Abstract
Strategies for enhancing protein degradation have been proposed for treating neurological diseases associated with a decline in proteasome activity. A proteasomal deubiquitinating enzyme that controls substrate entry into proteasomes, ubiquitin-specific protease 14 (USP14), is an attractive candidate for therapies that modulate proteasome activity. This report tests the validity of genetic and pharmacological tools to study USP14’s role in regulating protein abundance. Although previous studies implicated USP14 in the degradation of microtubule associate protein tau, tar DNA binding protein, and prion protein, the levels of these proteins were similar in our neurons cultured from wild type and USP14-deficient mice. Neither loss nor over-expression of USP14 affected the levels of these proteins in mice, implying that modifying the amount of USP14 is not sufficient to alter their steady-state levels. However, neuronal over-expression of a catalytic mutant of USP14 showed that manipulating USP14’s ubiquitin-hydrolase activity altered the levels of specific proteins in vivo. Although pharmacological inhibitors of USP14’s ubiquitin-hydrolase activity reduced microtubule associate protein tau, tar DNA binding protein, and prion protein in culture, the effect was similar in wild type and USP14-deficient neurons, thus impacting their use for specifically evaluating USP14 in a therapeutic manner. While examining how targeting USP14 may affect other proteins in vivo, this report showed that fatty acid synthase, v-rel reticuloendotheliosis viral oncogene homolog, CTNNB1, and synaptosome associated protein 23 are reduced in USP14-deficient mice; however, loss of USP14 differentially altered the levels of these proteins in the liver and brain. As such, it is critical to more thoroughly examine how inhibiting USP14 alters protein abundance to determine if targeting USP14 will be a beneficial strategy for treating neurodegenerative diseases.
Keywords: IU1, proteasome, USP14
1 ∣. INTRODUCTION
Protein turnover by the proteasome is essential for maintaining the appropriate levels of neuronal proteins, and mutations in components of the ubiquitin proteasome pathway have been implicated in several neurological disorders (Atkin & Paulson, 2014; Bingol & Sheng, 2011; George et al., 2018; Hegde & Upadhya, 2011; Im & Chung, 2016; Ristic et al., 2014; Upadhyay et al., 2017; Zheng et al., 2016). This tightly regulated process begins when proteins are tagged with ubiquitin and subsequently targeted to the proteasome (Glickman & Ciechanover, 2002). Dysfunction of the ubiquitin proteasome pathway has been attributed to the formation of protein aggregates within neurons and is thought to play an important role in disease progression (Bennett et al., 2007; Lam et al., 2000; McNaught et al., 2001; Miller et al., 2005; Shimura et al., 2001; Warrick et al., 2005; Zheng et al., 2016). As such, dissection of the molecular mechanisms controlling protein degradation has been an area of active research, with a particular focus on the development of targeted therapies to facilitate the clearance of toxic proteins. Since the buildup of aggregate-prone proteins is associated with a decrease in the activity of the proteasome (Im & Chung, 2016; McKinnon & Tabrizi, 2014; Saez & Vilchez, 2014), approaches aimed at enhancing proteasomal activity are attractive for the treatment of chronic neurodegenerative diseases associated with protein aggregation.
Studies in yeast and mammalian cell lines suggested that the proteasomal deubiquitinating enzyme USP14 acts to limit protein turnover by the proteasome (D'Arcy et al., 2011; Hanna et al., 2006; Lee et al., 2010). As a result, USP14’s catalytic activity was predicted to aid in rescuing proteins from destruction by disassembling ubiquitin side chains prior to the stable interaction of substrates with the proteasome. In support of that finding, a small molecule inhibitor of USP14, called IU1, which blocks the catalytic activity of USP14, was found to enhance the in vitro degradation of model proteasome substrates (Lee et al., 2010). Treatment of cell lines with IU1 also decreased the steady-state levels of aggregate-prone proteins, such as microtubule associate protein tau (MAPT), tar DNA binding protein (TARDBP), and prion protein (PRNP), that were ectopically expressed in the cell culture system (Homma et al., 2015; Lee et al., 2010). A variant of IU1, called IU1-47, which was 10-fold more potent in inhibiting USP14’s ubiquitin hydrolase activity, also enhanced degradation of MAPT in primary cortical neurons (Boselli et al., 2017). While these studies argue that targeting USP14 using IU1 or IU1-47 can alter protein turnover (Boselli et al., 2017; Homma et al., 2015; Lee et al., 2010), other studies questioned the ability of these inhibitors to reduce the levels of these substrates in a USP14-specific manner (Jin et al., 2012; Kiprowska et al., 2017; Ortuno et al., 2016). Although IU1 was not included in the study, a recent report noted that none of the eleven deubiquitinating enzyme inhibitors examined displayed strong selectivity toward a single deubiquitinating enzyme, and many unselectively inhibited all 32 deubiquitinating enzyme included in the study (Ritorto et al., 2014), indicating the difficulty associated with targeting a specific deubiquitinating enzyme in a therapeutic manner.
Our laboratory has previously generated mouse models for studying the effect of altering the levels and catalytic activity of USP14 in the nervous system (Crimmins et al., 2006; Marshall et al., 2013; Vaden et al., 2015; Wilson et al., 2002). In this study, we used these mouse models, as well as primary neurons cultured from these mice, to test how manipulating USP14 alters the expression of specific proteins in the nervous system. We also used these tools to expand on prior pharmacological investigations into the role USP14 plays in protein degradation by examining the effect of these inhibitors in USP14-deficient cortical neurons. In our studies, neither USP14 deficiency nor USP14 over-expression altered the steady-state levels of the MAPT, TARDBP, or PRNP proteins compared to what was observed in the hippocampi of wild type control mice. In contrast, transgenic over-expression of a catalytically-inactive form of USP14 in the nervous system reduced the levels of TARDBP and PRNP but did not alter the abundance of MAPT. When we used the USP14 inhibitors to pharmacologically block USP14’s ubiquitin hydrolase activity in cultured neurons, any effect that the inhibitors had on reducing the expression of the aggregate-prone proteins was similar in both the wild type and USP14-deficient neurons, suggesting a lack of specificity of the inhibitors for USP14. Our studies also revealed that loss of USP14 differentially affects the levels of specific proteins in the brain and liver. These findings indicate a complex role for USP14 in protein turnover and provide new insights on the ability to target USP14 to enhance protein turnover in the nervous system.
2 ∣. MATERIALS AND METHODS
2.1 ∣. Animals
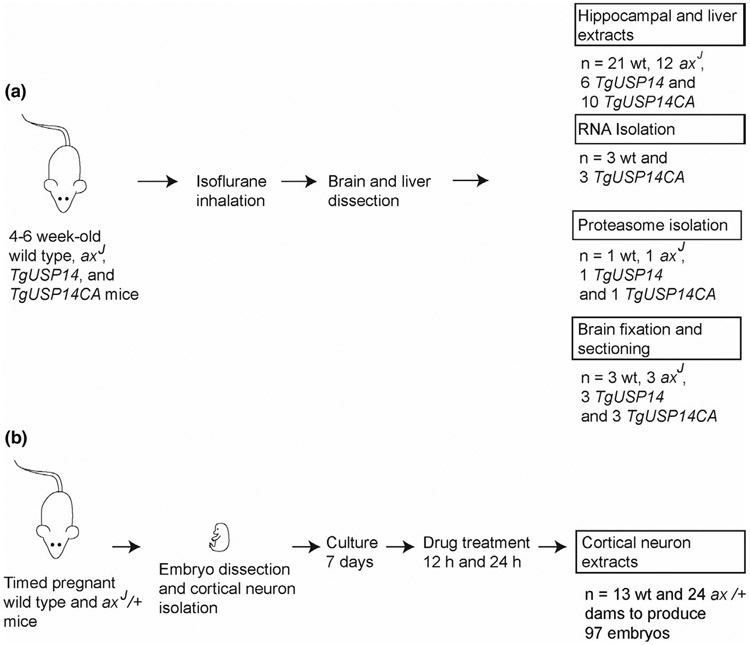
Wild type C57BL/6J mice and USP14-deficient axJ mice (Research Resource Identifier, RRID:MGI:5,527,108) were originally obtained from Jackson Laboratories, and transgenic mice expressing catalytically inactive USP14 (TgUsp14CA) or over-expressing USP14 (TgUsp14) were previously generated in our laboratory (Crimmins et al., 2006; Walters et al., 2014). All mouse strains have been maintained in our breeding colony at the University of Alabama at Birmingham, which is fully accredited by the Association for Assessment and Accreditation of Laboratory Animal Care International (A3255-01). All efforts were made to minimize animal suffering. All research was performed in compliance with the United States Animal Welfare Act and other federal statutes and regulations relating to animals, and all experiments involving mice adhered to the principles stated in the Guide for the Care and Use of Laboratory Animals, United States National Research Council. In addition, all experiments were carried out with the approval of the University of Alabama at Birmingham's Institutional Animal Care and Use Committee under animal protocol 20,457. To ensure that there was no gender bias, equal numbers of both female and male mice were used in these studies. The number of postnatal mice used for this study was 108, which includes 71 mice (28 wild type, 16 axJ, 10 TgUsp14, and 17 TgUsp14CA) that were used for protein analysis from hippocampal and liver extracts and 37 mice (13 wild type and 24 heterozygous axJ) from timed pregnancies that were used for obtaining cortical neurons (Figure 1). Cortical neurons were generated from 97 embryos. Animals were grouped solely by genotype, and no randomization was performed to allocate animals in this study. Mango Taq DNA polymerase (Bioline, Cat# BIO-21083) was used for all genotyping protocols according to the manufacturer's instructions. USP14 transgenic mice were genotyped using forward primer 5’-AAGGGGATAAAGAGAGGGGCTGAG-3’ and reverse primer 5’-TTTTTGTCTGGCTGGCTGGACTCC-3’ with thermocycling conditions of one cycle at 94°C for 2 min, 25 cycles at 94°C for 30 s, 58°C for 30 s and 72°C for 30 s, followed by a final cycle at 72°C for 7 min and then held at 4°C until resolved on a 1% agarose gel. The axJ mice were genotyped using forward primer 5’-TCA GAT TCA CTG CTA AGT CTT TTC-3’ and reverse primer 5’-AGC AAG AAA AGC AGG TGA GG-3’ with thermocycling conditions of one cycle at 94°C for 2 min, 30 cycles at 94°C for 30 s, 55°C for 30 s and 72°C for 30 s, followed by a final cycle at 72°C for 7 min and then held at 4°C until resolved on a 3% agarose gel.
FIGURE 1.
Experimental outline of animals used in these studies. (a) 71 mice at 4–6 weeks of age were used to generate hippocampal and liver extracts for immunoblotting, RNA, proteasomes, and brain sections for immunohistochemistry. (b) 37 timed pregnant mice were used for the production of cortical neuronal cultures for immunoblotting and neuronal viability assays. A total of 108 postnatal mice were used for the experiments described in this study
2.2 ∣. Isolation of proteins
Mice of the appropriate age were deeply anesthetized in 5% isoflurane prior to rapid decapitation. Hippocampi or livers were dissected and homogenized in modified RIPA buffer containing 50 mM Tris, pH 7.5, 150 mM NaCl, 5 mM MgCl2, 0.5 mM EGTA, 1 mM EDTA, 0.5% SDS, 1% Triton X-100, and 1% sodium deoxycholate. Complete protease inhibitor (ThermoFisher), phosphatase inhibitor cocktail III (ThermoFisher), iodoacetamide (Sigma), and n-ethylmaleimide (Sigma) were added to the homogenization buffer according to the manufacturer's instructions. Tissues were disrupted using a mechanical homogenizer. Following homogenization, samples were sonicated and centrifuged at 17,000 g for 10 min at 4°C to remove any insoluble material, and supernatants were stored at −20°C. Protein concentrations were determined using the BCA protein assay kit (ThermoFisher).
2.3 ∣. Immunoblotting
Proteins were resolved on either 12% or 4%–20% polyacrylamide gels and transferred onto nitrocellulose membranes. A solution of 2% bovine serum albumin (BSA) in tris-buffered saline with 0.1% Tween 20 (TBST) was used to block the membranes. Primary and horseradish peroxidase-conjugated secondary antibodies were diluted in a solution containing 0.5% BSA in TBST. Primary antibodies consisted of PRNP (RRID:AB_1128724), MAPT (RRID:AB_2232048), and glyceraldehyde 3-phosphate dehydrogenase (GAPDH; RRID:AB_632467) from Santa Cruz Biotech, β-Tubulin (RRID:AB_1157911) from Developmental Studies Hybridoma Bank, TARDBP (RRID:AB_2800143), CTNNB1 (RRID:AB_331149), ACTB (RRID:AB_2242334), and USP14 (RRID:AB_2242334) from Cell Signaling Technology, Ubiquitin from UAB Hybridoma Core, and fatty acid synthase (FASN) (RRID:AB_11156419), v-rel reticuloendotheliosis viral oncogene homolog (RELA) (RRID:AB_2178878), eukaryotic translation initiation factor 4A1 (EIF4A1) (RRID:AB_2692249), synaptosome associated protein 23 (SNAP23) (RRID:AB_2192025), and ubiquitin protein ligase E3A (UBE3A) (RRID:AB_2211801) from Thermo Fisher. All primary antibodies were used at a dilution of 1:1,000 except for the GAPDH, ACTB, and β-tubulin loading controls that were used at a dilution of 1:5,000. Horseradish peroxidase-conjugated secondary antibodies (Southern Biotechnology Associates) were used at a 1:10,000 dilution. SuperSignal West Pico Chemiluminescent Substrate (Thermo Scientific) was applied to each nitrocellulose membrane and allowed to incubate for 5 min before exposing to film. Blots were cropped to show reactive bands.
2.4 ∣. Cortical neuronal cell culture
Pregnant mice were deeply anesthetized with isoflurane prior to rapid decapitation. Embryos were isolated and placed in Hank's Balanced Salt Solution (HBSS) over ice. Brains of the embryos were removed, and neurons were prepared as described previously (Day et al., 2013). No blinding was performed during genotyping of embryos. Each embryo yielded approximately 5 × 106 cortical neurons, and embryos from identical genotypes were pooled to achieve the number of neurons needed for each experiment. The neurons were plated in a six-well dish at a density of 1 × 106 cells per well and incubated at 37°C in Neurobasal complete media that consists of 0.25% l-glutamine (ThermoFisher), 1% penicillin-streptomycin (ThermoFisher), and B-27 (ThermoFisher) in Neurobasal medium. Every 3 days, half of the media was replaced with fresh Neurobasal complete media to prevent stress to the neurons. Neuronal cultures were treated after 7 days in culture with either DMSO or the pharmacological inhibitor 3E,5E-bis[(4-nitrophenyl) methylene]-1-(1-oxo-2-propen-1-yl)-4-piperidinone (b-AP15), IU1, or IU1-47 at a concentration of either 10 μM or 50 μM. Neurons were treated for 12 or 24 hr prior to harvest. To harvest the neurons, the culture media was aspirated off, and cells were washed one time with 1× Phosphate buffered saline (PBS) and then harvested in modified RIPA buffer as stated above for isolation of proteins. Samples were removed from the plate, sonicated, and centrifuged at 17,000 g for 10 min at 4°C. The supernatants were removed and stored at −20°C. Protein concentrations were determined as stated above. A minimum of three neuronal preparations were prepared and analyzed for each experiment.
2.5 ∣. Neuronal viability
Survival data were generated for wild type and USP14-deficient neurons after 24 hr of treatment with either DMSO or the indicated pharmacological inhibitors. Drug-treated neurons were compared to DMSO-treated neurons, and the percentage of surviving cells was determined using trypan blue exclusion dye.
2.6 ∣. Quantitative PCR
Total RNA was isolated from the hippocampi of deeply anesthetized 4- to 6-week-old mice or from cultured neurons using RNA-STAT60 (Tel-Test) and 2 μg of total RNA was reverse transcribed using the Superscript VILO cDNA synthesis kit (ThermoFisher). Reverse transcription reactions were diluted 5-fold into water and 1 μl of the diluted reverse transcription reaction was used for qPCR analysis. Individual gene assays were purchased from Applied Biosystems and included Mapt (Mm00521988_m1), Tardbp (Mm012577504_g1), and Prnp (Mm00448389_m1) and Actb (Mm026195800_g1) which served as an internal control. The levels of all targets were determined by Δ/Δ Ct. qPCR results are shown as the average of three amplifications of cDNAs generated from three wild type and three TgUSP14CA mice. Each amplification was performed in triplicate.
2.7 ∣. USP14 inhibitors
The pharmacological inhibitors b-AP15 (Ubiquitin Proteasome Biotechnologies), IU1 (Sigma), and IU1-47 (Life Chemicals) were used in this study. Inhibitors were dissolved in anhydrous DMSO, aliquoted and stored at −80°C. Samples were briefly sonicated to ensure complete solubilization of the inhibitor before use and then diluted in Neurobasal complete media.
2.8 ∣. Isolation of ubiquitinated proteins
Cortical neuron lysates were prepared in 50 mM Tris (pH 7.5), 0.15 M NaCl, 1 mM EDTA, 1% NP-40, and 10% glycerol. A quantity of 500 μg of lysate was diluted into TBST, and 50 μl of pan-selective agarose-bound Tandem Ubiquitin-Binding Entities (TUBEs; Lifesensors; cat# UM401) were added to the lysate and incubated at 4°C on a rotator for 2 hr. Agarose beads were collected by centrifugation at 5,000 g at 4°C and washed three times with TBST. Ubiquitinated proteins were eluted from the TUBEs by the addition of 2× Laemmili buffer. Samples were heated to 100°C for 5 min and then separated on 4%–12% Bis-Tris polyacrylamide gels.
2.9 ∣. Hippocampal immunostaining and imaging
Hippocampal sections were prepared as described previously (Crimmins et al). Briefly, brains were rapidly dissected and submerged overnight in 4% paraformaldehyde in PBS and then embedded in paraffin. Brain sections from three mice per genotype were mounted onto glass slides and blocked in 10% normal goat serum, 1% BSA, and 0.1% Triton X-100. Primary and secondary antibodies were diluted in PBS containing 2% normal goat serum, 0.1% BSA, and 0.1% Triton X-100. Primary antibodies to TARDBP, PRNP, MAPT, beta-tubulin (TUBB), and CALB2 (RRID:AB_2619710) were diluted 1:200, and secondary antibodies (ThermoFisher Scientific, RRID:AB_ 2,534,744, RRID:AB_2534775, RRID:AB_429708 were diluted 1:500 in PBS containing 2% normal goat serum, 0.1% BSA, and 0.1% Triton X-100. Sections were washed three times with PBS containing 0.1% Triton X-100, and the DNA was stained with DAPI. Images were acquired using a Zeiss LSM-800 Airyscan confocal microscope.
2.10 ∣. Data analysis
A minimum of five hippocampal extracts from 4- to 6-week-old mice were analyzed for TARDBP, PRNP, and MAPT. This sample size was determined using the resource equation method since it was not possible to assume the effect size (Charan & Kantharia, 2013; Festing, 2006). These studies were not pre-registered, and no sample calculation was performed. Western blots were digitized, and band density was quantified with UN-SCAN-IT gel digitizing software (Silk Scientific, Inc.). Pixel totals were recorded and normalized to the level of GAPDH, β-Tubulin, or ACTB. Protein levels were reported as pixel density relative to wild type. No exclusion criteria were predetermined. Outliers were determined by performing the Grubbs’ test, and entries were excluded from the study if the results of the test concluded an alpha of < 0.05. Based on this criteria, one TgUSP14 protein extract and its wild type control was omitted from the analysis of the TARDBP and MAPT levels in Figure 2. These samples were not replaced with additional samples. The Student's unpaired t-test was performed to determine p-values for hippocampal immunoblot data, and the Welch's unequal variances t test was utilized to determine p-values for hippocampal qPCR data. For neuronal immunoblot data and neuronal viability assays, one-way ANOVA was followed by Holm-Sidak multiple comparisons post-test to determine p-values. The data were not assessed for normality. All data were analyzed in Prism Graphpad software by plotting the average ± the standard error of the mean.
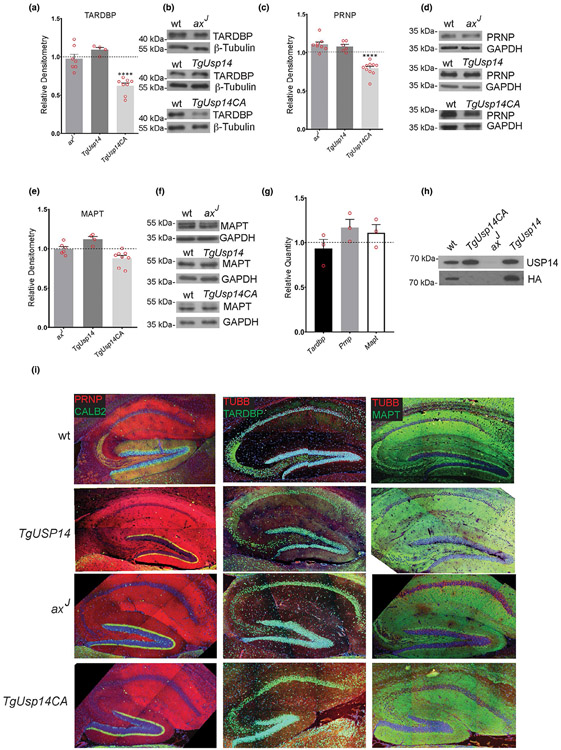
FIGURE 2.
Effect of altering the level or activity of ubiquitin-specific protease 14 (USP14) on steady-state levels of TARDBP, microtubule associate protein tau (MAPT), and prion protein (PRNP) proteins in mice. (a) Quantitation and (b) representative immunoblot of the steady-state levels of TARDBP in the hippocampi of 4- to 6-week-old wild type (wt), USP14-deficient (axJ), neuronally over-expressed USP14 (TgUSP14), and neuronally over-expressed, catalytically-inactive USP14 (TgUSP14CA) mice. (c) Quantitation and (d) representative immunoblot of the levels of PRNP in the hippocampi of 4- to 6-week-old wt, axJ, TgUSP14, TgUSP14CA mice. (e) Quantitation and (f) representative immunoblot of the levels of MAPT in the hippocampi of 4- to 6-week-old wt, axJ, TgUSP14, TgUSP14CA mice. For TARDBP analysis, n = 7 axJ, 4 TgUSP14, and eight TgUSP14CA extracts. For PRNP analysis, n = 8 axJ, 6 TgUSP14, and 10 TgUSP14CA extracts. For MAPT analysis, n = 6 axJ, 4 TgUSP14, and 8 TgUSP14CA extracts. An equivalent number of wild type extracts were analyzed for each protein. All quantitations are shown relative to wt control levels, and β-Tubulin or glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as a loading control. Data are shown as mean ± SEM. ****p < .0001. (g) Quantitative PCR depicting the levels of Tardbp, Prnp, and Mapt transcripts in the hippocampi of 5-week-old TgUSP14CA mice relative to wild type controls. n = 3. (h) Representative immunblot of HA-VME-Ub labeling assay of proteasomes from brains of wt, axJ, TgUSP14, and TgUSP14CA mice. Blots were probed with an anti-HA antibody to identify catalytically-active USP14 and an anti-USP14 antibody to measure total USP14 levels. (i) Expression patterns of PRNP, TARDBP, and MAPT in hippocampal sections from 6-week-old wt, TgUSP14, axJ, and TgUSP14CA mice. CALB2 (Calbindin 2) and tubulin beta chain (TUBB) were included as controls
3 ∣. RESULTS
3.1 ∣. Effect of altering the level and activity of USP14 on steady-state levels of aggregate-prone proteins in mice
Our previous studies showed that transgenic over-expression of either wild type or catalytically inactive USP14 in the nervous system of mice increases the level of proteasomal-bound USP14 (Crimmins et al., 2009; Vaden et al., 2015; Walters et al., 2014). In contrast, there is a 95% loss of USP14 on neuronal proteasomes in USP14-deficient axJ mice (Anderson et al., 2005). The present study utilized these mouse models to assess the role of USP14 in regulating the levels of three different proteins that are known to accumulate in various neurodegenerative diseases. Immunoblot analysis was used to measure the levels of MAPT, TARDBP, and PRNP in hippocampal extracts from either control C57BL/6J mice that express wild type levels of USP14, mice that over-express either USP14 (TgUsp14) or catalytically impaired USP14 (TgUsp14CA) in the nervous system, or axJ mice that are deficient for USP14 (Figure 2).
Altering the levels of USP14 in vivo either by over-expressing USP14 or by USP14 deficiency did not significantly affect the hippocampal levels of the TARDBP, PRNP, or MAPT proteins (Figure 2a-f). However, neuronal over-expression of a catalytically-inactive form of USP14 resulted in decreased steady-state levels of the TARDBP and PRNP proteins in the 4- to 6-week-old TgUsp14CA mice compared to the levels observed in the wild type controls (Figure 2a-d). This catalytic-inactivation of USP14 did not appear to have a global effect on all of the aggregate-prone proteins tested, as the hippocampal levels of MAPT were not significantly different between the TgUsp14CA and control mice (Figure 2e,f). When we investigated if the decreased levels of the TARDBP and PRNP proteins correlated with altered mRNA levels, qPCR analysis of reverse-transcribed hippocampal RNA did not reveal any significant differences in the levels of these mRNAs in the 5-week-old mice that expressed the catalytic-mutant of USP14 compared to controls (Figure 2g). As shown in our previous publications (Crimmins et al., 2009; Vaden et al., 2015; Walters et al., 2014), we observed an increase in the levels of USP14 bound to proteasomes isolated from the TgUSP14 and TgUSP14CA mice as well as a decrease in the levels of USP14 on the axJ proteasomes (Figure 2h).
To control for the possibility that the proteins analyzed in this study were insoluble in the conditions used for protein isolation, and that the immunoblots were therefore not reflective of their actual abundance, we also examined hippocampal sections of axJ, TgUsp14, TgUsp14CA, and wild type control mice for any focal protein accumulations that could be detected by immunohistochemistry. By this method, PRNP and MAPT staining was observed throughout the entire hippocampal section, and TARDBP was found predominantly in the nucleus in all of the sections examined (Figure 2i). Therefore, manipulating the level or catalytic activity of USP14 did not result in any detectable focal accumulations of PRNP, TARDBP, or MAPT in the hippocampi of the mice (Figure 2i).
3.2 ∣. Sensitivity of wild type cortical neurons to USP14 inhibitors
Several inhibitors have been developed to investigate the role of deubiquitinating enzymes in protein turnover. Inhibitors such as IU1, IU1-47, and b-AP15 have previously been used to target the catalytic activity of USP14 and study the role of USP14 in protein degradation (Boselli et al., 2017; D'Arcy et al., 2011; Lee et al., 2010; Ma et al., 2020; Xia et al., 2018; Yu et al., 2019; Yun et al., 2018). We therefore used these inhibitors to investigate if acute inhibition of USP14 could alter the levels of TARDBP, PRNP, and MAPT in cortical neurons. Although IU1 and IU1-47 are reported to be specific for USP14 (Boselli et al., 2017; Lee et al., 2010), b-AP15 is believed to inhibit the proteasomal deubiquitinating activity of both USP14 and UCH37 (D'Arcy et al., 2011).
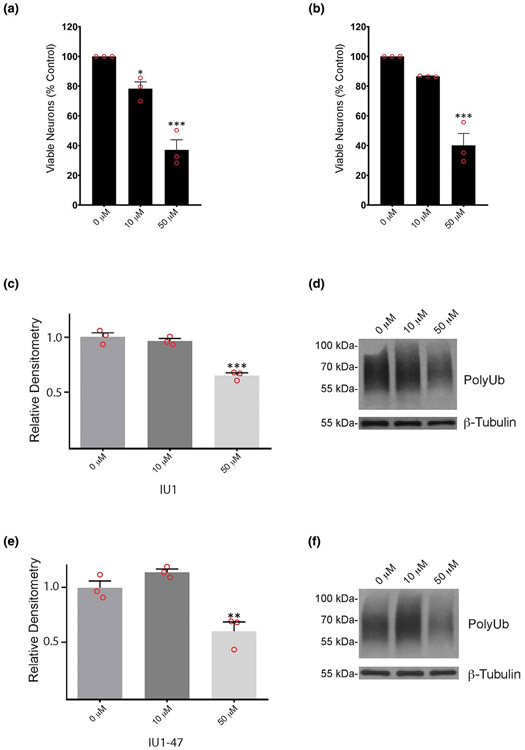
We first examined the sensitivity of wild type cortical neurons to b-AP15, IU1, and IU1-47 by measuring neuronal viability when the cells were exposed to either 10 or 50 μM concentrations of the inhibitors. Although studies have reported using these inhibitors at concentrations over 50 μM (Han et al., 2019; Li et al., 2019; Min et al., 2017; Zhou et al., 2019), all three of the inhibitors exhibited significant toxicity to the wild type cortical neurons at a concentration of 50 μM (Figure S1). Less than 50% of the cultured neurons were still viable after 24 hr of exposure to 50 μM of either IU1 or IU1-47, and only 10% of the cells were still viable after exposure to 50 μM of b-AP15 when compared to cells treated with the DMSO vehicle alone (Figure S1a-c). While exposure of the cultured neurons to a concentration of 10 μM of IU1 or IU1-47 for 24 hr was fairly well-tolerated (Figure S1b,c), neurons treated with 10 μM of b-AP15 showed a nearly 50% decrease in viability at 24 hr (Figure S1a).
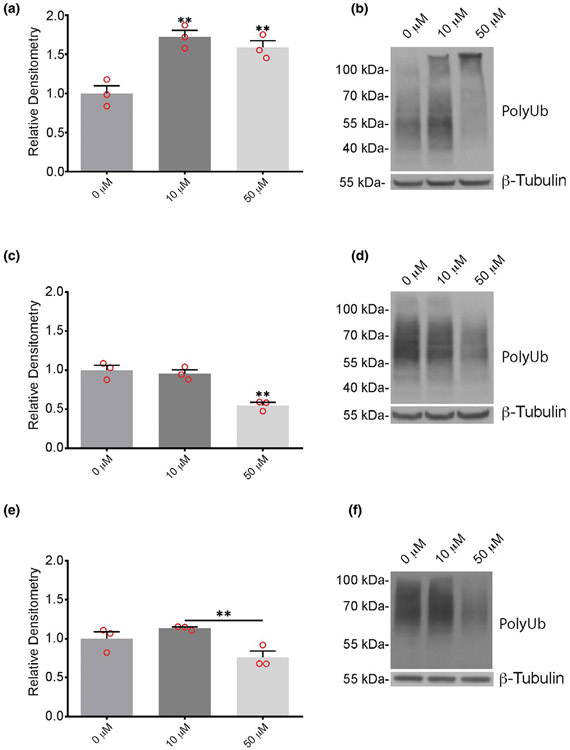
To determine if these inhibitors altered the level of ubiquitin protein conjugates in the neurons, protein extracts from wild type cortical neurons treated with DMSO, b-AP15, IU1, or IU1-47 were immunoblotted for ubiquitin. Although the cultures exhibited substantial cell loss after exposure to b-AP15, there was a significant increase in high-molecular weight ubiquitin-conjugates in the surviving neurons following treatment with either 10 or 50 μM of b-AP15 (Figure 3a,b). In contrast, there was a significant decrease in the ubiquitin conjugates in neurons treated with 50 μM of IU1 or IU1-47 (Figure 3c-f). No differences in ubiquitin conjugates were observed between neurons treated for 24 hr with 10 μM of either IU1 or IU1-47 compared to the DMSO vehicle alone (Figure 3c-f).
FIGURE 3.
Effect of pharmacological inhibitors on ubiquitin conjugates in cortical neurons from wild type mice. Quantitation and representative immunoblots of ubiquitin conjugates (PolyUb) in neurons after 24 hr treatment with 10 μM or 50 μM of (a, b) 3E,5E-bis[(4-nitrophenyl) methylene]-1-(1-oxo-2-propen-1-yl)-4-piperidinone, (c, d) IU1, or (e, f) IU1-47 showing dose-response effect on levels of poly-ubiquitin conjugates. Quantitations are shown relative to levels observed in neurons treated with DMSO alone (0 μM). β-tubulin was used as a loading control. n = 3 independent cortical neuron extracts. Data are shown as mean ± SEM. Significance is reported relative to treatment with DMSO, with the exception of IU1-47, which is shown relative to 10 μM inhibitor. **p < .01
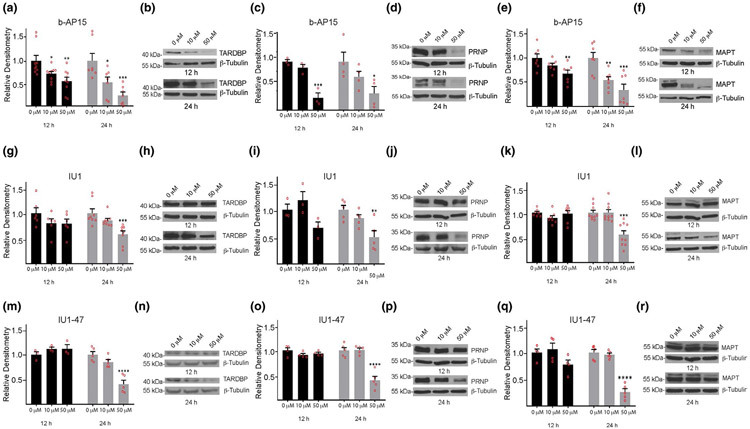
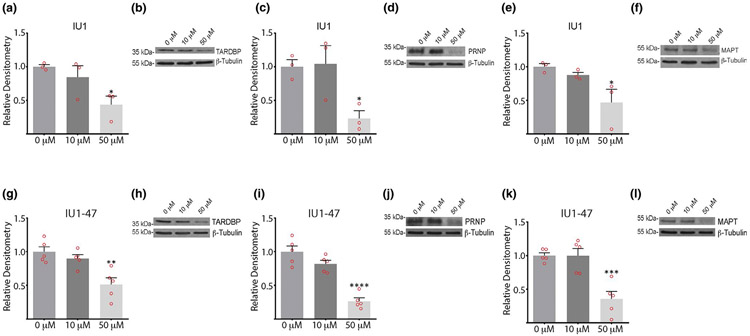
We next investigated if the inhibitors altered the abundance of the TARDBP, PRNP, or MAPT proteins. Cortical neurons from wild type mice were treated with either DMSO alone or with 10 or 50 μM of b-AP15, IU1, or IU1-47 for 12 or 24 hr, and the extracts were subjected to immunoblot analysis. Incubating the neurons with 10 μM b-AP15 for 12 hr resulted in a significant reduction in the steady-state level of TARDBP (Figure 4a,b). In contrast, wild type neurons treated with 10 μM b-AP15 for 12 hr did not show any significant changes in the levels of MAPT or PRNP compared to cells treated with DMSO alone (Figure 4c-f). After 24 hr in the presence of 10 μM b-AP15, the levels of both TARDBP and MAPT were significantly reduced compared to what was observed in the control neurons treated with DMSO (Figure 4a,b,e,f). Increasing the concentration of b-AP15 to 50 μM resulted in a significant reduction in all three proteins within 12 hr of exposure to the inhibitor (Figure 4a-f).
FIGURE 4.
Effect of 3E,5E-bis[(4-nitrophenyl) methylene]-1-(1-oxo-2-propen-1-yl)-4-piperidinone (b-AP15), IU1, and IU1-47 on the steady-state levels of aggregate-prone proteins in cortical neurons from wild type mice. Neurons were treated for 12 or 24 hr with 10 μM or 50 μM of inhibitor. Quantitation and representative immunoblots showing effect of b-AP15 on steady-state levels of (a, b) TARDBP, (c, d) prion protein (PRNP), and (e, f) microtubule associate protein tau (MAPT); IU1 on (g, h) TARDBP, (i, j) PRNP and (k, l) MAPT; and IU1-47 on (m, n) TARDBP, (o, p) PRNP, and (q, r) MAPT. Quantitations are shown relative to levels observed in neurons treated with DMSO alone (0 μM). β-tubulin was used as a loading control. Data are shown as mean ± SEM, and significance is reported relative to treatment with DMSO alone. For b-AP15: TARDBP, n = 9 at 12 hr and n = 7 at 24 hr; PRNP, n = 3 at 12 hr and n = 4 at 24 hr; MAPT, n = 7 at 12 hr and n = 7 at 24 hr. For IU1: TARDBP, n = 6 at 12 hr and n = 8 at 24 hr; PRNP, n = 3 at 12 hr and n = 5 at 24 hr; MAPT, n = 7 at 12 hr and n = 9 at 24 hr. For IU1-47: TARDBP, n = 3 at 12 hr and n = 5 at 24 hr; PRNP, n = 4 at 12 hr and n = 5 at 24 hr; MAPT, n = 4 at 12 hr and n = 5 at 24 hr, where each n represents an independent protein extract. *p < .05. **p < .01. ***p < .001. ****p < .0001
In contrast to what was observed for b-AP15, incubation of cortical neurons from wild type mice with 10 μM of IU1 did not significantly alter the levels of the TARDBP, PRNP, or MAPT proteins following 12 or 24 hr of exposure to the inhibitor (Figure 4g-l). Although 12 hr of exposure to 50 μM of IU1 was not sufficient to influence the steady-state levels of TARDBP, PRNP, or MAPT, increasing the incubation time to 24 hr with 50 μM of IU1 significantly decreased the levels of all three of the proteins in the cortical neurons from the wild type mice (Figure 4g-l). Despite the finding that IU1-47 is a more potent inhibitor of USP14’s ubiquitin hydrolase activity, the results were similar to those observed with IU1, with decreased expression of the TARDBP, PRNP, and MAPT proteins only apparent after 24 hr of exposure to the 50 μM concentration of IU1-47 (Figure 4m-r).
To determine if IU1 treatment affected the levels of ubiquitinated PRNP, TARDBP, and MAPT proteins, purified ubiquitinated proteins from wild type neurons treated with 50 μM IU1 for 24 hr were immunoblotted for PRNP, TARDBP, and MAPT (Figure S2). Similar to what was observed for the total steady-state levels of these proteins, we detected less ubiquitinated forms of PRNP, TARDBP, and MAPT in the cortical neurons, suggesting that these intermediate forms were unstable and were rapidly removed from the cells.
3.3 ∣. Effect of deubiquitinating enzyme inhibitors on cortical neurons from USP14-deficient axJ mice
When we examined the effect of the pharmacological inhibitors on cortical neurons from wild type mice, the inhibitors only seemed to reduce the levels of the TARDBP, PRNP, and MAPT proteins and alter the amount of ubiquitin conjugates under conditions that appeared to be toxic to the cells. Therefore, to investigate the selectivity of the inhibitors for USP14, we also examined the sensitivity of cortical neurons cultured from our USP14-deficient mice to either IU1 or IU1-47. Similar to what we observed for the cortical neurons isolated from wild type mice (Figure S1b,c), treatment of the neurons isolated from the axJ mice with 10 μM of either IU1 or IU1-47 for 24 hr reduced viability by approximately 20% compared to cells treated with DMSO alone (Figure 5a,b). Increasing the concentration of inhibitor to 50 μM also resulted in an approximately 60% decrease in cell viability after 24 hr compared to control axJ cortical neurons treated with DMSO (Figure 5a,b), indicating that the reduced viability observed with the inhibitors was not dependent on USP14. When we examined the effect of these inhibitors on the level of ubiquitin protein conjugates, similar to our findings with wild type neurons (Figure 3), we only observed a significant decrease in ubiquitin conjugates in the axJ neurons that were treated for 24 hr with 50 μM of IU1 or IU1-47 (Figure 5c-f). Since this decrease in ubiquitin conjugates was observed in both wild type and USP14-deficient cortical neurons, the effect of the inhibitors could not be attributed to changes in the activity of USP14.
FIGURE 5.
Effect of IU1 and IU1-47 on viability and level of ubiquitin conjugates in cortical neurons from ubiquitin-specific protease 14-deficient axJ mice. Viability of axJ cortical neurons treated for 24 hr with 10 μM or 50 μM of (a) IU1 or (b) IU1-47 shown relative to neurons treated with DMSO alone (0 μM). Quantitation and representative immunoblots showing the effect of (c, d) IU1 and (e, f) IU1-47 on the level of ubiquitin conjugates (PolyUb) in cortical neurons cultured from axJ mice. Neurons were treated with inhibitors for 24 hr, and quantitations are shown relative to levels observed in neurons treated with DMSO alone (0 μM). β-Tubulin was used as a loading control. Data are shown as mean ± SEM, and significance is reported relative to treatment with DMSO. n = 3. *p < .05. **p < .01. ***p < .001
To determine if the expression of the TARDBP, PRNP, and MAPT proteins in the cultured neurons replicated our in vivo findings in the hippocampi of mice, we also compared the levels of these proteins from cortical neurons cultured from the USP14-deficient axJ mice to those from wild type controls. Consistent with our previous results in the axJ mice (Crimmins et al., 2006), there was a 95% reduction in the level of USP14 in the axJ cortical neurons (Figure S3b). However, similar to what we observed in the hippocampi (Figure 2), this genetic reduction of USP14 was not sufficient to decrease the levels of the TARDBP, PRNP, or MAPT proteins, as the steady-state levels of these proteins were similar in the cortical neurons cultured from the USP14-deficient mice and the wild type controls (Figure S3).
To examine if IU1 and IU1-47 may be influencing the levels of the TARDBP, PRNP, and MAPT proteins independently of USP14, we also measured the steady-state levels of these proteins in cortical neurons from axJ mice treated for 24 hr with either 10 μM or 50 μM of the inhibitors. Similar to what we observed in the neurons cultured from wild type mice (Figure 4), incubation of cortical neurons from the USP14-deficient mice with 10 μM of IU1 or IU1-47 did not have a significant effect on the steady-state levels of the TARDBP, PRNP, or MAPT proteins (Figure 6). However, treatment of the neurons with 50 μM of IU1 or IU1-47 significantly reduced the levels of these proteins in the axJ neurons (Figure 6), indicating that this effect was not dependent on USP14. The effect of these inhibitors on cell viability, accumulation of ubiquitin conjugates, and the steady-state levels of the TARDBP, PRNP, and MAPT proteins therefore seems to be independent from their role of inhibiting the activity of USP14.
FIGURE 6.
Levels of aggregate-prone proteins in ubiquitin-specific protease 14-deficient cortical neurons treated with IU1 or IU1-47. Quantitation and representative immunoblots of the levels of (a, b) TARDBP, (c, d) prion protein (PRNP), and (e, f) microtubule associate protein tau (MAPT) in cortical neurons from axJ mice treated with 10 μM or 50 μM of IU1 for 24 hr. Quantitation and representative immunoblots of the levels of (g, h) TARDBP, (i, j) PRNP, and (k, l) MAPT in cortical neurons from axJ mice treated with 10 μM or 50 μM of IU1-47 for 24 hr. Quantitations are shown relative to levels observed in neurons treated with DMSO alone (0 μM). β-tubulin was used as a loading control. Data are shown as mean ± SEM. n = 3 for IU1 treatment and n = 5 for IU1-47 treatment. *p < .05. **p < .01. ***p < .001. ****p < .0001
3.4 ∣. USP14 deficiency alters the levels of potential USP14 substrates in a cell-type specific manner
A comprehensive proteome, ubiquitinome, and interactome analysis for USP14 substrates recently identified several proteins that are reduced in expression and display increased ubiquitination following knockdown of USP14 (Liu et al., 2018). These putative USP14 substrates were identified using an unbiased approach in HeLa cells and included FASN, RELA, CTNNB1, SNAP23, EIF4A1, and UBE3A (Liu et al., 2018). Since our studies are aimed at identifying how manipulating USP14 can affect the levels of other proteins and if this strategy can be used as a valid approach for therapeutic intervention in neurodegenerative diseases, we examined the effect of USP14 deficiency on the levels of these proteins in the hippocampi of mice. As the previous proteomics study used shRNA knockdown of USP14 in the liver to show that USP14 deficiency decreased the level of FASN (Liu et al., 2018), we also examined the level of these potential USP14 substrates in the liver of the axJ mice as a control for these studies.
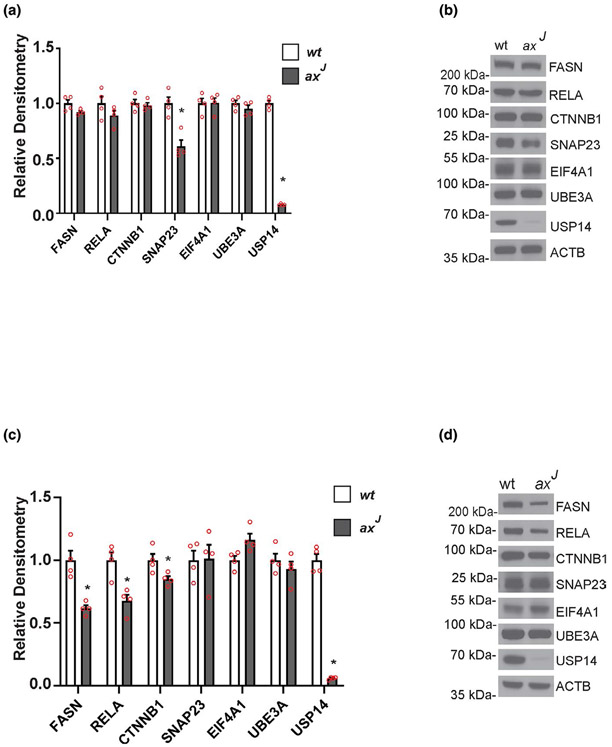
Comparison of hippocampal extracts from wild type and axJ mice demonstrated that loss of USP14 decreased expression of SNAP23 in the hippocampi of the axJ mice without affecting the steady-state levels of the FASN, RELA, CTNNB1, EIF4A1, or UBE3A proteins (Figure 7a,b). This reduction in SNAP23 levels was also observed in the USP14-deficient cortical neurons from the axJ mice (Figure S4). Contrary to what we saw in the hippocampi, loss of USP14 reduced the levels of FASN, RELA, and CTNNB1 but did not alter the level of SNAP23 in the livers of the axJ mice (Figure 7c,d). Although the proteomics study saw a decrease in EIF4A1 and UBE3A in the HeLa cells (Liu et al., 2018), loss of USP14 did not affect the levels of these proteins in the livers or hippocampi of the axJ mice (Figure 7), demonstrating that the effects of USP14 deficiency on protein expression vary between cells types.
FIGURE 7.
Tissue-specific effects of ubiquitin-specific protease 14 (USP14) deficiency on putative USP14 targets in vivo. Quantitation and representative immunoblots of (a, b) hippocampal and (c, d) liver extracts from 6-week-old wild type (wt) and USP-14-deficient axJ mice probed for USP14 and proteins that were previously reported as potential substrates for USP14 (Liu et al., 2018). β-tubulin was used as a loading control. Data are shown as mean ± SEM. n = 4 mice. *p < .05
4 ∣. DISCUSSION
Neurodegenerative diseases are comprised of a heterogeneous group of disorders that are characterized by the progressive degeneration of select populations of neurons. Many of these diseases are also accompanied by protein aggregation and are thus referred to collectively as proteinopathies (Golde et al., 2013; Taylor et al., 2002). Since the proteasome is a major regulator of protein abundance, enhancing proteasome function has been proposed as a way to reduce the expression of proteins that are believed to be critical drivers of human disease (Cromm & Crews, 2017). As regulators of proteasome function, proteasomal deubiquitinating enzymes are attractive candidates for therapeutic intervention, and several inhibitors have been developed to inactivate the ubiquitin hydrolase activity of these enzymes (Boselli et al., 2017; D'Arcy et al., 2011; Lee et al., 2010). However, a previous report demonstrated a lack of specificity for many deubiquitinating enzyme inhibitors (Ritorto et al., 2014), and there have been conflicting reports on the effect of blocking USP14’s ubiquitin hydrolase activity on protein abundance in a USP14-specific manner (Kiprowska et al., 2017; Ortuno et al., 2016). We therefore performed both genetic and pharmacological manipulation of USP14 in mice and in cortical neurons to investigate how altering the expression levels or ubiquitin hydrolase activity of USP14 affects the abundance of proteins in the nervous system.
Previous in vitro studies indicated that either loss of USP14 or inhibition of USP14’s catalytic activity increased the degradation of both model proteasome substrates and over-expressed TARDBP, PRNP, and MAPT proteins in cultured cells (Hanna et al., 2006; Homma et al., 2015; Lee et al., 2010; Lee et al., 2011). Therefore, it was expected that the levels of these putative USP14 substrates would be decreased in mice that were deficient for USP14 and in mice that expressed a ubiquitin-hydrolase-inactive version of USP14. While loss of USP14 did not significantly affect hippocampal expression of the TARDBP, PRNP, or MAPT proteins in the axJ mice compared to controls, over-expression of the ubiquitin-hydrolase inactive USP14 mutant in the TgUSP14CA mice decreased TARDBP and PRNP levels, indicating that USP14 can function in a catalytically independent manner to lower the expression of select proteins in the nervous system. Possible mechanisms for this catalytically-independent function of USP14 include the binding of the ubiquitin-like domain of USP14 to the proteasome and the subsequent interaction of USP14 with ubiquitinated proteins on the proteasome (Collins & Goldberg, 2020; Kim & Goldberg, 2018; Peth et al., 2009). Although disruptions in the stoichiometry of endogenous complexes have led to dominant-negative effects, (Clague et al., 2013; Lee et al., 2014; Liu et al., 2014;; Machida et al., 2009), the fact that the levels of TARDBP, PRNP, and MAPT were not altered by transgenically over-expressing wild type USP14 in the TgUSP14 mice shows that increasing the level of USP14 was not sufficient to disrupt the steady-state levels of these aggregate-prone proteins in the hippocampi of the mice. Further investigations will be required to determine the range of substrates affected by the changes in USP14’s ubiquitin hydrolase activity and to identify how the catalytic and non-catalytic activities of USP14 are coordinated to regulate protein degradation.
Our findings also suggest that USP14 is likely to be involved in the regulation of specific proteins in the nervous system, as we did not observe a significant difference in MAPT expression in the hippocampi of mice expressing the various USP14 constructs. This finding agrees with previous papers that have also used genetic inactivation of USP14 to question the role of USP14 in directly regulating the turnover of aggregate-prone proteins and found that there was no detectable change in the level of MAPT in both cell and animal models deficient for USP14 (Jin et al., 2012; Ortuno et al., 2016). While evidence exists that MAPT can be a substrate for the proteasome (Babu et al., 2005; Choi et al., 2016; Chu et al., 2016; Han et al., 2014; Petrucelli et al., 2004), other studies have suggested that MAPT turnover can also occur through a proteasome-independent pathway (Brown et al., 2005; Feuillette et al., 2005; Lee et al., 2013), such as the autophagic pathway (Dolan & Johnson, 2010; Hamano et al., 2008; Jimenez-Sanchez et al., 2012; Tang et al., 2019). Therefore, MAPT degradation likely occurs through both the proteasome and autophagy pathways, and the decision to utilize these pathways may reflect either constitutive or stress-induced MAPT degradation (Lee et al., 2013).
Since the metabolism of proteins may be inherently different in vivo than in vitro, this study also investigated if cortical neurons that are deficient for USP14 exhibited changes in the expression of these aggregate-prone proteins. Similar to our in vivo studies, we did not detect any significant difference in the steady state levels of the TARDBP, PRNP, or MAPT proteins in our USP14-deficient neurons compared to wild type control neurons. As it is possible that genetic compensation could have accounted for the absence of any effect on the expression of these proteins in the USP14-deficient neurons, we also investigated if acute pharmacological inhibition of USP14’s ubiquitin hydrolase activity altered the steady-state levels of the TARDBP, PRNP, or MAPT proteins. Treatment of cortical neurons from wild type mice with 50 μM of either IU1 or IU1-47 resulted in decreased expression of the TARDBP, PRNP, and MAPT proteins following 24 hr of exposure to the inhibitors. However, when we examined the effect these inhibitors had on the expression of these proteins in primary neurons cultured from our USP14-deficient mice, both control and USP14-deficient neurons treated with either IU1 or IU1-47 displayed similar losses in viability, reductions in steady-state levels of ubiquitin-protein conjugates, and decreases in the steady-state levels of the TARDBP, PRNP, and MAPT proteins. These findings suggest that the effects of IU1 and IU1-47 on the expression of these aggregate-prone proteins occurred in a manner that was not specific to USP14, and care must be taken when making conclusions about the use of these drugs to specifically target USP14.
Our studies on protein expression in the axJ mice point at the complex regulation of the ubiquitin proteasome system in vivo. The finding of reduced steady-state levels of FASN, RELA, and CTNNB1 in liver extracts from USP14-deficient mice suggests that USP14 is normally acting to limit the degradation of these proteins in the liver. However, USP14-deficiency did not affect the steady-state levels of these proteins in the hippocampus, demonstrating that USP14’s role in maintaining protein homeostasis can vary between cell types. Consistent with this observation, loss of USP14 alters the expression of SNAP23 in the hippocampus and in cortical neurons but not in the liver. These findings indicate that caution must be taken when targeting USP14 in vivo since it controls multiple cellular pathways in a tissue-specific manner. As USP14 has been shown to exist both on and off the proteasome (Anderson et al., 2005; Crimmins et al., 2006a; Xu et al., 2015), future studies will be required to determine if USP14 is regulating the abundance of its substrates when it is associated with the proteasome or if this regulation occurs through a proteasome-independent manner.
ACKNOWLEDGMENTS
This work was supported by the Department of Neurobiology, University of Alabama at Birmingham School of Medicine and by a grant from the National Institutes of Health, National Institute of Neurological Disorders and Stroke (NS110744).
All experiments were conducted in compliance with the ARRIVE guidelines.
Funding information
University of Alabama at Birmingham School of Medicine; National Institutes of Health, Grant/Award Number: NS110744; National Institute of Neurological Disorders and Stroke
Abbreviations:
- ACTB
beta-actin
- b-AP15
3E,5E-bis[(4-nitrophenyl) methylene]-1-(1-oxo-2-propen-1-yl)-4-piperidinone
- CTNNB1
beta catenin
- EIF4A1
eukaryotic translation initiation factor 4A1
- FASN
fatty acid synthase
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- IU1
1-[1-(4-fluorophenyl)-2,5-dimethyl-1H-pyrrol-3-yl]-2-(1-pyrrolidinyl)-ethanone
- IU1-47
1-[1-(4-chlorophenyl)-2,5-dimethyl-1H-pyrrol-3-yl]-2-(1-piperidinyl) ethanone
- MAPT
microtubule associate protein tau
- PRNP
prion protein
- RELA
v-rel reticuloendotheliosis viral oncogene homolog
- RRID
Research Resource Identifier
- SNAP23
synaptosome associated protein 23
- TARDBP
tar DNA binding protein
- TUBB
beta-tubulin
- UBE3A
ubiquitin protein ligase E3A
- USP14
ubiquitin specific protease 14
Footnotes
CONFLICTS OF INTEREST
The authors have no conflicts of interest to declare.
REFERENCES
- Anderson C, Crimmins S, Wilson JA, Korbel GA, Ploegh HL, & Wilson SM (2005). Loss of Usp14 results in reduced levels of ubiquitin in ataxia mice. Journal of Neurochemistry, 95, 724–731. 10.1111/j.1471-4159.2005.03409.x [DOI] [PubMed] [Google Scholar]
- Atkin G, & Paulson H (2014). Ubiquitin pathways in neurodegenerative disease. Frontiers in Molecular Neuroscience, 7, 63. 10.3389/fnmol.2014.00063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babu JR, Geetha T, & Wooten MW (2005). Sequestosome 1/p62 shuttles polyubiquitinated tau for proteasomal degradation. Journal of Neurochemistry, 94, 192–203. 10.1111/j.1471-4159.2005.03181.x [DOI] [PubMed] [Google Scholar]
- Bennett EJ, Shaler TA, Woodman B, Ryu K-Y, Zaitseva TS, Becker CH, Bates GP, Schulman H, & Kopito RR (2007). Global changes to the ubiquitin system in Huntington's disease. Nature, 448, 704–708. 10.1038/nature06022 [DOI] [PubMed] [Google Scholar]
- Bingol B, & Sheng M (2011). Deconstruction for reconstruction: The role of proteolysis in neural plasticity and disease. Neuron, 69, 22–32. 10.1016/j.neuron.2010.11.006 [DOI] [PubMed] [Google Scholar]
- Boselli M, Lee B-H, Robert J, Prado MA, Min S-W, Cheng C, Silva MC, Seong C, Elsasser S, Hatle KM, Gahman TC, Gygi SP, Haggarty SJ, Gan L, King RW, & Finley D (2017). An inhibitor of the proteasomal deubiquitinating enzyme USP14 induces tau elimination in cultured neurons. Journal of Biological Chemistry, M117, 815126. 10.1074/jbc.M117.815126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown MR, Bondada V, Keller JN, Thorpe J, & Geddes JW (2005). Proteasome or calpain inhibition does not alter cellular tau levels in neuroblastoma cells or primary neurons. Journal of Alzheimer's Disease, 7, 15–24. 10.3233/JAD-2005-7103 [DOI] [PubMed] [Google Scholar]
- Charan J, & Kantharia ND (2013). How to calculate sample size in animal studies? Journal of Pharmacology and Pharmacotherapeutics, 4, 303–306. 10.4103/0976-500X.119726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi WH, de Poot SAH, Lee JH, Kim JH, Han DH, Kim YK, Finley D, & Lee MJ (2016). Open-gate mutants of the mammalian proteasome show enhanced ubiquitin-conjugate degradation. Nature Communications, 7, 10963. 10.1038/ncomms10963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu T-T, Gao NA, Li Q-Q, Chen P-G, Yang X-F, Chen Y-X, Zhao Y-F, & Li Y-M (2016). Specific knockdown of endogenous tau protein by peptide-directed ubiquitin-proteasome degradation. Cell Chemical Biology, 23, 453–461. 10.1016/j.chembiol.2016.02.016 [DOI] [PubMed] [Google Scholar]
- Clague MJ, Barsukov I, Coulson JM, Liu H, Rigden DJ, & Urbe S (2013). Deubiquitylases from genes to organism. Physiological Reviews, 93, 1289–1315. [DOI] [PubMed] [Google Scholar]
- Collins GA, & Goldberg AL (2020). Proteins containing ubiquitin-like (Ubl) domains not only bind to 26S proteasomes but also induce their activation. Proceedings of the National Academy of Sciences of the United States of America, 117, 4664–4674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crimmins S, Jin Y, Wheeler C, Huffman AK, Chapman C, Dobrunz LE, Levey A, Roth KA, Wilson JA & Wilson SM (2006). Transgenic rescue of ataxia mice with neuronal-specific expression of ubiquitin-specific protease 14. Journal of Neuroscience, 26, 11423–11431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crimmins S, Sutovsky M, Chen PC, Huffman A, Wheeler C, Swing DA, Roth K, Wilson J, Sutovsky P & Wilson S (2009). Transgenic rescue of ataxia mice reveals a male-specific sterility defect. Developmental Biology, 325, 33–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cromm PM, & Crews CM (2017). The proteasome in modern drug discovery: Second life of a highly valuable drug target. ACS Central Science, 3, 830–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Arcy P, Brnjic S, Olofsson MH, Fryknäs M, Lindsten K, De Cesare M, Perego P, Sadeghi B, Hassan M, Larsson R & Linder S (2011). Inhibition of proteasome deubiquitinating activity as a new cancer therapy. Nature Medicine, 17, 1636–1640. [DOI] [PubMed] [Google Scholar]
- Day JJ, Childs D, Guzman-Karlsson MC, Kibe M, Moulden J, Song E, Tahir A and Sweatt JD (2013) DNA methylation regulates associative reward learning. Nature Neuroscience 16(10), 1445–1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolan PJ, & Johnson GV (2010). The role of tau kinases in Alzheimer's disease. Current Opinion in Drug Discovery and Development, 13, 595–603. [PMC free article] [PubMed] [Google Scholar]
- Festing MF (2006). Design and statistical methods in studies using animal models of development. ILAR Journal, 47, 5–14. 10.1093/ilar.47.1.5 [DOI] [PubMed] [Google Scholar]
- Feuillette S, Blard O, Lecourtois M, Frebourg T, Campion D, & Dumanchin C (2005). Tau is not normally degraded by the proteasome. Journal of Neuroscience Research, 80, 400–405. 10.1002/jnr.20414 [DOI] [PubMed] [Google Scholar]
- George AJ, Hoffiz YC, Charles AJ, Zhu Y, & Mabb AM (2018). A comprehensive Atlas of E3 ubiquitin ligase mutations in neurological disorders. Frontiers in Genetics, 9, 29. 10.3389/fgene.2018.00029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glickman MH, & Ciechanover A (2002). The ubiquitin-proteasome proteolytic pathway: Destruction for the sake of construction. Physiological Reviews, 82, 373–428. 10.1152/physrev.00027.2001 [DOI] [PubMed] [Google Scholar]
- Golde TE, Borchelt DR, Giasson BI, & Lewis J (2013). Thinking laterally about neurodegenerative proteinopathies. Journal of Clinical Investigation, 123, 1847–1855. 10.1172/JCI66029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamano T, Gendron TF, Causevic E, Yen S-H, Lin W-L, Isidoro C, DeTure M & Ko L-W (2008). Autophagic-lysosomal perturbation enhances tau aggregation in transfectants with induced wild-type tau expression. European Journal of Neuroscience, 27, 1119–1130. 10.1111/j.1460-9568.2008.06084.x [DOI] [PubMed] [Google Scholar]
- Han DH, Na H-K, Choi WH, Lee JH, Kim YK, Won C, Lee S-H, Kim KP, Kuret J, Min D-H, & Lee MJ (2014). Direct cellular delivery of human proteasomes to delay tau aggregation. Nature Communications, 5, 5633. 10.1038/ncomms6633 [DOI] [PubMed] [Google Scholar]
- Han KH, Kwak M, Lee TH, Park M-S, Jeong I-H, Kim MJ, Jin J-O, & Lee P-W (2019). USP14 inhibition regulates tumorigenesis by inducing autophagy in lung cancer in vitro. International Journal of Molecular Sciences, 20. 10.3390/ijms20215300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna J, Hathaway NA, Tone Y, Crosas B, Elsasser S, Kirkpatrick DS, Leggett DS, Gygi SP, King RW, & Finley D (2006). Deubiquitinating enzyme Ubp6 functions noncatalytically to delay proteasomal degradation. Cell, 127, 99–111. 10.1016/j.cell.2006.07.038 [DOI] [PubMed] [Google Scholar]
- Hegde AN, & Upadhya SC (2011). Role of ubiquitin-proteasome-mediated proteolysis in nervous system disease. Biochimica Et Biophysica Acta, 1809, 128–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homma T, Ishibashi D, Nakagaki T, Fuse T, Mori T, Satoh K, Atarashi R, Nishida N (2015). Ubiquitin-specific protease 14 modulates degradation of cellular prion protein. Scientific Reports, 5, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Im E, & Chung KC (2016). Precise assembly and regulation of 26S proteasome and correlation between proteasome dysfunction and neurodegenerative diseases. BMB Reports, 49, 459–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez-Sanchez M, Thomson F, Zavodszky E, & Rubinsztein DC (2012). Autophagy and polyglutamine diseases. Progress in Neurobiology, 97, 67–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin YN, Chen PC, Watson JA, Walters BJ, Phillips SE, Green K, Schmidt R, Wilson JA, Johnson GV, Roberson ED & Dobrunz LE (2012). Usp14 deficiency increases tau phosphorylation without altering tau degradation or causing tau-dependent deficits. PLoS One, 7, e47884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HT, & Goldberg AL (2018). UBL domain of Usp14 and other proteins stimulates proteasome activities and protein degradation in cells. Proceedings of the National Academy of Sciences of the United States of America, 115, E11642–E11650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiprowska MJ, Stepanova A, Todaro DR, Galkin A, Haas A, Wilson SM, & Figueiredo-Pereira ME (2017). Neurotoxic mechanisms by which the USP14 inhibitor IU1 depletes ubiquitinated proteins and Tau in rat cerebral cortical neurons: Relevance to Alzheimer's disease. Biochimica Et Biophysica Acta (BBA)-Molecular Basis of Disease, 1863, 1157–1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam YA, Pickart CM, Alban A, Landon M, Jamieson C, Ramage R, Mayer RJ & Layfield R (2000). Inhibition of the ubiquitin-proteasome system in Alzheimer's disease. Proceedings of the National Academy of Sciences of the United States of America, 97, 9902–9906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee B-H, Lee MJ, Park S, Oh D-C, Elsasser S, Chen PC, Gartner C, Dimova N, Hanna J, Gygi SP & Wilson SM (2010). Enhancement of proteasome activity by a small-molecule inhibitor of USP14. Nature, 467, 179–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JG, Kim W, Gygi S, & Ye Y (2014). Characterization of the deubiquitinating activity of USP19 and its role in endoplasmic reticulum-associated degradation. Journal of Biological Chemistry, 289, 3510–3517. 10.1074/jbc.M113.538934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MJ, Lee B-H, Hanna J, King RW, & Finley D (2011). Trimming of ubiquitin chains by proteasome-associated deubiquitinating enzymes. Molecular and Cellular Proteomics, 10(5), R110.003871. 10.1074/mcp.R110.003871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MJ, Lee JH, & Rubinsztein DC (2013). Tau degradation: The ubiquitin–proteasome system versus the autophagy-lysosome system. Progress in Neurobiology, 105, 49–59. 10.1016/j.pneurobio.2013.03.001 [DOI] [PubMed] [Google Scholar]
- Li H, Zhao Z, Ling J, Pan L, Zhao X, Zhu H, Yu J, Xie B, Shen J, & Chen W (2019). USP14 promotes K63-linked RIG-I deubiquitination and suppresses antiviral immune responses. European Journal of Immunology, 49, 42–53. 10.1002/eji.201847603 [DOI] [PubMed] [Google Scholar]
- Liu B, Jiang S, Li M, Xiong X, Zhu M, Li D, Zhao L, Qian L, Zhai L, Li J, Lu H, Sun S, Lin J, Lu Y, Li X, & Tan M (2018). Proteome-wide analysis of USP14 substrates revealed its role in hepatosteatosis via stabilization of FASN. Nature Communications, 9, 4770. 10.1038/s41467-018-07185-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Soetandyo N, Lee J-G, Liu L, Xu Y, Clemons WM, & Ye Y (2014). USP13 antagonizes gp78 to maintain functionality of a chaperone in ER-associated degradation. Elife, 3, e01369. 10.7554/eLife.01369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y-S, Wang X-F, Yu F, Wu T-M, Liu J-B, Zhang Y-J, Xia Q, Jiang Z-Y, Lin Q-L, & Fu DA (2020). Inhibition of USP14 and UCH37 deubiquitinating activity by b-AP15 as a potential therapy for tumors with p53 deficiency. Signal Transduction and Targeted Therapy, 5, 30. 10.1038/s41392-020-0143-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machida YJ, Machida Y, Vashisht AA, Wohlschlegel JA, & Dutta A (2009). The deubiquitinating enzyme BAP1 regulates cell growth via interaction with HCF-1. Journal of Biological Chemistry, 284, 34179–34188. 10.1074/jbc.M109.046755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall AG, Watson JA, Hallengren JJ, Walters BJ, Dobrunz LE, Francillon L, Wilson JA, Phillips SE, & Wilson SM (2013). Genetic background alters the severity and onset of neuromuscular disease caused by the loss of ubiquitin-specific protease 14 (usp14). PLoS One, 8, e84042. 10.1371/journal.pone.0084042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinnon C, & Tabrizi SJ (2014). The ubiquitin-proteasome system in neurodegeneration. Antioxidants and Redox Signaling, 21, 2302–2321. 10.1089/ars.2013.5802 [DOI] [PubMed] [Google Scholar]
- McNaught KSP, Olanow CW, Halliwell B, Isacson O, & Jenner P (2001). Failure of the ubiquitin–proteasome system in Parkinson's disease. Nature Reviews Neuroscience, 2, 589–594. [DOI] [PubMed] [Google Scholar]
- Miller VM, Nelson RF, Gouvion CM, Williams A, Rodriguez-Lebron E, Harper SQ, Davidson BL, Rebagliati MR & Paulson HL (2005). CHIP suppresses polyglutamine aggregation and toxicity in vitro and in vivo. Journal of Neuroscience, 25, 9152–9161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min JW, Lu L, Freeling JL, Martin DS, & Wang H (2017). USP14 inhibitor attenuates cerebral ischemia/reperfusion-induced neuronal injury in mice. Journal of Neurochemistry, 140, 826–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortuno D, Carlisle HJ, & Miller S (2016). Does inactivation of USP14 enhance degradation of proteasomal substrates that are associated with neurodegenerative diseases? F1000Res, 5, 137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peth A, Besche HC, & Goldberg AL (2009). Ubiquitinated proteins activate the proteasome by binding to Usp14/Ubp6, which causes 20S gate opening. Molecular Cell, 36, 794–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrucelli L, Dickson D, Kehoe K, Taylor J, Snyder H, Grover A, De Lucia M, McGowan E, Lewis J, Prihar G & Kim J (2004). CHIP and Hsp70 regulate tau ubiquitination, degradation and aggregation. Human Molecular Genetics, 13, 703–714. [DOI] [PubMed] [Google Scholar]
- Ristic G, Tsou W-L, & Todi SV (2014). An optimal ubiquitin-proteasome pathway in the nervous system: The role of deubiquitinating enzymes. Frontiers in Molecular Neuroscience, 7, 72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritorto MS, Ewan R, Perez-Oliva AB, Knebel A, Buhrlage SJ, Wightman M, Kelly SM, Wood NT, Virdee S, Gray NS & Morrice NA (2014). Screening of DUB activity and specificity by MALDI-TOF mass spectrometry. Nature Communications, 5, 4763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saez I, & Vilchez D (2014). The mechanistic links between proteasome activity, aging and age-related diseases. Current Genomics, 15, 38–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimura H, Schlossmacher MG, Hattori N, Frosch MP, Trockenbacher A, Schneider R, Mizuno Y, Kosik KS & Selkoe DJ (2001). Ubiquitination of a new form of alpha-synuclein by parkin from human brain: Implications for Parkinson's disease. Science, 293, 263–269. [DOI] [PubMed] [Google Scholar]
- Tang M, Harrison J, Deaton CA, & Johnson GVW (2019). Tau clearance mechanisms. Advances in Experimental Medicine and Biology, 1184, 57–68. [DOI] [PubMed] [Google Scholar]
- Taylor JP, Hardy J, & Fischbeck KH (2002). Toxic proteins in neurodegenerative disease. Science, 296, 1991–1995. [DOI] [PubMed] [Google Scholar]
- Upadhyay A, Joshi V, Amanullah A, Mishra R, Arora N, Prasad A, & Mishra A (2017). E3 ubiquitin ligases neurobiological mechanisms: development to degeneration. Frontiers in Molecular Neuroscience, 10, 151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaden JH, Bhattacharyya BJ, Chen P-C, Watson JA, Marshall AG, Phillips SE, Wilson JA, King GD, Miller RJ & Wilson SM (2015). Ubiquitin-specific protease 14 regulates c-Jun N-terminal kinase signaling at the neuromuscular junction. Molecular Neurodegeneration, 10, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters BJ, Hallengren JJ, Theile CS, Ploegh HL, Wilson SM, & Dobrunz LE (2014). A catalytic independent function of the deubiquitinating enzyme USP14 regulates hippocampal synaptic short-term plasticity and vesicle number. Journal of Physiology, 592, 571–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warrick JM, Morabito LM, Bilen J, Gordesky-Gold B, Faust LZ, Paulson HL, & Bonini NM (2005). Ataxin-3 suppresses polyglutamine neurodegeneration in Drosophila by a ubiquitin-associated mechanism. Molecular Cell, 18, 37–48. [DOI] [PubMed] [Google Scholar]
- Wilson SM, Bhattacharyya B, Rachel RA, Coppola V, Tessarollo L, Householder DB, Fletcher CF, Miller RJ, Copeland NG & Jenkins NA (2002). Synaptic defects in ataxia mice result from a mutation in Usp14, encoding a ubiquitin-specific protease. Nature Genetics, 32, 420–425. [DOI] [PubMed] [Google Scholar]
- Xia X, Liao Y, Guo Z, Li Y, Jiang L, Zhang F, Huang C, Liu Y, Wang X, Liu N & Liu J (2018). Targeting proteasome-associated deubiquitinases as a novel strategy for the treatment of estrogen receptor-positive breast cancer. Oncogenesis, 7, 75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D, Shan B, Lee BH, Zhu K, Zhang T, Sun H, Liu M, Shi L, Liang W, Qian L & Xiao J (2015). Phosphorylation and activation of ubiquitin-specific protease-14 by Akt regulates the ubiquitin-proteasome system. Elife, 4, e10510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Zhao Y, Fan Y, Chen Z, Li H, Lu J, Guo K, Woodfield SE, Vasudevan SA, Yang J & Nuchtern JG (2019). Inhibition of ubiquitin-specific protease 14 suppresses cell proliferation and synergizes with chemotherapeutic agents in neuroblastoma. Molecular Cancer Therapeutics, 18, 1045–1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun D, Zhuang Y, Kreutz MR, & Behnisch T (2018). The role of 19S proteasome associated deubiquitinases in activity-dependent hippocampal synaptic plasticity. Neuropharmacology, 133, 354–365. 10.1016/j.neuropharm.2018.01.043 [DOI] [PubMed] [Google Scholar]
- Zheng Q, Huang T, Zhang L, Zhou Y, Luo H, Xu H, & Wang X (2016). Dysregulation of ubiquitin-proteasome system in neurodegenerative diseases. Frontiers in Aging Neuroscience, 8, 303. 10.3389/fnagi.2016.00303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P, Xia D, Wang Y, Lv H, Wang Z, Xing M, Zhao Q, & Xu S (2019). Matrine derivate MASM protects murine MC3T3-E1 osteoblastic cells against dexamethasone-induced apoptosis via the regulation of USP14/p53. Artificial Cells, Nanomedicine, and Biotechnology, 47, 3720–3728. 10.1080/21691401.2019.1664563 [DOI] [PubMed] [Google Scholar]