Hu et al. demonstrate that transgenic expression of Glypican 4 (Gpc4) in the endoderm rescues convergence and extension (C&E) defects in all germ layers in gpc4−/− embryos. The rescue of mesoderm is mediated by Wnt5b and Wnt11f2 and depends on signaling filopodia.
Abstract
Glypicans influence signaling pathways by regulating morphogen trafficking and reception. However, the underlying mechanisms in vertebrates are poorly understood. In zebrafish, Glypican 4 (Gpc4) is required for convergence and extension (C&E) of both the mesoderm and endoderm. Here, we show that transgenic expression of GFP-Gpc4 in the endoderm of gpc4 mutants rescued C&E defects in all germ layers. The rescue of mesoderm was likely mediated by Wnt5b and Wnt11f2 and depended on signaling filopodia rather than on cleavage of the Gpc4 GPI anchor. Gpc4 bound both Wnt5b and Wnt11f2 and regulated formation of the filopodia that transport Wnt5b and Wnt11f2 to neighboring cells. Moreover, this rescue was suppressed by blocking signaling filopodia that extend from endodermal cells. Thus, GFP-Gpc4–labeled protrusions that emanated from endodermal cells transported Wnt5b and Wnt11f2 to other germ layers, rescuing the C&E defects caused by a gpc4 deficiency. Our study reveals a new mechanism that could explain in vivo morphogen distribution involving Gpc4.
Introduction
Glypicans (Gpc’s), members of the heparan sulfate proteoglycan family, are anchored to the external surface of the cell membrane by a C-terminal glycosylphosphatidylinositol (GPI). Gpc’s consist of a core protein that is covalently linked to glycosaminoglycan (GAG) heparan sulfate, a negatively charged moiety that interacts with numerous growth factors and morphogens such as Wnt, FGF, Bmp, and hedgehog (Hh). Thus, Gpc’s regulate a broad range of signaling pathways critical for animal development (Filmus et al., 2008; Lin, 2004; Poulain and Yost, 2015). Vertebrates have six GPC proteins (GPC1–GPC6), and in humans, mutations in the GPC3, GPC4, and GPC6 genes are associated with congenital diseases such as Simpson–Golabi–Behmel overgrowth syndrome (Amor et al., 2019; Campos-Xavier et al., 2009; Fico et al., 2011). Thus, it is important to understand how Gpc’s regulate the signaling pathways.
Gpc’s influence signaling in a variety of ways depending on cell type. Because Gpc’s bind to and interact with morphogens, they can control the diffusion or trafficking of morphogens and thus influence their distributions (Fico et al., 2011; Filmus et al., 2008). Cleavage of the GPI anchor results in the shedding of the Gpc into the extracellular environment, changing the morphogen concentration locally and in the distance (Kreuger et al., 2004). Gpc’s have also been shown to induce the endocytosis of morphogens to remove them from the cell surface (Capurro et al., 2008) or spread morphogens to neighboring cells via transcytosis (Callejo et al., 2011; Gallet et al., 2008), employ lipoprotein vesicles to transport morphogens to distant cells and release them there (Eugster et al., 2007; Panákova et al., 2005), and express in migrating cells to deliver morphogens to distant locations (Serralbo and Marcelle, 2014). Additionally, Gpc’s might facilitate morphogen transport by providing reservoirs of lipid moieties for solubilizing Wnt’s (McGough et al., 2020).
In addition to influencing morphogens, Gpc’s also act as coreceptors, stabilizing ligand–receptor interactions to enhance pathway activity (Kan et al., 1993; Yan et al., 2010), and as repressors, either competing with morphogens for receptor binding (Capurro et al., 2008) or recruiting a deacetylase to inhibit binding of a morphogen to its receptor (Kakugawa et al., 2015).
Recent studies indicate that morphogens can also be transported across distances by actin-based signaling filopodia known as cytonemes (González-Méndez et al., 2019; Ramírez-Weber and Kornberg, 1999). The Drosophila melanogaster Gpc’s Dally and Dally-like (Dlp) have been shown to coat cytonemes that transport Hh (González-Méndez et al., 2017), suggesting that Gpc’s play a role in cytoneme formation. Notably, in the zebrafish blastula, the formation of signaling filopodia that transport Wnt8a can be induced by noncanonical Wnt/planar cell polarity (Wnt/PCP) signaling (Mattes et al., 2018). However, it remains unknown if Gpc’s influence Wnt distribution by regulating the formation of filopodia.
In zebrafish and Xenopus laevis, Gpc4 was first identified as a positive modulator of Wnt11f2 in regulating mesodermal convergence and extension (C&E; Ohkawara et al., 2003; Topczewski et al., 2001), a process that establishes the animal body plan (Keller, 2002; Solnica-Krezel and Sepich, 2012). Later studies showed that Gpc4 contributes to many additional developmental processes by influencing Shh, BMP, and Wnt signaling (LeClair et al., 2009; Miles et al., 2017; Strate et al., 2015; Venero Galanternik et al., 2016). However, little is known about how Gpc4 affects morphogens in vivo.
Recently, we and others showed that Gpc4 is required for endoderm C&E in both the anterior and posterior regions (Hu et al., 2018; Miles et al., 2017). Thus, Gpc4 regulates the C&E of both mesoderm and endoderm. To investigate the cell autonomy of Gpc4 in the gut endoderm, we generated a transgenic line that expresses Gpc4 specifically in the endoderm. Intriguingly, our findings show that in gpc4−/− embryos, endodermal expression of GFP-Gpc4 not only completely rescued endodermal C&E defects but also partially, but significantly, rescued mesodermal C&E defects. These findings suggest that Gpc4 functions both within and outside the endoderm. Thus, our animal model provides a unique opportunity to explore the mechanisms underlying communication among germ layers. Our analyses also show that the mesodermal rescue was not due to Gpc4 cleavage at the GPI anchor. Instead, GFP-Gpc4 presented in the signaling filopodia of endodermal cells that transport Wnt5b and Wnt11f2 to neighboring cells was responsible. Thus, our study uncovers a new mechanism by which the contribution of Gpc4 to the formation of signaling filopodia accounts for its non–cell-autonomous functions.
Results
Endodermal expression of Gpc4 rescues C&E defects in all germ layers of gpc4−/− embryos
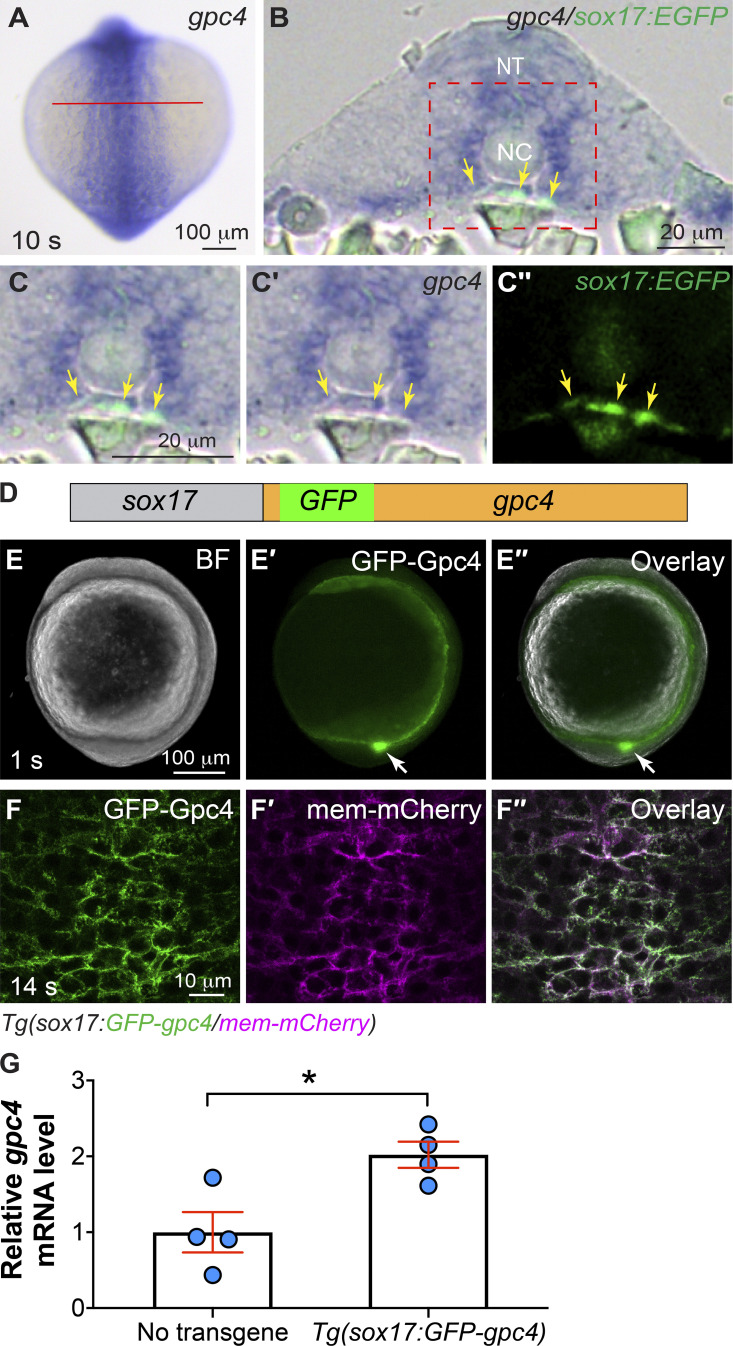
We found that gpc4 is expressed in the posterior endoderm (Fig. S1, A–C′′), from which the gut will develop. To evaluate gut formation in gpc4−/− embryos, we assessed the expression of foxa3 by whole-mount in situ hybridization (WISH). Compared with control (sibling) embryos, gpc4−/− embryos displayed an enlarged gut tube (Fig. 1 B versus A). To determine whether Gpc4 regulates morphogenesis of the gut endoderm cell autonomously, we generated transgenic line Tg(sox17:GFP-gpc4), in which expression of GFP-tagged Gpc4 is driven by the endoderm-specific promoter sox17 (Fig. S1 D; Woo et al., 2012). GFP was inserted immediately after the signal peptide of Gpc4 to avoid disrupting its membrane localization (Fig. S1 D). GFP-Gpc4 is functional, as injection of the encoding RNA rescued C&E defects in gpc4 mutants (Hu et al., 2018). Consistent with the expression pattern of sox17 (Aoki et al., 2002), in this line, GFP-Gpc4 signal was detected in the innermost tissue of the embryo, including in Kupffer’s vesicle (Fig. S1, E–E′′). Additionally, by crossing Tg(sox17:GFP-gpc4) into the Tg(sox17:mem-mCherry) line, in which mCherry is expressed in the plasma membrane of endodermal cells (Ye et al., 2015), we found that GFP-Gpc4 colocalized with mCherry (Fig. S1, F–F′′). Thus, GFP-Gpc4 is expressed in the endoderm. In this study, we used a line in which expression of GFP-Gpc4 was modest (gpc4 RNA levels were doubled relative to those in WT embryos; Fig. S1 G) and embryogenesis was normal.
Figure S1.
gpc4 is expressed in the posterior endoderm and transgenic lines that expresses Gpc4 in the endoderm. (A–C′′) Expression of gpc4 transcript in the posterior region of a Tg(sox17:EGFP) embryo at the 10 somite (s) stage, as detected by WISH. Cryosection was performed on the embryos after gpc4 WISH. (A) gpc4 expression in embryos. Posterior dorsal view, with anterior up. Red line shows the estimated plane for cross sectioning. (B–C′′) Transverse sections of the embryo. (C–C′′) Higher-magnification images of the region shown in red dashed box in B. (C) Overlay of WISH panel (C′) and anti-GFP IF staining panel (C′′). Yellow arrows, endoderm; NT, neural tube; NC, notochord. (D) Schematic depiction of the transgene sox17:GFP-Gpc4. GFP (green box) is inserted after the N-terminal signal peptide of Gpc4, and expression is driven by the endoderm-specific promoter sox17 (gray box). (E–E′′) Expression of transgenic GFP-Gpc4 at 1 somite (s) stage. (E) Bright-field image. (E′) Epifluorescence image of GFP expression. (E′′) Overlay of E and E′. White arrows, Kupffer’s vesicle. (F–F′′) A representative confocal z-stack image at 14 somite (s) stage, showing the expression of GFP-Gpc4 (F) and mem-mCherry (F′) on the plasma membranes of endodermal cells, and overlay of F and F′ (F′′). (G) Levels of gpc4 mRNA in embryos expressing the transgene relative to those that do not express it, as detected by qPCR at 3 somite (s) stage. Data are mean ± SEM. *, P < 0.05; unpaired Student's t test.
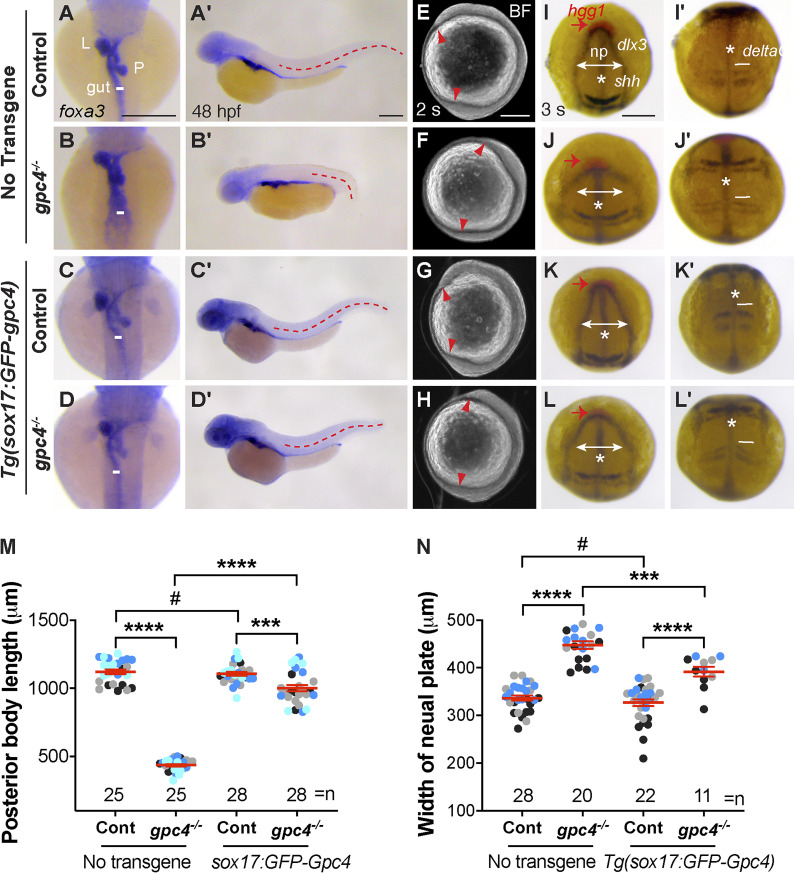
Figure 1.
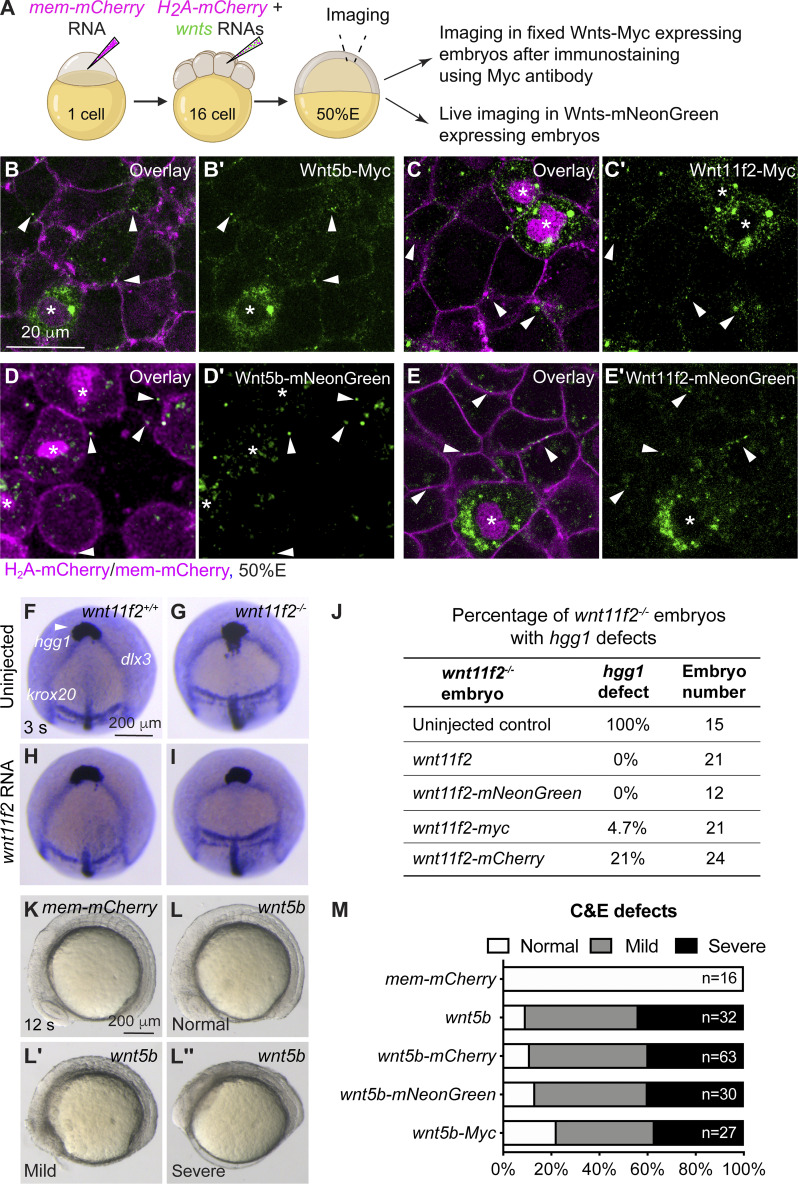
Endodermal expression of GFP-Gpc4 rescues C&E defects in all germ layers of gpc4−/− embryos. (A–D′) The expression of foxa3 as detected by WISH, showing the morphology of the gut, liver (L), and pancreas (P) in the indicated embryos. (A–D) Dorsal view. (A′–D′) Lateral view. White lines indicate the width of the gut tube, and lines in all embryos are equal in length; red dashed lines indicate posterior body length. (E–H) Bright-field images of the indicated embryos at 2 somite (s) stage. Red arrowheads point to the anterior and posterior points of the embryonic axis. Lateral view. (I–L′) Expression of hgg1 (red), dlx3, krox20, shh, and deltaC at 3 somite (s) stage, as detected by WISH. *, axial mesoderm (shh); np, neural plate (dlx3); red arrows indicate hgg1 expression; white lines with double arrows indicate the width of the neural plate, and white lines (deltaC) indicate the width of the first somite. All lines of each type are equal in length. (I–L) Dorsoanterior view. (I′–L′) Dorsal view. (M) Average posterior body length in embryos shown in A′–D′, from four independent experiments (represented by different color dots), with the number of embryos indicated. (N) Average width of the neural plate in embryos shown in I–L, from three independent experiments (represented by different color dots), with the number of embryos indicated. Data are mean ± SEM. #, P > 0.05; ***, P < 0.001; ****, P < 0.0001; unpaired Student's t test. Scale bars, 200 µm.
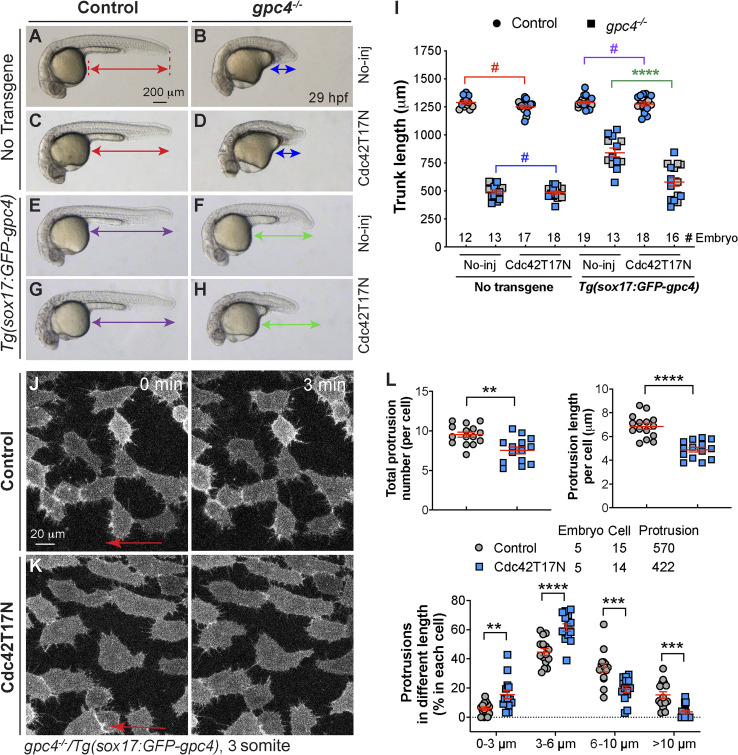
To determine whether endodermal defects in gpc4−/− embryos are due to gpc4 deficiency specifically in the endoderm, we generated the gpc4+/−/Tg(sox17:GFP-gpc4) line and determined whether the phenotypes in gpc4−/− embryos can be rescued by expressing GFP-Gpc4 in the endoderm. Strikingly, the gpc4−/− embryos derived from this line did not have the typical short body axis of these mutants. Further analyses revealed that endodermal expression of GFP-Gpc4 did not affect posterior body length in controls (Fig. 1, A′, C′, and M); it largely, but not completely, suppressed the shortening of posterior body length in gpc4−/− embryos (Fig. 1, B′, D′, and M), indicating that GFP-Gpc4 expression in the endoderm partially rescued the mesodermal C&E defects. Additionally, examination of foxa3 expression showed that the morphology of the gut tube and digestive organs in Tg(sox17:GFP-gpc4) embryos was normal (Fig. 1 C), suggesting that expressing GFP-Gpc4 in the endoderm in this line does not affect the normal development of the digestive system. Notably, in gpc4−/−/Tg(sox17:GFP-gpc4) embryos, the enlargement of the gut tube associated with the gpc4−/− genotype was absent (Fig. 1 D versus B). These data indicate that endodermal expression of Gpc4 rescues the C&E defects of both the mesoderm and endoderm in gpc4 mutants.
To determine when such rescue occurs, we examined embryos at the two-somite stage, when mesodermal C&E defects in gpc4−/− embryos are prominent (Topczewski et al., 2001). The body axes of gpc4−/−/Tg(sox17:GFP-gpc4) embryos were significantly longer than those of gpc4−/− embryos (Fig. 1 H versus F). Expression of tissue-specific markers of the neural plate (dlx3), notochord (shh), prechordal plate cells (hgg1), somites (deltaC), and rhombomeres 3 and 5 (krox20, for the purpose of staging) revealed that the broadening of the neural plate, notochord, and somites that is typically seen in gpc4−/− embryos was significantly suppressed (Fig. 1, I–L′ and N). Thus, the rescue of mesoderm and ectoderm defects by endodermal expression of GFP-Gpc4 was evident at early segmentation, suggesting that Gpc4 functions both within and outside of the endoderm.
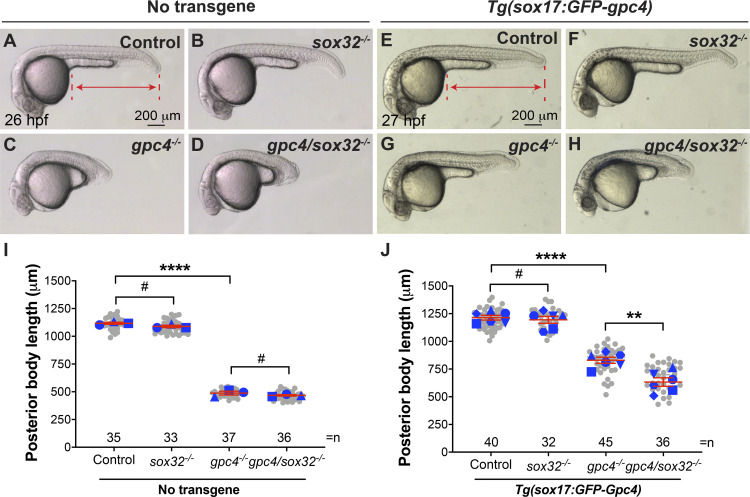
To exclude the possibility that the sox17 promoter induces expression of GFP-Gpc4 outside the endoderm, which could contribute to the observed rescue, we eliminated the formation of the endoderm. If the rescue is resulted from endodermal expression of Gpc4, then removal of the endoderm should abolish the rescue. The transcription factor sox32 is required for endoderm development (Alexander et al., 1999), and foxa3 expression in the endoderm is not detectable in sox32-deficient embryos (Stafford et al., 2006). Similar to earlier results (Alexander et al., 1999), we found sox32−/− embryos had relatively normal body length (Fig. 2 B versus A, I), and sox32 deficiency did not affect body length in gpc4−/− embryos (Fig. 2 D versus C, I ). These data suggest that the formation of the body axis does not rely on the endoderm. In the Tg(sox17:GFP-gpc4) background, sox32 deficiency did not affect body axis in control embryos (Fig. 2 F versus E, J) but led to significant shortening in gpc4−/− embryos (Fig. 2 H versus G, J). Thus, sox32 deficiency largely suppressed the rescue of body length, suggesting that the rescue is dependent on the endoderm.
Figure 2.
The rescue of C&E defects in gpc4 mutants by endodermal expression of GFP-Gpc4 is dependent on the endoderm. (A–H) Bright-field images of the indicated embryos. Lateral view. Lines with double arrows indicate posterior body length, and dashed lines represent points used to measure the posterior body length. (I) Posterior body length in embryos shown in A–D. (J) Posterior body length in embryos shown in E–H. Data from all embryos (gray circles) and separate experiments (different blue shapes) are superimposed, with the number of embryos indicated. Each blue shape represents the mean of data from one experiment. Statistical analyses were performed using the mean of the data from individual experiments. Data are mean ± SEM. #, P > 0.05; **, P < 0.01; ****, P < 0.0001; unpaired Student's t test.
The rescue of mesodermal C&E by endodermal expression of GFP-Gpc4 is partially mediated by Wnt5b and Wnt11f2
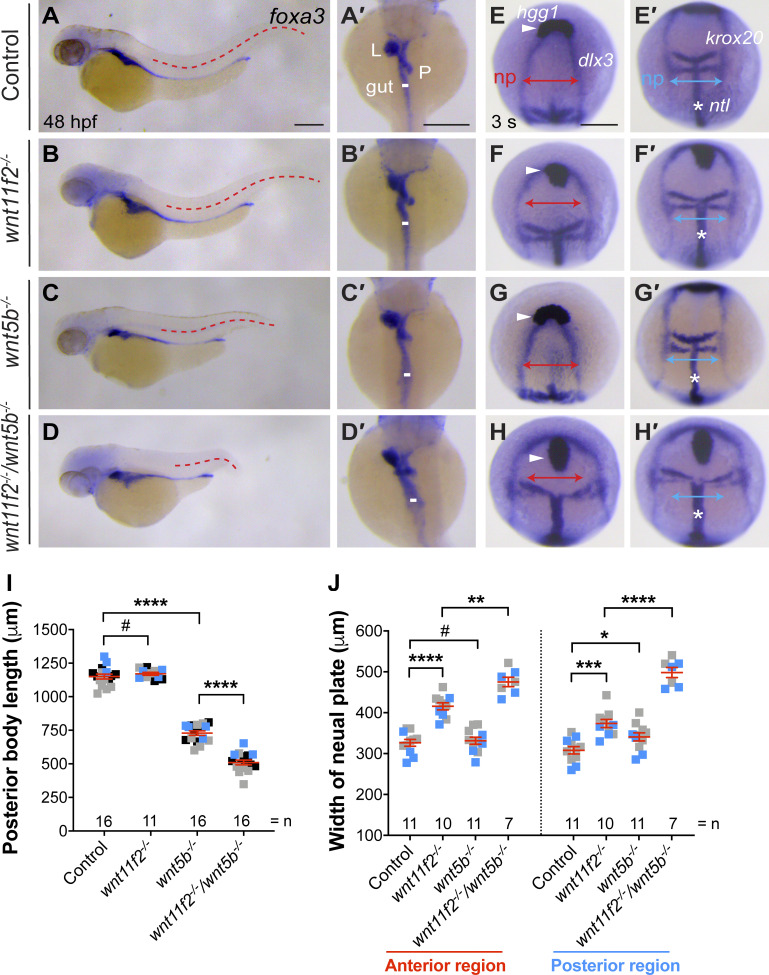
Since Gpc4 is known to regulate mesodermal C&E by interacting with Wnt11f2 (Ohkawara et al., 2003; Topczewski et al., 2001) and both Wnt5b and Wnt11f2 are involved in Wnt/PCP signaling (Kilian et al., 2003; Tada et al., 2002), we postulated that endodermal Gpc4 mediates the rescue of mesodermal C&E by influencing Wnt/PCP signaling. First, we tested the involvement of Wnt5b and Wnt11f2 in the morphogenesis of mesoderm and endoderm at 48 h postfertilization (hpf) in wnt11f2−/− and wnt5b−/− embryos. Assessment of posterior body length showed that wnt5b−/−, but not wnt11f2−/−, embryos had significantly shorter body axes than control siblings and that wnt11f2−/−/wnt5b−/− embryos had the shortest body axes (Fig. 3, A–D, and I). Similarly, the gut tube was normal in wnt11f2−/− embryos but slightly enlarged in wnt5b−/− embryos and significantly widened in wnt11f2−/−/wnt5b−/− embryos (Fig. 3, A′–D′). These data suggest that at day 2, Wnt5b, but not Wnt11f2, is required for elongation of the body axis and formation of the gut tube, but Wnt11f2 cooperates with Wnt5b to regulate endoderm morphogenesis.
Figure 3.
Synergistic C&E defects in mesoderm and endoderm of wnt5b and wnt11f2 mutants. (A–D′) foxa3 expression, as assessed by WISH. (A–D) Lateral view. (A′–D′) Dorsal view. Red dashed lines represent the length of the posterior body, and white lines indicate the width of the gut tube. All lines of the same type are equal in length. (E–H′) Expression of hgg1 (arrowheads), dlx3, krox20, and ntl (*) at 3 somite (s) stage, as detected by WISH. Red and blue lines with double arrows indicate the width of the neural plate (np) in the anterior and posterior regions. All lines of the same color are equal in length. (I) The posterior body length in embryos in A–D, from three independent experiments (represented by different color squares), with the number of embryos indicated. (J) Average width of the neural plate in the anterior and posterior regions in embryos shown in E–H′ from two independent experiments (represented by different color squares), with the number of embryos indicated. Data are mean ± SEM. #, P > 0.05; *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; unpaired Student's t test. Scale bars, 200 µm.
Given that the observed rescue occurred as early as 2-somite–3-somite stage (Fig. 1, E–L′), we tested if Wnt11f2 and Wnt5b regulate C&E at this stage. Consistent with published data (Heisenberg et al., 2000), we found that compared with controls, in wnt11f2−/− embryos, hgg1-expressing prechordal plate failed to migrate to the region anterior to dlx3-expressing neural plate, and the neural plate (dlx3) and notochord (ntl) were broader (Fig. 3, E–F′ and J). In wnt5b−/− embryos, the expression pattern of hgg, dlx3, and ntl in the anterior region appeared to be normal (Fig. 3, G and J), whereas dlx3-expressing neural plate was slightly wider in the posterior region (Fig. 3 G′ and J). In wnt11f2−/−/wnt5b−/− embryos, these defects were much more severe than those in the single mutants (Fig. 3, H, H′, and J), consistent with a previous report on effects at the tailbud stage (Kilian et al., 2003). These data indicate that at 3-somite stage, wnt11f2 is required for C&E of both the mesoderm and ectoderm in the anterior and posterior region and that wnt5b affects ectodermal C&E only in the posterior region. The nearly normal body length in wnt11f2−/− embryos and shorter body length in wnt5b−/− embryos at day 2 suggests that Wnt5b functions at a later stage of embryogenesis (segmentation) than Wnt11f2 (during gastrulation). However, consistent with what we found in day 2 embryos, Wnt5b and Wnt11f2 functioned synergistically in regulating the C&E of all germ layers at early segmentation. Therefore, both Wnt5b and Wnt11f2 might contribute to mesodermal rescue by endodermal expression of GFP-Gpc4 in gcp4−/− embryos.
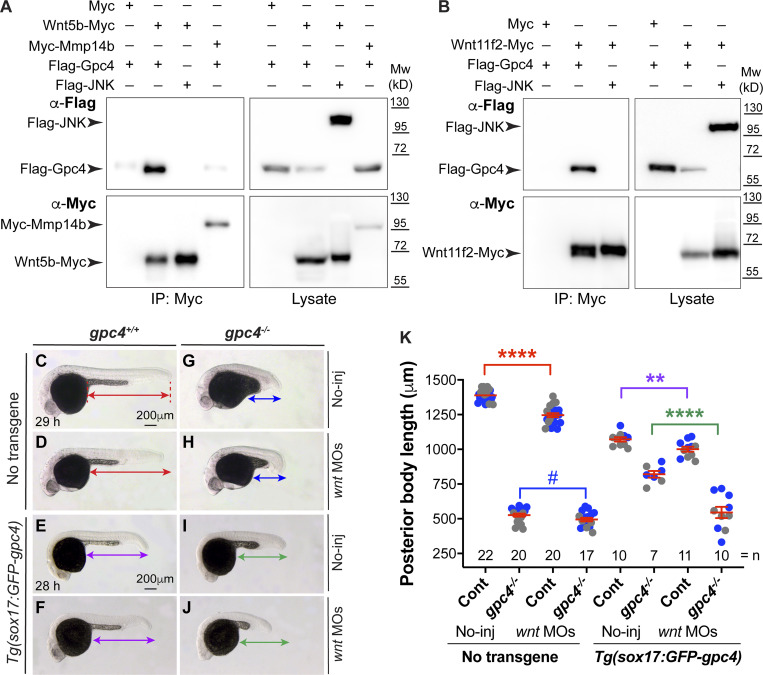
Gpc’s can bind morphogens to regulate their signaling. A previous study showed that Xenopus Gpc4 can physically bind Wnt5, Wnt8, and Wnt11 (Ohkawara et al., 2003). We further examined the ability of zebrafish Flag-Gpc4 to bind Wnt5b-Myc and Wnt11f2-Myc. First, we determined if these Wnt constructs are expressed normally and are functional. We conducted a blastula assay to assess the expression of Myc-tagged Wnt’s in vivo (Fig. S2 A). Briefly, we injected embryos at the one-cell stage with mem-mCherry RNA to label the plasma membrane of all cells. When embryos reached the 16-cell stage, one cell was coinjected with H2A-mCherry and wnt-Myc RNAs to express Myc-tagged Wnt’s and nuclei-mCherry (tracer) in a subset of cells. Embryos at 50% epiboly were fixed for Myc antibody immunostaining and subjected to confocal imaging. As shown in Fig. S2, B and C, punctate expression of Wnt proteins was observed in the extracellular space outside of the expressing cells (those with magenta nuclei), indicating that those Wnt’s were secreted from the Wnt-expressing cells. Furthermore, we found that injecting wnt11f2-Myc RNA rescued the C&E defects in wnt11f2−/− embryos (Fig. S2, F–J) and that overexpressing Wnt5b-Myc led to C&E defects (Fig. S2, K–M). These data suggest that our wnt constructs produce proteins that have the predicted subcellular localization patterns and are functional. Additionally, we found that Flag-Gpc4 is functional, as injection of the Flag-gpc4 RNA rescued the C&E defects in gpc4−/− embryos (Fig. S3, A–B′). Next, we transfected HEK293 cells with Flag-gpc4 or Flag-JNK (negative control) and wnt5b-Myc, wnt11f2-Myc, Myc, or Myc-mmp14b (Hu et al., 2018; the last two constructs were negative controls) and performed coimmunoprecipitation. Flag-Gpc4, but not Flag-JNK, was pulled down with Wnt5b-Myc or Wnt11f2-Myc, but not with Myc or Myc-Mmp14b (Fig. 4, A and B). Notably, Flag-Gpc4 was detected as a 60-kD band. This is consistent with previous findings that, in reducing gels, N-terminally tagged Dlp runs as a 50-kD band, although the C-terminally tagged Dlp runs as a smear (due to GAG modifications; Wang and Page-McCaw, 2014). Thus, zebrafish Gpc4 interacts physically with Wnt5b and Wnt11f2.
Figure S2.
Wnt11f2- and Wnt5b-tagged constructs are functional. (A) Schematic diagram illustrating mosaic labeling approach to assess the localization of tagged Wnt’s in vivo. At the 1-cell stage, embryos were injected with mem-mCherry RNA to label the plasma membrane of all cells; at the 16-cell stage, a single blastula cell was injected with RNAs encoding wnt5b or wnt11f2 (with distinct tags), together with H2A-mCherry, to express Wnt’s in a subset of cells (whose nuclei are labeled with mCherry). At 50% epiboly (50%E), embryos expressing Wnt-Myc were fixed for immunostaining using an anti–C-MYC antibody; and embryos expressing Wnt-mNeonGreen were subjected for live imaging. (B–E′) Confocal images of zebrafish embryos at 50%E, following mosaic injection. The expression of tagged Wnt constructs (green) is shown; all cells of the embryo are labeled with mem-mCherry (magenta). White arrowheads indicate Wnt-labeled puncta outside the expressing cells (asterisks). (B–C′) Confocal images showing the expression of Wnt5b-Myc (B and B′) and Wnt11f2-Myc (C and C′), as detected by immunostaining. (D–E′) Live confocal images showing the expression of Wnt5b-mNeonGreen (D and D′) and Wnt11f2-mNeonGreen (E and E′). (F-I) Expression of hgg1 (white arrowheads), dlx3, and krox20 at 3 somite (s) stage, as detected by WISH in control and wnt11f2−/− embryos (uninjected or injected with the wnt11f2 RNA). The expressing domain of hgg1 is lagging behind that of dlx3 in wnt11f2−/− embryos (G). Injecting wnt11f2 RNA did not impact hgg1 expression in control siblings (H) but largely rescued the defects of hgg1 expression pattern in wnt11f2−/− embryos (I). (J) Percentage of wnt11f2−/− control embryos and counterparts injected with RNAs encoding WT wnt11f2 or various tagged forms of wnt11f2 that display defects in hgg1 expression. (K–L′′) Bright-field images of live embryos, showing control WT embryos and counterparts injected with wnt5b RNA, showing C&E defects of different severity. 12 s, 12 somite stage. (M) Percentage of C&E defects of varying severity in embryos injected with RNAs encoding mem-mCherry or wnt5b or various tagged forms of wnt5b.
Figure S3.
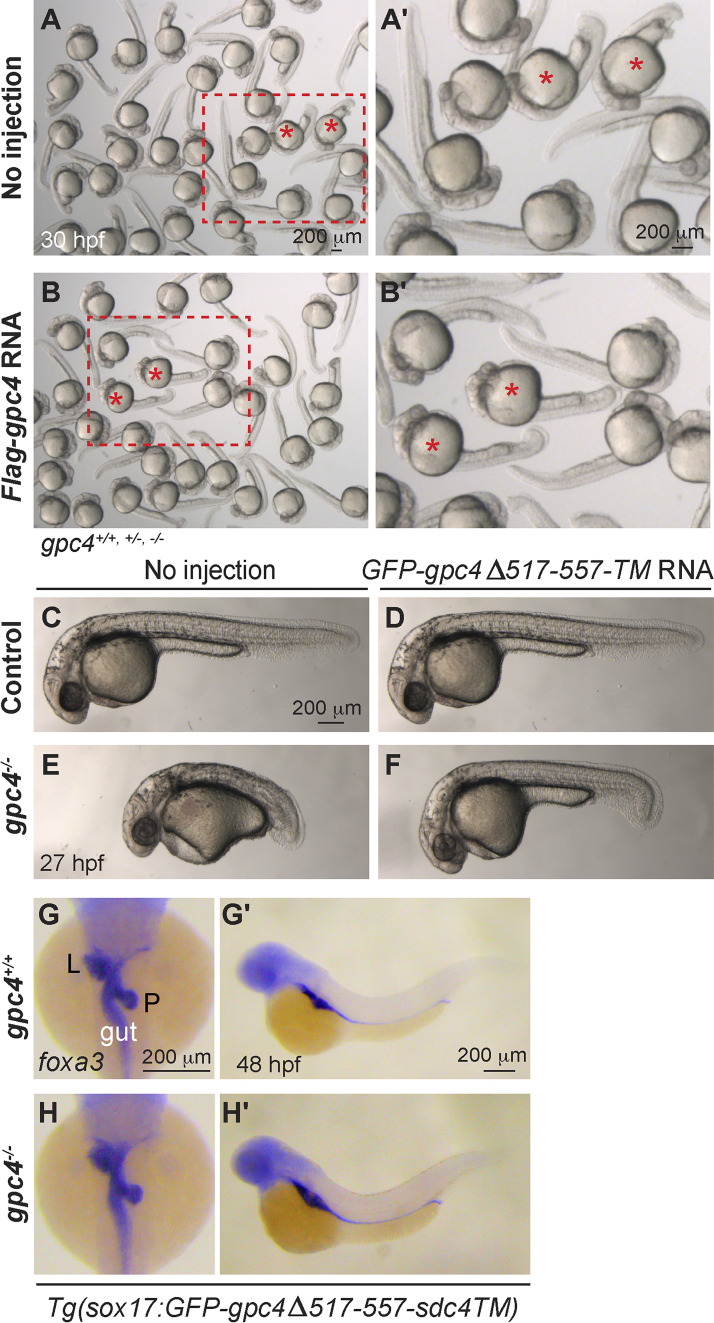
GPI cleavage of Gpc4 does not drive mesoderm rescue. (A–B′) Bright-field images of groups of embryos obtained from incrossing gpc4+/− fish, uninjected (controls; A), injected with Flag-gpc4 RNA (B). (A′ and B′) Magnified images from the rectangular areas outlined by red dashed lines in A and B. Red asterisks, gpc4−/− embryos exhibited shorter body axis (A and B), which was significantly rescued in those gpc4−/− embryos injected with Flag-gpc4 RNA (A′ and B′). (C–F) Bright-field images of the indicated embryos at 27 hpf. (G–H′) Expression of foxa3, as detected by WISH, in the indicated embryos at 48 hpf, showing the morphology of the gut, liver (L), and pancreas (P). (G and H) Dorsal view. (G′ and H′) Lateral view.
Figure 4.
The rescue of mesodermal C&E by endodermal expression of GFP-Gpc4 is mediated in part by Wnt5b and Wnt11f2. (A and B) Coimmunoprecipitation experiments were performed using lysates from HEK cells transfected with Myc or Myc-Mmp14b (negative controls), or Wnt5b-Myc or Wnt11f2-Myc and Flag-Gpc4 or Flag-JNK (negative controls) using anti–C-MYC antibody coupled to protein G magnetic beads. Western blotting of in both pellets (IP) and cell lysates (Lysate) was performed using anti-Flag and anti–C-MYC antibodies. Mw, molecular weight. (C–J) Bright-field images of the indicated control embryos and embryos injected with a subdose of MOs targeting wnt11f2/5b (5 and 1 ng). Lateral view. Lines with double arrows indicate posterior body length. Lines of the same color are equal in length. (K) Average posterior body length in embryos shown in C–J, from two independent experiments (represented by different color dots), with the number of embryos indicated. P values in different colors correspond to the embryos with posterior body length marked by the line of that color. Data are mean ± SEM. #, P > 0.05; **, P < 0.01; ****, P < 0.0001; unpaired Student's t test.
We next tested whether loss of wnt5b and wnt11f2 can suppress the rescue of C&E defects by endodermal expression of Gpc4. We reasoned that if the rescue of Gpc4 depends on these Wnt proteins, then embryos should be more sensitive to the suppression of their expression. Given that wnt5b/wnt11f2 double mutants have C&E defects, they cannot be used for this analysis. Thus, we injected embryos with morpholinos (MOs) targeting both wnt5b and wnt11f2 at subdoses that partially suppress their expression. Injecting control embryos with MOs at these doses led to a slight but significant reduction in posterior body length, regardless of whether GFP-Gpc4 was expressed in the endoderm (Fig. 4 C–F and K). Intriguingly, the injection of gpc4−/− embryos with these MOs did not affect the body length (Fig. 4 G, H, and K), but it suppressed the rescue caused by endodermal GFP-Gpc4 (Fig. 4, I–K). These results indicate that in gpc4−/− embryos, the rescue of mesodermal C&E defects induced by endodermal expression of GFP-Gpc4 is at least partially mediated by Wnt5b and Wnt11f2.
The ability of endodermal GFP-Gpc4 to rescue body length is not dependent on cleavage of the GPI anchor
We set out to identify the mechanisms whereby endodermal expression of GFP-Gpc4 exerts the rescue effect on other germ layers. GPI can be cleaved to release the attached proteins from the plasma membrane (Fujihara and Ikawa, 2016). For example, the Drosophila glypicans Dlp and Dally (Kreuger et al., 2004), as well as mammalian GPC3, GPC5, and GPC6 (Traister et al., 2008), can be cleaved from GPI. Similarly, Gpc4 was detected in the culture medium of zebrafish embryonic cells, and Gpc4 localization shifts from the membrane to the extracellular space in zebrafish embryos when GPI biosynthesis is disrupted (Shao et al., 2009). These findings suggest that Gpc4 can be released from the cell membrane and that the GPI anchor is critical for its attachment to the cell membrane. Thus, it is possible that GFP-Gpc4 expressed by the endoderm can be cleaved and released to the mesoderm and ectoderm to carry out its functions.
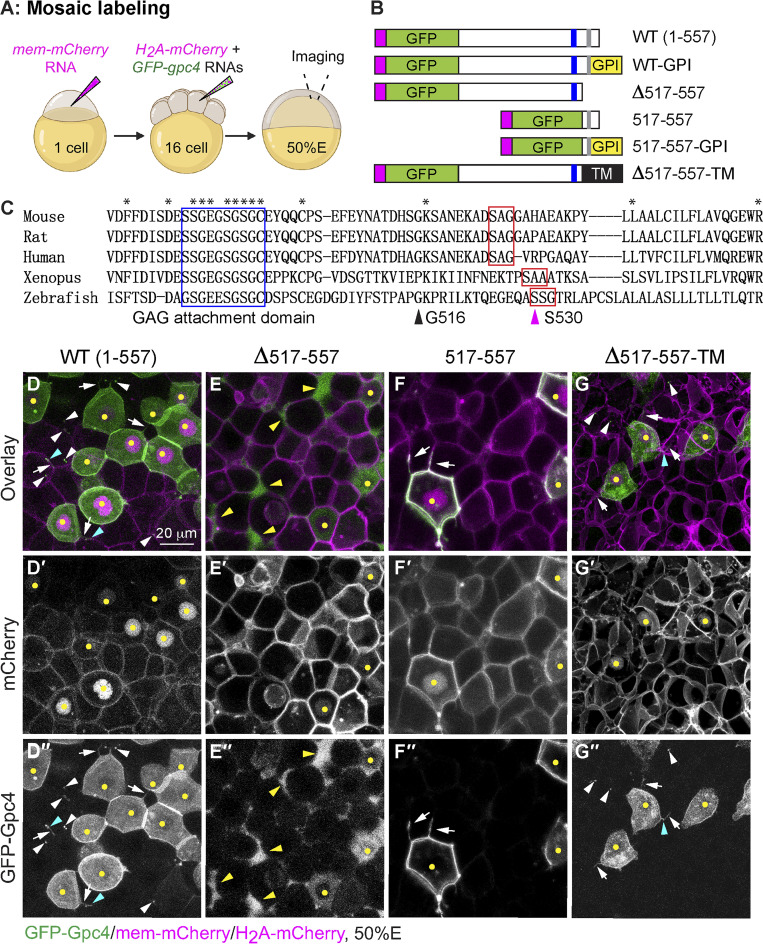
To test this hypothesis, we first assessed GFP-Gpc4 cleavage in vivo using the mosaic labeling approach that had been used to examine the expression of tagged Wnt’s (Fig. S2 A). As in the earlier experiment, embryos at the one-cell stage were injected with the mem-mCherry RNA to label the plasma membrane of all cells with mCherry. When these embryos reached the 16-cell stage, a single blastula cell in each was injected with the GFP-gpc4 and H2A-mCherry RNAs (Fig. 5 A). This approach resulted in a subset of cells in each embryo expressing GFP-Gpc4; these cells were labeled with nuclear mCherry. Live imaging of embryos at 50% epiboly showed that in the cells with mCherry-labeled nuclei, GFP-Gpc4 was present mainly on the plasma membrane; however, some GFP signal was also present in the cytosol, likely due to the overexpression or incomplete processing of proteins in this assay. Notably, GFP-Gpc4–expressing cells extended cellular protrusions (Fig. 5, D-D′′, white arrows), some of which were decorated with bright GFP puncta (Fig. 5, D–D′′, cyan arrowheads). In addition to nuclei-mCherry–labeled cells, cells distant from the site of expression harbored GFP-Gpc4 signal (Fig. 5, D–D′′, white arrowheads). These data suggest that GFP-Gpc4 can be delivered from cells that produce it to neighboring cells.
Figure 5.
GFP-Gpc4 mutant proteins localize to distinct sites in the zebrafish blastula. (A) Schematic diagram illustrating the mosaic labeling approach for examining the localization of GFP-Gpc4 in vivo. 50%E, 50% epiboly. (B) Schematic diagram of the constructs encoding WT GFP-gpc4 and various mutant forms of the protein. Magenta rectangle, the N-terminal signal peptide; GPI, GPI in the C terminus; blue rectangle, putative GAG attachment domain; gray rectangle, putative GPI attachment site; TM, transmembrane domain of Sdc4. (C) Alignment of C-terminal amino acids of Gpc4 from the indicated species. Asterisks, identical amino acids; blue rectangle, putative GAG attachment domain (G488-C497); red rectangles, putative conserved GPI attachment site and its adjacent residues; magenta arrowhead (at S530), putative GPI attachment site (⍵) in zebrafish; black arrowhead (at G516), the site of fusion to the TM domain. (D–G′′) Confocal images of blastulas, with all cells labeled with mem-mCherry (in magenta, gray in D′–G′) and a subset of cells colabeled with the indicated GFP-Gpc4 constructs (gray in D′′–G′′) and nuclear H2A-mCherry (yellow dots). White arrows, GFP-labeled protrusions; white arrowheads, punctate GFP signal outside of GFP-expressing cells; cyan arrowheads, punctate GFP signal on GFP-labeled protrusions; yellow arrowheads, GFP signal in the extracellular space outside GFP-expressing cells.
Next, we sought to identify the region of the putative GPI attachment signal in zebrafish Gpc4. During the posttranslational modification of GPI proteins, the N-terminal signal peptide (Fig. 5 B, magenta rectangle) guides Gpc4 preproproteins (WT [1–557]; Fig. 5 B) to the endoplasmic reticulum, where the GPI attachment signal peptide in the C terminus is cleaved and the GPI moiety is attached to the GPI attachment site (Fig. 5 B, gray rectangle; Kinoshita and Fujita, 2016). This GPI modification leads to loss of the C-terminal–most peptide but is attached with the GPI anchor (Fig. 5 B, WT-GPI). The GPI attachment signal contains the GPI attachment site, which is normally located between the GAG attachment domain and the C-terminal end. By comparing the C-terminal amino acids of zebrafish Gpc4 (I480-R557) with those of its mammalian and Xenopus orthologous, we found a putative GAG attachment domain (G488-C497) that contains conserved Ser-Gly dipeptide sites (Fig. 5 C, blue rectangle). The GPI attachment site (⍵) and its adjacent residues (⍵+1 and ⍵+2) have statistically conserved residues (Eisenhaber et al., 1998), with S occupying the ⍵ site 48% of the time; S, A, or G commonly occupying ⍵+1; and A or G occupying the ⍵+2 site 70% of the time. Using these criteria, we identified the putative conserved GPI attachment site and its adjacent residues (⍵, ⍵+1, and ⍵+2) of Gpc4: SSG (AA530-532) in zebrafish, SAG in mammals, and SAA in Xenopus (Fig. 5 C, red rectangles). To test the C-terminal region of zebrafish Gpc4 for the ability to anchor the protein to the membrane, we generated a series of C-terminal truncation mutants that lack the potential GPI attachment signal and assessed their expression patterns using the mosaic labeling approach described above. We found that Gpc4 lacking AA517-557 (Δ517–557; Fig. 5 B) failed to localize to the cell membrane and was present mainly in the extracellular space (Fig. 5, E–E′′). In contrast, GFP-Gpc4AA517-557, which consists of only AA517-557 and the N-terminal signal peptide of Gpc4 (517–557; Fig. 5 B) and was expected to be expressed on the cell membrane due to its ability to receive the GPI modification (517–557-GPI; Fig. 5 B), was expressed mainly on the cell membrane (Fig. 5, F–F′′). Thus, AA517-557 of Gpc4 encompasses the GPI attachment signal peptide, which is critical for the membrane anchor.
We postulated that if the rescue stems from the release of Gpc4 from the membrane after GPI cleavage, a Gpc4 that cannot be cleaved should not be able to rescue the mesodermal defects. Thus, we generated a chimeric construct, GFP-Gpc4Δ517-557-Sdc4TM (Δ517–557-TM; Fig. 5 B), in which GPI attachment signal peptide was replaced with the transmembrane (TM) domain of syndecan 4 (Sdc4), another heparan sulfate proteoglycan family member that is expressed on the cell membrane via its single TM domain (Lopes et al., 2006; Muñoz et al., 2006). In vivo localization revealed that GFP-Gpc4Δ517-557-Sdc4TM was mainly expressed on the cell membrane (Fig. 5, G–G′′). Thus, the TM domain restores membrane localization to GFP-Gpc4 lacking the GPI anchor. As expected, the cells expressing this TM construct extended GFP-labeled protrusions (Fig. 5, G–G′′, white arrows), some of which were decorated with GFP puncta (Fig. 5, G–G′′, cyan arrowheads). However, we also observed GFP punctae away from the TM construct–expressing cells (Fig. 5, G–G′′, white arrowheads), which could be due to the possibility that GFP-punctae was released from the protrusions to the extracellular space or the TM-construct might be cleaved at other regions rather than the GPI anchor. Furthermore, this chimeric construct was functional, as its overexpression rescued the shortened body length in gpc4−/− embryos (Fig. S3, C–F). We next generated transgenic line Tg(sox17:GFP-gpc4Δ517-557-sdc4TM), in which the construct that includes the TM domain but lacks the cleavable GPI signal is expressed specifically in the endoderm. Unexpectedly, like GFP-Gpc4, this chimeric protein rescued the C&E defects in both mesoderm and endoderm of gpc4−/− embryos, in spite of being expressed only in the endoderm (Fig. S3, G–H′ versus Fig. 1, A–D′). These data indicate that GPI cleavage of Gpc4 does not drive rescue of the mesoderm.
Gpc4 contributes to the formation of actin-based signaling filopodia that transport Wnt5b
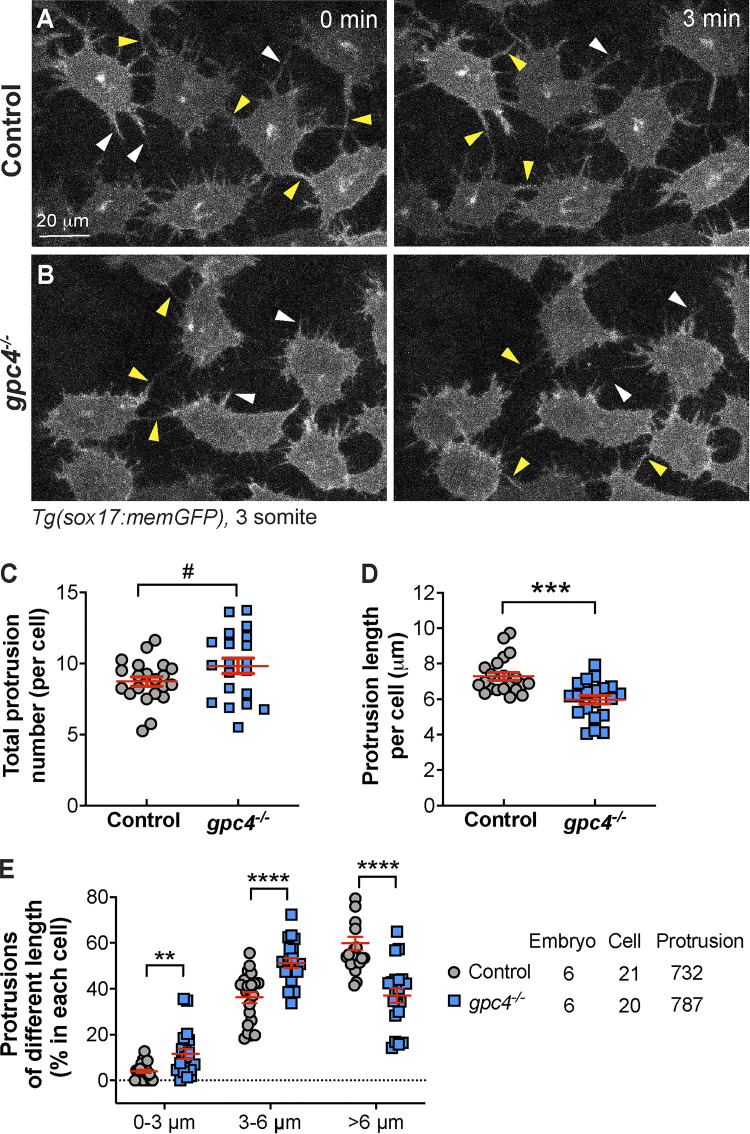
Some morphogens, including Wnt, can be transported between cells by specialized cell protrusions called signaling filopodia (Stanganello and Scholpp, 2016). To test if endodermal cells extend cellular protrusions and whether Gpc4 regulates the formation, we performed confocal time-lapse imaging of Tg(sox17:memGFP) embryos, in which the plasma membrane of endodermal cells is labeled with GFP. We found that endodermal cells extended robust finger-like protrusions; some of them extended to the space between endodermal cells (white arrowheads), and others reached the neighboring endodermal cells (yellow arrowheads; Fig. 6 A and Video 1). These findings suggest that these filopodia communicate with other cells. Notably, in gpc4−/− embryos, endodermal cells formed protrusions that might be too short to reach other cells (Fig. 6 B and Video 1). Quantification revealed that the total number of protrusions was comparable in gpc4−/− embryos and controls (Fig. 6 C), but the average length of the protrusions was shorter in the mutant embryos (Fig. 6 D). Specifically, the proportion of short protrusions (<6 µm) was significantly higher, while the portion of long protrusions (>6 µm) was significantly lower (Fig. 6 E) in the gpc4−/− embryos. These data suggest that Gpc4 is critical for generating long protrusions that enable communication between cells.
Figure 6.
Gpc4 is necessary for the formation of long endodermal protrusions. (A and B) Snapshots from confocal time-lapse imaging performed on the indicated embryos (Video 1). White arrowheads, protrusions in the space between endodermal cells; yellow arrowheads, protrusions that link neighboring endodermal cells. (C–E) The total number of protrusions (C), the length of the protrusion (D), and the percentages of protrusions of different lengths (grouped into 3-µm bins; E) in each endodermal cell. The number of embryos, cells, and protrusions analyzed is indicated. Data are mean ± SEM. #, P > 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; unpaired Student's t test.
Video 1.
Gpc4 is critical for the formation of endodermal protrusions. Confocal time-lapse experiments were performed on Tg(sox17:memGFP) control and gpc4−/− embryos at 3 somite stage using a Zeiss LSM880 confocal microscope with an LD C-Apo 40×/NA 1.1 water objective (shown in Fig. 6, A and B). Z-stacks of 13.5 µm were acquired at 1.5 µm intervals every 30 s using the following settings: zoom 1.0, 1,024 × 1,024 pixels, 9 speed, 4 averaging. The movie plays at five frames/s. White arrowheads, protrusions in the space between endodermal cells; yellow arrowheads, protrusions that link neighboring endodermal cells.
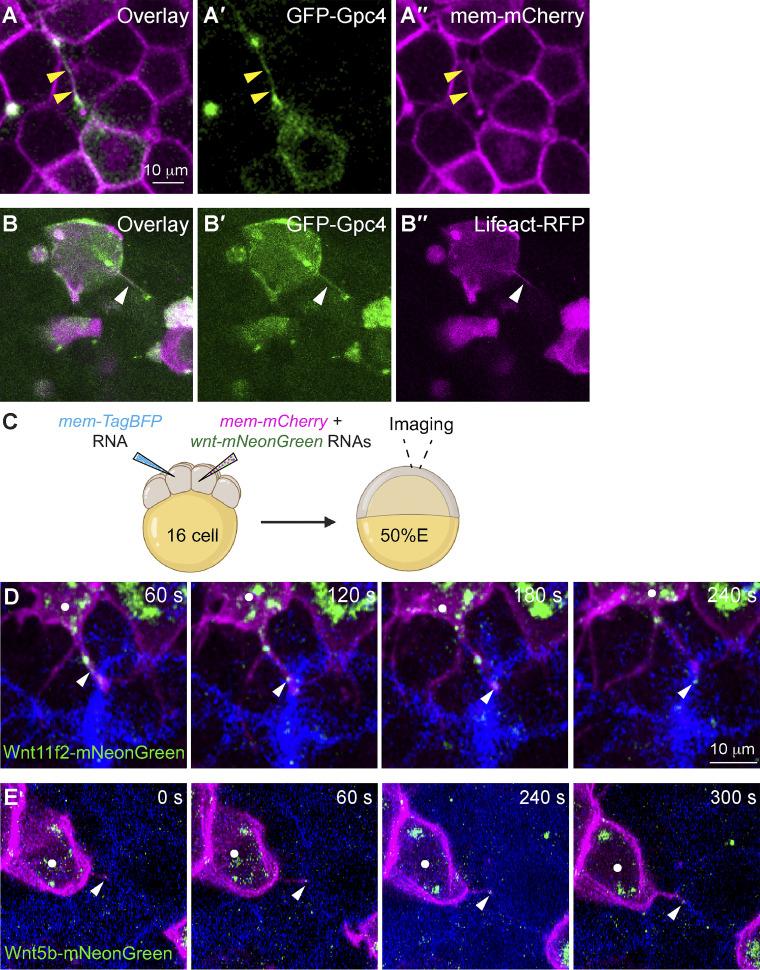
Signaling filopodia are actin-based structures that bind to and transport signaling molecules, enabling them to function at a distance from their site of expression (Kornberg and Roy, 2014; Mattila and Lappalainen, 2008). GFP-Gpc4 was mosaically expressed in a subset of zebrafish blastula cells. Their plasma membranes were labeled with mCherry, and their nuclei were labeled with H2A-mCherry. We found GFP-Gpc4–expressing cells extended GFP-positive protrusions (Fig. S4, A–A′′). To determine whether these GFP-Gpc4–labeled protrusions are also actin based, we performed mosaic injection with RNAs encoding GFP-gpc4 and Lifeact-RFP, an F-actin–binding protein that marks filopodia (Riedl et al., 2008). Live imaging showed that Lifeact-RFP illuminates GFP-Gpc4–labeled filopodia (Fig. S4, B–B′′). These results suggest that Gpc4-GFP–labeled filopodia are enriched with actin.
Figure S4.
Actin-based filopodia deliver Wnt proteins to neighboring cells. (A–A′′) Snapshots from confocal time-lapse imaging of zebrafish blastula cells (labeled with mem-mCherry, in magenta) showing a GFP-Gpc4 expressing cell extending a long cellular protrusion (yellow arrowheads). (B-B′′) Snapshots from confocal time-lapse imaging of zebrafish blastula cells, showing a GFP-Gpc4-labeled protrusion (white arrowheads) that is colabeled Lifeact-RFP (magenta). (C) Schematic diagram illustrating mosaic injection, with distinct cells of embryos at the 16-cell stage injected with specific sets of RNAs, as indicated. Confocal live imaging was performed, with a focus on the regions where the two populations of labeled cells were in close proximity. (D and E) Snapshots from confocal time-lapse imaging (Video 5) showing that mem-mCherry labeling protrusions extended from Wnts-mNeonGreen–expressing cells (white dots) transport Wnt11f2-mNeonGreen (D) or Wnt5b-mNeonGreen (E) to the neighboring BFP-expressing cells (Video 5). White arrowheads, Wnt-expressing puncta on protrusions.
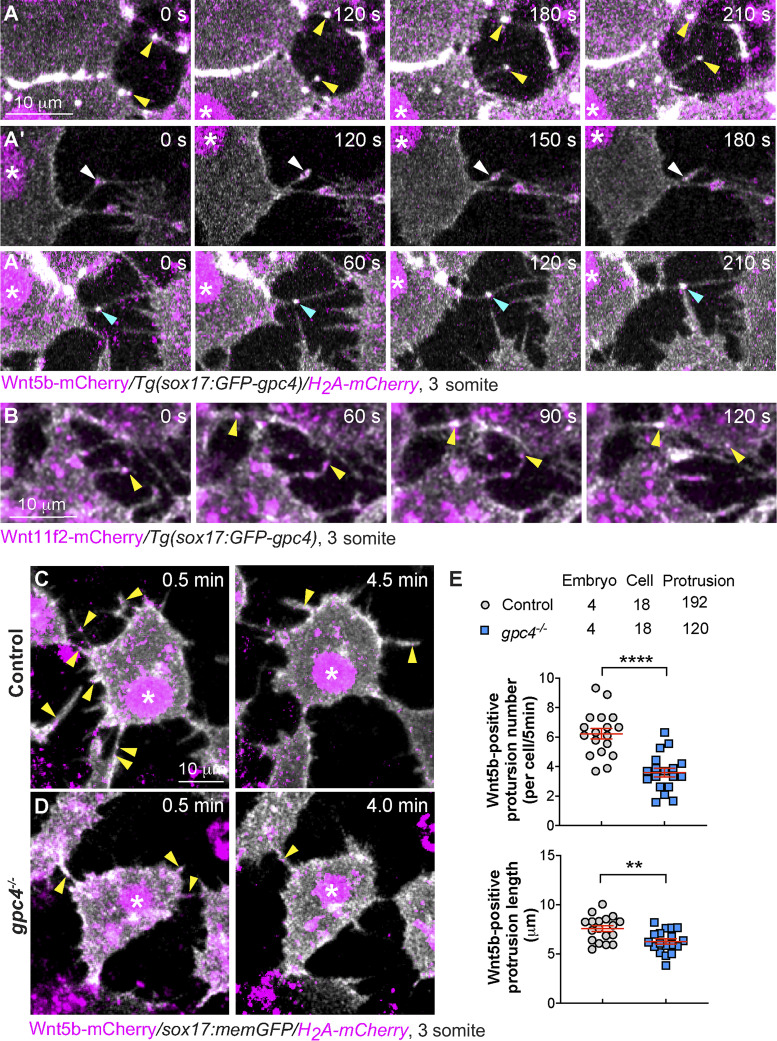
Signaling filopodia extending from zebrafish blastula cells were previously shown to transport Wnt8a (Stanganello et al., 2015). Our coimmunoprecipitation experiments showed that Gpc4 could physically bind Wnt5b and Wnt11f2 (Fig. 4, A and B); thus, we postulate that in Tg(sox17:GFP-gpc4) embryos, GFP-Gpc4–expressing endodermal cells can extend signaling filopodia that bind to and transport Wnt's, enabling these proteins to function outside the endoderm. To test this possibility, we injected Tg(sox17:GFP-gpc4/H2A-mCherry) embryos with a wnt5b-mCherry RNA and performed confocal time-lapse experiments. We found that, like memGFP-expressing endodermal cells (Fig. 6), GFP-Gpc4–expressing endodermal cells extended robust GFP-Gpc4–labeling filopodia, some of which having Wnt5b-mCherry puncta (Fig. 7 A). Notably, two types of Wnt5b-positive protrusions were observed: one extended to deliver Wnt5b out of the cells (yellow arrowheads), and the other retracted to carry Wnt5b back to cells (white arrowheads; Fig. 7, A–A′; and Video 2). In some cases, Wnt5b labeling was observed at protrusions that connect two adjacent endodermal cells (Fig. 7 A′′, cyan arrowheads; and Video 2). Similarly, we found that the GFP-Gpc4–labeled protrusions of endodermal cells transported Wnt11f2-mCherry (Fig. 7 B, yellow arrowheads; and Video 3). Next, we determined how Gpc4 affects Wnt5b transportation. We injected embryos obtained from incrossing gpc4+/−/Tg(sox17:memGFP/H2A-mCherry) fish with wnt5b-mCherry RNA and performed confocal time-lapse imaging to analyze Wnt5b-mCherry–labeled signaling protrusions extending from endodermal cells (Video 4). In gpc4−/− embryos, the Wnt5b-positive protrusions (yellow arrowheads) were shorter and fewer in number than those in control embryos (Fig. 7, C–E). These data indicate that GFP-Gpc4-labeled filopodia can transport Wnt5b and Wnt11f2 to other cells, and Gpc4 promotes the formation of signaling protrusions that deliver Wnt5b and Wnt11f2 to neighboring cells.
Figure 7.
Gpc4 is required for the formation of Wnt-positive filopodia. (A and B) Snapshots from confocal time-lapse images of Tg(sox17:GFP-gpc4/ H2A-mCherry) or Tg(sox17:GFP-gpc4) embryos injected with RNA encoding wnt5b-mCherry (A–A′′, Video 2) or wnt11f2-mCherry (B, Video 3), showing Wnt5b-mCherry or Wnt11f2-mCherry (in magenta) is present on GFP-Gpc4 labeled filopodia (in white). Asterisk, nucleus; yellow arrowheads, Wnt-mCherry at the extending protrusions; white arrowheads, Wnt-mCherry at the retracting protrusions; cyan arrowheads, Wnt-mCherry at protrusions from two cells merging or connected. (C and D) Snapshots from confocal time-lapse images of Tg(sox17:memGFP/H2A-mCherry) embryos injected with wnt5b-mCherry RNA in both control and gpc4−/− embryos (Video 4) showing Wnt5b-mCherry (in magenta, yellow arrowheads) is present on memGFP-labeled filopodia (in white). Asterisk, nucleus. (E) The number and length of protrusions positive for Wnt5b per endodermal cell during a 5-min window, in the indicated embryos, with the number of embryos, cells, and protrusions analyzed indicated. Data are mean ± SEM. **, P < 0.01; ****, P < 0.0001; unpaired Student's t test.
Video 2.
GFP-Gpc4–labeled protrusions emanated from endodermal cells transport Wnt5b-mCherry. Confocal time-lapse experiments were performed on Tg(sox17:GFP-gpc4) embryos injected with wnt5b-mCherry RNA at 3 somite stage using a Zeiss LSM880 confocal microscope with a LD C-Apo 40×/NA 1.1 water objective (shown in Fig. 7 A). Z-stacks of 10.2 µm were acquired at 0.6 µm intervals every 30 s using the following settings: Fast Airyscan mode, zoom 1.3, 1,024 × 1,024 pixels, 4 averaging. The movies were generated from four z-planes and are played at two frames/s. Yellow arrowheads, Wnt5b-mCherry–expressing puncta (in magenta) on the extending protrusions; white arrowheads, Wnt5b-mCherry on the retracting protrusions; cyan arrowheads, Wnt5b-mCherry at protrusions from two cells merging or connected.
Video 3.
GFP-Gpc4–labeled protrusions emanated from endodermal cells transport Wnt11f2-mCherry. Confocal time-lapse experiments were performed on Tg(sox17:GFP-gpc4) embryos wnt11f2-mCherry RNA at 3 somite stage using a Zeiss LSM880 confocal microscope with a LD C-Apo 40×/NA 1.1 water objective (shown in Fig. 7 B). Z-stacks of 14.45 µm were acquired at 0.85-µm intervals every 30 s using the following settings: Fast Airyscan mode, zoom 1.3, 1,024 × 1,024 pixels, 4 averaging. The movies were generated from five z-planes and are played at two frames/s. Yellow arrowheads, Wnt11f2-mCherry–expressing puncta (in magenta) on the extending protrusions.
Video 4.
Gpc4 is required for the formation of Wnt5b-labeled protrusions in endodermal cells. Confocal time-lapse experiments were performed on gpc4−/−/Tg(sox17:memGFP)/ Tg(sox17:H2AmCherry) embryos injected with Wnt5b-mCherry using a Zeiss LSM880 confocal microscope with a LD C-Apo 40×/NA 1.1 water objective (shown in Fig. 7, C and D). Z-stacks of 13.5 µm were acquired at 1.5-µm intervals every 30 s using the following settings: zoom 1.0, 1,024 × 1,024 pixels, 9 speed, 4 averaging. The movies were generated from four z-planes and are played at two frames/s. Yellow arrowheads, Wnt5b-mCherry–expressing puncta (in magenta) on memGFP-labeled filopodia in endoderm cells (in white).
Endodermal GFP-Gpc4–labeled signaling filopodia contribute to the rescue of mesodermal and ectodermal defects in gpc4−/− embryos
To further demonstrate that Wnt’s are delivered to neighboring cells on filopodia, we generated wnt11f2-mNeonGreen and wnt5b-mNeonGreen constructs for in vivo imaging. Like the wnt-Myc constructs, these mNeonGreen constructs displayed the predicted cellular localizations (Fig. S2, D–E′) and were functional (Fig. S2, J and M). We injected distinct cells of embryos at the 16-cell stage with RNAs encoding mem-mCherry and wnts-mNeonGreen and RNA encoding mem-TagBFP, respectively (Fig. S4 C), so that one subset of cells expressed mem-mCherry and Wnt11f2-mNeonGreen or Wnt5b-mNeonGreen and another subset expressed mem-TagBFP. We performed time-lapse imaging of embryos at 50% epiboly, focusing in the area where these two populations are in close proximity. We found that the cellular protrusions extending from mem-mCherry-expressing cells were able to transport Wnt11f2-mNeonGreen or Wnt5b-mNeonGreen to the neighboring BFP-expressing cells (Fig. S4, D and E; and Video 5). These results indicate that Wnt11f2-mNeonGreen and Wnt5b-mNeonGreen can be deposited from the producing cells to the receiving cells.
Video 5.
Mem-mCherry labeling protrusions from Wnt-mNeonGreen-expressing cells deliver Wn5b-mNeonGreen or Wn11f2-mNeonGreen to BFP-expressing receiving cells. Confocal time-lapse experiments were performed on WT embryos mosaically injected with various RNAs at 50% epiboly, using a Zeiss LSM880 confocal microscope with a LD C-Apo 40×/NA 1.1 water objective (shown in Fig. S4, C and D). Z-stacks of 22–41.25 µm were acquired at 1–1.65-µm intervals every 60 s using the following settings: Fast Airyscan mode, zoom 1.0–1.1, 1,064 × 1,064 to 1172 × 1172 pixels, 2 averaging. The movies were generated from two or three z-planes and are played at two frames/s. White arrowheads, Wnt-mNeonGreen–expressing puncta on mem-mCherry–labeled protrusions.
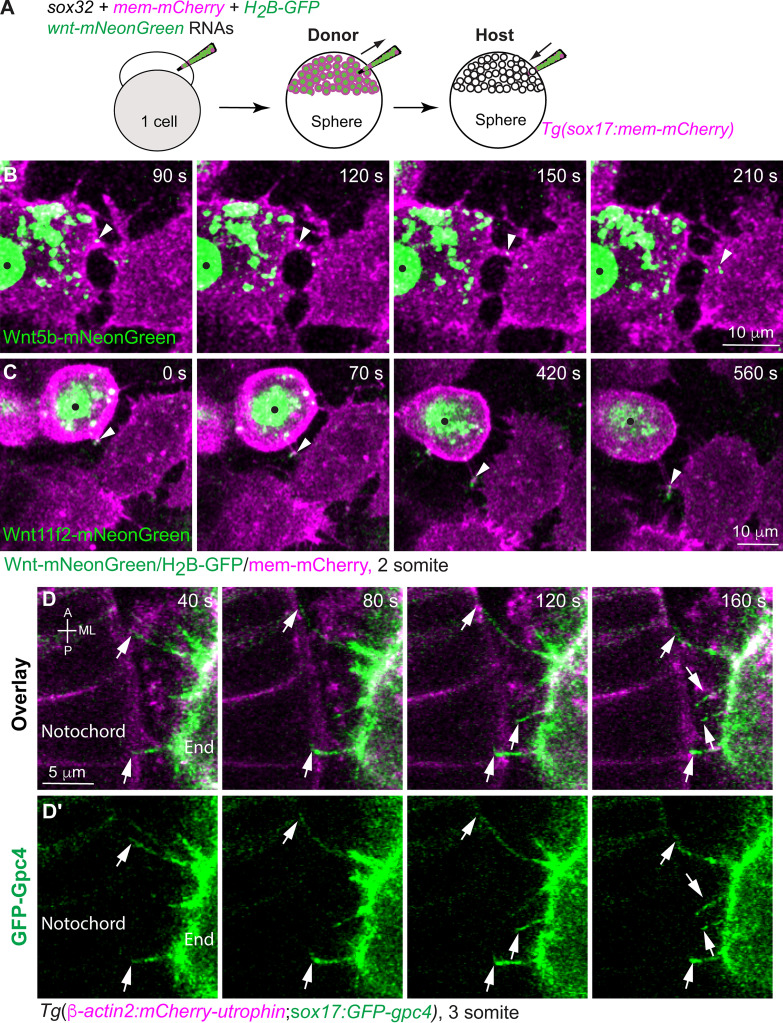
Additionally, we conducted endoderm transplantation to determine if Wnt’s can be delivered by endoderm cells. Briefly, donor embryos were injected with RNAs encoding mem-mCherry, H2B-GFP, wnt5b-mNeonGreen, or wnt11f2-mNeonGreen, and sox32 (to confer an endodermal identity to all cells). Thus, the donor endodermal cells, whose plasma membrane and nuclei were labeled with mCherry and GFP, respectively, expressed Wnt5b-mNeonGreen or Wnt11f2-mNeonGreen. Cells from the donor embryos were transplanted into Tg(sox17:mem-mCherry) embryos in which endodermal cells were labeled with mem-mCherry (Fig. 8 A). Like blastula cells, Wnt-expressing donor cells extended cellular protrusions and transported Wnt5b-mNeonGreen or Wnt11f2-mNeonGreen to the neighboring endodermal cells (Fig. 8, B and C; and Video 6). However, due to technical issues, we cannot label Wnt’s, endoderm, and mesoderm cells with three different fluorescent proteins. Nevertheless, by labeling endodermal cells and notochord, we were able to view the protrusions extending from endodermal cells making contact with notochord cells (Fig. 8 D, white arrows; and Video 7). These data indicate that endodermal cells directly contact neighboring mesoderm cells.
Figure 8.
Endodermal cells extend cellular protrusions to the neighboring endodermal and mesodermal cells. (A) Schematic illustrating endoderm transplantation, in which Wnts-mNeonGreen–expressing donor cells were transplanted into Tg(sox17:mem-mCherry) hosts. (B and C) Snapshots from confocal time-lapse imaging, showing that cellular protrusions extending from wnt5b-mNeonGreen–expressing (B) or wnt11r2-mNeonGreen–expressing donor endodermal cells (C; black dots) transport Wnt5b-mNeonGreen (B) or Wnt11f2-mNeonGreen (C) to neighboring endodermal cells (Video 6). Arrowheads, Wnt puncta on protrusions. (D–D′) Snapshots from confocal time-lapse imaging on Tg(β-actin2:mCherry-utrophin;sox17:GFP-gpc4) embryos in which the plasma membrane of notochord cells and endodermal cells was labeled with mCherry (Video 7). Images were taken on the region where endodermal cells and the notochord were in close proximity, showing that GFP-Gpc4–labeled protrusions from endodermal cells (D′, arrows) extended toward and contacted mCherry-Utrophin–expressing notochord cells (D). End, endoderm cell.
Video 6.
Protrusions from Wnt-mNeonGreen–expressing donor endodermal cells deliver Wn5b-mNeonGreen or Wn11f2-mNeonGreen to neighboring host endodermal cells. Confocal time-lapse experiments were performed on Tg(sox17:mem-mCherry) embryos with Wnt-expressing donor endodermal cells at 2 somite stage using a Zeiss LSM880 confocal microscope with a LD C-Apo 40×/NA 1.1 water objective (shown in Fig. 8, A–C). Z-stacks of 12.6–22.8 µm were acquired at 0.9–1.2-µm Z intervals every 25–35 s using the following settings: Fast Airyscan mode, zoom 1.2, 1,012 × 1,012 pixels, 4 averaging. The movies were generated from three or four z-planes and are played at two frames/s. White arrowheads, Wnt-mNeonGreen–expressing puncta on mem-mCherry–labeled protrusions extending from endodermal cells.
Video 7.
GFP-Gpc4–labeled protrusions extending from endodermal cells contact mCherry-utrophin-expressing notochord cells. Confocal time-lapse experiments were performed on Tg(β-actin2:mCherry-utrophin;sox17:GFP-gpc4) embryos at 3 somite stage using a Zeiss LSM880 confocal microscope with a LD C-Apo 40×/NA 1.1 water objective (shown in Fig. 8 D). Z-stacks of 28.5 µm were acquired at 1.5-µm intervals every 40 s using the following settings: Fast Airyscan, zoom 1.5, 1,012 × 1,012 pixels, 2 averaging. Five z-planes were stacked to generate the movie, which is played at two frames/s. White arrowheads, GFP-Gpc4–labeled protrusions from endodermal cells extending toward and contacting mCherry-utrophin–expressing notochord cells.
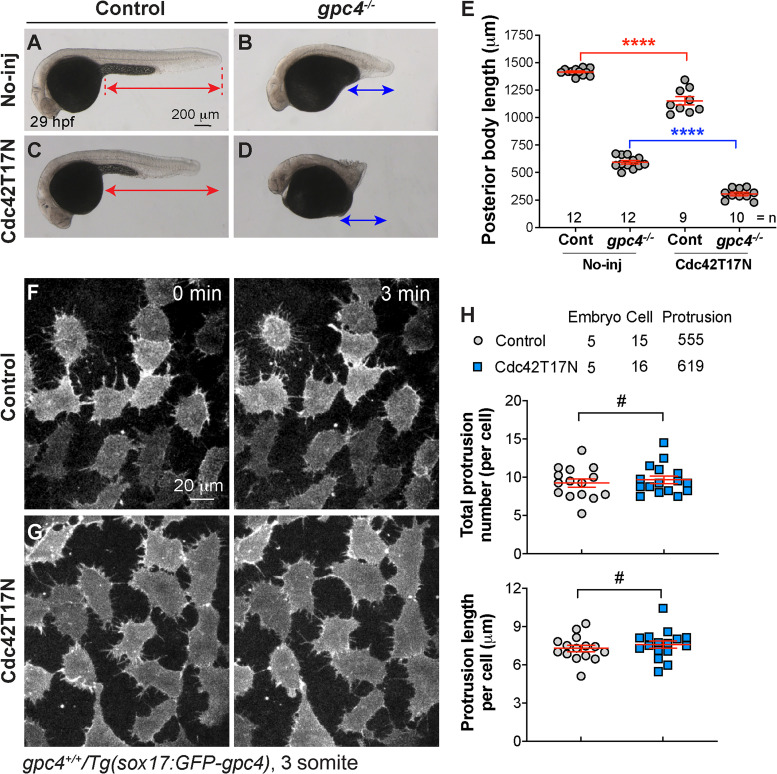
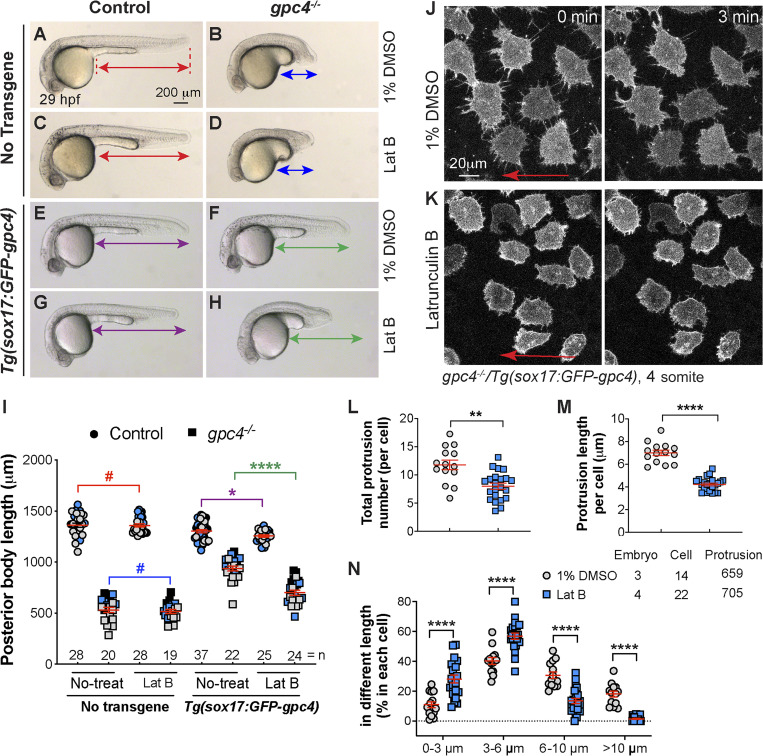
Next, we sought to determine whether signaling filopodia contribute to the rescue of mesoderm and ectoderm by endodermally expressed GFP-Gpc4 in gpc4 mutants. The small Rho GTPase Cdc42 is critical for the formation of filopodia in vitro (Kozma et al., 1995; Nobes and Hall, 1995). Recent studies in zebrafish showed that interference with Cdc42 activity by overexpression of dominant-negative Cdc42 (Cdc42T17N) prevents the formation of signaling filopodia in vivo (Cayuso et al., 2016; Stanganello et al., 2015). We tested whether such interference with Cdc42 activity could suppress the rescue of body length defects in the context of endodermal expression of Gpc4. Injection of control siblings and gpc4−/− embryos with a high dose (250 pg) of the cdc42T17N RNA impaired mesodermal C&E in both (Fig. S5, A–E), whereas injection of such embryos with a subdose (120 pg) of this RNA had little impact on the body axes (as shown in Fig. 9, A–D, and I). In the Tg(sox17:GFP-gpc4) background, injection of a subdose of the cdc42T17N RNA also had no effect on control embryos (Fig. 9 G versus E, I), but it significantly decreased posterior body length in the gpc4−/− embryos relative to that in the controls (Fig. 9 H versus F, I). Thus, inhibition of Cdc42 activity partially suppressed the rescue by endodermal expression of GFP-Gpc4 in gpc4−/− embryos. Furthermore, confocal live imaging showed that injection of a subdose of the cdc42T17N RNA had little effect on GFP-Gpc4–labeled cellular protrusions in WT embryos (Fig. S5, F–H) but produced shorter and fewer and filopodia in gpc4−/−/Tg(sox17:GFP-Gpc4) embryos than in uninjected counterparts (Fig. 9, J–L; and Video 8). Similar results were observed in embryos treated with a low dose of latrunculin B (Lat B), a well-characterized inhibitor of actin polymerization. In the no-transgene background, a subdose of Lat B did not affect posterior body length in either WT or gpc4−/− embryos (Fig. S6, A–D and I). In the Tg(sox17:GFP-gpc4) background, such treatment resulted in slight shortening of the posterior body in WT embryos (Fig. S6 G versus E, I) and more significant shortening in gpc4−/− embryos (Fig. S6 H versus F, I). In the latter case, the rescue effect of the transgene was abolished. As expected, this Lat B treatment also inhibited the formation of protrusions in gpc4−/−/Tg(sox17:GFP-gpc4) embryos (Fig. S6, J–N; and Video 9). Thus, the rescue of the body length in gpc4−/− embryos by endodermal expression of GFP-Gpc4 was suppressed by the expression of Cdc42T17N or Lat B treatment, likely due to the suppression of filopodia formation. Collectively, our data suggest that filopodia from GFP-Gpc4–expressing endodermal cells are critical for the observed rescue effects.
Figure S5.
Injection of cdc42T17N RNA at a high does causes short body axes in both control siblings and gpc4−/− embryos, and injection of a subdose does not disrupt the formation of protrusions in Tg(sox17:GFP-gpc4) embryos. (A–D) Bright-field images of the indicated embryos injected with a high dose of cdc42T17N RNA (250 pg). Lines with double arrows indicate length of the posterior body axis; lines of the same color are equal in length. (E) Average posterior body length in embryos shown in A–D. Colors of the P values correspond to the embryos in which the posterior body is marked with lines of the same color. (F and G) Snapshots from confocal time-lapse imaging performed on Tg(sox17:GFP-gpc4) embryos injected with a subdose of cdc42T17N RNA (120 pg). (H) Total number and length of protrusions in each endodermal cell of the embryos indicated shown in F and G. The number of embryos, cells, and protrusions analyzed is indicated. Data are mean ± SEM. #, P > 0.05; ****, P < 0.0001; unpaired Student's t tests.
Figure 9.
Suppression of GFP-Gpc4–labeled protrusions by expression of dnCdc42 partially blocks rescue by endodermal expression of Gpc4. (A–H) Bright-field images of the indicated embryos. Lines with double arrows show the length of the posterior body; lines of the same color are equal in length. (I) Average posterior body length in embryos shown in A–H from two independent experiments (represented by different color symbols), with the number of embryos indicated. Colored P values correspond to differences between the embryos in which the posterior body is marked with lines of the same color. (J and K) Snapshots from confocal time-lapse imaging performed on the indicated embryos (Video 8). Arrows indicate the direction of migration of the endodermal cells. (L) The total number of protrusions, the length of the protrusion, and the percentages of protrusions of different lengths (grouped into 3-µm bins) in each endodermal cell. The number of embryos, cells, and protrusions analyzed is indicated. Data are mean ± SEM. #, P > 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; unpaired Student's t test.
Video 8.
Formation of GFP-Gpc4 labeled protrusions in gpc4−/− embryos is suppressed by Cdc42T17N. Confocal time-lapse experiments were performed on gpc4−/−/Tg(sox17:GFP-gpc4) embryos at 3 somite stage using a Zeiss LSM880 confocal microscope with a LD C-Apo 40×/NA 1.1 water objective (shown in Fig. 9, J and K). Images were acquired at 30-s intervals using the following settings: zoom 1.0, 1,024 × 1,024 pixels, 9 speed, 4 averaging. The movie is played at five frames/s.
Figure S6.
Inhibition of actin polymerization by Lat B blocks the rescue mediated by endodermal expression of GFP-Gpc4. (A–H) Bright-field images of the indicated embryos. Lines with double arrows indicate length of the posterior body axis; lines of the same color are equal in length. (I) Average posterior body length in embryos shown in A–H, from three independent experiments (represented by different color symbols), with the number of embryos indicated. Colors of the P values correspond to the embryos in which the posterior body is marked with lines of the same color. (J and K) Snapshots from confocal time-lapse imaging performed on gpc4−/−/Tg(sox17:GFP-gpc4) embryos treated with 1% DMSO and Lat B (0.15 µg/ml; Video 9). Red arrows, direction of migration of the endodermal cells. (L–N) The total number of protrusions (L), the length of the protrusion (M), and the percentages of protrusions of different lengths (grouped into 3-µm bins; N) in each endodermal cell. The number of embryos, cells, and protrusions analyzed is indicated. Data are mean ± SEM. #, P > 0.05; *, P < 0.05; **, P < 0.01, ****, P < 0.0001; unpaired Student's t test.
Video 9.
GFP-Gpc4-labeled protrusions in in gpc4−/− embryos are suppressed by Lat B. Confocal time-lapse experiments were performed on gpc4−/−/Tg(sox17:GFP-gpc4) embryos at 4 somite stage using a Zeiss LSM880 confocal microscope with a LD C-Apo 40×/NA 1.1 water objective (shown in Fig. S6, J and K). Images were acquired at 30-s intervals using the following settings: zoom 1.0, 1,024 × 1,024 pixels, 9 speed, 4 averaging. The movie is played at five frames/s.
Endodermal expression of GFP-Gpc4 activates JNK to rescue mesodermal C&E
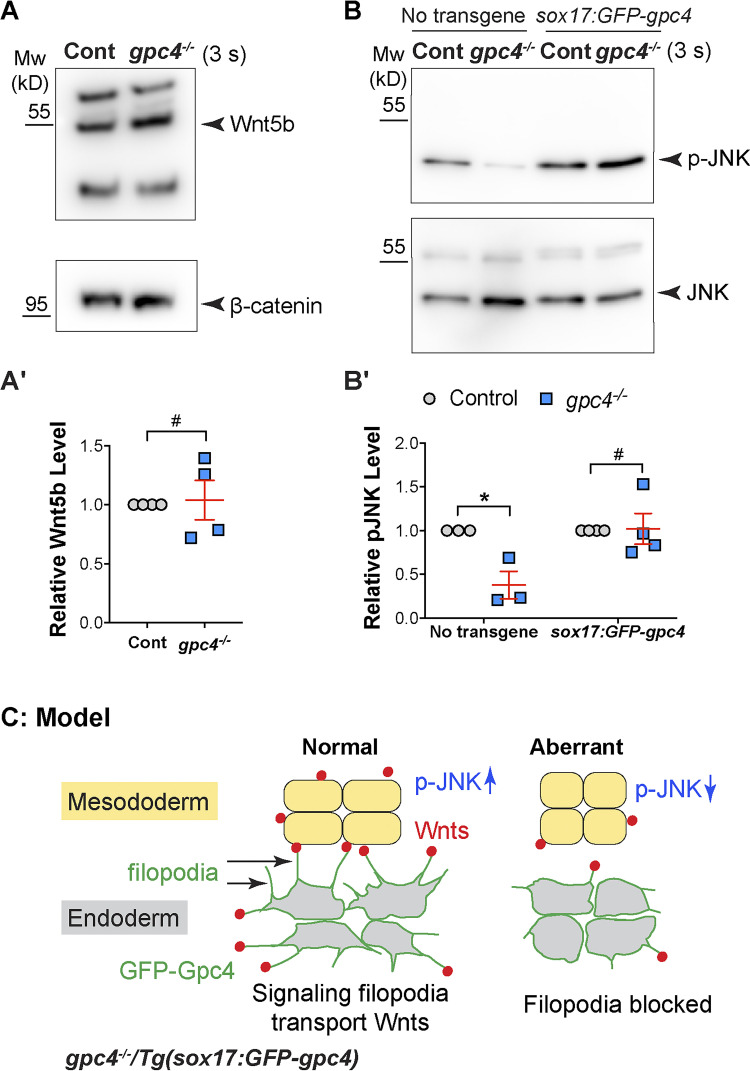
It has been shown that Gpc4 regulates mesodermal C&E by promoting Wnt/PCP signaling (Topczewski et al., 2001). Our results indicated that the delivery of Wnt5b and Wnt11f2 to the mesoderm by endodermal GFP-Gpc4 is responsible for the rescue of mesodermal C&E in gpc4−/− embryos. However, we cannot examine the expression patterns of endogenous Wnt5b and Wnt11f2 in embryos because we lack suitable antibodies. Using one antibody that detected zebrafish Wnt5b by Western blotting, we found that the expression of Wnt5b was not changed in gpc4−/− embryos (Fig. 10, A and A′), indicating that Gpc4 might affect the pattern, but not level, of Wnt5b expression. Next, we aimed to assess the effects of gpc4 deficiency on Wnt/PCP signaling. Wnt/PCP signaling can activate JNK by causing its phosphorylation (van Amerongen and Nusse, 2009; Yamanaka et al., 2002). Measurement of phosphorylated JNK (p-JNK) revealed that p-JNK expression was significantly reduced in gpc4−/− versus control embryos and that this reduction was reversed in gpc4−/−/Tg(sox17:GFP-gpc4) embryos (Fig. 10, B and B′). These data suggest that in gpc4−/− embryos, reduced Wnt/PCP signaling could be responsible for the C&E defects and that endodermal expression of GFP-Gpc4 restored Wnt/PCP signaling.
Figure 10.
Endodermal expression of GFP-Gpc4 restored JNK activation in gpc4 mutants. (A and B) The expression of Wnt5b and β-catenin (A), as well as p-JNK and JNK (B), as detected by Western blotting in the indicated embryos at 3 somite (s) stage from three or four independent experiments. (A′) Relative expression levels of Wnt5b versus β-catenin. (B′) Relative expression levels of p-JNK versus JNK. (C) Model of how endodermal GFP-Gpc4–labeled filopodia transport Wnt ligands to activate JNK in the mesoderm. Data are mean ± SEM. #, P > 0.05; *, P < 0.05; unpaired Student's t test.
Discussion
Our study leads us to propose a model whereby Gpc4 elicits its non–cell-autonomous functions by regulating the formation of signaling filopodia. In gpc4−/− embryos, endodermal filopodia that expressed Gpc4-GFP transported Wnt proteins to neighboring tissues to restore JNK activation and rescued mesodermal C&E defects; when the filopodia formation was blocked, these rescue effects were suppressed (Fig. 10 C).
Endodermal expression of GFP-Gpc4 partially rescues C&E defects in other germ layers
We provide multiple lines of evidence showing that in gpc4−/− embryos, expression of a GFP-gpc4 transgene specifically in the endoderm not only completely rescues C&E defects in this tissue but also partially rescues those in the mesoderm and ectoderm. We further show that this rescue stems from the endoderm, as depletion of the endoderm (sox32−/− background) abolished such rescue. Thus, Gpc4 functions both cell and non–cell autonomously. However, the fact that the phenotypic rescue in mesoderm and ectoderm was partial suggests that Gpc4 delivered from the endoderm is not able to play all of the roles of Gpc4 produced by those tissues.
Gpc proteins interact with signaling molecules to influence their pathways. In zebrafish, Gpc4 cooperates with Wnt11f2 to regulate mesodermal C&E (Topczewski et al., 2001), but the underlying mechanisms are not clear. Our study shows that Gpc4 can physically bind both Wnt5b and Wnt11f2 and that partial inhibiting the expression of both Wnt5b and Wnt11f2 using MOs at doses that do not cause significant defects in body length can suppress Gpc4-mediated rescue of mesodermal C&E defects in gpc4−/− embryos, suggesting that Gpc4 interacts with both Wnt5b and Wnt11f2 to influence mesodermal C&E defects. Furthermore, our finding that the total Wnt5b expression is not affected in gpc4−/− embryos suggests that Gpc4 instead interferes with the distribution and/or function of Wnt’s. Thus, it is possible that in gpc4−/− embryos the concentration of Wnt proteins in the mesoderm is reduced and that transportation of these proteins from the endoderm to the mesoderm helps restore the distribution of Wnt5b and thus also Wnt signaling. Our study does not shed light on the distribution of these endogenous ligands in vivo because of the failure of available antibodies to detect endogenous Wnt in whole-mount embryos; an alternative future approach will be the generation of knock-in reporter lines for wnt5b and wnt11f2. A second potential explanation for the reduced Wnt function observed in the absence of Gpc4 is that the latter serves as a coreceptor for Wnt’s (Franch-Marro et al., 2005) and that Gpc4 produced in the endoderm can compensate partly, but not completely, for its loss in the other germ layers. Future studies will test mesodermal expression of Gpc4 for the ability to rescue the endodermal defects in gpc4−/− embryos and also determine whether and how Gpc4 regulates the distributions and functions of Wnt proteins.
GPI cleavage is not necessary for the non–cell-autonomous functions of Gpc4
Gpc’s function both cell autonomously and non–cell autonomously. However, little is known about how they achieve their non–cell-autonomous functions. Our finding that endodermal expression of Gpc4 rescues the C&E defects in other germ layers provides an unprecedented opportunity to study the role of Gpc4 in communication among tissues.
The GPI anchors of Gpc’s can be cleaved to generate soluble forms of these proteins that can be released from the producing cells to the neighboring cells (Häcker et al., 2005; Lin, 2004). Our identification of Gpc4 fragment AA517-557 as a potential GPI anchor region is consistent with endodermal Gpc4 being cleaved and diffused to other germ layers. However, our surprising discovery that transgenic expression of even a membrane-bound form of Gpc4 in the endoderm rescued mesodermal C&E defects in gpc4−/− embryos suggests that cleavage of the GPI anchor is not necessary for its function in other germ layers. Two additional findings using a Gpc4 form that lacks the GPI anchor region (Gpc4Δ517–557) indicate that a membrane anchor is essential for effective Gpc4 function: Gpc4Δ517–557 failed to rescue gpc4−/− embryos, and Gpc4Δ517–557 overexpression caused C&E defects (data not shown). We speculate that when this form of the protein is present in the intracellular matrix, it interferes with the function of endogenous Gpc4 or Wnt/PCP signaling.
Endodermal, Gpc4-labeled signaling filopodia could be responsible for mesodermal C&E rescue
Accumulating evidence shows signaling molecules including Wnt’s can be delivered from one tissue to another one to exert their functions (González-Méndez et al., 2019; Kornberg and Roy, 2014; Stanganello and Scholpp, 2016). For example, recent studies revealed that zebrafish blastula cells can activate Wnt pathways in their neighbors by extending signaling filopodia that deliver Wnt8a (Stanganello et al., 2015). Our study builds on these findings, demonstrating that filopodia extended by blastula cells can bind to and deposit Wnt5b and Wnt11 to neighboring cells and that not only blastula cells but also endodermal cells extend signaling filopodia and rely on this ability for communication between tissues. These conclusions are supported by our findings that filopodia emanating from endodermal cells transported Wnt5b and Wnt11f2 to other endodermal cells and endodermal cells extended protrusions toward, and made contact with, neighboring notochord.
Studies in Drosophila showed that another Gpc protein, Dlp, decorates filopodia and is required for spreading of signaling filopodia (González-Méndez et al., 2017). A recent study showed that both Dlp and human GPC4 serve as reservoirs of lipid moieties, which are needed to solubilize and transport Wnt’s (McGough et al., 2020). However, whether Gpc’s regulate the distribution of Wnt’s by affecting signaling filopodia remained unknown. Our study provides evidence for a such a role for Gpc4 in transporting Wnt proteins. Our in vivo imaging shows that Gpc4 localizes to signaling filopodia that can bind and deliver Wnt5b-mCherry and Wnt11f2-mCherry to neighboring cells and that Gpc4 is required for the generation of long and productive protrusions. The discovery that Gpc4 could regulate Wnt distribution by participating in filopodia formation is consistent with a previous study indicating that Wnt/PCP signaling can induce filopodia formation in zebrafish embryos and a fibroblast (PAC2) line as well as in cancer cell lines and human organoids (Mattes et al., 2018). Thus, an involvement of Wnt/PCP signaling in the formation of signaling filopodia could contribute to many other developmental processes regulated by Wnt/PCP signaling.
In our study, the reduction in the proportion of long filopodia and number of mCherry-Wnt5b–bound filopodia in gpc4−/− mutant embryos could have reduced the efficiency of ligand delivery to distant sites. It is also possible that Gpc4 is needed to stabilize or elongate the protrusions. Intriguingly, in endodermal cells, GFP-Gpc4–labeled protrusions from endodermal cells not only extended and delivered Wnt5b to the neighboring cells but also retracted, in some cases bringing mCherry-Wnt5b back into the cell of origin. These behaviors of filopodia could potentially contribute to the regulation of local concentrations of morphogens, and perhaps gradients, in the tissues. Anchoring of Wnt proteins to filopodia could be one of the mechanisms whereby Gpc’s influence Wnt distribution. Given lack of Wnt antibodies that can detect endogenous proteins, the testing of this hypothesis will have to await the generation of appropriate knock-in lines.
Our data indicate that Wnt’s transported by endodermal signaling filopodia are likely responsible for the observed rescue. This is supported by the observation that the rescue was significantly impaired when the formation of filopodia was suppressed by either the expression of Cdc42T17N or treatment with Lat B. Our data also support the notion that Wnt/PCP signaling mediated by GFP-Gpc4 expression was responsible for the rescue, because JNK activation was restored in rescued embryos. Collectively, these findings reveal a novel mechanism whereby Gpc4 influences signaling pathways at a distance in vivo, i.e., by forming signaling filopodia to transport signaling molecules.
Materials and methods
Zebrafish strains and maintenance
Zebrafish were maintained according to animal protocols approved by the University of Iowa Animal Care and Use Committee. Embryos were obtained by natural spawning and staged according to morphological criteria or hpf at 28 or 32°C unless otherwise specified. The following zebrafish lines were used in this study: AB*/Tuebingen, Tg(sox17:mem-mCherry; Ye et al., 2015), gpc4/knypekfr6 (Topczewski et al., 2001), sox32/casanovas4 (Kikuchi et al., 2001), wnt11f2/silberblicktz216 (Heisenberg et al., 2000), and wnt5b/pipetailti265 (Hammerschmidt et al., 1996). To genotype mutants, PCR amplicons were amplified from genomic DNAs were digested with restriction enzymes for specific patterns. For gpc4fr6 mutants, an amplicon generated using the primers 5′-GACCAATCAAGGCTTATCTTC-3′ and 5′-AACTAACAATTAAGGAGGGCTA-3′ was digested with ClaI, producing 323-bp and 206-bp bands from WT embryos and a 529-bp band from mutants. For sox32s4 mutants, an amplicon generated from the primers 5′-TACATGCAAGAAGCAGAAAGACTACGGATCCAGG-3′ and 5′-ATGTTGCCTCGAAGTGGTATGATGAAGAGTGGTT-3′ was digested with KpnI, which produced a band at 271 bp from WT embryos and bands at 233 bp and 38 bp from the mutants. For wnt11f2tz216 mutants, an amplicon generated from the primers 5′-TAGTATTTGGGTGATTCCATTAGG-3′ and 5′-GTGGTTGAGGCTTTACCTGTCT-3′ was digested with FokI, which produced bands at 403 bp and 134 bp from WT embryos and a 537-bp band from the mutants. For wnt5bti265 mutants, an amplicon generated from the primers 5′-GTCTCTGGGCACCCAAGGCCGCCTATGC-3′ and 5′-CAAACTGGTCTACGAGTGACGTGCAGCGTTTGCTC-3′ was digested with XbaI, which produced a single band at 185 bp from WT embryos and bands at 147 bp and 38 bp from the mutants.
Sequence alignment
Alignment of C-terminal amino acids of Gpc4 from mouse (ENSMUST00000033450.2), rat (ENSRNOT00000003282.5), human (ENST00000370828.3), Xenopus (ENSXETT00000011898.2), and zebrafish (ENSDART00000026569.8) was performed in Clustal X, a multiple sequence alignment program (Thompson et al., 1997).
Plasmid constructs
To generate GFP-Gpc4 mutant constructs, we used GFP-gpc4/pCS2Dest plasmid as a template (this construct expresses Gpc4 in which EGFP is inserted after N-terminal signal peptide [AA1-23] of Gpc4; Hu et al., 2018). To generate GFP-gpc4-GPI (Δ24-516) that expresses the C-terminal GPI attachment signal (AA517-557) of Gpc4, an overlapping extension PCR strategy was used to amplify the coding sequences containing AA1-23 of Gpc4, EGFP, and AA517-557 of Gpc4. The resulting amplicon was digested by BstbI and XhoI and then cloned into pCS2Dest (from GFP-gpc4/pCS2Dest plasmid cut by BstbI and XhoI). To generate GFP-gpc4 (Δ517-557), primers containing BstbI and XhoI restriction enzyme sites were used to amply the region coding AA1-517; the resulting amplicons were cut using BstbI and XhoI and then cloned into the pCS2Dest. We also generated GFP-gpc4Δ517-557-sdc4TM/pCS2Dest construct that expresses Gpc4 AA1-517, the TM domain (T144-173) and a partial intracellular sequence (R174-L185) of zebrafish Syndecan4 (Sdc4; NM_001048149.1). The coding region of sdc4 that expresses AA144-185 was amplified from cDNAs obtained from 18s-zebrafish embryos using the following primers: 5′-CTATACCTGGTACAGAAGTGCTTGCAGCTGTT-3′ and 5′-TATACTCGAGTTACAGGTCGTAACTTCCTTCGTCT-3′ (the underlining indicates the SexAI and XhoI restriction sites, respectively). Sdc4 is shown to bind intracellular signaling molecules at the conserved cytosolic domain (G186-A201; Multhaupt et al., 2009). To avoid such interaction, we removed this domain and included only 12 amino acids of the cytosolic sequence (R174-L185) in our construct. The amplicon encoding sdc4TM (express AA144-185 of Sdc4) was digested with SexAI and XhoI and then cloned into GFP-gpc4/pCS2Dest plasmid following its digestion with SexAI and XhoI.
Flag-Gpc4/pCS2 is a construct (a gift from Dr. Jacek Topczewski, Northwestern University, Evanston, IL), which expresses Gpc4 in which the Flag epitope was inserted after its N-terminal signal peptide. To generate Lifeact-RFP/pCS2, the coding sequence of Lifeact-RFP was amplified from Abp140-17aaRuby-nos1-3′UTR/pCS2 (Kardash et al., 2010) and cloned into the pCS2 vector. To generate mem-TagBFP/pCS2Dest construct, a mem-TagBFP(CAAX)/pME was made from TagBFP/pME (a gift from Dr. Didier Stainier, Max Planck Institute, Munich, Germany) as a template. Primers containing the NcoI and BglII restriction enzyme sites was used to amplify the TagBFP sequences and the amplicons were digested and cloned into pME-EGFPCAAX cut by NcoI and BglII. The mem-TagBFP/pCS2Dest construct was generated by recombining mem-TagBFP/pME into a pCS2Dest vector using LR Clonase Enzyme mix (Invitrogen).
To generate zebrafish wnt5b/pCS2Dest and zebrafish wnt11f2/pCS2Dest, the coding regions of wnt5b and wnt11f2 were amplified using zebrafish wnt5b-Myc/pCS2 (Lin et al., 2010) and wnt11f2-Myc/pCS2 as the templates (both Wnt constructs are gifts from Dr. Diane Slusarski, The University of Iowa, Iowa City, IA), and cloned into pCS2Dest. To generate wnt5b-mNeonGreen/pCS2Dest, wnt11f2-mNeonGreen/pCS2Dest, and wnt11f2-mCherry/pCS2Dest constructs, an overlapping extension PCR strategy was used. Zebrafish wnt5b-Myc/pCS2, wnt11f2-Myc/pCS2, lamp1-mNeonGreen (Addgene; #98882), and mCherry/pME (Ye et al., 2015) were used as the templates. Overlapping primers containing a 39-bp DNA sequence (5′-GGCGGAGGTTCCGGAGGTGGCGGATCAGGAGGAGGTAGT-3′), which encodes a 13-aa linker peptide (GGGSGGGGSGGGS), were used to generate the sequences, resulting a protein product in which the 13-aa linker was inserted between the last coding amino acid of Wnt’s (K363 for Wnt5b and K353 for Wnt11f2) and the first amino acid of fluorescent proteins (V1 for both mNeonGreen and mCherry). The coding sequences was further amplified using the primers containing restriction enzyme sites of BstbI and XhoI (for wnt5b-mNeonGreen), or BstbI and SalI (for wnt5b-mNeonGreen and wnt11f2-mCherry). The amplicons were cloned into the pCS2Dest vector following its digestion with BstbI and XhoI. All the PCRs were performed using a Q5 high-fidelity DNA polymerase (New England Biolabs; M0491S). The constructs were confirmed correct by Sanger sequencing.
Generation of transgenic lines
Tg(sox17:memGFP/H2A-mCherry), Tg(sox17:GFP-gpc4), Tg(sox17:GFP-gpc4Δ517-557-sdc4TM), and Tg(β-actin2:mCherry-utrophin) were generated using a Tol2-based Multi-Site Gateway system (Invitrogen; Kwan et al., 2007; Villefranc et al., 2007). pME-GFP-Gpc4 was a gift from Dr. Jacek Topczewski (Northwestern University, Evanston, IL). The GFP-gpc4Δ517-557-TM/pME was generated by amplifying the coding sequence of GFP-Gpc4Δ517-557-TM from GFP-gpc4Δ517-557-TM/pCS2Dest plasmid (see above) using primers containing the attB sites, and the resulting PCR product was recombined into a pDONR221 vector using BP Clonase II Enzyme mix (Invitrogen). The GFP-gpc4Δ517-557-TM/pME, sox17/p5E (a 5′-entry vector containing a sox17 promoter was used to express genes specifically in the endoderm; Woo et al., 2012), p3E-polyA (a 3′-entry vector), and pDestTol2pA2 (a destination vector) were used for Multi-Site Gateway cloning. For generating Tg(β-actin2:mCherry-utrophin), β-actin2/p5E (a 5′-entry vector containing a ubiquitous promoter), mCherry/pME, and utrophin/p3E (a 3′-entry vector containing an actin-binding motif of human UTROPHIN gene, a gift from Dr. Woo; Woo et al., 2012), and pDestTol2pA2 (a destination vector) were used for Multi-Site Gateway cloning.
The cytoplasm of embryos at the one-cell stage was coinjected with the transgene plasmid DNA (40 pg) and the tol2 mRNA (25 pg). The injected embryos were screened for GFP expression in the endoderm, and those that were GFP positive were raised as F0 founders. The founders were then bred to AB* or Tuebingen WT fish to generate stable lines. For the genotyping of Tg(sox17:GFP-gpc4) and Tg(GFP-gpc4Δ517-557-sdc4TM) fish, an amplicon of 241 bp was generated using the primers 5′-TGTTTACAGTATGTATGTCTGTGGTGG-3′ (which targets the region that expresses the N-terminal signal peptide of Gpc4) and 5′-GTCAGGGTGGTCACGAGGG-3′ (which targets the open frame sequences of GFP).
RNA expression and MO injection
mRNA and MOs were injected into embryos at the one-cell stage at the doses indicated. Capped mRNAs were synthesized using the mMessage mMachine kit (Ambion) and were injected into one-cell embryos. RNAs encoding the following genes were used unless stated elsewhere: mem-mCherry (75 pg), H2A-mCherry (100 pg), H2B-GFP (40 pg), Lifeact-RFP (200 pg), mem-TagBFP (250 pg), sox32 (250 pg), wnt5b-mCherry (Lin et al., 2010; 120 pg for time-lapse experiments, 150 pg for overexpression experiments), wnt5b (150 pg), wnt5b-Myc (150 pg), wnt5b-mNeonGreen (150 pg), wnt11f2 (10 pg), wnt11f2-Myc (10 pg), wnt11f2-mCherry (120 pg for time-lapse experiments, 10 pg for rescue experiments), wnt11f2-mNeonGreen (120 pg for blastula assay, 10 pg for rescue experiments), cdc42T17N (Nobes and Hall, 1995; 120 pg), Flag-gpc4 (60 pg, for rescue experiment), GFP-gpc4Δ517-557-sdc4TM (60 pg for rescue experiment), GFP-gpc4, and GFP-gpc4 truncated constructs (200 pg each for in vivo blastula assay), Previously validated MO antisense oligonucleotides (MOs) targeting the following genes were used: sox32 (4 ng, 5′-CAGGGAGCATCCGGTCGAGATACAT-3′; Wong et al., 2012), wnt5b (1 ng, 5′-GCAAACACAATAATTTCTTACCACC-3′; Cirone et al., 2008), wnt11f2 (5 ng, 5′-ACTCCAGTGAAGTTTTTCCACAACG-3′; Muyskens and Kimmel, 2007), and p53 (1.5 ng, 5′-GCGCCATTGCTTTGCAAGAATTG-3′; Robu et al., 2007). All MOs were coinjected with the p53 MO to inhibit potential p53-dependent cell death induced by MO off-targeting effects (Robu et al., 2007).
RNA isolation and quantitative real-time PCR
RNAs were isolated from WT and Tg(sox17:GFP-gpc4) embryos (grouped by 20–30 embryos) at the 3-somite stage, and cDNAs were synthesized using the iScript Reverse Transcription kit (Bio-Rad Laboratories; #1708840). These cDNAs were then quantitated using real-time PCR and the iQ SYBR Green Supermix (Bio-Rad Laboratories; #1708880). The following primers were used to amplify gpc4 (5′-CAGCTCAAACCCTTCGGAGAC, 5′-CGCTACAGTACGGGCAGTATAACAT and eef1a1a (5′-GAGAAGTTCGAGAAGGAAGC, and 5′-CGTAGTATTTGCTGGTCTCG). Relative expression levels of gpc4 were determined by normalization to the expression to eef1a1a.
WISH and immunofluorescence (IF)
Digoxigenin-labeled antisense RNA probes targeting the following genes were synthesized by in vitro transcription: hgg1 (hatching gland 1, marks prechordal plate), dlx3 (distal-less homeobox 3b, marks neural plate boundary), shh (sonic hedgehog signaling molecule a, marks axial mesoderm; Marlow et al., 1998), foxa3 (forkhead box A3, marks the digestive system; Odenthal and Nüsslein-Volhard, 1998), deltaC (delta-like protein C, marks somites; Haddon et al., 1998), krox-20 (known as early growth response 2b, marks rhombomeres 3 and 5, for the staging purpose; Oxtoby and Jowett, 1993), ntl (tbxta, T-box transcription factor Ta, marks notochord; Schulte-Merker et al., 1994), and gpc4 (Topczewski et al., 2001).
For WISH, embryos were fixed in 4% paraformaldehyde (PFA) overnight at 4°C and washed with PBS/0.1% Tween (PBT). Embryos were then permeabilized by dipping into serial methanol solutions at increasing concentrations, for 5 min each, and stored in 100% methanol at −20°C until they were processed for WISH. They were then rehydrated by dipping into serial methanol dilutions of decreasing concentrations and treated with Proteinase K (10 µg/ml in PBS); times differed according to the age of the embryos (no treatment for embryos at 3-somite stage and 20 min for embryos at 2 dpf). Embryos were then refixed in 4% PFA for 20 min. Embryos were then washed in PBT, incubated in hybridization buffer (65% formamide, 5× saline sodium citrate [SSC], 50 mg/ml heparin, and 500 mg/ml tRNA in PBT) at 70°C for 3 h, and then incubated in hybridization buffer containing RNA probes at 70°C overnight. Embryos were then washed in serial dilutions of hybridization buffer without tRNA (HB−)/SSC at 70°C, as follows: 75% HB−/25% 2x SSC (15 min), 50% HB-/50% 2x SSC (15 min), 25% HB−/75% 2x SSC (15 min), 2x SSC (15 min), 0.05x SSC (2 × 30 min). Additional washes were performed at RT as follows: 75% 0.05x SSC/25% PBT (10 min), 50% 0.05x SSC/50% PBT (10 min), 25% 0.05x SSC/75% PBT (10 min), and PBT (10 min). Embryos were then incubated in blocking buffer (2% sheep serum and 2% BSA in PBT) for 3–4 h at RT and then in blocking buffer containing anti-Digoxigenin antibody (1:5,000; Roche; 11093274910) at 4°C overnight. After six washes in PBT (15 min each) at RT, the embryos were incubated in staining buffer (100 mM Tris-HCl, pH 9.5, 100 mM NaCl, and 50 mM MgCl2 in PBT) for 5 min (three times) and then in staining buffer containing the NBT/BCIP substrates (1:50; Roche; 11681451001) until the desired intensity was reached. For double ISH, a fluorescein-labeled hgg1 probe and an anti-fluorescein antibody (1:5000; Roche; 11426338910) were used; the signal was detected by Fast Red (Roche; 11496549001) staining. Staining was terminated by replacing the staining buffer with the stop solution (20 mM EDTA in PBS, pH 5.5).
For IF assay after gpc4 WISH, embryos were refixed in 4% PFA for 2 h at RT and then embedded in 1.2% agarose containing 5% sucrose. The agarose blocks were dehydrated in 30% sucrose and frozen in 2-methylbutane solution in a stainless steel beaker placed into liquid nitrogen (this makes it possible to freeze the agarose blocks within 1 min). The frozen agarose blocks were cryosectioned at 10 µm thickness using a Thermo Fisher Scientific Microm Cryostat. Sectioned slices were placed on a positively charged microscope slide and incubated with anti-GFP (1:300; sc-8334; Santa Cruz Biotechnology) and an Alexa Fluor 488–conjugated goat anti-mouse secondary antibody (1:200; Invitrogen; A-11001) in blocking solution (0.2% BSA and 2% FBS in PBS).
For IF staining using Myc antibody, embryos at 50% epiboly were dechorionated manually, fixed in 4% PFA, and permeabilized in PBS containing 0.5% Triton X-100. Embryos were then incubated with anti–C-MYC antibody (9E10; 1:200; Developmental Studies Hybridoma Bank; AB2266850) in blocking solution (0.5% BSA, 5% FBS, 2% DMSO, and 0.1% Triton X-100 in PBS) and an Alexa Fluor 488–conjugated goat anti-mouse secondary antibody (1:200; Invitrogen; A-11001). Embryos were counterstained with DAPI (0.2 µg/ml; Thermo Fisher Scientific; D1306) for 10 min and mounted in 2.5% methylcellulose on coverslides.
Cell transfection, immunoprecipitation, and Western blotting
HEK293 cells were transiently transfected with Myc/pCS2 or Myc-Mmp14b/pCS2 (Hu et al., 2018) or wnt5b-Myc/pCS2 (Lin et al., 2010) or wnt11f2-Myc/pCS2, Flag-gpc4/pCS2 Flag-JNK/pCDNA (human c-Jun N-terminal kinase, a gift from Ray Dunn; Young et al., 2014), using a GenJet Plus DNA in vitro Transfection Reagent (SignaGen Laboratories; SL100488) following the manufacturer’s protocol. 36 h after the transfection, the cells were collected in lysis buffer (150 mM NaCl, 50 mM Tris-HCl, pH 7.5, 5 mM EDTA, 0.5% NP-40, and 50 mM NaF; Ohkawara et al., 2003) containing protease inhibitors and subjected to sonication using an ultrasonic processor (Sonics & Materials; GE505) operated at five cycles of 1 s on, 10 s off. For immunoprecipitation, the cell lysates were incubated with Myc monoclonal antibody (9E10; Thermo Fisher Scientific; MA-1-980) coupled to protein G magnetic beads (Thermo Fisher Scientific; 88848) overnight at 4°C. The precipitates were collected using a magnetic separation rack (Thermo Fisher Scientific; CS15000) and washed with lysis buffer six times. Protein samples were resolved by SDS-PAGE and examined by Western blotting using an Amersham imager 600 detection system (GE Healthcare). The following antibodies were used for immunoblotting: anti-Flag M2 (1:2,000; Sigma-Aldrich; F3165), anti–C-MYC (9E10; 1:1,000; Developmental Studies Hybridoma Bank; AB2266850), goat anti-mouse IgG, and light chain–specific HRP conjugate (1:2,000; Jackson ImmunoResearch Laboratories; 115–035-174).
For Western blotting of embryo-derived proteins, embryos were manually dechorinated and pooled in deyolking buffer (55 mM NaCl, 1.8 mM KCl, and 1.25 mM NaHCO3; Link et al., 2006). The embryos were dissociated by pipetting with a 200-ml tip and vortexing for 30 s. The embryo solution was then centrifuged at 300 g for 30 s and washed with washing buffer (110 mM NaCl, 3.5 mM KCl, 10 mM Tris-HCl, pH 8.5, and 2.7 mM CaCl2) twice to remove the yolk. Embryonic cell pellets were collected by centrifuging at 300 g for 1 min and lysed in 2x SDS loading buffer (3 µl per embryo). Protease inhibitors (Roche; 05892970001) were added to the extraction and lysis buffers to prepare samples for p-JNK detection. Lysates representing the equivalent of 6–10 embryos were loaded into polyacrylamide gel wells for electrophoresis. The following antibodies were used for immunoblotting: anti-Wnt5b antibody (1:500; AnaSpec; AS-55880), anti–p-JNK (1:1,000; Cell Signaling Technology; #4668), anti-JNK (1:1,000; Cell Signaling Technology; #9252S), and anti–β-catenin (1:1,000; Sigma-Aldrich; C7207).
To quantify the intensity of bands in Western blotting assays, a “region of interest” tool in Fiji software was used. The region of the band (band) and a blank region right below the band (blank) were selected using a rectangle box, respectively. Mean gray value (MG) was measured in the selected areas. The intensity (I) of the individual band was calculated as MGband − MGblank. The “relative Wnt5b level” was obtained by dividing the IWnt5b by Iβ-catenin, while “the relative p-JNK level” was obtained by dividing the IpJNK by IJNK. The levels in the mutant embryos were calculated by normalizing that in the control embryos.
Endoderm transplantation
Endoderm transplantation was performed using a pneumatic microinjector (Narishige; 16375). Briefly, donor embryos at the one-cell stage were injected with RNAs encoding sox32 (250 pg, to confer an endodermal identity to all cells), mem-mCherry (120 pg), and H2B-GFP (40 pg, as lineage tracers), as well as wnt5b-mNeonGreen (300 pg) or wnt11-mNeonGreen (150 pg). At the sphere stage, 30–50 donor cells were transplanted into the host embryos along the blastoderm margin. Host embryos were screened for H2B-GFP labeling cells in the posterior endoderm before time-lapse imaging was initiated at 2-somite stage.
Lat B treatment
Embryos were dechorionated in glass dishes and treated with Lat B (Sigma-Aldrich; 428020), an inhibitor of actin polymerization, at a dose of 0.15 µg/ml (in 0.3x Danieau buffer with 1% DMSO) or with 0.3x Danieau buffer containing 1% DMSO. To evaluate its effects on body axis, embryos were treated from 80% epiboly to 29 hpf at 28°C and then subjected to bright-field imaging. To assess its effects on cellular protrusions, embryos were treated from 80% epiboly to 2-somite stage at 28°C and then washed thoroughly with 0.3x Danieau buffer before mounting for confocal time-lapse experiments.
Microscopy and image analysis
For still imaging, fixed or live embryos were mounted in 2.5% methylcellulose. Still epifluorescence images were acquired using a Leica DMI6000 microscope with a 5×/NA 0.15 or 10×/NA 0.3 objective. WISH and bright-field images were acquired using a Leica M165FC stereomicroscope with a Leica DFC290 color digital camera. All images were acquired using the Leica LAS X program. To image ISH sections, samples were mounted in 90% glycerol/PBS medium containing 0.2% propyl gallate and photographed using a Nikon Microphot-FX microscope and a Nikon Plan 20×/NA 0.5 objective, using the NIS-Elements acquisition program. Confocal images for the in vivo protein localization assay and fixed sample were taken on a laser-scanning confocal inverted microscope (Carl Zeiss; Zeiss LSM880) with EC Plan-Neo 40×/NA 1.3 oil or LD C-Apo 40×/NA 1.1 water objectives. Z-stacks were acquired at optimal intervals using the following settings: 1,024 × 1,024 pixel, 9 speed, 4 averaging.
For confocal time-lapse imaging, embryos were embedded in 0.7% (for embryos aged to <10 hpf) or 1% (for embryos aged >10 hpf) low-melting-point agarose using glass-bottom dishes, and images were taken at 28°C using a Zeiss LSM880 with a LD C-Apo 40×/NA 1.1 water objective and a temperature-controlled stage. Endoderm cells in the posterior region of embryos were focused for imaging cell protrusions. Confocal time-lapse imaging was performed using the regular or Fast Airyscan scanning mode. Regular scanning used the following settings: 1,024 × 1,024 pixels, 9 speed, 4 averaging. In most cases, z-stacks (13–15 µm) were acquired to cover the endoderm at 1- to 1.5-µm intervals. Images acquired using the Fast Airyscan scanning mode were subjected to deconvolution with the ZEN (Zeiss) software using the “Airyscan processing” tool and the default setting.
Images of the same type were acquired using the same settings, and all images were processed using Fiji software, edited, and compiled using Adobe Photoshop and Adobe Illustrator software. All analyses of the protrusions were repeated independently by different laboratory members in a blinded fashion. The average of two independent analyses is presented in the final results. To evaluate the anterior–posterior axis, we used an easy and accurate method to quantify the posterior body length, tracing from the starting point of the yolk extension to the tip of the tail using the segmented-line or straight-line tool in Fiji software. To assess the average number and length of cell protrusions in each cell, snapshots were taken from the confocal time-lapse movies every 3 min. The number and the length of the protrusions were qualified by Fiji software using a region of interest tool. The length of cell protrusion was measured from the starting point on cell membrane to the tip of the protrusion using a straight-line tool in Fiji software. Images of the region of interest were saved for validation. To assess the number of Wnt5b-binding cell protrusions, images of z-planes that cover the protrusions were stacked. The protrusions in each cell were manually tracked from 9-min movies at 30-s intervals. Images of each of the counted protrusions were saved for validation.
Statistical analysis
Data were compiled from two to three independent experiments and are presented as the mean ± SEM. Data distribution was assumed to be normal, but this was not formally tested. Statistical analyses were performed in GraphPad Prism (GraphPad Software) using unpaired two-tailed Student's t tests with unequal variance, and P < 0.05 was considered significant. Different symbols are used in the figures to show the different P values (#, P > 0.05; *, P < 0.05; **, P < 0.01; ****, P < 0.0001). The number of cells and embryos analyzed in each experiment is indicated in the figure legends.
Online supplemental materials
Fig. S1 shows that the gpc4 transcript is expressed in the posterior endoderm and GFP-Gpc4 is expressed on the membrane of endodermal cells in Tg(sox17:GFP-gpc4) embryos. It also shows the relative levels of gpc4 transcripts in Tg(sox17:GFP-gpc4) embryos. Fig. S2 shows that the Wnt11f2- and Wnt5b-tagged constructs displayed the predicted localization in the embryo and were functional. Fig. S3 shows that the Flag-Gpc4 construct is functional and that the expression of GFP-Gpc4Δ517-557-sdc4TM in the endoderm can rescue the C&E defects of mesoderm and endoderm in gpc4−/− embryos. Fig. S4 shows that GFP-Gpc4–expressing blastula cells extend GFP-positive protrusions colocalized with Lifeact-RFP and that Wnts-mNeonGreen–producing cells transport Wnt’s to mem-mTaqBFP–labeled receiving cells. Fig. S5 shows that injecting embryos with the cdc42T17N RNA at a high dose leads to C&E defects, whereas injecting them with this RNA at a subdose has little impact on cellular protrusions emanating from GFP-Gpc4–expressing endodermal cells. Fig. S6 shows that Lat B disrupts cellular protrusions extended from endodermal cells in gpc4−/−/ Tg(sox17:GFP-gpc4) embryos and blocks the rescue mediated by endodermal expression of GFP-Gpc4. Video 1 shows endodermal protrusions in gpc4−/− and their sibling control embryos. Video 2 shows GFP-Gpc4–labeled protrusions transport Wnt5b-mCherry in endoderm cells. Video 3 shows that GFP-Gpc4–labeled protrusions transport Wnt11f2-mCherry in endoderm cells. Video 4 shows that Wn5b-labeled endodermal protrusions are produced in gpc4−/− embryos and their WT sibling embryos. Video 5 shows that blastula cells deliver Wnts-mNeonGreen to the neighboring cells via protrusions. Video 6 shows that endoderm cells deliver Wnts-mNeonGreen to their neighboring cells via protrusions. Video 7 shows that GFP-Gpc4–labeled protrusions emanating from endoderm cells contact the adjacent notochord cells. Video 8 shows the impact that the cdc42T17N mRNA has on endodermal protrusions in gpc4−/−/Tg(sox17:GFP-gpc4) embryos. Video 9 shows that endodermal protrusions form in gpc4−/−/Tg(sox17:GFP-gpc4) embryos treated with Lat B or DMSO.
Supplementary Material
Acknowledgments
We are grateful to Jacek Topczewski (Northwestern University) for technical suggestions and providing the plasmid constructs and Diane Slusarski (University of Iowa), Bruce Appel (University of Colorado, Boulder, CO), Ray Dunn (Institute of Medical Biology, Singapore), Erez Raz (University of Münster, Münster, Germany), Stephanie Woo (University of California, Merced, Merced, CA), and Didier Stainier (Max Planck Institute) for providing the plasmid constructs. We thank other members in the laboratories of Drs. Fang Lin and Robert Cornell (The University of Iowa), as well as Lilianna Solnica-Krezel and Diane Sepich (Washington University in St. Louis), for their technical suggestions. We also thank Matthew Culver and Matthew Murry for excellent fish care and technical support.
This work was supported by the National Science Foundation (grant IOS-1354457 to F. Lin), the National Institutes of Health (grant 1R56DK123610-01 to F. Lin and R01 CA207889 to S. Chen), and the Department of Defense Breast Cancer Research Program breakthrough award level 2 (BC151478 to S. Chen).
The authors declare no competing financial interests.
Author contributions: B. Hu and F. Lin conceived the ideas and designed experiments; B. Hu, J.J. Rodriguez, A. Kakkerla Balaraju, Y. Gao, N.T. Nguyen, H. Steen, S. Suhaib, and S. Chen performed the experiments; B. Hu, J.J. Rodriguez, Y. Gao, and F. Lin analyzed the data; B. Hu and F. Lin wrote the manuscript.
References
- Alexander, J., Rothenberg M., Henry G.L., and Stainier D.Y.. 1999. casanova plays an early and essential role in endoderm formation in zebrafish. Dev. Biol. 215:343–357. 10.1006/dbio.1999.9441 [DOI] [PubMed] [Google Scholar]
- Amor, D.J., Stephenson S.E.M., Mustapha M., Mensah M.A., Ockeloen C.W., Lee W.S., Tankard R.M., Phelan D.G., Shinawi M., de Brouwer A.P.M., et al. 2019. Pathogenic Variants in GPC4 Cause Keipert Syndrome. Am. J. Hum. Genet. 104:914–924. 10.1016/j.ajhg.2019.02.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki, T.O., David N.B., Minchiotti G., Saint-Etienne L., Dickmeis T., Persico G.M., Strähle U., Mourrain P., and Rosa F.M.. 2002. Molecular integration of casanova in the Nodal signalling pathway controlling endoderm formation. Development. 129:275–286. 10.1242/dev.129.2.275 [DOI] [PubMed] [Google Scholar]
- Callejo, A., Bilioni A., Mollica E., Gorfinkiel N., Andrés G., Ibáñez C., Torroja C., Doglio L., Sierra J., and Guerrero I.. 2011. Dispatched mediates Hedgehog basolateral release to form the long-range morphogenetic gradient in the Drosophila wing disk epithelium. Proc. Natl. Acad. Sci. USA. 108:12591–12598. 10.1073/pnas.1106881108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos-Xavier, A.B., Martinet D., Bateman J., Belluoccio D., Rowley L., Tan T.Y., Baxová A., Gustavson K.H., Borochowitz Z.U., Innes A.M., et al. 2009. Mutations in the heparan-sulfate proteoglycan glypican 6 (GPC6) impair endochondral ossification and cause recessive omodysplasia. Am. J. Hum. Genet. 84:760–770. 10.1016/j.ajhg.2009.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capurro, M.I., Xu P., Shi W., Li F., Jia A., and Filmus J.. 2008. Glypican-3 inhibits Hedgehog signaling during development by competing with patched for Hedgehog binding. Dev. Cell. 14:700–711. 10.1016/j.devcel.2008.03.006 [DOI] [PubMed] [Google Scholar]
- Cayuso, J., Dzementsei A., Fischer J.C., Karemore G., Caviglia S., Bartholdson J., Wright G.J., and Ober E.A.. 2016. EphrinB1/EphB3b Coordinate Bidirectional Epithelial-Mesenchymal Interactions Controlling Liver Morphogenesis and Laterality. Dev. Cell. 39:316–328. 10.1016/j.devcel.2016.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirone, P., Lin S., Griesbach H.L., Zhang Y., Slusarski D.C., and Crews C.M.. 2008. A role for planar cell polarity signaling in angiogenesis. Angiogenesis. 11:347–360. 10.1007/s10456-008-9116-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenhaber, B., Bork P., and Eisenhaber F.. 1998. Sequence properties of GPI-anchored proteins near the omega-site: constraints for the polypeptide binding site of the putative transamidase. Protein Eng. 11:1155–1161. 10.1093/protein/11.12.1155 [DOI] [PubMed] [Google Scholar]
- Eugster, C., Panáková D., Mahmoud A., and Eaton S.. 2007. Lipoprotein-heparan sulfate interactions in the Hh pathway. Dev. Cell. 13:57–71. 10.1016/j.devcel.2007.04.019 [DOI] [PubMed] [Google Scholar]
- Fico, A., Maina F., and Dono R.. 2011. Fine-tuning of cell signaling by glypicans. Cell. Mol. Life Sci. 68:923–929. 10.1007/s00018-007-7471-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filmus, J., Capurro M., and Rast J.. 2008. Glypicans. Genome Biol. 9:224. 10.1186/gb-2008-9-5-224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franch-Marro, X., Marchand O., Piddini E., Ricardo S., Alexandre C., and Vincent J.P.. 2005. Glypicans shunt the Wingless signal between local signalling and further transport. Development. 132:659–666. 10.1242/dev.01639 [DOI] [PubMed] [Google Scholar]
- Fujihara, Y., and Ikawa M.. 2016. GPI-AP release in cellular, developmental, and reproductive biology. J. Lipid Res. 57:538–545. 10.1194/jlr.R063032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallet, A., Staccini-Lavenant L., and Thérond P.P.. 2008. Cellular trafficking of the glypican Dally-like is required for full-strength Hedgehog signaling and wingless transcytosis. Dev. Cell. 14:712–725. 10.1016/j.devcel.2008.03.001 [DOI] [PubMed] [Google Scholar]
- González-Méndez, L., Seijo-Barandiarán I., and Guerrero I.. 2017. Cytoneme-mediated cell-cell contacts for Hedgehog reception. eLife. 6:e24045. 10.7554/eLife.24045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Méndez, L., Gradilla A.C., and Guerrero I.. 2019. The cytoneme connection: direct long-distance signal transfer during development. Development. 146:dev174607. 10.1242/dev.174607 [DOI] [PubMed] [Google Scholar]
- Häcker, U., Nybakken K., and Perrimon N.. 2005. Heparan sulphate proteoglycans: the sweet side of development. Nat. Rev. Mol. Cell Biol. 6:530–541. 10.1038/nrm1681 [DOI] [PubMed] [Google Scholar]
- Haddon, C., Smithers L., Schneider-Maunoury S., Coche T., Henrique D., and Lewis J.. 1998. Multiple delta genes and lateral inhibition in zebrafish primary neurogenesis. Development. 125:359–370. 10.1242/dev.125.3.359 [DOI] [PubMed] [Google Scholar]
- Hammerschmidt, M., Pelegri F., Mullins M.C., Kane D.A., Brand M., van Eeden F.J., Furutani-Seiki M., Granato M., Haffter P., Heisenberg C.P., et al. 1996. Mutations affecting morphogenesis during gastrulation and tail formation in the zebrafish, Danio rerio. Development. 123:143–151. [DOI] [PubMed] [Google Scholar]
- Heisenberg, C.P., Tada M., Rauch G.J., Saúde L., Concha M.L., Geisler R., Stemple D.L., Smith J.C., and Wilson S.W.. 2000. Silberblick/Wnt11 mediates convergent extension movements during zebrafish gastrulation. Nature. 405:76–81. 10.1038/35011068 [DOI] [PubMed] [Google Scholar]
- Hu, B., Gao Y., Davies L., Woo S., Topczewski J., Jessen J.R., and Lin F.. 2018. Glypican 4 and Mmp14 interact in regulating the migration of anterior endodermal cells by limiting extracellular matrix deposition. Development. 145:dev.163303. 10.1242/dev.163303 [DOI] [PubMed] [Google Scholar]
- Kakugawa, S., Langton P.F., Zebisch M., Howell S., Chang T.H., Liu Y., Feizi T., Bineva G., O’Reilly N., Snijders A.P., et al. 2015. Notum deacylates Wnt proteins to suppress signalling activity. Nature. 519:187–192. 10.1038/nature14259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kan, M., Wang F., Xu J., Crabb J.W., Hou J., and McKeehan W.L.. 1993. An essential heparin-binding domain in the fibroblast growth factor receptor kinase. Science. 259:1918–1921. 10.1126/science.8456318 [DOI] [PubMed] [Google Scholar]
- Kardash, E., Reichman-Fried M., Maître J.L., Boldajipour B., Papusheva E., Messerschmidt E.M., Heisenberg C.P., and Raz E.. 2010. A role for Rho GTPases and cell-cell adhesion in single-cell motility in vivo. Nat. Cell Biol. 12:47–53: 1–11. 10.1038/ncb2003 [DOI] [PubMed] [Google Scholar]
- Keller, R. 2002. Shaping the vertebrate body plan by polarized embryonic cell movements. Science. 298:1950–1954. 10.1126/science.1079478 [DOI] [PubMed] [Google Scholar]
- Kikuchi, Y., Agathon A., Alexander J., Thisse C., Waldron S., Yelon D., Thisse B., and Stainier D.Y.. 2001. casanova encodes a novel Sox-related protein necessary and sufficient for early endoderm formation in zebrafish. Genes Dev. 15:1493–1505. 10.1101/gad.892301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilian, B., Mansukoski H., Barbosa F.C., Ulrich F., Tada M., and Heisenberg C.P.. 2003. The role of Ppt/Wnt5 in regulating cell shape and movement during zebrafish gastrulation. Mech. Dev. 120:467–476. 10.1016/S0925-4773(03)00004-2 [DOI] [PubMed] [Google Scholar]
- Kinoshita, T., and Fujita M.. 2016. Biosynthesis of GPI-anchored proteins: special emphasis on GPI lipid remodeling. J. Lipid Res. 57:6–24. 10.1194/jlr.R063313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornberg, T.B., and Roy S.. 2014. Cytonemes as specialized signaling filopodia. Development. 141:729–736. 10.1242/dev.086223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozma, R., Ahmed S., Best A., and Lim L.. 1995. The Ras-related protein Cdc42Hs and bradykinin promote formation of peripheral actin microspikes and filopodia in Swiss 3T3 fibroblasts. Mol. Cell. Biol. 15:1942–1952. 10.1128/MCB.15.4.1942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreuger, J., Perez L., Giraldez A.J., and Cohen S.M.. 2004. Opposing activities of Dally-like glypican at high and low levels of Wingless morphogen activity. Dev. Cell. 7:503–512. 10.1016/j.devcel.2004.08.005 [DOI] [PubMed] [Google Scholar]
- Kwan, K.M., Fujimoto E., Grabher C., Mangum B.D., Hardy M.E., Campbell D.S., Parant J.M., Yost H.J., Kanki J.P., and Chien C.B.. 2007. The Tol2kit: a multisite gateway-based construction kit for Tol2 transposon transgenesis constructs. Dev. Dyn. 236:3088–3099. 10.1002/dvdy.21343 [DOI] [PubMed] [Google Scholar]
- LeClair, E.E., Mui S.R., Huang A., Topczewska J.M., and Topczewski J.. 2009. Craniofacial skeletal defects of adult zebrafish Glypican 4 (knypek) mutants. Dev. Dyn. 238:2550–2563. 10.1002/dvdy.22086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, X. 2004. Functions of heparan sulfate proteoglycans in cell signaling during development. Development. 131:6009–6021. 10.1242/dev.01522 [DOI] [PubMed] [Google Scholar]
- Lin, S., Baye L.M., Westfall T.A., and Slusarski D.C.. 2010. Wnt5b-Ryk pathway provides directional signals to regulate gastrulation movement. J. Cell Biol. 190:263–278. 10.1083/jcb.200912128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Link, V., Shevchenko A., and Heisenberg C.P.. 2006. Proteomics of early zebrafish embryos. BMC Dev. Biol. 6:1. 10.1186/1471-213X-6-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes, C.C., Dietrich C.P., and Nader H.B.. 2006. Specific structural features of syndecans and heparan sulfate chains are needed for cell signaling. Braz. J. Med. Biol. Res. 39:157–167. 10.1590/S0100-879X2006000200001 [DOI] [PubMed] [Google Scholar]
- Marlow, F., Zwartkruis F., Malicki J., Neuhauss S.C., Abbas L., Weaver M., Driever W., and Solnica-Krezel L.. 1998. Functional interactions of genes mediating convergent extension, knypek and trilobite, during the partitioning of the eye primordium in zebrafish. Dev. Biol. 203:382–399. 10.1006/dbio.1998.9032 [DOI] [PubMed] [Google Scholar]
- Mattes, B., Dang Y., Greicius G., Kaufmann L.T., Prunsche B., Rosenbauer J., Stegmaier J., Mikut R., Özbek S., Nienhaus G.U., et al. 2018. Wnt/PCP controls spreading of Wnt/β-catenin signals by cytonemes in vertebrates. eLife. 7:e36953. 10.7554/eLife.36953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattila, P.K., and Lappalainen P.. 2008. Filopodia: molecular architecture and cellular functions. Nat. Rev. Mol. Cell Biol. 9:446–454. 10.1038/nrm2406 [DOI] [PubMed] [Google Scholar]
- McGough, I.J., Vecchia L., Bishop B., Malinauskas T., Beckett K., Joshi D., O’Reilly N., Siebold C., Jones E.Y., and Vincent J.P.. 2020. Glypicans shield the Wnt lipid moiety to enable signalling at a distance. Nature. 585:85–90. 10.1038/s41586-020-2498-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miles, L.B., Mizoguchi T., Kikuchi Y., and Verkade H.. 2017. A role for planar cell polarity during early endoderm morphogenesis. Biol. Open. 6:531–539. 10.1242/bio.021899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Multhaupt, H.A., Yoneda A., Whiteford J.R., Oh E.S., Lee W., and Couchman J.R.. 2009. Syndecan signaling: when, where and why? J. Physiol. Pharmacol. 60(Suppl 4):31–38. [PubMed] [Google Scholar]
- Muñoz, R., Moreno M., Oliva C., Orbenes C., and Larraín J.. 2006. Syndecan-4 regulates non-canonical Wnt signalling and is essential for convergent and extension movements in Xenopus embryos. Nat. Cell Biol. 8:492–500. 10.1038/ncb1399 [DOI] [PubMed] [Google Scholar]
- Muyskens, J.B., and Kimmel C.B.. 2007. Tbx16 cooperates with Wnt11 in assembling the zebrafish organizer. Mech. Dev. 124:35–42. 10.1016/j.mod.2006.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobes, C.D., and Hall A.. 1995. Rho, rac, and cdc42 GTPases regulate the assembly of multimolecular focal complexes associated with actin stress fibers, lamellipodia, and filopodia. Cell. 81:53–62. 10.1016/0092-8674(95)90370-4 [DOI] [PubMed] [Google Scholar]
- Odenthal, J., and Nüsslein-Volhard C.. 1998. fork head domain genes in zebrafish. Dev. Genes Evol. 208:245–258. 10.1007/s004270050179 [DOI] [PubMed] [Google Scholar]
- Ohkawara, B., Yamamoto T.S., Tada M., and Ueno N.. 2003. Role of glypican 4 in the regulation of convergent extension movements during gastrulation in Xenopus laevis. Development. 130:2129–2138. 10.1242/dev.00435 [DOI] [PubMed] [Google Scholar]
- Oxtoby, E., and Jowett T.. 1993. Cloning of the zebrafish krox-20 gene (krx-20) and its expression during hindbrain development. Nucleic Acids Res. 21:1087–1095. 10.1093/nar/21.5.1087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panáková, D., Sprong H., Marois E., Thiele C., and Eaton S.. 2005. Lipoprotein particles are required for Hedgehog and Wingless signalling. Nature. 435:58–65. 10.1038/nature03504 [DOI] [PubMed] [Google Scholar]
- Poulain, F.E., and Yost H.J.. 2015. Heparan sulfate proteoglycans: a sugar code for vertebrate development? Development. 142:3456–3467. 10.1242/dev.098178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez-Weber, F.A., and Kornberg T.B.. 1999. Cytonemes: cellular processes that project to the principal signaling center in Drosophila imaginal discs. Cell. 97:599–607. 10.1016/S0092-8674(00)80771-0 [DOI] [PubMed] [Google Scholar]
- Riedl, J., Crevenna A.H., Kessenbrock K., Yu J.H., Neukirchen D., Bista M., Bradke F., Jenne D., Holak T.A., Werb Z., et al. 2008. Lifeact: a versatile marker to visualize F-actin. Nat. Methods. 5:605–607. 10.1038/nmeth.1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robu, M.E., Larson J.D., Nasevicius A., Beiraghi S., Brenner C., Farber S.A., and Ekker S.C.. 2007. p53 activation by knockdown technologies. PLoS Genet. 3:e78. 10.1371/journal.pgen.0030078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulte-Merker, S., van Eeden F.J., Halpern M.E., Kimmel C.B., and Nüsslein-Volhard C.. 1994. no tail (ntl) is the zebrafish homologue of the mouse T (Brachyury) gene. Development. 120:1009–1015. 10.1242/dev.120.4.1009 [DOI] [PubMed] [Google Scholar]
- Serralbo, O., and Marcelle C.. 2014. Migrating cells mediate long-range WNT signaling. Development. 141:2057–2063. 10.1242/dev.107656 [DOI] [PubMed] [Google Scholar]
- Shao, M., Liu Z.Z., Wang C.D., Li H.Y., Carron C., Zhang H.W., and Shi D.L.. 2009. Down syndrome critical region protein 5 regulates membrane localization of Wnt receptors, Dishevelled stability and convergent extension in vertebrate embryos. Development. 136:2121–2131. 10.1242/dev.032649 [DOI] [PubMed] [Google Scholar]
- Solnica-Krezel, L., and Sepich D.S.. 2012. Gastrulation: making and shaping germ layers. Annu. Rev. Cell Dev. Biol. 28:687–717. 10.1146/annurev-cellbio-092910-154043 [DOI] [PubMed] [Google Scholar]
- Stafford, D., White R.J., Kinkel M.D., Linville A., Schilling T.F., and Prince V.E.. 2006. Retinoids signal directly to zebrafish endoderm to specify insulin-expressing beta-cells. Development. 133:949–956. 10.1242/dev.02263 [DOI] [PubMed] [Google Scholar]
- Stanganello, E., and Scholpp S.. 2016. Role of cytonemes in Wnt transport. J. Cell Sci. 129:665–672. [DOI] [PubMed] [Google Scholar]
- Stanganello, E., Hagemann A.I., Mattes B., Sinner C., Meyen D., Weber S., Schug A., Raz E., and Scholpp S.. 2015. Filopodia-based Wnt transport during vertebrate tissue patterning. Nat. Commun. 6:5846. 10.1038/ncomms6846 [DOI] [PubMed] [Google Scholar]
- Strate, I., Tessadori F., and Bakkers J.. 2015. Glypican4 promotes cardiac specification and differentiation by attenuating canonical Wnt and Bmp signaling. Development. 142:1767–1776. 10.1242/dev.113894 [DOI] [PubMed] [Google Scholar]
- Tada, M., Concha M.L., and Heisenberg C.P.. 2002. Non-canonical Wnt signalling and regulation of gastrulation movements. Semin. Cell Dev. Biol. 13:251–260. 10.1016/S1084-9521(02)00052-6 [DOI] [PubMed] [Google Scholar]
- Thompson, J.D., Gibson T.J., Plewniak F., Jeanmougin F., and Higgins D.G.. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876–4882. 10.1093/nar/25.24.4876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topczewski, J., Sepich D.S., Myers D.C., Walker C., Amores A., Lele Z., Hammerschmidt M., Postlethwait J., and Solnica-Krezel L.. 2001. The zebrafish glypican knypek controls cell polarity during gastrulation movements of convergent extension. Dev. Cell. 1:251–264. 10.1016/S1534-5807(01)00005-3 [DOI] [PubMed] [Google Scholar]
- Traister, A., Shi W., and Filmus J.. 2008. Mammalian Notum induces the release of glypicans and other GPI-anchored proteins from the cell surface. Biochem. J. 410:503–511. 10.1042/BJ20070511 [DOI] [PubMed] [Google Scholar]
- van Amerongen, R., and Nusse R.. 2009. Towards an integrated view of Wnt signaling in development. Development. 136:3205–3214. 10.1242/dev.033910 [DOI] [PubMed] [Google Scholar]
- Venero Galanternik, M., Lush M.E., and Piotrowski T.. 2016. Glypican4 modulates lateral line collective cell migration non cell-autonomously. Dev. Biol. 419:321–335. 10.1016/j.ydbio.2016.09.002 [DOI] [PubMed] [Google Scholar]
- Villefranc, J.A., Amigo J., and Lawson N.D.. 2007. Gateway compatible vectors for analysis of gene function in the zebrafish. Dev. Dyn. 236:3077–3087. 10.1002/dvdy.21354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, X., and Page-McCaw A.. 2014. A matrix metalloproteinase mediates long-distance attenuation of stem cell proliferation. J. Cell Biol. 206:923–936. 10.1083/jcb.201403084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong, K.S., Rehn K., Palencia-Desai S., Kohli V., Hunter W., Uhl J.D., Rost M.S., and Sumanas S.. 2012. Hedgehog signaling is required for differentiation of endocardial progenitors in zebrafish. Dev. Biol. 361:377–391. 10.1016/j.ydbio.2011.11.004 [DOI] [PubMed] [Google Scholar]
- Woo, S., Housley M.P., Weiner O.D., and Stainier D.Y.. 2012. Nodal signaling regulates endodermal cell motility and actin dynamics via Rac1 and Prex1. J. Cell Biol. 198:941–952. 10.1083/jcb.201203012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanaka, H., Moriguchi T., Masuyama N., Kusakabe M., Hanafusa H., Takada R., Takada S., and Nishida E.. 2002. JNK functions in the non-canonical Wnt pathway to regulate convergent extension movements in vertebrates. EMBO Rep. 3:69–75. 10.1093/embo-reports/kvf008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan, D., Wu Y., Yang Y., Belenkaya T.Y., Tang X., and Lin X.. 2010. The cell-surface proteins Dally-like and Ihog differentially regulate Hedgehog signaling strength and range during development. Development. 137:2033–2044. 10.1242/dev.045740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye, D., Xie H., Hu B., and Lin F.. 2015. Endoderm convergence controls subduction of the myocardial precursors during heart-tube formation. Development. 142:2928–2940. 10.1242/dev.113944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young, T., Poobalan Y., Tan E.K., Tao S., Ong S., Wehner P., Schwenty-Lara J., Lim C.Y., Sadasivam A., Lovatt M., et al. 2014. The PDZ domain protein Mcc is a novel effector of non-canonical Wnt signaling during convergence and extension in zebrafish. Development. 141:3505–3516. 10.1242/dev.114033 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.