ABSTRACT
Regeneration after peripheral nerve damage requires that axons re-grow to the correct target tissues in a process called target-specific regeneration. Although much is known about the mechanisms that promote axon re-growth, re-growing axons often fail to reach the correct targets, resulting in impaired nerve function. We know very little about how axons achieve target-specific regeneration, particularly in branched nerves that require distinct targeting decisions at branch points. The zebrafish vagus motor nerve is a branched nerve with a well-defined topographic organization. Here, we track regeneration of individual vagus axons after whole-nerve laser severing and find a robust capacity for target-specific, functional re-growth. We then develop a new single-cell chimera injury model for precise manipulation of axon-environment interactions and find that (1) the guidance mechanism used during regeneration is distinct from the nerve's developmental guidance mechanism, (2) target selection is specified by neurons' intrinsic memory of their position within the brain, and (3) targeting to a branch requires its pre-existing innervation. This work establishes the zebrafish vagus nerve as a tractable regeneration model and reveals the mechanistic basis of target-specific regeneration.
KEY WORDS: Axon guidance, Axon regeneration, Neuron identity, Topographic map, Vagus, Zebrafish
Summary: A new target-specific nerve regeneration model reveals that zebrafish vagus neurons rely on a cell-intrinsic memory of their positional identities to rebuild the topographic innervation pattern of the nerve robustly after injury.
INTRODUCTION
Peripheral nerve damage caused by injury, infection or disease is a common medical condition. Although humans possess some capacity to regenerate peripheral nerves, this process often fails or is incomplete, causing chronic deficits (Kuffler and Foy, 2020). Nerve injury generates axon ends that have lost synaptic connectivity with their target tissues. To restore functional connectivity, these axons must reprise two behaviors that are otherwise limited to their initial development. First, axons must re-establish a growth cone to promote their re-extension into the periphery (Leite et al., 2021). Second, re-extending axons must correctly interpret guidance cues along their growth trajectories in order to reach the appropriate target tissues, a process known as target-specific regeneration (Bolívar et al., 2020; Brushart, 1993; Koeberle and Bähr, 2004; Madison et al., 2007). This presents a unique challenge in the post-embryonic environment, which differs drastically in architecture and molecular composition from that of the embryo and may lack many of the growth and guidance factors that the axon relied upon during its initial development. However, the extent of these differences, their impacts on axon regeneration, and the mechanisms utilized by regenerating axons to confront these challenges are unclear. Recently, great strides have been made in identifying molecular pathways that promote axon re-growth after injury and in applying this knowledge to enhance regenerative axon growth in mammals (Anderson et al., 2018; Sun et al., 2011; Tedeschi et al., 2019). However, these interventions often fail to provide proper spatial guidance information, resulting in failure to re-innervate the correct targets (English, 2005; Luo et al., 2013). By contrast, zebrafish (Danio rerio) and other teleosts exhibit exceptional nerve regeneration; previous studies have reported 70-90% accuracy of axon re-targeting after injury (Isaacman-Beck et al., 2015; Lozano-Ortega et al., 2018; Stuermer and Easter, 1984). Although we know a good deal about the signals and mechanisms that guide axon targeting during embryonic development, we know very little about how regenerating axons navigate the post-developmental environment to ensure target-specific regeneration, particularly along complex paths that require distinct decision-making between multiple potential paths.
The conserved zebrafish vagus motor nerve (cranial motor nerve X) is a multifunctional branched nerve consisting of a single nucleus of neurons in the hindbrain that extend axons along a common fascicle before splitting into five distinct branches, four of which innervate muscles of posterior pharyngeal arches 4, 5, 6 and 7 (branches 4, 5, 6 and 7) and one of which extends posteriorly and innervates visceral organs (branch V) (Fig. 1A). The vagus nerve exhibits a conserved topographic organization along the anterior-posterior (A-P) axis, with more anterior neurons innervating more anterior branches (Fig. 1B) (Barsh et al., 2017; Bieger and Hopkins, 1987; Isabella et al., 2020). The mammalian vagus nerve regenerates poorly after injury (Peters et al., 2013; Phillips et al., 2003), which can result in loss of speech, difficulty swallowing, abnormal heart rate, and gastroparesis (Bunch et al., 2008; Vijayvargiya and Camilleri, 2019; Xie et al., 2020); therefore, it is important to understand how to improve this process.
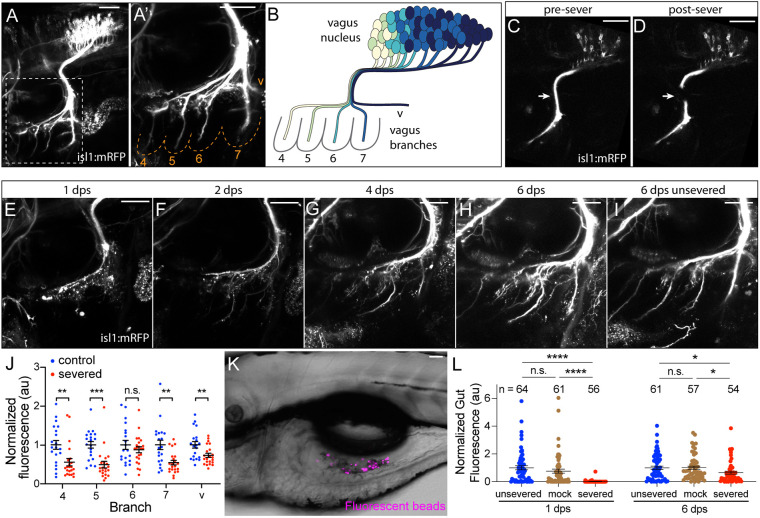
Fig. 1.
Structural and functional vagus nerve regeneration after laser axon severing. (A,A′) Zebrafish vagus nerve structure at 4 dpf. A′ shows an enlarged view of the boxed region in A. The five nerve branches are labeled. Dashed lines denote PAs. (B) A-P vagus nerve topography. Cells are color coded by branch. (C,D) Vagus nerve fascicle at 3 dpf immediately before (C) and after (D) laser severing; images taken from Movie 1. Arrows indicate sever site. (E-H) Time-lapse of vagus nerve regeneration after laser severing. At 1 dps (E), distal axon material is fragmented. See A′ for stage-matched unsevered control. By 2 dps (F), distal material is cleared and branches begin to re-grow. Between 4 dps (G) and 6 dps (H), branches complete re-growth. (I) Unsevered control at 6 dps. (J) Branch fluorescence intensity in control unsevered and severed larvae at 6 dps. All branches re-grow, but to varying extents. n=20 (control), 22 (severed). (K) Representative larval gut after eating paramecia containing fluorescent beads. (L) Gut fluorescence intensity after paramecia feeding in unsevered, bilaterally mock severed, and bilaterally severed larvae at 1 dps and 6 dps. In J and L, error bars represent mean with s.e.m. *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001; n.s., not significant (unpaired two-tailed t-test). (A,E-I) Maximum intensity projections. All images are lateral views with anterior to left. Scale bars: 50 µm.
Proper regeneration of the vagus nerve would require that each axon, upon reaching the peripheral branch point, accurately select between the five branches in order to maintain topographic organization, making it an excellent model in which to examine how axon targeting decisions are made during regeneration. Some topographically organized nerves, particularly those for which developmental axon targeting is guided by the stable expression of spatially patterned guidance cues, can re-deploy the same guidance cues used during their development (Rodger et al., 2004). We previously identified the mechanisms that guide topographic axon targeting during vagus development (Barsh et al., 2017; Isabella et al., 2020). Rather than relying on stable spatial guidance cues, vagus motor neurons use a highly dynamic process known as temporal matching, in which coordinated anterior-to-posterior expression wavefronts of the chemoattractant Hgf and its receptor, Met, in the pharyngeal arches and vagus motor neurons, respectively, drive temporally patterned topographic innervation of the pharyngeal arches by ensuring that motor neurons become competent to respond to pharyngeal arch chemoattraction (via Met expression) at the same time that their target pharyngeal arch becomes competent to receive them (via Hgf expression). Because temporal matching relies on the specific dynamic context of the embryo, it does not seem possible to re-use this mechanism during regeneration. The zebrafish vagus nerve thus provides an opportunity to discover de novo mechanisms of target-specific axon regeneration.
Here, we establish the larval zebrafish vagus nerve as a tractable model for target-specific axon regeneration. We establish two reproducible nerve injury models: a laser axon severing (Hu et al., 2018; Isaacman-Beck et al., 2015) model for whole-nerve injury and a novel single-cell chimera model for single-axon injury. We follow the re-growth of individual axons in both models and find that axon growth is properly guided to re-establish/maintain the topographic organization of the nerve during regeneration. We then use the single-cell chimera model to assess axon targeting under a series of environmental challenges, and find that (1) vagus motor axon targeting does not rely on the Hgf/Met developmental guidance signal; (2) larval vagus motor neurons possess an intrinsic knowledge of their target tissue based on a memory of their position in the vagus motor nucleus, and (3) re-growth of an axon to its target requires that pre-existing innervation of that target by other vagus axons. These results establish a conceptual basis for the mechanism by which regenerating motor axons make distinct guidance decisions to promote target-specific nerve regeneration.
RESULTS
Structural and functional vagus nerve regeneration after whole-nerve severing
By the beginning of the larval stage [3 days post-fertilization (dpf)], vagus motor neurogenesis has been completed (Barsh et al., 2017), and vagus axons have completed their developmental targeting to establish the five primary branches (Barsh et al., 2017) and begun to form neuromuscular synapses with their targets (Fig. S1A-D′). Although the vagus nerve appears to be fully ensheathed by Schwann cells at 2-3 dpf (Fig. S2A,A′,I,I′) (Cox et al., 2011), this appears to be transient: from 4-9 dpf we observe glia [marked by Tg(Sox10:mRFP)] and myelination [marked by Tg(mbp:eGFP-caax)] only in the region of the vagus fascicle proximal to where it turns ventrally out of the brain and not in the peripheral portions of the nerve (Fig. S2B-H′). We do not know whether the peripheral region becomes myelinated after 9 dpf. Overall, these data indicate that 3 dpf represents a sufficiently mature stage at which to examine regeneration.
To determine whether the vagus nerve can regenerate, we used a MicroPoint laser to sever the common fascicle of the vagus nerve at 3 dpf (Fig. 1C,D, Movie 1). This resulted in a rapid retraction of the severed ends away from the sever site (Movie 1). We then tracked nerve re-growth for 6 days after severing. In the first two days post-severing (dps), the axonal material distal to the cut site was fragmented (Fig. 1E) and cleared (Fig. 1F), indicative of Wallerian degeneration (Coleman and Freeman, 2010; Martin et al., 2010). Between 2 and 6 dps, axons re-grew into the denervated territory (Fig. 1F-H). Recovery of all five branches was observed in all animals, although the extent of that recovery varied, and branches often did not recover to a level equivalent to stage-matched unsevered controls (Fig. 1I,J). Regenerating axons also rebuilt neuromuscular synapses (Fig. S1E,E′). We observed no association of Schwann cells with regenerating vagus axons (Fig. S2I-J′), consistent with our observations that Schwann cells are normally absent from the peripheral vagus region after 3 dpf (Fig. S2B-D′).
Vagus motor innervation of pharyngeal and esophageal muscles is essential for swallowing (Erman et al., 2009; Finsterer and Grisold, 2015; Karpinski et al., 2014). To assess whether the regeneration we observed is sufficient to restore vagus function, we analyzed larval feeding behavior by incubating larvae for 1 h with paramecia containing fluorescent beads and measuring fluorescence in the gut (Fig. 1K) (Boyer et al., 2013). Because larvae do not begin to eat until 5 dpf, for this experiment we performed nerve severing at 4 dpf. Bilaterally severed larvae exhibited a virtually complete failure to eat at 1 dps relative to unsevered and mock-severed control animals (Fig. 1L). By 6 dps, however, feeding in these animals had recovered nearly to control levels (Fig. 1L), indicating substantial recovery of vagus function.
Target-specific vagus nerve regeneration after laser axon severing
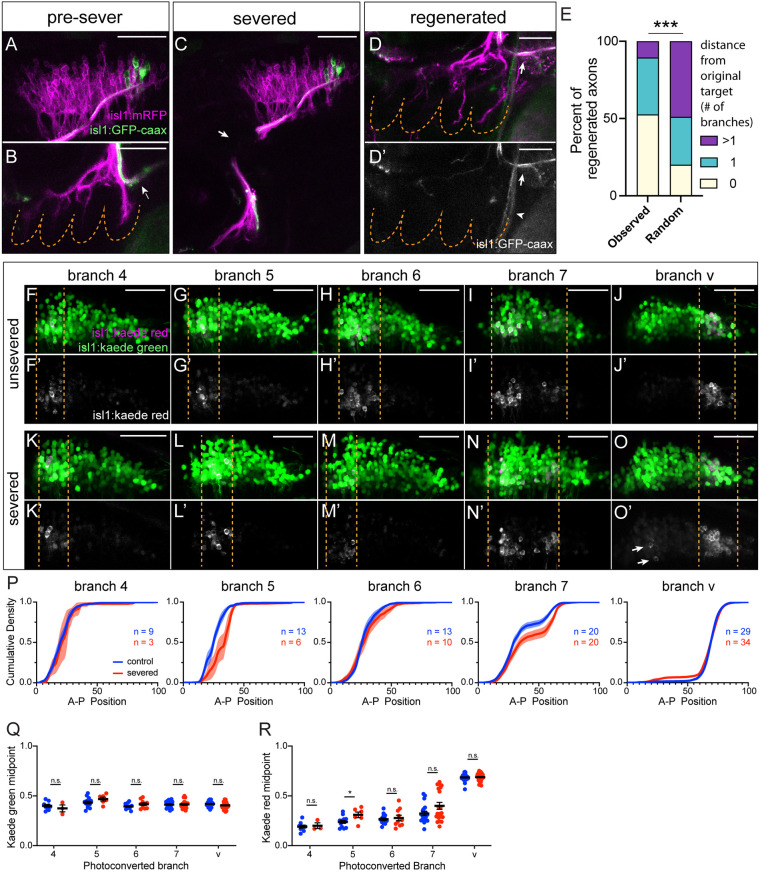
Re-establishing the correct topography is crucial to regeneration, as a scrambled map would likely have negative functional consequences. To test whether vagus axon regeneration is target specific, we combined our laser axon-severing experiment with sparse mosaic transgenic labeling of vagus neurons (Barsh et al., 2017). This allowed us to observe the innervation target of the same neuron(s) prior to severing at 3 dpf (Fig. 2A,B) and again after severing and regeneration (Fig. 2C-D′). We classified neurons by the distance (in number of branches) between their original branch and the branch they re-grew to, with 0 indicating re-targeting to the original branch (Fig. 2E) and found that 53% of axons returned to their original branch. However, because neurons at the same A-P position often innervate neighboring branches (Fig. 1B) (Barsh et al., 2017), re-growth of an axon to a branch that was one branch (score of 1) away from its original target would still, in almost all cases, represent topographically appropriate re-targeting. Re-growth to a branch greater than one branch (score of >1) away from the original target would, by contrast, almost always represent topographically inappropriate re-targeting. Combining the 0 and 1 categories, we found that 89% of axons exhibit topographically appropriate re-targeting. This distribution represents a statistically significant bias towards target-specific regeneration compared with expectations based on random targeting (Fig. 2E), and is similar to previous reports of target specificity in teleost fishes (Isaacman-Beck et al., 2015; Lozano-Ortega et al., 2018; Stuermer and Easter, 1984). Neurons showed an equivalent capacity for target-specific re-growth regardless of which branch they originally targeted (Fig. S3).
Fig. 2.
Target-specific vagus nerve regeneration after laser axon severing. (A,B) Sparsely labeled vagus neurons (green) (A) targeting branch v (arrow) (B) prior to severing at 3 dpf. (C) Severed vagus nerve at 3 dpf. Arrow indicates sever site. (D,D′) The same animal at 3 dps. Branches have regenerated, and green labeled axons (white in D′) have re-targeted to branch v (arrows). Arrowhead indicates autofluorescence of cleithrum bone. In B,D,D′, dashed lines denote PAs. (E) Frequency of re-targeting of sparsely labeled regenerated axons to a branch 0, 1 or >1 branches away from the original target, observed distribution (n=19) versus expected distribution assuming random branch selection. ***P=0.0004 (Chi-squared test). (F-O) Vagus topographic organization measured by cell backfills after photoconversion (magenta) of individual branches 4 (F,K), 5 (G,L), 6 (H,M), 7 (I,N) and v (J,O) in unsevered (F-J) and severed (K-O) larvae at 6 dps. (F′-O′) Photoconverted channel only. Dashed lines denote anterior and posterior boundaries of backfilled regions. Individual neurons with topographically incorrect re-targeting are occasionally observed in regenerated nerves (arrows in O′). (P) Anterior-to-posterior cumulative densities of Kaede red fluorescence in the vagus nucleus after photoconversion of each branch. Shaded area indicates s.e.m. (Q,R) Cumulative density midpoints of Kaede green fluorescence (Q) and Kaede red fluorescence (R) in the vagus nucleus after photoconversion of each branch. Error bars represent mean with s.e.m. *P<0.05; n.s., not significant (unpaired two-tailed t-test). All images are maximum intensity projections of lateral views with anterior to left. Scale bars: 50 µm.
As a second approach to assess the topography of the regenerated vagus nerve, we utilized a method we previously established to label the neurons that innervate individual branches by photoconversion of a branch and backfilling of photoconverted protein into the cell bodies (Isabella et al., 2020). We labeled the neurons innervating each branch in unsevered and severed Tg(isl1:Kaede)-expressing nerves at 6 dps (Fig. 2F-O′), and measured the anterior-to-posterior cumulative density of Kaede red fluorescence in the vagus nucleus (Fig. 2P-R). We found only very minor topographic differences between unsevered and regenerated nerves (Fig. 2P-R). Thus, regenerating vagus axons exhibit a very high degree of target specificity and re-establish topographic organization during regeneration.
Dynamics of single-cell chimera axon regeneration
Our results thus far suggest that each axon can distinguish between different targets and select the appropriate target for innervation. Fundamentally, this must require that vagus neurons have distinct molecular identities that allow them to interpret distinct environmental guidance signals along the path to each target. The capacity to experimentally perturb cell-environment interactions using the laser severing approach is limited. We therefore sought to establish a new and more experimentally powerful axon regeneration model. The generation of chimeras – composite animals containing cells from more than one individual – by cell transplantation is a classic embryological technique with two strengths that are well suited to this goal: the ability to track the behaviors and fates of specific cells and to manipulate the environmental context in which those cells act (Carmany-Rampey and Moens, 2006; Ribatti, 2019; Tam and Rossant, 2003). We previously transplanted developing vagus neurons in 1 dpf embryos (Barsh et al., 2017). Here, we established methods to transplant individual mature neurons from the vagus nucleus of Tg(isl1:GFP-caax) donor larvae into the vagus nucleus of Tg(isl1:mRFP) host larvae at 3 dpf (Fig. 3A). Crucially, this approach serves as a reproducible axon injury model – the donor neuron's original axon is torn off during transplantation and it must re-grow a new axon in the host.
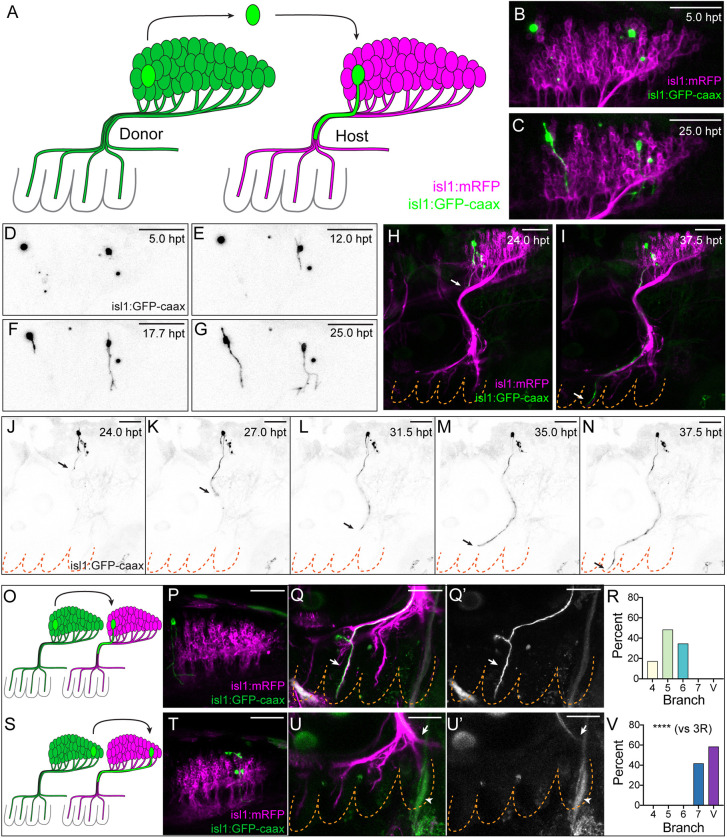
Fig. 3.
Target-specific axon regeneration in single-cell chimeras. (A) Transplantation scheme. Individual vagus neurons are transplanted from Tg(isl1:GFP-caax) donors to Tg(isl1:mRFP) hosts at 3 dpf and subsequently regenerate axons. (B-G) Dynamics of axon re-establishment in the first day after transplantation; images taken from Movie 2. (B) First frame (5 hpt). (C) Last frame (25 hpt). (D-G) Time-lapse of two GFP neurons regenerating axons after transplantation. (H-N) Dynamics of axon re-growth into the periphery during the second day after transplantation; images taken from Movie 3. (H) First frame (24 hpt). (I) Last frame (37.5 hpt). (J-N) Time-lapse of a GFP neuron re-extending an axon to branch 5 after transplantation. Images in B-G and H-N are from different animals. (O-V) Target-specific regeneration after neuron transplantation. (O) Homotopic A→A transplant schematic. (P-Q′) Representative A→A transplanted neuron (green, P) and its regenerated axon (green in Q and white in Q′) (arrows) in branch 5 at 3 dpt. (R) Branch distribution of regenerated A→A axons (n=29). (S) Homotopic P→P transplant schematic. (T-U′) Representative P→P transplanted neuron (green, T) and its regenerated axon (green in U and white in U′) (arrows) in branch v at 3 dpt. Arrowheads indicate autofluorescence of cleithrum bone. (V) Branch distribution of regenerated P→P axons (n=24). ****P=5.133e−15 [Fisher's exact test versus A→A (panel R)]. All images are maximum intensity projections of lateral views with anterior to left. Dashed lines denote PAs. Scale bars: 50 µm.
Axon regeneration after single-neuron transplantation is robust: 66% of transplants (n=453) resulted in the successful re-growth of an axon to one of the peripheral vagus branches. In nearly all remaining cases, the donor neuron died after being placed in the host, likely due to damage during transplantation, although in ∼2.5% of cases the transplanted neuron aberrantly extended an axon to somewhere other than the vagus branches.
The single-cell chimera regeneration model allows for high-resolution live imaging of single axon regeneration (Fig. 3B-G, Movie 2). Transplanted neurons initially rounded up (Fig. 3D, Movie 2). Beginning at ∼12 h post-transplantation (hpt), neurons exhibited multipolar protrusive activity (Fig. 3E-G, Movie 2). At ∼24 hpt, they extended an axon (Fig. 3G) that grew along the vagus fascicle to a peripheral branch (Fig. 3H-N, Movie 3). Most axons had extended to a branch by 2 days post-transplantation (dpt), and all completed their growth by 3 dpt.
Single-cell chimera axon regeneration is target specific
To assess the target specificity of axon regeneration in our single-cell chimeras, we performed homotopic transplants of anterior and posterior neurons, whereby neurons were placed in the same location in the host as they had occupied in the donor (denoted as A→A or P→P, respectively). ‘A’ corresponds to the anterior 20% of the vagus nucleus and ‘P’ corresponds to the posterior 40% of the vagus nucleus, by length. We then measured the branch that the regenerated axons innervated at 3 dpt. Although we could not measure the branch that the transplanted neuron originally innervated, we knew from our previously published topographic mapping data that A neurons exclusively innervate branches 4, 5 and 6, and P neurons exclusively innervate branches 7 and V (Fig. 1B) (Barsh et al., 2017), and could therefore determine whether re-targeting is topographically appropriate. We observed a perfect maintenance of topography during single-axon regeneration: A→A transplanted neurons re-grew exclusively to branches 4, 5 and 6 (Fig. 3O-R), and P→P transplanted neurons re-grew exclusively to branches 7 and V (Fig. 3S-V). Thus, consistent with our observations from the laser axon-severing experiments, vagus axon regeneration after neuron transplantation is target specific.
Target-specific regeneration does not require the embryonic guidance signal Hgf/Met
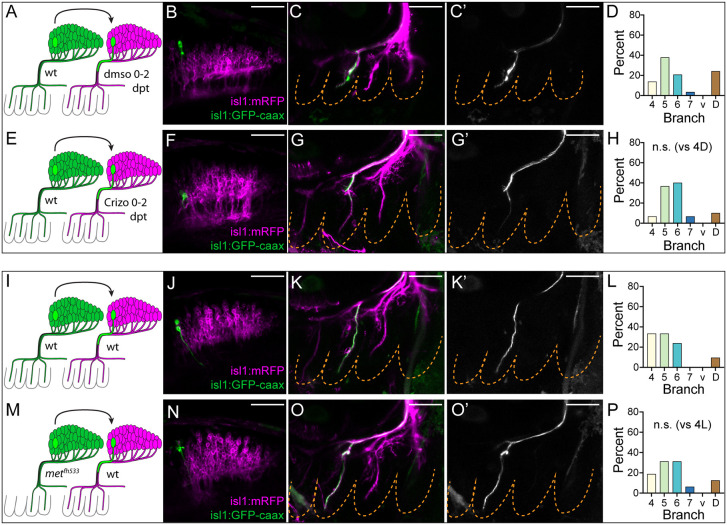
Some regenerating axons can reuse their developmental signaling mechanisms to guide re-growth (Rodger et al., 2004). However, because the embryonic mechanism promoting vagus topographic patterning requires that axon growth and guidance be precisely coordinated with the developmental progression of the pharyngeal arches (Barsh et al., 2017; Isabella et al., 2020), we hypothesize that this mechanism cannot be re-deployed to guide target-specific axon growth during regeneration. To determine whether regenerating axons utilize a different targeting mechanism, we examined whether Hgf/Met signaling, which constitutes the key chemical guidance cue during embryonic vagus axon targeting, influences axon targeting decisions during regeneration in the single-cell chimera model.
During embryonic development, the pharyngeal arch-derived chemoattractant Hgf signals to Met receptor-expressing vagus motor neurons to attract them sequentially to their targets. In embryos mutant for met or hgfa, or those treated with the Met inhibitor crizotinib (Crizo), vagus axons stalled at the choice point and, ultimately, failed to innervate anterior branches (branch 4 and sometimes branch 5) (Isabella et al., 2020). To examine whether Hgf/Met signaling influences targeting during regeneration, we employed two independent approaches to block Met function in regenerating neurons. First, we performed homotopic A→A transplants at 3 dpf and treated host animals with DMSO or 20 µM Crizo beginning immediately after transplantation until endpoint imaging (Fig. 4A-H). Second, we performed homotopic A→A transplants at 3 dpf from metfh533 mutant donors into wild-type hosts (metfh533 A→wt A) (Fig. 4I-P). Endpoint imaging was carried out at 2 dpt for these experiments because at that time point most but not all axons have completed their targeting (Fig. 4D), thereby allowing us to measure whether axons lacking Met function exhibited delayed axon extension in addition to altered targeting. Axons that had extended out of the brain but not past the choice point were categorized as ‘D’ for delayed in this analysis. In both experiments, regenerating axons lacking Met function had neither delayed axon outgrowth nor altered target selection compared with controls (Fig. 4D,H,L,P). Thus, the major signaling mechanism that guides vagus axon targeting during development is not involved in the guidance of regenerating vagus axons, which we conclude must instead rely on a regeneration-specific guidance mechanism.
Fig. 4.
Target-specific regeneration does not require the embryonic guidance signal Hgf/Met. (A-H) Met inhibition with crizotinib does not affect growth or targeting of regenerating axons. (A) Schematic for A→A transplants followed by DMSO treatment from 0 to 2 dpt. (B-C′) Representative A→A+DMSO 0-2 dpt transplanted neuron (green, B) and its regenerated axon (green in C and white in C′) in branch 5 at 2 dpt. (D) Branch distribution of regenerated A→A+DMSO 0-2 dpt axons (n=25). ‘D’ indicates axons that have extended out of the brain along the vagus fascicle but have not completed their targeting by 2 dpt. (E) Schematic for A→A transplants followed by Crizo treatment from 0 to 2 dpt. (F-G′) Representative A→A+Crizo 0-2 dpt transplanted neuron (green, F) and its regenerated axon (green in G and white in G′) in branch 5 at 2 dpt. (H) Branch distribution of regenerated A→A+Crizo 0-2 dpt axons (n=30). n.s., not significant. P=0.3486 [Fisher's exact test versus A→A+DMSO 0-2 dpt (panel D)]. (I-P) metfh533 mutant donor neurons show normal axon growth and targeting during regeneration. (I) Schematic for wt A→wt A transplants. (J-K′) Representative wt A→wt A transplanted neuron (green, J) and its regenerated axon (green in K and white in K′) in branch 5 at 2 dpt. (L) Branch distribution of regenerated wt A→wt A axons (n=21). ‘D’ indicates axons that have extended out of the brain along the vagus fascicle but have not completed their targeting by 2 dpt. (M) Schematic for metfh533 A→wt A transplants. (N-O′) Representative metfh533 A→wt A transplanted neuron (green, N) and its regenerated axon (green in O and white in O′) in branch 5 at 2 dpt. (P) Branch distribution of regenerated metfh533 A→wt A axons (n=16). n.s., not significant: P=0.7759 [Fisher's exact test versus wt →wt A (panel L)]. All images are maximum intensity projections of lateral views with anterior to left. Dashed lines denote PAs. Scale bars: 50 µm.
Intrinsic positional memory specifies target selection during regeneration
How does a neuron know which target to select during regeneration? This is clearly linked to A-P position, as A and P neurons reliably select different targets. During embryonic development, vagus neuron positional identity is determined by extrinsic signals in the hindbrain (Isabella et al., 2020). Larval neurons could similarly rely on extrinsic identity cues, or they could have an intrinsic molecular knowledge of their positional identity. To determine whether vagus neuron identity is extrinsically or intrinsically regulated during regeneration, we performed heterotopic transplants, in which neurons were placed in a different location in the host than they had occupied in the donor (A→P and P→A) and asked whether transplanted neurons selected targets consistent with their original position in the donor (indicative of intrinsic identity) or with their new position in the host (indicative of extrinsic identity). In both cases, axons re-grew to targets consistent with their original position in the donor (Fig. 5A-H). Although targeting was slightly more variable in these conditions than in the homotopic transplants, suggesting a modest environmental influence, we conclude that the identity of a regenerating neuron, defined here as its knowledge of which axon branch to target, is predominantly an intrinsic property of the cell.
Fig. 5.
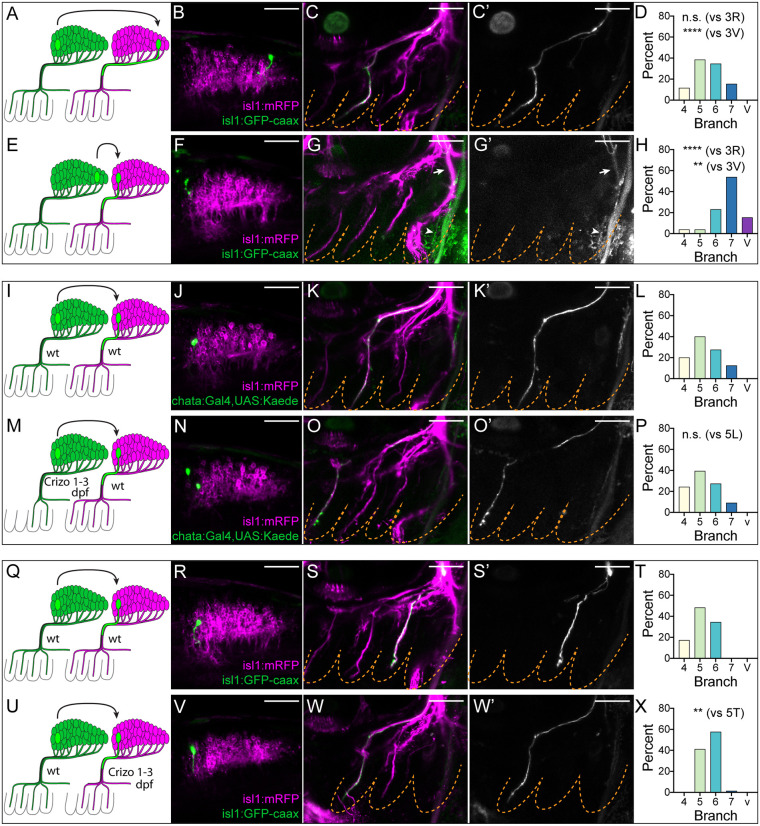
Target-specific regeneration is guided by intrinsic positional memory and pre-existing branch innervation. (A-H) Axon target selection during regeneration is intrinsically specified. (A) Heterotopic A→P transplant schematic. (B-C′) Representative A→P transplanted neuron (green, B) and its regenerated axon (green in C and white in C′) in branch 5 at 3 dpt. (D) Branch distribution of regenerated A→P axons (n=26). n.s., not significant: P=0.1839 [Fisher's exact test versus A→A (Fig. 3R)]; ****P=1.541e−10 [Fisher's exact test versus P→P (Fig. 3V)]. (E) Heterotopic P→A transplant schematic. (F-G′) Representative P→A transplanted neuron (green, F) and its regenerated axon (green in G and white in G′) in branch 7 at 3 dpt (arrows). Arrowheads indicate autofluorescence of cleithrum bone. (H) Branch distribution of regenerated P→A axons (n=26). ****P=3.075e−8 [Fisher's exact test versus A→A (Fig. 3R)]; **P=0.001256 [Fisher's exact test versus P→P (Fig. 3V)]. (I-P) Axon target selection during regeneration is specified by A-P position rather than by innervation history. (I) wt A→wt A transplant schematic. (J-K′) Representative wt A→wt A transplanted neuron (green, J) and its regenerated axon (green in K and white in K′) in branch 5 at 3 dpt. (L) Branch distribution of regenerated wt A→wt A axons (n=40). (M) Crizo 1-3 dpf A→wt A transplant schematic. (N-O′) Representative Crizo 1-3 dpf A→wt A transplanted neuron (green, N) and its regenerated axon (green in O and white in O′) in branch 4 at 3 dpt. (P) Branch distribution of regenerated Crizo 1-3 dpf A→wt A axons (n=33). n.s., not significant: P=0.9723 [Fisher's exact test versus wt A→wt A (panel L)]. (Q-X) Branch innervation during regeneration requires pre-existing innervation of that branch. (Q) wt A→wt A transplant schematic. (R-S′) Representative wt A→wt A transplanted neuron (green, R) and its regenerated axon (green in S and white in S′) in branch 6 at 3 dpt. (T) Branch distribution of regenerated wt A→wt A axons (n=29). Data shown in T are the same as presented in Fig. 3R. (U) wt A→Crizo 1-3 dpt A transplant schematic. (V-W′) Representative wt A→Crizo 1-3 dpt A transplanted neuron (green, V) and its regenerated axon (green in W and white in W′) in branch 5 at 3 dpt. Note the absence of host branch 4 (magenta). (X) Branch distribution of regenerated wt A→Crizo 1-3 dpt A axons (n=73). **P=0.001414 [Fisher's exact test versus wt A→wt A (panel T)]. All images are maximum intensity projections of lateral views with anterior to left. Scale bars: 50 µm.
The finding that a regenerating neuron can extend its axon to the correct target regardless of its position in the vagus nucleus suggests that the neuron possesses a molecular memory of its target. How is this memory conferred? There are two possible explanations: (1) the neuron remembers its original A-P position in the donor and selects an axon target consistent with that position during regeneration (Sperry, 1963); or (2) the neuron remembers which branch it had previously innervated in the donor and returns to that branch during regeneration. Distinguishing between these possibilities requires a donor larva in which A-P position and innervation target have been dissociated. We previously identified just such a condition: embryos treated with crizotinib from 24-72 h post-fertilization (Crizo 1-3 dpf) fail to generate branches 4 and often 5, and A neurons which would normally innervate branches 4 and 5, instead innervate branch 6 (Isabella et al., 2020). This condition does not affect the signals that regulate positional identity in the vagus nucleus. Therefore, by performing A→A transplants from Crizo 1-3 dpf donors lacking branches 4 and 5 into wild-type hosts (Crizo 1-3 dpf A→wt A), we could investigate whether transplanted neurons select targets consistent with their position (branches 4, 5 and 6) or with their innervation history (branch 6 only). Neurons transplanted from Crizo 1-3 dpf donors readily re-grew to branches 4 and 5 with the same frequency as controls (Fig. 5I-P), suggesting that the targeting history of a neuron does not determine which target it selects during regeneration. The metfh533 A→wt A transplants described in the last section (Fig. 4M-P) offer a conceptually similar test of this principle and showed the same result. We conclude that the intrinsic identity of vagus neurons in the larva is based on a memory of their A-P position within the hindbrain, rather than memory of their prior target.
Regeneration targeting requires pre-existing branch innervation
We next sought to understand how regenerating vagus axons are guided to the correct target tissues. This process is, by definition, extrinsically regulated: growing axons must interpret environmental cues along their growth trajectories to distinguish between potential paths. Two major processes that can guide axon growth have been established: (1) axons may sense chemoattractive and chemorepulsive cues to guide their growth (Chilton, 2006; Hindges et al., 2002; Kidd et al., 1998); and (2) axons may grow along a pre-existing physical scaffold, often in the form of other axons that have previously extended along their intended path (Bak and Fraser, 2003; Spead and Poulain, 2020; Treubert-Zimmermann et al., 2002). Although these processes are not mutually exclusive, a major distinguishing factor between them is the requirement for a pre-existing scaffold. The former process does not necessarily require such a scaffold and is the means by which ‘pioneer’ axons, which forge bravely into previously uninnervated territory, extend to their targets (Chilton, 2006; Hindges et al., 2002; Kidd et al., 1998). The latter process does require a pre-existing scaffold, and represents a means by which ‘follower’ axons, which extend along pioneer axons, reach their targets (Bak and Fraser, 2003; Spead and Poulain, 2020; Treubert-Zimmermann et al., 2002).
To determine whether regenerating vagus axons require a pre-existing scaffold, we performed the inverse of the experiment described in the last section: we homotopically transplanted wild-type A neurons into Crizo 1-3 dpf hosts, which were lacking branch 4 and 5 innervation (wt A→Crizo 1-3 dpf A). We then asked whether regenerating axons retained their ability to extend into all anterior branches (indicative of pioneering status) or whether they were constrained to branches with pre-existing innervation (indicative of follower status). Of note, although we screened for Crizo 1-3 dpf hosts lacking branches 4 and 5 prior to transplantation at 3 dpf, by 6 dpf (3 dpt) branch 5 was present in virtually all hosts, suggesting this branch can re-grow in the intervening period. However, branch 4 was still absent in all hosts; we therefore assessed whether axons could re-grow to branch 4 in this condition. Notably, unlike in control A→A transplants, in which ∼20% of neurons re-extended axons to branch 4 (Fig. 5Q-T), we did not observe a single instance of an axon re-growing to branch 4 in Crizo 1-3 dpf hosts (Fig. 5U-X). We therefore conclude that single regenerating axons require pre-existing innervation of their target.
DISCUSSION
We have established the larval zebrafish vagus nerve as a new model to further our understanding of the mechanisms guiding axon target selection during regeneration. We first combined sparse mosaic labeling with laser axon severing to show that peripheral vagus nerve branches regenerate after injury and re-establish both the function and the topographic organization of the nerve. We then developed a new single-cell chimera axon injury model, which we used to understand how individual regenerating neurons make axon targeting decisions under different environmental challenges. We find that regenerating axons use a targeting mechanism that does not utilize the developmental targeting machinery of the nerve, is driven by the neuron's memory of its position within the hindbrain, and requires that the target has pre-existing innervation. This work provides new insights into an understudied aspect of neuro-regeneration – the guidance of axons to the correct target tissues during re-growth.
Intrinsic regulation of larval neuron identity
We found that larval vagus neurons possess an intrinsic bias to re-innervate specific target branches during axon regeneration. We previously found that embryonic vagus neurons readily alter their target selection in response to environmental changes (Barsh et al., 2017), suggesting that identity, although initially plastic, becomes fixed sometime after the initial axon outgrowth of these neurons. We further found that the identity of a neuron is determined by its A-P position within the brain. Vagus spatial topography is not perfectly resolved: neurons that innervate branches 4, 5 and 6, for example, can be found at the same A-P position (Fig. 1B) (Barsh et al., 2017). We would therefore expect that a position-based identity mechanism may be insufficient to promote exact re-targeting (e.g. a branch 5 neuron returns to branch 5), but would be sufficient to promote topographically correct re-targeting, meaning that neurons re-grow to a branch that is correct for their position (e.g. a branch 5 neuron returns to branch 4, 5 or 6). Our findings are consistent with this model, as we observed that, after whole-nerve severing, although only about half of the regenerating axons showed exact re-targeting, almost all showed topographically correct re-targeting (Fig. 2E). The functional consequences of this inexactness are not clear – would a more exact re-targeting result in greater functional recovery? Or is a modest rearrangement of axons inconsequential? It is also possible that there are mechanisms to compensate for this inexactness, such as a subsequent rearrangement of dendritic connections. An experimental paradigm to measure precisely motor neuron activity in response to branch-specific stimuli will be important to address whether small-scale topographic rearrangements impact functional specificity after injury.
Environmental regulation of axon guidance during regeneration
Target specificity requires molecular differences between potential targets that growing axons can interpret. One potential source of guidance cues may be secreted chemoattractant and/or chemorepulsive ligands. The reliance of axon targeting on pre-existing innervation does not exclude the role of such agents, but is an important factor when considering what the nature of such cues might be. For instance, the axon could require a pre-existing scaffold as a generic growth substrate but use secreted chemical cues to distinguish between different paths. Or some material the presence of which relies on pre-existing motor innervation – be it extracellular matrix or the vagus sensory axons that also extend along the vagus fascicle (Cox et al., 2011) – could produce or localize secreted guidance cues. Schwann cells can provide guidance information in other regenerating nerves (Gonzalez and Allende, 2021; Isaacman-Beck et al., 2015), but their absence from the vagus nerve fascicle suggests that they are unlikely to contribute in this context. Lastly, the innervation of an arch could in itself alter the nature of the cues coming from that arch (Takeuchi et al., 2010).
Another potential source of target-specific molecular guidance information may be the growth substrate itself. It has been shown that follower axons can identify the correct path on which to grow by recognition of cell-surface molecules on pioneer axons (Raper et al., 1983; Spead and Poulain, 2020; Treubert-Zimmermann et al., 2002). In the context of the vagus nerve, motor or sensory axons extending to or from different peripheral branches could express distinct cell surface molecules that a regenerating axon could interpret to select the correct path to follow. Genetically identifying the molecular determinants of target-specific axon guidance, and the cells in which they are expressed, will be important to distinguish between the possible mechanisms discussed here, and will constitute important future work in this system.
Whole-nerve versus single-cell axon regeneration
An important and intriguing aspect of regeneration is that the starting point can vary greatly depending on the type and extent of the damage. Having the flexibility to rebuild appropriately from a variety of starting points is a particular challenge of regeneration. This fact is reflected in the varying types of nerve damage seen in humans, and the distinct therapeutic needs they exhibit. In cases of acute axonal injury, there is a major therapeutic focus on stimulating the re-growth and re-targeting of injured axons (Hutson and Di Giovanni, 2019; Lien et al., 2020). In cases such as neurodegenerative disease, including motor neuron diseases such as amyotrophic lateral sclerosis, in which neurons degenerate and die, there has been a focus on introducing new healthy neurons, either by stimulating neurogenesis or by stem cell therapies, and promoting their incorporation into existing circuits (Mazzini et al., 2018; Vasic et al., 2019). Understanding how regeneration can be flexibly achieved after varying types of damage is an important goal in the field.
In this study, we utilized two very different injury models: (1) laser axon severing, a model for severe acute nerve injury in which the entire vagus nerve is severed and axons must re-grow and re-establish the peripheral nerve branches and (2) single-cell chimeras, in which an introduced neuron must integrate itself into the existing nerve circuitry, which may serve as a valuable model for this aspect of neural cell replacement therapy. Although major biological differences exist between these two conditions, we observed fundamental similarities in how regeneration occurred. Likely the most significant difference is that laser axon severing results in a major peripheral wound; the axonal material distal to the wound is degraded in a process that may involve immune infiltration and remodeling of nerve support structures, as well as temporary denervation of peripheral target muscles, all of which could dramatically alter the molecular state of the pharyngeal region. In the chimeras, by contrast, although the transplanted neuron itself is injured, there is no peripheral wound in the host and peripheral innervation remains intact. Despite these differences, axons in both cases exhibited a dramatic capacity for target-specific regeneration, although this process was, perhaps unsurprisingly, slightly more error-prone after laser axon severing.
Our data also suggest potential differences in how regeneration may occur in these two conditions. For instance, we found that targeting of transplanted neurons requires pre-existing innervation. This may appear, at first glance, to contradict the whole-nerve severing data, as after whole-nerve severing axons are able to extend into regions lacking existing motor innervation. This may suggest that axons re-growing after whole-nerve severing possess a pioneering ability that axons of transplanted neurons do not. One potential explanation is that a neuron will pioneer into a new region if given no scaffold on which to grow, as after whole-nerve severing, but will preferentially follow an existing axon tract, even to a wrong target, if given that option, as seen after transplantation. Another possibility is that pioneering may be a low probability event; although we did not see it in the 73 single-cell transplants we performed, perhaps if enough axons are given enough time, as in the case of whole-nerve severing or in the case of spontaneous formation of branch 5 in the larva after Crizo 1-3 dpf treatment, one will manage the feat, and a single pioneer may be all that is needed to allow others to follow along. Alternatively, the relevant scaffold may indeed be present after laser axon severing. Although motor innervation is lost, it is likely that nerve support structures, such as extracellular matrix, as well as sensory axons that extend from pharyngeal arches to the brain, remain. These structures have been shown to promote axon growth and guidance in other contexts (Cox et al., 2011; Isaacman-Beck et al., 2015; Rosenberg et al., 2014; Roumazeilles et al., 2018). In the Crizo 1-3 dpf hosts used in the transplant experiment, however, such structures may not have been established in pharyngeal arch 4 owing to it never having received motor innervation (Cox et al., 2011). Although the full extent of the similarities and differences between these conditions remains to be examined, these complementary injury models provide a valuable opportunity to further examine nerve regeneration from distinct starting points.
Development versus regeneration in the vagus nerve
Regeneration represents, in large part, an application of developmental processes under unconventional circumstances. However, although development and regeneration share many conceptual and molecular characteristics, important contextual differences limit mechanistic transferability between them. Understanding what these differences are, how they arise, and how the organism adapts to them in order to promote successful regeneration, or fails to do so, is a key question in the field. There are very few nerve systems for which we understand the mechanisms of axon targeting in both development and regeneration. One such example is the optic nerve. During development, topographic retinal axon targeting is guided in large part by corresponding stable expression gradients of Eph receptors and Ephrin ligands in retinal ganglion cells and the optic tectum (Triplett and Feldheim, 2012). There is good evidence that these expression gradients can persist, or be re-established, in the adult, and may reprise their guidance functions after optic nerve injury (Bach et al., 2003; King et al., 2003; Koeberle and Bähr, 2004; Rodger et al., 2004). In contrast, we show here that larval vagus nerve regeneration relies on neither the guidance mechanism (temporal matching) nor the molecular machinery (Hgf/Met signaling) of the developmental targeting process, likely because of the tight temporal specificity of the embryonic mechanism. Thus, we conclude that the applicability of developmental targeting mechanisms to regeneration is dependent on the nature of those mechanisms, but that an inability to rely on the developmental mechanisms does not necessarily impair regeneration, as the animal can establish alternative targeting strategies. Why evolution might promote targeting mechanisms that may, as in the optic nerve, or may not, as in the vagus nerve, be transferable between development and regeneration, and how that relates to differences in regenerative capacity between species, is an open and intriguing question.
MATERIALS AND METHODS
Zebrafish care and maintenance
Danio rerio animals were raised at the Fred Hutchinson Cancer Research Center facility in accordance with institutional animal care and use committee (IACUC)-approved protocols. All experiments were carried out in accordance with IACUC standards. Fish were bred and maintained according to standard protocols (Westerfield, 2000). All experimental stages are noted in the figures and text. Sex is not a relevant biological variable in our experiments, which are carried out before sex is determined in zebrafish (Siegfried, 2010). Transgenic lines used in this study include: Tg(isl1:Kaede)ch103 (Barsh et al., 2017); Tg(isl1:mRFP)fh1 (Grant and Moens, 2010); Tg(isl1:EGFPCAAX)fh474 (Barsh et al., 2017); TgBAC(chata:GAL4-VP16)mpn202 (Förster et al., 2017); Tg(UAS-E1B:Kaede)s1999t (Scott et al., 2007); Tg(mbp:eGFP-caax)ue2 (Almeida et al., 2011); Tg(sox10:mRFP)vu234 (Kucenas et al., 2008); metfh533 (Isabella et al., 2020).
Drug treatments
Embryos were routinely treated with 0.2 mM N-phenylthiourea (Sigma-Aldrich, P7629) from 24 hpf to 72 hpf to block pigment formation. For embryonic crizotinib treatments, embryos were dechorionated and treated with 20 µM crizotinib (Selleck Chemicals, S1068) dissolved in DMSO (Sigma-Aldrich, 472301), or an equivalent concentration of DMSO alone (equaling 0.2% DMSO) as a control, in fish water from 24 hpf to 72 hpf. Crizotinib was washed out prior to transplantation. For larval crizotinib treatment, larvae were treated with 20 µM crizotinib or DMSO, as described above, in Ringers+1% Pen Strep (Gibco, 15140-122) from immediately after transplantation (∼72 hpf) until endpoint imaging at 2 dpt.
Mosaic transgenic labeling
Sparse transgenic labeling of neurons was carried out by injecting one-cell-stage embryos with 50 pg of Tg(isl1:GFP-caax)fh474 plasmid (Barsh et al., 2017) and 50 pg of Tol2 mRNA. Embryos were screened for sparse GFP expression at 3 dpf on a Zeiss AxioZoom.V16 microscope.
Fluorescence microscopy
Animals were anesthetized with 200 mg/l 3-aminobenzoic acid ethyl ester (MS-222) (Sigma-Aldrich, A5040), embedded laterally (unless otherwise noted) in 1.4% (for single time point imaging) or 0.7% (for time-lapse imaging) low-melt agarose (Invitrogen, 16520) and imaged live on a Zeiss LSM 700 inverted confocal microscope or Leica SP8 inverted confocal microscope. Animals were unmounted and recovered in standard fish water as needed after imaging. Images were processed in Fiji (Schindelin et al., 2012). For Kaede photoconversion on the LSM 700 microscope, a region of interest was drawn over an individual branch, and the region was photoconverted using 300 iterations of 405 nm laser at 9% power. At least 1 h elapsed between branch photoconversion and subsequent imaging of the vagus nucleus.
Whole-nerve laser axon severing
Larvae were mounted for standard imaging as described above. Larvae were visualized using a Leica SP8 inverted confocal microscope and the nerve severed using an attached MicroPoint laser (Andor) controlled by Metamorph software (Molecular Devices). For severing, a line of approximately ten points for MicroPoint illumination, spaced 10 pixels apart, was drawn over the fascicle in a perpendicular orientation in order to span the full width of the fascicle, in the region denoted in Fig. 1D. Each point was hit with two or three iterations of 65-80% laser power to induce severing. To confirm successful severing, time-lapse imaging was performed during the process to allow visualization of the rapid retraction of the severed ends that is stereotypical of a successful severing event (see Movie 1). For the feeding experiment, bilateral severing was achieved by repeating this process on both the left and right sides of the fish, and bilateral mock severing was achieved by performing the same severing protocol but aiming the MicroPoint laser just posterior to the vagus fascicle.
Vagus neuron transplantation
Larvae were anesthetized at 3 dpf with 200 mg/l MS-222 and screened for green or red fluorescence in vagus neurons prior to transplantation. For transplants involving Crizo 1-3 dpf treatment, larvae were screened at 3 dpf for the absence of branches 4 and 5. For Crizo 1-3 dpf donors, we used Tg(chata:Gal4), Tg(UAS:kaede) animals rather than Tg(isl1:GFP-caax) animals to aid in screening; although both of these lines express in vagus motor neurons, Tg(isl1:GFP-caax) also expresses in vagus sensory neurons, making it more difficult to screen for the absence of branch 4 and 5 innervation. A vertical column of one green- and three or four red-fluorescing 3 dpf larvae were mounted laterally in 1% low-melt agarose on a slide and immersed in normal Ringer's solution containing MESAB (Westerfield, 2000). A small wedge of agarose was cut out to expose the head, and the slide was mounted on a Zeiss AxioSkop fixed-stage microscope fitted with a 40× long working distance water-immersion lens. Wiretrol 10 µl capillary micropipettes (Drummond Scientific, 21-175B) were pulled into needles on a Flaming/Brown micropipette puller (Sutter Instruments), cut to a bore size of 10 µm, filled with mineral oil, and mounted on a transplantation rig consisting of an oil-controlled syringe and hydraulic micromanipulator (Kemp et al., 2009). The needle was inserted into the hindbrain at a 45° angle to the body axis at the level of the vagus nucleus from the dorsal side and GFP- or Kaede-expressing vagus neurons were removed from either the anterior 20% or posterior 40% of the nucleus. The needle was then similarly inserted into the vagus nucleus of each RFP-expressing animal in turn, and one to five green neurons were placed into the anterior 20% or posterior 40% of the nucleus. Following transplantation, larvae were unmounted from agarose and placed into normal Ringer's solution+1% Pen Strep (Gibco, 15140-122) until imaging.
Larval feeding assay
One liter of cultured paramecia was rinsed sequentially through 120 µm and 53 µm ‘plankton collector’ filters (Florida Aqua Farms) to remove large particulate matter, collected in a 10 µm filter, then resuspended in 100 ml water. Four microliters of Fluoresbrite Polychromatic Red Microspheres 2.0 µm (Polysciences, 19508-2) were added and the solution incubated 1 h to allow uptake of beads by paramecia. Paramecia were then again collected in the 10 µm filter, washed to remove excess microspheres, and resuspended in ∼4 ml water. Paramecia were added at a concentration of ∼100/ml to dishes containing zebrafish larvae at a concentration of four larvae per ml and incubated for 1 h. Larvae were then anesthetized and mounted laterally for imaging with their left sides facing the coverslip. The confocal pinhole was fully opened, and single confocal plane images of the larval gut were collected using a 10× objective. At 1 dps, larvae were unmounted after imaging and recovered in fish water to allow re-examination at 6 dps. Gut fluorescence was measured in Fiji using the ‘Raw Integrated Density’ measurement. Fluorescence intensity was normalized to the unsevered mean value for each time point.
Labeling of acetylcholine receptors at the neuromuscular synapse
Larvae were anesthetized and mounted dorsal side up in 1.4% low-melt-agarose, and a wedge of agarose was cut out to expose the head. Glass needles were prepared as for one-cell-stage embryo injections and loaded with 0.25 mg/ml Alexa 647-conjugated α-Bungarotoxin (Thermo Fisher, B35450) and 1 nl was injected into the forebrain ventricle. Animals were then unmounted, allowed to recover for ∼2 h, then remounted in the same orientation and imaged on an inverted Zeiss LSM700 confocal microscope.
Quantification and statistical analysis
Image analysis was performed in Fiji, and graphing and statistical analyses were performed in GraphPad Prism (t-tests) or R (Fisher's exact and Chi-squared tests), unless otherwise noted. Additional statistical information on each experiment is noted in figure legends.
Quantification of axon branch fluorescence intensity
At 6 dps (9 dpf), axon branches of severed and unsevered control animals were imaged using identical settings. For each animal, a 10-pixel-wide line was drawn perpendicularly over each of the five branches and the maximum fluorescence intensity measured in Fiji. Fluorescence intensity was normalized to the unsevered control mean value for each branch. Unpaired t-tests were performed to measure significance of difference in intensity between the groups for each branch.
Quantification of target-specific regeneration after laser axon severing
Larvae were screened for sparse GFP expression in vagus neurons at 3 dpf. The vagus nucleus and branches of mosaic transgenic animals were imaged at 3 dpf immediately prior to laser severing, and again after nerve regeneration (3-6 dps). For animals with innervation of a single branch by GFP axons both before severing and after regeneration, the innervated branch was manually determined. Innervation at each time point was determined blind relative to innervation status at the other time point. Animals were then categorized by the distance (in number of branches) between the original target branch and the regenerated target branch (0, 1 or >1). A chi-squared test was performed in R to determine whether the distribution of animals into these three categories differed significantly from the distribution expected by random re-targeting during regeneration, defined as a 20% probability of innervation for each branch after regeneration.
Quantification of regeneration after transplantation
The vagus nucleus and nerve branches of transplanted host larvae were imaged at 2 dpt or 3 dpt, as specified in the text. The A-P position of the transplanted neuron(s) within the nucleus was measured to confirm its correct placement during transplantation into either the anterior 20% or posterior 40% of the nucleus. The branch(es) innervated by green fluorescent axons was manually determined. In cases in which more than one branch was innervated by green axons in a given host (as a result of >1 cell having been transplanted), all innervated branches were counted. A Fisher's exact test was performed in R to determine whether the distribution of branches innervated by green fluorescent axons was significantly different between conditions.
Quantification of vagus nerve topography by branch-specific photoconversion
To determine the relative position of Kaede-red cell bodies within the vagus nucleus, we measured the cumulative density distribution of the converted Kaede signal along the AP axis of the vagus nucleus. First, to define the total length of the vagus nucleus, the most anterior and posterior positive pixels were identified after global thresholding of the maximum projection of the green (unconverted) Kaede image stack. Secondly, the red converted Kaede signal was binarized in 3D and the resulting binarized stack was subjected to two consecutive sum projections: the binarized stack was first sum-projected along the z-axis, and the resulting projected xy matrix was further sum-projected along the y-axis. The cumulative distribution of the resulting vector was computed from anterior to posterior and normalized to the length of the vagus nucleus defined by the green Kaede. The midpoint value, defined as the relative location of the median of this distribution, was then extracted. MATLAB R2019a was used to perform image segmentation and computation. Because in some animals a low signal-to-noise ratio prevented our script from discriminating between signal and background, after processing in MATLAB all data was blinded and we manually sorted out individuals in which the software had not properly discriminated between signal and background and did not include these in our analysis.
To graph the cumulative densities, we used a custom python script to transform the raw cumulative density data to fit a normalized vagus length spanning from 0 (anterior-most) to 100 (posterior-most). Resulting data were graphed in GraphPad Prism. Software generated for this analysis is available at https://github.com/MoensLab/Isabella_et_al_2021.
Supplementary Material
Acknowledgements
We thank Rachel Garcia for exceptional animal care, Dr Takuya Kaneko for advice and feedback, Dr Herwig Baier and Dr Sarah Kucenas for generously provided fish lines, Dr Lena Schroeder and Dr Peng Guo of the Fred Hutch Cellular Imaging Core for assistance with laser axon severing, and Dr Chad He for statistical assistance.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: A.J.I., C.B.M.; Methodology: A.J.I., J.D., C.B.M.; Software: J.D.; Formal analysis: A.J.I.; Investigation: A.J.I., J.A.S.; Writing - original draft: A.J.I.; Writing - review & editing: A.J.I., J.A.S., J.D., C.B.M.; Visualization: A.J.I., J.A.S.; Supervision: A.J.I., C.B.M.; Project administration: A.J.I., C.B.M.; Funding acquisition: A.J.I., C.B.M.
Funding
Funding for this project was provided by National Institutes of Health [R01 NS109425 to C.B.M., F32 HD096860 to A.J.I.]. Deposited in PMC for release after 12 months.
Peer review history
The peer review history is available online at https://journals.biologists.com/dev/article-lookup/doi/10.1242/dev.199706.
References
- Almeida, R. G., Czopka, T., Ffrench-Constant, C. and Lyons, D. A. (2011). Individual axons regulate the myelinating potential of single oligodendrocytes in vivo. Development 138, 4443-4450. 10.1242/dev.071001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson, M. A., O'Shea, T. M., Burda, J. E., Ao, Y., Barlatey, S. L., Bernstein, A. M., Kim, J. H., James, N. D., Rogers, A., Kato, B.et al. (2018). Required growth facilitators propel axon regeneration across complete spinal cord injury. Nature 561, 396-400. 10.1038/s41586-018-0467-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bach, H., Feldheim, D. A., Flanagan, J. G. and Scalia, F. (2003). Persistence of graded EphA/Ephrin-A expression in the adult frog visual system. J. Comp. Neurol. 467, 549-565. 10.1002/cne.10941 [DOI] [PubMed] [Google Scholar]
- Bak, M. and Fraser, S. E. (2003). Axon fasciculation and differences in midline kinetics between pioneer and follower axons within commissural fascicles. Development 130, 4999-5008. 10.1242/dev.00713 [DOI] [PubMed] [Google Scholar]
- Barsh, G. R., Isabella, A. J. and Moens, C. B. (2017). Vagus motor neuron topographic map determined by parallel mechanisms of hox5 expression and time of axon initiation. Curr. Biol. 27, 3812-3825.e3. 10.1016/j.cub.2017.11.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieger, D. and Hopkins, D. A. (1987). Viscerotopic representation of the upper alimentary tract in the medulla oblongata in the rat: the nucleus ambiguus. J. Comp. Neurol. 262, 546-562. 10.1002/cne.902620408 [DOI] [PubMed] [Google Scholar]
- Bolívar, S., Navarro, X. and Udina, E. (2020). Schwann cell role in selectivity of nerve regeneration. Cells 9, 2131. 10.3390/cells9092131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer, B., Ernest, S. and Rosa, F. (2013). Egr-1 induction provides a genetic response to food aversion in zebrafish. Front. Behav. Neurosci. 7, 1-12. 10.3389/fnbeh.2013.00051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brushart, T. M. E. (1993). Motor axons preferentially reinnervate motor pathways. J. Neurosci. 13, 2730-2738. 10.1523/JNEUROSCI.13-06-02730.1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunch, T. J., Ellenbogen, K. A., Packer, D. L. and Asirvatham, S. J. (2008). Vagus nerve injury after posterior atrial radiofrequency ablation. Hear. Rhythm 5, 1327-1330. 10.1016/j.hrthm.2008.05.014 [DOI] [PubMed] [Google Scholar]
- Carmany-Rampey, A. and Moens, C. B. (2006). Modern mosaic analysis in the zebrafish. Methods 39, 228-238. 10.1016/j.ymeth.2006.02.002 [DOI] [PubMed] [Google Scholar]
- Chilton, B. J. (2006). Molecular mechanisms of axon guidance. Dev. Biol. 292, 13-24. 10.1016/j.ydbio.2005.12.048 [DOI] [PubMed] [Google Scholar]
- Coleman, M. P. and Freeman, M. R. (2010). Wallerian degeneration, WldS, and Nmnat. Annu. Rev. Neurosci. 33, 245-267. 10.1146/annurev-neuro-060909-153248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox, J. A., LaMora, A., Johnson, S. L. and Voigt, M. M. (2011). Diverse mechanisms for assembly of branchiomeric nerves. Dev. Biol. 357, 305-317. 10.1016/j.ydbio.2011.06.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- English, A. W. (2005). Enhancing axon regeneration in peripheral nerves also increases functionally inappropriate reinnervation of targets. J. Comp. Neurol. 490, 427-441. 10.1002/cne.20678 [DOI] [PubMed] [Google Scholar]
- Erman, A. B., Kejner, A. E., Hogikyan, N. D. and Feldman, E. L. (2009). Disorders of cranial nerves IX and X. Semin. Neurol. 29, 85-92. 10.1055/s-0028-1124027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finsterer, J. and Grisold, W. (2015). Disorders of the lower cranial nerves. J. Neurosci. Rural Pract. 6, 377-391. 10.4103/0976-3147.158768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Förster, D., Arnold-Ammer, I., Laurell, E., Barker, A. J., Fernandes, A. M., Finger-Baier, K., Filosa, A., Helmbrecht, T. O., Kölsch, Y., Kühn, E.et al. (2017). Genetic targeting and anatomical registration of neuronal populations in the zebrafish brain with a new set of BAC transgenic tools. Sci. Rep. 7, 5230. 10.1038/s41598-017-04657-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez, D. and Allende, M. L. (2021). Current advances in comprehending dynamics of regenerating axons and axon–glia interactions after peripheral nerve injury in zebrafish. Int. J. Mol. Sci. 22, 2484. 10.3390/ijms22052484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant, P. K. and Moens, C. B. (2010). The neuroepithelial basement membrane serves as a boundary and a substrate for neuron migration in the zebrafish hindbrain. Neural Dev. 5, 9. 10.1186/1749-8104-5-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hindges, R., McLaughlin, T., Genoud, N., Henkemeyer, M. and O'Leary, D. D. M. (2002). EphB forward signaling controls directional branch extension and arborization required for dorsal-ventral retinotopic mapping. Neuron 35, 475-487. 10.1016/S0896-6273(02)00799-7 [DOI] [PubMed] [Google Scholar]
- Hu, B.-B., Chen, M., Huang, R.-C., Huang, Y.-B., Xu, Y., Yin, W., Li, L. and Hu, B. (2018). In vivo imaging of mauthner axon regeneration, remyelination and synapses re-establishment after laser axotomy in zebrafish larvae. Exp. Neurol. 300, 67-73. 10.1016/j.expneurol.2017.10.028 [DOI] [PubMed] [Google Scholar]
- Hutson, T. H. and Di Giovanni, S. (2019). The translational landscape in spinal cord injury: focus on neuroplasticity and regeneration. Nat. Rev. Neurol. 15, 732-745. 10.1038/s41582-019-0280-3 [DOI] [PubMed] [Google Scholar]
- Isaacman-Beck, J., Schneider, V., Franzini-Armstrong, C. and Granato, M. (2015). The lh3 glycosyltransferase directs target-selective peripheral nerve regeneration. Neuron 88, 691-703. 10.1016/j.neuron.2015.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isabella, A. J., Barsh, G. R., Stonick, J. A., Dubrulle, J. and Moens, C. B. (2020). Retinoic acid organizes the Zebrafish Vagus motor topographic map via spatiotemporal coordination of Hgf/Met signaling. Dev. Cell 53, 344-357.e5. 10.1016/j.devcel.2020.03.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpinski, B. A., Maynard, T. M., Fralish, M. S., Nuwayhid, S., Zohn, I. E., Moody, S. A. and LaMantia, A.-S. (2014). Dysphagia and disrupted cranial nerve development in a mouse model of DiGeorge (22q11) deletion syndrome. Dis. Model. Mech. 7, 245-257. 10.1242/dmm.012484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemp, H. A., Carmany-Rampey, A. and Moens, C. (2009). Generating chimeric zebrafish embryos by transplantation. J. Vis. Exp., 29, e1394. 10.3791/1394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidd, T., Brose, K., Mitchell, K. J., Fetter, R. D., Tessier-Lavigne, M., Goodman, C. S. and Tear, G. (1998). Roundabout controls axon crossing of the CNS midline and defines a novel subfamily of evolutionarily conserved guidance receptors. Cell 92, 205-215. 10.1016/S0092-8674(00)80915-0 [DOI] [PubMed] [Google Scholar]
- King, C. E., Wallace, A., Rodger, J., Bartlett, C., Beazley, L. D. and Dunlop, S. A. (2003). Transient up-regulation of retinal EphA3 and EphA5, but not ephrin-A2, coincides with re-establishment of a topographic map during optic nerve regeneration in goldfish. Exp. Neurol. 183, 593-599. 10.1016/S0014-4886(03)00211-5 [DOI] [PubMed] [Google Scholar]
- Koeberle, P. D. and Bähr, M. (2004). Growth and guidance cues for regenerating axons: where have they gone? J. Neurobiol. 59, 162-180. 10.1002/neu.10345 [DOI] [PubMed] [Google Scholar]
- Kucenas, S., Takada, N., Park, H.-C., Woodruff, E., Broadie, K. and Appel, B. (2008). CNS-derived glia ensheath peripheral nerves and mediate motor root development. Nat. Neurosci. 11, 143-151. 10.1038/nn2025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuffler, D. P. and Foy, C. (2020). Restoration of neurological function following peripheral nerve trauma. Int. J. Mol. Sci. 21, 1808. 10.3390/ijms21051808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leite, S. C., Pinto-Costa, R. and Sousa, M. M. (2021). Actin dynamics in the growth cone: a key player in axon regeneration. Curr. Opin. Neurobiol. 69, 11-18. 10.1016/j.conb.2020.11.015 [DOI] [PubMed] [Google Scholar]
- Lien, B. V., Brown, N. J., Ransom, S. C., Lehrich, B. M., Shahrestani, S., Tafreshi, A. R., Ransom, R. C. and Sahyouni, R. (2020). Enhancing peripheral nerve regeneration with neurotrophic factors and bioengineered scaffolds: A basic science and clinical perspective. J. Peripher. Nerv. Syst. 25, 320-334. 10.1111/jns.12414 [DOI] [PubMed] [Google Scholar]
- Lozano-Ortega, M., Valera, G., Xiao, Y., Faucherre, A. and López-Schier, H. (2018). Hair cell identity establishes labeled lines of directional mechanosensation. PLoS Biol. 16, 1-22. 10.1371/journal.pbio.2004404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo, X., Salgueiro, Y., Beckerman, S. R., Lemmon, V. P., Tsoulfas, P. and Park, K. K. (2013). Three-dimensional evaluation of retinal ganglion cell axon regeneration and pathfinding in whole mouse tissue after injury. Exp. Neurol. 247, 653-662. 10.1016/j.expneurol.2013.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madison, R. D., Robinson, G. A. and Chadaram, S. R. (2007). The specificity of motor neurone regeneration (preferential reinnervation). Acta Physiol. 189, 201-206. 10.1111/j.1748-1716.2006.01657.x [DOI] [PubMed] [Google Scholar]
- Martin, S. M., O'Brien, G. S., Portera-Cailliau, C. and Sagasti, A. (2010). Wallerian degeneration of zebrafish trigeminal axons in the skin is required for regeneration and developmental pruning. Development 137, 3985-3994. 10.1242/dev.053611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazzini, L., Ferrari, D., Andjus, P. R., Buzanska, L., Cantello, R., De Marchi, F., Gelati, M., Giniatullin, R., Glover, J. C., Grilli, M.et al. (2018). Advances in stem cell therapy for amyotrophic lateral sclerosis. Expert Opin. Biol. Ther. 18, 865-881. 10.1080/14712598.2018.1503248 [DOI] [PubMed] [Google Scholar]
- Peters, J. H., Gallaher, Z. R., Ryu, V. and Czaja, K. (2013). Withdrawal and restoration of central vagal afferents within the dorsal vagal complex following subdiaphragmatic vagotomy. J. Comp. Neurol. 521, 3584-3599. 10.1002/cne.23374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips, R. J., Baronowsky, E. A. and Powley, T. L. (2003). Long-term regeneration of abdominal vagus: efferents fail while afferents succeed. J. Comp. Neurol. 455, 222-237. 10.1002/cne.10470 [DOI] [PubMed] [Google Scholar]
- Raper, J. A., Bastiani, M. and Goodman, C. S. (1983). Pathfinding by neuronal growth cones in grasshopper embryos. II. Selection fasciculation onto specific axonal pathways. J. Neurosci. 3, 31-41. 10.1523/JNEUROSCI.03-01-00031.1983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribatti, D. (2019). Nicole Le Douarin and the use of quail-chick chimeras to study the developmental fate of neural crest and hematopoietic cells. Mech. Dev. 158, 103557. 10.1016/j.mod.2019.103557 [DOI] [PubMed] [Google Scholar]
- Rodger, J., Vitale, P. N., Tee, L. B. G., King, C. E., Bartlett, C. A., Fall, A., Brennan, C., O'Shea, J. E., Dunlop, S. A. and Beazley, L. D. (2004). EphA/ephrin-A interactions during optic nerve regeneration: restoration of topography and regulation of ephrin-A2 expression. Mol. Cell. Neurosci. 25, 56-68. 10.1016/j.mcn.2003.09.010 [DOI] [PubMed] [Google Scholar]
- Rosenberg, A. F., Isaacman-Beck, J., Franzini-Armstrong, C. and Granato, M. (2014). Schwann cells and deleted in colorectal carcinoma direct regenerating motor axons towards their original path. J. Neurosci. 34, 14668-14681. 10.1523/JNEUROSCI.2007-14.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roumazeilles, L., Dokalis, N., Kaulich, E. and Lelievre, V. (2018). It is all about the support — The role of the extracellular matrix in regenerating axon guidance. Cell Adhes. Migr. 12, 87-92. 10.1080/19336918.2017.1291481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin, J., Arganda-Carreras, I., Frise, E., Kaynig, V., Longair, M., Pietzsch, T., Preibisch, S., Rueden, C., Saalfeld, S., Schmid, B.et al. (2012). Fiji: An open-source platform for biological-image analysis. Nat. Methods 9, 676-682. 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott, E. K., Mason, L., Arrenberg, A. B., Ziv, L., Gosse, N. J., Xiao, T., Chi, N. C., Asakawa, K., Kawakami, K. and Baier, H. (2007). Targeting neural circuitry in zebrafish using GAL4 enhancer trapping. Nat. Methods 4, 323-326. 10.1038/nmeth1033 [DOI] [PubMed] [Google Scholar]
- Siegfried, K. R. (2010). In search of determinants: Gene expression during gonadal sex differentiation. J. Fish Biol. 76, 1879-1902. 10.1111/j.1095-8649.2010.02594.x [DOI] [PubMed] [Google Scholar]
- Spead, O. and Poulain, F. E. (2020). Trans-axonal signaling in neural circuit wiring. Int. J. Mol. Sci. 21, 5170. 10.3390/ijms21145170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperry, R. W. (1963). Chemoaffinity in the orderly growth of nerve fiber patterns and connections. Proc. Natl. Acad. Sci. USA 50, 703-710. 10.1073/pnas.50.4.703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuermer, C. A. O. and Easter, S. S. (1984). A comparison of the normal and regenerated retinotectal pathways of goldfish. J. Comp. Neurol. 223, 57-76. 10.1002/cne.902230106 [DOI] [PubMed] [Google Scholar]
- Sun, F., Park, K. K., Belin, S., Wang, D., Lu, T., Chen, G., Zhang, K., Yeung, C., Feng, G., Yankner, B. A.et al. (2011). Sustained axon regeneration induced by co-deletion of PTEN and SOCS3. Nature 480, 372-375. 10.1038/nature10594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi, H., Inokuchi, K., Aoki, M., Suto, F., Tsuboi, A., Matsuda, I., Suzuki, M., Aiba, A., Serizawa, S., Yoshihara, Y.et al. (2010). Sequential arrival and graded secretion of Sema3F by olfactory neuron axons specify map topography at the bulb. Cell 141, 1056-1067. 10.1016/j.cell.2010.04.041 [DOI] [PubMed] [Google Scholar]
- Tam, P. P. L. and Rossant, J. (2003). Mouse embryonic chimeras: tools for studying mammalian development. Development 130, 6155-6163. 10.1242/dev.00893 [DOI] [PubMed] [Google Scholar]
- Tedeschi, A., Dupraz, S., Curcio, M., Laskowski, C. J., Schaffran, B., Flynn, K. C., Santos, T. E., Stern, S., Hilton, B. J., Larson, M. J. E.et al. (2019). ADF/Cofilin-mediated actin turnover promotes axon regeneration in the adult CNS. Neuron 103, 1073-1085.e6. 10.1016/j.neuron.2019.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treubert-Zimmermann, U., Heyers, D. and Redies, C. (2002). Targeting axons to specific fiber tracts in vivo by altering cadherin expression. J. Neurosci. 22, 7617-7626. 10.1523/JNEUROSCI.22-17-07617.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triplett, J. W. and Feldheim, D. A. (2012). Eph and ephrin signaling in the formation of topographic maps. Semin. Cell Dev. Biol. 23, 7-15. 10.1016/j.semcdb.2011.10.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasic, V., Barth, K. and Schmidt, M. H. H. (2019). Neurodegeneration and neuro-regeneration— Alzheimer's disease and stem cell therapy. Int. J. Mol. Sci. 20, 4272. 10.3390/ijms20174272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijayvargiya, P. and Camilleri, M. (2019). Gastroparesis. In Essential Medical Disorders of the Stomach and Small Intestine (ed. Lacy B. E., DiBaise J. K., Pimentel M. and Ford A. C.), pp 23-50. Springer. 10.1007/978-3-030-01117-8_2 [DOI] [Google Scholar]
- Westerfield, M. (2000). The Zebrafish Book. A Guide for the Laboratory use of Zebrafish (Danio rerio), 4th edn. University of Oregon Press. [Google Scholar]
- Xie, Y., Schneider, K. J., Ali, S. A., Hogikyan, N. D., Feldman, E. L. and Brenner, M. J. (2020). Current landscape in motoneuron regeneration and reconstruction for motor cranial nerve injuries. Neural Regen. Res. 15, 1639-1649. 10.4103/1673-5374.276325 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.