Significance
This study demonstrates that underexpression of succinate dehydrogenase (SDH) subunits resulting in accumulation of oncogenic succinate is a common, adverse, epigenetic modulating feature in clear cell renal cell carcinoma (ccRCC), during pathogenesis and progression. The study sheds light on the mechanisms of down-regulation of SDH subunits in ccRCC and deciphers the consequent oncogenic effects. It shows that functional SDH deficiency is a common feature of ccRCC (∼80% of all kidney cancers), and not just limited to the 0.05 to 0.5% of kidney cancers with germline SDH mutations.
Keywords: succinate, succinate dehydrogenase, kidney cancer, TET-2
Abstract
Reduced succinate dehydrogenase (SDH) activity resulting in adverse succinate accumulation was previously considered relevant only in 0.05 to 0.5% of kidney cancers associated with germline SDH mutations. Here, we sought to examine a broader role for SDH loss in kidney cancer pathogenesis/progression. We report that underexpression of SDH subunits resulting in accumulation of oncogenic succinate is a common feature in clear cell renal cell carcinoma (ccRCC) (∼80% of all kidney cancers), with a marked adverse impact on survival in ccRCC patients (n = 516). We show that SDH down-regulation is a critical brake in the TCA cycle during ccRCC pathogenesis and progression. In exploring mechanisms of SDH down-regulation in ccRCC, we report that Von Hippel-Lindau loss-induced hypoxia-inducible factor–dependent up-regulation of miR-210 causes direct inhibition of the SDHD transcript. Moreover, shallow deletion of SDHB occurs in ∼20% of ccRCC. We then demonstrate that SDH loss-induced succinate accumulation contributes to adverse loss of 5-hydroxymethylcytosine, gain of 5-methylcytosine, and enhanced invasiveness in ccRCC via inhibition of ten-eleven translocation (TET)-2 activity. Intriguingly, binding affinity between the catalytic domain of recombinant TET-2 and succinate was found to be very low, suggesting that the mechanism of succinate-induced attenuation of TET-2 activity is likely via product inhibition rather than competitive inhibition. Finally, exogenous ascorbic acid, a TET-activating demethylating agent, led to reversal of the above oncogenic effects of succinate in ccRCC cells. Collectively, our study demonstrates that functional SDH deficiency is a common adverse feature of ccRCC and not just limited to the kidney cancers associated with germline SDH mutations.
Clear cell renal cell carcinoma (ccRCC) is by far the most common type of kidney cancer, accounting for ∼80% of all kidney cancers (1). Despite recent advances, metastatic ccRCC is a generally incurable malignancy, with a 5-y survival rate <20%, highlighting the need for further biologic and therapeutic insights in this disease.
The succinate dehydrogenase (SDH) complex is the only enzyme that is an integral component of both the TCA cycle and the Electron Transport Chain, thus playing an important role in oxidative phosphorylation. SDH converts succinate to fumarate in the TCA cycle. The complex is located in the inner mitochondrial membrane and is composed of four subunits: two hydrophilic subunits, SDHA and SDHB, and two hydrophobic membrane anchoring subunits, SDHC and SDHD.
In this study, using bioinformatic analyses of ccRCC TCGA (KIRC) data (transcriptome, methylome, and survival), ccRCC metabolomic repository, primary ccRCC tumors with adjacent normal renal tissue, and an array of mechanistic/representative experiments, we 1) investigated the expression of SDH subunits in ccRCC and the prognostic and functional consequences of loss of SDH in ccRCC, 2) shed light on the mechanisms of down-regulation of SDH in ccRCC, 3) established the role of succinate as an important epigenetic modulating oncometabolite in ccRCC pathogenesis and progression, and 4) determined the potential of ascorbic acid (AA) in reversing the oncogenic effects of succinate in ccRCC.
Results
SDH Subunits B, C, D Are Significantly Underexpressed in ccRCC and Associated with Markedly Worse Survival.
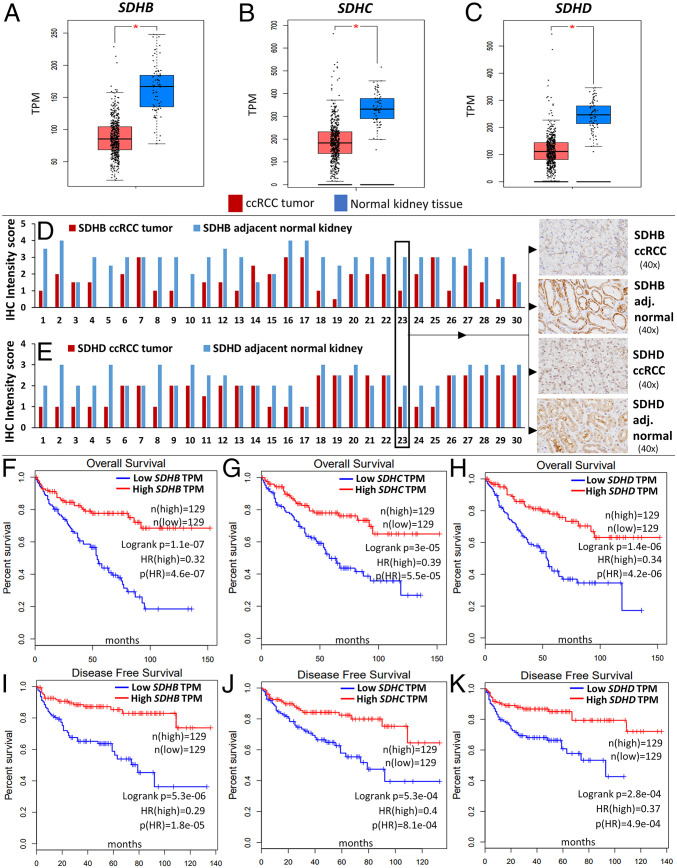
Analysis of the TCGA-KIRC dataset revealed that SDH subunits SDHB, SDHC, and SDHD are significantly down-regulated in ccRCC tumors (n = 573) compared to normal renal tissue (n = 72) (P < 0.01 for each subunit, Fig. 1 A–C). Paired ccRCC tumor–normal comparisons from TCGA-KIRC (n = 72) also revealed marked down-regulation of SDH subunits (SI Appendix, Fig. S1). SDHA expression in ccRCC, however, is not different from normal kidney tissue and does not impact survival (SI Appendix, Fig. S6).
Fig. 1.
SDH subunits (B, C, D) are significantly underexpressed in ccRCC and associated with markedly worse survival. (A–C) Analysis of the TCGA-KIRC dataset revealed that SDH subunits SDHB, SDHC, and SDHD are significantly down-regulated in ccRCC tumor (n = 573) compared to normal renal tissue (n = 72) (P < 0.05 for each subunit). Paired ccRCC tumor–normal comparisons are shown in SI Appendix, Fig. S1. (D, E) SDHB and SDHD Immunohistochemistry in primary ccRCC versus adjacent normal kidney tissue revealed down-regulation of both subunits in ccRCC at the protein level. Stain intensity was on a scale of 0 (negative) to 4+ (strongly positive). For both antibodies, the nonneoplastic tubular cells in the resection stained consistently stronger than the clear cell carcinoma cells (n = 30, paired t test P values of 1.4E-08 and 4.2E-06 for SDHB and SDHD, respectively). Representative IHC images are shown (case 23). Only two out of the 30 cases did not have a decrease in either SDHB or SDHD intensity score in ccRCC compared to adjacent normal kidney. (F–K) Survival analyses of the TCGA-KIRC dataset revealed a markedly worse overall survival (OS) (F–H) and disease-free survival (DFS) (I–K) with lower expression of SDHB, SDHC, and SDHD. The OS hazard ratio (HR) for high (n = 129) versus low (n = 129) SDHB, SDHC, and SDHD expression was 0.32, 0.39, and 0.34, respectively (high versus low quartiles, P < 0.001 for each). The DFS HR for high (n = 129) versus low (n = 129) SDHB, SDHC, and SDHD was 0.29, 0.4, and 0.37, respectively (high versus low quartiles, P < 0.001 for each). (Multivariate survival analysis adjusted for stage, grade, age, and sex also revealed a significant increase in survival with higher expression of SDH subunits—shown in SI Appendix, Fig. S5 A–D.)
We then investigated the protein expression levels of SDHB and SDHD in ccRCC tumors compared with adjacent normal tissue using immunohistochemistry. Consistent with the mRNA expression pattern, there was loss of both SDHB and SDHD in ccRCC compared to adjacent normal kidney tissue, as evidenced by the reduced intensity of immunostaining in ccRCC cells (n = 30 ccRCC tumors with paired normed tissue; paired t test P values < 0.001 for both SDHB and SDHD; Fig. 1 D and E and SI Appendix, Fig. S4 A and B). Only two out of the 30 cases did not have a decrease in either SDHB or SDHD intensity score in ccRCC compared to adjacent kidney. Mass spectrometry based proteomic profiling of ccRCC tumors (CPTAC samples) also showed a marked down-regulation of SDH subunits in ccRCC compared to normal kidney (SI Appendix, Figs. S2 and S3).
Survival analyses of the TCGA-KIRC dataset revealed a markedly worse overall survival (OS) and disease-free survival (DFS) with lower expression of SDHB, SDHC and SDHD. In univariate survival analysis, the OS Hazard Ratio (HR) for high (n = 129) versus low (n = 129) SDHB, SDHC and SDHD expression was 0.32, 0.39 and 0.34 respectively (high versus low quartiles, P < 0.001 for each, Fig. 1 F–H). The DFS HR for high (n = 129) versus low (n = 129) SDHB, SDHC and SDHD was 0.29, 0.4 and 0.37 respectively (high versus low quartiles, P < 0.001 for each, Fig. 1 I–K). Multivariate survival analysis adjusted for stage, grade, age, and sex also revealed a significant increase in survival with higher expression of SDH subunits (SI Appendix, Fig. S5 A–D). Furthermore, weaker immunostaining of SDHB has been shown to be adversely prognostic in ccRCC (2).
Down-regulation of SDH Is a Critical Brake in the TCA Cycle during ccRCC Pathogenesis and Progression.
In order to determine whether the down-regulation of SDH in ccRCC results in an accumulation of succinate, we analyzed the metabolomic repository of ccRCC (3). Primary ccRCC tumors (n = 138) had twofold higher succinate compared to adjacent normal kidney tissue (n = 138) (P < 0.001) (Fig. 2A). Furthermore, advanced (stage III/IV) ccRCC tumors (n = 90) had 1.6-fold higher succinate compared to early-stage (stage I/II) ccRCC tumors (n = 48) (P = 0.003) (Fig. 2B).
Fig. 2.
Down-regulation of SDH complex is a critical brake in Krebs cycle during ccRCC pathogenesis and progression. (A) Analysis of the ccRCC metabolite repository revealed that primary ccRCC tumors (n = 138) had twofold higher succinate compared to adjacent normal kidney tissue (n = 138) (adjusted P value = 6.9E-06). (B) Advanced (stage III/IV) ccRCC tumors (n = 90) had 1.6-fold higher succinate compared to early stage (stage I/II) ccRCC tumors (n = 48) (adjusted P = 0.003). (C, F) Fumarate hydratase and malate dehydrogenase are significantly lower in ccRCC tumor (n = 523) compared to normal renal tissue (n = 72) (KIRC TCGA, P < 0.01). (D, G) Despite lower fumarate hydratase and malate dehydrogenase, fumarate and malate are significantly lower in primary ccRCC tumor (n = 138) compared to adjacent normal renal tissue (n = 138) (0.3-fold and 0.5-fold, respectively; adjusted P = 1.2E-29 and 3.64E-21, respectively). (E, H) No significant difference in the malate and fumarate content between advanced (stage III/IV) ccRCC tumors (n = 90) and early stage (stage I/II) ccRCC tumors (n = 48). (I) Schematic representation depicting the accumulation of succinate with reduced SDH in ccRCC but reduced fumarate and malate despite reduced fumarate hydratase and malate dehydrogenase, highlighting that loss of SDH is a critical brake in Krebs cycle in ccRCC. [A, B, D, E, G, H have no error bars because the ccRCC metabolomic repository from which these data are derived (3) reports only average log2 fold change and adjusted P value for comparison between ccRCC and adjacent normal renal tissue for each metabolite]. (J, K) Knockdown of SDH subunits (SDHB, SDHD) in ccRCC cells (769P) resulted in accumulation of succinate and decrease in fumarate. (L–N) Simultaneous knockdown of SDH subunits (SDHB, SDHD), FH, and MDH2 (in 769P cells) resulted in accumulation of succinate but not of fumarate or malate. (Please see SI Appendix, Fig. S7 for knockdown of SDHB/SDHD/FH/MDH2.)
On the other hand, fumarate was significantly lower in primary ccRCC tumor (n = 138) compared to adjacent normal tissue (n = 138) (0.3-fold, P < 0.001, Fig. 2D), despite the fact that fumarate hydratase (FH) is significantly lower in ccRCC tumor (n = 523) compared to normal renal tissue (n = 72) (TCGA-KIRC, P < 0.01, Fig. 2C). Similarly, malate was significantly lower in primary ccRCC tumor (n = 138) compared to adjacent normal tissue (n = 138) (0.5-fold, P < 0.001, Fig. 2G), despite the fact that malate dehydrogenase (MDH) is significantly lower in ccRCC tumor (n = 523) compared to normal renal tissue (n = 72) (TCGA-KIRC, P < 0.01, Fig. 2F). Furthermore, there was no significant difference in the malate and fumarate content between advanced (stage III/IV) ccRCC tumors and early-stage (stage I/II) ccRCC tumors (Fig. 2 E and H).
Consistent with what is observed in ccRCC tumors, knockdown of SDHB and SDHD in ccRCC cells (769P) resulted in an expected increase in succinate and decrease in fumarate. Similarly, combined knockdown of SDHB, SDHD, FH, and MDH2 in 769P cells resulted in an increase in succinate but not of fumarate or malate (Fig. 2 J–N). Knockdown (fold change) of SDHB, SDHD, FH, and MDH2 is shown in SI Appendix, Fig. S7.
Put together, these data suggest that the down-regulation of SDH is a critical brake in the TCA cycle, preventing the conversion of succinate to fumarate and resulting in the accumulation of succinate. This accumulation of succinate is a feature of not just ccRCC pathogenesis (higher succinate in ccRCC tumor compared to adjacent normal) but also ccRCC progression (higher succinate in advanced stage compared to early-stage ccRCC).
Von Hippel-Lindau Loss Induced Hypoxia-Inducible Factor–Dependent Up-regulation of miR-210 in ccRCC Causes Direct Degradation of the SDHD Transcript.
Next, we sought to investigate the mechanism of SDH down-regulation in ccRCC. Initially, we studied the promoter (cytosine-phosphate-guanine) CpG island (CGI) methylation of SDH subunit genes in ccRCC versus normal kidney (TCGA; SI Appendix, Fig. S9). Although there was a statistically significant increase in promoter CGI methylation of SDHB, SDHD, and SDHC, the degree of increase in methylation was rather small and insufficient to explain the profound down-regulation of SDH subunits.
Therefore, we then investigated the possibility of microRNA (miRNA)-mediated down-regulation of SDH subunits. miR-210, a highly conserved miRNA, is known to be induced by hypoxia and co-ordinates various metabolic processes under hypoxic conditions. Furthermore, SDHD is a predicted target of this miRNA (mirDB-miRNA target prediction database). Given the Von Hippel-Lindau (VHL) loss induced pseudohypoxic signature of ccRCC, we hypothesized that miR-210 may play an important pathogenic role in ccRCC and may be involved in SDH down-regulation in this malignancy.
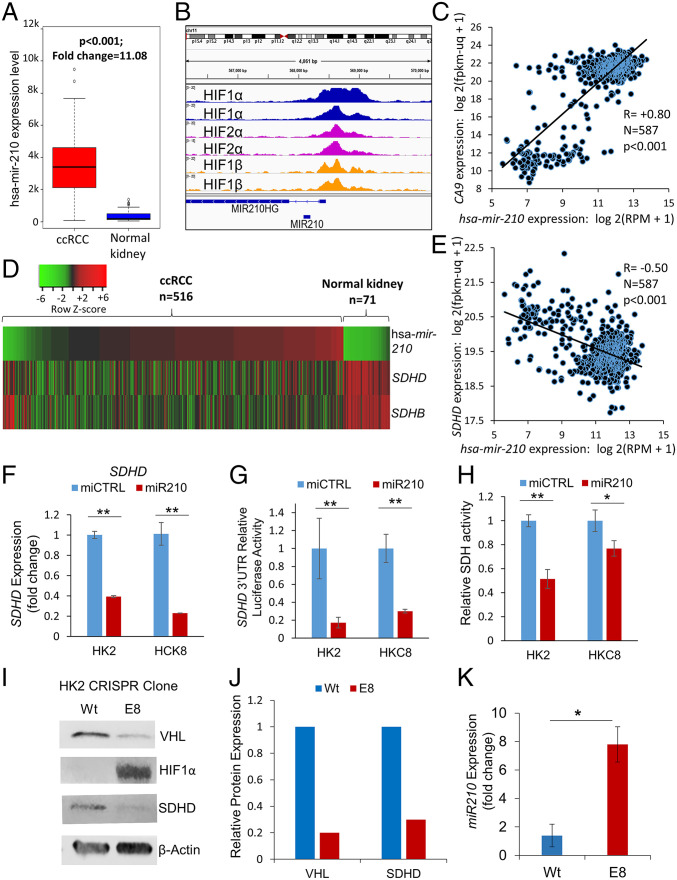
We found that miR-210 is the second-most up-regulated miRNA in ccRCC compared to normal kidney (11-fold, P < 0.001, TCGA, Fig. 3A). Paired ccRCC tumor–normal comparisons from TCGA-KIRC also revealed marked up-regulation of miR-210 (SI Appendix, Fig. S8). In the ccRCC cell line RCC-4, ChIP-seq (chromatin immunoprecipitation sequencing) revealed strong binding of hypoxia-inducible factor (HIF) 1α, HIF2α, and HIF1β to the miR-210 promoter region [Fig. 3B, data accessed through GSE120885 (4)]. Of all the miRNAs with a binding site for HIF2α in 786-O cells, the strength of binding of HIF2α on the miR-210 promoter is the highest by far [SI Appendix, Table S1, data accessed through GSE86092 (5)]. Furthermore, the HIF1α transcriptional target gene, CA9, strongly correlates with miR-210 expression in ccRCC (R = 0.80, P < 0.001, TCGA, Fig. 3C). We then mapped miR-210, SDHD and SDHB expression in ccRCC (n = 516) and normal kidney (n = 71) (TCGA, Fig. 3D). miR-210 expression had a significant negative correlation with SDHD (R = −0.5, P < 0.001, Fig. 3E) and with SDHB (R = −0.4, P < 0.001; SI Appendix, Fig. S10A).
Fig. 3.
VHL loss–induced HIF-dependent up-regulation of miR-210 in ccRCC causes direct degradation of the SDHD transcript in ccRCC. (A) miR-210 is markedly up-regulated in ccRCC compared to normal kidney (11-fold, P < 0.001, TCGA) (fold change = mean hsa-mir-210 expression levels in cancer samples/mean hsa-mir-210 expression levels in normal samples). (B) ChIP-seq for HIF1α, HIF2α, and HIF1β in the RCC4 cell line. HIF1α, HIF2α, and HIF1β show strong binding to the miR-210 promoter region. Data were accessed through GSE120885 (4). (C) The HIF1α transcriptional target gene, CA9, is strongly correlated with hsa-mir-210 expression in ccRCC (R = 0.80, TCGA). (D) Heatmap showing hsa-mir-210 expression along with SDHD and SDHB expression in ccRCC and normal kidney. (“n” as indicated, TCGA). (E) Marked inverse correlation between hsa-mir-210 expression and SDHD expression (R = −0.50, TCGA). (F) miR-210 or control miRNA were transfected into HK2 and HKC8 normal kidney cells. Cells were harvested 48 h posttransfection for SDHD gene expression. miR-210 significantly inhibited SDHD expression in both HK2 (P < 0.001) and HKC8 (P = 0.001) cell lines. (G) miR-210 or control miRNA were cotransfected with a SDHD 3′UTR reporter construct in HK2 and HKC8 cells, and luciferase activity was determined 48 h posttransfection. miR-210 inhibited SDHD 3′UTR activity in both HK2 (P = 0.004) and HKC8 (P = 0.002) cell lines. (H) miR-210 or control miRNA were transfected into HK2 and HKC8 normal kidney cells. SDH activity was measured 48 h posttransfection. miR-210 inhibited SDH activity in HK2 (P = 0.006) and HKC8 cell lines (P = 0.05). (I, J) CRISPR/Cas9 was used to generate a VHL knockdown clone, E8, from the HK2 cell line. VHL loss was validated by Western blot and Sanger sequencing (SI Appendix, Fig. S5). Western blot analysis showed marked elevation of HIF1α protein expression and down-regulation of SDHD in E8 compared to wt cells. (K) Knockdown of VHL in HK2 kidney cells results in up-regulation of hsa-miR-210-3p. (F–H, J, K) All data are indicated as means ± SE. P values for each pairwise comparison were derived from one-sided Student’s t test. *P ≤ 0.05; **P ≤ 0.01.
Next, we transfected miR-210 in immortalized human kidney cell lines HK2 and HKC8 and found that it led to marked down-regulation of SDHD expression in both HK2 (P < 0.001) and HKC8 (P < 0.001) cells (Fig. 3F). However, miR-210 transfection did not decrease SDHB expression (SI Appendix, Fig. S10B).
In order to determine whether SDHD down-regulation by miR-210 in these cell lines was through direct degradation of the SDHD transcript or an indirect mechanism, we cotransfected miR-210 (or control miRNA) with an SDHD 3′UTR reporter construct in HK2 and HKC8 cells, and luciferase activity was determined 48 h posttransfection. miR-210 inhibited SDHD 3′UTR activity in both HK2 (P = 0.004) and HKC8 (P = 0.002) cell lines (Fig. 3G), establishing that miR-210 directly degrades SDHD.
Next, we aimed to determine whether miR-210 decreased total SDH enzymatic activity. SDH activity was measured 48 h posttransfection of miR-210 or control miRNA into HK2 and HKC8 cells. As hypothesized, miR-210 inhibited SDH activity in both HK2 (P = 0.006) and HKC8 (P = 0.05) cells (Fig. 3H).
Finally, we aimed to determine whether loss of VHL was sufficient to induce up-regulation of miR-210 and down-regulation of SDHD. CRISPR mediated biallelic deletion of VHL (Fig. 3I) in the HK2 cell line (E8 clone) caused a marked increase in HIF1α expression (Fig. 3 I and J), concomitant with markedly increased miR-210 (>7-fold, Fig. 3K) and decreased SDHD expression (∼75%, Fig. 3 I and J) (VHL deletion confirmed with Sanger sequencing; SI Appendix, Fig. S11). Furthermore, CRISPR mediated monoallelic deletion of VHL in the HK2 cell line as well as a small interfering RNA (siRNA)–mediated VHL knockdown (∼60%) in the HKC-8 cell line, demonstrated consistent up-regulation of miR-210 (∼1.5-fold) and mild down-regulation of SDHD (SI Appendix, Fig. S12 A–C). The data on the biallelic and monoallelic VHL deleted clones and siRNA mediated VHL knockdown illustrate that the degree of VHL loss has a bearing on the degree of miR-210 up-regulation and loss of SDHD. This is consistent with the established fact that a single functional allele of VHL is sufficient to prevent RCC carcinogenesis and that a second hit of the preserved allele is required for the initiation of carcinogenesis.
In keeping with the findings above, in the ccRCC cell line 786-O cultured under long-term hypoxic conditions (1% O2, 3 mo), both an increased miR-210 expression as well as decreased SDHD/SDHB expression were observed [SI Appendix, Fig. S13, data accessed through GSE107848 (6)].
Interestingly, SDHB, located at 1p36, is deleted (shallow) in ∼20% of all ccRCC (TCGA-KIRC) (SI Appendix, Fig. S14A). This shallow deletion of SDHB is associated with lower expression of SDHB (SI Appendix, Fig. S14B), illustrating that the preserved allele does not fully compensate for the genetic loss. On the other hand, SDHD is deleted (shallow) in ∼5% and gained in ∼5% of all ccRCC. SDHC is deleted (shallow) in ∼6% and gained in ∼9% of all ccRCC.
Down-regulation of SDH Contributes to Adverse DNA Hypermethylation in ccRCC.
Next, we sought to investigate the mechanisms by which SDH down-regulation leads to adverse outcomes in ccRCC. Succinate has been shown to be an inhibitor of α-kg dependent dioxygenases, including the ten-eleven translocation (TET) enzymes (7–12). Furthermore, we have previously shown that aberrant DNA methylation and loss of hydroxymethylation are adverse prognostic factors in ccRCC (13, 14). Also, succinate has been shown to be an inhibitor of multiple α-kg dependent dioxygenases, including the TET enzymes (7). We therefore hypothesized that loss of SDH and subsequent accumulation of succinate may result in inhibition of TET enzyme activity and global regulatory hypermethylation, which may then play a key role in oncogenic pathways in ccRCC.
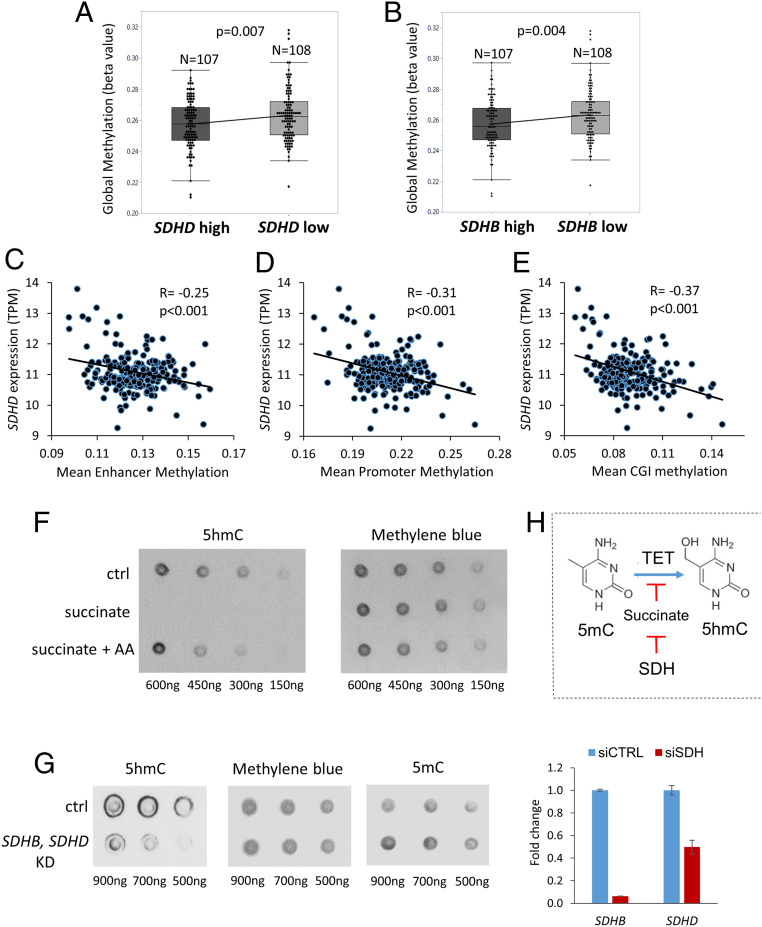
Analysis of 215 TCGA-KIRC cases with available methylation beta values and gene expression, revealed that lower SDHD expression (n = 108) was associated with higher global cytosine methylation compared to higher SDHD expression (n = 107) (P = 0.007, Fig. 4A). Similarly, lower SDHB expression was associated with higher global cytosine methylation compared to higher SDHB expression (n = 107) (P = 0.004, Fig. 4B).
Fig. 4.
Down-regulation of SDH contributes to adverse DNA hypermethylation in ccRCC. (A) Analysis of 215 KIRC TCGA cases with available methylation beta values and gene expression revealed that lower SDHD expression (n = 108) was associated with higher global cytosine methylation (mean beta values) compared to higher SDHD expression (n = 107) (P = 0.007). (B) Similarly, lower SDHB expression was associated with higher global cytosine methylation compared to higher SDHB expression (n = 107) (P = 0.004). (C–E) Analysis of specific regulatory regions of the genome revealed that SDHD expression had a significant inverse correlation with enhancer methylation (R = −0.25, P < 0.001), promoter methylation (R = −0.31, P < 0.001), and CGI methylation (R = −0.37, P < 0.001). (SI Appendix, Fig. S15: Similarly, SDHB expression also had a significant inverse correlation with enhancer methylation [R = −0.2, P = 0.003], promoter methylation [R = −0.26, P < 0.001], and CGI methylation [R = −0.26, P < 0.001].) (F) Dot blot showing exogenous succinate treatment (50 μM) resulting in marked loss of 5hmC in ccRCC cells and reversal with AA (a known TET-activating demethylating agent). (G) Knockdown of SDH subunits (B, D) in ccRCC cells (769P) resulted in loss of 5hmC and gain of 5mC. Fold change for knockdown of SDHB and SDHD is shown. (H) Schematic representation showing regulation (inhibition) of TET activity by succinate, which in turn is regulated by SDH.
Having determined the inverse relation between SDHB/SDHD expression with global cytosine methylation, we sought to correlate SDHB/SDHD expression with individual key regulatory regions of the genome-CGIs, promoters, and enhancers. SDHD expression had a significant inverse correlation with enhancer methylation (R = −0.25, P < 0.001), promoter methylation (R = −0.31, P < 0.001), and CGI methylation (R = −0.37, P < 0.001) (Fig. 4 C–E). Similarly, SDHB expression too had a significant inverse correlation with enhancer methylation (R = −0.2, P = 0.003), promoter methylation (R = −0.26, P < 0.001), and CGI methylation (R = −0.26, P < 0.001) (SI Appendix, Fig. S15).
Next, exogenous succinate treatment resulted in marked loss of 5hmC in ccRCC cells and reversal with AA, a TET-activating demethylating agent (dot blot, Fig. 4F). Furthermore, knockdown of SDH subunits (B, D) in ccRCC cells (769P) resulted in loss of 5hmC and gain of 5mC (dot blot, Fig. 4G). Simultaneous knockdown of SDH, FH, and MDH also increased global methylation in ccRCC cells (769P) (dot blot in SI Appendix, Fig. S16). Similarly, stable knockdown of SDHB in human embryonic kidney cells (HEK293T) has also been shown to markedly reduce ectopic TET-2 enzyme catalyzed 5hmC production by nearly 70% (7).
Given that deletions and underexpression of L2HGDH in ccRCC contributes to its hypermethylation via accumulation of L2HG, a TET-inhibiting oncometabolite (15–17), we sought to determine the relative contribution of low L2HGDH and SDH toward methylation in human ccRCC tumors. We stratified the 215 ccRCC cases into three subsets based on L2HGDH expression and correlated total methylation with L2HGDH and SDHB expression within each subset. We found that when the expression of L2HGDH is <7.38 TPM (i.e., lower third, n = 71), it is L2HGDH expression, and not SDHB expression, that has a significant inverse association with the mean total methylation. However, when the expression of L2HGDH is >7.38 TPM and <8.27 TPM (i.e., middle third, n = 71), and also when the expression of L2HGDH is >8.27 TPM (i.e., upper third, n = 73), it is SDHB expression, and not L2HGDH expression, that has a significant inverse association with the mean total methylation. (total n = 215; TCGA). (SI Appendix, Fig. S17 A–C)
Similarly, when the expression of L2HGDH is <7.38 TPM (i.e., lower third, n = 71), it is lower L2HGDH expression, and not lower SDHB expression, that is associated with significantly higher total methylation, higher promoter methylation, and higher CGI methylation. However, when the expression of L2HGDH is >7.38 TPM and <8.27 TPM (i.e., middle third, n = 71), and also when the expression of L2HGDH is >8.27 TPM (i.e., upper third, n = 73), it is lower SDHB expression, and not lower L2HGDH expression, that is associated with significantly higher total methylation, higher promoter methylation, and higher CGI methylation (“lower” and “higher” L2HGDH/SDHB separated by median within each group). Also, there is weak correlation between SDHB and L2HGDH expression in ccRCC (R = 0.18) (SI Appendix, Fig. S18 A–D).
Taken together, L2HG/L2HGDH impacts the methylation landscape of ccRCC predominantly when the expression of L2HGDH is low (genetic loss of L2HGDH is seen in ∼40% of all ccRCC as a part of 14q deletion, and shallow deletion of L2HGDH is associated with significantly lower L2HGDH levels). However, when L2HGDH expression is preserved, SDH expression impacts methylation landscape of ccRCC. The weak correlation between L2HGDH and SDHB in ccRCC further illustrates that the effect of loss of SDH (and accumulating succinate) on methylation in ccRCC is not related to L2HGDH expression. The data indicate that L2HG and succinate are the two major hypermethylating oncometabolites in ccRCC, likely with concurrent as well as independent influence.
Down-regulation of SDH Contributes to Enhanced Invasiveness in ccRCC.
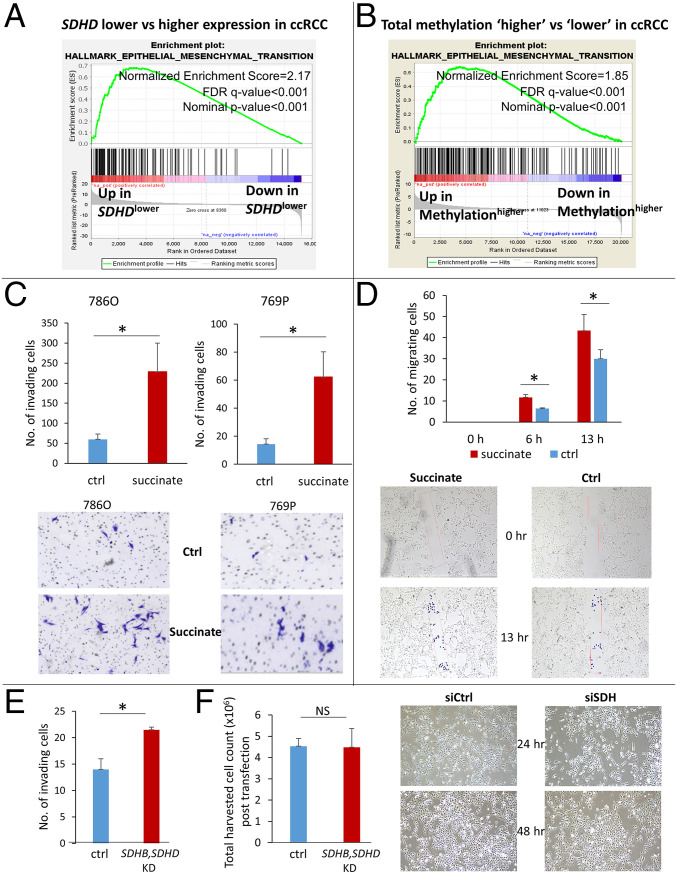
Next, we sought to determine the key oncogenic pathway enriched with both loss of SDH and gain of adverse methylation in ccRCC. We analyzed the TCGA-KIRC dataset with the Gene Set Enrichment Analysis (GSEA) software and found that lower SDHD is associated with marked enrichment of the epithelial mesenchymal transition (EMT) pathway, which aids invasion and metastasis (Normalized Enrichment Score: 2.17, nominal P value: 0.0, FDR q value: 0.0; Fig. 5A). In fact, EMT was the most positively enriched pathway in ccRCC tumors associated with lower SDHD expression. Similarly, total methylation, CGI methylation, promoter methylation, and enhancer methylation in ccRCC also strongly correlated with enrichment of the EMT pathway (Normalized Enrichment Score: 1.8, 2.1, and 1.5, respectively, Fig. 5B and SI Appendix Fig. S19 A–C). Expectedly, the EMT pathway is up-regulated in ccRCC compared to normal kidney, and a higher “EMT score” is associated with worse prognosis (SI Appendix, Figs. S20 and S21).
Fig. 5.
Down-regulation of SDH contributes to enhanced invasiveness in ccRCC. (A) Analysis of the KIRC TCGA with the GSEA revealed that lower SDHD is associated with marked enrichment of the Epithelial Mesenchymal Transition Pathway, known to aid invasion and metastasis (NES: 2.17; FDR q value: <0.001). EMT is the most positively enriched pathway in ccRCC tumors with lower SDHD expression. (B) Similarly, higher total methylation in ccRCC tumors is associated with marked enrichment of the EMT pathway (NES: 1.85; FDR q value: <0.001, KIRC TCGA). (C) Succinate treatment (50 μM) of ccRCC cell lines 786-O and 769P resulted in a significant increase in the invasiveness of ccRCC cells as determined by the Matrigel invasion assay (72-h time point, n = 3 for each cell line, data representing mean ± SEM, *P < 0.05). Representative pictures are shown. (D) The Scratch test revealed a significant increase in the migration of ccRCC cells with succinate (50 μM) treatment (n = 2, data representing mean ± SEM, *P < 0.05). Representative pictures shown for 0-, 6-, and 13-h time points. A rectangular area within the scratch (pink mark) was predefined for both succinate and control groups. During analysis, each cell that had invaded the rectangular area was highlighted with a blue dot using the ImageJ imaging software and counted digitally. (E) Knockdown of SDH subunits (SDHB, SDHD) in ccRCC cells (769P) resulted in increased invasiveness (72-h time point, n = 2, data representing mean ± SEM, *P < 0.05; fold change for knockdown of SDHB and SDHD shown in Fig. 4G). There was no difference in harvested cell counts between control and SDH knockdown after transfection in the ccRCC cells. (Please also see SI Appendix, Fig. S23. Knockdown of SDH in ccRCC cells 786-O also resulted in increased invasiveness.)
Having observed the marked enrichment of EMT pathway with lower SDH, we hypothesized that succinate accumulation from loss of SDH induces EMT phenotypic changes in RCC cells, aiding invasion, migration, and metastasis. Indeed, exogenous succinate treatment (50 μM) of ccRCC cell lines 786-O and 769P resulted in a significant increase in the invasiveness as determined by the Matrigel invasion assay (72-h time point, Fig. 5C). Furthermore, a Scratch assay also revealed a significant increase in the migration of ccRCC cells with succinate (50 μM) treatment (Fig. 5D). Prior to the invasion assay experiments with succinate, we performed the cell-viability assay in ccRCC cell line 769P with succinate (SI Appendix, Fig. S22) to determine the right concentration and time point for the invasion assay. At the 24-, 48-, and 72-h time points, there was no significant difference in cell viability with succinate treatment at 1 μM, 10 μM, and 100 μM concentrations. At the 96-h time point, there was slight decrease in viability with 100 μM concentration. We therefore chose the 50 μM concentration and a time point of 72 h for the invasion assay.
Furthermore, knockdown of SDH subunits (B, D) in ccRCC cells (769P) resulted in increased invasiveness (Fig. 5E). There was no difference in harvested cell counts between control and SDH knockdown posttransfection in the ccRCC cells, further indication of the remarkable ability of ccRCC cells to compensate for further down-regulation of oxidative phosphorylation. (Knockdown of the SDH complex also increased invasiveness in ccRCC cells 786-O; SI Appendix, Fig. S23.)
We found that in ccRCC, loss of SDH is significantly associated with loss of CDH1 (E-cadherin), an epithelial mark that is markedly underexpressed in this malignancy (with lower expression being associated with worse survival) (SI Appendix, Fig. S24A). The Pearson correlation R-value between the log2 expression of SDHB, SDHC, and SDHD and the log2 expression of CDH1 was 0.42, 0.56, and 0.4, respectively (P = 0 for each, SI Appendix, Fig. S24B). CDH1 is known to be suppressed by promoter methylation in ccRCC, with progressive loss of CDH1 expression and increase in promoter CpG methylation with higher grade in RCC (18). Succinate treatment of ccRCC cells (769P) resulted in a further increase in the hypermethylated fraction of CDH1 DNA and underexpression of CDH1, both of which were reversed with addition of AA (SI Appendix, Fig. S24C).
Succinate-Induced Enhanced Invasiveness in ccRCC Is Mediated by TET-2 Inhibition.
Next, we aimed to determine the key molecular mechanism by which succinate increases adverse methylation and enhances invasiveness in ccRCC specifically by studying its effect on the TET enzymes.
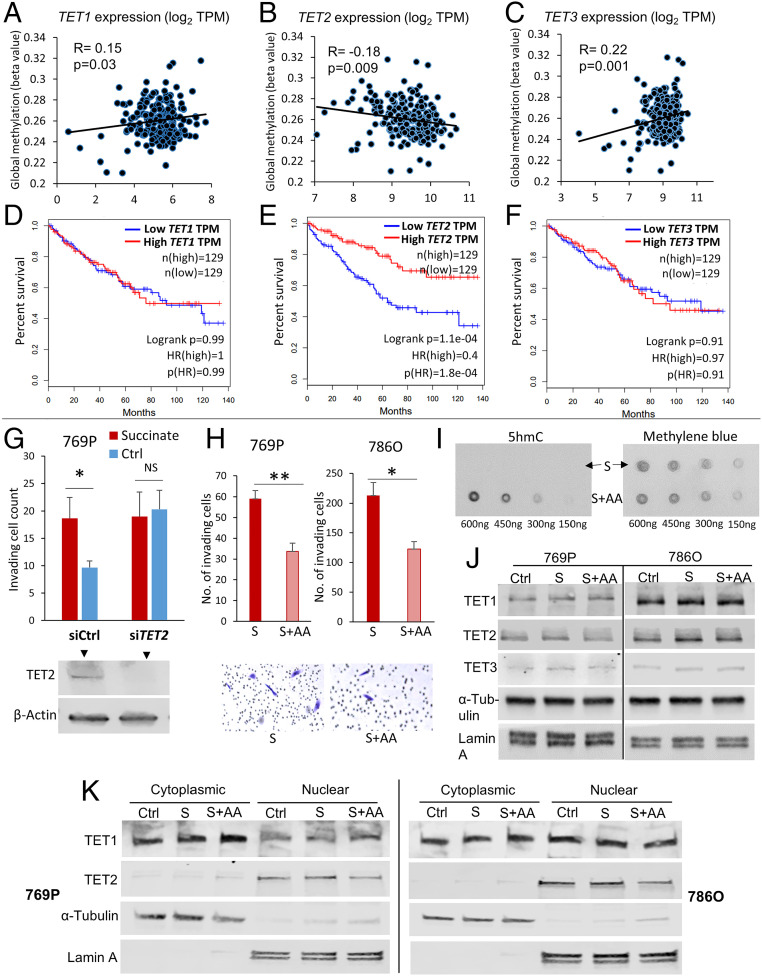
We found that in ccRCC, intriguingly, only TET2 (not TET1 or TET3) is negatively correlated with global methylation in ccRCC (Fig. 6 A–C) and significantly associated with adverse outcome (Fig. 6 D–F).
Fig. 6.
Succinate-induced enhanced invasiveness in ccRCC is mediated by TET-2 inhibition. (A–C) Only TET2, not TET1 or TET3, is negatively correlated with global cytosine methylation in ccRCC tumors (KIRC TCGA, n = 215). (Please also see SI Appendix, Fig. S25. Lower TET2 expression is significantly associated with higher global cytosine methylation in ccRCC tumors.) (D–F) In univariate survival analysis, only TET2, not TET1 or TET3, is significantly correlated with overall survival (OS) in ccRCC (KIRC TCGA, n = 215). Higher TET2 expression is significantly associated with improved OS in ccRCC. The OS HR for high (n = 129) versus low (n = 129) TET1, TET2, and TET3 was 1.0, 0.4, and 0.97, respectively (high versus low quartiles; log-rank P < 0.001 for TET2, although HR was not statistically significant in multivariate survival analysis adjusted for stage, grade, age, and sex). (G) Knockdown of TET2 resulted in abrogation of succinate-induced increase in invasiveness of ccRCC cells 769P. Knockdown of TET2, even without succinate treatment, increased invasiveness in comparison to control. (72-h time point, n = 2, *P < 0.05). (H, I) AA treatment with succinate significantly inhibited succinate-induced invasiveness in ccRCC cells (769P, 786-O) (72-h time point, n = 2 for each cell line, *P < 0.05; **P < 0.01). Representative pictures are shown. This inhibition of succinate-induced invasiveness was associated with a marked increase in 5hmC with AA treatment (dot blot for 769P shown in I; dot blot for 786-O shown in Fig. 4F). (J, K) Despite a marked change in 5hmC/5mC, there was no difference in the total protein expression of TET1, TET2, and TET3 with “succinate” or “succinate + AA” in the two ccRCC cell lines 769P and 786-O (J). Also, there was no difference in the localization of the TET proteins in the nucleus or cytoplasm with “succinate” or “succinate + AA.” Therefore, the loss of 5hmC with succinate and reversal with addition of AA is not due to any change in expression/localization of TETs but rather a change in TET activity. (These data, put together, strongly indicate that TET-2 inhibition-induced global regulatory DNA hypermethylation drives succinate-induced enhanced invasiveness in ccRCC. This was further supported by reversal of succinate-induced invasiveness of ccRCC cells by an archetypal DNA methyltransferase [DNMT1] inhibitor, Aza [SI Appendix, Fig. S27].)
Lower TET2 expression (n = 108) was associated with higher global cytosine methylation (mean beta value) compared to higher TET2 expression (n = 107) (P = 0.035; SI Appendix, Fig. S25). In univariate survival analysis, the OS HR for high (n = 129) versus low (n = 129) TET2 expression was 0.4 (TCGA-KIRC, high versus low quartiles, P < 0.001; Fig. 6E), although HR was not statistically significant in multivariate survival analysis adjusted for stage, grade, age, and sex.
We therefore hypothesized that TET-2 inhibition was mainly responsible for succinate-induced enhanced invasiveness. Knockdown of TET2 with siRNA (siTET2) resulted in abrogation of succinate-induced increase in invasiveness of ccRCC cells (769P). siTET2, even without succinate treatment, increased invasiveness in comparison to control (Fig. 6G). “Succinate” and “siTET2” increased global 5mC in ccRCC cells to a similar extent (∼2 to 2.5-fold). “Succinate+siTET2” did not increase global 5mC over “Succinate” alone, suggesting that succinate-induced increase in 5mC is largely mediated by TET-2 inhibition (SI Appendix, Fig. S26). Similarly, AA cotreatment with succinate significantly inhibited succinate-induced invasiveness of both ccRCC cell lines 769P (P = 0.004, 72-h time point) and 786-O (P = 0.01, 72-h time point) (Fig. 6H). This reversal of succinate-induced invasiveness by AA was associated with a dramatic increase in 5hmC levels as shown by the dot blot assay (Figs. 4F and 6I).
Next, we wanted to further confirm that the changes in 5hmC/5mC with succinate or AA were due to changes in TET enzymatic “activity” and not due to changes in expression or localization of TETs. Indeed, we found that there was no difference in the protein expression of TET-1, TET-2, and TET-3 either in the total or subcellular levels (nuclear and cytoplasmic) with “succinate” or “succinate + AA” in ccRCC cells (769P, 786-O) (Fig. 6 J and K). It was also interesting to note that in both cell lines, TET-2 was solely confined to the nucleus, whereas TET-1 was present more in the cytoplasm than nucleus (769P) or distributed equally (786-O).
Given that TET-2 inhibition-induced global regulatory DNA hypermethylation drives succinate-induced invasiveness, we hypothesized and demonstrated that succinate-induced invasiveness of ccRCC cells can also be reversed by an archetypal DNA methyltransferase inhibitor, azacytidine (Aza) (SI Appendix, Fig. S27A). This Aza-induced reversal of invasiveness was associated with an expected reduction in global methylation (SI Appendix, Fig. S27B).
Succinate-Induced Attenuation of TET-2 Activity Is Likely via Product Inhibition (Rather than Competitive Inhibition).
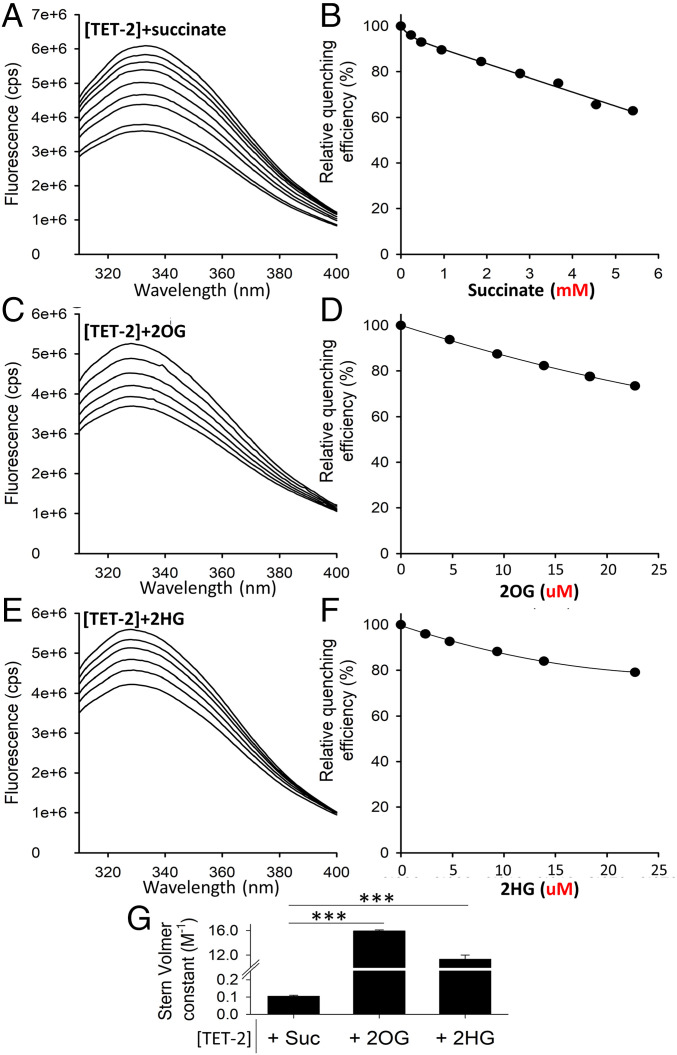
Having shown that succinate-induced enhanced invasiveness in ccRCC is mediated by TET-2 inhibition, we sought to determine the binding affinity of succinate with the catalytic domain of TET-2 using fluorescence spectroscopy and shed light on the biophysical mechanism of succinate-induced TET-2 inhibition (see Fluorescence Spectroscopy and Fig. 7: individual fluorescence spectra, relative quenching efficiency, and Stern-Volmer constants for TET-2+succinate, TET-2+2OG, and 2HG).
Fig. 7.
Succinate-induced decrease in TET-2 activity is likely via product inhibition (rather than competitive inhibition). Fluorescence quenching is a technique for measuring binding affinity between ligands and proteins. It is the decrease in quantum yield of fluorescence from a fluorophore, induced by molecular interactions with the quencher molecule(s). This experiment was undertaken to determine the binding affinity between succinate and the catalytic domain of recombinant human TET-2 protein (in comparison with the binding affinity of 2OG [natural cosubstrate] and 2HG [known competitive inhibitor] with the catalytic domain of TET-2). (A, C, E) Fluorescence spectra of 0.5 μM TET-2 are shown after excitation at 280 nm with increasing amounts of Succinate, 2OG, and 2HG (from top to bottom, respectively). (B, D, F) The relative fluorescence intensity at 328 nm is shown as a function of Succinate, 2OG, and 2HG. (Note the millimolar concentrations for succinate and micromolar concentrations for 2OG and 2HG in the relative quenching efficiency). (G) Comparison of the Stern-Volmer constants ± SEM obtained from the Stern-Volmer equation (Eq. 1 in Fluorescence Spectroscopy). Stern-Volmer constants ± SEM for [TET-2]+succinate, [TET-2]+2OG, and [TET-2]+2HG are 0.1 ± 0.0, 15.7 ± 0.1, and 12.2 ± 0.5, respectively. Put together, the binding affinity of succinate to the catalytic domain of TET-2 is less than a hundredth that of 2OG (natural cosubstrate) and 2HG (known competitive inhibitor), suggesting that the mechanism of succinate-induced inhibition of TET-2 is not via competitive inhibition but via product inhibition (each reaction of TET-2 utilizes 2OG as a cosubstrate and converts it to succinate).
We found that the binding affinity of succinate to the catalytic domain of TET-2 was less than a hundredth that of 2OG (natural cosubstrate) and 2HG (known competitive inhibitor), suggesting that the mechanism of succinate-induced inhibition of TET-2 is not via competitive inhibition but via product inhibition (each reaction of TET-2 utilizes 2OG as a cosubstrate and converts it to succinate).
Discussion
Previously, reduced SDH activity resulting in adverse succinate accumulation was thought to be relevant only to 0.05 to 0.5% of kidney cancers associated with germline SDH mutations. In this paper, we report that ccRCC (∼80% of all kidney cancers) is characterized by a marked loss of SDH subunits compared to normal renal tissue, and that this loss is associated with markedly worse overall and disease-free survival in a large cohort of ccRCC patients. Using the ccRCC metabolomic repository (3), we report that this loss is manifested by the accumulation of succinate during pathogenesis and progression in ccRCC tumors (loss of SDH complex prevents the conversion of succinate to fumarate).
While glucose oxidation is very low in ccRCC (19), the carbons of TCA cycle intermediates are predominantly derived from glutamine in this malignancy (20). Pyruvate derived from glucose via glycolysis needs the enzyme pyruvate dehydrogenase for conversion to acetyl-coa and subsequent entry into the TCA cycle. In clear cell renal cancer, the highly up-regulated HIFs (mainly secondary to biallelic VHL defects) increase the expression of pyruvate dehydrogenase kinase (a HIF-target gene), which phosphorylates and inhibits pyruvate dehydrogenase. Therefore, glucose derived pyruvate is unable to be converted to acetyl-coa for subsequent TCA cycle entry. However, acetyl-coa is an indispensable substrate for the synthesis of necessary macromolecules and is generated by HIF-induced reductive carboxylation of alpha ketoglutarate (2OG, 2-oxoglutarate) derived from glutamine (20–27). 2OG derived from glutamine, apart from contributing to acetyl-coa carbons, also functions as a cosubstrate for several 2OG-dependent enzymes (28, 29). Each reaction of each 2OG-dependent enzyme converts the cosubstrate 2OG into succinate (i.e., one of the reaction products) (29, 30). The only enzyme that converts succinate into fumarate is the SDH complex, which is markedly down-regulated. As a result, succinate accumulates and the amount of fumarate and malate formed is low (shown in Fig. 2 D and G) despite the expression of FH and malate dehydrogenase (MDH) being low. Succinate is higher in CCRC compared to normal kidney and also higher in late-stage ccRCC tumors compared to early-stage ccRCC tumors. Consistent with these findings, knockdown of SDH subunits in ccRCC cells resulted in an accumulation of succinate and a decrease in fumarate. Also, simultaneous knockdown of SDH subunits, FH, and MDH resulted in an accumulation of succinate but not of fumarate or malate. Collectively, these data indicate that down-regulation of the SDH complex (and resulting succinate accumulation) is a critical brake in the Krebs cycle during ccRCC pathogenesis and progression.
Next, we aimed to investigate the mechanism of SDH down-regulation in ccRCC. We found that VHL loss–induced HIF–dependent up-regulation of miR-210 in ccRCC causes direct inhibition of the SDHD transcript. We report that miR-210 is the second-most up-regulated miRNA in ccRCC, and of all the miRNAs with a binding site for HIF2α in ccRCC cell line 786-O, the strength of binding of HIF2α on the miR-210 promoter is the highest by a long margin. miR-210 expression had a strong negative correlation with SDH subunits in ccRCC (TCGA), which led us to hypothesize that it may be playing a causative role in SDH down-regulation. Furthermore, SDHD has been previously shown to be a target of miR-210 (31, 32). Indeed, miR-210 transfection of immortalized kidney cells led to marked down-regulation of SDHD expression (but not of SDHB expression) and was found to significantly inhibit SDHD 3′UTR activity. We then hypothesized, and demonstrated, using CRISPR as well as siRNA mediated knockdown that VHL loss (a pathognomonic feature of ccRCC carcinogenesis) is sufficient to induce miR-210 up-regulation and SDHD down-regulation in ccRCC carcinogenesis. Furthermore, we report that SDHB, located at 1p36, is deleted (shallow) in ∼20% of all ccRCC (TCGA-KIRC) and that shallow deletion is associated with lower expression. However, even with these data, the mechanisms governing the dramatic down-regulation of SDHB and SDHC (Fig. 1A) are not fully explained and could possibly be mediated by miRNAs other than miR-210. Even with SDHD, it is possible that other factors (apart from the HIF-miR-210 axis) may also influence its expression in ccRCC.
Next, we investigated the mechanisms by which SDH down-regulation leads to adverse outcomes in ccRCC. Succinate has been shown to be an inhibitor of α-kg dependent dioxygenases, including the TET enzymes (7–12). Furthermore, we have shown that aberrant DNA methylation and loss of hydroxymethylation are adverse prognostic factors in ccRCC (13, 14). We therefore hypothesized that loss of SDH and subsequent accumulation of succinate may result in inhibition of TET enzyme activity and global regulatory hypermethylation, which may then play a key role in oncogenic pathways. Indeed, we found that the loss of SDHD and SDHB in ccRCC significantly correlates with an increase in global genome-wide cytosine methylation. Exogenous succinate resulted in a marked loss of 5hmC in ccRCC cells and reversal with AA, a TET-activating demethylating agent. Similarly, knockdown of SDH subunits in ccRCC cells resulted in a loss of 5hmC and gain of 5mC, explaining the correlation between the loss of SDH subunits and gain of cytosine methylation in ccRCC tumors. Intriguingly, in ccRCC, only TET2, not TET1 or TET3, is negatively correlated with global methylation. We found that lower SDHD is associated with marked enrichment of the EMT pathway, which aids invasion and metastasis. Furthermore, global methylation in ccRCC strongly correlates with enrichment of the EMT pathway, providing a link between SDH loss–induced genome-wide methylation and enhancement of EMT in ccRCC tumors. We then hypothesized and demonstrated that succinate increases invasiveness and migratory ability of ccRCC cells and that succinate-induced invasiveness and hypermethylation is largely mediated by TET-2 inhibition. Finally, we show that the binding affinity of succinate to the catalytic domain of TET-2 is less than a hundredth that of 2OG (cosubstrate) and 2HG (known competitive inhibitor), suggesting that the mechanism of succinate-induced decrease in TET-2 activity is not via competitive inhibition but via product inhibition.
The relative contribution of loss of L2HGDH and SDH to the methylation landscape of human ccRCC tumors is an important consideration given the well-known impact of L2HGDH loss (and accumulation of L2HG) on ccRCC methylation reported by us and others (13, 16, 17, 33). Although L2HGDH had a significant inverse correlation with total methylation in ccRCC, it was by no means absolute, and there were several cases with high 5mC or loss of 5hmC with preserved/high L2HGDH expression (seen in figure 3 F and G of ref. 13). This led to the exploration of a second contributing hypermethylating oncometabolite inhibiting TET enzymes in ccRCC (given that there was no significant mutational or transcriptional inactivation of TET2 in ccRCC, and the protein expression of TET-2 revealed no difference between high-grade and low-grade ccRCC despite a marked difference in 5hmC levels between high-grade and low-grade ccRCC; figures 1 B and C and 3C of ref. 13). Now, having shown that succinate accumulates in ccRCC with down-regulation of SDH and contributes to its methylation landscape, we reasoned that the best method of assessing the relative contribution of low L2HGDH and SDH toward methylation in human ccRCC tumors would be to analyze the methylation beta values in a large sample size of ccRCC tumors in relation to the expression of SDH and L2HGDH. With stratification of 215 ccRCC cases (TCGA) into three subsets based on L2HGDH expression and correlation of methylation with L2HGDH and SDHB expression within each subset, we show that L2HG/L2HGDH impacts the methylation landscape of ccRCC predominantly when the expression of L2HGDH is low (genetic loss of L2HGDH is seen in ∼40% of all ccRCC as a part of 14q deletion, and shallow deletion of L2HGDH is associated with significantly lower L2HGDH levels). However, when L2HGDH expression is preserved, SDH expression impacts the methylation landscape of ccRCC. The relatively weak correlation between L2HGDH and SDHB in ccRCC provides further evidence that the effect of SDH loss on methylation in ccRCC is not related to L2HGDH expression. Together, the data indicate that L2HG and succinate are the two major hypermethylating oncometabolites in ccRCC, likely with concurrent as well as independent influence.
In summary, we show that a loss of SDH activity is a common feature in ccRCC, with adverse functional and prognostic implications. Succinate, which accumulates with the loss of SDH, is an important epigenetic modulating oncometabolite in ccRCC, contributing to the adverse methylation landscape and conferring enhanced invasive ability in ccRCC (Fig. 8). The 2016 WHO Classification of Renal Tumors (34) identified “SDH-deficient RCC” as a separate category, referring mainly to RCC that is associated with germline mutations in any of the SDH subunits. However, our findings reported here indicate that the term “SDH-deficient” is a misnomer because significant SDH subunit loss (and resulting accumulation of oncogenic succinate) is a characteristic adverse feature in ccRCC, which accounts for 75 to 80% of all kidney cancers (although the degree of functional SDH deficiency is likely lesser than that with germline SDH mutations). We therefore propose that the WHO “SDH-Deficient RCC” entity be renamed as “SDH germline mutation-associated RCC,” to avoid confusion in identifying the small group of RCC patients (0.05 to 0.5%) with germline mutations in SDH subunits, presenting at a median age of 35 y (much younger than that of ccRCC), and with characteristic pathologic findings of pale-to-eosinophilic cytoplasmic vacuoles and rare-to-occasional cytoplasmic inclusions (34–39).
Fig. 8.
Graphical abstract depicting the consequential adverse down-regulation of SDH in ccRCC and its central role in oxidative phosphorylation.
Methods
Cell Lines.
ccRCC cell lines 786-O and 769P were purchased from the American Type Culture Collection (ATCC). Cell line authentication was done at ATCC. Cells were cultured in RPMI-1640 media supplemented with 10% vol/vol fetal bovine serum (FBS) and 1% vol/vol Penicillin/Streptomycin. HK2 kidney cells were purchased from ATCC, and HKC8 cells were kindly provided by Lorainne Racusen (Johns Hopkins University, Baltimore, MD). Both cell lines were cultured in DMEM supplemented with 10% FBS and 1% Penicillin/Streptomycin.
Immunohistochemistry and Scoring (SDHB and SDHD).
Case selection.
Paraffin blocks containing ccRCC tumor and adjacent nonneoplastic carcinoma were selected for immunohistochemistry from nephrectomy and partial nephrectomy cases resected at Montefiore Medical Center between 2016 and 2019. All selected cases were diagnosed as clear cell carcinoma, WHO/International Society of Urological Pathologists grade 2 or 3, confined to the kidney, and had no sarcomatoid or rhabdoid features.
Immunohistochemistry.
Five-micrometer sections were cut from selected paraffin blocks, mounted on plus slides (Thermo Fisher Scientific), baked at 60 °C for 1 h, deparaffinized in xylene (2 × 5 min), rehydrated in 100% ethanol (2 × 2 min) followed by 95% ethanol (2 × 2 min), washed twice with deionized water (dH20), treated with 3% hydrogen peroxide for 10 min to quench endogenous peroxidase, and rinsed twice in dH20. Slides were microwaved in pH 6 antigen retrieval solution (1× Target Retrieval Solution, Agilent catalog no. S1699) to bring near to but not to boiling (∼1.5 min), placed in a steamer (Oster Model CKSTSTMD5-W) for 30 min, cooled at room temperature for 20 min, and washed twice in dH20 and then in PBS/0.05% Tween 20/1% BSA (PBSTB) for 5 min. Slides were wiped dry around but not on the tissue section, which was then surrounded by a hydrophobic barrier made with an ImmeEdge pen (Vector Labs catalog no. H-4000). Each section was treated with four drops of serum-free protein block (Agilent catalog no. X0909) for 20 min at room temperature and then removed by tapping the slide without washing, followed by incubation with 100 μL primary antibody (SDHB, Abcam no. ab14714 diluted 1:2,000 or SDHD, Abcam ab203199 diluted 1:200) in antibody diluent (Life Technologies catalog no. 003218) with 1% BSA in a slide box humidified using wet paper towels (Thermo Fisher Scientific catalog no. 03-448-1 or -2) for 30 min at room temperature, blotted with a paper towel, gently rinsed with two dips in PBS, and washed in PBSTB 1 × 5 min and then 2 × 1 min. Three drops per slide of secondary antibody (EnVision + labeled polymer-HRP anti-mouse, Dako K4001, or anti-rabbit, Dako K4003) were added and incubated for 30 min at room temperature, washed in PBSTB 1 × 5 min and then 2 × 1 min, and stained with diaminobenzidine (DAB; Cell Marque DAB Substrate Kit catalog no. 957D-60) for 4 min. Slides were then washed four times in tap water, counterstained with Harris hematoxylin (Leica catalog no. 3801561), blued with two dips in an ammonia solution (1:1,000 dilution of stock ammonium hydroxide; Thermo Fisher Scientific catalog no. A669S-500), washed once more in tap water, dehydrated through ethanol and xylene, and coverslipped with Cytoseal XYL (Thermo Scientific catalog no. 8312-4).
Evaluation of immunostaining.
Staining intensity was evaluated semiquantitatively on a scale of 0 to 4+. Groups of nonneoplastic tubule or carcinoma cells within contiguous areas were evaluated together since there was relatively little variation in staining (less than 1+ in the 0 to 4+ scale) among them (Fig. 1 D and E). Digital photography was performed with a Nikon Digisite DS-Fi3 microscope camera at a resolution of 2,880 × 2,048 pixels per image, using the same exposure and gain (contrast) settings for all slides. For both antibodies (SDHB and SDHD), the nonneoplastic tubular cells in the resection stained consistently stronger than the clear cell carcinoma cells. Stromal and other nonepithelial cells stained weakly or not at all with both antibodies.
CRISPR/Cas9 VHL Knockdown.
Three synthetic single-guide RNAs (sgRNAs) were designed targeting the first exon of VHL corresponding to the nucleotide sequences CGCGGAGGGAATGCCCCGGA, TGAAGAAGACGGCGGGGAGG, and GGAGGAACTGGGCGCCGAGG using the Synthego design tool (Synthego). Cas9 enzyme and sgRNAs were cotransfected using Lipofectamine CRISPRMAX (Invitrogen) following the manufacturer’s protocol (Synthego). Monoclonal cell populations were generated from the resulting transfection using limiting dilution and screened for knockdown efficiency by VHL Western blot and Sanger sequencing.
miRNA Transfection.
PremiRNA precursor molecules for hsa-miR-210-3p and negative control 1 (Ambion/Thermo Fisher Scientific) were transfected into HK2 and HKC8 cells at a final concentration of 30 nM using Lipofectamine 3000 (Invitrogen). All transfections were performed in triplicate and validated by qPCR for hsa-miR-210-3p.
siRNA Transfection.
HKC8 cells were transfected with siRNAs targeting VHL (Dharmacon, ON-TARGET Plus) and nontemplate control (Dharmacon) using Lipofectamine 3000. 769P and cells were transfected with siRNAs targeting SDHB, SDHD, FH, MDH2, or nontemplate control and 786-O cells with siRNAs targeting SDHB, SDHD, SDHA, SDHC, or nontemplate control (Dharmacon, ON-TARGET Plus) by electroporation. All cells were harvested at 48 h posttransfection for qPCR, methylation, and/or metabolite analyses. 769P and 786-O cells were plated for the invasion assay at 24 h posttransfection. All transfections were performed in triplicate.
Succinate, Fumarate, and Malate Metabolite Assays.
Colorimetric assay kits (Abcam) were used to quantify intracellular succinate (ab204718), fumarate (ab102516), and malate (ab83391) in cultured cells. Fresh cells were harvested and directly lysed and assayed following the manufacturer’s instructions. Standard curves were used to calculate nanomoles of each metabolite per 1 × 106 cells.
SDH Activity Assay.
At 48 h posttransfection with miRNA, cells were harvested and assayed immediately for SDH activity. SDH activity of transfected HK2 and HKC8 cells was determined using the Succinate Dehydrogenase Activity Colorimetric Assay Kit following the manufacturer’s instructions. The reaction was started by adding a blue colored artificial probe to accept electrons from the succinate to fumarate oxidation. Absorbance at 600 nm was recorded in kinetic mode at room temperature. The decrease in absorbance per unit time (slope) was compared to a standard curve obtained with known protein concentrations and reported proportional to SDH activity using the equation
, where is the nanomoles of protein reduced corresponding to the change in absorbance in the standard curve.
Immunoblotting.
Protein lysates were isolated from cells and run on 4 to 15% Tris⋅HCl poly-acrylamide gels. Proteins were transferred to nitrocellulose membranes and blocked with 1:1 Intercept PBS Blocking Buffer (LI-COR):PBS. Membranes were incubated in the following primary antibodies: HIF2α (Novus Biologicals NB100-122), VHL (Novus Biologicals, NB100-485), TET-2 (Cell Signaling Technology, 18950), TET-1 (Genetex, GTX124207), TET-3 (Genetex, GTX121453), HIF1α (Novus, NB100-105), SDHD (Abcam, ab189945), SDHB (Abcam, ab14714), Lamin A (Cell Signaling, 133A2), α-Tubulin (Cell Signaling, 2144S), and β-Actin (Novus Biologicals, NB600-501; AC-15). Proteins of interest were detected using IRDye 800CW and 680RD secondary antibodies (LI-COR) on a LI-COR Odyssey Fc Imaging System.
3-UTR Reporter Assay.
SDHD 3′UTR LightSwitch Luciferase Reporter Assay was purchased from Active Motif. Cells were cotransfected with 30 nM miRNA and 100 ng reporter using Lipofectamine 3000 in a 96-well plate. Luciferase activity was measured using the LightSwitch Luciferase Assay Reagent (Active Motif) on a SpectraMax M5 plate reader (Molecular Devices) following the manufacturer’s instructions.
ChIP-seq Analysis.
ChIP-seq data were downloaded from GSE34871 (40), GSE86092 (5), GSE67237 (41), and GSE120885 (4). ChIP-seq data were visualized in IGV (42). For analysis of HIF2α and HIF1α binding site distribution, ChIP-seq files were analyzed using ChIPSeeker (43).
RNA-seq Analysis.
RNA sequencing data were accessed from GSE1078484 (6) using GEO RNA-Seq Experiments Interactive Navigator [GREIN (44)].
Bioinformatic Analyses.
The Gene Expression Profiling Interactive Analysis (GEPIA) (45) was used for SDHA, SDHB, SDHC, SDHD, FH, and MDH expression comparison between ccRCC tumors and normal kidney (TCGA-KIRC dataset, Figs. 1 and 2); survival analyses with SDHA, SDHB, SDHC, SDHD, TET1, TET2, and TET3 expression (TCGA-KIRC dataset, Figs. 1 and 6 and SI Appendix, Fig. S6); and correlation between SDHB, SDHD, and SDHC expression with CDH1 in ccRCC tumors and normal kidney (TCGA-KIRC dataset, SI Appendix Fig. S24).
Methylation beta values for the available 215 ccRCC cases were downloaded from TCGA-KIRC, and correlation with expression of SDHD, SDHB, L2HGDH, TET1, TET2, and TET3 in those cases was studied with Excel (Figs. 4 A–E and 6 A–C and SI Appendix, Figs. S15, S17, and S18 A–C).
The TCGA Wanderer platform (46) was used for promoter CGI methylation of SDHB, SDHC, and SDHD comparison between ccRCC tumor and normal (TCGA-KIRC dataset, SI Appendix, Fig. S9)
The GSEA platform (47, 48) was used for Pathway Enrichment Analysis. ccRCC KIRC data were downloaded from TCGA. A ranked dataset comparing two groups (SDHD lower versus SDHD higher—Fig. 4A; total/CGI/promoter/enhancer methylation higher versus lower—Fig. 4B and SI Appendix, Fig. S19 A–C; tumor versus normal—SI Appendix, Fig. S20) was generated by calculating the product of the fold-change sign and −log(P value) for each gene. This ranked file was uploaded in the GSEA platform for analysis of pathways that were enriched for these comparisons.
The Xena (49) platform was used to access TCGA data for Fig. 3 C–E and SI Appendix, Fig. S10A. The Cancer Omics Atlas (50) platform was used to generate Fig. 3A. The UALCAN (51) platform was used for analysis of ccRCC samples from CPTAC (SI Appendix, Figs. S2 and S3). The CVCDAP (52) platform was used to perform multivariate survival analyses with TCGA-KIRC dataset (SI Appendix, Fig. S5 A–D).
Invasion Assay.
BD BioCoat Matrigel Invasion Chambers were used to assess the invasiveness of tumor cells following siRNA transfection or treated with control, succinate alone (50 μM), or together with AA (1 mM) or Aza (1 μM). A thin layer of Matrigel matrix at the bottom of each chamber serves as a reconstituted basement membrane in vitro, while the chemoattractant is present in the culture medium on the outside of the chamber. The manufacturer’s protocol to prepare the Matrigel chambers was followed, and the same number of cells per chamber (at least 20,000 cells) were plated in replicates. Incubation time of the cells in the Matrigel chambers was 72 h, after which the bottom of the chambers was fixed with buffered formaldehyde and stained with bromophenol blue. The number of invading cells were counted after removal of Matrigel layer and reported as mean ± SEM.
Scratch Assay.
Cells were grown to ∼60% confluence under each treatment condition (succinate and control) in 6-well plates. After 18 h treatment, scratches were made using a 200-μL pipet tip at least 5 mm apart on the cellular monolayer in each well. This was marked as t = 0 h point. The wells were quickly washed with PBS and replenished by fresh medium. Bright field images were captured at the indicated times as cells migrated toward the scratched area to close the wound. A fixed-dimensions rectangular field was demarcated using imaging software ImageJ such that the field contained no cell at t = 0 h. The number of cells migrating into the field were counted at each time point (with a blue dot over each migrated cell as shown in Fig. 5D) and reported.
EMT Score.
To avoid bias in the selection of markers, we used a previously defined EMT score (6), in which normalized expression values of mesenchymal markers (FN1, VIM, ZEB1, ZEB2, TWIST1, TWIST2, SNAI1, SNA2, and CDH2) are given a positive score and that of epithelial markers (CLDN4, CLDN7, TJP3, MUC1, and CDH1) are given a negative score. The TCGA-KIRC dataset was analyzed for these genes and the composite EMT score divided (based on median) into “high” and “low” EMT scores. These two groups were compared for overall survival.
Nuclear and Cytoplasmic Protein Extraction.
Nuclear and cytoplasmic protein fractions were isolated from cells using the NE-PER Nuclear and Cytoplasmic Extraction Kit (Thermo Fisher Scientific), according to the manufacturer’s instructions. Protein expression in isolated fractions was analyzed by Western blot.
Methylation PCR Assay.
Genomic DNA was extracted from ccRCC cells using the DNeasy Blood and Tissue Kit (Qiagen). Epitect Methyl II PCR Array System (Qiagen) was used to determine the CGI methylation status of CDH1 (E-Cadherin). At least 105 ng of total genomic DNA was used per sample. DNA was equally distributed among the parallel digests containing the mock, methylation-sensitive, methylation-dependent, and total digestion. Predesigned primers for CDH1 locus were obtained along with the kit and qPCR applied to each of the digests. Data analysis was carried out using the manufacturer’s protocol (analogous to ΔΔCt method in RT-PCR), which provides gene methylation extent of the total input genomic DNA as percentages of unmethylated, methylated, and hypermethylated fraction of input DNA.
Fluorescence Spectroscopy.
Fluorescence quenching is a technique for measuring binding affinity between ligands and proteins. It is the decrease in quantum yield of fluorescence from a fluorophore, induced by molecular interactions with the quencher molecule(s) (53). This experiment was undertaken to determine the binding affinity between succinate and the catalytic domain of TET-2 (in comparison with the binding affinity of 2OG [natural cosubstrate] and 2HG [known competitive inhibitor] with the catalytic domain of TET-2).
Recombinant TET-2 (1129-2002) protein that includes amino acids 1129-2002 of catalytic domain of human TET-2 protein was purchased from Abcam. The protein was expressed in Baculovirus-infected SF9 cells. SF9 cells were incubated with nickel beads, and the bound proteins were eluted with imidazole and resolved by SDS-PAGE (sodium dodecyl sulphate–polyacrylamide gel electrophoresis).
All fluorescence measurements were performed at 20 °C on a Horiba Jobin-Yvon Fluorolog 3 spectrofluorometer equipped with a Wavelength electronics Model LFI-3751 temperature controller. Protein fluorescence emission spectra of TET2 were averaged three times between 305 and 400 nm with excitation at 280 nm. The step width was 1 nm and the integration time 1 s.
TET2 protein solutions were prepared containing 0.5 µM protein in PBS buffer. Succinate (stock concentration = 100 mM), 2OG, and 2HG (stock concentration = 5.0 mM) in PBS were added stepwise to the solution, yielding final concentrations ranging from 0 to 5.4 mM for Succinate and 0 to 22.7 µM for 2OG and 2HG.
Quenching constants were obtained using the Stern-Volmer equation (54):
| [1] |
In this equation, KD is the quenching constant and [Q] is a defined concentration of succinate/2OG/2HG. F0 and F are the fluorescence intensities (at 328 nm) in absence and presence of succinate/2OG/2HG, respectively.
Quantitative Real-Time PCR.
At 48 h posttransfection with miRNA, cells were harvested for RNA extraction. Total RNA was isolated using miRNeasy kit (Qiagen) following the manufacturer’s instructions. cDNA was synthesized using SuperScript III (Invitrogen) and Taqman MicroRNA Reverse Transcription Kit (Thermo Fisher Scientific) for mRNA and miRNA analyses, respectively. Taqman primer probe assays (Thermo Fisher Scientific) were used for RNU6B (001093), hsa-miR-201-3p, SDHD (Hs00829723_g1), and SDHB (Hs00268117_m1). qPCR was performed for RNU6B, hsa-miR-201-3p, SDHD, and SDHB using TaqMan Universal Master Mix II (Thermo Fisher Scientific), following the manufacturer’s instructions. The relative expression of each gene was determined using the ΔΔCt normalized to RNU6B and GAPDH for miRNA and mRNA genes, respectively (Fig. 3 F and K and SI Appendix, Fig. S5B).
At 48 h posttransfection with siRNA, cells were harvested for RNA extraction. Total RNA was isolated using RNeasy kit (Qiagen) and cDNA synthesized using SuperScript III (Invitrogen). qPCR was performed using Taqman primer probe assays (Thermo Fisher Scientific) for SDHD (Hs00829723_g1), SDHB (Hs00268117_m1), SDHC (Hs01698067_s1), SDHA (Hs00188166_m1), FH (Hs00264683_m1), and MDH2 (Hs00938918_m1). The relative expression of each gene was determined using the ΔΔCt method normalized to GAPDH (SI Appendix, Figs. S3 and S18).
Total RNA was isolated from ccRCC cells (769P) using RNeasy Mini Kit (Qiagen), followed by first‐strand cDNA synthesis from 2 to 2.5 μg of RNA using SuperScript VILO (Invitrogen). Gene expression of CDH1 was measured via RT-PCR using forward (5′-GAA CAG CAC GTA CAC AGC CCT-3′) and reverse (5′-GCA GAA GTG TCC CTG TTC CAG-3′) primers. The expression values were normalized to GAPDH (forward primer: 5′-ACC CCT GGC CAA GGT CAT CCA-3′, reverse primer: 5′-ACA GTT TCC CGG AGG GGC CA-3′) (SI Appendix, Fig. S19C).
Dot Blot Assay for 5-hydroxymethylcytosine and 5-methylcytosine Analysis.
Genomic DNA from the indicated cell lines and treatment conditions was extracted. DNA was diluted in 8 µL water, denatured by adding 2 µL of 2 N NaOH/50 mM EDTA, and incubated for 10 min at 95 °C. Samples were quickly moved to ice followed by neutralization using 10 µL of ice cold 2 M ammonium acetate. Denatured DNA was manually spotted on a positively charged nylon membrane (Amersham Hybond N+). Hybridization was carried out by baking for 30 min at 80 °C. The membranes were blocked in 5% milk/TBST for 1 h followed by primary antibody blotting at 4 °C overnight incubation—1:2,000 rabbit anti-hmC (catalog no. 39069, Active Motif) or anti-5mC (catalog no. 39649, Active Motif). The next day, the blots were washed with TBST followed by incubation with secondary 1:2,000 anti-rabbit-HRP for 1 h, washed, and imaged using Western chemiluminescent reagents on a photographic X-ray film. Methylene blue stain (0.04% in 0.5 M sodium acetate, pH 5.2) was used as loading control. The photographic films and membrane with methylene blue spots were scanned as 8-bit black and white TIFF images on a scanner. Each spot was quantified using ImageJ by subtracting local background from the intensity readout from a fixed-size circle drawn around each spot.
Quantitative Global 5-methycytosine Assay (ELISA).
Genomic DNA was used to measure percent cytosine modification to 5-methylcytosine using a one-step ELISA (enzyme-linked immunosorbent assay) method (catalog no. 1030, Epigentek). Manufacturer’s protocol was followed to quantify the amount of modified cytosine residues in 50 ng of genomic DNA per treatment condition. Briefly, the protocol involves binding of a fixed amount of DNA to each well of a 96-well plate, followed by probing with the supplied anti-5mC antibody and colorimetric measurement at 450 nm representative of the concentration of modified DNA in the well. Readouts in duplicates were reported with SEM.
In Vitro AA Treatment.
Immediately prior to in vitro treatment with high dose L-AA (Sigma-Aldrich, catalog no. 50-81-7), cells were exposed to catalase to quench free radicals as previously published (13, 15). The 100 μg/mL catalase (Sigma) in 50 mM potassium phosphate was applied to all cells prior to treatment with or without 1 mM AA (Sigma). Both catalase and AA were prepared fresh for each experiment.
Statistical Analyses for Clinical Data and In Vitro Experiments.
A paired t test was used for statistical analyses in SI Appendix Fig. S2 (SDHB, SDHD IHC intensity scoring comparison between ccRCC and adjacent normal). A Log-rank test was used for survival analyses with the GEPIA platform. A one-sided Student’s t test was used for statistical analyses in SI Appendix, Fig. S4 (promoter CGI methylation comparison), Fig. 4 A and B, and SI Appendix Figs. S9B, S13 A–C, and S20.
A one-sided Student’s t test was used for statistical analyses in Figs. 2–7 and SI Appendix Figs. S7, S18, S19, and S22. Statistical significance was considered P < 0.05.
Study Approvals.
The use of ccRCC primary tumors and adjacent normal renal tissue for immunohistochemistry and the ccRCC xenograft study were approved by the Albert Einstein College of Medicine Institutional Review Board.
Supplementary Material
Acknowledgments
We acknowledge the ccRCC metabolomic repository generated by MSKCC (3) and made publicly available via supplemental attachments. The metabolite data in Fig. 2 A, B, D, E, G, and H of this manuscript were derived from analysis of this repository. We acknowledge The Cancer Genome Atlas (TCGA) (https://www.cancer.gov/tcga) (55). We also acknowledge interactive Web-based platforms for analysis of the TCGA database: GEPIA (45), Xena (49), TCGA Wanderer (46), The Cancer Omics Atlas (50), UALCAN (51), CVCDAP (52), and GSEA pathway analysis software (47). We acknowledge Albert Einstein Cancer Center core grant startup (to N.S.) 2P30CA013330-47; American Cancer Society Research Scholar Grant RSG-20-137-01 TBG (to N.S.); and 2017 American Society of Hematology Research Award (to N.S.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2106947118/-/DCSupplemental.
Data Availability
All study data are included in the article and/or SI Appendix.
References
- 1.Shenoy N., Pagliaro L., Sequential pathogenesis of metastatic VHL mutant clear cell renal cell carcinoma: Putting it together with a translational perspective. Ann. Oncol. 27, 1685–1695 (2016). [DOI] [PubMed] [Google Scholar]
- 2.Cornejo K. M., et al., Succinate dehydrogenase B: A new prognostic biomarker in clear cell renal cell carcinoma. Hum. Pathol. 46, 820–826 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hakimi A. A., et al., An integrated metabolic atlas of clear cell renal cell carcinoma. Cancer Cell 29, 104–116 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Smythies J. A., et al., Inherent DNA-binding specificities of the HIF-1α and HIF-2α transcription factors in chromatin. EMBO Rep. 20, e46401 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yao X., et al., VHL deficiency drives enhancer activation of oncogenes in clear cell renal cell carcinoma. Cancer Discov. 7, 1284–1305 (2017). [DOI] [PubMed] [Google Scholar]
- 6.Chen S. C., Chen F. W., Hsu Y. L., Kuo P. L., Systematic analysis of transcriptomic profile of renal cell carcinoma under long-term hypoxia using next-generation sequencing and bioinformatics. Int. J. Mol. Sci. 18, 2657 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xiao M., et al., Inhibition of α-KG-dependent histone and DNA demethylases by fumarate and succinate that are accumulated in mutations of FH and SDH tumor suppressors. Genes Dev. 26, 1326–1338 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Laukka T., et al., Fumarate and succinate regulate expression of hypoxia-inducible genes via TET enzymes. J. Biol. Chem. 291, 4256–4265 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mason E. F., Hornick J. L., Succinate dehydrogenase deficiency is associated with decreased 5-hydroxymethylcytosine production in gastrointestinal stromal tumors: Implications for mechanisms of tumorigenesis. Mod. Pathol. 26, 1492–1497 (2013). [DOI] [PubMed] [Google Scholar]
- 10.Carey B. W., Finley L. W., Cross J. R., Allis C. D., Thompson C. B., Intracellular α-ketoglutarate maintains the pluripotency of embryonic stem cells. Nature 518, 413–416 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schaefer I. M., Hornick J. L., Bovée J. V. M. G., The role of metabolic enzymes in mesenchymal tumors and tumor syndromes: Genetics, pathology, and molecular mechanisms. Lab. Invest. 98, 414–426 (2018). [DOI] [PubMed] [Google Scholar]
- 12.Hoekstra A. S., et al., Inactivation of SDH and FH cause loss of 5hmC and increased H3K9me3 in paraganglioma/pheochromocytoma and smooth muscle tumors. Oncotarget 6, 38777–38788 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shenoy N., et al., Ascorbic acid-induced TET activation mitigates adverse hydroxymethylcytosine loss in renal cell carcinoma. J. Clin. Invest. 129, 1612–1625 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hu C. Y., et al., Kidney cancer is characterized by aberrant methylation of tissue-specific enhancers that are prognostic for overall survival. Clin. Cancer Res. 20, 4349–4360 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shenoy N., et al., Upregulation of TET activity with ascorbic acid induces epigenetic modulation of lymphoma cells. Blood Cancer J. 7, e587 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shelar S., et al., Biochemical and epigenetic insights into L-2-hydroxyglutarate, a potential therapeutic target in renal cancer. Clin. Cancer Res. 24, 6433–6446 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shim E. H., et al., L-2-Hydroxyglutarate: An epigenetic modifier and putative oncometabolite in renal cancer. Cancer Discov. 4, 1290–1298 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nojima D., et al., CpG methylation of promoter region inactivates E-cadherin gene in renal cell carcinoma. Mol. Carcinog. 32, 19–27 (2001). [DOI] [PubMed] [Google Scholar]
- 19.Courtney K. D., et al., Isotope tracing of human clear cell renal cell carcinomas demonstrates suppressed glucose oxidation in vivo. Cell Metab. 28, 793–800.e2 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hoerner C. R., Chen V. J., Fan A. C., The ‘Achilles Heel’ of metabolism in renal cell carcinoma: Glutaminase inhibition as a rational treatment strategy. Kidney Cancer 3, 15–29 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gameiro P. A., et al., In vivo HIF-mediated reductive carboxylation is regulated by citrate levels and sensitizes VHL-deficient cells to glutamine deprivation. Cell Metab. 17, 372–385 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Okazaki A., et al., Glutaminase and poly(ADP-ribose) polymerase inhibitors suppress pyrimidine synthesis and VHL-deficient renal cancers. J. Clin. Invest. 127, 1631–1645 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wise D. R., et al., Hypoxia promotes isocitrate dehydrogenase-dependent carboxylation of α-ketoglutarate to citrate to support cell growth and viability. Proc. Natl. Acad. Sci. U.S.A. 108, 19611–19616 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sun R. C., Denko N. C., Hypoxic regulation of glutamine metabolism through HIF1 and SIAH2 supports lipid synthesis that is necessary for tumor growth. Cell Metab. 19, 285–292 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Semenza G. L., HIF-1: Upstream and downstream of cancer metabolism. Curr. Opin. Genet. Dev. 20, 51–56 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim J. W., Tchernyshyov I., Semenza G. L., Dang C. V., HIF-1-mediated expression of pyruvate dehydrogenase kinase: A metabolic switch required for cellular adaptation to hypoxia. Cell Metab. 3, 177–185 (2006). [DOI] [PubMed] [Google Scholar]
- 27.Kaelin W. G. Jr, The von Hippel-Lindau tumour suppressor protein: O2 sensing and cancer. Nat. Rev. Cancer 8, 865–873 (2008). [DOI] [PubMed] [Google Scholar]
- 28.Losman J. A., Koivunen P., Kaelin W. G. Jr, 2-Oxoglutarate-dependent dioxygenases in cancer. Nat. Rev. Cancer 20, 710–726 (2020). [DOI] [PubMed] [Google Scholar]
- 29.Islam M. S., Leissing T. M., Chowdhury R., Hopkinson R. J., Schofield C. J., 2-Oxoglutarate-dependent oxygenases. Annu. Rev. Biochem. 87, 585–620 (2018). [DOI] [PubMed] [Google Scholar]
- 30.Tretter L., Patocs A., Chinopoulos C., Succinate, an intermediate in metabolism, signal transduction, ROS, hypoxia, and tumorigenesis. Biochim. Biophys. Acta 1857, 1086–1101 (2016). [DOI] [PubMed] [Google Scholar]
- 31.Puisségur M. P., et al., miR-210 is overexpressed in late stages of lung cancer and mediates mitochondrial alterations associated with modulation of HIF-1 activity. Cell Death Differ. 18, 465–478 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Anton L., et al., HIF-1α stabilization increases miR-210 eliciting first trimester extravillous trophoblast mitochondrial dysfunction. Front. Physiol. 10, 699 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shenoy N., HIF1α is not a target of 14q deletion in clear cell renal cancer. Sci. Rep. 10, 17642 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moch H., Cubilla A. L., Humphrey P. A., Reuter V. E., Ulbright T. M., The 2016 WHO classification of tumours of the urinary system and male genital organs—Part A: Renal, penile, and testicular tumours. Eur. Urol. 70, 93–105 (2016). [DOI] [PubMed] [Google Scholar]
- 35.Udager A. M., Mehra R., Morphologic, molecular, and taxonomic evolution of renal cell carcinoma: A conceptual perspective with emphasis on updates to the 2016 World Health Organization Classification. Arch. Pathol. Lab. Med. 140, 1026–1037 (2016). [DOI] [PubMed] [Google Scholar]
- 36.Wang G., Rao P., Succinate dehydrogenase-deficient renal cell carcinoma: A short review. Arch. Pathol. Lab. Med. 142, 1284–1288 (2018). [DOI] [PubMed] [Google Scholar]
- 37.Gill A. J., et al., Succinate dehydrogenase (SDH)-deficient renal carcinoma: A morphologically distinct entity: A clinicopathologic series of 36 tumors from 27 patients. Am. J. Surg. Pathol. 38, 1588–1602 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Williamson S. R., et al., Succinate dehydrogenase-deficient renal cell carcinoma: Detailed characterization of 11 tumors defining a unique subtype of renal cell carcinoma. Mod. Pathol. 28, 80–94 (2015). [DOI] [PubMed] [Google Scholar]
- 39.Ricketts C. J., et al., Succinate dehydrogenase kidney cancer: An aggressive example of the Warburg effect in cancer. J. Urol. 188, 2063–2071 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schödel J., et al., Common genetic variants at the 11q13.3 renal cancer susceptibility locus influence binding of HIF to an enhancer of cyclin D1 expression. Nat. Genet. 44, 420–425 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Grampp S., et al., Multiple renal cancer susceptibility polymorphisms modulate the HIF pathway. PLoS Genet. 13, e1006872 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Robinson J. T., et al., Integrative genomics viewer. Nat. Biotechnol. 29, 24–26 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yu G., Wang L. G., He Q. Y., ChIPseeker: An R/Bioconductor package for ChIP peak annotation, comparison and visualization. Bioinformatics 31, 2382–2383 (2015). [DOI] [PubMed] [Google Scholar]
- 44.Mahi N. A., Najafabadi M. F., Pilarczyk M., Kouril M., Medvedovic M., GREIN: An interactive web platform for re-analyzing GEO RNA-seq data. Sci. Rep. 9, 7580 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tang Z., et al., GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 45 (W1), W98–W102 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Díez-Villanueva A., Mallona I., Peinado M. A., Wanderer, an interactive viewer to explore DNA methylation and gene expression data in human cancer. Epigenetics Chromatin 8, 22 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Subramanian A., et al., Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mootha V. K., et al., PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 34, 267–273 (2003). [DOI] [PubMed] [Google Scholar]
- 49.Goldman M. J., et al., Visualizing and interpreting cancer genomics data via the Xena platform. Nat. Biotechnol. 38, 675–678 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sun Q., Li M., Wang X., The Cancer Omics Atlas: An integrative resource for cancer omics annotations. BMC Med. Genomics 11, 63 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chandrashekar D. S., et al., UALCAN: A portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia 19, 649–658 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Guan X., et al., CVCDAP: An integrated platform for molecular and clinical analysis of cancer virtual cohorts. Nucleic Acids Res. 48, W463–W471 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lakowicz J., Principles of Fluorescence Spectroscopy (Springer, Boston, MA, 2006). [Google Scholar]
- 54.Lakowicz J. R. M., Barry R., Principles of fluorescence spectroscopy. J. Biomed. Opt. 13, 029901 (2008). [Google Scholar]
- 55.Weinstein J. N., et al., The Cancer Genome Atlas Pan-Cancer analysis project. Nat. Genet. 45, 1113–1120 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or SI Appendix.