Significance
Membrane proteins and their interactions are major drug targets because of their central roles in regulating cellular communication and signal transduction. Despite this, membrane receptors remain underrepresented in our knowledge base of protein–protein interactions because of the limitations of most available techniques. Here, we developed a vesicle-based membrane-protein display platform as a fast, reproducible, and broadly applicable method for unbiased receptor–ligand interaction discovery. Using this technology, we elucidated known and new single-pass transmembrane-binding partners for several immune checkpoint molecules as well as an architecturally distinct single-pass transmembrane protein that has resisted deorphanization by existing techniques. This method is well suited for extracellular protein interaction discovery for difficult-to-purify membrane proteins and whole vesicles.
Keywords: membrane protein, extracellular vesicle, high-throughput screening, receptor–ligand interaction
Abstract
Cell surface receptors are critical for cell signaling and constitute a quarter of all human genes. Despite their importance and abundance, receptor interaction networks remain understudied because of difficulties associated with maintaining membrane proteins in their native conformation and their typically weak interactions. To overcome these challenges, we developed an extracellular vesicle-based method for membrane protein display that enables purification-free and high-throughput detection of receptor–ligand interactions in membranes. We demonstrate that this platform is broadly applicable to a variety of membrane proteins, enabling enhanced detection of extracellular interactions over a wide range of binding affinities. We were able to recapitulate and expand the interactome for prominent members of the B7 family of immunoregulatory proteins such as PD-L1/CD274 and B7-H3/CD276. Moreover, when applied to the orphan cancer-associated fibroblast protein, LRRC15, we identified a membrane-dependent interaction with the tumor stroma marker TEM1/CD248. Furthermore, this platform enabled profiling of cellular receptors for target-expressing as well as endogenous extracellular vesicles. Overall, this study presents a sensitive and easy to use screening platform that bypasses membrane protein purification and enables characterization of interactomes for any cell surface–expressed target of interest in its native state.
Membrane proteins play an essential role in translating extracellular cues into intracellular responses. Their cell surface exposure, resulting in increased accessibility to therapeutic molecules, and their ability to orchestrate cellular behavior makes them attractive drug targets. Therefore, it is unsurprising that while they make up ∼30% of human genes, membrane proteins account for over 60% of all drug targets (1). However, progress on the characterization of membrane proteins and their interaction partners has lagged far behind that of cytoplasmic proteins (2). This gap is in part due to difficulties expressing membrane proteins in their native, active conformations and the scarcity of techniques that have sufficient sensitivity to detect the weak interactions common for membrane proteins (3, 4). Methods designed for general protein–protein interaction discovery require either well-folded purified protein [e.g., microarray- or plate-based screening methods (3, 5)] or strong interactions that can survive the extraction of proteins from membranes and washes [e.g., affinity purification–mass spectrometry (6)]. While some of the recently developed approaches enable the capture of many membrane protein interactions by strengthening weak interactions using multimerization (7, 8), they may still miss key interactions, because the proteins are removed from their native membrane contexts.
Physiological membranes contain a complex mix of lipids, proteins, and glycans that can participate in membrane protein interactions (9). In addition, membranes can strengthen individually weak protein–protein interactions by clustering, thereby increasing binding avidity, and facilitating receptor orientation (10–12). These membrane-dependent aspects of the receptor–ligand interactions remain a major bottleneck in the development of screening methods. Recent advances in proximity-based techniques allow detection of interactions in membranes and have been instrumental for the study of transient binders (13–16). However, they often struggle to distinguish direct interaction partners from nearby bystanders, typically focus on binding partners within the same cell (in cis), and are often incompatible with high-throughput studies. Alternative approaches such as nanodiscs and liposome particles enable membrane protein reconstitution and have been successfully employed to study challenging receptors (17, 18). However, these methods require protein purification, which can disrupt native folds and rarely account for potential protein or nonprotein cofactors. As a result, extracellular protein crosstalk remains remarkably underrepresented in existing datasets (5, 19). These limitations underscore the need for techniques specifically designed for the study of membrane proteins, with sufficient throughput and sensitivity for the characterization of receptor interactomes.
In this work, we combine the high throughput of a direct protein–protein interaction screen with the presentation of a target-of-interest in the context of a membrane using HIV gag-containing recombinant extracellular vesicles (rEVs). The rEVs contain proteins folded and inserted into their native membranes by the cell’s endogenous machinery, allowing them to maintain their native conformations and membrane cofactors. We show that rEVs provide a protein purification–free method for obtaining binding-competent receptors displayed in the context of the membrane. To take advantage of these secreted particles, we developed RDIMIS (Receptor-Display In Membranes Interaction Screen, pronounced like Artemis), a platform for membrane protein interaction discovery in high throughput. RDIMIS allows rapid, reproducible and unbiased identification of receptor interactomes by displaying proteins-of-interest on EVs that are screened against a comprehensive library of single-pass transmembrane (STM) ectodomains (20, 21). Applying this platform to four proteins central to cancer immunotherapy, the poliovirus receptor (PVR) and three members of the B7 family of immunoregulatory receptors, PD-L1/CD274/B7-H1, CD80/B7-1, CD276/B7-H3, we identified all expected binding partners alongside putative new ones. Furthermore, we successfully employed RDIMIS to study the hard-to-purify, orphan cancer-associated fibroblast (CAF) protein, LRRC15 (22). RDIMIS identified the tumor stroma marker TEM1/CD248/endosialin as a previously unknown receptor for LRRC15, an interaction that we confirmed using orthogonal methods. In addition to identifying interactions for proteins of interest, we show that RDIMIS enables the identification of vesicle-specific, receptor–agnostic binding partners, positioning this method as a powerful platform for the profiling of endogenous extracellular vesicles from diverse cellular sources.
In sum, this report presents a high-throughput and versatile platform with improved sensitivity that enables the characterization of human receptor interactomes for difficult-to-purify targets, critical for understanding cellular communication during homeostasis and disease.
Results
Extracellular Vesicles Are a Pragmatic Solution for Membrane Protein Display.
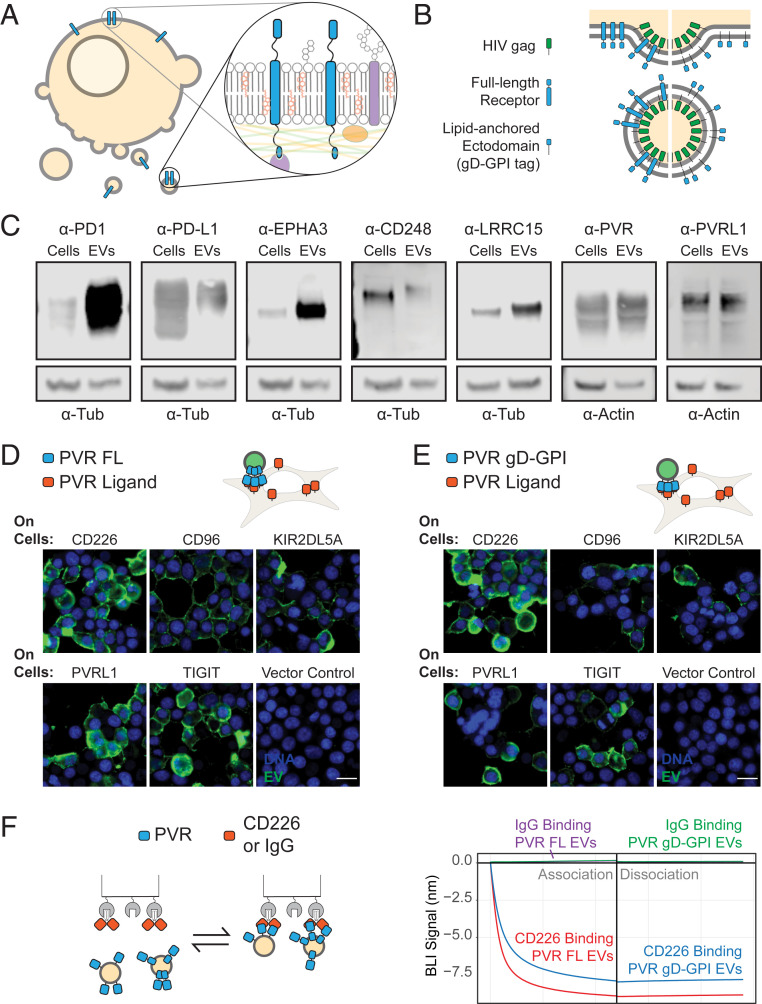
EVs incorporate a host of membrane-associated macromolecules (23), creating a microcosm of the cellular membrane environment (Fig. 1A). This environment often participates in membrane protein interactions, making EVs an ideal basis for a high-throughput and sensitive platform. However, some practical challenges had to be solved for EVs to be used for membrane protein interaction discovery. First, sensitive high-throughput screening requires large quantities of EVs. We therefore boosted EV production from the high–protein expression cell line, Expi293F (24, 25), by transfecting it with a HIV gag construct (26, 27) (Fig. 1B). This increased the 20- to 500-nm vesicle yield almost fourfold (SI Appendix, Fig. S1D). Second, cells release their EVs into a surrounding medium that contains a complex and variable background of other molecules. This complexity makes it challenging to get reproducible quantitative data from downstream assays. We therefore optimized a one-step, density-based EV purification (SI Appendix, Fig. S1A), which resulted in a pure and consistent population of rEVs free of aggregates (SI Appendix, Fig. S1B). Lastly, high-throughput screening necessarily shrinks the scale of each reaction, creating a challenge for robust and quantitative detection of bound rEVs. To maximize the sensitivity for detection, we fused Renilla luciferase (Rluc) to the gag protein employed to boost EV yields. This strategy resulted in robust luminescence signals linearly proportional to vesicle concentration over nearly three orders of magnitude (SI Appendix, Fig. S1E).
Fig. 1.
EVs enable STM protein display for receptor–ligand discovery. (A) EVs provide an endogenous membrane-like environment for receptors. (B) Screen-compatible rEVs can be generated by overexpressing HIV gag protein and a protein-of-interest either as a full-length protein or as a gD-GPI–tagged ectodomain. (C) Western blots of whole-cell lysates or EVs expressing different full-length, untagged receptors. (D) Full-length PVR expressing rEVs or (E) PVR gD-GPI EVs expressing gag-NeonGreen are incubated with cells transfected with known full-length PVR ligands, CD226, CD96, KIR2DL5A, PVRL1, and TIGIT. Green represents the direct fluorescence from EVs. (Scale bar, 20 µm.) (F) BLI experiment with either CD226-Fc or a control human IgG on the sensors dipped into 2.5 nM of rEVs expressing full-length PVR or gD-GPI ectodomains. The experimental conditions have been labeled in the figure to facilitate interpretation.
Next, to evaluate whether rEVs are broadly applicable as platforms for membrane protein display, we overexpressed a variety of full-length, untagged receptors in our rEV-producing cells. In all cases tested, the receptors were readily detectable in rEVs (Fig. 1C). To determine whether these receptors were accessible and binding competent, we carried out a proof-of-concept study using PVR, which binds a variety of cell surface–displayed proteins with different binding affinities (8). The rEVs displaying full-length PVR were isolated and tested for binding to these binding partners on cells. Notably, the PVR-EVs selectively bound to the surface of cells expressing PVR ligands, with negligible binding to cells transfected with an empty vector (Fig. 1D).
In parallel with our full-length receptor studies, we also designed a strategy to characterize the incorporation of different receptor ectodomains into rEVs without the need for receptor-specific antibodies. To do so, we fused a variety of receptor ectodomains to a glycoprotein D (gD) tag with a glycosylphosphatidylinositol (GPI) lipid anchor. While the GPI anchors the ectodomain to membranes, the gD epitope allows detection of all such constructs with a single anti-gD antibody. Importantly, when these proteins were tested for incorporation into vesicles, most of the receptors were readily detectable in rEVs using an anti-gD antibody (SI Appendix, Fig. S2 A–C). To determine whether this strategy resulted in binding-competent ectodomains, we applied it to PVR. Like full-length PVR rEVs, gD-GPI PVR rEVs selectively bound to cells expressing PVR ligands (Fig. 1E), suggesting that the tagged form is fully active. The gD tag also enabled detection of PVR receptors on the rEVs by electron microscopy (SI Appendix, Fig. S2B).
Next, we sought to assess whether receptor-expressing rEVs can bind tightly enough to survive the washing steps necessary for high-throughput assays while still enabling the detection of weak protein interactions. To this end, we determined the kinetics of binding for rEVs displaying either full-length PVR or gD-GPI–tagged PVR ectodomain to the known PVR ligand, CD226. Using the nondestructive biolayer interferometry (BLI) method, rEVs showed a strong negative signal for both PVR rEV types, as expected given the large size of the vesicles (28) (Fig. 1 F, Right). This was not observed when PVR rEVs were incubated with human IgG as a control, indicating that the signal represents a specific interaction. The rEVs showed slow dissociation over 10 min, indicative of a high-avidity interaction that make this approach suitable for high-throughput screening and the detection of weak protein interactions. Consistent with the cell-based binding assays (Fig. 1 D and E), the gD-GPI–tagged PVR rEVs showed a binding profile similar to the full-length PVR rEVs, further demonstrating that the engineered receptor is amenable for detection of in trans binding partners.
RDIMIS: A High-Throughput Platform for the Elucidation of Membrane Receptor Interactomes.
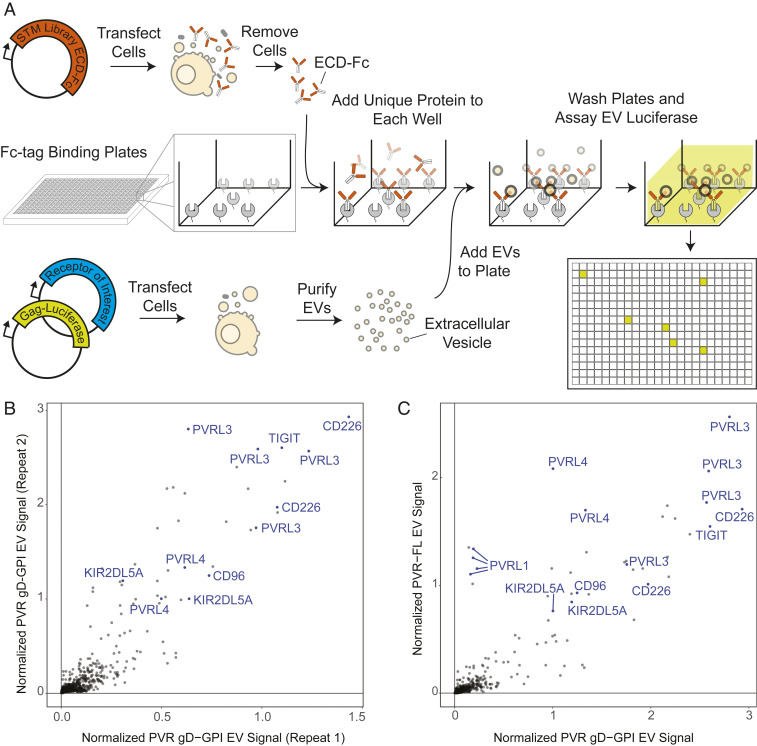
Having demonstrated that rEVs can display active receptors, we sought to use this strategy to enable high-throughput discovery of receptor–ligand interactions. To achieve unbiased binding partner profiling, we took advantage of our previously developed conditioned media library consisting of most human STM proteins expressed as ectodomain-Fc tag fusions (21, 29). To generate this library, Fc-tagged protein ectodomains were individually transfected in cells for expression and secretion as soluble proteins into the growth media. The conditioned media was then transferred to protein A–coated plates, with each well receiving a different ectodomain. This resulted in a collection of immobilized ectodomains suitable for high-throughput screening (21). In parallel, cell cultures were transiently transfected with the protein-of-interest alongside gag-Rluc for rEV isolation (Fig. 2A). The receptor-displaying rEVs were isolated using an optimized purification protocol that enabled rapid large-scale isolation of rEVs (SI Appendix, Fig. S1A). After rEVs were isolated, they were incubated with plates containing the STM protein library. The plates were thoroughly washed to remove unbound rEVs. The rEVs remaining in the wells are retained through an interaction between the protein displayed on the rEV and the STM protein immobilized on the well. Rluc substrate was then added to light up the wells at which an interaction had taken place (Fig. 2A).
Fig. 2.
A platform for detection of receptor–ligand interactions in the context of endogenous membranes. (A) Schematic of the RDIMIS workflow. The library of STM proteins, expressed as Fc-tagged ectodomains, are immobilized on plates. Independently, rEVs are isolated from the conditioned media of cells expressing the receptor-of-interest alongside gag-Rluc. Receptor-rEVs are screened against the collection of plate-bound STM proteins using a semiautomated workflow. Interactions between rEVs and ectodomains in the library are detected using luminescence. (B) Results show two independent RDIMIS screens for PVR gD-GPI rEVs. (C) RDIMIS screens for full-length (FL) PVR rEVs compared with repeat 2 from B.
We first tested this platform by studying the STM interactome of gD-GPI–tagged PVR ectodomain containing rEVs. Receptor display on the vesicles was confirmed using BLI, which showed robust binding of the rEVs to the gD antibody (Fig. 3A) or by Western blot (SI Appendix, Fig. S2C). Notably, RDIMIS identified all the expected PVR binding partners, CD96, CD226, PVRL3, PVRL4, and TIGIT as well as the recently described KIR2DL5A (8) (Fig. 2B, blue). Results were highly reproducible across two independent rEV and STM library preparations (Fig. 2B) (correlation coefficient of 0. 90) (SI Appendix, Fig. S5A). Moreover, the high scoring hits detected were virtually identical when RDIMIS was utilized to study full-length, untagged PVR in rEVs (Fig. 2C) (correlation coefficient of 0.88) (SI Appendix, Fig. S5A). Together, this further demonstrates that gD-GPI–tagged receptors expressed on rEVs allow detection of relevant ligands in trans, which in combination with the automated workflow developed, enables robust identification of membrane protein interactomes in an unbiased fashion and with enhanced sensitivity for detection of high- and low-affinity interactions.
Fig. 3.
RDIMIS enables identification of known interactions and putative binders for unrelated immune receptors-of-interest. (A) Expression and display of gD-GPI–tagged ectodomains on rEVs, detected by BLI. Anti-gD antibody was immobilized on the sensor and then incubated with rEVs expressing different gD-GPI tagged ectodomains. (B) RDIMIS screen for PD-L1 gD-GPI rEVs compared with the PVR gD-GPI rEV repeat 2 from Fig. 2A. (C) Overlap between the hits identified in this study for PD-L1 and PVR with interactions listed in the STRING database. (D) RDIMIS screen for CD80 gD-GPI rEVs compared with CD276 gD-GPI rEVs. (E) Overlap between the hits identified in this study for CD80 and CD276 with interactions listed in the STRING database. No experimentally verified interactions were listed in STRING for CD276. Screens are plotted against one another to differentiate receptor-specific hits (near either axis) from the generic vesicle binders (Fig. 6 and SI Appendix, Fig. S4) common between the screens. Hits that fall above the 98% quantile and show greater than a 4× screen-specific enrichment are labeled.
RDIMIS Identifies Putative Binders for Checkpoint Proteins.
To further benchmark the sensitivity of this technique, we applied our platform to three members of the B7 family of prominent immunoregulatory proteins, including the checkpoint inhibitor PD-L1. All three proteins were expressed as gD-GPI ectodomain fusions, allowing for their expression in vesicles to be monitored and directly compared. In all three rEVs, the tagged ectodomains bound to an anti-gD antibody at levels comparable to PVR gD-GPI rEVs when assayed by BLI (Fig. 3A) or by Western blot (SI Appendix, Fig. S2C). PD-L1 rEV screening results were compared with those from PVR rEV screens to identify receptor-specific interactions. Notably, RDIMIS using PD-L1 gD-GPI rEVs identified the known PD-L1 ligands, PDCD1 (PD1), EPHA3, CD80, and PDCD1LG2 (PD-L2) with high confidence (Fig. 3B and Dataset S1). Since the estimated affinity for the PD-L1/PD-L2 interaction is ∼10 µM (30), this further demonstrates that RDIMIS can identify biochemically challenging, weak interactions. Interestingly, a number of other high-scoring hits were identified (Fig. 3B). While IGF2R has been found to be broadly sticky in unrelated experiments (8), the other hits represent putative binders for PD-L1. To get a sense of the landscape of interactions that had been previously identified, we evaluated the overlap between our list of receptor-specific hits and experimentally supported interactions found in the STRING database (31) (Fig. 3C), a comprehensive repository for protein interactions. For a fair comparison, only interactions between proteins that were present in the STM library queried in this study were considered. Reassuringly, the most well-characterized interactions, those between PD-L1 and PD1 and CD80 and PD-L2, were represented in the STRING database. On the other hand, none of the remaining hits were found, suggesting that RDIMIS identified PD-L1 binding partners not represented in the STRING database.
To validate the sensitivity of the screens, we followed up on LRTM1, one of the putative PD-L1 binders with the lowest binding signal in our screens. Since we were not able to find commercially available protein for LRTM1, LRTM1 expressed on rEVs was used to study the interaction. PD-L1 ectodomains selectively bound to LRTM1 on rEVs presented as either a gD-GPI–tagged ectodomain or as the full-length protein (SI Appendix, Fig. S2D). LRTM1 rEVs also selectively bound to PD-L1 expressed on cells over cells transfected with a vector control (SI Appendix, Fig. S2E). Since PD1 is a known interaction partner of PD-L1 and the target of checkpoint blockade immunotherapy in cancer, we tested whether these interactions were competitive. Interestingly, increasing concentrations of recombinant PD1-Fc protein outcompeted LRTM1 vesicle binding in a concentration-dependent manner (SI Appendix, Fig. S2F), suggesting that the proteins bind to similar regions on PD-L1 and further supporting the specificity of the PD-L1/LRTM1 interaction.
To demonstrate the wide applicability of RDIMIS, the platform was applied to two additional membrane proteins, CD80 and CD276 (Fig. 3D and Dataset S1). Again, all the relevant partners were detected for both proteins, confirming the broad utility of this methodology to cell surface–expressed targets. For CD80, this included the well-described binders CD28, CTLA4, and PD-L1 as well as NGFR, recently suggested as an additional CD80 interactor (29). In the case of CD276, deorphanized only recently through advances in screening technology (8), RDIMIS captured the described interactor, IL20RA, PLA2R1, and MXRA5 which was found to be a nonspecific interactor (8) (Fig. 3D). Similar to PD-L1, an additional putative, receptor-specific binding partner, PLA2R1, was identified for CD276 that was previously not described in the literature or found in the STRING database (Fig. 3E). For all four proof-of-concept screens (PVR, PD-L1, CD80, and CD276), 11 out of 12 interactions in STRING were recovered showing that RDIMIS can capture the range of interactions that have been validated experimentally (Fig. 3 C and E). The sole exception was the CD80–CD86 interaction that remains controversial.
RDIMIS Enables the Deorphanization of a Cancer-Relevant Protein, LRRC15.
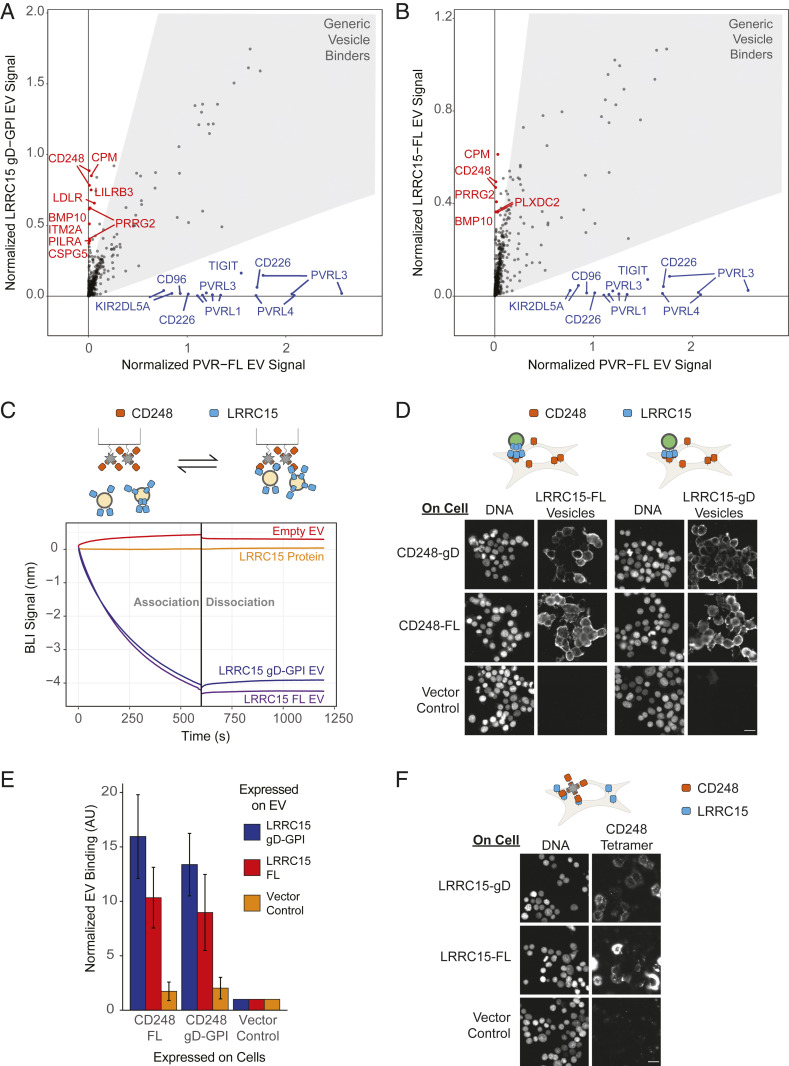
The performance of RDIMIS on the selected immunoregulatory proteins suggested that this platform may allow the deorphanization of challenging targets refractory to other biochemical screening approaches. As an example, we decided to apply RDIMIS to study the CAF protein, LRRC15, which recently emerged as a specific marker for CAFs associated with solid tumors (22). Despite its biological importance, no interaction partners have been identified, and thus, basic aspects of LRRC15 biology remained undefined. We first looked for LRRC15 interaction partners using an existing technology developed for extracellular protein–protein interactions, miniaturized AVEXIS (7, 21). Despite achieving high sensitivity through ectodomain pentamerization, no binding partners were identified for LRRC15 using this technique (SI Appendix, Fig. S6A). This may suggest that LRRC15 requires a more physiologically relevant setting for optimal activity. To test this hypothesis, LRRC15 was screened as gD-GPI (Fig. 4A and Dataset S1) and full-length (Fig. 4B and Dataset S1), protein-expressing rEVs using RDIMIS. Notably, both of these efforts identified similar sets of putative interactors for LRRC15, which were not described in the STRING database (SI Appendix, Fig. S6B).
Fig. 4.
CD248 is a membrane-facilitated binding partner for the orphan protein LRRC15. (A) RDIMIS results for LRRC15 gD-GPI or (B) LRRC15 full-length rEVs in which hits above the 98% quantile and show greater than 4× screen-specific enrichment are labeled. In both cases, results are compared with PVR screen results shown in Fig 2C. (C) Binding between CD248, expressed as a recombinant protein and immobilized on a sensor, and LRRC15-Fc (500 nM) or LRRC15 full-length or gD-GPI ectodomain expressing rEVs (0.25 mg/mL). (D) Full-length or gD-GPI CD248 was transiently expressed in cells. Cells were incubated with vesicles from cells transfected with gag-NeonGreen and full-length or gD-GPI LRRC15-expressing or an empty vector control. (E) Quantification of NeonGreen signal from D displayed as mean ± SE for three independent replicates. (F) Full-length or gD-GPI LRRC15 was transiently expressed in cells. CD248, as a tetramerized recombinant ectodomain, binding to the cell surface was determined using microscopy. (Scale bar, 20 µm.)
Since CD248 was a top scoring hit in both screens and its expression is up-regulated in tumor stroma (32–34) in which it may promote tumor growth (35), we focused on the LRRC15–CD248 interaction for further characterization. As a first step toward characterizing this protein pair, we assessed this interaction using biophysical and biochemical methods. First, miniature AVEXIS was performed using CD248-pentamerized ectodomains. Similar to LRRC15, no high-confidence hits were identified when CD248-pentamerized ectodomains were screened against the STM protein library (SI Appendix, Fig. S6C), which contained the LRRC15 ectodomain. Consistent with this result, no binding was observed between LRRC15 and CD248 recombinant proteins when the interaction was analyzed by either BLI (Fig. 4C) or surface plasmon resonance, even when experimental conditions to maximize sensitivity of detection were employed (SI Appendix, Fig. S6D). Given the lack of detectable binding between recombinant ectodomains, we tested this interaction in the context of a membrane. Even though BLI analysis did not detect binding between LRRC15 and CD248 as recombinant ectodomains, this interaction was readily detectable when LRRC15 was displayed on rEVs (Fig. 4C). Similarly, the LRRC15–CD248 interaction also occurred on the cell’s plasma membrane. LRRC15-expressing rEVs bound to CD248 overexpressed on the cell surface (Fig. 4D) over 10 times more than cells transfected with an empty vector control (Fig. 4E). Tetramerized CD248 recombinant protein also detectably bound to LRRC15-overexpressing cells but not to cells transfected with a vector control (Fig. 4F). These assays reinforced the notion that this interaction requires a membrane but not specifically rEVs and highlighted the sensitivity of our platform for detection of interactions that are refractory to other methodologies. To better understand the basis for the membrane dependence of this interaction, we altered rEV membranes either by disrupting them using Filipin III which form cholesterol ultrastructures (SI Appendix, Fig. S6 E–G) or by depleting membrane cholesterol with methyl-β-cyclodextrin (MβCD) (SI Appendix, Fig. S6 H–J) (36). We found that these cholesterol-disrupting reagents eliminated the binding of LRRC15 gD-GPI, and to a lesser extent LRRC15 full-length, expressing vesicles to CD248 monomer by BLI (SI Appendix, Fig. S6 E–G).
Though we showed that LRRC15 and CD248 can interact in a membrane-dependent manner, we did not know whether they were present in the same physiological environment. While LRRC15 and CD248 have been independently reported to be up-regulated in solid tumors (32, 37), it was unclear whether they were expressed in the same tumor samples. Bulk RNA sequencing (RNA-seq) data from The Cancer Genome Atlas (TCGA) for four different tumor indications showed significant correlations between the expression of CD248 and LRRC15 (Fig. 5 A and B and SI Appendix, Fig. S7 A and B). This correlation suggested that CD248 and LRRC15 are either found on the same cell type or that CD248 and LRRC15 were coregulated. To help answer that question, single-cell RNA-seq data from head and neck cancer patients was reanalyzed to highlight LRRC15 and CD248 expression (38). This analysis revealed that LRRC15 and CD248 are coexpressed on a subset of CAFs (co-occurrence score [Odds ratio] = 9.44) (Fig. 5 C and D), with CD248 showing a broader expression that encompasses all CAF and cancer-associated pericytes identified using markers such as DCN and RGS5 (SI Appendix, Fig. S7C).
Fig. 5.
LRRC15 and CD248 are coexpressed in tumors. (A) Bulk RNA-seq expression of LRRC15 (x-axis) and CD248 (y-axis) for head and neck squamous cell carcinoma and (B) breast-invasive carcinoma. Each point represents a single-patient sample. Spearman's rank correlation coefficient and significance values are given on the top right. (C and D) Uniform Manifold Approximation and Projection dimensionality reduction showing nontumor cells from single-cell RNA-seq data of head and neck cancer patients (39). Cells are colored by expression of (C) LRRC15 or (D) CD248.
Together, these results position RDIMIS as a robust method to identify new interactors for membrane proteins not amenable to technologies that rely on recombinant protein purification. Further, while it is unclear whether the interaction between LRRC15 and CD248 is occurring on the same cell or between cells, the above analysis suggests that these proteins have ample opportunity to interact in patient tumors, providing a potential biological context in which this interaction might be relevant.
RDIMIS Identifies Vesicle-Specific Binding Partners, Enabling the Profiling of Endogenous EVs.
Extracellular vesicles are increasingly recognized as critical mediators of cellular communication and have been implicated in disease progressions from cancer to infection (39). However, the molecular basis for much of their function and tissue-specific uptake has remained undiscovered. Since RDIMIS identifies all receptors that were able to retain rEVs in wells, this technique has the potential to profile binders for other molecules on the rEV surface, independent of the target-of-interest. Indeed, when batch-matched screens were plotted against each other (Figs. 3 B and D and 4 A and B) or against a screen in which cells were transfected with a vector control instead of a receptor-of-interest done independently (SI Appendix, Fig. S3 A–F), a number of high-confidence hits were common between the screens (gray shading in each plot). A consistent list of 80 common rEV binders emerged which we ranked by the average signal across all screens (SI Appendix, Table S1). To determine whether any biological pathways or functions were overrepresented, gene ontology (GO) enrichment analysis was performed. The molecular functions significantly enriched included carbohydrate, sulfur, and anion binding, all consistent with general binding to vesicles and cellular membranes (SI Appendix, Fig. S4A). In addition, GO analysis also indicated that rEV binders are involved in cell signaling, suggesting vesicle recognition by specific cellular receptors.
To generate a list of candidate cell–rEV, receptor–ligand pairs (SI Appendix, Fig. S4B), we cross-referenced our generic rEV binder list with the immunoglobulin superfamily interactome (29) and the STRING database (31). Using this data, we generated a network of putative EV receptor–ligand interactions (SI Appendix, Fig. S4C), showing vesicle-expressed proteins (blue) for the majority of the generic vesicle binders (green) identified using RDIMIS. To get a sense of whether these proteins are likely to be in the vesicles, we cross-referenced Cell Atlas expression data for human embryonic kidney 293 cells (HEK293) (40), the parent cell line for the Expi293F cells that we used to generate the rEVs. Since we found that the incorporation of proteins into the rEVs were typically correlated with their expression in the cells, we reasoned that a highly expressed protein was more likely to be responsible for the binding that we detected. Together, this network highlighted a series of likely rEV-expressed proteins for rEV binding receptors identified by RDIMIS, providing a putative map of protein–protein interactions that may regulate EV recognition and signaling.
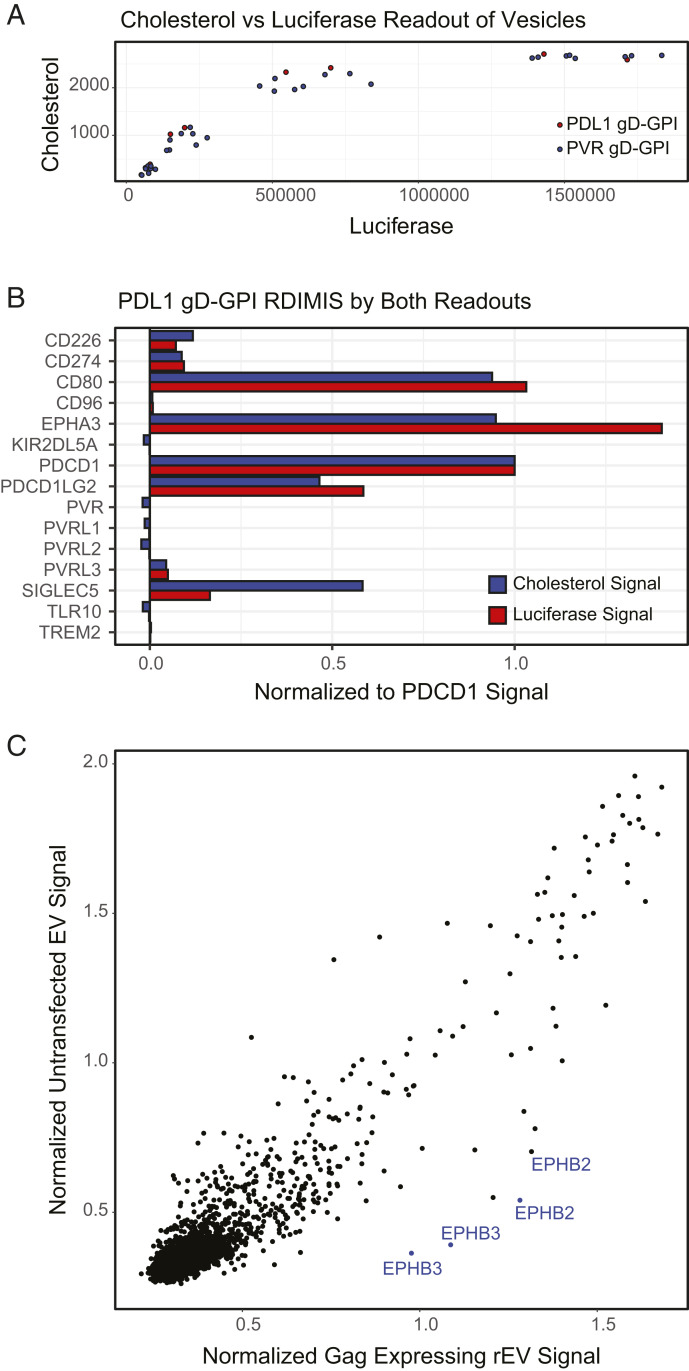
Having demonstrated that RDIMIS is a robust method to identify both vesicle-specific and target-of-interest–specific binding partners, we further developed the method to screen endogenous EVs devoid of any tags. Since cholesterol is a major component of the EV membrane (41), we used the Amplex Red Cholesterol assay that quantifies cholesterol levels. To ensure that this approach can detect EVs, we directly compared cholesterol readout to luciferase signal using our established rEVs. Luciferase signal and then cholesterol measurements were made for different dilutions of one batch of PD-L1 gD-GPI rEVs and three independent batches of PVR gD-GPI rEVs (Fig. 6A). The signal correlated well for the ranges of rEVs used for RDIMIS screening, though the cholesterol signal saturated at a lower concentration of rEVs. Next, we performed RDIMIS using PD-L1 gD-GPI rEVs at a small scale with a select set of receptors in our library (Fig. 6B). Importantly, both the luciferase and cholesterol signal were able to identify the known PD-L1 binders, CD80, EPHA3, PDCD1, and PDCD1LG2 as well as a generic rEV binder, SIGLEC5. Lastly, we performed two full RDIMIS screens using either rEVs that were transfected with a vector control but contained gag-Rluc or endogenous EVs harvested from untransfected cells (Fig. 6C). We found that both EV species behaved similarly (correlation coefficient of 0.92), showing that rEVs and naturally generated EVs from the same cell line have similar binding profiles. This analysis also suggests that similar predicted receptor–ligand interactions (SI Appendix, Fig. S4C) may participate in endogenous EV binding.
Fig. 6.
Unmodified vesicles can be profiled with RDIMIS using cholesterol as a readout. (A) PD-L1 gD-GPI or PVR gD-GPI rEVs containing gag-RLuc were diluted across a range of concentrations and then read out either by luciferase signal or using the AmplexRed Cholesterol Assay to measure cholesterol content of the solutions. (B) Small-batch RDIMIS with PD-L1 gD-GPI rEVs was performed and read out using either cholesterol or luciferase signal to compare the signal in a screening context. (C) RDIMIS was performed using cholesterol as a readout on rEVs expressing gag-Rluc (x-axis) or endogenous EVs harvested from untransfected cells (y-axis).
Discussion
In recent years, successes in both understanding and targeting membrane proteins and their interactions have fueled a multitude of drug development and exploratory research efforts across scientific disciplines. These efforts, however, have also continuously exposed the need for new specialized methods that specifically address membrane protein biology. While recent technological developments have significantly improved sensitivity for detection of interactions between these proteins, most of these techniques will inevitably miss specialized functions due to their membrane-embedded nature (2). The need for an in-membrane protein display method has spurred elegant solutions that take advantage of machinery from enveloped viruses to incorporate receptors into mammalian membranes. In particular, microarrays consisting of herpes simplex virions displaying different membrane proteins have been used successfully as a G protein–coupled receptor library for peptide ligand discovery (42, 43). A complementary approach that takes advantage of the HIV gag protein has previously been used to display multitransmembrane proteins for immunization and antibody generation (44), as well as antibody selection and ligand-binding characterization (45). RDIMIS builds on the use of HIV gag for rEV generation and developed the approach into a high-throughput workflow for the systematic query of interactions with human STM proteins. The result is a screening platform with the controllability, scalability, and high sensitivity of direct protein–protein interaction screens while also capturing the unique complexity of the cellular membrane.
While many molecular details remain ill-defined, it is clear that the different building blocks of membranes (i.e., carbohydrate groups, lipids, or other proteins) participate and influence receptor–ligand interactions. rEVs also contain a cytoplasmic luminal space with cytoskeletal elements that can play roles in higher-order complex formation (10, 23). In addition to capturing this complexity, the small size and stability of rEVs (41) make them an ideal vehicle for high-throughput screening in a physiologically relevant context. As such, the approach implemented in this study can be coupled to any library of choice, from high-coverage collections to more focused libraries such as receptor families or protein disease variants, enabling sensitive and relatively low resource-intensive identification of interactomes while maximizing query protein quality. Although broad and unbiased in nature, our work so far has been restricted to the identification of STM protein interactions and has not addressed interactions that involve other protein families. Therefore, RDIMIS could be further optimized to enable screening of multitransmembrane proteins or secreted factors. These aspects should be explored in future implementations of RDIMIS, as they represent an important fraction of the extracellular interactome that have not been addressed here.
Beyond the utilization of rEVs for the deorphanization of full-length receptors, we demonstrate that gD-GPI–tagged ectodomains can achieve results similar to full-length proteins and skirt the need for often elusive high-affinity antibodies to measure receptor incorporation. This strategy also enables the direct comparison of the interaction profiles of the ectodomain and full-length protein, quickly identifying interactions in which the transmembrane or cytoplasmic domains may play a role. Lastly, this tagging strategy can anchor nonmembrane proteins, providing a way to study extracellular matrix or secreted factors in proximity to membranes. Given the scalability of the method, and the ease of expression of receptors as gD-GPI–tagged ectodomains for rEV production, RDIMIS offer a platform for large-scale deorphanization of a relevant fraction of the extracellular proteome.
These advantages allowed us to identify an interaction between the cancer stroma markers LRRC15 and CD248, which we show is only measurable in the context of a membrane. There are several models that may explain the membrane dependence. The simplest model is that the LRRC15 ectodomain requires a membrane environment for proper folding. Interestingly, since both the full-length and gD-GPI–tagged ectodomain captured this interaction, the determinants responsible for this dependence may not be within the transmembrane domain or a precise spacing between the membrane and the ectodomain. Another plausible explanation is that the presence of the membrane promotes the formation of highly clustered arrays of receptors, increasing protein avidity beyond the tested pentamerization, to stabilize the interaction. This hypothesis is in part supported by the evidence that Filipin III and MβCD can disrupt this interaction. Alternatively, the LRRC15–CD248 interaction may depend on the recruitment of a yet unknown factor that promotes or stabilize the complex. While more work is needed to determine the nature of this membrane dependency, our study demonstrates that vesicle display provides increased sensitivity for detection of challenging protein interactions that have remained understudied by currently available methods. Similarly, although the functionality of the CD248–LRRC15 interaction remains unknown, our data indicate that these proteins are coexpressed in the tumor microenvironment, suggesting a possible regulatory function in solid tumors.
Beyond the identification of protein interactomes and the deorphanization of complex or hard-to-purify targets, this approach has wide applications in the study of human EV biology. For example, RDIMIS identified 80 vesicle-specific binders for a population of human cell line–derived EVs and showed that the interactome of recombinant and endogenously generated EVs were remarkably similar. This is consistent with previous works showing strong similarities between the proteomic and lipid compositions of recombinant and endogenous EVs (26, 46). Therefore, these binders may shed light on the receptor specificity of endogenous EVs and their potential signaling activity. The use of RDIMIS to analyze untagged EV using cholesterol content as a readout provides the foundation for endogenous vesicle profiling. This work sets the stage for future studies focused on disease-, tissue-, or cell-specific EVs and their roles in cellular communication or immune responses. As EVs are also being explored as avenues for drug delivery (47), RDIMIS can provide a way to address concerns about variability, immunogenicity, and off-target effects associated with EV complexity. Since this platform provides a unique tool to elucidate the players that influence EV functions at the molecular level, it can detect unexpected changes in the binding profile of EVs arising from the overexpression of target receptors, addition of therapeutics, or modifications of the cells generating the EVs.
All these possibilities arise because RDIMIS is a receptor–ligand discovery platform for unbiased STM interactome discovery of membrane proteins expressed in their native environment. The capture of membranes in high-throughput screens can quickly reveal new membrane-dependent modes of binding that has eluded researchers. This platform has broad applications that inform both basic and translational research endeavors.
Materials and Methods
Cell Culture.
HEK293T were cultured in Dulbecco's Modified Eagle Medium + GlutaMax supplemented with 10% fetal bovine serum and penicillin/streptomycin (Thermo Fisher Scientific). Expi293F were cultured in Expi293 expression media shaking at 150 rpm. All cells were cultured at 37 °C and 5% CO2.
Generation of Extracellular Vesicles and the STM Library.
HEK293T or Expi293F cells were transiently transfected with a protein-of-interest or an empty vector control and a plasmid expressing HIV gag fused to either Rluc (for screening) or mNeonGreen (for visualization). All plasmids were cloned into the expression vector pRK5 (Genentech). For vesicle harvesting, cells were removed by either spinning at 300 × g for 10 min followed by clearing at 2,000 × g for 20 min or filtered out. For full library screens, rEV expressions were done at a 1-L scale and grown for 7 d. Complete EDTA-Free Protease Inhibitor Mixture tablets (Roche) were added. Conditioned media was spun at 12,000 × g for 40 min to remove any remaining dense particulates and microvesicles. The supernatants were transferred to 70-mL polycarbonate ultracentrifuge tubes (Beckman Coulter). A total of 10 mL of 50% sucrose was layered from the bottom using a syringe and long needle, forming a sucrose cushion. Samples were spun at 100,000 × g for 90 min in a Ti-45 rotor. Vesicles were found to float on top of the sucrose. Media above the vesicle layer was aspirated. Cushion and vesicles were combined and diluted to 70 mL in new ultracentrifuge tubes using phosphate-buffered saline (PBS). Samples were spun at 100,000 × g, and the resulting pellet was dissolved in PBS. Halt Protease Inhibitor Mixture (Thermo Fisher Scientific) was added to 1×. All spins were done at 4 °C.
STM library generation was performed as in Martinez-Martin et al. (21).
Vesicle Characterization and Binding Measurements.
BLI measurements were done using an eight-channel Octet RED system (ForteBio). For PVR rEV binding, CD226-Fc (R&D Systems) or native human IgG (abcam) was loaded at 25 nM onto Anti-Human IgG Fc Capture Biosensors. For LRRC15 rEV binding, CD248 (R&D Systems) was biotinylated using the EZ-link Sulfo NHS-LC-LC-Biotin (Thermo Fisher) and cleaned up on Zeba desalting columns with 7K molecular weight cutoff (MWCO) (Thermo Fisher Scientific) or used as is. CD248 was loaded onto Streptavidin Biosensors at 25 nM or onto NiNTA Biosensors at 100 nM. LRRC15-Fc protein (Genentech) was provided at 500 nM. LRRC15 rEVs were provided at 0.25 mg/mL total protein (2.5 to 3.5 nM) for Fig. 4C or at 0.1 mg/mL for SI Appendix, Fig. S6 E–J. All measurements were done at 30 °C. Analysis was done on the Octet Data Analysis software. A PBS buffer control was subtracted to account for drift in the instrument unless the PBS curve is shown. Alignment was to baseline, and Savitzky–Golay filtering was performed. In screening cases, for gD-GPI vesicles, vesicles were measured at a total protein concentration of 0.1 mg/mL in PBS against 10 µg/mL mouse anti-gD antibody (Abcam) using Anti-Mouse IgG Fc Capture Biosensors (ForteBio) by BLI. Total protein concentrations of the vesicles samples were taken using the Quick Start Bradford Protein Assay Reagent (Bio-Rad). Vesicle particle numbers and concentrations were calculated from nanoparticle tracking analysis with NanoSight NTA (Malvern Panalytical). Vesicles at 0.1 mg/mL total protein were diluted 1,000× in PBS and run for five repeats of 1-min-long recordings using a 488-nm laser. Traces were analyzed using NTA 3.4 software, which provided a particle concentration that was used to calculate a molarity. Surface plasmon resonance was done using a Protein A Series S chip (GE Healthcare) on a Biacore 8K. LRRC15-Fc was loaded onto the chip at either 5 or 50 µg/mL.
Electron Microscopy.
The suspension of vesicles was adsorbed for 15 min to the surface of formvar and carbon coated transmission electron microscope (TEM) grids. Sample was stained with 2% phosphotungstic acid for rEV prep cleanup for 60 s and then air-dried. For gD epitope detection, vesicles were adsorbed for 30 min and blocked with Aurion Blocking Solution. Samples were then stained with mouse anti-gD (abcam) in the blocking solution and detected using goat anti-mouse 12-nm gold conjugate. Samples were then washed with PBS and water and stained with 1% uranyl acetate before being blotted and air dried. Imaging was done with a JEOL JEM-1400 TEM and a GATAN Ultrascan 1000 charge-coupled device camera at magnifications from 5,000× to 50,000×.
RDIMIS.
Vesicles were diluted into a final concentration of 0.03 to 0.05 mg/mL (as measured by Bradford) in 1× PBS + 0.49 mM MgCl2 + 0.9 mM CaCl2 (PCM) + 1% bovine serum albumin Fraction V (Sigma). Preparation of the human receptor library was performed using a robotic system consisting of liquid handling devices to allow for high-throughput analysis. Conditioned media containing Fc-tagged receptor ectodomains were dispensed directly into white 384-well protein A coated plates (Thermo Fisher Scientific) without adjusting concentrations (21). Plates were washed three times with PCM to remove unbound components. Vesicles were added to the plates and allowed to sit overnight at 4 °C. To prevent drying, 25 µL of PCM was added to the plates. For a positive control used for normalization, 25 µL of the same vesicle stocks used in the screens were added into the first column of each plate after all washing steps were completed. A total of 25 µL 1 µM coelantrazine h (Promega) in PCM was dispensed into the wells, incubated for 5 min, and read on a TECAN using 0.1 s of luminescence read time.
Western Blotting.
Protein concentrations were equalized in lysis buffer and diluted in lithium dodecyl sulfate Sample Buffer (Invitrogen) and Sample Reducing Agent (Invitrogen). Samples were loaded onto NuPAGE 4 to 12% Bis-Tris Protein Gels (Invitrogen) and transferred onto nitrocellulose using the iBlot Transfer system (Invitrogen). Blots were stained with anti-gD antibody (Abcam), rabbit anti-PD1 (Abcam), rabbit anti-PD-L1 (E1L3N, Cell Signaling), rabbit anti-EPHA3/4/5 (Cell Signaling), rabbit anti-TEM1 (Proteintech), rabbit anti-LRRC15 antibody (abcam), mouse anti-PVR (R&D Systems), mouse anti-Nectin1 (Thermo Fisher), mouse anti-alpha tubulin (Thermo Fisher), and rabbit anti-beta actin (Cell Signaling).
Immunofluorescence Validation of the LRRC15–CD248 Interaction.
HEK293T cells split into a 96-well SensoPlate (Greiner Bio-One) coated with 0.1 mg/mL Poly-D-Lysine (Gibco) for 30 min at 37 °C. Cells were transfected using LTX Reagent (Thermo Fisher Scientific) according to their specifications. For fluorescent rEV experiments, rEVs were harvested from Expi293F cells transiently cotransfected with gag-mNeonGreen and the protein-of-interest. They were purified by ultracentrifugation at 100,000 × g for 90 min after a 10-min spin at 300 × g and a 1-h spin at 3,000 × g to remove cells and debris. rEVs were incubated with cells for 30 min at 4 °C. Cells were washed with PBS and fixed using 4% paraformaldehyde for 10 min. LRRC15-Fc was generated at Genentech. CD248 protein (R&D Systems) was biotinylated using EZ-link Sulfo-NHS-LC-Biotin (Thermo Fisher Scientific), cleaned up on a Zeba 7K MWCO desalting column, and tetramerized using streptavidin-allophycocyanin (Agilent). DNA was stained with Hoechst 33342 (Tocris Bioscience).
Generic rEV Binder List Generation.
A cutoff was drawn at the 90% quantile, because the distribution of our data deviated from a normal distribution and had a long upper tail. This was done for each screen individually, and the final list was the list of genes that was common between all screens. To rank the genes, the intensity was averaged across all screens and sorted by that average. GO analysis was done through www.pantherdb.org using the PANTHER16.0 release overrepresentation test (48) on the list of generic vesicle binders. The reference list was a list of all genes in our STM library. We looked at the GO molecular function complete, biological process complete, and cellular component complete annotation data sets using the Fisher’s exact test.
Comparison with STRING Database Interactions.
Protein specific information was retrieved from the STRING database (https://string-db.org/). Only interactions with experiments or databases as “active interaction sources” were included. Only the subset of proteins represented in our STM library were kept for comparison. To generate a stringent list of hits for all screens, a cutoff was drawn at the 98% quantile for each screen, because the distribution of our data deviated from a normal distribution and had a long upper tail. In all screens, a receptor-specific hit was called if the signal in a particular screen was at least four times that of the other screen.
Single-Cell and Bulk RNA-Seq Data Analysis.
The coexpression of LRRC15 and CD248 in bulk RNA-seq samples across multiple cancer types was analyzed using gene expression profiling interactive analysis (49). Single-cell RNA-seq data from Head and Neck Cancer patients was processed and annotated as described previously (38). Odds ratios for co-occurrence of LRRC15 and CD248 expression (normalized gene expression > 0) in individual cells was calculated as described by Gao et al. (50).
Unlabeled Vesicle Detection Method.
Membrane cholesterol was detected using the Amplex Red Cholesterol Assay Kit (Invitrogen). Vesicles were screened according to the RDIMIS protocol, except rather than adding coelantrazine-h, plates were manually flicked dry and the Amplex Red Cholesterol Assay mix was added. For the titration against luciferase signal, four 3× serial dilutions were made. A total of 20 µL 0.5 µM Coelantrazine h (Promega) was added, incubated for 5 min, and read in the TECAN. A total of 20 µL the Amplex Red Cholesterol Assay mix was added to the wells, incubated for 1 h, and read. Luminescence was read out on a TECAN using 0.1 s of luminescence read time. Fluorescence was read out in a TECAN using an excitation of 560 nm and an emission wavelength of 590 nm. A blank well with just PBS and Coelantrazine h and Amplex Red Cholesterol Assay mix was also measured and the values subtracted from the signal. For PDL1 gD-GPI EV binding, wells were flicked dry between the Coelantrazine h and Amplex Red Cholesterol Assay.
Supplementary Material
Acknowledgments
We thank Genentech reviewers for critically reading the manuscript. We are deeply grateful for Jian Payandeh, J. T. Koerber, and Yonglian Sun for vesicle discussions as well as the gag-NeonGreen construct. We also thank Lovejit Singh, Zhong Rong Li, Isabelle Lehoux, Yvonne Franke, and Kristen Nailor for help with library construction and management of DNA reagents; Allison Gablin and Peter Harms for protein expression; Blair Wilson for expression analysis; and Andy Chang and Hei-Wai Lou for automation support. The results shown here are in part based upon data generated by the TCGA Research Network: https://www.cancer.gov/tcga.
Footnotes
Competing interest statement: All authors except C.M.A. are Genentech employees and own shares in the Roche/Genentech group.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2025451118/-/DCSupplemental.
Data Availability
All study data are included in the article and/or supporting information.
References
- 1.Santos R., et al., A comprehensive map of molecular drug targets. Nat. Rev. Drug Discov. 16, 19–34 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cao S., Martinez-Martin N., Unbiased Identification of Extracellular Protein – Protein Interactions for Drug Target and Biologic Drug Discovery (Intech Open, 2021). [Google Scholar]
- 3.Martinez-Martin N., Technologies for proteome-wide discovery of extracellular host-pathogen interactions. J. Immunol. Res. 2017, 2197615 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wright G. J., Signal initiation in biological systems: The properties and detection of transient extracellular protein interactions. Mol. Biosyst. 5, 1405–1412 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wright G. J., Martin S., Bushell K. M., Söllner C., High-throughput identification of transient extracellular protein interactions. Biochem. Soc. Trans. 38, 919–922 (2010). [DOI] [PubMed] [Google Scholar]
- 6.Huttlin E. L., et al., Dual proteome-scale networks reveal cell-specific remodeling of the human interactome. Cell 184, 3022–3040.e28 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bushell K. M., Söllner C., Schuster-Boeckler B., Bateman A., Wright G. J., Large-scale screening for novel low-affinity extracellular protein interactions. Genome Res. 18, 622–630 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Husain B., et al., A platform for extracellular interactome discovery identifies novel functional binding partners for the immune receptors B7-H3/CD276 and PVR/CD155. Mol. Cell. Proteomics 18, 2310–2323 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goñi F. M., The basic structure and dynamics of cell membranes: An update of the Singer-Nicolson model. Biochim. Biophys. Acta 1838, 1467–1476 (2014). [DOI] [PubMed] [Google Scholar]
- 10.Banjade S., Rosen M. K., Phase transitions of multivalent proteins can promote clustering of membrane receptors. eLife 3, 1–24 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Taylor M. J., Husain K., Gartner Z. J., Mayor S., Vale R. D., A DNA-based T cell receptor reveals a role for receptor clustering in ligand discrimination. Cell 169, 108–119.e20 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hu J., Lipowsky R., Weikl T. R., Binding constants of membrane-anchored receptors and ligands depend strongly on the nanoscale roughness of membranes. Proc. Natl. Acad. Sci. U.S.A. 110, 15283–15288 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Geri J. B., et al., Microenvironment mapping via Dexter energy transfer on immune cells. Science 367, 1091–1097 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li J., et al., Cell-surface proteomic profiling in the fly brain uncovers wiring regulators. Cell 180, 373–386.e15 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gingras A. C., Abe K. T., Raught B., Getting to know the neighborhood: Using proximity-dependent biotinylation to characterize protein complexes and map organelles. Curr. Opin. Chem. Biol. 48, 44–54 (2019). [DOI] [PubMed] [Google Scholar]
- 16.Frei A. P., Moest H., Novy K., Wollscheid B., Ligand-based receptor identification on living cells and tissues using TRICEPS. Nat. Protoc. 8, 1321–1336 (2013). [DOI] [PubMed] [Google Scholar]
- 17.Rouck J. E., Krapf J. E., Roy J., Huff H. C., Das A., Recent advances in nanodisc technology for membrane protein studies (2012-2017). FEBS Lett. 591, 2057–2088 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.De Franceschi N., et al., ProLIF – Quantitative integrin protein-protein interactions and synergistic membrane effects on proteoliposomes. J. Cell Sci. 132, jcs214270 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bausch-Fluck D., et al., The in silico human surfaceome. Proc. Natl. Acad. Sci. U.S.A. 115, E10988–E10997 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Czajkowsky D. M., Hu J., Shao Z., Pleass R. J., Fc-fusion proteins: New developments and future perspectives. EMBO Mol. Med. 4, 1015–1028 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martinez-Martin N., et al., An unbiased screen for human cytomegalovirus identifies neuropilin-2 as a central viral receptor. Cell 174, 1158–1171.e19 (2018). [DOI] [PubMed] [Google Scholar]
- 22.Dominguez C. X., et al., Single-cell RNA sequencing reveals stromal evolution into LRRC15+ myofibroblasts as a determinant of patient response to cancer immunotherapy. Cancer Discov. 10, 232–253 (2020). [DOI] [PubMed] [Google Scholar]
- 23.Keerthikumar S., et al., ExoCarta: A web-based compendium of exosomal cargo. J. Mol. Biol. 428, 688–692 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heath N., et al., Rapid isolation and enrichment of extracellular vesicle preparations using anion exchange chromatography. Sci. Rep. 8, 5730 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Arena T. A., Chou B., Harms P. D., Wong A. W., An anti-apoptotic HEK293 cell line provides a robust and high titer platform for transient protein expression in bioreactors. MAbs 11, 977–986 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Geeurickx E., et al., The generation and use of recombinant extracellular vesicles as biological reference material. Nat. Commun. 10, 3288 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cervera L., et al., Generation of HIV-1 Gag VLPs by transient transfection of HEK 293 suspension cell cultures using an optimized animal-derived component free medium. J. Biotechnol. 166, 152–165 (2013). [DOI] [PubMed] [Google Scholar]
- 28.Cameron C., et al., Octet potency assay: Development, qualification and validation strategies. https://www.fortebio.com/sites/default/files/en/assets/app-note/octet-potency-assay-development-qualification-and-validation-strategies.pdf. Accessed 9 September 2021.
- 29.Verschueren E., et al., The immunoglobulin superfamily receptome defines cancer-relevant networks associated with clinical outcome. Cell 182, 329–344.e19 (2020). [DOI] [PubMed] [Google Scholar]
- 30.Lee J. R., et al., Magneto-nanosensor platform for probing low-affinity protein-protein interactions and identification of a low-affinity PD-L1/PD-L2 interaction. Nat. Commun. 7, 12220 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Szklarczyk D., et al., STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 47, D607–D613 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rouleau C., et al., Endosialin protein expression and therapeutic target potential in human solid tumors: Sarcoma versus carcinoma. Clin. Cancer Res. 14, 7223–7236 (2008). [DOI] [PubMed] [Google Scholar]
- 33.Rouleau C., et al., Endosialin is expressed in high grade and advanced sarcomas: Evidence from clinical specimens and preclinical modeling. Int. J. Oncol. 39, 73–89 (2011). [DOI] [PubMed] [Google Scholar]
- 34.Teicher B. A., CD248: A therapeutic target in cancer and fibrotic diseases. Oncotarget 10, 993–1009 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maia M., et al., CD248 facilitates tumor growth via its cytoplasmic domain. BMC Cancer 2, 162 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Petro K. A., Dyer M. A., Yowler B. C., Schengrund C. L., Disruption of lipid rafts enhances activity of botulinum neurotoxin serotype A. Toxicon 48, 1035–1045 (2006). [DOI] [PubMed] [Google Scholar]
- 37.Purcell J. W., et al., LRRC15 is a novel mesenchymal protein and stromal target for antibody–drug conjugates. Cancer Res. 78, 4059–4072 (2018). [DOI] [PubMed] [Google Scholar]
- 38.Puram S. V., et al., Single-cell transcriptomic analysis of primary and metastatic tumor ecosystems in head and neck cancer. Cell 171, 1611–1624.e24 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schorey J. S., Bhatnagar S., Exosome function: From tumor immunology to pathogen biology. Traffic 9, 871–881 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thul P. J., et al., A subcellular map of the human proteome. Science 356, eaal3321 (2017). [DOI] [PubMed] [Google Scholar]
- 41.Colombo M., Raposo G., Théry C., Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu. Rev. Cell Dev. Biol. 30, 255–289 (2014). [DOI] [PubMed] [Google Scholar]
- 42.Hu S., et al., VirD: A virion display array for profiling functional membrane proteins. Anal. Chem. 85, 8046–8054 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Syu G. D., et al., Development and application of a high-content virion display human GPCR array. Nat. Commun. 10, 1997 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tucker D. F., et al., Isolation of state-dependent monoclonal antibodies against the 12-transmembrane domain glucose transporter 4 using virus-like particles. Proc. Natl. Acad. Sci. U.S.A. 115, E4990–E4999 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Willis S., et al., Virus-like particles as quantitative probes of membrane protein interactions. Biochemistry 47, 6988–6990 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lavado-García J., et al., Molecular characterization of the coproduced extracellular vesicles in HEK293 during virus-like particle production. J. Proteome Res. 19, 4516–4532 (2020). [DOI] [PubMed] [Google Scholar]
- 47.Vader P., Mol E. A., Pasterkamp G., Schiffelers R. M., Extracellular vesicles for drug delivery. Adv. Drug Deliv. Rev. 106, 148–156 (2016). [DOI] [PubMed] [Google Scholar]
- 48.Mi H., et al., PANTHER version 16: A revised family classification, tree-based classification tool, enhancer regions and extensive API. Nucleic Acids Res. 49, D394–D403 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tang Z., et al., GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 45, W98–W102 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gao J., et al., Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal complementary data sources and analysis options. Sci. Signal. 6, 1–20 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or supporting information.