Significance
Activation of plant innate immunity relies on the perception of microorganisms through elicitors. Rhamnolipids and their precursor HAAs are exoproducts produced by bacteria. They are involved in bacterial surface dissemination and biofilm development. As these compounds are released in the extracellular milieu, they have the potential to be perceived by the plant immune system. Our work shows that both compounds independently activate plant immunity. We demonstrate that HAAs are perceived by the receptor protein kinase LORE. By contrast, rhamnolipids are not sensed by LORE but activate a noncanonical immune response influenced by the sphingolipid composition of the plant plasma membrane. Thus, plants can sense bacterial molecules as well as their direct precursors to trigger distinct immune responses.
Keywords: plant immunity, rhamnolipids, HAA, Pseudomonas
Abstract
Plant innate immunity is activated upon perception of invasion pattern molecules by plant cell-surface immune receptors. Several bacteria of the genera Pseudomonas and Burkholderia produce rhamnolipids (RLs) from l-rhamnose and (R)-3-hydroxyalkanoate precursors (HAAs). RL and HAA secretion is required to modulate bacterial surface motility, biofilm development, and thus successful colonization of hosts. Here, we show that the lipidic secretome from the opportunistic pathogen Pseudomonas aeruginosa, mainly comprising RLs and HAAs, stimulates Arabidopsis immunity. We demonstrate that HAAs are sensed by the bulb-type lectin receptor kinase LIPOOLIGOSACCHARIDE-SPECIFIC REDUCED ELICITATION/S-DOMAIN-1-29 (LORE/SD1-29), which also mediates medium-chain 3-hydroxy fatty acid (mc-3-OH-FA) perception, in the plant Arabidopsis thaliana. HAA sensing induces canonical immune signaling and local resistance to plant pathogenic Pseudomonas infection. By contrast, RLs trigger an atypical immune response and resistance to Pseudomonas infection independent of LORE. Thus, the glycosyl moieties of RLs, although abolishing sensing by LORE, do not impair their ability to trigger plant defense. Moreover, our results show that the immune response triggered by RLs is affected by the sphingolipid composition of the plasma membrane. In conclusion, RLs and their precursors released by bacteria can both be perceived by plants but through distinct mechanisms.
Plant innate immunity activation relies on detection of invasion pattern (IP) molecules that are perceived by plant cells (1, 2). Non–self-recognition IPs include essential components of whole classes of microorganisms, such as fragments of flagellin, peptidoglycans, mc-3-OH-FAs from bacteria or fragments of chitin, and β-glucans from fungi and oomycetes, respectively (3, 4). Apoplastic IPs are sensed by plant plasma membrane–localized receptor kinases (RKs) or receptor-like proteins (RLPs) that function as pattern recognition receptors (PRRs) (5, 6). Activation of the immune response requires the recruitment of regulatory receptor kinases and receptor-like cytoplasmic kinases (RLCKs) by PRRs (7). Early cellular immune signaling of pattern-triggered immunity (PTI) includes ion-flux changes at the plasma membrane, rise in cytosolic Ca2+ levels, production of extracellular reactive oxygen species (ROS), and activation of mitogen-activated protein kinases (MAPKs) and/or Ca2+-dependent protein kinases (3, 8–10). Biosynthesis and mobilization of plant hormones, including salicylic acid, jasmonic acid, ethylene, abscisic acid and brassinosteroids, ultimately modulate plant resistance to phytopathogens (11–14).
Rhamnolipids (RLs) are extracellular amphiphilic metabolites produced by several bacteria, especially Pseudomonas and Burkholderia species (15–17). Acting as wetting agents, RLs are essential for bacterial surface dissemination called swarming motility and for normal biofilm development (18–20). These glycolipids are produced from l-rhamnose and 3-(3-hydroxyalkanoyloxy)alkanoic acid (HAA) precursors (15, 21). HAAs are synthesized by dimerization of (R)-3-hydroxyalkanoyl-CoA in Pseudomonas, forming congeners through the RhlA enzyme (21). The opportunistic plant pathogen Pseudomonas aeruginosa and the phytopathogen Pseudomonas syringae produce extracellular HAAs (16, 22–24). In P. syringae, HAA synthesis is coordinately regulated with the late-stage flagellar gene encoding flagellin (22). HAA and RL production is finely tuned and modulates the behavior of swarming migrating bacterial cells by acting as self-produced negative and positive chemotactic-like stimuli (25). RLs contribute to the alteration of the bacterial outer membrane composition, by shedding flagellin from the flagella (26) and by releasing lipopolysaccharides (LPS), resulting in an increased hydrophobicity of the bacterial cell surface (27). In mammalian cells, RLs produced by Burkholderia plantarii exhibit endotoxin-like properties similar to LPS, leading to the production of proinflammatory cytokines in human mononuclear cells (28, 29). They also subvert the host innate immune response through manipulation of the human beta-defensin-2 expression (30). Moreover, RLs from Burkholderia pseudomallei induce interferon gamma (IFN-γ)–dependent host immune response in goat (31).
In plants, RLs induce defense responses and resistance to biotrophic and necrotrophic pathogens (32, 33). They also contribute to the biocontrol activity of the plant beneficial bacterium P. aeruginosa PNA1 against oomycetes (17). Recently, it was reported that the bulb-type lectin receptor kinase LIPOOLIGOSACCHARIDE-SPECIFIC REDUCED ELICITATION/S-DOMAIN-1-29 (LORE/SD1-29) mediates medium-chain 3-hydroxy fatty acid (mc-3-OH-FA) sensing in Arabidopsis thaliana (hereafter, Arabidopsis) and that bacterial compounds comprising mc-3-OH-acyl building blocks including LPS and RLs do not stimulate LORE-dependent responses (34).
Here, we show that the lipidic secretome produced by P. aeruginosa (RL secretome), mostly composed of RLs and HAAs, induces Arabidopsis immunity. HAAs are perceived through the RK LORE. We demonstrate that, albeit not being sensed by LORE, RLs trigger an immune response characterized by an atypical defense signature. Altogether, our results demonstrate that RLs and their precursors produced by Pseudomonas bacteria stimulate the plant immune response by two distinct mechanisms.
Results
RL Secretome from Pseudomonas Induces Arabidopsis Immune Responses Partially Mediated by LORE.
Pseudomonas species, including opportunistic plant pathogenic or plant beneficial endophytic strains, release a mixture of RL congeners and HAA precursors, here collectively termed RL secretome (RLsec) (15, 25). High-performance liquid chromatography (HPLC)-tandem mass spectrometry (MS/MS) analyses of this RLsec from P. aeruginosa revealed the presence of mono-RLs and di-RLs at 50.9% and 44.9% of dry weight, respectively, and HAAs (3.8% of dry weight) (SI Appendix, Table S1 and Fig. S1). RLs comprising 10-carbon-long lipid tails, Rha-C10-C10 (α-l-rhamnopyranosyl-β-hydroxydecanoyl-β-hydroxydecanoate) and Rha-Rha-C10-C10 (α-l-rhamnopyranosyl-α-l-rhamnopyranosyl-β-hydroxydecanoyl-β-hydroxydecanoate), and C10-C10 [(R)-3-(((R)-3-hydroxydecanoyl)oxy)decanoate] HAAs were the most abundant molecules in this lipidic secretome (37.6, 33.1, and 2.1%, respectively). Notably, low amounts of free mc-3-OH-FAs (0.4% total), such as 3-OH-C8, 3-OH-C10, and 3-OH-C12, were also identified (SI Appendix, Table S1 and Fig. S1).
First, we monitored apoplastic ROS production triggered by RLsec in Arabidopsis (35). Wild-type (WT) plants challenged with RLsec displayed a transient extracellular ROS production, starting at 6 min and peaking at 15 min postelicitation (Fig. 1A). A robust ROS response was detected at concentrations of RLsec starting from 0.5 µg/mL (Fig. 1B and SI Appendix, Fig. S2). The ROS burst was dependent on the transmembrane NADPH oxidase RBOHD (36, 37) (Fig. 1C and SI Appendix, Fig. S3). RKs and RLPs mediate perception of IPs and early activation of PTI signaling (7). We monitored RLsec-triggered ROS production in Arabidopsis plants carrying loss-of-function mutations in genes encoding well-characterized RKs and RLPs fls2/efr-1 (38, 39), bak1-5, bkk1-1, bak1-5/bkk1-1 (40), bik1/pbl1 (41), cerk1-2 (42), sobir1-12, sobir1-13 (43), dorn1-1 (44), and lore-5 (45). RLsec-induced production of ROS was only reduced in lore-5 (Fig. 1C and SI Appendix, Fig. S3). Some IPs, including LPS extracts and mc-3-OH-FAs, were reported to induce a late ROS production in Arabidopsis (34, 46, 47). The late ROS response triggered by mc-3-OH-FAs was dependent on LORE (34). RLsec also induced a late and long-lasting ROS burst in Arabidopsis culminating at 6 to 8 h posttreatment (Fig. 2A), which was abolished in rbohD but not in lore-5 mutant plants (Fig. 2A).
Fig. 1.
RLsec activates LORE-dependent early immune-related responses in Arabidopsis. (A) Extracellular ROS production after treatment of WT leaf petioles with 50 µg/mL RLsec or EtOH as control. (B) Dose effect of RLsec on ROS production. ROS production measured after treatment of WT leaf petioles with the indicated concentrations of RLsec or EtOH as control. (C) ROS production measured after treatment of WT, fls2/efr-1, dorn1-1, bak1-5, bkk1-1, bak1-5/bkk1-1, cerk1-2, sobir1-12, sobir1-13, bik1/pbl1, lore-5, or rbohD leaf petioles with 50 µg/mL RLsec. (A–C) Data are mean ± SEM (n = 6). (A–C) Experiments have been realized three times with similar results.
Fig. 2.
RLsec activates LORE-independent responses in Arabidopsis. (A) Extracellular ROS production after treatment of WT, lore-5, and rbohD leaf petioles with 50 µg/mL RLsec or EtOH (control). ROS production was monitored over 720 min. Data are mean ± SEM (n = 6). Experiments have been realized three times with similar results. (B) WT (red) and lore-5 (blue) Arabidopsis leaves were treated with 50 µg/mL RLsec (triangle) or EtOH (square) (control) 48 h before infection. Pst titers were measured at 3 dpi. Data are individual data and mean (black line) (n = 5). Experiments have been realized three times with similar results. Letters represent results of pairwise Welch statistic test with Bonferroni correction with P > 0.05 (same letters) or P ≤ 0.05 (different letters).
Next, we tested whether RLsec induces local resistance to the hemibiotrophic phytopathogen P. syringae pathovar (pv.) tomato DC3000 (Pst) in Arabidopsis (48). RLsec pretreatment significantly enhanced resistance against Pst infection in WT and lore-5 plants (Fig. 2B). Taken together, our results show that RLsec induces immunity-related signaling events and disease resistance in Arabidopsis that are partially mediated by the bulb-type lectin RK LORE.
Pseudomonas HAAs and mc-3-OH-FAs from RLsec Trigger LORE-Dependent Arabidopsis Immunity.
By contrast to RLsec, purified RLs do not trigger LORE-dependent [Ca2+]cyt and early ROS signaling responses (34). Because RLsec contains significant amounts of HAAs, we investigated the role of these poorly studied compounds in RLsec-triggered immunity. We compared the responses to HAA with those to mc-3-OH-FAs, known to be sensed by LORE (34) and present in low amounts in RLsec (SI Appendix, Table S1). Side-by-side experiments with C10-C10 HAA purified from P. aeruginosa secretome and 3-OH-C10 revealed that both compounds induce [Ca2+]cyt signaling and ROS production in Arabidopsis plants in a dose-dependent manner (Fig. 3 A and B and SI Appendix, Figs. S4 and S5). As observed upon 3-OH-C10 elicitation, purified C10-C10–induced ROS response was impaired in rbohD and lore-5 mutants (Fig. 3C). Similarly, [Ca2+]cyt signaling triggered by C10-C10 was impaired in lore-5 (Fig. 3D). In addition, C10-C10 and 3-OH-C10 both triggered LORE-dependent MPK3 and MPK6 phosphorylation (SI Appendix, Fig. S6A). WT but not lore-5 mutant plants pretreated with C10-C10 or 3-OH-C10 displayed enhanced resistance against Pst (Fig. 3E). Similar to 3-OH-FAs (34), the acyl chain length of HAA affects its immune eliciting activity, as purified C14-C14 from B. glumae neither induced ROS production nor enhanced resistance to Pst in Arabidopsis plants (SI Appendix, Fig. S7 A and B).
Fig. 3.
Purified HAAs from P. aeruginosa trigger a LORE-dependent immune response in Arabidopsis. (A) Dose effect of 3-OH-C10 and C10-C10 purified from P. aeruginosa on ROS production by WT leaf petioles. EtOH was used as negative control. Data are mean ± SEM (n = 6). Experiments have been realized twice with similar results. (B) Maximum (max.) increases in [Ca2+]cyt in Arabidopsis Col-0AEQ seedlings treated with different concentrations of 3-OH-C10, C10-C10 purified from P. aeruginosa, or MeOH as control. Data are mean ± SD (n = 3). Experiments have been realized twice with similar results. (C) ROS production measured after treatment of WT, lore-5, or rbohD leaf petioles with 10 µM 3-OH-C10, 10 µM purified C10-C10, or EtOH as control. Data are mean ± SEM (n = 6). Experiments have been realized three times with similar results. (D) Max. increases in [Ca2+]cyt in Arabidopsis Col-0AEQ and lore-5AEQ seedlings treated with 5 µM 3-OH-C10 or purified C10-C10. Data are mean ± SD (n = 3). Experiments have been realized twice with similar results. For B and D, the same Col-0AEQ 5 µM data are presented (same experiments). (E) WT (red) and lore-5 (blue) Arabidopsis leaves were treated with 10 µM 3-OH-C10 (triangle), 10 µM purified C10-C10 (diamond), or EtOH (square) (control) 48 h before infection. Pst titers were measured at 3 dpi. Data are individual data and mean (black line) (n = 5). Experiments have been realized twice with similar results. Letters represent results of pairwise Welch statistic test with Bonferroni correction with P > 0.05 (same letters) or P ≤ 0.05 (different letters).
Trace amount of 3-OH-C10 was detected in C10-C10 purified from P. aeruginosa RLsec (SI Appendix, Table S2). To avoid any influence of potential contamination of HAAs with eliciting compounds related to purification procedure, we tested chemically synthesized C10-C10 for the ROS and [Ca2+]cyt responses. Synthetic C10-C10 triggered LORE-dependent [Ca2+]cyt signaling and ROS production in a dose-dependent manner (Fig. 4 A–C). WT plants pretreated with synthetic C10-C10 also displayed LORE-dependent enhanced resistance against Pst infection (Fig. 4D). LORE binds 3-OH-C10 through its extracellular domain (eLORE) (34). In a ligand depletion assay (SI Appendix, Fig. S8A), synthetic C10-C10 was depleted from the low-molecular-weight fraction after size-exclusion filtration when incubated with eLORE-mCherry fusion protein transiently expressed in the apoplast of Nicotiana benthamiana, but not upon incubation with an apoplastic mCherry control, similarly as observed for the 3-OH-C10 ligand (Fig. 4E and SI Appendix, Fig. S8) (84). This strongly suggests binding of C10-C10 to eLORE.
Fig. 4.
Synthetic HAAs trigger a LORE-dependent immune response in Arabidopsis. (A) Maximum (max.) increases in [Ca2+]cyt in Arabidopsis Col-0AEQ seedlings treated with different concentrations of 3-OH-C10, synthetic C10-C10, or MeOH. Data are mean ± SD (n = 3). Experiments have been realized twice with similar results. (B) Dose effect of synthetic C10-C10 on ROS production by WT leaf petioles. EtOH was used as negative control. Data are mean ± SEM (n = 6). Experiments have been realized twice with similar results. (C) Max. increases in [Ca2+]cyt in Arabidopsis Col-0AEQ and lore-5AEQ seedlings treated with 5 µM 3-OH-C10 or synthetic C10-C10. Data are mean ± SD (n = 3). Experiments have been realized twice with similar results. (D) WT (red) and lore-5 (blue) Arabidopsis leaves were treated with 10 µM 3-OH-C10 (triangle), 10 µM synthetic C10-C10 (diamond), or MeOH (square) (control) 48 h before infection. Pst titers were measured at 3 dpi. Data are individual data and mean (black line) (n = 5). Experiments have been realized twice with similar results. Letters represent data of pairwise Welch statistic test with Bonferroni correction with P > 0.05 (same letters) or P ≤ 0.05 (different letters). (E) Depletion of 1 µM synthetic C10-C10 or 100 nM 3-OH-C10 by concentrated apoplastic wash fluid from N. benthamiana containing LORE extracellular domain (eLORE)-mCherry fusion protein or apoplastic (apo)-mCherry as control as illustrated in SI Appendix, Fig. S8A. Unbound ligand content in low-molecular-weight filtrate was analyzed by [Ca2+]cyt measurements in LORE-overexpressing (red bars, gray triangles, n = 12) and lore-1 (blue bars, gray diamonds, n = 6) Arabidopsis seedlings. Data are mean ± SD and individual data of three pooled experiments. Letters represent data of pairwise Wilcoxon statistic test with Bonferroni correction with P > 0.05 (same letters) or P ≤ 0.05 (different letters).
Altogether, our results show that HAAs secreted by Pseudomonas are sensed by Arabidopsis through the bulb-type lectin RK LORE, activate canonical PTI-related immune responses, and provide resistance to bacterial infection.
RLs Trigger LORE-Independent Arabidopsis Immune Responses and Resistance to Pst.
To investigate whether RLs activate a LORE-independent immune response, we used purified Rha-Rha-C10-C10 and Rha-C10-C10, the most abundant molecules from P. aeruginosa RLsec. In Arabidopsis WT, both RL congeners induced a late and long-lasting ROS production but as observed previously (34), no early burst (Fig. 5A). As both RL congeners gave a similar ROS signature, we only used Rha-Rha-C10-C10 in the following experiments. The minimal concentration necessary to stimulate ROS production was 50 µM with an optimum at 100 µM (Fig. 5B). Late ROS production was compromised in rbohD but not in lore-5 mutants (Fig. 5C). Surprisingly, neither MPK3 nor MPK6 activation by Rha-Rha-C10-C10 was detectable over a 3-h time course (SI Appendix, Fig. S6 A and B). l-Rhamnose alone was inactive demonstrating that the lipid part of the RLs is necessary to trigger the immune response (Fig. 5A). Burkholderia species produce RL congeners with longer lipid chains than those produced by Pseudomonas (15). The RLsec from phytopathogenic Burkholderia glumae only contains congeners with fatty acid chain lengths varying from 12 to 16 carbons, in particular Rha-Rha-C14-C14 (49, 50). Challenge of Arabidopsis with purified Rha-Rha-C14-C14 from B. glumae did not trigger any ROS production (Fig. 5A) suggesting that the length of the fatty acid chain of RLs is critical for their eliciting activity. To determine whether RLs trigger local resistance to pathogenic Pseudomonas independent of LORE, plants were pretreated with 10 µM purified Rha-Rha-C10-C10 before Pst inoculation. WT plants displayed a significant enhanced resistance against Pst that was not compromised in lore-5 mutants (Fig. 5D).
Fig. 5.
Purified RLs trigger a LORE-independent Arabidopsis immune response. (A) Extracellular late ROS production after treatment of WT leaf petioles with 100 µM RLs, 100 µM L-rhamnose, or EtOH (control). Data are mean ± SEM (n = 6). (B) Dose effect of Rha-Rha-C10-C10 on late ROS production. ROS production measured after treatment of WT leaf petioles with the indicated concentrations of Rha-Rha-C10-C10 or EtOH (control). Data are mean ± SEM (n = 6). (C) Late ROS production measured after treatment of WT, lore-5, or rbohD leaf petioles with 100 µM Rha-Rha-C10-C10. Data are mean ± SEM (n = 6). (D) WT (red) and lore-5 (blue) Arabidopsis leaves were treated with 10 µM Rha-Rha-C10-C10 (triangle) or EtOH (square) (control) 48 h before infection. Pst titers were measured at 3 dpi. Data are individual data and mean (black line) (n = 5). Letters represent results of pairwise t statistic test with Bonferroni correction with P > 0.05 (same letters) or P ≤ 0.05 (different letters). (A–D) Experiments have been realized three times with similar results.
To get deeper insights into the mechanisms involved in RL sensing, we used Arabidopsis plants carrying loss-of-function mutations in genes encoding RK and RLPs but also plasma membrane channel mutants including quintuple mechano-sensitive channels of small conductance-like (msl4/5/6/9/10) and double mid1-complementing activity (mca1/2) channel mutants (51) that could monitor changes in membrane mechanical properties. None of these mutants were affected in the long-term ROS response (Fig. 6A). Glycosylinositol phosphorylceramide (GIPC) sphingolipids were recently involved in the sensing of microbial necrosis and ethylene-inducing peptide 1-like (NLP) proteins (52). We found that the fatty acid hydroxylase fah1/2 mutant that is disturbed in its complex sphingolipid composition (52) showed a reduced long-term ROS response (Fig. 6B). Ion leakage measurement confirmed that fah1/2 mutant plants were less affected than WT plants by RL treatment (Fig. 6C). fah1/2 plants are known to be more resistant to obligate biotrophic pathogens (85). We also observed that these mutants were more resistant to Pst (Fig. 6D). However, unlike in WT plants, challenge of fah1/2 with RLs did not trigger enhanced resistance to Pst in the mutants, confirming that RL-triggered responses are compromised in these plants (Fig. 6D). Ceramide synthase loh1 mutants are also impaired in GIPC levels but not in glucosyl ceramides (52). Interestingly, RL-triggered ROS production and ion leakage was unaltered in loh1 plants. Altogether, our results show that RLs activate an atypical immune response in Arabidopsis that is LORE independent but which is affected by the sphingolipid composition of the plasma membrane.
Fig. 6.
RL perception is impacted by plasma membrane sphingolipid composition. Extracellular late ROS production after treatment of (A) WT, cerk1-2, dorn1-1, sobir1-12, bak1-5/bkk1-1, bik1/pbl1, mca1/2, msl4/5/6/9/10 (msl), or rbohD, and (B) WT, loh1, or fah1/2 Arabidopsis leaf petioles with 100 µM Rha-Rha-C10-C10 or EtOH (control). Data are mean ± SEM (n = 6). Experiments have been realized three times with similar results. (C) Electrolyte leakage induced by 100 µM Rha-C10-C10 or EtOH (control) on WT, loh1, or fah1/2 Arabidopsis leaf discs 24 h posttreatment. Data are mean ± SEM (n = 6). Letters represent results of pairwise Wilcoxon Mann–Whitney U statistic test with P > 0.05 (same letters) or P ≤ 0.05 (different letters). Experiments have been realized twice with similar results. (D) WT (red) and fah1/2 (blue) Arabidopsis leaves were treated with 10 µM Rha-C10-C10 (triangle) or EtOH (square) (control) 48 h before infection. Pst titers were measured at 3 dpi. Data are individual data and mean (black line) (n = 5). Letters represent results of pairwise Welch statistic test with Bonferroni correction with P > 0.05 (same letters) or P ≤ 0.05 (different letters).
Discussion
In Pseudomonas and Burkholderia species, swarming motility is intimately related to the production of extracellular surface-active RLs and HAAs (22, 25, 53–55). In addition, RL production affects bacterial biofilm architecture and increases affinity of cells for initial adherence to surfaces through increasing the cell’s surface hydrophobicity (19, 56). These exoproducts are therefore at the frontline during host colonization. Our work demonstrates that both RLs and HAAs from the Pseudomonas lipidic secretome, referred to as RLsec here, are able to trigger Arabidopsis innate immunity by two distinct mechanisms.
We found that Pseudomonas RLs induce an atypical immune response. This response does not involve the RK LORE. Other bacterial compounds comprising mc-3-OH-acyl building blocks, but with large decorations including lipid A or LPS, lipopeptides, and N-acyl homoserine lactones, also do not trigger LORE-dependent immune responses (34). RLs are glycolipids made of l-rhamnose linked to an HAA lipid tail (15, 21). Therefore, we propose that glycosylation of HAAs abolishes their perception by LORE. Glycosylation is known to affect the perception of IPs. Glycosylation of the flagellin from Acidovorax avenae on Ser178 and Ser183 prevents its perception by rice cells (57). Similarly, unglycosylated flagellin from P. syringae pv. tabaci 6605 induces stronger defense responses in tobacco plants than glycosylated flagellin (58). In humans, glycosylation of Burkholderia cenocepacia flagellin significantly reduces its perception by epithelial cells (59).
We found that RL perception does not involve previously characterized RKs, RLPs, or mechanosensitive channels. However, the RL response is affected by alterations in sphingolipid synthesis suggesting a role of these key membrane lipids in RL-triggered immunity. Recently GIPCs, major structural components of the plant plasma membrane together with glucosylceramides (GlcCers), have been involved as receptors of cytotoxic NLPs (52). NLPs bind terminal monomeric hexose moieties of GIPCs. Only eudicot plants are sensing these NLPs through sphingolipid receptors. Insensitivity of monocots to NLPs is due to the length of the GIPC headgroup, consisting of three terminal hexoses compared to two in eudicots (52). fah1/2 mutants display reduced glycosylsphingolipid (GIPCs and GlcCers) content but also a lower level of ordered plasma membranes (52), suggesting that, similar to the NLP response, complex sphingolipids and/or ordered plasma membranes are necessary for the RL response. Unlike NLPs, RL responses were not significantly affected in loh1 mutant plants also suggesting that GlcCers more than GIPC could influence RL sensing (52). Surfactin and more recently synthetic RL bolaforms and synthetic glycolipids, also active in the micromolar range, have been proposed to directly interact with plasma membrane lipids (46, 60–62). Mono- and di-RLs from Pseudomonas interact with phospholipids in several synthetic membrane systems (63–66). In particular, RLs are able to fit into phospholipid bilayers of plant biomimetic plasma membranes (67). In this model, the rhamnose polar heads from RLs are located near the phosphate groups from phospholipids, and RL hydrophobic lipid tails are surrounded by the lipid chains from these phospholipids (67). The results obtained with these plant plasma membrane models suggest that the insertion of RLs into the lipid bilayer does not significantly affect lipid dynamics. The nature of the phytosterols could however influence the RL effect on plant plasma membrane destabilization. Subtle changes in lipid dynamics could then be linked to plant defense induction (67). Interestingly, RL bolaforms, like natural RLs, are inducing a noncanonical defense signature with a long-lasting oxidative burst without MPK3 or MPK6 activation (46). We showed that the late ROS response triggered by RLs and RL-bolaforms is fully dependent on RBOHD, which differs from the second ROS response observed after LPS treatment and produced by chloroplasts (47). The RL-related ROS response is not directly linked to Arabidopsis protection to Pst as some bolaforms can trigger this late ROS production without increasing resistance to Pst (46). However, the experiments from Luzuriaga-Loaiza et al. (46) and our results on natural RLs suggest that the late ROS response is closely linked to ion leakage and may therefore be involved in membrane destabilization. This atypical defense signature triggered by two structurally different RLs, displaying amphiphilic properties and biological activities at the micromolar range, may suggest a direct interaction of these molecules with plant plasma membrane lipids, similar to what has been shown on membrane models (46).
We also demonstrated that HAAs, found in large amount in Pseudomonas lipidic secretome, are IPs perceived by Arabidopsis. HAA sensing is mediated by LORE. HAAs, in the micromolar range, induce typical PTI responses including transient ROS production, [Ca2+]cyt signaling, and MPK3 and MPK6 phosphorylation in Arabidopsis. Interestingly, 3-OH-C10 activates similar responses but at concentrations 10 to 50 times lower. This is intriguing because HAAs are present in much larger quantities (more than 3%) compared to 3-OH-FAs (0.3%) in the lipidic secretome (SI Appendix, Table S1). This high amount of HAAs may compensate for their lower activity. RLs are activating an immune response at relatively high concentrations compared to both compounds. Interestingly, the RL concentration in the P. aeruginosa lipidic secretome is 10 to 100 times higher than HAAs and usually is in the millimolar range (23, 68). RLs are produced between 20 and 110 µM in vivo in mammals infected by P. aeruginosa, especially during cystic fibrosis disease (69–71). The high concentrations of RLs needed for plant elicitation are in the range of the concentrations produced by the bacteria.
Higher steric hindrance of HAA compared to 3-OH-FAs likely results in a lower affinity to the LORE receptor. Synthetic ethyl 3-hydroxydecanoate (Et-3-OH-C10:0) and n-butyl 3-hydroxydecanoate (nBut-3-OH-C10:0), which possess unbranched ester-bound carbon chains in place of the carboxyl group, also triggered LORE-dependent immune signaling in Arabidopsis, while 3-branched tert-butyl 3-hydroxydecanoate (tBut-3-OH-C10:0) was inactive (34). HAAs, possessing a 2-branched ester-bound headgroup, activate LORE signaling. The differences in efficacy could be explained by the different steric hindrance of the molecules. Alternatively, the additional carboxyl group could account for the LORE-eliciting activity of HAAs.
Pantoea, Dickeya, and Pseudomonas bacteria, in particular the well-known phytopathogen P. syringae, mainly produce HAAs containing 3-hydroxydecanoic acid (C10) tails (15, 22, 72). By contrast, Burkholderia species including the phytopathogenic bacterium B. glumae mainly produce HAAs comprising 3-hydroxytetradecanoic acid (C14) tails (49). Pseudomonas C10-containing HAAs activated Arabidopsis PTI whereas Burkholderia HAAs containing C14 fatty acid did not. Chain-length specificity was also observed for mc-3-OH-FA sensing by the LORE receptor with 3-OH-C10 representing the strongest immune elicitor (34). Thus, it could be hypothesized that Arabidopsis, and more generally Brassicaceae (73), are able to specifically recognize HAAs from specific bacterial species, among which several are plant opportunistic and phytopathogens (74–77). Interestingly, transcript profiles of the bean pathogen P. syringae pv. syringae B728a support a model in which leaf surface or epiphytic sites specifically favor swarming motility based on HAA surfactant production (55, 78). Low levels of HAAs contributing to motility are produced by these bacteria (22). HAA concentrations necessary to stimulate Arabidopsis innate immunity are in line with the concentration detected in RLsec and are produced by Pseudomonas (between 3 to 20% of the secretome) (23, 68, 79).
Low amounts of free mc-3-OH-FAs were found in RLsec from P. aeruginosa (SI Appendix, Table S1). In Pseudomonas, the outer membrane lipase PagL releases 3-OH-C10 during synthesis of penta-acylated lipid A (34). The further fate of this 3-OH-C10 is unknown. RLs are able to extract LPS from the outer membrane of P. aeruginosa (27). Conceivably, surface-active RLs, and presumably also HAAs, could release free 3-OH-C10, produced through PagL activity, along with LPS from the bacterial cell wall or outer membrane vesicles (27). Alternatively, degradation of HAAs/RLs in planta may also release free 3-OH-C10. Acyl carrier protein (ACP)- and coenzyme A (CoA)-bound mc-3-OH-FAs are precursors of HAA/RL synthesis (21). Upon bacterial cell lysis, enzymatic or nonenzymatic degradation processes may also generate free 3-OH-C10 from these precursors. In vivo, insights into IP release have been recently obtained for flagellin. The plant glycosidase BGAL1 facilitates the release of immunogenic peptides from glycosylated flagellin, upstream of cleavage by proteases (80). The pathogen may evade detection by altering flagellin glycosylation and inhibiting the plant glycosidase. Flagellin glycosylation increases its physical stability that could contribute to the nonliberation/recognition of the flg22 epitope (58, 81). RLs are able to shed flagellin from P. aeruginosa flagella (26), suggesting that these biosurfactants participate in the release of this and presumably other eliciting compounds.
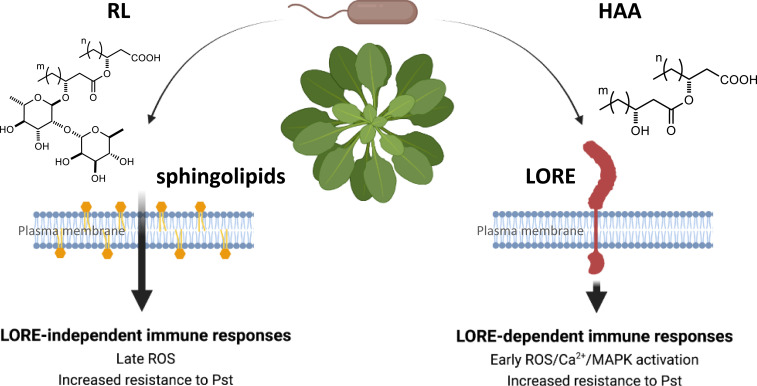
In conclusion, we hypothesize that when HAA- and RL-producing Pseudomonas colonize the leaf or root surface, they release RLs and HAAs, which are necessary for surface motility, biofilm development, and thus successful colonization. Whereas Arabidopsis senses HAAs and mc-3-OH-FAs through the bulb-type lectin receptor kinase LORE, RLs are perceived through a LORE-independent mechanism (Fig. 7). In addition to direct activation of a noncanonical defense response in plants, RLs, by releasing other IPs from bacteria, could orchestrate a node leading to strong activation of plant immunity.
Fig. 7.
Proposed model for HAA and rhamnolipid perception mechanisms in Arabidopsis and integrated immune responses. RL-producing bacteria excrete rhamnolipids and HAAs to form biofilm and for successful colonization. Arabidopsis perceives HAAs via LORE and rhamnolipids by a LORE-independent pathway affected by sphingolipids composition of the plant plasma membrane, resulting in the activation of efficient immune responses. Parts of this scheme were created with https://BioRender.com.
Methods
Molecules.
The P. aeruginosa lipidic secretome used in this study was obtained from Jeneil Biosurfactant Co. (JBR-599, lot no. 050629). Rha-Rha-C10-C10 and Rha-C10-C10 were purified from this lipidic secretome mixture, as previously described (33, 34). Rha-Rha-C14-C14 were purified from the B. glumae lipidic secretome (49). To obtain pure HAAs from P. aeruginosa or B. glumae, RLs were hydrolyzed using 1 M HCl in 1:1 dioxane–water boiling at reflux for 60 min. The mixture was extracted with ethyl acetate, and the extracts were dried over anhydrous Na2SO4. After filtration, the resulting extracts were then evaporated to dryness and resuspended in 2 mL methanol. HAAs were then isolated from digested mixture using flash chromatography on a Biotage Isorela One instrument with a SNAP Ultra C18 12 g column (Biotage) using an acetonitrile/water gradient at 12 mL/min flow rate. The elution was started with 0% acetonitrile for 4.5 min, and the acetonitrile concentration was raised to 100% over 28.2 min, followed by an isocratic elution of 100% acetonitrile for 13.3 min. The flash chromatography fraction containing the C10-C10 was further separated and purified using 0.25-mm thin-layer chromatographic plates (SiliCycle SilicaPlate F-254) and developed with n-hexanes-ethyl acetate-acetic acid (24:74:2). The bands were scraped from the plates, and the HAAs, including C10-C10, were extracted from the silica with chloroform-methanol (5:1). 3-OH-C10 was purchased from Sigma-Aldrich. All compounds were dissolved in ethanol or methanol as indicated to prepare stock solutions. Final aqueous compound dilutions were prepared freshly on the days of the experiment. Control solutions contained equal amounts of ethanol or methanol (0.05% for most experiments and not exceeding 0.5% for the highest concentrations tested). Chemical synthesis and HPLC purification of C10-C10 is described in Datasets S1 and S2.
LC-MS Analysis of HAAs.
Samples were prepared by diluting stock solutions using MeOH to final concentration of 50 parts per million (ppm). 16-Hydroxyhexadecanoic acid at 20 ppm was added to samples as internal standard (68). The analyses were performed with a Quattro II triple quadrupole mass spectrometer (Micromass) equipped with a Z-spray interface using electrospray ionization in negative mode. The capillary voltage was set at 3.5 kV and the cone voltage at 25 V. The source temperature was kept at 120 °C and the desolvation gas at 150 °C. The scanning mass range was from 130 to 930 Da. The instrument was interfaced to an HPLC (Waters 2795) equipped with a 100 × 4 mm internal diameter (i.d.) Luna Omega PS C18 reversed-phase column (particle size 5 µm) using a water-acetonitrile gradient with a constant 2 mmol/L−1 concentration of ammonium acetate (0.6 mL/min−1). Quantification of free 3-OH-C10 in purified C10-C10, Rha-Rha-C10-C10, Rha-C10-C10, Rha-Rha-C14-C14, or synthetic C10-C10 was performed as reported previously (34) and are presented in SI Appendix, Table S2.
Plant Material and Growth Conditions.
Arabidopsis thaliana ecotype Col-0 was used as WT parent for all experiments. Seeds from fls2/efr-1 (38, 39), bak1-5, bkk1-1, bak1-5/bkk1-1 (40), cerk1-2 (42), bik1/pbl1 (41), rbohD, msl4/5/6/9/10, and mca1/2 (51) were provided by C. Zipfel. Seeds from sobir1-12 and sobir1-13 (43) were provided by F. Brunner (Center for Plant Molecular Biology, University of Tübingen, Tübingen, PlantResponse). Seeds from sd1-29 (lore-5), Col-0AEQ, and lore-5AEQ were provided by S. Ranf (45). loh1 and fah1/2 seed (52) were provided by I. Feussner (University of Göttingen, Germany). dorn1-1 seeds (44) were obtained from NASC stock (SALK_042209). All mutants are in the Col-0 background. Plants were grown on soil in growth chambers at 20 °C, under 12-h light/12-h dark regime with fluorescent light of 150 µmol/m−2/s−1 and 60% relative humidity.
Extracellular ROS Production and Calcium Signaling.
ROS assays were performed on 4- to 6-wk-old Arabidopsis plants cultured on soil. Briefly, 5-mm-long petiole sections were cut and placed in 150 µL distilled water overnight in 96-wells plate (PerkinElmer) (46). Then, the protocol was conducted as previously described (82). Luminescence (relative light units, RLU) was measured every 2 min during 46 or 720 min with a Tecan Infinite F200 PRO (or a TECAN CM SPARK for SI Appendix, Fig. S6), Tecan. Total ROS production was calculated by summing RLU measured between 4 to 46 or 4 to 720 min after treatment. Control was realized on petioles of WT or mutant plants. [Ca2+]cyt measurements were done as previously described (34).
MAPK Phosphorylation Assays.
For MAPK phosphorylation assays, three leaf disks (9-mm diameter) were collected from 4- to 6-wk-old Arabidopsis plants grown on soil and incubated 8 h in distilled water. Leaf disks were mock treated or treated with different molecules. Fifteen minutes, 1 h, and 3 h after treatment, plant tissues were frozen in liquid nitrogen. To extract proteins, 60 mg leaf tissues were ground in a homogenizer Potter-Elvehjem with 60 µL extraction buffer (0.35 M Tris HCl [pH 6.8], 30% [volume(vol)/vol] glycerol, 10% [vol/vol] sodium dodecyl sulfate [SDS], 0.6 M dithiothreitol [DTT], 0.012% [weight/vol] bromophenol blue). Total protein extracts were denatured for 7 min at 95 °C, centrifuged at 11,000 g for 5 min, and 30 µL supernatant was separated by 12% SDS-polyacrylamide gel electrophoresis (PAGE). Phosphorylation state of MAPKs by immunobloting was performed as previously described (46).
Depletion Binding Assay.
Protein expression and ligand depletion were performed as described (84) with slight modifications. Briefly, LORE extracellular domain mCherry fusion protein (eLORE-mCherry) or mCherry control fused with N-terminal LORE signal peptide were transiently expressed in N. benthamiana leaf apoplast using a 1:1 mixture of Agrobacterium tumefaciens GV3101 carrying the respective expression plasmid or the p19 silencing suppressor (total optical density [OD]600 0.5). At 5 d postinoculation, N. benthamiana leaves were vacuum infiltrated with water, and apoplastic washing fluid (AWF) was collected by centrifugation (20 min, 800 g, 4 °C). AWF was cleared by centrifugation (20 min, 8,000 g, 4 °C), filter sterilized (pore size 0.22 μm), desalted (PD-10 Desalting Columns, Cytiva), and concentrated to a total protein concentration of 2 mg/mL (Vivaspin 20, 30,000 Da molecular weight [MW] cutoff, Sartorius). For ligand depletion assay, AWF concentrate was mixed in 9:1 ratio (vol:vol) with synthetic C10-C10 (50 μM) or 3-OH-C10 (5 μM) and incubated for 1 h at 4 °C on a rotator. Unbound ligands were separated from the mixture (Vivaspin 500, 30,000 Da MW cutoff, Sartorius). Content of unbound C10-C10 and 3-OH-C10 in filtrates was assessed by [Ca2+]cyt measurements using LORE-overexpressing (p35S:LORE/lore-1) and lore-1 seedlings with a filtrate:water ratio of 1:4 (vol:vol).
Conductivity Assay.
The assay was performed as described previously (83), with few modifications. Eight leaf discs of 6-mm diameter were incubated in distilled water overnight. One disk was transferred into a 1.5-mL tube containing fresh distilled water and the corresponding elicitor concentration or ethanol for control. Conductivity measurements (three to four replicates for each treatment) were then conducted using a B-771 LaquaTwin (Horiba) conductivity meter.
P. syringae Culture and Disease-Resistance Assays.
P. syringae pv. tomato strain DC3000 was grown at 28 °C under stirring in King’s B (KB) liquid medium supplemented with antibiotics: 50 µg/mL−1 rifampicin and 50 µg/mL−1 kanamycin. For local protection assays, 15 seeds were sown per pot and grown for 3 to 5 wk in soil. Plants were sprayed with molecules or ethanol as control and were placed for 2 d in high humidity atmosphere before infections. Plants were inoculated by spraying the leaves with 3 mL bacterial suspension at an OD600 of 0.01 (0.025% Silwet L-77, 10 mM MgCl2). For each experiment, five pots per conditions were used (n = 5). Quantification (colony forming units [CFU]) of in planta bacterial growth was performed 3 days post infection (dpi). To this end, all plant leaves from the same pot were harvested, weighed, and crushed in a mortar with 10 mL of 10 mM MgCl2, and serial dilutions were performed. For each dilution, 10 µL were dropped on KB plate supplemented with appropriate antibiotics. CFU were counted after 2 d of incubation at 28 °C. The number of bacteria per milligram of plants fresh mass was obtained with the following formula:
with N equal to CFU number, Vi the volume depot on plate, Vd the total volume, n the dilution number, and M the plants fresh mass.
Supplementary Material
Acknowledgments
We are thankful to Laetitia Parent and Sylvain Milot for technical support and Ralph Hückelhoven for critical discussion. This work was supported by grants from EliDeRham and Rhamnoprot (Région Grand Est). The project Rhamnoprot is cofunded by the European Union FEDER (Fonds Européens de Développement Régional) program. Financial support from the CNRS, the MESRI (Ministère de l’Enseignement Supérieur, de la Recherche et de l’Innovation), and the Federative Research Structure SFR Condorcet are gratefully acknowledged. Works on rhamnolipids in the E.D. and C.G. laboratories are funded by the Natural Sciences and Engineering Research Council of Canada (NSERC) through Discovery Grants RGPIN-2015-03931 and RGPIN-2020-06771 (to E.D.) and RGPIN-2016-04950 (to C.G.). Work in the S.R. laboratory is supported by the German Research Foundation (SFB924/TP-B10 and Emmy Noether programme RA2541/1). M.C. thanks The Natural Sciences and Engineering Research Council of Canada (NSERC) and Fonds de Recherche du Québec – Nature et Technologies (FRQNT) for M.Sc. and Ph.D. scholarships.
Footnotes
Competing interest statement: Technical University of Munich has filed a patent application to inventors A.K., C.D., T.H., and S.R.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2101366118/-/DCSupplemental.
Data Availability
All study data are included in the article and/or supporting information.
References
- 1.Cook D. E., Mesarich C. H., Thomma B. P., Understanding plant immunity as a surveillance system to detect invasion. Annu. Rev. Phytopathol. 53, 541–563 (2015). [DOI] [PubMed] [Google Scholar]
- 2.Kanyuka K., Rudd J. J., Cell surface immune receptors: The guardians of the plant’s extracellular spaces. Curr. Opin. Plant Biol. 50, 1–8 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boller T., Felix G., A renaissance of elicitors: Perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu. Rev. Plant Biol. 60, 379–406 (2009). [DOI] [PubMed] [Google Scholar]
- 4.Newman M. A., Sundelin T., Nielsen J. T., Erbs G., MAMP (microbe-associated molecular pattern) triggered immunity in plants. Front. Plant Sci. 4, 139 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boutrot F., Zipfel C., Function, discovery, and exploitation of plant pattern recognition receptors for broad-spectrum disease resistance. Annu. Rev. Phytopathol. 55, 257–286 (2017). [DOI] [PubMed] [Google Scholar]
- 6.Ranf S., Sensing of molecular patterns through cell surface immune receptors. Curr. Opin. Plant Biol. 38, 68–77 (2017). [DOI] [PubMed] [Google Scholar]
- 7.Couto D., Zipfel C., Regulation of pattern recognition receptor signalling in plants. Nat. Rev. Immunol. 16, 537–552 (2016). [DOI] [PubMed] [Google Scholar]
- 8.Bigeard J., Colcombet J., Hirt H., Signaling mechanisms in pattern-triggered immunity (PTI). Mol. Plant 8, 521–539 (2015). [DOI] [PubMed] [Google Scholar]
- 9.Garcia-Brugger A., et al., Early signaling events induced by elicitors of plant defenses. Mol. Plant Microbe Interact. 19, 711–724 (2006). [DOI] [PubMed] [Google Scholar]
- 10.Wu S., Shan L., He P., Microbial signature-triggered plant defense responses and early signaling mechanisms. Plant Sci. 228, 118–126 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.De Vleesschauwer D., Gheysen G., Höfte M., Hormone defense networking in rice: Tales from a different world. Trends Plant Sci. 18, 555–565 (2013). [DOI] [PubMed] [Google Scholar]
- 12.Glazebrook J., Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu. Rev. Phytopathol. 43, 205–227 (2005). [DOI] [PubMed] [Google Scholar]
- 13.Robert-Seilaniantz A., Grant M., Jones J. D., Hormone crosstalk in plant disease and defense: More than just jasmonate-salicylate antagonism. Annu. Rev. Phytopathol. 49, 317–343 (2011). [DOI] [PubMed] [Google Scholar]
- 14.Trdá L., et al., Perception of pathogenic or beneficial bacteria and their evasion of host immunity: Pattern recognition receptors in the frontline. Front. Plant Sci. 6, 219 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Abdel-Mawgoud A. M., Lépine F., Déziel E., Rhamnolipids: Diversity of structures, microbial origins and roles. Appl. Microbiol. Biotechnol. 86, 1323–1336 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Irorere V. U., Tripathi L., Marchant R., McClean S., Banat I. M., Microbial rhamnolipid production: A critical re-evaluation of published data and suggested future publication criteria. Appl. Microbiol. Biotechnol. 101, 3941–3951 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Perneel M., et al., Phenazines and biosurfactants interact in the biological control of soil-borne diseases caused by Pythium spp. Environ. Microbiol. 10, 778–788 (2008). [DOI] [PubMed] [Google Scholar]
- 18.Chrzanowski Ł., Ławniczak Ł., Czaczyk K., Why do microorganisms produce rhamnolipids? World J. Microbiol. Biotechnol. 28, 401–419 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nickzad A., Déziel E., The involvement of rhamnolipids in microbial cell adhesion and biofilm development–An approach for control? Lett. Appl. Microbiol. 58, 447–453 (2014). [DOI] [PubMed] [Google Scholar]
- 20.Vatsa P., Sanchez L., Clément C., Baillieul F., Dorey S., Rhamnolipid biosurfactants as new players in animal and plant defense against microbes. Int. J. Mol. Sci. 11, 5095–5108 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Abdel-Mawgoud A. M., Lépine F., Déziel E., A stereospecific pathway diverts β-oxidation intermediates to the biosynthesis of rhamnolipid biosurfactants. Chem. Biol. 21, 156–164 (2014). [DOI] [PubMed] [Google Scholar]
- 22.Burch A. Y., et al., Pseudomonas syringae coordinates production of a motility-enabling surfactant with flagellar assembly. J. Bacteriol. 194, 1287–1298 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Déziel E., Lépine F., Milot S., Villemur R., rhlA is required for the production of a novel biosurfactant promoting swarming motility in Pseudomonas aeruginosa: 3-(3-hydroxyalkanoyloxy)alkanoic acids (HAAs), the precursors of rhamnolipids. Microbiology (Reading) 149, 2005–2013 (2003). [DOI] [PubMed] [Google Scholar]
- 24.Plotnikova J. M., Rahme L. G., Ausubel F. M., Pathogenesis of the human opportunistic pathogen Pseudomonas aeruginosa PA14 in Arabidopsis. Plant Physiol. 124, 1766–1774 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tremblay J., Richardson A. P., Lépine F., Déziel E., Self-produced extracellular stimuli modulate the Pseudomonas aeruginosa swarming motility behaviour. Environ. Microbiol. 9, 2622–2630 (2007). [DOI] [PubMed] [Google Scholar]
- 26.Gerstel U., Czapp M., Bartels J., Schröder J. M., Rhamnolipid-induced shedding of flagellin from Pseudomonas aeruginosa provokes hBD-2 and IL-8 response in human keratinocytes. Cell. Microbiol. 11, 842–853 (2009). [DOI] [PubMed] [Google Scholar]
- 27.Al-Tahhan R. A., Sandrin T. R., Bodour A. A., Maier R. M., Rhamnolipid-induced removal of lipopolysaccharide from Pseudomonas aeruginosa: Effect on cell surface properties and interaction with hydrophobic substrates. Appl. Environ. Microbiol. 66, 3262–3268 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Andrä J., et al., Endotoxin-like properties of a rhamnolipid exotoxin from Burkholderia (Pseudomonas) plantarii: Immune cell stimulation and biophysical characterization. Biol. Chem. 387, 301–310 (2006). [DOI] [PubMed] [Google Scholar]
- 29.Bauer J., Brandenburg K., Zähringer U., Rademann J., Chemical synthesis of a glycolipid library by a solid-phase strategy allows elucidation of the structural specificity of immunostimulation by rhamnolipids. Chemistry 12, 7116–7124 (2006). [DOI] [PubMed] [Google Scholar]
- 30.Dössel J., Meyer-Hoffert U., Schröder J. M., Gerstel U., Pseudomonas aeruginosa-derived rhamnolipids subvert the host innate immune response through manipulation of the human beta-defensin-2 expression. Cell. Microbiol. 14, 1364–1375 (2012). [DOI] [PubMed] [Google Scholar]
- 31.Gonzalez-Juarrero M., et al., Polar lipids of Burkholderia pseudomallei induce different host immune responses. PLoS One 8, e80368 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sanchez L., et al., Rhamnolipids elicit defense responses and induce disease resistance against biotrophic, hemibiotrophic, and necrotrophic pathogens that require different signaling pathways in Arabidopsis and highlight a central role for salicylic acid. Plant Physiol. 160, 1630–1641 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Varnier A. L., et al., Bacterial rhamnolipids are novel MAMPs conferring resistance to Botrytis cinerea in grapevine. Plant Cell Environ. 32, 178–193 (2009). [DOI] [PubMed] [Google Scholar]
- 34.Kutschera A., et al., Bacterial medium-chain 3-hydroxy fatty acid metabolites trigger immunity in Arabidopsis plants. Science 364, 178–181 (2019). [DOI] [PubMed] [Google Scholar]
- 35.Qi J., Wang J., Gong Z., Zhou J. M., Apoplastic ROS signaling in plant immunity. Curr. Opin. Plant Biol. 38, 92–100 (2017). [DOI] [PubMed] [Google Scholar]
- 36.Kadota Y., Shirasu K., Zipfel C., Regulation of the NADPH oxidase RBOHD during plant immunity. Plant Cell Physiol. 56, 1472–1480 (2015). [DOI] [PubMed] [Google Scholar]
- 37.Torres M. A., Dangl J. L., Jones J. D., Arabidopsis gp91phox homologues AtrbohD and AtrbohF are required for accumulation of reactive oxygen intermediates in the plant defense response. Proc. Natl. Acad. Sci. U.S.A. 99, 517–522 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chinchilla D., Bauer Z., Regenass M., Boller T., Felix G., The Arabidopsis receptor kinase FLS2 binds flg22 and determines the specificity of flagellin perception. Plant Cell 18, 465–476 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zipfel C., et al., Perception of the bacterial PAMP EF-Tu by the receptor EFR restricts Agrobacterium-mediated transformation. Cell 125, 749–760 (2006). [DOI] [PubMed] [Google Scholar]
- 40.Roux M., et al., The Arabidopsis leucine-rich repeat receptor-like kinases BAK1/SERK3 and BKK1/SERK4 are required for innate immunity to hemibiotrophic and biotrophic pathogens. Plant Cell 23, 2440–2455 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li L., et al., The FLS2-associated kinase BIK1 directly phosphorylates the NADPH oxidase RbohD to control plant immunity. Cell Host Microbe 15, 329–338 (2014). [DOI] [PubMed] [Google Scholar]
- 42.Miya A., et al., CERK1, a LysM receptor kinase, is essential for chitin elicitor signaling in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 104, 19613–19618 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang W., et al., Arabidopsis receptor-like protein30 and receptor-like kinase suppressor of BIR1-1/EVERSHED mediate innate immunity to necrotrophic fungi. Plant Cell 25, 4227–4241 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Choi J., et al., Identification of a plant receptor for extracellular ATP. Science 343, 290–294 (2014). [DOI] [PubMed] [Google Scholar]
- 45.Ranf S., et al., A lectin S-domain receptor kinase mediates lipopolysaccharide sensing in Arabidopsis thaliana. Nat. Immunol. 16, 426–433 (2015). [DOI] [PubMed] [Google Scholar]
- 46.Luzuriaga-Loaiza W. P., et al., Synthetic rhamnolipid bolaforms trigger an innate immune response in Arabidopsis thaliana. Sci. Rep. 8, 8534 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shang-Guan K., et al., Lipopolysaccharides trigger two successive bursts of reactive oxygen species at distinct cellular locations. Plant Physiol. 176, 2543–2556 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xin X. F., He S. Y., Pseudomonas syringae pv. tomato DC3000: A model pathogen for probing disease susceptibility and hormone signaling in plants. Annu. Rev. Phytopathol. 51, 473–498 (2013). [DOI] [PubMed] [Google Scholar]
- 49.Costa S. G., Déziel E., Lépine F., Characterization of rhamnolipid production by Burkholderia glumae. Lett. Appl. Microbiol. 53, 620–627 (2011). [DOI] [PubMed] [Google Scholar]
- 50.Ham J. H., Melanson R. A., Rush M. C., Burkholderia glumae: Next major pathogen of rice? Mol. Plant Pathol. 12, 329–339 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stephan A. B., Kunz H. H., Yang E., Schroeder J. I., Rapid hyperosmotic-induced Ca2+ responses in Arabidopsis thaliana exhibit sensory potentiation and involvement of plastidial KEA transporters. Proc. Natl. Acad. Sci. U.S.A. 113, E5242–E5249 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lenarčič T., et al., Eudicot plant-specific sphingolipids determine host selectivity of microbial NLP cytolysins. Science 358, 1431–1434 (2017). [DOI] [PubMed] [Google Scholar]
- 53.Caiazza N. C., Shanks R. M., O’Toole G. A., Rhamnolipids modulate swarming motility patterns of Pseudomonas aeruginosa. J. Bacteriol. 187, 7351–7361 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nickzad A., Lépine F., Déziel E., Quorum sensing controls swarming motility of Burkholderia glumae through regulation of rhamnolipids. PLoS One 10, e0128509 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yu X., et al., Transcriptional responses of Pseudomonas syringae to growth in epiphytic versus apoplastic leaf sites. Proc. Natl. Acad. Sci. U.S.A. 110, E425–E434 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Davey M. E., Caiazza N. C., O’Toole G. A., Rhamnolipid surfactant production affects biofilm architecture in Pseudomonas aeruginosa PAO1. J. Bacteriol. 185, 1027–1036 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hirai H., et al., Glycosylation regulates specific induction of rice immune responses by Acidovorax avenae flagellin. J. Biol. Chem. 286, 25519–25530 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Taguchi F., et al., Glycosylation of flagellin from Pseudomonas syringae pv. tabaci 6605 contributes to evasion of host tobacco plant surveillance system. Physiol. Mol. Plant Pathol. 74, 11–17 (2009). [Google Scholar]
- 59.Hanuszkiewicz A., et al., Identification of the flagellin glycosylation system in Burkholderia cenocepacia and the contribution of glycosylated flagellin to evasion of human innate immune responses. J. Biol. Chem. 289, 19231–19244 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Henry G., Deleu M., Jourdan E., Thonart P., Ongena M., The bacterial lipopeptide surfactin targets the lipid fraction of the plant plasma membrane to trigger immune-related defence responses. Cell. Microbiol. 13, 1824–1837 (2011). [DOI] [PubMed] [Google Scholar]
- 61.Nasir M. N., et al., Differential interaction of synthetic glycolipids with biomimetic plasma membrane lipids correlates with the plant biological response. Langmuir 33, 9979–9987 (2017). [DOI] [PubMed] [Google Scholar]
- 62.Robineau M., et al., Synthetic mono-rhamnolipids display direct antifungal effects and trigger an innate immune response in tomato against Botrytis cinerea. Molecules 25, 3108 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Abbasi H., Noghabi K. A., Ortiz A., Interaction of a bacterial monorhamnolipid secreted by Pseudomonas aeruginosa MA01 with phosphatidylcholine model membranes. Chem. Phys. Lipids 165, 745–752 (2012). [DOI] [PubMed] [Google Scholar]
- 64.Aranda F. J., et al., Thermodynamics of the interaction of a dirhamnolipid biosurfactant secreted by Pseudomonas aeruginosa with phospholipid membranes. Langmuir 23, 2700–2705 (2007). [DOI] [PubMed] [Google Scholar]
- 65.Ortiz A., Aranda F. J., Teruel J. A., Interaction of dirhamnolipid biosurfactants with phospholipid membranes: A molecular level study. Adv. Exp. Med. Biol. 672, 42–53 (2010). [DOI] [PubMed] [Google Scholar]
- 66.Sánchez M., Aranda F. J., Teruel J. A., Ortiz A., Interaction of a bacterial dirhamnolipid with phosphatidylcholine membranes: A biophysical study. Chem. Phys. Lipids 161, 51–55 (2009). [DOI] [PubMed] [Google Scholar]
- 67.Monnier N., et al., Exploring the dual interaction of natural rhamnolipids with plant and fungal biomimetic plasma membranes through biophysical studies. Int. J. Mol. Sci. 20, 1009 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lépine F., Déziel E., Milot S., Villemur R., Liquid chromatographic/mass spectrometric detection of the 3-(3-hydroxyalkanoyloxy) alkanoic acid precursors of rhamnolipids in Pseudomonas aeruginosa cultures. J. Mass Spectrom. 37, 41–46 (2002). [DOI] [PubMed] [Google Scholar]
- 69.Kownatzki R., Tümmler B., Döring G., Rhamnolipid of Pseudomonas aeruginosa in sputum of cystic fibrosis patients. Lancet 1, 1026–1027 (1987). [DOI] [PubMed] [Google Scholar]
- 70.Read R. C., et al., Effect of Pseudomonas aeruginosa rhamnolipids on mucociliary transport and ciliary beating. J. Appl. Physiol. (1985) 72, 2271–2277 (1992). [DOI] [PubMed] [Google Scholar]
- 71.Somerville M., et al., Release of mucus glycoconjugates by Pseudomonas aeruginosa rhamnolipid into feline trachea in vivo and human bronchus in vitro. Am. J. Respir. Cell Mol. Biol. 6, 116–122 (1992). [DOI] [PubMed] [Google Scholar]
- 72.Germer A., et al., Exploiting the natural diversity of RhlA acyltransferases for the synthesis of the rhamnolipid precursor 3-(3-Hydroxyalkanoyloxy)alkanoic acid. Appl. Environ. Microbiol. 86, e02317–e02319 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ranf S., Immune sensing of lipopolysaccharide in plants and animals: Same but different. PLoS Pathog. 12, e1005596 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Compant S., Nowak J., Coenye T., Clément C., Ait Barka E., Diversity and occurrence of Burkholderia spp. in the natural environment. FEMS Microbiol. Rev. 32, 607–626 (2008). [DOI] [PubMed] [Google Scholar]
- 75.Kay E., Bertolla F., Vogel T. M., Simonet P., Opportunistic colonization of Ralstonia solanacearum-infected plants by Acinetobacter sp. and its natural competence development. Microb. Ecol. 43, 291–297 (2002). [DOI] [PubMed] [Google Scholar]
- 76.Silby M. W., Winstanley C., Godfrey S. A., Levy S. B., Jackson R. W., Pseudomonas genomes: Diverse and adaptable. FEMS Microbiol. Rev. 35, 652–680 (2011). [DOI] [PubMed] [Google Scholar]
- 77.Toth I. K., Pritchard L., Birch P. R. J., Comparative genomics reveals what makes an enterobacterial plant pathogen. Annu. Rev. Phytopathol. 44, 305–336 (2006). [DOI] [PubMed] [Google Scholar]
- 78.Yu X., et al., Transcriptional analysis of the global regulatory networks active in Pseudomonas syringae during leaf colonization. mBio 5, e01683–e14 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhu K., Rock C. O., RhlA converts beta-hydroxyacyl-acyl carrier protein intermediates in fatty acid synthesis to the beta-hydroxydecanoyl-beta-hydroxydecanoate component of rhamnolipids in Pseudomonas aeruginosa. J. Bacteriol. 190, 3147–3154 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Buscaill P., et al., Glycosidase and glycan polymorphism control hydrolytic release of immunogenic flagellin peptides. Science 364, eaav0748 (2019). [DOI] [PubMed] [Google Scholar]
- 81.Taguchi F., et al., Effects of glycosylation on swimming ability and flagellar polymorphic transformation in Pseudomonas syringae pv. tabaci 6605. J. Bacteriol. 190, 764–768 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Smith J. M., Heese A., Rapid bioassay to measure early reactive oxygen species production in Arabidopsis leave tissue in response to living Pseudomonas syringae. Plant Methods 10, 6 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Magnin-Robert M., et al., Modifications of sphingolipid content affect tolerance to hemibiotrophic and necrotrophic pathogens by modulating plant defense responses in Arabidopsis. Plant Physiol. 169, 2255–2274 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Shu L.-J., et al., Low cost, medium throughput depletion-binding assay for screening S-domain-receptor ligand interactions using in planta protein expression. bioRxiv [Preprint] (2021). 10.1101/2021.06.16.448648 (Accessed 18 June 2021). [DOI]
- 85.König S., et al., Arabidopsis mutants of sphingolipid fatty acid α-hydroxylases accumulate ceramides and salicylates. New Phytol. 196, 1086–1097 (2012). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or supporting information.