Abstract
Background
KRAS is one of the most frequently mutated oncogenes in colorectal cancer (CRC). Recently, a novel therapy targeting KRAS G12C mutation has demonstrated promising activities for corresponding advanced solid tumors, including metastatic CRC (mCRC). However, the prognostic impact of the KRAS G12C mutation remains unclear in patients with mCRC.
Materials and Methods
We retrospectively reviewed medical records of patients with mCRC who received first‐line chemotherapy between January 2005 and December 2017 at four large oncology facilities in Japan. Survival outcomes were compared between patients with KRAS G12C and those with non‐G12C mutations.
Results
Among 2,457 patients with mCRC, 1,632 met selection criteria, and of these, 696 had KRAS exon 2 mutations, including 45 with KRAS G12C mutation tumors. Patient characteristics were not significantly different between the KRAS G12C and non‐G12C groups. At a median follow‐up of 64.8 months, patients with the KRAS G12C mutation showed significantly shorter first‐line progression‐free survival (PFS; median, 9.4 vs. 10.8 months; p = .015) and overall survival (OS; median, 21.1 vs. 27.3 months; p = .015) than those with non‐G12C mutations. Multivariate analysis also showed that KRAS G12C mutation was significantly associated with shorter PFS (hazard ratio [HR], 1.43; 95% confidence interval [CI], 1.04–1.96, p = .030) and OS (HR, 1.42; 95% CI, 1.01–2.00; p = .044).
Conclusion
We demonstrate that, compared with non‐G12C mutations, KRAS G12C mutation is significantly correlated with shorter first‐line PFS and OS. These findings indicate the relevance of a stratified treatment targeting KRAS G12C mutation in mCRC.
Implications for Practice
Among patients with KRAS exon 2 mutated metastatic colorectal cancer (mCRC), median progression‐free survival (PFS) and overall survival (OS) were 9.4 and 21.1 months, respectively, for G12C mutation and 10.8 and 27.3 months, respectively, for patients with non‐G12C mutations, indicating significantly shorter PFS (hazard ratio [HR], 1.47; 95% confidence interval [CI], 1.08–2.01; p = .015) and OS (HR, 1.50; 95% CI, 1.08–2.08; p = .015) in patients with G12C mutation than in those with non‐G12C mutations. Furthermore, multivariate analysis showed that KRAS G12C mutation was independently associated with shorter first‐line PFS and OS. Thus, these findings underscore the relevance of a stratified treatment targeting KRAS G12C mutation in mCRC.
Keywords: KRAS, G12C, Colorectal cancer, Chemotherapy, Prognosis
Short abstract
KRAS is one of the most frequently mutated oncogenes in colorectal cancer. This article evaluates the prognostic impact of KRAS G12C mutation in metastatic colorectal cancer using real‐world data.
Introduction
The rat sarcoma (RAS) family of proto‐oncogenes, including Kirsten rat sarcoma (KRAS), neuroblastoma rat sarcoma (NRAS), and Harvey sarcoma, plays a central role in many human cancers [1], and KRAS is one of the most frequently mutated oncogenes in colorectal cancer (CRC). The various RAS mutation subtypes show specific functional profiles, and in cell lines, these mutation subtypes exhibit different patterns of transformation, aggressiveness, and/or drug response [2, 3, 4, 5]. RAS mutations are undruggable targets because of the presence of large, deep‐seated hydrophobic pockets in their molecular structure that are difficult to target with small‐molecule chemistry [6]; however, the development of small‐molecule inhibitors that selectively bind to a newly discovered allosteric regulatory site in the G12C KRAS mutant is underway [7].
A phase I trial of AMG 510, which is the first‐in‐class KRAS G12C inhibitor, achieved a partial response in 30% of patients with non‐small cell lung cancer and in 7.1% of patients with CRC [8]. Additionally, an ongoing clinical trial is testing a combination of the KRAS G12C inhibitor and anti–epidermal growth factor receptor (EGFR) antibody [9].
Thus, the prognostic impact of KRAS G12C mutation in metastatic CRC (mCRC) is important for the development of novel, clinically effective agents; however, there have been few reports on the prognostic impact of KRAS G12C mutation in mCRC. Therefore, the aim of this study was to evaluate the prognostic impact of KRAS G12C mutation in mCRC using real‐world data.
Materials and Methods
Data Acquisition
Data were collected from patients with mCRC who had undergone palliative chemotherapy at four centers from January 2005 to December 2017. Designated participating centers were large oncology facilities in Japan, namely, the National Cancer Center Hospital East (Kashiwa, Chiba, Japan), the Aichi Cancer Center (Nagoya, Aichi, Japan), the Shizuoka Cancer Center (Nagaizumi, Shizuoka, Japan), and the Hokkaido University Hospital (Sapporo, Hokkaido, Japan). All data were retrospectively collected from electronic medical records, and the eligibility criteria were as follows: (a) histologically proven colorectal adenocarcinoma, (b) RAS or BRAF mutation confirmed by polymerase chain reaction or next‐generation sequencing methods, (c) Eastern Cooperative Oncology Group performance status (ECOG PS) of 0–2, and (d) adequate organ function.
First‐line backbone chemotherapy regimens were divided into three groups: mono (5‐fluorouracil [5‐FU]/leucovorin [LV], capecitabine, tegafur‐uracil [UFT]/LV, or S‐1 monotherapy), doublet (oxaliplatin‐based regimens [FOLFOX, CAPOX, or SOX] or irinotecan‐based regimens [FOLFIRI, IRIS/SIRB, CAPIRI]), and triplet (FOLFOXIRI). Information on the history of bevacizumab administration was also obtained.
The study protocol was approved by the institutional review board of each institution and was carried out in accordance with the guidelines for biomedical research specified in the Declaration of Helsinki. The requirement for informed consent was waived because of the retrospective design of this study.
RAS/BRAFV600E Mutation Assessment
RAS and BRAF V600E mutation status were centrally assessed using polymerase chain reaction (PCR) kits, namely, TheraScreen K‐RAS Mutation Kit (QIAGEN, Germantown, MD), MuPACK KIT, MEBGEN RASKET KIT, and MEBGEN RASKET‐B KIT (Medical & Biological Laboratories, Tokyo, Japan) [10, 11, 12], and BRAF V600E status was centrally evaluated using a next‐generation sequencing method (Oncomine Cancer Research Panel or Oncomine Comprehensive Assay version 3; Thermo Fisher Scientific, Waltham, MA) as well as the PCR methods.
Statistical Analysis
Progression‐free survival (PFS) was defined as the time from first‐line chemotherapy initiation to disease progression or death from any cause. Overall survival (OS) was defined as the time from study treatment initiation to death from any cause. Both PFS and OS were calculated using the Kaplan‐Meier method, and the following pretreatment clinical data and baseline laboratory values were used as covariates, namely, age, gender, ECOG PS, primary tumor site (caecum, ascending colon, or transverse colon were classified as right‐sided, whereas those located in the splenic flexure, descending colon, sigmoid colon, or rectum were classified as left‐sided), surgery on the primary tumor, time of first metastasis (synchronous or metachronous), histology (well/moderately differentiated adenocarcinoma or poorly differentiated/mucinous adenocarcinoma), white blood cell count, serum albumin level, serum lactate dehydrogenase (LDH) level, serum C‐reactive protein level, metastatic tumor site (liver, lung, lymph node, or peritoneal dissemination), number of metastatic sites, and KRAS exon 2 mutation subtypes. Survival outcomes were compared between patients with KRAS G12C mutation and those with non‐G12C mutations.
Quantitative data are expressed as median and range, the cutoff value for LDH was set to the median, and the Glasgow prognostic score (GPS) was calculated based on data from previous reports [13]. The Mann‐Whitney U test or the Kruskal‐Wallis test was used to compare continuous variables, whereas Fisher's exact test or the χ2‐test was performed to compare categorical variables. Survival curves were estimated using the Kaplan‐Meier method and differences between the groups were tested by the log‐rank test. Hazard ratios (HRs) were estimated using the Cox proportional hazards model. PFS and OS were analyzed using univariate and multivariate Cox regression analyses. The backward method was used to select retained factors (p < .1) during multivariate analysis. All values of p < .05 were considered statistically significant, and all statistical analyses were performed using the statistical program R version 4.0.3 (R Foundation for Statistical Computing, Vienna, Austria).
Results
Patient Overview
Among 2,457 patients administered systemic chemotherapy for mCRC, we included 1,717 patients with known RAS status and clinical follow‐up data. BRAF V600E mutations and KRAS exon 2 mutations were identified in 43 patients and 702 patients, respectively, and the remaining 972 patients had tumors without KRAS exon 2 mutations. Among these, we excluded patients with KRAS co‐mutations (KRAS G13D and G13R [n = 4], KRAS G12V and G13D [n = 1], and KRAS G12A and G13D [n = 1]), KRAS exon 3/4 mutation (n = 45), and NRAS mutations (n = 34). Thus, 43, 893, and 696 patients were present in the BRAF V600E mutation, the KRAS exon 2 wild‐type, and the KRAS exon 2 mutation groups, respectively (Fig. 1). The distribution of KRAS exon 2 wild‐type and each combination of KRAS exon 2/BRAF V600E mutations is shown in Figure 2. KRAS exon 2 wild‐type accounted for 54.7% (893/1632), and the following six mutations accounted for the majority of KRAS exon 2 mutations: KRAS G12D (16.0%, 261/1632), G13D (9.8%,160/1632), G12V (9.3%, 151/1632), G12C (2.8%, 45/1632), G12S (2.2%, 36/1632), and G12A (1.9%, 31/1632). Characteristics of patients with KRAS G12C or non‐G12C mutations are summarized in Table 1. There was no significant difference between KRAS G12C and non‐G12C mutations; 5% of difference was observed in the following characteristics: gender, ECOG PS, surgery on primary tumor, time of first metastasis, liver metastasis, peritoneal dissemination, number of metastatic sites, and GPS. Characteristics of patients with each KRAS exon 2 mutation subtype are summarized in supplemental online Table 1, and there were no significant differences among the KRAS exon 2 mutation subtypes.
Figure 1.
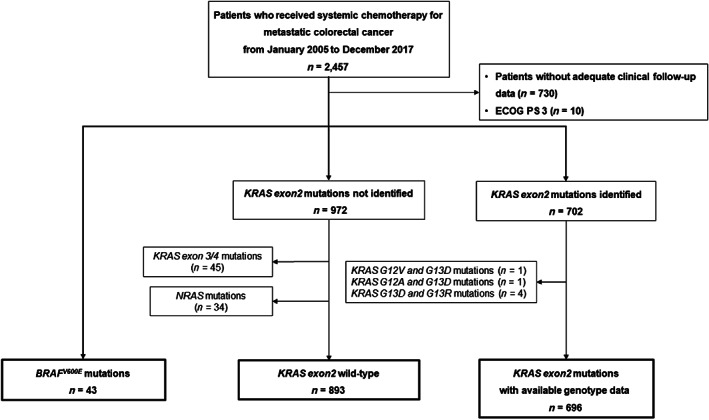
Patient selection flow diagram. Abbreviations: ECOG PS, Eastern Cooperative Oncology performance status; KRAS, Kirsten rat sarcoma; NRAS, neuroblastoma rat sarcoma.
Figure 2.
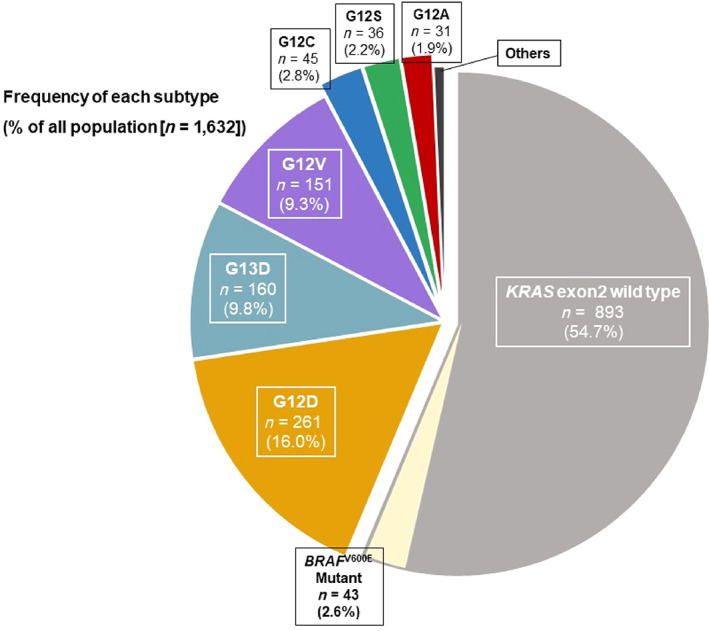
Frequency of each subtype (% of all population). KRAS exon 2 wild‐type was 56.3%, and the most prevalent mutations in KRAS exon 2 were G12D (16.3%), followed by G13D (10.1%), G12V (9.5%), G12C (2.8%), G12S (2.3%), and G12A (1.9%). Abbreviations: KRAS, Kirsten rat sarcoma.
Table 1.
Patient characteristics
| Characteristic | KRAS G12C mutations (n = 45), n (%) | KRAS non‐G12C mutations (n = 651), n (%) | p valuea |
|---|---|---|---|
| Age, years, median (range) | 65 (31–79) | 65 (24–88) | .709 |
| Age ≥65 years | 24 (53.3) | 339 (52.1) | .879 |
| Gender | .278 | ||
| Female | 24 (53.3) | 287 (44.1) | |
| Male | 21 (46.7) | 364 (55.9) | |
| ECOG PS | .113 | ||
| 0 | 31 (68.9) | 491 (75.4) | |
| 1 | 9 (20.0) | 133 (20.4) | |
| 2 | 5 (11.1) | 27 (4.1) | |
| Primary tumor location | .776 | ||
| Right | 16 (35.6) | 213 (32.7) | |
| Left | 29 (64.4) | 436 (67.0) | |
| Missing | 0 (0.0) | 2 (0.3) | |
| Surgery on primary tumor | .205 | ||
| Yes | 21 (46.7) | 240 (36.9) | |
| No | 24 (53.3) | 411 (63.1) | |
| Time of first metastasis | .075 | ||
| Metachronous | 10 (22.2) | 233 (35.8) | |
| Synchronous | 35 (77.8) | 418 (64.2) | |
| Histology | .706 | ||
| Well/mod | 40 (88.9) | 581 (89.2) | |
| Poor/muc | 4 (8.9) | 61 (9.4) | |
| Missing | 1 (2.2) | 9 (1.4) | |
| Metastatic sites | |||
| Liver | 30 (66.7) | 375 (57.6) | .275 |
| Lung | 18 (40.0) | 274 (42.1) | .876 |
| Peritoneal dissemination | 16 (35.6) | 150 (23.0) | .070 |
| Number of metastatic sites | .281 | ||
| 1 | 18 (40.0) | 319 (49.0) | |
| ≥2 | 27 (60.0) | 332 (51.0) | |
| Serum LDH, median (range), IU/L | 221 (140–1,459) | 210 (75–4,340) | .515 |
| GPS | .763 | ||
| 0 | 27 (60.0) | 427 (65.6) | |
| 1 | 10 (22.2) | 110 (16.9) | |
| 2 | 7 (15.6) | 105 (16.1) | |
| Missing | 1 (2.2) | 9 (1.4) | |
| First‐line backbone regimen | .357 | ||
| Monob | 2 (4.4) | 32 (4.9) | |
| Doubletc | 43 (95.6) | 591 (90.8) | |
| Tripletd | 0 (0.0) | 28 (4.3) | |
| First‐line bevacizumab | .843 | ||
| Yes | 36 (80.0) | 531 (81.6) | |
| No | 9 (20.0) | 120 (18.5) |
Values of p were calculated using Fisher's exact probability test for categorical variables.
Mono indicates 5‐fluorouracil (5‐FU)/leucovorin (LV), capecitabine, tegafur‐uracil (UFT)/LV, or S‐1 monotherapy.
Doublet indicates oxaliplatin‐based regimens (FOLFOX, CAPOX, and SOX) or irinotecan‐based regimens (FOLFIRI, IRIS [SIRB], CAPIRI).
Triplet indicates FOLFOXIRI.
Abbreviations: ECOG PS, Eastern Cooperative Oncology Group performance status; GPS, Glasgow prognostic score; KRAS, Kirsten rat sarcoma; LDH, lactate dehydrogenase; mod, moderately differentiated; muc, mucinous adenocarcinoma; por, poorly differentiated; well, well differentiated.
Survival in the Entire Population
Median follow‐up period was 64.8 months (95% confidence interval [CI], 59.6–71.1 months), and 1,243 (76.2%) patients had died. Median OS for the entire population was 29.4 months (95% CI, 27.9–30.9 months). Compared with patients with wild‐type KRAS exon 2, median OS was significantly shorter in patients with KRAS exon 2 mutations (HR, 1.23 [95% CI, 1.10–1.38], p < .001) and in those with BRAF V600E mutations (HR, 1.48 [95% CI, 1.25–1.75], p < .001) (Fig. 3). Furthermore, PFS and OS were not significantly influenced by genotype subgroups. The median PFS and OS of patients with KRAS exon 2 mutation subtypes ranged from 9.4 months (95% CI, 6.4–12.0) to 11.5 months (95% CI, 9.4–13.5) and from 21.1 months (95% CI, 12.8–29.4) to 29.8 months (20.5–41.9), respectively (supplemental online Figs. 1 and 2).
Figure 3.
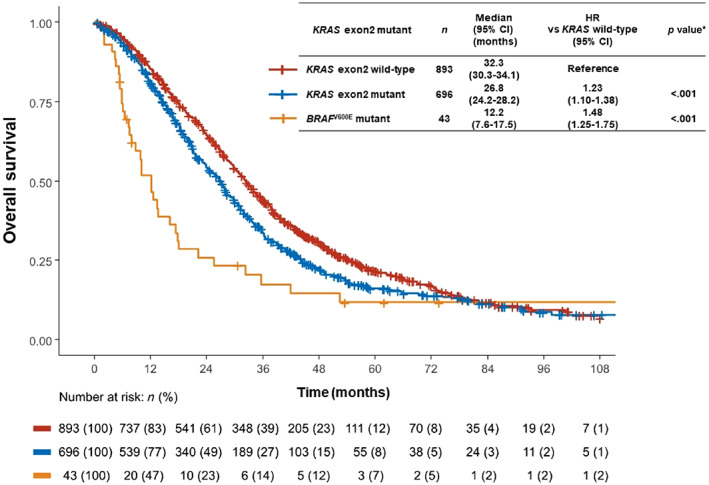
Kaplan‐Meier curves for overall survival (OS) in patients with KRAS wild‐type, KRAS exon 2 mutations, and BRAF V600E mutations. Compared with KRAS exon 2 wild‐type, the median OS was significantly shorter in patients with KRAS exon 2 mutations (HR, 1.23 [95% CI, 1.10–1.38], p < .001) and for those with BRAF V600E mutations (HR, 1.48 [95% CI, 1.25–1.75], p < .001). Abbreviations: CI, confidence interval; KRAS, Kirsten rat sarcoma; HR, hazard ratio.
Prognostic Impact of the KRAS G12C Mutation and Non‐G12C Mutations
Among patients with KRAS exon 2 mutations, those with the G12C mutation had significantly shorter PFS and OS than those with non‐G12C mutations (median PFS, 9.4 months [95% CI, 6.4–12.0] vs. 10.8 months [95% CI, 10.1–11.5]; HR, 1.47 [95% CI, 1.08–2.01], p = .015; median OS, 21.1 months [95% CI, 12.8–27.9] vs. 27.3 months [95% CI, 24.8–28.9]; HR, 1.50 [95% CI, 1.08–2.08], p = .015) (Fig. 4).
Figure 4.
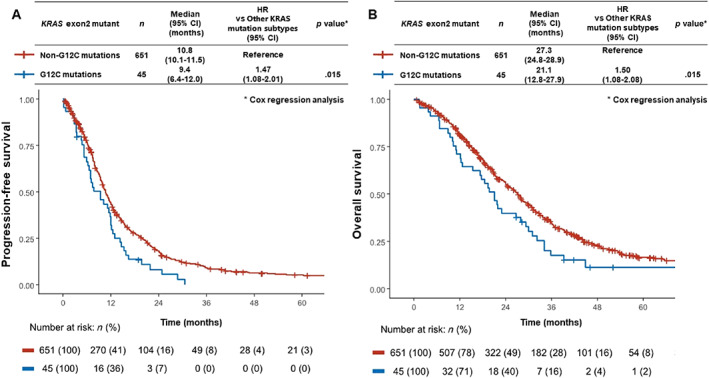
Kaplan‐Meier curves for progression‐free survival (PFS) and overall survival (OS) in KRAS G12C mutation versus non‐G12C mutations. (A): Median PFS in patients with KRAS G12C mutation was significantly shorter than that in patients with non‐G12C mutations (9.4 months [95% CI, 6.4–12.0] vs. 10.8 months [95% CI, 10.1–11.8]; HR, 1.47 [95% CI, 1.08–2.01], p = .015). (B): Median OS in patients with KRAS G12C mutation was significantly shorter than that in patients with non‐G12C mutations (21.1 months [95% CI, 12.8–27.9] vs. 27.3 months [95% CI, 24.8–28.9]; HR, 1.50 [95% CI, 1.08–2.08], p = .015).
Abbreviations: CI, confidence interval; KRAS, Kirsten rat sarcoma; HR, hazard ratio.
Table 2 shows the results of univariate and multivariate analyses for PFS in patients with KRAS exon 2 mutations. Multivariate analysis identified the following factors as significantly associated with PFS: KRAS G12C mutation (vs. non‐G12C mutation: HR, 1.43; 95% CI, 1.04–1.96; p = .030), surgery on primary tumor (yes vs. no: HR, 0.82; 95% CI, 0.69–0.99; p = .035), serum LDH (≥210 IU/L [median] vs. <210: HR, 1.24; 95% CI, 1.04–1.47; p = .014), GPS (1 or 2 vs. 0: HR, 1.53; 95% CI, 1.21–1.93, p < .001), number of metastatic organ sites (≥2 vs. 1: HR, 1.32; 95% CI, 1.12–1.55; p = .001), and first‐line bevacizumab (yes vs. no: HR, 0.57; 95% CI, 0.45–0.71; p < .001) (Table 2).
Table 2.
Univariate and multivariate analysis for progression‐free survival
| Category | Univariate | Multivariate | ||
|---|---|---|---|---|
| HR (95% CI) | p valuea | HR (95% CI) | p valuea | |
| KRAS exon 2 mutation: G12C vs. non‐G12C | 1.47 (1.08–2.01) | .015 | 1.43 (1.04–1.96) | .030 |
| Age: ≥65 vs. <65 years | 0.97 (0.83–1.14) | .724 | ||
| Gender: Male vs. female | 0.98 (0.83–1.15) | .794 | ||
| ECOG PS: 1 or 2 vs. 0 | 1.48 (1.24–1.78) | <.001 | 1.08 (0.88–1.33) | .443 |
| Primary tumor site: left vs. right | 0.93 (0.79–1.10) | .417 | ||
| Surgery on primary tumor: Yes vs. No | 0.69 (0.59–0.81) | <.001 | 0.82 (0.69–0.99) | .035 |
| Time of first metastasis: Synchronous vs. metachronous | 1.15 (0.97–1.35) | .113 | ||
| Histology: Poor/muc vs. well/mod | 1.17 (0.89–1.54) | .258 | ||
| Serum LDH, IU/L: ≥210 (median) vs. <210 | 1.50 (1.28–1.76) | <.001 | 1.24 (1.04–1.47) | .014 |
| GPS: 1 or 2 vs. 0 | 1.96 (1.58–2.44) | <.001 | 1.53 (1.21–1.93) | <.001 |
| Number of metastatic organ sites: ≥2 vs. 1 | 1.46 (1.25–1.71) | <.001 | 1.32 (1.12–1.55) | .001 |
| First‐line backbone regimen: Doubletb or tripletc vs. monod | 0.84 (0.57–1.22) | .355 | ||
| First‐line bevacizumab: Yes vs. no | 0.51 (0.42–0.63) | <.001 | 0.57 (0.45–0.71) | <.001 |
Values of p were calculated using the Cox proportional hazard model.
Doublet indicates oxaliplatin‐based regimens (FOLFOX, CAPOX, and SOX) or irinotecan‐based regimens (FOLFIRI, IRIS [SIRB], CAPIRI).
Triplet indicates FOLFOXIRI.
Mono indicates 5‐fluorouracil (5‐FU)/leucovorin (LV), capecitabine, tegafur‐uracil (UFT)/LV, or S‐1 monotherapy.
Abbreviations: CI, confidence interval; ECOG PS, Eastern Cooperative Oncology Group performance status; GPS, Glasgow prognostic score; HR, hazard ratio; KRAS, Kirsten rat sarcoma; LDH, lactate dehydrogenase; mod, moderately differentiated; muc, mucinous adenocarcinoma; poor, poorly differentiated; well, well differentiated.
Table 3 shows the results of univariate and multivariate analyses for OS in patients with KRAS exon 2 mutations. Here, multivariate analysis yielded significant association between OS and the following variables: KRAS G12C mutation (vs. non‐G12C mutation: HR, 1.42; 95% CI, 1.01–2.00; p = .044), ECOG PS (1 or 2 vs. 0: HR, 1.25; 95% CI, 1.00–1.57; p = .049), surgery on primary tumor (yes vs. no: HR, 0.75; 95% CI, 0.60–0.93; p = .010), histology (mucinous or poorly differentiated vs. well or moderately differentiated: HR, 1.44; 95% CI, 1.06–1.95; p = .021), serum LDH (≥210 IU/L [median] vs. <210: HR, 1.59; 95% CI, 1.31–1.92; p < .001), GPS (1 or 2 vs. 0: HR, 1.64; 95% CI, 1.27–2.12; p < .001), and number of metastatic organ sites (≥2 vs. 1: HR, 1.45; 95% CI, 1.21–1.74; p < .001) (Table 3).
Table 3.
Univariate and multivariate analysis for overall survival
| Category | Univariate | Multivariate | ||
|---|---|---|---|---|
| HR (95% CI) | p valuea | HR (95% CI) | p valuea | |
| KRAS exon 2 mutation: G12C vs. non‐G12C | 1.50 (1.08–2.08) | .015 | 1.42 (1.01–2.00) | .044 |
| Age: ≥65 vs. <65 years | 1.03 (0.87–1.22) | .725 | ||
| Gender: Male vs. female | 0.90 (0.76–1.07) | .219 | ||
| ECOG PS: 1 or 2 vs. 0 | 1.78 (1.46–2.17) | <.001 | 1.25 (1.00–1.57) | .049 |
| Primary tumor site: left vs. right | 0.82 (0.68–0.98) | .028 | 0.99 (0.82–1.21) | .976 |
| Surgery on primary tumor: Yes vs. no | 0.58 (0.48–0.69) | <.001 | 0.75 (0.60–0.93) | .010 |
| Time of first metastasis: Synchronous vs. metachronous | 1.47 (1.23–1.77) | <.001 | 0.95 (0.76–1.19) | .647 |
| Histology: por/muc vs. well/mod | 1.44 (1.07–1.94) | .016 | 1.44 (1.06–1.95) | .021 |
| Serum LDH, IU/L: ≥210 (median) vs. <210 | 1.93 (1.62–2.30) | <.001 | 1.59 (1.31–1.92) | <.001 |
| GPS: 1 or 2 vs. 0 | 2.31 (1.83–2.90) | <.001 | 1.64 (1.27–2.12) | <.001 |
| Number of metastatic organ site: ≥2 vs. 1 | 1.63 (1.37–1.94) | <.001 | 1.45 (1.21–1.74) | <.001 |
| First‐line backbone regimen: Doubletb or tripletc vs. monod | 0.60 (0.41–0.90) | .012 | 0.76 (0.50–1.17) | .219 |
| First‐line bevacizumab: Yes vs. no | 0.69 (0.55–0.86) | <.001 | 0.84 (0.66–1.08) | .174 |
Values of p were calculated using the Cox proportional hazard model.
Doublet indicates oxaliplatin‐based regimens (FOLFOX, CAPOX, and SOX) or irinotecan‐based regimens (FOLFIRI, IRIS [SIRB], CAPIRI).
Triplet indicates FOLFOXIRI.
Mono indicates 5‐fluorouracil (5‐FU)/leucovorin (LV), capecitabine, tegafur‐uracil (UFT)/LV, or S‐1 monotherapy.
Abbreviations: CI, confidence interval; ECOG PS, Eastern Cooperative Oncology Group performance status; GPS, Glasgow prognostic score; HR, hazard ratio; KRAS, Kirsten rat sarcoma; LDH, lactate dehydrogenase; mod, moderately differentiated; muc, mucinous adenocarcinoma; poor, poorly differentiated; well, well differentiated.
Furthermore, in the patients who received doublet or triplet chemotherapy, their characteristics were not significantly different between those with KRAS G12C and non‐G12C mutations (supplemental online Table 2). Also, consistently poor PFS and OS were observed in those with KRAS G12C mutation (supplemental online Fig. 3).
Discussion
Our study assessed the prognostic impact of KRAS G12C mutation in chemotherapy‐naïve patients with mCRC. Median OS for each type of mutation (i.e., KRAS exon 2 wild‐type, KRAS exon 2 mutant, and BRAF V600E mutant) was consistent with that reported previously [14, 15], indicating that our cohort was representative of the mCRC population and that the results presented here reflect prognosis in clinical practice. We modeled real‐world distributions of KRAS mutation subtypes and reveal poor PFS and OS in patients with KRAS G12C mutation compared with patients with non‐G12C mutations.
The overall prevalence of KRAS exon 2 mutations and that of the three major KRAS mutation subtypes, along with the targetable subtype KRAS G12C, obtained in this study is consistent with previous reports [16, 17, 18]. A recent analysis of the distribution of KRAS G12C mutation categorized according to race, gender, and cancer type identified a trend of greater prevalence among female patients, irrespective of their ethnicity; however, KRAS G12C mutation was less frequent in CRC compared with non‐small cell lung cancer (3.2% vs. 13.8%). This higher proportion of female patients is also consistent with the results from our study [18].
Although several reports have described the prognostic impact of KRAS mutation subtypes, most have focused on patients with non‐small cell lung cancer and/or nonmetastatic CRC [19, 20], and only a few studies have analyzed the prognostic impact of KRAS G12C mutation in mCRC [16, 21]. A pooled analysis by the Arbeitsgemeinschaft Internistische Onkologie [AIO] study group revealed that KRAS G12C mutation was significantly associated with lower OS compared with tumors with no KRAS mutations [16]; however, it must be noted here that this pooled analysis also included patients who had received a nonstandard treatment regimen, including FUFIRI, FUFOX, and mIROX as induction chemotherapies and bevacizumab monotherapy or observation alone as maintenance therapies in clinical trials [22, 23]. Schirripa et al. have recently reported poor prognosis in patients with mCRC and KRAS G12C mutation; however, the prevalence of KRAS G12C mutation was relatively high at 17.3%, and favorable outcomes, such as median OS greater than 35 months in patients with KRAS non‐G12C mutations, might not reflect real‐world clinical practice [21]. The strength of our study lies in validating poor prognosis in the presence of the KRAS G12C mutation with respect to PFS and OS using real‐world data obtained outside a clinical trial setting. We also included data from patients with mCRC receiving standard chemotherapies by performing a multivariate analysis that included important covariate factors.
A preclinical study has shown specific differences in intrinsic or GTPase Activating Protein‐mediated GTP hydrolysis among the KRAS mutation subtypes that result in differential activation of downstream effectors, such as the mitogen‐activated protein kinase [MAPK]/extracellular signal‐regulated kinase [ERK] pathway [24, 25]. Profiles of biochemical properties of KRAS mutations and G12C, G12D, and G13D mutants showed a high level of intrinsic GTPase activity compared with those of other KRAS mutations, such as G12A and G12V [5]. However, there are insufficient data on the reasons for the KRAS G12C mutation leading to poorer prognosis than non‐G12C mutations in mCRC; therefore, further investigations are needed to identify the precise mechanism underlying the observed poor prognosis.
Recently, two KRAS G12C inhibitors, AMG 510 and MRTX849, have been developed; both specifically bind to the mutant cysteine residue [7]. The first two trials with AMG 510 and MRTX849 revealed promising clinical results in non‐small cell lung cancer; however, the response in patients with CRC was unexpectedly limited [8, 26]. Indeed, rapid heterogeneous adaptation to conformation‐specific KRAS G12C inhibition has been pointed out as a mechanism of resistance to the KRAS G12C inhibitor, and importantly, this phenomenon could be reverted when therapy was combined with the EGFR inhibitor [27, 28]. Furthermore, a phase I study of a combination of AMG 510 plus EGFR inhibitor with or without chemotherapy is ongoing in patients with KRAS G12 mutant mCRC. Given the development and trial of novel targeted agents, our data would be an important source of reference on the real‐world distribution and prognostic impact of the KRAS G12C mutation in patients with mCRC.
Limitations to the present study need to be considered when interpreting these results. First, as previously noted, this was a nonrandomized retrospective study. Second, although our study included 1,632 patients with mCRC, the number of patients with the KRAS G12C mutation in tumors was limited. Nevertheless, taking into consideration that there are only a few reports that provide a detailed prognosis in KRAS G12C mutation, the data presented here can play an important role in the clinical development of KRAS G12C inhibitors. Third, all patients in our study were Asian. However, a recent report has revealed no difference in the distribution of KRAS G12C mutation in CRC between Asian and Western populations, unlike non‐small cell lung cancer [18]. Therefore, our result may be applied to Western populations as well as Asian populations.
Conclusion
We demonstrate that the KRAS G12C mutation is significantly correlated with a shorter PFS and OS compared with KRAS non‐G12C mutations in chemotherapy‐naïve patients with mCRC. These findings indicate the importance of individualized treatment that targets the KRAS G12C mutation.
Author Contributions
Conception/design: Keigo Chida, Daisuke Kotani, Toshiki Masuishi, Takayuki Yoshino
Provision of study material or patients: Keigo Chida, Daisuke Kotani, Toshiki Masuishi, Takeshi Kawakami, Yasuyuki Kawamoto, Kyoko Kato, Kunihiro Fushiki, Kentaro Sawada, Ryosuke Kumanishi, Hiromichi Shirasu, Yuki Matsubara, Satoshi Yuki, Yoshito Komatsu, Kentaro Yamazaki, Takayuki Yoshino
Collection and/or assembly of data: Keigo Chida, Toshiki Masuishi, Takeshi Kawakami, Yasuyuki Kawamoto, Kyoko Kato, Kunihiro Fushiki, Kentaro Sawada, Ryosuke Kumanishi, Hiromichi Shirasu, Yuki Matsubara
Data analysis and interpretation: Keigo Chida, Daisuke Kotani, Toshiki Masuishi, Takeshi Kawakami, Yasuyuki Kawamoto, Takayuki Yoshino
Manuscript writing: Keigo Chida, Daisuke Kotani, Takayuki Yoshino
Final approval of manuscript: Keigo Chida, Daisuke Kotani, Toshiki Masuishi, Takeshi Kawakami, Yasuyuki Kawamoto, Kyoko Kato, Kunihiro Fushiki, Kentaro Sawada, Ryosuke Kumanishi, Hiromichi Shirasu, Yuki Matsubara, Satoshi Yuki, Yoshito Komatsu, Kentaro Yamazaki, Takayuki Yoshino
Disclosures
Toshiki Masuishi: Takeda, Chugai, Merck Bio Pharma, Taiho, Bayer, Lilly Japan, Yakult Honsha, Bristol‐Myers Squibb, Ono, Sanofi (H), Merck Sharp & Dohme, Daiichi Sankyo, Ono, Novartis (RF); Takeshi Kawakami: Ono Pharmaceutical, Bristol‐Myers Squibb, Bayer (H); Kentaro Yamazaki: Daiichi Sankyo, Eli Lilly & Co., Yakult Honsha, Merck Serono, Bristol‐Myers Squibb, Ono Pharmaceutical, Merck Sharp & Dohme, Sanofi, Chugai Pharma, Takeda, Bayer, Taiho Pharmaceutical (H), Taiho Pharmaceutical (RF); Takayuki Yoshino: Taiho Pharmaceutical, Sumitomo Dainippon Pharma, Ono Pharmaceutical, Chugai Pharmaceutical, Amgen, PAREXEL International, Merck Sharp & Dohme, Daiichi Sankyo, Sanofi (RF). The other authors indicated no financial relationships.
(C/A) Consulting/advisory relationship; (RF) Research funding; (E) Employment; (ET) Expert testimony; (H) Honoraria received; (OI) Ownership interests; (IP) Intellectual property rights/inventor/patent holder; (SAB) Scientific advisory board
Supporting information
See http://www.TheOncologist.com for supplemental material available online.
Appendix S1. Figures.
Appendix S2. Tables.
Acknowledgments
The authors thank all patients and families for participation in the studies, as well as all involved study centers, colleagues, and nurses. The authors also would like to thank Enago (www.enago.jp) for the English language review.
Disclosures of potential conflicts of interest may be found at the end of this article.
No part of this article may be reproduced, stored, or transmitted in any form or for any means without the prior permission in writing from the copyright holder. For information on purchasing reprints contact commercialreprints@wiley.com.
References
- 1.Simanshu DK, Nissley DV, McCormick F. RAS proteins and their regulators in human disease. Cell 2017;170:17–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Guerrero S, Casanova I, Farré L et al. K‐ras codon 12 mutation induces higher level of resistance to apoptosis and predisposition to anchorage‐independent growth than codon 13 mutation or proto‐oncogene overexpression. Cancer Res 2000;60:6750–6756. [PubMed] [Google Scholar]
- 3.Messner I, Cadeddu G, Huckenbeck W et al. KRAS p.G13D mutations are associated with sensitivity to anti‐EGFR antibody treatment in colorectal cancer cell lines. J Cancer Res Clin 2012;139:201–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Garassino MC, Marabese M, Rusconi P et al. Different types of K‐Ras mutations could affect drug sensitivity and tumour behaviour in non‐small‐cell lung cancer. Ann Oncol 2011;22:235–237. [DOI] [PubMed] [Google Scholar]
- 5.Hunter JC, Manandhar A, Carrasco MA et al. Biochemical and structural analysis of common cancer‐associated KRAS mutations. Mol Cancer Res 2015;13:1325–1335. [DOI] [PubMed] [Google Scholar]
- 6.Cox AD, Fesik SW, Kimmelman AC et al. Drugging the undruggable RAS: Mission possible? Nat Rev Drug Discov 2014;13:828–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ostrem JM, Peters U, Sos ML et al. K‐Ras(G12C) inhibitors allosterically control GTP affinity and effector interactions. Nature 2013;503:548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hong DS, Fakih MG, Strickler JH et al. KRASG12C inhibition with sotorasib in advanced solid tumors. N Engl J Med 2020;383:1207–1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fakih M, Durm GA, Govindan R et al. Trial in progress: A phase Ib study of AMG 510, a specific and irreversible KRAS G12C inhibitor, in combination with other anticancer therapies in patients with advanced solid tumors harboring KRAS p.G12C mutation (CodeBreak 101). J Clin Oncol 2020;38(suppl 15):TPS3661a. [Google Scholar]
- 10.Kawazoe A, Shitara K, Fukuoka S et al. A retrospective observational study of clinicopathological features of KRAS, NRAS, BRAF and PIK3CA mutations in Japanese patients with metastatic colorectal cancer. BMC Cancer 2015;15:258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yoshino T, Muro K, Yamaguchi K et al. Clinical validation of a multiplex kit for RAS mutations in colorectal cancer: Results of the RASKET (RAS KEy Testing) prospective, multicenter study. EBioMedicine 2015;2:317–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Taniguchi H, Okamoto W, Muro K et al. Clinical validation of newly developed multiplex kit using Luminex xMAP technology for detecting simultaneous RAS and BRAF mutations in colorectal cancer: Results of the RASKET‐B study. Neoplasia 2018;20:1219–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu Y, He X, Pan J et al. Prognostic role of Glasgow prognostic score in patients with colorectal cancer: Evidence from population studies. Sci Rep 2017;7:6144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Phipps AI, Buchanan DD, Makar KW et al. KRAS‐mutation status in relation to colorectal cancer survival: The joint impact of correlated tumour markers. Br J Cancer 2013;108:1757–1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Richman SD, Seymour MT, Chambers P et al. KRAS and BRAF mutations in advanced colorectal cancer are associated with poor prognosis but do not preclude benefit from oxaliplatin or irinotecan: Results from the MRC FOCUS trial. J Clin Oncol 2009;27:5931–5937. [DOI] [PubMed] [Google Scholar]
- 16.Modest DP, Ricard I, Heinemann V et al. Outcome according to KRAS‐, NRAS‐ and BRAF‐mutation as well as KRAS mutation variants: Pooled analysis of five randomized trials in metastatic colorectal cancer by the AIO colorectal cancer study group. Ann Oncol 2016;27:1746–1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Neumann J, Zeindl‐Eberhart E, Kirchner T et al. Frequency and type of KRAS mutations in routine diagnostic analysis of metastatic colorectal cancer. Pathol Res Pract 2009;205:858–862. [DOI] [PubMed] [Google Scholar]
- 18.Nassar AH, Adib E, Kwiatkowski DJ. Distribution of KRAS G12C somatic mutations across race, sex, and cancer type. N Engl J Med 2021;382:185–187. [DOI] [PubMed] [Google Scholar]
- 19.Wiesweg M, Kasper S, Worm K et al. Impact of RAS mutation subtype on clinical outcome—a cross‐entity comparison of patients with advanced non‐small cell lung cancer and colorectal cancer. Oncogene 2019;38:2953–2966. [DOI] [PubMed] [Google Scholar]
- 20.Li W, Liu Y, Cai S et al. Not all mutations of KRAS predict poor prognosis in patients with colorectal cancer. Int J Clin Exp Pathol 2019;12:957–967. [PMC free article] [PubMed] [Google Scholar]
- 21.Schirripa M, Nappo F, Cremolini C et al. KRAS G12C metastatic colorectal cancer: Specific features of a new emerging target population. Clin Colorectal Cancer 2020;19:219–225. [DOI] [PubMed] [Google Scholar]
- 22.Venook AP, Niedzwiecki D, Lenz HJ et al. Effect of first‐line chemotherapy combined with cetuximab or bevacizumab on overall survival in patients with KRAS wild‐type advanced or metastatic colorectal cancer: A randomized clinical trial. JAMA 2017;317:2392–2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stintzing S, Modest DP, Rossius L et al. FOLFIRI plus cetuximab versus FOLFIRI plus bevacizumab for metastatic colorectal cancer (FIRE‐3): A post‐hoc analysis of tumour dynamics in the final RAS wild‐type subgroup of this randomised open‐label phase 3 trial. Lancet Oncol 2016;17:1426–1434. [DOI] [PubMed] [Google Scholar]
- 24.Smith MJ, Neel BG, Ikura M. NMR‐based functional profiling of RASopathies and oncogenic RAS mutations. Proc Natl Acad Sci USA 2013;110:4574–4579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Haigis KM. KRAS alleles: The devil is in the detail. Trends Cancer 2017;3:686–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hallin J, Engstrom LD, Hargis L et al. The KRASG12C inhibitor MRTX849 provides insight toward therapeutic susceptibility of KRAS‐mutant cancers in mouse models and patients. Cancer Discov 2020;10:54–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Amodio V, Yaeger R, Arcella P et al. EGFR blockade reverts resistance to KRASG12C inhibition in colorectal cancer. Cancer Discov 2020;10:1129–1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xue JY, Zhao Y, Aronowitz J et al. Rapid non‐uniform adaptation to conformation‐specific KRAS(G12C) inhibition. Nature 2020;577:421–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
See http://www.TheOncologist.com for supplemental material available online.
Appendix S1. Figures.
Appendix S2. Tables.


