The cystallization and structure determination of the armadillo repeat domain of Drosophila SARM1 are described. The phase problem was solved using the MIRAS technique with autoSHARP, combining diffraction data from native, selenomethionine-labelled and bromide-soaked crystals.
Keywords: armadillo repeat, Drosophila SARM1, nicotinamide mononucleotide, X-ray crystallography, multiple isomorphous replacement, anomalous scattering, phase combination
Abstract
The crystal structure determination of the armadillo repeat motif (ARM) domain of Drosophila SARM1 (dSARM1ARM) is described, which required the combination of a number of sources of phase information in order to obtain interpretable electron-density maps. SARM1 is a central executioner of programmed axon degeneration, a common feature of the early phase of many neurodegenerative diseases. SARM1 is held in the inactive state in healthy axons by its N-terminal auto-inhibitory ARM domain, and is activated to cleave NAD upon injury, triggering subsequent axon degeneration. To characterize the molecular mechanism of SARM1 activation, it was sought to determine the crystal structure of the SARM1 ARM domain. Here, the recombinant production and crystallization of dSARM1ARM is described, as well as the unconventional process used for structure determination. Crystals were obtained in the presence of NMN, a precursor of NAD and a potential activator of SARM1, only after in situ proteolysis of the N-terminal 63 residues. After molecular-replacement attempts failed, the crystal structure of dSARM1ARM was determined at 1.65 Å resolution using the MIRAS phasing technique with autoSHARP, combining data from native, selenomethionine-labelled and bromide-soaked crystals. The structure will further the understanding of SARM1 regulation.
1. Introduction
The protein SARM1 (sterile alpha and Toll/interleukin-1 receptor motif-containing 1) is a central executioner of injury-induced axon degeneration (Wallerian degeneration). Loss of SARM1 protects axons from degeneration for weeks after injury induced by axotomy or vincristine (Osterloh et al., 2012 ▸; Gerdts et al., 2013 ▸). In healthy axons, SARM1 is held in the inactive state by the N-terminal armadillo repeat motif (ARM) domain. Upon injury, this auto-inhibition is relieved, permitting the C-terminal TIR (Toll/interleukin-1 receptor) domains to hydrolyze nicotinamide adenine dinucleotide (NAD) into nicotinamide and either ADP-ribose (ADPR) or cyclic ADPR (cADPR) (Essuman et al., 2017 ▸; Horsefield et al., 2019 ▸). These changes in turn trigger an influx of Ca2+ into the cells, a corresponding loss of ATP and eventually axon degeneration (Loreto et al., 2015 ▸; Horsefield et al., 2019 ▸). Despite its important role in this process, the mechanism of SARM1 activation is poorly understood. Recently, it has been suggested that the accumulation of the NAD precursor nicotinamide mononucleotide (NMN) is a trigger of SARM1 activation, resulting in subsequent axon degeneration (Di Stefano et al., 2015 ▸). Therefore, we hypothesized that NMN may interact with the ARM domain and reverse the ARM domain-mediated auto-inhibition.
To determine crystal structures, the phase components of the structure factors, which are lost during diffraction data collection, need to be recovered. In the case where similar protein structures are available, one can use molecular replacement (Rossmann, 1990 ▸). Alternatively, one can locate the positions of heavy atoms (HAs) that are either incorporated into the crystals or already present within the macromolecule through techniques such as MIR (multiple isomorphous replacement), usually with the inclusion of the anomalous signal, and SAD (single-wavelength anomalous dispersion) (Vijayan & Ramaseshan, 2001 ▸).
We sought to determine the crystal structure of the NMN-bound ARM domain of Drosophila SARM1 (dSARM1ARM), which would greatly enhance our understanding of the mechanism of NMN-induced relief of ARM domain-mediated auto-inhibition in SARM1. We crystallized dSARM1ARM in the NMN-bound state. Although the crystals diffracted X-rays to high resolution (1.65 Å), attempts to determine the phases using the molecular-replacement technique were not successful. We then attempted SAD phasing using the anomalous signals from bromide or selenium, which were separately incorporated into the crystals. However, the anomalous signal present in either the Br-SAD or the Se-SAD data set was weak and initial phases could not be successfully estimated. The phase problem was eventually solved by employing the MIRAS (multiple isomorphous replacement with anomalous scattering) method with autoSHARP (Vonrhein et al., 2007 ▸), combining the data from native, selenomethionine (SeMet)-labelled and bromide-soaked crystals. Here, we report the protein production, crystallization and structure determination of dSARM1ARM, and present our experience as a case study of modern MIRAS phasing.
2. Materials and methods
2.1. Protein production
The cDNA encoding dSARM1ARM (residues 307–678; UniProtKB Q6IDD9) was codon-optimized for expression in Escherichia coli and was cloned into the pMCSG7 expression vector at the SspI site using the ligation-independent cloning technique (forward primer 5′-TACTTCCAATCCAATGCGAATGGACAGATGTTGAAGCTTGCGGATTTGAAATTAGACG-3′; reverse primer 5′-TTATCCACTTCCAATGTTACGTTTCCCCAATTAAGCGCAGCGCTTGGG-3′; Aslanidis & de Jong, 1990 ▸; Eschenfeldt et al., 2009 ▸). The plasmid was transformed into E. coli BL21 (DE3) (for the native protein) or B834 (DE3) (for the SeMet-labelled protein) cells by heat shock. The cells were grown on lysogeny broth (LB) agar plates containing 100 µg ml−1 ampicillin at 37°C overnight. Colonies were inoculated into 10 ml LB medium containing 100 µg ml−1 ampicillin and were incubated at 37°C and 225 rev min−1 overnight. To produce the native protein, 1 ml of the LB overnight culture of transformed E. coli BL21 (DE3) cells was inoculated into 1000 ml auto-induction medium (Studier, 2005 ▸) containing 100 µg ml−1 ampicillin and incubated at 37°C and 225 rev min−1 until the OD600 reached 0.8–1.0. The temperature was then decreased to 20°C for overnight protein expression. To produce the SeMet-labelled protein, 1 ml of the overnight LB culture of transformed E. coli B834 (DE3) cells was inoculated into 1000 ml M9 minimal medium containing 1× M9 salt (33.7 mM Na2HPO4, 22 mM KH2PO4, 8.55 mM NaCl, 9.35 mM NH4Cl), 1× trace elements solution (0.13 mM EDTA, 0.03 mM FeCl3, 6.2 µM ZnCl2, 0.76 µM CuCl2, 0.42 µM CoCl2, 1.62 µM H3BO3, 0.08 µM MnCl2), 0.4%(v/v) glucose, 1 mM MgSO4, 0.3 mM CaCl2, 1× BME vitamin solution (Sigma–Aldrich) and 100 µg ml−1 ampicillin. The bacteria were grown at 37°C and 22 rev min−1 until the OD600 reached 0.8–1.0. The temperature was then decreased to 20°C for a 30 min incubation. 1 ml 50 mg ml−1 SeMet (Sigma–Aldrich) was added to 1000 ml culture. Expression was induced by adding isopropyl β-d-1-thiogalactopyranoside (IPTG) to a final concentration of 1 mM and the cells were incubated overnight at 20°C and 225 rev min−1. The E. coli BL21 (DE3) or B834 (DE3) cells were harvested by centrifugation at 6000g for 20 min at 4°C and were treated identically in subsequent purification steps.
The harvested cells were resuspended in lysis buffer (50 mM HEPES pH 8.0, 500 mM NaCl, 30 mM imidazole, 1 mM DTT). Phenymethylsulfonyl fluoride was added to the cell suspension to a final concentration of 1 mM. The cells were subsequently lysed by sonication. The lysed cells were centrifuged at 15 300g for 40 min at 4°C. The supernatant was loaded onto a 5 ml HisTrap HP column (Cytiva) equilibrated with lysis buffer. The bound target protein was washed with 100 ml lysis buffer and eluted with elution buffer (50 mM HEPES pH 8.0, 500 mM NaCl, 300 mM imidazole, 1 mM DTT) on an ÄKTApurifier (Cytiva). Fractions containing dSARM1ARM were combined and incubated with Tobacco etch virus (TEV) protease (20:1 protein:TEV protease ratio) in SnakeSkin Dialysis Tubing (3.5 kDa molecular-weight cutoff; Thermo Fisher Scientific) and dialyzed against buffer consisting of 20 mM HEPES pH 8.0, 300 mM NaCl, 1 mM DTT at 4°C overnight. The His6-tag-removed protein was reloaded onto a 5 ml HisTrap HP column to remove uncleaved fusion protein and free His6 tag. The flowthrough was then collected, concentrated to a volume of 10 ml and injected onto a Superdex 75 HiLoad 26/600 column (Cytiva) equilibrated with gel-filtration buffer (10 mM HEPES pH 8.0, 150 mM NaCl, 1 mM DTT). The peak fractions containing pure dSARM1ARM were pooled, concentrated using a 30 kDa molecular-weight cutoff Amicon Ultra Centrifugal filter (Millipore), flash-frozen and stored at −80°C.
2.2. Crystallization
Prior to crystallization, dSARM1ARM protein (17 mg ml−1) was incubated with NMN in a 1:10 protein:compound molar ratio at 4°C overnight. Sparse-matrix protein crystallization screening was performed using the commercially available Index (Hampton Research), Combined Synergy (Hampton Research), PEG/Ion (Hampton Research), PEGRx (Hampton Research), JCSG+ (Molecular Dimensions), PACT premier (Molecular Dimensions), ProPlex (Molecular Dimensions) and ShotGun (Molecular Dimensions) screens. Crystallization trials were set up using a Mosquito liquid-handling robot (TTP LabTech) in a 96-well hanging-drop plate format with 100 nl protein solution and 100 nl reservoir solution per drop equilibrated against 75 µl reservoir solution at 20°C. To scale up the drop sizes, hanging drops consisting of 2 µl NMN-bound protein and 2 µl commercial reservoir solution were equilibrated against 500 µl homemade reservoir solution consisting of 0.1 M SPG buffer (succinic acid, sodium dihydrogen phosphate and glycine in a 2:7:7 molar ratio, pH 8.0) and 25%(w/v) PEG 1500 at 20°C. The SeMet-labelled crystals were produced using the same crystallization condition as for the native crystals. Crystals were observed in the crystallization drops after 3–5 days. SDS–PAGE and mass spectrometric analyses of these crystals were performed to ascertain the identity of the crystallized protein.
2.3. Diffraction data collection and processing
Prior to flash-cooling in liquid nitrogen, crystals of NMN-bound native dSARM1ARM and SeMet-labelled dSARM1ARM were cryoprotected in a cryoprotectant solution consisting of 0.1 M SPG buffer pH 8.0, 25%(w/v) PEG 1500, 25%(v/v) PEG 400. Crystals derivatized with bromide were prepared by soaking the native crystals in the mother liquor containing 0.5 M sodium bromide, 25%(v/v) PEG 400 for 2 min prior to flash-cooling. All data sets were collected on the MX2 beamline at the Australian Synchrotron using an EIGER X 16M detector (Aragão et al., 2018 ▸). The native data set was collected at a wavelength of 0.95372 Å, the SeMet-labelled data set was collected at the theoretical selenium absorption edge with a wavelength of 0.97857 Å and the bromide-soaked data set was collected at the theoretical bromine absorption edge with a wavelength of 0.91976 Å. Diffraction data from NMN-bound dSARM1ARM crystals (native, SeMet-labelled and bromide-soaked data) were processed and analyzed with autoPROC (Vonrhein et al., 2011 ▸). Initial phases were calculated using the MIRAS technique with autoSHARP (Vonrhein et al., 2007 ▸). The structure was refined using the strategy of TLS parameters with iterations of phenix.refine (Afonine et al., 2012 ▸) and manual model building in Coot (Emsley et al., 2010 ▸).
3. Results and discussion
The boundaries of the expression constructs for dSARM1ARM were determined based on sequence alignments among human, mouse, Caenorhabditis elegans and Drosophila SARM1, taking secondary-structure predictions into consideration. Small-scale expression tests were performed to identify constructs producing soluble target protein. Using the dSARM1ARM307–678 construct, we successfully expressed soluble dSARM1ARM protein, with a final yield of 10 mg per litre of bacterial culture.
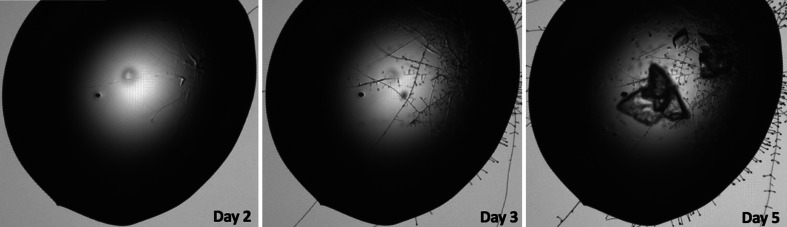
During sparse-matrix screening, we observed (after five days) the growth of a few crystals in 0.1 M SPG buffer pH 8.0, 25%(w/v) PEG 1500 (PACT premier condition A5) at 20°C. The crystals were chunky but irregular in shape (Fig. 1 ▸). However, despite our best efforts, we were not able to reproduce the NMN-bound dSARM1ARM crystals using homemade crystallization solutions. Importantly, we observed that what appeared to be fungal cells, possibly Penicillium, grew in the crystallization drop prior to the growth of the dSARM1ARM crystals (Fig. 1 ▸). SDS–PAGE analysis of the NMN-bound dSARM1ARM crystals, followed by mass spectrometry, indicated that the crystallized protein corresponded to residues 370–678 of SARM1 (i.e. lacking the N-terminal 63 amino acids). This led us to reason that partial proteolysis, mediated by proteases secreted by the fungal cells, was required for the crystallization of NMN-bound SARM1ARM. We therefore attempted in situ proteolysis using the proteases trypsin and chymotrypsin, but did not obtain any crystals. We also constructed dSARM1ARM370–678 with the hope of solving the crystal reproducibility issues. However, this construct failed to yield soluble protein. Crystals only grew when the original crystallization solution containing the fungal cells was added to the homemade crystallization solution. For these reasons, we had a limited number of crystals to optimize our diffraction experiments.
Figure 1.
Crystal growth of NMN-bound dSARM1ARM. Crystals were observed after five days in 0.1 M SPG buffer pH 8.0, 25%(w/v) PEG 1500 in the presence of fungal cells.
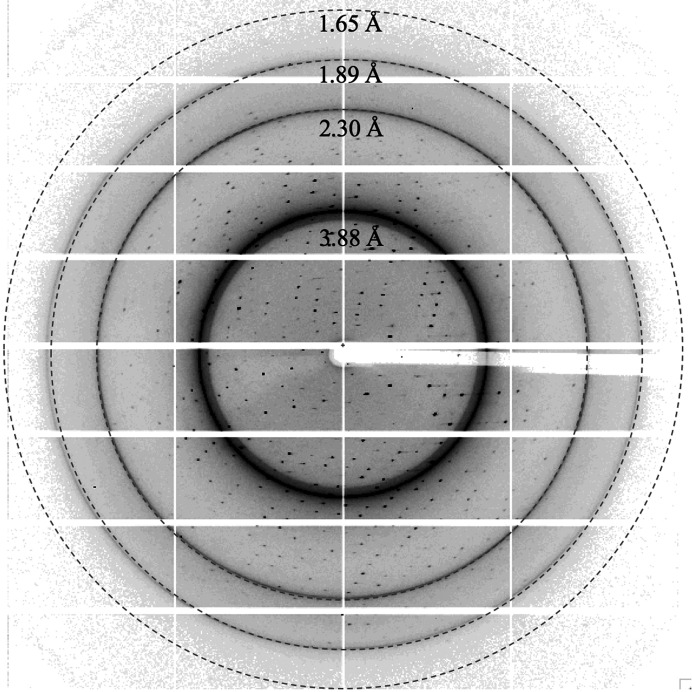
A native data set was collected from NMN-bound dSARM1ARM crystals on the MX2 beamline at the Australian Synchrotron. The data were initially processed using XDS (Kabsch, 2010 ▸) and AIMLESS (Evans & Murshudov, 2013 ▸). The crystals diffracted to ∼1.7 Å resolution, with strong ice rings appearing at ∼1.9, 2.3 and 3.9 Å (Table 1 ▸, Fig. 2 ▸). The crystal had the symmetry of space group P1 and was likely to contain two dSARM1ARM molecules in the asymmetric unit, with a Matthews coefficient of 2.07 Å3 Da−1 and a solvent content of 40%.
Table 1. Data-processing statistics from XDS and AIMLESS .
The statistics are based on the calculations from AIMLESS. Values in parentheses are for the highest resolution shell.
| Native | Bromide-soaked | SeMet-labelled | |
|---|---|---|---|
| Space group | P1 | P1 | P1 |
| a, b, c (Å) | 38.95, 50.82, 76.12 | 39.12, 51.15, 75.83 | 38.86, 50.28. 75.18 |
| α, β, γ (°) | 103.52, 101.89, 95.23 | 103.37, 101.51, 96.67 | 104.88, 101.37, 94.89 |
| Resolution (Å) | 48.9–1.74 (1.78–1.74) | 46.59–2.01 (2.06–2.01) | 48.1–4.18 (4.67–4.18) |
| R merge † | 0.058 (0.472) | 0.067 (0.621) | 0.036 (0.043) |
| R meas ‡ | 0.068 (0.554) | 0.077 (0.719) | 0.051 (0.060) |
| R p.i.m. § | 0.035 (0.288) | 0.038 (0.357) | 0.036 (0.043) |
| Mean I/σ(I) | 17.6 (3.4) | 17.8 (3.2) | 21.7 (19.9) |
| CC1/2 | 0.999 (0.931) | 0.999 (0.830) | 0.98 (0.96) |
| Total No. of observations | 3386169 (19534) | 284357 (19417) | 13961 (3827) |
| No. of unique observations | 54045 (2793) | 35879 (2561) | 3835 (1074) |
| Completeness (%) | 96.3 (89.2) | 97.9 (93.5) | 97.9 (96.8) |
| Multiplicity | 7.1 (7.0) | 7.9 (7.6) | 3.6 (3.6) |
| Anomalous completeness (%) | 95.4 (85.5) | 95.3 (88.5) | 92.6 (89.4) |
| Anomalous multiplicity | 3.5 (3.6) | 3.9 (3.9) | 1.8 (1.9) |
| DelAnom correlation between half-sets | −0.091 (0.076) | 0.398 (0.053) | 0.297 (0.183) |
| Mid-slope of anomalous normal probability | 0.956 | 1.173 | 1.392 |
Rmerge = \textstyle \sum_{hkl}\sum_{i}|I_{i}(hkl)- \langle I(hkl)\rangle|/\textstyle \sum_{hkl}\sum_{i}I_{i}(hkl).
Rmeas = \textstyle \sum_{hkl}\{N(hkl)/[N(hkl)-1]\}^{1/2}\sum_{i}|I_{i}(hkl)- \langle I(hkl)\rangle|/\textstyle \sum_{hkl}\sum_{i}I_{i}(hkl).
Rp.i.m. = \textstyle \sum_{hkl}\{1/[N(hkl)-1]\}^{1/2}\times\textstyle\sum_{i}|I_{i}(hkl)- \langle I(hkl)\rangle|/\textstyle \sum_{hkl}\sum_{i}I_{i}(hkl).
Figure 2.
A representative diffraction image of an NMN-bound dSARM1ARM crystal. The crystal diffracted X-rays to >1.65 Å resolution.
ARM domains are protein-interaction domains that are found in many proteins displaying diverse cellular roles from gene expression to cytoskeleton regulation (Coates, 2003 ▸). They typically consist of tandem repeats of armadillo motifs. One armadillo motif contains ∼42 residues, which fold into three α-helices (H1, H2 and H3). Stacking of these motifs forms a right-handed superhelix with an elongated concave surface, characterized by parallel H3 helices arranged in a ladder fashion (Coates, 2003 ▸). Initially, we attempted to solve the structure of dSARM1ARM by molecular replacement. We used the available ARM structures, such as importin-α (PDB entry 1ial; Kobe, 1999 ▸) and Vac8p (PDB entry 5xjg; Jeong et al., 2017 ▸), with various modifications, as search models within Phaser (McCoy et al., 2007 ▸). We attempted automated molecular replacement with MrBUMP (Keegan & Winn, 2008 ▸) and BALBES (Long et al., 2008 ▸), as well as ab initio macromolecular phasing with ARCIMBOLDO (Rodríguez et al., 2009 ▸). However, we did not obtain any clear solutions using these approaches. Post-mortem analysis of the structures revealed that the dSARM1ARM crystal structure adopted a curled conformation, which is drastically different from other existing ARM structures, with a root-mean-square deviation (r.m.s.d.) of the backbone Cα atoms of greater than 3.5 Å between them.
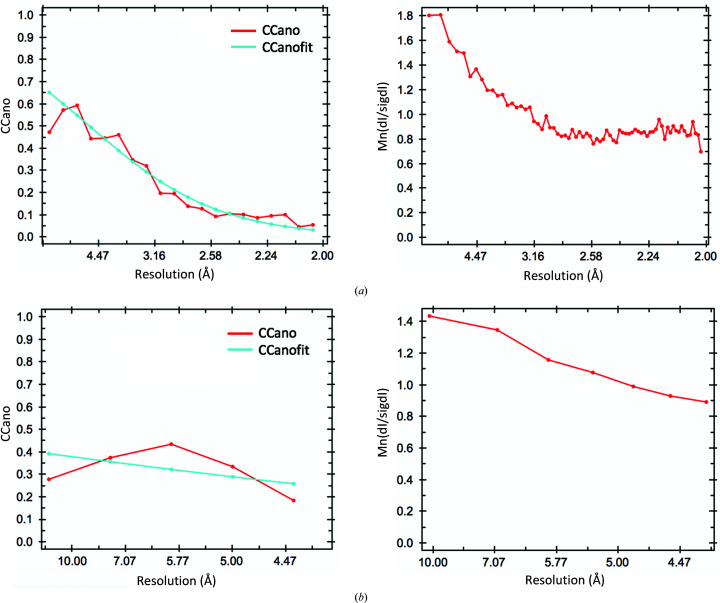
We alternatively sought to solve the phase problem by SAD phasing. To this end, we incorporated bromide (through soaking) and, separately, SeMet (during expression) into the protein crystals and collected anomalous data sets to ∼2.0 and 4.2 Å resolution, respectively, using X-ray wavelengths close to the absorption edges of the respective HAs (Table 1 ▸). Unfortunately, the crystals were prone to radiation damage, as demonstrated by the reduction in diffraction quality and a sharply decreasing Wilson B factor. Due to the low P1 symmetry, data sets with high multiplicity and therefore accurately determined anomalous differences, which are often required for successful SAD phasing, could not be acquired. Also, as we only had access to a limited number of crystals, merging multiple low-multiplicity data sets was not a viable option. The bromide data set had an anomalous multiplicity of ∼3.9, whereas the SeMet data set had an even lower anomalous multiplicity of merely 1.8. Data processing using XDS (Kabsch, 2010 ▸) and AIMLESS (Evans & Murshudov, 2013 ▸) indicated that the values of the mid-slope of anomalous normal probabilities of the bromide and SeMet data sets were 1.17 and 1.39, respectively, suggesting that detectable but weak anomalous signal was present in these two data sets. Using a CCano cutoff of 0.15, the detectable anomalous signals were up to 2.7 and 4.2 Å resolution for the bromide and SeMet data sets, respectively (Fig. 3 ▸), but subsequent searches for HAs in AutoSol within Phenix (Terwilliger et al., 2009 ▸) and CRANK2 within CCP4i2 (Skubák & Pannu, 2013 ▸; Potterton et al., 2018 ▸) invariably failed.
Figure 3.
Analysis of the anomalous signals using AIMLESS (manual). (a) CCano (left) and Mn(dI/sigdI) (right) as a function of resolution for the bromide data set. (b) CCano (left) and Mn(dI/sigdI) (right) as a function of resolution for the SeMet data set. The figures were automatically generated during manual data processing using AIMLESS.
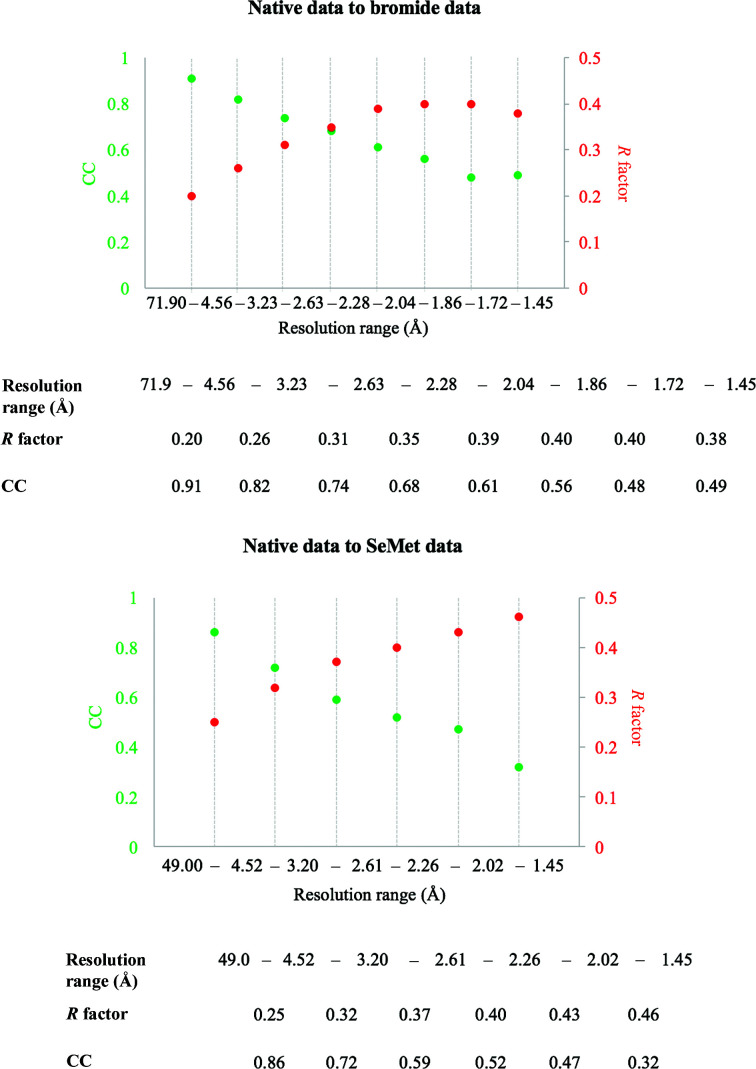
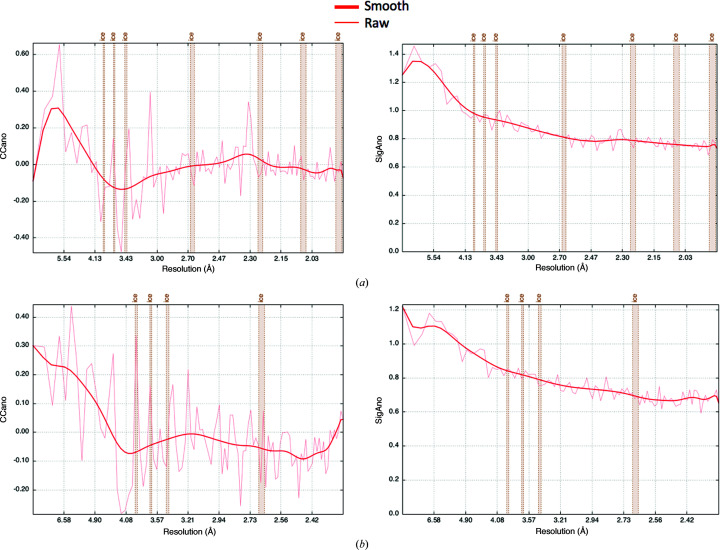
Fortunately, the bromide-soaked and SeMet-labelled crystals had similar unit-cell dimensions and the same space group as the native protein crystals (Table 1 ▸). Both HA data sets also shared relatively low R factors and a high correlation coefficient (CC) compared with the native data at low to intermediate resolutions (∼2.6 Å; Fig. 4 ▸), thus making MIRAS or SIRAS (single isomorphous replacement with anomalous scattering) phasing a possibility. During the 2019 CCP4/Shanghai Workshop we were able to use the autoPROC software package for data reprocessing (Vonrhein et al., 2011 ▸), which uses XDS for data processing (Kabsch, 2010 ▸), POINTLESS for space-group determination (Evans, 2006 ▸), AIMLESS for scaling (Evans & Murshudov, 2013 ▸) and STARANISO for analysis of diffraction anisotropy (Vonrhein et al., 2011 ▸), plus multiple additional tools for diffraction-image processing (Table 2 ▸, Fig. 5 ▸). Careful processing of the diffraction images is important, as diffraction outliers are damaging to the success of the HA search. We inspected the diffraction images to redefine accurate beam-stop masks for all of the data sets and subtracted ice rings from the diffraction data in autoPROC. In the SeMet data set, we observed four pixels with extremely high intensities, with coordinates (1675, 2512) at ∼6.2 Å, (5, 3182) at ∼1.6 Å, (21, 1668) at ∼1.7 Å and (1765, 2531) at ∼6.7 Å, respectively, that appeared in all diffraction images of the data set. Successful indexing of the SetMet data set was only achieved when these bad pixels were removed from the diffraction images using the image-analysis and inspection tool in autoPROC (aP_detect_damaged_pixels). We assume that these pixels accumulated damage over time and should therefore always be included in the detector pixel mask that defines inactive pixels and regions. Importantly, our SetMet data set also suffered from anisotropy, and within initial scaling using AIMLESS, an isotropic resolution cutoff of 4.2 Å was chosen. However, using STARANISO in autoPROC, the significant data that were previously excluded during scale determination in AIMLESS because of anisotropy could now be accounted for, extending the diffraction limit of the data set to 1.9 Å.
Figure 4.
Analysis of isomorphorism of the native, bromide-soaked and SeMet-labelled crystals. The statistics (R factor and CC on amplitudes) were generated within autoPROC using the native data as a reference (check_indexing -v).
Table 2. Data-processing statistics from autoPROC and refinement statistics from Phenix .
The statistics are based on the calculations from autoPROC and MolProbity. Values in parentheses are for the highest resolution shell.
| Native | Bromide-soaked | SeMet-labelled | |
|---|---|---|---|
| Data collection | |||
| Space group | P1 | P1 | P1 |
| a, b, c (Å) | 38.92, 50.79, 76.05 | 39.07, 51.08, 75.73 | 38.89, 50.31, 75.22 |
| α, β, γ (°) | 103.52, 101.90, 95.26 | 103.38, 101.52, 96.66 | 104.86, 101.36, 94.91 |
| Resolution (Å) | 71.9–1.46 (1.60–1.46) | 49.0–1.68 (1.82–1.68) | 48.1–1.89 (2.11–1.89) |
| R merge † | 0.05 (0.95) | 0.08 (1.77) | 0.07 (0.76) |
| R meas ‡ | 0.06 (1.15) | 0.09 (2.05) | 0.09 (1.08) |
| R p.i.m. § | 0.03 (0.63) | 0.04 (1.01) | 0.07 (0.76) |
| Mean I/σ(I) | 15.9 (1.6) | 13.9 (1.0) | 9.2 (1.2) |
| CC1/2 | 1.00 (0.63) | 1.00 (0.4) | 1.00 (0.47) |
| Total reflections | 456719 (20265) | 393818 (19421) | 86502 (4041) |
| Unique reflections | 64028 (3201) | 48934 (2447) | 24291 (1216) |
| Completeness (spherical) (%) | 67.2 (14.2) | 78.2 (19.1) | 56.6 (10.0) |
| Completeness (ellipsoidal) (%) | 83.5 (40.3) | 90.0 (44.7) | 85.3 (38.8) |
| Multiplicity | 7.1 (6.3) | 8.0 (7.9) | 3.6 (3.3) |
| Anomalous completeness (spherical) (%) | 66.5 (14.0) | 76.4 (18.5) | 55.8 (9.7) |
| Anomalous completeness (ellipsoidal) (%) | 82.7 (39.8) | 87.9 (43.4) | 84.1 (38.0) |
| Anomalous multiplicity | 3.6 (3.2) | 4.1 (4.1) | 1.8 (1.7) |
| Refinement | |||
| Resolution (Å) | 37.67–1.65 [native data set] | ||
| R work ¶ | 0.21 | ||
| R free ¶ | 0.23 | ||
| R.m.s.d., bonds (Å) | 0.002 | ||
| R.m.s.d., angles (°) | 0.47 | ||
| Ramachandran favoured (%) | 98.51 | ||
| Ramachandran outliers (%) | 0.33 | ||
| Rotamer outliers (%) | 0.19 | ||
| Clashscore | 1.64 | ||
| Average B factor (Å2) | 27.77 | ||
| Cβ outliers | 0 | ||
Rmerge = \textstyle \sum_{hkl}\sum_{i}|I_{i}(hkl)- \langle I(hkl)\rangle|/\textstyle \sum_{hkl}\sum_{i}I_{i}(hkl).
Rmeas = \textstyle \sum_{hkl}\{N(hkl)/[N(hkl)-1]\}^{1/2}\sum_{i}|I_{i}(hkl)- \langle I(hkl)\rangle|/\textstyle \sum_{hkl}\sum_{i}I_{i}(hkl).
Rp.i.m. = \textstyle \sum_{hkl}\{1/[N(hkl)-1]\}^{1/2}\textstyle\sum_{i}|I_{i}(hkl)- \langle I(hkl)\rangle|/\textstyle \sum_{hkl}\sum_{i}I_{i}(hkl).
Rwork = \textstyle \sum_{hkl}\big ||F_{\rm obs}|-|F_{\rm calc}|\big |/ \textstyle \sum_{hkl}|F_{\rm obs}|; R free is equivalent to R work, with 5% of data excluded from refinement process. |F obs| and |F calc| represent the observed and calculated structure-factor amplitudes, respectively.
Figure 5.
Analysis of the anomalous signals using autoPROC. (a) CCano (left) and SigAno (right) as a function of resolution for the bromide data set. (b) CCano (left) and SigAno (right) as a function of resolution for the SeMet data set. The figures were automatically generated during data processing with autoPROC. SigAno corresponds to Mn(dI/sigdI) in Fig. 3 ▸.
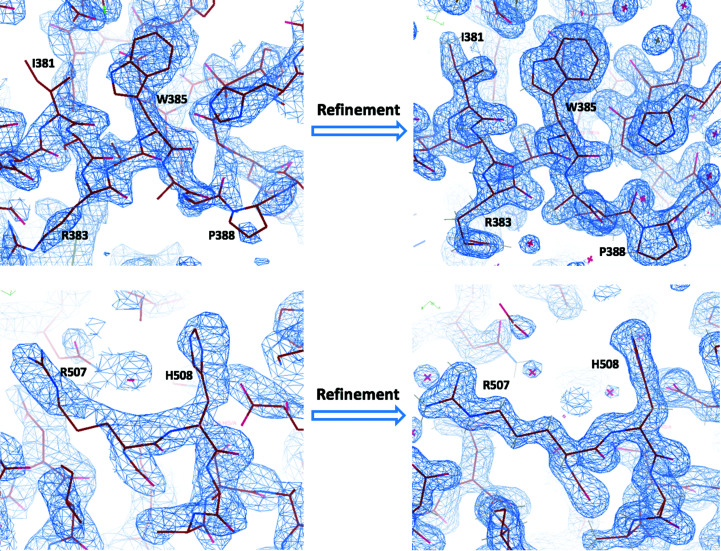
For MIRAS phasing and initial model building, we used autoSHARP (Vonrhein et al., 2007 ▸), which uses SHELXC/D for substructure determination (Schneider & Sheldrick, 2002 ▸), SHARP for HA refinement, phasing and substructure completion (de La Fortelle & Bricogne, 1997 ▸), SOLOMON for density modification (Abrahams & Leslie, 1996 ▸) and Buccaneer for automatic model building (Cowtan, 2006 ▸). After multiple attempts to choose an appropriate resolution cutoff to maximize the phasing power, phase determination was eventually performed using data with a resolution cutoff of 25–2.5 Å. The high-resolution cutoff seems slightly counterintuitive, because it is (i) much higher than the HA signal (isomorphous difference and anomalous signal) in the different data sets and (ii) lower than the overall diffraction limit of the available data sets. However, in cases where the difference between these two resolution limits (HA signal and overall) is rather large it is often beneficial to restrict the data to 2.5–3 Å resolution in the early stages of the structure-solution process. At this stage, one is mainly interested in achieving a successful substructure determination, a clear indication of the correct enantiomorph (during the density-modification step) and hopefully some meaningful secondary-structure elements resulting from the automatic model-building step. All of these can easily be achieved by using 2.5–3 Å resolution data. If even higher resolution data are used at this stage, the initial low-resolution phase information (which is expected to be poor) might be inadequate in helping density modification to bridge the large resolution range to the full limit of the available data, at least within the density-modification and phase-extension procedure using SOLOMON as implemented in autoSHARP. During HA detection, where combinations of SIR(AS) (native plus bromide or native plus SeMet data sets) and SAD (bromide or SeMet data set alone) were tried, autoSHARP calculates the CC between the observed and calculated normalized substructure amplitudes E. The solution with the highest CC(E), and therefore the initial substructure solution most likely to be successful, corresponded to SIRAS using the SeMet data set [CC(E) = 0.188 with four SeMet sites out of the expected ten sites in the asymmetric unit]. Using this solution as a starting point, SHARP within autoSHARP refined the coordinates, occupancy and temperature factors for the initial HA sites as well as scaling non-isomorphism parameters between the three data sets. It further detected ten bromide sites (overall phasing power of 0.216 for isomorphous differences and 0.483 for anomalous differences, with the phasing power dropping below 1 at 24.42 and 5.48 Å, resolution, respectively) and ten SeMet sites (overall phasing power of 0.136 for isomorphous differences and 0.299 for anomalous differences, with the phasing power dropping below 1 at 24.42 and 4.92 Å resolution, respectively). These values were consistent with the analysis of the anomalous signal from the data-processing stages and confirmed that the HA signal (and the initial maps computed with these phases) would present a rather low-resolution starting point for subsequent steps. A final set of phases was calculated in both hands and the most likely enantiomorph was determined by performing a single cycle of solvent flipping in SOLOMON as part of autoSHARP, suggesting that the correct phases were those from the inverted hand, based on its slightly higher score (a combination of the CC between observed E 2 values and the E 2 values of the modified map and the contrast in the assigned protein and solvent regions) of 0.1234 (two molecules in the asymmetric unit) compared with 0.1078 from the original hand. After multiple cycles of density modification to optimize the solvent content, the best density-modified map with a score of 1.8697 was finally handed over to Buccaneer, which managed to build a total of 614 residues in two chains (out of the expected 618 residues for a dimer in the asymmetric unit). Consistently, the ten SeMet sites, including the initial four sites during HA detection, have been shown to be consistent with the (Se)Met residues built by Buccaneer within the initial model. This was the final result of a fully automatic autoSHARP run (starting with the data sets, the sequence, information about the scattering properties of the heavy atoms in the different data sets and some indication of the expected number of bromide sites) and provided the starting point for subsequent steps. Further manual model building in Coot (Emsley et al., 2010 ▸) and refinement against the native data using phenix.refine (Afonine et al., 2012 ▸) improved the quality of the phases (Fig. 6 ▸). For refinement, we restricted the data resolution to 1.65 Å, as opposed to using the full resolution range of the anisotropically analysed STARANISO data. We observed that the data at 1.45–1.65 Å resolution contained greater than 40% phase error and less than 80% data completeness. Excluding the data at 1.45–1.65 Å resolution indeed resulted in a clearer overall electron-density map. The final structure was determined with R work and R free values of 0.21 and 0.23, respectively (Table 2 ▸).
Figure 6.
Representative electron-density maps, contoured at 1.5σ, before and after refinement. The panels on the left show the initial electron-density map (2F o − F c) calculated using the MIRAS-based phases from autoSHARP after density modification (SOLOMON) and model building (Buccaneer) at a resolution of 2.5 Å. The panels on the right show the electron-density map in the corresponding region after refinement with phenix.refine at 1.65 Å resolution.
4. Summary
In conclusion, we describe the crystal structure determination of dSARM1ARM as an illustrative example of how a number of technical difficulties in the process can be overcome. Multiple nonconventional steps were employed here. Firstly, in situ proteolysis allowed diffraction-quality crystals to be obtained in the first place. Secondly, after attempts to solve the phase problem by molecular replacement failed, heavy atoms were introduced by bromide soaking and SeMet incorporation. However, neither of these SAD data sets provided sufficient anomalous signal to obtain interpretable electron-density maps. The low symmetry of the crystal system provided little opportunity to collect high-multiplicity multiwavelength data sets around the Se and Br edges, which might have helped substructure detection within each bromide or SeMet data set. However, after careful data processing (manual beam-stop masking, exclusion of damaged pixels that are not yet in the detector mask and handling of ice rings) using autoPROC and combining the resulting improved data from the native, bromide-soaked and SeMet-labelled crystals, the MIRAS approach as implemented in autoSHARP led to interpretable electron-density maps and a clear initial starting model. The structure of NMN-bound dSARM1ARM displays a more compact conformation, differing from the canonical ARM domains, with an r.m.s.d. on backbone Cα atoms of over 3.5 Å. This could explain why molecular replacement failed to solve the phase problem. This structure will help us to understand the molecular mechanisms of regulation of SARM1, a protein with a central role in neurodegenerative disease. The biological implications of the structure are discussed in Figley et al. (2021 ▸).
Supplementary Material
PDB reference: armadillo repeat domain of Drosophila SARM1, 7lcz
Acknowledgments
We acknowledge the use of the University of Queensland Remote Operation Crystallization and X-ray (UQROCX) Facility at the Centre for Microscopy and Microanalysis and the support from the staff, Dr Gordon King and Karl Byriel. We acknowledge the use of the Australian Synchrotron MX beamlines, part of ANSTO, including the Australian Cancer Research Foundation detector, and the support of the staff. We also thank the CCP4/Shanghai 2019 workshop, their organisers, including CCP4, Dr Ruslan Sanishvili, National Facility for Protein Science in Shanghai, Shanghai Tech University iHuman Institute and Shanghai Synchrotron Radiation Facility, and Professor Gérard Bricogne from Global Phasing Ltd for the help with structure determination. We are grateful to Dr Gayle Petersen for diligent proofreading and discussion of the manuscript.
Funding Statement
This work was funded by National Health and Medical Research Council grants 1107804 , 1160570 , 1071659 , and 1108859 to Boštjan Kobe, Thomas Ve, Boštjan Kobe, Thomas Ve, Boštjan Kobe, and Thomas Ve; Australian Research Council grants FL180100109 and DE170100783 to Boštjan Kobe and Thomas Ve.
References
- Abrahams, J. P. & Leslie, A. G. W. (1996). Acta Cryst. D52, 30–42. [DOI] [PubMed]
- Afonine, P. V., Grosse-Kunstleve, R. W., Echols, N., Headd, J. J., Moriarty, N. W., Mustyakimov, M., Terwilliger, T. C., Urzhumtsev, A., Zwart, P. H. & Adams, P. D. (2012). Acta Cryst. D68, 352–367. [DOI] [PMC free article] [PubMed]
- Aragão, D., Aishima, J., Cherukuvada, H., Clarken, R., Clift, M., Cowieson, N. P., Ericsson, D. J., Gee, C. L., Macedo, S., Mudie, N., Panjikar, S., Price, J. R., Riboldi-Tunnicliffe, A., Rostan, R., Williamson, R. & Caradoc-Davies, T. T. (2018). J. Synchrotron Rad. 25, 885–891. [DOI] [PMC free article] [PubMed]
- Aslanidis, C. & de Jong, P. J. (1990). Nucleic Acids Res. 18, 6069–6074. [DOI] [PMC free article] [PubMed]
- Coates, J. C. (2003). Trends Cell Biol. 13, 463–471. [DOI] [PubMed]
- Cowtan, K. (2006). Acta Cryst. D62, 1002–1011. [DOI] [PubMed]
- Di Stefano, M., Nascimento-Ferreira, I., Orsomando, G., Mori, V., Gilley, J., Brown, R., Janeckova, L., Vargas, M. E., Worrell, L. A., Loreto, A., Tickle, J., Patrick, J., Webster, J. R. M., Marangoni, M., Carpi, F. M., Pucciarelli, S., Rossi, F., Meng, W., Sagasti, A., Ribchester, R. R., Magni, G., Coleman, M. P. & Conforti, L. (2015). Cell Death Differ. 22, 731–742. [DOI] [PMC free article] [PubMed]
- Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. (2010). Acta Cryst. D66, 486–501. [DOI] [PMC free article] [PubMed]
- Eschenfeldt, W. H., Lucy, S., Millard, C. S., Joachimiak, A. & Mark, I. D. (2009). Methods Mol. Biol. 498, 105–115. [DOI] [PMC free article] [PubMed]
- Essuman, K., Summers, D. W., Sasaki, Y., Mao, X., DiAntonio, A. & Milbrandt, J. (2017). Neuron, 93, 1334–1343. [DOI] [PMC free article] [PubMed]
- Evans, P. (2006). Acta Cryst. D62, 72–82. [DOI] [PubMed]
- Evans, P. R. & Murshudov, G. N. (2013). Acta Cryst. D69, 1204–1214. [DOI] [PMC free article] [PubMed]
- Figley, M. D., Gu, W., Nanson, J. D., Shi, Y., Sasaki, Y., Cunnea, K., Malde, A. K., Jia, X., Luo, Z., Saikot, F. K., Mosaiab, T., Masic, V., Holt, S., Hartley-Tassell, L., McGuinness, H. Y., Manik, M. K., Bosanac, T., Landsberg, M. J., Kerry, P. S., Mobli, M., Hughes, R. O., Milbrandt, J., Kobe, B., DiAntonio, A. & Ve, T. (2021). Neuron, 109, 1118–1136. [DOI] [PMC free article] [PubMed]
- Gerdts, J., Summers, D. W., Sasaki, Y., DiAntonio, A. & Milbrandt, J. (2013). J. Neurosci. 33, 13569–13580. [DOI] [PMC free article] [PubMed]
- Horsefield, S., Burdett, H., Zhang, X., Manik, M. K., Shi, Y., Chen, J., Qi, T., Gilley, J., Lai, J. S., Rank, M. X., Casey, L. W., Gu, W., Ericsson, D. J., Foley, G., Hughes, R. O., Bosanac, T., von Itzstein, M., Rathjen, J. P., Nanson, J. D., Boden, M., Dry, I. B., Williams, S. J., Staskawicz, B. J., Coleman, M. P., Ve, T., Dodds, P. N. & Kobe, B. (2019). Science, 365, 793–799. [DOI] [PubMed]
- Jeong, H., Park, J., Kim, H. I., Lee, M., Ko, Y. J., Lee, S., Jun, Y. & Lee, C. (2017). Proc. Natl Acad. Sci. USA, 114, E4539–E4548. [DOI] [PMC free article] [PubMed]
- Kabsch, W. (2010). Acta Cryst. D66, 125–132. [DOI] [PMC free article] [PubMed]
- Keegan, R. M. & Winn, M. D. (2008). Acta Cryst. D64, 119–124. [DOI] [PMC free article] [PubMed]
- Kobe, B. (1999). Nat. Struct. Biol. 6, 388–397. [DOI] [PubMed]
- La Fortelle, E. de & Bricogne, G. (1997). Methods Enzymol. 276, 472–494. [DOI] [PubMed]
- Long, F., Vagin, A. A., Young, P. & Murshudov, G. N. (2008). Acta Cryst. D64, 125–132. [DOI] [PMC free article] [PubMed]
- Loreto, A., Di Stefano, M., Gering, M. & Conforti, L. (2015). Cell. Rep. 13, 2539–2552. [DOI] [PubMed]
- McCoy, A. J., Grosse-Kunstleve, R. W., Adams, P. D., Winn, M. D., Storoni, L. C. & Read, R. J. (2007). J. Appl. Cryst. 40, 658–674. [DOI] [PMC free article] [PubMed]
- Osterloh, J. M., Yang, J., Rooney, T. M., Fox, A. N., Adalbert, R., Powell, E. H., Sheehan, A. E., Avery, M. A., Hackett, R., Logan, M. A., MacDonald, J. M., Ziegenfuss, J. S., Milde, S., Hou, Y. J., Nathan, C., Ding, A., Brown, R. H., Conforti, L., Coleman, M., Tessier-Lavigne, M., Züchner, S. & Freeman, M. R. (2012). Science, 337, 481–484. [DOI] [PMC free article] [PubMed]
- Potterton, L., Agirre, J., Ballard, C., Cowtan, K., Dodson, E., Evans, P. R., Jenkins, H. T., Keegan, R., Krissinel, E., Stevenson, K., Lebedev, A., McNicholas, S. J., Nicholls, R. A., Noble, M., Pannu, N. S., Roth, C., Sheldrick, G., Skubak, P., Turkenburg, J., Uski, V., von Delft, F., Waterman, D., Wilson, K., Winn, M. & Wojdyr, M. (2018). Acta Cryst. D74, 68–84. [DOI] [PMC free article] [PubMed]
- Rodríguez, D. D., Grosse, C., Himmel, S., González, C., de Ilarduya, I. M., Becker, S., Sheldrick, G. M. & Usón, I. (2009). Nat. Methods, 6, 651–653. [DOI] [PubMed]
- Rossmann, M. G. (1990). Acta Cryst. A46, 73–82. [DOI] [PubMed]
- Schneider, T. R. & Sheldrick, G. M. (2002). Acta Cryst. D58, 1772–1779. [DOI] [PubMed]
- Skubák, P. & Pannu, N. S. (2013). Nat. Commun. 4, 2777. [DOI] [PMC free article] [PubMed]
- Studier, F. W. (2005). Protein Expr. Purif. 41, 207–234. [DOI] [PubMed]
- Terwilliger, T. C., Adams, P. D., Read, R. J., McCoy, A. J., Moriarty, N. W., Grosse-Kunstleve, R. W., Afonine, P. V., Zwart, P. H. & Hung, L.-W. (2009). Acta Cryst. D65, 582–601. [DOI] [PMC free article] [PubMed]
- Vijayan, M. & Ramaseshan, S. (2001). International Tables for Crystallography, Vol. B, edited by U. Shmueli, pp. 264–275. Dordrecht: Springer.
- Vonrhein, C., Blanc, E., Roversi, P. & Bricogne, G. (2007). Methods Mol. Biol. 364, 215–230. [DOI] [PubMed]
- Vonrhein, C., Flensburg, C., Keller, P., Sharff, A., Smart, O., Paciorek, W., Womack, T. & Bricogne, G. (2011). Acta Cryst. D67, 293–302. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: armadillo repeat domain of Drosophila SARM1, 7lcz