Significance
Vaccines profit from the addition of adjuvants to better and more specifically initiate, amplify, and shape immune responses. Although the number of adjuvant candidates has steadily increased, peaking in the current SARS-CoV-2 pandemic, little is known about their inherent mode of action. Using human blood immune cells, we established a multilayer method to systematically assess the adjuvants’ effects on innate and adaptive immune cells. By employing a multiplex analysis with cells from 30 different donors, we determined important patterns of adjuvant function. Moreover, we demonstrate correlates of an antiviral immune response using a Toll-like receptor 7/8 ligand adjuvant and single-stranded RNA. This knowledge about adjuvants’ distinct immune signatures supports the selection of safe and effective adjuvants for future vaccines.
Keywords: adjuvants, mRNA vaccines, TLR, primary human cells
Abstract
The COVID-19 pandemic highlights the importance of efficient and safe vaccine development. Vaccine adjuvants are essential to boost and tailor the immune response to the corresponding pathogen. To allow for an educated selection, we assessed the effect of different adjuvants on human monocyte-derived dendritic cells (DCs) and their ability to polarize innate and adaptive immune responses. In contrast to commonly used adjuvants, such as aluminum hydroxide, Toll-like receptor (TLR) agonists induced robust phenotypic and functional DC maturation. In a DC-lymphocyte coculture system, we investigated the ensuing immune reactions. While monophosphoryl lipid A synthetic, a TLR4 ligand, induced checkpoint inhibitors indicative for immune exhaustion, the TLR7/8 agonist Resiquimod (R848) induced prominent type-1 interferon and interleukin 6 responses and robust CTL, B-cell, and NK-cell proliferation, which is particularly suited for antiviral immune responses. The recently licensed COVID-19 vaccines, BNT162b and mRNA-1273, are both based on single-stranded RNA. Indeed, we could confirm that the cytokine profile induced by lipid-complexed RNA was almost identical to the pattern induced by R848. Although this awaits further investigation, our results suggest that their efficacy involves the highly efficient antiviral response pattern stimulated by the RNAs’ TLR7/8 activation.
The current COVID-19 pandemic has unprecedently spurred vaccine development and has so far resulted in over 287 new vaccine candidates under evaluation (1–3).
In contrast to the vaccine formulations with inactivated or live-attenuated whole pathogens, the use of only purified antigens of the pathogen in today’s subunit vaccines is well tolerated and deemed to be safe. These antigens provide high specificity, but their low intrinsic immunogenicity requires the combined application with an adjuvant to activate the immune system (4). Adjuvants are not unified by structure, target, or mechanism of action (MoA), but they all shape, direct, and potentiate the immune response. The selection of an appropriate adjuvant is essential for vaccine efficacy (5).
Aluminum hydroxide [Al(OH)3] and MF59 belong to the first generation of adjuvants, but even though Al(OH)3 has a good safety profile, its MoA is still not fully understood (5, 6). Its limitation of stimulating a strong humoral response (7) has driven the development of second-generation adjuvants such as pattern recognition receptor (PRR) ligands or plant-derived compounds (e.g., saponins) to induce specific cell-mediated immune responses. Furthermore, the combination of different adjuvants like Alum and monophosphoryl lipid A (MPL) in the adjuvant system 04 (AS04) allowed for a reduction in antigen dose, faster production of antigen-specific antibodies, and longer duration of protective antibody titers compared to vaccines adjuvanted with aluminum salt alone (8).
In the case of AS04, efficiently activated dendritic cells (DCs) stimulate polyfunctional antigen-specific CD4+ T cells. This effect was primarily dependent on MPL, while Alum was shown to prolong the MPL-induced cytokine response at the injection site, which is required for recruitment of antigen-presenting cells (9–11).
Such designed adjuvant systems, containing different specific immune-modulating components, will be essential in current and future vaccine development. The risk for adverse effects, as observed upon Pandemrix vaccination, adjuvanted with an AS03 system (12, 13), needs to be as low as possible. In addition, adverse effects, which are potentially induced by adjuvants in healthy people, diminish the acceptance of vaccination within the population (14). Therefore, we need to increase our understanding of every distinct effect of single immunostimulatory components in adjuvants systems.
Supported by the Ministry of Health, our research group at the Federal Institute for Vaccines and Biomedicines set up an in-depth immunological analysis of potential immune-modulating components. Herein, we performed a side-by-side comparison of various adjuvants using a human primary immune cell-based in vitro assay composed of monocyte-derived DCs (moDCs) and autologous peripheral blood lymphocytes (PBLs). DCs are the main target of adjuvants and react by secretion of proinflammatory cytokines, increased uptake, and MHC presentation of antigens as well as an enhanced expression of the costimulatory proteins.
In our study, we aim to ascribe specific innate and adaptive immunogenic MoA to each adjuvant to facilitate the predictability of its potential to shape cellular immune responses and thus support the rational design of adjuvant-based vaccines. The adjuvant panel we investigated comprises second-generation and potential candidate adjuvants such as surface PRR ligands TDB (Mincle), Pam3CSK4 (Pam; TLR1/2), and MPL (TLR4); the endosomal PRR ligands Gardiquimod (GARD; TLR7), Imiquimod (IMQ; TLR7), and Resiquimod (R848; TLR7/8); and the saponin Quil A (Quil). In addition, first-generation adjuvants such as Al(OH)3 and an oil-in-water emulsion AddaVax (ADX) are included. In light of the current success of mRNA-based vaccines against SARS-CoV-2, we hypothesize that their excellent antiviral immune protection does not only rely on the produced antigen but also on TLR7/8-dependent adjuvanticity, and we added lipid-complexed RNA to investigate this accordingly.
In this study, several components were identified that induce full DC maturation, resulting in phenotypical and functional capacitation of DCs. In contrast to other single-component adjuvants, the TLR7 agonists GARD and IMQ only induced full DC maturation in the presence of PBLs. The tested adjuvants induced a highly specific response pattern, even when binding to similar or identical recognition receptors. It is the virtue of the approach taken in the current study to reveal the possibilities lying in the use of different agonists targeting the same receptor. Different components were additionally identified to have an antigen-dependent adjuvant effect on cells of the adaptive immune system. This effect was specific for the adjuvants Pam, MPL synthetic (MPL-s), and Al(OH)3, as they expanded FluM1-specific CD8+ T-cell populations in almost all of the tested donors. Using lipid-complexed single-stranded RNA as a surrogate for one of the recent mRNA vaccines, we could confirm that the cytokine profile induced by lipid-complexed RNA was almost identical to the pattern induced by synthetic TLR7/8 agonist R848. This supports the notion that the excellent antiviral immune protection that is observed with the innovative anti-SARS-CoV-2 RNA vaccines is in part due to the strong adjuvant activity of the single-stranded RNA component via TLR7/8.
Thus, clearly showing that even similar adjuvants induce different immunogenic profiles, our data will prove to be important for vaccine developers to tailor and modulate the envisaged immune response as well as for drug regulators to support their understanding of adjuvanticity in the process of authorization of both safe and effective new vaccines.
Results
Definition of Suitable Working Concentrations.
In a first step, a suitable working concentration for each of the 10 adjuvants was determined, as it was our particular concern to exclude any cytotoxic effects (SI Appendix, Table S1 and Fig. S1). Therefore, we analyzed the cytotoxicity of the different compounds by treating PBLs over 7 d. Concentrations that induced an increase of cell death (Annexin-V+/PI+ cells) to more than 25% compared to the unstimulated control were defined as unsuitable for this study. For adjuvants that induced lymphocyte proliferation, the proliferation-inducing capacity was also considered (SI Appendix, Fig. S1A). For all adjuvants, two concentrations were chosen: a lower concentration that barely showed any effect and a higher concentration that was well in the range of inducing cell proliferation but did not show cytotoxicity above the critical level, as described previously. Throughout the manuscript, the two conditions are termed “low” and “high” (SI Appendix, Table S1). We further evaluated the adjuvants for their endotoxin content by limulus amoebocyte lysate (LAL) assay (SI Appendix, Fig. S1B). The tests demonstrated that all adjuvants were negative for endotoxin, except for lipid A–containing adjuvants such as MPL-s and MPL-Salmonella minnesota R595 (MPL-SM) as well as LPS, as expected. Next, we investigated the pyrogenicity of the chosen adjuvant concentrations using the monocyte activation test (SI Appendix, Fig. S1C). We observed that stimulation even with high concentrations of TDB, Pam, GARD, IMQ, R848, and Al(OH)3 did not lead to interleukin (IL)-1β levels above the fever-inducing threshold (0.5 EU/mL of LPS) (15). By contrast, for MPL-s, MPL-SM, and LPS, only the lower working concentration resulted in IL-1β secretion below this threshold.
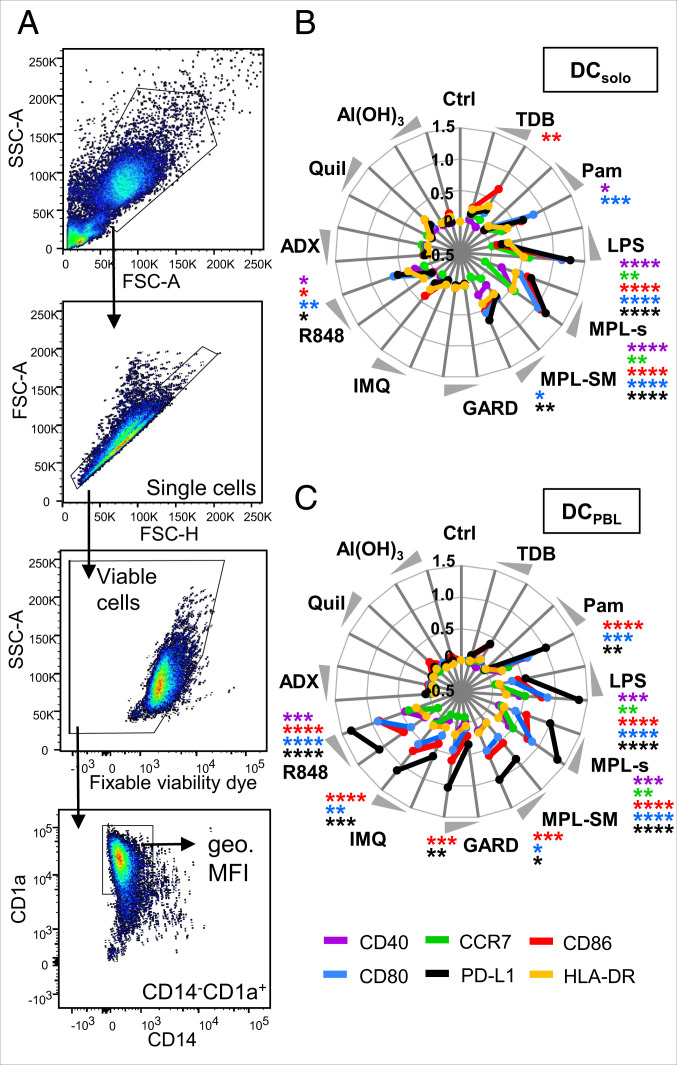
TLR Ligand Adjuvants Increase the Expression of Maturation Markers on DCs.
To assess the capacity of the adjuvants to induce DC maturation, we analyzed the expression of the maturation markers CD80, CD86, CD40, PD-L1, CCR7, and HLA-DR on the cell surface of moDCs after 24 h of stimulation with the respective adjuvant. The geometric mean fluorescence intensity of the different markers was assessed on viable CD14-CD1a+ DCs (Fig. 1A) when cultured either alone (DCsolo) or in the coculture with PBLs (DCPBL). For DCsolo, we observed a significant increased and concentration-dependent expression of two or more maturation markers upon stimulation with Pam, LPS, MPL-s, or R848 compared to the unstimulated control (Fig. 1B). The expression of the maturation markers on DCPBL was even more pronounced after Pam or R848 treatment (Fig. 1C). This synergistic effect of bystanding lymphocytes was not observed with the TLR4 ligands MPL-s, MPL-SM, and LPS. Except for PD-L1, levels of all maturation markers were either equal or lower. For the TLR7 ligands GARD and IMQ, no DC maturation was observed in the absence of PBLs, whereas high levels of CD80, CD86, and PD-L1 were induced under coculturing conditions (Fig. 1C and SI Appendix, Fig. S2). The indirect induction of these maturation markers seems to be in accordance with the fact that moDCs alone do not or very lowly express TLR7 (16). The observed elevated levels of CD40, CD80, CD86, and PD-L1 on DCsolo upon stimulation with the TLR7/8 ligand R848 suggests that these effects are TLR8-specific. Investigating receptor expression on protein level by flow cytometry or on mRNA level by PCR (SI Appendix, Table S3), we indeed detected less TLR7 in DCs compared to lymphocytes. Moreover, we found TLR8 to be expressed on DCs but not by PBLs (SI Appendix, Fig. S3 A–C). The adjuvants that do not interact with TLRs, like ADX, Quil, and Al(OH)3, induce no signs of DC maturation regardless of whether PBLs were present or not (Fig. 1 B and C).
Fig. 1.
TLR ligand adjuvants increase the expression of maturation markers on DCs. DCs or DC:PBL cocultures were stimulated for 24 h with the different adjuvants or left untreated. The expression of the maturation markers CD40, CCR7, CD86, CD80, PD-L1, and HLA-DR was assessed by flow cytometry. (A) Gating strategy to identify CD14−CD1a+ DCs. The geometric mean fluorescence intensity (geo. MFI) of single viable cells was analyzed. Respective isotype antibodies were used to determine background staining. (B and C) Radar plots showing the adjuvant-induced expression of maturation markers on DCsolo or DCPBL, respectively. Obtained geo. MFI values are displayed as fold change (compared to the respective unstimulated control) and transformed to log scale. Each maturation marker is represented as mean (n = 9 to 15 donors) in a colored line. Significant differences between the adjuvant-induced expression and the unstimulated control were analyzed using the Kruskal-Wallis test with Dunn’s correction for multiple comparisons on the nonnormalized data (*P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001). Statistics are depicted only for the high concentration of the adjuvant.
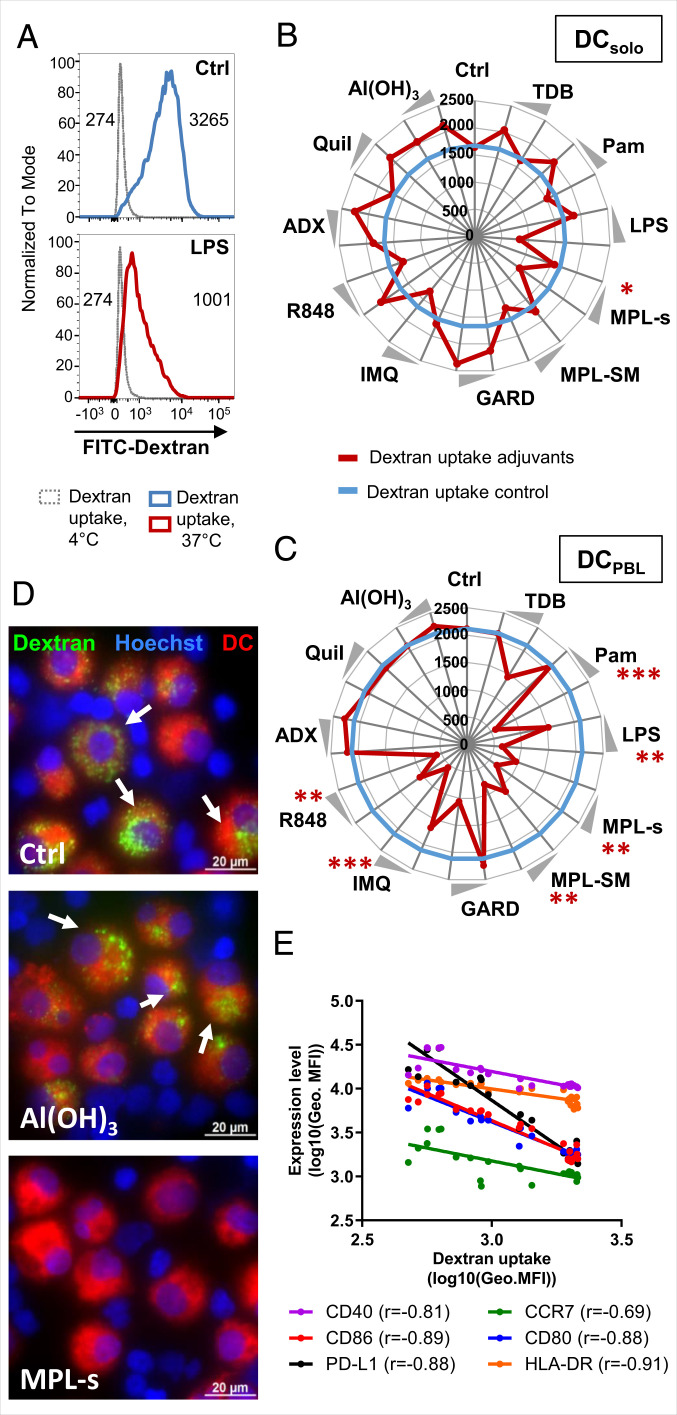
In Adjuvant-Stimulated DCs, Endocytic Activity Negatively Correlates with the Expression of Maturation Markers.
The down-regulation of endocytic activity is a hallmark of DC maturation. Hence, we investigated the functional maturation of the DCs by analyzing the endocytic uptake of fluorescein isothiocyanate (FITC)-labeled dextran using flow cytometry (Fig. 2A). For DCs alone, only LPS led to a significant decrease in the endocytic uptake of FITC-dextran compared to the unstimulated control (Fig. 2 A and B). With all other adjuvants, no effect on isolated DCs was observed, resulting in identical or even increased FITC-dextran uptake (Fig. 2B). By contrast, when DCs were coincubated with PBLs in the presence of Pam, LPS, MPL-s, MPL-SM, IMQ, or R848, the endocytic uptake of FITC-dextran was significantly reduced compared to the untreated coculture (Fig. 2 C and D). The physiochemical adjuvants like ADX, Quil A, and Al(OH)3 did not induce DC maturation and thus had no effect on the endocytic uptake of FITC-dextran regardless of whether PBLs were present or not (Fig. 2 C and D). Since we observed a clear dependency between the expression levels of maturation markers and the lack of endocytic activity of adjuvant-stimulated DCs (Fig. 2E), we calculated Spearman correlation coefficients. Indeed, HLA-DR, CD86, CD80, CD40, and PD-L1 showed a strong negative correlation, with values ranging from −0.81 to −0.91. These experiments show that the adjuvants Pam, GARD, IMQ, R848, MPL-s, and MPL-SM induce the development to fully mature DCs, which are phenotypically (expression of costimulatory and maturation markers) and functionally (low endocytic uptake capacity) different to immature DCs.
Fig. 2.
In adjuvant-stimulated DCs, endocytic activity negatively correlates with the expression of maturation markers. DCsolo or DCPBL cultures were stimulated with adjuvants for 24 h. Subsequently, 50 µg/mL FITC-dextran was added to the culture for another hour. After thorough washing, uptake of FITC-dextran in CD14−CD1a+DCs was measured as geometric mean fluorescence intensity (geo. MFI) in the unstimulated control or when stimulated with LPS. (A) Nonspecific binding of FITC-dextran at 4 °C served as control (dotted gray line). DC’s endocytic capacity of FITC-dextran was analyzed at 37 °C. Values within the histograms indicate the geo. MFI of the respective condition at 4 °C and 37 °C. (B and C) Radar plots showing the endocytic uptake of FITC-dextran in DCsolo or DCPBL, respectively. The blue solid line indicates the geo. MFI of FITC in the unstimulated controls. Adjuvant-induced changes of the endocytic capacity (geo. MFI of FITC) are shown in red. Both lines are represented as mean (n = 12) from at least three independent experiments. Statistical comparisons of the unstimulated control and the adjuvants were performed using the Kruskal-Wallis test with Dunn’s correction (*P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001). (D) Representative fluorescent microscopic pictures are shown for the unstimulated, Al(OH)3-stimulated, and MPL-s–stimulated DC:PBL cocultures (green: FITC-dextran, blue: Hoechst, red: DCs; scale bar: 20 µM). (E) Expression of the different maturation marker (geo. MFI) on DCPBL was correlated with the uptake of FITC-dextran (geo. MFI) using Spearman’s nonparametric rank correlation.
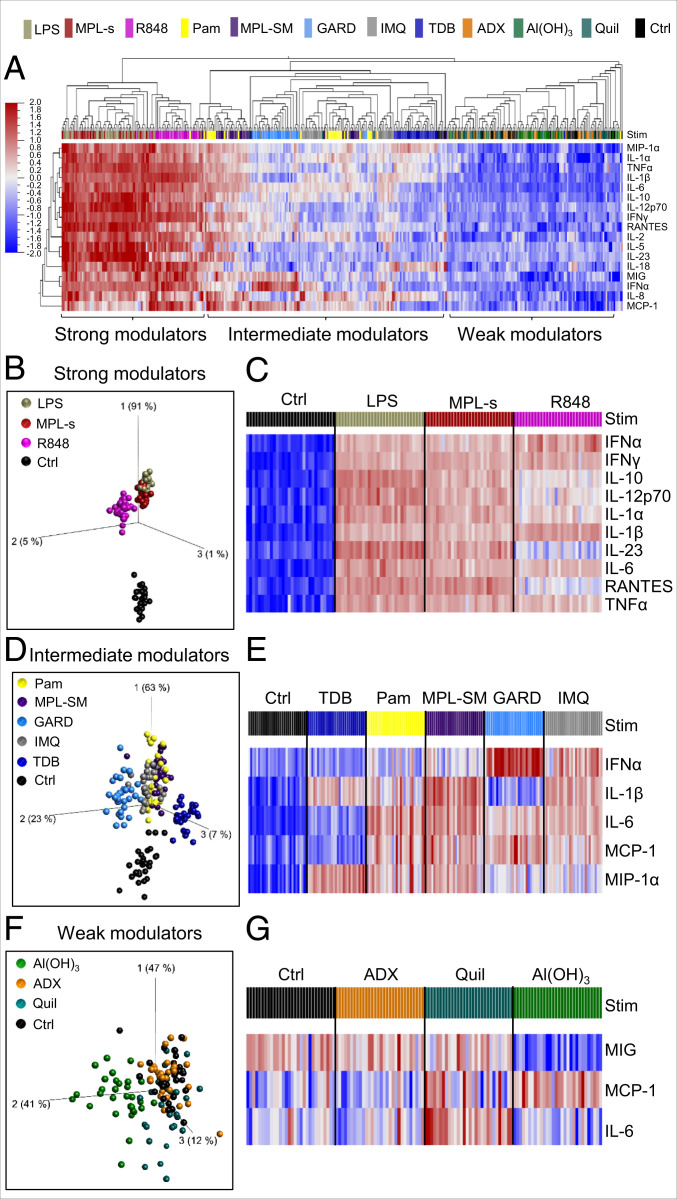
Adjuvants Separate into Strong, Intermediate, and Weak Immunomodulators Based on Their Induced Cytokine and Chemokine Expression Patterns.
To investigate the adjuvant-induced cytokine and chemokine response, we deliberately focused on the DC:PBL coculture system because it better reproduces the complex interplay between the immunomodulators and the cells of the innate and adaptive immune system. Hence, we collected culture supernatant of adjuvant-stimulated DC:PBL cultures from 30 healthy donors that were evenly distributed with respect to age and sex (15 male/female; 7 to 8 >40/<40 y of age). Using the Luminex xMAP technology, we performed multianalyte protein profiling and analyzed 25 cytokines and chemokines in total. The overall adjuvant-induced signature of four representative immunomodulators [LPS, Pam, Al(OH)3, and GARD] is exemplarily plotted in SI Appendix, Fig. S4. When stimulated with the positive control LPS, we observed a broad range of cytokine expression levels spanning up to 1,000-fold compared to the null response (e.g., IL-12p70, IL-6).
To better illustrate the individual characteristics of and the qualitative differences between the tested immunomodulators, we performed principal component analysis (PCA) using Qlucore Omics explorer 3.5 (17). The PCA revealed stimuli-specific clusters, with the first three PC vectors covering 76% of the total variance (SI Appendix, Fig. S5A). IL-10, IFNγ, TNFα, IL-6, and IL-12p70 predominantly contributed to the first vector. This axis alone accounted for 64% of the total variance, indicating that the adjuvants mainly segregate by the cytokines contributing to this axis (SI Appendix, Fig. S5 A and B). There was no evidence for sex- or age-specific responses (SI Appendix, Fig. S5C). To characterize the patterns of protein analytes induced by the different adjuvants, we performed hierarchical clustering with the focus on the 17 most differentially expressed proteins (Fig. 3A). This approach separated the adjuvants into three classes: strong, intermediate, and weak immunomodulators. The group of strong immunomodulators was defined by the high and simultaneous induction of all 17 cytokines. It comprised MPL-s, R848, and the positive control LPS. By performing PCA with this subset of strong immunomodulators, we observed that, expectedly, the TLR4-ligands MPL-s and LPS cluster together, indicating induction of a similar cytokine and chemokine response (Fig. 3B). Except for slightly lower levels of IL-10, IL-12p70, and IL-23 after MPL-s treatment, all other protein levels were highly comparable between the two adjuvants (Fig. 3C). R848 separated due to a lower expression of IL-23 and RANTES as well as higher protein levels of IFNα and IL-1β compared to LPS and MPL-s (Fig. 3C and SI Appendix, Fig. S6A). The group of intermediate immunomodulators is comprised of Pam, MPL-SM, GARD, IMQ, and TDB. GARD segregated from the other plotted adjuvants along PC vector 2 (Fig. 3D) and was characterized by a distinct high expression of IFNα and low levels of IL-1β (Fig. 3E and SI Appendix, Fig. S6B). TDB displayed low expression of IFNα, IL-6, and MCP-1 (Fig. 3D); however, within the group of intermediate immunomodulators, this adjuvant induced the highest expression levels of MIP-1α (Fig. 3E and SI Appendix, Fig. S6B). Among the weak immunomodulators, MIG, MCP-1, and IL-6 were the three cytokines that were most differentially expressed (SI Appendix, Fig. S6C). By PCA, we could identify a specific Al(OH)3 cluster that was characterized by lower levels of MIG and IL-6 along PC axis 2 and the highest induction of MCP-1 in this group (Fig. 3 F and G). By contrast, cytokine and chemokine expression profiles of ADX- or Quil A-treated cultures were statistically indistinguishable from the unstimulated control (Fig. 3 F and G).
Fig. 3.
Adjuvants separate into strong, intermediate, and weak immunomodulators based on their induced cytokine and chemokine expression patterns. Twenty-four hours after stimulating DC:PBL cocultures with the different adjuvants, supernatants were harvested and analyzed for cytokine and chemokine secretion (25 proteins in total) using the Luminex xMAP technology. (A) The dendogram, with each rectangle representing 1 of 30 tested donors stimulated with the high concentration of a specific adjuvant (color code on top of the heat map) shows the hierarchical clustering of the protein expression data. Red indicates high expression; blue indicates low expression. The analysis is based on the 17 most differentially induced proteins (cutoff value was determined by ANOVA, q-value < 1 × 10−50). Detailed analysis of cytokine and chemokine expression is shown for (B and C) strong modulators, (D and E) intermediate modulators and (F and G) weak modulators. (B, D, and F) PCA performed on the same data set but restricted to the strong, intermediate, or weak immunomodulators, respectively. Each dot represents a donor stimulated with the indicated adjuvant. The unstimulated control serves as base line. The cutoff q-value was defined by ANOVA and included the 10 (q < 1 × 10−25; B), 5 (q < 1 × 10−30; D), or 3 (q > 1 × 10−4; F) most differentially induced proteins. (C, E, and G) Heat map of the 10 (q-value < 1 × 10−58), 5 (q-value < 1 × 10−30), or 3 (q-value < 1 × 10−4) most differentially induced proteins within the respective immunomodulatory group.
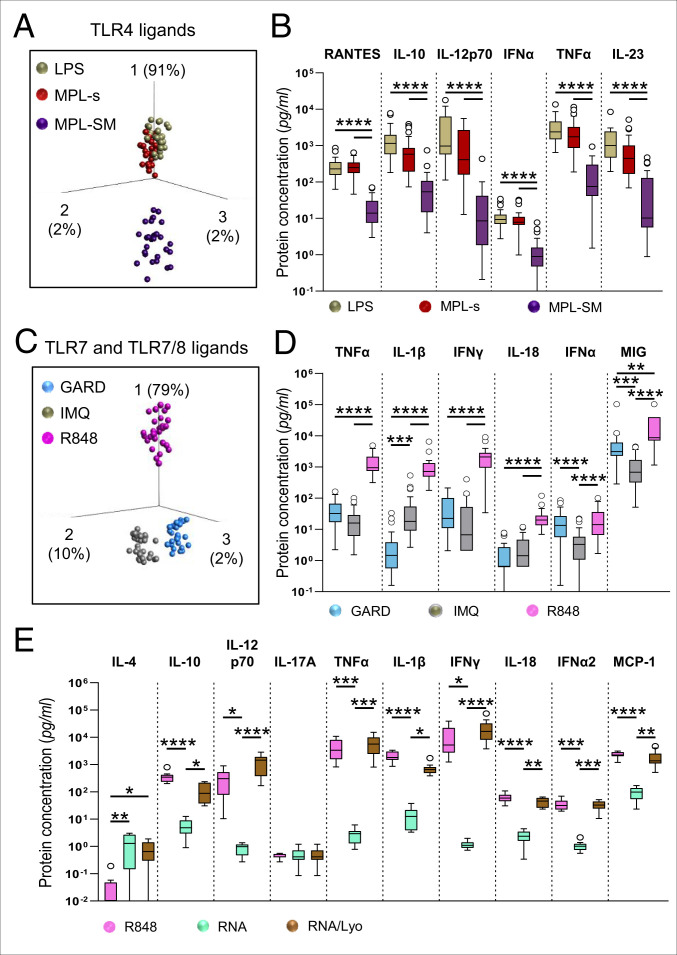
Despite Structure and Receptor Similarity, Individual TLR4 as well as Individual TLR7 or TLR7/8 Ligands Induce Distinct Cytokine and Chemokine Patterns.
To further dissect subtle differences between different immunomodulators targeting the same receptor, we compared the TLR4 ligands LPS, MPL-s, and MPL-SM by employing PCA (Fig. 4A). We selected the six most significantly regulated proteins with a q-value < 10 to 15, which allowed us to capture 95% of the measured variance in response to TLR4 stimulation. The TLR4 ligands strongly allocated along PCA vector 1 (91% of the total variance), indicating that most of the variance is due to the top listed proinflammatory cytokines of PCA vector 1, which are TNFα, IL-12p70, RANTES, IL-23, IL-10, and IFNα (SI Appendix, Fig. S7A). By applying an ANOVA test, these proteins were identified to be the most differentially induced proteins between LPS, MPL-s, and MPL-SM (Fig. 4B). The PCA plot and the absolute protein concentrations further demonstrated that LPS and MPL-s overlap in protein induction and concentration with no significant differences in protein levels (Fig. 4 A and B). However, overall MPL-s showed a trend of lower protein levels compared to LPS. In contrast to MPL-s and LPS, MPL-SM exhibited significantly lower protein levels for all six proteins shown here, leading to the clear separation in the PCA.
Fig. 4.
Despite structure and receptor similarity, individual TLR4 as well as individual TLR7 or TLR7/8 ligands induce distinct cytokine and chemokine patterns. Luminex data obtained from 30 donors was analyzed regarding differences within (A and B) TLR4- or (C and D) TLR7- and TLR7/8- stimulating adjuvants. (A and C) PCA on the TLR4 or TLR7 and TLR7/8 ligands including 12 or 14 of the 25 cytokines and chemokines, respectively (ANOVA, cutoff value q > 1 × 10−34 or q < 1 × 10−15, respectively). (B and D) The six most differentially induced proteins of the TLR4 or TLR7 and TLR7/8 ligands as defined by ANOVA, depicted as nontransformed and noncorrected data. (E) Analyte concentrations obtained by Legendplex for DC:PBL cocultures stimulated with either the high concentration of the TLR7/8 agonist R848 or the synthetic single-strand RNA preparations poly(U) (RNA) or poly(U)/Lyovec (RNA/Lyo) for 24 h. Statistical comparisons in B, D, and E were performed using the Kruskal-Wallis test with Dunn’s correction (*P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001). If not otherwise specified, comparison revealed no significance.
Interestingly, the two TLR7 ligands GARD and IMQ as well as the TLR7/8 ligand R848 could readily be segregated by PCA (Fig. 4C), with PCA vectors displaying 91% of the total variance. Although PCA vector 1 dominated with 79% of the total variance, the separation of GARD and IMQ was driven by PCA vector 2, corresponding to the differentially induced proteins IFNα, MIG, and IL-1β (Fig. 4 C and D and SI Appendix, Fig. S7B). In comparison to IMQ, IFNα and MIG protein levels were significantly elevated after GARD stimulation, whereas IL-1β expression was reduced. In comparison to GARD and IMQ, R848 showed overall significantly higher levels of TNFα, IL-1β, IFNγ, IL-18, and MIG. For IFNα, no significant difference in protein concentration between GARD and R848 was analyzed by Kruskal-Wallis test. A cell type known to secrete very high amounts of IFNα is plasmacytoid DCs (pDCs), which made up 0.38% to 0.60% of the viable cells in our 24 h coculture but strongly declined in viability until day 6 of coculture (0.04% to 0.11% of viable cells; SI Appendix, Fig. S7C). In summary, the overall higher cytokine and chemokine induction, except for IFNα, separated the TLR7/8 ligand R848 strongly from both TLR7 ligands GARD and IMQ on PCA vector 1 (SI Appendix, Fig. S7D and Fig. 4C), whereas GARD and IMQ’s separation is the result of varying IFNα and IL-1β protein levels (Fig. 4D).
Given the fact that potent antiviral mRNA vaccines have been developed during the current SARS-CoV-2 pandemic and that these mRNAs can display adjuvant function through binding to TLR7/8, we employed single-stranded polyuridylic acid [poly(U)] either naked (indicated as RNA) or in complex with the cationic lipid LyoVec (indicated as RNA/Lyo) for treatment of our DC:PBL coculture and measured the concentrations of a subset of cytokines and with a separate set of donors by Legendplex. While the naked RNA did not reveal significant stimulation of cytokine expression, probably due to its strongly impaired uptake into the cells, RNA/Lyo increased the secretion of the proinflammatory mediators IL-1β and TNFα as well as the immune cell recruiting or activating factors MCP-1, IL-12 p70, IFNα2, and IFNγ to a similar extent as R848 (Fig. 4E).
Adjuvants Can Induce the Proliferation of Different Lymphocyte Populations in an Antigen-Independent Manner.
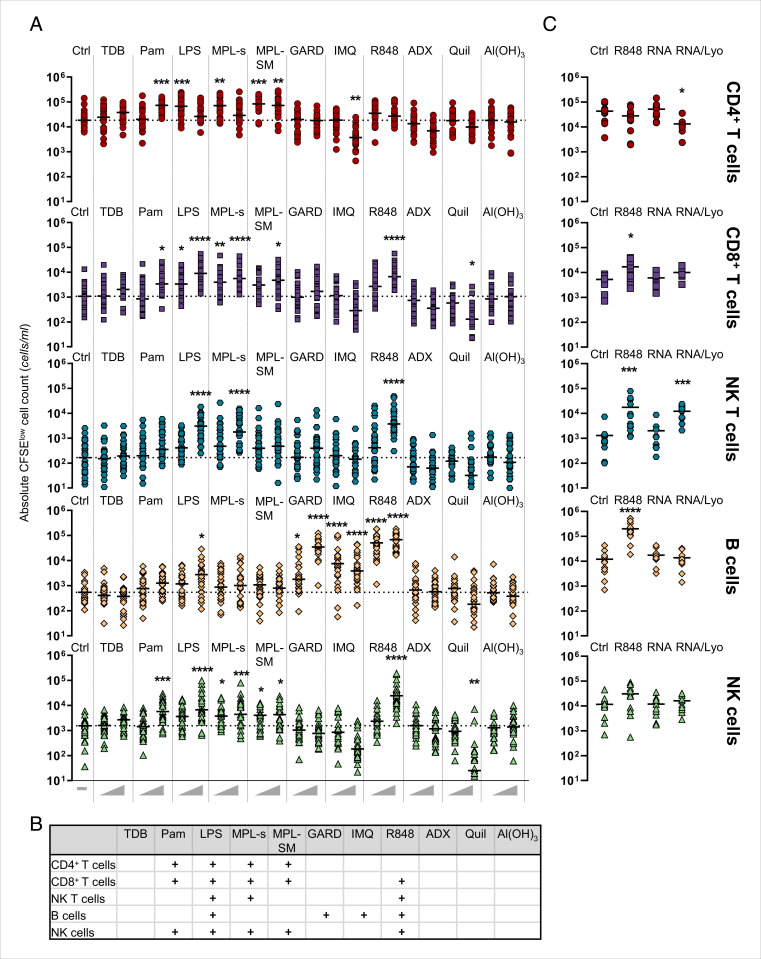
To assess the effect of adjuvant stimulation on the adaptive immune response, lymphocyte proliferation was investigated by carboxyfluorescein succinimidyl ester (CFSE) dilution after 6 d of coculture with DCs (SI Appendix, Fig. S8A). R848 was observed to induce the strongest proliferation of all adjuvants (mean ± SD: Ctrl: 7.4% ± 3.7%; LPS high: 22.6% ± 9% to 2%; R848 high: 36.5% ± 10.0%) (SI Appendix, Fig. S8B). In the case of TDB (high: 11.3% ± 5.9%), IMQ (high: 4.6% ± 4.1%), ADX (high: 3.3% ± 1.8%), Quil (high: 3.6% ± 2.2%), and Al(OH)3 (high: 5.5% ± 3.1%), no induction of lymphocyte proliferation compared to the unstimulated control was observed. In summary, with the exception of IMQ, all TLR ligands induced lymphocyte proliferation to different degrees. Using antibodies specific for CD3, CD4, CD8, CD56, and CD19, we could distinguish between CD4+, CD8+, and NKT as well as B and NK cell populations within the CFSE-low gate of proliferated lymphocytes (CFSElow cells). To address whether different immunomodulators induced adjuvant-specific proliferation profiles, we analyzed the absolute cell number for each lymphocyte population. Counting beads during sample acquisition were used to calculate cell counts (SI Appendix, Fig. S8A). A high concentration of the TLR2 ligand Pam led to the proliferation of both CD4+ and CD8+ T cells as well as NK cells (Fig. 5A). Within the group of TLR4 ligands, different proliferation profiles were found for the different adjuvants. LPS induced significant cell proliferation across all different lymphocyte populations. The proliferation pattern induced by MPL-s resembled that of LPS but showed little B-cell proliferation. Interestingly, we noted that high concentrations of LPS and MPL-s inhibited the proliferation of CD4+ T cells. This inhibition might be due to elevated levels of IL-10 that were secreted after 24 h in response to high concentrations of LPS or MPL-s (SI Appendix, Fig. S8C). By contrast, both concentrations of MPL-SM induced strong CD4+ T-cell proliferation. Accordingly, with both concentrations, comparably low IL-10 levels were observed. Also, in contrast to LPS and MPL-s, NKT cell levels were elevated upon MPL-SM stimulation but not significantly different to the unstimulated control using Kruskal-Wallis test (Fig. 5 A and B).
Fig. 5.
Adjuvants can induce the proliferation of different lymphocyte populations in an antigen-independent manner. (A) Immature dendritic cells (iDCs) were harvested on day 5 and cocultured with autologous PBLs at a 1:5 ratio. Both DCs and PBLs were labeled with CFSE prior to seeding. Subsequently, cells were stimulated with the different adjuvants or left untreated (Ctrl). After 6 d, PBLs were stained for CD3, CD4, CD8, CD19, and CD56, and proliferating cells were analyzed by flow cytometry. Counting beads were used during sample acquisition to allow for the calculation of absolute cell numbers. Aligned dot plot showing the individual values and median from 26 donors. Statistical significance obtained in A is summarized in B. + displays determined statistical significance regardless of the P value. (C) Experimental setup as in A but with data from 10 to 12 donors that were stimulated with the high concentration of R848 or the RNA stimuli (10 µg/mL). Statistical comparison of the unstimulated control and the adjuvants was performed using the Kruskal-Wallis test with Dunn’s correction (*P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001). If not otherwise specified, comparison revealed no significance.
All significant results are summarized in Fig. 5B. Likewise, the proliferation profiles varied between the TLR7 ligands GARD and IMQ and the TLR7/8 ligand R848. R848 stimulation led to the proliferation of CD8+ and NKT cells, B cells, and NK cells but not CD4+ T cells. Here, the induction of B, NK, and NKT cell proliferation was remarkable with respect to absolute cell counts as it exceeded even the numbers achieved with LPS stimulation. By contrast, GARD and IMQ only induced B-cell proliferation.
In this regard, we also tested the lymphocyte proliferation upon stimulation with the single-stranded RNA compounds (Fig. 5C). In line with the cytokine expression data obtained in Fig. 4E, stimulation of the coculture with the naked poly(U) RNA did not exceed the proliferation observed in the control condition. However, the LyoVec-complexed RNA modulated the proliferation of CD4+ and CD8+ T cells as well as NKT cells with the same potential as R848, whereas it did not stimulate the B-cell proliferation.
Notably, the high concentration of IMQ led to a decrease in lymphocyte proliferation (SI Appendix, Fig. S8B), which was probably caused by a significant drop in CD4+ T-cell count compared to the unstimulated control, which can be explained by TLR7-stimulated human T-cell anergy (18). A similar effect of reduced proliferation and a decrease in absolute cell counts of CD8+ and NKT cells was observed upon stimulation with Quil (SI Appendix, Fig. S8B and Fig. 5A). For TDB, ADX, and Al(OH)3, we did not observe any proliferative response of a distinct lymphocyte subset.
Adjuvants are mainly known to activate DCs, which in turn can stimulate the expansion of lymphocytes. Thus, it is not surprising that the proliferation of T and NK cells as well as the expression of early (CD69) or late (CD25) activation markers upon adjuvant treatment is much higher in a DC:PBL coculture than in a culture of only PBLs (SI Appendix, Fig. S9 A–C). In contrast, we obtained higher B-cell proliferation upon GARD or R848 treatment in the absence of DCs compared to the DC:PBL coculture (SI Appendix, Fig. S9A), suggesting a negatively regulating function of DCs in B-cell activation.
The Proliferation of Lymphocyte Subpopulations Can Be Linked to the Expression of Certain Cytokines and Chemokines.
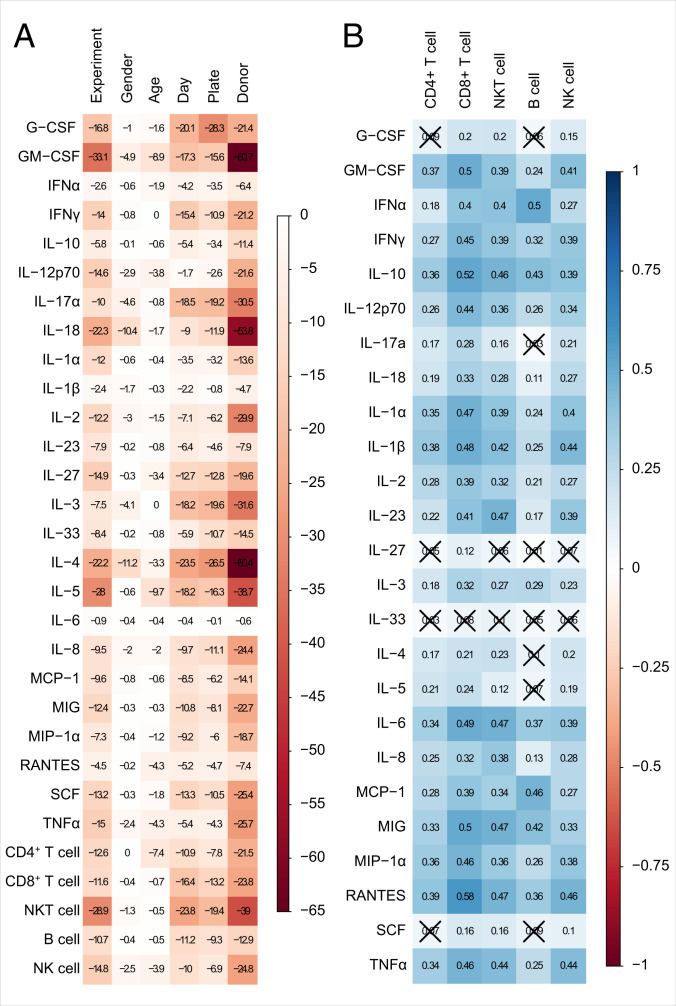
The use of the same donors for the analysis of the adjuvant-induced cytokine and chemokine signature as well as the proliferating lymphocyte population profile enabled us to investigate potential correlations between both data sets. The cytokine and chemokine data were assessed after 24 h of adjuvant stimulation and coculturing of DCs with autologous PBLs. Thus, we sought to correlate the cytokine and chemokine milieu at the starting point of the induction of lymphocyte proliferation with the resulting lymphocyte proliferation after 6 d. To this end, for each donor, we normalized the measured values of the cytokine and chemokine data set as well as the lymphocyte proliferation data set to their respective unstimulated control. Potential influences of the variables “experiment,” “sex of donor,” “age of donor,” “day of Luminex analysis,” “plate,” and “donor” on the levels of protein analytes and lymphocytes have been studied individually using the Kruskal-Wallis test (Fig. 6A). The logarithmized P values demonstrate a strong donor dependency, with high values especially for granulocyte-macrophage colony-stimulating factor (GM-CSF), IL-17A, IL-18, IL-2, IL-3, IL-4, IL-5, and NKT cells. In a next step, we determined the Spearman correlation of protein analytes and lymphocyte proliferation, which indicates an overall positive correlation (Fig. 6B). Although most of the values indicate only a weak correlation (around r = 0.3), the proliferation of distinct lymphocyte populations correlated moderately (around r = 0.5) to several cytokines. More precisely, CD8+ T-cell proliferation can be linked to the increased expression of multiple cytokines such as IFNy (r = 0.45), IL-10 (r = 0.52), IL-12p70 (r = 0.44), IL-1α/β (r = 0.47/0.48), IL-6 (r = 0.49), MIG (r = 0.5), MIP-1α (r = 0.46), RANTES (r = 0.58), and TNFα (r = 0.46). Also, NKT cell proliferation can be related to a cytokine pool out of IL-10 (r = 0.46), IL-23, IL-6, MIG, and RANTES (all r = 0.47). In contrast, the proliferation of B cells shows a moderate correlation to only two cytokines, namely IFNα (r = 0.5) and MCP-1 (r = 0.46), and NK cell proliferation only moderately correlates to RANTES (r = 0.46). Strikingly, CD4+ T-cell proliferation and the listed cytokines and chemokines correlate only weakly at the points of analysis. As cytokine secretion by various cell types over time relies on a broad range of crosstalk and feedback regulations, we used a second set of donors to compare the cytokine secretion on day 5 with the secretion after 24 h of adjuvant stimulation using Legendplex (SI Appendix, Fig. S10A). Interestingly, we found that concentrations of the proinflammatory mediators IL-1β and TNFα declined in the cell culture supernatants over time, while other proinflammatory mediators, e.g., IFNα and MCP-1, and cytokines that are predominantly expressed by T cells, e.g., IFNγ or IL-17A, showed a significant increase in various conditions. We then also assessed the correlation of lymphocyte proliferation on day 5 with the cytokines expressed after 24 h (SI Appendix, Fig. S10 B and C) or day 5 (SI Appendix, Fig. S10 D and E) of the coculture, again considering the variables day, plate, and donor (SI Appendix, Fig. S10 B and D). Linear relationships calculated for the cytokines measured after 24 h and lymphocyte proliferation on day 5 (SI Appendix, Fig. S10C) overall confirmed our initial correlation data shown in Fig. 6. However, as our second data set comprised a smaller sample size, only a few correlations reached significance. By contrast, cytokine concentrations measured on day 5 revealed a considerable correlation with proliferation, especially of CD8+ and NKT cells (SI Appendix, Fig. S10E), which was not so pronounced when looking at cytokines after 24 h even within the larger data set. Interestingly, while after 24 h, B-cell proliferation correlated best with IFNα and MCP-1 secretion, after 5 d, the strongest correlation was observed with IL-10.
Fig. 6.
The proliferation of lymphocyte subpopulations can be linked to the expression of certain cytokines and chemokines. Data sets of adjuvant-induced cytokine and chemokine secretion after 24 h (Figs. 3 and 4) and adjuvant-induced lymphocyte proliferation assessed after 6 d (Fig. 5) were linked using Spearman’s rank correlation. (A) The influence of several factors on the levels of cytokine and chemokine concentration as well as of absolute lymphocyte count been assessed individually by Kruskal-Wallis testing. The presented values are the common logarithms of the P values analyzed by the Kruskal-Wallis test. White color represents a high P value, whereas red color indicates a very low P value. All analyses are exploratory without corrections for multiple testing. (B) The correlation matrix is based on normalized values that were obtained by setting the unstimulated control for each donor to 1. Spearman correlation coefficients associated with a P value above 0.01 are crossed out. A value of 1 (blue) indicates a positive correlation, whereas a value of −1 (red) represents a negative correlation.
Pam, MPL-s, and Al(OH)3 Stimulate the Proliferation of FluM1-Specific CD8+ T Cells.
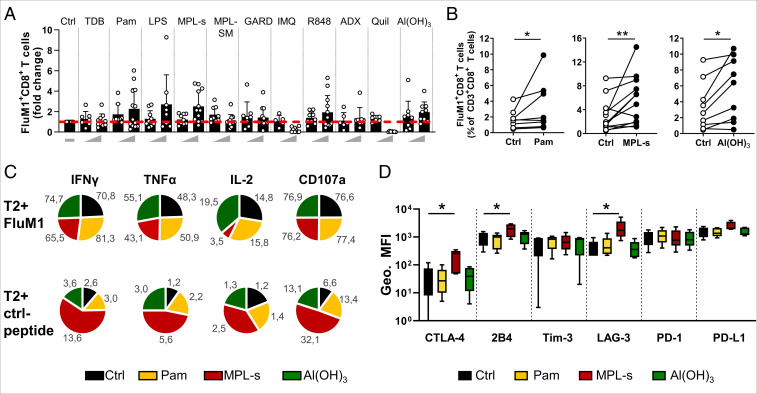
In a next step, we addressed the adjuvants’ influence on antigen-specific CD8+ T cells. To this end, CD8+ T cells were isolated and cocultured with DCs that had been pulsed with a peptide epitope of the influenza matrix protein (FluM158-66: GILGFVFTL). DCs that were not loaded with the FluM1-peptide served as control. After 6 to 7 d of costimulation with the different adjuvants, FluM1-specific CD8+ T cells were identified and analyzed using HLA tetramer-based flow cytometry. Since the allele is relatively frequent among Caucasian donors, we used HLA-A*0201 tetramers that readily bind the FluM158-66 peptide. Ex vivo, we found—in dependence of the respective donor—between 0.004% and 0.51% FluM1 tetramer-positive T cells within the CD8+ T-cell gate (SI Appendix, Fig. S11A). Irrespective of the added adjuvant, there was no change in the frequency of FluM1-specific T cells upon coculture with nonpulsed DCs (SI Appendix, Fig. S8B), whereas peptide-pulsed DCs expectedly led to a significant expansion of FluM1-specific CD8+ T cells (SI Appendix, Fig. S11B). Some of the tested immunomodulators were capable to boost this expansion (Fig. 7A). However, a significant enhancement of FluM1-specific CD8+ T-cell proliferation was only observed with Pam, MPL-s, and Al(OH)3 (Fig. 7B) These three components were therefore investigated in more detail. Interestingly, in the presence of high concentrations of IMQ and Quil, FluM1-specific CD8+ T-cell populations seemed to shrink rather than to expand, probably due to a general reduction of the CD8+ T-cell population, as observed in Fig. 5A.
Fig. 7.
Effects of Pam, MPL-s, and Al(OH)3 on the FluM1-specific CD8+ T-cell population and their polyfunctionality upon FluM1-restimulation. FluM1 peptide–loaded iDCs of HLA-A*0201+ donors and cocultured autologous CD8+ T cells were stimulated with the different adjuvants or left untreated. On day 6 or 7, CD8+ T cells were stained with peptide corresponding to MHC class I tetramer (GILGFVFTL HLA-A*0201-PE) and analyzed by flow cytometry. (A) Adjuvant-induced FluM1-specific CD8+ T-cell population (frequency of total CD3+CD8+ T cells) was normalized to its respective unstimulated control (dashed red line) (n = 7 to 12 donors). (B) For each donor, pairwise comparison of the unstimulated control and the adjuvant-stimulated conditions Al(OH)3 (10 µg/mL), Pam (100 pg/mL), and MPLs (1 µM) was performed using the Wilcoxon test (n = 8 to 11). (C) Cytokine expression upon restimulation of FluM1-specific CD8+ T cells using FluM1- or Ctrl peptide–loaded T2 cells. The mean frequency of total CD3+CD8+ T cells (%) is shown (n = 5 to 10). (D) Expression of the depicted exhaustion markers was assessed by flow cytometry (geometric MFI) on FluM1-specific CD8+ T cells, expanded with either Al(OH)3, Pam, or MPLs, or left untreated. Statistical differences were analyzed against the unstimulated control by Friedman test with Dunn’s correction for multiple comparisons (n = 5). P values *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001 were considered as statistically significant. Data are representative of at least three independent experiments.
Similarly to CD8+ T cells, we also observed adjuvant-dependent expansion of CD4+ T cells specific for tetanus toxoid peptide TT p2829-844. However, as the frequency of donors with the corresponding HLA-II allele is quite low within the population and there is a generally high variation in TT-specific CD4+ T-cell abundance between these donors, the results did not reach statistical significance. Nevertheless, we observed a clear trend of expanded antigen-specific CD4+ T cells with MPL-s–stimulated and peptide-pulsed DCs (SI Appendix, Fig. S11C).
Pam-Expanded and Al(OH)3-Expanded but Not MPL-s–Expanded FluM1-Specific T Cells Show a Polyfunctional Cytokine Secretion Profile upon a Second FluM1 Stimulus.
To assess whether adjuvant-expanded FluM1-specific CD8+ T cells would maintain their ability to exert pivotal effector functions, we analyzed their capacity to express multiple cytokines such as IFNγ, TNFα, and IL-2 upon restimulation with the FluM1 peptide. In addition, we tested whether the restimulation would lead to an increased surface expression of lysosomal-associated protein CD107a, which is used as a correlate to measure degranulation of cytolytic effector T cells. To this end, CD8+ T cells were harvested from the coculture with DCs and added in a 1:1 ratio to HLA*0201-positive TAP-deficient T2 cells that had been loaded with the FluM1 peptide or a nonrelated control peptide (NY-ESO-1). Here, the T2 cells served as peptide-presenting cells for restimulation of the CD8+ T cells. After 6 h of coculture, cells were stained for flow cytometric analysis. As outlined previously, we focused on the analysis of Pam-, MPL-s–, or Al(OH)3-expanded FluM1-specific T cells because only these three compounds consistently enhanced FluM1-specific CD8+ T-cell proliferation in almost all of the donors tested. As a reference, we analyzed the effector functions of FluM1-specific T cells that were expanded in the absence of adjuvant, and these cells maintained their specificity when presented to T2 cells loaded with the control peptide. By contrast, in response to their cognate peptide, the specific T cells expressed the three cytokines probed and expressed CD107a on their cell surface (Fig. 7C and SI Appendix, Fig. S12A). The expression levels were comparable to PMA/ionomycin stimulation of FluM1-specific T cells in the absence of the T2 cells and peptide stimulation, which served as nonspecific activation control (SI Appendix, Fig. S12A). Despite the accelerated expansion of FluM1-specific T cells in the presence of Pam, MPL-s, and Al(OH)3, T cells maintained their functional capacities. The majority of restimulated T cells expressed IFNγ, and while slightly fewer expressed TNFα, only a minority (20% to 30%) also expressed IL-2. CD107a was displayed on the cell surface of 80% to 90% of peptide- or PMA/ionomycin-stimulated cells. It is of note that upon expansion in the presence of MPL-s, less FluM1-specific T cells expressed IL-2 and TNFα when stimulated with PMA/ionomycin or FluM1 peptide (SI Appendix, Fig. S12A), and the number of IFNγ- and CD107a-expressing T cells was also slightly higher upon treatment with control peptide alone (Fig. 7C). This is presumably due to the strong adjuvant stimulus the T cells received during the expansion period. Since strong T-cell activation can go along with T-cell exhaustion, we evaluated the expression of inhibitory exhaustion markers on FluM1-expanded T cells. Here, we specifically observed elevated expression levels of LAG-3, 2B4, and CTLA-4 on MPL-s–expanded FluM1-specific T cells (Fig. 7D). While PD-1 and Tim-3 levels were not affected on MPL-s–expanded FluM1-specific T cells (Fig. 7D), they were elevated on the MPL-s–stimulated total CD8+ T-cell population (SI Appendix, Fig. S12B), which would be an indication for the strong adjuvant stimulus affecting the whole CD8+ T-cell population and not solely FluM1-specific T cells.
Discussion
Charles A. Janeway referred to adjuvants as the “immunologist’s dirty little secret.” Thirty years after Janeway’s bon mot and despite his founding work on the recognition of pathogen-associated molecular pattern (19), the highly complex effects of adjuvants on the immune system are still more an enigma to immunologists than a little secret at their service. However, a comprehensive understanding of their mode of action is a prerequisite for the intelligent design of modern vaccines. In view of the current COVID-19 pandemic, it is evident how important and life-saving the rapid development of a safe and efficacious vaccine against emerging pathogens can be. In the current study, we therefore aimed to systematically investigate the reaction pattern of human immune cells to a panel of candidate adjuvant substances and single-stranded RNA. The majority of the experiments were conducted in an antigen-independent manner to prove the sole adjuvant effect (20). Therefore, the first aspect of adjuvant characteristics we assessed was the changes in the functional phenotype of adjuvant-exposed DCs. In our study, isolated human moDCs responded in particular to TLR4, such as LPS and MPL-s as well as MPL-SM, the TLR2 agonist Pam, and also to the TLR7/8 ligand R848. Upon maturation, the cells shut down phagocytosis, as observed by strongly reduced FITC-dextran uptake and up-regulated CCR7, the homing receptor that guides mature DCs to the draining lymph node, as well as the costimulatory molecules CD80 and CD86, which are required for efficient T-cell priming (21). In contrast to the other substances, TLR4 and TLR2 agonists directly activated immature DCs, which is in line with the reported dominant expression of TLR4 and TLR2 on moDCs (22). Agonists triggering the endosomal TLR7 (GARD, IMQ) led to a DC maturation phenotype only when DCs were cocultivated with PBLs. This can be explained by the fact that immature moDCs do not or only very weakly express TLR7 (SI Appendix, Fig. S3), and thus, moDCs insufficiently respond to TLR7 ligand stimulation (23). However, as shown by Severa et al., TLR7 expression in moDCs can be elicited by type-I IFN, which is either generated in an autocrine manner upon TLR4 activation of the DCs or can be derived by a different (cellular) source (24). This highlights how the interaction with other immune cells has profound functional consequences on the entire cellular network.
Variation in vaccine efficacy within the population is known (e.g., influenza and hepatitis B as well as measles) and can be ascribed to intrinsic (immunogenetics, age, sex) and environmental factors affecting the individual’s immune response (25–27). Understanding this heterogeneity is of pivotal importance to rationalize adjuvant formulation strategies aiming to ensure broad vaccine efficacy. While we did not find distinct sex- or age-dependent specificities in the immune response to the tested adjuvants, significant variation between individual donors was clearly detectable.
Recently, the Milieu Intérieur Consortium analyzed the activation of innate immune cells of healthy donors after stimulation with 28 different conditions using a whole-blood assay and a multiplex cytokine readout. They suggested that the detected variations of the innate immune response explain differential outcomes of therapeutic interventions or vaccinations (28). We adapted the approach taken by the Milieu Intérieur study and employed the multiplex cytokine readout to investigate the cytokine pattern in the supernatant of cocultivated DCs. Using PCA, we identified specific cytokine and chemokine signatures for the various adjuvant candidates tested. In general, there was a broad up-regulation of various cytokines and chemokines in response to TLR4, TLR7/8, and TLR2 agonists. Over many decades, the potent immune stimulation exerted by these compounds has tempted vaccinologists to include them in vaccine formulations (29). However, it was only recently that compounds have been identified that combine the strong immune stimulation with a sufficient degree of tolerability (30, 31). Accordingly, we observed that MPL-SM, the compound that corresponds to detoxified monophosphoryl lipid A (MPLA) (32), and Pam showed a more sophisticated (intermediate) cytokine expression pattern compared to the strong immunomodulators LPS, MPL-s, or R848. In particular, the levels of IL-12 and IL-18, two cytokines that concertedly and strongly enhance proinflammatory responses (33, 34), were clearly lower.
Another interesting finding was the strong up-regulation of IFNα after stimulation with TLR7 and TLR7/8 agonists. In general, the source of TLR7-induced IFNα is not moDCs but rather pDCs (35). pDCs are able to produce 200 to 1,000-fold more IFNα than any other blood cell type (31), and thus, it is possible that IFNα levels measured in our Luminex analysis were strongly influenced by this pDC subtype. This is supported by the finding that about 0.5% viable pDCs were present in the PBL fraction after 24 h, which declined to 0.05% after 6 d of adjuvant stimulation (SI Appendix, Fig. S7C). An important finding was the induction of specific cytokine profiles by GARD, IMQ, and R848. In particular, the TLR7/8 ligand R848 segregated from the two TLR7 agonists but also GARD and IMQ showed clear distinctions. The highest cytokine levels were induced by R848, followed by GARD. However, although GARD induced higher levels in comparison to IMQ, the level of IMQ-induced IL-1β was superior in comparison to GARD. The differences in cytokine induction had significant implications for antigen-independent lymphocyte proliferation. The observation is intriguing because, in principle, all three compounds target the same receptors. However, it is conceivable that subtle differences in their TLR-binding account for the observed discrepancies: While R848 has been described to bind equally to both receptors (36–38), GARD and IMQ predominantly target TLR7. However, the binding to TLR7 is also different: both R848 and GARD are able to dimerize the receptor, but an N (Gardiquimod) to O (R848) atoms difference in the R1 moiety renders GARD less efficient. IMQ lacks that R1-moiety. Thus, it induces only very weak TLR7 dimerization (39).
The specific IL-17 induction in response to TLR ligands of bacterial origin is well in accordance with the presumed role of Th17 cells in orchestrating antibacterial immune responses. Under all conditions tested, the amount of IL-4 remained close to the detection limit (40). Together, this indicates that priming with the respective TLR ligands elicited a Th1 and/or Th17 profile but no Th2 responses.
When directly assessing the influence of the tested adjuvant components on the respective lymphocyte populations, we observed marked differences between the compounds, but they were all well in accordance with the induced cytokine pattern observed previously. One example was the strong proliferation of B lymphocytes in response to the TLR7 and TLR7/8 agonists. This was probably due to the concerted effect of IFNα secreted by dedicated innate immune cells, as discussed previously, and the observed DC-independent activation of B cells through R848, GARD, and IMQ (41). It is of note that pDC-derived IFNα has been described as a strong inducer of B-cell proliferation (42), which is in accordance with a very recent paper showing B-cell activation through IFNα (41). It remains to be defined whether TLR7/IFNα-activated B cells maintain a more autoreactive idiotype pattern, as recently suggested (43). Out of the TLR4 ligands, only LPS and MPL-s but not MPL-SM activated B and NKT cells. This might be attributed to the significantly higher cytokine levels of IL-23, IL-12p70, and TNFα when stimulating with MPL-s or LPS compared to MPL-SM. Along this line, Wang et al. recently demonstrated that various research-grade Salmonella enterica–derived MPLA preparations purchased from different companies showed varying efficacy on TLR4 stimulation and TNFα secretion in human THP-1 cells, whereas mouse RAW264.7 cells responded similarly to these compounds (44). The phenomenon of TLR-induced, antigen-independent lymphocyte expansion has been described before but has gained little interest in vaccine development so far. However, it is very well conceivable that it represents a very important adjuvant effect of single-component adjuvants.
While the antigen-independent expansion of lymphocyte populations was primarily observed with TLR7, TLR7/8, and TLR4 agonists, antigen-specific recall responses were also triggered in combination with classical adjuvants. Interestingly, in our model, mainly Al(OH)3, MPL-s, and Pam showed consistent FluM1 antigen-specific CD8+ T cell–activating properties throughout the tested donors. Similar results were observed for the proliferation of tetanus toxoid-specific CD4+ T cells that were most clearly stimulated by MPL-s. Our results are well in accordance with similar approaches in the mouse model (45–47). By contrast, CD8+ T-cell proliferation could not be detected in several clinical trials testing HB antigens or HIV-1 vaccine candidates adjuvanted with alum or the MPL-containing AS01 or AS04 (9, 47). These findings suggest that evoking antigen-specific CD8+ T cells in naïve adults with adjuvanted subunit vaccines is very complex and requires careful assessment.
Prolonged immune stimulation can cause T-cell exhaustion, which is characterized by the loss of robust effector function and up-regulation of various inhibitory receptors. Within the FluM1 adjuvant–expanded, tetramer-positive CD8+ T cells, only the immune stimulation with MPL-s induced the expression of inhibitory markers LAG-3, 2B4, and CTLA-4. PD-1 and Tim-3 were not induced. The intermediate or weak immunomodulators Pam or Al(OH)3, respectively, did not lead to elevated levels of inhibitory receptors. Whether adjuvant-induced T-cell exhaustion might already have been causative for the lower or inconsistent CD8+ proliferation upon treatment with one of the other adjuvants, especially LPS or R848, remains to be investigated.
Whether the observed differences in cytokine expression and lymphocyte proliferation translate into functional implications when individual adjuvant components are used in clinical trials is not clear at the moment. However, there are numerous publications indicating that commonly used preclinical animal models respond differently to TLR stimulation compared to humans (48, 49). Likewise, there are ample examples of experimental vaccines that conferred protection in animal models but failed when tested in humans (50–52). Bridging studies, like the human cell-based in vitro study described in the current manuscript, are therefore required to translate concepts of basic immunology into applied vaccinology (53). At the same time, it is obvious that our in vitro system is designed to study direct adjuvant effects on individual immune cell populations. It cannot reproduce effects that depend on tissue inflammation or the slow but continuous release of antigens from the adjuvant matrix. This is probably the reason why adjuvant substances, such as Al(OH)3 and ADX (formulation is similar to MF-59), which have been used over decades and stimulate a protective adaptive immune response in vivo, did not show a clear immune profile in the presented study. In particular, the weak effects of Al(OH)3 and ADX on DC maturation in our in vitro assay can be at least partially explained by the fact that these adjuvants were shown to instead act on macrophages and monocytes (54).
During the current SARS-CoV-2 pandemic, a variety of adjuvants are being screened for their potential application in various types of vaccines (55–57), and in this context, deep knowledge on adjuvant efficacy as reported here is essential. A recent World Health Organization draft landscape paper lists 287 SARS-CoV-2 vaccine candidates (3). In this list, prioritized vaccine candidates are based on protein subunits, vector platforms, such as recombinant adenoviruses, or mRNA constructs. It may very well be that the difficulty to select the most suitable adjuvant has hampered the development of adjuvanted SARS-CoV-2 subunit vaccines. However, in view of the data presented in the current study, it is of note that the two vaccines that were the first to be licensed by the US Food and Drug Administration and European Medicines Agency were mRNA vaccines (Comirnaty, COVID-19 Vaccine Moderna) (58–61). It is well known that mRNA vaccines activate TLR7 and TLR8 (62–64). Using lipid-complexed single-stranded RNA as a surrogate for mRNA vaccines, we could confirm TLR7/8-mediated induction of strongly antiviral cytokines. Furthermore, in response to lipid-formulated single-stranded RNA, we observed an antigen-independent proliferation of lymphocyte populations that are particularly required for virus control, such as CD8+ T cells, NKT cells, or NK cells. This inherent adjuvant activity of the single-stranded RNA is likely the key to the excellent efficacy of the recent COVID-19 mRNA vaccines.
Taken together, our study provides a comprehensive, comparative, multilayer analysis of the immunomodulatory functions of adjuvants and RNA molecules within a complex human cell-based in vitro system. The demonstration that even subtle differences in the receptor activation can alter the induced cytokine profiles and the responding cell populations may prove relevant for the careful preselection and design of future adjuvant systems.
Materials and Methods
Study Design.
The main objective of the study was to link specific innate and adaptive immunogenic modes of action to the adjuvant’s properties to facilitate the design of adjuvant-based vaccine and therapy formulations. Therefore, we analyzed and compared 10 single-component adjuvants in a human primary immune cell in vitro assay. We assessed the adjuvants’ capacities to stimulate DC maturation, cytokine and chemokine secretion, lymphocyte proliferation, and the boosting of antigen-specific T-cell populations. The samples were analyzed by flow cytometry, Luminex technology, and microscopy. Data were raised by a controlled laboratory experimental study.
The in vitro assay is based on human primary immune cells (DCs cocultured with autologous PBLs), which were isolated from buffy coats or whole blood. When using buffy coats, the inclusion criteria of donors are defined by DRK-Blutspendedienste on behalf of the hemotherapy guideline (§12a and 18 Transfusionsgesetz), assuming that only healthy subjects between 18 and 68 y of age were included in this study. Whole-blood donation was required from donors exhibiting an HLA-DRB1*11:01 phenotype and comprised a group of five healthy women ranging from 24 to 45 y of age. To study the cytokine- and chemokine-inducing signature of the adjuvants (Luminex multianalyte profiling), donors were selected by stratification, maintaining consistent sex and age distribution. The inclusion criteria were defined before donor enrollment for four groups (female/male; <40/>40 y of age) with seven to eight corresponding donors each. The selection of the buffy coats for this Luminex experiment was performed by DRK-Blutspendedienste. For all other experiments, buffy coats were selected randomly. HLA-A*02:01 donor positivity was assessed with antibody staining on six to eight ordered buffy coats (around 50% of them were HLA-A2 positive). Two to three positive donors were chosen randomly. If the phenotype of tested immature DCs was different from CD14−CD1a+CD209+ after harvest on day 5 to 6 or when lymphocytes proliferated >15% in the unstimulated control (without antigen) on day 6 to 7 of coculture with DCs, samples were excluded from experiments.
Immune cells isolated from the blood of donors were used to assess cytokine/chemokine levels with Luminex xMAP technology after stimulation with a total of 23 conditions (low and high concentration of 11 adjuvants, including the control condition “medium”). In order to be able to detect an effect size of 0.2 at a significance level of 0.05 and power of 95%, 30 donors were included in the study. The 30 replicates (donors) were analyzed in 15 independent experiments, with biological duplicates being measured and summarized as mean aggregated. For all other data sets, we performed at least three independent experiments with a total minimum of five donors. The assessment of experiments was not conducted in an anonymized manner.
Immunomodulators.
All immunomodulators listed in SI Appendix, Table S1 were purchased as VacciGrade products (Invivogen), which is a specific purity grade suitable for preclinical studies. Ultrapure Lipopolysaccharide from Escherichia coli 0111:B4 (LPS-EB) (Invivogen) served as positive control for immune induction. Concentrations of the adjuvants were determined by their cytotoxicity (Annexin-V/PI apoptosis staining kit, Miltenyi Biotec) and proliferation-inducing capacity (CellTrace Far Red Cell Proliferation Kit, Thermo Fisher Scientific) on lymphocytes at day 6 of the DC:PBL coculture (described in the Stimulation with the Immunomodulators section). Endotoxicity and pyrogenicity of the adjuvants were assessed using the LAL and monocyte activation test. The LAL test was based on the kinetic-turbidimetric method using Pyrotell-T lysate, which was solved in Pyrosol buffer (Associates of Cape Cod, Europe). Each sample underwent a product positivity test (spike) with 0.5 EU/mL LPS to exclude possible confounding factors within the sample. The determination of endotoxin amount was valid, if 50% to 200% of the spiked endotoxin was recovered. Turbidity was measured with ELx808 microplate reader (BioTek) at 340 nm. Endotoxin concentrations were calculated using the endotoxin standard Biological Reference Preparation (European pharmacopoeia reference standard, National Institute for Biological Standards and Control) standard curve. The monocyte activation test was performed as previously described (65, 66) on cryopreserved human whole blood and with an IL-β cytokine readout.
The TLR8 agonists ssPoly(U) Naked and ssPolyU/LyoVec were purchased from Invivogen and were applied to the cells in a concentration of 10 µg/mL.
Stimulation with the Immunomodulators.
DCs were harvested on day 5 to 6 of culture, and their immature phenotype CD14−CD1a+CD209+ (M5E2, Biolegend; HI149 and DCN46, both BD Bioscience) was confirmed by flow cytometry analysis. Autologous CD14− PBLs were thawed and added to DCs at a 5:1 ratio to achieve the DC:PBL coculture, which was then stimulated with the immunomodulators at the indicated concentrations (SI Appendix, Table S1).
Statistical Analysis and Data Visualization.
PCA, agglomerative hierarchical clustering, and heat maps were performed with Qlucore Omnics Explorer v3.5. Here, false discovery rate–adjusted ANOVA P values, called q-values, were used to define the cytokine cutoff of the data visualizations and to discriminate the most differentially induced proteins. Data were transformed prior to analysis by the software: logarithmized, mean-centered, and scaled to unit variance. The mean centering is in accordance with the paired structure of the data. Data were corrected for donor variation to observe adjuvant-specific effects. Dot plots/bar graphs, box plots, whisker plots, and two-way correlations were compiled using GraphPad Prism v8.3.1 or R v3.6.1 using the functions kruskal.test(), cor(), and cor.test() as well as the package corrplot for plotting correlation matrices v0. Radar plots and part-of-whole diagrams were drafted with Excel 2016. Statistical significance of antigen-specific experiments was determined by Wilcoxon matched-pairs signed rank test or Friedman test with Dunn’s correction. Spearman nonparametric ranks were used for correlation analysis. Further statistical analysis was performed using two-way ANOVA, with Sidak’s multiple comparison test or the Kruskal-Wallis test with Dunn’s multiple comparison correction (conducted as two-sided test with alpha = 0.05). P values <0.05 (*), P < 0.01 (**), P < 0.001 (***), and P < 0.0001 (****) were considered to be statistically significant.
Supplementary Material
Acknowledgments
We thank P. Windecker for excellent technical support in testing the adjuvants for the pyrogenic and endotoxic effects. We also thank S. Müller and H. Bönig (DRK-Blutspendedienste-Blutspendezentrum, Frankfurt a.M.) for their help in selecting the 30 buffy coats for the Luminex analysis according to the defined criteria and Z. Waibler and J. Kirberg (Paul-Ehrlich-Institut) for fruitful data discussions. This project was funded by the German Ministry of Health (BMG) to M.B. and G.v.Z.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2103651118/-/DCSupplemental.
Data Availability
All study data are included in the article and/or SI Appendix.
References
- 1.Koch T., Fathi A., Addo M. M., “The COVID-19 vaccine landscape” in Coronavirus Disease - COVID-19, Rezaei N., Ed. (Springer International Publishing AG, 2021), pp. 549–573. [DOI] [PubMed] [Google Scholar]
- 2.García-Montero C., et al., An updated review of SARS-CoV-2 vaccines and the importance of effective vaccination programs in pandemic times. Vaccines (Basel) 9, 9 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.World Health Organization , Data from “COVID-19 vaccine tracker and landscape.” World Health Organization. https://www.who.int/publications/m/item/draft-landscape-of-covid-19-candidate-vaccines). Accessed 6 November 2021.
- 4.Buonsanti C., D’Oro U., “Discovery of immune potentiators as vaccine adjuvants” in Immunopotentiators in Modern Vaccines, Schijns V. E. J. C., O’Hagan D. T., Eds. (Elsevier, 2017), pp. 85–104. [Google Scholar]
- 5.Reed S. G., Orr M. T., Fox C. B., Key roles of adjuvants in modern vaccines. Nat. Med. 19, 1597–1608 (2013). [DOI] [PubMed] [Google Scholar]
- 6.Marrack P., McKee A. S., Munks M. W., Towards an understanding of the adjuvant action of aluminium. Nat. Rev. Immunol. 9, 287–293 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moyer T. J., et al., Engineered immunogen binding to alum adjuvant enhances humoral immunity. Nat. Med. 26, 430–440 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garçon N., Segal L., Tavares F., Van Mechelen M., The safety evaluation of adjuvants during vaccine development: The AS04 experience. Vaccine 29, 4453–4459 (2011). [DOI] [PubMed] [Google Scholar]
- 9.Didierlaurent A. M., et al., AS04, an aluminum salt- and TLR4 agonist-based adjuvant system, induces a transient localized innate immune response leading to enhanced adaptive immunity. J. Immunol. 183, 6186–6197 (2009). [DOI] [PubMed] [Google Scholar]
- 10.De Mot L., et al., Transcriptional profiles of adjuvanted hepatitis B vaccines display variable interindividual homogeneity but a shared core signature. Sci. Transl. Med. 12, 12 (2020). [DOI] [PubMed] [Google Scholar]
- 11.Leroux-Roels G., et al., Impact of adjuvants on CD4(+) T cell and B cell responses to a protein antigen vaccine: Results from a phase II, randomized, multicenter trial. Clin. Immunol. 169, 16–27 (2016). [DOI] [PubMed] [Google Scholar]
- 12.Nohynek H., et al., AS03 adjuvanted AH1N1 vaccine associated with an abrupt increase in the incidence of childhood narcolepsy in Finland. PLoS One 7, e33536 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bardage C., et al., Neurological and autoimmune disorders after vaccination against pandemic influenza A (H1N1) with a monovalent adjuvanted vaccine: Population based cohort study in Stockholm, Sweden. BMJ 343, d5956 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.MacDonald N. E., Butler R., Dubé E., Addressing barriers to vaccine acceptance: An overview. Hum. Vaccin. Immunother. 14, 218–224 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schindler S., von Aulock S., Daneshian M., Hartung T., Development, validation and applications of the monocyte activation test for pyrogens based on human whole blood. ALTEX 26, 265–277 (2009). [DOI] [PubMed] [Google Scholar]
- 16.Osterlund P., et al., Gene expression and antiviral activity of alpha/beta interferons and interleukin-29 in virus-infected human myeloid dendritic cells. J. Virol. 79, 9608–9617 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dunn W. Jr, Burgun A., Krebs M.-O., Rance B., Exploring and visualizing multidimensional data in translational research platforms. Brief. Bioinform. 18, 1044–1056 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dominguez-Villar M., Gautron A.-S., de Marcken M., Keller M. J., Hafler D. A., TLR7 induces anergy in human CD4(+) T cells. Nat. Immunol. 16, 118–128 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gayed P. M., Toward a modern synthesis of immunity: Charles A. Janeway Jr. and the immunologist’s dirty little secret. Yale J. Biol. Med. 84, 131–138 (2011). [PMC free article] [PubMed] [Google Scholar]
- 20.Yang A. X., et al., Molecular characterization of antigen-peptide pulsed dendritic cells: Immature dendritic cells develop a distinct molecular profile when pulsed with antigen peptide. PLoS One 9, e86306 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schweitzer A. N., Borriello F., Wong R. C., Abbas A. K., Sharpe A. H., Role of costimulators in T cell differentiation: Studies using antigen-presenting cells lacking expression of CD80 or CD86. J. Immunol. 158, 2713–2722 (1997). [PubMed] [Google Scholar]
- 22.Jarrossay D., Napolitani G., Colonna M., Sallusto F., Lanzavecchia A., Specialization and complementarity in microbial molecule recognition by human myeloid and plasmacytoid dendritic cells. Eur. J. Immunol. 31, 3388–3393 (2001). [DOI] [PubMed] [Google Scholar]
- 23.Hackstein H., et al., The TLR7/8 ligand resiquimod targets monocyte-derived dendritic cell differentiation via TLR8 and augments functional dendritic cell generation. Cell. Immunol. 271, 401–412 (2011). [DOI] [PubMed] [Google Scholar]
- 24.Severa M., et al., Sensitization to TLR7 agonist in IFN-beta-preactivated dendritic cells. J. Immunol. 178, 6208–6216 (2007). [DOI] [PubMed] [Google Scholar]
- 25.Posteraro B., et al., The link between genetic variation and variability in vaccine responses: Systematic review and meta-analyses. Vaccine 32, 1661–1669 (2014). [DOI] [PubMed] [Google Scholar]
- 26.Thomas C., Moridani M., Interindividual variations in the efficacy and toxicity of vaccines. Toxicology 278, 204–210 (2010). [DOI] [PubMed] [Google Scholar]
- 27.White M. T., et al., A combined analysis of immunogenicity, antibody kinetics and vaccine efficacy from phase 2 trials of the RTS,S malaria vaccine. BMC Med. 12, 117 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Duffy D., et al., Functional analysis via standardized whole-blood stimulation systems defines the boundaries of a healthy immune response to complex stimuli. Immunity 40, 436–450 (2014). [DOI] [PubMed] [Google Scholar]
- 29.Skidmore B. J., Chiller J. M., Morrison D. C., Weigle W. O., Immunologic properties of bacterial lipopolysaccharide (LPS): Correlation between the mitogenic, adjuvant, and immunogenic activities. J. Immunol. 114, 770–775 (1975). [PubMed] [Google Scholar]
- 30.Casella C. R., Mitchell T. C., Putting endotoxin to work for us: Monophosphoryl lipid A as a safe and effective vaccine adjuvant. Cell. Mol. Life Sci. 65, 3231–3240 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Baldrick P., Richardson D., Elliott G., Wheeler A. W., Safety evaluation of monophosphoryl lipid A (MPL): An immunostimulatory adjuvant. Regul. Toxicol. Pharmacol. 35, 398–413 (2002). [DOI] [PubMed] [Google Scholar]
- 32.Fox C. B., Friede M., Reed S. G., Ireton G. C., Synthetic and natural TLR4 agonists as safe and effective vaccine adjuvants. Subcell. Biochem. 53, 303–321 (2010). [DOI] [PubMed] [Google Scholar]
- 33.Gracie J. A., Robertson S. E., McInnes I. B., Interleukin-18. J. Leukoc. Biol. 73, 213–224 (2003). [DOI] [PubMed] [Google Scholar]
- 34.Nakanishi K., Yoshimoto T., Tsutsui H., Okamura H., Interleukin-18 regulates both Th1 and Th2 responses. Annu. Rev. Immunol. 19, 423–474 (2001). [DOI] [PubMed] [Google Scholar]
- 35.Guiducci C., Coffman R. L., Barrat F. J., Signalling pathways leading to IFN-alpha production in human plasmacytoid dendritic cell and the possible use of agonists or antagonists of TLR7 and TLR9 in clinical indications. J. Intern. Med. 265, 43–57 (2009). [DOI] [PubMed] [Google Scholar]
- 36.de Marcken M., Dhaliwal K., Danielsen A. C., Gautron A. S., Dominguez-Villar M., TLR7 and TLR8 activate distinct pathways in monocytes during RNA virus infection. Sci. Signal. 12, 12 (2019). [DOI] [PubMed] [Google Scholar]
- 37.Berger M., et al., TLR8-driven IL-12-dependent reciprocal and synergistic activation of NK cells and monocytes by immunostimulatory RNA. J. Immunother. 32, 262–271 (2009). [DOI] [PubMed] [Google Scholar]
- 38.Bekeredjian-Ding I., et al., T cell-independent, TLR-induced IL-12p70 production in primary human monocytes. J. Immunol. 176, 7438–7446 (2006). [DOI] [PubMed] [Google Scholar]
- 39.Zhang Z., et al., Structural analyses of toll-like receptor 7 reveal detailed RNA sequence specificity and recognition mechanism of agonistic ligands. Cell Rep. 25, 3371–3381.e5 (2018). [DOI] [PubMed] [Google Scholar]
- 40.Lin Y., Slight S. R., Khader S. A., Th17 cytokines and vaccine-induced immunity. Semin. Immunopathol. 32, 79–90 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bender A. T., et al., TLR7 and TLR8 differentially activate the IRF and NF-κB pathways in specific cell types to promote inflammation. Immunohorizons 4, 93–107 (2020). [DOI] [PubMed] [Google Scholar]
- 42.Bekeredjian-Ding I. B., et al., Plasmacytoid dendritic cells control TLR7 sensitivity of naive B cells via type I IFN. J. Immunol. 174, 4043–4050 (2005). [DOI] [PubMed] [Google Scholar]
- 43.Simchoni N., Cunningham-Rundles C., TLR7- and TLR9-responsive human B cells share phenotypic and genetic characteristics. J. Immunol. 194, 3035–3044 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang Y.-Q., Bazin-Lee H., Evans J. T., Casella C. R., Mitchell T. C., MPL adjuvant contains competitive antagonists of human TLR4. Front. Immunol. 11, 577823 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Salerno F., Freen-van Heeren J. J., Guislain A., Nicolet B. P., Wolkers M. C., Costimulation through TLR2 drives polyfunctional CD8+ T cell responses. J. Immunol. 202, 714–723 (2019). [DOI] [PubMed] [Google Scholar]
- 46.Horrevorts S. K., et al., Toll-like receptor 4 triggering promotes cytosolic routing of DC-SIGN-targeted antigens for presentation on MHC class I. Front. Immunol. 9, 1231 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.MacLeod M. K. L., et al., Vaccine adjuvants aluminum and monophosphoryl lipid A provide distinct signals to generate protective cytotoxic memory CD8 T cells. Proc. Natl. Acad. Sci. U.S.A. 108, 7914–7919 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Davis H. L., Novel vaccines and adjuvant systems: The utility of animal models for predicting immunogenicity in humans. Hum. Vaccin. 4, 246–250 (2008). [DOI] [PubMed] [Google Scholar]
- 49.Barrat F. J., TLR8: No gain, no pain. J. Exp. Med. 215, 2964–2966 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Greek R., Animal models and the development of an HIV vaccine. J. AIDS Clinic Res. 01(2012). [Google Scholar]
- 51.Voss G., et al., Progress and challenges in TB vaccine development. F1000 Res. 7, 199 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jameson S. C., Masopust D., What is the predictive value of animal models for vaccine efficacy in humans? Reevaluating the potential of mouse models for the human immune system. Cold Spring Harb. Perspect. Biol. 10, 10 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Golding H., Khurana S., Zaitseva M., What is the predictive value of animal models for vaccine efficacy in humans? The importance of bridging studies and species-independent correlates of protection. Cold Spring Harb. Perspect. Biol. 10, 10 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cioncada R., et al., Vaccine adjuvant MF59 promotes the intranodal differentiation of antigen-loaded and activated monocyte-derived dendritic cells. PLoS One 12, e0185843 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gupta T., Gupta S. K., Potential adjuvants for the development of a SARS-CoV-2 vaccine based on experimental results from similar coronaviruses. Int. Immunopharmacol. 86, 106717 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Krammer F., SARS-CoV-2 vaccines in development. Nature 586, 516–527 (2020). [DOI] [PubMed] [Google Scholar]
- 57.Dong Y., Dai T., Wei Y., Zhang L., Zheng M., Zhou F., A systematic review of SARS-CoV-2 vaccine candidates. Sig. Transduct. Target Ther. 5, 237.(2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dooling K., et al., The advisory committee on immunization practices’ interim recommendation for allocating initial supplies of COVID-19 vaccine - United States, 2020. MMWR Morb. Mortal. Wkly. Rep. 69, 1857–1859 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ledford H., Moderna COVID vaccine becomes second to get US authorization. Nature, 10.1038/d41586-020-03593-7 (2020). [DOI] [PubMed] [Google Scholar]
- 60.Polack F. P., et al., Safety and efficacy of the BNT162b2 mRNA covid-19 vaccine. N. Engl. J. Med. 383, 2603–2615 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Baden L. R., et al., Efficacy and safety of the mRNA-1273 SARS-CoV-2 vaccine. N. Engl. J. Med. 384, 403–416 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fotin-Mleczek M., et al., Messenger RNA-based vaccines with dual activity induce balanced TLR-7 dependent adaptive immune responses and provide antitumor activity. J. Immunother. 34, 1–15 (2011). [DOI] [PubMed] [Google Scholar]
- 63.Kuhn A. N., et al., Determinants of intracellular RNA pharmacokinetics: Implications for RNA-based immunotherapeutics. RNA Biol. 8, 35–43 (2011). [DOI] [PubMed] [Google Scholar]
- 64.Pardi N., Hogan M. J., Porter F. W., Weissman D., mRNA vaccines - a new era in vaccinology. Nat. Rev. Drug Discov. 17, 261–279 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Montag T., et al., Safety testing of cell-based medicinal products: Opportunities for the monocyte activation test for pyrogens. ALTEX 24, 81–89 (2007). [DOI] [PubMed] [Google Scholar]
- 66.Schindler S., et al., International validation of pyrogen tests based on cryopreserved human primary blood cells. J. Immunol. Methods 316, 42–51 (2006). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or SI Appendix.