Summary
Next Generation Sequencing (NGS) is increasingly used in diagnostic centers for the assessment of genomic alterations to select patients for precision oncology. The Italian Society of Anatomic Pathology and Diagnostic Cytopathology (SIAPEC) through the Molecular Pathology and Predictive Medicine Study Group (PMMP) has been following the progressive development of centers that have adopted NGS technology in diagnostics over time. In July 2017, a study network on massive parallel sequencing was activated in Italy and recognized as the NGS SIAPeC National Network by the SIAPeC Scientific Society Board. Since then, activities have been implemented within the network that provide for alignment of laboratories through diagnostic concordance analysis and monitoring of centers adhering to the Network. Recently, considering the growing need for extended genomic analyses, the PMMP distributed a national survey to assess activities related to the use of genomic diagnostics in oncology within the NGS SIAPEC National Network.
Thirty centers participated in the survey. Eighty percent of the centers are laboratories within Pathology Departments. The distribution of laboratories in the country, the diagnostic laboratory/population ratio, the staff dedicated, the type and number of sequencing and mechatronics platforms available, the genomic panels utilized, and the type and number of diagnostic tests carried out in the last year in each center, are reported.
The centers were also asked whether they participated in a multidisciplinary Molecular Tumor Board (MTB) for management of patients. Thirty percent of the centers had a MTB that was ratified by regional decree. The professionals most frequently involved in the core team of the MTB are the pathologist, oncologist, molecular biologist, geneticist, pharmacologist, and bioinformatician.
The data from this survey indicate that NGS diagnostics in Italy is still heterogeneous in terms of geographical distribution and the characteristics of laboratories and diagnostic test performed. The implementation of activities that favors harmonization, the logistics and the convergence of biological material in reference centers for molecular analyses is a priority for the development of a functional laboratory network.
Key words: Molecular Pathology, Next generation Sequencing (NGS), Molecular Tumor Board (MTB)
Introduction
Precision oncology with targeted drugs requires accurate clinical evaluation of the patient and an in-depth morphological, immunophenotypic and molecular analysis of the tumor through a series of processes that require the involvement of different professionals working in a team 1,2.
Pathologists are increasingly involved in the molecular characterization of tumors. The current diagnostic needs in various oncological areas require broad spectrum analyses, which include information on the mutational pattern, DNA repair mechanisms, and immune response 3-6. These types of analyses can no longer be achieved with classical approaches, which are now impractical for reasons related to the costs of reagents, times needed for testing, and the scarcity of biological material.
Next Generation Sequencing (NGS), also referred to as Massive Parallel Sequencing (MPS), is increasingly used in diagnostic centers and is progressively replacing traditional methods of molecular analysis 7-11.
The activation of centers operating in the diagnostic field with NGS has required adaptations to new technologies over the years, with substantial investments and acquisition of new skills.
The Italian Society of Anatomic Pathology and Diagnostic Cytopathology (SIAPEC) through the Molecular Pathology and Predictive Medicine Study Group (PMMP) has been following the evolution of molecular diagnostic activities in Pathology centers operating in Italy for years. With the progressive increase of centers that have adopted NGS technology in diagnostics over time, a study network on massive parallel sequencing was activated in Italy and recognized as the NGS SIAPEC National Network by the SIAPeC Scientific Society Board on 5 July 2017.
Since then, activities have been implemented within the network that provide for alignment of laboratories through diagnostic concordance analysis and monitoring of centers adhering to the SIAPEC NGS National Network with dedicated questionnaires immediately prior to the annual meeting of the PMMP Group.
Considering the growing need for extended genomic analyses in Italy, and more specifically in the context of regional oncological networks, the scientific committee of the PMMP distributed a national survey to assess activities related to the use of genomic diagnostics in oncology within the SIAPEC-PMMP NGS National Network.
Matherial and methods
The SIAPEC-PMMP Study Group sent a survey to 30 centers belonging to the SIAPEC NGS National Network. The survey was conducted using a questionnaire to evaluate a series of parameters characterizing the center and the activities carried out.
The main points addressed in the questionnaire are as follows:
Name of the Center, Affiliation, Director, Manager.
Personnel at the center (Physicians, Molecular biologists, Laboratory technicians).
Equipment present (platforms for MPS, platforms for establishing genomic libraries).
Gene panels used (panel name, number of genes, nucleic acids required).
Quantification of the activities carried out (Diagnosis-Research) over the last year.
Main neoplastic pathologies examined.
Existence of a standardized report.
Laboratory report with multidisciplinary groups for selection of therapy in patients undergoing genomic analysis (Molecular Tumor Board).
Professionals involved in the Molecular Tumor Board.
Regional ratification of the reference Molecular Tumor Board.
The questionnaire is available as an Appendix.
The questionnaire was submitted to 30 centers in July-December 2020. Two months were initially granted (July-August 2020) to return the questionnaire. A total of 21 centers responded to the survey within that time. The results obtained were tabulated, anonymized, and analyzed. The preliminary data on the 21 centers were presented at the national meeting of the PMMP group, held virtually on 8 September 2020. Additional data were provided by the remaining centers in October-December 2020. The final data of the survey was tabulated in Excel and subjected to statistical analysis of frequency, T-test, and contingency tables. A P value < 0.05 was considered significant. The analysis was carried out using IBM® SPSS® software. The final data were presented to the scientific committee of the PMMP group, which met in plenary session on January 18, 2021.
Results
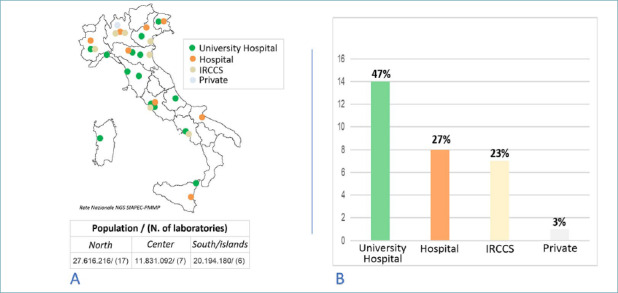
Within the SIAPEC-PMMP working group, 30 centers distributed throughout Italy as shown in Figure 1 (A) participated in the national survey; all centers answered all questions in the questionnaire.
Figure 1.
Type, number and distribution of centers within the SIAPEC NGS Network.
Seventeen centers are located in the north of the country, 7 in the center, and 6 in the south (including the main islands). Based on the geographical distribution of the Italian population (ISTAT data, 31 December 2019), the distribution of the laboratories was greater in the central north than in the south/islands. On average, the diagnostic laboratory/population ratio was 1/1.6 million in the north, 1/1.7 million in the center, and 1/3.4 million in the south/islands. Regarding the type of centers, 14 laboratories are located in Universities, 8 in large hospitals, 7 in Specialized care institutes (IRCCS), and one private center participated in the survey. Figure 1 (B) shows the types of centers and percentage distribution.
Twenty-four (80%) of the centers in the SIAPEC-PMMP NGS Network are laboratories specialized in molecular technologies within Pathology Departments, and the other centers are specialized in Clinical Pathology, Molecular Oncology and Molecular Diagnostics, which are strictly connected to the Pathology Departments. The staff dedicated to diagnostic activity in the centers was divided into 3 categories: Physicians, Molecular biologists, and Laboratory technicians.
As reported in Table I, in which the centers have been anonymized, on average about 3 personnel of each category are present in each center with a minimum of 3 and a maximum of 24; 30% of the centers (9 of 30) have fewer than 5 dedicated staff, while 12 (40%) have 10 or more personnel. Forthy percent of the centers reported having staff with bioinformatics skills.
Table I.
Number of personnel dedicated to diagnostic activity in each center.
| Center | Physicians | Biologists | Technicians | Total |
|---|---|---|---|---|
| 1 | 2 | 2 | 3 | 7 |
| 2 | 2 | 6 | 2 | 10 |
| 3 | 2 | 2 | 3 | 7 |
| 4 | 0 | 4 | 4 | 8 |
| 5 | 7 | 1 | 9 | 17 |
| 6 | 0 | 5 | 2 | 7 |
| 7 | 10 | 8 | 16 | 34 |
| 8 | 3 | 1 | 2 | 6 |
| 9 | 2 | 1 | 2 | 5 |
| 10 | 1 | 2 | 1 | 4 |
| 11 | 1 | 3 | 1 | 5 |
| 12 | 0 | 1 | 4 | 5 |
| 13 | 1 | 1 | 4 | 6 |
| 14 | 1 | 2 | 1 | 4 |
| 15 | 2 | 2 | 0 | 4 |
| 16 | 2 | 4 | 1 | 7 |
| 17 | 0 | 4 | 1 | 5 |
| 18 | 3 | 4 | 5 | 12 |
| 19 | 1 | 2 | 3 | 6 |
| 20 | 4 | 3 | 3 | 10 |
| 21 | 2 | 3 | 3 | 8 |
| 22 | 1 | 1 | 1 | 3 |
| 23 | 0 | 8 | 2 | 10 |
| 24 | 14 | 4 | 3 | 21 |
| 25 | 2 | 2 | 1 | 5 |
| 26 | 5 | 8 | 4 | 17 |
| 27 | 8 | 12 | 4 | 24 |
| 28 | 1 | 2 | 3 | 6 |
| 29 | 2 | 3 | 5 | 10 |
| 30 | 0 | 2 | 8 | 10 |
| Mean | 2.7 | 3.4 | 3.4 | 9.4 |
Regarding the number and type of NGS sequencers in the various centers, 1 sequencing platform is present in 33% of laboratories, 2 platforms in 30%, and 3 or more platforms in 37% of centers. Within the network there are 31 Illumina platforms, 28 IonTorrent platforms, and 4 Qiagen platforms.
Table II shows the survey data relating to the number and type of sequencing platforms present in the various centers. Turnover of the different platforms was high, as the majority of centers reported upgrades over the years. For the purposes of implementing NGS tests in clinical practice, the diffusion of automated systems for the preparation of genomic libraries is important, which considerably reduces the work needed by operators in the pre-analytical phase and minimizes human errors. Table II lists the instruments used for automation in the different centers. Some are an integral or complementary part of commercial platforms for NGS sequencing (e.g. Ion Chef for the Ion Torrent platform), while in other cases they are mechatronic systems that are adaptable to different sequencing platforms (Agilent, Hamilton Robotics, Masmec).
Table II.
Type and number of technological platforms in each center compared with the number of diagnostic NGS test performed in a year period (January-December 2019).
| Centers | Sequencing Platform | Genomic library preparation system | |
|---|---|---|---|
| Number | Type | ||
| 1 | 3 | MS/NS/GR | Magnis |
| 2 | 1 | S5 | N. 2 Ion Chef System |
| 3 | 2 | MS/NS | Microlab starlet |
| 4 | 1 | S5 | Ion Chef System |
| 5 | 3 | S5/PGM/HS | Ion Chef System |
| 6 | 3 | MS/NS/S5 | Ion Chef System - QIAgility |
| 7 | 2 | S5/MS | Ion Chef System |
| 8 | 1 | PGM | Ion Chef System |
| 9 | 3 | MS/PGM/S5 | NO |
| 10 | 1 | GR | Qiacube |
| 11 | 2 | GR | NO |
| 12 | 2 | MS/S5 | Ion Chef System - NGS Star |
| 13 | 3 | MS/NS/NoS | NO |
| 14 | 1 | PGM | NO |
| 15 | 2 | S5/PGM | Ion Chef System |
| 16 | 3 | MS/MS/NS | NGS Star |
| 17 | 1 | PGM | NO |
| 18 | 2 | S5/NS | Ion Chef System |
| 19 | 2 | MS/PGM | NO |
| 20 | 2 | S5/MS | NO |
| 21 | 1 | S5 | Ion Chef System |
| 22 | 3 | S5/MS/NS | Ion Chef System |
| 23 | 3 | S5/MS/NS | Ion Chef System - NGS Star |
| 24 | 1 | MS | NO |
| 25 | 1 | S5 | Ion Chef System |
| 26 | 4 | PGM/S5/S5/GN | N2. Ion Chef System |
| 27 | 2 | MS/NS | NO |
| 28 | 2 | MS/NS | NO |
| 29 | 3 | PGM/PGM/S5 | Ion Chef System - Magnis |
| 30 | 3 | MS/MS/MS | Omnia - Microlab starlet |
All data were collected relating to the genetic panels used in the various centers for NGS diagnostics, in order of frequency of use, with 6 possible opportunities for insertion in the questionnaire (from the panel of group 1, or first choice, to the panel of group 6).
Table III shows the data relating to the panels indicated by the various centers for group 1. These are panels with a number of genes that can be analyzed (from 2 to 80), with an average of 27 genes per panel; the table also shows the nucleic acids required to carry out the test.
Table III.
Characteristics of the genomic panels (Group 1) utilized in NGS diagnostics.
| Center | Type of panel: Group1 | Number of genes | Nucleic acid required |
|---|---|---|---|
| 1 | AIO ALL IN ONE | 22 | DNA |
| 2 | Oncomine™ Solid Tumour DNA | 22 | DNA |
| 3 | Myriapod NGS-LT 56G Onco Panel | 56 | DNA |
| 4 | Hotspot Cancer panel v2 | 50 | DNA |
| 5 | Oncomine Focus Assay | 80 | DNA/RNA |
| 6 | Archer FusionPlex Sarcoma Kit (Archer Dx) | 26 | DNA/RNA |
| 7 | ONCOMINE BRCA ASSAY | 2 | DNA |
| 8 | OST DNA | 22 | DNA |
| 9 | BRCA1/2 DEVYSER | 2 | DNA |
| 10 | Qiact BRCA UMI Panel | 5 | DNA |
| 11 | QIAact DNA AIT UMI panel | 30 | DNA |
| 12 | ONCOMINE UNIVERSAL DX | 56 | DNA/RNA |
| 13 | COLON-LUNG BENKIT panel | 5 | DNA |
| 14 | SiRe | 6 | DNA |
| 15 | AllRas | 4 | DNA |
| 16 | BRCA1/2 somatico mini-HRS (up-grade 2020 a Devyser BRCA) | 2 | DNA |
| 17 | Myriapod NGS-LT BRCA1-2 panel (HB: 2018/04) | 3 | DNA |
| 18 | Custom | 26 | DNA |
| 19 | Oncomine Focus Assay | 80 | DNA/RNA |
| 20 | Myriapod NGS-LT 56G Onco Panel | 56 | DNA |
| 21 | PAN-SOMATIC LAB-DEVELOPED 2.0 | 26 | DNA |
| 22 | BRCA1/2 DEVYSER | 2 | DNA |
| 23 | Oncomine Focus Assay | 52 | DNA/RNA |
| 24 | Myriapod NGS-LT 56G Onco Panel | 56 | DNA/RNA |
| 25 | CUSTOM | 25 | DNA |
| 26 | SirE | 7 | DNA |
| 27 | MYRIADPOD NGS SOLID TUMOR | 16 | DNA |
| 28 | Myriapod NGS-LT 56G Onco Panel | 56 | DNA |
| 29 | Oncomine BRCA | 2 | DNA |
| 30 | HEREDITARY CANCER SOLUTION™ | 26 | DNA |
The number of panels used in the different centers varies from 1 to 6, with 30 centers that reported a panel in group 1, while 22 centers also use a second panel, 17 centers use 3 panels, 9 use 4, 4 use 5, and 3 use 6. The number of genes in the panels reported in the different groups varied from 2 to 5.
As seen in Table IV, the average number of genes evaluable with the different panels progressively increases from 1st choice to 6th choice. It is clear that the 4th, 5th, and 6th choice panels, with an average of analyzable genes > 100, are used only by a limited number of centers with adequate technology and experience. For each panel, the dedicated commercial software was reported and, in the presence of bioinformatics skills in the center, the use of additional software for analysis of the results was also indicated. The data is available upon request to the SIAPEC Board.
Table IV.
Number of genes in the genomic panels utilized in NGS diagnostics.
| Panel: group 1 | Panel: group 2 | Panel: group 3 | Panel: group 4 | Panel: group 5 | Panel: group 6 | |
|---|---|---|---|---|---|---|
| Number of centers | 30 | 22 | 17 | 9 | 4 | 3 |
| Mean number of genes | 27 | 29 | 31 | 117 | 143 | 202 |
| Minimal number of genes | 2 | 1 | 2 | 9 | 7 | 17 |
| Maximum number of genes | 80 | 161 | 170 | 395 | 524 | 524 |
The centers also provided data on the tests performed in a year period (January-December 2019) for research or diagnostic activities. A total of 17,667 NGS tests were carried out (mean 654 tests per center), of which 6,386 were performed in the context of research activities (mean 237 tests per center), and 11,281 for diagnostic activity (mean 418 tests per center).
The number of tests carried out at the various centers was heterogeneous. There are 13 centers with high productivity (from 500 to 2500 tests/year), 6 centers with medium productivity (from 200 to 500 tests/year), and 11 centers that can be defined as having low activity or in a phase of implementation (< 200 tests/year). The data are reported in Table V.
Table V.
Number of NGS test performed in a year period (January-December 2019) in each center.
| Centers | Number of tests performed | Research tests | Diagnostic tests |
|---|---|---|---|
| 1 | 920 | 205 | 715 |
| 2 | 1700 | 500 | 1200 |
| 3 | 600 | 200 | 400 |
| 4 | In implementation | In implementation | In implementation |
| 5 | 60 | 25 | 35 |
| 6 | 145 | 85 | 60 |
| 7 | 486 | 288 | 198 |
| 8 | 620 | 90 | 530 |
| 9 | 100 | 40 | 60 |
| 10 | 150 | 0 | 150 |
| 11 | 460 | 100 | 360 |
| 12 | 250 | 100 | 150 |
| 13 | 405 | 200 | 205 |
| 14 | In implementation | In implementation | In implementation |
| 15 | 780 | 480 | 300 |
| 16 | 587 | 0 | 587 |
| 17 | 519 | 490 | 29 |
| 18 | 1650 | 150 | 1500 |
| 19 | 26 | 0 | 26 |
| 20 | 90 | 50 | 40 |
| 21 | 1359 | 0 | 1359 |
| 22 | 342 | 338 | 4 |
| 23 | 600 | 550 | 50 |
| 24 | In implementation | In implementation | In implementation |
| 25 | 120 | 120 | 0 |
| 26 | 2000 | 400 | 1600 |
| 27 | 220 | 200 | 20 |
| 28 | 175 | 150 | 25 |
| 29 | 2500 | 1625 | 875 |
| 30 | 803 | 0 | 803 |
| Total | 17667 (Mean: 654) | 6386 (Mean 237) | 11281 (Mean 418) |
A significant association emerged between diagnostic productivity and availability of platforms for automation of processes for sample preparation: centers with tools for automated processing of genomic libraries had an average annual diagnostic productivity of 502 cases compared to 78 cases in centers without automation (P = 0.026).
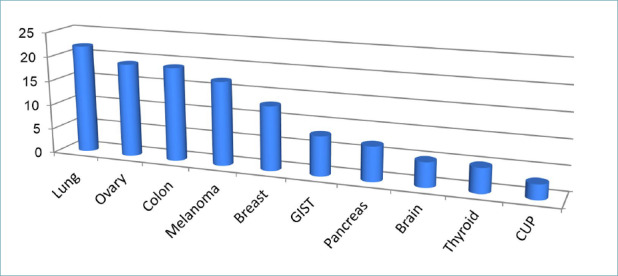
Centers were asked to indicate the oncological pathologies investigated by NGS diagnostics. The results of the survey showed that NGS tests were mainly used for the diagnosis of lung, ovarian, and colon cancers (with more than 15 centers involved), melanoma and breast cancers (over 10 centers), and to follow other oncological pathologies as shown in Figure 2.
Figure 2.
Type of solid tumors investigated by NGS in the centers that participated in the survey.
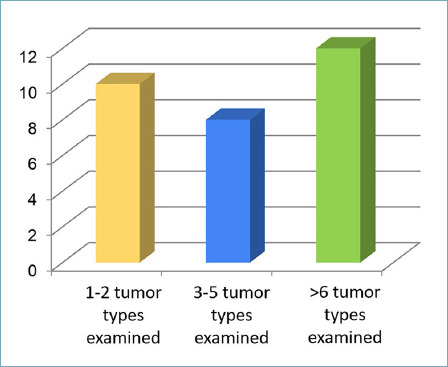
In all, 40% of centers applied diagnostic NGS to more than 6 different tumor types and 27% apply the technology to 3-5 tumor types, while 33% of centers carry out NGS focused on diagnosis of specific neoplastic forms (< 3 tumor types) (Fig. 3).
Figure 3.
Color Graph showing the diagnostic activity of centers categorized by the number of different tumor types investigated by NGS.
The survey also queried the availability of a reporting model in the center and the centers’ opinion on the utility of a standard model to be adopted at a national level. Twenty-three centers (77%) responded that they had a defined reporting model, while 97% referred that they would like to adhere to the adoption of a shared standardized model for reporting.
The centers were also asked whether they participated in a dedicated multidisciplinary group or “Molecular Tumor Board” (MTB) for management of patients.
Nineteen (63%) of the centers referred the existence of a multidisciplinary reference group with which to share diagnostic decisions based on cancer treatments. Multiple professionals are involved in MTB in the various centers. The most frequently reported professionals are the pathologist (100%), oncologist (100%), molecular biologist (79%), geneticist (63%), pharmacologist (37%), and bioinformatician (32%) as shown in Table VI.
Table VI.
Professionals included in the Molecular Tumor Boards activated in 19 centers.
| Centers | Professionals involved in a Molecular Tumor Board | |||||
|---|---|---|---|---|---|---|
| 1 | Oncologist | Pathologist | Molecular Biologist | |||
| 2 | Oncologist | Pathologist | Molecular Biologist | Geneticist | Farmacologist | Bioinformatician |
| 3 | Oncologist | Pathologist | Molecular Biologist | |||
| 4 | Oncologist | Pathologist | Molecular Biologist | |||
| 5 | Oncologist | Pathologist | Molecular Biologist | Geneticist | ||
| 6 | Oncologist | Pathologist | Geneticist | |||
| 7 | Oncologist | Pathologist | Molecular Biologist | |||
| 8 | Oncologist | Pathologist | Geneticist | |||
| 9 | Oncologist | Pathologist | Molecular Biologist | Geneticist | ||
| 10 | Oncologist | Pathologist | Molecular Biologist | Bioinformatician | ||
| 11 | Oncologist | Pathologist | Molecular Biologist | Geneticist | Bioinformatician | |
| 12 | Oncologist | Pathologist | Molecular Biologist | Geneticist | ||
| 13 | Oncologist | Pathologist | Molecular Biologist | Geneticist | Farmacologist | |
| 14 | Oncologist | Pathologist | Farmacologist | |||
| 15 | Oncologist | Pathologist | Molecular Biologist | |||
| 16 | Oncologist | Pathologist | Molecular Biologist | Geneticist | Farmacologist | Bioinformatician |
| 17 | Oncologist | Pathologist | Geneticist | Farmacologist | ||
| 18 | Oncologist | Pathologist | Molecular Biologist | Geneticist | Farmacologist | Bioinformatician |
| 19 | Oncologist | Pathologist | Molecular Biologist | Geneticist | Farmacologist | Bioinformatician |
These professionals are joined by others involved in instrumental diagnostics or in oncological treatments, which are reported in order of frequency: surgeon, radiologist, hematologist, radiotherapist, pulmonologist, endocrinologist, dermatologist, urologist, methodologist, psycho-oncologist. For 9 (47%) of the 19 centers that declared their participation in a MTB, the latter was ratified by a regional resolution. The regions that have taken action in this regard are: Campania, Lazio, Liguria, Lombardy, Sicily, Sardinia, Tuscany, and Veneto - eight regions in which a total of nine structures are located (two in Veneto). Some centers referred that they did not understand the difference between the Molecular Tumor Board and Multidisciplinary Oncology Group.
Discussion
The results of this national survey of centers that perform genomic tests in oncology using massive parallel sequencing have provided a heterogeneous and potentially rapidly evolving picture of the current situation. Overall, the data lead to interesting reflections on the rapid development of this new approach, on its main applications in diagnosis for a detailed characterization of oncological pathologies and suitability of targeted treatments, and on the progressive adaptations induced by new technology that will soon require regulatory interventions.
The survey assessed the type and number of network centers in Italy, their geographical distribution, and technological capabilities of the different laboratories. In 80% of cases, the centers participating in the survey were internal structures of Pathology Departments, with the remaining laboratories in close functional connection with Pathology Departments. Of the 30 centers that joined the NGS SIAPEC study network, more than half (57%) are located in the north of Italy and 80% in the center/north. This reflects the greater population density in these geographic areas compared to the south/islands. However, comparing the number of laboratories to the demographic distribution, in the center/north the density is almost homogeneous (one laboratory per 1.6-1.7 million inhabitants). In fact, in the south/islands, compared to the population there are about half of the number of laboratories present in the center/north. The distribution of laboratories in the center/north is in line with what has been repeatedly reported considering the ideal distribution of genomic diagnostic centers throughout the country 12,13. The number of laboratories in the south, on the other hand, represents a critical issue that should be overcome in a short time, particularly in some geographical areas.
From an instrumental point of view, all the centers, at the time of the survey, had one or more massive parallel sequencing instruments, mostly attributable to the two most widespread technologies available on the international market, i.e. Thermofisher and Illumina, with only a limited number of centers that adopted the Qiagen platform, which is currently being phased out. Two-thirds of laboratories have two or more platforms and most centers reported technological upgrades over a few years. This implies a significant economic commitment of laboratories to this new technology. Eight (80%) of the 10 centers equipped with a single platform opted for IonTorrent technology, while the multiple instruments in the various centers are mainly Illumina: this may depend on the technical characteristics of products, costs, and commercial policies. The survey also provided data on mechatronic systems for the automation of preparatory processes, and in particular with regards to establishing genomic libraries for massive parallel sequencing. Overall, the instrumental landscape is very heterogeneous and can be better interpreted only by evaluating the data relating to the instrumentation and diagnostic activity in the different centers (Tabs. II, V). Laboratories that have platforms for automation had significantly higher average diagnostic productivity than centers without automated platforms. This indicates the importance of automation of the pre-analytical phase which, in the case of massive parallel sequencing, is particularly lengthy and complex, with repetitive steps that can be associated with operator error, and therefore not highly suited for routine clinical activity in the absence of automated processes.
A further consideration concerns sequencing activities carried out within centers of the SIAPEC NGS National Network. As for most new technologies, next-generation sequencing was initially introduced in many laboratories for research purposes. With technological advances, both instrumental and in reagents used for sample preparation, the technology has become more reliable, progressively moving in the diagnostic field to provide new and important possibilities. At the time of the survey, 68% of NGS activity in the centers was devoted to oncology diagnostics and as many as 15 (50%) of the centers carry out more diagnostic activities than basic research. This suggests that within a few years massive parallel sequencing will have an important role as a diagnostic tool on the national territory.
Based on the numbers provided in terms of the annual diagnostic activity carried out, 13 laboratories handled a number of samples compatible with that reported for a reference center which must maximize resources 14. The other laboratories currently handle a much lower number of samples, while others have just recently been activated and diagnostic processes are still being implemented. The SIAPEC will continue to carry out annual surveys to monitor the activity of centers and encourage further development.
Overall, the data indicate that NGS technology is being used for genomic characterization of a wide range of neoplastic diseases and that the technology has a broad impact. The types of cancers most frequently investigated reflect what has been recently highlighted by the European Society of Medical Oncology 10. However, about one-third of the centers limit the technology to a few oncological pathologies, favoring lung, ovary, and colon cancers as well as melanoma, due to the greater complexity of genes that need to be examined (e.g. BRCA and related genes in ovarian cancer) and the need to simultaneously examine numerous markers that have entered routine clinical practice with limited quantities of biological material available for analysis (e.g. lung cancer, melanoma).
The personnel currently present in the diagnostic centers with higher throughput is on average adequate for the number of tests carried out and well represented in terms of professional categories among physicians, molecular biologists, and laboratory technicians. Personnel with specific bioinformatics skills are present in only 40% of centers, and their presence is not correlated with the number of exams performed. Evidently, the role of the bioinformatician is assumed by other personnel with more general skills, which is favored by the progressive development of dedicated software.
Of particular interest is the implementation of the Molecular Tumor Boards. These multidisciplinary groups are considered necessary for adequate and shared selection of patients to receive personalized oncological treatments after genomic characterization by NGS 12-19. About two-thirds of centers reported that they are involved in multidisciplinary working groups, or a Molecular Tumor Board which is dedicated to patient selection following NGS, but only 30% of the centers had a Molecular Tumor Board that was ratified by regional decree.
In general, the professionals most frequently involved in a Molecular Tumor Board are the oncologist, pathologist, molecular biologist, geneticist, pharmacologist, and bioinformatician.
This central “core team”, which represents the mainstay of the Molecular Tumor Board, associates less frequently with other professionals involved in the care pathway due to the specific oncological pathology in the various centers (hematologist, pulmonologist, endocrinologist, dermatologist, urologist). The results of the survey stress the need to better define the relationships between multidisciplinary cancer groups and Molecular Tumor Boards and to precisely outline their limits, specific functions, and activity.
The results of the survey are relative to the SIAPEC-PMMP study and monitoring network, which includes the vast majority of centers involved in NGS diagnostics in oncology that are active in Italy. Other centers, not operating within Pathology Departments or associated with them, were not included in the present analysis. It is therefore representative of the national situation, but not exhaustive. We believe, however, that only a few centers were excluded from the present survey.
The data from this survey indicate that diagnostic activity using massive parallel sequencing in Italy is carried out, but that it is heterogeneous in terms of geographical distribution and the characteristics of laboratories and activities performed. The diagnostic centers are still not fully structured within defined regional networks, which are not always connected to diagnostic-therapeutic pathways and molecular tumor boards, much less to a functional national network.
Broad-spectrum genomic characterization in oncology, based on massive parallel sequencing, is economically unsustainable in the absence of an economy of scale that provides for centralization of activities, adequate production processes, and automation. The implementation of a functional laboratory network that favors local logistics and the convergence of biological material in reference centers for molecular analyses is highly desirable. This, together with the management of data by molecular tumor boards, in close relationship with multidisciplinary oncology groups in well-defined diagnostic-therapeutic pathways, within regional oncological networks, will ensure a constant increase in quality, reduction in costs, and high levels of diagnostic and therapeutic appropriateness.
Figures and tables
References
- 1.Garraway LA.Genomics-driven oncology: framework for an emerging paradigm. J Clin Oncol 2013;31:1806-1814. https://doi.org/10.1200/JCO.2012.46.8934 10.1200/JCO.2012.46.8934 [DOI] [PubMed] [Google Scholar]
- 2.ÖzdoÐan M, Papadopoulou E, Tsoulos N, et al. BMC Med Genomics 2021;14:105. https://doi.org/10.1186/s12920-021-00952-9 10.1186/s12920-021-00952-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Giardina T, Robinson C, Grieu-Iacopetta F, et al. Implementation of next generation sequencing technology for somatic mutation detection in routine laboratory practice. Pathology 2018;50:389-401. https://doi.org/10.1016/j.pathol.2018.01.005 10.1016/j.pathol.2018.01.005 [DOI] [PubMed] [Google Scholar]
- 4.Luchini C, Bibeau F, Ligtenberg MJL, et al. ESMO recommendations on microsatellite instability testing for immunotherapy in cancer, and its relationship with PD-1/PD-L1 expression and tumour mutational burden: a systematic review-based approach. Ann Oncol 2019;30:1232-1243. https://doi.org/10.1093/annonc/mdz116 10.1093/annonc/mdz116 [DOI] [PubMed] [Google Scholar]
- 5.Alborelli I, Leonards K, Rothschild SI, et al. Tumor mutational burden assessed by targeted NGS predicts clinical benefit from immune checkpoint inhibitors in non-small cell lung cancer. J Pathol 2020;250:19-29. https://doi.org/10.1002/path.5344 10.1002/path.5344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rizvi H, Sanchez-Vega F, La K, et al. Molecular determinants of response to Anti-Programmed Cell Death (PD)-1 and AntiProgrammed Death-Ligand 1 (PD-L1) blockade in patients with non-small-cell lung cancer profiled with targeted next-generation sequencing. J Clin Oncol 2018;36:633-641. https://doi.org/10.1200/JCO.2017.75.3384 10.1200/JCO.2017.75.3384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Morganti S, Tarantino P, Ferraro E, et al. Next Generation Sequencing (NGS): a revolutionary technology in pharmacogenomics and personalized medicine in cancer. Adv Exp Med Biol 2019;1168:9-30. https://doi.org/10.1007/978-3-030-24100-1_2 10.1007/978-3-030-24100-1_2 [DOI] [PubMed] [Google Scholar]
- 8.Tan O, Shrestha R, Cunich M, et al. Application of next-generation sequencing to improve cancer management: A review of the clinical effectiveness and cost-effectiveness. Clin Genet 2018;93:533-544. https://doi.org/10.1111/cge.13199 10.1111/cge.13199 [DOI] [PubMed] [Google Scholar]
- 9.Deans ZC, Costa JL, Cree I, et al. Integration of next-generation sequencing in clinical diagnostic molecular pathology laboratories for analysis of solid tumours; an expert opinion on behalf of IQN Path ASBL. Virchows Arch. 2017;47:5-20. https://doi.org/10.1007/s00428-016-2025-7 10.1007/s00428-016-2025-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mosele F, Remon J, Mateo J, et al. Recommendations for the use of next-generation sequencing (NGS) for patients with metastatic cancers: a report from the ESMO Precision Medicine Working Group. Ann Oncol 2020;31:1491-1505. https://doi.org/10.1016/j.annonc.2020.07.014 10.1016/j.annonc.2020.07.014 [DOI] [PubMed] [Google Scholar]
- 11.van Krieken JH, Normanno N, Blackhall F, et al. Guideline on the requirements of external quality assessment programs in molecular pathology. Virchows Arch 2013;462:27-37. https://doi.org/10.1007/s00428-012-1354-4 10.1007/s00428-012-1354-4 [DOI] [PubMed] [Google Scholar]
- 12.Raccomandazioni AIOM 2020 - Tumor Board Molecolare. 19Novembre 2020. [Google Scholar]
- 13.Test Molecolari e Terapie Target in Oncologia. Atti del Workshop ISS-FICOG, 4 e 17 Novembre 2020. [Google Scholar]
- 14.Martini N, Marchetti P, Marchetti A, et al. Dall’Istologia al Target. Il futuro della Precision Medicine. I Quaderni di Medicina. Il Sole 24 ore, 2017. [Google Scholar]
- 15.VanderWalde A, Grothey A, Vaena D, et al. Establishment of a Molecular Tumor Board (MTB) and Uptake of Recommendations in a Community Setting. Pers Med 2020;10:252. https://doi.org/10.3390/jpm10040252 10.3390/jpm10040252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harada S, Arend R, Dai Q, Levesque JA, et al. Implementation and utilization of the molecular tumor board to guide precision medicine. Oncotarget 2017;8:57845-57854. https://doi.org/10.18632/oncotarget.18471 10.18632/oncotarget.18471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Walko C, Kiel PJ, Kolesar J.Am, et al. Precision medicine in oncology: New practice models and roles for oncology pharmacists. J Health Syst Pharm 2016;73:1935-1942. https://doi.org/10.2146/ajhp160211 10.2146/ajhp160211 [DOI] [PubMed] [Google Scholar]
- 18.Tafe LJ, Gorlov IP, de Abreu FB, et al. Implementation of a molecular tumor board: the impact on treatment decisions for 35 patients evaluated at Dartmouth-Hitchcock Medical Center. Oncologist 2015;20:1011-1018. https://doi.org/10.1634/theoncologist.2015-0097. https://doi.org/10.1634/theoncologist.2015-0097 10.1634/theoncologist.2015-0097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Patton EE, Mueller KL, Adams DJ, et al. Melanoma models for the next generation of therapies. Cancer Cell 2021:S15356108(21)00055-6. https://doi.org/10.1016/j.ccell.2021.01.011 10.1016/j.ccell.2021.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]


