Significance
Adult neurogenesis, which is known to be a heritable trait, is thought to be involved in learning, stress-related anxiety, and antidepressant action. In this study, we map genes influencing adult neurogenesis and identify a candidate gene, follistatin (Fst) for further study. By utilizing a brain-specific knockout and viral vector-mediated gene transfer, we reveal the importance of hippocampal FST in neurogenesis, learning, and synaptic plasticity. From RNA sequencing and chromatin immunoprecipitation experiments, we identify Asic4 as a critical downstream target gene regulated by FST. Our work demonstrates that FST functions in the hippocampus at least in part through regulating Asic4 expression. Overall, we illustrate the role of hippocampal Fst in learning and synaptic plasticity.
Keywords: adult neurogenesis, learning, follistatin, hippocampus, Asic4
Abstract
The biological mechanisms underpinning learning are unclear. Mounting evidence has suggested that adult hippocampal neurogenesis is involved although a causal relationship has not been well defined. Here, using high-resolution genetic mapping of adult neurogenesis, combined with sequencing information, we identify follistatin (Fst) and demonstrate its involvement in learning and adult neurogenesis. We confirmed that brain-specific Fst knockout (KO) mice exhibited decreased hippocampal neurogenesis and demonstrated that FST is critical for learning. Fst KO mice exhibit deficits in spatial learning, working memory, and long-term potentiation (LTP). In contrast, hippocampal overexpression of Fst in KO mice reversed these impairments. By utilizing RNA sequencing and chromatin immunoprecipitation, we identified Asic4 as a target gene regulated by FST and show that Asic4 plays a critical role in learning deficits caused by Fst deletion. Long-term overexpression of hippocampal Fst in C57BL/6 wild-type mice alleviates age-related decline in cognition, neurogenesis, and LTP. Collectively, our study reveals the functions for FST in adult neurogenesis and learning behaviors.
A wide variety of human disorders such as intellectual disability feature impairment of learning and memory. These conditions have a profound impact on quality of life and social functioning. Despite this, the biological mechanisms underpinning learning are not yet fully understood. However, mounting evidence has suggested that hippocampal neurogenesis is involved (1). Several publications report learning or emotional phenotypes in rodent models, which have little or no neurogenesis in adulthood (2–5), although these, and other findings, have been questioned (6). Despite the lack of consensus on the causal relationship about adult hippocampal neurogenesis on learning, it is possible that the same genes affect both neurogenesis and learning. Indeed, in mouse inbred strains, neurogenesis is genetically correlated with performance in spatial learning and memory tasks (7, 8), and spatial memory in rats is related to the levels of hippocampal neurogenesis (9).
We hypothesized that one way to identify genes that influence learning is to identify those that contribute to heritable variation in neurogenesis (10). In this study, we used genetic mapping data from heterogeneous stock (HS) mice to identify loci associated with neurogenesis (11). We increased mapping resolution further by the incorporation of sequence information. This technique has been shown to increase mapping resolution to the point of identifying causal variants (12). One of the target genes, Fst, is predominantly expressed in the cortex, olfactory bulb, and dentate gyrus. Interestingly, two of these regions are where adult neurogenesis occurs. Fst is known to encode the protein follistatin (FST), an activin-binding protein (13, 14), which neutralizes activin bioactivity (15). FST also binds to other members of the transforming growth factor-β superfamily but with a 10-fold lower affinity than for activin A (16). In the brain, activin has been shown to play a role in the maintenance of long-term memory (17). Despite numerous studies about the functions of FST in regulation of muscle growth (18) and energy metabolism (19), its roles in the brain are still unknown.
In this study, we used brain-specific Fst knockout (KO) mice to confirm its effect on neurogenesis, and we identified learning deficits in the Fst KO mice as well as deficits in long-term potentiation (LTP) through the regulation of acid-sensing ion channel 4 (ASIC4). Our study demonstrates the power of combining genetic mapping with functional work, and we provide insights into the role of FST in the hippocampus and its influence on learning.
Results
Genetic Mapping and Merge Analysis Identify Fst as a Candidate Gene Involved in Neurogenesis.
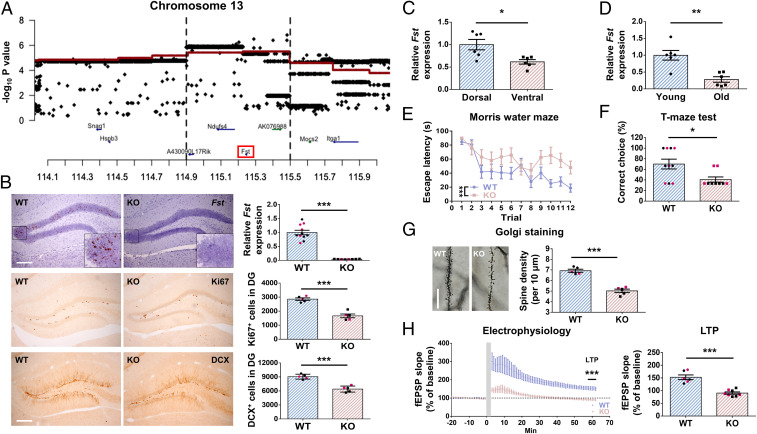
We mapped cellular proliferation in the subgranular zone of the dentate gyrus by counting the absolute number of Ki67-positive cells (a marker of cell proliferation) in 719 HS mice (11). We ran a genome-wide association test between genotypes from 13,495 markers and the number of Ki67 cells in each HS animal using a genomic relationship matrix to control for population structure (12). Applying a false discovery rate of 10% (logP of 5.1), we identified loci on chromosomes 1, 5, 6, 13, and 16. To reduce the interval containing the locus and to identify candidate genes, we incorporated all known sequence variants segregating at these loci and tested their likely involvement in adult neurogenesis using merge analysis (20). This method compares two models, one that tests association between a haplotype and the trait and the second that tests association between alleles at a marker and the trait. If the fit of the second model is worse than the first, then the marker can be excluded as causative for the association. Fig. 1A shows the result of the locus on chromosome 13 where single-marker results (black dots) exceed the haplotypic association (red line) at a region between 114.9 and 115.5 Mb. This region contains four genes.
Fig. 1.
Fst KO mice exhibit decreased hippocampal neurogenesis, impairment in learning, and synaptic plasticity. (A) Merge analysis identified candidate regions affecting adult neurogenesis. The figure shows a region (on chromosome 13) where there are genome-wide significant results from haplotypic analyses for association with adult neurogenesis (Ki67 counts) in HS mice. Each plot gives, on the vertical scale, the −log10 P values of association results for haplotypic (thick red lines) and single-marker analyses (black dots). (B) The expression of Fst in WT and KO hippocampus detected by in situ hybridization and real-time PCR. KO mice exhibit robust decreased Fst expression (P < 0.001, n = 8–11). Less Ki67 (P < 0.001, n = 5)– and DCX (P < 0.001, n = 5)–positive cells detected in the KO dentate gyrus compared to WT. (Scale bar, 200 µm.) (C) A greater expression of Fst in the dorsal than the ventral part of hippocampus in C57 mice (P = 0.01, n = 6). (D) The expression of hippocampal Fst is higher in young C57 mice (2 mo old) than aged mice (18 mo old) (P = 0.001, n = 6). (E) Fst KO mice spent more time finding the hidden platform than WT mice in a water maze learning test (P < 0.001, n = 9–11). (F) In the T-maze test, KO mice demonstrated a reduced percentage of correct choices (P = 0.03, n = 9–10). (G) Image and quantification of dendritic spine density in the dentate gyrus. Fst KO mice have significantly fewer dendritic spines in the dentate gyrus compared with WT (P < 0.001, n = 5). (Scale bar, 20 µm.) (H) High-frequency, stimulation-induced synaptic plasticity was recorded in the dentate gyrus region in WT and Fst KO mice (n = 6–8). The horizontal gray line indicates the average value of the normalized amplitude during the baseline period. fEPSP slope of LTP (55 to 60 min, P < 0.001) was dramatically reduced in the Fst KO group. Black dots signify males and red dots females.
We examined the expression profile of these genes as featured on the Allen Brain Atlas (https://portal.brain-map.org/), assuming that the genes involved in adult neurogenesis would be expressed in the adult dentate gyrus. We chose one gene, Fst, from the list of candidates for further analysis. Fst was chosen for three reasons. First, it features a clear expression profile in the dentate gyrus of the hippocampus, particularly in the dorsal hippocampus, which is involved to a greater extent in spatial learning (21). Second, we noted that overexpression of the human FST gene in transgenic mice is associated with reduced adult hippocampal neurogenesis (22). Third, activin A, to which FST has a high binding affinity, has been shown to play a role in learning (17). Despite these interesting features, the roles of endogenous Fst in the brain have not yet been studied.
To confirm the role of FST in adult hippocampal neurogenesis, we used nervous system–specific Fst conditional KO mice. This was done by crossing Fst floxed mice with Sox1::Cre mice, which specifically express Cre recombinase throughout the neural tube from E9.5 (23, 24). Fst KO mice exhibited no detectable Fst messenger RNA (mRNA) in the dentate gyrus of the hippocampus when assessed by in situ hybridization and real-time PCR (Fig. 1B), without a significant decrease in the expression of Fst in other peripheral tissues (SI Appendix, Fig. S1A). We found that Fst KO mice display a lower level of Ki67 (a cell proliferation marker), Tbr2, NeuroD, and DCX (markers for immature neurons) cell counts in the dentate gyrus compared to wild-type (WT) littermate controls (Fig. 1B and SI Appendix, Fig. S1B). These data suggest that FST plays a role in modulating adult hippocampal neurogenesis.
Altered Learning and Hippocampal Synaptic Plasticity in the Fst KO Mice.
We observed that Fst was more strongly expressed in the dorsal than ventral hippocampus. This was confirmed with real-time PCR (Fig. 1C). One feature of adult neurogenesis in rodents is that it decreases with age (25). We found that the expression of Fst was also decreased in aged mice (Fig. 1D). These results raise the possibility that FST plays a role in learning and memory. To investigate the relationship between FST and behavior, we subjected Fst KO and control mice to a battery of anxiety and learning behavior tests. This battery included open field, elevated-O-maze, light–dark box, T-maze, Morris water maze, contextual fear, and active avoidance. Our results showed no conclusive differences between the Fst KO and controls in three different anxiety tests (SI Appendix, Fig. S1 C–F). For learning and memory behaviors, Fst KO mice spent more time finding the hidden platform in a water maze (spatial learning), made fewer correct choices in a T-maze test (spatial working memory) (Fig. 1 E and F), and made fewer avoidance responses in the active avoidance test but displayed no difference in freezing time in the contextual fear-conditioning test (SI Appendix, Fig. S1 G and H). These results indicate that FST is essential for several types of learning.
It is widely believed that synaptic plasticity is the basis of learning and memory. To better understand the role of FST in synaptic plasticity, we measured dendritic spine density and hippocampal LTP of the dentate gyrus in Fst WT and KO mice. Our results show that Fst KO mice exhibit a lower level of dendritic spine density (Fig. 1G) and impairment in LTP (Fig. 1H). This suggests that mice without FST in the hippocampus display impairment of synaptic plasticity.
KO Mice with Hippocampal Fst Overexpression Reverses Impairment in Learning and Synaptic Plasticity.
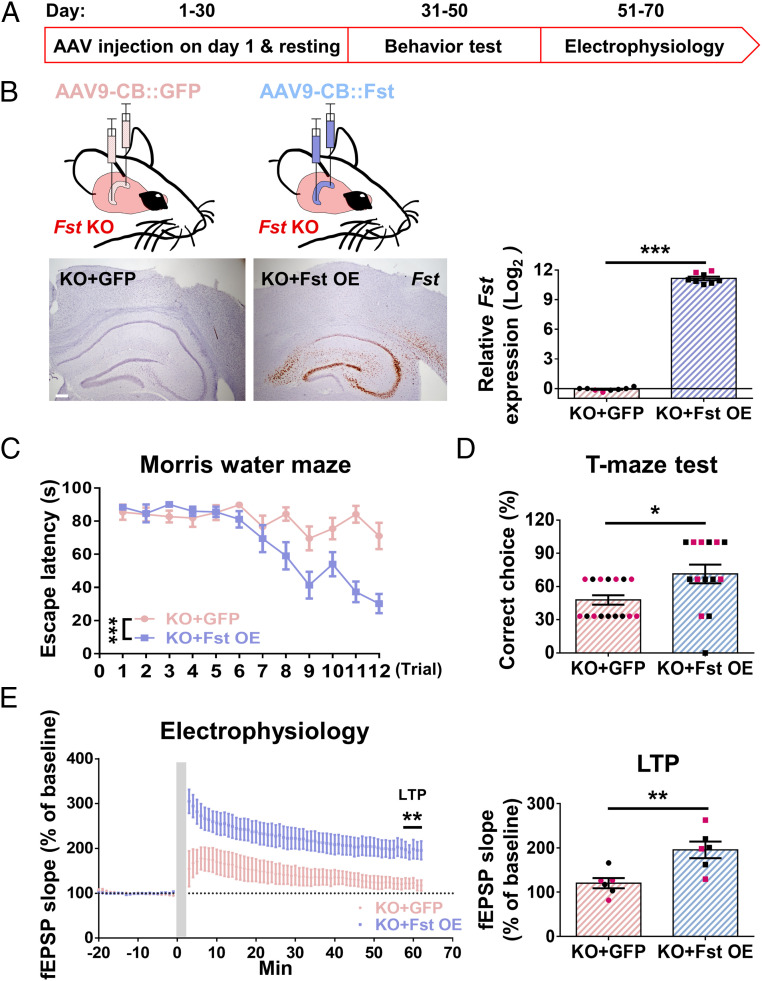
To examine whether the learning deficit of Fst KO mice can be rescued by increasing the expression of Fst in the hippocampus, we injected either AAV9-CB (chicken-beta actin)-Fst viral vector (Overexpression) or an AAV9-CB-GFP viral vector (Control) into the hippocampus of Fst KO mice. Fig. 2A shows the schematic of the experimental procedure.
Fig. 2.
Overexpression of hippocampal Fst reverses the impairment of learning and synaptic plasticity in KO mice. (A) Time course of the experimental procedure. (B) Illustration of intrahippocampal AAV-Fst or AAV-GFP injection in Fst KO mice. Image of Fst overexpression in the hippocampus 1 mo after the surgery by in situ hybridization (Left) and the confirmation of the Fst overexpression in the hippocampus by real-time PCR (n = 8 in each group) (Right). (Scale bar, 200 µm.) (C–E) Fst overexpression is sufficient to reverse the impairment in water maze learning (P < 0.001, n = 14–16) (C), T-maze (P = 0.03, n = 14–16) (D), and synaptic plasticity (LTP, P = 0.006, n = 6) (E). Black dots signify males and red dots females.
One month after surgery, we detected a robust increase of Fst expression in the hippocampus by in situ hybridization (Fig. 2B, Left) and by real-time PCR (Fig. 2B, Right). To examine the effect of Fst overexpression on behavior, 1 mo after surgery, we subjected all of the AAV-Fst– or AAV-GFP–injected Fst KO mice (Fstf/f-Sox1::Cre) to the battery of behavioral tests described previously. We detected no significant differences in any of the three anxiety tests (SI Appendix, Fig. S2 A–D). For learning behavior, Fst-overexpressing mice performed better in water maze training and T-maze (Fig. 2 C and D), but there was no significant difference in freezing time in contextual fear (SI Appendix, Fig. S2E) nor avoidance response in the active avoidance test (SI Appendix, Fig. S2F) compared to the AAV-GFP–injected control group. To further examine the effect of Fst overexpression in synaptic plasticity, we measured LTP using ex vivo electrophysiological recording. Our results reveal that Fst overexpression reversed the impairment of LTP (Fig. 2E). Our data demonstrates that some of the learning deficit (but not all) and impairment of LTP in Fst KO mice can be rescued by elevating hippocampal Fst expression.
Identification of Asic4 as a Target Gene Regulated by Fst Expression in the Hippocampus.
To search for downstream candidate genes which are regulated by FST and contribute to its effects on learning and LTP, we used RNA sequencing (RNA-seq) to define the differential gene expression list from WT versus Fst KO mice. We also compared the gene expression list from Fst KO with intrahippocampal AAV-GFP–injected mice versus AAV-Fst–injected mice. We then compared these gene lists and searched for the genes differentially expressed in opposite directions. There were 13 candidate genes in this list with a P value smaller than 0.01 (Fig. 3A, Left). Apart from Fst itself, only one gene, Asic4, had an adjusted P value less than or equal to 0.05. We further confirmed the expression pattern of Asic4 from RNA-seq analysis by using real-time PCR (Fig. 3A, Right).
Fig. 3.
Identifying downstream target genes of Fst. (A) Heatmap of the 13 most significant genes (P < 0.01) for which expression changes in opposite directions while comparing WT versus Fst KO and KO vs. Fst expression (n = 3 in each group) (Left). Confirmation of Asic4 expression in the dentate gyrus of the hippocampal formation by real-time PCR. WT versu Fst KO (P = 0.002, n = 9) (Upper right); Fst KO versus Fst overexpression (P < 0.001, n = 7) (Bottom right). Black dots signify males and red dots females. (B) Schematic diagram of ChIP analysis of H3K4me3 binding on chromatin region of the Asic4 promoters (Left). The extent of occupancy was compared between Control (KO+GFP)- and Fst-overexpressing (KO+Fst OE) hippocampus tissues. Each sample represents tissues merged from four different mice. To triplicate the experiment, a total of 12 mice were used in each group. We performed a logarithmic transformation of the ratio of the treatment sample to the reference control group; a ratio t test was used. The Asic4 promoter in the KO+Fst OE group underwent an increase in H3K4 trimethylation in region 1 (P = 0.06), region 2 (P = 0.002), and region 3 (P = 0.02) (Right). (C) Double in situ hybridization revealed that Fst and Asic4 were expressed in adjacent cells but not colocalized in the same cell in the dentate gyrus and cortex. (Scale bar, 200 µm.) (D) There is a dramatic decrease in the expression of hippocampal Fst in Fstf/f-CamKIIalpha::iCre mice (P < 0.001, n = 4, compared to their own WT littermates) but no difference in Fstf/f-Gad2::Cre mice (P = 0.29, n = 3, compared to their own WT littermate).
The up-regulated gene expression patterns shown in Fig. 3A suggested a positive effect of FST on the transcriptional rate of the Asic4 gene. To test this hypothesis, chromatin immunoprecipitation (ChIP) experiments were performed to monitor the epigenetic state of the Asic4 promoter (Fig. 3B; primer design shown in the left panel). For this purpose, we focused on the histone modification of trimethylated histone H3 lysine 4 (H3K4me3), as this represents a readout of the transcriptional activity and is detectable in the Asic4 promoter region (as shown by the The Encyclopedia of DNA Elements [ENCODE]/The University of California Santa Cruz [UCSC] Genome Browser). In line with the transcription activation effect of FST, H3K4me3 occupancy binding at the Asic4 promoter was elevated in the hippocampus tissues of KO mice with Fst overexpression as compared to the control (Fig. 3B, Right). Together, these data implied that the alteration of Asic4 expression is mediated at the chromatin level.
To better understand the relative location between Asic4- and Fst-expressing cells, we performed double-labeling in situ hybridization. Staining reveals that Asic4-expressing cells are not colocalized with Fst-expressing cells in either the dentate gyrus or cortex (Fig. 3C), suggesting that FST influences Asic4 gene expression in adjacent cells. Next, to determine whether Fst is expressed in excitatory or inhibitory neurons, we crossed Fst floxed mice with CamKIIalpha::iCre (Cre is expressed in excitatory neurons) or Gad2::Cre (Cre is expressed in inhibitory neurons) mice. We then assayed the hippocampal Fst expression in these tissue-specific Fst KO mice via real-time PCR. The results show that Fst is mainly expressed in excitatory neurons (Fig. 3D).
Overexpression of Asic4 in the Hippocampus of Fst KO Mice Rescues the Deficit in Learning and LTP.
To better understand the importance of Asci4 in Fst KO mice, we tested whether overexpressing Asic4 in the hippocampus of Fst KO mice could rescue the behavioral phenotype. We injected AAV-CB-Asic4 or AAV-CB-GFP control viral vector into the hippocampus of Fst KO mice. One month after surgery, the expression of Asic4 increased robustly in the hippocampus, as measured by in situ hybridization and real-time PCR (Fig. 4 A and B). We assayed all mice (Fst KO + AAV-GFP and Fst KO + AAV-Asic4) using our behavioral battery described above for the Fst WT vs. KO comparsion. We found no significant differences in the open field test and elevated-O-maze. However, in the light–dark box test, the Asic4 overexpression group spent more time in the light compartment compared to GFP control (SI Appendix, Fig. S3 A–D).
Fig. 4.
Overexpression of hippocampal Asic4 rescues the deficiency of learning and synaptic plasticity in Fst KO mice. (A) Time course of the experimental procedure. (B) Illustration of intrahippocampal AAV-GFP or AAV-Asic4 injection in Fst KO mice and image of Asic4 overexpression in the hippocampus by in situ hybridization (Bottom Left) and the expression of hippocampal Asic4 mRNA measured by real-time PCR (Bottom Right). (Scale bar, 200 µm.) (C–E) Asic4-overexpressed Fst KO mice exhibit better performance in water maze (P = 0.03, n = 16–18) (C), make more correct choices in T-maze (P = 0.02, n = 17–18) (D), and stronger synaptic plasticity (LTP, P = 0.001, n = 6–7) (E). Black dots signify males and red dots females.
For the learning and memory tests, Asic4-overexpressing Fst KO mice performed better in water maze training and made a higher percentage of correct choices in the T-maze compared to the GFP control (Fig. 4 C and D). However, there was no significant difference detected in the active avoidance test or contextual fear-conditioning test (SI Appendix, Fig. S3 E and F). To assess whether Asic4 overexpression can also reverse the characterization of LTP in Fst KO mice, we performed ex vivo electrophysiological recording. Our results show that Aisc4 overexpression can reverse the impairment of LTP in the Fst KO mice (Fig. 4E). Collectively, these data demonstrate that elevated expression of Asic4 in the hippocampus can rescue the deficits of learning behavior and LTP in the brain of Fst KO mice. This suggests that Fst regulates these functions of the hippocampus, at least in part, through Asic4 expression.
Long-Term Overexpression of Hippocampal Fst Improves Cognitive Behavior.
Having identified a role for FST in learning and LTP, we speculated whether increasing hippocampal Fst expression in C57BL/6 mice could improve learning performance. To study this, we injected AAV-GFP or AAV-Fst into the hippocampus of 2-mo-old male C57BL/6 mice. All mice were subjected to learning tests 1 mo after surgery (Fig. 5 A and B). However, our results show that there was only a minor difference in water maze training and no other obvious differences in all the other tests used (SI Appendix, Fig. S4).
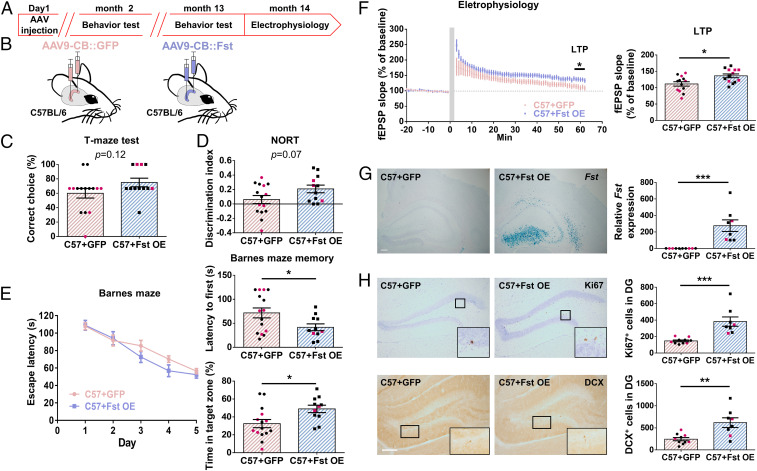
Fig. 5.
Mice with chronic hippocampal Fst expression enhance memory, LTP, and progenitor cell proliferation. (A) Time course of the experimental procedure. (B) Illustration of intrahippocampal AAV-GFP or AAV-Fst injection in C57BL/6 mice. (C) No significant difference in T-maze test (P = 0.12, n = 12–15). (D) There is a trend but no statistically significant difference in the discrimination index of the object recognition test (P = 0.07, n = 12–15). (E) There is no difference in the 5-d training for Barnes maze (P = 0.227, n = 11–15). Three days after the last training session, Fst-overexpressing mice spent less time finding the escape tunnel than control mice (P = 0.03) (Upper Right) and spent more time in the target zone (P = 0.01) (Bottom Right). (F) High-frequency, stimulation-induced synaptic plasticity was recorded in the dentate gyrus in Fst-overexpressing mice and control mice (n = 13 from five mice). The horizontal gray line indicates the average value of the normalized amplitude during the baseline period. HFS-induced LTP (P = 0.01) was significantly increased in the Fst-overexpressing mice. (G) Confirmation of hippocampal Fst overexpression by in situ hybridization and real-time PCR in mice 13 mo after AAV-Fst intrahippocampal injection (P < 0.001, n = 8–10) (Right). (H) For hippocampal neurogenesis (n = 8–10), there are increased Ki67 (P < 0.001)– and DCX (P = 0.002)–positive cells in the dentate gyrus of Fst-overexpressing mice 13 mo after AAV-Fst injection. (Scale bar, 200 µm.) Black dots signify males and red dots females.
To better understand whether long-term elevated expression of Fst plays a role in learning and memory performance, all these mice were subjected to behavioral tests again 12 mo later. The results showed no consistent outcome in the anxiety tests (no difference in the open field test and light–dark box, but Fst-overexpressing mice spent less time in the open arms in the elevated-O-maze test). There were also no significant differences in contextual fear, active avoidance (SI Appendix, Fig. S5 A–F), or the T-maze test (Fig. 5C) detected between C57BL/6 control and Fst-overexpressing mice. For the object recognition test, mice with Fst overexpression exhibited better performance, although this was not statistically significant (P = 0.07) (Fig. 5D). For the water maze test, we stopped the experiment, because a high percentage of these midage mice opted to float, not swim. Due to this, we decided to conduct a Barnes maze test, instead of the water maze, to assess spatial learning and memory. In this test, there was no significant difference observed in the 5 d of training. However, 3 d after the last training session, Fst-overexpressing mice exhibited better performance in the memory test (Fig. 5E). These results suggest that long-term elevated hippocampal Fst improves spatial memory and object recognition in mice.
We also found that mice with chronic overexpression of hippocampal Fst mice have greater LTP than control mice (Fig. 5F). We confirmed the robust increase in Fst expression and even higher level of Asic4 in Fst-overexpressing mice 14 mo after AAV-Fst injection (Fig. 5G and SI Appendix, Fig. S5G). With regards to hippocampal neurogenesis, there were more Ki67- and DCX-positive cells in Fst-overexpressing mice (Fig. 5H). Overall, these results reveal that chronic hippocampal Fst overexpression alleviates the decline of memory, LTP, and cell proliferation with age.
Sex Differences in Behaviors.
We explored whether sex differences in behavior contributed to our results. Results for males and females are shown in each figure (males are colored black and females colored red). We assessed differences between the sexes for each measure, testing in each case for the improvement of fit of a model containing sex over a model in which genotype predicted the behavior. Since this meant running 30 tests, we applied a Bonferroni-corrected threshold of 0.05/30 = 0.0167 as the 5% significance threshold. Only one result exceeded that threshold; conditioned fear tested at 12 mo in our analysis of Fst KO (P = 0.003, t = 4.193). Analysis of this measure for each sex yielded P = 0.504 for males and P = 0.274 for females; however, the small sample size and consequent low power means we cannot exclude the presence of a sex difference. We provide a summary of the sex difference analyses and the numbers of animals used for each experiment in SI Appendix, Table S1.
Discussion
In this study, we set out to identify genes involved in neurogenesis via genetic mapping in HS mice. The candidate gene, Fst, was identified and found to be expressed in the dentate gyrus of the hippocampus. We confirmed that FST regulates neurogenesis by using brain-specific Fst KO mice. From the Allen Brain Atlas, we noticed that Fst is expressed more intensely in the dorsal rather than the ventral hippocampus and confirmed this with real-time PCR. Furthermore, Fst was expressed more in young mice compared to older ones. These findings prompted us to investigate its role in learning and memory.
In our behavioral assays, we observed that Fst KO mice performed worse in water maze training, T-maze, and active avoidance tests. Our results also showed that the KO mice featured impairment of LTP and dendritic spine density. Conversely, overexpression of hippocampal Fst in the KO mice was shown to reverse some of the impairment of learning behavior and LTP. From the RNA-seq analysis, we identified Asic4 as a putative downstream target gene of FST. We subsequently confirmed that FST expression can regulate the promoter of Asic4. Next, we demonstrated that overexpression of hippocampal Asic4 in the Fst KO could rescue the learning behavior and LTP, suggesting that FST regulates hippocampal function via Asic4 expression.
A previous study has shown that mice with transgene-integrated human FST, driven by the CamKIIalpha promoter, exhibit severe impairment of hippocampal neurogenesis (22). In our study, we detected impaired adult neurogenesis in Fst brain-specific KO mice and increased neurogenesis in aged Fst-overexpressing mice. Although many factors may confound these apparently discordant results, the biggest difference is that in our study, we manipulated endogenous murine Fst expression in the brain. The results may be influenced by spatiotemporal differences in the CamKIIalpha overexpression versus our brain-specific KO mice. Furthermore, the human form of FST may function differently in the mouse brain to that of the endogenous FST. Our evidence presented here from hippocampal AAV-Fst–injected C57BL/6 mice suggests an important positive role for FST on neurogenesis. We found increased neurogenesis in mice 1 y after AAV-Fst injection, in contrast to the impaired neurogenesis of the brain-specific Fst KO. With regard to anxiety behavior, similar to the FST transgene-integrated mice (22), we did not detect a significant difference between WT and Fst KO mice. Similarly, we did not detect a significant difference in the AAV-Fst overexpression mice. These results suggest that FST does not play an essential role in anxiety.
Upon testing learning behavior in the Fst KO mice, we found deficits in water maze training, the T-maze, and active avoidance tests. Furthermore, we found that overexpression of Fst specifically in the hippocampus could rescue behavioral learning performance in both the water maze and T-maze. However, performance in the active avoidance test was not rescued. There are several potential explanations for this. First, FST may play a role in brain development. Consequently, overexpression of Fst in the adult mouse brain may not reverse avoidance behavior due to aberrant neurodevelopment. Second, other brain regions where Fst is expressed may be required for active avoidance.
Spatial learning has been shown to stimulate hippocampal neurogenesis (26) in some studies but not others (27). Some factors, such as running, stimulate adult neurogenesis and also increase performance in learning (28). However, whether spatial learning and memory requires adult newly formed neurons in hippocampus remains debatable (6). In this study, we did not intend to address whether the effect of FST on learning was due to hippocampal neurogenesis. Instead, we searched for downstream genes which may contribute to the effects of Fst on learning behavior and LTP. We identified the most likely target gene, Asic4, by RNA-seq. Asic4 shows strong homology to ASICs (29). It is expressed throughout the brain mainly in calretinin and/or vasoactive intestine peptide positive interneurons, oligodendrocyte precursor cells, and cerebellar granule cells (30). However, little is known about the function of Asic4.
We confirmed that Fst overexpression regulates the promoter region of Asic4. We also found that Asic4 is not colocalized with the expression of Fst, which is mainly expressed in excitatory neurons from our Fstf/f-CamKIIalpha::iCre KO result. This suggests that FST-expressing excitatory neurons have an influence on the expression of Asic4 in adjacent cells. This regulation may be a result of secretion of FST by these excitatory cells that directly influences Asic4-expressing cells. Alternatively, Asic4 may be regulated indirectly via other effects of FST on the excitatory cells. Furthermore, we overexpressed hippocampal Asic4 in Fst KO mice, and the impairment of learning and LTP was reversed. This suggests that the effects of FST on learning and LTP, at least partially, depend on the expression of Asic4. Here, we provide evidence that Asic4 is one of the downstream targets of FST.
Finally, we examined whether hippocampal Fst overexpression enhanced learning performance. The results demonstrate that short-term overexpression of Fst in the hippocampus of young mice does not change learning behavior. However, chronic Fst overexpression for more than 1 y resulted in better performance in object and spatial memory tests, greater LTP, and more progenitor proliferation in the dentate gyrus. This suggests that maintaining FST expression may contribute to protection from age-related cognitive decline.
In this study, we found little evidence for sex differences in the behavior that we tested in mice. However, others have reported such effects in rodents (31), and we are unable to exclude their contribution to our results. With our largest sample sizes (10 animals per group), we have only 40% power to detect a standardized mean difference of 0.8 (often regarded as a large effect by Cohen’s d measure). Sex differences are likely much smaller than this, so much larger sample sizes will be required to detect their presence.
In conclusion, we have identified a link between Fst and neurogenesis. Our results indicate that hippocampal FST plays a significant role in modulating neurogenesis, LTP, and learning. We further provide evidence that FST influences behavior via regulating Asic4 expression. Lastly, we have found that chronic Fst overexpression improves learning performance, LTP, and progenitor cell proliferation. Further understanding of the underlying cellular mechanisms will help the generation of management strategies in conditions such as intellectual disability, in which learning is impaired. Here, we suggest that brain FST is a potential target for drugs that influence learning and memory.
Materials and Methods
Animals.
Experiments were performed on 12-wk-old Fst KO mice and WT littermates, where possible with equal numbers of male and female animals (SI Appendix, Table S1). Fst with floxed alleles has been described previously (32). We crossed Fst floxed mice with Sox1::Cre mice (Access No. CDB0525K) to generate Fstf/f; Sox1::Cre mice (KO) and Fstf/f mice (WT) littermates were used in this study. Gad2::Cre transgenic mice were purchased from Jax Laboratory (Stock no. 004682 and 010802). CamKIIalpha::iCre mice were provided by laboratory of Che-Kun James Shen, Academia Sinica, Taiwan (33). C57BL/6 mice that were 12 wk-old were used for the hippocampal Fst overexpression experiment. Mice were bred in American Association for Accreditation of Laboratory Animal Care–certified, specific pathogen–free conditions. They were housed in a 12:12 h light:dark cycle at a temperature of 22 °C and a humidity level of 60 to 70%. Animals had ad libitum access to food and water. All procedures were carried out in accordance with the local regulations and approved by the Institutional Animal Care and Use Committee at Chang Gung University (Permit No.: CGU107-089).
Genetic Analysis in the HS Mice.
Association testing was carried out in a mixed-model framework, as described previously (12). In brief, the model used to test for association between the ancestral haplotypes segregating at a locus L and phenotypic variation was:
where yi is the phenotypic value of the mouse i, βc the regression coefficient of covariate c, and xic the value of the covariate c in mouse i. TLs is the deviation in phenotypic value that results from carrying one copy of a haplotype from strain s at locus L and PLi(s) the expected number of haplotypes of type s carried by mouse i at locus L output by HAPPY (34). ui and εi are random effects, with cov(ui, uj) = σg2Ki,j and cov(εi, εj) = σe2Ii,j, where σg2 and σe2 are estimated in the null model (no locus effect, TLs = 0) using the R package EMMA (35). K is the genetic covariance matrix and is estimated from the genome-wide genotypic data using identity by state (IBS, the proportion of shared alleles between any two animals). The IBS matrix was calculated using the R package EMMA (35). We implemented merge analysis (20) in a mixed-model framework as described (12). This test is applied at every variable site in the catalog of single-nucleotide variants that segregate between the eight HS founders. Because the two models (merged and unmerged) are nested, the best possible is obtained with the haplotype model. If the quantitative trait loci arises from variation at a single variant V, the fit of the merge model for variant V will be as good as the fit of the unmerged, and its significance will be greater due to the fewer number of degrees of freedom.
Behavioral Testing.
For behavioral tests, the movement of animals was recorded and tracked using a video recording system. Mice for conducting the behavioral analysis were randomized to a separate cage for testing, with bedding, food, and water before being transferred back to the home cage. To assess anxiety and cognitive abilities, mice were subjected to open field, elevated-O-maze, light–dark box, Morris water maze, object recognition, contextual fear-conditioning, and active avoidance tests. For the WT versus KO and KO versus Fst overexpression experiments, two cohorts of mice were used for each experiment: one cohort of mice for anxiety tests and one for learning tests. For Asic4 overexpression and Fst overexpression in C57 experiments, only one cohort of mice was used for behavior testing in each experiment. All the behavioral data were recorded and acquired automatically by Ethovision software. Some of these experimental details are described previously (36, 37).
Open Field.
Mice were allowed to explore for 5 min in an arena (radius = 45 cm, inner circle with radius = 30 cm). The time spent in the center zone and the exploration distance were measured.
Elevated-O-Maze.
The maze was a circle (radius = 55 cm) elevated 60 cm above the floor. The closed arms featured a 15-cm wall. Mice were first placed in the closed arm and allowed to freely move for 5 min under dim light. The time spent in the open arms and the exploration distance were measured.
Light–Dark Box.
At the start of the test, mice were put in the covered dark (black) box and permitted to move freely between the dark and the coverless light (white) box for 5 min. The time spent in light box and the number of light box entries were measured.
T-maze Test.
Before the test, all guillotine doors were raised. The mice were placed in the start area and allowed to choose a goal arm. A mouse was confined to the chosen arm and start area by quietly sliding the other door down. After 30 s, the mouse was removed. Each mouse repeated the experiment three times. We recorded the percentage of correct times each mouse explored the novel arm.
Morris Water Maze.
For training, mice were placed in the maze (radius = 60 cm) and allowed to swim/search for the hidden platform for a maximum of 90 s. The time taken to reach the platform was recorded. If the mice did not find the platform within 90 s, they were taken to the platform. This procedure was repeated three times over 3 d (12 trials in total). We then performed the memory test 7 d after training was complete, with the platform removed. We recorded the time and times that each mouse spent in the correct quadrant (target zone) and crossed the phantom platform location, respectively.
Barnes Maze.
For training, mice were placed in the maze (radius = 48 cm) and allowed to search for the escape tunnel for a maximum of 120 s. The time taken to reach the platform was recorded. If the mice did not find the escape tunnel within 120 s, they were guided to the escape tunnel. This procedure was repeated two times over 5 d (10 trials in total). We then performed the memory test 3 d after training was complete, with the escape tunnel removed. We recorded the latency and time that each mouse reached the phantom escape tunnel location and spent in the correct quadrant (target zone), respectively.
Contextual Fear-Conditioning.
Context-conditioned learning was assessed in a footshock chamber placed on a weight transducer (Panlab) and analyzed with PACKWIN software. Mice were allowed to explore the chamber for 3 min. At the end of the trial, an electric shock with current intensity of 0.75 mA was given through the underlying conducting rods. Mice were placed into the same footshock chamber the next day for 3 min, and the immobile time was recorded.
Active Avoidance.
Mice were placed in a two-compartment shuttle box equipped with a speaker and a light bulb in each compartment. Subjects were given conditioned stimuli (5 s of light and 8 kHz, 85-dB tone) followed by a unconditioned electric footshock (0.3 mA) from the underlying conducting rods. Once the mice moved to the other compartment or the cutoff time (10 s) was up, the unconditioned stimuli ceased. After a random intersession interval (range 3 to 10 s), the next session started. Mice repeated 50 sessions over 4 d (200 sessions in total). Avoidance number in each day was recorded.
Novel Object Recognition.
On day 1 (habituation), mice were put in the box (30 × 30 × 25 cm) without any object and permitted to explore for 10 min. On day 2 (training), we put four different objects in the box and let mice explore for 10 min. On day 3 (test), one of the four familiar objects was replaced with a novel object, and mice were permitted to explore for 10 min. The data were expressed by a discrimination index (D.I. = (tnovel − tfamiliar)/(tnovel + tfamiliar) × 100%).
Golgi Staining.
Brains were harvested and stained using the superGolgi Kit (Bioenno Tech) in accordance with the manufacturer's instructions. Brains were impregnated for 12 d following 2 d of dehydration in sucrose. Brains were later sectioned coronally at a thickness of 120 µm using a microtome. We calculated the spine density of granule cells in dentate gyrus. We selected 18 dendrites per animal and chose three different dendrites randomly per brain slice in both WT and KO mice. Each animal had a total of six brain slices analyzed.
Immunohistochemistry and Quantification.
Dissected brains were fixed in 4% paraformaldehyde overnight, following dehydration in 25% sucrose in phosphate-buffered saline (PBS). All sections for immunohistochemistry were sliced at a thickness of 40 µm. Brain sections were mounted on SuperFrost Plus slides (Thermo) and dried overnight. Slides were then incubated in 0.01 M citric acid buffer for 20 min at 95 °C, 3% H2O2 for 10 min, rinsed in PBS, and incubated overnight at room temperature in DCX antibody (1:250, Santa Cruz), Ki67 antibody (1:1,000, Vector Laboratory), Tbr2 antibody (1:1,000, Abcam), and NeuroD (1:1,000, Santa Cruz). Subsequently, we used a standard IgG ABC kit (Vector Laboratory) procedure according to the manufacturer’s instructions and incubated the slides for 5 to 10 min with a Sigma DAB (3,3′-diaminobenzidine) tablet.
For quantification, all slides were randomized and coded before quantitative analysis. Slides (half-brain) were examined under a 20× objective. Labeled cells were counted on every eighth section through the entire rostrocaudal extent of the granule cell layer (six sections per animal). The number of cells counted was then multiplied by 16 to obtain an estimate of the total number of positive cells in the dentate gyrus, as previously described (37).
In Situ Hybridization.
For Fst in situ hybridization, Fst probe (Cat No. 553211) and RNAscope 2.5 HD Brown Reagent Kit (Advanced Cell Diagnostics) were used, and staining was performed according to the product’s manual. Briefly, slides were incubated in 1× retrieval buffer for 5 min at 100 °C, 3% H2O2 for 10 min, and rinsed in 1× wash buffer. Slides were then incubated in protease for 20 min and, subsequently, target probe for 2 h at 40 °C. After a series of amplification, DAB was used to visualize the RNA. For Fst and Asic4 (Cat No. 511971-C2) double in situ hybridization, RNAscope 2.5 Duplex Detetcion Kit (Advanced Cell Diagnostics) was used, and the staining protocol followed the user manual.
Electrophysiology in Hippocampal Slices.
For LTP experiments, a bipolar stainless steel stimulating electrode (Frederick Haer Company, Bowdoinham) (10 Meg-ohm impedance) and a glass pipette filled with 3 M NaCl were positioned in the dentate gyrus. Stable baseline field excitatory postsynaptic potential (fEPSP) activity was evoked every 20 s for at least 20 min, high-frequency stimulation (HFS)–LTP was then induced using five trains of 100 pulses at 100 Hz with an inter train interval of 20 s. Electrophysiological traces were amplified with an amplifier (Multiclamp 700 B; Axon Instruments, Union City). All signals were low-pass–filtered at 1 kHz and digitized at 10 kHz using a CED Micro 1401 mKII interface (Cambridge Electronic Design). Data were collected using Signal software (Cambridge Electronic Design). Synaptic responses were normalized to the average of the baseline. LTP values were determined by averaging 5 min of normalized slope values at 55 to 60 min post-HFS. This protocol has been described previously (38).
Viral Vector Preparation and Intrahippocampal Injections.
For Fst and Asic4, overexpression and control vectors were derived from expressed mouse sequences. Briefly, total RNA was isolated from mouse hippocampal tissue, and complementary DNA (cDNA) was then made using SuperScript III reverse transcriptase. cDNA fragments that encoded FST (RefSeq: NM_001301373.1) or ASIC4 (RefSeq: NM_183022.3) were created by PCR and subcloned into the NotI site of aN AAV9 virus construct. Recombinant AAV9 vectors were produced by a standard triple-plasmid transfection method and purified by two rounds of CsCl centrifugation. The physical vector titres of AAVs were quantified using a real-time PCR method to measure the number of packaged vector genomes.
For the viral vector injections, surgery was performed under anesthesia. Mice received bilateral injections of viral vector into the hippocampus in a volume of 1.5 µL each injection side (2 × 1011 GC/mL for AAV9-Fst, 1.6 × 1012 GC/mL for AAV9-Asic4) under stereotaxic guidance. For hippocampal injections, there were four injection sites in each mouse (dorsal hippocampus: anterior–posterior [AP] −2.2 mm, medial–lateral [ML] ± 2.0 mm from bregma, and dorsal–ventral [DV] −1.8 mm from dura; ventral hippocampus: AP −3.0 mm, ML ± 3.0, and DV −3.3 mm). This protocol has been described previously (37).
RNA-Seq–Based Gene Expression and Analysis.
RNA from the dentate gyrus of the hippocampus was isolated using TRIzol reagent. Six mice were used in each group, and three independent samples were generated by pooling the tissue from two mice per sample. Following the manufacturer’s instructions, sequencing libraries were then generated using NEBNext Ultra RNA Library Prep Kit for Illumina. Using the Agilent Bioanalyzer 2100 system, library quality was then assessed. Subsequently, the clustering of the index-coded samples was performed on a cBot Cluster Generation System using HiSeq PE Cluster Kit cBot-HS (Illumina) according to the manufacturer’s instructions. The library preparations were then sequenced on an Illumina Hiseq platform. Paired-end reads of 125 bp/150 bp were generated.
High-throughput sequencing (HTSeq) version 0.6.1 was used to count the reads numbers mapped to each gene. Fragments per kilobase of exon per million reads mapped of each gene was calculated based on the length of the gene and reads count mapped to the gene (39). Using the DESeq R package (1.18.0), differential expression analysis of two groups was performed. The resulting P values were adjusted using the Benjamini and Hochberg’s approach for controlling the false discovery rate. Genes with an adjusted P value <0.05 found by DESeq were assigned as differentially expressed.
mRNA Quantification.
Hippocampal tissue was collected from WT and KO mice, then homogenized in TRIzol, followed by Phenol–Chloroform extraction of total RNA. cDNA was then made using SuperScript III reverse transcriptase (Invitrogen). Experiments were performed in duplicate. Gene expression levels were then calculated with the ΔΔCt method and normalized against a Gapdh control. Fst-Forward: TGCTGCTACTCTGCCAGTTC; Fst-Reverse: GTGCTGCAACACTCTTCCTTG, Asic4-Forward: CGCTATCCAGAGCCTGACAT; Asic4-Reverse: CCCTACTGGGCAGTGAACTC
ChIP.
ChIP assay was performed essentially as described previously (40). Hippocampus tissues obtained from adult male mice and homogenized in a 1% formaldehyde crosslinking solution for 15 min. Next, fixing was stopped by adding glycine solution (final concentration 0.125 M) to each reaction for 10 min. The homogenates were centrifuged at 500 × g for 6 min at 4 °C, and cell pellets were then resuspended in cell lysis buffer. Crosslinked chromatin was sonicated, precleared, and subjected to immunoprecipitation with 2.5 µg of H3K4me3 antibodies (with control IgG for mock immunoprecipitation). After extensive washes, immunocomplexes were treated with Proteinase K and de-crosslinked. The precipitated DNA as well as the input DNA (1/10 fragmented chromatin) was extracted, purified, and subjected to real-time PCR. Primers for different regions are region 1: Forward: CCTCACTAGGAGCCTGACCT; Reverse: GAGGGAGGGTCATCAGCGTA, region 2 Forward: CCCTTCCTTCCCTCCCTCAT, Reverse: CCCAGCTTCCTTGACTCCTG, region 3: Forward: TTCCACCTGGCCAATCTGAC, Reverse: CAGTGCGGTTGAGGATGTCT.
Statistical Analysis.
The mean ± SEM was determined for each group. Statistical analysis was performed using Graphpad Prism software. Data were analyzed via an ANOVA, Mann–Whitney U test (For T-maze), t test, or ratio t test as appropriate. Bonferroni method was performed when applicable. Differences were considered significant when P was less than 0.05. For sex differences in behaviors, we measured the significance of sex as the improvement in fit conferred by sex in a model in which genotype predicted the behavioral measure. The significance of the fixed effect sex was assessed using an approximation to the sequential F-test based on the Wald test (41). We fitted models by REML (Restricted maximum likelihood), using the lmer function from the R package lme4 (42).
Supplementary Material
Acknowledgments
We thank Dr. Cheng-Pu Sun (Academia Sinica, Taiwan) for his help on packing the AAV viral vector, Dr. Lichieh Julie Chu (Chang Gung University, Taiwan) for the heatmap graphical presentation of the figure, and Dr. Che-Kun James Shen (Academia Sinica, Taiwan) for providing CamKIIalpha::iCre mice. We thank Neuroscience Program of Academia Sinica Neuroscience Core Facility for technical support of electrophysiology (supported by grant AS-CFII-108-106) and the technical services provided by the Transgenic Mouse Model Core Facility of the National Core Facility for Biopharmaceuticals, Ministry of Science and Technology, Taiwan and the Animal Resources Laboratory of National Taiwan University Center of Genomic and Precision Medicine. We thank Dr. Andrew Edwards (National Health Service Greater Glasgow and Clyde, Scotland) for his helpful comments on this manuscript. The Fst mutant mice were created with the supported of the Eunice Kennedy Shriver National Institute of Child Health and Human Development (grant R01HD032067). This study was supported by grants from the Chang Gung Memorial Hospital (CMRPD1H0413), Healthy Aging Research Center (EMRPD1I0501), and the Ministry of Science and Technology of Taiwan (MOST 105-2320-B-182-040) (to G.-J.H.). This research was also supported by the “Molecular Medicine Research Center, Chang Gung University” (EMRPD1L0341) from The Featured Areas Research Center Program within the framework of the Higher Education Sprout Project by the Ministry of Education in Taiwan.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2109040118/-/DCSupplemental.
Data Availability
For the genetic mapping, the genotype and phenotype data available are on Heterogeneous stock (HS) mice webserver, http://mtweb.cs.ucl.ac.uk/HSMICE/GENOTYPES/ and http://mtweb.cs.ucl.ac.uk/HSMICE/PHENOTYPES/. The RNA-seq data are available on National Center for Biotechnology Information (GSE163449).
References
- 1.Anacker C., Hen R., Adult hippocampal neurogenesis and cognitive flexibility - Linking memory and mood. Nat. Rev. Neurosci. 18, 335–346 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shors T. J., et al., Neurogenesis in the adult is involved in the formation of trace memories. Nature 410, 372–376 (2001). [DOI] [PubMed] [Google Scholar]
- 3.Santarelli L., et al., Requirement of hippocampal neurogenesis for the behavioral effects of antidepressants. Science 301, 805–809 (2003). [DOI] [PubMed] [Google Scholar]
- 4.Saxe M. D., et al., Ablation of hippocampal neurogenesis impairs contextual fear conditioning and synaptic plasticity in the dentate gyrus. Proc. Natl. Acad. Sci. U.S.A. 103, 17501–17506 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Garthe A., Behr J., Kempermann G., Adult-generated hippocampal neurons allow the flexible use of spatially precise learning strategies. PLoS One 4, e5464 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Groves J. O., et al., Ablating adult neurogenesis in the rat has no effect on spatial processing: Evidence from a novel pharmacogenetic model. PLoS Genet. 9, e1003718 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kempermann G., Kuhn H. G., Gage F. H., Genetic influence on neurogenesis in the dentate gyrus of adult mice. Proc. Natl. Acad. Sci. U.S.A. 94, 10409–10414 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kempermann G., Gage F. H., Genetic determinants of adult hippocampal neurogenesis correlate with acquisition, but not probe trial performance, in the water maze task. Eur. J. Neurosci. 16, 129–136 (2002). [DOI] [PubMed] [Google Scholar]
- 9.Drapeau E., et al., Spatial memory performances of aged rats in the water maze predict levels of hippocampal neurogenesis. Proc. Natl. Acad. Sci. U.S.A. 100, 14385–14390 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kempermann G., Chesler E. J., Lu L., Williams R. W., Gage F. H., Natural variation and genetic covariance in adult hippocampal neurogenesis. Proc. Natl. Acad. Sci. U.S.A. 103, 780–785 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang G. J., et al., A genetic and functional relationship between T cells and cellular proliferation in the adult hippocampus. PLoS Biol. 8, e1000561 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baud A., et al., Combined sequence-based and genetic mapping analysis of complex traits in outbred rats. Nat. Genet. 45, 767–775 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nakamura T., et al., Activin-binding protein from rat ovary is follistatin. Science 247, 836–838 (1990). [DOI] [PubMed] [Google Scholar]
- 14.Ueno N., et al., Isolation and partial characterization of follistatin: A single-chain Mr 35,000 monomeric protein that inhibits the release of follicle-stimulating hormone. Proc. Natl. Acad. Sci. U.S.A. 84, 8282–8286 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Winter J. P., et al., Follistatins neutralize activin bioactivity by inhibition of activin binding to its type II receptors. Mol. Cell. Endocrinol. 116, 105–114 (1996). [DOI] [PubMed] [Google Scholar]
- 16.Hedger M. P., Winnall W. R., Phillips D. J., de Kretser D. M., The regulation and functions of activin and follistatin in inflammation and immunity. Vitam. Horm. 85, 255–297 (2011). [DOI] [PubMed] [Google Scholar]
- 17.Ageta H., et al., Activin plays a key role in the maintenance of long-term memory and late-LTP. Learn. Mem. 17, 176–185 (2010). [DOI] [PubMed] [Google Scholar]
- 18.Tsuchida K., The role of myostatin and bone morphogenetic proteins in muscular disorders. Expert Opin. Biol. Ther. 6, 147–154 (2006). [DOI] [PubMed] [Google Scholar]
- 19.Hansen J. S., Plomgaard P., Circulating follistatin in relation to energy metabolism. Mol. Cell. Endocrinol. 433, 87–93 (2016). [DOI] [PubMed] [Google Scholar]
- 20.Yalcin B., Flint J., Mott R., Using progenitor strain information to identify quantitative trait nucleotides in outbred mice. Genetics 171, 673–681 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bannerman D. M., et al., Double dissociation of function within the hippocampus: Spatial memory and hyponeophagia. Behav. Neurosci. 116, 884–901 (2002). [DOI] [PubMed] [Google Scholar]
- 22.Ageta H., et al., Activin in the brain modulates anxiety-related behavior and adult neurogenesis. PLoS One 3, e1869 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Takashima Y., et al., Neuroepithelial cells supply an initial transient wave of MSC differentiation. Cell 129, 1377–1388 (2007). [DOI] [PubMed] [Google Scholar]
- 24.Huang G. J., et al., Ectopic cerebellar cell migration causes maldevelopment of Purkinje cells and abnormal motor behaviour in Cxcr4 null mice. PLoS One 9, e86471 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Altman J., Das G. D., Autoradiographic and histological evidence of postnatal hippocampal neurogenesis in rats. J. Comp. Neurol. 124, 319–335 (1965). [DOI] [PubMed] [Google Scholar]
- 26.Gould E., Beylin A., Tanapat P., Reeves A., Shors T. J., Learning enhances adult neurogenesis in the hippocampal formation. Nat. Neurosci. 2, 260–265 (1999). [DOI] [PubMed] [Google Scholar]
- 27.van Praag H., Kempermann G., Gage F. H., Running increases cell proliferation and neurogenesis in the adult mouse dentate gyrus. Nat. Neurosci. 2, 266–270 (1999). [DOI] [PubMed] [Google Scholar]
- 28.van Praag H., Christie B. R., Sejnowski T. J., Gage F. H., Running enhances neurogenesis, learning, and long-term potentiation in mice. Proc. Natl. Acad. Sci. U.S.A. 96, 13427–13431 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gründer S., Geissler H. S., Bässler E. L., Ruppersberg J. P., A new member of acid-sensing ion channels from pituitary gland. Neuroreport 11, 1607–1611 (2000). [DOI] [PubMed] [Google Scholar]
- 30.Lin S. H., et al., Genetic mapping of ASIC4 and contrasting phenotype to ASIC1a in modulating innate fear and anxiety. Eur. J. Neurosci. 41, 1553–1568 (2015). [DOI] [PubMed] [Google Scholar]
- 31.Yagi S., Galea L. A. M., Sex differences in hippocampal cognition and neurogenesis. Neuropsychopharmacology 44, 200–213 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jorgez C. J., Klysik M., Jamin S. P., Behringer R. R., Matzuk M. M., Granulosa cell-specific inactivation of follistatin causes female fertility defects. Mol. Endocrinol. 18, 953–967 (2004). [DOI] [PubMed] [Google Scholar]
- 33.Casanova E., et al., A CamKIIalpha iCre BAC allows brain-specific gene inactivation. Genesis 31, 37–42 (2001). [DOI] [PubMed] [Google Scholar]
- 34.Mott R., Talbot C. J., Turri M. G., Collins A. C., Flint J., A method for fine mapping quantitative trait loci in outbred animal stocks. Proc. Natl. Acad. Sci. U.S.A. 97, 12649–12654 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kang H. M., et al., Efficient control of population structure in model organism association mapping. Genetics 178, 1709–1723 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chang S., Bok P., Sun C. P., Edwards A., Huang G. J., Neuropsin inactivation has protective effects against depressive-like behaviours and memory impairment induced by chronic stress. PLoS Genet. 12, e1006356 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chang S., et al., NPTX2 is a key component in the regulation of anxiety. Neuropsychopharmacology 43, 1943–1953 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu W. L., et al., The effect of ASIC3 knockout on corticostriatal circuit and mouse self-grooming behavior. Front. Cell. Neurosci. 13, 86 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Trapnell C., et al., Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28, 511–515 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Perna A., Alberi L. A., TF-ChIP method for tissue-specific gene targets. Front. Cell. Neurosci. 13, 95 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pinheiro J. C., Bates D. M., Mixed-Effects Models in S and S-PLUS (Springer, New York, 2000), pp. xvi, 528 pp. [Google Scholar]
- 42.R Core Team , R: A Language and Environment for Statistical Computing (R Foundation for Statistical Computing, Vienna, Austria, 2018). https://www.R-project.org/. Accessed 10 September 2021.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
For the genetic mapping, the genotype and phenotype data available are on Heterogeneous stock (HS) mice webserver, http://mtweb.cs.ucl.ac.uk/HSMICE/GENOTYPES/ and http://mtweb.cs.ucl.ac.uk/HSMICE/PHENOTYPES/. The RNA-seq data are available on National Center for Biotechnology Information (GSE163449).