Abstract
TFIIIC plays a key role in nucleating the assembly of the initiation factor TFIIIB on class III genes. We have characterized an essential gene, TFC8, encoding the 60-kDa polypeptide, τ60, present in affinity-purified TFIIIC. Hemagglutinin-tagged variants of τ60 were found to be part of TFIIIC-tDNA complexes and to reside at least in part in the downstream DNA-binding domain τB. Unexpectedly, the thermosensitive phenotype of N-terminally tagged τ60 was suppressed by overexpression of τ95, which belongs to the τA domain, and by two TFIIIB components, TATA-binding protein (TBP) and B"/TFIIIB90 (but not by TFIIIB70). Mutant TFIIIC was deficient in the activation of certain tRNA genes in vitro, and the transcription defect was selectively alleviated by increasing TBP concentration. Coimmunoprecipitation experiments support a direct interaction between TBP and τ60. It is suggested that τ60 links τA and τB domains and participates in TFIIIB assembly via its interaction with TBP.
The primary step in tRNA gene activation is the binding of TFIIIC to the A and B blocks of the intragenic promoter. Its main function is then to assemble the initiation factor TFIIIB upstream of the transcription start site (30). Yeast TFIIIC (τ factor) is a multisubunit protein of ca. 600 kDa organized in two large globular domains τA and τB of similar size and mass (10-nm diameter and ca. 300 kDa), each interacting with one promoter element, as visualized by electron microscopy (17, 53). Binding of τB to the B block is predominant and favors the binding of τA to the A block (4). TFIIIC-DNA interaction displays a remarkable adaptability to the variable A-B distances found in different tRNA genes (3). Affinity-purified Saccharomyces cerevisiae TFIIIC comprises six polypeptides of 138, 131, 95, 91, 60, and 55 kDa (6, 19, 47, 59). Gene cloning, protein-DNA cross-linking, mutagenesis, and protein-protein interaction studies provided a global view of the location and role of several TFIIIC subunits within the TFIIIC-DNA complex. τ138 and τ91 subunits reside in the τB domain and cooperate in downstream DNA binding (1, 10, 36, 37); τ95 and τ55 interact physically, belong to the τA domain, and are thought to participate in A block binding (7, 17, 40, 48, 59). τ131 (42) stands as the TFIIIC subunit responsible for TFIIIB assembly based on genetic evidence (49, 61), its upstream location (7), and its interaction with two TFIIIB components (14, 33, 51).
TFIIIB is not a stable molecular entity like TFIIIC. It can be resolved chromatographically into two fractions named B′ and B" (29). B′ comprises TATA-binding protein (TBP) and the TFIIB-related factor TFIIIB70/Brf1 (12, 16, 31, 39), while B" contains TFIIIB90 (32, 50, 51). The TFIIIC-dependent TFIIIB assembly on TATA-less class III genes is a multistep pathway that could be decomposed in vitro (29, 31) and reconstituted with recombinant TFIIIB components (32, 51). The order of interaction is TFIIIB70, then TBP, and then B", as shown by gel retardation and DNA photo-cross-linking (31). TBP stabilizes the weak interaction between TFIIIB70 and the TFIIIC-DNA complex but the complete upstream footprint and the characteristic stability of the TFIIIB-DNA complex requires the recruitment of B"/TFIIIB90 (29, 31). A cascade of conformational rearrangements at the protein and DNA levels are accompanying these assembly steps, as evidenced by successive changes in the accessibility of TFIIIB70, TBP, and τ131 to site-specific DNA cross-linking (31), by the DNA bending induced upon TFIIIB binding (11, 38, 46), and by the presence of a cryptic DNA binding domain in TFIIIB70 (24). τ131 appears to play the major role in positioning TFIIIB since it is the only TFIIIC subunit accessible to DNA cross-linking upstream of the start site (5, 7) and found to interact with TFIIIB70 (14, 33) and TFIIIB90 (51). TFIIIB can effect its own assembly onto the TATA-containing SNR6 gene through the interaction of TBP with the strong TATA box (27, 43, 45). Interestingly, Whitehall et al. (60) found that TBP could not discern the polarity of the TATA element and directed TFIIIB assembly in two orientations. However, in contrast to the TATA-dependent assembly, TFIIIC placed TFIIIB in the correct orientation. Since no TFIIIC component was known to interact with TBP, it was presumed that the unidirectional binding of TBP to the TATA box is dictated by the oriented interaction of TFIIIB70 with τ131 (60).
In the present work we have completed the characterization of TFIIIC components by cloning a yeast gene, named TFC8, that encodes the 60-kDa polypeptide. We present genetic and biochemical evidence that this component, named τ60, resides at least in part within the τB domain and participates in TFIIIB recruitment via TBP binding.
MATERIALS AND METHODS
Yeast strains, plasmids, and genetic techniques.
YNN281×YNN282 (21) was used for gene disruption. Cells were grown in standard rich medium (YPD) or minimal medium (Casamino acid medium). The plasmids used for suppression studies are described in Lefebvre et al. (37). Preparation of media, tetrad dissection, transformation of lithium acetate-treated cells, and plasmid shuffling by using 5-fluoro-orotic acid were performed by standard techniques (2).
Amino acid sequence determination.
The polypeptide components of purified TFIIIC preparation were separated by preparative electrophoresis on a 6% polyacrylamide–sodium dodecyl sulfate (SDS) gel (36). The gel was lightly stained with Coomassie blue. A gel slice containing the 60-kDa polypeptide was excised, crushed, and subjected to trypsin digestion. Preparation and analysis of tryptic peptides have been previously described (36, 59). One peptide sequence was partially determined (??TLYLTT[T/F]PT).
Cloning, construction of plasmids, and disruption of TFC8.
Two 32-mer oligonucleotides were used to amplify the open reading (ORF) frame of TFC8 by PCR on yeast genomic DNA. The resulting DNA fragment was then labeled with [α-32P]dCTP and used to screen the FL100 library (57) containing yeast genomic DNA fragments inserted into the pFL44L (2μ, URA3) plasmid (9). The pFL44L plasmid isolated from one of the hybridizing clones was found to contain a large DNA insert comprising the TFC8 gene and was named pCC12 (pFL44L-TFC8). The 2.3-kb SalI-FspI DNA fragment from pCC12 harboring the TFC8 gene was cloned into pUN45 creating the pYED1 plasmid. A 69-mer oligonucleotide was used to introduce a NdeI restriction site at the initiation codon of TFC8, followed by the sequence encoding the hemagglutinin (HA) epitope (YPYDVPDYA) derived from the influenza virus HA protein (62), by oligonucleotide-mediated mutagenesis on single-stranded pYED1 DNA by using a Muta-Gene kit (Bio-Rad), yielding the pYED2 plasmid (encoding HA Nter-τ60). The sequence encoding the HA epitope was also introduced before the stop codon of TFC8 by PCR-mediated mutagenesis with two oligonucleotides. One contained a SalI restriction site and nucleotides complementary to the upstream region of the TFC8 promoter, and the other harbored the sequence encoding the HA epitope, a NotI restriction site, and nucleotides complementary to the stop codon region of TFC8. After PCR amplification, the SalI-NotI DNA fragment was inserted into the corresponding sites of pYED1 to give pYED3 (encoding for HA Cter-τ60). The NdeI-BamHI DNA fragment from pYED2 was inserted into the corresponding sites of pET28c vector (Novagen), yielding pET60.
Disruption of the TFC8 gene was performed as previously described (8, 40). Two 55-mer oligonucleotides harboring sequences complementary to TFC8 and to the yeast HIS3 selectable marker were used to amplify by PCR an ∼1.1-kb DNA fragment containing the HIS3 gene flanked by TFC8 promoter and terminator sequences. The PCR-amplified DNA fragment was used to transform the strain YNN281×YNN282. The structure of several His+ diploids was verified by PCR analysis. To determine whether TFC8 was essential for growth, sporulation and dissection analysis were performed. The diploid His+ strain was also transformed with the pCC12 plasmid (pFL44L-TFC8) and sporulated. A His+ spore containing the pCC12 plasmid was chosen to give strain YCC8 used for plasmid shuffling.
Purification of TFIIIC.
TFIIIC was purified starting from ∼12 g of S. cerevisiae cells, using fast-protein liquid chromatography-grade resins. The preparation of the cell extract was done as described by Huet et al. (25). Crude extract was first diluted to 0.25 M ammonium sulfate (AS) with buffer I (20 mM Tris-HCl, pH 8.0; 0.5 mM EDTA; 10 mM β-mercaptoethanol; 10% [vol/vol] glycerol) and then loaded at 2.5 ml/min on a 25-ml heparin Hyper D (BioSepra) column previously equilibrated with buffer I (0.25 M AS). The resin was then washed at 5 ml/min with 250 ml of buffer I (0.35 M AS). A linear gradient of AS from 0.35 to 0.70 M in 180 ml of buffer I was then applied at 2.5 ml/min. Fractions (2 ml) were collected and assayed for TFIIIC-DNA binding activity. TFIIIC-containing fractions (0.45 to 0.55 M AS) were pooled and dialyzed against buffer I (0.07 M AS). Proteins were then loaded at 0.5 ml/min on a 1-ml MonoQ column (Pharmacia, Piscataway, N.J.) previously equilibrated with buffer I (0.07 M AS). The column was washed at 0.5 ml/min with 20 ml of buffer I (0.07 M AS). A linear gradient of AS from 0.07 to 0.4 M in 15 ml of buffer I was then applied at 0.5 ml/min. Fractions (200 μl) containing TFIIIC-DNA binding activity were eluted between 0.24 and 0.30 M AS. Based on Western blot experiments with anti-τ55 or anti-τ60 antibodies, the TFIIIC preparation from Ntag-τ60 mutant cells was found to contain half as much factor as the wild type.
Expression and purification of τ60 in Escherichia coli.
Recombinant Tfc8p tagged at its N-terminal end with six histidines and with the HA epitope (HA-rτ60) was expressed from plasmid pET60 in E. coli BL21 (pLysS). Cell culture, protein induction, and extract preparation were performed as described earlier (1). Crude cell extract was loaded at 1 ml/min on a 5 ml of Ni2+-charged HiTrap chelating column (Pharmacia) previously equilibrated with 20 mM HEPES (pH 7.8), 300 mM NaCl, and 10% glycerol containing 10 mM imidazole. Proteins were eluted by a linear gradient in 60 ml of the same buffer containing 20 to 270 mM imidazole at 1 ml/min. Fractions (1 ml) were assayed for HA-rτ60 by Western blotting with anti-τ60 antibodies. Fractions containing HA-rτ60 were eluted at concentrations between 160 and 190 mM imidazole.
Anti-τ60 polyclonal antibodies.
A 14-mer peptide (N-12-M) encompassing the C-terminal amino acid residues of τ60 was synthesized (Neosystem, Strasbourg, France) and conjugated to maleimide-activated keyhole limpet hemocyanin (KLH). N-12-M–KLH conjugate was then injected into rabbits for antibody production in three injections at about three week intervals.
Immunoprecipitation experiments.
rTBP alone (200 ng) or both rTBP (200 ng) and HA-rτ60 (650 ng) were preincubated for 90 min at 25°C in 40 μl of buffer A containing 20 mM HEPES (pH 8.0), 5 mM MgCl2, 1 mM dithiothreitol (DTT), 0.1 mM EDTA, 5% glycerol, 0.05% NP-40, and 110 mM KCl. Magnetic beads (Dynabeads/Dynal) coated with 12CA5 antibody were added to the mixtures. The beads were incubated with gentle shaking for 3 h at 10°C and then washed three times in buffer A. Bound proteins were eluted by boiling in Laemmli sample buffer, separated by SDS-polyacrylamide gel electrophoresis (PAGE) and analyzed by Western blotting by using a mixture of polyclonal antibodies directed to TBP and to τ60. Bound antibodies were revealed by using the Amersham ECL Kit. The polypeptides revealed by antibodies directed to TBP or τ60 were identified in independent experiments.
DNA binding and in vitro transcription assays.
TFIIIC-tDNA or τB-tDNA interaction was monitored by gel retardation analysis as described previously (25, 37). A 32P-labeled DNA fragment (3 to 10 fmol; 4,000 to 10,000 cpm) carrying the tRNA3Glu (198 bp) or the SUP4tRNATyr (375 bp) genes was incubated for 10 min at 25°C in a 15-μl reaction mixture containing 10 mM Tris-HCl (pH 8.0), 10% glycerol, bovine serum albumin, DNA competitor, and TFIIIC (MonoQ fraction). The final ammonium sulfate concentration was 75 mM. τB-tDNA complexes were obtained after a further 10 min incubation at 25°C of TFIIIC-tDNA complexes with 10 ng of α-chymotrypsin (Sigma). Digestion was stopped by addition of 1 ng of aprotinin (Sigma). Complexes were analyzed by nondenaturing gel electrophoresis (5% polyacrylamide).
Transcription mixtures (40 μl) contained 20 mM HEPES (pH 8.0); 5 mM MgCl2; 1 mM DTT; 0.1 mM EDTA; 5% glycerol; 8 U of RNasin (Amersham); 0.6 mM each ATP, GTP, and CTP; 0.03 mM UTP and 10 μCi of [32P]UTP; TFIIIB (370 ng of Cibacron-blue fraction) or rTBP (250 ng); rTFIIIB70 (1.2 μg) and partially purified B" fraction (1.8 μg); RNA polymerase III (Pol III; 50 ng); and TFIIIC (MonoQ fraction) as indicated. The final KCl concentration was 110 mM. After 10 min of preincubation at 25°C, transcription reaction was initiated by addition of 130 ng of plasmid DNA harboring different tRNA genes and allowed to proceed for 45 min at 25°C. Transcripts were analyzed by electrophoresis on an 8 M urea gel (6% polyacrylamide).
In vivo labeling of RNAs and Northern blot analysis.
RNA labeling was done with [3H]uracil with strains where the ura3 mutation was complemented by the URA3 plasmid pFL44L. Cells were exponentially grown in uracil-free Casamino acid medium supplemented with adenine (Casa+Ade) to an optical density of 0.4 at 600 nm. Then, 150 μCi of [3H]uracil was added to 10 ml of culture for 15 min. The cells were next harvested and chilled with 10 ml of ice-cold sterile water. RNAs were extracted as previously described (22, 52). Small RNA species were analyzed by loading and separating equal amounts of RNA (6 μg per lane) on a 7 M urea gel by electrophoresis (6% polyacrylamide).
RNA extraction and gel electrophoresis for Northern blot analysis were performed as described in the previous section. Electrophoretic transfer on nylon membrane (Bio-Rad apparatus), UV cross-linking DNA (Stratalinker apparatus), and hybridization with DNA probe in sodium phosphate buffer were carried out as previously described (15). The DNA probe used in the Northern blot experiment shown in Fig. 2B was a 327-bp 32P-labeled PCR fragment encompassing the yeast tRNA3Leu gene amplified from the pGE2 plasmid.
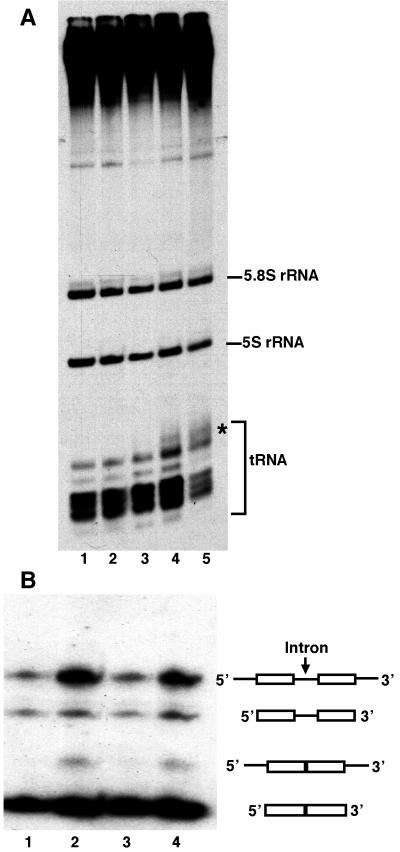
FIG. 2.
Comparison of in vivo tRNA synthesis in wild-type and mutant (Ntag-τ60) yeast strains. (A) Autoradiogram of the in vivo pulse-labeling experiments. [3H]uracil incorporation into wild-type and mutant strains was done for 15 min at 30°C (lane 1, wild-type; lane 2, mutant) or at 37°C after 6 h (lane 4, mutant) or 12 h (lane 3, wild-type; lane 5, mutant) of growth at 37°C. RNA species were analyzed by electrophoresis on a 7 M urea gel (6% polyacrylamide) by using equal amounts of RNA (6 μg per lane). The asterisk indicates the trailing of radioactive material above the tRNA species, suggesting a defect in tRNA maturation. (B) Northern blot analysis of tRNA precursors from wild-type and mutant cells with a labeled tDNA3Leu probe. RNA extraction was done after 8 h of growth at 37°C. Gel electrophoresis, electrophoretic transfer on nylon membrane, and hybridization with DNA probe was done as described in Materials and Methods. The positions of the primary transcript of tRNA3Leu, the 5′- and 3′-processed but nonspliced precursor, the spliced but 5′- and 3′-unprocessed precursor, and the mature tRNA3Leu are indicated on the right. Lanes 1 and 2, wild-type and mutant cells, respectively, containing the high-copy-number plasmid pFL44L; lane 3, mutant cells containing the high-copy-number plasmid pFL44L harboring RPR1 gene; lane 4, mutant cells containing pFL44L harboring SPT15 gene (TBP).
RESULTS
TFC8 encodes τ60, the 60-kDa subunit of yeast TFIIIC.
TFIIIC from S. cerevisiae comprises six polypeptides of 138, 131, 95, 91, 60, and 55 kDa. In order to clone the gene encoding the hypothetical τ60 subunit, the polypeptide was purified by SDS-PAGE and subjected to tryptic digestion. The sequence of only one peptide could be determined (??TLYLTT[T/F]PT). When compared to DNA sequences in databases by using the BLAST program, this peptide sequence was found with a slight variation (DGTLYLTTFPD) in a unique yeast hypothetical protein of 588 residues, with a theoretical molecular weight of 67,640 and an isoelectric point of 5.87. The gene encoding this protein is unique, maps on chromosome XVI (13), and shows no similarity to any sequences in the EMBL/GenBank data bank (S. cerevisiae, Caenorhabditis elegans, and current versions of Arabidopsis thaliana, Homo sapiens, and Schizosaccharomyces pombe genomic sequences). The gene, previously designated YPL007C, was named TFC8.
To investigate whether TFC8 is essential for growth, one chromosomal copy of the TFC8 gene was deleted in the diploid strain YNN281×YNN282 by replacing the ORF by the yeast HIS3 selectable marker. Sporulation and tetrad analysis revealed two viable His− spores and two nonviable spores, suggesting that TFC8 was an essential gene. To confirm this conclusion, the diploid His+ strain (which contains one copy of the TFC8 gene) was transformed with a 2μ plasmid, pCC12, harboring the TFC8 gene, and sporulated. Only the resulting His+ haploid strains containing the pCC12 plasmid with the TFC8 gene were able to grow. These results demonstrated that, like all of the previously characterized genes encoding the other TFIIIC subunits (1, 36, 40, 42, 48, 59), TFC8 is required for cell viability.
To demonstrate the presence of the TFC8 gene product in TFIIIC factor, the sequence encoding the HA epitope was fused to the 5′ or to the 3′ end of TFC8. A haploid strain which lacked the chromosomal TFC8 gene but expressed the HA C-terminally tagged τ60 (Ctag-τ60) grew normally. When the sequence encoding the HA epitope was fused just after the initiation codon, the growth of the haploid strain expressing the N-terminally tagged τ60 (Ntag-τ60) was slightly affected at 30°C (the mutant cells having a doubling time of 130 min in liquid medium, instead of 110 min for the wild-type strain). Furthermore, the Ntag-τ60 strain was temperature sensitive. At 37°C, the cells grew with an apparent doubling time of 190 min for approximately 16 to 18 h before cell death occurred.
TFIIIC factor was purified from these strains, as well as from a strain expressing an HA-tagged version of τ138 (36). Preformed TFIIIC-tDNA3Glu complexes were incubated for 30 min at 25°C with increasing concentrations of anti-HA monoclonal antibody and analyzed by gel retardation experiments (Fig. 1A). The anti-HA antibody clearly altered the migration rate of the Ctag-τ60 TFIIIC-tDNA3Glu complexes (compare lanes 6 and 7) to the same extent as HA-τ138-containing complexes (lanes 11 and 12). On the other hand, the migration rate of Ntag-τ60 TFIIIC-tDNA3Glu complexes was not altered (lanes 9 and 10), even in the presence of 1 μg or more of anti-HA antibody (data not shown), as if the HA epitope was buried and inaccessible to the antibody. Note also that, in contrast to previous results with TFIIIC containing HA-tagged τ131 (42) or τ138 subunits (see lanes 11 and 12 or reference 36), the binding of the anti-HA antibody to the Ctag-τ60 subunit appeared to cause some dissociation of the TFIIIC-tDNA3Glu complexes.
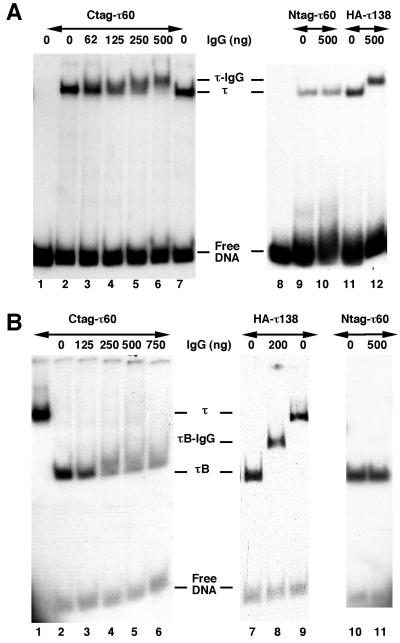
FIG. 1.
TFC8 encodes the 60-kDa subunit of yeast TFIIIC. (A) TFIIIC was purified from haploid strains expressing HA-tagged versions of TFC3 (HA-τ138) or TFC8 (Ctag-τ60 or Ntag-τ60, tagged at the C-terminal or N-terminal end of Tfc8p, respectively). TFIIIC was preincubated with a 32P-labeled DNA fragment containing the tRNA3Glu gene for 10 min at 25°C and then further incubated with various amounts of 12CA5 monoclonal antibody directed to the HA epitope for 30 min at 25°C. Protein-tDNA complexes were analyzed by gel retardation assay and revealed by autoradiography. Lanes 1 and 8, no TFIIIC, no antibody; lanes 2, 7, 9, or 11, no antibody; lanes 3 to 6, 10, and 12, addition of 12CA5 monoclonal antibody, as indicated. (B) τB, the protease-resistant domain of TFIIIC, HA-tagged on τ138 or τ60, was obtained after digestion of the TFIIIC-tDNA complex with 10 ng of α-chymotrypsin for 10 min at 25°C. Digestion was stopped with 1 ng of aprotinin before addition of the 12CA5 antibody. Left panel, C-terminally HA-tagged TFC8 gene (Ctag-τ60); right panel, HA-tagged TFC3 gene (HA-τ138) or N-terminally HA-tagged TFC8 gene (Ntag-τ60). Lanes 1 and 9, control TFIIIC-tDNA complex; lanes 2, 7, and 10, τB generated by proteolysis; lanes 3 to 6, 8, and 11, incubation of τB-tDNA complex with various amounts of 12CA5 monoclonal antibody, as indicated. τ, TFIIIC-tDNA complex; τ-IgG, immunoglobulin G (IgG)-TFIIIC-tDNA ternary complex; τB, τB-tDNA complex; τB-IgG, IgG-τB-tDNA ternary complex.
To gain some insight into the localization of τ60 within TFIIIC, we performed a limited proteolysis of TFIIIC-tDNA complexes by α-chymotrypsin. Limited proteolysis of yeast TFIIIC causes the separation of the transcription factor into two domains of ca. 300 kDa each, called τA and τB (44). The τB domain generated by proteolysis forms a stable complex with the B block that can be visualized in gel shift assays and supershifted by the addition of anti-τ138 polyclonal antibodies (19). A similar result was obtained in the present study as anti-HA antibodies neatly supershifted tagged-τ138 τB-tDNA complexes (Fig. 1B, lanes 7 and 8). When τB-tDNA complexes obtained after limited proteolysis of Ctag-τ60 TFIIIC-tDNA complexes were incubated with increasing amounts of anti-HA antibodies, no defined supershifted band of complex was observed, but the antibodies reduced the migration rate of τB-tDNA complexes, causing a marked trailing of the band (Fig. 1B, lanes 2 to 6). A weak accessibility or a partial proteolytic degradation of the HA epitope possibly caused some dissociation of the immune complex during electrophoresis. Such a phenomenon was not observed with Ntag-τ60 τB-tDNA complexes (lanes 10 and 11), nor with untagged τB-tDNA complexes (data not shown), whose yield and migration rate were unaffected by the monoclonal antibody. Altogether, these gel shift experiments indicated that the polypeptide encoded by TFC8 is part of TFIIIC and that at least its C-terminal end is located in the τB domain.
In vivo characterization of the Ntag-τ60 mutant.
We took advantage of the thermosensitive phenotype of the Ntag-τ60 mutant to explore the role of the TFC8 gene product in TFIIIC. The effect of the mutation on Pol III transcripts in vivo is shown in Fig. 2A. The wild-type and mutant cells were grown at 30°C and then shifted or not to nonpermissive temperature (37°C) for 6 or 12 h. The cells grown at 30 or 37°C were incubated at the same temperature with tritiated uracil for 15 min; the RNAs were then extracted, separated on a 7 M urea gel, and analyzed by autoradiography. No difference was observed in the RNA labeling pattern of the wild-type and mutant cells grown at 30°C (lanes 1 and 2). In contrast, when shifted for 6 or 12 h at 37°C, the mutation altered the labeling pattern of tRNAs. A smear of radioactivity was detected just above the labeled tRNA bands (lanes 4 and 5), which could correspond to partially matured tRNAs, and after 12 h the labeling of the tRNA bands was selectively reduced (lane 5). Clearly, the fusion of the HA epitope to τ60 at the N-terminal extremity disturbed the Pol III transcription system. Like in previous studies on different Pol III system mutants, the synthesis of 5S RNA did not decrease significantly (20, 41).
A similar tRNA maturation defect had previously been observed with thermosensitive mutants of other TFIIIC subunits, such as τ138 (37), τ131 (56) or, more recently, τ91 (1a). To explore a possible relationship between this maturation defect, TFIIIC function and the cell lethality phenotype, we performed a Northern blot analysis of various tRNA precursors. Total RNA was extracted from wild-type and mutant strains grown for 8 h at 37°C, separated by electrophoresis under denaturing conditions, transferred onto a membrane, and probed with various labeled tRNA genes (Fig. 2B). The wild-type and one mutant strain contained the multicopy plasmid pFL44L, whereas two other mutant strains contained pFL44L harboring either RPR1 or SPT15 (TBP). The precursor pattern of tRNA3Leu in the mutant was characterized by the presence of an extra RNA band absent in the wild-type extract (compare lanes 1 and 2). A similar result was obtained with labeled-SUP4tDNATyr and tDNA3Glu (data not shown). This additional RNA species has been previously observed in RNase P mutants and was shown to correspond to unmatured spliced tRNA (18, 35, 58). RNase P is an endonuclease that cleaves pre-tRNA substrates to yield a mature 5′ extremity. When the RNA component of RNaseP, RPR1, was overexpressed in the mutant strain, this anomalous RNA species disappeared (lane 3). However, the mutant cells overexpressing RPR1 retained the thermosensitive lethal phenotype (see Fig. 3). In contrast to RPR1, overexpression of TBP did not change the tRNA maturation pattern of the mutant (Fig. 2, lane 4) but suppressed the lethality (see below). A direct relationship between the maturation defect and the thermosensitive phenotype of the Ntag-τ60 mutant could therefore be excluded. The tRNA maturation problem was likely a consequence of the reduced synthesis of the class III RPR1 RNA, along with other class III transcripts (34).
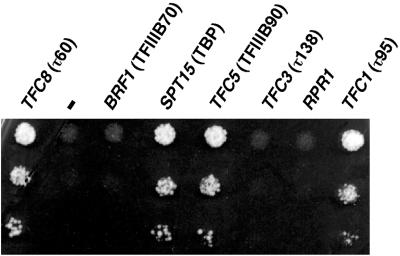
FIG. 3.
Suppressibility of Ntag-τ60 mutation by overexpression of Pol III-related genes. Stationary-phase cultures of mutant cells harboring different multicopy plasmids were diluted 10-, 102-, and 103-fold in water and spotted (5 μl) onto solid minimum medium (Casa+Ade plates). The plates were incubated for 4 days at 37°C. The different genes harbored on the high-copy-number vector pFL44L are indicated. −, plasmid without insert. RPR1 encodes the RNA cofactor of RNase P.
We looked for multicopy suppressors that could compensate for interaction defects within TFIIIC or the preinitiation complex. The Ntag-τ60 mutant cells were transformed with high-copy-number plasmids harboring different genes of the Pol III transcription system. A series of cell dilutions was plated on minimal medium and incubated at permissive or nonpermissive temperature. Among the TFIIIC genes, besides TFC8, only TFC1 (encoding τ95) acted as a multicopy suppressor (Fig. 3 and Table 1). None of the suppressors of tsv115 mutation in TFC3, such as RPR1, NOP1, and FHL1 (37), involved in the maturation of stable RNAs, were able to suppress the thermosensitive defect of the mutant cells (Table 1). Figure 3 shows that high dosage of two TFIIIB genes, TFC5 (TFIIIB90), and SPT15 (TBP), but not BRF1 (TFIIIB70), suppressed the lethal phenotype of Ntag-τ60 mutant cells on minimal medium at 37°C. Remarkably, only the overproduction of SPT15 (TBP) could restore cell growth on a rich YPD medium at the nonpermissive temperature (the suppression level was weaker under these conditions than the one observed on minimal medium, see Table 1).
TABLE 1.
Suppression of τ60 mutation by gene dosage
| Multicopy gene | Gene product | Suppression on:
|
|
|---|---|---|---|
| Minimal medium | Rich medium | ||
| TFC8 | τ60 | +++ | +++ |
| RPR1 | RNA component of RNase P (tRNA maturation) | − | − |
| NOP1 | Fibrillarin homolog (rRNA maturation) | − | − |
| FHL1 | Forkhead-like protein (rRNA maturation) | − | − |
| TFC3 | τ138, subunit of TFIIIC | − | − |
| TFC4 | τ131, subunit of TFIIIC | − | − |
| TFC1 | τ95, subunit of TFIIIC | ++ | − |
| TFC6 | τ91, subunit of TFIIIC | − | − |
| TFC7 | τ55, subunit of TFIIIC | − | − |
| BRF1 | Brf1/TFIIIB70, subunit of TFIIIB | − | − |
| SPT15 | TBP (TATA-binding protein), subunit of TFIIIB | ++ | + |
| TFC5 | B"/TFIIIB90, subunit of TFIIIB | ++ | − |
In vitro characterization of the Ntag-τ60 mutant.
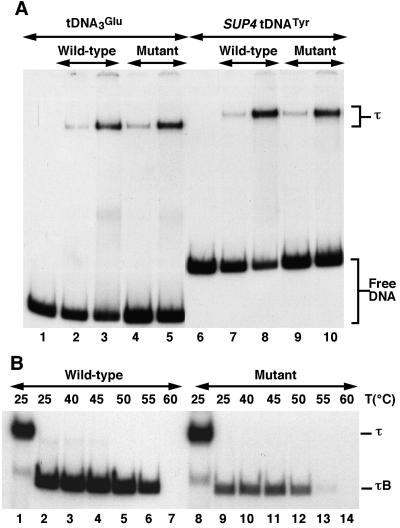
TFIIIC factor was partially purified from a wild-type strain and from the strain expressing the HA N-terminal version of τ60 to compare their DNA binding properties. TFIIIC fractions purified from the mutant strain were reproducibly found by Western blotting with anti-τ60 and anti-τ55 polyclonal antibodies (40) to contain half as much TFIIIC as wild-type fractions. Therefore, care was taken to use the same amount of wild-type or mutant TFIIIC in the following gel retardation experiments. Two labeled probes, tDNA3Glu or SUP4tDNATyr, were incubated with limiting amounts of wild-type or mutant TFIIIC. TFIIIC-tDNA complexes were analyzed by electrophoresis under nondenaturing conditions and revealed by autoradiography. Figure 4A shows that the yields of TFIIIC-DNA complexes formed with wild-type or mutant TFIIIC factor on tDNA3Glu or SUP4tDNATyr were very similar. In addition, wild-type and mutant TFIIIC-tDNA complex stabilities at various temperatures or ionic strengths were also indistinguishable (data not shown). In contrast, when TFIIIC-tDNA3Glu complexes were subjected to limited proteolysis, significantly fewer mutant τB-tDNA complexes were obtained compared to the wild type (Fig. 4B, compare lanes 2 and 9). Moreover, mutant τB-tDNA complexes were less stable than wild type at increasing temperatures. The amount of mutant τB-tDNA complexes decreased drastically after 10 min at 55°C to 5% of the initial control level at 25°C, while 60% of wild-type τB-tDNA complexes resisted under the same conditions (lanes 6 and 13), as determined by using a PhosphorImager with ImageQuant software (Molecular Dynamics). Whatever the structural defect causing the increased thermolability (either the presence of the HA epitope or some subtle difference in proteolysis indirectly caused by the presence of the epitope), these observations clearly revealed a defect at the τB level, enforcing the idea that the N-terminal part of τ60 belongs to the B block-binding domain.
FIG. 4.
DNA-binding properties of wild-type and mutant TFIIIC. (A) TFIIIC-tDNA complex formation on tRNA3Glu or SUP4tRNATyr genes. TFIIIC-DNA complexes were formed with Ntag-τ60 or wild-type TFIIIC preparations (MonoQ fraction) and analyzed by electrophoresis as described in Materials and Methods. To add similar amounts of wild-type and mutant TFIIIC (based on Western blot experiments), twice as much mutant protein fraction was added as follows: 25 ng (lanes 2 and 7) and 100 ng (lanes 3 and 8) of wild-type TFIIIC and 50 ng (lanes 4 and 9) and 200 ng (lanes 5 and 10) of mutant TFIIIC fraction. Lanes 1 to 5, tRNA3Glu gene; lanes 6 to 10, SUP4tRNATyr gene. (B) Effect of temperature on τB-tDNA complex stability. Preformed TFIIIC-tDNA3Glu complexes were digested with α-chymotrypsin to generate τB-DNA complexes, proteolytic digestion was stopped by addition of aprotinin, and the mixtures were then further incubated for 10 min at various temperatures as indicated. Lanes 1 and 8, wild-type and mutant TFIIIC-tDNA complexes, respectively; lanes 2 to 7, wild-type τB; lanes 9 to 14, mutant τB. τ, TFIIIC-tDNA complex; τB, τB-tDNA complex.
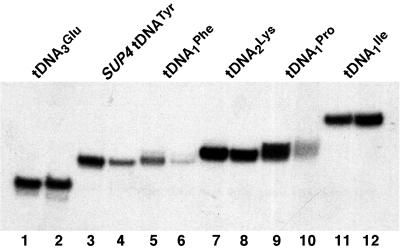
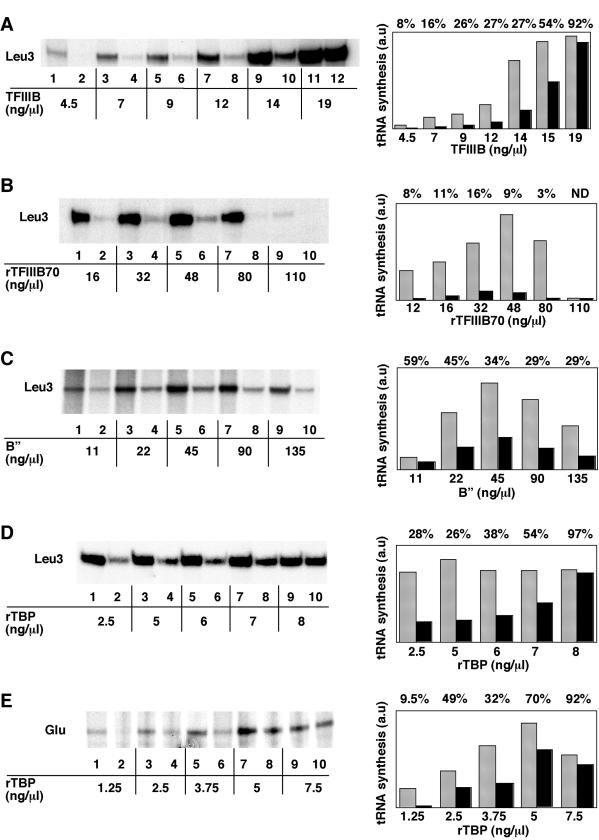
Wild-type and mutant TFIIIC were then compared for their ability to direct specific transcription of various tRNA genes in vitro. Identical amounts of both TFIIIC preparations, calibrated by Western blot analysis with anti-τ60 and anti-τ55 polyclonal antibodies and by gel retardation analysis with tDNA3Glu or SUP4tDNATyr probes, were used in reconstituted transcription assays in the presence of B" fraction, recombinant TBP (rTBP), TFIIIB70 (rTFIIIB70), and purified RNA Pol III. As shown in Fig. 5, the transcriptional activity of mutant TFIIIC was similar to that of the wild-type factor with tDNA3Glu, tDNA2Lys, or tDNA1Ile as templates (lanes 1 and 2, 7 and 8, and 11 and 12). Surprisingly, the transcriptional activity of the mutant factor was much impaired with other templates, such as SUP4tDNATyr, tDNA1Phe, or tDNA1Pro (lanes 3 to 6 and 9 and 10). However, as shown previously, the mutant and wild-type factor preparations bound similarly to tDNA3Glu or SUP4tDNATyr probes (see above). Since a difference in DNA-binding properties could not account for such a difference in transcription efficiency, we explored the ability of mutant TFIIIC to recruit TFIIIB. We used tDNA3Leu, which was, like SUP4tDNATyr, a poor template, with the mutant TFIIIC. Wild-type or mutant TFIIIC had also similar DNA-binding properties on tDNA3Leu (data not shown). Transcription was performed in the presence of increasing amounts of TFIIIB with the same amount of wild-type or mutant TFIIIC. Remarkably, as shown in Fig. 6A, high doses of TFIIIB fraction could compensate for the transcription defect of mutant TFIIIC. The relative transcriptional activity of mutant TFIIIC reached 92% of the wild-type level at 19 ng of TFIIIB per μl, a concentration at which TFIIIB was no longer limiting with wild-type TFIIIC (the maximum of tRNA synthesis in the presence of wild-type TFIIIC was reached at ca. 15 ng of TFIIIB per μl).
FIG. 5.
Specific transcription of various tRNA genes in the presence of wild-type or mutant TFIIIC. In vitro transcription was performed as described in Materials and Methods by using 100 and 200 ng of wild-type and Ntag-τ60 MonoQ-TFIIIC fraction (respectively), rTBP, rTFIIIB70, partially purified B" fraction, and RNA Pol III. The different plasmid templates are indicated. Odd lanes, wild-type TFIIIC. Even lanes, mutant TFIIIC.
FIG. 6.
Effects of TFIIIB, TFIIIB70, B", or TBP concentration on the transcription of tRNA3Leu or tRNA3Glu genes in the presence of wild-type or mutant TFIIIC. (A) Effect of TFIIIB. Transcription mixtures (40 μl) contained 100 ng of wild-type or 200 ng of Ntag-τ60 TFIIIC, 220 ng of plasmid DNA harboring the tRNA3Leu gene, RNA polymerase, and various concentrations of TFIIIB (Cibacron blue fraction) as indicated. (B) Effect of TFIIIB70. Transcription mixtures contained wild-type or mutant TFIIIC, plasmid DNA, and RNA polymerase as in panel A, 2.5 ng of rTBP per μl, 45 ng of partially purified B" fraction per μl, and various concentrations of rTFIIIB70 as indicated. (C) Effect of B". Transcription mixtures contained wild-type or mutant TFIIIC, plasmid DNA, and RNA polymerase as in panel A, 4 ng of rTBP per μl, 35 ng of rTFIIIB70 per μl, and various concentrations of partially purified B" fraction as indicated. (D) Effect of TBP. Lanes were as in other panels, with 35 ng of rTFIIIB70 per μl, 45 ng of partially purified B" fraction per μl, and various concentrations of rTBP as indicated. (E) Effect of TBP. Lanes are as described for panel D, except that plasmid DNA harboring the tRNA3Glu gene instead of the tRNA3Leu gene was used as the template. Transcripts were analyzed by electrophoresis and autoradiography (left panels). Odd lanes, wild-type TFIIIC; even lanes, mutant TFIIIC. Transcripts were quantified (right panels) by using a PhosphorImager and ImageQuant software (Molecular Dynamics). a.u., arbitrary units. Gray bars, wild-type TFIIIC; black bars, mutant TFIIIC. The relative transcription efficiencies of the mutant versus the wild-type TFIIIC are indicated above the bar graphs. ND, not determined. Two or three independent experiments were compiled for quantification.
To explore which TFIIIB component was critical for TFIIIB assembly by mutant TFIIIC, we performed multiple-round transcriptions with tDNA3Leu as template with various amounts of rTFIIIB70 (Fig. 6B), B" (Fig. 6C), or rTBP (Fig. 6D). Increasing the concentration of rTFIIIB70 did not correct the transcription defect of mutant TFIIIC (Fig. 6B). In a 12- to 48-ng/μl concentration range of rTFIIIB70 factor, tRNA synthesis increased similarly for both wild-type and mutant TFIIIC, and the relative transcriptional efficiency of mutant TFIIIC remained low and constant (8 to 16%). At a concentration of more than 48 ng/μl, rTFIIIB70 appeared to titrate a component of the transcription system, causing a dramatic decrease of tRNA synthesis directed by both wild-type and mutant TFIIIC. As shown in Fig. 6C, increasing the concentration of B" in a 11- to 45-ng/μl concentration range also did not correct the transcription of B" in a 11- to 45-ng/μl concentration range also did not correct the transcription defect of mutant TFIIIC. At a concentration of B" of >45 ng/μl, the transcription efficiency of both wild-type and mutant TFIIIC decreased similarly (Fig. 6C). The dosage-dependent effect of rTBP on transcriptional efficiency was totally different (Fig. 6D). In a concentration range of 2.5 to 8 ng of rTBP factor per μl, the transcriptional efficiency was optimal for wild-type TFIIIC. In contrast, with mutant TFIIIC, the tRNA synthesis rate increased with rTBP concentration to reach 97% of the wild-type level. This result indicated that rTBP was the limiting component for TFIIIB assembly directed by mutant TFIIIC, at least on tDNA3Leu.
The observation that several tRNA genes were normally transcribed in the presence of mutant TFIIIC (Fig. 5) suggested that these templates might have different TBP requirements. Indeed, we found that decreasing the TBP concentration revealed a defect of mutant TFIIIC in the transcription of tDNA3Glu template (Fig. 6E). Therefore, we inferred that the TBP concentration of our standard transcription assay was optimal for tDNA3Glu, tDNA2Lys, and tDNA1Ile but limiting with SUP4tDNATyr, tDNA1Phe, tDNA1Pro, and tDNA3Leu. The analysis of the promoter region of these genes did not disclose a feature within the TFIIIB binding region that might account for such a differential TBP requirement.
τ60 interacts with TBP.
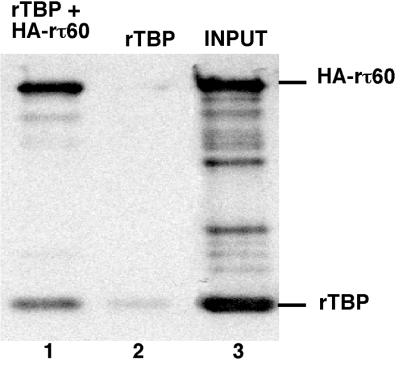
Two lines of evidence indicated that mutant TFIIIC was defective in TBP recruitment, both in vivo (the multicopy suppressor studies) and in vitro (the transcription experiments). We therefore explored the possibility that τ60 subunit might directly participate in TBP binding. The physical interaction between τ60 and TBP was investigated by coimmunoprecipitation experiments with recombinant proteins. rTBP (200 ng) was preincubated for 90 min at 25°C with HA-rτ60 (650 ng) in transcription buffer. The protein samples were then added to magnetic beads precoated with anti-HA antibodies, the beads were washed with the same buffer, and bound proteins were eluted with SDS or by competition with the HA peptide. The input and eluted proteins were subsequently analyzed by SDS-PAGE and immunoblotting with anti-TBP and anti-τ60 antibodies (Fig. 7). Compared to the low background level of TBP retained by the beads in the absence of τ60, a sevenfold-higher amount of TBP was bound upon preincubation with τ60, as quantified by using the ImageQuant software. A similar result was obtained when bound proteins were eluted by competition with the HA peptide or when the beads were washed extensively with the transcription buffer (data not shown). The background-corrected TBP/Tfc8 ratio in the eluted fraction has been quantified and corresponded to 0.56 (compared to 0.9 in the input), showing that each molecule of bound τ60 was specifically associated to 0.5 molecule of TBP. From that result, we tentatively estimate the Kd of TBP-τ60 interaction to be ca. 100 nM, which is indicative of a reasonably strong interaction. The same Kd was reported for TFIIIB70-TBP interaction by Sethy-Coraci et al. (55). These observations strongly suggested a direct interaction between the τ60 subunit and TBP. In a similar experiment, no physical interaction could be detected between rτ60 and rTFIIIB70: Western blot analysis revealed no significant difference in the amounts of rTFIIIB70 bound to the beads in the presence or absence of HA-rτ60 (data not shown).
FIG. 7.
Coimmunoprecipitation experiment. HA-tagged rτ60 (650 ng) was preincubated with rTBP (200 ng) for 1.5 h at 25°C and purified on magnetic beads coated with anti-HA antibodies. Bound proteins were eluted and analyzed by Western blotting with anti-τ60 and anti-TBP polyclonal antibodies. The positions of rTBP and of native HA-rτ60 are shown. The intermediate bands are specifically revealed by anti-τ60 antibodies. Lane 1, rTBP+HA-rτ60; lane 2, rTBP alone; lane 3, 25% of input rTBP+HA-rτ60 proteins.
DISCUSSION
We report here the characterization of the 60-kDa polypeptide present in affinity-purified TFIIIC. This polypeptide, named τ60, is shown to be an essential subunit of TFIIIC. The yeast TFIIIC factor, therefore, comprises six subunits, τ138, τ131, τ95, τ91, τ60, and τ55, all of which are essential for cell viability (Table 2). τ60 appears to reside, at least in part, in τB, the downstream DNA-binding domain. Paradoxically, however, analysis of a τ60 mutant indicated that τ60 plays a role in TFIIIB assembly via its interaction with TBP. The results suggest a network of interactions linking TFIIIC to TFIIIB components during preinitiation complex formation.
TABLE 2.
Subunits of yeast TFIIIC
| Subunit | Calculated mass (kDa) | Gene | Null phenotype | Location and proposed role in TFIIIC-DNA complex | Features | Source(s) or reference |
|---|---|---|---|---|---|---|
| τ138 | 132 | TFC3 | Lethal | In τB; B block binding | Two HMG domains | |
| τ131 | 120 | TFC4 | Lethal | Most 5′; TFIIIB assembly; interacts with TFIIIB70 and TFIIIB90 | 11 TPR motifs; phosphorylated; ortholog in H. sapiens, C. elegans, and K. lactis | 23, 41a |
| τ95 | 74 | TFC1 | Lethal | In τA; A block binding; interacts with τ55 | Ortholog in H. sapiens, D.m., C. elegans and S. pombe | 23, 27a |
| τ91 | 75 | TFC6 | Lethal | Most 3′; cooperates with τ138 in DNA binding | (Cys+His)-rich domain; ortholog in H. sapiens and S. pombe | 1a |
| τ60 | 68 | TFC8 | Lethal | In τB; TFIIIB assembly; interacts with TBP | ||
| τ55 | 49 | TFC7 | Lethal | In τA; interacts with τ95 | Chimeric protein |
The demonstration that τ60 is an integral subunit of TFIIIC rests on biochemical and genetic evidence. First, antibodies directed to C-terminally HA-tagged τ60 altered the migration of tagged TFIIIC-tDNA and τB-tDNA complexes. Second, a thermosensitive mutant TFIIIC (harboring N-terminally HA-tagged τ60) was affected in tRNA gene transcription in vivo and in vitro. Third, this in vivo phenotype was suppressed by overexpression of components of the class III transcription machinery (a TFIIIC subunit, τ95, and two components of the TFIIIB factor, TFIIIB90/B" and TBP). Fourth, the in vitro transcription defect was strongly alleviated by increasing the concentration of TFIIIB or TBP. Finally, like all of the subunits of TFIIIC previously characterized, the TFC8 gene encoding τ60 is essential for cell viability. Altogether, these results indicate that τ60 plays an important role in TFIIIC.
The location of TFIIIC subunits by site-specific DNA cross-linking has provided a broad view of the possible functions of the different polypeptides within the TFIIIC-DNA complex (Table 2), which are always in good agreement with other biochemical or genetic data (28). Unfortunately, τ60 was the only TFIIIC polypeptide that could not be cross-linked and located by this elegant and powerful approach (its subunit status could have been questioned for that reason). Two independent experiments suggested that τ60 resides within τB domain: τB-DNA complexes obtained by proteolysis of TFIIIC harboring HA-tagged τ60 were found to be more thermosensitive than untagged complexes or were recognized by antibodies directed to the HA epitope. Since, in these experiments, the HA tag was placed either at the N-terminal or at the C-terminal end of the polypeptide, we inferred that both extremities of τ60 resided in τB, together with τ138 (19) and τ91 (1). The total mass of these three polypeptides, 275 kDa, would correspond reasonably well to the mass of τB (∼300 kDa) visualized by scanning transmission electron microscopy over the B block (53). Note that the definition of τB by electron microscopy and by limited proteolysis are not necessarily equivalent. The protease-resistant form of τB observed by gel retardation assay may contain pieces of TFIIIC unrelated to the B-block-binding function. The participation of τ60 to τB (as defined by limited proteolysis and gel retardation assay) points to a role of this subunit in DNA binding. Indeed, the presence of the HA epitope at the N-terminal end of τ60 decreased the yield and weakened the stability of τB-DNA complex. However, mutant TFIIIC-DNA complex formation was not notably affected compared to the wild-type factor, even under unfavorable conditions, at high ionic strengths, or at high temperatures. Therefore, a role of τ60 in DNA binding is not excluded but is not strongly supported by the present data, in keeping with the lack of detectable τ60-DNA cross-linking (28). Schematic models of TFIIIC-DNA complexes based on site-specific cross-linking studies tentatively visualized the 60-kDa polypeptide as being unbound to DNA, connecting the τ95 and τ138 subunits, and overlapping the A block-B block interval (10). This proposal fits remarkably well with our present observations, since τ60 appears to reside at least partly in τB and possibly also contact τ95, as suggested by the in vivo suppression of τ60 mutant lethality by overexpression of τ95, although two-hybrid experiments did not confirm this interaction (data not shown).
In view of the position of τ60 in the TFIIIC-DNA complex, the effect of a τ60 mutation at the level of TFIIIB recruitment was rather unexpected. Since τ131 was the only subunit of TFIIIC extending upstream of the transcriptional start site (7), it has been assumed to be entirely responsible for TFIIIB assembly. Indeed, τ131 and no other TFIIIC subunit was found to interact with TFIIIB70/Brf1 and TFIIIB90/B" (14, 33, 51). Several lines of evidence, however, indicate that τ60 most likely also participates in TFIIIB recruitment: (i) the lethality of the Ntag-τ60 variant at a nonpermissive temperature was suppressed by overexpression of two TFIIIB components, TFIIIB90/B" and TBP (not by TFIIIB70 though); (ii) the in vitro transcription defect of the mutant TFIIIC was strongly alleviated by increasing TFIIIB concentration and more specifically by increasing TBP concentration (again not TFIIIB70 or B"); and (iii) τ60 was found to interact directly with TBP. The possibility of an indirect role of τ60 in TFIIIB recruitment could be envisioned in view of its possible interaction with τ95 that belongs to the τA domain. The τ60 mutation could perturb τ95-A block binding or affect indirectly, via τ95, TFIIIB assembly by τ131. If this were the case, one would expect a suppression of the in vivo or in vitro defects by increased TFIIIB70 dosage. Indeed, mutations in TFIIIC (37), in TBP (12, 16), or in promoter elements decreasing factor-DNA interaction (39) were always found to be best suppressed in vivo by overexpression of TFIIIB70. This makes sense since TFIIIB70 interacts with TFIIIC (14, 33), initiates TFIIIB assembly (31), and appears to be limiting for transcription both in whole-cell extracts and in vivo (39, 54, 55). Since increasing the TFIIIB70 concentration did not correct the TFIIIC mutant phenotype in vivo and in vitro, the rate-limiting step caused by the τ60 mutation was expected to follow TFIIIB70 recruitment step, i.e., the TBP recruitment step (31). Since it appears unlikely that the τ60 mutation would have an indirect effect on the recruitment of TBP by TFIIIB70, we rather favor the idea that τ60 is directly involved in TFIIIB recruitment by contacting TBP. The coimmunoprecipitation of TBP and τ60 comes in support of this proposal. The interaction of TBP with TFIIIC subunits has not been previously reported. However, Huet et al. (26) have noted that TBP slightly retarded the migration of TFIIIC-DNA complexes on a TATA-less gene. Since TFIIIC specifies the orientation of TFIIIB on the DNA (and thereby the direction of transcription) (60), the direct interaction of TFIIIC with TBP, via τ60, might well contribute to fix its correct orientation on the DNA. Alternatively, τ60-TBP interaction may not be involved in positioning TBP on the DNA but only represent a transient step facilitating the delivery of TBP to the TFIIIB70-TFIIIC-DNA complex.
ACKNOWLEDGMENTS
We are grateful to Christophe Carles, Michel Riva, Olivier Lefebvre, and Françoise Bouet for their help in peptide microsequence determination and to Jacques Grassi and Christophe Creminon for their help in raising polyclonal antibodies. We thank Emmanuel Favry for protein preparations and Olivier Lefebvre for advice on TFIIIC purification and immunoprecipitation experiments. We also thank Christian Marck and Hélène Dumay for providing plasmids harboring different tRNA genes and for helpful discussions and Olivier Lefebvre, Michel Riva, and Michel Werner for improving the manuscript.
E.D. was supported by postdoctoral fellowships from the Association pour la Recherche contre le Cancer, the Ligue Nationale contre le Cancer, and the Fondation pour la Recherche Médicale, and R.A. was supported by a postdoctoral fellowship from the European Union (Marie Curie Training and Mobility of Researchers).
REFERENCES
- 1.Arrebola R, Manaud N, Rozenfeld S, Marsolier M C, Lefebvre O, Carles C, Thuriaux P, Conesa C, Sentenac A. τ91, an essential subunit of yeast TFIIIC, cooperates with τ138 in DNA binding. Mol Cell Biol. 1998;18:1–9. doi: 10.1128/mcb.18.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1a.Arrebola, R. Unpublished data.
- 2.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1994. [Google Scholar]
- 3.Baker R E, Camier S, Sentenac A, Hall B D. Gene size differentially affects the binding of yeast transcription factor τ to two intragenic regions. Proc Natl Acad Sci USA. 1987;84:8768–8772. doi: 10.1073/pnas.84.24.8768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baker R E, Gabrielsen O S, Hall B D. Effects of tRNATyr point mutations on the binding of yeast RNA polymerase III transcription factor C. J Biol Chem. 1986;261:5275–5282. [PubMed] [Google Scholar]
- 5.Bartholomew B, Braun B R, Kassavetis G A, Geiduschek E P. Probing close DNA contacts of RNA polymerase III transcription complexes with the photoactive nucleoside 4-thiodeoxythymidine. J Biol Chem. 1994;269:18090–18095. [PubMed] [Google Scholar]
- 6.Bartholomew B, Kassavetis G A, Braun B R, Geiduschek E P. The subunit structure of Saccharomyces cerevisiae transcription factor IIIC probed with a novel photocrosslinking reagent. EMBO J. 1990;9:2197–2205. doi: 10.1002/j.1460-2075.1990.tb07389.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bartholomew B, Kassavetis G A, Geiduschek E P. Two components of Saccharomyces cerevisiae transcription factor IIIB (TFIIIB) are stereospecifically located upstream of a tRNA gene and interact with the second-largest subunit of TFIIIC. Mol Cell Biol. 1991;11:5181–5189. doi: 10.1128/mcb.11.10.5181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baudin A, Ozier-Kalogeropoulos O, Denouel A, Lacroute F, Cullin C. A simple and efficient method for direct gene deletion in Saccharomyces cerevisiae. Nucleic Acids Res. 1993;21:3329–3330. doi: 10.1093/nar/21.14.3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bonneaud N, Ozier-Kalogeropoulos O, Li G, Labouesse M, Minvielle-Sebastia L, Lacroute F. A family of low and high copy replicative, integrative and single-stranded S. cerevisiae/E. coli shuttle vectors. Yeast. 1991;7:609–615. doi: 10.1002/yea.320070609. [DOI] [PubMed] [Google Scholar]
- 10.Braun B R, Bartholomew B, Kassavetis G A, Geiduschek E P. Topography of transcription factor complexes on the Saccharomyces cerevisiae 5 S RNA gene. J Mol Biol. 1992;228:1063–1077. doi: 10.1016/0022-2836(92)90315-b. [DOI] [PubMed] [Google Scholar]
- 11.Braun B R, Kassavetis G A, Geiduschek E P. Bending of the Saccharomyces cerevisiae 5S rRNA gene in transcription factor complexes. J Biol Chem. 1992;267:22562–22569. [PubMed] [Google Scholar]
- 12.Buratowski S, Zhou H. A suppressor of TBP mutations encodes an RNA polymerase III transcription factor with homology to TFIIB. Cell. 1992;71:221–230. doi: 10.1016/0092-8674(92)90351-c. [DOI] [PubMed] [Google Scholar]
- 13.Bussey H, Storms R K, Ahmed A, Albermann K, Allen E, Ansorge W, Araujo R, Aparicio A, Barrell B, Badcock K, Benes V, Botstein D, Bowman S, Bruckner M, Carpenter J, Cherry J M, Chung E, Churcher C, Coster F, Davis K, Davis R W, Dietrich F S, Delius H, DiPaolo T, Hani J, et al. The nucleotide sequence of Saccharomyces cerevisiae chromosome XVI. Nature. 1997;387:103–105. [PubMed] [Google Scholar]
- 14.Chaussivert N, Conesa C, Shaaban S, Sentenac A. Complex interactions between yeast TFIIIB and TFIIIC. J Biol Chem. 1995;270:15353–15358. doi: 10.1074/jbc.270.25.15353. [DOI] [PubMed] [Google Scholar]
- 15.Church G M, Gilbert W. Genomic sequencing. Proc Natl Acad Sci USA. 1984;81:1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Colbert T, Hahn S. A yeast TFIIB-related factor involved in RNA polymerase III transcription. Genes Dev. 1992;6:1940–1949. doi: 10.1101/gad.6.10.1940. [DOI] [PubMed] [Google Scholar]
- 17.Conesa C, Swanson R N, Schultz P, Oudet P, Sentenac A. On the subunit composition, stoichiometry, and phosphorylation of the yeast transcription factor TFIIIC/τ. J Biol Chem. 1993;268:18047–18052. [PubMed] [Google Scholar]
- 18.Dichtl B, Tollervey D. Pop3p is essential for the activity of the RNase MRP and RNase P ribonucleoproteins in vivo. EMBO J. 1997;16:417–429. doi: 10.1093/emboj/16.2.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gabrielsen O S, Marzouki N, Ruet A, Sentenac A, Fromageot P. Two polypeptide chains in yeast transcription factor τ interact with DNA. J Biol Chem. 1989;264:7505–7511. [PubMed] [Google Scholar]
- 20.Gudenus R, Mariotte S, Moenne A, Ruet A, Mémet S, Buhler J-M, Sentenac A, Thuriaux P. Conditional mutants of RPC160, the gene encoding the largest subunit of RNA polymerase C in Saccharomyces cerevisiae. Genetics. 1988;119:517–526. doi: 10.1093/genetics/119.3.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heinemeyer W, Gruhler A, Mohrle V, Mahe Y, Wolf D H. PRE2, highly homologous to the human major histocompatibility complex-linked RING10 gene, codes for a yeast proteasome subunit necessary for chymotryptic activity and degradation of ubiquinated proteins. J Biol Chem. 1993;268:5115–5120. [PubMed] [Google Scholar]
- 22.Hermann-Le Denmat S, Werner M, Sentenac A, Thuriaux P. Suppression of yeast RNA polymerase III mutations by FHL1, a gene coding for a fork head protein involved in rRNA processing. Mol Cell Biol. 1994;14:2905–2913. doi: 10.1128/mcb.14.5.2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hsieh Y-J, Wang Z, Kovelman R, Roeder R G. Cloning and characterization of two evolutionarily conserved subunits (TFIIIC102 and TFIIIC63) of human TFIIIC and their involvement in functional interactions with TFIIIB and RNA polymerase III. Mol Cell Biol. 1999;19:4944–4952. doi: 10.1128/mcb.19.7.4944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huet J, Conesa C, Carles C, Sentenac A. A cryptic DNA binding domain at the COOH terminus of TFIIIB70 affects formation, stability, and function of preinitiation complexes. J Biol Chem. 1997;272:18341–18349. doi: 10.1074/jbc.272.29.18341. [DOI] [PubMed] [Google Scholar]
- 25.Huet J, Manaud N, Dieci G, Peyroche G, Conesa C, Lefebvre O, Ruet A, Riva M, Sentenac A. RNA polymerase III and class III transcription factors from Saccharomyces cerevisiae. Methods Enzymol. 1996;273:249–267. doi: 10.1016/s0076-6879(96)73024-0. [DOI] [PubMed] [Google Scholar]
- 26.Huet J, Sentenac A. The TATA-binding protein participates in TFIIIB assembly on tRNA genes. Nucleic Acids Res. 1992;20:6451–6454. doi: 10.1093/nar/20.24.6451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Joazeiro C A P, Kassavetis G A, Geiduschek E P. Identical components of yeast transcription factor IIIB are required and sufficient for transcription of TATA box-containing and TATA-less genes. Mol Cell Biol. 1994;14:2798–2808. doi: 10.1128/mcb.14.4.2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27a.Jourdain, S. Personal communication.
- 28.Kassavetis G A, Bardeleben C, Bartholomew B, Braun B R, Joazeiro C A P, Pisano M, Geiduschek E P. Transcription by RNA polymerase III. In: Conaway R C, Conaway J W, editors. Transcription: mechanisms and regulation. New York, N.Y: Raven Press, Ltd.; 1994. pp. 107–126. [Google Scholar]
- 29.Kassavetis G A, Bartholomew B, Blanco J A, Johnson T E, Geiduschek E P. Two essential components of the Saccharomyces cerevisiae transcription factor TFIIIB: transcription and DNA-binding properties. Proc Natl Acad Sci USA. 1991;88:7308–7312. doi: 10.1073/pnas.88.16.7308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kassavetis G A, Braun B R, Nguyen L H, Geiduschek E P. S. cerevisiae TFIIIB is the transcription initiation factor proper of RNA polymerase III, while TFIIIA and TFIIIC are assembly factors. Cell. 1990;60:235–245. doi: 10.1016/0092-8674(90)90739-2. [DOI] [PubMed] [Google Scholar]
- 31.Kassavetis G A, Joazeiro C A P, Pisano M, Geiduschek E P, Colbert T, Hahn S, Blanco J A. The role of the TATA-binding protein in the assembly and function of the multisubunit yeast RNA polymerase III transcription factor, TFIIIB. Cell. 1992;71:1055–1064. doi: 10.1016/0092-8674(92)90399-w. [DOI] [PubMed] [Google Scholar]
- 32.Kassavetis G A, Nguyen S T, Kobayashi R, Kumar A, Geiduschek E P, Pisano M. Cloning, expression, and function of TFC5, the gene encoding the B" component of the Saccharomyces cerevisiae RNA polymerase III transcription factor TFIIIB. Proc Natl Acad Sci USA. 1995;92:9786–9790. doi: 10.1073/pnas.92.21.9786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Khoo B, Brophy B, Jackson S P. Conserved functional domains of the RNA polymerase III general transcription factor BRF. Genes Dev. 1994;8:2879–2890. doi: 10.1101/gad.8.23.2879. [DOI] [PubMed] [Google Scholar]
- 34.Lee J Y, Evans C F, Engelke D R. Expression of RNase P RNA in Saccharomyces cerevisiae is controlled by an unusual RNA polymerase III promoter. Proc Natl Acad Sci USA. 1991;88:6986–6990. doi: 10.1073/pnas.88.16.6986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee J Y, Rohlman C E, Molony L A, Engelke D R. Characterization of RPR1, an essential gene encoding the RNA component of Saccharomyces cerevisiae nuclear RNase P. Mol Cell Biol. 1991;11:721–730. doi: 10.1128/mcb.11.2.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lefebvre O, Carles C, Conesa C, Swanson R N, Bouet F, Riva M, Sentenac A. TFC3: Gene encoding the B-block binding subunit of the yeast transcription factor TFIIIC. Proc Natl Acad Sci USA. 1992;89:10512–10516. doi: 10.1073/pnas.89.21.10512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lefebvre O, Rüth J, Sentenac A. A mutation in the largest subunit of yeast TFIIIC affects tRNA and 5 S RNA synthesis. Identification of two classes of suppressors. J Biol Chem. 1994;269:23374–23381. [PubMed] [Google Scholar]
- 38.Léveillard T, Kassavetis G A, Geiduschek E P. Saccharomyces cerevisiae transcription factors IIIB and IIIC bend the DNA of a tRNAGln gene. J Biol Chem. 1991;266:5162–5168. [PubMed] [Google Scholar]
- 39.López-De-León A, Librizzi M, Puglia K, Willis I. PCF4 encodes an RNA polymerase III transcription factor with homology to TFIIB. Cell. 1992;71:211–220. doi: 10.1016/0092-8674(92)90350-l. [DOI] [PubMed] [Google Scholar]
- 40.Manaud N, Arrebola R, Buffin-Meyer B, Lefebvre O, Voss H, Riva M, Conesa C, Sentenac A. A chimeric subunit of TFIIIC forms a subcomplex with τ95. Mol Cell Biol. 1998;18:3191–3200. doi: 10.1128/mcb.18.6.3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mann C, Micouin J Y, Chiannilkulchai N, Treich I, Buhler J-M, Sentenac A. RPC53 encodes a subunit of Saccharomyces cerevisiae RNA polymerase C (III) whose inactivation leads to a predominantly G1 arrest. Mol Cell Biol. 1992;12:4314–4326. doi: 10.1128/mcb.12.10.4314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41a.Marck, C. Personal communication.
- 42.Marck C, Lefebvre O, Carles C, Riva M, Chaussivert N, Ruet A, Sentenac A. The TFIIIB-assembling subunit of yeast transcription factor TFIIIC has both tetratricopeptide repeat and basic-helix-loop-helix motifs. Proc Natl Acad Sci USA. 1993;90:4027–4031. doi: 10.1073/pnas.90.9.4027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Margottin F, Dujardin G, Gérard M, Egly J M, Huet J, Sentenac A. Participation of the TATA factor in transcription of the yeast U6 gene by RNA polymerase C. Science. 1991;251:424–426. doi: 10.1126/science.1989075. [DOI] [PubMed] [Google Scholar]
- 44.Marzouki N, Camier S, Ruet A, Moenne A, Sentenac A. Selective proteolysis defines two DNA binding domains in yeast transcription factor τ. Nature. 1986;323:176–178. doi: 10.1038/323176a0. [DOI] [PubMed] [Google Scholar]
- 45.Moenne A, Camier S, Anderson G, Margottin F, Beggs J, Sentenac A. The U6 gene of Saccharomyces cerevisiae is transcribed by RNA polymerase C (III) in vivo and in vitro. EMBO J. 1990;9:271–277. doi: 10.1002/j.1460-2075.1990.tb08105.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pardee T S, Bangur C S, Ponticelli A S. The N-terminal region of yeast TFIIB contains two adjacent functional domains involved in stable RNA polymerase II binding and transcription start site selection. J Biol Chem. 1998;273:17859–64. doi: 10.1074/jbc.273.28.17859. [DOI] [PubMed] [Google Scholar]
- 47.Parsons M C, Weil P A. Purification and characterization of Saccharomyces cerevisiae transcription factor TFIIIC. Polypeptide composition defined with polyclonal antibodies. J Biol Chem. 1990;265:5095–5103. [PubMed] [Google Scholar]
- 48.Parsons M C, Weil P A. Cloning of TFC1, the Saccharomyces cerevisiae gene encoding the 95-kDa subunit of transcription factor TFIIIC. J Biol Chem. 1992;267:2894–2901. [PubMed] [Google Scholar]
- 49.Rameau R, Puglia K, Crowe A, Sethy I, Willis I M. A mutation in the second largest subunit of TFIIIC increases a rate-limiting step in transcription by RNA polymerase III. Mol Cell Biol. 1994;14:822–830. doi: 10.1128/mcb.14.1.822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Roberts S, Miller S J, Lane W S, Lee S, Hahn S. Cloning and functional characterization of the gene encoding the TFIIIB90 subunit of RNA polymerase III transcription factor TFIIIB. J Biol Chem. 1996;271:14903–14909. doi: 10.1074/jbc.271.25.14903. [DOI] [PubMed] [Google Scholar]
- 51.Rüth J, Conesa C, Dieci G, Lefebvre O, Düsterhöft A, Ottonello S, Sentenac A. A suppressor of mutations in the class III transcription system encodes a component of yeast TFIIIB. EMBO J. 1996;15:1941–1949. [PMC free article] [PubMed] [Google Scholar]
- 52.Schmitt M E, Brown T A, Trumpower B L. A rapid and simple method for preparation of RNA from Saccharomyces cerevisiae. Nucleic Acids Res. 1990;18:3091–3092. doi: 10.1093/nar/18.10.3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schultz P, Marzouki N, Marck C, Ruet A, Oudet P, Sentenac A. The two DNA-binding domains of yeast transcription factor τ as observed by scanning transmission electron microscopy. EMBO J. 1989;8:3815–3824. doi: 10.1002/j.1460-2075.1989.tb08559.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sethy I, Willis I M. Recessive mutations in the second largest subunit of TFIIIC suggest a new step in RNA polymerase III transcription. Gene Expr. 1995;5:35–47. [PMC free article] [PubMed] [Google Scholar]
- 55.Sethy-Coraci I, Moir R D, López-De-León A, Willis I M. A differential response of wild type and mutant promoters to TFIIIB70 overexpression in vivo and in vitro. Nucleic Acids Res. 1998;26:2344–52. doi: 10.1093/nar/26.10.2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Simos G, Tekotte H, Grosjean H, Segref A, Sharma K, Tollervey D, Hurt E C. Nuclear pore proteins are involved in the biogenesis of functional tRNA. EMBO J. 1996;15:2270–2284. [PMC free article] [PubMed] [Google Scholar]
- 57.Stettler S, Chiannilkulchai N, Hermann-Le Denmat S, Lalo D, Lacroute F, Sentenac A, Thuriaux P. A general suppressor of RNA polymerase I, II and III mutations in Saccharomyces cerevisiae. Mol Gen Genet. 1993;239:169–176. doi: 10.1007/BF00281615. [DOI] [PubMed] [Google Scholar]
- 58.Stolc V, Altman S. Rpp1, an essential protein subunit of nuclear RNase P required for processing of precursor tRNA and 35S precursor rRNA in Saccharomyces cerevisiae. Genes Dev. 1997;11:2414–2425. doi: 10.1101/gad.11.18.2414. . (Corrected and republished, 11:2926–2937.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Swanson R N, Conesa C, Lefebvre O, Carles C, Ruet A, Quemeneur E, Gagnon J, Sentenac A. Isolation of TFC1, a gene encoding one of two DNA-binding subunits of yeast transcription factor τ (TFIIIC) Proc Natl Acad Sci USA. 1991;88:4887–4891. doi: 10.1073/pnas.88.11.4887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Whitehall S K, Kassavetis G A, Geiduschek E P. The symmetry of the yeast U6 RNA gene’s TATA box and the orientation of the TATA-binding protein in yeast TFIIIB. Genes Dev. 1995;9:2974–2985. doi: 10.1101/gad.9.23.2974. [DOI] [PubMed] [Google Scholar]
- 61.Willis I, Schmidt P, Söll D. A selection for mutants of the RNA polymerase III transcription apparatus: PCF1 stimulates transcription of tRNA and 5S RNA genes. EMBO J. 1989;8:4281–4288. doi: 10.1002/j.1460-2075.1989.tb08614.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wilson I A, Niman H L, Houghten R A, Cherenson A R, Connolly M L, Lerner R A. The structure of an antigenic determinant in a protein. Cell. 1984;37:767–778. doi: 10.1016/0092-8674(84)90412-4. [DOI] [PubMed] [Google Scholar]