Significance
Wolbachia are maternally transmitted bacteria that infect most insects, making them the most common endosymbionts. Wolbachia achieved this status by manipulating host reproduction. For example, many Wolbachia cause cytoplasmic incompatibility (CI) that kills uninfected embryos. In females, Wolbachia can rescue CI, promoting their spread to high frequencies in host populations. CI strength varies in nature from weak to strong. Importantly, strong CI enables Wolbachia biocontrol strategies in mosquito systems, which protect millions of individuals from arboviruses. However, theory predicts that natural selection does not act to preserve genes that cause CI, suggesting mutations may disrupt it. We show that a single naturally observed mutation weakens CI by reducing deubiquitylation. These discoveries help elucidate the molecular basis of symbiont-induced reproductive manipulations.
Keywords: cytoplasmic incompatibility, Wolbachia, endosymbiosis, Drosophila, Ubiquitin
Abstract
Animals interact with microbes that affect their performance and fitness, including endosymbionts that reside inside their cells. Maternally transmitted Wolbachia bacteria are the most common known endosymbionts, in large part because of their manipulation of host reproduction. For example, many Wolbachia cause cytoplasmic incompatibility (CI) that reduces host embryonic viability when Wolbachia-modified sperm fertilize uninfected eggs. Operons termed cifs control CI, and a single factor (cifA) rescues it, providing Wolbachia-infected females a fitness advantage. Despite CI’s prevalence in nature, theory indicates that natural selection does not act to maintain CI, which varies widely in strength. Here, we investigate the genetic and functional basis of CI-strength variation observed among sister Wolbachia that infect Drosophila melanogaster subgroup hosts. We cloned, Sanger sequenced, and expressed cif repertoires from weak CI–causing wYak in Drosophila yakuba, revealing mutations suspected to weaken CI relative to model wMel in D. melanogaster. A single valine-to-leucine mutation within the deubiquitylating (DUB) domain of the wYak cifB homolog (cidB) ablates a CI-like phenotype in yeast. The same mutation reduces both DUB efficiency in vitro and transgenic CI strength in the fly, each by about twofold. Our results map hypomorphic transgenic CI to reduced DUB activity and indicate that deubiquitylation is central to CI induction in cid systems. We also characterize effects of other genetic variation distinguishing wMel-like cifs. Importantly, CI strength determines Wolbachia prevalence in natural systems and directly influences the efficacy of Wolbachia biocontrol strategies in transinfected mosquito systems. These approaches rely on strong CI to reduce human disease.
Many endosymbionts spread through host populations by manipulating their reproduction. For example, Rickettsiella (1), Mesenet (2), Cardinium (3), and Wolbachia (4) all cause cytoplasmic incompatibility (CI) that reduces the viability of uninfected host embryos fertilized by symbiont-modified sperm (5–8). CI is common among Wolbachia bacterial strains, being observed in at least 10 arthropod orders (6). CI strength influences Wolbachia prevalence, with stronger CI producing higher Wolbachia infection frequencies in host populations (8, 9). Indeed, CI contributes significantly to Wolbachia’s status as the most-common known endosymbionts in nature (10).
CI strength directly influences the efficacy of Wolbachia biocontrol programs, with vector-control groups relying on strong CI to either suppress mosquito populations (11, 12) or to transform them with pathogen-blocking Wolbachia like wMel that naturally infects Drosophila melanogaster (13–15). The World Health Organization recommends further developing these programs (16), which are currently protecting seven million people from disease with a goal of protecting half a billion by 2030 (14, 17).
Operons generally termed cifs control CI (cifA/B) (5, 18–22), and CI induction can be rescued by one factor (cifA) (5, 19, 23). Theory indicates that natural selection does not act to increase or maintain CI (24), which varies considerably among even very closely related Wolbachia (25–27), potentially due to mutational erosion of cifs (28). For example, CI strength differs significantly among model wMel from Drosophila melanogaster and closely related wMel-like Wolbachia in the Drosophila yakuba clade (wYak, wSan, and wTei) that wMel diverged from in only the last 30,000 y (25, 27, 29, 30). We sought to determine how much and why naturally observed mutations in wMel-like cifs influence CI strength.
Results
We cloned and Sanger sequenced the two divergent cif operons observed in wYak that infects D. yakuba in triplicate, confirming prior reports (29). Specifically, wYak has two pairs of cif loci: a Type 1 pair with a deubiquitylase (DUB) domain (cidwYak) that is homologous to cidwMel and a Type 4 pair with nuclease domains (cinwYak) that is homologous to cinwPip in wPip that infects Culex pipiens (29).
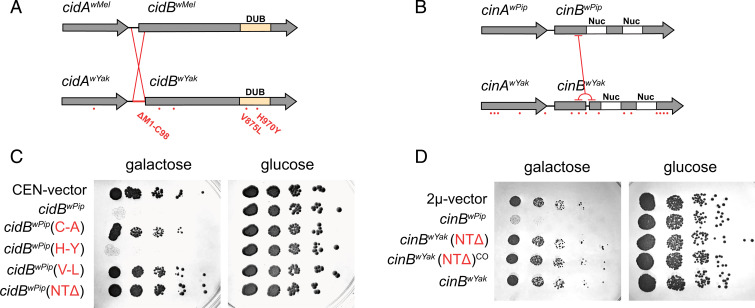
In search of sequence variation that differentiates wMel-like cifs, we aligned cidwYak to cidwMel and cinwYak to cinwPip (Fig. 1A) (29). In total, six coding permutations differentiate cidwMel from cidwYak, including five nonsynonymous mutations (one in cidAwYak and four in cidBwYak) and an inversion at a CACG palindrome on the 5′ end of cidBwYak that truncates CidBwYak (SI Appendix, Fig. S1). Two of the nonsynonymous mutations in cidBwYak fall within the DUB domain known to be catalytically active for CidBwMel (V875L, valine to leucine; H970Y, histidine to tyrosine) (19). cinwYak was differentiated from cinwPip by 16 coding permutations, including 15 nonsynonymous mutations (five in cinAwYak and 10 in cinBwYak), and a tandem duplication resulting in a frameshift and premature stop codon splitting CinBwYak into two parts (Fig. 1B). No mutations fell within the tandem nuclease domains (Nuc) known to be catalytically active for CinBwPip (31). For both CifBwYak copies, upstream methionines possibly initiate translation of N-terminally truncated CifB proteins.
Fig. 1.
Analysis of mutations in yeast model. Analysis of cif permutations in wYak, wMel, and wPip. Amino acid differences between homologs are highlighted with red dots for (A) CidwMel versus CidwYak and (B) CinwPip versus CinwYak. CidBwYak contains two mutations within the DUB domain and a large inversion. CinBwYak contains a tandem duplication yielding a frameshift and premature stop codon but no mutations in the nuclease (Nuc) domains (C). Serial dilutions of yeast-expressing constructs with analog mutations for cid (C) and cin (D). For cid, we express cidBwPip as in previous studies (19), since it produces a strong CI-like phenotype. Introducing (V-L) and (NTΔ) individually reduces yeast toxicity relative to wild-type control, cidBwPip. cidBwPip(C-A) is a catalytic inactive negative control. CEN-vector is an empty pRS416gal1 negative control. For cin, (NTΔ) beginning after the tandem duplication and the wild-type sequence containing the tandem duplication reduces yeast death relative to wild-type control, cinBwPip. (NTΔ) and (NTΔ)CO contain native sequence or codon optimized sequence, respectively. 2μ-vector is an empty pYes2 negative control.
We propose that naturally observed cif sequence variation, particularly in functional domains, contributes to CI-strength variation. To test this hypothesis, we first screened the effects of natural wYak cif mutations on a CI model in yeast (SI Appendix, Fig. S2). In this model, CifB proteins induce yeast toxicity, and CifA coexpression rescues it (19, 31, 32). Since catalytic inactive DUB mutants are unable to induce CI, we focused on wYak cif mutations found in the DUB domain (19). We also hypothesized that N-terminal truncations weaken CI, because this pattern is observed recurrently in putatively pseudogenized cifB genes (33, 34). In yeast serial dilutions, the wYak V875L (V-L) DUB mutation and a ΔM1-C98 N-terminal deletion (NTΔ) independently eliminated CidB-induced toxicity, while the H970Y (H-Y) DUB mutation did not differ from the CidB control (Fig. 1C) (phenotypic ablation can be attributed to reduced CidB deubiquitylation). For Cin, wild-type CinBwYak and an ΔM1-K126 N-terminal deletion (NTΔ) lacked CinB-induced toxicity (Fig. 1D and SI Appendix, Fig. S3). We conclude that wMel-like cif functions are disrupted by natural mutations in their functional domains and through truncation of N termini (NTΔ) of their CifB proteins. These variants observed in wYak cifs represent candidate mutations for involvement in CI strength variation.
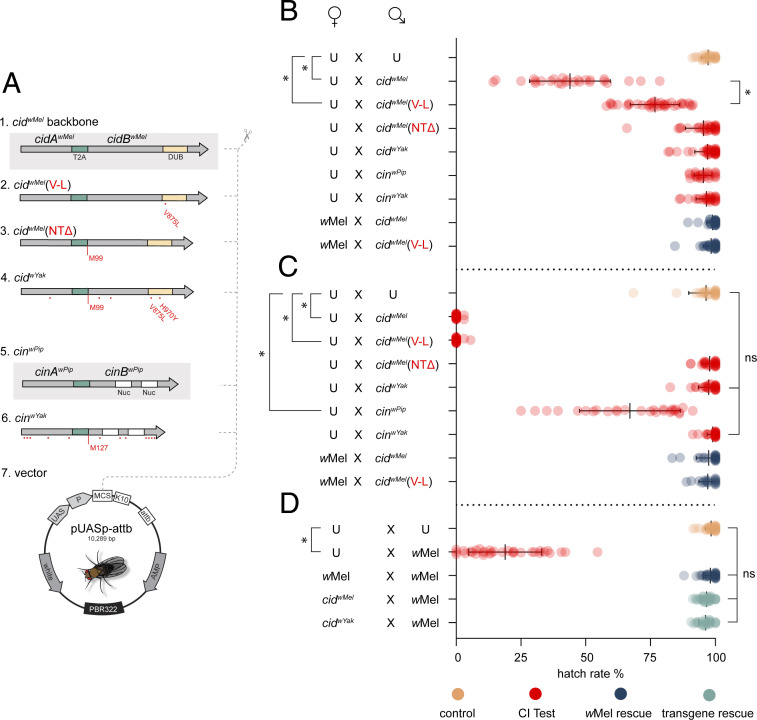
To test the hypothesis that (V-L) and (NTΔ) mutations observed in CidwYak reduce CidwMel CI strength, we next expressed transgenes in Wolbachia-free D. melanogaster males using the GAL4/UAS system (Fig. 2A). When driving transgenes with the nanos-GAL4-tubulin (NGT) driver in males (18, 32, 35), the CidwMel construct causes strong transgenic CI (hatch rate = 44% ± 16% SD, n = 26), measured as the percent of embryos that hatch from a mating pair’s clutch, relative to the hatch produced by uninfected controls (hatch rate = 97% ± 3% SD, n = 29) (Fig. 2B). As predicted, introducing the (V-L) mutation into CidwMel disrupts CI (hatch rate = 77% ± 10% SD, n = 36), reducing CI strength by an average of 2.6-fold (95% CI = 1.8- to 4.9-fold). This further suggests the DUB contributes to CI induction, as predicted (19). A maternal-cytoplasmic wMel infection fully rescues both CidwMel (99% ± 2% SD, n = 30) and weakened CidwMel(V-L) CI (98% ± 3% SD, n = 32) (Fig. 2B). The CidwMel(NTΔ) construct does not cause CI (hatch rate = 95% ± 7% SD, n = 31), indicating that the N terminus of CidB is also essential for CI. While wYak causes sporadic and weak CI in D. yakuba (27), neither the CidwYak (97% ± 5% SD, n = 36) nor the CinwYak (97% ± 5% SD, n = 36) complete constructs induce CI with the weak NGT driver (Fig. 2B). Therefore, we increased transgenic expression using the stronger Maternal Triple Driver (MTD) driver (21, 36) and repeated all crosses (Fig. 2C).
Fig. 2.
Analysis of mutations in a D. melanogaster model. (A) Design of transgene constructs. T2A is a viral sequence causing translation of two proteins. Red dots indicate amino acid changes. (M99) and (M127) label start codons after NTΔ. 1) positive CI control - the cidwMel backbone. 2) cidwMel (V-L) point mutant. 3) cidwMel (NTΔ) starting at M99. 4) cidwYak analog, in effect, wYak wild-type. 5) positive CI control, the cinwPip backbone. 6) cinwYak analog, in effect, wYak wild-type. 7) pUASp-attb insertion vector. (B) Transgenic CI with the weak NGT driver. Males expressing cidwMel cause strong CI relative to uninfected (U). Males expressing cidwMel(V-L) cause hypomorphic CI. wMel cytoplasmic infection rescues CI from males expressing cidwMel or cidwMel(V-L). (C) CI analysis with boosted expression using an MTD driver. Males expressing cidwMel or cidwMel(V-L) cause strong CI that cytoplasmic wMel infections in females fully rescue. Males expressing cinwPip cause weak CI. (D) Transgenic rescue. wMel cytoplasmic infection in males causes strong CI. wMel cytoplasmic infection in females rescues wMel-induced CI. Expression of operons cidwMel or cidwYak in females fully rescues wMel-induced CI. Raw hatch rates are plotted over means and SDs. (*) is P < 10−6, and (ns) is not significant. P values for B and C are calculated from one-tailed Wilcoxon tests with Bonferroni correction for multiple comparisons, while P values for D are calculated by a Kruskal–Wallis analysis with Dunn’s multiple comparison test.
Boosted CidwMel and CidwMel(V-L) expression yielded hatch rates near zero, demonstrating that hypomorphic CI produced by CidwMel(V-L) under the NGT driver increases to CidwMel intensity if sufficiently overexpressed (Fig. 2C). This is in contrast to the (NTΔ) mutant, in which CI is ablated even under boosted expression. These results suggest the (V-L) variant is a true enzymatic hypomorph.
Under similar conditions, our positive Cin control (CinwPip) induces weak CI, whereas neither CidwYak nor CinwYak complete constructs produce a phenotype. In addition, under the boosted driver, both transgenic phenotypes from CidwMel and CidwMel(V-L) are fully rescued by crossing to a maternal (cytoplasmic) wMel infection. Notably, while CidwYak cannot induce CI, when expressed in females, it fully rescues strong CI induced by a cytoplasmic wMel infection in young males (Fig. 2D). This agrees with theory and past results that indicate that selection does not act to maintain CI induction but does act to preserve rescue functions (24, 33, 34).
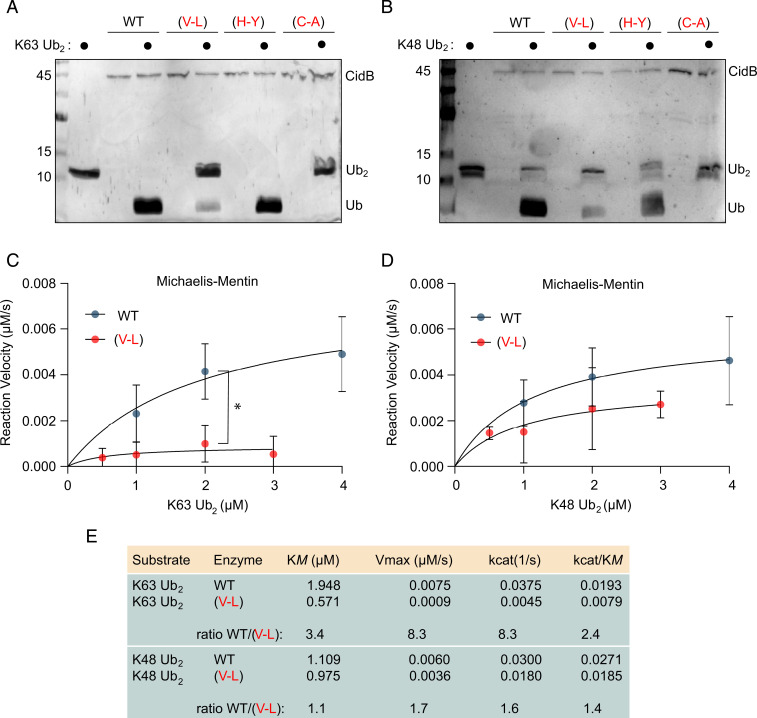
We hypothesized that variation in CI strength results from mutational effects on CidB’s DUB domain. To test this, we made His6-tagged recombinant-protein expression constructs to purify truncated CidBwMel DUB domains. These were then compared with constructs bearing amino acid substitutions found in wYak (H-Y; and V-L; in addition to a catalytic mutant C-A). DUBs were purified by affinity chromatography, quantified by densitometry, and subjected to downstream deubiquitylation kinetics assays (Fig. 3). Previous reports suggest that CidB preferentially cleaves lysine-63 (K63)-linked di-Ubiquitin as opposed to lysine-48 (K48) (19).
Fig. 3.
Ubiquitin cleavage assays with CidBwMel variants. CidBwMel proteins are truncated to the DUB domain (residues H717 through R1128). (V-L) and (H-Y) are substitutions found in CidBwYak. (C-A) is catalytic mutant negative control. Silver-stained SDS-PAGE analysis of 1-h digests with K63-linked di-Ubiquitin (Ub2) (A) and K48 linked Ub2 (B). The (V-L) mutant has reduced DUB activity compared to wild-type (WT). Markers are kDa. Michaelis–Mentin graphs showing reaction velocity versus substrate concentration from timed 5-min digests for K63 (C) and K48 (D). (V-L) reaction velocities are reduced compared to WT. At substrate concentrations of 2 μM, the difference is statistically significant (P = 0.008, unpaired t test with Welch’s correction). (E) Quantification of kinetic parameters for WT and (V-L) enzymes. Enzyme efficiency (kcat/KM) of (V-L) mutants are reduced 2.4-fold and 1.4-fold from the WT for K63 and K48, respectively.
Initial tests showed decreased deubiquitylation from the CidBwMel(V-L) when compared to CidBwMel and CidBwMel(H-Y) (Fig. 3A). We then repeated the tests with K48 linked di-Ubiquitin and poly-Ubiquitin substrates and observed the same patterns, though to a lesser degree (Fig. 3B and SI Appendix, Fig. S4). The K48 substrate data were more variable, and the divergences were not as strong compared to K63. Therefore, we quantified the hypomorphic activity of CidBwMel(V-L) using timed 5-min digests of both K63 and K48 linked di-Ubiquitin in conjunction with Michaelis–Mentin kinetics to measure the precise reduction in enzyme efficiency (Fig. 3 C–E). The (V-L) point mutation strongly reduces CidBwMel ability to bind to K63 (Fig. 3E), while the ability to bind to K48 is almost unchanged (minimal ratio change in KM, refer to value 1.1 in Fig. 3E). Similarly, the ability to cut K63 is more strongly impaired than the ability to cut K48 (comparing ratio changes in kcat values, refer to values 8.3 versus 1.6 in Fig. 3E). Overall, CidBwMel(V-L)’s enzyme efficiency is reduced 2.4-fold for the K63 substrate and 1.4-fold for K48. This indicates that CI is likely induced by an interaction involving K63 Ubiquitin linkages, because the CidBwMel(V-L) mutant also reduced transgenic CI penetrance. These data support a previous hypothesis that CI is induced by cleavage of K63-linked chains from an unknown substrate, perhaps P32 or Karyopherin-α (19, 32).
Discussion
In summary, while we do not know the order in which mutations distinguishing wMel-like cifs occurred, the wYak cid mutations that we predict to influence CI strength do, with one (V-L) reducing CidBwMel CI strength by an average of 2.6-fold and an N-terminal truncation ablating CidBwMel CI by a yet unknown mechanism. Parallel reduction in DUB activity on K63 by an average of 2.4-fold directly correlates CI induction to DUB function. These reductions cannot easily be explained away by reduced transgene expression or protein instability (SI Appendix, Figs. S5 and S6). The valine-to-leucine mutation is conservative, differing by only a single carbon. It is unlikely that the extra carbon unfolds CidB’s tertiary structure or causes significant instability. However, structural analysis will be needed to explore this.
Since wYak causes sporadic and weak CI (27) and because complete CidwYak and CinwYak did not individually induce CI here, our data suggest three possible hypotheses for the existence of relatively weak wYak CI: these operons interact to produce weak CI (20), expression from native Wolbachia contributes to and/or modulates cif-induced CI, and/or host genomes modulate CI strength (32, 37, 38), a pattern documented in natural D. yakuba clade crosses (27). Regardless, we predict that DUB contributions are significant, and our results demonstrate how single mutations may directly weaken CI through their effects on DUB enzymatic activity. Importantly, our discovery of a hypomorphic mutation that preferentially cleaves different Ubiquitin linkages will help identify the penultimate target of CI.
On what timescale is CI disrupted? Disruptions of cifB are prevalent within published genomes and appear to underlie a common process of pseudogenization (34), leading terminally toward rescue-only phenotypes as predicted by theory (24). This process appears to be concurrently playing out in wRi, wYak, and wMau. In these systems, wRi is an early stage example, still bearing strong CI (with 2/3 cifs putatively pseudogenized) (18, 20, 22, 34, 39); wYak is at an intermediate stage, with both cifs putatively pseudogenized (29), yet still capable of weak and sporadic CI (27); and wMau is at a terminal stage, retaining only rescue functions with cifB fully psuedogenized in the last few hundred thousand years (33). Our results demonstrate that individual mutations in cifs can influence the strength of CI caused by wMel-like Wolbachia that diverged in the last 30,000 y. Additional sampling of closely related Wolbachia that differ in CI phenotype is needed to better understand the timing of cif degradation, which is clearly relevant for Wolbachia biocontrol strategies in mosquito systems (11, 13, 40, 41).
Despite weak purifying selection, CI is incredibly common in nature (6). This motivates additional analyses of cif function, unrelated to CI, that might lead to selection pressures that indirectly preserve CI. The prevalence of CI-inducing strains in nature also provides the opportunity for them to move horizontally among host species (42), as demonstrated for wMel-like and wRi-like Wolbachia (26, 29, 43). As previously noted (34), such clade selection may also act on cif genes themselves, which are associated with mobile elements that mediate horizontal cif transfer among divergent genomes (18, 29, 44). Indeed, theoretical analysis supports the plausibility of this process of clade selection on Wolbachia and cifs to explain the contradiction of pervasive CI in nature and cif degradation.
Materials and Methods
DNA Manipulation.
Because Wolbachia cifs are prophage associated and typically flanked by transposable elements (18, 19, 29, 34), reliably assembling these fragmented and repetitive regions is improved with direct cloning (29, 34). To clone and sequence the native cif repertoire of wYak, we extracted genomic DNA from infected D. yakuba. DNA extraction was performed exactly as reported prior (19). In brief, we used a modified organic DNA extraction. A total 10 flies were placed in a tube with 200 µL lysis buffer-A (2% Triton X-100, 1% sodium dodecyl sulphate (SDS), 100 mM NaCl, 10mM Tris pH 8.0, and 1 mM ethylenediaminetetraacetic acid), 200 µL Phenol: Chloroform: Isoamyl alcohol (25:24:1), and ∼200 µL volume of acid-washed glass beads. Flies were homogenized by glass bead disruption in a Bead Ruptor Elite (OMNI International) for four cycles of 20 s on/ 20 s off. While on ice, 200 µL of dH2O was added to the mixture. Insoluble content was pelleted in a microcentrifuge at 13,000 rpm. DNA within the top aqueous phase (400 µL) was transferred into a fresh tube. DNA was precipitated by ethanol precipitation overnight at −20 °C after the addition of 800 µL (2 volumes) of 100% ethanol and 120 µL (= 10% total volume) 3M Sodium Acetate pH 5.4. Precipitate was pelleted, washed in 1 mL 70% ethanol, dried, then resuspended in 1 mL water; native cif operons were cloned by PCR using Phusion polymerase (New England Biolabs) into various yeast, Eshchericha coli, and Drosophila plasmids/vectors (for specifics, refer to construct and primer databases in Datasets S1 and S2). In general, various PCR amplicons produced throughout the study are visualized using 0.8 to 2% agarose gels with ethidium bromide. Mutants were made using site-directed mutagenesis or site-directed, ligase-independent mutagenesis (SLIM) as described previously (31, 32, 45). All construct inserts and variants were fully Sanger-sequenced to confirm DNA fidelity. Final plasmid constructs are transformed into Top10F’ E. coli by standard chemical transformation and stored in glycerol stocks at −80 °C.
Yeast.
We closely follow previous reports (19, 31, 32). Yeast strain W303-1a was used for all serial dilutions. Yeast were transformed using a standard Lithium acetate protocol (46). Wolbachia cifs were expressed under control of the GAL1 promoter in which an alternate sugar source induces expression (2% galactose) or represses expression (2% glucose). In our hands, CidBwPip has most consistently produced robust phenotypes in yeast. Because the mutated residues tested are conserved between CidBwPip and CidBwMel, we opted to use the CidBwPip as the most adept model. Yeast plasmid backbones were pRS416GAL1 (a low-copy centromeric [CEN] plasmid with URA3 selection) (47) and pYes2 (a high-copy 2µ plasmid with URA3 selection). Fivefold serial dilutions from a starting concentration of 0.1 optical density (OD600) were described previously (19). Experiments used solid, minimal, synthetic-defined media lacking uracil. Plates were placed at 34 to 35 °C for 3 d. All yeast serial dilutions show images representative of biological triplicate experiments.
Flies.
All lines were maintained on a 12:12 light:dark cycle at 23 °C on 10 mL of a standard media in vials. A previously described uninfected D. melanogaster stock wCS was used as our focal uninfected background (189) (19). A matching corresponding infected line was prior generated by crossing native wMel Wolbachia into the uninfected line (male wCS X female wMel-infected line) then backcrossing infected females into wCS males for greater than five generations (189wMel) (32). Our transgenic operon flies use the T2A peptide to cause translation of two protein products (19, 31, 32, 48). Backbone transgenic T2A operons from cidwMel, cinwPip, cidwYak, and cinwYak were codon-optimized and synthesized by Genscript. The cidwMel and cinwPip constructs were previously characterized and used as positive control backbones (31, 32). Mutants cidwMel(V-L) and cidwMel(NTΔ) were generated from synthesized backbones by subsequent site-directed mutagenesis and SLIM reactions. All fly construct inserts were subcloned into pUASp-attb26 vector (Fig. 2A) and validated by Sanger-sequencing. UAS transgenes were inserted via ΦC31 integrase into D.mel (BDSC 9744) on chromosome 3 (BestGene Inc.). Homozygous transgenic lines were confirmed by miniwhite selection (red eye phenotype). Two Gal4 lines were used to drive expression of transgenes: NGT (BDSC 4442) and MTD (BDSC 3777). F0 crosses were set up in vials with 10 mL of standard food and kept at 23 °C to generate flies for gene expression analysis and to be used as parents in F1 crosses for hatch rate assays. Multiple F0 crosses of Uninfected (189) or Infected (189wMel) females with GAL4 males produced our focal Uninfected (189/GAL4) or Infected (189wMel/GAL4) flies. At the same time, F0 crosses of GAL4 females with UAS males produced transgene-expressing (GAL4/UAS) flies.
Hatch Rate.
All F0 progeny were collected as virgins and aged to 4 to 5 d-old (except for Infected males used in the experiment for Fig. 2, which were only aged 18 to 24 h to ensure strong wMel CI). Age controlled GAL4/UAS flies were paired with either Infected (189wMel/GAL4) or Uninfected (189/GAL4) flies in vials containing spoons with cornmeal media and yeast paste. After 24 h, pairs were transferred to new spoons, and this process was repeated for up to 5 d. After flies were removed, embryos on each spoon were given 25 h at 23 °C to hatch. The hatch rate is the percentage of embryos that hatched into larvae. Only pairings that produced at least 15 eggs/day were included in analysis. To test for CI and Rescue in Fig. 2 B and C, we used one-tailed Wilcoxon tests with Bonferroni correction for multiple comparisons to compare egg hatch of all potential CI-causing crosses to a focal cross. To test for CI and Rescue in Fig. 2D, a Kruskal–Wallis analysis followed by a Dunn’s multiple comparison test is used. All experiments were carried out at 23 °C with a 12 light:12 dark photoperiod.
RNA Analysis.
To assay transgenic RNA expression levels under the various GAL4 drivers, transgene-expressing flies (that were siblings to those used in hatch rate assays) were collected. For each line, for each sex, eight samples of five abdomens were collected on the day that siblings were ovipositing for hatch rate assays. Abdomens were dissected in ice-cold phosphate-buffered saline and transferred to a nuclease-free 2.0-mL tube containing one 5-mm steel bead chilled in a dry ice/acetone bath. After five abdomens are added to a given tube, it is flash-frozen in liquid nitrogen and stored at −80 °C. Tissues were lysed by two cycles in a Tissue Lyser for 2 min at 25 Hz after adding 350 μL of TRIzol. RNA was then extracted from supernatant using the Direct-zol RNA MiniPrep Kit (Zymo) following the manufactures protocol without the on-column DNase step and eluting RNA in 35 μL of nuclease-free water. Copurifying DNA was degraded using the DNA-free kit (Ambion, Life Technologies). As a quality control for RNA, 2 μL is diluted 1:5, and this 10 μL RNA dilution is checked using the high sensitivity (HS) RNA kit (Qubit). DNA absence is further confirmed with a 28S PCR using GoTaq 2X Mastermix, and thermocycling conditions are the following: one cycle of 94 °C for 2 min, followed by 35 cycles of 94 °C for 30 s, 50 °C for 30 s, and 72 °C for 1 min, and one final cycle of 72 °C for 5 min. For qRT-PCR, RNA was diluted to 31.25 ng/μL, and 500 ng of total RNA was reverse-transcribed to complementary DNA with the SuperScript IV VILO Mastermix (Invitrogen). qPCR was performed on a Bio-Rad CFX-96 Real-Time System in triplicate using PowerUp SYBR Master Mix (Bio-Rad). Each transgene was compared to Drosophila host gene rp49. For details on primers, refer to Dataset S2. Fold expression of transgenes relative to rp49 was determined with 2-ΔΔCt. Each expression study was conducted once.
Cobalt Affinity Protein Purification.
A high-expressing truncated DUB domain from CidBwMel corresponding to residues H717 through R1128 of native sequence was cloned from genomic DNA into the pBadA (arabinose inducible, ampicillin selection) E.coli expression plasmid. A start codon and N-terminal His6 tag were inserted upstream (construct Cu31). Various mutants were built by site-directed mutagenesis off this plasmid backbone including (C-A), (V-L), and (H-Y). Plasmids were transformed by electroporation (Eporator, Eppendorf) into the arabinose-compatible expression strain BL21-AI (Thermo Fisher). For expression, a starter culture was inoculated into 8 L lysogeny broth (LB) ampicillin and shook at 37 °C. Cultures were induced at OD600 0.5 by addition of arabinose to final concentration of 0.02%. Temperature was then shifted to 18 °C, and cultures were shook all night long. Cultures were pelleted by centrifugation in a Sorval RC5B plus floor centrifuge using a SLA-3000 rotor at 16,9000 g for 10 mins. Cell pellets were placed on ice and resuspended in 5 mL binding wash buffer (50 mM sodium phosphate pH 8.0, 300 mM sodium chloride, 0.01% tween-20, and 10 mM Imidazole). To assist lysis, samples were incubated on ice with a pinch of egg white lysozyme (VWR) for 30 min. Samples were then passed twice through French press then centrifuged at ∼5,000 G for 6 h in a Heraeus Multifuge ×1R (Thermo Scientific) at 4 °C. Supernatants with soluble protein extracts were then passed twice through 0.45-μm syringe filters (Acrodisc, Pall) and transferred into a 15-mL conical tube with 2 mL HisPur cobalt beads (ThermoFisher). The tube was inverted at 4 °C with for 30 min. Contents were transferred to a disposable column and pushed through using a peristaltic pump. Column was then washed with 40 column volumes of binding wash buffer. Proteins were eluted off the column by addition of 1 mL elution buffer (50 mM sodium phosphate pH 8.0, 300 mM sodium chloride, 0.01% tween-20, and 300 mM Imidazole). The column was then capped and inverted for 15 min at 4 °C. Eluent was pushed through the column and collected into a fresh tube, followed by a final wash with 1 mL elution buffer yielding 2 mL total eluent. Samples were simultaneously concentrated and buffer-exchanged by centrifugation in a centrifugal filter (Amicon Ultra-4; 3K NMWL). Elution Buffer was exchanged to a storage buffer (50 mM Tris⋅HCl pH 7.6, 150 mM NaCl, and 1 mM dithiothreitol [DTT], 30% glycerol). Purified proteins were aliquoted into 1.5-mL tubes and stored at −80 °C. Protein yields were calculated by comparison to a standard bovine serum albumin curve using densitometry of Coomassie-stained SDS-polyacrylamide gel electrophoresis (PAGE) gels with the Gel Doc Ez Imager (Bio-Rad).
Ubiquitin Cleavage Assays.
Mono-Ubiquitin (Ub), di-Ubiquitin (Ub2), poly-Ubiquitin (all Boston Biochem), and purified CidB were thawed on ice. Ubiquitin digestion assays (total volume 20 µl) were mixed in Ubiquitin cleavage buffer (50 mM Tris HCL pH 7.6, 20 mM KCl, 5 mM MgCl2, and 1 mM DTT) to a final concentration of 0.2 µM enzyme and 2 µM substrate. Samples were incubated at 37 °C for 5 min on a rotator. Reactions were quenched by addition of 5 µl of 4× Laemmli SDS-PAGE buffer. Samples were warmed for 20 min on a hot plate at 40 °C (according to manufacturer specifications). Samples were run on 15% SDS-PAGE gels and stained using a standard silver stain kit (Fast Silver, G Biosciences). Michaelis–Menten kinetics were measured by holding enzyme concentration constant while varying substrate concentrations from 0.5 µM to 4 µM. These gels were silver-stained, and the change in concentration of mono-Ubiquitin [product] was compared to a standard mono-Ubiquitin curve using ImageJ software. Values were transferred to Graphpad Prism software, and plots were created using default Michaelis–Mentin algorithm analysis. Gels and kinetic data represent triplicate digests.
Supplementary Material
Acknowledgments
We thank Michael Turelli, Bill Sullivan, Mark Hochstrasser, Andrew Kern, Daniel Matute, and especially Dylan Shropshire for constructive comments on the manuscript. The editor and two anonymous reviewers also provided valuable comments. We also thank Judy Ronau for biochemical consultation, Kathleen Martin for the use of instruments, and the members of the B.S.C. laboratory for assistance with hatch rate analysis. Research reported in this publication was supported by the National Institute Of General Medical Sciences of the NIH under award number R35GM124701 to B.S.C., and by a US Department of Agriculture Hatch Grant (1015922) and Alabama Agricultural Experiment Station SEED grant to J.F.B. The University of Montana Genomics Core also provided support. Any opinions, findings, conclusions, or recommendations expressed in this material are those of the authors and do not necessarily reflect the views of the NIH.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2113271118/-/DCSupplemental.
Data Availability
All study data are included in the article and/or supporting information.
References
- 1.Rosenwald L. C., Sitvarin M. I., White J. A., Endosymbiotic Rickettsiella causes cytoplasmic incompatibility in a spider host. Proc. Biol. Sci. 287, 20201107 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Takano S. I., Gotoh Y., Hayashi T., “Candidatus Mesenet longicola”: Novel endosymbionts of Brontispa longissima that induce cytoplasmic incompatibility. Microb. Ecol., 10.1007/s00248-021-01686-y (2021). [DOI] [PubMed] [Google Scholar]
- 3.Hunter M. S., Perlman S. J., Kelly S. E., A bacterial symbiont in the Bacteroidetes induces cytoplasmic incompatibility in the parasitoid wasp Encarsia pergandiella. Proc. Biol. Sci. 270, 2185–2190 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yen J. H., Barr A. R., The etiological agent of cytoplasmic incompatibility in Culex pipiens. J. Invertebr. Pathol. 22, 242–250 (1973). [DOI] [PubMed] [Google Scholar]
- 5.Chen H., Zhang M., Hochstrasser M., The biochemistry of cytoplasmic incompatibility caused by endosymbiotic bacteria. Genes (Basel) 11, 852 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shropshire J. D., Leigh B., Bordenstein S. R., Symbiont-mediated cytoplasmic incompatibility: What have we learned in 50 years? eLife 9, e61989 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beckmann J. F., et al., The toxin-antidote model of cytoplasmic incompatibility: Genetics and evolutionary implications. Trends Genet. 35, 175–185 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hoffmann A. A., Turelli M., “Cytoplasmic incompatibility in insects” in Influential Passengers: Inherited Microorganisms and Arthropod Reproduction, O’Neill S. L., Hoffmann A. A., Werren J. H., Eds. (Oxford University Press, 1997), pp. 42–80. [Google Scholar]
- 9.Hoffmann A. A., Turelli M., Harshman L. G., Factors affecting the distribution of cytoplasmic incompatibility in Drosophila simulans. Genetics 126, 933–948 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zug R., Hammerstein P., Still a host of hosts for Wolbachia: Analysis of recent data suggests that 40% of terrestrial arthropod species are infected. PLoS One 7, e38544 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dobson S. L., Fox C. W., Jiggins F. M., The effect of Wolbachia-induced cytoplasmic incompatibility on host population size in natural and manipulated systems. Proc. Biol. Sci. 269, 437–445 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zheng X., et al., Incompatible and sterile insect techniques combined eliminate mosquitoes. Nature 572, 56–61 (2019). [DOI] [PubMed] [Google Scholar]
- 13.Hoffmann A. A., et al., Successful establishment of Wolbachia in Aedes populations to suppress dengue transmission. Nature 476, 454–457 (2011). [DOI] [PubMed] [Google Scholar]
- 14.O’Neill S. L., et al., Scaled deployment of Wolbachia to protect the community from dengue and other Aedes transmitted arboviruses. Gates Open Res. 2, 36 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.O’Neill S. L., The use of Wolbachia by the World Mosquito Program to interrupt transmission of Aedes aegypti transmitted viruses. Adv. Exp. Med. Biol. 1062, 355–360 (2018). [DOI] [PubMed] [Google Scholar]
- 16.World Health Organization , Thirteenth meeting of the WHO Vector Control Advisory Group (2021). https://www.who.int/publications/i/item/9789240021792. Accessed 22 March 2021.
- 17.Utarini A.et al.; AWED Study Group , Efficacy of Wolbachia-infected mosquito deployments for the control of dengue. N. Engl. J. Med. 384, 2177–2186 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.LePage D. P., et al., Prophage WO genes recapitulate and enhance Wolbachia-induced cytoplasmic incompatibility. Nature 543, 243–247 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Beckmann J. F., Ronau J. A., Hochstrasser M., A Wolbachia deubiquitylating enzyme induces cytoplasmic incompatibility. Nat. Microbiol. 2, 17007 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shropshire J. D., Rosenberg R., Bordenstein S. R., The impacts of cytoplasmic incompatibility factor (cifA and cifB) genetic variation on phenotypes. Genetics 217, 1–13 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shropshire J. D., Bordenstein S. R., Two-by-one model of cytoplasmic incompatibility: Synthetic recapitulation by transgenic expression of cifA and cifB in Drosophila. PLoS Genet. 15, e1008221 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Beckmann J. F., Fallon A. M., Detection of the Wolbachia protein WPIP0282 in mosquito spermathecae: Implications for cytoplasmic incompatibility. Insect Biochem. Mol. Biol. 43, 867–878 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shropshire J. D., On J., Layton E. M., Zhou H., Bordenstein S. R., One prophage WO gene rescues cytoplasmic incompatibility in Drosophila melanogaster. Proc. Natl. Acad. Sci. U.S.A. 115, 4987–4991 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Turelli M., Evolution of incompatibility-inducing microbes and their hosts. Evolution 48, 1500–1513 (1994). [DOI] [PubMed] [Google Scholar]
- 25.Martinez J., et al., Should symbionts be nice or selfish? Antiviral effects of Wolbachia are costly but reproductive parasitism is not. PLoS Pathog. 11, e1005021 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Turelli M., et al., Rapid global spread of wRi-like Wolbachia across multiple Drosophila. Curr. Biol. 28, 963–971.e8 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cooper B. S., Ginsberg P. S., Turelli M., Matute D. R., Wolbachia in the Drosophila yakuba complex: Pervasive frequency variation and weak cytoplasmic incompatibility, but no apparent effect on reproductive isolation. Genetics 205, 333–351 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Charlat S., Calmet C., Merçot H., On the mod resc model and the evolution of Wolbachia compatibility types. Genetics 159, 1415–1422 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cooper B. S., Vanderpool D., Conner W. R., Matute D. R., Turelli M., Wolbachia acquisition by Drosophila yakuba-clade hosts and transfer of incompatibility loci between distantly related Wolbachia. Genetics 212, 1399–1419 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zabalou S., et al., Multiple rescue factors within a Wolbachia strain. Genetics 178, 2145–2160 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen H., Ronau J. A., Beckmann J. F., Hochstrasser M., A Wolbachia nuclease and its binding partner provide a distinct mechanism for cytoplasmic incompatibility. Proc. Natl. Acad. Sci. U.S.A. 116, 22314–22321 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Beckmann J. F., Sharma G. D., Mendez L., Chen H., Hochstrasser M., The Wolbachia cytoplasmic incompatibility enzyme CidB targets nuclear import and protamine-histone exchange factors. eLife 8, e50026 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Meany M. K., et al., Loss of cytoplasmic incompatibility and minimal fecundity effects explain relatively low Wolbachia frequencies in Drosophila mauritiana. Evolution 73, 1278–1295 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Martinez J., Klasson L., Welch J. J., Jiggins F. M., Life and death of selfish genes: Comparative genomics reveals the dynamic evolution of cytoplasmic incompatibility. Mol. Biol. Evol. 38, 2–15 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shropshire J. D., Kalra M., Bordenstein S. R., Evolution-guided mutagenesis of the cytoplasmic incompatibility proteins: Identifying CifA’s complex functional repertoire and new essential regions in CifB. PLoS Pathog. 16, e1008794 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Petrella L. N., Smith-Leiker T., Cooley L., The Ovhts polyprotein is cleaved to produce fusome and ring canal proteins required for Drosophila oogenesis. Development 134, 703–712 (2007). [DOI] [PubMed] [Google Scholar]
- 37.Reynolds K. T., Hoffmann A. A., Male age, host effects and the weak expression or non-expression of cytoplasmic incompatibility in Drosophila strains infected by maternally transmitted Wolbachia. Genet. Res. 80, 79–87 (2002). [DOI] [PubMed] [Google Scholar]
- 38.Shropshire J. D., Hamant E., Cooper B. S., Male age and Wolbachia dynamics: Determining how fast and why bacterial densities and cytoplasmic incompatibility strengths vary. bioRxiv [Preprint] (2021). 10.1101/2021.06.01.446638. Accessed 1 June 2021. [DOI] [PMC free article] [PubMed]
- 39.Hoffmann A. A., Turelli M., Simmons G. M., Unidirectional incompatibility between populations of Drosophila simulans. Evolution 40, 692–701 (1986). [DOI] [PubMed] [Google Scholar]
- 40.Turelli M., Barton N. H., Deploying dengue-suppressing Wolbachia: Robust models predict slow but effective spatial spread in Aedes aegypti. Theor. Popul. Biol. 115, 45–60 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ross P. A., Ritchie S. A., Axford J. K., Hoffmann A. A., Loss of cytoplasmic incompatibility in Wolbachia-infected Aedes aegypti under field conditions. PLoS Negl. Trop. Dis. 13, e0007357 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hurst L. D., McVean G. T., Clade selection, reversible evolution and the persistence of selfish elements: The evolutionary dynamics of cytoplasmic incompatibility. Proc. R. Soc. Lond. B Biol. Sci. 263, 97–104 (1996). [Google Scholar]
- 43.Conner W. R., et al., Genome comparisons indicate recent transfer of wRi-like Wolbachia between sister species Drosophila suzukii and D. subpulchrella. Ecol. Evol. 7, 9391–9404 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gillespie J. J., et al., A tangled web: Origins of reproductive parasitism. Genome Biol. Evol. 10, 2292–2309 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chiu J., March P. E., Lee R., Tillett D., Site-directed, ligase-independent mutagenesis (SLIM): A single-tube methodology approaching 100% efficiency in 4 h. Nucleic Acids Res. 32, e174 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gietz R. D., Schiestl R. H., Quick and easy yeast transformation using the LiAc/SS carrier DNA/PEG method. Nat. Protoc. 2, 35–37 (2007). [DOI] [PubMed] [Google Scholar]
- 47.Mumberg D., Müller R., Funk M., Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene 156, 119–122 (1995). [DOI] [PubMed] [Google Scholar]
- 48.Diao F., White B. H., A novel approach for directing transgene expression in Drosophila: T2A-Gal4 in-frame fusion. Genetics 190, 1139–1144 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or supporting information.