Abstract
Objective
Recent studies using whole-body clock-disrupted animals identified a disruption in the circadian rhythm of the intestinal L-cell incretin hormone, glucagon-like peptide-1 (GLP-1). Although GLP-1 plays an essential role in metabolism through enhancement of both glucose-stimulated insulin secretion and satiety, recent evidence has also demonstrated its importance in regulating intestinal and microbial homeostasis. Therefore, using in vivo and in vitro models, this study assessed the role of the core circadian clock gene Arntl in the regulation of time-dependent GLP-1 secretion and its impact on the intestinal environment.
Methods
Oral glucose tolerance tests were conducted at zeitgeber time 2 and 14 in control and inducible Gcg-Arntl knockout (KO) mice. Colonic intraepithelial lymphocytes were isolated, mucosal gene expression analysis was conducted, and 16S rRNA gene sequencing of colonic feces as well as analysis of microbial metabolites were performed. Time-dependent GLP-1 secretion and transcriptomic analysis were conducted in murine (m) GLUTag L-cells following siRNA-mediated knockdown of Arntl.
Results
Gcg-Arntl KO mice displayed disrupted rhythmic release of GLP-1 associated with reduced secretion at the established peak time point. Analysis of the intestinal environment in KO mice revealed a decreased proportion of CD4+ intraepithelial lymphocytes in association with increased proinflammatory cytokine gene expression and increased colonic weight. Moreover, increased Actinobacteria within the colonic microbiome was found following L-cell Arntl disruption, as well as reductions in the microbial products, short chain fatty acids, and bile acids. Finally, siRNA-mediated knockdown of Arntl in mGLUTag L-cells resulted in both impaired time-dependent GLP-1 secretion and the disruption of pathways related to key cellular processes.
Conclusions
These data establish, for the first time, the essential role of Arntl in the intestinal L-cell in regulating time-dependent GLP-1 secretion. Furthermore, this study revealed the integral role of L-cell Arntl in mediating the intestinal environment, which ultimately may provide novel insight into the development of therapeutics for the treatment of intestinal and metabolic disorders.
Keywords: Circadian, Colon, GLP-1, GLP-2, Immunity, Inflammation, Microbiome
Highlights
-
•
L-cell Arntl is required for time-dependent GLP-1 secretion.
-
•
L-cell Arntl regulates metabolic as well as GLP-1 synthesis and exocytotic pathways.
-
•
Loss of L-cell Arntl results in altered colonic immune and microbial environments.
Abbreviations
- BAs
Bile acids
- IELs
Intraepithelial lymphocytes
- KD
Knockdown
- KO
Knockout
- OGTT
Oral glucose tolerance test
- SCFAs
Short-chain fatty acids
- ZT
Zeitgeber
1. Introduction
The incretin hormone glucagon-like peptide-1 (GLP-1) is secreted from enteroendocrine L-cells in response to nutrient intake [1,2]. The ability of GLP-1 to enhance both glucose-stimulated insulin secretion and satiety has led to the development of GLP-1 receptor agonists for the treatment of type 2 diabetes and obesity [1,2]. In addition to these well-established metabolic effects, recent studies have also implicated GLP-1 as an important mediator of the local intestinal environment through the regulation of inflammation, microbial composition, and mucosal mass [[3], [4], [5]]. Furthermore, intraepithelial lymphocytes (IELs) not only express the GLP-1 receptor, but also modulate systemic GLP-1 bioavailability [3,6]. Although GLP-1 secretion is known to be regulated by nutrient ingestion, circulating hormones, the parasympathetic nervous system, and microbial metabolites, recent studies have also demonstrated that the intestinal L-cell is under circadian control, with peak release of GLP-1 occurring at the onset of the dark/active period in rodents and differential secretion by time of day in humans [[7], [8], [9], [10], [11], [12]].
Circadian rhythms are endogenous biological rhythms with a phase of approximately 24 h. They have developed over evolutionary time as an adaptive response to diurnal variations in the environment, mainly the external light–dark cycle [[13], [14], [15]]. On the molecular level, these rhythms are generated through the heterodimerization and binding of BMAL1 (aryl hydrocarbon receptor nuclear translocator-like protein 1 (ARNTL) or brain and muscle (ARNT-Like 1)) and CLOCK to the E-box promoter elements of the Period (Per1/2/3) and Cryptochrome (Cry1/2) genes. PER1-3 and CRY1/2 then provide feedback to suppress BMAL1/CLOCK, thus forming a self-sustained autoregulatory transcriptional/translational feedback loop [[13], [14], [15]]. Although originally described in the suprachiasmatic nuclei of the hypothalamus, which has been termed the master clock, peripheral metabolic tissues, including the gastrointestinal tract, pancreatic α and β cells, hepatocytes, skeletal muscle, and adipose tissue have all been shown to have cell-autonomous circadian rhythms [[16], [17], [18], [19], [20], [21], [22], [23]]. Furthermore, circadian oscillations in the composition and function of the gut microbiome have been demonstrated and are critical for circadian gene rhythmicity in intestinal epithelial cells [[24], [25], [26], [27], [28]]. Consistent with these findings, whole-body clock knockout (KO) mouse models have been shown to develop metabolic abnormalities as well as microbial dysbiosis [17,25,[29], [30], [31], [32], [33]]. However, it is the generation of tissue-specific clock gene KO models that has provided invaluable insight into the role of the circadian clock in regulating distinct functions within these tissues [29,[34], [35], [36], [37], [38]].
Recent work using whole-body Arntl KO mice as well as primary intestinal cultures generated from these animals has implicated the circadian clock as a key determinant of the GLP-1 response to secretagogues [8,39]. However, the role of L-cell Arntl in mediating diurnal GLP-1 release and, thus, in establishing metabolic and intestinal homeostasis remains unknown. Herein, through inducible KO of Arntl in the intestinal L-cell using a Gcg-cre transgenic model, we demonstrate for the first time a crucial role for L-cell Arntl in regulating the time-dependent stimulation of GLP-1 secretion. Targeted disruption of Arntl also resulted in alterations in the intestinal, immune, and microbial environment within the mouse colon, suggesting a direct role for rhythmic L-cell secretion in mediating the intestinal environment. Arntl knockdown (KD) and subsequent GLP-1 secretion and transcriptomic analysis in the murine (m) GLUTag L-cell line provided additional mechanistic insight into the role of the core clock gene Arntl in regulating rhythmic L-cell secretion. Taken together, these data provide novel insight into the role and function of L-cell Arntl that may ultimately facilitate development of time-dependent treatments for metabolic and/or intestinal diseases.
2. Material and methods
2.1. Animal model
C57BL/6J Arntlflox/flox (Arntlfl/fl) mice that were originally purchased from Jackson Laboratories were crossed with C57BL/6J Gcg-CreERT2/+;Rosa26-GCamP3fl/fl mice (Gcg-CreERT2/+) [40] to generate Gcg-Arntl KO mice upon administration of tamoxifen (1 mg/100 μL sunflower oil i.p. for 5 days) [40,41]. The Gcg-cre transgene has previously been reported to be expressed in the intestinal L-cell, the pancreatic α-cell, and select neurons of the central nervous system [40]. Age (7–11 weeks)-, sex (male and female)-, and littermate-control (Gcg-CreERT2/+ and Arntlfl/fl mice with and without (i.e., vehicle alone) tamoxifen and Gcg-CreERT2/+; Arntlfl/fl mice without tamoxifen) mice were employed for all studies. All animals had free access to regular chow diet (2018 Teklad; Envigo) and water and were housed under a 12-hour light/12-hour dark cycle (with zeitgeber time (ZT) 0 being 06:00). All studies were conducted 7 days after the final tamoxifen injection to allow for an adequate wash-out period [40,42,43]. All animal work was approved by the Animal Care Committee at the University of Toronto and followed the guidelines set out by the Canadian Council on Animal Care.
2.2. Oral glucose tolerance tests (OGTTs)
OGTTs (5 g glucose/kg body weight) were conducted on 4-hour fasted mice at ZT2 (established trough of GLP-1 secretion) and ZT14 (established peak of GLP-1 secretion) with their basal blood glucose being measured prior to the oral gavage of glucose (t = 0 min) [8,40,41]. Tail vein blood was collected and blood glucose was measured using a OneTouch meter (LifeScan, Burnaby, BC, Canada) at t = 0, 10, and 60 min. Plasma GLP-1, insulin, and glucagon were analyzed at the same time points by MesoScale Discovery Assay, and plasma GIP was measured by Millipore ELISA.
2.3. Gravimetric, morphometric, and immunometric analyses
The colon was isolated and rinsed with ice-cold PBS. Weight and length were measured using a 2.8 g weight to provide constant tension. A 2-cm section of colon was collected into formalin for paraffin embedding, sectioning, and staining with hematoxylin & eosin (Pathology Research Program, University Health Network, Toronto, ON). Colon crypt depth was measured in blinded fashion using a Zeiss microscope with AxioVision software (Zeiss Microscopy Canada), with at least 20 well-oriented crypts measured from each mouse, to make n = 1. Crypt number was determined per intestinal cross-section by counting all well-oriented crypts in a blinded fashion, making n = 1 for each mouse.
Colonic sections were stained using anti-GLP-1 and anti-ARNTL primary antibodies (Abcam) with Alexa Fluor–coupled secondary antibodies (Table 1). GLP-1 staining was used to obtain the region of interest for quantification of ARNTL pixel fluorescence using a Nikon Swept Field Confocal microscope with NIS-Elements Imaging software (Nikon Corporation). Five cells were analyzed per mouse to make n = 1. The total number of L-cells was counted in 3 transverse colonic sections and normalized to one section per mouse to make n = 1.
Table 1.
List of Antibodies for Immunofluorescence.
| Primary Antibody | RRID | Secondary Antibody | RRID |
|---|---|---|---|
| Mouse anti-GLP-1 | AB_447455 | Anti-mouse IgG | AB_330924 |
| Rabbit anti-ARNTL | AB_10675117 | Anti-rabbit IgG | AB_2099233 |
2.4. Gene expression analyses
RNA was isolated from colonic mucosal scrapes using the RNeasy Plus Mini Kit (Qiagen Inc.) and cDNA was generated using 5x All-in-One RT Mastermix (Applied Biological Materials Inc.). Quantitative reverse-transcriptase polymerase chain reaction (qRT-PCR) was performed using the Taqman Gene Expression Assay (ThermoFisher) with primers (ThermoFisher) as listed in Table 2. Data were analyzed using the ΔΔCT method, with 18S as the internal control.
Table 2.
List of Primer Sequences.
| Target | Product Number |
|---|---|
| Tnf | Mm00443258_m1 |
| Ifng | Mm01168134_m1 |
| Il6 | Mm00446190_m1 |
| Il10 | Mm01288386_m1 |
| Tgfb | Mm01178820_m1 |
| 18S | Hs99999901_s1 |
2.5. Lymphocyte collection and flow cytometry
Colonic IELs were obtained as previously described [9]. In brief, the colon was divided into 2-cm segments and incubated with Hank's Balanced Salt Solution +5 mM dithiothreitol for mucus removal. After two consecutive 10 min incubations with 5 mM EDTA, IELs were collected. LIVE–DEAD fixable aqua dead cell stain (ThermoFisher) was used to exclude dead cells from single-cell suspensions. Prior to flow cytometric analysis, suspensions were incubated in flow cytometry staining buffer with Anti-Mo CD16/CD32 (ThermoFisher, Clone93) for the blocking of non-specific binding of antibodies to Fc receptors. Staining for cell surface antigens was conducted using the antibodies listed in Table 3, and fixation was carried out using the eBioscience Foxp3/Transcription Factor Staining Kit (ThermoFisher). Flow cytometry was conducted using the LSRFortessa Cell Analyzer (BD Biosciences) running FACSDiva acquisition software. Data were analyzed using FlowJo v10.
Table 3.
List of Antibodies for Cell Sorting.
| Target | Fluorophore |
|---|---|
| CD3 | PE-Cy7 |
| TCRb | eFluor 450 |
| CD8α | Alexa Fluor 700 |
| CD8β | APC |
| CD4 | APC-eFluor 780 |
2.6. Microbiome and microbial product analyses
Colonic fecal DNA was isolated using the DNeasy PowerSoil Kit (Qiagen Inc.), and 16S rRNA gene sequencing was performed by the Centre for the Analysis of Genome Evolution & Function (University of Toronto, Toronto, Ontario, Canada). Short-chain fatty acids (SCFAs) and bile acids (BAs) from the cecal feces were analyzed by gas chromatography–mass spectrometry and liquid chromatography–mass spectrometry (Biocrates), respectively, by the Analytical Facility for Bioactive Molecules at the Hospital for Sick Children (Toronto, Ontario, Canada).
16S rRNA gene sequencing data analysis was performed as previously described using R Statistical Programming language [9]. The Phyloseq package was used to read OTU counts and agglomerate them at the taxonomic levels of Family, Phylum, Genus, and Species. The ALDEx R package was used to compare relative abundances, estimating the technical variation inherent to high-throughput sequencing by Monte Carlo sampling from a Dirichlet distribution. Subsequently, ANOVA-like t-tests were performed with 250 Monte Carlo samples, with Benjamin Hochberg adjustment of p-values.
2.7. Cell culture
The colonic mGLUTag L-cell line was chosen as a model of the intestinal L-cell, as it has been demonstrated to exhibit a cell-autonomous circadian clock [7,8,39,44] and possess similarities to primary L-cells in terms of protein expression and regulation of GLP-1 secretion [45,46]. mGLUTag L-cells were grown in Dulbecco's modified Eagle's medium (DMEM, Gibco, Waltham, MA, USA) with 25 mmol/L glucose and 10% FBS and synchronized using a previously established protocol [7,8,39,40]. In brief, cells were incubated for 12 h with 0.5% FBS media to induce quiescence, after which they were treated with 20 μM forskolin (Sigma–Aldrich, Oakville, ON, Canada) for 1 h, with the media subsequently being changed to growing media for 8 h (established peak of GLP-1 secretion) and 20 h (established trough of GLP-1 secretion) [7,8,39].
Arntl KD was conducted using ON-TARGETplus siRNA for Arntl with ON-TARGETplus Non-targeting Pool scRNA as a control (Dharmacon Inc., Lafayette, CO, USA), based on a previously established protocol [8,40]. In short, a reverse-transfection protocol was utilized using the siRNA constructs in combination with Dharmafect3. Media for both the serum starvation (0.5% FBS), serum shock (10% FBS with forskolin), and all subsequent incubations included siRNA or scRNA in combination with Dharmafect3, as appropriate. Cells were then analyzed for GLP-1 secretion or extracted for RNA-sequencing analysis.
2.8. GLP-1 secretion assay
At 8 and 20 h post-synchronization, cells were incubated for 2 h in 0.5% DMEM containing vehicle or the known secretagogue, 10−7 M GIP [7,8,39,40,44]. Peptides in the media and cells were isolated by reversed-phase adsorption using C18 Sep-Pak cartridges (Waters Associates, Milford, MA, USA), and GLP-1 levels were measured using a Total GLP-1 Radioimmunoassay kit (GLP-1T-36HK, Millipore, Etobicoke, ON, Canada). GLP-1 secretion was represented as the percent of GLP-1 in the media over the total GLP-1 content (media + cells).
2.9. RNA sequencing
RNA was extracted using the RNeasy Plus Mini Kit (Qiagen), and sequencing was performed at the Donnelly Facility, University of Toronto. Samples were run in 50 base paired-end reads and sequencing data were then mapped using the Kallisto pseudo-alignment [47]. Count tables were assembled using the tximport [48] and normalized using the EdgeR R packages [49]. Genes with very low expression (<1) were removed, and the rest were normalized using the a sinh method. Differential expression analysis was performed using linear models through the R package limma [50] (Bioconductor) using an adjusted p-value of 0.01. Gene set enrichment analysis was done through the limma function camera using the Bader Lab gene set resource (http://download.baderlab.org/EM_Genesets).
Cytoscape plots were generated using EnrichmentMap [51,52], where nodes represent up- or downregulated pathways (green pathways are enriched to the scRNA condition, red are enriched to the siRNA condition). Lines connecting nodes (edges) were established through an overlap score based on the number of shared genes between the two pathways. Clusters of functionally related gene sets were then grouped and labelled.
2.10. Statistical analyses
Statistical analyses were conducted using GraphPad Prism. Data were analyzed for significance by 2-way ANOVA, followed by appropriate post hoc analysis for 3 or more groups or by Student's t-test for two groups. All data are expressed as mean ± SEM.
3. Results
3.1. L-cell Arntl is required for time-dependent L-cell secretion
Consistent with previous reports [8,9], time-dependent GLP-1 secretion in control animals aligned with the normal rodent feeding and fasting periods, with elevated GLP-1 secretion at ZT14 compared to ZT2 in response to identical glucose loads (Figure 1A). A 22% (p < 0.05) reduction in ARNTL fluorescent pixels was found in L-cells from Gcg-Arntl KO animals (2196 ± 192; n = 3) compared to control mice (2812 ± 235; n = 5), with no change in total L-cell number (114 ± 24 vs 126 ± 20, respectively). Gcg-Arntl KO animals demonstrated a loss of the glucose-stimulated GLP-1 secretory pattern in association with reduced secretion at the ZT14 time point following an OGTT, but did not display any changes in normal pattern of fasting GLP-1 levels (Figure 1A and S1A). Interestingly, the reduced GLP-1 secretion observed in KO mice at ZT14 was independent of changes in colonic mucosal proglucagon (Gcg) gene expression at that time point (Figure S1B). To verify the specificity of Arntl disruption of the L-cell within the intestine, the levels of the other incretin hormone, glucose-dependent insulinotropic polypeptide (GIP), were examined. As expected [9], control animals exhibited increased GIP secretion at ZT14 compared to ZT2, with no differences in either fasting levels or oral glucose-stimulated secretion being observed in Gcg-Arntl KO mice (Figure 1B and S1C). Notably, control mice did not display rhythmicity in glucagon levels between ZT2 and ZT14, likely a result of having only two sampling timepoints. However, consistent with the role of the circadian clock gene Arntl in pancreatic α-cell function [19] as well as with expression of the Gcg-cre transgene in these cells [40], glucagon levels were decreased in Gcg-Arntl KO mice at both ZT2 and ZT14 (Figure 1C). Consequently, 4-hour fasted Gcg-Arntl KO animals displayed hypoinsulinemia and normoglycemia, with no differences in insulin secretion following oral glucose administration and only a slight reduction in blood glucose at the ZT14 time point (Figures S1D–G). Finally, no differences were observed in body weight or in 4-hour food intake throughout the day/night cycle in the KO animals compared to controls, suggesting a lack of effect of Arntl disruption in Gcg-expressing neurons of the central nervous system (Figure 1D and S1H). Together, these data establish the Gcg-Arntl KO animals as a model of disrupted time-dependent L-cell secretion.
Figure 1.
L-cell Arntl KO mice display impairedtime-dependentGLP-1 secretion. Individual responses at ZT2 and ZT14 as well as the corresponding ΔAUCs for (A) GLP-1 and (B) GIP following an OGTT in control and Gcg-Arntl KO animals. (C) Glucagon levels at ZT2 and ZT14 and (D) 24-hour food intake in control and Gcg-Arntl KO animals. n = 7–8; ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001 (conducted by Student's t-test for individual responses and 2-way ANOVA for ΔAUCs). Control mice include Gcg-CreERT2/+ and Arntlfl/fl mice ± tamoxifen and Gcg-CreERT2/+; Arntlfl/fl mice without tamoxifen.
3.2. Disruption of L-cell Arntl alters the colonic immune environment
Colonic IEL cell populations were characterized given that the majority of murine L-cells are localized to the colon [41,53,54] and that the recent evidence depicts a tight interplay between GLP-1 and the intestinal immune environment [3,4,6]. No time-dependent differences were observed in the CD8αβ+, CD8αα+, and CD4+ IELs between ZT2 and ZT14 in control mice; however, while the CD8αβ+ and CD8αα+ cell populations did not differ between control and KO animals, Gcg-Arntl KO mice had a reduced proportion of the CD4+ cell population at the ZT14 time point (Figure 2A–C). Interestingly, while pro-inflammatory cytokine expression analyses demonstrated no differences in Tnf expression between control and KO mice, Ifng and Il6 were markedly elevated in KO animals at both ZT2 and ZT14 (Figure 2D–F). No changes were found in the time-dependent expression of the immunoregulatory, Tgfb, whereas Il10 expression was increased in KO mice at ZT2 (Figure 2G–H). Consistent with previous reports demonstrating increased intestinal weight in murine models of inflammation [55], colonic weight in Gcg-Arntl KO animals was significantly increased at both time points, independent of colonic length (Figure S2A–B). However, this change was not associated with alterations in either colonic crypt depth or number (Figure S2C–D).
Figure 2.
L-cell Arntl KO mice display a pro-inflammatory colonic environment. Proportion of (A) CD8αβ+, (B) CD8αα+, and (C) CD4+ on colonic TCRβ+ cells, as well as (D)Tnf, (E)Ifng, (F)Il6, (G)Tgfb, and (H)Il10 mRNA expression in colonic mucosa, at ZT2 and ZT14 in control and Gcg-Arntl KO animals. n = 7–8; ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001 (conducted by 2-way ANOVA). Control mice include Gcg-CreERT2/+ and Arntlfl/fl mice ± tamoxifen and Gcg-CreERT2/+; Arntlfl/fl mice without tamoxifen.
3.3. L-cell Arntl disruption alters the colonic microbiome
Given the known interplay between GLP-1, the host immune system, and the microbiome [3,4,6,9,56,57], 16S rRNA sequencing of colonic feces was conducted. Although no significant time-dependent changes were observed between ZT2 and ZT14, KO animals demonstrated a marked increase in the abundance of Actinobacteria and its major constituent family Bifidobacteriaceae at both time points (Figure 3A–C and S3A-B). The increase in Actinobacteria in the state of GLP-1 deficiency observed herein parallels the findings from GLP-1 receptor (GLP-1R) KO mice, which have also been reported to have elevated Actinobacteria [3]. To assess potential changes in microbial function, the major microbial products SCFAs and BAs were quantified in the cecal contents. Although total SCFA levels did not display any time-dependent changes, KO animals had reduced levels at ZT14, predominantly due to reduced acetic acid, in association with the reduced GLP-1 secretion at this time point (Figure 3D and S3C). In contrast, BA concentrations in control animals were largely elevated at ZT2 compared to ZT14, while KO mice displayed reduced levels of select BAs at both time points compared to controls (Figure 3E and S4D). These changes in microbially-generated products suggest not only compositional but also functional changes in the microbiome of L-cell Arntl-disrupted animals.
Figure 3.
L-cell Arntl KO mice display an altered microbial composition and function. Relative abundance of (A) phyla, (B) Actinobacteria, and (C)Bifidobacteriaceae in the colonic microbiomes of control and Gcg-Arntl KO mice at ZT2 and ZT14. Quantification of cecal (D) total short-chain fatty acids (SCFAs) and (E) bile acids (BAs) at ZT2 and ZT14 in control and Gcg-Arntl KO animals. n = 7–8; ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001 (conducted by 2-way ANOVA). Control mice include Gcg-CreERT2/+ and Arntlfl/fl mice ± tamoxifen and Gcg-CreERT2/+; Arntlfl/fl mice without tamoxifen.
3.4. Arntl knockdown in the mGLUTag L-cells results in impaired stimulation of GLP-1 secretion
To complement the in vivo findings of the role of L-cell Arntl in mediating time-dependent GLP-1 release, siRNA-mediated KD of Arntl was conducted in synchronized mGLUTag L-cells at the previously established peak (8 h) and trough (20 h) secretory time points [7,8,39,40,44]. scRNA treatment had no effect on cell synchronization, as shown by the previously established anti-phasic expression of Arntl and Per2 [7,8,39,44]. As expected, siRNA treatment resulted in decreased Arntl expression at both the 8- and 20-hour time points, with an associated loss of the temporal rhythm in Per2 (Figure 4A–B). GLP-1 secretion by scRNA-treated cells revealed the expected increase in the GLP-1 secretory response to GIP at 8 h in comparison to 20 h [7,8,39] (Figure 4C). Consistent with previous reports demonstrating that only stimulated GLP-1 release follows a circadian rhythm [7,8], vehicle-treated scRNA mGLUTag L-cells did not display time-dependent GLP-1 secretory differences between 8 and 20 h after cell synchronization. However, siRNA-mediated Arntl KD resulted in impaired GLP-1 secretion at the peak, but not at the trough time point, which also exhibited an unexpected increase in basal GLP-1 release (Figure 4C).
Figure 4.
Arntl KD in mGLUTag L-cells results in impaired GLP-1 release. (A)Arntl, (B)Per2 clock gene expression, and (C) vehicle- and 10−7 M GIP-stimulated GLP-1 secretion at 8 and 20 h after synchronization in mGLUTag L-cells treated with either scRNA or Arntl siRNA. n = 4–6; ∗p < 0.05, ∗∗∗p < 0.001 (conducted by 2-way ANOVA).
3.5. L-cell Arntl regulates key pathways involved in GLP-1 release
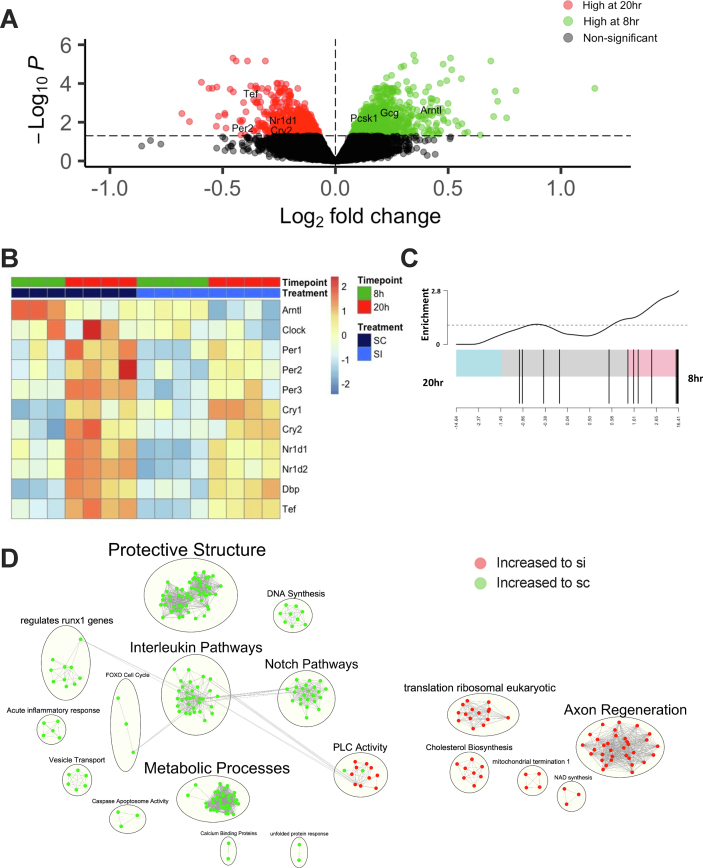
To provide mechanistic insight into the role of L-cell Arntl in regulating GLP-1 secretion, RNA-seq was conducted on synchronized control and clock-disrupted mGLUTag L-cells at the peak (8 h) and trough (20 h) time points of GLP-1 secretion. Following synchronization, as expected [7,8,39,44], control L-cells displayed rhythmicity in clock gene expression, with the positive arm of the clock being increased to the 8-hour time point and the negative arm of the clock largely increased to the 20-hour time point (Figure 5A–B). Interestingly, key genes involved in GLP-1 synthesis, including Gcg and Pcsk1 (Figure 5A), as well as GLP-1 and incretin synthesis and secretion gene sets, as illustrated by barcode plots, were upregulated to the 8-hour time point (Figure 5C, S4A and Supplementary Table 1). Furthermore, analysis of control cells between the peak and trough time points revealed an increase in peptide hormone metabolism pathways and an expected increase in the expression of key exocytotic proteins that are known regulators of L-cell secretion [8,40,41,58,59] (Figure S4B–C and Supplementary Table 1). Together, these data further confirm the role of the cell-autonomous clock in GLP-1 secretion under normal conditions.
Figure 5.
Arntl regulates key cellular pathwaysin mGLUTag L-cells. (A) Volcano plot of the transcriptome at the peak (8 h) and trough (20 h) of GLP-1 secretion in synchronized control mGLUTag L-cells. (B) Heat map depicting clock gene expression in scRNA- and Arntl siRNA-treated mGLUTag L-cells at 8 and 20 h after cell synchronization. (C) Representative GLP-1 processing gene set comparing control mGLUTag L-cells between the peak and trough GLP-1 secretory time points. (D) Network analysis comparing pathway enrichment between transcriptomes of scRNA- and Arntl siRNA-treated mGLUTag L-cells at the 8-hour (peak) secretory time point. Clusters of functionally related gene sets were grouped into nodes, with green and red indicating pathways that were relatively enriched in the scRNA-compared to siRNA-treated cells and vice versa, respectively; lines connecting the nodes indicate shared genes between the two pathways.
In contrast to the robust rhythmicity observed in normal cells, analysis of Arntl KD mGLUTag L-cells revealed significantly reduced expression of Arntl and disruption in the circadian clock machinery, as evidenced by an overall 27% reduction in amplitude (P < 0.05; calculated by determining the difference between peak and trough clock gene expression when comparing the scRNA and siRNA conditions) (Figure 5B and S4D). To further determine the molecular mechanism behind the impaired GLP-1 secretion observed at the 8-hour time point, pathway analysis of the transcriptome of scRNA- and siRNA-treated cells was performed. Compared to Arntl-disrupted L-cells, control cells had a marked increase in pathways related to metabolic processes as well as vesicular transport (Figure 5D and Supplementary Table 2), both of which are well-established regulators of GLP-1 release [8,39]. Interestingly, pathways related to immunity, such as interleukin signaling, as well as inflammatory responses were also dysregulated upon clock disruption (Figure 5D and Supplementary Table 2). Furthermore, Notch signaling was decreased in siRNA-treated cells (Figure 5D and Supplementary Table 2), suggesting a role for the circadian clock gene Arntl in the regulation of epithelial cell integrity [60]. Interestingly, a comparison of control and Arntl KD cells at the 20-hour time point revealed an increase in pathways related to the electron transport chain in the clock-disrupted cells (Figure S4E and Supplementary Table 3). Genes sets involved in the production of ATP (Fig. S4F and Supplementary Table 3) were also upregulated in the siRNA-treated cells. Given the known role of mitochondrial function in mediating GLP-1 release [39], this may provide a potential mechanism behind the increased basal levels of GLP-1 observed at the 20-hour time point in the Arntl KD cells.
4. Discussion
The intestinal L-cell has been shown to express a functional circadian clock [[7], [8], [9]]. While whole-body Arntl KO animals exhibit disrupted rhythmic GLP-1 release [8], use of Gcg-Arntl KO mice has now demonstrated the cell-autonomous role of the intestinal L-cell Arntl in time-dependent L-cell secretion. As in whole-body Arntl KO mice [8], Gcg-Arntl KO mice demonstrated a disrupted rhythm in GLP-1 secretion, with a dampened glucose-stimulated GLP-1 response occurring only at the established peak of L-cell secretion. Importantly, while whole-body Arntl KO animals exhibit temporal disruptions in food intake [61], a known zeitgeber for L-cell secretion [7], the Gcg-Arntl KO mice studied herein did not display any alterations in their 24-hour food intake patterns. Furthermore, primary small intestinal cultures generated from whole-body Arntl KO mice show increased Gcg gene expression [39], whereas colonic Gcg expression in Gcg-Arntl KO mice was decreased (at ZT2) despite decreased GLP-1 release in both models. Together, these findings highlight the importance of using cell-specific clock disruption models in the study of circadian L-cell secretion.
Given the known circadian rhythm of OGTT-induced GIP secretion [9] and the stimulatory effect of GIP on the L-cell [1,2], GIP levels were assessed and found to be unchanged in KO animals, confirming not only the specificity of the KO within the intestine to the L-cell, but also that the impaired GLP-1 secretion was a result of inherent L-cell Arntl disruption. Interestingly, while GLP-1 receptor (GLP-1R) KO mice have been shown to have a compensatory increase in GIP [62], normal levels of GIP were observed in the GLP-1–deficient Gcg-Arntl KO mice. These findings may indicate inherent differences in adaptation between chronic constitutive and short-term inducible KO models. However, despite decreased peak GLP-1 secretion, insulin levels following an OGTT were unchanged in the KO mice, in association with little impact on glycemia. This is likely due to Gcg-driven loss of Arntl in the pancreatic α-cells, as this circadian clock gene has been established as essential for coordinating glucagon release by these cells [19]. Therefore, the Gcg-Arntl KO mice may not be an ideal model for studying the role of L-cell Arntl and circadian GLP-1 release in 24-hour metabolic homeostasis. However, overall, these animals serve as an excellent model for determination of the localized role of diurnal L-cell secretion in the regulation of intestinal homeostasis.
Recent studies have identified the GLP-1R on IELs, which are key players in the prevention of intestinal inflammation [3,6]. Interestingly, while IEL GLP-1R expression has been demonstrated as essential in the regulation of GLP-1 bioavailability [6], no time-dependent differences were found in the proportion of colonic CD8αβ+, CD8αα+, and CD4+ IELs in control animals, thereby suggesting that temporal changes in circulating GLP-1 levels are not dependent on the binding of GLP-1 by this cell population. Given that recent studies have shown rhythmic changes in IEL cell populations within the small intestine [[63], [64], [65]], the lack of time-dependent differences in the colon suggests that these cell populations may be differentially regulated. Furthermore, lack of differences between ZT2 and ZT14 in the colonic IELs, despite a robust rhythm in GLP-1, indicates that GLP-1 is unlikely to act as a homing mechanism for these cells, despite their GLP-1R expression. Additionally, L-cell Arntl disruption resulted in a decrease in the proportion of colonic CD4+ IELs at the ZT14 time point, accompanied by decreased GLP-1 levels. This further negates the possible GLP-1-sequestering role of the GLP-1R expressed by these cells.
Interestingly, disruption of L-cell Arntl was associated with a marked increase in colonic mucosal pro-inflammatory cytokine expression. Given that depletion of CD4+ T cells has been identified as a marker of impaired mucosal immune function and epithelial integrity [66,67], the reduced proportion of this cell population in the Gcg-Arntl KO animals might therefore be a driver of the observed intestinal inflammation. Indeed, use of an intestinal-specific clock gene disruption model (i.e., driven by the villin promoter) demonstrated an essential role for the circadian clock gene RORα in attenuating intestinal epithelial inflammatory responses [34]. Although the intestinal L-cell only makes up approximately 0.5% of the intestinal epithelial cells [41,53,54], the data presented herein provides evidence for an important role of L-cell Arntl in mediating intestinal immune homeostasis, a key parameter in protection of the systemic circulation from both commensal and ingested pathogens.
In association with the pro-inflammatory environment in the Gcg-Arntl KO animals, colonic weight was increased, as has been previously shown in the dextran sulfate sodium-induced colitis model of inflammatory bowel disease [55]. The increase in colonic weight was independent of changes in either crypt number or crypt depth, suggesting that this was exclusive of the changes in L-cell secretion of either GLP-1, known to increase crypt fission [5], or the co-secreted hormone, GLP-2, which increases crypt cell proliferation [68]. However, consistent with the additional role of GLP-2 in maintaining intestinal barrier integrity [69], the presumed decrease in GLP-2 levels as a result of L-cell Arntl disruption may contribute to the proinflammatory environment within the colon of the Gcg-Arntl KO mice.
Consistent with the Gcg-Arntl KO mice displaying no differences in the proportion of colonic IELs between ZT2 and ZT14, the composition of the colonic microbiome also lacked any time-dependent variations. This is, however, unique to the control animals employed herein, as previous reports of rhythmic changes in colonic microbial composition have been demonstrated in other murine models [9,[24], [25], [26],28] and, therefore, speak to how both differences in strains and in vivaria (including diet) can dictate the outcome of microbial studies. Specifically, although recent studies have demonstrated that time-dependent changes in the species Akkermansia muciniphila parallel the GLP-1 secretory pattern in normal mice as well as in germ-free animals following recolonization [9], this was not observed in this study. Nonetheless, increases in the phyla Actinobacteria and the associated family Bifidobacteriaceae were observed in Gcg-Arntl KO animals, not only adding to the body of evidence supporting the role of the intestinal epithelial clock in establishing microbial composition [25], but also implicating L-cell Arntl specifically as a regulator of the colonic microbiome. Interestingly, whereas GLP-1 receptor KO animals have been reported to have elevated Actinobacteria, GLP-2 receptor KO mice exhibit decreased Actinobacteria [3,70]. Therefore, it is likely that the observed increase in Actinobacteria in the Gcg-Arntl KO animals represents a balance between decreased GLP-1 and GLP-2 co-release following L-cell Arntl disruption. Interestingly, increased abundance of Actinobacteria has been previously associated with enhanced proinflammatory cytokine expression [71] as well with inflammatory bowel disease [72], as observed in the Gcg-Arntl KO mice herein. Furthermore, the decreased levels of SCFAs and BAs observed in the Gcg-Arntl KO mice are consistent with their role in signaling to immune cells for intestinal immune regulation [73] as well as their established roles in stimulating L-cell secretion [74,75]. Together, these data provide further support for the role of the intestinal L-cell in maintaining intestinal immune and microbial homeostasis.
To provide mechanistic insight as to how L-cell Arntl disruption results in impaired rhythmic hormone secretion, the well-established mGLUTag L-cell line was utilized. Consistent with previous studies [7,8,39], control cells exhibited time-dependent GLP-1 release in response to the known L-cell secretagogue, GIP. Previous transcriptomic and proteomic analysis of mGLUTag L-cells identified increased expression of pathways related to vesicle-mediated transport at the peak of GLP-1 secretion [8]. Furthermore, analysis of primary L-cells revealed a similar increase in pathways related to SNARE-mediated exocytosis at the peak secretory time point [9]. Mechanistic studies using chromatin immunoprecipitation analysis further identified a direct role for L-cell ARNTL in regulating the time-dependent expression of key exocytotic proteins [8,40]. The demonstration of increased expression of pathways involved in hormone processing as well as elevated exocytotic protein expression at the peak GLP-1 secretion time point further adds to the body of evidence supporting the role of the circadian clock in regulating time-dependent GLP-1 release. These data are also consistent with the findings presented herein, whereby pathways related to vesicle transport were downregulated upon L-cell Arntl disruption. Interestingly, studies in the pancreatic α- and β-cells have demonstrated an essential role for Arntl in the regulation of glucagon and insulin secretion, respectively, with alterations in expression of the exocytotic SNARE protein genes observed in the β-cells [18,76]. In combination, data from these neuroendocrine cell types provide strong evidence for clock-controlled exocytosis, which is ultimately responsible for coordinating rhythmic hormone secretion.
Previous L-cell analyses have also identified pathways related to nutrient sensing and metabolic processes as important for L-cell secretion [8,9]. Furthermore, exposure to the saturated fatty acid, palmitate, impairs mGLUTag L-cell mitochondrial function, ATP production, and GLP-1 secretion in parallel [39]. Consistent with the notion of cellular metabolism being dependent on the circadian clock [39], Arntl KD also resulted in impaired L-cell metabolic processes in the mGLUTag L-cells. Finally, Arntl-disrupted mGLUTag L-cells were found to have dysregulated Notch signaling, which is a well-established pathway in the maintenance of intestinal epithelial integrity [60]. A recent study has demonstrated that Notch signaling is not essential for L-cell differentiation [77]; however, given the known role of intestinal epithelial Notch signaling in reducing intestinal inflammation [78], this may provide insight as to how L-cell Arntl disruption leads to impaired intestinal homeostasis in Gcg-Arntl KO mice.
5. Conclusions
Circadian disruption, induced by modern-day environmental factors such as shift work and jetlag, has been extensively linked to the development of metabolic diseases including type 2 diabetes and obesity [79]. Moreover, these circadian disruptors have been linked to impaired intestinal barrier integrity, microbial dysbiosis, and increased intestinal inflammation [80]. Clinically, circadian disruption is also implicated in the development and severity of inflammatory bowel disease [81]. Collectively, the results of the present study establish intestinal L-cell Arntl as essential in the regulation of key cellular pathways required for time-dependent stimulation of GLP-1 secretion and highlight the importance of the L-cell secretome for parameters of intestinal homeostasis. While this is the first time L-cell Arntl has been described as a regulator of the microbial and immune environment, the experimental model does have limitations that prevent complete mechanistic understanding of how L-cell Arntl facilities its novel role described herein, which could include clock-dependent and/or clock-independent mechanisms. Furthermore, although the decrease in ARNTL expression in the intestinal L-cell in Gcg-Arntl KO mice was 44% lower than we previously reported for STXBP1 using the same Gcg-cre model [40], this may be related to relative levels of expression of the two target genes and/or different efficiencies of the antisera used for the immunostaining. Additional analysis of ARNTL expression in pancreatic α-cells and Gcg-expressing neurons is warranted to provide a more complete characterization of this novel inducible KO mouse model. It must also be noted that the aberrant release of GLP-1 secretion following L-cell Arntl disruption suggests that L-cell secretory products contribute to the Gcg-Arntl KO mouse phenotype; however, further study is required to decipher the contribution made by each of the L-cell secretory products to circadian intestinal homeostasis, including not only GLP-1 and GLP-2, but also other GCG-derived peptides, including oxyntomodulin and co-expressed hormones such as peptide YY [82]. While transcriptomic analysis of the mGLUTag L-cells did provide mechanistic insight as to how altered L-cell function following Arntl knockdown may contribute to the in vivo phenotype observed in Gcg-Arntl KO mice, use of reporter animals to assess Arntl-disrupted primary L-cells would provide a further understanding of the circadian rhythm inherent to the L-cell. Furthermore, although the present study established the requirement of L-cell Arntl in time-dependent GLP-1 secretion, assessment of GLP-1 release throughout the 24-hour day in vivo and over 48 h in mGLUTag L-cells in vitro would be necessary to confirm the role of L-cell Arntl in the circadian secretion of GLP-1. Finally, the exact factors responsible for entraining the L-cell clock remain to be elucidated, including both diet and microbial metabolites. Nevertheless, these data collectively provide novel insight into both the regulation and role of time-dependent L-cell secretion. Establishing the requirement of the core clock gene Arntl within the L-cell in maintaining intestinal homeostasis means that therapeutic strategies aimed at increasing endogenous L-cell secretion to improve intestinal health can be developed for patients with intestinal and metabolic diseases.
Author contributions
S.E.M. researched and analyzed data and wrote the manuscript. A.M. researched data and reviewed and edited the manuscript. A.D.B. researched data and reviewed and edited the manuscript. A.W. analyzed data and reviewed and edited the manuscript. P.L.B. provided funding, reviewed and edited the manuscript, and is the guarantor of the work.
Acknowledgments
The authors are extremely grateful to S. Malekian Naeini and Dr. A. Hardy, University of Toronto, for the ARNTL immunostaining and analyses, Drs. F. Reimann and F. Gribble for the Gcg-CreERT2/+;Rosa26-GCamP3fl/fl mice, and N. Simard from the University of Toronto Faculty of Medicine Flow Cytometry Facility. S.E.M. was supported by graduate awards from the Ontario Graduate Scholarship program and the Banting and Best Diabetes Centre (University of Toronto); A.M. by graduate awards from the Canadian Institutes of Health Research, the Ontario Graduate Scholarship, the Banting and Best Diabetes Centre, and the Biological Rhythms Training Program (University of Toronto); A.D.B. by graduate awards from the Ontario Graduate Scholarship program and the Banting and Best Diabetes Centre (University of Toronto); and P.L.B. by a Tier I Canada Research Chair.
Operating support for this study was obtained (by P.L.B.) from the Canadian Institutes of Health Research (PJT-14853), and some of the equipment was provided by The 3D (Diet, Digestive Tract and Disease) Centre funded by the Canadian Foundation for Innovation and Ontario Research Fund (project numbers 19442 and 30961).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.molmet.2021.101340.
Conflict of interest
None declared.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Drucker D.J. Mechanisms of action and therapeutic application of glucagon-like peptide-1. Cell Metabolism. 2018;27(4):740–756. doi: 10.1016/j.cmet.2018.03.001. [DOI] [PubMed] [Google Scholar]
- 2.Müller T.D., Finan B., Bloom S.R., D'Alessio D., Drucker D.J., Flatt P.R. Glucagon-like peptide 1 (GLP-1) Molecular Metabalism. 2019;30:72–130. doi: 10.1016/j.molmet.2019.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yusta B., Baggio L.L., Koehler J., Holland D., Cao X., Pinnell L.J. GLP-1R agonists modulate enteric immune responses through the intestinal intraepithelial lymphocyte GLP-1R. Diabetes. 2015;64(7):2537–2549. doi: 10.2337/db14-1577. [DOI] [PubMed] [Google Scholar]
- 4.Lebrun L.J., Lenaerts K., Kiers D., Pais de Barros J.P., Le Guern N., Plesnik J. Enteroendocrine L cells sense LPS after gut barrier injury to enhance GLP-1 secretion. Cell Reports. 2017;21(5):1160–1168. doi: 10.1016/j.celrep.2017.10.008. [DOI] [PubMed] [Google Scholar]
- 5.Koehler J.A., Baggio L.L., Yusta B., Longuet C., Rowland K.J., Cao X. GLP-1R agonists promote normal and neoplastic intestinal growth through mechanisms requiring Fgf7. Cell Metabolism. 2015;21(3):379–391. doi: 10.1016/j.cmet.2015.02.005. [DOI] [PubMed] [Google Scholar]
- 6.He S., Kahles F., Rattik S., Nairz M., McAlpine C.S., Anzai A. Gut intraepithelial T cells calibrate metabolism and accelerate cardiovascular disease. Nature. 2019;566(7742):115–119. doi: 10.1038/s41586-018-0849-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gil-Lozano M., Mingomataj E.L., Wu W.K., Ridout S.A., Brubaker P.L. Circadian secretion of the intestinal hormone GLP-1 by the rodent L cell. Diabetes. 2014;63(11):3674–3685. doi: 10.2337/db13-1501. [DOI] [PubMed] [Google Scholar]
- 8.Biancolin A.D., Martchenko A., Mitova E., Gurges P., Michalchyshyn E., Chalmers J.A. The core clock gene, Bmal1, and its downstream target, the SNARE regulatory protein secretagogin, are necessary for circadian secretion of glucagon-like peptide-1. Molecular Metabolism. 2020;31:124–137. doi: 10.1016/j.molmet.2019.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martchenko S.E., Martchenko A., Cox B.J., Naismith K., Waller A., Gurges P. Circadian GLP-1 secretion in mice is dependent on the intestinal microbiome for maintenance of diurnal metabolic homeostasis. Diabetes. 2020;69(12):2589–2602. doi: 10.2337/db20-0262. [DOI] [PubMed] [Google Scholar]
- 10.Gil-Lozano M., Hunter P.M., Behan L.-A., Gladanac B., Casper R.F., Brubaker P.L. Short-term sleep deprivation with nocturnal light exposure alters time-dependent glucagon-like peptide-1 and insulin secretion in male volunteers. American Journal of Physiology. Endocrinology and Metabolism. 2016;310(1):41–50. doi: 10.1152/ajpendo.00298.2015. [DOI] [PubMed] [Google Scholar]
- 11.Lindgren O., Mari A., Deacon C.F., Carr R.D., Winzell M.S., Vikman J. Differential islet and incretin hormone responses in morning versus afternoon after standardized meal in healthy men. Journal of Clinical Endocrinology & Metabolism. 2009;94(8):2887e2892. doi: 10.1210/jc.2009-0366. [DOI] [PubMed] [Google Scholar]
- 12.Santiago J., Muñoz G., Jiménez Rodríguez D., José J., Morante H. Diurnal rhythms of plasma GLP-1 levels in normal and overweight/obese subjects: lack of effect of weight loss. Journal of Physiology & Biochemistry. 2015;71:17–28. doi: 10.1007/s13105-014-0375-7. [DOI] [PubMed] [Google Scholar]
- 13.Gerhart-Hines Z., Lazar M.A. Circadian metabolism in the light of evolution. Endocrine Reviews. 2015;36(3):289–304. doi: 10.1210/er.2015-1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bass J., Lazar M.A. Circadian time signatures of fitness and disease. Science. 2016;354(6315):994–999. doi: 10.1126/science.aah4965. [DOI] [PubMed] [Google Scholar]
- 15.Dibner C., Schibler U., Albrecht U. The mammalian circadian timing system: organization and coordination of central and peripheral clocks. Annual Review of Physiology. 2010;72(1):517–549. doi: 10.1146/annurev-physiol-021909-135821. [DOI] [PubMed] [Google Scholar]
- 16.Hoogerwerf W.A., Hellmich H.L., Cornélissen G., Halberg F., Shahinian V.B., Bostwick J. Clock gene expression in the murine gastrointestinal tract: endogenous rhythmicity and effects of a feeding regimen. Gastroenterology. 2007;133(4):1250e1260. doi: 10.1053/j.gastro.2007.07.009. [DOI] [PubMed] [Google Scholar]
- 17.Marcheva B., Ramsey K.M., Buhr E.D., Kobayashi Y., Su H., Ko C.H. Disruption of the clock components CLOCK and BMAL1 leads to hypoinsulinaemia and diabetes. Nature. 2010;466(7306):627e631. doi: 10.1038/nature09253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Perelis M., Marcheva B., Ramsey K.M., Schipma M.J., Hutchison A.L., Taguchi A. Pancreatic b cell enhancers regulate rhythmic transcription of genes controlling insulin secretion. Science. 2015;350(6261):aac4250. doi: 10.1126/science.aac4250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Petrenko V., Saini C., Giovannoni L., Gobet C., Sage D., Unser M. Pancreatic a- and b-cellular clocks have distinct molecular properties and impact on islet hormone secretion and gene expression. Genes & Development. 2017;31(4):383e398. doi: 10.1101/gad.290379.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rakshit K., Qian J., Ernst J., Matveyenko A.V. Circadian variation of the pancreatic islet transcriptome. Physiological Genomics. 2016;48(9):677–687. doi: 10.1152/physiolgenomics.00019.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rey G., Cesbron F., Rougemont J., Reinke H., Brunner M., Naef F. Genome-wide and phase-specific DNA-binding rhythms of BMAL1 control circadian output functions in mouse liver. PLoS Biology. 2011;9(2) doi: 10.1371/journal.pbio.1000595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McCarthy J.J., Andrews J.L., McDearmon E.L., Campbell K.S., Barber B.K., Miller B.H. Identification of the circadian transcriptome in adult mouse skeletal muscle. Physiological Genomics. 2007;31(1):86e95. doi: 10.1152/physiolgenomics.00066.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zvonic S., Ptitsyn A.A., Conrad S.A., Scott L.K., Floyd Z.E., Kilroy G. Characterization of peripheral circadian clocks in adipose tissues. Diabetes. 2006;55(4):962e970. doi: 10.2337/diabetes.55.04.06.db05-0873. [DOI] [PubMed] [Google Scholar]
- 24.Zarrinpar A., Chaix A., Yooseph S., Panda S. Diet and feeding pattern affect the diurnal dynamics of the gut microbiome. Cell Metabolism. 2014;20(6):1006–1017. doi: 10.1016/j.cmet.2014.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thaiss C.A., Zeevi D., Levy M., Zilberman-Schapira G., Suez J., Tengeler A.C. Transkingdom control of microbiota diurnal oscillations promotes metabolic homeostasis. Cell. 2014;159(3):514–529. doi: 10.1016/j.cell.2014.09.048. [DOI] [PubMed] [Google Scholar]
- 26.Thaiss C.A., Levy M., Korem T., Dohnalova L., Shapiro H., Jaitin D.A. Microbiota diurnal rhythmicity programs host transcriptome oscillations. Cell. 2016;167(6):1495–1510. doi: 10.1016/j.cell.2016.11.003. e12. [DOI] [PubMed] [Google Scholar]
- 27.Mukherji A., Kobiita A., Ye T., Chambon P. Homeostasis in intestinal epithelium is orchestrated by the circadian clock and microbiota cues transduced by TLRs. Cell. 2013;153(4):812–827. doi: 10.1016/j.cell.2013.04.020. [DOI] [PubMed] [Google Scholar]
- 28.Leone V., Gibbons S.M., Martinez K., Hutchison A.L., Huang E.Y., Cham C.M. Effects of diurnal variation of gut microbes and high-fat feeding on host circadian clock function and metabolism. Cell Host & Microbe. 2015;17(5):681–689. doi: 10.1016/j.chom.2015.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lamia K.A., Storch K.-F., Weitz C.J. Physiological significance of a peripheral tissue circadian clock. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(39):15172–15177. doi: 10.1073/pnas.0806717105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Andrews J.L., Zhang X., McCarthy J.J., McDearmon E.L., Hornberger T.A., Russell B. CLOCK and BMAL1 regulate MyoD and are necessary for maintenance of skeletal muscle phenotype and function. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(44):19090–19095. doi: 10.1073/pnas.1014523107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shimba S., Ishii N., Ohta Y., Ohno T., Watabe Y., Hayashi M. Brain and muscle Arnt-like protein-1 (BMAL1), a component of the molecular clock, regulates adipogenesis. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(34):12071–12076. doi: 10.1073/pnas.0502383102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang E.E., Liu Y., Dentin R., Pongsawakul P.Y., Liu A.C., Hirota T. Cryptochrome mediates circadian regulation of cAMP signaling and hepatic gluconeogenesis. Nature Medicine. 2010;16(10):1152–1156. doi: 10.1038/nm.2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lamia K.A., Papp S.J., Yu R.T., Barish G.D., Uhlenhaut N.H., Jonker J.W. Cryptochromes mediate rhythmic repression of the glucocorticoid receptor. Nature. 2011;480(7378):552–556. doi: 10.1038/nature10700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Oh S.K., Kim D., Kim K., Boo K., Yu Y.S., Kim I.S. RORα is crucial for attenuated inflammatory response to maintain intestinal homeostasis. Proceedings of the National Academy of Sciences of the United States of America. 2019;116(42):21140–21149. doi: 10.1073/pnas.1907595116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sadacca L.A., Lamia K.A., DeLemos A.S., Blum B., Weitz C.J. An intrinsic circadian clock of the pancreas is required for normal insulin release and glucose homeostasis in mice. Diabetologia. 2011;54(1):120–124. doi: 10.1007/s00125-010-1920-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rakshit K., Qian J., Gaonkar K.S., Dhawan S., Colwell C.S., Matveyenko A.V. Postnatal ontogenesis of the islet circadian clock plays a contributory role in β-cell maturation process. Diabetes. 2018;67(5):911–922. doi: 10.2337/db17-0850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dyar K.A., Ciciliot S., Wright L.E., Bienso R.S., Tagliazucchi G.M., Patel V.R. Muscle insulin sensitivity and glucose metabolism are controlled by the intrinsic muscle clock. Molecular Metabolism. 2014;3(1):29–41. doi: 10.1016/j.molmet.2013.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Paschos G.K., Ibrahim S., Song W.L., Kunieda T., Grant G., Reyes T.M. Obesity in mice with adipocyte-specific deletion of clock component Arntl. Nature Medicine. 2012;18(12):1768–1777. doi: 10.1038/nm.2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Martchenko A., Oh R.H., Wheeler S.E., Gurges P., Chalmers J.A., Brubaker P.L. Suppression of circadian secretion of glucagon-like peptide-1 by the saturated fatty acid, palmitate. Acta Physiologica. 2017;222(4) doi: 10.1111/apha.13007. [DOI] [PubMed] [Google Scholar]
- 40.Campbell J.R., Martchenko A., Sweeney M.E., Maalouf M.F., Psichas A., Gribble F.M. Essential role of syntaxin-binding protein-1 in the regulation of glucagon-like peptide-1 secretion. Endocrinology. 2020;161(5):bqaa039. doi: 10.1210/endocr/bqaa039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wheeler S.E., Stacey H.M., Nahaei Y., Hale S.J., Hardy A.B., Reimann F. The SNARE protein syntaxin-1a plays an essential role in biphasic exocytosis of the incretin hormone glucagon-like peptide 1. Diabetes. 2017;66(9):2327–2338. doi: 10.2337/db16-1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ceasrine A.M., Ruiz-Otero N., Lin E.E., Lumelsky D.N., Boehm E.D., Kuruvilla R. Tamoxifen improves glucose tolerance in a delivery-, Sex-, and strain-dependent manner in mice. Endocrinology. 2019;160(4):782–790. doi: 10.1210/en.2018-00985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bohin N., Carlson E.A., Samuelson L.C. Genome toxicity and impaired stem cell function after conditional activation of CreERT2 in the intestine. Stem Cell Reports. 2018;11(6):1337–1346. doi: 10.1016/j.stemcr.2018.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gil-Lozano M., Wu W.K., Martchenko A., Brubaker P.L. High-fat diet and palmitate alter the rhythmic secretion of glucagon-like peptide-1 by the rodent L-cell. Endocrinology. 2016;157(2):586–599. doi: 10.1210/en.2015-1732. [DOI] [PubMed] [Google Scholar]
- 45.Reimann F., Habib A.M., Tolhurst G., Parker H.E., Rogers G.J., Gribble F.M. Glucose sensing in L cells: a primary cell. Cell Metabolism. 2008;8(6):532–539. doi: 10.1016/j.cmet.2008.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brubaker P.L., Schloos J., Drucker D.J. Regulation of glucagon-like peptide-1 synthesis and secretion in the GLUTag enteroendocrine cell line. Endocrinology. 1998;139(10):4108–4114. doi: 10.1210/endo.139.10.6228. [DOI] [PubMed] [Google Scholar]
- 47.Bray N.L., Pimentel H., Melsted P., Pachter L. Near-optimal probabilistic RNA-seq quantification. Nature Biotechnology. 2016;34(5):525–527. doi: 10.1038/nbt.3519. [DOI] [PubMed] [Google Scholar]
- 48.Soneson C., Love M.I., Robinson M.D. Differential analyses for RNA-seq: transcript-level estimates improve gene-level inferences. F1000Research. 2015;4:1521. doi: 10.12688/f1000research.7563.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Robinson M.D., McCarthy D.J., Smyth G.K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Smyth G.K. Bioinformatics and computational biology solutions using R and bioconductor. Springer-Verlag; 2005. Limma: linear models for microarray data; pp. 397–420. [Google Scholar]
- 51.Merico D., Isserlin R., Stueker O., Emili A., Bader G.D. EnrichmentMap: a network-based method for gene-set enrichment visualization and interpretation. PloS One. 2010;5(11) doi: 10.1371/journal.pone.0013984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Reimand J., Isserlin R., Voisin V., Kucera M., Tannus-Lopes C., Rostamianfar A. Pathway enrichment analysis and visualization of omics data using g:Profiler, GSEA, Cytoscape and EnrichmentMap. Nature Protocols. 2019;14(2):482–517. doi: 10.1038/s41596-018-0103-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Egerod K.L., Engelstoft M.S., Grunddal K.V., Nohr M.K., Secher A., Sakata I. A major lineage of enteroendocrine cells coexpress CCK, secretin, GIP, GLP-1, PYY, and neurotensin but not somatostatin. Endocrinology. 2012;153(12):5782–5795. doi: 10.1210/en.2012-1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Grunddal K.V., Ratner C.F., Svendsen B., Sommer F., Engelstoft M.S., Madsen A.N. Neurotensin Is coexpressed, coreleased, and acts together with GLP-1 and PYY in enteroendocrine control of metabolism. Endocrinology. 2016;157(1):176–194. doi: 10.1210/en.2015-1600. [DOI] [PubMed] [Google Scholar]
- 55.Lo Sasso G., Phillips B.W., Sewer A., Battey J.N.D., Kondylis A., Talikka M. The reduction of DSS-induced colitis severity in mice exposed to cigarette smoke is linked to immune modulation and microbial shifts. Scientific Reports. 2020;10(1):3829. doi: 10.1038/s41598-020-60175-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zarrinpar A., Chaix A., Xu Z.Z., Chang M.W., Marotz C.A., Saghatelian A. Antibiotic-induced microbiome depletion alters metabolic homeostasis by affecting gut signaling and colonic metabolism. Nature Communications. 2018;9(1):2872. doi: 10.1038/s41467-018-05336-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Arora T., Akrami R., Pais R., Bergqvist L., Johansson B.R., Schwartz T.W. Microbial regulation of the L cell transcriptome. Scientific Reports. 2018;8(1):1207. doi: 10.1038/s41598-017-18079-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gustavsson N., Wang Y., Kang Y., Seah T., Chua S., Radda G.K. Synaptotagmin-7 as a positive regulator of glucose-induced glucagonlike peptide-1 secretion in mice. Diabetologia. 2011;54(7):1824–1830. doi: 10.1007/s00125-011-2119-3. [DOI] [PubMed] [Google Scholar]
- 59.Li S.K., Zhu D., Gaisano H.Y., Brubaker P.L. Role of vesicle-associated membrane protein 2 in exocytosis of glucagon-like peptide-1 from the murine intestinal L cell. Diabetologia. 2014;57(4):809–818. doi: 10.1007/s00125-013-3143-2. [DOI] [PubMed] [Google Scholar]
- 60.Noah T.K., Shroyer N.F. Notch in the intestine: regulation of homeostasis and pathogenesis. Annual Review of Physiology. 2013;75:263–288. doi: 10.1146/annurev-physiol-030212-183741. [DOI] [PubMed] [Google Scholar]
- 61.Laermans J., Vancleef L., Tack J., Depoortere I. Role of the clock gene Bmal1 and the gastric ghrelin-secreting cell in the circadian regulation of the ghrelin-GOAT system. Scientific Reports. 2015;5:16748. doi: 10.1038/srep16748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pederson R.A., Satkunarajah M., McIntosh C.H., Scrocchi L.A., Flamez D., Schuit F. Enhanced glucose-dependent insulinotropic polypeptide secretion and insulinotropic action in glucagon-like peptide 1 receptor -/- mice. Diabetes. 1998;47(7):1046–1052. doi: 10.2337/diabetes.47.7.1046. [DOI] [PubMed] [Google Scholar]
- 63.Suzuki H., Shibata S., Okutani T., Suzuki M., Nakayama M., Nishimura T. Diurnal changes in intraepithelial lymphocytes (IELs) in the small intestine of mice. Experimental Animals. 1999;48(2):115–118. doi: 10.1538/expanim.48.115. [DOI] [PubMed] [Google Scholar]
- 64.Tuganbaev T., Mor U., Bashiardes S., Liwinski T., Nobs S.P., Leshem A. Diet diurnally regulates small intestinal microbiome-epithelial-immune homeostasis and enteritis. Cell. 2020;182(6):1441–1459. doi: 10.1016/j.cell.2020.08.027. e21. [DOI] [PubMed] [Google Scholar]
- 65.Martchenko S.E., Prescott D., Martchenko A., Sweeney M.E., Philpott D.J., Brubaker P.L. 2021. Diurnal changes in the murine small intestine are disrupted by obesogenic western diet feeding and microbial dysbiosis. In revision. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gordon S.N., Cervasi B., Odorizzi P., Silverman R., Aberra F., Ginsberg G. Disruption of intestinal CD4+ T cell homeostasis is a key marker of systemic CD4+ T cell activation in HIV-infected individuals. The Journal of Immunology. 2010;185(9):5169–5179. doi: 10.4049/jimmunol.1001801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dondji B., Sun T., Bungiro R.D., Vermeire J.J., Harrison L.M., Bifulco C. CD4+ T cells mediate mucosal and systemic immune responses to experimental hookworm infection. Parasite Immunology. 2010;32(6):406–413. doi: 10.1111/j.1365-3024.2010.01204.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Drucker D.J., Ehrlich P., Asa S.L., Brubaker P.L. Induction of intestinal epithelial proliferation by glucagon-like peptide 2. Proceedings of the National Academy of Sciences of the United States of America. 1996;93(15):7911–7916. doi: 10.1073/pnas.93.15.7911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dong C.X., Zhao W., Solomon C., Rowland K.J., Ackerley C., Robine S. The intestinal epithelial insulin-like growth factor-1 receptor links glucagon-like peptide-2 action to gut barrier function. Endocrinology. 2014;155(2):370–379. doi: 10.1210/en.2013-1871. [DOI] [PubMed] [Google Scholar]
- 70.Lee S.J., Lee J., Li K.K., Holland D., Maughan H., Guttman D.S. Disruption of the murine Glp2r impairs paneth cell function and increases susceptibility to small bowel enteritis. Endocrinology. 2012;153(3):1141–1151. doi: 10.1210/en.2011-1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kim S.J., Kim S.E., Kim A.R., Kang S., Park M.Y., Sung M.K. Dietary fat intake and age modulate the composition of the gut microbiota and colonic inflammation in C57BL/6J mice. BMC Microbiology. 2019;19(1):193. doi: 10.1186/s12866-019-1557-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Alam M.T., Amos G.C.A., Murphy A.R.J., Murch S., Wellington E.M.H., Arasaradnam R.P. Microbial imbalance in inflammatory bowel disease patients at different taxonomic levels. Gut Pathogens. 2020;12:1. doi: 10.1186/s13099-019-0341-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.van der Hee B., Wells J.M. Microbial regulation of host physiology by short-chain fatty acids. Trends in Microbiology. 2021;S0966–842X(21) doi: 10.1016/j.tim.2021.02.001. 00035-4. [DOI] [PubMed] [Google Scholar]
- 74.Tolhurst G., Heffron H., Lam Y.S., Parker H.E., Habib A.M., Diakogiannaki E. Short-chain fatty acids stimulate glucagon-like peptide-1 secretion via the G-protein-coupled receptor FFAR2. Diabetes. 2012;61(2):364–371. doi: 10.2337/db11-1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Brighton C.A., Rievaj J., Kuhre R.E., Glass L.L., Schoonjans K., Holst J.J. Bile acids trigger GLP-1 release predominantly by accessing basolaterally located G protein-coupled bile acid receptors. Endocrinology. 2015;156(11):3961–3970. doi: 10.1210/en.2015-1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Petrenko V., Gandasi N.R., Sage D., Tengholm A., Barg S., Dibner C. In pancreatic islets from type 2 diabetes patients, the dampened circadian oscillators lead to reduced insulin and glucagon exocytosis. Proceedings of the National Academy of Sciences of the United States of America. 2020;117(5):2484–2495. doi: 10.1073/pnas.1916539117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Beumer J., Puschhof J., Bauzá-Martinez J., Martínez-Silgado A., Elmentaite R., James K.R. High-resolution mRNA and secretome atlas of human enteroendocrine cells. Cell. 2020;181(6):1291–1306. doi: 10.1016/j.cell.2020.04.036. e19. [DOI] [PubMed] [Google Scholar]
- 78.Obata Y., Takahashi D., Ebisawa M., Kakiguchi K., Yonemura S., Jinnohara T. Epithelial cell-intrinsic Notch signaling plays an essential role in the maintenance of gut immune homeostasis. The Journal of Immunology. 2012;188(5):2427–2436. doi: 10.4049/jimmunol.1101128. [DOI] [PubMed] [Google Scholar]
- 79.Fonken L.K., Nelson R.J. The effects of light at night on circadian clocks and metabolism. Endocrine Reviews. 2014;35(4):648–670. doi: 10.1210/er.2013-1051. [DOI] [PubMed] [Google Scholar]
- 80.Martchenko A., Martchenko S.E., Biancolin A.D., Brubaker P.L. Circadian rhythms and the gastrointestinal tract: relationship to metabolism and gut hormones. Endocrinology. 2020;161(12):bqaa167. doi: 10.1210/endocr/bqaa167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Voigt R.M., Forsyth C.B., Keshavarzian A. Circadian rhythms: a regulator of gastrointestinal health and dysfunction. Expert Review of Gastroenterology & Hepatology. 2019;13(5):411–424. doi: 10.1080/17474124.2019.1595588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Dong C.X., Brubaker P.L. Ghrelin, the proglucagon-derived peptides and peptide YY in nutrient homeostasis. Nature Reviews Gastroenterology & Hepatology. 2012;9(12):705–715. doi: 10.1038/nrgastro.2012.185. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.