Significance
Here, we describe inhibitors of the Plasmodium proteasome, an enzymatic complex that malaria parasites rely on to degrade proteins. Starting from inhibitors developed to treat cancer, derivatives were designed and synthesized with the aim of increasing potency against the Plasmodium proteasome and decreasing activity against the human enzyme. Biochemical and cellular assays identified compounds that exhibit selectivity and potency, both in vitro and in vivo, at different stages of the parasite’s lifecycle. Cryo-electron microscopy revealed that the inhibitors bind in a hydrophobic pocket that is structurally different in the human proteasome—underpinning their selectivity. The work will help develop antimalarial therapeutics, which are desperately needed to treat a disease that kills nearly half a million people annually.
Keywords: Plasmodium, proteasome, antimalarial drug, peptide boronate, cryo-EM
Abstract
The Plasmodium falciparum proteasome is a potential antimalarial drug target. We have identified a series of amino-amide boronates that are potent and specific inhibitors of the P. falciparum 20S proteasome (Pf20S) β5 active site and that exhibit fast-acting antimalarial activity. They selectively inhibit the growth of P. falciparum compared with a human cell line and exhibit high potency against field isolates of P. falciparum and Plasmodium vivax. They have a low propensity for development of resistance and possess liver stage and transmission-blocking activity. Exemplar compounds, MPI-5 and MPI-13, show potent activity against P. falciparum infections in a SCID mouse model with an oral dosing regimen that is well tolerated. We show that MPI-5 binds more strongly to Pf20S than to human constitutive 20S (Hs20Sc). Comparison of the cryo-electron microscopy (EM) structures of Pf20S and Hs20Sc in complex with MPI-5 and Pf20S in complex with the clinically used anti-cancer agent, bortezomib, reveal differences in binding modes that help to explain the selectivity. Together, this work provides insights into the 20S proteasome in P. falciparum, underpinning the design of potent and selective antimalarial proteasome inhibitors.
Each year, Plasmodium falciparum (Pf) malaria infects at least 200 million people and causes more than 400,000 deaths (1). Alarmingly, after a decade of gains, progress in tackling malaria has plateaued (1). A major concern is that current antimalarial control is highly dependent on artemisinin-based combination therapies, which are associated with a cure failure rate of ∼50% in some regions in Southeast Asia (2). A rise in resistance-associated mutations in Rwanda (3) may presage spread of resistance to Africa. New compounds with potent activity against all stages of the parasite cycle are needed to feed the antimalarial drug development pipeline.
As an organism that grows rapidly in oxidatively stressed niches, malaria parasites are particularly susceptible to compounds that compromise proteostasis. The Plasmodium proteasome is an experimentally validated drug target, with a number of studies showing that proteasome inhibitors rapidly kill parasites at different stages of development (4–12). Importantly, proteasome inhibitors strongly synergize artemisinin-mediated killing of Pf cultures (in vitro) and murine Plasmodium parasites (in vivo) (9–11).
The proteasome plays an important role in cellular homeostasis—degrading abnormal, damaged, and short-lived proteins. The inside of the barrel-shaped 20S proteasome complex has six proteolytic active sites, comprising two copies each of three distinct types, termed β1 (caspase-like), β2 (trypsin-like), and β5 (chymotrypsin-like). Previous work showed that inhibition of Pf β5 activity is crucial for parasite killing (11, 13). By contrast, inhibition of human constitutive β5 activity is tolerated in nonmalignant cells, if β2c activity is maintained, thereby providing a therapeutic window (7, 13, 14).
Medicines for Malaria Venture (MMV) has published Target Product Profiles for antimalarial compounds. They recommend that new compound classes show activity against all parasite stages and against drug-resistant parasites, preferably with a mechanism of action that is different from currently deployed compounds (15). New drugs should preferably act rapidly and exhibit pharmacokinetic (PK) properties that will enable complete parasite clearance, ideally with a single oral dose (15).
We previously reported a screen of a library of peptidyl boronic acids inhibitors of the human proteasome and identified hits that inhibit parasite growth. These dipeptide inhibitors show a range of selectivities for inhibition of the growth of Pf compared with human cell lines (11) and exhibit similar physicochemical properties to bortezomib (VELCADE), which is dosed weekly, intravenously, or subcutaneously, in the clinical treatment of multiple myeloma (16, 17). While dipeptide boronates are considered to have favorable safety and efficacy profiles in a cancer setting, enhanced selectivity and oral bioavailability is desired for the treatment of malaria.
Here, we describe a medicinal chemistry program that, coupled with structural biology and biochemical approaches, explores the potential of a series of amino-amide boronates (containing a single amide bond in the backbone) for development as antimalarials. We have identified compounds that exhibit rapid, potent, and selective action against Pf, including drug-resistant strains, and display oral efficacy in a mouse model of Pf malaria.
Results and Discussion
Design and Synthesis of a Series of Amino-Amide Boronates.
The Plasmodium proteasome is a well-validated target for multistage antimalarial activity. A major challenge for this target is to identify compounds that possess potent activity against malaria parasites while achieving a suitable level of selectivity versus the human proteasome and having physicochemical properties compatible with oral administration. In our previous study (11), we identified Malaria Proteasome Inhibitor-1 (MPI-1; Table 1) as an exemplar hit from a screen of a Takeda Pharmaceuticals peptide boronate library. MPI-1 has a dipeptide backbone, similar to that of bortezomib, and is not expected to have good oral PK properties. Here, we identified boronate inhibitors with a single amide-bond backbone, which in general have better physicochemical properties and, therefore, are expected to have greater permeability and oral bioavailability. We found that these compounds retained significant potency against Pf20S, and we generated a series of compounds to explore structure activity relationships for these Pf20S-targetted inhibitors (Table 1).
Table 1.
Inhibitory activities of selected compounds against purified 20S and against Pf and mammalian cancer cell lines

|
IC50 values for the compound set against Pf20S and Hs20S β5c/β5i activity. For Pf20S β5-selective compounds, MPI-5 to 14, enzyme assay IC50 values are the concentrations, resulting in 50% inhibition of the total inhibitable β5 activity. Data represent the mean ± SEM and n is the number of independent experiments. LD50 for toxicity against cell lines after 72 h. Data represent the mean ± SEM for three independent experiments, each performed in triplicate (3D7), and greater than two independent experiments (HepG2). BTZ = bortezomib.
Data from ref. 11.
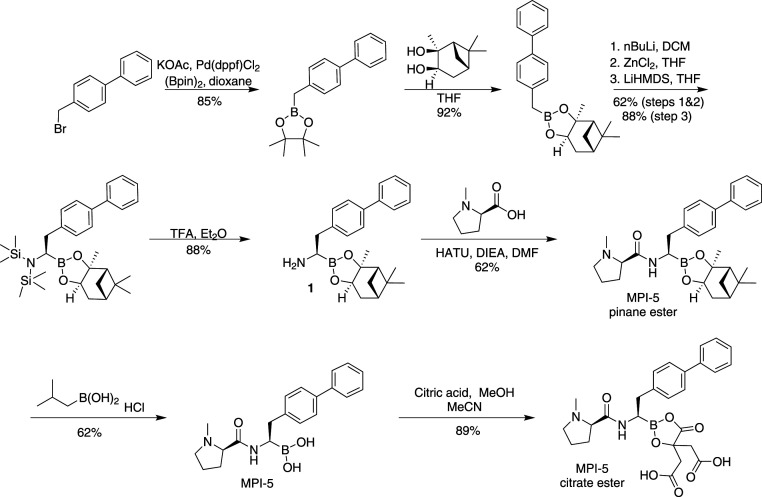
An exemplar synthesis of our boronate inhibitors is shown in Fig. 1. Biphenyl bromomethane was converted to the corresponding amino boronate pinane ester 1 in six steps (SI Appendix). Amide coupling resulted in the pinane ester boronate of MPI-5, which was used for our enzymatic and cellular assays as the pinane ester is readily hydrolyzed in aqueous media. For in vivo work, the citrate ester of MPI-5 was prepared in two steps, as shown (Fig. 1).
Fig. 1.
Synthesis of boronate analogs.
Optimization of the Assay for Inhibitors of Pf β5 Activity.
The ability of the compound set to inhibit the β5 subunit (chymotrypsin-like) activity of 20S purified from Pf and Homo sapiens (Hs) constitutive 20S was assessed based on cleavage of the fluorogenic peptide, amino-4-methylcoumarin (Ac-WLA-AMC), in the presence of human activator complex (PA28αβ) (11). In previous work, a Tris-based buffer has been employed for analysis of Pf20S β5 (10, 11), but incomplete inhibition of activity was often observed. We compared different buffers and found that a Hepes-based buffer (20 mM Hepes pH 7.4, 0.5 mM ethylenediaminetetraacetic acid [EDTA], and 0.01% bovine serum albumin [BSA]) enhances the potency of the inhibitor set at the Pf20S β5 active site and permits more complete inhibition (SI Appendix, Fig. S1 and Table S1). We employed the Hepes-based buffer for all further analyses of β5 activity. Where incomplete inhibition was still observed, we report the inhibition data as the concentration of compounds giving 50% inhibition of the total inhibitable β5 activity.
Biphenyl Substitution at P1 Is Associated with Enhanced Activity.
Activities of the compound set as inhibitors of substrate hydrolysis by Pf20S β5 and Hs20S β5c were compared with activities against Pf cultures (3D7 strain) and the HepG2 human cell line. As reported previously (11), bortezomib is twofold more potent as an inhibitor of Hsβ5c activity than of Pfβ5 activity (Table 1). Nonetheless, bortezomib inhibits growth of Pf more than the HepG2 cell line (selectivity index of 1.7-fold; Table 1). This difference is likely due to the particular sensitivity of Pf to inhibition of 20S β5 (7, 10).
In agreement with our previous report (11), MPI-1 is a much-less-efficient inhibitor of both Pfβ5 and Hsβ5c activity in comparison to bortezomib (Table 1). MPI-1 inhibits growth of Pf less potently; but shows much better cellular selectivity than bortezomib (Cellular selectivity index of 49-fold; Table 1).
As part of a synthetic campaign, we examined the effect of extending the aryl ring at P1 of MPI-1 to generate a biphenyl P1 substituent (MPI-6; Table 1). In the context of the dipeptide backbone, this was associated with greatly increased activity against Pfβ5 (50% inhibitory concentration [IC50] = 3 nM), and MPI-6 exhibited good selectivity compared with Hsβ5c (IC50 = 34 nM) (Table 1). As a consequence, MPI-6 exhibits ∼18-fold higher activity against 3D7 compared to MPI-1 and exhibits very good selectivity compared with HepG2 (323-fold; Table 1). This increased potency is associated with an increase in lipophilicity (AlogP; SI Appendix, Table S2) but also an increase in molecular mass. Building on the increased affinity offered by the biphenyl moiety, we sought to move to a single amide-containing backbone in an effort to generate a series with improved physicochemical properties and potentially good PK properties.
Amino-Amide Boronates Exhibit Enhanced Physicochemical Properties, Good Activity, and Specificity.
Among the various alterations explored, we found that removal of the terminal amide of MPI-1 and cyclization of the alanine side chain were both tolerated but did result in decreased enzymatic and cellular inhibition potency (MPI-7; Table 1). Potency and selectivity were improved by increasing the size of the P1 substituent from a phenyl to biphenyl moiety, yielding MPI-5 (Table 1) as a 5-nM inhibitor of Pfβ5 and a 21-nM inhibitor of 3D7 culture growth. MPI-5 exhibited 17-fold enzymatic selectivity and 125-fold cellular selectivity. We had identified a series of inhibitors to explore.
Having established increased potency with the P1 biphenyl substituent, we examined the effect of different substituents at both the P1 and P2 positions. Alteration of the terminal phenyl ring with substituents intended to increase solubility, such as pyridine (MPI-8) and pyrazole (MPI-9), did not improve either the enzymatic or cellular inhibition potency. Interestingly, although the observed cellular selectivity increased for MPI-8, the enzymatic selectivity did not.
We next explored the effect of modifications to the alkyl pyrrolidine on potency and selectivity. Increasing the ring size from pyrrolidine to piperidine (MPI-10) resulted in a decrease in selectivity with little change in enzyme potency; however, the potency against 3D7 increased—from 21 nM for MPI-5 to 9 nM for MPI-10. Opening the ring and introducing a methyl ether moiety to extend into the S3 pocket provided little improvement of either enzymatic or cellular potency and seemed to be less selective in cells (MPI-11 and MPI-12). We explored the effect of stereochemistry in the S2/S3 pocket with an N-isopropyl analog of the piperidine MPI-10 (MPI-13 and MPI-14). MPI-13 exhibited increased activity against both Hsβ5c and Pfβ5 compared with MPI-14 as well as improved biochemical selectivity (Table 1). It exhibited improved potency compared with both MPI-14 and MPI-5 against the Pf culture (Table 1).
In summary, each of the biphenyl amino-amide boronates exhibited enhanced activity against Pf 3D7 cultures compared with MPI-1, achieving values similar to or better than those for bortezomib, while the selectivity for Pf compared with HepG2 was greatly improved relative to bortezomib and MPI-1 (Table 1). A plot of activity in the Pfβ5 assay compared with the 3D7 assay reveals a moderate correlation (SI Appendix, Fig. S2A), suggesting that factors such as membrane permeability also affect cellular efficacy, as reported for inhibitors of the human proteasome (18).
Analysis of Activities against Different Constitutive and ImmunoProteasome Subunits.
We assessed the activity of selected inhibitors against the Pf β1 and β2 and Hs β1c and β2c subunits, using Ac-nLPnLD-AMC and Ac-WLR-AMC as substrates. While bortezomib exhibits moderate to strong inhibition of the β1 and β2 subunit activities of both Pf20S and Hs20Sc, all other compounds assessed exhibit no or very low inhibition of these activities (SI Appendix, Table S3). In immune cells, the three active constitutive proteasome subunits are replaced by “immuno” subunits to form immunoproteasomes. Activities against Hs20S β1i, β2i, and β5i were assessed as previously described (19). Our amino-amide proteasome inhibitors exhibited little activity against Hs20S β1i and β2i (SI Appendix, Table S3) but higher activity against Hs20S β5i (Table 1). It is important to note that selective inhibition of β5i by peptide boronates is not cytotoxic to immune cell lines (20); however, it can inhibit human T cell activation (21) and should be monitored during further development of this class of proteasome inhibitors.
We employed a fluorescent activity probe (BMV037) that contains an epoxyketone peptide scaffold based on the irreversible covalent inhibitor, carfilzomib (13), synthesized as previously described (11). Purified Pf20S was exposed to different peptide boronates (10 μM) before residual proteasome activity was detected by incorporation of BMV037. In the absence of inhibitor, the β5 and β2 subunits are labeled most efficiently, with some label incorporated into the β1 subunit (SI Appendix, Fig. S2B, left lane), consistent with our previous report (11). All of the inhibitors decrease labeling of the Pfβ5 subunit, consistent with preferential inhibition of the chymotrypsin-like activity (SI Appendix, Fig. S2B). An increase in labeling of the Pfβ2 subunit is observed, potentially due to a small allosteric activation of the Pfβ2 active sites when inhibitor is bound in the Pfβ5 active sites, as has been reported for the human enzyme (22).
Differences in Binding Kinetics Underpin Specificity.
Peptide boronates inhibit proteasome activity by binding into one or more of the enzyme’s six active sites. The binding is stabilized by formation of a coordinate covalent (dative) bond between the active site nucleophile (the Oγ atoms of the Thr1 residues in β1, β2, and β5 subunits) and the boron atom in the inhibitors (23–25). The formation of the dative bond is slow relative to the collision frequency giving rise to time-dependent (i.e., slow binding) behavior seen in progress curves of in vitro assays (SI Appendix, Fig. S2 C and D). On the other hand, once formed, the dative bonds can be quite stable resulting in long inhibitor residence times (several minutes to several hours) and potent proteasome inhibition.
We monitored progress curves of Ac-WLA-AMC hydrolysis (β5 activity) upon addition of inhibitors. The rate constant for approach to steady-state inhibition was measured as a function of inhibitor concentration for MPI-5 and the potent Hs20S inhibitor, bortezomib (SI Appendix, Figs. S2 C and D and S3 and Table S4). Inhibition of Hsβ5c by MPI-5 is weaker than by bortezomib, primarily because the dative bond formation is 17-fold slower (SI Appendix, Table S4). Additionally, the residence time of MPI-5 in the β5c active site of Hs20S is approximately one-half that of bortezomib (SI Appendix, Table S4). The differential kinetics parameters likely underpin the decreased toxicity of MPI-5 compared with bortezomib against human cell lines.
By contrast, the rate of dative bond formation for MPI-5 with Pf20S β5 is higher than for bortezomib and similar to that for bortezomib reacting with Hs20S β5c (SI Appendix, Figs. S2 C and D and S3 and Table S4). The data are consistent with the 14-fold-lower inhibition of Hs20S β5c activity and the 2.2-fold more potent inhibition of Pf20S β5 displayed by MPI-5 compared with bortezomib (Table 1), likely underpinning the 125-fold cellular selectivity (Table 1).
Exemplar Amino-Amide Boronates Exhibit a Fast Killing Rate, Cross-Species Anti-Plasmodial Activity, including against Drug-resistant Strains, Field Strains, and Extraerythrocytic and Transmissible Forms, as well as Synergy with Artemisinin.
Different members of the compound set, with a range of potencies, were assessed in a range of assays, based on availability of compound stocks and assay slots. We chose MPI-11 for a broader analysis of activity against different strains and at different life stages. We assessed the in vitro parasite reduction ratio using a standardized method (26). When employed at 10 times the inhibitory IC50 value, MPI-11 exhibited a fast killing rate, similar to that observed for artesunate (SI Appendix, Fig. S4A).
Demonstrating activity against known resistant parasites is critical when considering development of malaria therapeutic agents. MPI-11 was tested against various Pf laboratory strains to ensure activity against parasites resistant to historical antimalarial drugs (27). Encouragingly, full activity was maintained against all strains tested, irrespective of their resistance profile and geographical origin, with respect to the sensitive strain NF54 (SI Appendix, Table S5). Similarly, no cross-resistance was observed with representative antimalarial agents currently in development (SI Appendix, Table S5). MPI-10 showed good potency against Brazilian field isolates of Pf and Plasmodium vivax asexual blood stages (SI Appendix, Table S5). Activity against transmissible male gametes was assessed using the dual gamete formation assay (PfDGFA) (28). Treatment of mature (stage V) gametocytes with MPI-11 and MPI-13 inhibited male exflagellation (SI Appendix, Table S5). MPI-11 inhibited the exoerythrocytic stage of the rodent malaria parasite, Plasmodium berghei, in a human hepatoma cell line (29), as measured by decreases in sporozoite infection and liver schizont biomass, suggesting activity early in liver-stage development, as well as preventing hepatic merozoite formation. MPI-11 exhibited limited toxicity against HepG2 (SI Appendix, Table S5). MPI-11 also inhibited the development of Pf NF54 schizonts in primary human hepatocytes (30), and hepatic toxicity was not apparent (SI Appendix, Table S5).
Bortezomib and MPI-1 were previously shown to exhibit strong synergy with dihydroxyartemisinin (DHA) (9). Similarly, MPI-5 exerted a pronounced synergistic interaction with DHA against the early ring stages of the DHA-resistant, K13 mutant isolate, Cam3.IIR539T (SI Appendix, Fig. S4B).
Parasites Selected In Vitro for Resistance to MPI-12 Harbor a Pf20Sβ5 Met45Ile Mutation.
We exposed the Dd2-B2 clone (over a range of 2 × 105 to 6 × 107 parasites) at 4.2 times the MPI-12 IC50(72h) concentration (SI Appendix, SI Methods). No resistant parasites recrudesced. The selection was repeated with a 2 × 109 inoculum. Parasites recrudesced on day 20 and were cloned by limiting dilution. Dose–response profiling of four clones revealed IC50(72h) shifts of ∼14- to ∼19-fold (SI Appendix, Fig. S4C and Table S5). Whole-genome sequencing identified a Met45Ile mutation in the mature form of Pf20S β5 in all clones (SI Appendix, Tables S6 and S7). The same mutation was previously observed in parasites selected for resistance to bortezomib (11). No copy number variants were evident in any of the selected clones. The minimum inoculum for resistance that, for now, has been estimated to be more than 6 × 107 and less than 2 × 109 indicates a low propensity for resistance to MPI-12 (31). Despite the difficulty of obtaining resistant parasites, we observed no substantive difference in the growth rate of wild-type and Met45Ile mutant parasites over a period of 21 d, as measured using a mixed culture competition assay (32) (SI Appendix, Fig. S4E).
MPI-12 dose–response assays with Cam3.II and V1/S gene-edited lines expressing K13 wild-type or C580Y alleles (33) confirmed no impact of K13 C580Y-mediated artemisinin resistance on MPI-12 potency (SI Appendix, Fig. S4D and Table S5). We extended the susceptibility assays to include parasite lines resistant to the covalent vinyl sulfone proteasome inhibitors, WLL-vs. and WLW-vs. (10, 33). Cross-resistance was observed with V1/S parasites resistant to WLL-vs. and harboring the β6 A117V mutation, showing an eightfold IC50 increase relative to Dd2, indicating that mutations at the β5/β6 interface can interfere with the binding of covalent inhibitors of β5 activity. No cross-resistance was observed with Cam3.II parasites harboring the β5 A20S mutation. The β2 C31F mutant obtained following WLW-vs. selection in V1/S parasites showed a threefold lower IC50 for MPI-12 (SI Appendix, Fig. S4D and Table S5). This β2 C31F mutant was earlier observed to be hypersensitive to WLL-vs. (33).
Amino-Amide Boronates Exhibit Good Oral Bioavailability and Efficacy in an In Vivo Model of Pf Malaria.
Compounds from this series exhibit favorable physico-chemical and in vitro absorption, distribution, metabolism, and excretion (ADME) properties, including good kinetic solubility, Caco2 permeability, and metabolic stability (SI Appendix, Table S2). Four compounds from the series, with different substituents at the P2 position (MPI-5, MPI-10, MPI-11, and MPI-13), were profiled in rat PK studies (at 1 mg/kg per os [P.O.] and intravenously [IV] as the citrate esters). All four compounds showed promising bioavailability ranging from 50 to 100%, with Cmax values between 280 and 1,400 nM and clearance ranging from 25 to 42% of liver blood flow (SI Appendix, Fig. S5A and Table S8). The half-lives for these compounds were moderate (3.2 to 5.3 h) as were the volumes of distribution (1.3 to 4.2 L/kg). Excretion of the parent for all compounds in the 0- to 24-h urine sample was <1.56% of the dose, indicating that renal clearance is a minor clearance mechanism.
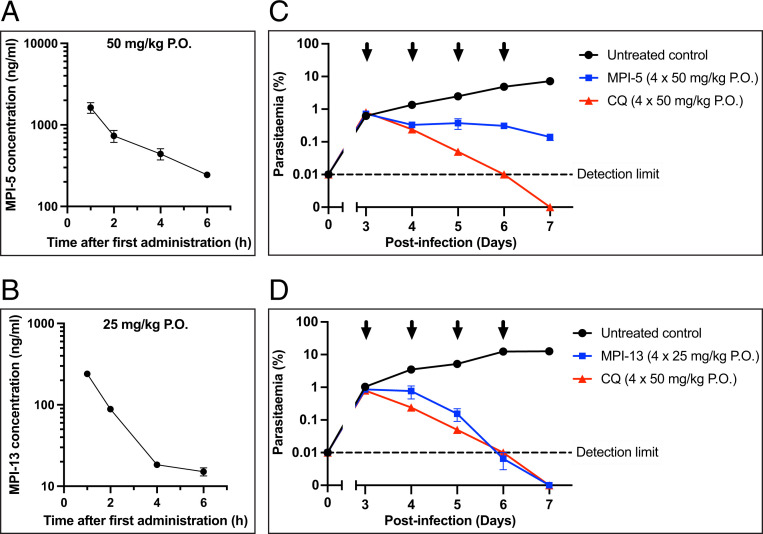
MPI-5 and MPI-13 were assessed for efficacy in vivo using a severe combined immune deficiency (SCID) mouse model of human Pf (NODscid/β2m−/−) in which mice are engrafted with human red blood cells (34). When dosed orally for four consecutive days at 50 mg/kg, MPI-5 demonstrated good oral exposure (4.6 μM [1,630 ng/mL] Cmax at 1 h; Fig. 2A and SI Appendix, Table S9) and good efficacy (∼98% parasite reduction in comparison to untreated control mice; Fig. 2B). When dosed orally at a lower dose (25 mg/kg QDx4), MPI-13 exhibited improved efficacy against Pf in the SCID mouse model (Fig. 2 C and D), despite a lower-observed exposure (609 nM [240 ng/mL] Cmax at 1 h).
Fig. 2.
Amino amide boronates exhibit potent activity against Pf in vivo after oral dosing. (A and B) PK profiles over the first day for SCID mice engrafted with human red blood cells infected with Pf following treatment with (A) MPI-5 at 50 mg/kg P.O. or (B) MPI-13 for 4 d at 25 mg/kg P.O. (C and D) Therapeutic efficacy of (C) MPI-5 and (D) MPI-13 in the SCID mouse Pf model, dosed at 50 and 25 mg/kg P.O., respectively, in comparison with gold standard antimalarial, chloroquine (CQ), dosed P.O. at 50 mg/kg.
The parasite clearance rate was similar to that for chloroquine, and parasitemia was reduced below the detection level in both mice at day 7. No apparent toxicity was observed. The blood concentrations for both compounds remain above the 50% lethal dose (LD50) values for MPI-5 and MPI-13 killing of 3D7 in vitro (21 nM and 11 nM, respectively; Table 1) for at least the first 6 h of treatment. The enhanced activity of MPI-13 compared with MPI-5 may in part reflect its higher cellular potency, although further work is needed to understand why MPI-13 achieves parasite clearance, while a low parasitemia (0.14%) persists following treatment with MPI-5.
We validated on-target activity by showing that MP1-5 and MP-13 exhibit similar levels of cross-resistance to the Met45Ile mutant line (SI Appendix, Fig. S5B) and have similar inhibitory effects on proteasome activity in Pf-infected red blood cells, as monitored using a degradation domain-linked (DD-GFP) reporter (11) (SI Appendix, Fig. S5C).
Cryo-electron microscopy (EM) Structures of Hs20S and Pf20S in Complex with MPI-5 and Bortezomib Reveal the Molecular Basis for Selectivity.
We determined the cryo-EM structures of Pf and human 20S proteasomes in complex with MPI-5 and bortezomib to understand the molecular basis for the enhanced specificity in the biochemical assays. Pf20S was purified from infected red blood cells using a method that incorporates a heparin-affinity chromatography step (35). Hs20Sc was characterized as described previously (36). Protein samples (0.15 mg/mL; ∼0.2 μM) were incubated with MPI-5 or bortezomib at a final concentration of 10 μM to maximize occupancy of the binding sites. The structures of Pf20S in complex with MPI-5 and bortezomib and Hs20S in complex with MPI-5 were solved to resolutions of 3.4 Å, 3.1 Å, and 3.4 Å, respectively (refer to SI Appendix, Fig. S6 A, D, and G for Fourier shell correlation [FSC] curves and SI Appendix, Table S10 for cryo-EM statistics; refer to SI Appendix, Fig. S6 B, E, and H for density maps).
In the Pf20S/MPI-5 and Hs20S/MPI-5 complexes, continuous density was observed extending from the N-terminal threonine residues of the β5 subunits, corresponding to the expected position of MPI-5 (SI Appendix, Fig. S6 C and F). No significant density was observed in the substrate binding pockets of the β1 and β2 active sites, suggesting low or no occupancy of these sites by the inhibitor. Superimposition of the apo-Pf20S structure with the inhibited MPI-5 complex structure revealed no significant global structural changes, with an overall Cα rmsd between the two structures of 0.56 Å. Similarly, no shifts in the positions of side chains surrounding the β5 active site are apparent upon binding of the inhibitor.
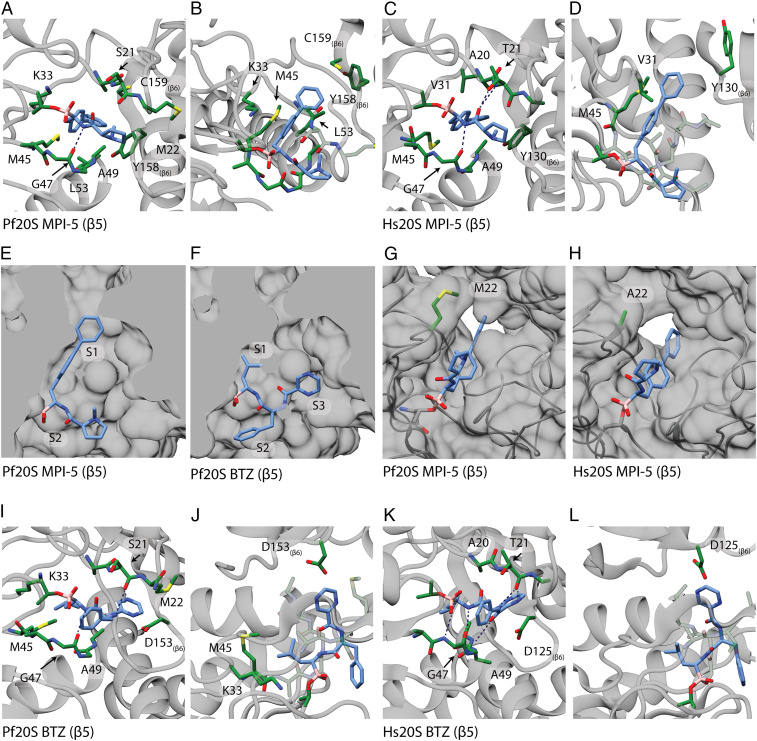
Apart from the covalent bond to the active site threonine, the interactions of MPI-5 with the Pf20S β5 active site appear to comprise predominantly hydrophobic interactions. These involve 11 residues lining the active site, particularly around the biphenyl rings (Fig. 3 A and B; refer to the LigPlot map in SI Appendix, Fig. S7A). The hydrophobic interactions are facilitated by the close fit of MPI-5 into the S1 binding pocket of the β5 active site (Fig. 3E and SI Appendix, Fig. S7 A and B). The Met22 sidechain forms a ledge over the S2 binding pocket and interacts with the methyl pyrrolidine group of MPI-5 (Fig. 3G). These hydrophobic interactions likely underpin the substantive gain in potency when comparing the biphenyl (e.g., MPI-5) and monophenyl (e.g., MPI-7) compounds. The inhibitor also makes a potential hydrogen bond to Gly47 (Fig. 3A and SI Appendix, Fig. S7A). The network of interactions with the Pf20S β5 active site appears to position MPI-5 favorably to form and maintain the covalent bond to the active site threonine. We note that the Met45 side chain runs parallel to and makes close hydrophobic interactions with the biphenyl rings of MPI-5 (Fig. 3B and SI Appendix, Fig. S7A). The importance of this residue is consistent with our finding that parasites selected for resistance to MPI-12 exhibit an amino acid change at this residue (Met45Ile). Modeling suggests that this change would significantly alter the shape of the S1 binding pocket (SI Appendix, Fig. S7B). We also note that Met22 and Met45 are replaced by Ser22 and Arg45 in the β1 active site and Glu22 and Gly45 in the β2 active site (SI Appendix, Fig. S8 E and M). These differences may underlie the specificity of MPI-5 for the β5 active site.
Fig. 3.
Structures of Pf20S and Hs20Sc β5 active sites with bound MPI-5 or bortezomib. Pf20S β5 (A and B) and Hs20S β5c (C and D) are shown as side (A and C) and top (B and D) views, with interacting residues (green) around the covalently bound MPI-5 (blue). (E and F) Cut-away showing solvent-accessible surface of the Pf20S β5 active site illustrating the different poses adopted by MPI-5 (E) and bortezomib (F). Substrate binding pockets, S1, S2, and S3 are marked. (G and H) Met22 creates a hydrophobic ledge over the ligand binding site in Pf20S β5, while Ala22 permits more solvent access in the Hs20S β5c active site around the covalently bound MPI-5. Pf20S β5 (I and J) and Hs20S β5c (K and L) are shown as side (I and K) and top (J and L) views ,with interacting residues (green) around the covalently bound bortezomib (blue).
MPI-5 is positioned in a similar orientation in the Hs20S β5c site (SI Appendix, Fig. S7D); however, it makes fewer close hydrophobic interactions with the β5c active site (SI Appendix, Fig. S7C), because of differences in amino acids lining the S1 pocket (Fig. 3 C and D) and the relatively open S2 pocket due to an alanine residue at position 22 (Fig. 3H) rather than methionine. Thus, MPI-5 achieves selectivity for the plasmodium enzyme via a decreased interaction network with the Hs20S β5c active site and absence of binding to Hs20S β1c and β2c active sites, while maintaining binding to Pf20S β5 via hydrophobic interactions.
For the Pf20S/bortezomib complex, continuous density extends from the N-terminal threonine residues of the β1, β2, and β5 subunits (SI Appendix, Figs. S6I and S8 D and L). Bortezomib appears to make hydrogen-bonding interactions with Thr21 and Gly47 in the Pf20S β5 subunit (Fig. 3 I and J and SI Appendix, Fig. S7E). Interactions with the S1 and S3 binding pockets (Fig. 3F) involve primarily the P1 leucine side chain and the P3 pyrazine group respectively, while the S2 binding pocket is shallower and more open, leaving the P2 phenylalanine side chain more solvent exposed (Fig. 3F). Thus, the network of hydrophobic interactions is smaller for bortezomib than for MPI-5 (Fig. 3 I and J and SI Appendix, Fig. S7E). This may decrease the bortezomib residence time and underpin the weaker inhibition of Pf20S β5 activity by bortezomib compared with MPI-5 (Table 1). We note that while the biphenyl group of the MPI-5 P1 residue penetrates deeply into the S1 pocket, the isopropyl group of the P1 residue of bortezomib is less engaged in this site and thus is less affected by the Arg45 /Gly47 residues in the S1 binding pockets of the β1 and β2 active sites. Nonetheless, the density is weaker at the Pf20S β1 and β2, suggesting reduced occupancy of bortezomib at these active sites relative to Pf20S β5.
We compared the interaction map for Pf20S/bortezomib with that for the published Hs20S/bortezomib structure (Protein Data Base [PDB] ID: 5LF3) (24, 37). The bortezomib binding pose in Hs20S β5c is similar to that for Pf20S β5, but a more extensive H-bond network (involving Thr21, Gly47, Ala49 and Thr1) appears to stabilize its binding to Hs20S β5c (Fig. 3 K and L and SI Appendix, Fig. S7 G and H). This is consistent with the lower kon value for MPI-5 and consequently much higher potency of inhibition of Hs20S β5c activity by bortezomib compared with MPI-5 (Table 1). Similarly, a network of potential H bonds appears to stabilize the binding of bortezomib into the Hs20S β1c and β2c active sites (SI Appendix, Fig. S8 F, H, N, and P), consistent with inhibition of these activities by bortezomib (SI Appendix, Table S3). We note that bortezomib binding to the Pf20S β1 and β2 subunits (SI Appendix, Fig. S8 A, C, I, and K) appears to be stabilized by fewer hydrogen bonds than for the Hs20S β1c and β2c subunits, consistent with the more potent activity of bortezomib against the human enzyme.
In summary, using a combination of structure-/activity-guided synthetic chemistry and cryo-EM–based structure determination, we have demonstrated that effective inhibition of Pf20S activity can be achieved by employing a biphenyl moiety at the P1 position of a peptide boronate, thereby exploiting a hydrophobic pocket in the Pf20S β5 active site. Selectivity is achieved because the Pf20S β5 pocket provides better complementarity to the biphenyl group than any of the Hs20S active sites. In particular, decreased activity against Hs20S β5c, coupled with no activity against Hs β2c, represents a signature that permits selective killing of Pf compared with a mammalian cell line. The enhanced affinity offered by the hydrophobic interactions enabled shortening of the peptide backbone, which resulted in oral bioavailability. We demonstrate that exemplar compounds exhibit efficacy in an in vivo model of Pf malaria. We show that this series of amino-amide boronates 1) has potent antimalarial activity across the parasite lifecycle, giving potential for use in TCP1, 4, and 5; 2) are fast-acting; 3) have equipotent activity against the major pathological strains (P. vivax and Pf); 4) have high selectivity for Pf cultures compared with a human cell line; 5) have good selectivity over the human enzyme; and 6) exhibit low propensity for resistance development compared to other classes of anti-malarial agents.
MPI-13 is an example of an orally bioavailable antimalarial proteasome inhibitor, with in vivo efficacy. Inhibition of human T cell activation should be monitored during further development of this class of proteasome inhibitors; and further improvement in in vivo half-life will be needed to permit single-dose efficacy, a highly desirable aspect of the MMV Target Product Profile. Nevertheless, the series demonstrates the potential for a proteasome inhibitor to be used in treatment, transmission-blocking, and chemoprophylaxis scenarios, especially when resistance to artemisinin limits the efficacy of existing combinations.
Methods
Proteasome and Mouse Studies.
Pf proteasome enrichment for use in the enzyme assays was as previously described (11). The Pf20S proteasome was subjected to additional chromatographic steps for cryo-EM studies (35). Proteasome activity assays were based on previously described methods (11). Pf Pf3D70087/N9 in NODscidIL2Rγnull mice engrafted with human red blood were infected intravenously with Pf (Pf3D70087/N9). Antimalarial efficacy of the proteasome inhibitors in vivo was assessed, following oral administration, using an adaptation of a previously described protocol (38).
Additional materials and methods are provided in SI Appendix, including information on chemical synthesis and characterization, analyses of proteasome activity, in vitro assays of potency against laboratory and field malaria parasites, gametocytes and liver stages, details of assays in the SCID mouse Pf model, analysis of propensity for resistance development, ADME/PK, cryo-EM, and structure determination.
Ethics Approvals.
Human biological samples were sourced ethically, and their research use was in accordance with the terms of the informed consent, as approved by the Ethics Committee of the Tropical Medicine Research Center (CAAE 58738416.1.0000.0011). All animal studies were ethically reviewed and carried out in accredited facilities in accordance with the relevant country’s directives and the institution’s Policy on the Care, Welfare and Treatment of Animals.
Supplementary Material
Acknowledgments
We thank the following colleagues: HepG2 toxicity assays: Lorna Campbell, Nicole Mutter, and Irene Hallyburton, University of Dundee; Anirban Koley, Pranab Dhar, and Partha Mukherjee, TCG Lifesciences;. Pf and P. vivax ex vivo patient isolate schizonts from the Brazilian Amazon: Dhelio B. Pereira and Carolina Bioni; liver-stage in vitro assays: Rianne van der Laak and Angelika Sturm, TropIQ Health Sciences; support with the SCID mouse and panel assays: Ursula Lehmann and Christian Scheurer, Swiss Tropical and Public Health Institute; support with the bioanalytical determination and PK evaluation in mouse blood samples: Anita Kress, Iris Barme, Mark Enzler, and Christoph Siethoff, Swiss BioQuant; assistance with whole-genome sequencing and analysis: Sachel Mok and Anne-Catrin Uhlemann, Columbia University; cryo-EM data acquisition: Andrew Leis, University of Melbourne; and useful advice: Steven Langston and Yongbo Hu, Takeda Pharmaceuticals. The cryo-EM work was performed in the Bio21 Ian Holmes Imaging Centre. We thank the Global Health Innovative Technology Fund (H2019-101), the Australian National Health and Medical Research Council, MMV, and Millennium Pharmaceuticals, a wholly owned subsidiary of Takeda Pharmaceuticals Company Limited, for research support. J.B. is supported by an Investigator Award from Wellcome (100993/B/13/Z) and MMV RD-08-2800. M.W.P. is a National Health and Medical Research Council of Australia Research Fellow (APP1117183) and Investigator (APP1194263). L.T. is supported by an Australian Research Council Laureate Fellowship. Funding from the Victorian Government Operational Infrastructure Support Scheme to St Vincent’s Institute is acknowledged. D.A.F. kindly acknowledges funding support from NIH (R33 AI127581). E.A.W. and S.O. are supported by MMV RD-12-0096. R.V.C.G. thanks the Sao Paulo Research Foundation (FAPESP), for funding the research (Centros de Pesquisa, Inovação e Difusão [CEPID] grants 2013/07600-3 and 2020/12904-5). A.C.C.A. and R.V.C.G. are supported by MMV RD-16-1066. The Talos Artica electron microscope was purchased with the help of an Australian Research Council Linkage Infrastructure, Equipment and Facilities grant led by M.W.P.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2107213118/-/DCSupplemental.
Data Availability
The following structures have been deposited in the PDB and Electron Microscopy Data Bank (EM-DB): Pf20S/bortezomib: 7LXT/EMD-23574; Pf20S/MPI-5: 7LXU/EMD-23575; and Hs20S/MPI-5: 7LXV/EMD-23576. All other study data are included in the article and/or supporting information.
Change History
September 24, 2021: The license for this article has been updated.
References
- 1.World Health Organisation , World Malaria Report 2020 (World Health Organization, 2020). [Google Scholar]
- 2.van der Pluijm R. W., et al., Determinants of dihydroartemisinin-piperaquine treatment failure in Plasmodium falciparum malaria in Cambodia, Thailand, and Vietnam: A prospective clinical, pharmacological, and genetic study. Lancet Infect. Dis. 19, 952–961 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bergmann C., et al., Increase in Kelch 13 Polymorphisms in Plasmodium falciparum, Southern Rwanda. Emerg. Infect. Dis. 27, 294–296 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li H., et al., Validation of the proteasome as a therapeutic target in Plasmodium using an epoxyketone inhibitor with parasite-specific toxicity. Chem. Biol. 19, 1535–1545 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tschan S., et al., Broad-spectrum antimalarial activity of peptido sulfonyl fluorides, a new class of proteasome inhibitors. Antimicrob. Agents Chemother. 57, 3576–3584 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kirkman L. A., et al., Antimalarial proteasome inhibitor reveals collateral sensitivity from intersubunit interactions and fitness cost of resistance. Proc. Natl. Acad. Sci. U.S.A. 115, E6863–E6870 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yoo E., et al., Defining the determinants of specificity of Plasmodium proteasome inhibitors. J. Am. Chem. Soc. 140, 11424–11437 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ng C. L., Fidock D. A., Bogyo M., Protein degradation systems as antimalarial therapeutic targets. Trends Parasitol. 33, 731–743 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dogovski C., et al., Targeting the cell stress response of Plasmodium falciparum to overcome artemisinin resistance. PLoS Biol. 13, e1002132 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li H., et al., Structure- and function-based design of Plasmodium-selective proteasome inhibitors. Nature 530, 233–236 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xie S. C., et al., Target validation and identification of novel boronate inhibitors of the Plasmodium falciparum proteasome. J. Med. Chem. 61, 10053–10066 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhan W., et al., Development of a highly selective Plasmodium falciparum proteasome inhibitor with anti-malaria activity in humanized mice. Angew. Chem. Int. Ed. Engl. 60, 9279–9283 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li H., et al., Assessing subunit dependency of the Plasmodium proteasome using small molecule inhibitors and active site probes. ACS Chem. Biol. 9, 1869–1876 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Britton M., et al., Selective inhibitor of proteasome’s caspase-like sites sensitizes cells to specific inhibition of chymotrypsin-like sites. Chem. Biol. 16, 1278–1289 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Burrows J. N., et al., New developments in anti-malarial target candidate and product profiles. Malar. J. 16, 26 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kreidenweiss A., Kremsner P. G., Mordmüller B., Comprehensive study of proteasome inhibitors against Plasmodium falciparum laboratory strains and field isolates from Gabon. Malar. J. 7, 187 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Scalzulli E., Grammatico S., Vozella F., Petrucci M. T., Proteasome inhibitors for the treatment of multiple myeloma. Expert Opin. Pharmacother. 19, 375–386 (2018). [DOI] [PubMed] [Google Scholar]
- 18.Kisselev A. F., Goldberg A. L., Proteasome inhibitors: From research tools to drug candidates. Chem. Biol. 8, 739–758 (2001). [DOI] [PubMed] [Google Scholar]
- 19.Blackburn C., et al., Characterization of a new series of non-covalent proteasome inhibitors with exquisite potency and selectivity for the 20S beta5-subunit. Biochem. J. 430, 461–476 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ladi E., et al., Design and evaluation of highly selective human immunoproteasome inhibitors reveal a compensatory process that preserves immune cell viability. J. Med. Chem. 62, 7032–7041 (2019). [DOI] [PubMed] [Google Scholar]
- 21.Zhan W., et al., Structure-activity relationships of noncovalent immunoproteasome β5i-selective dipeptides. J. Med. Chem. 63, 13103–13123 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lightcap E. S., et al., Proteasome inhibition measurements: Clinical application. Clin. Chem. 46, 673–683 (2000). [PubMed] [Google Scholar]
- 23.Adams J., et al., Potent and selective inhibitors of the proteasome: Dipeptidyl boronic acids. Bioorg. Med. Chem. Lett. 8, 333–338 (1998). [DOI] [PubMed] [Google Scholar]
- 24.Groll M., Berkers C. R., Ploegh H. L., Ovaa H., Crystal structure of the boronic acid-based proteasome inhibitor bortezomib in complex with the yeast 20S proteasome. Structure 14, 451–456 (2006). [DOI] [PubMed] [Google Scholar]
- 25.Kupperman E., et al., Evaluation of the proteasome inhibitor MLN9708 in preclinical models of human cancer. Cancer Res. 70, 1970–1980 (2010). [DOI] [PubMed] [Google Scholar]
- 26.Linares M., et al., Identifying rapidly parasiticidal anti-malarial drugs using a simple and reliable in vitro parasite viability fast assay. Malar. J. 14, 441 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Snyder C., Chollet J., Santo-Tomas J., Scheurer C., Wittlin S., In vitro and in vivo interaction of synthetic peroxide RBx11160 (OZ277) with piperaquine in Plasmodium models. Exp. Parasitol. 115, 296–300 (2007). [DOI] [PubMed] [Google Scholar]
- 28.Delves M. J., et al., A high throughput screen for next-generation leads targeting malaria parasite transmission. Nat. Commun. 9, 3805 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Swann J., et al., High-throughput luciferase-based assay for the discovery of therapeutics that prevent malaria. ACS Infect. Dis. 2, 281–293 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schalkwijk J., et al., Antimalarial pantothenamide metabolites target acetyl-coenzyme A biosynthesis in Plasmodium falciparum. Sci. Transl. Med. 11, eaas9917 (2019). [DOI] [PubMed] [Google Scholar]
- 31.Duffey M., et al., Assessing risks of Plasmodium falciparum resistance to select next-generation antimalarials. Trends Parasitol. 37, 709–721 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ross L. S., et al., Emerging Southeast Asian PfCRT mutations confer Plasmodium falciparum resistance to the first-line antimalarial piperaquine. Nat. Commun. 9, 3314 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stokes B. H., et al., Covalent Plasmodium falciparum-selective proteasome inhibitors exhibit a low propensity for generating resistance in vitro and synergize with multiple antimalarial agents. PLoS Pathog. 15, e1007722 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Angulo-Barturen I., et al., A murine model of falciparum-malaria by in vivo selection of competent strains in non-myelodepleted mice engrafted with human erythrocytes. PLoS One 3, e2252 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xie S. C., et al., The structure of the PA28-20S proteasome complex from Plasmodium falciparum and implications for proteostasis. Nat. Microbiol. 4, 1990–2000 (2019). [DOI] [PubMed] [Google Scholar]
- 36.Williamson M. J., et al., Comparison of biochemical and biological effects of ML858 (salinosporamide A) and bortezomib. Mol. Cancer Ther. 5, 3052–3061 (2006). [DOI] [PubMed] [Google Scholar]
- 37.Schrader J., et al., The inhibition mechanism of human 20S proteasomes enables next-generation inhibitor design. Science 353, 594–598 (2016). [DOI] [PubMed] [Google Scholar]
- 38.Jiménez-Díaz M. B., et al., Improved murine model of malaria using Plasmodium falciparum competent strains and non-myelodepleted NOD-scid IL2Rgammanull mice engrafted with human erythrocytes. Antimicrob. Agents Chemother. 53, 4533–4536 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The following structures have been deposited in the PDB and Electron Microscopy Data Bank (EM-DB): Pf20S/bortezomib: 7LXT/EMD-23574; Pf20S/MPI-5: 7LXU/EMD-23575; and Hs20S/MPI-5: 7LXV/EMD-23576. All other study data are included in the article and/or supporting information.