Abstract
The endocytic pathway is a system of dynamically communicating vesicles, known as early endosomes, that internalize, sort, and traffic nutrients, trophic factors, and signaling molecules to sites throughout the cell. In all eukaryotic cells, early endosome functions are regulated by rab5 activity, dependent upon its binding to GTP, whereas rab5 bound to GDP represents the biologically inactive form. An increasing number of neurodegenerative diseases are associated with endocytic dysfunction and, in the case of Alzheimer’s disease (AD) and Down syndrome (DS), an early appearing highly characteristic reflection of endocytic pathway dysfunction is an abnormal enlargement of rab5 positive endosomes. In AD and DS, endosome enlargement accompanying accelerated endocytosis and fusion, upregulated transcription of endocytosis-related genes, and aberrant signaling by endosomes are caused by pathological rab5 over-activation. In this chapter, we describe a battery of methods that have been used to assess Rab5 activation in models of AD/DS and are applicable to other cell and animal disease models. These methods include 1) fluorescence recovery after photobleaching (FRAP) assay; 2) quantitative measurement of endosome size by light, fluorescence and electron microscopy; 3) detection of GTP-rab5 by in situ immunocytochemistry in vitro and ex vivo; 4) immunoprecipitation and GTP-agarose pull-down assay; 5) biochemical detection of rab5 in endosome-enriched subcellular fractions obtained by OptiPrep™ density gradient centrifugation of mouse brain.
Keywords: Rab5 activity, GTP-binding, endosomes, endocytic dysfunction, neurodegeneration, Alzheimer’s disease, Down syndrome, endocytosis, GDI
1. Introduction
Rab5 is the master regulatory GTPase on early endosomes. Rab5 functions in endocytosis, endosome fusion, maturation, trafficking and signaling are dependent on its membrane localization and cycling between an active, GTP-bound and an inactive, GDP-bound state [1]. Rab5 membrane localization is regulated by guanyl nucleotide (GDP) dissociation inhibitors (GDIs) [2,3] and GDI displacement factors (GDFs) [4] whereas the activity of guanine nucleotide exchange factors (GEFs), e.g. Rabex5 [5], and GTPase-activating proteins (RabGAPs), e.g. RabGAP5 [6], facilitate rab5 activation and inactivation, respectively (Fig. 1). Upon activation, rab5 recruits effector proteins that mediate the various rab5 functions in the endocytic pathway. Pathological activation of rab5 drives endocytic dysfunction in Alzheimer’s disease (AD) and Down syndrome (DS) resulting in a signature enlargement of Rab5 positive early endosomes [7], accelerated endocytosis and fusion, in addition to several other downstream effects on endosome transport, signaling and neuronal survival [8].
Fig.1. Radb5 activation cycle.
Representative diagram showing some of the multiple components involved in Rab5 activation. Inactive GDP-bound Rab5 is present in the cytosol sequestered by GDI. In proximity of the target membrane, GDF may interact with Rab5 to facilitate GDI dissociation and Rab5 insertion into the membrane. Upon GDP dissociation, a GEF promotes GTP binding and subsequent activation of Rab5. In this active state Rab5 can interact with multiple effectors (not shown) to promote vesicle budding, trafficking, and fusion. Rab5 GTP hydrolysis is facilitated by a GAP, which converts Rab5 to its inactive GDP-bound state. Inactive rab5 can then be extracted from the membrane by GDI and recycled back to the cytosol. Inset, Under conditions promoting an increase in Rab5 expression (or transcription) or, as in the case of AD/DS, in the amyloid precursor protein (APP), rab5 undergoes hyper-activation, resulting in increased endocytosis and increased endosome fusion.
There are different methods to measure rab5 activation (e.g.[9-12]). The methods reported in this review have been validated by our group in models of AD and DS both in vitro [8,13,14] and in vivo [15-17] and allow either the direct or indirect determination of Rab5 activation. The indirect measures include analysis of the rate of rab5 dissociation from the endosomal membrane by fluorescence recovery after photobleaching (FRAP), which inversely correlates with rab5 activation state, and analyses of endosome number, size, morphology, which reflect endosomal enlargement, a robust immediate outcome of rab5-mediated endosome fusion. The direct measures are based on the use of a rab5-GTP-specific antibody for immunocytochemistry and immunoprecipitation and biochemical analysis of GTP-bound rab5 by GTP-agarose pull-down. We also describe an optimized method for subcellular fractionation using OptiPrep™ (iodixanol) [18] to efficiently separate and enrich different organelles, including early endosomes, from mouse brain tissue, which can be used for functional assays and further downstream applications. There is a growing interest in the study of endosome and rab5 due to the involvement of endocytic pathway abnormalities in diverse disease conditions [7,19-21]. The methods we report, therefore, represent a foundation to facilitate these expanding investigations.
2. Materials
Prepare all solutions by using ultrapure water (prepared using the Milli-Q Ultrapure Water Purification System) and analytical grade or the highest-grade reagents commercially available (unless otherwise specified).
2.1. Fluorescence Recovery After Photobleaching (FRAP) assay
Plasmids. GFP-rab5a wild-type (WT) construct is cloned from pHSV-Myc-rab5a plasmid. Dominant active GFP-rab5a Q79L and dominant negative GFP-rab5a S34N are generated by PCR based site-directed mutagenesis (Stratagene). The pEGFP-C1 vector is commercially available from Clontech.
Mouse neuroblastoma cell line. Neuro-2a cells (N2a cells are from ATCC, CCL-131).
Growth media: Dulbecco’s Minimal Essential media (DMEM) supplemented with 5 % Fetal Bovine Serum (Thermo Fisher Sci.,16000069) and 100 units/ml penicillin-streptomycin (Thermo Fisher Sci., 15140122).
Lipofectamine 2000 reagent and Opti-MEM reduced serum medium (Thermo Fisher Sci.).
Zeiss LSM520 confocal microscope (Zeiss). The microscope also has temperature and CO2 controller for live-imaging (Zeiss Pecon).
2.2. Quantitative measurement of endosome size changes
2.2.1. Rab5 positive endosome measurement in human fibroblasts
Primary human fibroblasts from Down syndrome patients (DS) and diploid age-matched controls (2N) (Coriell Cell Repositories, http://ccr.coriell.org) were used before reaching passage number 15.
Cell culture media: MEM (Thermo Fisher Sci., 10370021), plus 10% fetal bovine serum (Thermo Fisher Sci.,16000069) and 100 units/ml penicillin-streptomycin (Thermo Fisher Sci., 15140122).
Phosphate-buffered saline (PBS) 1X: 137 mM NaCl, 10 mM Na2HPO4, 1.8 mM KH2PO4, 2.7 mM KCl, adjust pH to 7.4.
Fixative solution: 4% PFA (Electron Microscopy Sciences, 15714) in PBS (PFA/PBS).
Antibody dilution buffer: 5% goat serum in PBS, plus 0.3% Triton X-100 (Sigma, T8787).
Primary antibodies: mouse anti-Rab5 (BD Biosciences, 610282); mouse anti-EEA1 (BD Biosciences, 610457).
Fluorescence-conjugated donkey anti-mouse Alexa-Fluor secondary antibodies (Thermo Fisher Sci.)
Fluoro-Gel mounting medium (Thermo Fisher Sci., 5024704).
LSM 510 Meta confocal microscope (Zeiss)
2.2.2. Rab5 positive endosome measurement in mouse brain sections
Anesthetic mix: ketamine (100 mg/kg BW) and xylazine (10 mg/kg BW) in PBS
Perfusion/fixation buffer solution: 4% paraformaldehyde (PFA) in 0.1M sodium cacodylate buffer, pH 7.4 (Electron Microscopy Sciences, 11652).
Vibratome (Leica VT1000 S).
Tris-buffered saline (TBS) 1X: 50 mM Tris-Cl, pH 7.5, 150mM NaCl, adjust pH to 7.5.
Endogenous peroxidase quenching buffer: 3% H2O2 and 10% methanol in TBS
Permeabilization/Antibody dilution buffer (A): 1% bovine serum albumin (BSA), 1% normal horse serum (NHS) and 0.4% Triton X-100.
Permeabilization/Antibody dilution buffer (B): 1% BSA, 1% NHS and 0.05% saponin.
Blocking buffer: 20% NHS in PBS.
Primary antibodies against Rab5: goat polyclonal anti-Rab5b (Santa Cruz Biotechnology Inc, SC-26569); rabbit polyclonal anti- Rab5a (Santa Cruz Biotechnology Inc, SC-309) and rabbit polyclonal anti-Rab5 (Abcam; 18211).
For DAB staining: biotinylated secondary antibodies (Vector Lab.,1:200-500); VECTASTAIN® ABC HRP Kit (Vector Lab, PK-4000); DAB (Vector Lab, SK-4100).
Permount (Fisher Sci, SP15-100).
Light microscope (Zeiss Axioskop II microscope).
For fluorescence labeling: fluorescence-conjugated donkey anti-goat and anti-rabbit Alexa-Fluor secondary antibodies (Thermo Fisher Sci.).
Fluoro-Gel mounting medium (Thermo Fisher Sci., 5024704).
Fisherbrand Superfrost Plus Microscope slides and glass coverslips (Fisher Sci.).
LSM880 laser scanning confocal microscope (ZEISS).
2.2.3. Electron Microscopy (EM) and post-embedding immuno-EM (iEM) from mouse brain
2.2.3.1. EM general Method
Anesthetic mix: as in 2.2.1
Perfusion/fixation buffer solution: 4% PFA, 2% glutaraldehyde in 0.1 M sodium cacodylate buffer, pH 7.4 (Electron Microscopy Sciences, 11652).
Vibratome (Leica VT1000 S).
1% osmium tetroxide.
Ascending ethanol series of 20%, 30%, 50%, 75%, 90% and 100%.
Spurr resin concentrations of 25%, 50%, 75% and 100%.
Aclar sheets (Electron Microscopy Sciences).
Ultramicrotome (Leica Reichert Ultracut S).
Nickel grids (200 mesh, Electron Microscopy Sciences).
Uranyl acetate (2%).
Lead citrate (3%).
Electron microscope (Thermo Fisher Talos L120C transmission electron microscope operating at 120kV).
2.2.3.2. Post-embedding iEM
Etching solution: 1% sodium metaperiodate in PBS followed
Double-distilled water (ddH2O).
Blocking buffer/Antibody dilution buffer: 1% BSA in PBS.
Primary antibodies: mouse anti-Rab5-GTP (NewEast Biosciences, 26911); rabbit anti-rab5 (Abcam, 18211).
Secondary antibodies: 5- and 20-nm gold-conjugated anti-rabbit and anti-mouse secondary antibodies, respectively (Aurion, 1:10).
2.3. Detection of GTP-Rab5
2.3.1. In situ immunofluorescence in vitro
Cell culture: murine neuroblastoma (N2a) cell line (ATCC, CCL-131), control or stably overexpressing human wild-type amyloid precursor protein (N2aAPP).
Growth media: Dulbecco’s Minimal Essential media (DMEM) supplemented with 5 % Fetal Bovine Serum and penicillin-streptomycin (as in 2.2.1) and 0.2 mg/ml G418 for selection (Millipore Sigma, G418-RO).
Dicer-substrate small interfering RNA (DsiRNA) targeting RabGAP5/Sgsm3 (IDT, mm.Ri.Sgsm3.13.1) and DsiRNA negative control (IDT, 51-01-14-03).
Lipofectamine 2000 and Opti-MEM reduced serum medium (Thermo Fisher Sci.).
Fixative solution: 4% PFA (Electron Microscopy Sciences, 15714) in PBS (PFA/PBS).
Blocking/Antibody dilution buffer: 2% NHS in PBS with 0.05% saponin.
Primary antibodies: mouse anti-Rab5-GTP (NewEast Biosciences, 26911); rabbit anti-RabGAP5 (Proteintech, 20825-AP).
Donkey anti-mouse and anti-rabbit Alexa Fluor secondary antibodies (ThermoFisher Scientific).
Fluoro-Gel mounting medium (Electron Microscopy Sciences).
Fisherbrand Superfrost Plus Microscope slides and glass coverslips (Fisher Scientific).
LSM880 laser scanning confocal microscope (ZEISS).
2.3.2. In situ immunofluorescence ex vivo
2.3.3. Immunoprecipitation from mouse brain tissue/subcellular fraction and Western blot
Anesthetic mix as in 2.2.2
Phosphate-buffered saline (PBS) for perfusion.
IP lysis/wash buffer: 50 mM Tris, pH 7.4, 150 mM NaCl, 1mM MgCl2, 1 mM EDTA, 1% Triton X-100, with protease and phosphatase inhibitors.
Method of choice for protein concentration assay (e.g. Bradford or Bicinchoninic acid).
Anti-Rab5-GTP (NewEast Biosciences, 26911).
PureProteome™ Protein A/G Magnetic beads (Millipore Sigma).
Bis (sulfosuccinimidyl) suberate (BS3) (Thermo Fisher Sci.).
Magnetic separator stand (Promega).
100 μM GTP-γ-S stock (NewEast Biosciences, 30302) in IP lysis/wash buffer.
2X Laemmli sample buffer.
Novex™ 4-20% Tris-Glycine gels (Thermo Fisher Sci.).
Nitrocellulose Membrane (Millipore).
Rabbit polyclonal anti-Rab5 primary antibody (Abcam; 18211).
HRP-conjugated goat anti-rabbit IgG (Jackson ImmunoResearch Laboratories).
ECL detection system (Invitrogen or Millipore Sigma).
2.3.4. Pull-down with GTP-Agarose from mouse brain tissue/subcellular fraction and Western Blot
Anesthetic mix and PBS for perfusion as in 2.3.3.
GTP-Agarose lysis/wash buffer: 50 mM Tris-HCl pH 7.5, 250 mM NaCl, 5 mM Mg acetate, 0.5% Triton X-100, and protease inhibitors.
GTP-agarose beads (Sigma Cat# G9768).
100 μM GTP-γ-S stock (NewEast Biosciences, 30302) in IP lysis/wash buffer.
Microcentrifuge (Eppendorf).
Method of choice for protein concentration assay (e.g. Bradford or Bicinchoninic acid).
For Western Blot analysis materials, primary and secondary antibodies see above (2.3.3., 7-12).
2.4. Endosome isolation by OptiPrep™ density gradient centrifugation from mouse brain tissue
Homogenization buffer: 0.25 M sucrose, 10 mM Tris, pH 7.4, 1 mM EDTA, protease and phosphatase inhibitor cocktail (Sigma).
Teflon-coated glass pestle (2 or 5 ml).
Method of choice for protein concentration assay (e.g. Bradford or Bicinchoninic acid).
50% (w/v) OptiPrep™ stock: 5 vol. of OptiPrep™ in 1 vol. of 0.25 M sucrose, 6mM EDTA, 60 mM Tris-HCl, pH 7.4.
OptiPrep™ gradient solutions at 5%, 10%, 12.5%, 14%, 15%, 20%, 25% obtained from 50% OptiPrep™ stock diluted with 1X homogenization buffer.
SW40 Rotor and ultracentrifuge (Beckman).
3. Methods
3.1. Fluorescence Recovery After Photobleaching (FRAP) assay
Transfection. Plasmid transfection in N2a cells is carried out using lipofectamine 2000. N2a cells are plated at 90-95% confluent at the time of transfection on the 35mm poly-lysine coated glass-bottom dish (BD bioscience) with 2ml of the growth media. Plasmids (4μg of each plasmid expressing GFP-rab5a WT, dominant active GFP-rab5a Q79L, or dominant negative GFP-rab5a S34N) are diluted in 250 μl of Opti-MEM medium (Invitrogen), and 4 μl lipofectamine 2000 (Invitrogen) is mixed in 250 μl of Opti-Mem medium, incubated for 5 minutes and combined with diluted plasmids (total 500 μl) for 20 minutes at room temperature (RT). Mixture is added into the culture, and cells are incubated overnight at 37 °C.
Photobleaching. In a FRAP experiment (Fig. 2), an area of the cell containing a fluorescently tagged protein is photobleached and the recovery of the fluorescence in the bleached region is monitored as the fluorescent protein replaces the bleached protein [22] (Fig. 2a).The GTP-loaded active form of rab5 associates with endosomal membranes while inactive rab5 (GDP-bound) is cytosolic [1]. Because the rate of dissociation of rab5 from endosomal membranes is determined by the conversion of the GTP-bound active to the GDP-bound inactive forms, determination of the rate of rab5 exchange from early endosomes indicates rab5 activation state (Fig. 2a). Dissociation rates can be calculated from the rate of fluorescence recovery after early endosomes expressing GFP-tagged rab5 have been photobleached [23]. Thus, FRAP is a useful tool to evaluate rab5 activity when GFP-tagged rab5 is introduced in cells. Here, FRAP analysis is carried out on a Zeiss LSM510 confocal microscope (Fig. 2b). GFP-rab5a is used for visualizing early endosomes. 24 hours post-transfection cells are imaged live using a 40X oil immersion objective and a zoom of 6 using a single line excitation at 488nm and emission BP 520-550 nm filter sets. A ROI (region of interest) is drawn around the GFP-rab5 positive puncta and laser transmission increased to 100%. Photobleaching results in roughly 70-80% loss of fluorescence in the bleached area
The recovery. The recovery of the GFP fluorescence in each condition is recorded with 20 time-lapse image series by scanning the whole cell at 15 second intervals. The cells are maintained at 37°C in the growth media on a heating stage (Zeiss Pecon) throughout the experiments.
Calculation. To calculate percent of recovery after photobleaching, intensity profiles for the bleach spot are calculated by Zeiss LSM510 image analysis software.
Fig.2. FRAP assay.
a, The principle of FRAP is represented by a schematic diagram of the cell containing GFP-rab5a-positive endosomes. The graph of FRAP analysis shows a schematic recovery curve that corresponds to the target endosome (1), photobleached endosome (2), and recovered endosome (3). The recovery curve provides an estimate for the overall fraction of the GFP-rab5a molecules that exhibit activation. b, Representative images and graphs of GFP-rab5a WT and Q79L FRAP analysis in N2a cells. WT rab5 shows marked increased recovery, while dominant active mutant of rab5a (Q79L) significantly reduces GFP-rab5a recovery after photobleaching, indicating persistent activation of rab5 GTPase (n=20 endosomes in each cell).
3.2. Quantitative measurement of endosome size changes
3.2.1. Rab5 positive endosome measurement in human fibroblasts
Human fibroblasts grown on glass coverslips in 12 well plate until they reached 70% confluency were gently washed with PBS and fixed in 4% PFA/PBS for 20 min.
After wash with PBS, anti-Rab5 (1:500) or anti-EEA1 (1:500) in antibody dilution buffer was added and incubated at 4°C overnight with gentle rocking.
After wash with PBS, fluorescence conjugated secondary anti-mouse antibodies (1:500) in antibody dilution buffer were added and incubated for 1-2 hr at room temperature.
Finally, the coverslips were washed, mounted, and images were taken at 40x magnifications, as previous described [14].
Confocal images were opened with Fiji-ImageJ (imagej.nih.gov/ij), threshold were set and individual cell was outlined. Image J will automatically set the scale unless the preference is to set the scale manually by Analyze-Set Scale. Use Analyze-Set Measurement to check Area, Integrated density, Area Fraction and other parameters as needed, limiting the analysis to threshold.
To separate coupled endosomes, use Process-Binary-Watershed.
Use Analyze-Analyze Particles to set the size range depending on the study (0.005 or 0.01 to infinity) and the method to visualize the results (e.g. check “display results” – to obtain individual particle measurements within the cell; check “summarize” – to obtain average particle measurements per cell) and/or to analyze the particles (e.g. “exclude on edges”, “include holes”). After clicking OK, separate windows will appear to show the result of individual endosomes inside the selected cell, as well as summary result, which will be both copied and pasted into a separate Excel work sheet for further data analysis.
Between 26-52 cells are evaluated for further statistical analysis. Results from rab5-positive or EEA1-positive endosomes can be expressed as average size, number and volume per cell, or relative number of rab5-positive or EEA1-positive endosomes per size bin (0.01-0.5 μm2, 0.51-1.4 μm2, 1.41-7.0 μm2) as previously described [14] [24] (also see Note3) (Fig. 3). Statistical analysis is performed with Prism-GraphPad with Student’t-test or ANOVA to determine significant difference between samples.
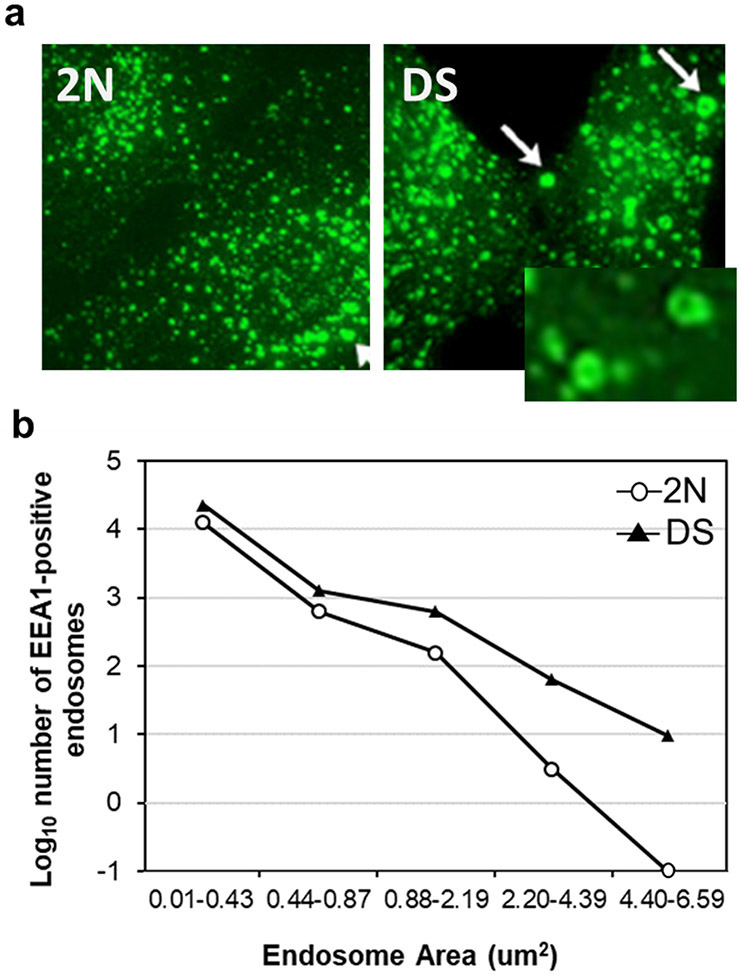
Fig.3. Early endosomes in DS.
a, Representative immunofluorescence micrographs of rab5-positive endosomes in DS and age-matched 2N controls. DS fibroblast show a striking enlargement in the size of rab5-positive endosomes compared to 2N. b, The numbers of EEA1-positive early endosomes in all size groups are also increased in the DS fibroblasts compared to 2N controls (total fibroblasts examined: 2N fibroblasts = 80; DS fibroblasts = 80).
3.2.2. Rab5 positive endosome measurement in mouse brain sections
Mice are anesthetized and transcardially perfused with perfusion/fixation buffer. Brains are removed and post-fixed in the same buffer overnight at 4°C for 48-72 hr.
Fixed brain are cut into 40 μm-thick coronal sections with a vibratome[15]. Regions of the medial septal nucleus (MSN), hippocampus including dentate gyrus, and neocortex are collected from each animal.
Brain sections are rinsed three times in TBS and incubated in endogenous peroxidase quenching solution for 30 min, room temperature (for DAB labeling), then rinsed three times with diluting buffer, before blocking 1 h at room temperature.
Various commercial antibodies against rab5 are used, including anti-Rab5b [16] in diluting buffer (A) and anti-Rab5a (1:100) and anti-Rab5 (1:500) [8] in diluting buffer (B) (see Note1 and Note2), and brain sections are incubated in the respective primary antibody solutions at 4 °C overnight with gentle rocking.
Sections are then incubated with biotinylated secondary antibodies for 30 min at room temperature for DAB or with donkey anti-goat (for Rab5b) or donkey anti-rabbit (Rab5a and Rab5) Alexa Fluor secondary antibodies (1:500) for 1-2 hr at room temperature.
For fluorescence labeling, cell nuclei are counterstained with DAPI (Thermo Fisher Sci, D1306), then brain sections are washed and mounted in Fluoro-Gel.
For DAB staining, brain sections are incubated with ABC solution for 1 hr at room temperature and DAB for 5-10 min (incubation time may vary depending on antibody and brain tissue), then washed and mount on to coated glass slide, air dry overnight, before going through the dehydrating step and mounting on glass coverslips with Permount.
A light microscope (Zeiss Axioskop II microscope) is used to collect images of DAB-stained sections with 100X magnifications while immunofluorescence images are captured at 40X and 3X zoom on top of 40X magnification with an oil immersion objective and a LSM880 laser scanning confocal microscope (ZEISS) at a resolution of 1024 x 1024 pixels as previously described [25].
Rab5-positive endosome number and size analysis in neurons are performed as detailed in 3.2.1 as well as described in previous publication[16,17] (see also Note3).
3.2.3. EM and post-embedding immunoEM from mouse brain
3.2.3.1. General EM Method
Anesthetize the mice with the anesthetic mix and perfuse them transcardially with perfusion-fixation buffer.
Collect the brain and store in the perfusion-fixation buffer at 4 °C 48-72 h.
Cut the brains into 80 μm-thick sagittal vibratome sections
Post-fix the sections in 1% osmium tetroxide for 30 min.
Dehydrate the sections in ascending ethanol solutions, 20%-90%, each for 30 min, followed by 100% for 1 hour.
Infiltrate the sections with increasing concentrations of Spurr resin, 25%-75% each for 1 hour and 100% overnight and flat embed them in Aclar sheets.
Cut out from the Aclar sheet regions of interest for ultrastructural analyses, (e.g. pre-frontal cortex and hippocampal CA1 and Dentate Gyrus areas).
Prepare 50 nm ultrathin sections using an ultramicrotome and place them on nickel grids.
Briefly stain the grids with uranyl acetate (2 %) and lead citrate (3%).
Image the material using an electron microscope (we use a Thermo Fisher Talos L120C transmission electron microscope operating at 120kV).
EM ultrastructural characterization. Endosomes are identified as vesicles with single limiting membranes and sparse intraluminal content[26] and most of them are relatively small, with an average diameter generally ranging between 100-150 nm[27]. Optimally, EM identification of early endosomes should be validated by immunoEM (see following). For endosome quantification, acquire approximately n=40-80 EM images at a direct magnification of 17,500x per mouse per genotype from a specific region of interest (e.g. the pyramidal cell layer V of the pre-frontal cortex), containing dendritic and synaptic profiles in the proximity (within 5-10 mm) of the neuronal soma. Glial cells are excluded on the basis of their morphology and chromatin patterns. Count the number of endosomes present in each field acquired per mouse and determine 1) the average vesicle diameter; 2) the average diameter size distributions falling within the following bins: 0-0.160 nm, 0.161-300 nm; > 300 nm; 3) the endosome area fraction relative to the total image area, using Fiji-ImageJ (imagej.nih.gov/ij) [17].
3.2.3.2. Post-embedding immuno-EM
Mount ultrathin sections prepared as described above (3.2.3.2, 1-8) on nickel grids and air-dry.
Proceed with etching 5 min with 1% sodium metaperiodate in PBS followed by 2 washes in filtered ddH2O.
Incubate sections in blocking solution (1% BSA in PBS) for 2 h at room temperature.
Incubate sections with primary antibodies (1:30) in a humidified chamber overnight at 4 °C.
Following 3x5 min washes in PBS, incubate sections with gold-conjugated anti-rabbit and anti-mouse secondary antibodies (1:10) for 2 h at room temperature.
Wash the grids several times and briefly stain with uranyl acetate and lead citrate before examination.
3.3. Detection of GTP-Rab5
3.3.1. In situ immunofluorescence in vitro
N2a cells and N2aAPP cells (see Note4) are seeded at a 70-80% confluency at the time of transfection on poly-lysine-coated glass coverslips placed at the bottom of a 12-well plate.
DsiRNA for RabGAP5/Sgsm3 or DsiRNA control are diluted in Opti-MEM medium and lipofectamine 2000 is mixed in Opti-Mem medium, incubated for 5 minutes and combined with diluted DsiRNAs for 20 minutes at room temperature. Mixture is added into the culture at a final 40 nM concentration and incubated with the cells for 4h at 37 °C, followed by addition of fresh DMEM for 48-72h. RabGAP5 is a rab5-specific GAP [6]. Here, silencing of Rab5GAP5 in N2a cells acts as a positive control for Rab5 activation.
Cells are gently washed with PBS and fixed in 4% PFA/PBS for 20 min.
After 3x5 min washes with PBS, cells are blocked with blocking/antibody dilution buffer for 30 min at room temperature.
Incubate cells with anti-Rab5-GTP (1:100) and anti-RabGAP5 (1:50) primary antibodies overnight at 4 °C, followed by 3x5 min washes with PBS and incubation with donkey anti-mouse and anti-rabbit Alexa Fluor secondary antibodies in antibody blocking/dilution buffer for 1 h at room temperature.
Coverslips are washed, mounted, and images are taken at 40x magnifications using a Zeiss LSM880 laser scanning confocal microscope (Fig.4).
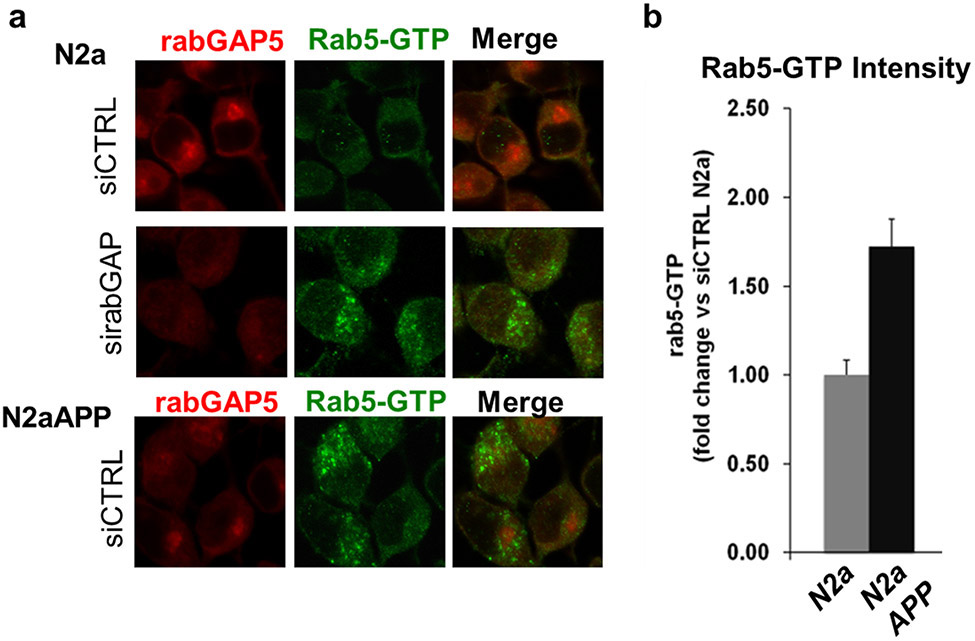
Fig. 4. Direct detection of rab5-GTP in vitro.
a, Representative immunofluorescence micrographs of N2a and N2aAPP cells double labeled with rabGAP5 antibody (red) and rab5-GTP antibody (green). rabGAP5 siRNA in N2a cells causes rab5 over-activation, as shown by the increased intensity of the rab5-GTP antibody. This level of activation is comparable to what is observed in N2aAPP cells (lower panel). b Quantification of rab5-GTP intensity in N2aAPP is approximately 2-fold greater than in N2a cells (total cells examined: N2a = 98; N2aAPP =101).
3.3.2. In situ immunofluorescence ex vivo
Mouse sections for immunofluorescence labeling are obtained as described in 3.2.2.
Sections are washed 3 x 10 min in permeabilization/antibody dilution buffer and blocked for 1 h at room temperature (see Note5).
Sections are incubated with anti-Rab5-GTP primary antibody in antibody dilution buffer (1:100) overnight at 4 °C, followed by incubation with donkey anti-mouse secondary antibodies (1:300) for 1 h at room temperature.
Following 10 min wash in TBS, cell nuclei are counterstained with DRAQ5 fluorescent probe for 5 min in PBS (1:2000).
Sections are then washed 3x10 min in TBS and mounted on glass coverslips using Fluoro-Gel.
Image acquisition settings and analysis of Rab5-GTP puncta are essentially the same as described in 3.2.1 and 3.2.2. In order to minimize analysis of non-specific signal, once the images are opened in Fiji/ImageJ, it is recommended to perform background subtraction using a rolling ball radius of 50 and apply the Yen’s method of image thresholding, before measuring Rab5-GTP puncta number, intensity, size and area fraction/cell (e.g. in the cortex) or area fraction/field (e.g in hippocampal CA1 subregion) [17].
3.3.3. Immunoprecipitation from mouse brain tissue/subcellular fraction and Western blot
Anesthetize the mice and transcardially perfuse using PBS.
Collect the mouse brain and proceed to sample preparation by dissecting a specific brain region or by obtaining a specific subcellular fraction of interest (e.g. hippocampal synaptosomes prepared as described by [28] (see Note6).
Homogenize the sample in 300 μl IP lysis/wash buffer, incubate on ice for 30 min and sonicate 3x 10 sec.
Save an aliquot as initial input (~10% vol/vol) and incubate ~ 0.3 mg of proteins in 300 μl of IP/wash buffer with anti-Rab5-GTP-, or Normal Mouse IgG-, crosslinked PureProteomeTM Protein A/G Magnetic beads overnight at 4°C with rotation (use at least 3-5 μg of antibody/reaction). Bis (sulfosuccinimidyl) suberate (BS3) is used as a cross-linking reagent, following the manufacturer’s instructions.
Carry out a separate reaction in the presence of 10 μM GTP-γ-S, a non-hydrolyzable G-protein-activating analog of GTP, at 30 °C for 30 min, prior to sample incubation with the respective beads, to control for specific binding (positive control).
Using a magnetic stand to allow bead-solution separation, wash the beads 3x with wash buffer, applying each time the magnet.
Resuspended the beads in 2X Laemmli sample buffer and boil 5 min at 95 °C.
Apply the magnet, collect the supernatant and load onto a 4-20% Tris-Glycine gel.
The amount of Rab5-GTP is determined following SDS-PAGE and blotting with an anti-Rab5 antibody (see Note7).
3.3.4. Pull-down with GTP-Agarose from mouse brain tissue/subcellular fraction and Western Blot
Anesthetize the mice and transcardially perfuse using PBS and proceed to sample preparation as described in 3.3.3 (see Note6).
Lyse the sample in 150- 300 μl GTP-Agarose lysis/wash buffer.
Centrifuge the lysates at 13,000 x g for 10 min at 4 °C and save an aliquot (10% vol-vol) of the supernatants as the loading control (input).
Incubate 0.5-1 mg of proteins with 300 μl of GTP-agarose beads for 4h at 4°C with rotation (incubation can be also extended overnight at 4°C if the reaction is carried out with lower protein amounts).
Carry out a separate reaction in the presence of 10 μM GTP-γ-S at 30 °C for 30 min, prior to sample incubation with the respective beads, to control for specific binding (in this case GTP-γ-S treatment acts as a negative control, by saturating the sample with GTP and preventing its binding to the GTP-agarose beads).
Pellet the beads by centrifugation at 10,000 x g for 2 min.
Wash the pellets 3x with GTP-Agarose lysis/wash buffer.
Resuspend the washed beads in 2X Laemmli Sample buffer, run the pulled-down samples along with the input on SDS-PAGE and blot with an anti-Rab5 antibody. The results are expressed as Rab5-GTP/total Rab5, where total Rab5 is the amount of Rab5 present in the input [17]. (see Note7 and Note8).
3.4. Endosome isolation by OptiPrep™ density gradient centrifugation from mouse brain tissue
Mice are anesthetized and transcardially perfused with saline (PBS) as in 3.3.3 and 3.3.4.
Collect the brain and use ½ for the subcellular fractionation (a specific brain regions can be dissected and used as well).
Quickly weight out the brain tissue and add 10x vol/brain weight homogenization buffer (e.g. for an average weight of 0.15-0.2 g use 1.5-2 ml of homogenization buffer).
Homogenize the tissue with 30-40 strokes of a Teflon-coated pestle in ice.
Centrifuge the samples at 1,000 x g for 20 min to pellet nuclei and unbroken tissue and obtain a post nuclear supernatant (PNS).
Collect the PNS and determine sample protein concentration (e.g. from one hemibrain homogenized in 1.4 ml of buffer the concentration of the PNS is about 7 mg/ml).
Adjust 5-6 mg of PNS in homogenization buffer to 25% OptiPrep™ adding an equal volume of 50% stock solution to a final volume of 2 ml. Keep an aliquot of the adjusted PNS as input (this will be mixed with Laemmli Sample Buffer followed by heat denaturation for western blot analysis).
Set up the gradient by loading the PNS in 25% OptiPrep™ at the bottom of a clear centrifuge tube and consecutively overlay with 1.5 ml of 20%, 15%, 14%, 12.5% and 10% and 5% OptiPrep™ solutions.
Load the tube on a SW 40 rotor and centrifuge overnight (18 h) at 100,000 x g at 4 °C.
After centrifugation carefully collect 22 x 0.5 ml aliquots from the top of the tube and analyze by Western Blot by loading equal volumes for each fraction collected, following resuspension in 5X Laemmli sample buffer and heat denaturation at 95 °C for 5 min.
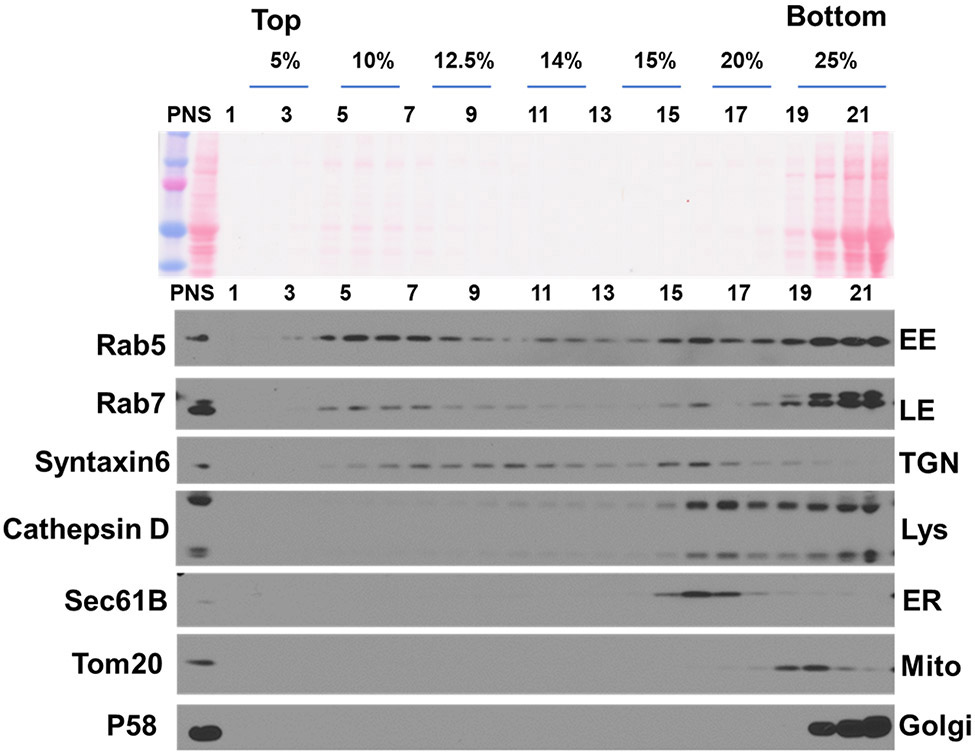
Probe the blots with primary antibodies against different subcellular compartments (Fig. 5), such as rab5 for early endosomes (Abcam); Rab7 (Abcam) for late endosomes; Cathepsin D (made in house) for lysosomes; Tom20 (Santa Cruz Biotechnology) for mitochondria; sec61B for endoplasmic reticulum (ER) (Proteintech); p58K for cis-Golgi (Sigma); syntaxin6 for trans-Golgi network (TGN) (Cell Signaling Technology). The following enrichment is expected at given Optiprep™ %: 5-10%, late endosomes; 10-12.5%, early endosomes; 12.5-15%, TGN; 15-20%, ER; 15-25%, lysosomes; 20-25%, mitochondria; 25%, Golgi. This method has been optimized from [18].
Fig. 5. Subcellular fractionation with Optiprep™.
a, Adult mouse brain homogenates were fractionated with an iodixanol step gradient and 22 fractions collected from the top. Equal volumes of each fraction (to show enrichment), along with the PNS input, are subject to Western blot analysis. Shown are the distributions of Rab5 (early endosome, EE), Rab7 (late endosome, LE), syntaxin6 (trans-golgi network, TGN), cathepsin D (lysosome, Lys), Sec61B (endoplasmic reticulum, ER), Tom20 (mitochondria, mito) and p58 (Golgi). Membrane proteins, weakly stained by Ponceau red, are enriched between 5-20 % Optiprep™).
Acknowledgements
We wish to thank Chitra Hindnavis for assistance with manuscript preparation. This work was supported by NIH P01AG017617 and NIH R01AG062376 to R.A.N.
Footnotes
Availability, specificity and quality of rab5 antibodies are critical to the study of endosomes. Most antibodies such as anti-Rab5 (BD Biosciences, 610281) and anti-EEA1 (BD Biosciences, 610457) are excellent for staining of cultured cell [14], but fail to work in mouse brain tissue section. The anti-Rab5b antibody (Santa Cruz Biotechnology Inc, SC-26569), which worked well in our previous studies [15,16], no longer exists. Finding and validating antibodies suitable for one’s specific needs is an ongoing priority task.
In order to detect vesicular/membrane-associated rab5, depending on the antibody and/or systems used (e.g cell culture vs. mouse tissue), it is of fundamental importance to determine the appropriate detergent type and concentration for sample permeabilization and antibody dilution. Non-ionic detergents such as Triton-X100 may extract proteins along with the lipids [29], resulting in poor staining. So, for instance, the anti-Rab5b antibody (Santa Cruz Biotechnology) works optimally in the presence of 0.4% Triton X-100, but it gives minimal signal with 0.05% saponin; on the other hand, saponin, which is more selective in targeting cholesterol [29], is preferred for the visualization of endosomes using Rab5a (Santa Cruz Biotechnology), Rab5-GTP (NewEast Biosciences) and Rab5 (Abcam) antibodies, whereas Triton X-100 results in a highly diffuse signal.
Size binning of Rab5-endosomes should be adjusted and set according to individual experimental conditions, including the cell type and antibody specificity. Presenting results in terms of a ratio to the experiment control instead of to absolute number is therefore recommended.
The stable N2aAPP line is generated after N2a cell transfection with a linearized pcDNA3-APP695 plasmid. Transfected cells are plated on 35mm dishes for 2 days and subcultured into 100mm dishes at a 1:10 dilution. Selection is obtained by cell incubation with G418 at 0.6mg/ml for two weeks as previously described [8].
Since the Rab5-GTP antibody is mouse monoclonal, the blocking reagent from the M.O.M kit (Vector Laboratories, BMK-2202) can be used to reduce the background when staining mouse tissue. However, it is recommended to use a custom-made antibody dilution buffer containing the appropriate detergent (e.g. 0.05% saponin), since other detergents eventually present in the commercial kit may affect the Rab5-GTP endosomal staining.
Synaptosomes can be isolated from mouse hippocampi, following homogenization in 250 μl of a sucrose solution (0.32 mol/L sucrose, 0.1 mmol/L CaCl2, 1 mmol/L MgCl2) with protease and phosphatase inhibitors [28]. After homogenization, samples are adjusted to 1.25 M and sequentially overlaid with 1.0 M Sucrose in 0.1 mM CaCl2 and homogenization buffer. Following sample centrifugation at 100,000 x for 3 h at 4 °C using a SW55Ti rotor (Beckman Coulter), the interface at 1.25-1.0 M sucrose, enriched in synaptosomes is diluted in ice-cold 0.1 mM CaCl2, centrifuged at 75,000 x g for 30 min at 4 °C in a SW55Ti rotor and the pellets washed in 0.1 mM CaCl2 and centrifuged again. The resultant washed synaptosome pellet is rich in pre-synaptic and post-synaptic membranes as well as in and synaptic soluble/vesicular components.
Since the Rab5-GTP antibody is “conformation-specific”, direct assessment of activated Rab5 by SDS-PAGE (or under denaturing conditions) cannot be performed.
In order to carry the Rab5 GTP-Agarose pull-down experiment successfully, the samples must be:1) fresh (do not freeze and thaw); 2) free of phosphatase inhibitors; 3) contain a cytosolic component, as factors regulating rab5 activation (GEF) and de-activation (GAP and GDI) are either cytosolic (GDI) or transiently associated with the endosomal membranes (GEF and GAP) [3]. Based on our experience, this assay would not work on isolated membranes.
References
- 1.Ullrich O, Horiuchi H, Bucci C, Zerial M (1994) Membrane association of Rab5 mediated by GDP-dissociation inhibitor and accompanied by GDP/GTP exchange. Nature 368 (6467):157–160. doi: 10.1038/368157a0 [DOI] [PubMed] [Google Scholar]
- 2.Felberbaum-Corti M, Van Der Goot FG, Gruenberg J (2003) Sliding doors: clathrin-coated pits or caveolae? Nature cell biology 5 (5):382–384. doi: 10.1038/ncb0503-382 [DOI] [PubMed] [Google Scholar]
- 3.Cavalli V, Vilbois F, Corti M, Marcote MJ, Tamura K, Karin M, Arkinstall S, Gruenberg J (2001) The stress-induced MAP kinase p38 regulates endocytic trafficking via the GDI:Rab5 complex. Mol Cell 7 (2):421–432 [DOI] [PubMed] [Google Scholar]
- 4.Dirac-Svejstrup AB, Sumizawa T, Pfeffer SR (1997) Identification of a GDI displacement factor that releases endosomal Rab GTPases from Rab-GDI. EMBO J 16 (3):465–472. doi: 10.1093/emboj/16.3.465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Horiuchi H, Giner A, Hoflack B, Zerial M (1995) A GDP/GTP exchange-stimulatory activity for the Rab5-RabGDI complex on clathrin-coated vesicles from bovine brain. J Biol Chem 270 (19):11257–11262. doi: 10.1074/jbc.270.19.11257 [DOI] [PubMed] [Google Scholar]
- 6.Haas AK, Fuchs E, Kopajtich R, Barr FA (2005) A GTPase-activating protein controls Rab5 function in endocytic trafficking. Nature cell biology 7 (9):887–893. doi: 10.1038/ncb1290 [DOI] [PubMed] [Google Scholar]
- 7.Nixon RA (2017) Amyloid precursor protein and endosomal-lysosomal dysfunction in Alzheimer's disease: inseparable partners in a multifactorial disease. FASEB J 31 (7):2729–2743. doi: 10.1096/fj.201700359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim S, Sato Y, Mohan PS, Peterhoff C, Pensalfini A, Rigoglioso A, Jiang Y, Nixon RA (2016) Evidence that the rab5 effector APPL1 mediates APP-betaCTF-induced dysfunction of endosomes in Down syndrome and Alzheimer's disease. Mol Psychiatry 21 (5):707–716. doi: 10.1038/mp.2015.97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bucci C, Parton RG, Mather IH, Stunnenberg H, Simons K, Hoflack B, Zerial M (1992) The small GTPase rab5 functions as a regulatory factor in the early endocytic pathway. Cell 70 (5):715–728 [DOI] [PubMed] [Google Scholar]
- 10.Gorvel JP, Chavrier P, Zerial M, Gruenberg J (1991) rab5 controls early endosome fusion in vitro. Cell 64 (5):915–925 [DOI] [PubMed] [Google Scholar]
- 11.Qi Y, Liang Z, Wang Z, Lu G, Li G (2015) Determination of Rab5 activity in the cell by effector pull-down assay. Methods Mol Biol 1298:259–270. doi: 10.1007/978-1-4939-2569-8_22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhan K, Xie H, Gall J, Ma M, Griesbeck O, Salehi A, Rao J (2011) Real-time imaging of Rab5 activity using a prequenched biosensor. ACS chemical biology 6 (7):692–699. doi: 10.1021/cb100377m [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cataldo AM, Mathews PM, Boiteau AB, Hassinger LC, Peterhoff CM, Jiang Y, Mullaney K, Neve RL, Gruenberg J, Nixon RA (2008) Down syndrome fibroblast model of Alzheimer-related endosome pathology: accelerated endocytosis promotes late endocytic defects. Am J Pathol 173 (2):370–384. doi: 10.2353/ajpath.2008.071053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jiang Y, Mullaney KA, Peterhoff CM, Che S, Schmidt SD, Boyer-Boiteau A, Ginsberg SD, Cataldo AM, Mathews PM, Nixon RA (2010) Alzheimer's-related endosome dysfunction in Down syndrome is Abeta-independent but requires APP and is reversed by BACE-1 inhibition. Proc Natl Acad Sci U S A 107 (4):1630–1635. doi: 10.1073/pnas.0908953107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cataldo AM, Petanceska S, Peterhoff CM, Terio NB, Epstein CJ, Villar A, Carlson EJ, Staufenbiel M, Nixon RA (2003) App gene dosage modulates endosomal abnormalities of Alzheimer's disease in a segmental trisomy 16 mouse model of down syndrome. J Neurosci 23 (17):6788–6792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jiang Y, Rigoglioso A, Peterhoff CM, Pawlik M, Sato Y, Bleiwas C, Stavrides P, Smiley JF, Ginsberg SD, Mathews PM, Levy E, Nixon RA (2016) Partial BACE1 reduction in a Down syndrome mouse model blocks Alzheimer-related endosomal anomalies and cholinergic neurodegeneration: role of APP-CTF. Neurobiol Aging 39:90–98. doi: 10.1016/j.neurobiolaging.2015.11.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pensalfini A, Seonil K, Subbanna S, Bleiwas C, Goulbourne CN, Stavrides P, Jiang Y, Lee J-H, Darji S, Pawlik M, Huo C, Berg M, Smiley J, Basavarajappa BS, Nixon RA (2019) Endosomal Dysfunction Induced by Directly Over-Activating Rab5 Recapitulates Prodromal and Neurodegenerative Features of Alzheimer's Disease. Cell Reports. doi: 10.2139/ssrn.3512167; SSRN: https://ssrn.com/abstract=3512167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee MS, Kao SC, Lemere CA, Xia W, Tseng HC, Zhou Y, Neve R, Ahlijanian MK, Tsai LH (2003) APP processing is regulated by cytoplasmic phosphorylation. J Cell Biol 163 (1):83–95. doi: 10.1083/jcb.200301115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shi MM, Shi CH, Xu YM (2017) Rab GTPases: The Key Players in the Molecular Pathway of Parkinson's Disease. Frontiers in cellular neuroscience 11:81. doi: 10.3389/fncel.2017.00081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Farg MA, Sundaramoorthy V, Sultana JM, Yang S, Atkinson RA, Levina V, Halloran MA, Gleeson PA, Blair IP, Soo KY, King AE, Atkin JD (2014) C9ORF72, implicated in amytrophic lateral sclerosis and frontotemporal dementia, regulates endosomal trafficking. Hum Mol Genet 23 (13):3579–3595. doi: 10.1093/hmg/ddu068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hadano S, Hand CK, Osuga H, Yanagisawa Y, Otomo A, Devon RS, Miyamoto N, Showguchi-Miyata J, Okada Y, Singaraja R, Figlewicz DA, Kwiatkowski T, Hosler BA, Sagie T, Skaug J, Nasir J, Brown RH Jr., Scherer SW, Rouleau GA, Hayden MR, Ikeda JE (2001) A gene encoding a putative GTPase regulator is mutated in familial amyotrophic lateral sclerosis 2. Nat Genet 29 (2):166–173. doi: 10.1038/ng1001-166 [DOI] [PubMed] [Google Scholar]
- 22.Axelrod D, Koppel DE, Schlessinger J, Elson E, Webb WW (1976) Mobility measurement by analysis of fluorescence photobleaching recovery kinetics. Biophysical journal 16 (9):1055–1069. doi: 10.1016/S0006-3495(76)85755-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vieira OV, Bucci C, Harrison RE, Trimble WS, Lanzetti L, Gruenberg J, Schreiber AD, Stahl PD, Grinstein S (2003) Modulation of Rab5 and Rab7 recruitment to phagosomes by phosphatidylinositol 3-kinase. Molecular and cellular biology 23 (7):2501–2514. doi: 10.1128/mcb.23.7.2501-2514.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cataldo AM, Mathews PM, Boiteau AB, Hassinger LC, Peterhoff CM, Jiang Y, Mullaney K, Neve RL, Gruenberg J, Nixon RA (2008) Down syndrome fibroblast model of Alzheimer-related endosome pathology. Accelerated endocytosis promotes late endocytic defects. Am J Pathol 173:370–384. doi:ajpath.2008.071053 [pii] 10.2353/ajpath.2008.071053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee JH, Rao MV, Yang DS, Stavrides P, Im E, Pensalfini A, Huo C, Sarkar P, Yoshimori T, Nixon RA (2018) Transgenic expression of a ratiometric autophagy probe specifically in neurons enables the interrogation of brain autophagy in vivo. Autophagy:1–15. doi: 10.1080/15548627.2018.1528812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gruenberg J, Griffiths G, Howell KE (1989) Characterization of the early endosome and putative endocytic carrier vesicles in vivo and with an assay of vesicle fusion in vitro. J Cell Biol 108 (4):1301–1316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huotari J, Helenius A (2011) Endosome maturation. EMBO J 30 (17):3481–3500. doi: 10.1038/emboj.2011.286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Louneva N, Cohen JW, Han LY, Talbot K, Wilson RS, Bennett DA, Trojanowski JQ, Arnold SE (2008) Caspase-3 is enriched in postsynaptic densities and increased in Alzheimer's disease. Am J Pathol 173 (5):1488–1495. doi: 10.2353/ajpath.2008.080434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oliver C, Jamur MC (2010) Immunocytochemical methods and protocols. Methods Mol Biol 588:iv–v. doi: 10.1007/978-1-59745-324-0 [DOI] [PubMed] [Google Scholar]