Abstract
Plasma cell‐free DNA (cfDNA) genotyping is an alternative to tissue genotyping, particularly when tissue specimens are insufficient or unavailable, and provides critical information that can be used to guide treatment decisions in managing patients with non‐small cell lung cancer (NSCLC). In this article, we review the evolution of plasma cfDNA genotyping from an emerging concept, through development of analytical methods, to its clinical applications as a standard‐of‐care tool in NSCLC.
The number of driver or resistance mutations recommended for testing in NSCLC continues to increase. Because of the expanding list of therapeutically relevant variants, comprehensive testing to investigate larger regions of multiple genes in a single run is often preferable and saves on time and cost, compared with performing serial single‐gene assays. Recent advances in nucleic acid next‐generation sequencing have led to a rapid expansion in cfDNA genotyping technologies. Analytic assays that have received regulatory approval are now routinely used as diagnostic companions in the setting of metastatic NSCLC. As the demand for plasma‐based technologies increases, more regulatory approvals of cfDNA genotyping assays are expected in the future.
Plasma cfDNA genotyping is currently aiding oncologists in the delivery of personalized care by facilitating matching of patients with targeted therapy and monitoring emergence of resistance to therapy in NSCLC. Further advances currently underway to increase assay sensitivity and specificity will potentially expand the use of plasma cfDNA genotyping in early cancer detection, monitoring response to therapy, detection of minimal residual disease, and measurement of tumor mutational burden in NSCLC.
Implications for Practice
Plasma cell‐free DNA (cfDNA) genotyping offers an alternative to tissue genotyping, particularly when tissue specimens are insufficient or unavailable. Advances in cfDNA genotyping technologies have led to analytic assays that are now routinely used to aid oncologists in the delivery of personalized care by facilitating matching of patients with targeted therapy and monitoring emergence of resistance to therapy. Further advances underway to increase assay sensitivity and specificity will potentially expand the use of plasma cfDNA genotyping in early cancer detection, monitoring response to therapy, detection of minimal residual disease, and evaluation of tumor mutational burden in non‐small cell lung cancer.
Keywords: Plasma cell‐free DNA genotyping, Circulating tumor DNA, Liquid biopsy, Non‐small cell lung cancer, Tissue biopsy
Short abstract
This article reviews the evolution of plasma cell‐free DNA genotyping from an emerging concept, through development of analytical methods, to its clinical applications as a standard‐of‐care tool in non‐small cell lung cancer.
Overview of Molecular Testing in Non‐Small Cell Lung Cancer
The diagnostic journey for lung cancer begins with identification of the carcinoma through pathological evaluation to classify the histologic subtype, imaging (including computed tomography [CT] and magnetic resonance imaging [MRI]) to determine the extent of disease, and molecular diagnostic testing to inform therapeutic strategies [1]. Targeted therapies have transformed the treatment landscape of non‐small cell lung cancer (NSCLC). Current clinical guidelines [1, 2, 3] recommend broad molecular diagnostic testing to identify driver mutations for which effective therapies are available, including alterations in epidermal growth factor receptor (EGFR), anaplastic lymphoma kinase (ALK), c‐ROS oncogene 1 (ROS1), B‐Raf proto‐oncogene (BRAF), MET proto‐oncogene (MET), RET proto‐oncogene (RET), and neurotrophic receptor tyrosine kinase (NTRK). The Kirsten rat sarcoma viral oncogene homolog (KRAS) gene mutation KRAS G12C is another oncogenic driver mutation that is now actionable based on the recent approval of a targeted therapy for KRAS G12C‐mutant NSCLC by the U.S. Food and Drug Administration (FDA) [4]. Another emerging actionable biomarker is ERBB2 (HER2) [1, 2, 3]. Guidelines also recommend testing for the expression level of tumor‐derived programmed cell death ligand 1 (PD‐L1) [1, 2, 3].
The number of driver or resistance mutations recommended to be tested in NSCLC continues to increase [5]. Because of the expanding list of therapeutic targets, broader comprehensive testing to investigate larger regions of multiple genes in a single run is preferable and saves time and cost, compared with performing serial single‐gene assays [5].
Genotyping has historically been performed using formalin‐fixed, paraffin‐embedded specimens from tissue biopsies [1, 2, 3], which often requires substantial amounts of tissue to achieve the minimum DNA or RNA input required for assay sensitivity. Small biopsy specimens with limited tumor cellularity might only be sufficient for morphological diagnosis, histologic subtype classification, and PD‐L1 staining, but the tissue might be insufficient in quantity and/or quality for genotyping. Although tissue genotyping is considered the gold standard for molecular profiling [3, 5, 6, 7], this is often not feasible for all patients and tumor types [8]. This is particularly relevant in lung cancer in which the tumor site may be difficult to access and obtaining adequate tissue specimens for comprehensive genotyping often necessitates invasive procedures. In NSCLC specifically, tissue biopsy has been shown to be inadequate for molecular testing [8, 9, 10].
Liquid biopsy involves the analysis of tumor‐derived material in body fluids, including blood, urine, saliva, pleural effusion, and cerebrospinal fluid [11, 12]. The most tested analyte in blood is plasma cell‐free DNA (cfDNA), composed of DNA fragments that are passively released from apoptotic or necrotic cells or released from digestion of cells by phagocytes [8, 13, 14, 15]. Plasma cfDNA contains circulating tumor DNA (ctDNA), which is a subset of DNA specifically shed from tumor cells [16]. The amount of ctDNA in plasma cfDNA varies depending on the tumor type, tumor stage and burden, tumor location, vascularization, apoptotic rate, metastatic potential of the cancer cells, and factors affecting the patient's blood volume [8, 15, 16]. The half‐life of ctDNA in the bloodstream varies from approximately 16 minutes to 2.5 hours, making ctDNA a “real‐time” molecular marker of disease [17, 18]. Other tested analytes in blood include tumor‐derived extracellular vesicles (EVs), circulating tumor cells (CTCs), messenger RNA (mRNA), and microRNA (miRNA). EVs are lipid bilayer–encapsulated vesicles of ~30 to 2,000 nm that contain DNA, RNA, proteins, and lipids from the tumor of origin and are believed to play a critical role in intercellular communication [19, 20, 21]. CTCs are detectable in some but not all cancers. Although EVs and CTCs are promising liquid biopsy analytes, they are not yet routinely used for clinical genotyping in NSCLC. mRNA and miRNA are currently still being evaluated as biomarkers in the research setting [22] and are not yet used for molecular testing in the clinic.
Plasma cfDNA genotyping technologies have recently advanced and increased the ability to identify oncogenic driver mutations. They are now routinely used to aid oncologists in the delivery of personalized care by facilitating matching of patients with targeted therapy and monitoring emergence of resistance to therapy. Tumor‐specific biomarkers that can be identified in plasma include somatic point mutations, insertions/deletions (indels), amplifications, gene fusions, mRNA splice variants, and tumor proteins [12]. In this article, we review the evolution of plasma cfDNA genotyping from an emerging concept, through development of analytical methods, to its clinical applications as a standard‐of‐care tool in NSCLC.
Evolution of Analytical Methods for cfDNA Genotyping
Advances in nucleic acid‐based cfDNA genotyping have led to the development of several analytical methods to identify somatic driver or resistance mutations (Table 1). These methods range from polymerase chain reaction (PCR)–based approaches to broader coverage next‐generation sequencing (NGS).
Table 1.
Analytical methods for plasma cfDNA genotyping
| Assay category | Assay technology/name (regulatory approval) | Analysis scale | Method | Limit of detection (as % of cfDNA) | Advantages |
|---|---|---|---|---|---|
| Quantitative PCR | qPCR | Single mutations or panels of known and well‐characterized mutations | Preferentially amplifies rare mutant DNA molecules | ~0.1%–1% |
Highly sensitive and specific. Low turnaround time. |
| cobas EGFR Mutation Test v2 (FDA and EMA approved) | |||||
| therascreen EGFR Plasma RGQ PCR kit | |||||
| PANAMutyper R EGFR kit | |||||
| Digital PCR | ddPCR | Single‐locus or multiplexed assays | Partitions target DNA into different reactions for massively parallel qPCR | ~0.04%–0.1% | Highly sensitive and specific. |
| Bio‐Rad QX200 ddPCR Dx system | |||||
| BEAMing | |||||
| OncoBEAM EGFR kit | |||||
| NGS whole genome and exome sequencing | Whole genome sequencing |
Genome wide Exome wide |
NGS of whole genome or whole exome |
~10% for whole genome ~5% for whole exome |
Evaluation of entire genome or exome can lead to discovery of new targets. Exome sequencing allows rapid aneuploidy assessment with lower cost than whole genome sequencing. Discovery of mechanisms of resistance. |
| Roche/454 | |||||
| Ion Torrent: Proton/PGM | |||||
| Illumina Sequencing (Solexa) | |||||
| SOLiD | |||||
| Hybrid capture‐based NGS | Targeted NGS sequencing | Targeted sequencing of captured regions of the genome | Subset of exome is hybridized to biotinylated probes and captured for NGS analysis | ~0.001%–0.5% |
Highly sensitive. Simultaneous detection of predetermined genes of interest. Comprehensive detection of known and unknown mutations. Lower cost and less bioinformatics data compared with whole genome sequencing. |
| Guardant360 CDx (FDA approved) | |||||
| FoundationOne Liquid CDx (FDA approved) | |||||
| Resolution ctDX Lung | |||||
| CAPP‐Seq | |||||
| TEC‐Seq | |||||
| Multiplex PCR‐based NGS | Targeted NGS sequencing | Targeted sequencing of predefined regions | PCR amplification enriches targets before NGS analysis | ~0.01–2.0% | Highly sensitive. |
| TAm‐Seq | |||||
| Enhanced TAm‐Seq | |||||
| Safe‐SeqS | |||||
| Natera | |||||
| Combination approaches (including DNA + biomarkers) | CAPP‐Seq + GRP | Single locus to genome wide | Combines different ctDNA detection methods, sometimes including protein biomarkers | Variable | Improved detection compared with standard ctDNA analysis alone in certain settings. |
| CancerSEEK | |||||
| UroSEEK |
Abbreviations: BEAMing, beads, emulsion, amplification and magnetics; CAPP‐Seq, cancer personalized profiling by deep sequencing; cfDNA, cell‐free DNA; ctDNA, circulating tumor DNA; ddPCR, digital droplet polymerase chain reaction; EGFR, epidermal growth factor receptor; EMA, European Medicines Agency; FDA, U.S. Food and Drug Administration; NGS, next‐generation sequencing; PGM, personal genome machine; PIK3CA, phosphatidylinositol‐4,5‐bisphosphate 3‐kinase catalytic subunit alpha; qPCR, quantitative polymerase chain reaction; Safe‐SeqS, Safe‐Sequencing System; SOLiD, sequencing by oligonucleotide ligation and detection; TAm‐Seq, tagged‐amplicon deep sequencing.
PCR‐based approaches can detect DNA alterations by amplifying small DNA regions of interest known as hotspots (Table 1). These assays can be used to detect driver mutations, such as those found in EGFR, KRAS, or BRAF [23, 24, 25]. The assays can also be used to identify the emergence of predefined treatment‐resistant clones in blood, often several weeks before imaging methods can confirm clinical progression, and to monitor emergence of specific mutations over the course of treatment [24, 26, 27, 28].
NGS, or high throughput sequencing or massively parallel sequencing, uses distinct approaches to the biochemistry of DNA sequencing to simultaneously perform millions of sequencing reactions. NGS is designed to investigate large regions of multiple genes in a single run and can detect somatic mutations, including single‐nucleotide variations (SNVs), copy number variations (CNVs), indels, and gene fusions [11, 26]. NGS platforms include whole genome sequencing, whole exome sequencing, hybrid capture panels, and amplicon sequencing panels (Table 1). The broad spectrum of genomic information from NGS surpasses that from PCR and fluorescence in situ hybridization techniques combined. However, the increased number of genes evaluable by NGS often reduces sensitivity, sometimes leading to the option of selecting PCR assays to maximize sensitivity for detection of a patient‐specific single‐nucleotide variant. More recently, cfDNA genotyping assays that combine PCR and NGS technologies have been developed to maximize on the advantages of both platforms.
FDA‐Approved Assays for Plasma cfDNA Genotyping in NSCLC
Several plasma cfDNA genotyping assays for NSCLC are in clinical practice, including PCR‐ and NGS‐based assays. Most are laboratory‐developed tests (LDTs) [29] and are performed in Clinical Laboratory Improvement Amendments (CLIA)–certified laboratories and monitored by the Centers for Medicare & Medicaid Services (CMS). As such, most tests in clinical practice are used in the market as LDTs, with a few that have received regulatory approval. A number of tests have received FDA approval as companion diagnostic (CDx) assays or have received FDA's breakthrough device designation.
Cobas EGFR Mutation Test
The cobas EGFR Mutation Test v2 (Roche Molecular Systems Inc., Pleasanton, CA; https://diagnostics.roche.com) was the first commercially available plasma‐genotyping test to receive FDA approval in June 2016 for the identification of patients with EGFR driver mutations who may benefit from treatment with tyrosine kinase inhibitors (TKIs) [30, 31]. The test kit is based on real‐time quantitative PCR (qPCR), which differs from classic PCR in that the intensity of the fluorescent light emitted by the probes is read every cycle, allowing for an estimate of the quantity of the loaded sample to be derived based on the number of cycles needed to obtain a threshold fluorescent signal [32]. The cobas EGFR assay is designed to detect G719X substitutions in exon 18, deletion mutations in exon 19, T790M and S768I substitutions and insertions in exon 20, and L858R and L861Q substitutions in exon 21 and is used as a CDx test for erlotinib [30]. Clinical trials in patients with advanced NSCLC showed that patients with exon 19 deletions or L858R substitutions in exon 21 who were treated with erlotinib as first‐line treatment were likely to experience clinical benefit compared with patients treated with chemotherapy [33, 34].
Guardant360 CDx
Guardant360 CDx (Guardant Health, Redwood City, CA; https://www.guardanthealth.com) is a commercially available targeted NGS panel that uses hybridization‐capture technology coupled with NGS. A subset of the exome or predetermined DNA sequences of interest are hybridized to biotinylated probes and captured using streptavidin, and then the captured DNA is sequenced by NGS. To date, Guardant360 CDx can detect ctDNA mutations (74 genes), amplifications (18 genes), fusions (6 genes), and indels (23 genes) [35, 36]. This includes detection of the National Comprehensive Cancer Network (NCCN) clinical guideline‐recommended biomarker mutations (EGFR, ALK, ROS1, BRAF, RET, MET, NTRK, KRAS, and ERBB2 [HER2]) for NSCLC. The FDA granted Guardant360 CDx breakthrough device status in January 2018 [37] and approved it for comprehensive genomic profiling in patients with any solid malignant neoplasm and as a CDx for the EGFR inhibitor osimertinib in August 2020 [38]. In May 2021, the FDA approved Guardant360 CDx for comprehensive genomic profiling in adult patients with KRAS G12C‐mutated locally advanced or metastatic NSCLC and as a CDx for the recently approved KRASG12C inhibitor sotorasib [4, 39].
In a prospective cohort study involving 323 patients with NSCLC [8], the addition of plasma cfDNA genotyping to tissue‐based genotyping markedly increased identification of targetable mutations, facilitating delivery of therapies that matched the patients’ DNA mutation. In 229 patients, actionable mutations were detected in 20.5% of patients with tissue genotyping alone, which increased to 35.8% with the use of Guardant360 CDx [8].
Guardant360 CDx was subsequently shown to have the same rate of biomarker detection as traditional tissue genotyping in the multi‐institution, head‐to‐head, Noninvasive versus Invasive Lung Evaluation (NILE) study [40] that analyzed 282 patients with newly diagnosed advanced NSCLC. Biomarkers were detected in a higher proportion of patients with plasma cfDNA genotyping than with tissue genotyping (77 patients, 27.3% vs. 60 patients, 21.3%; p < .0001), with the cfDNA genotyping results delivered significantly faster than the tissue genotyping results (median, 9 days vs. 15 days) [40]. A recent retrospective analysis of samples from the FLAURA [41] and AURA3 [42] studies also showed that Guardant360 CDx had similar diagnostic performance to the cobas EGFR Mutation Test in identifying patients with NSCLC who were positive for EGFR exon 19 deletions and T790M and L858R substitutions eligible for treatment with osimertinib [43].
FoundationOne Liquid CDx
FoundationOne Liquid CDx (Foundation Medicine, Cambridge, MA; https://www.foundationmedicine.com) is a commercially available targeted NGS panel that uses hybridization‐capture technology to detect more than 300 cancer‐related genes and multiple genomic signatures such as tumor mutational burden (TMB) and microsatellite instability. In August 2020, the FDA approved the FoundationOne Liquid CDx for comprehensive genomic profiling in patients with any solid tumor and for use as a CDx assay to identify patients with NSCLC who may benefit from treatment with three first‐line TKIs, gefitinib, osimertinib, and erlotinib [44]. Clinical validity of FoundationOne Liquid CDx as an aid in identifying patients with advanced NSCLC who may be eligible for treatment with the three first‐line TKIs was established through a noninferiority study that compared FoundationOne Liquid CDx with the cobas EGFR Mutation Test in identifying EGFR exon 19 deletion and EGFR exon 21 L858R substitutions involving 177 samples from patients with NSCLC [45].
Emerging Evidence to Support Use of Plasma cfDNA Genotyping in Oncology
Tissue genotyping, coupled with imaging (CT and MRI), remains the standard of care in oncology. Nevertheless, plasma cfDNA genotyping has notable advantages over tissue genotyping. Blood draws are less invasive and less risky, making plasma cfDNA genotyping more appealing to clinicians and patients. The turnaround time for plasma cfDNA genotyping is less than that for tissue genotyping, as the latter requires wait time for scheduling and performing the biopsy and tissue processing. cfDNA genotyping delivered results faster than tissue genotyping in the NILE study (median, 9 days vs. 15 days) [40]. In another study [46], the median time from pathologic diagnosis to delivery of genotyping results was 3 days with cfDNA plasma NGS performed before or in parallel with the diagnostic procedure versus 18 days with cfDNA plasma NGS and 35.5 days with tissue NGS, respectively, performed at the end of the diagnostic procedure per the routine standard of care. Technological advances have made it possible for clinicians to process blood on‐site or ship it to central laboratories for testing, limiting the earlier challenges of variable results because of sample collection and pretreatment conditions that have the potential to affect genotyping results. More importantly, plasma cfDNA genotyping provides information on the complete heterogeneity (both spatial and temporal) of the tumor as compared with a snapshot from a single needle biopsy. Furthermore, repeated sampling is more feasible with plasma cfDNA genotyping and allows real‐time monitoring of treatment efficacy, development of resistance, and cancer progression [11, 47].
A few studies have shown concordance between results obtained from plasma and tissue genotyping [48]. A study that evaluated orthogonal plasma and tissue genotyping using NGS‐based digital sequencing in >750 patients with solid tumors demonstrated high accuracy and specificity (>99% positive percent agreement and negative percent agreement and >92% positive predictive values) [35]. As already described in the section on Guardant360 CDx, the NILE study [40] demonstrated that Guardant360 CDx identified guideline‐recommended biomarkers at a rate at least as high as tissue genotyping in untreated metastatic NSCLCs, with high concordance, lower turnaround time, and higher biomarker discovery rate [40], and a prospective cohort study of patients with NSCLC [8] demonstrated that addition of plasma cfDNA genotyping to tissue genotyping markedly improved identification of targetable mutations, facilitating delivery of therapies that matched the patients’ DNA mutation. A retrospective subanalysis of the IFUM trial [49] that evaluated whether ctDNA could be used as a surrogate for determination of EGFR status using PCR to analyze exon 19 deletions, L858R mutation, and T790M mutation in paired tissue and plasma samples showed an agreement of 94.3% between 652 matched tumor and plasma samples, independent of mutation subtype; the test's sensitivity and specificity were 65.7% and 99.8%, respectively [49]. In a recent retrospective meta‐analysis of 25 studies involving 4,881 lung cancer cases [50], the sensitivity and specificity of EGFR mutation as detected by PCR genotyping of plasma cfDNA compared with matched tissue genotyping were 65.3% and 98.2%, respectively.
A recent position publication by the International Association for the Study of Lung Cancer [11] recommends implementation of plasma cfDNA genotyping in the clinic in a number of relevant therapeutic settings and provides algorithms to aid practicing oncologists in making treatment decisions for patients with advanced, treatment‐naïve NSCLC and patients with progressive or recurrent NSCLC. According to a previously published algorithm developed as a tool to aid in clinical decisions [51], plasma cfDNA genotyping can be used to test for detectable driver alterations if tissue is unavailable or inadequate for comprehensive genotyping or can be performed concurrently with tissue sequencing when tissue samples are available. Given the concordance between results obtained from plasma cfDNA and tissue genotyping [8, 35, 40, 49, 50] and the previously published recommendations [11, 51], a consideration is to use plasma cfDNA genotyping as the initial method to assess specimens from patients with NSCLC and provide critical information that could potentially be used to guide clinical decisions in some situations. In cases in which results from plasma cfDNA genotyping are inconclusive, tissue genotyping can then be performed. However, the approach of initially using plasma cfDNA genotyping alone and then using tissue genotyping only if results from plasma genotyping are negative is not currently considered standard of care; this approach should be reserved for special situations, such as in the context of acquired resistance or in selected cases at diagnosis [51].
Limitations of Plasma cfDNA Genotyping
Several limitations must be taken into consideration in the use of plasma cfDNA genotyping. For advanced NSCLC, the presence of cfDNA has been reported in approximately 85% of cases [52]. However, treatment‐naïve patients with slow‐growing tumors may be at risk of false‐negative findings [11]. A recent retrospective study that evaluated plasma samples from the FLAURA [41] and AURA3 [42] trials using the cobas EGFR Mutation Test demonstrated that EGFR exon 19 deletion and L858R and T790M mutations were not detected in plasma of a subset of patients identified to have EGFR‐mutant NSCLC by tissue genotyping as not all tumors shed detectable levels of mutated ctDNA into systemic circulation [43]. Patients who have visceral or extrathoracic disease are more likely to have detectable ctDNA in blood [8]. Because of this inherent limitation, it is recommended that, although positive findings from plasma cfDNA genotyping are clinically actionable, negative findings should be considered inconclusive and warrant tissue genotyping [11]. Another consideration in the context of NGS is the potential for false‐positive findings arising from somatic mutations in hematopoietic stem cells (clonal hematopoiesis of indeterminate potential; CHIP) [53, 54, 55]. Because most cfDNA fragments in blood originate from hematopoietic cells, these somatic mutations could be falsely identified as tumor‐specific mutations. However, most current algorithms filter out CHIP false positives [53, 56]. Additionally, plasma genotyping does not capture histologic transformation, which is a phenotypic switch between tumor histologies that can occur in patients with oncogene addicted NSCLC leading to resistance to therapy in the absence of significant changes in mutational profile seen in ctDNA [57, 58].
Reimbursement for plasma cfDNA genotyping is currently limited. CMS proposes that the evidence is sufficient to expand coverage of NGS as a diagnostic laboratory test when performed in a CLIA‐certified laboratory and in limited situations, restricting coverage to monitoring relapse, metastasis, or advanced stage III and IV cancers [59]. Accumulation of more extensive data from prospective trials of plasma cfDNA genotyping such as the NILE study [40], recommendations by clinical guidelines, and FDA approval of assays would likely increase the support of payors for plasma cfDNA genotyping and enable its broad adoption as standard of care in oncology.
Clinical Applications of Plasma cfDNA Genotyping
Current Clinical Applications of Plasma cfDNA Genotyping
Plasma cfDNA genotyping is routinely used in clinical practice as a complement to tissue genotyping to assist in guiding therapy at diagnosis and monitoring emergence of resistance to therapy [11] (Fig. 1).
Figure 1.
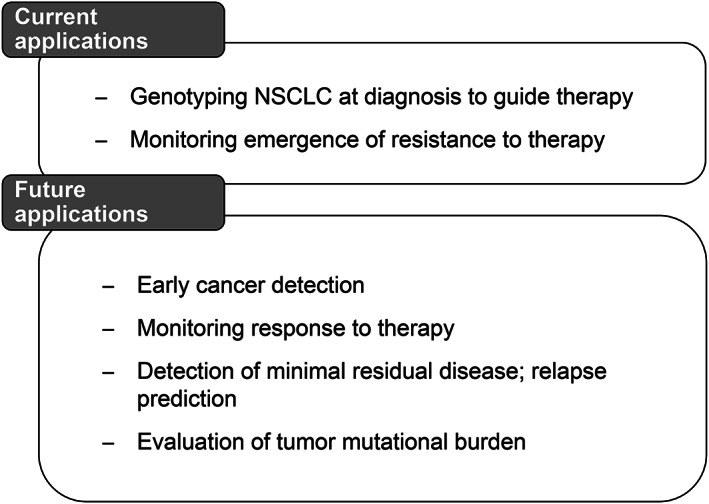
Broad potential application of plasma cell‐free DNA genotyping in NSCLC for precision medicine and improved patient care. Abbreviation: NSCLC, non‐small cell lung cancer.
Genotyping NSCLC at Diagnosis to Guide Therapy Decisions
In treatment‐naïve patients, plasma cfDNA genotyping should be considered at the time of initial diagnosis in all patients who need tumor genotyping and is particularly recommended when tumor tissue is scarce or unavailable or when a significant delay (typically ≥2 weeks) is expected in obtaining tumor tissue or tissue sequencing results [11]. Current clinical guidelines [1, 2, 3] recommend testing treatment‐naïve patients for known oncogenic driver alterations in EGFR, ALK, ROS1, BRAF, MET, RET, and NTRK, and if present, using one of several targeted therapies directed against the known gene mutations.
Monitoring Emergence of Resistance to Therapy
In addition to detecting mutations among treatment‐naïve patients, plasma cfDNA genotyping can also be used to track the emergence of resistance mutations during therapy. Patients with EGFR‐mutated or ALK‐rearranged NSCLCs treated with the indicated TKIs have been shown to develop resistance mutations associated with disease progression, and to benefit from successive lines of TKIs that can specifically target these new mutations [11]. Monitoring for the appearance of resistance mutations requires repeated tumor sampling to assess the mechanism of resistance, which requires performing serial biopsies at the site of tumor progression and is more suited to plasma versus tissue genotyping [11, 60].
After identification of the specific resistance mutation, the next step is matching the mutations to the most appropriate treatment agent [60]. Plasma cfDNA genotyping offers the advantage of early, noninvasive detection of resistance mutations. A comparison of plasma cfDNA genotyping and CT imaging in detecting early progression as indicated by the emergence of the T790M mutation showed plasma cfDNA genotyping detecting the mutation earlier than CT by 51 days in one study [61] and 103 days in another [62]. Of note, longitudinal monitoring of changes in ctDNA using plasma cfDNA genotyping is not a standard approach outside the research setting. Even if plasma cfDNA genotyping identifies resistance mutations that indicate early progression, the current clinical recommendation is to first confirm radiographic and clinical progression before modifying therapy. As such, plasma cfDNA genotyping results can be used to plan subsequent lines of therapy but not for making decisions to change therapy.
Future Clinical Applications of Plasma cfDNA Genotyping
Technical advances in plasma cfDNA genotyping to increase sensitivity and specificity are opening new potential applications including use in early cancer detection, monitoring response to therapy, detection of minimal residual disease (MRD), and evaluation of TMB [11, 63] (Fig. 1). These uses are likely to become part of routine clinical practice in the coming years.
Early Cancer Detection
Per NCCN guidelines [1], there is currently insufficient evidence to support use of plasma cfDNA genotyping for establishing primary lung cancer diagnosis as ctDNA shedding is low in early stages of NSCLC and ctDNA is not reliably detected with currently available technologies [26, 64, 65]. However, as sensitivity and specificity improve, plasma cfDNA genotyping could ultimately be used in NSCLC early detection and diagnosis. Leading technologies in this field have been granted FDA breakthrough device designation (Table 2) and include the GRAIL multicancer early detection platform (GRAIL; Menlo Park, CA; https://grail.com/), the CancerSEEK platform (Thrive Earlier Detection, Cambridge, MA; https://thrivedetect.com/) for cancer diagnosis, and the Ivy‐Gene CORE Test (Helio Health, Irvine, CA; https://www.heliohealth.com) for detecting variable ctDNA methylation patterns to confirm presence of early‐stage cancers. Another promising technology is the DELFI technology that specifically detects alternations in nucleosomal fragmentation profiles in plasma cfDNA from patients with cancer [66].
Table 2.
NSCLC plasma cfDNA genotyping assays that have received FDA approval or breakthrough device designation
| Kit/test | Company | Technology/application | FDA status |
|---|---|---|---|
| Targeted PCR‐based | |||
| cobas EGFR Mutation Test v2 | Roche Molecular Systems | RT‐PCR for detection of 42 gene mutations in EGFR | FDA approved in June 2016 |
| Multiplex NGS‐based | |||
| Guardant360 CDx | Guardant Health | NGS test that detects point mutations (74 genes), amplifications (18 genes), fusions (6 genes), and indels (23 genes) to guide treatment selection in non‐small‐cell lung carcinoma | FDA approved in August 2020; breakthrough device designation in May 2019 |
| FoundationOne Liquid CDx | Foundation Medicine | NGS test that detects over 300 cancer‐related genes including clinically relevant indels, substitutions, copy number variants, and selected genetic rearrangements in 70 oncogenes for companion diagnostics; detects multiple signatures including tumor mutational burden and microsatellite instability | FDA approved in August 2020; breakthrough device designation in April 2018 |
| Multicancer early detection test | GRAIL | NGS blood test analyzing ctDNA methylation patterns for detecting multiple cancer types | FDA breakthrough device designation in May 2019 |
| CancerSEEK | Thrive Earlier Detection | Multianalyte test that combines multiplexed PCR detection of mutations in ctDNA at 1,933 loci with measurements of validated protein biomarkers to diagnose eight common cancer types including breast, ovarian, and liver cancer | FDA breakthrough device designation in August 2018 |
| Ivy‐Gene CORE Test; Ivy‐Gene Dx Liver Test | Laboratory for Advanced Medicine | Analyzes presence of hyper‐methylated ctDNA from multiple gene targets to confirm the presence of breast, colon, liver, and lung cancers as early as stage I | FDA breakthrough device designation in September 2019 |
Abbreviations: CDx, companion diagnostics; cfDNA, cell‐free DNA; ctDNA, circulating tumor DNA; EGFR, epidermal growth factor receptor; FDA, U.S. Food and Drug Administration; NGS, next‐generation sequencing; NSCLC, non‐small cell lung cancer; PGM, personal genome machine; PCR, polymerase chain reaction; RT‐PCR, real‐time polymerase chain reaction.
The most advanced of the technologies in development is the GRAIL platform that employs DNA methylation signatures to detect early‐stage cancers. DNA methylation is a biological mechanism that controls genomic instructions that are carried out in the body [67]. The platform uses targeted bisulfite sequencing and machine learning to detect cfDNA methylation patterns and identify those that are abnormally methylated, with the additional ability of determining the tissue of origin of the ctDNA [68]. Use of this platform for population scale mass cancer screening is currently being validated in four clinical trials that enrolled a combined 180,000 participants in North America and the U.K. (CCGA [NCT02889978], STRIVE [NCT03085888], SUMMIT [NCT03934866], and PATHFINDER [NCT04241796]) [68].
Monitoring Response to Therapy
Tumors can change mutation patterns or acquire new mutations over time [69]. Pretreatment ctDNA levels have been shown to be prognostic and on‐treatment ctDNA levels to be predictive for patients with NSCLC receiving immune checkpoint inhibitors [70, 71]. In this context, plasma cfDNA genotyping to detect changes in ctDNA levels can be used to monitor the molecular make up of a patient's tumor over time. This has the potential of sparing patients from undergoing repeated invasive procedures to obtain tissue biopsy samples and enables monitoring of the patients’ state of disease systemically (as deciphered from biomarkers in the blood), and not just relying on results from a single, localized cell population represented by a needle biopsy [11].
Detection of Minimal Residual Disease
Detection of residual ctDNA following surgery or curative treatment can be used as a surrogate for MRD, predicting future relapse. ctDNA levels may be low following treatment and therefore require highly sensitive assays for detection. A few studies have demonstrated the ability of ctDNA to detect post‐treatment MRD, which was shown to be prognostic [72, 73]. The TRACERx study in early‐stage lung cancer [73] demonstrated that MRD indicative of recurrence could be detected in plasma at a median of 70 days before imaging‐confirmed relapse, suggesting the utility of plasma cfDNA genotyping in this setting. However, ctDNA analysis for MRD detection is not yet approved for clinical practice.
Evaluation of Tumor Mutational Burden
Studies have shown that an increase in somatic mutations present in tumor cells increases the potential for recognition of the tumor cells by the immune system [74]. The presence of mutations in the tumor generates antigens that are not expressed by normal cells (neoantigens), and the higher the TMB, the more the tumor is likely to be immunogenic [74]. Novel assays to measure TMB from blood have been developed, and these have demonstrated agreement of TMB derived from plasma and tissue genotyping [75, 76]. Additionally, evidence has emerged that a high TMB is associated with increased clinical activity of inhibitors of programmed cell death (PD) 1 and its ligand PD‐L1 in NSCLC [76, 77]. As such, plasma TMB has now become an important cfDNA genotyping marker and can potentially be used to stratify patients likely to respond to immunotherapy [63]. The FDA‐approved FoundationOne Liquid CDx includes TMB assessment as part of the platform.
Need for Prospective Trials to Assess the Clinical Utility of Plasma cfDNA Genotyping
Data from prospective clinical trials of plasma cfDNA genotyping are very limited, with clinical trial data available only from the NILE study [40]. Most current data supporting the clinical utility of plasma cfDNA genotyping comes from retrospective observational studies. Going forward, there is a need for more prospective clinical trials to evaluate use of plasma cfDNA genotyping in the clinic.
Conclusion
Plasma cfDNA genotyping has considerable potential in improving the management of patients with NSCLC as it offers an alternative when tissue biopsy specimens are insufficient or unfeasible. It also provides information on both spatial and temporal dynamic changes in tumor profiles that can be used to guide treatment decisions. Plasma cfDNA genotyping is currently being pursued in NSCLC, with some assays having received regulatory approval and having been put into clinical use; additional assays are in the development and validation stages. More regulatory approvals of plasma cfDNA genotyping assays are expected as the demand for plasma‐based technologies is increasing in oncology. Numerous studies have shown that plasma cfDNA genotyping is feasible in clinical practice. Broad adoption of plasma cfDNA genotyping as a standard‐of‐care tool in oncology practice depends on gathering prospective data to validate assays and identifying the most effective testing strategies to implement at different stages of NSCLC.
Plasma cfDNA genotyping has evolved from an emerging concept and is currently aiding oncologists in the delivery of personalized care by facilitating matching of patients with targeted therapy and monitoring emergence of resistance to therapy. Further advances currently underway to increase assay sensitivity and specificity will potentially expand the use of plasma cfDNA genotyping in early cancer detection, monitoring response to therapy, detection of MRD, and measurement of TMB in NSCLC.
Author Contributions
Conception/design: Jhanelle Gray, Jeffrey C. Thompson, Erica L. Carpenter, Ehab Elkhouly, Charu Aggarwal
Collection and/or assembly of data: Jhanelle Gray, Jeffrey C. Thompson, Erica L. Carpenter, Ehab Elkhouly, Charu Aggarwal
Data analysis and interpretation: Jhanelle Gray, Jeffrey C. Thompson, Erica L. Carpenter, Ehab Elkhouly, Charu Aggarwal
Manuscript writing: Jhanelle Gray, Jeffrey C. Thompson, Erica L. Carpenter, Ehab Elkhouly, Charu Aggarwal
Final approval of manuscript: Jhanelle Gray, Jeffrey C. Thompson, Erica L. Carpenter, Ehab Elkhouly, Charu Aggarwal
Disclosures
Jhanelle Gray: AstraZeneca, Boehringer Ingelheim, Bristol‐Myers Squibb, Genentech, GI Therapeutics, Merck, Pfizer, Novartis, Ludwig Institute of Cancer Research (RF); AstraZeneca, Blueprint Medicines, Bristol‐Myers Squibb, EMD Serono, Merck KGaA, Inivata, Merck, Novartis (C/A). Jeffrey C. Thompson: AstraZeneca, Guardant Health (SAB); Erica L. Carpenter: Janssen, Becton Dickinson, United Health Group, Merck (C/A); Guardant Health, Imedex, AstraZeneca, FoxChase Cancer Center, Bristol‐Myers Squibb (H); Menarini, VPS, American Gastroenterological Association, Imedex, AstraZeneca, Foundation Medicine, Gordon Conference, Americas Hepato‐Pancreato‐Biliary Association (Other—travel assistance). Ehab Elkhouly: Amgen (E), Amgen (OI). Charu Aggarwal: Merck, MacroGenics, Novartis, AstraZeneca (RF—institution); AstraZeneca, Blueprint, Celgene, Eli Lilly & Co., Merck, Daiichi Sankyo (SAB).
(C/A) Consulting/advisory relationship; (RF) Research funding; (E) Employment; (ET) Expert testimony; (H) Honoraria received; (OI) Ownership interests; (IP) Intellectual property rights/inventor/patent holder; (SAB) Scientific advisory board
Acknowledgments
We thank Byeong Yoon, Ph.D. (Amgen Inc.) for critical review of the manuscript and Martha Mutomba (on behalf of Amgen Inc.) for medical writing assistance. This study was supported by Amgen Inc. in the form of medical writing, submission fee, and open access publication.
Disclosures of potential conflicts of interest may be found at the end of this article.
References
- 1.NCCN clinical practice guidelines in oncology (NCCN Guidelines): Non‐Small Cell Lung Cancer. Version 2.2021. Plymouth Meeting, PA: National Comprehensive Cancer Center, 2021. Available at https://www.nccn.org/professionals/physician_gls/pdf/nscl.pdf. Accessed June 5, 2021. [Google Scholar]
- 2.Kalemkerian GP, Narula N, Kennedy EB et al. Molecular testing guideline for the selection of patients with lung cancer for treatment with targeted tyrosine kinase inhibitors: American Society of Clinical Oncology endorsement of the College of American Pathologists/International Association for the Study of Lung Cancer/Association for Molecular Pathology clinical practice guideline update. J Clin Oncol 2018;36:911–919. [DOI] [PubMed] [Google Scholar]
- 3.Lindeman NI, Cagle PT, Aisner DL et al. Updated molecular testing guideline for the selection of lung cancer patients for treatment with targeted tyrosine kinase inhibitors: Guideline from the College of American Pathologists, the International Association for the Study of Lung Cancer, and the Association for Molecular Pathology. Arch Pathol Lab Med 2018;142:321–346. [DOI] [PubMed] [Google Scholar]
- 4.Lumakras (sotorasib). Package insert. Available at https://www.accessdata.fda.gov/drugsatfda_docs/label/2021/214665s000lbl.pdf. Accessed June 5, 2021.
- 5.Pennell NA, Arcila ME, Gandara DR et al. Biomarker testing for patients with advanced non‐small cell lung cancer: Real‐world issues and tough choices. Am Soc Clin Oncol Educ Book 2019;39:531–542. [DOI] [PubMed] [Google Scholar]
- 6.Kim ES, Roy UB, Ersek JL et al. Updates regarding biomarker testing for non‐small cell lung cancer: Considerations from the National Lung Cancer Roundtable. J Thorac Oncol 2019;14:338–342. [DOI] [PubMed] [Google Scholar]
- 7.Lim C, Sekhon HS, Cutz JC et al. Improving molecular testing and personalized medicine in non‐small‐cell lung cancer in Ontario. Curr Oncol 2017;24:103–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aggarwal C, Thompson JC, Black TA et al. Clinical implications of plasma‐based genotyping with the delivery of personalized therapy in metastatic non‐small cell lung cancer. JAMA Oncol 2019;5:173–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lim C, Tsao MS, Le LW et al. Biomarker testing and time to treatment decision in patients with advanced nonsmall‐cell lung cancer. Ann Oncol 2015;26:1415–1421. [DOI] [PubMed] [Google Scholar]
- 10.Hellmann MD, Ciuleanu TE, Pluzanski A et al. Nivolumab plus ipilimumab in lung cancer with a high tumor mutational burden. N Engl J Med 2018;378:2093–2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rolfo C, Mack PC, Scagliotti GV et al. Liquid biopsy for advanced non‐small cell lung cancer (NSCLC): A statement paper from the IASLC. J Thorac Oncol 2018;13:1248–1268. [DOI] [PubMed] [Google Scholar]
- 12.Siravegna G, Marsoni S, Siena S et al. Integrating liquid biopsies into the management of cancer. Nat Rev Clin Oncol 2017;14:531–548. [DOI] [PubMed] [Google Scholar]
- 13.Crowley E, Di Nicolantonio F, Loupakis F et al. Liquid biopsy: Monitoring cancer‐genetics in the blood. Nat Rev Clin Oncol 2013;10:472–484. [DOI] [PubMed] [Google Scholar]
- 14.Jahr S, Hentze H, Englisch S et al. DNA fragments in the blood plasma of cancer patients: Quantitations and evidence for their origin from apoptotic and necrotic cells. Cancer Res 2001;61:1659–1665. [PubMed] [Google Scholar]
- 15.Kidess E, Jeffrey SS. Circulating tumor cells versus tumor‐derived cell‐free DNA: Rivals or partners in cancer care in the era of single‐cell analysis? Genome Med 2013;5:70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haber DA, Velculescu VE. Blood‐based analyses of cancer: Circulating tumor cells and circulating tumor DNA. Cancer Discov 2014;4:650–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Diehl F, Schmidt K, Choti MA et al. Circulating mutant DNA to assess tumor dynamics. Nat Med 2008;14:985–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lo YM, Zhang J, Leung TN et al. Rapid clearance of fetal DNA from maternal plasma. Am J Hum Genet 1999;64:218–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pasini L, Ulivi P. Liquid biopsy for the detection of resistance mechanisms in NSCLC: Comparison of different blood biomarkers. J Clin Med 2019;8:998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee JS, Hur JY, Kim IA et al. Liquid biopsy using the supernatant of a pleural effusion for EGFR genotyping in pulmonary adenocarcinoma patients: A comparison between cell‐free DNA and extracellular vesicle‐derived DNA. BMC Cancer 2018;18:1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Momen‐Heravi F, Getting SJ, Moschos SA. Extracellular vesicles and their nucleic acids for biomarker discovery. Pharmacol Ther 2018;192:170–187. [DOI] [PubMed] [Google Scholar]
- 22.Revelo AE, Martin A, Velasquez R et al. Liquid biopsy for lung cancers: An update on recent developments. Ann Transl Med 2019;7:349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guibert N, Pradines A, Casanova A et al. Detection and monitoring of the BRAF mutation in circulating tumor cells and circulating tumor DNA in BRAF‐mutated lung adenocarcinoma. J Thorac Oncol 2016;11:e109–e112. [DOI] [PubMed] [Google Scholar]
- 24.Oxnard GR, Paweletz CP, Kuang Y et al. Noninvasive detection of response and resistance in EGFR‐mutant lung cancer using quantitative next‐generation genotyping of cell‐free plasma DNA. Clin Cancer Res 2014;20:1698–1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guibert N, Pradines A, Farella M et al. Monitoring KRAS mutations in circulating DNA and tumor cells using digital droplet PCR during treatment of KRAS‐mutated lung adenocarcinoma. Lung Cancer 2016;100:1–4. [DOI] [PubMed] [Google Scholar]
- 26.Guibert N, Pradines A, Favre G et al. Current and future applications of liquid biopsy in nonsmall cell lung cancer from early to advanced stages. Eur Respir Rev 2020;29:190052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oxnard GR, Thress KS, Alden RS et al. Association between plasma genotyping and outcomes of treatment with osimertinib (AZD9291) in advanced non‐small‐cell lung cancer. J Clin Oncol 2016;34:3375–3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sacher AG, Alden RS, Oxnard GR. Early intervention in lung cancers with rapid plasma genotyping for EGFR and KRAS mutations‐Reply [letter]. JAMA Oncol 2016;2:1096–1097. [DOI] [PubMed] [Google Scholar]
- 29.Gatter K.FDA oversight of laboratory‐developed tests: Where are we now? Arch Pathol Lab Med 2017;141:746–748. [DOI] [PubMed] [Google Scholar]
- 30.cobas EGFR Mutation Test version 2 . Package insert. Available at https://www.accessdata.fda.gov/cdrh_docs/pdf12/P120019S007c.pdf. Accessed June 5, 2021.
- 31.Food and Drug Administration . cobas® EGFR Mutation Test v2. U.S. Food and Drug Administration Web site. Available at https://www.fda.gov/drugs/resources-information-approved-drugs/cobas-egfr-mutation-test-v2. Accessed June 5, 2021.
- 32.Schmittgen TD, Livak KJ. Analyzing real‐time PCR data by the comparative C(T) method. Nat Protoc 2008;3:1101–1108. [DOI] [PubMed] [Google Scholar]
- 33.Zhou C, Wu YL, Chen G et al. Erlotinib versus chemotherapy as first‐line treatment for patients with advanced EGFR mutation‐positive non‐small‐cell lung cancer (OPTIMAL, CTONG‐0802): A multicentre, open‐label, randomised, phase 3 study. Lancet Oncol 2011;12:735–742. [DOI] [PubMed] [Google Scholar]
- 34.Rosell R, Carcereny E, Gervais R et al. Erlotinib versus standard chemotherapy as first‐line treatment for European patients with advanced EGFR mutation‐positive non‐small‐cell lung cancer (EURTAC): A multicentre, open‐label, randomised phase 3 trial. Lancet Oncol 2012;13:239–246. [DOI] [PubMed] [Google Scholar]
- 35.Odegaard JI, Vincent JJ, Mortimer S et al. Validation of a plasma‐based comprehensive cancer genotyping assay utilizing orthogonal tissue‐ and plasma‐based methodologies. Clin Cancer Res 2018;24:3539–3549. [DOI] [PubMed] [Google Scholar]
- 36.Willis J, Lefterova MI, Artyomenko A et al. Validation of microsatellite instability detection using a comprehensive plasma‐based genotyping panel. Clin Cancer Res 2019;25:7035–7045. [DOI] [PubMed] [Google Scholar]
- 37.Food and Drug Administration . Summary of safety and effectiveness data: Guardant360 CDx. U.S. Food and Drug Administration Web site. Available at https://www.accessdata.fda.gov/cdrh_docs/pdf20/P200010B.pdf. Accessed June 5, 2021.
- 38.Guardant360 CDx . Package insert. Available at https://www.accessdata.fda.gov/cdrh_docs/pdf20/P200010C.pdf. Accessed June 5, 2021.
- 39.Food and Drug Administration . FDA approves first targeted therapy for lung cancer mutation previously considered resistant to drug therapy. News Release. FDA; May 28, 2021. Available at https://www.fda.gov/news‐events/press‐announcements/fda‐approves‐first‐targeted‐therapy‐lung‐cancer‐mutation‐previously‐considered‐resistant‐drug. Accessed June 5, 2021.
- 40.Leighl NB, Page RD, Raymond VM et al. Clinical utility of comprehensive cell‐free DNA analysis to identify genomic biomarkers in patients with newly diagnosed metastatic non‐small cell lung cancer. Clin Cancer Res 2019;25:4691–4700. [DOI] [PubMed] [Google Scholar]
- 41.Soria JC, Ohe Y, Vansteenkiste J et al. Osimertinib in untreated EGFR‐mutated advanced non‐small‐cell lung cancer. N Engl J Med 2018;378:113–125. [DOI] [PubMed] [Google Scholar]
- 42.Mok TS, Wu YL, Ahn MJ et al. Osimertinib or platinum‐pemetrexed in EGFR T790M‐positive lung cancer. N Engl J Med 2017;376:629–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gray JE, Han JY, Telaranta‐Keerie A et al. Clinical performance of a comprehensive novel liquid biopsy test for identifying non‐small cell lung cancer (NSCLC) patients for treatment with osimertinib. J Clin Oncol 2020;38(suppl 15):9553a. [Google Scholar]
- 44.Food and Drug Administration . FDA approves liquid biopsy next‐generation sequencing companion diagnostic test. U.S. Food and Drug Administration Web site, 2020. Available at https://www.fda.gov/drugs/drug-approvals-and-databases/fda-approves-liquid-biopsy-next-generation-sequencing-companion-diagnostic-test. Accessed June 5, 2021.
- 45.Woodhouse R, Li M, Hughes J et al. Clinical and analytical validation of FoundationOne Liquid CDx, a novel 324‐gene cfDNA‐based comprehensive genomic profiling assay for cancers of solid tumor origin. PLoS One 2020;15:e0237802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cheng ML, Milan MSD, Tamen RM et al. Plasma cfDNA genotyping in hospitalized patients with suspected metastatic NSCLC. JCO Precision Oncology 2021:726–732. [DOI] [PubMed] [Google Scholar]
- 47.Saarenheimo J, Eigeliene N, Andersen H et al. The value of liquid biopsies for guiding therapy decisions in non‐small cell lung cancer. Front Oncol 2019;9:129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thompson JC, Yee SS, Troxel AB et al. Detection of therapeutically targetable driver and resistance mutations in lung cancer patients by next‐generation sequencing of cell‐free circulating tumor DNA. Clin Cancer Res 2016;22:5772–5782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Douillard JY, Ostoros G, Cobo M et al. Gefitinib treatment in EGFR mutated Caucasian NSCLC: Circulating‐free tumor DNA as a surrogate for determination of EGFR status. J Thorac Oncol 2014;9:1345–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li C, He Q, Liang H et al. Diagnostic accuracy of droplet digital PCR and amplification refractory mutation system PCR for detecting EGFR mutation in cell‐free DNA of lung cancer: A meta‐analysis. Front Oncol 2020;10:290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Aggarwal C, Rolfo CD, Oxnard GR et al. Strategies for the successful implementation of plasma‐based NSCLC genotyping in clinical practice. Nat Rev Clin Oncol 2021;18:56–62. [DOI] [PubMed] [Google Scholar]
- 52.Zill OA, Banks KC, Fairclough SR et al. The landscape of actionable genomic alterations in cell‐free circulating tumor DNA from 21,807 advanced cancer patients. Clin Cancer Res 2018;24:3528–3538. [DOI] [PubMed] [Google Scholar]
- 53.Hu Y, Ulrich BC, Supplee J et al. False‐positive plasma genotyping due to clonal hematopoiesis. Clin Cancer Res 2018;24:4437–4443. [DOI] [PubMed] [Google Scholar]
- 54.Acuna‐Hidalgo R, Sengul H, Steehouwer M et al. Ultra‐sensitive sequencing identifies high prevalence of clonal hematopoiesis‐associated mutations throughout adult life. Am J Hum Genet 2017;101:50–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Steensma DP, Bejar R, Jaiswal S et al. Clonal hematopoiesis of indeterminate potential and its distinction from myelodysplastic syndromes. Blood 2015;126:9–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Razavi P, Li BT, Brown DN et al. High‐intensity sequencing reveals the sources of plasma circulating cell‐free DNA variants. Nat Med 2019;25:1928–1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schoenfeld AJ, Chan JM, Kubota D et al. Tumor analyses reveal squamous transformation and off‐target alterations as early resistance mechanisms to first‐line osimertinib in EGFR‐mutant lung cancer. Clin Cancer Res 2020;26:2654–2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Piotrowska Z, Hata AN. Resistance to first‐line osimertinib in EGFR‐mutant NSCLC: Tissue is the issue. Clin Cancer Res 2020;26:2441–2443. [DOI] [PubMed] [Google Scholar]
- 59.Centers for Medicare & Medicaid Services . Proposed decision memo for next‐generation sequencing (NGS) for Medicare beneficiaries with advanced cancer (CAG‐00450R). Centers for Medicare & Medicaid Services, 2019. Available at https://www.cms.gov/medicare-coverage-database/details/nca-proposed-decision-memo.aspx?NCAId=296. Accessed June 5, 2021.
- 60.Gainor JF, Dardaei L, Yoda S et al. Molecular mechanisms of resistance to first‐ and second‐generation ALK inhibitors in ALK‐rearranged lung cancer. Cancer Discov 2016;6:1118–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Provencio M, Torrente M, Calvo V et al. Dynamic circulating tumor DNA quantification for the individualization of non‐small‐cell lung cancer patients treatment. Oncotarget 2017;8:60291–60298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kim CG, Shim HS, Hong MH et al. Detection of activating and acquired resistant mutation in plasma from EGFR‐mutated NSCLC patients by peptide nucleic acid (PNA) clamping‐assisted fluorescence melting curve analysis. Oncotarget 2017;8:65111–65122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Aggarwal C, Thompson JC, Chien AL et al. Baseline plasma tumor mutation burden predicts response to pembrolizumab‐based therapy in patients with metastatic non‐small cell lung cancer. Clin Cancer Res 2020;26:2354–2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Newman AM, Bratman SV, To J et al. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat Med 2014;20:548–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cohen JD, Li L, Wang Y et al. Detection and localization of surgically resectable cancers with a multi‐analyte blood test. Science 2018;359:926–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cristiano S, Leal A, Phallen J et al. Genome‐wide cell‐free DNA fragmentation in patients with cancer. Nature 2019;570:385–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Huang CC, Du M, Wang L. Bioinformatics analysis for circulating cell‐free DNA in cancer. Cancers (Basel) 2019;11:805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Liu MC, Oxnard GR, Klein EA et al. Sensitive and specific multi‐cancer detection and localization using methylation signatures in cell‐free DNA. Ann Oncol 2020;31:745–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Vogelstein B, Papadopoulos N, Velculescu VE et al. Cancer genome landscapes. Science 2013;339:1546–1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nabet BY, Esfahani MS, Moding EJ et al. Noninvasive early identification of therapeutic benefit from immune checkpoint inhibition. Cell 2020;183:363–376.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhang Q, Luo J, Wu S et al. Prognostic and predictive impact of circulating tumor DNA in patients with advanced cancers treated with immune checkpoint blockade. Cancer Discov 2020;10:1842–1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Abbosh C, Birkbak NJ, Swanton C. Early stage NSCLC ‐ challenges to implementing ctDNA‐based screening and MRD detection. Nat Rev Clin Oncol 2018;15:577–586. [DOI] [PubMed] [Google Scholar]
- 73.Abbosh C, Birkbak NJ, Wilson GA et al. Phylogenetic ctDNA analysis depicts early‐stage lung cancer evolution. Nature 2017;545:446–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.El‐Deiry WS, Goldberg RM, Lenz HJ et al. The current state of molecular testing in the treatment of patients with solid tumors, 2019. CA Cancer J Clin 2019;69:305–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Parpart‐Li S, Bartlett B, Popoli M et al. The effect of preservative and temperature on the analysis of circulating tumor DNA. Clin Cancer Res 2017;23:2471–2477. [DOI] [PubMed] [Google Scholar]
- 76.Gandara DR, Paul SM, Kowanetz M et al. Blood‐based tumor mutational burden as a predictor of clinical benefit in non‐small‐cell lung cancer patients treated with atezolizumab. Nat Med 2018;24:1441–1448. [DOI] [PubMed] [Google Scholar]
- 77.Rizvi NA, Hellmann MD, Snyder A et al. Cancer immunology. Mutational landscape determines sensitivity to PD‐1 blockade in non‐small cell lung cancer. Science 2015;348:124–128. [DOI] [PMC free article] [PubMed] [Google Scholar]


