Abstract
Background
Preeclampsia (PE) is a condition of high blood pressure that is usually concurrent with proteinuria in pregnancy. PE complicates the management of both maternal and fetal health and contributes to most adverse pregnancy outcomes, but the mechanism underlying the development of PE remains unclear. In this study, we performed a case-control study to compare the gut microbiota of PE (n = 26), abnormal placental growth (APG, n = 25) and healthy pregnant women (n = 28) and analyzed the potential pathogenic role of gut microbiota in PE progression.
Results
The clinical pathophysiological state did not affect the bacterial diversity, while the compositions of the gut microbiota were significantly altered in both the PE and APG groups compared with healthy pregnant women. At the phylum level, TM7 was significantly increased in women with APG. Heterogeneity was observed at the genus level, especially in genera with positive LDA scores, suggesting the stage-dependent effect of gut microbiota on the development of PE. The beneficial bacterium Lactobacillus was markedly depleted in the PE and APG groups but was only correlated with blood pressure (BP) and proteinuria levels in the PE group. Two different bacterial taxa belonged to Lactobacillus showed different correlations (OTU255 and OTU784 were significantly related to PE and APG, respectively).
Conclusions
Our results indicated that shifts in the gut microbiota might occur from the early stages of the development of PE, which is of possible etiological and therapeutic importance.
Keywords: Gut microbiota, Preeclampsia, Placental growth factor, Pregnant women, Lactobacillus
Background
PE is a disorder of pregnancy characterized by the onset of hypertension and is usually concurrent with proteinuria in pregnancy [1]. Most PE occurs during the third trimester of pregnancy, and with generally worse outcomes when it occurs earlier [2, 3]. Since 2015, PE has become a leading cause of maternal and infant morbidity and mortality and affects 2 ~ 8% of all pregnancies worldwide annually [4, 5].
Although many studies have indicated that PE should be considered a subtype of metabolic disease [6, 7], the pathogenesis of PE has not been clarified, and there is no clear and effective clinical therapy. Some studies have suggested that placental obstruction, abnormal placentation, diffuse inflammatory response, vascular endothelial damage and environmental factors are possible etiologies of PE [1]. The most accepted pathogenesis of PE is placental dysplasia caused by anoxia, ischemia and hypoperfusion. In animal models, the increases in heterodimers formed by angiotensin II and bradykinin B2, two G protein-coupled receptors with opposing effects, can cause abnormal vasoconstriction, which triggers PE-like symptoms in pregnant mice [8]. However, the causes of the abovementioned abnormalities remain obscure; thus, pathogenetic heterogeneity complicates the management of both maternal and fetal health. For this reason, no reliable biomarkers or clinical tests can predict the emergence of PE in the earlier stages of pregnancy, and the only way to cure PE is termination [1].
A very encouraging and innovative hypothesis has recently been proposed whereby the mutual relationships between microbes and hosts could be an important factor affecting health [9]. Tens of trillions of microbes compose a complex ecosystem inhabiting mainly our intestine, also known as the gut microbiota [10], which has distinctive characteristics during pregnancy [11]. Numerous microbes in our gut help us digest complex and indigestible carbohydrates to produce beneficial metabolic output [12, 13]. Highly diverse bacterial-related proteins can maintain intestinal and immune homeostasis [14]. Dysbiosis in our gut is associated with metabolic disorders [15], immune system dysfunction [16], and even the development of obesity [17, 18] and diabetes [19, 20] of the host. However, the relationship between the gut microbiota and PE has remained largely unclarified until now. Some studies glimpsed the disturbance of both gut and placental microbiota in PE cohorts [21–23], but there has been a lack of stage-dependent and chronological analyses. In this study, healthy pregnant women and preeclamptic pregnant women were recruited. Because increasing evidence indicates that abnormal placentation is associated with PE [24] and that such abnormalities can often be predicted by clinical tests of placental growth factor (PIGF) [25] early in pregnancy, we also recruited 25 pregnant women with significantly decreased PIGF levels. Using 16S rRNA gene sequencing on stool samples, we revealed the gut microbiota profile of women with PE.
Results
Subject clinical features description
A total of 100 women were recruited, which included 21 healthy women (NW group), 28 healthy pregnant women (NP group), 25 pregnant women with abnormal placental growth (APG group, decreased PIGF Multiple of the Median, MoM, value) and 26 pregnant women with pre-eclampsia (PE group). There were no significant differences in age or body mass index (BMI) between the four groups. The gestational age of the APG group was significantly lower than those of the NP and PE groups. The setting of the APG group was more conducive to estimating gut microbiota changes at earlier gestational weeks, and the diastolic blood pressure (DBP), systolic blood pressure (SBP) and urinary protein concentration (UP) of the PE group were significantly higher than those of the other three groups (Table 1).
Table 1.
Summary of subject characteristics
| Groups | NW | NP | APG | PE |
|---|---|---|---|---|
| Subjects, n | 21 | 28 | 25 | 26 |
| Age range, years (SD) | 31.04 ± 3.73 | 29.03 ± 3.80 | 27.50 ± 0.57 | 29.23 ± 4.85 |
| gestational week (SD) | – | 36.50 ± 7.43 | 23.71 ± 5.80* | 36.73 ± 3.44 |
| BMI (SD) | 20.49 ± 1.30 | 21.87 ± 2.62 | 25.01 ± 3.55 | 24.27 ± 4.34 |
| SBP, mmHg (SD) | 110.95 ± 9.38 | 113.46 ± 8.79 | 115.00 ± 9.37 | 145.61 ± 10.44** |
| DBP, mmHg (SD) | 71.38 ± 5.62 | 73.21 ± 7.28 | 70.27 ± 6.99 | 95 ± 7.14** |
| Urine protein, scores (SD) | 0 | 0 | 0 | 1.57 ± 0.70** |
All values are mean ± standard deviation. An asterisk indicated a significant difference (* at p < .05, ** p < .01) between the NP and the labelled group. Paired t test followed by FDR correction
Effects of PE on the overall structure of the gut microbiota
To better analyze the reads, which ranged from 74,949 to 201,741 (approximately 124,761 clean reads per sample) obtained by fecal sample sequencing.
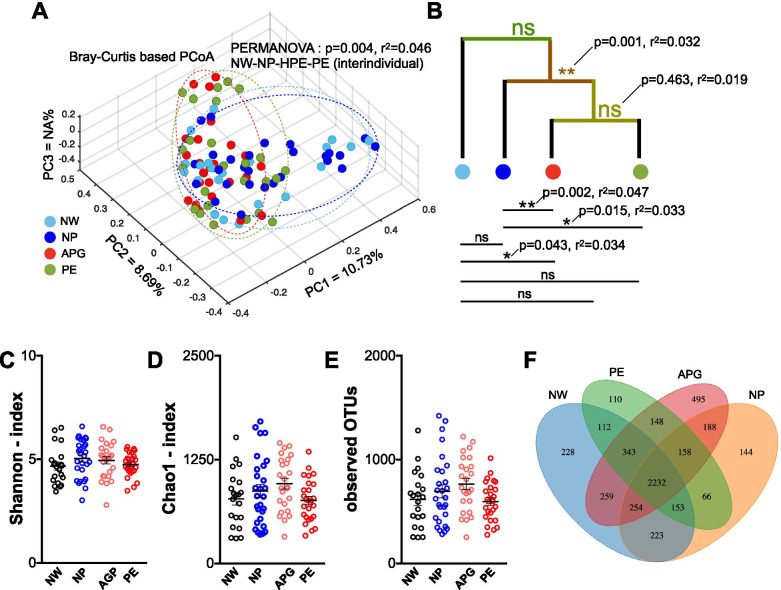
Compared with the NW group, principal component analysis (PCoA) based on the Bray-Curtis distance combined with PERMANOVA (also known as Adonis) test showed a significant difference (p = 0.043) in bacterial composition only to that of the APG group (Fig. 1A, B). As in the three groups of pregnant women, the gut microbiota compositions of the APG group, the PE group or all two groups of abnormal pregnancies (APG group plus PE group) were significantly shifted compared with that of the NP group (Adonis, p = 0.002, p = 0.015, p = 0.001, respectively) (Fig. 1B). Additionally, the PE group was significantly different from the NP group. The NW group showed no significant differences from the other groups when all the abnormally pregnant women were considered as a whole group. Although the pathophysiological state of pregnant women affected their gut microbiota, the alpha diversity, which included the Shannon and Chao1 indices, exhibited no statistical differences (Fig. 1C, D). The total number of OTUs did not differ significantly between any of the groups (Fig. 1E). For this analysis, only OTUs that are shared between 2 or more individuals within the group, were taken into account. A total of 2332 OTUs were observed in all four groups as the core shared features/OTUs (Fig. 1F), and the APG group harbored the highest number (495) of detectable unique OTUs.
Fig. 1.
Effects of PE on the Overall Structure of Gut Microbiota. (A, B) Principal coordinate analysis based on Bray-Curtis distance of the bacterial communities in NW, NP, APG and PE. The differences in beta diversity between each paired groups were tested by permutational multivariate analysis of variance (PERMANOVA, FDR was controlled at 5%). (C, D, E) The Shannon, Chao1 and observed OTUs estimates of fecal microbiota in each group. (F) Venn diagram of the numbers of the identified OTUs. Dots in box plot show values in each individual. An asterisk indicated a significant difference (* at p < .05, ** p < .01) between the labelled groups
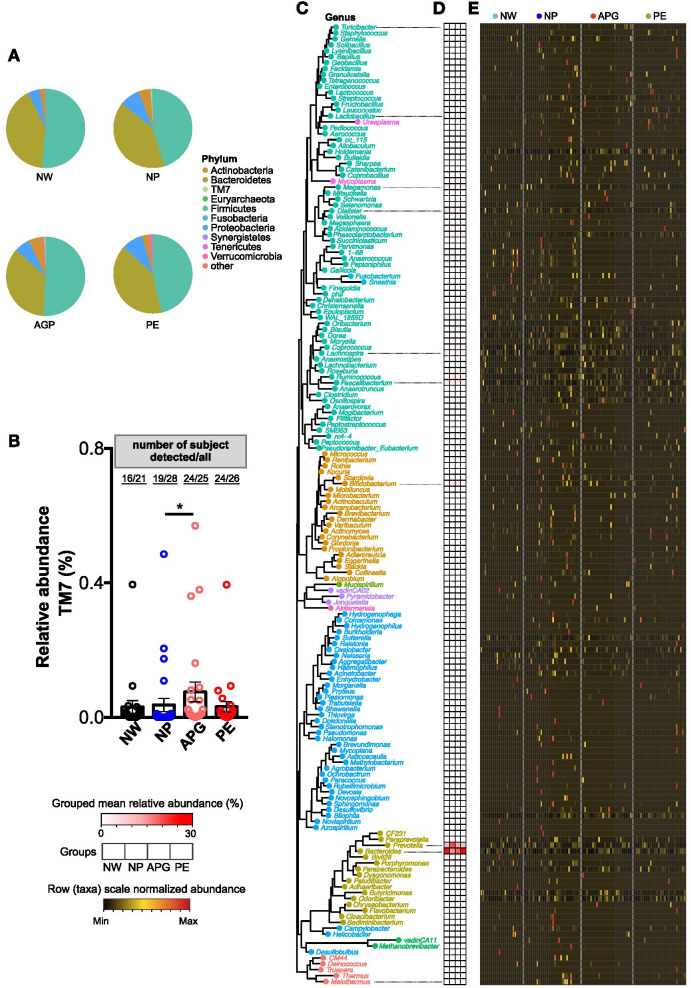
Gut microbiota compositional shifts in APG and PE at the phylum and genus level
As many other studies have indicated, the main microbial communities in the human gut microbiota mainly belong to Firmicutes, Bacteroidetes, Proteobacteria, Actinobacteria and Verrucomicrobia [26]. The same results were found in our data (Fig. 2A). At phylum level, only the percentage of TM7 was significantly increased in the APG group (Fig. 2B). Number of genera belonged to Firmicutes (68) was higher than that of Proteobacteria (40), Actinobacteria (22) and Bacteroidetes (16) (Fig. 2C). The most prevalent genera (at relative abundance level) detected in fecal samples were g_Prevotella, g_Bacteroides, g_Faecalibacterium, g_Lachnospira, g_Megamonas and g_Dialister across all subjects (Fig. 2D and E). Noteworthily, genera belonged to Proteobacteria and other rare phylum were more likely to be detected in NP subjects (Fig. 2C and E).
Fig. 2.
Gut microbiota compositional shifts in APG and PE at phylum and genus level. (A) Relative abundances of phyla in all the four groups. (B) Relative abundances of the phyla TM7 in samples. An asterisk indicated a significant difference (* at p < .05, ** p < .01) between the labelled paired groups. Paired t test followed by FDR correction. (C) Neighbour joining phylogenetic tree of all the 162 annotatable genera. Text indicated the generic name of each bacterial taxa (plot) and the colour of the text and plot represented the level of phylum of each genus. (D) Mean relative abundance of all genera in each group. (E) Normalized abundance of each genus in samples
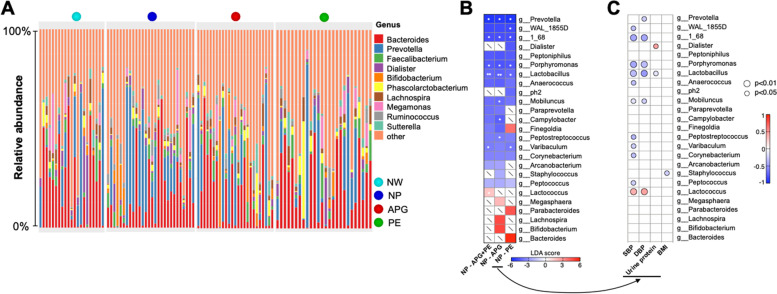
Disturbance of gut microbiota was associated with the clinical characteristics of PE
The composition of the gut microbiome at the genus level revealed by OTUs showed some more detailed changes in microbes. The compositions of the 10 most abundant genera in all subjects are shown in Fig. 3A. Apparently, g_Prevotella was the most common (detected in 19 subjects out of the 28 NP subjects) and prevalent (p = 0.017) genus in the NP group when compared with the NW group or any patient groups (Fig. 3A). To further identify the relationship between gut microbiota changes and the development of PE, linear discriminant analysis (LDA) was used to identify the core bacterial differences and revealed a total of ten genera with significant differences.
Fig. 3.
Disturbance of gut microbiota was associated with the development of PE. (A) Relative abundances of the most abundant ten genera in samples. (B) linear discriminant analysis effect size identified the most differentially abundant taxa between the selected two groups. The enriched taxa were indicated with a positive LDA score, and taxa enriched in NP have a negative score. Only taxa meeting an LDA significant threshold of > 3 were shown. The asterisk indicated a significant relative abundance change of the taxa (* at p < .05, ** p < .01) between the selected paired groups. Paired t test followed by FDR correction. (C) Correlations between the relative abundance of the selected genus and the clinical parameters
Specifically, the relative abundances of g_Prevotella, g_WAL_1855D, g_1_68, g_Porphyromonas, g_Varibaculum and g_Lactobacillus were significantly decreased in the PE group compared with the NP group (Fig. 3B). Between the APG and NP groups, g_Prevotella, g_1_68, g_Porphyromonas, g_Lactobacillus, g_Mobiluncus, g_Campylobacter and g_Peptostreptococcus were decreased significantly in the APG group (Fig. 3B). When considering the abnormally pregnant women as a whole group, 6 genera with significant differences were detected, whereby the significantly decreased abundance of g_Prevotella, g_1_68, g_Porphyromonas, g_Lactobacillus, and g_Varibaculum and the significantly increased abundance of g_Lactococcus (Fig. 3B) were compared to those of the NP group.
Obviously, g_Lactobacillus showed unanimous correlations with both blood pressure and UP (Fig. 3C). g_1_68, g_Porphyromonas, g_Mobiluncus, and g_Lactococcus were significantly correlated with SBP and DBP. Although there was no significant LDA difference in the PE group compared with the NP group (Fig. 3B), the relative abundance of g_Staphylococcus, a potential pathogen, was significantly (p = 0.029) inversely correlated with the BMI index in the NP and PE groups (Fig. 3C).
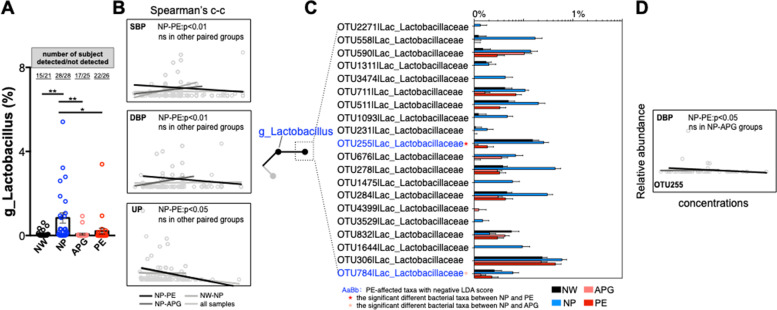
The loss of g_Lactobacilli is only related to the abnormal clinical indicators of pre-eclampsia patients
Although the abundance of g_Lactobacillus in human gut is very low, it has been regarded as the most common probiotic in keeping human health and has been used worldwide in food processing, drug development and clinical treatment over the past decades [27]. In our cohort, g_Lactobacillus was one of the most detectable genus in the four groups, rates of number of subject detected g_Lactobacillus /all ranged from 68 to 100% (Fig. 4A). The ratio of number of subject detected g_Lactobacillus /all and the relative abundance of g_Lactobacillus were significantly higher in the NP group than in any other group (Fig. 4A), implying that g_Lactobacillus may correlate with the health of pregnant women. Moreover, we analyzed the correlations between the relative abundance of g_Lactobacillus and blood pressure and UP in different grouping modes. The abundance of g_Lactobacillus was significantly (p = 0.0042, 0.0029 and 0.031, respectively) inversely correlated with SBP, DBP and UP in the NP and PE groups (Fig. 4B). However, in the NW and NP, NP and APG or all sample groups, no statistically significant correlations were found (Fig. 4B). We further found that 20 OTUs (bacterial taxa) belonged to g_Lactobacillus, and 2 OTUs, OTU255 and OTU784, showing significant differences (p = 0.017 and 0.023, respectively, and both of them had negative LDA scores) (Fig. 4C). The relative abundance of OTU255 was significantly decreased in the PE group compared with the NP group, and OTU784 was decreased with significance only between the APG and NP groups (Fig. 4C). Only OTU255 was significantly negatively related to DBP in the PE and NP groups (Fig. 4D). These results reveal profound changes in the intestinal microbiome structure of the APG and PE groups, indicating the importance of gut microbiota changes in the development of PE.
Fig. 4.
The loss of g_Lactobacilli is only related to the abnormal clinical indicators of Pre-eclampsia patients. (A) Relative abundances of g_Lactobacilli in samples. (B) The correlations between the relative abundance of g_Lactobacilli and the concentrations of SBP, DBP and UP. (C) The relative abundances of the OTUs belonged to the g_Lactobacilli. (D) The correlations between the relative abundance of OTU255 and the concentrations of DBP
Discussion
Pregnancy is one of the most special and vulnerable physiological states of humans under natural conditions. The maintenance of physical and mental health during pregnancy means a great deal to both maternal and fetal safety. Since PE was first described by Hippocrates [24], toxemia caused by external toxic substances, nutritional imbalance, genetic factors and placental dysfunction have been proposed as the pathogenesis of PE. The heterogeneity of etiology led to the lack of effective methods for both clinical prediction, prevention and intervention. We aimed to explore the potential role of the gut microbiota in the development of PE. To our knowledge, the cohort in this study was the first to include pregnant women with abnormal PIGF levels. Our results proposed that gut microbiota dysbiosis could play an important role in the development of PE in a chronological manner.
Microbial community disturbances in the gut [21–23] and the placenta [28] were previously detected in PE members, and using fecal microbiota transplantation, the gut–placenta axis was proposed [23]. Consistent with previous studies [23], we observed that the composition of the gut microbiota in the PE group was clearly and firmly shifted (Fig. 1A, B). Exclusively in our data, a more robust shift of the gut microbiota was also detected in the APG group (Fig. 1A, B). If we took all the abnormally pregnant women as a whole group, the gut microbiota was also significantly different from that of healthy women (Fig. 1B). Gestational age was one of the main factors affecting pregnant women’s gut microbiota [11, 21]. Interestingly, there was no difference between the PE and APG groups (Adonis, p = 0.463), even though a notable gestational age gap existed (Table 1), indicating that gut microbiota changes may have occurred in the early stage of pregnancy (considering the abnormal PIGF levels implied the abnormal pathophysiology of the pregnant women). Many diseases, such as obesity [17, 18], colorectal cancer [29] and arteriosclerosis [30], are accompanied by decreased gut microbiota diversity. A relatively high diversity of gut microbiota is considered a sign of health [31]. However, no significant differences in the Shannon and Chao1 indexes and the observed OTUs were observed (Fig. 1C, D).
The relative abundance of TM7 was significantly increased in the APG (Fig. 2B). The most prevalent genera were mainly belonged to Firmicutes and Bacteroidetes. Bacteria belonged to Firmicutes are believed to help us to improve the utilization of calories in food [32], and more prevalent in all the pregnant women group (NP, APG and PE) (Fig. 2E). However, more bacteria belonging to Bacteroidetes, which usually contain more gram-negative bacteria that can produce lipopolysaccharide [33, 34], were observed in the APG and PE (Fig. 2D, E).
More significant were the patterns of gut dysbiosis in the APG and PE. The relative abundance of g_Prevotella was significantly (p = 0.033 and 0.019, respectively) decreased (from ~ 20% in NP down to ~ 10% in APG and PE with significance). g_Prevotella is a main functional bacterial group in the human intestinal tract [35]. Recent studies have reported that g_Prevotella can use fiber and polysaccharides to produce short-chain fatty acids [36], such as butyrate, the main energy source of intestinal epithelial cells [37] and the regulator of colonic T cell differentiation [38]. At the same time, the colonization of g_Prevotella is also helpful to resist infections with pathogens [39, 40]. The beneficial effects of probiotics on human health have been widely studied. For pregnant women, the role of probiotics has not been fully revealed. g_Lactobacillus is the dominant bacteria in the female vagina for its viability in low pH and glycose condition [41] and has been considered a probiotic in keeping human health, which is associated with numerous beneficial outcomes [42, 43]. Although g_Lactobacillus is not the predominance genus in either male or female individuals gut [44], it is detectable in the maternity gut microbiota [21, 45]. In our cohort, we found that the g_Lactobacillus was detected, unexpectedly, in all of the NP subjects (Fig. 4A). Its relative abundance was significantly higher than that of any of the other three groups, implying that g_Lactobacillus may play an active role in healthy pregnancy (Fig. 4A). The abundance of g_Lactobacillus was significantly inversely related to SBP, DBP and the concentration of urine protein. Such microbiota shifts may be the result of physiological changes (such as hormone homeostasis) in the female body during pregnancy. For example, progesterone can be used as a nutrient to help the colonization and proliferation of Bifidobacteria in pregnant women’s feces [46]. A study of 33,399 primiparas showed that daily or weekly intake of Lactobacillus or their yogurt products during pregnancy significantly reduced the risk of PE [47]. Another study of 70,149 Norwegian mothers showed a significant correlation between supplementation of probiotic milk in late pregnancy and a lower risk of PE [48]. The loss of beneficial and functional genera may lead to the breeding of potential pathogens in both APG and PE, such as g_Lactococcus, and ultimately contribute to the development of PE. Correlations between clinical features and gut microbiota features provide potential evidence for future microbiota-targeted therapy, such as the reduction of the relative abundance of the SBP and (or) DBP positively related bacteria or treating potential probiotics (microbe inversely related to SBP and DBP) and (or) its corresponding prebiotics. In the future, probiotic supplementation and gut microbiota-targeted special diets may have therapeutic effects in improving or blocking the occurrence and development of preeclampsia and should be explored.
Conclusions
Taken together, we clearly revealed gut microbiota dysbiosis in PE and APG. This shift in the gut microbiota may occur from the early stages of the development of PE, and future longitudinal multiregion large-cohort studies are needed to constitute a microecological perspective for PE management.
Materials and methods
Study subject recruitment
All subjects were from the outpatient department or ward of Changsha Hospital for Maternal and Child Health Care, Hunan, China. The definition of PE was according to the diagnosis standards, and the subjects were screened by experienced clinicians. The diagnostic standards were as follows: hypertension (systolic blood pressure ≥ 140 mmHg and/or diastolic blood pressure ≥ 90 mmHg) and high urine protein (≥ 0.3 g/24 h urine collection or random dipstick reading urine protein positive). Additionally, PE women with systolic blood pressure ≥ 160 mmHg and/or diastolic blood pressure ≥ 110 mmHg, thrombocytopenia, impaired liver/kidney function, pulmonary edema or emerging nervous system abnormalities were clinically defined as severe. For the definition of patients with abnormal PIGF values in early and middle pregnancy, the values were obtained by normalizing the concentrations of PIGF with the median value in the localized database (anthropometry-matched pregnant women’s database), also known as the MoM value, in early and middle pregnancy. Patients with malignant tumors, depression, cardiovascular diseases, diabetes, chronic nephritis, liver and kidney function damage or immune system diseases were excluded.
Fecal sample collection and DNA extraction
Fecal samples of each subject were obtained at home. Fresh feces were immediately removed into our storage kit and transferred to a 4 °C domestic refrigerator. Within 1 day, the stool samples were sent to our outpatient clinic (4 °C) and stored at − 80 °C until further processing. The reagent for fecal collection and storage made it possible to deposit stools at ambient temperature for up to a month with minimal alterations when compared with deeply frozen samples (Zhejiang Hangzhou Equipment Preparation NO: 20190682, GUHE Laboratories, Hangzhou, China). Stool samples was excluded if any organic change is detected.
16S rRNA gene amplicon sequencing and analysis
Microbial genomic DNA was extracted from frozen fecal pellets using the e.Z.N.A. TM stool DNA kit (OMEGA Bio-Tek, USA) following the manufacturer’s instructions. The resultant DNA was quantified by Nanodrop and then stored at − 80 °C for further analysis. The V4 region of the 16S rRNA gene was PCR-amplified from microbial genomic DNA using primers (forward primer, 5′-GTG CCA GCM GCC GCG GTA A-3′; reverse primer, 5′-GGA CTA CHV GGG TWT CTA AT-3′). The PCR products were detected using dual-indexing amplification and sequencing approaches on the Illumina MiSeq platform. Briefly, raw sequences equipped with barcodes were assigned to their unique corresponding samples and identified as valid sequences. The following criteria were used for inferior sequence filtering: (i) sequences with a < 150-bp length or < 20 average Phred score and (ii) sequences that contained ambiguous bases or > 8-bp mononucleotide repeats, and the average number of clean reads from each sample was 124,761. Qualified paired-end reads were matched (blasted with each other), dereplicated (−-derep_fulllength), clustered (−-cluster_unoise) and chimeras filtered (−-uchime3_denovo) using VSEARCH (V2.4.4) against the SILVA138 database and then assembled into operational taxonomic units (OTUs) with sequence similarity ≥97% using the Quantitative Insights Into Microbial Ecology (QIIME2, v2020.6) pipeline. The OTUs containing less than 0.001% of total sequences across all samples were discarded. To minimize the difference in sequencing depth across samples, an averaged, rounded rarefied OTU table was generated by averaging 100 evenly resampled subsets under 90% of the minimum sequencing depth for further analysis. Measurements of OTU-level alpha diversity, such as the Chao1, Richness, abundance-based coverage estimator (ACE), Shannon and Simpson indices of each sample, were calculated using the OTU table in QIIME2. Maximum likelihood phylogenetic tree was constructed with phangorn (v.2.5.5) using a neighbour joining tree as the starting point.
Statistical analysis
Differences between two or more groups were evaluated by one-way ANOVA followed by Dunnett’s test or Fisher’s protected least significant difference test using SPSS 24.0 (SPSS, Chicago, Illinois). Values are expressed as the mean ± SEM. The significance of the differentiation of microbiota structure among groups was assessed by permutational multivariate analysis of variance (PERMANOVA) using the R package “vegan”.
Acknowledgements
The authors gratefully acknowledge all the participants and their families. We are particularly grateful to Hangzhou Guhe Info Co., Ltd., Hangzhou, China for its technical assistance and support. This study has been partly supported by the Scientific research project of Hunan Municipal Health Commission of China (award number: 20200421).
Abbreviations
- PE
Pre-eclampsia
- APG
Abnormal placental growth
- BP
Blood pressure
- PIGF
Placental growth factor
- BMI
Body mass index
- DBP
Diastolic blood pressure
- SBP
Systolic blood pressure
- UP
Urinary protein
- OTUs
Operational taxonomic units
- PCoA
Principal component analysis
- F/B
Firmicutes and Bacteroidetes
- LDA
Linear discriminant analysis
- LPS
Lipopolysaccharide
- SCFAs
Short chain fatty acids
Authors’ contributions
LH Huang, WT Yang, J He and Min Cai designed the research; L Li, Xin Zhang, Y Xu, Q Huang and GJ Luo programmed the task and collected the fecal samples and the clinical data; LH Huang, L Li, M Cai, ZZ Zeng and CY Jin analyzed the data and drafted the manuscript; JH Xiao, CY Jin and YX Jin revised the manuscript. All authors reviewed and approved the manuscript.
Funding
This study has been partly supported by the Scientific research project of Hunan Municipal Health Commission of China (award number: 20200421).
Availability of data and materials
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request. All 16S rRNA raw data have been submitted to China National GeneBank DataBase, CNGBdb (accession number, CNP0001512).
Declarations
Ethics approval and consent to participate
The study was approved by Research Ethics Committee of Changsha Hospital for Maternal and Child Health Care (NO: 2020005). Each participant provided written informed consent before any study procedure. All methods were performed in accordance with the relevant guidelines and regulations.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Lihui Huang, Min Cai and Li Li contributed equally to this work.
Contributor Information
Jun He, Email: junhe20210314@163.com.
Weitao Yang, Email: weitaoyang2021@163.com.
References
- 1.Mol BWJ, Roberts CT, Thangaratinam S, Magee LA, de Groot CJM, Hofmeyr GJ. Pre-eclampsia. Lancet. 2016;387(10022):999–1011. doi: 10.1016/S0140-6736(15)00070-7. [DOI] [PubMed] [Google Scholar]
- 2.Malik R, Kumar V. Hypertension in pregnancy. Adv Exp Med Biol. 2017;956:375–393. doi: 10.1007/5584_2016_150. [DOI] [PubMed] [Google Scholar]
- 3.Al-Jameil N, Aziz Khan F, Fareed Khan M, Tabassum H. A brief overview of preeclampsia. J Clin Med Res. 2014;6(1):1–7. doi: 10.4021/jocmr1682w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ghulmiyyah L, Sibai B. Maternal mortality from preeclampsia/eclampsia. Semin Perinatol. 2012;36(1):56–59. doi: 10.1053/j.semperi.2011.09.011. [DOI] [PubMed] [Google Scholar]
- 5.Brown MA, Magee LA, Kenny LC, Karumanchi SA, McCarthy FP, Saito S, et al. Hypertensive disorders of pregnancy: ISSHP classification, diagnosis, and management recommendations for international practice. Hypertension. 2018;72(1):24–43. doi: 10.1161/HYPERTENSIONAHA.117.10803. [DOI] [PubMed] [Google Scholar]
- 6.Jabalie G, Ahmadi M, Koushaeian L, Eghbal-Fard S, Mehdizadeh A, Kamrani A, et al. Metabolic syndrome mediates proinflammatory responses of inflammatory cells in preeclampsia. Am J Reprod Immunol. 2019;81(3):e13086. doi: 10.1111/aji.13086. [DOI] [PubMed] [Google Scholar]
- 7.Rodie VA, Freeman DJ, Sattar N, Greer IA. Pre-eclampsia and cardiovascular disease: metabolic syndrome of pregnancy? Atherosclerosis. 2004;175(2):189–202. doi: 10.1016/j.atherosclerosis.2004.01.038. [DOI] [PubMed] [Google Scholar]
- 8.Quitterer U, Fu X, Pohl A, Bayoumy KM, Langer A, AbdAlla S. Beta-Arrestin1 prevents preeclampsia by Downregulation of Mechanosensitive AT1-B2 receptor Heteromers. Cell. 2019;176(1–2):318–333. doi: 10.1016/j.cell.2018.10.050. [DOI] [PubMed] [Google Scholar]
- 9.Gilbert JA, Blaser MJ, Caporaso JG, Jansson JK, Lynch SV, Knight R. Current understanding of the human microbiome. Nat Med. 2018;24(4):392–400. doi: 10.1038/nm.4517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464(7285):59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Koren O, Goodrich JK, Cullender TC, Spor A, Laitinen K, Backhed HK, et al. Host remodeling of the gut microbiome and metabolic changes during pregnancy. Cell. 2012;150(3):470–480. doi: 10.1016/j.cell.2012.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.El Kaoutari A, Armougom F, Gordon JI, Raoult D, Henrissat B. The abundance and variety of carbohydrate-active enzymes in the human gut microbiota. Nat Rev Microbiol. 2013;11(7):497–504. doi: 10.1038/nrmicro3050. [DOI] [PubMed] [Google Scholar]
- 13.Wikoff WR, Anfora AT, Liu J, Schultz PG, Lesley SA, Peters EC, et al. Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. Proc Natl Acad Sci U S A. 2009;106(10):3698–3703. doi: 10.1073/pnas.0812874106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mukherji A, Kobiita A, Ye T, Chambon P. Homeostasis in intestinal epithelium is orchestrated by the circadian clock and microbiota cues transduced by TLRs. Cell. 2013;153(4):812–827. doi: 10.1016/j.cell.2013.04.020. [DOI] [PubMed] [Google Scholar]
- 15.Woting A, Blaut M. The intestinal microbiota in metabolic disease. Nutrients. 2016;8(4):202. doi: 10.3390/nu8040202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Round JL, Palm NW. Causal effects of the microbiota on immune-mediated diseases. Sci Immunol. 2018;3(20); doi: 10.1126/sciimmunol.aao1603. [DOI] [PubMed]
- 17.Ley RE, Backhed F, Turnbaugh P, Lozupone CA, Knight RD, Gordon JI. Obesity alters gut microbial ecology. Proc Natl Acad Sci U S A. 2005;102(31):11070–11075. doi: 10.1073/pnas.0504978102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ridaura VK, Faith JJ, Rey FE, Cheng J, Duncan AE, Kau AL, et al. Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science. 2013;341(6150):1241214. doi: 10.1126/science.1241214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Qin J, Li Y, Cai Z, Li S, Zhu J, Zhang F, et al. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature. 2012;490(7418):55–60. doi: 10.1038/nature11450. [DOI] [PubMed] [Google Scholar]
- 20.Hvistendahl M. My microbiome and me. Science. 2012;336(6086):1248–1250. doi: 10.1126/science.336.6086.1248. [DOI] [PubMed] [Google Scholar]
- 21.Goltsman DSA, Sun CL, Proctor DM, DiGiulio DB, Robaczewska A, Thomas BC, et al. Metagenomic analysis with strain-level resolution reveals fine-scale variation in the human pregnancy microbiome. Genome Res. 2018;28(10):1467–1480. doi: 10.1101/gr.236000.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu J, Yang H, Yin Z, Jiang X, Zhong H, Qiu D, et al. Remodeling of the gut microbiota and structural shifts in preeclampsia patients in South China. Eur J Clin Microbiol Infect Dis. 2017;36(4):713–719. doi: 10.1007/s10096-016-2853-z. [DOI] [PubMed] [Google Scholar]
- 23.Chen X, Li P, Liu M, Zheng H, He Y, Chen MX, et al. Gut dysbiosis induces the development of pre-eclampsia through bacterial translocation. Gut. 2020;69(3):513–522. doi: 10.1136/gutjnl-2019-319101. [DOI] [PubMed] [Google Scholar]
- 24.Phipps EA, Thadhani R, Benzing T, Karumanchi SA. Pre-eclampsia: pathogenesis, novel diagnostics and therapies. Nat Rev Nephrol. 2019;15(5):275–289. doi: 10.1038/s41581-019-0119-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Black C, Al-Amin A, Stolarek C, Kane SC, Rolnik DL, White A, et al. Midpregnancy prediction of pre-eclampsia using serum biomarkers sFlt-1 and PlGF. Pregnancy Hypertens. 2019;16:112–119. doi: 10.1016/j.preghy.2019.03.009. [DOI] [PubMed] [Google Scholar]
- 26.He Y, Wu W, Zheng HM, Li P, McDonald D, Sheng HF, et al. Regional variation limits applications of healthy gut microbiome reference ranges and disease models. Nat Med. 2018;24(10):1532–1535. doi: 10.1038/s41591-018-0164-x. [DOI] [PubMed] [Google Scholar]
- 27.Goh YJ, Klaenhammer TR. Genetic mechanisms of prebiotic oligosaccharide metabolism in probiotic microbes. Annu Rev Food Sci Technol. 2015;6:137–156. doi: 10.1146/annurev-food-022814-015706. [DOI] [PubMed] [Google Scholar]
- 28.Amarasekara R, Jayasekara RW, Senanayake H, Dissanayake VH. Microbiome of the placenta in pre-eclampsia supports the role of bacteria in the multifactorial cause of pre-eclampsia. J Obstet Gynaecol Res. 2015;41(5):662–669. doi: 10.1111/jog.12619. [DOI] [PubMed] [Google Scholar]
- 29.Thomas AM, Manghi P, Asnicar F, Pasolli E, Armanini F, Zolfo M, et al. Metagenomic analysis of colorectal cancer datasets identifies cross-cohort microbial diagnostic signatures and a link with choline degradation. Nat Med. 2019;25(4):667–678. doi: 10.1038/s41591-019-0405-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Menni C, Lin C, Cecelja M, Mangino M, Matey-Hernandez ML, Keehn L, et al. Gut microbial diversity is associated with lower arterial stiffness in women. Eur Heart J. 2018;39(25):2390–2397. doi: 10.1093/eurheartj/ehy226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bello MGD, Knight R, Gilbert JA, Blaser MJ. Preserving microbial diversity. Science. 2018;362(6410):33–34. doi: 10.1126/science.aau8816. [DOI] [PubMed] [Google Scholar]
- 32.Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444(7122):1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 33.Jandhyala SM, Talukdar R, Subramanyam C, Vuyyuru H, Sasikala M, Nageshwar RD. Role of the normal gut microbiota. World J Gastroenterol. 2015;21(29):8787–8803. doi: 10.3748/wjg.v21.i29.8787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vatanen T, Kostic AD, d'Hennezel E, Siljander H, Franzosa EA, Yassour M, et al. Variation in microbiome LPS immunogenicity contributes to autoimmunity in humans. Cell. 2016;165(4):842–853. doi: 10.1016/j.cell.2016.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Licht TR, Madsen B, Wilcks A. Selection of bacteria originating from a human intestinal microbiota in the gut of previously germ-free rats. FEMS Microbiol Lett. 2007;277(2):205–209. doi: 10.1111/j.1574-6968.2007.00962.x. [DOI] [PubMed] [Google Scholar]
- 36.Kovatcheva-Datchary P, Nilsson A, Akrami R, Lee YS, De Vadder F, Arora T, et al. Dietary Fiber-induced improvement in glucose metabolism is associated with increased abundance of Prevotella. Cell Metab. 2015;22(6):971–982. doi: 10.1016/j.cmet.2015.10.001. [DOI] [PubMed] [Google Scholar]
- 37.Wang RX, Lee JS, Campbell EL, Colgan SP. Microbiota-derived butyrate dynamically regulates intestinal homeostasis through regulation of actin-associated protein synaptopodin. Proc Natl Acad Sci U S A. 2020;117(21):11648–11657. doi: 10.1073/pnas.1917597117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Furusawa Y, Obata Y, Fukuda S, Endo TA, Nakato G, Takahashi D, et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature. 2013;504(7480):446–450. doi: 10.1038/nature12721. [DOI] [PubMed] [Google Scholar]
- 39.Tett A, Huang KD, Asnicar F, Fehlner-Peach H, Pasolli E, Karcher N, et al. The Prevotella copri complex comprises four distinct clades underrepresented in westernized populations. Cell Host Microbe. 2019;26(5):666–679. doi: 10.1016/j.chom.2019.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Leshem A, Liwinski T, Elinav E. Immune-microbiota interplay and colonization resistance in infection. Mol Cell. 2020;78(4):597–613. doi: 10.1016/j.molcel.2020.03.001. [DOI] [PubMed] [Google Scholar]
- 41.Anahtar MN, Gootenberg DB, Mitchell CM, Kwon DS. Cervicovaginal microbiota and reproductive health: the virtue of simplicity. Cell Host Microbe. 2018;23(2):159–168. doi: 10.1016/j.chom.2018.01.013. [DOI] [PubMed] [Google Scholar]
- 42.Nishida K, Sawada D, Kuwano Y, Tanaka H, Rokutan K. Health Benefits of Lactobacillus gasseri CP2305 Tablets in Young Adults Exposed to Chronic Stress: A Randomized, Double-Blind, Placebo-Controlled Study. Nutrients. 2019;11(8); doi: 10.3390/nu11081859. [DOI] [PMC free article] [PubMed]
- 43.Martin V, Cardenas N, Ocana S, Marin M, Arroyo R, Beltran D, et al. Rectal and Vaginal Eradication of Streptococcus agalactiae (GBS) in Pregnant Women by Using Lactobacillus salivarius CECT 9145, A Target-specific Probiotic Strain. Nutrients. 2019;11(4); doi: 10.3390/nu11040810. [DOI] [PMC free article] [PubMed]
- 44.Lloyd-Price J, Mahurkar A, Rahnavard G, Crabtree J, Orvis J, Hall AB, et al. Strains, functions and dynamics in the expanded human microbiome project. Nature. 2017;550(7674):61–66. doi: 10.1038/nature23889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang J, Zheng J, Shi W, Du N, Xu X, Zhang Y, et al. Dysbiosis of maternal and neonatal microbiota associated with gestational diabetes mellitus. Gut. 2018;67(9):1614–1625. doi: 10.1136/gutjnl-2018-315988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nuriel-Ohayon M, Neuman H, Ziv O, Belogolovski A, Barsheshet Y, Bloch N, et al. Progesterone increases Bifidobacterium relative abundance during late pregnancy. Cell Rep. 2019;27(3):730–736. doi: 10.1016/j.celrep.2019.03.075. [DOI] [PubMed] [Google Scholar]
- 47.Brantsaeter AL, Myhre R, Haugen M, Myking S, Sengpiel V, Magnus P, et al. Intake of probiotic food and risk of preeclampsia in primiparous women: the Norwegian mother and child cohort study. Am J Epidemiol. 2011;174(7):807–815. doi: 10.1093/aje/kwr168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nordqvist M, Jacobsson B, Brantsaeter AL, Myhre R, Nilsson S, Sengpiel V. Timing of probiotic milk consumption during pregnancy and effects on the incidence of preeclampsia and preterm delivery: a prospective observational cohort study in Norway. BMJ Open. 2018;8(1):e018021. doi: 10.1136/bmjopen-2017-018021. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request. All 16S rRNA raw data have been submitted to China National GeneBank DataBase, CNGBdb (accession number, CNP0001512).