Significance
Inactivating genetic variants in the Unfolded Protein Response (UPR) regulator Activating Transcription Factor 6 (ATF6) result in loss of color vision, which is linked to dysfunctional cone photoreceptors and the development of achromatopsia. The pathomechanims involving the lack of functional ATF6 remain unknown; retinal organoids from patient induced pluripotent stem cells and ATF6 null hESCs were generated to elucidate this question. Data from this study identified an unexpected UPR-independent role for ATF6 signaling during cone photoreceptor differentiation in human retinal development, identifying a pathogenic mechanism underlying ATF6-associated achromatopsia. It is believed that pharmacologic targeting of ATF6 may present a viable treatment option for color blinding diseases at early stages of retinal development.
Keywords: cone photoreceptors, retinal organoids, ATF6 signaling, achromatopsia, stem cell biology
Abstract
Endoplasmic reticulum (ER) stress and Unfolded Protein Response (UPR) signaling promote the pathology of many human diseases. Loss-of-function variants of the UPR regulator Activating Transcription Factor 6 (ATF6) cause severe congenital vision loss diseases such as achromatopsia by unclear pathomechanisms. To investigate this, we generated retinal organoids from achromatopsia patient induced pluripotent stem cells carrying ATF6 disease variants and from gene-edited ATF6 null hESCs. We found that achromatopsia patient and ATF6 null retinal organoids failed to form cone structures concomitant with loss of cone phototransduction gene expression, while rod photoreceptors developed normally. Adaptive optics retinal imaging of achromatopsia patients carrying ATF6 variants also showed absence of cone inner/outer segment structures but preserved rod structures, mirroring the defect in cone formation observed in our retinal organoids. These results establish that ATF6 is essential for human cone development. Interestingly, we find that a selective small molecule ATF6 signaling agonist restores the transcriptional activity of some ATF6 disease-causing variants and stimulates cone growth and gene expression in patient retinal organoids carrying these variants. These findings support that pharmacologic targeting of the ATF6 pathway can promote human cone development and should be further explored for blinding retinal diseases.
Activating Transcription Factor 6 (ATF6) encodes an endoplasmic reticulum (ER) resident type 2 transmembrane protein that controls a key signal transduction pathway of the mammalian Unfolded Protein Response (UPR) (1, 2). In response to pathologic or physiologic events that disrupt ER homeostasis, ATF6 migrates from the ER to the Golgi, where proteases cleave ATF6 to release the cytosolic, N-terminal ATF6 transcriptional activator fragment (3). The liberated ATF6 transcription factor enters the nucleus to induce ER chaperones and protein folding enzymes that increase the biosynthetic capacity of the ER, allowing the cell to adapt to episodes of ER stress (4, 5).
In people, variants in ATF6 cause heritable vision loss diseases such as achromatopsia (ACHM) and cone-rod dystrophy (6, 7). We previously found that disease-associated ATF6 variants impair transcriptional activity by interrupting essential steps in the ATF6 signal transduction mechanism (8). For example, Class 1 variants introduced missense changes in the ATF6 luminal domain that impeded ER-to-Golgi trafficking of ATF6, while Class 3 variants altered the ATF6 bZIP domain to prevent DNA binding by the cytosolic ATF6 transcriptional activator fragment (8). Visually impaired patients carry biallelic ATF6 disease alleles, while unaffected (normal vision) family members retain at least one copy of wild-type ATF6 (6, 8). To investigate the cellular basis by which ATF6 disease alleles cause vision loss, we generated induced pluripotent stem cells (iPSCs) from ACHM patients carrying biallelic ATF6 disease-causing variants c.1699T>A (p.Tyr567Asn, referred to ATF6[Y567N] hereinafter) and c.970C>T (p.Arg324Cys, referred to ATF6[R324C] hereinafter) and unaffected, normal vision family members (6, 8). We differentiated these ACHM and normal vision iPSCs into retinal organoids following established protocols and examined their photoreceptors (9).
Results
ATF6-Associated ACHM Patient Retinal Organoids Do Not Form Cone Structures.
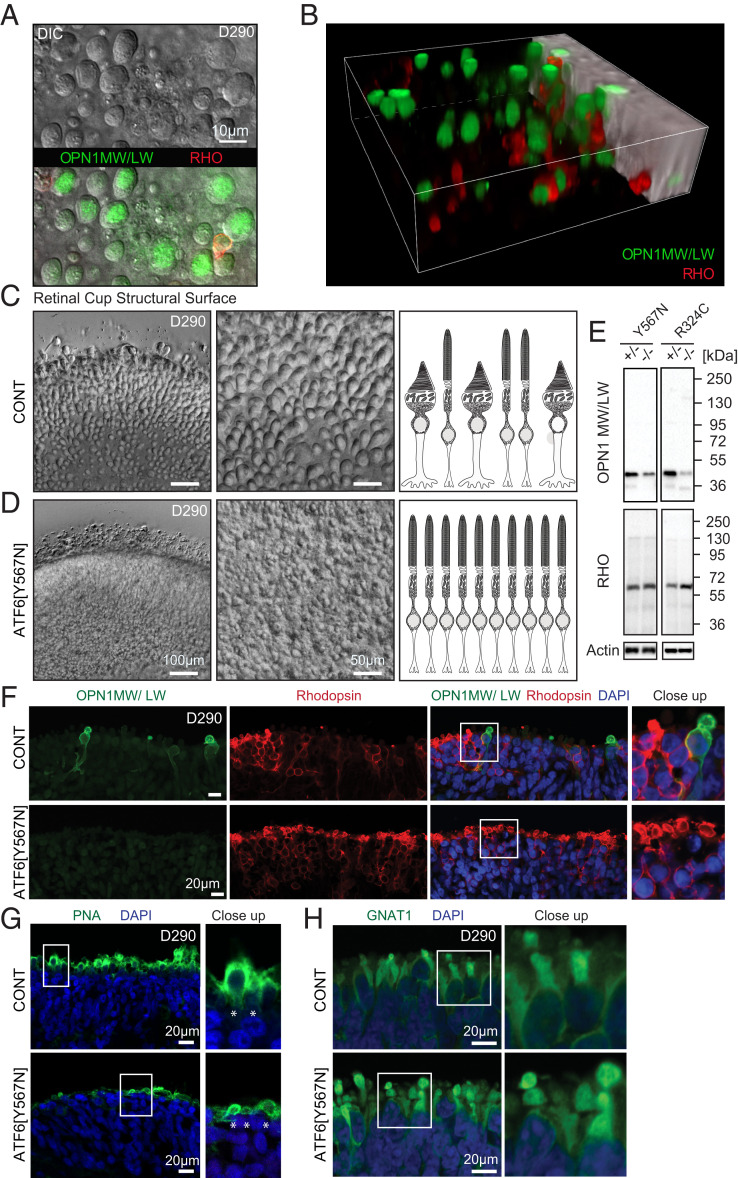
Developing photoreceptors in retinal organoids express cone/rod-specific genes and begin to show polarized cone/rod-like shapes (10–12). When we generated retinal organoids from normal vision patient iPSCs, we saw the emergence of round, ovoid protrusions on the surfaces of retinal organoids by ∼120 days (D) of differentiation that grew progressively larger and more bulbous with extended culturing as demonstrated at D290 (Fig. 1 A and B and Video S1). These ovoid protrusions contained cone opsins and excluded rhodopsin, which was confined to slimmer, cylindrical structures (Fig. 1 A and B and Video S1). These findings were consistent with prior studies showing the emergence of nascent cone and rod photoreceptors in retinal organoids (9, 12). These cone-like protrusions appeared on the surfaces of all retinal organoids developed from control normal vision patients (Fig. 1C and Video S2).
Fig. 1.
ATF6 ACHM patient retinal organoids lack cone structures and cone opsins. (A) Top-down view of DIC microscopy Z-stack series of RHO and cone opsin OPN1MW/LW-labeled retinal organoids shows large round structures on the surface of retinal organoids from normal vision patient control iPSCs (carrying wild-type ATF6). Superimposed immunofluorescence microscopy shows cone opsin OPN1MW/LW-labeling (green) colocalizing with round structures and exclusion of RHO labeling (red). (B) Snapshot from three-dimensional (3D) reconstruction video (Video S1) of confocal fluorescent microscopy images show that round structures on retinal organoid surfaces adopt ovoid morphologies in 3D encapsulating OPN1MW/LW (green) and excluding RHO (red). (C) Images were taken at the same optical magnification; digital zoom was used to demonstrate cellular details of the retinal organoid surface. Overview (Left) and enlargement for detailed view (Middle) DIC microscopy images show abundant cone structures on surface of normal vision patient retinal organoids (CONT). (D) ATF6[Y567N] patient retinal organoids (carrying biallelic inactivating ATF6 alleles) show absence of cone structures with retention of rod structures. Right cartoons in C and D summarize absence of cone structures in ATF6[Y567N] patient retinal organoids. (E) Protein lysates from ATF6[Y567N] and ATF6[R324C] ACHM and control patient retinal organoids (homozygous or heterozygous for ATF6 disease alleles as indicated) were immunoblotted for OPN1MW/LW or RHO. Actin was immunoblotted as a loading control. Protein lysates were combined from five to eight independent retinal organoids of each genotype, and representative immunoblots from three experimental replicates are shown. (F) Confocal fluorescence microscopy images show absence of OPN1MW/LW (green) and preservation of RHO (red) labeling in 290-d (D290)-old ATF6[Y567N] patient retinal organoids (Bottom Rows) compared to normal vision unaffected family member retinal organoid (CONT, Top Rows). DAPI (blue) identifies nuclei. (G) Confocal images show reduced expression of PNA (green) in ATF6[Y567N] (Bottom) retinal organoid compared to CONT (Top) patient retinal organoid. DAPI (blue) labels nuclei. (H) Confocal images show expression of rod transducin/GNAT1 (green) in both control normal vision (CONT, Top) and ATF6[Y567N] patient (Bottom) retinal organoids. For confocal microscopy studies, three independent retinal organoids were analyzed, and representative images are shown. Rhodopsin (RHO), green/red cone opsin (OPN1MW/LW), control (CONT).
Using this differentiation protocol, ACHM patient iPSCs (carrying biallelic ATF6[Y567N] disease variants) formed retinal organoids with no overt differences in differentiation efficiency, organoid size, or shape compared to control retinal organoids (SI Appendix, Fig. S1). However, bulbous protrusions were absent on the surfaces of ACHM patient retinal organoids, and instead, their surfaces had smoother contours (Fig. 1D and Video S3). These morphologic findings suggested a selective defect in cone cells in ACHM retinal organoids. To analyze photoreceptors in more detail, we prepared protein lysates from retinal organoids (generated from ACHM patient iPSCs carrying biallelic ATF6[Y567N] or ATF6[R324C] disease variants) and immunoblotted for cone (Fig. 1E, OPN1MW/LW) and rod (Fig. 1E, RHO) markers. We saw significantly reduced OPN1MW/LW protein levels in ACHM retinal organoids carrying biallelic ATF6 disease alleles compared to controls (Fig. 1E). Consistent with these biochemical findings, green/red cone opsin (OPN1MW/LW) protein expression was not detected by confocal immunofluorescence microscopy in nascent photoreceptors on sectioned ATF6[Y567N] patient retinal organoids compared to controls (Fig. 1F). Absence of cone-like structures and cone green/red opsin expression was observed in ACHM patient retinal organoids at all timepoints examined during retinal organoid differentiation (up to ∼D300). ATF6[Y567N] ACHM patient retinal organoids also showed significant loss of labeling by peanut agglutinin (PNA), a lectin that selectively labels cone outer segments (OS) (Fig. 1G, see asterisk), and interestingly, the remaining PNA staining in ATF6[Y567N] ACHM retinal organoids labeled flatter structures instead of the ovoid protrusions in control organoids (Fig. 1G, see asterisk in close-up). By contrast, ATF6[Y567N] ACHM patient retinal organoids formed abundant rod cells as determined by expression of rhodopsin and rod transducin (GNAT1) protein expression (Fig. 1 F and H). These retinal organoid findings suggested an early and selective defect in cone formation in ACHM patients carrying ATF6[Y567N] or ATF6[R324C] disease variants.
To further test if cone development in retinal organoids required ATF6, we next created isogenic ATF6 null hESCs by CRISPR-mediated indel introduced into exon 1 of the human ATF6 gene (ATF6ex1D/ex1D) in wild-type hESCs (SI Appendix, Fig. S2A). We confirmed complete loss of ATF6 protein in ATF6ex1D/ex1D cells and down-regulation of canonical downstream ATF6 transcriptional target genes such as GRP78/BiP/HSPA5 (SI Appendix, Fig. S2 B and C) (4, 5). When we differentiated isogenic ATF6+/+ and ATF6ex1D/ex1D hESCs into retinal organoids, we saw abundant ovoid conical structures on wild-type organoid surfaces but no ovoid protrusions on ATF6-null retinal organoids, similar to the phenotype observed in retinal organoids derived from the ATF6[Y567N] ACHM patients bearing inactivating ATF6 disease variants (SI Appendix, Fig. S2 D and E and Videos S4 and S5). These ATF6-null retinal organoid findings from isogenic hESCs were consistent with the cone defects observed in ATF6[Y567N] ACHM patient retinal organoids carrying loss-of-function ATF6 disease alleles.
ATF6-Associated ACHM Patients Do Not Have Cone Inner/Outer Segments.
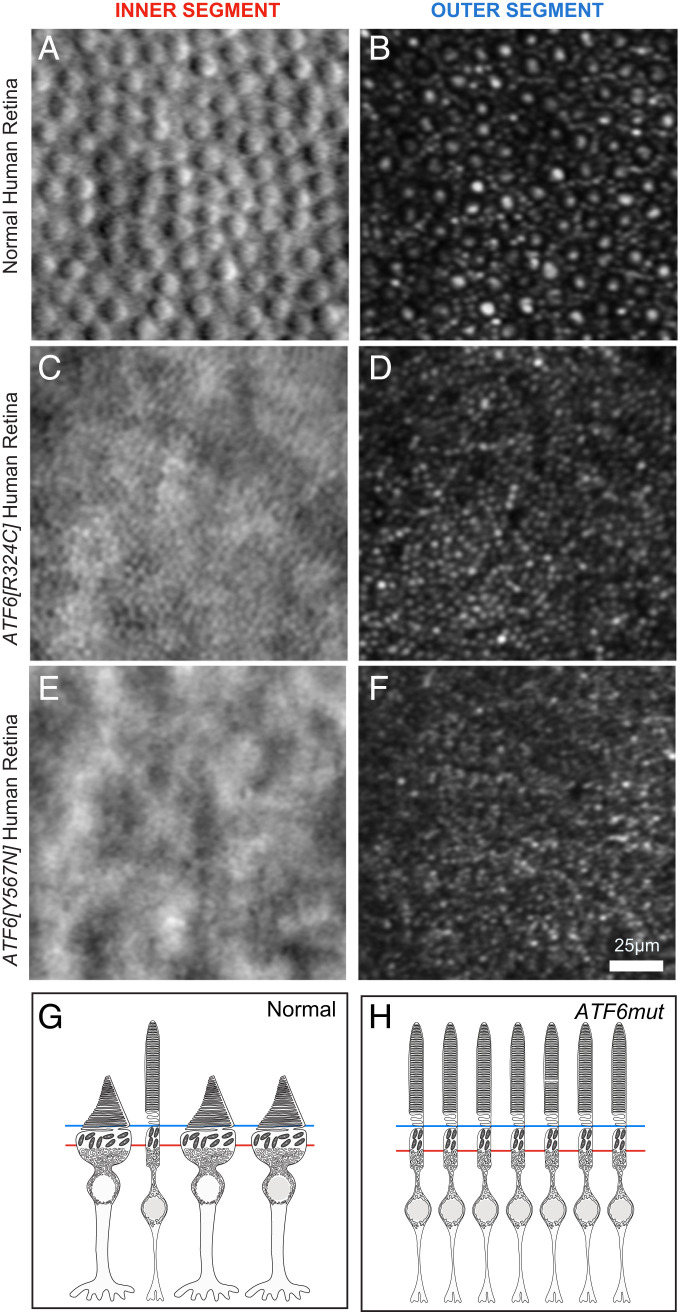
To determine if the defects in cone formation found in the retinal organoids occurred in ACHM patients, we examined adaptive optics scanning laser ophthalmoscopy (AOSLO) imaging data collected from the ACHM patients carrying the ATF6[Y567N] or ATF6[R324C] disease alleles who provided iPSCs for our retinal organoid studies (Fig. 1) (13). AOSLO enables noninvasive, imaging of the human retina at single-cell resolution (14). Cones and rods can be distinguished by cellular morphology in patients by AOSLO imaging of their retinas in planes orthogonal to the polarized photoreceptor cell bodies (15). Specifically, we examined patient AOSLO data collected at the levels of the photoreceptor OS and the photoreceptor inner segment (IS), subcellular regions of the photoreceptor that are markedly wider (conical) in mature cones compared to rods (16–18). In normal vision individuals carrying wild-type ATF6, AOSLO retinal imaging showed numerous large, circular, cone inner and outer segments (IS/OS) interspersed in a mosaic of slender rod IS/OS structures (Fig. 2 A and B). By contrast, AOSLO imaging of ACHM patients carrying biallelic inactivating ATF6 alleles showed near complete absence of cone IS/OS structures but retention of slender rod IS/OS structures (Fig. 2 C and D for ATF6[R324C] and Fig. 2 E and F for the ATF6[Y567N] patient). These patient findings are consistent with prior imaging studies of ACHM patients carrying other ATF6 disease alleles that showed absence of cone IS/OS structures with AOSLO imaging (13). Furthermore, longitudinal AOSLO imaging of ACHM patients at follow-up evaluations (up to 3-y duration) showed that the absence of cone IS/OS structures was a stationery phenotype (no episodes of growth/decay). These imaging findings identify absence of cone structures as a cellular defect in ACHM patients carrying inactivating ATF6 disease variants. These imaging findings support that the absence of conical protrusions on ATF6 mutant retinal organoids recapitulate the cellular defect found in ACHM patient retinas. More broadly, these imaging findings, coupled with the congenital onset of cone dysfunction in these patients, support a critical requirement for ATF6 in human cone photoreceptor development.
Fig. 2.
Adaptive optics imaging of ACHM patient retinas carrying inactivating ATF6 variants reveal absence of cone IS and OS. (A and B) Abundant cone IS (A) and OS (B) are identified in the retina of a normal vision patient (29-y-old female) (ATF6+/+) by split detector (A) and confocal (B) AOSLO (large round structures). (C and D) AOSLO of an ACHM patient (49-y-old female) carrying biallelic inactivating ATF6[R324C] variant shows absence of cone IS (C) and OS (D) but preservation of rod IS and OS. (E and F) AOSLO of an ACHM patient (25-y-old female) carrying biallelic inactivating ATF6[Y567N] variant shows absence of cone IS (E) and OS (F) but preservation of rod IS and OS. (G and H) Schematic cartoons model AOSLO images at the planes of the IS (red line) and OS (blue line) and summarize absence of cone structures in the ACHM patients carrying ATF6 mutations.
Disruption of Cone Opsin and Phototransduction Gene Expression in ACHM Patient ATF6[Y567N] Retinal Organoids.
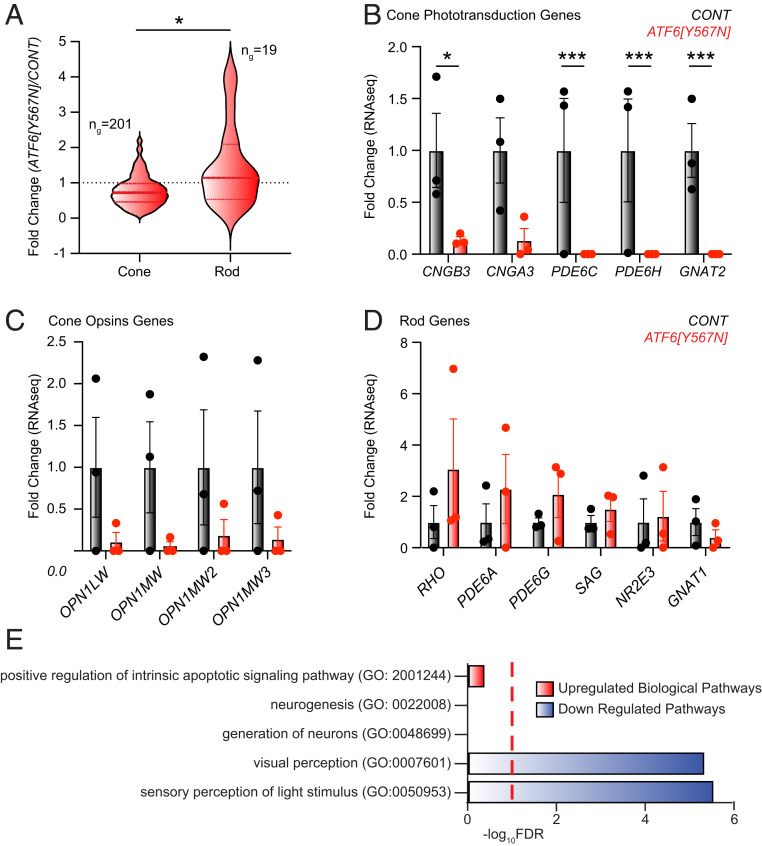
To further investigate the role of ATF6 in developing cones, we next performed bulk ribonucleic acid sequencing (RNA-seq) on retinal organoids prepared from ATF6[Y567N] ACHM patient iPSCs and normal vision iPSCs (Dataset S1). We then compared the expression of cone-specific and rod-specific genes, previously defined by human retina single-cell transcriptomic profiling (19), in these organoids (Dataset S2). The median expression of the cone-specific genes was moderately reduced in ACHM patient retinal organoids relative to control retinal organoids, affecting ∼70% of genes in the cone gene panel (Fig. 3A and Dataset S2). Furthermore, the expression of all cone phototransduction genes, including CNGB3, CNGA3, PDE6C, PDE6H, and GNAT2, was severely reduced in ACHM patient retinal organoids (Fig. 3B). Interestingly, inactivating variants of these cone phototransduction genes also cause ACHM, which is clinically indistinguishable from that observed in patients harboring disease-associated ATF6 alleles (6). Although more variable between individual replicates, cone visual pigment genes including red (OPN1LW) and green (OPN1MW) cone opsin also showed reduced expression in ATF6 mutant retinal organoids (Fig. 3C), consistent with the reduced expression of the green/ red cone opsins observed by immunoblotting and fluorescent microscopy (Fig. 1 E and F). By contrast, the median expression of the rod gene panel, including rhodopsin (RHO) and most rod phototransduction genes, was not significantly altered in ACHM patient retinal organoids (Fig. 3 A and D and Dataset S2). These findings suggested a striking defect in the expression of cone genes required for light detection and phototransduction in ATF6[Y567N] ACHM patient retinal organoids expressing loss-of-function ATF6.
Fig. 3.
Cone visual pigments and cone phototransduction gene expression are severely reduced in ACHM patient retinal organoids. (A) RNA-seq profiling reveals significant loss of cone photoreceptor transcripts in ACHM retinal organoids compared to control retinal organoids. Violin plots show expression levels of 201 cone photoreceptor genes and 19 rod photoreceptor genes identified by RNA-seq in ATF6[Y567N] ACHM retinal organoids, normalized to control retinal organoids. The thick, dashed horizontal line marks the median level of gene expression, and the thin horizontal lines delimit the upper and lower quartiles of genes in each violin plot. Cone- and rod-specific genes were defined in ref. 19. The complete gene sets are shown in Dataset S2. *P < 0.05 by Welch’s t test, t = 2.590. (B–D) Fold change of cone phototransduction gene transcripts (B), cone opsin (visual pigment) gene transcripts (C), and rod phototransduction gene transcripts (D) measured by RNA-seq from normal vision patient retinal organoids (gray bars) and ATF6[Y567N] ACHM patient retinal organoids (red bars). Individual replicates are shown, and error bars show mean ± SEM. ***P < 0.005, *P < 0.05 measured from DESEQ analysis (Dataset S1). (E) GO analysis identifies up-regulated (red) and down-regulated (blue) biological processes in RNA-seq transcriptional datasets from ATF6[Y567N] ACHM retinal organoids compared to control. Red dashed line indicates FDR = 0.1. The complete GO analysis is shown in Dataset S3.
Consistent with this, gene ontology (GO) analysis of the retinal organoid gene expression profiles also identified that the most significantly down-regulated GO biologic processes in ATF6[Y567N] ACHM patient mutant retinal organoids were visual perception and light detection (Fig. 3E and Dataset S3). Furthermore, we confirmed that the expression of cone-specific genes (Dataset S2) was reduced in ATF6[Y567N] patient retinal organoids by Gene Set Enrichment Analysis (GSEA; SI Appendix, Fig. S3A, false discovery rate [FDR] = 0.09). By contrast, genes involved in neurogenesis and neuronal development GO biological processes were not significantly changed in ACHM patient ATF6[Y567N] mutant retinal organoids (Fig. 3E and Dataset S3) but showed some reduction by targeted examination using GSEA (SI Appendix, Fig. S3 B and C, FDR = 0.1). Expression of CRX also showed reduction in ATF6[Y567N] ACHM patient retinal organoids, but other cone/rod photoreceptor cell fate genes were not significantly altered (SI Appendix, Fig. S3D) (20). These findings suggested that early aspects of photoreceptor cell fate commitment and identity specification were aberrant in ACHM patient retinal organoids in addition to the markedly negative effect on cone photodetection/transduction pathways.
GO and GSEA showed no significant change in apoptosis pathway gene expression (Fig. 3E and SI Appendix, Fig. S3E and Dataset S3). Furthermore, no increased expression of genes involved in ER stress-induced cell death including CHOP/DDIT3, DR5, CKB, and ATF4 was found in ATF6[Y567N] mutant retinal organoids (SI Appendix, Fig. S3F) (21–25). These results suggested that loss-of-function mutations in ATF6 did not trigger profound cell death in retinal organoids consistent with visual observations of no gross differences in organoid differentiation, growth, or appearance (SI Appendix, Fig. S1).
A Small Molecule ATF6 Agonist Rejuvenates Diseased Cone Photoreceptors.
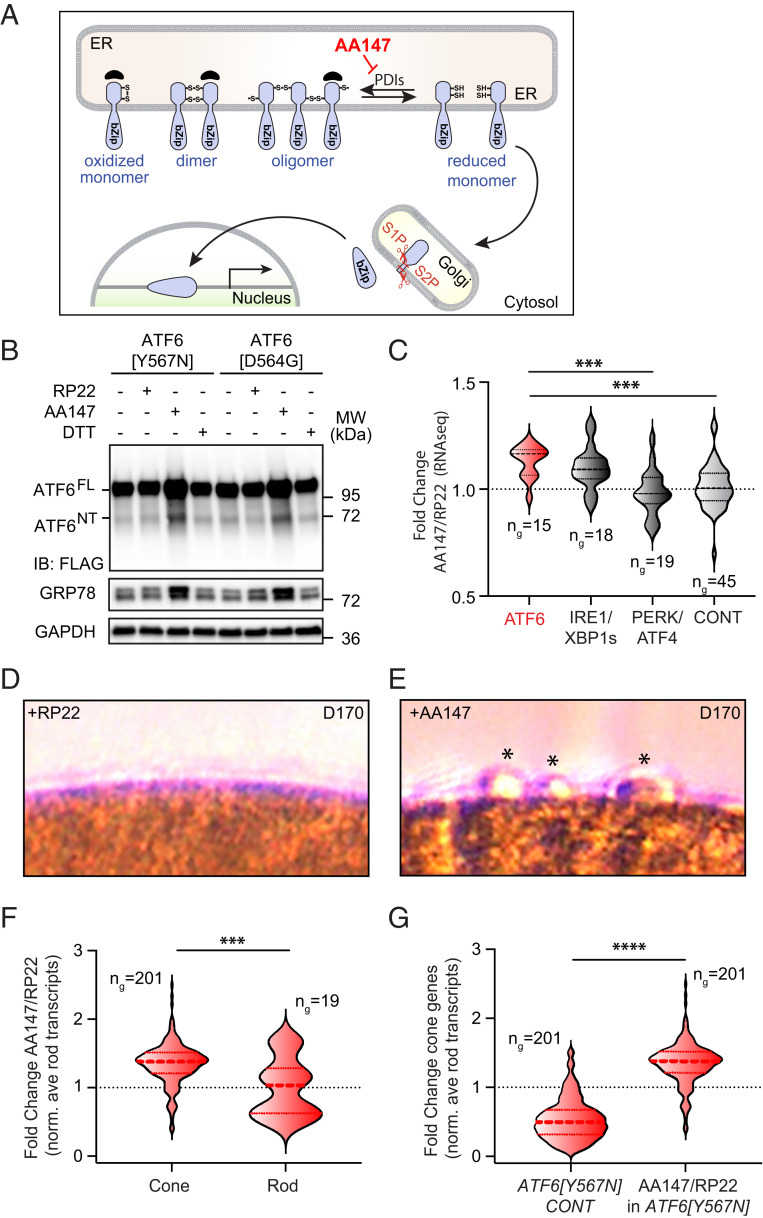
All ATF6 disease alleles identified to date impair ATF6 signaling (6–8, 26). Therefore, augmentation of ATF6 signaling could help patients with vision loss diseases arising from loss-of-function alleles. However, this strategy needs to be precisely tailored to the specific loss-of-function pathomechanisms caused by different ATF6 disease alleles (8). We previously identified soluble, nontoxic small molecules that increase ATF6 transcriptional signaling in cell culture, pluripotent stem cells, and mice by promoting ATF6 reduction and monomerization (Fig. 4A) (27–29). By increasing this pool of reduced ATF6 that exits the ER, these compounds increase the amount of proteolytically cleaved ATF6 transcriptional activator fragment available for downstream signaling (30). These small molecules offer an attractive chemical strategy to counter the molecular pathomechanism of Class 1 ATF6 disease alleles arising from decreased ATF6 release from the ER. To test this, we evaluated the efficacy of a lead small molecule ATF6 agonist, AA147, to increase transcriptional activity of the Class 1 ATF6 variants, Y567N and D564G, found in patients with ACHM and cone-rod dystrophy (6–8). Consistent with prior studies, treatment of HEK293 cells expressing the Class 1 ATF6 disease-associated proteins with the chemical ER stressor, dithiothreitol (DTT), did not induce production of the active ATF6 transcription factor (ATF6NT) or increase levels of the ATF6 target protein GRP78/BiP (Fig. 4B) (8). By contrast, media supplementation with AA147, but not the inactive AA147 analog RP22 (27), robustly restored production of the ATF6 transcriptional activator fragment from both Class 1 mutant proteins and increased protein levels of the ATF6 target protein, GRP78/BiP (Fig. 4B). These findings demonstrate that AA147 promotes transcriptional activity of Class 1 human ATF6 ACHM -associated disease variants.
Fig. 4.
Small molecule proteostasis agonist rescues ATF6 disease mutant transcriptional activity and promotes cone development in mutant retinal organoids. (A) Schematic cartoon shows mechanism of ATF6 signaling pathway activation by the small molecule proteostasis regulator AA147. AA147 inhibits the activity of protein disulfide isomerases (PDIs) to increase the population of trafficking competent, reduced ATF6 monomers in the ER. (B) HEK293 cells expressing FLAG-tagged ATF6 bearing [Y567N] or [D564G] disease mutations were cultured with AA147 (10 μM), RP22 (10 μM), DTT (2 mM), or DMSO solvent for 24 h, and protein lysates were immunoblotted for FLAG, GRP78, or GAPDH (loading control). Positions of full-length ATF6 (ATFFL) and the ATF6 transcriptional activator amino-terminal fragment (ATF6NT) proteins are shown. (C) Expression of ATF6 target genes, IRE1/XBP1s target genes, PERK/ATF4 target genes, and control genes measured by RNA-seq analysis of RNA isolated from pooled D170 retinal organoids (n = 2 for each condition) treated with AA147 (10 µM) or RP22 (10 µM) for 50 d. The thick, dashed horizontal line marks the mean, and the thin horizontal lines delimit the upper and lower quartiles of gene expression in each violin plot. The gene sets used were defined and analyzed as in ref. 31. Gene sets are shown in Dataset S5. ***P < 0.005 by Brown–Forsyth and Welch ANOVA. (D and E) DIC imaging shows presence of nascent conical protrusions (*) on ACHM [Y567N] variant patient retinal organoid surfaces after AA147 (10 μM) (E) but not RP22 (10 μM) treatment (D) for 50 d. (F) Comparison of the expression of cone-specific and rod-specific genes, measured by RNA-seq (Dataset S4) and normalized to the average rod-specific gene expression, in D170 ATF6[Y567N] retinal organoids treated for 50 d with AA147 (10 µM) relative to the inactive control compound RP22 (10 µM). Cone- and rod-specific genes were defined in ref. 19. Thick dashed line marks the mean, and the thin horizontal lines delimit the upper and lower quartiles of gene expression. The complete gene sets are shown in Dataset S6. ***P < 0.005, t = 3.421 measured by Welch’s t test. (G) The relative ratios of cone gene expression normalized to rod gene expression, measured by RNA-seq, in ATF6[Y567N] ACHM patient retinal organoids compared to normal vision patient control organoids (Left) and in AA147-treated compared to RP22-treated ACHM patient retinal organoids (Right). Thick dashed line marks the mean, and the thin horizontal lines delimit the upper and lower quartiles of gene expression. The complete gene sets are shown in Dataset S6. ****P < 0.0001. t = 26.80 by Welch’s t test.
Next, we examined the effects of AA147 in ACHM retinal organoids expressing biallelic copies of ATF6[Y567N]. We modified our retinal organoid differentiation protocol to incorporate AA147 or the inactive analog RP22 to the retinal organoids culture media from D120 of differentiation and then further differentiated these organoids in the presence of compounds for another D50 (SI Appendix, Fig. S4). RNA was isolated from pooled D170 ATF6[Y567N] organoids cultured in the presence of AA147 or RP22 (n = 2 organoids/condition). Gene expression was then analyzed by RNA-seq (Dataset S4). Using a targeted expression analysis (31), the expression of genes regulated by different UPR pathways (ATF6, IRE1/XBP1s, and PERK/ATF4) relative to a set of control genes were compared. We saw increased expression of ATF6 target genes in AA147-treated ATF6[Y567N] ACHM retinal organoids as compared to RP22-treated organoids (Fig. 4C and Dataset S5) (31). By contrast, we did not observe significant increases in expression of PERK/ATF4 target genes in AA147-treated organoids, indicating that this compound did not activate the entire UPR in retinal organoids (Fig. 4C and Dataset S5). We did observe a modest increase in XBP1s target gene expression in AA147-treated organoids (Fig. 4C and Dataset S5), reflecting the overlap between ATF6 and XBP1s transcriptional signaling previously described (27). However, the stronger induction of ATF6 target genes relative to XBP1s target genes in AA147-treated organoids is consistent with the preferential activation of ATF6 signaling previously reported for this compound (27, 28). Collectively, these results indicate that AA147 preferentially activates ATF6 signaling in retinal organoids and is consistent with results found in other cellular and in vivo models (27–29).
When we microscopically examined the surface morphology of ATF6[Y567N] ACHM retinal organoids at D170, we saw the emergence of ovoid protrusions on the surface of AA147-treated organoids, while RP22-treated ACHM organoids remained completely smooth (Fig. 4 D and E). These protrusions on D170 AA147-treated ATF6[Y567N] ACHM retinal organoids resembled smaller versions of the bulbous structures seen in the older D290 retinal organoids from normal vision patients and ATF6+/+ hESCs (Fig. 1C and SI Appendix, Fig. S2D) and suggested that AA147 reinstated cone photoreceptor development and maturation in an ACHM patient’s retinal organoids.
We then further defined the impact of AA147 on ACHM patient retinal organoids by comparing the relative expression of cone-specific and rod-specific genes in our RNA-seq analysis of RNA from pooled D170 ATF6[Y567N] retinal organoids prepared in the presence of AA147 or RP22 (Dataset S4). Interestingly, the expression of many cone transcripts, relative to rod transcripts, significantly increased in ATF6[Y567N] ACHM patient retinal organoids treated with AA147, as compared to those treated with RP22 (Fig. 4F and Dataset S6). This further indicates that AA147 rescues the deficiency in cone photoreceptor development in retinal organoids prepared from ACHM patients biallelic for ATF6[Y567N]. This can be further demonstrated by the ∼50% reduction in the relative expression of cone-specific genes, relative to rod-specific genes, in retinal organoids prepared from ACHM patients biallelic for ATF6[Y567N], as compared to organoids prepared from normal vision patients (Fig. 4G and Dataset S6). Treatment of ATF6[Y567N] ACHM patient retinal organoids with AA147 rescued this deficit. This indicates that pharmacologic strategies that rescue the transcriptional activity of Class 1 human ATF6 variants (e.g., Y567N) can stimulate cone photoreceptor development in ACHM patient retinal organoids biallelic for these types of disease-causing variants.
Discussion
ATF6 is a key transcriptional regulator of the UPR that ensures the ER organelle synthesizes high-quality proteins and lipids in human cells throughout life. However, it is unknown why inactivating variants of ATF6 cause vision loss in people. Here, we combine human stem cell retinal organoid models, high-resolution patient retinal imaging, and small molecule proteostasis modulators to show that ATF6 function is essential for human cone photoreceptor development. When ATF6 function is abrogated either with ATF6 disease variants or by indel insertion knockout, we find a selective disruption in cone development in retinal organoids and complete loss of cone polarized OS/IS structures in patient retinas. By contrast, rods develop in ACHM retinal organoids, and rod structures are present in adult ACHM patients carrying ATF6 disease alleles.
Why is ATF6 so critical for human cone development? ATF6−/− mouse fibroblasts and ACHM patient fibroblasts both show increased cell death in response to ER stress-inducing toxins (4, 5, 8), and ACHM cones and/or cone progenitor cells are also likely to be more sensitive to ER stress-inducing agents. However, ATF6−/− mice have normal cone function until relatively late in life, whereas patients carrying pathogenic human ATF6 variants have early (congenital) cone dysfunction (6), and patient retinal organoids also show developmental cone defects (Fig. 1). These may reflect intrinsic biologic differences in susceptibility to ER stress between murine and human cones associated with their significantly altered gene expression profiles (19). These may also reflect differences in environmental ER stressors encountered by murine and human cones, such as ambient light (32, 33), due to the nocturnal versus diurnal lifestyles of mice and people. Characterizing physiologic sources of ER stress in the developing human eye could provide insight into ATF6’s function in human cones. Importantly, we saw no induction of ER stress-induced cell death markers or cell death pathways in bulk RNA-Seq of ACHM retinal organoids, and this argues that ER stress occurs at sublethal levels during human retinal development.
Our retinal organoid studies identify a molecular strategy that may help diseased cones in ATF6-associated ACHM. In patients carrying Class 1 ATF6 disease variants, the full-length precursor ATF6 protein is trapped at the ER and unable to free its amino-terminal cytosolic transcriptional activator fragment. We show that we can overcome this defect using a small molecule that promotes ATF6 exit out of the ER. Using this strategy, we demonstrate that AA147 treatment restores production of the amino-terminal cytosolic ATF6 transcription factor fragment from Class 1 ATF6[Y567N] disease variants. This proteostasis modulator treatment results in up-regulation of an ATF6-regulated transcriptional expression program in retinal organoids carrying Class 1 ATF6[Y567N] variants and begins to stimulate cone gene expression and morphology in these ACHM patient retinal organoids. We anticipate even greater cone rescue in our retinal organoid model system can be achieved through optimization of small molecule proteostasis treatment conditions and timing and that this strategy will be effective for additional ATF6 variants that cause vision loss by an impaired intracellular trafficking pathomechanism.
Materials and Methods
Cell Culture.
Human somatic cell lines (fibroblasts and HEK293T cells) were maintained at 37 °C, 5% CO2 in Dulbecco’s modified Eagle medium (DMEM) (Mediatech), supplemented with 10% fetal bovine serum (FBS) (Mediatech), and 1% penicillin/streptomycin (Invitrogen). Primary human fibroblast cells were established from skin biopsies of ACHM patients expressing biallelic ATF6 disease alleles (including the R324C and Y567N missense variants) or unaffected family members retaining wild-type ATF6 as heterozygous carrier following institutionally approved protocols and deidentified as previously described (6–8). Fibroblasts were reprogrammed into iPSCs using the CytoTune-iPS 2.0 Sendai Reprogramming kit (Life Technologies), and three to five independent clones were created from each patient donor. All iPSC lines expressed pluripotency markers, had normal karyotypes, and were able to differentiate into all three germ layers using the embryoid body procedure. Early passage P35 hESC HUES9 clones were obtained from the Human Embryonic Stem Cell Core Facility at the Sanford Consortium for Regenerative Medicine at the University of California San Diego (UCSD). All iPSC and hESC lines were maintained on Corning Matrigel coated dishes (Corning Inc.) using mTESR1 medium (Stemcell Technologies) at 37 °C and 5% CO2. Medium was changed daily, and pluripotent stem cells were passaged every 5 to 7 d using ReLeSR medium (Stem Cell Technologies).
Generation of Retinal Organoids.
Pluripotent iPSCs and hESCs were differentiated into retinal organoids as previously described (ref. 9 and SI Appendix, Figs. S1 and S4B). In brief, pluripotent stem cells were dissociated from the culture dish at 75% confluency using dispase (day 0, d0). To induce aggregate formation, small cell clumps were cultured in suspension using mTESR1 media supplemented with 10 µM Blebbistatin (Sigma) for the first 24 h of differentiation. Early retinal differentiation was induced by transitioning the aggregates gradually to NIM1 media (DMEM/F12 [1:1], 1% N2 supplement [Invitrogen], 1× nonessential amino acids [NEAAs], and 2 mg/ml heparin [Sigma]). The ratios of media changes of mTESR1/10 µM Blebbistatin to Nim1 were 3:1 on d1; 1:1 on d2; and 100% Nim1 on d3. On d7, aggregates (∼20 aggregates/cm2) were plated on growth-factor-reduced Matrigel (BD Bioscience) coated dishes using Nim1 as culture media. Nim1 media was changed every 48 h. On d16, media was changed daily using Nim2 (DMEM/F12 [3:1] supplemented with 2% B27 [without vitamin A, Invitrogen], 1× NEAA, and 1% antibiotic–antimycotic [Gibco]). On the fourth week of differentiation, horseshoe-shaped neuronal retinas (NR) were manually collected using an inverted microscope, under a sterile tissue culture hood and a tungsten needle (SI Appendix, Fig. S1). To encourage the formation of retinal cups, collected NRs were cultured in suspension using Nim2 media at 37 °C and 5% CO2, media changes occurred twice a week. On d42, retinal cups were transitioned to long-term culture conditions using Nim3 media (DMEM/F12 [3:1] supplemented with 2% B27 [without vitamin A, Invitrogen], 1× NEAA and 1% antibiotic–antimycotic [Gibco], 10% fetal bovine serum [Gibco], 100 mM Taurine [Sigma], and 2mM GlutaMAX [Invitrogen]). To induce photoreceptor maturation, retinal cups were exposed to Nim3 media and 1 µM retinoic acid (RA, Sigma) from d70 to d98, the concentration of RA was reduced to 0.5 µM thereafter.
The retinal organoid data presented in this study derive from cell passages 10 to 21 iPSCs carrying the c.1699T>A (p.Tyr567Asn, referred to ATF6[Y567N]) and c.970C>T (p.Arg324Cys, referred to ATF6[R324C]) ATF6 variant as homozygous or heterozygous alleles. Passages for the transgenic lines HUES9, P35-45 and ATF6ex1D/ex1D, P8-15 were used. Dataset S7 summarizes cell lines and cones for this research study; no differences were found between iPSC or ESC clones of the same ATF6 genotype.
Generation of ATF6ex1D/ex1D HUES9 hESCs.
CRISPR/Cas9 editing of the ATF6 gene in hESCs was carried out by ThermoFisher Scientific Cell Model Services using the guide RNA (gRNA) “cgggctaaaaggtgactcca” to introduce indels into exon 1 of the human ATF6A gene. Sanger and next-generation sequencing (NGS) of isolated and expanded hESC clones revealed homozygous knockout clones (ATF6ex1D/ex1D). For Sanger sequencing, in brief, genomic DNA was isolated from hESC clones, and an ∼200- base pair (bp) fragment of genomic DNA encompassing the CRISPR/Cas9 editing site in the ATF6 gene was PCR amplified using forward primer, AAGTAGTTTGTCTTTACTAGGC, and reverse primer, TGAGTGGGATCTGAGAATGT, to confirm homozygous 1 to 2 bp indels in exon 1 of the 3 ATF6ex1D/ex1D clones. NGS of the parental unedited hESCs (ATF6+/+) and three expanded knockout clones (ATF6ex1D/ex1D) further revealed no off-target cleavage in gene-edited clones compared to parental clone.
All stem cell studies followed ethical guidelines with Embryonic Stem Cell Research Oversight (ESCRO) and Institutional Review Boards (IRB) approval.
Compounds.
ATF6-activating compound AA147 and its analog RP22 were prepared in DMSO as stock solution at 10 mM, and working aliquots were prepared and stored at −20 °C. The compound was used in cell culture medium at a final concentration of 10 μM. ER stress-inducing compound, DTT, (BioPioneer Inc.) was dissolved in water and added to the cell culture media at final concentration of 2 mM.
Molecular Biology.
Cells were lysed and total RNA collected using the RNeasy mini kit, according to manufacturer’s instructions (Qiagen). Messenger RNA (mRNA) was used for RNA-seq analysis or prepared for qRT-PCR using the iScript complementary DNA (cDNA) Synthesis Kit (Bio-Rad). cDNA was used as template in SYBR green qPCR supermix (Bio-Rad). PCR Primers used include the following: human RPL19, 5′-ATGTATCACAGCCTGTACCTG-3′ and 5′-TTCTTGGTCTCTTCCTCCTTG-3′; human BIP/GRP78, 5′-GCCTGTATTTCTAGACCTGCC-3′ and 5′-TTCATCTTGCCAGCCAGTTG-3′; human HERPUD1, 5′-AACGGCATGTTTTGCATCTG-3′ and 5′-GGGGAAGAAAGGTTCCGAAG-3′; human SEL1L, 5′-ATCTCCAAAAGGCAGCAAGC-3′ and 5′-TGGGAGAGCCTTCCTCAGTC-3′; and human EDEM1, 5′-TTCCCTCCTGGTGGAATTTG-3′ and 5′- AGGCCACTCTGCTTTCCAAC-3. RPL19 mRNA levels served as internal normalization standards. qPCR condition was 95 °C for 5 min, 95 °C for 10 s, 60 °C for 10 s, 72 °C for 10 s, with 40 cycles of amplification.
Immunoblotting Analysis.
HEK293 cells expressing wild-type or mutant ATF6 were lysed in sodium dodecyl sulfate (SDS) lysis buffer (2% SDS in phosphate-buffered saline (PBS) containing protease and phosphatase inhibitors [Thermo Scientific]). Protein concentrations of the total cell lysates were determined by bicinchoninic acid (BCA) protein assay (Pierce). Equal amounts of protein were loaded onto 10% or 4 to 15% Mini-PROTEAN TGX precast gels (Bio-Rad) and analyzed by Western blot. The following antibodies and dilutions were used: anti-FLAG at 1:5,000 (Sigma-Aldrich); anti-BiP/GRP78 (GeneTex) at 1:1,000; and anti-GAPDH (Santa Cruz Biotechnology) at 1:5,000. After overnight incubation with primary antibody, membranes were washed in PBS with 0.1% Tween-20, followed by incubation of a horseradish peroxidase–coupled secondary antibody (Cell Signaling). Immunoreactivity was detected using the SuperSignal West chemiluminescent substrate (Pierce). GAPDH levels were assessed as a loading control as indicated. Protein lysates from five to eight pooled retinal organoids per genotype were prepared and analyzed as described above. Primary antibodies for the protein detection of rhodopsin (mouse anti-RHO, Santa Cruz Biotechnologies), red and green opsin (rabbit anti-OPN1MW/ LW, EMD Millipore), and beta actin (mouse anti b-actin, Sigma) were used.
RNA-seq Analysis.
RNA-seq was performed as previously described (31). Briefly, RNA was isolated from individual D290 retinal organoids prepared from ATF6[Y567N] patient iPSCs, or normal vision patient iPSCs, or pooled D170 retinal organoids prepared from two independent retinal organoids treated with AA147 (10 µM) or RP22 (10 µM) for 50 d using the Qiagen RNAeasy Mini Kit according to the manufacturer’s instructions. RNA sequencing was performed by BGI Americas on the BGI proprietary platform (DNBseq), providing single-end 50-bp reads at 20 million reads per sample. Alignment of the sequencing data were performed using DNAstar Lasergene SeqManPro to the GRCh37.13 human genome reference assembly. Assembly data were then imported into ArrayStar 12.2 with QSeq (DNAStar Inc) to quantify the gene expression and normalized reads per kilobase million. Differential expression analysis and statistical significance calculations between conditions were assessed using DESeq in R using a standard negative binomial fit for the aligned counts data and are described relative to the indicated control (Datasets S1 and S4). For DESeq expression comparisons between retinal organoids prepared from ATF6[Y567N] patient iPSCs and normal vision patient iPSCs, standard DESeq arguments were used for dispersion calculation given the presence of biological replicates (method = “pooled,” sharingMode = “maximum”). For expression comparisons between ATF6[Y567N] patient retinal organoids treated with AA147 or RP22, modified DESeq arguments were utilized to account for single pooled populations for each condition (method = “blind,” sharingMODE = “fit-only”). This modified analysis assumes that the vast majority of transcript levels will be comparable among the two conditions and uses this information to perform the necessary variance estimation. This modification was performed as outlined in the DESeq user manual (https://rdrr.io/bioc/DESeq/f/inst/doc/DESeq.pdf). GO analysis was performed using Panther (http://geneontology.org; Dataset S3). GSEA was performed using denoted gene sets from GO on the GSEA platform (http://www.gsea-msigdb.org/gsea/index.jsp) (34, 35). Violin plots comparing mutant and control retinal organoids were generated using the fold change data from the differential expression analysis for human rod and cone genes (19) (Dataset S2). Violin plots demonstrating changes in UPR-associated gene expression were generated using the fold change data from differential expression analysis for described gene sets of UPR transcriptional targets (31) (Dataset S5). Violin plots comparing mutant retinal organoids treated with AA147 (10 µM) or the inactive analog, RP22 (10 µM), were generated using the normalized fold change data from the differential expression analysis for rod and cone genes, where fold change values of all genes of interest were normalized to the mean fold change of rod genes (Dataset S6).
Immunofluorescence and Confocal Microscopy.
Retinal organoids were washed three times in 1× PBS (without Mg2+/Ca2+) for 5 min, prior to fixing in 4% PFA for 45 min and gentle agitation at room temperature. After fixing, organoids were washed three times in 1× PBS (without Mg2+/Ca2+) for 5 min, followed by 30% (weight [wt]/[vol] volume) sucrose treatment for 48 h to allow organoids to settle down. Three to five retinal organoids from the same genotype were embedded in optimal cutting temperature compound (OCT) and cryosectioned in 10-µm sections (Leica CM 1950), slides were stored at −20 °C. Prepared slides were adjusted to room temperature and washed three times in 1× PBS (without Mg2+/Ca2+) for 5 min. Samples were blocked and permeabilized for at least 1 h in blocking buffer containing 1% (wt/vol) BSA/1× PBS (without Mg2+/Ca2+), 0.1% Triton X-100, and 5% goat serum. Primary antibody was prepared in blocking solution and incubated overnight at 4 °C with gentle agitation. The following primary antibodies were used: rabbit anti-GNAT1 (Sigma), mouse anti-rhodopsin (Santa Cruz Biotechnologies), and rabbit anti-cone opsin (OPN1MW/ LW; EMD Millipore), PNA Fluorescine (Vector). Slides were the washed six times in 1× PBS (without Mg2+/Ca2+) with 0.1% Triton X-100 for 5 min at room temperature with gentle agitation. Secondary antibody was prepared in blocking buffer and incubated for 1 h at room temperature. Secondary antibodies included Alexa546 goat anti-mouse (red) antibody (Molecular Probes) and Alexa488 goat anti-rabbit (green) antibody (Molecular Probes) used at 1:250 dilution. Slides were then washed six times in 1× PBS (without Mg2+/Ca2+) for 5 min at room temperature with gentle agitation followed by mounting samples with ProLong Gold antifade reagent with DAPI (Invitrogen). Images were collected with an Olympus FluoView-1000 confocal microscope and processed using Olympus FluoView version 2.0a Viewer software at UCSD, School of Medicine Microscopy Core.
Live Organoid Imaging and Video Preparation.
Three retinal organoids per genotype were placed in a chamber slide (Ibidi) without a lid and mounted in a stage top incubator (H301-K-FRAME with Koehler Lid, Okolab). The temperature (37 °C), CO2 (5%), and humidity (90%) were controlled by a H301-T-UNIT-BL-PLUS stage incubation system (Okolab), CO2-UNIT-BL gas controller (Okolab), and HM-ACTIVE Humidity Controller (Okolab). All images were collected on an Eclipse Ti2 inverted microscope (Nikon Instruments) equipped with a Plan Apo λ 20× numerical aperture (NA) 0.75 (Nikon Instruments). The differential interference contrast (DIC) mode used two polarizers, two Nomarski prims (N1 and 20×, Nikon Instruments), and diascopic light-emitted diode (LED) illumination. Images were acquired with a monochrome DS-Qi2 sCMOS camera (Nikon Instruments) controlled by NIS-Elements software (Nikon Instruments). Z-series optical sections were collected with a step size of 0.9 µm, using the Ti2 Z-Drive. The Z-series were processed using local intensity algorithm (25 0, radius = 5.46 µm), the focused image (balanced Z-map method) of the Z-series is displayed using NIS-Elements software (Nikon Instruments). To generate videos, a montage of the processed Z-series with 10 copies of the focused image is played back at 20 frames per second.
Fixed Retinal Organoid Imaging.
Whole organoids were stained using the same protocol as described for OCT embedded sections. After staining, the entire organoid was placed in a 35-mm round tissue culture dish without lid (FD35-100, World Precision Instruments). To stabilize whole organoids during images, organoids were maintained in NIM III media supplemented with 50% glycerol. All images were collected with an X-Light V2 LOV (Crestoptics, S.p.A.) with 50-µm pinholes spinning on an Eclipse Ti2 inverted microscope (Nikon Instruments) equipped with a Plan Apo VC 60xA WI NA 1.2 objective lens. The DIC mode used two polarizers, two Nomarski prims (N2 and 60xII, Nikon Instruments), and diascopic LED illumination. Cone opsin (OPN1MW/LW, EMD Millipore) was detected using Alexa Fluor 488 and was excited with the 473-nm line from a Celesta light engine (Lumencor); rod opsin (Santa Cruz Biotechnologies) was detected using Alexa Fluor 546 and was excited with the 545-nm line from a Celesta light engine (lumencor). The emission was collected using a penta dichroic mirror (FF421/491/567/659/776-Di01, Semrock) and a penta emission filter (FF01-441/511/593/684/817, Semrock). Images were acquired with an Iris 15 CMOS camera (Photometrics) controlled by NIS-Elements software (Nikon Instruments). Z-series optical sections were collected with a step size of 0.3 µm, using the Ti2 Z-Drive. The Z-series is processed with Fiji (36). For the DIC Z-series, a fast Fourier transform (FFT) bandpass filter (filtering large structures down to 40 pixels, small structures up to 2 pixels, and tolerance of direction 5%) was applied followed by an unsharp mask (with radius of 5 pixels and mask weight 0.60). For the green and red fluorescence Z-series, an FFT bandpass filter (filtering large structures down to 400 pixels, small structures up to 4 pixels, and tolerance of direction 5%) was applied followed by a background subtraction using a rolling ball (radius of 50 pixels). A three-dimensional image and movie were created using the Volume View and Movie Maker of the NIS-Elements Analysis software (Nikon Instruments).
AOSLO Retinal Imaging.
Informed consent was obtained from two patients with confirmed ATF6 disease-causing variants and a normal control. Prior to imaging, the combination of tropicamide (1%) and phenylephrine hydrochloride (2.5%) was used for cycloplegia and pupillary dilation. AOLSO videos were acquired and processed as previously described (13, 37). This study followed the tenets of the Declaration of Helsinki and was approved by the institutional review board at the Medical College of Wisconsin.
Statistical Analysis.
Student’s two-tailed t tests (for paired samples) were performed to determine P values for SI Appendix, Fig. S2B. A value of P < 0.05 was considered significant. **P < 0.05 and ***P < 0.005. For RNA-seq, statistical significance of differences in fold change expression cone phototransduction genes is reported as the P value for n = 3 independent organoids calculated by DESeq analysis (Fig. 3B). Gene sets comprising cone-specific and rod-specific genes (Figs. 3B and 4 F and G) were defined from single-nuclei RNA-seq analysis of 5,873 human retinal cells isolated from postmortem human donor eyes (19). Genes were assigned based on selective expression in cone or rod cells from this analysis. These gene sets are defined in Datasets S2 and S6. Gene sets comprising different stress-responsive signaling pathways (Fig. 4C) were selected and analyzed, as previously described (31). These gene sets are defined in Dataset S5. Statistical significance of differences in fold change expression values for gene sets were calculated using Brown–Forsyth and Welch ANOVA tests (Fig. 4C) or Welch t test (Figs. 3A and 4 F and G), as indicated. The adjusted P value for these tests is indicated in the figure and accompanying legend. All statistics were calculated in Graphpad Prism or DESeq in R.
Supplementary Material
Acknowledgments
We thank Eric Griffis of the Nikon Imaging Center at UCSD and Elise Heon, Yang Hu, Ajoy Vincent, Doug Vollrath, and Sui Wang for helpful feedback and sharing materials. This work was supported by NIH Grants R01AG046495, F31AG063489, R01EY027335, R01EY017607, R01NS088485, and P30NS047101; Veterans Affairs Merit Awards I01BX002284 and I01RX002340; California Institute for Regenerative Medicine DISC2-10973; and by grants from the NIH Research Biomedical Research Centre at Moorfields Eye Hospital National Health Service (NHS) Foundation Trust and University College London (UCL) Institute of Ophthalmology.
Footnotes
Competing interest statement: J.W.K. declares that he is a board member and shareholder of Proteostasis Therapeutics Inc., Protego BioPharma, and Yumanity, which may develop ATF6 activators to treat degenerative diseases, although not for stem cell–associated purposes at this time. R.L.W. is a shareholder and board member of Protego BioPharma. J.W.K. and R.L.W. are inventors on a patent describing the ATF6 activating compound used in this study. All other authors declare they have no competing interests.
This article is a PNAS Direct Submission. V.Y.A. is a guest editor invited by the Editorial Board.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2103196118/-/DCSupplemental.
Data Availability
The RNA-seq data have been deposited to the public National Center for Biotechnology Information Gene Expression Omnibus repository under the data identifier GSE153302. All other study data are included in the article and/or supporting information.
References
- 1.Walter P., Ron D., The unfolded protein response: From stress pathway to homeostatic regulation. Science 334, 1081–1086 (2011). [DOI] [PubMed] [Google Scholar]
- 2.Haze K., Yoshida H., Yanagi H., Yura T., Mori K., Mammalian transcription factor ATF6 is synthesized as a transmembrane protein and activated by proteolysis in response to endoplasmic reticulum stress. Mol. Biol. Cell 10, 3787–3799 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ye J., et al., ER stress induces cleavage of membrane-bound ATF6 by the same proteases that process SREBPs. Mol. Cell 6, 1355–1364 (2000). [DOI] [PubMed] [Google Scholar]
- 4.Wu J., et al., ATF6alpha optimizes long-term endoplasmic reticulum function to protect cells from chronic stress. Dev. Cell 13, 351–364 (2007). [DOI] [PubMed] [Google Scholar]
- 5.Yamamoto K., et al., Transcriptional induction of mammalian ER quality control proteins is mediated by single or combined action of ATF6α and XBP1. Dev. Cell 13, 365–376 (2007). [DOI] [PubMed] [Google Scholar]
- 6.Kohl S., et al., Mutations in the unfolded protein response regulator ATF6 cause the cone dysfunction disorder achromatopsia. Nat. Genet. 47, 757–765 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Skorczyk-Werner A., et al., Autosomal recessive cone-rod dystrophy can be caused by mutations in the ATF6 gene. Eur. J. Hum. Genet. 25, 1210–1216 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chiang W. C., et al., Achromatopsia mutations target sequential steps of ATF6 activation. Proc. Natl. Acad. Sci. U.S.A. 114, 400–405 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhong X., et al., Generation of three-dimensional retinal tissue with functional photoreceptors from human iPSCs. Nat. Commun. 5, 4047 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Parfitt D. A., et al., Identification and correction of mechanisms underlying inherited blindness in human iPSC-derived optic cups. Cell Stem Cell 18, 769–781 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wahlin K. J., et al., Photoreceptor outer segment-like structures in long-term 3D retinas from human pluripotent stem cells. Sci. Rep. 7, 766 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eldred K. C., et al., Thyroid hormone signaling specifies cone subtypes in human retinal organoids. Science 362, eaau6348 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mastey R. R., et al., Characterization of retinal structure in ATF6-associated achromatopsia. Invest. Ophthalmol. Vis. Sci. 60, 2631–2640 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wynne N., Carroll J., Duncan J. L., Promises and pitfalls of evaluating photoreceptor-based retinal disease with adaptive optics scanning light ophthalmoscopy (AOSLO). Prog. Retin. Eye Res. 83, 100920 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wells-Gray E. M., Choi S. S., Bries A., Doble N., Variation in rod and cone density from the fovea to the mid-periphery in healthy human retinas using adaptive optics scanning laser ophthalmoscopy. Eye (Lond.) 30, 1135–1143 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eckmiller M. S., Cone outer segment morphogenesis: Taper change and distal invaginations. J. Cell Biol. 105, 2267–2277 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mustafi D., Engel A. H., Palczewski K., Structure of cone photoreceptors. Prog. Retin. Eye Res. 28, 289–302 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goldberg A. F., Moritz O. L., Williams D. S., Molecular basis for photoreceptor outer segment architecture. Prog. Retin. Eye Res. 55, 52–81 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liang Q., et al., Single-nuclei RNA-seq on human retinal tissue provides improved transcriptome profiling. Nat. Commun. 10, 5743 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Swaroop A., Kim D., Forrest D., Transcriptional regulation of photoreceptor development and homeostasis in the mammalian retina. Nat. Rev. Neurosci. 11, 563–576 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zinszner H., et al., CHOP is implicated in programmed cell death in response to impaired function of the endoplasmic reticulum. Genes Dev. 12, 982–995 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lu M., et al., Opposing unfolded-protein-response signals converge on death receptor 5 to control apoptosis. Science 345, 98–101 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Han J., et al., ER-stress-induced transcriptional regulation increases protein synthesis leading to cell death. Nat. Cell Biol. 15, 481–490 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hiramatsu N., et al., PERK-mediated induction of microRNA-483 disrupts cellular ATP homeostasis during the unfolded protein response. J. Biol. Chem. 295, 237–249 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hiramatsu N., et al., Translational and posttranslational regulation of XIAP by eIF2α and ATF4 promotes ER stress-induced cell death during the unfolded protein response. Mol. Biol. Cell 25, 1411–1420 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee E. J., et al., Multiexon deletion alleles of ATF6 linked to achromatopsia. JCI Insight 5, e136041 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Plate L., et al., Small molecule proteostasis regulators that reprogram the ER to reduce extracellular protein aggregation. eLife 5, e15550 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Blackwood E. A., et al., Pharmacologic ATF6 activation confers global protection in widespread disease models by reprograming cellular proteostasis. Nat. Commun. 10, 187 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kroeger H., et al., The unfolded protein response regulator ATF6 promotes mesodermal differentiation. Sci. Signal. 11, eaan5785 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Paxman R., et al., Pharmacologic ATF6 activating compounds are metabolically activated to selectively modify endoplasmic reticulum proteins. eLife 7, e37168 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grandjean J. M. D., et al., Deconvoluting stress-responsive proteostasis signaling pathways for pharmacologic activation using targeted RNA sequencing. ACS Chem. Biol. 14, 784–795 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kroeger H., et al., Induction of endoplasmic reticulum stress genes, BiP and chop, in genetic and environmental models of retinal degeneration. Invest. Ophthalmol. Vis. Sci. 53, 7590–7599 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang L. P., Wu L. M., Guo X. J., Li Y., Tso M. O., Endoplasmic reticulum stress is activated in light-induced retinal degeneration. J. Neurosci. Res. 86, 910–919 (2008). [DOI] [PubMed] [Google Scholar]
- 34.Subramanian A., et al., Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liberzon A., et al., Molecular signatures database (MSigDB) 3.0. Bioinformatics 27, 1739–1740 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schindelin J., et al., Fiji: An open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Scoles D., et al., In vivo imaging of human cone photoreceptor inner segments. Invest. Ophthalmol. Vis. Sci. 55, 4244–4251 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA-seq data have been deposited to the public National Center for Biotechnology Information Gene Expression Omnibus repository under the data identifier GSE153302. All other study data are included in the article and/or supporting information.