLs were detected by IB with anti-HA or anti-Flag antibodies. E and F, retigabine deconjugated the SUMOylation of WT Kv7.2 and Kv7.3 to that of SUMO mutant Kv7.2 (E) and Kv7.3 (F). Indicated plasmids were transfected into HEK293T cells, followed by retigabine treatment, and the IP with anti-Flag from cell lysates were detected by IB with the anti-HA antibody. The WCLs were detected by IB with anti-HA or anti-Flag antibodies. G, retigabine decreased the interaction of Kv7.2 and Kv7.3 with CaM1. Indicated plasmids were transfected into HEK293T cells, followed by retigabine treatment, and the IP with anti-CaM from cell lysates were detected by IB with the anti-Flag antibody. The WCLs were detected by IB with anti-Flag, anti-CaM, or anti-Tubulin antibodies. IB, immunoblotting; IP, immunoprecipitates; SENP2, sentrin/SUMO-specific protease 2; SUMO, small ubiquitin-like modifier; WCLs, whole-cell lysates.
It is well known that retigabine prevents seizures by opening Kv7.2 and Kv7.3; however, whether SUMOylation is involved in this process is unknown. We cultured HEK293T cells and treated them with 10 μM retigabine for different time periods, and Western blotting showed that retigabine decreased the total level of SUMO1- and SUMO2-modified proteins, whereas the SUMO E2 UBC9 level showed no significant change (Fig. 7B). To further investigate whether retigabine regulates the transcription of SUMO or SUMO enzymes, HT22, PC12, and SH-SY5Y cells were treated with retigabine for 24 h, and real-time PCR indicated that SAE1, one of two vital SUMO E1 activating enzymes, was downregulated after retigabine treatment in all three neural cell lines (Fig. 7C). These results revealed that SUMOylation might be fine-tuned and regulated by retigabine through inhibition of the transcriptional level of SAE1.
We next detected the effect of retigabine on the SUMOylation of Kv7.2 and Kv7.3, and the results showed that retigabine significantly decreased the SUMOylation of both Kv7.2 and Kv7.3 (Fig. 7D), whereas retigabine showed no significant effect on the SUMO mutant Kv7.2 and Kv7.3 (Fig. 7, E and F). Because SUMOylation regulates the interaction between Kv7.2 or Kv7.3 and CaM1, we asked whether retigabine regulates the interaction between Kv7.2 or Kv7.3 and CaM1. Co-IP assay indicated that the interaction of Kv7.2 and Kv7.3 with CaM1 was significantly decreased after retigabine treatment (Fig. 7G). Taken together, these results showed that retigabine regulates the function of Kv7.2 and Kv7.3 by deconjugating Kv7.2 and Kv7.3 SUMOylation.
Discussion
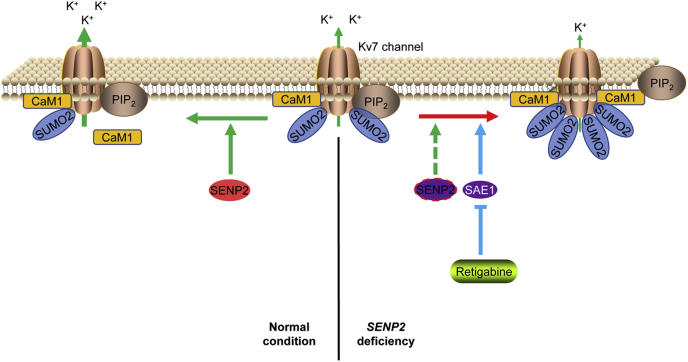
Our previous study showed that SENP2 deficiency leads to hyper-SUMOylation of Kv7.2/Kv7.3 and diminishes the hyperpolarizing M current, leading to neuronal hyperexcitability and seizures (6). In the present study, the SUMO sites of Kv7.2 and Kv7.3 were determined and mutated, and mutation of Kv7.2 and Kv7.3 SUMOylation sites increased the binding of Kv7.2 and Kv7.3 with CaM1 and affected the assembly of Kv7.2 and Kv7.3. The SENP2-deficient mice died between 6 and 8 weeks after birth, whereas chronic administration of retigabine prolonged their life span by almost 1 month via deSUMOylation of Kv7.2 and Kv7.3 (Fig. 8). These results indicated that SUMOylation of Kv7.2 and Kv7.3 is a novel mechanism to regulate the channel activity.
Figure 8.
Schematic model depicting the role of retigabine in the regulation of M currents via deSUMOylation of Kv7.2 and Kv7.3.
PIP2 interacts with voltage sensor elements and is required for opening and upregulates both the voltage sensitivity and current amplitude of the Kv7.2 and Kv7.3 channels (31). The distal C terminus has been proposed to be critical for the PIP2-dependent activation of Kv7 channels, and C terminus site mutation reduced channel activation by markedly decreasing channel opening (32, 33). Although residues in the transmembrane domain of Kv7.2 and Kv7.3 are likely involved in PIP2 interactions, we investigated the potential roles of SUMOylation of Kv7.2 and Kv7.3 C terminus in PIP2 interaction. SUMO2 fusion of the cytoplasmic tail of Kv7.2 and Kv7.3 decreased its binding with PIP2 (Fig. 3, C and D). These results indicated that SUMOylation of Kv7.2 and Kv7.3 decreased the binding of Kv7.2 and Kv7.3 with PIP2 and diminished the M current. These results were consistent with our previous study, which showed that SENP2 deficiency induced hyper-SUMOylation of Kv7.2 and Kv7.3 that diminished the M current (6). Thus, we identified that SUMOylation modulates Kv7.2 and Kv7.3 channel opening by regulating the binding of Kv7.2 with PIP2, which is an important mechanism to regulate the M current.
The distal Kv7 intracellular C-terminal domain endowed with two coiled coils appears to act as a module for proper channel heteromeric assembly and trafficking as well as a platform for interaction with several regulatory molecules (34, 35). CaM interacts with two noncontinuous ɑ-helical regions to affect the binding and function of Kv7 (7, 13, 36). It is interesting that the SUMOylation sites of Kv7.2 and Kv7.3 were also located in the C terminus (Fig. 1, D and E). In Kv7 channels, CaM1 binding and CaM1-dependent channel assembly were investigated after mutation of Kv7.2 and Kv7.3 SUMO sites. SUMOylation site mutation of Kv7.2 and Kv7.3 decreased the binding of the cytoplasmic tails of Kv7.2 and Kv7.3 with CaM1 (Fig. 4A), and Kv7.3 mutation decreased the heteromeric assembly of Kv7.2 and Kv7.3 (Fig. 5A). These data indicated that SUMOylation promoted the interaction of CaM1 with Kv7.2 and Kv7.3 but inhibited the assembly of Kv7.2 and Kv7.3, ultimately disrupting the normal function of the Kv7 channel.
Retigabine is a third-generation antiepileptic drug used in the treatment of partial-onset seizures (37). As a Kv7 channel opener, retigabine activates the Kv7.2-Kv7.5 channel by increasing the channel maximal opening probability and a hyperpolarization shift of the voltage dependence of channel activation (17). As expected, chronic administration of retigabine prolonged the mouse life span from 8 weeks to almost 12 weeks (Fig. 7A). Retigabine rescued the sudden death caused by hyper-SUMOylation of Kv7.2 and Kv7.3, and it was possible to ameliorate the function of the Kv7 channel through regulation of Kv7.2 and Kv7.3 SUMOylation. Treatment with retigabine decreased the level of the SUMO E1 SAE1 and decreased the SUMOylation levels of Kv7.2 and Kv7.3, which contributed to the opening of Kv7.2 and Kv7.3. Cryo-EM structural analysis showed that retigabine docked to its binding site in the pore of Kv7.2 and Kv7.4 (38). Our results indicated that the antiepileptic role of retigabine may partially occur via deconjugation of the SUMOylation of Kv7.2 and Kv7.3, which sheds light on a new mechanism of retigabine in the function of Kv7 channels. These results might give rise to new possibilities for the pharmaceutical and therapeutic mechanism of retigabine as an anticonvulsant for the treatment of epilepsy and sudden death.
In summary, SENP2 deconjugates the SUMOylation of Kv7.2 and Kv7.3, and SENP2 deficiency–induced hyper-SUMOylation of Kv7.2 and Kv7.3 affects their interaction with PIP2 and CaM1 and the assembly of Kv7.2 and Kv7.3. In SENP2-deficient mice, Kv7.2 and Kv7.3 are hyper-SUMOylated, which weakens the heteromeric assembly of Kv7.2 and Kv7.3 and ultimately causes seizures and sudden death. The Kv7 channel opener retigabine prolongs mouse life span by almost 1 month by decreasing the SUMOylation of Kv7.2 and Kv7.3 due to the decreased transcriptional level of the SUMO-activating enzyme SAE1. We have identified the mechanism whereby SUMOylation regulates the function of the Kv7 channel and retigabine prevents seizures through Kv7 SUMOylation. Balancing the SUMOylation of Kv7.2 and Kv7.3 through SUMO2 or SENP2 will be a novel therapeutic intervention for patients with SUDEP.
Experimental procedures
Mouse strains and animal care
SENP2-deficient mice were described previously (6). All animals were housed 2 to 5 per cage at room temperature (RT) (22 °C), with a 12-h light-dark cycle with ad libitum access to food and water. All animal studies were performed according to protocols approved by the Institutional Animal Care and Use Committee of Shaanxi Normal University, and all manipulations were conducted in consistence with established guidelines. For the rescue experiments, WT and SENP2-deficient mice were rested for 6 h and injected with either saline or atropine (1 mg/kg, Sigma), or retigabine (10 mg/kg, Selleck) with 20 mice in each group. The experiments were performed until all SENP2-deficient mice were dead.
Unilateral vagotomy
At the age of 5 weeks, WT and SENP2-deficient mice were randomized into sham and vagotomy groups with 20 mice in each group. Mice in the vagotomy group were anesthetized to make a 1-cm longitudinal midline cervical incision in the ventral region of the neck before blunt dissection. The overlying muscles and fascia were separated until the right vagus nerve was visible, and then, the right vagus nerve was isolated from the carotid artery and associated connective tissue bundle, ligated, and transected. The vagus was carefully stripped away from the carotid artery and lightly cutoff. Mice in the sham procedure group were anesthetized and had the vagus nerve isolated and left intact, but not cut, and the wound was closed and sutured. The experiments were performed until all SENP2-deficient mice were dead.
Cell culture and treatment
HEK293T, COS7, HT22, PC12, and SH-SY5Y cells were maintained at 37 °C in a 5% CO2 atmosphere. Cells were cultured in Dulbecco's modified Eagle's medium (Gibco) containing 10% fetal bovine serum (Gibco), 100 units/ml penicillin, and 100 μg/ml streptomycin. Cells were transiently transfected using Lipofectamine 3000 (Invitrogen), according to the manufacturer’s instructions. Cells were incubated with 10 μM retigabine or dimethyl sulfoxide, and the whole-cell lysates were analyzed by Western blotting.
Plasmid constructs
Eukaryotic expression plasmids, rat Flag-Kv7.2 and Flag-Kv7.3, were gifts from Dr K. George Chandy in University of California, Irvine. CaM1, Flag-CaM2, and CaM3 plasmids were provided by Dr Jichun Yang at Peking University. RGS-SENP2, RGS-SENP2 catalytic mutant, and HA-SUMO2 were previously described (6, 23, 24). Four SUMO mutant plasmids Flag-Kv7.2 K21R, Flag-Kv7.2 K634R, Flag-Kv7.3 K298R, and Flag-Kv7.3 K624R were generated using the QuikChange site-directed mutagenesis kit (TIANGEN). All plasmids were verified by DNA sequencing.
In vitro electrophysiological recordings
WT or SUMOylation site mutant Kv7.2 and Kv7.3 were cotransfected into HEK293T cells, and a single cell was selected for electrophysiological recording. M currents from transiently transfected HEK293T cells were recorded at RT (20–23 °C) 2 day after transfection, using the whole-cell configuration of the patch-clamp technique, with glass micropipettes of 4 to 8 MΩ resistance. The extracellular solution contained (in mM) the following chemicals: 150 NaCl, 2.5 CaCl2, 5 KCl, 1 MgCl2, 10 glucose, and 10 Hepes at pH 7.4. The intracellular solution contained (in mM) the following chemicals: 130 KCl, 5 MgCl2, 10 glucose, and 5 Hepes at pH 7.2. The Clampfit 11.0.3 software (Molecular Devices) was used for data acquisition and analysis. Cells were held at −80 mV and stepped from −80 to +30 mV for 200 ms in 10 mV increments with a 10-s interval between steps. Activation curves were constructed from the peak tail current evoked by a repolarization back to −80 mV after depolarizing voltage steps.
Protein–lipid overlay assay
Lipid–protein overlay assay was performed according to the protocol (39). Briefly, the plasmids inserted with the C-terminus of the WT or SUMO2-fusion Kv7.2 and Kv7.3 were constructed, and E. coli BL21 competent cells containing plasmids were grown in growth media with IPTG. The proteins containing MBP tag were purified with the chromatography column packed with amylose resin and concentrated. PIP strips were 2 × 6 cm hydrophobic membranes that have been spotted with 100 pM of all eight phosphoinositides and seven other biological important lipids (Echelon biosciences). PIP strips were blocked with 3% bovine serum albumin (Sigma) and gently agitated for 1 h at RT. The blocking buffer was discarded, and the strips were incubated with equal purified proteins overnight at 4 °C with gentle agitation. The strips were washed and incubated with anti-MBP antibody (NEB), followed by a secondary antibody coupled to peroxidase, and the interacted protein levels were detected with a chemo-luminescence system (Tanon) after additional washing steps.
Cell surface biotinylation assay
WT or SUMOylation site mutant Kv7.2 and Kv7.3 was transfected with or without SUMO2 into HEK293T cells. Forty-eight hours later, cells were washed twice with ice-cold PBS and treated with 1 mg/ml sulfosuccinimidyl 6-(biotinamido) hexanoate (Sulfo-NHS-Biotin, Thermo) in PBS for 40 min at 4 °C. After that, cells were washed with ice-cold buffer A (20 mM Tris HCl at pH 8.0 and 150 mM NaCl) and incubated in buffer A for 15 min at 25 °C. Cells were then collected by centrifuge and lysed in buffer B (10 mM Tris-HCl at pH 8.0, 150 mM NaCl, and 1% NP-40). Cell lysates were incubated with 100 μl 50% (v/v) NeutrAvidin agarose (Thermo) and rotated overnight at 4 °C. The agarose beads were then washed three times with buffer B. Biotinylated proteins bound to the beads were eluted by incubation with SDS-PAGE sample buffer at 37 °C for 30 min and analyzed by SDS-PAGE followed by Western blotting.
RNA isolation and real-time PCR
For quantitative analysis of gene expression, total RNA was extracted using RNeasy protocol (Qiagen) from cultured or treated cells. RNA was treated with DNase (Promega), and the concentration was measured by Nanodrop (Thermo). 1 μg total RNA was used to generate complementary DNA using the high-capacity cDNA reverse transcription protocol (Takara). Quantitative real-time PCR was then performed using reaction mixtures of cDNA, primers, and SYBR Green reagent (Takara) with the ABI StepOne system (PerkinElmer). PCR was done in triplicate, and SDs representing experimental errors were calculated. All data were analyzed using ABI PRISM SDS 2.0 software (PerkinElmer). The sequence of primers are as follows: SAE1-F: 5′-CAGTATGACCGACAGATCCGC-3′, R: 5′-GGCAACCTGAGCCTTTGATCT-3′; SAE2-F: 5′-CCACATCGACCTGATTGATCTG-3′, R: 5′-GGCAACCTGAGCCTTTGATCT-3′; UBC9-F: 5′-GGAGGAAGGACCACCCTTTTG-3′, R: 5′-GGATAGCGCACTCCCAGTT-3′; SUMO1-F: 5′-ATTGGACAGGATAGCAGTGAGA-3′, R: 5′-TCCCAGTTCTTTCGGAGTATGA-3′; SUMO2-F: 5′-AAGGAAGGAGTCAAGACTGAGAA-3′, R: 5′-CGGAATCTGATCTGCCTCATTG-3′; SENP2-F: 5′-CACCAAATGGAGCCTGATCT-3′, R: 5′-CTTCCGTTCTTCTGTCCTTCTC-3′.
Western blotting and immunoprecipitation
Whole mouse brain or transfected cells were extracted and subsequently homogenized on ice in RIPA lysis buffer with protease inhibitors (TargetMol). Total protein levels were quantified using the BCA assay (Pierce). Equal protein amounts were separated by electrophoresis and transferred to PVDF membranes by electroblotting. Membranes were blocked with 5% nonfat dried milk, incubated overnight with primary antibodies, including anti-UBC9 and anti-SENP2 (Santa Cruz), anti-SUMO1 (Cell Signaling Technology), anti-SUMO2 and anti-CaM (Abcam), and anti-Kv7.2 and anti-Kv7.3 (Alomone Labs) antibodies. The membrane was washed and incubated with the secondary antibody coupled to peroxidase, and protein levels were detected with the chemo-luminescence system (Tanon). For immunoprecipitation experiments, the brain or cell lysate was incubated overnight with antibodies. All incubations were performed at 4 °C with constant agitation. Antibody-bound protein complexes were captured by the addition of protein A/G agarose and incubated for another 2 h. Protein A/G agarose was pelleted by centrifugation, and the immunoprecipitated protein complex was eluted using SDS-PAGE sample buffer and Western blotting with antibodies.
Immunocytochemistry
Cells transfected with indicated plasmids were grown on cover slips. And 48 h later, cells were washed with PBS, fixed with 4% PFA (Sigma), and permeabilized with 0.5% Triton X-100 in PBS and then fixed with antibodies. Nonspecific antibody binding was minimized by treatment with 5% donkey serum in PBS for 30 min at RT. Primary antibodies were diluted in 0.1% Triton X-100 and incubated with the cells for 1 h at 37 °C. Cells were washed three times in PBS and then incubated for 1 h at RT with secondary antibodies conjugated to Cy3 or Cy5 fluorophores (Invitrogen). The cells were then washed three times in PBS and mounted using the anti-fade mounting solution (Dako) and then examined by confocal laser scanning microscopy (Zeiss).
Statistical analysis
All data are presented as the mean ± SD at least three separate experiments. Differences between groups were evaluated by one-way ANOVA followed by Dunnett’s or Tukey’s test or two-way ANOVA followed by Bonferroni’s test for multiple comparisons among more than two groups. For life span analysis, we performed the Log-rank and Mantel–Cox test by Primer-PriSM software. Statistical significance is defined as p < 0.05 (∗p < 0.05; ∗∗p < 0.01, ∗∗∗p < 0.001, and ∗∗∗∗p < 0.0001).
Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Conflict of interest
The authors declare that they have no conflict of interest with the contents of this article.
Acknowledgments
Author contributions
X. C., Y. Z., X. R., Q. S., Y. L., X. D., Yuanyuan Qin, X. Y., Z. X., and Y. S. investigation; H. W. and Yitao Qi conceptualization; Q. S., Y. L., and X. D. data curation; Yuanyuan Qin, Y. S., and Y. W. formal analysis; X. Y. methodology; Z. X. validation; Y. W., Z. B., E. T. H. Y., H. W., and Yitao Qi writing–review and editing; H. W. and Yitao Qi supervision; X. C., H. W., and Yitao Qi writing–original draft.
Funding and additional information
This work was supported by the National Natural Science Foundation of China (81671294 and 81870241 to Yitao Qi) and the Fundamental Research Funds for the Central Universities (GK201903066 to H. W.).
Edited by George DeMartino
Contributor Information
Hongmei Wu, Email: hq8479@snnu.edu.cn.
Yitao Qi, Email: qiyitao@snnu.edu.cn.
References
- 1.Goldman A.M., Glasscock E., Yoo J., Chen T.T., Klassen T.L., Noebels J.L. Arrhythmia in heart and brain: KCNQ1 mutations link epilepsy and sudden unexplained death. Sci. Transl. Med. 2009;1 doi: 10.1126/scitranslmed.3000289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Glasscock E., Yoo J.W., Chen T.T., Klassen T.L., Noebels J.L. Kv1.1 potassium channel deficiency reveals brain-driven cardiac dysfunction as a candidate mechanism for sudden unexplained death in epilepsy. J. Neurosci. 2010;30:5167–5175. doi: 10.1523/JNEUROSCI.5591-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sillanpaa M., Shinnar S. Long-term mortality in childhood-onset epilepsy. N. Engl. J. Med. 2010;363:2522–2529. doi: 10.1056/NEJMoa0911610. [DOI] [PubMed] [Google Scholar]
- 4.Massey C.A., Sowers L.P., Dlouhy B.J., Richerson G.B. Mechanisms of sudden unexpected death in epilepsy: The pathway to prevention. Nat. Rev. Neurol. 2014;10:271–282. doi: 10.1038/nrneurol.2014.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stewart M., Silverman J.B., Sundaram K., Kollmar R. Causes and effects contributing to sudden death in epilepsy and the rationale for prevention and intervention. Front. Neurol. 2020;11:765. doi: 10.3389/fneur.2020.00765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Qi Y., Wang J., Bomben V.C., Li D.P., Chen S.R., Sun H., Xi Y., Reed J.G., Cheng J., Pan H.L., Noebels J.L., Yeh E.T. Hyper-SUMOylation of the Kv7 potassium channel diminishes the M-current leading to seizures and sudden death. Neuron. 2014;83:1159–1171. doi: 10.1016/j.neuron.2014.07.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ambrosino P., Alaimo A., Bartollino S., Manocchio L., De Maria M., Mosca I., Gomis-Perez C., Alberdi A., Scambia G., Lesca G., Villarroel A., Taglialatela M., Soldovieri M.V. Epilepsy-causing mutations in Kv7.2 C-terminus affect binding and functional modulation by calmodulin. Biochim. Biophys. Acta. 2015;1852:1856–1866. doi: 10.1016/j.bbadis.2015.06.012. [DOI] [PubMed] [Google Scholar]
- 8.Ambrosino P., Freri E., Castellotti B., Soldovieri M.V., Mosca I., Manocchio L., Gellera C., Canafoglia L., Franceschetti S., Salis B., Iraci N., Miceli F., Ragona F., Granata T., DiFrancesco J.C. Kv7.3 compound heterozygous variants in early onset encephalopathy reveal additive contribution of C-terminal residues to PIP2-dependent K(+) channel gating. Mol. Neurobiol. 2018;55:7009–7024. doi: 10.1007/s12035-018-0883-5. [DOI] [PubMed] [Google Scholar]
- 9.Alaimo A., Villarroel A. Calmodulin: A multitasking protein in Kv7.2 potassium channel functions. Biomolecules. 2018;8:57. doi: 10.3390/biom8030057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang H., Craciun L.C., Mirshahi T., Rohacs T., Lopes C.M., Jin T., Logothetis D.E. PIP(2) activates KCNQ channels, and its hydrolysis underlies receptor-mediated inhibition of M currents. Neuron. 2003;37:963–975. doi: 10.1016/s0896-6273(03)00125-9. [DOI] [PubMed] [Google Scholar]
- 11.Bernardo-Seisdedos G., Nunez E., Gomis-Perez C., Malo C., Villarroel A., Millet O. Structural basis and energy landscape for the Ca(2+) gating and calmodulation of the Kv7.2 K(+) channel. Proc. Natl. Acad. Sci. U. S. A. 2018;115:2395–2400. doi: 10.1073/pnas.1800235115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu J., Qi Y., Zheng L., Cao Y., Wan L., Ye W., Fang L. Xinfeng capsule improves pulmonary function in ankylosing spondylitis patients via NF-KappaB-iNOS-NO signaling pathway. J. Tradit. Chin. Med. 2014;34:657–665. doi: 10.1016/s0254-6272(15)30079-0. [DOI] [PubMed] [Google Scholar]
- 13.Gomis-Perez C., Alaimo A., Fernandez-Orth J., Alberdi A., Aivar-Mateo P., Bernardo-Seisdedos G., Malo C., Areso P., Felipe A., Villarroel A. An unconventional calmodulin-anchoring site within the AB module of Kv7.2 channels. J. Cell Sci. 2015;128:3155–3163. doi: 10.1242/jcs.174128. [DOI] [PubMed] [Google Scholar]
- 14.Taylor K.C., Sanders C.R. Regulation of KCNQ/Kv7 family voltage-gated K(+) channels by lipids. Biochim. Biophys. Acta Biomembr. 2017;1859:586–597. doi: 10.1016/j.bbamem.2016.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nunez E., Muguruza-Montero A., Villarroel A. Atomistic insights of calmodulin gating of complete ion channels. Int. J. Mol. Sci. 2020;21:1285. doi: 10.3390/ijms21041285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sills G.J., Rogawski M.A. Mechanisms of action of currently used antiseizure drugs. Neuropharmacology. 2020;168:107966. doi: 10.1016/j.neuropharm.2020.107966. [DOI] [PubMed] [Google Scholar]
- 17.Tatulian L., Delmas P., Abogadie F.C., Brown D.A. Activation of expressed KCNQ potassium currents and native neuronal M-type potassium currents by the anti-convulsant drug retigabine. J. Neurosci. 2001;21:5535–5545. doi: 10.1523/JNEUROSCI.21-15-05535.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yeh E.T., Gong L., Kamitani T. Ubiquitin-like proteins: New wines in new bottles. Gene. 2000;248:1–14. doi: 10.1016/s0378-1119(00)00139-6. [DOI] [PubMed] [Google Scholar]
- 19.Yeh E.T. SUMOylation and de-SUMOylation: Wrestling with life's processes. J. Biol. Chem. 2009;284:8223–8227. doi: 10.1074/jbc.R800050200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chang H.M., Yeh E.T.H. SUMO: From bench to bedside. Physiol. Rev. 2020;100:1599–1619. doi: 10.1152/physrev.00025.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Long X., Zhao B., Lu W., Chen X., Yang X., Huang J., Zhang Y., An S., Qin Y., Xing Z., Shen Y., Wu H., Qi Y. The critical roles of the SUMO-specific protease SENP3 in human diseases and clinical implications. Front. Physiol. 2020;11:558220. doi: 10.3389/fphys.2020.558220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhao B., Zhang Z., Chen X., Shen Y., Qin Y., Yang X., Xing Z., Zhang S., Long X., Zhang Y., An S., Wu H., Qi Y. The important roles of protein SUMOylation in the occurrence and development of leukemia and clinical implications. J. Cell Physiol. 2021;236:3466–3480. doi: 10.1002/jcp.30143. [DOI] [PubMed] [Google Scholar]
- 23.Kang X., Qi Y., Zuo Y., Wang Q., Zou Y., Schwartz R.J., Cheng J., Yeh E.T. SUMO-specific protease 2 is essential for suppression of polycomb group protein-mediated gene silencing during embryonic development. Mol. Cell. 2010;38:191–201. doi: 10.1016/j.molcel.2010.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Qi Y., Zuo Y., Yeh E.T., Cheng J. An essential role of small ubiquitin-like modifier (SUMO)-specific protease 2 in myostatin expression and myogenesis. J. Biol. Chem. 2014;289:3288–3293. doi: 10.1074/jbc.M113.518282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu H., Chen X., Cheng J., Qi Y. SUMOylation and potassium channels: Links to epilepsy and sudden death. Adv. Protein Chem. Struct. Biol. 2016;103:295–321. doi: 10.1016/bs.apcsb.2015.11.009. [DOI] [PubMed] [Google Scholar]
- 26.Chen X., Zhang S., Huang J., Dong W., Xiao H., Shao H., Cheng J., Wu H., Qi Y. Hyper-SUMOylation of K(+) channels in sudden unexplained death in epilepsy: Isolation and primary culture of dissociated hippocampal neurons from newborn mice for subcellular localization. Methods Mol. Biol. 2018;1684:63–71. doi: 10.1007/978-1-4939-7362-0_6. [DOI] [PubMed] [Google Scholar]
- 27.Abidi A., Devaux J.J., Molinari F., Alcaraz G., Michon F.X., Sutera-Sardo J., Becq H., Lacoste C., Altuzarra C., Afenjar A., Mignot C., Doummar D., Isidor B., Guyen S.N., Colin E. A recurrent KCNQ2 pore mutation causing early onset epileptic encephalopathy has a moderate effect on M current but alters subcellular localization of Kv7 channels. Neurobiol. Dis. 2015;80:80–92. doi: 10.1016/j.nbd.2015.04.017. [DOI] [PubMed] [Google Scholar]
- 28.Manville R.W., Papanikolaou M., Abbott G.W. Direct neurotransmitter activation of voltage-gated potassium channels. Nat. Commun. 2018;9:1847. doi: 10.1038/s41467-018-04266-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim K.W., Kim K., Lee H., Suh B.C. Ethanol elevates excitability of superior cervical ganglion neurons by inhibiting Kv7 channels in a cell type-specific and PI(4,5)P2-dependent manner. Int. J. Mol. Sci. 2019;20:4419. doi: 10.3390/ijms20184419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Choveau F.S., De la Rosa V., Bierbower S.M., Hernandez C.C., Shapiro M.S. Phosphatidylinositol 4,5-bisphosphate (PIP2) regulates KCNQ3 K(+) channels by interacting with four cytoplasmic channel domains. J. Biol. Chem. 2018;293:19411–19428. doi: 10.1074/jbc.RA118.005401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang Q., Zhou P., Chen Z., Li M., Jiang H., Gao Z., Yang H. Dynamic PIP2 interactions with voltage sensor elements contribute to KCNQ2 channel gating. Proc. Natl. Acad. Sci. U. S. A. 2013;110:20093–20098. doi: 10.1073/pnas.1312483110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Telezhkin V., Brown D.A., Gibb A.J. Distinct subunit contributions to the activation of M-type potassium channels by PI(4,5)P2. J. Gen. Physiol. 2012;140:41–53. doi: 10.1085/jgp.201210796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brown D.A., Hughes S.A., Marsh S.J., Tinker A. Regulation of M(Kv7.2/7.3) channels in neurons by PIP(2) and products of PIP(2) hydrolysis: Significance for receptor-mediated inhibition. J. Physiol. 2007;582:917–925. doi: 10.1113/jphysiol.2007.132498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Haitin Y., Attali B. The C-terminus of Kv7 channels: A multifunctional module. J. Physiol. 2008;586:1803–1810. doi: 10.1113/jphysiol.2007.149187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Soldovieri M.V., Miceli F., Taglialatela M. Driving with no brakes: Molecular pathophysiology of Kv7 potassium channels. Physiology. 2011;26:365–376. doi: 10.1152/physiol.00009.2011. [DOI] [PubMed] [Google Scholar]
- 36.Yus-Najera E., Santana-Castro I., Villarroel A. The identification and characterization of a noncontinuous calmodulin-binding site in noninactivating voltage-dependent KCNQ potassium channels. J. Biol. Chem. 2002;277:28545–28553. doi: 10.1074/jbc.M204130200. [DOI] [PubMed] [Google Scholar]
- 37.Gunthorpe M.J., Large C.H., Sankar R. The mechanism of action of retigabine (ezogabine), a first-in-class K+ channel opener for the treatment of epilepsy. Epilepsia. 2012;53:412–424. doi: 10.1111/j.1528-1167.2011.03365.x. [DOI] [PubMed] [Google Scholar]
- 38.Li T., Wu K., Yue Z., Wang Y., Zhang F., Shen H. Structural basis for the modulation of human KCNQ4 by small-molecule drugs. Mol. Cell. 2021;81:25–37.e24. doi: 10.1016/j.molcel.2020.10.037. [DOI] [PubMed] [Google Scholar]
- 39.Thomas A.M., Tinker A. Determination of phosphoinositide binding to K(+) channel subunits using a protein-lipid overlay assay. Methods Mol. Biol. 2008;491:103–111. doi: 10.1007/978-1-59745-526-8_8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.