Abstract
PURPOSE:
To evaluate the diagnostic yield and clinical relevance of clinical genome sequencing (cGS) as a first genetic test for patients with suspected monogenic disorders.
METHODS:
We conducted a prospective randomized study with pediatric and adult patients recruited from genetics clinics at Massachusetts General Hospital who were undergoing planned genetic testing. Participants were randomized into two groups: standard-of-care genetic testing (SOC) only or SOC and cGS.
RESULTS:
Two hundred four participants were enrolled, 202 were randomized to one of the intervention arms, and 99 received cGS. In total, cGS returned 16 molecular diagnoses that fully or partially explained the indication for testing in 16 individuals (16.2% of the cohort, 95% confidence interval [CI] 8.9–23.4%), which was not significantly different from SOC (18.2%, 95% CI 10.6–25.8%, P = 0.71). An additional eight molecular diagnoses reported by cGS had uncertain relevance to the participant’s phenotype. Nevertheless, referring providers considered 20/24 total cGS molecular diagnoses (83%) to be explanatory for clinical features or worthy of additional workup.
CONCLUSION:
cGS is technically suitable as a first genetic test. In our cohort, diagnostic yield was not significantly different from SOC. Further studies addressing other variant types and implementation challenges are needed to support feasibility and utility of broad-scale cGS adoption.
INTRODUCTION
Currently, diagnostic standard-of-care (SOC) genetic testing practices are guided by specialty-based practice guidelines and clinical judgment.1–3 These practices may consist of a combination of methods such as karyotyping, chromosomal microarray analysis, single-gene analysis, and multigene panels.4 While high-coverage targeted sequencing technology has broadened the ability to assess and interpret the human genome, this approach has three key limitations. First, it requires that a set of genes be prespecified for each disease area; second, it limits the ability to reanalyze the data after new gene–disease associations are made; and third, it requires provider awareness and commercial availability of numerous disease-specific testing options.
In contrast to disease-focused genetic analysis, exome (ES) and genome sequencing (GS) have the potential to overcome the limitations of SOC and serve as effective diagnostic tools for rare genetic disorders.5–10 Furthermore, GS provides more uniform coverage of the genome, expands the scope of variants that can be identified based on documented medical and family history, and can reduce the number of genetic tests necessary to reach a diagnosis.11
In this prospective randomized study, we aimed to (1) assess diagnostic yield and clinical relevance of clinical genome sequencing (cGS) results across various disease phenotypes and ages at diagnostic evaluation, and (2) explore the challenges associated with implementing cGS as a diagnostic tool for patients with suspected genetic conditions. Here we report on diagnostic yield and clinical relevance of cGS as compared to SOC genetic tests.
MATERIALS AND METHODS
Study design and participants
Participants were recruited at the time of their clinical genetics evaluation at Massachusetts General Hospital (MGH) in Boston, Massachusetts between March 2018 and July 2019. Six MGH clinics participated in this study: the Cardiovascular Genetics Program, Medical Genetics and Metabolism Program (including the Diabetes Genetics Clinic), Ataxia Genetics Unit—Neurology, Gastrointestinal Cancer Program, Endocrine Tumor Genetics, and Pulmonary Genetics Clinic.
To be eligible for the study, patients were required to be pursuing a diagnostic genetic test at the time of enrollment; individuals were not eligible if they previously pursued genetic testing for the same indication. SOC testing was performed by reference laboratories or in-house at MGH. SOC laboratories were selected by the referring clinical team based on test availability and insurance coverage, among other reasons. Potential participants were identified through medical record review by a study coordinator and eligibility was confirmed by a study genetic counselor and the referring clinician. Given prior data on the utility of sequencing pediatric patients and their parents,12 patients under the age of 18 were offered enrollment as a family trio. Eligibility criteria are further described in Table S1. Consent sessions with a genetic counselor involved a discussion of study logistics, an overview of cGS, and potential results, which included both primary and nonprimary findings. Participants were allowed to opt out of receiving secondary findings in medically actionable genes recommended by the American College of Medical Genetics and Genomics (ACMG 59™).13 After enrollment, patient features were abstracted from electronic medical records (EMR) and recorded as Human Phenotype Ontology (HPO) terms using PhenoTips.14
Randomization was used as a strategy to avoid influencing the referring provider’s SOC approach and biasing patient choices for reflex testing. Enrolled participants were randomized 1:1 to receive only SOC or both SOC and cGS. Referring clinical providers, study staff members with patient interaction, and patients were blinded to randomization status until cGS report availability or three months after enrollment if randomized to the control arm. Block randomization stratified by clinic was implemented to ensure that a comparable proportion of individuals from each clinic received cGS. Participants enrolled as a trio were randomized independent of the clinic in which they were enrolled.
All participants were asked to complete two surveys—one at the time of enrollment and one after learning their randomization status and receiving cGS results. Survey questions and results will be described in a later paper.
Genome sequencing, analysis, and reporting
Genome sequencing was performed in the CLIA-certified, College of American Pathologists (CAP)–accredited Clinical Research Sequencing Platform at the Broad Institute of MIT and Harvard (Cambridge, MA; CLIA 22D2055652). All samples achieved a minimum coverage of 20 reads per base for >95% of the genome, with a minimum mean coverage of 30 reads per base.
The Partners Laboratory for Molecular Medicine (Cambridge, MA; CLIA 22D1005307) performed sequence realignment, variant calling, annotation, and reporting. Detailed analysis methods and reporting criteria are described in the Supplementary Methods and Fig. S1.
Molecular diagnosis and clinical relevance
In this study, sequencing results were categorized as a molecular diagnosis if they met all of the following criteria: (1) variant(s) classified as pathogenic (P) or likely pathogenic (LP), (2) variant(s) in genes with known disease association, and (3) variant(s) in allele states consistent with the inheritance pattern of the associated disorder. Molecular diagnoses were reported by cGS if they provided a full or partial explanation of the participant phenotype, or were predicted to cause a disease for which relevance to the participant’s phenotype could not be ruled out (uncertain). Further, phenotypic relevance of the molecular diagnoses was categorized as either primary (relevant to indication for SOC testing) or nonprimary (unrelated to the patient’s indication for SOC testing, but related to the patient’s family history, an additional phenotype identified upon EMR review, or ACMG 59™ secondary findings13).
The molecular diagnostic yield of SOC was compared to that of cGS for all patients who received both SOC and cGS reports. All molecular diagnoses on cGS were evaluated for clinical relevance. To assess clinical relevance, we evaluated if the result provided a diagnosis consistent with the patient’s reported phenotype and if the result informed medical management; clinical relevance was confirmed by the referring clinician.
Statistical analyses
Mean values between groups were compared using the two-sample t-test. Comparison of multiple values between the two study arms was performed using two-way analysis of variance (ANOVA). Diagnostic yields were compared using the two-sample test of proportions. Statistical significance threshold was set at ɑ= 0.05. All analyses were performed in Stata/IC 14.2.
RESULTS
Participant demographics, clinics of enrollment, and genetic test indications
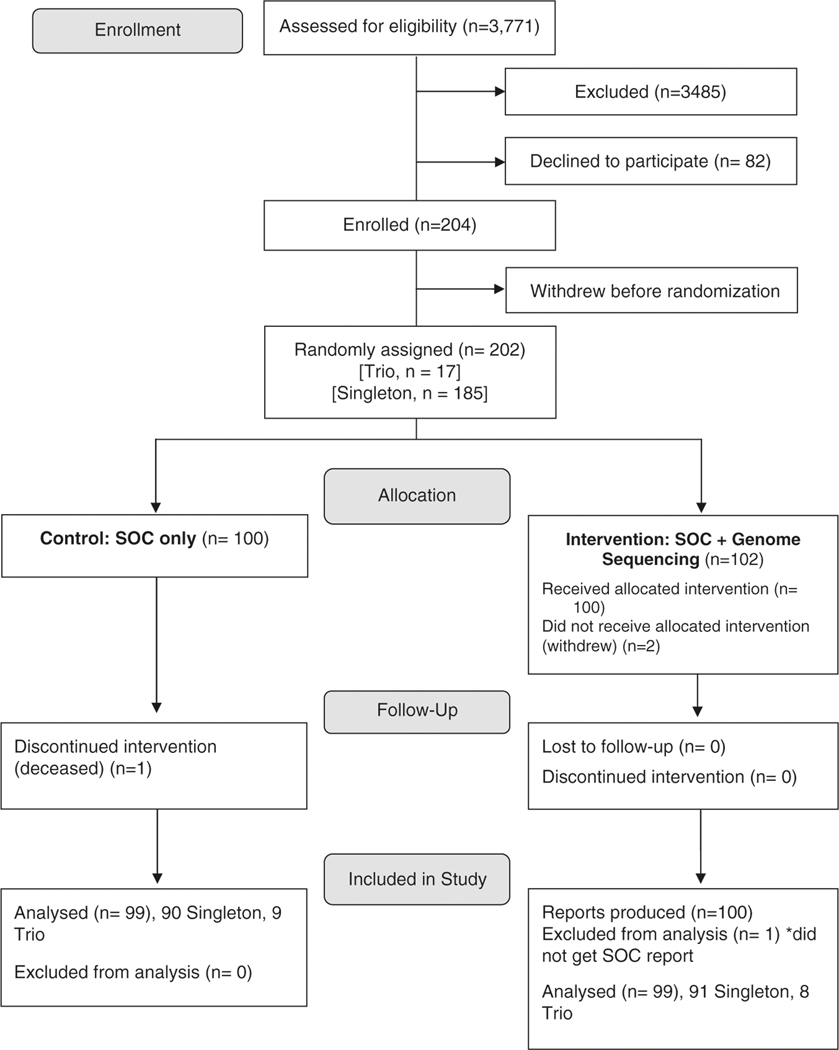
Between March 2018 and July 2019, 3,771 patients were evaluated by one of the six participating MGH genetics clinics; 204 patients were enrolled and 100 were randomized to receive cGS (Fig. 1, Fig. S2). One participant did not receive SOC due to insurance challenges and was removed from subsequent analysis—this resulted in 99 participants who received both SOC and cGS. The highest volume enrollment sites were the Cardiovascular Genetics Program (n = 69, 34%) and Medical Genetics and Metabolism Program (n = 60, 29%) (Table 1).
Fig. 1. Proband participant enrollment flowchart.
Two additional clinical genome sequencing (cGS) reports were produced for parents enrolled in a trio, but were not included in this diagram. SOC standard-of-care.
Table 1.
Participant characteristics and enrollment.
| Variable | Total cohort (n = 202)a | Control arm: SOC only (n = 100) | Intervention arm: SOC + cGS (n = 102)a | P valueb |
|---|---|---|---|---|
| Mean age, years (range) | 40 (2 months-81 years) | 42 (3 months-81 years) | 39 (2 months-78 years)c | 0.38 |
| Sex, n (%) | ||||
| Female | 110 (54%) | 52 (52%) | 57 (56%) | 0.58 |
| Male | 94 (46%) | 48 (48%) | 45 (44%) | |
| Race, n (%) | ||||
| White | 170(84%) | 86 (86%) | 84 (82%) | 0.96 |
| Asian | 6 (3%) | 3 (3%) | 3 (3%) | |
| Black or African American | 5 (2%) | 3 (3%) | 2 (2%) | |
| Unknown/not reported | 21 (10%) | 8 (8%) | 13 (13%) | |
| Ethnicity, n (%) | ||||
| NOT Hispanic/Latino | 172 (84%) | 87 (87%) | 83 (82%) | 0.67 |
| Hispanic/Latino | 7 (3%) | 3 (3%) | 4 (4%) | |
| Unknown/not reported | 25 (12%) | 10 (10%) | 15 (15%) | |
| Insurance, n (%) | ||||
| Private | 159 (78%) | 78 (78%) | 80 (78%) | 0.83 |
| Medicare | 41 (20%) | 19 (19%) | 21 (20%) | |
| Medicaid | 3 (1%) | 3 (3%) | 0 (0%) | |
| International | 1 (<1%) | 0 (0%) | 1 (1%) | |
| Clinic enrolled, n (% of total cohort) | ||||
| Cardiology | 69 (34%) | 35 (35%) | 34 (33%) | - |
| Medical Genetics | 60 (29%) | 29 (29%) | 29 (29%) | - |
| GI Cancer | 26 (13%) | 12 (12%) | 12 (13%) | - |
| Ataxia (Neurology) | 19 (9%) | 9 (9%) | 10 (10%) | - |
| Endocrine Tumor | 16 (8%) | 8 (8%) | 8 (8%) | - |
| Pulmonary | 14 (7%) | 7 (7%) | 7 (7%) | - |
| Human Phenotype Ontology (HPO) terms, n | ||||
| Average total | 6.14 | 6.16 | 6.11 | 0.85 |
| Primary indication | 3.67 | 3.76 | 3.57 | 0.66 |
| Nonprimary indication | 2.52 | 2.4 | 2.64 | 0.68 |
| Number of SOC genetic tests ordered, n (%) | ||||
| Average | 1.16 | 1.14 | 1.18 | 0.72 |
cGS clinical genome sequencing, GI gastrointestinal, SOC standard-of-care.
Two individuals enrolled and withdrew before randomization and two individuals withdrew after randomization; these individuals are not included in either in tervention arm of this table. Probands in a trio were randomized separately, but included in this table based on clinic of enrollment. Parents in a trio were not included in this table.
P value is for t-test comparing single values between the two study arms and analysis of variance (ANOVA) for comparing multiple values
Protocol deviation to enroll participant less than 3 months of age
The average age of the total cohort was 40.1 years, with 82% (n = 168) age 18 years or older. The majority of participants (82%) were White (Table 1). Seventeen of 36 pediatric probands were enrolled as a trio with both biological parents. The most common SOC test ordered was a multigene panel (n = 137, 65%). Eleven reference laboratories were used, which represented 96.6% of tests (Table 1, Fig. S3). The average number of HPO terms per participant was 6.14 (Table S2). No statistically significant differences in age, sex, race, ethnicity, insurance, HPO terms, or number of SOC tests ordered were observed between the control (SOC only) and intervention (SOC + cGS) groups (P values > 0.05, Table 1).
Molecular diagnostic yield: genome sequencing (cGS)
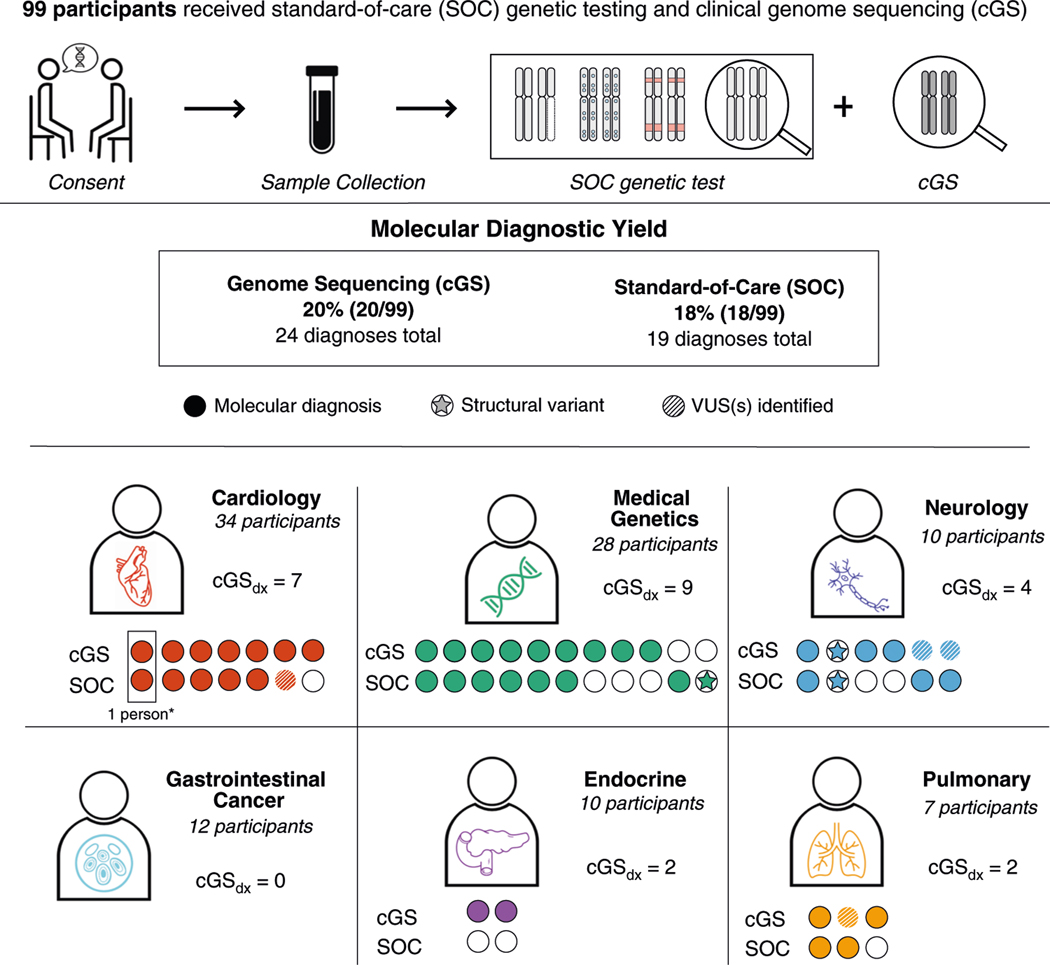
cGS identified molecular diagnoses in 20/99 participants. Some individuals received multiple diagnoses, yielding a total of 24 molecular diagnoses. Thirteen of these molecular diagnoses were full diagnoses that explained the participants’ primary indication for testing, and three were considered partial diagnoses that explained a portion of the phenotype (Table S3). The remaining diagnoses included one relevant to a family history of disease and seven uncertain diagnoses whose relevance to the participant’s phenotype was less clear but could not be ruled out. When considering only full and partial diagnoses, the molecular diagnostic yield of cGS was 16.2% (16/99, 95% CI 8.9–23.4%). Eighty-seven of 99 participants consented to receive secondary findings in the ACMG 59™ genes, but no returnable secondary findings were identified in this cohort.
When parsing by age group, 5/19 (26.3%) pediatric participants received molecular diagnoses (including 3 full, 1 partial, and 1 uncertain) and there was no significant difference in number of molecular diagnoses between singleton and family trio cGS (singleton: 27.3% [3/11], trio: 25% [2/8], P value > 0.05). Molecular diagnoses were identified in 20.0% of adult participants (16/80), ranging from 0% (0/12) in the Gastrointestinal Cancer Clinic to 40.0% (4/10) in the Ataxia Unit—Neurology (Fig. 2). When considering only full and partial diagnoses, 13.8% (11/80) of adults received molecular diagnoses from cGS (Table S3).
Fig. 2. Molecular diagnoses (probands only) made by clinical genome sequencing (cGS) and standard-of-care (SOC).
*A participant with multiple diagnoses is represented by more than one column. P/LP pathogenic/likely pathogenic, VUS variant of uncertain significance.
cGS technical sensitivity
All 27 of the P/LP variants reported by SOC were technically detected by cGS and filtered appropriately, corresponding to a sensitivity of 100% (Table S3, Table S4). These included 24 small (<20 bp) sequence variants and 3 copy-number variants (CNVs). Although analysis of repeat expansions (RE) and DNA methylation were included in some SOC tests, these variant types were not detected in our cohort.
Molecular diagnostic yield: cGS vs. SOC
SOC delivered a total of 19 molecular diagnoses in 18 individuals (Fig. 2)—a molecular diagnostic yield of 18.2% (18/99 participants, 95% CI −10.6–25.8%), which was not significantly different from cGS (P = 0.71). Similar to cGS, SOC diagnostic yield was lowest in the Gastrointestinal Cancer (0%; 0/12) and Endocrine Tumor (0%; 0/10) clinics, and highest in the Ataxia Unit—Neurology (30%; 3/10).
SOC reported 58.3% (14/24) of all molecular diagnoses reported by cGS. Additionally, one variant contributing to a cGS molecular diagnosis of MYH7-related hypertrophic cardiomyopathy was detected by SOC but classified as a variant of uncertain significance (VUS) (case 35CGS, Table S3). When disregarding this classification discrepancy, SOC reported 65.2% (15/24) of all cGS molecular diagnoses, and 87.5% (14/16) of cGS diagnoses that were categorized as full or partial (Table S3).
Among the nine molecular diagnoses reported only by cGS, none were considered to be full diagnoses (Table S3). However, two were partial diagnoses relevant to the primary indication for testing. In one case, the relevant gene was not analyzed by SOC testing (see case 65CGS vignette) and in the second case the molecular diagnosis was attributed to a variant that was detectable but not reported by SOC (see case 80CGS vignette). Two additional cGS-only diagnoses had uncertain relevance to the primary indication for testing but could not be ruled out as contributory (Table S3).
Case 65CGS:
A child presented to the Medical Genetics and Metabolism Program for evaluation due to delayed speech and language development and autistic behavior. At the time of her visit, three tests were ordered—fragile X, autism/ID panel, and microarray—all were negative. This patient was enrolled in the study as a family trio. cGS revealed two pathogenic GJB2 variants (p.Gly12ValfsX2 and p.Ser139Asn) confirmed in trans, suggesting a diagnosis of autosomal recessive deafness. This finding was considered a primary diagnosis given that hearing loss is frequently associated with delayed speech and language development. Upon review of this result with the family, it was uncovered that the patient had never undergone hearing evaluation.
The remaining five molecular diagnoses captured exclusively by cGS included four uncertain diagnoses with possible relevance to the probands’ nonprimary phenotypes, and one molecular diagnosis that was relevant to family history only (Table S3). Nonprimary phenotypes and family history were not the focus of SOC testing approaches. As a result, these genes were not included in the SOC tests.
It should be noted that five molecular diagnoses were made by SOC but not cGS (Fig. 2). Three of the diagnoses made only by SOC were the result of differential classification of variants that were reported by both methods (Table S3). For the remaining two cases (cases 204CGS, 187CGS), cGS detected but did not report the contributory variants since they were not highly relevant to the patient phenotype and were classified as VUS (Table S3).
cGS and SOC reports also differed in reporting of variants of uncertain significance. A total of 58 VUS were reported on SOC and/or cGS (Table S4). VUS identified exclusively by cGS in five participants prompted additional clinical workup (Fig. 3). Two case examples are described below—in both cases, familial testing was recommended to determine the phase of the identified variants; this testing was still pending at the time of this publication.
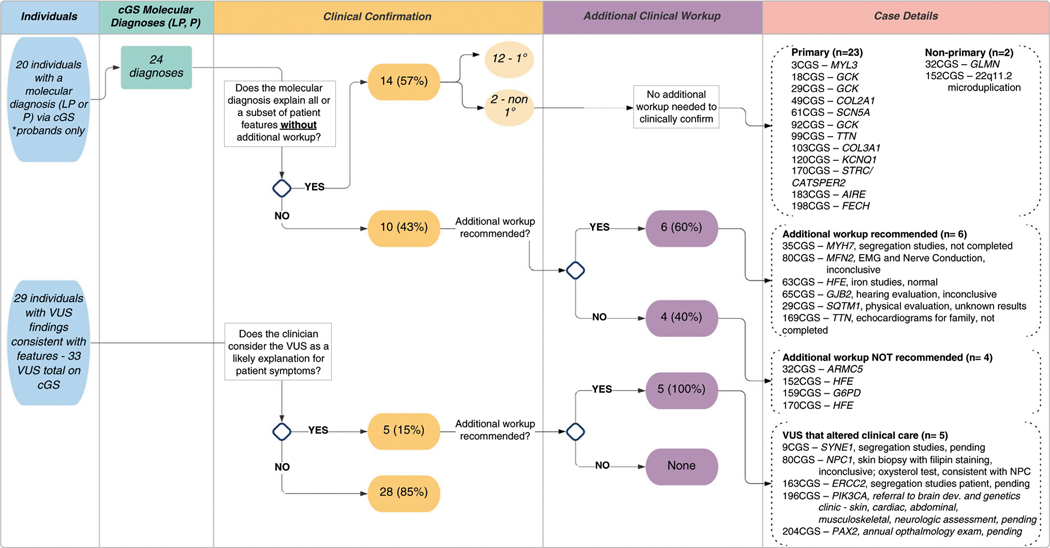
Fig. 3. Clinical relevance of clinical genome sequencing (cGS) molecular diagnoses and suspicious variant of uncertain significance (VUS) results. Each variant identified by cGS was reviewed for clinical relevance by the research team and referring clinical provider.
Column 1 is the number of individuals with a pathogenic/ likely pathogenic variant(s) or variant(s) of uncertain significance. Column 2 is the number of molecular diagnoses. Column 3 is an assessment of the degree to which the variant(s) identified explains patient features. Column 4 is an assessment of additional clinical workup needed to assess the significance of the variant(s) identified. Column 5 is the case identification number and corresponding gene of interest.
Case 9CGS:
Two VUS, c.3855C>T p.(Ile1285Ile) and c.2097 + 3_2097 + 15del p.(?), in the SYNE1 gene were detected by cGS in a proband referred for SOC RE testing based on his presentation of cerebellar atrophy, diplopia, and mild speech impairment. These phenotypes are consistent with a diagnosis of autosomal cerebellar ataxia type 1, which is caused by pathogenic variation in the SYNE1 gene. Given the close match in phenotype, and presence of two extremely rare variants, suspicion was higher for diagnostic relevance.
Case 163CGS:
In a proband with ataxia, abnormal magnetic resonance image (MRI), dysarthria, and a personal and family history of basal cell carcinoma, cGS identified one pathogenic variant (p.Arg616Pro) and one VUS (p.Gly413Val) in the ERCC2 gene, which is associated with a spectrum of autosomal recessive conditions including xeroderma pigmentosum. Notably, at least 25% of individuals with ERCC2-related disorders have progressive neurologic abnormalities, including ataxia and neurodegeneration in the cerebrum and cerebellum.
Clinical relevance and impact on management
Upon review of postclinic notes and/or discussions with referring providers, 14 of 24 (58%) cGS molecular diagnoses explained current clinical features or a subset of features without additional workup—12 were related to the primary indication for testing and 2 were related to nonprimary phenotypes (Fig. 3). Of the remaining ten cGS molecular diagnoses with unclear clinical relevance, referring providers recommended additional workup for six cases, including electromyography (EMG), hearing evaluation, and iron studies. Molecular diagnoses have not yet been clinically confirmed based on additional workup for these cases.
To further explore the medical importance of cGS results, we reviewed the relevance of clinically suspicious VUS findings. Despite uncertain variant pathogenicity, referring clinicians reported that they planned to change medical management and/or pursue additional workup for five patients with VUS reported by cGS (Fig. 3). To date, a diagnosis of Niemann–Pick type C was confirmed based on additional workup for one patient (case 80CGS).
Case 80CGS:
A female in her 40s presented to the Ataxia Unit for evaluation due to ataxia, cerebellar atrophy, dysphagia, and dysarthria. At the time of her visit, an autosomal dominant triplet repeat ataxia panel was ordered and was negative. Exome sequencing (ES) was pursued by the clinical team in follow-up to these results, which identified two variants (p.Gln438X, P and p.Phe68del, LP) in NPC1, suggesting a diagnosis of Niemann–Pick disease type C. In parallel, genome sequencing identified the same variants in NPC1; however, the variant classification differed (LP and VUS). Additionally, genome sequencing revealed a MFN2 variant (p. Arg707Trp, LP), suggesting a diagnosis of Charcot–Marie–Tooth type 2A. Follow-up with the ES laboratory revealed that the MFN2 variant was not reported due to a perceived lack of relevance to the patient phenotype. Upon review with the referring clinical team, additional workup was recommended, including: (1) skin biopsy with filipin staining to evaluate for Niemann–Pick disease type C—inconclusive (approximately 50% staining) and (2) electromyography and nerve conduction studies to evaluate for Charcot–Marie–Tooth type 2A, which were inconclusive. Subsequently, the patient received an oxysterol test, which was consistent with a diagnosis of Niemann–Pick disease type C. She is now taking miglustat to stabilize and slow progression of the disease.
cGS also confirmed one clinical diagnosis of hemochromatosis in a parent enrolled in this study as a part of a family trio. In total, 15 cGS molecular diagnoses were confirmed by clinical workup; 2 (170CGS parent, 32CGS) would not have been made by standard-of-care genetic test approaches.
DISCUSSION
Previous studies suggest that ES/GS be utilized as the first genetic test for individuals with suspected genetic disorders, citing increased diagnostic yield, reduced time to reach a diagnosis, and economic advantages over the SOC stepwise approach to genetic testing.15,16 While similar diagnostic yields have been reported for ES and GS,17 GS does offer added benefits, including more uniform sequencing coverage, greater power for structural variant (SV) analysis, and an expanded scope for future reanalysis as understanding of functional elements within noncoding regions improves.11 Given these benefits and the declining cost differential between ES and GS at our institution, we chose to test the utility of cGS as a firstline genetic test. In contrast to previous studies, which predominantly enrolled pediatric patients or focused on a specific disease area, this prospective study compares the diagnostic yield and clinical relevance of singleton and family trio cGS to that of SOC practices across age groups and medical specialties. An additional strength of this study was that genome analysis and interpretation were conducted within an integrated health-care setting, allowing for access to full medical records and collegial discussions about the significance of cGS results with referring providers.
cGS identified molecular diagnoses that fully or partially explained patient phenotypes in 16.2% (16/99) of our cohort; this yield was consistent with other studies that report diagnostic yields ranging from 14% to 76%.15,18,19 cGS detected all diagnostic variants reported by SOC, implying that cGS is sufficiently sensitive to replace SOC genetic testing. However, our study was limited by the narrow range of variant types detected. For example, no clinically suspicious SVs, mitochondrial variants, or deep noncoding variants (>50 bp from coding regions) were reported by either SOC or cGS, even though our genome analysis included these variant types. Similarly, important limitations of short-read NGS technology (e.g., detection of triplet repeat expansions) were not brought to light in this study, since the diagnostic variants identified by SOC included a limited number of variants for which cGS is expected to have reduced sensitivity. It is therefore important to note that cGS may not be an optimal firstline test for all clinical indications.
Specialized data processing algorithms have been developed to capture certain technically challenging variant types, including somatic mosaicism,20 repeat expansions,21 and recurrent variation in homologous regions.22 While they represent a promising new frontier in GS analysis, these algorithms have yet to see widespread clinical implementation and were not incorporated into the validated clinical pipeline used for our cGS analysis. As a result, low-level mosaic variants, repeat expansions, and variants in homologous regions were not comprehensively assessed in our study. While it will be important to determine whether the implementation of such algorithms improves cGS diagnostic yield, the results of SOC testing in this study, which included specialized assays for detecting these variant types, suggest that they may have a limited impact on yield in our cohort. The clinic with the highest proportion of cGS molecular diagnoses in this study was the adult Ataxia Unit, where all diagnoses were due to the identification of single-nucleotide variants (SNVs) or insertion/deletions in nonrepetitive regions. This was an unanticipated finding as 74% of the SOC genetic tests were ordered based on concern for a RE disorder (Fig. S3). However, none of the four cGS diagnoses from this clinic were considered to fully explain the indication for testing, and clinical follow-up did not support a contributory role for two cases. Nevertheless, cGS identified clinically suspicious VUS results not assessed by SOC in two additional participants from the Ataxia Unit (cases 9CGS, 163CGS), supporting other studies that suggest that ES/GS may improve diagnostic yield for adults with clinically heterogeneous cerebellar ataxias.23–26
This study revealed multiple sources of reporting differences between SOC and cGS that should be considered prior to adoption of ES or GS as a firstline test. First, the identification of diagnostic findings that partially explained participant phenotypes in genes that were omitted from the ordering provider’s SOC workup highlights the advantages of an unbiased approach to GS analysis, which has also been demonstrated in ES studies.5–9 However, cGS also revealed diagnoses that were unrelated to the patient’s primary indication for testing, which may be undesirable for some patients. Another source of reporting differences was due to discrepancies in variant classification,27 highlighting the importance of ongoing efforts to standardize classification criteria and support data sharing (ClinGen, https://clinicalgenome.org/; ClinVar, https://www.ncbi.nlm.nih.gov/clinvar/).
Laboratory reporting practices represented a key third source of discordance between cGS and SOC reports. Given the large number of variants identified by genomic sequencing methods, laboratories must define a subset of variants to analyze and report. Many laboratories restrict reporting to P and LP variants that match the patient phenotype or represent a medically actionable secondary finding.13,28,29 However, given that two molecular diagnoses reported as relevant to primary phenotypes by cGS were classified as irrelevant to the same phenotypes by SOC ES, this study highlights the subjective nature of “phenotypic match.” Additionally, while it is common practice for targeted sequencing tests to report variants classified as P, LP, or VUS, current guidelines for genomic sequencing suggest that VUS should only be reported in genes highly relevant to the patient phenotype. In following with this, several VUS included on SOC reports were excluded from reporting by cGS due to lack of phenotypic relevance. The observation of fewer reported VUS together with improved diagnostic yield suggests that more targeted genetic testing reports may be an unanticipated benefit of widespread implementation of cGS. Nevertheless, in accordance with other studies,30–32 our experience supports open communication between ordering providers and analysis teams to ensure that variants of interest to clinicians and patients are not omitted from reports.
This study did not address turnaround time (TAT), which is an important consideration for the feasibility of implementing cGS as a firstline test. cGS TAT was not informative in our study due to staffing levels that were not reflective of a typical diagnostic laboratory. Nevertheless, optimized sample preparation, sequencing, and data processing steps and artificial intelligence–assisted analyses have produced cGS TATs of less than 30 hours.33 While not necessary in many clinical contexts, the achievement of 30-hour TATs suggests that analysis infrastructure investments could make cGS TAT comparable to, or quicker than, existing SOC options.
Finally, we would be remiss not to note that this study was limited by multiple systemic barriers that impact access to and uptake of genetic services within a health-care system. A 2015 systematic review identified several obstacles, including lack of awareness of personal/patient risk factors, lack of knowledge of family medical history/lack of obtaining adequate family history, and lack of knowledge of genetic services.34 These factors influenced the patients identified and recruited for this study and negatively impacted participant diversity. Beyond access to genetic services, uptake of SOC appointments and testing was a barrier to participation. To participate in this study, individuals were required to attend an in-person appointment and pursue SOC at the time of enrollment. Given that 189 eligible patients did not attend their appointment and a portion of eligible patients deferred SOC genetic testing due to insurance coverage concerns, patients were likely excluded from the study due to challenges preventing them from traveling to an appointment as well as underlying insurance challenges imposed by the US health-care system (Fig. S2). Further, 176 eligible patients were excluded because they were not English-speaking, emphasizing the need for dedicated resources to support diverse populations in clinical care and research. Additionally, cGS in this study required a blood sample. Due to this requirement, we were limited in our ability to collect parental samples for trio GS when both parents were unable to come to clinic, often due to work, travel, and family-related obstacles. To equitably offer the most comprehensive cGS evaluation, resources are needed to develop methods that allow cGS to be run on saliva or buccal samples, which can be submitted remotely. Finally, this was a hospital-sponsored clinical research study. Most payers consider cGS to be investigational at this time and therefore efforts must be made to contract with insurance companies and conduct the necessary cost-effectiveness analyses needed to improve payer coverage of this test; doing so will make cGS accessible to more patients.
This study provides evidence that cGS is suitable as a firstline diagnostic genetic test, regardless of patient age or clinical specialty. However, metrics beyond diagnostic yield need to be considered prior to broad-scale implementation. Capturing the full scope of utility and feasibility, with a particular focus on payer coverage, will allow us to move towards equitable and scalable delivery models of genomic medicine.
Supplementary Material
ACKNOWLEDGEMENTS
This study was funded by the Department of Medicine at Massachusetts General Hospital. Illumina supplied a portion of the sequencing reagents to enable this study. M.U. was supported in part by NIDDK K23DK114551. We thank the patients and families for participating in this study. We are grateful to Stephanie Harris, Lauren O’Grady, Marcie Steeves, Jin Yun Helen Chen, Megan Hawley, Erica Blouch, Linda Rodgers, Kristen Shannon, David Sweetser, Paula Goldenberg, Frances High, Amel Karaa, Angela Lin, Stephanie Santoro, Steven Lubitz, Christopher Newton-Cheh, and Jeremy Schmahmann for referring patients to the study. We also thank Edyta Malolepsza, Harrison Brand, and members of Michael Talkowski’s laboratory for their assistance with SV calling and analysis.
Footnotes
DATA AVAILABILITY
Variants reported by cGS have been submitted to ClinVar. De-identified genomic and phenotype data will be made available on the National Human Genome Research Institute (NHGRI) AnVIL platform (pending approval of our application by AnVIL). Data access requests can be made per instructions here https://anvilproject.org/learn/accessing-data/requesting-data-access#accessing-controlled-access-data. For additional information, contact C.A.A-T. (ctse@mgh.harvard.edu).
ETHICS DECLARATION
All study participants provided written consent or assent. This study was completed as a demonstration project in the MGH Center for Genomic Medicine and was approved by the Massachusetts General Brigham Institutional Review Board. Additional information about the study can be found on Clinicaltrials.gov (NCT03829176).
COMPETING INTERESTS
The authors declare no competing interests.
ADDITIONAL INFORMATION
Supplementary information The online version contains supplementary material available at https://doi.org/10.1038/s41436s-021-01193-y.
REFERENCES
- 1.Ross LF, Saal HM, David KL & Anderson RR Technical report: ethical and policy issues in genetic testing and screening of children. Genet. Med 15, 234–245 (2013). [DOI] [PubMed] [Google Scholar]
- 2.Manning M. & Hudgins L. Array-based technology and recommendations for utilization in medical genetics practice for detection of chromosomal abnormalities. Genet. Med 12, 742–745 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.National Comprehensive Cancer Network. Genetic/familial high-risk assessment: breast, ovarian, and pancreatic (Version 1.2020). https://www.nccn.org/professionals/physician_gls/pdf/genetics_bop.pdf (2020). [DOI] [PubMed]
- 4.Xue Y, Ankala A, Wilcox WR & Hegde MR Solving the molecular diagnostic testing conundrum for Mendelian disorders in the era of next-generation sequencing: single-gene, gene panel, or exome/genome sequencing. Genet. Med 17, 444–451 (2015). [DOI] [PubMed] [Google Scholar]
- 5.Yang Y. et al. Molecular findings among patients referred for clinical whole-exome sequencing. JAMA. 312, 1870–1879 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee H. et al. Clinical exome sequencing for genetic identification of rare Mendelian disorders. JAMA. 312, 1880–1887 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vissers LELM et al. A clinical utility study of exome sequencing versus conventional genetic testing in pediatric neurology. Genet. Med 19, 1055–1063 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Ligt J. et al. Diagnostic exome sequencing in persons with severe intellectual disability. 10.1056/NEJMoa1206524 (2012). [DOI] [PubMed]
- 9.Stark Z. et al. A prospective evaluation of whole-exome sequencing as a first-tier molecular test in infants with suspected monogenic disorders. Genet. Med 18, 1090–1096 (2016). [DOI] [PubMed] [Google Scholar]
- 10.Lionel AC et al. Improved diagnostic yield compared with targeted gene sequencing panels suggests a role for whole-genome sequencing as a first-tier genetic test. Genet. Med 20, 435–443 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Meienberg J, Bruggmann R, Oexle K. & Matyas G. Clinical sequencing: is WGS the better WES? Hum. Genet 135, 359–362 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thiffault I. et al. Clinical genome sequencing in an unbiased pediatric cohort. Genet. Med 21, 303–310 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kalia SS et al. Recommendations for reporting of secondary findings in clinical exome and genome sequencing, 2016 update (ACMG SF v2.0): a policy statement of the American College of Medical Genetics and Genomics. Genet. Med 19, 249–255 (2017). [DOI] [PubMed] [Google Scholar]
- 14.Girdea M. et al. PhenoTips: patient phenotyping software for clinical and research use. Hum. Mutat 34, 1057–1065 (2013). [DOI] [PubMed] [Google Scholar]
- 15.Smith HS et al. Clinical application of genome and exome sequencing as a diagnostic tool for pediatric patients: a scoping review of the literature. Genet. Med 21, 3–16 (2019). [DOI] [PubMed] [Google Scholar]
- 16.Cirino Allison L. et al. A comparison of whole genome sequencing to multigene panel testing in hypertrophic cardiomyopathy patients. Circ. Cardiovasc. Genet 10, e001768 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kingsmore SF et al. A randomized, controlled trial of the analytic and diagnostic performance of singleton and trio, rapid genome and exome sequencing in ill infants. Am. J. Hum. Genet 105, 719–733 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Clark MM et al. Meta-analysis of the diagnostic and clinical utility of genome and exome sequencing and chromosomal microarray in children with suspected genetic diseases. npj Genomic Med. 10.1038/s41525-018-0053-8 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Scocchia A. et al. Clinical whole genome sequencing as a first-tier test at a resource-limited dysmorphology clinic in Mexico. npj Genomic Med 4, 1–12 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang AY et al. MosaicHunter: accurate detection of postzygotic single-nucleotide mosaicism through next-generation sequencing of unpaired, trio, and paired samples. Nucleic Acids Res. 45, e76 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dolzhenko E. et al. ExpansionHunter: a sequence-graph-based tool to analyze variation in short tandem repeat regions. Bioinformatics (Oxf.) 35, 4754–4756 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen X. et al. Spinal muscular atrophy diagnosis and carrier screening from genome sequencing data. Genet. Med 22, 945–953 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fogel BL et al. Exome sequencing in the clinical diagnosis of sporadic or familial cerebellar ataxia. JAMA Neurol. 71, 1237–1246 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pyle A. et al. Exome sequencing in undiagnosed inherited and sporadic ataxias. Brain. 138, 276–283 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kang C. et al. High degree of genetic heterogeneity for hereditary cerebellar ataxias in Australia. Cerebellum. 18, 137–146 (2019). [DOI] [PubMed] [Google Scholar]
- 26.Galatolo D, Tessa A, Filla A. & Santorelli FM Clinical application of next generation sequencing in hereditary spinocerebellar ataxia: increasing the diagnostic yield and broadening the ataxia-spasticity spectrum. A retrospective analysis. Neurogenetics. 19, 1–8 (2018). [DOI] [PubMed] [Google Scholar]
- 27.Richards S. et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med 17, 405–424 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rehm HL et al. ACMG clinical laboratory standards for next-generation sequencing. Genet. Med 15, 733–747 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hegde M. et al. Development and validation of clinical whole-exome and whole-genome sequencing for detection of germline variants in inherited disease. Arch. Pathol. Lab. Med 141, 798–805 (2017). [DOI] [PubMed] [Google Scholar]
- 30.Mak CC et al. Exome sequencing for paediatric-onset diseases: impact of the extensive involvement of medical geneticists in the diagnostic odyssey. npj Genomic Med. 3, 19 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Baldridge D. et al. The Exome Clinic and the role of medical genetics expertise in the interpretation of exome sequencing results. Genet. Med 19, 1040–1048 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vears DF, Elferink M, Kriek M, Borry P. & van Gassen KL Analysis of laboratory reporting practices using a quality assessment of a virtual patient. Genet. Med 30, 1–9 (2020). [DOI] [PubMed] [Google Scholar]
- 33.Clark MM et al. Diagnosis of genetic diseases in seriously ill children by rapid whole-genome sequencing and automated phenotyping and interpretation. Sci. Transl. Med 10.1126/scitranslmed.aat6177 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Delikurt T, Williamson GR, Anastasiadou V. & Skirton H. A systematic review of factors that act as barriers to patient referral to genetic services. Eur. J. Hum. Genet 23, 739–745 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.